Abstract
Macrophages derived from human induced pluripotent stem cells (iPSC) have the potential to enable the development of cell-based therapies for numerous disease conditions. We here provide a detailed protocol for the mass-production of iPSC-derived macrophages (iPSC-Mac) in scalable suspension culture on an orbital shaker or in stirred-tank bioreactors (STBRs). This strategy is straightforward, robust and characterized by the differentiation of primed iPSC-aggregates into so called “myeloid-cell-forming-complex (MCFC)” intermediates by means of a minimal cytokine cocktail. In contrast to the “batch-like differentiation approaches” established for other iPSC-derived lineages, MCFC-intermediates are stably maintained in suspension culture and continuously generate functional and highly pure iPSC-Mac. Employing a culture volume of 120 ml in the STBR platform, ~1-4 × 107 iPSC-Mac can be harvested at weekly intervals for several months. The STBR technology allows for real-time monitoring of crucial process parameters such as the biomass, pH, dissolved oxygen, as well as nutrition levels; the system also promotes systematic process development, optimization, and linear upscaling. The process duration, starting with the expansion of iPSC until the first iPSC-Mac harvest is 28 days. Successful application of the protocol requires expertise in pluripotent stem cell culture, differentiation and analytical methods, such as flow cytometry. Fundamental know-how in biotechnology is also advantageous to run the process in the STBR platform. The continuous, scalable production of well-defined iPSC-Mac populations is highly relevant to various fields ranging from developmental biology, immunology and cell therapies, to industrial applications for drug safety and discovery.
Keywords: iPSC, macrophages, hematopoiesis, suspension culture, bioreactor, upscaling, regenerative medicine
Introduction
Human iPSC-derivatives represent an attractive source of cells for regenerative medicine paving the way for the clinical translation of novel cell therapy products. To meet the growing demand for therapeutic iPSC-derived effector cells, efficient upscaling technologies are of central interest and 3D, suspension-based differentiation culture platforms, which are applicable to linear upscaling, have been introduced for several iPSC-derived cell types 1 . However, suitable systems for the scalable production of iPSC-derived hematopoietic cells are limited to a minority of hematopoietic cell types, and although clinically relevant numbers of iPSC-derived NK cells 2 or platelets 3 can be produced, methods to generate other hematopoietic derivatives from human iPSC are still lacking.
Within the hematopoietic system, one cell type of specific current interest are macrophages, including tissue-resident macrophages (TRMs), which are critically involved in organ function, tissue homeostasis and immune protection. While macrophages represent the first line of cellular immunity and respond to pathogen invasion with phagocytosis and activation of the immune response, they are also important for the termination of immune responses, tissue repair and homeostasis 4 . Moreover, each TRM population displays a unique gene expression profile and fulfills important organ-specific functions such as alveolar macrophages which break down surfactant material in the lungs 5 or Kupffer cells critically contributing to iron and cholesterol homeostasis in the liver 6 . Therefore, the absence or dysfunction of macrophages is associated with a variety of diseases ranging from immunodeficiencies, autoimmunity, neurodegenerative disorders, metabolic diseases to cancer 7 ; further highlighting macrophages as a highly interesting target for drug discovery and the development of novel cell-based therapies.
Given the broad field of applied macrophage research, advanced differentiation systems are needed to generate application-specific quantities of well-defined, functional macrophages. However, as of yet it remains challenging to derive these cells in suitable quantities and qualities 8 . Besides limitations of the availability of suitable healthy blood donors and patients, also the number of monocytes that can be isolated from a single apheresis is limited. Moreover, the pronounced batch-to-batch variability between monocyte-derived macrophages derived from different donors remains an obstacle for macrophage-based research in particular for potential cell-based therapies. In addition, monocytes/ macrophages derived from peripheral blood may not properly reflect the primitive origin of TRMs and thus may not represent an ideal model system to study TRM biology 9 . In this regard, human iPSCs offer an attractive cell source for the well-defined production of functional macrophages overcoming these restrictions in basic and applied research 9,10 .
Over the past years, a plethora of differentiation protocols to generate iPSC-derived macrophages (iPSC-Mac) have been described. The resulting cells have been shown to fulfill important functional requirements such as phagocytosis and antimicrobial activity in vitro, and, importantly, revealed the potential to adapt to a tissue-specific environment in vivo using immunodeficient mouse models 11,12 . As a current drawback however, most of these differentiation protocols are performed in 2D adherent-based, non-controlled cultivation platforms, which have substantial limitations for process upscaling and systematic optimization, limiting cell availability in a broader context.
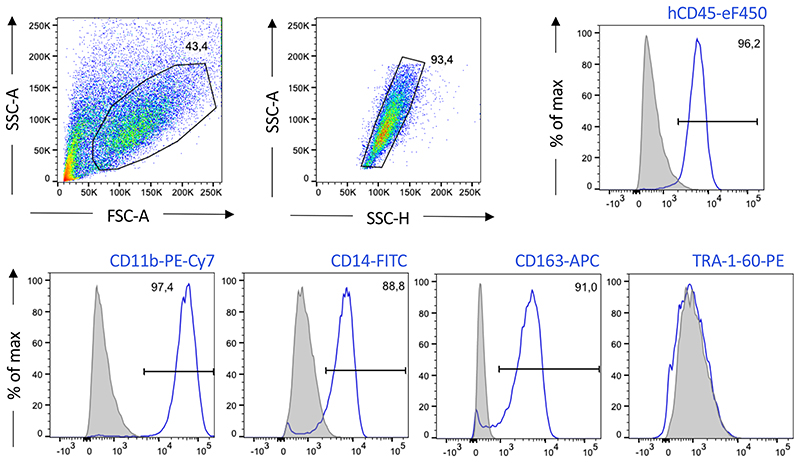
To overcome the current limitations for the derivation of myeloid lineages from human PSC, we recently established a strategy for the generation of iPSC-Mac using a scalable differentiation technique in 3D suspension culture. This approach is applicable to a broad spectrum of conventional culture vessels in laboratory scale as well as to industry-compatible STBR systems 13 . Here, we present a detailed step-by-step protocol describing the differentiation of iPSCs into “myeloid cell forming complexes (MCFC)” which can be maintained in suspension culture long-term to continuously produce iPSC-Mac. We detail the most critical steps to successfully apply this approach either in tissue culture plates agitated on orbital shakers or in STBR systems, thereby covering culture scales from 3 ml up to 150 ml. IPSC-Mac generated by these approaches represent a highly pure population characterized by a specific surface marker profile of CD45+/CD11b+/CD14+/CD163+/TRA-1-60- and display critical in vitro and in vivo functionality.
Development of the protocol
The emerging understanding of the in vivo functions and utility of TRMs recently promoted the use of in vitro generated iPSC-derived macrophages to study macrophage development, biology, and their role in the pathophysiology of certain diseases. A large number of recent studies employing iPSC-Mac have utilized protocols that are initiated by either an undirected differentiation or mesoderm priming of human iPSC, followed by a directed hematopoietic specification phase and allow for the continuous generation of macrophages with minimal cytokine supplementation (Interleukin 3 (IL-3), Macrophage colony-stimulating factor (M-CSF) and/or Granulocyte-macrophage colony-stimulating factor (GM-CSF)). The first description of this technique dates back to 2008, when Karlson and colleagues used human embryonic stem cells (ESCs) to generate macrophages 14 . Further advancements such as the use of defined, serum-free differentiation media have progressed this approach 15 . We made use of this system and established a flexible differentiation platform, which allows for the continuous production of iPSC-derived macrophages or granulocytes from “myeloid cell forming complex (MCFC)” intermediates, depending on the choice of cytokines 16 . Meanwhile, this protocol has been used in a number of studies related to hematopoietic development 17 or disease modeling of (rare) diseases such as chronic granulomatous disease (CGD) 18 , pulmonary alveolar proteinosis (PAP) 19 , mendelian susceptibility to mycobacterial disease (MSMD) 20,21 and others 22-26 . However, until recently, most approaches have been limited in scalability as they rely on a 2D adherent hematopoietic specification step(s), hampering the transition to GMP-compatible, iPSC-Mac production at a scale able to produce sufficient cells for clinical applications.
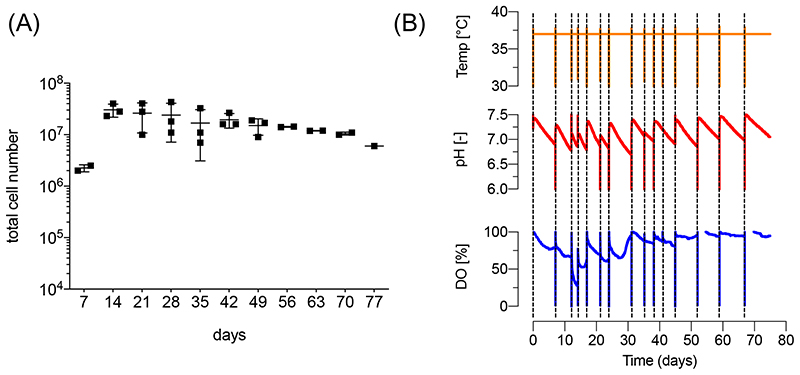
The backlog in manufacturing hematopoietic cell lineages is in clear contrast to the progress in other areas of iPSC bioprocessing, including the efficient high-density suspension culture possible for the pluripotent state 27,28 as well as the directed differentiation into functional mesodermal lineages such as cardiomyocytes 29,30 and endothelial cells 31 as well as endodermal derivatives 32 in controlled and scalable bioreactor systems. To close this gap, we aimed at adapting the advantages of controlled bioreactor technology to the derivation of myeloid cell types, while maintaining the unique feature of continuous cell production. As a first step, we established the differentiation process in small scale suspension culture using multi well-plates placed on an orbital shaker. We found that this simple-to-adapt approach is fully compatible with the formation of MCFCs, leading to the continuous production of iPSC-Mac with comparable efficiencies to the previously applied static 2D cultures. We consequently progressed to an STBR platform in 120 ml process scale. After fine tuning of the shear forces resulting from the impeller stirring, we were able to maintain MCFC intermediates in the bioreactor process and could demonstrate the continuous production of iPSC-Mac for up to 8 weeks 13 . Importantly, the resulting iPSC-Mac show high phenotypic and functional similarities to macrophages generated from peripheral blood. However, on the transcriptome level, these cells display a more primitive expression pattern in line with other studies 33 suggesting their suitability to study TRM biology. As a starting point for developing potential clinical applications, we demonstrated that the adoptive pulmonary transfer of bioreactor-derived iPSC-Mac reduced bacterial load and ameliorated disease pathophysiology in murine models for pulmonary Pseudomonas aeruginosa 13 or Staphylococcus aureus infections 34 .
Applications of the protocol
Macrophages are of central interest for various fields such as developmental biology or disease modeling studies. iPSC-Mac generated by our technique may also have potential for novel cell-based therapy strategies e.g. replacing (malfunctioning) tissue-resident macrophage populations 11,12 or immunotherapy approaches targeting infectious diseases 13 and beyond.
Given the high plasticity of macrophages, we hypothesize that the cells can also be directed to differentiate to numerous tissue-specific macrophage populations in vitro and in vivo thereby extending the application from lung diseases to other organs. For example iPSC-derived microglia were generated by transplantation of iPSC-Mac into the mouse brain by Fattorelli and colleagues 35 . Given the prominent role of microglia in the pathophysiology of neurodegenerative diseases, initial attempts have been made to develop a cell therapy approach for Metachromatic leukodystrophy (MLD) 36 suggesting future application of iPSC-Mac as a direct therapeutic intervention might be feasible. Our own data demonstrate similar plasticity of iPSC-derived macrophages generated with our platform 11 , suggesting cells generated by this protocol would facilitate the generation and study of various different TRM populations in vitro and in vivo. The field of macrophage-based cell therapeutics has been continuously growing over the past years, offering numerous opportunities for the application of iPSC-Mac. A potential future application of iPSC-Mac was introduced by S. Forbes and colleagues, who used adoptive transfer of autologous monocyte-derived macrophages to treat liver cirrhosis using a single dose of 107, 108 or up to 109 macrophages, depending on the yield of the apharesis 37,38 . Given the versatile roles of macrophages in cellular immunity, similar monocyte-derived macrophage transfer regimens are underway to target cancers 39 , and also to reduce immunosuppression in the field of solid organ transplantation, to treat chronic inflammatory conditions 40 or autoimmune diseases. As all these attempts rely on the use of defined macrophage populations, the technology of iPSC-Mac may represent an attractive tool to meet the growing demand for these cells.
In addition the possibility to introduce a plethora of genetic elements to iPSC and thereof derived macrophages (such as e.g. the conditional activation of a specific transcription factor that alters the macrophage phenotype) would allow the production of well-defined cell products for specific applications 41,42 . This off-the-shelf production could potentially overcome dose-limitations set by the availability of cells from an apheresis. The high quality and reproducibility of the cell product generated with our protocol also enables the iPSC-iMac to be used in drug development or (high throughput) drug screening.
Of note, while the production of macrophages is the focus of our work, our STBR-bases differentiation strategy can also be used to generate large quantities of other myeloid immune cells by simply switching the applied cytokine cocktail in the hematopoietic specification and production phase of the process (Figure 1). For example, iPSC-derived granulocytes can be generated when MCFCs are exposed to IL-3 and G-CSF 13,43 . Moreover, the continuous production of iPSC-derived erythrocytes using IL-3, SCF and EPO has been demonstrated in 2D cultures 44 . These results support the idea that a myeloid progenitor population develops and resides within the MCFC. This view is also supported by the observation that the cultivation of MCFCs with IL-3 only leads to the generation of CD45+/CD11b+/CD14-/CD163- myeloid progenitor cells, which can be further differentiated into macrophages, granulocytes or erythrocytes when exposed to specific lineage instructive cytokines 43 . Given this broad myeloid differentiation capacity, we propose that our differentiation strategy could also be applicable to generate e.g. iPSC-derived megakaryocytes or dendritic cells, thereby highlighting the overall potential and modular character of this seminal differentiation platform 16 .
