Abstract
Intracellular transport along microtubules enables cellular cargoes to efficiently reach the extremities of large, eukaryotic cells. While it would take more than 200 years for a small vesicle to diffuse from the cell body to the growing tip of a one-meter long axon, transport by a kinesin allows delivery in one week. It is clear from this example that the evolution of intracellular transport was tightly linked to the development of complex and macroscopic life forms. The human genome encodes 45 kinesins, 8 of those belonging to the family of kinesin-3 organelle transporters that are known to transport a variety of cargoes towards the plus end of microtubules. However, their mode of action, their tertiary structure, and regulation are controversial. In this review, we summarize the latest developments in our understanding of these fascinating molecular motors.
Keywords: molecular motors, microtubule-based transport, kinesin, autoinhibition, intracellular transport, Unc104/KIF1, cargo trafficking
Kinesins are molecular motors that step along microtubule tracks, thereby converting the chemical energy of one ATP per step into mechanical work. While moving along the microtubule, kinesins haul intracellular cargo such as chromosomes or mitochondria to achieve their correct positioning and transport secretory vesicles from the cell center to the cell cortex. Common to all kinesins is a structure that consists of a motor domain, a neck, and a tail. The motor domain combines both microtubule binding and ATPase activity. The ATP hydrolysis cycle is coupled to conformational changes within the motor and neck domains that result in forward movement of the tail-attached cargo. ATP turnover drives a sequence of conformational changes that cyclically change the microtubule binding affinity of the motor domains [1]. Kinesin motors exist in all eukaryotes and have been divided into 15 families based on the position and sequence homology of their motor domain [2, 3]. Amongst the 45 human kinesins, the largest family is the kinesin-3 family, a class of plus-end-directed transporters that have been implicated in the long-distance transport of vesicles and organelles in a variety of eukaryotic cells.
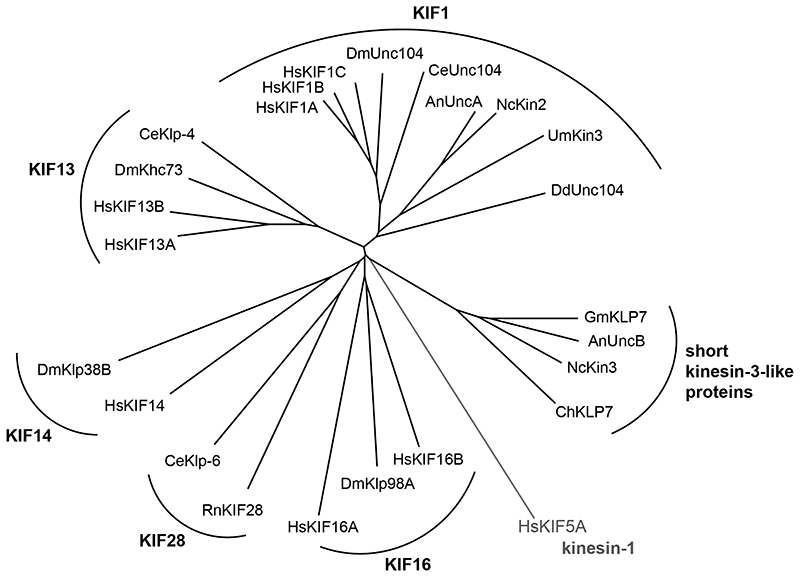
The founding member of the kinesin-3 family is Unc-104 from the nematode worm Caenorhabditis elegans. Mutations in Unc-104cause impaired transport of synaptic vesicles to the axon terminal and uncoordinated and slow movement of the nematode [4, 5]. Kinesin-3 family members have since been identified as fast organelle transporters in the amoeba Dictyostelium discoideum[6], as endosome transporters in fungi [7–9], and as transporters of vesicles, viral particles, and mitochondria in mammalian cells [10–17]. Kinesin-3s are thought to have been present in the last common eukaryotic ancestor, suggesting that cytoplasmic vesicle transport is evolutionarily ancient, even though today’s land plants lack kinesin-3s [3]. The kinesin-3 family comprises six subfamilies: the KIF1, KIF13, KIF14, KIF16, and KIF28 motors [18] plus a fungal-specific group of short kinesin-3-like proteins [19] (Fig. 1). While vertebrates usually have nine kinesin-3 genes with one to three of these representing each of the five major subfamilies [18], filamentous fungi usually have one KIF1 representative (Kin3 in Ustilago maydis, UncA in Aspergillus, and NKin2 in Neurospora crassa) plus one short kinesin-3-like protein (UncB in Aspergillus and NKin3 in Neurospora crassa) [7–9, 19]. Please see Fig. 1 for the phylogenetic relationship of kinesin-3 family members from vertebrates (Homo sapiens), insects (Drosophila melanogaster), worms (Caenorhabditis elegans), and several fungi, incorporating all motors mentioned in this review.
Fig. 1. Kinesin-3 tree.
Phylogenetic tree of all kinesin-3 family members from Homo sapiens (Hs), Drosophila melanogaster (Dm), Caenorhabditis elegans (Ce), Ustilago maydis (Um), Aspergillus nidulans (An), Neurospora crassa (Nc), and Dictyostelium discoideum (Dd). Selected kinesin-3 members from Rattus norvegicus (Rn), Gibberella moniliformis (Gm), and Cochliobolus heterostrophus (Ch) are also shown. Subfamilies are indicated in bold font. Human KIF5A, a kinesin-1, was used as root (shown in gray). To calculate tree information in Clustal Omega [130], kinesin motor domain sequences were aligned and cropped to a ~330-bp-long conserved region. The tree information was then used to generate a radial tree using T-REX tree viewer [131].
The number and variety of kinesin-3 motors in higher eukaryotes likely reflects the requirement for many different cargoes to be transported into different regions of the cell; thus, the different kinesin-3s are equipped with different specificities for both the cargo and the microtubule track and are activated by different mechanisms, as we will detail in the following sections.
Cellular Function and Human Disease
Kinesin-3-mediated transport is required for neuronal morphogenesis and function; mutations in any of the KIF1 motors KIF1A, KIF1B, or KIF1C cause neurological disorders, spastic paraplegia, or multiple sclerosis both in human patients and mouse models [13, 20–23]. DmUnc104 mutants also show defects in neuronal development, in particular in the morphogenesis of synaptic terminals and dendrites [24]. In fungi, transport of endosomes by kinesin-3 motors is required for optimal hyphal growth [9, 25]. Caenorhabditis elegans worms require axonal transport by Unc-104 for the coordination of their movement [5]. In addition, kinesin-3 motors have been shown to regulate signaling processes and the orderly progression of cell division. For example, KIF16A tethers the pericentriolar material (PCM) to the daughter centriole during mitosis, thereby preventing PCM fragmentation and enabling the formation of a bipolar mitotic spindle [26]; and KIF13A translocates a component of the cell abscission machinery to the spindle midzone, thereby controlling cytokinesis [27]. Likewise, deletion of the sole kinesin-3 in U. maydis results in a cell separation defect [7]. Important cargoes of kinesin-3 proteins are summarized in the table and range from mitochondria and viruses to vesicles containing a variety of receptors, pre-synaptic signaling proteins, microtubule regulators, and phospholipids [11, 12, 14, 15, 28–31]. It is becoming clear that the main function of kinesin-3 motors across species is the long-distance transport of membranous cargo. Kinesin-3 motors are exceptional in their high processivity, i.e. the distance they walk before falling off the microtubule track. This makes them particularly suited for long-haul tasks, and in the next section we will discuss the structure of kinesin-3 molecules and point out the features that underlie their properties.
Kinesin-3 cargoes. List of selected cargoes identified to be transported by kinesin-3 family members.
