Abstract
Background
African populations are experiencing health transitions due to rapid urbanization and international migration. However, the role of biological aging in this emerging burden of cardiometabolic diseases (CMD) among migrant and non-migrant Africans is unknown. We aimed to examine differences in epigenetic age acceleration (EAA) as measured by four clocks (Horvath, Hannum, PhenoAge and GrimAge) and their associations with cardiometabolic factors among migrant Ghanaians in Europe and non-migrant Ghanaians.
Methods
Genome-wide DNA methylation (DNAm) data of 712 Ghanaians from cross-sectional RODAM study were used to quantify EAA. We assessed correlation of DNAmAge measures with chronological age, and then performed linear regressions to determine associations of body mass index (BMI), fasting blood glucose (FBG), blood pressure, alcohol consumption, smoking, physical activity, and one-carbon metabolism nutrients with EAA among migrant and non-migrants. We replicated our findings among 172 rural-urban sibling pairs from India migration study and among 120 native South Africans from PURE-SA-NW study.
Findings
We found that Ghanaian migrants have lower EAA than non-migrants. Within migrants, higher FBG was positively associated with EAA measures. Within non-migrants, higher BMI, and Vitamin B9 (folate) intake were negatively associated with EAA measures. Our findings on FBG, BMI and folate were replicated in the independent cohorts.
Interpretation
Our study shows that migration is negatively associated with EAA among Ghanaians. Moreover, cardiometabolic factors are differentially associated with EAA within migrant and non-migrant subgroups. Our results call for context-based interventions for CMD among transitioning populations that account for effects of biological aging.
Funding
European Commission.
Introduction
African populations are experiencing epidemiological transitions due to rapid urbanization and international migration, often from low income to high income countries (HICs). 1,2 Changes in lifestyle factors upon migration (e.g. tobacco smoking, poor diet and physical inactivity) are prominent contributors to the emerging risk of cardiometabolic diseases (CMD). 3-5 While epigenetic aging has been linked to CMD and its risk factors in populations originating from HICs, little is known about the role of epigenetic aging in the emerging burden of CMD in transitioning African populations.
Epigenetic age, which is quantified using epigenetic clocks (also known as DNA methylation (DNAm) based age estimators) is a robust biomarker for chronologic age. 6 Epigenetic clocks comprise of a set of cytosine-phosphate-guanine sites (CpGs) coupled to mathematical algorithms that estimate age (in years) from a DNA source. 6 The difference between chronological age and epigenetic age in an individual is termed epigenetic age acceleration (EAA). 6 EAA can be used to predict all-cause mortality, life span, and incidence of chronic non-communicable diseases including CMD. 6-10 When quantified in blood, EAA can be divided into two forms; intrinsic and extrinsic. Intrinsic epigenetic age acceleration (IEAA) captures biological aging within each cell independently of proportions of naïve or senescent cytotoxic T cells, while extrinsic epigenetic age acceleration (EEAA) quantifies epigenetic aging in immune-related components. 6 Several epigenetic clocks have been developed to measure IEAA and EEAA. The most used are DNAm HorvathAge which measures IEAA, 11 DNAm HannumAge which measures EEAA, 12 DNAm PhenoAge which measures EEAA and includes clinical characteristics in its algorithm (i.e. chronological age, albumin, creatinine, glucose, CRP, lymphocyte percentage, mean cell volume, red blood cell distribution width, alkaline phosphatase, and white blood cell count), 9 and DNAm GrimAge which measures EEAA and incorporates chronological age, sex and seven plasma proteins in its algorithm (i.e. adrenomedullin, beta-2 macroglobulin, growth differentiation factor 15, Cystatin C, leptin, plasminogen activation inhibitor 1, tissue inhibitor metalloproteinase 1 and the amount of cigarettes smoked). 13
Previous studies have shown that physical activity, fruit and vegetable consumption, and education are negatively associated with EAA, while cardiometabolic traits such as obesity, diabetes and hypertension are positively associated with EAA. 7,14,15 Since DNAm that underlies EAA is affected by changes in lifestyle factors, and can also directly influence CMD, it is therefore important to understand the role of EAA in the emerging burden of CMD among transitioning African populations. 16 Considering that migrant (urbanizing) African populations have higher rates of CMD than their non-migrant counterparts, 17 we hypothesize that changes in lifestyle upon migration lead to accelerated biological aging of tissues (higher EAA), which in turn leads to higher CMD incidence. As such, we expect migrants and urbanizing populations to exhibit higher EAA than their non-migrant counterparts.
Using data from Research on Obesity and Diabetes Among Migrants (RODAM) study, we examined differences in EAAs and their associations with cardiometabolic and lifestyle factors among migrant Ghanaians residing in Europe and non-migrant Ghanaians residing in Ghana. Considering that maintenance of DNAm relies partly on one-carbon metabolism nutrients for transmethylation (folate, choline and betaine as methyl donors, vitamins B2, B6, and B12 as essential factors), we included dietary intake of one-carbon metabolism nutrients as part of our lifestyle factor measurements. 18
Methods
Study population
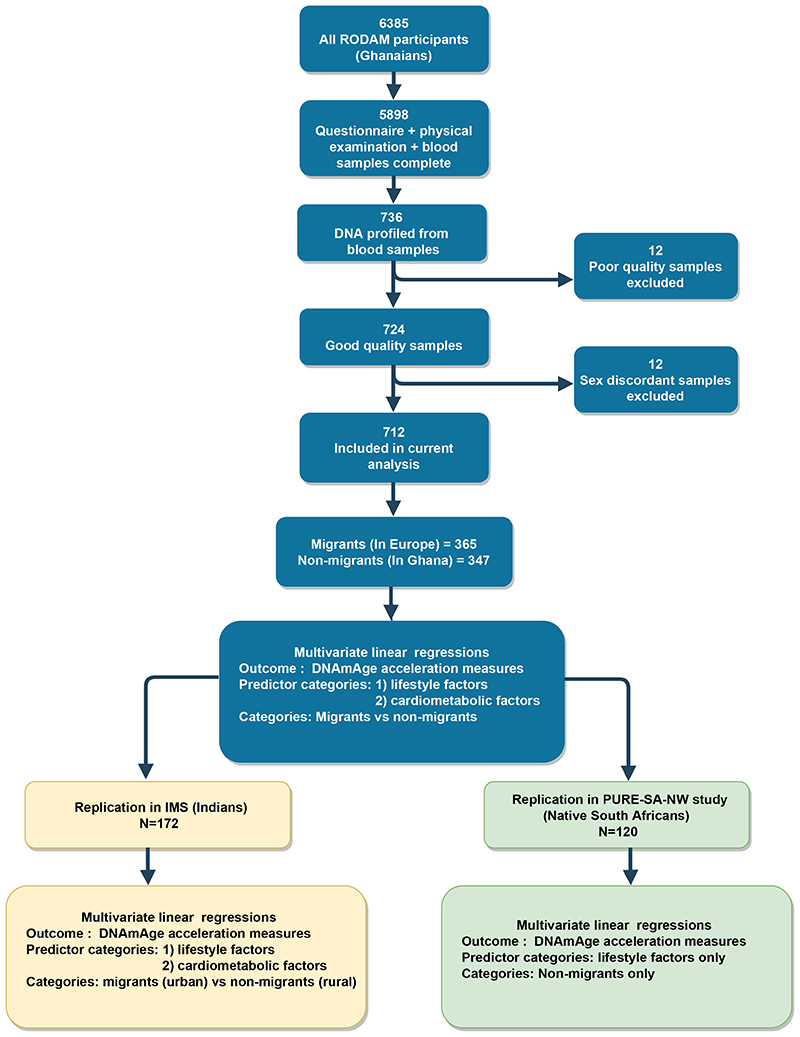
The cross-sectional RODAM study was initiated in 2012 to better understand the development of obesity and diabetes among African migrants at the phenotypic, epigenetic, and genetic levels. A detailed description of the study population is provided in appendix p 2. In brief, RODAM study enrolled 6385 non-migrant Ghanaian men and women living in Ghana, as well as first generation migrant Ghanaians residing in Europe. Participants were predominantly from Akan ethnic group. In Ghana, recruitment of urban participants was conducted in two cities (Kumasi and Obuasi), while recruitment in rural area was conducted in 15 villages in Ashanti region. In Europe, participants were recruited from cities of Amsterdam (Netherlands), Berlin (Germany) and London (UK). Participants were included if they were aged ≥ 25 years, had completed RODAM study questionnaire, were physically examined and had blood samples taken (figure 1).
Figure 1. Study flow chart.
