Mutations impairing early B cell development cause monogenic primary immunodeficiencies (PIDs) that manifest with markedly reduced or absent B cells, hypogammaglobulinemia, and recurrent bacterial infections from childhood. Approximately 85% of such patients have mutations in BTK, the gene responsible for X-linked agammaglobulinemia 1 . Current research focuses on patients with unknown genetic defects, because the identification of the causative genes will not only facilitate diagnosis of PIDs, but can also reveal new biological roles of the affected proteins in human B cell development and point at novel drug targets.
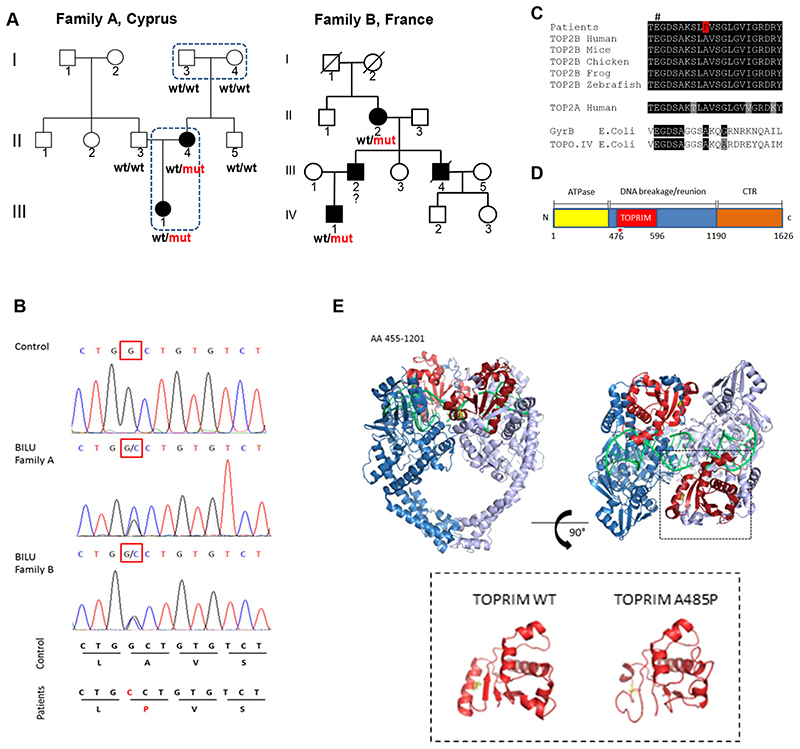
The autosomal dominant syndrome of unknown etiology called BILU (B-cell Immunodeficiency, Limb anomalies and Urogenital malformations) was previously described in two unrelated families originating from Cyprus and France 2,3 (families A and B, Figure 1A and Supplemental Tables 1 and >2). The BILU patients have absent or reduced B cells but normal T and myeloid cells 2,3 (Supplemental Figure 1). Here we studied these two families. Given that in family A both parents of patient II:4 are healthy, we hypothesized that the BILU syndrome was caused by a de novo mutation. We sequenced exomes of four subjects in family A (Figure 1A) and identified the only mutation in the coding part of the genome that was absent in healthy parents I:3 and I:4, appeared de novo in their affected daughter II:4 and then was transmitted to her affected daughter III:1. This heterozygous G>C mutation at the cDNA nucleotide 1,453 of the TOP2B gene (ENST00000435706) resulted in the alanine to proline substitution at position 485 (A485P) of topoisomerase 2β (TOP2B). Next, we used Sanger sequencing and found that exactly the same mutation was present in patients of family B (Figure 1B). Given that this mutation was never found in any healthy subject (e.g. absent from more than 90,000 subjects in the gnomAD database 4 ), but is present in the BILU patients from two unrelated families, this novel mutation is the cause of the BILU syndrome.
Figure 1. Novel dominant mutation A485P affects TOP2B catalytic site and causes the BILU syndrome.
