Abstract
Giant unilamellar vesicles (GUVs) are considered as the gold standard for assembling artificial cells from the bottom-up. In this study, we investigate the behavior of such biomimetic vesicles as they are subjected to mechanical compression. A novel microfluidic device is presented that comprises a trap to capture GUVs and a micro-stamp that is deflected downwards to mechanically compress the trapped vesicle. After characterization of the device, we show that single-phase GUVs can be controllably compressed to a high degree of deformation (D = 0.40) depending on the pressure applied to the micro-stamp. A permeation assay is implemented to show that vesicle bursting is prevented due to water efflux. Next, we mechanically compress GUVs with co-existing liquid-ordered and liquid-disordered membrane phases. Upon compression, we observe that the normally stable lipid domains reorganize themselves across the surface and fuse into larger domains. This phenomenon observed here in a model membrane system not only gives us insights into how multi-component membranes of artificial cells behave, but these observations may also have interesting consequences for the role of lipid rafts in biological cells that are subjected to compressive forces in a natural environment.
Keywords: artificial cells, giant unilamellar vesicles, microfluidics, mechanobiology, membrane domains, biomimetic, lipid rafts, bottom-up, synthetic biology
Introduction
In the quest for designing and realizing a fully functional minimal cell, there are two current approaches; the more known ‘top-down’ method where organisms are genetically modified, and the emerging field of ‘bottom-up’ synthetic biology where artificial cells are built-up from non-living components [1,2]. The latter approach has the potential to not only construct a minimal cell, but to go beyond our fundamental understanding of how cellular life exists as the completely artificial cells can be re-imagined to operate in a new manner not seen in nature and are therefore truly synthetic biology. However, suitable candidate materials must first be developed and in additional, the tools for handling them. Here we have chosen giant unilamellar vesicles (GUVs) as our artificial cells as they are considered the gold standard for cell models owning to their similar size to real cells and their membrane biocompatibility [3]. As part of normal physiological conditions, cells are subjected to external forces such as fluid shear stress osmotic pressure, gravity, surface topologies and mechanical loads.[4–7] It is well-known that these biomechanical forces can provoke responses in mechano-sensitive cells.[8,9] These can be immediate short term responses such as a change of membrane morphology[10] or membrane receptor activity,[11] but can also be long term effects such as cytoskeleton reorganization[7,12,13] or altered gene expression levels.[14]
When a eukaryotic cell is subjected to a mechanical load, the plasma membrane is the primary point of contact and the immediate result is a morphological deformation. Such prolonged forces may also result in a diffusion-based molecular reorganization (lipids, proteins, etc.) which occurs on a slower time-scale. Membrane proteins are a crucial part of the cell’s response to external stimuli, and in recent years so called ‘lipid rafts’ have been thought to play an important role in their spatial organization.[15–17] In the case of cell deformation, lipid rafts may be affected by changes in membrane curvature and surface tension which may in turn have consequences for membrane proteins and the resulting cellular response. In addition, understanding how membrane curvature couples to lipid composition is vital to understanding how cellular organelles are able to form complex structures.[18,19] Subjecting cells to mechanical forces is non-trivial, not only due to the difficulty in maintaining viability but also due to interpretation of data which may suffer interference from unknown sources [5,20]. Synthetic membranes systems such as giant unilamellar vesicles (GUVs) offer a reduced cell model, whereby individual membrane components can be isolated and studied without interference from the complexity of the natural cell. The simplest way to apply a force to a GUV is to unbalance the osmotic pressure between the lumen and the external solution. The resulting osmotic gradient can swell or deflate a vesicle which can change the membrane tension and therefore induce to lipid sorting and morphological changes.[21,22] Micropipette aspiration is a common technique used to spatially confine and conduct experiments on single giant vesicles. The method can be used to study the miscibility temperature dependence on membrane tension,[23] line-tensions,[24] bending moduli,[25] and can be combined with optically trapped beads to create membrane nanotubes for investigating membrane curvature.[26] Such nanotubes are an effective way of studying curvature driven lipid sorting or curvature sensing proteins.[27,28] This can either be performed by introducing membrane proteins to pre-existing nanotubes to observe binding[28,29] or by dynamically pulling nanotubes and observing the subsequent molecular reorganization.[26,27].
Hydrodynamic shear stress can also be used to deform giant vesicles either by using a counter-rotating Couette apparatus,[30] microfluidic channels[31–33] or by passing vesicles through channels of different cross-sectional areas.[34,35] Recently, we demonstrated that GUVs confined in microfluidic traps can be compressed by hydrodynamic forces.[33] However, the degree of deformation which can be achieved using hydrodynamic forces is limited and also results in unwanted shear force disruptions to the membrane.
Compression between two parallel plates has the advantage that the vesicle is free to expand (at least perpendicular to the applied forces) and as such, any changes to the membrane can easily be observed. In the literature, there are few experimental reports on GUVs subjected to parallel-plate compression. In 1981, Kwok and Evans investigated the effects of compressing large lecithin bilayer vesicles between a micro-pipette and a flat substrate to measure membrane thermoelastic properties.[36] Since then, theoretical approaches have been used to model liposomes compressed between plates,[37–39] and experimentally a tipless cantilever has been implemented by the Janshoff group to measure the mechanical response of GUVs compressed between parallel-plates.[40] Later, the same group used an AFM tip to apply indentations in GUVs.[41] Other methods to induce morphological changes in giant vesicles include contact induced vesicle deformation,[37,42,43] high-pressures,[44] dual optical beams,[45–47] optically trapped beads,[48] aqueous wetting,[49] light irradiation,[50–52] magnetic fields[53] and electric fields.[54–56]
Here, we report a microfluidic device with a novel integrated micro-stamp that is able to controllably compress and deform GUVs - a model cell system. First, we characterize the deflection of the micro-stamp as a function of externally applied pressure. Next, we apply the micro-stamp to pure 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) lipid GUVs to demonstrate the capability of the device to capture single vesicles and deform them. Finally, we introduce GUVs consisting of a ternary mixture of sphingomyelin (SM), DOPC, and cholesterol (Chol) that exhibit co-existing liquid ordered (lo) and disordered (ld) domains at room temperature. Upon compression, we observe induced reorganization and fusion of the lipid domains.