The chart depicts how the study population was arrived at, and how the study was performed with respect to discovery and replication.
Sample size justification
For the current analyses, we used a sub-sample of 736 participants from the RODAM study with DNAm data (figure 1). This epigenetics sub-sample was originally designed to detect 5% DNAm differences between participants with diabetes (n=265) compared to controls without diabetes (n=471), who were equally distributed between migrant and non-migrant groups (case-control design; power=0.80, α=0.05, appendix p 5). The final sample of 712 used in current analyses had passed DNAm quality control and was generally representative of the overall RODAM study population (figure 1, appendix p 7).
Ethical approval and consent to participate
Ethical approval was obtained from ethics committees of involved institutions in Ghana (Kwame Nkrumah University of Science & Technology: CHRPE/AP/200/12), Netherlands (Amsterdam University Medical Center: W12-062#12.17.0086), Germany (Charité University Berlin: EA1/307/12) and UK (London School of Hygiene & Tropical Medicine: 6208) before the start of data collection. 17 All participants gave written informed consent. 17
Phenotypic measurements
A standardized approach for questionnaires, anthropometric measurements and venipuncture samples was used across all study sites. A detailed description of phenotypic measurements, data handling and data entry procedures is provided in appendix p 8. For this study, the following measurements were obtained: chronological age, sex, location of residence, education, alcohol consumption, smoking, physical activity, dietary intake of Vitamin B2 (mg/day), Vitamin B6 (mg/day), Vitamin B9 (mcg/day), Vitamin B12 (mcg/day) and total energy intake (Kcal/day) from food frequency questionnaires, duration of stay in the host country among migrants, body mass index (BMI) in kg/m2, blood pressure (BP) in mmHg, fasting plasma glucose (FBG) in mmol/L and use of anti-diabetic medication. Education was categorized as follows; 1) none or elementary, 2) primary, 3) secondary, and 4) tertiary. Alcohol consumption was categorized as any or never consumption. Tobacco smoking was categorized into current, past, and never smokers. Physical activity was categorized into low, moderate, and high according to the global physical activity questionnaire. 17
DNAm processing, profiling, and quality control
Detailed procedures for DNAm processing, profiling, and quality control in RODAM study are also reported in appendix p 9. In brief, bisulfite conversion of DNA was conducted with Zymo EZ DNA Methylation™ kit. The converted DNA was amplified and hybridized on Infinium® HumanMethylation450 BeadChip, which quantifies DNAm levels of approximately 485,000 CpG sites. Quality control was performed using MethylAid package in R (version 1.4.0.). Functional normalization was applied using minfi package (version 3.1.0). 19 Probes annotated to X and Y chromosomes, known to involve cross hybridization or to involve single nucleotide polymorphisms with a minor allele frequency of ≥ 0.05 (5%) were removed from the dataset. 19 This resulted in a total set of 429,459 CpGs.
DNAmAge Calculation and Estimated blood cells
DNAmAges were calculated using online DNAmAge Calculator (http://dnamage.genetics.ucla.edu/) 20 from normalized DNAm data and cross checked via Bioconductor package Methylclock (version 0.5.0). 21 DNAm HorvathAge was estimated using 353 CpGs as specified in Horvath et al. 11 IEAA was obtained as residuals from the regression model that regresses HorvathAge on chronological age and blood cell counts. 11 DNAm Hannum Age was estimated using 71 CpGs as specified in Hannum et al. 12 EEAA was obtained as residuals from a regression model that regresses DNAm HannumAge aggregated with three blood cell components (naïve cytotoxic T cells, exhausted cytotoxic T cells, and plasmablasts) on chronological age. 12 DNAm PhenoAge was estimated using 513 CpGs as specified in Levine et al. 9 PhenoAge acceleration (PhenoAgeAccel) was obtained as residuals of the regression models that regress DNAm PhenoAge on chronological age without adjusting for blood cell counts. 9 DNAm GrimAge was estimated using 1030 CpG sites as specified by Lu et al. 13 GrimAge Acceleration (GrimAgeAccel) was obtained as residuals of the regression models that regress DNAm GrimAge on chronological age and sex, without adjusting for blood cell counts. Cell counts were estimated using a method developed by Houseman et al, implemented using Methylclock package. 22
Statistical analysis
Statistical analysis was carried out using R (version 4.0.2) and Bioconductor packages. There were no missing data in our sample. Descriptive statistics were presented as proportions for categorical variables, as means (with standard deviations) or as median (interquartile range) for skewed data. Pearson’s correlations were performed between chronological age and DNAmAges, as well as between the EAA measures. Linear regressions were performed between EAA measures (outcome variable) and CMD traits (smoking status, alcohol consumption, physical activity, vitamin intake, duration of stay in host country (migrants), BMI, FBP and BP) among migrants and non-migrants separately. Linear models were adjusted for chronological age, education, sex, smoking, alcohol consumption, physical activity, vitamin intake, total energy intake and duration of stay in host country among migrants.
Post-hoc analyses (appendix p 12-18 ): we performed post-hoc analyses to ascertain robustness our findings. First, we assessed whether associations of lifestyle and cardiometabolic factors with EAA measures were also apparent in combined sample of migrants and non-migrants. This would indicate that our findings were not substantially influenced by large differences in baseline characteristics between migrants and non-migrants. Second, we assessed whether associations between FBG and EAA were influenced by usage of anti-diabetic medications. Third, we aimed to replicate our findings among populations from Africa or other low- or middle-income countries (LMIC) where urbanization and health transitions are prominent. As such, we performed replication analysis among Indians from Indian Migration Study (IMS) and among native South Africans from Prospective Urban and Rural Epidemiology study’s South African, North West province cohort (PURE-SA-NW). 23,24 Emphasis was placed on consistency in direction of effects in replication analyses due to smaller sample sizes in replication cohorts (limited statistical power).
We aimed to minimize false positive findings in our study. Instead of utilizing standard multiple tests correction, we opted for other alternative ways because CMD risk factors have interconnected pathophysiological pathways and are complementary to each other in explaining our hypothesis. 25 For example, dietary intake, physical activity levels, obesity, and diabetes have interconnected pathophysiological pathways, 25 and moreover, changes in dietary intake, in physical activity and in BMI can together explain why migrants have higher rates of diabetes than non-migrants. 17 It is therefore possible to have statistically significant findings in all these CMD risk factors. We therefore opted out of standard multiple tests correction and considered detection of a similar effect in two or more EAA measures as a true finding in both the main and post-hoc analyses.
Role of the funding source
The study funder had no role in the study design, data collection, data analysis, data interpretation or writing of the report. The corresponding author had full access to all the data and the final responsibility to submit for publication.
Results
Between 2nd February 2012 and 30th September 2014, a total of 736 individuals participated in the RODAM epigenetics sub-study, of which 24 were excluded during DNAm quality control. Thus, 712 participants were included in the current analysis (figure 1). Of these; 365 were migrants and 347 were non-migrants. Mean chronological age of migrants was 50 (10) years and non-migrants was 52 (10) years. Mean DNAmAges were also lower in migrants compared to non-migrants. Migrants were better educated, smoked more and were more overweight than non-migrants. Generally, migrants had more male participants, higher BP, FBG and vitamin intake than non-migrants (table 1).
Table1. Baseline characteristics of Ghanaians participants in RODAM study.
