Genetic screening in a mouse model of biliary cancer revealed Pik3caH1047R activation but not oncogenic KrasG12D as critical for transformation and tumor formation and additionally reveals the context- and tissue-specific nature of oncogenic drivers.
Abstract
Biliary tract cancer ranks among the most lethal human malignancies, representing an unmet clinical need. Its abysmal prognosis is tied to an increasing incidence and a fundamental lack of mechanistic knowledge regarding the molecular basis of the disease. Here, we show that the Pdx1-positive extrahepatic biliary epithelium is highly susceptible toward transformation by activated PIK3CAH1047R but refractory to oncogenic KrasG12D. Using genome-wide transposon screens and genetic loss-of-function experiments, we discover context-dependent genetic interactions that drive extrahepatic cholangiocarcinoma (ECC) and show that PI3K signaling output strength and repression of the tumor suppressor p27Kip1 are critical context-specific determinants of tumor formation. This contrasts with the pancreas, where oncogenic Kras in concert with p53 loss is a key cancer driver. Notably, inactivation of p27Kip1 permits KrasG12D-driven ECC development. These studies provide a mechanistic link between PI3K signaling, tissue-specific tumor suppressor barriers, and ECC pathogenesis, and present a novel genetic model of autochthonous ECC and genes driving this highly lethal tumor subtype.
Significance:
We used the first genetically engineered mouse model for extrahepatic bile duct carcinoma to identify cancer genes by genome-wide transposon-based mutagenesis screening. Thereby, we show that PI3K signaling output strength and p27Kip1 function are critical determinants for context-specific ECC formation.
This article is highlighted in the In This Issue feature, p. 2945
INTRODUCTION
Adenocarcinomas of the bile duct express markers of cholangiocyte differentiation and are therefore termed cholangiocarcinomas (CC). They are classified into three subtypes, depending on their distinct anatomical location within the biliary tree: intrahepatic CC (ICC; comprising 10%–20% of all cases), and extrahepatic CC (ECC) with perihilar (PCC; 50%–60%) and distal (DCC; 20%–30%) location (1). This group of cancers is heterogeneous, and the anatomical subtypes differ not only in their anatomical site, but also in their epidemiology, pathogenesis, and genetic and clinical characteristics. This is reflected in different treatment options and therapeutic responses (1–3). CC develops from cholangiocytes or their progenitor cells lining the biliary tract, which arise from two distinct developmental origins. The intrahepatic biliary tree derives from liver progenitor cells, called hepatoblasts, which give rise to hepatocytes and intrahepatic cholangiocytes (4). The extrahepatic part of the biliary tree develops before the intrahepatic tract, and both systems merge at the level of the hepatic duct during embryonic development. Cholangiocytes lining the extrahepatic bile ducts share a common origin with the ventral pancreas, but not the liver. They derive from Pdx1-positive progenitor cells from the caudal part of the ventral foregut endoderm (4, 5).
The incidence of biliary tract cancer (BTC), which is highest in South/East Asia and South America, is increasing worldwide (1, 6). In the Western world, BTC accounts for 3% of all gastrointestinal tumors, with ECC being the most common type of BTC (1, 2). Despite modern multimodal treatment regimes, mortality rates of bile duct cancer have remained stubbornly unchanged for the last three decades, with approximately 10% of patients surviving 5 years (1–3, 7). This contrasts with other solid tumor entities such as colorectal cancers, where death rates have decreased more than 40% over the past 20 years (8). Surgical resection is the procedure of choice. However, early symptoms are unspecific and, by the time of diagnosis, most tumors are not surgically resectable. Consequently, there is an urgent need to identify and validate actionable cancer drivers as novel therapeutic targets.
Comprehensive molecular tumor profiling has greatly improved our understanding of the processes underlying tumorigenesis in the biliary tract (1, 3, 9–12). These studies revealed a subtype-associated prevalence of genomic alterations, methylation pattern, and gene-expression profiles in line with the observed clinicopathologic differences between ICC and ECC (2, 3, 11, 13). However, we are still far from understanding the nature of the described genetic alterations, their functional relevance, their downstream effectors, and their role in oncogenic networks. Specifically, mechanistic knowledge concerning true cancer drivers remains extremely poor for ECC, which is in part due to a lack of genetically defined animal models that accurately mimic this tumor subtype. Improving the survival of patients with ECC will therefore depend on unbiased functional systematic approaches in relevant model systems to identify the genes and pathways that drive the disease.
To this end, we generated a Cre/loxP-based genetically engineered mouse model (GEMM) of bile duct cancer with consequent extrahepatic localization and performed a forward genetic screen using the conditional piggyBac transposon mutagenesis system (14, 15). Analysis of recurrent transposon insertion sites identified numerous cancer genes and pathways, including known and new ECC-related genes, providing a unique resource for novel potential therapeutic targets. We used loss-of-function experiments in GEMMs to validate selected therapeutically tractable pathways and uncovered unexpected context-specific oncogenic networks, genetic dependencies, and tumor-suppressive functions in ECC development.
RESULTS
Pdx1-Cre Mediated Genetic Manipulation of the Extrahepatic Biliary Epithelium
To target the extrahepatic biliary epithelium, we used transgenic mice expressing Cre recombinase under the control of the murine Pdx1 promoter (ref. 16; Pdx1-Cre; Fig. 1A and B; Supplementary Fig. S1A). We evaluated the patterns of Pdx1-Cre transgene expression using two different Cre activatable reporter alleles. A lox–stop–lox (LSL) silenced fluorescent tdTomato reporter line (LSL-R26tdTomato/+; Fig. 1A; ref. 17) and a switchable floxed double-color fluorescent tdTomato-EGFP Cre reporter allele (R26mT-mG) that replaces the expression of tdTomato with EGFP after Cre-mediated recombination (Fig. 1B; ref. 18). Macroscopic fluorescence stereomicroscopy of Pdx1-Cre;LSL-R26tdTomato/+ mice revealed Cre-mediated recombination of the extrahepatic biliary tree, the pancreas, and duodenum in vivo, as evidenced by tdTomato expression (Fig. 1A). Using genetic Pdx1-Cre;R26mT-mG reporter animals, we confirmed Cre-mediated recombination specifically in the extrahepatic bile duct histologically, as shown by expression of membrane-tagged EGFP in CK19-positive biliary epithelial cells and peribiliary glands (Fig. 1B). In contrast, stromal cells remained unrecombined and expressed membrane-tagged tdTomato (Fig. 1B). No recombination was observed by fluorescence microscopy and genotyping PCR in liver, lung, kidney, heart, colon, brain, and spleen (Fig. 1; Supplementary Fig. S1A).
Figure 1.
Constitutive activation of the PI3K signaling pathway induces premalignant biliary intraepithelial neoplasia (BilIN). A, Left: genetic strategy and recombination scheme to analyze the patterns of Pdx1-Cre transgene expression using a conditional tdTomato reporter allele (LSL-R26tdTomato/+). Right: macroscopic fluorescence and white-light images of the Pdx1-Cre;LSL-R26tdTomato/+ reporter mouse. Visualization of tdTomato reveals reporter gene expression (red) in the pancreas, duodenum, gallbladder, and common bile duct (CBD). Scale bars, 1 cm. B, Left: genetic strategy and recombination scheme to analyze the patterns of Pdx1-Cre transgene expression using a switchable floxed double-color fluorescent tdTomato-EGFP Cre reporter line (R26mT-mG). Right panel, left image: representative confocal microscopic image of tdTomato (red color, non–Cre-recombined cells) and Cre-induced EGFP (green color) expression in the common bile duct. Note: Cre-mediated EGFP expression in the biliary epithelium and peribiliary glands, but not stromal cells. Nuclei were counterstained with TOPRO-3 (blue). Right image: representative immunofluorescence staining of CK19 (red color) of Pdx1-Cre;R26mT-mG/+ animal. Note: Blue color shows expression of tdTomato in unrecombined cells. Green color labels Cre-recombined cells that express EGFP. Colocalization of CK19 immunofluorescence staining (red) and EGFP expression (green) results in yellow color. Scale bars, 50 μm. C–E, Genetic strategy used to express oncogenic Pik3caH1047R (C), KrasG12D (D), or only Cre as control (E) in the common bile duct (top). Hematoxylin and eosin (H&E) staining and IHC analysis of PI3K/AKT pathway activation in the biliary epithelium of the common bile duct and different grades of dysplasia in BilIN (bottom). Scale bars, 50 μm for micrographs and 20 μm for insets.
Activation of Oncogenic Pik3caH1047R but Not KrasG12D Induces Premalignant Biliary Intraepithelial Neoplasia
Molecular profiling of human BTC revealed a large set of putative cancer drivers that are enriched for the RAS–RAF–MEK–ERK, PI3K–AKT–mTOR, TGFβ, and growth factor receptor signaling pathways, as well as epigenetic regulators and DNA damage response genes (1, 3, 10, 12, 13, 19–23). However, the functional consequences of these putative driver alterations and their relevance for ECC development have not been investigated in vivo. To test whether constitutive activation of oncogenic Kras or Pik3ca, which regulates two of the most frequently altered oncogenic pathways in BTC (3), is capable of transforming the extrahepatic biliary epithelium in vivo, we used the Pdx1-Cre model. We used an LSL-silenced oncogenic Pik3caH1047R allele as a knock-in at the Rosa26 locus (LSL-Pik3caH1047R/+ line; Fig. 1C; Supplementary Graphical Abstract and Supplementary Fig. S1A; ref. 24) to explore the mechanistic role of the PI3K–AKT–mTOR signaling pathway in ECC development. This mouse line carries a histidine (H) to arginine (R) mutation at codon 1047 (H1047R) within the highly conserved kinase domain of Pik3ca, resulting in increased catalytic activity and enhanced PI3K downstream signaling (24, 25). To achieve conditional activation of oncogenic Kras in the Pdx1-Cre lineage, we used a latent KrasG12D allele as a knock-in at the endogenous Kras locus (LSL-KrasG12D/+ line; Fig. 1D; Supplementary Fig. S1A and Supplementary Graphical Abstract; ref. 16). This mouse line carries a glycine (G) to aspartic acid (D) mutation at codon 12 (G12D) of Kras, which is refractory to GTPase-activating protein (GAP)–induced GTP hydrolysis, favoring an active state (26). Expression of mutationally activated Pik3caH1047R in Pdx1-Cre;LSL-Pik3caH1047R/+ mice revealed no overt phenotype after birth. However, adult animals developed an enlarged extrahepatic bile duct and biliary intraepithelial neoplasia (BilIN; Fig. 1C; Supplementary Fig. S1B), a precursor lesion of ECC (27) with complete penetrance. The grade of BilIN lesions increased over time; from low-grade BilINs in young animals to high-grade lesions in 9-month-old animals. In contrast to the Pik3caH1047R model, the extrahepatic bile duct was refractory toward neoplastic transformation by oncogenic KrasG12D (Fig. 1D; Supplementary Fig. S1B). Instead, KrasG12D expression in the Pdx1-Cre lineage induced only acinar–ductal metaplasia (ADM) and pancreatic intraepithelial neoplasia (PanIN), a precursor to invasive pancreatic ductal adenocarcinoma (PDAC). We observed similar lesions in a previous study in the pancreas after selective activation of Pik3caH1047R in pancreatic precursor cells using a Ptf1aCre line (24).
To compare Pik3ca expression levels between mutant and wild-type tissue and assess the activation status of the PI3K signaling pathway in Pik3caH1047R- and KrasG12D-mutant mice, we analyzed the common bile duct by IHC (Fig. 1C–E; Supplementary Fig. S1C). As expected, Pik3ca expression levels were moderately increased in the bile duct of Pik3caH1047R-mutant animals in comparison with wild-type mice (Supplementary Fig. S1C). In line with this, we observed activation of key downstream effectors of PI3K signaling, such as pAKT-T308, pAKT-S473, and pGSK3β-S9 (Fig. 1C). Interestingly, PI3K signaling is also activated in the extrahepatic bile duct of KrasG12D-mutant and wild-type mice, albeit to a lesser extent than in the wild-type (Fig. 1D and E). This indicates that signaling thresholds and tumor suppressor barriers might be important determinants of oncogenic transformation.
