Abstract
Metabolic characteristics of adult stem cells are distinct from their differentiated progeny, and cellular metabolism is emerging as a potential driver of cell fate conversions 1–4 . How these metabolic features are established remains unclear. Here, we identified inherited metabolism imposed by functionally distinct mitochondrial age-classes as a fate determinant in asymmetric division of epithelial stem-like cells. While chronologically old mitochondria support oxidative respiration, the electron transport chain of the new organelles is proteomically immature and they respire less. Upon cell division, selectively segregated mitochondrial age-classes elicit a metabolic bias in progeny cells, with oxidative energy metabolism promoting differentiation in cells inheriting old mitochondria. Cells inheriting newly synthesised mitochondria with low levels of Rieske iron-sulfur polypeptide 1 have a higher pentose phosphate pathway activity, which promotes de-novo purine biosynthesis and redox balance, and is required to maintain stemness during early fate determination after division. Our results demonstrate that fate decisions are susceptible to intrinsic metabolic bias imposed by selectively inherited mitochondria.
Cell fate changes in adult stem cells coincide with profound metabolic rewiring, and the importance of metabolic adaptation for successful differentiation has been demonstrated in a range of tissue-resident cell populations 5–9 . Metabolic rewiring correlates with changes in mitochondrial content and function 7,10,11 , and metabolism is emerging as an instructive signal for cell fate programs 12–14 . In particular, mitochondrial substrate use has the capacity to affect maintenance and function of adult stem cell populations 5,9,14–18 .
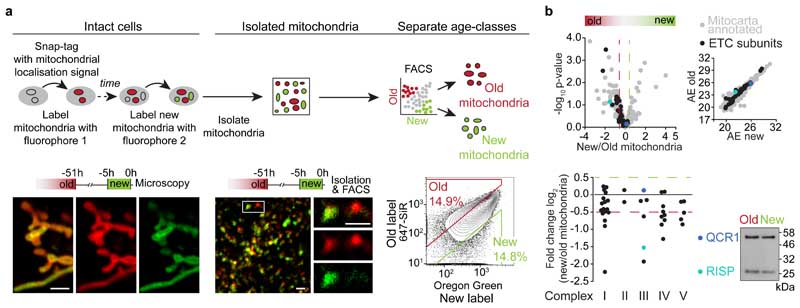
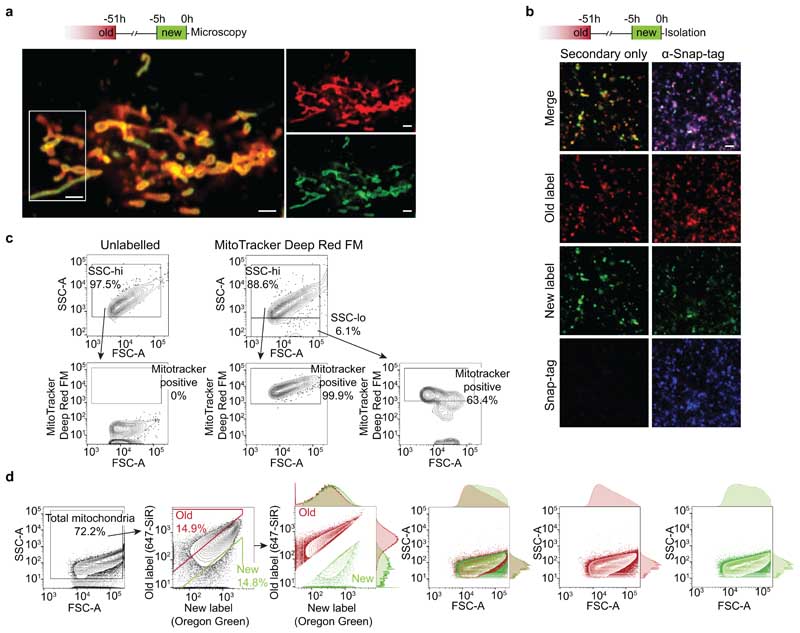
We have previously shown that human mammary epithelial cells (hMECs) contain a sub-population of stem-like cells (SLCs) 19,20 that apportion chronologically old mitochondria asymmetrically during cell division 21 . Progeny omitting old mitochondria maintain stem-like traits, whereas progeny inheriting old mitochondria differentiate 21 . To investigate how different mitochondrial age-classes influence the daughter cells after asymmetric divisions, we sequentially labelled hMECs that express Snap-tags 22 targeted to the outer mitochondrial membrane, and isolated their mitochondria (Fig. 1a). Labelling heterogeneity observed in isolated mitochondria corresponded to mitochondrial domains in cells before isolation (Fig. 1a, ED Fig. 1a, b). We selected a MitoTracker-positive mitochondrial population and used the enrichment for age-specific labels (ED Fig. 1c) for single-organelle FACS sorting to separate old and new mitochondria for proteomics (Fig. 1a, ED Fig. 1d).
Figure 1. Mitochondrial maturation with chronological age.
a, Experimental setup for age-specific mitochondrial labelling, isolation and separation of old and new mitochondria. Images show age-specifically labelled mitochondria in an intact living cell (left), after isolation (centre), and during age-selective single-organelle sorting (right). Microscopy images show a single confocal plane, and the FACS plot illustrates selection of mitochondria used for further analyses. Scale bars, 1.5 μm. b, Proteomic analysis of isolated old and new mitochondria (n = 3 isolations). Relative enrichment (log2 fold change) and average expression (AE) (upper) of mitochondrial proteins 23,24 in old and new mitochondria (shown in grey, with electron transport chain (ETC) subunits and selected proteins highlighted). Cut-off-values for enrichment, > 0.5 and < -0.5, were chosen based on the mean coefficient of variation for new and old mitochondrial samples (0.37 and 0.38, respectively). Relative enrichment (lower) of ETC subunits in old and new mitochondria, with proteins grouped by complex. Immunoblot validation is shown for QCR1 (unchanged) and RISP (enriched in old mitochondria). Statistical testing details in Supplementary Data Table 1. Numerical source data are available in Source Data Table 1 and unprocessed blots are available in Source Data Figure 1.
In addition to proteins annotated in MitoCarta 23,24 , both old and new mitochondrial samples contained proteins localised to other subcellular compartments (ED Fig. 2a, b, c, d). However, as virtually all events in the parent population were MitoTracker-positive (ED Fig. 1c), we concluded that detection of non-mitochondrially annotated proteins reflects inter-organelle interactions that support metabolic and functional regulation of cellular processes (reviewed in 25,26,27 ). Taken together, our approach yielded unique type of data, allowing us to analyse age-specific differences in composition and function between old and new mitochondria.
Among mitochondrial proteins, multiple inner membrane and matrix proteins were present at higher frequency in old mitochondria (ED Fig. 2e). Interestingly, electron transport chain (ETC) proteins were particularly enriched in old organelles (Fig. 1b), as well as ETC complex assembly factors, such as TIDC1, involved in complex I assembly (ED Fig. 2f, g) 28,29 . Multiple subunits of complexes III and IV were enriched in old mitochondria. These included the complex III catalytic subunit Rieske iron-sulfur polypeptide 1 (RISP) (Fig. 1b), which is among the last subunits to be incorporated to complex III (reviewed in 30 ). Notably, lack of RISP reduces complex III and IV quantity and activity in fibroblasts 31 , and affects hematopoietic stem cell differentiation 16 . Taken together, these data raised the possibility that old mitochondria contain a more functional ETC.
To further characterise old and new mitochondria, we analysed the lipid composition of age-selectively isolated organelles (ED Fig. 3a). Old mitochondria had higher levels of the key mitochondrial phospholipid cardiolipin (ED Fig. 3b), while overall phospholipid signature was similar to new mitochondria (ED Fig. 3c). Cardiolipin is synthesized in mitochondria, where it is enriched in the inner membrane to support ETC assembly and activity (reviewed in 32,33 ). Together with our proteomic analysis, these data suggest that our labelling strategy captures mitochondria at distinct stages of maturation.
As the molecular profiling suggested that old mitochondria support oxidative metabolism more readily than newly synthesised organelles, we assessed functional characteristics of mitochondrial age-classes by measuring their membrane potential. Old mitochondria had higher membrane potential than new mitochondria (Fig. 2a, ED Fig. 4a). Furthermore, chronologically old domains of the mitochondrial network showed high intensity of MitoSOX superoxide-probe labelling, reflecting complex I and III acivity 34 (Fig. 2b). This was in striking contrast to new mitochondrial domains, which showed low MitoSOX staining, indicating higher oxidative phosphorylation (Oxphos) activity in old mitochondria.
Figure 2. Age of inherited mitochondria predicts progeny cell metabolism.
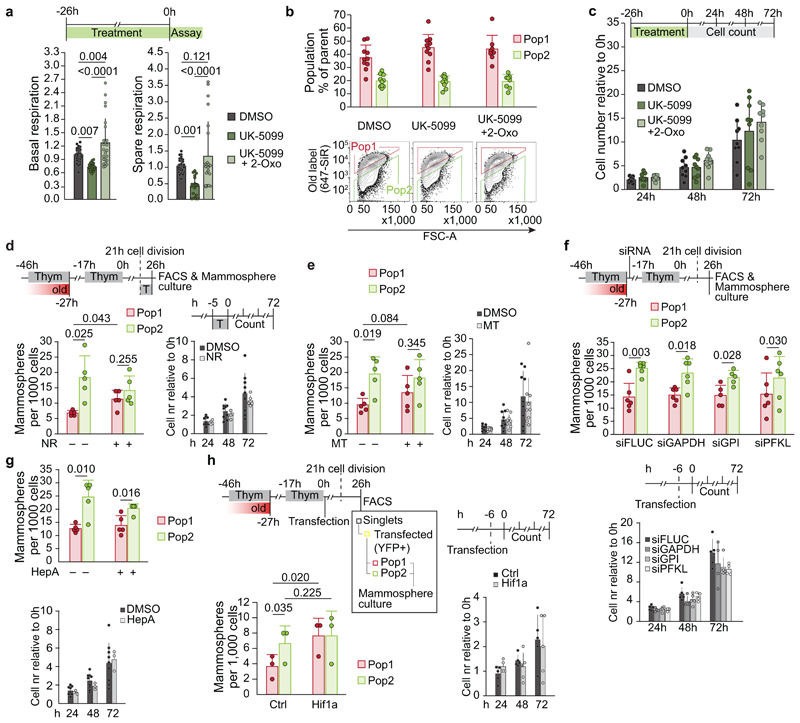
a, Membrane potential, representative TMRM FACS plots in isolated old and new mitochondria and quantitation of median fluorescence intensity (FI). Mean ± s.d. of three experiments. OG: Oregon Green b, Mitochondrial superoxide in mitochondrial domains enriched for old (arrow) or new (arrowhead) mitochondria label within intact cells. Maximum intensity projection of 1.2 μm (live cell imaging). Scale bars 1.5 μm. Quantitation of MitoSOX relative FI in individual mitochondria, 546 segmented mitochondrial domains from seven cells comparing mitochondria enriched for new (below first quartile of old/new ratio) and old label (above third quartile). c, Old mitochondria and MitoSOX during cytokinesis. Maximum intensity projection of two planes (live cell imaging). Scale bar 5 μm. Distribution of total fluorescence as fraction per cell within a cell pair (P1 and P2) in division from 58 pairs in five experiments. 25 pairs were asymmetric for of old mitochondria inheritance. Asymmetry cut-off: 1.5-fold enrichment of old mitochondria (P1/P2 ratio > 60/40 %). d, Oxygen consumption rate (OCR) of Pop1 and Pop2 cells FACS-sorted by inheritance of old mitochondria. Representative FACS plot and OCR curves from one experiment (Pop1 seven, Pop2 five repeats). Bar graphs show data (pmol/min) from three experiments (Pop1 14, Pop2 13 repeats). Mean ± s.d.. e, Redox balance in Pop1 and Pop2 cells FACS-sorted by inheritance of old mitochondria. NAD+/NADH and GSSG/GSH ratio in Pop1 cells compared to Pop2 cells from the same experiment, mean ± s.d. of five (NAD+/NADH) and seven (GSSG/GSH) biological replicates. f, Mammosphere formation (left) after treatment with UK-5099 for 26h alone or in combination with 2-Oxo, or DMSO prior to FACS-sorting by inheritance of old mitochondria. Mean ± s.d. of UK-5099 18, UK-5099 + 2-Oxo 11 and DMSO 13 biological replicates. Representative OCR curves (right) for cells treated with UK-5099 (seven replicates), UK-5099 and 2-Oxo (eleven replicates), or DMSO (seven replicates) from two experiments. Treatments were maintained during the assay. Mean ± s.d. UK-5099: 10 μM, 2-Oxo: 400 μM. Statistical testing details in Supplementary Data Table 1. Numerical source data are available in Source Data Table 2.
