Abstract
Size at birth is known to be influenced by various fetal and maternal factors including genetic effects. South Asians have a high burden of low birthweight and cardiometabolic diseases, yet studies of common genetic variations underpinning these phenotypes are lacking. We generated independent, weighted fetal genetic score (fGS) and maternal genetic score (mGS) from 196 birthweight-associated variants identified in Europeans and conducted association analysis with various fetal birth parameters and anthropometric and cardiometabolic traits measured at different follow-up stages (5-6 years’ intervals) from seven Indian and Bangladeshi cohorts of South Asian ancestry. The results from above cohorts were compared with South Asians in UK BioBank and The Exeter Family Study of Childhood Health, a European ancestry cohort. Birthweight increased by 50.7g and 33.6g per standard deviation of fGS (p = 9.1×10-11) and mGS (p = 0.003) respectively in South Asians. A relatively weaker maternal genetic score effect compared to Europeans indicates possible different intrauterine exposures between Europeans and South Asians. Birthweight was strongly associated with body size in both childhood and adolescence (p = 3×10-5 − 1.9×10-51), however, fetal genetic score was associated with body size in childhood only (p < 0.01) and with head circumference, fasting glucose and triglycerides in adults (p < 0.01). The substantially smaller newborn size in South Asians with comparable fetal genetic effect to Europeans on birthweight suggests a significant role of factors related to fetal growth that were not captured by the present genetic scores. These factors may include different environmental exposures, maternal body size, health and nutritional status etc. Persistent influence of genetic loci on size at birth and adult metabolic syndrome in our study supports a common genetic mechanism partly explaining associations between early development and later cardiometabolic health in various populations, despite marked differences in phenotypic and environmental factors in South Asians.
Keywords: Birthweight, anthropometric traits, association, cardiometabolic risk, DOHaD, fetal genetic score, maternal genetic score, South Asian populations
Abbreviations
- DOHaD
Developmental Origins of Health and Disease
- EAF
Effect allele frequency
- EFSOCH
The Exeter Family Study of Childhood Health
- EGG
Early Growth Genetics
- fGS
Fetal genetic score
- GDM
Gestational diabetes mellitus
- GIFTS
Genomic and lIfestyle predictors of Fetal ouTcomeS
- GWASs
Genome-wide association studies
- MBRC
Mysore Birth Records Cohort
- mGS
Maternal genetic score
- MMNP
Mumbai Maternal Nutritional Project
- PMNS
Pune Maternal Nutrition Study
- PS
Parthenon Study
- SEM
Structural equation modelling
- UK-Bang
London UK Bangladeshi cohort
- UKBB
UK Biobank
- UKBB-SAS
UK Bio Bank South Asian Subjects
- WP2
GIFTS Work Package 2
- WP3
GIFTS Work Package 3
Introduction
Size at birth is a summary measure for intrauterine nutrition, growth and development (1; 2). It is influenced by genetic and environmental factors, and in clinical practice helps predict neonatal wellbeing (3; 4). Several longitudinal population-based studies both in higher and lower-middle-income countries including India have demonstrated a correlation between birth size (both small and large) and future risk of cardiometabolic diseases (1; 2; 5–8). This led to the ‘Fetal Programming’ or Developmental Origins of Health and Disease (DOHaD) hypothesis which proposes that the intrauterine environment (meaning maternal diet, smoking, etc) drives fetal growth and also affects the development of metabolic organs, setting up later risk of disease (1; 2). Up to one third of South Asians living in the Indian sub-continent are born low birthweight (9). They also have a high prevalence of type 2 diabetes and cardiovascular diseases and develop these conditions at a younger age and a lower BMI than Europeans (10). Understanding the genetic determinants of neonatal size and their association with later phenotypes may provide important insights into mechanisms of how fetal growth and development relate to later risk of cardiometabolic diseases in various ancestral groups with different environmental exposures.
Large-scale genome-wide association studies (GWASs), mostly in individuals of European ancestry, including participants from the Early Growth Genetics (EGG) consortium and the UK Biobank (UKBB) have identified several genetic variants associated with birthweight (11-15). These genetic associations include (i) direct effects, where the fetus’s own genotype influences its birthweight, (ii) indirect effects of the maternal genotype which influence birthweight via the intrauterine environment, and (iii) those which have a combination of direct fetal and indirect maternal effects (11; 15). A recent study in Europeans reported 209 conditionally independent GWAS significant genetic variants at 190 independent loci that were associated with birthweight and explained 7% of birthweight variance (fetal genotype 6%, maternal genotype 2%, and covariance -0.5%) further confirming the relatively weaker effect of maternal genetics than fetal genetics (15). It further partitioned the genetic effects on birthweight into fetal and maternal effects using structure equation model (SEM) and also demonstrated their association with various cardiometabolic traits. Genetic risk score is one of the approaches to summarise the genetic effects of multiple risk genes on a given trait such as birthweight. Based on the observations that fetal genetic score (fGS) for birthweight is negatively associated with adult BP, lipids, glucose and insulin levels, and insulin resistance, Warrington et al. concluded that common genetic variants contribute to the observed associations between lower birthweight and later cardiometabolic disease. This is something akin to the ‘Fetal Insulin Hypothesis’ first set out by Hatterseley et al. (16), which purports that the same genotype at a variant can influence birthweight and later cardiometabolic risk.
The dual burden of low birthweight and cardiometabolic diseases in South Asians and the fact that South Asians, especially those living in lower and middle income countries are not well represented in the majority of GWAS studies demands investigating genetic variants associated with fetal development, and how they relate to later cardiometabolic traits (17–19). Here, we studied associations of the weighted genetic scores with birth size in ~1900 mother-offspring pairs from South Asian birth cohorts in India, Bangladesh and UK. Association analysis was also conducted with body size and cardiometabolic traits among children, adolescents and adults using available follow-up data from Indian cohorts. Overall, the study has tried to answer two questions: (1) are fetal and maternal genetic scores related to newborn size in South Asians in the same way as in Europeans and (2) do the genetic scores related to birthweight influence cardiometabolic risk in a direction that would support a genetic contribution to the birthweight-cardiometabolic diseases link in the South Asian population?
Research Design and Methods
Study participants
The participants in this study were mother-child pairs from different prospective birth cohort studies from India, Bangladesh and UK. The Indian cohorts comprise the Pune Maternal Nutrition Study (PMNS), Parthenon Study (PS), Mumbai Maternal Nutritional Project (MMNP) and Mysore Birth Records Cohort (MBRC). The individuals from PMNS and MMNP are Indo-Europeans, and those from the PS and MBRC are Dravidians, the two major ethnic populations in the Indian sub-continent (20; 21). Informed consent was obtained from all participants following the guidelines of Indian Council of Medical Research, Govt. of India, New Delhi. The Bangladeshi cohorts were from a sub-study of a prospective multi-center European Union FP7 project GIFTS (Genomic and lIfestyle predictors of Fetal ouTcome relevant to diabetes and obesity and their relevance to prevention strategies in South Asian people) consisting of work package (WP2), work package (WP3) and London UK Bangladeshi cohort (UK-Bang) that was conducted following appropriate Institutional Review Board approval.
Pune Maternal Nutrition Study (PMNS)
The PMNS cohort, based in six rural villages near Pune in Western India, was established in 1993 to examine the relationship of maternal health and nutrition during pregnancy to fetal growth and development, and future cardiometabolic risk (22). Women were recruited pre-conceptionally. A 75gm oral glucose tolerance test was carried out at 28 weeks’ gestation in pregnancy and GDM was diagnosed based on then prevalent WHO guidelines. Gestational age was based on last menstrual period dates (recorded every month during the pre-conception period) unless it differed from early (<20 weeks’ gestation) ultrasound scan dating by 2 weeks or more, in which case the latter was used. Detailed new born anthropometry was carried out by trained research staff within 72 hours of birth. Multiple follow-up studies have been conducted starting from pre-pregnancy, during pregnancy, at birth, early childhood, adolescence and young adulthood and detailed anthropometric and biochemical data have been collected. At 6 years of age, we measured anthropometry, resting systolic and diastolic blood pressure, plasma glucose and insulin (fasting and after an oral glucose load) and fasting lipids (triglycerides and LDL- and HDL-cholesterol). At 12 years, detailed anthropometry, and measurements of blood pressure, fasting glucose, insulin and lipids were repeated. At both time points, the same measurements were carried out in both parents. We have used these data in the current study. The DNA samples isolated from the 6 years follow up stage were used for genotyping.
Parthenon Study (PS)
The Parthenon study (PS) was established in 1997-98 in Mysore, South India, to examine the long-term effects of maternal glucose tolerance and nutritional status during pregnancy on cardiovascular risk factors and cognition in the offspring (23). Women (<32 weeks’ gestation) were recruited in the antenatal clinic of the Holdsworth Memorial Hospital, Mysore. Gestational age was assessed using last menstrual period dates collected at recruitment. A 100gm oral glucose tolerance test was carried out at 28-32 weeks’ gestation and GDM was diagnosed based on Carpenter and Coustan criteria (24). Detailed newborn anthropometry was carried out by trained research staff within 72 hours of birth. At 5 and 13.5 years of age, we measured anthropometry, resting systolic and diastolic blood pressure, plasma fasting glucose and insulin) and fasting lipids (triglycerides and LDL- and HDL-cholesterol). At 5 years, the same measurements were carried out in their mothers and only fasting glucose and insulin in the fathers. These data were used in this study. Genotyping was performed on the DNA samples isolated from the 5 years follow up stage blood samples.
Mumbai Maternal Nutritional Project (MMNP)
The Mumbai Maternal Nutrition Project was a randomised controlled trial, set up in 2006 among women living in slums in the city of Mumbai, Western India with the objective to test whether improving women’s dietary micronutrient quality before and during conception improves birthweight and other related outcomes (25). Women were recruited before conception. As in the PMNS, gestational age was assessed using a combination of last menstrual period dates (which were collected monthly during the pre-conceptional period) and ultrasound scans conducted before 20 weeks’ gestation. A 75g oral glucose tolerance test was carried out at 28-32 weeks’ gestation and GDM was diagnosed based on revised WHO 1999 guidelines. Trained research staff carried out newborn anthropometry within 10 days of birth. In the current study, we have used the child phenotype data at birth (anthropometry) and in early childhood (5-7-year follow-up), when detailed anthropometry, systolic and diastolic blood pressure, fasting and post-load glucose and insulin, and fasting LDL- and HDL-cholesterol and triglycerides were measured (26). Maternal anthropometry, blood pressure and fasting plasma glucose and insulin concentrations were also measured at this follow-up. Genomic DNA isolated from blood samples at the same stage were used for genotyping.