| Non-migrants
1
(n=347) |
Migrants
2
(n=365) |
|
|---|---|---|
| Aging estimators, mean (SD) | ||
| Chronological Age 3 | 52.36(9.84) | 49.9(9.76) |
| DNA HorvathAge 4 | 50.78(10.04) | 48(9.62) |
| DNAm HannumAge 5 | 44.01(9.93) | 39.82(8.96) |
| DNAm PhenoAge 6 | 41.2(10.77) | 35.68(10.07) |
| DNAM GrimAge 7 | 39.35(8.72) | 37.07(8.33) |
| IEAA 8 | 0.35(6.14) | -0.34(5.88) |
| EEAA 9 | 0.90(5.26) | -0.86(5.09) |
| PhenoAgeAccel 10 | 1.77(6.9) | -1.68(6.41) |
| GrimAgeAccel 11 | 0.19(3.79) | -0.18(3.85) |
| Other Demographic factors | ||
| Female, n (%) | 243(70.02) | 166(45.47) |
| Education, n (%) | ||
| No education | 160 (46.11) | 79(21.64) |
| Primary school | 136(39.19) | 154(42.19) |
| Secondary school | 38(10.96) | 78(21.37) |
| Tertiary | 13(3.74) | 54(14.79) |
| Behavior related factors | ||
| Any Alcohol consumption, n (%) | 121 (34.90) | 152(41.60) |
| Smoking, n (%) | ||
| Current | 5(1.44) | 18(4.93) |
| Never | 309(89.04) | 316(86.58) |
| Past | 33(9.52) | 31(8.49) |
| Physical activity levels, n (%) 12 | ||
| Low | 131(37.75) | 137(37.53) |
| Moderate | 65(18.73) | 86(23.56) |
| High | 151(43.52) | 142(38.90) |
| Total Energy intake, Kcal/day 13 | 2386.24(824.96) | 2894.10 (1113.31) |
| Length of stay for migrants, years 14 | NA | 20.39(12.30-25.35) |
| One carbon metabolism nutrient intake, mean (SD) 15 | ||
| Vitamin B2 (riboflavin, mg/day) | 1.29(0.51) | 2.24(1.60) |
| Vitamin B6 (mg/day) | 2.32(0.80) | 3.01(1.29) |
| Vitamin B9 (folate; µg /day) | 311.56(113.22) | 445.31(203.87) |
| Vitamin B12 (cyanocobalamin; µg /day) 14 | 5.92(3.81-10.17) | 12.82(7.19-34.43) |
| Cardio-metabolic Factors, mean (SD) | ||
| Body mass index (kg/m2) | 24.74(5.59) | 28.18(4.92) |
| Systolic blood pressure (mmHg) | 130.91(22.27) | 136.77(18.44) |
| Diastolic blood pressure (mmHg) | 80.8(12.32) | 84.85(11.44) |
| Fasting blood glucose (mmol/L) 14 | 5.13(4.73-6.18) | 5.26(4.87-7.61) |
| Use of medication | ||
| Use of anti-diabetic medication, n (%) 16 | 23(6.62) | 37(10.14) |
Ghanaians living in rural and urban Ghana were categorised as non-migrants
Ghanaians living in Amsterdam, Berlin and London were categorised as migrants.
Age provided by the participant during questionnaire interviews
DNA methylation age obtained using the Horvath clock
DNA methylation age obtained using the Hannum clock
DNA methylation age obtained using the PhenoAge clock
DNA methylation age obtained using GrimAge clock
Intrinsic epigenetic age acceleration (within each cell) obtained using Horvath clock
Extrinsic epigenetic age acceleration (between different cells) using Hannum clock
Epigenetic age acceleration incorporating clinical traits obtained using the PhenoAge clock
Epigenetic age acceleration incorporating plasma proteins obtained using GrimAge clock
Physical activity categorised according to the global physical activity questionnaire (GPAQ) criteria
Total energy intakes obtained by food frequency questionnaires in Kcal/day
Data presented as medians, interquartile range.
One carbon metabolism nutrients intake obtained via food frequency questionnaires.
Use of medications to treat diabetes (oral and injectables).
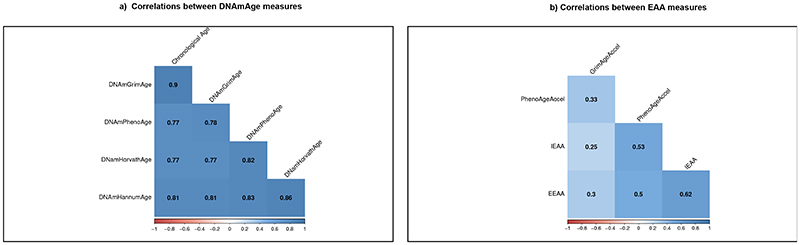
Chronological age positively correlated with all four DNAmAges; r = 0.77 for DNAm HorvathAge, 0.81 for DNAm HannumAge, 0.77 for DNAm PhenoAge and 0.90 for DNAm GrimAge (figure 2). These correlations are comparable to those of previous studies and validate utility of DNAmAge estimators in this study. 9,14 Correlations among EAA measurements were weaker with correlation coefficients ranging from 0.25 (IEAA vs GrimAgeAccel) to 0.62 (IEAA and EEAA). These lower correlations between EAA measures have been previously observed and represent uniqueness of each of the four EAA measures. 14
Figure 2. Correlations between DNAmAge measures and between EAA measures in the RODAM study.
The figure depicts Pearson’s correlations between DNAmAge measures and between EAA measures.
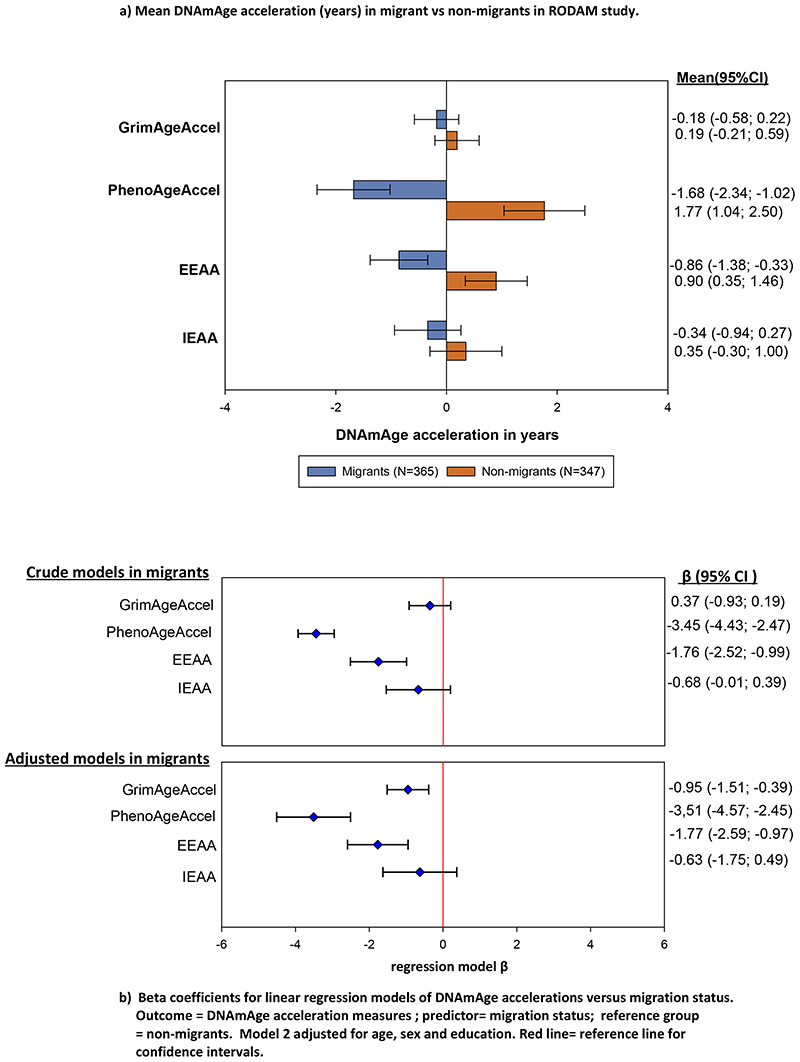
We found that migrants had generally lower EAA compared to non-migrants (IEAA = –0.34 vs 0.35; EEAA = –0.86 vs 0.90, PhenoAgeAccel = –1.68 vs 1.77, and GrimAgeAccel = –0.18 vs 0.19 years; table 1, figure 3). Migration status was also negatively associated with EEAA, PhenoAgeAccel and GrimAgeAccel after adjusting for age, sex, and education (figure 3). However, duration of stay in the host country among migrants was not associated with any EAA measure (table 2, figure 4 & 5).
Figure 3. Migration status and EAA Measures in RODAM study.
The plot depicts mean (SD) epigenetic age acceleration (IEAA, EEAA, PhenoAgeAccel and GrimAgeAccel) in migrants compared to non-migrants. Additionally, the plot also depicts regression model β with 95% confidence intervals for the associations between migration status and four EAA Measures in RODAM study. Red line = reference line for confidence intervals. Abbreviations: IEAA= Intrinsic epigenetic age acceleration, EEAA= extrinsic epigenetic age acceleration, PhenoAge Accel= Pheno Age Acceleration, GrimAge Accel= Grim Age Acceleration.
