Abstract
The combination of computational design and directed evolution could offer a general strategy to create enzymes with new functions. To date, this approach has delivered enzymes for a handful of model reactions. Here we show that new catalytic mechanisms can be engineered into proteins to accelerate more challenging chemical transformations. Evolutionary optimization of a primitive design afforded an efficient and enantioselective enzyme (BH32.14) for the Morita-Baylis-Hillman (MBH) reaction. BH32.14 is suitable for preparative scale transformations, accepts a broad range of aldehyde and enone coupling partners, and is able to promote selective mono-functionalizations of dialdehydes. Crystallographic, biochemical and computational studies reveal that BH32.14 operates via a sophisticated catalytic mechanism comprising a His23 nucleophile paired with a judiciously positioned Arg124. This catalytic arginine shuttles between conformational states to stabilize multiple oxyanion intermediates and serves as a genetically encoded surrogate of privileged bidentate hydrogen bonding catalysts (e.g. thioureas). This study demonstrates that elaborate catalytic devices can be built from scratch to promote demanding multi-step processes not observed in Nature.
Introduction
The ability to reliably design enzymes would have significant impacts across the chemical industry, allowing the rapid delivery of new biocatalysts in response to diverse societal challenges. Computational enzyme design is conceptually similar to catalytic antibody technology; in that it aims to generate protein catalysts based on fundamental principles of transition state stabilization. 1 However, in principle computational design offers a far more flexible approach, as it is not limited to the antibody fold or reliant on the availability of an imperfect transition state mimic. Thus far, computational algorithms have enabled the design of primitive catalysts for a handful of transformations that have been optimized through laboratory evolution to deliver enzymes with efficiencies approaching natural systems. 2–7 Naturally, early efforts targeted simple model transformations previously achieved with catalytic antibodies. 8–10 For example, highly effective antibodies and enzymes have been created for the retro-aldol reaction, 3,7,10 which employ reactive lysine residues reminiscent of natural type I aldolases. If we are to establish ‘bottom up’ enzyme design as a reliable source of biocatalysts for practical applications, we must now move beyond the functional capabilities of antibodies and develop enzymes for complex bimolecular chemical processes.
The Morita-Baylis-Hillman (MBH) reaction (Figure 1A), involving the coupling of activated alkenes (e.g. α,β-unsaturated carbonyl compounds) with carbon electrophiles (e.g. aldehydes), is an iconic transformation in organic synthesis. 11–13 This transformation provides a versatile and atom economical approach to generate densely functionalized chiral building blocks for synthesis. MBH reactions are typically promoted by small catalytic nucleophiles such as 1,4-diazabicyclo[2.2.2]octane (DABCO), 4-dimethylaminopyridine (DMAP) and imidazole. Enantioselective versions of the MBH reaction have been developed by employing catalytic peptides, quinidine derivatives, chiral DMAP surrogates or by pairing catalytic nucleophiles with chiral hydrogen bond donors such as thioureas. 13,14 Although good selectivity can be achieved in favourable cases, in other instances achieving high levels of stereo-control remains a considerable challenge (e.g. with substrates 1 and 2 used in this study). 15,16 Despite the great synthetic potential, the practical utility of MBH reactions is further compromised by the low efficiencies achieved by existing catalytic systems, which results in prolonged reaction times and the requirement for high catalyst loadings. Given the enormous rate accelerations achieved by enzymes, biological catalysts could offer a plausible solution to these long-standing challenges. However there are no catalytic antibodies or known natural enzymes for the MBH reaction, although thymidylate synthase that catalyses the conversion of deoxyuridine monophosphate to deoxythymidine monophosphate operates along similar mechanistic principles. 17 Only extremely low levels of promiscuous MBH activity have been reported with a handful of proteins, 18–20 with unspecific protein catalysis implicated in a number of these cases.
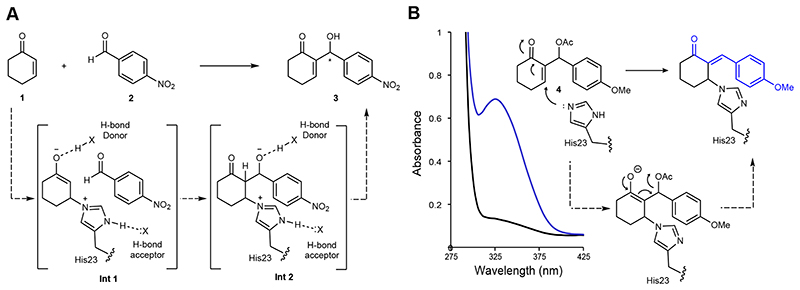
Figure 1. A designed enzyme for the Morita-Baylis-Hillman (MBH) reaction and the development of a dual function mechanistic inhibitor.
A) Chemical scheme of the MBH reaction between 1 and 2 catalysed by the computationally designed enzyme BH32. 21 Intended design features include a His23 nucleophile positioned by a hydrogen bond acceptor, and hydrogen bond donors for oxyanion stabilization. B) Chemical scheme of BH32 inhibition with the mechanistic inhibitor 4. Addition of the His23 nucleophile is followed by E1cB elimination of the acetoxy group, generating a conjugated π-system, which can be monitored by an increase in absorbance at 325 nm. The stereochemistry of the exocyclic double bond in the inhibited complex is unknown. The wavelength scan shows the spectral changes that occur when BH32 (25 μM) is incubated with inhibitor 4 (250 μM) for 40 minutes.
Results and Discussion
In the absence of a suitable natural enzyme, we selected a primitive computationally designed protein for the MBH reaction (BH32), as a starting template for evolutionary optimization. 21 BH32 utilizes a histidine nucleophile (His23) built into the cap domain of haloacid dehalogenase from Pyrococcus horikoshii by introducing 12 active site mutations predicted by the Rosetta software suite. Other intended design features (Figure 1A) include: Glu46 to position and activate the His23 catalytic nucleophile; an aromatic aldehyde binding pocket shaped by Phe132 and Leu10; Gln128 to serve as an oxyanion-hole to stabilize the first covalent enzyme-substrate intermediate formed upon reaction with 2-cyclohexen-1-one (Int 1); and an ordered water molecule bound through Ser22 designed to stabilize Int 2, formed following C-C bond formation, through hydrogen bonding. Despite intensive design efforts, BH32 promotes the coupling of 2-cyclohexen-1-one (1) with 4-nitrobenzaldehyde (2) with extremely low activity (k cat = 0.13 ± 0.01 hr-1). This activity is dependent on the His23 nucleophile, however comparison of the design model with the structurally characterized protein reveals discrepancies, which compromise the intended design features and likely contribute towards low efficiency. For example, key designed residues including His23 & Ser22 adopt altered conformational states from those predicted by design, while a shift of helix 126-132 alters the position of the designed Gln128 oxyanion hole.
Evolution of an efficient and selective MBHase
A high-throughput spectrophotometric assay was developed to allow rapid evaluation of large libraries of BH32 derivatives during evolution. This was achieved by modifying the MBH product 3 through acetylation of the C3 secondary alcohol to generate a dual function mechanistic inhibitor and spectroscopic probe (4) (Figure 1B). Upon reaction with the His23 nucleophile, it was expected that E1cB elimination of the acetoxy group would generate a stable, conjugated product suitable for spectrophotometric detection. Indeed, incubation of BH32 with 4 resulted in the anticipated time resolved spectral changes. The spectroscopic changes were made more pronounced to improve assay sensitivity, by introducing a 4-OMe substituent to increase the degree of conjugation upon inhibition. Formation of a stable 1:1 protein-inhibitor complex was confirmed by MS analysis of the intact protein (Supplementary Table 1). No spectral or mass changes were observed with the BH32 His23Ala variant, confirming the His23 catalytic nucleophile as the site of covalent attachment. Although this assay doesn’t report on overall catalytic turnover, we anticipated that it would provide a useful tool for MBHase engineering capable of reporting on key features of MBH catalysis, including nucleophilicity of His23, stabilization of C1 oxyanion intermediates and shape complementarity between the active site and the MBH product.
The evolutionary strategy used for BH32 optimization included both local and global mutagenesis (Supplementary Figure 1): error-prone PCR was used to target the entire gene sequence leading to the identification of ‘hot spots’ that were further interrogated as small focused libraries (Rounds 1-4, 10-11 & 13-14); iterative cassette mutagenesis was used to individually randomize up to twenty positions per round (Rounds 5-6, 8-9 & 12); and combinatorial active site saturation mutagenesis (CASTing) was used to target active site residues (Round 2 & 7). Beneficial diversity identified during each round was combined by DNA shuffling. The aforementioned spectrophotometric assay was used to rapidly evaluate libraries during rounds 1-8 (Figure 2C). The most active (ca. 1%) clones identified during each round were further evaluated as purified enzymes for MBH activity using HPLC analysis. The correlation between increased rate of reaction with inhibitor 4 and improving MBH activity was generally excellent. While the reaction of BH32 with inhibitor 4 takes > 60 mins to reach completion, under identical conditions selective modification of BH32.8 is essentially complete within 1.5 minutes (Figure 2C and Supplementary Table 1). To further refine the catalytic mechanism, BH32.8 was subjected to an additional four rounds of evolution using a HPLC assay to monitor the formation of MBH product 3 catalysed by individual variants arrayed as cell lysates in 96-well plates.
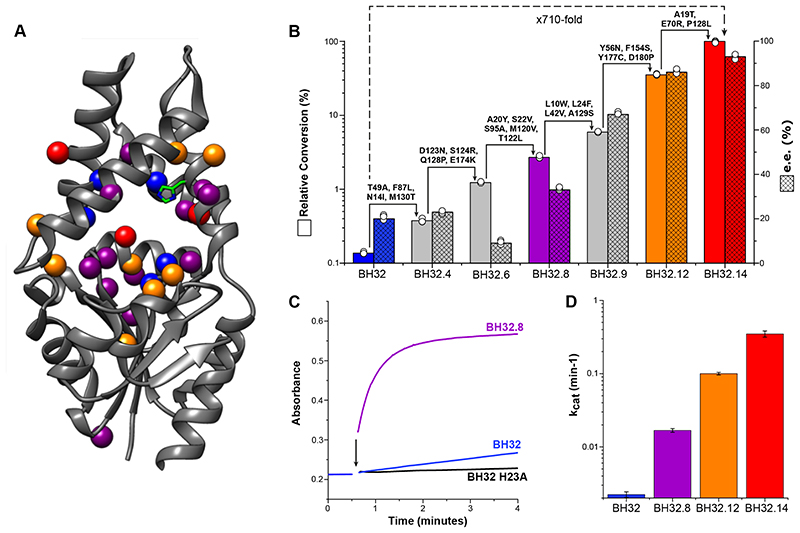
Figure 2. Characterization of BH32, BH32.14 and selected variants.
A) Structure showing the amino acid positions mutated in BH32.14 (represented as spheres) mapped onto the structure of haloacid dehalogenase from Pyrococcus horikoshii (PDB code: 1X42). Mutations introduced during rounds 1-8 are shown in purple, rounds 9-12 in orange and rounds 13-14 in red. Position Asn14 was mutated twice (round 3 & 4) and is shown in purple, position Pro128 was mutated twice (round 5 & 14) and is shown in red. Computationally designed residues remaining following evolution are shown in blue. The His23 catalytic nucleophile is shown in stick representation. B) Bar chart showing the mean relative conversion (solid colour) and enantiomeric excess (hatched) achieved by selected variants along the evolutionary trajectory. Biotransformations were performed using 1 (3 mM), 2 (0.6 mM) and enzyme (18 μM), and analysed following 4.5 hours incubation (see Supplementary Table 2 for conversion and selectivity data). Error bars represent the standard deviation of measurements made in triplicate. C) Time course for the inhibition of BH32.8 (25 μM, purple), BH32 (25 μM, blue) and BH32 H23A (25 μM, black) with inhibitor 4 (250 μM). The black arrow indicates the time of protein addition. D) Bar chart comparing the turnover number (k cat) of BH32 (blue), BH32.8 (purple), BH32.12 (orange) and BH32.14 (red) for the MBH reaction between 1 and 2. Steady state kinetic data (average of measurements made in triplicate at each substrate concentration) were fitted globally using a kinetic model for two substrates with randomly ordered binding to extract kinetic constants and associated errors (shown as error bars). Representative Michaelis-Menten plots at fixed concentrations of 1 or 2 are also shown in Supplementary Figure 2.
