Abstract
We introduce pathway complexity on a multicomponent systems level in chemically fueled transient DNA polymerization systems, achieving autonomous evolution over multiple structural dynamic steady states from monomers to dimers, oligomers of dimers to randomized polymer structures before being ultimately degraded back to monomers once the fuel is consumed. The enabling key principle is to design monomer species having kinetically selected molecular recognition in the structure-forming step and which are reconfigured in an enzymatic reaction network. This non-equilibrium systems chemistry approach to pathway complexity provides new conceptual insights in fuel-driven automatons and autonomous materials design.
Nature relies on non-equilibrium structures and processes to perform work by dissipating energy stored in ATP or GTP. 1–4 Pathway complexity plays a pivotal role to associate those dynamic structures and thus realize a living system. For instance, intracellular signaling networks involve multiple steps of ATP/GTP-powered phosphorylation and enzyme activation for downstream regulations. 5–8 Thus far, synthetic systems with fuel-driven transient properties are developed in the fields of systems chemistry, materials science and synthetic biology. 4, 9–12 Synthetic systems requiring continuous energy influx have extended the features and functions of present-day functional materials. 13–14
Most presently existing fuel-driven systems show single trajectories of their non-equilibrium states, such as helical structure switching, 15 supramolecular polymerization, 14, 16 nanoreactors, 13, 17 transient hydrogels, 18–19 and photonic materials. 20 On the contrary, in classical supra-molecular chemistry, pathway complexity is regularly exploited to transiently pass energetically downhill from metastable structures to equilibrium. 21–22 A relevant example showing multiple states in an autonomous fashion is the case of supramolecular helicity switching coupled to hydrolysis of environmental ATP to ADP, AMP, and to adenosine. 15 In directly chemically driven systems, Boekhoven and coworkers recently showed that small fuel amounts allowed the formation of reversibly assembling transient clusters, while large fuel amounts lead to a trapping of aggregates in a non-transient fashion. 23 Critically, pathway-controlled uphill fuel-driven systems involving multiple building blocks with an autonomous structural reconfiguration of multiple dynamic transient states have not been shown by any previous report. Such a concept would however be very valuable to promote a further integration of systems chemistry concepts with reaction networks and dissipative and transient structure formation.
The difficulty in implementing such a concept in synthetic system is related to the need for highest levels of programmability and ability for deterministic autonomous reconfiguration leading to multiple transient dynamic steady-state (DySS) structures, which is a profound challenge in supramolecular chemistry. 24–26 On the contrary, DNA shows great potential to program reaction networks and pathways for self-assemblies in a systems chemistry approach. 27–29
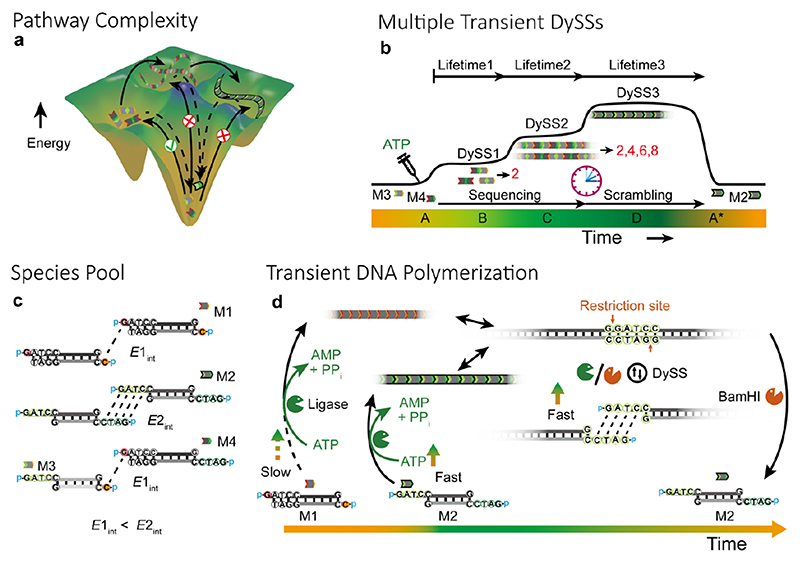
Herein, we demonstrate pathway complexity in ATP-fueled transient DNA polymerizations from a species pool realizing autonomous and transient multiple DySS structures (Figure 1a,b). Our systems build on our recently reported ATP-fueled DySS polymerization of DNA nanostructures using an uphill-driven dynamic covalent DNA bond orchestrated by an enzymatic reaction network (ERN) of concurrent ligation and cleavage of double-stranded DNA (dsDNA) building blocks (Figure 1d). 30 Importantly, the molecular recognition for ligation can be programmed by sticky-end length, enabling distinct complementary recognition pairs and kinetically selected pathways in the starting species pool.
