Abstract
Proteases are among the key regulators of most forms of programmed cell death (PCD) in animals. Also in plants, many PCD processes have been associated with protease expression or activation. However, the functional evidence of the roles and actual modes of action of plant proteases in PCD remains surprisingly limited. In this review, we give an update on protease involvement in the context of developmentally regulated plant PCD. To illustrate the diversity of protease functions, we focus on several prominent developmental PCD processes, including xylem and tapetum maturation, suspensor elimination, endosperm degradation and seed coat formation, as well as plant senescence processes. Despite the substantial advance in the field, protease functions are still often only correlatively linked to developmental PCD, and the specific molecular roles of proteases in many developmental PCD processes remain to be elucidated.
Keywords: plant, programmed cell death (PCD), developmental PCD (dPCD), protease, development, protein degradation
Abbreviations
- PCD
Programmed cell death
- dPCD
Developmentally controlled PCD
- ePCD
Environmentally triggered PCD
- ER
Endoplasmic reticulum
- GFP
Green fluorescent protein
- PLCPs
Papain-like cysteine proteases
- ROS
Reactive oxygen species
- SI
Self-incompatibility or Self-incompatible
- SI-PCD
Self-incompatibility induced programmed cell death
- TUNEL
Terminal deoxynucleotidyl transferase dUTP nick end labeling
- TE
Tracheary element cell in the xylem
- VPEs
Vacuolar processing enzymes
1. Introduction
Programmed cell death (PCD) designates different forms of genetically encoded, tightly controlled processes to eliminate no longer needed, damaged or harmful cells in a targeted fashion. There is a plethora of different PCD pathways, and many of them are vital for the development and fitness of animals and plants. In plants, PCD occurs in the course of regular development (dPCD), as well as a part of various stress responses to the biotic and abiotic environment (ePCD) (Daneva et al., 2016; Huysmans et al., 2017).
During plant development, a surprising number of – partly still putative – dPCD processes have been recognized in different stages of vegetative and reproductive development (Van Hautegem et al., 2015). A paradigm for dPCD in plants occurs during the development of the water-conducting xylem network, whose efficiency relies on the formation of hollow xylem cells that die and become functional in water transport (Escamez and Tuominen, 2014). A precise control of xylem cell death in early vascular plants was a key evolutionary prerequisite for the successful colonialization of terrestrial habitats. Another well-established example of dPCD is the degeneration of the tapetum layer in the floral anther. Several mutants have been characterized in which tapetum differentiation and degeneration is altered, which almost invariably leads to abortion of pollen development and thus male sterility (Wilson and Zhang, 2009). A form of dPCD also disposes of embryonic suspensor cells, which has been shown to be important for embryo development in some plant species (Smertenko and Bozhkov, 2014). After seedling germination, the organ size of the root cap in Arabidopsis (Arabidopsis thaliana) is controlled by a precisely timed dPCD event, which is important for optimal root growth and the patterning of the root system (Fendrych et al., 2014; Xuan et al., 2016). Finally, reproductive self-incompatibility (SI) to avoid inbreeding relies on the specific execution of a cell death program in incompatible self-pollen in some plant taxa (Eaves et al., 2014) (see also article in this Special Issue by Wang et al.). These selected examples of dPCD processes in plants highlight the importance of PCD for successful plant growth and reproduction. Notably, PCD has to be tightly controlled in any context, as ectopic or untimely cellular death can be as detrimental to plant development as the delay or lack of PCD.
Despite substantial advances achieved over the last two decades in the discovery and characterization of plant PCD processes, we still lack a coherent mechanistic understanding of dPCD pathways. In many cases there seems to be a strong link between tissue differentiation and execution of PCD as the ultimate differentiation step (Figure 1). A number of transcription factors have been identified in the coordination of differentiation and dPCD preparation(Cubria-Radio and Nowack, 2019; Van Durme and Nowack, 2016), emphasizing the importance of transcriptionally regulated dPCD-associated genes as a starting point for the generation of research hypotheses. In a large-scale meta-analysis of commonly regulated genes in diverse PCD contexts, we identified a core set of dPCD-associated genes (Olvera-Carrillo et al., 2015). Among these, there is a substantial number encoding hydrolases (Table 1). Though transcriptional reporters based on some of these genes provide a convenient tool for dPCD research (Olvera-Carrillo et al., 2015), there is still little insight into the mechanisms of action of most of the corresponding proteins. Despite substantial efforts, unveiling targets and precise functions of proteases remains a challenging task (Sueldo and van der Hoorn, 2017).
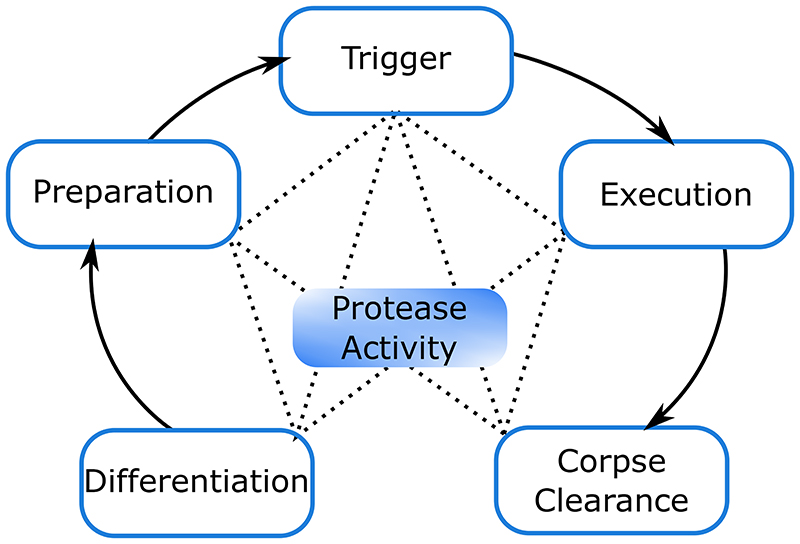
Figure 1. Hypothetical regulatory network of the plant dPCD process.
Coordinated with cellular differentiation, dPCD competency is achieved at least partly on the transcriptional level. A range of PCD-associated genes are upregulated, and their proteins accumulate in cells that are preparing to undergo PCD. Once competent, a hypothetical trigger starts the PCD execution process. Finally, partial or complete corpse clearance is achieved by activity of degrading enzymes that are activated during PCD and complete corpse clearance post mortem. Conceivably, proteases might play roles in all of these steps, but so far only few protease functions in dPCD have been characterized in detail.
Table 1. Proteases commonly upregulated during Arabidopsis dPCD (Olvera-Carrillo et al., 2015).
| AGI | Name | Annotation | Co-expressed with* | Upregulated in dPCD** |
|---|---|---|---|---|
| AT5G04200 | AtMC9 | Metacaspase 9 | 7 genes | b.c. |
| AT4G04460 | PASPA3 | Saposin-like aspartyl protease family protein | 6 genes | 12 conditions |
| AT3G45010 | SCPL48 | Serine carboxypeptidase-like 48 | 5 genes | 12 conditions |
| AT4G35350 | XCP1 | Xylem cysteine peptidase 1 | 4 genes | b.c. |
| AT2G25940 | αVPE | Alpha-vacuolar processing enzyme | 3 genes | 8 conditions |
| AT1G20850 | XCP2 | Xylem cysteine peptidase 2 | 3 genes | b.c. |
| AT1G23460 | n.n. | Pectin lyase-like superfamily protein | 3 genes | b.c. |
| AT1G63120 | ATRBL2 | RHOMBOID-like 2 | 3 genes | b.c. |
| AT5G50260 | CEP1 | Cysteine proteinases superfamily protein | 2 genes | b.c. |
| AT1G01900 | ATSBT1.1 | Subtilase family protein | n.a. | 8 conditions |
based on a Genevestigator co-expression analysis with other dPCD-associated genes
dPCD conditions covered in meta-analysis
n.n. no name
n.a. not applicable
b.c. below cutoff value of 8 conditions
As described in more detail in other articles of this Special Issue, proteases (also called peptidases or proteinases) are enzymes that cleave target proteins, generally via hydrolysis-driven proteolysis. There are hundreds of protease-encoding genes in the genomes of multicellular organisms, for instance, there are more than 800 in Arabidopsis (van der Hoorn, 2008). This vast number of proteases can be grouped according to their catalytic residues as aspartic-, cysteine-, serine-, threonine-, glutamic-, asparagine-, or metalloproteases (Oda, 2012). In the MEROPS database (http://merops.sanger.ac.uk/, (Rawlings et al., 2010)), proteases are grouped in families (based on amino-acid sequence similarity) and in clans (based on tertiary structure), with some groups comprising over 100 individual members.
Proteases likely originated in the earliest protein-producing organisms as a result of the need to recycle amino acids (Poręba et al., 2013). In addition to this ancestral housekeeping function, proteases have adopted novel functions as posttranslational modifiers, thus becoming key components in the regulation of biological processes. Hydrolytic cleavage irreversibly alters target proteins; they can be activated or inactivated, stabilized or destabilized, their structure can be modified, or targets can be re-localized to different subcellular compartments (Schaller, 2004). Owing to this versatile regulatory capacity, proteases have been identified as regulators of a plethora of biological processes, including, but not restricted to, many steps of plant vegetative and reproductive development, as well as the orchestration of local and systemic defense responses (van der Hoorn, 2008).
Proteases have also been implicated in the regulation of many plant PCD processes. In fact, proteases are key regulators and executors of several cell death processes in animal systems, including apoptosis (caspase-mediated PCD) and pyroptosis (caspase-1 dependent proinflammatory PCD) (Fink and Cookson, 2005). In these contexts, the most prominent proteases are cysteine-dependent, aspartate-specific proteases (caspases). The activation of initiator caspases occurs both in the extrinsic and the intrinsic apoptosis pathway and leads to the activation of executioner caspases. Thus, caspases form a proteolytic cascade that eventually leads to the direct or indirect cleavage of hundreds of target proteins (Poręba et al., 2013). Although a few key caspase targets have been identified, it seems that the concerted cleavage of many proteins is decisive for the execution of caspase-mediated cell death (Julien and Wells, 2017). More recently, alternative caspase-independent pathways have been described. Necroptosis, for instance, does not require caspase activity to be initiated; on the contrary, caspase-8 is an important inhibitor of necroptosis (Fuchslocher Chico et al., 2017). On the other hand, caspases have also been implicated in other, non-PCD-related cellular remodeling events (Julien and Wells, 2017).
Plant scientists started to investigate the role of proteases in cell death regulation over two decades ago. In several plant PCD systems, specific activity probes reported the presence of caspase-like activities, and, in some of these systems, chemical caspase inhibitors were shown to alter PCD progression (Bonneau et al., 2008). The quest for caspase homologs in plants took a turn with the publication of whole genome sequences, which revealed that caspases are not conserved outside animals (Uren et al., 2000), leading to the study of the structurally related metacaspases and paracaspases, grouped in the same family (C14) and clan (CD) of cysteine proteases. Metacaspases are subdivided into types I, II, and III, depending on their overall structure. Only type I and II metacaspases are found in plants. Type I metacaspases may contain an N-terminal extension, whereas type II metacaspases lack extra N-terminal motifs and generally have a longer linker region between the p10 and p20 motifs. The diversity of caspase homologs has been recently reviewed (Klemenčič and Funk, 2018). Although metacaspases became prime candidates in the search of functional caspase analogs in plants, it was finally shown that plant metacaspases lack aspartate specificity, and rather cleave after basic arginine or lysine residues (Vercammen et al., 2004). Hence, although some metacaspases have been implicated in types of plant PCD (Minina et al., 2017), they cannot be directly responsible for the observed caspase-like activities. Instead, several non-C14 family proteins have been shown to generate caspase-like activities in the plant PCD context, among them vacuolar processing enzymes (VPEs, (Hatsugai et al., 2015), phytaspases (Galiullina et al., 2015; Reichardt et al., 2018), cathepsin B (Cai et al., 2018), and subunits of the 26S-proteasome (Hatsugai et al., 2009).
In this review, we will explore the roles of proteases that have been implicated in the regulation or progression of developmentally regulated PCD processes in plants. We will focus on dPCD occurring in the xylem, the tapetum, the embryonic suspensor, the endosperm and the seed coat, and finally in senescing plant organs (Figure 2). Other PCD processes, including the Arabidopsis root cap turnover (Huysmans et al., 2018; Kumpf and Nowack, 2015; Xuan et al., 2016), lace plant (Aponogeton madagascariensis) leaf perforation (Dauphinee et al., 2014; Lord et al., 2013), or root aerenchyma formation (Takahashi et al., 2015; Yamauchi et al., 2016) will not be included, as there is still very little functional data available on the proteases involved in these processes. Although in many dPCD processes, proteases and protease activities have been found to be associated with different stages of cell death progression, evidence of the precise function and the specific mode of action of these proteases is rare. We attempted not only to summarize up-to-date information on dPCD proteases, but also to point out areas in which we still have to make further progress to understand dPCD regulation and execution. Of note, the roles of proteases in the context of plant immunity- and stress-related PCD processes will not be covered, and we refer to a number of excellent reviews focusing on these topics that have been recently published (Balakireva and Zamyatnin, 2018; Hou et al., 2018; Thomas and van der Hoorn, 2018).
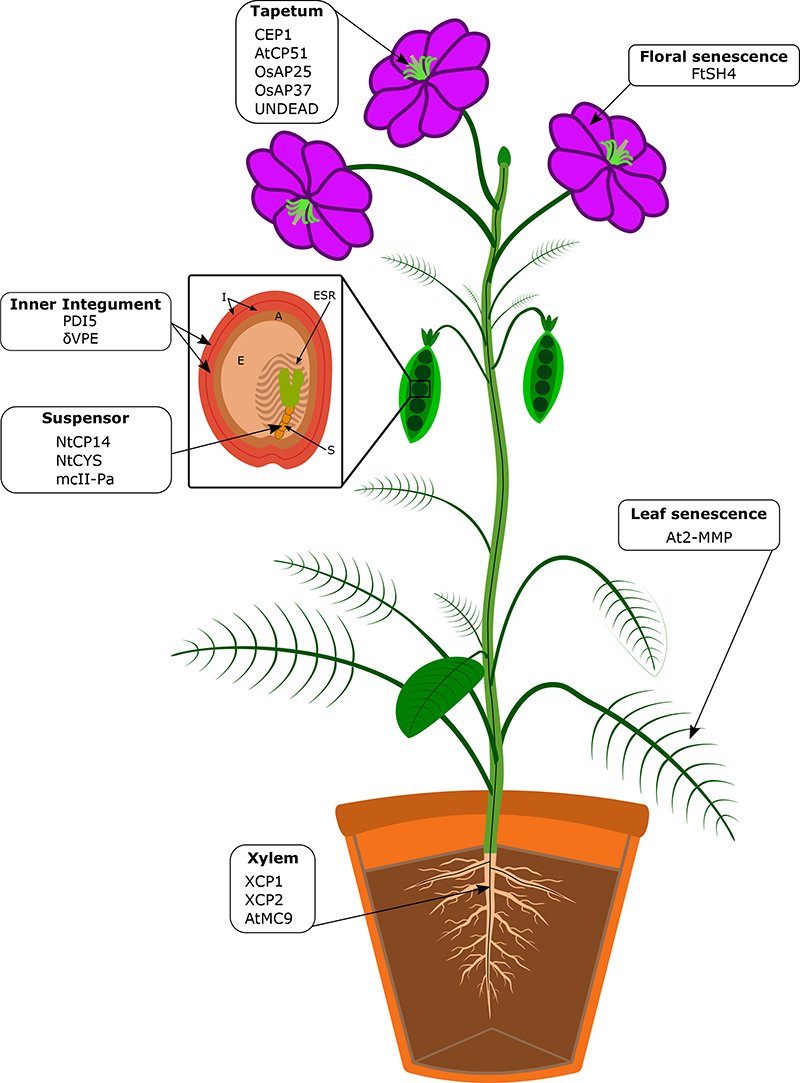
Figure 2. Proteases implicated in selected PCD processes occurring during plant development.
