Abstract
Automated ultrasound (US)-probe movement guidance is desirable to assist inexperienced human operators during obstetric US scanning. In this paper, we present a new visual-assisted probe movement technique using automated landmark retrieval for assistive obstetric US scanning. In a first step, a set of landmarks is constructed uniformly around a virtual 3D fetal model. Then, during obstetric scanning, a deep neural network (DNN) model locates the nearest landmark through descriptor search between the current observation and landmarks. The global position cues are visualised in real-time on a monitor to assist the human operator in probe movement. A Transformer-VLAD network is proposed to learn a global descriptor to represent each US image. This method abandons the need for deep parameter regression to enhance the generalization ability of the network. To avoid prohibitively expensive human annotation, anchor-positive-negative US image-pairs are automatically constructed through a KD-tree search of 3D probe positions. This leads to an end-to-end network trained in a self-supervised way through contrastive learning.
Keywords: obstetric US, probe guidance, landmark retrieval
1. Introduction
Motivation
Obstetric US scanning is known to be highly experienced-operator dependent. Simplifying US to be more accessible to non-expert operators is a recognized priority for wider deployment of US in clinical practice. Automatic probe movement guidance may assist less-experienced operators to perform scanning more confidently, and widen the use of US in existing and new areas of clinical medicine. Our target is to develop automated machine learning (ML)-based visual interventions to provide helpful visualization cues for guiding an inexperienced operator using US scanning as an exemplar. In this case the target end-user might be a sonographer trainee, midwife, emergency medicine doctors, or primary care practitioners for instance.
Automatic ML-based probe movement guidance to assist a human operator (rather than a robot) is currently an open research problem. Two recent methods [6][3] propose to predict control parameters of probe movement such as translation distance and rotation degree. Li et al. [6] propose an Iterative Transformation Network (ITN) to automatically detect 2D standard planes from a pre-scanned 3D US volume. The CNN-based ITN learns to predict the parameters of the geometric transformation required to move the current plane towards the position/orientation of the 2D standard plane in the 3D volume. Droste et al. [3] develop a real-time probe rotation guidance algorithm using US images with Inertial Measurement Unit (IMU) signals for obstetric scanning. The proposed deep multi-modality model predicts both the rotation towards the standard plane position, and the next rotation that an expert operator might perform.
These control parameter prediction style methods are best suited for guiding a robot agent rather than a human. There is a parallel here with the self-driving vehicle literature where, for example, the most efficient way to assist a person driving is via real-time GPS localization visualization, while control parameter (steering wheel and accelerator) prediction is more useful for a self-driving car. We are therefore interested in discovering whether a similar visual intervention such as [11] can assist obstetric US scanning.
Grimwood et al. [5] formulate the probe guidance problem as a high-level command classification problem during prostate external beam radiotherapy using US images and transducer pose signals. The proposed CNN-RNN based classification network predictss 6 different high-level guidance cues i.e. outside prostate, prostate periphery, prostate centre for position and move left, move right, stop for direction to recommend probe adjustments. However, training this classification network requires a large number of expensive ground-truth annotated by physicists and radiotherapy practitioners.
From a technical viewpoint, deep regression-based methods such as [6][3] take advantage of the powerful non-linearity of a DNN to regress the control parameters from the collected data. These methods leverage the DNN to learn to overfit on the training data of some specific users, so these methods naturally lack generalization ability, as mentioned in [8], for real clinical applications.