Figure 1. Pathway complexity in ATP-powered transient multi-state systems.
(a) Schematic illustration of pathway complexity in ATP-driven DNA polymerization. (b) ATP-driven autonomous multiple transient DySSs. (c) Building blocks with varied length of sticky ends. (d) Schematic illustration of transient polymerization of 1 nt and 4 nt sticky-end dsDNA tiles. Note the reconfiguration of the 1 nt ends into 4 nt ends.
Our species pool contains monomers with 1 nucleotide (nt) and 4 nt sticky ends and combinations of those (Figure 1c). The 1 nt sticky ends in M1 have lower stickiness and thus slower ligation than the 4 nt sticky ends in M2. M1 and M2 have orthogonal molecular recognitions. The tiles with combined 1 nt and 4 nt sticky ends, M3 and M4, serve as the key for pathway complexity in the system due to the kinetically selective sequential ligation of these sticky ends. Subsequently, uniformization of the sticky ends occurs in the ERN due to BamHI restriction.
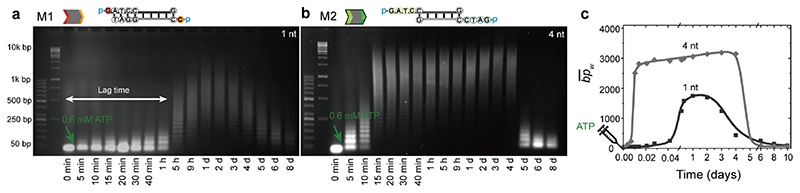
We first demonstrate lag times for the DySS DNA polymers by shortening the ligatable sticky ends from previously 30 used 4 nt to 1 nt (M1) (Figure 1d; Figure S1). Agarose gel electrophoresis (AGE) analysis shows the length distributions of the DySS DNA polymers. M2 displays a fast and direct growth phase. The chains rapidly grow to ca. 800 bp within 10 min and reach a DySS plateau at ca. 9000 bp after 15 min (Figure 2c; Figure S2). In striking contrast, the transient polymerization of M1 does not show any significant chain elongation for the first hour (Figure 2a). This system can only enter the growth phase after substantial completion of the M1 conversion phase, during which the 1 nt sticky-end overhangs are increasingly converted to 4 nt sticky-end overhangs to promote ligation efficiency. This is increasingly achieved after ca. 1 h of transient polymerization, yet, only after 5 h, there is an obvious gel band shift to higher molecular weights. After four days, the DySS DNA polymers are degraded and both systems decompose to the monomer state; M2 in both cases. The DySS DNA polymers from M1 show ca. 1000 bp lower average mass-weighted chain length ) than the DNA polymers from M2 (Figure 2b,d), which is attributed to a continuous evolution of M1 to M2 at the DySS and thus slowed down ligation. In addition, a 2 nt or 3 nt overhang cannot induce an obvious lag time for the transient DNA polymerization (Figure S3).
Figure 2. Sticky-end length controlled lag time in ATP-driven DNA assembly.
(a,b) Time-dependent AGE and (c) development of for the transient polymerization of M1 and M2 by fueling with 0.6 mM ATP. Lines are guides to the eye.
