Abstract
OBJECTIVE
Maturity-onset diabetes of the young (MODY) is a rare monogenic form of diabetes. In 2009, >80% of U.K. cases were estimated to be misdiagnosed. Since then, there have been a number of initiatives to improve the awareness and detection of MODY, including education initiatives (Genetic Diabetes Nurse [GDN] project), the MODY probability calculator, and targeted next-generation sequencing (tNGS). We examined how the estimated prevalence of MODY and other forms of monogenic diabetes diagnosed outside the neonatal period has changed over time and how the initiatives have impacted case finding.
RESEARCH DESIGN AND METHODS
U.K. referrals for genetic testing for monogenic diabetes diagnosed >1 year of age from 1 January 1996 to 31 December 2019 were examined. Positive test rates were compared for referrals reporting GDN involvement/MODY calculator use with those that did not.
RESULTS
A diagnosis of monogenic diabetes was confirmed in 3,860 individuals, more than threefold higher than 2009 (1 January 1996 to 28 February 2009, n = 1,177). Median age at diagnosis in probands was 21 years. GDN involvement was reported in 21% of referrals; these referrals had a higher positive test rate than those without GDN involvement (32% vs. 23%, P < 0.001). MODY calculator usage was indicated in 74% of eligible referrals since 2014; these referrals had a higher positive test rate than those not using the calculator (33% vs. 25%, P = 0.001). Four hundred ten (10.6%) cases were identified through tNGS. Monogenic diabetes prevalence was estimated to be 248 cases/million (double that estimated in 2009 because of increased case finding).
CONCLUSIONS
Since 2009, referral rates and case diagnosis have increased threefold. This is likely to be the consequence of tNGS, GDN education, and use of the MODY calculator.
Introduction
Maturity-onset diabetes of the young (MODY) is a rare, young-onset, monogenic form of diabetes. Identifying MODY is crucial for the patient, as a correct diagnosis can inform optimal treatment, long-term complication risk, risk to other family members, and other aspects of clinical care, such as pregnancy management (1).
Based on population screening studies, MODY has been estimated to account for 1–4% of pediatric and young adult diabetes cases (2–7), varying depending on the genes tested, how pathogenicity of variants is determined, age-group of the cohort, and screening criteria chosen. In practice, however, referral of patients for diagnostic genetic testing for MODY is often less systematic and based on clinician opinion, so many of these cases are missed. In the U.K., 2009 data (published in 2010) estimated that >80% of MODY cases were misdiagnosed, with significant regional variation in referral rates (8).
Since 2009, more resources have been put into the recognition and awareness of MODY. Education initiatives, such as the Genetic Diabetes Nurse (GDN) project in the U.K., have been set up to raise awareness and support local clinicians and patients and their families with testing and changes to treatment and management following a genetic diagnosis (9). The MODY calculator (https://www.diabetesgenes.org/exeter-diabetes-app) has been developed as a free-to-use clinical tool, accessible worldwide, that provides the probability of a patient having MODY, on the basis of their clinical features, to help clinicians make decisions on which patients to refer for diagnostic genetic testing (10). In addition, targeted next-generation sequencing (tNGS) allows all potential MODY genes and additional monogenic diabetes genes to be sequenced in parallel, meaning a greater chance of identifying mutations, particularly in rarer genes, compared with traditional Sanger sequencing, which is limited to testing the common MODY genes in series unless a specific phenotype is recognized (11).
To date, studies have examined the prevalence of MODY but have not considered how the estimated prevalence may have changed over time as efforts to raise awareness and detection of MODY have improved. We have assessed the change in estimated prevalence of MODY, as well as other monogenic causes for diabetes diagnosed outside the neonatal period, over time at a national level in the U.K. We examined all referrals to the two laboratories responsible for all diagnostic genetic testing for monogenic diabetes in the U.K. and the potential impact of three initiatives (the GDN project, the MODY calculator, and tNGS) on improving the identification of patients with MODY in routine clinical practice.
Research Design and Methods
We examined data on referrals and cases from the two laboratories in the U.K. responsible for all national diagnostic genetic testing for monogenic diabetes. The MODY diagnostic service within the Exeter Genomics Laboratory at the Royal Devon and Exeter National Health Service (NHS) Foundation Trust provides monogenic diabetes testing for England, Wales, and Northern Ireland. Referral details of all patients who undergo monogenic diabetes testing at the Exeter Genomics Laboratory are recorded within an in-house database. Scotland has offered a separate service for Scottish residents since 2016 through the East of Scotland Regional Genetics Service and provided data on referrals and cases for this study.