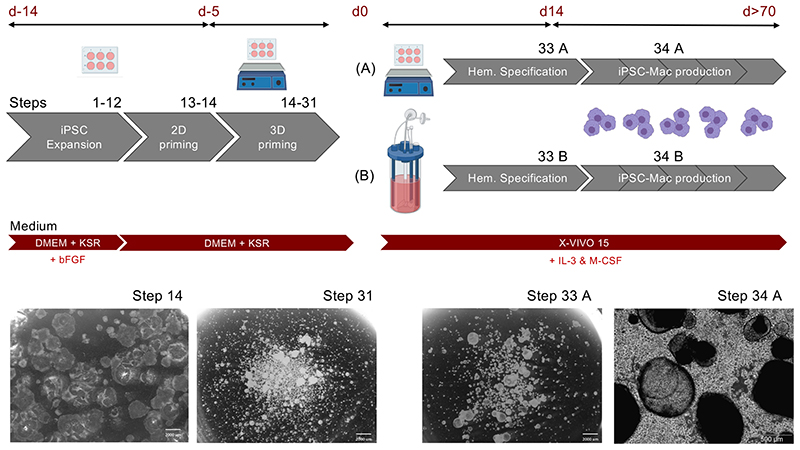
Figure 1. Schematic representation of the differentiation platform.
Human iPSC are expanded in 2D adherent cultures using DMEM+KSR+bFGF (“Full iPSC medium”), followed by a 2D priming phase in DMEM+KSR (“minimal iPSC medium), both performed on murine embryonic fibroblasts (MEFs) without intermediate passaging. Subsequently, fragmented colonies are transferred to 3D suspension culture in a 6-well plate on an orbital shaker for 3D priming in minimal iPSC medium. After 5 days, the largest primed aggregates are selected by defined sedimentation and transferred (A) to a new 6-well suspension culture plate and maintained on the orbital shaker in a culture volume of 3 ml/well or (B) to a stirred tank bioreactor in a culture volume of 120 ml. By cultivation of primed aggregates in X-VIVO15 medium supplemented with IL-3 and M-CSF for 14 days (Hematopoietic specification) myeloid cell forming complexes (MCFCs) develop. From day 14 onwards, iPSC-derived macrophages can be harvested in parallel to the medium change, while MCFCs remain in the process. Abbreviations: iPSC: Induced pluripotent stem cell, DMEM: Dulbecco’s Modified Eagle’s Medium, KSR: Knock our serum replacement, bFGF: basic fibroblast growth factor, IL-3: Interleukin 3, M-CSF: Macrophage colony stimulating factor. Scale bars: 2000 μm/ magnification 6.1x and 500 μm, magnification 40x.
Comparison with other methods
Most hematopoietic differentiation protocols for human pluripotent stem cells (PSCs) are either based on co-culture systems with murine stromal cells or the formation of primed 3D-aggregate structures, (historically) often referred to as embryoid bodies (EBs). Co-culture based protocols often include an intermediate step of generating stem/progenitor cells that are purified from the differentiation cultures and can be further directed into the target cells i.e. macrophages 45 . As an alternative, other protocols rely on the genetic manipulation of PSC to foster the hematopoietic program and accelerate the generation of hematopoietic target cells 46,47 . However, these systems lack scalability due to the required purification steps and/or adherence-based cultivation conditions. In contrast, our 3D-suspension STBR based technique provides a scalable and continuous production platform for the generation of highly pure iPSC-derived macrophages with in vitro and in vivo functionality. Moreover, this protocol is robust and can be used to generate several macrophage harvests, thus increasing the overall cell yields compared to alternative individual differentiation approaches. If bioreactor technology is used, this allows the monitoring and control of numerous process parameters as well as supports the development of automated and continuous feeding strategies that facilitate upscaling into larger bioreactor platforms 28 .
Experimental design
As schematically presented in Figure 1, our protocol starts with the cultivation of undifferentiated iPSC on murine embryonic fibroblasts (MEFs). For differentiation, we typically expand the culture to several 6-well tissue culture plates and culture the iPSC for 5 days in “full iPSC-medium” (see Reagent Setup). After 5 days, the medium is changed to “minimal iPSC-medium” (2D pre-priming). After 9 days, we detach the iPSC colonies by enzymatic treatment with collagenase IV (day -5), followed by transition into suspension plates on an orbital shaker in priming medium for 5 days (d 0) to induce undirected differentiation and mesoderm formation (3D priming). Subsequently, primed iPSC aggregates are selected by defined sedimentation, transferred to X-VIVO15 medium, and subsequently differentiation cultures are continued in (A) a suspension plate on an orbital shaker or (B) a stirred tank bioreactor (Figure 1). Hematopoietic specification in the defined differentiation medium X-VIVO15 supplemented only with IL-3 and M-CSF leads to the differentiation of primed aggregates into MCFCs (hematopoietic specification), which can be maintained in the process and start to produce iPSC-derived macrophages from day 14 onwards.
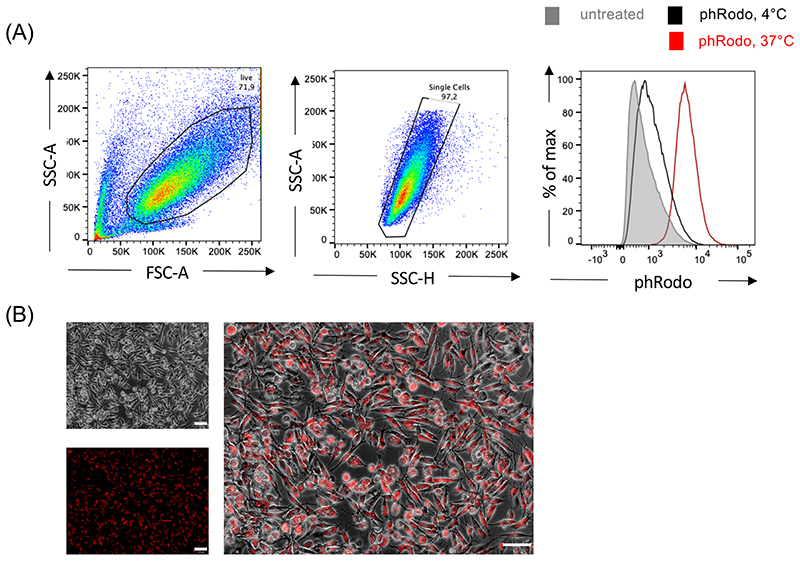
The iPSC-Mac can be harvested in the “macrophage production phase” from the medium in parallel to the weekly medium change after controlled sedimentation of the MCFCs. When undertaking small scale differentiation, one well of a 6-well tissue culture plate (culture volume 3 ml) yields approx. 0.4-1x106 iPSC-Mac/week, whereas in the bioreactor platform in 120 ml culture volume the generation of approx. 10-40x106 iPSC-Mac/week is expected; and the harvest, irrespectively of the process scale, can be continued for up to 11 weeks. Importantly, the harvested cell population retains a high purity so there is no need for further enrichment strategies. The cells maintain functionality in vitro and in vivo. Flow cytometry-based analysis of CD45, CD11b, CD14 and CD163 expression enables monitoring of the identity of the generated iPSC-Macs. Macrophage specific functions such as the phagocytosis of fluorescence labeled bioparticles (e.g. pHrodo™ Red E. coli BioParticles™ (ThermoFisher Scientific)) should also be tested in a functional control stage of quality assurance. A 2D terminal differentiation phase in tissue culture plates can be added if experimentally required for downstream applications.
Expertise needed to implement the protocol
All key steps of the small-scale protocol are easy-to-handle and can be implemented by a trained cell scientist. In contrast, set-up, handling and operation of the bioreactor platform requires specific expertise in bioprocessing. If basic understanding of the bioreactor system-specific properties is required, technical support can be requested from the bioreactor system vendor. It is important users understand how to assemble the reactor, maintain sterility, calibrate the sensors and handle the reactor vessel under a sterile hood. After gaining familiarity with these technical features, a scientist trained in cell culture technologies should be able to successfully follow and apply the detailed, step-wise procedures described in this protocol.
Current limitations
Fully defined, xeno-free culture conditions have gained increasing attention as they represent an important prerequisite for the envisioned clinical translation of iPSC-based cell therapies. While our protocol employs a defined and potentially GMP-compatible differentiation medium (X-VIVO15), the first steps of iPSC maintenance and expansion described here are based on murine embryonic fibroblasts and “classical” iPSC medium containing complex serum replacement supplement. However, former studies using an adherent-based approach of this differentiation strategy 33,48 as well as own unpublished data suggest that the protocol can also be implemented when using feeder free culture conditions. Yet, when using feeder free cultures as a starting point, supplementation of mesoderm priming cytokines such as BMP4, SCF and VEGF seems to be necessary to ensure differentiation. Given the heterogeneity seen between the proliferation and differentiation potential of different iPSC lines, the protocol may require further line-specific adaptations.
Another practical issue is that when using this protocol iPSC differentiate as multi-cellular aggregates. This makes assessment of the exact cell numbers present at each differentiation stage challenging as precise cell counts cannot easily be obtained. We provide detailed microscopic pictures of every process step to facilitate the monitoring of experimental progress. Recent studies show that the directed differentiation of iPSCs in 3D e.g. to generate tissue-specific organoids, can closely resemble key aspects of native organ development 49 . Our MCFCs also recapitulate important steps of embryonic hematopoietic development 17 and apparently seem to maintain a progenitor cell population that can give rise to macrophages over a prolonged culture period. Thus, it seems likely that differentiation as multi-cellular aggregates is required to generate macrophages long term in culture.
Although this protocol includes in-process monitoring of dissolved oxygen (DO) and pH when the STBR platform is used, this can be further advanced by establishing feedback-based control loops to keep these parameters stable. This can be achieved by perfusion-based feeding (enabled by the STBR technology), allowing for the continuous media replacement and the addition of supplements without process interruption. Applying this technique for expanding human PSCs at the pluripotent state, we recently demonstrated improved process efficiency, resulting in 10-fold higher cell yields (per given process scale) and the parallel reduction of (costly) media consumption by 75% 28 . Moreover, implementing monitoring and controlling key metabolites such as the glucose-, lactate- and glutamine-concentrations, which can be specifically tailored to the needs of hematopoietic cells, could potentially stabilize the differentiation process, increase cell yields, and prolong the stable production of macrophages (and other blood cell types generated by an adapted version of this protocol). Such optimization might be required to enable generation of sufficient cells for treatment of patients. For example cell doses of 107-109 macrophages per patient are currently used to treat liver cirrhosis 38 . Given the current process efficiency, weekly production of 108-109 iPSC-Mac seems feasible if the protocol is applied on a production scale of 0.6-6 l. Such production scales are generally feasible using currently available GMP-compliant bioreactor technologies. In addition to further upscaling, cryopreservation of iPSC-Mac is an option to enable larger cell doses to be collected and pooled from repetitive harvests. Freezing enables the required quality control of cell products to be undertaken fully prior to use of the product. Whereas cryopreservation of macrophages has been proven challenging, some recent publications describe the efficient cryopreservation of macrophages, including from iPSC 50 . Ensuring STBR-derived iPSC-Mac can be cryopreserved will be an important future step required for clinical translation. As an alternative to cryopreservation, short term storage of iPSC-Mac in a spinner flask has recently been described 48 , which may be included in this protocol alongside other strategies that enable iPSC-Mac to be stored for limited periods of time.
Materials
Biological Materials
Human pluripotent stem cells
We used two human induced pluripotent stem cell lines derived from mobilized peripheral blood CD34+ cells (hCD34iPSC11 51 /MHHi015-B: https://hpscreg.eu/cell-line/MHHi015-B) and hCD34iPSC16 19 /MHHi015-A: https://hpscreg.eu/cell-line/MHHi015-A) to establish this protocol, which can be obtained from our laboratory upon request. CRITICAL Both of the iPSC lines used to establish this protocol were derived from female donors. In principle, any human iPSC line, regardless of the origin, reprogramming method, sex or donor age can be used in this protocol. Using the small-scale version of the protocol, we and collaborators efficiently used iPSC lines from either male and female donors, which were derived from fibroblast or hematopoietic cells, and which have been reprogrammed either by lenti- or Sendai viral systems 11,17,20,41,52-54 . However, as cell production efficiencies can vary between iPSC lines, we recommend evaluating several lines whilst establishing the protocol.
CRITICAL The iPSC lines used should undergo regular quality control such as karyotype analysis or Comparative Genomic Hybridization (aCGH), mycoplasma test, as well as flow cytometric analysis for expression of pluripotency markers (e.g. TRA-1-60, SSEA-4) 19,51 .
! CAUTION Before working with human PSCs, ensure National regulations will be followed, required institutional ethics and review board approval is obtained and any individual funding agency requirements are adhered to.
CAUTION The generation and the use of human iPSCs described here was approved by the local ethical committee (Approval numbers: 1427-2012, 2127-2014, 1303-2012).
Murine Embryonic Fibroblast
The growth of undifferentiated iPSCs in culture was supported by murine embryonic fibroblast (MEF) which were derived from CF-1 strain. Suitable primary MEFs can be purchased from Merck (EmbryoMax® Primary Mouse Embryonic Fibroblasts, Cat. # PMEF-CFL-P1). Further information about seeding, expanding and irradiating MEFs can be found in the Reagent set up section entitled “Preparation of irradiated murine embryonic fibroblasts”.
Reagents
Cell culture components
KnockOut-DMEM (1×), 500 ml (Thermo Fisher Scientific_Gibco, cat. no. 10829-018)
RPMI 1640 Medium (1×), 500 ml (Thermo Fisher Scientific_Gibco, cat. no. 21875-034)
X-VIVO 15 medium, 500 ml (Lonza, cat.no. BE02-060F)
DMEM, low glucose, DMEM 1g/l D-Glucose; 500 ml; (Thermo Fisher Scientific_Gibco, cat. no. 31885-023)
KnockOut™ Serum Replacement, 500 ml (Thermo Fisher Scientific_Gibco, cat no. 10828-028)
Fetal Bovine Serum (FBS), 500 ml (Merck, cat. no. S0615)
DPBS (1×) w/o CaCl2, MgCl2; pH 7.0 - 7.3, 500 ml (Thermo Fisher Scientific_Gibco, cat. no.14190-094)
Ethylenediamine tetraacetic acid disodium salt dehydrate (EDTA), 250g (Carl Roth, cat. no.8043.1)
Rho Kinase Inhibitor (RI) Y-27632 (Tocris, cat. no. 1254)
L-Glutamin 200 mM (100×), 100 ml (Thermo Fisher Scientific_Gibco, cat. no. 25030-024)
Albumin Fraction V, 50g (Carl Roth, cat. no. 8076.2)
Non-essential amino acids (100×), MEM-NEAA, 100ml (Thermo Fisher Scientific_Gibco, cat. no.11140-035)
-
2-Mercaptoethanol 50 mM, 10 ml (Thermo Fisher Scientific_Gibco, cat. no. 31350-010)
! CAUTION This reagent is toxic.
