Abstract
Toehold-mediated DNA strand displacement (DSD) is a powerful strategy to engineer dynamics in DNA-based devices and for molecular computing. However, facile strategies for autonomously self-resettable DSD cascades with programmable lifetimes are still missing. Here, we concatenate an ATP-powered ligation/restriction network with toehold-mediated DSD reactions realizing ATP-driven transient DSD with self-resetting behavior. The ATP-fueled ligation biases strand displacement reactions by increasing the toehold length and increasing the local concentration by covalent fixation, while the concurrent endonuclease restriction eliminates this bias allowing to reset the system. The lifetimes and adaptive dynamic steady states for the DSD are engineered by the ATP-fueled uphill-driven non-equilibrium ligation/restriction system. By programming the toeholds for downstream DSD reaction networks, we realize ATP-fueled transient DSD cascades. Higher level of fuel-driven automaton is achieved by combining two sub-systems for the transient DSD, where the expelled strands are each other’s input strands and the restriction-induced double-stranded DNA melting resets the systems.
Keywords: Systems Chemistry, Chemical Reaction Network, DNA strand displacement, Feedback Systems, Non-equilibrium Self-Assembly
Introduction
Biological systems implement complex molecular networks to execute signal transduction and regulate system transformation. 1–2 This typically involves the coordination and concatenation of multiple networks able to process chemically or physically distinct signals to allow for communication and for making sense of diverse signals. 3–4 Such biological systems inspire researchers to pursue regulated self-assemblies for complex molecular construction and dynamic behavior, which expand the capability of molecular nanotechnology and advance the development of artificial molecular machinery and automatons. 5–13
DNA has emerged as a powerful material for structural nanoscience by exploiting programmable Watson-Crick base pairing. 14–15 Towards implementation of dynamics in autonomous systems, chemical reaction networks (CRNs) including toehold-mediated DNA strand displacement (DSD) circuits, DNA/RNA transcription machinery, polymerase/exonuclease/nickase (PEN) reactions, and ATP-driven ligation/restriction networks have been developed. 7, 11, 16–26 Increasingly complex behaviors have been shown individually by exploiting energy to modulate energy landscapes, but, new synergetic behavior can be in reach by combination of them. DSD has given rise to molecular actuators, 18, 27–28 soft robots, 29–30 and logic circuits capable of computing. 31 The use of toehold-mediated DSD allows to temporally and spatially reproduce reconfigurable and autonomous DNA nanostructures. 32–33 However, to date, most of the DSD reactions operate unidirectionally via kinetically and thermodynamically controlled DNA hybridization, in which thermodynamic equilibration drives the structural evolution. Reversibility in DSD has been realized by Winfree and coworkers through equal rate of toehold exchange. 34 This strategy has been used for DNA catalyzed reaction networks to amplify nucleic acid signals, digital signal restoration, and neural-network-like computations. 35–36
However, it is unfortunately hard to reset those DSD systems. Fluidic mixing is often used to introduce recovery strands to reset a DSD system, 27 which is, however, non-robust and wasteful of materials. Strand displacement oscillators have been prepared by CRNs consisting of three autocatalytic reactions in cyclic competition, 10, 37 which, however, continuously consume helper strands and accumulate waste. Shih and coworker reported a strategy to reset DSDs using temperature cycling, 38 where DSD cascades were favored at low temperatures while rebinding of recovery strands was favored at higher temperatures. For such systems, however, an automated operation with programmable lifetimes is not achievable. To engineer transient DSD reactions, earlier on Li and coworker developed adenosine-driven transient double-stranded DNA dehybridization using an adenosine-binding aptamer and adenosine deaminase, 39 and recently Ricci and coworkers used disulfide bond embedded single-stranded DNA as a redox input to engineer transient DNA-based nanostructures in the presence of a reducing agent (tris(2-carboxyethyl)phosphine). 9 Unfortunately, those transient DSD systems either rely on fixed sequences from aptamers or are restricted to specific chemical modifications (disulfide bond), and multiple layers of DSD cascades are hard to achieve. We hypothesized that new resetting behavior allowing for complicated DSD cascades could be encoded by combing energetically downhill DSDs with a fuel-driven CRN with an energetically uphill transient component that has a chemical clock behavior with an internal fuel-dependent lifetime reset function.
In the present study, we demonstrate a strategy to program fully self-resettable and autonomous DSDs by combination with an ATP-dependent DNA clock, where ATP-powered ligation using T4 DNA ligase induces forward DSDs, while the endonuclease restriction induces duplex dissociation followed by backward DSDs, whereby the system returns to its original state. By further incorporating conventional toehold-mediated DSD reaction networks with the ATP-driven transient DSD, we realize ATP-fueled transient DSD cascades. Critically, due to the flexible programmability of the molecular recognition sequences located in the sticky ends for ligation in the ATP-driven network and of the DNA sequences for DSD, it is possible to further achieve ATP-fueled transient feedback DSD systems by programming the expelled strands from two subsystems to be each other’s input strands.
