Abstract
The biological basis of male-female brain differences has been difficult to elucidate in humans. The most striking morphological difference is size, with males having on average a larger brain than females 1,2 , yet a mechanistic understanding of how this difference arises remains to be elucidated. Here, we use brain organoids 3 to demonstrate that while sex chromosomal complement has no observable effect on neurogenesis, sex steroids, namely androgens, lead to increased proliferation of cortical progenitors and an increased neurogenic pool. Transcriptomic analysis and functional studies demonstrate downstream effects on HDAC activity and mTOR pathway. Finally, we show that androgens specifically increase neurogenic output of excitatory neuronal progenitors, while inhibitory neuronal progenitors are not increased. These findings uncover a hitherto unknown role for androgens in regulating excitatory neuron number and represent a first step towards understanding the origin of human sex-related brain differences.
Introduction
Sex-based brain differences are controversial, yet negating their existence has detrimental consequences for all sexes and genders, and for developing accurate diagnostic tools and effective drugs. One of the most striking differences is divergent susceptibility to certain neurological conditions 4,5 , for example, autism spectrum disorder (ASD) 6 and schizophrenia 7 , pointing to differences in brain structure and/or function. Indeed, decades of research in model organisms has revealed numerous sex-specific morphological and behavioural differences 8–10 . However, the presence of sex-specific characteristics in humans has remained a topic of heated debate, particularly since such differences are not dimorphic, but rather a continuum with substantial overlap 1 . Nonetheless, quantitative differences in size are well established, with adult males having on average larger brain volume, increased neuronal density 11 and increased grey matter in several brain regions 1,2 . The neonatal presence of size differences, even after correcting for birth weight 12 , speaks to a developmental origin. Yet, how these size differences arise remains to be determined.
Sexual differentiation has two main sources: cell-intrinsic chromosomal complement and circulating sex hormones 13 . The effects of sex hormones on brain development have been studied more extensively in mice 14 , where testosterone has its masculinising effects through its conversion to estradiol. Thus, counter intuitively, it is estradiol, and not testosterone, which masculinises the mouse brain 15 . However, studies in nonhuman primates point to divergent mechanisms 16 , and the binary nature of rodent brain sexualisation is strikingly different from humans. Furthermore, studies have mainly focused on the sexually dimorphic nuclei, but the underlying mechanism for the most striking difference in humans, brain size, remains to be examined.
Results
Androgens increase basal progenitors
To gain an understanding of developmental events that may lead to male-female differences in the human context, we used cerebral organoids, which provide a reliable model of progenitor behaviour and neurogenesis 3 We compared male and female organoids and assessed neural progenitor responses to sex hormones (Fig. 1a, Extended Data Fig. 1-2). We saw no differences in these measures between organoids made from female (XX) and male (XY) cell lines, suggesting intrinsic sex chromosomal makeup does not influence forebrain neurogenesis, at least in this model system.
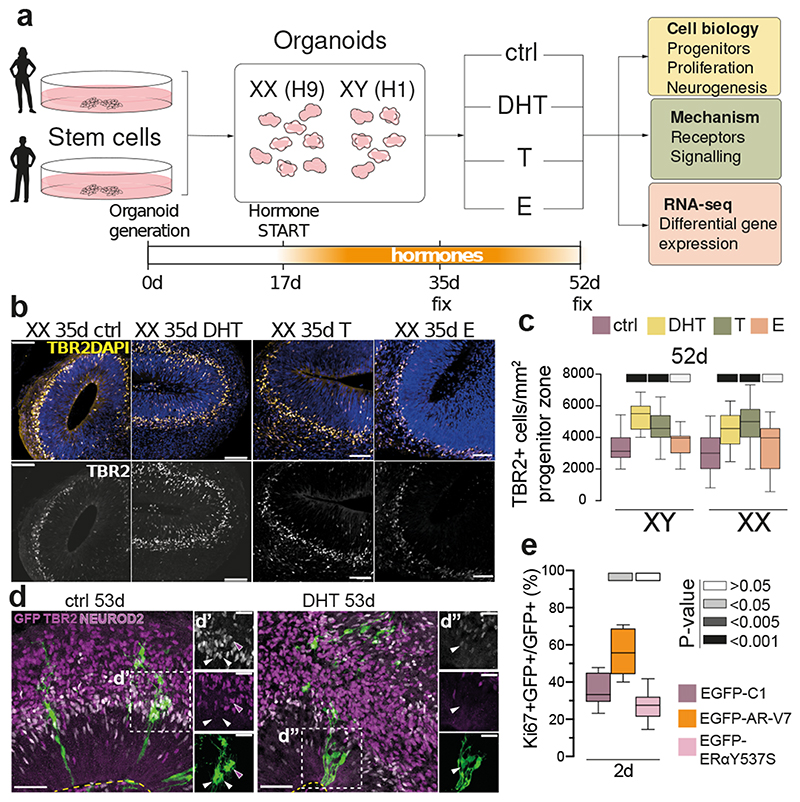
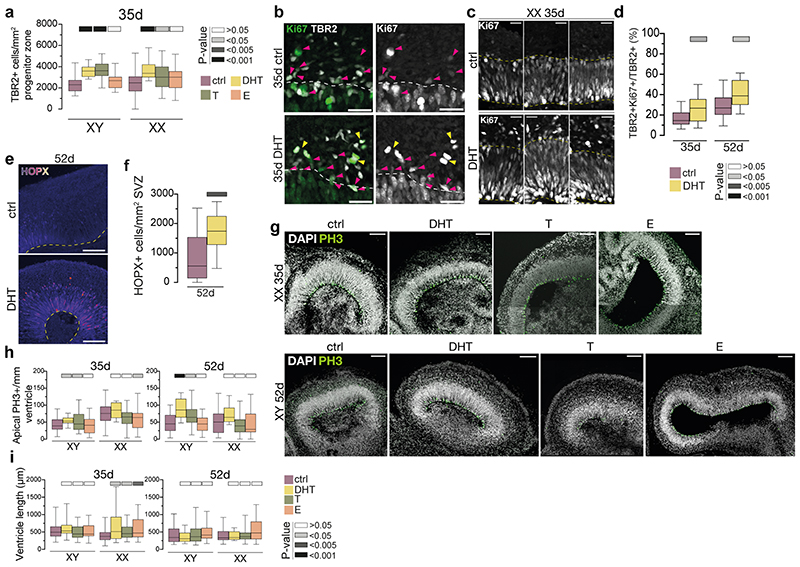
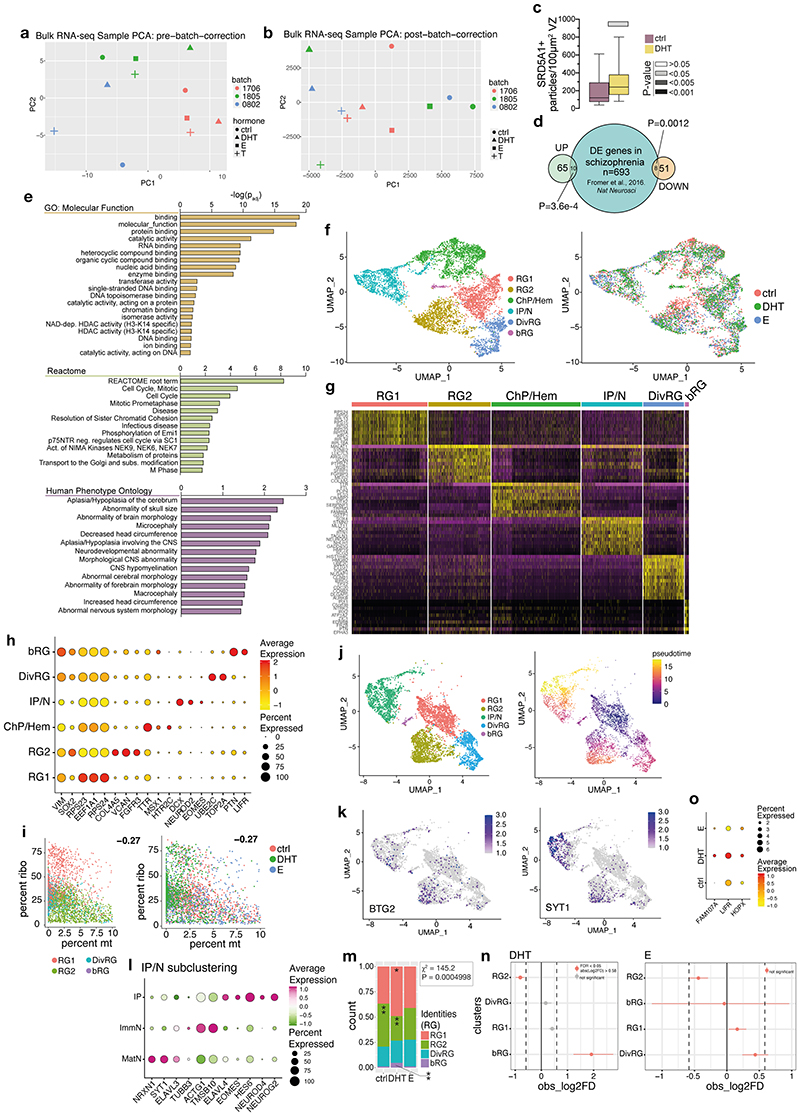
Figure 1. Androgens lead to expanded basal progenitors through increased radial glial stem cell proliferation.
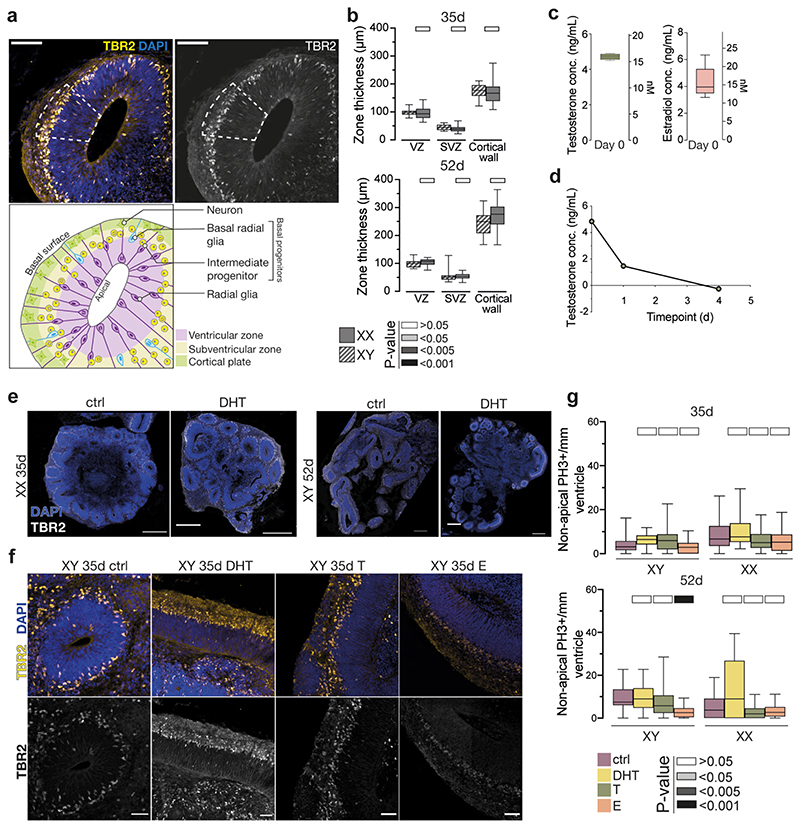
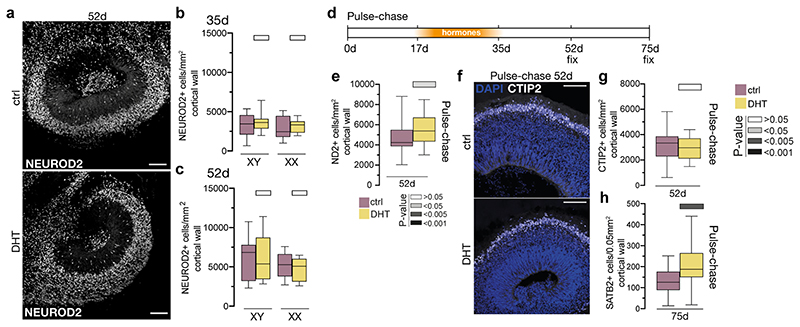
a) Schematic of the project workflow and treatment timeline. b) Female (XX) treated organoids stained for the intermediate progenitor marker TBR2 and DAPI. c) Quantification of TBR2+ cells per mm2 progenitor zone (see Extended Data Fig. 1a). Grey bars refer to P values as in panel e. d) Treated XX organoids injected with Sendai emGFP at 45d, fixed at 53d and stained for GFP, TBR2 and NEUROD2. Yellow dashed lines indicate ventricular surface. White arrowheads: TBR2/GFP double positive cells. Magenta arrowhead: NEUROD2/GFP double positive cell. Note increased GFP+ cells in the VZ upon DHT treatment. e) Quantification of GFP+ cells co-staining for the proliferation marker Ki67 at 2 days post electroporation of indicated constructs in XX organoids. Scale bars: b), d) 50μm, d’), d”) 25μm. See Methods for details of statistics and Supplementary Table 5 for details of n numbers.
Next, we exposed brain organoids to the androgens testosterone (T) and dihydrotestosterone (DHT), and to estradiol (E) (Fig. 1a, see Methods). DHT is a more potent androgen and cannot be aromatised into E, as opposed to T 17 . In both male and female organoids, androgens (DHT or T) led to an increase in intermediate progenitors, which are basally located cells that produce excitatory cortical neurons (Fig. 1b, c, Extended Data Fig. 2a). This was not seen upon treatment with E. Staining for the proliferative marker Ki67 revealed an increased proportion of intermediate progenitors that remained proliferative upon DHT treatment (Extended Data Fig. 2b-d). In addition, we observed an increase in the number of another basal progenitor type known as basal radial glia 18 in DHT-treated organoids (Extended Data Fig. 2e, f). Taken together, these findings suggest that androgens increase the numbers and proliferation of basal progenitors.