Penicillin Streptomycin,100 ml (Thermo Fisher Scientific_Gibco, cat. no. 15140-122)
Collagenase type IV, 1g (Thermo Fisher Scientific_Gibco, cat. no. 17104-019)
Trypsin-EDTA (0.05%), phenol red (Thermo Fisher Scientific_Gibco, cat. no. 25300062)
Ampuwa, sterile pyrogen-free water, 1000 ml (Fresenius Kabi, cat. no. 108813)
Cytokines
Recombinant Human M-CSF (Peprotech cat.no. 300-25)
Recombinant Human IL-3 (Peprotech cat.no. 200-03)
Recombinant Human FGF-basic (bFGF) (Peprotech, cat.no. 100-18B)
CRITICAL: In our lab, most of the differentiations were performed using cytokines purchased from Peprotech. As it is likely there are differences in cytokine activity amongst cytokines produced by different vendors, we recommend Peprotech cytokines when following this protocol. Of note, cytokines purchased from Miltenyi Biotec have been applied in successful differentiation runs as well, using the same concentrations, so can be used as an alternative.
Equipment
15 and 50 ml conical tubes (Sarstedt, cat. nos. 62.554.502 and 62.547.254)
5, 10, 25 and 50 ml serological pipettes (Sarstedt, cat nos. 86.1253.001, 86.1254.001, 86.1685.001, and 86.1689.001)
Serological Pipette Controller - Pipetus (Hirschmann Laborgeräte, cat. no. 9907200)
1000 μl, 200 μl, 20 μl, 10 μl sterile filter pipette tips (Sarstedt cat. nos. 70.3050.255, 70.760.211, 70.760.213, 70.1130.210)
6-well tissue culture dishes (Biochrom, cat-no. P 92406) and (NUNC, cat.no. 140675)
Glass beads, Ø1.7-2.1mm (Carl Roth, cat.no. A556.1)
PluriStrainer 70μm (PluriSelect, cat. no. 43-50070-01)
0,22 μm Vacuum Filtration “rapid”-Filtermaxx 250 ml (TPP, cat. no. P 99250)
Syringe filters 0,22 μm (TPP, cat. no. P 99722)
Glass Pasteur pipette (sterilized at 180 °C for >3 h; Brand, cat. no. 747715)
Disposable syringes, Omnifix 5 ml with Luer-lock (B. Braun Melsungen, cat. no. 4617053V)
Disposable syringes, Omnifix 50 ml with Luer-lock (B. Braun Melsungen, cat. no. 8728810F)
Technical Buffer pH 4.01 (WTW, cat. no. 108 800)
Technical Buffer pH 7.00 (WTW, cat. no. 108 802)
Hamilton Storage Solution, 500 ml (Hamilton, cat. No. 238931)
Terg-a-zyme® enzyme detergent pack (Sigma-Aldrich, cat. no. Z273287-1EA)
Orbital shaker Celltron (Infors, cat. no. 6922)
Humidified incubator, model MCO-18AIC, 37 °C, 5% CO2 (SANYO, cat. no. SA-MCO18)
Water bath GFL TYP 1083 (Omnilab, cat. no. DE 67770231)
Cell culture cabinet/laminar flow, HERAsafe KS 12 (Thermo Scientific, cat. no. 107922482
Benchtop centrifuge, Heraeus Multifuge 3SR+ (Thermo Scientific, cat. no. 75004371)
DASbox Mini bioreactor system for cell culture applications (Eppendorf, cat. no. 76DX04CC)
DASbox Mini bioreactor vessel for cell culture applications (Eppendorf, cat. no. 76DS0250ODSS)
DASware® control (Eppendorf, cat. no. 78600167)
Stereo microscope (Leica S9i, SZX16-ILLT, Olympus)
Inverted fluorescence & phase contrast microscope (IX71, Olympus)
Hemocytometer, Neubauer improved (Carl Roth, cat. no. T728.1)
Sampling valve (Eppendorf, cat. no. 78200077)
Mohr pinchcock clamp (Carl Roth, cat. no. KY01.1)
DASGIP Dip Tube Pg 13.5, 220 mm (Eppendorf, cat. no. 76DGDT220)
Female Luer-lock connector (Carl Roth, cat. no. CT62.1)
Male Luer-lock plug (Carl Roth, cat. no. CT70.1)
Silicone tubing, inner diameter (i.d.) 1.0 mm, outer diameter (o.d.) 3.0 mm (Carl Roth, cat. no. HC61.1) hose reduction piece (Carl Roth, cat. no. CT46.1)
Silicone tubing, inner diameter (i.d.) 4.8 mm (Watson-Marlow, cat. no. 913.A048.024)
DASbox exhaust system (Eppendorf, cat. no. 76DXOFF)
DASbox exhaust condenser, Peltier (Eppendorf, cat. no. 76DXCOND)
Polytetrafluoroethylene (PTFE) membrane inline vent filter; pore size 0.2 μm (mdi Membrane Technologies, cat. no. ITFX0801BBXX109)
EasyFerm Bio PHI K8 120 (Hamilton, cat. no. 243632-1513)
OxyFerm FDA 225, 215 mm (Hamilton, cat. no. 237452)
DASGIP compression fitting, for outer diameter (o.d.) 12 mm, w/Pg 13.5 male thread (Eppendorf, cat. no. 76DGCF12)
DASbox overhead drive (Eppendorf, cat. no. 76DXOHD)
Impeller: Pitched-Blade Impeller, 8-blade, 60° pitch, stainless steel, O.D. 34 mm, I.D. 5 mm (Eppendorf, cat. no. 78100604)
Holding Sleeve, for 8-blade impeller, stainless steel with set screw, I.D. 8 mm, for shaft with O.D. 5 mm (Eppendorf, cat. no. 78100595)
Sigmacote (Sigma-Aldrich, cat. no. SL2)
Reagent Setup
Minimal iPSC Medium
To prepare 500 ml minimal iPSC medium, add the following supplements to 384 ml KnockOut DMEM: 100 ml Knockout Serum Replacement (final concentration 20% vol/vol), 5 ml L-glutamine (final concentration 2 mM), 5 ml nonessential amino acid (final concentration 1% vol/vol), 5 ml Penicillin/Streptomycin (final concentration 1% vol/vol), and 1 ml β-mercaptoethanol (final concentration 0.2% vol/vol). Filter the medium using 0.22 μm Stericup vacuum filtration system. iPSC medium can be stored at 4 °C for up to two weeks.
Full iPSC Medium
To prepare full iPSC medium, add bFGF to minimal iPSC medium to give a final concentration of 10 ng/ml. Full iPSC medium can be stored at 4 °C for up to 1 week.
CRITICAL To ensure the activity of bFGF over time, avoid repeated warming of the bFGF iPSC medium.
MEF Medium
To prepare MEF medium, combine 450 ml DMEM-Low Glucose (1 g/L) with 50 ml fetal bovine serum (final concentration 10% vol/vol), 5 ml nonessential amino acid (final concentration 1% vol/vol), 5 ml Penicillin/Streptomycin (final concentration 1% vol/vol), and 1 ml β-mercaptoethanol (final concentration 0.2% vol/vol). MEF medium can be stored at 4 °C for up to two weeks.
CRITICAL If DMEM-Low Glucose medium is not already supplemented with L-Glutamine, add 5 ml L-glutamine to the final concentration of 2 mM.
Complete X-VIVO 15 Medium
To prepare complete X-VIVO 15 medium, supplement 500 ml X-VIVO 15 medium with 5 ml L-glutamine (final concentration 2 mM), 5 ml Penicillin/Streptomycin (final concentration 1% vol/vol), and 1 ml β-mercaptoethanol (final concentration 0.2% vol/vol). X-VIVO 15 medium can be stored at 4 °C for up to one month.
Differentiation Medium I
Prepare differentiation medium I by adding M-CSF (final concentration of 50 ng/ml) and IL-3 (final concentration of 25 ng/ml).
CRITICAL Add cytokines immediately before use to the volume of medium needed, and do not store cytokine-supplemented medium.
Complete RPMI Medium
To prepare complete RPMI medium, supplement 445 ml RPMI 1640 medium with 50 ml fetal bovine serum (final concentration 10% vol/vol), and 5 ml Penicillin/Streptomycin (final concentration 1% vol/vol). Complete RPMI medium can be stored at 4 °C for up to one month. CRITICAL If RPMI 1640 medium is not supplemented with L-Glutamine, add 5 ml L-glutamine to the final concentration of 2 mM.
Differentiation Medium II
Prepare differentiation medium II by adding M-CSF (final concentration of 50 ng/ml). CRITICAL Add cytokines immediately before use to the volume of medium needed. Do not store cytokine-supplemented medium.
Collagenase type IV
To prepare 25000 U/ml stock solution (100×) of collagenase type IV, dissolve 1 g of collagenase IV powder in an appropriate volume of Hank’s Balanced Salt Solution (HBSS) with Ca2+ and Mg2+. Vortex to ensure it is completely dissolved and filter-sterile the solution through a 0.22 μm syringe filter. Divide the stock solution into 1 ml aliquots and store them at -20 °C for up to one year. Prepare working solution by diluting the 1 ml stock solution in 100 ml KO DMEM medium. Aliquot the working solution and store at -20 °C for up to 2 months.
! CAUTION Collagenase type IV may cause allergy or asthma symptoms or breathing difficulties if inhaled. Avoid breathing dust/fume/gas/mist/vapours/spray. Wear personal respiratory protection.
Rho-kinase inhibitor (RI) Y-27632
To prepare a 10 mM stock solution (1000×), dissolve 10 mg of Y-27632 powder in 3.12 ml of pure water. Filter-sterile the solution using a syringe filter, prepare small aliquots and store them at -20 °C for up to 3 months. Dilute 10 mM ROCK inhibitor in cell culture medium to achieve a final concentration of 10 μM.
CRITICAL avoid repeated freeze-thaw cycles.
! CAUTION Y27632 is toxic. Avoid ingestion, inhalation and skin contact. Wear protective gloves and eye protection when you are preparing the stock solution.
bFGF
Dissolve 100 μg of bFGF in 200 μl Tris HCl 5 mM (pH 7.5) to prepare 500 ng/ml stock solution. Dilute 50 μl of stock solution in 1.2 ml of 0.1% bovine serum albumin (BSA) solution in PBS to prepare 20 μg/ml working solution (2000×). Aliquots can be stored at -20 °C for up to 6 months.
CRITICAL Avoid repeated freeze-thaw cycles and thawing aliquot in temperatures above room temperature (20–25 °C).
M-CSF and IL-3
To prepare 100 μg/ml stock solution (2000× and 4000×, respectively), dissolve 100 μg lyophilized cytokine in 1 ml 0.1% bovine serum albumin (BSA) solution in PBS. Aliquots can be store at -20 °C for up to 6 months.
CRITICAL Avoid freeze-thaw cycles. Avoid thawing aliquot in temperatures above room temperature (20–25 °C).
Gelatin solution
To prepare gelatin solution (final concentration 0.1% wt/vol), dissolve 0.5 g in 500 ml sterile pyrogen-free water. 0.1% gelatin solution can be stored at 4°C for up to 2 months.
Preparation of irradiated Murine Embryonic Fibroblasts (MEFs)
CRITICAL MEF cells can be purchased from different vendors (e.g. Merck, see “biological material”) or can be directly isolated from mouse embryos.
Thaw the cryovial of frozen non-irradiated MEFs.
Seed 0.4- 0.5 × 104 cells/cm 2 in gelatin-coated 15 cm dishes.
Place the cells in a 37 °C incubator at 5% CO2.
Passage every 3-4 days. To passage, first remove the old medium, then add 4 ml trypsin-EDTA, and incubate at 37 °C for 3 min. Stop trypsin reaction by adding 8 ml MEF medium.
Collect cell suspension in a conical tube, and centrifuge the cell suspension at 300 × g for 5 min at room temperature.
Expand cells by plating at a ratio of 1:3- 1:4 on gelatin-coated 15 cm dishes.
Continue expanding the cells up to passage 6 by repeating steps 4-6.
After expansion, collect all cells in 30 ml of MEF medium in a 50 ml conical tube and inactivate MEFs with gamma irradiation at 661,6 keV.
Freeze MEFs in appropriately sized aliquots, e.g. as 1 x 106 cells to be used for one 6- well tissue culture plate or 3 x 106 cells to be used for three 6-well tissue culture plates.
▲ CRITICAL STEP Evaluate each MEF batch to ensure they enable hiPSC to retain pluripotency. We recommend testing MEFs provided by other vendors or prepared in house by comparing them with the EmbryoMax ® primary MEFs provided by Merck (Cat. # PMEF-CFL-P1)
Seeding irradiated MEFs for iPSC culture
Coat 6-well tissue culture plates with pre-warmed 0.1% gelatin solution and incubate for at least 20 min at room temperature.
For each 6-well tissue culture plate, defrost a vial containing 1 × 106 y-irradiated MEFs and resuspend cells in 12 ml of MEF medium.
Aspirate gelatin from the 6-well tissue culture plates and distribute cells across a 6-well tissue culture plate, (~ 1.6 × 105 cells/well, 2ml medium per well). Distribute MEFs evenly with a back/forth and right/left movement of the plate and place in a CO2 incubator at 37 °C.
CRITICAL Due to batch-to-batch differences in MEF viability and quality, different seeding densities need to be tested to reach the optimal density required for iPSCs maintenance. Each batch should be tested before use.
Procedure
Passaging of human iPSCs on murine embryonic fibroblasts (MEFs) TIMING ~ 1 h
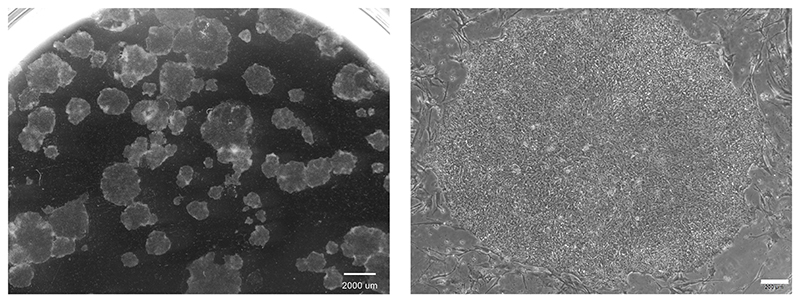
CRITICAL Human iPSCs should be passaged when colonies are reaching > 1500 μm diameter and the culture is semi-confluent (see Figure 2).
