Abstract
Genetics has recently benefited from the genome engineering revolution: genes can be knocked out, knocked down, or activated more easily than ever before. This range of genetic manipulations has also provided a range of outcomes, sometimes contradictory. But how much interesting biology hides within these discrepancies? Recent studies have shown that genetic compensation can be activated by some gene perturbations and not others, hinting that this phenomenon might skew our understanding of the genotype–phenotype relationship. We review the main findings regarding transcriptional adaptation, a newly discovered form of genetic compensation, and discuss their possible implications for establishing and analyzing animal and plant models to study gene function. We also touch upon how this new knowledge could benefit our understanding of disease-causing mutations and help explain cases of low penetrance or variable expressivity in human genetics.
From Classical to Molecular Genetics
When Gregor Mendel was trying to understand how traits are passed from one generation to the next, a phenomenon well known to farmers since ancient times, he first needed to establish the concept of factors (nowadays called genes) that control individual traits. He also coined the terms dominant (see Glossary) and recessive to explain the way alleles interact to produce the phenotypic outcomes he was studying. The resulting Mendelian laws of inheritance, which founded the field of genetics, were such an intellectual leap that it took decades until their importance was appreciated.
Glossary
- Allele
a version of a gene.
- Antisense oligonucleotides
modified or synthetic nucleic acid, or nucleic acid like, molecules complementary to endogenous RNAs that are employed to modify gene expression. RNA hairpins and double-stranded RNA molecules fall into the broader category of antisense technology but utilize the endogenous RNAi pathway.
- Dominant
an allele whose presence in the heterozygous state dictates the phenotype.
- Expressivity
the degree/severity of phenotype manifestation observed in individuals carrying the same mutation(s).
- Genetic compensation
the phenomenon whereby the effect of a deleterious mutation is buffered by the genome.
- Genetic redundancy
the phenomenon whereby two genes contribute to the same biological process such that inactivation of either gene is not disruptive.
- Homologous gene
a gene in the genome of a different species that has a shared evolutionary ancestry.
- Hypomorphic allele
also known as a hypomorph, a version of a gene that retains some wild-type function.
- Morpholino
a synthetic antisense molecule composed of a methylenemorpholine ring backbone and phosphorodiamidate-linked nucleic acid bases.
- Mutagenesis screen
the systematic use of mutagens to induce genomic alterations and recover phenotypes.
- Nonsense-mediated mRNA decay (NMD)
a cellular quality-control pathway that identifies and degrades mRNAs which contain a premature termination codon.
- Penetrance
the percentage of individuals carrying a particular allele that display a phenotype related to this allele.
- Recessive
an allele that dictates the phenotype only when in the homozygous state.
- Reverse genetics
the introduction of a mutation into a gene of interest to analyze the resulting phenotype.
- Small molecules
compounds of low molecular weight, usually less than 900 Da.
- Transcriptional adaptation
modulation of the transcriptome of a cell due to a mutation in a gene, independent of the mutation’s effect on the encoded protein.
Eventually, seminal work from many scientists confirmed Mendel’s laws and expanded his findings, leading to the description of many naturally occurring phenotypes and the establishment of genetic model organisms. Furthermore, random mutagenesis screens granted access to an even wider spectrum of phenotypes under laboratory conditions [1–3]. Until the advent of transposable element mutagenesis and gene trapping, approaches that facilitate the mapping of genomic disruptions and the isolation of the affected genes, most studies were restricted to phenotype characterization [2–5]. All this knowledge was later exploited by new tools and techniques with the emergence of molecular biology. For example, the large collection of P-element insertions in Drosophila allowed selective mutagenesis by mobilizing transposable elements close to or within specific genes, and methods such as TILLING (Targeting Induced Local Lesions In Genomes) allowed the identification of mutations in specific genes from a random pool [4–7]. Furthermore, although whole-genome sequencing helped to isolate the causative mutations for well-characterized phenotypes, it also provided a long list of annotated genes that were not linked to any phenotype.
This situation led to the explosion of reverse genetics, and with it the need to develop tools and methodologies to mutate specific genes. One technique, gene targeting through homologous recombination, is a powerful but slow and laborious technique that is mostly limited to models where embryonic stem cell technology is available; therefore, complementary strategies that focused on perturbing gene function became more prevalent. Targeting gene products (RNAs or proteins) to ablate their function became synonymous to mutating a gene. Small molecules can bind to enzymes or receptors and block their function [8], and antisense oligonucleotides can bind to and degrade RNA molecules, or inhibit their translation or splicing [9–12]. However, unlike small molecules, little prior knowledge other than gene sequence is necessary for the design and use of antisense approaches. Thus, their versatility made them very popular with animal models, especially where efficient gene targeting was not available, including worms, flies, fish, frogs, and chickens [12–14].