(A) Two families with the BILU syndrome: ○ and □-unaffected, ● and ■-affected; wt-wild-type allele, mut - A485P mutation in TOP2B. Exome sequencing was performed in four subjects shown by dotted lines. (B) Electrophoregrams of the TOP2B genomic DNA sequence showing the same mutation in patients from the two BILU families. (C) Multispecies protein sequence alignment of type II topoisomerases. # indicates a conserved glutamate essential for TOP2B activity. (D) Schematic representation of domains of the TOP2B protein. The A485P mutation in the TOPRIM domain is shown by a red star. The TOPRIM domain is part of the DNA gate that catalyzes DNA cleavage and re-ligation. (E) Structure of the TOP2B homodimer (amino acid residues 455-1201) in complex with DNA (green); the TOPRIM domain (red) and the alanine at codon 485 (yellow) (upper panel). I-TASSER-modeled structures of the TOP2B TOPRIM domain; WT – wild-type (lower panel).
TOP2B is a type II topoisomerase, an enzyme that generates transient DNA double strand breaks (DSBs) and solves topological problems during replication and transcription, e.g. removes DNA supercoils, knots and catenanes 5 . TOP2B and the other human type II topoisomerase 2α (TOP2A) can make active homodimers and heterodimers 6,7 . First, we studied how the newly discovered A485P mutation interfered with the structure of the TOP2B protein. The mutation affects alanine that is conserved in eukaryotic and even in prokaryotic type II topoisomerases (Figure 1C). Its substitution with proline is predicted to destabilize an α helix within the TOPRIM domain (Figure 1D, E) that is essential for the catalytic activity of the TOP2B protein 8,9 .
Since BILU patients have no or few B cells in the blood, we studied T cells, skin fibroblasts and induced pluripotent stem cells (iPSCs) and found reduced amounts of the TOP2B protein in patients’ cells in comparison to cells of healthy controls (Figure 2A). We then used CRISPR-Cas9 to knock-out TOP2B in HEK-293 cells and expressed in these cells the wild-type and mutant proteins, TOP2BWT and TOP2BA485P. We found a low molecular weight product of TOP2BA485P degradation, suggesting that the mutant protein is less stable than wild-type TOP2B (Supplemental Figure 2). Co-immunoprecipitation showed that both TOP2BWT and TOP2BA485P interact with endogenous TOP2A (Supplemental Figure 2).
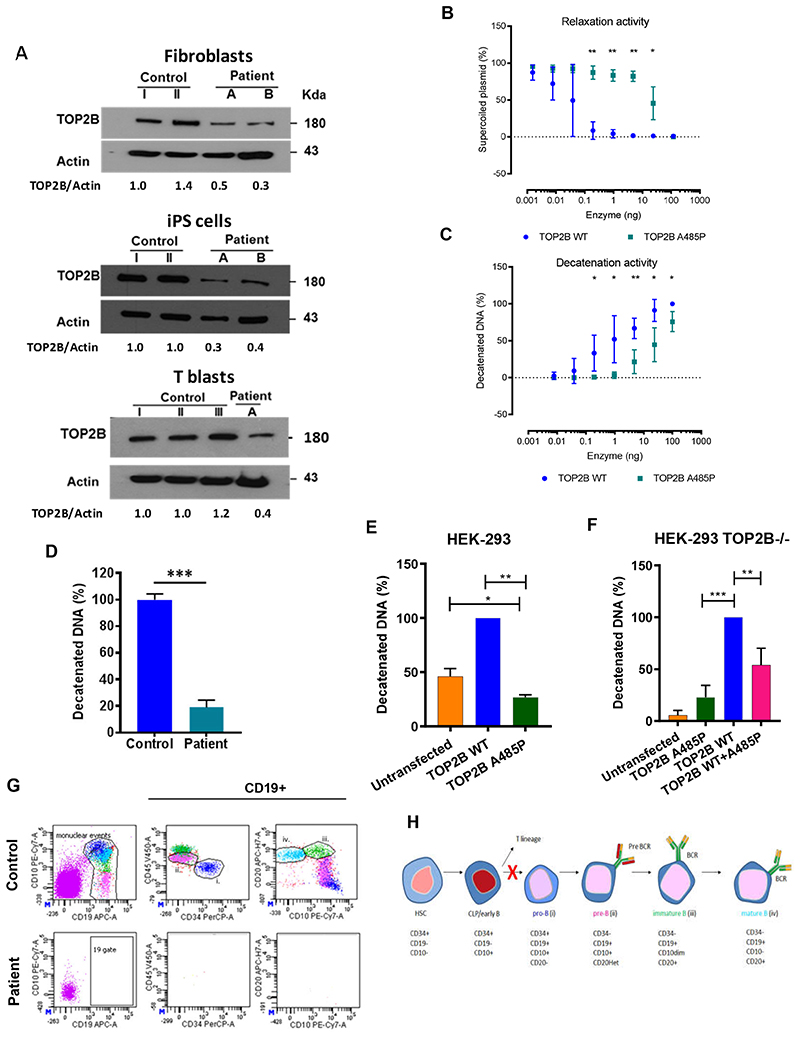
Figure 2. Mutation A485P reduces expression and activity of the TOP2B protein and impairs early B cell development.