Contribution
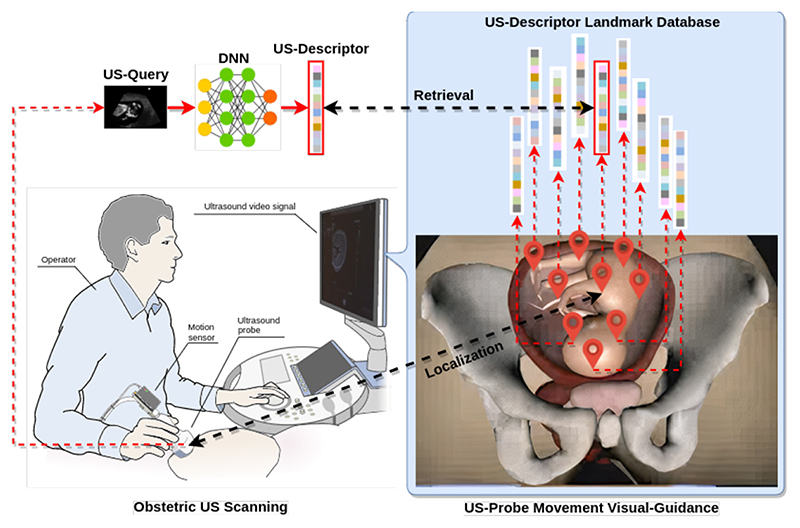
In this paper we propose a landmark retrieval-based method as a visual-assisted intervention to guide US-probe movement as shown in Fig.1. The goal is to provide global position visualization cues to the operator during US scanning. To be specific, we firstly construct a set of landmarks uniformly around a virtual fetal model. Each landmark stores a data-pair of information: the 3D position relative to the virtual fetal model, and the global descriptor of the US image captured at this position. During US scanning, the network transforms the current observed US image to a global descriptor, and then retrieves the landmark dataset to locate the nearest landmark through descriptor search. The nearest landmark provides the relative 3D position between the probe and the virtual fetal model in 3D space. This global position visualization is displayed on the monitor in real-time as visual guidance to assist the operator.
Fig. 1. Overview of landmark retrieval-based US-probe movement guidance.
This descriptor learning-based landmark retrieval method abandons any need for deep parameter regression, which can avoid the degeneration of network generalization ability. The proposed method is trained end-to-end in a self-supervised way without any expensive human expert annotation. The main contributions are: 1) we formulate US-probe movement guidance as a landmark retrieval problem through learned descriptor search; 2) a Transformer-VLAD network is proposed to learn a generalized descriptor for automatic landmark retrieval; 3) the descriptor learning is achieved by contrastive learning using self-constructed anchor-positive-negative US image-pairs.
2. Methodology
Overview
Building on the representation ability of DNN, we cast US-probe movement guidance as landmark retrieval. The query, i.e. current observed US image, at an unknown position is used to visually search a landmark dataset. The positions of top-ranked landmarks are used as suggestions for the query position. It is achieved by designing a DNN, i.e. Transformer-VLAD, to extract a global descriptor given an US image for visual search. During inference, only the descriptor of the query US image is computed online, while the other descriptors of the landmarks are computed once offline and stored in memory, thus enabling a real-time performance (0.01s on NVIDIA TITAN RTX). The visual search is performed by finding the nearest landmarks to the query. This can be achieved through fast approximate nearest neighbour search by sorting landmarks according to the Euclidean distance between the learned descriptors.
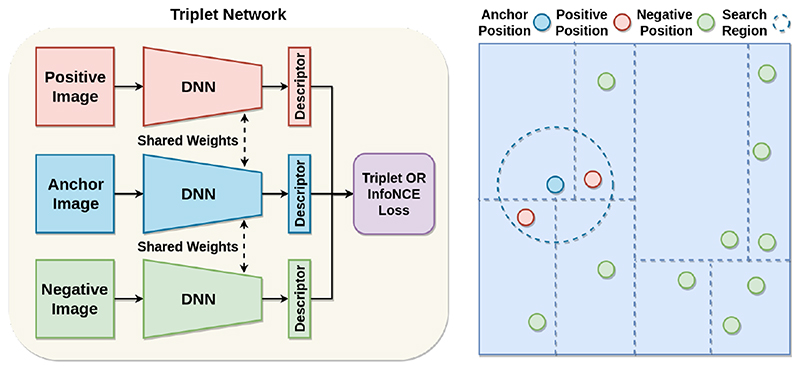
The proposed Transformer-VLAD network is a typical triplet network which is a variation of a Siamese network, as shown in Fig.2 left. It utilizes a triplet of images, including anchor, positive and negative US images in network training. The triplet network simultaneously minimizes the feature distance between the anchor and positive US image-pair, and maximizes the feature distance between the anchor and negative US image-pair through contrastive learning. The anchor-positive-negative triplet data is automatically constructed according to the KD-tree based probe 3D position without use of human annotation. Hence, the whole network is trained end-to-end in a self-supervised way.
Fig. 2. (left) The triplet network architecture. (right) The probe position KD-tree based anchor-positive-negative triplet data construction.