Figure 2. Morphology of human iPSCs in 2D culture.
hCD34iPSC16 were maintained on murine embyonic fibroblasts. Pictures were taken after 7 days in culture. Scale bars: left: 2000 μm, magnification 6.1x and right: 200 μm, magnification 40x.
1-2 days before passaging iPSCs, seed γ-irradiated MEFs in 6-well tissue culture plates as described in Reagent Setup (“Seeding irradiated MEFs for iPSC culture” section). ? TROUBLESHOOTING
On day of passage, obtain a 6-well tissue culture plate of semi-confluent iPSC culture maintained on MEFs in full iPSC culture medium containing 20% Knock-Out Serum Replacement and 10 ng/ml bFGF.? TROUBLESHOOTING
Aspirate medium and wash cells once with pre-warmed PBS without Ca2+ and Mg2+ (1 ml per well).
Aspirate PBS, add 500 μl pre-warmed Collagenase type IV per well and incubate at 37°C for 30-40 min.
-
Add 1 ml minimal iPSC medium per well and carefully break down the colonies by gentle pipetting up and down 2-3 times using a 1 ml tip to detach the colonies as small cell aggregates.
▲ CRITICAL STEP When detaching the colonies, gently pipette the medium a max. of 3-4 times to obtain iPSC aggregates of an appropriate size. If colonies do not detach completely, transfer the cell suspension into a conical tube, and repeat step 5. Alternatively, use autoclaved glass beads (Ø 1.7-2.1 mm) for mechanical dissociation. To do this, add sufficient beads to cover approximately half the surface of each well. Tap or move the plate slowly until all cells are detached. Use a 1 ml tip to collect only the cell suspension. Since the beads are larger than the tip head, they cannot pass through the tip and will remain in the well and only colony fragments will be collected. Rinse the beads 1-2 times with PBS to collect all colony fragments. Transfer the cell suspension to a conical tube to collect colony fragments.
? TROUBLESHOOTING
After transferring the cell suspension to a conical tube, centrifuge cell clumps at 100 x g for 3 min at room temperature.
Aspirate the supernatant without disturbing the pellet and carefully resuspend the cells in full iPSC medium using a 1 ml tip. Add 1 ml of medium for each well detached from a 6-well plate.
-
Gently resuspend cell clumps in the medium using a 1 ml tip to reach a homogenous suspension.
▲ CRITICAL STEP Resuspension of the pellet should be performed very gently and be limited to prevent further dissociation of the colony fragments and generation of single cells.
Take the pre-seeded MEFs from the incubator, aspirate the old medium, wash with 2 ml pre-warmed PBS/well and add 2 ml full iPSC medium.
-
Add an appropriate volume of homogenously resuspended iPSC to each well, distribute the aggregates evenly with a back/forth and right/left movement of the plate and place the plate in the incubator at 37 °C, 5% CO2.
▲ CRITICAL STEP For maintenance, passage a semi-confluent iPSC culture in a ratio of 1:20 to 1:40. Different splitting ratios may need to be tested for individual iPSC lines to reach optimal cell growth and confluency (see Figure 2).
Maintenance of iPSCs on MEFs TIMING 7 days
-
11
Feed the iPSC culture 1-2 days after seeding. Aspirate old medium, wash the cells with 1 ml pre-warmed PBS/well and add 2 ml full iPSC medium.
-
12
From day 4 onwards change 3-4 ml medium/well every two days and passage the iPSC culture every 7 days by following steps 1-10.
▲ CRITICAL STEP Observe the iPSC under a microscope before medium change. If differentiation occurs, differentiated colonies should be eliminated during maintenance.
Expansion & pre-priming of human iPSC in preparation of macrophage differentiation TIMING 9 days
CRITICAL For small-scale differentiation in 6-well tissue culture plates on an orbital shaker, expand iPSC culture in at least 3 wells of a 6-well tissue culture plate to generate 1 well of primed aggregates within 3 ml culture volume. For large-scale differentiation using the bioreactor with a culture volume of 120 ml, expand the iPSC culture to 18 × 6-well plates (90 wells, see table 1).
-
13
Use a semi-confluent iPSCs maintenance culture, preferably large colonies, and expand the iPSC to the required number of plates following steps 1-11.
-
14
After 5 days of culture, replace the full iPSC medium with minimal iPSC medium (3- 4ml/well) to induce a 2D pre-priming. Keep the iPSC culture in minimal iPSC medium for an additional 4 days without passaging (total culture period 9 days). On day 7 of the total culture period, remove the medium and add 3-4 ml fresh minimal iPSC medium/well.
▲ CRITICAL STEP The timing of pre-priming may need to be adapted for individual iPSC lines and can be initiated between day 3 and 7. Smaller colonies or cultures already showing some spontaneous differentiation should be kept longer in complete iPSC medium (initiation of pre-priming on day 5-7), whereas large colonies of 1000-2000 μm and cultures without spontaneous differentiations can be subjected to pre-priming between day 3-5.
Table 1. Specifications for small- and large-scale hematopoietic differentiation.
| No. of wells iPSC expansion & 2D priming | No. wells 3D priming | No. of wells hem. specification & iPSC-Mac production | volume (ml) |
|---|---|---|---|
| 3 | 1 | 1 to 2 | 3 to 6 |
| 18 | 6 | 6 to 12 | 18 to 36 |
| 90 | 30 | Bioreactor | 90 to 150 |
Preparation of 3D priming cultures TIMING ~ 1-3 h (depending on scale)
-
15
Obtain semi confluent pre-primed iPSC cultures with large colonies.
▲ CRITICAL STEP The quality of the iPSC colonies is important for the generation of primed aggregates. The colonies should be quite large and > 1500 μm diameter (if necessary cultivation time to obtain these colonies can be up to 10 days). Note that pre-priming can induce some spontaneous differentiation of the colonies.
-
16
Aspirate old medium and rinse cells with PBS. Add 500 μl pre-warmed Collagenase IV per well and place in the incubator at 37 °C for 30-40 min.
-
17
Add 1 ml minimal iPSC medium per well.
-
18
Tilt the plate, pipette perpendicular up and down 2-3 times using a 1 ml tip until some colonies are detached as cell aggregates. Transfer the suspension to a 50 ml conical tube. Repeat gentle pipetting and transfer remaining colony fragments into the conical tube until all colonies are detached.
▲ CRITICAL STEP The success in generating high-quality primed aggregates highly depends on the size of cell aggregates. Therefore, minimize pipetting times and remove colonies fragments from wells to the conical tube directly after detachment.
? TROUBLESHOOTING
-
19
Centrifuge cell clumps at 100 × g for 3 min at room temperature. Aspirate the supernatant without disturbing the pellet and carefully resuspend the pellet in minimal iPSC medium containing 10μM ROCK inhibitor. Add 4 ml of medium for 3 detached wells of a 6-well suspension culture plate.
-
20
Gently pipette the clumps up and down in a 10 ml serological pipette 1-2 times to generate a homogenous suspension. Transfer 4 ml of the cell suspension to one well of a 6-well suspension culture plate (three wells of iPSC culture are transferred to one well of 3D priming culture, see table 1). When processing a larger number of plates, gently invert the tube during pipetting or gently pipette up and down with the serological pipette to remain a homogenous suspension of iPSC aggregates.
-
21
Place the plate on an orbital shaker installed in the 37 °C, 5% CO2 incubator (this is day -5, see Figure 1).
▲ CRITICAL STEP The optimal speed of the orbital shaker (shaking throw 25 mm) is 85 rpm. An incorrect shaker speed can impact the development of primed aggregates, moreover higher or lower shaker speed can affect the size of primed aggregates. If using another shaker model, the speed has to be adjusted carefully.
? TROUBLESHOOTING
Generation of primed aggregates and induction of differentiation TIMING 5 days
-
22
Observe the plate on day 1 and 2 after transition into suspension culture to check aggregates are developing (Figure 3A).
-
23
Refresh ~70-80% of the medium on day 2 of 3D priming. Carefully tilt the plate by 4060°, let the aggregates settle to the edge and remove 3 ml of medium from the top of each well using a 1 ml tip. Add 3 ml of minimal iPSC medium or ROCK inhibitor to each well.
-
24
Place the plate back on the orbital shaker installed in the 37 °C, 5% CO2 incubator. CRITICAL STEP If planning to use a bioreactor, note that assembly and preparation of the bioreactor (BOX 1) should be started at least 2 days before inoculation of the bioreactor on day 5, i.e. on day 3 of priming.
-
25
Feed the primed aggregates by refreshing ~80% of the medium on day 4.
▲ CRITICAL STEP When changing medium, tilt the plate by 40-60° to accumulate primed aggregates at the edge of the well. Cystic and translucent primed aggregates specifically observed towards the end of 3D priming (day 4-5) require more time to settle down. Do not use the vacuum aspirator to remove the old medium as this might result in the loss of aggregates. If the pH drops too low (indicated by a change of the medium to yellow color), change the medium of the 3D priming culture daily from day 2 onwards.
-
26
Continue the culture till day 5.
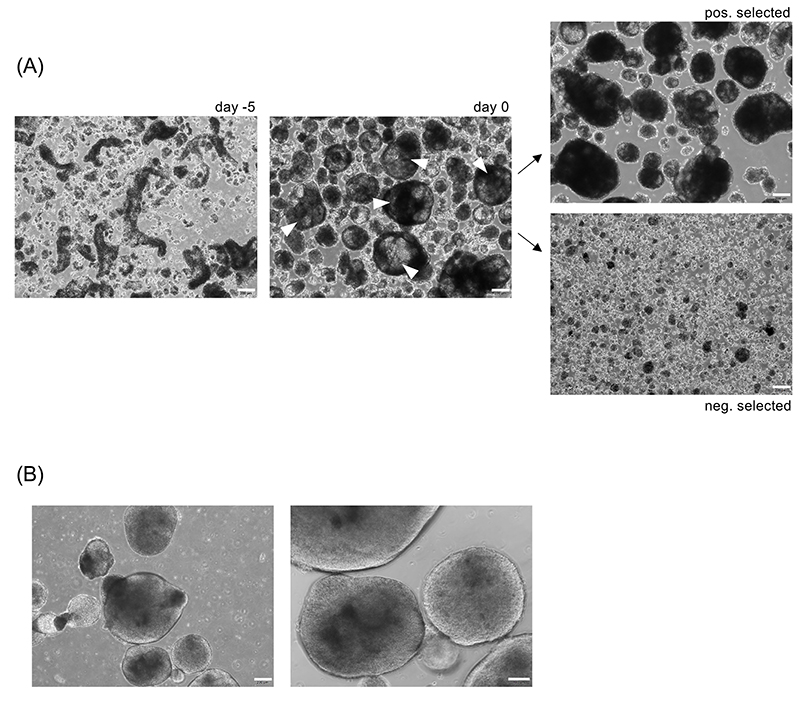
Figure 3. Morphology of primed aggregates.
(A) Colony fragments of hCD34iPSC16 (left picture, day -5) were cultured in 6-well suspension culture plate on an orbital shaker in priming medium for 5 days. White error heads indicate primed aggregates with optimal morphology which is defined by a size of > 300-500 μm diameter, the presence of some cystic structures within the aggregates and a translucent appearance (middle picture, day 0). The largest primed aggregates were selected by defined sedimentation. Upper image shows larger, positively selected primed aggregates; lower image shows smaller, negatively selected aggregates. Scale bar: 200 μm, magnification 40x. (B) Primed aggregates with sub-optimal morphology appearing round and dense. Scale bar 200 μm, 40x magnification and 100 μm, 100x magnification.
Box 1. Preparation of the bioreactor • TIMING 5 h (plus overnight vessel coating plus overnight DO sensor polarization).
Vessel coating
-
1
Make sure that the glass vessel as well as the installations are completely clean and dry. (If not follow cleaning instructions in BOX 2).
-
2
Siliconize the bioreactor glass vessel under the flow hood by repeatedly pipetting 1 mL of Sigmacote solution to the sides and bottom of the vessel until no solution is left. Keep the vessel at RT overnight.
-
3
On the next day: Rinse the bioreactor vessel thoroughly with water to remove any residues of Sigmacote.
! CAUTION Sigmacote is harmful if swallowed or inhaled. Wear protective gloves and work under the fume or flow hood.
▲ CRITICAL STEP Non-coated areas of the inner vessel wall could cause unwanted attachment of cells.
Head plate configuration (shown in Figure 4C)
-
4
To enable sampling without process interruption, add a piece of silicone tubing (i.d. 1.0 mm) with a Mohr pinchcock clamp, a female Luer-Lock connector and a Luer-Lock sampling valve to the sample port. Fix connections with cable ties.
-
5
Exhaust and inlet air filter by first adding a piece of silicone tubing (i.d. 4.8 mm) with a vent filter to the exit port of the exhaust gas line. Assemble a piece of small silicone tubing (i.d. 1.0 mm) with a piece of larger silicone tubing (i.d. 4.8 mm) with the help of a hose reduction piece. Attach a vent filter to the larger tube and connect everything to the headspace gassing port (for overlay gassing), Fix connections with cable ties; make sure that the filters are free.
Impeller installation
-
6
Make sure that the impeller is completely clean.
-
7
Mount the impeller (8-blade impeller, 60° pitch) on the lowest position of the impeller shaft and tighten with the help of the holding sleeve (shown in Figure 4B and 4D). Make sure that the impeller is properly fixed to prevent damage to sensors.
pH sensor calibration
-
8
Remove pH sensor from storage solution and check that the pH sensor’s diaphragm is undamaged.
-
9
Attach the probe to the cable of the bioreactor station.
-
10
Rinse the sensor with pure water and carefully dry with a tissue without rubbing at the diaphragm. Place the pH and temperature sensor into pH 7.00 calibration buffer. Make sure that the pH sensor diaphragm is completely submerged.
-
11
Wait until the pH reading is stable, and reset the calibration offset value at the bioreactor control unit. The raw offset value should be 0+/-25 mV.
-
12
Repeat steps 10 - 11 with a pH 4.01 calibration buffer. Wait until the pH reading is stable, and calibrate the slope value.
▲ CRITICAL STEP pH measurement is temperature dependent, therefore correct temperature measurement is necessary. To ensure correct calibration, use fresh buffers. For high precision of pH measurement recalibrate pH sensors by additional offline measurement.
? TROUBLESHOOTING
Bioreactor sterilization
-
13
Cover the bottom of the vessel with pure water to prevent pH probes drying out during steam autoclaving.
