Abstract
Using mass spectrometry we have detected both assembly intermediates and the final product, the T = 3 viral capsid, during reassembly of the RNA bacteriophage MS2. Assembly is only efficient when both types of quasiequivalent coat protein dimer seen in the final capsid are present in solution. NMR experiments confirm that interconversion of these conformers is allosterically regulated by sequence-specific binding of a short RNA stem−loop. Isotope pulse−chase experiments confirm that all intermediates observed are competent for further coat protein addition, i.e., they are all on the pathway to capsid formation, and that the unit of capsid growth is a coat protein dimer. The major intermediate species are dominated by stoichiometries derived from formation of the particle threefold axis, implying that there is a defined pathway toward the T = 3 shell. These results provide the first experimental evidence for a detailed mechanistic explanation of the regulation of quasiequivalent capsid assembly. They suggest a direct role for the encapsidated RNA in assembly in vivo, which is consistent with the structure of the genomic RNA within wild-type phage.
Keywords: virus assembly, RNA−protein interaction, mass spectrometry
Introduction
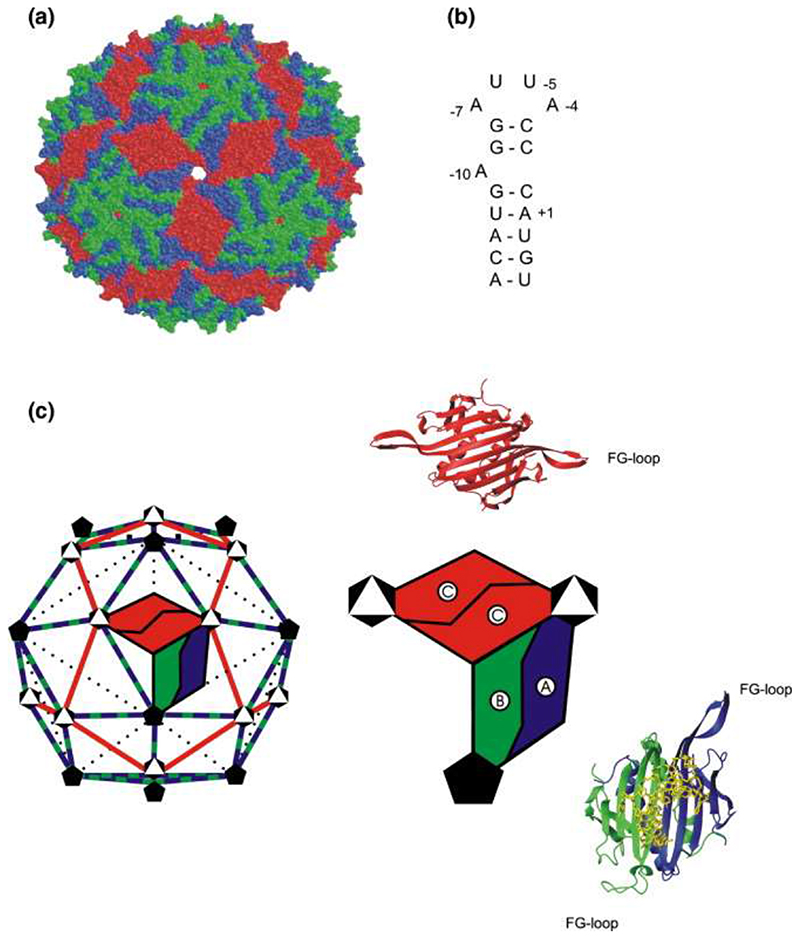
Simple spherical viruses consisting of a nucleic acid genome enclosed in a protein shell composed of multiple copies of one, or very few, coat protein (CP) subunits generally have isometric capsids. The positioning of the CP subunits is predictable from the allowed tessellations of a sphere. 1,2 We studied the RNA bacteriophage MS2, which conforms to a T = 3 shell in the Caspar−Klug nomenclature. The triangulation number (T = 3) implies that the phage CP subunit can exist in three quasiequivalent conformations (A, B, and C), allowing 180 copies of the subunit to create an enclosed capsid. This is indeed the case (Figure 1); the CP subunits form interdigitated noncovalent dimers (CP2) that act as the basic capsid building blocks composed of A/B or C/C conformers. 3,4 Within each monomer, these conformers are defined by the orientation of the loop of the polypeptide connecting the F and G β-strands, with A and C subunits having extended loops while B subunit loops fold back toward the globular core of the protein (Figure 1(c)). In the complete capsid, A/B dimers surround the particle fivefold axes with a ring of B-type loops, while A- and C-type loops alternate around the threefold axes.
Figure 1. The structures and solution components associated with MS2 bacteriophage assembly.
(a) A space-filling representation of the X-ray structure of the T = 3 capsid of wild-type MS23 (Protein Data Bank 2MS2). (b) The sequence and secondary structure of the TR RNA. (c) The distribution of symmetry axes (black and white symbols) within the icosahedral surface lattice of the MS2 capsid and the arrangement of the quasiequivalent dimers, AB (blue/green) and CC (red), within it. Alongside are ribbon models for the structures of each, with the A/B dimer bound to TR RNA, and an enlarged cartoon of their relationship within the capsid.
A major unresolved problem in structural virology is understanding the detailed molecular mechanism(s) that gives rise to the assembly of capsids of the correct size and symmetry. Various proposals for the formation of T = 3 shells based on the initial formation of three 5 or fivefold 6 assembly initiation complexes have been made. However, capsid assembly is spontaneous and usually very rapid, with the result that it has been technically difficult to isolate and characterize intermediates beyond these “initiation complexes” on the pathway to the final products. 7,8 The MS2 system is an ideal model for investigating such phenomena due to the extensive biochemical and structural information available. 3, 9–13 Capsid reassembly can be triggered in vitro by a sequence-specific RNA− protein interaction between a CP2 and an RNA stem−loop (TR) of just 19 nt 14,15 that encompasses the start codon of the viral replicase on the RNA genome (Figure 1). RNA−protein binding thus achieves two functions via a single molecular recognition event: translational repression of replicase and creation of an assembly competent complex on viral RNA.
We used electrospray ionization-mass spectrometry (ESI-MS) to investigate the mechanism of capsid reassembly and here show for the first time the time-dependent formation of both intermediates and the final product in a single assay. ESI-MS has proven to be a powerful technique for the analysis of noncovalently bound protein complexes because the mild ionization conditions are able to preserve often fragile macromolecular structures, thus permitting mass analysis of the intact complex 16,17 Our analysis of the yield and kinetics of the assembly of both intermediates and the final capsid leads to one of the first mechanistic descriptions of the control of quasiequivalent interactions during virus assembly, in this case via an allosteric switch controlled by interaction with phage RNA.
Results and Discussion
An RNA-mediated conformational change
The reassembly experiments were analyzed using a customized nanoelectrospray ionization−time-of-flight instrument with an extended m/z range (up to m/z 60,000), collisional cooling capabilities, 18–21 and a temperature-controlled, automated injection and ionization system (see Materials and Methods). Capsid reassembly in vitro was achieved by raising the pH of a solution of acid-disassembled coat protein in the presence of TR RNAs. 22,23 Working in solutions of final pH 5.2−5.7 and in buffers compatible with mass spectrometry slows the reassembly process, allowing us to observe early and intermediate stages.
In our ESI-MS experiments the possible molecular components in the reassembly reaction consist of the following: the CP; the CP dimer (CP2); the TR RNA (TR), the RNA−protein dimer complex (CP2:TR), and various higher-order species. Systematic analysis of reassembly conditions by gel filtration-light-scattering prior to the MS assay allowed us to choose conditions for the reaction that slow the production of the final product, the T = 3 capsid. These conditions favor very high overall yields of the desired end product and essentially eliminate the production of mis-structured aggregates that could otherwise complicate the analysis.
