Abstract
Idiopathic hypogonadotropic hypogonadism (IHH) is a rare genetic condition characterized by absent puberty and infertility due to gonadotropin-releasing hormone (GnRH) deficiency. IHH can be accompanied by normal (normosmic IHH, nIHH) or compromised olfaction (Kallmann syndrome, KS). Several semaphorins have been shown to be potent modulators of the GnRH, olfactory and vomeronasal system development. Using exome sequencing, we screened 216 IHH patients and identified 10 ultra-rare missense variants in SEMA3F and PLXNA3 in 15 patients, corresponding to 6.9% of our study cohort. Most of these variants are predicted to affect SEMA3F secretion or signaling activity based on predictive algorithms and in vitro functional assays. We also demonstrated the expression of SEMA3F, and of its obligatory holoreceptors, PlexinAs, along the GnRH migratory route in human fetuses. We report that SEMA3F signaling insufficiency contributes to the pathogenesis of IHH.
Keywords: Hypogonadotropic hypogonadism, puberty, PLXNA3, SEMA3F
Idiopathic Hypogonadotropic Hypogonadism (IHH) is a rare genetic disorder characterized by complete or partial pubertal failure caused by gonadotropin-releasing hormone (GnRH) deficiency. According to olfactory function, IHH is divided into two major forms, normal sense of smell (normosmic IHH, nIHH) and inability to smell, anosmia, defined as Kallmann syndrome, KS. However, this distinction is often blurred, probably reflecting the wealth of patho-physiological mechanisms and their variety of combinations in individual cases. Up to the present, nearly 50 genes have been reported to be associated with IHH 1 , which account for nearly 50% all cases and thus suggests that other associated genes remain to be discovered. To date, the genes implicated in IHH impact GnRH neuron ontogenesis, GnRH neuron migration and/or axon growth, GnRH secretion, and/or gonadotrope function. GnRH–secreting neurons are unique neuroendocrine cells as they originate in the nasal placode, outside the central nervous system, during embryonic development, and migrate to the hypothalamus along the vomeronasal and terminal nerves (VNN, TN) 2 . This process is evolutionarily conserved and follows a similar spatio-temporal pattern in all mammals 2 , including humans 3,4 . Maldevelopment of this neuroendocrine system results in hypothalamic hypogonadotropic hypogonadism. Unraveling new genetic pathways involved in the development or function of GnRH neurons is relevant for understanding the basis of pathogenesis leading to IHH in humans.
Proper navigation of growing axons and neurons during embryonic development depend on the action of guidance cues, which include semaphorins, a large and diverse family of secreted and membrane-associated proteins 5 . Correct targeting of GnRH neurons and olfactory/vomeronasal projections have been shown to depend on the orchestrated action of this family of guidance cues 6 . Mutations in members of class-3 semaphorins, SEMA3A, SEMA3E and SEMA3G, have been associated with IHH 7–9 . To exert its functions, SEMA3s bind to Neuropilin co-receptors (NRP1 and NRP2) in hetero-complexes with PlexinA1-4 (PLXNA1-4) receptors to activate plexin signal-transduction 10 . Loss of function of Plxna1, Nrp1 and Nrp2 have been linked to defective GnRH neuron development in mice 9,11–13 and nonsynonymous heterozygous variants in PLXNA1 and NRP2 have been identified in KS individuals 12 .
