Summary
Multivalency-driven liquid-liquid phase separation (LLPS) is essential in biomolecular condensates to facilitate spatiotemporal regulation of biological functions. Providing programmable model systems would help to better understand the LLPS processes in biology, and furnish new types of compartmentalized synthetic reaction crucibles that exploit biological principles. Herein, we demonstrate a concept for programming LLPS using transient multivalency between ATP-driven sequence-defined functionalized nucleic acid polymers (SfNAPs), which serve as simple models for membrane-less organelles. The ATP-driven SfNAPs are transiently formed by an enzymatic reaction network (ERN) of concurrent ATP-powered DNA ligation and DNA restriction. The lifetimes can be programmed by the ATP concentration, which manifests on the LLPS length scale in tunable lifetimes for the all-DNA coacervates. Critically, the prominent programmability of the DNA-based building blocks allows to encode distinct molecular recognitions for multiple multivalent systems, enabling sorted LLPS and thus multicomponent DNA coacervates, reminiscent of the diverse membraneless organelles in biological systems. The ATP-driven coacervates are capable for multivalent trapping of micron-scale colloids and biomolecules to generate functions as emphasized for rate enhancements in enzymatic cascades. This work supports ATP-driven multivalent coacervation as a valuable mechanism for dynamic multicomponent and function biomolecular condensate mimics and for autonomous materials design in general.
Keywords: coacervates, liquid-liquid phase separation, multivalency, synthetic biology, protocells
Introduction
Membrane-less biomolecular condensates are ubiquitous in biological cells and are important for spatiotemporal organization of subcellular processes. 1–3 These biomolecular condensates function to concentrate proteins and nucleic acids, and provide diverse biological functions such as RNA metabolism, ribosome biogenesis, and signal transduction. 2, 4–6 Many of them show liquid-like properties such as rapid condensation and dissolution, fusion and wetting. 7–8 For instance, the nucleoli, Cajal bodies, and nuclear speckles in the nucleus can vary their size, number, and composition over time and can reversibly appear and disappear during mitosis. 9–10 Although all mechanisms for the dynamic changes remain elusive, multivalent interactions between the (bio)macromolecular components are a key driving principle in their dynamic regulations. 6, 11–13 Inspired by this natural multivalency-driven liquid-liquid phase separation (LLPS), biomolecular recognition units have for instance been encoded into pairs of engineered proteins, which then show the capability for phase transition upon mixing, leading to micrometer-sized liquid droplets. 6
Aside from biomolecular recognition, numerous strategies such as charge-charge, dipole-dipole, cation-π, and π-π interactions have been developed to construct related, in parts fully synthetic, coacervates and complex coacervates. 14–18 Recently, such coacervates have regained interest as artificial membrane-less organelles, which could act as open reactors with facile exchanges of matter. 19–22 Specifically, polyelectrolyte complexation is intriguing for facile and versatile coacervate fabrication. Examples include carboxymethyl dextran sodium salt/poly-L-lysine (PLL), 20 oppositely charged amylose derivatives, 23 ATP/poly(diallyldimethyl ammonium chloride), 24 ATP/PLL, 25 and others. 26–31 The unstable membrane-less coacervates can be stabilized by interfacial self-assembly of synthetic surfactants and stoichiometry mismatch of the charges. 23
To allow external control mechanisms, various chemical signals and physical fields have been used to reversibly assemble polyelectrolyte-based coacervates. 32–36 Mann and co-workers reported reversible coacervation via alternately bubbling CO2 and NH3 to ATP/PLL coacervates to regulate the PLL charge. 37 Keating and co-workers investigated the switching of RNA/peptide coacervates by changing the peptide phosphorylation state using a kinase/phosphatase pair. 38 Moreover, ATP/PLL coacervates were coupled to an enzymatic reaction network (ERN) that transiently generates ATP as one component of the coacervate. 39 Towards active matter coacervates, Huck and coworkers reported a GTP-driven polymerization of FtsZ protein within coacervates for dynamic coacervate deformation and fission. 40 In summary, although great progress has been made in coacervates as mimics for membrane-less organelles and for protocell research, most coacervates rely on polyelectrolyte interactions, whereas highly programmable molecular recognition for the formation of LLPS has scarcely been addressed. 6 Such highly programmable interactions would however contribute fundamentally relevant model systems to quantitatively understand kinetics and thermodynamics of LLPS, provide means to modulate the dynamics and selective uptake, and, if orthogonal interactions were available, also spatially sorted dynamic artificial membrane-less organelles could be prepared. Additionally, the emerging concepts of non-equilibrium chemically fueled systems empowered by chemical reaction networks can contribute new features such as transience and dynamics. 41–43
In the DNA nanoscience arena, droplets made of DNA could feature outstanding programmability and molecular recognition. 44–47 In terms of DNA droplets, DNA nanostars have been investigated for controllable liquid properties and macromolecular structures. 48–50 Mao and coworkers reported all-DNA droplets by condensation of palindromic domains in single-stranded DNA (ssDNA), 51 which however showed ill-defined structures. Moreover, we previously reported pathway-controlled formation of membrane-less all-DNA droplets with well-defined structures using temperature-induced LLPS of poly(adenine)-rich ssDNA strands and subsequent kinetic trapping of the coacervates during cooling – again using palindromic domains. 52 Both strategies need high temperature annealing to form the droplets, which is incompatible with applications in a biological system, involving for instance the encapsulation of enzymes or other temperature-sensitive components. Although first DNA temperature-annealing derived droplets exist, exploiting distinct molecular recognition for constructing multicomponent DNA-based droplets is uncharted territory, and the potential of DNA for regulating dynamics (eg. reversible condensation and dissociation, and transient coacervation) and for realizing isothermal LLPS to open possibilities to encapsulate temperature-sensitive components such as enzymes or proteins in general has not been addressed.
Herein, we report ATP-driven membrane-less all-DNA coacervates, which form through multivalency-driven LLPS of programmable sequence-defined functionalized nucleic acid polymers (SfNAPs) orchestrated by an ERN of concurrent ATP-powered ligation (using T4 DNA ligase) and endonuclease-controlled restriction (using BsaI). Using orthogonally polymerizing SfNAPs system (due to distinct molecular recognition in the main chain of various building blocks), we further demonstrate the self-sorting of LLPS for multicomponent coacervates in the same system. To showcase functions of the coacervates, we reveal transient entrapment and release of functionalized micron-scale colloids and functional DNA polymers within the ATP-driven all-DNA coacervates. Additionally, glucose oxidase (GOx) and horseradish peroxidase (HPR) fusion proteins bound to streptavidin can be encapsulated in the coacervates by anchoring onto biotinylated coacervate-forming DNA SfNAPs. An improved catalytic activity of the GOx/HPR enzyme cascade is found within the membrane-less compartments with a macromolecularly crowded DNA environment.