| Motor | Cargo | Cell type | Reference |
|---|---|---|---|
| KIF1A | tyrosine kinase A receptor (TrkA) | mouse dorsal root ganglion neurons | [32] |
| synaptotagmin and synaptophysin | rat spinal nerves (cauda equina) | [14] | |
| dense core vesicles (DCVs) | rat primary hippocampal neurons | [11] | |
| beta secretase-1 (BACE-1) | mouse SCG neurons | [33] | |
| AMPA receptors | Rat brain | [34] | |
| KIF1B | mitochondria | mouse Neuro2a cells | [12] |
| SCG10 / Stathmin-2 | sensory axons in zebrafish | [28] | |
| lysosomes | Cos7 African green monkey fibroblast cells | [35] | |
| KIF1C | α5β1-integrin | RPE1 human epithelial cell line | [15] |
| KIF13A | serotonin type 1A receptor | mouse hippocampal neurons | [36] |
| viral matrix proteins | Huh7 human hepatoma cell line | [29] | |
| mannose-6-phosphate receptors (MPRs) | MDCK canine epithelial cell line | [37] | |
| FYVE-CENT | HeLa human cervical cancer cell line | [27] | |
| KIF13B | human discs large (hDlg) tumor suppressor | in vitro reconstitution with purified human KIF13B | [38] |
| PtdIns(3,4,5)P3-containing vesicles | rat PC12 cells and in vitro reconstitution | [30] | |
| vascular endothelial growth factor receptor 2 (VEGFR2) | human umbilical vein endothelial cells (HUVECs) | [39] | |
| transient receptor potential vanilloid 1 (TRPV1) | CHO cells, rat dorsal root ganglion neurons | [40] | |
| KIF16B | fibroblast growth factor receptor (FGFR) | mouse embryonic stem cells | [31] |
| Kin3 | early endosomes | Ustilago maydis | [41] |
| Nkin2 / Nkin3 | mitochondria | Neurospora crassa | [19] |
| UncA | early endosomes | Aspergillus nidulans | [9] |
| Unc-104 | presynaptic vesicles | Caenorhabditis elegans | [4] |
| KLP-6 | mitochondria | Caenorhabditis elegans Neuro2 cells | [42] |
| polycystins LOV-1 and PKD-2 | Caenorhabditis elegans | [43] | |
| KLP-4 | GLR-1 glutamate receptors | Caenorhabditis elegans | [44] |
Structure of Kinesin-3 Motors
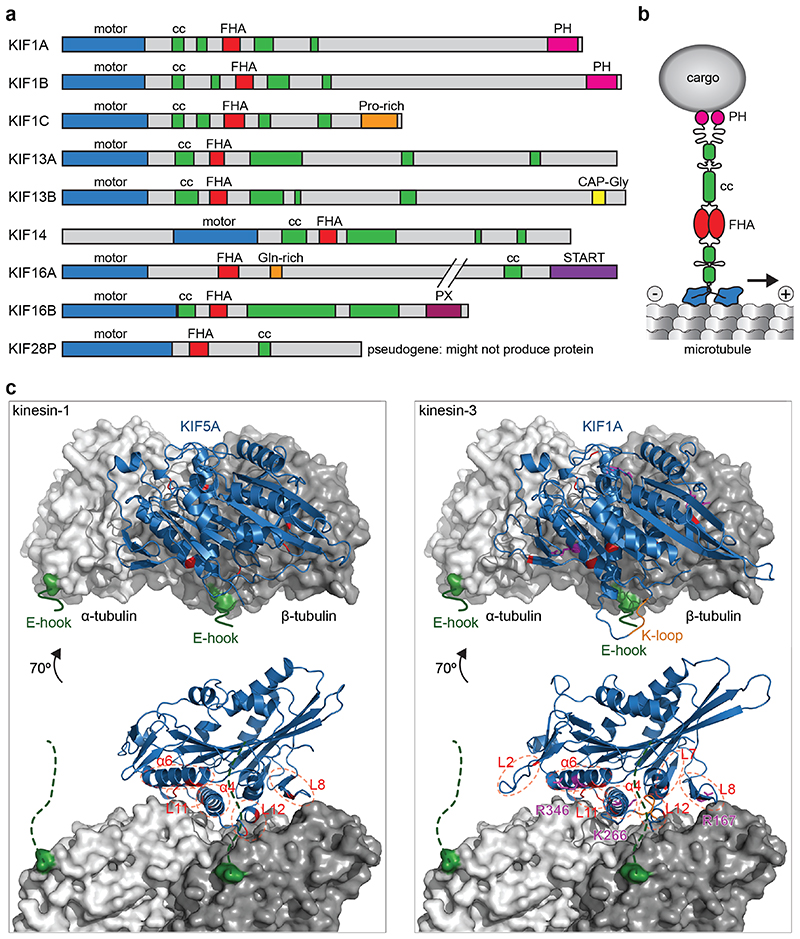
All kinesin motors that walk towards the plus end of microtubules have their motor domain at the N-terminus of the molecule. This is also true for kinesin-3 family motors (Fig. 2a). What sets kinesin-3 motors apart from other kinesins is the organization of the neck region, which contains a β-sheet as well as a helix [18], and the presence of a forkhead-associated (FHA) domain [45] in the tail. In addition to the FHA domain, the tail region contains several short coiled-coils and diverse protein and lipid interaction domains that mediate binding to cargo and regulators (Fig. 2, a and b). In this section, we will discuss kinesin-3 specific features of each region, those that are common to most kinesin-3 motors and those that give a motor unique properties.
Fig. 2. Structure of kinesin-3 motors.
a) Primary structure of human kinesin-3 members with characteristic N-terminal motor domain, FHA domain, and tail with several short coiled-coil (CC) regions in addition to a variety of protein or lipid interaction motifs. b) Schematic representation of a dimeric kinesin-3 motor and its interaction with the microtubule surface as well as a cargo vesicle. c) Structural model of kinesin motor domains binding to the microtubule (one αβ - tubulin heterodimer shown, in gray). The flexible C-terminal tubulin tails (E-hooks) are indicated in green. Key regions of the kinesin motor domain (blue) that contribute to interaction with microtubules are highlighted in red for both KIF5A, a kinesin-1, and KIF1A, a kinesin-3. Key residues that were shown to contribute to 10-fold higher processivity of kinesin-3 are shown in magenta [53, 54]. PDB accession numbers: 4UXP and 4UXY.
The motor domain binds to the microtubule, and the energy from ATP hydrolysis is used to produce directional movement [46, 47]. A characteristic feature of the kinesin-3 family is the presence of a stretch of positively charged lysine residues designated as the K-loop in loop 12 of the motor domain. This loop is ideally positioned so that it can contact the negatively charged glutamate-rich (E-hook) C-terminal tail of β-tubulin (Fig. 2c). The K-loop was proposed to enable processive motion by monomeric KIF1A by mediating diffusive interaction to microtubules throughout the ATPase cycle [48, 49]. However, while the K-loop in KIF1, KIF13, and KIF16 has been shown to increase microtubule affinity [50–52], an increase in processivity could not be attributed to the K-loop when these motors are working as dimers [50]. Instead, the K-loop increases the microtubule-binding rate and enables kinesin-3 motors to effectively work in teams [50, 51]. Recent comparative high-resolution cryoelectron microscopy structures of kinesin-1 (KIF5A) and kinesin-3 (KIF1A) motor domains bound to microtubules in different nucleotide states paired with molecular dynamics simulations ascertained which family-specific residue changes result in the 200-fold increased affinity of kinesin-3 motors to microtubules relative to kinesin-1 [53, 54]. These residues reside in loops L2, L7, L8, L11, L12, and α-helices α4 and α6 (Fig. 2c). Thus, the contribution of multiple sites increases kinesin-3s’ interaction surface with microtubules and results in a large effect on affinity. This increased affinity then increases the processivity of dimeric kinesin-3 motors [54]. Key residues that result in a 10-fold increased processivity of kinesin-3 versus kinesin-1 are Arg167 in loop 8, Lys266 in loop 11, and Arg346 in α-helix 6 of KIF1A (Fig. 2c) [53].
Coiled coils are important structural features that mediate motor dimerization [55]. Kinesin-3 motors tend not to contain the extended coiled coils that are typical for the tails of other kinesins, but instead contain several smaller predicted coiled-coil regions scattered along the tail (Fig. 2a). It is presently unclear whether all of these contribute to dimer formation. So far, the only direct test of this was performed with the fourth coiled-coil domain of KIF1C, which is sufficient to drive dimerization in a yeast-two-hybrid assay [56]. In KIF1A, KIF13A, and KIF13B, the coiled-coil domains seem to interfere with dimerization. It has been shown that instead, the neck coil alone efficiently dimerizes these motors [57, 58].
FHA domains are small protein modules that recognize phosphothreonine epitopes on proteins and mediate protein–protein interactions [59, 60]. FHA domains have been found in more than 200 different proteins with diverse cellular functions such as transcription, DNA repair, and protein degradation [61]. Besides fulfilling a structural role in kinesin-3 proteins, the FHA domain also confers specific cargo interactions. For example, the FHA domain of KIF13B medicates binding to its cargo transient receptor potential vanilloid 1 (TRPV1). Interestingly, this interaction depends on phosphorylation of KIF13B at T506 in the FHA domain by cyclin-dependent kinase 5 (Cdk-5) [40]. A point mutation that is likely to alter the folding of the FHA domain of KIF1C causes a change in the susceptibility of mice to anthrax lethal toxin, further demonstrating the functional importance of the domain [61, 62].
Several kinesin-3 tails contain domains that allow direct interaction with membranes, e.g. KIF16A contains a START/lipid sterol-binding domain at the C-terminus [26]. KIF1A and KIF1B have a pleckstrin homology (PH) domain that is important for binding cargo vesicles [63], probably through specific interaction with phosphatidylinositol4,5-bisphosphate (PtdIns(4,5)-P2) [64]. KIF16B possesses a phosphoinositide-binding structural domain (PX), which binds to PtdIns(3,4,5)P3 and is involved in the trafficking of early endosomes [65, 66].
