Abstract
Tissue maintenance and repair depend on the integrated activity of multiple cell types 1 . Whereas the contributions of epithelial 2,3 , immune 4,5 and stromal cells 6,7 in intestinal tissue integrity are well understood, the role of intrinsic neuroglia networks remains largely unknown. Here, we uncover pivotal roles of enteric glial cells (EGCs) in intestinal homeostasis, immunity and tissue repair. We demonstrate that infection of mice with Heligmosomoides polygyrus leads to enteric gliosis and upregulation of an interferon gamma (IFN-γ) gene signature. IFN-γ-dependent gene modules were also induced in EGCs from inflammatory bowel disease patients 8 . Single-cell transcriptomics of the tunica muscularis (TM) showed that glia-specific abrogation of IFN-γ signaling leads to tissue-wide activation of pro-inflammatory transcriptional programs. In addition, disruption of the IFN-γ-EGC signaling axis enhanced the inflammatory and granulomatous response of the TM to helminths. Mechanistically, we show that upregulation of Cxcl10 is an early immediate response of EGCs to IFN-γ signaling and provide evidence that this chemokine and the downstream amplification of IFN-γ signaling in the TM are required for a measured inflammatory response to helminths and resolution of granulomatous pathology. Our study demonstrates that IFN-γ signaling in enteric glia is central to intestinal homeostasis and reveals critical roles of the IFN-γ-EGC-Cxcl10 axis in immune response and tissue repair following infectious challenge.
Helminth-induced gliosis of EGCs
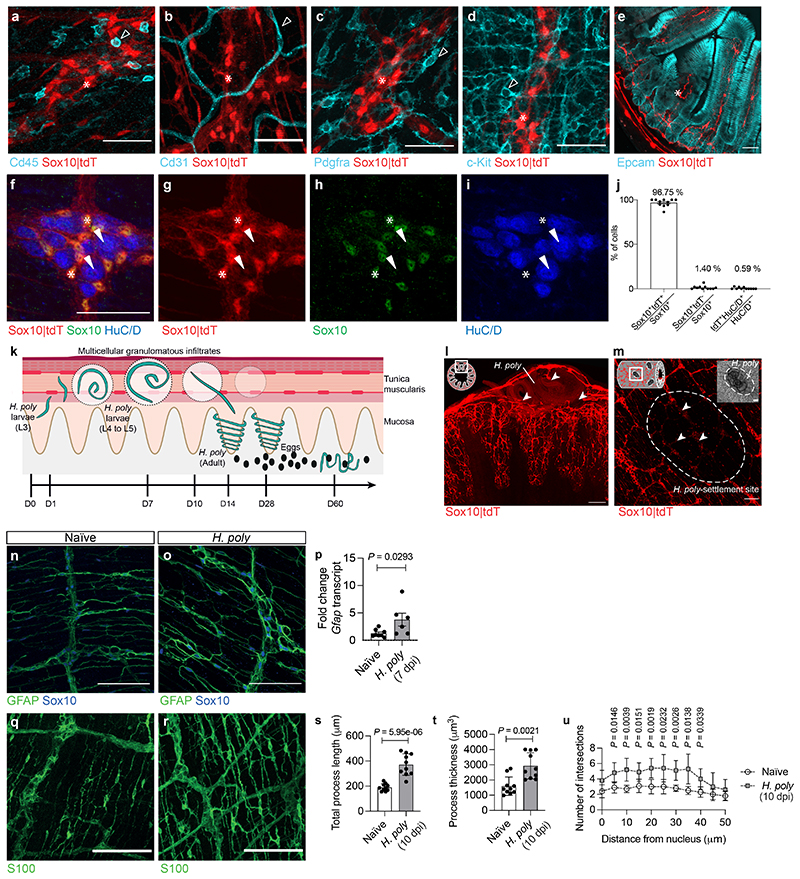
To investigate how EGCs respond to pathogens, Sox10CreERT2;Rosa26tdTomato mice 9 (Sox10|tdT) expressing the fluorescent reporter tdTomato (tdT) in enteric glia (Extended Data Fig. 1a-j), were infected with Heligmosomoides polygyrus (H. poly). L3 H. poly larvae settle in the TM of the duodenum, where they elicit local tissue inflammation and multicellular granulomatous infiltrates 10 (Extended Data Fig. 1k). At 7 days post infection (dpi) tdT signal was reduced at the sites of worm settlement, indicating local EGC network damage (Extended Data Fig. 1l, m). However, the overall expression of GFAP and the length, thickness and density of glial processes were increased (Extended Data Fig. 1n-u). The proliferation marker Ki67 was upregulated in interganglionic EGCs 11 at the vicinity of H. poly, with kinetics correlating with worm residence in the TM 10 (Fig. 1a-d). Thus, intestinal helminth infections induce proliferation and reactive gliosis of EGCs.
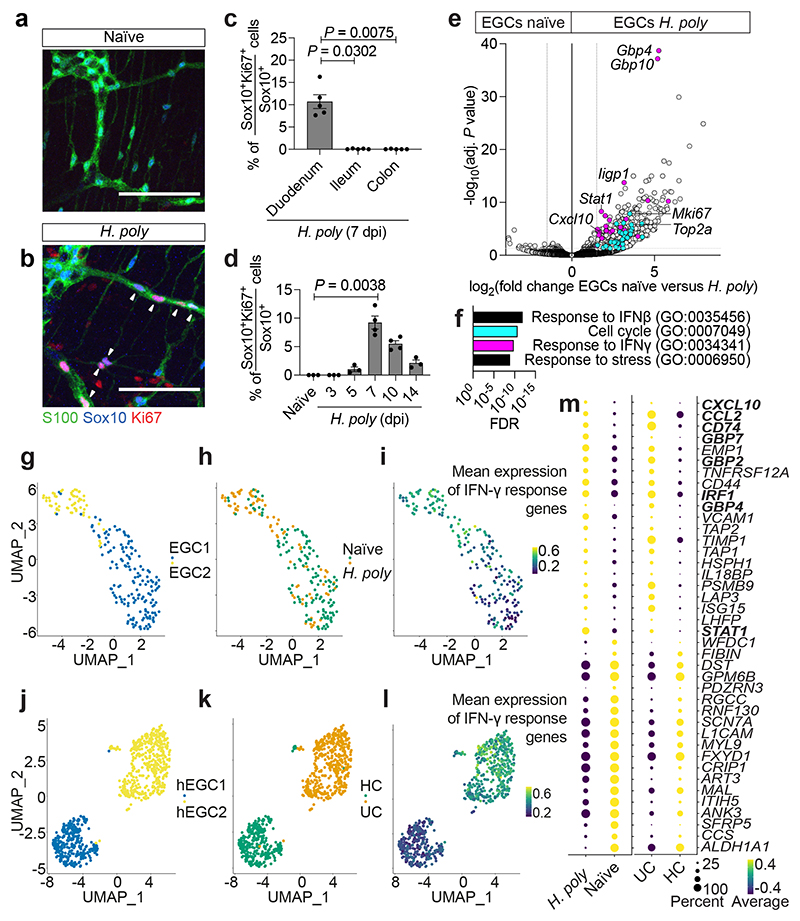
Figure 1. Inflammatory injury induces IFN-γ signature in EGCs.
(a, b) TM preparations from the duodenum of naïve (a) and H. poly-infected (b) animals (7 dpi) immunostained for S100, Sox10 and Ki67. Arrowheads: Ki67+ EGCs. (c) Quantification of Ki67+ EGCs in the duodenum, ileum and colon of H. poly-infected animals (n=4). (d) Time-course analysis of EGC-proliferation after H. poly infection (nnaïve,3dpi,5dpi,14dpi= 3, n7dpi,10dpi= 4, data from one experiment, data at 7 dpi representative of 3 experiments). (e) Volcano plot showing mean log2-transformed fold change and significance (-log10(adjusted P value)) of differentially expressed genes in tdT+ cells from naïve and H. poly-infected Sox10|tdT animals (7 dpi). Coloured dots: genes associated with the GO terms “cell cycle” (turquoise) and “response to IFN-γ” (magenta). n=4. (f) Top GO terms associated with the differentially expressed genes in EGCs from H. poly infected mice. “Cell cycle” and “response to IFN-γ” terms colours correspond to colour of genes highlighted in (g). (g-l) UMAP of sequenced EGCs from the TM of naïve and H. poly infected mice (g-i) or human healthy colon (HC) and Ulcerative Colitis colon (UC) (j-l) (human dataset, GSE114374 8 ). Cells are color-coded according to Louvain clusters (g, j), experimental (h) or disease condition (k) and mean scaled expression of IFN-γ response-associated genes (i, l; selection based on GO term “response to IFN-γ”). (m) Intersection of genes differentially expressed by mouse (left, naïve versus H. poly-infected mice) and human (right, HC versus UC patients) EGCs. Bold: IFN-γ-target genes. Kruskal-Wallis test (c, d). Mean±SEM (c, d). Scale bars: 100 μm
Inflammation-induced IFN-γ-EGC signalling
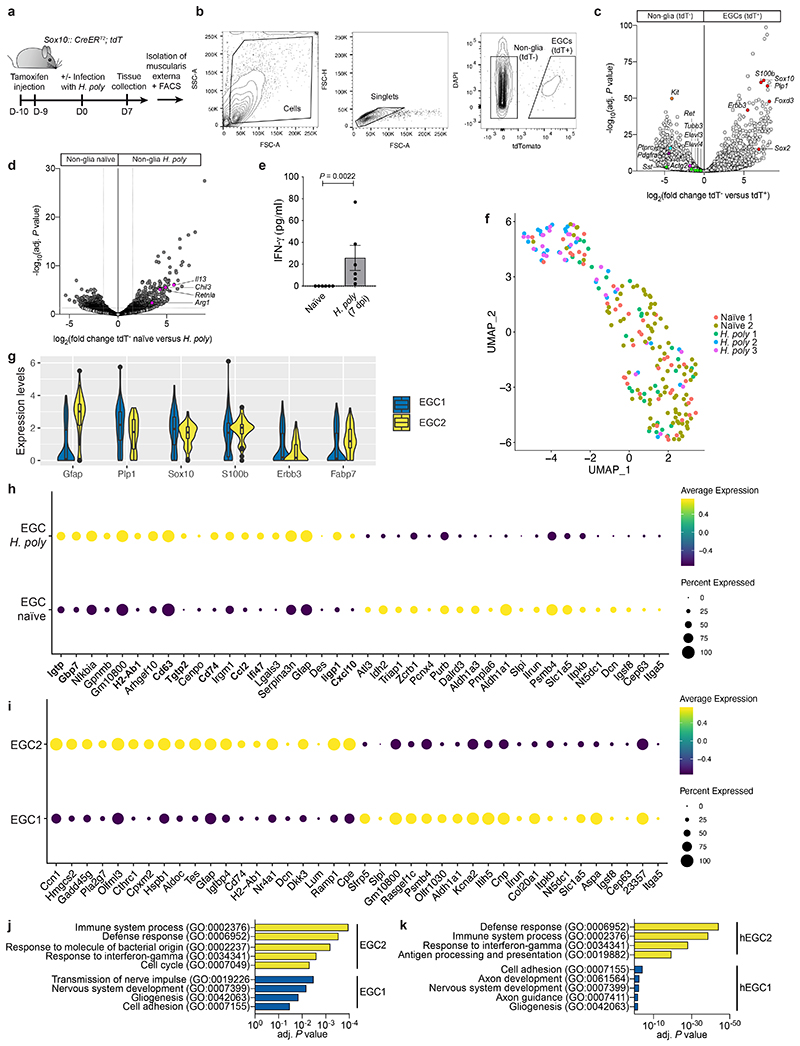
To identify helminth-activated transcriptional programmes in EGCs, we performed bulk RNA sequencing (RNAseq) of the TM from naïve and H. poly infected (7 dpi) Sox10|tdT mice (Extended Data Fig. 2a, b). As expected, EGC markers (Sox10, Plp1, S100b, Foxd3, Erbb3, Sox2) 12,13 were expressed by tdT+ cells, while tdT- cells expressed markers of neurons (Ret, Tubb3, Sst, Elavl3, Elavl4), immune cells (Ptprc), interstitial cells of Cajal (ICCs, Kit), smooth muscle cells (Actg2) and fibroblasts (Pdgfra) (Extended Data Fig. 2c). Consistent with the robust TH2 response associated with helminths 10 , alternative macrophage markers (Arg1, Retnla, Chil3) and TH2 cytokines (Il13) were upregulated in tdT- cells from H. poly- infected mice (Extended Data Fig. 2d). Surprisingly, in addition to cell cycle genes, EGCs from infected animals were enriched for GO terms for interferon and stress response and upregulated IFN-γ target genes (Gbp4, Gbp10, Iigp1, Stat1, Cxcl10) (Fig. 1e, f). Accordingly, increased levels of IFN-γ were detected in the TM of H. poly-infected mice at 7 dpi (Extended Data Fig. 2e). To characterize the response of EGCs at single cell resolution, we performed single-cell RNAseq (scRNAseq) of tdT+ cells from the TM of naïve and H. poly-infected Sox10|tdT mice. We identified two clusters, EGC1 and EGC2 (Fig. 1g), expressing Plp1, Sox10, S100b, Erbb3, Fabp7 and Gfap (Extended Data Fig. 2f, g). Similar to bulk RNAseq, EGCs from H. poly-infected mice upregulated Gfap and IFN-γ target genes (Extended Data Fig. 2h), and were enriched for the “IFN-γ response” gene set (padj = 0.005, normalized enrichment score (NES)=2.88). Interestingly, a higher percentage of EGCs from infected mice were localised in EGC2 (46.99% versus 14.18%; Fisher exact test, p-value=2.28-07) (Fig. 1h), suggesting that EGC2 represents an activated cell state. Accordingly, EGC2 expressed higher levels of Gfap and were enriched for immune cell activation GO terms, including “response to IFN-γ” (Fig. 1i; Extended Data Fig. 2i, j). Therefore, the reactive state adopted by EGCs following H. poly infection is characterized by induction of IFN-γ gene signature.