Results and discussion
General concept for ATP-driven molecular recognition and multicomponent all-DNA coacervates
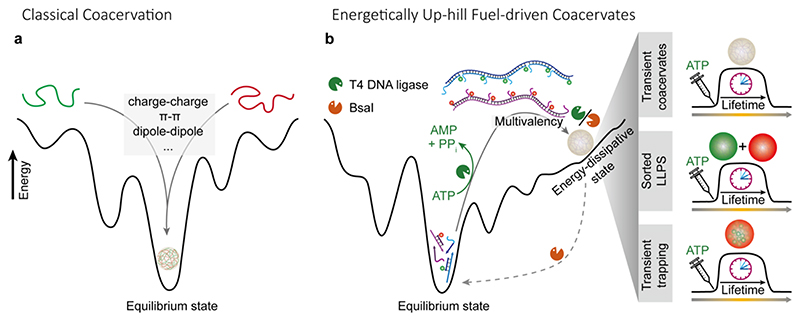
We first highlight the differences in free energy between classical coacervates and chemically fueled coacervates. Classical coacervation based on attractive interactions between two polymers occurs in an energetically down-hill process by formation of enthalpic interactions and entropically favored release of water (and counterions for polyelectrolytes). 18 The resulting coacervate droplets sit near the global minimum in the energy landscape (Figure 1a). In our design, we implement an overall energetically up-hill system to obtain the coacervates in a dynamic and energy-dissipative state (Figure 1b). It builds on our recently introduced systems of ATP-driven transient DNA polymerization and SfNAPs with transient generation of multivalent interactions with colloid objects. 53–55 We hypothesized that two SfNAPs with complementary side chain interactions could allow to establish membrane-less coacervates by LLPS, providing a programmable mimicry of biomolecular condensates driven by multivalency. The injection of ATP to a system containing small building blocks with complementary sticky ends and the enzymes (T4 DNA ligase and BsaI restriction enzyme) allows to drive the system isothermally into a dynamic steady state (DySS) with temporal SfNAP formation. More details on the ERN are below in Figure 3a. The ATP is consumed in the process and the energy is transiently transferred into DNA phosphodiester bonds, resulting in an up-hill driven dynamic covalent bond with an average steady-state chain length. Under the conditions of the ERN, the phosphodiester bond is unstable (ΔG = -5.3 kcal/mol 56 ). Constant ATP-dissipating bond shuffling occurs in the energy-driven steady state. The up-hill and down-hill reactions are not each other’s inverse and are separate kinetically tunable reactions. The network is reminiscent of non-equilibrium steady state phosphorylation/dephosphorylation networks without any outside regulator. 57–61 This qualifies the system as a chemically fueled non-equilibrium system, in which the ATP concentration majorly sets the lifetime, and the antagonistic enzymes engineer the transduction kinetics and dynamics of the system. 53
Figure 1. ATP-driven LLPS by programmable interactions in multivalent SfNAPs with complementary side chain interactions.
(a) Schematic illustration of the energy landscape for classical coacervation processes. (b) ATP-driven transient SfNAPs to engineer transient all-DNA coacervates, sorted LLPS coacervates system, and transient trapping of secondary components. The dynamic coacervate structures sit at an energy-dissipative state out of the global minimum in the energy landscape. Transient lifetimes occur because the ATP fuel is consumed as detailed in the ERN in Figure 3a.
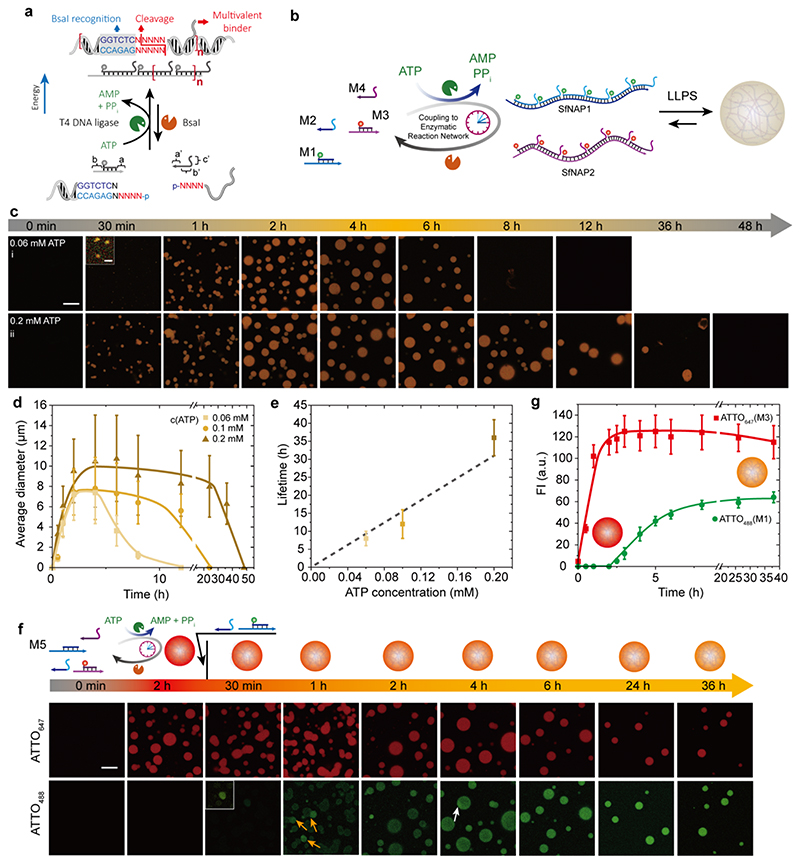
Figure 3. ATP-driven transient all-DNA coacervates with programmable lifetimes.
(a) Schematic illustration of the ERN furnishing an ATP-driven transient SfNAP. (b) Schematic illustration of ATP-driven membrane-less all-DNA coacervates via LLPS driven by transient multivalency of orthogonal SfNAPs. (c) Time-dependent CLSM measurements for transient all-DNA coacervates with programmable lifetimes by fueling with varied concentration of ATP (0.06 and 0.2 mM). CLSM measurements were conducted consecutively with the same microscopy settings. Scale bar = 20 μm; insert = 2 μm. (d) Time-dependent size of the transient coacervates with programmable lifetimes. Error bars are standard deviations from three random fields of view of duplicate experiments. Lines are guides to the eye. (e) Lifetimes are controlled by the ATP concentrations. Error bars are standard deviations of duplicate measurements. Line is a guide to the eye. (f) Tile integration and dynamization during a running transient coacervates system. The tile with ATTO488 was added 2 h after starting the initial system. CLSM before and after adding the ATTO488 tile confirm the integration and reconfiguration. Scale bar = 20 μm. (g) Time-dependent fluorescence intensities. Error bars are standard deviations from three random fields of view of duplicate experiments. Lines are guides to the eye. Conditions for (b): 37°C, 0.01 mM M1, 0.012 mM M2, 0.01 mM M3, 0.012 mM M4, 0.92 WU/μL T4 DNA ligase, 1.0 U/μL BsaI, and varying amounts of ATP, under orbital shaking at 80 rpm. Conditions for (f) 37°C, 0.01 mM M5, 0.012 mM M2, 0.01 mM M3, 0.012 mM M4, 0.92 WU/μL T4 DNA ligase, 1.0 U/μL BsaI, and 0.2 mM of ATP, under orbital shaking at 80 rpm, followed by addition of another 0.002 mM M1 and 0.0024 mM M2 after 2 h running of the transient coacervation reaction.
Since the side chains on the SfNAPs are independent from the ERN, they are freely programmable with regard to their sequences and thus hybridization strength. Additionally, since BsaI is a class IIS endonuclease with separated domains for recognition and restriction, a careful selection of the ligation parts for the main chain and of the side chain interaction sites on various DNA building blocks allows running multiple coacervating systems in parallel and achieving sorted LLPS and multicomponent coacervates. 4 nucleotides (nt) of the sticky end overhangs at the restriction site can be freely programmed in BsaI, enabling in principle 4 4 = 256 orthogonal interactions in multicomponent systems. Importantly, the multivalency-driven coacervates are able to trap multivalent binder functionalized objects, such as micron-scale colloids and catalytic enzymes (Figure 1).
