Abstract
Clathrin-mediated endocytosis (CME) is a highly conserved and essential cellular process in eukaryotic cells. Although classical genetics has been used to understand CME, its highly dynamic and vital nature is difficult to approach with such tools. In contrast, small molecules can acutely and reversibly perturb CME, however, the few chemical CME inhibitors that have been applied to plants are ineffective or show undesirable side effects. Here, we identify the previously described endosidin9 (ES9) as an inhibitor of clathrin heavy chain (CHC) function in both Arabidopsis and human cells through affinity-based target isolation, in vitro binding studies and X-ray crystallography. Moreover, we present a chemically improved ES9 analog, ES9-17, which lacks the undesirable side effects of ES9, while retaining the ability to target CHC. ES9 and ES9-17 have expanded the chemical toolbox to probe CHC function, and present chemical scaffolds for further design of more specific and potent CHC inhibitors across different systems.
Clathrin-mediated endocytosis (CME) is a major route for internalization of plasma membrane (PM) proteins and molecules from the extracellular enviroment 1,2 , but its dynamic and essential nature makes it difficult to dissect using classical genetics approaches. Chemical inhibitors of endocytosis are an attractive alternative to the current methods for disrupting protein functions. However, despite the extensive structural and biochemical knowledge about CME in eukaryotic cells 3 , the development of chemicals that interfere with this process is still at a relatively early stage. Until now, a few small molecules were shown to target the CME machinery in mammalian, yeast or plant systems 4 . Some of the most commonly used small-molecule CME inhibitors in mammalian systems are Pitstop2 (ref. 5), targeting the N-terminal domain (nTD) of the clathrin heavy chain (CHC), Dynasore 6 and the Dynasore-based series of small molecules called Dyngo 7 , both affecting the dynamin function. Recently, a natural product, Ikarugamycin, has been used to inhibit CME in different systems, but neither its potency nor specificity toward CME have been extensively examined 8 . Since none of the above mentioned molecules showed consistent effects in plant cells, plant cell biology has taken advantage of TyrphostinA23 (TyrA23), a CME-inhibiting small molecule 9 . However, TyrA23 has recently been described as a protonophore in Arabidopsis thaliana, and its inhibition of endocytosis was shown to occur through non-specific cytoplasmic acidification 9 . Therefore, CME research in plants would benefit from novel, potent small molecule inhibitors to dissect endocytosis to improve our understanding of the many physiological processes that rely on it.
Previously, the small molecule endosidin9 (ES9) was identified as an endocytosis inhibitor in different model systems 9 . Although ES9 is primarily a protonophore, its observed interference with CME did not seem to originate solely from cytosol acidification. For example, in Drosophila melanogaster, ES9 blocked synaptic vesicle recycling, closely mimicking the phenotype in mutants defective in clathrin or dynamin functions, while in Arabidopsis, ES9 was found to retain its ability to inhibit endocytosis at an increased apoplastic pH, in marked contrast to other protonophores such as TyrA23 (ref. 9). These results suggested that, despite its protonophore activity, ES9 might target proteins involved in CME. Here, we identified CHC as the protein target of ES9. Structural activity relation (SAR) analysis of ES9 aimed at disconnecting the protonophoric and CME-inhibiting activities discovered the improved ES9-17 analog, which inhibited endocytosis, but without its former protonophore activity. In vitro target validation strategies, including cellular thermal shift assays (CETSA) 10 and Drug Affinity Responsive Target Stability (DARTS) 11 further confirmed CHC as a target of ES9-17. Altogether ES9 and ES9-17 expand the current chemical toolbox for CME inhibition and present promising scaffolds for further development of chemical probes targeting CHC across different systems.
Results
Affinity purification identifies Arabidopsis thaliana CHC1 as potential ES9 binder
Although the small molecule ES9 (1) had been previously characterized as a protonophore and an unspecific inhibitor of CME 9 , not all ES9-induced phenotypes were explained by its protonophore activity, implying that ES9 might inhibit CME through a direct interaction with its machinery 9 . To explore this possibility, we performed a SAR analysis of ES9 to identify a suitable position for derivatization with a linker and a biotin tag for carrying out an affinity-based target identification (Supplementary Fig. 1a; Supplementary Note 1). A small collection of commercially available and in house synthesized ES9 analogs were investigated for their ability to inhibit the uptake of the lipophilic styryl endocytic tracer dye N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl)hexatrienyl)pyridinium dibromide (FM4-64) 12 in Arabidopsis root epidermal cells at a concentration of 50 μM and upon 30 min treatment (Supplementary Fig. 1a–c). The partial structure or possible hydrolysis products ES9-3 (2) and ES9-7 (3) lacked activity in this endocytosis inhibition assay. Similarly, methylation of the acidic NH position of ES9-6 (4) resulted in FM4-64 uptake and, thus, loss of activity. Replacement of the bulky bromine substituent on the thiophene ring by a hydrogen produced a simplified analog ES9-2 (5) that retained activity, hinting at a suitable anchoring place that can tolerate the introduction of a linker moiety for a biotin label attachment. Replacement of the thiophene ring itself by its phenyl bioisostere gave the active phenylsulfonamide analog ES9-8 (6) that readily allowed the further derivatization of the bromine-occupied position. Thus, a hydroxyl-terminated linker was introduced at this position giving the analog ES9-9 (7) which indeed retained its activity in the endocytosis inhibition assay. Next, a biotin tag was introduced to generate a biotin-coupled variant of the active ES9-9, named ES9-10 (8). However, ES9-10 lost the ability to inhibit FM4-64 uptake, likely because ES9-10 was prevented from entering the cell by either the biotin moiety requiring transport 13 or the cell wall. A linker-biotin probe without the ES9 moiety, ES9-13 (9) (Supplementary Fig. 1a–c), was included as a negative control in further pull-down experiments.
To identify possible targets of ES9, we prepared protein extracts for affinity purification from PSB-D wild type Arabidopsis cell cultures. Proteins bound to ES9-10 or ES9-13 were analyzed by means of mass spectrometry. In total, three biological replicates were examined for the ES9-10 affinity purification and the ES9-13 control, each represented by three technical repeats. Proteins were listed based on the number of times they were identified in the different biological and technical replicates, and based on the number of peptide-to-spectrum matches. To reduce the number of the putative ES9-10 interactors we filtered the obtained data as previously described 14,15 . Only proteins found more than once in the three biological replicates in the ES9-10 pull-down experiments, and not in the ES9-13, were considered as potential hits, resulting in a list of 11 candidates (Supplementary Data Set 1), among which the Arabidopsis thaliana CHC1 isoform is an essential CME component 16 .
ES9 binds CHC
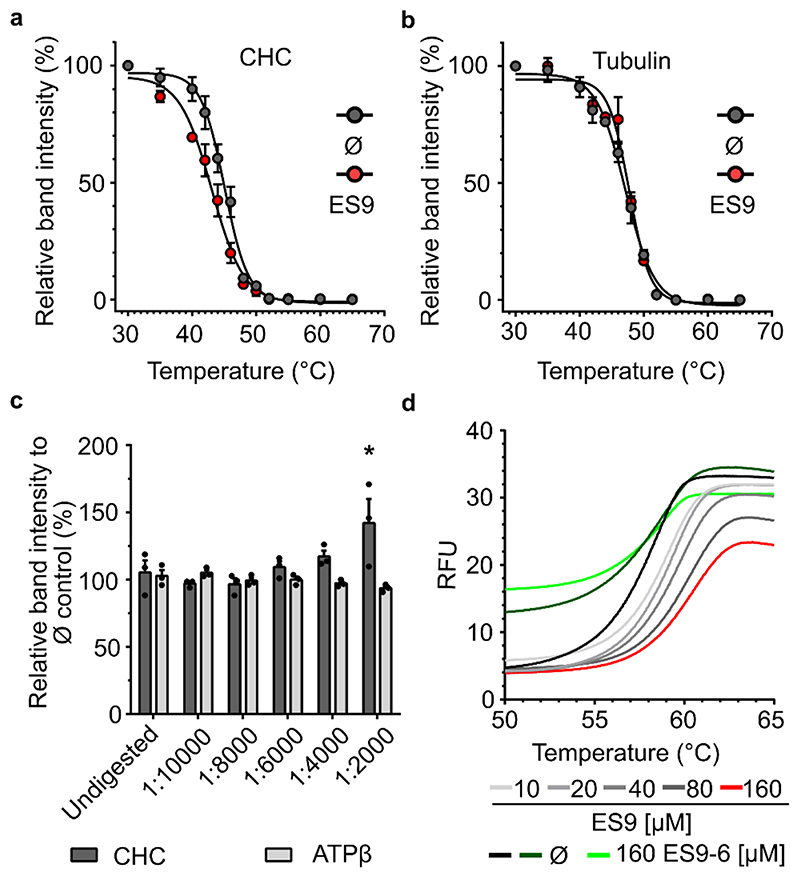
Although we cannot exclude that other proteins identified in the ES9-10 pull-down might play a role in CME, we chose to focus on CHC, as CHC1 was found as one of the top candidates, and has a well described role in CME. To validate the possible interaction between ES9 and CHC1 in Arabidopsis, we employed the cellular thermal shift assay (CETSA) that monitors target engagement based on small molecule-induced changes in the thermal stability of protein targets 10 . The aggregation temperature (Tagg) of CHC was assessed through immunoblot analysis using anti-CHC antibody, which cross-reacts with the two Arabidopsis CHC isoforms, CHC1 (AT3G11130) and CHC2 (AT3G08530), in Arabidopsis cell culture lysates treated with DMSO for 30 min and heated for 2 min at 12 different temperature points (30-65°C). Thermal denaturation of CHC under control (DMSO) conditions indicated a Tagg of 45±0.23°C (mean ± standard error of the mean, SEM) (Fig. 1a). For further isothermal dose-response fingerprint (ITDRFCETSA) experiments of CHC in the presence of ES9, we set the temperature at 46°C to ensure a sufficient shift in the denaturation temperature and determined the half maximum effective concentration (EC50) to be 120.5±1.09 μM (mean±SEM) (Supplementary Fig. 2a). In contrast, the highly abundant protein ATP synthaseβ (ATPβ), chosen as a control, was largely unaffected, even at high compound concentrations. Although the EC50 for the thermal denaturation of CHC in the presence of ES9 was 120 μM, a higher concentration of 250 μM ES9 was used for CETSA to achieve sufficiently sized Tagg shifts. The presence of 250 μM ES9 for 30 min and heating for 2 min at 12 different temperature points (30-65°C) generated a lower Tagg of 43 ±0.29°C (mean ± SEM) than that of the vehicle control (45±0.23°C, mean±SEM), thus resulting in a Tagg shift of 2°C (Fig. 1a). Thermal denaturation of the control proteins, tubulin and ATPβ, showed negligible Tagg shifts in the presence of either 250 μM ES9 or the vehicle (Fig. 1b; Supplementary Fig. 2b). Moreover, in the presence of the inactive analog ES9-6 (250 μM), the Tagg for CHC did not differ when lysates were either treated with DMSO (44.76±0.47°C) or ES9-6 (44.49±0.27°C) (mean±SEM) (Supplementary Fig. 2c). The results obtained with CETSA were further corroborated by means of DARTS 11 study, used to determine the potential of ES9 to protect CHC from protease digestion (Fig. 1c). In the presence of ES9 (250 μM), CHC was significantly stabilized at one pronase concentration when compared to the DMSO-treated control samples. Such stabilization was not observed for the control protein ATPβ (Fig. 1c). Taken together, our data indicate that CHC likely is the target of ES9 in Arabidopsis cells.