Together, these functional studies increased our understanding of the role of different factors during development and organogenesis. In addition, the extensive use of RNAi in Drosophila, Caenorhabditis elegans, and less popular model organisms such as sea urchin and Parhyale has driven many important discoveries ranging from evolutionary developmental biology to behavior [15–19]. Furthermore, large-scale in vitro siRNA screens have helped identify new drug targets for regulating cell growth and viability in disease-relevant contexts [20]. However, these approaches also suffer from variability and off-target effects, and are not easily applicable at later developmental stages or to study regeneration or aging [21].
The Targeted Mutagenesis Revolution and Mutants on Demand
Cells target proteins to specific regions of their genome to regulate gene transcription, as well as DNA replication and repair. Inspired by mechanisms found in nature, scientists have developed tools to guide DNA nucleases to specific genes and activate error-prone DNA-repair mechanisms, hoping to inactivate parts of the genome. Although early versions of these tools such as zinc-finger nucleases (ZFNs) were somewhat inefficient and challenging to assemble, transcription Activator-Like Effector Nucleases (TALENs) made genome-engineering technology more widely available [22]. The latest development came from repurposing prokaryotic nucleases that are part of a defense system against viruses, known as the CRISPR/Cas system [23–25].
Very much as the introduction of genome sequencing led to an abundance of new genes, modern genome-engineering tools have offered an abundance of mutant alleles. This technological revolution shifted the bottleneck of reverse genetics from targeting and knocking out candidate genes to identifying and analyzing the phenotypic outcome of the new mutant alleles. Genetics had entered the ‘mutants on demand’ era – any gene could now be mutated in the laboratory. The first engineered zebrafish mutant phenotypes were reported in 2008, describing developmental defects such as no tail, pigmentation loss, and vascular malformations [26,27]. Other more technical reports focused primarily on the spectrum and prevalence of mutations induced at the DNA level, and did not include in-depth phenotypic analysis [28,29].
Discrepancies in the Field of Genetics and Genetic Compensation
Because negative results tend to be under-represented in the scientific literature, it is reasonable to assume that the initial publications on engineered alleles did not paint the whole picture regarding how often mutations failed to produce a clear phenotype. Failure to identify a phenotype could be because the gene is not involved in the biological process examined and/or because subtle phenotypes were not detected. Candidate genes are often selected based on their tissue-specific expression pattern or their expression dynamics during a given biological process. Even when information about the gene product is available or mutations in a homologous gene are described in other model systems, or are implicated in human disease, lack of a phenotype could be attributed to hypomorphic alleles, genetic redundancy, or merely differences between evolutionarily distant species.
Since the early 1980s, zebrafish (Danio rerio) has emerged as a powerful genetic model organism to study vertebrate development, organogenesis, and regeneration. Its fast ex utero development and embryo transparency were key factors in establishing zebrafish as a genetic model. In addition to spontaneous mutations [30], large forward-genetic screens have provided a wealth of mutants with phenotypes in early embryogenesis, vascular development, and behavior, to name only a few [31]. Even so, zebrafish research has especially benefited from the introduction of antisense technology, mainly in the form of morpholinos, that allowed knocking down virtually any target mRNA, either by inhibiting its translation or inducing its mis-splicing [12].
During the approximately two decades of extensive morpholino use in zebrafish, >5000 genes have been targeted, and on average two morpholinos have been designed for each gene (zfin.org). As with other antisense technology, concerns about the off-target effects of morpholinos led to the early publication of good practices for morpholino use [32]. These concerns were renewed with the advent of modern genetic engineering tools, such as TALENs, that were easy to design and were thus broadly implemented in zebrafish laboratories. The massive shift to mutant generation and analysis, which started gaining momentum in 2012, revealed that many genes lacked observable phenotypes when mutated, despite previous data based on morpholino antisense approaches.
Luckily, these anecdotal reports were consolidated in 2015 in an extensive analysis by Kok et al. alerting the community about the poor correlation between morpholino-induced and mutant phenotypes in zebrafish [33,34]. Around 80% of the mutants analyzed by Kok et al. did not exhibit the morphant phenotype [33]. In one example, the authors generated a zebrafish line with a deletion of the noncoding gene megamind and failed to recover the previously reported hydrocephaly phenotype [35]. More importantly, they showed that injection of the megamind morpholino in the same line, which also lacks the morpholino binding site, still caused hydrocephaly, showing that this phenotype was due to off-target effects. Of note, the original megamind study included three different morpholinos against that gene, mismatch morpholino controls, and rescue experiments [35]. Despite these measures, the morpholino-based conclusions of this study were now being questioned.