Other kinesin-3 tails contain protein interaction domains, such as a CAP-Gly domain at the C-terminus of KIF13B. CAP-Gly domains bind to sequence motifs at the C-terminus of tubulin and EBs, zinc-finger motifs, and proline rich sequences [67]. KIF1C possesses a proline-rich region at the C-terminus. Proline-rich regions play a structural role and also act as binding sites for protein interaction [68]. In the case of KIF1C, this domain mediates several protein interactions, including the cargo adapter protein BICDR1, 14-3-3 proteins, and Rab6 [56, 69, 70].
Surprisingly, a monomeric motor construct of KIF1A has been observed to undergo processive plus-end directed movement along microtubules [48]. This is thought to be possible due to the presence of the K-loop and a stable microtubule interaction surface that persists throughout the ATPase cycle (Fig. 2c) [49, 54]. However, monomeric KIF1A only moves very slowly (0.15μm/s) and weakly (~0.15pN) along microtubules, while multiple KIF1A motors transport cargo at 1.5 μm/s[14, 71]. Teams of 10 monomeric KIF1A motors have been proposed to become approximately 100-fold stronger than a single monomeric motor [72]; however, experimental data on the force generation of kinesin-3 teams are lacking. There is evidence suggesting that kinesin-3 motors exist as inactive monomers in cells until activated by dimerization [58, 73–75]. Other studies suggest that KIF1A motors are dimeric in vivo, but in an autoinhibited state until activated by cargo binding [6, 57]. Thus, the extent to which individual kinesin-3 family members exist as monomers or dimers in cells remains to be elucidated. However, it is clear that a single monomeric motor cannot achieve the high processivity of kinesin-3 mediated cargo transport observed in cells. Thus, these would need either to work in teams formed by recruitment of several monomeric motors to the same cargo, or to form dimers.
Mechanism of Autoinhibition
Early work and biochemical characterization of conventional kinesin revealed that the molecule exists in two conformations: a folded inactive conformation and an extended active one [76, 77]. A small peptide region in the tail of kinesin-1 binds to the motor domain to inhibit it [78–80]. While kinesin-3 motors do not contain such an extensive coiled-coil region with a hinge that allows neat folding and unfolding of the tail, inactive kinesin-3 motors have been shown to adopt a compact conformation with a crumpled tail [81], which probably extends when activated and/or under load. That the pool of motors exists in an autoinhibited state in cells is important because in the absence of cargo, motor activity needs to be tightly regulated to avoid microtubule crowding and futile ATP consumption.
Currently, there are two models of autoinhibition that apply to kinesin-3 motors. In the monomer–dimer switch model, intramolecular interactions involving neck and tail regions hold some kinesin-3 motors in a monomeric, inactive state. Upon activation, these motors dimerize with their neck coil and tail regions undergoing intermolecular interactions. In the alternative tail block model, the motors are stable dimers, but regions of the tail interact with the motor or neck domains and interfere with motor activity until cargo binding occupies the tail region and releases the motor. Evidence exists for both models, and the picture emerging is that different kinesin-3 motors might use either or a combination of both of these methods of autoinhibition.
Most KIF1 and KIF13 motors are thought to undergo a monomer–dimer switch. Consistently with an autoinhibited state, the full length CeUnc-104 and MmKIF1A are inactive in motility assays [14, 57]. As a monomeric motor domain construct of KIF1A could produce some directional motion by itself and work as a processive motor when dimerized artificially [48, 75], regions of the neck or tail interfere with motor activity. Indeed, in Unc-104, the two neck helices can form an intramolecular coiled-coil, thereby inhibiting the ATPase and microtubule binding cycle of the motor and holding the motor in a monomeric state [73]. The neck helices can also form an intermolecular coiled-coil, thereby enabling the switch from monomer to dimer, which is required to obtain a processive Unc-104 motor [73]. In MmKIF1A, a similar switch through intra- and intermolecular coiled-coil formation is proposed to occur between the neck coil region and the first coiled-coil domain (CC1). Surprisingly, the truncation of the entire tail results in processive dimeric motors of KIF1A, KIF13A, and KIF13B, while all longer constructs containing CC1 result in monomers that only show diffusive movement [57, 58]. If autoinhibition is prevented by deletion of the flexible hinge between the neck helices in C. elegans Unc-104, the motility of the motor in vitro is unperturbed, but transgenic worms show severe defects in the coordination of their movement [73]. Likewise, mutations in the CC1 segment of KIF1A result in activation of the motor [82, 83]. In the KIF13 subfamily, a proline residue at the junction of neck coil and CC1 provides the flexibility to enable CC1 to fold back and interact with the neck coil. Deletion of this proline residue results in dimerization via the neck coil domains and active, processive motors [58, 84]. Control of the autoinhibited state of the KIF1A motor might also involve the FHA domain and the following coiled coil CC2. A tandem construct of CC1 and FHA domains forms a very stable dimer. Furthermore, the dimerization of CC1–FHA sequesters the CC1 region and makes it unavailable for the autoinhibition of the neck coil region [82]. Also, CC2 can fold back to interact with the FHA domain, which disrupts the motor activity [57]. Disruption of the CC1–FHA dimer severely impairs synaptic vesicle transport and locomotion in C. elegans worms, suggesting that robust dimerization is crucially important for KIF1A function in vivo [83].
Evidence for a tail-block mechanism exists for KIF13B and KIF16B. In KIF16B, microtubule binding is inhibited by the interaction of the second and third coiledcoil with the motor domain in an ATP-dependent manner. This tail-mediated inhibition is important for the correct localization of early endosomes to somatodendritic regions in neurons and the recycling of AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolpropionate) and NGF (nerve growth factor) receptors [85]. An interaction of a tail domain with the motor domain also contributes to the autoinhibition of KIF13B [38, 86]. Upon phosphorylation of KIF13B close to its C-terminus by Par1b/MARK2 (microtubule affinity-regulating kinase), 14-3-3β binds and promotes the intramolecular interaction of KIF13B motor and tail domains. This in turn negatively regulates KIF13B microtubule binding, resulting in the dispersal of the motor in the cytoplasm and a reduction in cell protrusion and axon formation [86]. In addition, KIF1C, which is known to exist as a stable dimer, interacts with 14-3-3 proteins in a phosphorylation-dependent manner [56]. However, whether this mediates an autoinhibitory tail–motor interaction similarly to KIF13B remains to be elucidated.
Taken together, these data suggest specific autoinhibition mechanisms for each kinesin-3 family member. These might require different interaction partners to achieve release from autoinhibition and activate the motors for transport of specific cargoes.
Activation By Cargo Interaction
Many kinesins are activated upon cargo binding. Full-length KIF13B, also known as guanylate kinase-associated kinesin (GAKIN), exists in an autoinhibited state in solution. It is activated by the direct binding of its cargo, human disc large (hDlg) tumor suppressor [38]. In contrast to KIF1A, full-length KIF13B is active in a gliding assay. This could be because the binding of the C-terminus to the glass surface might mimic the cargo-bound state, thus relieving autoinhibition [38]. In contrast, KIF16B is a monomer in the cytoplasm and dimerizes at the cargo surface. The localized dimerization of KIF16B on early endosomes has been directly observed using Förster resonance energy transfer (FRET) in live cells [58]. Thus, these examples support the idea that due to the diverse cargo binding tail, the different kinesin-3 family members use diverse means of autoinhibition and cargo-dependent release of inhibition, involving changes in the dimerization status for some members and competitive binding of a peptide region that weakly interacts with the motor domain for others. The mechanisms of cargo-mediated activation thus require elucidation for each family member.
While some motors bind their cargo directly, often cargo adapter proteins mediate both the motor activation and cargo recruitment. For C. elegans kinesin-3 motor Unc-104, a number of adapter proteins are known that are involved in cargo loading; a bimolecular fluorescence complementation assay (BiFC) was employed to show that binding of different adapters Unc-16 (JIP3), DNC-1 (DCTN-1/Glued), and SYD-2 (Liprin-α) to Unc-104 results in translocation to different subcellular compartments in neuronal cells. This suggests that adapter proteins can recruit the motor to their cargo and steer their transport [16, 87]. Further, binding of LIN-2 (CASK) and SYD-2 was shown to positively regulate the Unc-104 motor by increasing its velocity, and binding of LIN-2 also increased run lengths. The cargo transport of synaptobrevin-1 (SNB-1) was markedly reduced in the neurons of LIN-2 knockout worms, implying that LIN-2 is an activator of Unc-104 motor [88]. In Ustilago maydis,the cargo adapter Hook protein (Hok1) mediates the recruitment of Kin3 and dynein to early endosomes and regulates bidirectional motility. Hok1 releases Kin3, and this allows for dynein to bind and drive the subsequent change in directionality [41]. Like Kin3, KIF1C binds to another dynein adapter protein, Bicaudal-D-related protein 1 (BICDR-1) [69]. BICDR-1 also binds Rab6A vesicles, thus linking both motors to secretory vesicles and controlling the bidirectional vesicle transport in developing neurons [69]. Centaurin α1 (CENTA1) acts as a cargo adapter for KIF13B and recruits the motor to PtdIns(3,4)P2/PtdIns(3,4,5)P3-containing vesicles [30, 89]. CENTA1 contains two PH domains that bind the headgroups of phosphoinositides, and PH1 also directly binds the FHA domain of KIF13B in a phosphorylation-independent manner [89]. As KIF13B FHA simultaneously interacts with the ArfGAP domain of a second CENTA1 molecule, CENTA1–KIF13B form a heterotetrameric transport complex for PtdIns(3,4,5)P3-rich vesicles [30, 89].