The most active variant to emerge following 14 rounds of evolution (BH32.14) contains 24 mutations (Figure 2A). The relative activities of variants along the evolutionary trajectory were compared in biotransformations of 1 (3 mM) and 2 (0.6 mM) using 3 mol% catalyst (Figure 2B). The starting variant BH32 gave ~0.08 % conversion to product (3) after 4.5 hours (Supplementary Table 2) which increased to only 0.4% following incubation for 22 h (Table 1, Entry 1). Furthermore, product formation was accompanied by a substantial proportion of a competing aldol by-product S1 (5:1 ratio of 3:S1, Supplementary Figure 3). In contrast, under identical reaction conditions BH32.14 afforded 3 as the sole product in 58% (Table 1, Entry 2) and 83% conversion (Table 1, Entry 3) after 4.5 hours and 22 hours, respectively. For comparison, with commonly employed small molecule catalytic nucleophiles imidazole, DMAP and DABCO, low but detectable conversions (<0.5%) are only achieved using high catalyst loadings and prolonged reaction times (Table 1, Entry 7-9). This analysis shows how steady improvements across the evolutionary trajectory have afforded an efficient MBHase that is >710-fold more active than the starting design under these assay conditions. This improvement in catalytic performance is primarily achieved through a substantial 160-fold increase in turnover number (k cat = 0.35 ± 0.03 min-1 and 0.13 ± 0.01 hr-1 for BH32.14 and BH32, respectively, Figure 2D). Although at this stage we cannot differentiate between random and ordered binding mechanisms operating in BH32 and its evolved variants, the magnitude of these k cat values is largely insensitive to the kinetic model used (Supplementary Table 3). Evolution has also resulted in modest improvements in apparent K M values for both the enone and aldehyde coupling partners (Kapp [1] = 2.6 ± 0.4 mM and 8.0 ± 1.0 mM, Kapp [2] = 1.1 ± 0.2 mM and 1.8 ± 0.2 mM for BH32.14 and BH32, respectively), meaning that substrate saturation is more readily achieved in the evolved BH32.14 variant (Supplementary Figure 2 and Supplementary Table 3).
Table 1. The MBH reactions of 1 and 2 catalysed by small molecule nucleophiles, BH32, BH32.14 and selected variants.
| Entry | Catalyst | Catalyst Loading (mol%) | Time (h) | Conversion (%) |
|---|---|---|---|---|
| 1 | BH32 | 3 | 22 | <0.5 |
| 2 | BH32.14 | 3 | 4.5 | 58 |
| 3 | BH32.14 | 3 | 22 | 83 |
| 4 | BH32.14 His23Ala | 3 | 4.5 | <0.5 |
| 5 | BH32.14 Arg124Ala | 3 | 4.5 | <1 |
| 6 | BH32.14 Trp10Ala | 3 | 4.5 | 3.8 |
| 7 | Imidazole | 167 | 22 | <0.5 |
| 8 | DMAP | 167 | 22 | <0.5 |
| 9 | DABCO | 167 | 22 | <0.5 |
| 10 | BH32.14 | 0.5 | 19 | 94 |
Entries 1-9 were carried out using 1 (3 mM) and 2 (0.6 mM) in PBS (pH 7.4) with 3% DMSO as a cosolvent. Conversion to product was determined by HPLC analysis. Entry 10 was performed on a preparative scale using 1 (50 mM) and 2 (10 mM) in PBS (pH 7.4) with 20% DMSO as a cosolvent. Conversion to product was determined by HPLC analysis. The final product was isolated in 88% yield following extraction into organic solvent and chromatographic purification.
Although we didn’t explicitly select for improvements in enantioselectivity, evolution led to substantial selectivity gains, with reactions catalysed by BH32 and BH32.14 affording (R)-3 in 20% and 93% e.e., respectively (Figure 2B and Supplementary Table 2). Kinetic improvements were evidently achieved by an increasingly precise recognition of the rate-limiting transition state(s) leading to the formation of the (R)-configured product. This selectivity compares favourably with the modest selectivities achieved for the synthesis of 3 by chiral small molecule catalysts. 15,16 We also explored the utility of BH32.14 for preparative scale biotransformations (Table 1, Entry 10). Significantly, while the enzyme is rather intolerant to elevated MeCN concentrations, it readily tolerates 20 % DMSO as an organic cosolvent, which allows reactions to be performed with increased substrate loadings (10 mM concentration of 2). High conversions (94%) and isolated yields (88%) are obtained within 19 h using only 0.5% catalyst loading to allow synthesis of several hundred milligrams of MBH product 3 (Supplementary Figure 4). Reactions using 0.1% catalyst loading show that the enzyme is able to achieve >530 turnovers (Supplementary Figure 5). Interestingly, BH32.14 displays reduced, but appreciable levels of activity in up to 40% DMSO (Supplementary Table 4) and up to 50 °C (Supplementary Figure 6), highlighting the potential for further reaction intensification in the future. The high degree of solvent tolerance and mutational flexibility of the BH32 scaffold (12% of the protein was mutated during computational design and evolution) is noteworthy, and likely reflects the stability of the starting scaffold, which originates from a hyperthermophilic organism that grows optimally at 98 °C. 22,23
Synthetic utility of engineered MBHases
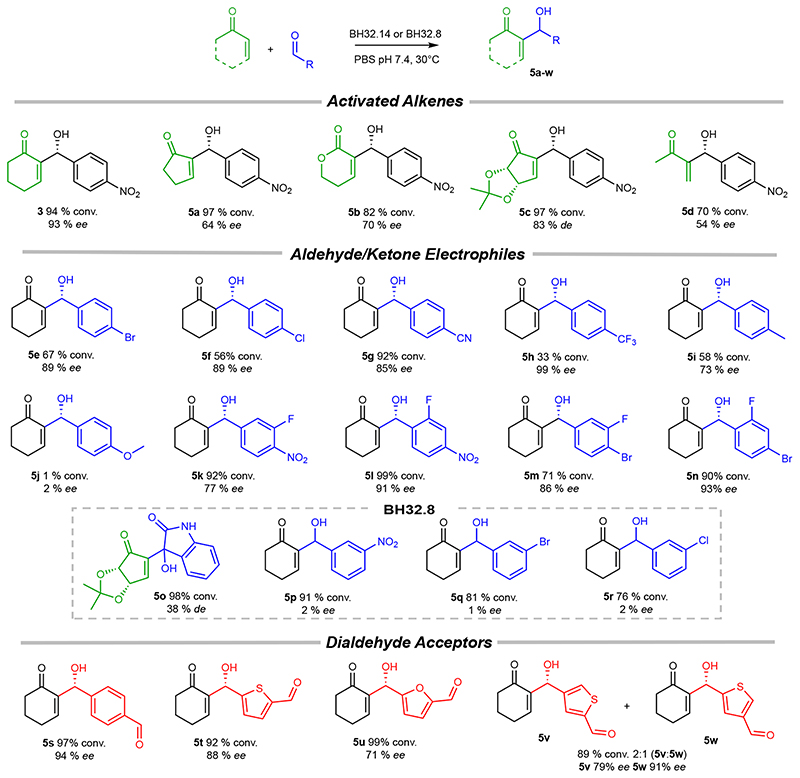
To further explore synthetic utility, BH32.14 was evaluated for activity towards a range of activated alkene and aldehyde/ketone coupling partners, leading to the synthesis of structurally diverse MBH products 5a-w (Figure 3). In addition to cyclohexenone 1, BH32.14 displays high levels of activity in MBH reactions with cyclopentenone, functionalized cyclopentenones and methyl vinyl ketone. Testament to its catalytic power, BH32.14 can even promote enantioselective transformations of unsaturated lactones (product 5b), which are notoriously challenging as MBH substrates. 24,25 BH32.14 also accepts a range of mono- and di-substituted aromatic aldehydes as substrates, although has a clear preference for para-substituted derivatives. While a modestly electron-donating p-Me substituent is well tolerated (5i), introduction of a strongly donating p-OMe group leads to a significant reduction in activity (5j). The aliphatic aldehydes heptanal, acetaldehyde and cyclohexanecarboxyaldehyde are not substrates for BH32.14 (data not shown). Compared with BH32.14, the less highly evolved BH32.8 accepts a broader range of substituents at the 2-and 3-positions, including the activated ketone istatin (product 5o), albeit with reduced efficiency and minimal selectivity. These results suggests BH32.14 has been highly specialized through evolution to operate efficiently and selectively on specific classes of substrate, whereas its ancestor BH32.8 is more promiscuous and thus provides an attractive starting template for engineering MBHases to produce a broader range of target structures. Similar correlations between improving efficiency and increased substrate specificity have been observed previously during evolutionary optimization of designed enzymes. 6 These trends can be attributed to improved shape complementarity between the target substrate(s) and the engineered active site, which aids efficient catalysis by maximising productive interactions with the transition states but makes the active site less tolerant of non-native reaction partners. We also recognized the opportunity to perform selective monofunctionalizations of aromatic dialdehydes. Indeed, biotransformations of terephthaldehyde, thiophene-2,5-dicarboxaldehyde and furan-2,5-dicarboxaldehyde proceed with high conversions to chiral monofunctionalized MBH adducts (5s-w). The reaction of cyclohexenone with the unsymmetrical dialdehyde, thiophene-2,4-dicarboxaldehyde delivered enantioenriched monosubstituted products 5v and 5w with modest regiocontrol, which could likely be improved through additional rounds of evolution.
Figure 3. Substrate scope of engineered MBHases.
BH32.14 tolerates a range of activated alkenes, aldehydes and dialdehydes as substrates, leading to the production of densely functionalized MBH adducts with high conversions and selectivities. The less highly evolved BH32.8 accepts a broader range of substituents at the 2- and 3- positions, albeit with reduced efficiency and minimal enantioselectivity. Reported conversions and selectivities are the mean of biotransformations performed in triplicate. The stereochemistry of 5a-i, k-o & 5s-w are assigned by analogy to the (R)-3 product formed in BH32.14 mediated biotransformations. Biotransformations were performed using aldehyde/ketone (10 mM), activated alkene (50 mM for 3, 5a-c, 5f-h & 5j-w, 100 mM for 5d-e & 5i) and catalyst (0.5-5 mol%). Specific reaction conditions for the synthesis of 3 & 5a-w are presented in Supplementary Table 5.
Structural characterization and computational modelling
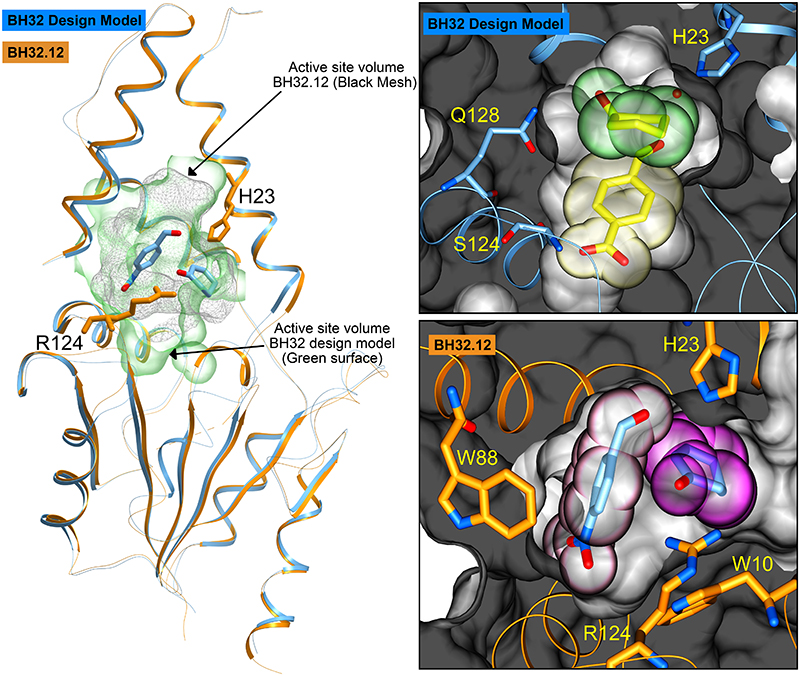
To understand the origins of improved efficiency following evolution, crystal structures of several evolved BH32 variants were solved for comparison to the original design. The most active variant, BH32.14, proved challenging to crystallize; however we were able to solve the apo structure of BH32.12, which contains 21 out of 24 mutations present in BH32.14, to 2.3 Å resolution (Supplementary Table 7). Thus far, we have been unable to obtain BH32.12 structures with substrate, product or inhibitor bound. The BH32.12 and BH32 apo-structures superimpose well, with a root-mean-square-deviation of 0.8 Å. However evolution has resulted in extensive remodelling of the active site, including a ~30% reduction in volume (Figure 4). The designed His23 nucleophile has been preserved and is essential for catalytic activity in BH32.14 (Table 1, Entry 4). In contrast, Gln128 and Ser22 intended to stabilize key oxyanion intermediates through hydrogen bonding interactions, have been abandoned during evolution. Instead, an active site Arg124 has emerged which is critical for effective catalysis (Table 1, Entry 5). This catalytic residue emerged in the early rounds of evolutionary optimization, and in the crystal structure of BH32.6 (resolution 1.5 Å, Supplementary Table 7) adopts a conformation which places the guanidinium motif in close proximity to the amide side chain of the originally designed Gln128. However, subsequent active site remodelling resulted in a substantial repositioning of the arginine side chain leading to a ~5 Å displacement of the guanidinium ion in BH32.12 (Supplementary Figure 7). The originally designed aldehyde binding site is occluded in BH32.12 by residues Trp10, Leu122, Arg124 & Ser129 which emerged during evolution. Molecular docking of substrates 1 and 2 reveals the presence of a new aldehyde binding pocket shaped by Trp10, Val22, Ile26, Leu64, Trp88, Ser91, Leu92, Phe132, Arg124 & Ser129, and a 2-cyclohexen-1-one binding mode suitable for nucleophilic attack by His23 (Figure 4).