We examined all U.K. patients with diabetes diagnosed ≥1 year of age who were referred to these services for monogenic diabetes testing from 1 January 1996 to 31 December 2019, 10 years and 10 months after the previous study that reported patients detected on 28 February 2009. In the previous study, >99% of the patients reported had mutations in the four most common MODY genes (GCK, HNF1A, HNF4A, and HNF1B). For this previous study, they were tested by Sanger sequencing and multiplex ligation-dependent probe amplification (for gene deletions), so testing typically would be initially based on phenotype and tested serially rather than in parallel. In 2013, tNGS was introduced to the Exeter Genomics Laboratory as a routine testing option where multiple known monogenic diabetes genes are tested in parallel. Here, in line with updated testing criteria, we tested cases for variants (including gene deletions) in 28 known monogenic diabetes genes (ABCC8, CEL, CISD2, GATA4, GATA6, GCK, HNF1A, HNF1B, HNF4A, INS, INSR, KCNJ11, LMNA, MAFA, NEUROD1, PAX6, PCBD1, PDX1, PLIN1, POLD1, PPARG, RFX6, SLC19A2, SLC29A3, TRMT10A, WFS1, ZBTB20, and ZFP57) and the mitochondrial DNA variant m.3243A>G by tNGS as described previously (11). The classification of variant pathogenicity in both laboratories was assessed in line with the 2015 American College of Genetics and Genomics guidelines (12) or prior to these being introduced in 2017, according to an in-house framework. Cases with variants of uncertain clinical significance are not included. Only referrals for genetic testing where the patient presented with diabetes were included, so HNF1B-associated renal disease and HNF4A hyperinsulinism were excluded from the data cohort.
Case and referral rates were calculated by region. Regional and national population data from mid-2019 was obtained from the U.K. Office for National Statistics (13). Because of local research interest, the Exeter region (EX postal code area, population size 547,511 by U.K. Census 2011 [14]) has the highest referral rate and so was used to calculate a minimum prevalence of monogenic diabetes and estimate the number of missing cases in the U.K.
GDNs
The GDN project was set up in 2002 and has provided training for 62 diabetes nurses to gain specialist knowledge in genetic forms of diabetes. GDN involvement in a referral is recorded on the Exeter Genomic Laboratory’s patient diagnostic referral form and recorded in the referrals database. This information is not available for the Scottish laboratory. Positive test rates of GDN and non-GDN–associated referrals were compared (using χ2 test), as well as number of family members followed. Regional GDN activity was calculated based on GDN person-time in a post (e.g., two GDNs in a post for 12 months would be classified as 24 months of GDN person-time). Regional GDN activity was correlated with cases identified per million population, as summarized using Pearson correlation coefficients.
MODY Calculator
The MODY calculator was launched in 2012. Since August 2014, referring clinicians to the Exeter Genomics Laboratory have been reporting on the diagnostic referral form whether they have used the MODY calculator. This information is not available for referrals to the Scottish laboratory. The number of eligible referrals for White patients diagnosed between ages 1 and 35 years (as the calculator has not been validated for use in other races/ethnicities and older ages) that reported using the MODY calculator were recorded, and positive test rates were compared between those that reported using the calculator and those that did not using χ2 test.
Ethics Committee Approval
This project analyzed anonymized data based on referrals to the MODY diagnostic services in Exeter and Scotland. Only aggregate data were shared between sites. On the Integrated Research Application System application form, this work was classified as research limited to secondary use of information previously collected in the course of normal care (without an intention to use it for research at the time of collection), which generally is excluded from research ethics committee review provided that the patients or service users are not identifiable to the research team carrying out the research. The U.K. Health Research Authority’s online tool for assessment of need for ethical approval was completed and confirmed that research ethics committee review was not needed.
Results
Overview
MODY Cases and Referrals Across the U.K.