Pdx1-Cre–mediated activation of oncogenic Pik3caH1047R or KrasG12D in the pancreas resulted in very similar patterns of ADM and PanIN induction (Supplementary Fig. S2A and S2B). Surrogate markers of PI3K signaling were almost identical in Pik3caH1047R- and KrasG12D-driven tumors, as shown by phospho-specific pAKT-T308, pAKT-S473, and pGSK3β-S9 stainings, indicating an important role of this pathway for PDAC formation (Supplementary Fig. S2B and S2C). Previous studies validated its essential role for PanIN and PDAC development by genetic loss-of-function experiments in KrasG12D-driven GEMMs (24, 28).
To assess the signaling output of the canonical MAPK pathway, we analyzed ERK 1/2 activation using a phosphorylation-specific antibody. MAPK pathway signaling was decreased in the bile duct of the Pik3caH1047R model compared with wild-type animals. In the pancreas, we observed strong ERK 1/2 activation in KrasG12D-driven PanINs and PDAC, whereas Pik3caH1047R-induced tumors showed lower phosphorylation levels (Supplementary Fig. S2B–S2D).
Taken together, our data demonstrate that activation of oncogenic Pik3caH1047R in the extrahepatic bile duct models the full spectrum of human BilIN precursor lesion progression. In addition, they uncover fundamental different context-specific oncogenic vulnerabilities of the extrahepatic bile duct and the pancreas, although both organs originate from the same Pdx1-positive precursor cell lineage.
Expression of Mutant Pik3caH1047R Induces Invasive ECC
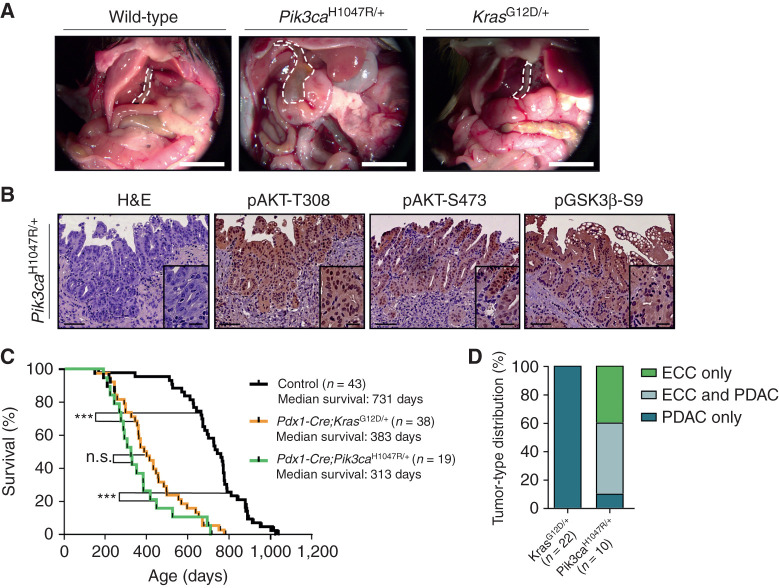
To test whether Pik3caH1047R is capable of inducing ECC, we aged Pdx1-Cre;LSL-Pik3caH1047R/+ mice. Within 800 days, 90% of the Pik3caH1047R-mutant animals in the tumor watch cohort developed extrahepatic bile duct cancer (Fig. 2A–D; Supplementary Graphical Abstract) displaying strong PI3K/AKT pathway activation, as demonstrated by key downstream surrogates of PI3K signaling: AKT-T308/S473 and GSK3β-S9 phosphorylation (Fig. 2B). ECCs recapitulated the full spectrum of the human disease, ranging from well-differentiated, stroma-rich ductal adenocarcinomas to more undifferentiated tumors (Fig. 2B; Supplementary Fig. S2; refs. 27, 29). Notably, at the time of necropsy, all animals developing ECC displayed cholestasis with bile duct obstruction and jaundice. This caused termination of the experiment as humane endpoint. Thus, the new Pdx1-Cre;LSL-Pik3caH1047R/+ model faithfully mimics human ECC; from precursor lesion formation and progression to full—blown bile duct–obstructing invasive cancers, with a characteristic strong desmoplastic stromal reaction (27, 29).
Figure 2.
Expression of oncogenic Pik3caH1047R but not KrasG12D induces ECC. A, Representative in situ images of 12-month-old wild-type (control), Pdx1-Cre;LSL-Pik3caH1047R/+, and Pdx1-Cre;LSL-KrasG12D/+ mice. The common bile duct is outlined by a white dashed line. Scale bars, 1 cm. B, Representative hematoxylin and eosin (H&E) stainings and IHC analyses of PI3K/AKT pathway activation in the common bile duct of aged Pdx1-Cre;LSL-Pik3caH1047R/+ mice with invasive ECC. Scale bars, 50 μm for micrographs and 20 μm for insets. C, Kaplan–Meier survival curves of the indicated genotypes (n.s., not significant; ***, P < 0.001, log-rank test). D, Tumor-type distribution in percentage according to histologic analysis of the extrahepatic bile duct and the pancreas from Pdx1-Cre;LSL-KrasG12D/+ and Pdx1-Cre;LSL-Pik3caH1047R/+ mice.
Comparison of tumor formation of Pik3caH1047R- and KrasG12D-mutant animals revealed striking differences. Although survival times were nearly identical, KrasG12D-mutant mice developed exclusively invasive tumors of the pancreas, recapitulating PDAC, but not BTC (Fig. 2C and D; Supplementary Fig. S3A and Supplementary Graphical Abstract). We never observed high-grade BilIN lesions or invasive ECC in KrasG12D-mutant animals aged up to 800 days. This unexpected resistance of the extrahepatic biliary epithelium against oncogenic transformation by KrasG12D contrasts with other tissue types, such as the lung or the intrahepatic biliary tract, which are highly susceptible to KrasG12D-induced tumorigenesis (24, 30, 31).
To assess tumor development in other Pdx1-Cre recombined organs in Pik3caH1047R-mutant mice (see Fig. 1A; Supplementary Fig. S1A), we performed a comprehensive macroscopic and microscopic histopathologic analysis and observed PDAC development in 60%, hyperplasia of the duodenal mucosa in 100%, and duodenal adenoma formation in 10% of the investigated double-transgenic animals (Fig. 2D; Supplementary Fig. S3A–S3C). Histopathologic analyses revealed that Pik3caH1047R-induced tumors were indistinguishable from KrasG12D-driven PDAC and showed the full spectrum of the disease, including ADM and PanIN lesions (Fig. 2; Supplementary Fig. S3A).
Together, these data support the notion that PI3K signaling is an important functional driver of ECC development and suggest that distinct context-specific tumor suppressor barriers and/or signaling thresholds are operative in the pancreas and the extrahepatic bile duct. This is remarkable due to the shared developmental origin of the pancreas and the extrahepatic bile duct and, therefore, a common cell of origin of both tissue types.
Oncogenic PI3K Signaling Activates a Senescence Program in the Extrahepatic Bile Duct That Is Independent of the p53 Pathway
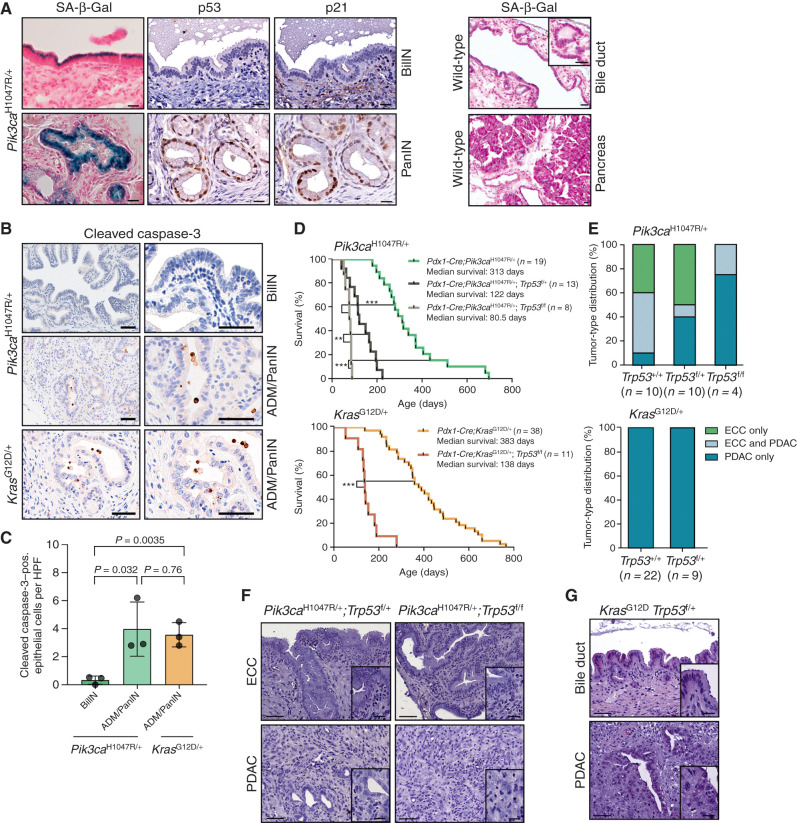
We next analyzed tissue-specific tumor-suppressive mechanisms in the Pik3caH1047R model. Expression of oncogenic Pik3caH1047R induced a senescence program in the extrahepatic biliary epithelium, similar to the activation of oncogenic KrasG12D in the pancreas (Fig. 3A; Supplementary Graphical Abstract; ref. 32). Low-grade BilIN lesions but not wild-type epithelium stained positive for senescence-associated β-galactosidase (SA-β-Gal), which has been shown to be a reliable oncogene-induced senescence (OIS) biomarker (Fig. 3A; ref. 33). OIS can be induced by the tumor suppressor p53 and its target gene p21Cip1. Interestingly, and in contrast to Pik3caH1047R- and KrasG12D-induced ADM and PanINs in the pancreas, BilIN lesions lack activation of the canonical p53–p21Cip1 senescence pathway (Fig. 3A) and display significantly reduced levels of apoptotic cell death (Fig. 3B and C). Previously, it has been shown that OIS and apoptotic cell death are triggered by the p53 network in the pancreas (32, 34). The absence of p53 induction and the significantly lower levels of apoptosis in the extrahepatic bile duct versus the pancreas upon Pik3caH1047R activation indicate fundamental context-dependent differences in the genetic networks and tumor suppressor barriers that prevent tumor formation in both organs (Supplementary Graphical Abstract).
Figure 3.
Oncogenic PI3K signaling induces senescence in the extrahepatic bile duct that is independent of the Trp53 pathway. A, Left, representative SA-β-Gal staining, and p53 and p21 IHC of BilIN (top) and PanIN (bottom) lesions of a Pdx1-Cre;Pik3caH1047R/+ mouse. Right, representative SA-β-Gal staining of wild-type extrahepatic bile duct and pancreas. Scale bars, 20 μm. B, Representative IHC cleaved caspase-3 staining of BilIN and ADM/PanIN of a Pdx1-Cre;LSL-Pik3caH1047R/+ mouse and an ADM/PanIN of Pdx1-Cre;LSL-KrasG12D/+ mouse. Scale bars, 50 μm. C, Quantification of cleaved caspase-3–positive epithelial cells in BilIN and ADM/PanIN lesions of the indicated genotypes. Each dot represents one animal (mean ± SD; P values are indicated, two-tailed Student t test). HPF, high-power field; pos., positive. D, Kaplan–Meier survival curves of the indicated genotypes (**, P < 0.01; ***, P < 0.001, log-rank test). E, Top: tumor-type distribution according to histologic analysis of the extrahepatic bile duct and pancreas of Pdx1-Cre;LSL-Pik3caH1047R/+, Pdx1-Cre;LSL-Pik3caH1047R/+;p53f/+, and Pdx1-Cre;LSL-Pik3caH1047R/+;p53f/f mice. Note: Reduced ECC fraction in mice with the Pdx1-Cre;Pik3caH1047R/+;p53f/f genotype (P = 0.02, Fisher exact test). Bottom: tumor-type distribution according to histologic analysis of the extrahepatic bile duct and pancreas of Pdx1-Cre;LSL-KrasG12D/+ and Pdx1-Cre;LSL-KrasG12D/+;p53f/+ mice. F, Representative hematoxylin and eosin (H&E) staining of ECC and PDAC of Pdx1-Cre;LSL-Pik3caH1047R/+;p53f/+ and Pdx1-Cre;LSL-Pik3caH1047R/+;p53f/f mice. G, Representative H&E staining of the common bile duct and PDAC of a Pdx1-Cre;LSL-KrasG12D/+;p53f/+ mouse. Scale bars, 50 μm for micrographs and 20 μm for insets. The Pdx1-Cre;Pik3caH1047R/+ and Pdx1-Cre;LSL-KrasG12D/+ cohorts shown in D and E are the same as those shown in Fig. 2C and D.