We next asked whether mitochondrial age-specific characteristics are retained during cell division, and may therefore impact daughter cells immediately after their age-selective apportioning in asymmetric cell divisions 21 . To this end, we analysed mitochondrial Oxphos activity by superoxide production during cytokinesis, and found that daughter cells inheriting the majority of old mitochondria (Progeny 1, P1) had significantly higher activity than their corresponding P2 daughter cells inheriting fewer old mitochondria (Fig. 2c). This was elicited by the mitochondrial composition and not quantity, as total mitochondrial content - measured by mtDNA copy number and mitochondrial protein levels - was similar in both cell types (ED Fig. 4b, c).
Mitochondrial bioenergetics is a central regulator of cellular metabolism and redox maintenance 5,14,35 . To test if the age-selective inheritance of functionally distinct mitochondria defines the cell-level bioenergetics of the daughter cells, we sorted cells five hours after synchronised division based on age-selective mitochondrial inheritance, and assessed their respiration. We designated cells enriched for old mitochondria as Population 1 (Pop1), and cells omitting old mitochondria as Population 2 (Pop2) (Fig. 2d, ED Fig. 5a). Consistent with the inheritance of old mitochondria enriched for ETC subunits, Pop1 cells showed higher basal respiration and spare respiratory capacity than Pop2 cells (Fig. 2d). In line with their higher respiration and superoxide production, the redox balance indicated by NAD+/NADH and GSSG/GSH-ratios was shifted towards oxidized forms in Pop1 cells (Fig. 2e). Further highlighting mitochondrial functional differences while suggesting no primary role for ROS-induced protein damage 36 , total cellular levels of reactive oxygen species (ROS) in recently divided cells correlated with the age of mitochondria they had inherited (ED Fig. 5b), but levels of protein carbonylation, an indicator of ROS induced oxidative protein damage, were comparable (ED Fig. 5c). Taken together, these data are in line with our findings on age-dependent functional differences of mitochondria, and with the immature respiratory activity of newly synthesised mitochondria.
Mammosphere formation provides an in vitro assay for self-renewal and stem-like growth of HMECs 37 , and as we showed previously, Pop2 cells form more mammospheres than Pop1 cells 21 (Fig. 2f). To test whether old mitochondria with highly active respiration determine cell fate via mitochondrial metabolism, we reduced respiration (Fig. 2f, ED Fig. 6a) by treating the cells with the mitochondrial pyruvate carrier (MPC) inhibitor UK-5099. Importantly, UK-5099 abolished the difference in stem-like properties between Pop1 and Pop2 cells, and the effect was nullified by rescuing respiration with simultaneous administration of an α-ketoglutarate precursor (2-Oxo) (Fig. 2f and ED Fig. 6a). As neither treatment affected cell proliferation or apportioning of old mitochondria (ED Fig. 6b, c), these findings indicate that metabolism acts as a fate determinant, and active respiration induces differentiation in our system.
Given the difference in redox balance between Pop1 and Pop2 cells (Fig. 2e), we next studied how redox capacity impacts stem cell traits. We pulse-treated cells with the NAD+ precursor NR or mitochondrial superoxide scavenger MitoTEMPO shortly after division. Both NR and MitoTEMPO have previously been shown to increase self-renewal 18,38 . While both treatments increased mammosphere formation of Pop1 cells (ED Fig. 6d, e), neither of them nullified the difference between Pop1 and Pop2 cells. Moreover, inhibition or depletion of glycolytic enzymes did not abrogate the difference in mammosphere formation between Pop1 and Pop2 cells, indicating that glycolytic flux per se was not causal for the difference in stem cell traits or redox balance (ED Fig. 6f, g). In contrast, the more general metabolic adaptation promoted by Hif1α overexpression nullified differences in stemness (ED Fig. 6h), suggesting involvement of other pathways beyond those influenced by redox.
To more specifically probe the metabolic pathways that respond to age-selectively segregated mitochondria, we performed isotopic tracing on Pop1 and Pop2 cells. To accurately address the primary metabolic profile of the cell populations after asymmetric division, we administered a short pulse of U-13C glucose close to the division time and assessed levels of 13C-labelled intermediates of central carbon metabolism pathways (Fig. 3a, ED Fig. 7a). Consistent with their lower basal and spare respiration (Fig. 2d), Pop2 cells contained less 13C-labelled Tricarboxylic Acid (TCA) cycle metabolites, especially fumarate and malate (Fig. 3a). Cells with such reduction in mitochondrial metabolism are often active in aerobic glycolysis increasing lactate production 39 . In line with this, UK-5099 treatment increased the NAD+/NADH ratio and lactate secretion (Fig. 3b, ED Fig.7b). In contrast, Pop2 cells surprisingly produced significantly lower levels of intracellular and secreted lactate than Pop1 cells (Fig. 3a,c). Taken together, UK-5099 treatment partially phenocopied the stem cell traits of Pop2 cells (Fig. 2f), but did not fully recapitulate the metabolic features of Pop2 cells. Consequently, our data suggested that stem-like Pop2 cells may use glycolysis for metabolite supply rather than for ATP-production.
Figure 3. Inherited metabolic cell fate bias is dependent on the pentose phosphate pathway.
a-b LC-MS/MS analysis of TCA cycle (1st turn) and glycolysis metabolites tracing from a one or two-hour pulse of U-13C glucose. Data shown as log2 metabolite levels (peak area/relative cell number) in a, Pop2/Pop1 cells (four biological repeats per time point) from the same experiment. Green lines indicate mean, b, cells treated with 10 μM UK-5099 for 24h (three biological replicates) relative to the mean of DMSO treated samples (four biological replicates). Black lines indicate mean. Empty circles denote metabolites undetected or below background level. Schematics show experimental strategy and 13C-labelling patterns in the TCA cycle and in glycolysis. c, Cumulative lactate production during the first 20 hours after FACS-sorting. Data are mean ± s.d. of six biological replicates. d, IMP M+5 relative to Ru5P M+5 derived from the pentose phosphate pathway in Pop1 and Pop2 cells. Data are mean ± s.d of eight biological replicates. e, LC-MS/MS analysis of M+1 and M+2 lactate in media secreted from Pop1 and Pop2 cells treated with a one-hour pulse of 1,2-13C glucose after FACS-sorting. Ratio, mean ± s.d., of M+1 to M+2 isotopomers derived via the pentose phosphate pathway (M+1) or directly from glycolysis (M+2) in six biological replicates. f, Mammosphere forming capacity of cells pre-treated with a five-hour pulse of PPP inhibitor 10 μM (6-AN) or DMSO. Data are mean ± s.d. of four biological replicates. g, Reconstitution efficiency (left) of the mammary epithelium in cleared fat pads transplanted with MMECs pre-treated with 500 μM 6-AN or DMSO for 24 hours before transplantation. Bar graphs show reconstitution efficiency as percentage of glands reaching 25, 50, 75, or 100 percent reconstitution. n shows the number of successful transplants out of 14 mice transplanted with DMSO- and 6-AN-treated cells to contralateral sides: 5/14 and 8/14, not significantly different. Whole-mounts of carmine-alumn stained mammary glands (right) with 100 (DMSO) and 25 (6-AN) percent reconstitution. Scale bar is 5mm. Statistical testing details in Supplementary Data Table 1. Numerical source data are available in Source Data Table 3.
Given the unique metabolic features of Pop1 and Pop2 cells, we asked if these cells invest carbon differently into selected metabolic pathways. Here, we used labelling ratios of key metabolites 16,40,41 as a comparable readout of pathway activity between the cell populations (ED Fig. 7c). In this analysis, the most striking difference was a significantly higher ratio between M+5 labelled inosine monophosphate (IMP) and Ribulose-5-phosphate (Ru5P) in Pop2 cells, representing shunting of carbons from the oxidative branch of the pentose phosphate pathway (PPP) to de novo purine biosynthesis (Fig. 3d). As expected, 6-phospho-gluconate levels were elevated and subsequently, the Ru5P M+5/6PG M+6- and IMP M+5/Ru5P M+5-ratios were reduced by inhibiting the key PPP enzyme 6-phospho-gluconate dehydrogenase with 6-aminonicotinamide (6-AN) 42 (ED Fig. 7d). Further validating the higher relative PPP activity in Pop2 cells, isotope tracing from 1,2-13C glucose immediately after division showed that lactate secreted by Pop2 cells had a significantly higher M+1/M+2 isotopomer ratio 43,44 than lactate from Pop1 cells (Fig. 3e).
The immediate presence of metabolic differences after asymmetric cell division implied that the metabolic features associated with daughter cell fate existed already upon cytokinesis. Metabolic bias imposed by age-selectively inherited mitochondria may therefore be one of the chronologically first determinants of cell fate after division. To test this, we assessed the role of high PPP activity in maintaining stemness of Pop2 cells immediately after cell division. We restricted 6-AN treatment to a five-hour window following synchronised cell division, and analysed effects on stem-like growth 10-12 days later. Strikingly, the transient and partial PPP inhibition with a low dose of 6-AN (ED Fig. 8a) reduced mammosphere formation selectively in Pop2 cells without effects on cell survival or oxidative or glycolytic metabolism (Fig. 3f, ED Fig. 8b, c). Similarly, inhibition of the first step of glycolysis, also required for PPP activity, by 2-Deoxy-D-Glucose (2-DG) at a dose that had no effect on cell survival, or glycolytic metabolism (ED Fig. 8d-f), reduced Pop2 cell mammosphere formation to a level comparable with Pop1 cells (ED Fig. 9a, b).
