Preface
The function of neuronal circuits relies on the properties of individual neuronal cells and their synapses. We propose that a substantial degree of synapse formation and function are instructed by molecular codes resulting from transcriptional programs. Recent studies on the Neurexin protein family and its ligands provide fundamental insight into how synapses are assembled and remodeled, how synaptic properties are specified, and how single gene mutations associated with neurodevelopmental and psychiatric disorders might modify the operation of neuronal circuits and behavior. In this review, we first summarize insights into Neurexin function obtained from various model organisms. We then discuss mechanisms and logic of the cell type-specific regulation of Neurexin isoforms, in particular at the level of alternative mRNA splicing. Finally, we propose a conceptual framework for how combinations of synaptic protein isoforms act as “senders” and “readers” to instruct synapse formation and the acquisition of cell type- and synapse-specific functional properties.
Introduction
Nervous systems represent remarkable examples of a highly organized tissue with an abundance of specialized cells joint into an intricate structure. During development, neuronal connectivity arises from a series of steps, including cell specification, migration, targeted growth, synapse formation, and remodeling. Spontaneous activity and sensory experience propagated through the developing networks play a significant role in organizing aspects of neuronal wiring. However, many fundamental steps of neuronal morphogenesis and synapse formation proceed normally even in the absence of neurotransmission 1-3 . Thus, genetically encoded programs are thought to orchestrate key aspects of the timing and dynamics of neuronal growth and nervous system wiring 4-7 . Cell surface adhesion and signaling molecules significantly contribute to all of these developmental steps. Thus, each neuronal cell type carries an array of cues linked to cellular origin and cell fate that is integral to its developmental specification. While signaling processes, neuronal activity, and disease states may shift these codes, there are constraints that restrict this plasticity, thereby maintaining cell type-specific properties. One critical, and extensively studied process in nervous system development is the selective growth and targeting of neurites, which encompasses axon guidance and synaptic specificity 7,8 . The present review aims to discuss a second key aspect of neuronal wiring: the molecular principles of neuronal synapse formation and the specification of synapse function. We will use the Neurexins, one class of cell adhesion molecules, to illustrate fundamental principles of this process that likely apply to many other adhesion systems operating at neuronal synapses.
Adhesive modules for synapse assembly
Synaptic differentiation relies on a large number of synaptic adhesion and signalling molecules with so-called synaptogenic properties, that is, the ability of an isolated factor to trigger a substantial degree of the synaptic differentiation process. When presented in non-neuronal cells or on synthetic surfaces, synaptogenic proteins nucleate the formation of functional pre- or postsynaptic assemblies 9-12 . For example, postsynaptic adhesion molecules of the Neuroligin family trigger the assembly of functional presynaptic terminals in axons through interaction with their receptor Neurexin 9,10 (Fig. 1a). Conversely, Neurexin-mediated clustering of Neuroligins triggers the recruitment of NMDA-type glutamate receptors and scaffolding molecules 12,13 . This early cell biological analysis uncovered fundamental activities of Neurexin proteins and their ligands. Subsequent genetic studies then probed the functional consequences of inactivating Neurexin genes in various model organisms (see below). What makes the roles for Neurexins in this process so fascinating is twofold. First, the Neurexin gene family encodes a vast array of distinct transcript isoforms generated from multiple genes (Nrxn1, Nrxn2, Nrxn3), alternative promoters (α, β, γ), and extensive alternative splicing, with individual isoforms linked to specific neuronal cell types. Second, Neurexins serve as presynaptic receptors for several structurally unrelated extracellular binding partners, indicating that they represent a hub for presynaptic organization (Fig.1b). For example, Neurexins are presynaptic receptors for the secreted protein Cerebellin-1 (CBLN1), the transmembrane proteins Neuroligin 1-4, α-Dystroglycan, Leucine-rich repeat transmembrane proteins (LRRTM1,2,3,4), and Calsyntenin-3 14-17 . Recent reviews provided a comprehensive summary of this array of Neurexin ligands 6,18 . In the present article, we focus on the contribution of alternative splicing of Neurexins in controlling such interactions and on the interplay of multiple synaptic recognition systems at neuronal synapses. In the following, we will first discuss genetic studies in various model organisms where many or most Neurexin isoforms are ablated.
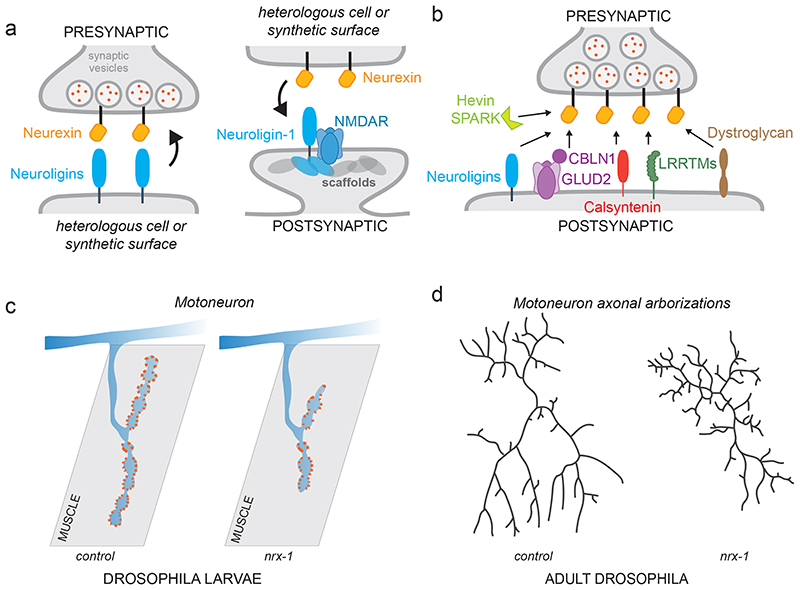
Figure 1. Synaptogenic function of Neurexins.
a) Illustration of bi-directional synapse-organizing activity of Neurexins. Presentation of Neurexin proteins or their ligands on synthetic surfaces in vitro and overexpression of Neurexin ligands in vitro and in vivo drives assembly of pre- and postsynaptic structures, respectively.
b) Neurexin isoforms interact with a large array of structurally unrelated extracellular binding partners. Only a selection of ligands is displayed in this simplified schematic. Depending on cellular context, several ligands can be co-expressed at single synapses or can be differentially expressed across neuronal cell populations.
c) Example for loss-of-function phenotype resulting from loss of Neurexin (nrx-1 mutant) at the neuromuscular junction of Drosophila larvae (adapted from Li et al. 27 ), synaptic release sites are marked in orange).
d) Illustration of contribution of Neurexin-Neuroligin adhesion system to growth of axonal arborisations of the motoneurons that innervate the abdominal pleural muscles of adult Drosophila (adapted from Constance et al. 29 ).
Neurexin and neuronal connectivity
Genetic loss-of-function studies highlight critical roles for Neurexin proteins at synapses in vivo. Initial work emphasized functional alterations in synaptic transmission, in particular calcium-dependent neurotransmitter release 19,20 . Consistent with the synaptogenic activity of Neurexins, a large body of genetic studies also support an evolutionary conserved role for Neurexin proteins in structural synapse assembly. In vivo models present with a wide array of phenotypes, depending on the cellular context and the Neurexin isoforms and/or genes ablated. Early studies in Neurexin-1,2,3α triple knock-out mice uncovered that inhibitory synapse density in the brainstem is reduced by 50%, whereas the density of excitatory synapses is unchanged at birth 19 . In the nematode Caenorhabditis elegans, loss of one particular Neurexin isoform, γ-Neurexin, diminishes synapse numbers in the DA9 motoneuron supporting critical functions for this form in synapse formation 21 . A broader nrx-1 mutation results in a loss of postsynaptic neurotransmitter receptors from synapses and loss of spine-like protrusions from the postsynaptic neuron 22,23 . In another class of C.elegans neurons that display experience-dependent and sexually dimorphic plasticity, synapse rearrangements are impaired in nrx-1 mutants 24 . In the fruit fly Drosophila melanogaster, mutations in Neurexin and its ligand Neuroligin, result in severe loss of neuromuscular synaptic release sites in larvae 25-27 (Fig.1c). Importantly, impairing Neurexin- Neuroligin adhesion also modifies the growth of axonal and dendritic arbors in some model organisms. Timelapse imaging experiments in developing tadpoles suggest that adhesion through Neurexin and its ligand, Neuroligin, confer transient morphological stabilization of dendritic contacts 28 . Similarly, the growth and arborization of Drosophila motoneuron axons during metamorphosis is disrupted in Neurexin-deficient fruit flies (Fig.1d) 29 . While such macroscopic alterations in neuronal arborizations have not been reported in mice, the roles for Neurexins in synapse assembly are conserved from invertebrates to mammals.
Two aspects have significantly delayed the emergence of the present picture for Neurexin functions: first, in the mammalian system, the phenotypic space explored in in vivo studies was quite limited. While work in invertebrate model systems has long explored synapse formation between genetically-defined cell types, this approach has only been implemented in mammalian systems this past decade. Second, unlike the vertebrate neuromuscular junction, where synaptic differentiation relies heavily on one primary signaling system 30 , central synapses engage a complex combination of signals. This cooperation between multiple trans-synaptic signals greatly complicates generalizing conclusions from individual genetic experiments. The same Neurexin mutation can result in very different phenotypes when analysed in different cell types. For example, mutation of Caenorhabditis elegans nrx-1 severely disrupts AChR clusters in DD GABA neurons but not in muscle 23 . In mice, the conditional ablation of all Neurexin isoforms in somatostatin-versus parvalbumin-positive interneurons exhibit very different phenotypes: Mutant parvalbumin-positive interneurons severely reduce synapse formation on principal neurons in the medial prefrontal cortex. By contrast, the number of synapses formed by Neurexin-deficient somatostatin-positive interneurons in the same region is unchanged 31 . However, somatostatin-positive interneurons show altered voltage-gated calcium channel function and defects in neurotransmitter release 31 . The reasons for such disparate observations are likely manifold. First, many studies examine mutations in cells without knowing the expression of the disrupted Nrxn gene, its transcript isoforms, and paralogues. Second, there is an array of additional presynaptic receptors unrelated to Neurexins that contribute to synapse assembly 32,33 . Third, different neuronal cell types express different Neurexin isoforms, generated from alternative promoters (like the α, β, γ forms) and modified through extensive alternative splicing. Notably, such Neurexin isoforms differentially interact with selective synaptic ligands. Thus, deletions of individual Neurexin genes precipitates impairment of different receptor-ligand modules in different cell types. This complexity most likely underlies the diversity of phenotypes reported in previous studies.
Molecular diversity of Neurexins
Combinations of genomic and proteomic features, including posttranslational modifications, impart Neurexins with numerous adhesive motifs that underlie low- and high-affinity interactions. These structural motifs - acting individually or cooperatively - recruit macromolecular complexes that span the synaptic cleft and coordinate bidirectional signalling and organization.