To elucidate the role of the p53 pathway in ECC and PDAC formation functionally, we inactivated Trp53 genetically using floxed Trp53 mice (35). Strikingly, at the humane endpoint, we observed a significantly reduced survival of Trp53-deleted animals and a shift of the tumor spectrum from ECC to PDAC in the Pik3caH1047R model, whereas KrasG12D;Trp53f/+ animals again developed PDAC only (Fig. 3D–G; Supplementary Graphical Abstract). To investigate the timing and order of BilIN/PanIN and ECC/PDAC formation in this model, we analyzed Pik3caH1047R;Trp53f/+ mice without clinical signs of cancer development at two selected early time points. This revealed widespread occurrence of low- and high-grade BilIN lesions along the extrahepatic bile duct already in 1.5-month-old animals, whereas only a few ADMs and even less low-grade PanIN lesions were present in the pancreas (Supplementary Fig. S4A). Nevertheless, most Trp53-deleted animals at the endpoint displayed PDAC and BilINs, but not invasive ECC (Fig. 3E). Only one mouse at an age of 3 months developed invasive PDAC and ECC in parallel in the same animal (Supplementary Fig. S4B). This indicates that BilIN development may precede PanINs; however, deletion of Trp53 accelerates progression of low-grade PanIN precursor lesions to invasive pancreatic cancer, leading to a substantial shift in the tumor spectrum toward PDAC. These findings support the view of tissue-specific tumor-suppressive functions of p53 being a pivotal tumor barrier in the pancreas (Supplementary Graphical Abstract).
Identification of Cancer Genes in the Extrahepatic Biliary Tract by a PiggyBac Transposon Mutagenesis Screen
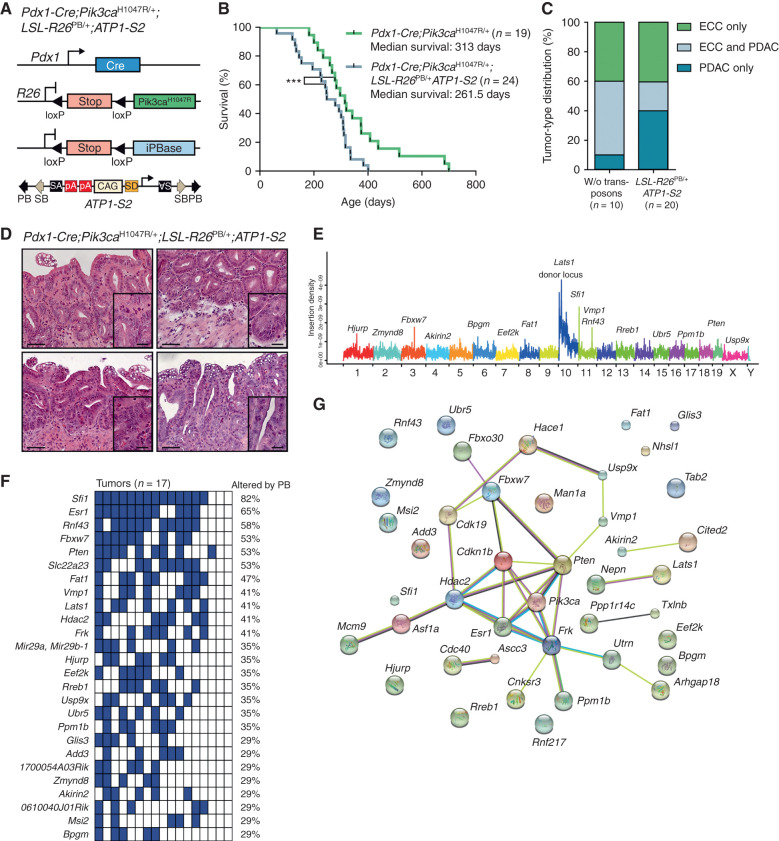
To study cooperating genetic events in extrahepatic bile duct cancer, we used a previously developed conditional piggyBac transposon–based somatic insertional mutagenesis system in mice with activated Pik3caH1047R (14). To this end, we crossed an LSL-silenced piggyBac transposase allele (LSL-R26PB) with ATP1-S2 mice, which possess mutagenic ATP1 transposons (14, 15). Compound mutant mice were then crossed to the Pdx1-Cre;LSL-Pik3caH1047R/+ line (Fig. 4A). To study ECC formation, we aged quadruple-mutant mice and observed at the humane endpoint that piggyBac mutagenesis led to an acceleration of Pik3ca-induced cancer development, reduced survival, and a shift in the tumor spectrum toward PDAC (Fig. 4B–D).
Figure 4.
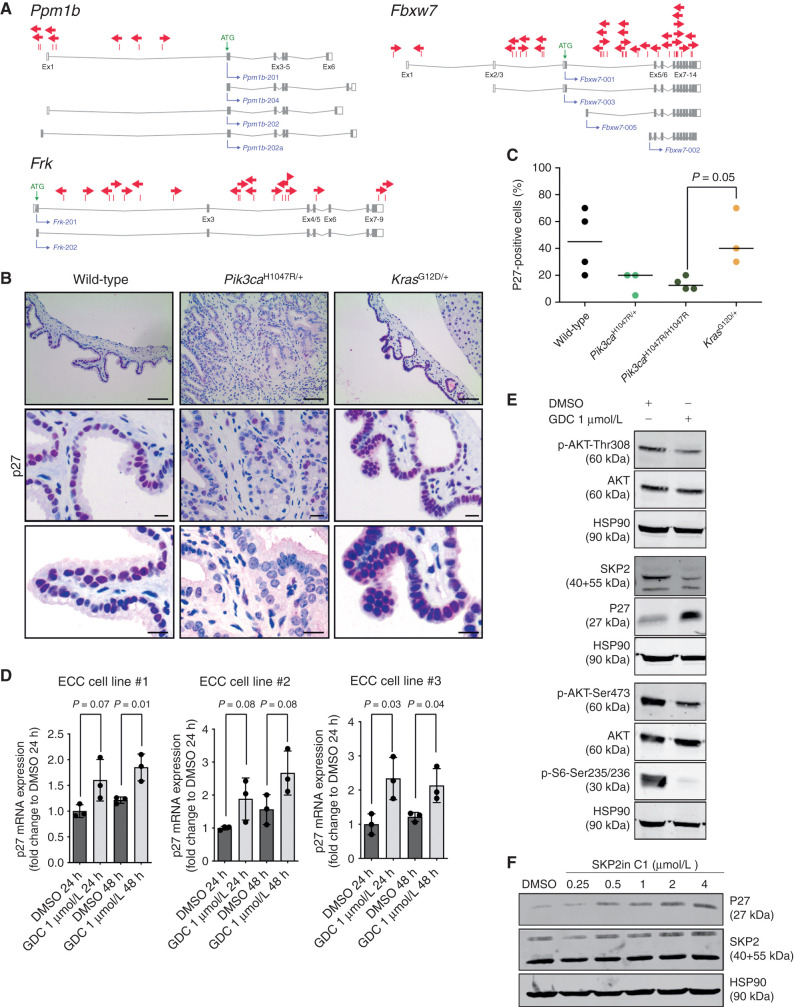
Identification of cancer genes in the extrahepatic biliary tract by a piggyBac transposon mutagenesis screen. A, Genetic strategy used to activate ATP1-S2 transposons by the piggyBac (PB) transposase in a Pdx1-Cre;LSL-Pik3caH1047R/+-mutant background. The ATP1-S2 mouse line carries 20 copies of the transposon construct on chromosome 10, which can be mobilized by the piggyBac transposase. Unidirectional integration of the transposon can lead to gene activation through its CAG promoter. Gene inactivation is independent of transposon orientation. CAG, CAG promoter; iPBase, insect version of the piggyBac transposase; pA, poly adenylation site; SA, splice acceptor; SD, splice donor. B, Kaplan–Meier survival curves of the indicated genotypes (***, P < 0.001, log-rank test). The Pdx1-Cre;Pik3caH1047R/+ cohort is the same as that shown in Fig. 2C. C, Tumor-type distribution according to histologic analysis of the extrahepatic bile duct and pancreas from Pdx1-Cre;LSL-Pik3caH1047R/+ and Pdx1-Cre;LSL-Pik3caH1047R/+;LSL-R26PB;ATP1-S2 mice. W/o, without. D, Representative hematoxylin and eosin stainings of four individual piggyBac-induced ECC of Pdx1-Cre;LSL-Pik3caH1047R/+;LSL-R26PB;ATP1-S2 mice. Scale bars, 50 μm for micrographs and 20 μm for insets. E, Genome-wide representation of transposon insertion densities in ECCs (pooled data from 17 tumors). Selected CIS genes are depicted. Chromosomes are labeled by different colors, and chromosome number is indicated on the x-axis. The transposon donor locus is on chromosome 10. F, Co-occurrence analysis of the CIS identified by TAPDANCE analysis in 17 tumors. Each column represents one tumor where insertions in the respective genes are indicated in blue. The fraction of tumors carrying an insertion in the respective genes is given as percentage. G, Network of protein interactions between CIS genes generated by STRING analysis (38). Each network node represents one protein. Interactions are marked by lines in different colors (green, neighborhood evidence; purple, experimental evidence; light blue, database evidence; black, coexpression evidence). Cdkn1b, p27Kip1.
To identify transposon insertions, we performed quantitative insertion site sequencing (QISeq) of ECCs as described previously (14). We identified 38,391 nonredundant integrations with a minimal read coverage of 2 (which was set as the threshold for insertion calling). To identify genomic regions that have been hit by transposons more frequently than expected by chance, we carried out TAPDANCE (Transposon Annotation Poisson Distribution Association Network Connectivity Environment) statistical analysis (36), which identified 90 common insertion sites (CIS; Supplementary Table S1). Figure 4E shows the genome-wide distribution of these insertions and indicates CIS genes within some of the transposon insertion density peaks. Among the 90 TAPDANCE CISs, we identified four main gene clusters: (i) insertions into important regulators of PI3K signaling, such as Pten, Lats1, and Frk; (ii) CIS in genes that have been reported to be direct or indirect regulators of p27Kip1 (Cdkn1b) protein abundance, such as Fbxw7 and Ppm1b; (iii) regulators of cell cycle and DNA replication, such as Sfi1, Hdac2, and Hjurp; and (iv) genes of the ubiquitin proteasome system, such as Usp9x, Fbxo30, and Ubr5 (Fig. 4F). Most of the identified genes have not been implicated in epithelial cancer development but are interesting candidates for further analysis. Vmp1, for example, is a downstream target of the PI3K–Akt pathway and implicated in autophagy (37).
To identify significantly enriched interaction networks in ECC, we used the STRING database (http://string-db.org; ref. 38). Thereby, we confirmed enrichment of CIS gene interactions with Pik3ca/Pten and p27Kip1 (Cdkn1b) in ECC (P = 5.43e−06; Fig. 4G). Cross-species comparison of the top 24 CISs of the transposon screen with recurrent genetic aberrations found in human bile duct cancer revealed an overlap of 15 altered genes, including RNF43, FBXW7, PTEN, FAT1, HDAC2, USP9X, and UBR5 (Supplementary Fig. S5).
The main insertional mutagenesis hits, and their function as direct or indirect regulators of PI3K signaling, p27Kip1 abundance, and cell-cycle progression are summarized in Supplementary Fig. S6.