Next, we analysed the transcriptomes of enteric glia from healthy colon (HC) and ulcerative colitis colon (UC) 8 . EGCs were distributed into two distinct clusters, hEGC1 and hEGC2 (Fig. 1j), which shared GO terms with mouse EGC1 and EGC2, respectively (Extended Data Fig. 2j, k). Virtually all EGCs from UC samples clustered in hEGC2 (Fig. 1k), suggesting that inflammatory gut pathology also leads to EGC activation. In support of this, the most significant term enriched in UC EGCs was “IFN-γ response” (padj=0.01, NES=2.98), while plotting “ IFN-γ response” genes onto the UMAP indicated increased response of UC EGCs to IFN-γ (Fig. 1l). Finally, EGCs from UC patients and helminth-infected mice shared IFN-γ module genes (Fig. 1m). Therefore, the IFN-γ response of EGCs to infectious and inflammatory gut pathologies is conserved in mammals and can be elicited throughout the gut.
IFN-γ-EGC signaling in tissue repair
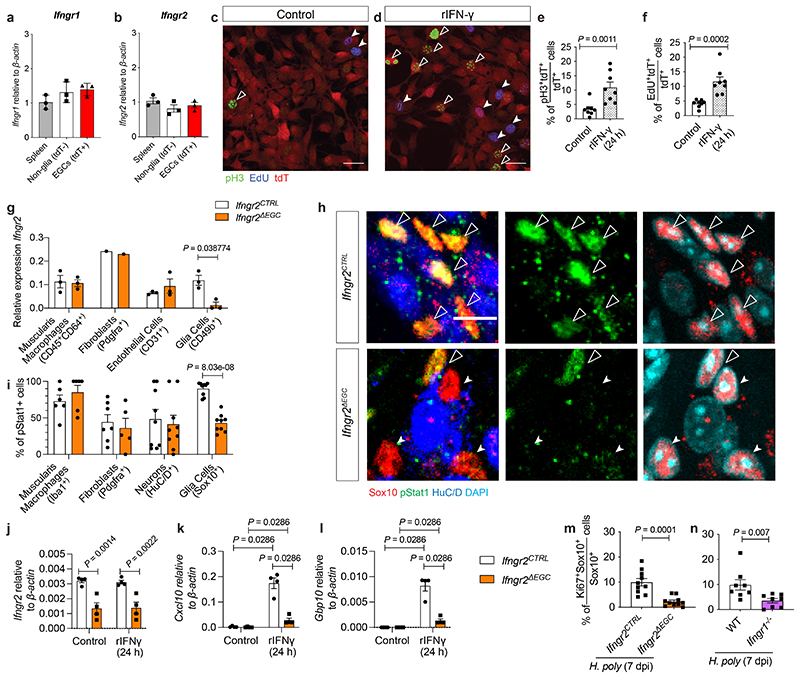
Next, we examined the cell autonomous effect of IFN-γ signalling on enteric glia. EGCs expressed Ifngr1 and Ifngr2 (which encode the IFN-γ receptor heterodimer 14 ) (Extended Data Fig. 3a, b) and were induced to proliferate in culture by IFN-γ (Extended data Fig. 3c-f). Also, by comparing the response of EGCs from Sox10CreERT2;Ifngr2fl/fl 15 (Ifngr2ΔEGC ) and Sox10CreERT2;Ifngr2fl/+ (Ifngr2CTRL ) mice to IFN-γ, we demonstrated that conditional ablation of Ifngr2 from enteric glia diminished the activation of STAT1 and the upregulation of Cxcl10 and Gbp10 (Extended data Fig. 3g-l). Finally, H. poly infection of Ifngr2ΔEGC and Ifngr1-/- 16 mice led to reduced proliferation of TM EGCs (Extended Data Fig. 3m, n). Therefore, helminth infection activates a novel IFN-γ-EGC signalling axis in the mammalian intestine.
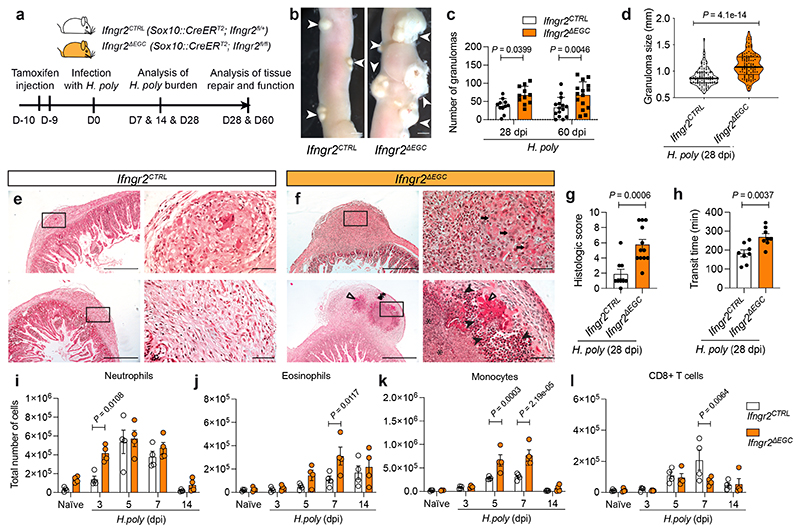
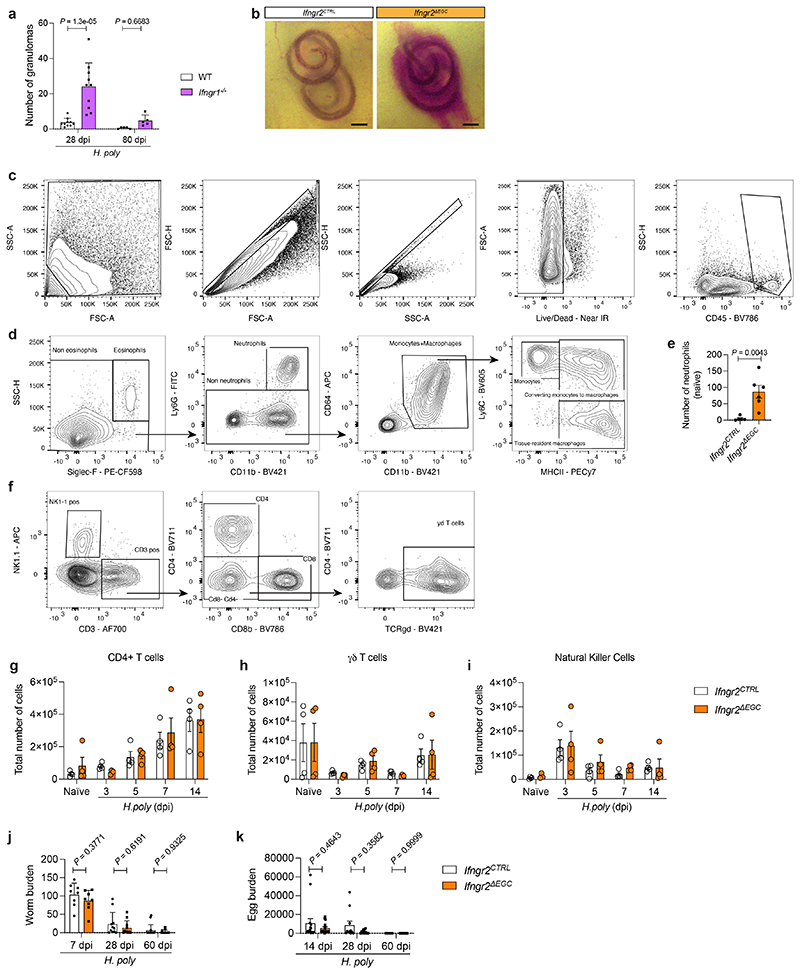
According to earlier reports which implicate IFN-γ-signalling in tissue repair 17,18 , we found that H. poly-infected Ifngr1-/- mice exhibited increased number and delayed resolution of granulomas (Extended Data Fig. 4a). To specifically examine the role of the IFN-γ-EGC signalling axis in helminth-induced inflammatory gut pathology, we infected Ifngr2CTRL and Ifngr2ΔEGC mice with H. poly (Fig. 2a). Strikingly, at 7 dpi, H. poly sites in Ifngr2ΔEGC mice were characterized by bleeding (Extended Data Fig. 4b), while at 28 dpi we recorded a higher number and size of granulomas, which persisted well after worms had migrated to the intestinal lumen (Fig. 2b-d). H. poly-infected Ifngr2ΔEGC were also characterised by pyogranulomatous enteritis and serositis and showed abnormal gut peristalsis (Fig. 2e-h). By comparing the immune cell profiles of TM from H. poly-infected Ifngr2ΔEGC and Ifngr2CTRL mice at 7 dpi, we detected increased accumulation of inflammatory cells in H. poly-infected Ifngr2ΔEGC mice (Fig. 2i-k; Extended data Fig. 4c, d). Interestingly, in contrast to eosinophils and monocytes, increased recruitment of neutrophils in Ifngr2ΔEGC gut was restricted to the early post-infection stages (Fig. 2i) and was evident even in uninfected animals (Extended Data Fig. 4e), suggesting a role of the IFN-γ-EGC signalling axis in immune homeostasis of TM. Notably, despite a comparable initial increase, CD8+ T cells were significantly reduced in H. poly-infected Ifngr2ΔEGC mice at 7 dpi (Fig. 2l), suggesting lack of cues that sustain this cell population. Finally, we detected no difference in the number of CD4+, γδT and NK cells or the worm or egg burden between the two genotypes (Extended Data Fig. 4f-k). Together, these analyses demonstrate that IFN-γ-mediated activation of EGCs in the TM regulates the immune response and repair of tissue damage associated with H. poly infection.
Figure 2. IFN-γ-signaling in EGCs spromotes intestinal tissue repair following helminth infection.
(a) Experimental strategy. (b-g) Characterisation of the granulomatous response of intestines from H. poly-infected Ifngr2CTRL and Ifngr2ΔEGC mice. Representative images of the small intestine at 28 dpi (b); quantification of the number (c, n28dpi=11, n60dpi=15) and size (d, n=11) of granulomas; representative H&E stained sections through granulomas (e, f; arrows: aggregates of macrophages; filled arrowheads: viable and degenerate neutrophils and eosinophils; empty arrowheads: dense eosinophilic fibrillar material, major basic protein; asterisks: basophilic karyorrhectic and eosinophilic cytoplasmic debris, necrosis); histology severity score (g, nCTRL=9, nΔEGC=12). Two experiments. (h) Intestinal transit time at 60 dpi; nCTRL=8, nΔEGC=7. Two experiments. (i-l) Flow cytometric quantification of neutrophils (i), eosinophils (j), monocytes (k) and CD8+ T cells (l) at indicated time-points in the TM of H. poly-infected Ifngr2ΔEGC and Ifngr2CTRL mice. (n=4, one experiment, data on day 7 representative of 3 experiments. Two-tailed Mann-Whitney test (d, g), unpaired two-tailed t-test (h), Two-way ANOVA with Sidak’s multiple comparison test (c, i-l). Mean±SEM (c, g, h, i, j, k, l). Scale bars: c: 0.5 cm, e, f: 500 μm, (insets: 50 μm).
Tissue-wide effects of IFN-γ-EGC axis
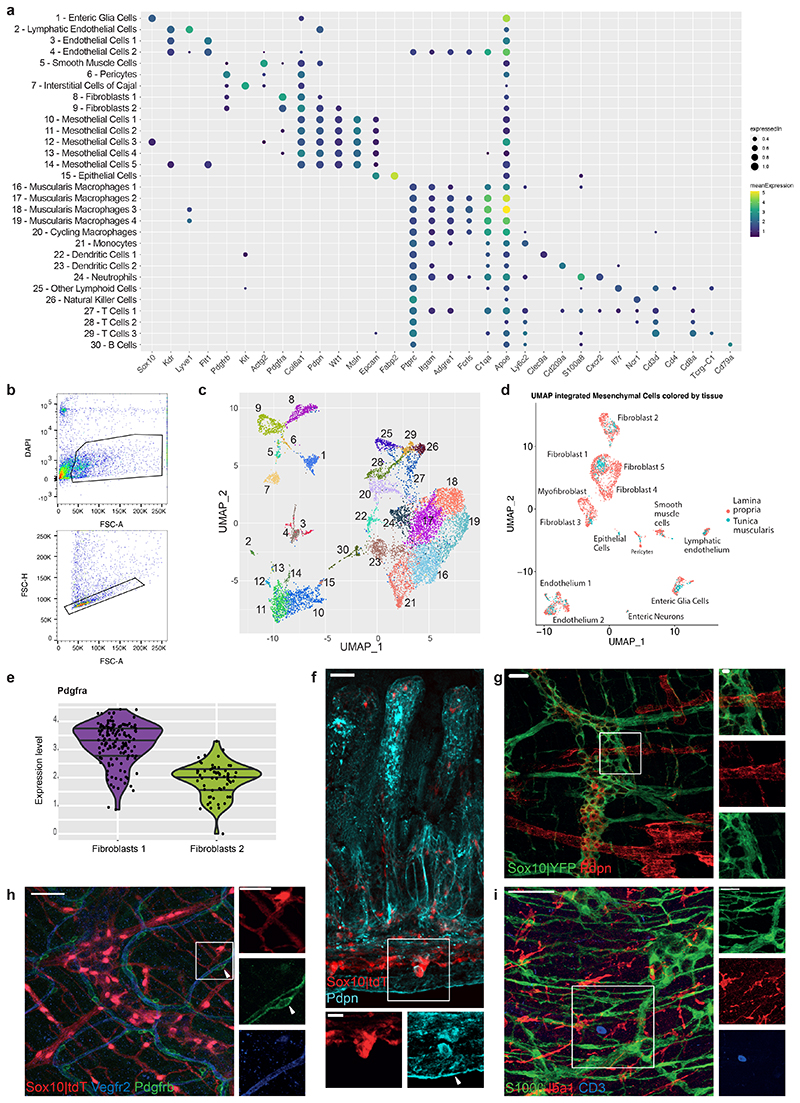
To understand mechanistically how the IFN-γ-EGC axis regulates the intestinal response to helminth infection, we sequenced 35733 cells from the TM of naïve and H. poly-infected (7 dpi) Ifngr2CTRL and Ifngr2ΔEGC mice (Fig. 3a and Extended Data Fig. 5a-c). Based on well-established markers, we identified EGCs (Sox10), smooth muscle cells (Actg2) and ICCs (Kit) (Extended Data Fig. 5a, c and Extended Data Fig. 1d). We also identified Pdgfrα+ fibroblasts with gene expression profile similar to their counterparts in the mucosa 19 (Extended Data Fig. 5a, c-e and Extended Data Fig. 1c), mesothelial cells (Wt1, Msln, Pdpn), which have key roles in peritoneal homeostasis 20 (Extended Data Fig. 5a, c, f) and cell clusters associated with the vascular network, such as lymphatic endothelial cells (Lyve1, Pdpn), endothelial cells (Kdr, encoding Vegfr2), pericytes (Pdgfrb), and various types of immune cells, such as microglia-like macrophages (Fcrls, C1qa and Apoe), monocytes, dendritic cells, NK cells and T cells (Extended Data Fig. 5a, c, g-i).