Three very large genes encode mammalian Neurexin1, -2, and -3 (1.0, 0.1, 1.6 Mb in mice; 1.1,0.1,1.8 Mb in human). Invertebrates, such as Caenorhabditis elegans and Drosophila melanogaster, possess a single Neurexin ortholog 34,35 . Each mammalian Neurexin gene contains two promoters that produce a long, α- and a shorter, β-Neurexin pre-mRNA, which encode proteins of approximately 1500 and 450 amino acids (Fig. 2a). For mouse Nrxn1 an additional, very short γ-isoform is generated from a third, internal promoter 36 , and an orthologous γ-isoform is reported in Caenorhabditis elegans 21 . We refer to these transcripts (irrespective of their further modification by alternative splicing as “primary Neurexin transcripts”). Differential usage of α-, β-, γ-promoters in the three mammalian Neurexin paralogues drive highly divergent levels of the primary Neurexin transcripts across neuronal cell types. For example, mouse hippocampal CA3 pyramidal neurons express high levels of Nrxn1β, whereas the same transcript is very low in CA1 pyramidal cells 37-39 . In the mouse neocortex, GABAergic SST-interneurons express threefold higher levels of Nrxn3α transcripts as compared to layer 4 pyramidal cells 39 .
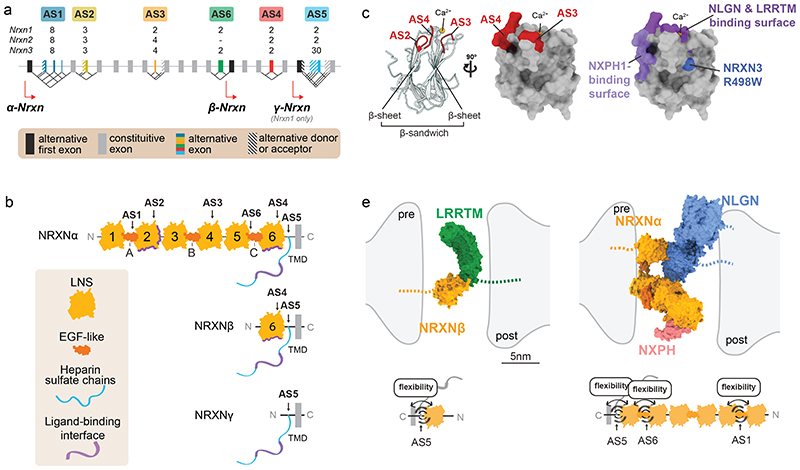
Figure 2. Molecular and structural features of Neurexin isoform diversity.
a) Schematic illustrating the alternatively spliced segments of mouse Neurexin genes. Mouse Neurexin transcript isoforms are generated from three genes (Nrxn1 = 1.1Mb, Nrxn2 = 0.1Mb, Nrxn3 = 1.8Mb; note that given the big differences in gene sizes, the exons and introns are not drawn to scale), each containing up to three alternative promoters (α, β and γ) and exhibiting extensive alternative splicing at six alternatively spliced segments (AS1-6). Individual segments can contain single alternative cassette exons (e.g. AS4, AS6) or consist of complex combinations of alternative splice donor and acceptor sites (e.g. AS1, AS2, AS3, AS5). Numbers (2,3,4,8,30) depict counts of potential splice variations generated at each segment. Alternative exons are illustrated in color, constitutive exons in grey, alternative donor or acceptor sites are striped.
b) Alternative promoters and alternative mRNA splicing result in Neurexin protein isoforms that share transmembrane domain (TMD) and cytoplasmic sequences but differ in their extracellular protein sequences. The extracellular sequences are composed of three major elements: Laminin-Neurexin-Sex hormone-binding globulin domains (LNS), epidermal growth factor-like domains (EGF), and attachment sites for heparan sulfates (HS). The largest Neurexin proteins are the NRXNα forms composed of six LNS domains (LNS 1-6), three interposed EGF domains (EGF A-C) and the HS attachment sites. Interaction surfaces for ligands are marked with purple lines. The NRXNβ forms contain a single LNS domain and HS attachment sites whereas the NRXNγ is the smallest form lacking LNS and EGF domains.
c) Mapping of alternatively spliced segments (AS2, AS3, AS4), ligand-binding domains, and sequence variants on a prototypical LNS domain: (left) ribbon diagram and positions of alternatively spliced segments, (middle) view of a 90-degree rotation and surface representation LNS domain with mapped alternatively spliced segments, (right) surface representation as in middle panel highlighting the position of ligand-binding domains and the naturally occurring R498W variant in NRXN3α which has been linked to behavioral alterations in mice 129 .
d) Illustration of approximate sizes and hypothetical conformation of adhesion molecule complexes in the synaptic cleft: β-Neurexin (orange, left panel) with LRRTM2 (green), and α-Neurexin (orange, right panel) with Neurexophilin (NXPH 1, pink) and Neuroligin (NLGN, blue). Structural models of the extracellular domains were drawn with ChimeraX 1.0 from the following Protein Data Bank IDs: 3POY 48 , 3B3Q 53 , 6PNP 50 , 5Z8Y 139 . The position of stalk, transmembrane and cytoplasmic sequences is indicated as dashed lines. Diagrams at the bottom display positions within these structures where alternative splicing at AS1, AS6 and AS5 in NRXN β (left) and NRXN α (right) modifies the flexibility of the extracellular domains in the synaptic cleft
Besides the use of these alternative promoters and corresponding transcription start sites, extensive diversification of Neurexin transcripts is further driven by alternative splicing. Thus far, up to six alternatively spliced segments (AS1-6) - some containing multiple alternative splice acceptor and donor splice sites - exist in primary Neurexin transcripts (Fig.2b). The combinatorial usage of alternative promoters and alternative splice sites has the potential to yield >12’000 Neurexin transcript isoforms in mice. Long-read, singlemolecule PacBio sequencing studies experimentally confirmed the presence of hundreds of Neurexin transcript isoforms in the mouse brain 40,41 . Interestingly, the relative usage and combination of alternative splice insertions are conserved between rodents and humans, evidenced by the analysis of postmortem human brain samples and hIPSC-derived neuronal preparations 42 . These transcriptomic studies provide a basis for interpreting the function of Neurexin diversity. However, one caveat is that transcript levels are not sufficiently informative regarding Neurexin protein isoform expression as multiple Neurexin gene products undergo further control at the level of mRNA translation 40,43 . Advances in targeted proteomics should clarify the accurate relative and absolute quantification of protein abundance, even for peptides derived from specific splice insertions 44 .
Structure of macromolecular assemblies
The defining feature of all Neurexins (except for the non-canonical NRXN1γ) is the presence of extracellular Laminin Neurexin Sex hormone-binding globulin (LNS) domains. α-Neurexins present six LNS domains interspersed by single EGF domains in the extracellular region (Fig.2b). These alternating repeats tether to the cell surface via a rigid and extensively O-linked glycosylated stalk and a single transmembrane domain 45,46 . The short intracellular tail contains interaction sites for cytoskeletal adaptors (Protein 4.1) and a C-terminal PDZ-binding motif 47 . The smaller β-NRXN proteins have a short, unique N-terminal sequence but are otherwise identical to α-NRXN beginning at the sixth LNS domain. γ-Nrxn transcripts encode a truncated isoform that lacks the extracellular LNS and EGF structured domains, yet retains a transmembrane and intracellular tail 36 .
Remarkably, despite having low sequence identity (20%) between each other, crystal structures from α-Neurexin1 LNS domains 2-6 reveal high structural homology 48 . The architectural prototype of an LNS domain consists of a β sandwich – two slightly curved β sheets juxtaposed, forming a ‘lens-like’ structure (Fig.2c). Importantly, at the rim of this β- sandwich are calcium and ligand-binding sites. A ligand-binding surface emerges from the folds that connect the two β sheets and is subject to alteration by AS2, AS3, and AS4, and accordingly is referred to as the hypervariable domain 49 . A one-quarter turn along the rim of the β-sandwich of LNS2 reveals an additional ligand-binding surface 50 . Exon inclusion or exclusion of Neurexin pre-mRNA encoding the hypervariable domain alters the length of the folds at the rim, modifies the binding surface topography, and regulates assorted low- and high-affinity interactions of Neurexins. Interestingly, an analogous surface of LNS-containing proteins agrin and laminin also confers ligand-binding specificity at the mammalian neuromuscular junction 51 .
Extensive studies of the LNS domain of β-NRXN1 (also NRXN1α LNS6) reveal critical structural elements for binding to Neuroligins 52-56 . The β loops of the Neurexin hypervariable domain clasp a single calcium ion creating an electropositive surface for binding to a complementary electro-negative surface on Neuroligin. Neurexin ligand selectivity also relies on the accessibility of ligand-binding surfaces, ligand concentration, and identity of the splice isoform. This dynamic balance is best documented in the interaction of β-NRXN and Neuroligin 57,58 . β-NRXN1,-2, and -3 bind to all Neuroligins with nanomolar affinity in a splice form-dependent manner 58 . Two general conclusions emerged from these in vitro binding assays: (1) the presence of AS4 insertions in β-NRXN1, and -2 diminishes affinity to Neuroligin; (2) by contrast the presence of AS4 insertions in β-NRXN3 increases affinity to Neuroligin. LRRTM1/2 also exhibits AS4- isoform-dependent binding at the same Neurexin LNS6 site, whereas Neurexins containing the alternative insertions at AS4 bind to CBLN 15,59 . These examples highlight the combinatorial and competitive activities of Neurexin-ligand interactions.
In addition to these protein-protein interaction sites, some interactions of Neurexins with ligands involve interactions with carbohydrate moieties on the Neurexins. The juxtamembrane region of α-, β-, and γ-Neurexins contain heparan sulfate carbohydrate structures that provide an additional interaction site for postsynaptic ligands such as LRRTMs and Neuroligins (Fig.2) 60,61 . LRRTM1/2 and Neuroligins require cooperative binding to Neurexin LNS6 and the carbohydrate chains for macromolecular assembly, whereas LRRTM3/4 only requires the carbohydrate structures and thus can act through γ- Neurexin isoforms which lack LNS domains. Interestingly, the identity of heparan sulfate proteoglycan structures is controlled by a series of cellular enzymes that produce cell type-specific carbohydrate modifications. Individual glycosyltransferases, sulfotransferases, and epimerases have emerged as critical regulators for neuronal development and wiring 62 . Thus, the molecular diversity of Neurexins generated at the level of alternative splicing may be complemented by a “glycan code” generated by differential heparan sulfate modifications.
The larger α-Neurexin isoforms, which are more abundant at the protein level 44 , interact with additional extracellular ligands. Thus, Calsyntenin-3 and α-Dystroglycan, two postsynaptic proteins at GABAergic synapses, interact only with α- but not β-Neurexin isoforms 16,63 . To accommodate the large, α-Neurexin in the narrow synaptic cleft, flexibility in the linker regions connecting the LNS domains bend the large, extracellular domain to fit in the 25nm synaptic cleft 46,48,64 . On the other hand, the short length of β-Neurexin is not constrained by its confirmation by the narrow synaptic cleft. Indeed, β-Neurexin and Neuroligin expressed in heterologous cells form a lattice-like sheet spanning the length of cell-to-cell contacts. By contrast, α-Neurexin-Neuroligin interactions fail to recruit widespread lateral assemblies in this assay. Thus, there might be an isoform-specific constraint on the macromolecular assembly of adhesion complexes within the limits of the synaptic cleft 65 . Moreover, while alternative splicing of Neurexin AS2, AS3, or AS4 tunes the affinity of Neurexin to ligands, the splice insertions at AS1, AS5, and AS6 – regions that encode for the linker regions – adjust the interdomain length (Fig.2d) 40,48 . Shortening or lengthening of these linker regions constrains the configuration of α-Neurexin and modulates the exposure of the ligand-binding domains. Ultimately, adjustments in the ligand-binding interface and changes in interdomain flexibility may govern the high and low-affinity interactions of Neurexins with their ligands.