Functional In Vivo Validation of a Context-Specific Oncogenic Pten–Pik3ca Signaling Node in ECC
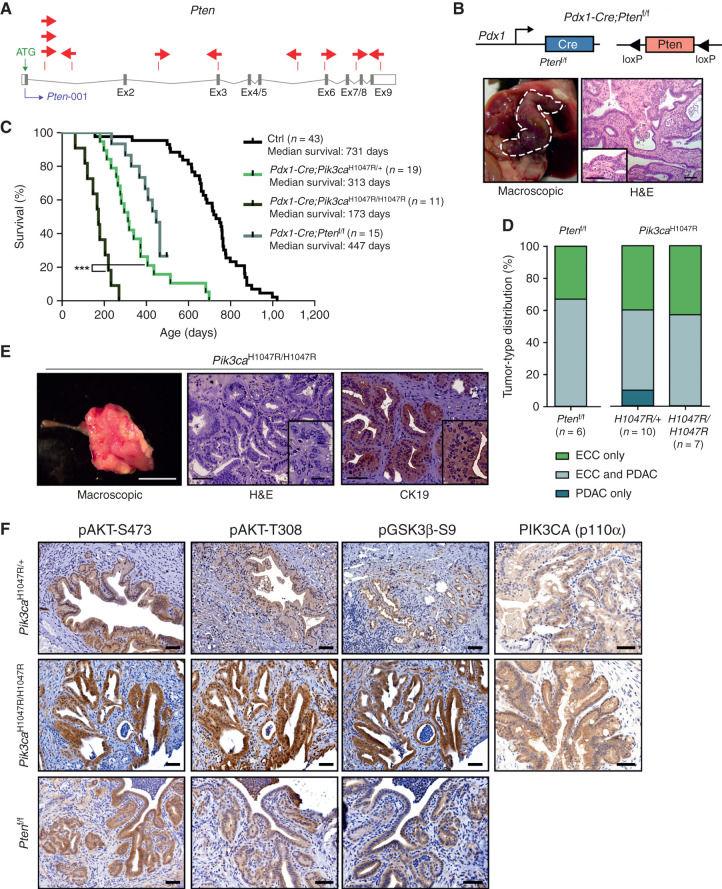
More than 50% of the ECC tumors (9/17) had insertions in Pten. The position and orientation of the ATP1-S2 transposon, which carries a CAG promoter, a splice donor, bidirectional SV40 polyadenylation signals, and two splice acceptors, can be used to predict whether it is likely to drive or disrupt gene transcription (14, 15). Disruption of transcription may occur if the transposon inserts into a gene with no CAG promoter relative to the direction of transcription; in this situation, the gene is a putative tumor suppressor gene (TSG). Transcriptional activation may occur if the transposon is sense-oriented and upstream of the translation start site, supporting gene expression from the unidirectional CAG promoter. Analysis of transposon insertion position and orientation in the Pten locus revealed a random distribution and sense or antisense orientation, suggesting that gene function is disrupted (Fig. 5A).
Figure 5.
Functional validation of transposon integrations: PI3K signaling output is a critical determinant of ECC development. A,piggyBac insertion patterns in Pten. Pten possesses one protein-coding isoform consisting of nine exons. Each arrow represents one insertion and indicates the orientation of the CAG promoter that was introduced into the transposon. B, Top: genetic strategy used to inactivate Pten in the Pdx1 linage using a Pdx1-Cre line. Bottom: representative macroscopic picture (left, bile duct depicted by a white dashed line) and hematoxylin and eosin (H&E)–stained tissue section (right) of an ECC from a Pdx1-Cre;Ptenf/f mouse. Scale bars, 50 μm for micrographs and 50 μm for insets. C, Kaplan–Meier survival curves of the indicated genotypes (***, P < 0.001, log-rank test). The Pdx1-Cre;Pik3caH1047R/+ and the control cohort are the same as those shown in Fig. 2C. D, Tumor-type distribution according to histologic analysis of the bile duct and pancreas from Pdx1-Cre;Ptenf/f, Pdx1-Cre;LSL-Pik3caH1047R/+, and Pdx1-Cre;LSL-Pik3caH1047R/H1047R mice. The Pdx1-Cre;Pik3caH1047R/+ cohort is the same as shown in Fig. 2D.E, Left: representative macroscopic picture of an ECC from a Pdx1-Cre;LSL-Pik3caH1047R/H1047R mouse. Scale bar, 1 cm. Middle and right: representative H&E-stained (middle) and IHC CK19–stained (right) tissue section of an ECC from a Pdx1-Cre;Pik3caH1047R/H1047R mouse. Scale bars, 50 μm for micrographs and 20 μm for insets. F, Representative H&E stainings and IHC analyses of PI3K/AKT pathway activation and Pik3ca (p110α) expression in the common bile duct of Pdx1-Cre;LSL-Pik3caH1047R/+, Pdx1-Cre;LSL-Pik3caH1047R/H1047R, and Pdx1-Cre;Ptenf/f mice. Scale bars, 50 μm.
To functionally validate Pten as a TSG in ECC, we used a genetic loss-of-function approach and inactivated Pten in the Pdx1 lineage using floxed Pten mice (Ptenf/f; ref. 39). Pdx1-Cre mediated deletion of Pten-induced extrahepatic bile duct cancer formation in all animals of the tumor watch cohort analyzed so far (Fig. 5B–D). These findings confirm PI3K signaling as a driver of ECC development in an independent genetic model and validate our transposon screen functionally.
The inactivation of PTEN argues for the need to select for aberrant enrichment of PI3K signaling during tumor evolution. This note is underscored by the observation that several CISs in the Pik3caH1047R model, such as Lats1 and Frk/Rak, have been shown to regulate the PI3K pathway (refs. 40, 41; Fig. 4F and G). To test genetically whether a threshold effect of PI3K signaling output is indeed a critical determinant of ECC formation in vivo, we increased the Pik3caH1047R gene dose and used homozygous Pik3caH1047R-mutant mice, which express the oncogene from both alleles (LSL-Pik3caH1047R/H1047R). This increased Pik3ca expression levels, PI3K signaling output, and animal mortality; shortened survival; and further shifted the tumor spectrum from PDAC toward ECC (Fig. 5C–F).
Taken together, these results provide mechanistic in vivo evidence that the canonical PI3K-dependent signaling pathway drives ECC formation in a gene dose–dependent manner, arguing that a certain level of PI3K signaling output strength is important for cancer formation in the extrahepatic bile duct.
p27Kip1 Is a Tissue-Specific Barrier of Tumor Evolution
Combining data from STRING analysis (http://string-db.org) of the piggyBac insertional mutagenesis screen and correlative data from literature reviews suggests that p27Kip1 (Cdkn1b) is an important barrier in ECC tumorigenesis and a target of several of the identified CIS genes (Fig. 4G; Supplementary Fig. S6). Overactivation of the PI3K signaling pathway through dysregulated Pik3ca and/or Pten loss, as well as alterations in Fbxw7, Ppm1b, and Frk, has been shown to regulate p27Kip1 protein expression directly or indirectly (41–50). Fbxw7, the substrate recognition component of an SCF (SKP1–CUL1–F-box protein) E3 ubiquitin–ligase complex, for example, is inactivated as a putative TSG by missense mutations in 5% to 15% of patients with bile duct cancer (3, 51), and deletion of Fbxw7 results in decreased p27Kip1 protein abundance (43). Ppmb1b, a magnesium-dependent protein phosphatase, is another positive regulator of p27Kip1 abundance (50), eventually via dephosphorylation of p27Kip1 at threonine 187 to remove a signal for proteasomal degradation, as described for Ppmb1h, another member of the Ppmb1 family (52).
As demonstrated for Pten (Fig. 5A), the transposon insertion pattern in Fbxw7, Ppm1b, and Frk/Rak (Fig. 6A) revealed random distribution across each gene and no orientation bias, predicting that inactivation of these TSGs and positive regulators of p27Kip1 are the mechanisms of cancer promotion. Because p27Kip1 levels are significantly decreased or absent specifically in patients with ECC (53), these observations suggest that p27Kip1 might be an important roadblock for extrahepatic biliary cancer formation in mice and men. To test this hypothesis, we analyzed p27Kip1 expression levels in the extrahepatic bile duct epithelium by IHC in wild-type, Pik3caH1047R, and KrasG12D-mutant animals. In contrast to oncogenic KrasG12D, p27Kip1 expression was selectively and significantly downregulated by Pik3caH1047R (Fig. 6B and C), consistent with previous reports showing that activated PI3K signaling impairs p27Kip1 expression in different tissue types in vitro and in vivo (42, 54, 55). To validate these findings, we analyzed the effect of PI3K inhibition on p27Kip1 expression levels using primary low-passaged ECC cells derived from the Pik3caH1047R model. Treatment with the potent and selective pan-class I PI3K inhibitor GDC-0941 blocked protein abundance of SKP2 and surrogates of PI3K signaling, such as Akt-T308, Akt-S473, and p70-S6 kinase1-S235/236 phosphorylation, and induced p27Kip1 expression at the mRNA and protein levels (Fig. 6D and E). To investigate whether SKP2 regulates p27Kip1 protein abundance in our system, we treated primary low-passaged ECC cells with a specific SKP2 inhibitor (SKP2in C1), which blocks SKP2 substrate binding. This increased p27Kip1 protein expression levels in a dose-dependent manner, providing additional insights into the mechanism of PI3K-mediated p27Kip1 downregulation (Fig. 6F).
Figure 6.
p27Kip1 (Cdkn1b) expression is downregulated by mutant PIK3CAH1047R. A,piggyBac insertion patterns for selected CIS genes implicated in p27Kip1 (Cdkn1b) regulation. Ppmb1b and Fbxw7 possess four protein-coding transcripts. Frk has two protein-coding isoforms consisting of eight and nine exons. Each arrow represents one insertion and indicates the orientation of the CAG promoter that was introduced into the transposon. B, Representative IHC analysis of p27Kip1 expression in the common bile duct of 6-month-old wild-type, Pdx1-Cre;LSL-Pik3caH1047R/+, and Pdx1-Cre;LSL-KrasG12D/+ mice. Scale bars, 80 μm (top) and 20 μm (middle and bottom). IHC staining was performed using 3-amino-9-ethylcarbazole (AEC) as chromogen, resulting in a pink color of positively stained cells. C, Quantification of p27Kip1-positive bile duct epithelial cells in 6-month-old wild-type, Pdx1-Cre;LSL-Pik3caH1047R/+, Pdx1-Cre;LSL-Pik3caH1047R/H1047R, and Pdx1-Cre;LSL-KrasG12D/+ mice (n = 3–4 animals per genotype). Each dot represents one animal; the horizontal line represents the mean (P = 0.05, Pdx1-Cre;LSL-Pik3caH1047R/H1047R vs. Pdx1-Cre;LSL-KrasG12D/+, Wilcoxon rank sum test). D, qRT-PCR analysis of p27Kip1 mRNA expression in three primary murine ECC cell lines from Pdx1-Cre;LSL-Pik3caH1047R/+ mice treated with either vehicle (DMSO) or 1 μmol/L GDC-0941 (GDC) for 24 or 48 hours (h) as indicated. Data are shown as fold change to DMSO treatment (n = 3; mean + SD; P values are indicated, Student t test). E, Immunoblot analysis of PI3K/AKT pathway activation and p27Kip1 expression in primary murine ECC cell line #1 treated with either vehicle (DMSO) or 1 μmol/L GDC-0941 for 48 hours. Hsp90α/β was used as loading control. F, Immunoblot analysis of p27Kip1 expression in primary murine ECC cell line #1 treated with either vehicle (DMSO) or the indicated concentrations of the SKP2 inhibitor SKP2in C1 for 48 hours. Hsp90α/β was used as loading control.