Multivalency-guided LLPS of complementary SfNAPs for membrane-less all-DNA coacervates
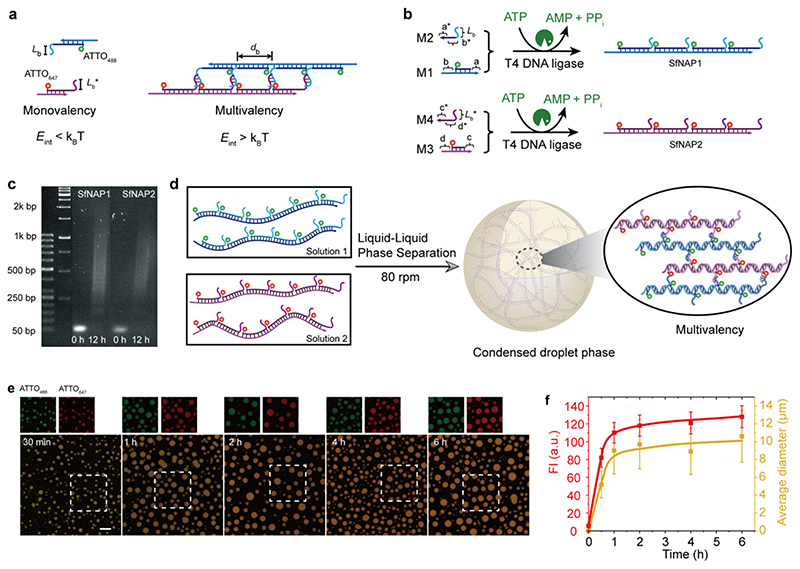
For proof-of-concept, we discuss the static multivalency-guided LLPS of two SfNAPs with complementary side chains before discussing the dynamic ATP-fueled transient systems. Under appropriate enthalpic balance, efficient cooperative multivalent binding between the side chains of two SfNAPs occurs only in the polymer state, whereas binding is absent in the monomer state. This is a function of the interaction length of the complementary part (L b) and the distance between the side chains (d b; Figure 2a). Based on preliminary screening, we found that a L b of 8 base pairs (bp; CGAATAGA/TCTATTCG (T m = 33 °C) requires a d b of ca. 38 bp or smaller to realize efficient multivalency in the ATP-driven SfNAP state. These conditions set the starting point here. We synthesized two orthogonal SfNAPs, SfNAP1 and SfNAP2, by separate ligation of M1/M2 and M3/M4, whereby M2 and M4 carry the side chains with complementary interactions (Figure 2b, sequences in Supplementary Table 1). ATTO488 and ATTO647 dyes in M1 and M3 fluorescently label both SfNAPs for confocal laser scanning microscopy (CLSM). Agarose gel electrophoresis (AGE) yields polymer length distributions of the SfNAPs with a growth up to 1500 bps (Figure 2c).
Figure 2. Multivalency-driven LLPS for membrane-less all-DNA coacervates.
(a) Monovalent vs. multivalent binding with programmable interaction strength, L b, and branch distance, d b. (b) Schematic illustration of ATP-driven multiple SfNAPs. (c) AGE for the polymerizations at start and end (12 h; 2 wt. %, 90 V, 2 h). (d) Schematic illustration of multivalency-guided LLPS of two SfNAPs with complementary side chain interactions for all-DNA coacervate formation. (e) Time-dependent CLSM of DNA coacervates. Scale bar = 20 μm. (f) Time-dependent fluorescence intensities of ATTO647 and average diameters for the coacervates. Error bars are standard deviations from three random fields of view of duplicate experiments (ca. 300 coacervates). Lines are guides to the eye.
Afterwards, we investigated the coacervation of SfNAP1 and SfNAP2 (10 μM for each species regarding amounts of repeating units) by mixing and shaking using an orbital shaker at 80 revolutions per minute (rpm) at 37°C. CLSM in Figure 2e indeed confirms the formation of spherical all-DNA coacervates. After 30 min, DNA coacervates with a number average diameter (D n) of ca. 5.2 μm are present, and the D n increases to ca. 10.5 μm at 1 h, reaching a balanced state for LLPS at ca. 1h. Both fluorophores are homogeneously distributed in the coacervates confirming intimate mixing of the SfNAPs (Figure 2e). Note that control experiments of monomer mixtures or the individual SfNAPs do not give any structure in CLSM, because the multivalent interactions are absent. Herein, we realize LLPS for membrane-less coacervates in a non-protein based synthetic system via DNA-based multivalent interaction.
ATP-driven transient all-DNA coacervates with programmable lifetimes
In the above investigation, the distinct difference between LLPS for the mixed SfNAPs and absence of LLPS for the monomer tiles is the key to regulate the temporal behavior of the all-DNA coacervates. To realize an energy-dissipative coacervation system, we combined the SfNAPs with the full ERN featuring concurrent ligation/polymerization and restriction/depolymerization (Figure 3a,b). Typical experiments were conducted using ca. 0.01 mM of each DNA tile (M1-M4), 0.92 WU/μL T4 DNA ligase, 1.0 U/μL BsaI, and varied ATP concentrations at 37°C. CLSM measurements at different time points reveal the transient nature of the ATP-driven all-DNA coacervates regarding their structural evolution (Figure 3c; Figure S1). Before ATP injection, the system is a fully homogeneous solution. The injection of 0.06 mM ATP drives the multicomponent system into the SfNAP state with dynamic sub-unit exchange, and the build-up of the multivalency between the SfNAPs induces LLPS (Figure 3c-i). After 30 min, some small DNA coacervates with a D n ≈ 1 μm are observed under CLSM, for which the fluorescence intensity (FI) is very weak. Subsequently, further growth occurs and a relatively steady plateau with a D n ≈ 7 μm is reached after ca. 2 h (Figure 3d). Concurrently, the averaged fluorescence intensities for the coacervates also drastically increase to a DySS (Figure S2). The constant D n in the plateau is obtained because of slight orbital shaking to mitigate coalescence. After the ATP is consumed, the DNA coacervates disappear, because the long SfNAPs are degraded back to monomeric building blocks by BsaI. The coacervate lifetime is tunable from 6 hours to 1.5 days by changing the ATP concentration from 0.06 to 0.2 mM (Figure 3e), which indicates the tiles produced after endonuclease restriction are continue to be active and recycled in the ERN. The kinetics of coacervate formation and the coacervate size increase mildly with the ATP concentration, which is due to a faster polymerization process as ATP is a co-factor in the ligation and provides ionic strength to promote DNA interactions (Figure 3c-ii,d; Figure S1). AGE analyses of the transient SfNAPs in these systems confirms similar lifetimes for the molecular and coacervate levels. Higher ATP concentrations indeed lead to faster polymerization of transient SfNAPs to promote the kinetics for LLPS (Figure S3). Interestingly, selected coacervates degrade from the inside and end up being hollow capsules during the degradation. This confirms the entrapment and operation of the enzymes inside the coacervates (Figure S1). Additionally, in the dynamic steady state (when the system is still rich in ATP), no capsule formation is observed. This indicates that in the coacervates the kinetic for ligation is faster than restriction to maintain the structure. Consequently, the polymers can still be dynamically extended when in the coacervate phase if excess ATP and ligation overhangs are present.
It is also worth noting that orbital shaking is crucial for controlling the dynamic property of the coacervates. Without any shaking, the coacervates sediment and merge together to form random aggregates (Figure S4d), while strong shaking (eg. at 400 rpm) leads to smaller coacervates with D n from several hundred nanometers to ca. 1 μm (Figure S4b,c). This demonstrates that fusion and fission occur during shaking as a dynamic system feature. We also characterized the liquid property of fully suspended coacervates by real-time fusion experiments. Upon contacting, two coacervates merge into one large coacervate, which verifies the liquid-like nature of the DNA coacervates (Figure S5 and Supplementary Video 1). The droplets can wet the glass surface (Figure S4d), but after wetting the droplets cannot be recovered by further shaking. Hence, the orbital shaking can also mitigate the wetting of the droplets on the glass surface.