-
14
Make sure that all O-rings (underside of the head plate, pH and DO probe) are undamaged
-
15
Screw the bioreactor’s head plate onto the coated vessel. At this point, make sure that the assembled bioreactor looks like Figure 4A.
-
16
Check whether the membrane of the DO sensor is clean and undamaged.
-
17
Screw sensors into the respective head space ports and cover their plugs with caps.
▲ CRITICAL STEP The DO probe is not directly screwed into the head plate but rather fixed with the DASGIP compression fitting. When attaching the DO probe, the lowest point of the probe should end slightly above the impeller to ensure a more homogenous aggregation. If the probe is too low, the impeller can be damaged by the probe (shown in Figure 4D).
-
18
Wrap vent filters with aluminium to protect from humidity. Close silicone tubing of sampling port with the Mohr pinchcock clamp.
-
19
Autoclave bioreactor for 20 min at 120 °C.
-
20
Place sterilized bioreactor under the laminar flow and fill vessel with 100 ml PBS.
▲ CRITICAL STEP Bioreactor should be filled with medium or PBS as soon as possible to avoid draining of the pH sensor’s diaphragm.
-
21
Place the bioreactor vessel into the bioreactor station and connect to the overhead drive, exhaust condenser and temperature sensor.
Dissolved oxygen sensor calibration
-
22
Connect gas supply of the bioreactor station with the inlet filter and start overlay gassing with 21% O2 (and 5% CO2, only necessary when calibration is conducted in culture medium). Start stirring and temperature control at process conditions (stirring at 50 rpm, 37 °C). Attach the probe to the cable of the bioreactor station and polarize the sensor for at least 6 h or overnight.
-
23
Perform a one-point slope calibration at 100% DO. The slope is expected to be between 85% - 105%.
▲ CRITICAL STEP For highly accurate DO measurement, additionally apply gassing with pure N2 until the PBS is saturated followed by zero point calibration.
? TROUBLESHOOTING
Selection of primed aggregates TIMING ~ 30-60 min
CRITICAL Primed aggregates should be selected and transferred to hematopoietic specification after 5 days (day 0, see Figure 1).
-
27
Prepare Differentiation medium I (3 ml/well for small scale and 120 ml for bioreactor scale).
-
28
Observe quality and quantity of generated, primed aggregates under the microscope.
▲ CRITICAL STEP The majority of primed aggregates should be large (> 300-500 μm diameter), cystic and translucent at this stage (see Figure 3A). Typically, primed aggregates generated in one well of 6-well plate can be transferred to a culture volume of 3-6 ml (see table 1).
? TROUBLESHOOTING
-
29
Transfer all primed aggregates and medium to 15 or 50 ml conical tubes using a 1 ml tip or 10 ml serological pipette, respectively.
-
30
Allow the primed aggregates to sediment by gravity for 3-4 min.
-
31
After sedimentation of the largest primed aggregates, slowly aspirate the supernatant using a 10 or 25 ml serological pipette.
▲ CRITICAL STEP Sedimentation by gravity allows separation of larger and smaller aggregates. The sedimentation time may need to be adapted depending on the size and quality of aggregates. Monitor under the microscope whether any large and cystic aggregates remain in the supernatant. If this is the case, increase sedimentation time until all big and cystic aggregates settle.
-
32
Carefully aspirate the remaining supernatant without disturbing sedimented aggregates.
Hematopoietic specification TIMING 14 days
-
33
For hematopoietic specification in small-scale differentiation in 6-well suspension plates on an orbital shaker follow option A. If aiming for large-scale differentiation in the bioreactor follow option B.
(A) Small scale hematopoietic specification in 6-well suspension plates TIMING ~ 30 min to set up cultures and ~14 days of culture
Preparing small scale differentiation cultures
Prepare appropriate amount of differentiation medium I (3 ml/differentiation well).
Add 2 ml of differentiation medium I to each well of a 6-well suspension plate.
Carefully resuspend selected, primed aggregates in 1 ml Differentiation medium I per well using a 5 or 10 ml serological pipette and gently pipette up and down using the serological pipette 2-3 times to generate a homogenous suspension.
Add 1 ml of the aggregate suspension to each well of the differentiation plate using a 1 ml tip to achieve a final culture volume of 3 ml/well. To achieve a consistent distribution of primed aggregates between the wells, resuspend the aggregates in suspension by regular trembling the tube.
-
Place the differentiation plate on the orbital shaker installed in the 37 °C, 5% CO2 incubator (this is day 0 of differentiation)
▲ CRITICAL STEP The optimal speed of the orbital shaker (shaking throw 25mm) is 85 rpm.
Hematopoietic specification in small scale
-
vi
Refresh ~ 80% of the medium at day 7 of differentiation. Carefully tilt the plate by 40-60°, let the developing MCFCs settle to the edge and remove 2.5 ml of medium from the top of each well using a 1 ml tip. Add 2.5 ml of fresh differentiation medium I to each well.
▲ CRITICAL STEP It is important not to remove the cystic MCFCs during the medium change. Therefore, the process should be performed using a 1 ml pipette.
(B) Large scale hematopoietic specification in STBR TIMING ~ 1 h to set up cultures (plus preparation of the bioreactor) plus ~14 days of culture
Preparing large scale differentiation cultures
Assemble and prepare the DASbox Mini bioreactor according to instructions in BOX 1 (shown in Figure 4, all ensuring it is clean (cleaning instructions are given in BOX 2). Start at least two days before inoculation of the bioreactor as the vessel needs to be coated and the DO sensor calibration requires overnight (> 6 h) sensor polarization.
Prepare 120 ml Differentiation medium I
Pool the selected, primed aggregates from different 50ml tubes and resuspend in 20 ml Differentiation medium I using a 25 ml serological pipette.
Detach the bioreactor from the bioreactor station and place it under laminar flow. Open the bioreactor and hold the head plate in one hand while whilst adding 100 ml Differentiation medium I with your other hand.
-
Add 20 ml of primed aggregates in suspension from step (ii) into the bioreactor vessel.
TROUBLESHOOTING
Place the head plate back on the vessel and screw it tight.
-
Place the bioreactor vessel into the bioreactor station and connect temperature, pH and DO sensors as well as overhead drive, exhaust condenser and overlay gas supply. Start stirring (50 rpm, up-flow [counter clockwise with the here described impeller]), overlay gassing (3 sl/h with 21% O2 and 5% CO2), heating (37 °C) as well as on-line recording of temperature, pH and DO in the software.
▲ CRITICAL STEP Make sure the temperature sensor is connected before starting temperature control to exclude system overheating.
TROUBLESHOOTING
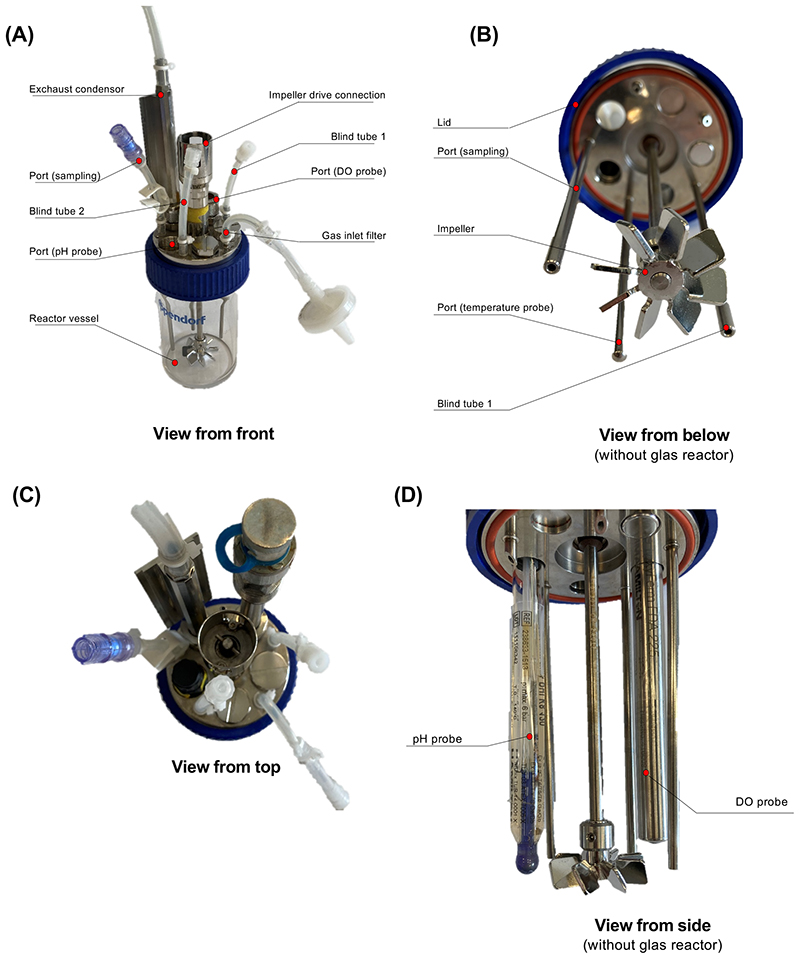
Figure 4. Photography of the assembled bioreactor (related to BOX1).
(A) Assembled Bioreactor. (B) Bottom view of the bioreactor. Positioning of the Impeller at the lowest position of the impeller shaft. (C) Headplate configuration with pH and DO probes. (D) Side view of the headplate configuration to highlight the positioning of the DO probe
Box 2. Cleaning of the bioreactor.
• TIMING 1 h (plus overnight incubation)
-
1
Disconnect bioreactor from the bioreactor station.
-
2
Fill the bioreactor vessel with 250 ml of Terg-a-zyme® enzyme detergent solution and close the vessel via the head plate to place the impeller and the inner installations into the disinfectant solution. Place the bioreactor in the bioreactor station again and connect the overhead stirrer and the temperature probe. Set the temperature to 37 °C and the stirring rate to 150 rpm. Discard the disinfectant solution afterwards.
-
3
Rinse the sample option tubing thoroughly with enzyme detergent solution and incubate for 24 h.
-
4
Wash and rinse three times with pure water after incubation.
-
5
Unscrew DO and pH sensors off the head plate. Place pH sensor back into the storage solution.
-
6
Rinse the bioreactor vessel and inner installations with 70% ethanol.
-
7
Repeat steps 4| + 6| twice. Ultimately wash the bioreactor vessel and inner installations thoroughly with pure water and pat dry.
▲ CRITICAL STEP Visually check that the bioreactor’s vessel, inner installations and tubing are free of any culture or cleaning residues after these cleaning steps.
Hematopoietic specification in STBR
-
viii
After 7 days of culture, stop temperature control, stirring and gassing, detach the bioreactor from the bioreactor station and place it under the laminar flow.
-
ix
Let the primed aggregates settle for 3 minutes
-
x
Open the bioreactor and hold the head plate in one hand while with the other remove 100 ml supernatant using a 50 ml serological pipette. About 20mL medium with MCFCs will remain in the bioreactor vessel.
-
xi
Carefully add 100 ml of differentiation medium I to the bioreactor
-
xii
Place the head plate back on the vessel and screw it tight.
-
xiii
Place the bioreactor vessel into the bioreactor station and connect temperature, pH and DO sensors as well as overhead drive, exhaust condenser and overlay gas supply. Restart temperature control, stirring and gassing.
Macrophage production TIMING 7 days (per harvest)
-
34
For macrophage production in small-scale differentiation in 6-well suspension plates on an orbital shaker follow option A. If aiming for large-scale differentiation in the bioreactor follow option B.
(A) Harvest of iPSC-derived macrophages from 6-well suspension plates (continuously every 7 days) TIMING ~ 1 hour
Prepare the required amount of Differentiation medium I (2.5 ml/well).
Remove the differentiation plate from the incubator and monitor the plate under the microscope for macrophage production (Figure 5).
Create short, circular motions in the plate to accumulate all MCFCs in the middle of the well.
Slowly tilt the plate by 40-60°, and let the MCFCs to settle to the edge of the well.
Carefully collect the 2.5 ml medium including iPSC-Mac from the top of the well using 1 ml tip. Pass the cells through a 70 μm cell strainer, and collect them in a conical tube.
Centrifuge iPSC-Mac at 300 × g for 5 min at room temperature.
-
Resuspend iPSC-Mac in PBS (0.5 ml/well) and count the cells using a hemocytometer and TrypanBlue live/dead staining
TROUBLESHOOTING
Figure 5. Production of iPSC-derived macrophages by myeloid cell forming complexes (MCFCs).
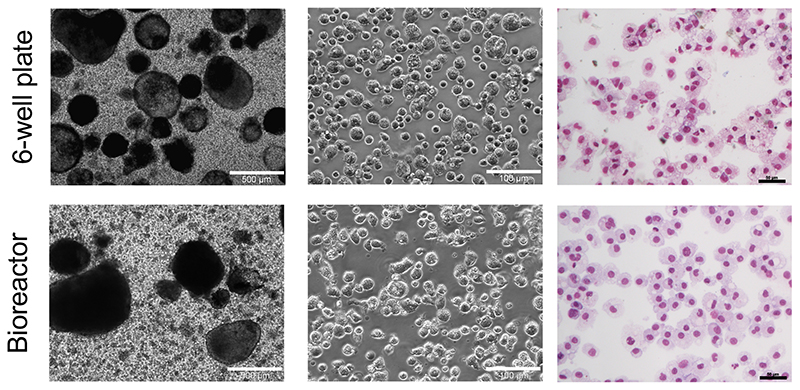
From left to right: Bright field microscopy of MCFCs producing iPSC-Mac in 6-well plates (upper row) and the bioreactor platform (lower row) (Scale bar: 500 μm (magnification 40x) and 100 μm (magnification 100x)). May-Grünwald-Giemsa stained cytospin preparations of generated iPSC-Mac (Scale bar: 50 μm, magnification 200x).
(B) Harvest of iPSC-derived macrophages from STBR (continuously every 7 days) TIMING ~ 1hour
▲ CRITICAL STEP Monitor macrophage production (Figure 5) before detaching the bioreactor as described in BOX 3.
Box 3. Sampling from the bioreactor.
• TIMING 30 min.