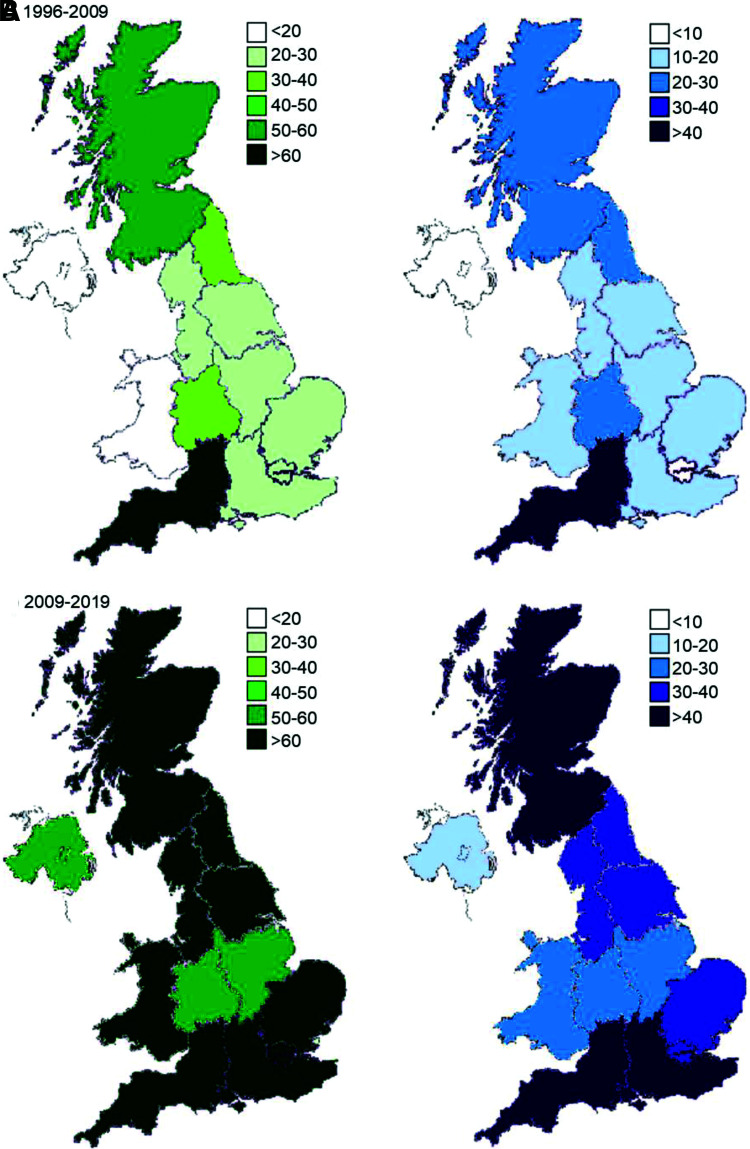
A total of 3,860 cases of monogenic diabetes outside the neonatal period were genetically confirmed in the U.K. in 2019 (Table 1) compared with 1,177 in 2009, a 3.3-fold increase in 10.83 years. The number of cases and referrals has increased across all regions since the 2010 article (8) (Fig. 1). There was still considerable regional variation in the number of cases identified across the U.K. (see Supplementary Fig. 1 for maps with revised scales), ranging from 24.3 per million of the population in Northern Ireland to 113.3 in South West England (Table 1). The number of cases identified was highly correlated to the proband referral rate of a region (r = 0.9). Scotland and South West England had the highest rates of proband referrals in the U.K. (both ∼250 cases per million population) (Table 1). Northern Ireland maintained the lowest rate of referral at 67.1 referrals per million of the population. The majority of cases were diagnosed in adults: 63% (1,924 of 3,035) reported in individuals >18 years of age (median 21 years [interquartile range 15, 31]).
Table 1.
Regional variation in referrals for genetic testing for MODY and cases of diabetes with a confirmed diagnosis of MODY from 1 January 1996 to 31 December 2019
| Probands | ||||||||
|---|---|---|---|---|---|---|---|---|
| Country/region | Population | Referrals, n | Referrals/ million | With mutation, n | Positive test rate, %† | Relatives with diabetes with mutation, n | Total cases,* n | Total cases/ million |
| Scotland | 5,463,300 | 1,369 | 250.6 | 265 | 19.4 | 178 | 443 | 81.1 |
| Wales | 3,152,879 | 255 | 80.9 | 69 | 27.1 | 60 | 129 | 40.9 |
| Northern Ireland | 1,893,667 | 127 | 67.1 | 23 | 18.1 | 23 | 46 | 24.3 |
| England | 56,286,961 | 6,781 | 120.5 | 1,724 | 25.4 | 1,479 | 3,203 | 56.9 |
| English regions | ||||||||
| East | 6,236,072 | 571 | 91.6 | 154 | 27.0 | 145 | 299 | 47.9 |
| South East | 9,180,135 | 1,163 | 126.7 | 346 | 29.8 | 280 | 626 | 68.2 |
| South West | 5,624,696 | 1,402 | 249.3 | 312 | 22.3 | 325 | 637 | 113.3 |
| London | 8,961,989 | 1,019 | 113.7 | 246 | 24.1 | 145 | 391 | 43.6 |
| West Midlands | 5,934,037 | 461 | 77.7 | 120 | 26.0 | 122 | 242 | 40.8 |
| East Midlands | 4,835,928 | 393 | 81.3 | 115 | 29.3 | 99 | 214 | 44.3 |
| Yorkshire/Humber | 5,502,967 | 543 | 98.7 | 153 | 28.2 | 121 | 274 | 49.8 |
| North East | 2,669,941 | 448 | 167.8 | 92 | 20.5 | 59 | 151 | 56.6 |
| North West | 7,341,196 | 781 | 106.4 | 186 | 23.8 | 183 | 369 | 50.3 |
| Unknown | 5 | 2 | 37 | 39 | ||||
| U.K. total | 66,796,807 | 8,537 | 127.8 | 2,083 | 24.4 | 1,777 | 3,860 | 57.8 |
Proband referral rates and confirmed cases were calculated per million of regional population.