We further probed the internal dynamics of the coacervates in the ATP-driven steady state to detect the efficiency of component diffusion in the coacervates. We added differently labeled ATTO488-M1 monomers to a running system based on non-fluorescent SfNAP1 (M2/M5) and ATTO647-labeled SfNAP2 (M3/M4). ATTO488-M1 shares the same sequence with M5 to co-integrate with SfNAP1. Before M1 addition, only red fluorescence can be observed, because the coacervates are only labeled by ATTO647 on SfNAP2 (Figure 3f). After the addition of ATTO488-M1, green ATTO488 fluorescence gradually arises in the coacervates (Figure 3f), while the red ATTO647 fluorescence intensity remains unchanged (Figure 3g). Interestingly, for the first 4 h after the addition of M1, the uptake and distribution of ATTO488-M1 to the coacervates is not homogeneous. Coacervates with distinctly brighter fluorescence can be observed at e.g. 1h. This is due to slow mixing when adding the ATTO488-M1 and preferential uptake. More interestingly, at 4h, patches of brighter fluorescence can be observed in otherwise spherical coacervates (white arrow). These arise from fusion events (visible e.g. at 1h, orange arrows) and subsequent recircularization of the coacervates to minimize interfacial energy (Figure 3f). These patches disappear within 6 h and fully homogeneous coacervates are obtained. This confirms the dynamic chain shuffling inside the coacervates and deep integration of secondary tiles. Such a covalent integration of a new monomer species would be impossible in a static non-driven system as the DNA bonds would be static. This clearly emphasizes an advantage towards quick structural and functional adaptation due to operating the structures in a DySS with constant bond shuffling. 62
ATP-driven transient multiple sorted membrane-less coacervates
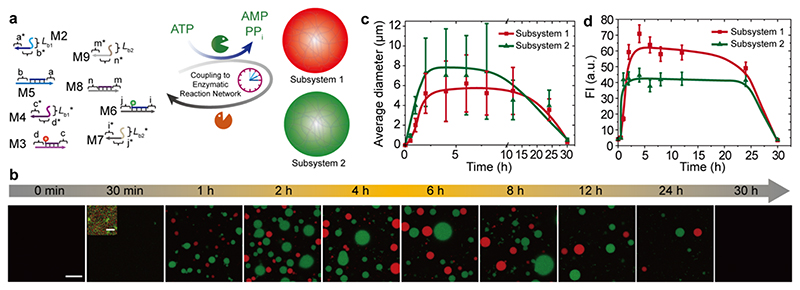
The programmability of the ligation-restriction position and of the complementary side chains on the SfNAPs opens the possibility to potentially run multiple LLPS systems in parallel. Figure 4a shows such a design, in which 4 distinct SfNAPs can be temporarily programmed in the ERN, where two SfNAPs subsets feature orthogonal multivalent interactions (L 1b/L 1b * and L 2b/L 2b *) to form self-sorted LLPS. Subsystem 1 consists of M5/M2 and ATTO647-M3/M4. Subsystem 2 consists of ATTO488-M6/M7 and M8/M9. The fluorescent labels allow distinguishing the self-sorted LLPS. Time-dependent CLSM indeed confirms orthogonal coacervate formation and the transient nature of both ATP-driven multicomponent coacervates (Figure 4). The transient behavior is analogous to the single-pair coacervates and spatially resolved fluorescence analysis in the CLSM shows absence of crosstalk. The absence of crosstalk here is attributed to the absence of attractive specific multivalent interactions between unlike coacervates, that otherwise possess repulsive electrostatic interactions due to their charged nature. This also confirms that the multivalent interaction among the coacervates from the same species facilitate the fusion of contacting coacervates (Figure S5 and Supplementary Video 1). This demonstrates that ATP-driven sorted LLPS can be achieved by defining molecular recognition events to form the ATP-driven SfNAPs and to guide the multivalent interactions between them. Although collision events happen between the two sorted populations during operation, those do not suffice for a merging and fusion into common droplets. A crosstalk such as in common polyelectrolyte coacervates using non-specific interactions can be prevented. 23 Such multicomponent membrane-less coacervates are reminiscent of diverse membrane-less organelles driven by multivalent recognitions in a biological system, and are a clear advantage provided by the high programmability of DNA and of our ATP-driven ERN using BsaI as class IIS endonuclease. 2
Figure 4. Self-sorting ATP-driven transient multicomponent membrane-less coacervates.
(a) Schematic illustration of ATP-driven multiple coacervation systems. Subsystem one is formed by M2 - M5 and subsystem two is formed by M6 - M9. (b) Time-dependent CLSM measurements of the transient multiple coacervates. CLSM measurements were conducted consecutively with the same microscopy settings. Scale bar = 20 μm; insert = 4 μm. (c) Time-dependent average diameters of the transient coacervates for both systems. Lines are guides to the eye. Error bars are standard deviation from three random fields of view of duplicate experiments. (d) Time-dependent fluorescence intensities of the transient coacervates for both systems. Error bars are standard deviations from three random fields of view of duplicate experiments. Lines are guides to the eye. Conditions: 37°C, 0.01 mM M5, 0.012 mM M2, 0.01 mM M3, 0.012 mM M4, 0.01 mM M6, 0.012 mM M7, 0.01 mM M8, 0.012 mM M9, 0.92 WU/μL T4 DNA ligase, 1.0 U/μL BsaI, and 0.12 mM ATP, under orbital shaking at 80 rpm.
Transient trapping and releasing for temporal superstructures
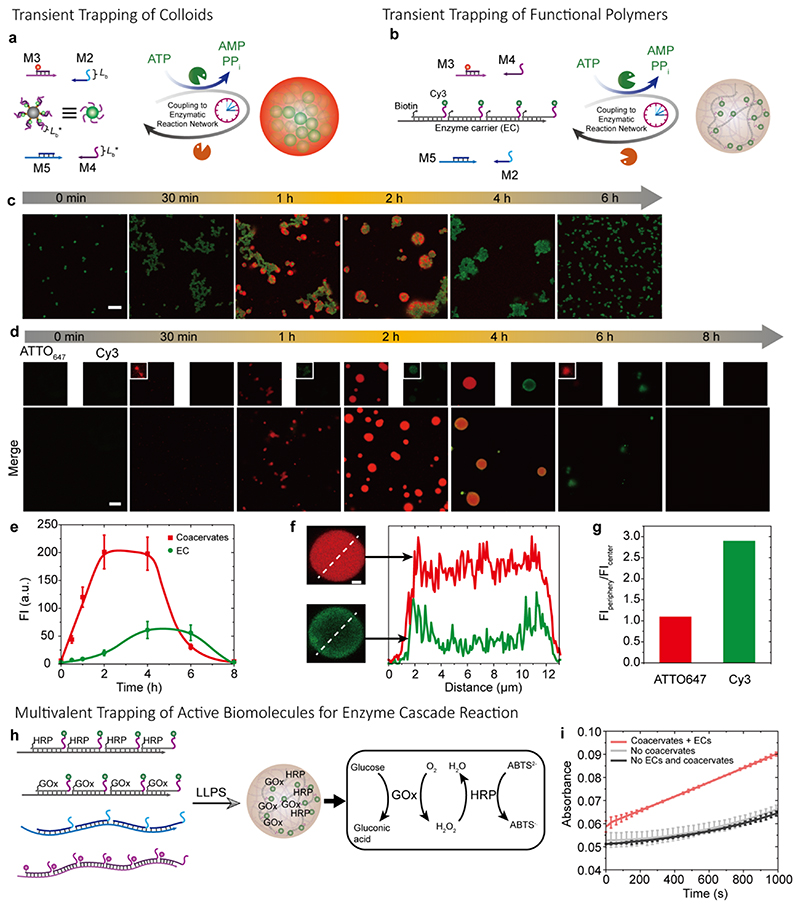
We surmised that the multivalent recognition patterns driving the LLPS would provide a generic platform to entrap objects functionalized with complementary ssDNA. Figure 5a shows a first example by transiently trapping ssDNA-functionalized micron-sized colloids (D = 1 μm) in the DNA coacervates. For clear visualization, the colloids and coacervates are labeled with ATTO488 and ATTO647, respectively. Time-dependent CLSM visualizes the transient all-DNA coacervate formation and concurrent colloid aggregation, engulfment and restructuring. Before ATP injection, the colloids are freely dispersed in the solution (Figure 5c). The addition of ATP drives the formation of two orthogonal SfNAPs with complementary side chain interactions. On the one hand this leads to LLPS between SfNAP1 and SfNAP2. From above (Figure 2), we know that this process takes up to 30 min to lead to visible structure formation. This is due to the fact that both SfNAPs first need to build up long chains for efficient interaction. On the other hand, the non-fluorescent SfNAP1 (M5 and M2) can interact with the already present multivalent recognition pattern on the colloids. This process is faster (less than 10 min) and hence colloid assembly happens prior to the formation of any obvious DNA coacervates. After 30 min, clear fractal aggregates of the colloids (reminiscent of diffusion-controlled aggregation) are visible. After 1 h, the coacervates become clearly visible (red fluorescent) and nucleate largely on these structures. Engulfment and encapsulation happen due to the competition of multivalent interactions and the dynamics of the system (Figure 5c). Additionally, the dynamics lead to some level of reorientation from the more fractal aggregation to a more spherical superstructure. Interestingly, in the disassembly phase, after ATP is consumed (4 h), the degradation of the coacervates is much faster than the colloidal disassembly. We attribute this to the lower BsaI accessibility of the SfNPAs between the colloid aggregates. After 6 h, all colloid aggregates largely re-disperse in the solution (Figure 5c). It is worth noting that by using lower concentration of functionalized colloids (10 % of the one above), smaller colloid aggregates can be obtained and the fractal aggregation in the initial state is absent (Figure S6). This is due to the fact that lower particle concentrations allow significantly less collisions needed for assembly, and the assembly time scale is more synchronized with the LLPS of both SfNAPs.