Results and discussion
General concept for ATP-driven transient DNA strand displacement circuitry
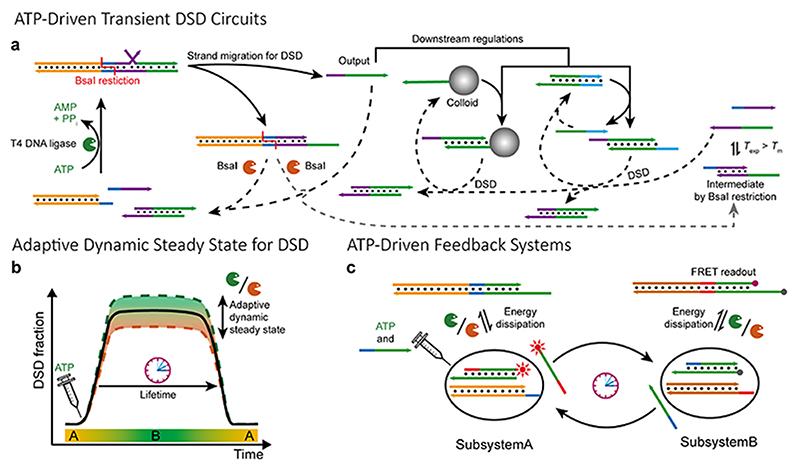
In our design, the ATP-powered ligation of the DNA species using T4 DNA ligase induces forward DSDs, while the concurrent endonuclease restriction induces duplex dissociation triggering backward DSDs (Figure 1a). Intermediately released strands (output) can target single-stranded DNA modified colloids for transient functionalization or regulate multiple layers of downstream DSD reactions via toehold-mediated DSD. The DSD is maintained in an ATP-driven dynamic steady state (DySS) through an enzymatic reaction network (ERN) of ligation and restriction, and the combined systems are bound in their behavior to the consumption of ATP in the ERN, leading to programmable lifetimes. The adaptive DySS properties (frequency for ligation/cutting and fractional degree of DSD) for the system are mainly regulated by the kinetic balance and total concentration of both enzymes (Figure 1b). 23 Aside from the transient DSD circuitry, fuel-driven automatons can be achieved by feedback-controlled DSD systems, where the expelled strands are each other’s input strands and the restriction-induced dsDNA melting triggers the recovery of the systems (Figure 1c).
Figure 1. Chemical fuel-driven transient DSD circuitry.
(a) Overview of ATP-fueled transient DSD circuits. Experimental temperature (T exp) = 37.0 °C. Melting temperature (T m) of the Intermediate (right) by BsaI restriction = 30.8 °C. The 3’ ends are marked with an arrow. (b) Adaptive DySS properties of ATP-fueled DSD, which is regulated by the kinetics of both enzymes. (c) Schematic illustration of ATP-fueled automaton for feedback component exchange between two subsystems. Forward reactions are in solid lines and backward resetting reactions are in dashed lines.
ATP-fueled transient DNA strand displacement reaction
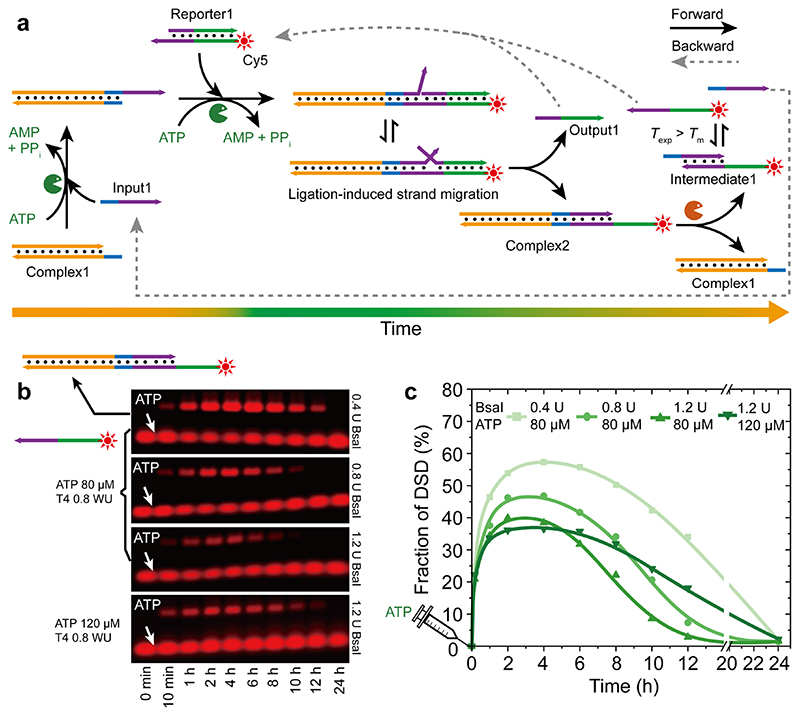
Figure 2a illustrates a proof-of-concept design for an ATP-powered transient DSD. The system contains a dsDNA Complex1, a single-stranded DNA (ssDNA) input (Input1), and a dsDNA reporter (Reporter1). Reporter1 carries a 4 nucleotide (nt) toehold and the length of the violet dsDNA part is 5 nt. The green part of the reporter is 8 nt (details of the sequences in Supplementary Table 1). Without ATP, no DSD happens between Input1 and Reporter1 due to uncompetitive toehold (4 nt violet vs. 8 nt green). However, after adding ATP, Input1 is covalently ligated to Complex1, which itself triggers subsequent ligation with the toehold in Reporter1. This second step ligation activates the strand displacement, because the original 4 nt toehold interaction between Input1 and Reporter1 is now increased by the whole length of Complex1 to 29 nt due to covalent fixation. Consequently, strand migration happens and Output1 is expelled from Reporter1. Complex1 changes to Complex2. Simultaneously, BsaI endonuclease is able to cleave the newly formed Complex2 to regenerate Complex1 and a dsDNA Intermediate1 which dissociates into two ssDNA due to its low T m (Figure S1) and thereby reproduces Input1, as well as Reporter1 by rehybridzation with Output1.
