Summary
Gene activation requires the cooperative activity of multiple transcription factors at cis-regulatory elements. Yet, most transcription factors have short residence time, questioning the requirement of their physical co-occupancy on DNA to achieve cooperativity. Here, we present a DNA footprinting method that detects individual molecular interactions of transcription factors and nucleosomes with DNA in vivo. We apply this strategy to quantify the simultaneous binding of multiple transcription factors on single DNA molecules at mouse cis-regulatory elements. Analysis of the binary occupancy patterns at thousands of motif combinations reveals that high DNA co-occupancy occurs for most types of transcription factors, in absence of direct physical interaction, at sites of competition with nucleosomes. Perturbation of pairwise interactions demonstrates the function of molecular co-occupancy in binding cooperativity. Our results reveal the interactions regulating cis-regulatory elements at molecular resolution and identify DNA co-occupancy as a widespread cooperativity mechanism used by transcription factors to remodel chromatin.
Keywords: Gene regulation, enhancers, chromatin, genomics, transcription factor cooperativity
Introduction
The binding of transcription factors (TFs) translates the regulatory information contained in cis-regulatory elements (CREs) into gene expression patterns. Upon binding, TFs activate or repress transcription by recruiting protein complexes that modulate the activity of RNA Polymerase II (Pol II) at promoters of genes. The DNA binding domains of TFs recognize 6-25bp DNA sequence motifs with low specificity (Inukai et al., 2017). Thus, each individual TF has millions of theoretical recognition sequences in mammalian genomes, few of which are observed to be bound in vivo (Neph et al., 2012). The affinity of a TF for its motif does not explain its genome occupancy; instead, combinatorial action of multiple TFs likely shapes the precise control of binding at CREs (Gerstein et al., 2012; Iwafuchi-Doi and Zaret, 2014; Yan et al., 2013).
Evidence that TFs may collaborate to bind CREs came from the observation that certain key TFs tend to frequently bind the same set of CREs in the genome (Junion et al., 2012; Siersbæk et al., 2014; Tijssen et al., 2011). This dependency was further demonstrated by deleting individual TFs or their motifs and measuring the effect on the binding of other TFs at target CREs (Junion et al., 2012; Siersbæk et al., 2014). Complementary evidence for binding dependency comes from comparative genomics showing that binding of a given TF can correlate with changes in the genotype affecting neighbouring binding motifs (He et al., 2011; Kilpinen et al., 2013; Stefflova et al., 2013). These studies have established cooperativity as a prevalent mechanism explaining TF binding at CREs, yet lack the necessary resolution to determine the precise mechanisms of TF cooperativity at specific loci.
Several mechanisms have been described to explain the binding cooperativity of TFs (reviewed in (Deplancke et al., 2016; Inukai et al., 2017; Morgunova and Taipale, 2017; Reiter et al., 2017; Spitz and Furlong, 2012)). Cooperativity was shown to occur through direct protein-protein interactions between TFs (TF-TF). For instance, TFs such as Myc/Max or NFY undergo physical interactions that combine different TF DNA binding domains to increase their binding affinity and motif specificity (Amoutzias et al., 2008; Luna-Zurita et al., 2016). A systematic study has estimated the existence of >800 interactions between TFs, largely expanding the binding repertoire of the ~2000 known human TFs (Ravasi et al., 2010). Cooperativity has also been observed in the absence of direct TF-TF interactions through changes of the local DNA structure upon binding (Ibarra et al., 2020; Jolma et al., 2015). Finally, passive TF cooperativity has been proposed to occur through the competition of TFs with nucleosomes (Klemm et al., 2019; Spitz and Furlong, 2012). At some CREs, binding of a single TF might be insufficient to displace nucleosomes; instead, collective binding of multiple TFs may be required (Mirny, 2010; Polach and Widom, 1995, 1996; Wasson and Hartemink, 2009). Collective TF binding has been observed when TFs compete with nucleosomes to activate genes (Miller and Widom, 2003) as well as in vivo, using artificial systems (Adams and Workman, 1995; Pettersson and Schaffner, 1990; Vashee et al., 1998).
An important open question is whether simultaneous co-occupancy of DNA by multiple TFs is required for binding cooperativity. Most TFs have short residence times on DNA (Agarwal et al., 2017; Arnold et al., 2013a; Gebhardt et al., 2013; Sung et al., 2014) and binding cooperativity was observed in absence of physical co-occupancy for some TFs (Voss et al., 2011). Addressing this question requires direct quantification of how frequently two TFs co-occupy the same DNA molecule in vivo. Current methods to measure TF binding, such as Chromatin-Immunoprecipitation (ChIP) are based on enrichment of TF-bound DNA fragments. These approaches precisely resolve the binding of individual TFs but, they lose information on the co-occurrence of binding at neighbouring sites. To overcome these limitations, we recently developed a Single Molecule Footprinting (SMF) approach for Drosophila melanogaster genomes. SMF uses recombinant enzymes to methylate accessible regions of the genome. Its combination with bisulfite-sequencing allows to quantify the footprints created by DNA-bound proteins on individual molecules (Krebs et al., 2017).
Here, we adapt SMF for mammalian genomes and demonstrate that the assay resolves TF binding and nucleosome occupancy (NO) at single molecule resolution. We show that SMF allows the simultaneous quantification of multiple TF binding events on single DNA molecules, enabling us to systematically quantify the frequency of co-occupancy for thousands of TF pairs across the genome. Analysis of these molecular TF occupancy patterns reveals that simultaneous binding is largely independent of the identity of the TFs involved, does not require a strict organization of motifs and is prevalent at regions having high nucleosome occupancy. Reduction of TF concentration using siRNAs indicate that co-occupancy is a mechanism used by TFs to bind their cognate motifs. Altogether, our data comprehensively identify TF interactions at CREs at molecular resolution and show that DNA co-occupancy is widespread at sites of TF cooperativity.
Results
Single Molecule Footprinting of the mouse genome
Quantification of genomic binding events by SMF requires the sequencing of a large number of DNA molecules encompassing the binding regions of the factor of interest. Generation of high coverage SMF datasets is challenging in mammalian genomes, which are a factor of twenty bigger than Drosophila genomes. To overcome this bottleneck, we took advantage of the fact that TF binding is mostly restricted to CREs, which represent only a small fraction of the genome (Stamatoyannopoulos et al., 2012). We employed DNA capture to enrich for CREs prior to sequencing (Figure 1A). We used a library of RNA baits tiling 297,000 regions (~2% of the genome), covering a large fraction of CREs accessible in mouse embryonic stem cells (mESCs) (59.7% of open regions as detected by DNAse-seq).
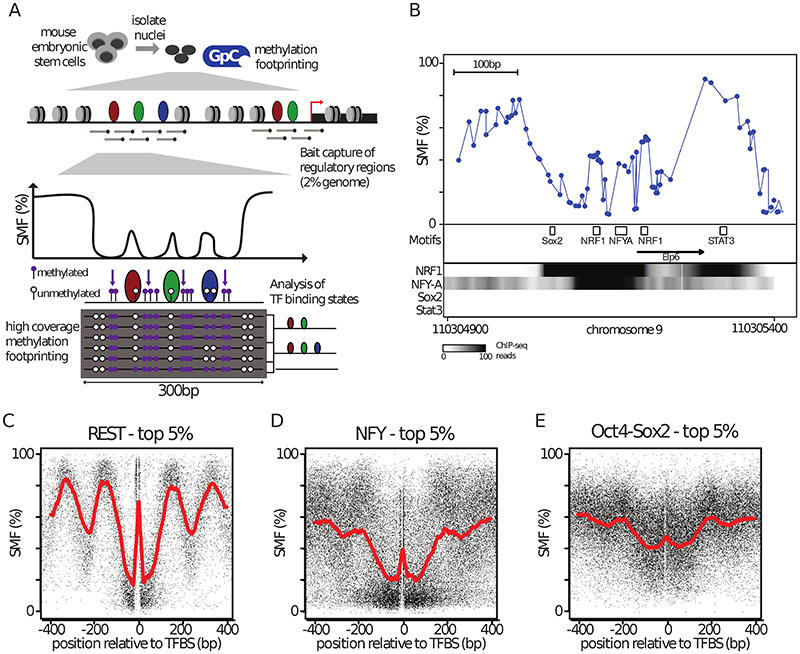
Figure 1. Methylation Footprinting detects Transcription Factors in mouse embryonic stem cells.
(A) Overview of Single Molecule Footprinting (SMF) for mammalian genomes. Nuclei are isolated and incubated with a recombinant cytosine methyl-transferase that targets GCs (M.CviPI) which are distinct from CGs that are endogenously methylated. To generate data with a genomics coverage compatible with single molecule interpretation, the resulting methylated DNA is subjected to capture using probes targeting ~60% of the Cis-Regulatory Elements (CREs) used in mESCs. Methylation is detected by bisulfite sequencing using long reads (300bp), enabling the detection and analysis of multiple footprints on a single molecule. (B) SMF pattern at the active promoter of the Elp6 gene revealing short footprints (<20bp) at binding sites of the activators NRF1 and NFY. Shown is the inverse frequency of methylation (1-methylation (%)) (blue line). Black boxes represent the location of consensus motifs for TFs. Black arrow indicates the transcriptional start site. Read counts for ChIP-seq of the respective TFs are shown as intensity heatmap (datasets as indicated), revealing strong binding of NFYA and NRF1 but no detectable binding for Sox2 and Stat3. (C-E) TF binding creates discrete footprints detectable by SMF. Composite profile of SMF signal at various (C, D, E) bound TF motifs (top 5% of the respective TF ChIP-seq) as indicated. Shown is the footprinting frequency (1-methylation [%]) of individual cytosines (black dots). See also Figure S1.
Unlike Drosophila, mammalian genomes have endogenous DNA methylation in CG context (Stadler et al., 2011), preventing the use of exogenous methyl-transferases targeting this sequence context and thus, reducing the spatial resolution of SMF. We took two complementary approaches to avoid interference of endogenous DNA methylation with SMF. First, we used the methyltransferase M.CviPI which only methylates GC dinucleotides, therefore compromising on the spatial resolution of the assay (median distance of ~14bp). Second, we leveraged the ability of mESCs to proliferate in absence of DNA methylation (Tsumura et al., 2006). We used an isogenic mESC line depleted for all three DNA methyl-transferases (DNMT TKO), which shows only discrete changes in chromatin accessibility and gene expression (Domcke et al., 2015). This line enables the use of GC as well as CG methyl-transferases for footprinting, significantly increasing the spatial resolution (up to 7bp). However, such analysis is limited to the stem cell state (Sakaue et al., 2010).
We generated high coverage SMF datasets in wild-type (WT) and DNMT TKO mESCs with highly reproducible methylation footprints between biological replicates (R>0.90, Figure S1A, B). Moreover, footprinting levels were in close agreement between WT and DNMT TKO cells (R=0.90, Figure S1C), consistent with previous observations (Domcke et al., 2015). The high capture efficiency (>70% of reads were within bait regions) achieved coverage levels that allowed data interpretation at the single molecule resolution for 78 807 CREs in mESCs.