Protein levels of key PPP enzymes were not different between Pop1 and Pop2 cells (ED Fig. 9c) and overexpression of G6PD did not increase stemness of Pop1 cells (ED Fig. 9d), supporting the primary role of mitochondrial segregation in driving the metabolic divergence. In this light, the shunt to purine synthesis found in Pop2 cells was interesting, as de novo purine synthesis is tightly linked with mitochondria via the folate pathway providing crucial cytoplasmic one-carbon units 45 . Inhibition of dihydrofolate reductase (DHFR) by methotrexate (Mtx) resulted in reduced stemness selectively of Pop2 cells (ED Fig. 9e), suggesting that de novo purine synthesis is indeed another stemness-supporting feature of Pop2 cells and possibly links the mitochondria-driven metabolic bias after asymmetric cell division with PPP activity.
Unperturbed PPP activity was a requirement for HMECs to maintain stem-like properties after asymmetric division. To address the role of the PPP in mammary epithelial cell stemness in a more physiological setting, we assessed the effect of 6-AN-induced PPP disturbance on stemness in primary murine mammary epithelial cells (MMECs). We extended the pulse treatment to 24 hours due to the non-synchronous cell cycle of isolated MMECs. Similarly to non-synchronised bulk HMECs (ED Fig. 9f, g), isolated MMECs pre-treated in 2D culture with 6-AN formed fewer mammospheres than control cells, while cell viability was not affected (ED Fig. 9h). We transplanted MMECs pre-treated with 6-AN or DMSO into cleared mammary fat pads and assessed their ability to reconstitute the mammary gland in vivo. 6-AN significantly reduced reconstitution efficiency, with only 1/8 successful transplants yielding a fully reconstituted gland, whereas DMSO-treated cells reconstituted the gland fully in 4/5 successful transplants (Fig. 3g), further emphasizing metabolism and the PPP upstream of cell fate decisions.
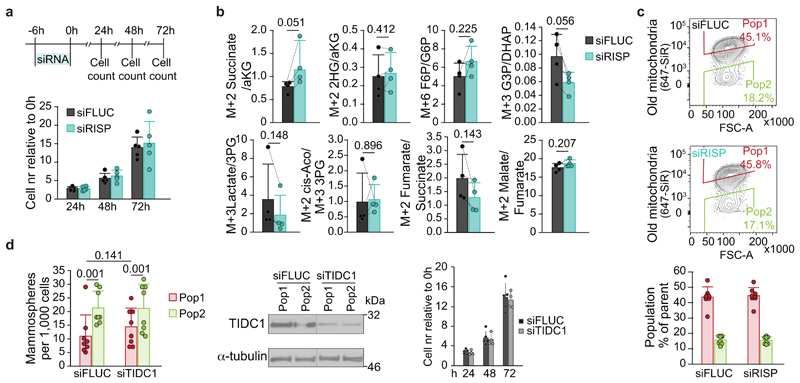
Our data indicated that selectively inherited mitochondria impose a metabolic bias on progeny cells and acts as a cue for cell fate determination. To directly address whether differences between the mitochondrial age-classes are sufficient to induce this divergence of metabolism and cell fate, we returned to the proteomic analysis of new and old mitochondria (Fig. 1). We focused on ETC subunits, as many were significantly enriched in old mitochondria, and high mitochondrial respiration entailed lower mammosphere forming capacity (Fig. 2). Among the candidates, complex III subunit RISP was particularly interesting, as it is essential for hematopoietic stem cell (HSC) function by balancing self-renewal and differentiation 16 . We first analysed RISP localization in old and new domains of the mitochondrial network. Corroborating our proteomic analysis (Fig. 1b), RISP was localized preferentially to old mitochondrial domains (Fig. 4a, b). Next, we reduced RISP expression moderately with siRNA (Fig. 4c), mimicking the RISP protein level difference observed between old and new mitochondria (Fig.1b). Partial depletion of RISP did not affect cell proliferation (ED Fig. 10a) in contrast to the complete knockout of RISP in HSCs reported previously 16 . As expected, RISP depletion resulted in reduction of respiratory capacity with the most pronounced effect on spare respiratory capacity, indicating that RISP levels are rate-limiting for respiration (Fig. 4d). Similarly to Pop2 cells with fewer old mitochondria, the reduced respiration was coupled with a modest decrease in 13C-labelled TCA cycle metabolites shortly after cell division (Fig. 4e). RISP depletion also led to slightly increased M+2 succinate/aKG ratio (ED Fig. 10b), consistent with a previous study 16 but not M+2 2HG/aKG ratio, likely due to partial RISP-depletion mimicking the difference between mitochondrial age-classes. Interestingly, partial RISP-depletion largely phenocopied the metabolic status of Pop2 cells (Fig. 3a, ED Fig. 7c), resulting in decreased lactate M+3 levels and altered ratios of 13C-labelled metabolites such as M+3 G3P/DHAP (Fig. 4e, ED Fig. 10b). Importantly, the M+5 IMP/Ru5P ratio, indicative of PPP activity towards de-novo purine synthesis, was significantly increased upon RISP depletion (Fig. 4e). Together, these results indicate that partial RISP depletion induced a Pop2-like metabolic phenotype in HMECs, suggesting that low RISP levels support stemness. Consistent with this notion, RISP knockdown increased mammosphere formation capacity especially in Pop1 cells (Fig. 4f, ED Fig. 10c). Highlighting the specific role of RISP in maintaining Pop1 cell identity, knockdown of the complex I assembly factor TIDC1 (ED Fig. 2f) did not affect Pop1 mammosphere formation (ED Fig. 10d). Taken together, our data confirmed that cell fate decisions can be regulated by qualitative differences in the mitochondrial proteome, which can emerge through selective inheritance of mitochondrial age classes during asymmetric cell divisions.
Figure 4. RISP is required for oxidative metabolism and reduced stemness of cells inheriting more old mitochondria.
a, Microscopy images show RISP immunofluorescent staining in mitochondrial domains enriched for old or new label. A representative area of a confocal image (maximum intensity projection) is shown. Scale bar, four left panels 5 μm, four right panels 2 μm. b, Quantitation of immunofluorescent staining for QCR1 and RISP in mitochondrial domains enriched for old or new label within intact cells. Pooled analysis of segmented mitochondrial domains from 4 cells each, comparing relative staining intensity in mitochondria enriched for new (below first quartile of old/new label ratio) and old label (above third quartile). c, Western blot showing RISP expression in Pop1 and Pop2 cells, one out of eight biological replicates presented also in f. Cells treated with the indicated siRNAs two days prior to FACS-sorting by inheritance of old mitochondria according to the schematic. d, Oxygen consumption rate (OCR) relative to cell number for cells treated with the indicated siRNAs. Representative OCR curves from one experiment (siFLUC: six, siRISP: five repeats), and spare respiratory capacity, pmol/min/relative cell number, (siFLUC: 25 repeats, siRISP: 28 repeats from four experiments). Data are mean ± s.d.. e, LC-MS/MS analysis of TCA cycle (1st turn) and glycolysis metabolites tracing from a two-hour pulse of U-13C glucose in cells transfected with the indicated siRNAs. Data shown as log2 metabolite levels (left) in siRISP cells relative to siFLUC cells from the same experiment. Blue lines indicate mean of four biological replicates. IMP M+5 relative to Ru5P M+5 (right) derived from the pentose phosphate pathway in cells transfected with siRNA for RISP or FLUC. Data are mean ± s.d. of four experiments. f, Mammosphere forming capacity of Pop1 and Pop2 cells treated with indicated siRNAs two days prior to FACS-sorting by inheritance of old mitochondria according to the schematic on top. Data are mean ± s.d. of eight biological replicates. Details of statistical analysis in Supplementary Data Table 1. Numerical source data are available in Source Data Table 4 and unprocessed blots are available in Source Data Figure 4.
Our data indicate that modulation of metabolism can bias fate of adult stem cells and alter the balance between stem cell self-renewal and differentiation. We propose that selectively and asymmetrically inherited mitochondria impose a metabolic bias on progeny cells, which acts as a cue at a branch point in cell fate determination after cell division. Metabolism of inherited mitochondria can act as a cell-intrinsic determinant of fate decisions shortly after cell division. This mechanism is in line with recent evidence on metabolism driving cell fate and differentiation 12–14 , rather than being a consequence or precondition for either maintenance of stemness or differentiation. Further studies are needed to assess whether fate determination through inherited metabolic bias has a role in other stem cell populations. While proliferating stem cells are highly dependent on the supply of metabolic intermediates for biosynthetic reactions, also quiescent stem cells in different tissues undergo a metabolic switch upon activation 4 . Considering recent findings on the role of mitochondria and mitochondrial metabolism in stem cell expansion 1,3,5,7,8,12–17 , it will be interesting to determine whether selective segregation of mitochondria balances regeneration and maintenance of stem cell pools in vivo.
Methods
Ethical considerations and animals
Animal studies were approved by the National Animal Ethics Committee of Finland (ELLA) and conducted with the support of HiLIFE Laboratory Animal Centre Core Facility, University of Helsinki, Finland, under institutional guidelines. The licencse numbers covering all related work: ESAVI-18179-2020, ESAVI-7011-2019. Female C57BL6/Rcc mice were obtained from Envigo. Mice were housed in germ-free conditions, with Tapvei aspen bedding and nesting material. The light/dark cycle was 12 hours, and the temperature maintained at 21-22°C and humidity at 40-60%. The mice had ad libitum access to food (Teklad 2916) and water.
Cell culture
Human mammary epithelial cell (HMEC) line FL2, stem-like cells (SLCs) identified in cultures of immortalized human mammary epithelial cells 19 expressing Snap-tag on the outer mitochondrial membrane (Snap-Omp25 21 ) was maintained in MEGM (Lonza, CC-3153) or phenol-red free media base (Lonza, CC-3150) for imaging.
Snap labelling and cell synchronisation
Cells were synchronised with a double thymidine block 21 . After incubation in 2 mM thymidine (Sigma-Aldrich, T1895) for 19h, PBS washes, collection using trypsin-EDTA (Thermo Fisher Scientific, 15400054), and centrifugation, cells were treated with Snap-Cell substrates to label old mitochondria, washed, and plated on cell culture dishes, or poly-L-lysine (Merck Millipore, A-005-C, final concentration 1 μg/ml) coated 35 mm glass bottom dishes (MatTek, P35G-1.5-14-C). After 10 hours, thymidine was added for 17 hours, cells washed and returned to MEGM. Cells divided 21-26h after the second thymidine release 21 .
Snap-Cell Block or Snap-Cell substrates (New England Biolabs) were applied for 30min at 37°C as indicated, diluted in full MEGM with 0.5% BSA (Thermo Fisher Scientific, 15260037) at 5-10μM (Snap-Cell Block S9106S), 1.5-3μM (Snap-Cell 647-SiR S9102S), 3μM (Snap-Cell TMR-Star S9105S), 5μM (Snap-Cell 430 S9109S), 5μM (Snap-Cell Oregon Green S9104S).
Live imaging
Cells were maintained at 37°C and 5% CO2 (see Supplementary Data Table 2). Live cell Airyscan superresolution microscopy used a Zeiss LSM 880 confocal microscope with Airyscan detector and 63× Plan-Apochromat oil objective, NA 1.4. Sequential excitation of fluorophores was used, with 488nm laser line for Snap-Cell Oregon Green and MitoSOX, and 633nm laser for Snap-Cell 647-SiR, with appropriate dual bandpass filters. The Airyscan detector was adjusted regularly, and images were Airyscan processed in 2D using the Zeiss Zen 2.3 software package (Carl Zeiss Microscopy).