Results and Discussion
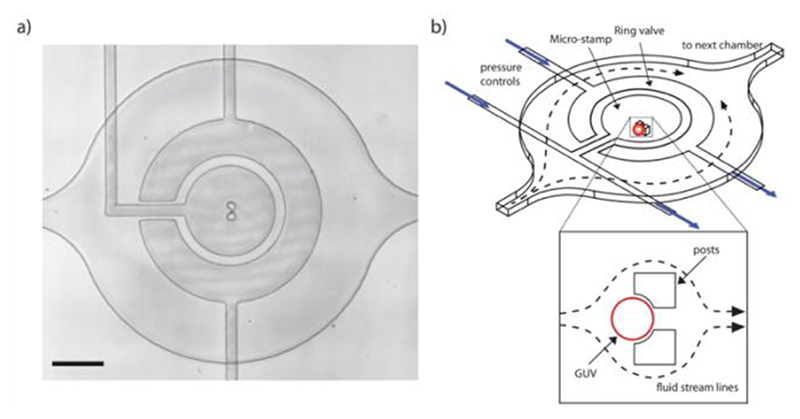
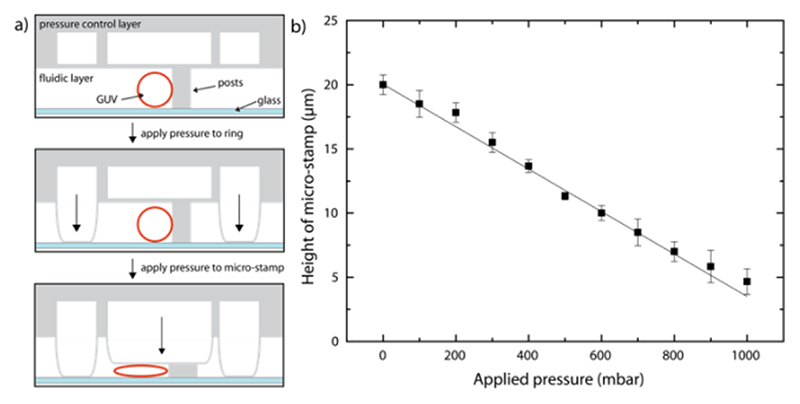
Microfluidic device design & micro-stamp function. The device was fabricated using PDMS and consists of two layers containing microfluidic channels. The lower fluidic layer is designed to contain the GUVs while the upper layer serves as a control layer for the micro-stamps and ring-valves. There is an array of 60 chambers in the lower layer each containing a pair of PDMS posts designed to trap a single vesicle underneath a micro-stamp (Figure 1 & and Figure S1). A ring-valve and a micro-stamp are positioned above each chamber (within the upper layer). Once actuated with 3 bars of pressure, the purpose of the ring-valve (Figure 2a) is to block unwanted flow at the posts and therefore prevent shear stress on the vesicle membrane. The micro-stamp can then be controllably deflected (0 – 1000 mbar) to deform the GUVs as shown schematically in Figure 2a. In order to calibrate the movement of the micro-stamp, the device was filled with calcein (0.1 mM) in water and the applied pressure was increased up to 1000 mbar in 100 mbar steps. The deflection of the micro-stamp (at the center of the chamber) was analyzed using confocal microscopy (Figure S2) and the resulting linear relationship (Figure 2b) demonstrates the controllability and reproducibility of the device. This ensures the same GUV compression is reached for each specific pressure. Note that the flexible PDMS posts of the trap are also compressed by the micro-stamp preventing it from reaching the glass surface but still allowing the GUVs to be deformed to a height of 4.7 ± 1.0 μm.
Figure 1. Design of the microfluidic device.
a) Bright-field image of a single chamber showing alignment of the micro-stamp above the trapping posts. Scale bar: 100 μm. b) Schematic showing a single chamber with a ring-valve and a micro-stamp. Insert: Once a GUV is trapped the fluid flow is diverted around the posts.
Figure 2. Operation of the device.
a) Side view (x-z) schematic showing how the control layer, situated above the fluidic layer, can be pressurized to lower the ring-valve or micro-stamps. Top: A single GUV is trapped at the posts. Middle: The ring-valve is actuated. Bottom: The micro-stamp is lowered to compress the GUV. b) The micro-stamp height can be precisely controlled by increasing the applied pressure in the control layer. The height of the micro-stamp above the glass coverslip is measured in front of the posts in the center where GUVs are captured. Note that the center of the stamp is prevented from making contact with the glass due to the presence of the PDMS posts. Averages are from three different chambers and errors bars taken from the standard deviation.
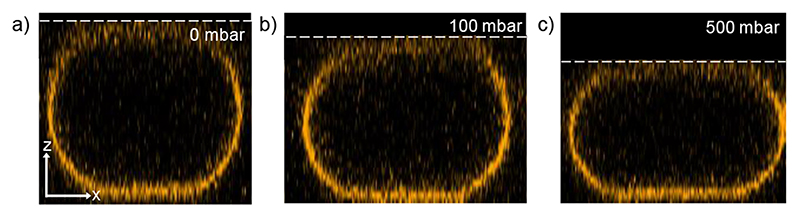
Compression of single-phase GUVs. To demonstrate the ability of the device to deform GUVs, vesicles composed of DOPC (with 0.1 mol% DiI) were introduced into the device (see Methods). First the ring-valve is actuated to eliminate shear stress and then the micro-stamp is deflected to compress the GUV. Figure 3 shows a side view of GUV (reconstructed from confocal optical slices) acquired without (0 bar) and with compression (0.5 bar) from the micro-stamp. This revealed the natural spherical shape becomes an oblate truncated spheroid under the micro-stamp. After the ring-valve is actuated and the flow is stopped, the vesicles are no longer in contact with the PDMS posts (not visible). To assess the compression, we define the degree of deformation (D) using Equation 1, [37]
| (1) |
where W and H are the diameter and height of the vesicle respectively.
Figure 3. Confocal renderings of a pure DOPC GUV under compression.
Side view (x-z) with micro-stamp pressures of a) 0 mbar (D = 0.13), b) 100 mbar (D = 0.28) and x) 500 mbar (D = 0.40). Scale bar arrows: 5 μm.