Table 2. Lifestyle factors, cardiometabolic traits and epigenetic age acceleration in migrant and non-migrant Ghanaians in RODAM study.
| IEAA (Horvath) 1 | EEAA (Hannum) 2 | PhenoAgeAccel 3 | GrimAgeAccel 4 | |||||
|---|---|---|---|---|---|---|---|---|
| Non-migrants
5
(N=347) |
Migrants
6
(N=365) |
Non-migrants
5
(N=347) |
Migrants
6
(N=365) |
Non-migrants
5
(N=347) |
Migrants
6
(N=365) |
Non-migrants
5
(N=347) |
Migrants
6
(N=365) |
|
| β (95% CI) | β (95% CI) | β (95% CI) | β (95% CI) | β (95% CI) | β (95% CI) | β (95% CI) | β (95% CI) | |
| Lifestyle factors | ||||||||
| Alcohol consumption | ||||||||
| No | ref | ref | ref | ref | ref | ref | ref | ref |
| Yes (crude) | –0.77 (–2.13;0.59) | –1.07 (–2.29;0.16) | –0.82 (–1.99;0.34) | –0.39 (–1.46;0.67) | –0.37 (–1.90;1.16) | –0.38 (–1.72;0.96) | –0.58 (–0.28;1.39) | 0.64 (–0.17;1.44) |
| Yes (adjusted) | –0.92 (–2.32;0.48) | –0.01 (–1.71;1.70) | –1.03 (–2.23;0.18) | –0.27 (–1.70;1.16) | –0.74 (–2.31;0.84) | –0.08 (–1.86;1.71) | –0.17 (–0.97;0.63) | 0.44 (–0.33;1.22) |
| Physical activity levels 7 | ||||||||
| Low | ref | ref | ref | ref | ref | ref | ref | ref |
| Moderate (crude) | –0.77 (–2.60;1.07) | –0.27 (–1.86;1.32) | –0.20 (–1.78;1.37) | –0.90 (–2.28;0.48) | 0.96 (–1.10;3.03) | –0.37 (–2.10;1.35) | 0.74 (–0.38;1.86) | –0.29 (–1.34;0.74) |
| Moderate (adjusted) | –0.78 (–2.67;1.11) | 0.64 (–2.82;2.82) | –0.18 (–1.81;1.44) | –0.33 (–2.16;1.49) | 0.71 (–1.41;2.85) | 0.12 (–2.16;2.41) | –0.13 (–1.21;0.04) | –0.52 (–1.49;0.44) |
| High (crude) | –0.67 (–2.12;0.77) | –1.16 (–2.54;0.22) | –0.674 (–1.91;0.56) | –0.71 (–1.92;0.48) | 0.41 (–1.21;2.04) | –1.91 (–3.41;0.41) | –0.53 (–1.43;0.35) | 0.49 (–0.41;1.39) |
| High (adjusted) | –0.33 (–1.85;1.20) | –0.19 (–1.69;1.69) | –0.347 (–1.66;0.96) | –0.21 (–1.79;1.36) | 0.55 (–1.16;2.27) | –2.16 (–4.13; –0.20) | –1.12 (–1.98; 0.25) | –0.15 (–1.00;0.69) |
| Tobacco smoking | ||||||||
| Past (crude) | 1.59 (–0.61;3.79) | –0.35 (–0.72;0.03) | 0.62 (–1.28;2.52) | –0.13 (–2.02;1.75) | –0.11 (–2.60;2.37) | –1.67 (–4.05;0.70) | 3.14 (1.81;4.49) | 1.89 (0.52;3.26) |
| Past (adjusted) | –3.15 (–9.09;2.79) | –0.26 (–0.42;0.42) | –1.33 (–6.44;3.78) | 3.11 (–0.79;7.02) | –7.07 (–13.76; –0.40) | 1.44 (–3.4;6.32) | 0.51 (–2.86;3.89) | –2.09 (–4.22;0.02) |
| Current (crude) | 5.01 (–0.40;10.43) | –0.03 (–2.83;2.78) | 1.07 (–3.60;5.75) | –1.91 (–4.33;0.52) | 5.62 (–0.48;11.73) | –0.24 (–3.30;2.81) | 1.83 (–1.43;5.19) | 4.56 (2.79;6.32) |
| Current (adjusted) | –5.13 (–10.61;0.35) | –1.39 (–5.19;5.19) | –1.34 (–6.06;3.37) | 2.91 (–0.28;6.09) | –6.31 (–12.47; –0.15) | 1.29 (–2.68;5.28) | –1.26 (–4.38;1.85) | –3.60 (–5.36; –1.80) |
| Length of stay in Europe among migrants | ||||||||
| Length of stay (crude) | NA | –0.01 (–0.07;0.05) | NA | –0.03 (–0.07;0.01) | NA | –0.04 (–0.14;0.06) | NA | 0.01 (–0.04;0.05) |
| Length of stay (adjusted) | NA | 0.04 (–0.02;0.12) | NA | 0.04 (–0.03;0.11) | NA | 0.05 (–0.04;0.13) | NA | 0.03 (–0.02;0.08) |
| One carbon metabolism nutrients 7 | ||||||||
| Vit B2 (crude) | –1.37 (–2.63;0.12) | –0.35 (–0.72;0.03) | –0.585 (–1.67;0.50) | –0.13 (–0.47;0.19) | –1.59 (–3.01; –0.18) | 0.07 (–0.34;0.49) | –0.54 (–1.32;0.24) | 0.09 (–0.15;0.33) |
| Vit B2 (adjusted) | –1.06 (–3.39;1.27) | –0.26 (–0.94;0.42) | –1.92 (–3.94;0.08) | –0.12 (–0.69;0.45) | –3.09 (–5.72; –0.47) | –0.29 (–1.01;0.41) | –1.51 (–2.75;0.27) | –0.06 (–0.39;0.26) |
| Vit B6 (crude) | –0.69 (–1.50;0.12) | –0.14 (–0.61;0.33) | 0.01 (–0.69;0.70) | –0.05 (–0.46;0.36) | –0.43 (–1.35;0.47) | 0.46 (–0.05;0.97) | –0.10 (–0.60;0.41) | 0.10 (–0.20;0.49) |
| Vit B6 (adjusted) | –1.31 (–3.78;1.15) | 0.66 (–2.18;2.18) | –0.59 (–2.72;1.52) | –0.08 (–1.36;1.19) | –2.13 (–4.90;0.64) | 0.96 (–0.63;2.56) | –0.93 (–2.26;0.39) | 0.39 (–0.32;1.03) |
| Vit B9 (crude) | 0.00 (–0.01;0.00) | 0.00 (0.00;0.00) | 0.02 (0.00;0.01) | –0.01 (0.00;0.00) | 0.01 (–0.01;0.01) | 0.00 (0.00;0.01) | 0.01 (–0.02;0.05) | 0.00 (–0.02;0.02) |
| Vit B9 (adjusted) | –0.01 (0.00;0.03) | 0.00 (–0.01;0.01) | –0.01 (–0.02; –0.001) | –0.01 (–0.01;0.00) | –0.02 (0.04; –0.001) | –0.00 (–0.01;0.01) | -0.01 (–0.03; –0.02) | 0.00 (–0.04;0.03) |
| Vit B12 (crude) | 0.00 (–0.04;0.03) | 0.04 (–0.03;0.10) | 0.02 (–0.01;0.05) | –0.01 (–0.06;0.05) | –0.01 (–0.04;0.04) | 0.08 (0.00;0.14) | –0.01 (–0.02;0.02) | –0.02 (–0.01; –0.03) |
| Vit B12 (adjusted) | 0.00 (–0.04;0.04) | 0.05 (–0.20;0.31) | 0.03 (;0.001;0.06) | 0.14 (–0.09;0.36) | 0.01 (–0.03;0.05) | 0.13 (–0.15;0.41) | –0.01 (–0.03;0.02) | 0.02 (–0.02;0.06) |
| Cardiometabolic factors | ||||||||
| BMI (crude) | –0.15 (–0.27; –0.04) | –0.04 (–0.16;0.09) | –0.16 (–0.26; –0.07) | –0.03 (–0.14;0.07) | –0.22 (–0.35; –0.10) | –0.13 (–0.26;0.01) | –0.16 (–0.23; –0.09) | –0.09 (–0.17;0.02) |
| BMI (adjusted) | –0.14 (–0.26; –0.02) | 0.00 (–0.14;0.14) | –0.14 (–0.25; –0.04) | 0.00 (–0.11;0.12) | –0.21 (–0.34; –0.07) | –0.13 (–0.27;0.01) | –0.11 (–0.18; –0.04) | –0.01 (–0.09;0.07) |
| SBP (crude) | 0.01 (–0.03;0.04) | –0.01 (–0.05;0.02) | 0.01 (–0.01;0.04) | 0.00 (–0.03;0.03) | –0.01 (–0.04;0.02) | 0.01 (–0.03;0.05) | 0.00 (–0.01;0.02) | 0.02 (–0.01;0.04) |
| SBP (adjusted) | 0.00 (–0.03;0.03) | –0.02 (–0.02;0.02) | 0.00 (–0.02;0.03) | 0.00 (–0.03;0.03) | –0.02 (–0.05;0.02) | 0.02 (–0.02;0.06) | –0.01 (–0.02;0.02) | 0.01 (–0.01;0.03) |
| DBP (crude) | –0.02 (–0.08;0.03) | –0.02 (–0.07;0.03) | 0.02 (–0.03;0.06) | 0.01 (–0.04;0.06) | –0.04 (–0.10;0.02) | 0.00 (–0.06–0.06) | 0.01 (–0.02;0.04) | 0.07 (0.03;0.11) |
| DBP (adjusted) | –0.03 (–0.09;0.02) | –0.04 (–0.02;0.02) | 0.01 (–0.04;0.05) | –0.01 (–0.06;0.05) | –0.05 (–0.11;0.02) | 0.01 (–0.06–0.07) | –0.01 (–0.04;0.02) | 0.03 (–0.01;0.06) |
| FBG (crude) | 0.12 (–0.03;0.28) | 0.29 (0.01;0.58) | 0.10 (–0.03;0.23) | 0.32 (0.08;0.57) | 0.15 (–0.03;0.32) | 0.37 (0.05–0.68) | 0.15 (0.06;0.25) | 0.29 (0.11;0.48) |
| FBG (adjusted) | 0.12 (–0.04;0.28) | 0.30 (0.01;0.59) | 0.08 (–0.06;0.22) | 0.31 (0.05;0.56) | 0.14 (–0.0;0.32) | 0.39 (0.07;0.71) | 0.12 (0.03;0.21) | 0.18 (0.01;0.34) |
The table depicts linear regression model β with 95% confidence intervals. Crude = crude linear regression model, adjusted= fully adjusted linear regression model for age, sex, education, smoking, physical activity, alcohol intake, total energy intake, and duration of stay in host countries for migrants, respectively.