Probands and relatives.
Minimum percent positive test rate based on number of probands with mutations identified per all proband referrals for region specified.
Figure 1.
Maps of the U.K. showing regional variations for referrals for MODY testing (per million population) (in green) and cases diagnosed with monogenic diabetes outside the neonatal period (per million population) (in blue) for 1996 to 28 February 2009 (A) and 1 March 2009 to 31 December 2019 (B). Same scale as in original 2010 study (8) used for both.
Yearly Referrals and Cases
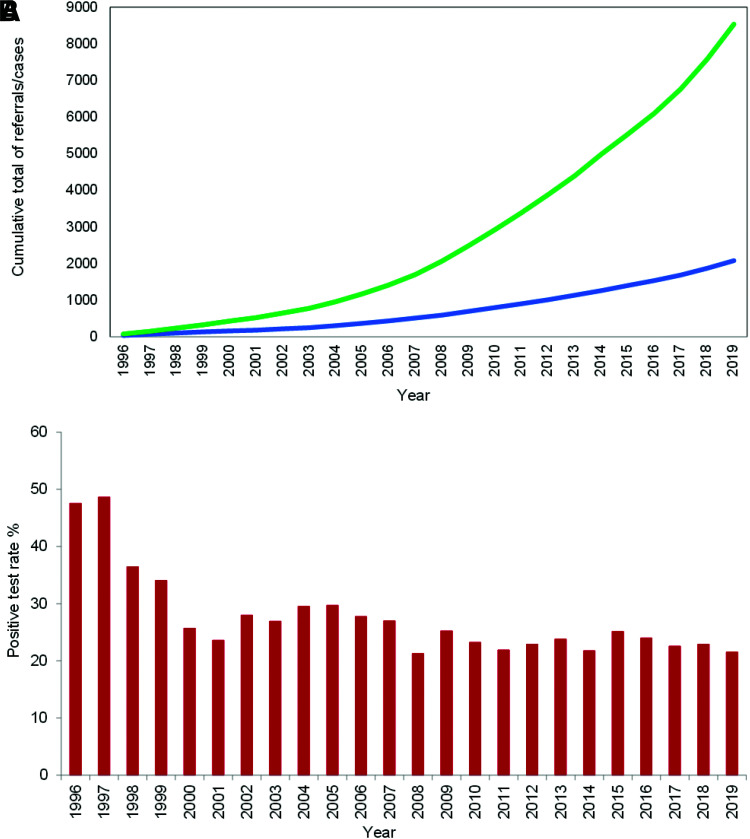
Referrals and confirmed cases of monogenic diabetes have been increasing yearly (Fig. 2A), whereas the positive test rate for probands has been stable at ∼23% (Fig. 2B). Since the 2010 report (8), there has been a fourfold increase in the number of probands who have been referred for testing (n = 8,537 vs. 2,072), and the number of confirmed cases in the U.K. has more than tripled (3,860 vs. 1,177, including family members; 2,083 vs. 564, for probands alone).
Figure 2.
A: Cumulative frequency of proband referrals (green) and proband cases (blue) of monogenic diabetes from 1996 to 2019 at the Exeter Genomics Laboratory. B: Yearly proband positive test rate of monogenic diabetes. Positive test rate was calculated as the proportion of positive cases of all referrals in that year.