Figure 1. ES9 binds clathrin heavy chain.
(a) and (b) Protein extracts from Arabidopsis PSB-D cell cultures were treated with ES9 (250 μM) or DMSO (Ø) for 30 min and the thermal denaturation curves for endogenous clathrin heavy chain (CHC) (a) and tubulin (b) were recorded across 12 temperature points (30-65°C). The relative band intensity compared to the lowest temperature (30°C) sample was measured by Western blot with anti-CHC and anti-tubulin antibodies. Error bars in (a) and (b) indicate standard error of the mean (SEM) of three biological replicates. For the uncropped blots, see Supplementary Fig. 9. (c) Protein extracts from Arabidopsis PSB-D cell cultures were incubated with ES9 (250 μM) or DMSO (Ø) for 30 min and digested with different concentrations of pronase. An undigested sample was included as control. The relative band intensity compared to the DMSO control was measured by Western blot with anti-CHC and anti-ATP synthase subunitβ (ATPβ) antibodies. Error bars indicate SEM, individual data points are shown. *P<0.05, for a one-way analysis of variance (ANOVA) test with a Dunnett’s multiple comparisons test, and compared to the undigested control; (n=3); n, biological replicates. For the uncropped blots, see Supplementary Fig. 11. (d) Changes in the thermodynamic stability of the Arabidopsis CHC1 N-terminal domain in the presence of different concentrations of ES9 and the inactive analog ES9-6. Controls are respective to ES9 and ES9-6 treatments. The data shown are representative of three experiments. RFU, relative fluorescence units.
ES9 binds the N-terminal domain (nTD) of CHC
In an attempt to predict the binding site of ES9 on CHC, ES9 was docked to the only available structures of the human CHC1 nTD in a complex with either Pitstop1 (pdb2XZG) or Pitstop2 (pdb4G55) 5 . The obtained prediction with pdb4G55 suggested that the binding site for ES9 on Arabidopsis CHC1 might be the same as that for Pitstop2 in human CHC1 (Supplementary Fig. 3a). Residues Arg64, Phe91 and Gln89 in the binding site were set as flexible during docking and the first nine predicted positions were all similar, except for minor reorientations of the flexible residues. To confirm this prediction, the highly homologous β-propeller nTDs of the human CHC1 and the two Arabidopsis CHC1 and CHC2 isoforms (Supplementary Fig. 3b) were produced and the respective protein-ES9 interactions were analyzed in vitro by means of differential scanning fluorimetry (DSF). Upon ES9 binding to the nTD of the Arabidopsis CHC1, DSF detected a shift in the thermostability of CHC1, resulting in a change in its melting temperature (ΔTm) (Fig. 1d; Supplementary Table 1). In contrast, the thermal denaturation curve for the inactive analog ES9-6 (160 μM) overlapped with that of the (DMSO) control (Fig. 1d). The protein stability of the nTD in the presence of ES9, assessed as ΔTm, was similar for the CHC1 and CHC2 of Arabidopsis and the human CHC1 (Supplementary Fig. 2d,e; Supplementary Table 1). Altogether, our results demonstrate that ES9 binds to the nTD of CHC both in Arabidopsis and in human.
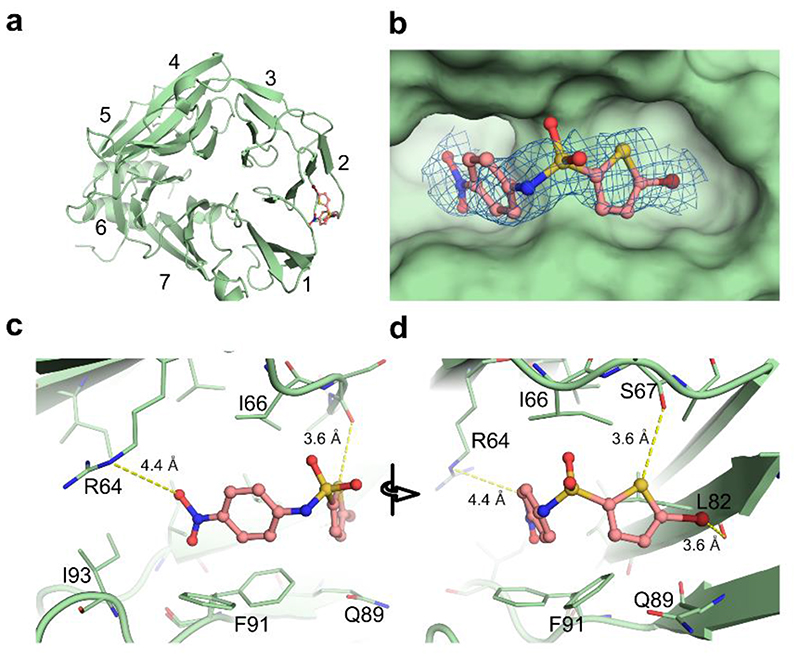
To understand the molecular basis of the CME inhibition, the binding site of ES9 in the nTD of CHC was mapped by determining the structure of the human nTD of CHC1 in a complex with ES9 by protein X-ray crystallography (Fig. 2a). Crystals with the clathrin nTD diffracted up to a resolution of 1.6Å and the structure was solved by molecular replacement with the CHC1 nTD as a search model (Supplementary Table 2). The CHC1 nTD forms a seven-bladed β-propeller, each with four antiparallel strands. The electron density for ES9 could be identified between the first and second blades (Fig. 2b). Modeling of ES9 into this electron density revealed that the ES9-binding site on the CHC nTD overlaps with the binding site for the CME adaptor proteins harboring a clathrin box motif 17 . ES9 is positioned in the cavity formed by six amino acids (Arg64, Ile93, Phe91, Gln89, Leu 82, and Ile66). The conformation of ES9 is stabilized by electrostatic interactions between its benzonitro moiety and Arg64, and between its thiophene group and the carboxyl oxygen of Ser67 (Fig. 2c,d). All the six amino acids involved in binding of ES9 to nTD are conserved between human CHC1 and Arabidopsis CHC1 and CHC2 (Supplementary Fig. 3b). Thus, the crystal structure analysis corroborated the previous docking predictions that ES9 targets the nTD of CHC in a similar fashion as the Pitstop2 (ref. 5).
Figure 2. ES9 binds the terminal domain of clathrin heavy chain.
(a) The structure of the human clathrin heavy chain1 (CHC1) N-terminal domain (nTD) (green cartoon) is shown in complex with ES9 (salmon) (b). Omitted electron density (2Fo–Fc) of ES9, contoured at 1σ level, is observed inside the hydrophobic clathrin box, which is located between the first and second blades of the nTD. (c) The interaction distances between the nTD residues and ES9 are shown with yellow dashes. The zoom-in view of the benzonitro moiety of ES9 indicates that the oxygen of the nitro group interacts electrostatically with R64. (d) A 60-degree rotated view of ES9 focuses on the bromothiophene moiety. Whereas the sulfur atom of the thiophene group is shown in close contact to the carboxyl oxygen of S67, the bromide atom points toward the carboxyl oxygen of L82 by a σ hole.