Regulation By Rab GTPases
The members of the Rab family of GTPases are known to control the localization of vesicles/organelles in a nucleotide-dependent manner. Rab proteins act at all stages including vesicle formation, motility, and tethering of vesicles to the designated compartment [90]. Rab GTPases exist in either GTP or GDP bound states, and are activated by GEFs (GTP/GDP exchange factors) and switched off by GAPs (GTPase activating factors) [91]. Once activated, the Rab proteins bind to vesicles that are translocated to the destination compartment, where they dock and fuse. The Rab proteins are then recycled back via a cytosolic intermediate [92]. KIF1A and KIF1Bβ both transport Rab3-coated vesicles in the axon. Rab3 is a synaptic vesicle protein that controls the exocytosis of synaptic vesicles [93–95]. It has been found that DENN/MADD (differentially expressed in normal and neoplastic cells/MAP kinase activating death domain), a GEF for Rab3, binds to Rab3 and the tail domain of KIF1A and KIF1Bβ and is thought to mediate the transport to the axon terminal while maintaining Rab3 in the GTP-bound form [13].
Rab6 binds to KIF1C at two sites, to the motor domain and near the C-terminus. Rab6 binding to the motor domain disrupts the motor’s ability to bind microtubules [70], while the binding to the C-terminus might activate cargo loading and relief from autoinhibition. Secretory Rab6 vesicles are transported bidirectionally, and it is thought that the dual ability of Rab6 to activate and inhibit KIF1C might regulate the directional switch. KIF1C also transports Rab11-positive vesicles for the recycling of integrins [15]. Whether Rab11 is directly involved in controlling the activity of KIF1C is yet unclear.
KIF13A binds to the active GTP-bound recycling endosomes associated with Rab11 and controls endosomal sorting and recycling of endosomal cargo [96]. KIF16B transports Rab5-positive early endosomes and Rab14-positive vesicles in non-neuronal cells [31, 66]. Also, the Neurospora kinesin-3 NKin2 colocalizes with the Rab5 GTPase YPT52 [8]. DmKlp98A interacts with Rab14 and Atg8 (autophagic-vesicle associated protein). This allows the motor to transport endocytic and autophagic vesicles [97]. To what extent these Rabs affect kinesin remains to be understood.
Specificity for a Subset of Microtubule Tracks
The microtubule tracks on which kinesin motors walk are not uniform. Depending on the cell type or its differentiation status, cells express different tubulin isoforms, accumulate microtubules with different posttranslational modifications, and also express different microtubule-associated proteins (MAPs) that decorate the microtubules. Kinesins are known to be sensitive to both changes to tubulin and MAP composition.
Tubulin undergoes a diverse range of chemical modifications known as posttranslational modifications after polymerization into microtubules. These modifications mainly occur on the C-terminal tails of both α- and β-tubulin and include the removal of terminal amino acids, such as detyrosination, and the addition of polyglutamate and polyglycine side chains [98–100]. Considering that the kinesin-3-specific K-loop is thought to interact with the C-terminal tail of β-tubulin (Fig. 2b), it is expected that changes in this region would impact kinesin-3 binding. Further modification at other sites of tubulin have been described, such as the acetylation of K40 in α-tubulin and phosphorylation of tubulin at various sites [101]. These modifications may change the stability of microtubules and act as signposts for motor transport by selectively increasing or decreasing the affinity of certain motors to the microtubule [102]. In line with this idea, knockdown of polyglutamylase PGs1 in ROSA22 mice decreases the localization of KIF1A to neurites [103]. Further, the ciliary localization of kinesin-3 KLP-6 in C. elegans is positively regulated by tubulin deglutamylase CCPP-1 [104]. However, in COS cells, the truncated, constitutively active KIF1A(1-393) was a non-selective motor [105]. The fungal kinesin-3 UncA from A. nidulans has been reported to selectively walk on detyrosinated microtubules, and the tail is necessary and sufficient for this recognition [9, 106]. Also, the N. crassa kinesin-3 NKin2 preferentially binds to a subset of microtubules [8]. However, this feature is not conserved in all fungi, as Kin3 from U. maydis uses all microtubules equally [107]. Like the finding in COS cells, the negative result could be due to the lack of modified microtubules in these cells rather than a different property of the motor, and this would require further investigation to elucidate. The subcellular localization of KIF1C is regulated by acetylation in primary human macrophages in a way that suggests that tubulin acetylation is a negative signal for KIF1C transport [108]. Likewise, KIF1Bβ and KIF1A have been reported to drive lysosomal transport preferentially along tyrosinated (i.e. non-modified) microtubules [109]. These data suggest that most kinesin-3 motors are sensitive to tubulin posttranslational modifications, but with different preferences.
MAPs regulate the assembly and disassembly kinetics of microtubules as well as the interactions of motors with microtubules [110, 111]. Lattice-decorating MAPs such as the neuronal protein tau regulate the attachment rate and can act as roadblocks that affect motors differently, depending on their ability to take side or backwards steps to circumvent the roadblock [112–114]. MAP4, which is a tau-related protein in non-neuronal cells, negatively regulates force generation and transport by dynein, but it positively regulates kinesin-based movement [115, 116]. Thus, MAPs can regulate microtubule-based transport directionality and access of motors to microtubules.
For kinesin-3, MAPs known to regulate the motor include doublecortin-like kinase-1 (DCLK-1), which regulates KIF1 transport of dense core vesicles (DCVs) along dendrites in neurons. DCLK-1 specifically binds to microtubules in dendrites, which acts as a positive signal to promote dendritic transport of KIF1 cargoes. In the absence of DCLK-1, KIF1 motors predominantly transport DCVs into the axon [117]. In C. elegans, the retrograde motility of Unc-104 was affected in tau/PTL-1 (protein with tau-like repeats) knockout worms. Unc-104 usually moves bidirectionally, but in the absence of PTL-1 the motor travels preferentially in anterograde direction [118]. It is thought that kinesin-3 motors cooperate with dynein for bidirectional motility, so whether PTL-1 affects Unc-104 directly or negatively regulates dynein to cause the observed phenotype remains to be elucidated. The microtubule plus-end tracking protein CLASP is required to stimulate the trafficking of KIF1C [119]. KIF1C has also been described to move with growing microtubule plus ends in cells [120]. This could be either due to the preference for unmodified (i.e. freshly assembled) microtubules [108], or due to its fast transport speed and thus ability to catch up with the growing microtubule end [51], or due to its interaction with CLASP [119].
Cooperation of Motors
Kinesin-3s have been implicated in the bidirectional transport of cargo. This means that when a specific kinesin-3 is inhibited or depleted, the transport of its cargo both towards the plus and the minus end of the microtubule is impaired [15, 118, 121]. It has been suggested that kinesin-3 cooperates with dynein in the bidirectional transport of cargoes, but the mechanism underlying the mutual activation of these opposite-polarity motors remains to be elucidated [122]. It has been suggested that cooperation depends on the opposing force generated, resulting in a mechanical activation [121]. Other proposed models include a steric inhibition mechanism whereby the direct interaction of the opposing motor or accessory protein relieves autoinhibition, and a microtubule tethering mechanism whereby the opposing motor is in a weakly bound state and acts as a processivity factor [122]. This is different to the idea of tug-of-war that has been proposed and reconstituted for kinesin-1 and dynein-mediated transport, where the motors pull against each other and the strongest team wins, i.e. the number of motors of each type loaded to a cargo molecule and the force that each motor can produce determine the net movement of the cargo [123, 124]. Potential linkers to facilitatecooperation of dynein and kinesin-3 include Hook and Bicaudal, cargo adapter proteins that have been identified to interact with both dynein and kinesin-3 tail domains [41, 69, 125, 126]. Interestingly, the presence of BICD2 increases the force generation and processivity of dynein/dynactin [127, 128], demonstrating that these cargo adapter proteins regulate motor activity and could act as switches to control transport directionality within a complex containing two opposing motors. Other control mechanisms could come from accessory proteins such as kinesin-binding protein (KBP), which has been shown to stimulate KIF1B, but inhibit KIF1A-mediated bidirectional transport [28, 129]. If the activity of such regulatory proteins were spatially controlled, this would enable directional switching of transport complexes in the presence of opposing motors.
