Abstract
Lactate is an abundant oncometabolite in the tumor environment. In prostate cancer (PCa), cancer-associated fibroblasts are major contributors of secreted lactate, which can be taken up by cancer cells to sustain mitochondrial metabolism. However, how lactate impacts transcriptional regulation in tumors has yet to be fully elucidated. Here, we describe a mechanism by which CAF-secreted lactate is able to increase the expression of genes involved in lipid metabolism in PCa cells. This regulation enhanced intracellular lipid accumulation in lipid droplets (LD) and provided acetyl moieties for histone acetylation, establishing a regulatory loop between metabolites and epigenetic modification. Inhibition of this loop by targeting the bromodomain and extraterminal (BET) protein family of histone acetylation readers suppressed the expression of perilipin-2 (PLIN2), a crucial component of LDs, disrupting lactate-dependent lipid metabolic rewiring. Inhibition of this CAF-induced metabolic-epigenetic regulatory loop in vivo reduced growth and metastasis of prostate cancer cells, demonstrating its translational relevance as a therapeutic target in PCa. Clinically, PLIN2 expression was elevated in tumors with a higher Gleason grade and in castration resistant prostate cancer compared to primary PCa. Overall, these findings show that lactate has both a metabolic and an epigenetic role in promoting PCa progression.
Keywords: histone acetylation, cancer-associated fibroblasts, lactate metabolism, lipid droplets, prostate cancer
Introduction
Collaborative interactions between neoplastic and stromal cells dictate cancer initiation and progression in a plethora of tumors (1). Cancer cells generally exhibit profound alterations in their metabolism, in particular they reveal a high degree of flexibility in the use of nutrients available in the tumor microenvironment often provided by corrupted stromal cells. We and others have demonstrated that lactate, which is abundant in several tumors (2–4) influence the metabolism of cancer cells. In several cancer models, the secretion of lactate as the by-product of fermentative metabolism of a given cell population (i.e. hypoxic cancer cells (5,6); endothelial cells (6), cancer-associated fibroblasts (CAFs) (7)) is functional to sustain the metabolism of the oxidative counterpart, such as cancer cells. In keeping, environmental lactate is preferentially uploaded by lung, prostate and pancreatic cancer cells (7–9) and exploited to sustain catabolic pathways. However, the role of lactate is not limited to sustain the oxidative metabolism (7). Indeed, we have recently demonstrated that the CAF-secreted lactate supports prostate cancer (PCa) progression by acting as an immunosuppressive agent (10) and by promoting a SIRT1-dependent PGC-1α activation and subsequent enhancement of mitochondrial mass and activity (11). Both lactate-mediated effects foster PCa cell invasiveness and malignancy in vivo. Interestingly, enhanced lactate transport and predominant usage of its carbons moieties correlate with a high likelihood of distant metastasis and worse outcome in cancer patients (8).
Several evidence point out that metabolites can be substrates of enzymes that carry out post-translational modifications thereby serving as regulators of gene expression and impacting on cell function (12). In addition to the role of other tricarboxylic acid (TCA) cycle intermediates to regulate histone and DNA methylation (13), acetyl-CoA is a critical metabolite within tumor metabolic pathways as it acts as fatty acid (FA) and cholesterol biosynthetic intermediate as well as the universal donor of acetyl groups, thereby supporting tumor cell TCA cycle and contributing to the histone acetylation, a key event in the epigenetic regulation of gene transcription (14). As lipid‐derived acetyl‐CoA is a major source of carbons for histone acetylation (15), alterations in lipid metabolism (either in anabolic or catabolic processes) are emerging as recurrent traits in several cancers (16). The stromal-driven metabolic and epigenetic regulation is therefore an interesting area of research in oncology as it could unveil innovative vulnerabilities with therapeutic implications.