Identification of nonprotonophoric ES9 analog
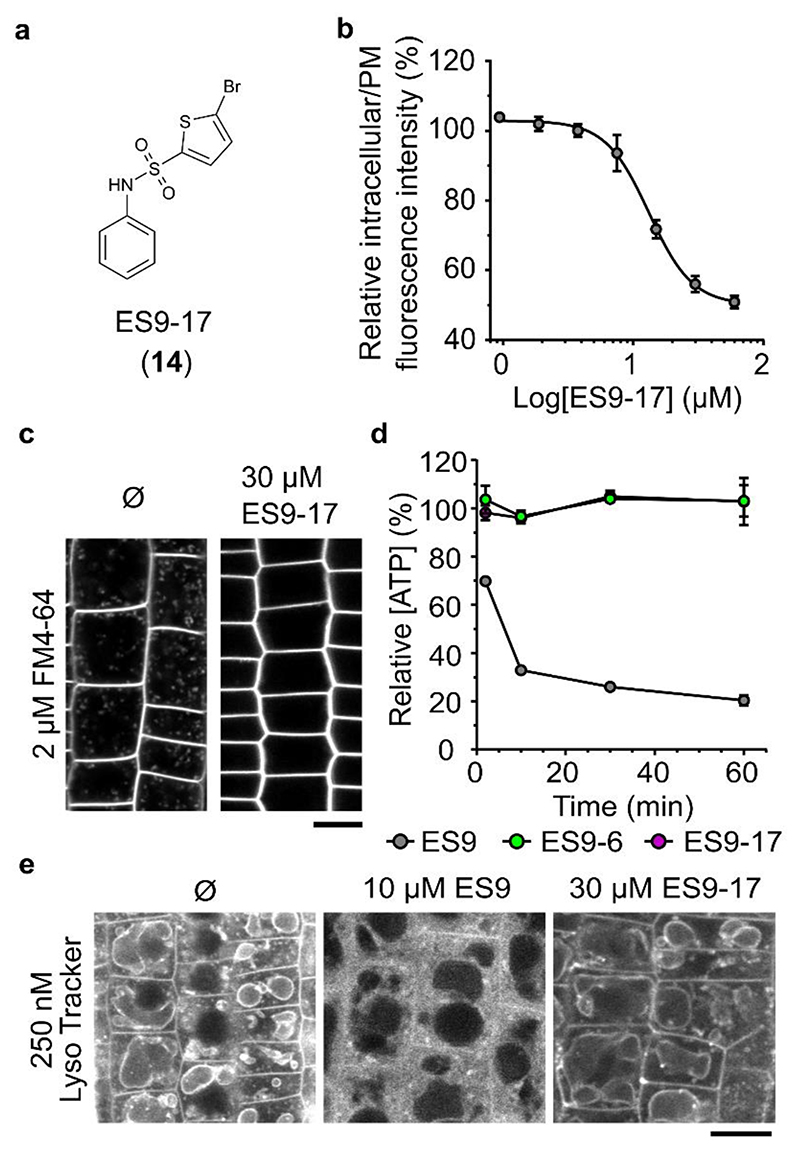
Although ES9 binds CHC, its protonophore activity 9 limits the use of this small molecule as a specific CME inhibitor. With the aim to uncouple CHC binding of ES9 from its protonophore activity, we carried out a focused SAR analysis to identify ES9 analogs that lack protonophore activity, but retain the ability to inhibit FM4-64 uptake (Supplementary Fig. 4a; Supplementary Note 1). As a weak acid that gives a delocalized anionic charge via electronic conjugation within a large hydrophobic compound is a known molecular architecture that allows protonophore activity, resulting in lipohilic anions 18 , we attempted to influence the acidity of the sulfonamide of ES9. For several analogs with expected differences in acidity and/or charge delocalization, we assessed both the FM4-64 uptake inhibition and the impact on the mitochondrial membrane potential as visualized with MitoTracker Red CM-H2XRos dye 19 in Arabidopsis root epidermal cells 9 when applied at concentration of 50 μM (Supplementary Fig. 4b–d). Transfer of the nitro group from the para to the meta position in ES9-29 (10) is expected to make the sulfonamide less acidic, while preserving specific contacts with target biomolecules. However, ES9-29 retained its protonophore activity, indicating that the sulfonamide was still fairly acidic. Other substitutions on the phenyl ring, like in ES9-15 (11) and ES9-16 (12) led to loss of the FM-64 uptake inhibition activity. Altogether, elimination of the nitro group in ES9 should strongly shift the equilibrium toward the protonated form, and inhibit resonance of the charge to the phenyl ring but it could affect the binding affinities for biomolecular targets. In fact, we found that ES9-14 (13) (Supplementary Fig. 4a) and ES9-17 (14) (Fig. 3a) both remained active with respect to FM4-64 uptake inhibition and did not abolish MitoTracker Red CM-H2XRos staining of the mitochondria at 50 μM (Supplementary Fig. 4b–d). Although, ES9-17 inhibited the FM4-64 uptake in Arabidopsis root epidermal cells with an EC50 of 13 μM (Fig. 3b), a concentration of 30 μM was used for further cellular analysis to ensure a substantial reduction in endocytosis (Fig. 3c). In addition, CME inhibition with 30 μM ES9-17 proved to be reversible, as FM4-64 uptake was clearly recovered after 120 min washout with control medium in two independent experiments (Supplementary Fig. 4e).
Figure 3. ES9-17 is not a protonophore.
(a) Structure of ES9-17 (14). (b) Dose-response of ES9-17-mediated FM4-64 uptake inhibition. The data are plotted as the ratio of the intracellular over the plasma membrane (PM) FM4-64 fluorescence intensities. The EC50 is 13 μM. At least 3 cells were measured per seedling, n=6 seedlings per treatment. (c) Strongly reduced FM4-64 uptake (2 μM, 30 min) in Arabidopsis root epidermal cells in the presence of ES9-17 (30 μM) when compared to the DMSO (Ø) control. The samples were pretreated with either ES9-17 (30 μM) or DMSO for 30 min. (d) Measurements of the ATP concentration in wild type Arabidopsis PSB-D cell cultures treated for 30 min with ES9 (10 μM), ES9-17 (30 μM) and ES9-6 (50 μM), showing ATP concentration relative to the DMSO control. Error bars indicate standard error of the mean of three biological replicates. (e) Confocal images of Arabidopsis epidermal root cells stained with Lyso Tracker Red DND 99 (30 min) and treated additionally for 30 min with DMSO (Ø), ES9 (10 μM) or ES9-17 (30 μM). Scale bars, 10 μm.
To rule out the protonophore activity of ES9-17, we assessed its capacity to affect cellular ATP in dark-grown Arabidopsis PSB-D cell cultures when used at 30 μM together with ES9 (10 μM) and the inactive analog ES9-6 (50 μM). As expected and in contrast to the sharp ATP decrease after treatment with 10 μM ES9 (ref. 9), neither 30 μM of ES9-17 nor 50 μM ES9-6 depleted ATP (Fig. 3d). All small molecules did not interfere with increasing fluorescein diacetate (FDA) fluorescence over time, indicating that they had no cytotoxic properties (Supplementary Fig. 5a). Previously, the protonophore activity of ES9 was shown to be unspecific to the mitochondria, because ES9 acidified the cytoplasm probably through dissipation of the proton gradients over the PM 9 . Therefore, we assessed whether ES9-17 affected the cytoplasmic pH. Arabidopsis seedlings were preincubated with Lyso Tracker Red DND 99 (ref. 20) to label membranes delineating acidic compartments, followed by a 30-min incubation with DMSO, 10 μM ES9 or 30 μM ES9-17 (Fig. 3e). As anticipated, 10 μM of ES9 relocated Lyso Tracker Red DND 99 predominantly to the cytosol when compared to the DMSO control, whereas 30 μM of ES9-17 failed to induce a similar staining pattern. Furthermore, unlike ES9, application of ES9-17 (30 μM for 30 min) did not compromise the motility of the actin cytoskeleton (ABD2 of Fimbrin-GFP) 21 , microtubules (GFP-MAP4) 22 , the Golgi (ST-mRFP) 23 and the trans-Golgi network (TGN)/early endosome (EE) compartments (VHA-a1-GFP) 24 (Supplemental Fig. 5b–e).
ES9-17 is a CME inhibitor
To characterize the potential of ES9-17 as a CME inhibitor, we evaluated the internalization of several PM-localized cargos subjected to CME in Arabidopsis and in human HeLa cells. We assessed whether CME-mediated uptake of the brassinosteroid (BR) receptor 25 BR INSENSITIVE1 (BRI1) would be affected by ES9-17. Arabidopsis seedlings expressing GFP-tagged BRI1 (BRI1::BRI1-GFP) (ref. 26) were pretreated for 1 h with 50 μM cycloheximide (CHX) to reduce the amount of newly synthesized proteins, followed by treatments with DMSO or ES9-17 (30 μM) for 30 min and then with 50 μM Brefeldin A (BFA) and FM4-64 for 30 min. In the presence of ES9-17, BRI1-GFP as well as FM4-64 failed to label the BFA bodies (Fig. 4a), hinting at an inhibition of uptake from the PM. The lack of BRI1-GFP-labeled BFA bodies was not due to a limitation of BFA body formation, because BFA bodies marked by the TGN/EE marker VHAa1-GFP 24 were formed (Supplementary Fig. 6a). Furthermore, fluorescently labeled Alexa fluor 674 castasterone (AFCS), which binds to BRI1 and consequently enters the cell through CME 25 , failed to stain the vacuole in root epidermal cells when applied to Arabidopsis seedlings expressing BRI1-GFP 26 after pretreatment with 30 μM ES9-17 for 30 min (Supplementary Fig. 6b). To corroborate the CME inhibition by ES9-17, we examined another PM-localized cargo, which is subjected to CME, the leucine-rich repeat receptor kinase PEP RECEPTOR1 (PEPR1) 27 . Treatment of Arabidopsis root cells expressing RPS5A::PEPRI-GFP with 100 nM of the Pep1 peptide for 10 s after pretreatment with DMSO for 30 min induced PEPR1-GFP internalization (Fig. 4b). However, when roots were pretreated with 30 μM ES9-17 for 30 min and elicited with 1 μM Pep1, PEPR1-GFP was still predominantly localized to the PM even 90 min after the elicitation, implying an inhibitory effect of ES9-17 on the CME-mediated PEPR1 uptake (Fig. 4b).
Figure 4. ES9-17 is a CME inhibitor.
(a) ES9-17 inhibited the recruitment of the plasma membrane-localized BRI1-GFP to the Brefeldin A (BFA) body. Samples were pretreated with 50 μM cycloheximide (CHX) for 1 h and treated with either DMSO (Ø) or ES9-17 (30 μM) for 30 min, followed by a combined application of FM4-64 (2 μM) and BFA (50 μM) for an additional 30 min in the presence of the inhibitor. Scale bar, 10 μm. (b) Confocal images of Arabidopsis root epidermal cells expressing PEPR1-GFP. Seedlings were treated with ES9-17 (30 μM) or DMSO (Ø) for 30 min, followed by elicitation with 100 nM Pep1. The internalization of PEPR1-GFP was followed until 90 min post elicitation. Scale bar, 20 μm. (c) Histograms representing the measured endocytic foci life time of clathrin light chain1 (CLC1)-GFP (CLC1::CLC1-GFP/Col-0) in the presence of ES9-17 (30 μM) or DMSO (Ø), n=1276 and 1069 measurements, from 10 and 4 seedlings respectively. Kymographs represent a line trace (horizontal axis) over a time period (vertical axis), taken from a time lapse (2 f/sec) of Arabidopsis root epidermal cells, that illustrate the life times of endocytic foci labeled by CHC1-GFP. Scale bar, 10 sec.
To link the reduction of cargo internalization caused by ES9-17 to its possible effects on PM recruitment of the CME machinery we evaluated the dynamic behavior of clathrin, CLC1-GFP 9 and CHC1-GFP 9 , and one representative of the CME adaptor proteins in plants, the TPLATE muniscin-like (TML) subunit of the TPLATE complex (TPC) 28 in root epidermal cells of Arabidopsis after ES9-17 application. In the presence of DMSO, the majority of the PM-localized foci had an average residence life time of 31.98 sec, 44.62 s and 36.72 s for CLC1-GFP, CHC1-GFP and TML-GFP, respectively (Fig. 4c; Supplementary Fig. 6c, d). When seedlings were treated with 30 μM ES9-17, the residence life time of CLC1, CHC1 and TML in the PM increased substantially with an average of 73.99 s, 83.55 s and 68.66 s, respectively.
