Abstract
Oncogene-induced replication stress characterizes many aggressive cancers. Several treatments are being developed that target replication stress, however, identification of tumors with high levels of replication stress remains challenging.
We describe a gene expression signature of oncogene-induced replication stress. A panel of triple-negative breast cancer (TNBC) and non-transformed cell lines were engineered to overexpress CDC25A, CCNE1 or MYC, which resulted in slower replication kinetics. RNA sequencing analysis revealed a set of 52 commonly upregulated genes. In parallel, mRNA expression analysis of patient-derived tumor samples (TCGA, n=10,592) also revealed differential gene expression in tumors with amplification of oncogenes that trigger replication stress (CDC25A, CCNE1, MYC, CCND1, MYB, MOS, KRAS, ERBB2, and E2F1). Upon integration, we identified a six-gene signature of oncogene-induced replication stress (NAT10, DDX27, ZNF48, C8ORF33, MOCS3, and MPP6). Immunohistochemical analysis of NAT10 in breast cancer samples (n=330) showed strong correlation with expression of phospho-RPA (R=0.451, p=1.82x10-20) and γH2AX (R=0.304, p=2.95x10-9). Finally, we applied our oncogene-induced replication stress signature to patient samples from TCGA (n=8,862) and GEO (n=13,912) to define the levels of replication stress across 27 tumor subtypes, identifying diffuse large B cell lymphoma, ovarian cancer, TNBC and colorectal carcinoma as cancer subtypes with high levels of oncogene-induced replication stress.
Introduction
Breast cancers can be classified based on the expression of the estrogen receptor (ER), the progesterone receptor (PR) and the human epidermal growth factor receptor 2 (HER2). Breast cancers that lack expression of the ER, PR and HER2 are called ‘triple-negative breast cancer’ (TNBC), and are characterized by profound genomic instability 1,2 . This phenomenon is characterized by continuous gains and losses of chromosomal fragments and complex genomic rearrangements. This genomic instability underlies the rapid acquisition of genomic aberrations that drive therapy failure 3 . Finding new treatment options for genomically unstable cancers is not only relevant for TNBC, but also other hard-to-treat cancers with extensive genomic instability, including high-grade serous ovarian cancer (HGSOC) 4 . Genomic instability is observed in multiple aggressive cancer subtypes and is associated with the inability of cancer cells to faithfully repair DNA damage. An important source of the DNA lesions that fuels genomic instability is replication stress 5,6 .
During S-phase of the cell cycle, all DNA must be replicated in a coordinated manner, which is initiated at genomic loci called 'replication origins' 7 . Replication origins firing adheres to a temporally-controlled program, which prevents exhaustion of the nucleotide pool and warrants the availability of essential components of the replication machinery 8 . DNA replication can be challenged in various ways, which is collectively referred to as replication stress. An important cause of replication stress is the uncoordinated initiation of origin firing due to oncogene activation 5,9,10 . Consequently, oncogene activation leads to depletion of the nucleotide pool and collisions of the replisome with the transcription machinery, resulting in slowing or complete stalling of replication forks 5,8,11 . When stalled replication forks are not resolved in time, they can collapse and cause DNA double-strand breaks (DSBs) and lead to genomic instability.
Several oncogenes have been linked to induction of replication stress, many of them leading to aberrant activation of CDK2. For instance, overexpression of the CDK2-binding partner Cyclin E1 (CCNE1) or the CDK2-activating phosphatase CDC25A leads to replication slow-down and reversal of replication forks 12 . In line with this notion, amplification of CCNE1 has been proposed as a biomarker for tumors with high levels of replication stress 13 . However, multiple other oncogenic events beyond CCNE1 amplification also can lead to replication stress, including overexpression of MYC 14,15 , MOS 16 , E2F1 10 , or expression of the E6/E7 HPV oncoproteins 8 . Currently, there is no uniform way to determine oncogene-induced replication stress levels in cancer samples.
Identification of cancers with high levels of replication stress is increasingly relevant because several drugs have been developed that specifically target tumor cells with high levels of replication stress. Inhibition of WEE1 has been shown to have therapeutic efficacy in HGSOC, which is considered as a prototypical tumor type with high levels of replication stress 17 . Interestingly, patients with the largest decrease in tumor size upon WEE1 inhibitor treatment showed enrichment for CCNE1 amplification 17 . In line with this notion, overexpression of CCNE1 sensitized TNBC cell lines to WEE1 inhibition 18,19 . In good agreement with these data, an unbiased genomic screen identified regulators of CDK2 as determinants of WEE1 inhibitor sensitivity 20 . Mechanistically, WEE1 inhibition is thought to exacerbate levels of replication stress further, while it inactivates the G2/M cell cycle checkpoint, driving cells into mitotic catastrophe 21 . Similarly, inhibition of the ATR or CHK1 checkpoint kinases has been shown to preferentially target tumor cells with molecular characteristics that are associated with replication stress 22,23 .
It is currently unclear how patients can be optimally selected for treatment with agents that target replication stress. To this end, we performed gene expression profiling of cell lines with oncogene-induced replication. Further refinement with expression profiles of patient derived tumor samples yielded a gene expression signature of replication stress, which allowed us to describe a pan-cancer landscape of oncogene-induced replication stress.
