A loss-of-function CRISPR screen in human CTC-derived xenografts identifies genes critical for individual steps of the metastatic cascade, suggesting novel drivers and treatment opportunities for metastatic breast cancers.
Abstract
Blood-borne metastasis of breast cancer involves a series of tightly regulated sequential steps, including the growth of a primary tumor lesion, intravasation of circulating tumor cells (CTC), and adaptation in various distant metastatic sites. The genes orchestrating each of these steps are poorly understood in physiologically relevant contexts, owing to the rarity of experimental models that faithfully recapitulate the biology, growth kinetics, and tropism of human breast cancer. Here, we conducted an in vivo loss-of-function CRISPR screen in newly derived CTC xenografts, unique in their ability to spontaneously mirror the human disease, and identified specific genetic dependencies for each step of the metastatic process. Validation experiments revealed sensitivities to inhibitors that are already available, such as PLK1 inhibitors, to prevent CTC intravasation. Together, these findings present a new tool to reclassify driver genes involved in the spread of human cancer, providing insights into the biology of metastasis and paving the way to test targeted treatment approaches.
Significance:
A loss-of-function CRISPR screen in human CTC-derived xenografts identifies genes critical for individual steps of the metastatic cascade, suggesting novel drivers and treatment opportunities for metastatic breast cancers.
Introduction
The vast majority of cancer-related deaths is due to the clinical manifestation of metastasis and the inability to prevent or block their progression (1). Distant metastases result from circulating tumor cells (CTC) that are shed from primary or secondary tumors into the bloodstream, survive within the circulation and reach distant sites, where they extravasate and establish cancerous deposits (2, 3). CTCs are found as single cells or as multicellular aggregates in circulation, with the latter displaying an elevated metastasis-forming capability (4–10). In breast cancer, CTCs typically seed metastasis in organs such as the bone, brain, lung, liver, and lymph nodes (11, 12). Generally, treatments tailored to prevent or subvert a metastatic disease are lacking, as the genes that orchestrate the metastatic cascade in physiological contexts are poorly understood. Widely used models to study metastasis typically fail to precisely recapitulate the human disease. For instance, most patient-derived xenografts obtained from primary tumor biopsies display a preferential tropism for visceral organs (e.g., liver and lungs), whereas human breast cancer metastasis occurs most often within the bone (13, 14). This dichotomy underlies the need to first develop new patient-derived models that mirror the human disease, and then to exploit them to investigate metastasis-relevant phenomena with functional assays.
Recent advances in CRISPR-based genetic screens demonstrated the suitability of this technology for unbiased identification of disease-relevant genes, including those that are involved in cancer biology (15–21). Yet, key players in cancer cells that influence individual steps of the metastatic cascade are poorly understood in clinically relevant contexts, and in particular in freshly isolated primary cancer cells. Here, we set out to perform an in vivo loss-of-function CRISPR-Cas9 screen in human CTC-derived xenografts (CDX)—mirroring the metastatic pattern of the patient of origin—to pinpoint genes required for primary tumor growth, intravasation of single and clustered CTCs, as well as spontaneous metastasis to bone, brain, liver, and lymph nodes. We provide evidence of the suitability of human CTC-derived xenografts—combined with unbiased loss-of-function screens—to mimic human cancer progression and to highlight metastasis-relevant genes with potential for therapy development.
Materials and Methods
Human blood sample collection
After obtaining written informed consent from patients, blood specimens were taken in EDTA vacutainers at the University Hospital Basel. The study was performed in compliance with the Declaration of Helsinki, and received ethical approval from the Ethics Committee northwest/central Switzerland (EKNZ BASEC #2016–00067 and #2020–00014).
CTC capture and identification
Blood from BR16 patient was drawn in EDTA vacutainers and processed within 1 hour from blood draw through a Parsortix GEN3D6.5 Cell Separation Cassette (Angle Europe). Captured CTCs were stained directly within the Parsortix cassette with EpCAM-AF488 (Cell Signaling Technology, CST5198), HER2-AF488 (#324410, BioLegend), EGFR-FITC (GeneTex, GTX11400), and CD45-BV605 (BioLegend, 304042; anti-human) antibodies. Subsequently, captured CTCs were released and cultured as described below.
Cell culture
MDA-MB-231-LM2 (LM2) human breast cancer cells were obtained from Dr. Joan Massagué (Memorial Sloan Kettering Cancer Center, New York, NY) and grown in DMEM medium (Invitrogen, 11330–057) supplemented with 10% FBS (Invitrogen, 10500064) and 1x antibiotic/antimycotic (Invitrogen, 15240062) in a humidified incubator at 37°C with 20% O2 and 5% CO2. For passaging, LM2 cells were washed once with D-PBS (Invitrogen, 14190169) and detached using 0.25% Trypsin (Invitrogen, 25200056). Human CTC-derived BR16 cells and their Cas9-GFP-expressing derivatives were maintained under hypoxic conditions (5% O2) in ultra-low attachment (ULA) 6-well plates (Corning, 3471-COR), T-75 flasks (Corning, CLS3814) or CellSTACK (Corning, CLS3303). CTC growth medium containing 20 ng/mL recombinant HER2 (Gibco, PHG0313), 20 ng/mL recombinant human FGF (Gibco, 100–18B), 1x B27 supplement (Invitrogen, 17504–044), and 1x antibiotic-antimycotic (Invitrogen, 15240062) in RPMI-1640 Medium (Invitrogen, 52400–025) was added every third day.
Generation of Cas9-GFP–expressing human CTC-derived BR16 and LM2 cells
MDA-MB-231 (LM2) and human CTC-derived cell line (BR16) were transduced with the Cas9-NLS-FLAG-2A-EGFP (Cas9-GFP) lentivirus at a low multiplicity of infection (MOI < 1). GFP-positive cells were sorted either with the ARIAIII (BD Biosciences) for LM2 cells or with the Influx Cell Sorter (BD Biosciences) for BR16 cells, and individual GFP-positive single cells were seeded in 96-well plates and cultured as clonal cell lines. Multiple clones were established and Cas9 expression was confirmed by Western blot using antibodies against Cas9 (BioLegend, 844301). Clonal cell lines with 100% GFP-positive cells and detectable Cas9 protein expression were kept, whereas those with lower GFP expression or lower Cas9 expression were discarded. On the basis of this, we generated LM2-Cas9-GFP-C18, LM2-Cas9-GFP-C32, BR16-Cas9-GFP-A2, and BR16-Cas9-GFP-H3 cell lines. In addition, all Cas9-expressing cell lines were transduced with lentiviruses carrying EF1a-LUC-RFP plasmid (Biosettia, GlowCell-15–1) for in vivo tracking through bioluminescence imaging.
Knockout efficiency test for Cas9-GFP–expressing BR16 and LM2 cells
To test whether gene amplification interferes with the knockout efficiency of the CRISPR/Cas9 system, BR16-Cas9-GFP-A2/H3 and LM2-Cas9-GFP-C18/C32 cells were transduced with lentiviruses carrying hPGK-RFP plasmid (Addgene, 26001) at increasing MOI. Subsequently, 3 different sgRNAs targeting the RFP gene were transduced at low MOI (<0.4). After seven days of puromycin selection, the percentage of RFP-positive cells was measured using the CytoFLEX (Beckman Coulter) and analyzed using FlowJo software. All sequences for protospacer oligos used for RFP knockout are listed in Supplementary Table S1.
Viral production of genome-wide sgRNA library (GeCKOv2)
To produce virus, hGeCKOa and hGeCKOb plasmid pools (Addgene, #1000000049) were co-transfected into HEK293T cells with lentiviral packaging plasmids pMDLG, pMD2G and pRSV-Rev (Addgene #12251, #12259, #12253). HEK293T cells were cultured in DMEM medium (as described above) and seeded in CellStacks (Corning, CLS3313) the day before transfection. For transfection of one CellStack, 40 μg hGeCKOa+b, 40 μg pMDL.G, 20 μg pMD2.G, and 14.4 μg pRSV-Rev were mixed and diluted with ddH2O to a total volume of 1,580 μL. 1 mol/L CaCl2 (Sigma-Aldrich, C7902) was added dropwise to the plasmid-mix during vortexing with low speed. CaCl2-plasmid-mix was then transferred dropwise to 1,580 μL 2xHBS during vortexing and incubated for 15 minutes at room temperature. Subsequently, 500 mL DMEM medium was mixed with 2,250 μL chloroquine (Sigma-Aldrich, C6628) and the CaCl2-HBS-plasmid-mix and added to the cells. Medium was exchanged with freshly prepared DMEM medium 6 hours after transfection. Viral particles were harvested after 72 hours later by filtering the viral supernatant through a 0.22-μm Steriflip-GP filter (Sigma-Aldrich, Z660493) and immediately snap-frozen in liquid nitrogen and stored at −80°C until usage.
Genome-wide sgRNA library transduction of Cas9-GFP–expressing CTC-derived BR16 cells
Each batch of virus was titrated by transduction of 1.2 × 105 BR16-Cas9-GFP-A2 or BR16-Cas9-GFP-H3 cells per well in a 6-well plate with different dilutions of the virus in each well. For each viral concentration, two replicate wells were seeded. After 24 hours, medium containing 2 μg/mL puromycin (Invivogen, ant-pr-1) was exchanged in one of the replicate wells. A non-virus control was always included and untransduced control cells did not survive 72 hours puromycin treatment. After puromycin selection, live and dead cells were counted, to identify the viral volume that resulted in 30% of cells surviving in puromycin-containing medium, corresponding to an MOI of 0.3 and a single-infection percentage (SIP) of 83% if the infection events are considered as independently-occurring. The SIP was calculated directly from the puromycin survival (Psurvival) as described previously (15). For each transduction replicate of the GeCKOv2 (a+b), a total of 1.2 × 108 cells were infected at MOI = 0.3, and cultured under puromycin at 2 μg/mL for seven days. After this selection, 6 × 107 cells were spun down at 800 rpm for 6 minutes and used for genomic DNA extraction, whereas the remaining cells were used for transplantation in mice.