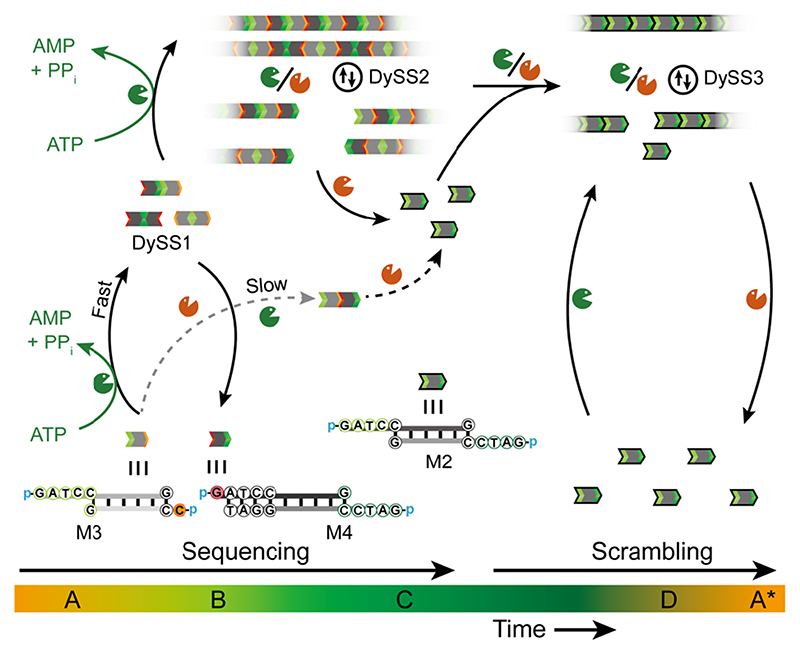
Next we discuss higher levels of pathway complexity of the fuel-driven self-assembly by designing and using two asymmetric dsDNA tiles bearing 1 nt and 4 nt overhangs on their terminal parts, M3 and M4 (Figure 3). Due to the lag time for 1 nt overhang ligation, first, a dimer state (DySS1) forms by the ligation of the 4 nt overhangs. Subsequently, oligomers of dimers (DySS2) form by the polymerization of dimers via the ligation of 1 nt overhangs, being representative of a different fuel-driven uphill localized minimum in the energy landscape. Afterwards, M3 and M4 evolve in the ERN to standard tiles with 4 nt overhang due to cycles of dynamic ligation and cleavage. Thus, the sequenced DNA oligomers with even numbers of monomers from DySS1 and DySS2 are scrambled in this long-lasting conversion phase, enabling the formation of DNA polymers with randomized numbers of initial monomers in DySS3. Different DySSs only indicate the majority states of the system. All DNA polymers degrade to 4 nt overhang dsDNA tiles after the ATP is consumed. Hence, this concept demonstrates pathway-controlled sequencing and scrambling that leads to the formation of multiple transient DySSs.
Figure 3. Schematic illustration of pathway-controlled sequencing and scrambling for autonomous multiple transient DySSs.
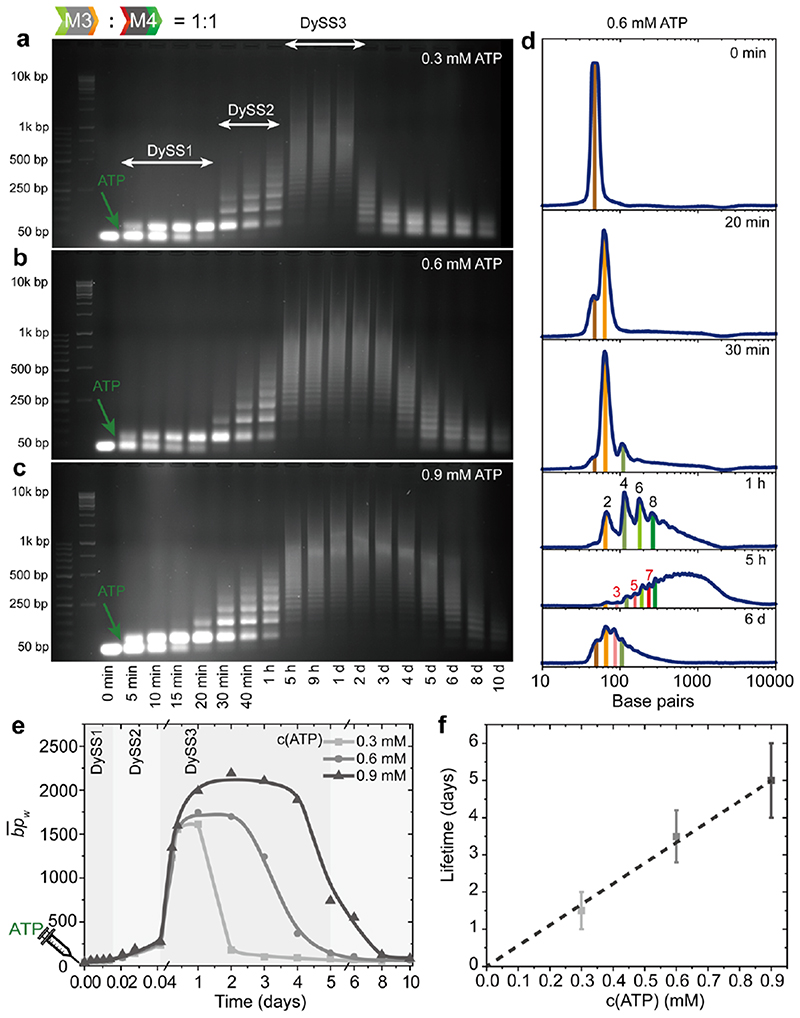
Indeed, AGE visualizes the transient multi-state nature of the systems fueled by 0.3, 0.6 and 0.9 mM ATP with three consecutive and different DySSs (Figure 4a-c). For instance, by fueling with 0.3 mM ATP, the system consecutively stays at DySS1 and DySS2, each for ca. 30 min, and then autonomously enters DySS3. Gray scale profiles extracted from AGE at 0.6 mM ATP show more direct evidence for the sequential sequencing and scrambling effects (Figure 4d). Below 30 min, the gel band only shifts to the position for dimers. Then