For spinning disk confocal imaging, cells were imaged on a 3i Marianas platform with a Zeiss Axio Observer Z1, Optovar 1×, 1.6× and 2.5×, with a Yokogawa CSU-X1 M1 spinning disk, Sutter LB-10W fast 10-position filter wheel with filters: 445/45, 525/30, 617/73, and Andor Neo sCMOS camera using a 63× Alpha Plan-Apochromat oil objective, NA 1.46 (Carl Zeiss Microscopy), using Slidebook 5.5 acquisition software (3i) and lasers: Violet (solid state 405nm/100mW), Blue (solid state 488nm/150mW), and Lime (solid state 561nm/50mW).
An inverted Nikon widefield system with Andor Zyla 4.2+ camera, external filter wheel and SpectraX LED was used for widefield imaging. Images were acquired using a 60× water objective, NA 1.20, and NIS-Elements 4.5 software.
FACS-sorting of cells
HMECs were synchronised and Snap-labelled as described above. Cells were collected and sorted on a BD FACSAria II or FACSAria fusion sorter (lasers: near UV 375nm, blue 488nm, red 633nm; or violet 405nm, blue 488nm, yellow-green 561nm, red 633nm) with an 85μm or 100μm nozzle (FSC 1.0x or 1.5x ND filter) using BD FACSDiva software (versions 7 and 8). Snap-Cell Oregon Green was detected in the GFP/FITC channel, Snap-Cell 647-SiR in the APC/A-647 channel. FlowJo V10 analysis software (FlowJo LLC) was used for further data analysis. Plots are presented as contour plots (5 % probability contouring threshold, with outliers).
Pharmacological treatments and transfections
For pharmacological treatments, cells were synchronised and Snap-labelled as described above. After the second thymidine release, or 21h later, pharmacological treatments: 10μM, 50μM, or 1mM 2-Deoxy-D-glucose (2-DG, Sigma-Aldrich, D6134), 10μM 6-Aminonicotinamide (6-AN, Cayman Chemical Company, 10009315); 10μM UK-5099 (R&D Systems, 4186); 400μM dimethyl 2-oxoglutarate (2-Oxo, Sigma-Aldrich, 349631), 300 nM Heptelidic acid (AdipoGen AGCN20118C250), 100 μM Nicotinamide riboside (NR, Cayman Chemical Company, 23132), 250nM MitoTEMPO (Sigma-Aldrich, SML0737) or 10nM Methotrexate (Mtx, Abcam, ab142445) were added, and maintained until cells were detached for FACS-sorting 26h after release. Control samples were untreated, or supplemented with dimethyl sulfoxate (DMSO, 1:1,000, Sigma-Aldrich, D2650).
For transfections, cells were synchronised and Snap-labelled as described above. After the second thymidine release, cells were transfected in antibiotic-free MEGM with FuGene 6 Transfection Reagent (Promega, E2691) according to manufacturer’s instructions for 6h and plasmids at a total concentration of 1.9 μg/ml, or after labelling of old mitochondria, cells were plated in antibiotic-free MEGM and transfected with DharmaFECT1 Transfection Reagent (PerkinElmer) according to manufacturer’s instructions for 6h and siRNAs at a final concentration of 18nM. Plasmids: pYFP_C1, FLAG-HA-pcDNA3.1-(Addgene #52535 1:5), HA-HIF1alpha-pcDNA3 (Addgene #18949), G6PD/pRK5 (Addgene #41521) and pRK5-control (modified G6PD/pRK5). SiRNAs: MISSION esiRNA targeting FLUC (Sigma-Aldrich, EHUFLUC), siGENOME Human UQCRFS1 (RISP) siRNA-SMARTpool (PerkinElmer, 7386), MISSION esiRNA targeting human TIMMDC1 (Sigma-Aldrich EHU129821), MISSION esiRNA targeting human PFKL (Sigma-Aldrich, EHU05887), MISSION esiRNA targeting human GPI (Sigma-Aldrich EHU078901), MISSION esiRNA targeting human GAPDH (Sigma-Aldrich EHU146741).
Mammosphere culture
Cells were sorted into MEGM and plated into mammosphere culture in MEGM media containing 1% methylcellulose (15 cP, Sigma-Aldrich, M7027) on 96-well Ultra Low Attachment Microplates (Corning, 3474) at 250 or 500 cells per well, 4-8 technical replicates per sample. Mammospheres were counted 10-12 days later.
For analysis of mammosphere formation in bulk HMECs, cells were pre-treated for 24h in 2D culture before collection, and plated into mammosphere culture at 300 cells per well, 4-8 replicates per group. Mammospheres were counted 10-12 days later.
For pre-treatment with 2-DG, cells were maintained in media supplemented with 2-DG (Sigma-Aldrich, D6134) for at least 5 days before start of synchronisation, and until FACS-sorting. Before FACS-sorting, cells were labelled with 100nM MitoTracker Deep Red FM (Thermo Fisher Scientific, M22426), diluted in MEGM, for 15 minutes.
Mitochondrial isolation
Mitochondria were isolated using the Focus SubCell kit (G-Biosciences, 786-260). After centrifugation, cells were resuspended in cold Buffer I (provided with the kit). Cells were lysed using a Potter-Elvehjem homogenizer (Sigma-Aldrich, P7734), Buffer II added, and nuclei and intact cells were pelleted at 700g for 10 minutes. The supernatant was then centrifuged again at 12,000g for 15 min, pelleted mitochondria resuspended in Mitochondrial Storage Buffer (MSB) supplied with the kit and kept on ice.
Single-organelle FACS-sorting
For separation of mitochondrial age-classes, mitochondrial fractions were sorted on a BD FACSAria II sorter (lasers: near UV 375nm, blue 488 nm, red 633nm; or violet 405nm, blue 488nm, yellow-green 561nm, red 633 nm) with a 70 μm nozzle (FSC 0.5× or 1.0× ND filter). Snap-Cell Oregon Green was detected in the GFP/FITC channel, and Snap-Cell 647-SiR in the APC/A-647-channel, using BD FACSDiva software versions 7 and 8. FlowJo V10 analysis software (FlowJo LLC) was used for data analysis. Plots shown are contour plots (5 % probability contouring threshold, with outliers).
Mitochondria were FACS-sorted and pelleted at 21,000g for 20 min. Supernatant was removed and mitochondrial pellets were snap-frozen in liquid nitrogen or on dry ice, or lysed for Western blotting.
To determine gating, separate samples of isolated mitochondria were incubated on ice for 30 min with MitoTracker Deep Red FM (40nM in MSB) before analysis on a BD FACSAria II sorter (lasers: blue 488nm, red 633nm, near UV 375nm), using the APC-channel.
Flow cytometric analysis of isolated mitochondria
Isolated mitochondria were supplemented with tetramethylrhodamine methyl ester (TMRM; Thermo Fisher Scientific, T668, 2nM in MSB) for 10min on ice before analysis on a BD LSRFortessa with BD FACSDiva software and violet 405nm, blue 488nm, yellow-green561nm and red 640nm lasers. Snap-Cell Oregon Green was detected in the GFP/FITC channel, Snap-Cell 647-SiR in the APC/A-647-channel, and TMRM in the PE-channel. FlowJo V10 analysis software (FlowJo LLC) was used. Plots are contour plots (5 % probability contouring threshold, with outliers). Median TMRM fluorescence intensity, normalised to the parent population, was analysed.
Immunofluorescent staining of isolated mitochondria
Mitochondria sedimented on poly-L-lysine (Merck Millipore, A-005-C) coated cover glasses (VWR International, 631-0150) for 30min at 37°C were fixed with methanol for 8min at -20°C. Samples were blocked with 10% FBS (Thermo Fisher Scientific, 10270106) in PBS, incubated with anti-Snap tag antibody (New England Biolabs, P9310S, 1:400) in PBS with 1% BSA (Biowest, P6154) and 0.05% Tween-20 (Sigma-Aldrich) for 1h at room temperature (RT), washed and incubated with Alexa fluor goat anti-rabbit 594 secondary antibody (Thermo Fisher Scientific, A11012, 1:600) for 30min at RT. Cover glasses were washed, mounted with fluorescence mounting medium (Dako, S302380-2), and imaged on a Leica TCS SP5 confocal system with a Leica DM5000 with an HCX APO 63× glycerol objective, NA 1.3, using Leica Application Suite LAS X 2.7.3. See Supplementary Data Table 2.
Immunofluorescent staining of cells
Cells on poly-L-lysine (Merck Millipore, A-005-C) coated cover glasses (VWR International, 631-0150) were fixed in 2% PFA for 5min at +37°C. Samples were permeabilized in PBS with 0.1% Triton-X100, 1% BSA in for 10min at 4°C, blocked with 10% FBS (Thermo Fisher Scientific, 10270106) in PBS for 1 h at RT, incubated with primary antibody Anti-UQCRFS1 [EPR16288] (Abcam, ab191078, rabbit, 1:50) or anti-UQCRC1 (Abcam, ab125882, rabbit, 1:50), in 1% BSA (Biowest, P6154), 0.05 % Tween-20 (Sigma-Aldrich) in PBS for 1h at RT, washed, incubated with Alexa fluor 488 Goat anti-Rabbit IgG, Thermo Fisher Scientific, A-11008, 1:400 for 30min at RT, washed, and mounted with fluorescence mounting medium (Dako, S302380-2). For imaging settings, see Supplementary Data Table 2.
Mitochondrial domains were outlined manually based on Snap-labels. For analysis of fluorescence intensities using ImageJ 1.51 (NIH), confocal stacks for slices of 0.25μm were collapsed using maximum intensity projection. Mean fluorescence intensity was measured for each domain, and background was subtracted. Staining intensity was compared between mitochondrial domain, and background subtracted.
Proteomic analysis of old and new mitochondria
Sample preparation – In solution rapigest digestion and Oasis clean-up
Mitochondria were resuspended in 0.4% (w/v) RapiGest SF Surfactant (Waters, 186001861) in 5mM ammonium bicarbonate (Carl Roth) and heated at 95°C for 10min. After a short cool-down, 10M Urea (Sigma-Aldrich) in 250mM ammonium bicarbonate was added to 4M Urea, and lysates sonicated in a Bioruptor Plus (Diagenode, 10 cycles with 1min on, 30s off with high intensity at 20°C). Lysates were clarified by centrifugation for 5min at 14,000g, reduced by incubating at 37°C for 30min with 10mM DTT (Dithiothreitol; Carl Roth), and cysteines alkylated with 15mM iodacetamide (Sigma-Aldrich, I1149) for 30min in the dark. Proteins were digested for 4h at 37°C using 1:100 (w/w) LysC (Wako Chemicals, 125-05061), samples diluted to 1.5M Urea with HPLC grade water (Sigma-Aldrich, 270733), and digested with 1:100 (w/w) trypsin (Promega Biotech, V5111) for 16h at 37°C.