Here we demonstrate that CAF-secreted lactate induces a lipid metabolic reprogramming that culminates into the formation and mobilization of lipid droplets (LDs), concurring to enhance the invasive abilities of PCa cells. In particular, we documented that lactate-dependent lipid metabolic reprogramming is responsible for epigenetic modifications in aggressive PCa. Crucially, targeting the bromodomain and extraterminal (BET) proteins family of acetylation readers, blocks the lactate-induced epigenetic and metabolic rewiring, ultimately leading to the impairment of PCa metastases.
Methods
Cell lines
Human PCa cells (DU145, RRID:CVCL_0105; PC3, RRID:CVCL_0035; 22Rv1, RRID:CVCL_1045; and LNCaP, RRID:CVCL_4783) were obtained from ATCC. Human prostate fibroblasts (healthy prostate fibroblasts – HPFs – isolated from benign prostate hyperplasia and CAFs) were isolated from surgical explants after patient informed consent, according to the Ethics Committee of the Azienda Ospedaliera Universitaria Careggi (Florence, Italy). Clinicopathological analysis to discriminate hyperplasia from carcinoma was performed according to the presence/absence of benign markers (CK5/6, p63, HMWCK 34βE12) and the preoperative PSA levels. CAFs specimens were collected and selected from patients according to the Gleason score, PSA levels, positivity for specific marker (i.e.FAP) (Table1). All cells were maintained in DMEM (#ECB7501L; Euroclone) supplemented with 10% fetal bovine serum (#ECB4004L; Euroclone), 2 mM L-Glutamine and 1% penicillin/streptomycin, except for PC3 or LNCaP that were maintained in Nutrient Mixture F-12 Ham (#N4888; Merck Millipore) and RPMI (#ECB2000L). All cell lines were maintained at 37°C and 5% CO2 and were routinely tested for mycoplasma contamination with the MycoAlert™ Mycoplasma Detection kit (#LT07-318; Lonza). DU145 shBRD4 cells were previously described (17). Cell lines and primary cells once thawed were kept in culture for a maximum of 20 and 12 passages, respectively.
Table 1. Source of CAFs.
| SAMPLE | AGE | PSA (ng/mL) | Gleason score | FAP |
|---|---|---|---|---|
| Patient #1 | 70 | 9,62 | 4+5 | |
| Patient #2 | 73 | 1,01 | 3+4 | |
| Patient #3 | 72 | 3,4 | 4+5 | |
| Patient #4 | 74 | 5,68 | 3+4 | ++ |
| Patient #5 | 72 | 14,58 | 4+4 | ++ |
| Patient #6 | 70 | 10,6 | 4+5 | |
| Patient #7 | 71 | 6,1 | 3+3 | ++ |
| Patient #8 | 72 | 9,39 | 4+5 | |
| Patient #9 | 73 | 13,3 | 3+3 | -/+ |
| Patient #10 | 75 | 7,18 | 4+5 | |
| Patient #11 | 71 | 8,93 | 3+4 | |
| Patient #12 | 73 | 17,76 | 3+4 | |
| Patient #13 | 71 | 7,18 | 3+4 | |
| Patient #14 | 70 | 16,4 | 4+4 | |
| Patient #15 | 71 | 10,6 | 3+4 | |
| Patient #16 | 71 | 5,04 | 4+5 | ++ |
| Patient #17 | 74 | 4,95 | 3+3 | ++ |
| Patient #18 | 70 | 10,31 | 3+3 |
Conditioned media from fibroblasts
HPFs and CAFs were grown to sub-confluence and treated for 48 hours with serum-free medium to obtain the corresponding Conditioned Media (CM) which is filtered and used fresh or stored for further analysis.