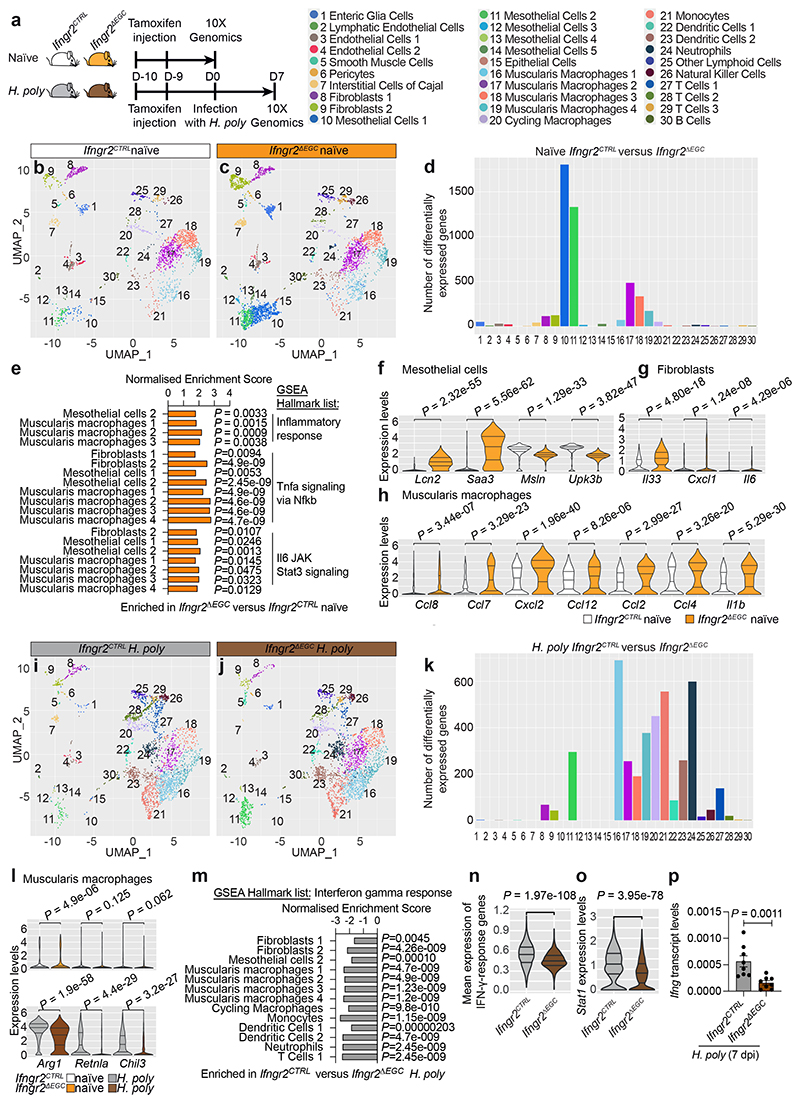
Figure 3. Tissue-wide regulation of immune homeostasis and immune responses in the TM by the IFN-γ-EGC signalling axis.
(a) Genotypes and experimental treatment of mice (color-coded) used for scRNAseq of the TM. (b, c) UMAP of TM cells from naïve Ifngr2CTRL (b) and Ifngr2ΔEGC (c) mice. Identity of cell clusters is indicated (left). (d) Quantification of differentially expressed genes in clusters from naïve Ifngr2CTRL versus Ifngr2ΔEGC mice. (e) GSEA showing selected significant hallmark list terms for inflammatory pathways in the indicated cell clusters from naïve Ifngr2CTRL and Ifngr2ΔEGC mice. (f-h) Violin plot quantification of selected differentially expressed genes in mesothelial cells (f), fibroblasts (g) and muscularis macrophages (h) from naïve Ifngr2CTRL and Ifngr2ΔEGC mice. (i, j) UMAP of TM cells from H. poly-infected Ifngr2CTRL (i) and Ifngr2ΔEGC (j) mice. (k) Quantification of differentially expressed genes in cell clusters from H. poly-infected Ifngr2CTRL versus Ifngr2ΔEGC mice. (l) Violin plot quantification of expression of Arg1, Retnla and Chil3 in muscularis macrophages from naïve and H. poly-infected Ifngr2CTRL and Ifngr2ΔEGC mice. (m) GSEA showing selected significant hallmark list term for IFN-γ response in the indicated cell clusters from H. poly-infected Ifngr2CTRL and Ifngr2ΔEGC mice. (n, o) Violin plot of mean scaled expression of IFN-γ response-associated genes (from the GO term “response to IFN-γ”; n) and Stat1 (o) across all UMAP clusters. (p) qRT-PCR of Ifng in H. poly-infected TM (7 dpi). Mean±SEM. n=8, two experiments. Wilcoxon test (f, g, h, l, n, o), two-tailed Mann-Whitney test (p).
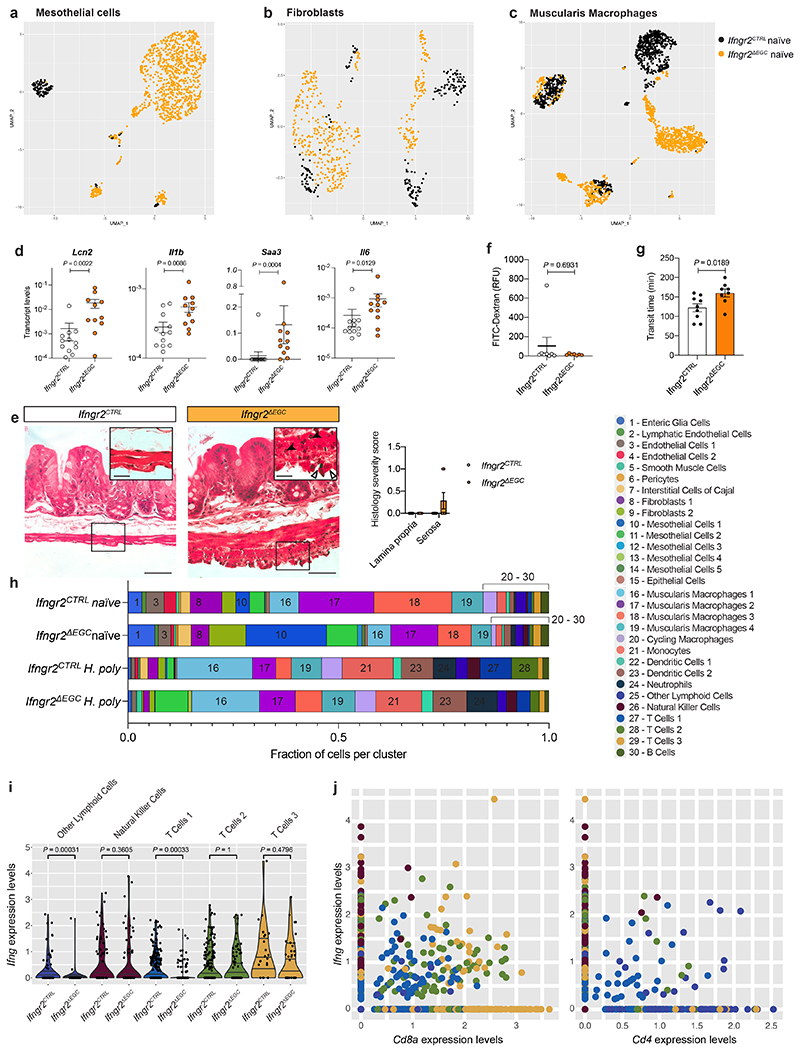
To understand the homeostatic roles of the IFN-γ-EGC signalling axis on the TM, we compared cell clusters from naïve Ifngr2CTRL and Ifngr2ΔEGC mice (Fig. 3b, c). Surprisingly, several clusters, such as mesothelial cells, macrophages and fibroblasts exhibited high numbers of differentially expressed genes (Fig. 3d) and were enriched for pro-inflammatory hallmark terms in Ifngr2ΔEGC mice (Fig. 3e). They also exhibited genotype-dependent shifts in UMAP position (Extended Data Fig. 6a-c) and upregulated genes encoding inflammatory markers and pro-inflammatory chemokines, such as Lcn2 and Saa3 in mesothelial cells, Il33, Cxcl1 and Il6 in fibroblasts and Ccl8, Ccl7, Cxcl2, Ccl12, Ccl4 and Il1b in muscularis macrophages (Fig. 3f-h; Extended data Fig. 6d). Furthermore, 25% (2/8) of naïve Ifngr2ΔEGC mice showed signs of inflammation in the TM, such as thickening of the muscular layer, serositis and activation of mesothelial cells (Extended data Fig. 6e). In contrast, mucosal histology and epithelial barrier function were normal (Extended data Fig. 6f). Ifngr2ΔEGC mice also showed significant increase in intestinal transit time (Extended data Fig. 6g). Finally, expression of the mesothelial cell differentiation markers Msln and Upk3b 20 was reduced in Ifngr2ΔEGC mice (Fig. 3f), suggesting mesothelial-to-mesenchymal transition associated with impaired tissue repair and fibrosis 21 . Therefore, selective abrogation of IFN-γ signalling in enteric glia is sufficient to activate pro-inflammatory programs in cell types critical for intestinal homeostasis and pathology.
To examine the role of IFN-γ-EGC signalling in the response of host tissue to helminths, we compared the cellular and transcriptional landscape of the TM from H. poly-infected Ifngr2CTRL and Ifngr2ΔEGC mice. As expected 10 , both sets of animals showed increased representation of diverse immune cell types (compare Fig. 3, i and j to b and c, respectively; Extended Data Fig. 6h) and induction of TH2 response genes (Arg1, Retnla and Chil3). However, the TH2 response in Ifngr2ΔEGC mice was markedly weaker relative to Ifngr2CTRL controls (Fig. 3l). Furthermore, GSEA analysis on clusters with the largest number of differentially expressed genes (Fig. 3k), showed that the hallmark term “interferon gamma response” was enriched in H. poly-infected Ifngr2CTRL mice (Fig. 3m), while Ifng expression in the TM and the mean expression of IFN-γ target genes, including Stat1, across UMAP clusters, were reduced in Ifngr2ΔEGC animals (Fig. 3n-p). The diminished IFN-γ response likely resulted from reduced expression of Ifng and reduced number of IFN-γ+ cell types, such as CD8+ T cells (Fig. 2l; Extended data Fig. 6i, j). Collectively, our analyses demonstrate that glia-specific ablation of IFN-γ signalling impairs the amplification of IFN-γ and tissue healing responses associated with helminth infection.