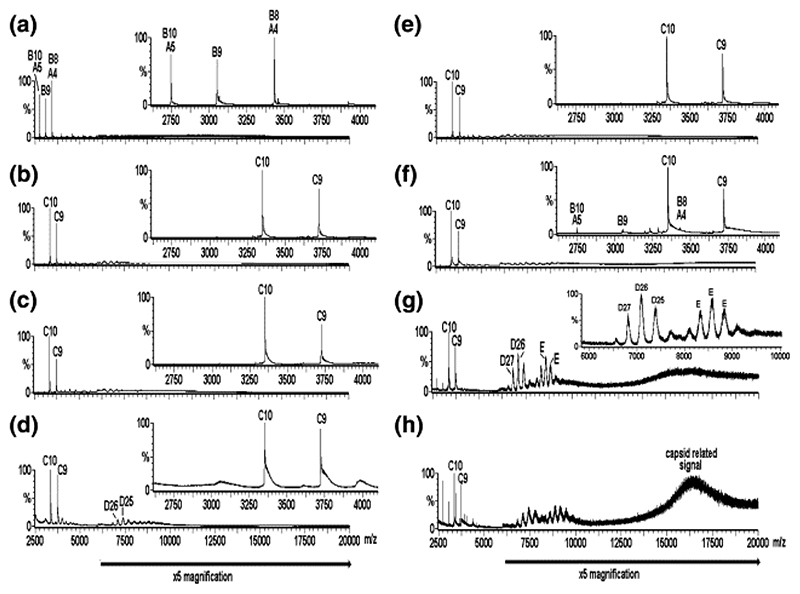
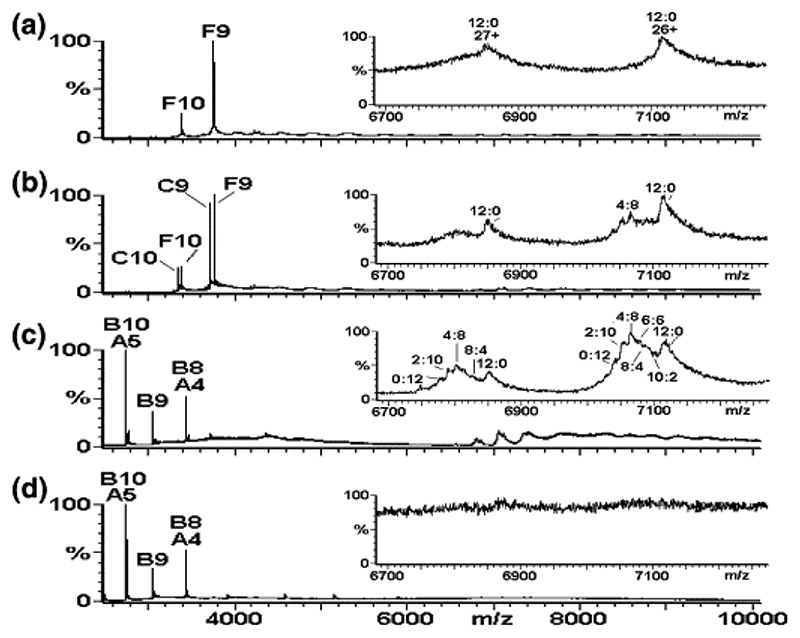
At pH ∼ 3.0 the CP obtained by acid disassembly of recombinant T = 3 capsids 24 shows a spectrum dominated by a mixture of CP (13.7 kDa) and CP2 (Figure 2(a); components labeled A and B, respectively), even at concentrations well above the dissociation constant for dimer formation determined at neutral pH, suggesting that the CP2 is not completely stable at this pH, at least in the gas phase of the mass spectrometer. These major species are accompanied by minor species with masses consistent with the CP trimer, tetramer, and pentamer (Figure 2(a); unlabeled, minor peaks (< 1%) in the range m/z 4000−7000). The addition of TR to a solution of coat protein at a final pH ∼ 5.2−5.7 and at a stoichiometry of 1:1 (CP2 to TR) changes the spectrum radically, resulting in the complete disappearance of CP, CP2, and the other minor species and the emergence of the complex CP2:TR of mass 33.5 kDa (Figure 2(b); component C). The presence of this complex as the major species implies that all of the CP2 binds TR RNA and is thus stabilized. The narrow ESI charge state distribution (9+−10+ ions) of the complex reflects its well-defined structure. This species is accompanied by masses consistent with trace amounts of higher-order oligomers in the region m/z 4000−10,000, which do not appear to alter significantly in concentration over time (Figure 2(c)) until after some 5 h, when certain higher-order species increase slightly in intensity (e.g., component D in Figure 2(d); see below for an explanation).
Figure 2.
Virus capsid reassembly monitored over time by real-time nanoESI-MS. The spectra were acquired over the range m/z 500−30,000 for samples in ammonium acetate (40 mM) at pH 5.2−5.7. The components are labeled as follows: A = CP; B = CP2; C = CP2:TR; D = 182.7 kDa, assigned as [3(CP2:TR)+3CP2], and E = ∼ 288−300 kDa, unassigned. The number immediately following each letter is the charge state of those particular ions, i.e., [M+ nH]n+. The spectra are as follows: (a) CP alone (8 μM); pH 3.2; (b)−(d) are for reassembly at pH 5.2−5.7 at a stoichiometry of CP2 to TR 1:1 (8 μM:8 μM), at 1, 90, 300 min, respectively. Spectrum (e) is the 1:1 pre-equilibrated starting point for reassembly at a stoichiometry of CP2 to TR 2:1 (16 μM:8 μM), spectra (f)−(h), at 1, 120, and 180 min, respectively. Note that spectra (b) and (e) are essentially equivalent. The bars below the spectra indicate a magnification factor of 5 for all ions above m/z 6000 to enhance clarity. The insets in (a)− (f) highlight the range m/z 2600−4100 to enhance clarity in the region containing ions arising from components A, B, and C. The inset in (g) highlights the range m/z 6000−10,000 to enhance clarity of the ions arising from components D and E and is normalized to the D26 ions, which are ∼10% of the intensity of the full spectrum.