Results
Overexpression of CDC25A, CCNE1, or MYC results in replication stress
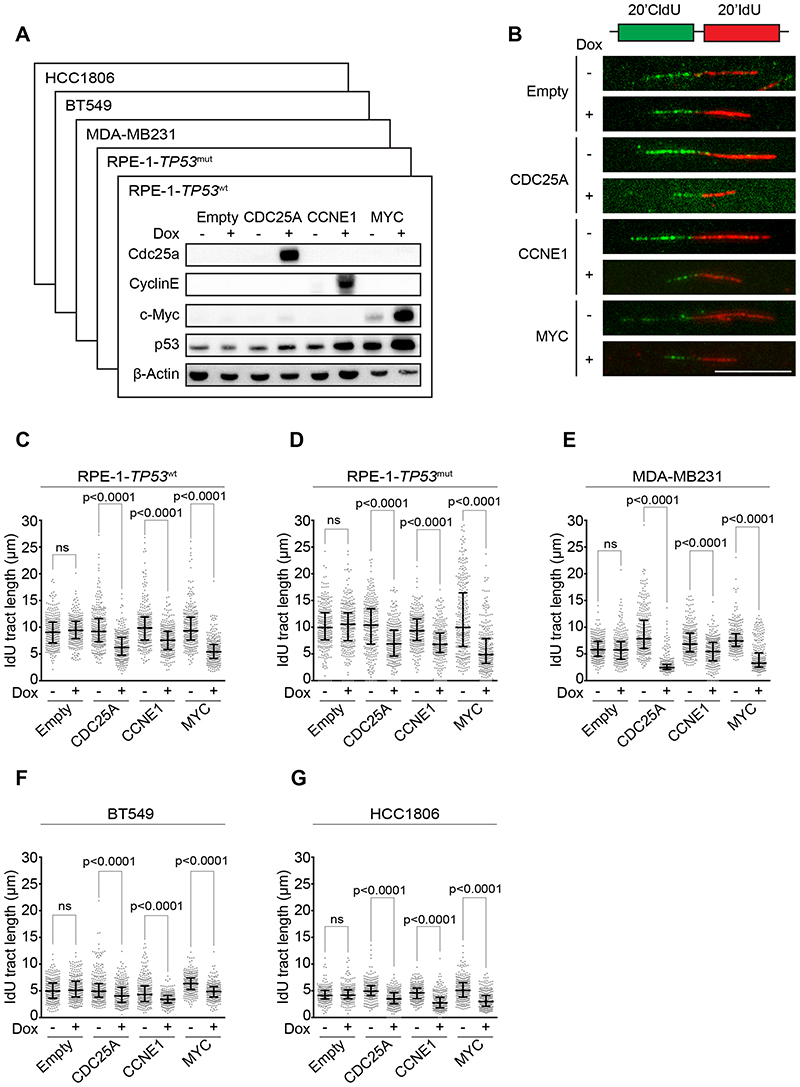
To develop an mRNA expression-based signature for oncogene-induced replication stress, we engineered a panel of cell lines to overexpress CDC25A, CCNE1, or MYC in a doxycycline-inducible manner. This cell line panel included non-transformed human retina epithelial (RPE1) cell lines, either with wild type TP53 (RPE1-TP53 wt) or a derivative in which TP53 was mutated using CRISPR-Cas9 (RPE1-TP53 mut), and TNBC cell lines MDA-MB-231, BT549 and HCC-1806 (Fig. 1A, Suppl. Fig. 1A-D). To validate that oncogene induction indeed affected DNA replication, DNA fiber analysis was performed (Fig. 1B). A severe reduction in DNA synthesis velocity was observed upon induction of CDC25A, CCNE1, or MYC, as measured by IdU fiber tract lengths in RPE1-TP53 wt cells (32%, 23% and 42% decrease in CDC25A, CCNE1, and MYC overexpressing cells versus controls, respectively, Fig. 1C). Similar decreases in DNA replication dynamics were observed in RPE1-TP53 mut cells (34%, 27%, and 51% decrease in CDC25A, CCNE1 and MYC overexpressing cells versus controls, respectively, Fig. 1D), indicating that these effects are independent of TP53 status. Subsequently, the impact of oncogene expression on DNA synthesis was analyzed in TNBC cell lines. Again, we consistently observed shortening of IdU tract lengths in MDA-MB-231, BT549 and HCC-1806 cell lines upon doxycycline-induced overexpression of CDC25A, CCNE1 or MYC (Fig. 1E-G), but not in empty vector controls (Fig. 1C-G). Taken together, these data indicate that induction of CDC25A, CCNE1, or MYC results in replication stress in cancer cells, as well as in untransformed cell lines, independently of TP53 status.
Figure 1. Overexpression of CDC25A, CCNE1 or MYC leads to replication stress.
(A) Indicated RPE1-TP53 wt cell lines were treated with doxycycline for 48 hours. Immunoblotting was performed for CDC25A, CCNE1, p53, and β-Actin. (B) Cells were treated with doxycycline as described for panel A and subsequently pulse-labeled for 20 minutes with CldU (25 μM) and 20 minutes with IdU (250 μM). Representative DNA fibers from RPE1-TP53 wt cells are shown. Scale bar represents 10 μm. (C-G) Quantification of IdU DNA fiber lengths, as described in panel B. Per condition, 300 fibers from RPE1-TP53 wt cells were analyzed. P values were calculated using the Mann-Whitney U test for RPE1-TP53 wt (panel C), RPE1-TP53 mut (panel D), MDA-MB-231 (panel E), BT549 (panel F) and HCC1806 (panel G).