With the rapid evolution of the CRISPR/Cas technology, and trust in antisense technology challenged [36], the era of morpholino use seemed to be over. Although mutants are seen as the gold standard, complementary approaches such as morpholinos still have their advantages: in prescreening candidates before investing time and effort to raise engineered mutants, knocking down genes in different genetic backgrounds, and complying with the increasing needs and regulations regarding animal experimentation and welfare. Good practices should of course also be used when engineering mutants. Analyzing several different independent mutant alleles of each gene to avoid off-target effects, and assessing the strength of each mutant allele to avoid hypomorphs, are points to consider when designing and engineering genetic loss-of-function models.
Faced with this transition from morpholino-based experiments to generating and analyzing mutants, it would first be important to carefully assess the strengths and weaknesses of each approach. To this end, the endothelial enriched epidermal growth factor like 7 (egfl7) gene, which encodes an extracellular matrix protein, was analyzed in detail using both approaches. This gene had been previously implicated, using morpholino knockdowns, in vascular tube formation [37,38], but engineered zebrafish and mouse mutants for this gene did not exhibit a vascular phenotype [39,40]. Furthermore, morpholino efficiency and zebrafish mutant allele strength had been evaluated through RNA and/or protein levels in a tagged eglf7 zebrafish line and in cell culture experiments [39]. These results indicated that the egfl7 morpholino injections efficiently targeted the egfl7 mRNA, and that the engineered egfl7 mutation disrupted the expression of full-length Egfl7 protein.
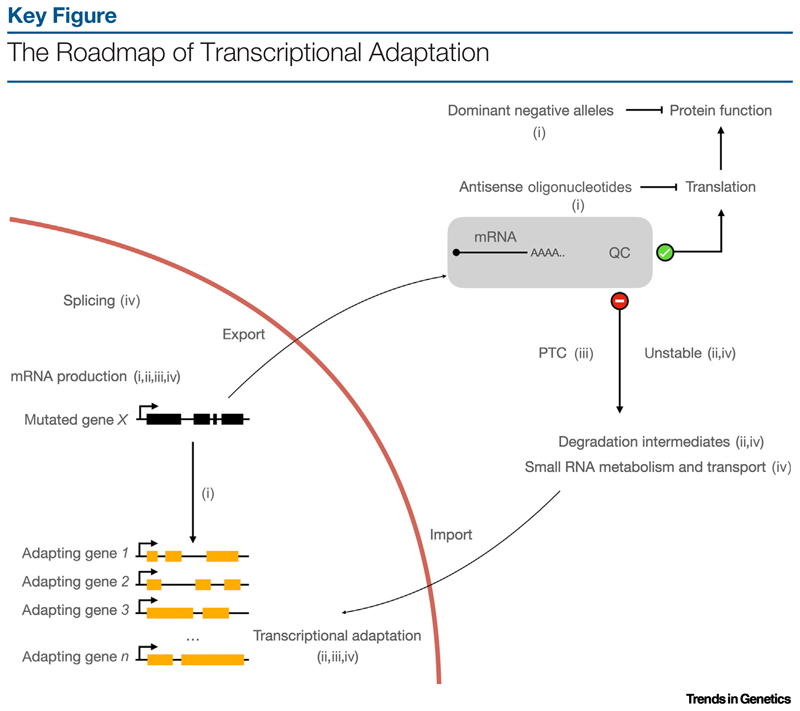
Similarly to the megamind study [35], the key experiment to test the two approaches – morpholinos and genetic mutants – was to inject the egfl7 morpholino into the egfl7 mutants, and determine which phenotypic outcome prevailed – the vascular phenotype of the morphants or the lack of phenotype of the mutants [35,39]. A morphant phenotype in these embryos would suggest that the egfl7 morpholino eliminated residual eglf7 activity in the mutants or affected non-specific targets. A lack of phenotype, on the other hand, would imply that vascular development in egfl7 mutants was independent of Egfl7, and that any off-targets of the egfl7 morpholino did not cause vascular phenotypes under these conditions. To ensure a blinded experimental setup, the authors injected embryos from egfl7 heterozygous intercrosses; siblings served as an internal control. The injected embryos exhibiting vascular defects were sorted and genotyped, leading to the observation that the mutants were strongly under-represented in that population. Taken together, these results led to the hypothesis that the phenotypic differences between egfl7 mutants and morphants were not caused by nonspecific effects of the morpholino injections or residual Egfl7 activity in mutants. Instead, there was a fundamental difference in the way in which embryos responded to a mutation in the egfl7 locus versus inhibition of egfl7 mRNA translation. How could cells overcome loss of egfl7 in one case and not the other? Does the egfl7 mutation somehow protect from egfl7 loss produced by morpholino knockdown? By comparing the transcriptomes and proteomes of mutant and morphant embryos, one might identify the changes responsible for the different phenotypic outcomes. A set of genes of the emilin family, which encodes extracellular proteins, were in fact found to be upregulated in mutants, but not in morphants. Because emilin proteins share domains with EGFL7, it was hypothesized that their increased expression could compensate for the loss of Egfl7. Indeed, this hypothesis was supported by rescue experiments in which egfl7 morphants, which do not upregulate emilin genes, displayed only mild vascular phenotypes after Emilin mRNA injections. These and other data led the authors to propose a new mode of genetic compensation, whereby cells can upregulate particular genes when they harbor a mutation in their genome, but fail to do so when challenged by knockdown through morpholino antisense technology [39] (Figure 1, Key Figure).