Cell type-specific Neurexin isoforms
The molecular diversification of the Neurexin transcripts and the selective biochemical interactions of the resulting proteins raises the question of whether these proteins contribute to some form of molecular code that specifies aspects of neuronal wiring. The key elements to consider the coding power of such a system are: (1) the number of distinct recognition tags or “senders” (i.e., Neurexin protein isoforms generated), (2) the number of biochemical interaction partners that detect/distinguish or “read” these protein isoforms, and (3) the spatial logic of how such tags and “readers” array over neuronal cell types. Quantitative single-molecule sequencing of full-length Nrxn1 transcripts uncovered a large number of highly represented transcript isoforms in heterogenous brain tissue but a much more narrow isoform complement in a purified neuronal cell population 40 . Similarly, alternative exon and splice site choices at individual alternatively spliced segments display cell type-specific regulation of individual splicing decisions. For example, the relative abundance of exon usage at several alternatively spliced segments differs between parvalbumin- and CCK-positive interneuron populations 39,66,67 . Moreover, differential usage of combinatorial alternative splicing profiles of Nrxn1 and -3 or alternative splicing at AS3, correlate with the developmental origin of interneurons 67 . Quantitative assessments of the absolute usage of alternative exons uncovered that hippocampal CA3/CA1 pyramidal neurons and parvalbumin-positive interneurons contain different pools of Nrxn1 AS6 and Nrxn2 AS2. Hippocampal excitatory neurons contain substantially higher amounts of AS4- (exon lacking) than AS4+ (exon containing) transcript isoforms for all three Nrxn genes, whereas the higher AS4 inclusion rates can be observed in parvalbumin-positive interneurons 37 . A similar trend is observed for alternative splicing at AS4 of Nrxn3 in somatostatin-positive interneurons 37,68 . Conditional ablation of the Nrxn1 and 3 AS4 alternative exons in parvalbumin-positive neurons results in elevated hippocampal network activity, and impaired performance in a learning task 37 – a first demonstration that this cell type-specific isoform regulation is indeed essential for circuit function. A recent single-cell study on somatostatin-positive interneurons residing in the stratum oriens of the hippocampus, further revealed that neurons with similar electrophysiological properties exhibit similar expression of Nrxn1 and 3 splice isoforms 68 . In aggregate, these studies establish that Nrxn isoform repertoires link to neuronal cell type identity. This raises the question of how these repertoires are generated, whether they are dynamically regulated, and what specific aspect of neuronal function do such isoforms instruct?
Regulation of Neurexin splice isoforms
Alternative splicing is a highly dynamic process which is guided by cis-acting RNA sequence elements such as donor and acceptor sites, branchpoint and polypyrimidine tract for spliceosome assembly (Fig3a) 69,70 . Trans-acting factors such as RNA-binding proteins bind intronic or exonic sequence elements in the pre-mRNAs and bias the choice of splice donor and acceptor sites, resulting in inclusion or skipping of alternative sequence elements 70-72 (Fig.3a). Several classes of RNA binding proteins are implicated in the generation of cell type-specific Neurexin repertoires. In cultured rat neurons, the polypyrimidine tract binding protein PTBP2 modifies exon skipping at Nrxn2 AS4 73 . Genome-wide screens for transcript isoform alterations in mouse mutants for the RNA-binding proteins NOVA2, PTBP2 and RBFOX1 uncover modifications in Nrxn transcripts at several segments 74-76 , however, the functional consequences of these alterations are unknown.
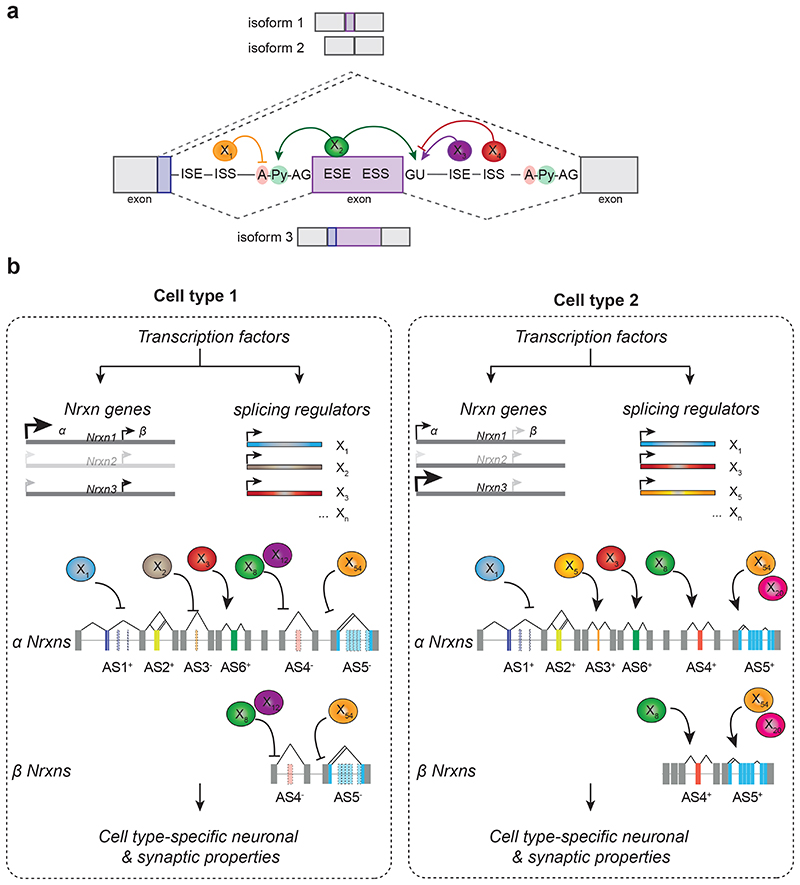
Figure 3. Control of synapse specification by alternative splicing programs.
a) Schematic illustrating the control of alternative splicing by RNA motifs and trans-acting factors (coloured spheres X1, X2, X3…). The displayed alternatively spliced segment contains an alternative splice donor site in the upstream exon (depicted as grey and blue boxes on left), followed by a cassette exon (purple box) and a downstream constitutive exon (grey box, right). Exons can contain exonic splicing enhancers (ESE) and exonic splicing silencers (ESS). The intronic between the exon boxes contain RNA motifs that act as intronic splicing enhancers (ISE) and silencers (ISS). GU marks the 5’ splice site, A-Py-AG marks the branchpoint and Polypyrimidine tract followed by the terminal AG sequence in the intron. Motifs recruit trans-acting RNA-binding proteins (depicted as colored spheres) which either promote or suppress usage of individual splice donor acceptor sites, resulting in inclusion or skipping of the alternative cassette exon (isoform 1 and isoform 2). Usage of the alternative donor site in the first exon, results in a third transcript isoform (isoform3).
b) Intersection of Nrxn transcriptional programs and the combinatorial action of splicing regulators. The illustration depicts two hypothetical cell types (cell type 1 and cell type 2) that produce different Neurexin transcript repertoires. Cell type-specific transcription from promoters/enhancers (arrowheads) drives the differential transcription of the primary transcripts from the Nrxn genes (e.g. Cell type 1 transcribes high levels of NRXN1α whereas Cell type 2 transcribes high levels of NRXN3α). In addition, each expresses a specific battery of splicing regulators (Cell type 1: X1,X2, X3,…; Cell type 2: X1, X3, X5,…). The intersection of these splicing regulators (colored spheres) with the primary Nrxn transcripts then produces the cell type-specific Neurexin isoforms (e.g. β Nrxn AS4-AS5- in cell type 1 and β Nrxn AS4+AS5+ in cell type 2) 76,79,89 .
Probably the best-characterized regulators of Neurexin alternative splicing are members of the STAR (signal transduction activators of RNA) protein family. These proteins are defined by an evolutionarily conserved RNA binding domain of approximately 200 amino acids. In mice, there are 5 STAR family proteins, the splicing factors SF1, Quaking, and SAM68, SLM1, and SLM2 – the latter three are closely related paralogues (encoded by the genes Khdrbs1,2,3) 77,78 . SAM68, SLM1, SLM2 directly bind RNA recognition motifs in introns flanking the highly conserved alternative exon at AS4 of the Neurexin pre-mRNAs 79-81 . This binding promotes skipping of the alternative exon at AS4. Indeed, a close correlation exists between AS4+ (exon containing) and AS4- Neurexin isoforms and the absence and presence, respectively, of SLM1 or SLM2 in neuronal cell types 37,80,81 . Alternative splicing regulation at AS4 is particularly interesting as the 30 amino acids encoded by the alternative exon strongly impact affinities for Neurexin ligands 17,59,82,83 . For example, AS4 containing Neurexins (NRXNAS4+) in cerebellar granule cell axons form tripartite complexes with the extracellular scaffolding protein CBLN1 and the postsynaptic receptor GLUD2 expressed in Purkinje cells. Mutation of any of the three components of this tripartite complex impairs presynaptic differentiation and results in a severe reduction in synapse assembly and density 59,84,85 . SLM1 and SLM2 proteins exhibit highly selective, mutually exclusive expression in neuronal cell types 80,86,87 . In the mouse hippocampus, SLM2 protein expression is primarily restricted to principal cells of the pyramidal cell layer (CA1, CA2, CA3) and subsets of somatostatin- and VIP-positive interneurons. In SLM2KO hippocampus, there is a highly selective loss of Nrxn1,2,3 AS4 insertions 87-89 . Conversely, ectopic expression of SLM1 or SLM2 in cells that do not endogenously express either of these proteins results in the generation of AS4- isoforms 37,80 . Thus, single, cell type-specific RNA binding proteins selectively instruct alternative splicing at one of the six alternatively spliced segments of Neurexin genes.
Detailed functional analysis of Slm2KO mice, as well as conditional mutations of AS4, uncovered that Neurexin splice variants in hippocampal CA3 pyramidal cells control postsynaptic properties of Schaffer collateral synapses. In Slm2KO hippocampi, macroscopic neuronal morphology and density of Schaffer collateral synapses is normal. However, SLM2-deficiency selectively elevates AMPA-receptor (GluA1) surface expression, leading to increased evoked glutamatergic transmission and impaired Schaffer collateral long-term potentiation (LTP). Selective genetic restoration of the NRXNAS4- splice isoform (which is lost in Slm2KO mice) restores normal levels of GluA1, partially restores LTP, and rescues behavioural alterations in the Slm2KO mice 89 . A germline mutation in Nrxn3 that constitutively includes the AS4 exon reduces AMPAR surface expression and impairs long-term potentiation (LTP) in a subset of subicular neurons 90 . Interestingly, the postsynaptic modifications that result from altered presynaptic Nrxn3 isoforms suggest the disruption of a transsynaptic link. The exact mechanisms resulting in altered postsynaptic properties remain to be worked out. However, the phenotypes in Slm2KO and Nrxn3AS4 mice are consistent with shifted ligand affinities of AS4- versus AS4+ splice variants for the interaction with postsynaptic Neuroligins and LRRTMs. Indeed, NRXNAS4+ isoforms display reduced affinity for postsynaptic Neuroligins 13,57,58 , and Neurexin-Neuroligin1/3 interactions are disrupted in Slm2KO hippocampus 89 . In Nrxn3AS4+ mice, expression of the postsynaptic NrxnAS4- ligand LRRTM2 is reduced 90 . Given that mutations in these postsynaptic Neurexin-ligands likewise disrupt LTP and synaptic transmission 91-93 , their altered interactions with presynaptic Neurexin isoforms may be responsible for aspects of synaptic dysfunction.