Gene expression profiling of cell line models with oncogene-induced replications stress
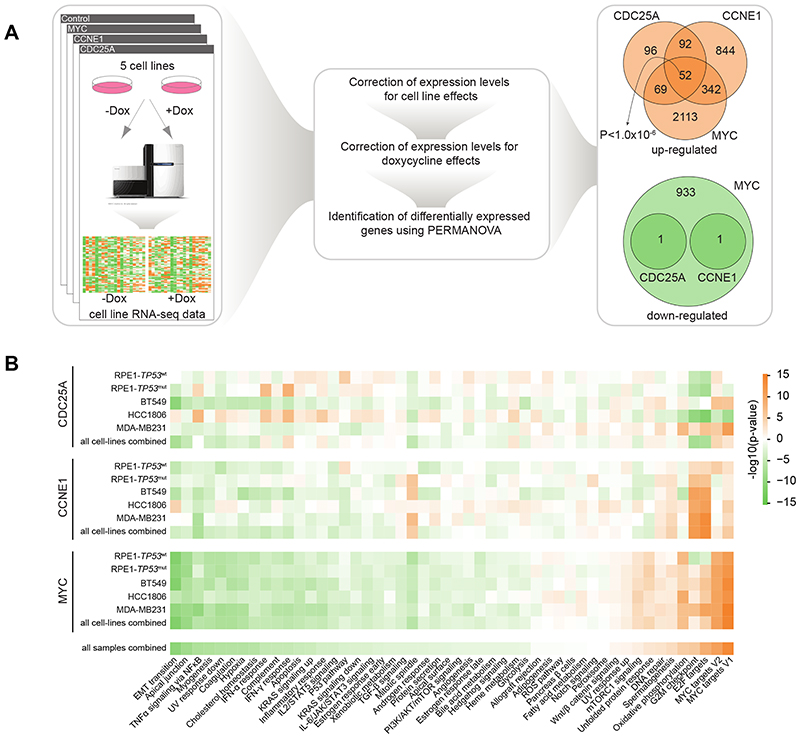
To map the transcriptional consequences of overexpression of CDC25A, CCNE1 and MYC, RNA sequencing of RPE1-TP53 wt, RPE1-TP53 mut, MDA-MB-231, BT549, and HCC-1806, cells was performed both at 48 hours and at 120 hours after oncogene induction to capture gene expression changes provoked by replication stress (Fig. 2A). Subsequently, all cell lines and genetic perturbations were analyzed in a pooled fashion. To this end, the pooled RNAseq dataset was first normalized to remove any possible effects of doxycycline treatment in control cell lines as well as cell line-specific effects (see Supplementary Methods for details). Subsequently, permutational multivariate analysis of variance (PERMANOVA) was performed to identify genes that were differentially expressed upon overexpression of each oncogene (CDC25A, CCNE1, and MYC) across cell lines (Fig. 2B). Induction of CCNE1 or CDC25A led to significantly differentially upregulated genes (n=1,330 and n=309 respectively; p<0.01), with only 2 genes being downregulated (Fig. 2A, right panel). In contrast, MYC overexpression resulted in a substantial differential gene expression, involving both downregulation (n=2,576) and upregulation (n=935) of gene expression (Fig. 2A, right panel). Interestingly, expression of 52 genes was found to be commonly upregulated in response to induction of CCNE1, CDC25A, or MYC (Fig. 2A, right panel). Of note, because of the relatively limited number of cell line samples, we did not correct for multiple testing, as we wanted to keep our type II error low in this step of the discovery phase.
Figure 2. Overexpression of CDC25A, CCNE1, or MYC leads to upregulation of 52 genes.
(A) RNAseq data of 5 cell lines were corrected for cell line-specific and doxycycline treatment effects on gene expression. Subsequently, PERMANOVA was used to identify common differentially expressed genes upon oncogene expression. 52 genes were found to be commonly upregulated whereas no genes were found to be commonly downregulated in response to induction of CCNE1, CDC25A or MYC (B) Gene Set Enrichment Analysis using two-sided Welch’s t-test for the MSigDB Hallmark collection. An orange box indicates enrichment for upregulated genes due to overexpression of oncogenes in corresponding cell lines, and a green box indicates enrichment for downregulated genes.
Importantly, gene-set enrichment analysis (GSEA) on metrics obtained from PERMANOVA (see Supplementary Methods for details) revealed that common biological pathways were affected upon induction of CCNE1, CDC25A, or MYC (Fig. 2B), with strong upregulation in expression of MYC targets, and genes involved in cell cycle control and oxidative phosphorylation (Fig. 2B). In contrast, expression of genes related to biological pathways involved in cellular morphology and inflammatory signaling were commonly downregulated (Fig. 2B). Taken together, these results indicate that oncogene overexpression induces distinct yet overlapping gene expression changes, affecting common biological pathways.
Common differential gene expression upon oncogene overexpression between in vitro models and patient samples
To investigate whether the differential gene expression we observed in our cell line models overlaps with patient-derived tumor samples with amplification of these oncogenes, we retrieved copy number data and mRNA expression data from The Cancer Genome Atlas (TCGA) (Fig. 3A). TCGA samples were classified into two groups based on the copy number of each of the oncogenes (‘amplified’ versus ‘neutral’). After that, genes that were significantly differentially expressed upon amplification of each oncogene (CDC25A, CCNE1, or MYC) were identified (Fig. 3A). In tumor samples with amplification of the three oncogenes, expression of 720 common genes was significantly upregulated (permutation test: p<1.0x10-6), and that of 597 genes was down-regulated (permutation test: p<1.0x10-6). GSEA revealed strong upregulation in expression of genes related to MYC targets, cell cycle control, and oxidative phosphorylation (Fig. 3B). In contrast, expression of genes related to immune signatures was commonly downregulated in samples with oncogene amplification (Fig. 3B). The majority of the enrichments in the TCGA data were similar to those obtained by differential expression analysis from the cell line data. Of note, two genesets (i.e., “allograft rejection” and “IL-6/JAK/STAT3 signaling”) were only significantly enriched in the analysis with patient-derived tumor samples. This is in line with immune activities in the patient samples not being reflected in cell line models.
