Abstract
Viruses are ubiquitous pathogens of global impact. Prompted by the hypothesis that their earliest progenitors recruited host proteins for virion formation, we have used stringent laboratory evolution to convert a bacterial enzyme lacking affinity for nucleic acids into an artificial nucleocapsid that efficiently packages and protects multiple copies of its own encoding mRNA. Revealing remarkable convergence on the molecular hallmarks of natural viruses, the accompanying changes refashioned the protein building blocks into a 240-subunit icosahedral capsid impermeable to nucleases, while emergence of a robust RNA stem-loop packaging cassette ensured high encapsidation yields and specificity. In addition to evincing a plausible evolutionary pathway for primordial viruses, these findings highlight practical strategies for developing non-viral carriers for diverse vaccine and delivery applications.
Understanding the origins and evolutionary trajectories of viruses is a fundamental scientific challenge ( 1 ). Even the simplest virions, optimized for genome propagation over billions of years of evolution, require co-assembly of many copies of a single protein with an RNA or DNA molecule to afford a closed-shell container of defined size, shape, and symmetry. Strategies for excluding competing host nucleic acids and protecting the viral genome from nucleases are also needed. While recreating such properties in non-viral containers is challenging ( 2 – 6 ), capsids generated by bottom-up design are promising as customizable tools for delivery and display ( 7 – 9 ).
Previous efforts to produce artificial nucleocapsids that encapsulate their own genetic information have utilized natural and computationally designed protein cages possessing engineered cationic interiors ( 5 , 6 ). However, even after directed evolution only ~10% of the resulting particles contained the full-length target RNA, underscoring the difficulties associated with packaging and protecting nucleic acids in a cell. In addition to competition from abundant host nucleic acids, genome degradation by cellular RNases is problematic owing to slow assembly, cage dynamics and/or porosity. Here we show that complementary adaptations of cargo and container can be harnessed to address these challenges and recapitulate the structural and packaging properties of natural viruses.
Our starting point was a previously evolved nucleocapsid, derived from Aquifex aeolicus lumazine synthase (AaLS), a bacterial enzyme that naturally forms 60-subunit nanocontainers but has no inherent affinity for nucleic acids ( 10 ). AaLS was redesigned by circular permutation and appending the arginine-rich peptide λN+, which tightly binds an RNA stem-loop called BoxB ( 11 , 12 ) (Fig. S1A). The resulting nucleocapsid variant, NC-1, was subsequently evolved via intermediate NC-2 to NC-3 by selecting for variants that capture capsid-encoding mRNA transcripts flanked by BoxB tags. Nevertheless, only one in eight of the NC-3 capsids packaged the full-length RNA genome ( 6 ).
To improve NC-3’s packaging properties, we mutagenized its gene by error-prone PCR and subjected the library to three cycles of expression, purification, and nuclease challenge, followed by re-amplification of the surviving mRNA. Selection stringency was steadily increased in each cycle by decreasing nuclease size (60 kDa benzonase ➞ 14 kDa RNase A ➞ 11 kDa RNase T1) and extending nuclease exposure from 1 to 4 hours. This strategy ensured 1) efficient assembly of RNA-containing capsids, 2) protection of the cargo from nucleases, and 3) enrichment of variants that package the full-length mRNA (Fig. 1A). The best variant, NC-4, had nine new mutations, three of which were silent (Fig. S2).
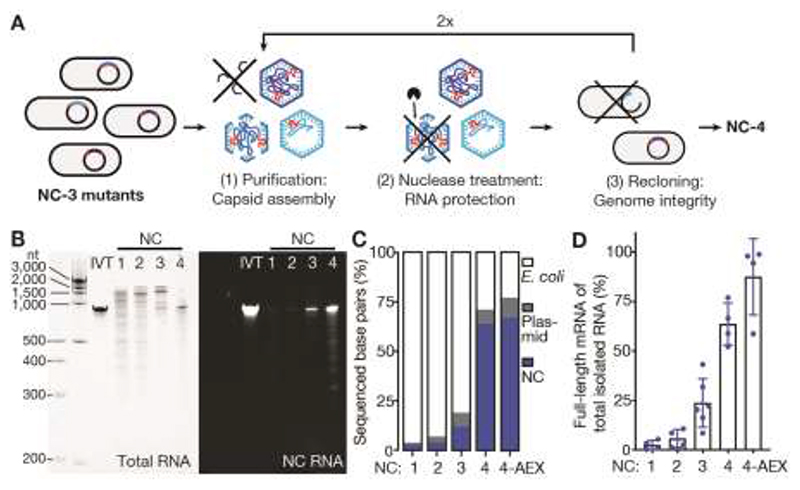
Fig. 1. NC-4 packages its genome with high selectivity.