Figure 2. ATP-fueled transient DSD with transient formation of Output1.
(a) Schematic illustration for coupling ATP-driven ligation/restriction ERNs with DSD. T exp = 37.0 °C, T m of Intermediate1 = 34.0 °C. (b) AGE analyses of DSD with time for different sets of enzyme and ATP concentrations. (c) Fraction of DSD for the systems for different concentrations of BsaI and ATP. Lines are guides to the eye. Most relevant concentrations are indicated within the Figure. Concentration of T4 DNA ligase is 0.8 WU. Concentration of each DNA species is 10 μM.
The experimental systems were set at 10 μM of each species, 0.8 Weiss units/μL (WU) T4 DNA ligase, and varied concentrations of ATP and BsaI. Agarose gel electrophoresis (AGE) analyses of time-dependent aliquots of the systems give insights into the transient nature of the systems regarding their fractions of DSD (Figure 2b). To visualize and quantify the key component via AGE, Cy5 was attached to Reporter1 that allows separation in AGE when transformed to Complex2. The intensity ratio between the Complex2 and all fluorescent DNA species was used to calculate the DSD fraction, where the amount of intermediate1 is not considered as it can quickly rehybridize with Output1 to regenerate Reporter1. Figure 2b displays AGE analyses for different BsaI concentrations. A transient Cy5-containing, high molecular weight species of Complex2 indicative of the DSD reaction is observed at shorter migration distance.
The degradation of the transient Complex2 occurs earlier for increased BsaI concentration and changes from ca. 10, 6, and 4 h for BsaI concentration 0.4, 0.8, and 1.2 U, respectively. This is due to the larger transduction speed of ATP by a higher BsaI concentration (Figure 2b). Additional, by increasing BsaI from 0.4 to 1.2 U, the fraction of DSD (calculated by the yield of Complex2) at the DySS decreases from ca. 57 % to 38 %, because a higher BsaI concentration shifts the balance of the average degree of ligation of the T4 DNA ligase/BsaI ERN to the side of the cut species (Figure 2c). 23 Common to all systems is the fact that they reach their DySS plateaus at around 2 h.
An increase of the ATP concentration from 80 to 120 μM at constant 1.2 U BsaI extends the lifetime from ca. 8 to 12 h (Figure 2b), yet, the fraction of DSD in the fueled state remains on a similar plateau. To better understand the similar plateau height, it is important to consider the ATP concentration in relation to the DNA concentration. One full junction ligation by T4 DNA ligase requires two ATP molecules. In the systems discussed here, the injected ATP is eight or twelve folds compared to the DNA substrates. This significant excess also explains, why there is no significant effect of the ATP concentration on the DySS regarding the fraction of DSD, because an effect on the DySS plateau would only appear for much lower stochiometric ratios, i.e. insufficient ATP. 23 Hence, the plateau of the adaptive DySS is majorly set by the kinetics of both enzymes (their ratio) in the presence of an excess ATP. Additionally, both enzymes are sufficiently stable in the investigated time regime. 23–24 For general consideration, it is also important to mention that such ATP-driven system can be refueled by a fresh batch of ATP 23–24 due to the cyclic nature of ERN with repeated activation and deactivation.
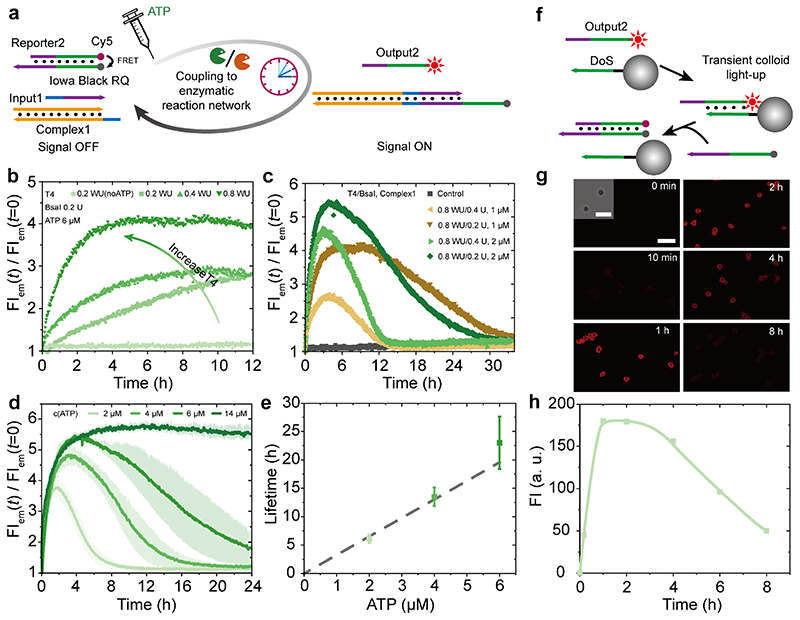
Although AGE gives detailed insights into the formation of different species, it is not an ideal method for determining the lifetime due to limited temporal resolution. Therefore, we will next use FRET readout to more accurately monitor the DSD reactions below. To this end, we added a FRET pair to Reporter2 for in-situ readout of the DSD via a fluorescence spectroscopy and for visualization of downstream reactions at lower DNA concentrations (1 μM of all DNA species; Figure 3a). Before adding ATP, no DSD occurs in the system and the fluorophore and quencher in Reporter2 are in close proximity, leading to the quenching of any excited state of the fluorophore. After addition of ATP, Complex1 is consecutively ligated with Input1 and Reporter2, triggering the release of Output2 from Reporter2, and, thus, an increase of the fluorescence intensity (FI) is observed. A higher FI indicates a higher fraction of DSD. After ATP is running out, the system returns back to its original state with a quenched fluorescence.