Intrinsic epigenetic age acceleration (within each cell) obtained using Horvath clock
Extrinsic epigenetic age acceleration (between different cells) using Hannum clock
Epigenetic age acceleration incorporating clinical traits obtained using the PhenoAge clock
Epigenetic age acceleration incorporating plasma proteins obtained using GrimAge clock
Ghanaians living in rural and urban Ghana were categorised as non-migrants
Ghanaians living in Amsterdam, Berlin and London were categorised as migrants.
Levels of physical activity categorised according to GPAQ criteria. Abbreviations: Length stay= duration of stay in Europe for migrants, Vit= Vitamin, Vitamin intake was measured in mg/day for Vitamin B2 and Vitamin B6, while vitamin B9 and Vitamin B12 were measured in mcg/day, BMI= body mass index (kg/m2), SBP= systolic blood pressure (mmHg), DBP= diastolic blood pressure (mmHg), FBG= fasting blood glucose (mmol/L). Highlighted results are the statistically significant results in the adjusted linear regression models.
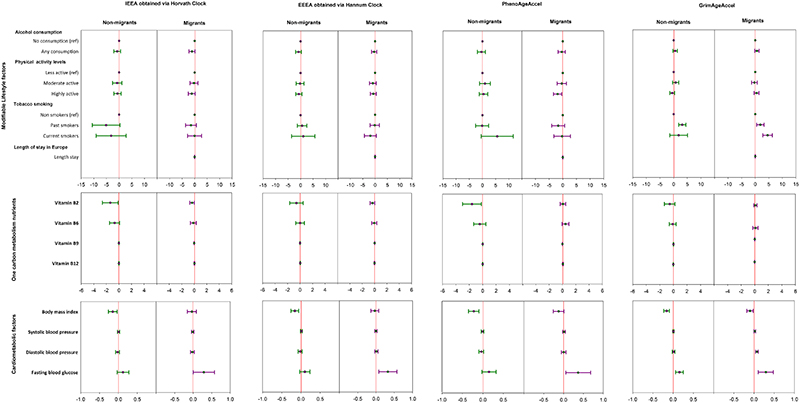
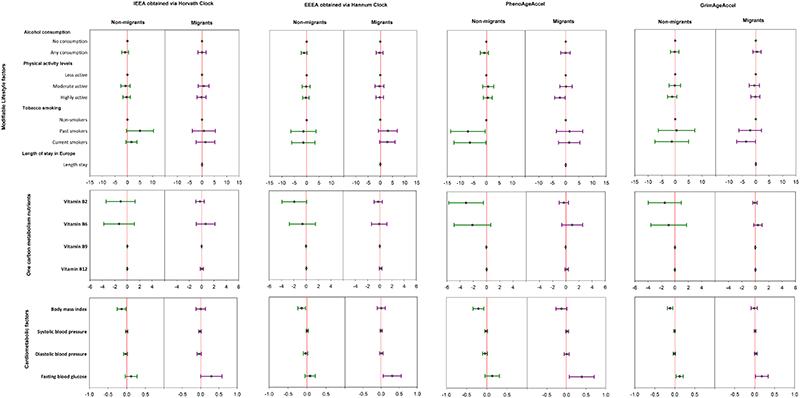
Figure 4. Forest plots of crude regression model β (and 95% confidence intervals) relating EAA measures to cardiometabolic related traits in the RODAM study.
The plot depicts regression model β with 95% confidence intervals in linear regression models adjusted for age, sex, education, smoking, physical activity, alcohol intake, total energy intake, and duration of stay in host countries for migrants, respectively. N=365 for migrants and 347 for non-migrants. Red line = reference line for confidence intervals. Abbreviations: IEAA= Intrinsic epigenetic age acceleration, EEAA= extrinsic epigenetic age acceleration, PhenoAge Accel= Pheno Age Acceleration, GrimAge Accel= Grim Age Acceleration. ref= reference, physical activity activity levels categorized according to Global Physical Activity Questionnaire (GPAQ) criteria. Vitamin intake was measured in mg/day for Vitamin B6 and Vitamin B12, while vitamin B9 and Vitamin B12 were measured in mcg/day. body mass index in kg/m2, fasting blood glucose in mmol/L, blood pressure in mmHg, Length stay= duration of stay in Europe for migrants. Reference (comparison) groups for modifiable risk factors: smoking = non-smokers, alcohol consumption = no (never) alcohol consumption, physical activity= less physically active.
Figure 5. Forest plots of adjusted regression model β (and 95% confidence intervals) relating EAA measures to cardiometabolic related traits in the RODAM study.
The plot depicts regression model β with 95% confidence intervals in linear regression models adjusted for age, sex, education, smoking, physical activity, alcohol intake, total energy intake, and duration of stay in host countries for migrants, respectively. N=365 for migrants and 347 for non-migrants. Red line = reference line for confidence intervals. Abbreviations: IEAA= Intrinsic epigenetic age acceleration, EEAA= extrinsic epigenetic age acceleration, PhenoAge Accel= Pheno Age Acceleration, GrimAge Accel= Grim Age Acceleration. ref= reference, physical activity activity levels categorized according to Global Physical Activity Questionnaire (GPAQ) criteria. Vitamin intake was measured in mg/day for Vitamin B6 and Vitamin B12, while vitamin B9 and Vitamin B12 were measured in mcg/day. body mass index in kg/m2, fasting blood glucose in mmol/L, blood pressure in mmHg, Length stay= duration of stay in Europe for migrants. Reference (comparison) groups for modifiable risk factors: smoking = non-smokers, alcohol consumption = no (never) alcohol consumption, physical activity = less physically active
When investigating modifiable lifestyle risk factors (table 2, figure 4&5), adjusted linear regression models showed that alcohol consumption was not associated with any EAA measure in either migrants or non-migrants. Current tobacco smoking was negatively associated with GrimAgeAccel among migrants, while current and past tobacco smoking were negatively associated with PhenoAgeAccel among non-migrants. Higher physical activity levels were negatively associated with PhenoAgeAccel among migrants but not non-migrants.
When investigating one-carbon metabolism nutrients (table 2, figure 4&5), adjusted linear regression models showed that higher Vitamin B2 (riboflavin) intake was negatively associated with PhenoAgeAccel among non-migrants but not among migrants. Higher Vitamin B9 (folate) intake was negatively associated with EEAA, PhenoAgeAccel and GrimAgeAccel among non-migrants but not among migrants. Such associations were not observed for Vitamin B6 (pyridoxine) and Vitamin B12 (cobalamin) in both migrants and non-migrants.
When investigating cardiometabolic factors (table 2, figure 4&5), adjusted linear regression models showed that higher BMI was negatively associated with all EAA measures among non-migrants. Such associations were not observed among migrants. Higher FBG was positively associated with all EAA measures among migrants. Higher FBG was also positively associated with GrimAgeAccel among non-migrants. Systolic and diastolic blood pressure were not associated with any EAA measure, regardless of migration status.