The spectra are significantly different, however, when reactions are performed at a final stoichiometry of 2:1 CP2 to TR (Figure 2(f)-(h)), by allowing pre-equilibration of the complex (1:1, CP2:TR) for 10 min (Figure 2(e)) followed by the addition of an equal aliquot of CP2 lacking RNA (Figure 2(f)). The proportion of the higher-order species increases rapidly and dramatically and is not consistent with a simple mass action effect (Figure 2, cf. (c) and (g)). This effect was not seen when equal aliquots of CP2 and TR were added to 1:1 reactions. Several higher-order species are clearly visible in the 2:1 reaction, some of which we are able to assign to unambiguous stoichiometries, as well as weaker species whose signals are too broad to measure accurately. For the mass assignment, the measured masses, based on a minimum of three adjacent charge states, were compared with the calculated masses of all possible multiples of CP and TR. A significant higher mass species of 182.7 kDa is consistent with the complex [3(CP2:TR) + 3CP2] (Figure 2(g) and (h); component D). In addition to CP, CP2, CP2:TR, and [3(CP2:TR) + 3CP2] (components A−D, respectively), other, minor, higher-order species consistent with [CP2:TR+ CP] (mass 47 kDa), [2(CP2:TR)] (mass 67 kDa), and [3(CP2:TR) + CP2 + CP] (mass 142 kDa) were unambiguously identified (Figure 2; not labeled). Other species of masses > 200 kDa (e.g., component(s) E, mass ∼ 288−300 kDa) were more difficult to identify due to low-intensity broad peaks, which made unambiguous charge state assignment difficult and hence gave rise to the possibility of several theoretically compatible stoichiometries. The presence of complexes with CP monomer components may arise because of the presence of the monomer under the starting conditions. Over time the peaks corresponding to all of these species decreased in intensity. In contrast to the time-course for the 1:1 CP2 to TR reaction (Figure 2(b)-(d)), the higher-order aggregates (m/z 4000−10,000) are accompanied by broad unresolved higher mass to charge signals (> m/z 14000) visible in the spectra for the 2:1 reaction from the earliest time-point sampled (∼ 1 min; Figure 2(f)-(h)). Such broad peaks have been mass assigned to the MS2 T = 3 capsid by others 25 and are also seen when we analyze the intact recombinant capsid. 24
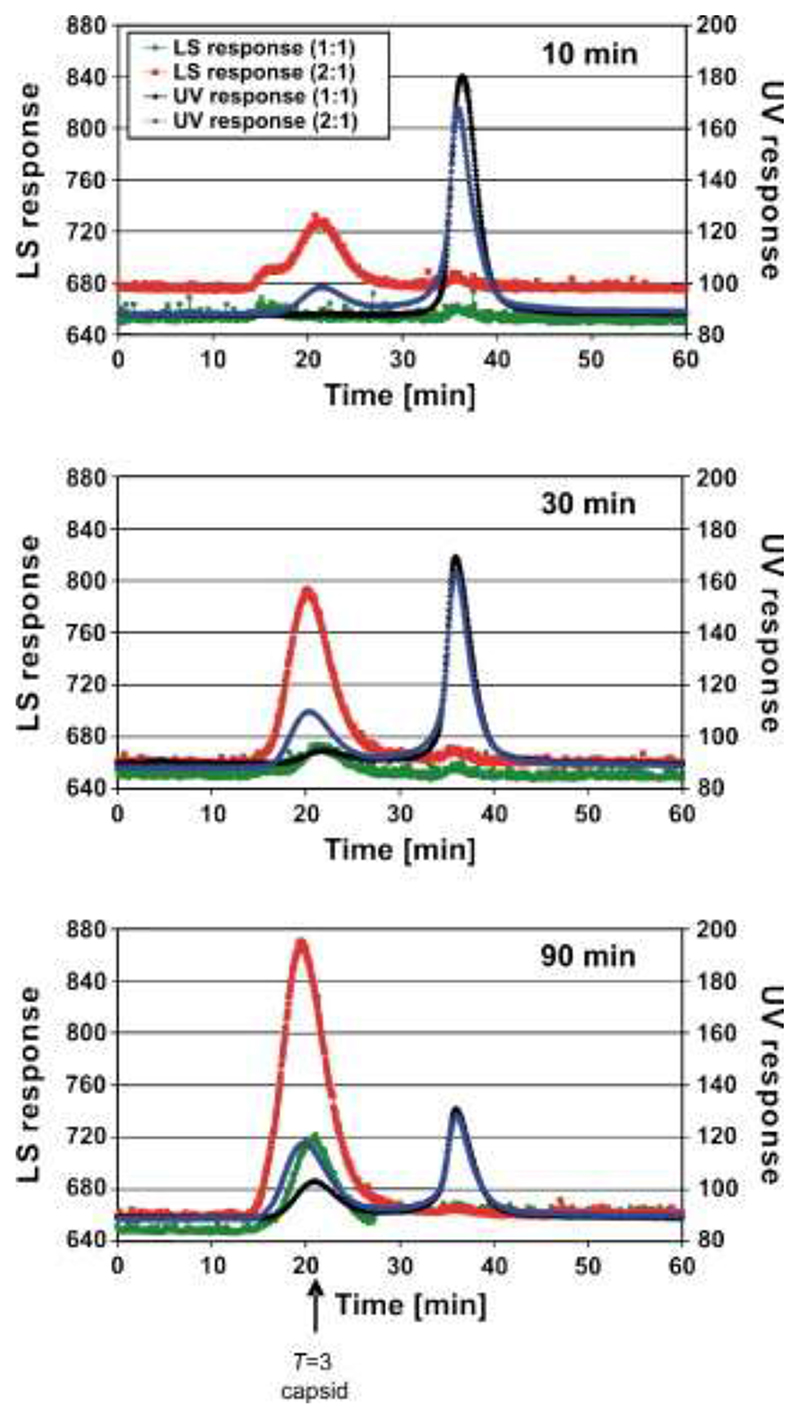
To confirm that the high mass peaks corresponded to the formation of T = 3 shells, identical reassembly reactions were analyzed by gel filtration-light-scattering (Figure 3). The 1:1 reaction forms capsids only very slowly, whereas the 2:1 equivalent has capsids present from the earliest time-points. These data imply that the small amounts of reassembly intermediates seen in Figure 2(b)-(d) are caused by a limiting concentration of an RNA-free CP component in the assembly pathway. Increasing the concentration of CP2 triggers a dramatic increase in the efficiency of assembly of the higher-order species observed in Figure 2(f)-(h). Further increasing the concentration of RNA-free CP subunits in the MS assay (to a 4:1 CP2 to TR ratio) results in more rapid loss of these higher-order species, including the CP2:TR complex, from the m/z spectrum, suggesting that they do participate in capsid formation rather than being simply MS artifacts. The spectrum is then dominated by peaks originating from CP monomer and dimer, together with a trace of trimer (data not shown). A reasonable inference from these effects is that the RNA-free coat protein added to the preformed CP2:TR complex is in a different quasiequivalent conformation from the one in the complex. Because both A/B and C/C dimers are required to build the T = 3 capsid, mixing these conformers at high concentration allows the assembly reaction to proceed efficiently. The 1:1 CP2:TR mixture also makes T = 3 shells, although under these conditions the RNA-free sample does not. Because the life-time of the CP2:TR is 42 s, 26,27 both types of dimer are available in the 1:1 reaction but in these reactions the concentration of the RNA-free form would be limiting.
Figure 3.
Gel filtration-light-scattering assays of capsid reassembly. MS2 CP2 and TR were mixed in 40 mM ammonium acetate, pH 5.2−5.7, to form a 1:1 reaction, incubated at 4 °C for 10, 30, or 90 min, and then loaded onto a Sepharose 6 gel filtration column equilibrated in 50 mM Tris−acetate, pH 7.4, and eluted at 0.36 ml/min. The outflow from the column was analyzed simultaneously via UV absorbance (black trace) and light-scattering (green trace). Similar reactions were then set up, preincubated at a molar ratio of 1:1 for 10 min, and then an additional aliquot of protein was added to create the 2:1 reaction. The traces are shown as UV absorbance (blue trace) and light-scattering (red trace). The position at which authentic T = 3 capsids elute is marked with an arrow; the peak at ∼35 min corresponds to the starting materials and smaller complexes.