In this representative example, D was initially 0.13 due to the vesicle’s spherical diameter being slightly larger than the 20 μm channel height. After 0.5 bar was applied, D increased to 0.40. This is comparable to other methods of deforming GUVs such as optical stretching[46] or electro-deformation[57] which can achieve D~0.17 and ~0.33 respectively. Vesicles which had an initial diameter close to that of the channel height showed similar results. Here the compressed GUVs were prepared without buffers to ensure that water can permeate across the membrane [58]. However, when vesicles were prepared in the presence of a sugar solution (900 mM sucrose), they burst after a few minutes of compression (Figure S3). This is because the sugar osmotically balances the pressure across the membrane and prohibits water permeation. As the GUVs become more truncated with compression, the surface to volume ratio increases. Because the volume is held constant, the membrane is stretched beyond the maximum limit of 3-5% area increase and leads to vesicle rupture [36,59,60]. This highlights the need to exclude sugar solutions (typically used for GUV preparations) to allow water permeation when deforming GUVs to such high degrees.
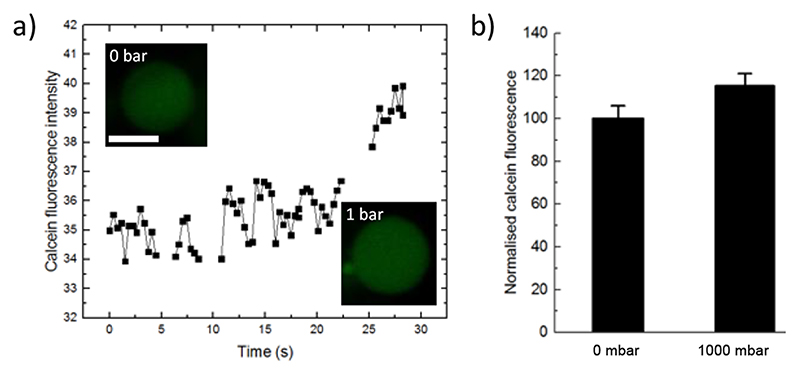
A permeation assay was performed to further investigate the flow of water across the membrane. Pure DOPC GUVs are prepared in 20 μM calcein, trapped in the posts and then flushed with water to remove external calcein. The ring-valve was lowered and a confocal time series was acquired. The data shown in Figure 4 shows the internal calcein fluorescence intensity of an exemplary GUV as it was compressed by the stamp resulting in ~11% increase in diameter. Here, the average fluorescence intensity of encapsulated calcein dye increased by 15.3 ± 5.8 % revealing a volume decrease due to the efflux of water and concentration of the dye. This data agrees with the above and shows that water is forced out of the GUVs to prevent the membrane surface area exceeding an increase of 3-5 %.[36,59]
Figure 4. Permeation assay.
a) Plot of the internal calcein fluorescence intensity of an exemplary GUV as it is compressed by the stamp (1000 mbar). Note that gaps in the data are when the GUV had to be re-centred in relation to the objective focal plane. Inserts show confocal images before and after compression. Scale bar: 5 μm. b) The average calcein fluorescence was measured before and 3 min after compression (n=6), and mean intensity increase was 15.3 ± 5.8 %. Error bars are taken from the standard deviations.
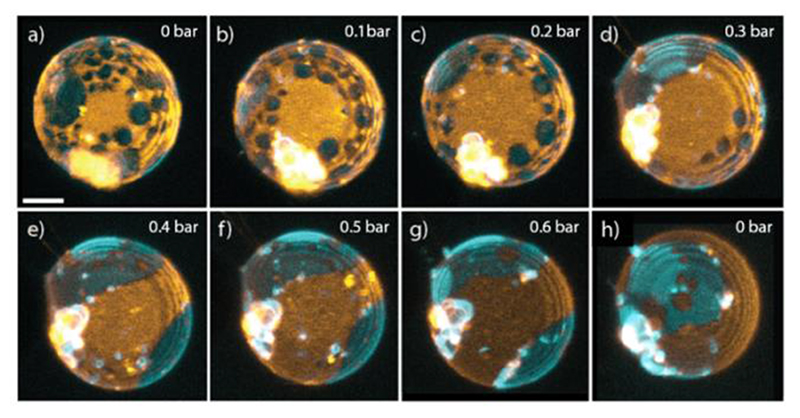
Compression induced domain fusion. To study the effects of compression on membrane phase-separated domains, we used GUVs with a lipid composition of SM/DOPC/Chol 4:4:2 which lies inside the lo/ld fluid co-existence region.[61–63] This would allow us to observe any changes in the lateral lipid organization that may occur as a result of compression. We included 0.1 mol% of the fluorescent dyes NAP and DiI which partition into the liquid ordered (cholesterol-rich) and liquid disordered phases respectively and therefore allow visualization of membrane domains. Confocal images were acquired with increasing amounts of compression as well as after releasing the pressure. Figure 5 shows 3-D renderings of a typical phase-separated vesicle under compression. In most cases, when a high degree of compression was achieved, the appearance of the domains changed as the lipids re-organized across the surface. For the GUV shown in Figure 5, the circular lo domains, which constituted the minority phase (in terms of area fraction), appeared to migrate to the vesicle equator and fuse together into larger patches.
Figure 5. Compression dependent domain fusion of a phase-separated GUVs, SM/DOPC/Chol 4:4:2.
a) to g) 3-D renderings when increasing the applied pressure from 0 mbar to 600 mbar. h) Releasing the pressure and returning the GUV to its original spherical shape. Scale bar: 5 μm. DiI (orange) and NAP (cyan) partition into the ld and lo phases respectively. Note that from from g) to h) the vesicle has undergone a rotation.