Figure 1.
Data from key studies {(i), Rossi et al. [39]; (ii), El-Brolosy et al. [48]; (iii), Ma et al. [49]; (iv), Serobyan et al. [52]} have helped build a basic framework for transcriptional adaptation: a frameshift mutation in gene X can activate the transcription of similar genes in trans [39,48,49,52]. This activation is not dependent on loss of protein activity as transcriptional [39,48,49,52] or translational [39] inhibition of gene X, or dominant negative alleles [39], do not trigger this response. The mRNA quality control (QC) mechanism of the cell determines whether an mRNA is used for protein production or is recycled [48,49,52]. If not used for translation, mRNAs can also enter the transcriptional adaptation pathway, either repurposed as long noncoding RNAs [49] or by contributing small degradation intermediates [48,52]. Different processes have been implicated in transcriptional adaptation, either upstream or downstream of the QC step [48,49,52]. Better understanding of the crosstalk between these processes will help explain different transcriptional adaptation responses and allow modulation of this pathway. Abbreviation: PTC, premature termination codon.
Transcriptional Adaptation beyond Zebrafish
The observation of genetic compensation is not new. Protein feedback loops have been described in bacteria and yeast, allowing the utilization of alternative biochemical pathways [41]. Such a response is somewhat linked to the idea of genetic robustness: biological systems that are less sensitive to genetic changes should be favored by natural selection [42]. Nevertheless, the cellular response described in zebrafish seemed to be inherently different. An additional layer of transcriptional regulation was activated upstream of protein function, a notion also supported by the use of a dominant negative allele [38]. To distinguish this special mode of genetic compensation from others, it was called transcriptional adaptation [43]. Furthermore, even though the lack of a phenotype in egfl7 mutants helped to identify this phenomenon, it would be a mistake to think of it as a purposeful response to compensate for gene loss. After all, it was also shown that zebrafish vegfaa mutants, like their respective morphants, exhibit severe vascular hypoplasia despite showing transcriptional differences compared to vegfaa morphants: the upregulation of vegfab observed in vegfaa mutants is not sufficient to compensate for the loss of Vegfaa. Accordingly, vegfab mRNA injections cannot rescue vegfaa morphants or mutants [44], indicating that vegfaa and vegfab are not functionally redundant. In this example, transcriptional adaptation, at least the upregulation of vegfab, does not modify the mutant phenotype.
Two main questions arose from this seminal work – how prevalent is this phenomenon of transcriptional adaptation, and what is the molecular mechanism that triggers this response? [43]. Many reports have since implicated transcriptional adaptation to explain phenotypic differences between mutants and morphants in zebrafish, but only a few have performed careful analysis to exclude other possible reasons for these discrepancies, for example, hypomorphic alleles and/or off-target effects of the morpholinos [45–47]. Moreover, a major challenge when studying potential examples of transcriptional adaptation is to distinguish the gene(s) responding to the genomic mutation (hereafter named adapting genes) from expression changes caused by loss of protein function. Therefore, most efforts to identify adapting genes have so far focused on paralogs; however, responses from other genes should not be excluded.