PCD processes are an integral part of vegetative and reproductive plant development. They occur during xylem and tapetum differentiation, in various tissues during seed development and germination, and during plant organ senescence. We indicate proteases that have been implicated with aspects of these cell death processes in the individual tissues. For some of the PCD processes listed, no functional data for protease involvement exists to date. Abbreviations are: A, aleurone layer; E, endosperm; ESR, embryo-surrounding region of the endosperm; I, seed integuments; S, suspensor.
2. Proteases in xylem PCD
Long distance water transport in vascular plants is facilitated by the xylem. In angiosperms, the xylem is composed of an interconnected network of tracheary elements (TEs) and tracheids, as well as parenchyma and fiber cells. The differentiation of TEs and tracheids involves extensive secondary cell wall deposition and lignification. PCD and the ensuing post-mortem degradation of the protoplast, while maintaining the reinforced cell walls, is essential for the final function of the xylem as an efficient water transport system (Daneva et al., 2016). The progression of PCD in TEs is marked by the release of vacuolar contents caused by tonoplast rupture, triggering the deterioration of the cytoplasm, the dismantling of the endomembrane system, and mitochondrial degradation (Groover et al., 1997; Kuriyama, 1999; Yu et al., 2002).
Xylem-expressed proteases have been identified and implicated in xylem differentiation and PCD in different plant systems (Böllhoner et al., 2018; Hao et al., 2008; Liu et al., 2015; Obudulu et al., 2016; Petzold et al., 2012; Sueldo and van der Hoorn, 2017; Zhang et al., 2015). In fact, xylem differentiation and PCD are tightly coordinated. VND6 (VASCULAR-RELATED NAC-DOMAIN 6) and VND7 are two central NAC transcription factors implicated in xylem differentiation in Arabidopsis and poplar (Populus spec.) (Nakaba et al., 2015; Ohashi-Ito et al., 2010; Yamaguchi et al., 2010). Closely related NAC transcription factors have been implicated in xylem development of other species, including gymnosperms and non-vascular plants (Duval et al., 2014; Laubscher et al., 2018; Xu et al., 2014), suggesting a high degree of evolutionary conservation of xylem differentiation. Although the ectopic overexpression of VND6 or VND7 leads to the expression of several genes believed to control differentiation and PCD, among them several proteases (Ohashi-Ito et al., 2010; Valdivia et al., 2013; Yamaguchi et al., 2010; Yamaguchi et al., 2011; Zhong et al., 2010), only few xylem proteases have been functionally investigated in detail to date.
XCP1 (XYLEM CYSTEINE PROTEASE 1) and XCP2 have been identified as xylem-expressed papain-like cysteine proteases (PLCPs) that are produced in preparation for PCD, accumulate in the vacuolar lumen and are to a smaller extent associated with the endoplasmic reticulum (ER) and Golgi of Arabidopsis TEs (Avci et al., 2008; Zhao et al., 2000). Although simultaneous deletion of both gene functions does not lead to developmental disturbances or PCD impairment, transmission electron microscopy showed a delay in the PCD-associated cell clearance (Avci et al., 2008). Consistent with this role in Arabidopsis, Tr-cp14, a cysteine protease with homology to XCP1 and XCP2, has been identified in white clover (Trifolium repens) and shown to accumulate in the ER prior to vacuolar collapse (Mulisch et al., 2013). Although accumulation of XCPs seems to be part of xylem differentiation, their activation seems to be confined to precise developmental stages. Despite the transcriptional accumulation of PsgXCP1, PsgXCP2A, and PsgXCP2B in poplar, ray parenchyma cells (radially arranged cells in woody tissues) remained alive for extended periods of time, even after secondary cell wall deposition (Nakaba et al., 2015). Despite the fact that ray parenchyma cells might follow differentiation programs distinct from TEs, this finding suggests that XCP proteases have to be post-transcriptionally activated in order to affect post-mortem clearance.
In poplar, the activity of the proteasome has been linked to the caspase 3-like activity observed during TE differentiation and PCD (Han et al., 2012). Although chemical inhibition of proteasome activity by clasto-lactacystin β-lactone in Zinnia (Zinnia elegans) and Arabidopsis xylogenic cell cultures suppressed PCD, it was difficult to separate this effect from a post-mortem clearance effect or a potential disruption of early xylem differentiation (Han et al., 2012; Woffenden et al., 1998). Reinforcing the complexity of separating the potential pro-PCD effect of proteases from a role in early differentiation, chemical caspase inhibitors have been shown to affect the kinetics of TE differentiation in Zinnia cell cultures, but not to block PCD itself (Twumasi et al., 2010).
Albeit not exerting caspase-like activity, metacaspases have been implicated in xylem PCD. In Arabidopsis, an atmc9 mutant displays delayed xylem post-mortem corpse clearance, but PCD onset is not affected. Although post-mortem clearance is eventually achieved in triple atmc9 xcp1 xcp2 mutants, activity profiling of PLCPs in whole-plant extracts showed overall reduction in PLCP activity in the triple mutants in a manner that cannot be explained by atmc9 or xcp1 xcp2 mutations alone (Bollhöner et al., 2013), pointing to an interplay between the different proteolytic activities. AtMC9 has been localized to nucleocytoplasm, the apoplast, and the vacuolar lumen (Böllhoner et al., 2018; Escamez et al., 2016; Tsiatsiani et al., 2013; Vercammen et al., 2006) (Figure 3). Further supporting a post-mortem role, the acidic pH optimum for the cytoplasmic-localized AtMC9 might only be achieved after vacuolar collapse as a committing step toward PCD (Bollhöner et al., 2013).. Whereas the lowering of cytoplasmic pH might enhance the activity of the cytoplasmic pool of AtMC9 without necessarily changing its abundance, the apoplastic pool might be independently regulated by the abundance of apoplastic Serpin1 (Vercammen et al., 2006). Serpin1 is a suicide protease inhibitor that has also been localized to the cytoplasm and shown to interact with other proteases such as the PLCP RESPONSIVE TO DISSECATION 21 (RD21), likely after vacuolar collapse as RD21 accumulates in the vacuole (Lampl et al., 2013; Lema Asqui et al., 2018). In agreement with a non-essential role of MC9 in xylem PCD, the downregulation of putative MC9 poplar homologs, PttMC13 and PttMC14, is not sufficient to alter xylem differentiation (Böllhoner et al., 2018). However, the activity of PttMC13 and PttMC14 coincides with the processing of RD21, again suggesting a participation of metacaspase activity in a proteolytic cascade after vacuolar collapse potentially aiding in corpse clearance (Böllhoner et al., 2018). Similarly, downregulation of AtMC9 in Arabidopsis xylogenic cell cultures fails to alter the timing or extent of TE-PCD, though in accordance with the atmc9 mutant, post-mortem autolysis is affected. Interestingly, AtMC9 downregulation causes an accumulation of autophagic bodies in TE-cell vacuoles, and is associated with an increased death rate of parenchymatic non-TE cells (Escamez et al., 2016). Non-TE cells do not express AtMC9 in these cell cultures and, in contrast to TE-cells, survive xylogenic induction. Concomitant downregulation of AtMC9 and the autophagy-related gene ATG2 in TE cells alleviates the ectopic cell death phenotype, suggesting that the level of autophagy in TE cells influences the survival rate of neighboring non-TE cells (Escamez et al., 2016).
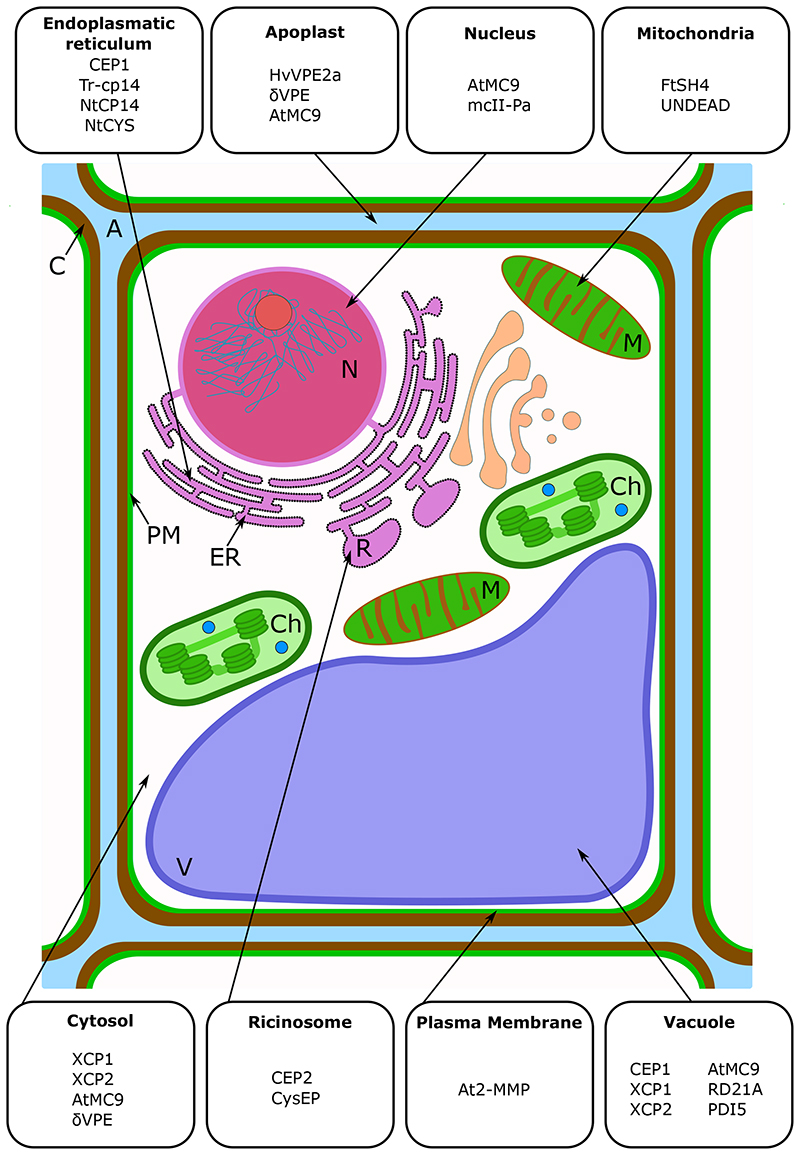
Figure 3. Selection of dPCD-associated proteases with known or predicted subcellular localizations.
Many dPCD-associated proteases accumulate as inactive proenzymes or in specific subcellular compartments before they are activated. Especially the vacuole, but also the ER and ER-derived vesicles can serve as storage space for inactive proteases. Once PCD execution is triggered, the breakup of cellular compartments is thought to release these proteases into the same compartments as their targets. The specific chemical environment of subcellular compartments before and after compartment breakup can be an additional cue to activate PCD-associated proteases. Abbreviations are: A, apoplast; C, cell wall; Ch, chloroplast; ER, endoplasmic reticulum; M, mitochondrion; N, nucleus; PM, plasma membrane; R, ricinosome; V, vacuole.
Although xylem PCD is one of the most intensively studied dPCD processes, and many proteases and protease activities have been linked to xylem PCD, our knowledge of roles and mechanistic details of plant protease functions in this process remains limited. For instance, although a general program of xylem differentiation including upregulation of NAC transcription factors with subsequent upregulation of protease genes has been observed for the different species studied, the subcellular localization and target identification of these proteases need better elucidation. Moreover, with most of the phenotypes of analyzed mutants being related with post-mortem degradation, the role of proteases in earlier TE PCD steps remains to be unraveled.
3. Proteases in tapetum PCD
Pollen development is essential for successful sexual reproduction in plants. Pollen formation takes place within the anthers and is dependent on the proper development of the tapetum, a layer of cells lining the anther locule (Daneva et al., 2016). Pollen development and pollen exine (tough outer walls of pollen grains) formation depend on correct tapetum differentiation, which induces dPCD of tapetal cells at a specific stage of pollen development (Ariizumi and Toriyama, 2011; Plackett et al., 2011). Prior to dPCD, sporopollenin components are exported from intact tapetum cells, whereas the pollen coat is deposited later when tapetum dPCD occurs and their cellular content is released in the locule cavity in which the pollen grains develop. The precise timing of differentiation and PCD is crucial, as precocious or delayed tapetum differentiation and PCD almost invariably result in male sterility (Bedinger, 1992; Kawanabe et al., 2006; Lam et al., 2000). Nevertheless, as differentiation and degeneration of the tapetum is tightly linked, it is difficult to distinguish if tapetum differentiation and early secretion or later dPCD-related defects affect pollen development in mutant plants. In this context, several protease activities have been associated with tapetum development (Hierl et al., 2012; Omidvar et al., 2017; PHan et al., 2012; Sheoran et al., 2009; Shukla et al., 2016; Solís et al., 2014), but few of them have been functionally analyzed in detail.
Arabidopsis CYSTEINE ENDOPEPTIDASE 1 (CEP1), a KDEL-tailed cysteine protease from the PLCP family, has been identified as a key element of tapetum PCD (Zhang et al., 2014). CEP1 is expressed in the tapetum from stage 5 to 11 of anther development. CEP1 is transported in vesicles as an inactive proenzyme to the vacuole, where maturation occurs, and the active form is released to the cytosol after vacuolar rupture. Knock-out of CEP1 leads to abnormal and delayed tapetal PCD, and reduction in pollen fertility. Conversely, the overexpression of CEP1 under the 35S promoter causes premature tapetal PCD and, as in the cep1 mutant, a reduction in the number of viable pollen grains (Zhang et al., 2014). Interestingly, transcriptome analyses of cep1 mutant flower buds revealed the upregulation of hundreds of genes in comparison to the wild type already in early stages of anther development before tapetum PCD (Zhang et al., 2014), suggesting an additional role for CEP1 in early tapetum differentiation.