Among SEMA3s, there is semaphorin-3F (SEMA3F), a secreted protein that serves as a guidance cue to repel late-arriving olfactory axons that express neuropilin-2 (Nrp2) receptor to the olfactory bulbs 14 . SEMA3F, PLXNA3, and NRP2 have a spatio-temporal expression consistent with a possible role on the regulation of the GnRH system, olfactory, and vomeronasal development 3,13,14
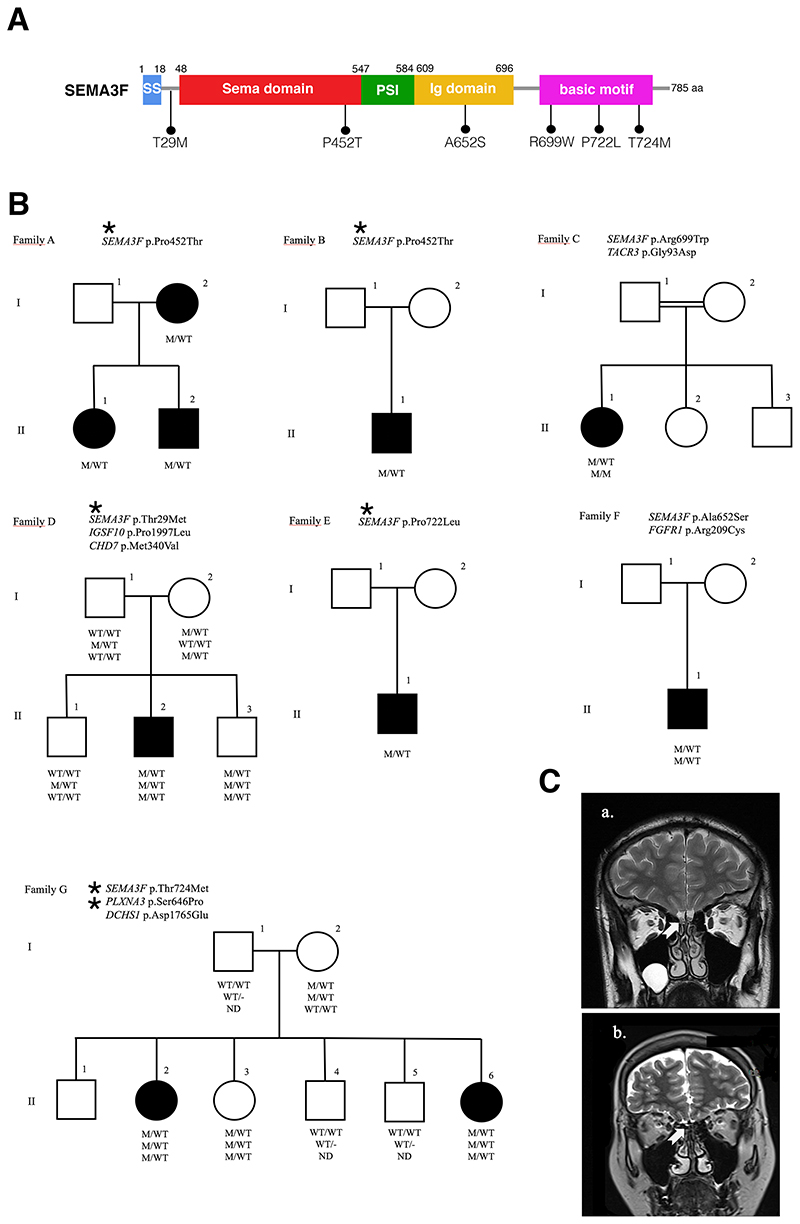
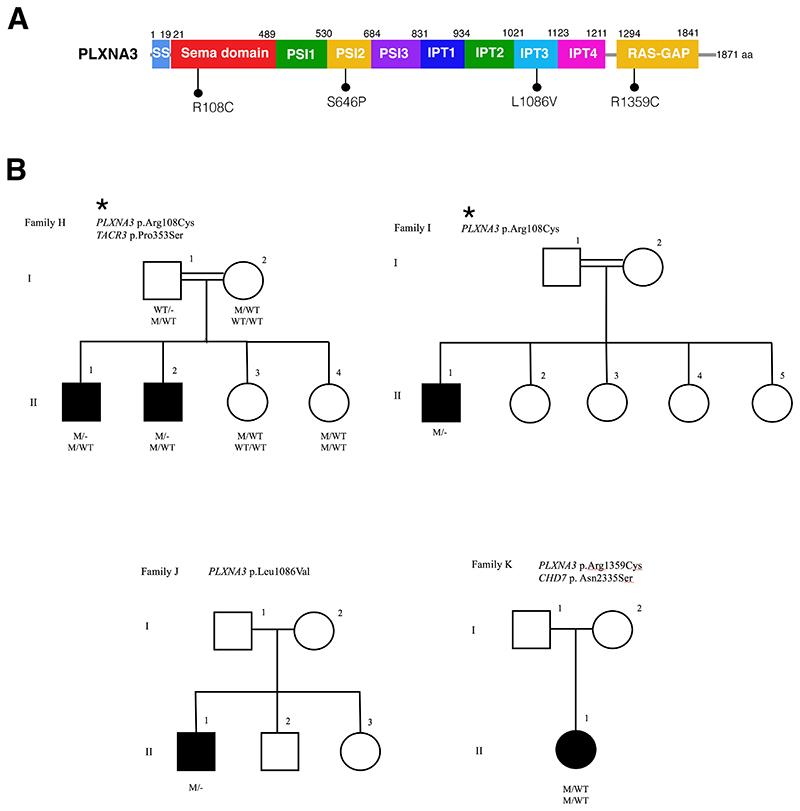
In this study, we performed exome sequencing in 216 well-phenotyped IHH/KS patients, and identified heterozygous or hemizygous missense variants in SEMA3F (HGNC:10728) and PLXNA3 (HGNC:9101) genes respectively in 15 patients from 11 unrelated families. Clinical and molecular genetic characteristics of the patients and their alterations are shown in Table 1 and Supplementary Table 1, respectively. The location of the variants on SEMA3F and PLXNA3 gene diagrams and the pedigrees are depicted in Figures 1 and 2. We found six rare SEMA3F variants located in critical regions for the proper protein function or its dimerization (Figure 1A). We identified one variant (T29M) located close to the signal sequence (SS) of the protein, one (P452T) located within the SEMA domain, one (A652S) in the Ig domain and three (R699W, P722L, T724M) located in the basic motif of the protein (Figure 1A). Seven IHH patients carried variants in PLXNA3 respectively in the SEMA binding domain (R108C), in the Plexin-Semaphorin-Integrin (PSI2) domain (S646P) and in the Ig domain (IPT3 region) (L1086V), and RAS-GAP (R1359C), which suggest that these mutants are likely to affect ligand and co-receptor interactions (Figure 2A).
Table 1. Clinical characteristics of individuals with SEMA3F and PLXNA3 mutations. M, male; F, female; NA, Not available.
| Family/in dividual no | Gene/variant | Age at diagnosis | Sex | Obesity | Olfaction | Reproductive phenotype |
|---|---|---|---|---|---|---|
| A I-1 | SEMA3F p.Pro452Thr | 28 | F | No | Anosmic | No delayed puberty or infertility reported |
| A II-1 | SEMA3F p.Pro452Thr | 20 | F | No | Anosmic | Absent puberty, primary amenorrhea |
| A II-2 | SEMA3F p.Pro452Thr | 12 | M | Obese | Anosmic | Absent puberty, cryptorchidism |
| B II-1 | SEMA3F p.Pro452Thr | 10.5 | M | Overweight | Normosmic | Cryptorchidism |
| C II-1 | SEMA3F p.Arg699Trp | 16 | F | No | Normosmic | Absent puberty, primary amenorrhea |
| D II-2 | SEMA3F p.Thr29Met | 17 | M | Obese | Hyposmic | Absent puberty, cryptorchidism |
| E II-1 | SEMA3F p.Pro722Leu | 14 | M | Obese | Normosmic | Absent puberty |
| F I-1 | SEMA3F p.Ala652Ser | 35 | M | No | Hyposmic | Absent puberty, micropenis, infertility |
| G II-3 | SEMA3F p.Thr724Met PLXNA3 p.Ser646Pro | 20 | F | No | Normosmic | Primary amenorrhea, Delayed menarche at age 16 |
| G II-6 | SEMA3F p.Thr724Met PLXNA3 p.Ser646Pro | 14 | F | No | Normosmic | Absent puberty |
| H II-1 | PLXNA3 p.Arg108Cys | 19 | M | No | Normosmic | Absent puberty |
| H II-2 | PLXNA3 p.Arg108Cys | 17 | M | No | Normosmic | Absent puberty |
| I II-1 | PLXNA3 p.Arg108Cys | 21 | M | No | Hyposmic | Absent puberty, cryptorchidism |
| J II-1 | PLXNA3 p.Leu1086Val | 14 | M | Overweight | Normosmic | Absent puberty, micropenis |
| K II-1 | PLXNA3 p.Arg1359Cys | 18 | F | Obese | Normosmic | Absent puberty, primary amenorrhea |
Figure 1. SEMA3F Mutations in the etiology of idiopathic hypogonadotropic hypogonadism.