Detection of TF binding using SMF
When inspecting SMF signal at CREs, we observed discrete footprints (<25bp) around TF motifs (as exemplified in the Elp6 promoter, Figure 1B). These motifs are recognized by TFs that have ChIP-seq enrichments at respective regions (Figure 1B), suggesting that TF binding results in footprints detectable by SMF, consistent with previous observations (Abdulhay et al., 2020; Gal-Yam et al., 2006; Kelly et al., 2012; Levo et al., 2017; Shipony et al., 2020; Stergachis et al., 2020). This prompted us to assess whether footprints at TF motifs can be found genome-wide and ask whether their presence is consistent with orthogonal measures of TF binding. We used published TF ChIP-seq datasets (Supplementary Table 1) to identify the subset of motifs that are bound by their TF genome-wide. We plotted the SMF signal around these motifs and found that some TFs, such as the transcriptional repressor RE1-Silencing Transcription factor (REST), create short footprints over their bound sites (Figure 1C), in contrast to unbound motifs (Figure S1F). These footprints are directly flanked by highly accessible regions and larger periodic footprints consistent with nucleosomal phasing (Figure 1C). We also detected footprints at the binding sites of activators, such as NFY (Figure 1D, Figure S1G) or Oct4-Sox2 (Figure 1E, Figure S1H). A systematic assessment of footprints at bound motifs revealed that many TFs create footprints detectable by SMF (Figure S1J). These vary in size (~15-30bp) and intensity, suggesting that TFs occupy DNA at different frequencies. We conclude that methylation footprinting has the sensitivity and the resolution to detect TF binding at CREs.
Heterogeneity of occupancy at TF binding regions
Competition between TFs and nucleosomes is assumed to be a critical determinant of TF binding (Iwafuchi-Doi and Zaret, 2014; Morgunova and Taipale, 2017). To determine the fraction of molecules bound by TFs or nucleosomes at individual TF motifs, we adapted the single molecule classification strategy we developed to study GTF binding at promoters (Krebs et al., 2017). We combined the accessibility information at the TF binding location with its flanking regions to discriminate short footprints, presumably created by TFs, from longer nucleosomal footprints (Figure S2A). We collected methylation information in a 15bp window around TF motifs to avoid restricting our analysis to TFs having GCs in their recognition motifs. For each individual molecule, the algorithm collects binarized methylation within three bins, creating 8 (23) possible combinations (see Methods). We further grouped these patterns into three binding states, separating molecules showing a short footprint at the TF motif, fully accessible molecules (no detectible binding at the motif), and molecules showing large nucleosomal footprints (Figure S2B, C).
We applied single molecule quantification of TF binding to the 7383 REST motifs targeted by our capture method. In the GC methyltransferase treated WT ESC, we reproducibly quantified the binding frequency of REST for >77% of the 1357 motifs that contain informative GCs within all three collection bins (Figure S2D). The high percentage of recovery confirms the efficiency of our DNA capture strategy.
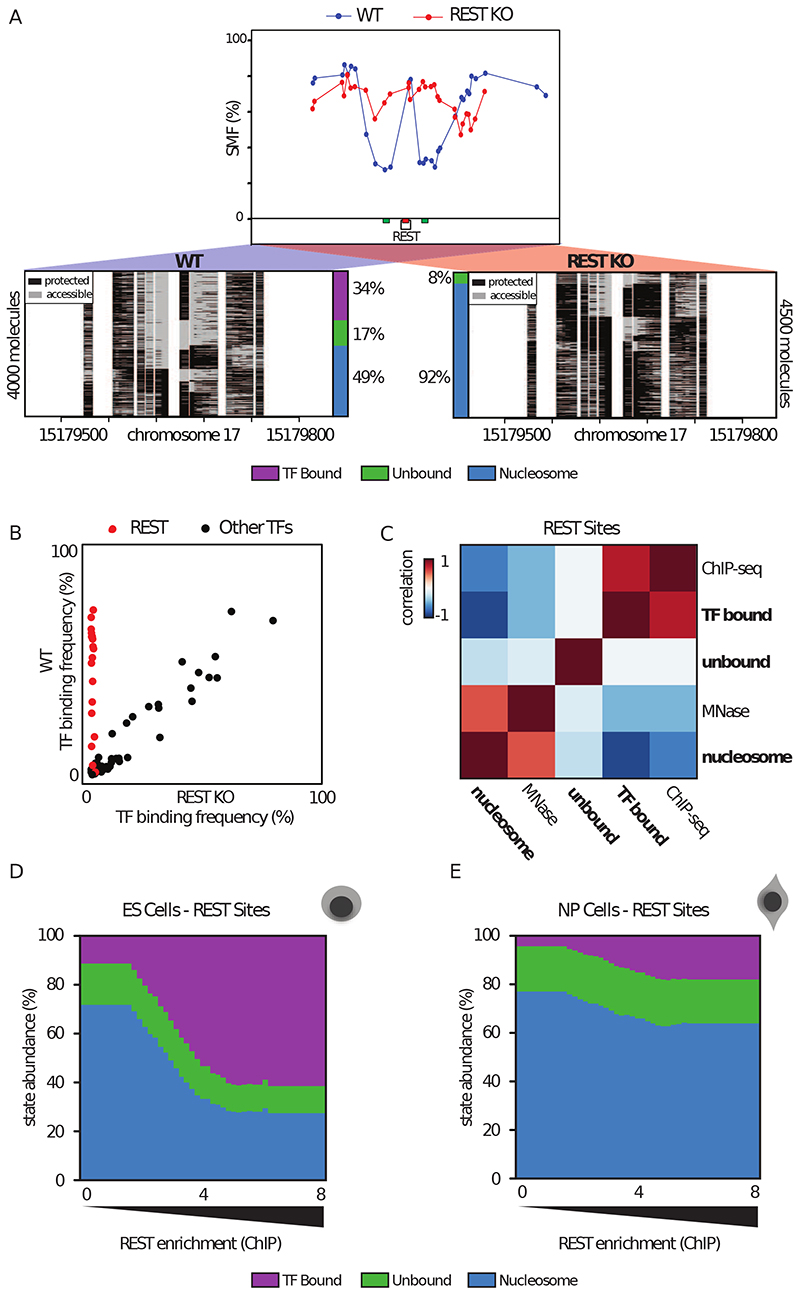
To analyse single molecule accessibility, we stacked the sequencing reads covering each locus and sorted them according to their binding states (Figure 2A). When inspecting these data, we observed considerable heterogeneity in the accessibility patterns of bound REST motifs. For instance, even at a highly bound site only 34% of the DNA molecules showed a short footprint at the motif potentially created by REST (Figure 2A, left panel). Other molecules were accessible (unbound, 17%) or harboured larger footprints compatible with nucleosomal occupancy (49%). If these short footprints are created by REST, genetic deletion of the TF should abolish them. We compared REST single molecule profiles with those obtained in cells genetically depleted for REST (REST-KO) (Chen et al., 1998) (Figure S2E). In the absence of REST, the discrete footprint at the binding site disappeared and almost all molecules showed the large footprints assigned to nucleosomes (92%) (Figure 2A, right panel). A complete loss of the TF bound molecules was observed for all 16 REST binding sites tested (Figure 2B), while the binding frequencies of sites bound by other factors remained unchanged (Figure 2B; Tables S2, S3 for binding site selection). Together, these suggest that the footprints observed at REST motifs are created by the binding of the transcription factor REST.
Figure 2. Quantification of TF binding frequency at single DNA molecule resolution.
(A) Single-locus examples of a REST bound region in wild type (WT, blue line and dots) versus REST knock out (REST-KO, red line and dots) mESCs. Shown are average methylation levels (top panel, blue dots connected by a blue line) and single-molecule stacks (bottom panels) measured by targeted amplicon bisulfite sequencing, sorted into three states using the classification algorithm. Informative cytosines (GpC) are represented by vertical stripes (methylated Cs, accessible, light gray; unmethylated Cs, protected, black), separated by a blank space. The vertical side bar depicts the frequency of each state. Color legend for the states is given on the bottom of the panel (purple: TF bound; green: accessible; blue: nucleosome occupied). The percentages of molecules in each state are indicated on the right side of the plot. The number of sequenced molecules is displayed on the side and varies as a function of the coverage of individual samples. Collection bins are indicated on x-axis (red: TF bin; green: Flank bins). (B) Scatter plot comparing TF binding frequencies in WT and REST KO mESC at 16 REST motifs (red) and motifs of other TFs (black) covered by 96 amplicons using targeted SMF. (C) Global relationship between state frequencies and independent bulk measurements of TF and nucleosome occupancies at REST motifs, depicted as a heatmap of similarity (Pearson correlation). States separate into two groups that either correlate with occupancy by the TF (ChIP-seq) or nucleosomes (MNase-seq) illustrating accurate state quantification. (D-E) Distribution of state frequencies in mESC (D) and neuronal progenitors (NP) (E) as a function of REST occupancy as determined by ChIP-seq. Cumulative bar plot depicting the distribution of state frequencies. TF motifs were binned based on REST enrichment in mESCs (log2 ChIP-seq) and the median frequency of each state was calculated within each bin. The frequency of each state is color coded as in Figure 2A. TF occupancy changes across loci and between cell types are tightly coupled with nucleosome occupancy. See also Figure S2.
If SMF accurately quantifies TF binding and nucleosome occupancy, then the frequency of the TF footprints should scale with TF and NO as determined by bulk assays. For all captured REST motifs, we analysed how the frequencies of different states as measured by SMF scale with REST ChIP-seq and MNase-seq signal at those respective motifs. We found that across all motifs, the TF bound state strongly correlated with REST ChIP-seq signal (R=0.77, Figure 2C, Figure S2C). Conversely, frequency of the nucleosome occupied state showed a correlation with NO at the TF binding site as measured by MNase-seq (R=0.53, Figure 2C). We conclude that SMF simultaneously quantifies TF and nucleosome occupancy, revealing the heterogeneity of binding patterns at CREs.
Co-variation of nucleosome and TF binding at REST binding sites
The heterogeneity observed in SMF patterns implies that within a cell population, most REST motifs can either be occupied by REST or by nucleosomes, suggesting a competition between these two states. If so, variations in TF binding and NO levels should co-vary when compared across these regions. To test this idea, we analysed the relationship between TF and NO as a function of REST binding intensity (Figure 2D). The frequency of TF bound molecules grew with increasing ChIP-seq enrichment, with up to ~60% occupancy for the top TF bound sites. Increases in TF occupancy were accompanied by a proportional decrease in NO (Figure 2D). Overall, nucleosomes occupy 20-60% of the REST bound sites (Figure 2D), implying that at any given time only a fraction of the cells undergo TF binding at a particular binding site.