Individual knockout of candidate genes in Cas9-GFP–expressing LM2 and BR16 cells
sgRNAs targeting individual candidate genes were cloned into the lentiGuide-Puro plasmid (Addgene, 52963). Lentiviral particles carrying individual sgRNAs were produced by transfection of HEK293T cells applying the same procedure as described previously for the hGeCKOv2 (a+b) library lentivirus production. After harvest, BR16-Cas9-GFP-A2 and LM2-Cas9-GFP-C32 were transduced with individual sgRNAs at an MOI = 0.3. 24 hours after transduction, cells were maintained under puromycin selection for seven days. After selection, a portion of cells, each carrying a single sgRNA, was collected for RNA isolation using RNeasy Mini Kit (Qiagen, 74104), following the manufacturer's instructions and subsequently used for cDNA production and RT-qPCR analysis. The remaining portion of cells was directly used for transplantation in mice. All sequences for sgRNA oligos used for individual knockout cell lines are listed in Supplementary Table S1.
Real-time qPCR
Total RNA (1 μg) was treated with DNaseI (Promega, M6101), then reverse-transcribed using Superscript II (Thermo Fisher Scientific, 18064014), dNTPs (Invitorgen, 10297–018), and Random Hexamers (Invitrogen, C118A). Transcripts were quantified by real-time PCR using SYBR Green PCR MasterMix (Invitrogen, 4367659). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used for normalization. PCR assays were performed in duplicates, and the percentage of remaining gene expression was calculated using the comparative Ct method (ΔΔ Ct). Primers used for RT-qPCR are listed in Supplementary Table S5.
Cluster size measurement
Upon generation of individual loss-of-function BR16-Cas9-GFP-A2 cells, 40 μL of cell suspension was transferred into a 96-well black/clear tissue culture treated plate (BD Falcon, 353219) and stained for 1 hour at 37°C with a final concentration of 4 mmol/L Hoechst 34580 (Invitrogen, H21486), 2 mmol/L TMRM (Invitrogen, T668), and 4 mmol/L TOTO-3 (Invitrogen, T3604). For each plate, a negative control (clusters of BR16-Cas9-GFP-A2 cells harboring nontargeting sgRNA) and a positive control (40-μm filtered BR16-Cas9-GFP-A2 cells) were included. All measurements were done in quadruplicates. Plates were scanned using Operetta High Content Imaging System (PerkinElmer) and CTC cluster size analysis was performed using Columbus Image Data Storage and Analysis System (PerkinElmer) as previously described (6).
Mouse experiments
All mouse experiments were carried out in agreement with institutional and cantonal guidelines (approved mouse protocols #2781 and #3053, cantonal veterinary office of Basel-City). Maximal approved tumor volumes of 2,800 mm3 were never exceeded. Eight-to-10-week-old female NSG (NOD-scid-Il2rgnull) mice (The Jackson Laboratory) were anesthetized with isoflurane and injected with either 1 × 106 BR16, 1 × 107 untransduced or hGeCKOv2 (a+b)-transduced BR16-Cas9-GFP-A2 or BR16-Cas9-GFP-H3, 1 × 106 BR16-Cas9-GFP-A2 or LM2-Cas9-GFP-C32 individual loss-of-function cells into the right mammary fat pad. In all cases, before injection, cells were resuspended in 100 μL of 1:1 PBS (Invitrogen, 14190169) and Cultrex PathClear Reduced Growth Factor Basement Membrane Extract (R&D Biosystems, 3533–010–02). Primary tumors, single CTCs, CTC clusters, and organs with metastasis were sampled 22–28 weeks upon injection for NSG/BR16, NSG/BR16-Cas9-GFP-A2 or NSG/BR16-Cas9-GFP-H3 and eight weeks upon injection for NSG/LM2-Cas9-C32-individual loss-of-function. For RNA-seq of cells originating from metastatic lesions, 7 × 105 cells were injected intravenously in NSG mice and organs with metastasis were sampled 12–14 weeks upon injection. For PLK1 KO seeding capacity evaluation, 2 × 105 LM2-Cas9-GFP-C32 cells expressing a PLK1 or non-targeting sgRNAs were injected intravenously via the tail vein of NSG mice and metastatic growth was followed over time via bioluminescence imaging. For all orthotopic intracranial-injections, NSG mice at age between 7 and 10 weeks were anaesthetized by inhalation of isoflurane and positioned in a stereotaxic apparatus (David Kopf Instruments). For the stereotactic intracranial injection, the surgical site was shaved and prepared with 70% ethyl alcohol. A midline incision was made and a 1-mm diameter hole, centered 2-mm posterior to the coronal suture and 2-mm lateral to the sagittal suture, was drilled. Two microliters of PBS containing 2.0 × 105 MDA-MB-231-GFP-Luc cells or 4 mL of PBS containing 4.5 × 105 BR16-RFP-Luc cells were injected into the striatum (0-mm anteroposterior, 2.5-mm lateral to the bregma, and 4-mm below the surface of the skull) using a Hamilton syringe (Hamilton). All bioluminescence imaging of primary tumors and organs with metastasis was conducted with IVIS Lumina II (PerkinElmer). Blood with CTCs was obtained by cardiac puncture and up to 1 mL was collected and applied through the Parsortix (Angle) device. CTCs from all xenograft models were identified through ectopic GFP and RFP signals via fluorescence microscopy. For PLK1 inhibitor treatment, BI2536 (MCE) was dissolved in DMSO, diluted in 0.9% NaCl + 0.1N HCl and administered intravenously at 40 mg/kg for two consecutive days per week for one (BR16-GFP-Luc bearing mice) or two (MDA-MB-231-GFP-Luc bearing mice) cycles. For HDAC inhibitor treatment, belinostat (MCE) was dissolved in DMSO and diluted in L-arginine (100 mg/mL) in ddH2O and administered intraperitoneally at 80 mg/kg for five consecutive days per week for three cycles. For Rho GTPases inhibitor treatment, rhosin (MCE) was dissolved in DMSO and diluted in 0.9% NaCl solution and administered intraperitoneally at 20 mg/kg once per day, each day for three weeks.
Setup of sgRNA minipool library (intravasation sgRNA library) for validation
The intravasation sgRNA minipool library comprises 54 sgRNAs from 9 hit genes identified with the genome-wide pooled screen and 50 nontargeting control sgRNA (Supplementary Table S9). The 9 hit genes were selected upon the following criteria: LogFC > −3, P < 0.05, expression in patient-derived CTCs, correlation of their gene expression with worst overall survival (OS), observation of depletion in CTCs in at least 4 mice and they possessing an extracellular domain or are secreted or having a kinase domain. The sgRNA minipool library was synthesized by GenScript and amplified following the procedure previously shown in Joung and colleagues (22). Following amplification, the minipool library was sequenced to verify full representation of total sgRNA complexity and lentivirus was generated with the same procedure as for the hGeckov2 library described above. As before, sgRNA minipool library viruses were titered by transduction of 5 × 105 LM2-Cas9-GFP-C32 cells in a ULA 6-well plate or 10-cm dish, respectively, with increasing volume of lentivirus (including no virus control). Subsequent selection with 1 μg/mL puromycin (Sigma) for seven days identified the viral volume and number of cells to achieve an MOI of 0.2. Representation for the minipool screen was above 1,000-fold. Using viral volumes resulting in an MOI of 0.2, LM2-Cas9-GFP-C32 were transduced and selected for 7 days with 1 μg/mL puromycin. Subsequently, cell samples were taken for genomic extraction and the rest of the cells was used for transplantation into the mammary fat pad of NSG mice (1 × 106 cells per injection). After 8–10 weeks after transplantation, mice were sacrificed, primary tumors were dissected and single CTCs and CTC clusters were isolated from mouse blood and subsequently genomic DNA from all samples was isolated as described above. Subsequent next-generation sequencing (NGS) was used to readout sgNRA representation as described below.
Flow cytometry–based separation of single CTCs and CTC clusters
Flow cytometry–based CTC separation was performed using the BD Influx cell sorter equipped with a 200-μm nozzle and the BD FACS Software. To identify the sample pressure that does not break CTC clusters during the separation process, as well as to define the gating strategy to efficiently separate single cells from clustered cells, BR16-GFP single cells were mixed with BR16-RFP clustered cells in a ratio of 1:1 and directly applied to the Influx cell sorter. RFP-positive cells found in the single cell sort and that could only be derived from broken clustered cells were quantified using fluorescence microscopy (Leica DMI4000). With this method, we identified a sample pressure of ≤5 PSI to be optimal for the separation of single and clustered CTCs with our protocol. These parameters were applied for all following separations of single CTCs versus CTC clusters originating from xenograft models used in this study.
Genomic DNA extraction from cells, CTCs, and xenograft tissues
For gDNA extraction of xenograft primary tumors and organs with metastasis, samples were first homogenized in lysing matrix tubes (MP Bioscience, 116925500) using the FastPrep Instrument (MP Bioscience). Of note, samples were corresponding to the entire tumor tissue (primary and/or metastatic) as well as the entire CTC population. Subsequently, homogenized tissue samples or 6 × 107 cells were transferred in a 50-mL conical tube and incubated with 18 mL of lysis buffer (50 mmol/L Tris, 50 mmol/L EDTA, 1% SDS, pH 8) supplemented with 100 μL of 20 mg/mL Proteinase K (Qiagen, 19131) at 55°C overnight. Afterwards, 100 μL of 100 mg/mL RNase A (Qiagen, 19101) was added to the lysed sample, inverted 30 times and incubated at 37°C for 30 minutes. Subsequently, samples were immediately put on ice before addition of 6 mL pre-chilled 7.5 mol/L ammonium acetate (Sigma, A1542). The samples were then thoroughly vortexed and then centrifuged at 4,400 × g for 10 minutes. After centrifugation, the supernatant was carefully decanted into a new 50 mL conical tube without transferring the pellet. To precipitate genomic DNA, 18 mL of 100% 2-propanol (Sigma-Aldrich, 59304) was added to each tube, inverted at least 50 times and centrifuged at 4,400 × g for 10 minutes. After decanting the supernatant, 12 mL of freshly prepared 70% ethanol (Sigma-Aldrich, 51976) was added; the tubes were inverted at least 10 times, and then centrifuged at 4,200 × g for 4 minutes. The supernatant was then discarded. After air drying for 20 minutes under a PCR cabinet with laminar air flow, 1.5 mL nuclease-free H2O (Thermo Fisher Scientific, AM9932) was added and the tubes were incubated at 65°C for 1 hour to dissolve the gDNA. gDNA from CTC samples was extracted using the QIAmp DNA Micro Kit (Qiagen, 56304), following the manufacturer's instructions. The gDNA concentration was measured using a Nanodrop (ThermoFisher) and stored at −20°C until usage.
