Abstract
Structural Maintenance of Chromosomes (SMC) protein complexes play key roles in the three-dimensional organization of genomes in all kingdoms of life. Recent insights from chromosome contact mapping experiments and single-molecule imaging assays suggest that these complexes achieve distinct cellular functions by extruding large loops of DNA while they move along the chromatin fiber. In this short review, we summarize recent insights into the molecular architecture of these unconventional DNA motor complexes, their interaction with their DNA substrates, and the remarkable dynamic changes they can undergo during their ATPase reaction cycle.
Introduction
A multitude of genomic functions – ranging from the control of gene expression and the repair of DNA damage to the compaction of individual chromatin fibers into mitotic chromosomes – require a series of complex rearrangements in the spatial organization of the chromatin fiber. These structural changes in chromosome architecture are governed by ubiquitous multi-subunit protein complexes of the SMC family, which can be subdivided into three major categories both in prokaryotes (MukBEF, MksBEF, SMC–ScpAB) and eukaryotes (cohesin, condensin and Smc5/6) [1]. The eukaryotic SMC complexes fulfill specific functions during different stages of the cell cycle: cohesin maintains chromatin domains during interphase and holds together sister chromatids once they emerge from the DNA replication fork [2]; condensin organizes mitotic and meiotic chromosomes into the iconic cylindrical structures described decades ago [3]; Smc5/6 controls DNA superhelicity and plays a role in DNA damage repair [4].
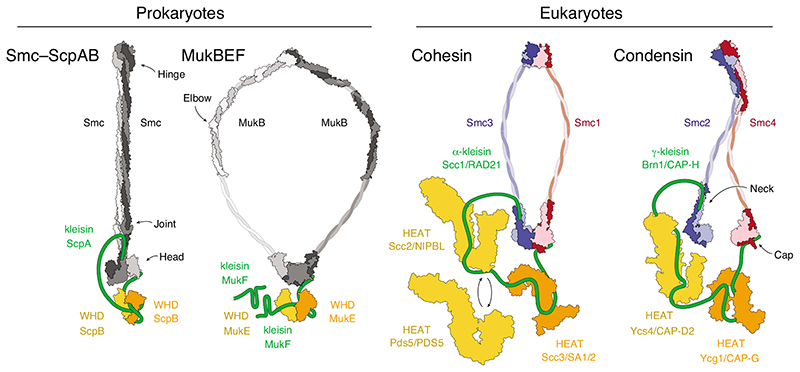
Despite their discrete roles, SMC complexes share a common three-dimensional architecture. At their core is a pair of ~50-nm-long intra-molecular coiled-coil SMC subunits that dimerize via a globular ‘hinge’ domain at one end of the coils. The amino and carboxy termini brought together at the other ends of the coils fold into ATP-Binding-Cassette (ABC) ATPase ‘head’ domains (Figure 1). The two ATPase heads of an SMC dimer are able to engage with each other upon binding a pair of ATP molecules between the Walker A/B motifs of one head and the conserved ABC signature motifs of the opposite head, whereas nucleotide hydrolysis and release are thought to again drive the head apart. The SMC heads are furthermore connected by a subunit of the kleisin protein family, which binds via a helical domain located at its amino terminus to the coiled-coil ‘neck’ region immediately adjacent to one head and via a winged-helix domain located at its carboxy terminus to the opposite ‘cap’ surface of the opposite head. The kleisin recruits additional subunits to the complex that are either composed of tandem winged-helix domains (in MukBEF, MksBEF, SMC–ScpAB and Smc5/6) or of helical HEAT (Huntingtin, EF3, PP2A, TOR1) repeats (in cohesin and condensin) [5].
Figure 1. Architecture of SMC protein complexes.
Cartoon representations of available subunit structures of prokaryotic Smc–ScpAB and MukBEF complexes and eukaryotic cohesin and condensin complexes. Available structural models are indicated for the Pyrocuccus furiosus Smc dimer (light/dark grey) in the rod-shaped, nucleotide-free state [42] and Geobacillus stearothermophilus ScpB (orange/yellow; pdb: 3W6J) [28]; the Escherichia coli MukB dimer (light/dark grey) hinge (pdb: 3IBP) [53] and coiled-coil elbow (pdb: 6H2X) [44] and Haemophilus ducreyi head domains (pdb: 3EUK) in the nucleotide-bound state and E. coli MukE (orange/yellow; pdb: 3EUH) [54]; the cohesin Smc1–Smc3 dimer (red/blue) Mus musculus hinge (pdb: 2WD5) [32] and Saccharomyces cerevisiae/Chaetomium thermophilum heads (pdb: 6QPW) in the nucleotide-bound state [31] and C. thermophilum Scc2 (yellow; pdb: 5T8V) [55], Lachancea thermotolerans Pds5 (yellow; pdb: 5F0O) [56] and Zygosaccharomyces rouxii Scc3 (orange; pdb: 4UVK) [57]; the condensin Smc2–Smc4 dimer (blue/red) S. cerevisiae hinge (pdb: 4RSI) [43] and C. thermophilum heads (pdb: 6QJ1 and 6QJ2) in the nucleotide-free state and Ycs4 (yellow; pdb: 6QJ3) [49] and S. cerevisiae Ycg1 (orange; pdb: 5OQQ) [20]. The cartoons indicate different coiled-coil conformations that are presumably accessible by all complexes. Note that MukBEF complexes can form dimers of dimers via the amino terminus of the MukF kleisin subunit, indicated by the presence of a second MukF subunit.