Mysore Birth Records Cohort (MBRC)
The MBRC is a retrospective birth cohort of urban men and women born at the CSI Holdsworth Memorial Hospital during 1934-55 (27). They were recruited for the first time as adults (mean age 47 years) in 1993-95 and cardiometabolic risk factors were measured (7). Birthweight, length and head circumference were obtained from their mothers’ obstetric records. We have included the anthropometric data at birth and cardiometabolic parameters measured between 40 and 70 years during 2013-2017. Gestational age was missing in the majority of subjects and gestational diabetes status was not available. Since maternal DNA samples were not available, the analyses were restricted to the association of fetal genetic score and their birth measures and later life outcomes. GIFTS Dhaka Bangladeshi cohorts (WP2 and WP3)
WP2 samples were collected between 2011 and 2012 in Dhaka, Bangladesh from women attending the Maternal and Child Health Training Institute, a tertiary Government hospital for antenatal care and registration in Dhaka. Primigravid pregnant women who were in the first trimester of their pregnancy (≤ 14 week gestation), with a singleton pregnancy conceived naturally and who were willing to participate in the study were included in an observational study during pregnancy and immediately post-partum after written consent (28). GDM was diagnosed based on revised WHO 1999 guidelines. Women with a prior history of type 2 diabetes, or gestational diabetes or pregnancy induced hypertension were excluded. The aim of WP2 was to establish the methods and feasibility of recruitment and follow-up for an interventional study (WP3). WP3 samples were collected between 2014 and 2015 in Dhaka, Bangladesh from pregnant women attending MCHTI who consented to an open-label micro-nutrient supplement trial of vitamin D and vitamin B12 supplementation (29). All consenting women eligible under the WP2 criteria were included in the study and samples were collected from mother and baby under the same sampling frame as WP2. Women who were diagnosed later in pregnancy with GDM remained in the study.
London UK Bangladeshi cohort (UK-Bang)
The cohort was set up between 2012-2015 as an exploratory observational study of gestational diabetes and its consequences on offspring. Pregnant women of Bangladeshi origin were recruited from the Royal London Hospital antenatal clinics at 28 weeks gestation at the time of 75 gm OGTT. GDM was diagnosed based on Revised WHO, 1999 guidelines. Women were recruited during routine antenatal care and enriched for the presence of GDM. Women with multiple pregnancies, pre-existing or overt type 1 or type 2 diabetes were excluded. Gestational age was based on ultrasound scan dating. Detailed new born anthropometry was carried out by trained research staff within 72 hours of birth.
The Exeter Family Study of Childhood Health (EFSOCH)
EFSOCH is a prospective study of children born between 2000 and 2004, and their parents, from a geographically defined region of Exeter, UK. All women gave informed consent and ethical approval was obtained from the local review committee. Details of study protocol, including measurement of birthweight, are described in Knight et al (30). Maternal and paternal DNA samples were extracted from parental blood samples obtained at the study visit (when the women were 28 weeks pregnant), and offspring DNA was obtained from cord blood at birth. Genotyping and imputation of EFSOCH samples has been described previously (31).
UK Bio Bank South Asian participants (UKBB-SAS)
The UK Biobank phenotype preparation has been described in detail elsewhere (15). Briefly, a total of 280,315 participants reported their own birthweight in kilograms and 216,839 women reported the birthweight of their first child on at least one assessment centre visit. Multiple birth were excluded where reported. In the absence of gestational data, participants with birthweight values <2.5kg or >4.5kg were considered pre-term births and excluded. In addition to the genotype quality control metrics performed centrally by the UK Biobank, we defined a subset of “South Asian” ancestry samples (32). To do this, we generated ancestry informative principal components (PCs) in the 1000 genomes samples. The UK Biobank samples were then projected into this PC space using the SNP loadings obtained from the principal components analysis using the 1000 genomes samples. The UK Biobank participants’ ancestry was classified using K-means clustering centred on the three main 1000 genomes populations (European, African, and South Asian). Those clustering with the South Asian cluster were classified as having South Asian ancestry.
Inclusion and exclusion criteria, and phenotype measurements
In all the cohorts, the association analysis was restricted to individuals with both genotype and phenotype data available. The anthropometric measurements at birth were conducted within 72 hours after birth, and babies with congenital defects were excluded from the analysis. Twins and babies born lesser than 37 weeks of gestational age (9-14%) were excluded from the association analysis at birth. For anthropometric and cardiometabolic analysis at follow up stages during childhood and adolescence, we included all the individuals with phenotype-genotype data available irrespective of their gestational age at birth. For adults, phenotypes data were taken from the follow up stages as PMNS mother at 6 years, PMNS fathers at 12 years, PS mother and father at 5 years, MMNP mother at 7 years, and MBRC at the latest follow up during 2013-2017. Anthropometric measurements at birth and follow up stages were conducted using standard methods. Body fat percentage was measured by whole-body dual energy X-ray absorptiometry (DEXA) scans. Biochemical measurements were conducted from fasting plasma samples using standard methods. Plasma glucose was measured by the glucose oxidase peroxidase method, plasma insulin was measured using Delfia technique. Insulin resistance was calculated using the homeostatic model assessment of insulin resistance (HOMA-IR). Plasma lipid levels including total cholesterol, triglycerides, high density lipoprotein (HDL) and low density lipoprotein (LDL) cholesterols were measured by standard enzymatic methods. Individuals with missing phenotype were excluded from the analysis of the particular trait.
Genotyping and imputation QCs
For Indian cohorts, genome-wide genotyping were performed using Affymetrix Genome-Wide Human SNP Array 6.0 for fathers of PMNS cohort; Illumina Infinium Human CoreExome-24 array for children and mothers of PMNS and PS cohorts; and Illumina Infinium Global Screening Array for children and mothers of MMNP, fathers of PS and individuals of MBRC cohorts. Individuals with genotyping call rate ≤ 95% and SNPs with call rate ≤ 95% and Hardy Weinberg equilibrium P ≤10-6 were removed. Genome-wide imputation was performed by using IMPUTEv2 software (https://mathgen.stats.ox.ac.uk/impute/impute_v2.html) and 1000 Genome Phase 3 as reference panel and SNPs with imputation info score ≤ 0.4 were removed. The genome-wide genotyping for the children and mothers of all the Bangladeshi cohorts were performed using Illumina Infinium Global Screening Array and genome-wide imputation using HRC imputation panel.
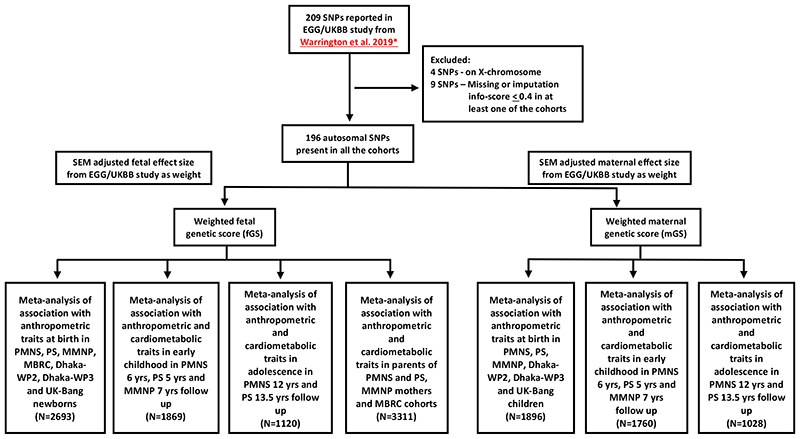
Selection of genetic variants and calculation of weighted genetic scores
The scheme for selecting SNPs for the calculation of birthweight genetic score is shown in Figure 1. Of the 205 autosomal SNPs reported as associated with birthweight in Warrington et al., 9 SNPs were excluded due to either being missing or having an imputation info score less than 0.4 in at least one of the cohorts (15). Finally, 196 autosomal SNPs were used for generating weighted fetal genetic score (fGS) and maternal genetic score (mGS). Details of the 196 SNPs were provided in Supplementary Table 1. The SNP weights for generating the fGS and mGS were taken from the SEM adjusted effect estimates of the fetal and maternal effects respectively from the recent GWAS of birthweight from the EGG/UKBB consortium (Supplementary Table 1) (15). The SEM estimates associations of both maternal and fetal scores with birthweight while accounting for the relationship between fetal and maternal genotypes, thereby producing independent estimates of the fetal and maternal genetic effects on birthweight. The weighted genetic score was calculated using the following formula:
Figure 1. Flow chart showing the overall study design including SNP selection, generation of weighted fetal and maternal genetic scores, association analysis and final meta-analyses at different stages of follow-up.
SNP, single nucleotide polymorphism; SEM, structure equation model; EGG, Early Growth Genetics Consortium; UKBB, UK Biobank; PMNS, Pune Maternal Nutrition Study; PS, Parthenon Study; MMNP, Mumbai Maternal Nutrition Project; MBRC, Mysore Birth Records Cohort; Dhaka-WP2, Work Package 2 of GIFTS; Dhaka-WP3, Work Package 3 of GIFTS; UK-Bang, London UK Bangladeshi cohort.
*, Warrington NM, et al. 2019 (15)
Where βn is the weight of SNPn taken from the EGG/UKBB birthweight GWAS, nSNPs is the number of SNP available (n=196), and −βn is the sum total weight of all 196 SNPs.
We identified independent genetic variants from the 196 SNPs used above by looking at pairwise linkage disequilibrium (r2<0.01) in a window of 1000kb in the 1000 Genome Phase 3 reference panel and freshly conducted association analysis with birthweight.
