Abstract
Cytokinetic membrane abscission is a spatially and temporally regulated process that requires ESCRT (endosomal sorting complexes required for transport)-–dependent control of membrane remodeling at the midbody, a subcellular organelle that defines the cleavage site. Alteration of ESCRT function can lead to cataract, but the underlying mechanism and its relation to cytokinesis are unclear. We found a lens-specific cytokinetic process that required phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2α (PI3K-C2α), its lipid product PI(3,4)P2 (phosphatidylinositol 3,4-bisphosphate), and the PI(3,4)P2-–binding ESCRT-II subunit VPS36 (vacuolar protein-sorting-–associated protein 36). Loss of each of these components led to impaired cytokinesis, triggering premature senescence in the lens of fish, mice, and humans. Thus, an evolutionarily conserved pathway underlies the cell type-–specific control of cytokinesis that helps to prevent early onset cataract by protecting from senescence.
Indroduction
Cytokinesis is the final step of mitosis, driving the physical separation of the two daughter cells, in which an organelle called midbody orchestrates the cleavage of the intercellular bridge. At the midbody, several cytokinesis-associated events take place, — including cytoskeleton rearrangements, cell cycle regulation, membrane traffic, and plasma membrane remodelling —that progressively constrict the intercellular bridge until cleavage, in a process called abscission (1).
The ESCRT (endosomal sorting complexes required for transport) machinery directs a conserved membrane cleavage reaction that is important in multiple cellular processes, such as multivesicular endosome (MVE) formation, virus budding, plasma membrane repair, and, importantly, cytokinetic abscission. All of these processes share a similar topology of budding away from the cytosol, starting with the recruitment of early ESCRT components (ESCRT-0, -I and –-II) to the designated membrane (2). The final step of membrane fission relates to the self-polymerization and remodeling of the ESCRT-III subunits into helical filaments on the inner side of the membrane (3). Cytokinetic abscission has been thought to require only ESCRT-I, ESCRT-III, and the ESCRT-associated ALG2-interacting protein X (ALIX) that serves to recruit ESCRT-III in abscission. Nonetheless, mice that lack ALIX show reduced brain size without affecting other tissues, indicating the presence of redundant pathways that enable cytokinesis in non-neuronal cells (4–7). Consistent with this view, increasing evidence suggests that recruitment of the ESCRT-III component charged multivesicular body protein 4B (CHMP4B) to the midbody involves not only ALIX, but also other parallel and additional pathways controlled by the ESCRT-I and -II cascade (8–11). However, the precise mechanism underlying ESCRT-II recruitment to the midbody and the resulting ESCRT-III accumulation remains incompletely understood (1).
During cargo sorting and budding at endosomes, ESCRTs are targeted to the membrane through multiple low-affinity interactions with 3-phosphoinositides (12–14). Phosphoinositides are produced on intracellular membranes and at the cytosolic face of the plasma membrane, where they serve as docking sites for proteins that govern diverse processes, including endocytosis, intracellular signaling, and vesicular trafficking (1). Whether a similar phosphoinositide-based mechanism directs ESCRT recruitment to the abscission site during cytokinesis is unknown.
Cytokinetic defects can end up in binucleation, and the ensuing tetraploidization can lead to senescence. Abnormal development of the lens as well as late-onset cataracts of the elderly are linked to premature cellular senescence (15), and cytokinetic defects may trigger these abnormalities (16). In addition, loss of members of the ESCRT machinery, such as CHMP4B and VPS4, determines early cataract onset in vivo (17–19), thus suggesting that perturbation of ESCRT-mediated cytokinesis can disturb the appropriate organization of lens epithelial cells. Thus, a connection between ESCRT-mediated abscission, senescence, and cataract development can be envisioned, but molecular details of this cascade of events are still poorly defined.
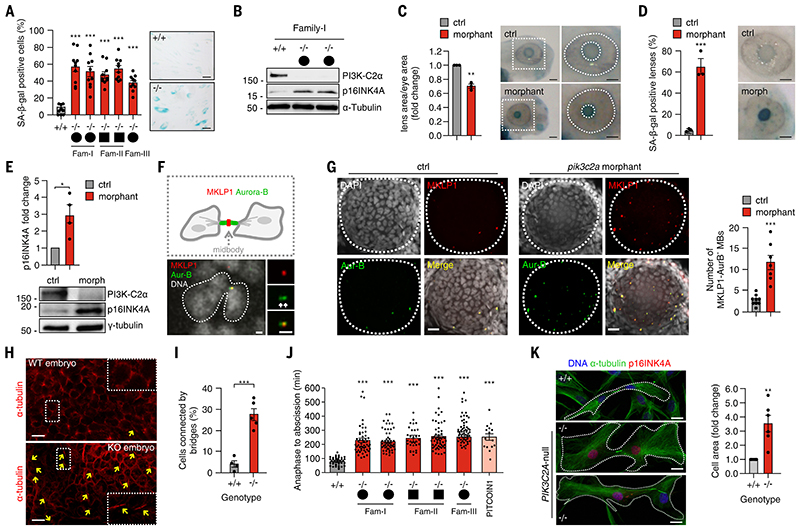
Senescence and premature aging in patients carrying null mutations of PIK3C2A
PIK3C2A-(phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2A)–null patients have congenital syndromic features reminiscent of premature aging, including early onset of cataract and secondary glaucoma (20). To investigate a possible role of PIK3C2A in premature senescence, we analysed markers such as senescence-associated β-galactosidase (SA-β-gal) and p16INK4A (21) in PIK3C2A-null fibroblasts from patients and their controls. While less than 10% of wild-type cells scored positive, more than 50% of PIK3C2-null fibroblasts from different families displayed pronounced SA-β-gal activity (Fig. 1A and fig.S1A). Similar results were observed in Pik3c2a−/− primary mouse embryonic fibroblasts (MEFs) (fig. S1B) and in human lens epithelial cells in which PIK3C2A was suppressed (HLE-B3) (fig. S1C). After 2 weeks of culture, expression of senescence markers [p16INK4A, p21, BCL2/BAX ratio, and senescence-associated secretory phenotype (SASP)] were significantly higher in PIK3C2A-null patients’ fibroblasts than in wildtype controls (Fig. 1B and fig. S1D), (3.02-fold induction ± 0.34, n = 5 replicates, p = 0.0013; fig. S1, E- to G). Similar results were observed in PI3K-C2α-–depleted HLE-B3 cells (fig. S1, E-, F, and H) (3.31-fold induction ± 0.23, n = 3 replicates, p < 0.0001; fig. S1I). Next, a selective inhibitor for PI3K-C2α (PITCOIN1) was tested on human fibroblasts and found to significantly induce p16INK4A expression (fig. S1J), confirming that loss of the lipid products of this kinase could induce senescence.
Figure 1. Loss of PI3K-C2α induces defective abscission and early senescence.
A) Quantification and representative images of SA-b-gal–positive fibroblasts derived from Family I, II, and III after 2 weeks in culture. +/+ genotypes are shown in the chart as pulled together. (B) Immunoblot analysis of p16INK4A protein level in fibroblasts derived from Family I after 2 weeks in culture. (C) Quantification and representative images of the ratio between lens and eye size in control and pik3c2a morphant 72 hours post-fertilization (hpf) zebrafish embryos. (D) Quantification and representative images of SA-b-gal intensity on the lens of control and pik3c2a morphant 72-hpf embryos. (E) Immunoblot analysis of p16INK4A and PI3K-C2α in control and pik3c2a morphant embryos (n = 4 pools of 15 embryos each). (F and G) Confocal images of whole-mount immunofluorescence performed on 72-hpf embryos lens using MKLP1 (red) and Aur-B (green) antibodies to stain midbody and TO-PRO-3 (gray) to stain nuclei. (H) Immunofluorescence of wild-type and Pik3c2a−/− embryo sections by using a-tubulin to mark intercellular bridges connecting cells in cytokinesis. (I) Quantification of the number of cells connected by bridges (%). n = 6 fields in at least four independent experiments. (J) Time-lapse analysis of the time required to progress from anaphase to cytokinesis in fibroblasts derived from patients with homozygous deletion of PI3K-C2α or in control fibroblasts treated with PITCOIN1. (K) (Left) Immunofluorescence of p16INK4A (red), DNA (blue), and a-tubulin (green) in wild-type and PI3K-C2α-null fibroblast. (Right) Quantification of cell area in control and PIK3C2A-null fibroblasts. If not previously specified, all results are shown as mean or representative picture of at least three independent experiments ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
Loss of PI3K-C2α induces early senescence, defective lens development, and cataracts in vivo
To test whether PI3K-C2α depletion causes premature senescence in the eye lens, we focused on zebrafish in which pik3c2a was suppressed (fig. S2A) that develop to term and recapitulate the human phenotype more faithfully than do Pik3c2a−/− mice, which die in utero (22). The lens size was 25% smaller in pik3c2a morphants than in controls (Fig. 1C). We also found a significantly higher number of SA-β-gal –positive lenses in pik3c2a-suppressed fish than in controls (65.5 versus 5.1%, respectively); (Fig. 1D and fig. S2B). Levels of other senescence markers (p16INK4A, p21, BCL2/BAX ratio, and SASP) were also increased in pik3c2a morphants (Fig. 1E and fig. S2, C to E). Similarly, p16INK4A was elevated in wildtype embryos treated with PITCOIN1 (fig. S2F).
To further investigate whether pik3c2a loss in adult fish caused premature lens cell senescence and cataracts akin to PIK3C2A-null patients, we generated and examined compound heterozygotes from two zebrafish strains carrying distinct null mutations in the pik3c2a gene. The frequency of genotypes in the offspring from pik3c2a+/− intercrosses followed the expected Mendelian ratio (table S1), and the gross morphology of the mutants was indistinguishable from that of controls. All compound heterozygotes carrying the two distinct null alleles (n = 118) displayed lenticular abnormalities (P < 0.005, two-tailed Fisher’s exact test), consisting in circular cataract and posterior lenticonus (fig. S3A; and movie S1), like what is observed in PIK3C2A-null patients (fig. S3B) (20).
To challenge these findings in a different model organism, we analyzed the eye of a mouse homozygous for a Pik3c2a hypomorphic allele (Pik3c2ahypo/hypo) (23) and found reduced eye size (fig. S3C). Pik3c2ahypo/hypo lenses displayed reduced numbers of KI-67-–positive nuclei and increased p16INK4A expression compared with those of controls (fig. S3, D, and E). To analyze whether this could occur independently of a developmental defect, loss ofPI3K-C2α in the adult eye by from Cre-mediate gene deletion induced a significant up-regulation of p16INK4A and other senescence markers (p21, BCL2/BAX ratio, and SASP) in the lens (fig. S4, A- to D), leading to overt cataracts within 3 months (fig. S4E). This was consistent with cataract patients, where PIK3C2A lens expression is significantly lower in individuals with age-related cataracts than in age-matched controls (36.7% ± 5.2 reduction; P = 0.0001) (24). Thus, loss of PI3K-C2α in vertebrates can induce early senescence, defective lens development, and cataracts.
Cytokinesis defects in cells lacking PI3K-C2α
Abnormal cell division and subsequent tetraploidization can lead, in the lens epithelium, to senescence and cataract formation (15, 16). We reverted to zebrafish morphants as well as PITCOIN1-treated embryos and analyzed tetraploidy by means of flow cytometry in dissociated cells of the developing lens (fig. S5, A to C). The number of cells in the G2/M phase of the cell cycle was substantially increased (2-twofold difference between controls and either pik3c2a morphants or PITCOIN1-treated embryos), indicating increased number of cells stalled in late cytokinesis (25). In agreement with this hypothesis, immunofluorescence analysis revealed a threefold enrichment of cells connected by a cytokinetic bridge positive for markers of late cytokinesis, such as Aurora B and MKLP1 (Fig. 1, F, and G, and fig. S5, D, and E). Cells connected by intercellular bridges were also observed in embryonic day 11.5 (E11.5) Pik3c2a−/− mouse embryos (Fig. 1, H, and I). Furthermore, time-lapse analysis of the progression from anaphase onset to abscission showed that Pik3c2a−/− MEFs were delayed in the completion of cytokinesis (fig. S5, F, and G, and Movie S2). Whereas post-anaphase furrowing, and intercellular bridge establishment proceeded normally (fig. S5, F, and G, and movie S2), cytokinesis failure and refusion appeared more frequently (fig. S5, H, and I).