Kinesin-3 molecules are important cargo transporters in neuronal cells that show a number of remarkable features. Their high affinity to the microtubule surface in all nucleotide states makes them highly processive motors, ideally suited to drive long-distance transport in neuronal cells. Their processivity is so high that even monomeric motor domain constructs show some directional motion. The activity of kinesin-3 motors is tightly regulated, in some motors via a switch from monomer to dimer, in others via autoinhibitory interactions of motor and tail domains. These interactions are relieved by cargo recruitment or regulated by kinases. Kinesin-3 motors are sensitive to the changes in the microtubule track and follow signposting modifications such as posttranslational modifications of tubulin or MAP decoration. Finally, kinesin-3 motors cooperate with dynein to bring about bidirectional transport of cargo. Many of these fascinating features remain to be understood mechanistically. Future work will illuminate this problem and enable us to appreciate kinesin-3 motor physiology, including the causation of the disease states arising from mutated kinesin-3 motors.
Acknowledgments
We thank Kristen Verhey (University of Michigan) and Carolyn Moores (Birkbeck) for useful discussions about kinesin-3 motors, and Andrew McAinsh and Rob Cross for critical reading of the manuscript.
A. S. is a Prize Fellow of the Lister Institute of Preventive Medicine and a Wellcome Trust Investigator (200870/Z/16/Z). N. S. is funded by a Chancellor’s International PhD Scholarship of the University of Warwick.
References
- 1.Cross RA. Review: Mechanochemistry of the kinesin-1 ATPase. Biopolymers. 2016;105:476–482. doi: 10.1002/bip.22862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, Malmberg RL, et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wickstead B, Gull K, Richards TA. Patterns of kinesin evolution reveal a complex ancestral eukaryote with a multifunctional cytoskeleton. BMC Evol Biol. 2010;10:110. doi: 10.1186/1471-2148-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall DH, Hedgecock EM. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C.elegans . Cell. 1991;65:837–847. doi: 10.1016/0092-8674(91)90391-b. [DOI] [PubMed] [Google Scholar]
- 5.Otsuka AJ, Jeyaprakash A, Garcia-Anoveros J, Tang LZ, Fisk G, Hartshorne T, Franco R, Born T. The C. elegans unc-104 gene encodes a putative kinesin heavy chain-like protein. Neuron. 1991;6:113–122. doi: 10.1016/0896-6273(91)90126-k. [DOI] [PubMed] [Google Scholar]
- 6.Pollock N, De Hostos EL, Turck CW, Vale RD. Reconstitution of membrane transport powered by a novel dimeric kinesin motor of the Unc104/KIF1A family purified from Dictyostelium . J Cell Biol. 1999;147:493–506. doi: 10.1083/jcb.147.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wedlich-Soldner R. A balance of KIF1A-like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis . EMBO J. 2002;21:2946–2957. doi: 10.1093/emboj/cdf296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seidel C, Moreno-Velasquez SD, Riquelme M, Fischer R. Neurospora crassa NKIN2, a kinesin-3 motor, transports early endosomes and is required for polarized growth, Eukaryot . Cell. 2013;12:1020–1032. doi: 10.1128/EC.00081-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zekert N, Fischer R. The Aspergillus nidulans kinesin-3 UncA motor moves vesicles along a subpopulation of microtubules. Mol Biol Cell. 2009;20:673–684. doi: 10.1091/mbc.E08-07-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kratchmarov R, Kramer T, Greco TM, Taylor MP, Ch’ng TH, Cristea IM, Enquist LW. Glycoproteins gE and gI are required for efficient KIF1A-dependent anterograde axonal transport of alphaherpesvirus particles in neurons. J Virol. 2013;87:9431–9440. doi: 10.1128/JVI.01317-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo KY, Kuzmin A, Unger SM, Petersen JD, Silverman MA. KIF1A is the primary anterograde motor protein required for the axonal transport of dense-core vesicles in cultured hippocampal neurons. Neurosci Lett. 2011;491:168–173. doi: 10.1016/j.neulet.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 12.Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell. 1994;79:1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 13.Niwa S, Tanaka Y, Hirokawa N. KIF1Bbeta- and KIF1A-mediated axonal transport of presynaptic regulator Rab3 occurs in a GTP-dependent manner through DENN/MADD. Nat Cell Biol. 2008;10:1269–1279. doi: 10.1038/ncb1785. [DOI] [PubMed] [Google Scholar]
- 14.Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81:769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- 15.Theisen U, Straube E, Straube A. Directional persistence of migrating cells requires Kif1C-mediated stabilization of trailing adhesions. Dev Cell. 2012;23:1153–1166. doi: 10.1016/j.devcel.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Wagner OI, Esposito A, Kohler B, Chen CW, Shen CP, Wu GH, Butkevich E, Mandalapu S, Wenzel D, Wouters FS, Klopfenstein DR. Synaptic scaffolding protein SYD-2 clusters and activates kinesin-3 UNC-104 in C. elegans . Proc Natl Acad Sci USA. 2009;106:19605–19610. doi: 10.1073/pnas.0902949106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wozniak MJ, Melzer M, Dorner C, Haring HU, Lammers R. The novel protein KBP regulates mitochondria localization by interaction with a kinesin-like protein. BMC Cell Biol. 2005;6:35. doi: 10.1186/1471-2121-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Fuchs F. Role of Unc104/KIF1-related motor proteins in mitochondrial transport in neurospora crassa. Mol Biol Cell. 2004;16:153–161. doi: 10.1091/mbc.E04-05-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dor T, Cinnamon Y, Raymond L, Shaag A, Bouslam N, Bouhouche A, Gaussen M, Meyer V, Durr A, Brice A, Benomar A, et al. KIF1C mutations in two families with hereditary spastic paraparesis and cerebellar dysfunction. J Med Genet. 2014;51:137–142. doi: 10.1136/jmedgenet-2013-102012. [DOI] [PubMed] [Google Scholar]
- 21.Yonekawa Y, Harada A, Okada Y, Funakoshi T, Kanai Y, Takei Y, Terada S, Noda T, Hirokawa N. Defect in synaptic vesicle precursor transport and neuronal cell death in KIF1A motor protein-deficient mice. J Cell Biol. 1998;141:431–441. doi: 10.1083/jcb.141.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caballero Oteyza A, Battaloglu E, Ocek L, Lindig T, Reichbauer J, Rebelo AP, Gonzalez MA, Zorlu Y, Ozes B, Timmann D, Bender B, et al. Motor protein mutations cause a new form of hereditary spastic paraplegia. Neurology. 2014;82:2007–2016. doi: 10.1212/WNL.0000000000000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aulchenko YS, Hoppenbrouwers IA, Ramagopalan SV, Broer L, Jafari N, Hillert J, Link J, Lundstrom W, Greiner E, Dessa Sadovnick A, Goossens D, et al. Genetic variation in the KIF1B locus influences susceptibility to multiple sclerosis. Nat Genet. 2008;40:1402–1403. doi: 10.1038/ng.251. [DOI] [PubMed] [Google Scholar]
- 24.Kern JV, Zhang YV, Kramer S, Brenman JE, Rasse TM. The kinesin-3, Unc-104 regulates dendrite morphogenesis and synaptic development in Drosophila . Genetics. 2013;195:59–72. doi: 10.1534/genetics.113.151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lenz JH, Schuchardt I, Straube A, Steinberg G. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 2006;25:2275–2286. doi: 10.1038/sj.emboj.7601119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torres JZ, Summers MK, Peterson D, Brauer MJ, Lee J, Senese S, Gholkar AA, Lo YC, Lei X, Jung K, Anderson DC, et al. The STARD9/Kif16a kinesin associates with mitotic microtubules and regulates spindle pole assembly. Cell. 2011;147:1309–1323. doi: 10.1016/j.cell.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sagona AP, Nezis IP, Pedersen NM, Liestol K, Poulton J, Rusten TE, Skotheim RI, Raiborg C, Stenmark H. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat Cell Biol. 2010;12:362–371. doi: 10.1038/ncb2036. [DOI] [PubMed] [Google Scholar]
- 28.Drerup CM, Lusk S, Nechiporuk A. Kif1B interacts with KBP to promote axon elongation by localizing a microtubule regulator to growth cones. J Neurosci. 2016;36:7014–7026. doi: 10.1523/JNEUROSCI.0054-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fehling SK, Noda T, Maisner A, Lamp B, Conzelmann KK, Kawaoka Y, Klenk HD, Garten W, Strecker T. The microtubule motor protein KIF13A is involved in intracellular trafficking of the Lassa virus matrix protein Z. Cell Microbiol. 2013;15:315–334. doi: 10.1111/cmi.12095. [DOI] [PubMed] [Google Scholar]