Deep sequencing and sgRNA library readout
The sgRNA library readout was performed using the protocol previously described in Chen and colleagues (15). In brief, a two-step PCR was performed, where the first PCR (PCR#1) amplifies a broad region containing the sgRNA sequence using specific primers that recognize the plenti-Guide-PURO vector (Addgene, 52963). For PCR#1, the amplification parameters are: 98°C for 30 seconds, 24 cycles of (98°C for 1 s, 62°C for 5 s, 72°C for 35 s), and 72°C for 1 minute. In each PCR#1 reaction for primary tumor, organs with metastasis and cells, 1.5 μg of gDNA was used. For CTC samples, 20 μL of eluted gDNA for each PCR#1 reaction was used. To ensure full capture of sgRNA complexity for each sample, the appropriate number of PCR#1 was calculated as described previously in Chen and colleagues (15). All PCR#1 reactions for each individual sample were pooled and 10 μL were used for each individual PCR#2 reaction. PCR#2 was conducted using barcoded PCR primers, including Illumina adapters P5 and P7. All PCR#1 and PCR#2 reactions were performed using Phusion Flash High Fidelity Master Mix (Thermo Fisher Scientific, F548L). PCR#2 products were pooled and gel purified from a self-casted 1.5% ultra-pure agarose gel (Invitrogen, 16500500) using the QIAquick Gel Extraction Kit (Qiagen, 28706). The purified libraries from each biological sample were quantified with dsDNA High-Sensitivity Qubit (Life Technologies). Diluted libraries were pooled according to the number of necessary reads for each sample and sequenced together with 20% PhiX DNA on the Illumina NextSeq 500 sequencer.
Immunofluorescence staining
Primary tumors and organs with metastasis were fixed for 24 hours in 4% PFA at 4°C. The tissue pieces were then cut, embedded in 4% low-gelling temperature agarose, and subsequently sectioned (50–100-μm-thick sections) using a vibratome (Leica VT1200 S). All steps were performed at room temperature with gentle rocking in double side adhesive silicon chambers (Grace Biolabs) glued on glass slides. Sections were blocked and permeabilized with TBS (final concentration 0.1 mol/L Tris, 0.15 mol/L NaCl, pH 7.5) containing 0.05% Tween-20, 20% DMSO, 1% Triton X 100 and 10% donkey serum (Jackson Immuno Research) for a minimum of 2 hours. This buffer was also used to dilute all primary and secondary antibodies. Primary antibodies were applied overnight. Secondary antibodies were applied for 2 hours. After extensive washing (with TBST), sections were mounted in home-made mounting medium (80% glycerol in TBS containing 0.2 mol/L N-propyl gallate, pH 8.5) using size 1.5 coverslips. Primary human cytokeratin (hCK; Milteny Biotec, 130–112–931), laminin (Novus Biologicals, NB300–144), CD31 (R&D Systems, AF3628), Ki67 (Invitrogen, 14–5698–82), glial fibrillary acidic protein (GFAP; Invitrogen, PA1–10019) antibodies and secondary IgG-Cy3 (Jackson ImmunoResearch, 712–165–153), IgG-594 (Invitrogen, SA5–10028), IgG-594 (Invitrogen, SA5–10040), IgG-555 (Invitrogen, A-31572), IgG-647 (Invitrogen, A-21447), and IgG-488 (709–545–149) antibodies were used.
Cytospin and staining of BR16-Cas9-GFP-A2 PLK1 knockout cells
A total of 2 × 103–5 × 103 BR16-Cas9-GFP-A2 cells transduced with either nontargeting control sgRNA or sgRNAs against PLK1 were spun down on a coated cytoslide (Shandon) at 500 rpm for 4 minutes. Subsequently, cells were fixed with 4% PFA solution for 15 minutes at room temperature. Slides with fixed cells were washed with PBS. Then cells were blocked for 1 hour at room temperature in blocking buffer [10% donkey serum (Millipore, S30), 1% BSA in H2O (Sigma, 05482), 0.5% Triton X-100 (Sigma, 93443), and 0.01% sodium azide (Sigma, S2002)]. Cells were washed with PBS after blocking and then incubated with primary antibodies in 3% donkey diluent buffer [3% donkey serum (Millipore, S30), 1% BSA in H2O (Sigma, 05482), 0.5% Triton X-100 (Sigma, 93443) and 0.01% sodium azide (Sigma, S2002)] for 1 hour at room temperature. Afterwards, cells were washed with PBS and then incubated with secondary antibodies that were diluted in 3% donkey buffer for 1 hour at room temperature. Again, cells were washed 2x with PBS before they were stained for DAPI and mounted using Prolong Gold. 3 hours after DAPI staining, cells were imaged using Leica DMI 4000 microscope. The following antibodies were used: Ki67 (Abcam, ab15580), cleaved caspase-3 (Cell Signaling Technology, CS9664S), EpCAM (Cell Signaling Technology, 5198), vimentin (Abcam, ab92547), rabbit polyclonal AF647 IgG (Thermo Fisher Scientific, A-31573).
Confocal microscopy
Confocal microscopy was performed on a Leica TCS SP8 microscope equipped with three photomultiplier tubes, two HyD detectors, three lasers ([405 nm, Argon Laser (458, 476, 488, 496, and 514 nm)] and a white light laser using Leica type G immersion liquid and a ×20 glycerol immersion lens (NA 0.75) as previously described (23). All scans were acquired at 20°C–25°C, 400 Hz, in the bidirectional mode, with z-spacing of 2.5 at 1,024 × 1,024 pixel resolution. Images were acquired in 8 bit. For signal acquisition, only HyD detectors were used.
sgRNA analysis
Sequencing reads were demultiplexed and trimmed using cutadapt (v1.16). Only reads with a minimum quality of 20 in both ends and 20nt, corresponding to the sgRNA spacer sequences length, were retained. Final reads were mapped to the human GeCKO library using bowtie2 (v2.3.4.2), allowing for one mismatch. For distribution and complexity analysis, only sgRNA spacers with at least five assigned reads were analyzed. To correct for differences in library size and count distribution, counts were normalized using median ratio method (24). To identify depleted genes between two conditions, sgRNA counts were aggregated at gene level. Genes detected in at least 3 mice with ≥ 30 counts in one of the conditions were analyzed with DESeq2 (25). Genes with a log2-fold change (LFC) ≤ −3 and adjusted P ≤ 0.01 were defined as dropouts. LFCs of −30 represent cases with 0 counts in all the replicates of one of the groups. In those cases, the manual calculation of the fold change will give us a result of Infinity, which is assigned with a predetermined large value of 30 to the LFC. To assess whether the genes were expressed in CTCs, normalized gene expression data were downloaded from GSE111065 (6). A gene was considered expressed if the average counts per million (cpm) was larger than zero in each specific sample group (e.g., CTC clusters of BR16-CDX or patients). The effect of each gene on OS in patients with breast cancer from The Cancer Genome Atlas (TCGA) assessed using precomputed Cox proportional hazards regression analysis downloaded from OncoLnc (http://www.oncolnc.org). For organotropism analysis, a list of organ-depleted sgRNA was obtained by comparing the abundances in each specific organ against other metastatic sites. For each mouse, dropouts were defined as absent in the query organ (0 counts) and detected in the other organs (reference organs) with an average of at least 30 counts. Dropouts across replicates were defined as sgRNAs depleted in a minimum of 3 mice and in at least 80% of cases, using as denominator the number of replicates with an average count of ≥30 in the query or the reference organs. Enriched sgRNA were defined in the same fashion than depleted but considering only sgRNAs with at least 30 counts in query organ and absent (0 counts) in all reference organs. Finally, genes with a minimum of 2 sgRNA depleted and 0 enriched were considered as organ-specific dropouts. For minipool libraries, raw reads were processed as described previously for GeCKO library. Then, the read counts of sgRNAs were first normalized at mouse level by the size factor derived from non-targeting control guides. Then, a mouse specific LFC was calculated only for guides with more than 30 read counts in the control condition. The LFC of each sgRNA across all mice was tested for negative selection against the population of nontargeting control sgRNAs using a one-sided Mann–Whitney U test. The median LFC by guide was normalized by subtracting the median LFC in nontargeting control guides. Then sgRNA results were aggregated at gene level by combining the P values using the Fisher method, adjusting by multiple comparisons using Benjamini–Hochberg (BH) procedure and using the median of normalized LFC. Genes with a Padjusted value of ≤ 0.05, at least 3 sgRNAs with LFC < 0, and a median LFC < 0 were categorized as validated.
RNA sequencing analysis
Quality assessment for RNA sequencing data was performed using FastQC (v.0.11.4), FastQ Screen (v.0.11.4), and visualized with MultiQC (v.1.7; ref. 26). Adaptor sequences, first 9 base pairs and low-quality ends were removed with Trim Galore (v.0.4.2). To eliminate any residual contamination from mouse RNA, trimmed reads were aligned to human (GRCh38) and mouse (GRCm38) genome references using STAR (v.2.5.2a; ref. 27) and ambiguous reads were removed using Disambiguate (28). Gene level expression was quantified with Salmon (29) using Ensembl transcripts (release 89). Samples were retained for further analyses if they had at least 10,000 genes with non-zero expression, less than 90% of counts from the 100 most abundant features and having a minority of reads mapping to mitochondrial genes. Size factor normalization was performed using the TrimmedMean of M values method (30) and the mouse of origin effect was removed using the removeBatchEffect function from Limma bioconductor package (31).
Survival analysis
Metastasis-free survival (MFS) analysis was performed on METABRIC molecular dataset downloaded from Rueda and colleagues (32). From the 582 patients who had a distant relapse we only include the 414 patients with a complete description of all recurrences. The final dataset consists of 1,736 patients with breast cancer, of which, 232 showing distant recurrences to bone, liver, brain or lymph nodes before 5 years. Signature scores were assigned to each patient by calculating the mean of the gene-level standardized expression (z-scores) across the genes found depleted in metastases of the CRISPR–CDX–BR16 model. Survival was assessed via univariate Cox proportional hazards regression models, using the “coxph” function of the R survival package. For visualization, the scores were divided into quartiles and compared using the Kaplan–Meier method.
Data analysis
Data analysis, statistical testing, and visualization were conducted in GraphPad Prism (v.8.2.1.), R (version 3.6.0; R Foundation for Statistical Computing), and bioconductor (v.3.9; ref. 33). Figure legends describe the statistical approach used for each analysis.