This enables sampling via the self-sealing sample valve from the bioreactor without interrupting the process:
-
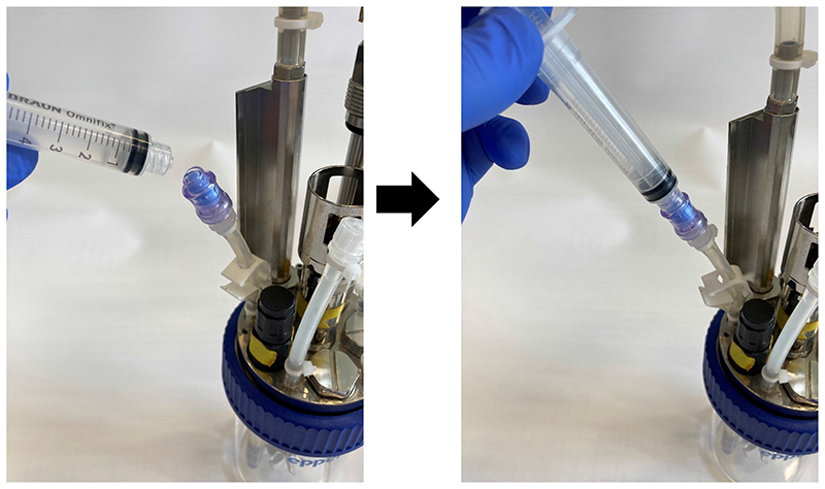
1
Remove the cap from the valve and spray with 70% ethanol.
-
2
Connect a sterile Luer-Lock syringe to the valve (shown in the images below), create an under pressure and open the Mohr pinchcock clamp of the sampling tube. Carefully draw up ~1.0 ml culture into the syringe as clearance volume. Close the clamp before unplugging the syringe and discard the clearance volume.
-
3
Connect a second sterile Luer-Lock syringe to the valve and draw in ~2.5 ml culture suspension as described in (step 2). Transfer the sample into one well of a 6-well plate.
-
4
Spray the valve with 70% ethanol and cover the sampling valve with the cap.
Sampling from the bioreactor.
After 4 days of macrophage production culture, control the pH value of the bioreactor. If the pH has dropped to 6.7 or lower, please follow steps ii.-v. If the pH is >6.7, continue with step vi.
Stop temperature control, stirring and gassing, detach the bioreactor from the bioreactor station and place it under the laminar flow.
Open the bioreactor and hold the head plate in one hand while with the other add additional 30 ml Differentiation medium I.
Place the head plate back on the vessel and screw it tight.
Place the bioreactor vessel into the bioreactor station and connect temperature, pH and DO sensors as well as overhead drive, exhaust condenser and overlay gas supply. Restart temperature control, stirring and gassing.
After 7 days, stop temperature control, stirring and gassing, detach the bioreactor from the bioreactor station and place it under the laminar flow.
Let the MCFCs settle for 3 minutes.
-
Open the bioreactor and hold the head plate in one hand while with the other pipet 130 ml of the medium containing iPSC-Mac through a 70 μm cell strainer into three 50 ml conical tubes. About 20 mL medium with MCFCs will remain in the bioreactor vessel.
▲ CRITICAL STEP If no additional medium was added on day 4, only remove 100 ml medium from the bioreactor.
Invert the strainer and flush remaining MCFCs with 25 ml fresh Differentiation medium I back into the bioreactor. Carefully add 75 ml Differentiation medium I to the vessel.
Place the head plate back on the vessel and screw it tight.
Place the bioreactor vessel into the bioreactor station and connect temperature, pH and DO sensors as well as overhead drive, exhaust condenser and overlay gas supply. Restart temperature control, stirring and gassing.
Centrifuge iPSC-Mac at 300 × g for 5 min at room temperature.
-
Resuspend iPSC-Mac in 10 ml PBS and count the cells using a hemocytometer and TrypanBlue live/dead staining
TROUBLESHOOTING
OPTIONAL: Terminal Differentiation of Macrophage TIMING ~ 7 days
CRITICAL For further maturation/maintenance of macrophages, freshly harvested iPSC-derived macrophages can be cultured in differentiation medium II for up to 7 days.
-
35
Prepare the required amount of Differentiation medium II (1 ml/ 0.5×106 cells).
-
36
Centrifuge the desired quantity of iPSC-Mac derived from step 34 A vii. or 34 B xiii. at 300 × g for 5 min at room temperature.
-
37
Resuspend the cells in Differentiation medium II (1 ml/0.5×106 cells), and seed 5 × 105 cells/ cm 2 of a tissue culture plate (1.25×106/well of a 6-well plate), and place in a CO2 incubator at 37 °C.
▲ CRITICAL STEP Macrophages should attach to plate within 24 hours after seeding.
-
38
Feed the cells with fresh Differentiation medium II 4 days after seeding. Carefully remove the old medium and add 2.5 ml fresh Differentiation medium II. Matured iPSC-Mac are ready for use in further experiments after 4-7 days.
Timing
Passaging of iPSCs on murine embryonic fibroblasts (MEFs) (steps 1-10) TIMING ~ 1 h Maintenance of iPSCs on MEFs (steps 11-12) TIMING 7 days
Expansion & pre-priming of iPSC in preparation of macrophage differentiation (steps 13-14) TIMING 9 days
Preparation of 3D priming cultures TIMING ~ 1-3 h (steps 15-21) (depending on scale) Generation of primed aggregates and induction of differentiation (steps 22-26) TIMING 5 days Selection of primed aggregates (steps 27-32)TIMING ~ 30-60 min
Hematopoietic specification (step 33) TIMING 14 days
-
(A)
Small scale hematopoietic specification in 6-well suspension plates TIMING ~ 30 min to set up cultures and ~14 days of culture
-
(B)
Large scale hematopoietic specification in STBR TIMING ~ 1 h to set up cultures (plus preparation of the bioreactor) plus ~14 days of culture
Macrophage production (step 34) TIMING 7 days
-
(A)
Harvest of iPSC-derived macrophages from 6-well suspension plates (continuously every 7 days) TIMING ~ 1 hour
-
(B)
Harvest of iPSC-derived macrophages from STBR (continuously every 7 days) TIMING ~ 1hour
OPTIONAL: Terminal Differentiation of Macrophage (steps 35-38) TIMING ~ 7 days
BOX 1 | Preparation of the bioreactor TIMING 5 h (plus overnight vessel coating plus overnight DO sensor polarization)
BOX 2 | Cleaning of the bioreactor TIMING 1 h (plus overnight incubation)
BOX 3 | Sampling from the stirred tank bioreactor TIMING 30min
BOX 4 | Quality control of iPSC-derived macrophages by flow cytometry TIMING 3 h
Box 4. I Quality control of iPSC-derived macrophages by flow cytometry • TIMING 3 h.
Additional Materials Required
Antibodies (see Table 3 for antibodies we have successfully used).
Flow Cytometer, Cytoflex S V4-B4-R3-12 (Beckman Coulter Life sciences, cat. no. C01161)
FACS buffer: Add 10 ml FBS (final concentration 2% vol/vol), and 1 ml EDTA (1 mM) to 500 ml PBS and store at 4 °C. FACS buffer can be stored at 4 °C for up to 2 months
Procedure
-
1
Transfer 1 × 105 iPSC-Mac into three 15 ml conical tubes and centrifuge at 300 × g for 5 min at room temperature.
-
2
Resuspend each cell pellet in 100 μl cold FACS buffer.
PAUSE POINT: Cells can be kept on ice for up to 2 hours before staining.
-
3
Add 1 μl of fc blocking Ab to the cell suspension and incubate for 20 min at 4 °C.
-
4Add 1-2 μl of appropriate antibodies, as indicated in the table below, to the tubes and incubate for 30-45 min at 4 °C in the dark. Table 3 contains details of the antibodies we have successfully used.
Tube Condition Antibodies 1 unstained None 2 isotype IgG1 κ-eF450, IgG1 κ-FITC, IgG1 κ-APC, IgG1 κ-PE-Cy7, IgM-PE 3 stained CD45-eF450, CD14 FITC, CD163-APC, CD11b PE-Cy7, TRA-1-60-PE -
5
Add 1ml FACS buffer per sample, centrifuge at 300 × g for 5 min at 4 °C and resuspend the pellet in 100-200 μl FACS buffer.
PAUSE POINT: Cells can be kept on ice or at 4 °C in the dark for up to 3 hours before measurement.
-
6
Analyze the samples using a suitable flow cytometer (e.g., Cytoflex, Beckman Coulter or LSRII BD Bioscience). FlowJo (TreeStar) can be used for further analysis.
CRITICAL: As spillover of the fluorochromes is expected, prepare single stained controls for each antibody using either iPSC-Mac or compensation beads (e.g. OneComp eBeads™ Compensation Beads, ThermoFisher) and apply proper compensation.
BOX 5 | Evaluating the phagocytosis potential of iPSC-derived macrophages TIMING 6 h
Box 5. I Evaluating the phagocytosis potential of iPSC-derived macrophages • TIMING 7-9 h.
Additional Materials required
pHrodo™ Red E. coli BioParticles™ (TermoFisher Scientific, cat. no. P35361)
Procedure
-
Prepare pHrodo™ Red E. coli BioParticles™ (TermoFisher Scientific) according to the manufacturer’s instructions. Pipette 2 ml of Phenol red-free RPMI 1640 medium and 40 μl of HEPES into the vial containing lyophilized pHrodo™ Red E. coli BioParticles™. Resuspend by using a sonicator or alternatively vortex the solution for 5 min at maximum speed.
PAUSEPOINT The solution can be kept in light-protected condition in a 4 °C fridge for up to six months.
-
Seed 1-2 × 105 iPSC-Mac into 2 wells of two 24-well plate in 500 μl complete RPMI 1640 medium. Allow the cells to settle and adhere to the plate for at least 4 hours in a cell culture incubator with 5% CO2 at 37 °C.
PAUSE POINT: Cells can be cultured for up to 3 days before analysis.
CRITICAL STEP: Two 24-well plates are required as the control plate will be incubated at 4 °C.
After the cells have adhered, remove the culture medium. Replace the culture medium with 500 μl complete phenol-red free RPMI 1640 medium.
Pre-cool the cells allocated to 4 °C in a fridge.
Add 10 μl of the prepared pHrodo™ BioParticles®suspension to each experimental well.
-
Protect the plate from light and centrifuge the bioparticles on top of cells at 500xg for
3 min.
Place the plates in a 37 °C incubator or 4 °C fridge for 2-4 hours.
Detect the phagocytic cells via flow cytometry or fluorescent microscopy after 4 hours.
TROUBLESHOOTING
Troubleshooting advice can be found in Table 2. General guidance on issues related to setting-up the bioreactor system were described before 30 . More specific issues specifically related to the generation of iPSC-Mac in the STBR platform are outlined in Table 2 below.
Table 2. Troubleshooting.
| Step | Problem | Possible reason | Possible solution |
|---|---|---|---|
| iPSC cutivation and expansion | |||
| 1 | Poor quality of MEF cells | Poor quality of MEF stock and / or MEF culture handling | Test different fetal calf serum (FSC) vendors/ batches for good support of MEF culture |
| Do not expand MEFs for more than 5-6 passages | |||
| Make sure tissue culture plates are properly coated with gelatin | |||
| Purchase or create new MEF stock | |||
| 2 | iPSC colonies lose their well-defined edges and start to differentiate | Insufficient bFGF activity | Try to increase bFGF concentration to 20 ng/ml, or higher, depending on the vendor |
| Purchase new bFGF stock. Biological activity of your bFGF batch can be tested as described 55 | |||
| 5 | Poor detachment of iPSCs colonies after treatment with Collagenase IV | Insufficient incubation time and/or poor Collagenase IV activity | Increase Collagenase IV concentration or purchase new stock |
| Extend incubation time for an additional 20 min | |||
| Use glass beads for mechanical dissociation (see step 5 description) | |||
| 3D priming | |||
| 28 | Primed aggregates are too big | Improper orbital shaker speed | Monitor orbital shaker speed and carefully increase the speed depending on the throw of your shaker model |
| Improper volume of culture medium during the generation of primed aggregates | Do not exceed a maximum of 4 ml medium per well of a 6-well plate | ||
| Clumping of aggregates during priming phase | Mechanically dissociate big clumps on day 1 or 2 of 3D priming carefully using a 1 ml pipette tip | ||
| Primed aggregates are too small | Small iPSC colonies | Seed iPSC at lower density during passaging and increase the time between passages to allow colonies to grow larger (>1500 um) | |
| Poorly detachment of iPSC colonies from the culture plate | See Step 5 in this table | ||
| Too harsh pipetting | Pipette gently max. 3-4 times up and down with a 1ml pipette tip. If further pipetting is needed to detach colonies, use fresh medium | ||
| Orbital shaker speed is too high | Monitor orbital shaker speed and carefully decrease the speed depending on the throw of your shaker model | ||
| Improper volume of culture medium during 3D priming | Do not use less than 3 ml medium per well of a 6-well plate | ||
| Primed aggregates appear round and dense (see Figure 3B), and do not show the morphology depicted in Figure 3A | Improper differentiation/ mesoderm formation | Ensure proper quality of iPSC culture following steps 1, 2 and 5 in this table | |
| Reduce bFGF concentration to 5 ng/ml during maintenance of your iPSC culture | |||
| Prolong iPSC culture in minimal iPSC medium (2D priming) before the initiation of 3D priming | |||
| Extend 3D priming to 7 days | |||
| Add mesoderm priming cytokines (BMP4, SCF, VEGF) during the 3D priming to promote mesodermal differentiation 33 especially when starting with feeder-free cultures | |||
| Macrophage Production | |||
| 34 (A) and (B) | No or low Macrophage production | Inefficient pre-priming; poor quality of MCFC | Follow trouble shooting advise of steps 1 – 27 listed in this table |
| Harvested cells contain a large fraction of small and CD45+/CD11b+/CD14- /CD163- cells | Insufficient cytokine concentration | Reduce the number of MCFCs/mL | |
| Check origin and quality of your cytokine stocks; eventually increase the concertation of cytokines | |||
| Inefficient differentiation | Include a terminal differentiation step (35-38) to further mature myeloid progenitors to iPSC-Mac | ||
| Bioreactor process-related issues: | |||
| 33 and 34 B | Undesired MCFC sedimentation at the bottom of the vessel during the process that may induce MCFC fusion disrupting production of iPSC-Mac | Improper/ low stirring speed | Adapt / increase the stirring speed; typical range to be tested: 55 – 65 rpm |
| Improper/ too high impeller positioning | Modify / adjust the position following description in BOX 1, step 7, Figure 4B and D | ||
| Different impeller type/geometry used than described in BOX 1, step 7 | Modify the stirring speed such that the primed aggregates / MCFC are properly maintained in suspension but without disrupting MCFCs. Range to be tested: ~45 – 65 rpm (also depending on impeller type used) | ||
| MCFC disruption resulting into the formation of smaller fragments and loss of iPSC-Mac production | Improper/ high stirring speed | Adapt / increase the stirring speed; typical range to be tested: 45 – 55 rpm | |
| Different impeller type/geometry used than described in BOX 1, step 7 | Modify the stirring speed such that the primed aggregates / MCFC are properly maintained in suspension but without “disrupting” MCFCs. Use recommended impeller type/geometry | ||
| Decreasing number of MCFCs after each harvest | MCFCs do not sediment properly | Increase the sedimentation time in step 33 B xiii for up to 8 min | |
| MCFCs get stuck in the cell strainer | Carefully evaluate the cell strainer for remaining MCFCs after flushing, eventually flush again | ||
| BOX 1 | pH drop below 6.7 between independent harvests | Improper density e.g. too many MCFCs per culture medium volume | Reduce the density of MCFCs e.g., by adding additional medium / process volume within the bioreactorlimited range of ~120 – 200 ml or by removing primed aggregates / MCFCs from the process |
| No (proper) signal of the bioprocess sensors for pH and oxygen | Cable of respective sensors not tightly connected | Ensure proper connection closely following vendors instructions | |
| Contacts of sensors and sensor cables might be wet (e.g. in consequences to sterilization) | Dry the contacts and reconnect | ||
| pH sensor is broken due to extended exposure time without liquid submersion. Electrolyte of the DO sensor depleted after autoclaving | Sensor to be replaced, recalibrated and exchange under the flow hood | ||
| Replace membrane and electrolyte following product instructions. Remember to autoclave the bioreactor again before usage | |||
Anticipated results
To perform macrophage differentiation in suspension culture, as a first step primed aggregates are generated by cultivating fragmented iPSC colonies in a 6-well suspension plate placed on an orbital shaker. After 24 hours, the fragments should have formed aggregates of different sizes. After 5 days in 3D priming culture, large (~300-500 μm diameter) primed aggregates comprising some cystic structures and appearing translucent should have formed (Figure 3A). If the primed aggregates do not show the desired size distribution and morphology, respective trouble shooting measures outlined in table 2 should be followed. While the translucent morphology with some cystic structures within the aggregate is an important quality criterion at this stage, the formation of some budding cystic structures is optional here and can also happen later during the hematopoietic specification phase. During the subsequent hematopoietic specification period, primed aggregates typically become more cystic (including some budding cystic structures) and develop into MCFCs (Figure 5). Between day 14 and day 21, we have observed that most iPSC lines tested by us start to produce iPSC-Mac, which appear as large round cells (approx. 20 μm diameter) under the microscope (Figure 5). Typically, upon the first medium change (day 7/week 1), only low numbers of iPSC-Mac are present (Figure 6A). However, during subsequent weekly harvests, one well of a 6-well suspension plate typically yields ~0.4-1 × 106 iPSC-Mac/week and the bioreactor platform at 120 ml culture scale typically generates approximately 10-40 × 106 iPSC-Mac/week (Figure 6A) when operating at optimal conditions. Once initiated, the production phase typically extends for up to three months. Although we did not observe a reduced cell quality when the quantity of the production drops, we typically discontinue the cultures when the macrophage yield drops below 10 × 106 in the bioreactor platform or below 300,000 for the 6-well culture. Generated iPSC-Mac should be large cells (diameter approx. 20 um) with vacuoles and some filopodia visible on microscopic evaluation. Furthermore, cytospin preparations can confirm typical macrophage morphology such as a large cytoplasm filled with vacuoles (Figure 5). Flow cytometry should be used to more specifically investigate the macrophage identity and functionality. If the protocol is implemented correctly, an iPSC-Mac population harvested from our protocol should comprise >90% CD45+ and CD11b+ and >80% CD14+ and CD163+ cells after the correct pre-gating for viable cells (see Figure 7 and BOX 4). More than 80% of the analyzed cells should demonstrate efficient phagocytosis of pHrodo™ BioParticles (see Figure 8 and BOX 5). Live cell imaging can be applied as an alternative strategy to study the rate of phagocytosis 56,57 .