Figure 4. Crystal structures of BH32 and BH32.12.
A ribbon representation of the superimposed coordinates of the BH32 design model (blue) and the evolved variant BH32.12 (orange). The His23 nucleophile and catalytic Arg124 from BH32.12 are shown in orange stick representation. Substrates docked into BH32.12 are shown in all atom coloured stick representation. The active site surface volume of the BH32 design model is shown as a green transparency whilst the equivalent active site surface volume for BH32.12 is shown as a grey mesh. The right hand panels show a close-up representation of the active sites. The top panel shows the substrate binding pocket of BH32 design model highlighting the spatial arrangement of the designed residues Gln128, Ser124 and His23. The protein backbone is shown in ribbon representation (blue) with the protein surface shown in grey. The substrates are derived from the original composite transition state model 21 with the aldehyde shown in stick representation and transparent yellow CPK spheres. 2-Cyclohexen-1-one is also shown in stick representation with accompanying transparent green CPK spheres. The bottom panel shows the aldehyde and enone binding pocket of BH32.12 highlighting the spatial arrangement of key residues Trp88, Trp10, Arg124 and His23 (stick representation - orange carbon atoms). The protein backbone is shown in ribbon representation (orange) with the protein surface shown in grey. The substrate positions depicted are those obtained from initial docking studies prior to DFT calculations. The aldehyde substrate is shown in stick representation with accompanying transparent pink CPK spheres. 2-Cyclohexen-1-one is shown in stick representation with transparent magenta CPK spheres.
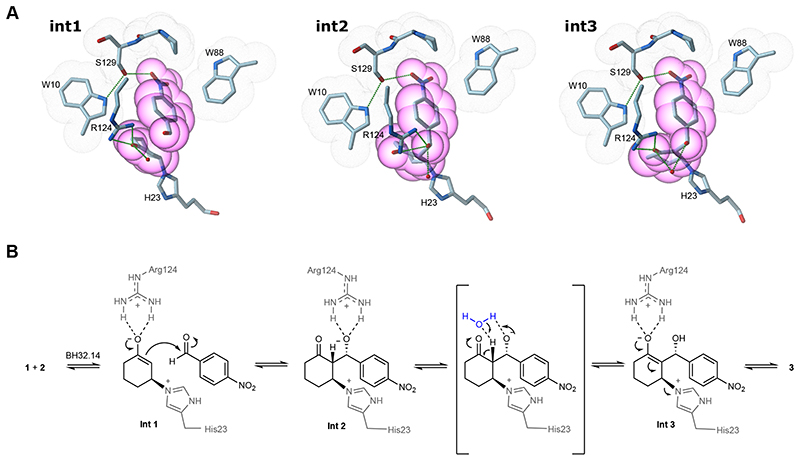
To shed light on the catalytic mechanism, an active site DFT ‘cluster’ model comprising 273 atoms was constructed from the BH32.12 crystal structure and docked substrates, with peripheral atoms fixed to maintain the crystal structure geometry (Supplementary Figure 8). A water molecule was also included in the calculations to facilitate proton transfer in the third chemical step. Relaxed potential energy scans were performed to model each step of the proposed mechanism (Supplementary Figures 9 and 10). During energy minimisation of the reactant state, the aromatic aldehyde forms an edge to face π-stacking interaction with Trp88. The modelling suggests that aldehyde binding is further supported by a hydrogen bonding network involving the para-nitro substituent, Ser129 and Trp10, two residues that emerged during the latter stages of evolution. Trp10 lies in close proximity to Arg124 and likely plays an important role in positioning the guanadinium side chain (Figure 4 and Supplementary Figure 7). A Trp10Ala substitution in BH32.14 leads to a substantial reduction in activity (Table 1, Entry 6), underscoring the importance of this residue to the catalytic mechanism. Nucleophilic addition of His23 to the si-face of 1 generates the first oxyanion intermediate (Int1) which is stabilized by a bidentate hydrogen bond to the side chain of Arg124. Similar interactions have been observed in natural 26 and de novo hydrolases, 27 as well as catalytic antibodies, 28–30 and are the hallmark of small molecule hydrogen bonding catalysts such as thiourea and guanidinium ions. 31,32 While BH32.14 reacts rapidly with inhibitor 4, this activity is dramatically reduced upon mutation of Arg124 to Ala (Supplementary Figure 11), supporting the role of this residue in stabilizing oxyanion intermediates at C1. Consistent with its role as the catalytic nucleophile, activity with inhibitor 4 is abolished in BH32.14 His23Ala. DFT modelling shows that inhibitor 4 is well accommodated in the active site with His23 poised for nucleophilic attack and Arg124 suitably positioned for oxyanion stabilization (Supplementary Figure 12). The enolate of Int1 is well positioned for subsequent addition to aldehyde 2. Diastereoselective carbon-carbon bond formation generates a second oxyanion intermediate (Int2) with (R)-configuration at the C3-position, consistent with the stereochemical outcome of the MBH reactions observed experimentally, and proceeds with transfer of negative charge to the C3-oxygen (Figure 5). Arg124 is ideally positioned to support this charge transfer, and the calculations suggest that this residue can shuttle between bidentate hydrogen bonding modes to stabilize both Int1 and Int2, via a bridging mode at the transition state involving a single hydrogen bond to each oxygen atom. Consistent with previous models of thiourea promoted MBH reactions, 33 the third chemical step involves water mediated proton transfer from C2 to the C3 alkoxide, leading to the generation of a third oxyanion intermediate (Int 3) which again can be stabilized by bidentate hydrogen bonding to Arg124. The final chemical step involves elimination of the His23 nucleophile to generate Morita-Baylis-Hillman product (R)-3. Our calculations suggest that the third chemical step is rate limiting (or partially rate limiting) (Supplementary Figure 9), with an energy barrier of 61.9 kJmol-1 compared with barriers of 50.1 kJmol-1 and 47.4 kJmol-1 for steps 1 and 4, respectively. If this were the case, we would expect to observe a solvent kinetic isotope effect (SKIE) as an exchangeable proton is transferred in step 3. Comparison of biotransformations with BH32.14 performed in H2O and D2O reveals a modest SKIE of 1.4 ± 0.09 (Supplementary Figure 13), indicating that proton transfer is involved in the rate-limiting step as suggested by the modelling. Interestingly, no SKIE is observed in biotransformations with BH32 or BH32.8, suggesting that the nature of the rate-limiting step has changed during the course of evolution.
Figure 5. Proposed catalytic mechanism of an engineered MBHase.
A) DFT states for intermediates 1, 2 & 3 are shown in all atom coloured stick representation. The images presented show only a subset of the atoms used in the full DFT calculation for greater visual clarity. Atoms derived from several residues that form close packing interactions with substrates 1 and 2 during catalysis (Trp10, Trp88, Arg124 & Ser129) are highlighted with grey dot surfaces. Heavy atoms from the substrates are highlighted with magenta transparent CPK spheres. Atoms from the His23 nucleophile and catalytic Arg124 are shown in all atom coloured stick representation. The hydrogen bonding network that emerged during evolution between Trp10, Ser129 and the aldehyde substrate are shown as black dashed lines. In addition, hydrogen bonds between Arg124 and each intermediate state are shown. B) The BH32.14 catalytic mechanism showing the role played by Arg124 in stabilizing three intermediates covalently bound through His23. A catalytic water molecule is shown in blue to facilitate proton transfer from C2 to the C3 alkoxide.
Conclusions
This study provides a remarkable glimpse into the evolution of new catalytic mechanisms in protein active sites, where multiple components work in concert to accelerate valuable and demanding chemical transformations. The development of BH32.14 now allows us to add MBHases to the restricted repertoire of biocatalysts available for C-C bond forming reactions. High-throughput enzyme engineering was facilitated by the development of a spectrophotometric assay, based on an irreversible inhibition reaction with mechanistic similarities to MBH transformations. We anticipate that similar assays will prove valuable for identifying highly active biocatalysts for the MBH reaction or mechanistically related transformations in the future. Beyond its synthetic utility, analysis of the BH32.14 evolution offers valuable lessons to improve enzyme design protocols, to facilitate the design of more efficient catalysts for a broader range of transformations in the future. BH32.14 employs a nucleophilic His23 and a multi-functional Arg124 to accelerate the MBH reaction, in a catalytic mechanism with strong similarities to that observed with small molecule systems. 31,32 This suggests that in cases where no natural enzyme or catalytic antibodies are available to guide the design of new active site arrangements, we should turn to small molecule catalysis for inspiration. In particular, the observation that an appropriately positioned arginine can serve as a genetically encoded surrogate of privileged bidendate hydrogen bonding catalysts (e.g. thioureas) should inspire the design of biocatalysts for a broad range of non-biological transformations. 31,32,34,35 Arginine was not considered as a hydrogen bond donor in the original BH32 design constraints and is typically avoided as a catalytic motif during enzyme design due to its high degree of conformational flexibility, which makes accurate positioning of the guanidinium side chain challenging. However, our study shows that in BH32.14 this conformational flexibility is advantageous, with Arg124 providing a highly effective and economical means of stabilizing multiple oxyanion intermediates and transition states. While natural enzymes are inherently dynamic molecules, existing enzyme design protocols generate a static model of the protein-transition state complex. For chemical transformations achieved with catalytic antibodies, it seems reasonable to assume that these single state design protocols could deliver enzymes with comparable or improved efficiencies to antibodies. However, our results suggest that for complex transformations with several high-energy transition states, multi-state enzyme design protocols 36,37 will be required to account for exchanges between conformational states along the reaction coordinate. The development of these next generation enzyme design methodologies is now underway, using the evolutionary optimization of the BH32.14 catalytic mechanism as inspiration.
Methods
Materials
All chemicals and biological materials were obtained from commercial suppliers. Lysozyme, DNase I and kanamycin were purchased from Sigma-Aldrich; polymyxin B sulfate from AlfaAesar; LB agar, LB media, 2×YT media and arabinose from Formedium; Escherichia coli (E. coli) 5α, Q5 DNA polymerase, T4 DNA ligase and restriction enzymes from New England BioLabs; and oligonucleotides were synthesized by Integrated DNA Technologies.
Construction of pBbE8k_BH32 and variants
The original BH32 design 20 was modified to introduce C186A and C212A mutations to avoid any non-specific alkylations of these positions. These substitutions had no effects on MBH activity. The C186A and C212A double mutant is referred to as BH32 throughout this study. BH32 was subcloned, using NdeI and XhoI restriction sites, into a pBbE8k vector 38 modified to include a 6×His tag or Strep-tag following the XhoI restriction site to yield pBbE8k_BH32_6His or pBbE8k_BH32_Strep. The H23A mutation was introduced into the pBbE8k_BH32 constructs using QuikChange site-directed mutagenesis.
Protein production and purification
For expression of BH32 and variants, chemically competent E. coli 5α were transformed with the relevant pBbE8k_BH32 constructs. Single colonies of freshly transformed cells were cultured for 18 h in 10 mL LB medium containing 25 μg mL−1 kanamycin. Starter cultures (500 μL) were used to inoculate 50 mL 2×YT medium supplemented with 25 μg mL−1 kanamycin. Cultures were grown at 37 °C, 200 r.p.m. to an optical density at 600 nm (OD600) of around 0.5. Protein expression was induced with the addition of L-arabinose to a final concentration of 10 mM. Induced cultures were incubated for 20 h at 25 °C and the cells were subsequently collected by centrifugation (3,220g for 10 min). For His-tagged variants, pelleted cells were resuspended in lysis buffer (50 mM HEPES, 300 mM NaCl, pH 7.5 containing 20 mM imidazole) and lysed by sonication. Cell lysates were cleared by centrifugation (27,216g for 30 min), and supernatants were subjected to affinity chromatography using Ni-NTA Agarose (Qiagen). Purified protein was eluted using 50 mM HEPES, 300 mM NaCl, pH 7.5 containing 250 mM imidazole. For Strep-tagged variants, pelleted cells were resuspended in Buffer NP (50 mM NaH2PO4, 300 mM NaCl, pH 8) and lysed by sonication. Cell lysates were cleared by centrifugation (27,216g for 30 min), supernatants were subjected to a Strep-Tactin® Superflow Plus resin (Qiagen) and purified protein was eluted using 50 mM NaH2PO4, 300 mM NaCl, 2.5 mM desthiobiotin at pH 8.0. Proteins were desalted using 10DG desalting columns (Bio-Rad) with PBS pH 7.4 and analysed by SDS-PAGE. Proteins were further purified by size-exclusion chromatography using a Superdex 200 column (GE Healthcare) in PBS pH 7.4. Proteins were aliquoted, flash-frozen in liquid nitrogen and stored at −80 °C. Protein concentrations were determined by measuring the absorbance at 280 nm and assuming an extinction coefficient of 31400 M−1 cm−1 for BH32-BH32.7, 32890 M−1 cm−1 for BH32.8, 38390 M−1 cm−1 for BH32.9-BH32.11, and 35410 M−1 cm−1 for BH32.12-BH32.14.