- 30.Horiguchi K, Hanada T, Fukui Y, Chishti AH. Transport of PIP3 by GAKIN, a kinesin-3 family protein, regulates neuronal cell polarity. J Cell Biol. 2006;174:425–436. doi: 10.1083/jcb.200604031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ueno H, Huang X, Tanaka Y, Hirokawa N. KIF16B/Rab14 molecular motor complex is critical for early embryonic development by transporting FGF receptor. Dev Cell. 2011;20:60–71. doi: 10.1016/j.devcel.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka Y, Niwa S, Dong M, Farkhondeh A, Wang L, Zhou R, Hirokawa N. The molecular motor KIF1A transports the TrkA neurotrophin receptor and is essential for sensory neuron survival and function. Neuron. 2016;90:1215–1229. doi: 10.1016/j.neuron.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Hung CO, Coleman MP. KIF1A mediates axonal transport of BACE1 and identification of independently moving cargoes in living SCG neurons. Traffic. 2016;17:1155–1167. doi: 10.1111/tra.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin H, Wyszynski M, Huh KH, Valtschanoff JG, Lee JR, Ko J, Streuli M, Weinberg RJ, Sheng M, Kim E. Association of the kinesin motor KIF1A with the multimodular protein liprin-alpha. J Biol Chem. 2003;278:11393–11401. doi: 10.1074/jbc.M211874200. [DOI] [PubMed] [Google Scholar]
- 35.Matsushita M, Tanaka S, Nakamura N, Inoue H, Kanazawa H. A novel kinesin-like protein, KIF1Bbeta3 is involved in the movement of lysosomes to the cell periphery in non-neuronal cells. Traffic. 2004;5:140–151. doi: 10.1111/j.1600-0854.2003.00165.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhou R, Niwa S, Guillaud L, Tong Y, Hirokawa N. A molecular motor, KIF13A, controls anxiety by transporting the serotonin type 1A receptor. Cell Rep. 2013;3:509–519. doi: 10.1016/j.celrep.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 37.Nakagawa T, Setou M, Seog D, Ogasawara K, Dohmae N, Takio K, Hirokawa N. A novel motor, KIF13A, transports mannose-6-phosphate receptor to plasma membrane through direct interaction with AP-1 complex. Cell. 2000;103:569–581. doi: 10.1016/s0092-8674(00)00161-6. [DOI] [PubMed] [Google Scholar]
- 38.Yamada KH, Hanada T, Chishti AH. The effector domain of human Dlg tumor suppressor acts as a switch that relieves autoinhibition of kinesin-3 motor GAKIN/KIF13B. Biochemistry. 2007;46:10039–10045. doi: 10.1021/bi701169w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamada KH, Nakajima Y, Geyer M, Wary KK, Ushio-Fukai M, Komarova Y, Malik AB. KIF13B regulates angiogenesis through Golgi to plasma membrane trafficking of VEGFR2. J Cell Sci. 2014;127:4518–4530. doi: 10.1242/jcs.156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xing BM, Yang YR, Du JX, Chen HJ, Qi C, Huang ZH, Zhang Y, Wang Y. Cyclin-dependent kinase 5 controls TRPV1 membrane trafficking and the heat sensitivity of nociceptors through KIF13B. J Neurosci. 2012;32:14709–14721. doi: 10.1523/JNEUROSCI.1634-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bielska E, Schuster M, Roger Y, Berepiki A, Soanes DM, Talbot NJ, Steinberg G. Hook is an adapter that coordinates kinesin-3 and dynein cargo attachment on early endosomes. J Cell Biol. 2014;204:989–1007. doi: 10.1083/jcb.201309022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka K, Sugiura Y, Ichishita R, Mihara K, Oka T. KLP6: a newly identified kinesin that regulates the morphology and transport of mitochondria in neuronal cells. J Cell Sci. 2011;124:2457–2465. doi: 10.1242/jcs.086470. [DOI] [PubMed] [Google Scholar]
- 43.Peden EM, Barr MM. The KLP-6 kinesin is required for male mating behaviors and polycystin localization in Caenorhabditis elegans . Curr Biol. 2005;15:394–404. doi: 10.1016/j.cub.2004.12.073. [DOI] [PubMed] [Google Scholar]
- 44.Monteiro MI, Ahlawat S, Kowalski JR, Malkin E, Koushika SP, Juo P. The kinesin-3 family motor KLP-4 regulates anterograde trafficking of GLR-1 glutamate receptors in the ventral nerve cord of Caenorhabditis elegans. Mol Biol Cell. 2012;23:3647–3662. doi: 10.1091/mbc.E12-04-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westerholm-Parvinen A, Vernos I, Serrano L. Kinesin subfamily UNC104 contains a FHA domain: boundaries and physicochemical characterization. FEBS Lett. 2000;486:285–290. doi: 10.1016/s0014-5793(00)02310-3. [DOI] [PubMed] [Google Scholar]
- 46.Endow SA. Determinants of molecular motor directionality. Nat Cell Biol. 1999;1:E163–167. doi: 10.1038/14113. [DOI] [PubMed] [Google Scholar]
- 47.Vale RD, Case R, Sablin E, Hart C, Fletterick R. Searching for kinesin’s mechanical amplifier. Philos Trans R Soc Lond B Biol Sci. 2000;355:449–457. doi: 10.1098/rstb.2000.0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okada Y, Hirokawa N. A processive single-headed motor: kinesin superfamily protein KIF1A. Science. 1999;283:1152–1157. doi: 10.1126/science.283.5405.1152. [DOI] [PubMed] [Google Scholar]
- 49.Okada Y, Hirokawa N. Mechanism of the single-headed processivity: diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc Natl Acad Sci USA. 2000;97:640–645. doi: 10.1073/pnas.97.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soppina V, Verhey KJ. The family-specific K-loop influences the microtubule on-rate but not the superprocessivity of kinesin-3 motors, Mol. Biol Cell. 2014;25:2161–2170. doi: 10.1091/mbc.E14-01-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rogers KR, Weiss S, Crevel I, Brophy PJ, Geeves M, Cross R. KIF1D is a fast non-processive kinesin that demonstrates novel K-loop-dependent mechanochemistry. EMBO J. 2001;20:5101–5113. doi: 10.1093/emboj/20.18.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsushita M, Yamamoto R, Mitsui K, Kanazawa H. Altered motor activity of alternative splice variants of the mammalian kinesin-3 protein KIF1B. Traffic. 2009;10:1647–1654. doi: 10.1111/j.1600-0854.2009.00975.x. [DOI] [PubMed] [Google Scholar]
- 53.Scarabelli G, Soppina V, Yao XQ, Atherton J, Moores CA, Verhey KJ, Grant BJ. Mapping the processivity determinants of the kinesin-3 motor domain. Biophys J. 2015;109:1537–1540. doi: 10.1016/j.bpj.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Atherton J, Farabella I, Yu IM, Rosenfeld SS, Houdusse A, Topf M, Moores CA. Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins. Elife. 2014;3:e03680. doi: 10.7554/eLife.03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peckham M. Coiled coils and SAH domains in cytoskeletal molecular motors. Biochem Soc Trans. 2011;39:1142–1148. doi: 10.1042/BST0391142. [DOI] [PubMed] [Google Scholar]
- 56.Dorner C, Ullrich A, Haring HU, Lammers R. The kinesin-like motor protein KIF1C occurs in intact cells as a dimer and associates with proteins of the 14-3-3 family. J Biol Chem. 1999;274:33654–33660. doi: 10.1074/jbc.274.47.33654. [DOI] [PubMed] [Google Scholar]
- 57.Hammond JW, Cai D, Blasius TL, Li Z, Jiang Y, Jih GT, Meyhofer E, Verhey KJ. Mammalian kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 2009;7:e72. doi: 10.1371/journal.pbio.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soppina V, Norris SR, Dizaji AS, Kortus M, Veatch S, Peckham M, Verhey KJ. Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proc Natl Acad Sci USA. 2014;111:5562–5567. doi: 10.1073/pnas.1400759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J, Lee GI, Van Doren SR, Walker JC. The FHA domain mediates phosphoprotein interactions. J Cell Sci. 2000;113(Pt. 23):4143–4149. doi: 10.1242/jcs.113.23.4143. [DOI] [PubMed] [Google Scholar]
- 60.Hammet A, Pike BL, Mcnees CJ, Conlan LA, Tenis N, Heierhorst J. FHA domains as phospho-threonine binding modules in cell signaling. IUBMB Life. 2003;55:23–27. doi: 10.1080/1521654031000070636. [DOI] [PubMed] [Google Scholar]
- 61.Durocher D, Jackson SP. The FHA domain. FEBS Lett. 2002;513:58–66. doi: 10.1016/s0014-5793(01)03294-x. [DOI] [PubMed] [Google Scholar]
- 62.Watters JW, Dewar K, Lehoczky J, Boyartchuk V, Dietrich WF. Kif1C, a kinesin-like motor protein, mediates mouse macrophage resistance to anthrax lethal factor. Curr Biol. 2001;11:1503–1511. doi: 10.1016/s0960-9822(01)00476-6. [DOI] [PubMed] [Google Scholar]