Increased radial glial proliferation
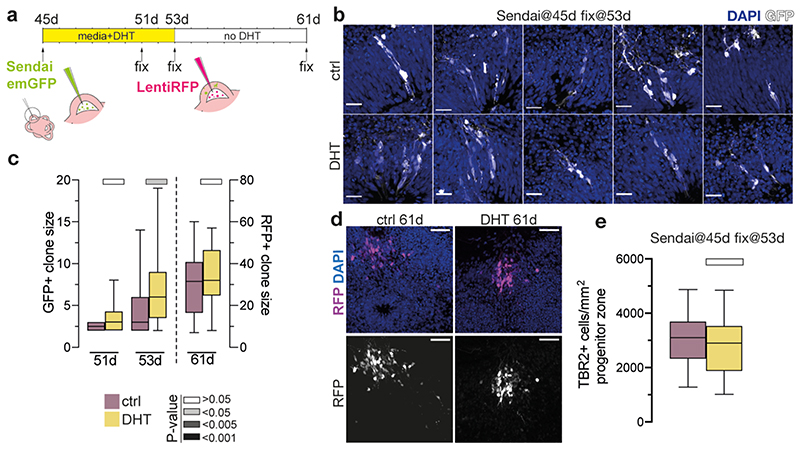
We next examined the neural stem cells, called radial glia, that give rise to basal progenitors, revealing features suggestive of increased proliferation (Extended Data Fig. 2g-i). Lineage tracing using viral sparse labelling revealed increased radial glial clone size upon 8 days of DHT treatment (Fig. 1d, Extended Data Fig. 3). Removal of the hormone and labelling with a different virus resulted in newly-labelled clones of comparable size in control and previously DHT-treated organoids (Extended Data Fig. 3). These results indicate that androgen acts specifically and reversibly to promote proliferative divisions of radial glial stem cells.
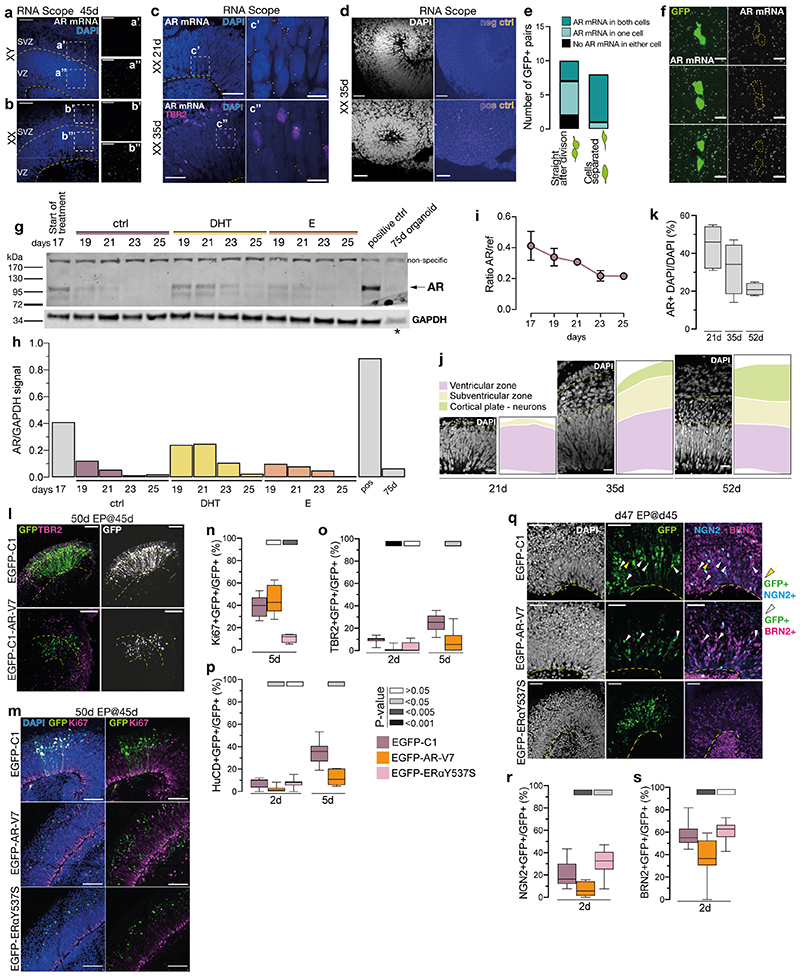
Upon binding androgen, androgen receptor (AR) translocates to the nucleus where it acts as a transcription factor 19 . Fluorescent in situ hybridization with RNAScope 20 for AR mRNA revealed widespread signal in radial glia, but not in more superficial zones containing differentiated cells (Extended Data Figure 4a-d). The distribution of the signal within radial glia varied depending on cell cycle stage (Extended Data Fig. 4e-f). Western blot (Extended Data Fig. 4g, h) and quantitative droplet digital PCR (Extended Data Fig. 4i) revealed a decline in AR over time, likely due to increased numbers of differentiated cells (Extended Data Fig. 4j) not expressing AR and hence a decrease in the proportion of AR-expressing cells over time (Extended Data Fig. 4k). In contrast, organoids exposed to DHT showed a more sustained expression of AR protein (Extended Data Fig. 4g, h) in accordance with previous observations 21 .
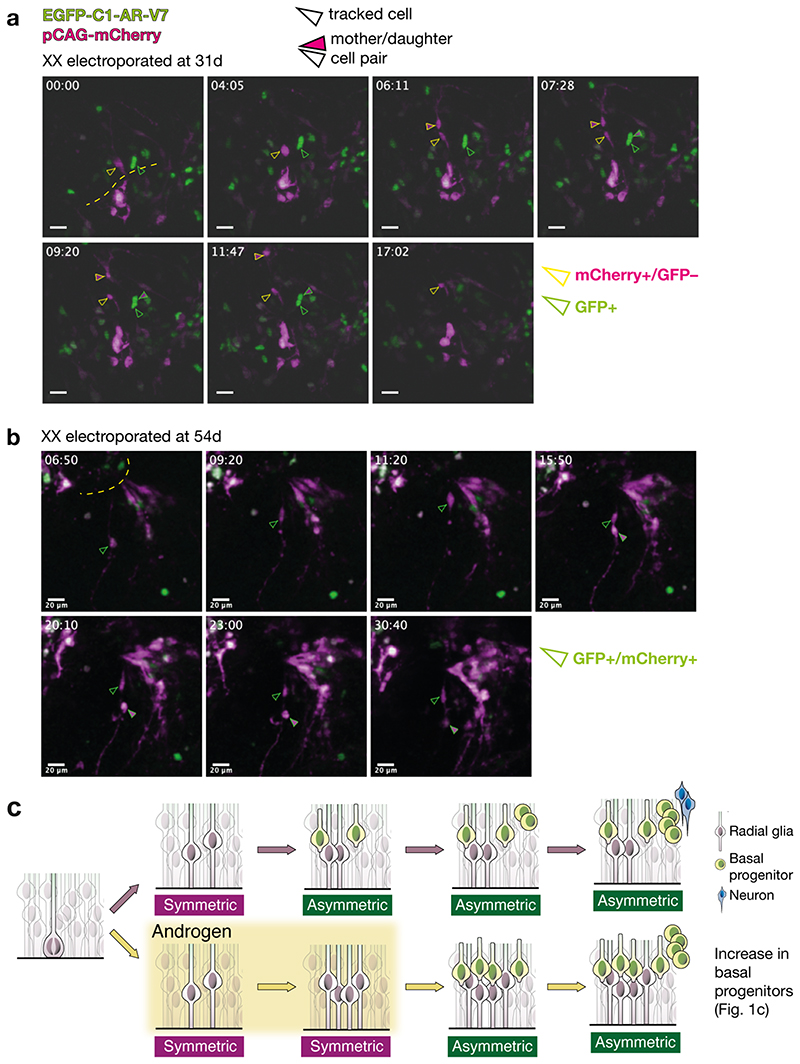
We next introduced a GFP-tagged constitutively active AR, AR-V7 22 , which showed the expected nuclear localisation with most cells remaining in the radial glial ventricular zone, whereas in control organoids, GFP-labelled cells were distributed in both ventricular and more differentiated basal compartments (Extended Data Fig. 4l). Ki67 staining, a marker of cycling cells (Extended Data Fig. 4m, n), revealed a ~60% increase in proliferation (Fig. 1e), while differentiation markers were decreased in cells expressing AR-V7 (Extended Data Fig. 4o-s). To test the specificity of this effect, we expressed a constitutively active form of another sex hormone receptor, estrogen receptor alpha (ERα) 23 . Unlike AR-V7, expression of GFP tagged constitutively active ERαY537S revealed proliferation status similar to control (Fig. 1e, Extended Data Fig. 4m-s). Live imaging revealed symmetric proliferative divisions of GFP tagged AR-V7 positive cells in contrast to the asymmetric differentiative divisions of control cells (Extended Data Fig. 5a, b, Supplementary Video 1-3). Taken together, these findings reveal a proliferative action of AR signalling on cortical progenitors leading to their increased numbers as well as increased progeny upon release from signalling (Extended Data Fig. 5c).
Androgen induced gene expression changes
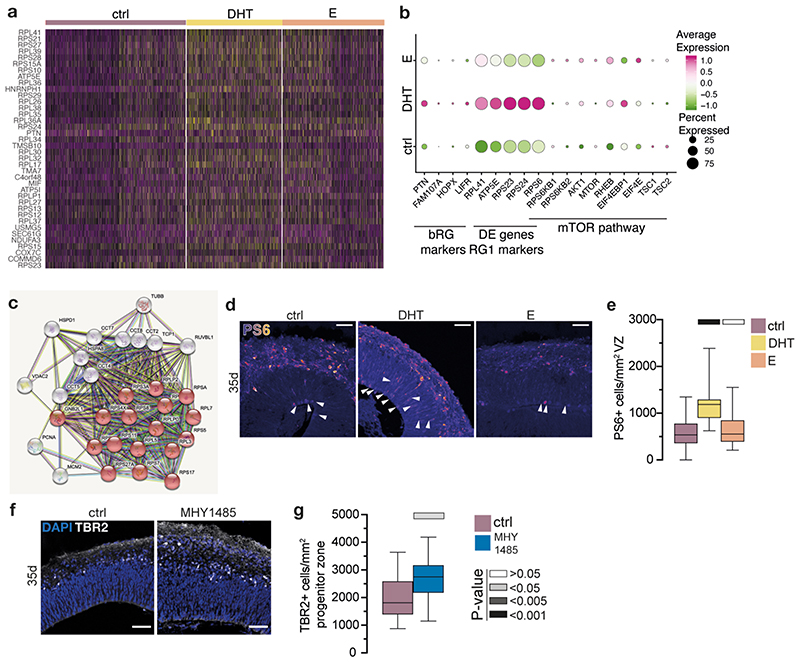
Androgen induced changes in gene expression have previously been reported to be subtle 21 . Therefore, to capture the downstream response with enough transcriptomic coverage we performed bulk RNA-seq on control and treated organoids (Extended Data Fig. 6a, b). To detect differentially expressed genes (DEGs), we employed a new method 24 designed to ensure high sensitivity while controlling for false discovery rate (see Methods). We identified 65 up-regulated and 51 down-regulated genes upon androgen treatment (Fig. 2a, Supplementary Table 1). Up-regulated genes included several with roles in neural development: HDAC2 25 , HDAC3 26 , YBX1 27 , EML1 28 . It was encouraging to find SRD5A1, the enzyme that converts T into DHT 17 , upregulated, which we validated by immunostaining (Extended Data Fig. 6c). Several of the DEGs were associated with ASD 29 (MCM4, PHB) and/or schizophrenia (PARK2 30 , SELENBP1 31 , GABBR1 32 , SPTLC2 33 ) (Fig. 2a, Extended Data Fig. 6d). Gene ontology 34 analysis of downregulated genes showed very low enrichment scores, while upregulated genes were enriched in terms associated with brain size, developmental disorders, and chromatin and HDAC activity (Extended Data Fig. 6e).
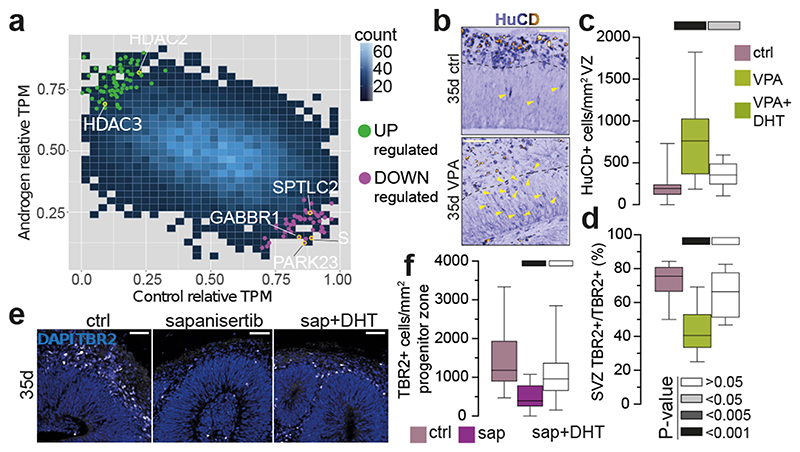
Figure 2. Transcriptomics reveal involvement of HDACs and mTOR in androgen-mediated phenotype.
a) Normalised TPM values from bulk RNA-seq plotted for control and androgen treatment. DEGs detected using Delboy 24 . Green: upregulated DEGs, magenta: downregulated DEGs. Blue shade indicates data density (count). b) HuC/D immunostaining in treated XX 35d organoids. Yellow arrowheads: HuC/D+ cells in the VZ. c) Quantification of HuC/D+ cells per mm2 VZ in treated XX 35d organoids. d) Percentage of TBR2+ cells in the SVZ, out of all TBR2+ cells, for XX 35d treated organoids. e) Immunostaining for TBR2 and DAPI in control, sapanisertib and sapanisertib+DHT treated 35d XX organoids. f) Quantification of TBR2+ cells per mm2 progenitor layer in control, sapanisertib and sapanisertib+DHT treated 35d XX organoids. Scale bars: b), e) 50μm. See Methods for details of statistics and Supplementary Table 5 for details of n numbers.