(A) Western blots showing expression of the TOP2B protein in primary dermal fibroblasts, induced pluripotent stem cells (iPSCs) dedifferentiated from dermal fibroblasts and T cell blasts derived from PBMC. Patients: A is III.1 in family A; B is IV.1 in family B. Fold change of band densitometry is shown. (B) Relaxation of negatively-supercoiled DNA by the purified recombinant TOP2BWT and TOP2BA485P proteins. (C) Decatenation of kinetoplast DNA by the purified recombinant TOP2BWT and TOP2BA485P proteins. (D) Decatenation of kinetoplast DNA by nuclear extracts from T cell blasts. (E, F) Decatenation of kinetoplast DNA by nuclear extracts from wild-type HEK-293 cells (E) or TOP2B-knockout HEK-293 cell (F), untransfected or transfected with plasmids encoding TOP2BWT and TOP2BA485P. P-values were calculated using two-tailed (E) and one-tailed (F) paired t-tests. Graphs show averages ± SD. * P < 0.05, ** P < 0.01, *** P < 0.001. (G) Bone marrow immunophenotyping of the BILU patient and a healthy unrelated control showing pro-B (i), pre-B (ii), immature B (iii) and mature B (iv) cells. (H) B cell development stages; red X shows defect in the BILU patients. HSC - hematopoietic stem cell, CLP - common lymphoid progenitor.
To find out if the mutation affects enzymatic activity, we then studied purified recombinant full-length TOP2BWT and TOP2BA485P proteins using DNA relaxation and decatenation in vitro assays 10 . We found that enzymatic activities of TOP2BA485P were reduced more than 10 fold (Figure 2B, C and Supplemental Figure 3A, B). In human cells, DNA relaxation can be performed by both type I and type II topoisomerases, whilst DNA decatenation is performed only by type II topoisomerases TOP2B and TOP2A. Therefore, we studied cell lysates using only the decatenation assay. The decatenation activity of lysed patient’s T cell blasts was reduced (Figure 2D and Supplemental Figure 4). Likewise, the decatenation activity of lysed wild-type HEK-293 cells transiently expressing TOP2BA485P was lower than the activity of cells expressing TOP2BWT (Figure 2E and Supplemental Figure 5). Interestingly, it was also lower than the decatenation activity of untransfected wildtype HEK-293 cells that expressed only endogenous TOP2B, as well as endogenous TOP2A (Figure 2E and Supplemental Figure 5). Similarly, in TOP2B-knockout HEK-293 cells co-expression of TOP2BWT and TOP2BA485P proteins resulted in lower decatenation activity than the expression of TOP2BWT alone (Figure 2F and Supplemental Figure 6). These results indicate that mutant TOP2BA485P protein not only itself has reduced intrinsic enzymatic activity, but also exerts a dominant negative effect on the activities of wild-type TOP2B and TOP2A. Thus, our experimental data are consistent with the dominant negative impact of the TOP2B mutation that causes BILU syndrome, rather than with haploinsufficiency. Moreover, haploinsufficiency of TOP2B is an unlikely causative mechanism of BILU because multiple subjects with various heterozygous loss-of-function TOP2B mutations have been detected in population cohorts (e.g. the gnomAD database 4 has 42 such unaffected subjects; https://gnomad.broadinstitute.org/).
Recently, other dominant mutations affecting the TOPRIM domain of TOP2B have been shown to cause Hoffman syndrome that is characterized by B cell deficiency, limb abnormalities and facial dysmorphism 11 (Supplemental Tables 1 and 2). Our results indicate that BILU and Hoffman syndromes are manifestations of the same disease, TOP2B deficiency. Importantly, these findings demonstrate a previously unknown critical role of TOP2B in B cell development.