Statistical analysis and power calculation
Birthweight and other birth measures were transformed to standardized Z-scores (Z-score = (value − mean)/standard deviation). Association analysis was performed by linear regression, using Z- scores as the dependent variables and weighted genetic score as the independent variable, adjusted for the child’s sex and gestational age. The models were as follows:
Power calculations were conducted to estimate the probable association observable in our analysis with a sample size of 2693 individuals of South Asian ancestry. If the birthweight SNPs explain equal variance in South Asians to that explained in Europeans (6% and 2% for fGS and mGS respectively) (Warrington et al, 2019), we would have > 99% power to see an association with the fGS and 98% power with the mGS at α = 0.05. However, it is likely that due to differing linkage disequilibrium between marker SNPs and underlying causal genetic variants, genetic variants identified in GWAS samples that were largely of European ancestry may explain less variation in non-European samples. Therefore, assuming that the genetic scores explain only 75% of the European ancestry variation in South Asian ancestry individuals, we would still have 99% and 83% power for fGS and mGS respectively to detect an association with birthweight.
Association analysis of the anthropometric and cardiometabolic phenotype data acquired during follow-up at childhood and adolescence was performed by linear regression, using log10..transformed standardized Z-scores as the dependent variables and weighted genetic score as an independent variable, adjusted for sex and age. Imputed genotype data from parents of children in the PMNS and PS, mothers of children in MMNP, and men and women in MBRC were utilized for investigating the effect of the genetic risk scores on adult anthropometric and cardiometabolic phenotypes. BMI was included as an additional covariate for the cardiometabolic traits. The models were as follows:
The association analyses for birthweight and other birth measures and for anthropometric and cardiometabolic traits were conducted independently for each cohort and fixed effect inverse variance weighted meta-analysis (using the metan command in STATA) was performed to combine the final results. A total of 57 tests in the three stages (childhood, adolescence and adulthood) were conducted and the significance level was set at p < 0.001 (α < 0.05/57 tests) to allow for multiple testing.
Results
Clinical and demographic characteristics of study participants
Newborn measurements, maternal details and phenotypes at different follow-up stages are shown in Table 1 and Supplementary Tables 2, 3, 4 and 5. The mean birthweight of term babies in different cohorts ranged between 2.64 and 3.12 kg. Within the cohorts of South Asian ancestry, babies born in India and Bangladesh were comparatively smaller, whereas Bangladeshi babies born in UK from the UK-Bang and the UKBB-SAS were relatively larger (Supplementary Table 2 and 3). Birthweight was much higher in the European babies as observed in the EFSOCH (Table 1). Boys were bigger than girls across all the cohorts. In contrast, sum of skin-fold thickness, a measure of adiposity, was greater in girls. Amongst all the cohorts, PMNS mothers living in rural India were the thinnest (mean BMI = 18.0 kg/m2) whereas Bangladeshi mothers living in the UK (UK-Bang) were the heaviest (mean BMI = 26.2 kg/m2). Mean BMI in the mothers from the other cohorts were in the normal range, between 20.3 to 23.6 kg/m2. The percentage of mothers with gestational diabetes mellitus (GDM) was higher in the Bangladeshi cohorts (UK-Bang = 50%, WP2 = 24.5% and WP3 = 25.8%), whereas, in the Indian cohorts, it was 0.6%, 6.1% and 6.9% in PMNS, PS and MMNP respectively. The UK-Bang cohort was positively selected to have higher rates of GDM than the underlying population, but the high rates of GDM in the Bangladeshi Dhaka WP2 and WP3 cohorts represent the high rates of GDM in the community. The mothers of MBRC individuals were not tested for diabetes. Principal Components Analysis did not reveal any evidence of population stratification within the cohorts (The data can be made available on request).
Table 1. Maternal and newborn details in the study cohorts, and fetal and maternal genetic scores for the South Asian and European cohorts.
| Traits | PMNS (N=515) | PS (N=511) | MMNP (N=466) | MBRC (N=684) | Dhaka-WP2 (N=53) | Dhaka-WP3 (N=314) | UK-Bang (N=150) | UKBB-SAS* (N=2732) | EFSOCH* (N=674) |
|---|---|---|---|---|---|---|---|---|---|
| Birthweight (kg) | 2.68 (0.34) | 2.91 (0.41) | 2.64 (0.37) | 2.76 (0.42) | 2.90 (0.38) | 2.84 (0.42) | 3.12 (0.45) | 3.10 (0.68) | 3.52 (0.47) |
| Birth length (cm) | 47.8 (1.97) | 48.8 (2.11) | 48.2 (2.26) | 48.0 (2.95) | 46.2 (2.56) | 49.6 (2.60) | 46.6 (2.03) | NA | 50.3 (2.12) |
| Ponderal index (kg/m3) | 24.5 (2.44) | 25.0 (2.75) | 23.6 (2.60) | 25.3 (4.85) | 29.5 (4.42) | 23.3 (3.50) | 28.9 (4.27) | NA | 27.7 (2.58) |
| Head circumference (cm) | 33.1 (1.24) | 33.9 (1.28) | 33.2 (1.20) | 35.6 (1.58) | 33.4 (1.39) | 33.0 (2.40) | 33.6 (1.31) | NA | 35.2 (1.26) |
| Chest circumference (cm) | 31.2 (1.59) | 32.0 (1.64) | 30.9 (1.75) | NA | NA | NA | 33.4 (1.97) | NA | 34.2 (1.86) |
| Abdomen circumference (cm) | 28.7 (1.91) | 30.0 (1.92) | 28.4 (2.08) | NA | NA | NA | 31.4 (2.56) | NA | NA |
| Mid-upper arm circumference (cm) | 9.7 (0.88) | 10.4 (0.92) | 9.7 (0.82) | NA | 9.9 (0.71) | 10.2 (2.09) | 10.9 (2.13) | NA | 11.1 (0.90) |
| Triceps skinfold (mm) | 4.3 (0.87) | 4.3 (0.90) | 4.2 (1.05) | NA | NA | NA | 5.0 (1.93) | NA | 4.86 (1.08) |
| Subscapular skinfold (mm) | 4.2 (0.89) | 4.5 (0.91) | 4.2 (0.99) | NA | NA | NA | 5.3 (1.87) | NA | 4.87 (1.08) |
| Gestational age (weeks) | 39.0 (1.06) | 39.5 (1.14) | 39.3 (1.17) | NA | 40.3 (1.17) | 39.2 (1.53) | 40.0 (3.44) | NA | 40.1 (1.22) |
| Maternal Age (years) | 21.4 (3.56) | 23.8 (4.24) | 24.8 (3.83) | NA | 19.9 (2.45) | 22.7 (4.29) | 29.7 (5.40) | NA | 30.5 (5.19) |
| Maternal Height (cm) | 152.1(4.9) | 154.5(5.4) | 151.3(5.4) | NA | 151.1 (5.8) | 150.9 (5.7) | 156.0 (5.8) | NA | 165.0 (6.3) |
| Maternal BMI (kg/m2) | 18.0 (1.9) | 23.6 (3.55) | 20.3 (3.67) | NA | 20.6 (3.40) | 22.7 (4.03) | 26.2 (4.34) | NA | 24.0 (4.34) |
| Maternal GDM status [n (%)] | 3 (0.6) | 31 (6.1) | 32 (6.9) | NA | 13 (24.5) | 81 (25.8) | 75 (50.0) | NA | NA |
| Year of birth | 1994-95 | 1998-99 | 2006-12 | 1934-66 | 2011-12 | 2015-16 | 2011-15 | 1934-70 | 2000-04 |
| Fetal Genetic Score | 191.0 (9.0) | 191.0 (9.6) | 189.0 (9.4) | 189.0 (9.6) | 191.0 (8.1) | 188.0 (9.4) | 188.0 (9.3) | 192.0 (9.9) | 192.0 (9.8) |
| Maternal Genetic Score | 215.0 (10.3) | 215.0 (10.4) | 215.0 (10.5) | NA | 218.0 (10.2) | 217.0 (10.2) | 216.0 (9.3) | 214.8 (11.0) | 214.0 (10.8) |
All values are mean (SD); N, subjects included in this study; SD, standard deviation; GDM, Gestational diabetes mellitus; PMNS, Pune Maternal Nutrition Study; PS, Parthenon Study; MMNP, Mumbai Maternal Nutrition Project; MBRC, Mysore Birth Records Cohort; Dhaka-WP2, Work Package 2 of GIFTS; Dhaka-WP3, Work Package 3 of GIFTS; UK-Bang, London UK Bangladeshi cohort; UKBB-SAS, UK Biobank South Asian component; EFSOCH, The Exeter Family Study of Childhood Health study; *, Not used for meta-analysis. Fetal and maternal genetic scores were calculated from 196 birthweight-associated variants in children and mothers, respectively.
Association of genetic scores with birthweight and other birth measures
The effect allele frequencies (EAFs) of 196 SNPs were similar in all seven South Asian cohorts, except two outliers, one each in the MBRC (rs2306547) and GIFTS (rs9851257) cohorts (Supplementary Figure 1A and Supplementary Table 1). Although, the EAFs at several SNPs varied considerably between South Asians and the EGG/UKBB subjects (Supplementary Figure 1B and Supplementary Table 1), mean values for both fGS and mGS in South Asian cohorts were similar to those in the European cohort, EFSOCH (Table 1).