Single cell RNA sequencing (scRNA-seq) at the same stage identified 6 major clusters (Extended Data Fig. 6f-h), including 4 subtypes of radial glia: dividing (DivRG), basal radial glia (bRG), a cluster (RG1) with high levels of ribosomal biogenesis transcripts but low mitochondrial counts (Extended Data Fig. 6g, i) and another (RG2) expressing extracellular matrix factors, growth factor receptors, and maturation markers (Extended Data Fig. 6g, Supplementary Table 2). Pseudotime analysis demonstrated a progression from RG1 to RG2 to IP/N (intermediate progenitor/neurons) (Extended Data Fig. 6j), suggesting RG2 represented more committed radial glia (Extended Data Fig. 6k). The IP/N cluster contained both intermediate progenitors and neuronal populations, which separated upon further subclustering (Extended Data Fig. 6l). While there was very good overlap between different organoids across treatments (Extended Data Fig. 6f), indicating reproducibility, DHT treated organoids displayed increased RG1 and bRG clusters (Extended Data Fig. 6m, n), consistent with increased radial glial proliferation and increased basal progenitors (Extended Data Fig. 2e, f, 6o). Differences in other RG clusters were not detected but this may relate to statistical power and the numbers of cells sequenced.
Downstream HDACs affect differentiation
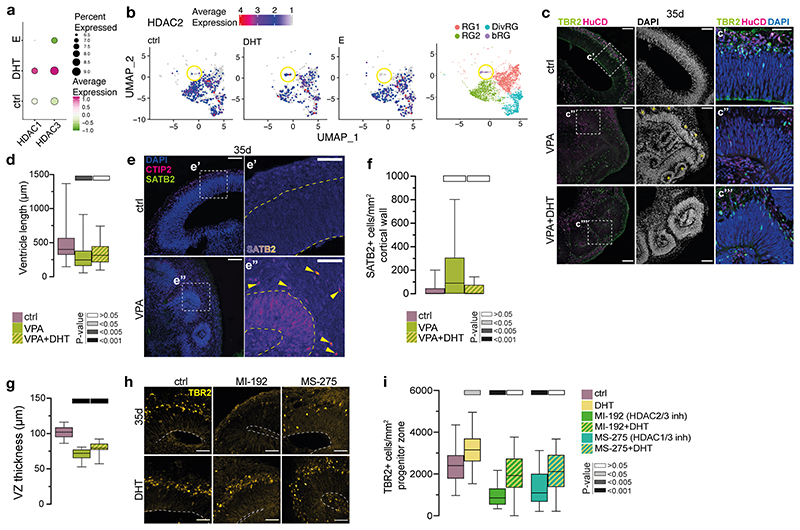
We examined levels of DEGs identified by bulk RNA-seq in the context of cellular subtypes afforded by scRNA-seq, revealing increased HDAC1 and 3 upon DHT treatment (Extended Data Fig. 7a), as well as increased HDAC2 in bRG cells (Extended Data Fig. 7b). We tested for a functional interaction between HDACs and androgens by treating organoids with valproic acid (VPA), a potent inhibitor of HDACs 35 . VPA-treated organoids had smaller ventricles and increased neurons in the ventricular zone (Fig. 2b, c, Extended Data Fig. 7c, d), as well as premature production of later-born upper layer neurons (Extended Data Fig. 7e, f). VPA-treated organoids also exhibited a thinner ventricular zone (Extended Data Fig. 7g) and reduced intermediate progenitors (Fig. 2d). While administration of DHT together with VPA did not rescue the thickness of the ventricular zone (Extended Data Fig. 7g), it did rescue the reduced ventricular length and returned intermediate progenitor distribution to control levels (Fig. 2d, Extended Data Fig. 7d). Treatments with specific inhibitors MI192 36 (HDAC2/3 inhibitor) and MS275 37 (HDAC1/3 inhibitor) provoked a similar response (Extended Data Fig. 7h, i). These results indicate that HDACs, like androgens, inhibit differentiation, and the ability of DHT to rescue phenotypes also affected by androgen signalling suggests these pathways act together in cortical neurogenesis.
Biogenesis and mTOR signalling
We next performed differential gene expression analysis in our scRNA-seq dataset, identifying 38 transcripts that were increased in DHT treated organoids (Supplementary Table 3). Among these was a marker of bRGs, PTN 18 , consistent with increased bRG cells, as well as ribosome biogenesis genes overlapping with markers of RG1 cells, consistent with their increase in DHT treated organoids (Extended Data Fig. 8a, b). Re-examination of bulk RNA-seq for interaction partners 38 also confirmed these ribosomal gene enrichments (Extended Data Fig. 8c), as well as several DEGs directly involved in translation or ribosomal complex assembly (EIF2B3, RPL6, PES1, RSL24D1, EEF1A1). An increase in biogenesis and the presence of certain metabolic markers (ATP5E, ATP5I, USMG5, NDUFA3) could be indicative of increased mTOR signalling 39 . Indeed, we observed an increase in some individual mTOR-related transcripts in scRNA-seq (Extended Data Fig. 8b).
mTOR signalling is well described in neural progenitor cells where it is involved in brain size determination 40 . Upon mTOR activation, S6 ribosomal protein becomes phosphorylated 41 . Staining for phosphorylated S6 (PS6) in DHT treated organoids revealed increased PS6+ radial glia (Extended Data Fig. 8d, e). Sapanisertib, an inhibitor of both mTORC1 and mTORC2 42 , caused a significant decrease in the number of intermediate progenitors, which was rescued by the addition of DHT (Fig. 2e, f). Conversely, activation of mTOR signalling by an mTOR signalling activator, MHY1485 43 , phenocopied the effect of androgens by increasing the number of intermediate progenitors (Extended Data Fig. 8f, g). These drug interaction experiments thus confirmed the role of HDACs and mTOR signalling in the androgen-induced increase in progenitors.
Inhibitory neurogenesis response
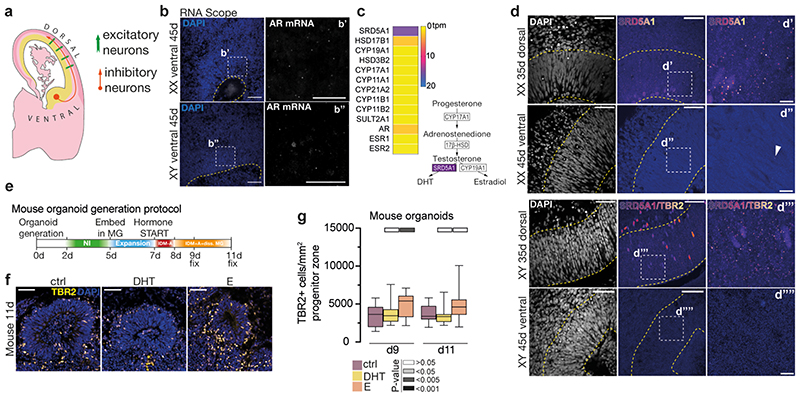
There is increasing evidence in both ASD and schizophrenia for an imbalance in excitatory and inhibitory neurons 44 , raising the question of whether androgens may also regulate inhibitory neurogenesis. Because inhibitory neurons are generated outside the cortex, in the ventral forebrain 45 (Extended Data Fig. 9a), we ventralised organoids and treated with sex steroid hormones, revealing no change or a slight decrease in ventral intermediate progenitors upon DHT treatment (Fig. 3a, b) while E treatment had no effect. RNAScope demonstrated much lower levels of AR in ventral forebrain (Extended data Fig. 9b). A hallmark of androgen-responsive cells is expression of SRD5A1 46 . Bulk RNA-seq and immunostaining showed expression of SRD5A1 in dorsal organoids (Extended Data Fig. 9c, d), whereas ventral organoids had almost no SRD5A1 staining (Extended Data Fig. 9d). Together, these data indicate lower androgen responsiveness in inhibitory neural progenitors.
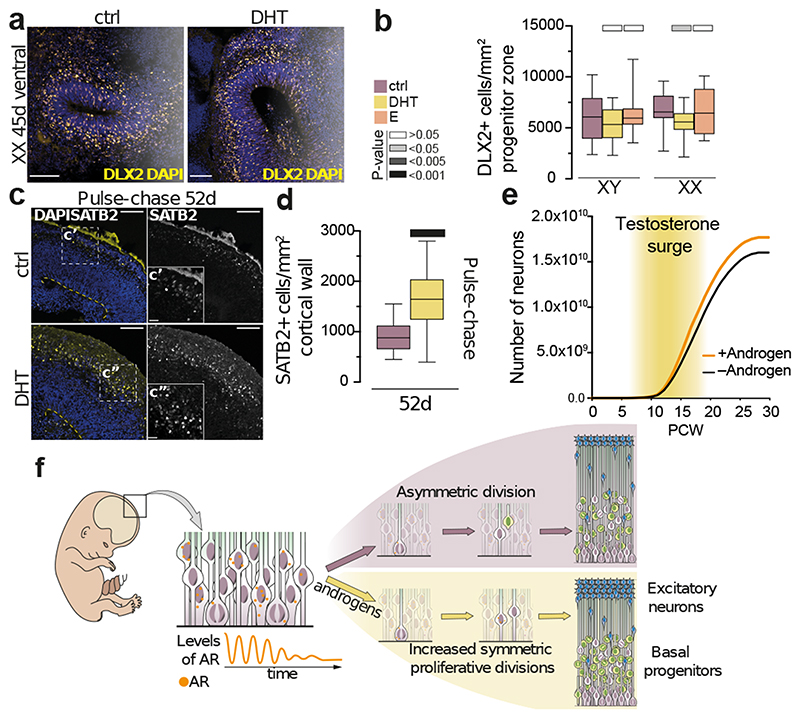
Figure 3. Specific increase in excitatory neurogenesis upon androgen surge.
a) Immunostaining for ventral intermediate progenitor marker DLX2 and DAPI on XX 45d ventralised organoids. b) DLX2+ cells per mm2 progenitor layer for treated XX and XY organoids. c) SATB2 immunostaining and DAPI after pulse-chase treatment. Yellow dashed line indicates the ventricular surface. d) SATB2+ cells per mm2 cortical wall after pulse-chase treatment. e) Growth curve of the predicted effect of androgens on neuron number (9.4% increase). f) Proposed model: Radial glia express androgen receptor intermittently and decreasing over time. Androgens act on radial glia to increase proliferation, increasing the stem cell pool. As AR and the fetal surge drop over time, these effects weaken and the increased radial glia give rise to increased basal progenitors and finally to increased excitatory neurons. Scale bars: a) 50μm, c) 100μm, c’) c”) 25μm. See Methods for details of statistics and Supplementary Table 5 for details of n numbers.
Interestingly, we observed no expression of aromatase (CYP19A1), an enzyme that converts testosterone to estrogen 17 (Extended Data Fig. 9c), or estrogen receptors (ESR1 and ESR2), supporting a nonresponsive state to estrogen as we observed, and in contrast to the mouse 15,47 . The differences between these observations in human tissue and previous studies in mice prompted us to explore the potential evolutionary divergence between these two species using organoids. We observed that in mouse organoids, it was E, and not DHT that caused an increase in intermediate progenitors (Extended Data Fig. 9e-g), in contrast to human.
Androgens increase neuron number
The enrichment for ontology terms associated with brain size (Extended Data Fig. 6e), as well as the fact that males on average exhibit a larger brain size, pointed to a role for androgens on neuron number. With constant androgen treatment, we did not observe any change in neuronal numbers despite the increase in basal progenitors (Extended Data Fig. 10a-c). We hypothesised that, due to the continuous supply of androgens, the progenitors were maintained in a proliferative state. We therefore performed a pulse-chase experiment, not unlike the fetal testosterone surge, in which organoids were subjected to DHT for 18 days, followed by 17 days without DHT (Extended Data Fig. 10d) resulting in a subtle, but significant increase in neurons (Extended Data Fig. 10e). Staining for subsets of neurons revealed no change in early-born deep-layer neurons (Extended Data Fig. 10f, g), but an increase in later-born upper-layer neurons (Fig. 3c, d), which persisted if organoids were left to develop further (Extended Data Fig. 10h). This subtype specificity may be due to the timing of hormone treatment, with the androgen pulse preventing differentiation during deep-layer neurogenesis, while androgen withdrawal during upper-layer neurogenesis would enable progenitor “release” from the proliferative state.
Growth curve modelling to predict what effect these measured differences in neurogenesis would have on excitatory neuron number revealed a predicted overall increase of 9.4% (Fig. 3e), similar to the measured average increase of 9.8% in males 2 . These findings demonstrate that a transient androgen-induced switch towards more proliferative divisions of neural progenitors would result in a later increase in excitatory cortical neuron numbers.
Discussion
Because sex-based brain differences in humans have remained controversial, we sought to provide some clarity to this issue from a cell biological and mechanistic standpoint. Our findings reveal a shift towards proliferative divisions of radial glial stem cells upon androgen exposure, leading to an eventual increase in neuron numbers in both male and female brain organoids (Fig. 3f). This reveals a primarily cell-extrinsic driven sex difference determined by the presence of androgens, which would fit with the non-binary nature of brain sex-related phenotypes 48 , since female fetuses also possess low levels of androgens coming from the developing adrenal gland and potentially from the placenta 49 . The subtle, cell stage-dependent, and dynamic nature of the androgen response calls for future studies, especially in the context of different brain areas where females exhibit a larger size 2 .