We have recently started to understand more about transcriptional adaptation, guided by two studies focusing on the mechanistic underpinnings of this process in zebrafish, and also expanding some of the studies for the first time in mouse cells [48,49]. The authors analyzed different zebrafish and mouse cell line mutants, and found a correlation between alleles that harbor a premature termination codon (PTC) and the upregulation of adapting genes. Further analyses found that reduced mutant mRNA levels were predictive of whether the adapting genes were upregulated [48]. These two sets of data pointed to the importance of the mRNA surveillance machineries including nonsense-mediated mRNA decay (NMD), an mRNA quality-control mechanism that clears defective transcripts [50]. Recognition and degradation of error-containing mRNAs is classically thought to protect from the accumulation of nonfunctional or even toxic translation products. The new findings on transcriptional adaptation indicate that, during this process, mutant mRNAs can also be repurposed to activate the transcription of adapting genes. How this activation occurs is currently not understood. For example, even though the two studies independently recognize the importance of the mutant mRNA in activating transcriptional adaptation, each favors a slightly different model. The main debate lies with whether the recognition of the PTC-bearing mRNA is followed by degradation and repurposing of decay intermediates for gene regulation [48,51], or whether transcriptional adaptation represents a parallel pathway in which mRNAs bearing PTCs evade degradation and participate as long or even full-length transcripts in gene expression regulation [49] (Figure 1).
More insights came from the first transcriptional adaptation work in the nematode C. elegans. The authors established and used two gene pairs to further dissect the transcriptional adaptation pathway by performing a targeted RNAi screen [52]. Briefly, act-5 and unc-89 mutations were found to upregulate act-3 and sax-3 expression, respectively. Interestingly, knocking down SMG-6, the only known endonuclease implicated in NMD, restores mutant act-5 and adapting act-3 mRNA levels [52]. Although this result argues that, at least, the initial endonucleolytic cleavage of the mutant mRNA is necessary for transcriptional adaptation, differences between gene models cannot be excluded. Knocking down SMG-4, another member of the NMD pathway which lies upstream of SMG-6, abolishes transcriptional adaptation in the unc-89/sax-3 model, whereas SMG-6 knockdown has no effect in this model [52]. C. elegans smg-4, the ortholog of yeast UPF3, has two homologs in zebrafish and mice. The differential involvement of the two upf3 members has been proposed to be decisive for sorting the PTC-containing mRNAs into the degradation or transcriptional adaptation pathways in zebrafish [49]. It is thus possible that different genes or alleles utilize somewhat different factors to activate transcriptional adaptation. These differences also emphasize the value of establishing and studying several transcriptional adaptation models to grasp the general and particular rules underlying this phenomenon.
Modern tools enable us to assess gene function in a fast and efficient manner. Genetic screens for the activation or suppression of transcriptional adaptation can help identify new genes and assemble the pathways that regulate this process. The targeted RNAi screen in C. elegans has already shown the strength of such an approach [52]. In addition to ‘expected’ findings, such as the involvement of NMD-related factors, this screen identified factors involved in mRNA splicing and small RNA biogenesis. The most interesting result, however, was that loss of some factors involved in small RNA biogenesis and transport, including the Argonaute proteins ERGO-1 and NRDE-3, the RNA-dependent RNA polymerase RRF-3, and the RNase DCR-1, blocked activation of transcriptional adaptation without restoring mutant RNA levels [52]. Irrespective of what template these factors use, full-length PTC-containing mRNAs or their degradation products, or even some derivatives, their involvement is essential to integrate the transcriptional adaptation pathway into the cellular gene expression machinery (Figure 1).
Genetic Models and Transcriptional Adaptation
The ability to generate genetic models on demand entails a larger responsibility. Although random mutagenesis cannot be controlled, the decision on how to perturb gene function now depends on the scientific question and prior knowledge. For example, if the goal is to create human disease models, one might decide to engineer known or suspected pathological mutations. However, when the goal is less well defined, for instance when investigating gene function, exploiting the error-prone nonhomologous end-joining (NHEJ) machinery after DNA cleavage is a straightforward way to insert frameshift mutations and disrupt the amino acid sequence of the protein product. The aim in such cases is to isolate complete knockout alleles, and hypomorphic alleles (encoding a partially functional protein or leading to genetic compensation by activating transcriptional adaptation) must therefore be identified and excluded.
If transcriptional adaptation is a concern, for example, in cases where multiple paralogs are present, and with our current understanding of how this pathway is activated, generating unstable (e.g., PTC-containing) alleles should be avoided. Deletion of single or multiple exons is a very popular strategy, especially when generating conditional alleles [53]. However, such deletions can also generate PTC-containing transcripts, which could trigger transcriptional adaptation. Alternatives include selecting, whenever possible, in-frame deletions when using NHEJ, and floxing exons whose excision maintains the reading frame of the gene. In both strategies, targeting functionally important domains, or evolutionarily conserved regions, a proxy for low tolerance to mutations, is more likely to lead to an inactive protein product.