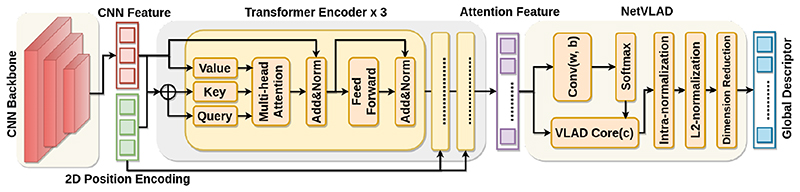
The more detailed architecture inside the Transformer-VLAD network is illustrated in Fig.3. It consists of three components: feature extraction (left), Transformer (middle) and NetVLAD (right). The feature extraction is composed of a series of convolution stacks to extract the local feature representation from the US image. The Transformer includes three transformer encoder stacks in series with 2D position encoding, enabling co-contextual information extraction from a set of feature representations. The NetVLAD is a differentiable version of the vector of locally aggregated descriptors (VLAD), which aggregates a set of local descriptors and generate one global descriptor.
Fig. 3. The Transformer-VLAD network architecture.
Local Feature Extraction
We employ VGG-16 per-trained on the ImageNet dataset as a backbone to extract local features. This CNN-backbone transforms the initial US image 𝐼 US ∈ ℝ1×H0×W 0 to a lower-resolution feature map 𝐹0 ∈ ℝ D×H×W , where H0 = 400, W 0 = 274, D = 512 and H,W = H 0/32, W 0/32. So each pixel feature representation in the final feature map represents a 32 × 32 US patch in the original US image. Finally, we collapse the feature map 𝐹0 into a one-dimensional sentence-like feature vector 𝐹 ∈ ℝ D×H·W as input to the Transformer.
Contextual Feature Extraction
Given the feature vector 𝐹 ∈ ℝ D×H·W , the Transformer [9] extracts the contextual cues within the CNN feature representations to generate a new feature vector 𝐴 ∈ ℝ D×H·W . The Transformer consists of three encoders, and each of which is composed of a series of modules, i.e. multi-head self-attention (MHSA), feed-forward network (FFN) and layer normalization (LN). Each encoder can be stacked on top of each other multiple times.
Because the Transformer architecture is permutation-invariant, we supplement it with fixed positional encodings 𝑃 ∈ ℝ D×H·W that are added to each encoder. Specifically, 𝑃 is a sinusoidal positional encoding following [7]. We add 𝑃 to the query Q and key K without value V in each MHSA to maintain the position information of the feature representation,
| (1) |
Then the Transformer encoder can learn a co-contextual message Attn captured by the MHSA mechanism,
| (2) |
where Qi , Ki , Vi stand for ith head of queries, keys, values of the feature representation respectively, and direfers to the dimension of queries. In this implementation, an eight head attention (i.e. i = 1, 2,…, 8) is adopted to enhance the discriminativeness of the feature attributes. The MSHA mechanism automatically builds the connections between the current representation and the other salient representations within the sentence-like feature vector.
Finally, the attentional representation 𝐴 can be obtained as,
| (3) |
where FFN contains two fully connected layers. This procedure is performed three times in the three encoders, and the position encoding is inputted to the MHSA in each encoder.
Feature Aggregation
In order to improve permutation invariance of the feature representation 𝐴 ∈ ℝ D×H·W , we adopt NetVLAD [1] rather than a bag-of-visual-words or max-pooling operation. It is designed to aggregate a set of local descriptors 𝐴 ={xi} to generate one global descriptor 𝒱0 ∈ ℝ D×K , where K = 64 denotes the number of cluster centers {ck },
| (4) |
Here xi (d) denotes the dth dimension of the ith descriptor, and ck (d) denotes dth dimension of the kth cluster center. {w k }, {bk } and {ck } are the trainable parameters of the kth cluster. In contrast to conventional VLAD, the parameters of NetVLAD, especially the assignment score αk (xi) of the descriptor xi to kth cluster center, are learned through an end-to-end training.
NetVLAD records statistical information with respect to local signatures and sums the differences between these signatures and their respective cluster. To avoid computationally expensive descriptor search, we use a fully connected layer to compress the high-dimensional descriptor 𝑉0 ∈ ℝ D×K into a compact descriptor 𝑉 ∈ ℝ4096.