Although the structures of most of their subunits have now been resolved to near-atomic detail, it still remains mysterious how SMC protein complexes engage with their chromatin substrates and carry out their functions. In this brief review, we focus our discussion on the current understanding of the physical interactions between SMC complexes and DNA and speculate how these interactions might mediate diverse biological functions.
Chromosome organization by DNA loop extrusion
A unifying mechanism for the seemingly different functions of SMC protein complexes might be their ability to generate large DNA loops. The idea that SMC complexes act as a pair of bidirectional DNA motors that move into opposite directions on the same DNA and thereby extrude a DNA loop elegantly explains several features of chromosome contact frequency maps that were obtained from high-throughput sequencing techniques in vertebrate cells. These features include the co-localization of cohesin and the transcriptional repressor CTCF bound to DNA sequences that are oriented in a convergent (i.e. head-to-head) orientation with respect to one another at the bases of DNA loops during interphase [6,7] and the folding of mitotic chromosomes by condensin into arrays of loops with an average size of ~80 kilobase pairs [8,9]. The alignment of the left and right arms of circular bacterial chromosomes can similarly be rationalized by Smc–ScpAB complexes that zip up both chromosome arms into loops as they translocate along them [10,11].
In single-molecule imaging experiments, purified yeast condensin complexes [12] or human cohesin complexes [13,14] were indeed able to track along tens of kilobase pairs and progressively extrude DNA into loops. Unlike cohesin, condensin reeled in the DNA double helix in a strictly unidirectional manner, which suggests that condensin must possess two principally distinct DNA-binding elements: an ‘anchor’ element that stably holds on to the DNA segment the complex initially bound to and a ‘motor’ element that translocates along a second segment of the same DNA. Such an asymmetric motor could then be converted into a symmetric, bidirectional motor by the dimerization of two complexes in a back-to-back orientation, as has been suggested for human cohesin [14] or the Escherichia coli MukBEF complex [15]. Computational simulations likewise suggest that conversion of condensin into a bidirectional motor might be necessary to achieve efficient compaction of mitotic chromosomes [16]. Asymmetric motors that can traverse each other, as recently observed for condensin [17], provide an alternative solution for closing gaps between DNA loops that would otherwise have been left uncompacted. Yet another possibility foresees that asymmetric motors might be able to switch directionality without releasing the DNA loop they had created to achieve bidirectional loop extrusion. To gain insights into the DNA loop extrusion mechanism, understanding how SMC complexes contact DNA and how conformational changes turn static complexes into DNA motors will hence be crucial.
DNA anchor elements in SMC complexes
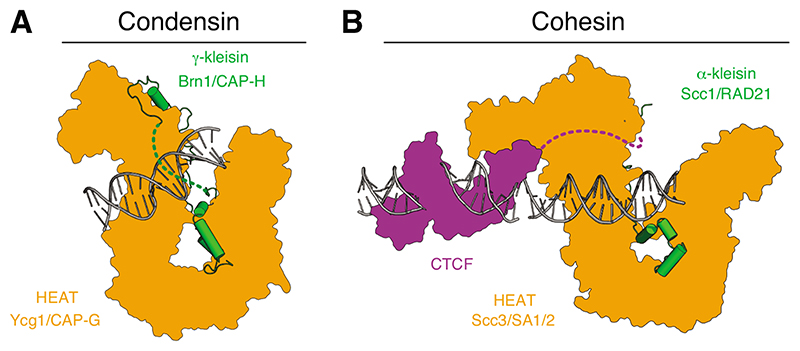
Whereas the identity of the DNA motor element(s) in condensin and other SMC complexes remains unknown (see below), it is likely that a positively charged groove, which is formed when the Ycg1/CAP-G HEAT-repeat subunit of condensin engages the Brn1/CAP-H kleisin subunit (Figure 2A), serves as an anchor element [18–20]. Co-crystal structures revealed that DNA is bound in the groove exclusively via backbone contacts with residues from both the kleisin and HEAT-repeat subunits, and is furthermore locked into place by its entrapment within a ‘safety belt’ peptide loop of the kleisin [20]. Loosening of this safety belt by mutation causes condensin to slip while it extrudes DNA loops, which is consistent with the proposal that this part of the complex functions as a DNA anchor [12].
Figure 2. DNA anchor domains of eukaryotic SMC complexes.