In rodents, androgens are thought to primarily act through their conversion to estrogen during brain development, and in line with this, our mouse organoid assays revealed no effect of DHT while estrogen led to increased basal progenitors. This discrepancy with the human data points to primate-rodent divergence as further supported by nonhuman primate studies 16 . However, further studies of the role of estrogen itself on other stages or processes during human brain development are warranted.
Downstream of androgens, our functional studies point to HDAC and mTOR signaling in mediating the effect on neural progenitors. HDAC and mTOR activity have been linked to ASD and schizophrenia 50,51 , both of which also exhibit a sex-bias and present with sex-differential manifestations. Interestingly, we observed a specific effect for androgens on excitatory, but not inhibitory neurogenesis, pointing to a role in regulating the balance of excitatory to inhibitory neurons. Along these lines, there is some evidence that ASD and schizophrenia may share a pathophysiological mechanism of excitatory to inhibitory neuron imbalance 44 . Thus, our findings are able to consolidate these various observations and point to androgens as a potential factor in these conditions as well as revealing a role in neuron number, which would impact brain size.
Methods
Human stem cell maintenance and organoid generation
Male (XY) and female (XX) stem cells (male - H1 (WiCell WA01) and female - H9 (WiCell WA09)) were approved for use in the project by the UK Stem Cell Bank Steering Committee. Cells were cultured on Matrigel® (Corning, 356234) – coated plates and maintained in StemFlex™ medium (Thermo Fisher, A3349401). We used the commercial kit (STEMdiff™ Cerebral Organoid Kit, StemCell Technologies 08570) with previously described modifications to increase embryoid body surface area 52 , namely microscaffolds (18000 cells seeded) or smaller EBs (2000 cells seeded). Organoids were transferred to the orbital shaker at 57 rpm (f25 cm) at 15d (±1 day, depending on the morphology). Matrigel® was removed manually between days 14-17, after visual inspection. Organoids were transferred to IDM+A supplemented with diluted Matrigel® on day 30 and kept at those conditions until fixation. Ventral organoids were generated as previously described 53 , with ventralizing factors (2.5 μM IWP-2, Sigma, I0536; 100 nM SAG, Sigma, 566661) supplemented in the neural induction (NI) medium for 2 days.
Human hormone and drug treatments
All steroid hormones were diluted in 96% ethanol (EtOH) to obtain stock solution. Organoids were treated with 30 nM or 100 nM dihydrotestosterone (DHT, Sigma A8380-1G), 100 nM testosterone (Sigma, T1500), 100 nM estradiol (active form of estrogen) (Sigma, E1024) and 100 nM progesterone (Sigma, P8783). 96% ethanol was used as a vehicle control (1.4 μl of 1:10 absolute ethanol (EtOH) in PBS to 5mL of media, which does not impact the normal development of the neuroepithelium). The treatments started on day 17 of the protocol. Treatments were started at day 17 when organoids morphologically resemble the time point when the male human testes begin to produce the testosterone surge (~8 gestational weeks) 54 . A low dose of hormones (~20 nM) did not yield an observable phenotype. However, with an increased amount of 100 nM as used in previously published reports 21 , we measured by ELISA an actual concentration of ~16nM, which is within the physiological range detected during the fetal testosterone surge 55 (Extended Data Fig. 1c). This decrease is likely due to the known affinity of steroid hormones for plastic, and thus sequestration in the culture vessel plasticware 56 , as well as short hormone half-life 57 , which was similarly observed in this context (Extended Data Fig. 1d). We therefore continued with this dose, except in the case of DHT where a lower dose was used due to its increased potency. Organoids exposed to 100 nM DHT for prolonged periods of time exhibited impaired morphology at later stages (increased cell death and general cytoarchitectural disturbances, data not shown), so we reduced the amount of DHT to 30 nM. This amount was chosen because the potency of DHT is roughly three times that of testosterone 58 , meaning any effects caused by 100 nM T and 30 nM DHT would be comparable.
Hormone treatments were administered twice a week, with changing of the media. Organoids were collected for analysis at 35 (period of extensive deep-layer neuron production), 52 days (production of upper-layer neurons) and 75 days (late time point for pulse-chase experiments). Ventral organoids were collected at 45 days, as a mid-point between the time points analysed for dorsal organoids. Manipulations (Sendai injections, electroporations, see below) were performed at 45 days, due to younger organoids’ ventricles being difficult to access. After establishing the phenotype in both cell lines, and since we observed no significant difference between cell lines in their response to different hormones (two-way ANOVA P=0.23), we focused on the female cell line for subsequent experiments, as it showed greater batch-to-batch consistency.
For mechanistic experiments of the androgen-induced phenotype and the investigation of candidate genes/pathways, we have omitted the treatments with T, as in our system, both T and DHT provoked a very similar response.
The application of progesterone to the organoids did not yield any observable phenotype (data not shown). This also eliminated the possibility that progesterone present in the commercially available media (present at 20 nM in N2 and B27 supplements, which are routinely added to the organoid media) might have an effect on organoid development.
For continuous hormone treatments (Sendai/Lenti virus labelling experiments), hormone treatments started immediately after labelling (day 30 or day 45) and were administered every day with the change of media.
All drug treatments were started at 17d and continued through until 35d, with fresh drug supplementation twice a week during media change. DHT was added at 30 nM in the drug+DHT experiment. Organoids were collected for analysis at 35d. The following drugs were used: valproic acid (VPA) (sodium valproate; Stratech, S1168-SEL-200mg; used at 0.7 mM, which is close to therapeutic doses 59 ), MS275 (Selleckchem 209783-80-2, used at 200 nM), MI-192 (Tocris 5647, dissolved in DMSO and used at 200 nM), sapanisertib (Selleckchem 1224844-38-5, dissolved in DMSO and used at 100 nM), MHY1485 (Sigma, SML0810, used at 2 μM).
The pulse-chase experiment was started at 17 days, and hormones were removed from the media at 35 days. Organoids were then grown in IDM+A with dissolved Matrigel® and fixed at 52 and 75 days.
ELISA
ELISA (enzyme-linked immunosorbent assay) kits for testosterone (Abcam, ab108666) and estradiol (R&D Systems, KGE014) were performed as per manufacturer’s instructions. The day of the hormone addition was designated as d0. Media samples were collected ~10’ after the initial addition of hormones and over 4 days, at a similar time of the day from organoid cultures containing 2 organoids per dish. Absorbance was measured using CLARIOstar (BMG Labtech).
Mouse stem cell maintenance, organoid generation
Mouse embryonic stem cells (ES-E14TG2a, ATCC CRL-1821) were cultured on Martigel®-coated plates in Stemflex™ medium,with 1μg/mL recombinant mouse LIF (R&D Systems, 8878-LF-025). We used the commercial kit, as in the generation of human organoids, with the following modifications: day 0 - organoids generation, day 2 - transfer to neural induction (NI) medium, day 5 - embedding in Matrigel in Expansion medium, day 7 - detailed removal of Matrigel; transfer organoids to IDM-A, day 8 - transfer organoids to IDM+A+dissolved Matrigel; shaker. Mouse organoids were treated with hormones at the same concentrations as the human ones. Treatments started at day 7 (start of neurogenesis), and collected for analyses at days 9 and 11 (Extended Data Fig. 9e).
Sendai and Lenti virus labelling
Organoids were labelled using a CytoTune™ EmGFP Sendai Fluorescence Reporter (ThermoFisher, A16519). The stock solution of the virus was diluted with PBS to 1:10 and kept on ice. The solution was visualized with Fast Green FCF (Sigma, F7252) and was delivered to the ventricles at 45d using a pulled capillary. Organoids were immediately subjected to hormone treatment. The media was changed every day. Organoids were collected for analysis at 6- and 8-days post-labelling. At 8 days post labelling, some of the organoids were injected with LentiBrite™ RFP Control Lentiviral Biosensor (Millipore, 17-10409), using the same procedure as with EmGFP Sendai virus. These organoids were kept in IDM+A with dissolved Matrigel® for a further 8 days and then fixed for clonal analysis.
Plasmids
Constitutively active androgen receptor (EGFP-C1-AR-V7) 22 was obtained from Michael Mancini and Marco Marcelli (Addgene #86856). A plasmid containing estrogen receptor alpha (pEGFP-C1-ERα) was obtained from Michael Mancini (Addgene #28230) and mutated into a constitutively active form (pEGFP-C1-ERαY537S) using the Q5® Site-Directed Mutagenesis Kit (New England Biolabs, E054S).
Electroporations
Organoids were injected and electroporated at 45d as described previously 3 with the following parameters: voltage - 40 mV, duration – 50 ms, pulses – 5, interval – 1.0 s. All plasmids were used at 1 μg/mL. EGFP-C1 plasmid was used as a control. Organoids were collected 2- and 5-days post-electroporation and media was changed at 2 days post-electroporation.
Organoid fixation and processing
Organoids were collected at different stages and fixed in 4% PFA in phosphate buffer (PB) for 1 hour at room temperature (RT). After washing in PBS for 2 minutes (2’), organoids were infiltrated with 30% sucrose in PB at 4°C until the tissue sank. After embedding in 7.5% gelatin/30% sucrose in PB and freezing in isopentane at -40°C, tissue blocks were stored at -80°C until sectioning. Organoids were sectioned using a cryostat (Leica, CM1950) at 20 μm and sections were collected on coated slides (ThermoFisher Superfrost® Plus, J1800AMNZ). Sections were stored at -20°C until further processing.
Immunohistochemistry
Immunostaining was performed as follows: Slides were briefly washed in PBS. Antigen retrieval (if necessary) was performed by using 0.01 M sodium citrate, pH6.0. Slides were submerged in the antigen retrieval solution and heated for one hour at 70°C in a water bath. After antigen retrieval, slides were left to cool for 20 minutes at RT. After washing with PBS, sections were blocked with 0.25% Triton-X 100 with 4% donkey serum in PBS (permeabilization buffer) for 1-2 hours at RT. Primary antibodies were diluted in 0.1% Triton-X 100 with 4% donkey serum in PBS (blocking buffer) and left overnight at 4°C. The following primary antibodies were used: rat anti-histone H3 (phospho S28) (Abcam, ab10543, 1:500), rat anti-CTIP2 (Abcam, ab18465, 1:200), mouse anti-HuC/D (LifeTech, A21271, 1:200), mouse anti-Ki67 (Dako, MIB-1, 1:100), mouse anti-DLX2 (Santa Cruz, sc-393879, 1:200), mouse anti-SATB2 (Abcam, ab51502, 1:50), rabbit anti-NEUROD2 (Abcam, ab104430, 1:200), rabbit anti-Ki67 (Abcam, ab15580, 1:200), rabbit anti-NGN2 (Cell Signalling Technologies, D2R3D, 1:200), rabbit anti-HOPX (FL-73) (Santa Cruz, sc-30216, 1:100), rabbit anti-PS6 (Cell Signallin, 2211S, 1:200), chicken anti-GFP (Thermo Fisher, A10262, 1:500), goat anti-SRD5A1 (Abcam, ab110123, 1:200), goat anti-BRN2 (Santa Cruz, sc-6029, 1:200), sheep anti-TBR2 (R&D Systems, AF6166, 1:200).
The next day, sections were washed with PBS and secondary antibodies coupled to Alexa Fluor fluorescent dyes (ThermoFisher Scientific) were added (diluted in blocking buffer 1:1000) in combination with DAPI (1:1000). Sections were incubated for 1 hour at RT. After PBS washes, slides were mounted in ProLong™ Diamond (ThermoFisher, P36961), left to dry at RT in the dark, and stored at 4°C after imaging.
AR in situ hybridisation (RNAScope)
AR mRNA was detected using the RNAscope® Fluorescent Multiplex Reagent Kit (ACD, 320850). The probe used was Hs-AR-O2 (ACD, 522941). The slides were dehydrated using a sequence of increasing concentrations of EtOH (50-100%). Dehydrated sections were air-dried for 5’ at RT. Heat mediated target retrieval was performed in Target retrieval solution for 2’ at 95°C. Diluted protease III (1:15) was applied and incubated for 15’ at 40°C in a humidified chamber. After washing 3 times in PBS, pre-heated probe was applied to the slides and hybridised for 2 hours at 40°C in a humidified chamber. After hybridisation, the slides were washed in wash buffer twice. AMP 1-FL was applied and incubated for 30’ at 40°C. Sections were washed twice for 2’ in 1x Wash buffer at RT. AMP 2-FL, AMP 3-FL and AMP 4-FL were individually added to the slides and incubated for 15’, 30’ and 30’ at 40°C, respectively, with wash steps in 1x Wash buffer in between (twice for 2’ at RT). The slides were washed 3 times in MilliQ water. Permeabilisation buffer (0.4% Triton-X, 4% normal donkey serum in PBS) was applied and incubated for 1 hour at RT. Primary antibody (diluted in blocking buffer) was incubated over night at RT in a humidified chamber. The following day, slides were washed 3 times for 5’ in PBS. Secondary antibody and DAPI were incubated for 2 hours at RT. Slides were washed in PBS 3 times for 5’ and mounted in ProLong™ Diamond.
For analysis of AR mRNA after cell division, XX organoids were injected with EmGFP Sendai Fluorescence Reporter at 50d as described above to trace individual radial glia stem cells and their progeny. Organoids were fixed at 54d and submitted to the RNA Scope protocol. The AR mRNA signal was analysed only in well-defined GFP+ pairs of cells. Only AR mRNA puncta that were inside the GFP signal were taken into consideration.