A. The mutations are depicted on the functional gene diagram of SEMA3F. B. The pedigrees of seven families with SEMA3F mutations are shown. Affected males and females are represented by black squares and black circles respectively. White square symbols indicate unaffected male family members, white circle symbols represent unaffected female family members, and the double line indicates consanguinity. Under each symbol are the genotypes in the same order as the gene and variantdescriptions, with WT and M denoting wild type and mutant, respectively. ND: Not determined. C. The T2-weighted magnetic resonance imaging (MRI) from a healthy control (a) and from patient DII-2 (b). The arrows point to normal (a) and aplastic olfactory bulbs (b).
Figure 2. PLXNA3 mutations in patients with idiopathic hypogonadotropic hypogonadism.
A. The diagram of PLXNA3 showing the positions of the missense mutations found in patients. B. The pedigrees of the three families with PLXNA3 mutations are shown. Note that patients in Family G in Figure 1 also have PLXNA3 mutations in addition to SEMA3F mutations. Affected males and females are represented by black squares and black circles respectively. White square symbols indicate unaffected male family members, white circle symbols represent unaffected female family members, and the double line indicates consanguinity. Under each symbol are the genotypes in the same order as the gene and variantdescriptions, with WT and M denoting wild type and mutant, respectively.
In order to avoid harmless polymorphisms, in addition to quality and read depth filtering, we filtered the protein altering heterozygous/hemizygous variants by setting a threshold occurrence of 1:10.000. These 10 variants were either not seen or extremely rare with a minor allele frequency <0.0001 in the two largest reference population databases, TOPMed and gnomAD. Similarly, these variants were not reported in a much smaller but regional database, the Greater Middle East Variome (GME). Sanger sequencing confirmed the presence of these variants. No variant was seen in ClinVar. These variants were all classified as variants of uncertain significance (VUS) by ACMG/AMP classification 15 .
In the etiology of IHH, almost all causative gene alterations show phenotypic variability and segregations in families are often irregular as in some of the pedigrees in this study. Oligogenic etiology 16 and clinical reversibility 17 , among others, are well recognized in this condition to explain these complex pedigrees. In these situations, in order to identify novel puberty genes, strategies such as burden testing should be employed. In fact, in the recent discovery of several IHH genes, burden tests were routinely carried out 18,19 . Also, burden testing, as aggregate variant analysis in case-control studies, was recognized by the authoritative, Clinical Genome Resource (ClinGen), which sets up evidence-based guidelines in establishing new gene-disease associations, as comparably valuable to linkage analysis 20 . To determine if putative pathogenic SEMA3F signaling gene variants are more enriched in the disease cohort than in the control population, we performed gene-based burden tests. Working on the same disease as in this study with a comparable patient population, Guo et al. developed a gene-based burden test using gnomAD and TOPMed as control populations to their IHH cases 21 . Emulating that study, we did a statistical comparison of occurrences of rare (<1:10.000) protein-truncating and missense alleles with a CADD score >20 using Fisher exact test. We found that rare putatively harmful SEMA3F or PLXNA3 variants were statistically more enriched in our patient cohort compared to the TOPMed database representing the general (presumably healthy) population (Supplementary Table 1). Specifically, our individual gene-based burden analyses revealed the following p values and odds ratios: SEMA3F (p = 0.005) with an odds ratio 4.51 (1.86-10.97 95% CI) and PLXNA3 (p = 0.019) with an odds ratio 2.47 (1.02-6.01 95% CI). Patients in this study were recruited in Turkey. The SNP profile in the general Turkish population was previously reported to be comparable to that of Europeans 22 . Although the largest ancestral group in both gnomAD and TOPMed is non-Finnish Europeans, both databases are considerably represented by other groups. It has been recognized that testing across mix ancestries even further increases statistical power, attesting to the validity of our comparisons 21 . In summary, these burden testing results provide strong support to the contention that variants in SEMA3F signaling contribute to IHH.