Even though PTC-containing mRNAs have received most attention to date when investigating transcriptional adaptation, other aberrant mRNAs, which are recognized by other cellular quality-control mechanisms, could also contribute to transcriptional adaptation or similar processes. Some of the alleles studied are predicted to generate unstable mRNAs owing to the formation of strong secondary structures or the lack of a stop codon [39,48]. Blocking transcription through CRISPR interference (CRISPRi) or by deleting promoters or whole genes does not lead to transcriptional adaptation [39,48,49]. Likewise, tissues that do not express the mutant gene do not activate adapting genes [52]. It is thus expected that RNA-less alleles are a good way to avoid transcriptional adaptation. Nevertheless, such extreme modifications of the genome should be carried out with caution because removal of noncoding RNAs or unforeseen regulatory elements of nearby genes could lead to incorrect conclusions regarding the function of the mutated gene.
As we start to understand more about the ways in which different mutations are interpreted by the cellular machinery, we will be able to make better use of the available genome-engineering tools and design new ways to modify and dissect gene function. Current tools for targeting the genome still need improvement. Increasing sequencing power now allows us to identify off-target effects with better resolution than ever before. For example, although initial experiments on tolerance of mismatches in guide sequences identified a seed region in which mutations abrogated targeting in vitro [23], we now know that many in vivo off-targets do not follow this rule [54,55]. For these reasons, and because all tools have drawbacks (some known and quantifiable, others unknown), it is essential to use orthogonal approaches to challenge results and avoid confirmation bias.
Human Genetics and Transcriptional Adaptation
The human genome project was a milestone for human genetics. Soon after its completion, however, it became evident that this effort was only the first step towards understanding the information stored in our genome and how this information is used and interpreted by cells. Moreover, in the field of genetics, discovery of gene function is driven by differences between genotypes, and thus projects that sample the diversity of the human genome are providing us with invaluable information about variants that could be linked to susceptibility to specific diseases. Building on this information, genome-wide association studies (GWAS) have been an important method to identify possible causative mutations. However, in the absence of direct experimental evidence, most conclusions from GWAS are based on correlations.
Penetrance and expressivity are two terms used by geneticists to describe the black box that lies between genotype and phenotype. It is remarkable that, even without knowing the underlying causes of the complete lack, or variable severity, of an expected phenotype, scientists conceived and quantified these concepts. It is now widely accepted that possible causes for incomplete penetrance and variable expressivity are genetic background (modifier genes) and environmental factors. However, identifying modifier genes has proven to be a major challenge.
Transcriptional adaptation could be seen as a disruptive phenomenon when trying to understand genotype–phenotype relationships. Changes in gene expression caused by transcriptional adaptation can reduce the severity of the expected phenotype, as seen with egfl7 mutants in zebrafish and Actin mutants in zebrafish, mouse cells, and worms [39,46,48,52]. In other cases, for example, in zebrafish vegfaa mutants, transcriptional adaptation does not have an obvious effect on the phenotype [39,44]. It cannot be excluded that these changes in gene expression could also have detrimental effects on cellular fitness. For example, Lgr6 knockout mice are predisposed to squamous cell carcinoma owing to upregulation of Lgr5, which is not observed with short hairpin RNA knockdown of Lgr6 [56]. Similarly, in another report in zebrafish, while marcksb morphants exhibit early patterning defects due to decreased bone morphogenetic protein (BMP) signaling, mutants overcompensate by upregulating MARCKS-family members, and display increased BMP signaling, leading to a mild ventralization phenotype [57]. Thus, transcriptional adaptation can have beneficial as well as detrimental effects.
In light of these findings, factors involved in transcriptional adaptation constitute excellent modifier gene candidates. Variations in the protein sequence or expression levels of these molecules could influence the ability of the cell to regulate the expression of adapting genes, among others. For example, differences in NMD activity have been proposed to affect human disease, and both interindividual variability and cross-tissue variability have been reported in NMD [58–60]. Moreover, an aging-related decrease in NMD activity has been observed in C. elegans [61]. Similarly, the activity levels of splicing and small RNA biogenesis factors, that are also linked to transcriptional adaptation [52], could potentially influence the outcome of the response to various mutant alleles.
Concluding Remarks
Basic research provides the foundation to understand how our world works. It also allows us to use this knowledge to build more complex hypotheses and find alternative avenues to solve problems. In addition to correcting genetic mutations through gene therapy, manipulating modifier genes or pathways to stimulate the cells’ own mechanisms to compensate for the loss of protein function could be a way to tackle genetic diseases and alleviate symptoms (see Outstanding Questions). Although transcriptional adaptation has not yet been reported in human cells, recent reports in mouse cells and C. elegans [39,52], and the implication of conserved cellular pathways, suggest that this phenomenon is likely to be widespread. These first descriptions of transcriptional adaptation have also reshaped our view of different gene perturbation strategies, and additional exciting discoveries clearly lie ahead.