Like fish and mice, PIK3C2A-null patients’ cells (20), showed a significantly delayed abscission (Fig. 1J) and binucleation (fig. S5J). Mutant cells still connected with the intercellular bridge resulted positive for the senescence marker p16INK4A (Fig. 1K and fig. S6, A and C). In addition, mutant daughter cells that were stalled in cytokinesis exhibited p16INK4A positivity in only one of the two nuclei, indicating that the increase in p16INK4A expression did not occur post mitotically after mitosis but appeared in cells enduring delayed abscission (Fig. 1K and fig. S6A, lower bottom right panel). In agreement, cell flattening that is typical of senescent cells occurred in p16INK4A-positive PIK3C2A-null cells that did not complete cytokinesis (Fig. 1K, right panel). This phenotype was recapitulated in PIK3C2A-depleted HLE-B3 cells (fig. S6, B and D), as well as in cells treated with an inhibitor of cytokinesis [Paprotrain (26] (fig. S6E). Thus, data from humans, mice, and cultured cells indicate that senescence in cells or tissues that lack PI3K-C2α function could be a direct consequence of defective and/ or delayed cytokinesis.
PI3K-C2α localizes at the midbody and directs abscission
To dissect the molecular mechanism underlying the requirement for PI3K-C2α to complete cytokinesis and to protect cells from senescence, we used HeLa cells, in which depletion of PI3K-C2α delayed abscission (fig. S7, A- to E). Localization of endogenous PI3K-C2α during cytokinesis was analyzed by means of immunofluorescence and found in a characteristic ring-like arrangement at the midbody. PI3K-C2α appeared to be embraced by intercellular-bridge microtubules, indicating that PI3K-C2α was associated with the midbody ring (Fig. 2A and figs. S7, F, top, and G, and S8A). Similar results were seen in cells transfected with a green fluorescent protein (GFP)-–tagged PI3K-C2α (figs. S7, F, bottom, and S8, B andC, and movie S3). Both transfected GFP-–PI3K-C2α and the endogenous protein were absent from the cleavage furrow during early cytokinesis but became enriched at the midbody during late stages of cytokinesis (fig. S7F). Thus, PI3K-C2α localizes to the midbody and controls late cytokinetic abscission.
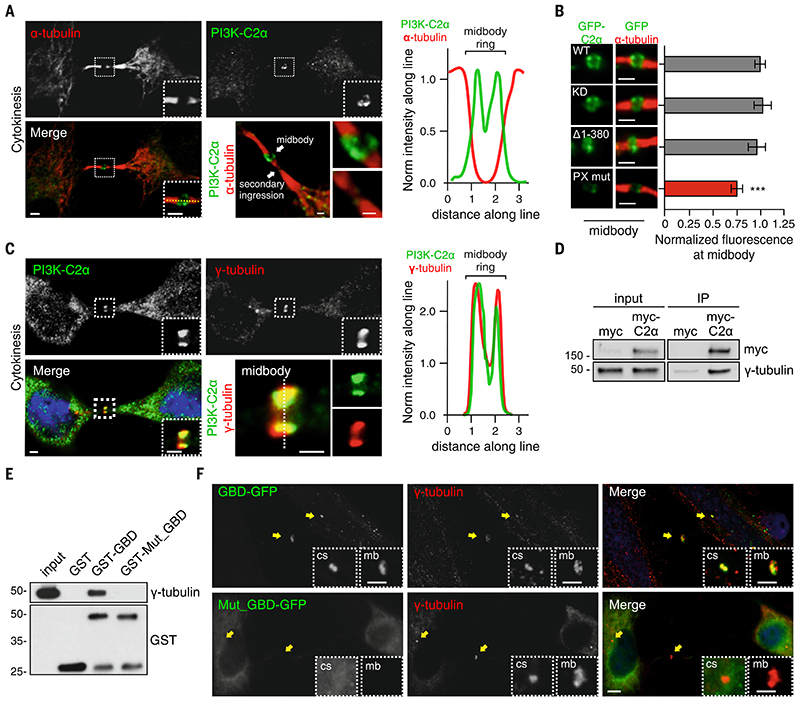
Figure 2. PI3K-C2α localizes to midbody through the PX- and the g-tubulin–binding domain.
A) Confocal images of HeLa cells stained for PI3K-C2α (green) and a-tubulin (red) (left) during cytokinesis and (right) fluorescence intensity along the line showing localization of PI3K-C2α at midbody. (B) Hela cells transfected with wild-type, kinase inactive (KD), clathrin-binding deletion (D1-380), and PX-binding mutant PI3K-C2α GFP-tagged. Immunofluorescence staining by using antibody to GFP showing (left) the enrichment of the different PI3K-C2α constructs and (right) their quantification at the midbody. (C) Confocal images of (left) HeLa cells stained for PI3K-C2α (green), 4′,6-diamidino-2-phenylindole (DAPI) (blue), and g-tubulin (red) and (right) fluorescence intensity along the line showing colocalization between PI3K-C2α and g-tubulin. (D) Immunoprecipitation of myc-PI3K-C2α from cells synchronized in cytokinesis. (E) Pull-down experiment by using GST-GBD and GST-GBDQ1022A-T1025A-S1081A from cells synchronized in cytokinesis. (F) Immunofluorescence staining of GFP-GBD and GFP-GBDQ1022A-T1025A-S1081A (green) with g-tubulin (red) during cytokinesis. Enlarged section shows colocalization with centrosome and midbody. If not previously specified, all results are shown as mean or representative picture of at least three independent experiments ± SEM. ***P < 0.001.
Coincidental binding of γ-tubulin and PI(4,5)P2 localizes PI3K-C2α at the midbody
During the furrowing process, PI(4,5)P2 progressively accumulates at the membrane surrounding the furrow (27). The PX domain of PI3K-C2α specifically binds PI(4,5)P2 with high affinity and is required for the activation of PI3K-C2α catalytic activity (28). We thus analyzed the localization in cytokinesis of a PI3K-C2α PX-domain mutant as well as a kinase inactive (KD) form, a clathrin-binding domain mutant (Δ1-380), and a GFP-tagged wild-type protein and found that only the PX-binding mutant reduced PI3K-C2α localization (Fig. 2B and fig. S9A).
Previous proximity biotinylation experiments identified PI3K-C2α as a putative γ-tubulin interactor (29). In agreement, we found γ-tubulin to be enriched in an unbiased proteomic analysis of PI3K-C2α interactors (table S2). Given the specific localization of γ-tubulin at the midbody (30), we tested its possible function in recruiting PI3K-C2α during abscission by analyzing the localization of PI3K-C2α and γ-tubulin. As expected, both proteins were enriched at the midbody where they colocalized (Fig. 2C). Immunoprecipitation from cells synchronized in cytokinesis of myc-PI3K-C2α together with γ-tubulin revealed that the two proteins interact during abscission (Fig. 2D). A docking model of the PI3K-C2α and γ-tubulin complex showed a region comprising the C-terminal part of the helical domain of PI3K-C2α fitting to the γ-tubulin crystal structure (fig. S9, B- to D). We created an enhance GFP (EGFP) fusion protein between the C-terminal part of the PI3K-C2α helical domain [referred to hereafter as γ-tubulin binding domain (GBD), amino acids 981- to 120)] and analyzed its intracellular localization by means of immunofluorescence. EGFP-GBD displayed complete co-localization with γ-tubulin in HeLa cells during interphase (fig. S9, E, and F). Pull-down assays by using GST-GBD showed that the GBD is sufficient to precipitate γ-tubulin from cells synchronized in cytokinesis (Fig. 2E). Last, mutation of three residues within the γ-tubulin interaction site (Q1022A, T1025A, and S1081A) inhibited γ-tubulin binding (Fig. 2F) as well as colocalization at the centrosome and midbody (Fig. 2G). These data demonstrate that coincidental binding of PI(4,5)P2 and γ-tubulin drives PI3K-C2α localization.
PI3K-C2α produces PI(3,4)P2 at the midbody
To characterize the role of PI3K-C2α at the midbody, we explored the function of its kinase activity during cytokinesis. Delayed abscission in HeLa cells was rescued through re-expression of a small interfering RNA (siRNA) –resistant wild-type PI3K-C2α, whereas add-back of the KD form had no effect (Fig. 3A and fig. S10A), demonstrating that cytokinesis completion requires PI3K-C2α kinase activity at the midbody.
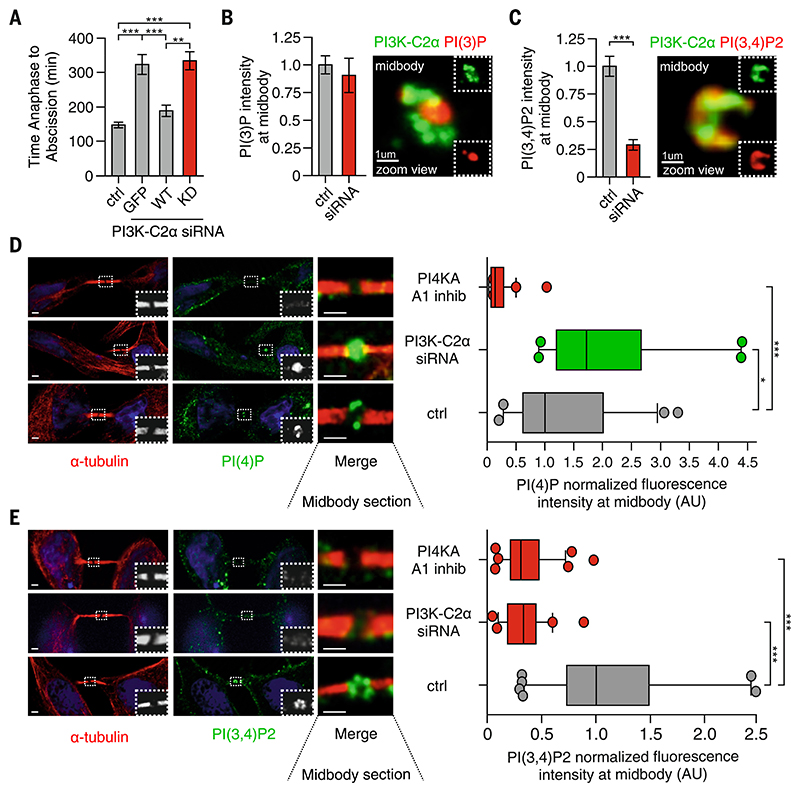
Figure 3. PI3K-C2α produces PI(3,4)P2 at the midbody ring.
(A) Time-lapse microscopy measuring the time required to progress from anaphase to abscission in HeLa cells treated with siRNA for PI3K-C2α alone or expressing GFP and siRNA-resistant GFP–wild-type (WT) or GFP-kinase inactive (KD) form of PI3K-C2α. n ≥ 25 cells imaged. (B) (Left) Quantification of PI(3)P levels at midbody by means of immunofluorescence staining (n ≥100 cells) and (right) confocal image of 3D reconstructed midbody stained for PI(3)P (red) and PI3K-C2α (green). (C) (Left) Quantification of PI(3,4)P2 levels at midbody by immunofluorescence staining (n ≥100 cells) and (right) confocal image of 3D reconstructed midbody stained for PI(3,4)P2 (red) and PI3K-C2α (green). (D) Quantification of PI(4)P levels at midbody in control, A1 inhibitor, and siRNA-treated cells. n ≥100 cells, mean ± SD. (E) Quantification of PI(3,4)P2 levels at midbody in control, A1 inhibitor, and siRNA-treated cells. n ≥100 cells, mean ± SD. If not previously specified, all results are shown as mean or representative picture of at least three independent experiments ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
The scaffold function of PI3K-C2α is involved in spindle organization at metaphase, and lack of PI3K-C2α causes aberrant chromosome congression (31). In line with these findings, loss of PI3K-C2α increased the amount of LAP-2 –positive lagging chromosomes (fig. S10B), which, when entrapped at the bridge, cause cytokinetic delay. To exclude a role of the scaffold function, we re-expressed a kinase-dead version of PI3K-C2α that allows proper chromosome congression but fails to produce PI(3,4)P2 (31). The kinase-dead mutant was able to abolish LAP2-positive bridges but not delayed cytokinesis, thus excluding that defective abscission relies on the role of PI3K-C2α in metaphase (fig. S10B).
PI3K-C2α has been reported to produce phosphatidylinositol 3-phosphate [PI(3)P] on endosomes and PI(3,4)P2 at the plasma membrane (22). To address the identity of the PI3K-C2α lipid product at the midbody, we analyzed the localization of 3-phosphoinositides during cytokinesis. PI(3,4,5)P3, mainly produced by class I PI3Ks, was almost undetectable at the midbody (fig. S11A). As expected (32), PI(3)P was enriched at the midbody (fig. S11B), and no significant changes in its levels were observed upon PI3K-C2α depletion (Fig. 3B). PI(3)P mainly appeared inside the intercellular bridge (Fig. 3B), differently from the ring-like distribution of PI3K-C2α. By contrast, PI3K-C2α localization matched that of PI(3,4)P2 (Fig. 3C; and fig. S11, D, and E), and downregulation of PI3K-C2α resulted in a 73% reduction in PI(3,4)P2 abundance, whereas levels of PI(3)P remained unchanged (Fig. 3, B and C).