Family Member Referrals
Most probands (54%) had at least one family member referred (median three per family). Of the 3,190 U.K. family members, 1,821 were recorded as having diabetes on the request form and 86% (1,562 of 1,821) of these patients were diagnosed with monogenic diabetes.
GDN Network
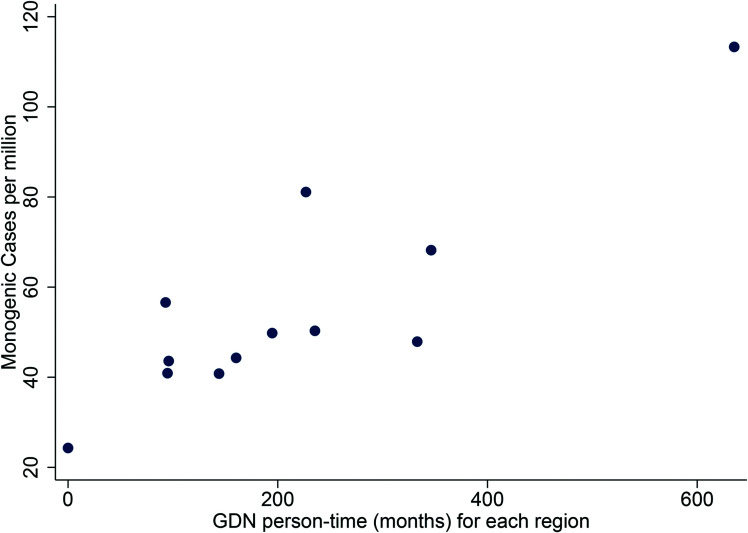
GDN involvement was recorded for 21% (1,821 of 7,981) of proband referrals to the Exeter Genomics Laboratory, ranging from 8% to 38% across the various regions (Supplementary Table 1 and Supplementary Fig. 2). Referrals associated with a GDN had a higher positive test rate than those without (32% vs. 23%, P < 0.001). Proband cases referred by a GDN (588 of 2,011, 29%) were more likely to have additional family members referred for testing compared with those with no GDN involvement (58% vs. 51%, P = 0.007). Regions with higher GDN activity (as measured by person-time in a post) were associated with a higher number of confirmed cases of monogenic diabetes (r = 0.86, P < 0.001) (Fig. 3).
Figure 3.
Association between GDN involvement and confirmed cases across the U.K. Each point represents a region of the U.K., with GDN person-time in months on the x-axis against cases diagnosed with confirmed MODY per million population on the y-axis. Association determined using Pearson correlation coefficients (r = 0.86, P < 0.001).
MODY Calculator
There were 1,657 referrals to the Exeter Genomics Laboratory from August 2014 (the date when recording on the diagnostic referral forms began) to 31 December 2019 that were eligible for the MODY calculator (diagnosed with diabetes between the ages of 1 and 35 years and White race). Of these diagnostic request forms, 1,224 (74%) reported using the MODY calculator, ranging from 56% to 87% across the various regions (Supplementary Table 1 and Supplementary Fig. 2). Referrals reporting a MODY probability score had a higher positive test rate than those without (33% vs. 25%, P = 0.001).
To What Extent Has the Testing of Additional Genes Improved the Number of Cases Found?
Supplementary Fig. 3 shows the distribution of genetic causes found for the 3,860 confirmed monogenic diabetes cases in the U.K. The four most common genes (GCK, HNF1A, HNF4A, and HNF1B) accounted for 89.4% of cases. In the original study, 1,177 cases of MODY were identified, with >99% having mutations in GCK, HNF1A, HNF4A, and HNF1B. The use of tNGS greatly increased the testing of all genes but particularly for the rarer causes. A total of 410 (10.6%) of the 3,860 cases had causes other than the four most common MODY genes detected through tNGS. The full breakdown of genetic causes identified is given in Supplementary Table 2. Twenty-two rarer causes were found, of which the most common were the mitochondrial DNA variant m.3243A>G (4.5%) and mutations in the ABCC8 gene (1.8%). Forty patients had biallelic mutations causing a recessively inherited subtype of monogenic diabetes. Of these, four were homozygous for a mutation as a result of known consanguinity.