The coupling between the states suggests a possible competition between NO and REST at its binding sites. To further test this possibility, we investigated how binding states redistribute upon perturbation of REST expression levels. During the early stages of neuronal differentiation, REST expression levels are reduced by ~4-fold (Figure S2F) without significant redistribution of REST target sites (Arnold et al., 2013b). We generated a SMF dataset in in vitro derived neuronal progenitors (NPs) (Bibel et al., 2004) (Figure S1D, E). Upon reduction of REST expression, we observed a global decrease of ~4-fold in REST occupancy at its motifs (Figure 2E), indicating that REST abundance correlates with its binding frequency in the cell population. We also observed a concomitant increase in NO at REST sites (Figure 2E), whereas there was very little effect on CTCF binding frequencies at CTCF binding regions in NPs (Figure S2G, H, I). This co-variation in TF and NO suggests that the binding heterogeneity observed at REST sites could be the result of the competition between the TF and nucleosomes for DNA occupancy.
TFs adopt various strategies to compete with nucleosomes for DNA occupancy
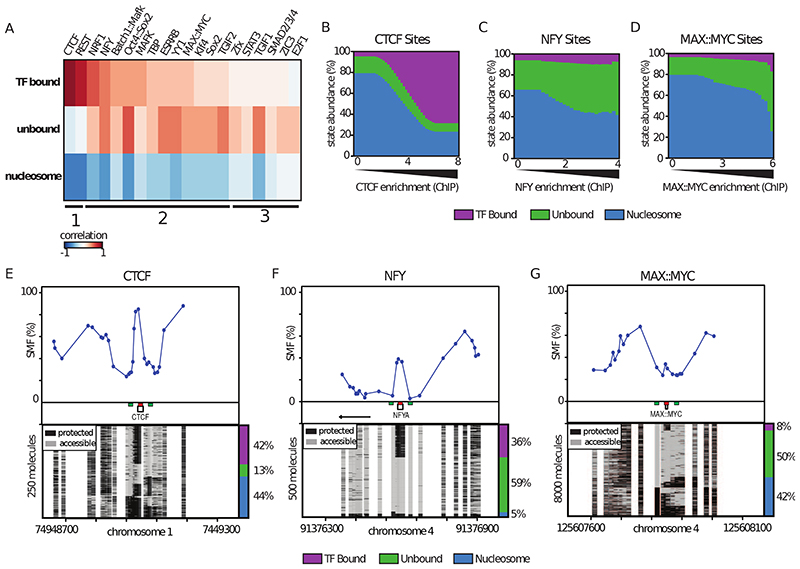
A small number of TFs are able to bind their target sites in isolation (i.e. CTCF), but most bind within larger clusters of motifs at CREs. To understand how this functional diversity influences the single molecule occupancy of TFs, and how it relates to alterations of chromatin structure, we classified the footprint patterns around all binding sites for which binding motifs are known (~500 from JASPAR (Mathelier et al., 2015)). We subsequently selected the motifs for which ChIP-seq data were available in mESCs (Table S1), and obtained genome-wide TF binding frequencies for 20 TFs that were highly correlated between biological replicates (Figure S3). For these TFs, we asked if the frequency of unbound, TF bound, or nucleosome occupied DNA molecules correlates with TF enrichment as measured by ChIP-seq (Figure 3A). We observed that CTCF has very similar binding characteristics to REST (Figure 3A; category 1), where molecular occupancy by the TF correlates with ChIP-seq enrichment (Figure 3A). For these two factors, we observed that DNA molecules are bound by either the TF or nucleosomes with a constant small fraction (~10%) of unbound DNA molecules (Figure 3B, E).
Figure 3. Identification of the strategies used by TFs to outcompete nucleosomes.
(A) Global relationship between SMF state frequencies and TF occupancy as determined by ChIP-seq at TF motifs. Heatmap of similarity (Pearson correlation) between the frequency of states (y-axis label) at specific TF motifs and ChIP-seq enrichment of the matching TF (x-axis label). Columns are ranked by similarity between the TF bound state and ChIP-seq. The bottom lines indicate groups of TFs sharing similar profiles. (B-D) Distribution of state frequencies as a function of TF occupancy as determined by ChIP-seq for (B) CTCF, (C) NFY, (D) MAX. Cumulative bar plot depicting the distribution of state frequencies. TF motifs were binned based on their respective ChIP-seq enrichment in mESCs (log2), and the median frequency of each state was calculated within each bin. The frequency of each state is color coded as in Figure 2A, color legend for the states is given on the bottom of the panels. (E-G) Single-locus examples of a (E) CTCF, (F) NFY, (G) MAX bound region mESCs. Shown are average methylation levels (top panel, blue dots connected by a blue line) and single-molecule stacks measured by targeted amplicon bisulfite sequencing, sorted into three states using the classification algorithm (methylated Cs, accessible, light gray; unmethylated Cs, protected, black). Collection bins are indicated on x-axis (red: TF bin; green: Flank bins). The vertical side bar depicts the frequency of each state. A color legend for the states is given on the bottom of the panels. The percentages of molecules in each state are indicated on the right side of the plot.
For a majority of the other TFs, the occurrence of TF footprints also scaled with ChIP-seq enrichments, but had lower maximal frequencies of TF-bound molecules (Figure 3A, C, D). In this category, a large fraction of DNA molecules was unbound (Figure 3A; category 2), as exemplified for the activators NFY or MAX (Figure 3C-D, F-G). For a smaller set of factors, we observed a good correlation between unbound DNA molecules and ChIP-seq enrichments with very infrequent TF-bound molecules (Figure 3A; category 3).
The degree of anti-correlation between the nucleosome-occupied fraction and ChIP-seq enrichments is lower for categories 2 and 3, compared to category 1 (Figure 3A). For factors in categories 2 and 3, a significant fraction of DNA molecules is unbound, even at low ChIP-seq enrichments (exemplified in Figure 3C, F). The frequency of these unbound DNA molecules increases at higher ChIP-seq enrichment. Together, these suggest that the competition between TFs and nucleosomes can only be partially explained by the binding of individual factors at these regions (Figure 2D, Figure 3B). This is consistent with the idea that many of these binding events occur within clusters of motifs that may collectively outcompete nucleosomes. We conclude that many of the tested TFs create footprints quantifiable at the single molecule level, but that the range of binding frequencies vary between TFs.
Quantification of the molecular co-occupancy of TFs
Having established the ability of SMF to quantify the binding of TFs, we developed a strategy to quantify their degree of molecular co-occupancy (see methods section, Figure S4A). Here, used the dual enzyme footprinting dataset generated in DNMT TKO cells, which enables quantification of approximately five times more binding events than in WT cells (Figure S4B). We did not observe major differences in TF binding frequencies when comparing DNMT TKO cells to WT cells (R=0.94, Figure S4C). In our analysis, we distinguished co-occupancy at dimeric motifs (Figure 4A) from TFs occupying distinct motifs lying in the vicinity of each other (15-140bp, Figure 4B). When applying this strategy to the TFs creating quantifiable footprints by SMF (Figure 3A), we obtained reproducible, high confidence co-occupancy measurements for 1238 TF dimers and 381 TF pairs (Figure S4B, C).
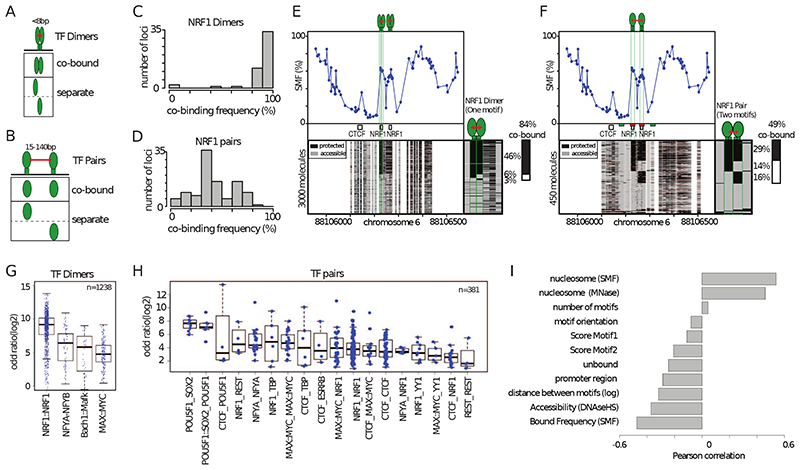
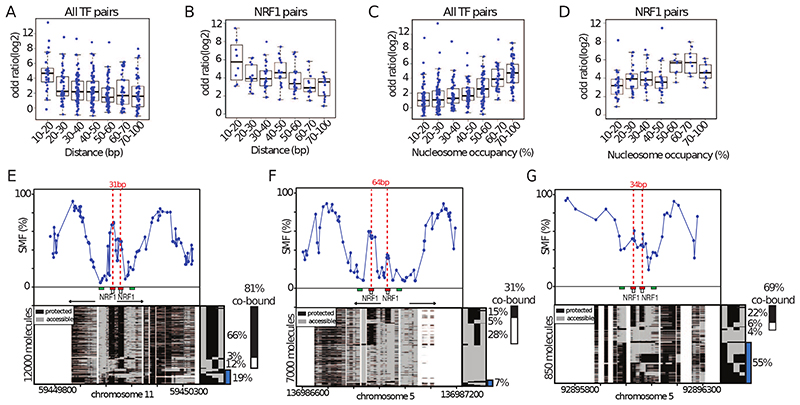
Figure 4. High TF co-occupancy does not rely on a precise motif arrangement.
(A) Footprints for dimeric TFs occur nearby (<8bp) on two halves of the same recognition motif. Schematic representation of the theoretical binding states expected to occur at such motifs, bound by TF dimers. (B) Footprints for neighboring binding sites occurs at separate recognition motifs within the same CRE (<140bp). Schematic representation of the theoretical binding states expected to occur at a pair of neighboring motifs. (C) Histogram depicting the percentage of dimeric co-occupancy at NRF1 motifs. (D) Histogram depicting the percentage of co-occupancy at neighboring pairs of NRF1 motifs. (E, F) Single-locus example of a region bound by NRF1 at two neighboring recognition motifs. Analysis of the degree of TF co-occupancy (E) within the NRF1 dimeric motif and (F) between neighboring motif pairs. Shown are average methylation levels (top panel, blue dots connected by a blue line) and single-molecule stacks measured by targeted amplicon bisulfite sequencing (bottom left), sorted into four states using the co-occupancy classification algorithm (methylated Cs, accessible, light gray; unmethylated Cs, protected, black). The states heatmap (bottom right, binarized heatmap) depicts the occupancy states for each of the protein analyzed in the pairs. The percentages of molecules where individual or co-binding are observed is indicated on the right side of the plot. The fraction of co-bound molecules within all bound molecules is indicated on top of a side bar depicting the co-bound frequency. Collection bins are indicated on x-axis (F) (red: TF bins; green: Flank bins). (G-H) Half motifs are frequently co-bound by TF dimers while motif pairs show highly variable degrees of co-occupancy. Degree of co-occupancy observed at (G) half-motifs bound by TF dimers, and (H) at neighboring motifs bound by TF pairs. Boxplot summarizing the log2 odds ratios of a Fischer’s exact test for each pair of TFs analyzed. Blue dots represent individual TF pairs. (I) Analysis of the determinants of TF co-occupancy. Barplot depicting the Pearson correlation coefficient between the degree of co-occupancy of TF pairs and various genomic features (y axis label). See also Figure S4.