Data availability
Raw sequencing reads and the matrix of read counts from RNA-seq have been deposited in the Gene Expression Omnibus (GEO, NCBI) with accession number GSE138813.
Results
Generation and characterization of CTC-derived xenografts
We first established a human breast CTC-derived xenograft model (CDX-BR16) by mammary fat pad injection of CTCs obtained from the blood of a patient with progressing luminal breast cancer with overt metastasis to the bone, brain, liver and lymph nodes and refractory to endocrine therapies (Fig. 1A; Supplementary Fig. S1A and S1B). Of note, somatic mutational analysis of the BR16 CTC-derived cell line confirmed the presence of 95.8% of mutations that were identified in freshly isolated CTCs of the same patient; although clonal mutations are identified as expected either in homozygosity (variant allele frequency close to 100%) or heterozygosity (variant allele frequency around 50%), the majority of mutations (>74%) are present below a variant allele frequency of 20%, highlighting the maintenance of the original heterogeneity of BR16 CTCs (Supplementary Fig. S1C). Primary tumor development in mice followed a slow growth kinetic—as typically observed for human breast tumors—and resulted in tumors with a mean size of 212.4 mm3 (5 months after injection) with analogous histological features compared with the patient's original tumor tissue (Supplementary Fig. S1D and S1E). Importantly, despite the small primary tumor size, CDX-BR16 mice spontaneously developed single and clustered CTCs, as well as proliferative metastatic deposits in bone, brain, liver and lymph nodes, as highlighted by staining with EpCAM, HER2 and EGFR antibodies (CTCs) or human cytokeratin (primary tumor and all metastases; Fig. 1B and C; Supplementary Fig. S1F).
Figure 1.
Generation and characterization of a CTC-derived xenograft model. A, Schematic of the establishment of human CTC-derived xenografts (CDX) from a patient with breast cancer (BR16). CTCs from BR16 patient blood were isolated, expanded ex vivo, and injected in the mammary fat pad of NSG mice. Upon tumor development, spontaneously formed CTCs seed metastases to defined organs, mirroring the metastatic pattern of the patient-of-origin. B, Representative pictures of primary tumor, single CTCs, and CTC clusters from the BR16-CDX model. The primary tumor was stained for human pan-cytokeratin (hCK; green), laminin (turquoise), CD31 (red), Ki67 (purple), and DAPI (nuclei; white), whereas single CTCs and CTC clusters were live-stained for epithelial cell adhesion molecule (EpCAM), HER2, and EGFR (green) within the microfluidic cassette, directly after capture. C, Representative pictures of spontaneous bone, brain, liver and lymph node metastasis of BR16-CDX mice, stained for hCK (green), laminin (turquoise), glial fibrillary acidic protein (GFAP; turquoise), CD31 (red), Ki67 (purple), and DAPI (white).
In vivo loss-of-function screen in CTC-derived xenografts
We then used this model to engineer an in vivo genome-wide loss-of-function CRISPR screen to identify genes that are required for (i) primary tumor growth, (ii) generation of single CTCs and CTC clusters, and (iii) organotropism of disseminated breast cancer cells to brain, bone, liver and lymph nodes. Particularly, we transduced BR16 cells with lentiviruses carrying Cas9-GFP (MOI<1) and selected two clones with positive Cas9-GFP protein expression (namely, BR16-Cas9-GFP-A2 and BR16-Cas9-GFP-H3; Supplementary Fig. S2A). We then assessed the efficiency of genome editing in both clonal cell lines by transduction of a hPGK-H2B-RFP vector at increasing MOI (ranging from 0.3 to 4, aiming to mimic gene amplification events that are typical in breast cancer (34, 35), followed by transduction of a nontargeting sgRNA control or three different sgRNAs targeting RFP (Supplementary Table S1) and subsequent FACS analysis to determine the remaining percentage of RFP-positive cells (Supplementary Fig. S2B). Both clonal BR16 lines transduced with RFP-targeting sgRNAs showed significant decrease of RFP positive cells in all conditions (Supplementary Fig. S2C and S2D), highlighting the efficiency of gene editing in our primary, patient-derived CTC model even in the presence of multiple copies of the same gene. We next transduced BR16-Cas9-GFP-A2 and BR16-Cas9-GFP-H3 clones with a pooled genome-wide human sgRNA library (hGeCKOv2a+b; ref. 36) in four independent infection replicates, each comprising the full library complexity of 122,411 sgRNAs at 1,000 cells per sgRNA (6 sgRNAs per gene, 4 sgRNAs per microRNA precursor, and 1,000 control sgRNAs). Upon seven days of puromycin selection, mutant cell pools were injected into the mammary fat pad of 70 NSG mice (5 to 10 mice per infection replicate) to create a CRISPR–CDX–model (Fig. 2A). As control, untransduced BR16-Cas9-GFP-A2 and BR16-Cas9-GFP-H3 cells were injected into 5 mice each. Primary tumors of mice harboring the sgRNA library grew with a similar rate yet reached a later endpoint compared with the control group, suggesting intra-tumor selection processes, as expected (Supplementary Fig. S3A), whereas the abundance of single and clustered CTCs as well as the metastatic pattern to brain, bone, liver, and lymph nodes were unaltered (Supplementary Fig. S3B–S3D). We then harvested the primary tumors, CTCs and metastatic organs from each mouse at 22–28 weeks upon injection and processed them for genomic DNA extraction and next-generation barcode sequencing (NGS) to quantitate sgRNA representation at different steps of the metastatic cascade (Supplementary Fig. S4A). Particularly, to analyze the sgRNA representation in both single CTCs and CTC clusters, we established a FACS-based separation procedure upon microfluidic CTC enrichment, using ultra-low sample pressure (<5 PSI) and an optimized gating strategy based on multicolor spike-in experimental controls, resulting in high-purity single CTCs (>94%) and CTC cluster (100%) isolation (Supplementary Fig. S4B and S4C). We then determined the sgRNA representation in all CRISPR–CDX-derived samples (n = 475), aiming to identify selectively depleted sgRNAs across different clones and infection replicates, pinpointing genes that are potentially required for individual steps of the metastatic cascade and for organotropism in breast cancer. Of note, sgRNA complexity in BR16 cells (before injection into mice) showed high concordance between all infection replicates [median Spearman correlation coefficient (ρ) = 0.82, interquartile range (IQR) = 0.15, n = 8; Supplementary Fig. S5A]. After 22–28 weeks of spontaneous tumor progression and in vivo sgRNAs selection, despite expected mouse-to-mouse variations and random sgRNA drift during the time required for efficient metastatic colonization, we still observed concordance between individual mice concerning primary tumors (median ρ = 0.3, IQR = 0.09, n = 69; Supplementary Fig. S5B), single and clustered CTCs (median ρ single CTC = 0.37, IQR = 0.18, n = 55; median ρ CTC cluster = 0.34, IQR = 0.19, n = 55; Supplementary Fig. S5C and S5D) as well as metastasis (median ρ Bone = 0.50, IQR = 0.11, n = 69; median ρ Brain = 0.49, IQR = 0.37, n = 69; median ρ Liver = 0.46, IQR = 0.50, n = 69; median ρ Lymph node = 0.48, IQR = 0.12, n = 69; Supplementary Fig. S5E–S5H). We then compared the overall number of detected sgRNAs in all samples, and found that although cells (one-week after sgRNA library transduction, before injection in mice) are characterized by a nearly full complexity (a mean of 115,142 detected sgRNAs, corresponding to 94% of the initial pool), sgRNA representation is decreased in early primary tumors (12 weeks in vivo), highlighting an ongoing selection process, as expected (Fig. 2B; Supplementary Fig. S6A). Despite this large bottleneck, we identified an average number of 3.93 (sd = 1.43) remaining sgRNAs per gene in late primary tumors (Supplementary Fig. S6B). Using gene ontology (GO) analysis, we found that sgRNAs with significantly decreased abundance in early primary tumor compared with cells comprise genes involved in essential cellular processes, such as RNA processing, ribosomal proteins, transcription regulation, as well as genes involved in proliferation, such as spindle chromosome attachment, kinetochore assembly and centrosome duplication, highlighting the expected selection processes during primary tumor development (Supplementary Fig. S7A and S7B; Supplementary Table S2). sgRNA representation analysis confirmed stable complexity levels in corresponding late primary tumors (22–28 weeks in vivo), which then decreased in corresponding CTCs and metastases, suggesting a further major selection step at the time of CTC intravasation (Fig. 2B) and consistent with the expectation that only a minority of tumor cells are capable to intravasate. Of note, as internal control, we observed that the number of detected sgRNAs in CTCs did not exceed the number of sequenced cells, suggesting that virtually all cancer cells contain one sgRNA, as initially projected (Supplementary Fig. S6C). In addition, when calculating sgRNA distribution across all samples (Gini index), we observe a more balanced sgRNA distribution in CTCs compared with all metastatic sites, arguing for further organ-specific selection processes downstream of the circulatory system (Supplementary Fig. S6D). This notion is confirmed at the level of individual mice when interrogating the dynamics of representative sgRNAs across the entire metastatic process, that is, highlighting the occurrence of sgRNA dropouts and enrichments in a site-specific fashion (Fig. 2C). Most importantly, when assessing overall sgRNA correlation and visualizing it through unsupervised hierarchical clustering, we note that samples strongly cluster by tissue type, independently of the specific clone, mouse or infection replicate, clearly pointing toward the occurrence of defined biological processes as the major driving force that shapes sgRNA complexity (Fig. 2D). Moreover, when comparing the sgRNA correlation (average Spearman ρ) of samples derived from the same tissue (i.e., within tissue) and of samples from different tissue types (i.e., between tissue), we observe that similarity is higher in samples from the same origin (Supplementary Fig. S6E). Thus, our CRISPR–CDX model provides a framework to interrogate biology-driven genetic dependencies of individual steps of the metastatic cascade in models that spontaneously recapitulate a human metastatic disease. Although this approach is strictly related to the specific genetic background of the donor patient, it provides a clinically relevant resource to interrogate metastasis vulnerabilities.
Figure 2.