Since ES9 was found to bind both Arabidopsis and human CHC, we assessed if ES9-17 could inhibit transferrin uptake in HeLa cells, a well-known clathrin-mediated process 29 . Treatment of HeLa cells with 30 μM ES9-17 for 30 min appeared to reduce the uptake of transferrin, but not to an extent as observed with 20 μM Pitstop2 (Supplementary Fig. 7a). In parallel with the inhibitory treatments, a cell proliferation assay ascertained that the resulting inhibition of the transferrin uptake was not due to the cytotoxicity of the compounds (Supplementary Fig. 7b). In an effort to compare ES9-17 with Pitstop2 we sought to assess Pitstop2 activity in Arabidopsis but failed to detect CME inhibition. Pitstop2 did not block FM4-64 uptake when applied at 30 μM for 30 min (Supplementary Fig. 7c), and similarly failed to inhibit the internalization of the CME cargo, BRI1-GFP in the presence of BFA (Supplementary Fig. 7d). Higher concentrations of Pitstop2 appeared to affect overall FM4-64 fluorescence, but did not result in uptake inhibition (Supplementary Fig. 7e). Moreover, the CETSA thermal denaturation curves for CHC and ATPβ in the presence of either DMSO (Ø) or 250 μM Pitstop2 were very similar. The Tagg with DMSO (Ø) and Pitstop2 for CHC was 44.57±0.29°C and 44.5±0.34°C (mean±SEM), respectively and 55.03±1.41°C and 54.63±1.05°C (mean±SEM) for ATPβ, respectively (Supplementary Fig. 7f). Taken together, ES9-17 affected dynamic behavior of core components of CME, and the uptake of different cargoes. In addition, ES9-17 affected transferrin uptake in HeLa cells, similar to Pitstop2, while Pitstop2 proved to be inactive in Arabidopsis with respect to CME inhibition.
ES9-17 targets CHC
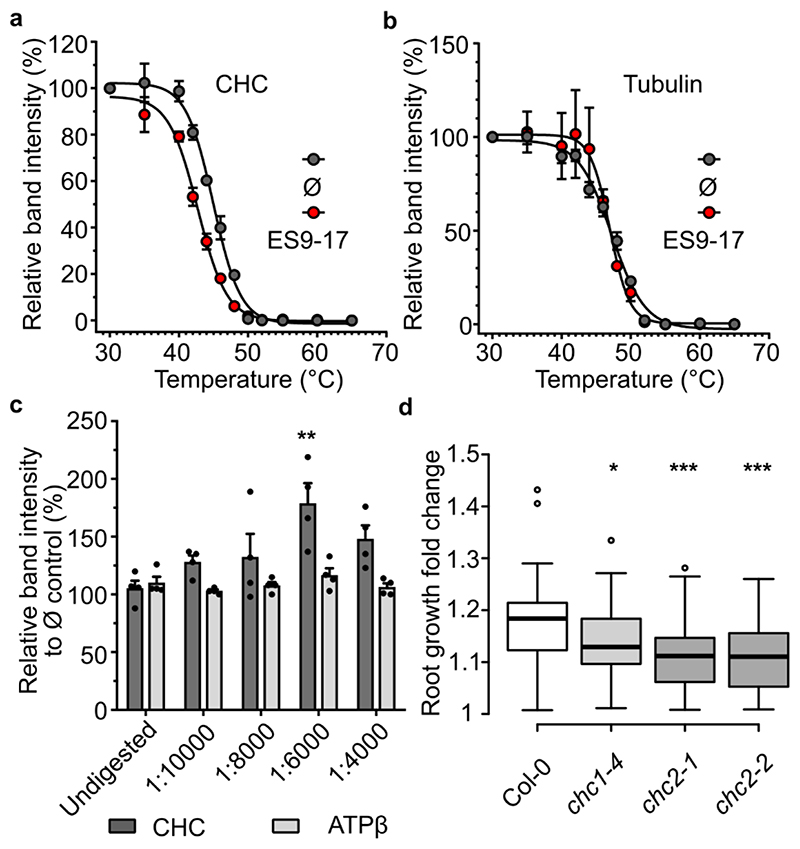
As ES9-17 inhibited several clathrin-dependent processes and the parent molecule ES9 was found to bind CHC, we hypothesized that CHC might also be the target of ES9-17. To test this hypothesis, we made use of the target validation approaches CETSA and DARTS (Fig. 5a). ITDRFCETSA analysis at 46°C indicated an EC50 of 123±1.13 μM (mean±SEM) for ES9-17 (Supplementary Fig. 8a), which is very close to the EC50 observed for ES9. Similarly, as observed for ES9, a shift in Tagg for CHC was detected in the presence of ES9-17. Arabidopsis cell culture lysates treated with DMSO (Ø) for 30 min and heated for 2 min at 12 different temperature points (30-65°C) resulted in a Tagg for CHC of 44.96±0.2°C (mean±SEM), whereas in the presence of 250 μM ES9-17 the Tagg was 42.71±0.2°C (mean±SEM), thus generating a Tagg shift of 2.2°C (Fig. 5a). In contrast, ES9-17 failed to induce a shift in Tagg for the control proteins, tubulin and ATPβ (Fig. 5b; Supplementary Fig. 8b). These results were supported by the DARTS assay, in which cell lysates were treated with ES9-17 (250 μM) at particular dilutions of pronase (Fig. 5c). At pronase concentrations of 1:6000 and 1:4000, a 2-fold and 1.5-fold stabilization was observed for CHC in ES9-17-treated cell lysates, respectively, of which only the 1/6000 dilution was found to be statistically significant (one-way ANOVA with Dunett’s multiple comparisons test), whereas the control proteins, ATPβ (Fig. 5c) and selected TPC 28 and adaptor protein complex-2 (AP-2) subunits 30 , were not stabilized (Supplementary Fig. 8c–e). To further strengthen the specificity of ES9-17, we assessed its possible interaction with the coatomer subunit γ (AT4G34450, γ-COP), since this protein ranked second in the ES9 affinity purification (Supplementary Data Set 1). The DARTS assay with ES9-17 failed to demonstrate significant protection of the γ-COP from protease digestion (Supplementary Fig. 8f), concluding that ES9-17 does not bind γ-COP. Previously we observed that ES9 affects Golgi cisternae morphology and induces the formation of Golgi-endoplasmic reticulum (ER) hybrid compartments 9 indicative of a non-functional coat protein I (COPI) trafficking. In contrast, transmission electron microscopy (TEM) analysis of Arabidopsis Col-0 root tips treated with ES9-17 (30 μM) did not reveal striking changes in Golgi and ER morphology (Supplementary Fig. 8g), though occasionally ES9-17 appeared to induce larger multivesicular bodies (Supplementary Fig. 8g, white triangle). Altogether our results showed that ES9-17 did not affect the COPI function in Arabidopsis.
Figure 5. ES9-17 targets clathrin heavy chain.
(a) and (b) Protein extracts from Arabidopsis PSB-D cell cultures were treated with ES9-17 (250 μM) or DMSO (Ø) for 30 min. The thermal denaturation curves for endogenous clathrin heavy chain (CHC) (a) and tubulin (b) were observed across 12 temperature points (30-65°C). The relative band intensity compared to the lowest temperature (30°C) sample was measured by Western blot using anti-CHC and anti-tubulin antibodies. Error bars in (a) and (b) indicate standard error of the mean (SEM) of three biological replicates. For the uncropped blots, see Supplementary Fig. 9. (c) Protein extracts from Arabidopsis PSB-D cell cultures were incubated with ES9-17 (250 μM) or DMSO (Ø) for 30 min and digested with different concentrations of pronase. An undigested sample was included as a control. The relative band intensity compared to the DMSO control was measured by Western blot with anti-CHC and anti-ATP synthase subunitβ (ATPβ) antibodies. Error bars indicate SEM, individual data points are shown. **P<0.01 for a one-way analysis of variance (ANOVA) test with a Dunnett’s multiple comparisons test, and compared to the undigested control. n=4 biological replicates. For the uncropped blots, see Supplementary Fig. 12. (d) Boxplot representation of the root growth fold change over 48 h for wild type Arabidopsis Col-0 seedlings treated with ES9-17 (12 μM). Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by the R software, outliers are represented by dots. (n=80); n, number of seedlings in 3 independent experiments. *P<0.05, ***P<0.001 for a one-way analysis of variance (ANOVA) test with a Dunnett’s multiple comparisons test.
To confirm the results obtained with ES9-17 in CETSA and DARTS assays, we assessed genetically whether ES9-17 interacts with CHC. In contrast to ES9, of which the protonophore activity prevents meaningful genetic studies, ES9-17 lacks the protonophore characteristics and, hence, can be used to evaluate the increased sensitivity or resistance of mutant alleles. Double knockout mutant lines in CHC cannot be used, because CHC has an essential function. Therefore, several single mutant alleles for CHC1 and CHC2 (ref. 16) were examined for their higher sensitivity to ES9-17 than that of the wild type. Primary root growth for 5-day-old Arabidopsis (accession Columbia-0) seedlings was inhibited with an EC50 of 9 μM (Supplementary Fig. 8h). To assure a sufficient root growth inhibition, we set the concentration of ES9-17 at 12 μM. Seedlings of chc1-4 (Supplementary Fig. 8i), chc2-1 and chc2-2 (ref. 16) mutant lines displayed a significantly increased sensitivity to ES9-17 compared to the Col-0 seedlings (one-way ANOVA with Dunett’s multiple comparisons test) (Fig. 5d). Root growth did not significantly vary for the different mutant alleles when compared to Col-0, when grown under control conditions (Supplementary Fig. 8j) (one-way ANOVA with Dunett’s multiple comparisons test). Taken together and similar to the results obtained with ES9, the results obtained with CETSA, DARTS and CHC mutant alleles point toward CHC as a protein target of ES9-17 in Arabidopsis.