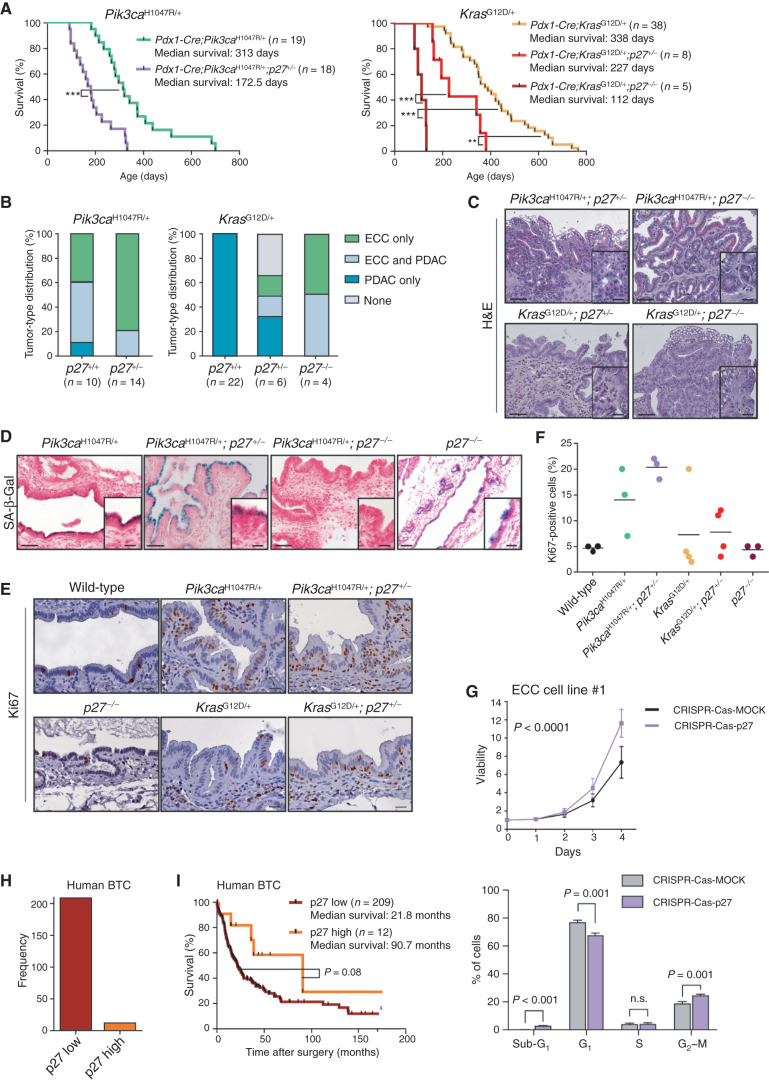
These data demonstrate that oncogenic PI3K signaling is an important regulator of p27Kip1 mRNA and protein abundance in the extrahepatic bile duct and ECC cells. To functionally analyze the role of reduced p27Kip1 expression for ECC formation in the extrahepatic bile duct in vivo, we used p27Kip1 knockout mice (56). Genetic deletion of one p27Kip1 allele in the Pik3caH1047R-driven model dramatically accelerated tumor formation, shortened mouse survival to a median of 172.5 days, and further shifted the tumor spectrum from PDAC toward ECC (Fig. 7A and B). All animals in the tumor watch cohort developed invasive ECCs with a desmoplastic stromal reaction (Fig. 7C).
Figure 7.
p27Kip1 is a context-specific roadblock for Kras-induced ECC formation. A, Kaplan–Meier survival curves of the indicated genotypes (***, P < 0.001; **, P < 0.01, log-rank test). B, Tumor type distribution according to histologic analysis of the extrahepatic bile duct and pancreas from Pdx1-Cre;LSL-Pik3caH1047R/+ and Pdx1-Cre;LSL-Pik3caH1047R/+;p27+/− mice (left) and Pdx1-Cre;LSL-KrasG12D/+, Pdx1-Cre;LSL-KrasG12D/+;p27+/−, and Pdx1-Cre;LSL-KrasG12D/+;p27−/− animals (right). Significant increase of ECC development in mice with the Pdx1-Cre;LSL-KrasG12D/+;p27−/− genotype (P < 0.0001, Fisher exact test). C, Representative hematoxylin and eosin (H&E) staining of the common bile duct of aged Pdx1-Cre;LSL-Pik3caH1047R/+;p27+/−, Pdx1-Cre;LSL-Pik3caH1047R/+;p27−/−, Pdx1-Cre;LSL-KrasG12D/+;p27+/−, and Pdx1-Cre;LSL-KrasG12D/+;p27−/− mice. D, Representative SA-β-Gal staining of the common bile duct of Pdx1-Cre;LSL-Pik3caH1047R/+, Pdx1-Cre;LSL-Pik3caH1047R/+;p27+/−, Pdx1-Cre;LSL-Pik3caH1047R/+;p27−/−, and p27−/− mice. C and D, Scale bars, 50 μm for micrographs and 20 μm for insets. E, Representative images of Ki67-stained common bile duct tissue sections of 3-month-old wild-type (control), Pdx1-Cre;LSL-Pik3caH1047R/+, Pdx1-Cre;LSL-Pik3caH1047R/+;p27+/−, p27−/−, Pdx1-Cre;LSL-KrasG12D/+, and Pdx1-Cre;LSL-KrasG12D/+;p27+/− mice. Scale bars, 20 μm. F, Quantification of Ki67-positive biliary epithelial cells of indicated genotypes. Each dot represents one animal, and the horizontal line represents the mean. G, Top: viability assay of primary murine ECC cell line #1 from a Pdx1-Cre;LSL-Pik3caH1047R/+ mouse after CRISPR-Cas9–mediated deletion of p27Kip1 (Cdkn1b). Cells were transfected with either Cas9-sgRNA-p27Kip1 targeting Cdkn1b (CRISPR-Cas-p27) or a MOCK Cas9-sgRNA-MOCK vector, selected using puromycin, and cell viability was measured in triplicate using the MTT assay (n = 4 independent experiments; mean ± SD; P < 0.0001, two-way ANOVA). Bottom: FACS-based cell-cycle analysis of the cells shown in the top panel. The P values were determined with multiple t tests and Benjamini correction and are shown in the figure. n.s., not significant. H, Quantification of p27Kip1 expression by IHC of 221 surgically resected human BTC specimens. I, Kaplan–Meier survival curves of patients with BTC with low or high p27Kip1 protein abundance (P = 0.08, log-rank test). The Pdx1-Cre;Pik3caH1047R/+ and Pdx1-Cre;LSL-KrasG12D/+ cohorts shown in A and B are the same as those shown in Fig. 2C and D.
Expression of oncogenic KrasG12D showed no effect on p27Kip1 protein abundance in the extrahepatic bile duct and failed to induce ECC formation (Figs. 1, 2, 6B; Supplementary Fig. S1B). To test the hypothesis of a causal relation of p27Kip1 as a tissue-specific roadblock for Kras-induced tumor formation in biliary epithelium, we deleted p27Kip1 in the Pdx1-Cre;KrasG12D-driven model genetically. Inactivation of one p27Kip1 allele induced high-grade dysplastic lesions in the extrahepatic bile duct as well as ECC formation in some animals, a phenotype never observed in KrasG12D mice with intact p27Kip1. This reduced median survival of KrasG12D/+;p27Kip1+/− mice significantly to 227 days compared with KrasG12D/+ animals with a median survival of 383 days. Homozygous p27Kip1 deletion led to ECC formation in all animals analyzed and further reduced median survival to 112 days (Fig. 7A–C).
Overall, these data validate at the genetic level the notion that p27Kip1 is a tissue- and context-specific tumor suppressor in the extrahepatic bile duct, and contrasts with the pancreas, where p53 represents a critical tumor-suppressive barrier; furthermore, they support the view that downregulation of p27Kip1 is important for ECC initiation and essential for KRAS-induced transformation of the extrahepatic bile duct (Supplementary Graphical Abstract).
To investigate the transcriptional programs involved in the context-dependent regulation of the p27Kip1 network, we performed RNA-sequencing (RNA-seq) analysis of Pik3caH1047R- and KrasG12D-driven low-passaged primary ECC and PDAC cell cultures treated for 48 hours with the PI3K inhibitor GDC-0941 or DMSO as control. We identified profound differences between Pik3caH1047R- and KrasG12D-driven tumors. Whereas blockade of PI3K signaling induced significant changes to the transcriptional program of Pik3caH1047R-driven ECCs and PDACs (146 and 142 differentially expressed genes in ECC and PDAC, respectively), it had only minor effects on the transcriptional output of KrasG12D-induced PDAC (22 differentially expressed genes; Supplementary Fig. S7A and S7B).
Pathway analysis of the significantly regulated genes revealed substantial changes in RNA binding, processing, splicing, and metabolism, as well as E2F targets in Pik3caH1047R-driven ECCs (top 10 pathways ranked by FDRq value shown in Supplementary Fig. S7C). In contrast, Pik3caH1047R-driven PDAC showed mainly changes of metabolic pathways and the mitochondrion upon PI3K pathway perturbation. E2F transcription factors (TF) have previously been shown to regulate p27Kip1 abundance via the transcriptional regulation of SKP2. Therefore, the observed E2F target gene signature provides evidence that a PI3K-induced E2F node regulates p27Kip1 abundance via SKP2, which is downregulated at the mRNA and protein levels upon GDC-0941 treatment (Fig. 6E; Supplementary Fig. S7D) and regulates p27Kip1 protein abundance in ECC cells (Fig. 6F). Accordingly, the Biocarta p27 pathway is significantly altered in Pik3caH1047R-driven ECC cells upon GDC-0941 treatment (Supplementary Fig. S7D). For Kras-driven PDAC, we could not detect any significantly altered pathway, because of the low number of significantly differentially expressed genes. These findings support the view that PI3K pathway activation by oncogenic KrasG12D does not reach a critical threshold that is necessary to alter the p27 network and downregulate p27Kip1 protein abundance, which is necessary for ECC induction.
To gain more insights into the context-specific transcriptional control of the p27Kip1 network under basal conditions, we analyzed the top 10 differentially targeting TFs in Pik3caH1047R-driven ECC and KrasG12D-induced PDAC. We observed fundamental differences in p27Kip1 network regulation and identified context-specific TFs that control the p27Kip1 tumor suppressor node specifically in ECC (Supplementary Fig. S7E).
To analyze p27Kip1 function in the extrahepatic biliary epithelium, we investigated early events of tumorigenesis. Surprisingly, p27Kip1 inactivation triggered senescence of the normal biliary epithelium in the absence of an oncogene. In contrast, it bypassed OIS in the context of Pik3caH1047R-induced early neoplastic BilIN lesions (Fig. 7D; Supplementary Graphical Abstract). Importantly, these BilIN lesions showed concomitant increased proliferation rates as evidenced by Ki67 IHC in situ (Fig. 7E and F). CRISPR-Cas9–mediated deletion of p27Kip1 in primary low-passaged ECC cells derived from the Pik3caH1047R model induced cell proliferation and cell-cycle progression (Fig. 7G; Supplementary Fig. S8A and S8B).
These results provide evidence that canonical p27Kip1 tumor suppressor functions, which regulate G0 to M phase cell-cycle transitions, contribute significantly to the observed effects, although the importance of additional and alternate effectors of p27Kip1 cannot be completely excluded (42).
To further substantiate our hypothesis that p27Kip1 is also a roadblock in human BTC, we analyzed p27Kip1 expression in 221 primary surgically resected tumor specimens. In accordance with the murine in vivo studies, we observed weak or absent p27Kip1 expression in nearly all human BTC (weak or absent p27Kip1: n = 209; strong p27Kip1: n = 12) specimens (Fig. 7H). In addition, tumors with weak or absent p27Kip1 expression were associated with poor survival. Kaplan–Meier curves comparing p27Kip1 expression with survival showed a trend toward separation (P = 0.08, log-rank test; Fig. 7I). This is in accordance with previous studies reporting that p27Kip1 protein abundance is an independent predictor of survival for patients with CC (53, 57–59).
DISCUSSION
There have been no major improvements in the treatment of ECC in the last three decades, and no effective targeted therapies are available (1–3). Next-generation sequencing and molecular tumor profiling uncovered a plethora of potentially actionable molecular alterations associated with ECC malignancy (3, 13, 19–23). However, despite these exciting new data describing the genomic landscape of ECC, we are far from understanding the functional consequences of these genetic alterations and the underlying tumor biology. This is due to (i) the inherent limitations of current cancer genome analysis to distinguish driver and passenger mutations and to predict the functional consequences of missense mutations, (ii) the often descriptive nature of such data, (iii) the complexity of signaling processes and tumor suppressor barriers downstream of altered genes, (iv) context-specific differences between different cancer entities, (v) tumor heterogeneity, and (vi) a lack of understanding of signals from the tumor micro- and macroenvironment. Thus, there is an urgent need for complementary functional studies using informative and predictive model systems, which fill the gap between the genetic landscape and the largely unknown biology of ECC (2, 3). This offers the best means of identifying novel therapeutic approaches for ECC that are robust and effective in the clinic (60, 61).
GEMMs have been developed for diverse cancer types in the last two decades that faithfully recapitulate the histologic, molecular, genetic, and clinical hallmarks of the human disease (60). They are often based on the Cre/loxP system and tissue-specific expression of oncogenes and/or deletion of TSGs (62). Such conditional models have provided important information about the natural biology of many cancer types and revealed potential targets for early detection and therapeutic intervention (61, 63). Accordingly, they have become one of the gold standards for preclinical investigation of novel lead compounds and therapeutic regimes (60, 61).