Loss Function
After getting the query, positive, negative global descriptors 𝑉 q , 𝑉 pos , 𝑉 neg of the triplet data from the Transformer-VLAD network, we explore both Triplet loss and InfoNCE loss to train the network through contrastive learning. The contrastive learning aims to push representations of positive pairs closer together, while representations of negative pairs are pushed farther with each other. Triplet loss requires the positive pairs to be closer than the negative pairs by a fixed margin δ given the same anchor,
| (5) |
where δ = 0.3 and dis(·,·) denotes the Euclidean distance. InfoNCE loss formulates it as a dictionary look-up task using cross-entropy to measure the descriptor similarity from the similar/dissimilar date-pairs,
| (6) |
where τ = 0:5 is a temperature hyper-parameter.
3. Experiments
Data Acquisition and Processing
Data acquired in this work came from a ScanTrainer Simulator1 (Intelligent Ultrasound Ltd). This realistic simulator is based on real clinical 3D US volumes and allows a user to learn how to acquire 2D images for a virtual patient. In this case, we captured the 2D US image with the corresponding 6DoF probe pose during virtual obstetric scanning from the simulator. We collected a large number of obstetric US images of 2nd (20 weeks) and 3rd trimester (28 weeks) scans from different subjects in the simulator library of examples. We acquired 535,775 US images with the corresponding 6DoF probe pose captured to construct the anchor-positive-negative triple data for the network training. We constructed 5 landmark-query testing cases for the 2nd trimester scanning, and also 5 landmark-query testing cases for the 3rd trimester scanning. For the landmark setting, we firstly collected a very large number of US images with probe poses. Then, a spatially distributed sampling was used to generate 400 evenly distributed landmarks in 3D space, which can not only transform an arbitrary size position to a fixed number of positions, but also simultaneously preserve structural information within the original positions. The query number of 5 test cases in the 2nd trimester are 1471, 927, 1295, 934, 1031 respectively, and that of the 3rd trimester are 1027, 936, 813, 830, 818 respectively. Note the training and testing data are collected from different cases/women of 2nd and 3rd trimester scans.
Self-supervised Network Training
To avoid expensive human annotation, we construct anchor-positive-negative triple data using KD-tree searching according to the 3D probe position, as shown in Fig. 2 right. To be specific, we extract the 3D probe position from the US-probe data-pair to build a KD-tree. Each node in the KD-tree stores the corresponding US image. Given an anchor US image, we set the search radius to 15mm for the KD-tree search region. The US images located inside the search region are assigned as positive US images related to the anchor image, while those outside US images are assigned as negative US images. In this case, the anchor-positive-negative triple data is constructed automatically from data itself without human annotation so that the network is trained end-to-end in a self-supervised way. The hyper-parameter 15mm is empirically set according to the number of landmarks and 3D volume i.e. density of landmarks. It can be adjusted according to specific clinical tasks.
Performance Evaluation
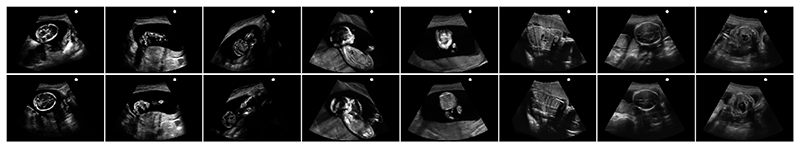
A standard evaluation procedure of image retrieval is employed for performance evaluation. Specifically, the query image is deemed correctly retrieved if at least one of the topN retrieved landmarks is within 15mm from the probe position of the query. The percentage of correctly retrieved queries (recall) is calculated for different values of N, i.e. recall@topN number(%). Some selected examples of query US images and successfully retrieved top1 US landmarks are shown in the Fig.4. We can see the successfully retrieved top1 landmark has very similar appearance to the query US image.
Fig. 4.
Selected examples of query US images (first row) and successfully retrieved top1 US landmarks (second row). The images in the first 5 columns are captured from 2nd trimester, and the last 3 columns are captured from 3rd trimester.