PI3K-C2α generates PI(3,4)P2 starting from PI(4)P (33), suggesting that PI4K may synthetize PI(4)P at the midbody. Because PI4KA is well known to be active at the plasma membrane (34), we treated cells with the PI4KA-specific inhibitor A1 and found it to abolish PI(4)P at the midbody (Fig. 3D). Conversely, depletion of PI3K-C2α significantly increased PI(4)P at the midbody (Fig. 3D) and reduced PI(3,4)P2 to the same extent of PI4KA inhibition (Fig. 3E), thus indicating that PI3K-C2α uses PI(4)P as a substrate.
PI(3,4)P2 locally produced by PI3K-C2α controls the recruitment of ESCRT-II/VPS36 to the midbody
Cytokinetic abscission relies on a cascade of interactions in which ESCRT-I binds to ALIX, which eventually recruits ESCRT-III (4, 6). Unexpectedly and in contrast to this canonical model, cataracts have been observed in humans and mice harbouring inactivating mutations of the ESCRT-III component CHMP4B (17), but not in ALIX-null mice (7). We hypothesized that PI3K-C2α might be a key component of an ALIX-independent pathway that leads to cytokinetic abscission that is of importance in the lens. The lens epithelium of both fish and mice, although expressing high levels of PI3K-C2α, appeared nearly devoid of ALIX (Fig. 4, A and B, and fig. S12A).
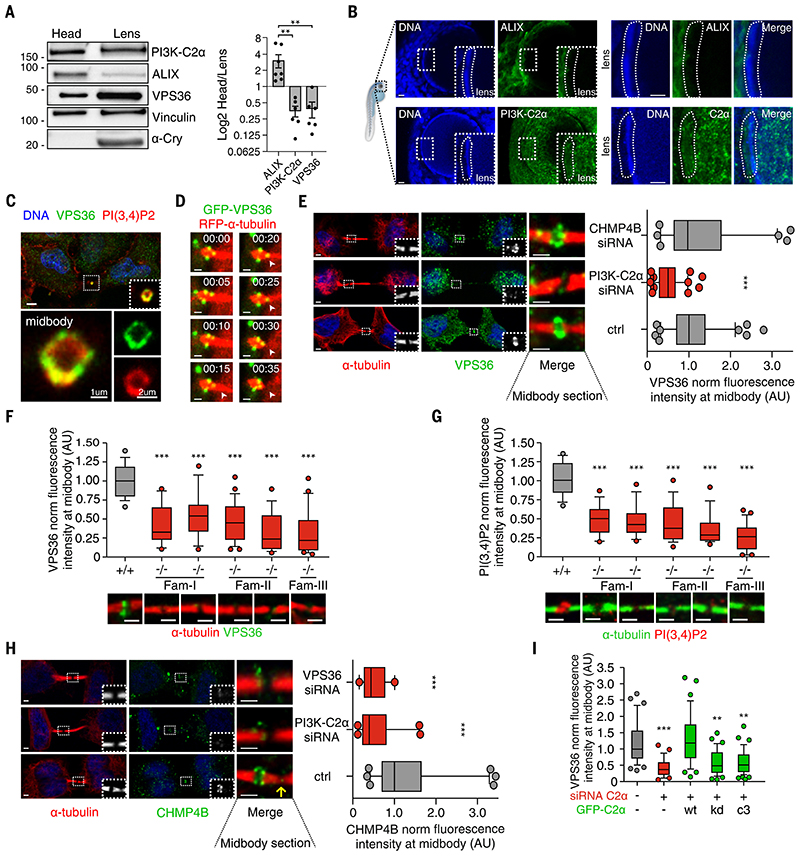
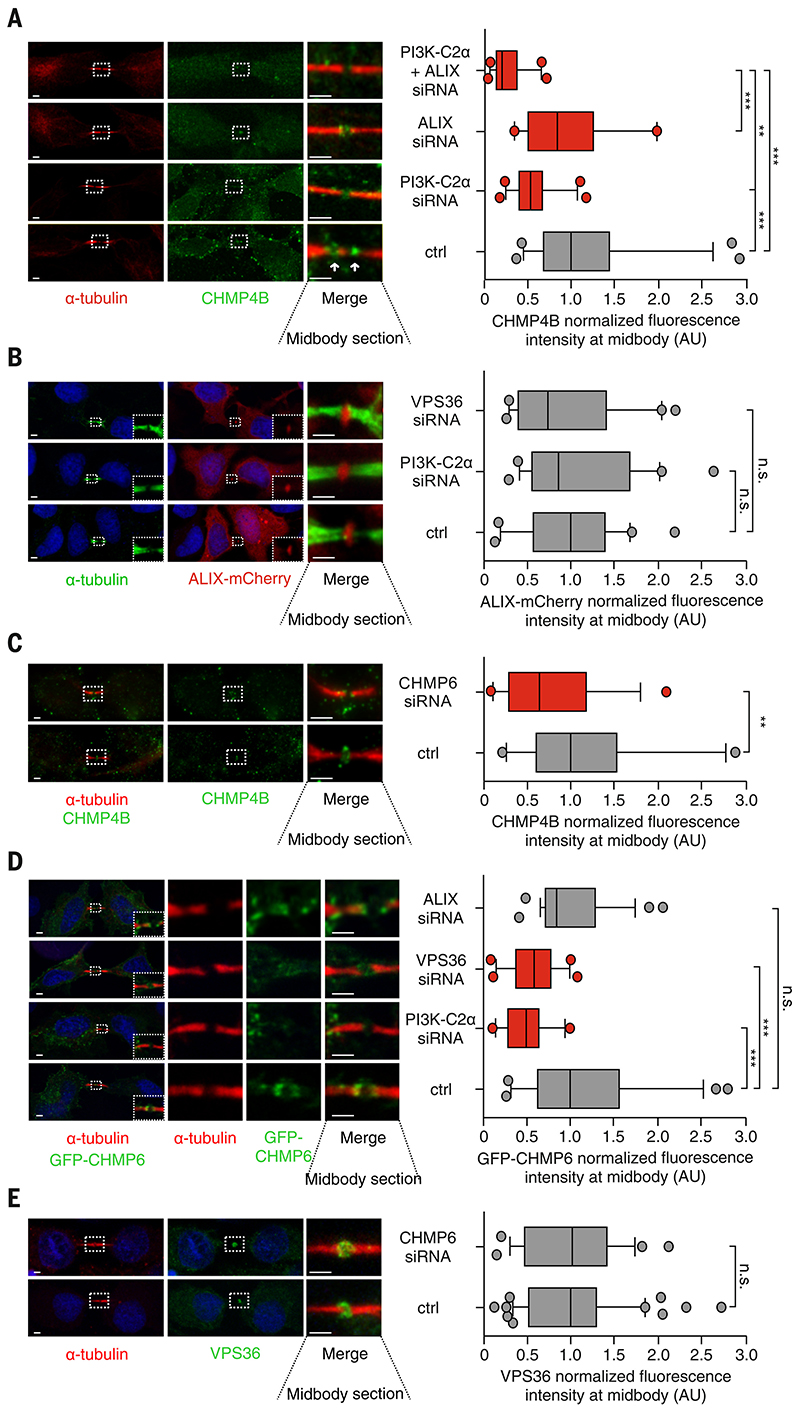
Figure 4. PI3K-C2α/VPS36 axis is an ALIX alternative pathway to recruit CHMP4B to the midbody in lens.
(A) Immunoblot analysis and protein quantification showing ALIX/PI3K-C2α expression ratio in zebrafish lens. (B) Immunofluorescence showing (top) ALIX or (bottom) PI3K-C2α expression in 72-hpf zebrafish lens. (C) Immunofluorescence staining showing colocalization between endogenous VPS36 and PI(3,4)P2 at the midbody ring. (D) Snapshots taken from time-lapse imaging of GFP-VPS36 during cytokinesis in HeLa cells stably expressing red fluorescent protein (RFP)–a-tubulin. Arrowheads indicate enrichment of VPS36 at the abscission site. (E) Quantification of VPS36 levels at midbody in control and siRNA-treated cells. n ≥ 200 cells, mean ± SD. (F and G) Immunofluorescence staining and quantification of endogenous (F) VPS36 and (G) PI(3,4)P2 levels at the midbody in fibroblasts derived from patients with homozygous deletion of PI3K-C2α. +/+ genotypes were pulled together. n ≥ 50 cells, mean ± SD. (H) Quantification of CHMP4B levels at midbody in control and siRNA-treated HeLa cells. n ≥ 120 cells, mean ± SD. (I) Quantification of VPS36 levels at midbody in control and siRNA treated cells upon transfection of siRNA-resistant WT, KD, and PI(3)P–producing (C3) forms of PI3K-C2α. n ≥ 30 cells, mean ± SD. If not previously specified, all results are shown as mean or representative picture of at least three independent experiments ± SEM. **P < 0.01; ***P < 0.001.
These data led us to hypothesize that, in cells expressing low or negligible amounts of ALIX, cytokinetic abscission depends on PI3K-C2α. We further reasoned that even in cells that express ALIX, such as HeLa cells, the PI3K-C2α pathway may act synergistically with ALIX to drive completion of cytokinesis. Thus, we tested whether PI3K-C2α–-derived PI(3,4)P2 could recruit effector proteins linking ESCRT-III to the cytokinetic bridge. Among the known ESCRT components, only VPS36, a core subunit of ESCRT-II, binds PI(3,4)P2 (13). Immunofluorescence staining of endogenous VPS36 showed that, during cytokinesis, this protein was enriched at the midbody, where it co-localized with PI(3,4)P2 in ring-like structures (Fig. 4C). Like PI3K-C2α, VPS36 was absent from the cleavage furrow but was enriched at the midbody during late cytokinesis (fig. S12, B- to D). Time-lapse imaging of living cells confirmed the enrichment of GFP-VPS36 at the midbody during abscission (Fig. 4D and movie S4). VPS36 suppression caused delayed cytokinesis (fig. S13A) and refusion during abscission like that observed in cells lacking PI3K-C2α (fig. S13B).
To dissect the order of events during cytokinetic abscission, we analyzed the effects of the depletion of either PI3K-C2α or VPS36 on the localization of the various components of the ESCRT machinery. Suppression of either PI3K-C2α or VPS36 had no effect on TSG101 (ESCRT-I) localization at the midbody (fig. S13C). Similarly, down-modulation of TSG101 did not lead to changes in either PI3K-C2α or PI(3,4)P2 amounts. Thus, PI3K-C2α acts downstream of TSG101 in an ESCRT-I – independent manner (fig. S13, D and E). Conversely, suppression of PI3K-C2α, but not class III PI3K inhibition, resulted in a significantly reduced recruitment of VPS36 to the midbody (Fig. 4E and fig. S13F, respectively). These findings were confirmed in PIK3C2A-null human fibroblasts that showed a >50% reduction in VPS36 and PI(3,4)P2 levels at the midbody as well as elongated intercellular bridges (Fig. 4, F, and G and fig. S14A). Removal of TSG101 together with PI3K-C2α synergized and almost completely abolished VPS36 recruitment to the midbody (fig. S13G). Thus, PI3K-C2α–-derived PI(3,4)P2 facilitates VPS36 recruitment and ESCRT-III assembly. In agreement, PI4KA inhibition reduced the ESCRT-III subunit CHMP4B at the midbody (fig. S14B), but suppression of CHMP4B had no effect on VPS36 localization (Fig. 4E), confirming that VPS36 is recruited before ESCRT-III during abscission. At the midbody, depletion of either PI3K-C2α or VPS36 reduced CHMP4B enrichment (Fig. 4H), but overexpression of VPS36-mCherry increased CHMP4B-GFP localization (fig. S15A). Thus, VPS36/ESCRT-II participates in the hierarchical recruitment of ESCRT-III during abscission. Midbody fractionation showed that PI3K-C2α loss led to significantly reduced levels of VPS36 and CHMP4B (fig. S15, B and C) but not of the ESCRT-I subunit TSG101 and Citron kinase (fig. S15, B to D).
Last, VPS36 localization at the midbody in PI3K-C2α–suppressed cells was completely restored by a wild-type PI3K-C2α but neither by the KD nor by the C3 mutant that produces PI(3)P only (Fig. 4I and fig. S15E) (33, 35). Thus, PI(3,4)P2 produced by PI3K-C2α controls VPS36 localization, contributing to the midbody enrichment of CHMP4B driving cytokinesis.