Inhibition of BH32 variants
Proteins were inhibited under two different reaction conditions (Figure 2C and Supplementary Figure 11): For comparison of BH32, BH32 H23A and BH32.8, proteins (25 μM) were inhibited following incubation with 4-methoxyphenyl-(6-oxocyclohex-1-en-1-yl)methyl acetate, 4 (250 μM) in PBS pH 7.4 with 3% (v/v) acetonitrile at room temperature. Formation of the covalently modified protein was monitored spectrophotometrically at 325 nm (Figure 2C). Samples were diluted in PBS pH 7.4, excess inhibitor was removed using a Vivaspin® 10k MWCO (Sartorius) and inhibited proteins were characterized by mass spectrometry (Supplementary Table 1). Under these conditions, inhibition of BH32.12 and BH32.14 was too rapid to be monitored spectrophotometrically. To allow comparison of BH32, BH32 H23A, BH32.8, BH32.12, BH32.14, BH32.14 H23A and BH32.14 R124A (Supplementary Figure 11), enzyme variants (10 μM) were inhibited following incubation with 4 (25 μM) in PBS pH 7.4 with 3% (v/v) acetonitrile at room temperature.
Mass spectrometry
Purified protein samples were buffer exchanged into 0.1% acetic acid using a 10k MWCO Vivaspin (Sartorius) and diluted to a final concentration of 0.5 mg mL−1. Mass spectrometry was performed using a 1200 series Agilent LC, with a 5 μL injection into 5% acetonitrile (with 0.1% formic acid) and desalted inline for 1 min. Protein was eluted over 1 min using 95% acetonitrile with 5% water. The resulting multiply charged spectrum was analysed using an Agilent QTOF 6510 and deconvoluted using Agilent MassHunter Software.
Library construction
Primer sequences used to generate DNA libraries are shown in Supplementary Table 9.
Rounds 1, 3, 10 & 13: random mutagenesis using error-prone PCR
Random libraries were generated by error-prone PCR of the entire gene using a JBS Error-Prone Kit (Jena Bioscience) according to the manufacturer’s protocol. PCR conditions were adjusted to generate an average of 2.5 mutations per gene. The linear library fragments and the modified pBbE8k vector were digested using NdeI and XhoI endonucleases, gel-purified and subsequently ligated using T4 DNA ligase. Variants with improved activity were sequenced and identified ‘hot spots’ were subsequently individually randomized by saturation mutagenesis.
Rounds 2, 4-9, 11-12 & 14: saturation mutagenesis
Positions were either individually randomized using NNK codon degeneracy (Rounds 2, 4-6, 8-9, 11-12 &14), or randomized in pairs using the 22-codon trick 39 (Rounds 2 & 7). DNA libraries were constructed by overlap extension PCR and genes were cloned as described above.
Shuffling by overlap extension PCR
After each round of evolution, beneficial diversity was combined by DNA shuffling of fragments generated by overlap extension PCR. Primers were designed that encoded either the parent amino acid or the identified mutation. These primers were used to generate short fragments (up to 6) which were gel-purified and mixed appropriately in overlap extension PCR to generate genes containing all possible combinations of mutations. Genes were cloned as described above.
Library screening
For protein expression and screening, all transfer and aliquotting steps were performed using Hamilton liquid-handling robots. Chemically competent E. coli 5α cells were transformed with the ligated libraries. Freshly transformed clones were used to inoculate 180 μL of 2xYT medium supplemented with 25 μg mL-1 kanamycin in Corning® Costar® 96-well microtitre round bottom plates. For reference, each plate contained 6 freshly transformed clones of the parent template and 2 clones containing an empty pBbE8k vector. Plates were incubated overnight at 30 °C, 80 % humidity in a shaking incubator at 900 r.p.m. 20 μL of overnight culture was used to inoculate 480 μL 2xYT medium supplemented with 25 μg mL-1 kanamycin. The cultures were incubated at 30 °C, 80 % humidity with shaking at 900 r.p.m. until an OD600 of about 0.5 was reached, and L-arabinose was added to a final concentration of 10 mM. Induced plates were incubated for 20 h at 30 °C, 80 % humidity with shaking at 900 r.p.m. Cells were harvested by centrifugation at 2,900 g for 5 min. The supernatant was discarded and the pelleted cells were resuspended in 400 μL of lysis buffer (PBS pH 7.4 buffer supplemented with 1.0 mg mL-1 lysozyme, 0.5 mg mL-1 polymixin B and 10 μg mL-1 DNase I) and incubated for 2 h at 30 °C, 80 % humidity with shaking at 900 r.p.m. Cell debris was removed by centrifugation at 2,900 g for 5 min.
Rounds 1-8
100 μL Clarified lysate was transferred to 96-well microtitre plates containing 80 μL PBS buffer pH 7.4. Reactions were initiated with the addition of 20 μL inhibitor 4 (Rounds 1-4: 250 μM final concentration, Rounds 5-6: 100 μM final concentration) in PBS pH 7.4 containing acetonitrile (3% (v/v) final concentration). Inhibition was monitored spectrophotometrically at 325 nm, over 15 minutes using a CLARIOstar plate reader (BMG Labtech). Reaction rates of individual variants were normalized to the average of the 6 parent clones.
Rounds 8-14
75 μL Clarified lysate was transferred to 96-well polypropylene microtitre plates and the reaction was initiated with the addition of 25 μL assay mix containing 4-nitrobenzaldehyde (0.6 mM final concentration) and 2-cyclohexen-1-one (3 mM final concentration) in PBS pH 7.4. Reactions were heat-sealed and incubated for 18 h at 30 °C, 80 % humidity with shaking at 900 r.p.m. Reactions were quenched with the addition of 100 μL acetonitrile and incubated for a further 2 h at 30 °C, 80 % humidity with shaking at 900 r.p.m. Precipitated proteins were removed by centrifugation at 2,900g for 10 min. 100 μL of the clarified reaction was transferred to 96-well polypropylene microtitre plates and heat-sealed with pierceable foil. Reactions were evaluated by HPLC analysis as described below.
Following each round, the most active variants were rescreened as purified proteins using the HPLC assay. Proteins were produced and purified as described above, however starter cultures were inoculated from glycerol stocks prepared from the original overnight cultures.
Kinetic characterization
Initial velocity (V 0) vs [4-nitrobenzaldehyde] kinetic data were measured using strep-tagged purified enzyme (60 μM BH32, 40 μM BH32.8, 10 μM BH32.12 & BH32.14), a fixed concentration of 1 (25 mM) and varying concentrations of 2 (0.25-2 mM for BH32 and 0.1-2 mM for BH32.8, BH32.12 & BH32.14). Reactions were performed in PBS pH 7.4 with 3% acetonitrile and were incubated at 30 °C with shaking (800 r.p.m.). BH32.14 & BH32.12 catalyzed reactions were sampled at 15 minute intervals, BH32.8 was sampled at 35, 80, 120, 150, 180, 225, 270 minutes, and BH32 was sampled at 24 h, 40 h, 49 h, 68 h and 94 h. V 0 vs [2-cyclohexen-1-one] kinetic data were measured using a fixed concentration of 2 (2 mM) and varying concentrations of 1 (1-25 mM for BH32, BH32.8 & BH32.12, and 0.5-25 mM for BH32.14) using the enzyme concentrations and buffer conditions described above. Samples were quenched with 1 volume of acetonitrile and analysed by HPLC as described below (see chromatographic analysis).
Linear fits of conversion vs time allowed determination of V 0 at each condition (Supplementary Figure 2). The combined V 0 vs [4-nitrobenzaldehyde] and V 0 vs [2-cyclohexen-1-one] steady state kinetic data were fitted globally using both the random order binding model (Eq. 1) and a two-substrate ordered binding kinetic model (Eq. 2): v = k cat[E][A][B]/((K mA + [A]) (K mB + [B]))(Eq. 1); v = kcat [E][A][B]/([A][B]+K mA [B] + K mB [A] + K sA K mB)(Eq. 2). Where k cat has its usual meaning, [E] is the total enzyme concentration, [A] and [B] are the initial 2-cyclohexen-1-one and 4-nitrobenzaldehyde concentrations respectively, K mA and K mB are the corresponding apparent Michaelis constants, and K sA is the dissociation constant for the enzyme-substrate A complex. Note that Eq. 1 and Eq. 2 are equivalent when K sA = K mA. Global fitting was performed with shared k cat, K mA, K mB and K sA values (i.e. as a 3D surface fit). Kinetic constants are shown in Supplementary Table 3.
Kinetic solvent isotope effects
KSIE experiments were performed in PBS (pH/pD 7.4) (Supplementary Figure 13). Deuterated buffers were prepared using 99.9% D2O with pD adjusted according to the following relationship: pD = pHobs + 0.38. To compare the activity of BH32, BH32.8 and BH32.14, analytical scale biotransformations were performed using 1 (25 mM), 2 (2 mM) and the relevant biocatalyst (50 μM BH32, 30 μM BH32.8, 10 μM BH32.14) in both deuterated and non-deuterated PBS buffer with 3 % MeCN as a cosolvent. Reactions were performed in triplicate. Reactions in deuterated buffer contained <0.5% H2O final concentration. All reactions were incubated at 25 °C with shaking (800 r.p.m.) with samples taken every 10 minutes for 1 hour with BH32.8 and BH32.14 and at 15, 21, 24, 40 and 46 hours for BH32. For HPLC analysis, reactions were quenched by the addition of 1 volume of acetonitrile. Samples were vortexed and precipitated proteins removed by centrifugation (14,000 g for 5 minutes).
BH32.14 total turnover numbers
Total turnover numbers achieved by BH32.14 were determined as follows: BH32.14 (0.5 or 0.1 mol%) catalyzed biotransformations were performed in glass vials using 1 (50 mM) and 2 (10 mM) in PBS (pH 7.4) with 20 % DMSO cosolvent (Supplementary Figure 5). Reactions were incubated at 25 °C with shaking (300 r.p.m.) and samples were taken at 3.0, 28.5, 52.5, 70.25, 100, 159 and 187 hours. For HPLC analysis, reactions were quenched at the stated time points with the addition of 1 volume acetonitrile. Samples were vortexed and precipitated proteins were removed by centrifugation (14,000 g for 5 minutes).
General procedure for analytical scale biotransformations
To compare the activity of BH32, BH32.14 and its variants (Table 1), analytical scale biotransformations were performed using 1 (3 mM), 2 (0.6 mM) and the relevant biocatalyst (18 μM) in PBS (pH 7.4) with 3 % DMSO as a cosolvent. For comparison, reactions were also performed at varying concentrations of small molecule catalytic nucleophiles imidazole, DMAP and DABCO (18 μM, 60 μM and 1 mM). For reaction conditions used in the preparation of MBH adducts 5a-w (Figure 3) see Supplementary Table 5.
For HPLC analysis, reactions were quenched at the stated time points with the addition of 1 volume acetonitrile. Samples were vortexed and precipitated proteins were removed by centrifugation (14,000 g for 5 minutes). For SFC analysis, the substrates and products were extracted with 3 volumes of ethyl acetate. Precipitated proteins were cleared by centrifugation (14,000 g for 5 minutes), the organic phase was separated and directly injected onto the SFC.
BH32.14 cosolvent tolerance and temperature profile
To investigate cosolvent tolerance, analytical scale biotransformations were performed using 1 (50 mM), 2 (10 mM) and BH32.14 (50 μM) in PBS (pH 7.4) with either 20 %, 30 % and 40 % of DMSO or MeCN as a cosolvent (Supplementary Table 4). All reactions were incubated at 25 °C with shaking (800 r.p.m.) for 5 hours. To evaluate the activity of BH32.14 at elevated temperatures (Supplementary Figure 6) analytical scale biotransformations were performed using 1 (50 mM), 2 (10 mM) and BH32.14 (50 μM) in PBS (pH 7.4) with 20 % DMSO as a cosolvent. Enzyme solutions were pre-incubated at the required temperature (25-85 °C at 5 °C intervals) for 15 minutes prior to initiation of the reaction by addition of substrate. Reactions were quenched after 5 hours with the addition of 1 volume MeCN and analysed by HPLC (see below).
Chromatographic analysis
HPLC analysis was performed on a 1290 Infinity II Agilent LC system with a Kinetex® 5 μm XB-C18 100 Å LC Column, 50 x 2.1 mm (Phenomenex). For library screening an isocratic method using 25% acetonitrile in water at 1 mL min-1 for 1.5 minutes was used. For substrate profiling reactions, substrates and products (5a-w) were eluted over 20 minutes using a gradient of 5-95% acetonitrile in water at 1 mL min-1. Peaks were assigned by comparison to chemically synthesized standards and the peak areas were integrated using Agilent OpenLab software. The extinction coefficients used to calculate the conversion are reported in Supplementary Table 5.