(A) Cartoon representations of the budding yeast cohesin Ycg1–Brn1 subcomplex bound to 18 base pairs (bp) double-stranded DNA (dsDNA) (pdb: 5OQP) [20]. A dotted line indicates the ‘safety belt’ peptide loop. (B) Cartoon model of the human cohesin SA2–SCC1 subcomplex (pdb: 6QNY) [22] bound to 19 bp dsDNA (positioned based on the structure of the yeast Scc3– Scc1 subcomplex, pdb: 6H8Q) [21] and the zinc-fingers 2–7 of human CTCF bound to 23 bp dsDNA (pdb 5T0U) [58]. A dotted line indicates a linker to a peptide motif that binds to the ‘conserved essential surface’ region on the backside of SA2.
Although the homologous Scc3/SA2 HEAT-repeat subunit of cohesin binds DNA at a similar position of its U-shaped architecture [21], amino acid sequence analysis suggests that the cohesin Scc1/RAD21 kleisin lacks a safety belt loop [20]. It is tempting to speculate that the absence of a safety belt might allow cohesin to frequently switch its directionality to produce symmetric DNA loops over a period of time. Cohesin nevertheless becomes stably anchored to DNA when it encounters the transcriptional repressor CTCF, which uses a hydrophobic peptide motif close to its amino terminus to bind to a conserved patch on the Scc3/SA2 HEAT-repeat subunit [22] (Figure 2B). The proposal that this peptide motif might only be accessible when cohesin approaches from the 3’ end of the CTCF-binding sequence provides a simple explanation for the accumulation of cohesin specifically at CTCF sequences that are in a convergent orientation [23,24]. In this case, the CTCF–SA2 complex would function as a DNA anchor element analogous to the Ycg1/CAP-G anchor found in condensin, yet one that targets cohesin to specific positions in the genome. Whether SMC–kleisin complexes that contain tandem winged-helix subunits in place of HEAT-repeat subunits, such as Smc–ScpAB or Smc5/6, possess a comparable DNA anchor element remains unknown.
Potential DNA motor elements in SMC complexes
It seems reasonable to assume that any DNA motor element must require the presence of at least two distinct DNA binding sites that alternate in their contact with the DNA double helix in a manner that is controlled by cycles of ATP binding and hydrolysis by the SMC heads. Where might such DNA binding sites be located?
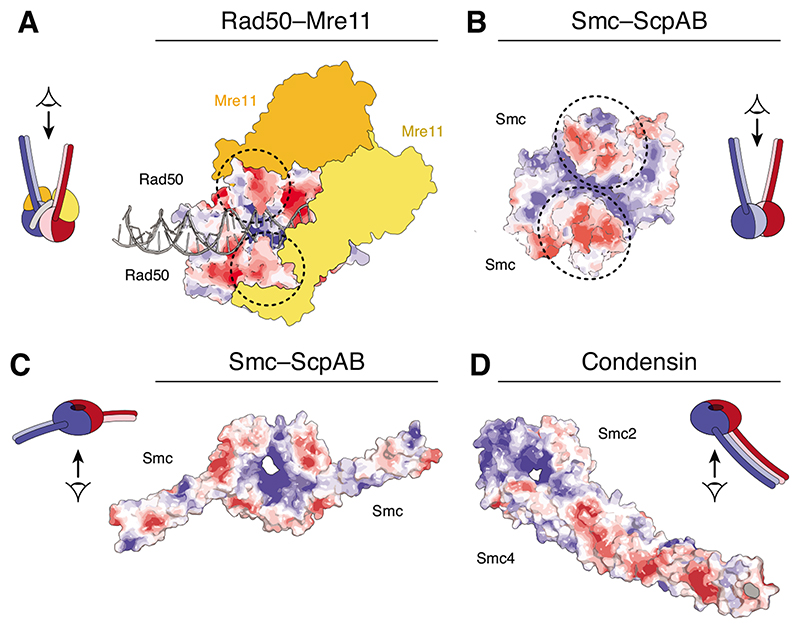
The SMC ATPase heads themselves are excellent candidates for creating a transient DNA binding site. Crystal and cryo-electron microscopy (cryo-EM) structures of the SMC-related Rad50–Mre11 DNA damage repair complex reveal binding of a DNA double helix along a positively charged cleft that forms between the coiled coils of the engaged Rad50 ATPase heads, with the Mre11 endonuclease closing off one side of the cleft to lock DNA in place [25–27] (Figure 3A). A similar cleft can be detected in the crystal structure of dimerized prokaryotic SMC heads [28,29] (Figure 3B) and mutation of positively charged side chains of surrounding residues reduces the affinity of the ATP-engaged head dimer for DNA [30]. Whether the same cleft accommodates DNA in eukaryotic SMC complexes is less clear, since crystal structures of ATP-engaged head heterodimers are not yet available and a recent Smc1–Smc3 head cryo-EM structure did not resolve the positions of several central residues of the cleft [31]. In either case, the formation of a composite positively charged cleft upon nucleotide-dependent ATPase head engagement would provide a simple control mechanism for the creation and dissolution of a transient DNA binding site during the SMC catalytic cycle.
Figure 3. Potential DNA binding surfaces on the SMC heads.