Cell treatments and transfection
Cell treatments were performed in HPF- or CAF-CM or in serum-free culture medium supplemented with 15 mM lactic acid. Unless specified otherwise, all reagents were obtained from Merck Millipore. The following inhibitors were used in this study: 40 μM MCT1 inhibitor AR-C155858 (#4960; Tocris BioScience), 25 μM ATGListatin, 40 μM Etomoxir, 10 μM SB-204990, 100 nM I-BET762 (I-BET762) inhibitor, 50-100 nM Simvastatin. Cells were transfected with a pool of siRNA targeting human ACLY and PLIN2 (esiRNA #EHU081921 and #EHU105291; Merck Millipore) using Lipofectamine™ RNAiMAX transfection reagent (#13778150; Thermo Fisher), according to manufacturer’s instructions. shPLIN2 cells were obtained by infecting with lentiviral particles (PLIN2 #TL314932V, OriGene Technologies Inc., Rockville, MD, USA) for 24-48h. Fresh medium containing puromycin (1 μg/mL) (#P8833; Merck Millipore) was added after 24 h and cells were selected as stable pools before use.
Lipid droplets staining
Glass coverslip-plated cells were stained with BODIPY 493/503 (#D3922; Thermo Fisher Scientific) for 15 min at 37°C and then fixed with 4% paraformaldehyde (PFA) for 10 min. For nuclei staining, PFA-fixed cells were incubated with DAPI (#D3571, Thermo Fisher) for 10 min at room temperature. Sample images were acquired using TCS SP8 microscope (Leica Microsystems) with LAS-AF image acquisition software. The quantification of lipid droplets was performed using FiJi software: an outline was drawn around each cell and spots were counted in at least 50 cells derived from three representative 63x images from 3 independent experiments.
RNA extraction and qRT-PCR
Total RNA was extracted using RNeasy Kit (#74104; Qiagen) and cDNA synthesis was performed using iScript™ cDNA Synthesis Kit (#1708891; Bio-Rad). qRT-PCR was carried out by CFX96 Touch Real-Time PCR Detection System (Bio-Rad) using TaqMan assays (Applied Biosystems). The following probes were used in this study: PLIN2 (#Hs00605340_m1, Life Technologies). Data were normalized on HPRT1 (#Hs02800695_m1, Life Technologies).
Western blot analysis
Protein samples were denatured for 10 min at 95°C with Laemmli sample buffer and protein concentration was measured by BCA kit. Samples (20-25 μg) were then loaded on 4-20% acrylamide precast SDS-PAGE gels (Bio-Rad #456-8096) and then transferred on PVDF membrane by Trans-Blot Turbo Transfer Pack (Bio-Rad #1704157). The membranes were activated with methanol and then incubated with the appropriate antibodies overnight at 4°C. The following primary antibodies used in this study were applied in 5% milk overnight at 4°C: rabbit anti-H3K27Ac (1:1000; #8173, RRID:AB_10949503), anti-H3K9Ac (1:1000; #9649), anti-ACC (1:500; #3676) and anti-pACC (1:1000; #11818, RRID:AB_2687505), mouse anti-H3 (1:2000; #4499, RRID:AB_10544537) (Cell Signalling Technology), mouse anti-ACLY (1:1000; MA5-17027, RRID:AB_2538499; Thermo Fisher Scientific), mouse anti-FASN (1:1000; sc-48357, RRID:AB_627584), mouse anti-pACLY (1:1000; sc-374647, RRID:AB_10988728; Santa Cruz Biotechnology) and anti-actin (1:1000; sc-47778, RRID:AB_2714189; Santa Cruz Biotechnology), rabbit anti-PLIN2 (1:500; ab78920, RRID:AB_2040415; Abcam).