For ECC research, many sophisticated tools of fundamental importance, such as conditional GEMMs, have not been developed so far. This represents a major bottleneck, which is rate limiting for preclinical research and explains in part the poor molecular understanding of this devastating disease (1, 12, 19). We addressed this limitation and developed novel GEMMs of oncogenic KRAS- and PI3K-driven ECC subtypes that exhibit hallmarks of the human disease—from multistage BilIN precursor lesion progression to invasive stroma-rich ductal adenocarcinomas (see Supplementary Graphical Abstract for an overview of the models).
The pathogenesis of human ECC is a complex process involving alterations in the activity of multiple oncogenes and TSGs and their downstream effector pathways (1, 3, 64). Of these, activation of RAS and PI3K signaling pathways represents the most frequent events (3). Our new autochthonous ECC models offer for the first time the possibility to (i) study the biology of these molecular cancer subtypes and their associated pathways, (ii) discover and validate therapeutic targets, and (iii) provide the context for preclinical trials to find new ways to treat ECC subtypes and identify biomarkers of response and resistance.
We used the novel Pik3caH1047R-driven GEMM of ECC to perform a large genome-wide unbiased functional transposon-based mutagenesis screen to discover cooperating cancer genes in ECC. Using the piggyBac transposition system, we identified many novel cancer genes and cancer-driving networks. Some of them specifically feed into the PI3k pathway, such as inactivation of Pten or Lats1, or drive inactivation of the tumor suppressor p27Kip1 (Cdkn1b), which we identified as a major roadblock for ECC pathogenesis. Subsequently, we validated the essential role of PI3K signaling output strength and p27Kip1 downregulation for ECC formation in vivo using KrasG12D- and Pik3caH1047R-driven GEMMs with genetic alteration in these pathways (please see Supplementary Graphical Abstract for an overview of the tissue-specific tumor suppressor barriers of ECC formation).
p27 Kip1 is a cyclin-dependent kinase (CDK) inhibitor and one of the most frequently misregulated proteins in human cancers. It is an atypical TSG, as it is rarely mutated or deleted; rather, p27 protein levels are reduced or the protein is mislocalized, which promotes uncontrolled cell growth (42). Our data suggest that a certain level of PI3K output is necessary to block p27Kip1 expression to a level that allows extrahepatic bile duct cancer formation. To test this hypothesis, we overactivated the PI3K pathway in vivo and observed decreased p27Kip1 levels and accelerated ECC formation. In line with this, genetic manipulation of p27Kip1 (Cdkn1b) expression using knockout mice validated its role as a roadblock for ECC pathogenesis in the PI3K-driven model. In contrast to Pik3caH1047R, activation of oncogenic KrasG12D induced tumor development only in the pancreas but completely failed to drive ECC. However, deletion of p27Kip1 (Cdkn1b) enabled Kras-driven tumorigenesis in the extrahepatic bile duct with high penetrance. Because the TSG p27Kip1 is tightly linked to PI3K signaling (42, 54, 55), our genetic in vivo data show for the first time that a PI3K-p27Kip1 node is an essential determinant of PI3K- as well as Kras-driven ECC development (see Supplementary Graphical Abstract). This has important therapeutic implications, because several pharmacologic inhibitors directed against the PI3K pathway are under clinical investigation and drugs that combat p27Kip1 dysfunction are currently under development (48, 65).
Our data allow for two important conclusions. First, the response and vulnerability to distinct oncogenic drivers, allowing or preventing cancer development at different anatomical sites, are strictly tissue-specific, even if the different tissue types have a common developmental origin and indistinguishable morphology, as shown for the extrahepatic bile duct and the pancreatic duct. Second, distinct tumor suppressor barriers and signaling thresholds are operative in different tissue types even in the presence of the same initial oncogenic lesion as shown for KrasG12D and p27Kip1 in the extrahepatic bile duct and the pancreas. Thus, our results demonstrate that tissue-specific mechanisms contribute significantly to the context-specific oncogenic potency of a cancer driver and support the view that each tissue has its own unique signaling requirement during oncogene-induced transformation. This provides in vivo evidence for the rationale to investigate KRAS- and PI3K-driven oncogenic pathways in a strictly tissue- and context-specific manner to characterize the relevant nodes engaged in different tumor entities. Tissue context is an important determinant of treatment response and resistance. Therefore, the advance in our understanding of tissue-specific effects of oncogenic Kras and PI3K signaling presented here will affect clinical practice in the future because it clearly shows that results obtained in one tumor entity cannot simply be extrapolated to another.
In conclusion, our findings establish cell context–specific functions for KRAS and PI3K signaling and engagement of p27Kip1 in transformation of the extrahepatic bile duct that are independent of the p53 pathway. They also suggest that the role of oncogenic drivers varies depending on the cell and tissue type and microenvironment. Our findings underscore that tissue context is an important determinant of tumorigenesis. A deeper understanding of the molecular mechanisms involved in ECC pathogenesis and tumor biology will provide novel therapeutic targets and biomarkers of response essentially needed for clinical decision-making and the design of novel and improved next-generation targeted treatment regimens for patients with ECC.
METHODS
Mouse Strains and Tumor Models
Pdx1-Cre (16), LSL-R26tdTomato/+ (17), R26mT-mG/+ (18), LSL-Pik3caH1047R/+ (24), LSL-KrasG12D/+ (16), Trp53f/f (35), p27−/− (56), LSL-R26PB/+ (14), ATP1-S2 (15), and Ptenf/+ (39) alleles have been previously described. All animal studies were conducted in compliance with European guidelines for the care and use of laboratory animals and were approved by the Institutional Animal Care and Use Committees of Technische Universität München, Regierung von Oberbayern, and UK Home Office. Animals on a mixed C57Bl6/129S6 genetic background were intercrossed to obtain the desired genotype, and both male and female mice with the correct genotype were aged to assess cancer formation and survival of the animals. Wild-type and animals containing only Cre recombinase served as controls as indicated in the figure legends. Sequence information of genotyping PCR primers is given in the Supplementary Methods.
Materials
Cell culture reagents were obtained from Thermo Fisher Scientific. Primers were made by Eurofins Genomics, and restriction endonucleases were obtained from New England Biolabs. The Escherichia coli strain Stbl3 (Thermo Fisher Scientific) was used for transformation and plasmid amplification. The PI3K inhibitor GDC-0941 free base was purchased from LC Laboratories. The SKP2 inhibitor SKP2in C1 was purchased from Selleckchem.
White Light and Fluorescence Stereomicroscopy
Macroscopic pathologic analysis of murine tissues including ECC and PDAC was performed, and white-light and fluorescence pictures were taken using a Zeiss Stemi 11 fluorescence stereomicroscope (Zeiss).
Histopathology and IHC
Mouse tissue specimens were fixed in 4% buffered formalin overnight, embedded in paraffin, and sectioned (2 μm thick). Murine PanIN lesions and PDAC were graded according to the established nomenclature (66). Murine BilIN were graded in analogy to the human classification (27, 67). The normal common bile duct in mice is morphologically similar to its human counterpart. It is lined by a single layer of uniform cuboidal cells and accompanied by small peribiliary glands. Proliferative lesions with a flat or micropapillary structure composed of tall columnar cells with some loss of cellular polarity and/or nuclear pseudostratification with mild nuclear abnormalities (i.e., slight hyperchromasia or irregular nuclear membrane) were categorized as low-grade BilIN. High-grade BilIN were defined as lesions with micropapillary architecture and diffuse loss of cellular polarity, “budding off” of single epithelial cells into the lumen and severe cellular and nuclear pleomorphia. By definition, invasion through the basement membrane had to be absent in these strictly intraepithelial lesions. The examiner was blinded to the genotype of the animals.
For immunodetection, formalin-fixed, paraffin-embedded tissue sections were dewaxed, rehydrated, and placed in a microwave (10 minutes, 600 watt) in unmasking solution H-3300 (Vector Laboratories) to recover antigens. Sections were incubated with primary pERK (p44/42; #4376; 1:1,000; RRID:AB_331772), cleaved caspase-3 (#9664; 1:150; RRID:AB_2070042), PIK3CA (p110α; #4249; 1:400; RRID:AB_2165248), pAKT-T308 (#2965; 1:50; RRID:AB_2255933), pAKT-S473 (#4060; 1:50; RRID:AB_2315049), pGSK3β-S9 (#9323; 1:100; RRID:AB_2115201; all from Cell Signaling Technology), p21 (sc-397-G; 1:50; RRID:AB_632127; Santa Cruz Biotechnology), p53 (CM5; 1:300; RRID:AB_2744683; Novocastra/Leica Mikrosysteme), p27Kip1 (ab62364; 1:500; RRID:AB_944575; Abcam), and Ki67 (SP6; 1:50; RRID:AB_2722785; DCS Diagnostics) antibodies followed by a secondary antibody conjugated to biotin (Vector Laboratories). Antibody detection of pERK, cleaved caspase-3, and PIK3CA (p110α) was performed with the Bond Polymer Refine Detection Kit (Leica). Images were acquired using Leica AT2 Scanner (Leica). Quantification of p27Kip1-, cleaved caspase-3–, and Ki67-positive cells was performed by exclusively counting epithelial cells in at least 10 high-power fields in at least three animals, and the examiner was blinded to the genotype of the animals.
SA-β-Gal Staining
SA-β-Gal staining of cryosections was performed with the Senescence β-Galactosidase Staining Kit in accordance with the manufacturer's protocol (Cell Signaling Technology). Counterstaining was carried out with nuclear fast red. Images were acquired using Aperio Versa Scanner (Leica).
Immunofluorescence and Confocal Laser-Scanning Microscopy
Tissues were fixed in 4% buffered formalin for 2 hours, dehydrated at 4°C (15% sucrose in PBS for 4 hours; 30% sucrose in PBS overnight), and embedded in Tissue-Tek (Sakura) before being frozen in liquid nitrogen. 20-μm thick frozen sections were post-fixed for 1 minute in 4% buffered formalin, washed twice in PBS, and incubated for 1 hour in PBS with 3% (w/v) bovine serum albumin (BSA), 1% (w/v) saponin, and 1% (v/v) Triton-X 100. Subsequently, slides were incubated with CK19 primary antibody (ab52625, 1:100, RRID:AB_2281020, Abcam) and a DyLight 680–conjugated secondary antibody (#5366, 1:100, RRID:AB_10693812, Cell Signaling Technology). Nuclei were counterstained with TOPRO-3-iodide (1:1,000, Thermo Fisher Scientific). Sections were examined on a Leica TCS SP5 DMI 6000 CS confocal laser-scanning microscope using a 40/1.25 oil-immersion objective (Leica Microsystems).
Human BTC Tissue Samples
This study conformed to the Declaration of Helsinki and was approved by the ethics committee of the Universität Heidelberg. Written informed consent was obtained from all patients. The use of the tissue for this study was approved by the institutional ethics committee (206/05). Our cohort comprised 221 patients with CC. All tumors were resected between 1995 and 2010 at the University Hospital in Heidelberg, Germany. Neoadjuvant radiotherapy and/or chemotherapy was not applied in any of the patients. One hundred twenty-three (55.7%) patients were male, and 98 (44.3%) were female. Mean age at diagnosis was 64.7 years (range, 33.5–91.6 years). Twenty-seven (12.2%) tumors were classified as pT1, 116 (52.5%) as pT2, 61 (27.6%) as pT3, and 17 (7.7%) as pT4. One hundred fifty-one (68.3%) of the patients received concomitant lymph node resection. Of these, 83 (55%) were pN1 and 68 (45%) were pN0. Nineteen (8.6%) patients had distant metastasis at the time of resection. Fifty-seven (25.8%) of the tumors were ICCs, 94 (42.5%) were ECCs, and 70 (31.7%) were gallbladder carcinomas. Follow-up data were available for 191 patients (86.4%). The mean follow-up time of patients alive at the endpoint of analysis was 45.7 months. One hundred twenty-five (65.4%) patients died during follow-up.
Evaluation of the Human BTC Cohort
From all 221 CCs, 3-μm sections were cut and stained with hematoxylin and eosin. Representative areas from the tumor center and invasive margins were marked by two pathologists with a special expertise in CC pathology (B. Goeppert and W. Weichert). For each case, tumor tissue cores (1.5 mm diameter) from the selected representative tumor areas were punched out of the sample tissue blocks and embedded into a new paraffin array block using a tissue microarrayer (Beecher Instruments). For the detection of p27Kip1, a monoclonal mouse IgG antibody directed against p27Kip1 [BD Biosciences; clone: 57/Kip1/p27 (RUO); #610242; 1:100 dilution, RRID:AB_397637] was used. Bound antibody was detected with the Dako K5003 AEC kit (Dako). To exclude cases with only faint and focal putatively biologically irrelevant p27Kip1 expression from the p27Kip1 “positive” group, cases were only scored as positive when a clearly distinguishable nuclear signal of at least moderate intensity was seen in more than 50% of tumor cells.