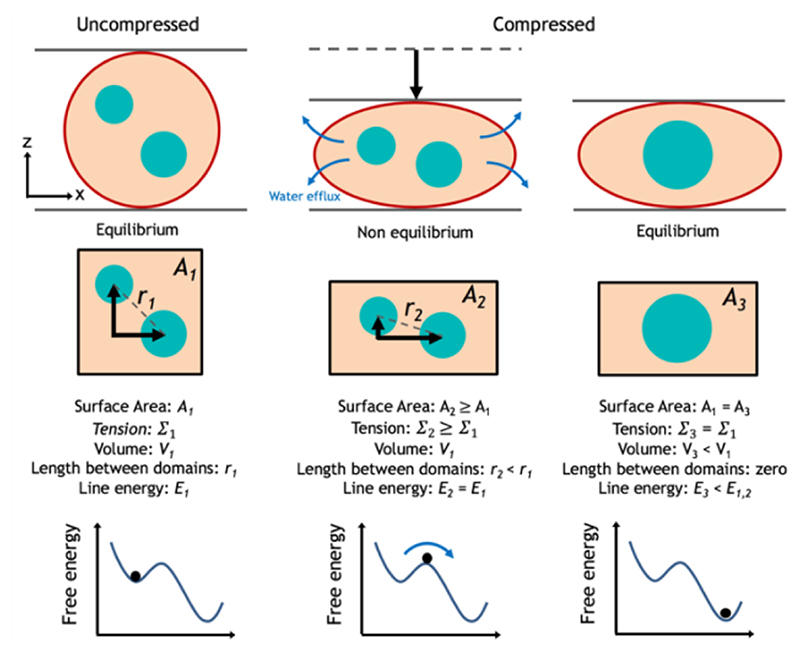
We frequently observed domain fusion with compression and in order to explain this we must first consider the geometry of the vesicles. Under this mechanical force they experience a compression in one axial dimension and expansion in the other. As we do not observe rupture, we again assume that the total surface area can only expand by a maximum of 3-5%. Therefore, they undergo this extension and compression at a near constant area [64]. This unidirectional compression favors re-distribution of the fluidic membrane (i.e. via mass transport) reducing the distance between a population of the circular domains and increasing the probability of contact and fusion (see Figure 6 for a schematic description). We must also consider the vesicles in terms of their free energy which is the sum of membrane tension, bending energy, and the interfacial energy of the line tension.[65,66] Let us first consider the relationship between membrane tension Σ and surface area A given by,
| (2) |
where k_A is the compressibility modulus and A_0 is the initial area. Here, an initially spherical GUV has zero membrane tension [36,67] but there must be a short period (tens of seconds [59]) in which the area and tension increases before the water finishes permeating across the membrane and the internal and external hydrostatic pressures reach equilibrium (Figure 6). A change in lateral membrane tension can result in a change in the critical temperature and hence line tension between domains.[23,68–71] In response, there are two ways in which the total global free energy can be reduced again: fusion or budding of domains. [66,72,73] In both cases the perimeter interface between the two phases is minimized and the total line energy and hence global energy is reduced. Our observations of domain fusion can therefore also be understood if the transient tension increase induced by compression increases the critical temperature which implies an increase in line tension. Such an increase in line tension therefore favors fusion of domains and reduces the total interfacial line energy. It should be noted that lo and ld domains are known to have different bending rigidities but these differences are not dominant in determining the global vesicle shapes.[74] Contributions from bending energies are expected to be small compared to the cost of increasing the membrane tension or interfacial line energy. Indeed, we also observe the migration of ld domains to the equatorial region in other examples (see Figure S4). This can be explained due to the geometry of the system and the protrusion of the budding domains. At the micro-stamp and glass coverslip, the membrane is unnaturally flattened and therefore prohibits the formation of buds from the vesicle surface. Domains which are in the minority (in terms of area fraction) will prefer to bud and therefore migrate to the equatorial regions (more visible in Figure S4). Due to population heterogeneities, vesicles produced with the same lipid mixtures can have either lo or ld minority domains and explains why we observe different equatorial migrations for different GUVs. We note that normal lipid movement can be affected when in contact with a surface as previously observed, [33] but here this is minimized by the use of surface passivation by BSA coating. Any small unwanted surface effects may have prevented fusion of domains in the contact zones.
Figure 6. Description of domain fusion as a result of compression.
The uncompressed GUV is at equilibrium with zero membrane tension (Σ). As the compression is initially applied, the area per lipid and lateral membrane tension increase temporally. As the volume (V) is reduced, water is forced out of the vesicle until it reaches equilibrium in the final state and the tension returns to zero. While the final vesicle surface area (A) has not changed compared to the uncompressed state, the domains are now more restricted in one axial dimension due to the new geometry of the compressed oblate vesicle causing a reduction in the inter-domain distance (r) for a population. Moreover, during the transient non-equilibrium intermediate state, the increased membrane tension favours domain fusion in order to decrease the total line energy (E) and lower the total free energy.
In a unique event, we performed the compression on a phase-separated vesicle which was already partially deformed when it was flushed into the microfluidic channels (due to its large spherical diameter > 20 μm). Surprisingly, smaller ld domains began bud and finally completely detached from the main GUV with increasing compression (Figure S5). Both of these events are consistent with our observations of domain fusion as they all serve to reduce the total interfacial line energy of the system.[66,67,72,74–76]
Finally, when the pressure was released from the micro-stamp the vesicles returned to their original spherical shape but the original domain appearance was not recovered (Figure 4h). Fused domains are more energetically favorable due to the reduction in interfacial line energy (E) which serves to keep the vesicle in a lower global energetic state (Figure 6). Spontaneous fusion of the small ld domains in Figure 4a was mostly likely prevented due to ‘trapped coarsening’[77] which keeps the vesicle in a metastable state that can persist for days.[21] A control experiment was performed, where confocal stacks were recorded at various time intervals (3 – 6 min) of an uncompressed phase-separated vesicle inside a device (see Figure S6). The data shows that no re-organization of the domains occurred and that the observations presented here are the result of compression and not, for example, an imaging artifact.[21] It should be noted that reorganization of lipid domains has been observed when GUVs adhere to a substrate[78] or to each other,[79] but here the surfaces of the microfluidic device are coated with BSA to prevent any contribution from adhesive forces. Changes in osmotic pressures,[21] buffer asymmetry,[80] and protein binding[81] can also affect the appearance of domains, but the external solution in our setup remains constant.