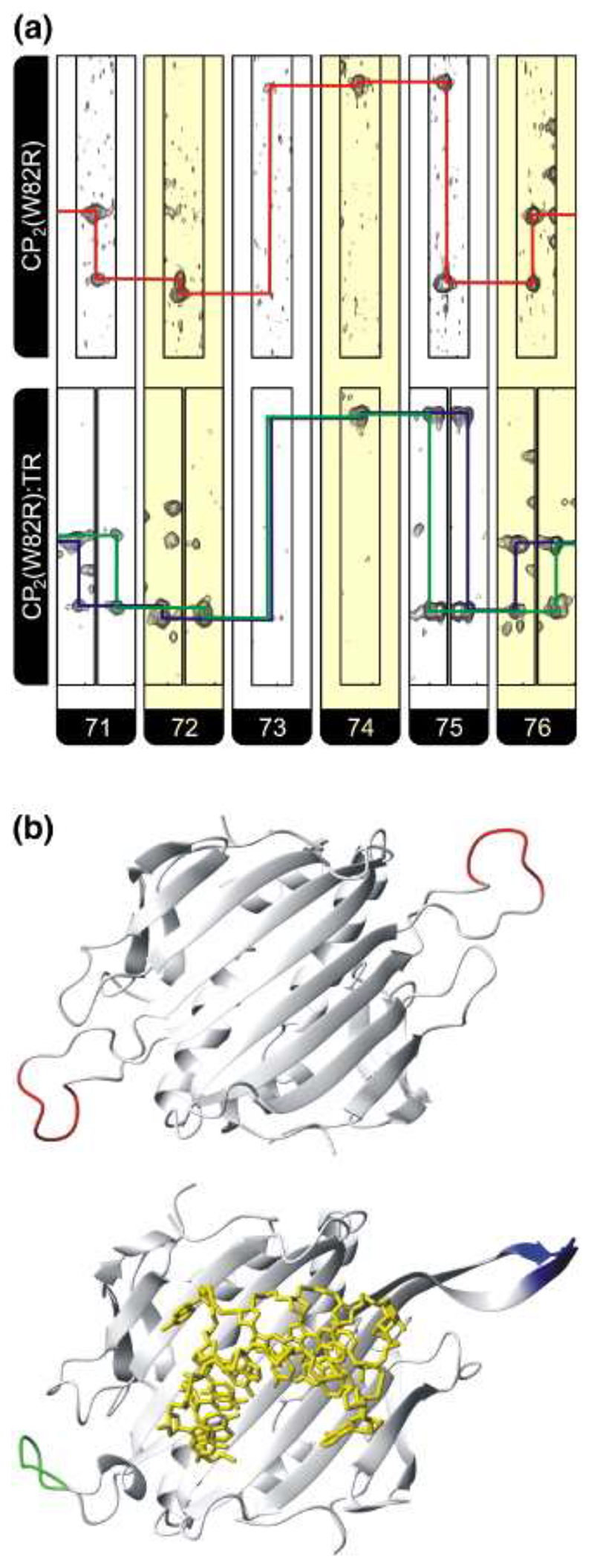
To probe whether TR binding leads to conformational change within the CP subunit, using NMR we carried out measurements with a mutant CP subunit (W82R) that does not assemble beyond dimer but does bind TR RNA with the same affinity as the wild-type (unpublished results). Residue 82 lies outside both the FG-loop and the TR binding site. The backbone resonances from the 1H, 13C, 15N triple resonance NMR spectrum of the apo-CP2(W82R) were analyzed with 87% of the backbone NH resonances being assigned, including virtually all the nonproline resonances in the FG loop region. The full details of these experiments will be published elsewhere. The degeneracy of all pairs of backbone resonances present in apo-CP2(W82R) resulting from pairs of symmetry-related atoms in the two subunits of the dimer, encompassing the FG loop, show that in the absence of RNA the protein subunits are equivalent on the NMR chemical shift timescale (Figure 4). Furthermore, 1H−15N heteronuclear nuclear Overhauser effect spectra (data not shown) show considerable motion within the FG loop on picosecond to nanosecond timescales, indicating that the chemical shift equivalence seen in these regions results from an average of a large number of conformers.
Figure 4. Observed changes in the chemical shifts for the FG loop residues 71−76 in both apo and RNA-bound complexes.
(a) Strips, alternately colored white and yellow for each residue, from HNCA spectra of CP2(W82R) (top) and CP2(W82R):TR (bottom). The assignment pathway for CP2(W82R) is shown in red while the split pathways for the complex are in blue and green. Note that no assignment of resonances to specific FG loop conformations is made or assumed. (b) Cartoon representation of (top) the backbone of CP2(W82R) showing the position of residues 71−76 in red for the two CP subunits derived from the crystal structure of this mutant 38 (Protein Data Bank 1MSC) and (bottom) the equivalent view for a wild-type A/B dimer (green/blue) bound to TR RNA (yellow sticks), which is the conformation we assume is closest to that present in solution.
The addition of TR to the labeled sample leads to a change in the spectrum of CP2(W82R), with resonances for the apo-CP2 state disappearing and pairs of peaks appearing at perturbed positions, which are associated with the [CP2(W82R):TR] complex. The data show that the complex is in slow exchange with the apo state, as expected for a RNA−protein complex with a nanomolar binding affinity. The sequential assignment for the backbone HN, N, and Cα resonances of residues 71−76 were made from HNCA, HncoCA, and 1H−15N heteronuclear single-quantum coherence spectra and the position of the fragment within the CP2(W82R) primary sequence assured by the presence of distinctive Cα resonances for residues 73 and 74, which are a GG pair, one of only two such pairs within the primary sequence of the CP. Pairs of resonances can be seen for residues 71, 72, 75, and 76, which are clearly delineated by shift differences in the 1H+15N dimensions (Figure 4). Comparison of the observed differences in shifts for the FG loop 1H amide resonances were made with those predicted by the Heigh Mallion ring current model using the program MOLMOL and ring current intensities suitable for RNA. 28,29 These calculations show that the observed changes in shifts in the presence and absence of TR are approximately an order of magnitude greater than that expected from theory (data not shown), confirming that the observed shifts are not caused by RNA binding per se but arise either from RNA-induced changes in the conformations of the loops or from fixing of a defined conformational state from within an equilibrium of states present in the apo protein. In summary, binding of TR results in conformational changes involving the FG loops and creates asymmetry in the complex. We propose that the conformations of the loops in the RNA-bound dimer are likely to be closer to those in an A/B coat protein dimer, compared to C/C-like or ill-defined conformers in the apo protein.
Why then does the W82R mutant fail to assemble beyond the TR:CP2 complex? To address this question we carried out an ESI-MS chasing experiment with combinations of wild-type and W82R CP. The W82R mutant dimer forms a CP2(W82R):TR complex when mixed 1:1 with TR with very little evidence of any higher-order species. Interestingly, the charge state distribution of the CP2(W82R):TR complex was slightly but noticeably wider (n = 7+−10+) than that of the wild-type complex (n = 9+−10+) analyzed under the same buffer and instrumental conditions, indicating a more dynamic structure (data not shown). Upon the addition of wild-type CP2 to the preformed CP2(W82R):TR complex, both the CP2:TR and the CP2(W82R):TR complexes were detected with no higher-order species visible. The reverse experiment, i.e., preforming the wild-type CP2:TR complex (with its associated dimer, trimer, and traces of other higher-order species) followed by the addition of CP2(W82R), resulted again in the observation of only the CP2:TR and CP2(W82R):TR complexes, in this case with the concomitant disappearance of the higher-order species (data not shown). There were no signals corresponding to a mixed CP-CP(W82R):TR complex. These results are consistent with our expectation that W82R can bind TR normally but is unable to support further interactions with other coat protein dimers, thus acting as an inhibitor of reassembly.
It is a reasonable assumption that the RNA-induced conformational change in the FG loops observed with W82R protein also occurs with the wild-type coat protein. This RNA-mediated allosteric effect at the FG loops explains the dependency of the kinetics and yield of assembly products on the presence of RNA-free and RNA-bound forms of the coat protein dimer.