Chiral analysis was performed using an SFC 1290 Infinity II Agilent system. Enantiomers of the MBH product 3 were separated using a Daicel 80S82 CHIRALPAK ® IA-3 SFC column, 3 mm, 50 mm, 3 μm, and an isocratic method with 35% methanol in CO2 at 1 mL min-1 for 1 minute. For substrate profiling reactions a range of different SFC methods were used and these are summarized in Supplementary Table 6. For MBH adducts 5c-d, j, l, n-o, r & t the major stereoisomer formed in the biotransformation eluted first; for MBH adducts 3, 5a-b, e-i, k, m, p-q, s & u-w the major stereoisomer formed in the biotransformation eluted second. Peaks were assigned by comparison to chemically synthesized standards and peak areas were integrated using Agilent OpenLabs software.
Preparative scale biotransformation
A preparative scale biotransformation was performed using 1 (50 mM), 2 (10 mM), strep-tag purified BH32.14 (50 μM) in PBS (pH 7.4, 72 mL) with 20 % DMSO (18 mL) as a cosolvent. The reaction was incubated at 30 °C with shaking at 100 r.p.m. for 19 hours. An aliquot (100 μL) was removed and quenched with acetonitrile for HPLC analysis, which showed the reaction had proceeded to 94 % conversion. The reaction mixture was extracted with ethyl acetate (2 x 400 mL), dried over MgSO4, filtered and the solvent was removed in vacuo. The crude product (Supplementary Figure 4) was purified by flash column chromatography (5:1 cyclohexane:ethyl acetate) to give 2-(hydroxy(4-nitrophenyl)methyl)cyclohex-2-en-1-one, 3 as a light yellow solid (197 mg, 88%). Spectral data is consistent with literature values. 40 δH (400 MHz, CDCl3): 8.21 (m, 2H), 7.56 (m, 2H), 6.82 (t, J = 4.1 Hz, 1H), 5.62 (d, J = 6.1 Hz, 1 H), 3.51 (d, J = 6.0 Hz, 1H), 2.46 (m, 4H), 2.03 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 200.1, 149.3, 148.1, 147.2, 140.2, 127.1, 123.5, 72.0, 38.4, 25.8, 22.3. ESI+ m/z = 270 ([M +Na]+,100)
Preparation of product standards 3 and S1 41
4-Nitrobenzaldehyde (1.5 g, 10 mmol), 2-cyclohexen-1-one (970 μL, 10 mmol) and imidazole (681 mg, 10 mmol) were stirred in 1M NaHCO3 (40 mL) and THF (10 mL) for 40 hours at room temperature. The reaction was acidified with 1M HCl and extracted with ethyl acetate (150 mL). The organic layer was dried over MgSO4, filtered and the solvent was removed in vacuo. The reaction yielded a mixture of the Morita-Baylis-Hillman adduct 3 and aldol adduct S1 which were separated by flash column chromatography (5:1 cyclohexane:ethyl acetate).
2-(hydroxy(4-nitrophenyl)methyl)cyclohex-2-en-1-one 3 (562 mg, 23%). See characterization data above.
6-(hydroxy(4-nitrophenyl)methyl)cyclohex-2-en-1-one S1 (180 mg, 7%) as a 4:1 mixture of diastereoisomers. Spectral data is consistent with literature values. 41 δH (400 MHz, CDCl3): 8.26-8.20 (m, 2H), 7.57-7.51 (m, 2H), 7.10-6.97 (m, 1H), 6.13-6.08 (m, 1H), 5.70 (d, J = 2.3 Hz, 1Hmaj), 4.99 (d, J = 8.7 Hz, 1Hmin), 4.95 (br s, OHmin), 2.95 (br s, OHmaj), 2.72-2.65 (m, 1Hmaj), 2.62-2.53 (m, 1Hmin), 2.48-2.25 (m, 2H), 2.06-1.93 (m, 1H), 1.57-1.46 (m, 1H). ESI+ m/z = 270 ([M +Na]+,100)
Preparation of 5j and inhibitor 4
Anisaldehyde (1.36 g, 10 mmol), 2-cyclohexen-1-one (970 μL, 10 mmol) and imidazole (681 mg, 10 mmol) were stirred in 1M NaHCO3 (40 mL) and THF (10 mL) 48 hours at room temperature. The reaction was acidified with 1M HCl and extracted with ethyl acetate (150 mL). The organic layer was dried over MgSO4, filtered and the solvent was removed in vacuo. The product was purified by flash column chromatography (5:1 cyclohexane:ethyl acetate) to yield 5j (440 mg, 19%). Spectral data is consistent with literature values. 42 1H NMR (400 MHz, CDCl3) δ 7.30 – 7.25 (m, 2H), 6.90 – 6.85 (m, 2H), 6.74 (t, J = 4.2 Hz, 1H), 5.51 (s, 1H), 3.80 (s, 3H), 3.35 (br s, 1H), 2.48 – 2.42 (m, 2H), 2.42 – 2.35 (m, 2H), 2.03 – 1.96 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 200.4, 158.9, 147.0, 141.1, 133.8, 127.7, 113.7, 72.0, 55.2, 38.5, 25.7, 22.5. ESI+ m/z = 255 ([M +Na]+,100) 5j (50 mg, 0.25 mmol) and acetic anhydride (50 μL) were stirred in pyridine (1 mL) overnight at room temperature. The reaction was diluted in ethyl acetate (30 mL) and washed with 1M NaHCO3 (2 x 10 mL), 10% CuSO4 (2 x 10 mL) and then brine (10 mL). The organic layer was dried over MgSO4, filtered and the solvent removed in vacuo. The product was purified by flash column chromatography (5:1 cyclohexane: ethyl acetate) to give the product (4-methoxyphenyl)(6-oxocyclohex-1-en-1-yl)methyl acetate, 4 (49 mg, 84%). 1H NMR (400 MHz, CDCl3) δ 7.30 – 7.24 (m, 2H), 6.96 (t, J = 4.3 Hz, 1H), 6.87 – 6.82 (m, 2H), 6.67 (s, 1H), 3.78 (s, 3H), 2.50 – 2.33 (m, 4H), 2.07 (s, 3H), 2.05 – 1.90 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 197.0, 169.5, 159.3, 145.3, 138.9, 130.9, 128.6, 113.7, 71.4, 55.2, 38.3, 25.7, 22.5, 21.2. ESI+ m/z = 297 ([M +Na]+,100)
Preparation of chiral standards
The enantiomers of 3 were separated by preparative chiral HPLC by Reach Separations (Nottingham) to afford (R)-3 (99.5% e.e) and (S)-3 (99.9% e.e) as white solids. The absolute stereochemistry was determined by measuring the optical rotation ((R)-3 (-52.5°) and (S)-3 (+50.0°) at 0.008 g mL-1 in DCM at 27 °C) and comparison to literature values. 43
General procedure for the preparation of racemic product standards (5a, c, e-I, k-n & p-w)
Arylaldehyde (3.3 mmol, 1.0 equiv), cyclic enone (3.3 mmol, 1.0 equiv) and imidazole (227 mg, 3.3 mmol, 1.0 equiv) were stirred in 1M NaHCO3 (13.3 mL) and THF (3.3 mL) for 24h at room temperature. The reaction was acidified with 1M HCl and extracted with ethyl acetate (3 x 50 mL). The organic layer was dried over MgSO4, filtered and the solvent removed in vacuo to give the crude product.
2-((4-nitrophenyl)(hydroxy)methyl)cyclopent-2-en-1-one (5a)
Crude product purified by flash chromatography (2:1 cyclohexane:ethyl acetate) to give the product as a yellow solid (62 mg, 8%). Spectral data is consistent with literature values. 15 1H NMR (400 MHz, CDCl3) δ 8.25-8.20 (m, 2H), 7.62-7.57 (m, 2H), 7.29 (td, J = 2.8, 1.2 Hz, 1H), 5.68 (s, 1H), 3.56 (s, 1H), 2.67-2.61 (m, 2H), 2.52-2.46 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 209.5, 159.9, 148.6, 147.9, 146.8, 127.2, 123.9, 69.3, 35.3, 26.9. ESI+ m/z = 216.0657 ([M -OH]+, 100).
5-(hydroxy(4-nitrophenyl)methyl)-2,2-dimethyl-3a,6a-dihydro-4H-cyclopenta[d][1,3]dioxol-4-one (5c)
Crude product purified by flash chromatography (3:1 cyclohexane:ethyl acetate) to give the two diastereomeric products. Stereoisomer 1 eluted first and was formed as a colourless oil (96 mg, 10%). 1H NMR (400 MHz, CDCl3) δ 8.24-8.18 (m, 2H), 7.64-7.57 (m, 2H), 7.40-7.35 (m, 1H), 5.67 (d, J = 4.2 Hz, 1H), 5.21 (dd, J = 5.4, 2.4 Hz, 1H), 4.50 (d, J = 5.4 Hz, 1H), 3.05 (d, J = 4.4 Hz, 1H), 1.40 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 201.9, 153.7, 147.9, 147.5, 147.4, 127.3, 124.0, 115.9, 77.8, 77.0, 68.6, 27.6, 26.2. ESI-m/z = 304.0844 ([M -H]-, 80), 288.08916 (27), 246.0421 (100). Stereoisomer 2 was formed as a colourless oil (75 mg, 8%). 1H NMR (400 MHz, CDCl3) δ 8.25-8.19 (m, 2H), 7.61-7.56 (m, 2H), 7.27-7.24 (m, 1H), 5.69 (m, 1H), 5.22 (ddd, J = 5.5, 2.4, 1.1 Hz, 1H), 4.55 (d, J = 5.4 Hz, 1H), 1.42 (s, 3H), 1.38 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 201.7, 153.8, 147.8, 147.0, 147.0, 127.3, 123.9, 115.7, 77.7, 76.8, 68.5, 27.5, 26.1. ESI-m/z = 304. 0842 ([M -H]-, 85), 288.0892 (33), 246.0423 (100).
2-((4-bromophenyl)(hydroxy)methyl)cyclohex-2-en-1-one (5e)
Crude product was purified by flash chromatography (4:1 cyclohexane:ethyl acetate) to give the product as a white solid (128 mg, 14%). Spectral data is consistent with literature values. 15 1H NMR (400 MHz, CDCl3) δ 7.49-7.44 (m, 2H), 7.25-7.21 (m, 2H), 6.73 (t, J = 4.2 Hz, 1H), 5.50 (s, 1H), 2.56-2.28 (m, 4H), 2.09-1.89 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 200.5, 147.7, 140.9, 140.8, 131.6, 128.3, 121.5, 72.3, 38.7, 25.9, 22.6. ESI+ m/z = 265.0070 ([M 81Br -OH]+, 100), 263.0090 ([M 79Br -OH]+, 91).
2-((4-chlorophenyl)(hydroxy)methyl)cyclohex-2-en-1-one (5f)
Crude product purified by flash chromatography (4:1 cyclohexane:ethyl acetate) to give the product as a white solid (114 mg, 15 %). Spectral data is consistent with literature values. 15 1H NMR (400 MHz, CDCl3) δ 7.30-7.22 (m, 4H), 6.74 (t, J = 4.3 Hz, 1H), 5.48 (s, 1H), 3.46 (br s, 1H), 2.45-2.32 (m, 4H), 1.95 (app quint, J = 6.3 Hz, 2H). 13C NMR (101 MHz, CDCl3) δ 200.4, 147.6, 140.8, 140.4, 133.2, 128.5, 127.9, 71.8, 38.5, 25.8, 22.5. ESI+ m/z = 221.0564 ([M 37Cl -OH]+, 30), 219.0591 ([M 35Cl -OH]+, 100).
2-((4-cyanophenyl)(hydroxy)methyl)cyclohex-2-en-1-one (5g)
Crude product purified by flash chromatography (3:1 cyclohexane:ethyl acetate) to give the product as a colourless oil (115 mg, 15%). Spectral data is consistent with literature values. 44 1H NMR (400 MHz, CDCl3) δ 7.62 (d, J = 8.3 Hz, 2H), 7.48 (d, J = 8.1 Hz, 2H), 6.79 (t, J = 4.2 Hz, 1H), 5.55 (d, J = 5.9 Hz, 1H), 3.51 (d, J = 5.9 Hz, 1H), 2.48-2.38 (m, 4H), 2.05-1.96 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 200.1, 148.0, 147.5, 140.4, 132.5, 127.1, 118.9, 111.3, 72.0, 38.5, 25.8, 22.5. ESI+ m/z = 210.0922 ([M -OH]+, 100).
2-((4-(trifluoromethyl)phenyl)(hydroxy)methyl)cyclohex-2-en-1-one (5h)
Crude product purified by flash chromatography (4:1 cyclohexane:ethyl acetate) to give the product as a colourless oil (196 mg, 22%). Spectral data is consistent with literature values. 45 1H NMR (400 MHz, CDCl3) δ 7.58 (d, J = 8.1 Hz, 2H), 7.47 (d, J = 8.2 Hz, 2H), 6.77 (t, J = 4.3 Hz, 1H), 5.57 (d, J = 5.4 Hz, 1H), 3.59 (d, J = 5.7 Hz, 1H), 2.48-2.37 (m, 4H), 2.04-1.95 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 200.4, 147.9, 145.9, 140.7, 129.6 (q, J = 32.3 Hz), 126.8, 125.4 (q, J = 3.8 Hz), 122.8, 72.4, 38.6, 25.9, 22.6. ESI+ m/z = 253.0853 ([M - OH]+, 100).