Prevalence
In 2009, we estimated that there were 108 cases of monogenic diabetes per million population, which would lead to an estimated 7,214 cases in the U.K. population. Now with 3,860 cases identified, only 46% would be missing. However, the finding of new cases suggests that this past prevalence was an underestimate. An updated prevalence of monogenic diabetes in the U.K. was calculated based on data from the Exeter area where there is the most testing and the most awareness (as defined by the EX postal code region of the U.K., population size 547,511). One hundred thirty-six cases of monogenic diabetes have been identified in the EX postal code region (105 having mutations in genes included in the original study), leading to a prevalence of 248 cases per million population. Based on this prevalence and extrapolation to the whole U.K. population, this would suggest that there is a minimum of 16,566 cases of monogenic diabetes in the U.K. Of these estimated total U.K. cases, 3,860 (23%) have been identified through genetic testing, suggesting that 77% remain undiagnosed.
Conclusions
Over the past 10 years, the referral rates and detection of monogenic diabetes cases outside the neonatal period has improved across the U.K., with more than a threefold increase in the number of cases detected since 2009. The referral rate has consistently increased, and we have shown that measures such as the GDNs and MODY calculator have led to better positive test rates.
There have been a number of population-based research studies that aimed to determine the prevalence of MODY, but these have been largely carried out in pediatric cohorts (2–6). In contrast, our study examined monogenic diabetes cases identified through all routine diagnostic referrals at a national level and, importantly, that had no restriction on age. More than one-half of the cases in our cohort were diagnosed in adults. Therefore, our study gives a clear indication of not only the minimum prevalence but also the extent to which patients with monogenic diabetes may be misdiagnosed in routine clinical practice across all ages.
Our minimum prevalence estimate is higher than previously thought, and this reflects both the continuing increase in referral rates and the introduction of tNGS. The more we have looked for cases, the more we have found, and the more we have realized are still missing. Our data suggest that despite the improved detection of MODY across the U.K., more than three-quarters of U.K. cases are still misdiagnosed (as type 1 or 2 diabetes), and considerable regional variation in referral rates remains. There are many possible reasons for this variability in use of genetic testing for monogenic diabetes, but historically, this could be due to the funding model for testing (£650 per test, recharged to the requesting organization for referrals to the Exeter Genomics Laboratory), meaning that certain organizations may be more willing than others to pay for this service. However, in Scotland, funding has always been central, and a change to central funding for genomic testing was introduced in England during 2020/2021, which is predicted to increase future referral rates. A further reason for regional variability in referral rates could be differences in awareness of MODY/monogenic diabetes across the country. The GDN project has improved awareness, with a correlation between regional GDN time in a post and cases in that region, but the reach of GDNs is still limited in certain areas.
Both GDN involvement and use of the MODY calculator were found to be frequently indicated on diagnostic referral forms. In both cases, the use of these initiatives was associated with a better positive test rate, suggesting that they are helpful in targeting testing at more appropriate patients and ensuring that resources are not unnecessarily used for those highly unlikely to have a monogenic cause for diabetes. The GDN project has shown how increased awareness, education, and support for identifying patients with monogenic diabetes can have real benefits and is a model that could be adopted in other countries around the world. The MODY calculator can be easily accessed online and on smartphones for free worldwide. Both can be helpful to clinicians who may not have specialist training in monogenic diabetes and are faced with what can be a challenging decision on whether to make a referral for diagnostic molecular genetic testing.
In addition to the initiatives aimed at clinicians to improve referral rates for MODY and monogenic diabetes, since 2013, tNGS has helped to ensure that more cases are found at the diagnostic testing stage. More than 10% of cases had rarer monogenic causes identified through tNGS, which is in contrast to the <1% identified in our previous report (8) where we performed testing using Sanger sequencing of specific genes.
Clinical Implications
The fact that 77% of monogenic cases outside the neonatal period are still estimated to be misdiagnosed indicates that further work is crucially needed to identify these missing cases. The initiatives that we described to improve awareness and diagnosis are easy to implement, but to encourage wide use, these approaches need to be introduced into guidelines and taken up at a national level to have real impact. Use of the MODY calculator is now proposed in the NHS’s National Genomic Test Directory’s Testing Criteria for Rare and Inherited Disease (15), and such criteria could be introduced in other diagnostic laboratories worldwide. We are now working with NHS England and the regional Genomic Medicine Service Alliances on plans to train and identify a monogenic diabetes lead consultant and diabetes specialist nurse in every NHS Trust through a targeted approach and virtual training. In addition to the initiatives we have described, screening approaches using C-peptide and islet autoantibodies are helpful in insulin-treated patients (6,7). In line with this, national C-peptide screening in Scotland has recently been introduced, which will help not only with the diagnosis of MODY but also with better classification of diabetes more broadly, and the impact of this at a population level will be of considerable interest.