The pathway to T = 3 shell formation
To confirm that the intermediates seen in the MS spectra were on-pathway to capsid formation and to determine the mass step size of capsid growth, we repeated the reassembly assay using isotopically labeled wild-type CP. The CP2:TR complex was preformed as above with a 1:1 ratio of reactants but using 15N-labeled CP2 (Figure 5(a), component F). The intermediate complex [3(CP2:TR) + 3CP2] discussed above was also visible in low yield in the region m/z 6700−7200, as expected. The measured molecular mass of this component indicated a totally 15N-labeled [3(CP2:TR) + 3CP2] complex, i.e., with all 12 CP subunits 15N-labeled, and hence is labeled 12:0 (15N to 14N). RNA-free 14N-CP2 was then added to make the 2:1 reaction (Figure 5(b)). There were then two peaks of approximately equivalent intensity corresponding to 14N and 15N-labeled CP2:TR complexes (components C and F), but there is no evidence for a heterodimer complex, indicating that while the TR molecule can dissociate from the CP2:TR complex and reassociate with a different CP2, the dimer does not dissociate to monomer under these conditions. Inspection of the region m/z 6700−7200 containing the multiply charged peaks for the complex [3(CP2:TR) + 3CP2] in the 2:1 reaction reveals that the 15N-labeled complex peaks are still present (Figure 5(b), inset, labeled 12:0) with additional peaks of the same charge state but with lower m/z values, i.e., of lower molecular mass. Further addition of 14N-CP2 (15N to 14N ratio now 1:3) causes these peaks to intensify (Figure 5(c)) and finally (15N to 14N ratio now 1:4) disappear (Figure 5(d)), as expected from the previous experiments. The masses of these components indicated the presence of complexes consisting of 15N to 14N CP in the ratios of 10:2, 8:4, 6:6, 4:8, 2:10, and 0:12 subunits, respectively, i.e., equivalent to all stoichiometries differing by dimer addition. The mass differences between these species consolidate the assignment of this complex and confirm the presence of 12 CP molecules. Again, no evidence was found for complexes of this stoichiometry with mixed 15N:14N CP2 units. The predominance of the 15N to 14N 4:8 ratio species in Figure 5(c) is consistent with the ratio of 15N to 14N CP present in this particular reassembly reaction (i.e., 1:3).
Figure 5.
Mapping the capsid reassembly pathway using isotopic chase experiments. Virus capsid reassembly from 15N-labeled CP2 chased by adding increasing quantities of 14N-CP2, with each reaction monitored by real-time nanoESI-MS at t = 1 min. The spectra were acquired over the range m/z 500−10,000 for samples in 40 mM ammonium acetate, pH 5.2−5.7. The larger panels show the range m/z 2500−10,000 and the components are labeled as follows: A = CP; B = CP2; C = CP2:TR; F = 15N-CP2:TR. The number immediately following each letter is the charge state of those particular ions. Spectra are (a) 15N-CP2 to TR 1:1 (8 μM:8 μM), t = 1 min; (b) 15N-CP2 to TR 1:1 (8 μM:8 μM) with 14N-CP2 (8 μM) added, t = 1 min; (c) 15N-CP2 to TR 1:1 (8 μM:8 μM) with 14N-CP2 (24 μM) added, t = 1 min; (d) 15N-CP2 to TR 1:1 (8 μM:8 μM) with 14N-CP2 (32 μM) added, t = 1 min. Inset spectra highlight the range m/z 6700−7200, showing the 26+ and 27+ charge states of the major reassembly intermediate [3(CP2:TR)+3CP2]. The peaks are labeled with the ratio of 15N:14N CP contained in the species, i.e., 12:0, 10:2, 8:4, 6:6, 4:8, 2:10, and 0:12.
Thus, it appears that the TR in the reassembly reaction rapidly equilibrates between the 14N and 15N-labeled coat protein dimer subunits, presumably because of the short half-life of the CP2:TR complex. 26,27 As the initially formed [3(CP2:TR) + 3CP2] 15N-labeled complexes continue on the assembly pathway to form capsids, new [3(CP2:TR) + 3CP2] complexes are formed from a mixture of the remaining 15N-labeled CP2 species and the added 14N material. Gradually, the percentage of all 15N-labeled [3(CP2:TR) + 3CP2] decreases with increasing CP2 concentration, while the mixed 14N:15N (Figure 5(b) and (c)), and eventually all 14N complexes, become predominant. Eventually, the addition of further CP2 dimers results in the disappearance of this complex, as well as other higher-order species, as capsid formation progresses (Figure 5(d)). One prediction of this interpretation is that performing the experiment in the opposite direction, i.e., preforming the complex with 14N CP2 and chasing with 15N-labeled material, should produce an equivalent, but mirror image, spectrum. This is indeed the case (see Supplementary Information).
The data confirm that the intermediate species detected in the MS spectra are competent to bind additional coat proteins and are therefore likely to be on the capsid assembly pathway. The unit of capsid growth is clearly a coat protein dimer under these conditions. Although some intermediate species are visible for extended periods during the assembly process, there does not appear to be a significant build-up of partially formed structures that later coalesce. This last observation is consistent with cryo-EM reconstructions of species formed in reassembly reactions that show particles of increasing completeness over time. 23
The results lead directly to a molecular mechanism for T = 3 capsid formation (Figure 6). RNA-free CP2 in solution has a preponderant, quasiequivalent conformation distinct from that created when TR binds, both of which are needed to allow the capsid to be built. This is consistent with the previous observation that some CP2 subunits will not bind TR until they have undergone a conformational change. 12 The TR binding site lies entirely within a CP2 and it makes distinct contacts with each monomer, suggesting that it might bind preferentially to an asymmetric dimer, e.g., an A/B unit. However, the NMR data from the apo-CP2 suggest that the dynamic motions of the FG loop regions also propagate along the attached F and G β-strands and overlap the RNA binding site, providing a potentially direct mechanism to couple RNA binding and FG loop conformational preference. A 1:1 reassembly reaction would thus be dominated by A/B-like species, whereas the 2:1 reactions described above would, in addition, contain more symmetrical dimers but with very dynamic FG loop regions. These could be C/C-like. Formation of A/B dimers would in principle permit formation of a capsid fivefold axis via interacting B-type loops. Such dimers could also contribute to the formation of a threefold axis by the A conformers interacting with C/C dimers. The stimulatory effect of adding RNA-free CP2 to the preformed CP2:TR suggests that formation of particle threefold axes is the dominant pathway toward T = 3 shell formation under these conditions. This is consistent with the fact that none of the assigned mass peaks in the spectra corresponds to a fivefold intermediate, 5(CP2:TR); rather, they are essentially all consistent with the threefold stoichiometry, [3(CP2:TR) + 3CP2], implying that there is a unique assembly pathway. Formation of the threefold complex determines that assembly yields a T = 3 rather than a T = 1 capsid; the latter would require only complexes with fivefold stoichiometries.
Figure 6.
Model for the initiation of T = 3 capsid assembly. A pair of chemically equivalent (C/C-like) subunits in solution (top left, shaded pink) binds a TR RNA, resulting in a conformational switch into an A/B-like asymmetric dimer (shaded green and blue). This complex binds an additional C/C-like dimer, creating the first higher-order intermediate on the pathway to capsid formation. A variety of subpathways involving the addition of coat protein dimers (arrows) can then generate the major assigned intermediate encompassing the first threefold axis of the virus particle. We have mass assignments for all the cartoon species shown in the diagram, as well as a number of components that may be in the subassembly pathway, such as 2[CP2:TR]+2CP2 & 3[CP2:TR]+2CP2+CP. These latter species are not always present in the reactions and when present are in trace amounts.