ChIP and ChIP-qPCR
ChIP experiments were performed by using Pierce™ Magnetic ChIP Kit (#26157; Thermo Fisher Scientific). Briefly, cells were crosslinked with 1% formaldehyde for 10 min followed by quenching with 0.125 M Glycine for 5 min. Post-wash, 3x106 cells were scraped and resuspended in 900 μL of nuclear lysis buffer, incubated on ice for 20 min and disrupted in a Q500 sonicator (Qsonica) at 4°C (30 sec on/off pulses). After verification on agarose gel of the right size of chromatin shearing (approximately 200-400 bp), the samples were centrifuged, and supernatant collected in a fresh tube after removal of 10% input samples. Chromatin was incubated with H3K27Ac antibody (or corresponding IgG) overnight at 4°C. Next day A/G magnetic beads were added, and incubation continued for an additional 4 hours, followed by sequential washes. In parallel with the input samples, the eluted chromatin was subjected to RNase treatment, reverse crosslinked, followed by Proteinase K digestion for 4 hours heated at 65°C to reverse the cross-linking. DNA was recovered by applying the samples on provided columns. The final DNA was dissolved in 50 μL nuclease-free water for ChIP-qPCR. The primers used for evaluating PLIN2 promoter in SYBR-Green qPCR are forward 5’-GCTAGTCAGGTTGGAAAGTAAGC-3’ and reverse 5’-GCTGCCAAATCTTAGAATGAGG-3’.
Invasion assay
Invasion assay was performed using 8-μm-pore transwell (#3428; Corning) coated with reconstituted Matrigel (#356334; BD Biosciences). Cells were seeded (1x 105/well), in the upper chamber of the transwell in serum-free medium and allowed to invade for 16 hours toward complete medium. Air-dried membranes were stained with DiffQuick solution (#726443; BD Bioscience) and invasion was evaluated by counting invading cells to the lower surface of the filters (six randomly chosen fields).
Radioactive assays
To analyze the incorporation of radioactive lactate into lipids or histones, culture media were changed to a serum-free medium and then supplemented with 1 μCi 14C-lactic acid (Perkin Elmer #NEC599050UC) 4 hours prior to the end of the experiment. For radioactive incorporation into lipids, cells were washed three times in ice cold PBS and then lysed in methanol. Samples were first resuspended in 4 volumes of a CHCl3:MeOH (1:1) solution and then an additional volume of dH2O was added. The solution was then centrifuged at 1000 rpm for 5 min at room temperature. The lower phase was collected, transferred to a scintillation vial and counted on the scintillation counter. For radioactive incorporation into histones, cells were lysed in cold NIB buffer (5 mM MgCl2, 1 mM CaCl2, 15 mM Tris-HCl, 60 mM KCl, 15 mM NaCl, 250 mM sucrose, 10 mM sodium butyrate, 1 mM DTT, protease inhibitors) supplemented with 0.2% NP-40 and nuclei were harvested by stroking 3 times every 10 min with 20-Gauge needle. Samples were centrifuged (3000 rpm for 5 min at 4°C) and pellet was resuspended in H2SO4. Samples were centrifuged (13000 rpm 10 min at 4°C) and the upper aqueous phase was collected and resuspended with 33% TCA. The resulting pellet was washed twice with 100% acetone and allowed to dry. The pellet was resuspended in Laemmli sample buffer and radioactivity signal was measured normalized on protein content.
Seahorse XFe96 MitoStress Test
Cells were seeded in XFe96 cell culture plate with 2.5x104 cells per well in full medium and incubated overnight at 37°C. 24 hours post-seeding cells were treated as indicated. Oxygen consumption rate (OCR) was real-time measured with Seahorse XFe96 plate reader (Agilent Technologies) in basal conditions and upon the addiction of compounds interfering with mitochondrial respiration: oligomycin (1.5 μM), FCCP (1 μM) and rotenone/antimycin A (0.5 μM). Protein quantification was used to normalize the results. Basal respiration is calculated as last rate measurement before injection – non-mitochondrial respiration rate. Maximal respiration is calculated as the maximum rate measurement after FCCP injection – non-mitochondrial respiration.