- 63.Xue X, Jaulin F, Espenel C, Kreitzer G. PH-domain-dependent selective transport of p75 by kinesin-3 family motors in non-polarized MDCK cells. J Cell Sci. 2010;123:1732–1741. doi: 10.1242/jcs.056366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klopfenstein DR, Tomishige M, Stuurman N, Vale RD. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 2002;109:347–358. doi: 10.1016/s0092-8674(02)00708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blatner NR, Wilson MI, Lei C, Hong W, Murray D, Williams RL, Cho W. The structural basis of novel endosome anchoring activity of KIF16B kinesin. EMBO J. 2007;26:3709–3719. doi: 10.1038/sj.emboj.7601800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoepfner S, Severin F, Cabezas A, Habermann B, Runge A, Gillooly D, Stenmark H, Zerial M. Modulation of receptor recycling and degradation by the endosomal kinesin KIF16B. Cell. 2005;121:437–450. doi: 10.1016/j.cell.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 67.Steinmetz MO, Akhmanova A. Capturing protein tails by CAP-Gly domains. Trends Biochem Sci. 2008;33:535–545. doi: 10.1016/j.tibs.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 68.Williamson MP. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297(Pt. 2):249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schlager MA, Kapitein LC, Grigoriev I, Burzynski GM, Wulf PS, Keijzer N, De Graaff E, Fukuda M, Shepherd IT, Akhmanova A, Hoogenraad CC. Pericentrosomal targeting of Rab6 secretory vesicles by Bicaudal-D-related protein 1 (BICDR-1) regulates neuritogenesis. EMBO J. 2010;29:1637–1651. doi: 10.1038/emboj.2010.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee PL, Ohlson MB, Pfeffer SR. Rab6 regulation of the kinesin family KIF1C motor domain contributes to Golgi tethering. Elife. 2015;4 doi: 10.7554/eLife.06029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Okada Y, Higuchi H, Hirokawa N. Processivity of the single-headed kinesin KIF1A through biased binding to tubulin. Nature. 2003;424:574–577. doi: 10.1038/nature01804. [DOI] [PubMed] [Google Scholar]
- 72.Oriola D, Casademunt J. Cooperative force generation of KIF1A Brownian motors. Phys Rev Lett. 2013;111:048103. doi: 10.1103/PhysRevLett.111.048103. [DOI] [PubMed] [Google Scholar]
- 73.Al-Bassam J, Cui Y, Klopfenstein D, Carragher BO, Vale RD, Milligan RA. Distinct conformations of the kinesin Unc104 neck regulate a monomer to dimer motor transition. J Cell Biol. 2003;163:743–753. doi: 10.1083/jcb.200308020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rashid DJ, Bononi J, Tripet BP, Hodges RS, Pierce DW. Monomeric and dimeric states exhibited by the kinesin-related motor protein KIF1A. J Pept Res. 2005;65:538–549. doi: 10.1111/j.1399-3011.2005.00255.x. [DOI] [PubMed] [Google Scholar]
- 75.Tomishige M, Klopfenstein DR, Vale RD. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 2002;297:2263–2267. doi: 10.1126/science.1073386. [DOI] [PubMed] [Google Scholar]
- 76.Hirokawa N, Pfister KK, Yorifuji H, Wagner MC, Brady ST, Bloom GS. Submolecular domains of bovine brain kinesin identified by electron microscopy and monoclonal antibody decoration. Cell. 1989;56:867–878. doi: 10.1016/0092-8674(89)90691-0. [DOI] [PubMed] [Google Scholar]
- 77.Hackney DD, Levitt JD, Suhan J. Kinesin undergoes a 9 S to 6 S conformational transition. J Biol Chem. 1992;267:8696–8701. [PubMed] [Google Scholar]
- 78.Friedman DS, Vale RD. Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat Cell Biol. 1999;1:293–297. doi: 10.1038/13008. [DOI] [PubMed] [Google Scholar]
- 79.Stock MF, Guerrero J, Cobb B, Eggers CT, Huang TG, Li X, Hackney DD. Formation of the compact confomer of kinesin requires a COOH-terminal heavy chain domain and inhibits microtubule-stimulated ATPase activity. J Biol Chem. 1999;274:14617–14623. doi: 10.1074/jbc.274.21.14617. [DOI] [PubMed] [Google Scholar]
- 80.Coy DL, Hancock WO, Wagenbach M, Howard J. Kinesin’s tail domain is an inhibitory regulator of the motor domain. Nat Cell Biol. 1999;1:288–292. doi: 10.1038/13001. [DOI] [PubMed] [Google Scholar]
- 81.Hirokawa N, Noda Y. Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol Rev. 2008;88:1089–1118. doi: 10.1152/physrev.00023.2007. [DOI] [PubMed] [Google Scholar]
- 82.Huo L, Yue Y, Ren J, Yu J, Liu J, Yu Y, Ye F, Xu T, Zhang M, Feng W. The CC1-FHA tandem as a central hub for controlling the dimerization and activation of kinesin-3 KIF1A. Structure. 2012;20:1550–1561. doi: 10.1016/j.str.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 83.Yue Y, Sheng Y, Zhang HN, Yu Y, Huo L, Feng W, Xu T. The CC1-FHA dimer is essential for KIF1A-mediated axonal transport of synaptic vesicles in C. elegans . Biochem Biophys Res Commun. 2013;435:441–446. doi: 10.1016/j.bbrc.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 84.Ren J, Huo L, Wang W, Zhang Y, Li W, Lou J, Xu T, Feng W. Structural correlation of the neck coil with the coiled-coil (CC1)-forkhead-associated (FHA) tandem for active kinesin-3 KIF13A. J Biol Chem. 2016;291:3581–3594. doi: 10.1074/jbc.M115.689091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Farkhondeh A, Niwa S, Takei Y, Hirokawa N. Characterizing KIF16B in neurons reveals a novel intramolecular “stalk inhibition” mechanism that regulates its capacity to potentiate the selective somatodendritic localization of early endosomes. J Neurosci. 2015;35:5067–5086. doi: 10.1523/JNEUROSCI.4240-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yoshimura Y, Terabayashi T, Miki H. Par1b/MARK2 phosphorylates kinesin-like motor protein GAKIN/KIF13B to regulate axon formation. Mol Cell Biol. 2010;30:2206–2219. doi: 10.1128/MCB.01181-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hsu CC, Moncaleano JD, Wagner OI. Sub-cellular distribution of UNC-104(KIF1A) upon binding to adaptors as UNC-16(JIP3), DNC-1(DCTN1/Glued) and SYD-2(Liprin-alpha) in C. elegans neurons. Neuroscience. 2011;176:39–52. doi: 10.1016/j.neuroscience.2010.12.044. [DOI] [PubMed] [Google Scholar]
- 88.Wu GH, Muthaiyan Shanmugam M, Bhan P, Huang YH, Wagner OI. Identification and characterization of LIN-2(CASK) as a regulator of kinesin-3 UNC-104(KIF1A) motility and clustering in neurons. Traffic. 2016;17:891–907. doi: 10.1111/tra.12413. [DOI] [PubMed] [Google Scholar]
- 89.Tong Y, Tempel W, Wang H, Yamada K, Shen L, Senisterra GA, Mackenzie F, Chishti AH, Park HW. Phosphorylation-independent dual-site binding of the FHA domain of KIF13 mediates phosphoinositide transport via centaurin alpha1. Proc Natl Acad Sci USA. 2010;107:20346–20351. doi: 10.1073/pnas.1009008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Novick P, Zerial M. The diversity of Rab proteins in vesicle transport. Curr Opin Cell Biol. 1997;9:496–504. doi: 10.1016/s0955-0674(97)80025-7. [DOI] [PubMed] [Google Scholar]
- 92.Zerial M, Mcbride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 93.Fischer Von Mollard G, Sudhof TC, Jahn R. A small GTP-binding protein dissociates from synaptic vesicles during exocytosis. Nature. 1991;349:79–81. doi: 10.1038/349079a0. [DOI] [PubMed] [Google Scholar]
- 94.Stettler O, Moya KL, Zahraoui A, Tavitian B. Developmental changes in the localization of the synaptic vesicle protein rab3A in rat brain. Neuroscience. 1994;62:587–600. doi: 10.1016/0306-4522(94)90391-3. [DOI] [PubMed] [Google Scholar]
- 95.Schluter OM, Schmitz F, Jahn R, Rosenmund C, Sudhof TC. A complete genetic analysis of neuronal Rab3 function. J Neurosci. 2004;24:6629–6637. doi: 10.1523/JNEUROSCI.1610-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Delevoye C, Miserey-Lenkei S, Montagnac G, Gilles-Marsens F, Paul-Gilloteaux P, Giordano F, Waharte F, Marks MS, Goud B, Raposo G. Recycling endosome tubule morphogenesis from sorting endosomes requires the kinesin motor KIF13A. Cell Rep. 2014;6:445–454. doi: 10.1016/j.celrep.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mauvezin C, Neisch AL, Ayala CI, Kim J, Beltrame A, Braden CR, Gardner MK, Hays TS, Neufeld TP. Coordination of autophagosome-lysosome fusion and transport by a Klp98A–Rab14 complex in Drosophila. J Cell Sci. 2016;129:971–982. doi: 10.1242/jcs.175224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hallak ME, Rodriguez JA, Barra HS, Caputto R. Release of tyrosine from tyrosinated tubulin. Some common factors that affect this process and the assembly of tubulin. FEBS Lett. 1977;73:147–150. doi: 10.1016/0014-5793(77)80968-x. [DOI] [PubMed] [Google Scholar]