In vivo loss-of-function genome-wide CRISPR screen in human CTC-derived xenografts. A, Schematic of in vivo genome-wide loss-of-function CRISPR screen in CRISPR-CDX-BR16 mice. BR16-Cas9-GFP cells were transduced with hGeCKOv2 sgRNA library and injected in the mammary fat pad of NSG mice. After 22–28 weeks, primary tumor, single CTCs, CTC clusters, and organs with metastasis were isolated and subjected to NGS for sgRNA readout. B, Violin plot showing the number of detected sgRNAs (threshold ≥ 5 reads per sgRNA) in cells before injection, early primary tumors (12 weeks after transplantation), late primary tumors, single CTCs, CTC clusters, and organs with metastasis (bone, brain, liver, lymph node; all 22–28 weeks after transplantation); n = 8 for cells, n = 10 for early primary tumor, n = 69 for late primary tumor, n = 55 for single CTCs, n = 59 for CTC clusters, n = 69 for bone, n = 68 for brain, n = 68 liver, and n = 69 for lymph nodes. C, Representative plots showing sgRNA population dynamics along the metastatic process in individual mice. Colored areas represent counts per million (CPM) of sgRNAs detected in multiple sample categories. D, Unsupervised hierarchical clustering of the pairwise Spearman correlation of log2 sgRNA normalized counts from cells before injection (cells), late primary tumor, single CTCs, CTC clusters, and organs with metastasis (bone, brain, liver, and lymph node), showing high concordance by sample type.
Loss-of-function of IL18R1, ITGA2, CSNK1A1L, and CSNK2A2 decreases CTC cluster generation and metastatic burden in vivo
We next investigated sgRNA dropouts comparing single CTCs and CTC clusters, focusing on those sgRNAs that are found in single CTCs but are selectively depleted in CTC clusters, that is, aiming at the identification of genes whose knockout would interfere with CTC clusters generation (Fig. 3A). We then evaluated differential abundance at gene-level using DESeq2, considering only those sgRNAs that have at least 30 reads in single CTCs or CTC clusters (Fig. 3B). To reduce the impact of outliers, the differential abundance analysis was followed by a heuristic approach that was validated using cross-validation analysis based on random sampling. We divided mice into two nonoverlapping random groups (50 times for each analysis) and analyzed which proportion of the genes that were identified in each subgroup was overlapping with our previously identified genes. This analysis shows high consistency of the identified genes and of the fold-change (log2) in each of the 50 iterations between the two subgroups (Supplementary Fig. S8A and S8B). We found 365 genes specifically depleted in CTC clusters (Fig. 3B; Supplementary Table S3), of which, 256 were also found to be expressed in CTC clusters of patients with breast cancer (n = 6 patients, n = 13 CTC clusters) and from the CDX models (n = 47 from BR16-CDX and n = 10 from LM2-CDX; Supplementary Table S4; ref. 6). Among these, we identified 44 genes coding for proteins that could be targetable pharmacologically, that is, having either a kinase domain, an extracellular domain or that are secreted in the extracellular compartment (Fig. 3B). We then selected four candidates with extracellular domain (CDH7, IL18R1, ITGA2, IL1RAPL1) and two candidates with a kinase domain (CSNK1A1L, CSNK2A2) for further validation. Although the candidates with extracellular domain were prioritized on the basis of their biological function (e.g., part of a well-defined cell–cell junction complex or receptors for targetable cytokines), candidate kinases were chosen on the basis of the co-occurrence of several members of the same family within the 44 short-listed genes, yet with unknown biological function in the metastatic process. We then generated individual loss-of-function BR16-Cas9-GFP-A2 cells using two sgRNAs for each of the candidates (Supplementary Tables S1; Supplementary Fig. S9A) and measured CTC cluster size in suspension cultures with a dedicated assay (6), comparing the results to BR16-Cas9-GFP-A2 cells expressing a nontargeting sgRNA (Fig. 3C). In line with our in vivo screen results, knockout of all candidates individually yielded a significant reduction in CTC cluster size compared with controls, without affecting cellular viability (>79%; Fig. 3D; Supplementary Fig. S10). We next sought to extend our findings to an independent model and generated Cas9-GFP–expressing human breast cancer MDA-MB-231 cells (lung metastatic variant, referred to as LM2; ref. 37), which we further validated for their knockout efficiency (Supplementary Fig. S2A, S2B, and S2D). We then transduced LM2-Cas9-GFP-C32 cells with individual sgRNAs targeting candidate genes (Supplementary Fig. S9B) and injected them in the mammary fat pad of NSG mice, aiming to assess primary tumor development and spontaneous generation of single and clustered CTCs, as well as metastasis formation (Fig. 3E). Mice transplanted with either individual knockout cells or cells harboring a nontargeting sgRNA developed primary tumors with resembling growth kinetics and tumor size (Supplementary Fig. S9C). Remarkably, individual gene knockouts highlighted a general trend for all genes toward reduction of CTC cluster-shedding rate (Fig. 3F), in line with our in vitro assay. Of note, IL18R1, ITGA2, CSNK1A1L, and CSNK2A2 stood out as targetable genes whose knockout most significantly reduced CTC cluster formation as well as spontaneous metastasis in vivo (Fig. 3F–H), and rescue experiments with ectopic re-expression of these genes confirmed their requirement for the observed phenotype (Supplementary Fig. S11A–S11C). Thus, our genome-wide loss-of-function approach followed by individual gene validation enabled the identification of new genes involved in CTC cluster formation in the tested models, whose targeting leads to metastasis reduction.
Figure 3.
Identification of targetable genes required for CTC cluster generation. A, Schematic of the experimental design. sgRNA that are enriched in single CTCs but depleted in CTC clusters are identified, highlighting genes that are required for CTC cluster formation. B, Plot showing, at gene-level, the fold change and the average of normalized read counts over all CTC samples. The counts for each gene were obtained through sgRNA counts aggregation (see Materials and Methods). Depleted genes (≤ −3 log2-fold change; adjusted P ≤ 0.01) coding for proteins possessing an extracellular domain are shown in purple, kinases are shown in red, and others are shown in gray. Gene names highlighted with a border were selected for validation. C, Schematic describing in vitro validation of candidate genes by generation of BR16-Cas9-GFP-A2 individual loss-of-function cell lines and subsequent cluster size measurement with a dedicated assay. D, Plot showing the mean cluster size (area in μm2) of BR16-Cas9-GFP-A2 cells that were filtered through a 40-μm cell strainer (positive control; n = 12), transduced with a nontargeting sgRNA (negative control; n = 12), or transduced with sgRNAs targeting individual candidate genes (n = 4). The heatmap shows the mean percentage (%) of viability in each condition compared with negative control cells. E, Schematic showing the experimental design for in vivo validation of candidate genes. LM2-Cas9-GFP-C32 cells carrying sgRNAs targeting individual candidate genes were injected in the mammary fat pad of NSG mice. Upon tumor growth, the percentage of single CTCs and CTC clusters as well as the lung metastasis burden were assessed. F, Bar plot showing the percentage (%) of single CTCs and CTC clusters in mice injected with LM2-Cas9-GFP-C32 transduced either with nontargeting control sgRNA or with individual sgRNAs targeting candidate genes (n = 4). G, Plot showing the metastatic index of LM2-Cas9-GFP-C32 xenografts expressing a nontargeting control sgRNA or individual sgRNAs targeting candidate genes (n = 3–4). H, Representative images of the bioluminescence signal measured in the lungs of LM2-Cas9-GFP-C32 xenografts expressing a nontargeting control sgRNA or individual sgRNAs targeting candidate genes. *, P < 0.05 by two-tailed Student t test; ns, not significant. Error bars, SEM.
PLK1 is required for intravasation of both single CTCs and CTC clusters
We then sought to determine genes that are required for intravasation of all CTCs by identifying sgRNAs that are present in the primary tumor but depleted in both single CTCs and CTC clusters (Fig. 4A). For this purpose, we first calculated the log median of normalized read counts for all CTC samples and primary tumors, considering only those sgRNAs that have at least 30 reads, and then defined differential abundance (fold-change; Fig. 4B). We found sgRNAs specifically depleted in both single and clustered CTCs compared with the primary tumor, corresponding to 3,100 genes (Supplementary Table S6), of which, 2,137 are also expressed in CTCs of patients with breast cancer (n = 10 patients, n = 41 CTCs) and from the BR16 CDX model (n = 78 CTCs; Supplementary Table S7; ref. 6). We further stratified this gene list by asking if high expression of these genes in the primary tumor of patients with breast cancer from the TCGA database would confer a reduced OS. Among 2,136 genes, the expression of 109 genes correlates with a worse survival (adjusted P < 0.05; Supplementary Table S8). Among these, we identified 26 targetable genes, that is, having either a kinase domain, an extracellular domain or that are secreted in the extracellular compartment (Fig. 4B). We then extended these findings to an independent in vivo model, reasoning that validation would be imperative in this case given a potentially high false-positive rate when comparing sgRNA dropout rates in populations of cells with inherent differences in overall cell number (i.e., CTCs versus primary tumors). Thus, we conducted a sgRNA minipool screen in LM2-Cas9-GFP-C32, including 9 hit genes (PLK1, PANX1, HPSE, PAK6, IL27, PCDHGA1, PCDHGA2, BAMBI and IYD; represented by 6 sgRNAs for each gene and selected on the basis of the same parameters described above) that were significantly depleted in single CTCs and CTC clusters and 50 non-targeting sgRNAs, resulting in a total library complexity of 104 sgRNAs (Fig. 4C; Supplementary Table S9). Interestingly, 5 out of 9 (55.5%) of our identified candidate genes were still significantly depleted in CTCs compared with primary tumor (PLK1, PANX1, HPSE, PAK6, and IL27), highlighting the reproducibility of our in vivo loss-of-function screen (Fig. 4D). Importantly, only 2/50 (4%) nontargeting sgRNAs were significantly depleted in CTCs compared with primary tumor, suggesting a low false-positive rate (Fig. 4D). We selected two candidates with an extracellular domain that were not validated (PCDHGA1, PCDHGA2) and one kinase that was validated (PLK1) in our minipool experiment as significant hits, and further generated individual loss-of-function LM2-Cas9-GFP-C32 cells using two sgRNAs per candidate gene (Supplementary Table S1; Supplementary Fig. S12A). We then injected these cells into the mammary fat pad of NSG mice, for subsequent quantification of primary tumor growth, CTCs and metastasis. We found no difference in primary tumor growth kinetics or size at the time of CTC isolation in any of the knockouts (Supplementary Fig. S12B), and in agreement with minipool validation results, depletion of PCDHGA1 and its paralog PCDHGA2 did not affect CTC counts, ratios, nor metastasis formation (Fig. 4E; Supplementary Fig. S12C and S12D). Yet, knockout of PLK1 led to a remarkable reduction of both single CTCs and CTC clusters as well as metastasis formation in vivo (Fig. 4E; Supplementary Fig. S12C and S12D). As PLK1 has been proposed to play a role in mitosis, EMT and its dysfunction can promote malignant proliferation (38), we further investigated whether decreased CTC numbers are related to proliferation, apoptosis or impaired DNA integrity. We found no difference in the mean percentage of Ki67-, cleaved caspase-3-, Annexin V-, vimentin- and EpCAM-positive cells between nontargeting control and PLK1 knockout BR16-Cas9-GFP-A2 cells in vitro (Supplementary Fig. S13A–S13H), and DNA integrity was also not impaired (Supplementary Fig. S13I), in line with previous reports highlighting PLK1 addiction for standard (serum-dependent) cell lines in vitro, yet not confirmed in primary cells (39). Similarly, in LM2-Cas9-GFP-C32 xenografts, we found no difference in the mean percent of Ki67-, cleaved caspase-3-positive, vimentin- or EpCAM-positive cells within primary tumors in the presence or absence of a PLK1 sgRNAs (Supplementary Fig. S14A–S14F). Rescue of PLK1 in knockout cell lines restored intravasation of both single CTCs and CTC clusters without affecting tumor growth rate (Supplementary Fig. S14G–S14I). To investigate whether PLK1 knockout impairs lung colonization, we injected NSG mice with LM2-GFP-Luc cells expressing PLK1 or non-targeting sgRNAs via the tail vein and measured metastatic outgrowth in the lungs over time. PLK1 depletion did not affect the growth of metastatic foci in the lungs (Supplementary Fig. S14J and S14K). To explore the clinical applicability of our results, we treated LM2-GFP-Luc and BR16-RFP-Luc xenografts with the PLK1 inhibitor BI 2536. Consistently, although no significant effect was observed at the level of the primary tumor, we detect a sharp decrease in both single and clustered CTCs upon treatment with BI 2536 (Fig. 4F and G; Supplementary Fig. S15A and S15B). Furthermore, using the METABRIC dataset (32, 40), we find that high expression of PLK1 in the primary tumor is linked to reduced MFS in patients with breast cancer (Supplementary Fig. S15C). Altogether, our approach provides new means to identify genes that promote intravasation of both single and clustered CTCs in their physiological setting, such as PLK1, whose suppression reduces overall CTC shedding and metastasis in the tested models.