Cxcl10: an effector of IFN-γ-EGC axis
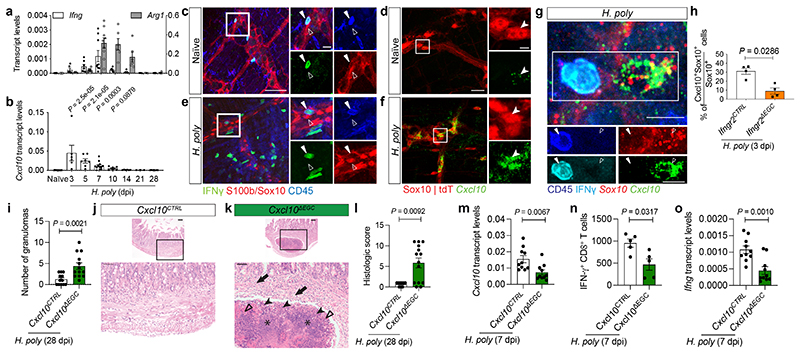
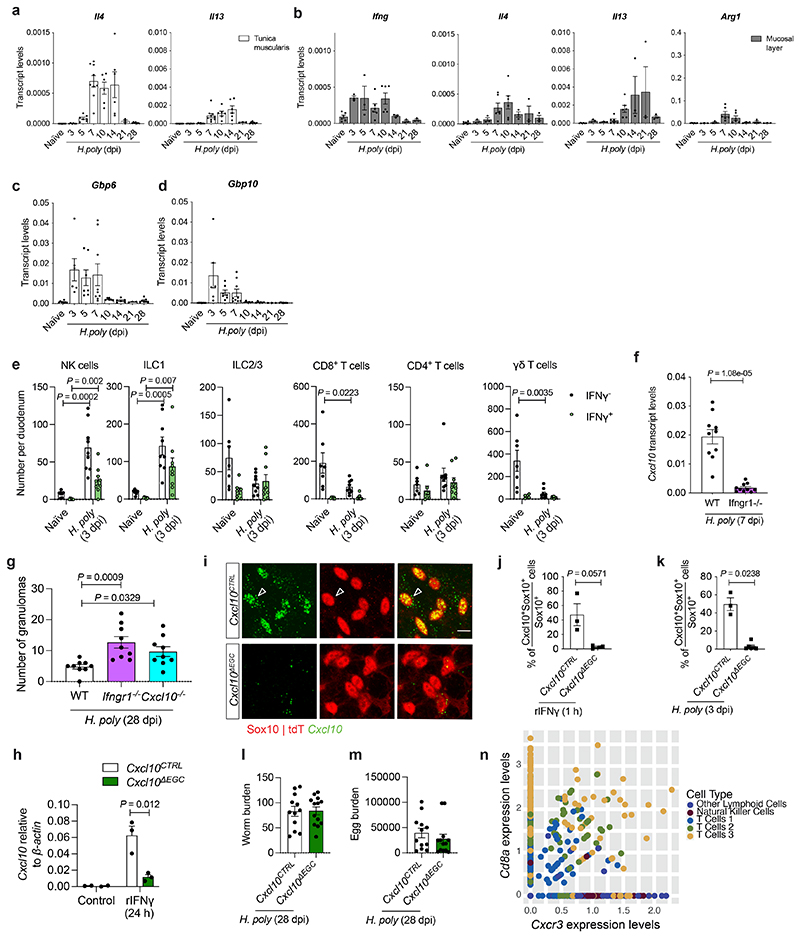
Our experiments so far suggest that IFN-γ-producing immune cells recruited in the TM shortly after helminth invasion activate effector mechanisms in EGCs that calibrate local immune responses and promote tissue repair. In support of this idea, H. poly-induced upregulation of Ifng in mucosa and TM preceded the induction of TH2 marker genes (Arg1, Il4 and Il13) (Fig. 4a and Extended Data Fig. 7a, b). In addition, upregulation of the IFN-γ targets Cxcl10, Gbp6 and Gbp10, had already peaked at 3 dpi (Fig. 4b and Extended Data Fig. 7c, d). At this state, the main sources of IFN-γ were NK cells (Extended data Fig. 4i) and ILC1 cells (Extended data Fig. 7e). Immunostaining of TM preparations from Yeti reporter mice 22 for GFP and CD45 showed that GFP+CD45+ cells were closely associated with the EGC network of the myenteric plexus (Fig. 4c). Interestingly, low levels of Cxcl10 expression was detected in a subset of Sox10+ EGCs (Fig. 4d), suggesting that TM glia respond to basal levels of IFN-γ in steady state animals. In contrast, H. poly infection led to recruitment of IFN-γ producing cells, many of which were embedded within the glial cell network (Fig. 4e), and upregulation of Cxcl10 in EGCs (Fig. 4f). The functional link between IFN-γ producing haematopoietic cells and EGCs was further supported by the requirement of IFN-γ receptor for the upregulation of Cxcl10 in TM (Extended data Fig. 7f), the close physical association between CD45+IFN-γ+ and Sox10+Cxcl10+ cells (Fig. 4g), and the significant reduction of Cxcl10+ EGCs in Ifngr2ΔEGC animals (Fig. 4h). To examine whether Cxcl10 is an effector chemokine downstream of the IFN-γ-EGC signalling axis promoting tissue healing in the context of helminth infections, we first analysed the small intestine of H. poly-infected mice homozygous for a null mutation of Cxcl10 (Cxcl10-/- ) 23 . Similar to Ifngr1-/- and Ifngr2ΔEGC mice, the number of granulomas at 28 dpi was higher in Cxcl10-/- mice relative to Cxcl10+/+ controls (Extended Data Fig. 7g), supporting a critical role of Cxcl10 in repair of H. poly-induced tissue damage. To test directly the role of EGC-produced Cxcl10, we used the Cxcl10fl allele 24 to generate Sox10CreERT2;Cxcl10fl/fl (Cxcl10ΔEGC ; carrying a conditional deletion of Cxcl10 in EGCs) and Sox10CreERT2;Cxcl10fl/+ (Cxcl10CTRL ; control) mice (Extended data Fig. 7h-k). Strikingly, similar to Ifngr2ΔEGC mice, the size and number of granulomas in H. poly-infected Cxcl10ΔEGC animals at 28 dpi was significantly higher relative to Cxcl10CTRL controls (Fig. 4i), while histological analysis revealed increased inflammatory infiltrates in Cxcl10ΔEGC mice (Fig. 4j-l). However, worm and egg burden were similar between genotypes (Extended data Fig. 7l, m). The similar granulomatous phenotype of Ifngr2ΔEGC and Cxcl10ΔEGC mice, together with the widespread expression of the Cxcl10 receptor-encoding gene Cxcr3 in IFN-γ-producing immune cells at 7 dpi (Extended data Fig. 7n), suggested that reduced levels of Cxcl10 in the TM of H. poly-infected Cxcl10ΔEGC mice (Fig. 4m) fail to sustain robust levels of IFN-γ production in the TM of H.poly-infected animals. In support of this idea, the number of IFN-γ+CD8+ cells and expression of Ifng were reduced in Cxcl10ΔEGC animals (Fig. 4n, o). Together, these findings demonstrate that Cxcl10 is part of the IFN-γ induced effector mechanisms that mediate the tissue healing effects of EGCs in the TM.
Figure 4. Early activation of IFN-γ-EGC-Cxcl10 signalling regulates tissue repair after helminth infection.
(a, b)qRT-PCR kinetics of Ifng and Arg1 (a) and Cxcl10 (b) in H. poly-infected TM. Mean±SEM, a: nnaïve==8, n3dpi,7dpi,10dpi,14dpi=6, n5dpi,28dpi=7, n21dpi=3; b: nnaïve=11, n3dpi=6, n7dpi=12. Two experiments. (c-f) TM from naïve (c, d) and H. poly-infected (3 dpi) (e, f) Yeti (c, e) and Sox10|tdT (d, f) mice immunostained for GFP, Sox10, S100β and CD45 (c, e) or hybridised for Cxcl10 (d, f). Combined and single-spectrum images of open squares shown on the right (c, e: empty and filled arrowheads indicate an EGC and an IFN-γ-producing GFP+ cell, respectively, n=4; d, f: arrowhead indicates an EGC; n=5). (g) Combined GFP/CD45 immunostaining and Sox10/Cxcl10 hybridisation of TM from H. poly-infected (3 dpi) Yeti mice. Combined and single-spectrum images of open squares at the bottom. (h) Quantification of Cxcl10+ TM EGCs of H. poly-infected (3 dpi) Ifngr2CTRL and Ifngr2ΔEGC mice; n=4. (i-l) Granulomatous response of H. poly-infected (28 dpi) intestine from Cxcl10CTRL and Cxcl10ΔEGC mice. Quantification of granulomas (i, nCTRL =12, nΔEGC=13); sections through granulomas (j, k; arrows: high numbers of neutrophils, eosinophils with scattered lymphocytes within the submucosa; filled arrowheads: viable and degenerate neutrophils and eosinophils; empty arrowheads: dense eosinophilic fibrillar material-major basic protein; asterisks: basophilic karyorrhectic and eosinophilic cytoplasmic debris-necrosis), histology severity score (l, nCTRL =9, nΔEGC=14). Two experiments. (m, o) Quantification of Cxcl10 (m) and Ifng (o) induction in H. poly-infected (7 dpi) TM of Cxcl10CTRL and Cxcl10ΔEGC mice (n=10; two experiments). (n) Quantification of TM IFN-γ+CD8+ T cells after H. poly-infection (7dpi); n=5. Representative of two experiments. Two-tailed Mann-Whitney test (h, l, n). Unpaired two-tailed t-test (i, m, o).Two-way ANOVA with Sidak’s multiple comparison (b). Mean±SEM (a, b, h, i, l-o). Scale bars: c, e: 15 μm (insets 5 μm);d, f: 50 μm (insets 10 μm); g: 10 μm; j, k: 100 μm (insets 50 μm).
Discussion
Collectively, our study demonstrates that IFN-γ signalling in enteric glia promotes immune homeostasis and tissue repair following pathogen-induced intestinal damage. Identification of the IFN-γ-EGC signalling module uncovers a fundamental regulatory mechanism in which immune signals maintain and restore tissue integrity by regulating the transcriptional output of a neuroectodermal lineage. Therefore, by monitoring the levels of IFN-γ at steady state, EGCs regulate healthy intestinal dynamics while inflammation-induced upregulation of this cytokine activates a cascade of immunomodulators that co-ordinate tissue repair. Although the full range of effector mechanisms is unknown, the demonstration that Cxcl10 is a critical mediator of tissue repair downstream of IFN-γ-induced activation of EGCs highlights the paracrine engagement of these cells with multiple immune cell lineages that are responsive to this and potentially other glia-derived cytokines. Together with recent studies 17,18 , our findings reveal a fundamental role of IFN-γ in coordinating tissue responses to pathogens throughout the gut wall. Many of the lineages downstream of the IFN-γ-EGC signalling axis (mesothelial cells, fibroblasts, immune cells) play crucial roles in the pathogenesis of gastrointestinal disorders, including inflammatory bowel disease and tumour formation 2–8 . This, together with the demonstration that EGCs from the colon of UC patients 8 share an IFN-γ response signature with helminth-infected mouse EGCs and the fact that GWAS and expression analysis have associated elevated Cxcl10 levels with increased inflammatory bowel risk 25,26 , suggests that mammalian enteric glia are implicated in the pathogenesis of common gastrointestinal disorders and thus represent a potential therapeutic target.
Methods
Data reporting
No statistical methods were used to predetermine sample size. Wherever possible investigators were blinded to allocation during experiments and outcome assessment. Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Mice
All mouse procedures were carried out at The Francis Crick Institute in accordance with the regulatory standards of the UK Home Office (ASPA 1986) and the ARRIVE guidelines and approved by the local Animal Welfare and Ethics Review Body (AWERB).
Mice were appropriately bred and housed under specific pathogen-free (SPF) conditions in individually ventilated cages on a 12/12 hour light/dark cycle at ambient temperature (19°C-21°C) and humidity (45-55 %). Standard food and water were provided ad libitum. For all experiments, mice were randomly assigned to control and treatment groups, which were coded until the experiment was finished and data was analyzed. Control and treatment groups were co-housed and littermate controls were used. Age-matched mice aged between 2 and 4 months were used for each experiment and cohorts were of mixed genders. C57BL/6J wild-type (WT), B6.Ifngr1tm1Agt (Ifngr1-/- , MGI: 1857286 16 ), B6.129S4-Cxcl10tm1Adl/J (Cxcl10-/- , MGI: 2180678 23 ) and B6.129P2-Ifngtm1Lky (YFP-enhanced transcript for IFN-γ, Yeti, MGI: 3665254 22 ) were used. The genetic background of other lines used in this study is considered mixed: Sox10CreERT2 ;Rosa26tdTomato (B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze, MGI:3809524), Ifngr2CTRL (Sox10-CreERT2;Ifngr2+/fl , Ifngr2tm1.1Lflu , MGI: 6402706 15 ) and Ifngr2ΔEGC (Sox10-CreERT2;Ifngr2fl/fl ) combined with Gt(ROSA)26Sortm1(EYFP)Cos (MGI: 2449038) or Rosa26tdTomato mice. Cxcl10ΔCTRL (Sox10-CreERT2;Cxcl10+/fl , Cxcl10tm1.1Dple, MGI: 6357982 24 ) and Cxcl10ΔEGC (Sox10-CreERT2;Cxcl10fl/fl ) combined with Rosa26tdTomato. Genotyping was carried out by Transnetyx.
Cre-mediated allele recombination
Labelling of EGCs with tdTomato and conditional knock-out of Ifngr2 and Cxcl10 using the Sox10-CreERT2 driver was performed using intraperitoneal injections or oral gavage of a 20 mg/mL tamoxifen (Sigma, T5648) solution in corn oil (Sigma, C8267) and ethanol at a ratio of 9:1. Animals were injected on two consecutive days or gavaged on three consecutive days with 0.1 mg/g tamoxifen per body weight 10-14 days prior to analysis of naïve animals or H. poly infection.
H. poly infection
Mice were infected with 200 H. poly third-stage (L3) larvae by oral gavage (200 μl), which were obtained from fecal cultures of H. poly-infected mice. H. poly egg counts were performed on fecal pellets using a McMaster 2 cell counter (Hawksley). The number of adult worms in the intestinal wall at 7 dpi, the number of adult worms in the intestinal lumen and the number of macroscopically visible granulomas were counted in a blinded manner. The size of granulomas per animal was averaged from ≥ 10 granulomas per intestine.
Total transit time assay
Mice were individually placed into cages devoid of bedding and fasted for 1 hour before oral gavage with 300 μL of 6% (w/v) carmine red dye (Sigma-Aldrich, C1022-25G) and 0.5% (w/v) methylcellulose (Sigma, M0512-500G) in dH20. Animals were returned to their individual cages with access to water and food. The total intestinal transit time was calculated by the time from gavage until the extrusion of the first red-coloured pellet.