Results from post-hoc analyses are presented in appendix p 20-37 and summarized in appendix p 38. In brief, our findings on BMI, FBG and folate in the main analyses were also apparent in the combined sample of migrants and non-migrants (appendix p 20). Our results on FBG in the main analyses were also observable in linear regression models excluding participants taking anti-diabetic medications (appendix p 21). Our findings on BMI, FBG and folate in main analyses were also detectable in the independent replication cohorts (appendix p 22-39).
Discussion
In this analysis of EAA and CMD risk factors among migrant and non-migrant Ghanaians, we have found that migration status was negatively associated with EAA measures. Within migrants, FBG was positively associated with EAA measures (independent of use of anti-diabetic medications). Within non-migrants, BMI was negatively associated with EAA measures. The findings among Ghanaians also demonstrate a role of folate, a one-carbon metabolism nutrient, whereby higher folate intake was negatively associated with EAA measures among non-migrants. Our findings on FBG, BMI and folate among Ghanaians were observable in the combined migrant and non-migrant sample and were also successfully replicated in independent cohorts.
Our results among Ghanaians suggest that the interplay between migration, EAA and CMD is not as we hypothesized. Based on findings from HICs where lifestyle factors (tobacco smoking, unhealthy diets and increased alcohol intake) and cardiometabolic factors (obesity, diabetes and hypertension) were positively associated with EAA, we had hypothesized that changes in lifestyle upon migration would lead to accelerated biological aging of tissues, which would in turn be associated with CMD. 7,14,15 This would explain why migrant and urban populations have higher rates of obesity, hypertension and diabetes than non-migrant or rural populations. 16,17,23 However, we found in our current study that migration was negatively associated with EAA measures among Ghanaians (even after adjusting for potential confounders), despite their increased risk of CMD. Nevertheless, Horvath et al. reported a similar observation of decreased EAA with urbanization or migration among African hunter gatherers and Mexicans. 26 In that study, African hunter gatherers living in forests had higher EEAA compared to those who had shifted to an agriculturist life, while Mexicans born outside of the USA (but living in USA) had higher EEAA than Mexicans born in the USA (also living in USA). 26 Since all measures of EAA have been shown to predict all-cause mortality, our findings of lower EAA in migrants could be related to, at least in part, the mortality advantage observed among migrant and urbanizing populations (“healthy migrant effect”). 27 The “healthy migrant effect” describes an empirically observed mortality advantage of migrants relative to the remaining population in the native country, but also to the majority in the host country. Since previous studies have shown that migrants have lower all-cause mortality compared to non-migrant populations even with higher rates of CMD derived from multiple factors including better access to health services, 27 our findings seem to provide some molecular evidence for that mortality advantage observed among some migrants. 27
We found that FBG was positively associated with all EAA measures among Ghanaian migrants independent of anti-diabetic medication usage. Considering that migrants have higher rates of diabetes than non-migrants, this finding was expected. Epigenetic aging represents tissue aging, thus higher EAA was expected among individuals with higher FBG since glucose homeostasis mechanisms are impaired. Moreover, previous studies in HICs have also shown a positive relationship between diabetes and higher EAA. 7,14,15
We found that BMI was negatively associated with all EAA measures among non-migrant Ghanaians. This inverse relationship between BMI and EAA is in direct contrast to previous studies from HICs that found positive associations between obesity and EAA measures. 7,14,15 The explanations for these inconsistent results are unclear. However, a recent study undertaken in rural South Africa has also shown that overweight/obese individuals have a lower risk of all-cause mortality than those with a normal BMI after adjusting for potential confounders. 28 In that study, the protective effect of overweight and mild obesity was best demonstrated for infectious causes of death. 28 Putting our study and the South African study together, it is possible that both these findings point to the protective effects of BMI (overweight and mild obesity) on mortality in environments where the infectious disease (inflammatory) load is high. For instance, infectious diseases and high chronic inflammatory load are common among less urbanized populations (non-migrant Ghanaians in our case, as compared to migrants). Such a lifetime of diverse pathogen stresses, elevated inflammation and extensive immune activation are known to rapidly deplete naïve CD4+ T cells and lead to greater expression of exhausted T cells (rapid immunoscence), and eventually to increase EAA (as also seen among non-migrant Ghanaians in our study). 26 These ongoing environmental insults may result in increased nutritional needs for cell repair. 29 As such, it is possible that individuals with a nutritional reserve (moderately higher BMI) have better capacity for cell repair than lean individuals in contexts with a high load of environmental insults, hence the lower EAA.
Our study demonstrated a role for folate as a one-carbon metabolism nutrient. DNAm is dependent upon one-carbon pathways, which enables transmethylation reactions to occur. 19 Therefore, dietary intake of one-carbon metabolism nutrients could influence EAA. 19 Our findings on folate further support our hypothesis on BMI that EAA in environments with high inflammatory load depends on nutritional reserve for ongoing cell repair to these environmental insults (i.e. nutritional reserve → better cell repair → lower EAA). For instance, we found in the RODAM study that higher Vitamin B9 (folate) intake was negatively associated with EAA measures among non-migrant Ghanaians after adjusting for confounders. Folate plays a crucial role in cell repair (especially DNA repair). 30-32 It is essential for the de novo synthesis of purines and pyrimidines, which are required during the replication and repair of DNA, 31 Thus, similarly to BMI, presence of this one-carbon metabolism nutrient could possibly enhance the capacity for cell repair in contexts with a high load of inflammation, thereby decreasing EAA among persons with higher intake of folate compared to those with deficiencies.
Our goal was to validate our findings in independent cohorts. One thing that was clear was the scarcity of epigenetic data in populations from LMIC, especially cohorts examining effects of migration from LMIC to HIC in homogenous populations. Subsequently, we utilized the IMS and PURE-SA-NW cohorts to replicate our findings. 23,24 Although the IMS (Indians) investigated rural-to-urban migration, and was in an entirely different ethnic group, changes in CMD risk factors at the phenotypic level have been shown to be similar whether migrating from rural-to-urban areas within-country, 16 or internationally from LMIC to HIC (though effect sizes may differ). 17 Moreover, migrants and non-migrants in IMS were matched by age range and sex, which minimized differences in baseline characteristics, which can confound migration studies. 23 Associations of migration status and EAA did not pass the replication criteria in IMS, which could be due to a broad range of factors including type of migration (rural-to-urban vs LMIC-to- HIC), ethnicity (genetic influences on the epigenome) and other environmental factors (e.g. pollution, etc.) not shared between Ghanaians and Indians. 33 Despite the unsuccessful replication of migration status among Indians, one thing that was clear was the mirroring of EAA in migrants and non-migrants. For instance, when the mean IEAA was -0.30 years in migrants, it would mirror in non-migrants as 0.30 years. This was observed in all EAA measures, as well as in the RODAM study. This finding is interesting because determination of EAA in one group (let us say migrants) could possibly predict an opposite EAA effect in the other group (homogenous group of non-migrants). The effects of FBG, BMI and folate observed among Ghanaian migrant and non-migrants were also detected in the IMS. This shared finding between Ghanaians and Indians could point to the fact that these specific effects are similar across populations undergoing health transitions irrespective of type of migration and ethnicity.
The PURE-SA-NW cohort (native South Africans) was not the most ideal cohort to replicate our findings due to factors such as lack of comparison groups and lack of participants with higher BMI and FBG. 24 However, this was the only African cohort with data on DNA methylation and CMD risk factors. Despite the challenges, we believed that the effects of lifestyle factors on EAA among native South Africans residing in Africa would be still captured. These effects would then in turn be compared to those of Ghanaians also residing in Africa (non-migrants). We subsequently found that higher folate intake was negatively associated with EAA measures as had been observed in non-migrant Ghanaians. This finding validated the role of folate in EAA for populations residing in Africa in general.
Our findings have the potential to improve population health. For example, further studies could determine a cut-off point at which higher BMI has a benefit on biological aging (improved life expectancy) and yet minimizes the risk of other CMDs in environments with a high inflammatory (infectious disease) load. Moreover, with high rates of chronic undernutrition in less urbanized populations, further studies could determine whether supplementation of one-carbon metabolism nutrients like folate (i.e., correcting vitamin deficiencies) can reverse EAA in these environments. More importantly, there have been recent breakthroughs in reversal of biological clocks. 34 Such future treatments will be handy for specific group of migrants and non-migrants that exhibit higher EAA.