Interestingly, selective manipulation of AS4 alternative splice insertions in Nrxn1 and Nrxn3 differentially modify postsynaptic NMDAR- and AMPAR-mediated transmission 94 . Constitutive misexpression of Nrxn1AS4+ enhances NMDAR-mediated responses at hippocampal CA1-subiculum synapses, whereas Nrxn3AS4+ misexpression suppress AMPAR-mediated currents 94 . Whether these differential phenotypes are a consequence of different Nrxn1 and Nrxn3 expression levels in CA pyramidal cells or unique properties of the proteins derived from either gene is unknown. However, these findings raise the possibility that AS4 isoforms derived from two Nrxn paralogues may exhibit non-overlapping functions 94 .
In aggregate, these studies demonstrate that cell type-specific RNA binding proteins drive highly selective regulation of Neurexin alternative splicing. This regulation establishes cell type-specific molecular Neurexin isoform repertoires that engage in trans-synaptic interactions with dedicated receptors in the postsynaptic cell, thereby shaping fundamental synaptic properties. While this concept is best analysed for the alternative splicing factor SLM2 and the AS4 alternative exons, it may extend to the other Neurexin alternatively spliced segments. At Nrxn3 AS5, there are multiple alternative splice acceptor sites in the downstream exon of the segment (sometimes referred to as exon 25a,b,c). The amino acids encoded by Nrxn3 exon 25b confer binding of the Nrxn 3 isoform to the extracellular linker proteins C1QL2 and C1QL3, which in turn mediate the formation of a tri-partite, trans-synaptic complex with postsynaptic kainate receptors (GluK2 and 4) at hippocampal mossy fiber synapses 95 . Trans-acting factors regulating this splicing event remain to be identified.
While splicing choices link to cell identity, they also underlie dynamic regulation in response to neuronal signalling. Previous studies demonstrate shifts in alternative splicing regulation by strong pharmacological or electrical stimulation 96,97 . In Neurexins, such paradigms shift alternative exon incorporation at several alternatively spliced segments 81,98 . In the mouse cerebellum, this phenomenon requires calcium influx, calmodulin-dependent kinase IV, and the broadly expressed STAR protein, SAM68 81 . In granule cells of the mouse dentate gyrus, recall of a contextual fear memory triggers the inclusion of the alternative exon at Nrxn1AS4 . Interestingly, this shift in alternative splicing requires HDAC2 and is controlled by a selective histone modification (H3K9me3) in the Nrxn1 gene. This modification is thought to control memory stability, by temporarily shifting Nrxn1 alternative splicing at AS4, and ultimately, contributing to re-wiring of dentate granule cell synapses to support learning 99 . Yet another level of regulation is the proteolytic cleavage of Neurexins, resulting in shedding of the extracellular domain of the protein 100,101 . The physiological contexts and functional relevance of these modes of regulation remain to be explored. However, these mechanistically diverse modes of transcriptional, post-transcriptional, and proteolytic regulation highlight the perplexing complexity of Neurexin cell biology and pose challenges for linking cell type-specific repertoires to synaptic function.
Cell type logic of splicing regulators
Recent genome-wide studies on mRNA transcript isoforms and targets of RNA binding proteins provide insight into the complex logic of neuronal cell type-specific alternative splicing 39,75,76 . Several alternative transcript programs are linked to neuronal cell type, and some alternative splicing regulators increase in expression upon the commitment to a postmitotic fate 102 . Examining RNA binding protein expression across neuronal cell types highlights a remarkable range of cell type selectivity: some splicing regulators are expressed in essentially all neurons, and many of them are “neuron-specific”, i.e. largely not expressed in non-neuronal cells. These broadly expressed splicing regulators include the widely studied NOVA, PTBPs or RBFOX proteins. Other splicing regulators such as SLM1 and SLM2 exhibit a much more selective expression in a subset of neuronal cell types (Fig.3b). Intriguing, the selectively expressed RNA-binding protein SLM2 controls alternative splicing choices of only a few genes, acting as a highly targeted regulator of a small number of synaptic proteins 88,89 . Both SAM68 and SLM2 bind the same consensus motif, however, SLM2 but not SAM68 regulates alternative splicing of Nrxn2 at AS4 in vivo 80 . This selective activity is dependent on the abundance of binding sites flanking the alternative exon 79 . Furthermore, the specificity of RNA recognition by SLM2 and SAM68 may be modified by their ability to homo- and heterodimerize 78,80 . Thus, these different modes of action provide additional flexibility for the STAR family of proteins to generate neuronal cell class-specific synaptic properties (Fig. 3b X8+X12) 88,89 . By contrast, broadly expressed RNA-binding proteins tend to regulate alternative splicing events in mRNAs from hundreds of genes, possibly generating cell type-specific outcomes by coordinate and/or competitive action of multiple trans-acting factors on a single RNA segment. For example, NOVA2-dependent intron retention in hundreds of transcripts sequesters the trans-acting splicing factor PTBP2 75 . Remarkably, NOVA2 regulates diverse target transcripts in different cell populations, demonstrating that selectivity can emerge from a complex, cell type-specific interplay of splicing regulators. Moreover, selective alternative splicing decisions also arise from histone modifications and alterations in transcriptional kinetics 103 .
Acquiring a specific complement of RNA binding proteins during synaptic terminal differentiation may act much like the terminal selector genes for transcriptional cell type specification. Terminal selector genes have been postulated to be transcription factors which regulate the expression of genes required to give neurons their unique identity 4 . Congruent with this hypothesis, the combinatorial expression of RNA binding proteins in individual neuronal cell classes could ultimately instruct the generation of a cell type-specific complement of Neurexin alternative splice isoforms and their unique functions (Fig.3b). While this review article focuses on the Neurexin gene family, a similar molecular logic likely applies to other neuronal recognition and synaptic proteins 104 . Detailed profiling of transcript isoforms across genetically defined neuronal cell populations uncovered hundreds of differentially regulated alternative splicing events in the mouse neocortex and hippocampus. Furthermore, gene expression analysis of 52 bona fide splicing regulators highlighted broad, overlapping versus highly restricted expression within neuronal cell classes 39 . This supports the possibility that combinatorial expression of RNA binding proteins provides different classes of neurons with unique compositions of synaptic proteins and function (Fig 3b). Studies on additional neuronal recognition systems, such as the receptor protein tyrosine phosphatases, uncovered alternative splice insertions that modify molecular interfaces in trans-synaptic receptor-ligand interactions 33,104,105 . Thus, the principles discussed here for the generation and synaptic function of Neurexins likely extend broadly to the control of neuronal recognition (Fig.3b).
Circuits and disorders
From human genetic studies, the Neurexins, and predominantly NRXN1, emerge as significant risk genes for a wide range of neurodevelopmental, psychiatric, neurological and neuropsychological phenotypes 106-108 . Likewise, Neurexin gene mutations are associated with Schizophrenia 109 , Autism 110 , Tourettes Syndrome 111 , Nicotine dependence 112 , developmental delay, dysmorphic features and infantile epileptic encephalopathy 107,113 . Many of these deletions span promoter and initial exons of the NRXN1 gene (2p16.3). Transcriptomic studies in IPSC-derived neurons carrying such mutations uncover significantly reduced NRXN1α transcripts. Moreover, de-novo expression of isoforms divergent from the repertoire in neurotypical controls might occur 42 . Heterozygous exonic deletions do not appear to be fully penetrant as rearrangements in the NRXN1 gene are frequent in the control population 114 , and NRXN1 deletions are frequently inherited from a healthy parent 115 . However, such mono-allelic NRXN1 deletion carriers may share common alterations in anxiety, intelligence, and impulsivity, which go undiagnosed without an indepth evaluation 116 . Notably, a small number of bi-allelic NRXN1 mutations result in a severe mental retardation syndrome, which phenotypically overlaps with Pitt-Hopkins syndrome, an autism-like developmental disorder with variable characteristics 107,117,118 .
In some cases, specific NRXN sequence variants may elevate risk to certain disorders. However, considering the wide range of neurodevelopmental conditions observed in individuals with NRXN mutations, it is more likely that alterations in NRXN gene expression alter neurodevelopmental trajectories which – depending on genetic background and the environmental conditions - precipitate diverse phenotypes. It is widely appreciated that many symptoms are comorbid with multiple neurodevelopmental disorders, such as attention-deficit/hyperactivity, tic disorder, developmental coordination disorder, and autism. For clinical evaluations, it is encouraged - in particular for young children - to focus on impairments in specific domains, such as communication and language, motor coordination, attention, mood, and sleep, rather than to separate patients into discrete disorders. This is conceptualized in “ESSENCE” (Early Symptomatic Syndromes Eliciting Neurodevelopmental Clinical Examinations) 119 .
The significant disease association with the human NRXN genes has spurred efforts to obtain insights into how Neurexin mutations impact neuronal circuits and behaviour. It should be noted that some components of the Neurexin adhesion systems are also expressed in non-neuronal cells (Box 1). However, studies modelling the impact of disease-associated NRXN mutations have largely focused on synaptic phenotypes. Invertebrate systems have provided important opportunities for probing common cellular nodes modified by various risk gene mutations 118,120 . In mice, global Nrxn1α knock-out results in multiple behavioural alterations. These include impaired nest building, decreases in prepulse inhibition of startle responses, and an improvement in motor learning 121 . Interestingly, some phenotypes are sex-specific: Male homozygous Nrxn1α knock-out mice exhibit increased aggressive behaviors. Male heterozygous Nrxn1α knock-out mice show increased novelty responses as assessed by locomotor activity in a new environment and enhanced habituation upon repeated exposure to this environment 122 . In rats, non-social deficits, such as hyperactivity and deficits in instrumental and spatial learning tasks, result from Nrxn1α-deficiency 123 . Nrxn2α homozygous and heterozygous knock-out mice exhibit diminished social approach and social novelty responses in behavioral assays but also impaired recognition of novel objects 124-126 , indicating social and broader cognitive deficits.
Considering that these behavioral observations were made in global, constitutive knock-out mice, linking such phenotypes to selective developmental, circuit, and synaptic functions of the Neurexin proteins is difficult. Reversible overexpression studies using dominant-negative mutant Nrxn1 isoforms support the notion that behavioral phenotypes may result from dysfunction, rather than irreversible mis-wiring of circuits during development 127 . For Nrxn3, a requirement for Neurexin function in somatostatin interneurons in the anterior cingulate cortex affects empathy in conditional mutant mice. Empathy is a key element of social interactions, and the loss of empathy is an important feature of autism spectrum disorders and psychiatric conditions 128 . Mice carrying a single nucleotide polymorphism (SNP) in Nrxn3, which results in a single amino acid change (R498W), increase observational fear in a behavioral task 129 . In this task, an observer mouse adopts a conditioned context-dependent freezing response after observing a second mouse receiving repetitive foot shocks. Given that human performance in a similar paradigm correlates with metrics of empathy, this task is thought to assess an evolutionarily conserved aspect of empathy 130 . Conditional deletion of Nrxn3 in somatostatin-positive interneurons of the anterior cingulate cortex impairs synaptic transmission from these GABAergic neurons and elevates freezing responses, whereas activation of the same neuronal population suppresses them 129 . The R498W variant maps to the third LNS domain of Nrxn3α (see Fig.2c). This region may confer a Ca2+-mediated conformational switch for ligand binding 48,64 . However, ligands that contribute to the differential function of the Nrxn3 R498W protein remain to be uncovered. Nevertheless, these studies illustrate that Neurexin functions – and likely specifically the synaptic recognition codes controlled by the Neurexins - are not a cell biological detail but are fundamental for nervous system operation, behavior, and neurodevelopmental disorders.