Figure 6. Harvest efficiency and process monitoring of the STBR production platform.
(A) iPSC-Mac production efficiencies of three independent bioreactor runs over a period of up to 11 weeks (individual values, mean +/- SD, n=3). (B) Data from one continuous process monitoring (temperature, pH, and dissolved oxygen (DO) level) for the entire cultivation phase of one 77-day bioreactor run.
Figure 7. Analysis of iPSC-derived macrophages by flow cytometry (related to BOX 4).
Representative gating strategy for the analysis of iPSC-Mac freshly harvested from the bioreactor by flow cytometry. Pre-gating on viable cells by FSC/SSC properties was followed by a single-cell gating step (SSC-A vs. SSC-H). Subsequently, expression of CD45, CD11b, CD14 and CD163 was analyzed. Grey histograms represent unstained controls (30.056 cells analyzed), blue lines respective surface marker staining (33.799 cells analyzed). Flow cytometry was performed with a Cytoflex S (Beckman Coulter) and data was analyzed using FlowJo (BD Bioscience). Underlying flow cytometry data sets can be accessed under http://flowrepository.org/, Repository IDs: FR-FCM-Z4WE)
Figure 8. Phagocytosis of bioparticles by iPSC-derived macrophages (related to BOX 5).
To evaluate the functionality of iPSC-Mac, cells were incubated with pHrodo™ Red E. coli BioParticles™ for 4 hours at 4 or 37 °C. (A) Flow cytometric analysis. Pre-gating on viable cells by FSC/SSC properties was followed by a single-cell gating step (SSC-A vs. SSC-H). Untreated iPSC-Mac (grey filled, 143.913 cells), iPSC-Mac incubated with bioparticles at 4 °C (black line, 100.785 cells) and iPSC-Mac incubated with bioparticles at 37 °C (70.466 cells) were analyzed. Flow cytometry was performed with a Cytoflex S (Beckman Coulter) and data was analyzed using FlowJo (BD Bioscience). Underlying flow cytometry data sets can be accessed under http://flowrepository.org/, Repository IDs: FR-FCM-Z4WC). (B) Fluorescence microscopy of iPSC-Mac incubated with pHrodo™ Red E. coli BioParticles™ at 37 °C (scale bar: 100 μm, magnification 200x).
We did not observe differences in these quality criteria for iPSC-Mac generated by the two production platforms described here. Moreover, the expression pattern of the described surface markers as well the functional features of iPSC-Mac should remain stable and essentially unchanged across the entire harvest period i.e. for up to 3 months.
Editorial summary.
iPSC-derived macrophages (iPSC-Mac) are mass-produced in a scalable suspension culture on an orbital shaker or in a stirred-tank bioreactor, and can be harvested at weekly intervals for several months.
Tweet.
Mass production of iPSC-derived #macrophages in bioreactors, enabling innovative immunotherapies @natureprotocols
Cover teaser.
iPSC-derived macrophage production
Table 3. Antibodies.
| Antigen | Conjugated | Host | Isotype | Clone | Supplier | Cat. no. | RRID | Dilution factor |
|---|---|---|---|---|---|---|---|---|
| CD11b | PE-Cy7 | Mouse | lgG1 k | ICRF45 | eBioscience | 17-0118-42 | AB_2016659 | 1:100 |
| CD14 | FITC | Mouse | lgG1 k | 61D3 | eBioscience | 11-0149-41 | AB_10597445 | 1:50 |
| CD45 | eFluor 450 | Mouse | lgG1 k | HI30 | eBioscience | 48-0459-41 | AB_2016610 | 1:50 |
| CD163 | APC | Mouse | lgG1 k | eBioGHI/61 | eBioscience | 17-1639-41 | AB_2573I67 | 1:100 |
| TRA-1-60 | PE | Mouse | IgM | Tra1-60-R | Biolegend | 330609 | AB_1279447 | 1:100 |
| lgG1 k | APC | Mouse | - | P3.6.2.8.1 | eBioscience | 17-4714-81 | AB_763650 | 1:200 |
| lgG1 k | FITC | Mouse | - | P3.6.2.8.1 | eBioscience | 11-4714-41 | AB_10598647 | 1:50 |
| lgG1 k | PE-Cy7 | Mouse | - | P3.6.2.8.1 | eBioscience | 25-4714-80 | AB_6579I4 | 1:100 |
| lgG1 k | eFluor 450 | Mouse | - | P3.6.2.8.1 | eBioscience | 48-4714-82 | AB_1271992 | 1:100 |
| IgM | PE | Mouse | - | MM-30 | Biolegend | 401611 | AB_2892810 | 1:400 |
Acknowledgments
The authors thank Doreen Kloos, Theresa Buchegger, Luisa Bach for technical support. The work received funding from the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG): Cluster of Excellence REBIRTH EXC 62/3 (R.Z. and N.L.), ZW64/4-1, ZW 64/4-2, KFO311 / ZW64/7-1 (R.Z.); the German Ministry for Education and Science (Bundesministerium für Bildung und Forschung, BMBF): 01EK1602A (R.Z. and N.L.) 13N14086, 01EK1601A, 655 13XP5092B, 031L0249 (R.Z.), “Förderung aus Mitteln des Niedersächsischen Vorab” (grant: ZN3340) (R.Z. and N.L.), the Else Kröner-Fresenius-Stiftung (EKFS; 2016_A146) (M.A.). The work also received funding from Hannover Medical School Transplantation Center (Tx Center) (N.L.) and the European Society of Clinical Microbiology and Infectious Diseases (ESCMID)(N.L.). This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No. 852178).
Footnotes
Author contributions
M.A., H.K., R.Z. and N.L. developed the protocol; M.A., A.R.H. and F.M. performed the experiments and analyzed the data; M.A., A.R.H., F.M., M.C.O., R.Z. and N.L wrote the manuscript; all authors approved the final paper.
Competing Interests
This work is included in a patent application. M.A., H.K., R.Z., and N.L. are authors of the patent application (European patent application number PCT/EP2018/061574) entitled “Stem-cell derived myeloid cells, generation and use thereof”. The priority date of the application is 04.05.2017. H.K. is employee of Novo Nordisk A/S, Måløv, Denmark and N.L. and R.Z. receive funding from Novo Nordisk A/S, Måløv, Denmark.
Data availability statement
The raw data for Figures 7 and 8 are available at http://flowrepository.org/ (Repository IDs: FR-FCM-Z4WC (Figure 7) and FR-FCM-Z4WE (Figure 8)). The raw data for Figure 6 is available as source data.