Another cysteine protease, AtCP51, has been identified as being expressed during anther development (Yang et al., 2014). Its expression was observed in different parts of the anther with strong promoter activity detected in tapetal cells and microspores. Contrarily to CEP1, AtCP51 seems to have a pro-survival function, because silencing of AtCP51 by RNAi results in male sterility as a result of early tapetum degradation. Homologs of AtCP51 have been found in various plant species such as OsCP1 in rice (Oryza sativa) and NtCP56 in tobacco (Nicotiana tabacum) (Yang et al., 2014). Analysis of promoter activity suggested OsCP1 expression in late anther development, including pollen and tapetum. The oscp1 mutant shows overall delay in plant growth, and abnormal pollen development with reduced male fertility (Lee et al., 2004). However, whether OsCP1 controls pollen viability directly or via tapetum cell death has not been established. OsCP1 together with Osc6, encoding a putative protease inhibitor, have been shown to be directly regulated by the tapetum-specific transcription factor TDR (Tapetum Degeneration Retardation). Knock-out of TDR results in a delayed tapetum degeneration in rice, characterized by lower OsCP1 and Osc6 levels (Li et al., 2006), confirming the link between OsCP1 expression and tapetal PCD.
Next to cysteine proteases, aspartic proteases have been implicated in PCD processes throughout plant development, including tapetum PCD (Gao et al., 2017a; Gao et al., 2017b; Ge et al., 2005). Two aspartic proteases in rice, OsAP25 and OsAP37, have been found to be under direct transcriptional control of ETERNAL TAPETUM1 (EAT1) (Niu et al., 2013). EAT1 is a tapetum-expressed basic helix-loop-helix transcription factor and EAT1 knock-out leads to a delay in PCD, causing male sterility. Both OsAP25 and OsAP37 are strongly downregulated in eat1 mutants, and transient expression of both proteases causes ectopic cell death in Nicotiana benthamiana leaves, which was blocked in presence of the aspartic protease inhibitor pepstatin A. These results suggest that the protease activities of OsAP25 and OsAP37 are necessary and sufficient to drive cells into PCD. (Niu et al., 2013)
In contrast to OsAP25 and OsAP37, the Arabidopsis aspartic protease UNDEAD is a negative regulator of tapetum PCD. UNDEAD expression is controlled by the tapetum transcription factor AtMYB80, a key regulator of tapetum differentiation and PCD (Phan et al., 2011). Promoter activity analysis showed expression in the tapetum throughout the development of anthers, but mainly in young floral stages. The presence of a putative mitochondrial targeting signal suggests that UNDEAD might be localized to, and act in, mitochondria (Figure 3). RNAi-mediated knock-down of UNDEAD mimics the myb80 phenotype of premature tapetal degeneration, characterized by an early onset of chromatin degradation in tapetum cells (Phan et al., 2011). These results establish UNDEAD as a pro-survival protease that fine-tunes the timing of tapetum PCD downstream of MYB80. It is tempting to speculate that UNDEAD targets and inactivates mitochondria-localized pro-PCD factors, whose premature accumulation leads to early cell death in myb80 and undead mutants. Future studies with UNDEAD overexpression might help to elucidate its pro-survival role downstream of MYB80.
4. Proteases in seed development-related PCD
The plant seed is a complex organ consisting of both zygotic fertilization products (embryo and endosperm) as well as maternally derived tissues (seed integuments, nucellus). While the embryo develops to carry on life to a new sporophytic generation, all other seed tissues die at specific stages during seed development (Daneva et al., 2016; López-Fernández and Maldonado, 2015). In the following section, we will review the role of proteases in cell death processes occurring in the different seed compartments during seed development and germination.
4. 1 Proteases in embryonic suspensor PCD
The embryonic suspensor is formed after the asymmetric division of the zygote. The terminal cell gives rise to the embryo proper, the basal cell to the suspensor cells. The suspensor promotes early embryo growth and undergoes a form of dPCD when the embryo enters the maturation phase (Kawashima and Goldberg, 2010; López-Fernández and Maldonado, 2015; Smertenko and Bozhkov, 2014). Although suspensor dPCD has only been investigated and described in few species, it is clear that correct PCD progression in suspensor cells can be critical for embryo development (Daneva et al., 2016).
One of the major model systems to study suspensor dPCD is the suspensor of somatically generated embryos of Norway spruce (Picea abies). In this in-vitro system, the large spruce suspensor is easily accessible for biochemical, pharmacological, and genetic approaches (Filonova et al., 2008). Caspase-like VEIDase activity has been detected specifically in embryos at the onset of suspensor PCD, and its inhibition has been found to disturb embryo development by interfering with suspensor differentiation (Bozhkov et al., 2004). In this context, a type II metacaspase, mcII-Pa, has been identified as being expressed in suspensor cells that are committed to PCD (Suarez et al., 2004). While silencing of mcII-Pa leads to a reduced caspase-like activity on a VEID-based substrate (Suarez et al., 2004), it has been shown that mcII-Pa, like other metacaspases, does not possess an intrinsic caspase activity (Bozhkov et al., 2005). Nonetheless, mcII-Pa has been described to localize to the nucleus in dying cells (Figure 3), recombinant mcII-Pa is able to promote the degradation of nuclei in suspensor cell extracts, and its silencing reduces the frequency of DNA degradation as seen by TUNEL staining (Bozhkov et al., 2005; Suarez et al., 2004). A recent RNA-sequencing-based transcriptomics analysis revealed that several dPCD-associated genes are expressed in suspensor cells of Norway spruce. Those include the transcription factors XND1 and ANAC075, as well as putative spruce homologs of MC9, CEP1, and cathepsin B-like genes (Reza et al., 2018).
An interplay between mcII-pa and autophagy during the development of suspensor cells has been observed in Norway spruce, pointing to a regulatory role for protease activities. The downregulation of autophagy has been shown to phenocopy embryo and suspensor development defects and leads to aberrant suspensor cell differentiation with divergent cell death (Minina et al., 2013). Moreover, the downregulation of mcII-Pa leads to reduced autophagy, potentially reducing the cytoprotective activity of the latter in the rapidly differentiating suspensor cells. Although the mechanisms for the action of mcII-Pa in autophagy are not yet clear, a role for this metacaspase in the inactivation of an autophagy repressor has been suggested by Minina and colleagues (2014).
In tobacco, the differentiation and final PCD of suspensor cells have been linked to the opposing activity of NtCP14, a cathepsin H-like PLCP, and the interacting cystatin protease inhibitor NtCYS (Zhao et al., 2013). While upregulation of NtCP14 is sufficient to induce embryonic cell death and abortion, both overexpression of NtCYS or silencing of NtCP14 leads to a delay in the onset of suspensor PCD. However, downregulation of NtCYS also leads to increased caspase 1-, 3-, and 6-like activities that do not seem to be caused by NtCP14, suggesting other targets or indirect effects (Zhao et al., 2013). NtCYS has also been shown to localize, together with NtCP14, to the ER (Zhao et al., 2013), suggesting either a subcellular confinement of its action to the ER or the existence of roles that are only fulfilled after the release of ER content (Figure 3). Nine other cystatins have been identified in tobacco and their differential localizations among the cytoplasm, ER, and vacuolar lumen can be a hint at different targets and processes (Zhao et al., 2014). Interestingly, the control of suspensor PCD by NtCYS in tobacco interspecific hybrids is controlled in a parent-of-origin dependent fashion. The levels of the maternally, but not the paternally, inherited allele are controlled to ensure the correct timing of suspensor PCD (Luo et al., 2016).
In contrast to the spruce tobacco suspensors, little is known about the regulation of dPCD in the Arabidopsis 7- to 9-cell suspensor (Peng and Sun, 2018), and even less about the role of proteases in this process. CEP1 is expressed in suspensor cells prior to PCD execution where it localizes to subdomains of the ER (Zhou et al., 2016). This localization prompts the hypothesis that CEP1 is stored in these structures until triggering of PCD leads to its release, unleashing its proteolytic activity (Figure 3). However, its actual role in the dying suspensor remains uncertain, as no suspensor phenotype has been identified in the respective mutants (Zhou et al., 2016). KISS OF DEATH (KOD), a PCD-inducing peptide expressed in Arabidopsis suspensor cells, has been shown to be involved in suspensor PCD. Loss-of-function kod mutants have a reduced proportion of suspensor cells initiating PCD. In agreement with a pro-PCD role, ectopic overexpression of KOD in both tobacco leaves and Arabidopsis seedlings is sufficient to induce cell death. Interestingly, the expression of KOD leads to increased caspase 3-like activity, and co-expression of the pan-caspase inhibitor p35 with KOD causes a reduction of cell death (Blanvillain et al., 2011). These data suggest that the enhancement of caspase-like activity is crucial for KOD-induced death, though the mechanism of peptide signaling in the regulation of proteolytic activity is yet poorly understood.
Further suggesting a role for proteases in suspensor development and its final PCD, caspase-like activities have also been observed in the large suspensor of runner bean (Phaseolus coccineus) (Lombardi et al., 2007). Altogether, an enhancement of proteolytic activity during suspensor development culminating in PCD seems to be a general phenomenon affecting embryo development.
4.2. Proteases in endosperm PCD
Next to the embryo, the endosperm is the second fertilization product generated by the double fertilization process in seed plants. In contrast to the embryo, however, the endosperm does not survive past the seed stage. Nevertheless, the endosperm has important functions as hybridization barrier and as nutritive tissue for the embryo during seed development and germination (Becraft and Gutierrez-Marcos, 2012; Lafon-Placette and Köhler, 2016). Because of its role as nutrient storage in mature cereal seeds, several cereal species have been domesticated over the past millennia and now form the most important source of carbohydrate nutrition for humankind.
Several instances of dPCD terminate the life of different endosperm tissues at specific stages of seed development (Costa et al., 2004; Daneva et al., 2016; López-Fernández and Maldonado, 2015). In many plant species, the endosperm adjacent to the developing embryo will disappear, providing space for the growing embryo (Sabelli and Larkins, 2009). In cereals, most of the nutrients for seedling germination are stored in the starchy endosperm, which undergoes PCD after seed filling is completed (Sabelli and Larkins, 2009). In contrast to lytic endosperm death, starchy endosperm death preserves the intact starch-filled cell corpses until seed germination. Unlike the central endosperm, the peripheral aleurone layer of the endosperm shows similarities to the embryo in that both survive desiccation during seed maturation (Becraft and Gutierrez-Marcos, 2012). In cereals, the aleurone organizes nutrient remobilization during germination by secreting lytic enzymes into the dead mass of the starchy endosperm (Domínguez and Cejudo, 2014). When this function has been fulfilled, the aleurone as last surviving endosperm tissue will in turn undergo cell death (Bethke et al., 2007; Sabelli and Larkins, 2009). Though many protease genes and specific protease activities have been found to be expressed in association with endosperm degeneration (Domínguez and Cejudo, 1999; Domínguez et al., 2002; Rocha et al., 2013; Sreenivasulu et al., 2006; Szewińska et al., 2016; Tran et al., 2014), our knowledge about functional aspects of these proteases is still very restricted.
During lytic endosperm breakdown in the embryo-surrounding region of Arabidopsis, the bHLH transcription factor ZHOUPI (ZOU) appears to have a central role, as zou mutant endosperm persists, severely impeding embryo growth (Waters et al., 2013). The action of ZOU appears to result in a weakening of endosperm cell walls, allowing the growing embryo to exert mechanical pressure on the endosperm cells (Fourquin et al., 2016), causing either a passive disruption, or a mechanically triggered active PCD process in the embryo-surrounding endosperm. The expression of several proteases depends on ZOU, including the PCD-associated PUTATIVE ASPARTIC PROTEINASE A3 (PASPA3) and the subtilisin-like protease ABNORMAL LEAF SHAPE1 (ALE1) (Fourquin et al., 2016; Yang et al., 2008). Although the functional role of PASPA3 is still unknown, ALE1 is not involved in endosperm breakdown, but in the epidermal surface formation of the developing embryo (Yang et al., 2008). Recently, putative ZOU orthologs have been described in other species in association with endosperm development (Dou et al., 2018; Feng et al., 2018; Zhang et al., 2017b), but whether the role of ZOU in endosperm breakdown is conserved in all plant clades is unclear.
During starchy endosperm PCD in cereal seeds, the pattern of cell death progression varies (Becraft and Gutierrez-Marcos, 2012). In wheat, a rather stochastic cell death pattern has been observed (Young and Gallie, 1999), whereas maize (Zea mays) and rice endosperm cell death is initiated in the central part and progresses outward (Kobayashi et al., 2013; Young et al., 1997). In several cases, caspase-like activities have been detected in association with different instances of starchy endosperm cell death. For example, an increase in several caspase-like activities has been detected in the maturing endosperm of barley (Hordeum vulgare) and found to coincide with the expression of protease-encoding genes such as HvVPE1 and HvPhS1, and with nuclear DNA degradation in the starchy endosperm (Tran et al., 2014). Previously, VEIDase (caspase 6-like) activity has been detected, though preferentially in young, rapidly developing tissues of the barley caryopsis, not corresponding to the onset of DNA degradation (Borén et al., 2006). Also in the starchy endosperm of rice, VEIDase activity has been detected prior to the onset of endosperm degradation (Kobayashi et al., 2013). In the later stage of starchy endosperm development, several protease-encoding genes are upregulated. Endosperm-expressed proteases include metalloproteases and subunits of the 20S/26S proteasome complex. Interestingly, serine and aspartic protease transcripts that are present in pericarp tissues undergoing cell death are not expressed in the starchy endosperm (Sreenivasulu et al., 2006), suggesting that the preservation of starchy endosperm corpses might depend on the lack of protease activity. Specific protease activity might be further regulated by protease inhibitors that are expressed in the endosperm, but not in the degenerating pericarp (Sreenivasulu et al., 2006). Despite these expression data, to date there is still little knowledge about the role of proteases in the direct regulation of cell death in the starchy endosperm in cereals.