Conclusions
We have presented a novel approach to investigate mechanical compression on model cells using a microfluidic device with an integrated micro-stamp. Our method allows for observations of vesicles far from their natural spherical equilibrium state, i.e. a large degree of deformation. We applied the micro-stamp to single component vesicles and demonstrated that water efflux prevents membrane rupture by reducing the internal volume. When multicomponent phase separated GUVs were compressed, we observed the reorganization and subsequent fusion of membrane domains. A unidirectional compression causes lipid transport which reduces inter-domain distances and promotes an increase in the probability of domain contact and fusion. The findings can also be understood by considering that the total global energy is altered by a temporary increase in the lateral membrane tension (before water efflux and equilibrium). Chen and Santore also note the near-zero tension of vesicles without osmotic balance in pure water [58]. Domain fusion therefore serves to reduce to total energy of the system by minimizing the interfacial line energies. To investigate these effects further, it would be desirable to develop a calibration system that enables the determination of the final applied force imposed on the vesicles by the micro-stamp. Moreover, we hope that researchers will implement simulations of the dynamic compression induced by our system to gain further insights into the relationship between compression, membrane tension, and domain organization.
Our method has several advantages but the main being that due to the mechanical trapping of the vesicles there is no need to adhere them to a surface which is often done with cells [20]. Adhesion of GUVs to substrate can introduce unwanted and uncontrolled membrane tensions which can lead to alterations of domain patterns [82]. This makes our device universally applicable for any soft object (water in oils droplets, aqueous droplets, cells, lipid vesicles, polymersomes, proteinosomes, etc.) in which compressive forces should be applied.
Here we applied the micro-stamp to a biomimetic membrane system, but the results could have a wider implication for how biological cells respond to compressive mechanical forces. By observing vesicles exhibiting micrometer sized membrane domains, we can speculate about the role of so called ‘lipid rafts’ in cell membranes under compression. We have shown that compression has a significant effect on phase-separated membranes, demonstrating the fuse of domains as a direct result. This behavior would be challenging to observe in cellular membranes, but if lipid rafts were to exist, one could imagine that the dramatic changes observed in our model system might have implications in real cells. In particular, lipid rafts are thought to play a role in membrane protein sorting. Therefore, compressive mechanical forces on cells may result in membrane protein organization via remodeling of the lipids themselves. Interestingly, the formation of cholesterol-rich domains was recently observed in red blood cells upon mechanical deformation[20].
Finally, recent interest in the development of minimal cells using the bottom-up approach will inevitably lead to the construction of minimal tissues systems [1]. Such assemblies are expected to shed light on the emergence and function of early proto-tissue systems (or the first multicellular organisms or colonies of cells). While some reports have focused on aggregation [83] of or biomolecular transfer [84] between collections of GUVs, no work as ever investigated how environmental factors effects such systems. External triggers in the early Earth will have surely been essential in the evolution of proto-cell behavior. For this reason it will be interesting to observe how multiple vesicle systems respond to external factors. Not only compression as shown here on single artificial cells, but with osmotic gradients, changes in heat, and also applied shear stresses. We are therefore developing systems to model proto-tissues using high capacity microfluidic vesicle capture with this future goal in mind [85,86].
Experimental Section
Chemicals. Naphtho(2,3-a)pyrene (NAP) was purchased from TCI (Germany). 1,1’-Dioctadecyl-3,3,3’,3’-Tetramethylindocarbocyanine Perchlorate (DiI) was obtained from Invitrogen. Sphingomyelin (egg chicken, SM), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and cholesterol (Chol) were purchased from Avanti Polar Lipids (Alabama, USA). Methanol and chloroform were from Acros Organics. PDMS and curing agent (Sylgard 184) were purchased from Dow Corning (MI, USA). SU-8 2015 photoresist was obtained from Microchem Corp (MA, USA). Bovine serum albumin (BSA), sucrose and calcein were purchased from Sigma Aldrich. mr-Dev 600 developer was obtained from micro resist technology (Germany). 1H,1H,2H,2H-Perfluorodecyltrichlorosilane was purchased from ABCR-Chemicals (Germany).
Microfluidic device fabrication. Standard 100 mm diameter silicon wafers (Si-Mat, Germany) were used to fabricate the master molds. First, the wafers were dehydrated for 10 min at 200 °C and then spin coated with SU-8 2015 for 30 s at 2100 rpm. Afterwards, a soft bake was performed for 240 s 95°C, then the wafers were exposed with 150 mJ/cm2 at 365 nm on a MA 6 mask aligner (Süss MicroTec, Germany) through a transparency photomask (Micro Lithography Services, UK). After a post exposure bake was conducted at 95 °C for 300 s, the wafers were developed using mr-Dev 600 developer for 4 min. Finally, a hard bake was performed for at least 3 hr at 200°C. To prevent adhesion of the PDMS to the wafer surface, they were treated with a 1H,1H,2H,2H-Perfluorodecyltrichlorosilane atmosphere at 100 mbar for 12 hr. The PDMS part of the device consists of a lower fluidic layer and an upper pressure control layer. Oligomer and curing agent were mixed in a ratio of 10:1. For the upper layer (feature height 20 μm), the PDMS mixture was poured to a height of 5 mm and cured for 3 h at 80 °C. Holes for the pressured nitrogen were punched using a 1 mm outer diameter biopsy puncher (Miltex, PA, USA). For the lower layer (feature height 20 μm), the mixture was spin-coated to a height of 40 μm at 2000 rpm and cured for 1 h at 80 °C. Both layers were treated with an air plasma in a custom-built chamber and bonded together under a microscope for alignment of upper and lower channels. After heating the two bonded parts for 2 hr at 80 °C, fluidic access holes were punched with a 1.5 mm outer diameter biopsy puncher (Miltex, PA, USA). The final device was completed by bonding the PDMS section to a glass coverslip using a plasma cleaner (PDC-32G, Harrick, NY, USA).
GUV preparation. GUVs exhibiting domains were prepared by electroformation in a custom-built temperature controlled chamber. Specific ratios of SM, DOPC and cholesterol were dissolved in chloroform/methanol (9:1 v/v) at a concentration of 1 mM with 1 μM DiI and NAP. Approximately 2 μl of the mixture was deposited onto a conductive indium tin oxide (ITO) slide (15-25 Ω/sq, Sigma-Aldrich) in 12 different locations (for multiple lipid ratios) separated by a 1.5 mm thick silicone rubber spacer and dried in a vacuum overnight. Millipore filtered water was then used to hydrate the lipid films and the 12 chambers were sealed using a second ITO coated slide. A custom-built heating device set the chamber temperature to 60°C, and 1 V at 10 Hz was applied for 4 h using a function generator (HMF2525, Hameg) to form GUVs. The voltage was turned off and the device was cooled to room temperature. GUVs were subsequently detached from the ITO surface by applying 1.5 V at 3 Hz for 30 min, harvested by gentle pipetting of the solution, and used on the same day.