- 99.Edde B, Rossier J, Le Caer JP, Desbruyeres E, Gros F, Denoulet P. Posttranslational glutamylation of alpha-tubulin. Science. 1990;247:83–85. doi: 10.1126/science.1967194. [DOI] [PubMed] [Google Scholar]
- 100.Redeker V, Levilliers N, Schmitter JM, Le Caer JP, Rossier J, Adoutte A, Bre MH. Polyglycylation of tubulin: a posttranslational modification in axonemal microtubules. Science. 1994;266:1688–1691. doi: 10.1126/science.7992051. [DOI] [PubMed] [Google Scholar]
- 101.L’hernault SW, Rosenbaum JL. Chlamydomonas alpha-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry. 1985;24:473–478. doi: 10.1021/bi00323a034. [DOI] [PubMed] [Google Scholar]
- 102.Janke C. The tubulin code: molecular components, readout mechanisms, and functions. J Cell Biol. 2014;206:461–472. doi: 10.1083/jcb.201406055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ikegami K, Heier RL, Taruishi M, Takagi H, Mukai M, Shimma S, Taira S, Hatanaka K, Morone N, Yao I, Campbell PK, et al. Loss of alpha-tubulin polyglutamylation in ROSA22 mice is associated with abnormal targeting of KIF1A and modulated synaptic function. Proc Natl Acad Sci USA. 2007;104:3213–3218. doi: 10.1073/pnas.0611547104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.O’hagan R, Piasecki BP, Silva M, Phirke P, Nguyen KC, Hall DH, Swoboda P, Barr MM. The tubulin deglutamylase CCPP-1 regulates the function and stability of sensory cilia in C. elegans . Curr Biol. 2011;21:1685–1694. doi: 10.1016/j.cub.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cai D, Mcewen DP, Martens JR, Meyhofer E, Verhey KJ. Single molecule imaging reveals differences in microtubule track selection between kinesin motors. PLoS Biol. 2009;7:e1000216. doi: 10.1371/journal.pbio.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Seidel C, Zekert N, Fischer R. The Aspergillus nidulans kinesin-3 tail is necessary and sufficient to recognize modified microtubules. PLoS One. 2012;7:e30976. doi: 10.1371/journal.pone.0030976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Steinberg G. Kinesin-3 in the basidiomycete Ustilago maydis transports organelles along the entire microtubule array. Fungal Genet Biol. 2015;74:59–61. doi: 10.1016/j.fgb.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 108.Bhuwania R, Castro-Castro A, Linder S. Microtubule acetylation regulates dynamics of KIF1C-powered vesicles and contact of microtubule plus ends with podosomes. Eur J Cell Biol. 2014;93:424–437. doi: 10.1016/j.ejcb.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 109.Guardia CM, Farias GG, Jia R, Pu J, Bonifacino JS. BORC functions upstream of kinesins 1 and 3 to coordinate regional movement of lysosomes along different microtubule tracks. Cell Rep. 2016;17:1950–1961. doi: 10.1016/j.celrep.2016.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Van Der Vaart B, Akhmanova A, Straube A. Regulation of microtubule dynamic instability. Biochem Soc Trans. 2009;37:1007–1013. doi: 10.1042/BST0371007. [DOI] [PubMed] [Google Scholar]
- 111.Atherton J, Houdusse A, Moores C. MAPping out distribution routes for kinesin couriers. Biol Cell. 2013;105:465–487. doi: 10.1111/boc.201300012. [DOI] [PubMed] [Google Scholar]
- 112.Schneider R, Korten T, Walter Wilhelm J, Diez S. Kinesin-1 motors can circumvent permanent roadblocks by side-shifting to neighboring protofilaments. Biophys J. 2015;108:2249–2257. doi: 10.1016/j.bpj.2015.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dixit R, Ross JL, Goldman YE, Holzbaur EL. Differential regulation of dynein and kinesin motor proteins by tau. Science. 2008;319:1086–1089. doi: 10.1126/science.1152993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Seitz A, Kojima H, Oiwa K, Mandelkow EM, Song YH, Mandelkow E. Single-molecule investigation of the interference between kinesin, tau and MAP2c. EMBO J. 2002;21:4896–4905. doi: 10.1093/emboj/cdf503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Samora CP, Mogessie B, Conway L, Ross JL, Straube A, Mcainsh AD. MAP4 and CLASP1 operate as a safety mechanism to maintain a stable spindle position in mitosis. Nat Cell Biol. 2011;13:1040–1050. doi: 10.1038/ncb2297. [DOI] [PubMed] [Google Scholar]
- 116.Semenova I, Ikeda K, Resaul K, Kraikivski P, Aguiar M, Gygi S, Zaliapin I, Cowan A, Rodionov V. Regulation of microtubule-based transport by MAP4. Mol Biol Cell. 2014;25:3119–3132. doi: 10.1091/mbc.E14-01-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lipka J, Kapitein LC, Jaworski J, Hoogenraad CC. Microtubule-binding protein doublecortin-like kinase 1 (DCLK1) guides kinesin-3-mediated cargo transport to dendrites. EMBO J. 2016;35:302–318. doi: 10.15252/embj.201592929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tien NW, Wu GH, Hsu CC, Chang CY, Wagner OI. Tau/PTL-1 associates with kinesin-3 KIF1A/UNC-104 and affects the motor’s motility characteristics in C. elegans neurons. Neurobiol Dis. 2011;43:495–506. doi: 10.1016/j.nbd.2011.04.023. [DOI] [PubMed] [Google Scholar]
- 119.Efimova N, Grimaldi A, Bachmann A, Frye K, Zhu X, Feoktistov A, Straube A, Kaverina I. Podosome-regulating kinesin KIF1C translocates to the cell periphery in a CLASP-dependent manner. J Cell Sci. 2014;127:5179–5188. doi: 10.1242/jcs.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kopp P, Lammers R, Aepfelbacher M, Woehlke G, Rudel T, Machuy N, Steffen W, Linder S. The kinesin KIF1C and microtubule plus ends regulate podosome dynamics in macrophages. Mol Biol Cell. 2006;17:2811–2823. doi: 10.1091/mbc.E05-11-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ally S, Larson AG, Barlan K, Rice SE, Gelfand VI. Opposite-polarity motors activate one another to trigger cargo transport in live cells. J Cell Biol. 2009;187:1071–1082. doi: 10.1083/jcb.200908075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hancock WO. Bidirectional cargo transport: moving beyond tug of war. Nat Rev Mol Cell Biol. 2014;15:615–628. doi: 10.1038/nrm3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Amrute-Nayak M, Bullock SL. Single-molecule assays reveal that RNA localization signals regulate dynein-dynactin copy number on individual transcript cargoes. Nat Cell Biol. 2012;14:416–423. doi: 10.1038/ncb2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Derr ND, Goodman BS, Jungmann R, Leschziner AE, Shih WM, Reck-Peterson SL. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science. 2012;338:662–665. doi: 10.1126/science.1226734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, Grigoriev I, Engelsma D, Haasdijk ED, Keijzer N, Demmers J, et al. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 2010;8:e1000350. doi: 10.1371/journal.pbio.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fu MM, Holzbaur EL. Integrated regulation of motor-driven organelle transport by scaffolding proteins. Trends Cell Biol. 2014;24:564–574. doi: 10.1016/j.tcb.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Belyy V, Schlager MA, Foster H, Reimer AE, Carter AP, Yildiz A. The mammalian dynein-dynactin complex is a strong opponent to kinesin in a tug-of-war competition. Nat Cell Biol. 2016;18:1018–1024. doi: 10.1038/ncb3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schlager MA, Serra-Marques A, Grigoriev I, Gumy LF, Esteves Da Silva M, Wulf PS, Akhmanova A, Hoogenraad CC. Bicaudal d-family adaptor proteins control the velocity of dynein-based movements. Cell Rep. 2014;8:1248–1256. doi: 10.1016/j.celrep.2014.07.052. [DOI] [PubMed] [Google Scholar]
- 129.Kevenaar JT, Bianchi S, Van Spronsen M, Olieric N, Lipka J, Frias CP, Mikhaylova M, Harterink M, Keijzer N, Wulf PS, Hilbert M, et al. Kinesin-binding protein controls microtubule dynamics and cargo trafficking by regulating kinesin motor activity. Curr Biol. 2016;26:849–861. doi: 10.1016/j.cub.2016.01.048. [DOI] [PubMed] [Google Scholar]
- 130.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, Mcwilliam H, Remmert M, Soding J, Thompson JD, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Boc A, Diallo AB, Makarenkov V. T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012;40:W573–579. doi: 10.1093/nar/gks485. [DOI] [PMC free article] [PubMed] [Google Scholar]