Discussion
In plants, CME is one of the most studied pathways for internalization of membrane-associated and soluble cargos from the PM and the extracellular environment 1 . As loss-of-function, CME mutants are often lethal or have no phenotypes because of gene redundancy 16 , methods to perturb CME largely rely on the use of inducible expression of mutant forms of critical proteins involved in endocytosis, such as clathrin and auxillin 31,32 , and siRNA-mediated depletion of adaptor proteins 28 . The drawbacks of such approaches are the low gene induction efficiency, the construct silencing and the considerable time needed to deplete existing complexes of these proteins in a cell (2-5 days), while the cell may adapt and even alter its gene expression, without certitude that only CME is impacted. Prolonged loss of a protein, such as clathrin, might impair post-Golgi trafficking 33 , with possible defects in secretion and vacuolar targeting as a consequence. Therefore, application of small molecule inhibitors of CME combined with live-cell imaging can greatly facilitate studies of the CME machinery and dynamics. An attractive feature of chemical inhibitors is that they can be applied acutely to reveal the direct block of a particular process and that their effect is reversible 34 .
Over the years, different small molecule effectors of endocytosis and endosomal function in Arabidopsis have been described, including Secdin 15 and several compounds from the endosidin (ES) series 35–38 . The identification and characterization of the CME inhibitor ES9 (ref. 9) revealed that the mode of action of this compound is mediated via its mitochondrial uncoupling and protonophore activities, therefore causing ATP depletion and cytoplasmic acidification. This mode of action appeared to be shared with the until then, most commonly used plant CME inhibitor, TyrA23. Therefore, in contrast to mammalian systems, in which the small CME inhibitors Pistop2 and Dynasore are well established 5,6 , plants lack small molecules that specifically target the CME machinery. While Dynasore has been used as a CME inhibitor in plants 39 , reports on the activity of Pistop2 in plant cells are lacking. Our results revealed that Pitstop2 is inactive as a CME inhibitor in Arabidopsis. This dissonance in activity behavior can be rationalized using structural considerations based on a key amino acid difference between Arabidopsis CHC1/CHC2 and human CHC1, and in light of the structurally characterized binding pocket of Pitstop2 in human CHC1. The hallmark of Pitstop2 binding by human CHC1 centers on the snug accommodation of its naphthalene ring by an evenly distributed cushion of β-branched residues (Ile52, Ile66, Ile80, Ile93 and Val50) 5 . However, Arabidopsis CHC1 and CHC2 display a Leu at position 80 (Supplementary Fig. 3b), which would be expected to introduce an irreconcilable steric hindrance that would prevent Pitstop2 binding.
Even though ES9 acts as a protonophore, evidence suggested that ES9 may have CME inhibiting effects beyond those resulting from cytoplasmic acidification 9 , prompting an affinity-based target identification strategy that identified the Arabidopsis CHC1 as a possible ES9-binding protein. Interestingly, both CHC1 and CHC2 equally interacted with ES9 in subsequent in vitro target validation strategies, such as CETSA, DARTS, and DSF, despite the fact that only CHC1 was found as a meaningful ES9 interactor and CHC2 was detected in the background lists (Supplementary Data Set 1). The interaction with CHC was further confirmed by the crystal structure of the human nTD of CHC1 with ES9, revealing that ES9 binds the same clathrin box as the Pitstop2 molecule 5 , positioned between blades 1 and 2 of the nTD β-propeller.
Regardless of the binding of ES9 to CHC, its usefulness as a specific CME inhibitor is compromised by its pronounced protonophore characteristics. Here we have described ES9-17 as an improved analog of ES9, lacking the nitro group, that was capable of inhibiting CME without protonophore activity. Docking predictions and the crystal structure of the nTD of human CHC1 in complex with ES9 suggested that the nitro group establishes an electrostatic interaction with Arg64. As a consequence, ES9-17 might bind the nTD of CHC in a different than ES9 orientation and affinity (Supplementary Fig. 3a). Nevertheless, ES9-17 inhibited the uptake of FM4-64 and of several CME cargos in Arabidopsis, validating ES9-17 as an inhibitor of endocytosis. Moreover, and similar to the previously reported CME inhibitors from the Pitstop family 5 , ES9-17 increased the dwell time of the endocytic foci at the PM, in contrast to the general freeze of CME dynamics observed with ES9 (ref. 9). In addition, ES9-17 did not acidify the cytoplasm and, thus, did not hamper endomembrane compartment motility and cytoplasmic streaming, as visualized by the TGN/EE, Golgi and cytoskeleton dynamics and the overall Golgi, TGN/EE and BFA body morphology.
The target validation strategy for ES9-17 indicated binding to CHC in CETSA and DARTS assays similar to what we describe for ES9. Both the temperature shift in the CHC denaturation and the EC50 concentration for ES9-17 and ES9 were essentially the same, as was the protection from protease digestion in the DARTS assay, strongly suggesting that ES9-17 also targets CHC. Currently we can only speculate why we observed a sizable difference in EC50 values for FM4-64 inhibition and in vitro validation strategies such as CETSA for both ES9 and ES9-17. Possibly, accessory proteins present in a biological context might increase affinity, but equally, in vitro conditions might increase the apparent EC50 values. Furthermore, ES9 appeared to be able to inhibit FM4-64 uptake with a higher potency (EC50 of 5.16 μM) 9 than that of ES9-17 (EC50 of 13 μM) likely due to being both a protonophore and a CHC inhibitor.
Notably, Golgi morphology in presence of ES9-17 appeared the same as in the control, unlike ES9, which induces substantial morphological changes in the Golgi 9 . These morphological changes are in part the result of the protonophore characteristics but are possibly also the result of targeting the γ-COP subunit of the coatomer complex, which ranked second after CHC in the ES9 affinity purification list. The lack of γ-COP protection from protease digestion in presence of ES9-17, as observed with the DARTS assay, further highlight the specificity of ES9-17 in terms of CME inhibition. Together with the genetic data, our results make for compelling evidence that ES9-17 is a specific CME inhibitor in Arabidopsis.
With ES9-17, several new opportunities arise in the quest to understand clathrin-mediated trafficking. For example, ES9 and ES9-17 offer a chemical scaffold to further improve on and identify more potent inhibitors of CHC function. In doing so we would further increase our understanding of the molecular aspects of CHC function in Arabidopsis. As we established that ES9 binds the same pocket as Pitstop2, it is reasonable to assume that the mode of ES9 inhibition would be similar to that of Pitstop2, that is, interfering with the recruitment of accessory proteins harboring a clathrin box motif 5 . Furthermore, although the core function of CHC in CME is known, we have little understanding of the specific roles of CHC1 and CHC2 in Arabidopsis. Rendering either CHC1 or CHC2 insensitive to ES9-17, yet still biologically functional, might help to deconvolve their function. The ability to selectively inhibit CHC1 or CHC2 function might help to highlight their different functions in the endomembrane system, or at the tissue and developmental level.
In summary, with ES9-17 we have presented for the first time a small molecule inhibitor of CME in Arabidopsis. While its activity is not limited to Arabidopsis, other inhibitors of CME, such as Pitstop2, lack activity in Arabidopsis, making ES9-17 to our knowledge the only small molecule probe for CME in Arabidopsis, which in addition is chemically different from the Pitstop2 family. It therefore allows to dynamically and reversibly inhibit CME pharmacologically, and together with ES9 offers a scaffold and platform for the further rational development of additional ES9 analogs. Such analogs might possess an increased affinity toward CHC or allow the distinction between CHC1 and CHC2, and serve as a platform to allow detailed dissection of the CHC function in Arabidopsis and other systems, both in a CME context and beyond.
Methods
Methods, including statements of data availability and any associated accession codes and references, are available in the online version of the paper.
Methods
Plant material and growth conditions
Arabidopsis thaliana (L.) Heyhn. (accession Columbia-0 [Col-0]) seedlings and other lines were stratified for 2 days at 4°C and grown vertically on agar plates containing half-strength Murashige and Skoog (½MS) medium supplemented with 1% (w/v) sucrose for 5 days at 22°C in a 16-h/8-h light/dark cycle, prior to use. The Arabidopsis mutants and transgenic lines used were: chc2-1 (ref. 16), chc2-2 (ref. 16), TML::TML-GFP/tml-1 (ref. 28), CLC1::CLC1-GFP/Col-0 (ref. 9), RPS5A::CHC1-GFP/Col-0 (ref. 9), VHA-a1::VHA-a1-GFP/Col-0 (ref. 24), 35S::ST-mRFP/Col-0 (ref. 23), RPS5A::PEPR1-GFP/pepr1pepr2 (ref. 27), BRI1::BRI1-GFP/Col-0 (ref. 26) 35S::GFP-MAP4/Ler 22 and 35S::Fimbrin-GFP/Col-0 (ref. 21). The T-DNA insertion line chc1-4 (SALK_110063) was obtained from the Nottingham Arabidopsis Stock Centre. The T-DAN insertion of chc1-4 was confirmed by PCR by using left primer flanking SALK insertion (5’-CAAGTGACGCATCACAACATG-3’), right primer (5’-ACCATTGCTCAAAACATACGC-3’) and T-DNA specific primer, LBA1 (5’-TGGTTCACGTAGTGGGCCATC-3’) (Supplementary Fig. 8i).
Generation of hemagglutin (HA)-tagged AP2S and AP2M constructs and Arabidopsis PSB-D cell culture transformation
Entry clones pDONRRP4-1R-RPS5A 9 , pDONRRP2R-P3-HAstop and pDONR211-AP2S (AT1G47830) or pDONR211-AP2M 30 (AT5G46630) were used together with pH7m34GW in multisite Gateway reactions (Life Technologies) to generate the RPS5A::AP2S-HA and RPS5A::AP2M-HA constructs. The AP2M-HA and AP2S-HA expression vectors were used to transform Agrobacterium tumefaciens C58. The PSB-D wild type cell cultures were subsequently transformed as described 14 .