2-(hydroxy(p-tolyl)methyl)cyclohex-2-en-1-one (5i)
Crude product purified by flash chromatography (3:1 cyclohexane:ethyl acetate) to give the product as a colourless oil (112mg, 16%). Spectral data is consistent with literature values. 46 1H NMR (400 MHz, CDCl3) δ 7.24 (d, J = 8.1 Hz, 2H), 7.15 (d, J = 7.9 Hz, 2H), 6.74 (t, J = 4.2 Hz, 1H), 5.52 (s, 1H), 3.36 (br s, 1H), 2.48 – 2.42 (m, 2H), 2.42 – 2.35 (m, 2H), 2.34 (s, 3H), 2.04 – 1.95 (m, 2H). 13C NMR (126 MHz, CDCl3) δ 200.6, 147.4, 141.3, 138.8, 137.3, 129.2, 126.5, 72.6, 38.8, 25.9, 22.7, 21.3. ESI+ m/z = 239.1050 ([M +Na]+, 16), 199.1125 ([M -OH]+, 100).
2-((3-fluoro-4-nitrophenyl)(hydroxy)methyl)cyclohex-2-en-1-one (5k)
Crude product purified by flash chromatography (4:1 cyclohexane:ethyl acetate) to give the product as a white solid (53 mg, 6%). 1H NMR (400 MHz, CDCl3) δ 8.02 (dd, J = 8.5, 7.5 Hz, 1H), 7.34 (dd, J = 11.8, 1.8 Hz, 1H), 7.31-7.23 (m, 1H), 6.87 (t, J = 4.2 Hz, 1H), 5.54 (s, 1H), 2.54-2.37 (m, 4H), 2.02 (app quint, J = 6.2 Hz, 2H). 13C NMR (101 MHz, CDCl3) δ 200.1, 155.7 (d, J = 265.2 Hz), 151.6 (d, J = 7.7 Hz), 148.7, 139.9, 136.4 (d, J = 7.2 Hz), 126.2 (d, J = 2.7 Hz), 122.2 (d, J = 3.8 Hz), 116.2 (d, J = 21.9 Hz), 71.8 (d, J = 1.4 Hz), 38.5, 25.9, 22.5. ESI+ m/z = 248.0697 ([M -OH]+, 100).
2-((2-fluoro-4-nitrophenyl)(hydroxy)methyl)cyclohex-2-en-1-one (5l)
Crude product purified by flash chromatography (4:1 cyclohexane:ethyl acetate) to give the product as a white solid (90 mg, 10%). 1H NMR (400 MHz, CDCl3) δ 8.08 (dd, J = 8.6, 1.9 Hz, 1H), 7.89 (dd, J = 9.8, 2.2 Hz, 1H), 7.83-7.77 (m, 1H), 6.78-6.72 (m, 1H), 5.81 (d, J = 6.7 Hz, 1H), 3.92 (d, J = 6.3 Hz, 1H), 2.55-2.37 (m, 4H), 2.08-1.93 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 200.7, 159.0 (d, J = 251.0 Hz), 148.5 (d, J = 1.8 Hz), 148.1, 138.6, 136.5 (d, J = 13.3 Hz), 129.2 (d, J = 4.4 Hz), 119.5 (d, J = 3.7 Hz), 111.2 (d, J = 26.9 Hz), 67.6 (d, J = 2.5 Hz), 38.5, 25.9, 22.4. ESI+ m/z = 248.0717 ([M -OH]+, 100).
2-((3-fluoro-4-bromophenyl)(hydroxy)methyl)cyclohex-2-en-1-one (5m)
Crude product purified by flash chromatography (4:1 cyclohexane:ethyl acetate) to give the product as a colourless oil (43 mg, 4%). 1H NMR (400 MHz, CDCl3) δ 7.48 (dd, J = 8.3, 7.0 Hz, 1H), 7.16-7.11 (m, 1H), 7.01 (dd, J = 8.2, 2.0 Hz, 1H), 6.79 (t, J = 4.1, 1H), 5.46 (s, 1H), 3.50 (br s, 1H), 2.49-2.35 (m, 4H), 2.06-1.92 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 200.3, 159.1 (d, J = 247.4 Hz), 147.9, 144.1 (d, J = 6.2 Hz), 140.4, 133.3, 123.3 (d, J = 3.3 Hz), 114.7 (d, J = 23.1 Hz), 107.8 (d, J = 20.9 Hz), 71.7 (d, J = 1.6 Hz), 38.6, 25.9, 22.5. ESI+ m/z = 282.9955 ([M81Br -OH]+, 95), 280.9975 ([M79Br -OH]+, 100).
2-((2-fluoro-4-bromophenyl)(hydroxy)methyl)cyclohex-2-en-1-one (5n)
Crude product purified by flash chromatography (4:1 cyclohexane:ethyl acetate) to give the product as a colourless oil (68 mg, 7%). 1H NMR (400 MHz, CDCl3) δ 7.46-7.40 (m, 1H), 7.32 (dd, J = 8.4, 1.9 Hz, 1H), 7.20 (dd, J = 9.7, 1.9 Hz, 1H), 6.71-6.66 (m, 1H), 5.74 (s, 1H), 3.80 (s, 1H), 2.50-2.35 (m, 4H), 2.05-1.93 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 200.8, 159.5 (d, J = 251.1 Hz), 147.8, 139.3, 129.6 (d, J = 4.7 Hz), 127.9 (d, J = 13.3 Hz), 127.7 (d, J = 3.6 Hz), 121.6 (d, J = 9.5 Hz), 118.9 (d, J = 24.9 Hz), 67.1 (d, J = 2.9 Hz), 38.6, 25.9, 22.5. ESI+ m/z = 282.9962 ([M81Br -OH]+, 99), 280.9981 ([M79Br -OH]+, 100).
2-((3-nitrophenyl)(hydroxy)methyl)cyclohex-2-en-1-one (5p)
Crude product purified by flash chromatography (4:1 cyclohexane:ethyl acetate) to give the product as a colourless oil (132 mg, 16%). Spectral data consistent with literature values. 15 1H NMR (400 MHz, CDCl3) δ 8.21 – 8.18 (m, 1H), 8.09 (dd, J = 8.3, 1.2 Hz, 1H), 7.70 (d, J = 7.6 Hz, 1H), 7.49 (t, J = 7.9 Hz, 1H), 6.86 (t, J = 4.2 Hz, 1H), 5.58 (d, J = 5.7 Hz, 1H), 3.67 (d, J = 5.8 Hz, 1H), 2.48 – 2.37 (m, 4H), 2.00 (app quint, J = 6.3 Hz, 2H). 13C NMR (101 MHz, CDCl3) δ 200.2, 148.4, 148.2, 144.4, 140.3, 132.7, 129.3, 122.5, 121.4, 71.9, 38.5, 25.9, 22.5. ESI+ m/z = 230.08221 ([M -OH]+, 100).
2-((3-bromophenyl)(hydroxy)methyl)cyclohex-2-en-1-one (5q)
Crude product purified by flash chromatography (4:1 cyclohexane:ethyl acetate) to give the product as a colourless oil (67 mg, 7%). Spectral data is consistent with literature values. 15 1H NMR (400 MHz, CDCl3) δ 7.55-7.47 (m, 1H), 7.42-7.36 (m, 1H), 7.31-7.26 (m, 1H), 7.20 (t, J = 7.8 Hz, 1H), 6.76 (t, J = 4.2 Hz, 1H), 5.50 (d, J = 5.3 Hz, 1H), 3.48 (d, J = 5.6 Hz, 1H), 2.48-2.38 (m, 4H), 2.04-1.96 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 200.4, 147.9, 144.3, 140.7, 130.7, 130.0, 129.6, 125.2, 122.7, 72.2, 38.6, 25.9, 22.6. ESI+ m/z = 265.0051 ([M 81Br -OH]+, 100), 263.0070 ([M 79Br -OH]+, 94).
2-((3-chlorophenyl)(hydroxy)methyl)cyclohex-2-en-1-one (5r)
Crude product purified by flash chromatography (4:1 cyclohexane:ethyl acetate) to give the product as a colourless oil (152 mg, 20%). Spectral data consistent is with literature values. 15 1H NMR (400 MHz, CDCl3) δ 7.37-7.34 (m, 1H), 7.30-7.21 (m, 3H), 6.75 (t, J = 4.2 Hz, 1H), 5.51 (d, J = 5.6 Hz, 1H), 3.46 (d, J = 5.6 Hz, 1H), 2.49-2.38 (m, 4H), 2.05-1.97 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 200.5, 147.9, 143.9, 140.7, 134.5, 129.7, 127.8, 126.7, 124.8, 72.3, 38.7, 25.9, 22.6. ESI+ m/z = 221.0549 ([M 37Cl -OH]+, 27), 219.0575 ([M 35Cl -OH]+, 100).
4-(hydroxy(6-oxocyclohex-1-en-1-yl)methyl)benzaldehyde (5s)
Crude product purified by flash chromatography (3:1 cyclohexane:ethyl acetate) to give the product as a colourless oil (183 mg, 24%). 1H NMR (400 MHz, CDCl3) δ 9.96 (s, 1H), 7.82 (d, J = 8.0 Hz, 2H), 7.51 (d, J = 7.9 Hz, 2H), 6.80 (t, J = 4.2 Hz, 1H), 5.58 (d, J = 5.5 Hz, 1H), 3.67 (d, J = 5.6 Hz, 1H), 2.50-2.30 (m, 4H), 2.07-1.90 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 200.2, 192.1, 148.9, 147.9, 140.6, 135.7, 129.9, 127.0, 72.2, 38.5, 25.9, 22.5. ESI+ m/z = 213.0923 ([M -OH]+, 20), 185.0976 ([M -OH -CO]+, 100).
5-(hydroxy(6-oxocyclohex-1-en-1-yl)methyl)thiophene-2-carbaldehyde (5t)
Crude product purified by flash chromatography (2:1 cyclohexane:ethyl acetate) to give the product as a yellow solid (73 mg, 9%). 1H NMR (400 MHz, CDCl3) δ 9.84 (s, 1H), 7.63 (d, J = 3.8 Hz, 1H), 7.08 (dd, J = 3.8, 1.0 Hz, 1H), 6.99 (t, J = 4.2 Hz, 1H), 5.66 (m, 1H), 3.01 (br s, 1H), 2.54-2.40 (m, 4H), 2.13-1.96 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 200.3, 183.1, 158.1, 148.5, 142.9, 139.5, 136.7, 125.4, 70.5, 38.6, 25.9, 22.5. ESI+ m/z = 237.0596 ([M + H]+, 33), 219.0491 ([M -OH]+, 100), 191.0540 ([M -OH -CO]+, 21).
5-(hydroxy(6-oxocyclohex-1-en-1-yl)methyl)furan-2-carbaldehyde (5u)
Crude product purified by flash chromatography (2:1 cyclohexane:ethyl acetate) to give the product as a yellow solid (98 mg, 13%). 1H NMR (400 MHz, CDCl3) δ 9.55 (s, 1H), 7.20 (d, J = 3.6 Hz, 1H), 6.99 (t, J = 4.1 Hz, 1H), 6.53 (d, J = 3.6 Hz, 1H), 5.51 (s, 1H), 2.53-2.36 (m, 4H), 2.02 (app quint, J = 6.2 Hz, 2H). 13C NMR (101 MHz, CDCl3) δ 200.2, 177.7, 161.8, 152.3, 149.4, 137.4, 122.9, 109.8, 68.2, 38.4, 25.9, 22.4. ESI+ m/z = 243.0642 ([M +Na]+, 78), 203.0716 ([M -OH]+, 100), 175.0765 ([M -OH -CO]+, 87).
4-(hydroxy(6-oxocyclohex-1-en-1-yl)methyl)thiophene-2-carbaldehyde (5v) and 5-(hydroxy(6-oxocyclohex-1-en-1-yl)methyl)thiophene-3-carbaldehyde (5w)
Regioisomers 5v and 5w were produced in the same reaction and separated by flash chromatography (3:1 cyclohexane:ethyl acetate). The isomers were assigned based on the chemical shifts of closely related regioisomeric structures.47-48 5v was obtained as a yellow oil (10 mg, 2%). 1H NMR (400 MHz, CDCl3) δ 9.88 (d, J = 1.2 Hz, 1H), 7.69 (d, J = 1.5 Hz, 1H), 7.64 (m, 1H), 6.86 (t, J = 4.2 Hz, 1H), 5.57 (s, 1H), 2.54-2.39 (m, 4H), 2.09-1.96 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 200.49, 183.12, 147.70, 145.20, 144.2, 140.23, 135.10, 131.11, 69.70, 38.65, 25.89, 22.59. ESI+ m/z = 219.0490 ([M -OH]+, 100). 5w was obtained as a yellow oil (23 mg, 3%). 1H NMR (400 MHz, CDCl3) δ 9.81 (s, 1H), 8.02 (d, J = 1.3 Hz, 1H), 7.28-7.24 (m, 1H), 6.96 (t, J = 4.0 Hz, 1H), 5.63 (br s, 1H), 2.54-2.38 (m, 4H), 2.08-1.99 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 200.5, 185.3, 149.7, 148.4, 142.8, 139.4, 136.9, 121.4, 70.2, 38.6, 25.9, 22.5. ESI+ m/z = 219.0491 ([M -OH]+, 100), 191.0540 ([M -OH -CO]+, 15).