References
- 1.Kempf H, Zweigerdt R. Scalable Cardiac Differentiation of Pluripotent Stem Cells Using Specific Growth Factors and Small Molecules. Adv Biochem Eng Biotechnol. 2018;163:39–69. doi: 10.1007/10_2017_30. [DOI] [PubMed] [Google Scholar]
- 2.Knorr DA, Ni Z, Hermanson D, Hexum MK, Bendzick L, Cooper LJ, Lee DA, Kaufman DS. Clinical-scale derivation of natural killer cells from human pluripotent stem cells for cancer therapy. Stem Cells Transl Med. 2013;2:274–283. doi: 10.5966/sctm.2012-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito Y, Nakamura S, Sugimoto N, Shigemori T, Kato Y, Ohno M, Sakuma S, Ito K, Kumon H, Hirose H, Okamoto H, et al. Turbulence Activates Platelet Biogenesis to Enable Clinical Scale Ex Vivo Production. Cell. 2018;174:636–648.:e618. doi: 10.1016/j.cell.2018.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hetzel M, Ackermann M, Lachmann N. Beyond “Big Eaters”: The Versatile Role of Alveolar Macrophages in Health and Disease. Int J Mol Sci. 2021;22 doi: 10.3390/ijms22073308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singanayagam A, Triantafyllou E. Macrophages in Chronic Liver Failure: Diversity, Plasticity and Therapeutic Targeting. Front Immunol. 2021;12:661182. doi: 10.3389/fimmu.2021.661182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ginhoux F, Guilliams M. Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity. 2016;44:439–449. doi: 10.1016/j.immuni.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 8.Haake K, Ackermann M, Lachmann N. Concise Review: Towards the Clinical Translation of Induced Pluripotent Stem Cell-Derived Blood Cells-Ready for Take-Off. Stem Cells Transl Med. 2019;8:332–339. doi: 10.1002/sctm.18-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee CZW, Kozaki T, Ginhoux F. Studying tissue macrophages in vitro: are iPSC-derived cells the answer? Nat Rev Immunol. 2018;18:716–725. doi: 10.1038/s41577-018-0054-y. [DOI] [PubMed] [Google Scholar]
- 10.Ackermann M, Dragon AC, Lachmann N. The Immune-Modulatory Properties of iPSC-Derived Antigen-Presenting Cells. Transfus Med Hemother. 2020;47:444–453. doi: 10.1159/000512721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Happle C, Lachmann N, Ackermann M, Mirenska A, Göhring G, Thomay K, Mucci A, Hetzel M, Glomb T, Suzuki T, Chalk C, et al. Pulmonary Transplantation of Human Induced Pluripotent Stem Cell-derived Macrophages Ameliorates Pulmonary Alveolar Proteinosis. Am J Respir Crit Care Med. 2018;198:350–360. doi: 10.1164/rccm.201708-1562OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu R, Li X, Boreland AJ, Posyton A, Kwan K, Hart RP, Jiang P. Human iPSC-derived mature microglia retain their identity and functionally integrate in the chimeric mouse brain. Nat Commun. 2020;11:1577. doi: 10.1038/s41467-020-15411-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ackermann M, Kempf H, Hetzel M, Hesse C, Hashtchin AR, Brinkert K, Schott JW, Haake K, Kühnel MP, Glage S, Figueiredo C, et al. Bioreactor-based mass production of human iPSC-derived macrophages enables immunotherapies against bacterial airway infections. Nat Commun. 2018;9:5088. doi: 10.1038/s41467-018-07570-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlsson KR, Cowley S, Martinez FO, Shaw M, Minger SL, James W. Homogeneous monocytes and macrophages from human embryonic stem cells following coculture-free differentiation in M-CSF and IL-3. Exp Hematol. 2008;36:1167–1175. doi: 10.1016/j.exphem.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Wilgenburg B, Browne C, Vowles J, Cowley SA. Efficient, long term production of monocyte-derived macrophages from human pluripotent stem cells under partly-defined and fully-defined conditions. PLoS One. 2013;8:e71098. doi: 10.1371/journal.pone.0071098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lachmann N, Ackermann M, Frenzel E, Liebhaber S, Brennig S, Happle C, Hoffmann D, Klimenkova O, Lüttge D, Buchegger T, Kühnel MP, et al. Large-scale hematopoietic differentiation of human induced pluripotent stem cells provides granulocytes or macrophages for cell replacement therapies. Stem Cell Reports. 2015;4:282–296. doi: 10.1016/j.stemcr.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ackermann M, Haake K, Kempf H, Kaschutnig P, Weiss AC, Nguyen AHH, Abeln M, Merkert S, Kühnel MP, Hartmann D, Jonigk D, et al. A 3D iPSC-differentiation model identifies interleukin-3 as a regulator of early human hematopoietic specification. Haematologica. 2021;106:1354–1367. doi: 10.3324/haematol.2019.228064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dreyer AK, Hoffmann D, Lachmann N, Ackermann M, Steinemann D, Timm B, Siler U, Reichenbach J, Grez M, Moritz T, Schambach A, et al. TALEN-mediated functional correction of X-linked chronic granulomatous disease in patient-derived induced pluripotent stem cells. Biomaterials. 2015;69:191–200. doi: 10.1016/j.biomaterials.2015.07.057. [DOI] [PubMed] [Google Scholar]
- 19.Lachmann N, Happle C, Ackermann M, Lüttge D, Wetzke M, Merkert S, Hetzel M, Kensah G, Jara-Avaca M, Mucci A, Skuljec J, et al. Gene correction of human induced pluripotent stem cells repairs the cellular phenotype in pulmonary alveolar proteinosis. Am J Respir Crit Care Med. 2014;189:167–182. doi: 10.1164/rccm.201306-1012OC. [DOI] [PubMed] [Google Scholar]
- 20.Haake K, Neehus AL, Buchegger T, Kühnel MP, Blank P, Philipp F, Oleaga-Quintas C, Schulz A, Grimley M, Goethe R, Jonigk D, et al. Patient iPSC-Derived Macrophages to Study Inborn Errors of the IFN-γ Responsive Pathway. Cells. 2020;9 doi: 10.3390/cells9020483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neehus AL, Lam J, Haake K, Merkert S, Schmidt N, Mucci A, Ackermann M, Schubert M, Happle C, Kühnel MP, Blank P, et al. Impaired IFNγ-Signaling and Mycobacterial Clearance in IFNγR1-Deficient Human iPSC-Derived Macrophages. Stem Cell Reports. 2018;10:7–16. doi: 10.1016/j.stemcr.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dannenmann B, Klimiankou M, Oswald B, Solovyeva A, Mardan J, Nasri M, Ritter M, Zahabi A, Arreba-Tutusaus P, Mir P, Stein F, et al. iPSC modeling of stage-specific leukemogenesis reveals BAALC as a key oncogene in severe congenital neutropenia. Cell Stem Cell. 2021 doi: 10.1016/j.stem.2021.03.023. [DOI] [PubMed] [Google Scholar]
- 23.Dannenmann B, Zahabi A, Mir P, Oswald B, Bernhard R, Klimiankou M, Morishima T, Schulze-Osthoff K, Zeidler C, Kanz L, Lachmann N, et al. Human iPSC-based model of severe congenital neutropenia reveals elevated UPR and DNA damage in CD34(+) cells preceding leukemic transformation. Exp Hematol. 2019;71:51–60. doi: 10.1016/j.exphem.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Le Voyer T, Neehus AL, Yang R, Ogishi M, Rosain J, Alroqi F, Alshalan M, Blumental S, Al Ali F, Khan T, Ata M, et al. Inherited deficiency of stress granule ZNFX1 in patients with monocytosis and mycobacterial disease. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2102804118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makaryan V, Kelley ML, Fletcher B, Bolyard AA, Aprikyan AA, Dale DC. Elastase inhibitors as potential therapies for ELANE-associated neutropenia. J Leukoc Biol. 2017;102:1143–1151. doi: 10.1189/jlb.5A1016-445R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pittermann E, Lachmann N, MacLean G, Emmrich S, Ackermann M, Göhring G, Schlegelberger B, Welte K, Schambach A, Heckl D, Orkin SH, et al. Gene correction of HAX1 reversed Kostmann disease phenotype in patient-specific induced pluripotent stem cells. Blood Adv. 2017;1:903–914. doi: 10.1182/bloodadvances.2016003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kropp C, Kempf H, Halloin C, Robles-Diaz D, Franke A, Scheper T, Kinast K, Knorpp T, Joos TO, Haverich A, Martin U, et al. Impact of Feeding Strategies on the Scalable Expansion of Human Pluripotent Stem Cells in Single-Use Stirred Tank Bioreactors. Stem Cells Transl Med. 2016;5:1289–1301. doi: 10.5966/sctm.2015-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manstein F, Ullmann K, Kropp C, Halloin C, Triebert W, Franke A, Farr CM, Sahabian A, Haase A, Breitkreuz Y, Peitz M, et al. High density bioprocessing of human pluripotent stem cells by metabolic control and in silico modeling. Stem Cells Transl Med. 2021 doi: 10.1002/sctm.20-0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halloin C, Schwanke K, Löbel W, Franke A, Szepes M, Biswanath S, Wunderlich S, Merkert S, Weber N, Osten F, de la Roche J, et al. Continuous WNT Control Enables Advanced hPSC Cardiac Processing and Prognostic Surface Marker Identification in Chemically Defined Suspension Culture. Stem Cell Reports. 2019;13:775. doi: 10.1016/j.stemcr.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kempf H, Kropp C, Olmer R, Martin U, Zweigerdt R. Cardiac differentiation of human pluripotent stem cells in scalable suspension culture. Nat Protoc. 2015;10:1345–1361. doi: 10.1038/nprot.2015.089. [DOI] [PubMed] [Google Scholar]
- 31.Olmer R, Engels L, Usman A, Menke S, Malik MNH, Pessler F, Göhring G, Bornhorst D, Bolten S, Abdelilah-Seyfried S, Scheper T, et al. Differentiation of Human Pluripotent Stem Cells into Functional Endothelial Cells in Scalable Suspension Culture. Stem Cell Reports. 2018;10:1657–1672. doi: 10.1016/j.stemcr.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sahabian A, Dahlmann J, Martin U, Olmer R. Production and cryopreservation of definitive endoderm from human pluripotent stem cells under defined and scalable culture conditions. Nat Protoc. 2021;16:1581–1599. doi: 10.1038/s41596-020-00470-5. [DOI] [PubMed] [Google Scholar]
- 33.Buchrieser J, James W, Moore MD. Human Induced Pluripotent Stem Cell-Derived Macrophages Share Ontogeny with MYB-Independent Tissue-Resident Macrophages. Stem Cell Reports. 2017;8:334–345. doi: 10.1016/j.stemcr.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rafiei Hashtchin A, Fehlhaber B, Hetzel M, Manstein F, Stalp JL, Glage S, Abeln M, Zweigerdt R, Munder A, Viemann D, Ackermann M, et al. Human iPSC-derived macrophages for efficient Staphylococcus aureus clearance in a murine pulmonary infection model. Blood Adv. 2021 doi: 10.1182/bloodadvances.2021004853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fattorelli N, Martinez-Muriana A, Wolfs L, Geric I, De Strooper B, Mancuso R. Stem-cell-derived human microglia transplanted into mouse brain to study human disease. Nat Protoc. 2021;16:1013–1033. doi: 10.1038/s41596-020-00447-4. [DOI] [PubMed] [Google Scholar]
- 36.Capotondo A, Milazzo R, Garcia-Manteiga JM, Cavalca E, Montepeloso A, Garrison BS, Peviani M, Rossi DJ, Biffi A. Intracerebroventricular delivery of hematopoietic progenitors results in rapid and robust engraftment of microglia-like cells. Sci Adv. 2017;3:e1701211. doi: 10.1126/sciadv.1701211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bird TG, Lu WY, Boulter L, Gordon-Keylock S, Ridgway RA, Williams MJ, Taube J, Thomas JA, Wojtacha D, Gambardella A, Sansom OJ, et al. Bone marrow injection stimulates hepatic ductular reactions in the absence of injury via macrophage-mediated TWEAK signaling. Proc Natl Acad Sci U S A. 2013;110:6542–6547. doi: 10.1073/pnas.1302168110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moroni F, Dwyer BJ, Graham C, Pass C, Bailey L, Ritchie L, Mitchell D, Glover A, Laurie A, Doig S, Hargreaves E, et al. Safety profile of autologous macrophage therapy for liver cirrhosis. Nat Med. 2019;25:1560–1565. doi: 10.1038/s41591-019-0599-8. [DOI] [PubMed] [Google Scholar]
- 39.Klichinsky M, Ruella M, Shestova O, Lu XM, Best A, Zeeman M, Schmierer M, Gabrusiewicz K, Anderson NR, Petty NE, Cummins KD, et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat Biotechnol. 2020;38:947–953. doi: 10.1038/s41587-020-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ackermann M, Mucci A, McCabe A, Frei S, Wright K, Snapper SB, Lachmann N, Williams DA, Brendel C. Restored macrophage function ameliorates disease pathophysiology in a mouse model for IL10 receptor deficient very early onset inflammatory bowel disease. J Crohns Colitis. 2021 doi: 10.1093/ecco-jcc/jjab031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ackermann M, Kuhn A, Kunkiel J, Merkert S, Martin U, Moritz T, Lachmann N. Ex vivo Generation of Genetically Modified Macrophages from Human Induced Pluripotent Stem Cells. Transfus Med Hemother. 2017;44:135–142. doi: 10.1159/000477129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopez-Yrigoyen M, Yang CT, Fidanza A, Cassetta L, Taylor AH, McCahill A, Sellink E, von Lindern M, van den Akker E, Mountford JC, Pollard JW, et al. Genetic programming of macrophages generates an in vitro model for the human erythroid island niche. Nat Commun. 2019;10:881. doi: 10.1038/s41467-019-08705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ackermann M, Haake K, Kempf H, Kaschutnig P, Weiss AC, Nguyen AHH, Abeln M, Merkert S, Kühnel MP, Hartmann D, Jonigk D, et al. A 3D iPSC-differentiation model identifies interleukin-3 as a regulator of early human hematopoietic specification. Haematologica. 2020 doi: 10.3324/haematol.2019.228064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernecker C, Ackermann M, Lachmann N, Rohrhofer L, Zaehres H, Araúzo-Bravo MJ, van den Akker E, Schlenke P, Dorn I. Enhanced Ex Vivo Generation of Erythroid Cells from Human Induced Pluripotent Stem Cells in a Simplified Cell Culture System with Low Cytokine Support. Stem Cells Dev. 2019;28:1540–1551. doi: 10.1089/scd.2019.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi KD, Vodyanik M, Slukvin II. Hematopoietic differentiation and production of mature myeloid cells from human pluripotent stem cells. Nat Protoc. 2011;6:296–313. doi: 10.1038/nprot.2010.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doulatov S, Vo LT, Chou SS, Kim PG, Arora N, Li H, Hadland BK, Bernstein ID, Collins JJ, Zon LI, Daley GQ. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell. 2013;13:459–470. doi: 10.1016/j.stem.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vo LT, Kinney MA, Liu X, Zhang Y, Barragan J, Sousa PM, Jha DK, Han A, Cesana M, Shao Z, North TE, et al. Regulation of embryonic haematopoietic multipotency by EZH1. Nature. 2018;553:506–510. doi: 10.1038/nature25435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gutbier S, Wanke F, Dahm N, Rümmelin A, Zimmermann S, Christensen K, Köchl F, Rautanen A, Hatje K, Geering B, Zhang JD, et al. Large-Scale Production of Human iPSC-Derived Macrophages for Drug Screening. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21134808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 50.Cao X, Yakala GK, van den Hil FE, Cochrane A, Mummery CL, Orlova VV. Differentiation and Functional Comparison of Monocytes and Macrophages from hiPSCs with Peripheral Blood Derivatives. Stem Cell Reports. 2019;12:1282–1297. doi: 10.1016/j.stemcr.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ackermann M, Lachmann N, Hartung S, Eggenschwiler R, Pfaff N, Happle C, Mucci A, Göhring G, Niemann H, Hansen G, Schambach A, et al. Promoter and lineage independent anti-silencing activity of the A2 ubiquitous chromatin opening element for optimized human pluripotent stem cell-based gene therapy. Biomaterials. 2014;35:1531–1542. doi: 10.1016/j.biomaterials.2013.11.024. [DOI] [PubMed] [Google Scholar]
- 52.Hong D, Ding J, Li O, He Q, Ke M, Zhu M, Liu L, Ou WB, He Y, Wu Y. Human-induced pluripotent stem cell-derived macrophages and their immunological function in response to tuberculosis infection. Stem Cell Res Ther. 2018;9:49. doi: 10.1186/s13287-018-0800-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lopez-Yrigoyen M, Fidanza A, Cassetta L, Axton RA, Taylor AH, Meseguer-Ripolles J, Tsakiridis A, Wilson V, Hay DC, Pollard JW, Forrester LM. A human iPSC line capable of differentiating into functional macrophages expressing ZsGreen: a tool for the study and in vivo tracking of therapeutic cells. Philos Trans R Soc Lond B Biol Sci. 2018;373 doi: 10.1098/rstb.2017.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neehus AL, Moriya K, Nieto-Patlán A, Le Voyer T, Lévy R, Özen A, Karakoc-Aydiner E, Baris S, Yildiran A, Altundag E, Roynard M, et al. Impaired respiratory burst contributes to infections in PKCδ-deficient patients. J Exp Med. 2021;218 doi: 10.1084/jem.20210501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuo HH, Gao X, DeKeyser JM, Fetterman KA, Pinheiro EA, Weddle CJ, Fonoudi H, Orman MV, Romero-Tejeda M, Jouni M, Blancard M, et al. Negligible-Cost and Weekend-Free Chemically Defined Human iPSC Culture. Stem Cell Reports. 2020;14:256–270. doi: 10.1016/j.stemcr.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishida T, Fujihara N, Nishimura T, Funabashi H, Hirota R, Ikeda T, Kuroda A. Live-cell imaging of macrophage phagocytosis of asbestos fibers under fluorescence microscopy. Genes Environ. 2019;41:14. doi: 10.1186/s41021-019-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lewis LE, Bain JM, Okai B, Gow NA, Erwig LP. Live-cell video microscopy of fungal pathogen phagocytosis. J Vis Exp. 2013 doi: 10.3791/50196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data for Figures 7 and 8 are available at http://flowrepository.org/ (Repository IDs: FR-FCM-Z4WC (Figure 7) and FR-FCM-Z4WE (Figure 8)). The raw data for Figure 6 is available as source data.