Figure 3. Common differential gene expression of 6 genes upon oncogene overexpression between in vitro models and patient samples.
(A) TCGA RNAseq samples were queried for amplification of CCNE1, CDC25A or MYC. Expression of 720 genes was found to be commonly upregulated, whereas 597 genes were found to be commonly downregulated in response to amplification of CCNE1, CDC25A or MYC. (B) Gene Set Enrichment Analysis using two-sided Welch’s t-test for the MSigDB Hallmark collection. An orange box indicates enrichment for upregulated genes due to overexpression of oncogenes in corresponding cell lines, whereas a green box indicates enrichment for downregulated genes. (C) TCGA RNAseq samples were queried for amplification of CCND1, MYB, MOS, KRAS, ERBB2 and E2F1. The resulting commonly upregulated genes were overlaid with upregulated genes identified upon overexpression of CCNE1, CDC25A or MYC in cell lines and upregulated genes in TCGA samples with CCNE1, CDC25A or MYC amplifications, resulting in 6 commonly upregulated genes (D) Co-functionality analysis of commonly upregulated genes using the GenetICA algorithm.
Since replication stress can also be caused by oncogenes beyond MYC, CCNE1, CDC25A and to increase stringency in obtaining a replication stress signature, we additionally identified commonly upregulated genes in TCGA tumor samples overexpressing other oncogenes that have been associated with replication stress, including CCND1, MYB, MOS, KRAS, ERBB2, and E2F1 10,16,24–29 (Fig. 3C). P-values from the differential expression analysis using the TCGA data were not corrected for multiple testing to keep the type II error low. Analysis of overlap between genes that were commonly upregulated upon oncogenes expression in cell line models (n=52, Fig. 2A) with genes upregulated in patient-derived tumor samples revealed six genes (i.e., NAT10, DDX27, ZNF48, C8ORF33, MOCS3, and MPP6) (Fig. 3C, D and Supplementary Fig. 2). For cross validation, we also conducted differential expression analysis using the ‘limma’ package, and found that these 6 commonly upregulated genes were also part of 104 commonly upregulated genes upon overexpression or amplification of oncogenes in cell line dataset or TCGA dataset respectively (see Supplementary Fig. 3). Subsequently, we investigated if these genes were differentially expressed in the TCGA dataset by using PERMANOVA. We found that the 6 signature genes are significantly upregulated in all these conditions with a p-value cutoff of 0.01, except MPP6 in the condition of CDC25A amplification, which showed borderline significance (p= 0.0112) (see Supplementary Fig. 4). Co-functionality analysis of commonly upregulated genes using the GenetICA algorithm 30 (see Supplementary Methods for details) pointed at roles for these genes in ncRNA processing, DNA repair, and ribosome biogenesis (Fig. 3D).
NAT10 protein expression is associated with markers of replication stress in breast cancer samples
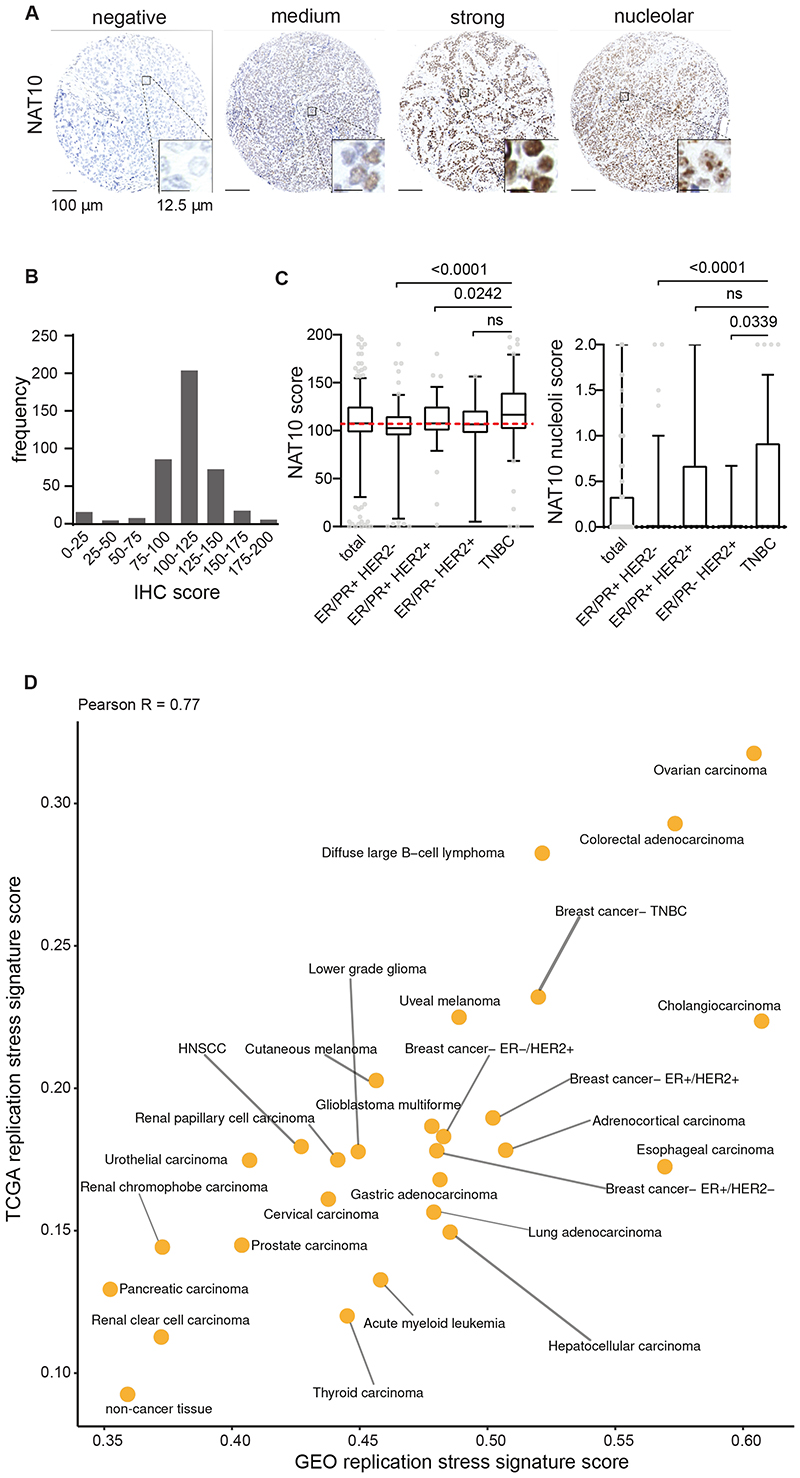
To validate the mRNA-based replication stress signature, we selected NAT10 for immunohistochemical analysis. The acetyltransferase NAT10 (N-Acetyltransferase-10) has previously been implicated in regulation of various processes, including regulation of the DNA damage response 31,32 and regulation of translation 33,34 . NAT10 is a nuclear protein, predominantly localized to the nucleolus 35 . Immunohistochemical analysis of NAT10 staining in a series of breast cancer tissues (n=410), confirmed nuclear localization of NAT10, with nucleolar expression in a subset of cancer samples (Fig. 4A, Supplementary Fig. 5). Importantly, a significant variation in protein expression was observed, with triple-negative breast cancer samples showing the highest NAT10 expression levels (Fig. 4B). Of note, absence or presence of nucleolar localization was not different between breast cancer subgroups (Fig. 4B).