In cereals and other taxa, the aleurone endosperm layer is the only endosperm tissue that survives seed development. In developing seeds of wheat (Triticum aestivum), both the embryo and the aleurone express a cystatin protease inhibitor named WC5 (Wheat Cystatin 5) (Corre-Menguy et al., 2002), possibly to protect these tissues from death-associated protease activities. Nevertheless, the aleurone finally dies during germination. Aleurone PCD is a well-established process, characterized by an accumulation of reactive oxygen species and the occurrence of internucleosomal DNA fragmentation (Domínguez and Cejudo, 2014). Before dying, the aleurone secretes large quantities of enzymes, including proteases, to mobilize the storage starch and proteins in the dead bulk of the starchy endosperm (Domínguez and Cejudo, 1999; Martinez et al., 2009). Recently, the N-end rule pathway controlling targeted proteolysis has been implicated in the storage reserve mobilization process in Arabidopsis (Zhang et al., 2018). However, proteases involved in aleurone PCD remain little investigated. In dying barley aleurone cells, a spectrum of nuclease and protease activities has been described (Fath et al., 2000). The aleurone-expressed wheat carboxypeptidase CPIII has also been found to be associated with xylem TE differentiation and PCD in wheat seedlings, suggesting a common PCD-relevant function in both tissues (Domínguez et al., 2002). However, whether CPIII is functionally involved in xylem cell death and might have other roles than the mobilization of endosperm proteins during germination, remains to be investigated.
In other plant taxa, for instance, castor bean (Ricinus communis), the bulk of the endosperm undergoes cell death only during germination (Schmid et al., 1999). In castor bean, endopeptidases with a C-terminal KDEL retention motif have been identified in specific ER-derived compartments, dubbed ricinosomes (Schmid et al., 1998; Schmid et al., 2001). Ricinosomes accumulate the KDEL-proteases and release them during PCD into the cytoplasm, where they become activated by cleavage of a pro-peptide (Schmid et al., 1999). Similar KDEL-proteases, CEP1, CEP2, and CEP3, have also been identified in Arabidopsis as being expressed in association with dPCD (Hierl et al., 2014; Zhou et al., 2016) and involved in pathogen defense (Höwing et al., 2017; Höwing et al., 2014), though their precise mode of action remains to be elucidated. CEP2 was found to accumulate in ricinosome-like vesicles and to be activated by a pH-dependent proteolytic step (Hierl et al., 2014). Similarly, in the endosperm of germinating tomato (Solanum lycopersicum) seeds, ricinosome-like structures and KDEL-tailed cysteine protease activities have been detected (Trobacher et al., 2013).
Although there is ample evidence of protease expression and activity during the different cell death and degradation processes that occur in the endosperm, it remains challenging to define the precise mechanism of protease action in control of the cell death process.
4.3. Proteases in seed integument and nucellus PCD
The seed integuments are a multi-layered structure of maternal tissue that surrounds the developing embryo and endosperm in seed plants. During seed development and maturation, the different layers follow individual differentiation programs (Beeckman et al., 2000). All cell layers eventually undergo dPCD, leading to the formation of the seed coat or testa, a tough protective layer surrounding the mature embryo (Daneva et al., 2016). Further to its protective role, the seed coat stores an abundance of different proteins, including proteases, that might fulfil important functions during seed dormancy, protection, and germination (Raviv et al., 2017).
Accumulation and upregulation of proteases concomitant with the development of the seed coat and PCD have been reported for different species. Several proteases are upregulated during seed coat formation in the oil-producing tree Jatropha curcas, including cysteine and serine proteases, and subtilases (Rocha et al., 2013; Soares et al., 2017). In tomato, five VPE genes, named SlVPE1 through SlVPE5, have been identified. Based on phylogenetic and expression analysis, SlVPE1 and SlVPE2 are expressed in the seed coat, similarly to δVPE in Arabidopsis. However, even with the downregulation of all five SlVPEs by RNAi, the seed coat is able to differentiate normally (Ariizumi et al., 2011). In rapeseed (Brassica napus), BnCysP1, a putative cysteine protease that lacks a KDEL motif and is structurally related to the leaf senescence-associated genes BnSAG-12-1 and BnSAG-12- 1, has been identified to be expressed only in the inner integument prior to cell death (Obermeier et al., 2009; Wan et al., 2002).
In Arabidopsis, the seed coat develops from five integument layers that differentiate and finally undergo cell death in a specific succession (Haughn and Chaudhury, 2005). The Arabidopsis genome codes for four different VPE genes, with δVPE being specifically expressed in the inner integument (Nakaune et al., 2005). While the other three Arabidopsis VPEs (αVPE, βVPE, and γVPE) have been shown to localize to vacuoles, δVPE was found in aggregates between the cell wall and plasma membrane of shrinking integument cells (Figure 3). In the δvpe-1 mutant, PCD characterized by nuclear degradation and cellular breakdown is delayed. However, mature δvpe-1 seeds show a similar appearance, dormancy, and germination rate as wild-type seeds. Using a YVAD-based substrate, δVPE has been shown to have caspase 1-like activity (Nakaune et al., 2005). In the Arabidopsis seed coat, loss of function of the also inner integument-accumulated PDI5 (Protein Disulfide Isomerase 5), a Cys protease inhibitor, causes premature PCD of the inner seed coat layer and loss of embryo viability (Andème Ondzighi et al., 2008). The pdi5 mutant phenotype suggests that the regulation of proteolytic activity during seed coat development is crucial for proper seed set. PDI5 has been shown to interact with the Cys proteases RD21, CP43, and CP19 in integument cells. Both RD21 and PDI5 traffic together through the endomembrane system towards the vacuole, where these accumulate. Accumulation of PDI5 has been observed in flowers, stems, leaves, siliques, and immature seeds (Andème Ondzighi et al., 2008). The reduction of PDI5 levels prior to the onset of PCD might indicate that PDI5 has a pro-survival role by maintaining the suppression of proteolytic activity to avoid premature seed coat PCD.
The nucellus is a tissue of maternal origin that gives rise to and supports the early female gametophyte and developing seed, and in many species degenerates through PCD (Lu and Magnani, 2018). In aubergine (Solanum melongena), a putative cysteine protease similar to tobacco’s CYP-8 has been identified and is expressed in nucellar cells as well as in several other tissue types that undergo PCD (Xu and Chye, 1999). In castor bean, the expression of CysEP, a cysteine protease found in ricinosomes in senescing tissues, coincides with nucellar PCD (Greenwood et al., 2005). In cardoon (Cynara cardunculus), aspartic proteases named cardosins accumulate in flowers. These are traditionally used for cheese manufacture in the Iberian Peninsula. The nucellus of cardoon has been shown by TUNEL staining to undergo PCD and also to accumulate cardosin B (Figueiredo et al., 2006). This protease localizes to the extracellular space in the stigma and the transmitting tissue in the style during floral development (Vieira et al., 2001). In quinoa (Chenopodium quinoa), the nucellus gives rise to a layer of nutritive tissue called perisperm. It consists of dead, starch-filled cells and takes over the role of the nutritive endosperm in some angiosperms. In the forming perisperm, caspase 6-like activity has been found when starch accumulation starts and few cells already show signs of nuclear degradation (López-Fernández and Maldonado, 2013). In the degenerating nucellus of chayote (Sechium edule), caspase 1-, 3-, and 6-like activities have been found to be upregulated during cell death (Lombardi et al., 2007). The nucellus has also been shown to produce nitric oxide and hydrogen peroxide during its development, and exogenous applications of these compounds enhances the observed caspase-like activity and cell death (Lombardi et al., 2010).
In barley, seven VPE genes, named HvVPE1 through HvVPE7, have been found to be expressed in developing grains. HvVPE2a and HvVPE2b, as well as two phytaspases HvPhS1 and HvPhS2, and the aspartic protease nucellin are expressed during preparation and initiation of PCD in the nucellus (Chen and Foolad, 1997; Radchuk et al., 2011; Tran et al., 2014). Interestingly, HvVPE2a/nucellain has been found to localize to the cell wall (Linnestad et al., 1998), complicating the understanding of its role in pre-mortem intracellular events (Figure 3). In rice, suppression of the transcription factor MADS29 leads to the downregulation of PCD-related genes in the nucellar projection, among them one encoding a Cys protease, causing defective nucellar PCD and abnormal seed development with shrunken seeds containing abnormal starch granules (Yin and Xue, 2012). These results suggest a role for transcriptionally regulated proteolytic activity in the developmentally controlled degradation of the nucellus. The investigation of PCD progression in many plant tissues, including the nucellus, is hampered by the low accessibility of the tissue. To circumvent this issue, a system using maize nucellus protoplasts has been developed to allow the rapid manipulation of the cellular environment. In this system, overexpression of MADS29 leads to the upregulation of Cys proteases, as was observed in rice (Chen et al., 2015), indicating that nucellar protoplasts might be promising to aid gene discovery and characterization in nucellar dPCD.
While the current studies showcase a significant and developmentally controlled proteolytic activity during seed coat and nucellus development, functional links between these activities and the occurring PCD are still limited. Further studies will be necessary to untangle the roles of the identified proteases to understand if their activity is responsible for triggering or executing cell death, or whether these are responsible primarily for post-mortem corpse degradation activities.
5. Proteases in senescence-induced PCD
Senescence is an integrative final stage of plant development that can also be induced or accelerated by a variety of environmental stresses. Senescence can occur on the level of individual organs, but can also encompass the entire plant at the end of its life cycle. Of interest, leaf and flower senescence are tightly controlled and are crucial for the degradation of high-weight macromolecules and their remobilization to actively growing parts of the plant (Diaz-Mendoza et al., 2016).
5.1. Leaf senescence
Leaf senescence is an intensely studied phenomenon in which a large number of genes have been implicated (Li et al., 2017). Proteolysis is among the most important catabolic processes occurring during leaf senescence and is central in the recycling of nitrogen compounds. Accordingly, genes encoding proteases are among the most upregulated genes during leaf senescence (Guo et al., 2004). Although some proteases, like SENESCENCE-ASSOCIATED GENE 12 (SAG12), represent important genetic markers for senescence, their actual functions often remain unknown, likely due to extensive genetic or functional redundancy (Pružinská et al., 2017). Generally, it is assumed that protein breakdown in senescing leaves involves protease activities in different cellular compartments, maximizing the catabolism of proteins into transportable units (Diaz-Mendoza et al., 2016; Schippers et al., 2015).
Although often used interchangeably, leaf senescence and PCD do not refer to the same process. Whereas nutrient mobilization is an active cellular program depending on cellular viability, PCD marks the endpoint of the senescence process (Thomas, 2013). As such, senescence-induced cell death needs to be tightly coordinated with the catabolic senescence processes to optimize recuperation of nutrients (Kim et al., 2016), with nuclei and mitochondria remaining active until the very late stages of senescence (Lim et al., 2007). Levels of the phytohormone salicylic acid (SA) increase during senescence. SAG12 expression and senescence-associated cell death are delayed in the SA signaling mutant pad4, indicating that SA has a role in the control of gene expression during developmental senescence, and that protease activity downstream of SA can have an effect on late senescence and cell death (Morris et al., 2000).
Several protease-encoding genes have been shown to be differentially regulated during leaf senescence (Bhalerao et al., 2003; Gregersen and Holm, 2007; Guo et al., 2004; Kinoshita et al., 1999; Parrott et al., 2010; Roberts et al., 2012). However, untangling proteases that act primarily on nutrient mobilization from those acting on the initiation and execution of senescence-induced PCD remains problematic. While the role of proteolysis in nutrient mobilization has been extensively reviewed (Diaz-Mendoza et al., 2016; Roberts et al., 2012; Schippers et al., 2015), here we focus on the few instances in which proteases have been implicated in senescence-associated cell death.
One approach to identify PCD-regulating genes expressed during plant organ senescence is to identify genes that are also expressed in other, senescence-unrelated PCD processes. A meta-analysis of different PCD-associated transcriptional signatures revealed the upregulation of senescence-associated genes also during osmotic stresses, the hypersensitive response, and cell-differentiation induced PCD (Olvera-Carrillo et al., 2015). Thus, especially proteases that are co-regulated during leaf senescence and in cell types that undergo differentiation-induced death might represent interesting candidates for death-promoting proteases during senescence. For instance, SERINE CARBOXYPEPTIDASE-LIKE48 (SCPL48), PASPA3, and some VPEs are upregulated in organ senescence as well as diverse cell types that undergo cell death as final differentiation step (Olvera-Carrillo et al., 2015).
Additionally, several proteases linked with senescence-induced PCD have been reported in different protease groups. The cucumber (Cucumis sativus) metalloprotease Cs1-MMP is expressed in the final stage of cotyledon senescence just prior to DNA fragmentation (Delorme et al., 2000). This late upregulation suggests a role in PCD regulation rather than in nutrient recycling. Additionally, the expression of the At2-MMP metalloprotease in Arabidopsis increases with organ age and senescence, and senescence-induced PCD is accelerated in at2-mmp-1 mutants (Golldack et al., 2002). Similarly, mutants of the mitochondrial metalloprotease FtSH4 display early leaf senescence, higher autophagic activity and cell death, suggesting a pro-survival function for FtSH4 and At2-MMP (Zhang et al., 2017a). Naturally, the effects of these loss-of-function mutants might be pleiotropic and the influence on leaf viability might be only indirect.
Next to protease activity, also autophagy has been involved in nutrient remobilization during leaf senescence (Havé et al., 2017). Interestingly, the autophagy-deficient atg5 mutant has been found to display higher levels of protease activity, which was attributed to the proteasome, and cytosolic and vacuolar cysteine proteases (Havé et al., 2018). Although these might act directly in promoting cell death, it is possible that these activities are back-up proteolytic systems to compensate for the absence of canonical autophagy.
5.2. Floral organ senescence
Leaf and floral senescence share the upregulation of PCD- and senescence-associated genes (Trivellini et al., 2016). In comparison with leaf senescence, floral senescence might be more dependent on developmental than on environmental stimuli. For example, the lifespan of the stigma is tightly controlled by NAC transcription factors, which induce PCD to terminate flower receptivity for pollen (Gao et al., 2018). Also, pollination itself is an important trigger for floral organ senescence, often mediated by the phytohormone ethylene (Rogers, 2006, 2013).
Ageing floral organs display certain PCD hallmarks, including degradation of total protein content, DNA fragmentation, vacuolar rupture and protoplast shrinkage (Battelli et al., 2011). The activity of serine-, cysteine-, and metalloproteases, and of KDEL-like and caspase-like proteases has been shown throughout floral senescence (Battelli et al., 2014; Pak and Van Doorn, 2005; Tripathi et al., 2009; Zhou et al., 2016), but functional evidence for involvement in PCD is still limited. For instance, senescing lily (Lilium longiflorum) flowers develop caspase-like activities during late senescence, and YVADase, VEIDase, and DEVDase inhibitors strongly decrease this activity (Battelli et al., 2011). However, none of the three inhibitors affect flower longevity or the progression of senescence(Battelli et al., 2011). In another study on lily flowers, signs of PCD including decreasing protein content and DNA degradation have been described in mesophyll and epidermal cells (Mochizuki-Kawai et al., 2015). During early senescence, mesophyll cells show an upregulation of the KDEL-tailed cysteine protease LoCYP and display signs of PCD. Only during later flower senescence, epidermal cells start to undergo PCD, with the simultaneous upregulation of the lily SAG12 homolog, LoSAG12 (Mochizuki-Kawai et al., 2015).