Chip preparation and operation. To prevent adhesion of the vesicle to the PDMS or glass surfaces, 2% (w/v) BSA solution in Millipore filtered water was centrifuged into the fluidic layer and incubated for 1 h. At the same time, Millipore filtered water was centrifuged into the upper layer. Centrifugation ensures that fluids fill the entire device and no air pockets remain trapped in the micro-channels. A syringe-pump (neMESYS, Cetoni, Germany), was subsequently used to exchange the BSA solution in the lower layer with Millipore filtered water (20 μl/min). The same pump was used to draw the GUV solution through the fluid channels during the experiments (10 μl/min). All experiments were performed at room temperature (22 - 23 ⍰C). Once more than 55 out of 60 traps were filled with GUVs, the ring-valves in the upper layer were actuated by applying 3 bar of pressure (using an in-house nitrogen line) using a custom-built pressure control instrument. The hydraulic pressure deforms the 20 μm of PDMS between the two layers so that it makes contact with the glass bottom to form an open ring shape around the posts and GUV. The purpose of which is to prevent shear stress during the experiments. Additionally, a time period of 30 min is given before performing the experiments to ensure the domains are stable. Deformation of the GUVs was achieved in a similar manner by applying 0 to 1000 mbar to the micro-stamp in the upper layer using a pressure control system (MFCS-FLEX, Fluigent, France). Typical steps of 100 - 200 mbar were implemented with time intervals of 3 - 4 mins between steps. The final assembled device with connected tubing can be found in Figure S1b.
Microscopy. Wide-field microscopy was achieved using an inverted microscope (IX70, Olympus, Japan) with an EMCCD camera (iXon DV887, Andor, Germany). GUVs were imaged by optically sectioned microscopy using a confocal laser scanning microscope (Axiovert 200M, Zeiss, Germany) with a 63x/1.4 NA oil immersion objective lens. NAP fluorescence was recorded using a 458 nm Argon ion laser line, an HFT 458/561 dichroic, and a 505-550 nm band-pass filter. DiI fluorescence was recorded using a 561nm-diode laser, an HFT 458/561 dichroic, and a 575 nm long-pass filter.
Image analysis. Image analysis and 3-D rendering of confocal data sets were performed using Zen (Zeiss, Germany). To account for sample drift during image acquisition, alignment of the z-stacks was achieved using AutoAligner (Bitplane AG, Switzerland). Wide-field images were processed using ImageJ.
Supplementary Material
Acknowledgements
Thanks to Reinhard Lipowsky, Jaime Agudo-Canalejo, Jan Steinkühler, and Tripta Bhatia for fruitful discussions. Financial support by the European Research Commission (ERC Consolidator Grant No. 681587 “HybCell”) is gratefully acknowledged. TR acknowledges support from the MaxSynBio consortium, which is jointly funded by the Federal Ministry of Education and Research of Germany and the Max Planck Society.
References
- [1].Schwille P, Spatz J, Landfester K, Bodenschatz E, Herminghaus S, Sourjik V, Erb TJ, Bastiaens P, Lipowsky R, Hyman A, et al. Angew Chemie Int Ed. 2018;57:13382–13392. doi: 10.1002/anie.201802288. [DOI] [PubMed] [Google Scholar]
- [2].Göpfrich K, Platzman I, Spatz JP. Trends Biotechnol. 2018;36:938–951. doi: 10.1016/j.tibtech.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Walde P, Cosentino K, Engel H, Stano P. ChemBioChem. 2010;11:848–865. doi: 10.1002/cbic.201000010. [DOI] [PubMed] [Google Scholar]
- [4].Discher DE, Janmey P, Wang Y-L. Science. 2005;310:1139–43. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- [5].Kurth F, Eyer K, Franco-Obregón A, Dittrich PS. Curr Opin Chem Biol. 2012;16:400–408. doi: 10.1016/j.cbpa.2012.03.014. [DOI] [PubMed] [Google Scholar]
- [6].Hamill OP, Martinac B. Physiol Rev. 2001;81:685–740. doi: 10.1152/physrev.2001.81.2.685. [DOI] [PubMed] [Google Scholar]
- [7].Fletcher DA, Mullins RD. Nature. 2010;463:485–92. doi: 10.1038/nature08908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Janmey PA, McCulloch CA. Annu Rev Biomed Eng. 2007;9:1–34. doi: 10.1146/annurev.bioeng.9.060906.151927. [DOI] [PubMed] [Google Scholar]
- [9].Schaffhauser DS, Andrini O, Ghezzi C, Schaffner M, Forster IC, Egli M. Methods. 2011:320–322. [Google Scholar]
- [10].Vogel V, Sheetz M. Nat Rev Mol Cell Biol. 2006;7:265–75. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- [11].Árnadóttir J, Chalfie M, Arnadóttir J, Chalfie M. Annu Rev Biophys. 2010;39:111–137. doi: 10.1146/annurev.biophys.37.032807.125836. [DOI] [PubMed] [Google Scholar]
- [12].Loufrani L, Henrion D. Med Biol Eng Comput. 2008;46:451–60. doi: 10.1007/s11517-008-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pollard TD, Cooper JA. Science. 2009;326:1208–12. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chan MWC, Hinz B, McCulloch CA. Mechanical Induction of Gene Expression in Connective Tissue Cells. 2010 doi: 10.1016/S0091-679X(10)98008-4. [DOI] [PubMed] [Google Scholar]