Figure 4. Immunohistochemical analysis of NAT10 in breast cancer patients and the landscape of oncogene-induced replication stress across cancer types.
(A) Representative staining of NAT10 in breast cancer patient samples (n=410). Scale bar represents 100 μm. (B, C) Patients from the combined cohort (n=410) and breast cancer subgroups ER/PR+HER2- (n=164), ER/PR+HER2+ (n=95), ER/PR-HER2+ (n=21) and TNBC (n=130) were analyzed. Tumor tissue was immunohistochemically scored for expression of NAT10 and NAT10 nucleoli. indicated P values were calculated using Mann-Whitney U test. Interquartile ranges are displayed, dashed red line represents the median score from the combined cohort and outliers are shown as dots. (D) Scatter plot of replication stress signature for 27 cancer types as well as non-cancerous tissues of patient samples from TCGA (y axis) and GEO (x axis).
For this breast cancer cohort, we previously reported the presence of p-RPA, a marker of replication stress, and γH2AX, which reflects DNA breaks, a possible result of stalled replication forks collapse 36 . In line with NAT10 expression being part of the oncogene-induced replication stress signature, Spearman correlation analysis showed an association of NAT10 expression with both p-RPA (R=0.451, p=1.82x10-20) and γH2AX (R=0.304, p=2.95x10-9) (Supplemental Table 1A). Subgroup analysis showed that the most significant correlations between NAT10 expression and pRPA or γH2AX were observed in ER/PR-/HER2+ and TNBC samples (Supplemental Table 1A).
Next, we tested whether NAT10 expression was also correlated to expression of two oncogenes that are frequently amplified in breast cancer, Cyclin E1 and MYC. NAT10 expression was positively correlated to expression of both Cyclin E1 (R=0.303, p=1.86x10-9) and MYC (R=0.264, p=1.45x10-7) (Supplemental Table 1B). Again, associations were most significant in ER/PR-/HER2+ and TNBC samples (Supplemental Table 1B). Combined, these analyses validated NAT10 as part of our oncogene-induced replication stress signature, which is associated with markers of replication stress as well as expression of oncogenes, which are known to induce replication stress.
Landscape of replication stress across tumor types
To investigate the landscape of oncogene-induced replication stress across cancer types, we used the six-gene signature of oncogene-induced replication stress, as a proxy for oncogene-induced replication stress levels. We applied our signature to RNAseq expression data of 8,862 samples retrieved from TCGA (Supplemental Table 2, Supplemental Dataset 1). This dataset represents 27 cancer subtypes as well as non-cancer tissues, and displayed large differences in the oncogene-induced replication stress signature score, with diffuse large B cell lymphoma (DLBCL), ovarian cancer and colorectal carcinoma showing highest scores (Supplemental Fig. 5). in line with expectations, normal tissues were among the tissue types with lowest scores (Supplemental Fig. 6). These observations are in line with previous reports on these cancer subtypes 15,37,38 . To validate the landscape of oncogene-induced replication stress levels in an independent dataset, we retrieved microarray mRNA expression data of 13,912 patient-derived samples from the GEO database (Fig. 4D, Supplemental Table 2, Supplemental Dataset 2). A high concordance between the tumor type replication stress levels in TCGA and GEO was observed (Pearson R=0.77), indicating that our oncogene-induced replication stress signature captures the level of oncogene-induced replication stress also in a platform-independent fashion.
Discussion
In the present study, we examined transcriptional changes that go along with oncogene-induced replication stress in cell line models and tumor samples to build a gene expression signature of oncogene-induced replication stress. Analysis of a panel of TNBC and non-transformed RPE1 cell lines, combined with analysis of a large dataset of patient-derived cancer samples, yielded a six-gene signature of oncogene-induced replication stress.