VPS36-dependent CHMP4B recruitment to the midbody through an ALIX-independent pathway
Our results pointed to two mechanisms for recruitment of ESCRT-III at the midbody: one depending on ALIX (12, 25) and the other on PI3K-C2α/ESCRT-II (fig. S15F). In agreement with this hypothesis, we observed a 16, 49, and 76% reduction in CHMP4B levels at the midbody upon suppression of either ALIX or PI3K-C2α or their concomitant depletion, respectively (Fig. 5A). No changes in ALIX localization at the midbody could be observed after loss of either PI3K-C2α or VPS36 (Fig. 5B), suggesting that ALIX and PI3K-C2α/VPS36 operate independently of each other (fig. S15F). Thus, CHMP4B can associate with the midbody through either ALIX (ALIX pathway) or PI3K-C2α/ESCRT-II (fig. S15F). Furthermore, the complex of VPS36 and VPS22, anchored at the midbody membrane, can recruit CHMP6, eventually promoting the addition of CHMP4B (fig. S15F) (8). Consistent with this model, suppression of CHMP6 resulted in a nearly 50% reduction in CHMP4B levels at the midbody (Fig. 5C). Furthermore, CHMP6 recruitment was decreased after suppression of either PI3K-C2α or VPS36 but not after suppression of ALIX (Fig. 5D). Conversely, removal of CHMP6 did not impair VPS36 recruitment (Fig. 5E). Thus, a pathway anchoring CHMP4B/ ESCRT III to the midbody plasma membrane acts in parallel to ALIX and depends on PI3K-C2α/PI(3,4)2, VPS36, and CHMP6 (fig. S15F).
Figure 5. VPS36 recruits CHMP4B to the midbody through ALIX parallel pathway.
(A) Quantification of CHMP4B levels at midbody in HeLa cells treated with control siRNA or siRNA targeting PI3K-C2α, ALIX, or PI3K-C2α and ALIX. n ≥ 50 cells, mean ± SD. (B) Quantification of ALIX levels at midbody in HeLa cells transfected with ALIX-Cherry and treated with control or siRNA targeting PI3K-C2α or VPS36. n ≥ 60 cells, mean ± SD. (C) Quantification of CHMP4B levels at midbody in HeLa cells treated with control or siRNA targeting CHMP6. n ≥ 45 cells, mean ± SD. (D) Quantification of CHMP6 levels at midbody in HeLa cells transfected with GFP-CHMP6 and treated with control or siRNA targeting PI3K-C2α, VPS36, or ALIX. n ≥ 50 cells, mean ± SD. (E) Quantification of VPS36 levels at midbody in HeLa cells treated with control or siRNA targeting CHMP6. n ≥ 40 cells. If not previously specified, all results are shown as mean or representative picture of at least three independent experiments. (n.s. = not significant; **P < 0.01; ***P < 0.001).
Binding of VPS36 to PI(3,4)P2 is required for VPS36 localization to the midbody
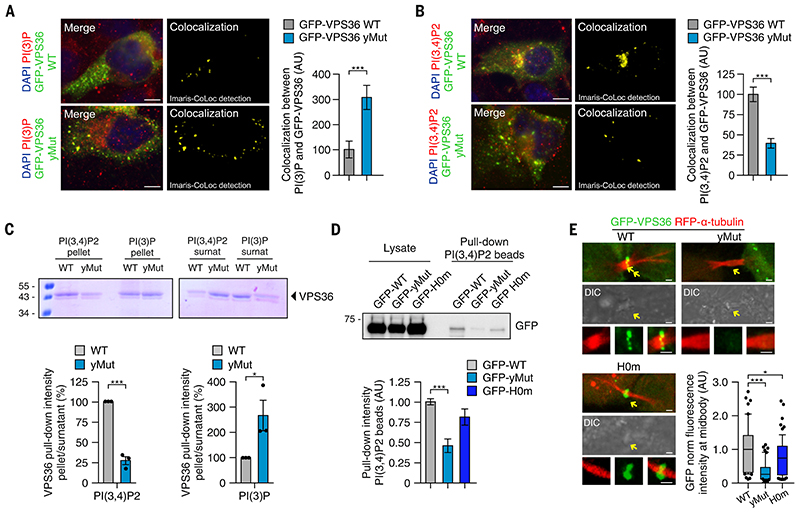
VPS36 function is highly conserved during evolution, but its structure substantially diverged from yeast to mammals, with multiple changes in phosphoinositide binding selectivity (13, 14, 36, 37). The yeast GRAM-like ubiquitin binding in EAP45 (GLUE)-domain in VPS36 shows specificity toward PI(3)P, whereas the mammalian GLUE-domain binds strongly but relatively promiscuously to phosphoinositides, including PI(3,4)P2 (13, 14). The molecular explanation for this evolutionary change is the presence of additional charged residues in mammalian VPS36 flanking the charged residues responsible for PI(3)P binding in yeast (36). We mutated all three of the additional basic residues from the mammalian GLUE -domain, partially reestablishing the yeast PI-binding sequence (fig. S16A). Mutant mammalian VPS36 (yMut-VPS36) showed a punctate staining in cells during interphase that resembled that of wild-type VPS36 localization (fig. S16B). Similar to yeast VPS36, yMut-VPS36 showed a threefold increase in co-localization with PI(3)P-positive vesicles (Fig. 6A). Conversely, reduced co-localization of yMut-VPS36 with PI(3,4)P2 was observed (Fig. 6B). In agreement, liposome binding assays showed reduced binding of yMut-VPS36 to PI(3,4)P2 and increased association with PI(3)P (Fig. 6C and F fig. S16C). Furthermore, a pull-down assay by using PI(3,4)P2-–coated beads revealed a reduced ability of yMut-VPS36 to bind to PI(3,4)P2 (Fig. 6D). The previously reported H0m mutant of VPS36 (14) that lacks binding to ESCRT-I retained its interaction with PI(3,4)P2 (Fig. 6D).
Figure 6. PI(3,4)P2 is required for VPS36 recruitment to the midbody.
(A and B) (Left) Immunofluorescence and (right) quantification by using Imaris software of the colocalization between GFP-VPS36 WT or GFP-VPS36-yMut with either (A) PI(3)P or (B) PI(3,4)P2. (C) Lipid sedimentation assay showing binding of WT and yMut VPS36 to PI(3,4)P2 and PI(3)P. (D) Pull-down experiment by using PI(3,4)P2–coated beads and lysates from cells expressing WT, yMut, or H0m VPS36. (E) Time-lapse of HeLa cells stably expressing RFP–a-tubulin, showing localization of WT, H0m, or yMut VPS36 enrichment at the midbody during abscission. Quantification reports the average fluorescence intensity observed in n ≥ 60 cells and normalized on total GFP fluorescence inside each cell (mean ± SD). If not previously specified, all results are shown as mean or representative picture of at least three independent experiments ± SEM. *P < 0.05; ***P < 0.001.
Considering that suppression of PI3K-C2α decreased PI(3,4)P2 and VPS36 enrichment at the midbody, we tested whether the reduced binding observed for yMut-VPS36 to PI(3,4)P2 might be sufficient to prevent its localization to the abscission site. Live cell imaging of GFP-H0m-VPS36 and GFP-yMut-VPS36 showed a 24 and 72% reduction of their localization at the midbody, respectively (Fig. 6E, fig. S16D, and movies S5 to S7). Thus, PI(3,4)P2 provides a spatial cue that enhances VPS36 localization.
Loss of either PI3K-C2α or VPS36 impairs secondary ingression formation
Defective ESCRT assembly during cytokinesis often results in impaired secondary ingression formation, which is the final cause of delayed abscission (38, 39). Suppression of either PI3K-C2α or VPS36 led to stable and elongated intercellular bridges, and reduced percentage of cells displaying a secondary ingression at one or both sides, respectively (fig. S17, A and B). Although control HeLa cells formed the secondary ingression at an expected distance from the midbody of 0.77 μm (38)(fig. S17C) (38), loss of either PI3K-C2α or VPS36 led to an average of 0.90μm and 0.88 μm, respectively (fig. S17C). Thus, loss of PI3K-C2α impairs secondary ingression formation.
Recruitment of ESCRT machinery to the midbody causes deformation of the secondary ingression site into wavy patterns of the membrane at the intercellular bridge (40). To support the role of PI3K-C2α and VPS36 in the assembly of the ESCRT machinery at the midbody, a kymograph analysis of the sequential wavy constrictions occurring at the membrane flanking the midbody was analyzed by use of live cell imaging. Depletion of either PI3K-C2α or VPS36 caused a significant elongation of the intercellular bridge, likely because of the stretching movement of the two dividing cells (fig. S17D). These intercellular bridges were very stable, with significantly fewer constriction events compared to with that of control cells (fig. S17, E and F). No differences in localization at the intercellular bridge of other secondary ingression site effectors, such as FIP3 and F-actin, were observed (fig. S17, G and H).
Loss of VPS36 induces cytokinesis defects and early senescence in the lens
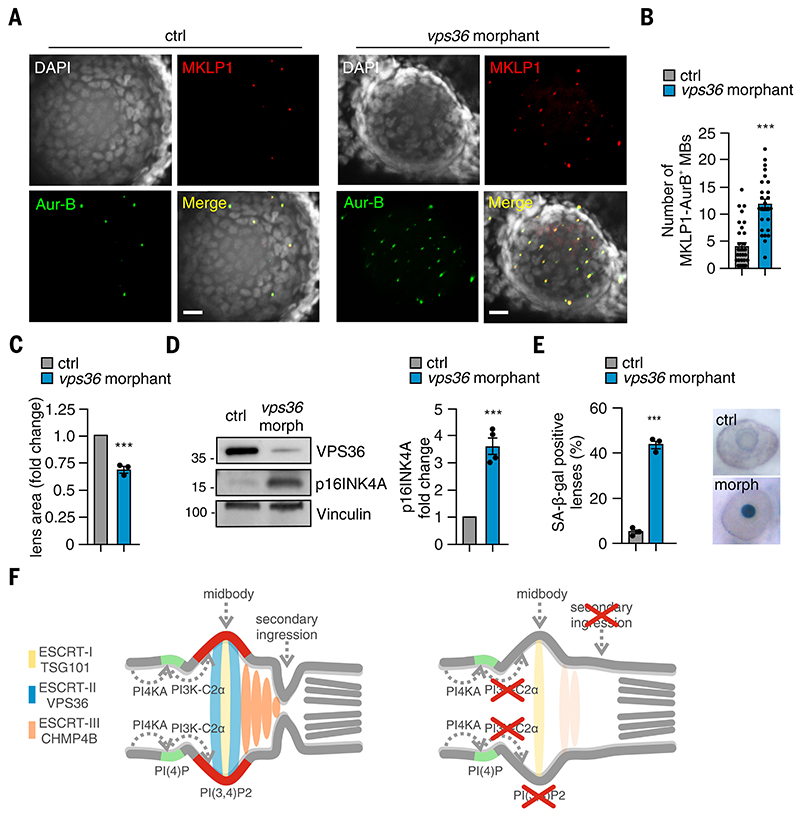
Our data suggest that loss of VPS36 should phenocopy the lack of PI3K-C2α. Analysis of cytokinesis in the lens of zebrafish embryos in which vps36 was suppressed (fig. S18A) showed that MKLP1-positive midbodies, double stained with either Aurora B (Fig. 7A) or α-tubulin (fig. S5, D and E), were 3-fold more abundant than in controls (Fig. 7B and fig. S5E). Like with pik3c2a suppression, this was not due to increased proliferation, because the lens area in vps36 morphants was 31% smaller than in wild-type controls (Fig. 7C). Consistent with this idea, a reduced number of phospho-H3 –positive cells was detected in the lens of vps36 or pik3c2a morphants (fig. S18B), and reduced EdU incorporation was found in HLE-B3–suppressed cells as well as after treatment with the cytokinesis inhibitor Paprotrain (fig. S18C). Furthermore, expression of senescence markers (p16INK4A, SA-βGal, p21, BCL2/BAX ratio, and SASP) was more abundant in the eyes of vps36 morphants than in wild-type controls (Fig. 7, D and E, and fig. S19, A to C). Similar results were observed in VPS36-deficient HLE-B3 cells, where p16INK4A (fig. S19D) and SA-βGal (fig. S19E) were increased, with p16INK4A appearing in the nuclei of cells blocked in cytokinesis (fig. S19F). Appearance of senescence markers (SA-β-gal, p16INK4A, and p21), previously observed in pik3c2a morphants (Fig. 1, C to E, and fig. S2, C to E), was prevented by a morpholino-resistant wild-type pik3c2a mRNA but not by the KD or by the C3 pik3c2a mutants (35)(fig. S19, G to I) (35). Thus, PI3K-C2α kinase activity specifically linked to cytokinesis is involved in protection from senescence. Last, we tested whether cytokinesis-interfering inhibitors targeting Aurora-B (ZM447439), Myosin-II (blebbistatin), and MKLP2 (Paprotrain) induced senescence. Like in vps36 morphants, up-regulation of senescence markers (SA-βGal, p16INK4A, p21, and SASP) was observed (fig. S19, J, and K), indicating that, although potentially perturbing distinct biological functions, the common effect of delaying cytokinesis is likely the cause of senescence.