at 1 h, the gray scale profile shows sharp peaks for oligomers of dimers. After 5 h, new peaks for the oligomers with odd numbers of monomers appear between the peaks for oligomers of dimers, and elongation into polymers takes place. The system fueled with 0.3 and 0.6 mM ATP show a plateau of at ca. 1700 bp. Increasing the ATP concentration to 0.9 mM further promotes the by ca. 400 bp in a sustained DySS polymerization (Figure 4e). The lifetimes for DySS1 and DySS2 do not significantly depend on the fuel level, because there is a significant excess of ATP in the beginning, yet the lifetime for DySS3 is almost doubled when increasing the ATP level from 0.3 mM to 0.6 mM. Further increasing the ATP concentration to 0.9 mM only slightly shortens the lifetime for DySS1 by 5 min, but the lifetime for the whole process, which is defined by the point where the declines below half of the of the DySS plateau, is further increased to ca. 5 days (Figure 4f). The reason for the insensitivity of the lifetimes of DySS1 and DySS2 to the ATP concentration is because the sequencing process is programmed by the lag time of 1 nt overhang ligation and evolution of M3 and M4 to M2 in the ERN, which mainly correlates to the BamHI cleavage kinetics in the ERN (independent of ATP). The linear evolution of the lifetimes is indicative of a relatively stable enzymatic activity with limited side product (AMP, PPi) inhibition, which we discuss in Figure S4 and Supplementary Note1. 30–33
Figure 4. ATP-powered autonomous multiple transient DySS structures.
(a-c) Time-dependent AGE for the transient DySS polymerizations of M3 and M4 by fueling with (a) 0.3, (b) 0.6, and (c) 0.9 mM ATP. (d) Gray scale profiles from AGE for the system in (b). (e) development with time for the systems (a-c). Lines are guides to the eye. (f) Lifetime control via ATP concentration. Error bars are standard deviations of duplicate measurements.