In six of the 11 kindreds (54%), there was at least one more gene known to be associated with IHH (oligogenecity) 1,16 . This high rate is consistent with our recent observation of the increased detection of oligogenecity in the etiology of IHH 23 , which was earlier reported to be 10-20% 16 . This probably reflects increased number of recognized genes for IHH as well as recent diagnostic popularity of exome sequencing. Overall, the pattern of inheritance among the SEMA3F pedigrees is consistent with an autosomal dominant inheritance of each variant with variable penetrance and expressivity, irrespective of the inheritance whether it is familial or sporadic, which was repeatedly observed in recent IHH gene discoveries 18 The inheritance of PLXNA3 variants are consistent with X-linked recessive pattern. With the exception of one SEMA3F (p.Pro452Thr) and one PLXNA3 (p.Arg108Cys) variants, all other variants were encountered once. Notably, two sisters in family G had variants in both SEMA3F and PLXNA3, which were both inherited from their apparently unaffected mother, but their phenotypes were not more severe, in fact, these sisters had a lighter phenotype, one of the sisters having only delayed menarche. Overall, six of 15 (40%) patients (five of them with SEMA3F variants) had compromised olfaction. In two of them, brain MRIs were obtained, which showed hypoplastic/aplastic olfactory bulbs. Figure 1C depicts the absent olfactory bulbs in Case D-II-2 due to p.Thr29Met in SEMA3F. These data are consistent with previous rodent studies showing the role of SEMA3F in the proper projections and fasciculations of the olfactory nerve and thus in the development of the olfactory bulb 14 . Interestingly, the same SEMA3F variant (Pro452Thr) was associated with normosmia or anosmia in two different families (Family A and B respectively). On the other hand, male patients in both families had cryptorchidism, indicating the presence of severe congenital hypogonadism. In congenital IHH, fetal pituitary gonadotropin secretion is low, leading to inadequate testosterone levels in fetal serum. As the testicular descent and growth of phallus are androgen-dependent, consequently, boys with severe IHH present with micropenis, cryptorchidism, and hypospadias at birth 24 . Again, there was a discrepancy in olfactory function in patients carrying the same PLXNA3 variant (p.Arg108Cys) in two different kindreds (Family H and I, Figure 2A, B). These observations suggest that there is a complex input to the olfactory phenotype. To date, all Semaphorin signaling gene variant discoveries were carried out solely in the KS patients cohorts 9,12,25 . This may have obscured the breadth of the phenotypic spectrum associated with these versatile family of molecules. Our population, on the contrary, is made up of all-inclusive IHH patients regardless of their olfactory functions. Therefore, our study has an unrestricted ability to observe all possible phenotypes. Case in point, in a previous screening study of PLXNA1 mutant cases in the same cohort as in this study, we found nine cases, among which only one third had compromised olfaction 23 . In contrast, the seminal publication on this gene was from an all anosmic (KS) cohort 12 .
In order to test the functionality of the variants identified in KS and nIHH probands, we transiently transfected HEK293T cells with plasmids encoding the human wild-type proteins (SEMA3F and PLXNA3) or the corresponding variants and investigated whether the SEMA3F secretory capacity or PLXNA3 protein maturation and trafficking of transfected cells was affected. Because mutations in genes such as TAC3 and FGFR1 have been shown to be highly penetrant and to, most likely, primarily drive the IHH phenotype 26 , we opted not to test functionally the SEMA3F variants segregating with mutations in one of the aforementioned genes.
Western blot analysis revealed normal protein content of SEMA3F variants in whole cell lysates (Supplementary Figure 1A, B). By contrast, SEMA3Fs harboring the P452T, T29M, and T724M missense variants showed impaired SEMA3F secretion, as shown by western blot analysis of the conditioned media (Supplementary Figure 1A, C). Since SEMA3F is a bifunctional guidance molecule that can exert both axon-repulsive and -attractive effects depending on its gradient and on receptors’ composition on target cells 27 , future studies will be aimed at understanding how different SEMA3F concentrations and different receptors’ complexes may affect GnRH neuronal migration and/or olfactory and vomeronasal axon orientation. We also tested the effect of the PLXNA3 variants (with the exception of R1359C, which was detected while preparing the manuscript) on receptor synthesis by western-blot analyses on transfected HEK293T cells producing PLXNA3 WT, PLXNA3 S646P and PLXNA3 R108C (Supplementary Figure 2). Immunoblots on cell lysates from HEK293T cells transfected with PLXNA3 WT or with the mutant proteins revealed the expression of a band for PLXNA3 at the expected molecular weight (220 kDa), which was absent in mock-transfected cells (Supplementary Figure 2A). However, cells expressing the mutant PLXNA3s presented two isoforms, likely to be due to altered post-translational modifications (Supplementary Figure 2A). We next immunostained transfected HEK293 cells for PLXNA3 and Calnexin, a marker of the endoplasmic reticulum (ER). WT PLXNA3 was predominantly localized on the plasma membrane (Supplementary Figure 2B). In contrast, S646P, but not R108C, variant showed PLXNA3 localization exclusively in the ER (Supplementary Figure 2B, 2C), indicating that the variant S646P disrupts cell-surface localization of PLXNA3. Overall, these data revealed the existence of two PLXNA3 isoforms associated with the expression of the mutants, which are likely due to altered glycosylation. Moreover, the observation that PLXNA3 S646P variant was retained within the ER suggests that these variants may affect the processing and/or maturation of the glycosylation of the receptor with a subsequent accumulation of PLXNA3 in the ER.