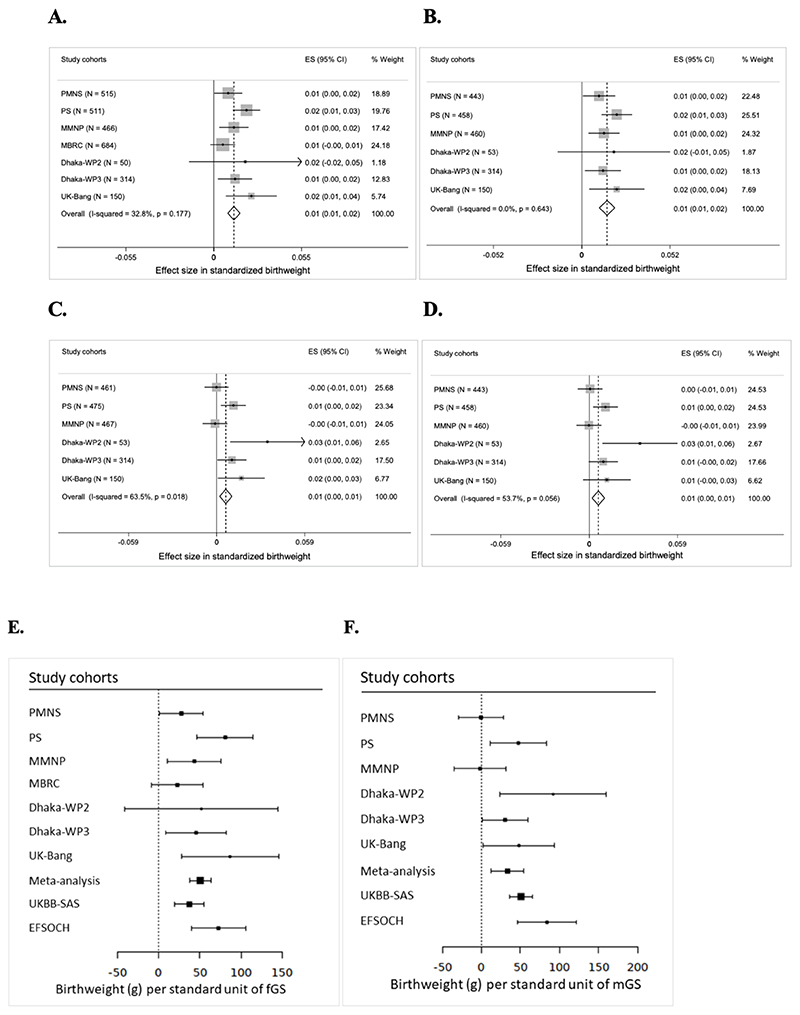
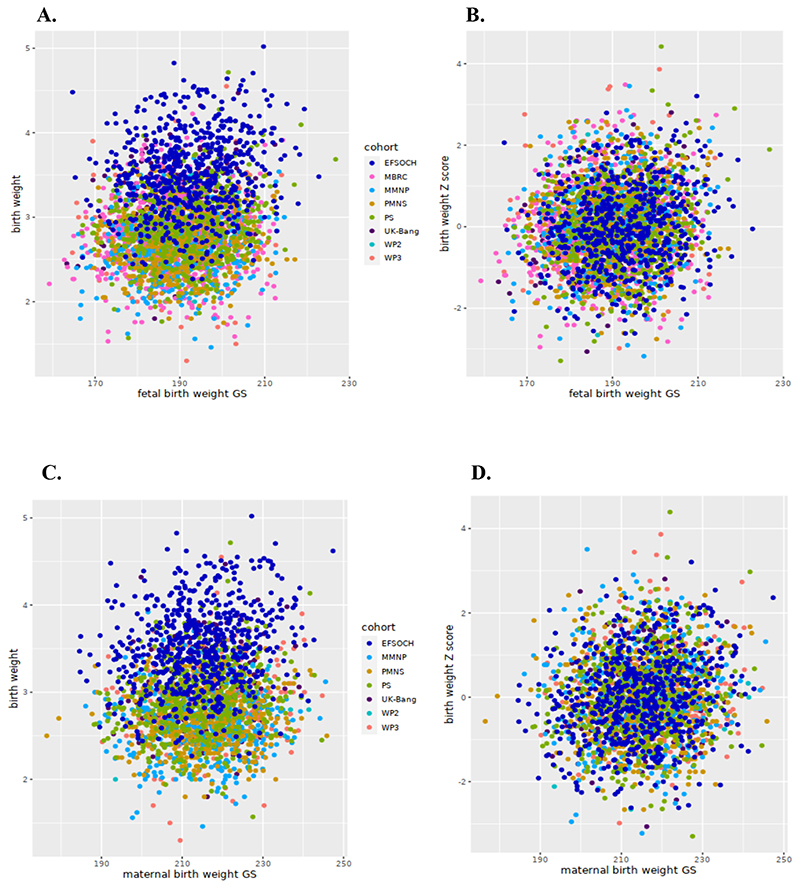
We noted that the fGS calculated from 196 SNPs was strongly associated with birthweight in South Asians (Table 2). The meta-analysis of the South Asian cohorts showed a 0.013 SD higher birthweight per 1 unit higher fGS, adjusted for the child’s sex and gestational age (p = 9.1×10-11) (Figure 2A and Table 2). This is equivalent to 50.7 g of birthweight per SD unit of fGS (Figure 2E). The strength of association was only partially attenuated after additional adjustment for the mGS (Effect = 0.015 SD, p = 1.1×10-10) (Figure 2B and Table 2). The mGS was also directly associated with offspring birthweight although compared to the fGS, the effect size was smaller (effect = 0.006 SD, p = 0.003). This is equivalent to 33.6 g of birthweight per SD unit of mGS and adjustment for fGS made little difference (effect = 0.006 SD; p = 0.004) (Figures 2C, 2D and 2F, Table 2). Analyses of only Indians and only Bangladeshis showed consistent and overlapping effect sizes in the fGS association analysis, but the mGS association with birthweight was largely driven by the Bangladeshi cohorts (Supplementary Tables 8 and 9). Since GDM is associated with excess fetal growth, we repeated association analysis after the exclusion of offspring of GDM women and observed similar associations (effect = 0.010; p = 5.1×10-8 for the fGS and effect = 0.005; p = 0.011 for the mGS) (Supplementary Tables 6 and 7). A plot of fGS versus birthweight showed that for each fGS, birthweight was substantially smaller in the South Asians (Figures 3A and 3B). Similar observations were noted for the association of mGS with birthweight (Figures 3C and 3D). The effect sizes of the fGS on birthweight in the South Asian cohorts was comparable to the same in EFSOCH (n = 674) and also with South Asians in the UK Biobank study (UKBB-SAS; n = 2732) (p = 0.17; p = 0.23 respectively) (Figure 2E). Similarly, the association between mGS and offspring birthweight in our study was similar to that observed in UKBB-SAS (p = 0.93). However, we noted a statistically significant smaller effect size of mGS among all the South Asian cohorts combined than in EFSOCH (p = 0.048) (Figure 2F). The fGS was also positively associated with other birth measures; no associations were seen with the mGS (Table 3). Respective adjustments for mGS and fGS did not substantially change the strength of these associations (Supplementary Table 10). Further, sensitivity analysis using 167 LD-pruned SNPs (after exclusion of 29 SNPs with an r2>0.01 with other variants from the list of 196 SNPs) did not make any significant changes in the strength of association (Supplementary Tables 11-13).
Table 2. Associations of fetal genetic score with own birthweight and maternal genetic score with its offspring birthweight in South Asian cohorts.
| Cohort | fGS adjusted for sex and GA* | fGS adjusted for sex, GA and mGS† | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | Effect | L95 | U95 | P | N | Effec | L95 | U95 | P | |
| PMNS | 515 | 0.009 | 0.000 | 0.018 | 0.042 | 443 | 0.010 | 0.001 | 0.020 | 0.040 |
| PS | 511 | 0.021 | 0.012 | 0.029 | 3.8×10-6 | 458 | 0.021 | 0.012 | 0.030 | 1.0×10-5 |
| MMNPt | 466 | 0.013 | 0.003 | 0.022 | 0.007 | 460 | 0.013 | 0.004 | 0.022 | 0.006 |
| MBRC§ | 684 | 0.006 | -0.002 | 0.013 | 0.154 | NA | NA | NA | NA | NA |
| Dhaka-WP2 | 53 | 0.020 | -0.015 | 0.055 | 0.277 | 53 | 0.019 | -0.014 | 0.052 | 0.269 |
| Dhaka-WP3 | 314 | 0.013 | 0.003 | 0.024 | 0.015 | 314 | 0.013 | 0.002 | 0.023 | 0.022 |
| UK-Bang | 150 | 0.024 | 0.008 | 0.040 | 0.004 | 150 | 0.021 | 0.004 | 0.037 | 0.015 |
| Meta-analysis | 2693 | 0.013 | 0.009 | 0.017 | 9.1×10 11 | 1878 | 0.015 | 0.01 | 0.020 | 1.1×10 10 |
| mGS adjusted for sex and GA ‖ | mGS adjusted for sex, GA and fGS ¶ | |||||||||
| N | Effect | L95 | U95 | P | N | Effec | L95 | U95 | P | |
| PMNS | 461 | 0.000 | -0.008 | 0.008 | 0.976 | 443 | 0.001 | -0.008 | 0.009 | 0.876 |
| PS | 475 | 0.011 | 0.003 | 0.020 | 0.013 | 458 | 0.011 | 0.003 | 0.019 | 0.011 |
| MMNPt | 467 | -0.001 | -0.009 | 0.007 | 0.804 | 460 | 0.000 | -0.009 | 0.008 | 0.957 |
| Dhaka-WP2 | 53 | 0.034 | 0.009 | 0.059 | 0.011 | 53 | 0.034 | 0.009 | 0.059 | 0.011 |
| Dhaka-WP3 | 314 | 0.010 | 0.001 | 0.020 | 0.040 | 314 | 0.009 | 0.000 | 0.019 | 0.060 |
| UK-Bang | 150 | 0.016 | 0.001 | 0.032 | 0.041 | 150 | 0.012 | -0.004 | 0.028 | 0.150 |
| Metaanalysis | 1920 | 0.006 | 0.002 | 0.010 | 0.003 | 1878 | 0.006 | 0.002 | 0.010 | 0.004 |
Association analysis was performed using linear regression with standardized birthweight adjusted for sex and gestational age as the dependent variable for each cohort separately and finally the summary results were meta-analyzed. t, In MMNP, allocation group was additionally adjusted for, and §, in MBRC only sex was adjusted for, since gestational age data was not available for the majority of the sample. The effect size is in standard deviation units of birthweight per unit change in genetic score. The standard deviation of birthweight in kg in all these cohorts ranged from 0.34 to 0.45 kg. N, number of term babies; GA, gestational age; I2, heterogeneity; Het-P, P value for heterozygosity; P, P value; fGS, fetal genetic score; mGS, maternal genetic score; GA, gestational age. PMNS, Pune Maternal Nutrition Study; PS, Parthenon Study; MMNP, Mumbai Maternal Nutrition Project; MBRC, Mysore Birth Records Cohort; Dhaka-WP2, Work Package 2 of GIFTS; Dhaka-WP3, Work Package 3 of GIFTS; UK-Bang, London UK Bangladeshi cohort.
For fGS, *, I2 = 32.8 and Het-P = 0.177; †, I2= 0 and Het-P = 0.643
For mGS, ‖, I2= 63.5 and Het-P = 0.018; ¶, I2=53.7 and Het-P = 0.056.