Synthesis of 3-(hydroxy(4-nitrophenyl)methyl)-5,6-dihydro-2H-pyran-2-one (5b)
4-Nitrobenzaldehyde (151 mg, 1.0 mmol, 1.0 equiv) and 5,6-dihydro-pyran-2-one (0.1 mL, 1.1 mmol, 1.1 equiv) were stirred under nitrogen in dichloromethane (4.0 mL) and cooled to 0 °C. A solution of diethylaluminium iodide (0.9 M solution in toluene, 1.3 mL, 1.2 mmol, 1.2 equiv) was added dropwise and the resulting brown mixture was stirred for 24h at room temperature. The reaction was quenched with saturated NaHCO3 (2.0 mL). The dichloromethane layer was separated and the aqueous layer was extracted with dichloromethane (3 x 10 mL). The combined organic layers were dried over MgSO4, filtered and the solvent was removed in vacuo. The crude product was purified by flash column chromatography (1:1 cyclohexane:ethyl acetate) to give the product as an orange solid (23 mg, 9%). Spectral data is consistent with literature values. 24 1H NMR (400 MHz, CDCl3) δ 8.27-8.15 (m, 2H), 7.64-7.54 (m, 2H), 6.77 (t, J = 4.3 Hz, 1H), 5.66 (d, J = 4.9 Hz, 1H), 4.44-4.33 (m, 2H), 3.63 (d, J = 5.5 Hz, 1H), 2.62-2.47 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 164.5, 148.3, 147.6, 141.8, 134.2, 127.5, 123.8, 71.8, 66.5, 24.3. ESI-m/z = 248.0613 ([M -H]-, 100).
Synthesis of 3-(hydroxy(4-nitrophenyl)methyl)but-3-en-2-one (5d)
4-Nitrobenzaldehyde (604mg, 4.0 mmol, 1.0 equiv), DABCO (291 mg, 2.6 mmol, 0.65 equiv), but-3-en-2-one (433 μL, 5.2 mmol, 1.3 equiv) were added to dichloromethane (8 mL) and stirred for 20h at room temperature. The reaction mixture was diluted in dichloromethane (50 mL) and washed with 10% HCl (2 x 30 mL). The organic layer was dried over MgSO4, filtered and the solvent was removed in vacuo. The crude product was purified by flash column chromatography (2:1 cyclohexane:ethyl acetate) to give the product as a white solid (133mg, 15%). Spectral data is consistent with literature values. 49 1H NMR (400 MHz, CDCl3) δ 8.22 – 8.18 (m, 2H), 7.57 – 7.53 (m, 2H), 6.26 (d, J = 1.1 Hz, 1H), 6.03 (d, J = 1.1 Hz, 1H), 5.68 (s, 1H), 2.36 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 200.2, 149.1, 149.0, 147.5, 127.9, 127.4, 123.8, 72.5, 26.5. ESI+ m/z = 220.0714 ([M -H]-, 100), 204.07552 (25).
Preparative biotransformation for the synthesis of 3-(2,2-dimethyl-4-oxo-3α,6α-dihydro-4H-cyclopenta[δ][1,3]dioxol-5-yl)-3-hydroxyindolin-2-one (5o)
A preparative scale biotransformation (10 mL) was performed using (3αS,6αS)-2,2-dimethyl-3α,6α-dihydro-4H-cyclopenta[δ][1,3]dioxol-4-one (50 mM), isatin (10 mM), His-tag purified BH32.8 (60 μM) in PBS (pH 7.0, 8 mL) with DMSO (2 mL) as a cosolvent. The reaction was incubated at 30 °C with shaking at 180 r.p.m. for 1.5 hours. The reaction mixture was extracted with ethyl acetate (2 x 15 mL), dried over MgSO4, filtered and the solvent removed in vacuo. The crude product was purified by flash chromatography (2:1 cyclohexane:ethyl acetate) to give two diastereomeric products. Stereoisomer 1 eluted first as a yellow oil (10 mg, 33%). 1H NMR (400 MHz, MeOD) δ 7.86 (d, J = 2.5 Hz, 1H), 7.25 (t, J = 7.8 Hz, 1H), 7.12 (d, J = 7.4 Hz, 1H), 6.96 (t, J = 7.5 Hz, 1H), 6.91 (d, J = 7.8 Hz, 1H), 5.31 (dd, J = 5.5, 2.5 Hz, 1H), 4.44 (d, J = 5.5 Hz, 1H), 1.37 (s, 3H), 1.34 (s, 3H). 13C NMR (101 MHz, MeOD) δ 202.2, 178.6, 156.9, 147.6, 143.9, 131.2, 131.0, 125.4, 123.6, 116.2, 111.4, 79.4, 78.2, 75.5, 28.2, 27.0. ESI+ m/z = 324.0842 ([M +Na]+, 100). Stereoisomer 2 eluted second as a yellow oil (3 mg, 10%). 1H NMR (400 MHz, MeOD) δ 7.86 (d, J = 2.4 Hz, 1H), 7.27 (t, J = 7.7 Hz, 1H), 7.10 (d, J = 7.4 Hz, 1H), 6.98 (t, J = 7.5 Hz, 1H), 6.92 (d, J = 7.7 Hz, 1H), 5.32 (dd, J = 5.4, 2.5 Hz, 1H), 4.47 (d, J = 5.4 Hz, 1H), 1.36 (s, 3H), 1.30 (s, 3H). 13C NMR (101 MHz, MeOD) δ 201.3, 179.1, 156.7, 147.3, 143.8, 131.3, 130.9, 125.2, 123.6, 116.2, 111.6, 79.3, 78.3, 75.4, 28.3, 26.7. ESI+ m/z = 324.0844 ([M +Na]+, 100).
Crystallization, refinement and model building
Crystals of BH32.6, BH32.7 and BH32.12 variants were prepared by mixing 200 nL of 20 mg mL−1 protein in PBS buffer pH 7.4 with equal volumes of precipitant. All trials were conducted by sitting-drop vapour diffusion and incubated at 4 °C. Protein crystallization conditions are given in Supplementary Table 8. BH32.6 and BH32.7 crystals were flash cooled in liquid nitrogen whilst BH32.12 crystals were first cryoprotected by the addition of 10 % PEG 200 to the mother liquor prior to flash cooling. Data were collected from single crystals at Diamond Light Source and subsequently scaled and reduced with Xia2. 50 Preliminary phasing was performed by molecular replacement in Phaser using a search model derived from wild-type BH32 (PDB code: 3U26). Iterative cycles of rebuilding and refinement were performed in COOT 51 and Phenix.refine, 52 respectively. Structure validation with MolProbity and PDBREDO were integrated into the iterative rebuild and refinement process. Complete data collection and refinement statistics can be found in Supplementary Table 7. Coordinates and structure factors have been deposited in the Protein Data Bank under accession numbers 6Z1K, 7O1D & 6Z1L. All figures and surface representations were generated in ICM Pro (Molsoft).
Molecular Docking
To generate starting structures for DFT modelling, molecular docking was performed with Autodock Vina, 53 using AutoDockTools 54 to assign hydrogen atoms to the crystal structure and generate input files. The protein was kept rigid during docking but all relevant substrate bonds were rotatable. For the product, the highest-scoring pose was selected. For the reactant state, 2-cyclohexen-1-one was docked first and the pose which showed the best overlap with the corresponding fragment of the docked product was selected (this had an estimated binding energy only 0.2 kcal mol-1 higher than the lowest energy pose, which was very similar but rotated around the keto group). The aldehyde was then docked into the 2-cyclohexen-1-one bound structure, and again the binding mode with the best overlap to the corresponding fragment in the docked product was selected (this had an estimated binding energy of 0.8 kcal mol-1 higher than the lowest binding mode).
DFT Modelling
A cluster model was constructed using the docked conformations for the substrates, including the first-shell amino acids around the substrates (Trp10, Ile14, Ala19, Val22, His23, Ile26, Leu64, Trp88, Ser91, Leu92, Ala95, Arg124, Phe128, Ser129, Phe132). Residues were truncated at the Cβ unless their backbone atoms are in near contact with the substrates or involved in hydrogen bonding with the substrates or other amino acids in the cluster model. A water molecule was included in the calculation to facilitate the third chemical step (Int2 to Int3). The models contained a total of 273 atoms as shown in Supplementary Figure 8. For DFT modelling of the inhibitor 4-enzyme complex, the energy minimised product state was modified to remove the water molecule and to introduce the acetoxy group present in inhibitor 4, followed by energy minimization. The coordinates for all energy minimized ground and transition state structures are provided as supplementary files. Calculations were performed using Gaussian16 revision A03, 55 using the B3LYP functional 56 and the 6-31G(d,p) basis sets 57–58 for all atoms except for the Arg124 guanidino nitrogens and the C=O groups of 2-cyclohexen-1-one and aldehyde, for which 6-311+G(d,p) was used. Grimme’s D3 dispersion correction with Becke-Johnson damping was employed 59 and implicit solvation was treated using the polarizable conductor calculation model (CPCM) 60–61 with a dielectric constant ε = 5.7 to mimic the enzyme environment. 62–63 For comparison, single-point energy calculations were then performed with ε = 80. All models have a net charge of +1 (from Arg124) and spin multiplicity 1. Sixteen peripheral atoms were kept fixed during the calculations: the Cα of residues whose backbone was included, and Cβ for residues truncated at Cβ (atom numbers 16, 26, 31, 36, 43, 52, 54, 57, 60, 71, 77, 85, 89, 97, 104, 245). Each chemical step was modelled by performing a relaxed potential energy scan of the making/breaking bond for steps 1, 2 and 4. For step 3 (the proton transfer) a simple reaction coordinate, z = R(C-H) - R(O-H) was scanned where R is distance, C and H are the carbon and hydrogen atoms of the breaking bond and O is the oxygen atom of the water molecule. In each case, the transition state was selected as the structure with the highest potential energy along the scanned coordinate, to within ±0.03 Å along the breaking/forming bond or z.
Supplementary Material
Acknowledgements
We acknowledge the Biotechnology and Biological Sciences Research Council (David Phillips Fellowship BB/M027023/1 to A.P.G.), the European Research Council (ERC Starter Grant, no. 757991 to A.P.G.) and the UK Research and Innovation Council (Future Leader Fellowship MR/T041722/1 to S.L.L.). We thank the Faculty of Science and Engineering (University of Manchester) for the award of a Presidential Fellowship to S.L.L. A.E.C. was supported by a BBSRC Industrial CASE PhD studentship (BB/S507040/1) supported by GSK. We are grateful to Diamond Light Source for time on beamlines i03 and i04 under proposals MX17773-33 & MX17773-74, and to Manchester SYNBIOCHEM Centre (BB/M017702/1), the Future Biomanufacturing Hub (EP/S01778X/1) and the Henry Royce Institute for Advanced Materials (funded through EPSRC grants EP/R00661X/1, EP/S019367/1, EP/P025021/1 and EP/P025498/1) for access to their facilities, and to M. Dunstan (Manchester Institute of Biotechnology) for guidance on automating directed evolution workflows. We thank R. Spiess and R. Sung (Manchester Institute of Biotechnology) for acquiring protein mass spectra and for assistance in HPLC method development, and Reach Separations (Nottingham) for supplying individual enantiomers of the MBH adduct 3. The authors acknowledge the assistance given by IT Services and the use of the Computational Shared Facility at The University of Manchester.
Footnotes
Author Contributions
R.C carried out molecular biology, protein production, purification, crystallization & kinetic characterization, and directed evolution experiments. A.E.C and R.C carried out organic synthesis and substrate profiling of BH32 variants. R.C., A.J.B. and A.E.C. developed spectrophotometric assays and performed enzyme-inhibition experiments. L.J. and S.H. carried out molecular docking and DFT calculations. S.H. interpreted and analysed kinetic data. C.L. interpreted, analysed and presented structural data. D.B. provided the BH32 design model. All authors discussed the results and participated in writing the manuscript. A.P.G. and S.L directed the research.
Competing Interests
The authors declare no competing interests.
Data Availability Statement
Coordinates and structure factors have been deposited in the Protein Data Bank under accession numbers 6Z1K, 7O1D and 6Z1L. The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary Information files.