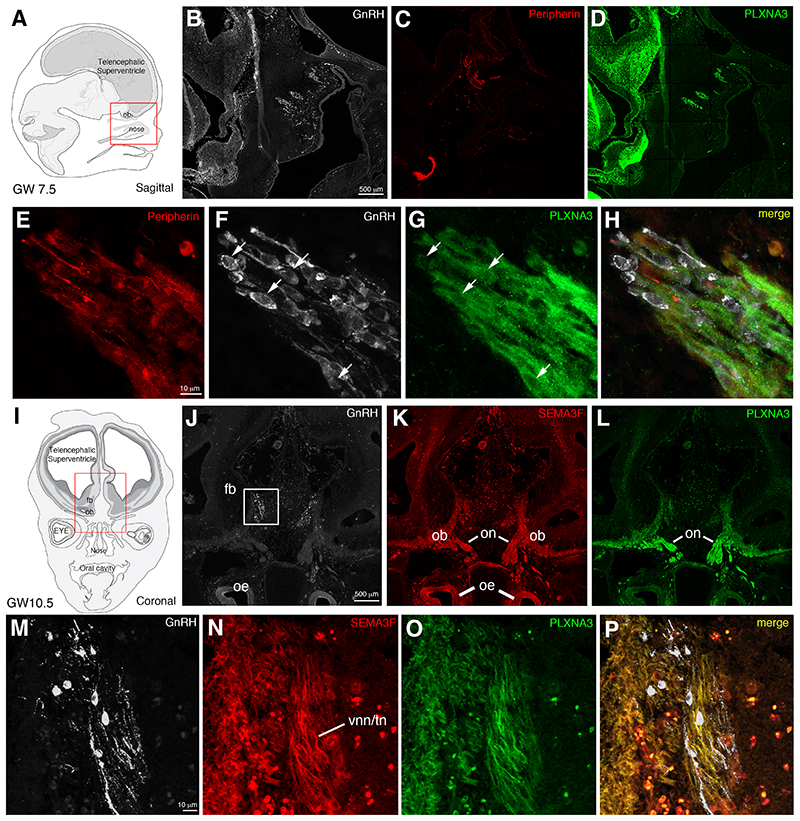
With the exception of NRP2 3 , expression of SEMA3F signaling pathway has never been investigated in the developing human GnRH and olfactory system. We, thus, evaluated the expression pattern of SEMA3F and PLXNA1-A4 receptors in sagittal and coronal sections of human fetal heads (gestational weeks post amenorrhea: GW, GW7.5 and GW10.5), together with the expression of GnRH and peripherin, a marker of the developing olfactory and vomeronasal fibers 3,28 (Figures 3, 4). At GW7.5, the great majority of GnRH neurons migrate in chains across the nasal septum along vomeronasal/terminal peripherin-positive fibers directed toward the forebrain (Figure 3 B, C, E, H), in agreement with our previous immunohistochemical study 3 . PLXNA3 expression was found along the vomeronasal and terminal nerves (Figure 3 D-H), as well as in migratory GnRH neurons (see arrows in Figure 3 F, G). At GW11, about 80% of GnRH neurons have been reported to enter the brain and migrate toward their final target areas 3 . Our immunofluorescence analysis of a GW10.5 fetus indeed revealed robust GnRH-immunoreactivity in migratory cells invading the developing forebrain (Figure 3 I, J, M). Similarly to the mouse olfactory system, where SEMA3F is secreted by early-arriving olfactory axons and deposited at the anterodorsal olfactory bulb (OB) 14 , in GW10.5 human fetuses, SEMA3F is expressed along the olfactory nerve (on), by the developing olfactory epithelium (oe) and by the vomeronasal and terminal nerves (VNN and TN) entering the brain (Figure 3 K, N). SEMA3F expression was absent in human migratory GnRH neurons (Figure 3 M, N). At this fetal stage, like at GW7.5, PLXNA3 is expressed along the vomeronasal and terminal nerves, which also express its ligand (Figure 3 L-P). However, at GW10.5 GnRH neurons are PLXNA3-negative (Figure 3 M, O, P), thus suggesting that PLXNA3 could be downregulated in GnRH neurons that entered the brain compartment.
Figure 3. Semaphorin 3F and Plexin A3 are expressed along the migratory route of GnRH neurons in human fetuses.
Fluorescent IHC was performed to assess SEMA3F and PLXNA3 expression both in the nasal region (A) and at the nasal/forebrain junction (I) during the early fetal development. (B-D) Representative immunostaining for GnRH, Peripherin and Plexin A3 on sagittal sections of a GW7.5 human fetus. (E-H) Magnification of the boxed area in B, revealing that PLXNA3 is found in the peripherin-positive olfactory/vomeronasal scaffold, as well as in GnRH neurons (arrows) during their migration through the nasal compartment. (J-L) Representative pictures of immunolabelings for GnRH, PLXNA3 and SEMA3F on coronal sections of a GW10.5 human fetus. (M-P) Magnification of the boxed area in J. The olfactory and vomeronasal/terminal nerves still show immunoreactivity for PLXNA3 at the nose/forebrain junction. SEMA3F is strongly expressed in this region and detected along the PLXNA3-positive nerves. GnRH neurons migrating inside the forebrain do not longer express PLXNA3. fb, forebrain; ob, olfactory bulbs; oe, olfactory epithelium; on, olfactory nerves; vnn/tn, vomeronasal/terminal nerves. Scale bars: B, J = 500 μm; E, M = 10 μm.
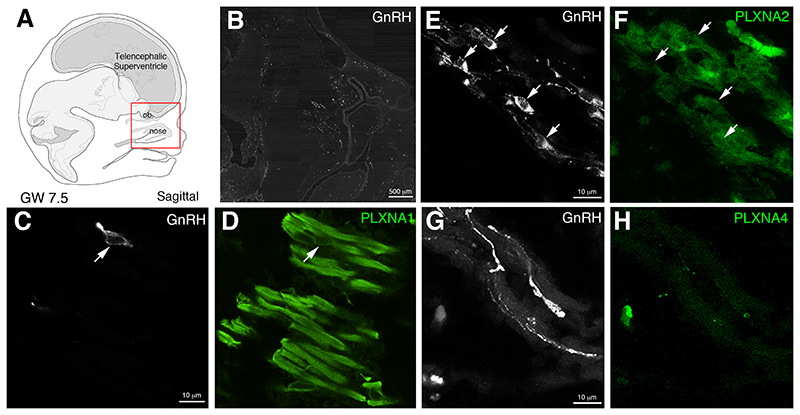
Figure 4. PLXNA1 and PLXNA2 could also contribute to SEMA3F signaling in the human fetal nose.
Since SEMA3F can signal not only through PLXNA3, but through the PLXNA1-A4 family, additional stainings were performed to check for PLXNA1-A4 expression in the nasal compartment of a GW7.5 human fetus. In addition to PLXNA3, migrating GnRH neurons and the olfactory/vomeronasal scaffold also express PLXNA1 (C-D) while PLXNA2 was detected in GnRH neurons and other cells of the migratory mass, but not in the olfactory/vomeronasal projections (E-F). However, PLXNA4 immunoreactivity was only found in scattered cells, distant from the migratory route of GNRH neurons (G-H). ob, olfactory bulbs. Scale bars: B = 500 μm; E,C,G = 10 μm.