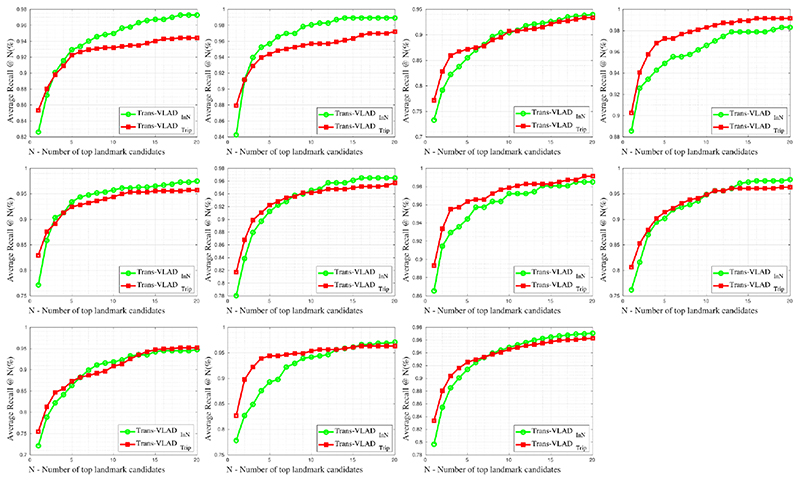
The recall@number(%) of each test case for the 2nd and 3rd trimester cases are given in the Table 1. We can see that the contrastive learning with Triplet loss achieves better performance than with InfoNCE loss. Performance for the 2nd trimester cases is slightly better than that for the 3rd trimester cases. A performance comparison with baselines using ablation analysis is provided in the Table 2. Compared with Transformer-VLAD, there is no Transformer sub-network in the baseline NetVLAD[1]. The baselines Transformer-Max and Transformer-TEN replace differentiable VLAD with Max-pooling and DeepTEN[10] respectively for the local feature aggregation. The baseline ViT[2][4]-VLAD uses a pure Transformer encoder instead of a CNN backbone operated on a sequence of 16 × 16 image patches. Note the latest research [4] achieves SOAT performance on the public benchmarks of natural image retrieval and we replaces its Max-pooling with VLAD to get better results in our dataset. We can see the that performance difference between the compared methods decreases as the number N increases. The Transformer sub-network improves performance compared with its absence. The VLAD-pooling outperforms Max-pooling significantly when the dataset does not achieve the large-scale level as public benchmark dataset. The VLAD-pooling provides slightly performance improvement comparing with DeepTEN-pooling due to their similar mechanism. We also find that the CNN backbone achieves better performance than a patch-style Transformer for US image retrieval although ViT[2][4] has attained excellent results compared to CNN on some public benchmarks. We also report the average recall curves from top1 to top20 landmark candidates for all 2nd and 3rd trimester test cases in the Fig.5. The sub-figures from left to right, top to bottom refer to the recall curves of case1,2,3,4,5 for 2nd trimester, and case1,2,3,4,5 for 3rd trimester, and the average recall curve of all 10 test cases (last sub-figure).
Table 1. Performance comparison on different test cases with different losses.
| Test cases | Trans-VLAD Trip | Trans-VLAD InN | ||||
|---|---|---|---|---|---|---|
| r@1 | r@5 | r@10 | r@1 | r@5 | r@10 | |
| case1 Sec | 85.3 | 92.3 | 93.4 | 82.6 | 92.9 | 94.5 |
| case2 Sec | 87.9 | 94.4 | 95.7 | 84.3 | 95.7 | 98.0 |
| case3 Sec | 77.2 | 87.2 | 90.7 | 73.3 | 85.5 | 90.4 |
| case4 Sec | 90.3 | 96.8 | 98.1 | 88.6 | 95.6 | 97.8 |
| case5 Sec | 83.0 | 92.8 | 94.4 | 77.1 | 93.4 | 95.7 |
| case1 Thi | 81.7 | 92.2 | 94.1 | 78.0 | 91.2 | 94.6 |
| case2 Thi | 89.3 | 96.4 | 97.9 | 86.5 | 94.4 | 97.2 |
| case3 Thi | 80.6 | 91.4 | 94.8 | 76.2 | 90.2 | 94.8 |
| case4 Thi | 75.5 | 87.3 | 89.7 | 72.1 | 86.3 | 91.1 |
| case5 Thi | 82.8 | 94.4 | 95.3 | 77.8 | 90.3 | 94.1 |
| Average | 83.4 | 92.5 | 94.4 | 79.7 | 91.6 | 94.8 |
Trans signifies Transformer. r@N signifies recall@number(%). Trip and InN signify Triplet and InfoNCE loss. Sec and Thi signify 2nd and 3rd Trimester.