Solutions were acidified with 10% trifluoroacetic acid (Biosolve, 20234131) and desalted with Oasis HLB μElution Plate 30μm (Waters, 186001828BA) in the presence of a slow vacuum. The columns were conditioned with 3x100μl solvent B (80% acetonitrile; 0.05% formic acid; Sigma Aldrich, 34998; Biosolve, 6914143) and equilibrated with 3x100 μl solvent A (0.05% formic acid in HPLC grade water). Samples were loaded, washed 3 times with 100μl solvent A, and eluted into PCR tubes with 50μl solvent B. Eluates were dried in a speed vacuum centrifuge (Eppendorf) and dissolved in 5% acetonitrile, 95% HPLC grade water, with 0.1% formic acid to a peptide concentration of 1μg/μl before analysis by LC-MS/MS.
LC-MS/MS
Peptides were separated using the nanoAcquity UPLC system (Waters) with trapping (nanoAcquity Symmetry C18, 5μm, 180μm x 20mm) and an analytical column (nanoAcquity BEH C18, 1.7μm, 75μm x 250mm). The outlet of the analytical column was coupled to an Orbitrap Fusion Lumos (Thermo Fisher Scientific) using the Proxeon nanospray source. Solvent A was water, 0.1% formic acid and solvent B was acetonitrile, 0.1% formic acid. The samples (500ng) were loaded with a constant flow of solvent A at 5μl/min onto the trapping column. Trapping time was 6min. Peptides were eluted via the analytical column with a constant flow of 0.3μl/min. After isocratic flow for 5min with 5% B, the elution step proceeded, with the percentage of solvent B increasing in a linear fashion from 5% to 22% in 105 min, then to 32% in 10 min and finally to 50% in a further 1min. Column clean-up to 85% B followed before re-equilibration to 5% B. Total runtime was 155min. The peptides were introduced into the mass spectrometer via a Pico-Tip Emitter 360μm OD x 20μm ID; 10μm tip (New Objective) and a spray voltage of 2.2kV was applied. The capillary temperature was set at 300°C. The RF lens was set to 30. Full scan MS spectra with mass range 375-1500m/z (using quadrupole isolation) were acquired in profile mode in the Orbitrap with resolution of 60,000. The filling time was set at maximum of 50ms with limitation of 2x 105 ions. The “Top Speed” method was employed to take the maximum number of precursor ions (with an intensity threshold of 5x 104) from the full scan MS for fragmentation (using HCD collision energy, 30%) and quadrupole isolation (1.4Da window) and measurement in the Orbitrap, with a cycle time of 3s. The MIPS (monoisotopic precursor selection) peptide algorithm was employed. MS/MS data were acquired in centroid mode in the Orbitrap, with a resolution of 15,000 and a fixed first mass of 120m/z. The filling time was set at maximum of 22ms with limitation of 1x 105 ions. Only multiply charged (2+ - 7+) precursor ions were selected for MS/MS. Dynamic exclusion was employed with maximum retention period of 15s and relative mass window of 10ppm. Isotopes were excluded. In order to improve the mass accuracy, a lock mass correction using a background ion (m/z 445.12003) was applied. For data acquisition and processing of the raw data, Xcalibur 4.0 (Thermo Fisher Scientific) and Tune version 2.0 were used.
Data analysis
Raw files were searched using the Andromeda search engine 46 included in MaxQuant (version 1.5.3.28) 47 . The data were searched against a human database (Swiss-Prot entries of the Uniprot KB database release 2016_01, 20198 entries) with a list of common contaminants appended. The data were searched with the following modifications: Carbamidomethyl (C) (Fixed) and Oxidation (M) and Acetyl (Protein N-term) (variable). The mass error tolerance for the full scan MS spectra was set at 20ppm and for the MS/MS spectra at 0.5Da. A maximum of 2 missed cleavages were allowed. Peptide and protein level 1% FDR were applied using a target-decoy strategy 48 . Only proteins identified by at least two unique peptides were retained for further analysis. LFQ (label free quantification) intensity values were exported from the MaxQuant output table and used to perform quantitative analysis using in house written R (version 3.4.0) procedures. Data were processed using the R package MSnbase 49 and data imputation was performed using imputeLCMD. A mixed strategy based on the definition of Missing At Random (MAR) and Missing Not At Random (MNAR) values was used for data imputation. MNAR were defined as values that were (i) missing in 3 out of 3, or 2 out of 3 biological replicates in one sample group, and (ii) present in all the 3 biological replicates in the second sample group. Because of their non-random distribution across samples, these values were considered as underlying biological difference between sample groups. MNAR values were computed using the method “MinDet” by replacing values with minimal values observed in the sample. MAR were consequently defined as values that were missing in 1 out of 3 biological replicates per sample group. MAR values were imputed based on the method “knn” (k-nearest neighbors) 49 . All the other cases (e.g., protein groups that had less than 2 values in both sample groups) were filtered out. Following data imputation, data were quantile normalized and differential protein abundance was evaluated using the limma package 50 . False discovery rate was estimated using fdrtool 51 .
Mass spectrometry of lipids
Lysates of new and old mitochondria from 4 individual FACS sorts were pooled. Mitochondrial samples, MEGM media sample and whole cell sample were frozen on dry ice, lipids extracted according to the method of Folch 52 and stored at -80°C. The solvent was evaporated, lipid residue dissolved in methanol/chloroform (2:1, v/v), spiked with a cocktail of 13 internal standards (including representatives for each PL class detected), and supplemented with 1% NH4OH before direct infusion of the sample solution into the ESI source of a triple quadrupole mass spectrometer, Agilent 6490 Triple Quad LC/MS (with iFunnel technology; Agilent Technologies). In addition to positive and negative ion mode MS detections, PL class specific MS/MS detection modes were used with information on equipment and ion source settings 53 . Spectra were processed by MassHunter Workstation Qualitative Analysis B.06.00 software (Agilent Technologies) and the concentrations of individual PL species quantified using internal standards and software Lipid Mass Spectrum Analysis (LIMSA, v.1.0) 54 . Concentration data were converted to PL class composition (as molar %).
Gas chromatography of fatty acids
Fatty acid compositions of cell culture medium, cells and mitochondria were studied by gas chromatography after conversion of lipid acyl chains and free fatty acids to methyl ester derivatives. Fatty acid methyl esters were quantified by using FID detection (Shimadzu GC-2010 Plus, the FID response of which has a large linear range) and identified by using MS detection (Shimadzu GCMS-QP2010 Ultra, recording electron impact mass spectra) 53 .
Western blotting
Samples were lysed in radioimmunoprecipitation assay (RIPA) buffer (150mM NaCl, 20mM Tris pH 7.5, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, 1% Triton X-100) with protease inhibitor (Abcam, ab201116 or Thermo Fisher Scientific, 87786), supernatant processed after centrifugation at 12,000g for 5min. 250,000 - 1,000,000 sorted mitochondria or 50,000 cells were loaded per lane.
For the Bolt system (Thermo Fisher Scientific), samples were denatured with Bolt LDS Sample Buffer (B0007), Sample Reducing Agent (B0009), and heated to 70°C for 10min. Samples were resolved on 4-12% Bis-Tris Plus Gels (BG04120BOX) and transferred to 0.2μM nitrocellulose membranes (Bio-Rad, 1620112) using Bolt MES SDS Running Buffer (Thermo Fisher Scientific, B0002) and Transfer Buffer (BT00061). For the Bio-Rad Mini-PROTEAN II system, samples were heated to 95°C for 10min in Laemmli buffer (62.5mM Tris-HCl, 2% SDS, 20% (v/v) glycerol, 0.01% bromophenol blue (Sigma-Aldrich)), separated on 4-20% Tris-Glycine Gels (Bio-Rad, 4561096) in Tris/ Glycine Buffer (Bio-Rad, 1610734) with 0.1% SDS, and transferred to 0.2μM nitrocellulose membranes (Bio-Rad 1620112) using Tris-Glycine buffer (Bio-Rad, 1610734) with 20% methanol and 0.05% SDS.
Membranes were incubated with blocking buffer (0.1% BSA, 5% milk, 0.1% Tween-20 in TBS) for 1h at RT, followed by primary antibodies Anti-UQCRFS1 [EPR16288] (Abcam, ab191078, rabbit, 1:1,000), anti-UQCRC1 (Abcam, ab125882, rabbit, 1:1,000), anti-TIMMDC1 [EPR11808] (Abcam, ab171949, rabbit, 1:700), anti-OPA1 (Thermo Fisher Scientific, MA5-16149, mouse, 1:700), anti-α-tubulin (Cell Signaling Technology, DM1A, 1:2,000), anti-G6PDH (Abcam, ab133525, 1:2,000), anti-PGD (Abcam, ab129199, 1:2,000), anti-Transaldolase (Abcam, ab137629, 1:1,000), anti-PGC1α (Sigma-Aldrich, T1202, 1:500), anti-HSP60 (Santa Cruz, sc-1052, 1:1,000), anti-MTCO2 (COXII, Gene Tex EPR3314/Abcam ab79393, 1:500), anti-GAPDH (Cell Signal, 14C10, 1:1,000), anti-Vinculin (Sigma-Aldrich, V9264, 1:10,000), anti-β-actin (Abcam, ab8227, rabbit, 1:4,000) in blocking solution, at 4 °C overnight, washed in TBS-T (0.05% Tween-20 in TBS), and incubated with appropriate HRP-conjugated secondary antibodies (anti-mouse IgG HRP-linked antibody, Cell Signaling Technology, 7076S, 1:1,000; or anti-rabbit IgG HRP-linked antibody, Sigma-Aldrich, A0545, 1:1,000-3,000 or anti-goat IgG HRP-linked antibody, Agilent, P044901, 1:1,000-3,000), in blocking solution, for 1h at RT, washed in TBS-T and detected using Thermo Fisher Scientific Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific, 10005943) and Fujifilm Super RX films or a Molecular Imager ChemiDoc XRS+ with ImageLab (Bio-Rad).
MitoSOX imaging
For analysis of MitoSOX fluorescence intensity in mitochondrial domains, cells were plated on MatTek glass bottom dishes. After Snap-labelling, samples were loaded with MitoSOX (Thermo Fisher Scientific, M36008, 5μM in PBS or phenol red free MEGM) 15min before start of imaging on a 3i Marianas platform (see above).
Images were deconvolved using AutoQuant X3 software (Bitplane) with the settings: theoretical PSF, Medium Noise Level (Noise Value 20), 10 iterations. ImageJ 1.51 (NIH) was used for image analysis. In maximum intensity projections of six confocal planes close to the attachment surface of the cells, domains of the mitochondrial network were outlined manually using Snap-labels. Mean fluorescence intensities for Snap-labels and TMRM were measured, and background (mean intensity outside of the cell in the same image) subtracted. Old-to-new-label ratio for individual mitochondrial domains was calculated and divided by the average ratio for each cell for normalisation. Mitochondria were grouped by enrichment of old and new labels (enriched for new label: old-to-new label ratio smaller than first quartile of all mitochondrial domains analysed; intermediate ratio: between first and third quartile; enriched for old label: higher than third quartile). Relative MitoSOX fluorescence intensity was calculated for individual mitochondrial domains by dividing background corrected fluorescence intensity by average intensity per cell.
For analysis of MitoSOX in cells, synchronised HMECs were Snap-labelled, and 21h after release from the second thymidine, loaded with MitoSOX (Thermo Fisher Scientific, M36008, 1-5μM final concentration in PBS or phenol red free MEGM), and cell division pairs were imaged starting 15min later on a 3i Marianas platform or an inverted Nikon widefield system (see above).