Framework for synaptic action modules
A particular challenge in defining cellular Neurexin functions and predicting the impact of mutations on neuronal circuit function arise from extensive multiplexing at the biochemical level. A single presynaptic Neurexin isoform can recruit fundamentally different postsynaptic ligands. At a single synapse, multiple ligands compete for interaction with a limited pool of Neurexin molecules. Moreover, numerous Neurexin paralogues cooperate with various additional, independent trans-synaptic systems localized at the same synaptic contact. This complexity demands significant caution when interpreting loss-of-function studies. As discussed above, the same Neurexin loss-of-function manipulation applied in different cellular contexts results in widely differing phenotypes, ranging from a substantial loss of synaptic structures and entire axonal branches to the comparably subtle impairment of one or multiple synaptic ion channels. Such context-dependent synaptic phenotypes are not unique to the Neurexin gene family and have been reported for other trans-synaptic adhesion systems like the type III mGluR – Elfn 131-133 or receptor protein tyrosine phosphatase complexes 134,135 .
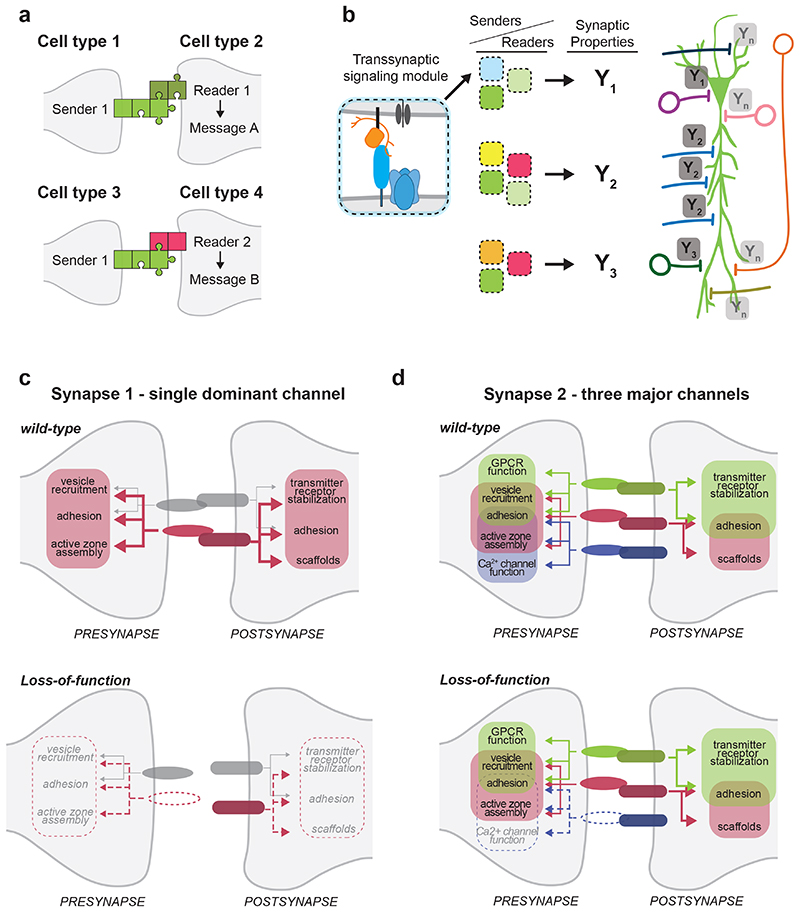
We propose that the combinatorial actions of synaptic adhesion and signaling proteins (Neurexins and other protein families) can be rationalized as modules for nucleating synaptic structures, scaffolding proteins, and ion channels. Multiple modules can be present at single synapses and contain overlapping components (Fig.4) 136-138 . These trans-synaptic recognition and synapse-organizing systems can be conceptualized as “senders” and “readers” arrayed across neuronal populations (Fig.5a). Upon fate specification, each neuronal cell type contains a set of cues – or a molecular code - that is integral to its neuronal identity. We postulate that this code instructs, but also constrains cellular interactions, and thereby, directs aspects of neuronal wiring and plasticity, thereby maintaining cell type-specific properties and circuit function (Fig.5b). Importantly, the messages conveyed by a particular sender (e.g., a specific Neurexin isoform) are strongly context-dependent. Thus, the nucleation of a trans-synaptic module largely depends on the molecular repertoire of “readers” available (Fig.5a) and may even rely on certain extracellular proteins being absent from a particular synaptic site. A second critical parameter is the number of trans-synaptic communication channels. Some synapses with little demand for plasticity and extensive neuromodulation may heavily rely on a few trans-synaptic channels, or even just a single dominant sender-reader pair (Fig. 5c). In such cases, loss of any of the core components results in a substantial dissociation of synaptic contacts, for example loss of the Neurexin-CBLN1 link at cerebellar parallel fiber synapses 84,85 or the Elfn-mGluR6 link in photoreceptor synapses 131 . At synapses with multiple prominent trans-synaptic channels, the same mutation may modestly destabilize a particular neurotransmitter receptor recruited by the sender or reader – however, a second trans-synaptic channel would take over additional functions and maintain the overall structural integrity of the synapse (Fig.5d).
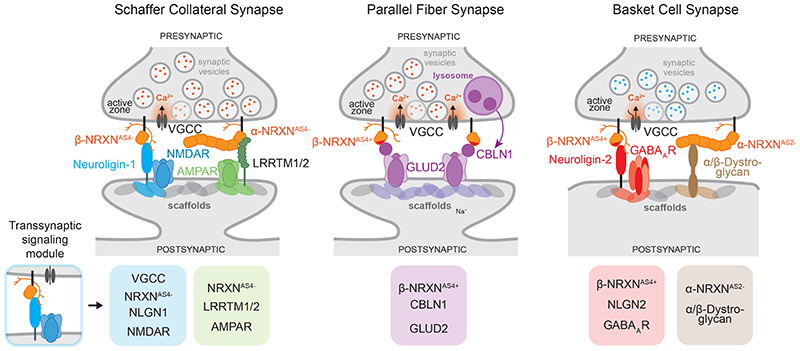
Figure 4. Examples of synaptic interaction modules nucleated by Neurexin proteins.
Simplified model illustrating trans-synaptic interaction modules assembled around presynaptic Neurexin protein isoforms. Amongst other components, CA3-CA1 Schaffer collateral synapses in the mouse hippocampus contain trans-synaptic NRXNAS4--NLGN1 and NRXN-LRRTM complexes which recruit postsynaptic NMDA and AMPA-type glutamate receptors. Note that LRRTM proteins bind α and β Neurexins – but for simplicity only the interaction with α is depicted here. Cerebellar parallel fiber synapses largely rely on a single trans-synaptic module consisting of NRXNAS4+ isoforms, extracellular CBLN1 proteins and the postsynaptic receptor GLUD2. CBLN1 is secreted from lysosome-like carrier vesicles 140 . Note that CBLN1 interacts with α and β Neurexins – but for simplicity only the interaction with β is depicted here. GABAergic basket cell synapses in the mouse hippocampus contain modules of NRXNAS4+ and NRXNAS2- isoforms linking to postsynaptic NLGN2 or α/β Dystroglycan proteins, respectively. Red dots within synaptic vesicles indicate the neurotransmitter glutamate, blue dots represent GABA. One major contribution of Neurexins at synapses is the incorporation of functional voltage-gated calcium channels (VGCC) at synapses, facilitating the calcium-dependent release of synaptic vesicles 19,142,143 . The lower row displays illustrations of individual trans-synaptic modules present at the respective synapses.
Figure 5. Context-dependent functions of synaptic interaction modules.
a) Model for the action of synaptic interaction modules. Cell types express specific complements of “senders” (light green) and “readers” (dark green and red), such as Neurexin splice isoforms and corresponding binding partners. The impact on synapse assembly and functional specification (“message”) depends on the cell type-specific abundance of corresponding senders and readers (dark green and red). Cell type 1 and cell type 3 both express the same sender but because their downstream synaptic partners cell types 2 and 4 express different readers (reader 1 and 2, respectively), the messages will be different (message A and message B, respectively). Thus, the same presynaptic sender can convey divergent messages at different synapses.
b) Across the surface of an individual neuron (here an illustration of a pyramidal cell), the combinations of axonal senders and dendritic readers create trans-synaptic modules (depicted as blue, green, light green boxes etc.) which represent a molecular code. This code sets synapse-specific properties (Y1, Y2, Y3, .. Yn; the blue labels represent glutamatergic synapses, the red labels GABAergic synapses) and, thereby, input integration in the postsynaptic cell.
c) Synapses across the central nervous system employ various numbers of trans-synaptic modules that can be viewed as trans-synaptic communication channels (displayed in different colors). Some synapses, rely on a single dominant channel (here depicted in red), which has a major contribution to synapse assembly, stability and functional properties (“Synapse 1” - an example for this would be parallel fiber synapses in the cerebellum which rely on the NRXN-CBLN1-GLUD2 module 59,85,141 ). Loss of a single presynaptic sender (illustrated in the lower panel, e.g. CBLN1) results in disruption of the trans-synaptic module and substantial disruption of synapse formation and function, despite the presence of an additional, minor channel (depicted in grey).
d) Other synapses (“Synapse 2”) contain multiple prominent trans-synaptic modules (here depicted in red, green, and purple) – likely to afford a larger dynamic range of plasticity. These modules drive overlapping elements of synaptic differentiation. For example, the green module drives bi-directional adhesion, presynaptic vesicle recruitment, presynaptic GPCR function, and post-synaptic stabilization of neurotransmitter receptors, whereas the purple module controls adhesion, active zone assembly and calcium channel function. Loss of a single presynaptic sender (the purple module) results in a loss of presynaptic calcium channel function but active zone assembly and adhesion are maintained by the overlapping red and green modules at this synapse. See references for examples on Ca2+ channel function 31,142,143 .
Such complex systems likely evolved for CNS synaptogenesis as they render a synaptic contact more tunable, providing a high degree of freedom to control plasticity of individual synaptic sites – but, at the same time they provide constraints for wiring in highly complex circuits. By integrating the observations made in reductionist biochemical and in vitro systems, across vertebrate and invertebrate systems, and loss-of-function studies in multiple cell types, we can define action modules for synaptic adhesion molecules. A key question for the future will be to explore how molecular codes and activity-dependent mechanisms intersect to shape circuitry during development. There is mounting evidence that synaptic transmission per se is not required for a significant degree of neuronal wiring and cell type-specific connectivity. Thus, activity-independent mechanisms generate an initial blueprint of neuronal circuits. However, within one molecularly and anatomically recognizable cell type, subpopulations of cells are recruited to represent or encode unique aspects of the external world, such as direction-selective cells in the visual cortex, “reward” cells in the cerebellum, or place cells in the hippocampus. Neuronal activity and synaptic plasticity mechanisms play a major role in establishing such neuronal ensembles – and future work may elucidate how molecular recognition systems constrain and execute such key steps of circuit assembly.
TOC Summary.
In this review, Scheiffele and colleagues discuss the molecular basis of transsynaptic signaling by Neurexins. Linking cell type-specific alternative splicing to the combinatorial action of Neurexin isoforms, this review synthesizes a mechanistic framework for Neurexins functions in synapse organization.
Box 1. Non-neuronal and non-synaptic roles for Neurexin complexes.