Figure 4.
Identification of genes required for intravasation. A, Schematic of the experimental design. sgRNA that are present in the primary tumor but absent in all CTCs (both single CTCs and CTC clusters) are identified, highlighting genes that are required for CTC intravasation. B, Plot showing the fold change (log2) of depleted genes in single CTCs and CTC clusters when compared with late primary tumor. Dotted lines indicate a threshold for significant depletion (≤ −3 LFC; P ≤ 0.01). Genes coding for proteins possessing an extracellular domain are shown in purple, kinases are shown in red, and others are shown in gray. Gene names highlighted with a border were selected for validation. C, Schematic showing the experimental design for in vivo validation of candidate genes using a minipool sgRNA library (intravasation sgRNA library) comprising six sgRNAs per hit gene and 50 nontargeting control sgRNAs. After generation of a LM2-Cas9-GFP-C32 mutant cell pool, cells were injected into the mammary fat pad of NSG mice. Upon tumor growth, sgRNA complexity of primary tumors, single and clustered CTCs was assessed by isolation of gDNA and subsequent NGS. D, Volcano plot showing the median of normalized LFC and the Padjusted value for individual hit genes and non-targeting control sgRNAs between primary tumors and all CTCs (single and clustered CTCs) of mice injected with cells carrying the intravasation sgRNA minipool (n = 30). Genes with an adjusted P ≤ 0.05, at least 3 sgRNAs with LFC <0, and a median LFC <0 are labeled. E, Bar plot showing the ratio of single CTCs and CTC clusters in mice injected with LM2-Cas9-GFP-C32 cells transduced with individual sgRNAs targeting candidate genes compared with nontargeting control sgRNA (n = 4; *, P < 0.05 by two-tailed Student t test). F, Plot showing the primary tumor growth curve of LM2-GFP-Luc xenografts. n = 4; error bars, SEM. Mice were injected intravenously with either vehicle (0.9% NaCl + 0.1N HCL) or 40 mg/kg BI2536 for two consecutive days a week, for a total of two cycles (green arrows); ns, not significant by two-tailed Student t test. G, The plots show the mean number of single CTCs (left) and CTC clusters (right) per mL of blood isolated from LM2-GFP-Luc xenografts treated with either vehicle (0.9% NaCl + 0.1N HCL) or 40 mg/kg BI2536. n = 4; The P value is shown in the graph by two-sided Student t test.
CTC metastatic signature correlates with decreased MFS in patients with breast cancer
We next compared sgRNA representation in brain, bone, liver, and lymph node metastases, aiming to gain insights into sgRNAs that selectively dropped out in a specific metastatic site, highlighting genes that might be required for organotropism of breast cancer cells (Fig. 5A). With this approach, we identified selective dropouts for 68 genes in bone, 45 genes in liver, 180 genes in brain, and 98 genes in lymph node metastasis, some of which are selective to each site and some others are shared with other sites (Fig. 5B; Supplementary Table S10). Furthermore, we asked whether the same genes are also expressed in metastatic lesions in bone, brain, liver and lymph nodes of the BR16-CDX model, by means of RNA-sequencing of directly isolated GFP-positive cells from the respective metastatic organs. We find that 25/68 genes in bone, 22/45 genes in liver, 67/180 genes in brain, and 39/98 genes in lymph node metastasis are indeed expressed at the RNA level in the respective organs (Fig. 5C; Supplementary Table S11), suggesting their possible involvement in metastatic outgrowth and tumor maintenance within their respective site. In contrast, genes that are not expressed at the RNA levels could highlight their possible involvement in the extravasation process, rather than their activity during actual metastasis outgrowth. On the basis of the expressed genes, we generated a “metastasis signature” comprising a total of 114 genes resulting from the union of the 4 sets of genes. We next attempted to validate these findings directly in patient samples. Although data on metastatic tropism of patient samples are rarely available from curated datasets, we took a dual approach to assess whether our findings are relevant in a clinical context. First, we evaluated our “metastasis signature” in the METABRIC (32, 40) molecular dataset containing gene expression profiles of 1,736 breast cancer patients with long-term follow-up, including MFS, and determined patients with low (Q1) or high (Q4) expression of the signature genes (Supplementary Fig. S16A). Accordingly, we found that a high “metastasis signature” score has an inverse association with the lower-risk integrative cluster (IC) 4 ER+ (Padj < 0.001), IC 3 (Padj = 0.01), IC 7 (Padj = 0.007), and a positive association with the poor-prognosis IC 5 (Padj = 0.001), IC 1 (Padj = 0.01), and IC 10 (Padj < 0.001; Supplementary Fig. S16B). Furthermore, because the development of brain metastasis is considered to be among the most lethal metastatic events in breast cancer (41), we identified three targets among “metastasis signature” genes that were selectively depleted in brain metastases and for which inhibitors were already available, corresponding to class II histone deacetylase 4 (HDAC4) and two Rho-GTPases RhoA and RhoC. We then tested the effects of belinostat (HDAC class I and II inhibitor) and rhosin (RhoA/RhoC GTPases inhibitor) on brain metastases derived from intracranial injection of parental MDA-MB-231 and BR16 cells in NSG mice. Remarkably, brain metastasis was significantly reduced upon treatment with either belinostat or rhosin in both models (Fig. 5D–G). Hence, our in vivo CRISPR-based analysis allowed us to identify candidate genes involved in orchestrating organ-specific metastasis, whose expression correlates with a reduced MFS in patients with breast cancer and whose inhibition reduces brain metastasis in vivo.
Figure 5.
Identification of genes required for organ-specific metastasis and treatment of brain metastasis. A, Schematic of the experimental design. sgRNAs that are specifically depleted in individual metastatic organs are identified, highlighting genes that are required for organ-specific metastasis. B, UpSetR plot showing the intersection of gene sets that are depleted in liver (n = 45), bone (n = 68), lymph node (n = 98), and brain (n = 180). C, Heatmap of expression (transcripts per million) of genes that were depleted in bone (n = 3), brain (n = 9), liver (n = 3), and lymph node (n = 11) metastases in the BR16–CDX model. D, Plot showing the luminescence signal (ph/sec/cm2/sr) over time from NSG mice intracranially injected with either 2.0 × 105 MDA-MB-231-GFP-Luc (top) or 4.5 × 105 BR16-RFP-Luc (bottom) cells and treated either with vehicle (l-arginine (100 mg/mL) in ddH2O) or 80 mg/kg belinostat for five consecutive days a week, for a total of three cycles. Black arrow, treatment start. n = 3 (vehicle), n = 5 (belinostat). E, Representative images of the bioluminescence signal measured from brains of NSG mice intracranially injected with either MDA-MB-231-GFP-Luc (top) or BR16-GFP-Luc (bottom) cells and treated either with vehicle [l-arginine (100 mg/mL) in ddH2O] or 80 mg/kg belinostat. F, Plot showing the luminescence signal (ph/sec/cm2/sr) over time from NSG mice intracranially injected with 2.0 × 105 MDA-MB-231-GFP-Luc (top) or 4.5 × 105 BR16-RFP-Luc (bottom) cells and treated with either vehicle (0.9% NaCl) or 20 mg/kg rhosin each day, for a total of three weeks. Black arrow, treatment start. n = 3 (vehicle); n = 5 (rhosin). G, Representative images of the bioluminescence signal measured from brains of NSG mice intracranially injected with either MDA-MB-231-GFP-Luc (top) or BR16-GFP-Luc (bottom) cells and treated either with vehicle (0.9% NaCl) or 20 mg/kg rhosin. *, P < 0.05 by Student t test; error bars, SEM.