Figure 2. Meta-analysis of associations of fetal genetic score with birthweight in South Asian populations and comparison with European cohorts. Panel A-D: Fetal genetic score with birthweight.
(A) Fetal genetic score adjusted for sex and gestational age; (B) Fetal genetic score adjusted for sex, gestational age and maternal genetic score; (C) Maternal genetic score adjusted for sex and gestational age and (D) Maternal genetic score adjusted for sex, gestational age and fetal genetic score. The X-axis indicates the effect size for standardized birthweight per unit of weighted genetic score. In MMNP, allocation group was additionally adjusted for and in MBRC, only sex was adjusted for, since gestational data was not available for the majority of the samples. Panel E-F: Comparison between South Asians and European cohorts. (E) Weighted fetal genetic score and (F) Weighted maternal genetic score. The X-axis indicates the effect size for birthweight in gram (g) per standardized weighted genetic score. PMNS, Pune Maternal Nutrition Study; PS, Parthenon Study; MBRC, Mysore Birth Records Cohort; MMNP, Mumbai Maternal Nutrition Project; Dhaka-WP2, Work Package 2 of GIFTS; Dhaka- WP3, Work Package 3 of GIFTS; UK-Bang, London UK Bangladeshi cohort; UK Biobank South Asian component (UKBB-SAS); EFSOCH, The Exeter Family Study of Childhood Health; fGS, fetal genetic score; mGS, weighted maternal genetic score; ES, effect size; CI, confidence interval; I2, heterogeneity; p, p- value. Heterogeneity p value for fGS is 0.1777 and for mGS is 0.0046.
Figure 3. Scatter plot comparing the correlation between birthweight and fetal genetic score and maternal genetic score in South Asian and European cohorts. Panel A-B: birthweight and fetal genetic score (fGS).
A, indicates absolute birthweight and fGS; B, shows the same between cohort-specific birthweight Z-scores and fGS; Panel C-D: birthweight and maternal genetic score (mGS). C, indicates absolute birthweight and mGS and D, shows the same between cohort-specific birthweight Z-scores and mGS.
South Asian cohorts include PMNS, Pune Maternal Nutrition Study; PS, Parthenon Study; MMNP, Mumbai Maternal Nutrition Project; MBRC, Mysore Birth Records Cohort; Dhaka- WP2, Work Package 2 of GIFTS; Dhaka-WP3, Work Package 3 of GIFTS; UK-Bang, London UK Bangladeshi cohort) while EFSOCH (The Exeter Family Study of Childhood Health) is the European Cohort.
Table 3. Associations of fetal and maternal genetic scores with other birth measures in South Asian populations.
| Trait | fGS adjusted for sex and gestational age* | mGS adjusted for sex and gestational age* | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Effect | L95 | U95 | P | I2 | Het-P | N | Effect | L95 | U95 | P | I2 | Het-P | |
| Birth length (Z) | 2544 | 0.004 | 0.000 | 0.009 | 0.048 | 44.1 | 0.097 | 1820 | 0.003 | -0.002 | 0.008 | 0.153 | 42.5 | 0.122 |
| Ponderal Index (Z) | 2517 | 0.009 | 0.004 | 0.013 | 2.1×10-4 | 28.3 | 0.213 | 1796 | 0.000 | -0.004 | 0.006 | 0.906 | 14.3 | 0.323 |
| Head circumference (Z) | 2564 | 0.005 | 0.000 | 0.009 | 0.030 | 48.0 | 0.073 | 1844 | 0.002 | -0.002 | 0.007 | 0.425 | 0 | 0.741 |
| Chest circumference (Z) | 1586 | 0.012 | 0.007 | 0.017 | 8.2×10-6 | 23.1 | 0.273 | 1477 | 0.002 | -0.002 | 0.007 | 0.383 | 3.7 | 0.374 |
| Abdominal circumference (Z) | 1586 | 0.014 | 0.008 | 0.019 | 3.4×10-7 | 68.5 | 0.023 | 1477 | 0.002 | -0.003 | 0.007 | 0.554 | 62.0 | 0.048 |
| Mid-upper arm circumference (Z) | 1953 | 0.014 | 0.009 | 0.019 | 1.3×10-7 | 0 | 0.485 | 1844 | 0.005 | 0.000 | 0.010 | 0.045 | 0 | 0.982 |
| Triceps skinfold (Z) | 1564 | 0.013 | 0.007 | 0.018 | 3.6×10-6 | 44.6 | 0.144 | 1455 | 0.003 | -0.001 | 0.009 | 0.181 | 61.7 | 0.050 |
| Subscapular skinfold (Z) | 1563 | 0.012 | 0.006 | 0.017 | 2.4×10-5 | 42.3 | 0.158 | 1454 | 0.003 | -0.002 | 0.008 | 0.260 | 25.7 | 0.258 |
Association analysis was performed using linear regression with standardized birth measures adjusted for sex and gestational age as the dependent variables for each cohort independently and finally the summary results were meta-analyzed. The effect size is in standard deviation units of the birth measure per unit change in genetic score. The South Asian populations include PMNS, Pune Maternal Nutrition Study; PS, Parthenon Study; MMNP, Mumbai Maternal Nutrition Project from India; MBRC, Mysore Birth Records Cohort; Dhaka-WP2 of GIFTS; Dhaka-WP3 of GIFTS; UK-Bang, London UK Bangladeshi cohort. *, In MMNP, allocation group was additionally adjusted for, and in MBRC only sex was adjusted for since gestational age data was not available for the majority of the sample. N, number of term babies; L95, U95, 95% confidence interval; I2, heterogeneity; Het-P, P value for heterozygosity; P, P value; fGS, fetal genetic score; mGS, maternal genetic score. The N was different for each trait due to missingness of some phenotype data in MBRC, Dhaka-WP2 and Dhaka-WP3.
Associations of birthweight and fetal genetic score with anthropometric and cardiometabolic traits in follow-up stages
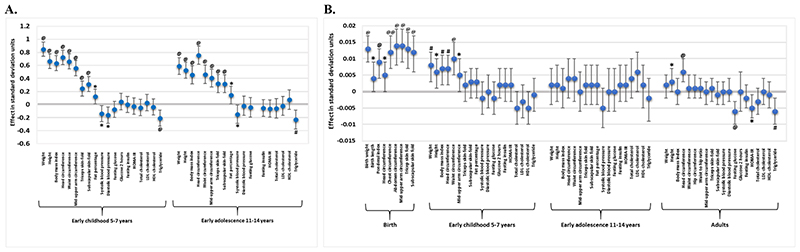
The associations of birthweight and the fGS with later anthropometric and cardiometabolic traits in early childhood and early adolescence were investigated in the Indian cohorts only, since they had longitudinal follow-up data. Birthweight was strongly positively associated with all anthropometric traits in childhood (5-7 years; p = 3×10-5 − 1.9×10-51) and adolescence (11-14 years; p = 5.7×10-6 − 8.1×10-27) (Figure 4A; Supplementary Table 14). It also showed strong evidence of a negative association with triglycerides levels in childhood (p = 9.8×10-4) and a weak association in adolescence (p = 0.002). We observed a negative association with SBP and DBP and a positive association with fat percentage both in childhood and adolescence but these did not pass the Bonferroni-corrected threshold of p < 0.001 (Figure 4A; Supplementary Table 14). Similar to birthweight, a higher fGS was associated with larger body size in childhood (Table 3). We observed a strong positive association of the fGS with waist circumference (effect = 0.01 SD per standard unit, p = 5.7×10-5) but the associations with other anthropometric parameters including weight, height, BMI, head circumference and mid-upper arm circumference were weaker (p = 0.017 − 0.001) and did not pass the multiple testing threshold of p < 0.001 (Table 4; Figure 4B)]. No evidence of associations between fGS and anthropometric traits were detected in adolescents. The fGS was not associated with any of the cardiometabolic parameters in children or in adolescents (Table 4) and mGS had no association with any anthropometric and cardiometabolic parameters in children or in adolescents (Supplementary Table 15).
Figure 4. Birthweight and fetal genetic score associations with various anthropometric and cardiometabolic traits at different follow-up stages in the Indian cohorts. (A) Birthweight (B) fetal genetic score.
The X-axis shows anthropometric and cardiometabolic traits at different stages of follow-up including birth, early childhood, early adolescence and adults. The Y-axis indicates the effect size in standard deviation units. HOMA-IR, Homeostasis Model Assessment of Insulin Resistance; LDL, low density lipoprotein; HDL, high density lipoprotein. ‘Early childhood 5-7 years’ included children from Pune Maternal Nutrition Study at 6 yrs, Parthenon Study at 5 yrs, and Mumbai Maternal Nutrition Project at 7 yrs of age whereas adolescents from Pune Maternal Nutrition Study at 12 yrs and Parthenon Study at 13.5 yrs formed the group ‘Early adolescence 12-14 years’. ‘Adults’ consisted of parents from Pune Maternal Nutrition Study and Parthenon Study, mothers from Mumbai Maternal Nutrition Project, and individuals from Mysore Birth Records Cohort. @, P-value ≤ 0.001; #, P-value ≤ 0.01; *, P-value ≤ 0.05.