(A) Laboratory evolution: a library of NC-3 mutants generated by error-prone PCR was expressed in Escherichia coli and purified by affinity and size exclusion chromatography. This step recovers assembled capsids. The purified capsid library was then treated with nucleases to enrich for capsids that protect their RNA cargo. Finally, the RNA was extracted from capsids, reverse-transcribed, and re-cloned into the original expression vector. This step selects for capsids that contain full-length genomes. (B) Denaturing PAGE (5%) of NC-1 to NC-4 stained for total RNA with GelRed (left) and the fluorogenic dye DFHBI-1T (right), which selectively binds the broccoli aptamer present in the 5’ - and 3’-untranslated regions of the mRNA genome (NC RNA). IVT, in vitro-transcribed reference mRNA. (C) RNA identities and their relative abundance were determined by Oxford Nanopore Sequencing ( 32 ) for all four capsids, including anion-exchanged NC-4 (4-AEX), and assigned to three main categories: bacterial RNA (E. coli), nucleocapsid mRNA (NC), and RNA originating from other plasmid-associated genes (plasmid). The encapsulated E. coli genes are primarily rRNA (Fig. S3). (D) The fraction of total extracted RNA corresponding to the full-length mRNA genome was determined by real-time quantitative PCR (mean of at least two biological replicates, each measured in two separate laboratories, error bars represent the standard deviation of the mean).
After optimizing protein production and purification, we compared NC-4 to its precursors. Particle heterogeneity decreased notably over the course of evolution from NC-1 so that NC-4 assembles into homogeneous capsids (Fig. S1B,C), with protein yields after purification (~35 mg/L medium) that increased by an order of magnitude in the last evolutionary step. Additionally, nuclease resistance steadily improved. NC-1 RNA is almost completely degraded upon treatment with either benzonase or RNase A, whereas NC-2 protects small amounts of full-length mRNA from benzonase but not RNase A (Fig. S1D). In contrast, both NC-3 and NC-4 protect most of their encapsidated RNA from both nucleases (Fig. S1D,E). Importantly, NC-4 also packages its own full-length mRNA with improved specificity. While earlier generations encapsidate a broad size range of RNA species (400–2000 nt), NC-4 binds one major species corresponding to the 863 nt-long capsid mRNA (Fig. 1B, left). Long-read direct cDNA sequencing confirmed the decrease in encapsidated host RNA (Fig. 1C), which was largely ribosomal (Fig. S3). The simultaneous increase in genome packaging efficiency over the four generations is clearly evident in gels stained with the fluorogenic dye DFHBI-1T, which binds the Broccoli aptamers ( 13 ) introduced with the BoxB tags (Fig. 1B, right) ( 6 ).
The fraction of full-length genome relative to total encapsidated RNA was quantified by real-time PCR to be (2±2)% for NC-1, (6±5)% for NC-2, (24±12)% for NC-3 and (64±11)% for NC-4 (Fig. 1D). When NC-4 was further purified by ion-exchange chromatography to remove incomplete or poorly-assembled capsids, (87±19)% of the RNA corresponded to the full-length genome. Given the total number of encapsidated nucleotides (~2500), NC-4 packages on average 2.5 full-length mRNAs per capsid, a dramatic improvement compared to its precursors and other artificial nucleocapsids ( 5 , 6 ). This packaging capacity suggests that the evolved capsid could readily accommodate substantially longer RNAs, such as more complex genomes or large RNA molecules of medical interest.
Improved genome packaging and protection were accompanied by major structural transformations. The cavity of the starting 16 nm diameter AaLS scaffold is too small to package an 863 nt-long RNA ( 2 , 6 ). However, addition of the λN+ peptide to circularly permuted AaLS afforded expanded capsids with diameters in the 20–30 nm range, which were subsequently evolved toward uniform ~30 nm diameter particles (Fig. S1C). To elucidate the nature of these changes, we turned to cryo-EM.