Highlight.
Different strategies to perturb gene function can produce different outcomes due to biological reasons.
Transcription of some mutant mRNA species can affect gene expression at distant loci, a phenomenon we call transcriptional adaptation.
Transcriptional adaptation can sometimes upregulate genes that compensate for the loss of the mutant gene function, thereby masking the expected mutant phenotype.
The mechanisms of transcriptional adaptation remain poorly understood, and our ability to modulate this phenomenon could be used to uncover or alleviate phenotypes.
Outstanding Questions.
Do we need so many different ways to perturb gene function? Are not all approaches equivalent? What can we learn from using different approaches?
Can some presumably null alleles behave as hypomorphs despite the lack of protein function? Does this imply that protein-coding genes can also have protein-independent functions?
Can differences in how tissues or individuals respond to mutations, independently of their outcome on protein function, affect the resulting phenotype? Can we mobilize similar mechanisms to strengthen or weaken the phenotype of an allele as needed?
Acknowledgements
We thank M.A. El-Brolosy, G. Jakutis, Z. Jiang, A. Rossi, V. Serobyan, and J.M. Welker for discussion and comments on the manuscript; research in the laboratory of D.Y.R.S. is supported by the Max Planck Society, the EU (European Research Council AdG project ZMOD 694455), the Deutsche Forschungs Gemeinschaft (DFG), and the Leducq Foundation. Z.K. is supported by ETHZ.
References
- 1.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Driever W, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- 3.Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet. 2002;3:176–188. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- 4.O’Kane CJ, Gehring WJ. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci U S A. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartenstein V, Jan YN. Studying Drosophila embryogenesis with P-lacZ enhancer trap lines. Roux Arch Dev Biol. 1992;201:194–220. doi: 10.1007/BF00188752. [DOI] [PubMed] [Google Scholar]
- 6.McCallum CM, et al. Targeted screening for induced mutations. Nat Biotechnol. 2000;18:455–457. doi: 10.1038/74542. [DOI] [PubMed] [Google Scholar]
- 7.Bentley A, et al. Targeted recovery of mutations in Drosophila. Genetics. 2000;156:1169–1173. doi: 10.1093/genetics/156.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 9.Bennett CF. Therapeutic antisense oligonucleotides are coming of age. Annu Rev Med. 2019;70:307–321. doi: 10.1146/annurev-med-041217-010829. [DOI] [PubMed] [Google Scholar]
- 10.Kennerdell JR, Carthew RW. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- 11.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 12.Moulton JD, Yan YL. Using morpholinos to control gene expression. Curr Protoc Mol Biol Chapter. 2008;26:2681–268. doi: 10.1002/0471142727.mb2608s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohr SE. RNAi screening in Drosophila cells and in vivo. Methods (San Diego, Calif) 2014;68:82–88. doi: 10.1016/j.ymeth.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods (San Diego, Calif) 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 15.Firnhaber C, Hammarlund M. Neuron-specific feeding RNAi in C. elegans and its use in a screen for essential genes required for GABA neuron function. PLoS Genet. 2013;9:e1003921. doi: 10.1371/journal.pgen.1003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nix P, et al. Axon regeneration genes identified by RNAi screening in C. elegans. J Neurosci. 2014;34:629–645. doi: 10.1523/JNEUROSCI.3859-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng F, et al. TRAP-seq profiling and RNAi-based genetic screens identify conserved glial genes required for adult Drosophila behavior. Front Mol Neurosci. 2016;9:146. doi: 10.3389/fnmol.2016.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peter I, et al. Predictive computation of genomic logic processing functions in embryonic development. Proc Natl Acad Sci U S A. 2012;109:16434–16442. doi: 10.1073/pnas.1207852109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liubicich D, et al. Knockdown of Parhyale ultrabithorax recapitulates evolutionary changes in crustacean appendage morphology. Proc Natl Acad Sci U S A. 2009;106:13892–13896. doi: 10.1073/pnas.0903105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perwitasari O, et al. siRNA genome acreening approaches to therapeutic drug repositioning. Pharmaceuticals. 2013;6:124–160. doi: 10.3390/ph6020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fedorov Y, et al. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12:1188–1196. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaj T, et al. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doyon Y, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng X, et al. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foley JE, et al. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by oligomerized pool engineering (OPEN) PLoS One. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sander JD, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reischauer S, et al. Cloche is a bHLH-PAS transcription factor that drives haemato-vascular specification. Nature. 2016;535:294–298. doi: 10.1038/nature18614. [DOI] [PubMed] [Google Scholar]