Figure 5. Multivalency-driven superstructures and catalytically active coacervates.
(a) Schematic illustration of ATP-driven transient coacervates for temporal trapping of micron-sized colloids. (b) Schematic illustration for ATP-driven transient trapping of functional DNA polymers in the DNA coacervates via multivalency. (c) Time-dependent CLSM of the transient colloid assembly and transient coacervates. CLSM measurements were conducted consecutively with the same microscopy settings. Scale bar = 10 μm. (d) Time-dependent CLSM measurements of the transient coacervates with transient trapping and releasing functional DNA polymers via transient multivalency. CLSM measurements were conducted consecutively with the same microscopy settings. Scale bar = 10 μm. (e) Time-dependent fluorescence intensities for the transient coacervates and entrapped fluorescent DNA polymers. Error bars are standard deviations from three random fields of view of duplicate experiments. Lines are guides to the eye. (f) Fluorescence intensity profiles for coacervates and entrapped functional DNA polymers at 4 h. Scale bar = 2 μm. (g) Quantified fluorescence intensity (FI) ratios of the FIperiphery to the FIcenter of the coacervates. (h) Schematic illustration of GOx and HRP encapsulation in the coacervates. (i) Catalytic activity of the multienzyme complex inside the coacervates in comparison to free enzymes and enzymes on enzyme carriers (EC) but without coacervates. Reactions monitored by absorbance at 414 nm. Error bars are standard deviations of duplicate measurements. Conditions for (a): 37°C, 0.01 mM M5, 0.012 mM M2, 0.01 mM M3, 0.012 mM M4, ca. 4.68-6.68×107 beads/mL, 0.92 WU/μL T4 DNA ligase, 1.0 U/μL BsaI, and 0.06 mM ATP, under orbital shaking at 80 rpm. Conditions for (b): 37°C, 0.01 mM M5, 0.012 mM M2, 0.01 mM M3, 0.012 mM M4, 0.92 WU/μL T4 DNA ligase, 1.0 U/μL BsaI, ca. inert functional polymers of 2 nM repeating units, and 0.06 mM ATP, under orbital shaking at 80 rpm. Conditions for (h): 37°C, 0.5 nM GOx on enzyme carriers, 0.5 nM HRP on enzyme carriers, 2 mM ABTS2-, SfNAP1 and SfNAP2 of 0.008 mM repeating units, under orbital shaking at 97 rpm. After coacervating for 2 h, 1 mM glucose was added, and absorbance at 414 nm was measured.
Aside from trapping colloids in the coacervates for superstructure formation, we further show the encapsulation of functional polymers inside the coacervates (Figure 5b,d-g). Typical experiments were performed by running the above ATTO647-labeled M3/M4-M5/M2 coacervates (red channel) together with a Cy3 and biotin-labeled static Cy3/biotin-SfNAP (green channel) without recognition sequence for BsaI, bearing one of the complementary multivalent binders. The Cy3/biotin-SfNAPs is only present in 0.02 mol% of the coacervate SfNAPs. Time-dependent CLSM images visualize the transient trapping and release of the Cy3-SfNAPs (Figure 5d). Both fluorescence channels show a transient increase of the intensities, yet there is some relevant delay for the Cy3/biotin-SfNAPs (Figure 5e). The uptake is slightly delayed due to the small quantity compared to the coacervate-forming SfNAPs. Additionally, CLSM shows a homogeneous distribution of the static Cy3-SfNAP polymers in the coacervates for the first 2 h. Yet, after 4 h, when degradation sets in, the Cy3-SfNAP is clearly more visible at the periphery of the coacervates (Figure 5f,g). This is because, when ATP is substantially consumed, there is a degradation of the SfNAP1 (bearing the same multivalent binder as the static Cy3/biotin-SfNAPs), and its decreased multivalent binding to the coacervates accumulates some Cy3/biotin-SfNAPs at the periphery of the coacervates.
One of the key aspects of LLPS is to provide spatially confined reaction compartments for catalysis with local enrichment and macromolecular crowding. To demonstrate a proof-of-concept catalytic reaction, we used the biotin motifs on the Cy3/biotin-SfNAPs to encapsulate streptavidin(StAv)-conjugated biomolecules, more precisely StAv-based fusion proteins of GOx and HRP, which can form an enzymatic cascade (Figure S7, Supplementary Note 1). In this enzyme cascade system, GOx firstly catalyzes oxidization of glucose and the generated H2O2 is used by HRP to oxidize 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS2–) into ABTS–., which can be traced colorimetrically at 414 nm. Here, static coacervates were applied to investigate the catalytic efficacy of the enzyme cascade reaction of GOx and HRP in the macromolecularly crowded coacervate environment (Figure 5h). Non-fluorescent SfNAP1 and ATTO647-SfNAP2 (8 μM for each species regarding amounts of repeating units) were mixed together with 0.5 nM GOx and HRP (separately pre-anchored to Cy3/biotin-SfNAPs), and 2 mM ABTS2-. After 2 h coacervation via orbital shaking at 97 rpm, 1 mM glucose was added and the catalytic efficacy was monitored at 414 nm absorbance. We find a significantly increased catalytic efficiency for the enzyme cascade encapsulated in the DNA coacervates as compared to free enzymes or enzymes immobilized on the DNA polymers (EC; enzyme carrier; Figure 5i). This demonstrates first of all efficient diffusion of educts and products within the coacervates and, secondly, that the coacervates provide a propitious environment for the catalysis, which is due to molecular crowding and spatial proximity effect. 63 The results underscore the potential for using the multivalency guided all-DNA coacervates as reactors for versatile reactions.