6. Conclusion
Based on the enormous regulatory potential of targeted protein modification and degradation by proteases, it comes as no surprise that proteases are involved in the regulation of most biological processes, including PCD. A major question is, however, if there are plant proteases that are similarly central to plant PCD as caspases are to apoptosis and other animal cell death types, or whether proteolytic events merely accompany plant PCD without being central for its actual regulation.
In this review, we aim to provide an overview of existing data on proteases in the context of dPCD and cover a selection of relatively well-described PCD processes in which the functional involvement of proteases has been demonstrated (Figure 2). (Dauphinee et al., 2014; Huysmans et al., 2018; Kumpf and Nowack, 2015; Lord et al., 2013; Takahashi et al., 2015; Xuan et al., 2016; Yamauchi et al., 2016)But even in the intensively studied PCD model systems, most available data are still associative in that they describe the expression of protease-encoding genes or activity of proteases in connection with PCD processes. Only in a limited number of cases functional data exist that allow to draw causal relationships between proteases and/or protease activity and dPCD (Table 2).
Table 2. A selection of proteases implicated in dPCD processes.
| Tissue/Process | Name | Species | MEROPS clan/family/ID | Catalytic type | Localization | Mode of action | Reference |
|---|---|---|---|---|---|---|---|
| Xylem | XCP1 | At | CA/C1/C01.065 | Cysteine | Vacuole and to a lesser extend in cytoplasm | Post-mortem clearance | (Avci et al., 2008) |
| XCP2 | At | CA/C1/C01.065 | Cysteine | Vacuole and to a lesser extend in cytoplasm | Post-mortem clearance | (Avci et al., 2008) | |
| Tr-cp14 | Tr | CA/C1/C01.065 | Cysteine | ER | n.d. | (Mulisch et al., 2013) | |
| AtMC9 | At | CD/C14/C14.034 | Cysteine | Nucleocytoplasm, apoplast, and vacuole | Post-mortem clearance | (Vercammen et al., 2006); (Escamez et al., 2016); (Bollhöner et al., 2013) | |
| Tapetum | CEP1 | At | CA/C1/A03 | Cysteine | Vacuole | Pro-PCD | (Zhang et al., 2014) |
| AtCP51 | At | CA/C1/A26 | Cysteine | n.d. | Pro-survival | (Yang et al., 2014) | |
| OsAP25 | Os | AA/A1/UPW | Aspartic | n.d. | Pro-PCD | (Niu et al., 2013) | |
| OsAP37 | Os | AA/A1/UPW | Aspartic | n.d. | Pro-PCD | (Niu et al., 2013) | |
| UNDEAD | At | AA/A1/A01.A49 | Aspartic | Predicted mitochondrial | Pro-survival | (Phan et al., 2011) | |
| Suspensor | CEP1 | At | CA/C1/C01.A03 | Cysteine | Subdomains of ER | n.d. | (Zhou et al., 2016) |
| NtCP14 | Nt | CA/C1/C01.A13 | Cysteine | ER | Pro-PCD | (Zhao et al., 2013) | |
| NtCYS | Nt | n.p. | inhibitor | ER | Pro-survival | (Zhao et al., 2013) | |
| mcII-Pa | Pa | CD / C14 / C14.036 | Cysteine | Nucleus | Promotes nuclei degradation | (Bozhkov et al., 2005) | |
| Endosperm | CEP2 | At | CA/C1/C01.A01 | Cysteine | Ricinosome-like vesicles | n.d. | (Hierl et al., 2014) |
| Inner integument | RD21A | At | CA/C1/C01.064 | Cysteine | Vacuole | n.d. | (Andème Ondzighi et al., 2008) |
| PDI5 | At | n.p. | inhibitor | Vacuole | Pro-survival | (Andème Ondzighi et al., 2008) | |
| δVPE | At | CD/C13/C13.A01 | Cysteine | Cytosol, Apoplast | Pro-PCD | (Nakaune et al., 2005) | |
| Nucellus | CysEP | Rc | CA/C1/C01.010 | Cysteine | Ricinosomes | n.d. | (Greenwood et al., 2005) |
| HvVPE2a | Hv | CD/C13/C13.001 | Cysteine | Apoplast | n.d. | (Linnestad et al., 1998) | |
| Leaf senescence | At2-MMP | At | MA/M10/M10.012 | Metallo | Predicted plasma membrane | Pro-survival | (Golldack et al., 2002) |
| Floral senescence | FtSH4 | At | MA/M41/M41.004 | Metallo | Mitochondria | Pro-survival | (Zhang et al., 2017a) |
n.d. not determined; n.p. not present; At - Arabidopsis thaliana; Tr - Trifolium repens; Os - Oryza sativa; Nt - Nicotiana tabacum; Pa - Picea abies; Rc - Ricinus communis; Hv - Hordeum vulgare
The determination of the actual mode of action of PCD-associated proteases remains a key challenge. VPEs for instance have been implicated in tonoplast breakdown as a committing step in PCD (Hatsugai et al., 2015), but how tonoplast disintegration is achieved remains unresolved. A substantial limitation might be presented by the genetic and functional redundancy of proteases – many plant proteases exist in large families and even higher-order mutants might fail to produce phenotypes that would allow an inference of the proteases’ function. The development and application of synthetic protease inhibitors have the potential to overcome such redundancy. However, the limited and often overestimated selectivity can restrict the utility of chemical approaches (Poręba et al., 2013). A way forward would consist in comprehensive approaches combining pharmacology and reverse genetics. Protein localization is key to protease function, especially regarding the sequestration of inactive zymogens and the re-localization of activated proteases (Figure 3). So far, only a few publications describe the dynamic subcellular localization of proteases, for instance by fusion proteins that take activation cleavage sites into account (Hierl et al., 2014). Additionally, strategies to identify the protease targets in quantitative degradomics studies (Demir et al., 2018; Huesgen and Overall, 2012) will be an important path to a functional understanding of the role proteases play in the control and execution of dPCD in plants.
One sentence abstract.
We review recent developments on the role of proteases in key developmental programmed cell death contexts, and discuss their modes of action.
Acknowledgements
We thank all members of the PCD lab at the VIB-UGENT Center for Plant Systems Biology for vivid discussions and critical feedback. We also thank Annick Bleys for help with preparing the manuscript. We gratefully acknowledge funding by the ERC StG PROCELLDEATH (Project 639234) to M.K.N. We apologize to the colleagues whose papers we were not able to cite due to space restrictions.
Contributor Information
Rafael Andrade Buono, Email: rabuo@psb.vib-ugent.be.
Roman Hudecek, Email: rohud@psb.vib-ugent.be.
Moritz K. Nowack, Email: moritz.nowack@vib.be.
References
- Andème Ondzighi C, Christopher DA, Cho EJ, Chang S-C, Staehelin LA. Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds. Plant Cell. 2008;20:2205–2220. doi: 10.1105/tpc.108.058339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Higuchi K, Arakaki S, Sano T, Asamizu E, Ezura H. Genetic suppression analysis in novel vacuolar processing enzymes reveals their roles in controlling sugar accumulation in tomato fruits. Journal of Experimental Botany. 2011;62:2773–2786. doi: 10.1093/jxb/erq451. [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Toriyama K. Genetic regulation of sporopollenin synthesis and pollen exine development. Annual Review of Plant Biology. 2011;62:437–460. doi: 10.1146/annurev-arplant-042809-112312. [DOI] [PubMed] [Google Scholar]
- Avci U, Earl Petzold H, Ismail IO, Beers EP, Haigler CH. Cysteine proteases XCP1 and XCP2 aid micro-autolysis within the intact central vacuole during xylogenesis in Arabidopsis roots. Plant Journal. 2008;56:303–315. doi: 10.1111/j.1365-313X.2008.03592.x. [DOI] [PubMed] [Google Scholar]
- Balakireva AV, Zamyatnin AA. Indispensable role of proteases in plant innate immunity. International Journal of Molecular Sciences. 2018;19:629. doi: 10.3390/ijms19020629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battelli R, Lombardi L, Picciarelli P, Lorenzi R, Frigerio L, Rogers HJ. Expression and localisation of a senescence-associated KDEL-cysteine protease from Lilium longiflorum tepals. Plant Science. 2014;214:38–46. doi: 10.1016/j.plantsci.2013.09.011. [DOI] [PubMed] [Google Scholar]
- Battelli R, Lombardi L, Rogers HJ, Picciarelli P, Lorenzi R, Ceccarelli N. Changes in ultrastructure, protease and caspase-like activities during flower senescence in Lilium longiflorum . Plant Science. 2011;180:716–725. doi: 10.1016/j.plantsci.2011.01.024. [DOI] [PubMed] [Google Scholar]
- Becraft PW, Gutierrez-Marcos J. Endosperm development: dynamic processes and cellular innovations underlying sibling altruism. Wiley Interdisciplinary Reviews: Developmental Biology. 2012;1:579–593. doi: 10.1002/wdev.31. [DOI] [PubMed] [Google Scholar]
- Bedinger P. The remarkable biology of pollen. Plant Cell. 1992;4:879–887. doi: 10.1105/tpc.4.8.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman T, De Rycke R, Viane R, Inzé D. Histological study of seed coat development in Arabidopsis thaliana . Journal of Plant Research. 2000;113:139–148. [Google Scholar]
- Bethke PC, Libourel IGL, Aoyama N, Chung Y-Y, Still DW, Jones RL. The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiology. 2007;143:1173–1188. doi: 10.1104/pp.106.093435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao R, Keskitalo J, Sterky F, Erlandsson R, Björkbacka H, Birve SJ, Karlsson J, Gardeström P, Gustafsson P, Lundeberg J, Jansson S. Gene expression in autumn leaves. Plant Physiology. 2003;131:430–442. doi: 10.1104/pp.012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanvillain R, Young B, Cai Y-m, Hecht V, Varoquaux F, Delorme V, Lancelin J-M, Delseny M, Gallois P. The Arabidopsis peptide kiss of death is an inducer of programmed cell death. EMBO Journal. 2011;30:1173–1183. doi: 10.1038/emboj.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böllhoner B, Jokipii-Lukkari S, Bygdell J, Stael S, Adriasola M, Muñiz L, Van Breusegem F, Ezcurra I, Wingsle G, Tuominen H. The function of two type II metacaspases in woody tissues of Populus trees. New Phytologist. 2018;217:1551–1565. doi: 10.1111/nph.14945. [DOI] [PubMed] [Google Scholar]
- Bollhöner B, Zhang B, Stael S, Denancé N, Overmyer K, Goffner D, Van Breusegem F, Tuominen H. Post mortem function of AtMC9 in xylem vessel elements. New Phytologist. 2013;200:498–510. doi: 10.1111/nph.12387. [DOI] [PubMed] [Google Scholar]
- Bonneau L, Ge Y, Drury GE, Gallois P. What happened to plant caspases? Journal of Experimental Botany. 2008;59:491–499. doi: 10.1093/jxb/erm352. [DOI] [PubMed] [Google Scholar]
- Borén M, Höglund AS, Bozhkov P, Jansson C. Developmental regulation of a VEIDase caspaselike proteolytic activity in barley caryopsis. Journal of Experimental Botany. 2006;57:3747–3753. doi: 10.1093/jxb/erl136. [DOI] [PubMed] [Google Scholar]
- Bozhkov PV, Filonova LH, Suarez MF, Helmersson A, Smertenko AP, Zhivotovsky B, von Arnold S. VEIDase is a principal caspase-like activity involved in plant programmed cell death and essential for embryonic pattern formation. Cell Death and Differentiation. 2004;11:175–182. doi: 10.1038/sj.cdd.4401330. [DOI] [PubMed] [Google Scholar]
- Bozhkov PV, Suarez MF, Filonova LH, Daniel G, Zamyatnin AA, Jr, Rodriguez-Nieto S, Zhivotovsky B, Smertenko A. Cysteine protease mcII-Pa executes programmed cell death during plant embryogenesis. Proceedings of the Institution of Mechanical Engineers Part A-Journal of Power and Energy. 2005;102:14463–14468. doi: 10.1073/pnas.0506948102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y-M, Yu J, Ge Y, Mironov A, Gallois P. Two proteases with caspase-3-like activity, cathepsin B and proteasome, antagonistically control ER-stress-induced programmed cell death in Arabidopsis. New Phytologist. 2018;218:1143–1155. doi: 10.1111/nph.14676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Foolad MR. Molecular organization of a gene in barley which encodes a protein similar to aspartic protease and its specific expression in nucellar cells during degeneration. Plant Molecular Biology. 1997;35:821–831. doi: 10.1023/a:1005833207707. [DOI] [PubMed] [Google Scholar]
- Chen J, Yi Q, Song Q, Gu Y, Zhang J, Hu Y, Liu H, Liu Y, Yu G, Huang Y. A highly efficient maize nucellus protoplast system for transient gene expression and studying programmed cell death-related processes. Plant Cell Reports. 2015;34:1239–1251. doi: 10.1007/s00299-015-1783-z. [DOI] [PubMed] [Google Scholar]
- Corre-Menguy F, Cejudo FJ, Mazubert C, Vidal J, Lelandais-Brière C, Torres G, Rode A, Hartmann C. Characterization of the expression of a wheat cystatin gene during caryopsis development. Plant Molecular Biology. 2002;50:687–698. doi: 10.1023/a:1019906031305. [DOI] [PubMed] [Google Scholar]
- Costa LM, Gutièrrez-Marcos JF, Dickinson HG. More than a yolk: the short life and complex times of the plant endosperm. Trends in Plant Science. 2004;9:507–514. doi: 10.1016/j.tplants.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Cubria-Radio M, Nowack MK. Transcriptional networks orchestrating programmed cell death during plant development. Curr Top Dev Biol. 2019;131:161–184. doi: 10.1016/bs.ctdb.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneva A, Gao Z, Van Durme M, Nowack MK. Functions and regulation of programmed cell death in plant development. Annual Review of Cell and Developmental Biology. 2016;32:441–468. doi: 10.1146/annurev-cellbio-111315-124915. [DOI] [PubMed] [Google Scholar]
- Dauphinee AN, Warner TS, Gunawardena AHLAN. A comparison of induced and developmental cell death morphologies in lace plant Aponogeton madagascariensis leaves. BMC Plant Biology. 2014;14:389. doi: 10.1186/s12870-014-0389-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme VGR, McCabe PF, Kim D-J, Leaver CJ. A matrix metalloproteinase gene is expressed at the boundary of senescence and programmed cell death in cucumber. Plant Physiology. 2000;123:917–927. doi: 10.1104/pp.123.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir F, Niedermaier S, Villamor JG, Huesgen PF. Quantitative proteomics in plant protease substrate identification. New Phytologist. 2018;218:936–943. doi: 10.1111/nph.14587. [DOI] [PubMed] [Google Scholar]