Figure 7. Loss of VPS36 impairs abscission and leads to senescence in vitro and in vivo.
(A) Confocal images of whole-mount immunofluorescence performed on 72-hpf embryos lens by using MKLP1 (red) and Aur-B (green) antibodies to stain midbody and TO-PRO-3 (gray) to stain nuclei. (B) Quantification of number of MKLP1 and Aur-B–positive midbody in 72-hpf embryos lens. (C) Measure of lens area in control and vps36 morphants. (D) Immunoblot analysis and quantification of p16INK4A in control and vps36 morphant 72-hpf embryos. n = 4 pools of 15 embryos each. (E) Quantification and representative images of SA-b-gal intensity on the lens of control and morphant (vps36) 72-hpf embryos. (F) During cytokinesis, PI3K-C2α produced PI(3,4)P2 (red) at the midbody by converting PI(4)P (light blue) synthetized by PI4KA. PI(3,4)P2 triggers the recruitment and the stabilization of the ESCRT-II subunits VPS36 (green), which in turn contributed to the accumulation of ESCRT-III CHMP4B at the midbody. When PI3K-C2α is lost, the secondary ingression where abscission occurs does not form properly, resulting in impaired abscission. This leads to defective cytokinesis and early onset of senescence, which is particularly evident in the lens epithelium. If not previously specified, all results are shown as mean or representative picture of at least three independent experiments ± SEM. ***P < 0.001.
Discussion
Signs of premature aging, —including high prevalence of stroke, bone fragility, and cataract, —are a major phenotypical feature of PIK3C2A-null patients (20). Previous reports identify PIK3C2A within the evolutionarily conserved “CellAge” gene network in 24 different mammalian species that, when suppressed, leads to senescence (41). In line with these findings, in this work in fish, mice, and humans the loss of PI3K-C2α was directly linked to senescence, with a particularly high penetrance in the eye lens. The lack of the lipid product generated by PI3K-C2α appeared as the driver of a cytokinetic defect, leading to premature up-regulation of senescence markers in the crystallin. Loss of other enzymes that, like PI3K-C2α, control phosphoinositide turnover can lead to cataract development. For example, inactivating mutations in the two phosphatases OCRL and INPP5K result in early-onset cataracts (42, 43). Both enzymes remove the phosphate in position 5 of PI(4,5)P2 to generate PI(4)P, the main substrate of PI3K-C2α, indicating that a specific phosphoinositide shift critically protects lens cells from degenerative senescence.
Our observations in different vertebrate species converge on the notion that, at the midbody, PI3K-C2α orchestrates the assembly of the ESCRT machinery required for abscission through its lipid product PI(3,4)P2. The synthesis of specific phosphoinositide species is one of the mechanisms adopted by cells to selectively recruit proteins in defined cellular regions (44). Our observations suggest that binding to specific membrane components contributes to the precise positioning of the ESCRT III complex required to complete abscission. The recruitment of ESCRT-III at the midbody depends on ALIX (4), but the presence of other mechanisms that trigger ESCRT III-–mediated abscission have been observed (9, 11). For example, ESCRT-III recruitment to the midbody can alternatively be mediated by the ESCRT-I and -II axis (8–10)and by the localization of the VPS22 subunit of ESCRT-II at the midbody, through the interaction with the ESCRT-I subunit TSG101 (8, 9). ALIX itself is also localized at the midbody by both TSG101 association and the binding to a complex that contains syntenin, bridging ESCRT III to a transmembrane syndecan-4 molecule (11). However, how the core ESCRT-II subunit, VPS36, could be recruited through this alternative pathway has remained elusive. In light of the inability of VPS22 to bind a select lipid (14) and thus localize to a specific subcellular domain, other ESCRT-II components must be involved in the targeting to the midbody-encasing plasma membrane. Our data demonstrate that VPS36 localizes to the midbody through the detection of a confined PI(3,4)P2 pool.
Although the function of VPS36 is generally conserved throughout evolution, it substantially differs from yeast to mammals regarding the selective binding to phosphoinositides. A study based on comparison of the intact and GLUE-domain – deleted ESCRT-II complexes further demonstrates that the human GLUE-domain binds with high affinity not only to PI(3)P, but also to a variety of other phosphoinositide isomers (14). In mammalian cells, PI(3)P is never found on the plasma membrane (34), where ESCRT polymerizes (38), thus indicating that another phosphoinositide must be involved in the localization at the midbody-enclosing membrane. This likely explains why the removal of PI(3,4)P2 through the down-modulation of PI3K-C2α can lead to a substantial reduction in VPS36 amounts at the midbody. This modification might have evolved to support specific functions acting in parallel or in place of the ALIX pathway.
The finding that PI3K-C2α and ALIX are not always co-expressed, indicates three modalities of ESCRT recruitment in abscission that can rely on either one alone or both of the two proteins together. In most cells, simultaneous presence of the two pathways allows partial compensation, and, in cells such as fibroblasts and HeLa, the effect of the loss of PI3K-C2α is mitigated by ALIX activity, as suggested by limited CHMP4B localization at the midbody in the absence of PI3K-C2α. Nonetheless, the compensatory effect of ALIX is incomplete because delayed cytokinesis and senescence are still observed in PIK3C2A-null fibroblasts and HeLa cells, thus indicating a potential synergy between the two pathways. In the lens, where ALIX is substantially less expressed than in other tissues, cells mainly depend on PI3K-C2α. Consistent with this idea, ALIX-deficient mice do not show either eye abnormalities or premature aging (7), but loss-of-function mutations in ESCRT-III components, such as CHMP4B (17, 45, 46) and VPS4 (18, 19), lead to altered cell division and premature aging with early-onset cataract. Other alterations of cytokinesis unrelated to defective ESCRT-III can equally result in the development of cataracts. For example, cataract can follow a dysfunction in lens cytoskeletal components acting in cytokinesis, such as vimentin mutation (47). The finding that loss of either PI3K-C2α or VPS36 leads to the same cytokinetic defect, and the ensuing cellular senescence indicate that, of the various functions that these two proteins play in the cell, their common and epistatic role in cytokinesis links them to senescence prevention. Our data that point to disturbance of cell division as a main driver of cataract development consolidate the causative link between aberrant cytokinesis and senescence. Defective cytokinesis has been generally associated with senescence through the activation of the senescence program following after tetraploidization (48). However, our findings that p16INK4A starts to be expressed by cells blocked before abscission indicate that the senescence program could also be triggered by altered ESCRT function before tetraploidization, likely to prevent cell cycle re-entry after refusion.
We defined a non-redundant molecular link between highly localized phosphoinositide signals and ESCRT assembly at the abscission site, together with an evolutionarily conserved mechanism that connects cytokinesis failure to senescence and eventually determines cataract, one of the most common conditions of the elderly population worldwide (Fig. 7F).
Materials and methods
Cell lines and primary cultures
HeLa and HEK293T, were purchased from ATCC (without further authentication) and cultured in DMEM GlutaMAXTM medium supplied with 10% fetal bovine serum (FBS) and 1% Penicillin-Streptomycin (10,000 U/mL). Cell lines used in this paper are not listed in the database of commonly misidentified cell lines maintained by ICLAC. Mouse embryonic fibroblasts (MEFs) were obtained as previously described (35). Fibroblast from patients were obtained and cultured as previously described (20). HLE-B3 cells were purchased from ATCC and cultured in MEM-a supplied with 20% fetal bovine serum (FBS), 1% Penicillin-Streptomycin (10,000 U/mL) and GlutaMAX. Cell lines were routinely tested formycoplasma contamination.
Protein analysis
Cells and tissues were homogenized in lysis buffer (120 mM NaCl, 50 mM Tris-HCl pH=8, 1% Triton X-100) supplemented with 25x protease inhibitor cocktail (Roche), 50mMsodium fluoride and 1 mM sodium orthovanadate. Lysates were cleared by centrifugation at 13,000 rpm for 15 min at 4°C. Protein concentration was determined by Bradford method and supernatants were analyzed for immunoblotting or for immunoprecipitation (IP) with the indicated antibodies. Membranes probed with the indicated antibodies were then incubated with HRP conjugated secondary antibodies (antimouse used 1:10000, anti-rabbit 1:5000, Sigma) and developed with enhanced chemiluminescence (ECL, BD). For IP assays, 1 mg of precleared extracts were incubated with 1 mg of the indicated antibody at 4°C on a rotating rack. After 1.5 hours, 15 ml of protein G-Sepharose (Amersham Biosciences, Buckinghamshire, UK) were added for 30 min. Samples were collected by centrifugation (13000 rpm 1 min) and washed six-times with lysis buffer. Bound protein complexes were then eluted by adding 30 ml Laemmli sample buffer. For pull-down experiment, HEK293T cells homogenized in lysis buffer (120 mM NaCl, 50 mM Tris-HCl pH = 8, 1% Triton X-100) supplemented with 25x protease inhibitor cocktail (Roche), 50mM sodium fluoride and 1 mM sodium orthovanadate. Lysates were cleared by centrifugation at 13,000 rpm for 15min at 4°C. 1 mg GST-GBDor 140 ng GST was incubated with 15 ml of protein G-Sepharose for 1 hour at 4°C in 1mg of cell lysate. Beads were washed four times with 1 ml of reaction buffer and analyzed by immunoblotting after the addition of 30 ml of Laemmli buffer. Bound protein complexes were then eluted by adding 30 ml Laemmli sample buffer.
Antibodies
Anti-PI3K-C2α (#611046, BD Transduction Laboratories; #22028-1-AP, Proteintech; a rabbit polyclonal develop by VH (33)), anti-GFP (gift fromEmilia Turco, University of Turin, Italy), anti a-tubulin (#2125, Cell Signaling; #3873, Cell Signaling), anti Myc-tag (#2276, Cell Signaling), anti TSG101 (GTX70255), anti PI(3)P (Z-P003, Echelon), anti PI(3,4)P2 (Z-P034b, Echelon), anti PI(4)P (Z-P004, Echelon), anti PI(3,4,5)P3 (Z-P345B, Echelon) anti CHMP4B (ab105767, Abcam; sc82556 (C12), Santa Cruz Biotechnology), anti VPS36 (ab76331, Abcam; ab247016, Abcam; #043947, Sigma), anti GFP (ab291, Abcam), anti g-tubulin (#T5326, GTU-88 Sigma), anti MKLP-1 (sc-869, Santa Cruz), anti E-Cadherin (4A2, Cell Signaling), anti-ALIX (#92880, Cell Signaling; #634502, BioLegend), anti aACrystallin (#PA1-009, Thermo Fisher), anti-b Crystallin (Santa Cruz Biotechnology sc-376006), anti p16INK4A (#SAB4500072, Sigma; 10883-1-AP, Proteintech; #80772, Cell Signaling), anti p21 (Cell Signaling, #2947), anti-Aurora B (GTX132702,GeneTex; BDbioscience, #611082), anti Phospho-Histone H3 (Ser10) (#9701. Cell Signaling), anti GAPDH (Cell Signaling, #5174), anti Vdac (Cell Signaling, #4866), anti-Vinculin (gift from Emilia Turco, University of Turin, Italy), anti-Citron Kinase (Transduction Laboratories, BD Biosciences, Franklin Lakes, NJ), anti-ALIX and CHMP4B (gift from Harald Stenmark, Institute for Cancer Research, The Norwegian Radium Hospital, Oslo, Norway).