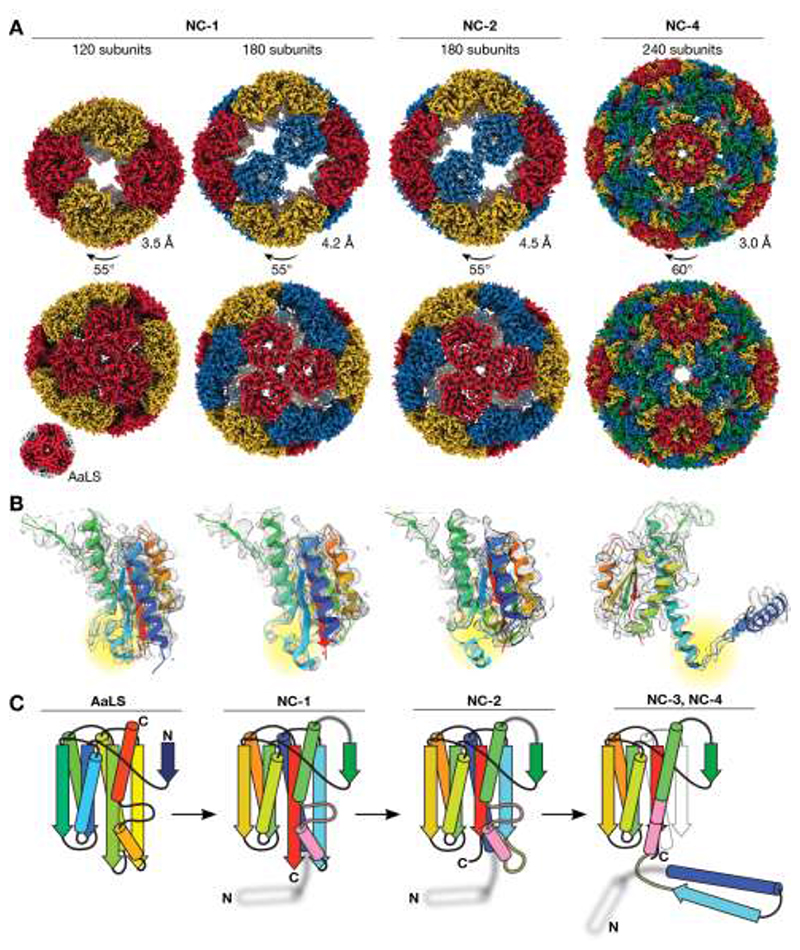
Characterization of the initial NC-1 design revealed a range of assemblies of varying size and shape (Fig. S4A,B). Although particle heterogeneity and aggregation complicated single-particle reconstruction, two expanded structures with tetrahedral symmetry were successfully obtained (Fig. 2A). Like the wild-type protein, both are composed entirely of canonical lumazine synthase pentamers (Fig. 3A), but they possess large, keyhole-shaped pores (~4 nm wide) through which nucleases could diffuse. One capsid is a 180-mer (Fig. S4C–F, Table S1) that closely resembles a previously characterized AaLS variant possessing a negatively charged lumen ( 14 , 15 ). The other NC-1 structure is an unprecedented 120-mer (Fig. S4G–I, Table S1). It features wild-type-like pentamer-pentamer interactions as well as inter-pentamer contacts characteristic of its 180-mer sibling (Fig. 2A). At the monomer level, the major deviation from the AaLS fold is seen in a short helix (residues 67–74) and adjacent loop (residues 75–81) (Fig. 2B,C). In AaLS, this region is involved in lumenal interactions between the pentameric building blocks at the threefold-symmetry axes. In NC-1 chains that are not involved in wild-type-like pentamer-pentamer contacts, this loop assumes altered conformations and is resolved to lower local resolution (Fig. 2B,C, Fig. S4E,H).
Fig. 2. Structural evolution towards virus-like nucleocapsids.
(A) Maps are shown for the tetrahedrally symmetric NC-1 and NC-2 structures with symmetry-related pentamers in the same color, and the icosahedral T=4 NC-4 capsid with the four quasi-equivalent chains highlighted in different colors. The lower resolution NC-3 capsid (7.0 Å, Fig. S6) resembles NC-4. Wild-type AaLS ( 10 ) is shown for comparison (not to scale). Resolutions were estimated by Fourier shell correlation (0.143 threshold). (B) Fits of single chains (rainbow; N-terminus to C-terminus from blue to red) in the electron density of the capsids above show the evolution of the monomer. Residues 66 to 81 are highlighted (yellow). Clear density is seen for this segment in NC-1 protomers involved in AaLS-like inter-pentamer contacts. In other chains, as in the 180-mer NC-1 structure, this region is less well resolved. In NC-2 the nearby beta-sheet is also perturbed, further enhancing the flexibility of this region. In NC-3 and NC-4, this segment rearranges into an extended helix that supports the domain swap. (C) Rainbow-colored models depict the changes in the protein fold. The helix (67–74) and loop (75–81) that undergo a major rearrangement are colored in pink, and the hinge loop (62–66) in yellow; the structurally unresolved RNA-binding peptide is depicted as a blurry white helix.
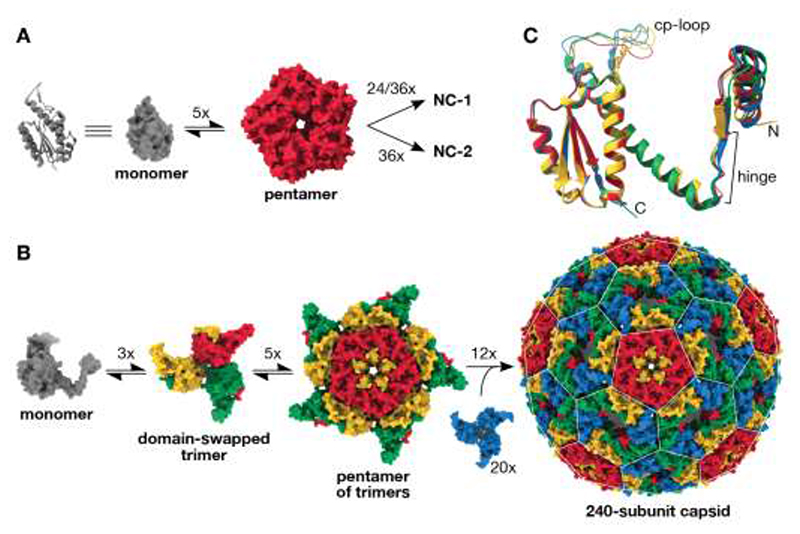
Fig. 3. Virus-like architecture by protomer reorganization.