- Diaz-Mendoza M, Velasco-Arroyo B, Santamaria ME, González-Melendi P, Martinez M, Diaz I. Plant senescence and proteolysis: two processes with one destiny. Genetics and Molecular Biology. 2016;39:329–338. doi: 10.1590/1678-4685-GMB-2016-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez F, Cejudo FJ. Patterns of starchy endosperm acidification and protease gene expression in wheat grains following germination. Plant Physiology. 1999;119:81–87. doi: 10.1104/pp.119.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez F, Cejudo FJ. Programmed cell death (PCD): an essential process of cereal seed development and germination. Frontiers in Plant Science. 2014;5:366. doi: 10.3389/fpls.2014.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez F, González M, Cejudo FJ. A germination-related gene encoding a serine carboxypeptidase is expressed during the differentiation of the vascular tissue in wheat grains and seedlings. Planta. 2002;215:727–734. doi: 10.1007/s00425-002-0809-2. [DOI] [PubMed] [Google Scholar]
- Dou M, Zhang Y, Yang S, Feng X. Identification of ZHOUPI orthologs in rice involved in endosperm development and cuticle formation. Frontiers in Plant Science. 2018;9:223. doi: 10.3389/fpls.2018.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval I, Lachance D, Giguère I, Bomal C, Morency M-J, Pelletier G, Boyle B, MacKay JJ, Séguin A. Large-scale screening of transcription factor–promoter interactions in spruce reveals a transcriptional network involved in vascular development. Journal of Experimental Botany. 2014;65:2319–2333. doi: 10.1093/jxb/eru116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves DJ, Flores-Ortiz C, Haque T, Lin Z, Teng N, Franklin-Tong VE. Self-incompatibility in Papaver advances in integrating the signalling network. Biochemical Society Transactions. 2014;42:370–376. doi: 10.1042/BST20130248. [DOI] [PubMed] [Google Scholar]
- Escamez S, André D, Zhang B, Bollhöner B, Pesquet E, Tuominen H. METACASPASE9 modulates autophagy to confine cell death to the target cells during Arabidopsis vascular xylem differentiation. Biology Open. 2016;5:122–129. doi: 10.1242/bio.015529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamez S, Tuominen H. Programmes of cell death and autolysis in tracheary elements: when a suicidal cell arranges its own corpse removal. Journal of Experimental Botany. 2014;65:1313–1321. doi: 10.1093/jxb/eru057. [DOI] [PubMed] [Google Scholar]
- Fath A, Bethke P, Lonsdale J, Meza-Romero R, Jones R. Programmed cell death in cereal aleurone. Plant Molecular Biology. 2000;44:255–266. doi: 10.1023/a:1026584207243. [DOI] [PubMed] [Google Scholar]
- Fendrych M, Van Hautegem T, Van Durme M, Olvera-Carrillo Y, Huysmans M, Karimi M, Lippens S, Guérin CJ, Krebs M, Schumacher K, Nowack MK. Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis . Current Biology. 2014;24:931–940. doi: 10.1016/j.cub.2014.03.025. [DOI] [PubMed] [Google Scholar]
- Feng F, Qi W, Lv Y, Yan S, Xu L, Yang W, Yuan Y, Chen Y, Zhao H, Song R. OPAQUE11 is a central hub of the regulatory network for maize endosperm development and nutrient metabolism. Plant Cell. 2018;30:375–396. doi: 10.1105/tpc.17.00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo R, Duarte P, Pereira S, Pissarra J. The embryo sac of Cynara cardunculus ultrastructure of the development and localisation of the aspartic proteinase cardosin B. Sexual Plant Reproduction. 2006;19:93–101. [Google Scholar]
- Filonova LH, Suárez MF, Bozhkov PV. Detection of programmed cell death in plant embryos. Methods in Molecular Biology. 2008;427:173–179. doi: 10.1007/978-1-59745-273-1_14. [DOI] [PubMed] [Google Scholar]
- Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infection and Immunity. 2005;73:1907–1916. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourquin C, Beauzamy L, Chamot S, Creff A, Goodrich J, Boudaoud A, Ingram G. Mechanical stress mediated by both endosperm softening and embryo growth underlies endosperm elimination in Arabidopsis seeds. Development. 2016;143:3300–3305. doi: 10.1242/dev.137224. [DOI] [PubMed] [Google Scholar]
- Fuchslocher Chico J, Saggau C, Adam D. Proteolytic control of regulated necrosis. Biochimica et Biophysica Acta - Molecular Cell Research. 2017;1864:2147–2161. doi: 10.1016/j.bbamcr.2017.05.025. [DOI] [PubMed] [Google Scholar]
- Galiullina RA, Kasperkiewicz P, Chichkova NV, Szalek A, Serebryakova MV, Poreba M, Drag M, Vartapetian AB. Substrate specificity and possible heterologous targets of phytaspase, a plant cell death protease. Journal of Biological Chemistry. 2015;290:24806–24815. doi: 10.1074/jbc.M115.675819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Li R, Guo Y. Arabidopsis aspartic proteases A36 and A39 play roles in plant reproduction. Plant Signaling & Behavior. 2017a;12:e1304343. doi: 10.1080/15592324.2017.1304343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Zhang Y, Wang W, Zhao K, Liu C, Bai L, Li R, Guo Y. Two membrane-anchored aspartic proteases contribute to pollen and ovule development. Plant Physiology. 2017b;173:219–239. doi: 10.1104/pp.16.01719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Daneva A, Salanenka Y, Van Durme M, Huysmans M, Lin Z, De Winter F, Vanneste S, Karimi M, Van de Velde J, Vandepoele K, et al. KIRA1 and ORESARA1 terminate flower receptivity by promoting cell death in the stigma of Arabidopsis . Nature Plants. 2018;4:365–375. doi: 10.1038/s41477-018-0160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Dietrich C, Matsuno M, Li G, Berg H, Xia Y. An Arabidopsis aspartic protease functions as an anti-cell-death component in reproduction and embryogenesis. EMBO Reports. 2005;6:282–288. doi: 10.1038/sj.embor.7400357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D, Popova OV, Dietz K-J. Mutation of the matrix metalloproteinase At2-MMP inhibits growth and causes late flowering and early senescence in Arabidopsis . Journal of Biological Chemistry. 2002;277:5541–5547. doi: 10.1074/jbc.M106197200. [DOI] [PubMed] [Google Scholar]
- Greenwood JS, Helm M, Gietl C. Ricinosomes and endosperm transfer cell structure in programmed cell death of the nucellus during Ricinus seed development. Proceedings of the Institution of Mechanical Engineers Part A-Journal of Power and Energy. 2005;102:2238–2243. doi: 10.1073/pnas.0409429102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen PL, Holm PB. Transcriptome analysis of senescence in the flag leaf of wheat Triticum aestivum L.) Plant Biotechnology Journal. 2007;5:192–206. doi: 10.1111/j.1467-7652.2006.00232.x. [DOI] [PubMed] [Google Scholar]
- Groover A, DeWitt N, Heidel A, Jones A. Programmed cell death of plant tracheary elements differentiating in vitro. Protoplasma. 1997;196:197–211. [Google Scholar]
- Guo Y, Cai Z, Gan S. Transcriptome of Arabidopsis leaf senescence. Plant, Cell & Environment. 2004;27:521–549. [Google Scholar]
- Han J-J, Lin W, Oda Y, Cui K-M, Fukuda H, He X-Q. The proteasome is responsible for caspase-3-like activity during xylem development. Plant Journal. 2012;72:129–141. doi: 10.1111/j.1365-313X.2012.05070.x. [DOI] [PubMed] [Google Scholar]
- Hao X, Qian J, Xu S, Song X, Zhu J. Location of caspase 3-like protease in the development of sieve element and tracheary element of stem in Cucurbita moschata . Journal of Integrative Plant Biology. 2008;50:1499–1507. doi: 10.1111/j.1744-7909.2008.00719.x. [DOI] [PubMed] [Google Scholar]
- Hatsugai N, Iwasaki S, Tamura K, Kondo M, Fuji K, Ogasawara K, Nishimura M, Hara-Nishimura I. A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes & Development. 2009;23:2496–2506. doi: 10.1101/gad.1825209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsugai N, Yamada K, Goto-Yamada S, Hara-Nishimura I. Vacuolar processing enzyme in plant programmed cell death. Frontiers in Plant Science. 2015;6:234. doi: 10.3389/fpls.2015.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn G, Chaudhury A. Genetic analysis of seed coat development in Arabidopsis . Trends in Plant Science. 2005;10:472–477. doi: 10.1016/j.tplants.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Havé M, Balliau T, Cottyn-Boitte B, Dérond E, Cueff G, Soulay F, Lornac A, Reichman P, Dissmeyer N, Avice J-C, Gallois P, et al. Increases in activity of proteasome and papain-like cysteine protease in Arabidopsis autophagy mutants: back-up compensatory effect or cell-death promoting effect? Journal of Experimental Botany. 2018;69:1369–1385. doi: 10.1093/jxb/erx482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havé M, Marmagne A, Chardon F, Masclaux-Daubresse C. Nitrogen remobilization during leaf senescence: lessons from Arabidopsis to crops. Journal of Experimental Botany. 2017;68:2513–2529. doi: 10.1093/jxb/erw365. [DOI] [PubMed] [Google Scholar]
- Hierl G, Höwing T, Isono E, Lottspeich F, Gietl C. Ex vivo processing for maturation of Arabidopsis KDEL-tailed cysteine endopeptidase 2 (AtCEP2) pro-enzyme and its storage in endoplasmic reticulum derived organelles. Plant Molecular Biology. 2014;84:605–620. doi: 10.1007/s11103-013-0157-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierl G, Vothknecht U, Gietl C. Programmed cell death in Ricinus and Arabidopsis the function of KDEL cysteine peptidases in development. Physiologia Plantarum. 2012;145:103–113. doi: 10.1111/j.1399-3054.2012.01580.x. [DOI] [PubMed] [Google Scholar]
- Hou S, Jamieson P, He P. The cloak, dagger, and shield: proteases in plant-pathogen interactions. Biochemical Journal. 2018;475:2491–2509. doi: 10.1042/BCJ20170781. [DOI] [PubMed] [Google Scholar]
- Höwing T, Dann M, Hoefle C, Hückelhoven R, Gietl C. Involvement of Arabidopsis thaliana endoplasmic reticulum KDEL-tailed cysteine endopeptidase 1 (AtCEP1) in powdery mildew-induced and AtCPR5-controlled cell death. PLoS ONE. 2017;12:e0183870. doi: 10.1371/journal.pone.0183870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höwing T, Huesmann C, Hoefle C, Nagel M-K, Isono E, Hückelhoven R, Gietl C. Endoplasmic reticulum KDEL-tailed cysteine endopeptidase 1 of Arabidopsis (AtCEP1) is involved in pathogen defense. Frontiers in Plant Science. 2014;5:58. doi: 10.3389/fpls.2014.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesgen PF, Overall CM. N- and C-terminal degradomics: new approaches to reveal biological roles for plant proteases from substrate identification. Physiologia Plantarum. 2012;145:5–17. doi: 10.1111/j.1399-3054.2011.01536.x. [DOI] [PubMed] [Google Scholar]
- Huysmans M, Buono RA, Skorzinski N, Cubria Radio M, De Winter F, Parizot B, Mertens J, Karimi M, Fendrych M, Nowack MK. NAC transcription factors ANAC087 and ANAC046 control distinct aspects of programmed cell death in the Arabidopsis columella and lateral root cap. Plant Cell. 2018;30:2197–2213. doi: 10.1105/tpc.18.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysmans M, Lema AS, Coll NS, Nowack MK. Dying two deaths — programmed cell death regulation in development and disease. Current Opinion in Plant Biology. 2017;35:37–44. doi: 10.1016/j.pbi.2016.11.005. [DOI] [PubMed] [Google Scholar]
- Julien O, Wells JA. Caspases and their substrates. Cell Death and Differentiation. 2017;24:1380–1389. doi: 10.1038/cdd.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanabe T, Ariizumi T, Kawai-Yamada M, Uchimiya H, Toriyama K. Abolition of the tapetum suicide program ruins microsporogenesis. Plant & Cell Physiology. 2006;47:784–787. doi: 10.1093/pcp/pcj039. [DOI] [PubMed] [Google Scholar]
- Kawashima T, Goldberg RB. The suspensor: not just suspending the embryo. Trends in Plant Science. 2010;15:23–30. doi: 10.1016/j.tplants.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Kim J, Woo HR, Nam HG. Toward systems understanding of leaf senescence: an integrated multi-omics perspective on leaf senescence research. Molecular Plant. 2016;9:813–825. doi: 10.1016/j.molp.2016.04.017. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Yamada K, Hiraiwa N, Kondo M, Nishimura M, Hara-Nishimura I. Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetative tissues during senescence and under various stressed conditions. Plant Journal. 1999;19:43–53. doi: 10.1046/j.1365-313x.1999.00497.x. [DOI] [PubMed] [Google Scholar]
- Klemenčič M, Funk C. Structural and functional diversity of caspase homologues in nonmetazoan organisms. Protoplasma. 2018;255:387–397. doi: 10.1007/s00709-017-1145-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Ikeda TM, Nagata K. Spatial and temporal progress of programmed cell death in the developing starchy endosperm of rice. Planta. 2013;237:1393–1400. doi: 10.1007/s00425-013-1854-8. [DOI] [PubMed] [Google Scholar]
- Kumpf RP, Nowack MK. The root cap: a short story of life and death. Journal of Experimental Botany. 2015;66:5651–5662. doi: 10.1093/jxb/erv295. [DOI] [PubMed] [Google Scholar]
- Kuriyama H. Loss of tonoplast integrity programmed in tracheary element differentiation. Plant Physiology. 1999;121:763–774. doi: 10.1104/pp.121.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon-Placette C, Köhler C. Endosperm-based postzygotic hybridization barriers: developmental mechanisms and evolutionary drivers. Molecular Ecology. 2016;25:2620–2629. doi: 10.1111/mec.13552. [DOI] [PubMed] [Google Scholar]
- Lam E, Fukuda H, Greenberg J. Programmed cell death in higher plants. Springer Science & Business Media; New York: 2000. [Google Scholar]
- Lampl N, Alkan N, Davydov O, Fluhr R. Set-point control of RD21 protease activity by AtSerpin1 controls cell death in Arabidopsis. Plant Journal. 2013;74:498–510. doi: 10.1111/tpj.12141. [DOI] [PubMed] [Google Scholar]
- Laubscher M, Brown K, Tonfack LB, Myburg AA, Mizrachi E, Hussey SG. Temporal analysis of Arabidopsis genes activated by Eucalyptus grandis NAC transcription factors associated with xylem fibre and vessel development. Scientific Reports. 2018;8:10983. doi: 10.1038/s41598-018-29278-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Jung K-H, An GH, Chung Y-Y. Isolation and characterization of a rice cysteine protease gene, OSCP1 using T-DNA gene-trap system. Plant Molecular Biology. 2004;54:755–765. doi: 10.1023/B:PLAN.0000040904.15329.29. [DOI] [PubMed] [Google Scholar]