Discussion
We have demonstrated, for the first time, the multivalency-driven LLPS of two orthogonal SfNAPs for artificial membrane-less organelles, serving as a model system to understand the multivalency-driven LLPS in biological systems. Energetically up-hill driven coacervates have been realized by coupling the SfNAPs-forming building blocks to an ERN of ATP-powered ligation and BsaI-controlled restriction, rendering transient multivalency-driven all-DNA coacervates showing programmable lifetimes by ATP fuel level. By encoding molecular recognitions both in sticky ends and short side chains on various building blocks, we further demonstrated self-sorting of the LLPS for multicomponent coacervates in the same system, reminiscent of the diverse membrane-less organelles in biological systems.
Critically, the molecular recognition during LLPS allows for multivalent trapping of complementary ssDNA functionalized objects, and the isothermal LLPS opens possibilities to encapsulate temperature-sensitive components such as enzymes or proteins in general. We proved this by trapping micron-scale colloids for temporal formation of hybrid superstructures and trapping catalytically active biomolecules, GOx and HRP, for improved efficiency of the enzyme cascade.
Looking to the future, we foresee that this transient multivalency driven LLPS provides completely new insights for significantly and swiftly advancing the fields of life-like materials, systems chemistry, and synthetic biology, as well as for better understanding some principles of the dynamic structures in living systems. We believe that more external and autonomous control mechanisms can simply be encoded by providing light-switchable and reconfigurable components. 36, 54, 64 However, oscillatory behavior is hard to achieve in our coacervate system, as the reversible switches in the ERN cannot be as swift as the systems using photo-switchable chromophores. We envision in particular that the concept of transient multivalency driven temporal coacervates will allow to investigate biocatalytic reactions inside the coacervates, dynamic condensation of proteins and nucleic acids for programmable biofunctions, and to program synthetic cells with hierarchical dynamic structures, enabling applications in biomedical engineering.
Experimental Procedures
Resource Availability
Lead Contact Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Andreas Walther (Andreas.walther@makro.uni-freiburg.de)
Materials Availability
This study did not generate new unique reagents.
Syntheses of sequence-defined functionalized nucleic acid polymers (SfNAPs)
dsDNA tiles M1 and M3 were first annealed from complementary ssDNA, D1/D2, and D4/D5, respectively (see sequences in Supplementary Table 1). The syntheses of SfNAP1 and SfNAP2 followed with the same protocol. Briefly, the experiments were performed at 37°C in 1× NEB CutSmart buffer (50 mM potassium acetate, 20 mM tris-acetate, 10 mM magnesium acetate, 100 μg/mL BSA) containing 0.02 mM M1 (or M3), 0.024 mM M2 (or M4), 0.46 Weiss Units (WU)/μL T4 DNA ligase as well as 0.25 mM ATP, and the reactions were run for 12 h (the addition of a bit more tile M2 or M4 helps to suppress e.g. slight pipetting errors, ensuring efficient polymerizations). For analyses of the SfNAPs, 6 μL aliquots of the reaction solutions were quenched by 8 μL of the quenching buffer. Afterwards, all quenched samples were analyzed by agarose gel electrophoresis (AGE) (2 wt.% agarose gel in TAE buffer) using 90 V for 2 h run time. The results were recorded by a transilluminator (UVsolo touch Gel electrophoresis documentation system, Analytik Jena).
Liquid-liquid phase separation (LLPS) of two orthogonal SfNAPs
The above obtained SfNAP1 and SfNAP2 solutions were mixed with equal volumes (60 μL in total) in a well of an imaging plate at 37°C under orbital shaking at 80 revolutions per minute (rpm). The temperature was controlled by a Hokai-Hit Stage Top Incubator System. The mixed solution was sealed with a layer of hexadecane (40 μL) to prevent evaporation. At different time intervals, confocal laser scanning microscopy (CLSM) was applied to characterize the coacervates formed through multivaleny-driven LLPS. 63× water immersion objective was used throughout this study. CLSM measurements were conducted consecutively with the same microscopy settings. The DNA coacervates were quantified by ImageJ with regard to their size and fluorescent intensity over time. Briefly, the size and average fluorescent intensity of each individual coacervate were measured manually by line and circle selections, respectively. The line was drawn through the center of the coacervate and the circle selection covers at least 80 % of the area for the whole coacervate. Small and irregular aggregates with very faint fluorescence and less than 1 μm diameter were not counted. For the autonomous process below, the small coacervates with less than 1 μm diameter were also counted at 30 min due to their overall very small size.
ATP-fueled transient membrane-less all-DNA coacervates
All the coacervation experiments for CLSM imaging were performed using 60 μL aqueous solutions sealed by 40 μL hexadecane. Typical experiments were conducted in 1× NEB CutSmart buffer at 37°C containing 0.01 mM M1, 0.012 mM M2, 0.01 mM M3, 0.012 mM M4, 0.92 WU/μL T4 DNA ligase, 1.0 U/μL BsaI as well as 0.06, 0.1 or 0.2 mM ATP, under orbital shaking at 80 rpm. The reaction solution was sealed in a well of an imaging plate by a layer of hexadecane. At different time intervals, CLSM was applied to monitor the DNA coacervates. It is worth noting that for the measurements at 30 min, the solutions were set still for ca. 5 min prior the measurements to let the coacervates sediment a bit and facilitate the nucleation. For the AGE analyses of the time-dependent distributions of the SfNAPs, the experiments were performed with same conditions as above. At different time intervals, 6 μL aliquots of the reaction solutions were quenched by 8 μL of the quenching buffer, and analyzed by AGE (see above). To investigate the effects of shaking on the coacervates, the experiments were conducted similar to above. Two groups of experiments were fueled with 0.06 mM ATP and performed under orbital shaking at 400 rpm and without shaking, respectively. CLSM was implemented to characterize the coacervates, and CLSM measurements were conducted consecutively with the same microscopy settings.
Supplementary Material
Supplemental Information can be found online at https://www.cell.com/chem/ or from the author.
Acknowledgments
We acknowledge support by the European Research Council starting Grant (TimeProSAMat) Agreement 677960, as well as from the DFG Cluster of Excellence livMatS “Living, Adaptive and Energy-Autonomous Materials Systems”, EXC-2193/1 - 390951807.
Footnotes
AUTHOR CONTRIBUTIONS
J.D. and A.W. conceived the project. J.D. designed and performed all the experiments. A.W. designed experiments and supervised the project. J.D. and A.W. analyzed the data and wrote the manuscript.
DECLARATION OF INTERESTS
The authors declare no competing interests.
Data and Code Availability
This study did not generate or analyze datasets or code.