Table 4. Meta-analysis of associations of fetal genetic score with anthropometric and cardiometabolic traits in early childhood, adolescence and adults in Indians.
| Traits | Children | Adolescents | Adults | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Effect | P | I2 | Het-P | N | Effect | P | I2 | Het-P | N | Effect | P | I2 | Het-P | |
| Weight (Z) | 1866 | 0.008 | 0.001 | 0 | 0.830 | 1120 | 0.002 | 0.592 | 0 | 0.641 | 3311 | 0.002 | 0.341 | 0 | 0.698 |
| Height (Z) | 1865 | 0.006 | 0.017 | 0 | 0.846 | 1120 | 0.002 | 0.437 | 0 | 0.889 | 3307 | 0.003 | 0.037 | 0 | 0.574 |
| Body mass index (Z) | 1865 | 0.007 | 0.007 | 0 | 0.666 | 1120 | 0.001 | 0.844 | 0 | 0.581 | 3306 | 0.000 | 0.977 | 0 | 0.438 |
| Head circumference (Z) | 1866 | 0.007 | 0.003 | 0 | 0.999 | 1115 | 0.004 | 0.223 | 0 | 0.633 | 3256 | 0.006 | 5.5×10-4 | 32.2 | 0.194 |
| Waist circumference (Z) | 1864 | 0.010 | 5.5×10-5 | 0 | 0.463 | 1096 | 0.004 | 0.254 | 0 | 0.918 | 3251 | 0.001 | 0.528 | 13.8 | 0.326 |
| Hip circumference (Z) | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 3256 | 0.001 | 0.456 | 0 | 0.680 |
| Waist to hip ratio (Z) | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 3247 | 0.001 | 0.603 | 9.8 | 0.353 |
| Mid upper arm circumference (Z) | 1865 | 0.005 | 0.032 | 0 | 0.705 | 1112 | 0.000 | 0.976 | 0 | 0.595 | 3258 | 0.000 | 0.852 | 0 | 0.645 |
| Triceps skinfold (Z) | 1865 | 0.002 | 0.511 | 0 | 0.760 | 1114 | 0.002 | 0.487 | 0 | 0.790 | 3259 | 0.001 | 0.748 | 0 | 0.725 |
| Subscapular skinfold (Z) | 1865 | 0.003 | 0.280 | 52.2 | 0.123 | 1113 | 0.002 | 0.603 | 0 | 0.825 | 3238 | -0.001 | 0.673 | 0 | 0.926 |
| Fat percentage (Z) | 1860 | 0.003 | 0.254 | 50.3 | 0.133 | 1085 | 0.002 | 0.475 | 45.8 | 0.174 | NA | NA | NA | NA | NA |
| Systolic blood pressure (Z)* | 1847 | -0.002 | 0.411 | 0 | 0.410 | 1102 | -0.005 | 0.112 | 88.6 | 0.003 | 3081 | 0.000 | 0.801 | 0 | 0.454 |
| Diastolic blood pressure (Z)* | 1848 | 0.000 | 0.989 | 0 | 0.765 | 1102 | 0.000 | 0.904 | 92.4 | 0.000 | 3082 | 0.000 | 0.922 | 0 | 0.467 |
| Fasting glucose (Z)* | 1840 | -0.002 | 0.483 | 0 | 0.497 | 1110 | 0.000 | 0.908 | 92.8 | 0.000 | 2601 | -0.006 | 9.3×10-4 | 30.5 | 0.218 |
| 120 minutes glucose (Z)* | 1809 | 0.002 | 0.321 | 0 | 0.434 | NA | NA | NA | NA | NA | 1320 | 0.000 | 0.905 | 0 | 0.707 |
| Fasting insulin (Z)* | 1831 | 0.002 | 0.369 | 18.9 | 0.291 | 1111 | 0.002 | 0.463 | 47.7 | 0.167 | 2596 | -0.002 | 0.359 | 0 | 0.823 |
| HOMA-IR (Z)* | 1756 | 0.002 | 0.401 | 0 | 0.997 | 1110 | 0.002 | 0.407 | 74.4 | 0.048 | 2432 | -0.005 | 0.022 | 0 | 0.802 |
| Total cholesterol (Z)* | 1838 | -0.005 | 0.050 | 50.7 | 0.131 | 1111 | 0.004 | 0.224 | 0 | 0.488 | 2601 | -0.003 | 0.118 | 0 | 0.968 |
| LDL-cholesterol (Z)* | 1847 | -0.003 | 0.280 | 52.9 | 0.119 | 1111 | 0.006 | 0.070 | 0 | 0.676 | 2600 | -0.001 | 0.594 | 0 | 0.957 |
| HDL cholesterol (Z)* | 1849 | -0.005 | 0.059 | 0 | 0.513 | 1111 | 0.002 | 0.632 | 0 | 0.631 | 2584 | 0.000 | 0.867 | 0 | 0.809 |
| Triglycerides (Z)* | 1838 | -0.001 | 0.666 | 0 | 0.668 | 1111 | -0.002 | 0.440 | 37.8 | 0.205 | 2601 | -0.006 | 0.002 | 0 | 0.673 |
Association analysis was performed using linear regression with standardized log10 transformed traits as the dependent variable for each cohort independently and finally the summary results were meta-analyzed. Age and sex were included as covariates in the regression model for all traits; BMI was additionally included as a covariate for analysis of traits marked with an asterisk (*). Allocation group was additionally adjusted for in MMNP. Meta-analysis for children included those from Pune Maternal Nutrition Study at 6 yrs, Parthenon Study at 5 yrs, and Mumbai Maternal Nutrition Project at 7 yrs of age; for adolescents from Pune Maternal Nutrition Study at 12 yrs and Parthenon Study at 13.5 yrs; and for adults from parents from Pune Maternal Nutrition Study and Parthenon Study, mothers from Mumbai Maternal Nutrition Project, and individuals from Mysore Birth Records Cohort; P, P value; I2, heterogeneity; Het-P, P value for heterozygosity; SNP, single nucleotide polymorphism; HOMA-IR, homeostasis model assessment of insulin resistance, LDL, low density lipoprotein; HDL, high density lipoprotein, NA, not available. Those passing the Bonferroni corrected P≤0.001 were considered as statistically significant.
Using data on parents of children in the PMNS and PS, men and women in the MBRC and mothers in the MMNP cohort, we investigated the influence of fGS on anthropometric and cardiometabolic traits in adults (Figure 4B, Table 4). The fGS showed a strong positive association with head circumference (effect = 0.006; p = 5.5×10-4) and a statistically insignificant positive association with adult height (effect = 0.002; p = 0.037) (Table 4; Figure 4B). It was also negatively associated with fasting glucose (effect = -0.006; p = 9.3×10-4) and showed a weak negative association with HOMA-IR and triglycerides (p = 0.022 and 2.0×10-3 respectively). The direction of associations was the same as the genome-wide correlations reported in Europeans (p range, 0.002 − 5.5×10-4) (Figure 4B; Table 4) [14]. No evidence of association was noted between fGS and other anthropometric and cardiometabolic traits in adults (p > 0.05) (Table 4).
Discussion
In this study which included four Indian and three Bangladeshi cohorts from both the Indian subcontinent and the UK, we investigated whether the genetic variants identified in a GWAS of birthweight in Europeans also influence birth size in South Asians (Warrington et al, 2019) (15). We further investigated whether the same genetic variants (either fetal variants that directly influence birthweight, or those in the mother that act indirectly via the intrauterine environment) were associated with anthropometric and cardiometabolic parameters measured during childhood, adolescence and adulthood. We observed strong positive associations of fetal genetic score with birthweight and other birth measurements in these populations of South Asian ancestry despite a large variation in maternal BMI and fetal birthweight. While birthweight positively predicted body size in both children and adolescents, fGS did so only in children but not in adolescents. We also noted a strong association of birthweight with plasma triglycerides levels both in children and adolescents, but fGS was not related to any of the child/adolescent cardiometabolic outcomes. However, fGS was inversely associated with plasma glucose and triglycerides in adults. Maternal genetic score was weakly positively linked to birthweight and was unrelated to body size and cardiometabolic traits in both children and adolescents. Our study thus reports a strong association of fGS and relatively weak association of mGS with birthweight and other birth measures in a non-European population. Further, the genetic constitution of the fetus at specific variants influences body size and the data from the adults suggest that it contributes to future cardiometabolic risk in Indians. Overall, it provides support to the observational association between low birth size and non-communicable diseases like type 2 diabetes and cardiovascular diseases in South Asians. Follow up studies on a larger sample size will be required to answer our second research question (is the birthweight−cardiometabolic risk association explained by shared genetic variants) with confidence.
Most genetic studies associating early life parameters with future risk of cardiometabolic disorders have been conducted in Europeans. As far as we are aware, this is first such analysis in South Asians. We found similar associations of fGS generated using weights from European studies with birth size in a consortium of seven birth cohorts of South Asian ancestry comprising Indian and Bangladeshi mother-child pairs. This was despite a wide variability in birthweight and maternal BMI within the South Asian cohorts and significant differences in the EAFs of many of the birthweight associated variants between the EGG/UKBB and the South Asian subjects. Despite similar fGS association with birthweight as in Europeans, the newborn size of South Asian babies was substantially smaller indicating a significant role of factors not captured by the genetic score on fetal growth. These factors may include different environmental exposures, maternal body size, health and nutritional status etc. We noted an increase of 50.7g of birthweight per SD of fGS which is consistent with the observation in the UKBB-SAS and is marginally smaller than in EFSOCH, examples of South Asian and European ancestry cohorts respectively. The significant association of fGS with body size at birth persisted even after adjustment for mGS, indicating that the genetic effect is not significantly influenced by aspects of the intrauterine environment predicted by the genetic variants used in this study. This is further supported by a similar strength of association after exclusion of children born to GDM mothers which suggests that the fetal genetic effects are independent of maternal diabetes status during pregnancy. The similar association for fGS with birthweight observed between South Asian and European ancestry individuals in this study suggests that although it is difficult to conclude at individual variant level, there are likely common genetic pathways for fetal growth and development in both ancestry groups. Although mGS was relatively weakly associated with fetal birthweight, the association was unaffected by the fetus’s own genotype suggesting that the maternal genetic effect on birthweight was mediated through intrauterine environment. The weaker association of mGS is not unexpected given the lower proportion of variance explained in birthweight by the mGS (~2%) compared to fGS (~6%). Thus, birthweight (body size) is an outcome of the baby’s genetic constitution and an influence of the intrauterine environment, partly determined by the mother’s genotype. However, with the exception of a small number of variants that are known to influence fasting glucose levels, it is largely unclear which intrauterine exposures are influenced by which genetic variants used in the study, making it difficult to dissect their individual role. It was interesting to note that the influence of the maternal genetic score on birthweight varied considerably amongst the cohorts investigated in this study (heterogeneity p = 0.018). This heterogeneity in effect estimates could be driven by ethnicity, maternal BMI, height and nutritional status, socio-economic status, and GDM status; this needs further investigation.