Our mRNA-based signature points towards ovarian cancers, colorectal cancers, DLBCLs, TNBCs, cholangiocarcinomas, and esophageal carcinomas having high levels of replication stress. Among these are cancer subtypes that were previously described as prototypical cancers with high levels of replication stress. Specifically, HGSOCs almost invariably have TP53 mutations (96%), frequently contain high levels of somatic copy number alterations and structural variations, and often have amplification of MYC (>30%) and CCNE1 (>20%) 4 , both of which have been linked to replication stress and genomic instability 6,12 . Likewise, TNBCs show biological similarities with HGSOC and also frequently contain somatic TP53 mutations as well as amplification of MYC and CCNE1 1,2 . In good agreement with these data, our immunohistochemical analysis of TNBC samples revealed high levels of p-RPA and γH2AX, markers of single-stranded DNA and DNA breaks, which have associated with replication stress 36 .
Cholangiocarcinoma was among the highest-ranked cancer subtypes based on our replication stress signature. These bile duct cancers have not previously been linked to genomic features associated with replication stress. However, recent studies described recurrent alterations in the proto-oncogene CCND1, cell cycle regulatory gene CDKN2A as well as the chromatin remodelers ARID1A, IDH1/2 and PBRM1 39 , which could underlie DNA replication perturbations 25,40–43 . Of note, mixed type hepatocellular-cholangiocarcinoma was described to share genetic features with hepatocellular carcinoma 44 , including CCNE1 amplification, which are causally implicated in hepatocellular carcinogenesis 13 .
Also, diffuse large B-cell lymphoma (DLBCL) showed a high score using our gene expression signature. This finding is in line with observations of MYC amplification, ARID1A mutation and CDKN2A/B deletion in DLBCL, with accompanying sensitivity to inhibitors of replication checkpoint kinases 15,45,46 . Although colorectal cancer (CRC) is not commonly regarded as a tumor type with high levels of oncogene-induced replication stress, CRC scored high in our classifier. This observation is strengthened by earlier observations that CRC subgroups in which pRB is inactivated show activation of the DNA damage response 10,47 .Additionally, the chromosomal instability that characterizes CRCs was shown to be linked to replication stress, as judged by slower replication fork progression, and increased numbers of ultrafine anaphase bridges and 53BP1 bodies in G1 cells 37 .
A gene expression signature may aid in patient selection for drugs that target cancers with high levels of replication stress. Early clinical trials evaluating inhibitors of the ATR, WEE1, and CHK1 checkpoint kinases have shown promising results 17,48,49 , but not all patients respond and biomarkers defining optimal patient subgroups has been challenging. For instance, TP53 mutation status was used to select patients for Wee1 inhibitor treatment, but additional features are needed to define patients who will likely benefit 17,50 . Interestingly, CCNE1 amplification was among the genetic features that appeared enriched in patients responding to Wee1 inhibitor treatment 17 . focused analysis of oncogene-induced replication stress in these clinical trials is warranted to test whether a more optimal patient selection is achievable.
Materials and Methods
Methodology is described in the Supplementary Methods document.
Supplementary Material
Acknowledgments
We thank members of the Medical Oncology laboratory for fruitful discussions.
Funding
This work was financially supported by grant from the Netherlands Organization for Scientific Research (NWO-VIDI #917.13334 to M.A.T.M.v.V. and NWO-VENI #916-16025 to R.S.N.F.), the Dutch Cancer Society (RUG 2013-5960 to R.S.N.F) and from the European Research Council (ERC-Consolidator grant “TENSION” to M.A.T.M.v.V.).
Footnotes
Author contribution
R.S.N.F. and M.A.T.M.v.V. conceived and supervised the study. S.G.L. performed in vitro studies. S.G.L, A.B, K.K. and R.S.N.F. generated and analyzed RNAseq data. A.K. and R.S.N.F. conducted computational analyses. M.E. and B.v.d.V. conducted immunohistochemical analysis of patient samples. S.G.L, A.B, R.S.N.F. and M.A.T.M.v.V. wrote the manuscript. All authors provided feedback and approved the manuscript.
Conflict of Interest
M.A.T.M.v.V. has acted on the Scientific Advisory Board of Repare Therapeutics, which is unrelated to this work. The other authors declare no conflict of interest.
Data and code availability
All sequencing data have been deposited at the GEO archive of NCBI (identifier GSE185512). All code has been made available through GitHub: (https://github.com/arkajyotibhattacharya/Replication_stress_signature).