RNA sequencing analysis
The RNA-seq analysis was performed to investigate the expression profiles of DU145 cells in 4 conditions: untreated cells, cells exposed to lactate, cells treated with I-BET762, cells exposed to lactate and treated with I-BET762. For each condition, four technical replicates were produced. Total RNA was extracted using RNeasy Kit (#74104, Qiagen) according to manufacturer’s instructions. RNA quantity and quality were evaluated with a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific). RNA sequencing for all experiments was performed at the Institute of Oncology Research using Next Ultra II Directional RNA Library Prep Kit for Illumina starting from 800 ng of total RNA each sample and sequenced on the Illumina NextSeq500 with single-end, 75 bp long reads.
Alignment and quantification
The read quality was accessed through the FastQC tool (18) (18) (RRID:SCR_014583). Reads were aligned to the reference genome using STAR 2.6.1. along with the release of human genome assembly (GRCh38.p12,v29) (19). Nucleotide sequences of the GRCh38.p12 genome contain all regions, including reference chromosomes, scaffolds, assembly patches and haplotypes. The reads were quantified at the gene level using GTF Gencode annotations (Comprehensive gene annotation).
Downstream transcriptomic analysis
All the following bioinformatic analyses were performed in R environment (20) (20).
Outlier identification
Before the downstream analysis a preliminary analysis aimed to investigate the conformity of the data was performed. One sample from lactate group completely diverged from the other replicates of the same group and from all the other samples. Network-based approach proposed by Oldham et al. (21) was adopted to determine if this sample should be discarded from the further analyses. The selected criterium to consider a sample an outlier is: standardized sample connectivity lower than -2 and since in the replicate 1 of lactate group this value is equal to -3, this sample was removed (Refer to the pdf attachment “Outlier Analysis” for the images and more details about the network-based approach proposed by Oldham at al.).
Gene expression levels and differential expression results (baseMean, log2FoldChange, Wald statistic, p-value) of the considered comparisons (Lactate vs Untreated, Lactate+I-BET762 vs Lactate) were retrieved according to DESeq pipeline (22). The p-value threshold was set at 0.05 and at 0.1 for Lactate vs Untreated and Lactate+I-BET762 vs Lactate comparisons respectively. Enrichment analysis was performed through the cameraPR function from Limma package (23) (RRID:SCR_010943): the accounted genes are the ones that are significantly differentially expressed and are weighted (statistic parameter of cameraPR function) according to the Wald statistic value obtained from the differential expression analysis. The p-value cut-off was set at 0.05. The Benjamini-Hochberg multiple correction method was applied to all multiple tests. The enrichment plots were produced through the “GSEAPreranked” tool (24): for the enrichment statistic the exploited weights were the Wald statistic values. Volcano plot, heatmap and violin plot images were made with pheatmap (25) (25) and ggplot2 R packages (26) (RRID:SCR_014601).
Network analysis
The network analysis was performed using Cytoscape_v3.8.2 (27) (RRID:SCR_003032). The network was constructed considering a confidence cutoff equal to 0.4 and a maximum additional interactor equal to 0. The graph nodes were represented by the genes resulted significantly differentially expressed with a log2FoldChange lower than -0.5 or higher thn 0.5 and the edges (connections between nodes) were established according to the functional interactions between the corresponding proteins that are collected in the STRING database (28) (RRID:SCR_005223). The network parameters (betweenness centrality, closeness centrality, degree) were computed through the “NetworkAnalyzer” tool of Cytoscape.
Metabolomics
Gas Chromatography–Mass Spectrometry (GC–MS)
For SCAN mode, 1x106 cells were collected and subjected to extraction using a mixture of CHCl3:MeOH:H2O (1:1:1), before quenching with ice-cold MeOH:H2O (1:1) containing norvaline, used as internal standard. Snap frozen tissue samples were minced and subjected to the same extraction mixture. CHCl3 was then added, and the samples were vortexed at 4°C for 30 min, centrifuged at 3000g for 10 min, and the aqueous phase was collected and allowed to evaporate at room temperature. Dried polar metabolites were dissolved in 60μL of 2% methoxyamine hydrochloride in pyridine (Thermo Fisher), and held at 30°C for 2 hours. After dissolution and reaction, 90μL N-Trimethylsilyl-N-methyl trifluoroacetamide (MSTFA) + 1% trimethylchlorosilane TMCS were added and samples were incubated at 37°C for 60 min. Gas chromatographic runs were performed with helium as carrier gas at 0.6mL/min. The split inlet temperature was set to 250°C and the injection volume to 1μL. A split ratio of 1:10 was used. The GC oven temperature ramp was from 60 to 325°C at 10°C/min. The data acquisition rate was 10Hz. For the Quadrupole, an Electron Ionization (EI) source (70eV) was used, and full-scan spectra (mass range from 50 to 600) were recorded in the positive ion mode. The ion source and transfer line temperatures were set, respectively, to 250 and 290°C.