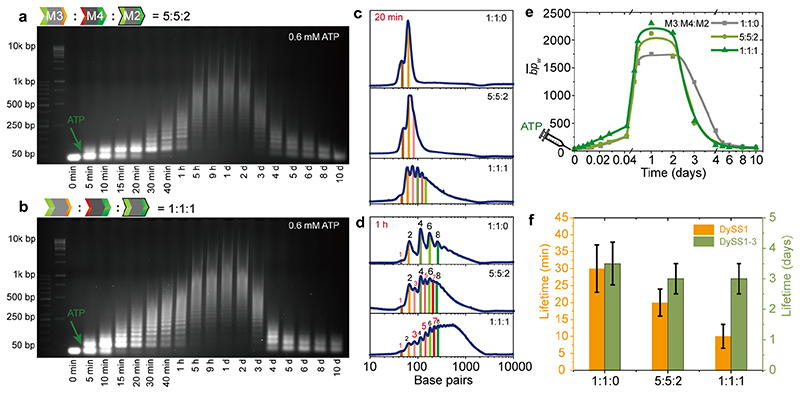
Next we show how to disrupt the transient sequencing in binary systems by adding M2 to the reach a ternary M2/M3/M4 system. Compared to the system without M2 (M3:M4:M2 = 1:1:0, Figure 4b), the introduction of 16.7 mol% M2 (5:5:2) allows DySS2 and DySS3 to compete with each other to a greater extent and DySS2 is greatly suppressed. The lifetime of DySS1 decreases by 10 min and a swift transition into polymers occurs (Figure 5a). Although the introduced M2 can promote the ligation, M3 and M4 in the system act as terminators for the ligation of M2, hence hindering the formation of longer oligomers. After 20 min, the increasing reconfiguration of 1 nt overhangs to 4 nt overhangs and free M2 lead to elongated oligomers. Gel bands corresponding to DySS2 and DySS3 overlap with each other in the time region from 30 min to 1 h. A further increase of M2 to 33.3 mol% significantly speeds up the polymerization process (Figure 5b) and the lifetime of DySS1 is further decreased by 20 min. A rather smooth but much slower evolution into the DySS3 occurs as compared to for instance using M2 alone (Figure 2b; Figure S2b).The gray scale plots at 20 min visualize the chain elongation efficiency with increased M2 ratio (Figure 5c). After the dimer state, the polymerized DNA oligomers from M3, M4, and M2 show both even and odd number of monomers. The signals of DNA oligomers with odd number of monomers increase with the M2 content (Figure 5d). The addition of M2 also promotes the to higher values in a sustained DySS polymerization as well as fast polymerization after DySS1 (Figure 5e).
Figure 5. Disruption of programmable transient multi-states.
(a-b) Time-dependent AGE for transient polymerizations of M3, M4, and M2 with a ratio of (a) 5:5:2 and (b) 1:1:1:1 by fueling with 0.6 mM ATP. (c-d) Gray scale profiles from AGE at (c) 20 min and (d) 1 h for transient polymerization of M3, M4, and M2 with varied ratios (0.6 mM ATP). (e) development with time for the systems of varied ratios of M3, M4, and M2. Lines are guides to the eye. (f) Lifetime control in the disrupted systems for DySS1 and DySS1-3. Error bars are standard deviations of duplicate measurements.
A comparison of the lifetimes of the DySS1 shows an inverse linear relationship with the M2 concentration (Figure 5f). However, the whole lifetimes for the autonomous multi-state transition lifecycle are mainly correlated to the ATP fuel level. The system without M2 shows a slightly longer lifetime due to the slower overall ligation kinetics related to the relatively higher ratio of 1 nt sticky-end overhangs (Figure 5f).
In summary, we introduced an energetically uphill driven autonomous switching of multiple transient DySS structures in a fuel-driven system, whereby multiple building blocks from a species pool are reconfigured in a pre-programmable fashion. This multicomponent systems chemistry approach to chemically fueled pathway complexity contrasts earlier work on classical supramolecular pathway complexity (oriented towards equilibrium) and comparably simple one-component chemically fueled systems that only have limited ability for structural reconfiguration. This strategy allows to reconfigure building blocks to approach complex trajectories in space and time. The autonomous reconfiguration mechanism shows general potential in the assembly of versatile DNA functionalized building blocks, such as nano/micro particles, DNA origami, and proteins. It also serves as an inspiring starting point to think how fully synthetic systems with similar ability for reconfiguration could be designed. It paves new avenues for non-equilibrium systems with structurally evolving multiple transient DySS states for higher levels of functions in emerging chemically fueled non-equilibrium systems, automatons and dynamic molecular materials.
Supplementary Material
Acknowledgment
We acknowledge support by the European Research Council starting Grant (TimeProSAMat) Agreement 677960. We thank L. Heinen for establishing the ATP-driven DNA polymerization system and F. Lossada for the help with Blender.
Footnotes
Notes
The authors declare no competing financial interests.