Intestinal paracellular permeability assay
Mice were fasted for 4 hours and then administered 600 μg/g per body weight fluorescein isothiocyanate-conjugated (FITC)-dextran (Sigma-Aldrich, 46944) by oral gavage before culling. The concentration of FITC dextran in plasma was determined with a TECAN Spark Multimode Microplate Reader and represents a measure for paracellular permeability of the intestinal epithelium.
Isolation of the tunica muscularis (TM)
The small intestines from Sox10|tdT mice were dissected in ice-cold PBS, and the TM was removed by peeling as previously described 11,27 . The remainder of the tissue containing the lamina propria and the epithelial layer was termed mucosal layer.
Immunohistochemistry of the TM and cultured primary EGCs
Adult TM preparations were fixed for 2 hours in 4% paraformaldehyde at 4°C. Whole gut segments were fixed overnight in 4% paraformaldehyde at 4°C. EGC cultures were fixed for 30 mins at room temperature (RT). After fixation, samples were washed three times in PBS at RT and processed for immunohistochemistry. Whole gut segments were embedded in 4% low-melting temperature agarose (Invitrogen) and transversal sections of 60 μm were obtained with a Leica vibratome. Samples were blocked with 10% donkey serum/1% Triton-X100 for TM samples and 10% donkey serum/0.3% Triton-X100 for cultured cells for 2 hours at room temperature before being incubated in primary antibodies overnight at RT, washed three times with 1% donkey serum/0.3% Triton-X100 and incubated in secondary antibodies at RT. For anti-Ki67 antibody staining an antigen-retrival step for 10 minutes at 80°C in 10 mM sodium citrate was performed prior to blocking and staining. For pStat1 staining, samples were treated with ice-cold methanol for 10 minutes followed by three washes in PBS prior to blocking and staining. Primary antibodies rabbit anti-pStat1 (Tyr701) (1:400, Cell Signaling Technology, 9167), chick anti-GFP (1:300, Abcam, ab13970), rat anti-CD45 (1:100, Biolegend, 103101), rabbit anti-CD45 (1:100, Abcam, ab10558), rat anti-Ep-Cam Biotin (1:100, Biolegend, 118212), goat anti-cKit (1:100, R&D Systems, AF1356), rat anti-CD31 (1:100, BDPharmingen, 550274), mouse anti-HuC/D (1:400, Life Technologies, A-21271), mouse anti-Ki67 (1:200, BD Biosciences, 550609); goat anti-Sox10 (1:200, Santa-Cruz Biotechnology, sc17342); rabbit anti-Sox10 (1:200, Proteintech, 10422-1-AP), chicken anti-GFAP (1:500, Abcam, ab4674), rabbit anti-S100β (1:500, Proteintech, 15146-1-AP), rabbit anti-S100 (1:500, DAKO, z0311); rabbit phosphor histone 3 (1:500, Millipore, 06-570), goat anti-Iba1 (1:200, Abcam, Ab5076), Armenian hamster anti-Cd3-ε (1:200, Santa Cruz, sc-18871), goat anti-Pdgfra (1:200, R&D Systems, AF1062), mouse anti-Vegfr2 (1:200, & Systems, AF644), rat anti-Pdgfrb (1:100, eBioscience, clone APB5, 14-1402-81), Syrian hamster anti-Pdpn PE (1:50, Biolegend, clone 8.1.1, 127407), Syrian hamster anti-Pdpn purified (1:50, Biolegend, clone 8.1.1, 127403) and secondary antibodies donkey anti-chicken Alexa Fluor (AF) 488 (1:500, Jackson Immuno Research, 703-545-155), donkey anti-mouse AF 568 (1:500, Invitrogen, A10037); goat anti-rat (1:500, Invitrogen, A-11077), donkey anti-mouse AF 647 (1:500, Invitrogen, A-31571), donkey anti-goat AF 647 (1:500, Invitrogen, A21447); donkey anti-goat AF 405 (1:500, Invitrogen, A48259), donkey anti-rabbit AF 488 (1:500, Invitrogen, A21206), donkey anti-goat AF 568 (1:500, Invitrogen, A-11057), donkey anti-hamster AF 647 (1:500, Invitrogen, A21451), donkey anti-rat AF 488 (1:500, Invitrogen, A21208), donkey anti-rat AF 647 (1:500, Invitrogen, A48272), streptavidin 405 (1:300, Invitrogen, S32351); were diluted in 1% donkey serum/0.3% Triton-X100 for TM samples and 1% donkey serum/0.1% Triton-X100 for cultured EGCs. Cultured EGCs were co-stained with the Click-iT EdU Alexa Fluor 647 kit according to the manufacturer’s instructions (Thermofisher, C10340). After staining, tissues were extensively washed in 0.3% PBT containing 1 μg/ml 4',6-Diamidino-2-Phenylindole (DAPI, Thermo Fisher Scientific, D1306) and flat-mounted onto glass slides using Vectashield™ mounting medium (Vector Labs, H-1200).
Fluorescence in situ hybridization
Fluorescence in situ hybridization on the duodenal myenteric plexus was carried out using the Advanced Cell Diagnostics RNAscope® Fluorescent Multiplex Kit (ACD, 320850) according to the manufacturer’s instructions. After fixation O/N, the TM layer was dehydrated by serial ethanol treatments and treated with RNAscope® Protease III for 30 min at RT. Tissue was then incubated with fluorescent probes, 3-Plex Positive Control Probe, 3-Plex Negative Control Probe or customized probes (RNAscope® Probe - Mm-Cxcl10, 408921, RNAscope® Probe - Mm-Sox10, 435931-C3) at 40°C O/N. Following hybridization, tissue was washed twice with wash buffer and then subjected to sequential hybridization with pre-amplifier, amplifier DNA (Amp1-FL, Amp 2-FL and Amp 3-FL) and fluorophore (Amp 4 Alt A-FL) at 40°C for 30 min, 15 min, 30 min and 15 min, respectively. After hybridization, tissues were counterstained with DAPI and mounted on Superfrost Plus Adhesion Microscope Slides (ThermoFisher Scientific, 10149870) using VectaMount Permanent Mounting Medium (ACD, 321584).
Histopathology
For histopathological analysis, tissue samples were fixed in 10% neutral buffered formalin, embedded in paraffin wax, sectioned at 4 μm, and stained with haematoxylin and eosin (H&E). Histopathological examination was performed in a blinded manner by a board-certified veterinary pathologist (ASB). Sections from small intestine were examined and in each case a descriptive and quantitative assessment of the granulomatous inflammation present was provided. For analysis of naïve Ifngr2CTRL and Ifngr2ΔEGC animals after tamoxifen induction the following score was used to assess inflammation in the lamina propria/ mucosa (epithelium hyperplasia and goblet cell depletion: 0 = not present; 1 = mild; 2 = moderate; 3 = severe; lamina propria inflammation: 0 = not present; 1 = mild; 2 = moderate; 3 = severe; affected area: 0 = none, 1 = up to 25%, 2 = 25-50%, 3 = more than 50%) and the TM (serosal inflammation, 0 = not present; 1 = mild; 2 = moderate; 3 = severe) while for animals at 28 days after H. poly infection the following criteria to assess granulomatous inflammation were scored (Inflammation: 0 = not present; 1 = mild; 2 = moderate; 3 = severe; presence of granulomas: 0 = not present; 1 = mild; 2 = moderate; 3 = severe; size of granulomas: 0 = not present; 1 = mild; 2 = moderate; 3 = severe; necrosis, 0 = not present, 1 = present).
Microscopy and image analysis
Confocal images were acquired using an upright Olympus Confocal Laser Scanning Microscope FV3000 run by the FV31S-SW software using standard excitation and emission filters for visualizing DAPI, Alexa Fluor 488, Alexa Fluor 568, and Alexa Fluor 647. Images were processed using Fiji ImageJ (2.0.0-rc-68/1.52h) 28 . Cells were manually counted by confirming the presence of DAPI in single z-stacks per acquired image for whole mount TM layers and in maximum projections for EGC cultures using the Cell Counter plugin in Fiji ImageJ software. For H. poly infection experiments, 3 images of random areas within a 1000 μm zone bordering the site of H. poly settlement were acquired per animal, while for cultured EGCs 8 random images were acquired per well. Changes in glial morphology (10 cells were traced) were quantified using Z-stacks of confocal images of S100-immunostained preparations of the TM with the FIJI- Image J Simple Neurite Tracer plugin as previously described 29 . The total process length was quantified by the sum of the length of individual paths, process thickness was estimated from S100β-thickness provided by “fill out” analysis function, and morphology complexity was estimated from the number of intersections at each radial distance from the starting point provided by the Sholl analysis. Final figures were composed using Adobe Illustrator CC (Adobe Systems).
Cell culture of whole gut tissue and primary EGCs
For culture of whole gut tissues, small intestine regions were dissected, cut into 2 cm pieces and cultured in DMEM/F12 supplemented with penicillin-streptomycin and 10% FBS and cultured at 37°C for 1 hour in the presence of IFN-γ (rIFN-γ, 100 ng/ml, R&D Systems, 485-MI). Tissues were then removed, washed and the TM isolated and fixed. For isolation of primary EGCs, The TM was isolated, minced and dissociated as previously described, with some modifications 11 . Briefly, isolated TM samples were washed thoroughly in DMEM/F12 media containing 2% FBS (Sigma-Aldrich), 100 U/ml Penicillin/Streptomycin (P/S) (Thermo Fisher Scientific) and 12.5 mM HEPES (Thermo Fisher Scientific). Washed TM samples were then minced using fine scissors and digested in the buffer as described above containing 0.5 mg/ml Type 1 collagenase (Sigma Aldrich) and 0.5 mg/ml DNase I (Sigma-Aldrich), incubated in a shaker at 37°C for 30 mins. Following, trituration of the digest using an unpolished glass pasteur pipette was carried out. Triturated samples were then washed twice with DMEM/F12 media containing 10% FBS with P/S before seeding. Samples derived from the entire length of a mouse’s small intestines were used to seed two 60 mm (Corning) dishes coated with fibronectin (20 μg/ml; Sigma-Aldrich) and cultured overnight in DMEM/F12 media containing 10% FBS and P/S. On the next day, the culture media was switched to serum-free media made up of: DMEM/F12 containing 1% N2 supplement (Thermo Fisher Scientific), 1% G5 supplement (Thermo Fisher Scientific) and 50 ng/mL NGF 7s (Thermo Fisher Scientific). Half of the proliferative media were exchanged with fresh media on day 3 to ensure robust proliferation of glial cells. On day 4, cells were trypsinized using TrypLE express (Thermo Fisher Scientific), followed by the removal of remaining tissue clumps using a 40 μm cell strainer (Thermo Fisher Scientific) to obtain a single cell suspension. tdTomato positive cells were FACS sorted and seeded at 40,000 cells per well in a 48-well format, pre-coated with fibronectin for subsequent experiments. Cells were treated with recombinant IFN-γ (rIFN-γ, 100 ng/ml, R&D Systems, 485-MI). In some cases, cultured cells were exposed to 5-Ethynyl-2'-deoxyuridine (EdU) at 10 μM for an hour before fixation to label proliferating cells. Cells were consistently maintained at an atmosphere of 5% CO2.
Isolation of RNA and real-time PCR
RNA from TM and submucosal preparations, FACS-isolated EGCs, cultured EGCs and spleen was extracted using Trizol™-LS (Thermofisher, 10296010) and the PureLink™ RNA Micro Kit (Life Technologies, 12183016) according to the manufacturer’s instructions. cDNA was synthesized using the High-Capacity cDNA Archive Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. qRT-PCR was performed with 2% of complementary DNA (cDNA) generated using Taqman fast universal 2 x PCR Master Mix (Thermo Fisher Scientific) and Taqman primer and probes assays (all Thermo Fisher Scientific) for: β-actin (Mm02619580_g1), Gfap (Mm01253033_m1), Ifng (Mm00801778_m1), Il4 (Mm00445259_m1), Il13 (Mm00434205_g1), Cxcl10 (Mm00445235_m1), Arg1 (Mm00475988_m1), Ifngr1 (Mm00599890_m1), Ifngr2 (Mm00492626_m1), Ifngr2 (Mm01210592_m1), Lcn2 (Mm01324470_m1), Gbp6 (Mm00843395_m1), Gbp10 (Mm03647514_m1), Il6 (Mm00446190_m1), Il1b (Mm01336189_m1), Saa3 (Mm00441203_m1). qRT-PCR reactions were reactions were analysed using an Applied Biosystems Quantstudio 7 Flex Real-Time PCR System. Ct values obtained are normalized to β-actin and are either calibrated to the median control sample for relative quantification or presented as relative expression using the standard ΔΔCt method.
ELISA analysis
TM samples were prepared as described above and cultured in DMEM/F12 supplemented with penicillin-streptomycin and 10% FBS and cultured at 37°C overnight. Interferon-γ (IFN-γ) in supernatants was measured by ELISA (Quantikine® ELISA, R&D Systems, Inc) according to the manufacturer’s instructions. The optical density was read at 450 nm on a microplate reader (Bio-Rad).