- [15].Lingwood D, Simons K. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- [16].Simons K, Ikonen E. Nature. 1997;387:569–72. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- [17].Brown DA, London E. Biochem Biophys Res Commun. 1997;240:1–7. doi: 10.1006/bbrc.1997.7575. [DOI] [PubMed] [Google Scholar]
- [18].Shibata Y, Hu J, Kozlov MM, Rapoport TA. Annu Rev Cell Dev Biol. 2009;25:329–354. doi: 10.1146/annurev.cellbio.042308.113324. [DOI] [PubMed] [Google Scholar]
- [19].van Meer G, Voelker DR, Feigenson GW. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Leonard C, Conrard L, Guthmann M, Pollet H, Carquin M, Vermylen C, Gailly P, Van Der Smissen P, Mingeot-Leclercq MP, Tyteca D. Sci Rep. 2017;7:4264. doi: 10.1038/s41598-017-04388-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Morales-Penningston NF, Wu J, Farkas ER, Goh SL, Konyakhina TM, Zheng JY, Webb WW, Feigenson GW. Biochim Biophys Acta - Biomembr. 2010;1798:1324–1332. doi: 10.1016/j.bbamem.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].la Oglecka K, Sanborn J, Parikh AN, Kraut RS. Front Physiol. 2012 MAY;3 doi: 10.3389/fphys.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Portet T, Gordon SE, Keller SL. Biophys J. 2012;103 doi: 10.1016/j.bpj.2012.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tian A, Johnson C, Wang W, Baumgart T. Phys Rev Lett. 2007;98 doi: 10.1103/PhysRevLett.98.208102. [DOI] [PubMed] [Google Scholar]
- [25].Rawicz W, Olbrich KC, McIntosh T, Needham D, Evans E, Nagle JF. Biophys J. 2000;79:328–39. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Callan-Jones A, Sorre B, Bassereau P. Cold Spring Harb Perspect Biol. 2011;3:1–14. doi: 10.1101/cshperspect.a004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sorre B, Callan-Jones A, Manneville J-B, Nassoy P, Joanny J-F, Prost J, Goud B, Bassereau P. Proc Natl Acad Sci U S A. 2009;106:5622–6. doi: 10.1073/pnas.0811243106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sorre B, Callan-Jones A, Manzi J, Goud B, Prost J, Bassereau P, Roux A. Proc Natl Acad Sci U S A. 2012;109:173–178. doi: 10.1073/pnas.1103594108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Prévost C, Zhao H, Manzi J, Lemichez E, Lappalainen P, Callan-Jones A, Bassereau P. Nat Commun. 2015 Jun;:8529. doi: 10.1038/ncomms9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].de Haas KH, van den Ende D, Blom C, Altena EG, Beukema GJ, Mellema J. 1998. http://oasc12039.247realmedia.com/RealMedia/ads/click_lx.ads/www.aip.org/pt/adcenter/pdfcover_test/L-37/1156540762/x01/AIP-PT/MCL_RSIArticleDL_030817/RSI-Banner-March17.gif/434f71374e315a556e61414141774c75?x . [DOI]
- [31].Kantsler V, Segre E, Steinberg V. Phys Rev Lett. 2007;99 doi: 10.1103/PhysRevLett.99.178102. [DOI] [PubMed] [Google Scholar]
- [32].Deschamps J, Kantsler V, Segre E, Steinberg V. Proc Natl Acad Sci U S A. 2009;106:11444–7. doi: 10.1073/pnas.0902657106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sturzenegger F, Robinson T, Hess D, Dittrich PS. Soft Matter. 2016;12:5072–5076. doi: 10.1039/c6sm00049e. [DOI] [PubMed] [Google Scholar]
- [34].Noguchi H, Gompper G, Schmid L, Wixforth A, Franke T. Europhys Lett. 2010;89:1–6.:28002 [Google Scholar]
- [35].Coupier G, Farutin A, Minetti C, Podgorski T, Misbah C. Phys Rev Lett. 2012;108 doi: 10.1103/PhysRevLett.108.178106. [DOI] [PubMed] [Google Scholar]
- [36].Kwok R, Evans E. Biophys J. 1981;35:637–652. doi: 10.1016/S0006-3495(81)84817-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Foo JJ, Chan V, Liu KK. J Biomech. 2006;39:2338–2343. doi: 10.1016/j.jbiomech.2005.07.008. [DOI] [PubMed] [Google Scholar]
- [38].Parker KH, Winlove CP. Biophys J. 1999;77:3096–3107. doi: 10.1016/S0006-3495(99)77140-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Preston SP, Jensen OE, Richardson G, Mech QJ. Appl Math. 2008;61:1–24. [Google Scholar]
- [40].Schäfer E, Kliesch T-T, Janshoff A. Langmuir. 2013;29:10463–10474. doi: 10.1021/la401969t. [DOI] [PubMed] [Google Scholar]
- [41].Schäfer E, Vache M, Kliesch T-T, Janshoff A. Soft Matter. 2015;11:4487–4495. doi: 10.1039/c5sm00191a. [DOI] [PubMed] [Google Scholar]
- [42].Steinkühler J, Agudo-Canalejo J, Lipowsky R, Dimova R. Biophys J. 2016;111:1454–1464. doi: 10.1016/j.bpj.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Aoki PHB, Schroder AP, Constantino CJL, Marques CM. Soft Matter. 2015;11:5995–5998. doi: 10.1039/c5sm01019e. [DOI] [PubMed] [Google Scholar]
- [44].Nicolini C, Celli A, Gratton E, Winter R. Biophys J. 2006;91:2936–2942. doi: 10.1529/biophysj.106.088716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Solmaz ME, Biswas R, Sankhagowit S, Thompson JR, Mejia Ca, Malmstadt N, Povinelli ML. Biomed Opt Express. 2012;3:2419. doi: 10.1364/BOE.3.002419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Solmaz ME, Sankhagowit S, Biswas R, a Mejia C, Povinelli ML, Malmstadt N. RSC Adv. 2013;3:16632–16638. doi: 10.1039/C3RA42510J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Delabre U, Feld K, Crespo E, Whyte G, Sykes C, Seifert U, Guck J. Soft Matter. 2015;11:6075–6088. doi: 10.1039/c5sm00562k. [DOI] [PubMed] [Google Scholar]
- [48].Poole C, Losert W. Methods Mol Biol. 2007;400:389–404. doi: 10.1007/978-1-59745-519-0_26. [DOI] [PubMed] [Google Scholar]