Surface charge potential representations of (A) Rad50 homodimer heads bound to 31 bp dsDNA and the Mre11 dimer (yellow/orange) (pdb: 6S85) [25], (B) Geobacillus stearothermophilus SMC head homodimer (pdb 5H68) [59] and (C) a chimeric Saccharomyces cerevisiae/Chaetomium thermophilum Smc1–Smc3 head heterodimer (pdb: 6QPW) [31] viewed from in-between the coiled coils. Surface charge scale ranges from – 5 kT/e (red) to +5 kT/e (blue). Circles indicate the positions of the coiled-coil stems.
A second candidate for creating a DNA-binding interface is the SMC hinge dimerization domain. Crystal structures of hinge dimers from different origins revealed that the central channel of the doughnut-shaped domain is consistently positively charged [32,33]. Mutation of basic residues within this channel prevent DNA entrapment by cohesin complexes [34]. Moreover, mutation of basic residues at the outer surface reduces DNA binding by prokaryotic SMCs in vitro [35,36]. These results suggest that the hinge domain must make an important contribution to the function of SMC complexes, presumably through its ability to contact DNA. However, the majority of isolated hinge domains assayed for DNA binding in vitro displayed considerably greater affinities for single-stranded than for double-stranded DNA [33,36–38]. Even more surprising, SMC complexes were still able to support rapid cell divisions in Bacillus subtilis even when their hinge dimerization domains had been replaced by the structurally distinct zinc-hook dimerization domain of Rad50 [39]. It therefore remains questionable whether SMC hinge domain could be a crucial part of any DNA motor element.
Conformational changes that could drive DNA loop extrusion
Comparison of ATP hydrolysis rates measured in ensemble assays and DNA-loop extrusion speeds measured in single-molecule experiments suggests that cohesin and condensin might be able to take steps that are in the range of the entire length of these complexes – 50 nm or more – in a single reaction cycle [12,13]. If this comparison were accurate, SMC complexes must be able to undergo extensive conformational changes that allow them to move in such large steps along DNA and, more importantly, entire chromatin fibers, by a mechanism that is remarkably different from those used by well-characterized DNA motor proteins, like DNA helicases or translocases [40].
It seems plausible that any large-scale movements originate in the properties of the SMC coiled coils. When imaged in solution by real-time atomic force microscopy, the coiled coils of condensin are frequently separated and display a large degree of elasticity. This flexibility allows them to bend into a ‘butterfly’-like shape that brings together head and hinge regions [41]. In contrast, data from electron and dry atomic force microscopy, protein cross-linking experiments and crystal structures imply that two SMC coiled coils frequently align into stiff rods along almost their entire lengths [42,43] and that such rods can sharply fold back at an ‘elbow’ region to bring head and hinge domains into proximity [44,45]. The rod-shaped coiled-coil conformation might hence be less rigid than initially anticipated, which is also consistent with recent molecular dynamics simulations [46].
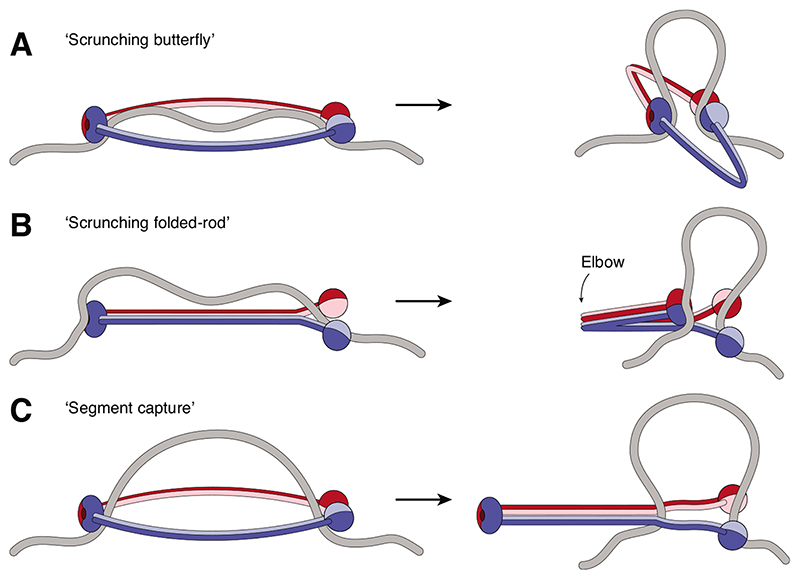
Bending of the coiled coils, irrespective of whether they are split apart or aligned into a rod, might provide a mechanism to bring together two otherwise distant DNA-binding sites at the head and hinge domains and thereby bend the segment of DNA between them into a loop [12,40,47,48] (Figures 4A and B).It is conceivable that such a ‘scrunching’-type motion might be powered by the SMC ATPase heads to drive the DNA motor activity. Comparison of structures of nucleotide-bound and nucleotide-free condensin and cohesin SMC heads indeed suggests that nucleotide binding to one of the two head domains results in a reorientation of the attached coiled coil and disengagement of the kleisin amino terminus from the coiled-coil neck region [31,49]. Whether this structural transition directly affects DNA binding, possibly by creating additional DNA binding interfaces at the coiled coil, like the one observed in a recent Rad50 crystal structure [50], is currently unclear. Scrunching-type models fall, however, short in explaining why bending of the coiled coils could never be observed in electron micrographs of Bacillus subtilis SMC complexes [43], why the hinge domain can be replaced, or why the coiled coils can be shortened or extended by one superhelical period without interfering with SMC function in this species [39].