Discussion
The study of spontaneous breast cancer metastasis in vivo with patient-derived material has evidenced an exaggerated proclivity for these models to metastasize to visceral organs, whereas the major metastatic site in patients with breast cancer is the bone. This discrepancy has interfered with our ability to study (and challenge) the molecular drivers of metastasis in clinically relevant contexts and further, it has limited our ability to faithfully recapitulate human breast cancer progression with patient-derived xenografts. Our study provides evidence that when using metastasis-derived CTCs (rather than primary tumor cells) as the source of transplanted cells, consistent and abundant metastatic outgrowth occurs in the bone, brain, liver and lymph node, mirroring the disease status of the original patient at the moment of CTC isolation. Although additional models will be needed to increase the repertoire of CTC-derived xenografts (many of which are now available in a number of different laboratories worldwide), this current tool already allowed us to interrogate the metastatic process in great detail with loss-of-function approaches.
We applied a two-step, CRISPR-based procedure to identify metastasis-relevant genes during disease progression in vivo. We first transduced breast CTC-derived cells with a genome-wide sgRNA library and transplanted them orthotopically in vivo, where we observed spontaneous sgRNA dropout all along metastatic progression. The pattern of sgRNA complexity at different stages clearly highlighted that metastasis occurs through various bottlenecks, as expected, yet allowed us to infer the identity of genes that could play a role in specific aspects, such as CTC cluster formation, CTC intravasation, and organ-specific metastatic spread. Given the chosen experimental design and inherent bottlenecks of the metastatic cascade, we performed a second step to validate candidate genes by means of mini-pool or individual sgRNA knockouts in an independent breast cancer model. Results of this approach allowed us to identify previously unappreciated genes whose expression regulates CTC cluster formation (such as casein kinases and interleukin 18 receptor), as well as genes that regulate the intravasation of both single and clustered CTCs (such as PLK1) and organ-specific metastasis (such as HDAC and Rho GTPases in brain metastasis).
In the context of CTC intravasation, pharmacological inhibition of PLK1 mirrors the results obtained with Cas9-mediated gene knockout. Particularly, in vivo, PLK1 appears to be critical for the entry of cancer cells into the bloodstream as opposed to a role in tumor growth or metastatic colonization. Given that PLK1 inhibitors are available, future trials will be needed to confirm this aspect, yet adopting endpoints that consider its role in metastatic dissemination, rather than tumor growth. In regard to brain metastasis, we observe that pharmacological inhibition of HDAC or RhoA/C GTPases leads to a reduction in their growth rate, suggesting novel therapeutic opportunities. Broadly, our study provides a novel CTC-based tool to study spontaneous breast cancer metastasis in a highly clinically relevant fashion. Using this model in combination with a CRISPR-based approach, we demonstrate the involvement of specific (targetable) players of the metastatic spread of cancer with potential clinical applicability.
Authors' Disclosures
S. Gkountela reports personal fees from Novartis Pharma AG outside the submitted work. N. Aceto reports grants and personal fees from Novartis, and grants from Roche, and grants and personal fees from Lilly, Basilea Pharmaceutica Ltd., and personal fees from Merck, Swiss Re, Thermo Fisher Scientific, and grants and personal fees from Angle plc., Tethis Spa, and personal fees from Bracco Suisse during the conduct of the study; as well as other support from Novartis outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
The authors thank all patients that donated blood for the study as well as all involved clinicians and study nurses. The authors thank Dr. J. Massagué (Memorial Sloan Kettering Cancer Center, New York, New York) for donating cell lines. They thank all members of the Aceto laboratory for feedback and discussions. They thank the Genomics Facility Basel (D-BSSE of the ETH Zürich) for performing next-generation sequencing. The authors thank Dr. T. Lopes and Emmanuel Traunecker of the flow cytometer core (DBM of University of Basel) for help with CTC sorting experiments. The authors thank Dr. T. Heye (University Hospital Basel) for providing CT scan images. Calculations were performed at sciCORE (http://scicore.unibas.ch/) scientific computing center at the University of Basel. C. Giachino acknowledges funding by the Swiss Cancer League (KLS-4518-08-2018). Research in the Aceto laboratory is supported by the European Research Council (#101001652), the European Union (801159-B2B), the Swiss National Science Foundation (PP00P3_190077), the Swiss Cancer League (KLS-4834-08-2019), the Basel Cancer League (KLbB-4763-02-2019), the two Cantons of Basel through the ETH Zürich (PMB-01-16), the University of Basel and the ETH Zurich, Switzerland. Z. Diamantopoulou is a H2020 Marie Skłodowska-Curie Actions (#101028567) fellow.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
M.C. Scheidmann: Investigation. F. Castro-Giner: Investigation. K. Strittmatter: Investigation. I. Krol: Investigation. A. Paasinen-Sohns: Investigation. R. Scherrer: Investigation. C. Donato: Investigation. S. Gkountela: Investigation. B.M. Szczerba: Investigation. Z. Diamantopoulou: Investigation. S. Muenst: Investigation. T. Vlajnic: Investigation. L. Kunz: Investigation. M. Vetter: Resources. C. Rochlitz: Resources. V. Taylor: Resources. C. Giachino: Resources, funding acquisition, investigation, methodology. T. Schroeder: Resources. R.J. Platt: Resources. N. Aceto: Conceptualization, resources, supervision, funding acquisition, investigation.
References
- 1. Nguyen DX, Bos PD, Massagué J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer 2009;9:274–84. [DOI] [PubMed] [Google Scholar]
- 2. Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer 2014;14:623–31. [DOI] [PubMed] [Google Scholar]
- 3. Krebs MG, Metcalf RL, Carter L, Brady G, Blackhall FH, Dive C. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat Rev Clin Oncol 2014;11:129–44. [DOI] [PubMed] [Google Scholar]
- 4. Liotta LA, Saidel MG, Kleinerman J. The significance of hematogenous tumor cell clumps in the metastatic process. Cancer Res 1976;36:889–94. [PubMed] [Google Scholar]
- 5. Molnar B, Ladanyi A, Tanko L, Sréter L, Tulassay Z. Circulating tumor cell clusters in the peripheral blood of colorectal cancer patients. Clin Cancer Res 2001;7:4080–5. [PubMed] [Google Scholar]
- 6. Gkountela S, Castro-Giner F, Szczerba BM, Vetter M, Landin J, Scherrer R, et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell 2019;176:98–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Szczerba BM, Castro-Giner F, Vetter M, Krol I, Gkountela S, Landin J, et al. Neutrophils escort circulating tumour cells to enable cell-cycle progression. Nature 2019;566:553–7. [DOI] [PubMed] [Google Scholar]
- 8. Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014;158:1110–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang C, Mu Z, Chervoneva I, Austin L, Ye Z, Rossi G, et al. Longitudinally collected CTCs and CTC-clusters and clinical outcomes of metastatic breast cancer. Breast Cancer Res Treat 2017;161:83–94. [DOI] [PubMed] [Google Scholar]
- 10. Donato C, Kunz L, Castro-Giner F, Paasinen-Sohns A, Strittmatter K, Szczerba BM, et al. Hypoxia triggers the intravasation of clustered circulating tumor cells. Cell Rep 2020;32:108105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kennecke H, Yerushalmi R, Woods R, Cheang MCU, Voduc D, Speers CH, et al. Metastatic behavior of breast cancer subtypes. J Clin Oncol 2010;28:3271–7. [DOI] [PubMed] [Google Scholar]
- 12. Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell 2011;147:275–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weilbaecher KN, Guise TA, McCauley LK. Cancer to bone: a fatal attraction. Nat Rev Cancer 2011;11:411–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sowder ME, Johnson RW. Bone as a preferential site for metastasis. JBMR Plus 2019;3:e10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 2015;160:1246–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Katigbak A, Cencic R, Robert F, Sénécha P, Scuoppo C, Pelletier J. A CRISPR/Cas9 functional screen identifies rare tumor suppressors. Sci Rep 2016;6:38968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kodama M, Kodama T, Newberg JY, Katayama H, Kobayashi M, Hanash SM, et al. In vivo loss-of-function screens identify KPNB1 as a new druggable oncogene in epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2017;114:E7301–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 2017;547:413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yau EH, Kummetha IR, Lichinchi G, Tang R, Zhang Y, Rana TM. Genome-wide CRISPR screen for essential cell growth mediators in mutant KRAS colorectal cancers. Cancer Res 2017;77:6330–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang G, Chow RD, Ye L, Guzman CD, Dai X, Dong MB, et al. Mapping a functional cancer genome atlas of tumor suppressors in mouse liver using AAV-CRISPR–mediated direct in vivo screening. Sci Adv 2018;4:eaao5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chow RD, Guzman CD, Wang G, Schmidt F, Youngblood MW, Ye L, et al. AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat Neurosci 2017;20:1329–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc 2017;12:828–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Coutu DL, Kokkaliaris KD, Kunz L, Schroeder T. Multicolor quantitative confocal imaging cytometry. Nat Methods 2018;15:39–46. [DOI] [PubMed] [Google Scholar]
- 24. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016;32:3047–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ahdesmäki MJ, Gray SR, Johnson JH, Lai Z. Disambiguate: an open-source application for disambiguating two species in next-generation sequencing data from grafted samples. F1000Res. 2016;5:2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 2017;14:417–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rueda OM, Sammut S-J, Seoane JA, Chin S-F, Caswell-Jin JL, Callari M, et al. Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature 2019;567:399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 2015;12:115–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kadota M, Sato M, Duncan B, Ooshima A, Yang HH, Diaz-Meyer N, et al. Identification of novel gene amplifications in breast cancer and coexistence of gene amplification with an activating mutation of PIK3CA. Cancer Res 2009;69:7357–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Holst F, Stahl PR, Ruiz C, Hellwinkel O, Jehan Z, Wendland M, et al. Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat Genet 2007;39:655–60. [DOI] [PubMed] [Google Scholar]
- 36. Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014;11:783–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature 2005;436:518–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu Z, Sun Q, Wang X. PLK1, a potential target for cancer therapy. Transl Oncol 2017;10:22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Raab M, Kappel S, Krämer A, Sanhaji M, Matthess Y, Kurunci-Csacsko E, et al. Toxicity modelling of Plk1-targeted therapies in genetically engineered mice and cultured primary mammalian cells. Nat Commun 2011;2:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer. Nat Rev Dis Primers. 2019;5:66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing reads and the matrix of read counts from RNA-seq have been deposited in the Gene Expression Omnibus (GEO, NCBI) with accession number GSE138813.