TM tissue digestion and fluorescence activated cell sorting (FACS)
TM preparations were dissociated into single cell suspensions using two different protocols. For EGC isolation for qRT-PCR, bulk and scRNAseq, TM preparations were digested for 15 mins at 37°C with HBSS (Gibco, 14170112) containing 10 mg/mL collagenase IV (Worthington Biochemicals, CLS-4) followed by a wash step in ice-cold HBSS and subsequent incubation with 1U papain (Worthington Biochemicals, PAP2) for 5 mins at 37°C. Samples were then washed and resuspended in L15 medium without phenol red (Gibco, 21083027) containing 1% P/S (Gibco, 15140122), 1 mg/ml BSA (Sigma, A9418), 10 mM Hepes pH 7.4 (Gibco, 15630106), 10 % Biowhittaker water (Lonza, BE17-724F) and 400 U DNAse I (Grade II; Roche, 10104159001), filtered through a 70 μm and a 40 μm strainer and subjected to FACS on a FACS Aria (BD Biosciences) using DAPI (1 μg/ml) for live/ dead cell discrimination. For qRT-PCR and bulk sequencing, cells were directly sorted into Trizol™-LS. For isolation of TM cell populations for scRNAseq and for flow cytometry analysis of immune cell populations, TM preparations were digested for 20 mins at 37°C with HBSS containing 0.075 mg/mL Liberase TL (Sigma, 5401020001) followed by manual dissociation using pipetting. Samples were washed, filtered through a 100 μm strained and either directly subjected to FACS using DAPI (1 μg/ml) for live/ dead cell discrimination and sorted into Trizol™-LS or incubated with antibodies for immunophenotyping.
Flow cytometry/immunophenotyping
For the immunophenotyping, TM preparations were dissociated using Liberase TL as described above. Single cell suspensions were then counted and pre-incubated with antibodies against: CD16/CD32 for FcγR-blocking (1:200; clone 2.4G2, BD Bioscience, 553142), followed by incubation with different antibody panels. Innate immune cell panel: Live/Dead marker (1:1000, Thermofisher Scientific, L10119), CD64 (1:200, APC, clone X54-5/7.1, BD Bioscience, 558539), MHCII (1:300, PE-Cy7, clone M5/114.15.2, eBioscience, 25-5231-80), Ly6C (1:200, BV605, clone HK1.4, Biolegend, 128035), Ly6G (1:200, FITC, clone 1A8, BD Bioscience, 551460), CD11c (1:200, PE, clone N418, eBioscience, 12-0114083), CD45 (1:300, BV786, clone 30-F11, BD Bioscience, 564225), CD11b (1:200, BV421, clone M1/70, Biolegend, 101235), Siglec-F (1:100, PE-CF598, clone E50-2440, BD Bioscience, 562757). NK/ T-cell panel: Live/Dead marker (1:1000, Thermofisher Scientific, L10119), CD8b (1:200, BV786, clone H35-17.2, BD Bioscience, 740952), CD4 (1:200, BV711, clone RM4-5, Biolegend, 100594), CD3 (1:100, AF700, clone 17-A2, Biolegend, 100216), TCR γ/δ (1:100, BV421, clone GL3, Biolegend, 118119), CD8a (1:200, FITC, clone 53-6.7, eBioscience, 11-0081-85), CD8b (1:200, BV786, H35-17.2, 740952), CD45.2 (1:200, PE/Cy7, clone 104, Biolegend, 109830), Nkp46 (1:200, PE, clone 29A1.4, Biolegend, 137604), NK1.1 (1:100, APC, clone PK136, Biolegend, 108710). Intracellular NK/ ILC/T-cell panel: Live/Dead marker (1:1000, Thermofisher Scientific, L10119), CD45 (1:200, AF700, clone 30-F11, Biolegend, 103128), TCR β (1:100, FITC, clone RR8-1, BD Bioscience, 553277), TCR γ/δ (1:100, BV421, clone GL3, Biolegend, 118119), CD4 (1:200, BV786, clone RM4-5, BD Bioscience, 563727), Cd8a (1:200, PECy7, clone 53-6.7, Biolegend, 100722), NK1.1 (1:100, APC, clone PK136, Biolegend, 108710), CD127 (1:100, BV605, clone A7R34, Biolegend, 135025), Thy1.1/CD90 (1:100, BV650, clone OX-7, Biolegend, 202533), CD3 (1:100, Biotin, clone 145-2C11, Biolegend, 100304), CD11c (1:100, Biotin, clone N418, Biolegend, 117304), CD11b (1:100, Biotin, clone M1/70, Biolegend, 101203), CD19 (1:100, Biotin, clone 6D5, Biolegend, 115504), CD5 (1:100, Biotin, clone 53-6.7, Biolegend, 100704), TER-119 (1:100, Biotin, clone TER-119, Biolegend, 116204), Gr1 (1:100, Biotin, clone RB6-8C5, Biolegend, 108404), IFNγ (1:100, PE, clone XMG1.2, BD Bioscience, 554412), T-bet (1:100, clone o4-46, BD Bioscience, 563320). Mesenchymal-/Immune cell panel: Live/Dead marker (1:1000, Thermofisher Scientific, L10119), CD45 (1:200, BV786, clone 30-F11, BD Biosciences, 564335), CD64 (1:200, PE-Cy7, clone X54-5/7.1, Biolegend, 139314), CD31 (1:200, BV421, clone 390, Biolegend, 102424), Pdgfra (1:200, APC, clone APA5, Biolegend, 135908), Cd49b (1:200, FITC, clone DX5, Invitrogen, 11-5971-82). The stained samples were analyzed using a FACS FORTESSA (BD Bioscience), and the obtained raw data were further analyzed using FlowJo software version 10.3 and 10.7 (Tomy Digital Biology). Different cell populations were identified by the following general gating strategy: single cell selection (FSC-A vs FSC-H and SSC-A vs SSC-H), dead cell discrimination (Live/Dead cell marker-) and hematopoietic cell selection (CD45+) followed by the expression of the following surface markers: eosinophils (CD11b+, Siglec-F+, SSC-H), neutrophils (CD11b+, Siglec-F-, Ly6G+), monocytes (CD11b+, MHCII-, Ly6C+), tissue-resident macrophages (CD11b+, CD64+, MHCII+, Ly6C-), NK cells (CD3-, NK1.1+, Nkp46+), CD4 T-cells (CD3+, CD4+), CD8 T-cells (CD3+, CD8a+, CD8b+), TCR γ/δ T-cells (CD3+, TCR γ/δ+). For quantification of IFN-γ+-cells in Extended Data Fig. 7e, cells were gated on single, live CD45+ cells, gated on IFN-γ- and IFN-γ+-cells, followed by gating on γ/δ T-cells (TCR γ/δ+), CD4 T-cells (TCR β+, CD4+), CD8 T-cells (TCR β +, CD8+), NK cells (TCR γ/δ-, TCR β-, Nk1.1+, Thy1.2-), ILC1 (TCR γ/δ-, TCR β-, NK1.1+, Thy1.2+) and ILC2/3 (TCR γ/δ-, TCR β-, Nk1.1-, Thy1.2+). Mesenchymal-/Immune cell panel: after single cell selection and dead cell discrimination the following populations were isolated using FACS: muscularis macrophages (CD45+, CD64+), fibroblasts (Pdgfra+), endothelial cells (CD31+), EGCs (CD45-, CD49b+).The total number of cells was estimated either by the cell count multiplied by the percentage of live cells or using CountBrightTM Absolute Counting Beads (Invitrogen, C36950).
RNA-sequencing
Bulk sequencing
RNA from FACS-sorted EGCs was isolated using PureLink RNA Micro Kit (Invitrogen, 12183016) according to the manufacturer’s instructions. Total RNA samples were converted to cDNA libraries using Ovation RNA-Seq System V2 (NuGEN Technologies -7102-A01) according to manufacturer’s instructions. After the amplification procedure Qubit dsDNA HS Assay kit (ThermoFisher - Invitrogen - Q32854) was used to measure the cDNA concentration. Sample volume of 15 μl of SPIA cDNA was sheared to a size of 200 bp by sonication (Covaris – S220/E220 Focused-ultrasonicator). The Ovation Ultralow System V2 1-16 (NuGEN - PN: 0344NB-A01) was used to produce libraries from fragmented double stranded cDNA. Then the fragment ends are repaired before ligation of sequencing adaptors, followed by PCR amplification and library purification. The quality and the molarity of the libraries were checked using Agilent TapeStation, (HS D1000 Reagent - 5067-5585, HS D1000 Screentape - 5067-5584). The final libraries were pooled and sequenced on the HiSeq 4000.
Fluidigm scRNAseq
We isolated cells from five biological samples (2x naïve and 3x H. poly infected animals (7 dpi)) from both male and female Sox10|tdT adult mice at 24 hours after tamoxifen induction. TM samples and isolation of tdT+ cells using FACS were prepared as described above. The quantity and quality of a single cell suspension was assessed using the Eve Cell Counter and trypan blue. 200-1000 cells were loaded into a Fluidigm C1 Single Cell Auto Prep IFC for mRNA-seq for use with the Fluidigm C1 system. After the cell load program cells were imagined using the JuliStage to identify wells that contain 1 single cell. The SMART-Seq v4 Ultra Low Input RNA Kit for the Fluidigm C1 System, IFCs was used to reverse transcribe and amplify cDNA. cDNA was quantified using the Glomax (Promega). Libraries were prepared from cDNA generated from single cells using the Nextera XT kit from Illumina. Final libraries were pooled and sequenced on the HiSeq 4000.
10x scRNAseq
We isolated cells from two experimental replicates (1: one mouse per sample, 2: a pool of three mice per sample) for each condition (naïve Ifngr2CTRL , naïve Ifngr2ΔEGC , 7 days of H. poly infection Ifngr2CTRL, 7 days of H. poly infection Ifngr2 ΔEGC ) from both males and females at 10 days after tamoxifen induction. TM samples and isolation of live (DAPI-) cells using FACS were prepared as described above. Cells were quantified using the Eve Automatic Cell Counter and trypan blue. Approximately 10 000 cells were loaded into a 10x Chromium Chip G (10x 3’ v3.1) and partitioned into GEMs using the 10x Chromium Controller. cDNA was generated using the manufacturers protocol and quantified using the Agilent TapeStation (D5000 ScreenTape). Libraries were prepared according to the manufacturers guidelines and QC’d using the Agilent TapeStation and the High Sensitivity dsDNA Qubit. The final libraries were pooled and sequenced using the HiSeq 4000.
Bioinformatic analysis
Bulk RNAseq
For the bioinformatics analysis, the ‘Trim Galore!’ utility version 0.4.2 was used to remove sequencing adaptors and to quality trim individual reads with the q-parameter set to 20. The sequencing reads were then aligned to the mouse genome and transcriptome (Ensembl GRCm38 release-89) using RSEM version 1.3.0 in conjunction with the STAR aligner version. Sequencing quality of individual samples was assessed using FASTQC version 1.11.5 and RNA-SeQC version 1.1.8. Differential gene expression was determined using the R-Bioconductor package DESeq2 version 1.24.0.
Despite the high degree of enrichment of EGCs-specific genes in RNAseq samples isolated from tdT+ populations (Extended Data Fig. 2c), we found a small increase in some immune cell gene contaminants (Ptprc, Itgam), likely reflecting nonspecific adherence of some immune cells to EGCs during FACS isolation. Importantly, there was a significant increase in these transcripts after H. poly infection (log2(fold change EGCs H. poly versus naïve): Ptprc=3.659, Itgam=2.926; P-valueadj.(EGCs infected versus naïve): Ptprc=6.12-9, Itgam=1.26-5), possibly due to the overwhelming immune response associated with these treatments. We devised a selection strategy that we applied to EGC-RNAseq samples isolated from tdT+ populations, to determine genes that were specifically induced in EGCs and that could not be accounted for by potential immune cell contamination: log2(fold change EGCs treated versus untreated) > 1.5 & P-valueadj.(EGCs treated versus untreated) < 0.05 & log2(fold change EGCs treated versus Non-EGCs treated) > -1. Predicted mouse IFN targets were determined using the related GO term "response to IFN-γ" (GO:0034341) and the website Interferome (http://interferome.its.monash.edu.au/interferome) with default settings.
scRNAseq (Fluidigm)
Nextera adapter sequences were removed from reads using Cutadapt 30 . Reads were aligned to the Ensembl GRCm38 genome using STAR 31 . Gene counts were obtained using the featureCounts program from Subread. Quality control, normalisation and downstream analysis were performed using the software Seurat 32,33 . Cells expressing more than 2000 genes and less than 10 percent mitochondrial reads were retained for further analysis. Contaminating macrophages (n=7) were removed based on the expression of Itgam (> 10 counts), and contaminating neurons (n=95) were removed based on expression of Elavl4 (> 30 counts). Genes detected in fewer than 3 cells or with total counts < 10 were omitted from further analysis. The dimension of the final data matrix was 18643 genes by 217 cells (134 cells from naïve mice and 83 from H. poly infected mice). Counts were normalized to ten thousand counts per cell, natural-log transformed using log1p, and scaled. Highly variable genes were identified using Seurat, and the top 1000 used for dimensionality reduction and clustering 32 . The first 4 principal components were selected as input for dimensionality reduction using the UMAP algorithm and Louvain clustering. These were selected visually (inspecting heat maps) based on the presence of clear associated patterns of gene expression. Louvain clustering was performed at a resolution of 0.1, as this resolution allows for the detection of clusters with distinct properties according to downstream analysis. Visualisation of batches on the resulting UMAP reveals that cells do not cluster according to batch and that any batch effects are unlikely to obscure the underlying biology (Extended Data Fig. 2f).