- Lema Asqui S, Vercammen D, Serrano I, Valls M, Rivas S, Van Breusegem F, Conlon FL, Dangl JL, Coll NS. AtSERPIN1 is an inhibitor of the metacaspase AtMC1-mediated cell death and autocatalytic processing in planta . New Phytologist. 2018;218:1156–1166. doi: 10.1111/nph.14446. [DOI] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, Wen TQ, et al. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell. 2006;18:2999–3014. doi: 10.1105/tpc.106.044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhao Y, Liu X, Jiang Z, Peng J, Jin J, Guo H, Luo J. Construction of the leaf senescence database and functional assessment of senescence-associated genes. Methods in Molecular Biology. 2017;1533:315–333. doi: 10.1007/978-1-4939-6658-5_19. [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. Leaf senescence. Annual Review of Plant Biology. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- Linnestad C, Doan DNP, Brown RC, Lemmon BE, Meyer DJ, Jung R, Olsen O-A. Nucellain, a barley homolog of the dicot vacuolar-processing protease, is localized in nucellar cell walls. Plant Physiology. 1998;118:1169–1180. doi: 10.1104/pp.118.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hai G, Wang C, Cao S, Xu W, Jia Z, Yang C, Wang JP, Dai S, Cheng Y. Comparative proteomic analysis of Populus trichocarpa early stem from primary to secondary growth. Journal of Proteomics. 2015;126:94–108. doi: 10.1016/j.jprot.2015.05.032. [DOI] [PubMed] [Google Scholar]
- Lombardi L, Casani S, Ceccarelli N, Galleschi L, Picciarelli P, Lorenzi R. Programmed cell death of the nucellus during Sechium edule Sw. seed development is associated with activation of caspaselike proteases. Journal of Experimental Botany. 2007;58:2949–2958. doi: 10.1093/jxb/erm137. [DOI] [PubMed] [Google Scholar]
- Lombardi L, Ceccarelli N, Picciarelli P, Sorce C, Lorenzi R. Nitric oxide and hydrogen peroxide involvement during programmed cell death of Sechium edule nucellus. Physiologia Plantarum. 2010;140:89–102. doi: 10.1111/j.1399-3054.2010.01381.x. [DOI] [PubMed] [Google Scholar]
- López-Fernández MP, Maldonado S. Programmed cell death during quinoa perisperm development. Journal of Experimental Botany. 2013;64:3313–3325. doi: 10.1093/jxb/ert170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Fernández MP, Maldonado S. Programmed cell death in seeds of angiosperms. Journal of Integrative Plant Biology. 2015;57:996–1002. doi: 10.1111/jipb.12367. [DOI] [PubMed] [Google Scholar]
- Lord CEN, Dauphinee AN, Watts RL, Gunawardena AHLAN. Unveiling interactions among mitochondria, caspase-like proteases, and the actin cytoskeleton during plant programmed cell death (PCD) PLoS ONE. 2013;8:e57110. doi: 10.1371/journal.pone.0057110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Magnani E. Seed tissue and nutrient partitioning, a case for the nucellus. Plant Reproduction. 2018;31:309–317. doi: 10.1007/s00497-018-0338-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo A, Zhao P, Zhang L-Y, Sun M-X. Initiation of programmed cell death in the suspensor is predominantly regulated maternally in a tobacco hybrid. Scientific Reports. 2016;6:29467. doi: 10.1038/srep29467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M, Cambra I, Carrillo L, Diaz-Mendoza M, Diaz I. Characterization of the entire cystatin gene family in barley and their target cathepsin L-like cysteine-proteases, partners in the hordein mobilization during seed germination. Plant Physiology. 2009;151:1531–1545. doi: 10.1104/pp.109.146019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minina EA, Coll NS, Tuominen H, Bozhkov PV. Metacaspases versus caspases in development and cell fate regulation. Cell Death and Differentiation. 2017;24:1314–1325. doi: 10.1038/cdd.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minina EA, Filonova LH, Fukada K, Savenkov EI, Gogvadze V, Clapham D, Sanchez-Vera V, Suarez MF, Zhivotovsky B, Daniel G, Smertenko A, et al. Autophagy and metacaspase determine the mode of cell death in plants. Journal of Cell Biology. 2013;203:917–927. doi: 10.1083/jcb.201307082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minina EA, Smertenko AP, Bozhkov PV. Vacuolar cell death in plants: metacaspase releases the brakes on autophagy. Autophagy. 2014;10:928–929. doi: 10.4161/auto.28236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki-Kawai H, Niki T, Shibuya K, Ichimura K. Programmed cell death progresses differentially in epidermal and mesophyll cells of lily petals. PLoS ONE. 2015;10:e0143502. doi: 10.1371/journal.pone.0143502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K, MacKerness SA-H, Page T, John CF, Murphy AM, Carr JP, Buchanan-Wollaston V. Salicylic acid has a role in regulating gene expression during leaf senescence. Plant Journal. 2000;23:677–685. doi: 10.1046/j.1365-313x.2000.00836.x. [DOI] [PubMed] [Google Scholar]
- Mulisch M, Asp T, Krupinska K, Hollmann J, Holm PB. The Tr-cp 14 cysteine protease in white clover Trifolium repens is localized to the endoplasmic reticulum and is associated with programmed cell death during development of tracheary elements. Protoplasma. 2013;250:623–629. doi: 10.1007/s00709-012-0427-1. [DOI] [PubMed] [Google Scholar]
- Nakaba S, Takata N, Yoshida M, Funada R. Continuous expression of genes for xylem cysteine peptidases in long-lived ray parenchyma cells in Populus . Plant Biotechnology. 2015;32:21–29. [Google Scholar]
- Nakaune S, Yamada K, Kondo M, Kato T, Tabata S, Nishimura M, Hara-Nishimura I. A vacuolar processing enzyme, δVPE, is involved in seed coat formation at the early stage of seed development. Plant Cell. 2005;17:876–887. doi: 10.1105/tpc.104.026872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu N, Liang W, Yang X, Jin W, Wilson ZA, Hu J, Zhang D. EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nature Communications. 2013;4:1445. doi: 10.1038/ncomms2396. [DOI] [PubMed] [Google Scholar]
- Obermeier C, Hosseini B, Friedt W, Snowdon R. Gene expression profiling via LongSAGE in a non-model plant species: a case study in seeds of Brassica napus . BMC Genomics. 2009;10:295. doi: 10.1186/1471-2164-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obudulu O, Bygdell J, Sundberg B, Moritz T, Hvidsten TR, Trygg J, Wingsle G. Quantitative proteomics reveals protein profiles underlying major transitions in aspen wood development. BMC Genomics. 2016;17:119. doi: 10.1186/s12864-016-2458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K. New families of carboxyl peptidases: serine-carboxyl peptidases and glutamic peptidases. Journal of Biochemistry. 2012;151:13–25. doi: 10.1093/jb/mvr129. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Oda Y, Fukuda H. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell. 2010;22:3461–3473. doi: 10.1105/tpc.110.075036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olvera-Carrillo Y, Van Bel M, Van Hautegem T, Fendrych M, Huysmans M, Simaskova M, van Durme M, Buscaill P, Rivas S, Coll NS, Coppens F, et al. A conserved core of programmed cell death indicator genes discriminates developmentally and environmentally induced programmed cell death in plants. Plant Physiology. 2015;169:2684–2699. doi: 10.1104/pp.15.00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidvar V, Mohorianu I, Dalmay T, Zheng Y, Fei Z, Pucci A, Mazzucato A, Večeřová V, Sedlářova M, Fellner M. Transcriptional regulation of male-sterility in 7B-1 male-sterile tomato mutant. PLoS ONE. 2017;12:e0170715. doi: 10.1371/journal.pone.0170715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak C, Van Doorn WG. Delay of Iris flower senescence by protease inhibitors. New Phytologist. 2005;165:473–480. doi: 10.1111/j.1469-8137.2004.01226.x. [DOI] [PubMed] [Google Scholar]
- Parrott DL, Martin JM, Fischer AM. Analysis of barley Hordeum vulgare leaf senescence and protease gene expression: a family C1A cysteine protease is specifically induced under conditions characterized by high carbohydrate, but low to moderate nitrogen levels. New Phytologist. 2010;187:313–331. doi: 10.1111/j.1469-8137.2010.03278.x. [DOI] [PubMed] [Google Scholar]
- Peng X, Sun M-X. The suspensor as a model system to study the mechanism of cell fate specification during early embryogenesis. Plant Reproduction. 2018;31:59–65. doi: 10.1007/s00497-018-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold HE, Zhao M, Beers EP. Expression and functions of proteases in vascular tissues. Physiologia Plantarum. 2012;145:121–129. doi: 10.1111/j.1399-3054.2011.01538.x. [DOI] [PubMed] [Google Scholar]
- Phan HA, Iacuone S, Li SF, Parish RW. The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana . Plant Cell. 2011;23:2209–2224. doi: 10.1105/tpc.110.082651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan HA, Li SF, Parish RW. MYB80, a regulator of tapetal and pollen development, is functionally conserved in crops. Plant Molecular Biology. 2012;78:171–183. doi: 10.1007/s11103-011-9855-0. [DOI] [PubMed] [Google Scholar]
- Plackett ARG, Thomas SG, Wilson ZA, Hedden P. Gibberellin control of stamen development: a fertile field. Trends in Plant Science. 2011;16:568–578. doi: 10.1016/j.tplants.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Poręba M, Stróżyk A, Salvesen GS, Drąg M. Caspase substrates and inhibitors. Cold Spring Harbor Perspectives in Biology. 2013;5:a008680. doi: 10.1101/cshperspect.a008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pružinská A, Shindo T, Niessen S, Kaschani F, Tóth R, Millar AH, van der Hoorn RAL. Major Cys protease activities are not essential for senescence in individually darkened Arabidopsis leaves. BMC Plant Biology. 2017;17:4. doi: 10.1186/s12870-016-0955-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk V, Weier D, Radchuk R, Weschke W, Weber H. Development of maternal seed tissue in barley is mediated by regulated cell expansion and cell disintegration and coordinated with endosperm growth. Journal of Experimental Botany. 2011;62:1217–1227. doi: 10.1093/jxb/erq348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raviv B, Aghajanyan L, Granot G, Makover V, Frenkel O, Gutterman Y, Grafi G. The dead seed coat functions as a long-term storage for active hydrolytic enzymes. PLoS ONE. 2017;12:e0181102. doi: 10.1371/journal.pone.0181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Research. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt S, Repper D, Tuzhikov AI, Galiullina RA, Planas-Marquès M, Chichkova NV, Vartapetian AB, Stintzi A, Schaller A. The tomato subtilase family includes several cell death-related proteinases with caspase specificity. Scientific Reports. 2018;8:10531. doi: 10.1038/s41598-018-28769-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reza SH, Delhomme N, Street NR, Ramachandran P, Dalman K, Nilsson O, Minina EA, Bozhkov PV. Transcriptome analysis of embryonic domains in Norway spruce reveals potential regulators of suspensor cell death. PLoS ONE. 2018;13:e0192945. doi: 10.1371/journal.pone.0192945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts IN, Caputo C, Criado MV, Funk C. Senescence-associated proteases in plants. Physiologia Plantarum. 2012;145:130–139. doi: 10.1111/j.1399-3054.2012.01574.x. [DOI] [PubMed] [Google Scholar]
- Rocha AJ, Soares EL, Costa JH, Costa WLG, Soares AA, Nogueira FCS, Domont GB, Campos FAP. Differential expression of cysteine peptidase genes in the inner integument and endosperm of developing seeds of Jatropha curcas L. (Euphorbiaceae) Plant Science. 2013;213:30–37. doi: 10.1016/j.plantsci.2013.08.009. [DOI] [PubMed] [Google Scholar]
- Rogers HJ. Programmed cell death in floral organs: how and why do flowers die? Annals of Botany. 2006;97:309–315. doi: 10.1093/aob/mcj051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HJ. From models to ornamentals: how is flower senescence regulated? Plant Molecular Biology. 2013;82:563–574. doi: 10.1007/s11103-012-9968-0. [DOI] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA. The development of endosperm in grasses. Plant Physiology. 2009;149:14–26. doi: 10.1104/pp.108.129437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A. A cut above the rest: the regulatory function of plant proteases. Planta. 2004;220:183–197. doi: 10.1007/s00425-004-1407-2. [DOI] [PubMed] [Google Scholar]
- Schippers JHM, Schmidt R, Wagstaff C, Jing H-C. Living to die and dying to live: the survival strategy behind leaf senescence. Plant Physiology. 2015;169:914–930. doi: 10.1104/pp.15.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Simpson D, Gietl C. Programmed cell death in castor bean endosperm is associated with the accumulation and release of a cysteine endopeptidase from ricinosomes. Proceedings of the Institution of Mechanical Engineers Part A-Journal of Power and Energy. 1999;96:14159–14164. doi: 10.1073/pnas.96.24.14159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Simpson D, Kalousek F, Gietl C. A cysteine endopeptidase with a C-terminal KDEL motif isolated from castor bean endosperm is a marker enzyme for the ricinosome, a putative lytic compartment. Planta. 1998;206:466–475. doi: 10.1007/s004250050423. [DOI] [PubMed] [Google Scholar]
- Schmid M, Simpson DJ, Sarioglu H, Lottspeich F, Gietl C. The ricinosomes of senescing plant tissue bud from the endoplasmic reticulum. Proceedings of the Institution of Mechanical Engineers Part A-Journal of Power and Energy. 2001;98:5353–5358. doi: 10.1073/pnas.061038298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheoran IS, Ross ARS, Olson DJH, Sawhney VK. Differential expression of proteins in the wild type and 7B-1 male-sterile mutant anthers of tomato Solanum lycopersicum a proteomic analysis. Journal of Proteomics. 2009;71:624–636. doi: 10.1016/j.jprot.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Shukla P, Subhashini M, Singh NK, Ahmed I, Trishla S, Kirti PB. Targeted expression of cystatin restores fertility in cysteine protease induced male sterile tobacco plants. Plant Science. 2016;246:52–61. doi: 10.1016/j.plantsci.2016.02.010. [DOI] [PubMed] [Google Scholar]