References
- 1.Van den Heuvel MG, Dekker C. Motor Proteins at Work for Nanotechnology. Science. 2007;317(5836):333–336. doi: 10.1126/science.1139570. [DOI] [PubMed] [Google Scholar]
- 2.Pollard TD. The Cytoskeleton, Cellular Motility and the Reductionist Agenda. Nature. 2003;422(6933):741. doi: 10.1038/nature01598. [DOI] [PubMed] [Google Scholar]
- 3.Mitchison T, Kirschner M. Dynamic Instability of Microtubule Growth. Nature. 1984;312(5991):237. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 4.Merindol R, Walther A. Materials Learning from Life: Concepts for Active, Adaptive and Autonomous Molecular Systems. Chem Soc Rev. 2017;46(18):5588–5619. doi: 10.1039/c6cs00738d. [DOI] [PubMed] [Google Scholar]
- 5.Taylor MJ, Husain K, Gartner ZJ, Mayor S, Vale RD. A DNA-based T Cell Receptor Reveals a Role for Receptor Clustering in Ligand Discrimination. Cell. 2017;169(1):108–119.:e20. doi: 10.1016/j.cell.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li L, Yang J, Wang J, Kopeček Ji. Amplification of Cd20 Cross-linking in Rituximab-resistant B-lymphoma Cells Enhances Apoptosis Induction by Drug-free Macromolecular Therapeutics. ACS Nano. 2018;12(4):3658–3670. doi: 10.1021/acsnano.8b00797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaudry D, Stork P, Lazarovici P, Eiden L. Signaling Pathways for PC12 Cell Differentiation: Making the Right Connections. Science. 2002;296(5573):1648–1649. doi: 10.1126/science.1071552. [DOI] [PubMed] [Google Scholar]
- 8.Salazar-Roa M, Malumbres M. Fueling the Cell Division Cycle. Trends Cell Biol. 2017;27(1):69–81. doi: 10.1016/j.tcb.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Grzybowski BA, Huck WT. The Nanotechnology of Life-inspired Systems. Nat Nanotechnol. 2016;11(7):585. doi: 10.1038/nnano.2016.116. [DOI] [PubMed] [Google Scholar]
- 10.van Rossum SA, Tena-Solsona M, van Esch JH, Eelkema R, Boekhoven J. Dissipative out-of-equilibrium Assembly of Man-made Supramolecular Materials. Chem Soc Rev. 2017;46(18):5519–5535. doi: 10.1039/c7cs00246g. [DOI] [PubMed] [Google Scholar]
- 11.Sorrenti A, Leira-Iglesias J, Markvoort AJ, de Greef TF, Hermans TM. Non-equilibrium Supramolecular Polymerization. Chem Soc Rev. 2017;46(18):5476–5490. doi: 10.1039/c7cs00121e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattia E, Otto S. Supramolecular Systems Chemistry. Nat Nanotechnol. 2015;10(2):111. doi: 10.1038/nnano.2014.337. [DOI] [PubMed] [Google Scholar]
- 13.Maiti S, Fortunati I, Ferrante C, Scrimin P, Prins LJ. Dissipative Self-assembly of Vesicular Nanoreactors. Nat Chem. 2016;8(7):725. doi: 10.1038/nchem.2511. [DOI] [PubMed] [Google Scholar]
- 14.Sorrenti A, Leira-Iglesias J, Sato A, Hermans TM. Non-equilibrium Steady States in Supramolecular Polymerization. Nat Commun. 2017;8:15899. doi: 10.1038/ncomms15899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhiman S, Jain A, Kumar M, George SJ. Adenosine-phosphate-fueled, Temporally Programmed Supramolecular Polymers with Multiple Transient States. J Am Chem Soc. 2017;139(46):16568–16575. doi: 10.1021/jacs.7b07469. [DOI] [PubMed] [Google Scholar]
- 16.Mishra A, Korlepara DB, Kumar M, Jain A, Jonnalagadda N, Bejagam KK, Balasubramanian S, George SJ. Biomimetic Temporal Self-assembly Via Fuel-driven Controlled Supramolecular Polymerization. Nat Commun. 2018;9(1):1295. doi: 10.1038/s41467-018-03542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pezzato C, Prins LJ. Transient Signal Generation in a Self-assembled Nanosystem Fueled by Atp. Nat Commun. 2015;6:7790. doi: 10.1038/ncomms8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heuser T, Weyandt E, Walther A. Biocatalytic Feedback-driven Temporal Programming of Self - regulating Peptide Hydrogels. Angew Chem Int Ed. 2015;54(45):13258–13262. doi: 10.1002/anie.201505013. [DOI] [PubMed] [Google Scholar]