Gene silencing and inhibitors
Plasmids containing shRNA sequences (arrest GIPZ lentiviral shRNA) against PIK3C2A were purchased fromThermo Scientific and used to generate lentiviral particles to stably infect cell lines as previously described (35). The Sh1 (V2LMM_73461) target sequence was 5′-GGCAAGATATGTTAGCTTT-3′, and the Sh2 (V2LMM_66190) target sequence was 5′-CAAAGTTTCTTTAACTCT-3′. Cells at early passages after infection (second to third) were used for experiments. siRNA treatment against PIK3C2A was previously described (33). siRNA against VPS36 was purchased by Thermo Fischer (cd: 134802 20nM). siRNA against OCRL was purchased by Thermo Fischer (cd: 104448 20nM). siRNA against CHMP6 was purchased by Sigma (esiRNA human CHMP6 EHU144401). The sequence of siRNA against TSG101 was 5′-CCUCCAGUCUUCUCUCGUCUU-3′. The sequence of siRNA against CHMP4B was 5′-AUCGAUAAAGUUGAUGAGUUAUUUU-3′. The sequence of siRNA against ALIX was 5′-CCUGGAUAAUGAUGAAGGA-3′. The sequence of siRNA against VPS36 was 5′-AAGUGAAUGCCAAUAUGAA-3′. VPS34-IN1 was purchased by Sigma. Paprotrain was purchased from Abcam. Blebbistatin was purchased from Sigma. ZM 447439 was purchased from TOCRIS (#2458). The PI4K A1 inhibitor was used as described (49). For experiments in zebrafish, Tuebingen zygotes were treated with 40 mM PITCOIN1 in standard fish water. Medium was changed every 24 hours, embryos were sacrificed at 72 hpf for further analysis. For fibroblasts, 60 mM PITCOIN1 was added to normal culture medium every 24 hours for a maximum of four days.
Plasmids
Wild-type PI3K-C2α-GFP, kinase inactive (KD) PI3K-C2α-GFP, clathrin-binding deleted PI3KC2a-GFP (D1–380) were previously described (31, 33). PI3K-C2α-Myc construct was generated by cutting and pasting equivalent fragments from the PI3K-C2α-GFP mutants into pCDNA3.1 plasmid. HumanVPS36 andCHMP4B were obtained by RLW and inserted in pEGFP empty vector. For Cherry-VPS36 plasmid generation, EGFP was replaced with Cherry by directly cloning Cherry sequence using restriction enzymes. GST-GBD and GFP-GBD were obtained by PCR amplification from WT PI3K-C2α of the predicted sequence and by cloning in either pGEX 5x1 or pEGFP-C2. GST-GBDQ1022A-T1025A-S1081A and GFPGBDQ1022A-T1025A-S1081A were obtained by site-directed mutagenesis using QuikChange II Site-Directed Mutagenesis Kit (Agilent). GFP-VPS36 H0m and GFP-VPS36 yMut were generated by QuikChange II Site-Directed Mutagenesis Kit (Agilent). Recombinant VPS36 (WT and yMut), VPS25 and VPS22 were generated as described (14). mCherry-hALIX was a gift from James Hurley (Addgene plasmid #21504; http://n2t.net/addgene:21504; RRID: Addgene_21504). CHMP6_GFP was a gift from Daniel Gerlich (Addgene plasmid # 31806; http://n2t.net/addgene:31806 ; RRID:Addgene_31806). All constructs were verified by restriction digest and automated DNA sequencing. HEK293T cells and HeLa cells were transfected by lipofection using Lipofectamine® 2000 (Life Technologies) or X-tremeGENE™ HP DNA Transfection Reagent (Roche), according to the manufacturer’s instructions. For silencing experiments, HeLa and HLE-B3 cells were transfected with 25 nMsiRNAs for 48-72 hours using X-tremeGENE 360 Transfection Reagent (Roche) or Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer’s instructions.
Lipid sedimentation assay and pull-down using PI(3,4)P2-coated beads
Lipid sedimentation assay was performed in accordance with what previously described in Im et al. 2008 (14). Pull-down using PI(3,4)P2-coated beads was performed using cell lysate of HeLa cells transfected with the indicated plasmid, in accordance with manufacturer instructions (Echelon, P-B034a).
Immunofluorescence
Immunofluorescence was performed by ice cold methanol or PFA fixation of MEFs, human fibroblasts, HLE-B3 and HeLa cells followed by standard procedures (31, 35). Cells were stained with DAPI, TO-PRO-3 iodide (Thermo Fischer) or Sytox Green (Thermo Fischer) and examined with either Zeiss Observer-Z1 microscope, equipped with the Apotome, Leica TCS-II SP5 or Leica TSC-II SP8 confocal microscope. Zebrafish embryos were fixed overnight in PFA 4%, nucleiwere stainedwith TO-PRO-3 iodide (Thermo Fisher). Whole mounted embryos were examined with Leica TSC-II SP5 or Leica TSC-II SP8 confocal microscope. Raw images were digitally processed only to normalize the background and enhance the contrast. Z-stacks were acquired and processed with the Maximum Projection tool. 3D morphometric measurement and reconstruction was performed with Imaris (BitPlane, Zurich, Switzerland). Fluorescence intensity was calculated with ImageJ tools, modifying existing protocols (50), (https://theolb.readthedocs.io/en/latest/imaging/measuring-cell-fluorescenceusing-imagej.html) and as outlined in fig. S20. Whiskers plots show either 90/10, 95/5 or 97.5/2.5 percentile, and the median in the center line.
Time-lapse microscopy
For measurement of the time required to progress from anaphase onset to abscission, cells were taken in a humidified chamber, at 37°C and 5% CO2 and imaged every 4 min in accordance with what previously described (31). For time-lapse of GFP-VPS36, Cherry-VPS36 and GFP-CHMP4B, either Zeiss Observer-Z1 microscope or Leica TSC-II SP8 confocal microscope were used and HeLa cells were imaged every 30 s for at least 10 min. For time-lapse of GFP-PI3K-C2α, Leica TSC-II SP8 confocal microscope was used, and HeLa cells were imaged every 2 min for at least 2 hours.
Cell synchronization and Midbody purification
Cell synchronization and midbody purification were adapted from Capalbo et al, 2019 and Skop et al, 2004. Briefly, to synchronize cells in cytokinesis we used a thymidine-nocodazole block and release procedures. To purify midbodies, 5 × 107 HeLa or HEK-293T cells were used. Cells were synchronized using the thymidine-nocodazole block. After nocodazole washout cells were incubated for 2 h in fresh medium containing 10 mMMG132 (Sigma-Aldrich) to further increase the effectiveness of the synchronization, and then incubated at 37 °C for 60 min after release from MG132. Before collection, 5 mg/ml taxol (Sigma-Aldrich) was added to the medium for 4min to stabilize microtubules in vivo. Cells were then transferred into a Corning 50 ml centrifuge tube and collected by centrifugation at 200 × g for 5min. Cells were then washed once in H2O and gently resuspended in 25 ml of swelling solution (1 mM PIPES pH 7.0, 1 mM MgCl2, 5 mg/ml taxol and Roche Complete Protease Inhibitors) and centrifuged at 200 × g for 5 min. The cell pellet was then resuspended in 50 ml of lysis buffer (1 mM PIPES pH 7, 1 mMEGTA, 1% NP-40, 5 mg/ml taxol, 3 U/ml DNAse I, 10 mg/ml RNAse A, 1 U/ml micrococcal nuclease, and Roche Complete Protease Inhibitors) and vortexed vigorously. After the addition of 0.3 volumes of cold 50mM 2-(Nmopholino) ethanesulfonic acid (MES) pH 6.3, the sample was incubated on ice for 20 min and then centrifuged at 200 × g for 10 min at 4 °C. The supernatant was transferred to a new tube and centrifuged at 650 × g for 20min at 4 °C to pellet midbodies. The midbody pellet was then resuspended in 4 ml of 50mM MES pH 6.3 and centrifuged through a 25ml glycerol cushion (40%[w/v] glycerol diluted in 50mM MES pH 6.3) at 2800 × g for 45 min at 4 °C. After removal of the glycerol cushion, the midbody pellet was washed with 2 ml of 50 mM MES pH 6.3, transferred to a 15 ml conical tube and centrifuged at 2800 × g for 20 min at 4 °C. After removing as much liquid as possible, the midbody pellet was resuspended in laemmli buffer for immunoblot analysis.
Zebrafish strains and treatments
All procedures using zebrafish (Danio rerio) were authorized by the Ethical Committee of the University of Torino and the Italian Ministry of Health. The wild-type fish strain Tuebingen was used. Adult fish were routinely maintained under a 14h light and 10h dark photoperiod at approximately 28°C, bred and genotyped according to standard procedures. Allelic transmission followed the expected mendelian ratios. Eggs were generated by natural mating, and following fertilization were collected, treated and maintained under a 12h light and 12h dark photoperiod, incubated at 28°C. Embryos and adult fish were sacrificed with a tricaine overdose. Embryos with the alleles sa10124/+ and sa12328/+ were created as part of the Zebrafish Mutation Project (51) and were obtained by in vitro fertilization from frozen sperm samples by the Zebrafish International Resource Center. The alleles sa10124 and sa12328 encode nonsense mutations in PI3K-C2α at amino acids 585 and 1236, respectively, that were confirmed by Sanger sequencing. Both nonsense mutations occurred prior to the end of the catalytic domain and were thus predicted to encode null alleles. We generated homozygous pik3-c2asa12328/sa12328 and pik3c2asa10124/sa10124 zebrafish, as well as compound heterozygous pik3c2asa12328/sa10124 mutants by intercrossing heterozygous adults. As these alleles were generated by random ENU mutagenesis (51), analysis of the compound heterozygous pik3-c2asa12328/sa10124 mutants minimized the likelihood of homozygosity for any unlinked ENU-induced mutations. For inhibitor treatment, zebrafish embryos were treated from 48hpf to 72hpf with blebbistatin (B05660, Sigma-Aldrich), paprotrain (512533, Sigma-Aldrich) and ZM447439 (189410, Sigma-Aldrich) at a concentration of 5 mM, 100 mMand 40 mM respectively.
Cataract formation in zebrafish
Adult zebrafish were anesthetized with tricaine and examined. Coaxial illumination using a Leica M841 surgical microscope was used to visualize lenticular defects. Digital video recordings were made using a Panasonic GPUS932AHDcamera systemand later reviewed by an ophthalmologist without knowledge of the genotype of each animal. Optical sectioning by changing the z-axis focus was used to aid in the identification of cataracts. All zebrafish examined for cataracts were offspring of a cross between pik3c2asa10124/+ and pik3c2asa12328/+ fish.
Morpholino microinjection
AntisenseMorpholino Oligonucleotides against pik3c2a, vps36 and scrambled control were purchased from GeneTools (LLC, Philomat, OR, USA). The pik3c2a-MO sequence was 5′-TATGTGGGCCATGGTGTCAGCTCT -3′. The vps36-MO sequence was 5′-CATTTGTCCACATAAATCGGTCCAT-3′. The scrambled control-MO sequence was 5′-CCTCTTACCTCAGTTACAATTTATA-3′ Tuebingen zygotes were collected at 1-cell stage and injected under stereological examination with 400 mM of pik3c2a-MO, vps36-MOor scrambled control-MO in the presence of Phenol Red for subsequent selection of the injected embryos.
Rescue experiments in zebrafish
For the rescue experiment, plasmids containing a GFP-tagged zebrafish transcript variant of pik3c2a (XM_021467821.1), modified with silent mutations in the region complementary to morpholino oligonucleotide, was constructed and packaged by VectorBuilder. For the mutagenesis, morpholino-resistant plasmids were obtained by site-directed mutagenesis using QuikChange II Site-Directed Mutagenesis Kit (Agilent) as previously described (33, 35). Capped mRNA was transcribed with mMESSAGE mMACHINE T7 Transcription Kit (Invitrogen). Zebrafish embryos were then injected at 1-cell-stage with morpholino oligonucleotides and mRNA (500 ng/mL). Positive embryos were selected based on GFP-fluorescence.
Lens size measurement
Zebrafish embryos were treated with 0.003% 1-phenyl-2-thiourea (#P7629, Sigma) at 12 hpf to prevent formation of melanin pigment. Picture of fixed72hpf larvaewere takenunder stereological examination using fixed magnification. Images were analyzed using ImageJ tools. Lens size was quantified as ratio between lens and eye diameters.
SA-b-galactosidase assay
Cells or embryos were stained using Senescence Cells Histochemical Staining Kit (#CS00, Sigma), according to the manufacturer’s instructions. Cell were fixed 1h RT, washed 3 times and incubated 3 hours in b-Gal staining solution at 37° in absence of CO2. Zebrafish embryos were treated with 0.003% 1-phenyl-2-thiourea (#P7629, Sigma) at 12hpf to prevent formation of melanin pigment, then 72 hpf larvae were fixed at 4° overnight, washed 3 times with PBS and incubated in b-Gal staining solution at 37° for 12 hours in absence of CO2.