For selected ion monitoring (SIM) mode MS analysis, cells were scraped in 80% methanol and phase separation was achieved by centrifugation at 4°C. The methanol-water phase containing polar metabolites was separated and dried using a vacuum concentrator. Dried polar metabolites were dissolved in 20μL of 2% methoxyamine hydrochloride in pyridine (Pierce-Thermo Fisher) and held at 37°C for 2 hours. After dissolution and reaction, 80μL N-tert-Butyldimethylsilyl-N-methyltrifluoroacetamide with 1% tert-Butyldimethylchlorosilane MBTSTFA + 1% TBDMCS (Thermo Fisher) was added and samples were incubated at 60°C for 60 min. Gas chromatographic runs were performed with helium as carrier gas at 0.6 mL/min. The split inlet temperature was set to 250°C and the injection volume of 1 μL. The GC oven temperature ramp was from 70 to 280°C. The first temperature ramp was from 70°C to 140°C at 3°C/min. The second temperature ramp was from 140°C to 180°C at 1°C/min. Finally, the latest temperature ramp was from 180 to 280°C at 3°C/min. For the Quadrupole, an EI source (70eV) was used. The ion source and transfer line temperatures were set, respectively, to 250 and 290°C. For the determination of relative metabolite abundances, the integrated signal of all ions for each metabolite fragment was normalized by the signal from norvaline, weight of tissue samples and cell number.
13C tracing experiments
For 13C-carbon incorporation from lactate in metabolites, cells were incubated for 24 hours with labelled lactate (confirmation of steady state). For mass spectrometry analysis cells were scraped in 80% methanol and phase separation was achieved by centrifugation at 4°C. The methanol-water phase containing polar metabolites was separated and dried using a vacuum concentrator. Dried polar metabolites were dissolved in 10μL of 2% methoxyamine hydrochloride in pyridine (Pierce, Thermo Fisher Scientific) and held at 37°C for 2 h. After dissolution and reaction, 50μL N-tert-Butyldimethylsilyl-N-methyltrifluoroacetamide with 1% tert-Butyldimethylchlorosilane MBTSTFA + 1% TBDMCS (Thermo Fisher Scientific) was added and samples were incubated at 60°C for 60 min. Gas chromatographic runs were performed with helium as carrier gas at 0.6 mL/min. The split inlet temperature was set to 250°C and the injection volume of 1μL. The GC oven temperature ramp was from 70 to 280°C. The first temperature ramp was from 70 to 140°C at 3°C/min. The second temperature ramp was from 140 to 180°C at 1 °C/min. Finally, the latest temperature ramp was from 180 to 280°C at 3°C/min. For the Quadrupole, an EI source (70 eV) was used. The ion source and transfer line temperatures were set, respectively, to 250 and 290°C. For the determination of relative isotopomers abundances, the integrated signal of all ions for each isotopomers was normalized by the signal from norvaline and the per cell number. The measured distributions of mass isotopomers were corrected for natural abundance of 13C using IsoCor software (29).
Acetyl-CoA quantification
Acetyl-CoA analyses were performed by liquid chromatography-mass spectrometry (AB SCIEX QTRAP 5500 LC-MS/MS; Agilent) following the procedures as previously described (30).