Many active promoters harbour clustered motifs for the transcriptional activator NRF1, which binds a tandem repeat recognition sequence as a homodimer. We analysed whether NRF1 monomers have different co-occupancy properties when binding the two halves of the NRF1 motif as homodimers (Figure 4A) compared binding a pair of NRF1 motifs (Figure 4B). We observed nearly systematic co-occurrence of footprints at the NRF1 half-sites (Figure 4C). This high co-occurrence of footprints at NRF1 half sites is not simply explained by the close proximity of the collection bins (4bp) (Figure S4F and G). The frequencies of co-occupancy observed between neighbouring NRF1 motifs ranged from very low (<20%) to levels comparable to those between dimeric half-sites (>80%) (Figure 4D). This is also evident when analysing co-occupancy within half-sites (85%, Figure 4E) and between two NRF1 binding sites (48%, Figure 4F) of a single locus. Together, these data provide evidence that NRF1 binds as an obligatory homodimer in vivo and suggests that binding dependency between neighbouring NRF1 binding sites varies substantially from one CRE to another.
SMF is performed on permeabilized nuclei, in the absence of protein-DNA cross-linking. The residence times for TFs in living cells range from a few seconds to a couple of minutes, which are much shorter than the time nuclei are incubated with methyltransferases in vitro. Thus, it is possible that the co-occupancy patterns detected by SMF could reflect TF retention on chromatin in vitro, rather than binding dependencies between TFs occurring in vivo. To address this question, we developed an independent SMF protocol where we fixed protein-DNA interactions in vivo using formaldehyde prior to methylation footprinting (X-link SMF, see methods section). We observed minimal differences in TF binding frequencies when comparing SMF data between native and crosslinked conditions (Figure S4D). Moreover, TF co-occupancy profiles of individual loci were very similar at the single molecule level (Figure S4E, F). Together, these findings suggest that co-binding frequencies between TFs is highly variable at CREs.
Co-occupancy is largely independent of TF identity
To test if co-occupancy patterns are specific to the type of TFs involved, we compared the results obtained for NRF1 with those for other pairs of TF dimers or neighbouring binding motifs. For each binding event, we tested whether the observed TF co-occupancy exceeds the one expected by chance. Consistent with our observations for NRF1, we observed that co-occupancy at the half-sites of dimeric motifs is often higher than expected by chance (Figure 4G). However, none of the tested cases showed a systematic co-occupancy that would suggest an obligatory dimerization at comparable levels to what was observed for NRF1 dimers (Figure 4G). For example, we observed that the tandem E-box motif bound by the transcriptional activators Myc and Max showed varying degrees of co-occupancy across the genome (Figure 4G) and we could identify Myc-Max bound regions where >80% is monomeric (Figure S4F). Our data argues that dimerization is not a prerequisite for binding for most of the tested dimeric factors.
The degree of co-occupancy between pairs of TFs that bind distinct motifs (Figure 4H) is on average lower than the one we observed for TF dimers (Figure 4G). Grouping the data according to the identity of the TFs involved reveals that certain pair types tend to have higher co-occupancy than others (i.e. Sox2-Oct4, Figure 4H). However, we also observed a broad distribution of co-occupancies across the same TF-TF pair at different loci. For instance, we observed NRF1-NRF1 pairs with over 80% of co-occupancy at certain sites, but also lower than 20% co-occupancy at other sites (Figure 4D, H). Thus, certain TF combinations tend to co-occupy DNA more frequently than others. However, most of this variation cannot be explained by the identity of the TFs involved.
Co-occupancy decreases as a function of distance between motifs
Most of the tested pairs of TFs co-occupy DNA at high frequency at a subset of their binding loci. Thus, increased co-occupancy is unlikely to require specific TF-TF interaction domains. To identify potential determinants of the variation observed in TF co-occupancy patterns, we tested how much motif organisation (i.e. number of motifs, their score, distance and orientation) and the binding frequencies of TFs and nucleosomes predict the observed levels of co-occupancy (Figure 4I). Several of the tested parameters show a significant correlation with the degree of TF co-occupancy, including the inter-motif distance, NO at the binding region, and TF occupancy level (Figure 4I). Interestingly, the precise organisation of motifs in the CRE does not appear to be necessary for high co-occupancy. For instance, high co-occupancy does not require precise orientation of the motifs (Figure 4I). Moreover, we did not find strong evidence for an effect of the number of TF motifs in the cluster (Figure 4I, Figure S5A-C).
The degree of co-occupancy between TFs decreases as a function of the distance between their binding sites (Figure 4I, Figure 5A). Nearby binding events (within 20bp) show greater co-occupancy (Figure 5A). The frequencies of co-occupancy exponentially decrease with increasing inter-motif distance, with close to random distribution of binding for sites located at greater than 70bp apart (Figure 5A). Similar correlations were seen when considering TF identity (Figure 5B). A large majority of NRF1-NRF1 pairs were frequently co-bound when located at <40bp distance (Figure 5E), whereas co-occupancy was significantly reduced when the sites were located further apart (Figure 5F). However, while distance is an important determinant of co-occupancy, it only partially explains the co-occupancy levels observed at intermediate distances (20-70bp, Figure 5A, B). In summary, co-occupancy between TFs increases with proximity but this does not rely on precise spacing or orientation of the motifs.
Figure 5. Increased TF co-occupancy occurs at nucleosome occupied regions.
(A-B) Co-occupancy at pairs of motifs decrease as a function of genomic distance. Boxplot depicting the degree of co-occupancy as a function of the genomic distance between the TF motifs for (A) all analyzed TFs and (B) pairs of NRF1 motifs. Shown are odds ratios of a Fisher’s exact test (log2). (C-D) Co-occupancy between pairs of motifs is increased at regions having high nucleosomal occupancy. Boxplot depicting the degree of co-occupancy as a function of the fractions of molecules occupied by nucleosomes for (A) all analyzed TFs and (B) pairs of NRF1 sites. (E-F) Comparison of single loci harboring (E) nearby or (F) distant tandem NRF1 motifs. Same representation as in Figure 4F. Blue bar depicts the fraction of nucleosome occupied molecules (G) Example of a NRF1 bound locus having high nucleosome occupancy. Same representation as in Figure 5E. See also Figure S5.
TF co-occupancy is high at nucleosome occupied regions
Another predictor of TF co-occupancy is the fraction of DNA molecules occupied by nucleosomes at the same locus (Figure 4I). Conversely, the sum of the molecules bound by either of the two TFs is anti-correlated with co-occupancy (Figure 4I). This suggests that regions of high co-occupancy are sites that are only partially occupied by TFs with incomplete opening of the chromatin. Global analysis of SMF data reveals an increase in TF co-occupancy for regions, which display high frequencies (> 50%) of nucleosome binding (Figure 5C). While this observation is independent of TF identity (Figure 5C), it is also evident when analysing NRF1 pairs only (Figure 5D). This effect can be exemplified by comparing two loci bound by the same combination of TFs, with different degrees of NO. A locus with low NO (7% - Figure 5F) tends to have lower TF co-occupancy than a locus with high NO (55% - Figure 5G). When considered together, distance between binding sites and NO account for a larger fraction of the variance (41%) than when considered individually (10% and 29%, respectively), explaining why co-occupancy can be observed at regions in absence of high NO when inter-motif distance is short (Figure 5E). Regions of high NO are likely to be regions where TFs and nucleosomes are actively competing for DNA binding. In these cases, binding of individual TFs would be insufficient to overcome the energetic costs of outcompeting nucleosome binding, leading to a requirement for simultaneous DNA co-occupancy of multiple TFs at these loci.
TF co-occupancy identifies cooperativity between TFs
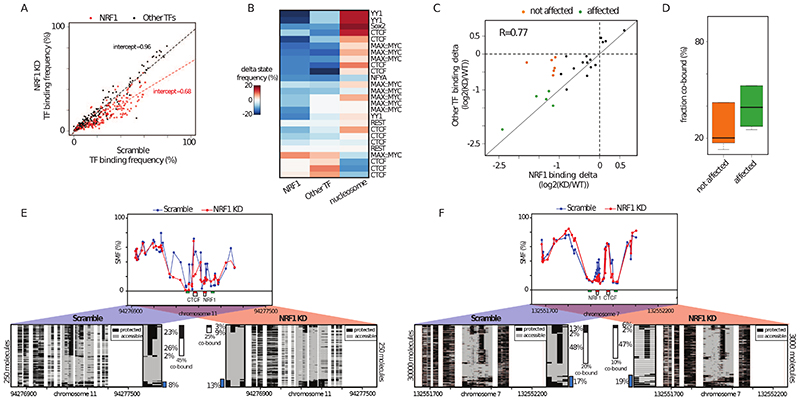
The high co-occupancy observed between certain TF pairs suggests the existence of a dependency between these factors for binding. If correct, perturbation of the binding of TFs with high frequency of co-occupancy should impact the binding of their partners. To test this hypothesis, we performed siRNA knock-down (KD) of NRF1. NRF1 has a broad range of co-occupancy frequencies and is involved in heterologous pairs with many other TFs (Figure 4H). Upon KD, we observed a reduction in NRF1 protein levels (Figure S6A) and an average decrease of about one third of NRF1 occupancy at all its binding sites (intercept=0,68, Figure 6A), in contrast with the binding frequencies of other TFs, which are generally unaffected (Figure 6A). At NRF1 motifs, we observed a gain of nucleosome-occupied DNA molecules, which corresponded to the loss of NRF1 binding at the respective site (Figure S6B).
Figure 6. TF co-occupancy is a mechanism of TF binding cooperativity.
(A) Specific reduction of TF footprints at NRF1 motifs upon NRF1 knock down (KD). Scatterplot depicting the binding frequency of TFs upon NRF1 KD. Binding frequency at NRF1 motifs is consistently decreased (red dots) while other TFs are mostly unaffected (black dots). Dotted lines represent a linear regression fitted to NRF1 (red) and the other TFs (black). A proportional loss of 30% is observed at NRF1 binding motifs. (B) Decrease in NRF1 binding affects binding of heterologous factors at neighboring motifs. Heatmap depicting the changes in binding frequencies for NRF1-containing heterologous motif pairs. Shown is the difference in TF binding frequency and NO between WT and NRF1 KD. The identity of the second TF is indicated on the row labels of the heatmap. The rows were grouped using k-means clustering. (C) Binding changes are correlated at most NRF1-containing heterologous TF pairs. Scatterplot comparing the loss at NRF1 binding motifs with the one at neighboring heterologous factors. A fraction of the TFs involved in heterologous pairs with NRF1 have correlated reduction of their occupancy upon NRF1 KD (green dots), while other are not affected (orange dots). (D) Binding frequency is decreased upon KD for TFs having high co-occupancy with NRF1. Boxplot depicting the frequency of co-occupancy for TFs that are not affected (orange) or strongly reduced (green) by NRF1 KD. Categories are similar to Figure 6C. (E-F) Single-locus examples of CTCF bound regions where (E) high or (F) low binding cooperativity with NRF1 is observed. Shown is the average SMF signal in mESCs treated with scramble (top panel, blue dots connected by a blue line) or NRF1 siRNA (red dots connected by a red line). Same representation as in Figure 4F. Blue bar depicts the fraction of nucleosome occupied molecules. See also Figure S6.