Cell pairs were identified and cells outlined manually in bright-field images. For analysis of fluorescence intensities using ImageJ 1.51 (NIH), confocal stacks of entire cells were collapsed using maximum intensity projection. Integrated density (IntDen) was measured for each cell. The corrected total cell fluorescence (CTCF) value was calculated as IntDen-(cell area×mean background fluorescence intensity). For each channel, the sum of CTCF values per cell pair was calculated. Relative fluorescence intensities for each cell are expressed as percentage of the sum for each division pair. Additional images were acquired with airyscan superresolution microscopy (described above, see Supplementary Data Table 2).
Immunofluorescent detection of protein carbonylation
The OxyBlot Protein Oxidation Detection Kit (Merck Millipore, S7150) was adapted for immunofluorescence 36,55 . Synchronised and Snap-labelled HMECs on glass bottom dishes, treated with 100nM antimycin A (Sigma-Aldrich, A8674) as indicated, were fixed with 95% ethanol for 13min at -20°C, and permeabilised with 0.25% Triton X-100 (Sigma-Aldrich) in PBS for 20min at RT. Carbonyl groups were derivatised to 2,4-dinitrophenylhydrazone with 2,4-dinitrophenylhydrazine solution (OxyBlot kit) for 15 min at RT, or treated with derivatisation control solution. Neutralisation solution was added and non-specific antigens were blocked with 10% FBS (Thermo Fisher Scientific, 10270106) in PBS for 1h at RT. Samples were incubated with primary antibody against the derivatized DNP moiety (Rabbit anti-DNP, 1:80, OxyBlot kit), in PBS with 1% BSA (Biowest, P6154) and 0.05% Tween-20 (Sigma-Aldrich), or buffer for controls, for 1h at RT, washed with 0.05% Tween-20 in PBS, and incubatedwith secondary antibody (Alexa fluor 488 Goat anti-Rabbit IgG, Thermo Fisher Scientific, A-11008, 1:400) for 1h at RT. Samples were washed with PBS, and stored in PBS until imaging on a 3i Marianas platform (See section ”live imaging”, Supplementary Data Table 2).
ImageJ 1.51 (NIH) was used for image analysis. Confocal stacks of entire cells were collapsed using maximum intensity projection, cells were outlined manually in bright-field images, integrated density (IntDen) per cell was measured for each fluorescent channel, and the corrected total cell fluorescence (CTCF) calculated as described above, and OxyBlot CTCF values normalised to cell area. P1 progeny was defined as the cell with the higher CTCF for old mitochondrial label, P2 with lower CTCF.
Flow cytometric analysis of CellROX
Synchronised and Snap-labelled cells were loaded with 2.5μM CellROX Deep Red Reagent (Thermo Fisher Scientific, C10422) for 30min at 37°C, washed, trypsinised, collected, and analysed on a BD FACSAria II sorter (lasers: Near UV 375nm, Blue 488nm, Red 633nm; or Violet 405nm, Blue 488nm, Yellow-Green 561nm, Red 633nm) with a 85μm or 100μm nozzle (FSC 1.0× or 1.5× ND filter). Instruments were equipped with BD FACSDiva software, and FlowJo V10 analysis software (FlowJo LLC) was used for data analysis. Snap-Cell Oregon Green (old mitochondria) was detected in the GFP/FITC channel, CellROX in the A-647/APC channel.
Mitochondrial DNA copy number
Pop1 and Pop2 cells were sorted, pelleted and resuspended in lysis buffer for DNA extraction using Zymo Microprep DNA kit according to manufacturer’s instructions. Mitochondrial DNA copy number was determined by qPCR using two sets of primers to detect nuclear and mitochondrial genomes. Primers for mitochondrial Cytb gene were 5′-GCCTGCCTGATCCTCCAAAT-3′ and 5′-AAGGTAGCGGATGATTCAGCC-3′, and for nuclear APP gene 5′-TGTGTGCTCTCCCAGGTCTA-3′ and 5′-CAGTTCTGGATGGTCACTGG-3′. Targets were amplified in separate wells in duplicates using Power SYBR Green PCR master mix (Applied Biosystems). Mean values were used for analysis and results are shown as mtDNA/nDNA.
Seahorse XF Mito Stress Test
Mitochondrial activity was assessed using the Seahorse XF Mito Stress Test kit (Agilent Technologies, 103015-100) in a Seahorse XFe96 Flux Analyser with Seahorse Wave 2.4 software (Agilent Technologies). Synchronized and labelled cells were FACS-sorted into Seahorse assay medium (sodium bicarbonate-free DMEM with phenol red, 10mM glucose, 2mM glutamine, 0.11g/l sodium pyruvate; all from Sigma-Aldrich), plated on poly-L-lysine-coated (Merck Millipore, A-005-C) XFe96 well plates (Agilent Technologies, 102416-100) at 60,000 cells per well, and incubated in a non-CO2 incubator at 37 °C for 1h. Basal oxygen consumption (OCR) was recorded 3 times (3min recording and 3min mixing cycles), followed by sequential injections of 1μM oligomycin, 1μM FCCP and 0.5μM rotenone and antimycin A (Mito Stress Test kit). OCR was recorded in 3 cycles of 3min mixing followed by 3min recording after injection of each drug. For basal OCR, non-mitochondrial OCR (recorded after rotenone and antimycin A injection) was subtracted from the last OCR reading before oligomycin injection. For spare respiratory capacity, basal OCR was subtracted from maximal respiration (OCR after FCCP).
Bulk HMECs were plated on XFe96 well plates at 25,000 cells per well and cultured in MEGM overnight. Pharmacological treatments were added 22 or 5h before the assay, or cells were transfected with siRNAs 48h before the assay (see section “Pharmacological treatments and transfections”). Cells were trypsinised and replated on poly-L-lysine-coated (Merck Millipore, A-005-C) XFe96 well plates (Agilent Technologies, 102416-100), and analysed as described above. Before the assay, medium was changed to sodium bicarbonate-free DMEM-based assay medium with phenol red (10mM glucose, 2mM glutamine, 0.11g/l sodium pyruvate, all from Sigma-Aldrich), and treatments as indicated, and the cells were equilibrated for 30min in a non-CO2 incubator at 37°C.
Oxygen consumption rate data of siRNA-treated cells were corrected for cell numbers based on thresholded bright field images captured in the centre of each well before the assay in a Nikon ECLIPSE Ts2R-FL microscope with Infinity 3 Lumenera camera and 4x objective. Images within the same assay were batch-analysed using ImageJ 1.51 (NIH). After background correction, images were processed with the Auto-Local Threshold function (Bernsen method; radius 15), and watershed correction. Cells were counted in the same ROI using the analyze particles function with particle size >10μm.
Seahorse XF Glycolytic Rate Assay
For Seahorse XF Glycolytic Rate Assay (Agilent Technologies) cells were plated and treated as described for the Mito Stress Test. Medium was changed to a phenol-red and sodium bicarbonate-free assay medium (5mM HEPES (Agilent Technologies), 10mM glucose, 2mM glutamine, 0.11g/l sodium pyruvate (all from Sigma-Aldrich) and treatments as indicated). Cells were equilibrated for 30min in a non-CO2 incubator at 37°C. Extracellular acidification rate (ECAR) and OCR were measured in a Seahorse XFe96 Flux Analyser with Seahorse Wave 2.4 software in 3 cycles of 3min mixing and 3min recording sequential injection of 0.55μM rotenone (Sigma-Aldrich, 45656) and antimycin A (Sigma-Aldrich, A8674), followed by 6.25 mM2-DG (Sigma-Aldrich, D6134). Recordings were performed 3 and 5 times after the two injections, following the same mixing and measuring cycles. Glycolytic proton efflux rate (GlycoPER) was calculated using the Seahorse Glycolysis rate assay Report Generator (Agilent Technologies).
NAD+/NADH measurement
Total NAD+ and NADH were measured using the NAD+/NADH assay kit (Abcam, ab65348) according to manufacturer’s instructions. Cells were synchronised and Snap-labelled (see above). 400,000 FACS-sorted HMECs were collected in MEGM, pelleted, washed with cold PBS, pelleted and resuspended in 400μl extraction buffer (NAD+/NADH assay kit), with protease inhibitor (Abcam, ab201116). Pharmacologically treated cells were trypsinised, collected, washed and lysed with repeated freeze-thaw cycles on dry ice. Samples deproteinized using 10kDa spin columns (Abcam, ab93349) were divided into samples for measurement of total NAD+ and NADH (following decomposition of NAD+ at 60°C for 30min). Total amounts of NAD+ (NADt) and NADH were calculated based on absorbance at OD 450nm and NAD+/NADH ratio calculated as (NADt – NADH) / NADH.
Lactate measurement
HMECs were plated on 12-well plates (Sarstedt), treated as indicated, and media samples taken from the same wells at indicated time points after addition of treatments. Alternatively, cells were sorted into MEGM media and plated on 48-well plates (Sarstedt). Media samples for lactate measurement were taken at indicated time points. After centrifugation at 13,000g for 10min, lactate content in the supernatant was measured using the Lactate Assay Kit (Sigma-Aldrich, MAK064) according to manufacturer’s instructions. Calculations were based on absorbance measured at OD 570 nm.
Enzymatic assay of reduced and oxidized glutathione
Cells were synchronised, Snap-labelled and FACS-sorted as described above. Cells were resuspended in pre-warmed (45-60°C) extraction buffer containing 0.1N HCl and 0.1% Triton X-100, using 150μl extraction buffer per 400,000 cells, boiled at 100°C for 2min, cooled on ice for 2min and centrifuged at 14 000g for 15min at 4°C. For reduced glutathione measurement, 1 -methyl-4-vinyl-pyridinium trifluoromethane sulfonate (M4VP) was added to the supernatant in the presence of Na2CO3 56 . Glutathione (CS0260) dilutions in 0.1N HCl were used as standards. To 10μl of the sample or standard loaded onto 96 well-plate, 150μl of assay buffer (100mM PBS, 1mM EDTA, 0.04mg/ml DTNB, 0.2U/ml glutathione reductase (Sigma, G3664)) and 50μl of NADPH (Sigma, N7505, 33mg/ml in 100mM PBS with 1mM EDTA) were added, incubated at RT for 10min, and absorbance was measured at 412nm. Values were normalized to protein concentrations measured by BCA Protein Assay Kit (Thermo Fisher Scientific, 23225).