Figure 4. Possible DNA motor movements of SMC complexes.
(A) Extended ring-shaped SMCs might bind to DNA via their head and hinge domains and DNA loops might be formed when the coiled-coils bend. (B) Rod-shaped SMCs might bind DNA as in (A) and then fold at their elbow region to create and extend a DNA loop. (C) Ring-shaped SMC complexes might capture a DNA loop within their lumen and then zip up their coiled coils from hinge to heads to move the loop into a new compartment created by the kleisin subunit (not depicted). In all models, the bases of the DNA loop would need to be held in place when the reaction cycle resets.
An alternative model stems from the finding that, in the reconstituted structure of the rod-shaped B. subtilis SMC dimer, the ATPase head domains tilt away from each other in an orientation that prevents nucleotide binding [42]. Protein cross-linking data support a similar orientation of the Smc1 and Smc3 heads of cohesin [51]. Bringing the heads into a position that enables them to sandwich ATP between their Walker A/B and ABC signature motifs requires a major re-orientation in the heads, which is thought to drive the opening of the rod-shaped coils into an open ring-shaped conformation [43]. If a DNA segment could enter the open ring structure in the ATP-engaged conformation and subsequent head disengagement would cause the coiled coils to zip up from the hinge, then this movement could drive any DNA loop entrapped between the coiled coils towards the heads (Figure 4C) and into a separate compartment formed by the connection of the latter with the kleisin subunit [52]. Such a ‘segment capture’ model would not require a strong DNA binding site at the SMC hinge, but bears the challenge that DNA would somehow need to be bent and fed into the coiled-coil interspace. Independent of whether the DNA translocation step is based on a scrunching-type movement or follows the segment capture model, future experiments will need to address whether, and if so how, re-arrangements in the coiled coils are linked to DNA movement and loop extrusion.
Conclusions
SMC protein complexes have emerged as the universal key players in the functional organization of genomes. Their common working principle presumably depends on the extrusion of large chromatin loops by their processive movement along the DNA double helix, but the molecular basis of their DNA motor activity has remained largely mysterious. Uncovering how these exceptional machines move DNA and entire chromatin fibers will require the unambiguous identification of the parts of the complexes that interact with DNA and of the conformational changes that are essential to generate the large steps inferred from biophysical measurements. Structural insights into the architecture of SMC holo complexes, by themselves and when bound to DNA and/or chromatin, as well as new techniques that can capture the dynamic changes in these structures, will undoubtedly provide unique new insights into one of the last few unsolved puzzles of chromosome biology.
Highlights.
Protein complexes of the SMC family organize chromosomes by extruding large DNA loops.
SMC-mediated loop extrusion presumably requires DNA anchor and motor elements.
Changes in coiled-coil conformations might appose distant DNA binding sites during DNA translocation.
Acknowledgements
We are grateful to Markus Hassler and Indra Shaltiel for comments on the manuscript. S.D. acknowledges support from a Darwin Trust Fellowship. Work in the Haering group is funded by the European Research Council (ERC, grant number 681365), the German Research Foundation (DFG, grant number HA5853/4-1), the European Molecular Biology Laboratory (EMBL) and the University of Würzburg.