We observed that upon KD, the majority of TF binding events involved in a heterologous pair with NRF1 showed reduced TF binding (Figure 6B). Concomitant to this loss, we observed a proportional increase in NO at some of these sites (Figure 6B). NRF1 KD affected diverse transcriptional activators but also a subset of the binding sites of the insulator CTCF. We observed a good correlation between loss of NRF1 binding and reduction in binding of the heterologous TF (Figure 6C), suggesting that this reduction is a direct effect of the decrease in NRF1 binding. However, we could identify a set of TF binding events that were less affected by the reduction of NRF1 binding in their vicinity (Figure 6C – orange dots). These less affected TF binding events tend to have a lower frequency of co-occupancy with NRF1 than those showing a substantial reduction upon NRF1 KD (Figure 6D). This is further evident when comparing the important decrease in CTCF binding frequency at a locus where NRF1 co-occupies 46% of its bound molecules (Figure 6E) with the limited changes observed at a locus where it only co-occupies 22% CTCF’s bound molecules (Figure 6E). The loss of binding is not coupled with nucleosomal gain at this locus (Figure 6E, blue bars), suggesting that additional TFs may determine NO at these sites. Thus, high TF co-occupancy is consistent with cooperativity between NRF1 and its partners at these sites. Together, these results demonstrate the potency of molecular co-occupancy to identify cooperative events at CREs.
Modeling co-occupancy in nucleosome-mediated cooperativity
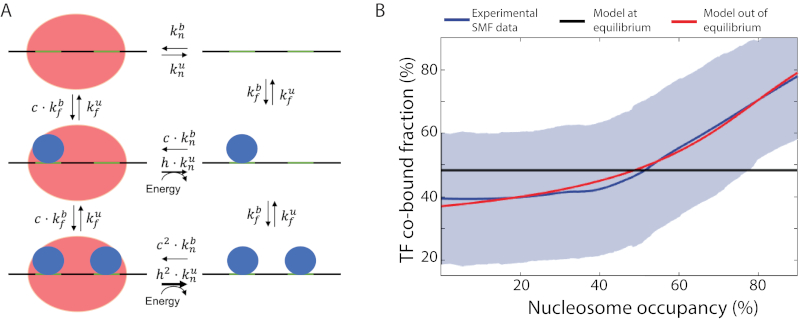
Our observations suggest a model in which increased TF co-occupancy is required to open chromatin at sites of competition with nucleosomes. We then asked if our data can be recapitulated by predictions of the nucleosome-mediated cooperativity model as proposed by (Mirny, 2010)(Figure S7A). This model assumes that the system is at equilibrium, implying that every reaction in the process is balanced by its reverse counterpart. This assumes for example, a free exchange between nucleosomes and TFs with no external energy flux (see Methods). Interestingly, we found that this model does not predict the increase in co-binding frequencies that we observed at sites having higher NO (Figure 7B). To reconcile our observations with this model, we explored the possibility that binding energies of TFs alone are insufficient to explain competition with nucleosomes. We tested the prediction of an extended model that does not assume an equilibrium between TF and nucleosome-bound states (Figure 7A). This model speculates that binding of TFs promotes nucleosome removal and their co-binding does so more efficiently (Figure 7A, larger arrows). We find that such a model predicts that the fraction of TF co-bound state scales with NO, recapitulating our experimental observations (Figure 7B). We conclude that our findings are compatible with the nucleosome-mediated TF cooperativity hypothesis, yet our data suggest that enzymatic activity such as ATP-dependent remodeling may be involved for TFs to co-occupy DNA.
Figure 7. Modelling TF co-binding as a function of nucleosome occupancy.
(A) Diagram representing an extension of the nucleosome-mediated TF cooperativity model in an out-of-equilibrium state, on an exemplary locus with two TF motifs. Diagrams show the reactions described by the model where parameters and describe the binding and unbinding rates for TFs, respectively and parameters and describe these rates for nucleosomes. This model assumes that TF (co-)occupancy provides a competitive advantage against nucleosome binding, as illustrated by a lack of equilibrium (energy influx) and described by the parameter h. (B) The out-of-equilibrium model accurately predicts experimental observations made with SMF. The fraction of co-bound molecules was plotted as a function of nucleosome occupancy. Shown are experimental observations in bait-capture SMF data (smoothed average, blue straight line; standard deviation, blue shadow), the imputed results of nucleosome-mediated cooperativity model in equilibrium (black line) and out of equilibrium (red straight line). NO is shown as the fraction of molecules with nucleosome binding in SMF data. See also Figure S7.
Discussion
Here, we demonstrate that SMF can quantify the binding of multiple TFs and nucleosomes at mouse CREs. We use this property to systematically quantify the degree of co-occupancy of neighbouring TFs. We find widespread evidence of co-occupancy for most of the tested TFs and demonstrate that high co-occupancy identifies loci with binding cooperativity between TFs. Our data suggest that TF co-occupancy is an important component of the mechanism used by TFs to outcompete nucleosomes in order to access their binding sites.
We detected quantifiable footprints for more than half of the TFs we tested in mESCs. Differences of in vivo footprint patterns between TFs has been previously reported for other DNA footprinting technologies, such as ATAC-seq or DNase-seq (Karabacak Calviello et al., 2019; Neph et al., 2012) and may result from differences in the residence time of TFs on DNA (Agarwal et al., 2017; Arnold et al., 2013a; Gebhardt et al., 2013; Sung et al., 2014). For instance, the insulator CTCF occupies a majority of its target molecules, in contrast to the activator Sox2-Oct4 that rarely occupies more than 10% of the molecules. This is in line with CTCF’s binding kinetics that are an order of magnitude slower than the kinetics of Oct4-Sox2 (Agarwal et al., 2017; Chen et al., 2014). Moreover, we observed through the examples of REST and NRF1, that binding frequencies are altered by TF concentration changes in the nucleus. Thus, differences in expression levels of TFs is likely to also contribute to the observed occupancy differences.
The unique ability of SMF to detect the binding of multiple TFs on single DNA molecules enabled us to determine their frequency of co-occupancy at CREs. As expected, we found that the frequency of co-occupancy is increased at known dimeric sites where TF-TF interactions are expected to occur. However, for most of the known TF dimers tested, dimerization is not obligatory for in vivo binding, suggesting that sequential monomer binding is the binding mode used by many TF dimers. Our data would for instance suggest that Myc-Max dimer formation preferentially occurs through a sequential monomer binding pathway in agreement with in vitro evidence (Kohler et al., 2002); possibly through initial binding of Max, followed by Myc stabilisation (Beaulieu et al., 2020).
Interestingly, we also observed comparably high frequency of co-occupancy for TFs that are unlikely to physically interact when these are binding at regions having high nucleosome occupancy. This shows that TF-TF interactions are not mandatory for their high co-occupancy, but are likely to contribute to the stabilisation of TF bound complexes in vivo. These observations make predictions on how TF binding motifs are organised at CREs (Spitz and Furlong, 2012) and has implications for the models of enhancer activity (Arnosti and Kulkarni, 2005; Spitz and Furlong, 2012). Our data argue that strict motif organisation is dispensable for TF cooperativity at most CREs but that TF-TF interactions stabilize binding at dimeric motifs. This agrees with a ‘billboard’ model for CREs, where motif organisation is generally flexible with the occurrence of enhanced cooperativity modules requiring stricter motif organisation.
Our data suggest that a fraction of DNA co-occupancy events is dictated by the competition between TFs and nucleosomes at CREs. We find that TF co-occupancy is particularly frequent at CREs with high levels of nucleosome binding. TF co-occupancy is independent of a precise arrangement of binding sites, but is exponentially reduced with genomic distance between the motifs. To explain our data, we propose an extension of the model of nucleosome-mediated cooperativity as proposed by (Mirny, 2010). This model no longer assumes a reversible equilibrium between TF binding and nucleosome occupancy. Instead, it postulates that binding of TFs actively promotes nucleosome removal, which could for instance be mediated through the recruitment of ATP-dependent remodelling complexes. We anticipate that the single molecule approach developed in this study, combined with perturbations in cis and trans, will enable to test this hypothesis and reveal mechanisms controlling TF binding at CREs.
STAR Methods
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Arnaud Krebs (arnaud.krebs@embl.de).
Materials Availability
This study did not generate new unique reagents.
Experimental model and subject details
Wild-type ES 159 and DNA methylation-null ES (DNMT TKO) cells were grown on gelatin-coated 10cm dishes, using Dulbecco’s Modified Eagle Medium (DMEM), supplemented with 15% FBS, LIF, 2-Mercaptoethanol, 1mM L-Glutamine and 1x non-essential amino acids (ThermoFisher 11140050) with regular splitting of cells.
Single Molecule Footprinting
Footprinting protocol was adapted from Krebs et al 2017 and optimized for mESC. Per reaction, 250.000 mESC (ES 159 or DNMT TKO) were trypsinized, washed in ice-cold PBS and re-suspended in ice-cold hypertonic lysis buffer (10mM Tris (pH = 7.4), 10mM NaCl, 3mM Mgcl2, 0.1mM EDTA, 0.5% NP40). Cells were incubated 10 minutes on ice, releasing nuclei and span down, pelleting the nuclei. Nuclei were washed with SMF Wash Buffer (10mM Tris (pH = 7.4), 10mM NaCl, 3mM Mgcl2, 0.1mM EDTA) and re-suspended in 1x M.CviPI reaction buffer (50mM Tris (pH 8.5), 50mM NaCl, 10mM DTT). The nuclei were then incubated with 200U of M.CviPI (NEB-M0227L) at 37°C for 7.5 minutes (in presence of 0.6mM SAM, and 300mM Sucrose). The reaction was supplemented with 100U of M.CviPI and 128pmol of SAM before a second incubation round of 37°C for 7.5 minutes. For ES samples, reactions were stopped at this point by adding a SDS containing buffer (20mM Tris, 600mM NaCl, 1%SDS 10mM EDTA). For DNMT TKO samples, where dual enzyme footprinting was applied, 10mM MgCl2, 128pmol of SAM and 60U of M.SssI (NEB- M0226L) were added for a third incubation round of 37°C for 7.5 minutes followed by stoppage of the reaction by adding the same SDS containing buffer. For all samples, material was digested with Proteinase K (200 mg/ml) overnight at 55°C, followed by phenol/chloroform purification of DNA, which was resuspended in water and treated with RNase A at for 1 hour 60°C and quantified using Qubit 1x dsDNA High Sensitivity kit.