Biological implications
The mechanism described above immediately begs an obvious question: what triggers the C/C-like dimer in solution to adopt the A/B conformer at appropriate positions in the growing capsid? In vivo there is only one copy of the TR stem−loop per genome and there are no obvious candidates for closely related sequences/structures in the remainder of the phage RNA. However, we have shown that very many RNA stem−loop variants can bind at the TR binding site. Indeed, we have characterized over 30 such species by X-ray crystallography. 13,30 In such experiments preformed capsids are soaked with RNAs that penetrate into the interior of the particles via the pores at the symmetry axes, from where they can bind to all possible binding sites within the interior, including both A/B and C/C conformers. The orientation of the RNA at C/C dimers is degenerate but it is unique at A/B, allowing the details of the RNA−protein complex to be determined. These results suggest that the difference in RNA affinity between A/B and C/C dimers in a capsid is probably modest, although for many of the RNA variants we have studied the measured in vitro affinity for isolated dimers is very low. Note that the recombinant T = 3 shells used to generate the CP2 starting material here were described previously as “empty,” because they appear to lack any MS2 RNA or other long RNA sequences. 24 They are not, however, free of cellular RNA fragments, which may serve as the triggers of quasiequivalent switching when the recombinant protein is expressed at high levels.
It appears, therefore, that if there were suitably oriented genomic sequences/structures outside the TR region, coat protein dimers adding to a growing shell could bind them, thus fixing the appropriate quasiequivalent conformation. Cryo-EM reconstruction of the intact phage reveals the existence of such ordered RNA segments located underneath the protein shell, 31 supporting this idea. We have also shown that RNAs that encompass the TR sequence but have 5′ or 3′ genomic extensions appear to alter the efficiency of capsid formation. 23 This mechanism may well play a direct role in the life-cycle of the phage. The TR sequence is a transient signal that is only generated when ribosomes read through the coat protein gene, disrupting a long-range base-pairing interaction, the Min Jou sequence 32 . Binding of a CP2 to the TR stem−loop would lead to translational repression of the replicase cistron and simultaneously mark the MS2 genome for packaging into capsids. Many of the genomes marked in this way, however, might only be partially replicated and it might be inappropriate to begin assembly immediately after the repression complex forms. If protein-free genomic RNAs present higher affinity binding sites for CP2 at low concentrations, this would allow all progeny phage genomes to complete replication and carry an assembly initiation species. As the concentration of CP2 rises to levels in excess of the genomic RNA copy number, these C/C-like species can begin the assembly process by interacting with the A/B dimer bound at TR. Presumably, the additional A/B dimers required to complete assembly would then be triggered to form at appropriate points by interactions with genomic sequences outside the TR region, where both RNA− protein and protein−protein binding energy can contribute to the affinity of the interaction.
The pathway to capsid formation described above corresponds to the mechanism of autostery first described by Caspar, in which free subunits are held in an assembly-incompetent state until they make contact with the growing viral shell or another viral ligand. 33 In many other viral systems quasiequivalent conformer formation is dominated by alteration of the orientation of distinct protein domains or extended “arm” regions. 5 Many such proteins share structurally conserved protein domains and these have been co-opted by viruses from diverse sources from bacteria to humans. 34 The RNA phage coat proteins, in contrast, contain only a single domain and lack such extended arms. They form a unique protein fold that has to date no other viral homologues. This isolation may be because in these cases assembly is so dependent on interactions with the viral genome that it places such strict evolutionary constraints on the genomic sequence that the fold has not been used elsewhere. A related issue is the conformation of the phage genome during the assembly process. Does CP2 binding to the growing shell provide the driving force for genome folding and condensation or does the protein bind to a prefolded RNA? AFM measurements on a number of other single-stranded RNA viruses suggest that the genomic RNAs can fold stably into a compact structure with dimensions similar to that of the interior of the final protein shell, suggesting that the latter proposal is more likely. 35
The data presented here demonstrate the power of mass spectrometry to dissect the molecular details of a viral assembly pathway and gain insight into MS2 phage capsid construction by the identification of on-pathway intermediates. The ability to monitor mass and intensity changes in co-existing, noncovalently bound, high mass oligomeric species in real time is a vital asset for these studies. Such information is potentially very important for designing novel inhibitors of viral life-cycles, e.g., in the RNA phage, reagents that block the conformational changes required to create the A/B dimer would be potent inhibitors of assembly. Additionally, there are growing applications of virus-like particles in therapeutic and bionanotechnologic applications in which fundamental information on assembly mechanisms would be extremely useful. 36,37 ESI-MS can clearly contribute significantly to interdisciplinary studies of such systems.
Materials and Methods
Preparation of coat proteins and TR RNA
Proteins
Wild-type recombinant coat protein in the form of T = 3 shells lacking specifically bound RNA were prepared by overexpression in Escherichia coli as described previously. 24 These capsids were purified and then disassembled coat protein dimers isolated by treatment with glacial acetic acid, followed by exchange into 20 mM acetic acid. 12,22
NMR experiments were carried out on samples of the MS2 mutant coat protein, W82R, that does not assemble beyond the dimer. 38 W82R protein was purified by successive chromatography over diethylaminoethanol and sulphopropyl exchange media following the methodology described previously 38 with slight modifications. Following elution from the sulphopropyl column in 20 mM phosphate, pH 5.8, all samples were treated with 10 μL/ml of 2 M Tris, pH 8.5, to prevent crystallization and were then dialyzed into 50 mM Tris, pH 8.5, 100 mM KCl, 10 mM DTT, and 20 mg/l complete protease inhibitor for long-term storage at 4 °C. The purity and identity of the protein samples were assessed by SDS−PAGE, ESI-MS, and N-terminal sequencing of the first eight residues in the protein.
Isotopically labeled reagents were purchased from (Spectra Stable Isotopes Inc., Columbia, MD, USA), Goss Scientific Instruments (Great Baddow, Essex, UK), or Cambridge Isotope Laboratories (Andover, MA, USA). W82R samples were prepared by growing cells in M9 medium prepared with 50% D2O/50% H2O (v/v) and supplemented with 40 mg/l FeCl3 and 1% (v/v) 100× BME vitamin solution. Isotopes were introduced into the growth medium by the addition of 4 g/l of uniformly 13C-labeled 50% 2H-random-labeled glucose and 1 g/l of 15NH4Cl. Plasmid selection during expression was maintained using 100 μg/l carbenicillin. Cells were grown to an A600 of 0.8 starting from a labeled starter culture grown overnight and were induced with 0.1 mM isopropyl-β-𝒟-thiogalactopyranoside and harvested after a further 8 h of growth.
15N-labeled wild-type MS2 coat protein was prepared identically with wild-type protein, with the exception that the cells were grown in 15NH4Cl-containing minimal medium.
RNAs
Synthetic oligonucleotides encompassing the TR sequence (Figure 1) or the −5C variant were prepared as described previously. 39 The −5C variant binds to the coat protein dimer significantly more tightly than the wild-type sequence, with half-lives of > 300 and 42 s, respectively. 27 The crystal structure of the −5C variant bound to the coat protein in a preformed capsid10 shows that this effect is likely the result of an additional intramolecular hydrogen bond within the RNA, from the exocyclic amino group of C to the phosphodiester bridging the − 6 and − 7 positions. This presumably stabilizes the conformation of the RNA in solution closer to that resulting from protein binding reducing the free energy cost of distortion. There are no differences in contacts to the protein. Therefore, the − 5C variant was used in the NMR experiments.
Reassembly reactions
Reaction conditions
Assembly reactions were carried out in 40 mM ammonium acetate, pH 6.8, at 8 °C. Initially, CP2 was mixed with TR to a final concentration of 8 μM, and further aliquots of CP2 were added as indicated. The concentration of the CP2 solution was calculated from UV absorbance, and the sample was passed through a 0.2-μm syringe filter prior to measurement.