A correct genetic diagnosis is crucial and can significantly improve patients’ quality of life. The specific genetic cause can inform the most appropriate treatment: Insulin injections are essential for patients with type 1 diabetes (the most common young-onset form of diabetes), whereas patients with the most common forms of monogenic diabetes can be treated with an oral-based sulfonylurea (as for HNF1A/HNF4A MODY) (16–18) or require no pharmacological treatment (as for GCK MODY) (19). Testing is also important because a positive result has implications for family members or future offspring who may also require treatment and can inform future complication risk (20,21) and pregnancy management (22,23).
Limitations
This study focused on data collected from an internal database that has been transcribed from referral forms. Clinical information can be missing on referral forms, meaning that we may have missed some patients, but this is likely to be minimal because the key criteria of diabetes status and country of origin are nearly always reported, particularly for probands. Our assessment of the likely impact of GDN involvement or use of the MODY calculator is limited by the data available. Both are indicated by a box on the diagnostic request form, and this can also be missed. Furthermore, we have no way of knowing of any potential patients who may not have been referred following either of these initiatives. Referrals were not tested for all genes, and not all were tested using tNGS, typically in cases where a specific gene is suspected (24). This means that we cannot rule out that cases without a positive test result in the database do not have an as-yet undiagnosed genetic cause.
In conclusion, since 2009, referral rates and the number of cases diagnosed with monogenic diabetes outside the neonatal period have increased more than threefold. Improvements in referrals and diagnosis of cases are likely to be due to the introduction of better education and awareness through initiatives such as GDNs, the MODY calculator, and tNGS of more genes. Despite this, 77% of cases are still estimated to be undiagnosed or misdiagnosed in the U.K., and wide variation in referral rates exists across the country. Thus, further work in disseminating knowledge is needed to ensure that more patients obtain the right diagnosis and the optimal care for their diabetes.
Article Information
Funding. M.H.S. is a National Institute for Health Research (NIHR) senior nurse and midwife research leader. M.H.S., A.TH., and B.M.S. are core members of the NIHR Exeter Clinical Research Facility, which is a partnership between the University of Exeter Medical School College of Medicine and Health and Royal Devon and Exeter NHS Foundation Trust. The GDNs were supported with funding from Health Education England and from the Scottish Government.
The views expressed in this article are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care.
Duality of Interest. No conflicts of interest relevant to this article were reported.
Author Contributions. L.P. collated data, analyzed data, and drafted the manuscript. L.P. and B.M.S. verified the underlying data. K.C.C. runs the Exeter monogenic diabetes diagnostic service and helped with access to data and writing of the manuscript. M.H.S. reviewed and edited the manuscript. J.M. collated data from the Scottish monogenic diabetes diagnostic service and reviewed and edited the manuscript. E.R.P. provided data from the Scottish monogenic diabetes diagnostic service and reviewed and edited the manuscript. S.E. led the genomics testing laboratory in Exeter and helped to review and draft the manuscript. A.T.H. helped to design the study and review and draft the manuscript. B.M.S. designed the study, analyzed data, and helped with the writing of the manuscript. B.M.S. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.17493602.
References
- 1. Thanabalasingham G, Owen KR. Diagnosis and management of maturity onset diabetes of the young (MODY). BMJ 2011;343:d6044. [DOI] [PubMed] [Google Scholar]
- 2. Carlsson A, Shepherd M, Ellard S, et al. Absence of islet autoantibodies and modestly raised glucose values at diabetes diagnosis should lead to testing for MODY: lessons from a 5-year pediatric Swedish national cohort study. Diabetes Care 2020;43:82–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johansson BB, Irgens HU, Molnes J, et al. Targeted next-generation sequencing reveals MODY in up to 6.5% of antibody-negative diabetes cases listed in the Norwegian Childhood Diabetes Registry. Diabetologia 2017;60:625–635 [DOI] [PubMed] [Google Scholar]
- 4. Johnson SR, Ellis JJ, Leo PJ, et al. Comprehensive genetic screening: the prevalence of maturity-onset diabetes of the young gene variants in a population-based childhood diabetes cohort. Pediatr Diabetes 2019;20:57–64 [DOI] [PubMed] [Google Scholar]