Metabolomics
For isotopic tracing experiments, HMECs were synchronised and Snap-labelled as described above. Dividing cells were treated with 16mM U-13C glucose (Cambridge Isotope Laboratories, CLM-1396-1) in MEGM for 1-2h before sorting. Sorted cells were washed in cold PBS and metabolites extracted in ice cold 80% acetonitrile and vortexed for 5s. Samples were centrifuged at 15,800g for10min at +4°C and supernatant stored in liquid nitrogen. Samples were analyzed on a Thermo Q Exactive Focus Quadrupole Orbitrap mass spectrometer coupled with a Thermo Dionex UltiMate 3000 HPLC system (Thermo Fisher Scientific, Inc.). The HPLC was equipped with a hydrophilic ZIC-pHILIC column (150× 2.1mm, 5μm) with a ZIC-pHILIC guard column (20× 2.1mm, 5μm, Merck Sequant). 5μl sample was injected into the LC-MS after quality controls in randomized order having every 10th samples as blank. A linear solvent gradient was applied in decreasing organic solvent (80–35%, 16min) at 0.15ml/min flow rate and 45°C column oven temperature. Mobile phases were: aqueous 200mmol/l ammonium bicarbonate solution (pH 9.3, adjusted with 25% ammonium hydroxide), 100% acetonitrile, 100% water. Ammonium bicarbonate solution was kept at 10% throughout the run resulting in steady 20mmol/l concentration. Metabolites were analyzed using a MS with a heated electrospray ionization (H-ESI) source using polarity switching and following settings: resolution of 70,000 at m/z of 200, spray voltages: 3400 V for positive and 3000 V for negative mode, sheath gas: 28 arbitrary units (AU), and auxiliary gas: 8AU, vaporizer temperature: 280°C, ion transfer tube temperature: 300°C. The instrument was controlled with the Xcalibur 4.1.31.9 software (Thermo Scientific). Metabolite peaks were confirmed using commercial standards (Sigma-Aldrich). Data quality was monitored throughout the run using inhouse quality control cell line extracted similarly to other samples. After final peak integration with TraceFinder 4.1 SP2 software (Thermo Scientific), peak area data was exported as excel files. Metabolite peak areas were corrected for background by subtracting the peak areas observed in an extraction buffer sample and the corrected values normalized to cell number.
For pharmacological treatments, HMECs were synchronised and supplemented with 10μM UK-5099 after second thymidine release or 10, 50 or 200μM 6-AN 21h after release, or DMSO as control, and with 16mM U-13C glucose for the last 2h before FACS-sorting 26h after release. Sorted cells were washed in cold PBS and metabolites extracted in 30μl ice cold 80% acetonitrile and processed for LC-MS/MS analysis as above.
For RISP depletion, HMECs were synchronised and transfected with siRNA targeting RISP or FLUC after release from the first thymidine, (see “Pharmacological treatments and transfections”) and treated with 16mM U-13C glucose for 2h before FACS-sorting at 26h after second thymidine release. Sorted cells were washed in cold PBS and metabolites extracted in 30μl ice cold 80% acetonitrile and processed for LC-MS/MS analysis as above.
For isotopic tracing from 1,2-13C glucose, HMECs were synchronized and Snap-labelled (see above), FACS-sorted at 23h after second thymidine release into MEGM, pelleted at 300g for 10min at RT, washed, pelleted and resuspended in assay medium (glucose-, pyruvate- and glutamine-free DMEM (Agilent, 103334-100) with 2 mM HEPES and Lonza bullet kit supplements (Lonza, CC-4136)) and 8mM 1,2-13C glucose (Cambridge Isotope Laboratories, CLM-504-0.25). Cells were seeded on poly-L-lysine-coated (Merck Millipore, A-005-C, 1μg/ml) 12-well plates in a total volume of 800μl. 20μl medium were collected 0 and 1h later in 480μl ice cold 80% acetonitrile buffer. Samples were centrifuged for 10min at 15,800g at +4°and stored at -80°C until analysis as above. All data were corrected for 13C natural abundance using the IsoCor method 57 and the ratio of M+1 to M+2 lactate was calculated from the corrected peak areas.
Population doubling dynamics
HMECs were plated on 24-well plates (Sarstedt) one day before the assay and treated as indicated. At 0h, cells were washed and returned to MEGM. Cells were counted in phase-contrast images of the same field of view acquired at 0h, 24h, 48h, and 72h using a Nikon Eclipse Ts2 microscope with Nikon CFI Ph1 ADL 10× objective, NA 0.25, using the ImageJ Cell Counter plugin (NIH). Cell numbers were normalized to 0h for each field of view. Where indicated, images were acquired and processed similarly after treatment addition at 0h.
For treatment with 1mM 2-DG, cells plated on a 24-well plate (Thermo Fisher Scientific) were mounted on a Cell-IQ cell culture platform equipped with Nikon CFI Plan Achromat 10× objective, NA 0.25 and Imagen Retiga EXi camera (Chip-Man Technologies), using Cell-IQ Imagen 4.1.0, and treatments added at the start of imaging.
Primary cell isolation
Primary mammary epithelial cells (MMECs) were isolated from 10-16 week old virgin female mice C57BL6/Rcc mice (obtained from Envigo and housed as described above) 58 . Mammary glands #3-#5 were dissected, the lymph node in #4 glands removed, and the glands finely chopped. Tissue was incubated with 0.01mg of Collagenase A (Sigma-Aldrich, C5138) per 1g of tissue in DMEM/F12 (Thermo Fisher Scientific, 12634010) containing 2.5% FCS (Thermo Fisher Scientific, 10270106), 5μg/ml insulin (Sigma-Aldrich, I-9278), 50μg/ml gentamycin (Duchefa Biochemie, G0124) and 2mM glutamine (Sigma-Aldrich, G8540) in an environmental shaker at 37°C at 120 rpm for 2-2.5h. The cell suspension was centrifuged at 400g for 10min and pulse centrifuged 3-5 times at 400g to obtain a preparation free of other cells than MMEC organoids. Organoids were trypsinised with 0.05% Trypsin-EDTA (BD Biosciences and Thermo Fisher Scientific) for 5-10min, drained through a 70μm cell strainer (BD Biosciences) and resuspended in MMEC growth media (DMEM/F12 media, Thermo Fisher Scientific, 12634010) containing 5μg/ml insulin (Sigma-Aldrich, I-9278), 1μg/ml hydrocortisone (Sigma-Aldrich, H-4001), 10ng/ml mouse EGF (R&D Systems, 2028-EG-200), 2mM glutamine (Sigma-Aldrich, G8540), 50μg/ml gentamycin (Duchefa Biochemie, G0124) and penicillin and streptomycin (Orion and Sigma-Aldrich) and 10% FCS (Thermo Fisher Scientific, 10270106).
Primary cell culture and treatments
MMECs were seeded on 12-well plates (Greiner Bio-One) precoated with 50 μg/ml of Laminin-111 (BioLamina, LN111-02) at 4°C overnight, and incubated for 24h in MMEC growth media. MMECs were treated with 500μM 6-AN (Cayman Chemical Company, 10009315), or DMSO (Sigma-Aldrich, D2650) for 24h and used to assess cell viability, or trypsinised, counted and plated in mammosphere culture or used in fat pad transplantations. Live and Dead Cell Assay (Abcam, ab115347) was used for cell viability studies according to manufacturer’s instructions. Samples were imaged using a Nikon Eclipse Ts2 microscope with CFI Ph1 ADL 20× objective, NA 0.4, and CoolLED pE-300 white light source and DS-Qi1 camera (Nikon), and cells were counted using ImageJ 1.51 (NIH) in 2-3 images per group. For mammosphere culture, cells were plated in 1% methylcellulose (15 cP, Sigma-Aldrich, M7027) in MMEC growth media on 96-well low adhesion plates (Corning, 3474) at 1,000 cells per well. Mammospheres were counted 14 days after plating.
Fat pad transplantation
MMECs for transplantation 58 were isolated and treated as described above. 14 3-week-old female C57BL6/Rcc mice (obtained from Envigo and housed as described above) were anesthetised using 2-2.5 % Isoflurane (Baxter) and the anterior part of #4 mammary gland, lymph node and bridge to #5 gland were surgically removed. Pre-treated MMECs were injected into the remaining fat pad at 105 cells/gland in 10μl volume using a Hamilton syringe. Mice were injected with control and treated MMECs into contralateral glands. Wounds were sutured using wound clips (Autoclip Physicians kit, BD Biosciences, 427638), which were removed 1 week later. The fat pad transplanted mice were sacrificed 8 weeks after transplantation.
Wholemount stainings
#4 inguinal mammary glands were fixed in 4 % PFA overnight, stained for several hours in carmine-alumn staining solution (2% w/v Carmine, Sigma-Aldrich; 5% w/v Aluminium Potassium Sulfate, Sigma-Aldrich) until the desired color had developed, mounted to glass coverslips and imaged using a Leica S9i stereo microscope with 10 MP CMOS-camera.
Statistics and reproducibility
GraphPad Prism 7, Microsoft Excel XLSTAT 2017.6, and RStudio 3.4.0 were used for data analysis. Data are presented as mean ± standard deviation (s.d.) where indicated. One-way ANOVA with two-sided Dunnett’s post hoc test was applied for comparison of means of multiple groups to the respective controls, or two-tailed Student’s t-test for comparisons of means of two data sets, using paired or unpaired comparison as appropriate, where indicated. Fisher’s exact test was used to assess differences in starting frequencies in mammary fat pad reconstitution assay. Multinomial goodness of fit test using the Monte Carlo method was used to compare distributions of mammary fat pad reconstitution efficiency (with four degrees of freedom, categories were 0, 25, 50, 75 and 100% reconstituted). Alpha was 0.05 for analysis of statistical significance. See Supplementary Data Table 1 for exact P-values and sample sizes.
For analysis of mammary fat pad reconstitution, investigators were blinded to group allocation during data collection and analysis (imaging and counting of stained glands by blinded observers). Mammosphere experiments were regularly validated by blinded investigators by repeating data collection and/or analysis from the same samples or a subset of samples used by the main investigators for analysis. When possible, sample and measurement order was randomized for other experiments. No statistical methods were used to predetermine sample size. Sample sizes were estimated empirically on the basis of pilot experiments and previously performed experiments with similar setup to provide sufficient sample sizes for statistical analysis. No data was removed from the analysis, with the exception of Seahorse experiments where technically inadequate (negative values) and outliers (technical replicates with cutoff: more than 2 standard deviations from the mean for all technical replicates from the same experiment). Experiments were repeated multiple times. The number of biological and technical replicates are reported in the legend of each figure.
Extended Data
Extended Data Figure 1.
Extended Data Figure 2.
Extended Data Figure 3.
Extended Data Figure 4.
Extended Data Figure 5.
Extended Data Figure 6.
Extended Data Figure 7.
Extended Data Figure 8.
Extended Data Figure 9.
Extended Data Figure 10.
Supplementary Material
Acknowledgements
The authors thank Jenny Bärlund and Maija Simula for technical assistance, and all members of the Katajisto laboratory for discussion and comments, Åsa-Lena Dackland at the FACS facility at the Department of Laboratory Medicine at Karolinska Institutet, Juhana Juutila at the Centre of Excellence in Stem Cell Metabolism Metabolomics facility, University of Helsinki and Marja Lohela at the Biomedicum Imaging Unit at the University of Helsinki. Part of this study was performed at the Live Cell Imaging facility, Karolinska Institutet, Sweden, and at the Light Microscopy Unit at the Institute of Biotechnology, University of Helsinki. Anas Kamleh (Thermo Fisher Scientific), Gillian Mackay and Karen Vousden (Beatson Institute) are acknowledged for their support on metabolomics set-up. This study was funded by grants from European Research Council (ERC, #677809), Academy of Finland (#266869, #304591, #312436), Knut and Alice Wallenberg Foundation (KAW 2014.0207), Swedish Research Council 2018-03078, Cancerfonden 190634, Center for Innovative Medicine (CIMED), Sigrid Juselius Foundation, Cancer Society of Finland, Doctoral Programme in Biomedicine at the University of Helsinki (J.D.), Alfred Kordelin Foundation (J.D.), Finnish Cultural Foundation (J.D.).