Figure 3. ATP-fueled transient DSD monitored by FRET and its use for transient colloid surface functionalization.
(a) Schematic illustration of transient DSD with FRET pair for in-situ DSD readout. (b) Tuning the adaptive DSD output and speed by increasing the concentration of T4 DNA ligase from 0.2 to 0.8 WU while keeping 0.2 U BsaI, 6 μM ATP, and 1 μM each DNA species constant. (c) Increasing of the Complex1 concentration and increasing of BsaI lead to higher DSD output and speed up the ATP consumption, respectively. Complex1 was increased from 1 to 2 μM, and BsaI was varied from 0.2 to 0.4 U, while 6 μM ATP and 0.8 WU T4 DNA ligase were kept constant. (d-e) Programmable lifetimes seen by time-dependent FI at different ATP concentrations. ATP concentration was varied from 14 to 2 μM, while 2 μM Complex1, 0.8 WU T4 DNA ligase, and 0.2 U BsaI were kept constant. Shaded areas correspond to the standard deviation of duplicate measurements. (f) Schematic illustration of transient colloid functionalization by Output2 from the transient DSD. T exp = 37.0 °C, T m of Intermediate = 30.8 °C (see also Figure 1). (g) Time-dependent confocal images for the colloids for transient colloid functionalization via transient DSD using 2 μM ATP, 2 μM Complex1, 0.8 WU T4 DNA ligase, and 0.2 U BsaI supplemented with ca. 4.68-6.68×107 beads/mL of ssDNA-modified colloids. All scale bars = 5 μm. (h) Time-dependent FI of the colloids. The line is a guide to the eye.
We first focused on tuning the DSD level by variation of the T4 DNA ligase content. By increasing the T4 DNA ligase from 0.2 to 0.8 WU (while keeping 0.2 U BsaI and 6 μM ATP constant), a faster DSD and an increased fraction of DSD are observed by the faster and higher FI increase (Figure 3b). Hence, at fixed concentrations of DNA species, ATP and BsaI, an increase of the T4 DNA ligase concentration gives rise to a higher fraction of DSD by shifting the dynamic balance in the ERN to higher degrees of ligation. Note that here we only focus on the “early stages” of the transient behavior. In order to bring the transient lifetimes into an experimentally more accessible regime (< days), we furthermore doubled the concentration of Complex1 to 2 μM and also increased the BsaI concentration. This indeed shortens the lifetimes, and gratifyingly 2 μM Complex1 also leads to consistently higher FI and thus higher fractions of DSD (both at 0.2 and 0.4 U BsaI; Figure 3c). More specifically, by fixing the concentrations of ATP, T4 DNA ligase and DNA species, an increase of the BsaI concentration from 0.2 to 0.4 U leads to a significantly lower fraction of DSD and a shorter lifetime. In contrast, when keeping the concentrations of T4 DNA ligase, BsaI, and ATP constant, a doubling of the concentration of Complex1 leads to a significantly higher FI, and, thus, to a higher fraction of DSD, because higher Complex1 concentrations promote the T4 DNA ligase-mediated ligation and deliver more ligated species needed to trigger the DSD. The lifetime of the transient DSD is not sensitive to the Complex1 concentration, because the endonuclease restriction is majorly the rate-limiting step in the ERN governing the ATP-driven lifetime behavior.
We judged the conditions of 0.8 WU T4 DNA ligase, 0.2 U BsaI, and 2 μM Complex1 to provide a good experimental time and DSD space, and fixed these conditions to investigate ATP-programmable lifetime of the transient DSD. Indeed when decreasing the ATP concentration from 6 to 2 μM, the lifetimes for the transient DSD similarly shorten from ca. 23 to 6 h, as calculated from the point where the FI decreases to 20 % of its maximum increased value (Figure 3d,e). The close-to-linear trend in Figure 3e simply reflects the fuel-dependent transduction kinetics. A decrease of the transient FI maximum occurs at concentrations of 2 μM ATP, because ATP becomes a limiting factor in coupling the ATP-driven ERN to the DSD, as one DSD reaction requires two ligation steps where each ligation consumes one molecule of ATP (Figure 2a). This is relevant when the Complex1 concentration of the ERN is of similar concentration (also 2 μM as in these experiments). The slower decrease of the FI at higher ATP concentration is because there is not enough ATP to maintain the DSD at the highest fraction of DSD for extended times, and hence ATP becomes a limiting factor, leading to continuously lower fractions of DSD during the FI decrease phase. Additionally, even though the enzymes are sufficiently stable in the system (see e.g. also transient curves of 30 h in Figure 3c), some loss of activity of BsaI can contribute to an elongated decreases of the FI. 24 It is worth noting that 6 μM ATP is sufficient to drive the DSD to the fueled DySS of the ERN, but a further increase of the ATP concentration cannot induce significantly higher FI (Figure 3d).