- Smertenko A, Bozhkov PV. Somatic embryogenesis: life and death processes during apical-basal patterning. Journal of Experimental Botany. 2014;65:1343–1360. doi: 10.1093/jxb/eru005. [DOI] [PubMed] [Google Scholar]
- Soares EL, Lima MLB, Nascimento JRS, Soares AA, Coutinho IAC, Campos FAP. Seed development of Jatropha curcas L. (Euphorbiaceae): integrating anatomical, ultrastructural and molecular studies. Plant Cell Reports. 2017;36:1707–1716. doi: 10.1007/s00299-017-2184-2. [DOI] [PubMed] [Google Scholar]
- Solís M-T, Chakrabarti N, Corredor E, Cortés-Eslava J, Rodríguez-Serrano M, Biggiogera M, Risueño MC, Testillano PS. Epigenetic changes accompany developmental programmed cell death in tapetum cells. Plant & Cell Physiology. 2014;55:16–29. doi: 10.1093/pcp/pct152. [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Radchuk V, Strickert M, Miersch O, Weschke W, Wobus U. Gene expression patterns reveal tissue-specific signaling networks controlling programmed cell death and ABA-regulated maturation in developing barley seeds. Plant Journal. 2006;47:310–327. doi: 10.1111/j.1365-313X.2006.02789.x. [DOI] [PubMed] [Google Scholar]
- Suarez MF, Filonova LH, Smertenko A, Savenkov EI, Clapham DH, von Arnold S, Zhivotovsky B, Bozhkov PV. Metacaspase-dependent programmed cell death is essential for plant embryogenesis. Current Biology. 2004;14:R339–R340. doi: 10.1016/j.cub.2004.04.019. [DOI] [PubMed] [Google Scholar]
- Sueldo DJ, van der Hoorn RAL. Plant life needs cell death, but does plant cell death need Cys proteases? FEBS Journal. 2017;284:1577–1585. doi: 10.1111/febs.14034. [DOI] [PubMed] [Google Scholar]
- Szewińska J, Simińska J, Bielawski W. The roles of cysteine proteases and phytocystatins in development and germination of cereal seeds. Journal of Plant Physiology. 2016;207:10–21. doi: 10.1016/j.jplph.2016.09.008. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Yamauchi T, Rajhi I, Nishizawa NK, Nakazono M. Transcript profiles in cortical cells of maize primary root during ethylene-induced lysigenous aerenchyma formation under aerobic conditions. Annals of Botany. 2015;115:879–894. doi: 10.1093/aob/mcv018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas EL, van der Hoorn RAL. Ten prominent host proteases in plant-pathogen interactions. International Journal of Molecular Sciences. 2018;19:639. doi: 10.3390/ijms19020639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H. Senescence, ageing and death of the whole plant. New Phytologist. 2013;197:696–711. doi: 10.1111/nph.12047. [DOI] [PubMed] [Google Scholar]
- Tran V, Weier D, Radchuk R, Thiel J, Radchuk V. Caspase-like activities accompany programmed cell death events in developing barley grains. PLoS ONE. 2014;9:e109426. doi: 10.1371/journal.pone.0109426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi SK, Singh AP, Sane AP, Nath P. Transcriptional activation of a 37 kDa ethylene responsive cysteine protease gene, RbCP1 is associated with protein degradation during petal abscission in rose. Journal of Experimental Botany. 2009;60:2035–2044. doi: 10.1093/jxb/erp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivellini A, Cocetta G, Hunter DA, Vernieri P, Ferrante A. Spatial and temporal transcriptome changes occurring during flower opening and senescence of the ephemeral hibiscus flower, Hibiscus rosa-sinensis . Journal of Experimental Botany. 2016;67:5919–5931. doi: 10.1093/jxb/erw295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobacher CP, Senatore A, Holley C, Greenwood JS. Induction of a ricinosomal-protease and programmed cell death in tomato endosperm by gibberellic acid. Planta. 2013;237:665–679. doi: 10.1007/s00425-012-1780-1. [DOI] [PubMed] [Google Scholar]
- Tsiatsiani L, Timmerman E, De Bock P-J, Vercammen D, Stael S, van de Cotte B, Staes A, Goethals M, Beunens T, Van Damme P, Gevaert K, et al. The Arabidopsis metacaspase9 degradome. Plant Cell. 2013;25:2831–2847. doi: 10.1105/tpc.113.115287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twumasi P, Iakimova ET, Qian T, van Ieperen W, Schel JHN, Emons AMC, van Kooten O, Woltering EJ. Caspase inhibitors affect the kinetics and dimensions of tracheary elements in xylogenic Zinnia Zinnia elegans cell cultures. BMC Plant Biology. 2010;10:162. doi: 10.1186/1471-2229-10-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren AG, O’Rourke K, Aravind L, Pisabarro MT, Seshagiri S, Koonin EV, Dixit VM. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Molecular Cell. 2000;6:961–967. doi: 10.1016/s1097-2765(00)00094-0. [DOI] [PubMed] [Google Scholar]
- Valdivia ER, Herrera MT, Gianzo C, Fidalgo J, Revilla G, Zarra I, Sampedro J. Regulation of secondary wall synthesis and cell death by NAC transcription factors in the monocot Brachypodium distachyon . Journal of Experimental Botany. 2013;64:1333–1343. doi: 10.1093/jxb/ers394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn RAL. Plant proteases: from phenotypes to molecular mechanisms. Annual Review of Plant Biology. 2008;59:191–223. doi: 10.1146/annurev.arplant.59.032607.092835. [DOI] [PubMed] [Google Scholar]
- Van Durme M, Nowack MK. Mechanisms of developmentally controlled cell death in plants. Current Opinion in Plant Biology. 2016;29:29–37. doi: 10.1016/j.pbi.2015.10.013. [DOI] [PubMed] [Google Scholar]
- Van Hautegem T, Waters AJ, Goodrich J, Nowack MK. Only in dying, life: programmed cell death during plant development. Trends in Plant Science. 2015;20:102–113. doi: 10.1016/j.tplants.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Vercammen D, Belenghi B, van de Cotte B, Beunens T, Gavigan J-A, De Rycke R, Brackenier A, Inzé D, Harris JL, Van Breusegem F. Serpin1 of Arabidopsis thaliana is a suicide inhibitor for metacaspase 9. Journal of Molecular Biology. 2006;364:625–636. doi: 10.1016/j.jmb.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Vercammen D, van de Cotte B, De Jaeger G, Eeckhout D, Casteels P, Vandepoele K, Vandenberghe I, Van Beeumen J, Inzé D, Van Breusegem F. Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. Journal of Biological Chemistry. 2004;279:45329–45336. doi: 10.1074/jbc.M406329200. [DOI] [PubMed] [Google Scholar]
- Vieira M, Pissarr J, Veríssimo P, Castanheira P, Costa Y, Pires E, Faro C. Molecular cloning and characterization of cDNA encoding cardosin B, an aspartic proteinase accumulating extracellularly in the transmitting tissue of Cynara cardunculus L. Plant Molecular Biology. 2001;45:529–539. doi: 10.1023/a:1010675015318. [DOI] [PubMed] [Google Scholar]
- Wan L, Xia Q, Qiu X, Selvaraj G. Early stages of seed development in Brassica napus a seed coat-specific cysteine proteinase associated with programmed cell death of the inner integument. Plant Journal. 2002;30:1–10. doi: 10.1046/j.1365-313x.2002.01262.x. [DOI] [PubMed] [Google Scholar]
- Waters A, Creff A, Goodrich J, Ingram G. “What we’ve got here is failure to communicate”: Zou mutants and endosperm cell death in seed development. Plant Signaling & Behavior. 2013;8:e24368. doi: 10.4161/psb.24368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ZA, Zhang D-B. From Arabidopsis to rice: pathways in pollen development. Journal of Experimental Botany. 2009;60:1479–1492. doi: 10.1093/jxb/erp095. [DOI] [PubMed] [Google Scholar]
- Woffenden BJ, Freeman TB, Beers EP. Proteasome inhibitors prevent tracheary element differentiation in zinnia mesophyll cell cultures. Plant Physiology. 1998;118:419–430. doi: 10.1104/pp.118.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Ohtani M, Yamaguchi M, Toyooka K, Wakazaki M, Sato M, Kubo M, Nakano Y, Sano R, Hiwatashi Y, Murata T, et al. Contribution of NAC transcription factors to plant adaptation to land. Science. 2014;343:1505–1508. doi: 10.1126/science.1248417. [DOI] [PubMed] [Google Scholar]
- Xu F-X, Chye M-L. Expression of cysteine proteinase during developmental events associated with programmed cell death in brinjal. Plant Journal. 1999;17:321–327. doi: 10.1046/j.1365-313x.1999.00370.x. [DOI] [PubMed] [Google Scholar]
- Xuan W, Band LR, Kumpf RP, Van Damme D, Parizot B, De Rop G, Opdenacker D, Möller BK, Skorzinski N, Njo MF, De Rybel B, et al. Cyclic programmed cell death stimulates hormone signaling and root development in Arabidopsis . Science. 2016;351:384–387. doi: 10.1126/science.aad2776. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Goué N, Igarashi H, Ohtani M, Nakano Y, Mortimer JC, Nishikubo N, Kubo M, Katayama Y, Kakegawa K, Dupree P, et al. VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiology. 2010;153:906–914. doi: 10.1104/pp.110.154013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Mitsuda N, Ohtani M, Ohme-Takagi M, Kato K, Demura T. VASCULAR-RELATED NAC-DOMAIN 7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant Journal. 2011;66:579–590. doi: 10.1111/j.1365-313X.2011.04514.x. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Tanaka A, Mori H, Takamure I, Kato K, Nakazono M. Ethylene-dependent aerenchyma formation in adventitious roots is regulated differently in rice and maize. Plant, Cell & Environment. 2016;39:2145–2157. doi: 10.1111/pce.12766. [DOI] [PubMed] [Google Scholar]
- Yang S, Johnston N, Talideh E, Mitchell S, Jeffree C, Goodrich J, Ingram G. The endospermspecific ZHOUPI gene of Arabidopsis thaliana regulates endosperm breakdown and embryonic epidermal development. Development. 2008;135:3501–3509. doi: 10.1242/dev.026708. [DOI] [PubMed] [Google Scholar]
- Yang Y, Dong C, Yu J, Shi L, Tong C, Li Z, Huang J, Liu S. Cysteine Protease 51 CP51 an antherspecific cysteine protease gene, is essential for pollen exine formation in Arabidopsis . Plant Cell, Tissue and Organ Culture. 2014;119:383–397. [Google Scholar]
- Yin L-L, Xue H-W. The MADS29 transcription factor regulates the degradation of the nucellus and the nucellar projection during rice seed development. Plant Cell. 2012;24:1049–1065. doi: 10.1105/tpc.111.094854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TE, Gallie DR. Analysis of programmed cell death in wheat endosperm reveals differences in endosperm development between cereals. Plant Molecular Biology. 1999;39:915–926. doi: 10.1023/a:1006134027834. [DOI] [PubMed] [Google Scholar]
- Young TE, Gallie DR, DeMason DA. Ethylene-mediated programmed cell death during maize endosperm development of wild-type and shrunken2 genotypes. Plant Physiology. 1997;115:737–751. doi: 10.1104/pp.115.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X-H, Perdue TD, Heimer YM, Jones AM. Mitochondrial involvement in tracheary element programmed cell death. Cell Death and Differentiation. 2002;9:189–198. doi: 10.1038/sj.cdd.4400940. [DOI] [PubMed] [Google Scholar]
- Zhang D, Liu D, Lv X, Wang Y, Xun Z, Liu Z, Li F, Lu H. The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis . Plant Cell. 2014;26:2939–2961. doi: 10.1105/tpc.114.127282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gannon L, Hassall KL, Deery MJ, Gibbs DJ, Holdsworth MJ, van der Hoorn RAL, Lilley KS, Theodoulou FL. N-terminomics reveals control of Arabidopsis seed storage proteins and proteases by the Arg/N-end rule pathway. New Phytologist. 2018;218:1106–1126. doi: 10.1111/nph.14909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Li C, Wang R, Chen Y, Shu S, Huang R, Zhang D, Li J, Xiao S, Yao N, Yang C. The Arabidopsis mitochondrial protease FtSH4 is involved in leaf senescence via regulation of WRKY-dependent salicylic acid accumulation and signaling. Plant Physiology. 2017a;173:2294–2307. doi: 10.1104/pp.16.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li X, Yang S, Feng X. Identification of ZOUPI orthologs in soybean potentially involved in endosperm breakdown and embryogenic development. Frontiers in Plant Science. 2017b;8:139. doi: 10.3389/fpls.2017.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xin W, Wang S, Zhang X, Dai H, Sun R, Frazier T, Zhang B, Wang Q. Xylem sap in cotton contains proteins that contribute to environmental stress response and cell wall development. Functional & Integrative Genomics. 2015;15:17–26. doi: 10.1007/s10142-014-0395-y. [DOI] [PubMed] [Google Scholar]
- Zhao C, Johnson BJ, Kositsup B, Beers EP. Exploiting secondary growth in Arabidopsis. Construction of xylem and bark cDNA libraries and cloning of three xylem endopeptidases. Plant Physiology. 2000;123:1185–1196. doi: 10.1104/pp.123.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Zhou X-m, Zhang L-y, Wang W, Ma L-g, Yang L-b, Peng X-b, Bozhkov PV, Sun M-x. A bipartite molecular module controls cell death activation in the Basal cell lineage of plant embryos. PLoS Biology. 2013;11:e1001655. doi: 10.1371/journal.pbio.1001655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Zhou X-m, Zou J, Wang W, Wang L, Peng X-b, Sun M-x. Comprehensive analysis of cystatin family genes suggests their putative functions in sexual reproduction, embryogenesis, and seed formation. Journal of Experimental Botany. 2014;65:5093–5107. doi: 10.1093/jxb/eru274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye Z-H. Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis . Molecular Plant. 2010;3:1087–1103. doi: 10.1093/mp/ssq062. [DOI] [PubMed] [Google Scholar]
- Zhou L-Z, Howing T, Muller B, Hammes UZ, Gietl C, Dresselhaus T. Expression analysis of KDEL-CysEPs programmed cell death markers during reproduction in Arabidopsis . Plant Reproduction. 2016;29:265–272. doi: 10.1007/s00497-016-0288-4. [DOI] [PubMed] [Google Scholar]