Marker genes for each cluster were detected using a Wilcoxon test in Seurat. Differential expression between glia from control and H. poly infected mice was also performed in Seurat using the MAST package 34 and the formula ~condition + nFeature_RNA. Functional analysis was performed on markers detected for each cluster (adjusted p-value < 0.01) using g:Profiler 35 , accessed programmatically using the gprofiler python module. Gene set enrichment analysis was also performed using the full set of differentially expressed genes between the two clusters using the R package fgsea 36 . Here hallmark gene sets were used and converted to mouse genes using a mapping obtained from HomoloGene database 37 .
scRNAseq (10X
Fastq files were aligned to the mm10 genome and assembled into cell feature matrices using the 10X Cell Ranger software suite. Further computations were carried out in R 38 . The filtered cell feature matrices were assembled into a Seurat object using published steps 39 for each of the four cases Ifngr2CTRL naïve, Ifngr2CTRL H. poly, Ifngr2ΔEGC naïve and Ifngr2ΔEGC H. poly (two replicates each case). Visual inspection led us to apply quality control cutoffs to each of these Seurat objects of percent.mt < 10 and nCountRNA ≥ 5000. Each of these four Seurat objects was normalized (ten thousand counts per cell, natural-log transformed using log1p) and the four were then combined using Seurat’s FindIntegrationAnchors() and IntegrateData() functions. Data for the resulting Seurat object was scaled using ScaleData(). Cell types were determined by an iterative process which started by applying Louvain clustering of the cells using Seurat’s FindNeighbors() function using principal components (PCs) 1 through 20, and FindClusters() function with resolution 0.9. To discover the group marker genes for each cluster, we then applied the FindMarkers() function to these clusters with using the default Wilcoxon test and only.pos=TRUE and for each cluster queried those genes with p_val_adj ≥ 0.05. The resulting genes were used to manually curate an assessment of the biological type of the cells in each cluster. In this way, we were able to arrive at the cell classification used for further downstream analyses. During this process, a small number of cells were discarded from further evaluation as their gene expression profiles, i.e. the expression of multiple canonical markers across different cell types, suggested they were doublets. Lists of differentially expressed genes between experimental conditions were found using Seurat’s FindMarkers() with the parameter test='MAST' 34 and the formula ~condition. We confirmed the difference in the number of differentially expressed genes by subsampling to ensure that the difference in the number of differentially expressed genes was due to clear transcriptomic differences and not due to differences in statistical power due to the different number of cells within the clusters. For each comparison to be made we considered the clusters in the two conditions and randomly subsampled to 100 cells (larger cluster) and to 50 cells (smaller cluster). In addition, the UMI count was taken into account as a latent variable (~condition+nUMI). The count matrices for the analysis of single-cell transcriptomes of the mouse (GSE142431 19 ) and human (GSE114374 8 ) mesenchyme were obtained from GEO. Individual datasets were combined using Seurat’s FindIntegrationAnchors() and IntegrateData() functions. Highly variable genes were identified using Seurat, and the top 2000 used for dimensionality reduction and clustering. The top 20 PCs were chosen for as input for dimensionality reduction using the UMAP algorithm and Louvain clustering. Subclustering of human glia was performed based on the selection of the cluster that expressed S100b, Sox10 and Plp1. The resolution for the FindClusters() function was set to 0.1. Gene set enrichment analysis was performed comparing the full list of differentially expressed genes, not filtered by p_val_adj against the hallmark gene sets using the fgsea package 36 . We used ggplot2 40 for many of the figures. Additional packages used in plotting include ggsignif 41 , ggrepel 42 and ggsci 43 .
Statistics
Bioinformatic statistical analysis was carried out in R 38 . All other statistical analysis was carried out using GraphPad Prism 9.0 software (GraphPad software, CA, USA). Normality distribution was tested with the D'Agostino–Pearson omnibus test. When comparing two groups unpaired two-tailed t-tests (followed by Welch’s correction test for non-equal standard deviations) and two-tailed Mann-Whitney tests were used for parametric and non-parametric datasets respectively. When comparing more than two groups, one-way ANOVA followed by Tukey’s or Bonferroni’s multiple comparison test and Kruskal Wallis test followed by a Dunn’s multiple comparison test were used for parametric and non-parametric datasetsrespectively. Two-way ANOVA was used to compare time-course curves followed by Sidak’s multiple comparison test to determine the significance. P-values of less than 0.05 were deemed statistically significant and all P values are denoted in the figures. The nature of the entity of n is defined as individual animals unless otherwise indicated and all experimental replicates are biological.
Extended Data
Extended Data Figure 1. Intestinal helminth infection induces ENS injury and gliosis.
(a-i) TM preparations (a-d, f-i) and cross-section (e) from the duodenum of Sox10|tdT mice immunostained for CD45 (a), CD31 (b), Pdgfra (c), c-Kit (d), Epcam (e), Sox10 (f, h) and HuC/D (f, i). Indicated are cells negative for tdT (empty arrowheads), tdT+ EGCs (asterisks) and neurons (arrowheads). g, h and i show single spectrum images of f. n=3. (j) Quantification of tdT+ cells expressing Sox10 (EGCs) and HuC/D (neurons) (n=11 field of views, representative of 3 experiments). (k) Schematic of intestinal cross-section illustrating the organisation of the EGC-network and the life-cycleof H. poly. L3 H. poly larvae penetrate the duodenum mucosa and settle in the TM, eliciting local tissue damage, inflammation and formation of granulomatous infiltrates. 10 days later they emerge as adult worms into the lumen where they mate and produce eggs. (l, m) Cross-section (l) and whole-mount view (m) of the duodenum of H. poly-infected Sox10|tdT mice (7 dpi). Schematics (top left) show the orientation of the images. Arrowheads point to H. poly settlement sites. n=4. (n, o, q, r) TM preparations from the duodenum of naïve and H. poly-infected (7 dpi n, o; 10 dpi q, r) animals immunostained for GFAP and Sox10 (n, o) and S100 (q, r).n=5. (p) Quantification (qRT-PCR) of Gfap transcripts in the TM of naïve and H. poly-infected mice (7 dpi). (nNaïve=8, n H.poly =6). 2 experiments. (s-u) Quantification of S100+ type III EGC morphology including total process length (s) (n=10), process thickness (t) (n=10) and Scholl analysis for EGC process branching (u) (n=5). 2 experiments. Two-tailed Mann-Whitney test (p, t), unpairedtwo-tailed t-test (s), Two-way ANOVA with multiple comparisons (u). Mean±SEM. Scale bars: a-i: 50 μm; l, m, n, o, q, r:100 μm, insets: 12.5 μm
Extended Data Figure 2. Transcriptomic analysis of H. poly-infected TM.
(a) Experimental design for bulk RNAseq of EGCs from the TM of naïve and H. poly-infected (7 dpi) Sox10|tdT mice. tdT+ and tdT- cell populations of dissociated TM were separated using FACS and subjected to RNAseq. (b) Sorting strategy for tdT+ EGCs and tdT- non-glia cells. (c, d) Volcano plot showing mean log2-transformed fold change (x-axis) and significance (-log10(adjusted Pvalue)) of differentially expressed genes between tdT- and tdT+ cells from naïve mice (c) and in tdT- cells from naïve and H. poly-infected animals (d). Coloured dots in (c) indicate genes specific to EGCs (Sox10, Plp1, S100b, Foxd3, Erbb3, Sox2; red), enteric neurons (Ret, Tubb3, Sst, Elavl3, Elavl4; green), immune cells (Ptprc; cyan), interstitial cells of Cajal (Kit; orange), smooth muscle cells (Acgt2; pink), fibroblasts (Pdgfra; purple) and in (d) genes specific to type II immune response (Arg1, Retnla, Chil3) and TH2 cytokines (Il13).n=4. (e) Quantification of IFN-γ in the TM of naïve and H. poly-infected animals (7 dpi). Mean±SD; n=6. Two-tailed Mann-Whitney test. (f) UMAP of sequenced EGCs from the TM of naïve and H. poly infected mice. Cells are color-coded according to experimental batches. (g) Violin plots showing normalized expression of representative EGC marker genes in EGC1 and EGC2 clusters in Fig. 1g. (h, i) Top 20 up-regulated and top 20 down-regulated genes in EGCs from H. poly-infected mice (h) and in EGC2 relative to EGC1 (i). IFN-γ-target genes shown in bold (h). Dot size indicates proportion of expressing cells and fill colour indicates mean normalized, centred and scaled expression level. (j, k) GO terms significantly enriched among the differentially expressed genes in EGC1 and EGC2 clusters shown in Fig. 1g (j) and in hEGC1 and hEGC2 clusters shown in Fig. 1j (k).
Extended Data Figure 3. Cell autonomous activation of EGCs by IFN-γ.
(a, b) qRT-PCR analysis of Ifngr1 (a) and Ifngr2 (b) transcript levels from spleen cells and FACS-isolated EGCs (tdT+) and non-glia cells (tdT-) from Sox10|tdT mice. n=3. (c, d) Cultures of FACS-isolated EGCs from Sox10|tdT mice in the absence (c) or presence (d) of IFN-γ immunostained for pH3 (green) and labelled with EdU (blue). Scale bars: 100 μm. (e, f) Quantification of pH3+ (e) and EdU+ (f) EGCs (tdT+ cells) in the cultures shown in c and d, respectively. n=8 field of views from each of 3 experiments. (g) qRT-PCR analysis of Ifngr2 transcript levels in muscularis macrophages, fibroblasts, endothelial cells and EGCs FACS-isolated from the TM of Ifngr2CTRL and Ifngr2ΔEGC mice. n=3. (h) Images of IFN-γ-treated (1 hour) myenteric plexus preparations from the duodenum of Ifngr2CTRL and Ifngr2ΔEGC mice immunostained for pStat1, Sox10 and HuC/D and counterstained for DAPI. Indicated are pStat1+ EGCs (empty arrowheads), pStat1- EGCs (filled arrowheads). Scale bar = 10 μm. (i) Quantification of pStat1+ muscularis macrophages, fibroblasts, neurons and EGCs in IFN-γ-treated (1 hour) TM preparations from Ifngr2CTRL and Ifngr2ΔEGC mice. n=8. (j-l) qRT-PCR analysis of Ifngr2, Cxcl10 and Gbp10 transcript levels from rIFN-γ treated EGCs isolated from Ifngr2CTRL and Ifngr2ΔEGC mice. n=4. (m) Quantification of Ki67+ EGCs in the TM of H. poly-infected Ifngr2CTRL and Ifngr2ΔEGC mice at 7 dpi (n=10). (n) Quantification of Ki67+ EGCs in the TM of WT and Ifngr1-/- mice (n=8). 2 experiments (m, n). Two-tailed Mann-Whitney test (e, f, n, o), unpaired two-tailed t-test (g, i, j, k, l, m). Mean±SEM
Extended Data Figure 4. Characterisation of mice with glia-specific deletion of IFN-γ signalling.
(a) Quantification of granulomas in the small intestine at 28 dpi (n=10) and 80 dpi (n=5) WT and Ifngr1 -/- mice (2 experiments). (b) Representative images of H. poly settlement sites in Ifngr2CTRL and Ifngr2ΔEGC gut at 7 dpi. Note bleeding in Ifngr2ΔEGC mice (65.2±3.14 versus 31.8±1.56 in Ifngr2CTRL mice). n=20 animals analysed. (c, d, f) Flow cytometry gating strategy to immunophenotype the TM of naïve and H. poly-infected Ifngr2CTRL andIfngr2ΔEGC mice showing debris exclusion and doublet discrimination, selection of live immune cells (c), followed by gating of myeloid cells (d) or NK-/ T cells (f). (e) Flow cytometry quantification of neutrophils in the TM of naïve Ifngr2CTRL and Ifngr2ΔEGC mice. (nCTRL=5, nΔEGC=6). (g-i) Flow cytometry quantification of CD4 and γδT cells and NK cells at indicated time-points after H. poly infection within the TM of Ifngr2CTRL and Ifngr2ΔEGC mice. (n=4, data from 1 experiment). (j) Quantification of worms in the TM at 7 dpi (nCTRL&ΔEGC=8) or recovered from the intestinal tract at 28 (nCTRL&ΔEGC=11) and 60 days post-infection (nCTRL&ΔEGC=15). Mean±SD. Two-way Anova. (k) H. poly egg burden at 14 (nCTRL=15, nΔEGC=16), 28 (nCTRL&ΔEGC=11) and 60 dpi (nCTRL&ΔEGC=9). Two-tailed Mann-Whitney Test (e), Two-way Anova (a, g-k). Mean±SD (a, j, k), Mean±SEM (e, g-i).
Extended Data Figure 5. Cellular atlas of small intestine TM.
(a) Mean normalized expression of representative marker genes and proportion of expressing cells (indicated by dot size) in the cell clusters shown in (c). Clusters are labelled with post facto annotation based on known markers. (b) Sorting strategy for live TM cells. (c) UMAP of all sequenced cells from small intestine TM. The numbers of clusters in c correspond to the numbers in a. (d) UMAP analysis of integrated scRNAseq datasets of mesenchymal cells from the lamina propria (GSE142431) 19 and the TM (present study; Fig. 3a, b). Annotation of cellular clusters matches those reported by Roulis et al. 19 on the basis of respective marker genes. Note that cell populations from the TM overlap with those from the lamina propria. (e) Violin plot quantification of Pdgfra expression per single cell highlighting that, similar to those in the lamina propria 21 , TM fibroblasts are divided into Pdgfrahigh and Pdgfralow cells. (f-i) Representative images of cross-section (f) or TM preparations (g-i) from the small intestine of Sox10|tdT mice immunostained for Pdpn to identify mesothelial cells (f, arrowhead) and lymphatic endothelial cells (g), Vegfr2 and Pdgfrb to identify endothelial cells and pericytes (arrowhead), respectively (h), and Cd3 and Iba1 to identify T cells and macrophages, respectively (i). Note the small number of T cells in TM relative to macrophages. Scale bars: 50 μm, insets: 25 μm. Images representative of n=5 animals analysed.
Extended Data Figure 6. Glia-specific ablation of IFN-γ signalling induces a tissue-wide pro-inflammatory state of TM at steady state and modulates the response to H. poly infection.