Imaging and image analysis
Imaging was performed either by using a Pannoramic Confocal slide scanner, or Zeiss 710, 780UV or 880 confocal microscopes. Objectives used: 10×, 20×, 40× and 63×. Images were taken as either Z-stacks, or individual planes. Quantifications of data from all systems were compared to ensure reproducibility of the data. Images obtained were processed using Fiji 60 . For Figure 2c, HuC/D staining was visualized using the “ICA” lookup table. For Extended Data Fig. 2d, HOPX staining was visualised using the Fire lookup table.
Quantifications
All quantifications were done using Fiji. Manual cell counting was performed by a custom plug-in, written by Johannes Schindelin. Only ventricles on the outside of the organoid were analysed. Ventricles of perpendicular cutting angle (visible apical and basal sides), appropriate morphology (distinct progenitor and neuronal layers, see below) and healthy appearance were chosen for quantifications.
Cell type, morphology and proliferation quantifications
The ventricular length was defined as an uninterrupted stretch of apical surface populated by radial glia, as shown by phospho-histone H3 (PH3) or a radial glia marker staining (PAX6 or SOX2, not shown). The ventricular zone (VZ) was defined as a layer of cells directly abutting the ventricle, with a distinctive radial organisation of nuclei and high density of cells, as revealed by DAPI staining. The subventricular zone (SVZ) was defined as a zone basally adjacent to the VZ, with cells expressing TBR2 and nuclei of a rounder shape. Progenitor zone was defined as the combined area of the ventricular (VZ) and subventricular zone (SVZ) expressing the progenitor marker TBR2. The cortical wall was defined as the area between the apical and the basal side of the cortical lobule. The cortical plate was recognised as a zone at the basal side of the cortical wall containing neurons arranged in an ordered fashion. Regions of interest (ROI) were designated following the tissue’s radial organisation, except for the pulse-chase 75d time point. Due to a lack of ventricles, ROIs at 75d were designated as a square (223.6 μm × 223.6 μm), located at the basal-most portion of the cortical wall.
All counts were normalized per mm2, unless indicated otherwise. Counts were done without using pseudocolour. All quantifications were done on a minimum of three independent organoid batches, with a minimum of 3 ventricles from 2 different organoids per treatment, per batch. In the electroporation experiments quantifications were done on a minimum of two ventricles per batch, per treatment, due to low levels of expression of particularly EGFP-C1-ERαY537S plasmid. Exact numbers of batches and ventricles analysed can be found in Supplementary Table 5. Only live cells were counted. When quantifying multiple stainings, the cell was scored as double positive only if the signal from both channels analysed clearly corresponded in shape.
Analysis of cell number quantification was performed using GraphPad Prism 8 software. Statistical analyses of different cortical wall zones (Fig. 1b) were done as pairwise comparison of the measurements from the XX and XY cell lines (unpaired non-parametric Mann-Whitney test, two-tailed). Statistical analyses of control versus hormone (or drug) treatments were done as multiple pairwise analyses, using unpaired non-parametric tests (Mann-Whitney, two-tailed) with significance threshold of P<0.05.
Clonal analysis
Only cells labelled GFP- (labelled with CytoTune™ EmGFP Sendai Fluorescence Reporter) or RFP-positive (labelled with LentiBrite™ RFP Control Lentiviral Biosensor) were analysed. During imaging, special care was taken to collect all the GFP- or RFP-expressing cells from the 20 μm section in the form of a Z-stack. Only clones consisting of >2 cells were taken into account, to ensure that the cell had divided upon receiving the virus. Only isolated clones were counted, in order to minimize the probability of several founder cells being infected upon the delivery of the virus, and their clonal progeny intermingling. Measurements were done on a minimum of 3 independent organoid batches, with a minimum of 2 organoids per timepoint, with a minimum of 5 clones analysed per timepoint, per treatment, per batch.
Quantification of SRD5A1 puncta
After immunostaining with anti-SRD5A1 antibody, images were obtained using a 63´ objective, with exactly the same laser power, exposure time and digital gain for both the control and DHT-treated sections. A square measuring 100 μm×100 μm was designated in the VZ. Threshold was adjusted in order to remove any immunostaining background which was easily discernible from the SRD5A1 signal. SRD5A1 puncta were then automatically counted using the “Analyze particles” option.
Quantification of AR mRNA-positive nuclei
Quantifications were done on sections of 21, 35 and 52d old organoids, after detection of AR mRNA with RNA Scope (see above). All healthy nuclei marked by DAPI were counted in a provisional unit of cortical wall from the apical to the basal surface. A nucleus was considered AR+ (containing AR mRNA) if there was at least one AR punctum inside the DAPI signal, or directly abutting the nucleus.
Image Quantification Statistics
Quantifications are represented as box and whisker plots. The body of the boxplot represents the 25th to the 75th percentile of the data. The centre line represents the median. Whiskers represent the minimum and maximum. Test for significance on image quantification values was performed using Mann-Whitney (two tailed) comparison of treatment to control, and P values are indicated in figures by bars of different shades of grey as indicated. For exact sample size for each experiment, see Supplementary Table 5.
Live imaging
Organoids were electroporated at day 31 or 54, as described above. Plasmids containing pCAG-mCherry and EGFP-C1-AR-V7 were mixed to 300 ng/mL and 1 μg/mL, respectively, and delivered to the ventricles. Electroporated organoids were transferred to IDM+A+Matrigel® medium for several hours for recovery. After that, organoids were embedded in 3% Low melting agarose (Sigma, A9414) in HBSS (1×) (Gibco, 14025092), left to settle and sectioned on the vibratome at 300 μm. Sections were placed on Millicell Organotypic Cell Culture Inserts (Merck, PICM0RG50) and supplemented with 800 μl serum-free media 61 . After 24 hours at 37°C, slices were imaged using Zeiss 880 Airy Scan, with the following parametres: 20 ´ objective, FAST scanning mode, 1072x1072 pixels, scanning interval of 10 - 11.45 minutes, total imaging time >25 hours. Movies were processed using Zeiss ZEN software and Fiji.
Western blot
Organoids for Western blot were treated with 30 nM DHT or 100 nM E every day, starting from 17d and collected every two days. Organoids (7 to 10 per condition) were washed twice in PBS prior to snap-freezing in liquid nitrogen. XX stem cells were used as a positive control, and XX 75d organoids as a negative (due to very low AR expression at this stage). Organoids and cells were lysed in modified RIPA buffer (mRIPA: 1% Triton-X, 0.1% SDS, 150mM NaCl, 50mM Tris pH7.4, 2mM EDTA, 12mM sodium deoxycholate) supplemented with protease (Thermo Fisher, 78430) and phosphatase (Sigma-Aldrich, 4906845001) inhibitors. Quick Start Bradford Dye Reagent (Bio-Rad, 5000205) assay was used to determine protein concentration. 20 μg of total protein was loaded for every sample except for the negative control where 10 μg of protein were loaded (indicated by an asterisk in Extended Data Fig. 4g). Samples were run on SDS polyacrylamide gel electrophoresis (SDS-PAGE) 4-20% Tris glycine gel at 90 V in MOPS-SDS buffer and transferred overnight at 30 V to Amersham Hybond P 0.45 PVDF blotting membranes (GE Healthcare, 10600023). Membranes were blocked in 5% BSA for 2 hours at RT and incubated overnight with the following primary antibodies, rabbit anti-AR (Cell Signalling, D6F11, 1:2000), mouse anti-GAPDH (Abcam, ab8245, 1:2000). Membranes were washed in TBST buffer prior to incubation with secondary fluorophore conjugated antibodies 1:2000 for 1h at RT (goat anti-mouse DyLight 680 Invitrogen A21058, goat anti-rabbit DyLight 800 CST 5151). Membranes were washed in TBST and imaged using a Li-COR Odyssey CLx Infrared Imaging System.
Droplet digital (dd) PCR
RNA was isolated from organoids collected at 19-25 days by using RNeasy Mini kit (Qiagen, 74104). Remaining DNA was removed with Invitrogen™ TURBO DNA-free™ kit (Invitrogen, AM1907). cDNA was produced using SuperScript™ III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen, 11752-050). PCR was performed as per manufacturer’s instructions, using the ddPCR Supermix for Probes (Bio-Rad, 1863026). 10ng of cDNA was used for the reaction. EIF2B2 was used as reference. Primers used: AR_F GGTGAGCAGAGTGCCCTATC, AR_R GCAGTCTCCAAACGCATGTC (FAM), EIF2B2_F TCCGGGAGGAGTATGGCAG, EIF2B2_R AATGGAAGCTGAAATCCTCG (HEX). Droplets were generated and read using the QX200 Droplet Digital PCR System (Bio-Rad, 1864001).
RNA-Seq library preparation and sequencing
Organoids were generated from the XX cell line and treated as described above (hormone treatment started at 17d). Organoids were collected at 35d, snap-frozen in liquid nitrogen and stored at -80°C until processing. Two to three individual organoids were collected per treatment. Total RNA from individual organoids was isolated using the RNeasy Mini Kit (Qiagen, 74104). Genomic DNA was removed using the Invitrogen™ TURBO DNA-free™ kit (Invitrogen, AM1907). After isolation, total RNA from individual organoids of the same treatment was pooled for each library preparation. RNA quality was assessed using Bioanalyzer (Agilent, G2939BA). Libraries were prepared using the NEBNext™ Ultra II DNA Library Prep Kit for Illumina™ (New England BioLabs, E7645) with starting input of 500ng of total RNA per sample. Ribosomal RNA was removed and mRNA isolated using the NEBNext™ Poly(A) mRNA Magnetic Isolation Module (New England BioLabs, E7490). During library preparation, AMPure XP Beads (Beckman Coulter, A63881) were used for reaction clean ups. NEBNext™ Multiplex Oligos (dual index primers) (New England BioLabs, E7600) were used in the PCR enrichment step, with 11 cycles. Library quality was assessed by Bioanalyzer and the concentration was determined using the NEBNext™ Library Quant Kit for Illumina™ (New England BioLabs, E7630). Libraries were pooled and sequenced on the HiSeq4000 machine (Illumina) at 50 base pair length, single ended, to a minimum of 30 million reads per library.
Bulk RNA-seq data analysis
FASTQ sequence read files were aligned to the hg38 transcriptome (with decoy sequences) and quantified using salmon v0.14.1 62 applying both a sequence bias correction and a GC bias correction with a mean fragment length of 300 bp and a standard deviation of 50 bp. Salmon quantification files containing Transcripts Per Million (TPM) estimates were imported into R using tximport v1.16.1 and DESeq2 v1.28.1 63 . Prior to analysis, a non-parametric empirical Bayesian batch-correction was applied to the data using the function ‘ComBat’ in sva v3.36.0 64 (Extended Data Fig. 6a, b), and only genes with >4 TPM on average per sample (6 androgen (DHT/T), 3 control) were retained for downstream analysis.
Differential expression analysis was conducted using a novel approach 24 that employs a logistic elastic-net regression implemented in the R package glmnet v4.0-2 65 using an alpha of 0.5. The response variable was taken to be the sample hormone status (Androgen or Control), and the independent variable was the transcript abundance estimates for individual genes. To determine the value of lambda – the parameter controlling the strength of the regression penalty - for selecting differentially expressed genes, cross-validation was performed using the function ‘cv.glmnet’ and the ‘type.measure’ argument set to ‘deviance’. The value of lambda that minimised the deviance (a measure of goodness-of-fit) of the model was selected, and genes with a non-zero regression coefficient at the selected lambda value were considered to be differentially expressed, which included both up- and down-regulated genes. Log-fold change estimates for the selected genes were estimated using DESeq2.
Validation of the logistic elastic-net model was carried out using the same input dataset in which a ComBat batch-correction was applied to the treatment variable (Androgen or Control) to remove signal from the dataset. Then, a known amount of log-fold change signal was added to a random sample of genes using the binomial-thinning approach implemented in the R package seqgendiff v1.2.2 66 using the function ‘thin_diff’ and randomly selecting 6 of the samples to be androgen samples and 3 to be controls. To determine the number of genes to add signal to, the R package locfdr v1.1-8 was used to fit a mixture model to DESeq2 raw p-values derived from the original unmodified dataset. This package enables an estimate of the total number of non-null genes in the dataset – in this case 362. ‘locfdr’ was also used to infer the distribution of log-fold changes for non-null genes using DESeq2 log-fold change estimates derived from the original unmodified dataset. 362 log-fold changes were then randomly sampled from this distribution and paired with 362 randomly sampled genes. Both DESeq2 and the elastic-net models were then used to infer differential expression in this modified dataset and both the sensitivity and precision of each method was calculated. While DESeq2 achieved 100% precision, it had a sensitivity of just 2% (genes with an adjusted p-value <0.1 were considered differentially expressed). In contrast, the elastic-net method achieved a sensitivity of 20% (72 correctly called genes) with a precision of 96% - all three false positives (out of 75 hits) had absolute log fold changes below 0.31 whereas 75% of the 72 true positive hits had an absolute log fold change above 0.37. In addition, false negatives had a significantly lower log fold change than true positives (Wilcoxon rank sum test, p = 3e-13). A reproducible R markdown script and data can be accessed at https://github.com/alextkalinka/hormone-DE-analysis.
Single cell RNA sequencing and analysis
Single cell dissociation was performed on a total of 6 XX (H9) organoids, 2 control mock treated, 2 treated with DHT (30 nM) from day 17 to 35, and two treated with E (100 nM) from day 17 to 35. Samples were collected on day 35 and incubated in 1ml of Accumax (Sigma, A7089) with 400 μg DNase I and 15 μM actinomycin D at 37°C for 20 min with gentle agitation. At 5 min intervals, the sample tubes were flicked, and after 15 min pipetted 10 times, followed by a final pipetting 10 times at 20 min. Clumps were allowed to settle and supernatant collected, to which 100 μl media was added to inactivate Accumax. Samples were then spun at 300xg for 5min. Dead Cell Removal kit and MACS column (Miltenyi, 130-090-101) were used to remove dead cells before another spin at 300g for 5min. Cells were resuspended in an appropriate volume of 0.04% BSA in PBS and loaded on the 10X Chromium (10X Genomics).