Electrospray ionization-mass spectrometry
Samples were analyzed by positive ionization nanoelectrospray using an LCT Premier Mass Spectrometer (Waters Corp., Manchester, UK) with collisional cooling capabilities equipped with a NanoMate (Advion, Inc., Ithaca, NY, USA) temperature-controlled automated sample handling and ionization interface, which was thermostated at 8 °C. A capillary voltage of 1.9 kV was set with a nitrogen gas flow of 0.5 psi for sample introduction and ionization. The sampling cone voltage was optimized at 80 V, ion guide 1 at 130 V, and aperture 1 at 60 V. The analyzer pressure was 7.2 × 10-7 mBar and the pressure in the transfer optics region was 4.9 × 10-3 mBar. Data were acquired over the range m/z 500−30,000 and data processing was performed using the MassLynx software supplied with the mass spectrometer. An external calibration using CsI clusters was applied to the data.
Gel filtration-light-scattering assays
For assays of reassembly by gel filtration, reactions at differing stoichiometries were set up with slight variations on the protocol for mass spectrometry. For convenience, the concentration of CP2 in the 1:1 reaction was 10 and 20 μM for the 1:2 sample, with the final TR concentration being 10 μM for both. The final 100-μl reaction volume was made up with assembly buffer (40 mM ammonium acetate, pH 6.8), and the samples were incubated on ice for various times until injection onto a Tricon high-performance column of Sepharose 6 10/300 GL (GE Healthcare Bio-Sciences AB, Sweden) thermostated at 12 °C. The eluant was 50 mM Tris acetate, pH 7.4 (chosen to avoid degassing problems with ammonium acetate), at a flow rate 0.36 ml/min.
The outflow of the column was monitored for UV absorption and for light-scattering using a static light-scattering detector PD 2000 (Precision Detectors, Inc., Bellingham, Mass., USA).
NMR data collection
Conditions providing optimum solubility of CP2 were determined from a set of hanging drop trials 40 in a range of buffers at a pH between 6.0 and 7.0 at 25 °C. Suitable solution conditions were thus determined to be 1% v/v glycerol, 100 mM Bis−Tris, pH 7.0, 100 mM KCl. In addition, the buffer also contained 10% D2O, 2 mg/ml complete protease inhibitor, 10 mM NaN3, 2 mM EDTA, 1 mM DTT, and 0.05 mM sodium 2,2-dimethyl-2-silapentane-5-sulfonate. Samples were introduced into the required solution conditions and concentrated using Vivaspin centrifugal concentrators (Sartorius, Ltd., Epsom, UK). All experiments were carried out in Shigemi microcells (Shigemi, Inc. Allison Park, PA, USA) containing 250−300 μl of solution with concentrations of CP2(W82R) between 0.5 and 1.0 mM. NMR experiments were measured on a Unity Inova 750-MHz NMR spectrometer (Varian, Inc., Palo Alto, CA USA) equipped with four radiofrequency channels and a triple-resonance (1H−15N−13C) room temperature probe with Z axis magnetic field gradients. The set of triple-resonance NMR experiments applied to the CP2 complex consisted of 1H−15N heteronuclear single-quantum coherence, HNCA, HNcoCA, HNCO, HNcaCO, HNcaCB, and HNcocaCB spectra, which were acquired using Varian Biopack pulse sequences. In the case of the HNCA, HNcoCA, HncaCB, and HNcocaCB experiments the pulse sequences were modified 41 so as to maintain optimal transverse relaxation optimized spectroscpy effects in partially deuterated proteins during the Cα/β chemical shift evolution period. The HNcoCA sequence was modified to have a Cα selective inversion pulse. 1H−15N heteronuclear nuclear Overhauser effect spectra were acquired on the CP2 dimer using a Varian Unity Inova 600-MHz NMR spectrometer equipped with four radiofrequency channels and a triple-resonance (1H−15N−13C) probe with Z axis magnetic field gradients.
In contrast to the experiments measured on CP2 the, HNCA and HNcoCA spectra of the CP2:TR complex were measured using a Varian cryogenic probe with receiver coils and preamplifiers cooled to − 253 °C. The limited assignment of the CP2:TR complex used spectra from 1H−15N heteronuclear single-quantum coherence, HNCA, and HNcoCA experiments. The buffer for the CP2:TR samples used for NMR spectroscopy was supplemented with 800 U RNAsin (Promega, Southampton, UK), which was brought to the final solution conditions using a Vivaspin centrifugal concentrator. Solutions of the CP2:TR complex were prepared by addition of a slight excess of TR to CP2 as determined by the loss of signals from the apo CP2 dimer.
All experiments were measured with the use of transverse relaxation optimized spectroscopy to reduce T2 relaxation effects and improve dispersion. Data processing and analysis were carried out using NMRPipe 42 and Nmrview 43 on computers running under Linux. Segments of assigned residues from CP2 were mapped to the primary sequence of the protein using the program Mapper2. 44 1H, 13C, and 15N chemical shifts were referenced directly (1H) and indirectly (13C, 15N) to sodium 2,2-dimethyl-2-silapentane-5-sulfonate.
Acknowledgements
We thank Drs Hugo Lago and Andrew Smith for preliminary mass spectrometry and help with data collection, respectively, and Dr Abdul Rashid for production of some of the materials used in these experiments. We also thank Professors Sheena Radford (Leeds), Bentley Fane (Arizona), and Jon King (Cambridge, Mass.) and Dr Reidun Twarock (York) for their helpful discussions of the results and the manuscript. This work was supported in part by the Leverhulme Trust, the Wellcome Trust, and the UK BBSRC. OR is a Wellcome Trust-funded PhD student.
References
- 1.Caspar D, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 2.Twarock R. A tiling approach to virus capsid assembly explaining a structural puzzle in virology. J Theoret Biol. 2004;226:477–482. doi: 10.1016/j.jtbi.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Valegård K, Liljas L, Fridborg K, Unge T. The three-dimensional structure of the bacterial virus MS2. Nature. 1990;345:36–41. doi: 10.1038/345036a0. [DOI] [PubMed] [Google Scholar]
- 4.Golmohammadi R, Valegård K, Fridborg K, Liljas L. Allosteric Conformer Switching in Virus Assembly. The refined structure of bacteriophage MS2 at 2.8 Å resolution. J Mol Biol. 1993;234:620–639. doi: 10.1006/jmbi.1993.1616. [DOI] [PubMed] [Google Scholar]
- 5.Sorger PK, Stockley P, Harrison S. Structure and assembly of turnip crinkle virus. 2. Mechanism of Reassembly in vitro. J Mol Biol. 1986;191:639–658. doi: 10.1016/0022-2836(86)90451-1. [DOI] [PubMed] [Google Scholar]
- 6.Rossmann MG, Abad-Zapatero C, Erickson JW, Savithri HS. RNA-protein interactions in some small plant viruses. J Biomol Struct Dyn. 1983;1:565–579. doi: 10.1080/07391102.1983.10507462. [DOI] [PubMed] [Google Scholar]
- 7.Rossmann MG, Johnson JE. Icosahedral RNA virus structure. Annu Rev Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 8.Zlotnick A, Aldrich R, Johnson JM, Ceres P, Young MJ. Mechanism of capsid assembly for an icosahedral plant virus. Virology. 2000;277:450–456. doi: 10.1006/viro.2000.0619. [DOI] [PubMed] [Google Scholar]
- 9.Witherell GW, Gott JM, Uhlenbeck OC. Specific interaction between RNA phage coat proteins and RNA. Prog Nucl Acid Res Mol Biol. 1991;40:185–220. doi: 10.1016/s0079-6603(08)60842-9. [DOI] [PubMed] [Google Scholar]