Genome-wide studies have established a robust association between fetal genetic score and later cardiometabolic risk including glycaemic and lipid parameters in Europeans (13; 15). An important feature of our study is that we have been able to independently compare associations of birthweight and birthweight-associated genetic variants with later anthropometric and cardiometabolic traits. Birthweight showed a strong positive association with body composition, and an inverse association with blood triglycerides concentrations in both childhood and adolescence. Fetal genetic score explains only about 6% of the variance in birthweight in European individuals (15) and considering equal effect of fetal genetic score on birthweight in South Asians as in Europeans, it is worth noting that a positive association with body size in childhood and height and head circumference in adults was observed. Effect estimates of fGS with other anthropometric traits was directionally consistent with the direct effect of birthweight; a lack of strong association may be due to a relatively smaller sample size and the smaller effect size compared to the birthweight itself. Absence of association between fGS and any of the traits during adolescence is consistent with findings from even larger studies that have found little evidence of influence of fetal birthweight variants on BMI beyond early childhood (33). Similar to our study, previous studies have demonstrated a pattern of positive genetic correlations with birthweight, and 22 with childhood and adulthood height (13; 15). The fact that the fetus’s genotype at birthweight-associated genetic variants also influenced plasma glucose and triglycerides in adulthood is consistent with the fetal insulin hypothesis, which proposes that birthweight and later cardiometabolic risk are two effects of the same genotype (34). Our findings need to be replicated in larger independent studies of South Asian subjects. Further understanding of the link between birthweight and future cardiometabolic risk will be possible as we understand the exact role of each genetic variant, whether it operates directly or indirectly through its effects on intrauterine environment.
Our study has several strengths and a few limitations. This is the first study exploring the influence of fetal and/or maternal genotype on birth size and their role in future cardiometabolic risk in South Asians. We combined diverse cohorts from India (including both Indo-European and Dravidian ethnicity) and from Bangladesh (local and migrants to the UK), hence the observations can be considered representative of South Asians. The greatest strength of the study is availability of mother-child pairs and anthropometric and cardiometabolic traits in early childhood and adolescence and hence the conclusions drawn from these prospective cohorts are robust. The limitations of the study include a relatively small sample size although assuming equal variance explained by these SNPs in Europeans, our study in South Asians had > 99% and 98% power to detect association of fGS and mGS with birthweight respectively. Lack of adult phenotype data in children of these cohorts is another limitation, but we have partly circumvented this issue by using the genotype and phenotype data from parents of the children in the Indian cohorts. However, lack of birth size and maternal genotype data for these parents did not allow us to study the maternal influence in this group. The availability of a genetic score specific to individuals of South Asian ancestry would also allow us to further investigate the difference in association of mGS with birthweight compared to European ancestry individuals observed here, helping to disentangle environmental effects from those expected from a GS which may not capture the same underlying genetic associations in different ancestry groups.
The observations made in this study are important because the sub-continent is facing the twin burden of poor fetal health and an emerging epidemic of type 2 diabetes and cardiovascular diseases (9; 35; 36). This has been linked to unique phenotypic features, environmental exposures, and a different genetic makeup of South Asians compared to Europeans (17–21). However, this study suggests that the genetic contribution to birth size is largely similar to that in the Europeans, and that other factors may be responsible for the thin-fat phenotype of South Asians which predisposes them to a higher risk of diabetes and related disorders compared to Caucasians. The validation of genetic associations with birthweight in populations of two ancestries, Europeans and South Asians provides a hint that there may be common pathways affecting fetal development which can be influenced by different environmental exposures.
To conclude, we report the associations of genetic scores identified in Europeans with size at birth in participants of South Asian ancestry. However, fetal genetic score is known to explain only about 6% variability in birthweight in Europeans. Interestingly, despite similar association of fetal genetic scores with birthweight as in Europeans, South Asians have a considerably lower birthweight. This indicates a significant role of other factors on fetal growth such as different environmental exposures which are not captured by the genetic variants included in the present study. These genetic loci also influenced early childhood body size and were associated with fasting glucose and triglycerides levels in adults, suggesting that common genetic variants explain part of the association between birth size and adult metabolic syndrome. This supports the “fetal insulin hypothesis” but also highlights an important interaction with environment (16; 34). Lack of association between fetal genetic scores and cardiometabolic traits in the children and adolescents deserves more exploration. Further, birthweight-fetal genotype associations were consistent across all cohorts, association of fetal birthweight with maternal genotype showed heterogeneity between cohorts. This may be related to differences in maternal size, glycemia and socio-economic status and needs further research.
Supplementary Material
Acknowledgements
We sincerely acknowledge the unflinching support of individuals who voluntarily participated in the study and continued to attend regular follow-ups. S.S.N. and A.D. are grateful to the Council of Scientific and Industrial Research (CSIR Mission Mode Project; HCP0008) and A.S. to the Indian Council for Medical Research (ICMR-CAR; GAP0504), Govt of India for their fellowship. We acknowledge the Genetic Laboratory, Erasmus MC, University Medical Centre Rotterdam for the help with genotyping of the Bangladeshi cohorts. Thanks are also due to the UK Medical Research Council Clinical Research Training Fellowship (No. G0800441) to S.F. G.R.C. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
This manuscript was accepted for publication in Diabetes on 19 January, 2022. The final version of the paper will be available on the Diabetes website at https://doi.org/10.2337/db21-0479.
Funding
The Pune Maternal Nutrition Study, Parthenon Study, Mumbai Maternal Nutrition Project and Mysore Birth Records Study were funded by the Medical Research Council (UK), Wellcome Trust (UK), Parthenon Trust (Switzerland) and Newton Fund. The GIFTS and London Bangladeshi cohorts were supported by the MRC [Clinical Research Training Fellowship (G0800441)] and the European Union (FP7 EU grant: 83599025). MMNP was supported by the Wellcome Trust, Parthenon Trust, ICICI Bank Ltd., Mumbai, the UK Medical Research Council (MRC) and the UK Department for International Development (DFID) under the MRC/DFID Concordat. Children’s follow-up was funded by MRC (MR/M005186/1). MBRC studies were also supported by an early career fellowship to Murali Krishna by Welcome DBT India Alliance. Genotyping for the MMNP mother and children (The EMPHASIS study) is jointly funded by MRC, DFID and the Department of Biotechnology (DBT), Ministry of Science and Technology, India under the Newton Fund initiative (MRC grant no.: MR/N006208/1 and DBT grant no.: BT/IN/DBT-MRC/DFID/24/GRC/2015-16). High throughput genotyping of the mother-child pairs from PMNS was funded through the GIFTS European Union (FP7 EU grant: 83599025); for all other cohorts, the genetic analysis was funded by the Council of Scientific and Industrial Research (CSIR), Ministry of Science and Technology, Government of India, through its Network projects. RMF and RNB are supported by Sir Henry Dale Fellowship (Wellcome Trust and Royal Society Grant: WT104150). Both authors would like to acknowledge the use of the University of Exeter High-Performance Computing (HPC) facility in carrying out this work. This research has been conducted using the UK Biobank Resource under application number 7036. The Exeter Family Study of Childhood Health (EFSOCH) was supported by South West NHS Research and Development, Exeter NHS Research and Development, the Darlington Trust and the Peninsula National Institute of Health Research (NIHR) Clinical Research Facility at the University of Exeter. This project is supported by NIHR which is a partnership between the University of Exeter Medical School College of Medicine and Health, and Royal Devon and Exeter NHS Foundation Trust. The views expressed in this paper are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care. Genotyping of the EFSOCH study samples was funded by the Wellcome Trust and Royal Society grant 104150/Z/14/Z.
This research was funded, in part, by the Wellcome Trust (grant WT220390). A CC BY or equivalent licence is applied to author accepted manuscript arising from this submission, in accordance with the grant’s open access conditions.
The funding bodies played no role in the design of the study and collection, analysis, interpretation of data or writing of the manuscript.
Footnotes
Authors’ contributions
G.R.C., C.S.Y., G.A.H., C.H.D.F., S.F. and R.M.F. conceptualised and contributed to the study design; collated and interpreted overall results from various cohorts in the study. G.V.K., K.K., S.A.S., R.D.P., M.K., C.D.G., C.S.Y. and C.H.D.F. are coordinators for various Indian cohorts and played important role in the follow-up and acquisition of phenotype data at different stages. G.R.C. supervised the overall Indian cohort studies. S.F., G.A.H. are the lead supervisor of UK cohort while A.H. and A.K.A.K. managed the Bangladeshi cohort studies. B.W.B. oversaw data collection and phenotyping of subjects in Bangladeshi cohorts. B.A.K. carried out sample collection and phenotyping in the EFSOCH cohort. I.D.M. provided technical support in DNA isolation and quality control analysis in Indian cohorts. S.S.N. and A.D. performed high throughput genotyping of Indian cohorts while B.O., Z.H. T.M.F. and R.M.F. were responsible for preparing samples and genotyping in the Bangladeshi and EFSOCH cohorts. S.S.N., A.D., A.S. cleaned Indian cohorts’ genotype data and generated imputed genotypes whereas R.N.B. performed quality control and imputation of the Bangladeshi and EFSOCH cohort genotype data. A.R.W. defined the South Asian samples of the UK Biobank dataset using ancestry principal components. S.S.N. and R.N.B. performed the central analysis and wrote the first draft of the manuscript. All authors have contributed to manuscript writing, provided critical inputs and approved the final version of the manuscript.
Competing Interests
The authors have no competing interests to declare.
Data and Resource Availability
The datasets and generated during and/or analyzed during the current study are available upon reasonable request. Researchers interested in accessing the data are expected to send a reasonable request by sending an email to the contact authors as detailed below.
Indian cohorts (PMNS, PS, MMNP and MBRC): Giriraj R Chandak at chandakgrc@ccmb.res.in EFSOCH: The Exeter Clinical Research Facility at crf@exeter.ac.uk
GIFTS (WP2 & WP3) and UK Bang cohorts: Graham A Hitman at g.a.hitman@qmul.ac.uk UK-biobank data - https://www.ukbiobank.ac.uk/using-the-resource/ [ukbiobank.ac.uk] No applicable resources were generated or analyzed during the current study.