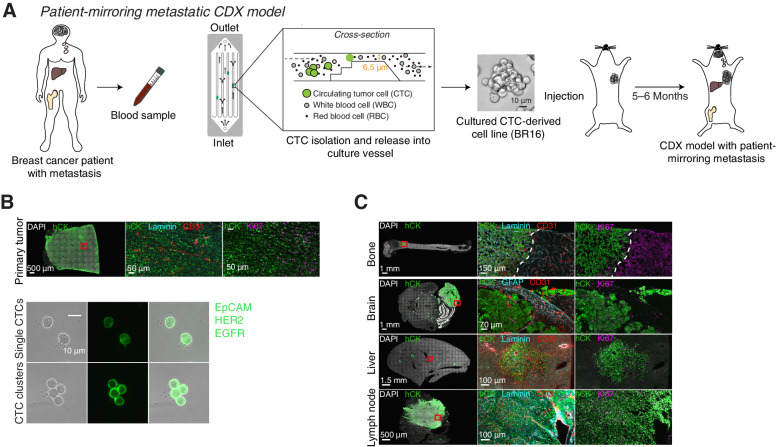
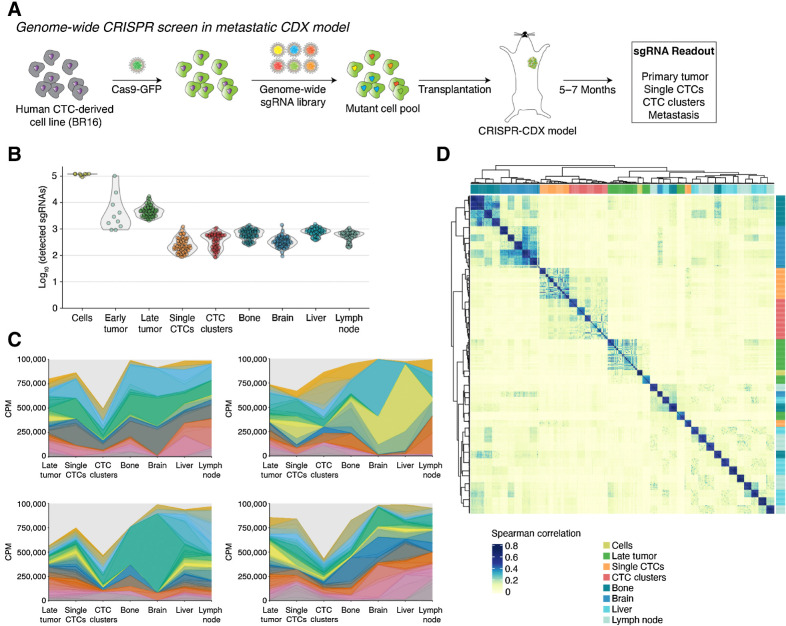
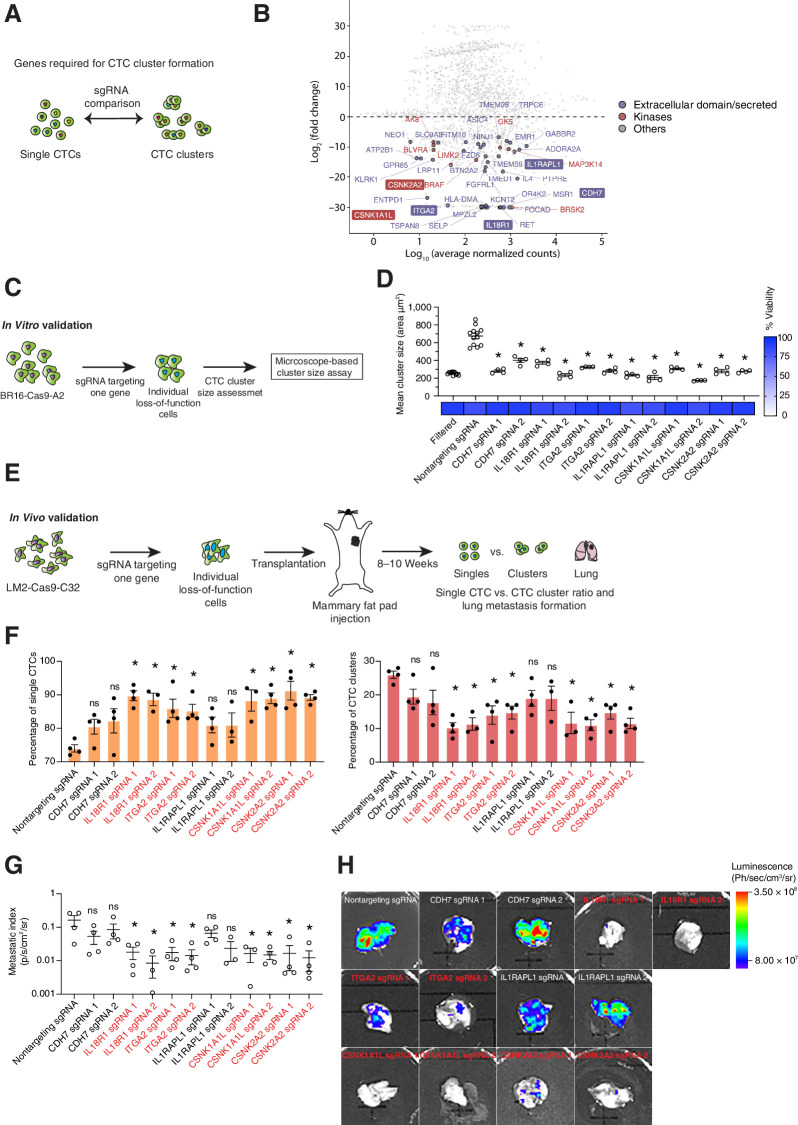
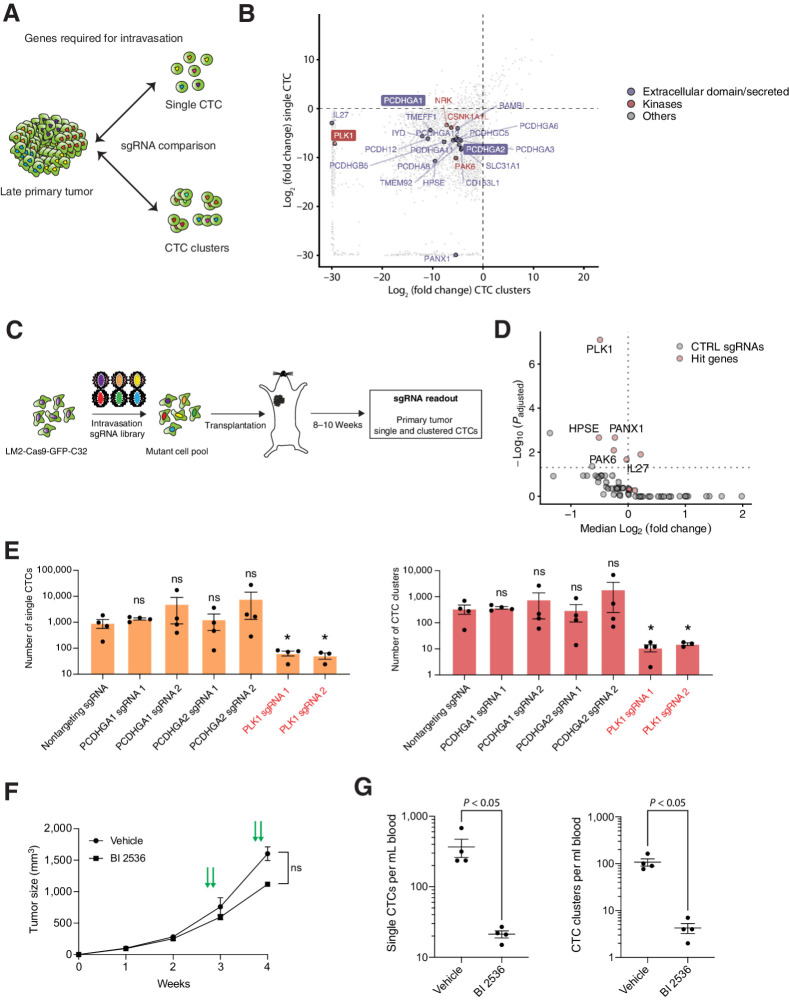
![Figure 5. Identification of genes required for organ-specific metastasis and treatment of brain metastasis. A, Schematic of the experimental design. sgRNAs that are specifically depleted in individual metastatic organs are identified, highlighting genes that are required for organ-specific metastasis. B, UpSetR plot showing the intersection of gene sets that are depleted in liver (n = 45), bone (n = 68), lymph node (n = 98), and brain (n = 180). C, Heatmap of expression (transcripts per million) of genes that were depleted in bone (n = 3), brain (n = 9), liver (n = 3), and lymph node (n = 11) metastases in the BR16–CDX model. D, Plot showing the luminescence signal (ph/sec/cm2/sr) over time from NSG mice intracranially injected with either 2.0 × 105 MDA-MB-231-GFP-Luc (top) or 4.5 × 105 BR16-RFP-Luc (bottom) cells and treated either with vehicle (l-arginine (100 mg/mL) in ddH2O) or 80 mg/kg belinostat for five consecutive days a week, for a total of three cycles. Black arrow, treatment start. n = 3 (vehicle), n = 5 (belinostat). E, Representative images of the bioluminescence signal measured from brains of NSG mice intracranially injected with either MDA-MB-231-GFP-Luc (top) or BR16-GFP-Luc (bottom) cells and treated either with vehicle [l-arginine (100 mg/mL) in ddH2O] or 80 mg/kg belinostat. F, Plot showing the luminescence signal (ph/sec/cm2/sr) over time from NSG mice intracranially injected with 2.0 × 105 MDA-MB-231-GFP-Luc (top) or 4.5 × 105 BR16-RFP-Luc (bottom) cells and treated with either vehicle (0.9% NaCl) or 20 mg/kg rhosin each day, for a total of three weeks. Black arrow, treatment start. n = 3 (vehicle); n = 5 (rhosin). G, Representative images of the bioluminescence signal measured from brains of NSG mice intracranially injected with either MDA-MB-231-GFP-Luc (top) or BR16-GFP-Luc (bottom) cells and treated either with vehicle (0.9% NaCl) or 20 mg/kg rhosin. *, P < 0.05 by Student t test; error bars, SEM.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/429b/9359744/fb2e08e932e0/681fig5.jpg)