Chemical treatments, chemical labeling and imaging in Arabidopsis
ES9 and analogs were acquired through Chembridge (http://www.chembridge.com/) or synthesized. They were dissolved in DMSO (Sigma-Aldrich) and 50°mM stocks were stored at -20°C in glass vials. ES9-17 was freshly prepared from lyophilized powder (stored at -20°C) prior to use. Brefeldin A, cycloheximide and Pitstop2 (Sigma-Aldrich) were dissolved in DMSO. Endocytosis was visualized with 2 μM FM4-64 (Life technologies). Washout experiments involved a 30 min pretreatment of seedlings in liquid ½ MS medium with ES9-17 (30 μM), followed by an additional 30 min treatment in presence of FM4-64 (2 μM). Subsequent substitution of treatment medium with medium without treatment, but with FM4-64, constituted the washout. Seedlings were imaged at indicated time points. The experiment was performed twice independently. Repeat 1 was washed with regular ½ MS and FM4-64 (2 μM), while repeat 2 was washed with ½ MS plus DMSO and FM4-64 (2 μM). Staining and imaging of mitochondria and acidic compartments in the seedlings treated with the small molecules was performed as described previously 9 . To observe the internalization of AFCS, seedlings expressing BRI1-GFP were treated with ES9-17 for 30 min followed by 20 μM of AFCS for 20 min. Seedlings were then quickly washed and mounted for microscopic observations for 20 min. To follow the PEPR1-GFP internalization, seedlings expressing PEPR1-GFP were treated with ES9-17 for 30 min and FM4-64 (2 μM) was added for further 15 min. Seedlings were then elicited with Pep1 100 nM and imaged at different time points. Imaging for AFCS uptake and PEPR1 internalization was performed with an SP8 confocal laser scanning microscope with 40×water immersion lens. Life-time of endocytic foci were measured in seedlings upon treatment with ES9-17/DMSO for 30 min as described previously 9 . Seedlings expressing VHA-a1-GFP and ST-mRFP were treated with or without drug for 30 min followed by time lapse images with 6 time points for 90 s, 10 s interval. Images were taken with Olympus FV10 ASW confocal laser scanning microscope with a 60× water immersion lens (NA 1.2) and 3× digital zoom.
The cytoskeletal dynamics of Arabidopsis root cells with and without drug treatments were performed with the Perkin Elmer Spinning disc using a 60x water corrected Plan Apo (NA 1.2) objective. Time lapse series were taken for 12 min, 5 time points per minute (MAP4) or 5 min, 1 time points per second (Fimbrin). Images were processed by means of the ImageJ (Fiji) software package. More specifically, for the MAP4 and Fimbrin superposed multi-color images, the background was subtracted using a rolling ball radius of 50 pixels and a walking average of 4 was subsequently applied to the time series. Colored projections were generated by superposing six different time points spread evenly over the duration of the acquisition. The six time points were merged using the merge channels tool of Fiji where the grey channel was left blank. All experiments described for Pitstop2 in Arabidopsis were performed as described for ES9-17. For the quantification of the cytosolic/PM signal intensity ratio, non-saturated images were converted in ImageJ to 8-bit and regions of interest (ROIs) were selected based on the PM or cytosol localization. Histograms listing all intensity values per ROI were generated and the averages of the 100 most intense pixels were used for calculations. Three cells were quantified per seedling and averaged.
ATP measurements
ATP and FDA measurements in three-day-old wild type PSB-D Arabidopsis cell cultures were performed as described previously 9 .
Affinity purification with biotinylated small molecules
PSB-D wild type cell cultures were harvested by separating them from the medium, flash frozen in liquid nitrogen and ground with a Retsch® MM400. Cell material was weighed and extraction buffer (EB) (50 mM Tris-HCl, pH 8, 150 mM NaCl, 0.1% NP-40 [Sigma-Aldrich] with 1 tablet/10 mL cOmplete ULTRA protease inhibitor cocktail, EDTA free [Roche]) was added in 2:1 ratio (200 μl extraction buffer for 100 mg material). The protein concentration was determined with the Bradford method (Quick start Bradford 1× Dye reagent [Bio-Rad]). Lysate was prepared by removing endogenous biotin, with Streptavidin Sepharose High Performance beads (GE Healthcare; hereafter referred to as beads). Washed beads (50 μl, and washed 3× with EB) were applied to 1-ml lysate and incubated at 4°C for 1 h on a rotary wheel. A second batch of beads was washed 3x with EB, and biotinylated small molecules (2 μl of 50 mM stock per 50 μl beads) were added with the last wash. The mixture was left at room temperature for 15 min, supernatant of the last wash removed, and transferred to 4°C until further use. Supernatant constituting the biotin cleared lysate was collected and added to beads incubated with the biotinylated small molecules. The subsequent mixture was incubated for a minimum of 2 h, or overnight at 4°C on a rotary wheel, followed by centrifugation whereafter the mixture was spun down (4°C), the supernatant removed, and beads were washed 3 times (50 mM Tris-HCl, pH 8, 150 mM NaCl). Appropriate amounts of the LDS sample buffer (4× LDS sample buffer supplemented with 10× sample reducing agent; Novex, Life Technologies) were added and samples were incubated for 10 min at 70°C, followed by a 2-min centrifugation at maximum speed (18 000 x g). The resulting supernatant was collected and run on 4-12% Bis-Tris protein gels with MES buffer (NuPage Novex gels and buffer from Life technologies). Gels were stained with SYPRO Ruby protein gel stain (Molecular probes, Invitrogen) and stained gel regions were excised and cut in approximately 3 pieces before submission to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
LC–MS/MS Analysis
Gel pieces containing the proteins of interest were cut from the gel and transferred to Biopure® Eppendorf tubes (Eppendorf AG, Hamburg, Germany). After two consecutive 15-min wash steps with water/acetonitrile (1/1, v/v) (both HPLC analytical; Mallinckrodt Baker B.V., Deventer, The Netherlands), the gel pieces were completely dried in a centrifugal vacuum concentrator, subsequently rehydrated in 10 μl of a 0.02 μg/μl of a sequencing-grade modified trypsin stock solution (Promega Corporation) and, completely submerged in freshly prepared 50 mM ammonium bicarbonate solution. The overnight digestion at 37°C was stopped by acidification with TFA. After centrifugation (16 000 x g for 5 min), the peptide mixture was removed from the gel pieces, transferred to a new Eppendorf tube, completely dried in a centrifugal vacuum concentrator and redissolved in 20 μl of 0.1% (w/v) TFA in water/acetonitrile (98/2, v/v) (loading solvent). 10 μl of the 20 μl obtained peptide mixtures mixtures were introduced into an LC-MS/MS system through an Ultimate 3,000 RSLC nano LC (Thermo Fisher Scientific) in-line connected to a Q Exactive mass spectrometer (Thermo Fisher Scientific). The sample mixture was first loaded on an in-house made trapping column (100 μm internal diameter [I.D.] × 20 mm, 5 μm beads C18 Reprosil-HD; Dr. Maisch, Ammerbuch-Entringen, Germany). After flushing from the trapping column, the sample was loaded on an in-house made analytical column (75 μm I.D. × 150 mm, 5 μm beads C18 Reprosil-HD; Dr. Maisch) packed in the needle (PicoFrit SELF/P PicoTip emitter, PF360-75-15-N-5; New Objective, Woburn, MA, USA). Peptides were loaded with loading solvent (0.1% (v/v) TFA in water/acetonitrile, 2/98 (v/v)) and separated with a linear gradient from 98% solvent A’ (0.1% (v/v) formic acid in water) to 40% solvent B′ (0.1% (v/v) formic acid in water/acetonitrile, 20/80 (v/v)) in 30 min at a flow rate of 300 nL/min, followed by a 5-min wash, reaching 99% solvent B’. Two packing and two analytical columns were configured in tandem LC mode. Switching between two flow paths, an analysis and a regeneration flow path, allows column washing and re-equilibration off-line; thus, while one column is re-equilibrated, the system injects a sample on the other column. The mass spectrometer was operated in data-dependent, positive ionization mode, automatically switching between MS and MS/MS acquisition for the 10 most abundant peaks in a given MS spectrum. The source voltage was 3.4 kV and the capillary temperature was 275°C. One MS1 scan (m/z 400-2,000, AGC target 3 × 106 ions, maximum ion injection time 80 ms) acquired at a resolution of 70,000 (at 200 m/z) was followed by up to 10 MS/MS scans (resolution 17,500 at 200 m/z) of the most intense ions fulfilling predefined selection criteria (AGC target 5 × 104 ions, maximum ion injection time 60 ms, isolation window 2 Da, fixed first mass 140 m/z, spectrum data type: centroid, underfill ratio 2%, intensity threshold 1.7×E4, exclusion of unassigned, 1, 5-8, >8 charged precursors, peptide match preferred, exclude isotopes on, dynamic exclusion time 20 sec). The HCD collision energy was set at 25% Normalized Collision Energy and the polydimethylcyclosiloxane background ion at 445.120025 Da was used for internal calibration (lock mass).
From the MS/MS data in each LC run, Mascot Generic Files (mgf) were created with the Mascot Distiller software (version 2.4.3.3; Matrix Science). These peak lists were then searched with the Mascot search engine and the Mascot Daemon interface (version 2.4, Matrix Science). Spectra were searched against the TAIR10 database. Variable modifications were set to pyro-glutamate formation of amino-terminal glutamine, acetylation of the protein N-terminus, methionine oxidation and propionamide cysteine formation. Mass tolerance on precursor ions was set to ± 10 ppm (with the Mascot C13 option set at 1) and on fragment ions to 20 mmu. The instrument setting was on ESI-QUAD. The enzyme was set to trypsin/P, allowing one missed cleavage, whereas cleavage was allowed also when lysine or arginine was followed by proline. Only peptides that were ranked first and scored above the threshold score, set at 99% confidence, were withheld. All data were managed by ms_lims 40 and analyzed with R (http://www.R-project.org) embedded in KNIME. The data were filtered by removing all peptides smaller than eight amino acids and only the proteins containing at least two peptides in one of the experiments were taken into account for data analysis.