While the vast majority of research focus on the roles for Neurexins at synapses, recent evidence highlights potential functions of these adhesion molecules at other cellular junctions. A starting point for such investigations were transcriptomic and in situ hybridization studies that detected significant Neurexin mRNA expression in nonneuronal cells, including astrocytes, oligodendrocytes, and oligodendrocyte precursor cells 36,144,145 . Moreover, also major Neurexin ligands, such as the Neuroligins were detected in non-neuronal cells. For example, gliomas have been reported to produce significant levels of Neuroligin-3 protein 146-148 . Interestingly, the proliferation of glioma cells is elevated by electrical signalling that may involve synapse-like structures formed by axons onto the Neuroligin-expressing glioma cells as a critical pathological feature. An example for further extrasynaptic roles for Neurexins are interactions between neuronal Neurexins and the Neurexin ligand Neuroligin-2 in astrocytes 149 . These interactions were proposed to contribute to the elaboration of astrocytic morphology in the developing neocortex of mice. Moreover, the extracellular matrix proteins Hevin, SPARC, and Thrombospondin, secreted from astrocytes, have been suggested to directly bind to Neurexins and their postsynaptic ligands 150,151 , thereby reconfiguring their availability for adhesive interactions in the synaptic cleft. Collectively, these recent findings indicate a physiologically relevant “re-purposing” of Neurexins and their ligands to organize cellular interactions beyond synaptic neuron-neuron contacts.
Glossary.
- Posttranslational modifications
Enzymatic chemical modifications of specific amino acids in a protein that occur in the cell after or during mRNA translation (e.g. through phosphorylation, glycosylation, acetylation etc.).
- Structural motifs
A structurally conserved building block or “supersecondary structure” which appears in a variety of protein molecules that may or may not be functionally related.
- Alternative splicing
Process in which exons of an mRNA are assembled in multiple different (alternative) ways to yield multiple different versions of a final mRNA molecule that may contain different RNA regulatory motifs or encode alternative protein forms.
- Isoforms
Variants of an mRNA transcript or protein generated from a single gene but differing in sequence (e.g. resulting from alternative promoters or from alternative splicing).
Acknowledgements
A.M.G. was financially supported by an advanced EMBO long-term fellowship. L.T. was supported by the Boehringer Ingelheim Fonds and the Doris Dietschy and Denise Dietschy- Frick-Stiftung. Work in the laboratory of P.S. is supported by the Swiss National Science Foundation, a European Research Council Advanced Grant (SPLICECODE), EU-AIMS and AIMS-2-TRIALS which are supported by the Innovative Medicines Initiatives from the European Commission. This Joint Undertaking receives support from the European Union's Horizon 2020 research and innovation programme and EFPIA and AUTISM SPEAKS, Autistica, SFARI.
References
- 1.Lu W, Bushong EA, Shih TP, Ellisman MH, Nicoll RA. The cell-autonomous role of excitatory synaptic transmission in the regulation of neuronal structure and function. Neuron. 2013;78:433–439. doi: 10.1016/j.neuron.2013.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sigler A, et al. Formation and Maintenance of Functional Spines in the Absence of Presynaptic Glutamate Release. Neuron. 2017;94:304–311.:e304. doi: 10.1016/j.neuron.2017.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sando R, et al. Assembly of Excitatory Synapses in the Absence of Glutamatergic Neurotransmission. Neuron. 2017;94:312–321.:e313. doi: 10.1016/j.neuron.2017.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hobert O. Terminal Selectors of Neuronal Identity. Curr Top Dev Biol. 2016;116:455–475. doi: 10.1016/bs.ctdb.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Hassan BA, Hiesinger PR. Beyond Molecular Codes: Simple Rules to Wire Complex Brains. Cell. 2015;163:285–291. doi: 10.1016/j.cell.2015.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sudhof TC. Synaptic Neurexin Complexes: A Molecular Code for the Logic of Neural Circuits. Cell. 2017;171:745–769. doi: 10.1016/j.cell.2017.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanes JR, Zipursky SL. Synaptic Specificity, Recognition Molecules, and Assembly of Neural Circuits. Cell. 2020;181:536–556. doi: 10.1016/j.cell.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Stoeckli ET. Understanding axon guidance: are we nearly there yet? Development (Cambridge, England) 2018;145 doi: 10.1242/dev.151415. [DOI] [PubMed] [Google Scholar]
- 9.Dean C, et al. Neurexin mediates the assembly of presynaptic terminals. Nat Neurosci. 2003;6:708–716. doi: 10.1038/nn1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 11.Dalva MB, et al. EphB receptors interact with NMDA receptors and regulate excitatory synapse formation. Cell. 2000;103:945–956. doi: 10.1016/s0092-8674(00)00197-5. [DOI] [PubMed] [Google Scholar]
- 12.Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chih B, Gollan L, Scheiffele P. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 2006;51:171–178. doi: 10.1016/j.neuron.2006.06.005. [ These two studies uncovered regulation of Neurexin synaptogenic activity by alternative splicing ] [DOI] [PubMed] [Google Scholar]
- 14.de Wit J, et al. LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron. 2009;64:799–806. doi: 10.1016/j.neuron.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko J, Fuccillo MV, Malenka RC, Sudhof TC. LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron. 2009;64:791–798. doi: 10.1016/j.neuron.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pettem KL, et al. The specific alpha-neurexin interactor calsyntenin-3 promotes excitatory and inhibitory synapse development. Neuron. 2013;80:113–128. doi: 10.1016/j.neuron.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siddiqui TJ, Pancaroglu R, Kang Y, Rooyakkers A, Craig AM. LRRTMs and neuroligins bind neurexins with a differential code to cooperate in glutamate synapse development. J Neurosci. 2010;30:7495–7506. doi: 10.1523/JNEUROSCI.0470-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Wit J, Ghosh A. Specification of synaptic connectivity by cell surface interactions. Nat Rev Neurosci. 2016;17:22–35. doi: 10.1038/nrn.2015.3. [DOI] [PubMed] [Google Scholar]
- 19.Missler M, et al. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423:939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- 20.Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurshan PT, et al. gamma-Neurexin and Frizzled Mediate Parallel Synapse Assembly Pathways Antagonized by Receptor Endocytosis. Neuron. 2018;100:150–166.:e154. doi: 10.1016/j.neuron.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maro GS, et al. MADD-4/Punctin and Neurexin Organize C. elegans GABAergic Postsynapses through Neuroligin. Neuron. 2015;86:1420–1432. doi: 10.1016/j.neuron.2015.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philbrook A, et al. Neurexin directs partner-specific synaptic connectivity in C. elegans. eLife. 2018;7 doi: 10.7554/eLife.35692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hart MP, Hobert O. Neurexin controls plasticity of a mature, sexually dimorphic neuron. Nature. 2018;553:165–170. doi: 10.1038/nature25192. [ This study uncovered a key role for Neurexin in structural plasticity in C.elegans .] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banovic D, et al. Drosophila neuroligin 1 promotes growth and postsynaptic differentiation at glutamatergic neuromuscular junctions. Neuron. 2010;66:724–738. doi: 10.1016/j.neuron.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 26.Chen YC, et al. Drosophila neuroligin 2 is required presynaptically and postsynaptically for proper synaptic differentiation and synaptic transmission. J Neurosci. 2012;32:16018–16030. doi: 10.1523/JNEUROSCI.1685-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Ashley J, Budnik V, Bhat MA. Crucial role of Drosophila neurexin in proper active zone apposition to postsynaptic densities, synaptic growth, and synaptic transmission. Neuron. 2007;55:741–755. doi: 10.1016/j.neuron.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen SX, Tari PK, She K, Haas K. Neurexin-neuroligin cell adhesion complexes contribute to synaptotropic dendritogenesis via growth stabilization mechanisms in vivo. Neuron. 2010;67:967–983. doi: 10.1016/j.neuron.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 29.Constance WD, et al. Neurexin and Neuroligin-based adhesion complexes drive axonal arborisation growth independent of synaptic activity. eLife. 2018;7 doi: 10.7554/eLife.31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burden SJ, Huijbers MG, Remedio L. Fundamental Molecules and Mechanisms for Forming and Maintaining Neuromuscular Synapses. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19020490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen LY, Jiang M, Zhang B, Gokce O, Sudhof TC. Conditional Deletion of All Neurexins Defines Diversity of Essential Synaptic Organizer Functions for Neurexins. Neuron. 2017;94:611–625.:e614. doi: 10.1016/j.neuron.2017.04.011. [ This study highlighted cell type-specific alterations in synapse formation and synaptic function resulting from genetic Neurexin deletion .] [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Takahashi H, Craig AM. Protein tyrosine phosphatases PTPdelta, PTPsigma, and LAR: presynaptic hubs for synapse organization. Trends in neurosciences. 2013;36:522–534. doi: 10.1016/j.tins.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, et al. Splicing-Dependent Trans-synaptic SALM3-LAR-RPTP Interactions Regulate Excitatory Synapse Development and Locomotion. Cell reports. 2015;12:1618–1630. doi: 10.1016/j.celrep.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haklai-Topper L, et al. The neurexin superfamily of Caenorhabditis elegans. Gene Expr Patterns. 2011;11:144–150. doi: 10.1016/j.gep.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knight D, Xie W, Boulianne GL. Neurexins and neuroligins: recent insights from invertebrates. Mol Neurobiol. 2011;44:426–440. doi: 10.1007/s12035-011-8213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan Q, et al. Systematic discovery of regulated and conserved alternative exons in the mammalian brain reveals NMD modulating chromatin regulators. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:1502849112. doi: 10.1073/pnas.1502849112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen TM, et al. An alternative splicing switch shapes neurexin repertoires in principal neurons versus interneurons in the mouse hippocampus. eLife. 2016;5 doi: 10.7554/eLife.22757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ullrich B, Ushkaryov YA, Sudhof TC. Cartography of neurexins: more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 1995;14:497–507. doi: 10.1016/0896-6273(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 39.Furlanis E, Traunmuller L, Fucile G, Scheiffele P. Landscape of ribosome-engaged transcript isoforms reveals extensive neuronal-cell-class-specific alternative splicing programs. Nature neuroscience. 2019;22:1709–1717. doi: 10.1038/s41593-019-0465-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schreiner D, et al. Targeted Combinatorial Alternative Splicing Generates Brain Region-Specific Repertoires of Neurexins. Neuron. 2014 doi: 10.1016/j.neuron.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 41.Treutlein B, Gokce O, Quake SR, Südhof TC. Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E1291–1299. doi: 10.1073/pnas.1403244111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flaherty E, et al. Neuronal impact of patient-specific aberrant NRXN1alpha splicing. Nat Genet. 2019;51:1679–1690. doi: 10.1038/s41588-019-0539-z. [ These three studies reported numbers and cell type-selectivity of Neurexin splice isoforms in rodent and human neurons by long-read mRNA sequencing .] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding X, et al. Translational Inhibition of alpha-Neurexin 2. Sci Rep. 2020;10:3403. doi: 10.1038/s41598-020-60289-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schreiner D, Simicevic J, Ahrné E, Schmidt A, Scheiffele P. Quantitative isoform-profiling of highly diversified recognition molecules. eLife. 2015;4:1–17. doi: 10.7554/eLife.07794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ushkaryov YA, Petrenko AG, Geppert M, Sudhof TC. Neurexins: synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science (New York, N Y. 1992;257:50–56. doi: 10.1126/science.1621094. [DOI] [PubMed] [Google Scholar]