References
- 1.Hilvert D. Design of protein catalysts. Annu Rev Biochem. 2013;82:447–470. doi: 10.1146/annurev-biochem-072611-101825. [DOI] [PubMed] [Google Scholar]
- 2.Röthlisberger D, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 3.Jiang L, et al. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel JB, et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science. 2010;329:309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eiben CB, et al. Increased Diels-Alderase activity through backbone remodelling guided by Foldit players. Nat Biotechnol. 2012;30:190–192. doi: 10.1038/nbt.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomberg R, et al. Precision is essential for efficient catalysis in an evolved Kemp eliminase. Nature. 2013;503:418–421. doi: 10.1038/nature12623. [DOI] [PubMed] [Google Scholar]
- 7.Obexer R, et al. Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase. Nat Chem. 2017;9:50–56. doi: 10.1038/nchem.2596. [DOI] [PubMed] [Google Scholar]
- 8.Gouverneur VE, et al. Control of the exo and endo pathways of the Diels-Alder reaction by antibody catalysis. Science. 1993;262:204–208. doi: 10.1126/science.8211138. [DOI] [PubMed] [Google Scholar]
- 9.Stewart JD, Benkovic SJ. Transition-state stabilization as a measure of the efficiency of antibody catalysis. Nature. 1995;375:388–391. doi: 10.1038/375388a0. [DOI] [PubMed] [Google Scholar]
- 10.Barbas CF, III, et al. Immune versus natural selection: antibody aldolases with enzymic rates but broader scope. Science. 1997;278:2085–2092. doi: 10.1126/science.278.5346.2085. [DOI] [PubMed] [Google Scholar]
- 11.Basavaiah D, Rao AJ, Satyanarayana T. Recent advances in the Baylis-Hillman reaction and applications. Chem Rev. 2003;103:811–892. doi: 10.1021/cr010043d. [DOI] [PubMed] [Google Scholar]
- 12.Basavaiah D, Reddy BS, Badsara SS. Recent contributions from the Baylis-Hillman Reaction to Organic Chemistry. Chem Rev. 2010;9:5447–5674. doi: 10.1021/cr900291g. [DOI] [PubMed] [Google Scholar]
- 13.Wei Y, Shi M. Recent advances in organocatalytic asymmetric Morita-Baylis-Hillman/aza-Morita-Baylis-Hillman Reactions. Chem Rev. 2013;113:6659–6690. doi: 10.1021/cr300192h. [DOI] [PubMed] [Google Scholar]
- 14.Metrano AJ, et al. Asymmetric catalysis mediated by synthetic peptides, version 2.0: expansion of scope and mechanisms. Chem Rev. 2020;120:11479–11615. doi: 10.1021/acs.chemrev.0c00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi M, Liu X-G. Asymmetric Morita-Baylis-Hillman reaction of arylaldehydes with 2-cyclohexen-1-one and 2-cyclopenten-1-one catalyzed by chiral bis(thio)urea and DABCO. Org Lett. 2008;10:1043–1046. doi: 10.1021/ol7028806. [DOI] [PubMed] [Google Scholar]
- 16.Nakayama Y, Gotanda T, Ito K. Asymmetric Morita-Baylis-Hillman reactions of 2-cyclohexen-1-one catalyzed by chiral biaryl-based bis(thiourea) organocatalysts. Tetrahedron Lett. 2011;52:6234–6237. [Google Scholar]
- 17.Islam Z, Strutzenberg TS, Gurevic I, Kohen A. Concerted versus stepwise mechanism in thymidylate synthase. J Am Chem Soc. 2014;136:9850–9853. doi: 10.1021/ja504341g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reetz MT, Mondière R, Carballeira JD. Enzyme promiscuity: first protein-catalyzed Morita-Baylis-Hillman reaction. Tetrahedron Lett. 2007;48:1679–1681. [Google Scholar]
- 19.López-Iglesias M, Busto E, Gotor V, Gotor-Fernández V. Use of protease from Bacillus licheniformis as promiscuous catalyst for organic synthesis: applications in C-C and C-N bond formation reactions. Adv Synth Catal. 2011;353:2345–2353. [Google Scholar]
- 20.Joshi PN, Purushottam L, Das NK, Mukherjee S, Rai V. Protein self-assembly induces promiscuous nucleophilic biocatalysis in Morita-Baylis-Hillman (MBH) reaction. RSC Adv. 2016;6:208–211. [Google Scholar]
- 21.Bjelic S, et al. Computational design of enone-binding proteins with catalytic activity for the Morita-Baylis-Hillman reaction. ACS Chem Biol. 2013;8:749–757. doi: 10.1021/cb3006227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Prod Nat Acad Sci. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol. 2009;19:596–604. doi: 10.1016/j.sbi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Aggarwal VK, Emme I, Fulford SY. Correlation between pK a and reactivity of quinuclidine-based catalysts in the Baylis−Hillman reaction: Discovery of quinuclidine as optimum catalyst leading to substantial enhancement of scope. J Org Chem. 2003;3:692–700. doi: 10.1021/jo026671s. [DOI] [PubMed] [Google Scholar]
- 25.Karur S, Hardin J, Headley A, Li G. A novel approach to Morita-Baylis-Hillman (MBH) lactones via the Lewis acid-promoted couplings of α,β-unsaturated lactone with aldehydes. Tettrahedron Lett. 2003;44:2991–2994. [Google Scholar]
- 26.Phillips MA, Fletterick R, Rutter WJ. Arginine 127 stabilizes the transition state in carboxypeptidase. J Biol Chem. 1990;265:20692–20698. [PubMed] [Google Scholar]
- 27.Studer S, et al. Evolution of a highly active and enantiospecific metalloenzyme from short peptides. Science. 2018;362:1285–1288. doi: 10.1126/science.aau3744. [DOI] [PubMed] [Google Scholar]
- 28.Janda KD, Schloeder D, Benkovic SJ, Lerner RA. Induction of an antibody that catalyzes the hydrolysis of an amide bond. Science. 1988;241:1188–1191. doi: 10.1126/science.3413482. [DOI] [PubMed] [Google Scholar]
- 29.Thayer MM, et al. Structural basis for amide hydrolysis catalyzed by the 43C9 antibody. J Mol Biol. 1993;291:329–345. doi: 10.1006/jmbi.1999.2960. [DOI] [PubMed] [Google Scholar]
- 30.Roberts VA, Stewart J, Benkovic SJ, Getzoff ED. Catalytic antibody model and mutagenesis implicate arginine in transition-state stabilization. J Mol Biol. 1994;235:1098–1116. doi: 10.1006/jmbi.1994.1060. [DOI] [PubMed] [Google Scholar]
- 31.Doyle AG, Jacobsen EN. Small-molecule H-bond donors in asymmetric catalysis. Chem Rev. 2007;107:5713–5743. doi: 10.1021/cr068373r. [DOI] [PubMed] [Google Scholar]
- 32.Taylor MS, Jacobsen EN. Asymmetric catalysis by chiral hydrogen-bond donors. Angew Chem Int Ed. 2006;45:1520–1543. doi: 10.1002/anie.200503132. [DOI] [PubMed] [Google Scholar]
- 33.Amarante GW, et al. Brønsted acid catalyzed Morita-Baylis-Hillman reaction: a new mechanistic view for thioureas revealed by ESI‐MS(/MS) monitoring and DFT calculations. Chem Eur J. 2009;15:12460–12469. doi: 10.1002/chem.200900966. [DOI] [PubMed] [Google Scholar]
- 34.Knowles RR, Lin S, Jacobsen EN. Enantioselective thiourea-catalyzed cationic polycyclizations. J Am Chem Soc. 2010;132:5030–5032. doi: 10.1021/ja101256v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park Y, et al. Macrocyclic bis-thioureas catalyze stereospecific glycosylation reactions. Science. 2017;355:162–166. doi: 10.1126/science.aal1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.St-Jacques AD, Eyahpaise M-EC, Chica RA. Computational design of multisubstrate enzyme specificity. ACS Catal. 2019;9:5480–5485. [Google Scholar]
- 37.Davey JA, Damry AM, Goto NK, Chica RA. Rational design of proteins that exchange on functional timescales. Nat Chem Biol. 2017;13:1280–1285. doi: 10.1038/nchembio.2503. [DOI] [PubMed] [Google Scholar]
- 38.Lee TS, et al. BglBrick vectors and datasheets: A synthetic biology platform for gene expression. J Biol Eng. 2011;5:12. doi: 10.1186/1754-1611-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kille S, et al. Reducing codon redundancy and screening effort of combinatorial protein libraries created by saturation mutagenesis. ACS Synth Biol. 2013;2:83–92. doi: 10.1021/sb300037w. [DOI] [PubMed] [Google Scholar]
- 40.Luo S, Wang PG, Cheng J-P. Remarkable rate acceleration of imidazole-promoted Baylis-Hillman reaction involving cyclic enones in basic water solution. J Org Chem. 2004;69:555–558. doi: 10.1021/jo035345p. [DOI] [PubMed] [Google Scholar]
- 41.Kataoka T, Iwama T, Tsujiyama S, Iwamura T, Watanabe S. The Chalcogeno-Baylis-Hillman reaction: a new preparation of allylic alcohols from aldehydes and electron-deficient alkenes. Tetrahedron. 1998;54:11813–11824. [Google Scholar]
- 42.Vazquez-Chavez J, et al. Effect of chiral N-substituents with methyl and trifluoromethyl groups on the catalytic performance of mono- and bifunctional thioureas. Org Biomol Chem. 2019;17:10045–10051. doi: 10.1039/c9ob01893j. [DOI] [PubMed] [Google Scholar]
- 43.Wang F, et al. A highly efficient kinetic resolution of Morita-Baylis-Hillman adducts achieved by N-Ar axially chiral Pd-complexes catalyzed asymmetric allylation. Chem Commun. 2011;47:12813–12815. doi: 10.1039/c1cc15543a. [DOI] [PubMed] [Google Scholar]
- 44.Kwong CK-W, Huang R, Zhang M, Shi M, Toy PH. Bifunctional polymeric organocatalysts and their application in the cooperative catalysis of Morita-Baylis-Hillman reaction. Chemistry. 2007;13:2369–2376. doi: 10.1002/chem.200601197. [DOI] [PubMed] [Google Scholar]
- 45.Li G, Wei H-X, Gao JJ, Caputo TD. TiCl4-mediated Baylis-Hillman and aldol reactions without the direct use of a Lewis base. Tetrahedron Lett. 2000;41:1–5. [Google Scholar]
- 46.Yang J, et al. Endohedral functionalized cage as a tool to create frustrated lewis pairs. Angew Chem Int Ed. 2018;57:14212–14215. doi: 10.1002/anie.201808291. [DOI] [PubMed] [Google Scholar]
- 47.Venable JD, et al. Preparation and Biological Evaluation of Indole, Benzimidazole, and Thienopyrrole Piperazine Carboxamides:? Potent Human Histamine H4 Antagonists. J Med Chem. 2005;48:8289–8298. doi: 10.1021/jm0502081. [DOI] [PubMed] [Google Scholar]
- 48.Comins DL, Killpack MO. Lithiation of heterocycles directed by-amino alkoxides. J Org Chem. 1987;52:104–109. [Google Scholar]
- 49.Coelho F, et al. Ultrasound in Baylis-Hillman reactions with aliphatic and aromatic aldehydes: scope and limitations. Tetrahedron. 2002;58:7437–7447. [Google Scholar]
- 50.Winter G, et al. DIALS: implementation and evaluation of a new integration package. Acta Crystallogr D. 2018;74:85–97. doi: 10.1107/S2059798317017235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystalogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–67. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morris GM, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–91. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frisch MJ, et al. Gaussian 16 Rev A03. Wallingford CT: 2016. [Google Scholar]
- 56.Becke AD. Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 57.Hehre W, Ditchfield R, Pople J. Further extensions of gaussian-type basis sets for use in molecular orbital studies of organic molecules. J Chem Phys. 1972;56:2257–2261. [Google Scholar]
- 58.Francl M, et al. Self-consistent molecular orbital methods. XXIII. A polarization-type basis set for 2nd-row elements. J Chem Phys. 1982;77:3654–3665. [Google Scholar]
- 59.Grimme S, Ehrlich S, Goerigk L. Effect of the damping function in dispersion corrected density functional theory. J Comput Chem. 2011;32:1456–65. doi: 10.1002/jcc.21759. [DOI] [PubMed] [Google Scholar]
- 60.Barone V, Cossi M. Quantum calculation of molecular energies and energy gradients in solution by a conductor solvent model. J Phys Chem. 1998;102:1995–2001. [Google Scholar]
- 61.Cossi M, Rega N, Scalmani G, Barone V. Energies, structures, and electronic properties of molecules in solution with the C‐PCM solvation model. J Comput Chem. 2003;24:669–681. doi: 10.1002/jcc.10189. [DOI] [PubMed] [Google Scholar]
- 62.Shaik S, Kumar D, de Visser SP, Altun A, Thiel W. Theoretical perspective on the structure and mechanism of cytochrome P450 enzymes. Chem Rev. 2005;105:2279–2328. doi: 10.1021/cr030722j. [DOI] [PubMed] [Google Scholar]
- 63.Heyes DJ, Sakuma M, de Visser SP, Scrutton NS. Nuclear quantum tunneling in the light-activated enzyme protochlorophyllide oxidoreductase. J Biol Chem. 2009;284:3762–3767. doi: 10.1074/jbc.M808548200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors have been deposited in the Protein Data Bank under accession numbers 6Z1K, 7O1D and 6Z1L. The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary Information files.