(A) The assembly of 120- and 180-subunit NC-1 and NC-2 cages from monomers (cartoon and surface shown in grey) presumably proceeds via AaLS-like pentamers. (B) Based on the assembly mechanisms of other viral capsids( 33 ), the T=4 capsids likely arise from domain-swapped trimeric building blocks that further combine into pentamers. Combining the latter with additional domain-swapped trimers (blue) would afford the complete 240-subunit capsid. The pentagonal and hexagonal faces of the icosahedrally symmetric capsid are highlighted by a white lattice. (C) Assembly of the T=4 icosahedral NC-3 and NC-4 structures requires the subunits to adopt different, quasi-equivalent conformations. An overlay of the four quasi-equivalent chains of NC-4, colored as in panel b, shows that the hinge region provides flexibility for subtle adjustments in the relative orientation of the flanking segments. Additional differences are visible in the poorly resolved surface loop introduced by circular permutation (cp-loop), which interacts with the neighboring subunit in both pentamers and hexamers via a single short beta strand.
The second-generation variant NC-2, obtained after benzonase challenge, is also polymorphic and aggregation-prone. Several distinct morphologies were identified by 2D-classification (Fig. S5), one of which was reconstructed as a tetrahedrally symmetric 180-mer (4.5 Å, Fig. 2A, Table S1) that superimposes on the analogous NC-1 structure. Four mutations (I58V, G61D, V62I, and I191F) shorten two strands of the core beta-sheet and, indirectly, further increase disorder in neighboring residues 66–81 (Fig. 2B,C). These changes likely disfavor wild type-like pentamer-pentamer interactions, explaining the absence of smaller capsids with more tightly packed capsomers. Structural heterogeneity and particle aggregation precluded reconstruction of additional structures that may contribute to the benzonase-resistance phenotype.
The ability of NC-3 and NC-4 to protect their cargo from RNases significantly smaller than the pores in the parental structures suggests a novel solution to nuclease resistance. In fact, three-dimensional reconstructions of NC-3 (7.0 Å) and NC-4 (3.0 Å) (Fig. S6, Table S1) yielded superimposable structures that are markedly different from any previously characterized AaLS derivative (Fig. 2A). Both capsids form icosahedrally symmetric 240-mers that feature smaller pores (~2.5 nm) than their progenitors. The pentagonal vertices align with AaLS pentamers, and are surrounded by 30 hexagonal patches (Fig. 3B). This architecture is typical of T=4 virus capsids, in which a single protein chain assumes four similar, quasi-equivalent conformations, repeated with icosahedral symmetry to afford a closed container with increased volume ( 16 ).
The most striking feature of our evolved cages is a 3D-domain swap ( 17 ), which links neighboring monomers and reorganizes the structure into trimeric building blocks (Fig. 3B). As reported for some viral capsids ( 18 – 21 ), such interlacing may enhance particle stability. This rearrangement was made possible by a hinge around residues 62–66, which permits dissociation of the N-terminal helix and strand of each subunit from the core, allowing it to dock onto a neighboring subunit in the trimeric capsomer. An elongated alpha-helix extends C-terminally from this hinge, formed by fusing the short helix (residues 67–74) to the following helix by ordering of the intervening loop (residues 75–81) (Fig. 2C). Slight variations in the hinge angles allow the subunits to occupy four quasi-equivalent positions within the expanded icosahedral lattice (Fig. 3C) ( 22 ). Such flexible elements might similarly be exploited for the rational design of large (T>1) capsid assemblies from a single protein chain, an as yet unmet challenge due to the difficulty of designing proteins capable of adopting several distinct conformations.
The smaller pores in the NC-3 and NC-4 shells provide a compelling explanation for nuclease resistance. The structurally unresolved λN+ peptides, which line the lumenal edge of these openings, likely further restrict access to the cage interior. Nevertheless, the superimposable structures do not account for the differences in packaging efficiency between NC-3 and NC-4. Although a lysine to arginine mutation that appeared in the λN+ peptide of NC-4 is known to increase affinity to the BoxB tags ~3-fold ( 11 ), the effects of reverting this mutation are modest (Fig. S7), indicating that other factors must be at play.
Some viruses that package single-stranded RNA genomes utilize multiple stem-loop packaging signals to ensure cargo specificity and orchestrate capsid assembly within the crowded confines of the cell ( 23 ). Could the evolution of additional RNA packaging signals in the NC-4 genome explain its superiority to NC-3? Besides the originally introduced BoxB tags ( 6 ), BB1 and BB2, both genomes have 37 BoxB-like URxRxRR (R = purine) and URxR sequences ( 24 ) (Table S2). In order to determine whether any of these serve as packaging signals, we used synchrotron X-ray footprinting (XRF). Synchrotron radiation generates hydroxyl radicals, which cleave the RNA backbone. Because base-pairing and contact with protein decrease local cleavage propensity, XRF provides a means to map intermolecular interactions and RNA secondary structure ( 25 ).