- 46.Comoletti D, et al. The macromolecular architecture of extracellular domain of alphaNRXN1: domain organization, flexibility, and insights into trans-synaptic disposition. Structure. 2010;18:1044–1053. doi: 10.1016/j.str.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biederer T, Sudhof TC. CASK and protein 4.1 support F-actin nucleation on neurexins. J Biol Chem. 2001;276:47869–47876. doi: 10.1074/jbc.M105287200. [DOI] [PubMed] [Google Scholar]
- 48.Miller MT, et al. The crystal structure of the alpha-neurexin-1 extracellular region reveals a hinge point for mediating synaptic adhesion and function. Structure. 2011;19:767–778. doi: 10.1016/j.str.2011.03.011. [ This study and ref. 64 uncovered the structural organization of the Neurexin alpha extracellular domain .] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheckler LR, Henry L, Sugita S, Sudhof TC, Rudenko G. Crystal structure of the second LNS/LG domain from neurexin 1alpha: Ca2+ binding and the effects of alternative splicing. J Biol Chem. 2006;281:22896–22905. doi: 10.1074/jbc.M603464200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson SC, et al. Structures of neurexophilin-neurexin complexes reveal a regulatory mechanism of alternative splicing. EMBO J. 2019;38:e101603. doi: 10.15252/embj.2019101603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rudenko G, Hohenester E, Muller YA. LG/LNS domains: multiple functions – one business end? Trends in biochemical sciences. 2001;26:363–368. doi: 10.1016/s0968-0004(01)01832-1. [DOI] [PubMed] [Google Scholar]
- 52.Arac D, et al. Structures of neuroligin-1 and the neuroligin-1/neurexin-1 beta complex reveal specific protein-protein and protein-Ca2+ interactions. Neuron. 2007;56:992–1003. doi: 10.1016/j.neuron.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 53.Chen X, Liu H, Shim AH, Focia PJ, He X. Structural basis for synaptic adhesion mediated by neuroligin-neurexin interactions. Nat Struct Mol Biol. 2008;15:50–56. doi: 10.1038/nsmb1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fabrichny IP, et al. Structural analysis of the synaptic protein neuroligin and its beta-neurexin complex: determinants for folding and cell adhesion. Neuron. 2007;56:979–991. doi: 10.1016/j.neuron.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koehnke J, et al. Crystal structure of the extracellular cholinesterase-like domain from neuroligin-2. Proc Natl Acad Sci U S A. 2008;105:1873–1878. doi: 10.1073/pnas.0711701105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koehnke J, et al. Crystal Structures of beta-Neurexin 1 and beta-Neurexin 2 Ectodomains and Dynamics of Splice Insertion Sequence 4. Structure. 2008;16:410–421. doi: 10.1016/j.str.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boucard AA, Chubykin AA, Comoletti D, Taylor P, Sudhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 2005;48:229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 58.Koehnke J, et al. Splice form dependence of beta-neurexin/neuroligin binding interactions. Neuron. 2010;67:61–74. doi: 10.1016/j.neuron.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uemura T, et al. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–1079. doi: 10.1016/j.cell.2010.04.035. [ This study discovered a tripartite synaptic complex consisting of Neurexins, Cerebellins and the postsynaptic receptor GLUD2 .] [DOI] [PubMed] [Google Scholar]
- 60.Zhang P, et al. Heparan Sulfate Organizes Neuronal Synapses through Neurexin Partnerships. Cell. 2018;174:1450–1464.:e1423. doi: 10.1016/j.cell.2018.07.002. [ This work revealed a major contribution of Neurexin carbohydrate modifications to the assembly of synaptic protein complexes .] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roppongi RT, et al. LRRTMs Organize Synapses through Differential Engagement of Neurexin and PTPsigma. Neuron. 2020;106:108–125.:e112. doi: 10.1016/j.neuron.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 62.Bulow HE, Hobert O. The molecular diversity of glycosaminoglycans shapes animal development. Annu Rev Cell Dev Biol. 2006;22:375–407. doi: 10.1146/annurev.cellbio.22.010605.093433. [DOI] [PubMed] [Google Scholar]
- 63.Sugita S, et al. A stoichiometric complex of neurexins and dystroglycan in brain. The Journal of cell biology. 2001;154:435–445. doi: 10.1083/jcb.200105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen F, Venugopal V, Murray B, Rudenko G. The structure of neurexin 1alpha reveals features promoting a role as synaptic organizer. Structure. 2011;19:779–789. doi: 10.1016/j.str.2011.03.012. [ This study and ref. 48 uncovered the structural organization of the Neurexin alpha extracellular domain .] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tanaka H, et al. Higher-order architecture of cell adhesion mediated by polymorphic synaptic adhesion molecules neurexin and neuroligin. Cell reports. 2012;2:101–110. doi: 10.1016/j.celrep.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 66.Fuccillo MV, et al. Single-Cell mRNA Profiling Reveals Cell-Type-Specific Expression of Neurexin Isoforms. Neuron. 2015;87:326–340. doi: 10.1016/j.neuron.2015.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lukacsovich D, et al. Single-Cell RNA-Seq Reveals Developmental Origins and Ontogenetic Stability of Neurexin Alternative Splicing Profiles. Cell reports. 2019;27:3752–3759.:e3754. doi: 10.1016/j.celrep.2019.05.090. [DOI] [PubMed] [Google Scholar]
- 68.Winterer J, et al. Single-cell RNA-Seq characterization of anatomically identified OLM interneurons in different transgenic mouse lines. Eur J Neurosci. 2019;50:3750–3771. doi: 10.1111/ejn.14549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neugebauer KM. Nascent RNA and the Coordination of Splicing with Transcription. Cold Spring Harbor perspectives in biology. 2019;11 doi: 10.1101/cshperspect.a032227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Raj B, Blencowe BJ. Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron. 2015;87:14–27. doi: 10.1016/j.neuron.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 72.Darnell RB. RNA protein interaction in neurons. Annu Rev Neurosci. 2013;36:243–270. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Resnick M, Segall A, G GR, Lupowitz Z, Zisapel N. Alternative splicing of neurexins: a role for neuronal polypyrimidine tract binding protein. Neuroscience letters. 2008;439:235–240. doi: 10.1016/j.neulet.2008.05.034. [DOI] [PubMed] [Google Scholar]
- 74.Gehman LT, et al. The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes Dev. 2012;26:445–460. doi: 10.1101/gad.182477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saito Y, et al. Differential NOVA2-Mediated Splicing in Excitatory and Inhibitory Neurons Regulates Cortical Development and Cerebellar Function. Neuron. 2019;101:707–720.:e705. doi: 10.1016/j.neuron.2018.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wamsley B, et al. Rbfox1 Mediates Cell-type-Specific Splicing in Cortical Interneurons. Neuron. 2018 doi: 10.1016/j.neuron.2018.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Di Fruscio M, Chen T, Richard S. Characterization of Sam68-like mammalian proteins SLM-1 and SLM-2: SLM-1 is a Src substrate during mitosis. Proc Natl Acad Sci U S A. 1999;96:2710–2715. doi: 10.1073/pnas.96.6.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Feracci M, et al. Structural basis of RNA recognition and dimerization by the STAR proteins T-STAR and Sam68. Nat Commun. 2016;7:10355. doi: 10.1038/ncomms10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Danilenko M, et al. Binding site density enables paralog-specific activity of SLM2 and Sam68 proteins in Neurexin2 AS4 splicing control. Nucleic acids research. 2017;45:4120–4130. doi: 10.1093/nar/gkw1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Iijima T, Iijima Y, Witte H, Scheiffele P. Neuronal cell type-specific alternative splicing is regulated by the KH domain protein SLM1. The Journal of cell biology. 2014;204:331–342. doi: 10.1083/jcb.201310136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Iijima T, et al. SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell. 2011;147:1601–1614. doi: 10.1016/j.cell.2011.11.028. [ This study discovered the regulation of neuronal activity-dependent alternative splicing of Neurexins by the STAR-family RNA binding protein SAM68 .] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Elegheert J, et al. Structural basis for integration of GluD receptors within synaptic organizer complexes. Science (New York, N Y. 2016;353:295–299. doi: 10.1126/science.aae0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matsuda K, Yuzaki M. Cbln family proteins promote synapse formation by regulating distinct neurexin signaling pathways in various brain regions. Eur J Neurosci. 2011;33:1447–1461. doi: 10.1111/j.1460-9568.2011.07638.x. [DOI] [PubMed] [Google Scholar]
- 84.Ito-Ishida A, et al. Cbln1 regulates rapid formation and maintenance of excitatory synapses in mature cerebellar Purkinje cells in vitro and in vivo. J Neurosci. 2008;28:5920–5930. doi: 10.1523/JNEUROSCI.1030-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Matsuda K, et al. Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science. 2010;328:363–368. doi: 10.1126/science.1185152. [ This studies and ref. 141 demonstrate critical in vivo functions of the Neurexin – Cerebellin – GLUD2 complex in cerebellar synapse formation and maintenance .] [DOI] [PubMed] [Google Scholar]
- 86.Stoss O, et al. p59(fyn)-mediated phosphorylation regulates the activity of the tissuespecific splicing factor rSLM-1. Mol Cell Neurosci. 2004;27:8–21. doi: 10.1016/j.mcn.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 87.Traunmüller L, Bornmann C, Scheiffele P. Alternative splicing coupled nonsense-mediated decay generates neuronal cell type-specific expression of SLM proteins. J Neurosci. 2014;34:16755–16761. doi: 10.1523/JNEUROSCI.3395-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ehrmann I, et al. The tissue-specific RNA binding protein T-STAR controls regional splicing patterns of neurexin pre-mRNAs in the brain. PLoS Genet. 2013;9:e1003474. doi: 10.1371/journal.pgen.1003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Traunmüller L, Gomez AM, Nguyen T-M, Scheiffele P. Control of neuronal synapse specification by highly dedicated alternative splicing program. Science. 2016;352:982–986. doi: 10.1126/science.aaf2397. [DOI] [PubMed] [Google Scholar]
- 90.Aoto J, Martinelli DC, Malenka RC, Tabuchi K, Südhof TC. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell. 2013;154:75–88. doi: 10.1016/j.cell.2013.05.060. [ These two studies demonstrated a major impact of Neurexin alternative splicing on glutamatergic synapse function .] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chubykin AA, et al. Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron. 2007;54:919–931. doi: 10.1016/j.neuron.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shipman SL, Nicoll RA. A subtype-specific function for the extracellular domain of neuroligin 1 in hippocampal LTP. Neuron. 2012;76:309–316. doi: 10.1016/j.neuron.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Soler-Llavina GJ, et al. Leucine-rich repeat transmembrane proteins are essential for maintenance of long-term potentiation. Neuron. 2013;79:439–446. doi: 10.1016/j.neuron.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dai J, Aoto J, Sudhof TC. Alternative Splicing of Presynaptic Neurexins Differentially Controls Postsynaptic NMDA and AMPA Receptor Responses. Neuron. 2019;102:993–1008.:e1005. doi: 10.1016/j.neuron.2019.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Matsuda K, et al. Transsynaptic Modulation of Kainate Receptor Functions by C1q-like Proteins. Neuron. 2016;90:752–767. doi: 10.1016/j.neuron.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 96.Quesnel-Vallieres M, et al. Misregulation of an Activity-Dependent Splicing Network as a Common Mechanism Underlying Autism Spectrum Disorders. Mol Cell. 2016;64:1023–1034. doi: 10.1016/j.molcel.2016.11.033. [DOI] [PubMed] [Google Scholar]