(a-c) UMAP representation of mesothelial cells (a), fibroblasts (b) and muscularis macrophages (c) from naïve Ifngr2CTRL (black) and Ifngr2ΔEGC (orange) mice. (d) qRT-PCR analysis of Lcn2, Il1b, Saa3 and Il6 transcript levels in the TM from naïve Ifngr2CTRL and Ifngr2ΔEGC mice. nCTRL=12, nΔEGC=11. (e) Representative H&E stained intestinal cross-sections from naïve Ifngr2CTRL and Ifngr2ΔEGC mice. Empty and filled arrowheads in inset highlight reactive mesothelial cells and eosinophils, respectively. Scale bars = 50 μm, insets: 20 μm. Shown also is histology severity score (right) assessing inflammation in the lamina propria and tunica muscularis from naïve Ifngr2CTRL and Ifngr2ΔEGC mice. n=8. 2 experiments. (f) Intestinal paracellular permeability in naïve Ifngr2CTRL and Ifngr2ΔEGC mice. nCTRL=8, nΔEGC=7 (2 independent experiments). (g) Whole intestinal transit time in naïveIfngr2CTRL and Ifngr2ΔEGC mice. nCTRL=9, nΔEGC=8. (h) Fraction of cells per cluster in naïve and H. poly-infected Ifngr2CTRL and Ifngr2ΔEGC mice. (i) Violin plot visualisation of Ifng expression levels per single cell in indicated cell clusters. (j) Dot plot quantification of expression levels of Ifng vs Cd8a (left panel) and Ifng vs Cd4 (right panel) in the Other lymphoid cells, NK cells, T Cells 1, T Cells 2 and T Cells 3 clusters, indicating that Cd8a T cells are a major source of Ifng in the TM of H. poly-infected mice at 7 dpi. Two-tailed Mann-Whitney test (d, f). Unpaired two-tailed t-test (g). Mean±SEM.
Extended Data Figure 7. Activation of the IFN-γ-Cxcl10 axis in TM precedes type II immune response and promotes tissue repair after helminth infection.
(a, b) qRT-PCR analysis of Il4 and Il13 transcripts in TM (a; nnaïve=8, n3dpi,7dpi,10dpi,14dpi=6, n5dpi,28dpi=7, n21dpi=3) and Ifng, Il4, Il13 and Arg1 transcripts in mucosa (b; nnaïve,7dpi,10dpi=6, n3dpi,5dpi,14dpi,21dpi,28dpi=3) of small intestine following H. poly infection. 2 experiments. (c, d) qRT-PCR time-course analysis of Gbp6 (c) and Gbp10 (d) transcript levels in TM after H. poly infection.nnaïve=8, n3 dpi,7 dpi,10 dpi,14 dpi=6, n5 dpi,28 dpi=7, n21 dpi=3. 2 experiments. (e) Flow cytometric quantification of IFN-γ-producing cells in the TM of H. poly-infected WT mice (3 dpi). nnaïve=7, n3dpi=9. 2 experiments. (f) qRT-PCR analysis of Cxcl10 transcripts in TM from H. poly-infected (7 dpi) WT and Ifngr1 -/- mice. n=10. 2 experiments. (g) Quantification of granulomas in the small intestine H. poly-infected (28 dpi) Ifngr1 -/- and Cxcl10 -/- mice. n=9. 2 experiments. (h) qRT-PCR analysis of Cxcl10 transcripts from rIFN-γ-treated cultures (24 hours) of EGCs from Cxcl10CTRL and Cxcl10ΔEGC mice. nControl=2, nrIFN-γ=3. (i) In situ hybridization for Cxcl10 in IFN-γ-treated (1 hour) myenteric plexus from the duodenum of Cxcl10CTRL and Cxcl10ΔEGC mice. Cxcl10+ EGCs indicated by empty arrowhead. Scale bar = 10 μm. (j) Quantification of Cxcl10+ EGCs in IFN-γ-treated (1 hour) TM preparations from Cxcl10CTRL and Cxcl10ΔEGC mice. nCTRL=3, nΔEGC=4. (k) Quantification of Cxcl10+ EGCs in the TM of H. poly-infected Cxcl10CTRL and Cxcl10ΔEGC mice (3 dpi). nCTRL =3, nΔEGC=6. (l, m) Quantification of adult worms (l) and eggs (m) in small intestine from H. poly-infected Cxcl10CTRL and Cxcl10ΔEGC mice (28 dpi). nCTRL=12, nΔEGC=13. 2 experiments. (n) Dot plot analysis of Cxcr3 vs Cd8a expression indicating that Cd8+ T cells in TM express Cxcr3. Two-tailed Mann-Whitney test (f, j, k). Kruskal-Wallis test (g). Unpaired two-tailed t-test (e, h). Mean±SEM.
Acknowledgements
We thank the Crick Science Technology Platforms (STPs) for expert support, in particular the Biological Research Facility, the Flow Cytometry STP, the Advanced Sequencing Facility and the Experimental Histopathology Laboratory; C. Minutti for useful advice, discussions and insightful comments on the manuscript and all members of the Pachnis laboratory for insightful comments on the manuscript. We thank Werner Müller for providing the Ifngr2 floxed mice. This work was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001128, FC001159), the UK Medical Research Council (FC001128, FC001159), and the Wellcome Trust ((FC001128, FC001159). V.P. acknowledges additional funding from BBSRC (BB/L022974) and the Wellcome Trust (212300/Z/18/Z). B.S. was also funded by the Wellcome Trust (210556/Z/18/Z).
Footnotes
Author contributions
F.P. and V.P. conceived the study and together with B.S. and M.S.W. designed the experiments. S.H.C., C.H.C., S.S., M.R., E.M.A., A.C.B.-F, R.L and L.J.E. helped with the experiments. S.H.C. performed cell culture of primary EGCs, M.S. and A.L. performed scRNAseq bioinformatic analysis, S.B. performed bulk RNAseq bioinformatics analysis, A.C.B.-F, S.S. and B.G.-C. performed immunohistochemistry of the TM, B.G.-C. and C.H.C. designed, performed and analysed immunophenotyping experiments, K.S. designed some of the immunophenotyping experiments, A.S.-B. performed the histopathology examination. V.P. and F.P. wrote the manuscript with help from B.S., and contributions from all authors.
The authors declare no competing financial interest.
Data availability
The. fastq files, Seurat objects, count matrices and associated meta data are publicly available in the Gene Expression Omnibus repository with the accession numbers GSE182708 and GSE182715 for the bulk and scRNAseq of naïve and H. poly-infected EGCs, respectively and GSE182506 for the scRNAseq of the naïve and H. poly-infected TM of Ifngr2CTRL and Ifngr2ΔEGC mice. All datasets are available to search on https://biologic.crick.ac.uk/ENS/EGCinflammation. Source data for all graphs plotted in figures and extended data figures that have associated raw data are provided with the paper. Data for all graphs associated with the scRNAseq analysis are available at GitHub (https://github.com/michaeldshapiro/RegulationOfIntestinalImmuneHomeostasis).
Code availability
The code used for all scRNAseq analysis is available at GitHub (https://github.com/michaeldshapiro/RegulationOfIntestinalImmuneHomeostasis).
References
- 1.Kotas ME, Medzhitov R. Homeostasis, inflammation, and disease susceptibility. Cell. 2015;160:816–827. doi: 10.1016/j.cell.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 3.Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010;10:131–144. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]
- 4.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 5.Klose CS, Artis D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat Immunol. 2016;17:765–774. doi: 10.1038/ni.3489. [DOI] [PubMed] [Google Scholar]
- 6.Roulis M, Flavell RA. Fibroblasts and myofibroblasts of the intestinal lamina propria in physiology and disease. Differentiation. 2016;92:116–131. doi: 10.1016/j.diff.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Koliaraki V, Prados A, Armaka M, Kollias G. The mesenchymal context in inflammation, immunity and cancer. Nat Immunol. 2020;21:974–982. doi: 10.1038/s41590-020-0741-2. [DOI] [PubMed] [Google Scholar]
- 8.Kinchen J, et al. Structural Remodeling of the Human Colonic Mesenchyme in Inflammatory Bowel Disease. Cell. 2018;175:372–386.:e317. doi: 10.1016/j.cell.2018.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laranjeira C, et al. Glial cells in the mouse enteric nervous system can undergo neurogenesis in response to injury. J Clin Invest. 2011;121:3412–3424. doi: 10.1172/JCI58200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reynolds LA, Filbey KJ, Maizels RM. Immunity to the model intestinal helminth parasite Heligmosomoides polygyrus. Semin Immunopathol. 2012;34:829–846. doi: 10.1007/s00281-012-0347-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boesmans W, Lasrado R, Vanden Berghe P, Pachnis V. Heterogeneity and phenotypic plasticity of glial cells in the mammalian enteric nervous system. Glia. 2015;63:229–241. doi: 10.1002/glia.22746. [DOI] [PubMed] [Google Scholar]
- 12.Heanue TA, Pachnis V. Prospective identification and isolation of enteric nervous system progenitors using Sox2. Stem Cells. 2011;29:128–140. doi: 10.1002/stem.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lasrado R, et al. Lineage-dependent spatial and functional organization of the mammalian enteric nervous system. Science. 2017;356:722–726. doi: 10.1126/science.aam7511. [DOI] [PubMed] [Google Scholar]
- 14.Bach EA, Aguet M, Schreiber RD. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol. 1997;15:563–591. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- 15.Lee HM, et al. IFNgamma signaling endows DCs with the capacity to control type I inflammation during parasitic infection through promoting T-bet+ regulatory T cells. PLoS Pathog. 2015;11:e1004635. doi: 10.1371/journal.ppat.1004635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang S, et al. Immune response in mice that lack the interferon-gamma receptor. Science. 1993;259:1742–1745. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- 17.Gentile ME, et al. NK cell recruitment limits tissue damage during an enteric helminth infection. Mucosal Immunol. 2019 doi: 10.1038/s41385-019-0231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nusse YM, et al. Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature. 2018;559:109–113. doi: 10.1038/s41586-018-0257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roulis M, et al. Paracrine orchestration of intestinal tumorigenesis by a mesenchymal niche. Nature. 2020;580:524–529. doi: 10.1038/s41586-020-2166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rinkevich Y, et al. Identification and prospective isolation of a mesothelial precursor lineage giving rise to smooth muscle cells and fibroblasts for mammalian internal organs, and their vasculature. Nat Cell Biol. 2012;14:1251–1260. doi: 10.1038/ncb2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mutsaers SE, et al. Mesothelial cells in tissue repair and fibrosis. Front Pharmacol. 2015;6:113. doi: 10.3389/fphar.2015.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stetson DB, et al. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J Exp Med. 2003;198:1069–1076. doi: 10.1084/jem.20030630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dufour JH, et al. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J Immunol. 2002;168:3195–3204. doi: 10.4049/jimmunol.168.7.3195. [DOI] [PubMed] [Google Scholar]
- 24.Mills Ko E, et al. Deletion of astroglial CXCL10 delays clinical onset but does not affect progressive axon loss in a murine autoimmune multiple sclerosis model. J Neuroinflammation. 2014;11:105. doi: 10.1186/1742-2094-11-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostvik AE, et al. Enhanced expression of CXCL10 in inflammatory bowel disease: potential role of mucosal Toll-like receptor 3 stimulation. Inflamm Bowel Dis. 2013;19:265–274. doi: 10.1002/ibd.23034. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, et al. An Atlas of Genetic Variation Linking Pathogen-Induced Cellular Traits to Human Disease. Cell Host Microbe. 2018;24:308–323.:e306. doi: 10.1016/j.chom.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sasselli V, et al. Planar cell polarity genes control the connectivity of enteric neurons. J Clin Invest. 2013;123:1763–1772. doi: 10.1172/JCI66759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tavares G, et al. Employing an open-source tool to assess astrocyte tridimensional structure. Brain Struct Funct. 2017;222:1989–1999. doi: 10.1007/s00429-016-1316-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011;17:3. doi: 10.14806/ej.17.1.200. 2011. [DOI] [Google Scholar]
- 31.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su S, et al. CellBench: R/Bioconductor software for comparing single-cell RNA-seq analysis methods. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finak G, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reimand J, et al. g:Profiler-a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Res. 2016;44:W83–89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sergushichev A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv. 2016 doi: 10.1101/060012. [DOI] [Google Scholar]
- 37.Coordinators, N.R. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018;46:D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2019. [Google Scholar]
- 39.Stuart T, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.:e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York: 2016. [Google Scholar]
- 41.ggsignif: Significance Brackets for 'ggplot2'. 2019 [Google Scholar]
- 42.ggrepel: Automatically Position Non-Overlapping Text Labels with 'ggplot2'. 2020 [Google Scholar]
- 43.ggsci: Scientific Journal and Sci-Fi Themed Color Palettes for 'ggplot2'. 2018 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The. fastq files, Seurat objects, count matrices and associated meta data are publicly available in the Gene Expression Omnibus repository with the accession numbers GSE182708 and GSE182715 for the bulk and scRNAseq of naïve and H. poly-infected EGCs, respectively and GSE182506 for the scRNAseq of the naïve and H. poly-infected TM of Ifngr2CTRL and Ifngr2ΔEGC mice. All datasets are available to search on https://biologic.crick.ac.uk/ENS/EGCinflammation. Source data for all graphs plotted in figures and extended data figures that have associated raw data are provided with the paper. Data for all graphs associated with the scRNAseq analysis are available at GitHub (https://github.com/michaeldshapiro/RegulationOfIntestinalImmuneHomeostasis).
The code used for all scRNAseq analysis is available at GitHub (https://github.com/michaeldshapiro/RegulationOfIntestinalImmuneHomeostasis).