Footprints for packaged NC-3 and NC-4 RNA show that only BB1, BB2, and 11 out of 37 BoxB-like motifs exhibit low XRF reactivity (Table S2). Furthermore, XRF-informed prediction of RNA secondary structure ensembles ( 26 ) indicates that only seven of these motifs (BB1, BB2, and potential packaging signals PS1–5) are presented as stem-loops with significant frequency (Fig. S8A, S9A). Assuming that interactions with the λN+ peptides stabilize the stem-loops, comparison of their display frequency in encapsulated versus free RNA pinpoints which of these motifs might serve as packaging signals.
In NC-3, the secondary structure predictions (Fig. 4A,C,E, Fig. S8) indicate that the original high-affinity BoxB tags are more frequently displayed as stem-loops in free transcripts than in capsids (96% vs. 63% for BB1 and 75% vs. 52% for BB2). Although the five lower affinity PS1–5 motifs are displayed more frequently upon encapsulation, their broad distribution, coupled with modest display of the high-affinity tags, contrasts with natural viruses, which appear to utilize narrow clusters of packaging signals surrounding an efficiently displayed, high-affinity stem-loop to initiate capsid assembly ( 23 ). The lack of robust assembly instructions may explain why 72% of the RNA packaged in NC-3 is ribosomal. Ribosomal RNA is compact, abundant and also possesses multiple BoxB-like signals (Fig. S3C,D), which may allow it to function as an alternative nucleation hub for capsid assembly.
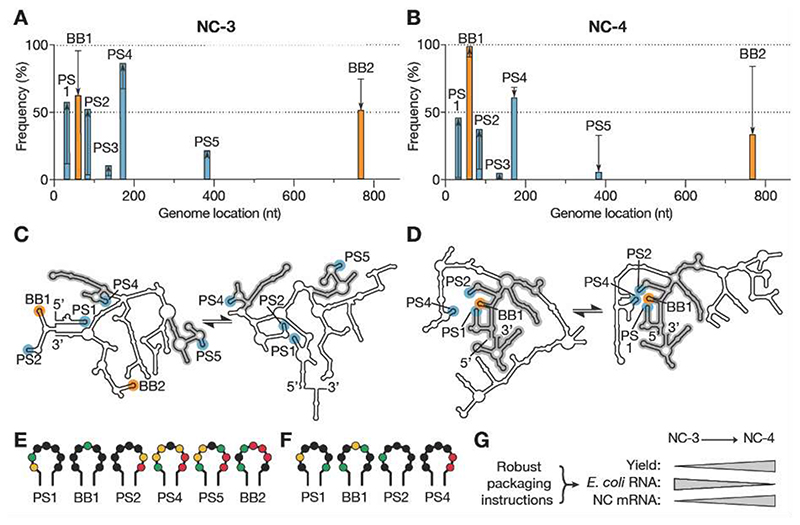
Fig. 4. Virus-like genome packaging mediated by packaging signals.
(A,B) XRF reactivities were used to calculate how frequently the seven packaging signal candidates occur in stem-loops in the NC-3 (A) and NC-4 (B) mRNA genomes; 1000 sample folds were generated for each of 1116 combinations of reactivity offsetting and scaling factors (see Figs. S8,S9). Cumulative display frequencies of the motifs as stem-loop are plotted against genome position for the packaged transcripts (bars), with the high-affinity BoxB tags highlighted in orange, BoxB-like PS1–5 motifs in blue, and the respective in vitro-transcribed RNA indicated by black lines; arrows show the increase or decrease observed upon packaging. (C,D) Two consensus folds predicted for packaged NC-3 and NC-4 mRNA (see also Figs. S8,S9). Secondary structure features shared between the respective folds are highlighted in grey. These structures indicate more extensive fold conservation in NC-4, as well as more robust display of a packaging cassette comprising PS1, BB1, PS2, and PS4, than in NC-3. (E,F) Reactivities of the URxRxRR motifs displayed in the packaged NC-3 (E) and NC-4 (F) RNA folds depicted in panels (C) and (D), respectively. Reactivity follows the order: red (high)➞ yellow➞ green➞ black (low). The four packaging signal candidates in NC-4 show low reactivities, consistent with protection by capsid protein. (G) The evolution of a packaging cassette that steers efficient capsid assembly around the target RNA provides a compelling explanation for the improved properties of NC-4 compared to NC-3.