- 10.Valegård K, Murray JB, Stockley PG, Stonehouse NJ, Liljas L. Crystal structure of a bacteriophage RNA coat protein operator system. Nature. 1994;371:623–626. doi: 10.1038/371623a0. [DOI] [PubMed] [Google Scholar]
- 11.Stockley PG, Stonehouse NJ, Valegård K. Molecular mechanism of RNA phage morphogenesis. Int J Biochem. 1994;26:1249–1260. doi: 10.1016/0020-711x(94)90094-9. [DOI] [PubMed] [Google Scholar]
- 12.Lago H, Parrott AM, Moss T, Stonehouse NJ, Stockley PG. Probing the kinetics of formation of the bacteriophage MS2 translational operator complex: identification of a protein conformer unable to bind RNA. J Mol Biol. 2001;305:1131–1144. doi: 10.1006/jmbi.2000.4355. [DOI] [PubMed] [Google Scholar]
- 13.Horn WT, Tars K, Grahn E, Helgstrand C, Baron AJ. Lago, et al. Structural basis of RNA binding discrimination between bacteriophages Qβ and MS2. Structure. 2006;14:487–495. doi: 10.1016/j.str.2005.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beckett D, Uhlenbeck OC. Ribonucleoprotein complexes of R17 coat protein and a translational operator analog. J Mol Biol. 1988;204:927–938. doi: 10.1016/0022-2836(88)90052-6. [DOI] [PubMed] [Google Scholar]
- 15.Beckett D, Wu H-N, Uhlenbeck OC. Roles of operator and non-operator RNA sequences in bacteriophage R17 capsid assembly. J Mol Biol. 1988;204:939–947. doi: 10.1016/0022-2836(88)90053-8. [DOI] [PubMed] [Google Scholar]
- 16.Loo JA. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 17.Ashcroft AE. Recent developments in electrospray ionisation mass spectrometry: noncovalently bound protein complexes. Nat Prod Rep. 2005;22:452–464. doi: 10.1039/b417724j. [DOI] [PubMed] [Google Scholar]
- 18.Krutchinsky AN, Chernushevich IV, Spicer VL, Ens W, Standing KG. Collisional damping interface for an electrospray ionization time-of-flight mass spectrometer. J Am Soc Mass Spectrom. 1998;9:569–579. [Google Scholar]
- 19.Chernushevich IV, Thomson BA. Collisional cooling of large ions in electrospray mass spectrometry. Anal Chem. 2004;76:1754–1760. doi: 10.1021/ac035406j. [DOI] [PubMed] [Google Scholar]
- 20.Van Berkel WJH, Van den Heuvel RHH, Versluis C, Heck AJR. Detection of intact megaDalton protein assemblies of vanillylalcohol oxidase by mass spectrometry. Protein Sci. 2000;9:435–439. doi: 10.1110/ps.9.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobott F, Hernandez H, McCammon MG, Tito MA, Robinson CV. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal Chem. 2002;74:1402–1407. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- 22.Sugiyama T, Nakada D. Control of translation of MS2 RNA cistrons by MS2 coat protein. Biochemistry. 1967;57:1750–1774. doi: 10.1073/pnas.57.6.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stockley PG, Ashcroft AE, Francese S, Thompson GS, Ranson NA, Smith AM, et al. Dissecting the fine details of assembly of a T=3 phage capsid. J Theoret Med. 2005;6:119–125. [Google Scholar]
- 24.Mastico RA, Talbot SJ, Stockley PG. Multiple presentation of foreign peptides on the surface of an RNA-free spherical bacteriophage capsid. J Gen Virol. 1993;74:541–548. doi: 10.1099/0022-1317-74-4-541. [DOI] [PubMed] [Google Scholar]
- 25.Tito MA, Tars K, Valegard J, Hajdu J, Robinson CV. Electrospray time-of-flight mass spectrometry of the intact MS2 virus capsid. J Am Chem Soc. 2000;122:3550–3551. [Google Scholar]
- 26.Carey J, Uhlenbeck OC. Kinetic and thermodynamic characterization of the R17 coat protein-ribonucleic acid interaction. Biochemistry. 1983;22:2610–2615. doi: 10.1021/bi00280a003. [DOI] [PubMed] [Google Scholar]
- 27.Talbot SJ, Goodman S, Bates SRE, Fishwick CWG, Stockley PG. Use of synthetic oligoribonucleotides to probe RNA-protein interactions in the MS2 translational operator complex. Nucl Acids Res. 1990;18:3521–3528. doi: 10.1093/nar/18.12.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 29.Case DA. Calibration of ring-current effects in proteins and nucleic acids. J Biomol NMR. 1995;6:341–346. doi: 10.1007/BF00197633. [DOI] [PubMed] [Google Scholar]
- 30.Valegård K, Murray JB, Stonehouse NJ, van den Worm S, Stockley PG, Liljas L. The three dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence specific protein-RNA interactions. J Mol Biol. 1997;270:724–738. doi: 10.1006/jmbi.1997.1144. [DOI] [PubMed] [Google Scholar]
- 31.Koning R, van denWorm S, Plaisier JR, van Duin J, Pieter Abrahams J, Koerten H. Visualization by cryo-electron microscopy of genomic RNA that binds to the protein capsid inside bacteriophage MS2. J Mol Biol. 2003;332:415–422. doi: 10.1016/s0022-2836(03)00846-5. [DOI] [PubMed] [Google Scholar]
- 32.Berkhout B, van Duin J. Mechanism of translational coupling between coat protein and replicase genes of RNA bacteriophage MS2. Nucl Acids Res. 1985;13:6955–6967. doi: 10.1093/nar/13.19.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caspar D. Movement and self-control in protein assemblies. Quasi-equivalence revisited. Biophys J. 1980;32:103–138. doi: 10.1016/S0006-3495(80)84929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struct Biol. 2005;15:655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 35.Kuznetsov YG, Daijogo S, Zhou J, Semler BL, McPherson A. Atomic force microscopy analysis of icosahedral virus RNA. J Mol Biol. 2005;347:41–52. doi: 10.1016/j.jmb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 36.Wu M, Brown WL, Stockley PG. Cellspecific delivery of bacteriophage-encapsidated ricin- A chain. Bioconjug Chem. 1995;6:587–595. doi: 10.1021/bc00035a013. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Lin TW, Tang L, Johnson JE, Finn MG. Icosahedral virus particles as addressable nanoscale building blocks. Angew Chem Int Ed Engl. 2002;41:459–462. doi: 10.1002/1521-3773(20020201)41:3<459::aid-anie459>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 38.Ni CZ, Kodandapani R, Wickersham J, Peabody DS, Ely KR, Syed R. Crystal structure of the MS2 coat protein dimer: implications for RNA binding and virus assembly. Structure. 1995;3:255–263. doi: 10.1016/S0969-2126(01)00156-3. [DOI] [PubMed] [Google Scholar]
- 39.Murray JB, Collier AK, Arnold JRP. A general purification procedure for chemically synthesised oligoribonucleotides. Anal Biochem. 1994;218:177–184. doi: 10.1006/abio.1994.1157. [DOI] [PubMed] [Google Scholar]
- 40.Lepre CA, Moore JM. Microdrop screening: a rapid method to optimize solvent conditions for NMR spectroscopy of proteins. J Biomol NMR. 1998;12:493–499. doi: 10.1023/a:1008353000679. [DOI] [PubMed] [Google Scholar]
- 41.Eletsky A, Kienhofer A, Pervushin K. TROSY NMR with partially deuterated proteins. J Biomol NMR. 2001;2:177–180. doi: 10.1023/a:1011265430149. [DOI] [PubMed] [Google Scholar]
- 42.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe—a multidimensional spectral processing system based on unix pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 43.Johnson BA, Blevins RA. NMR View—a computer-program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 44.Güntert P, Salzmann M, Braun D, Wuthrich K. Sequence-specific NMR assignment of proteins by global fragment mapping with the program MAPPER. J Biomol NMR. 2000;18:129–137. doi: 10.1023/a:1008318805889. [DOI] [PubMed] [Google Scholar]