CETSA
The protocol is largely based on the previously published procedure 10 with minor adjustments. Wild type or transgenic Arabidopsis PSB-D cell cultures were harvested, flash frozen in liquid nitrogen and ground with a Retsch® MM400. Cell material was added at a ratio of 2:1 in extraction buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 0.1% NP-40 with 1 tablet/10 mL cOmplete ULTRA protease inhibitor cocktail, EDTA free (Roche)). Cell material was allowed to thaw on ice for 15 min with occasional mixing. Samples were centrifuged for 30 min (18 000 x g at 4°C. Lysates were pooled per cell type and the protein concentration was determined with the Bradford method (Quick start Bradford 1× Dye reagent (Bio-Rad)). Pooled lysates were treated with small molecules or mock (DMSO) and incubated for 30 min at room temperature on a rotary wheel. The treated lysates were aliquoted in 60-μl fractions in PCR tubes, treated for 2 min at 12 temperature points (30, 35, 40, 42, 44, 46, 48, 50, 52, 55, 60, and 65°C) in a Bio-Rad thermal cycler, allowed to cool down and centrifuged (18,000 x g) for 30 min at 4°C. The supernatant (50 μl) was taken and processed for standard Western blot analysis to detect the proteins. All antibodies were diluted in TBS-T with 5% (w/v) skimmed milk. Anti-CHC (1/3.000, AS10 690 Agrisera) anti-AtpB (1/4.000, AS05085 Agrisera) were detected with horseradish peroxidase (HRP)-linked anti-rabbit IgG (1/10.000, GE Healthcare), anti-tubulin (1/15.000) with HRP-linkked anti-mouse IgG (1/10.000, GE Healthcare), anti-TPLATE (1/1.000) and HA with anti-HA-HRP (1/4.000, Abcam). Blots were developed with Western Lightning® Plus–ECL, Enhanced Chemiluminescence Substrate (Perkin-Helmer) and imaged with a Bio-Rad ChemiDoc XRS+ molecular imager. Band intensity was measured with the Bio-Rad Image Lab software package.
DARTS
The DARTS protocol was adopted as previously described 11,15 . Lysates were prepared as described for CETSA. Lysates treated with ES9-17 (250 μM) or mock (DMSO) were incubated for 30 min at room temperature. After incubation, lysates were split into equal aliquots for pronase (Roche, 10 165 921 001) digestion. Pronases were diluted from a 10 mg/ml stock solution in dH2O. The 1/100 starting dilution of pronase that was used for all the subsequent dilutions as indicated was obtained by dissolving 12.5 μl pronase stock solution in 87.5 μl 1× TNC buffer (500 mM Tris-HCl, pH 8, 500 mM NaCl, 100 mM CaCl2, 10× stock). All dilutions were prepared with 1× TNC buffer. Digestion (30 min) was started with 1-min intervals and stopped by addition of the sample buffer at a 1× final concentration (4× LDS sample buffer with 10× sample reducing agent; Novex, Life Technologies) in the same sequence as the digestion had been started. Western blotting, protein detection and quantification were as described for CETSA. The anti-γ-COP (anti-Sec21p) (1/1.000, AS08327, Agrisera) was detected with anti-rabbit IgG, HRP-linked antibody (1/10.000, GE Healthcare).
Molecular docking
Water and all ligands of the human clathrin nTD crystal structure pdb-entry 4G55 in complex with Pitstop2 (ref. 5) were manually deleted from the pdb-text file. The emptied structure was subjected to a local minimization with the GROMOS96 (43B1 parameter set) 41 implementation within the Swiss-PdbViewer 42 , and polar hydrogens were added. The ES9 ligand was drawn three-dimensionally with Avogadro 1.1.1 (ref. 43) and was minimized with the built-in MMFF94s force field 44 . The AutoDockTools 1.5.4 suite 45 was used for pdbqt-format preparations of proteins and ligands. Dockings were done with AutoDock-Vina 1.1.0 (ref. 46) with exhaustiveness set at 64; residues Arg64, Phe91 and Gln89 were set as flexible. The grid-box size was x = 20, y = 22 and z = 20 Å, centered at x = 50.0, y = -10.2 and z = 24.5. PyMOL (Molecular Graphics System, Version 1.7 Schrödinger, LLC) was used for visualization.
Cloning, expression and purification of human nTD CHC1 and Arabidopsis nTD CHC1/2
Arabidopsis nTD CHC1 (residues 1-377), CHC2 (residues 1-378) and human nTD CHC (residues 1-363) were cloned into the pGEX4T-1 vector and transformed into the competent Escherichia coli BL21 (DE3) cells. Transformed cells were cultured in Luria-Bertani medium supplemented with carbenicillin (100 μg/ml)at 37°C until an O.D. of 0.6 was reached and expression was induced by the addition of 1 mM isopropyl-1-thio-D-galactopyranoside (IPTG). Cell cultures were kept for another 4 h at 37°C. Next, cells were harvested by centrifugation at 6000 x g,, resuspended in lysis buffer (10 mM Tris-HCl, pH 8.3, 500 mM NaCl, 10% (w/v) glycerol and 5 mM DTT) supplemented with protease inhibitors (Roche) and lysed by sonication. The proteins were purified by affinity purification using a GSTrap FF 1-ml column. The column was first equilibrated with equilibration buffer (10 mM sodium phosphate buffer, 150 mM NaCl, pH 7.5) and the sample applied to the GSTrap column. The column was subsequently washed with equilibration buffer and the bound protein was eluted with the elution buffer (10 mM NaH2PO4, 150 mM NaCl, 10 mM reduced glutathione, pH 7.5) and collected in 1-ml aliquots. The GST tag was cleaved by overnight thrombin digest at 20° C, after which the sample was again applied to a GSTrap column to remove the cleaved GST as well as uncleaved GST-CHC. The flow through was collected and further polished by size exclusion chromatography with a SD75 16/600 column (GE Healthcare) equilibrated on HBS (20mM HEPES pH 7.4 150mM NaCl). Protein concentration (A280) was measured with a Nanodrop 1000 (Thermo Scientific) and aliquots at a concentration of 2 mg/ml were stored at –80°C.
X-ray data collection
Purification and crystallization of nTD were carried out as previously described 5 . X-ray data were collected at beamline BL14.2 at BESSY-II (Berlin, Germany) and processed in XDS and Xscale 47 . Data collection statistics are shown in Supplementary Table 3. The phase problem was solved by molecular replacement using phaser 48 using the 1.7 Å structure of nTD (PDB ID:4G55) as a model. All water molecules and ligand atoms were omitted from the starting model. Subsequent cycles of refinement to 1.6 Å resolution were performed in PHENIX 49 . Structure file of ES9 was generated using the Dundee PRODRG2 server 50 and manually fitted to the electron density. All structural figures were produced with PyMOL (Molecular Graphics System, Version 1.7 Schrödinger, LLC). The data were deposited in the PDB under ID: xxxx.
Differential scanning fluorimetry (DSF) assay
For the DSF assay, the Light cycler 480, Real-time PCR system (Roche) was used as described previously 49 . The purified nTD of human CHC1 and Arabidopsis CHC1/2 was diluted in a buffer containing 20 mM Hepes, pH 7.4, 150 mM NaCl. Each well of a 96-well microplate contained a concentration of 1.6 μM protein, 5x Sypro Orange (Invitrogen), 2.5 μl of compound and buffer up to a total volume of 25 μl. Thermal scanning 10 to 95°C at 1.5°C/min) was done with a real-time PCR setup and the fluorescence intensity was measured every 10 sec. The software Light cycler 480Sw 1.5.1 was utilized for calculating melting temperature.
Transferrin uptake in HeLa cell cultures
Maintenance and imaging of HeLa cells for transferrin uptake was performed as described previously 9 .
Cell viability assay
For the WST-1 assay, HeLa cells were grown in a 96-well plate and were incubated with the compounds for 30 min followed by addition of 10 μl WST-1 reagent (Sigma-Aldrich) to 100 μl of medium in a 96-well plate. Absorbance was measured at 450 nm versus a 690 nm reference by means of a plate reader.
Root Growth Assay
Seeds were sown on ½ MS solid medium, stratified for 2 days at 4 °C in the dark, and placed vertically in the light. At 5 days after germination, seedlings were transferred to solid ½ MS medium without sucrose supplemented with 100 mM D-sorbitol with ES9-17 or DMSO and incubated for another 2 days, after which the plates were scanned and root growth was measured. For measurements, scanned images were processed and evaluated with ImageJ. Fold change in root growth was measured as a ratio of root length on 2 days after treatment with ES9-17/DMSO to the root length at the start of the treatment.
TEM
Five-days-old Arabidopsis thaliana Col-0 seedlings, grown on solid ½ MS medium, were treated by immersing them in liquid ½ MS supplemented with DMSO and 30 μM ES9 17 for 30 min. Root tips were subsequently excised, immersed in 20% (w/v) BSA and frozen immediately in a high-pressure freezer (Leica EM ICE; Leica Microsystems, Vienna, Austria). Freeze substitution was carried out using a Leica EM AFS (Leica Microsystems) in dry acetone containing 1% (w/v) OsO4 and 0.5% glutaraldehyde over a 4-days period as follows: -90°C for 54 h, 2°C per hour increase for 15 h, -60°C for 8 h, 2°C per hour increase for 15 h, and -30°C for 8 h. Samples were then slowly warmed up to 4°C, rinsed 3 times with acetone for 20 min each time and infiltrated stepwise over 3 days at 4°C in Spurr’s resin and embedded in capsules. The polymerization was performed at 70°C for 16 h. Ultrathin sections were made using an ultra-microtome (Leica EM UC6) and post-stained in in a Leica EM AC20 for 40 min in uranyl acetate at 20°C and for 10 min in lead stain at 20°C. Sections were collected on formvar-coated copper slot grids. Grids were viewed with a JEM 1400plus transmission electron microscope (JEOL, Tokyo, Japan) operating at 60 kV.
Statistical tests and generation of graphs
All statistical tests and graphs other than boxplots were done and generated with Graphpad Prism 6. Dose-response curves with a log-transformed x-axis were generated using nonlinear regression with a log(inhibitor) vs. response model, and setting a variable slope (four parameters) and top and bottom constraints to 1 and 0 respectively. Other dose-response and CETSA curves were generated with a Boltzmann sigmoid equation with top and bottom constraints set to 1 and 0 respectively, when applicable. Boxplots were generated with the online tool BoxPlotR (http://boxplot.tyerslab.com/) from the Tyers and Rappsilber laboratories.
Life sciences reporting summary
Further information on experimental design and reagents is available in the Life Sciences Reporting Summary.
Supplementary Material
Any supplementary information, chemical compound information and source data are available in the online version of the paper.