In order to assess the expression pattern of the other PLXN receptors through which SEMA3F can possibly signal, we next performed double-immunofluorescence staining of sagittal sections of GW7.5 fetuses using antibodies raised against GnRH and PLXNA1, PLXNA2 and PLXN4. This analysis revealed that GnRH neurons migrating in the nasal compartment express PLXNA1 and PLXNA2 (Figure 4 A-F; arrows) but not PLXNA4 (Figure 4 G, H). Also, PLXNA1 was found to be expressed by the vomeronasal/terminal nerves that form the migratory scaffold for GnRH neurons (Figure 4 D). GnRH neurons migrate together with a heterogeneous coalescence of placode-derived and neural crest-derived migratory cells and olfactory axons, collectively called the ‘migratory mass’ (MM) 29 . At this stage, we observed a mixed mass of GnRH neurons and other cells migrating across the nasal mesenchyme towards the telencephalon expressing PLXNA2 (Figure 4 E, F). Overall, these results show that SEMA3F and PLXNA3 are both expressed along the olfactory nerve (on) and along the intracranial projection of the vomeronasal nerve/terminal nerve (vnn/tn). Our immohistochemical data also indicated that the signal-transducing receptors PLXNA1-A3 are expressed in early migratory GnRH neurons, therefore suggesting that SEMA3F signaling through PLXNA1-A3 could be involved in the guidance of GnRH neurons and of olfactory and vomeronasal nerve fibers in humans.
The central message of this paper is that SEMA3F signaling is necessary for correct human puberty onset and reproduction. To substantiate this claim, we present clinical, molecular genetic, and in vitro functional data from 15 patients who belonged to 11 independent families. These patients all presented with pubertal failure and were clinically diagnosed with IHH or KS. Our study reveals that 6.9% of probands with nIHH/KS harbor variants in SEMA3F and/or PLXNA3. This indicates that genes in the SEMA3F signaling pathway are among the most frequently mutated ones in congenital GnRH deficiency, together with FGFR1 and CHD7 1 .
With regards to any previous implication of SEMA3F in human puberty/reproduction, in a large genome-wide association study, an SNP near SEMA3F was found to be strongly correlated with “age at first birth,” which indicates earlier puberty timing and reproductive success 30 , suggesting the relevance of SEMA3F to reproductive function in humans.
To support further our message, we provide unequivocal human embryonic data showing the expression of SEMA3F along the developing human GnRH migratory pathway. Moreover, our data provide compelling evidence that all the receptors required for the SEMA3F signaling, including PLXNA3, a key component of the SEMA3F holoreceptor complex 31 , are expressed by the human GnRH and olfactory/vomeronasal systems. Based on the expression of SEMA3F signaling pathway in the nasal compartment of early human fetuses, and on the chemotactic role of this semaphorin reported in other species 31 , our data raise the hypotheses that SEMA3F/PLXNA3 could regulate GnRH neuronal migration and olfactory/vomeronasal axonal elongation in humans. Notably, our data indicated that the signal-transducing receptors PLXNA1-A3 are expressed in early migratory GnRH neurons. This is consistent with a recent investigation highlighting cooperation of Plxna1 and Plxna3 in the formation of the GnRH migratory scaffold in mice, based on which the human orthologue of Plxna3 (i.e., PLXNA3), like PLXNA1, was proposed to be a candidate gene for mutation screening in patient with KS 13 .
The existing literature indicates that the SEMA3F signals through Nrp2/Plxna3 holoreceptor complex, while Sema3a signaling occurs mainly via Nrp1/Plxna1 6,31 . Importantly, Nrp2 and its ligand, SEMA3F, are expressed by olfactory sensory neurons in rodents in a complementary manner that is important for establishing olfactory map topography 14 . Despite these evidences, based on typical GnRH migration patterns and expected numbers of GnRH neurons in the Sema3f deficient mice, this guidance molecule was reported to be dispensable for GnRH neuron migration 32 . However, pubertal and adult reproductive phenotypes of this mouse model, such as the timing of puberty onset, first estrus, fertility index, litter size, etc. have not been described. Besides, speciesspecific differences in Sema3s functions between mice and humans may exist. In fact, significant species differences, even with well-established reproductive genes including TAC3/TACR3 and KISS1/KISS1R, have been previously documented. Specifically, patients bearing mutations in TAC3 and TACR3 26 have sexual infantilism and infertility due to GnRH deficiency. In contrast, a recent transgenic Tacr3(-/-) mouse model demonstrated that normal sexual maturation occurs in these mice, albeit some significant reproductive defects are evident in adulthood 33 . Likewise, while humans with KISS1 34 and KISS1R 35 mutations typically suffer from pubertal failure and infertility, a considerable proportion of female Kiss1 and Kiss1r knockout mice exhibit normal estrous cyclicity, and some male Kiss1- and Kiss1r-null mice exhibit spermatogenesis 36 . These species differences underscore the importance of human studies in puberty/reproduction research.
Finally, a recent work provided compelling evidence that SEMA3s-mediated signaling drives the development of hypothalamic melanocortin circuits and that mutations in these pathways cause obesity 37 . Interestingly, the hypothalamic pro-opiomelanocortin (POMC)-expressing neurons have been repeatedly observed to send projections to and be in close contact with kisspeptin/neurokinin B/dynorphin (KNDy) cells 38,39 . A plethora of investigations have recently identified the role of the hypothalamic KNDy neuronal population as the GnRH pulse generator 38–40 . Importantly, in the study of van der Klaauw and colleagues, one of the patients harboring a PLXNA3 variant (D1710N) had hypogonadotropic hypogonadism in addition to obesity 37 . However, since the majority of the probands in that study were pre-pubertal age children, the true prevalence of hypogonadotropic hypogonadism among variant carriers may be underestimated 37 . Although we noted in this study that a significant proportion of the patients (40%) was obese, SEMA3F/PLXNA3 variants were not statistically more enriched in the subset of obese IHH patients than in our general IHH cohort, where 56 patients (26.0%) featured overweight or obesity (p = 0.24). It would be interesting in the future to carefully look at larger and multiple IHH cohorts and determine whether statistical significance in the frequency of obesity or metabolic disorders could emerge when analyzing larger populations and probands carrying variants in SEMA3F signaling pathway.