- 97.Xie J, Black DL. A CaMK IV responsive RNA element mediates depolarization-induced alternative splicing of ion channels. Nature. 2001;410:936–939. doi: 10.1038/35073593. [DOI] [PubMed] [Google Scholar]
- 98.Rozic-Kotliroff G, Zisapel N. Ca2+ -dependent splicing of neurexin IIalpha. Biochem Biophys Res Commun. 2007;352:226–230. doi: 10.1016/j.bbrc.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 99.Ding X, et al. Activity-induced histone modifications govern Neurexin-1 mRNA splicing and memory preservation. Nature neuroscience. 2017;20:690–699. doi: 10.1038/nn.4536. [DOI] [PubMed] [Google Scholar]
- 100.Saura CA, Servian-Morilla E, Scholl FG. Presenilin/gamma-secretase regulates neurexin processing at synapses. PLoS One. 2011;6:e19430. doi: 10.1371/journal.pone.0019430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Servian-Morilla E, et al. Proteolytic Processing of Neurexins by Presenilins Sustains Synaptic Vesicle Release. J Neurosci. 2018;38:901–917. doi: 10.1523/JNEUROSCI.1357-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lim L, et al. Optimization of interneuron function by direct coupling of cell migration and axonal targeting. Nature neuroscience. 2018;21:920–931. doi: 10.1038/s41593-018-0162-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kornblihtt AR. Transcriptional control of alternative splicing along time: ideas change, experiments remain. RNA. 2015;21:670–672. doi: 10.1261/rna.051151.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Furlanis E, Scheiffele P. Regulation of neuronal differentiation, function, and plasticity by alternative splicing. Annual Review of Cell and Developmental Biology. 2018;34:451–469. doi: 10.1146/annurev-cellbio-100617-062826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tanabe Y, et al. IgSF21 promotes differentiation of inhibitory synapses via binding to neurexin2alpha. Nat Commun. 2017;8:408. doi: 10.1038/s41467-017-00333-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hu Z, Xiao X, Zhang Z, Li M. Genetic insights and neurobiological implications from NRXN1 in neuropsychiatric disorders. Molecular psychiatry. 2019;24:1400–1414. doi: 10.1038/s41380-019-0438-9. [DOI] [PubMed] [Google Scholar]
- 107.Castronovo P, et al. Phenotypic spectrum of NRXN1 mono- and bi-allelic deficiency: A systematic review. Clinical genetics. 2020;97:125–137. doi: 10.1111/cge.13537. [DOI] [PubMed] [Google Scholar]
- 108.Cosemans N, et al. The clinical relevance of intragenic NRXN1 deletions. Journal of medical genetics. 2020;57:347–355. doi: 10.1136/jmedgenet-2019-106448. [DOI] [PubMed] [Google Scholar]
- 109.Rujescu D, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vaags AK, et al. Rare deletions at the neurexin 3 locus in autism spectrum disorder. Am J Hum Genet. 2012;90:133–141. doi: 10.1016/j.ajhg.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang AY, et al. Rare Copy Number Variants in NRXN1 and CNTN6 Increase Risk for Tourette Syndrome. Neuron. 2017;94:1101–1111.:e1107. doi: 10.1016/j.neuron.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nussbaum J, et al. Significant association of the neurexin-1 gene (NRXN1) with nicotine dependence in European- and African-American smokers. Hum Mol Genet. 2008;17:1569–1577. doi: 10.1093/hmg/ddn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rochtus AM, et al. Mutations in NRXN1 and NRXN2 in a patient with early-onset epileptic encephalopathy and respiratory depression. Cold Spring Harb Mol Case Stud. 2019;5 doi: 10.1101/mcs.a003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bena F, et al. Molecular and clinical characterization of 25 individuals with exonic deletions of NRXN1 and comprehensive review of the literature. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:388–403. doi: 10.1002/ajmg.b.32148. [DOI] [PubMed] [Google Scholar]
- 116.Vinas-Jornet M, et al. A common cognitive, psychiatric, and dysmorphic phenotype in carriers of NRXN1 deletion. Mol Genet Genomic Med. 2014;2:512–521. doi: 10.1002/mgg3.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Harrison V, et al. Compound heterozygous deletion of NRXN1 causing severe developmental delay with early onset epilepsy in two sisters. Am J Med Genet A. 2011;155A:2826–2831. doi: 10.1002/ajmg.a.34255. [DOI] [PubMed] [Google Scholar]
- 118.Zweier C, et al. CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am J Hum Genet. 2009;85:655–666. doi: 10.1016/j.ajhg.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gillberg C. The ESSENCE in child psychiatry: Early Symptomatic Syndromes Eliciting Neurodevelopmental Clinical Examinations. Res Dev Disabil. 2010;31:1543–1551. doi: 10.1016/j.ridd.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 120.Tong XJ, Hu Z, Liu Y, Anderson D, Kaplan JM. A network of autism linked genes stabilizes two pools of synaptic GABA(A) receptors. eLife. 2015;4:e09648. doi: 10.7554/eLife.09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci U S A. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Laarakker MC, Reinders NR, Bruining H, Ophoff RA, Kas MJ. Sexdependent novelty response in neurexin-1alpha mutant mice. PLoS One. 2012;7:e31503. doi: 10.1371/journal.pone.0031503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Esclassan F, Francois J, Phillips KG, Loomis S, Gilmour G. Phenotypic characterization of nonsocial behavioral impairment in neurexin 1alpha knockout rats. Behav Neurosci. 2015;129:74–85. doi: 10.1037/bne0000024. [DOI] [PubMed] [Google Scholar]
- 124.Dachtler J, et al. Deletion of alpha-neurexin II results in autism-related behaviors in mice. Transl Psychiatry. 2014;4:e484. doi: 10.1038/tp.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dachtler J, et al. Heterozygous deletion of alpha-neurexin I or alpha-neurexin II results in behaviors relevant to autism and schizophrenia. Behav Neurosci. 2015;129:765–776. doi: 10.1037/bne0000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Born G, et al. Genetic targeting of NRXN2 in mice unveils role in excitatory cortical synapse function and social behaviors. Front Synaptic Neurosci. 2015;7:3. doi: 10.3389/fnsyn.2015.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rabaneda LG, Robles-Lanuza E, Nieto-Gonzalez JL, Scholl FG. Neurexin dysfunction in adult neurons results in autistic-like behavior in mice. Cell reports. 2014;8:338–346. doi: 10.1016/j.celrep.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 128.Greenberg DM, Warrier V, Allison C, Baron-Cohen S. Testing the Empathizing-Systemizing theory of sex differences and the Extreme Male Brain theory of autism in half a million people. Proc Natl Acad Sci U S A. 2018;115:12152–12157. doi: 10.1073/pnas.1811032115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Keum S, et al. A Missense Variant at the Nrxn3 Locus Enhances Empathy Fear in the Mouse. Neuron. 2018;98:588–601.:e585. doi: 10.1016/j.neuron.2018.03.041. [ This study reports the critical contribution of a Nrxn3 sequence variant in mice to cortical circuit function and emotional behavior .] [DOI] [PubMed] [Google Scholar]
- 130.Panksepp J, Panksepp JB. Toward a cross-species understanding of empathy. Trends in neurosciences. 2013;36:489–496. doi: 10.1016/j.tins.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cao Y, et al. Mechanism for Selective Synaptic Wiring of Rod Photoreceptors into the Retinal Circuitry and Its Role in Vision. Neuron. 2015;87:1248–1260. doi: 10.1016/j.neuron.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sylwestrak EL, Ghosh A. Elfn1 regulates target-specific release probability at CA1-interneuron synapses. Science (New York, NY. 2012;338:536–540. doi: 10.1126/science.1222482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tomioka NH, et al. Elfn1 recruits presynaptic mGluR7 in trans and its loss results in seizures. Nat Commun. 2014;5:4501. doi: 10.1038/ncomms5501. [DOI] [PubMed] [Google Scholar]
- 134.Park H, et al. Splice-dependent trans-synaptic PTPdelta-IL1RAPL1 interaction regulates synapse formation and non-REM sleep. EMBO J. 2020;39:e104150. doi: 10.15252/embj.2019104150. [ This work uncovers a key role for an alternative exon in the regulation of transsynaptic interactions, synaptic transmission and mouse behavior .] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kim K, et al. Presynaptic PTPsigma regulates postsynaptic NMDA receptor function through direct adhesion-independent mechanisms. eLife. 2020;9 doi: 10.7554/eLife.54224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shen K, Scheiffele P. Genetics and cell biology of building specific synaptic connectivity. Annu Rev Neurosci. 2010;33:473–507. doi: 10.1146/annurev.neuro.051508.135302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kim B, Emmons SW. Multiple conserved cell adhesion protein interactions mediate neural wiring of a sensory circuit in C. elegans. eLife. 2017;6 doi: 10.7554/eLife.29257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schroeder A, et al. A Modular Organization of LRR Protein-Mediated Synaptic Adhesion Defines Synapse Identity. Neuron. 2018;99:329–344.:e327. doi: 10.1016/j.neuron.2018.06.026. [DOI] [PubMed] [Google Scholar]
- 139.Yamagata A, et al. Structural insights into modulation and selectivity of transsynaptic neurexin-LRRTM interaction. Nat Commun. 2018;9:3964. doi: 10.1038/s41467-018-06333-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ibata K, et al. Activity-Dependent Secretion of Synaptic Organizer Cbln1 from Lysosomes in Granule Cell Axons. Neuron. 2019;102:1184–1198.:e1110. doi: 10.1016/j.neuron.2019.03.044. [DOI] [PubMed] [Google Scholar]
- 141.Ito-Ishida A, et al. Presynaptically released Cbln1 induces dynamic axonal structural changes by interacting with GluD2 during cerebellar synapse formation. Developmental Cell. 2012 doi: 10.1016/j.neuron.2012.07.027. [ This studies and ref. 85 demonstrate critical in vivo functions of the Neurexin – Cerebellin – GLUD2 complex in cerebellar synapse formation and maintenance .] [DOI] [PubMed] [Google Scholar]
- 142.Brockhaus J, et al. alpha-Neurexins Together with alpha2delta-1 Auxiliary Subunits Regulate Ca(2+) Influx through Cav2.1 Channels. J Neurosci. 2018;38:8277–8294. doi: 10.1523/JNEUROSCI.0511-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tong XJ, et al. Retrograde Synaptic Inhibition Is Mediated by alpha-Neurexin Binding to the alpha2delta Subunits of N-Type Calcium Channels. Neuron. 2017;95:326–340.:e325. doi: 10.1016/j.neuron.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hrvatin S, et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nature neuroscience. 2018;21:120–129. doi: 10.1038/s41593-017-0029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Uchigashima M, Cheung A, Suh J, Watanabe M, Futai K. Differential expression of neurexin genes in the mouse brain. The Journal of comparative neurology. 2019;527:1940–1965. doi: 10.1002/cne.24664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Venkatesh HS, et al. Targeting neuronal activity-regulated neuroligin-3 dependency in high-grade glioma. Nature. 2017;549:533–537. doi: 10.1038/nature24014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Venkataramani V, et al. Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature. 2019;573:532–538. doi: 10.1038/s41586-019-1564-x. [DOI] [PubMed] [Google Scholar]
- 148.Zeng Q, et al. Synaptic proximity enables NMDAR signalling to promote brain metastasis. Nature. 2019;573:526–531. doi: 10.1038/s41586-019-1576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Stogsdill JA, et al. Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature. 2017;551:192–197. doi: 10.1038/nature24638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Singh SK, et al. Astrocytes Assemble Thalamocortical Synapses by Bridging NRX1alpha and NL1 via Hevin. Cell. 2016;164:183–196. doi: 10.1016/j.cell.2015.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xu J, Xiao N, Xia J. Thrombospondin 1 accelerates synaptogenesis in hippocampal neurons through neuroligin 1. Nature neuroscience. 2009 doi: 10.1038/nn.2459. [DOI] [PubMed] [Google Scholar]