Acknowledgements
We thank Steffen Vanneste for fruitful discussions, Rahul Kumar for providing the pDONR211-AP2S plasmid, Daniel Martinez Molina for help with the CETSA protocol and Martine De Cock for help in preparing the manuscript. This work was supported by the Research Foundation-Flanders (project no. G022516N to E.R. and the joint project G0E5718N to E.R. and J.F.), the European Research Council (ERC Co T-Rex grant 682436 to D.V.D), the Deutsche Forschungsgemeinschaft (TRR186/A08 to V.H.), the Agency for Innovation by Science and Technology (IWT) for a postdoctoral (K.M.) and predoctoral (W.D. and S.D.M) fellowship, the China Science Council for a predoctoral fellowship (Q.L.), the joint research projects (VS.025.13N and VS.095.16N) within the framework of cooperation between the Research Foundation-Flanders and the Bulgarian Academy of Sciences (K.M.) and the Belgian Science Policy Office (BELSPO) for a postdoctoral fellowship to non-EU researchers (I.S.).
Footnotes
Author contributions
W.D. and E.R. initiated the work; W.D., I.S., and E.R. designed the experiments; W.D., I.S., B.D., A.M., and J.W. did SAR; W.D., K.M., A.S. and K.G. did affinity purification and MS analysis; H.B. and V.H. performed the X-ray crystallography; I.S., S.D.M. and S.S. did in vitro binding assay; W.N. did molecular docking; W.D., Q.L and I.S. did CETSA; W.D., I.S. and Q.L. did DARTS; I.S., D.V.S., E.M., and D.V.D. did imaging and data analysis; I.S. did cloning and generated transgenic Arabidopsis cell cultures; W.D., I.S., A.D., and D.A. did ATP measurements; M.V. and J.F. contributed the HeLa cells assays; K. Y. generated the TPLATE antibody; R.D.R and Q.L. did TEM; W.D., I.S. and E.R. wrote the manuscript. All authors commented on the results and the manuscript.
Competing financial interests
The authors declare no competing financial interests: details accompany the online version of the paper.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Reynolds GD, Wang C, Pan J, Bednarek SY. Inroads into internalization: five years of endocytic exploration. Plant Physiol. 2018;176:208–218. doi: 10.1104/pp.17.01117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaksonen M, Roux A. Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2018;19:313–326. doi: 10.1038/nrm.2017.132. [DOI] [PubMed] [Google Scholar]
- 3.Mettlen M, Chen P-H, Srinivasan S, Danuser G, Schmid SL. Regulation of clathrin-mediated endocytosis. Annu Rev Biochem. 2018;87:871–896. doi: 10.1146/annurev-biochem-062917-012644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mishev K, Dejonghe W, Russinova E. Small molecules for dissecting endomembrane trafficking: a cross-systems view. Chem Biol. 2013;20:475–486. doi: 10.1016/j.chembiol.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 5.von Kleist L, et al. Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell. 2011;146:471–484. doi: 10.1016/j.cell.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 6.Macia E, et al. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 7.McCluskey A, et al. Building a better dynasore: the Dyngo compounds potently inhibit dynamin and endocytosis. Traffic. 2013;14:1272–1289. doi: 10.1111/tra.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elkin SR, et al. Ikarugamycin: a natural product inhibitor of clathrin-mediated endocytosis. Traffic. 2016;17:1139–1149. doi: 10.1111/tra.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dejonghe W, et al. Mitochondrial uncouplers inhibit clathrin-mediated endocytosis largely through cytoplasmic acidification. Nat Commun. 2016;7:11710. doi: 10.1038/ncomms11710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jafari R, et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc. 2014;9:2100. doi: 10.1038/nprot.2014.138. [DOI] [PubMed] [Google Scholar]
- 11.Lomenick B, et al. Target identification using drug affinity responsive target stability (DARTS) Proc Natl Acad Sci USA. 2009;106:21984–21989. doi: 10.1073/pnas.0910040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jelínková A, et al. Probing plant membranes with FM dyes: tracking, dragging or blocking? Plant J. 2010;61:883–892. doi: 10.1111/j.1365-313X.2009.04102.x. [DOI] [PubMed] [Google Scholar]
- 13.Ludwig A, Stolz J, Sauer N. Plant sucrose-H+ symporters mediate the transport of vitamin H. Plant J. 2000;24:503–509. doi: 10.1046/j.1365-313x.2000.00900.x. [DOI] [PubMed] [Google Scholar]
- 14.Van Leene J, et al. An improved toolbox to unravel the plant cellular machinery by tandem affinity purification of Arabidopsis protein complexes. Nat Protoc. 2015;10:169–187. doi: 10.1038/nprot.2014.199. [DOI] [PubMed] [Google Scholar]
- 15.Mishev K, et al. Nonselective chemical inhibition of Sec7 domain-containing ARF GTPase exchange factors. Plant Cell. 2018;30:2573–2593. doi: 10.1105/tpc.18.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitakura S, et al. Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis . Plant Cell. 2011;23:1920–1931. doi: 10.1105/tpc.111.083030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popova NV, Deyev IE, Petrenko AG. Clathrin-mediated endocytosis and adaptor proteins. Acta Naturae. 2013;5:62–73. [PMC free article] [PubMed] [Google Scholar]
- 18.Onizuka S, Ikewaki N, Shiraishi S. A mechanism by which propofol induces cytotoxicity. J Drug Metab Toxicol. 2017;8:230. [Google Scholar]
- 19.Poot M, et al. Analysis of mitochondrial morphology and function with novel fixable fluorescent stains. J Histochem Cytochem. 1996;44:1363–1372. doi: 10.1177/44.12.8985128. [DOI] [PubMed] [Google Scholar]
- 20.Dolman NJ, Kilgore JA, Davidson MW. A review of reagents for fluorescence microscopy of cellular compartments and structures, Part I: BacMam labeling and reagents for vesicular structures. Curr Protoc Cytom. 2013;65:12.30.1–12.30.27. doi: 10.1002/0471142956.cy1230s65. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y-S, Yoo C-M, Blancaflor EB. Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C- and N-termini of the fimbrin actin-binding domain 2. New Phytol. 2008;177:525–536. doi: 10.1111/j.1469-8137.2007.02261.x. [DOI] [PubMed] [Google Scholar]
- 22.Marc J, et al. GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell. 1998;10:1927–1940. doi: 10.1105/tpc.10.11.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teh O-k, Moore I. An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature. 2007;448:493–496. doi: 10.1038/nature06023. [DOI] [PubMed] [Google Scholar]
- 24.Dettmer J, Hong-Hermesdorf A, Stierhof Y-D, Schumacher K. Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis . Plant Cell. 2006;18:715–730. doi: 10.1105/tpc.105.037978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irani NG, et al. Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat Chem Biol. 2012;8:583–589. doi: 10.1038/nchembio.958. [DOI] [PubMed] [Google Scholar]
- 26.Friedrichsen DM, Joazeiro CAP, Li J, Hunter T, Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123:1247–1255. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ortiz-Morea FA, et al. Danger-associated peptide signaling in Arabidopsis requires clathrin. Proc Natl Acad Sci USA. 2016;113:11028–11033. doi: 10.1073/pnas.1605588113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gadeyne A, et al. The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell. 2014;156:691–704. doi: 10.1016/j.cell.2014.01.039. [DOI] [PubMed] [Google Scholar]
- 29.Kaksonen M, Roux A. Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2018;19:313–326. doi: 10.1038/nrm.2017.132. [DOI] [PubMed] [Google Scholar]
- 30.Di Rubbo S, et al. The clathrin adaptor complex AP-2 mediates endocytosis of brassinosteroid insensitive1 in Arabidopsis . Plant Cell. 2013;25:2986–2997. doi: 10.1105/tpc.113.114058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dhonukshe P, et al. Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis . Curr Biol. 2007;17:520–527. doi: 10.1016/j.cub.2007.01.052. [DOI] [PubMed] [Google Scholar]
- 32.Adamowski M, et al. A functional study of AUXILIN-LIKE1 and 2, two putative clathrin uncoating factors in Arabidopsis. Plant Cell. 2018;30:700–716. doi: 10.1105/tpc.17.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson DG, Pimpl P. Clathrin and post-Golgi trafficking: a very complicated issue. Trends in Plant Science. 2014;19:134–139. doi: 10.1016/j.tplants.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 34.Hicks GR, Raikhel NV. Small molecules present large opportunities in plant biology. Annu Rev Plant Biol. 2012;63:261–282. doi: 10.1146/annurev-arplant-042811-105456. [DOI] [PubMed] [Google Scholar]
- 35.Drakakaki G, et al. Clusters of bioactive compounds target dynamic endomembrane networks in vivo. Proc Natl Acad Sci USA. 2011;108:17850–17855. doi: 10.1073/pnas.1108581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang C, et al. Endosidin2 targets conserved exocyst complex subunit EXO70 to inhibit exocytosis. Proc Natl Acad Sci USA. 2016;113:E41–E50. doi: 10.1073/pnas.1521248112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kania U, et al. Endosidin 4 inhibitor targets the SEC7 1 domain-type ARF-GEFs and interferes with 2 subcellular trafficking in eukaryotes. Plant Cell. 2018;30:2553–2572. doi: 10.1105/tpc.18.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li R, et al. Different endomembrane trafficking pathways establish apical and basal polarities. Plant Cell. 2017;29:90–108. doi: 10.1105/tpc.16.00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharfman M, et al. Endosomal signaling of the tomato leucine-rich repeat receptor-like protein LeEix2. Plant J. 2011;68:413–423. doi: 10.1111/j.1365-313X.2011.04696.x. [DOI] [PubMed] [Google Scholar]
- 40.Helsens K, et al. ms_lims, a simple yet powerful open source laboratory information management system for MS-driven proteomics. Proteomics. 2010;10:1261–4. doi: 10.1002/pmic.200900409. [DOI] [PubMed] [Google Scholar]
- 41.Scott WRP, et al. The GROMOS biomolecular simulation program package. J Phys Chem A. 1999;103:3596–3607. [Google Scholar]
- 42.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 43.Hanwell MD, et al. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminformatics. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halgren TA. MMFF VI. MMFF94s option for energy minimization studies. J Comput Chem. 1999;20:720–729. doi: 10.1002/(SICI)1096-987X(199905)20:7<720::AID-JCC7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 45.Sanner MF. Python: a programming language for software integration and development. J Mol Graph Model. 1999;17:57–61. [PubMed] [Google Scholar]
- 46.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schüttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 51.Huynh K, Partch CL. Analysis of protein stability and ligand interactions by thermal shift assay. Curr Protoc Protein Sci. 2015;79:28.9.1–28.9.14. doi: 10.1002/0471140864.ps2809s79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.