References
- 1.Barker DJ. Fetal origins of coronary heart disease. BMJ. 1995;311:171–174. doi: 10.1136/bmj.311.6998.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker DJ, Gluckman PD, Godfrey KM, Harding JE, Owens JA, Robinson JS. Fetal nutrition and cardiovascular disease in adult life. Lancet. 1993;341:938–941. doi: 10.1016/0140-6736(93)91224-a. [DOI] [PubMed] [Google Scholar]
- 3.Godfrey KM, Barker DJ. Fetal nutrition and adult disease. Am J Clin Nutr. 2000;71:1344S–1352S. doi: 10.1093/ajcn/71.5.1344s. [DOI] [PubMed] [Google Scholar]
- 4.Wells JC, Sharp G, Steer PJ, Leon DA. Paternal and maternal influences on differences in birth weight between Europeans and Indians born in the UK. PLoS One. 2013;8:e61116. doi: 10.1371/journal.pone.0061116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fall CH, Stein CE, Kumaran K, Cox V, Osmond C, Barker DJ, Hales CN. Size at birth, maternal weight, and type 2 diabetes in South India. Diabet Med. 1998;15:220–227. doi: 10.1002/(SICI)1096-9136(199803)15:3<220::AID-DIA544>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 6.Mi J, Law C, Zhang KL, Osmond C, Stein C, Barker D. Effects of infant birthweight and maternal body mass index in pregnancy on components of the insulin resistance syndrome in China. Ann Intern Med. 2000;132:253–260. doi: 10.7326/0003-4819-132-4-200002150-00002. [DOI] [PubMed] [Google Scholar]
- 7.Stein CE, Fall CH, Kumaran K, Osmond C, Cox V, Barker DJ. Fetal growth and coronary heart disease in south India. Lancet. 1996;348:1269–1273. doi: 10.1016/s0140-6736(96)04547-3. [DOI] [PubMed] [Google Scholar]
- 8.Yajnik CS. Early life origins of insulin resistance and type 2 diabetes in India and other Asian countries. J Nutr. 2004;134:205–210. doi: 10.1093/jn/134.1.205. [DOI] [PubMed] [Google Scholar]
- 9.World Health Organization & United Nations Children’s Fund (UNICEF) Low Birthweight: Country, regional and global estimates. 2004. https://apps.who.int/iris/handle/10665/43184 .
- 10.Ramachandran A, Snehalatha C, Viswanathan V, Viswanathan M, Haffner SM. Risk of noninsulin dependent diabetes mellitus conferred by obesity and central adiposity in different ethnic groups: a comparative analysis between Asian Indians, Mexican Americans and Whites. Diabetes Res Clin Pract. 1997;36:121–125. doi: 10.1016/s0168-8227(97)00040-5. [DOI] [PubMed] [Google Scholar]
- 11.Beaumont RN, Warrington NM, Cavadino A, Tyrrell J, Nodzenski M, Horikoshi M, Geller F, Myhre R, Richmond RC, Paternoster L, Bradfield JP, et al. Genome-wide association study of offspring birth weight in 86 577 women identifies five novel loci and highlights maternal genetic effects that are independent of fetal genetics. Hum Mol Genet. 2018;27:742–756. doi: 10.1093/hmg/ddx429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freathy RM, Mook-Kanamori DO, Sovio U, Prokopenko I, Timpson NJ, Berry DJ, Warrington NM, Widen E, Hottenga JJ, Kaakinen M, Lange LA, et al. Variants in ADCY5 and near CCNL1 are associated with fetal growth and birth weight. Nat Genet. 2010;42:430–435. doi: 10.1038/ng.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horikoshi M, Beaumont RN, Day FR, Warrington NM, Kooijman MN, Fernandez-Tajes J, Feenstra B, van Zuydam NR, Gaulton KJ, Grarup N, Bradfield JP, et al. Genome-wide associations for birth weight and correlations with adult disease. Nature. 2016;538:248–252. doi: 10.1038/nature19806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Early Growth Genetics C. Horikoshi M, Yaghootkar H, Mook-Kanamori DO, Sovio U, Taal HR, Hennig BJ, Bradfield JP, St Pourcain B, Evans DM, Charoen P, et al. New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nat Genet. 2013;45:76–82. doi: 10.1038/ng.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warrington NM, Beaumont RN, Horikoshi M, Day FR, Helgeland O, Laurin C, Bacelis J, Peng S, Hao K, Feenstra B, Wood AR, et al. Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat Genet. 2019;51:804–814. doi: 10.1038/s41588-019-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hattersley AT, Tooke JE. The fetal insulin hypothesis: an alternative explanation of the association of low birthweight with diabetes and vascular disease. Lancet. 1999;353:1789–1792. doi: 10.1016/S0140-6736(98)07546-1. [DOI] [PubMed] [Google Scholar]
- 17.Krishnaveni GV, Yajnik CS. Developmental origins of diabetes-an Indian perspective. Eur J Clin Nutr. 2017;71:865–869. doi: 10.1038/ejcn.2017.87. [DOI] [PubMed] [Google Scholar]
- 18.Wells JC, Pomeroy E, Walimbe SR, Popkin BM, Yajnik CS. The Elevated Susceptibility to Diabetes in India: An Evolutionary Perspective. Front Public Health. 2016;4:145. doi: 10.3389/fpubh.2016.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yajnik CS, Fall CH, Coyaji KJ, Hirve SS, Rao S, Barker DJ, Joglekar C, Kellingray S. Neonatal anthropometry: the thin-fat Indian baby. The Pune Maternal Nutrition Study. Int J Obes Relat Metab Disord. 2003;27:173–180. doi: 10.1038/sj.ijo.802219. [DOI] [PubMed] [Google Scholar]
- 20.Basu A, Sarkar-Roy N, Majumder PP. Genomic reconstruction of the history of extant populations of India reveals five distinct ancestral components and a complex structure. Proc Natl Acad Sci U S A. 2016;113:1594–1599. doi: 10.1073/pnas.1513197113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao S, Yajnik CS, Kanade A, Fall CH, Margetts BM, Jackson AA, Shier R, Joshi S, Rege S, Lubree H, Desai B. Intake of micronutrient-rich foods in rural Indian mothers is associated with the size of their babies at birth: Pune Maternal Nutrition Study. J Nutr. 2001;131:1217–1224. doi: 10.1093/jn/131.4.1217. [DOI] [PubMed] [Google Scholar]
- 23.Krishnaveni GV, Veena SR, Hill JC, Karat SC, Fall CH. Cohort profile: Mysore parthenon birth cohort. Int J Epidemiol. 2015;44:28–36. doi: 10.1093/ije/dyu050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carpenter MW, Coustan DR. Criteria for screening tests for gestational diabetes. Am J Obstet Gynecol. 1982;144:768–773. doi: 10.1016/0002-9378(82)90349-0. [DOI] [PubMed] [Google Scholar]
- 25.Potdar RD, Sahariah SA, Gandhi M, Kehoe SH, Brown N, Sane H, Dayama M, Jha S, Lawande A, Coakley PJ, Marley-Zagar E, et al. Improving women’s diet quality preconceptionally and during gestation: effects on birth weight and prevalence of low birth weight--a randomized controlled efficacy trial in India (Mumbai Maternal Nutrition Project) Am J Clin Nutr. 2014;100:1257–1268. doi: 10.3945/ajcn.114.084921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saffari A, Shrestha S, Issarapu P, Sajjadi S, Betts M, Sahariah SA, Tomar AS, James P, Dedaniya A, Yadav DK, Kumaran K, et al. Effect of maternal preconceptional and pregnancy micronutrient interventions on children’s DNA methylation: Findings from the EMPHASIS study. Am J Clin Nutr. 2020;112:1099–1113. doi: 10.1093/ajcn/nqaa193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krishna M, Kalyanaraman K, Veena SR, Krishanveni GV, Karat SC, Cox V, Coakley P, Nagaraj K, Stein C, Paul B, Prince M, et al. Cohort Profile: The 1934-66 Mysore Birth Records Cohort in South India. Int J Epidemiol. 2015;44:1833–1841. doi: 10.1093/ije/dyv176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhowmik B, Siddique T, Majumder A, Mdala I, Hossain IA, Hassan Z, Jahan I, Moreira N, Alim A, Basit A, Hitman GA, et al. Maternal BMI and nutritional status in early pregnancy and its impact on neonatal outcomes at birth in Bangladesh. BMC Pregnancy Childbirth. 2019;19:413. doi: 10.1186/s12884-019-2571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhowmik B, Siddiquee T, Mdala I, Quamrun Nesa L, Jahan Shelly S, Hassan Z, Moreira N, Jahan I, Azad Khan AK, Hitman GA, Hussain A. Vitamin D3 and B12 supplementation in pregnancy. Diabetes Res Clin Pract. 2021;174:108728. doi: 10.1016/j.diabres.2021.108728. [DOI] [PubMed] [Google Scholar]
- 30.Knight B, Shields BM, Hattersley AT. The Exeter Family Study of Childhood Health (EFSOCH): study protocol and methodology. Paediatr Perinat Epidemiol. 2006;20:172–179. doi: 10.1111/j.1365-3016.2006.00701.x. [DOI] [PubMed] [Google Scholar]
- 31.Hughes AE, Nodzenski M, Beaumont RN, Talbot O, Shields BM, Scholtens DM, Knight BA, Lowe WL, Jr, Hattersley AT, Freathy RM. Fetal Genotype and Maternal Glucose Have Independent and Additive Effects on Birth Weight. Diabetes. 2018;67:1024–1029. doi: 10.2337/db17-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, Cortes A, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cousminer DL, Freathy RM. Genetics of early growth traits. Hum Mol Genet. 2020;29:R66–R72. doi: 10.1093/hmg/ddaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hattersley AT, Beards F, Ballantyne E, Appleton M, Harvey R, Ellard S. Mutations in the glucokinase gene of the fetus result in reduced birth weight. Nat Genet. 1998;19:268–270. doi: 10.1038/953. [DOI] [PubMed] [Google Scholar]
- 35.International Diabetes Federation: IDF report 7th edition. 2015. http://www.diabetesatlas.org .
- 36.Prabhakaran D, Jeemon P, Roy A. Cardiovascular Diseases in India: Current Epidemiology and Future Directions. Circulation. 2016;133:1605–1620. doi: 10.1161/CIRCULATIONAHA.114.008729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets and generated during and/or analyzed during the current study are available upon reasonable request. Researchers interested in accessing the data are expected to send a reasonable request by sending an email to the contact authors as detailed below.
Indian cohorts (PMNS, PS, MMNP and MBRC): Giriraj R Chandak at chandakgrc@ccmb.res.in EFSOCH: The Exeter Clinical Research Facility at crf@exeter.ac.uk
GIFTS (WP2 & WP3) and UK Bang cohorts: Graham A Hitman at g.a.hitman@qmul.ac.uk UK-biobank data - https://www.ukbiobank.ac.uk/using-the-resource/ [ukbiobank.ac.uk] No applicable resources were generated or analyzed during the current study.