Single cell RNA-seq libraries were prepared according to manufacturer’s instructions using the 10X Genomics Chromium Single Cell 3’ Library & Gel Bead Kit v3.1 (10X Genomics) workflow. The Chromium Controller was run according to the protocol producing the single cell gel beads in emulsion mixture. Reverse transcriptase reaction and subsequent amplification was carried out on a C1000 Touch Thermal Cycler (Biorad) and libraries were quality tested using a 2100 Bioanalyzer Instrument (Agilent). Samples were pooled together and sequenced on one lane of SP flowcell of the Novaseq sequencer (Illumina). Raw sequencing output was converted to the standard fastq format by bcl2fastq, before running through CellRanger Count (version 3.1.0, 10X Genomics) which also performed read alignment using STAR with GRCh38 human reference genome. QC output revealed 1,708 cells, with mean reads per cell of 54,907 and median UMI counts per cell of 3,926 for Control sample 1; 1,432 cells, with mean reads per cell of 47,496 and median UMI counts per cell of 1,708 for Control sample 2; 557 cells, with mean reads per cell of 107,192 and median UMI counts per cell of 1,587 for DHT sample 1; 1,386 cells, with mean reads per cell of 48,845 and median UMI counts per cell of 1,471 for DHT sample 2; 1,179 cells, with mean reads per cell of 46,140 and median UMI counts per cell of 1,130 for E sample 1; 1,005 cells, with mean reads per cell of 68,252 and median UMI counts per cell of 2,934 for E sample 2. The filtered feature barcode matrix for each sample was then analysed further with the Seurat v3 R package 67 by first merging the matrices, followed by removal of cells with mitochondrial percentage >10%, and removal of likely doublets based on nCount_RNA, resulting in a total dataset of 6,899 cells. Data were then normalised for read depth across cells, scaling the data, and variable feature finding using SCTransform. Mitochondrial mapping percentage was regressed out during normalization to remove this confounding source of variation. Unbiased clustering was performed by principal component analysis (PCA), followed by FindNeighbors, FindClusters, and UMAP dimensionality reduction visualization. Clusters were identified based on the top 10 differentially expressed genes in each cluster, as well as analysis of known marker genes. To statistically test the significance of observed distribution of RG cell clusters in treatments and controls, a chi-square test was performed. For the overall distribution, the test was performed with Monte Carlo simulation, and for each individual value chi-square value and calculated p-value is reported in Supplementary Table 4. A further test for significant difference in distribution between treatments and control was performed as described in https://github.com/rpolicastro/scProportionTest in which a permutation test is used to calculate p-value for each cluster in treatment compared with control and confidence intervals returned using bootstrapping. For further analysis, IP/N and RG cell clusters were subsetted, which was used for pseudotime analysis by converting the seurat object to a cell data set for analysis in Monocle3 68,69 using SeuratWrappers, and pseudotime was mapped onto the seurat UMAP. IP/N cluster was also further subclustered as above with increased resolution. For comparison across treatments, differential gene expression was performed on RG clusters by first assigning cluster ID to treatment status, which was added to metadata, followed by FindMarkers in DHT compared to Control.
Growth curve modeling
Mathematical modeling of cell numbers used a standard growth curve equation to calculate the number of radial glia over time, which was used to predict numbers of neurons. In order to more accurately model the trajectory of neurogenesis, several assumptions were taken from the literature, as detailed below, and the mathematical model was applied equally to both control and androgen exposure. The only difference was the addition of a coefficient reflecting the increase in neurogenesis seen in organoids treated with androgen (Fig. 3e). This was only applied during the fetal stage when the testosterone surge has been reported at its highest (post-conception weeks (pcw) 8 to 16) 46 . Otherwise, the same modeling was performed for both conditions.
The following growth curve equation was first used to calculate radial glial growth during the first 7 weeks:
where pt is the number of radial glial progenitors at time t in days, and f is the number of divisions in one day. The following assumptions were applied in order to more accurately model the trajectory: Neural development begins at embryonic disk stage at day 18 with 1 progenitor; 18.831 hour cell cycle until week 4 when tNE starts 70 ; 22.099 hour cell cycle during tNE until onset of neurogenesis at week 5. During early neurogenesis (weeks 5 and 6) 48 hour cell cycle and 9/17 (0.53) divisions are symmetric proliferative according to previous studies 71 while the rest of the divisions (0.47) are asymmetric direct neurogenic. Thus, from week 5-6, number of neurons was calculated as follows:
where nt is the number of neurons at time t in days.
From week 7 on, radial glial divisions were calculated as occurring every 4 days, and were calculated according to the above growth curve where time t is a multiple of four days, for simplicity. The following assumptions were then applied: Weeks 7-10: deep layer neurogenesis, 9/17 divisions are symmetric, the rest are asymmetric giving rise to one RG and one BP, which gives rise to 2 neurons; Weeks 11-14: upper layer neurogenesis, symmetric divisions steadily decrease to 1/19, asymmetric giving rise to one RG and one BP, which generates 2 neurons; Weeks 15-16: upper layer neurogenesis, symmetric divisions drop to 0, asymmetric giving rise to one RG and one BP, which makes 2 neurons; Weeks 17-18: upper layer neurogenesis, all divisions are asymmetric giving rise to one RG and one BP, which makes 2 neurons; Weeks 19-27: asymmetric divisions giving rise to BP decrease (being replaced by gliogenic divisions) steadily to 0, BPs give rise to 2 neurons; Weeks 28 onwards: cortical excitatory neurogenesis is complete.
BPs were calculated accordingly:
where bt is the number of BPs at time t, and a is the ratio of RG asymmetric divisions. Neurons from week 7 on were calculated according to the following:
Finally, the only difference between −Androgen and +Androgen was the inclusion of a coefficient to reflect the increased neurogenesis as measured in organoids during the fetal surge. Thus, from weeks 8 to 16, numbers of neurons in +Androgen model was calculated according to the following equation:
where At is the number of neurons in +Androgen at time t, Δnt is the increase in the number of neurons in −Androgen between time t and t − 1 and 1.272 is the measured increased neurogenesis upon androgen treatment (Fig. 3e). This equation applied only during the fetal surge, from 8 to 16 weeks, and otherwise ΔAt equaled Δnt .
Overall, the model revealed an increase in neurons in +Androgen of 9.4%, which was consistent and independent of the arbitrary values otherwise used to generate the growth curves.
Extended Data
Extended Data Figure 1. Treatment and phenotypic characterization of cerebral organoids.
a) Illustration of the determination of the progenitor zone for quantifications. The progenitor zone (VZ+SVZ) was determined based on immunostaining and histological landmarks (see Methods). Immunostaining of XX 35d organoids stained for TBR2 (yellow, white), co-stained with DAPI (blue). Dashed line indicates the progenitor zone. Schematic of an organoid ventricle with different progenitor populations of interest. b) Quantification of thicknesses of different morphological zones between male (XY) and female (XX) brain organoids at 35d (above) and 52d (below). Significance values (Mann-Whitney, two tailed) of the measurements from XX organoids, as compared to XY organoids, are indicated by different shades of grey. VZ-ventricular zone, SVZ-subventricular zone. c) Actual measured concentration of testosterone (left) and estradiol (right) in the media after the addition of 100nM (calculated) on day 0, as measured by ELISA assay. d) Actual measured concentration of testosterone over 4 days of organoid culture after addition of 100nM (calculated) on day 0, as measured by ELISA. e) Sections of whole XX organoids at 35d (left), and XY organoids at 52d (right), stained for TBR2 (white). Co-stained with DAPI (blue). f) XY 35d organoids stained for TBR2 (yellow, white), co-stained with DAPI (blue), treated with DHT, T or E. g) Quantification of non-apical mitotic, PH3+ cells in XY and XX, 35d and 52d organoids, treated with DHT, T and E. Scale bars: e) 500μm, a), f) 100 μm. See Methods for details of statistics and Supplementary Table 5 for details of n numbers.
Extended Data Figure 2. Basal progenitors are increased upon androgen treatment.
a) Quantification of TBR2+ cells per mm2 progenitor zone (see Extended Data Fig. 1a) at 35d. b) Image of the ventricular zone/subventricular zone (VZ/SVZ) border in XX 35d organoids stained for Ki67 (green, white) and TBR2 (white). Cells positive for both Ki67 and TBR2 are indicated with magenta arrowheads. Double positive mitotic cells are indicated with yellow arrowheads. A white dashed line delineates VZ/SVZ border with the ventricular zone below. c) Immunostaining for proliferation marker Ki67 (white) on XX 35d control (left) and DHT treated (right) organoids. Dashed yellow lines represent the apical surface (bottom line) and the VZ/SVZ boundary. Note the difference in Ki67+ cells in the SVZ of DHT-treated organoids. d) Quantification of Ki67/TBR2 double positive cells, out of all TBR2+ cells, in XX 35d and 52d organoids. e) Immunostaining for HOPX (fire LUT) of XX 52d organoids. Yellow dashed line indicates the ventricular surface. Note the HOPX signal in radial glia. Images are single, 1.2 μm optical planes. f) Quantification of HOPX+ basal radial glia per mm2 SVZ of XX 52d organoids. g) Immunostaining for phosphorylated histone H3 (PH3) (green) on XX 35d and XY 52d old organoids. Co-stained with DAPI (white). h) Quantification of apical mitotic cells (PH3+), normalized per mm length of ventricle in male (XY) and female (XX) organoids, at 35d and 52d, treated with DHT, T and E. i) Quantification of ventricular length in male (XY) and female (XX) organoids, at 35d and 52d, treated with DHT, T and E. Scale bars: g) 100 μm, e) 50μm, b), c) 25μm. See Methods for details of statistics and Supplementary Table 5 for details of n numbers.
Extended Data Figure 3. Clonal labelling reveals increased radial glial proliferation upon androgen treatment.
a) Timeline and schematic of the Sendai/Lenti virus lineage tracing and analysis, with virus encoding emGFP injected at day 45 and virus encoding RFP injected at day 53. b) Representative images of GFP-labelled clones (white) 8 days post Sendai emGFP labelling in control (above) and DHT-treated (below) XX organoids. DAPI is in blue. c) Quantification of GFP+ clone size (left side) at 51d and 53d and RFP+ clone size at 61d. d) XX control and DHT organoids injected with Lenti-RFP at 53d, and fixed at 61d. RFP (magenta). Co-stained with DAPI (blue). e) Quantification of TBR2+ cells per mm2 of progenitor layer in control and DHT-treated XX organoids, at 8 days post-labelling with Sendai emGFP. Note that TBR2+ intermediate progenitors were not yet increased at this time point, as the radial glia which produce them were still in the VZ. Scale bars: c) 50μm, a) 25μm. See Methods for details of statistics and Supplementary Table 5 for details of n numbers.
Extended Data Figure 4. Androgen receptor activity in radial glia promotes their proliferation rather than differentiation.
a) RNA Scope (fluorescent in situ) for AR of XY organoids at 45d. Single AR mRNA puncta (white). Co-stained with DAPI (blue). Yellow dashed lines demarcate ventricular zone (VZ) and the subventricular zone (SVZ). a’) Portion of the VZ from a. a”) Portion of the SVZ from a. b) RNA Scope for AR of XX organoids at 45d. Single AR mRNA puncta (white). Co-stained with DAPI (blue). b’) Portion of the VZ from b. b”) Portion of the SVZ from b. Note the difference in the amount of AR mRNA in the VZ and SVZ in both cell lines. c) RNA Scope for AR of XX 21d (top) and 35d (bottom) organoids. Single AR mRNA puncta (white). Immunostaining for TBR2 (magenta). Co-stained with DAPI (blue). Yellow dashed line demarcates the apical surface c’) Portion of the ventricular zone (VZ) from XX 21d organoid. c”) Portion of the VZ from XX 35d organoid. d) RNA Scope negative (above) and positive (below) control (fire look up table) in female (XX) 35d organoids. DAPI is in white. e) Quantification of AR mRNA distribution in GFP+ pairs of cells, depending on the stage of cell division. f) RNA Scope of androgen receptor (AR) mRNA (white puncta), together with GFP signal (green) from EmGFP Sendai Fluorescence Reporter showing examples of three daughter cell pairs. Yellow dashed lines indicate GFP+ cell body. g) Western blot for AR on control, DHT and E treated organoids at 17-25 days. AR specific band is predicted to be ~110kDa. Note the increased AR signal in DHT-treated organoids. Asterisk indicates a lower amount of protein loaded for the 75d organoid lane (see Methods). For gel source data, see Supplementary Figure 1. h) Quantification of levels of AR protein from g), normalised by GAPDH expression. i) ddPCR results showing the decrease in AR transcription between days 17 and 25. ref = housekeeping gene EIF2B2. j) Representative images of the cortical wall at 21, 35 and 52d, with DAPI-labelled nuclei visible, showing the relative reduction in the radial glial progenitor layer (ventricular zone - VZ) and an increase in the thickness of the neuronal layer over time. k) Quantification of the percentage (%) of cells (AR+ DAPI), containing AR mRNA puncta (as detected by RNA Scope), out of all cells (DAPI) at 21, 35 and 52d. Cells counted: 21d – 1222, 35d – 1067, 52d – 1201. Co-stained with DAPI (white). l) XX organoids electroporated (EP) at 45d and fixed at 50d. EGFP-C1: control; EGFP-C1-AR-V7: constitutively active AR. Immunostaining for GFP and TBR2. Yellow dashed lines demarcate the apical and basal boundaries of the VZ. Note increased GFP+ nuclei in the VZ in EGFP-C1-AR-V7. m) Immunostaining for GFP (green) and Ki67 (magenta) on XX 50d organoids, electroporated at 45d. Co-stained with DAPI (blue). EGFP-C1: control plasmid; EGFP-C1-AR-V7: plasmid expressing constitutively active AR; EGFP-C1-ERaY537S: plasmid expressing constitutively active ERa. n) Quantification of the proportion of GFP+ cells co-staining for the proliferation marker Ki67 at 5 days post electroporation in XX organoids electroporated with the indicated plasmid at 45d. o) Quantification of the proportion of GFP+ cells co-staining for the intermediate progenitor marker TBR2 at 2- and 5 days post electroporation in XX organoids electroporated at 45d. At 5 days post electroporation, most cells electroporated with EGFP-C1-ERaY537S died indicating a later effect on cell survival. p) Quantification of the proportion of GFP+ cells co-staining for the neuronal marker HuC/D 2- and 5 days post electroporation in XX organoids electroporated at 45d. At 5 days post electroporation, most cells electroporated with EGFP-C1-ERaY537S died. q) XX 47d organoids, electroporated at 45d. Immunostaining for GFP (green), NGN2 (cyan) and BRN2 (magenta). Co-stained with DAPI (white). Yellow arrowheads: GFP+/NGN2+ cells. White arrowheads: GFP+/BRN2+ cells. Yellow dashed line demarcates the apical surface. Note the increased expression of differentiation markers upon expression of ERaY537S perhaps indicating a premature cell cycle exit and relating to the cell death observed. r) Quantification of the proportion of GFP+ cells co-staining for the neurogenic marker NGN2 2 days post electroporation in XX organoids electroporated at 45d. s) Quantification of the proportion of GFP+ cells co-staining for the upper layer neurogenesis marker BRN2 2 days post electroporation in XX organoids electroporated at 45d. Scale bars: a), b), l), m) 50μm, c), d), q) 25μm, a’), a”), b’), b”), j) 20μm, c’), c”), f) 5μm. See Methods for details of statistics and Supplementary Table 5 for details of n numbers.