In NC-4, four of the seven potential packaging signals are significantly populated as stem loops in packaged genomes (PS1, BB1, PS2, PS4) and all are clustered at the 5’-end of the transcript. Notably, BB1 is displayed in 99% of all packaged RNA folds (Fig. 4B,D, Fig. S9). The low reactivities observed for the four URxR sub-motifs within the capsid (Fig. 4F) imply that they are in contact with protein. Robust display of a high-affinity packaging signal within a cassette of lower affinity motifs (PS1, PS2, and PS4) is reminiscent of nucleation complexes found in Satellite Tobacco Necrosis Virus ( 27 ), MS2 phage ( 28 ), and Hepatitis B Virus ( 29 ). This finding suggests that NC-4 similarly evolved a key hallmark of RNA packaging signal-mediated assembly. Genome-encoded packaging instructions likely foster selective RNA encapsulation as well as rapid, efficient capsid assembly ( 30 ), providing a compelling explanation for the improved properties of the evolved cage (Fig. 4G). Encapsulation of alternative or longer, more complex genomes may similarly benefit from optimization of RNA sequence and structure.
Successful conversion of a bacterial enzyme into a nucleocapsid that packages and protects its own encoding mRNA with high efficiency and selectivity shows how primordial self-replicators could have recruited host proteins for virion formation ( 1 ). The convergence on structural properties characteristic of natural RNA viruses through co-evolution of capsid and cargo is striking. Introduction of destabilizing mutations into the starting protein was key to the dramatic remodeling of the protein shell, providing the molecular heterogeneity needed to depart from the initial, energetically stable, architectural solution and converge on a regular, 240-subunit, closed-shell icosahedral assembly. At the same time, evolution of multiple RNA packaging motifs that can cooperatively bind the coat proteins likely guided specificity and efficient assembly. While such constructs are themselves attractive as customizable and potentially safe alternatives to natural viruses for gene delivery and vaccine applications, the lessons learned from their evolution may also inform ongoing efforts to tailor the properties of natural viruses for more effective gene therapy ( 31 ).
Supplementary Material
Acknowledgements
We acknowledge technical support from the Functional Genomics Center Zurich, Miroslav Peterek and Peter Tittmann (Scientific Center for Optical and Electron Microscopy, ETH Zurich), Daniel Böhringer (Cryo-EM Knowledge Hub, ETH Zurich) for help with cryoEM Data collection and elaboration, and Oliver Allemann for help with optimization of nucleocapsid expression and purification. We thank DNA Sequencing & Services (MRC I PPU, School of Life Sciences, University of Dundee, Scotland, www.dnaseq.co.uk) for DNA sequencing.
Funding
DH thanks the Swiss National Science Foundation and the ETH Zurich for financial support. RT acknowledges funding via an EPSRC Established Career Fellowship (EP/R023204/1) and a Royal Society Wolfson Fellowship (RSWF/R1/180009). RT & PGS acknowledge support from a Joint Wellcome Trust Investigator Award (110145 & 110146), and PGS also thanks the NSLS-II at the Brookhaven Synchrotron for the award of slots for data collection at Beamline 17-BM, together with Erik Farquhar for his expert assistance during these visits. NT acknowledges support from the Human Frontier Science Program (LT000426/2015-L). AS is the recipient of a Marie Skłodowska-Curie Individual Fellowship (LEVERAGE mRNA).
Footnotes
Author contributions: NT performed laboratory evolution, AS, ST, and NT biochemical characterization of nucleocapsids, and ST cryo-EM analysis with help from ML. APS, NP, and EW performed, and RJB, SC, PGS, and RT analyzed X-ray footprinting experiments. ST, AS, and DH wrote the manuscript with input from all other authors.
Competing interests: Authors declare no competing interests.
Data and materials availability
The principal data supporting the findings of this study are provided in the figures and Supplementary Information. Additional data that support the findings of this study are available from the corresponding author upon request. Cryo-EM maps and atomic models have been deposited in the Electron Microscopy Data Bank (EMDB) and wwPDB, respectively, with the following accession codes: EMDB-11631 and PDB 7A4F (NC-1 120-mer), EMDB-11632 and PDB 7A4G (NC-1 180-mer), EMDB-11633 and PDB 7A4H (NC-2), EMDB-11634 and PDB 7A4I (NC-3), and EMDB-11635 and PDB 7A4J (NC-4).