Bait-capture of footprinted DNA
Genome-wide data were obtained using Agilent SureSelectXT Mouse Methyl-Seq Kit. The library preparations for the genome-wide data were performed according to the SureSelect XT Mouse Methyl-Seq Kit Enrichment System for Illumina Multiplexed Sequencing Library protocol (Agilent Technologies, Santa Clara CA, Version E0, April 2018). A total of 3 μg of footprinted DNA was used as input for bait capture, according to the company’s specifications. (# 5190-4836). DNA was first sonicated using a Covaris S220 sonicator (Covaris, Woburn, MA) (duty factor 10%, intensity 4 and 200 cycles/burst for 100 seconds duration time) to obtain products of 200-300 bp. DNA was then end-repaired, A-tailed and ligated with methylated adapters to create a pre-capture DNA library. Adapter-ligated libraries were purified using (0.65X) AMPure XP beads (Beckman Coulter, Inc., Brea, CA, USA) then quality and quantity of libraries were determined by bioanalyzer using DNA high sensitivity chip (Agilent). Next, 350 ng of each library was hybridized with the SureSelect Mouse methyl-seq capture library at 65°C for 16 hours. Hybridized products were purified by capture with Dynabeads MyOne Streptavidin T1 magnetic beads (ThermoFisher Scientific, Waltham, MA)and then subjected to bisulfite conversion using EZ DNA Methylation- Gold Kit (Zymo Research D5005)kit according to manufacturer’s protocol. As described in manufacturer’s protocol (Agilent SureSelectXT Mouse Methyl-Seq Kit), bisulfite converted libraries were PCR-amplified for 8 cycles with supplied universal primers and purified using AMPure XP beads. Captured libraries were indexed by PCR for another 6 cycles, using supplied indexes for downstream multiplexing. Quality of libraries were ensured with an Agilent Bioanalyzer using DNA High Sensitivity chip prior to pooling for sequencing. Supplied primers and recommended amplification parameters of the manufacturer were used throughout library preparation.
Single Molecule Footprinting of crosslinked chromatin
ES cells grown under standard conditions were washed once with PBS and supplemented with fresh ES medium 2 hours prior to fixation. After 2 hours, cells were washed twice with room temperature PBS, all liquid was carefully removed and 10ml of crosslinking medium (1% formaldehyde in DMEM) was added to plates, followed by a 10 minutes incubation on a shaker at room temperature. A replicate plate was spared from fixation and trypsinized to count the number of cells per plate. After 10 minutes, crosslinking was stopped by addition of 500μl of 2.5M Glycin per plate, followed by a 10 minutes incubation on a shaker at 4°C. All liquid was removed and 1ml ice cold PBS was added, followed by collection of cells using scraping. Fixed material was centrifuged at 600g for 5 minutes at 4°C, resuspended in 1ml detergent containing Paro buffer (0.25% Triton X-100, 10mM Tris pH 8, 10mM EDTA pH 8, 0.5mM EGTA) and incubated on ice for 10 minutes. Material was spun down again, resuspended in SMF wash buffer (see above), followed by another centrifugation resuspension in 1ml SMF wash buffer. Using cell count measures, material equating to 0.5 million cells were aliquoted into a new tube, spun down and resuspended in 300μl 1x GC Buffer (50mM Tris (pH 8.5), 50mM NaCl, 10mM DTT) and transferred to a Bioruptor Pico tube, followed by sonication at Bioruptor Pico instrument for 10 cycles at 15 secs ON / 90 secs OFF settings. M.CviPI reaction mixture (50mM Tris (pH 8.5), 50mM NaCl, 10mM DTT, 640μM SAM, 0.3M Sucrose) was prepared and used to equilibrate a Slide-A-Lyzer MINI Dialysis Device (0.5 mL, cutoff 3.5K MW, Thermo #88400). Sonicated chromatin was loaded into the dialysis cuvette, which was set up in a 15ml Falcon tube filled with M.CviPI reaction mixture. 100U of M.CviPI was added to the dialysis cuvette, followed by incubation at a 37°C water bath for 4 hours . Reaction was replenished with 20U of M.CviPI and 4μl of 32mM SAM after every hour. After 4 hours (3 replenishments), 10mM MgCl2 was added to the reaction mixture, SAM was replenished and 40U of S.ssI was added to the dialysis cuvette, followed by 2 hours of incubation at 37°C, where SAM and S.ssI were replenished after 1 hour. After a total of 6 hours of footprinting, reactions were stopped by addition of a SDS-containing buffer (20mM Tris, 600mM NaCl, 1%SDS, 10mM EDTA), RNase A and incubated at 60°C overnight. Proteinase K was added to the samples, which were then incubated at 65°C for 2 hours, followed by phenol/chloroform extraction of the footprinted DNA.
Amplicon bisulfite sequencing
To confirm the footprints we observe in bait-capture data, we employed a PCR-based strategy for target enrichment after SMF. This allows for the generation of high coverage SMF data for regions of interest in a cost- and time-effective manner. When picking targets, we focused on TFs with a consensus motif and a matching ChIP-seq signal (16 TFs in total, Table S2). These targets encompass sites where TFs bind in isolation or not bind at all (Table S2, Plate 1) as well as sites where binding is observed for multiple TFs (Table S2, Plate 2 and Plate 3). We ensured to pick regions with varying features such as NO and distance between the motifs. We laid a focus on capturing a high number of sites for TFs which were perturbed throughout this study, i.e REST and NRF1, while also profiling a sufficient number of sites for other studied TFs. Overall, we target 96 single binding sites (Plate 1) and 143 unique TF clusters, containing 481 motifs (Plates 2, 3).
A custom R script was used to design primers against in silico bisulfite converted templates, using Primer3 with slight modifications. Products ranged from 200 to 500bp in size with majority of amplicons being over 450bp (TableS3). Primers were commercially synthesized and reconstituted in 96-well plates. DNA material obtained from one footprinting reaction (~1μg) was bisulfite converted following standard Epitect bisulfite conversion kit protocol (Qiagen 59124). Entire material of one conversion was equally distributed to a 96-well plate and amplified via PCR using KAPA HiFi Uracil+ Kit (Roche) in a total volume of 16μl, with 625nM primers (forward and reverse combined) under following cycling conditions: 95°C for 3 minutes, 35 cycles of 20 seconds at 98°C, 30 seconds at 56°C, 60 seconds at 72°C, followed by 5 minutes at 72°C. 10μl of each reaction was pooled together and subjected to 0.8x AMPureXP bead purification (Beckman Coulter - A63880). 1μg of purified DNA was used for each amplicon bisulfite sample and sequencing libraries were prepared using the NEBNext Ultra II Kit (E7645). Up to 12 libraries were multiplexed using NEBNext Multiplex Oligos (E7335) and sequenced on a MiSeq instrument, generating 250bp paired-end reads. All targeted amplification experiments have been performed in duplicates.
siRNA transfection and TF knockdown
Dharmacon ON-TARGETplus siRNAs against CTCF, NRF1, MAX and NFYA were ordered as SMARTpools, consisting of 4 different siRNA sequences targeting each mRNA, and resuspended in sterile water. As control, ON-TARGETplus non-targeting control pool (Horizon Discovery, D-001810-10) was used at the same concentration. Following guidelines of the manufacturer for ES cells, DharmaFECT 1 (Horizon Discovery, T-2001) was used as transfection reagent. Transfection was performed in gelatinized 6-well plates (Falcon # 353934) seeded with 250.000 DNMT TKO ES cells, according to the instructions of the manufacturer, with a final siRNA concentration of 25nM and a final DharmaFECT volume of 4ul in each well with a total volume of 2ml Opti-MEM reduced serum medium (ThermoFisher 51985034). 24 hours post-transfection, siRNA medium was exchanged with standard ES medium. After a recovery period of 24 hours in standard medium, cells were supplemented again with the transfection mixture for further knockdown of the targets. After a total treatment of 72 hours, cells were pooled in two independent technical replicates, counted and aliquoted for footprinting according to standard SMF procedure as well as RNA- and protein isolation.
Confirmation of siRNA knockdowns via qPCR and Western blots
siRNA transfected cells were pelleted and resuspended in TRIzol reagent (ThermoFisher 15596026) or Protein Extraction Buffer (1% Triton-X-100, 150mM NaCl, 50mM Tris pH=8) for RNA and protein purification, respectively. Upon purification, RNA concentration was measured used Qubit RNA High Sensitivity Kit and 500ng RNA was reverse transcribed using SuperScript IV (ThermoFisher 18091050) with random hexamer priming. cDNA was diluted 20-fold in PCR-grade water and subjected to real-time quantitative PCR (qPCR) using SYBR Green PCR Master Mix (ThermoFisher 4309155) with gene specific, intron-spanning primers. Primers were checked for quantitative range with dilution series, and knockdown data was analyzed using DeltaDeltaCT method.
Western blots were used to confirm the knockdowns on protein expression level. 5ug total protein from transfected cells were subjected to SDS-PAGE (12% Gels) and transferred onto a nitrocellulose membrane at 150V for 1 hour. Transfer was confirmed with Ponceau staining and blots were incubated with primary antibodies overnight. NRF1 antibody (Abcam ab55744) was used at 1:1000 dilution and CTCF antibody (Sigma 07-729) was used at 1:2000 dilution in TBS-T. Blots were washed twice, incubated with secondary antibodies for 1 hour at room temperature and imaged using at a Bio-Rad gel documentation system. Consequently, blots were washed with TBS-T, incubated with tubulin primary antibody for loading control (1:1000 dilution) and imaged in the same manner.
Quantification and statistical analysis
Alignment and data extraction
Data alignment and methylation call
SMF data were processed as previously described (Krebs et al., 2017). Briefly, raw sequence files were pre-processed using Trimmomatic to remove Illumina adaptor sequences, remove low quality reads and trim low quality bases. The trimmed reads were then aligned using QuasR (using Bowtie as an aligner) (Gaidatzis et al., 2015) against a bisulfite index of the Mus Musculus genome (BSgenome.Mmusculus.UCSC.mm10). For other datasets (ChIP-seq, RNA-seq, MNase-seq), reads were aligned using QuasR against the Mus Musculus genome (BSgenome.Mmusculus.UCSC.mm10). Context independent cytosine methylation call was performed using QuasR. Custom R functions were developed to determine context dependent (CG, GC) average methylation. Methylation has been called genome wide for Cs covered at least 10 times.
Single molecule methylation call
Single molecule C methylation extraction was performed using QuasR (Gaidatzis et al., 2015). Custom R functions have been developed to determine nucleotide context and sort the molecules according to their methylation pattern using a molecular classifier. In order to ensure uniqueness of the molecules, we filtered PCR duplicates using Picard (Broad Institute, 2009) selecting reads with unique start and end mapping positions.
Mapping of TF motifs
Position weight matrices (PWM) for vertebrates TFs were downloaded from the Jaspar database (Mathelier et al., 2015) and mapped over non repetitive regions of the mouse genome (BSgenome.Mmusculus.UCSC.mm10) using the matchPWM function of the Biostrings package. PWM with a score of 10 or above were retained for single molecule TF classification.