References
- 1.Yatskevich S, Rhodes J, Nasmyth K. Organization of Chromosomal DNA by SMC Complexes. Annu Rev Genet. 2019;53:445–482. doi: 10.1146/annurev-genet-112618-043633. [DOI] [PubMed] [Google Scholar]
- 2.Nishiyama T. Cohesion and cohesin-dependent chromatin organization. Curr Opin Cell Biol. 2019;58:8–14. doi: 10.1016/j.ceb.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Paul MR, Hochwagen A, Ercan S. Condensin action and compaction. Curr Genet. 2019;65:407–415. doi: 10.1007/s00294-018-0899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aragon L. The Smc5/6 Complex: New and Old Functions of the Enigmatic Long-Distance Relative. Annu Rev Genet. 2018;52:89–107. doi: 10.1146/annurev-genet-120417-031353. [DOI] [PubMed] [Google Scholar]
- 5.Wells JN, Gligoris TG, Nasmyth KA, Marsh JA. Evolution of condensin and cohesin complexes driven by replacement of Kite by Hawk proteins. Curr Biol. 2017;27:R17–R18. doi: 10.1016/j.cub.2016.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goloborodko A, Imakaev MV, Marko JF, Mirny L. Compaction and segregation of sister chromatids via active loop extrusion. Elife. 2016;5 doi: 10.7554/eLife.14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibcus JH, Samejima K, Goloborodko A, Samejima I, Naumova N, Nuebler J, Kanemaki MT, Xie L, Paulson JR, Earnshaw WC, et al. A pathway for mitotic chromosome formation. Science. 2018;359 doi: 10.1126/science.aao6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Brandao HB, Le TB, Laub MT, Rudner DZ. Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science. 2017;355:524–527. doi: 10.1126/science.aai8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran NT, Laub MT, Le TBK. SMC Progressively Aligns Chromosomal Arms in Caulobacter crescentus but Is Antagonized by Convergent Transcription. Cell Rep. 2017;20:2057–2071. doi: 10.1016/j.celrep.2017.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganji M, Shaltiel IA, Bisht S, Kim E, Kalichava A, Haering CH, Dekker C. Real-time imaging of DNA loop extrusion by condensin. Science. 2018;360:102–105. doi: 10.1126/science.aar7831. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Single-molecule experiments visualize, for the first time, DNA-loop extrusion activity by an SMC protein complex and suggest that a single yeast condensin complex extrudes DNA unidirectionally in ~50 nm steps
- 13.Davidson IF, Bauer B, Goetz D, Tang W, Wutz G, Peters JM. DNA loop extrusion by human cohesin. Science. 2019;366:1338–1345. doi: 10.1126/science.aaz3418. [DOI] [PubMed] [Google Scholar]; ** Single-molecule experiments reveal bidirectional DNA loop extrusion by monomeric human cohesin complexes, even when all three interfaces in the Smc1–Smc3–kleisin ring are covalently linked
- 14.Kim Y, Shi Z, Zhang H, Finkelstein IJ, Yu H. Human cohesin compacts DNA by loop extrusion. Science. 2019;366:1345–1349. doi: 10.1126/science.aaz4475. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Single-molecule imaging demonstrates the compaction of DNA loops by dimeric human cohesin complexes
- 15.Badrinarayanan A, Reyes-Lamothe R, Uphoff S, Leake MC, Sherratt DJ. In vivo architecture and action of bacterial structural maintenance of chromosome proteins. Science. 2012;338:528–531. doi: 10.1126/science.1227126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banigan EJ, Mirny LA. Limits of Chromosome Compaction by Loop-Extruding Motors. Physical Review X. 2019;9 [Google Scholar]; * Banigan and Mirny discuss the theoretical limits of unidirectional loop extrusion and presents computational simulations that support the effectiveness of bidirectional loop extrusion
- 17.Kim E, Kerssemakers J, Shaltiel IA, Haering CH, Dekker C. DNA-loop extruding condensin complexes can traverse one another. Nature. 2020;579:438–442. doi: 10.1038/s41586-020-2067-5. [DOI] [PubMed] [Google Scholar]; * Real-time single-molecule imaging reveals that two DNA-loop extruding condensin complexes can cross each other, providing a possible explanation for the efficient compaction of chromosomes by unidirectional loop extrusion
- 18.Manalastas-Cantos K, Kschonsak M, Haering CH, Svergun DI. Solution structure and flexibility of the condensin HEAT-repeat subunit Ycg1. J Biol Chem. 2019;294:13822–13829. doi: 10.1074/jbc.RA119.008661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hara K, Kinoshita K, Migita T, Murakami K, Shimizu K, Takeuchi K, Hirano T, Hashimoto H. Structural basis of HEAT-kleisin interactions in the human condensin I subcomplex. EMBO Rep. 2019;20 doi: 10.15252/embr.201847183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kschonsak M, Merkel F, Bisht S, Metz J, Rybin V, Hassler M, Haering CH. Structural Basis for a Safety-Belt Mechanism That Anchors Condensin to Chromosomes. Cell. 2017;171:588–600.:e524. doi: 10.1016/j.cell.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Muir KW, Bowler MW, Metz J, Haering CH, Panne D. Structural basis for Scc3-dependent cohesin recruitment to chromatin. Elife. 