Mapping of Transposon Insertion Sequences to the Mouse Genome and Identification of CISs
Genomic DNA was isolated from fresh-frozen ECC samples with the QIAamp DNA Mini Kit according to the manufacturer's protocol. Amplification of the transposon insertion sites and next-generation sequencing on an Illumina MySeq desktop sequencer was performed as described (14). Transposon insertion sites with a coverage of two or more reads were subjected to TAPDANCE analysis to identify nonredundant CISs (36). Network analysis was conducted with STRING (38).
Generation and Cultivation of Primary Low-Passaged Dispersed Murine ECC and PDAC Cell Cultures
Primary dispersed murine ECC and PDAC cell cultures were established from Pdx1-Cre;LSL-Pik3caH1047R/+ and Pdx1-Cre;LSL-KrasG12D/+ mice, and cultivated as previously described (68). Cell viability was quantified by MTT assay using the substrate 3-[4,5-dimethythiazol-2-yl]-2,5-diphenyltetrazolium bromide (Sigma-Aldrich Chemie). All cell lines were authenticated by genotyping and tested negative for Mycoplasma contamination by an inhouse PCR-based Mycoplasma detection assay every two months. The low-passaged cell cultures were cultivated for a maximum of 20 passages and typically used for the experiments between passages 5 and 10.
Drug Treatment
Cells were seeded in a 10-cm dish and treated 24 hours later with GDC-0941 or SKP2in C1 at the indicated concentrations or vehicle for 24 or 48 hours as indicated. The medium containing either the drug or the vehicle was changed after 24 hours.
Gene-Expression Profiling and Analysis
RNA was isolated with the RNeasy kit (Qiagen) from 80% confluent primary cells and immediately transferred into RLT buffer (Qiagen) containing β-mercaptoethanol.
RNA-seq library preparation was performed as previously described (69). Analyses were carried out using R version 3.6.2 and Bioconductor version 3.1. Differential gene-expression analysis was performed using DESeq2 version 1.26.0. A gene was considered to be differentially expressed with a Benjamini–Hochberg adjusted P value of 0.05 and an absolute fold change >0.5. The overrepresentation analysis was performed with MSigDB v7.1 gene sets provided by the Broad Institute, Massachusetts Institute of Technology, and Harvard University.
RNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-10515.
To investigate the tissue-specific regulatory networks of the p27Kip1 pathway, we used PANDA. PANDA integrates multiple types of data including protein–protein interactions (PPI) and gene expression to infer interaction between TFs and their target genes (70). We used the netZooR implementation of PANDA (version 0.9) to infer two separate regulatory networks for Pik3caH1047R-driven ECC and KrasG12D-driven PDAC. For each network, we used the same TF/target prior and PPI networks and normalized tissue-specific gene-expression data. The TF/gene priori was downloaded from https://sites.google.com/a/channing.harvard.edu/kimberlyglass/tools/resources. The PPI network was downloaded from STRINGdb, integrated in the netZooR library. The resulting networks allowed us to analyze the behavior of 441 TFs, >18,000 genes, and more than 8 million weighted edges between them. The visualization of the p27Kip1-related subnetworks was done in Cytoscape (v3.8.2, https://cytoscape.org).
CRISPR-Cas9–Mediated p27Kip1 Deletion in Primary Murine ECC Cell Lines
Two different sgRNAs targeting exon 1 of the Cdkn1b (p27Kip1) gene were designed using the online tool CRISPR Design (http://crispr.mit.edu/job/4733254127165968,1.8.2014). The Cas9 expression vector pSpCas9(BB)-2A-Puro (pX459; Addgene plasmid #48139; www.addgene.org) was used to insert either the annealed sgRNA1 or sgRNA2 both targeting Cdkn1b (p27Kip1). Sequence information of the sgRNAs is provided in the Supplementary Methods. Primary low-passaged murine ECC cells were cotransfected with the two modified versions of pX459 containing either sgRNA1 or sgRNA2 using Lipofectamine 2000 reagent according to the manufacturer's protocol (Invitrogen). For MOCK cells, cell lines were transfected with a modified version of the Cas9 nickase expression vector pSpCas9n(BB)-2A-Puro (pX462; Addgene plasmid #48141; www.addgene.org) containing only sgRNA1. Transfected cells were selected 24 hours later by addition of puromycin to the culture medium (2.5 μg/mL). Cell viability of passaged cell lines was determined daily for 96 hours by MTT assay, or cell proliferation was assessed by cell counting.
Quantitative Reverse-Transcriptase PCR
Total RNA was isolated from primary ECC cell lines using the RNeasy kit (Qiagen) following the manufacturer's instructions and reverse transcribed (Thermo Fisher Scientific). Relative mRNA quantification using cyclophilin as reference gene was performed by real-time PCR analysis (TaqMan, PE Applied Biosystems). Sequence information of the qRT-PCR primers is provided in Supplementary Methods.
Cell-Cycle Analysis by Flow Cytometry
To synchronize the cell cultures, cells were seeded in 10-cm dishes in growth medium with 10% FCS overnight. The cultures were then washed by PBS and changed to serum-free medium. After 24 hours of serum starvation, the cells were passaged and released into cell cycle by the addition of serum for 6 hours. Cells were trypsinized and washed two times with PBS followed by fixation in 1 mL cold 70% EtOH. The EtOH was removed after 24 hours, and cells were washed with PBS. Cells were stained with DAPI (Sigma, 1:6,000) and acquired using Cytoflex LX (Beckman Coulter). Flow cytometry data were analyzed using FlowJo software (Version 10.6.2).
Preparation of Total Cell Lysates and Immunoblotting
Whole-cell lysates were prepared, and isolated proteins were separated in SDS-polyacrylamide gels, transferred to polyvinylidene difluoride membranes, and incubated at 4°C overnight with the following primary antibodies: pAKT-T308 (#2965, RRID:AB_2255933), pAKT-S473 (#4060, RRID:AB_2315049), pan-AKT (#4691, RRID:AB_915783), p-S6-S235/236 (#4858, RRID:AB_916156; all from Cell Signaling Technology) and p27Kip1 (sc528, RRID:AB_632129), Skp2 (sc7164, RRID:AB_2187650), and Hsp90 alpha/beta (sc13119, RRID:AB_675659; all from Santa Cruz Biotechnology). Primary antibodies were followed by Alexa680-coupled secondary antibodies (Thermo Fisher Scientific) and detected by the Odyssey Infrared Imaging System (Licor).
Data Analysis
Unless otherwise indicated, all data were determined from at least three independent experiments and expressed as mean values ± SEM. GraphPad Prism (versions 5 and 8) was used for graphical presentation of the data and calculation of statistical tests except for the Wilcoxon rank sum test. The Wilcoxon rank sum test was computed in R Language. Statistical tests and P values are provided in the figure legends. Values of P = 0.05 or less were considered to be statistically significant.
Authors' Disclosures
K. Steiger reports other support from Roche and TRIMT outside the submitted work, as well as a patent for radiopharmaceutical issued. S. Roessler reports grants from German Cancer Aid, German Research Foundation, and PSC Partners Seeking a Cure Foundation outside the submitted work. A. Bradley reports other support from Kymab outside the submitted work. M. Reichert reports personal fees from Roche, Celgene, and Falk outside the submitted work. W. Weichert reports personal fees from Roche, MSD, Bristol Myers Squibb, AstraZeneca, Pfizer, Merck, Lilly, Boehringer, Novartis, Takeda, Bayer, Amgen, Astellas, Eisai, Illumina, Siemens, Agilent, ADC, GlaxoSmithKline, and Molecular Health and grants from Roche, MSD, Bristol Myers Squibb, and AstraZeneca outside the submitted work. O.J. Sansom reports grants from Novartis, AstraZeneca, and Cancer Research Technology; personal fees from Boehringer Ingelheim; and other support from Ionctura outside the submitted work. J.P. Morton reports grants from CRUK during the conduct of the study, as well as grants from Cancer Research UK, Pancreatic Cancer UK, and Medical Research Council outside the submitted work. G. Schneider reports grants from Deutsche Forschungsgemeinschaft and German Cancer Consortium (DKTK) during the conduct of the study. D. Saur reports grants from Deutsche Forschungsgemeinschaft (DFG SA 1374/4-3 and SFB 1321 Project-ID 329628492), European Research Council, and German Cancer Consortium during the conduct of the study, as well as personal fees and nonfinancial support from Novartis and Amgen, and nonfinancial support from Johnson & Johnson outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Supplementary graphical abstract, eight Supplemental Figures, Supplementary Methods
Common insertion sites of piggyBac transposon mutagenesis screen.
Acknowledgments
We would like to thank Dr. J. Roberts, Dr. T. Jacks, Dr. J. Jonkers, Dr. A. Berns, Dr. A. Lowy, Dr. T. Mak, Dr. L. Luo, Dr. H. Zeng, and Dr. D. Tuveson for providing transgenic animals and J. Götzfried, M. Mielke, A. Hering, and O. Seelbach for excellent technical assistance. This work was supported by funding from Deutsche Forschungs-gemeinschaft (DFG SA 1374/4-3 to D. Saur, SFB 1321 Project-ID 329628492, P06, P11, P13 to M. Reichert, M. Schmidt-Supprian, G. Schneider, and D. Saur, and S1 to W. Weichert and K. Steiger), SFB 1371 Project-ID 395357507 P12 to D. Saur), the German Cancer Consortium (DKTK to K. Steiger, M. Schmidt-Supprian, W. Weichert, R. Rad, G. Schneider, and D. Saur), the European Research Council (ERC CoG No. 648521 to D. Saur), Cancer Research UK (CRUK) core funding to the CRUK Beatson Institute (A17196), and CRUK core funding to O.J. Sansom (A21139).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors' Contributions
C. Falcomatà: Data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. S. Bärthel: Data curation, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. A. Ulrich: Formal analysis, investigation, methodology. S. Diersch: Investigation, methodology. C. Veltkamp: Investigation. L. Rad: Conceptualization, formal analysis, supervision, investigation, methodology. F. Boniolo: Data curation, software, formal analysis, investigation, visualization. M. Solar: Resources, investigation. K. Steiger: Formal analysis. B. Seidler: Conceptualization, resources, formal analysis, validation, investigation. M. Zukowska: Resources, investigation. J. Madej: Resources. M. Wang: Investigation. R. Öllinger: Resources, data curation, formal analysis, investigation. R. Maresch: Resources, investigation. M. Barenboim: Software, formal analysis. S. Eser: Investigation. M. Tschurtschenthaler: Investigation. A. Mehrabi: Resources. S. Roessler: Resources, data curation, formal analysis. B. Goeppert: Resources, data curation, investigation. A. Kind: Writing–original draft. A. Schnieke: Resources, supervision, writing–original draft. M.S. Robles: Resources, funding acquisition, validation, investigation. A. Bradley: Resources. R.M. Schmid: Resources. M. Schmidt-Supprian: Resources, funding acquisition, validation, investigation. M. Reichert: Resources, investigation. W. Weichert: Resources, investigation. O.J. Sansom: Resources, data curation, investigation. J.P. Morton: Resources, data curation, investigation. R. Rad: Resources, software, formal analysis, supervision, validation, investigation, methodology. G. Schneider: Conceptualization, resources, supervision, funding acquisition, validation, investigation, writing–review and editing. D. Saur: Conceptualization, resources, formal analysis, supervision, funding acquisition, validation, investigation, methodology, writing–original draft, writing–review and editing.