Flow cytometry analysis
Heads from 72 hpf zebrafish embryos were disaggregated by mechanical homogenization at 30°C in 0.25% trypsin containing 5 mg/mL collagenase. Enzymatic reaction was blocked by addition of DMED medium containing 10% FBS. Disaggregation products were washed in PBS and fixed by dropwise addition of absolute ethanol while vortexing reaching a final concentration of 70% ethanol, cells were incubated 30 min in ice. After further PBS washes, cells were resuspended in PIstaining solution containing 25 mg/mL propidium iodide (P4864, Sigma), 0.05% Triton-X 100 and 0.1 mg/mL RNAse in PBS and incubated at 37°C for 40min. Samples were washed in PBS and analyzed using BD FACSVerse© flow cytometer. Cell cycle was analyzed quantifying incorporated propidium iodide using FCSalyzer ver.0.9.22-alpha (http://sourceforge.net/projects/FCSalyzer).
Quantitative RT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA). cDNA was synthesized from 1000 ng of total RNA using cDNA reverse transcription kits (Applied Biosystems, Foster City, CA). Relative mRNA level was analyzed by real time PCR (ABI 7900HT FAST Real-Time PCR system, Applied Biosystems, Foster City, CA) with SYBR Green master mix (Applied Biosystems). GAPDH gene was used as housekeeping control. The primers are listed in Table S3.
EdU assay
The EdU staining on cells was done using Click-iT® EdU Alexa Fluor® 647 HCS Assay (C10419, Invitrogen). Cells were incubated for 60’ in complete medium supplemented with 10 mM EdU. Cells labeling, fixation and detection was done following manufacturer’s instructions.
Immunohistochemistry
Immunohistochemistry was performed on formalin-fixed, paraffin-embedded tissue sections after hydration. After blocking in TBS with 0.1% Triton-X 100, 5% BSA and 1% goat serum, primary antibody incubation was carried overnight in in humified chamber at 4°C, dilutions followed manufacturer’s instructions. The Novolink Polymer detection system (Leica) was used for visualization, as described inmanufacturer’s instructions. Slides were then stained with hematoxylin and eosin then dehydrated, cleared and cover slipped.
Mouse strains and treatments
All mice were from C57/Bl6 background. They were born healthy and according to mendelian ratio. Animals were kept in SPF rooms in temperature and light controlled environment and fed standard chow diet ad libitum and had free access to water. All the animal use followed institutional animal welfare guidelines and legislation, as approved by the local Animal Ethics Committee (Comitato di Bioetica e Valutazione, Torino, Italy). Pik3c2ahypo/hypo were generated at Lexicon Pharmaceuticals by gene trapping as described (23). To generate Pik3c2afl/fl mice, Pik3c2atm1a(EUCOMM)Hmgu mouse embryonic stem cells (clone HEPD0636_4_E04) were received from the International Mouse Phenotyping Consortium and used to generate chimeric mice. Pik3cafl/fl mice were finally obtained from germ-line transmitters intercrossed with transgenic mice expressing the Flp recombinase under the b-actin promoter [B6.Cg-Tg(ACTFLPe)9205Dym/J, Jackson Laboratory] and eventually eliminating lacZ/neomycin cassettes from the Pik3c2a locus. Subsequently, Pik3c2afl/fl mice were crossed with Cre transgenic mice [B6.Cg-Tg(CAG-cre/Esr1*)5Amc/J, Jackson Laboratory] to generate Pik3c2afl/fl;Cre+ and Pik3c2afl/fl;Cre- mice. Cre recombinase was activated using 50 mg/kg of tamoxifen (Sigma-Aldrich) in corn oil, via a daily eye drop administration for 14 days. Pik3c2afl/fl; Cre- mice received the same tamoxifen treatment and were used as experimental controls.
Computational modelling
Structure predictions were performed using the I-Tasser server without additional constraints or templates (52). The following amino acid sequence of the helical domain of PI3KC2a was used (Uniprot id: O00443, aa 861-1397), helical domain (aa. 861-1037) and PI3K/PI4K domain (aa. 1133-1397) are underlined while predicted sequence (P sequence - aa. 980-1203) is in bold: QHNLETLENDIKGKLLDILHKDSSLGLSKEDKAFLWEKRYYCFKHPNCLPKILASAPNWKWVNLAKTYSLLHQWPALYPLIALELLDSKFADQEVRSLAVTWIEAISDDELTDLLPQ FVQALKYEIYLNSSLVQFLLSRALGNIQIAHNLYWLLKDALHDVQFSTRYEHVLGALLSVGGKRLREELLKQTKLVQLLGGVAEKVRQASGSARQVVLQRSMERVQSFFQKNKCRLPLKPSLVAKELNIKSCSFFSSNAVPLKVTMVNADPMGEEINVMFKVGEDLRQDMLALQMIKIMDKIWLKEGLDLRMVIFKCLSTGRDRGMVELVPASDTLRKIQVEYGVT GSFKDKPLAEWLRKYNPSEEEYEKASENFIYSCAGCCVATYVLGICDRHNDNIMLRSTGHMFHIDFGKFLGHAQMFGSFKRDRAPFVLTSDMAYVINGGEKPTIRFQLFVD LCCQAYNLIRKQTNLFLNLLSLMIPSGLPELTSIQDLKYVRDALQPQTTDAEATIFFTRLIESSLGSIATKFNFFIHNLAQLR. Based on amino acid sequence, three dimensional atomicmodels were generated frommultiple threading alignment and iterative structural assembly simulations. The output fromthe server run contained full-length secondary and tertiary structure predictions. An estimate of accuracy of the predictions is provided based on the confidence score of the modeling (52). The TM-scores and root mean square deviations (RMSDs) of the generated structures with respect to the template structures were calculated. Crystal structure of human g-tubulin was obtained from PDB deposited under ID: 3CB2 (resolved at 2.3 Å) (53). PI3K-C2α/g-tubulin interaction was studied using High Ambiguity Driven biomolecular DOCK-ing (HADDOCK) webserver (haddock.science.uu.nl).
HADDOCK webserver offers protein–protein ab initio dockings based on random patch definition and center of mass restraints. Possible residues of interfaces in each structure were predicted using the Consensus Prediction Of Interface Residues in Transient complexes facility and integrated intoHADDOCK. Then, two hundred complexes were generated by the HADDOCK program and clustered. Selection of the best complex was based on cluster size, HADDOCK score and electrostatic energy. Among the ten best clusters, we selected a cluster that was the second most populated cluster but was characterized by highest HADDOCK score and lowest electrostatic energy. PISA was used to analyze the protein-protein docking and binding interfaces. The illustrations and visualizations of the 3D models were produced in PyMOL (version 1.7.1.7).
Proteomic analysis of PI3K-C2α
HEK293 cells (0.8 × 106) were transfected with eGFP (mock), eGFP-PI3K-C2α (full length), or eGFP-PI3K-C2α DNT (amino acids 377-1686, i.e., a mutant lacking the clathrin binding N-terminal domain) using calcium-phosphate. 16 hours post-transfection cells were harvested and washed twice with ice-cold PBS. The cells were resuspended with 1ml lysis buffer (20mM HEPES-NaOH pH 7.4, 130 mM NaCl, 10 mM NaF, 1 tablet/50 ml EDTA-free protease inhibitor cocktail, 0.05% Saponin). Cell debris was removed by centrifugation at 16,000 × g for 10 min. Cell lysates containing 1.5 mg of total protein were used for affinity-purification using 10 ml of GFP-Trap_MA beads (ChromoTek). Samples were incubated while rotating for 1h at 4°C. The beads were washed 3x with lysis buffer. Bound proteins were eluted with 2x SDS-PAGE loading buffer and separated by 4-12% SDS-PAGE. Each lane was cut into 12 slices, before in-gel digestion with trypsin was performed. Tryptic peptides were analyzed by reversed-phase capillary liquid chromatography system connected to an Orbitrap Elite mass spectrometer (ThermoScientific). The identification and label-free quantification of proteins was performed by MaxQuant software (Version 1.5.2.8). The LFQ intensities of the identified proteins were used to identify PI3K-C2α-associated proteins.
Development of PI3K-C2α inhibitor
We capitalized on the availability of active recombinant PI3K-C2α (Wang, Lo et al., 2018) to conduct high-throughput screening of > 37,000 small molecules from an in-house library followed by iterative rounds of medicinal chemistry optimization and selectivity profiling. These experiments identified a potent PI3K-C2α-selective small molecule inhibitor that we named PITCOIN1 (for phosphatidylinositol three-kinase class two inhibitor 1). PITCOIN1 inhibits PI3K-C2α with an IC50 of 95 nM but is inactive against class I PI3Ks or Vps34 and does not display off-target activity toward a panel of > 100 kinases including related lipid kinases. Details are reported in a separate study in preparation.
General experimental approaches
All the statistical details of experiments can be found in the figure legends, in the Results and the Methods sections, including the statistical tests used, exact value of n, what n represents (cells, experiments) and precision measures (mean, median, SD, SEM, confidence intervals). No statistical methods were used to predetermine sample size. No samples or data points were excluded from the reported analyses. Samples were not randomized to experimental groups. Sample size was determined on the basis of our previous studies (Franco et al., 2014, Franco et al., 2015). The investigators were not blinded to allocation during experiments and outcome assessment.
Supplementary Material
Acknowledgments
We acknowledge M. Gai and the Open Lab of Advance Microscopy (OLMA@MBC) for technical assistance. Funding: This work was supported by Associazione Italiana Ricerca Cancro (AIRC; 21875 to E.H.), Leducq Foundation (19CVD02 to E.H.), PRIN (20177E9EPY to E.H.), Fondazione Ricerca Molinette (to E.H.), Fondazione Pezcoller-SIC “Patrizia Coser” (to F.G.), Fondazione Italiana per la Ricerca sul Cancro (FIRC 22558 to J.P.M. and FIRC 22248 to M.C.D.S.), Deutsche Forschungsgemeinschaft (DFG) (TRR186/A08 to V.H. and SCHU 3314/1-1 to M.S.), NeuroCure Cluster of Excellence (Charité Universitätsmedizin Berlin, 10117 Berlin, Germany to V.H.), National Institute of Diabetes and Digestive and Kidney Diseases (DK119305 to D.A.B.), and Sigma Xi research foundation (to A.C.). Author contributions: F.G. designed and performed research, analyzed data, and wrote the manuscript; L.P., H.L., I.C., S.J.C., A.M., P.K., A.C., J.F., A.Y., W.-T.L., N.T.S., M.C.D.S, and J.P.M. performed research and analyzed data; O.V., D.R.P., H.N.B., D.T., R.L.W., M.T., T.B., B.M.D., M.S., M.S.W., A.G., M.M., B.D.P., V.H, E.B, G.R.M., and D.A.B. provided critical reagents and supervised the work; E.H. designed the research, supervised the work and wrote the manuscript. Competing interests: E.H. and A.G. are cofounder and board member of Kither Biotech, a pharmaceutical product company developing PI3K inhibitors for the treatment of respiratory diseases not in conflict with statements made in this article. The other authors declare no conflict of interest. Data and materials availability: All data are available in the manuscript or the supplementary materials. PITCOIN1 is subject to a European patent application, and its distribution is dependent on completion of a materials transfer agreement with the FMP.