Additionally, we exploited the transient DSD generation of Output2 in a system fueled by 2 μM ATP to showcase the transient reconfiguration of the surface functionality of added ssDNA-functionalized colloids that can hybridize with Output2 (Figure 3f; ca. 4.68-6.68×107 beads/mL, and 3.34×10-9 μM ssDNA on colloidal surfaces in total). Indeed as expected, the colloids are lightened up transiently, with a stable FI after ca. 1h, that decays over time (Figure 3g,h). This simple downstream functionalization demonstrates that the transient Ouput2 can be utilized. The timescale correspond to the molecular network characteristics in Figure 3d.
ATP-fueled transient DNA strand displacement cascades
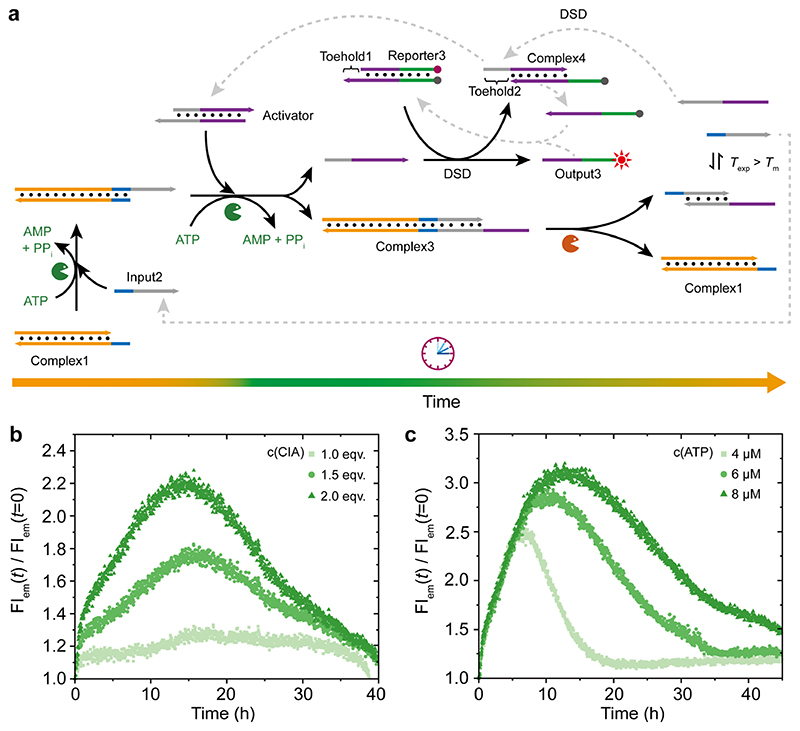
Importantly, downstream toehold-mediated DSD processes can be incorporated into the above transient DSD to realize transient DSD cascades, and thus realize autonomously resettable DSD systems on a circuitry level. Figure 4a shows such a design, where we incorporated an Activator between Complex1 and the Output3 that is generated by two consecutive DSDs. The first DSD is conditional on the ATP-powered ligation, while the second DSD uses classical toehold-mediated strand displacement utilizing Toehold1. The process can be tracked by the FI of Output3 due to the FRET pair located at Reporter3. Simultaneously, the restriction of Complex3 produces Complex1 and a thermodynamically unstable dsDNA that dissociates into two ssDNA due to melting. One ssDNA is simply the original Input2, while the other ssDNA is able break Complex4 by DSD utilizing Toehold2 to induce the backward reaction. This process regenerates the Activator and the released ssDNA rehybridizes with Output3 to regenerate Reporter3. Hence, once ATP is consumed, the system returns to its original state.
Figure 4. ATP-fueled transient DSD circuitry by combining an ATP-fueled DSD with a classical toehold-mediated DSD downstream.
(a) Schematic illustration of the molecular mechanism for the ATP-fueled DSD cascade circuitry. T exp = 37.0 °C, T m of the Intermediate by BsaI restriction = 30.8 °C. (b) Adaptive DySS for transient DSD by changing the concentration of CIA from 1 to 2 μM and keeping 0.1 WU T4 DNA ligase, 0.4 U BsaI, and 6 μM ATP constant. (c) ATP-dependent lifetime for the transient DSD circuitry using 2 μM CIA, 0.2 WU T4 DNA ligase, and 0.4 U BsaI, and varying ATP concentration from 4 to 8 μM. Note that increased T4 DNA ligase was used to speed up the process.