Footnotes
Author contributions
P.K., J.D., E.K. A.A and J.I.E. conceived and designed experiments. J.D., E.K. A.A., J.I.E., S.G., S.K., E.S.S., R.T.M., K.A. Y.Y.and H.B. performed experiments. N.G. and A.O. performed proteomic analysis. R.K. performed lipidomic analysis. E.K., A.A. and A.N. performed metabolomics data analysis. J.D., E.K. and P.K. wrote the manuscript with input from S.G., H.T., V.H., T.O. and all co-authors. Correspondence and requests for materials should be addressed to P.K. (pekka.katajisto@helsinki.fi).
Competing interests
The authors declare no competing interests.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD010667. 59 Proteomics data were searched against a human database (Swiss-Prot entries of the Uniprot KB database release 2016_01, 20198 entries) with a list of common contaminants appended, for details see section “Proteomic analysis of old and new mitochondria”. The datasets generated and analysed in this study are included as Extended Data and Supplementary information and Source Data. Other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
To facilitate further analysis of the proteomics data, the code is available from the authors in a ready to implement form per request.
References
- 1.Folmes CDL, Dzeja PP, Nelson TJ, Terzic A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell. 2012;11:596–606. doi: 10.1016/j.stem.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vacanti NM, Metallo CM. Exploring metabolic pathways that contribute to the stem cell phenotype. Biochim Biophys Acta - Gen Subj. 2013;1830:2361–2369. doi: 10.1016/j.bbagen.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Chandel NS, Jasper H, Ho TT, Passegué E. Metabolic regulation of stem cell function in tissue homeostasis and organismal ageing. Nat Cell Biol. 2016;18:823–832. doi: 10.1038/ncb3385. [DOI] [PubMed] [Google Scholar]
- 4.Shyh-Chang N, Ng HH. The metabolic programming of stem cells. Genes Dev. 2017;31:336–346. doi: 10.1101/gad.293167.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ron-Harel N, et al. Mitochondrial Biogenesis and Proteome Remodeling Promote One-Carbon Metabolism for T Cell Activation. Cell Metab. 2016;24:104–117. doi: 10.1016/j.cmet.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tormos KV, et al. Mitochondrial complex III ROS regulate adipocyte differentiation. Cell Metab. 2011;14:537–544. doi: 10.1016/j.cmet.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jang K-J, et al. Mitochondrial function provides instructive signals for activation-induced B-cell fates. Nat Commun. 2015;6:6750. doi: 10.1038/ncomms7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu W-M, et al. Metabolic regulation by the mitochondrial phosphatase PTPMT1 is required for hematopoietic stem cell differentiation. Cell Stem Cell. 2013;12:62–74. doi: 10.1016/j.stem.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knobloch M, et al. A Fatty Acid Oxidation-Dependent Metabolic Shift Regulates Adult Neural Stem Cell Activity. Cell Rep. 2017;20:2144–2155. doi: 10.1016/j.celrep.2017.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Almeida MJ, Luchsinger LL, Corrigan DJ, Williams LJ, Snoeck H-W. Dye-Independent Methods Reveal Elevated Mitochondrial Mass in Hematopoietic Stem Cells. Cell Stem Cell. 2017;21:725–729.:e4. doi: 10.1016/j.stem.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams WC, et al. Anabolism-Associated Mitochondrial Stasis Driving Lymphocyte Differentiation over Self-Renewal. Cell Rep. 2016;17:3142–3152. doi: 10.1016/j.celrep.2016.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito K, et al. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18:1350–1358. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryall JG, et al. The NAD+-dependent sirt1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell. 2015;16:171–183. doi: 10.1016/j.stem.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schell JC, et al. Control of intestinal stem cell function and proliferation by mitochondrial pyruvate metabolism. Nat Cell Biol. 2017;19:1027–1036. doi: 10.1038/ncb3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito K, et al. Self-renewal of a purified Tie2+hematopoietic stem cell population relies on mitochondrial clearance. Science (80-) 2016;354:1156–1160. doi: 10.1126/science.aaf5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ansó E, et al. The mitochondrial respiratory chain is essential for haematopoietic stem cell function. Nat Cell Biol. 2017;19:614–625. doi: 10.1038/ncb3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buck MDDD, et al. Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming. Cell. 2016;166:63–76. doi: 10.1016/j.cell.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khacho M, et al. Mitochondrial Dynamics Impacts Stem Cell Identity and Fate Decisions by Regulating a Nuclear Transcriptional Program. Cell Stem Cell. 2015;19:232–247. doi: 10.1016/j.stem.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 19.Chaffer CL, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raouf A, et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell. 2008;3:109–118. doi: 10.1016/j.stem.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 21.Katajisto P, et al. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science. 2015;348:340–3. doi: 10.1126/science.1260384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keppler A, et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 23.Pagliarini DJ, et al. A Mitochondrial Protein Compendium Elucidates Complex I Disease Biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hung V, et al. Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. Elife. 2017;6:1–38. doi: 10.7554/eLife.24463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottschling DE, Nyström T. The Upsides and Downsides of Organelle Interconnectivity. Cell. 2017;169:24–34. doi: 10.1016/j.cell.2017.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murley A, Nunnari J. The Emerging Network of Mitochondria-Organelle Contacts. Mol Cell. 2016;61:648–653. doi: 10.1016/j.molcel.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guarani V, et al. TIMMDC1/C3orf1 Functions as a Membrane-Embedded Mitochondrial Complex I Assembly Factor through Association with the MCIA Complex. Mol Cell Biol. 2014;34:847–861. doi: 10.1128/MCB.01551-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guerrero-Castillo S, et al. The Assembly Pathway of Mitochondrial Respiratory Chain Complex I. Cell Metab. 2017;25:128–139. doi: 10.1016/j.cmet.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Fernández-Vizarra E, Zeviani M. Nuclear gene mutations as the cause of mitochondrial complex III deficiency. Front Genet. 2015;6:1–11. doi: 10.3389/fgene.2015.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diaz F, Enríquez JA, Moraes CT. Cells Lacking Rieske Iron-Sulfur Protein Have a Reactive Oxygen Species-Associated Decrease in Respiratory Complexes I and IV. Mol Cell Biol. 2012;32:415–429. doi: 10.1128/MCB.06051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paradies G, Paradies V, De Benedictis V, Ruggiero FM, Petrosillo G. Functional role of cardiolipin in mitochondrial bioenergetics. Biochim Biophys Acta -Bioenerg. 2014;1837:408–417. doi: 10.1016/j.bbabio.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Mejia EM, Hatch GM. Mitochondrial phospholipids: role in mitochondrial function. J Bioenerg Biomembr. 2016;48:99–112. doi: 10.1007/s10863-015-9601-4. [DOI] [PubMed] [Google Scholar]
- 34.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patten DA, et al. OPA1-dependent cristae modulation is essential for cellular adaptation to metabolic demand. EMBO J. 2014;33:2676–2691. doi: 10.15252/embj.201488349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aguilaniu H, Gustafsson L, Rigoulet M, Nyström T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science (80-) 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 37.Shaw FL, et al. A Detailed Mammosphere Assay Protocol for the Quantification of Breast Stem Cell Activity. J Mammary Gland Biol Neoplasia. 2012;17:111–117. doi: 10.1007/s10911-012-9255-3. [DOI] [PubMed] [Google Scholar]
- 38.Vannini N, et al. The NAD-Booster Nicotinamide Riboside Potently Stimulates Hematopoiesis through Increased Mitochondrial Clearance. Cell Stem Cell. 2019;24:405–418.:e7. doi: 10.1016/j.stem.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 39.Sanderson SM, Locasale JW. Revisiting the Warburg Effect: Some Tumors Hold Their Breath. Cell Metabolism. 2018;28:669–670. doi: 10.1016/j.cmet.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fendt SM, et al. Reductive glutamine metabolism is a function of the alpha-ketoglutarate to citrate ratio in cells. Nat Commun. 2013;4:2236. doi: 10.1038/ncomms3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faubert B, et al. Lactate Metabolism in Human Lung Tumors. Cell. 2017;171:358–371.:e9. doi: 10.1016/j.cell.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan J, et al. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jalloh I, et al. Glycolysis and the pentose phosphate pathway after human traumatic brain injury: Microdialysis studies using 1,2-13C2 glucose. J Cereb Blood Flow Metab. 2015;35:111–120. doi: 10.1038/jcbfm.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miccheli A, et al. Metabolic profiling by 13C-NMR spectroscopy: [1,2-13C2]glucose reveals a heterogeneous metabolism in human leukemia T cells. Biochimie. 2006;88:437–448. doi: 10.1016/j.biochi.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 45.Ducker GS, et al. Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell Metab. 2016;23:1140–1153. doi: 10.1016/j.cmet.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox J, et al. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 47.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 48.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 49.Gatto L, Lilley KS. MSnbase-an R/Bioconductor package for isobaric tagged mass spectrometry data visualization, processing and quantitation. Bioinformatics. 2012;28:288–289. doi: 10.1093/bioinformatics/btr645. [DOI] [PubMed] [Google Scholar]
- 50.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47–e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strimmer K. A unified approach to false discovery rate estimation. BMC Bioinformatics. 2008;9:303. doi: 10.1186/1471-2105-9-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Folch J, Lees M, Sloane Stanley G. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 53.Tigistu-Sahle F, et al. Metabolism and phospholipid assembly of polyunsaturated fatty acids in human bone marrow mesenchymal stromal cells. J Lipid Res. 2017;58:92–110. doi: 10.1194/jlr.M070680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haimi P, Uphoff A, Hermansson M, Somerharju P. Software tools for analysis of mass spectrometric lipidome data. Anal Chem. 2006;78:8324–8331. doi: 10.1021/ac061390w. [DOI] [PubMed] [Google Scholar]
- 55.Hernebring M, Brolen G, Aguilaniu H, Semb H, Nystrom T. Elimination of damaged proteins during differentiation of embryonic stem cells. Proc Natl Acad Sci. 2006;103:7700–7705. doi: 10.1073/pnas.0510944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaik IH, Mehvar R. Rapid determination of reduced and oxidized glutathione levels using a new thiol-masking reagent and the enzymatic recycling method: Application to the rat liver and bile samples. Anal Bioanal Chem. 2006;385:105–113. doi: 10.1007/s00216-006-0375-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Millard P, Letisse F, Sokol S, Portais JC. IsoCor: Correcting MS data in isotope labeling experiments. Bioinformatics. 2012;28:1294–1296. doi: 10.1093/bioinformatics/bts127. [DOI] [PubMed] [Google Scholar]
- 58.Partanen JI, et al. Tumor suppressor function of Liver kinase B1 (Lkb1) is linked to regulation of epithelial integrity. Proc Natl Acad Sci. 2012;109:E388–E397. doi: 10.1073/pnas.1120421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD010667. 59 Proteomics data were searched against a human database (Swiss-Prot entries of the Uniprot KB database release 2016_01, 20198 entries) with a list of common contaminants appended, for details see section “Proteomic analysis of old and new mitochondria”. The datasets generated and analysed in this study are included as Extended Data and Supplementary information and Source Data. Other data supporting the findings of this study are available from the corresponding author on reasonable request.