2018;7 doi: 10.7554/eLife.38356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Haarhuis JHI, Sedeno Cacciatore A, Oldenkamp R, van Ruiten MS, Willems L, Teunissen H, Muir KW, de Wit E, Rowland BD, et al. The structural basis for cohesin-CTCF-anchored loops. Nature. 2020;578:472–476. doi: 10.1038/s41586-019-1910-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Wit E, Vos ES, Holwerda SJ, Valdes-Quezada C, Verstegen MJ, Teunissen H, Splinter E, Wijchers PJ, Krijger PH, de Laat W. CTCF Binding Polarity Determines Chromatin Looping. Mol Cell. 2015;60:676–684. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 25.Kashammer L, Saathoff JH, Lammens K, Gut F, Bartho J, Alt A, Kessler B, Hopfner KP. Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex. Mol Cell. 2019;76:382–394.:e386. doi: 10.1016/j.molcel.2019.07.035. [DOI] [PubMed] [Google Scholar]; ** Cryo-EM structures of the Rad50–Mre11 active site complex at different functional states provide a mechanism for DNA binding at the nucleotide-engaged ATPase heads
- 26.Seifert FU, Lammens K, Stoehr G, Kessler B, Hopfner KP. Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50. EMBO J. 2016;35:759–772. doi: 10.15252/embj.201592934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Sung S, Kim Y, Li F, Gwon G, Jo A, Kim AK, Kim T, Song OK, Lee SE, et al. ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex. EMBO J. 2016;35:743–758. doi: 10.15252/embj.201592462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamada K, Miyata M, Hirano T. Molecular basis of SMC ATPase activation: role of internal structural changes of the regulatory subcomplex ScpAB. Structure. 2013;21:581–594. doi: 10.1016/j.str.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 29.Lammens A, Schele A, Hopfner KP. Structural biochemistry of ATP-driven dimerization and DNA-stimulated activation of SMC ATPases. Curr Biol. 2004;14:1778–1782. doi: 10.1016/j.cub.2004.09.044. [DOI] [PubMed] [Google Scholar]
- 30.Vazquez Nunez R, Ruiz Avila LB, Gruber S. Transient DNA Occupancy of the SMC Interarm Space in Prokaryotic Condensin. Mol Cell. 2019;75:209–223.:e206. doi: 10.1016/j.molcel.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Gruber and colleagues provide in vivo and in vitro evidence for DNA binding by prokaryotic SMC head domains and supports the topological entrapment in alternating compartments of the SMC–ScpAB complex
- 31.Muir KW, Li Y, Weis F, Panne D. The structure of the cohesin ATPase elucidates the mechanism of SMC-kleisin ring opening. Nat Struct Mol Biol. 2020;27:233–239. doi: 10.1038/s41594-020-0379-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; * The first high-resolution cryo-EM structure of nucleotide-engaged cohesin Smc1–Smc3 ATPase heads puts forward a mechanism for kleisin disengagement from the Smc3 coiled coil
- 32.Kurze A, Michie KA, Dixon SE, Mishra A, Itoh T, Khalid S, Strmecki L, Shirahige K, Haering CH, Lowe J, et al. A positively charged channel within the Smc1/Smc3 hinge required for sister chromatid cohesion. EMBO J. 2011;30:364–378. doi: 10.1038/emboj.2010.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alt A, Dang HQ, Wells OS, Polo LM, Smith MA, McGregor GA, Welte T, Lehmann AR, Pearl LH, Murray JM, et al. Specialized interfaces of Smc5/6 control hinge stability and DNA association. Nat Commun. 2017;8:14011. doi: 10.1038/ncomms14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srinivasan M, Scheinost JC, Petela NJ, Gligoris TG, Wissler M, Ogushi S, Collier JE, Voulgaris M, Kurze A, Chan KL, et al. The Cohesin Ring Uses Its Hinge to Organize DNA Using Non-topological as well as Topological Mechanisms. Cell. 2018;173:1508–1519.:e1518. doi: 10.1016/j.cell.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirano M, Hirano T. Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J. 2002;21:5733–5744. doi: 10.1093/emboj/cdf575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griese JJ, Hopfner KP. Structure and DNA-binding activity of the Pyrococcus furiosus SMC protein hinge domain. Proteins. 2011;79:558–568. doi: 10.1002/prot.22903. [DOI] [PubMed] [Google Scholar]
- 37.Griese JJ, Witte G, Hopfner KP. Structure and DNA binding activity of the mouse condensin hinge domain highlight common and diverse features of SMC proteins. Nucleic Acids Res. 2010;38:3454–3465. doi: 10.1093/nar/gkq038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piazza I, Rutkowska A, Ori A, Walczak M, Metz J, Pelechano V, Beck M, Haering CH. Association of condensin with chromosomes depends on DNA binding by its HEATrepeat subunits. Nat Struct Mol Biol. 2014;21:560–568. doi: 10.1038/nsmb.2831. [DOI] [PubMed] [Google Scholar]