Extended Data Figure 5. Live imaging of radial glial division modes upon activation and androgen signaling.
a) Still images from live imaging of organoids electroporated at 31d and imaged beginning at 2 days post-electroporation (see Methods for details of image acquisition). pCAG-mCherry+ cells are shown in magenta, and EGFP-C1-AR-V7+ are shown in green. Green arrowheads: GFP+ cells. Yellow arrowheads: mCherry+/GFP– cells. Filled in arrowheads: one of the tracked daughter cells. a) Comparison of the behaviour of mCherry+ and GFP+ only cells at 33d. Basal surface is up. mCherry+ only cell, upon division (06:11) produces two daughter cells, one of which migrates basally, indicating a more differentiated identity, while the other stays in the ventricular zone (VZ), representing an example of asymmetric division. GFP+ cell divides (07:28), but both of the daughter cells continue to reside in the VZ. b) Still images from live imaging of organoids electroporated at 54d and imaged at 56d. Basal surface is down. After division (15:50), both of the daughter cells remain in the VZ. Yellow dashed lines demarcate the apical surfaces. Time scale: hours:minutes. c) Schematic of the proposed mechanism leading to increased basal progenitors. Radial glia can divide symmetrically, increasing their numbers, or asymmetrically, generating one radial glia and one basal progenitor. Application of androgen supports symmetric, proliferative divisions, thus increasing the size of radial glial clones. After androgen is withdrawn, these radial glia can start producing basal progenitors in increased numbers, as shown in Fig. 1b, c. Basal progenitors represented as a single population (yellow cells), for clarity. Scale bars: a), b) 20μm. See Methods for details of statistics and Supplementary Table 5 for details of n numbers.
Extended Data Figure 6. Transcriptomics reveals key signaling pathways and cell types.
a) Principle component analysis (PCA) plots of XX 35d bulk RNA-seq, uncorrected for batch effects. b) Principle component analysis (PCA) plots of XX 35d bulk RNA-seq, corrected for batch effects using the R package ComBat. Filtering of lowly expressed genes was performed prior to batch correction, resulting in different PC value scales between a and b. See Methods. c) Quantification of SRD5A1, an upregulated DEG under androgen treatment. SRD5A1 was quantified as the number of SRD5A1+ puncta per 10μm2 of the ventricular zone (VZ) in XX 35d control and DHT-treated organoids. d) Venn diagram representing the overlap of up- and downregulated DEGs in androgen treated XX 35d organoids with genes differentially expressed in schizophrenia patients 72 . P values represent significance of a hypergeometric test of the intersection with up- and downregulated genes (P=3.6E-4 and P=0.0012, respectively). DE: differentially expressed. e) Top significant GO 34 term enrichments in XX 35d androgen upregulated genes. f) Left, UMAP plot showing different clusters identified by scRNA-seq in 35d old treated and control XX organoids. Right, UMAP plot showing the distribution of cells belonging to control, DHT and E treatments. Note the high degree of overlap indicating reproducibility. g) Heatmap showing scaled expression levels of cluster specific genes identified through differential gene expression analysis across clusters. The top 10 genes ordered by average log fold change are shown for each cluster. h) Dot plot showing the relative expression of cell type-specific markers. i) Feature scatter plot for percent ribosome (percent.ribo) by percent mitochondrial (percent.mt) reads. Left, plotted data points grouped by RG subcluster. Right, plotted data points grouped by treatment status. -0.27 refers to the correlation coefficient. j) Pseudotime analysis using Monocle3 mapped onto the reclustered seurat neural clusters (all clusters except ChP/Hem) UMAP. k) Feature plot of the neurogenic marker BTG2 (Tis21) and more mature marker synaptotagmin (SYT1) which was also identified as upregulated in RG2 cluster. l) Dot plot of the subclustering of the IP/N cell population showing the relative expression of cell type-specific markers and clear separation of different cell subpopulations. ImmN = immature neurons, MatN = maturing neurons. m) Stacked bar plot of radial glial (RG) cluster proportions detected by scRNA-seq at 35d. Upper right: chi-square with Monte Carlo simulation for overall distribution (Supplementary Table 4). Note increased RG1 and bRG, but decrease in more committed RG2, with no significant difference compared with expected counts in total RG (combined four RG clusters) and non-RG clusters (IP/N and ChP/Hem) in DHT treatment as determined by chi-square test (not shown) indicating specificity for RG subclusters. *P<0.05 and **P<0.00005 n) Point range plot of Monte Carlo permutation test of statistical significance between cell cluster proportions in DHT compared with control (left) and E compared with control (right) displaying bootstrapped confidence intervals for the difference in cluster proportion (reported as observed log2 fold-change) (see Methods). o) Dot plot showing the relative expression of bRG markers in different treatments, detected by scRNA-seq, in 35d old organoids. See Methods for details of statistics and Supplementary Table 5 for details of n numbers.
Extended data Figure 7. HDAC activity interacts with androgen signalling to influence progenitor behaviour.
a) Dot plot showing the relative expression of HDACs in different treatments, detected by scRNA-seq, in 35d old organoids. b) UMAP plots showing the relative expression of HDAC2 in cells from control, DHT and E treated organoids. Right - UMAP plot showing the radial glia clusters identified by scRNA-seq. Yellow circle demarcates the bRG subpopulation. Note the increased expression of HDAC2 in DHT bRG cluster. c) Immunostaining for TBR2 (green) and HuC/D (magenta) in XX 35d organoids, treated with VPA from 17d. Co-stained with DAPI (white, blue). c’) c”) c”’) insets from c), as indicated. Yellow asterisks indicate neural rosettes forming above the VZ in some VPA-treated organoids. d) Quantification of ventricular length for XX 35d old organoids treated with VPA and VPA+DHT. e) Immunostaining for CTIP2 (magenta), and SATB2 (green, white) on XX 35d control and VPA-treated organoids. Co-stained with DAPI (blue). e’), e”) insets from e) showing just SATB2 in fire look-up table. e”) Note the SATB2+ cells (yellow arrowheads) and SATB2+ staining present in the VZ in VPA-treated organoids. Yellow dashed lines demarcate the apical and basal boundaries of the VZ. f) Quantification of SATB2+ cells in the cortical wall of control, VPA- and VPA+DHT-treated organoids in XX organoids at 35d. g) Quantification of the thickness of VZ for XX 35d organoids treated with VPA and VPA+DHT. h) Immunostaining for TBR2 (yellow) in 35d old XX organoids with the following treatments: DHT, MI-192 (HDAC2/3 inhibitor), MI-192+DHT, MS-275 (HDAC1/3 inhibitor), MS-275+DHT. White dashed line demarcates the apical surface. i) Quantification of TBR2+ cells per mm2 of the control and the following treatments: DHT, MI-192 (HDAC2/3 inhibitor), MI-192+DHT, MS-275 (HDAC1/3 inhibitor), MS-275+DHT in 35d old XX organoids. Scale bar: c) 100 μm, c’), c”), c”’), e), h) 50μm, e’), e”) 20μm. See Methods for details of statistics and Supplementary Table 5 for details of n numbers.
Extended Data Figure 8. Downstream mTOR activity influences progenitor expansion.
a) Heatmap showing scaled expression of differentially expressed genes in DHT-treated compared to control organoids, detected by scRNAseq. Cell identities were assigned to their treatment group and differential expression analysis performed in Seurat as performed for comparison across clusters (see Methods). b) Dotplot showing the relative expression of bRG markers, overlapping markers of RG1 cells and DE transcripts in DHT, and genes of the mTOR pathway in cells from control, DHT and E treated organoids, detected by scRNAseq. c) Protein interaction network of upregulated DEG directly-interacting genes, obtained by bulk RNA seq. Translation/ribosome biogenesis genes are in red. d) Immunostaining for phosphorylated S6 (PS6) (fire look-up table), an indicator of mTOR activity, in 35d old XX organoids, treated with DHT and E. White arrowheads indicate PS6+ cell bodies in the ventricular zone (VZ). e) Quantification of PS6+ cells in the VZ of control, DHT- and E-treated 35d old XX organoids. f) Immunostaining for TBR2 (white) in control and MHY-1485-treated 35d XX organoids. Costained with DAPI (blue). g) Quantification of TBR2+ cells per mm2 progenitor layer in control and MHY-1485-treated 35d XX organoids. Scale bars: d), f) 50μm. See Methods for details of statistics and Supplementary Table 5 for details of n numbers.
Extended Data Figure 9. Ventral progenitors and mouse organoids exhibit differential responses to those of human excitatory neurogenic progenitors.
a) Schematic of neurogenesis in the human developing brain at ~12 gestation weeks. Excitatory neurons (green arrows) are born within the dorsal cortex and migrate basally to form the cortical plate. Inhibitory neurons (orange arrow) are born in the ventral telencephalon and migrate towards the dorsal side to incorporate themselves into the cortical plate. Yellow: progenitor zone, dark pink: cortical plate. b) RNA Scope (fluorescent in situ hybridization) for AR mRNA (white) in XX and XY 45d old ventral organoids. Co-stained with DAPI (blue). b’) and b”) - portions of the VZ, as indicated. Note the decreased levels compared with Extended Data Fig. 4a. c) Above: Heatmap of steroidogenic enzyme expression from XX 35d bulk RNA-seq. Values are in tpm. Below: Schematic of the steroidogenic pathway. Purple – SRD5A1. ESR1, ESR2-estrogen receptors a and b, respectively. d) Comparison of SRD5A1 (fire LUT) immunostaining on XX and XY dorsal 35d and ventral 45d organoids. Both anti-SRD5A1 and anti-TBR2 primary antibodies were used in XY samples, and the secondary antibody recognised both primary antibodies but are discernible by their different subcellular localisations. Co-stained with DAPI (white). Yellow dashed lines demarcate the apical and basal boundaries of the VZ. d’) Portion of the VZ in XX dorsal 35d organoids stained for SRD5A1. Note the SRD5A1+ puncta. d”) Portion of the VZ in XX ventral 45d organoids stained for SRD5A1. d”’) Portion of the VZ in XY dorsal 35d organoids stained for SRD5A1. Note the SRD5A1+ puncta. d””) Portion of the VZ in XY ventral 45d organoids stained for SRD5A1. White arrowhead - SRD5A1+ punctum. e) Timeline of the mouse organoid generation protocol. See Methods for details. f) Immunostaining for TBR2 (yellow) on 11d old mouse organoids in control, DHT- and E- treated organoids. Costained with DAPI (blue). g) Quantification of TBR2+ cells per mm2 of progenitor layer in mouse organoids at 9d and 11d. Scale bars: b), d), f) 50μm, b’), b”) 20μm, d’), d”), d”’), d””) 10μm. See Methods for details of statistics and Supplementary Table 5 for details of n numbers.
Extended Data Figure 10. Excitatory neurons are increased upon release of androgen signaling.
a) Immunostaining of XX 52d continuously treated control and DHT organoids stained for NEUROD2 (white). b) Quantification of NEUROD2+ cells per mm2 of cortical wall at 35 days, in XY and XX organoids. c) Quantification of NEUROD2+ cells per mm2 of cortical wall at 52 days, in XY and XX organoids. d) Timeline of the pulse-chase experiment. Hormones were administered between 17-35d, then removed. Organoids were fixed at 52d and 75d. e) NEUROD2+ cells per mm2 cortical wall after pulse-chase treatment. f) Immunostaining for CTIP2 (white) of XX 52d organoids after pulse-chase treatment (control and DHT). Co-stained with DAPI (blue). g) Quantification of CTIP2+ cells per mm2 cortical wall after pulse-chase treatment. h) Quantification of SATB2+ cells per 0.5mm2 cortical wall at 75d after pulse-chase treatment. Scale bars: a) 100μm, f) 50μm. See Methods for details of statistics and Supplementary Table 5 for details of n numbers.