References
- 1.Gulluni F, Martini M, Hirsch E. Cytokinetic abscission: Phosphoinositides and ESCRTs direct the final cut. J Cell Biochem. 2017;118:3561–3568. doi: 10.1002/jcb.26066. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt O, Teis D. The ESCRT machinery. Curr Biol. 2012;22:R116–R120. doi: 10.1016/j.cub.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adell MAY, Teis D. Assembly and disassembly of the ESCRT-III membrane scission complex. FEBS Lett. 2011;585:3191–3196. doi: 10.1016/j.febslet.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlton JG, Agromayor M, Martin-Serrano J. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc Natl Acad Sci USA. 2008;105:10541–10546. doi: 10.1073/pnas.0802008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhutta MS, McInerny CJ, Gould GW. ESCRT function in cytokinesis: Location, dynamics and regulation by mitotic kinases. Int J Mol Sci. 2014;15:21723–21739. doi: 10.3390/ijms151221723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurley JH. ESCRTs are everywhere. EMBO J. 2015;34:2398–2407. doi: 10.15252/embj.201592484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laporte MH, Chatellard C, Vauchez V, Hemming FJ, Deloulme J-C, Vossier F, Blot B, Fraboulet S, Sadoul R. Alix is required during development for normal growth of the mouse brain. Sci Rep. 2017;7:44767. doi: 10.1038/srep44767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christ L, Raiborg C, Wenzel EM, Campsteijn C, Stenmark H. Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem Sci. 2017;42:42–56. doi: 10.1016/j.tibs.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 9.Christ L, Wenzel EM, Liestøl K, Raiborg C, Campsteijn C, Stenmark H. ALIX and ESCRT-I/II function as parallel ESCRT-III recruiters in cytokinetic abscission. J Cell Biol. 2016;212:499–513. doi: 10.1083/jcb.201507009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goliand I, Nachmias D, Gershony O, Elia N. Inhibition of ESCRT-II-CHMP6 interactions impedes cytokinetic abscission and leads to cell death. Mol Biol Cell. 2014;25:3740–3748. doi: 10.1091/mbc.E14-08-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Addi C, Presle A, Frémont S, Cuvelier F, Rocancourt M, Milin F, Schmutz S, Chamot-Rooke J, Douché T, Duchateau M, Giai Gianetto Q, et al. The Flemmingsome reveals an ESCRT-to-membrane coupling via ALIX/syntenin/syndecan-4 required for completion ofcytokinesis. Nat Commun. 2020;11:1941. doi: 10.1038/s41467-020-15205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurley JH. The ESCRT complexes. Crit Rev Biochem Mol Biol. 2010;45:463–487. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teo H, Gill DJ, Sun J, Perisic O, Veprintsev DB, Vallis Y, Emr SD, Williams RL. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell. 2006;125:99–111. doi: 10.1016/j.cell.2006.01.047. [DOI] [PubMed] [Google Scholar]
- 14.Im YJ, Hurley JH. Integrated structural model and membrane targeting mechanism of the human ESCRT-II complex. Dev Cell. 2008;14:902–913. doi: 10.1016/j.devcel.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sreekumar PG, Hinton DR, Kannan R. The emerging role of senescence in ocular disease. Oxid Med Cell Longev. 2020:2583601. doi: 10.1155/2020/2583601. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuyama M, Tanaka H, Inoko A, Goto H, Yonemura S, Kobori K, Hayashi Y, Kondo E, Itohara S, Izawa I, Inagaki M. Defect of mitotic vimentin phosphorylation causes microophthalmia and cataract via aneuploidy and senescence in lens epithelial cells. J Biol Chem. 2013;288:35626–35635. doi: 10.1074/jbc.M113.514737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Bennett TM, Shiels A. A charged multivesicular body protein (CHMP4B) is required for lens growth and differentiation. Differentiation. 2019;109:16–27. doi: 10.1016/j.diff.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodger C, Flex E, Allison RJ, Sanchis-Juan A, Hasenahuer MA, Cecchetti S, French CE, Edgar JR, Carpentieri G, Ciolfi A, Pantaleoni F, et al. Genomics England Research Consortium, De novoVPS4A mutations cause multisystem disease with abnormal neurodevelopment. Am J Hum Genet. 2020;107:1129–1148. doi: 10.1016/j.ajhg.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seu KG, Trump LR, Emberesh S, Lorsbach RB, Johnson C, Meznarich J, Underhill HR, Chou ST, Sakthivel H, Nassar NN, Seu KJ, et al. VPS4A mutations in humans cause syndromic congenital dyserythropoietic anemia due to cytokinesis and trafficking defects. Am J Hum Genet. 2020;107:1149–1156. doi: 10.1016/j.ajhg.2020.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiosano D, Baris HN, Chen A, Hitzert MM, Schueler M, Gulluni F, Wiesener A, Bergua A, Mory A, Copeland B, Gleeson JG, et al. Mutations in PIK3C2A cause syndromic short stature, skeletal abnormalities, and cataracts associated with ciliary dysfunction. PLOS Genet. 2019;15:e1008088. doi: 10.1371/journal.pgen.1008088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorgoulis V, Adams PD, Alimonti A, Bennett DC, Bischof O, Bishop C, Campisi J, Collado M, Evangelou K, Ferbeyre G, Gil J, et al. Cellular senescence: Defining a path forward. Cell. 2019;179:813–827. doi: 10.1016/j.cell.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Gulluni F, De Santis MC, Margaria JP, Martini M, Hirsch E. Class II PI3K functions in cell biology and disease. Trends Cell Biol. 2019;29:339–359. doi: 10.1016/j.tcb.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Harris DP, Vogel P, Wims M, Moberg K, Humphries J, Jhaver KG, DaCosta CM, Shadoan MK, Xu N, Hansen GM, Balakrishnan S, et al. Requirement for class II phosphoinositide 3-kinase C2α in maintenance of glomerular structure and function. Mol Cell Biol. 2011;31:63–80. doi: 10.1128/MCB.00468-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodes JD, Lott MC, Russell SL, Moulton V, Sanderson J, Wormstone IM, Broadway DC. Activation of the innate immune response and interferon signalling in myotonic dystrophy type 1 and type 2 cataracts. Hum Mol Genet. 2012;21:852–862. doi: 10.1093/hmg/ddr515. [DOI] [PubMed] [Google Scholar]
- 25.Morita E, Sandrin V, Chung H-Y, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tcherniuk S, Skoufias DA, Labriere C, Rath O, Gueritte F, Guillou C, Kozielski F. Relocation of Aurora B and survivin from centromeres to the central spindle impaired by a kinesin-specific MKLP-2 inhibitor. Angew Chem Int Ed. 2010;49:8228–8231. doi: 10.1002/anie.201003254. [DOI] [PubMed] [Google Scholar]
- 27.Brill JA, Wong R, Wilde A. Phosphoinositide function in cytokinesis. Curr Biol. 2011;21:R930–R934. doi: 10.1016/j.cub.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Lo W-T, Vujičić Žagar A, Gulluni F, Lehmann M, Scapozza L, Haucke V, Vadas O. Autoregulation of class II α PI3K activity by its lipid-binding PX-C2 domain module. Mol Cell. 2018;71:343–351.:e4. doi: 10.1016/j.molcel.2018.06.042. [DOI] [PubMed] [Google Scholar]
- 29.Gupta GD, Coyaud É, Gonçalves J, Mojarad BA, Liu Y, Wu Q, Gheiratmand L, Comartin D, Tkach JM, Cheung SWT, Bashkurov M, et al. A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell. 2015;163:1484–1499. doi: 10.1016/j.cell.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fanarraga ML, Bellido J, Jaén C, Villegas JC, Zabala JC. TBCD links centriologenesis, spindle microtubule dynamics, and midbody abscission in human cells. PLOS ONE. 2010;5:e8846. doi: 10.1371/journal.pone.0008846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gulluni F, Martini M, De Santis MC, Campa CC, Ghigo A, Margaria JP, Ciraolo E, Franco I, Ala U, Annaratone L, Disalvatore D, et al. Mitotic spindle assembly and genomic stability in breast cancer require PI3K-C2α scaffolding function. Cancer Cell. 2017;32:444–459.:e7. doi: 10.1016/j.ccell.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Sagona AP, Nezis IP, Pedersen NM, Liestøl K, Poulton J, Rusten TE, Skotheim RI, Raiborg C, Stenmark H. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat Cell Biol. 2010;12:362–371. doi: 10.1038/ncb2036. [DOI] [PubMed] [Google Scholar]
- 33.Posor Y, Eichhorn-Gruenig M, Puchkov D, Schöneberg J, Ullrich A, Lampe A, Müller R, Zarbakhsh S, Gulluni F, Hirsch E, Krauss M, et al. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. 2013;499:233–237. doi: 10.1038/nature12360. [DOI] [PubMed] [Google Scholar]
- 34.Balla T. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franco I, Gulluni F, Campa CC, Costa C, Margaria JP, Ciraolo E, Martini M, Monteyne D, De Luca E, Germena G, Posor Y, et al. PI3K class II α controls spatially restricted endosomal PtdIns3P and Rab11 activation to promote primary cilium function. Dev Cell. 2014;28:647–658. doi: 10.1016/j.devcel.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alam SL, Langelier C, Whitby FG, Koirala S, Robinson H, Hill CP, Sundquist WI. Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain. Nat Struct Mol Biol. 2006;13:1029–1030. doi: 10.1038/nsmb1160. [DOI] [PubMed] [Google Scholar]
- 37.Hirano S, Suzuki N, Slagsvold T, Kawasaki M, Trambaiolo D, Kato R, Stenmark H, Wakatsuki S. Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain. Nat Struct Mol Biol. 2006;13:1031–1032. doi: 10.1038/nsmb1163. [DOI] [PubMed] [Google Scholar]
- 38.Elia N, Fabrikant G, Kozlov MM, Lippincott-Schwartz J. Computational model of cytokinetic abscission driven by ESCRT-III polymerization and remodeling. Biophys J. 2012;102:2309–2320. doi: 10.1016/j.bpj.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mierzwa B, Gerlich DW. Cytokinetic abscission: Molecular mechanisms and temporal control. Dev Cell. 2014;31:525–538. doi: 10.1016/j.devcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 40.Schiel JA, Simon GC, Zaharris C, Weisz J, Castle D, Wu CC, Prekeris R. FIP3-endosome-dependent formation of the secondary ingression mediates ESCRT-III recruitment during cytokinesis. Nat Cell Biol. 2012;14:1068–1078. doi: 10.1038/ncb2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Avelar RA, Ortega JG, Tacutu R, Tyler EJ, Bennett D, Binetti P, Budovsky A, Chatsirisupachai K, Johnson E, Murray A, Shields S, et al. A multidimensional systems biology analysis of cellular senescence in aging and disease. Genome Biol. 2020;21:91. doi: 10.1186/s13059-020-01990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Matteis MA, Staiano L, Emma F, Devuyst O. The 5-phosphatase OCRL in Lowe syndrome and Dent disease 2. Nat Rev Nephrol. 2017;13:455–470. doi: 10.1038/nrneph.2017.83. [DOI] [PubMed] [Google Scholar]
- 43.Wiessner M, Roos A, Munn CJ, Viswanathan R, Whyte T, Cox D, Schoser B, Sewry C, Roper H, Phadke R, Marini Bettolo C, et al. Mutations in INPP5K, encoding a phosphoinositide 5-phosphatase, cause congenital muscular dystrophy with cataracts and mild cognitive impairment. Am J Hum Genet. 2017;100:523–536. doi: 10.1016/j.ajhg.2017.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cauvin C, Echard A. Phosphoinositides: Lipids with informative heads and mastermind functions in cell division. Biochim Biophys Acta. 2015;1851:832–843. doi: 10.1016/j.bbalip.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 45.Sagona AP, Nezis IP, Stenmark H. Association of CHMP4B and autophagy with micronuclei: Implications for cataract formation. BioMed Res Int. 2014:974393. doi: 10.1155/2014/974393. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alfred V, Vaccari T. When membranes need an ESCRT: Endosomal sorting and membrane remodelling in health and disease. Swiss Med Wkly. 2016;146:w14347. doi: 10.4414/smw.2016.14347. [DOI] [PubMed] [Google Scholar]
- 47.Goto H, Inagaki M. New insights into roles of intermediate filament phosphorylation and progeria pathogenesis. IUBMB Life. 2014;66:195–200. doi: 10.1002/iub.1260. [DOI] [PubMed] [Google Scholar]
- 48.Panopoulos A, Pacios-Bras C, Choi J, Yenjerla M, Sussman MA, Fotedar R, Margolis RL. Failure of cell cleavage induces senescence in tetraploid primary cells. Mol Biol Cell. 2014;25:3105–3118. doi: 10.1091/mbc.E14-03-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bojjireddy N, Botyanszki J, Hammond G, Creech D, Peterson R, Kemp DC, Snead M, Brown R, Morrison A, Wilson S, Harrison S, et al. Pharmacological and genetic targeting of the PI4KA enzyme reveals its important role in maintaining plasma membrane phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate levels. J Biol Chem. 2014;289:6120–6132. doi: 10.1074/jbc.M113.531426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thoresen SB, Campsteijn C, Vietri M, Schink KO, Liestøl K, Andersen JS, Raiborg C, Stenmark H. ANCHR mediates aurora-B-dependent abscission checkpoint control through retention of VPS4. Nat Cell Biol. 2014;16:550–560. doi: 10.1038/ncb2959. [DOI] [PubMed] [Google Scholar]
- 51.Kettleborough RN, Busch-Nentwich EM, Harvey SA, Dooley CM, de Bruijn E, van Eeden F, Sealy I, White RJ, Herd C, Nijman IJ, Fényes F, et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496:494–497. doi: 10.1038/nature11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rice LM, Montabana EA, Agard DA. The lattice as allosteric effector: Structural studies of alphabeta-and γ-tubulin clarify the role of GTP in microtubule assembly. Proc Natl Acad Sci USA. 2008;105:5378–5383. doi: 10.1073/pnas.0801155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.