References
- 1.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerlinger M, Swanton C. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br J Cancer. 2010;103:1139–43. doi: 10.1038/sj.bjc.6605912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 7.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–74. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 8.Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–46. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–5. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 10.Bartkova J, Horejsí Z, Koed K, Krämer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 11.Macheret M, Halazonetis TD. Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature. 2018;555:112–116. doi: 10.1038/nature25507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neelsen KJ, Zanini IMY, Herrador R, Lopes M. Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J Cell Biol. 2013;200:699–708. doi: 10.1083/jcb.201212058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aziz K, Limzerwala JF, Sturmlechner I, Hurley E, Zhang C, Jeganathan KB, et al. Ccne1 Overexpression Causes Chromosome Instability in Liver Cells and Liver Tumor Development in Mice. Gastroenterology. 2019;157:210–226.:e12. doi: 10.1053/j.gastro.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.López-Contreras AJ, Gutierrez-Martinez P, Specks J, Rodrigo-Perez S, Fernandez-Capetillo O. An extra allele of Chk1 limits oncogene-induced replicative stress and promotes transformation. J Exp Med. 2012;209:455–61. doi: 10.1084/jem.20112147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young LA, O’Connor LO, de Renty C, Veldman-Jones MH, Dorval T, Wilson Z, et al. Differential Activity of ATR and WEE1 Inhibitors in a Highly Sensitive Subpopulation of DLBCL Linked to Replication Stress. Cancer Res. 2019;79:3762–3775. doi: 10.1158/0008-5472.CAN-18-2480. [DOI] [PubMed] [Google Scholar]
- 16.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–7. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 17.Leijen S, van Geel RMJM, Sonke GS, de Jong D, Rosenberg EH, Marchetti S, et al. Phase II Study of WEE1 Inhibitor AZD1775 Plus Carboplatin in Patients With TP53-Mutated Ovarian Cancer Refractory or Resistant to First-Line Therapy Within 3 Months. J Clin Oncol. 2016;34:4354–4361. doi: 10.1200/JCO.2016.67.5942. [DOI] [PubMed] [Google Scholar]
- 18.Chen X, Low K-H, Alexander A, Jiang Y, Karakas C, Hess KR, et al. Cyclin E Overexpression Sensitizes Triple-Negative Breast Cancer to Wee1 Kinase Inhibition. Clin Cancer Res. 2018;24:6594–6610. doi: 10.1158/1078-0432.CCR-18-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kok YP, Guerrero Llobet S, Schoonen PM, Everts M, Bhattacharya A, Fehrmann RSN, et al. Overexpression of Cyclin E1 or Cdc25A leads to replication stress, mitotic aberrancies, and increased sensitivity to replication checkpoint inhibitors. Oncogenesis. 2020;9:88. doi: 10.1038/s41389-020-00270-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heijink AM, Blomen VA, Bisteau X, Degener F, Matsushita FY, Kaldis P, et al. A haploid genetic screen identifies the G1/S regulatory machinery as a determinant of Wee1 inhibitor sensitivity. Proc Natl Acad Sci U S A. 2015;112:15160–5. doi: 10.1073/pnas.1505283112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aarts M, Sharpe R, Garcia-Murillas I, Gevensleben H, Hurd MS, Shumway SD, et al. Forced mitotic entry of S-phase cells as a therapeutic strategy induced by inhibition of WEE1. Cancer Discov. 2012;2:524–39. doi: 10.1158/2159-8290.CD-11-0320. [DOI] [PubMed] [Google Scholar]
- 22.Wengner AM, Siemeister G, Lücking U, Lefranc J, Wortmann L, Lienau P, et al. The Novel ATR Inhibitor BAY 1895344 Is Efficacious as Monotherapy and Combined with DNA Damage-Inducing or Repair-Compromising Therapies in Preclinical Cancer Models. Mol Cancer Ther. 2020;19:26–38. doi: 10.1158/1535-7163.MCT-19-0019. [DOI] [PubMed] [Google Scholar]
- 23.Ferrao PT, Bukczynska EP, Johnstone RW, McArthur GA. Efficacy of CHK inhibitors as single agents in MYC-driven lymphoma cells. Oncogene. 2012;31:1661–72. doi: 10.1038/onc.2011.358. [DOI] [PubMed] [Google Scholar]
- 24.Shimura T, Ochiai Y, Noma N, Oikawa T, Sano Y, Fukumoto M. Cyclin D1 overexpression perturbs DNA replication and induces replication-associated DNA double-strand breaks in acquired radioresistant cells. Cell Cycle. 2013;12:773–82. doi: 10.4161/cc.23719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aggarwal P, Lessie MD, Lin DI, Pontano L, Gladden AB, Nuskey B, et al. Nuclear accumulation of cyclin D1 during S phase inhibits Cul4-dependent Cdt1 proteolysis and triggers p53-dependent DNA rereplication. Genes Dev. 2007;21:2908–22. doi: 10.1101/gad.1586007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maya-Mendoza A, Ostrakova J, Kosar M, Hall A, Duskova P, Mistrik M, et al. Myc and Ras oncogenes engage different energy metabolism programs and evoke distinct patterns of oxidative and DNA replication stress. Mol Oncol. 2015;9:601–16. doi: 10.1016/j.molonc.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyperreplication. Nature. 2006;444:638–42. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 28.Carlos AR, Escandell JM, Kotsantis P, Suwaki N, Bouwman P, Badie S, et al. ARF triggers senescence in Brca2-deficient cells by altering the spectrum of p53 transcriptional targets. Nat Commun. 2013;4:2697. doi: 10.1038/ncomms3697. [DOI] [PubMed] [Google Scholar]