Given that MAX and MYC both bind the palindromic E-box sequence and can’t be teased apart from one another in SMF data, we merged all instances for these TFs into MAX::MYC and excluded them from orientation analysis. Conversely, Sox2 and Pou5f1::Sox2 correspond to two separate motifs, mapping to distinct sequences in the genome. Therefore, we kept the instances separate for these TFs and analysed them as different TF binding events.
TF footprint quantification
A molecular classifier has been developed to quantify the binding of individual TFs. It extracts methylation for every read in 3 bins designed around the TFs motifs [-35:-25], [-7:7], [25:35] (Figure S2A). Bins were designed to distinguish short footprints created by the binding of TFs from larger footprints created by nucleosomes occupancy. All reads were aligned relative to the TF motifs and methylation was extracted for every molecule in each bin. Methylation was binarized in each bin, creating a 3 bits vector classifying the state of every molecule among 2^3=8 theoretical possibilities (Figure S2B). In cases where a bin contains multiple informative cytosines, methylation was averaged and rounded up (distribution of methylation in bins is bimodal). These include a TF bound pattern (101); a fully accessible pattern (111); patterns which may indicate the presence of nucleosomes at various position (Figure S2B). For the analysis of particular TF binding event, all reads that cover the 3 respective collection bins were probed with the single molecule classifier, revealing the frequencies of different binding states at the target region. Coverage cutoffs of 40 reads have been used for genome wide analysis and amplicon bisulfite sequencing.
A second molecular classifier has been developed to quantify the frequency of co-occupancy at neighboring motifs (Figure S4A). This strategy first identifies the positions of neighboring motifs at a genomic locus and classifies these loci into TF clusters, based on the number of motifs present (n). Then, n+2 bins are designed per cluster, one around each motif [-7:7] and two that flank the cluster [-35:-25+, *25:35+ from the most 5’ and 3’ motifs respectively (Figure S4A). For every read covering a cluster, binary methylation information was extracted from the n+2 bins. For each TF in the cluster, we defined the TF bound state as a combination of a footprint at their motif and accessibility at the flanks of the cluster (Figure S4A). Since the molecular classifier requires to have informative Cs in each of the bins, increasing the genomic density of the accessibility measures should enhance the number of clusters that can be analyzed with this strategy. We were indeed able to analyze five times more TF clusters when combining information of GCs and CGs from the tandem footprinting dataset in methylation null ESCs (Figure S4B, C). Importantly, SMF signal (Figure S1C – R=0.90) and TF binding frequencies are extremely comparable in WT and DNMT TKO mESCs (Figure S4C, R=0.94).
Data analysis
All analyses were performed using R- Bioconductor.
Comparison with external datasets
Data collection was performed using the qCount function of QuasR (Gaidatzis et al., 2015). Reads were counted in a window around each TF motif [-100:100] using the qCount function of QuasR (Gaidatzis et al., 2015) and enrichment over input were derived. Correlations were calculated on log2 transformed data.
Identification of dependencies between TFs
For each analyzed TF pair, states frequency was computed (Bound-Bound, Bound-Unbound, Unbound-Bound, Unbound-Unbound) and to build a contingency table. A Fischer’s exact test was performed on the contingency table representing each TF pair, with the null hypothesis that the binding states of two TFs are not dependent (odds ratio equals 1). The output of this test is an odds ratio, representing the likelihood that TF1 and TF2 will be in the same state (e.g. both bound) over the likelihood that TF1 and TF2 will be in different states. Fischer’s test also calculates an associated p-value with the odd’s ratios. Here, we report the odd’s ratios of TF pairs as a proxy for dependency in their binding. The co-bound fraction was calculated for each TF in the pair individually by dividing the number of co-bound molecules by the total number of bound molecules by the TF.
Kinetic model of TF and nucleosome binding process
Nucleosome-mediated cooperativity (NMC) model
The NMC model proposed by (Mirny, 2010) considers a DNA region containing n binding sites for a transcription factor (TF). In its simplest version, all binding sites have equal affinity which can be described with the binding and unbinding rates and . The DNA region can be in two states: occupied by a nucleosome (N) or free (F). The transition between these two states is govern by the binding and unbinding rates and . Importantly, the model assumes that binding sites can be bound by TFs when DNA is wrapped around a nucleosome with an affinity reduction that is tuned with a parameter c (Figure S7). To allow comparison with the pairwise analysis of TF co-occupancy in SMF data, we considered a DNA region with two binding sites that can be bound with equal affinities leading to a system with 6 states whose probabilities are denoted as: N 0, N 1, N 2, F 0, F 1 and F 2 where the subscript indicates the number of occupied binding sites. Then, assuming that every reaction in the system is at equilibrium the probabilities must hold the following detailed balance equations:
The above-mentioned equations depend only on the ratio of binding and unbind rates allowing a simplification of the model by introducing the following the dimensionless parameters: , the equilibrium stability of the nucleosome in the absence of TFs, and , the effective concentration of the TF. Assuming that the system is closed, i.e. , the system of equations can be solved leading to the following expressions for the NO (p) and the fraction of co-binding (f):
The NO can be tuned between [0%, 100%] by the parameter L.
Out-of-equilibrium kinetic (OEK) model
The NMC model was extended by breaking the equilibrium introducing an unbalanced reaction of nucleosome removal upon TF binding. The strength of this reaction is controlled by the dimensionless parameter h (see Fig 1). As detailed balance is not anymore obeyed the probability of each state is described by the following kinetic model:
To obtain the steady state solution, we set the equations above equal to zero and solved the resulting system of algebraical equations using Mathematica. As a result, the NO and the fraction of co-binding are nonlinear functions of the parameters L, α, c, h and that represents the ratio of binding stability between the nucleosome and the TF:
Notice that for h = 1 we obtain the same results than with the NMC model.
Fitting the model to the observed fraction of co-binding
We fitted the OEK model to the observed NO and fraction of co-binding using the method of least squares. To do so, we assume that all pairs share common average parameters α, c, h and q and the NO in each pair is determined by a different nucleosome stability (i.e. different L’s). The fitted common parameters were α = 1.2, c = 0.7, h = 10 and q = 0.5. This model is a first approximation on how the NMC model can be extended to be able to explain the observed changes in TF co-occupancy. Further work will be required for a full quantitative understanding of mechanisms underlying collective TF binding genome-wide.
Supplementary Material
Acknowledgments
The authors are grateful to Ferris Jung, Leslie Horner, Christiane Wirbelauer for technical assistance, Bernd Klaus, Lukas Burger and Charles Girardot for for computational and statistical support. The authors are grateful to Ralph Grand, Duncan Odom and members of the Krebs laboratory for helpful discussions and comments on the manuscript. Authors are thankful to Veronique Juvin (SciArtWork) for support with artwork and Guy Riddihough (LifeScienceEditors) for manuscript editing. Research in the laboratory of A.R.K is supported by core funding of the European Molecular Biology Laboratory, Deutche Forschung Gemenischaft (KR 5247/1-1) and a Swiss National Fund Ambizione grant (PZOOP3_161493). Salary of D.I was supported by a Swiss National Fund Ambizione grant (PZOOP3_161493). Research in the laboratory of D.S. is supported by the Novartis Research Foundation, the European Research Council (ERC) under the European Union’s Horizon research and innovation program (Grant agreement no. 667951) and the Swiss National Sciences Foundation.
Footnotes
Contributions
A.R.K designed the study. A.R.K and C.S wrote the manuscript. C.S designed and performed the experiments with the help of R.K, D.I, L.V and V.B for Illumina sequencing developments and library preparation. A.R.K supervised conduction of the experiments with help of D.S. C.S. and A.R.K analyzed the data with help of G.B. N.M wrote the mathematical model with the help of C.S and A.R.K. All authors discussed the results and commented on the manuscript.
Declaration of interest
The authors declare no conflicts of interest.
Data and Code Availability
The raw and processed sequencing data generated in this study can be accessed at ArrayExpress under following accessions: E-MTAB-9123 and E-MTAB-9033. Custom scripts generated for this study can be publicly accessed under the following link: https://github.com/Krebslabrep/Soenmezer_2020_SMF.
References
- Abdulhay NJ, McNally CP, Hsieh LJ, Kasinathan S, Keith A, Estes LS, Karimzadeh M, Underwood JG, Goodarzi H, Narlikar GJ, et al. Massively multiplex singlemolecule oligonucleosome footprinting. BioRxiv. 2020:2020.05.20.105379. doi: 10.7554/eLife.59404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams CC, Workman JL. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol Cell Biol. 1995;15:1405–1421. doi: 10.1128/mcb.15.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal H, Reisser M, Wortmann C, Gebhardt JCM. Direct Observation of Cell-Cycle-Dependent Interactions between CTCF and Chromatin. Biophys J. 2017 doi: 10.1016/j.bpj.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoutzias GD, Robertson DL, Van de Peer Y, Oliver SG. Choose your partners: dimerization in eukaryotic transcription factors. Trends Biochem Sci. 2008;33:220–229. doi: 10.1016/j.tibs.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Stelzer C, Bory ŁM, Rath M, Stark A. Genome-Wide Quantitative Enhancer Activity Maps Identified by STARR-seq. 2013a doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- Arnold P, Schöler A, Pachkov M, Balwierz PJ, Jøsrgensen H, Stadler MB, van Nimwegen E, Schübeler D. Modeling of epigenome dynamics identifies transcription factors that mediate Polycomb targeting. Genome Res. 2013b;23:60–73. doi: 10.1101/gr.142661.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnosti DN, Kulkarni MM. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J Cell Biochem. 2005;94:890–898. doi: 10.1002/jcb.20352. [DOI] [PubMed] [Google Scholar]
- Barisic D, Stadler MB, Iurlaro M, Schübeler D. Mammalian ISWI and SWI/SNF selectively mediate binding of distinct transcription factors. Nature. 2019;569:136–140. doi: 10.1038/s41586-019-1115-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu ME, Castillo F, Soucek L. Structural and Biophysical Insights into the Function of the Intrinsically Disordered Myc Oncoprotein. Cells. 2020;9:1–27. doi: 10.3390/cells9041038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibel M, Richter J, Schrenk K, Tucker KL, Staiger V, Korte M, Goetz M, Barde YA. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat Neurosci. 2004;7:1003–1009. doi: 10.1038/nn1301. [DOI] [PubMed] [Google Scholar]
- Broad Institute. Picard Tools - By Broad Institute. 2009.
- Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014 doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998 doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- Crocker J, Abe N, Rinaldi L, McGregor AP, Frankel N, Wang S, Alsawadi A, Valenti P, Plaza S, Payre F, et al. Low affinity binding site clusters confer HOX specificity and regulatory robustness. Cell. 2015;160:191–203. doi: 10.1016/j.cell.2014.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Alpern D, Gardeux V. The Genetics of Transcription Factor DNA Binding Variation. Cell. 2016 doi: 10.1016/j.cell.2016.07.012. [DOI] [PubMed] [Google Scholar]
- Domcke S, Bardet AF, Adrian Ginno P, Hartl D, Burger L, Schübeler D. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 2015;528:575–579. doi: 10.1038/nature16462. [DOI] [PubMed] [Google Scholar]
- Echigoya K, Koyama M, Negishi L, Takizawa Y, Mizukami Y, Shimabayashi H, Kuroda A, Kurumizaka H. Nucleosome binding by the pioneer transcription factor OCT4. Sci Rep. 2020;10:11832. doi: 10.1038/s41598-020-68850-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley EK, Olson KM, Zhang W, Brandt AJ, Rokhsar DS, Levine MS. Suboptimization of developmental enhancers. Science (80- ) 2015;350:325–328. doi: 10.1126/science.aac6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidatzis D, Lerch A, Hahne F, Stadler MB. QuasR: quantification and annotation of short reads in R. Bioinformatics. 2015;31:1130–1132. doi: 10.1093/bioinformatics/btu781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam EN, Jeong S, Tanay A, Egger G, Lee AS, Jones PA. Constitutive nucleosome depletion and ordered factor assembly at the GRP78 promoter revealed by single molecule footprinting. PLoS Genet. 2006;2:e160. doi: 10.1371/journal.pgen.0020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt JCM, Suter DM, Roy R, Zhao ZW, Chapman AR, Basu S, Maniatis T, Xie XS. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat Methods. 2013;10:421–426. doi: 10.1038/nmeth.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012 doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Bardet AF, Patton B, Purvis J, Johnston J, Paulson A, Gogol M, Stark A, Zeitlinger J. High conservation of transcription factor binding and evidence for combinatorial regulation across six Drosophila species. Nat Genet. 2011 doi: 10.1038/ng.808. [DOI] [PubMed] [Google Scholar]
- Ibarra IL, Hollmann NM, Klaus B, Augsten S, Velten B, Hennig J, Zaugg JB. Mechanistic insights into transcription factor cooperativity and its impact on proteinphenotype interactions. Nat Commun. 2020;11 doi: 10.1038/s41467-019-13888-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai S, Kock KH, Bulyk ML. ScienceDirect Transcription factor - DNA binding : beyond binding site motifs. Curr Opin Genet Dev. 2017;43:110–119. doi: 10.1016/j.gde.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Zaret KKS. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature. 2015;527:384–388. doi: 10.1038/nature15518. [DOI] [PubMed] [Google Scholar]
- Joseph SR, Pálfy M, Hilbert L, Kumar M, Karschau J, Zaburdaev V, Shevchenko A, Vastenhouw NL. Competition between histone and transcription factor binding regulates the onset of transcription in zebrafish embryos. Elife. 2017;6 doi: 10.7554/eLife.23326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junion G, Spivakov M, Girardot C, Braun M, Gustafson EH, Birney E, Furlong EEM. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell. 2012 doi: 10.1016/j.cell.2012.01.030. [DOI] [PubMed] [Google Scholar]
- Karabacak Calviello A, Hirsekorn A, Wurmus R, Yusuf D, Ohler U. Reproducible inference of transcription factor footprints in ATAC-seq and DNase-seq datasets using protocol-specific bias modeling. Genome Biol. 2019;20:1–13. doi: 10.1186/s13059-019-1654-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TK, Liu Y, Lay FD, Liang G, Berman BP, Jones PA. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 2012;22:2497–2506. doi: 10.1101/gr.143008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H, Waszak SM, Gschwind AR, Raghav SK, Witwicki RM, Orioli A, Migliavacca E, Wiederkehr M, Gutierrez-Arcelus M, Panousis NI, et al. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science. 2013;342(6159):744–747. doi: 10.1126/science.1242463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm SL, Shipony Z, Greenleaf WJ. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet. 2019;1 doi: 10.1038/s41576-018-0089-8. [DOI] [PubMed] [Google Scholar]
- Kohler JJ, Metallo SJ, Schneider TL, Schepartz A. DNA specificity enhanced by sequential binding of protein monomers. Proc Natl Acad Sci. 2002;96:11735–11739. doi: 10.1073/pnas.96.21.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs AR, Imanci D, Hoerner L, Gaidatzis D, Burger L, Schübeler D. Genome-wide Single-Molecule Footprinting Reveals High RNA Polymerase II Turnover at Paused Promoters. Mol Cell. 2017;67:411–422.:e4. doi: 10.1016/j.molcel.2017.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levo M, Avnit-Sagi T, Lotan-Pompan M, Kalma Y, Weinberger A, Yakhini Z, Segal E. Systematic Investigation of Transcription Factor Activity in the Context of Chromatin Using Massively Parallel Binding and Expression Assays. Mol Cell. 2017;65:604–617.:e6. doi: 10.1016/j.molcel.2017.01.007. [DOI] [PubMed] [Google Scholar]
- Luna-Zurita L, Stirnimann CU, Glatt S, Kaynak BL, Thomas S, Baudin F, Samee MAH, He D, Small EM, Mileikovsky M, et al. Complex Interdependence Regulates Heterotypic Transcription Factor Distribution and Coordinates Cardiogenesis. Cell. 2016;164:999–1014. doi: 10.1016/j.cell.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Fornes O, Arenillas DJ, Chen C, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, et al. JASPAR 2016: a major expansion and update of the openaccess database of transcription factor binding profiles. Nucleic Acids Res. 2015;44:gkv1176. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Widom J. Collaborative Competition Mechanism for Gene Activation In Vivo. Mol Cell Biol. 2003;23:1623–1632. doi: 10.1128/MCB.23.5.1623-1632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA. Nucleosome-mediated cooperativity between transcription factors. Proc Natl Acad Sci U S A. 2010;107:22534–22539. doi: 10.1073/pnas.0913805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgunova E, Taipale J. Structural perspective of cooperative transcription factor binding. Curr Opin Struct Biol. 2017;47:1–8. doi: 10.1016/j.sbi.2017.03.006. [DOI] [PubMed] [Google Scholar]
- Nabilsi NH, Deleyrolle LP, Darst RP, Riva A, Reynolds BA, Kladde MP. Multiplex mapping of chromatin accessibility and DNA methylation within targeted single molecules identifies epigenetic heterogeneity in neural stem cells and glioblastoma. Genome Res. 2014;24:329–339. doi: 10.1101/gr.161737.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh KS, Ha J, Baek S, Sung MH. XL-DNase-seq: Improved footprinting of dynamic transcription factors. Epigenetics and Chromatin. 2019;12:1–12. doi: 10.1186/s13072-019-0277-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson M, Schaffner W. Synergistic activation of transcription by multiple binding sites for NF-κB even in absence of co-operative factor binding to DNA. J Mol Biol. 1990;214:373–380. doi: 10.1016/0022-2836(90)90187-q. [DOI] [PubMed] [Google Scholar]
- Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: A dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- Polach KJ, Widom J. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J Mol Biol. 1996;258:800–812. doi: 10.1006/jmbi.1996.0288. [DOI] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, et al. An Atlas of Combinatorial Transcriptional Regulation in Mouse and Man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter F, Wienerroither S, Stark A. ScienceDirect Combinatorial function of transcription factors and cofactors. Curr Opin Genet Dev. 2017;43:73–81. doi: 10.1016/j.gde.2016.12.007. [DOI] [PubMed] [Google Scholar]
- Sakaue M, Ohta H, Kumaki Y, Oda M, Sakaide Y, Matsuoka C, Yamagiwa A, Niwa H, Wakayama T, Okano M. DNA methylation is dispensable for the growth and survival of the extraembryonic lineages. Curr Biol. 2010 doi: 10.1016/j.cub.2010.06.050. [DOI] [PubMed] [Google Scholar]
- Shipony Z, Marinov GK, Swaffer MP, Sinnott-Armstrong NA, Skotheim JM, Kundaje A, Greenleaf WJ. Long-range single-molecule mapping of chromatin accessibility in eukaryotes. Nat Methods. 2020 doi: 10.1038/s41592-019-0730-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbæk R, Rabiee A, Nielsen R, Sidoli S, Traynor S, Loft A, Poulsen LLC, Rogowska-Wrzesinska A, Jensen ON, Mandrup S. Transcription Factor Cooperativity in Early Adipogenic Hotspots and Super-Enhancers. Cell Rep. 2014;7:1443–1455. doi: 10.1016/j.celrep.2014.04.042. [DOI] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015 doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, Canfield T, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefflova K, Thybert D, Wilson MD, Streeter I, Aleksic J, Karagianni P, Brazma A, Adams DJ, Talianidis I, Marioni JC, et al. Cooperativity and rapid evolution of cobound transcription factors in closely related mammals. Cell. 2013;154 doi: 10.1016/j.cell.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis AB, Debo BM, Haugen E, Churchman LS, Stamatoyannopoulos JA. Single-molecule regulatory architectures captured by chromatin fiber sequencing. Science. 2020;368:1449–1454. doi: 10.1126/science.aaz1646. [DOI] [PubMed] [Google Scholar]
- Sung MH, Guertin MJ, Baek S, Hager GL. DNase footprint signatures are dictated by factor dynamics and DNA sequence. Mol Cell. 2014;56:275–285. doi: 10.1016/j.molcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Supplemental Data Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Reactions. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thanos D, Maniatis T. Virus induction of human IFN-b gene expression requires the assembly of an enhancosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- Tijssen MR, Cvejic A, Joshi A, Hannah RL, Ferreira R, Forrai A, Bellissimo DC, Oram SH, Smethurst PA, Wilson NK, et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1 FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev Cell. 2011;20:597–609. doi: 10.1016/j.devcel.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes to Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- Vashee S, Melchert K, Ding WV, Johnstont SA, Kodadek T. Evidence for two modes of cooperative DNA binding in vivo that do not involve, direct protein-protein interactions. Curr Biol. 1998;8:452–458. doi: 10.1016/s0960-9822(98)70179-4. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010 doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J, Lazar J, Sandstrom R, Halow J, Lee K, Bates D, Diegel M, Dunn D, Neri F, Haugen E, et al. Global reference mapping and dynamics of human transcription factor footprints. BioRxiv. 2020:2020.01.31.927798. doi: 10.1038/s41586-020-2528-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, Schiltz RL, Sung M-H, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin XY, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson T, Hartemink AJ. An ensemble model of competitive multi-factor binding of the genome. Genome Res. 2009;19:2101–2112. doi: 10.1101/gr.093450.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, Schmierer B, Jolma A, Kivioja T, Taipale M, et al. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell. 2013 doi: 10.1016/j.cell.2013.07.034. [DOI] [PubMed] [Google Scholar]
- Zhu F, Farnung L, Kaasinen E, Sahu B, Yin Y, Wei B, Dodonova SO, Nitta KR, Morgunova E, Taipale M, et al. The interaction landscape between transcription factors and the nucleosome. Nature. 2018;562:76–81. doi: 10.1038/s41586-018-0549-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw and processed sequencing data generated in this study can be accessed at ArrayExpress under following accessions: E-MTAB-9123 and E-MTAB-9033. Custom scripts generated for this study can be publicly accessed under the following link: https://github.com/Krebslabrep/Soenmezer_2020_SMF.