- 39.Burmann F, Basfeld A, Vazquez Nunez R, Diebold-Durand ML, Wilhelm L, Gruber S. Tuned SMC Arms Drive Chromosomal Loading of Prokaryotic Condensin. Mol Cell. 2017;65:861–872.:e869. doi: 10.1016/j.molcel.2017.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hassler M, Shaltiel IA, Haering CH. Towards a Unified Model of SMC Complex Function. Curr Biol. 2018;28:R1266–R1281. doi: 10.1016/j.cub.2018.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eeftens JM, Katan AJ, Kschonsak M, Hassler M, de Wilde L, Dief EM, Haering CH, Dekker C. Condensin Smc2-Smc4 Dimers Are Flexible and Dynamic. Cell Rep. 2016;14:1813–1818. doi: 10.1016/j.celrep.2016.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diebold-Durand ML, Lee H, Ruiz Avila LB, Noh H, Shin HC, Im H, Bock FP, Burmann F, Durand A, Basfeld A, et al. Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization. Mol Cell. 2017;67:334–347.:e335. doi: 10.1016/j.molcel.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soh YM, Burmann F, Shin HC, Oda T, Jin KS, Toseland CP, Kim C, Lee H, Kim SJ, Kong MS, et al. Molecular basis for SMC rod formation and its dissolution upon DNA binding. Mol Cell. 2015;57:290–303. doi: 10.1016/j.molcel.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burmann F, Lee BG, Than T, Sinn L, O’Reilly FJ, Yatskevich S, Rappsilber J, Hu B, Nasmyth K, Lowe J. A folded conformation of MukBEF and cohesin. Nat Struct Mol Biol. 2019;26:227–236. doi: 10.1038/s41594-019-0196-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Negative stain electron microscopy, protein cross-linking and crystallography reveal an ‘elbow’ region in SMC coiled coils that gives rise to a folded rod-shaped conformation
- 45.Yoshimura SH, Hizume K, Murakami A, Sutani T, Takeyasu K, Yanagida M. Condensin architecture and interaction with DNA: regulatory non-SMC subunits bind to the head of SMC heterodimer. Curr Biol. 2002;12:508–513. doi: 10.1016/s0960-9822(02)00719-4. [DOI] [PubMed] [Google Scholar]
- 46.Krepel D, Davtyan A, Schafer NP, Wolynes PG, Onuchic JN. Braiding topology and the energy landscape of chromosome organization proteins. Proc Natl Acad Sci U S A. 2020;117:1468–1477. doi: 10.1073/pnas.1917750117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terakawa T, Bisht S, Eeftens JM, Dekker C, Haering CH, Greene EC. The condensin complex is a mechanochemical motor that translocates along DNA. Science. 2017;358:672–676. doi: 10.1126/science.aan6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sedeno Cacciatore A, Rowland BD. Loop formation by SMC complexes: turning heads, bending elbows, and fixed anchors. Curr Opin Genet Dev. 2019;55:11–18. doi: 10.1016/j.gde.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 49.Hassler M, Shaltiel IA, Kschonsak M, Simon B, Merkel F, Tharichen L, Bailey HJ, Macosek J, Bravo S, Metz J, et al. Structural Basis of an Asymmetric Condensin ATPase Cycle. Mol Cell. 2019;74:1175–1188.:e1179. doi: 10.1016/j.molcel.2019.03.037. ** A set of crystal structures and biochemical reconstitution experiments describe a sequence of structural transitions at the SMC ATPase core during its reaction cycle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rojowska A, Lammens K, Seifert FU, Direnberger C, Feldmann H, Hopfner KP. Structure of the Rad50 DNA double-strand break repair protein in complex with DNA. EMBO J. 2014;33:2847–2859. doi: 10.15252/embj.201488889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chapard C, Jones R, van Oepen T, Scheinost JC, Nasmyth K. Sister DNA Entrapment between Juxtaposed Smc Heads and Kleisin of the Cohesin Complex. Mol Cell. 2019;75:224–237.:e225. doi: 10.1016/j.molcel.2019.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marko JF, De Los Rios P, Barducci A, Gruber S. DNA-segment-capture model for loop extrusion by structural maintenance of chromosome (SMC) protein complexes. Nucleic Acids Res. 2019;47:6956–6972. doi: 10.1093/nar/gkz497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Schoeffler AJ, Berger JM, Oakley MG. The crystal structure of the hinge domain of the Escherichia coli structural maintenance of chromosomes protein MukB. J Mol Biol. 2010;395:11–19. doi: 10.1016/j.jmb.2009.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woo JS, Lim JH, Shin HC, Suh MK, Ku B, Lee KH, Joo K, Robinson H, Lee J, Park SY, et al. Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell. 2009;136:85–96. doi: 10.1016/j.cell.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 55.Kikuchi S, Borek DM, Otwinowski Z, Tomchick DR, Yu H. Crystal structure of the cohesin loader Scc2 and insight into cohesinopathy. Proc Natl Acad Sci U S A. 2016;113:12444–12449. doi: 10.1073/pnas.1611333113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee BG, Roig MB, Jansma M, Petela N, Metson J, Nasmyth K, Lowe J. Crystal Structure of the Cohesin Gatekeeper Pds5 and in Complex with Kleisin Scc1. Cell Rep. 2016;14:2108–2115. doi: 10.1016/j.celrep.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roig MB, Lowe J, Chan KL, Beckouet F, Metson J, Nasmyth K. Structure and function of cohesin’s Scc3/SA regulatory subunit. FEBS Lett. 2014;588:3692–3702. doi: 10.1016/j.febslet.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hashimoto H, Wang D, Horton JR, Zhang X, Corces VG, Cheng X. Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA. Mol Cell. 2017;66:711–720.:e713. doi: 10.1016/j.molcel.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamada K, Su’etsugu M, Takada H, Miyata M, Hirano T. Overall Shapes of the SMC-ScpAB Complex Are Determined by Balance between Constraint and Relaxation of Its Structural Parts. Structure. 2017;25:603–616.:e604. doi: 10.1016/j.str.2017.02.008. [DOI] [PubMed] [Google Scholar]