The experimental systems were set at constant concentrations of T4 DNA ligase (0.1 WU), BsaI (0.4 U), ATP (6 μM), and reporter2 (1 μM). Here, a very low concentration of T4 DNA ligase was used to emphasize the effects of the substrates (Complex1, Input2, and Activator) on the transient DSD cascades. Since we have demonstrated above that the DNA species involved in the ligation steps for inducing the DSD can speed up the kinetics and increase the fraction for the DSD at higher concentrations, here the concentrations for Complex1, Input2, and Activator (CIA) were set at the same stoichiometry and were tuned from 1 to 2 μM. Importantly, since the output signal is regulated by toehold-mediated DSD triggered by ssDNA released from the upstream ATP-fueled transient ligation-induced DSD, a higher concentration of the ssDNA strands released from the dsDNA Activator via a higher concentration of CIA speeds up the downstream toehold-mediated DSD (Figure 4b). As expected, higher FIs and thus higher fractions of DSD are obtained at a higher concentration of CIA, as significantly accelerated by faster ligation and a higher concentration of accumulated ssDNA strands released from the dsDNA Activator. Due to the very low concentration of T4 DNA ligase, it is quite time-consuming for the systems to reach the peaks of the DSD cascades (ca. 13 h). An increase of T4 DNA ligase concentration can shorten the time needed for reaching the peak for the DSD cascades, which will be shown below in Figure 4c.
To further program the lifetime of the DSD cascades via different ATP concentration, we used a 2 fold excess of CIA and doubled the T4 DNA ligase concentration to further speed up the ligation and get more continuous increase of the FIs. By changing the ATP concentration from 4 to 8 μM, the lifetime for the transient DSD circuitry changes from ca. 18 to 45 h (Figure 4c), in line with expectations for a fuel-driven system. Compared to the system using 0.1 WU T4 DNA ligase and 2 μM CIA in Figure 4b, here the system with doubled T4 DNA ligase fueled by 6 μM ATP shows a higher FI and the time needed to reach the peak of DSD cascades is shortened by ca. 4 h.
ATP-fueled feedback systems
Going beyond the ATP-fueled DSD cascades, we further implement a fuel-driven automaton exploiting the above developed transient DSD strategy as a first examples of increased systems complexity. To this end, we intertwine two transient DSD subsystems by their input and output information so that they can communicate (Figure 5a,b). Briefly, Input3 for SubsystemA triggers the production of Output4 via ATP-fueled DSD. This Output4 functions as the input for SubsystemB, giving rise to the formation of Output5, which is indeed identical to Input3. Hence, once the system is injected with ATP, autonomous feedback trade-off happens between these two subsystems (Figure 5a,b). Once ATP is consumed, the BsaI controlled restriction resets the systems.
Building on the conditions established above, we designed a system containing 1 μM Complex1 and Complex5, Reporter4, and Reporter5, 0.8 WU T4 DNA ligase, and 6 μM ATP. We first investigated the transient automation via 0.2 U BsaI and different amount of Input3 (0.4, 0.2, and 0.1 μM). Overall, the transient FI readout allows to identify the transient feedback molecular system (Figure 5c). Consistently, there is a transient decrease of the FI, which – importantly – can only be explained by the fact that Output4 is transiently incorporated into SubsystemB, where it is temporally quenched due to the ligation with Complex5 and the quencher-containing Reporter5. When Complex5 is absent, there is no FI change during automation of SubsystemA (no feedback from Sub-systemB) (Figure S2). While Input3 concentrations of 0.4 and 0.2 μM show good recovery of the transient fluorescence quenching, an Input3 concentration of 0.1 μM shows some lack of recovery. This is because the ligation between Complex1 and Input3 is significantly slower at 0.1 μM Output3, while the more significant decrease of FI by 0.1 μM is due to the leakage of Reporter5 (Figure S3), where the released Output4 can quickly ligate with Complex5 together with the quencher strand dissociated from Reporter5. Hence, the slower kinetics for the FI decrease is attributed to system optimization for less leakage. Admittedly, it is challenging to program such feedback systems with highly associated input and output strands and also thermodynamically unstable short duplexes after restriction. We found that the leakage can be significantly decreased by higher concentration of Input3 (Figure S3), which explains the better recovery. Higher concentration of Input3 induces less quenching for the transient feedback systems, because the formation of the FRET pair is majorly set by the kinetically slower ligation-induced strand migration on the activated Complex5, and strand expulsion, but not by the ligation of the strands from leakage. Moreover, by increasing the BsaI concentration to 0.4 U, a smaller FI decrease is obtained due to the enzymatically controlled adaptive DySS for the ATP-driven ERN and the transient DSD that shifts the ligation balance to the restricted species (Figure 5c and 5d).
Conclusion
Taken together, we introduced the concatenation of an ATP-driven ligation/restriction network with DSD reaction networks that allows for ATP-fueled transient and reversible DSD with timed operation. The key step relies on an ATP-fueled ligation that biases strand displacement reactions in a transient fashion by temporally increasing the toehold length and increasing the local concentration by covalent fixation. By concurrent cleavage using the restriction enzyme, this bias can be removed allowing for the overall reversibility of the DSD. The DSD part follows the lifetime considerations imposed by the chemically fueled uphill-driven non-equilibrium ATP ligation/restriction system. Adaptive DySS properties regarding the fraction of DSD can be regulated by the kinetics of both enzymes and the concentration of ligation template and input trigger.