- 5. Pihoker C, Gilliam LK, Ellard S, et al.; SEARCH for Diabetes in Youth Study Group . Prevalence, characteristics and clinical diagnosis of maturity onset diabetes of the young due to mutations in HNF1A, HNF4A, and glucokinase: results from the SEARCH for Diabetes in Youth. J Clin Endocrinol Metab 2013;98:4055–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shepherd M, Shields B, Hammersley S, et al.; UNITED Team . Systematic population screening, using biomarkers and genetic testing, identifies 2.5% of the U.K. pediatric diabetes population with monogenic diabetes. Diabetes Care 2016;39:1879–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shields BM, Shepherd M, Hudson M, et al.; UNITED study team . Population-based assessment of a biomarker-based screening pathway to aid diagnosis of monogenic diabetes in young-onset patients. Diabetes Care 2017;40:1017–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shields BM, Hicks S, Shepherd MH, Colclough K, Hattersley AT, Ellard S. Maturity-onset diabetes of the young (MODY): how many cases are we missing? Diabetologia 2010;53:2504–2508 [DOI] [PubMed] [Google Scholar]
- 9. Shepherd M, Colclough K, Ellard S, Hattersley AT. Ten years of the National Genetic Diabetes Nurse Network: a model for the translation of genetic information into clinical care. Clin Med (Lond) 2014;14:117–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shields BM, McDonald TJ, Ellard S, Campbell MJ, Hyde C, Hattersley AT. The development and validation of a clinical prediction model to determine the probability of MODY in patients with young-onset diabetes. Diabetologia 2012;55:1265–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ellard S, Lango Allen H, De Franco E, et al. Improved genetic testing for monogenic diabetes using targeted next-generation sequencing. Diabetologia 2013;56:1958–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Richards S, Aziz N, Bale S, et al.; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Office for National Statistics . Estimates of the population for the UK, England and Wales, Scotland and Northern Ireland. Accessed 10 February 2021. Available from https://www.ons.gov.uk/peoplepopulationandcommunity/populationandmigration/populationestimates/datasets/populationestimatesforukenglandandwalesscotlandandnorthernireland
- 14. Office for National Statistics . Usual resident population. Accessed 18 January 2022. Available from https://www.nomisweb.co.uk/census/2011/ks101ew
- 15. National Health Service . National Genomic Test Directory: testing criteria for rare and inherited disease. Accessed 18 January 2022. Available from https://www.england.nhs.uk/wp-content/uploads/2018/08/rare-and-inherited-disease-eligibility-criteria-v2.pdf
- 16. Pearson ER, Starkey BJ, Powell RJ, Gribble FM, Clark PM, Hattersley AT. Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet 2003;362:1275–1281 [DOI] [PubMed] [Google Scholar]
- 17. Shepherd M, Shields B, Ellard S, Rubio-Cabezas O, Hattersley AT. A genetic diagnosis of HNF1A diabetes alters treatment and improves glycaemic control in the majority of insulin-treated patients. Diabet Med 2009;26:437–441 [DOI] [PubMed] [Google Scholar]
- 18. Hattersley AT, Greeley SAW, Polak M, et al. ISPAD clinical practice consensus guidelines 2018: the diagnosis and management of monogenic diabetes in children and adolescents. Pediatr Diabetes 2018;19(Suppl. 27):47–63 [DOI] [PubMed] [Google Scholar]
- 19. Stride A, Shields B, Gill-Carey O, et al. Cross-sectional and longitudinal studies suggest pharmacological treatment used in patients with glucokinase mutations does not alter glycaemia. Diabetologia 2014;57:54–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Steele AM, Shields BM, Wensley KJ, Colclough K, Ellard S, Hattersley AT. Prevalence of vascular complications among patients with glucokinase mutations and prolonged, mild hyperglycemia. JAMA 2014;311:279–286 [DOI] [PubMed] [Google Scholar]
- 21. Steele AM, Shields BM, Shepherd M, Ellard S, Hattersley AT, Pearson ER. Increased all-cause and cardiovascular mortality in monogenic diabetes as a result of mutations in the HNF1A gene. Diabet Med 2010;27:157–161 [DOI] [PubMed] [Google Scholar]
- 22. Dickens LT, Naylor RN. Clinical management of women with monogenic diabetes during pregnancy. Curr Diab Rep 2018;18:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shepherd M, Brook AJ, Chakera AJ, Hattersley AT. Management of sulfonylurea-treated monogenic diabetes in pregnancy: implications of placental glibenclamide transfer. Diabet Med 2017;34:1332–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ellard S, Bellanné-Chantelot C; European Molecular Genetics Quality Network (EMQN) MODY group . Best practice guidelines for the molecular genetic diagnosis of maturity-onset diabetes of the young. Diabetologia 2008;51:546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]