References and Notes
- 1.Krupovic M, Dolja VV, Koonin EV. Origin of viruses: primordial replicators recruiting capsids from hosts. Nat Rev Microbiol. 2019;17:449–458. doi: 10.1038/s41579-019-0205-6. [DOI] [PubMed] [Google Scholar]
- 2.Azuma Y, Edwardson TGW, Terasaka N, Hilvert D. Modular protein cages for size-selective RNA packaging in vivo. J Am Chem Soc. 2018;140:566–569. doi: 10.1021/jacs.7b10798. [DOI] [PubMed] [Google Scholar]
- 3.Edwardson TGW, Mori T, Hilvert D. Rational engineering of a designed protein cage for siRNA delivery. J Am Chem Soc. 2018;140:10439–10442. doi: 10.1021/jacs.8b06442. [DOI] [PubMed] [Google Scholar]
- 4.Lilavivat S, Sardar D, Jana S, Thomas GC, Woycechowsky KJ. In vivo encapsulation of nucleic acids using an engineered nonviral protein capsid. J Am Chem Soc. 2012;134:13152–13155. doi: 10.1021/ja302743g. [DOI] [PubMed] [Google Scholar]
- 5.Butterfield GL, Lajoie MJ, Gustafson HH, Sellers DL, Nattermann U, Ellis D, Bale JB, Ke S, Lenz GH, Yehdego A, Ravichandran R, et al. Evolution of a designed protein assembly encapsulating its own RNA genome. Nature. 2017;552:415–420. doi: 10.1038/nature25157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terasaka N, Azuma Y, Hilvert D. Laboratory evolution of virus-like nucleocapsids from nonviral protein cages. Proc Natl Acad Sci USA. 2018;115:5432–5437. doi: 10.1073/pnas.1800527115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwardson TGW, Hilvert D. Virus-Inspired Function in Engineered Protein Cages. J Am Chem Soc. 2019;141:9432–9443. doi: 10.1021/jacs.9b03705. [DOI] [PubMed] [Google Scholar]
- 8.Cannon KA, Ochoa JM, Yeates TO. High-symmetry protein assemblies: patterns and emerging applications. Curr Opin Struct Biol. 2019;55:77–84. doi: 10.1016/j.sbi.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Walls AC, Fiala B, Schäfer A, Wrenn S, Pham MN, Murphy M, Tse LV, Shehata L, O’Connor MA, Chen C, Navarro MJ, et al. Elicitation of Potent Neutralizing Antibody Responses by Designed Protein Nanoparticle Vaccines for SARS-CoV-2. Cell. 2020;183:1367–1382. doi: 10.1016/j.cell.2020.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ladenstein R, Fischer M, Bacher A. The lumazine synthase/riboflavin synthase complex: shapes and functions of a highly variable enzyme system. FEBS J. 2013;280:2537–2563. doi: 10.1111/febs.12255. [DOI] [PubMed] [Google Scholar]
- 11.Austin RJ, Xia T, Ren J, Takahashi TT, Roberts RW. Designed arginine-rich RNA-binding peptides with picomolar affinity. J Am Chem Soc. 2002;124:10966–10967. doi: 10.1021/ja026610b. [DOI] [PubMed] [Google Scholar]
- 12.Di Tomasso G, Lampron P, Dagenais P, Omichinski JG, Legault P. The ARiBo tag: A reliable tool for affinity purification of RNAs under native conditions. Nucleic Acids Res. 2011;39:e18. doi: 10.1093/nar/gkq1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filonov GS, Kam CW, Song W, Jaffrey SR. In-gel imaging of RNA processing using broccoli reveals optimal aptamer expression strategies. Chem Biol. 2015;22:649–660. doi: 10.1016/j.chembiol.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasaki E, Böhringer D, Van De Waterbeemd M, Leibundgut M, Zschoche R, Heck AJRR, Ban N, Hilvert D. Structure and assembly of scalable porous protein cages. Nat Commun. 2017;8:14663. doi: 10.1038/ncomms14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seebeck FP, Woycechowsky KJ, Zhuang W, Rabe JP, Hilvert D. A simple tagging system for protein encapsulation. J Am Chem Soc. 2006;128:4516–4517. doi: 10.1021/ja058363s. [DOI] [PubMed] [Google Scholar]
- 16.Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 17.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: A mechanism for oligomer assembly. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu C, Liljas L, Opalka N, Brugidou C, Yeager M, Beachy RN, Fauquet CM, Johnson JE, Lin T. 3D domain swapping modulates the stability of members of an icosahedral virus group. Structure. 2000;8:1095–1103. doi: 10.1016/s0969-2126(00)00508-6. [DOI] [PubMed] [Google Scholar]
- 19.Sun Z, El Omari K, Sun X, Ilca SL, Kotecha A, Stuart DI, Poranen MM, Huiskonen JT. Double-stranded RNA virus outer shell assembly by bona fide domain-swapping. Nat Commun. 2017;8:14814. doi: 10.1038/ncomms14814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sánchez-Eugenia R, Durana A, López-Marijuan I, Marti GA, Guérin DMA. X-ray structure of Triatoma virus empty capsid: Insights into the mechanism of uncoating and RNA release in dicistroviruses. J Gen Virol. 2016;97:2769–2779. doi: 10.1099/jgv.0.000580. [DOI] [PubMed] [Google Scholar]