References
- 1.Shin Y, Brangwynne CP. Liquid Phase Condensation in Cell Physiology and Disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 2.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular Condensates: Organizers of Cellular Biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Case LB, Zhang X, Ditlev JA, Rosen MK. Stoichiometry Controls Activity of Phase-Separated Clusters of Actin Signaling Proteins. Science. 2019;363:1093–1097. doi: 10.1126/science.aau6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X-H, Chavali PL, Pancsa R, Chavali S, Babu MM. Function and Regulation of Phase-Separated Biological Condensates. Biochemistry. 2018;57:2452–2461. doi: 10.1021/acs.biochem.7b01228. [DOI] [PubMed] [Google Scholar]
- 5.Strzyz P. Phase Separation Tunes Signal Transduction. Nat Rev Mol Cell Biol. 2019;20:263. doi: 10.1038/s41580-019-0121-7. [DOI] [PubMed] [Google Scholar]
- 6.Li P, Banjade S, Cheng H-C, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF. Phase Transitions in the Assembly of Multivalent Signalling Proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Jülicher F, Hyman AA. Germline P Granules Are Liquid Droplets That Localize by Controlled Dissolution/Condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 8.Brangwynne CP, Mitchison TJ, Hyman AA. Active Liquid-Like Behavior of Nucleoli Determines Their Size and Shape in Xenopus Laevis Oocytes. Proc Natl Acad Sci USA. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mao YS, Zhang B, Spector DL. Biogenesis and Function of Nuclear Bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dundr M. Nuclear Bodies: Multifunctional Companions of the Genome. Curr Opin Cell Biol. 2012;24:415–422. doi: 10.1016/j.ceb.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD. Phase Transition of a Disordered Nuage Protein Generates Environmentally Responsive Membraneless Organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernandez-Verdun D. Assembly and Disassembly of the Nucleolus During the Cell Cycle. Nucleus. 2011;2:189–194. doi: 10.4161/nucl.2.3.16246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matera AG, Terns RM, Terns MP. Non-Coding RNAs: Lessons from the Small Nuclear and Small Nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 14.Martin N. Dynamic Synthetic Cells Based on Liquid-Liquid Phase Separation. ChemBioChem. 2019;20:2553–2568. doi: 10.1002/cbic.201900183. [DOI] [PubMed] [Google Scholar]
- 15.Tang TD, Hak CRC, Thompson AJ, Kuimova MK, Williams D, Perriman AW, Mann S. Fatty Acid Membrane Assembly on Coacervate Microdroplets as a Step Towards a Hybrid Protocell Model. Nat Chem. 2014;6:527–533. doi: 10.1038/nchem.1921. [DOI] [PubMed] [Google Scholar]
- 16.Vieregg JR, Lueckheide M, Marciel AB, Leon L, Bologna AJ, Rivera JR, Tirrell MV. Oligonucleotide–Peptide Complexes: Phase Control by Hybridization. J Am Chem Soc. 2018;140:1632–1638. doi: 10.1021/jacs.7b03567. [DOI] [PubMed] [Google Scholar]
- 17.Adhikari S, Prabhu VM, Muthukumar M. Lower Critical Solution Temperature Behavior in Polyelectrolyte Complex Coacervates. Macromolecules. 2019;52:6998–7004. [PMC free article] [PubMed] [Google Scholar]
- 18.Priftis D, Tirrell M. Phase Behaviour and Complex Coacervation of Aqueous Polypeptide Solutions. Soft Matter. 2012;8:9396–9405. [Google Scholar]
- 19.Poudyal RR, Guth-Metzler RM, Veenis AJ, Frankel EA, Keating CD, Bevilacqua PC. Template-Directed Rna Polymerization and Enhanced Ribozyme Catalysis inside Membraneless Compartments Formed by Coacervates. Nat Commun. 2019;10 doi: 10.1038/s41467-019-08353-4. 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drobot B, Iglesias-Artola JM, Le Vay K, Mayr V, Kar M, Kreysing M, Mutschler H, Tang TD. Compartmentalised RNA Catalysis in Membrane-Free Coacervate Protocells. Nat Commun. 2018;9:3643. doi: 10.1038/s41467-018-06072-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schoonen L, van Hest JC. Compartmentalization Approaches in Soft Matter Science: From Nanoreactor Development to Organelle Mimics. Adv Mater. 2016;28:1109–1128. doi: 10.1002/adma.201502389. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Liu X, Abdelmohsen LK, Williams DS, Huang X. Spatial Organization in Proteinaceous Membrane-Stabilized Coacervate Protocells. Small. 2019;15:1902893. doi: 10.1002/smll.201902893. [DOI] [PubMed] [Google Scholar]
- 23.Mason AF, Buddingh’ BC, Williams DS, van Hest JC. Hierarchical Self-Assembly of a Copolymer-Stabilized Coacervate Protocell. J Am Chem Soc. 2017;139:17309–17312. doi: 10.1021/jacs.7b10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams DS, Patil AJ, Mann S. Spontaneous Structuration in Coacervate-Based Protocells by Polyoxometalate-Mediated Membrane Assembly. Small. 2014;10:1830–1840. doi: 10.1002/smll.201303654. [DOI] [PubMed] [Google Scholar]
- 25.Deshpande S, Brandenburg F, Lau A, Last MG, Spoelstra WK, Reese L, Wunnava S, Dogterom M, Dekker C. Spatiotemporal Control of Coacervate Formation within Liposomes. Nat Commun. 2019;10:1800. doi: 10.1038/s41467-019-09855-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang L-W, Lytle TK, Radhakrishna M, Madinya JJ, Vélez J, Sing CE, Perry SL. Sequence and Entropy-Based Control of Complex Coacervates. Nat Commun. 2017;8:1273. doi: 10.1038/s41467-017-01249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Black KA, Priftis D, Perry SL, Yip J, Byun WY, Tirrell M. Protein Encapsulation Via Polypeptide Complex Coacervation. ACS Macro Lett. 2014;3:1088–1091. doi: 10.1021/mz500529v. [DOI] [PubMed] [Google Scholar]
- 28.Priftis D, Leon L, Song Z, Perry SL, Margossian KO, Tropnikova A, Cheng J, Tirrell M. Self-Assembly of α-Helical Polypeptides Driven by Complex Coacervation. Angew Chem, Int Ed. 2015;127:11280–11284. doi: 10.1002/anie.201504861. [DOI] [PubMed] [Google Scholar]
- 29.Fan Y, Tang S, Thomas EL, Olsen BD. Responsive Block Copolymer Photonics Triggered by Protein–Polyelectrolyte Coacervation. ACS Nano. 2014;8:11467–11473. doi: 10.1021/nn504565r. [DOI] [PubMed] [Google Scholar]
- 30.Jeon O, Wolfson DW, Alsberg E. In-Situ Formation of Growth-Factor-Loaded Coacervate Microparticle-Embedded Hydrogels for Directing Encapsulated Stem Cell Fate. Adv Mater. 2015;27:2216–2223. doi: 10.1002/adma.201405337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cinar H, Cinar S, Chan HS, Winter R. Pressure-Induced Dissolution and Reentrant Formation of Condensed, Liquid–Liquid Phase-Separated Elastomeric α-Elastin. Chem: Eur J. 2018;24:8286–8291. doi: 10.1002/chem.201801643. [DOI] [PubMed] [Google Scholar]
- 32.Yang S, Li B, Wu C, Xu W, Tu M, Yan Y, Huang J, Drechsler M, Granick S, Jiang L. Steering Coacervation by a Pair of Broad-Spectrum Regulators. ACS Nano. 2019;13:2420–2426. doi: 10.1021/acsnano.8b09332. [DOI] [PubMed] [Google Scholar]