Table 2. Performance comparison with baselines using ablation analysis.
| Method | r@1 | r@5 | r@10 |
|---|---|---|---|
| Trans-VLAD | 83.4 | 92.5 | 94.4 |
| NetVLAD[1] | 80.2 | 90.9 | 93.5 |
| ViT[2][4]-VLAD | 81.8 | 91.8 | 93.8 |
| Trans-Max | 77.2 | 87.8 | 92.1 |
| Trans-TEN[10] | 82.5 | 91.5 | 94.0 |
Trans signifies Transformer. Max signifies Max-pooling operation.
Fig. 5. The average recall curves from top1 to top20 landmark candidates for all 2nd and 3rd trimester test cases.
4. Conclusions
In this paper, we present a novel visual-assisted US-probe movement guidance method using landmark retrieval aimed at assisting inexperienced operators to scan. In contrast to conventional control parameter prediction methods, we design a global position visualization intervention which is more intuitive and suitable for the human operator. We also proposed a Transformer-VLAD network to learn a generalized descriptor of each US image to achieve automated landmark retrieval. To avoid the need for expensive human annotation, we take advantage of the 3D probe position to construct anchor-positive-negative US image-pairs automatically for contrastive learning. The experiments demonstrate the potential of the proposed method to simulate realistic imaging acquisitions.
Supplementary Material
Acknowledgments
We acknowledge the ERC (ERC-ADG-2015 694581, project PULSE), the EP-SRC (EP/MO13774/1, EP/R013853/1), and the NIHR Biomedical Research Centre funding scheme.
Footnotes
References
- 1.Arandjelovic R, Gronat P, Torii A, Pajdla T, Sivic J. Netvlad: Cnn architecture for weakly supervised place recognition; Proceedings of the IEEE conference on computer vision and pattern recognition; 2016. pp. 5297–5307. [DOI] [PubMed] [Google Scholar]
- 2.Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S, et al. An image is worth 16x16 words: Transformers for image recognition at scale; International Conference on Learning Representations; 2021. [Google Scholar]
- 3.Droste R, Drukker L, Papageorghiou AT, Noble JA. Medical Image Computing and Computer-Assisted Intervention. LNCS, Springer; 2020. Automatic probe movement guidance for freehand obstetric ultrasound. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-Nouby A, Neverova N, Laptev I, Jégou H. Training vision transformers for image retrieval. arXiv preprint arXiv. 2021:2102.05644 [Google Scholar]
- 5.Grimwood A, McNair H, Hu Y, Bonmati E, Barratt D, Harris EJ. Assisted probe positioning for ultrasound guided radiotherapy using image sequence classification; International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer; 2020. pp. 544–552. [Google Scholar]
- 6.Li Y, Khanal B, Hou B, Alansary A, Cerrolaza JJ, Sinclair M, Matthew J, Gupta C, Knight C, Kainz B, et al. Standard plane detection in 3d fetal ultrasound using an iterative transformation network; International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer; 2018. pp. 392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parmar N, Vaswani A, Uszkoreit J, Kaiser L, Shazeer N, Ku A, Tran D. Image transformer; International Conference on Machine Learning; 2018. 40554064 [Google Scholar]
- 8.Sattler T, Zhou Q, Pollefeys M, Leal-Taixe L. Understanding the limitations of cnn-based absolute camera pose regression; Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2019. pp. 3302–3312. [Google Scholar]
- 9.Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser L, Polosukhin I. Attention is all you need. Advances in neural information processing systems. 2017:5998–6008. [Google Scholar]
- 10.Zhang H, Xue J, Dana K. Deep ten: Texture encoding network; Proceedings of the IEEE conference on computer vision and pattern recognition; 2017. pp. 708–717. [Google Scholar]
- 11.Zhao C, Shen M, Sun L, Yang GZ. Generative localization with uncertainty estimation through video-ct data for bronchoscopic biopsy. IEEE Robotics and Automation Letters. 2019;5(1):258–265. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.