- [49].Li Y, Kusumaatmaja H, Lipowsky R, Dimova R. J Phys Chem B. 2012;116:1819–1823. doi: 10.1021/jp211850t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Riske KA, Sudbrack TP, Archilha NL, Uchoa AF, Schroder AP, Marques CM, Baptista MS, Itri R. Biophys J. 2009;97:1362–1370. doi: 10.1016/j.bpj.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sankhagowit S, Wu SH, Biswas R, Riche CT, Povinelli ML, Malmstadt N. Biochim Biophys Acta-Biomembr. 2014;1838:2615–2624. doi: 10.1016/j.bbamem.2014.06.020. [DOI] [PubMed] [Google Scholar]
- [52].Brückner E, Sonntag AP, Rehage H. 2001 doi: 10.1021/LA000256I. [DOI] [Google Scholar]
- [53].Bacri J-C, Cabuil V, Cebers A, Menager C, Perzynski R. Eur Lett. 1996;33:235–240. [Google Scholar]
- [54].Winterhalter M, Helfrich W, Colloid J. Interface Sci. 1988;122:583–586. [Google Scholar]
- [55].Dimova R, Riske KA, Aranda S, Bezlyepkina N, Knorr RL, Lipowsky R. Soft Matter. 2007;3:817–827. doi: 10.1039/b703580b. [DOI] [PubMed] [Google Scholar]
- [56].Korlach J, Reichle C, Müller T, Schnelle T, Webb WW. Biophys J. 2005;89:554–562. doi: 10.1529/biophysj.104.050401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kummrow M, Helfrich W. Phys Rev A. 1991;44:8356–8360. doi: 10.1103/physreva.44.8356. [DOI] [PubMed] [Google Scholar]
- [58].Chen D, Santore MM. Langmuir. 2014;30:9484–9493. doi: 10.1021/la502089t. [DOI] [PubMed] [Google Scholar]
- [59].Olbrich K, Rawicz W, Needham D, Evans E. doi: 10.1016/S0006-3495(00)76294-1. n.d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Emami S, Su W-C, Purushothaman S, Ngassam VN, Parikh AN. Biophys J. 2018;115:1942–1955. doi: 10.1016/j.bpj.2018.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Veatch SL, Keller SL. Phys Rev Lett. 2005;94 doi: 10.1103/PhysRevLett.94.148101. [DOI] [PubMed] [Google Scholar]
- [62].Bezlyepkina N, Gracià RS, Shchelokovskyy P, Lipowsky R, Dimova R. Biophys J. 2013;104:1456–1464. doi: 10.1016/j.bpj.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ionova IV, Livshits VA, Marsh D. Biophys J. 2012;102:1856–65. doi: 10.1016/j.bpj.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Evans EA, Skalak R. Mechanics and Thermodynamics of Biomembranes. CRC Press; 2018. [Google Scholar]
- [65].Rim JE, Ursell TS, Phillips R, Klug WS. Phys Rev Lett. 2011;106:057801. doi: 10.1103/PhysRevLett.106.057801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Lipowsky R. J Phys II. 1992;2:1825–1840. [Google Scholar]
- [67].Sackmann E, Käs J, Rädler J. Phys Scr. 1993;T49A:111–118. [Google Scholar]
- [68].Akimov SA, Kuzmin PI, Zimmerberg J, Cohen FS. Phys Rev E. 2007;75:011919. doi: 10.1103/PhysRevE.75.011919. [DOI] [PubMed] [Google Scholar]
- [69].Lipowsky R, Rouhiparkouhi T, Discher DE, Weikl TR. doi: 10.1039/c3sm50712b. n.d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ayuyan AG, Cohen FS. Biophys J. 2008;94:2654–2666. doi: 10.1529/biophysj.107.118596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Hamada T, Kishimoto Y, Nagasaki T, Takagi M. Soft Matter. 2011;7:9061 [Google Scholar]
- [72].Jülicher F, Lipowsky R. Phys Rev Lett. 1993;70:2964–2967. doi: 10.1103/PhysRevLett.70.2964. [DOI] [PubMed] [Google Scholar]
- [73].Ursell TS, Klug WS, Phillips R. PNAS. 2009;106:13301–13306. doi: 10.1073/pnas.0903825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Baumgart T, Hess ST, Webb WW. Nature. 2003;425:821–824. doi: 10.1038/nature02013. [DOI] [PubMed] [Google Scholar]
- [75].Allain J-M, Ben Amar M. Eur Phys J E. 2006;20:409–420. doi: 10.1140/epje/i2006-10030-4. [DOI] [PubMed] [Google Scholar]
- [76].Döbereiner HG, Käs J, Noppl D, Sprenger I, Sackmann E. Biophys J. 1993;65:1396–403. doi: 10.1016/S0006-3495(93)81203-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Yanagisawa M, Imai M, Masui T, Komura S, Ohta T. Biophys J. 2007;92:115–125. doi: 10.1529/biophysj.106.087494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Sarmento MJ, Prieto M, Fernandes F. Biochim Biophys Acta-Biomembr. 2012;1818:2605–2615. doi: 10.1016/j.bbamem.2012.05.028. [DOI] [PubMed] [Google Scholar]
- [79].Gordon VD, Deserno M, Andrew CMJ, Egelhaaf SU, Poon WCK. EPL (Europhysics Lett) 2008;84:48003 [Google Scholar]
- [80].Kubsch B, Robinson T, Lipowsky R, Dimova R. Biophys J. 2016;110:2581–2584. doi: 10.1016/j.bpj.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Pataraia S, Liu Y, Lipowsky R, Dimova R. Biochim Biophys Acta - Biomembr. 2014;1838:2036–2045. doi: 10.1016/j.bbamem.2014.04.019. [DOI] [PubMed] [Google Scholar]
- [82].Sarmento MJ, Prieto M, Fernandes F. Biochim Biophys Acta - Biomembr. 2012;1818:2605–2615. doi: 10.1016/j.bbamem.2012.05.028. [DOI] [PubMed] [Google Scholar]
- [83].de Souza TP, Bossa GV, Stano P, Steiniger F, May S, Luisi PL, Fahr A. Phys Chem Chem Phys. 2017;19:20082–20092. doi: 10.1039/c7cp03751a. [DOI] [PubMed] [Google Scholar]
- [84].Litschel T, Ganzinger KA, Movinkel T, Heymann M, Robinson T, Mutschler H, Schwille P. New J Phys. 2018;20:055008 [Google Scholar]
- [85].Yandrapalli N, Robinson T. Lab Chip. 2019;19:626–633. doi: 10.1039/c8lc01275j. [DOI] [PubMed] [Google Scholar]
- [86].Robinson T. Adv Biosyst. 2019:1800318. doi: 10.1002/adbi.201800318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.