In summary, we provide clinical, genetic, molecular/cellular, and developmental evidence to implicate variants in SEMA3F signaling in the etiology of IHH. We propose that this phenotype could be exerted via a direct deleterious effect of these mutants on the GnRH neuronal migration and potentially a harmful impact of SEMA3F/PLXNA3 mutants on the development of hypothalamic melanocortin circuits involved in the regulation of energy homeostasis, which are also known to influence GnRH secretion and reproduction. Each of these events or a combination could hence negatively affect the development and function of the HPG axis. Further studies aimed at addressing the contribution of SEMA3F and PLXNA3 in each one of the above-mentioned developmental processes may uncover new mechanisms underlying human disorders characterized by central hypothalamic dysfunction and infertility.
Supplementary Material
Acknowledgements
We thank the midwives of the Gynecology Department, Jeanne de Flandre Hospital of Lille (Centre d’Orthogénie), France, for their kind assistance and support; M Tardivel and A Bongiovanni (BICeL core microscopy facility of the Lille University School of Medicine) for expert technical assistance. The authors acknowledge support of the Inserm Cross-Cutting Scientific Program (HuDeCA). This work was supported by the Institut National de la Santé et de la Recherche Médicale (INSERM), France [grant number U1172], by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (ERC-2016-CoG to P.G. grant agreement n° 725149/REPRODAMH), Agence Nationale de la Recherche (ANR), France (grant number ANR-18-CE14-0017-02 to P.G.). This study was supported by a start-up grant (DN00305) by UMMC to AKT. This work was supported by the Cukurova University scientific research project number 11364.
Footnotes
Conflict of interests statement: The authors declare no conflict of interests.
Declaration of interests The authors declare no competing interests.
Accessions: The following variants with associated clinical information were submitted to Clin Var: SEMA3F p.Pro452Thr, p.Arg699Trp, p.Thr29Met, p.Pro722Leu, p.Ala652Ser, p.Thr724Met. PLXNA3 p.Ser646Pro, p.Arg108Cys, p.Leu1086Val, p.Arg1359Cys.
Web Resources: HGNC, https://www.genenames.org/
GenBank, https://www.ncbi.nlm.nih.gov/genbank/
TOPMed, https://bravo.sph.umich.edu/freeze5/hg38/
gnomAD Browser, https://gnomad.broadinstitute.org/
Greater Middle East Variom Project (GME), http://igm.ucsd.edu/gme/data-browser.php
InterVar http://wintervar.wglab.org/
ClinVar https://www.ncbi.nlm.nih.gov/clinvar/
Combined Annotation Dependent Depletion (CADD), https://cadd.gs.washington.edu
MutationTaster, http://www.mutationtaster.org
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
References
- 1.Topaloglu AK. Update on the Genetics of Idiopathic Hypogonadotropic Hypogonadism. J Clin Res Pediatr Endocrinol. 2017;9:113–122. doi: 10.4274/jcrpe.2017.S010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wray S, Grant P, Gainer H. Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc Natl Acad Sci U S A. 1989;86:8132–6. doi: 10.1073/pnas.86.20.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casoni F, et al. Development of the neurons controlling fertility in humans: new insights from 3D imaging and transparent fetal brains. Development. 2016;143:3969–3981. doi: 10.1242/dev.139444. [DOI] [PubMed] [Google Scholar]
- 4.Schwanzel-Fukuda M, et al. Migration of luteinizing hormone-releasing hormone (LHRH) neurons in early human embryos. J Comp Neurol. 1996;366:547–57. doi: 10.1002/(SICI)1096-9861(19960311)366:3<547::AID-CNE12>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 5.Pasterkamp RJ. Getting neural circuits into shape with semaphorins. Nat Rev Neurosci. 2012;13:605–18. doi: 10.1038/nrn3302. [DOI] [PubMed] [Google Scholar]
- 6.Giacobini P. Shaping the Reproductive System: Role of Semaphorins in GnRH Development and Function. Neuroendocrinology. 2015 doi: 10.1159/000431021. [DOI] [PubMed] [Google Scholar]
- 7.Oleari R, et al. A novel SEMA3G mutation in two siblings affected by syndromic GnRH deficiency. Neuroendocrinology. 2020 doi: 10.1159/000508375. [DOI] [PubMed] [Google Scholar]
- 8.Cariboni A, et al. Dysfunctional SEMA3E signaling underlies gonadotropin-releasing hormone neuron deficiency in Kallmann syndrome. J Clin Invest. 2015;125:2413–28. doi: 10.1172/JCI78448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanchate NK, et al. SEMA3A, a gene involved in axonal pathfinding, is mutated in patients with Kallmann syndrome. PLoS Genet. 2012;8:e1002896. doi: 10.1371/journal.pgen.1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janssen BJ, et al. Neuropilins lock secreted semaphorins onto plexins in a ternary signaling complex. Nat Struct Mol Biol. 2012;19:1293–9. doi: 10.1038/nsmb.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cariboni A, et al. Neuropilins and their ligands are important in the migration of gonadotropin-releasing hormone neurons. J Neurosci. 2007;27:2387–95. doi: 10.1523/JNEUROSCI.5075-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcos S, et al. Defective signaling through plexin-A1 compromises the development of the peripheral olfactory system and neuroendocrine reproductive axis in mice. Hum Mol Genet. 2017;26:2006–2017. doi: 10.1093/hmg/ddx080. [DOI] [PubMed] [Google Scholar]
- 13.Oleari R, et al. PLXNA1 and PLXNA3 cooperate to pattern the nasal axons that guide gonadotropin-releasing hormone neurons. Development. 2019;146 doi: 10.1242/dev.176461. [DOI] [PubMed] [Google Scholar]
- 14.Takeuchi H, et al. Sequential arrival and graded secretion of Sema3F by olfactory neuron axons specify map topography at the bulb. Cell. 2010;141:1056–67. doi: 10.1016/j.cell.2010.04.041. [DOI] [PubMed] [Google Scholar]