The most important strength of our study is that we attempted replication in independent cohorts, which validates our findings. Second, the migration models utilized in this study are powerful because they allow for investigation of changing environment exposures while controlling for ethnicity and early life exposure. However, our study is not without limitations. First, we used a sub-sample of the overall RODAM study, which could have introduced selection bias. Nevertheless, our epigenetic sub-sample was generally representative of the overall RODAM population (appendix p 7). Second, there were large baseline differences between migrants and non-migrants, which could confound our results. Although these differences represented the real-life contrasts between migrants and non-migrants (whereby migrants are mainly young, with poor lifestyle and cardiometabolic profiles compared to non-migrants), we minimized confounding by performing sensitivity analyses in the combined migrant and non-migrant sample, where the effect would be largely free of sub-group baseline differences (appendix p 20-21). Third, socio economic status (SES) is known to influence BMI and overall health status. As such, our findings could also be possibly influenced by SES. For instance, the lower EAA seen in migrants could be because of migrants having better SES than non-migrants, while lower EAA associated with higher BMI among non-migrants might also reflect the higher SES among those with higher BMI. We minimized this SES confounding by adjusting for education. Although education has been shown to be a powerful predictor for SES, 35 it does not capture the full spectrum of SES. It is therefore possible that other components of SES such as income levels and occupation might still influence our findings. Since currently there is no data on how SES affects EAA in Africans or other populations from LMIC, further studies are needed to evaluate in detail the effects of other indicators of SES on EAA in these populations. Fourth, although we adjusted for age and sex, we cannot rule the possibility of residual confounding. Nevertheless, successful replication of most of our findings in IMS where confounding was minimized by matching migrants and non-migrants by age and sex, further substantiated our findings. 24 Fourth, we introduced criteria to minimize false positives. Some positive findings on alcohol consumption, smoking, physical activity, and vitamin intake which did not pass the criteria (were observed in only one EAA measure), might have been true findings. However, application of a criteria to minimize false positives in our study enabled us to identify effects that were consistent across most EAA measures. Lastly, vitamin intake was calculated from FFQs, relying on participants ability to recall well frequency and number of consumed foods, which might have introduced recall bias.
In conclusion, our study among Ghanaians shows that migration is negatively associated with EAA. Moreover, CMD traits, are differentially associated with EAA within migrant and non-migrant subgroups. Many of such associations are also apparent in other ethnic groups. There is potential to harness the EAA effects of BMI, and folate intake to improve life expectancy in the rural areas where nutritional deficiencies and infectious diseases might be highly prevalent. Our study therefore calls for interventions that consider the effects of biological aging (EAA) in the prevention and treatment of CMD among Africans and other LMIC populations.
Supplementary Material
Research in Context.
Evidence before this study
We searched PubMed for articles published between September 2010 and September 2020 using the MESH search terms transients and migrants, urbanization, hypertension, diabetes mellitus type 2, DNA methylation, body mass index, biological clocks, Africa and developing countries. We did not find any study that had investigated the relationship between biological aging (epigenetic age acceleration) and cardiometabolic traits such as obesity, diabetes and hypertension in Africa or any other low-income countries.
Added value of this study
We provide evidence for the role of epigenetic age acceleration (EAA) in the growing burden of obesity, diabetes and hypertension in populations undergoing epidemiological (health) transitions. We use powerful migration models that allow investigation of changing environmental exposures while controlling for ethnicity and early life exposure. We also assess for the first time ever, how one-carbon metabolism nutrients can influence EAA. We find that transitioning Ghanaian populations (migrants) have lower EAA than their non-migrant counterparts. We find among migrants that higher fasting blood glucose (diabetes) is positively associated with EAA. We also find that higher BMI (overweight and mild obesity) and intake of folate are negatively associated with EAA in non-migrants, which could possibly be linked to the nutritional reserve that is required to repair the deleterious effects of chronic inflammation in the human body. Our results on overweight, diabetes and folate intake were successfully replicated in independent cohorts of Indians and native South Africans, which shows that most of the relationships between EAA and cardiometabolic factors are similar across populations experiencing epidemiological transitions.
Implications of all the available evidence
Our results show that EAA is negatively associated with epidemiological (health) transitions among Ghanaians. Clearly, the relationship between EAA and cardiometabolic factors depends on whether a population is urbanizing or not, as well as on the prevalent environment exposures like infectious disease and chronic inflammation. There is potential to harness the EAA effects of BMI, and folate intake to improve life expectancy in the rural areas where nutritional deficiencies and infectious diseases might be highly prevalent. Our study calls for interventions that consider the effects of biological aging (EAA) in the prevention and treatment of cardiometabolic diseases among Africans and other populations undergoing epidemiological (health) transitions.
Acknowledgements
The authors are very grateful to the Ghanaian, Indian and Batswana volunteers participating in RODAM, IMS and PURE-SA-NW studies, respectively. We also thank all research staff who were involved in all these studies. This work was supported by the European Commission under the Framework Program (Grant Number: 278901) and European Research Council Consolidation (Grant Number: 772244). FPC is supported by the Erasmus Mundus Joint Doctorate Program of the European Union through the Amsterdam Institute of Global Health and Development (AIGHD) [grant agreement 2015–1595]. KACM. and AA are supported by the Intramural Research Program of the National Institutes of Health in the Center for Research on Genomics and Global Health (CRGGH). The CRGGH is supported by the National Human Genome Research Institute, the National Institute of Diabetes and Digestive and Kidney Diseases, the Center for Information Technology, and the Office of the Director at the National Institutes of Health (1ZIAHG200362). HRE works in the Medical Research Council Integrative Epidemiology Unit (MRC-IEU) at the University of Bristol, which is financially supported by the Medical Research Council (MRC) and the University of Bristol (MC_UU_00011/5). HTC is supported by a grant from the Novo Nordisk Foundation Challenge Program: Harnessing the Power of Big Data to Address the Societal Challenge of Aging (NNF17OC0027812).
Declarations
Author contributions
FPC, PH and CA conceived and designed the study. FPC, HRE, HTC analysed the data. PH, CA, GKW and MP verified the underlying data. FPC, wrote the paper with PH and CA. All authors FPC, PH, HRE, HTC, JRG, GKW, KACM, ARM, AA, ID, SB, KKG, AA, MP, MMAMM and CA participated in interpreting the data, drafting the article, or revising it critically for content.
Declarations of interests
We declare no competing interests.
Contributor Information
Felix P Chilunga, Department of Public Health, Amsterdam Public Health Research Institute, Amsterdam University Medical Centres, University of Amsterdam, Amsterdam, The Netherlands.
Peter Henneman, Department of Clinical Genetics, Amsterdam Reproduction & Development research institute, Amsterdam University Medical Centres, University of Amsterdam, Amsterdam, The Netherlands.
Hannah R Elliott, MRC Integrative Epidemiology Unit, University of Bristol, Bristol, United Kingdom; Department of population health sciences, Bristol Medical School, University of Bristol, Bristol, United Kingdom.
H Toinét Cronjé, Section of Epidemiology, Department of Public Health, University of Copenhagen, Copenhagen, Denmark; Centre of Excellence for Nutrition, North-West University, Potchefstroom, South Africa.
Gagandeep K Walia, Public Health Foundation of India, New Delhi, India.
Karlijn A.C Meeks, Center for Research on Genomics and Global Health, National Human Genome Research Institute, National Institutes of Health, Bethesda, Maryland, United States of America.
Ana Requena-Mendez, Barcelona Institute for Global Health (ISGlobal), Barcelona, Spain; Department of Global Public Health, Karolinska Institutet, Solna, Sweden.
Andrea Venema, Department of Clinical Genetics, Amsterdam Reproduction & Development research institute, Amsterdam University Medical Centres, University of Amsterdam, Amsterdam, The Netherlands.
Silver Bahendeka, Department of Medicine, MKPGMS-Uganda Martyrs University, Kampala, Uganda.
Ina Danquah, Heidelberg Institute of Global Health (HIGH), Universitätsklinikum Heidelberg, Heidelberg, Germany.
Adebowale Adeyemo, Center for Research on Genomics and Global Health, National Human Genome Research Institute, National Institutes of Health, Bethesda, Maryland, United States of America.
Kerstin Klipstein-Grobusch, Julius Global Health, Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherland; Division of Epidemiology and Biostatistics, School of Public Health, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa.
Prof Marlien Pieters, Medical Research Council Unit for Hypertension and Cardiovascular Disease, North-West University, Potchefstroom, South Africa.
Prof Marcels M.A.M Mannens, Department of Clinical Genetics, Amsterdam Reproduction & Development research institute, Amsterdam University Medical Centres, University of Amsterdam, Amsterdam, The Netherlands.