- 29.Kanu N, Cerone MA, Goh G, Zalmas L-P, Bartkova J, Dietzen M, et al. DNA replication stress mediates APOBEC3 family mutagenesis in breast cancer. Genome Biol. 2016;17:185. doi: 10.1186/s13059-016-1042-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urzúa-Traslaviña CG, Leeuwenburgh VC, Bhattacharya A, Loipfinger S, van Vugt MATM, de Vries EGE, et al. Improving gene function predictions using independent transcriptional components. Nat Commun. 2021;12:1464. doi: 10.1038/s41467-021-21671-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H-Y, Liu Y-Y, Yang F, Zhang L, Zhang F-L, Hu X, et al. Acetylation of MORC2 by NAT10 regulates cell-cycle checkpoint control and resistance to DNA-damaging chemotherapy and radiotherapy in breast cancer. Nucleic Acids Res. 2020;48:3638–3656. doi: 10.1093/nar/gkaa130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Tan Y, Zhang C, Zhang Y, Zhang L, Ren P, et al. NAT10 regulates p53 activation through acetylating p53 at K120 and ubiquitinating Mdm2. EMBO Rep. 2016;17:349–66. doi: 10.15252/embr.201540505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, et al. Acetylation of Cytidine in mRNA Promotes Translation Efficiency. Cell. 2018;175:1872–1886.:e24. doi: 10.1016/j.cell.2018.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dominissini D, Rechavi G. N4-acetylation of Cytidine in mRNA by NAT10 Regulates Stability and Translation. Cell. 2018;175:1725–1727. doi: 10.1016/j.cell.2018.11.037. [DOI] [PubMed] [Google Scholar]
- 35.Shen Q, Zheng X, McNutt MA, Guang L, Sun Y, Wang J, et al. NAT10, a nucleolar protein, localizes to the midbody and regulates cytokinesis and acetylation of microtubules. Exp Cell Res. 2009;315:1653–67. doi: 10.1016/j.yexcr.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Guerrero Llobet S, van der Vegt B, Jongeneel E, Bense RD, Zwager MC, Schröder CP, et al. Cyclin E expression is associated with high levels of replication stress in triplenegative breast cancer. NPJ breast cancer. 2020;6:40. doi: 10.1038/s41523-020-00181-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calzetta NL, González Besteiro MA, Gottifredi V. Mus81-Eme1-dependent aberrant processing of DNA replication intermediates in mitosis impairs genome integrity. Sci Adv. 2020;6:492–496. doi: 10.1126/sciadv.abc8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pillay N, Tighe A, Nelson L, Littler S, Coulson-Gilmer C, Bah N, et al. DNA Replication Vulnerabilities Render Ovarian Cancer Cells Sensitive to Poly(ADP-Ribose) Glycohydrolase Inhibitors. Cancer Cell. 2019;35:519–533.:e8. doi: 10.1016/j.ccell.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farshidfar F, Zheng S, Gingras M-C, Newton Y, Shih J, Robertson AG, et al. Integrative Genomic Analysis of Cholangiocarcinoma Identifies Distinct IDH-Mutant Molecular Profiles. Cell Rep. 2017;18:2780–2794. doi: 10.1016/j.celrep.2017.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gadhikar MA, Zhang J, Shen L, Rao X, Wang J, Zhao M, et al. CDKN2A/p16 Deletion in Head and Neck Cancer Cells Is Associated with CDK2 Activation, Replication Stress, and Vulnerability to CHK1 Inhibition. Cancer Res. 2018;78:781–797. doi: 10.1158/0008-5472.CAN-17-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williamson CT, Miller R, Pemberton HN, Jones SE, Campbell J, Konde A, et al. ATR inhibitors as a synthetic lethal therapy for tumours deficient in ARID1A. Nat Commun. 2016;7:13837. doi: 10.1038/ncomms13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inoue S, Li WY, Tseng A, Beerman I, Elia AJ, Bendall SC, et al. Mutant IDH1 Downregulates ATM and Alters DNA Repair and Sensitivity to DNA Damage Independent of TET2. Cancer Cell. 2016;30:337–348. doi: 10.1016/j.ccell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Espana-Agusti J, Warren A, Chew SK, Adams DJ, Matakidou A. Loss of PBRM1 rescues VHL dependent replication stress to promote renal carcinogenesis. Nat Commun. 2017;8:2026. doi: 10.1038/s41467-017-02245-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joseph NM, Tsokos CG, Umetsu SE, Shain AH, Kelley RK, Onodera C, et al. Genomic profiling of combined hepatocellular-cholangiocarcinoma reveals similar genetics to hepatocellular carcinoma. J Pathol. 2019;248:164–178. doi: 10.1002/path.5243. [DOI] [PubMed] [Google Scholar]
- 45.de Jong MRW, Visser L, Huls G, Diepstra A, van Vugt M, Ammatuna E, et al. Identification of relevant drugable targets in diffuse large B-cell lymphoma using a genome-wide unbiased CD20 guilt-by association approach. PLoS One. 2018;13:e0193098. doi: 10.1371/journal.pone.0193098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell. 2017;171:481–494.:e15. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tort F, Bartkova J, Sehested M, Orntoft T, Lukas J, Bartek J. Retinoblastoma pathway defects show differential ability to activate the constitutive DNA damage response in human tumorigenesis. Cancer Res. 2006;66:10258–63. doi: 10.1158/0008-5472.CAN-06-2178. [DOI] [PubMed] [Google Scholar]
- 48.Konstantinopoulos PA, Cheng S-C, Wahner Hendrickson AE, Penson RT, Schumer ST, Doyle LA, et al. Berzosertib plus gemcitabine versus gemcitabine alone in platinum-resistant high-grade serous ovarian cancer: a multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020;21:957–968. doi: 10.1016/S1470-2045(20)30180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Italiano A, Infante JR, Shapiro GI, Moore KN, LoRusso PM, Hamilton E, et al. Phase I study of the checkpoint kinase 1 inhibitor GDC-0575 in combination with gemcitabine in patients with refractory solid tumors. Ann Oncol Off J Eur Soc Med Oncol. 2018;29:1304–1311. doi: 10.1093/annonc/mdy076. [DOI] [PubMed] [Google Scholar]
- 50.Rajeshkumar NV, De Oliveira E, Ottenhof N, Watters J, Brooks D, Demuth T, et al. MK-1775, a potent Wee1 inhibitor, synergizes with gemcitabine to achieve tumor regressions, selectively in p53-deficient pancreatic cancer xenografts. Clin Cancer Res. 2011;17:2799–806. doi: 10.1158/1078-0432.CCR-10-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data have been deposited at the GEO archive of NCBI (identifier GSE185512). All code has been made available through GitHub: (https://github.com/arkajyotibhattacharya/Replication_stress_signature).