- 15.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sykiotis GP, et al. Oligogenic basis of isolated gonadotropin-releasing hormone deficiency. Proc Natl Acad Sci U S A. 2010;107:15140–4. doi: 10.1073/pnas.1009622107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sidhoum VF, et al. Reversal and relapse of hypogonadotropic hypogonadism: resilience and fragility of the reproductive neuroendocrine system. J Clin Endocrinol Metab. 2014;99:861–70. doi: 10.1210/jc.2013-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouilly J, et al. DCC/NTN1 complex mutations in patients with congenital hypogonadotropic hypogonadism impair GnRH neuron development. Hum Mol Genet. 2018;27:359–372. doi: 10.1093/hmg/ddx408. [DOI] [PubMed] [Google Scholar]
- 19.Messina A, et al. Neuron-Derived Neurotrophic Factor Is Mutated in Congenital Hypogonadotropic Hypogonadism. Am J Hum Genet. 2020;106:58–70. doi: 10.1016/j.ajhg.2019.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strande NT, et al. Evaluating the Clinical Validity of Gene-Disease Associations: An Evidence-Based Framework Developed by the Clinical Genome Resource. Am J Hum Genet. 2017;100:895–906. doi: 10.1016/j.ajhg.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo MH, Plummer L, Chan YM, Hirschhorn JN, Lippincott MF. Burden Testing of Rare Variants Identified through Exome Sequencing via Publicly Available Control Data. Am J Hum Genet. 2018;103:522–534. doi: 10.1016/j.ajhg.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alkan C, et al. Whole genome sequencing of Turkish genomes reveals functional private alleles and impact of genetic interactions with Europe, Asia and Africa. BMC Genomics. 2014;15:963. doi: 10.1186/1471-2164-15-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotan LD, et al. Prevalence and associated phenotypes of PLXNA1 variants in normosmic and anosmic idiopathic hypogonadotropic hypogonadism. Clin Genet. 2019;95:320–324. doi: 10.1111/cge.13482. [DOI] [PubMed] [Google Scholar]
- 24.Pitteloud N, et al. Predictors of outcome of long-term GnRH therapy in men with idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2002;87:4128–36. doi: 10.1210/jc.2002-020518. [DOI] [PubMed] [Google Scholar]
- 25.Young J, et al. SEMA3A deletion in a family with Kallmann syndrome validates the role of semaphorin 3A in human puberty and olfactory system development. Hum Reprod. 2012;27:1460–5. doi: 10.1093/humrep/des022. [DOI] [PubMed] [Google Scholar]
- 26.Topaloglu AK, et al. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat Genet. 2009;41:354–358. doi: 10.1038/ng.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolk SM, et al. Semaphorin 3F is a bifunctional guidance cue for dopaminergic axons and controls their fasciculation, channeling, rostral growth, and intracortical targeting. J Neurosci. 2009;29:12542–57. doi: 10.1523/JNEUROSCI.2521-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fueshko S, Wray S. LHRH cells migrate on peripherin fibers in embryonic olfactory explant cultures: an in vitro model for neurophilic neuronal migration. Dev Biol. 1994;166:331–48. doi: 10.1006/dbio.1994.1319. [DOI] [PubMed] [Google Scholar]
- 29.Valverde F, Santacana M, Heredia M. Formation of an olfactory glomerulus: morphological aspects of development and organization. Neuroscience. 1992;49:255–75. doi: 10.1016/0306-4522(92)90094-i. [DOI] [PubMed] [Google Scholar]
- 30.Day FR, et al. Physical and neurobehavioral determinants of reproductive onset and success. Nat Genet. 2016;48:617–623. doi: 10.1038/ng.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma A, Verhaagen J, Harvey AR. Receptor complexes for each of the Class 3 Semaphorins. Front Cell Neurosci. 2012;6:28. doi: 10.3389/fncel.2012.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cariboni A, et al. Defective gonadotropin-releasing hormone neuron migration in mice lacking SEMA3A signalling through NRP1 and NRP2: implications for the aetiology of hypogonadotropic hypogonadism. Hum Mol Genet. 2011;20:336–44. doi: 10.1093/hmg/ddq468. [DOI] [PubMed] [Google Scholar]
- 33.Yang JJ, Caligioni CS, Chan YM, Seminara SB. Uncovering novel reproductive defects in neurokinin B receptor null mice: closing the gap between mice and men. Endocrinology. 2012;153:1498–508. doi: 10.1210/en.2011-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Topaloglu AK, et al. Inactivating KISS1 mutation and hypogonadotropic hypogonadism. N Engl J Med. 2012;366:629–35. doi: 10.1056/NEJMoa1111184. [DOI] [PubMed] [Google Scholar]
- 35.Seminara SB, et al. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–27. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 36.Lapatto R, et al. Kiss1-/-mice exhibit more variable hypogonadism than Gpr54-/-mice. Endocrinology. 2007;148:4927–36. doi: 10.1210/en.2007-0078. [DOI] [PubMed] [Google Scholar]
- 37.van der Klaauw AA, et al. Human Semaphorin 3 Variants Link Melanocortin Circuit Development and Energy Balance. Cell. 2019;176:729–742.:e18. doi: 10.1016/j.cell.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manfredi-Lozano M, Roa J, Tena-Sempere M. Connecting metabolism and gonadal function: Novel central neuropeptide pathways involved in the metabolic control of puberty and fertility. Front Neuroendocrinol. 2018;48:37–49. doi: 10.1016/j.yfrne.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 39.Moore AM, Coolen LM, Lehman MN. Kisspeptin/Neurokinin B/Dynorphin (KNDy) cells as integrators of diverse internal and external cues: evidence from viral-based monosynaptic tracttracing in mice. Sci Rep. 2019;9:14768. doi: 10.1038/s41598-019-51201-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeo SH, Kyle V, Blouet C, Jones S, Colledge WH. Mapping neuronal inputs to Kiss1 neurons in the arcuate nucleus of the mouse. PLoS One. 2019;14:e0213927. doi: 10.1371/journal.pone.0213927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.