- 21.Squires G, Pous J, Agirre J, Rozas-Dennis GS, Costabel MD, Marti GA, Navaza J, Bressanelli S, Guérin DMA, Rey FA. Structure of the Triatoma virus capsid. Acta Crystallogr Sect D Biol Crystallogr. 2013;69:1026–1037. doi: 10.1107/S0907444913004617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonjack-Shterengartz M, Avnir D. The enigma of the near-symmetry of proteins: Domain swapping. PLoS One. 2017;12:e0180030. doi: 10.1371/journal.pone.0180030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Twarock R, Stockley PG. RNA-mediated virus assembly: Mechanisms and consequences for viral evolution and therapy. Annu Rev Biophys. 2019;48:495–514. doi: 10.1146/annurev-biophys-052118-115611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Legault P, Li J, Mogridge J, Kay LE, Greenblatt J. NMR structure of the bacteriophage λ N peptide/boxB RNA complex: Recognition of a GNRA fold by an arginine-rich motif. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- 25.Hao Y, Bohon J, Hulscher R, Rappé MC, Gupta S, Adilakshmi T, Woodson SA. Time-resolved hydroxyl radical footprinting of RNA with X-rays. Curr Protoc Nucleic Acid Chem. 2018;73:e52. doi: 10.1002/cpnc.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandler-Bostock R, Bingham R, Clark S, Scott AP, Wroblewski E, Barker A, White SJ, Dykeman EJ, Mata CP, Bohon J, Farquhar E, et al. RNA X-ray footprinting reveals an in vivo determinant of viral infectivity. In preparation [Google Scholar]
- 27.Ford RJ, Barker AM, Bakker SE, Coutts RH, Ranson NA, Phillips SEV, Pearson AR, Stockley PG. Sequence-specific, RNA-protein interactions overcome electrostatic barriers preventing assembly of satellite tobacco necrosis virus coat protein. J Mol Biol. 2013;425:1050–1064. doi: 10.1016/j.jmb.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rolfsson Ó, Middleton S, Manfield IW, White SJ, Fan B, Vaughan R, Ranson NA, Dykeman E, Twarock R, Ford J, Kao CC, et al. Direct evidence for packaging signal-mediated assembly of bacteriophage MS2. J Mol Biol. 2016;428:431–448. doi: 10.1016/j.jmb.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel N, White SJ, Thompson RF, Bingham R, Weiß EU, Maskell DP, Zlotnick A, Dykeman EC, Tuma R, Twarock R, Ranson NA, et al. The HBV RNA pre-genome encodes specific motifs that mediate interactions with the viral core protein that promote nucleocapsid assembly. Nat Microbiol. 2017;2:17098. doi: 10.1038/nmicrobiol.2017.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dykeman EC, Stockley PG, Twarock R. Solving a Levinthal’s paradox for virus assembly identifies a unique antiviral strategy. Proc Natl Acad Sci USA. 2014;111:5361–5366. doi: 10.1073/pnas.1319479111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C, Samulski RJ. Engineering adeno-associated virus vectors for gene therapy. Nat Rev Genet. 2020;21:255–272. doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- 32.Jain M, Olsen HE, Paten B, Akeson M. The Oxford Nanopore MinION: Delivery of nanopore sequencing to the genomics community. Genome Biol. 2016;17:239. doi: 10.1186/s13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perlmutter JD, Hagan MF. Mechanisms of virus assembly. Annu Rev Phys Chem. 2015;66:217–239. doi: 10.1146/annurev-physchem-040214-121637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porterfield JZ, Zlotnick A. A simple and general method for determining the protein and nucleic acid content of viruses by UV absorbance. Virology. 2010;407:281–288. doi: 10.1016/j.virol.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. In: The Proteomics Protocols Handbook. Walker JM, editor. Vol. 112. Humana Press; Totowa, NJ: 2005. pp. 571–607. [Google Scholar]
- 36.Greisch JF, Tamara S, Scheltema RA, Maxwell HWR, Fagerlund RD, Fineran PC, Tetter S, Hilvert D, Heck AJR. Expanding the mass range for UVPD-based native top-down mass spectrometry. Chem Sci. 2019;10:7163–7171. doi: 10.1039/c9sc01857c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beck T, Tetter S, Künzle M, Hilvert D. Construction of Matryoshka-type structures from supercharged protein nanocages. Angew Chem Int Ed. 2015;54:937–940. doi: 10.1002/anie.201408677. [DOI] [PubMed] [Google Scholar]
- 38.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJH, Lindahl E, Scheres SHW. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, Moriarty NW, et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr Sect D Struct Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karabiber F, McGinnis JL, Favorov OV, Weeks KM. QuShape: Rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA. 2013;19:63–73. doi: 10.1261/rna.036327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding Y, Chan CY, Lawrence CE. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004;32:W135–W141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The principal data supporting the findings of this study are provided in the figures and Supplementary Information. Additional data that support the findings of this study are available from the corresponding author upon request. Cryo-EM maps and atomic models have been deposited in the Electron Microscopy Data Bank (EMDB) and wwPDB, respectively, with the following accession codes: EMDB-11631 and PDB 7A4F (NC-1 120-mer), EMDB-11632 and PDB 7A4G (NC-1 180-mer), EMDB-11633 and PDB 7A4H (NC-2), EMDB-11634 and PDB 7A4I (NC-3), and EMDB-11635 and PDB 7A4J (NC-4).