This strategy paves a new way for fuel-driven non-equilibrium transient DSD as well as DSD-programmed self-assemblies for more complex system behavior. Importantly, this fuel-driven transient DSD can be further combined with classical toehold-mediated DSD to realize transient and self-resettable DSD circuits, which has not been possible by previous DSD system. Moreover, this fuel-driven transient DSD strategy can be evolved to program ATP-driven automatons, which use two communicating ATP-driven DSD subsystems, that exchange information via output/input couples that are each other’s inverse. We envision that our ATP-fueled transient DSD system shows great potential in DNA computing and for devices for information processing.
Supplementary Material
Acknowledgment
We acknowledge support by the European Research Council starting Grant (TimeProSAMat) Agreement 677960, as well as from the DFG Cluster of Excellence livMatS “Living, Adaptive and Energy-Autonomous Materials Systems”, EXC-2193/1 - 390951807.
Footnotes
Notes
The authors declare no competing financial interests.
References
- 1.Kiel C, Yus E, Serrano L. Engineering Signal Transduction Pathways. Cell. 2010;140(1):33–47. doi: 10.1016/j.cell.2009.12.028. [DOI] [PubMed] [Google Scholar]
- 2.Mann S. Life as a Nanoscale Phenomenon. Angew Chem Int Ed. 2008;47(29):5306–5320. doi: 10.1002/anie.200705538. [DOI] [PubMed] [Google Scholar]
- 3.Leiman P, Kanamaru S, Mesyanzhinov V, Arisaka F, Rossmann M. Structure and Morphogenesis of Bacteriophage T4. Cell Mol Life Sci. 2003;60(11):2356–2370. doi: 10.1007/s00018-003-3072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walther A. From Responsive to Adaptive and Interactive Materials and Materials Systems: A Roadmap. Adv Mater. 2019;32(20):1905111. doi: 10.1002/adma.201905111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benenson Y, Paz-Elizur T, Adar R, Keinan E, Livneh Z, Shapiro E. Programmable and Autonomous Computing Machine Made of Biomolecules. Nature. 2001;414(6862):430–434. doi: 10.1038/35106533. [DOI] [PubMed] [Google Scholar]
- 6.Deng J, Walther A. Pathway Complexity in Fuel-Driven DNA Nanostructures with Autonomous Reconfiguration of Multiple Dynamic Steady States. J Am Chem Soc. 2020;142(2):685–689. doi: 10.1021/jacs.9b11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green LN, Subramanian HK, Mardanlou V, Kim J, Hariadi RF, Franco E. Autonomous Dynamic Control of DNA Nanostructure Self-Assembly. Nat Chem. 2019;11(6):510–520. doi: 10.1038/s41557-019-0251-8. [DOI] [PubMed] [Google Scholar]
- 8.Deng J, Bezold D, Jessen H, Walther A. Multiple Light Control Mechanisms in ATP-Fueled Non-Equilibrium DNA Systems. Angew Chem Int Ed. 2020;59(29):12084–12092. doi: 10.1002/anie.202003102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Del Grosso E, Prins L, Ricci F. Transient DNA-Based Nanostructures Controlled by Redox Inputs. Angew Chem Int Ed. 2020;59(32):13238–13245. doi: 10.1002/anie.202002180. [DOI] [PubMed] [Google Scholar]
- 10.Srinivas N, Parkin J, Seelig G, Winfree E, Soloveichik D. Enzyme-Free Nucleic Acid Dynamical Systems. Science. 2017;358(6369):eaal2052. doi: 10.1126/science.aal2052. [DOI] [PubMed] [Google Scholar]
- 11.Su H, Xu J, Wang Q, Wang F, Zhou X. High-Efficiency and Integrable DNA Arithmetic and Logic System Based on Strand Displacement Synthesis. Nat Commun. 2019;10(1):5390. doi: 10.1038/s41467-019-13310-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng J, Walther A. ATP-Responsive and ATP-Fueled Self-Assembling Systems and Materials. Adv Mater. 2020;32(42):2002629. doi: 10.1002/adma.202002629. [DOI] [PubMed] [Google Scholar]
- 13.Mishra A, Dhiman S, George SJ. ATP-Driven Synthetic Supramolecular Assemblies: From ATP as a Template to Fuel. Angew Chem Int Ed. 2020 doi: 10.1002/anie.202006614. [DOI] [PubMed] [Google Scholar]
- 14.Chen RP, Blackstock D, Sun Q, Chen W. Dynamic Protein Assembly by Programmable DNA Strand Displacement. Nat Chem. 2018;10(4):474–481. doi: 10.1038/s41557-018-0016-9. [DOI] [PubMed] [Google Scholar]
- 15.Aldaye FA, Palmer AL, Sleiman HF. Assembling Materials with DNA as the Guide. Science. 2008;321(5897):1795–1799. doi: 10.1126/science.1154533. [DOI] [PubMed] [Google Scholar]
- 16.Wang ZG, Elbaz J, Willner I. A Dynamically Programmed DNA Transporter. Angew Chem Int Ed. 2012;51(18):4322–4326. doi: 10.1002/anie.201107855. [DOI] [PubMed] [Google Scholar]
- 17.Seelig G, Yurke B, Winfree E. Catalyzed Relaxation of a Metastable DNA Fuel. J Am Chem Soc. 2006;128(37):12211–12220. doi: 10.1021/ja0635635. [DOI] [PubMed] [Google Scholar]