Prof Charles Agyemang, Department of Public Health, Amsterdam Public Health Research Institute, Amsterdam University Medical Centres, University of Amsterdam, Amsterdam, The Netherlands.
Availability of data and materials
Individual participant data from the RODAM study used in the current analyses will be deposited to the European Genome-phenome Archive (EGA; https://ega-archive.org/) by June 2021 in a deidentified or anonymized format. The study protocol and statistical analysis plans were previously published (doi: 10.1136/bmjopen-2014-004877). Data will be shared with bona fide researchers submitting a research proposal and requesting access to data. Data will be made available for analyses as approved by the data access committee. Data from IMS is available upon request, details can be found on the study webpage https://apcaps.lshtm.ac.uk/relatedstudies/ims/, while from PURE-SA-NW is available with permission of the Health Research Ethics Committee of the North-West University and the principal investigator of the PURE-SA-NW study, Prof. I.M. Kruger (lanthe.kruger@nwu.ac.za) and MP (marlien.pieters@nwu.ac.za)
References
- 1.Rechel B, Mladovsky P, Ingleby D, Mackenbach JP, McKee M. Migration and health in an increasingly diverse Europe. The Lancet. 2013;381(9873):1235–45. doi: 10.1016/S0140-6736(12)62086-8. [DOI] [PubMed] [Google Scholar]
- 2.Bickler SW, Wang A, Amin S, et al. Urbanization in Sub-Saharan Africa: Declining Rates of Chronic and Recurrent Infection and Their Possible Role in the Origins of Non-communicable Diseases. World Journal of Surgery. 2017:1–12. doi: 10.1007/s00268-017-4389-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim SS, Vos T, Flaxman AD, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. The lancet. 2012;380(9859):2224–60. doi: 10.1016/S0140-6736(12)61766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imamura F, Micha R, Khatibzadeh S, et al. Dietary quality among men and women in 187 countries in 1990 and 2010: a systematic assessment. The lancet global health. 2015;3(3):e132–e42. doi: 10.1016/S2214-109X(14)70381-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collaborators GRF. Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet (London, England) 2015;386(10010):2287. doi: 10.1016/S0140-6736(15)00128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nature Reviews Genetics. 2018;19(6):371. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- 7.Quach A, Levine ME, Tanaka T, et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY) 2017;9(2):419. doi: 10.18632/aging.101168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine ME, Hosgood HD, Chen B, Absher D, Assimes T, Horvath S. DNA methylation age of blood predicts future onset of lung cancer in the women’s health initiative. Aging (Albany NY) 2015;7(9):690. doi: 10.18632/aging.100809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 2018;10(4):573. doi: 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marioni RE, Shah S, McRae AF, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome biology. 2015;16(1):1–12. doi: 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath S. DNA methylation age of human tissues and cell types. Genome biology. 2013;14(10):3156. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Molecular cell. 2013;49(2):359–67. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu AT, Quach A, Wilson JG, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 2019;11(2):303. doi: 10.18632/aging.101684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao W, Ammous F, Ratliff S, et al. Education and Lifestyle Factors Are Associated with DNA Methylation Clocks in Older African Americans. International journal of environmental research and public health. 2019;16(17):3141. doi: 10.3390/ijerph16173141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiorito G, McCrory C, Robinson O, et al. Socioeconomic position, lifestyle habits and biomarkers of epigenetic aging: a multi-cohort analysis. Aging (Albany NY) 2019;11(7):2045. doi: 10.18632/aging.101900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chilunga FP, Musicha C, Tafatatha T, et al. Investigating associations between rural-to-urban migration and cardiometabolic disease in Malawi: a population-level study. International journal of epidemiology. 2019;48(6):1850–62. doi: 10.1093/ije/dyz198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agyemang C, Meeks K, Beune E, et al. Obesity and type 2 diabetes in sub-Saharan Africans–Is the burden in today’s Africa similar to African migrants in Europe? The RODAM study. BMC medicine. 2016;14(1):166. doi: 10.1186/s12916-016-0709-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson OS, Sant KE, Dolinoy DC. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. The Journal of nutritional biochemistry. 2012;23(8):853–9. doi: 10.1016/j.jnutbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–9. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horvath S. DNA Methylation Age Calculator. 2021. [accessed 15 February 2021]. http://dnamage.genetics.ucla.edu/
- 21.Alfonso G, Gonzalez JR. Bayesian neural networks for the optimisation of biological clocks in humans. bioRxiv. 2020 [Google Scholar]
- 22.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics. 2012;13(1):86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebrahim S, Kinra S, Bowen L, et al. The effect of rural-to-urban migration on obesity and diabetes in India: a cross-sectional study. PLoS Med. 2010;7(4):e1000268. doi: 10.1371/journal.pmed.1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cronjé HT, Elliott HR, Nienaber-Rousseau C, Pieters M. Replication and expansion of epigenome-wide association literature in a black South African population. Clinical Epigenetics. 2020;12(1):1–13. doi: 10.1186/s13148-019-0805-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lechner K, von Schacky C, McKenzie AL, et al. Lifestyle factors and high-risk atherosclerosis: Pathways and mechanisms beyond traditional risk factors. European journal of preventive cardiology. 2020;27(4):394–406. doi: 10.1177/2047487319869400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horvath S, Gurven M, Levine ME, et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome biology. 2016;17(1):171. doi: 10.1186/s13059-016-1030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallace M, Khlat M, Guillot M. Mortality advantage among migrants according to duration of stay in France, 2004–2014. BMC public health. 2019;19(1):1–9. doi: 10.1186/s12889-019-6652-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manne-Goehler J, Baisley K, Vandormael A, et al. BMI and All-Cause Mortality in a Population-Based Cohort in Rural South Africa. Obesity. 2020 doi: 10.1002/oby.23005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarry-Adkins J, Chen J, Smith N, Jones R, Cherif H, Ozanne S. Poor maternal nutrition followed by accelerated postnatal growth leads to telomere shortening and increased markers of cell senescence in rat islets. The FASEB Journal. 2009;23(5):1521–8. doi: 10.1096/fj.08-122796. [DOI] [PubMed] [Google Scholar]
- 30.Duthie SJ, Narayanan S, Blum S, Pirie L, Brand GM. Folate deficiency in vitro induces uracil misincorporation and DNA hypomethylation and inhibits DNA excision repair in immortalized normal human colon epithelial cells. Nutrition and cancer. 2000;37(2):245–51. doi: 10.1207/S15327914NC372_18. [DOI] [PubMed] [Google Scholar]
- 31.Duthie SJ, Hawdon A. DNA instability (strand breakage, uracil misincorporation, and defective repair) is increased by folic acid depletion in human lymphocytes in vitro. The FASEB Journal. 1998;12(14):1491–7. [PubMed] [Google Scholar]
- 32.Manthey KC, Rodriguez-Melendez R, Hoi JT, Zempleni J. Riboflavin deficiency causes protein and DNA damage in HepG2 cells, triggering arrest in G1 phase of the cell cycle. The Journal of nutritional biochemistry. 2006;17(4):250–6. doi: 10.1016/j.jnutbio.2005.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pena MSB, Rollins A. Environmental exposures and cardiovascular disease: a challenge for health and development in low-and middle-income countries. Cardiology clinics. 2017;35(1):71–86. doi: 10.1016/j.ccl.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu Y, Brommer B, Tian X, et al. Reprogramming to recover youthful epigenetic information and restore vision. 2020;588(7836):124–9. doi: 10.1038/s41586-020-2975-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winkleby MA, Jatulis DE, Frank E, Fortmann SP. Socioeconomic status and health: how education, income, and occupation contribute to risk factors for cardiovascular disease. American journal of public health. 1992;82(6):816–20. doi: 10.2105/ajph.82.6.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Individual participant data from the RODAM study used in the current analyses will be deposited to the European Genome-phenome Archive (EGA; https://ega-archive.org/) by June 2021 in a deidentified or anonymized format. The study protocol and statistical analysis plans were previously published (doi: 10.1136/bmjopen-2014-004877). Data will be shared with bona fide researchers submitting a research proposal and requesting access to data. Data will be made available for analyses as approved by the data access committee. Data from IMS is available upon request, details can be found on the study webpage https://apcaps.lshtm.ac.uk/relatedstudies/ims/, while from PURE-SA-NW is available with permission of the Health Research Ethics Committee of the North-West University and the principal investigator of the PURE-SA-NW study, Prof. I.M. Kruger (lanthe.kruger@nwu.ac.za) and MP (marlien.pieters@nwu.ac.za)