References
- 1. Banales JM, Marin JJG, Lamarca A, Rodrigues PM, Khan SA, Roberts LRet al. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol 2020;17:557–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology 2013;145:1215–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato Met al. Genomic spectra of biliary tract cancer. Nat Genet 2015;47:1003–10. [DOI] [PubMed] [Google Scholar]
- 4. Zong Y, Stanger BZ. Molecular mechanisms of bile duct development. Int J Biochem Cell Biol 2011;43:257–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Spence JR, Lange AW, Lin S-CJ, Kaestner KH, Lowy AM, Kim Iet al. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Dev Cell 2009;17:62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- 7. Tyson GL, El-Serag HB. Risk factors for cholangiocarcinoma. Hepatology 2011;54:173–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 9. Tian W, Hu W, Shi X, Liu P, Ma X, Zhao Wet al. Comprehensive genomic profile of cholangiocarcinomas in China. Oncol Lett 2020;19:3101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lowery MA, Ptashkin R, Jordan E, Berger MF, Zehir A, Capanu Met al. Comprehensive molecular profiling of intrahepatic and extrahepatic cholangiocarcinomas: potential targets for intervention. Clin Cancer Res 2018;24:4154–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fujimoto A, Furuta M, Shiraishi Y, Gotoh K, Kawakami Y, Arihiro Ket al. Whole-genome mutational landscape of liver cancers displaying biliary phenotype reveals hepatitis impact and molecular diversity. Nat Commun 2015;6:6120. [DOI] [PubMed] [Google Scholar]
- 12. Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet 2014;383:2168–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goeppert B, Konermann C, Schmidt CR, Bogatyrova O, Geiselhart L, Ernst Cet al. Global alterations of DNA methylation in cholangiocarcinoma target the Wnt signaling pathway. Hepatology 2014;59:544–54. [DOI] [PubMed] [Google Scholar]
- 14. Rad R, Rad L, Wang W, Strong A, Ponstingl H, Bronner IFet al. A conditional piggyBac transposition system for genetic screening in mice identifies oncogenic networks in pancreatic cancer. Nat Genet 2015;47:47–56. [DOI] [PubMed] [Google Scholar]
- 15. Rad R, Rad L, Wang W, Cadinanos J, Vassiliou G, Rice Set al. PiggyBac transposon mutagenesis: a tool for cancer gene discovery in mice. Science 2010;330:1104–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MAet al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 2003;4:437–50. [DOI] [PubMed] [Google Scholar]
- 17. Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu Het al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 2010;13:133–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis 2007;45:593–605. [DOI] [PubMed] [Google Scholar]
- 19. Chong DQ, Zhu AX. The landscape of targeted therapies for cholangiocarcinoma: current status and emerging targets. Oncotarget 2016;7:46750–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rizvi S, Borad MJ, Patel T, Gores GJ. Cholangiocarcinoma: molecular pathways and therapeutic opportunities. Semin Liver Dis 2014;34:456–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deshpande V, Nduaguba A, Zimmerman SM, Kehoe SM, Macconaill LE, Lauwers GYet al. Mutational profiling reveals PIK3CA mutations in gallbladder carcinoma. BMC Cancer 2011;11:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yoo KH, Kim NKD, Kwon WI, Lee C, Kim SY, Jang Jet al. Genomic alterations in biliary tract cancer using targeted sequencing. Transl Oncol 2016;9:173–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zou S, Li J, Zhou H, Frech C, Jiang X, Chu JSCet al. Mutational landscape of intrahepatic cholangiocarcinoma. Nat Commun 2014;5:5696. [DOI] [PubMed] [Google Scholar]
- 24. Eser S, Reiff N, Messer M, Seidler B, Gottschalk K, Dobler Met al. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell 2013;23:406–20. [DOI] [PubMed] [Google Scholar]
- 25. Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer 2005;5:921–9. [DOI] [PubMed] [Google Scholar]
- 26. Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer 2011;11:761–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zen Y, Adsay NV, Bardadin K, Colombari R, Ferrell L, Haga Het al. Biliary intraepithelial neoplasia: an international interobserver agreement study and proposal for diagnostic criteria. Mod Pathol 2007;20:701–9. [DOI] [PubMed] [Google Scholar]
- 28. Baer R, Cintas C, Dufresne M, Cassant-Sourdy S, Schönhuber N, Planque Let al. Pancreatic cell plasticity and cancer initiation induced by oncogenic Kras is completely dependent on wild-type PI 3-kinase p110α. Genes Dev 2014;28:2621–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Esposito I, Schirmacher P. Pathological aspects of cholangiocarcinoma. HPB (Oxford) 2008;10:83–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O'Dell MR, Huang JL, Whitney-Miller CL, Deshpande V, Rothberg P, Grose Vet al. Kras(G12D) and p53 mutation cause primary intrahepatic cholangiocarcinoma. Cancer Res 2012;72:1557–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya Ret al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 2001;15:3243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle Bet al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci U S A 2010;107:246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Caldwell ME, DeNicola GM, Martins CP, Jacobetz MA, Maitra A, Hruban RHet al. Cellular features of senescence during the evolution of human and murine ductal pancreatic cancer. Oncogene 2012;31:1599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schofield HK, Zeller J, Espinoza C, Halbrook CJ, del Vecchio A, Magnuson Bet al. Mutant p53R270H drives altered metabolism and increased invasion in pancreatic ductal adenocarcinoma. JCI Insight 2018;3:e97422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet 2001;29:418–25. [DOI] [PubMed] [Google Scholar]
- 36. Sarver AL, Erdman J, Starr T, Largaespada DA, Silverstein KAT. TAPDANCE: an automated tool to identify and annotate transposon insertion CISs and associations between CISs from next generation sequence data. BMC Bioinformatics 2012;13:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lo Re AE, Fernandez-Barrena MG, Almada LL, Mills LD, Elsawa SF, Lund Get al. Novel AKT1-GLI3-VMP1 pathway mediates KRAS oncogene-induced autophagy in cancer cells. J Biol Chem 2012;287:25325–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas Jet al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Suzuki A, Yamaguchi MT, Ohteki T, Sasaki T, Kaisho T, Kimura Yet al. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity 2001;14:523–34. [DOI] [PubMed] [Google Scholar]
- 40. Tumaneng K, Schlegelmilch K, Russell RC, Yimlamai D, Basnet H, Mahadevan Net al. YAP mediates crosstalk between the Hippo and PI(3)K-TOR pathways by suppressing PTEN via miR-29. Nat Cell Biol 2012;14:1322–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yim EK, Peng G, Dai H, Hu R, Li K, Lu Yet al. Rak functions as a tumor suppressor by regulating PTEN protein stability and function. Cancer Cell 2009;15:304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat Rev Cancer 2008;8:253–67. [DOI] [PubMed] [Google Scholar]
- 43. Masuda K, Ishikawa Y, Onoyama I, Unno M, de Alboran IM, Nakayama KIet al. Complex regulation of cell-cycle inhibitors by Fbxw7 in mouse embryonic fibroblasts. Oncogene 2010;29:1798–809. [DOI] [PubMed] [Google Scholar]
- 44. Lee YH, Seo D, Choi KJ, Andersen JB, Won MA, Kitade Met al. Antitumor effects in hepatocarcinoma of isoform-selective inhibition of HDAC2. Cancer Res 2014;74:4752–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CHet al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature 2010;464:374–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lin HK, Wang G, Chen Z, Teruya-Feldstein J, Liu Y, Chan CHet al. Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of Skp2 by Akt/PKB. Nat Cell Biol 2009;11:420–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol 2010;11:9–22. [DOI] [PubMed] [Google Scholar]
- 48. Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov 2014;13:140–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pang M, Ma L, Liu N, Ponnusamy M, Zhao TC, Yan Het al. Histone deacetylase 1/2 mediates proliferation of renal interstitial fibroblasts and expression of cell cycle proteins. J Cell Biochem 2011;112:2138–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang J, Yuan D, Li J, Zheng S, Wang B. miR-186 downregulates protein phosphatase PPM1B in bladder cancer and mediates G1-S phase transition. Tumour Biol 2016;37:4331–41. [DOI] [PubMed] [Google Scholar]
- 51. Churi CR, Shroff R, Wang Y, Rashid A, Kang HC, Weatherly Jet al. Mutation profiling in cholangiocarcinoma: prognostic and therapeutic implications. PLoS One 2014;9:e115383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee-Hoeflich ST, Pham TQ, Dowbenko D, Munroe X, Lee J, Li Let al. PPM1H is a p27 phosphatase implicated in trastuzumab resistance. Cancer Discov 2011;1:326–37. [DOI] [PubMed] [Google Scholar]
- 53. Jarnagin WR, Klimstra DS, Hezel M, Gonen M, Fong Y, Roggin Ket al. Differential cell cycle-regulatory protein expression in biliary tract adenocarcinoma: correlation with anatomic site, pathologic variables, and clinical outcome. J Clin Oncol 2006;24:1152–60. [DOI] [PubMed] [Google Scholar]
- 54. Diersch S, Wenzel P, Szameitat M, Eser P, Paul MC, Seidler Bet al. Efemp1 and p27(Kip1) modulate responsiveness of pancreatic cancer cells towards a dual PI3K/mTOR inhibitor in preclinical models. Oncotarget 2013;4:277–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kelly-Spratt KS, Philipp-Staheli J, Gurley KE, Hoon-Kim K, Knoblaugh S, Kemp CJ. Inhibition of PI-3K restores nuclear p27Kip1 expression in a mouse model of Kras-driven lung cancer. Oncogene 2009;28:3652–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fero ML, Rivkin M, Tasch M, Porter P, Carow CE, Firpo Eet al. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell 1996;85:733–44. [DOI] [PubMed] [Google Scholar]
- 57. Fiorentino M, Altimari A, D'Errico A, Gabusi E, Chieco P, Masetti Met al. Low p27 expression is an independent predictor of survival for patients with either hilar or peripheral intrahepatic cholangiocarcinoma. Clin Cancer Res 2001;7:3994–9. [PubMed] [Google Scholar]
- 58. Jones RP, Bird NTE, Smith RA, Palmer DH, Fenwick SW, Poston GJet al. Prognostic molecular markers in resected extrahepatic biliary tract cancers; a systematic review and meta-analysis of immunohistochemically detected biomarkers. Biomark Med 2015;9:763–75. [DOI] [PubMed] [Google Scholar]
- 59. Ruys AT, Groot Koerkamp B, Wiggers JK, Klümpen HJ, ten Kate FJ, van Gulik TM. Prognostic biomarkers in patients with resected cholangiocarcinoma: a systematic review and meta-analysis. Ann Surg Oncol 2014;21:487–500. [DOI] [PubMed] [Google Scholar]
- 60. Schneider G, Schmidt-Supprian M, Rad R, Saur D. Tissue-specific tumorigenesis: context matters. Nat Rev Cancer 2017;17:239–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tuveson D, Hanahan D. Translational medicine: cancer lessons from mice to humans. Nature 2011;471:316–7. [DOI] [PubMed] [Google Scholar]
- 62. Jonkers J, Berns A. Conditional mouse models of sporadic cancer. Nat Rev Cancer 2002;2:251–65. [DOI] [PubMed] [Google Scholar]
- 63. Eser S, Messer M, Eser P, von Werder A, Seidler B, Bajbouj Met al. In vivo diagnosis of murine pancreatic intraepithelial neoplasia and early-stage pancreatic cancer by molecular imaging. Proc Natl Acad Sci U S A 2011;108:9945–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lee H, Wang K, Johnson A, Jones DM, Ali SM, Elvin JAet al. Comprehensive genomic profiling of extrahepatic cholangiocarcinoma reveals a long tail of therapeutic targets. J Clin Pathol 2016;69:403–8. [DOI] [PubMed] [Google Scholar]
- 65. Huang X, Dixit VM. Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res 2016;26:484–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hruban RH, Adsay NV, Albores-Saavedra J, Anver MR, Biankin AV, Boivin GPet al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: consensus report and recommendations. Cancer Res 2006;66:95–106. [DOI] [PubMed] [Google Scholar]
- 67. Albores-Saavedra J, Henson DE, Klimstra DS. Tumors of the gallbladder, extrahepatic bile ducts, and Vaterian system. Silver Spring (MD)American Registry of Pathology; 2015. [Google Scholar]
- 68. von Burstin J, Eser S, Paul MC, Seidler B, Brandl M, Messer Met al. E-cadherin regulates metastasis of pancreatic cancer in vivo and is suppressed by a SNAIL/HDAC1/HDAC2 repressor complex. Gastroenterology 2009;137:361–71. [DOI] [PubMed] [Google Scholar]
- 69. Mueller S, Engleitner T, Maresch R, Zukowska M, Lange S, Kaltenbacher Tet al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature 2018;554:62–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Glass K, Huttenhower C, Quackenbush J, Yuan GC. Passing messages between biological networks to refine predicted interactions. PLoS One 2013;8:e64832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary graphical abstract, eight Supplemental Figures, Supplementary Methods
Common insertion sites of piggyBac transposon mutagenesis screen.