The developmental defect leading to B-cell deficiency in patients with BILU and Hoffman syndromes has not been investigated previously. To reveal the affected stage of B cell development, we studied a bone marrow aspirate of the BILU patient II:4 from family A using multicolor flow cytometry and found a complete absence of any CD19+ cells, including pro-B, pre-B, immature and mature B cells, but normal T, NK and myeloid cell lineages (Figure 2G and Supplemental Figure 7). This finding for the first time shows that TOP2B is critical during the earliest stages of B cell lineage after the Common Lymphoid Progenitor (CLP) stage (Figure 2H). The early block in B cell development distinguishes patients with TOP2B deficiency from most other B-cell immunodeficiencies that either impair later stages of B cell differentiation resulting in the accumulation of CD19+ pro-B cells (e.g. BTK deficiency) or affect multiple hematopoietic cell lineages (e.g. ADA or GATA2 deficiencies) 12 . Rather, it is reminiscent of the early block of B cell differentiation seen in patients with dominant TCF3 mutations and recessive PIK3R1 mutations 13,14 . Nevertheless, in TOP2B deficiency this block is leaky, because immunoglobulins and small numbers of B cells have been found in peripheral blood of several patients (Supplemental Table 2).
TOP2B had been shown to produce signaling-induced DSBs at gene promoters 15–18 and was involved in activation of transcription 16–18 , transcription of long genes 19 , as well as formation and maintenance of topologically associated domains and chromatin loops 20,21 . While these TOP2B functions may contribute to the B-cell developmental defect, the exact molecular mechanism affecting specifically B cells, but not other immune cell lineages, remains unclear.
TOP2B and TOP2A are the targets of the anti-cancer drug etoposide that traps these enzymes in a complex with cleaved DNA, which eventually leads cells to apoptosis. Etoposide, in combination with clofarabine and cyclophosphamide, had been used for chemotherapy of acute lymphoblastic leukemia (ALL) 22–24 . Interestingly, in pediatric patients with refractory/multiple relapse ALL, this chemotherapy regimen was found to be more effective against B-cell precursor ALL than T-cell ALL 23 . This clinical observation is consistent with the particularly important role of TOP2B in B cell precursors, rather than in T cell lineage. Thus, etoposide, as well as other inhibitors of type II topoisomerases, may be especially effective for the treatment of B-cell malignancies.
Supplementary Material
Acknowledgements
We would like to acknowledge the important contribution of Dr. David Webster, who worked with the BILU patients, but, sadly, passed away before this manuscript was completed. S.N. was supported by the Wellcome Trust Senior Research Fellowship (095198/Z/10/Z), ERC Starting grant (260477), ERC Advanced grant (832721), MRC grant (MR/M012328) and the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre. O.P. was supported by the Marie Skłodowska-Curie Individual Fellowship (657633). AC was supported by Wellcome Clinical Research Career Development Fellowships (103413/Z/13/Z, 206618/Z/17/Z). S.K. is a Centre National de la Recherche Scientifique staff researcher. Authors declare no competing interests.
Footnotes
Author Contributions
S.N. conceived the study, identified the TOP2B mutation, planned the experiments and analyzed the data. O.P. planned and performed the experiments and analyzed the data with the help from A.C., D.E., M.M., A.A., J.C., E.B., and D.C.-L. S.I. analyzed bone marrow aspirate of the BILU patient. A.S.A analyzed lymphocytes in peripheral blood of the BILU patients. V.P. performed bioinformatics analysis of the exome data. S.O.B., S.K., A.D., O.H., C.P. and A.F. looked after the BILU patients and collected clinical data. K.O. participated in the planning of experiments and analyzed the data.