- 33.Deshpande S, Brandenburg F, Lau A, Last MG, Spoelstra WK, Reese L, Wunnava S, Dogterom M, Dekker C. Spatiotemporal Control of Coacervate Formation within Liposomes. Nat Commun. 2019;10:1–11. doi: 10.1038/s41467-019-09855-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuster BS, Reed EH, Parthasarathy R, Jahnke CN, Caldwell RM, Bermudez JG, Ramage H, Good MC, Hammer DA. Controllable Protein Phase Separation and Modular Recruitment to Form Responsive Membraneless Organelles. Nat Commun. 2018;9:2985. doi: 10.1038/s41467-018-05403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin N, Douliez J-P, Qiao Y, Booth R, Li M, Mann S. Antagonistic Chemical Coupling in Self-Reconfigurable Host–Guest Protocells. Nat Commun. 2018;9:3652. doi: 10.1038/s41467-018-06087-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin N, Tian L, Spencer D, Coutable-Pennarun A, Anderson JR, Mann S. Photoswitchable Phase Separation and Oligonucleotide Trafficking in DNA Coacervate Microdroplets. Angew Chem Int Ed. 2019;58:14594–14598. doi: 10.1002/anie.201909228. [DOI] [PubMed] [Google Scholar]
- 37.Koga S, Williams DS, Perriman AW, Mann S. Peptide–Nucleotide Microdroplets as a Step Towards a Membrane-Free Protocell Model. Nat Chem. 2011;3:720–724. doi: 10.1038/nchem.1110. [DOI] [PubMed] [Google Scholar]
- 38.Aumiller WM, Jr, Keating CD. Phosphorylation-Mediated Rna/Peptide Complex Coacervation as a Model for Intracellular Liquid Organelles. Nat Chem. 2016;8:129–137. doi: 10.1038/nchem.2414. [DOI] [PubMed] [Google Scholar]
- 39.Nakashima KK, Baaij JF, Spruijt E. Reversible Generation of Coacervate Droplets in an Enzymatic Network. Soft Matter. 2018;14:361–367. doi: 10.1039/c7sm01897e. [DOI] [PubMed] [Google Scholar]
- 40.Te Brinke E, Groen J, Herrmann A, Heus HA, Rivas G, Spruijt E, Huck WT. Dissipative Adaptation in Driven Self-Assembly Leading to Self-Dividing Fibrils. Nat Nanotechnol. 2018;13:849–855. doi: 10.1038/s41565-018-0192-1. [DOI] [PubMed] [Google Scholar]
- 41.Rieß B, Grötsch RK, Boekhoven J. The Design of Dissipative Molecular Assemblies Driven by Chemical Reaction Cycles. Chem. 2020;6:552–578. [Google Scholar]
- 42.Deng J, Walther A. ATP-Responsive and ATP-Fueled Self-Assembling Systems and Materials. Adv Mater. 2020 doi: 10.1002/adma.202002629. [DOI] [PubMed] [Google Scholar]
- 43.Sun M, Deng J, Walther A. Polymer Transformers: Interdigitating Reaction Networks of Fueled Monomer Species to Reconfigure Functional Polymer States. Angew Chem Int Ed. 2020 doi: 10.1002/anie.202006526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang DY, Seelig G. Dynamic DNA Nanotechnology Using Strand-Displacement Reactions. Nat Chem. 2011;3:103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- 45.Qian L, Winfree E. Scaling up Digital Circuit Computation with DNA Strand Displacement Cascades. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 46.Song T, Shah S, Bui H, Garg S, Eshra A, Fu D, Yang M, Mokhtar R, Reif J. Programming DNA-Based Biomolecular Reaction Networks on Cancer Cell Membranes. J Am Chem Soc. 2019;141:16539–16543. doi: 10.1021/jacs.9b05598. [DOI] [PubMed] [Google Scholar]
- 47.Kim Y-Y, Bang Y, Lee A-H, Song Y-K. Multivalent Traptavidin–DNA Conjugates for the Programmable Assembly of Nanostructures. ACS Nano. 2019;13:1183–1194. doi: 10.1021/acsnano.8b06170. [DOI] [PubMed] [Google Scholar]
- 48.Sato Y, Sakamoto T, Takinoue M. Sequence-Based Engineering of Dynamic Functions of Micrometer-Sized DNA Droplets. Sci Adv. 2020;6:eaba3471. doi: 10.1126/sciadv.aba3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen DT, Jeon B-j, Abraham GR, Saleh OA. Length-Dependence and Spatial Structure of DNA Partitioning into a DNA Liquid. Langmuir. 2019;35:14849–14854. doi: 10.1021/acs.langmuir.9b02098. [DOI] [PubMed] [Google Scholar]
- 50.Jeon B-j, Nguyen DT, Abraham GR, Conrad N, Fygenson DK, Saleh OA. Salt-Dependent Properties of a Coacervate-Like, Self-Assembled DNA Liquid. Soft Matter. 2018;14:7009–7015. doi: 10.1039/c8sm01085d. [DOI] [PubMed] [Google Scholar]
- 51.Zeng J, Fu W, Qi Z, Zhu Q, He H, Huang C, Zuo H, Mao C. Self-Assembly of Microparticles by Supramolecular Homopolymerization of One Component DNA Molecule. Small. 2019;15:1805552. doi: 10.1002/smll.201805552. [DOI] [PubMed] [Google Scholar]
- 52.Merindol R, Loescher S, Samanta A, Walther A. Pathway-Controlled Formation of Mesostructured All-DNA Colloids and Superstructures. Nat Nanotechnol. 2018;13:730–738. doi: 10.1038/s41565-018-0168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heinen L, Walther A. Programmable Dynamic Steady States in Atp-Driven Nonequilibrium DNA Systems. Sci Adv. 2019;5:eaaw0590. doi: 10.1126/sciadv.aaw0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deng J, Walther A. Pathway Complexity in Fuel-Driven DNA Nanostructures with Autonomous Reconfiguration of Multiple Dynamic Steady States. J Am Chem Soc. 2020;142:685–689. doi: 10.1021/jacs.9b11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deng J, Walther A. ATP-Powered Molecular Recognition to Engineer Transient Multivalency and Self-Sorting 4d Hierarchical Systems. Nat Commun. 2020;11:3658. doi: 10.1038/s41467-020-17479-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dickson KS, Burns CM, Richardson JP. Determination of the Free-Energy Change for Repair of a DNA Phosphodiester Bond. J Biol Chem. 2000;275:15828–15831. doi: 10.1074/jbc.M910044199. [DOI] [PubMed] [Google Scholar]
- 57.Ge H, Qian H. Dissipation, Generalized Free Energy, and a Self-Consistent Nonequilibrium Thermodynamics of Chemically Driven Open Subsystems. Phys Rev E. 2013;87:062125. doi: 10.1103/PhysRevE.87.062125. [DOI] [PubMed] [Google Scholar]
- 58.Qian H. Open-System Nonequilibrium Steady State: Statistical Thermodynamics, Fluctuations, and Chemical Oscillations. J Phys Chem B. 2006;110:15063–15074. doi: 10.1021/jp061858z. [DOI] [PubMed] [Google Scholar]
- 59.Qian H, Kjelstrup S, Kolomeisky AB, Bedeaux D. Entropy Production in Mesoscopic Stochastic Thermodynamics: Nonequilibrium Kinetic Cycles Driven by Chemical Potentials, Temperatures, and Mechanical Forces. J Phys: Condens Matter. 2016;28:153004. doi: 10.1088/0953-8984/28/15/153004. [DOI] [PubMed] [Google Scholar]
- 60.Qian H, Roy S. An Information Theoretical Analysis of Kinase Activated Phosphorylation Dephosphorylation Cycle. IEEE Trans Nanobioscience. 2012;11:289–295. doi: 10.1109/TNB.2011.2182658. [DOI] [PubMed] [Google Scholar]
- 61.Qian H, Bishop LM. The Chemical Master Equation Approach to Nonequilibrium Steady-State of Open Biochemical Systems: Linear Single-Molecule Enzyme Kinetics and Nonlinear Biochemical Reaction Networks. Int J Mol Sci. 2010;11:3472–3500. doi: 10.3390/ijms11093472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walther A. From Responsive to Adaptive and Interactive Materials and Materials Systems: A Roadmap. Adv Mater. 2019;32:1905111. doi: 10.1002/adma.201905111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ellis RJ. Macromolecular Crowding: Obvious but Underappreciated. Trends Biochem Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 64.Deng J, Bezold D, Jessen HJ, Walther A. Multiple Light Control Mechanisms in Atp-Fueled Non-Equilibrium DNA Systems. Angew Chem Int Ed. 2020;59:12084–12092. doi: 10.1002/anie.202003102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate or analyze datasets or code.