Supplementary Material
Acknowledgements
We would like to thank Deepak Srivastava, Sean Munro and Manu Hegde for helpful comments. We would like to thank the MRC LMB Light Microscopy facility and Bioinformatics (Paula Freire-Pritchett), as well as the CRUK Genomics facility. We would like to thank Michael Mancini, Marco Marcelli and Elizabeth Wilson for depositing their plasmids in Addgene. We thank the other members of the Lancaster lab and the MRC LMB Cell biology division for fruitful discussions. Work in the Lancaster lab is supported by the Medical Research Council (MC_UP_1201/9) and the European Research Council (ERC STG 757710).
Footnotes
Author contributions
I.K. conceived the study, performed experiments, analysed data and wrote the paper. I.C. performed experiments and analysed the data. L.P. performed experiments. A.T.K. performed bioinformatics analysis. M.A.L. supervised the study, analysed data and co-wrote the paper.
Conflict of interest
M.A.L. is an inventor on several patents related to cerebral organoids, is co-founder and scientific advisory board member of a:head bio, and scientific advisory board member of the Roche Institute for Translational Bioengineering.
Data Availability Statement
All sequencing data (bulk and single-cell) have been deposited on NCBI Gene Expression Omnibus (GEO) under accession GSE187877. The following open access databases were used in this study: SZDB: A Database for Schizophrenia Genetic Research (szdb.org), SFARI gene (https://gene.sfari.org/).
Code Availability
Delboy code is available on https://github.com/alextkalinka/delboy.
References
- 1.Ritchie SJ, et al. Sex Differences in the Adult Human Brain: Evidence from 5216 UK Biobank Participants. Cereb Cortex. 2018;28:2959–2975. doi: 10.1093/cercor/bhy109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruigrok ANV, et al. A meta-analysis of sex differences in human brain structure. Neurosci Biobehav Rev. 2014;39:34–50. doi: 10.1016/j.neubiorev.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green T, Flash S, Reiss AL. Sex differences in psychiatric disorders: what we can learn from sex chromosome aneuploidies. Neuropsychopharmacol. 2018;44:9–21. doi: 10.1038/s41386-018-0153-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCarthy MM. Multifaceted origins of sex differences in the brain. Philosophical Transactions Royal Soc B Biological Sci. 2016;371:20150106. doi: 10.1098/rstb.2015.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loomes R, Hull L, Mandy WPL. What Is the Male-to-Female Ratio in Autism Spectrum Disorder? A Systematic Review and Meta-Analysis. J Am Acad Child Adolesc Psychiatry. 2017;56:466–474. doi: 10.1016/j.jaac.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 7.Abel KM, Drake R, Goldstein JM. Sex differences in schizophrenia. Int Rev Psychiatr. 2010;22:417–428. doi: 10.3109/09540261.2010.515205. [DOI] [PubMed] [Google Scholar]
- 8.Hines M. Neuroscience and sex/gender: Looking back and looking forward. J Neurosci. 2019;40:0750-19 [Google Scholar]
- 9.McCarthy MM, Arnold AP. Reframing sexual differentiation of the brain. Nat Neurosci. 2011;14:677. doi: 10.1038/nn.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodson JC, Gorski RA. In: Sexual Differentiation of the Brain. Matsumoto A, editor. CRC Press; 1999. Structural Sex Differences in the Mammalian Brain: Reconsidering the Male/Female Dichotomy. [Google Scholar]
- 11.Rabinowicz T, Dean DE, Petetot JM-C, de Courten-Myers GM. Gender Differences in the Human Cerebral Cortex: More Neurons in Males; More Processes in Females. J Child Neurol. 1999;14:98–107. doi: 10.1177/088307389901400207. [DOI] [PubMed] [Google Scholar]
- 12.Knickmeyer RC, et al. Impact of Sex and Gonadal Steroids on Neonatal Brain Structure. Cereb Cortex. 2014;24:2721–2731. doi: 10.1093/cercor/bht125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnold AP. A general theory of sexual differentiation. J Neurosci Res. 2017;95:291–300. doi: 10.1002/jnr.23884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold AP. The organizational–activational hypothesis as the foundation for a unified theory of sexual differentiation of all mammalian tissues. Horm Behav. 2009;55:570–578. doi: 10.1016/j.yhbeh.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarthy MM. Estradiol and the Developing Brain. Physiol Rev. 2008;88:91–134. doi: 10.1152/physrev.00010.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallen K. Hormonal influences on sexually differentiated behavior in nonhuman primates. Front Neuroendocrin. 2005;26:7–26. doi: 10.1016/j.yfrne.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Miller WL, Auchus RJ. The Molecular Biology, Biochemistry, and Physiology of Human Steroidogenesis and Its Disorders. Endocr Rev. 2011;32:81–151. doi: 10.1210/er.2010-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollen AA, et al. Molecular Identity of Human Outer Radial Glia during Cortical Development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davey RA, Grossmann M. Androgen Receptor Structure, Function and Biology: From Bench to Bedside. Clin Biochem Rev Australian Assoc Clin Biochem. 2016;37:3–15. [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagnostics Jmd. 2012;14:22–9. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quartier A, et al. Genes and Pathways Regulated by Androgens in Human Neural Cells, Potential Candidates for the Male Excess in Autism Spectrum Disorder. Biol Psychiat. 2018;84:239–252. doi: 10.1016/j.biopsych.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Qu Y, et al. Constitutively Active AR-V7 Plays an Essential Role in the Development and Progression of Castration-Resistant Prostate Cancer. Sci Rep-uk. 2015;5:7654. doi: 10.1038/srep07654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeselsohn R, et al. Emergence of Constitutively Active Estrogen Receptor-Mutations in Pretreated Advanced Estrogen Receptor-Positive Breast Cancer. Clin Cancer Res. 2014;20:1757–1767. doi: 10.1158/1078-0432.CCR-13-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalinka AT. Improving the sensitivity of differential-expression analyses for under-powered RNA-seq experiments. bioRxiv. 2020 doi: 10.1101/2020.10.15.340737. [DOI] [Google Scholar]
- 25.Tang T, et al. HDAC1 and HDAC2 Regulate Intermediate Progenitor Positioning to Safeguard Neocortical Development. Neuron. 2019;101:1117–1133.:e5. doi: 10.1016/j.neuron.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Li L, Jin J, Yang X-J. Histone deacetylase 3 governs perinatal cerebral development via neural stem and progenitor cells. Iscience. 2019;20:148–167. doi: 10.1016/j.isci.2019.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dey A, et al. YB-1 is elevated in medulloblastoma and drives proliferation in Sonic hedgehog-dependent cerebellar granule neuron progenitor cells and medulloblastoma cells. Oncogene. 2016;35:4256–4268. doi: 10.1038/onc.2015.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kielar M, et al. Mutations in Eml1 lead to ectopic progenitors and neuronal heterotopia in mouse and human. Nat Neurosci. 2014;17:923–933. doi: 10.1038/nn.3729. [DOI] [PubMed] [Google Scholar]
- 29.Abrahams BS, et al. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol Autism. 2013;4:36–36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim TJ, Kim TJ, Lee H, Kim YE, Jeon BS. A case of Parkin disease (PARK2) with schizophrenia: Evidence of regional selectivity. Clin Neurol Neurosur. 2014;126:35–7. doi: 10.1016/j.clineuro.2014.07.034. [DOI] [PubMed] [Google Scholar]
- 31.Udawela M, et al. SELENBP1 expression in the prefrontal cortex of subjects with schizophrenia. Transl Psychiat. 2015;5:e615. doi: 10.1038/tp.2015.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fatemi SH, Folsom TD, Thuras PD. Deficits in GABA(B) receptor system in schizophrenia and mood disorders: a postmortem study. Schizophr Res. 2011;128:37–43. doi: 10.1016/j.schres.2010.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narayan S, Head SR, Gilmartin TJ, Dean B, Thomas EA. Evidence for disruption of sphingolipid metabolism in schizophrenia. J Neurosci Res. 2009;87:278–88. doi: 10.1002/jnr.21822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raudvere U, et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Göttlicher M, et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. Embo J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boissinot M, et al. Induction of differentiation and apoptosis in leukaemic cell lines by the novel benzamide family histone deacetylase 2 and 3 inhibitor MI-192. Leukemia Res. 2012;36:1304–1310. doi: 10.1016/j.leukres.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Saito A, et al. A synthetic inhibitor of histone deacetylase, MS-27-275, with marked in vivo antitumor activity against human tumors. Proc National Acad Sci. 1999;96:4592–4597. doi: 10.1073/pnas.96.8.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexeyenko A, et al. Comparative interactomics with Funcoup 2.0. Nucleic Acids Res. 2012;40:D821–D828. doi: 10.1093/nar/gkr1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 40.Kim W-Y. Brain size is controlled by the mammalian target of rapamycin (mTOR) in mice. Commun Integr Biology. 2015;8:e994377. doi: 10.4161/19420889.2014.994377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 42.Voss MH, et al. Phase 1 study of mTORC1/2 inhibitor sapanisertib (TAK-228) in advanced solid tumours, with an expansion phase in renal, endometrial or bladder cancer. Brit J Cancer. 2020;123:1590–1598. doi: 10.1038/s41416-020-01041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi YJ, et al. Inhibitory Effect of mTOR Activator MHY1485 on Autophagy: Suppression of Lysosomal Fusion. Plos One. 2012;7:e43418. doi: 10.1371/journal.pone.0043418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sohal VS, Rubenstein JLR. Excitation-inhibition balance as a framework for investigating mechanisms in neuropsychiatric disorders. Mol Psychiatr. 2019;24:1248–1257. doi: 10.1038/s41380-019-0426-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marín O, Müller U. Lineage origins of GABAergic versus glutamatergic neurons in the neocortex. Curr Opin Neurobiol. 2014;26:132–141. doi: 10.1016/j.conb.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Shaughnessy PJ, et al. Alternative (backdoor) androgen production and masculinization in the human fetus. Plos Biol. 2019;17:e3000002. doi: 10.1371/journal.pbio.3000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martíez-Cerdeño V, Noctor SC, Kriegstein AR. Estradiol stimulates progenitor cell division in the ventricular and subventricular zones of the embryonic neocortex. Eur J Neurosci. 2006;24:3475–3488. doi: 10.1111/j.1460-9568.2006.05239.x. [DOI] [PubMed] [Google Scholar]
- 48.Eliot L, Ahmed A, Khan H, Patel J. Dump the “dimorphism”: Comprehensive synthesis of human brain studies reveals few male-female differences beyond size. Neurosci Biobehav Rev. 2021;125:667–697. doi: 10.1016/j.neubiorev.2021.02.026. [DOI] [PubMed] [Google Scholar]
- 49.Knickmeyer RC, Baron-Cohen S. Fetal testosterone and sex differences. Early Hum Dev. 2006;82:755–760. doi: 10.1016/j.earlhumdev.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 50.Bahari-Javan S, et al. HDAC1 links early life stress to schizophrenia-like phenotypes. Proc National Acad Sci. 2017;114:E4686–E4694. doi: 10.1073/pnas.1613842114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryskalin L, Limanaqi F, Frati A, Busceti CL, Fornai F. mTOR-Related Brain Dysfunctions in Neuropsychiatric Disorders. Int J Mol Sci. 2018;19:2226. doi: 10.3390/ijms19082226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lancaster MA, et al. Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol. 2017;35:659. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bagley JA, Reumann D, Bian S, Lévi-Strauss J, Knoblich JA. Fused cerebral organoids model interactions between brain regions. Nat Methods. 2017;14:743–751. doi: 10.1038/nmeth.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hines M, Constantinescu M, Spencer D. Early androgen exposure and human gender development. Biol Sex Differ. 2015;6:3. doi: 10.1186/s13293-015-0022-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reyes FI, Boroditsky RS, Winter JDS, Faiman C. Studies on Human Sexual Development. II. Fetal and MaternalSerum Gonadotropin and Sex Steroid Concentrations. J Clin Endocrinol Metabolism. 1974;38:612–617. doi: 10.1210/jcem-38-4-612. [DOI] [PubMed] [Google Scholar]
- 56.McManus JM, Sharifi N. Structure-dependent retention of steroid hormones by common laboratory materials. J Steroid Biochem Mol Biology. 2020;198:105572. doi: 10.1016/j.jsbmb.2019.105572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shoskes JJ, Wilson MK, Spinner ML. Pharmacology of testosterone replacement therapy preparations. Transl Androl Urology. 2016;5:834–843. doi: 10.21037/tau.2016.07.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wright AS, Thomas LN, Douglas RC, Lazier CB, Rittmaster RS. Relative potency of testosterone and dihydrotestosterone in preventing atrophy and apoptosis in the prostate of the castrated rat. J Clin Invest. 1996;98:2558–2563. doi: 10.1172/JCI119074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iacopino F, et al. Valproic acid activity in androgen-sensitive and-insensitive human prostate cancer cells. Int J Oncol. 1992;32:1293–1303. doi: 10.3892/ijo_32_6_1293. [DOI] [PubMed] [Google Scholar]
- 60.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giandomenico SL, et al. Cerebral organoids at the air–liquid interface generate diverse nerve tracts with functional output. Nat Neurosci. 2019;22:669–679. doi: 10.1038/s41593-019-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinform Oxf Engl. 2012;28:882–3. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 66.Gerard D. Data-based RNA-seq simulations by binomial thinning. Bmc Bioinformatics. 2020;21:206. doi: 10.1186/s12859-020-3450-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stuart T, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.:e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cao J, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benito-Kwiecinski S, et al. An early cell shape transition drives evolutionary expansion of the human forebrain. Cell. 2021 doi: 10.1016/j.cell.2021.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Noctor SC, Martínez-Cerdeño V, Kriegstein AR. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J Comp Neurol. 2008;508:28–44. doi: 10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fromer M, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data (bulk and single-cell) have been deposited on NCBI Gene Expression Omnibus (GEO) under accession GSE187877. The following open access databases were used in this study: SZDB: A Database for Schizophrenia Genetic Research (szdb.org), SFARI gene (https://gene.sfari.org/).
Delboy code is available on https://github.com/alextkalinka/delboy.