- 19.Heinen L, Heuser T, Steinschulte A, Walther A. Antagonistic Enzymes in a Biocatalytic pH Feedback System Program Autonomous DNA Hydrogel Life Cycles. Nano Lett. 2017;17(8):4989–4995. doi: 10.1021/acs.nanolett.7b02165. [DOI] [PubMed] [Google Scholar]
- 20.Heuser T, Merindol R, Loescher S, Klaus A, Walther A. Photonic Devices out of Equilibrium: Transient Memory, Signal Propagation, and Sensing. Adv Mater. 2017;29(17):1606842. doi: 10.1002/adma.201606842. [DOI] [PubMed] [Google Scholar]
- 21.Heinen L, Walther A. Celebrating Soft Matter’s 10th Anniversary: Approaches to Program the Time Domain of Self-assemblies. Soft Matter. 2015;11(40):7857–7866. doi: 10.1039/c5sm01660f. [DOI] [PubMed] [Google Scholar]
- 22.Madan I, Buh J, Baranov VV, Kabanov VV, Mrzel A, Mihailovic D. Nonequilibrium Optical Control of Dynamical States in Superconducting Nanowire Circuits. Sci Adv. 2018;4(3):eaao0043. doi: 10.1126/sciadv.aao0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grötsch RK, Wanzke C, Speckbacher M, Angi A, Rieger B, Boekhoven J. Pathway Dependence in the Fuel-driven Dissipative Selfassembly of Nanoparticles. J Am Chem Soc. 2019;141(25):9872–9878. doi: 10.1021/jacs.9b02004. [DOI] [PubMed] [Google Scholar]
- 24.Dong B, Liu L, Hu C. ATP-driven Temporal Control over Structure Switching of Polymeric Micelles. Biomacromolecules. 2018;19(9):3659–3668. doi: 10.1021/acs.biomac.8b00769. [DOI] [PubMed] [Google Scholar]
- 25.della Sala F, Maiti S, Bonanni A, Scrimin P, Prins LJ. Fuel-selective Transient Activation of Nanosystems for Signal Generation. Angew Chem Int Ed. 2018;57(6):1611–1615. doi: 10.1002/anie.201711964. [DOI] [PubMed] [Google Scholar]
- 26.Hao X, Chen L, Sang W, Yan Q. Periodically Self-pulsating Microcapsule as Programmed Microseparator Via ATP-regulated Energy Dissipation. Adv Sci. 2018;5(3):1700591. doi: 10.1002/advs.201700591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian L, Winfree E. Scaling up Digital Circuit Computation with DNA Strand Displacement Cascades. Science. 2011;332(6034):1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 28.Qian L, Winfree E, Bruck J. Neural Network Computation with DNA Strand Displacement Cascades. Nature. 2011;475(7356):368. doi: 10.1038/nature10262. [DOI] [PubMed] [Google Scholar]
- 29.Song T, Shah S, Bui H, Garg S, Eshra A, Fu D, Yang M, Mokhtar R, Reif J. Programming DNA-Based Biomolecular Reaction Networks on Cancer Cell Membranes. J Am Chem Soc. 2019;141(42):16539–16543. doi: 10.1021/jacs.9b05598. [DOI] [PubMed] [Google Scholar]
- 30.Heinen L, Walther A. Programmable Dynamic Steady States in Atp-driven Nonequilibrium DNA Systems. Sci Adv. 2019;5(7):eaaw0590. doi: 10.1126/sciadv.aaw0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cherepanov AV, De Vries S. Binding of Nucleotides by T4 DNA Ligase and T4 Rna Ligase: Optical Absorbance and Fluorescence Studies. Biophys J. 2001;81(6):3545–3559. doi: 10.1016/S0006-3495(01)75985-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhat R, Grossman L. Purification and Properties of Two DNA Ligases from Human Placenta. Arch Biochem Biophys. 1986;244(2):801–812. doi: 10.1016/0003-9861(86)90649-1. [DOI] [PubMed] [Google Scholar]
- 33.Huang Y, Zhao S, Shi M, Chen J, Chen Z-F, Liang H. Intermolecular and Intramolecular Quencher Based Quantum Dot Nanoprobes for Multiplexed Detection of Endonuclease Activity and Inhibition. Anal Chem. 2011;83(23):8913–8918. doi: 10.1021/ac2013114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.