- 31.Nüsslein-Volhard C. The zebrafish issue of development. Development (Cambridge, England) 2012;139:4099–4103. doi: 10.1242/dev.085217. [DOI] [PubMed] [Google Scholar]
- 32.Eisen JS, Smith JC. Controlling morpholino experiments: don’t stop making antisense. Development (Cambridge, England) 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- 33.Kok F, et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell. 2015;32:97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stainier D, et al. Making sense of anti-sense data. Dev Cell. 2015;32:7–8. doi: 10.1016/j.devcel.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 35.Ulitsky I, et al. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stainier D, et al. Guidelines for morpholino use in zebrafish. PLoS Genet. 2017;13:e1007000. doi: 10.1371/journal.pgen.1007000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker LH, et al. The endothelial-cell-derived secreted factor Egfl7 regulates vascular tube formation. Nature. 2004;428:754–758. doi: 10.1038/nature02416. [DOI] [PubMed] [Google Scholar]
- 38.Charpentier MS, et al. CASZ1 promotes vascular assembly and morphogenesis through the direct regulation of an EGFL7/RhoA-mediated pathway. Dev Cell. 2013;25:132–143. doi: 10.1016/j.devcel.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rossi A, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- 40.Kuhnert F, et al. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development (Cambridge, England) 2008;135:3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- 41.Mitrophanov AY, Groisman EA. Positive feedback in cellular control systems. BioEssays. 2008;30:542–555. doi: 10.1002/bies.20769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitano H. Biological robustness. Nat Rev Genet. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- 43.El-Brolosy MA, Stainier D. Genetic compensation: a phenomenon in search of mechanisms. PLoS Genet. 2017;13:e1006780. doi: 10.1371/journal.pgen.1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossi A, et al. Regulation of Vegf signaling by natural and synthetic ligands. Blood. 2016;128:2359–2366. doi: 10.1182/blood-2016-04-711192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu P, et al. Short body length phenotype is compensated by the upregulation of nidogen family members in a deleterious nid1a mutation of zebrafish. J Genet Genomics. 2017;44:553–556. doi: 10.1016/j.jgg.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 46.Sztal TE, et al. Genetic compensation triggered by actin mutation prevents the muscle damage caused by loss of actin protein. PLoS Genet. 2018;14:e1007212. doi: 10.1371/journal.pgen.1007212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heng J, et al. Rab5c-mediated endocytic trafficking regulates hematopoietic stem and progenitor cell development via Notch and AKT signaling. PLoS Biol. 2020;18:e3000696. doi: 10.1371/journal.pbio.3000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El-Brolosy MA, et al. Genetic compensation triggered by mutant mRNA degradation. Nature. 2019;568:193–197. doi: 10.1038/s41586-019-1064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma Z, et al. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature. 2019;568:259–263. doi: 10.1038/s41586-019-1057-y. [DOI] [PubMed] [Google Scholar]
- 50.Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16:665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 51.Haimovich G, et al. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 2013;153:1000–1011. doi: 10.1016/j.cell.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 52.Serobyan V, et al. Transcriptional adaptation in Caenorhabditis elegans. eLife. 2020;9:e50014. doi: 10.7554/eLife.50014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang H, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wienert B, et al. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science. 2019;364:286–289. doi: 10.1126/science.aav9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang PY, et al. Lgr6 is a stem cell marker in mouse skin squamous cell carcinoma. Nat Genet. 2017;49:1624–1632. doi: 10.1038/ng.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ye D, et al. Marcksb plays a key role in the secretory pathway of zebrafish Bmp2b. PLoS Genet. 2019;15:e1008306. doi: 10.1371/journal.pgen.1008306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang L, et al. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol Cell. 2011;43:950–961. doi: 10.1016/j.molcel.2011.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nguyen LS, et al. Nonsense-mediated mRNA decay: inter-individual variability and human disease. Neurosci Biobehav Rev. 2014;46:175–186. doi: 10.1016/j.neubiorev.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lindeboom R, et al. The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy. Nat Genet. 2019;51:1645–1651. doi: 10.1038/s41588-019-0517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Son HG, et al. RNA surveillance via nonsense-mediated mRNA decay is crucial for longevity in daf-2/insulin/IGF-1 mutant C. elegans. Nat Commun. 2017;8:14749. doi: 10.1038/ncomms14749. [DOI] [PMC free article] [PubMed] [Google Scholar]