References
- 1.Conley ME, Dobbs AK, Farmer DM, et al. Primary B cell immunodeficiencies: comparisons and contrasts. Annu Rev Immunol. 2009;27:199–227. doi: 10.1146/annurev.immunol.021908.132649. [DOI] [PubMed] [Google Scholar]
- 2.Edery P, Le Deist F, Briard ML, et al. B cell immunodeficiency, distal limb abnormalities, and urogenital malformations in a three generation family: a novel autosomal dominant syndrome? J Med Genet. 2001;38(7):488–493. doi: 10.1136/jmg.38.7.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tischkowitz M, Goodman F, Koliou M, et al. Autosomal dominant B-cell immunodeficiency, distal limb anomalies and urogenital malformations (BILU syndrome) - report of a second family. Clin Genet. 2004;66(6):550–555. doi: 10.1111/j.1399-0004.2004.00349.x. [DOI] [PubMed] [Google Scholar]
- 4.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pommier Y, Sun Y, Huang SN, Nitiss JL. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol. 2016;17(11):703–721. doi: 10.1038/nrm.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biersack H, Jensen S, Gromova I, Nielsen IS, Westergaard O, Andersen AH. Active heterodimers are formed from human DNA topoisomerase II alpha and II beta isoforms. Proc Natl Acad Sci U S A. 1996;93(16):8288–8293. doi: 10.1073/pnas.93.16.8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gromova I, Biersack H, Jensen S, Nielsen OF, Westergaard O, Andersen AH. Characterization of DNA topoisomerase II alpha/beta heterodimers in HeLa cells. Biochemistry. 1998;37(47):16645–16652. doi: 10.1021/bi981391l. [DOI] [PubMed] [Google Scholar]
- 8.Aravind L, Leipe DD, Koonin EV. Toprim--a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26(18):4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450(7173):1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 10.Nitiss JL, Soans E, Rogojina A, Seth A, Mishina M. Topoisomerase assays. Curr Protoc Pharmacol. 2012 doi: 10.1002/0471141755.ph0303s57. Chapter 3:Unit 3.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broderick L, Yost S, Li D, et al. Mutations in topoisomerase IIbeta result in a B cell immunodeficiency. Nat Commun. 2019;10(1):3644. doi: 10.1038/s41467-019-11570-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durandy A, Kracker S, Fischer A. Primary antibody deficiencies. Nat Rev Immunol. 2013;13(7):519–533. doi: 10.1038/nri3466. [DOI] [PubMed] [Google Scholar]
- 13.Boisson B, Wang YD, Bosompem A, et al. A recurrent dominant negative E47 mutation causes agammaglobulinemia and BCR-B cells. J Clin Invest. 2013;123(11):4781–4785. doi: 10.1172/JCI71927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conley ME, Dobbs AK, Quintana AM, et al. Agammaglobulinemia and absent B lineage cells in a patient lacking the p85alpha subunit of PI3K. J Exp Med. 2012;209(3):463–470. doi: 10.1084/jem.20112533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haffner MC, Aryee MJ, Toubaji A, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42(8):668–675. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ju BG, Lunyak VV, Perissi V, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312(5781):1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 17.Madabhushi R, Gao F, Pfenning AR, et al. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell. 2015;161(7):1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trotter KW, King HA, Archer TK. Glucocorticoid Receptor Transcriptional Activation via the BRG1-Dependent Recruitment of TOP2beta and Ku70/86. Mol Cell Biol. 2015;35(16):2799–2817. doi: 10.1128/MCB.00230-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King IF, Yandava CN, Mabb AM, et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013;501(7465):58–62. doi: 10.1038/nature12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uuskula-Reimand L, Hou H, Samavarchi-Tehrani P, et al. Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biol. 2016;17(1):182. doi: 10.1186/s13059-016-1043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canela A, Maman Y, Jung S, et al. Genome Organization Drives Chromosome Fragility. Cell. 2017;170(3):507–521. doi: 10.1016/j.cell.2017.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hijiya N, Gaynon P, Barry E, et al. A multi-center phase I study of clofarabine, etoposide and cyclophosphamide in combination in pediatric patients with refractory or relapsed acute leukemia. Leukemia. 2009;23(12):2259–2264. doi: 10.1038/leu.2009.185. [DOI] [PubMed] [Google Scholar]
- 23.Locatelli F, Testi AM, Bernardo ME, et al. Clofarabine, cyclophosphamide and etoposide as single-course re-induction therapy for children with refractory/multiple relapsed acute lymphoblastic leukaemia. Br J Haematol. 2009;147(3):371–378. doi: 10.1111/j.1365-2141.2009.07882.x. [DOI] [PubMed] [Google Scholar]
- 24.Hijiya N, Thomson B, Isakoff MS, et al. Phase 2 trial of clofarabine in combination with etoposide and cyclophosphamide in pediatric patients with refractory or relapsed acute lymphoblastic leukemia. Blood. 2011;118(23):6043–6049. doi: 10.1182/blood-2011-08-374710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.