- 18.Zhang DY, Seelig G. Dynamic DNA Nanotechnology Using Strand-Displacement Reactions. Nat Chem. 2011;3(2):103. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- 19.Shah S, Wee J, Song T, Ceze L, Strauss K, Chen Y-J, Reif J. Using Strand Displacing Polymerase to Program Chemical Reaction Networks. J Am Chem Soc. 2020;142(21):9587–9593. doi: 10.1021/jacs.0c02240. [DOI] [PubMed] [Google Scholar]
- 20.Agarwal S, Franco E. Enzyme-Driven Assembly and Disassembly of Hybrid DNA–RNA Nanotubes. J Am Chem Soc. 2019;141(19):7831–7841. doi: 10.1021/jacs.9b01550. [DOI] [PubMed] [Google Scholar]
- 21.Montagne K, Plasson R, Sakai Y, Fujii T, Rondelez Y. Programming an in Vitro DNA Oscillator Using a Molecular Networking Strategy. Mol Syst Biol. 2011;7(1):466. doi: 10.1038/msb.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baccouche A, Montagne K, Padirac A, Fujii T, Rondelez Y. Dynamic DNA-Toolbox Reaction Circuits: A Walkthrough. Methods. 2014;67(2):234–249. doi: 10.1016/j.ymeth.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 23.Heinen L, Walther A. Programmable Dynamic Steady States in ATP-Driven Nonequilibrium DNA Systems. Sci Adv. 2019;5(7):eaaw0590. doi: 10.1126/sciadv.aaw0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng J, Walther A. ATP-Powered Molecular Recognition to Engineer Transient Multivalency and Self-Sorting 4D Hierarchical Systems. Nat Commun. 2020;11(1):3658. doi: 10.1038/s41467-020-17479-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun M, Deng J, Walther A. Polymer Transformers: Interdigitating Reaction Networks of Fueled Monomer Species to Reconfigure Functional Polymer States. Angew Chem Int Ed. 2020;59(41):18161–18165. doi: 10.1002/anie.202006526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng J, Walther A. Programmable ATP-Fueled DNA Coacervates by Transient Liquid-Liquid Phase Separation. Chem. 2020 doi: 10.1016/j.chempr.2020.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. A DNA-Fuelled Molecular Machine Made of DNA. Nature. 2000;406(6796):605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 28.Yan H, Zhang X, Shen Z, Seeman NC. A Robust DNA Mechanical Device Controlled by Hybridization Topology. Nature. 2002;415(6867):62–65. doi: 10.1038/415062a. [DOI] [PubMed] [Google Scholar]
- 29.Fern J, Schulman R. Modular DNA Strand-Displacement Controllers for Directing Material Expansion. Nat Commun. 2018;9(1):3766. doi: 10.1038/s41467-018-06218-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cangialosi A, Yoon C, Liu J, Huang Q, Guo J, Nguyen TD, Gracias DH, Schulman R. DNA Sequence–Directed Shape Change of Photopatterned Hydrogels Via High-Degree Swelling. Science. 2017;357(6356):1126–1130. doi: 10.1126/science.aan3925. [DOI] [PubMed] [Google Scholar]
- 31.Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-Free Nucleic Acid Logic Circuits. Science. 2006;314(5805):1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 32.Wei B, Ong LL, Chen J, Jaffe AS, Yin P. Complex Reconfiguration of DNA Nanostructures. Angew Chem Int Ed. 2014;126(29):7605–7609. doi: 10.1002/anie.201402437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joesaar A, Yang S, Bögels B, van der Linden A, Pieters P, Kumar BP, Dalchau N, Phillips A, Mann S, de Greef TF. DNA-Based Communication in Populations of Synthetic Protocells. Nat Nanotechnol. 2019;14(4):369–378. doi: 10.1038/s41565-019-0399-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang DY, Winfree E. Control of DNA Strand Displacement Kinetics Using Toehold Exchange. J Am Chem Soc. 2009;131(47):17303–17314. doi: 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]
- 35.Qian L, Winfree E. Scaling up Digital Circuit Computation with DNA Strand Displacement Cascades. Science. 2011;332(6034):1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 36.Qian L, Winfree E, Bruck J. Neural Network Computation with DNA Strand Displacement Cascades. Nature. 2011;475(7356):368–372. doi: 10.1038/nature10262. [DOI] [PubMed] [Google Scholar]
- 37.Soloveichik D, Seelig G, Winfree E. DNA as a Universal Substrate for Chemical Kinetics. Proc Natl Acad Sci USA. 2010;107(12):5393–5398. doi: 10.1073/pnas.0909380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hahn J, Shih WM. Thermal Cycling of DNA Devices Via Associative Strand Displacement. Nucleic Acids Res. 2019;47(20):10968–10975. doi: 10.1093/nar/gkz844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nutiu R, Li Y. A DNA-Protein Nanoengine for “on-Demand” Release and Precise Delivery of Molecules. Angew Chem Int Ed. 2005;44(34):5464–5467. doi: 10.1002/anie.200501214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.