Histology, IHC and Oil Red staining
Individual specimens from human or mouse tumors/lungs were dissected and fixed in 4% PFA for histology and IHC analysis, or were cryo-embedded in OCT compound (Leica Biosystems) for Oil Red staining. Frozen tissues were stained by Oil Red working solution as previously described (31). For prostate tissue analyses, we used a tissue microarray (#PR633, US Biomax). Sections were stained with PLIN2 (rabbit; HPA016607, RRID:AB_2669403; Merck Millipore, 1:250) and MCT1 (mouse ab90582; RRID:AB_2050317; Abcam, 1:200). Immunohistochemistry was performed using the Leica BOND-MAX™ automated system (Leica Microsystems). Slides were developed with 3′3-diaminobenzidine or Fast Red (Leica Microsystems) and counterstained with hematoxylin. Staining intensity for MCT1 was analyzed independently by at least two researchers and given a score of 0, 1+, 2+, 3+. 5-μm lungs sections were cut and stained using hematoxylin and eosin (HE) for pathological assessment. The lung tumor metastatization was confirmed by the presence of panCytokeratin+ epithelial human cells (mouse ab86734, RRID:AB_10674321; Abcam; 1:200). The specificity of the antibodies was established using a negative and positive human tissue samples. For antibodies used on mouse tissues, the blocking Mouse on Mouse (M.O.M.) basic kit (Vector Laboratories, BMK-2202) was used according to the manufacturer’s datasheet. Images were acquired by using a slide scanner (Aperio LV1; Leica Biosystems) and analyzed with software ImageScope (RRID:SCR_020993).
Animal experiments
6-8 weeks-old male SCID mice (NOD.CB17-Prkdcscid/NCrHsd, Envigo) were housed and used at the Centro Stabulazione Animali da Laboratorio (CESAL, Florence, Italy) under sterile conditions with ad libitum access to food and water. For experimental metastasis assay, 1x106 DU145 cells – pre-treated with HPF- or CAF-CM or lactate for 48h – in 100 μL of PBS were intravenously injected into the tail of mice. Where indicated, mice were randomized into six groups and i.p. treated with 10 mg/Kg I-BET762 inhibitor or vehicle (water) every day. Number of metastases was determined 8 weeks post-injection using hematoxylin/eosin (HE) staining on lungs of sacrificed mice. To assess primary tumor growth of control/shPLIN2 DU145 cells or the effect of BET inhibition, 1×106 cells were resuspended in 100 μl growth-factor reduced Matrigel (#A1413201; ThermoFisher) and subcutaneously co-injected with 5x105 HPFs or CAFs into the flanks of 6-week-old male athymic mice (Hsd:Athymic Nude-Foxn1nu, Envigo). 1×106 cells without fibroblasts were injected for the lactate group (i.p. 1g/kg/daily). For drug treatment, when tumor volume reached 100 mm3, mice (n=5) were randomly assigned to receive vehicle or I-BET762 (10 mg/kg/daily, i.p.). Tumors were measured every 4 days with a digital caliper and the tumor volume was calculated using the following formula: V = (length × width2)/2. After 12 weeks, mice were sacrificed, and the lungs and tumors were harvested, formalin-fixed and paraffin-embedded for immunohistochemical analysis. Animal work was carried out under the Project license n.762/2018 and was approved by the Ministero della Salute. All the procedures were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee of University of Florence.
Statistical analysis
Bioinformatic analysis was performed using patient raw data from the web-based interface Cancertool (32). Statistical analyses were performed using GraphPad Prism 8. Two-tailed unpaired Student t-test, one-way or two-way ANOVA tests (Tukey’s post hoc test) were used where appropriate. For all statistical analyses, the expected variance was similar between the groups that were compared, and significance was accepted at the 95% confidence level (*p< 0.05, **p < 0.01, ***p < 0.001).
Results
1. CAF-reprogrammed PCa cells exhibit a lactate-dependent lipid anabolism
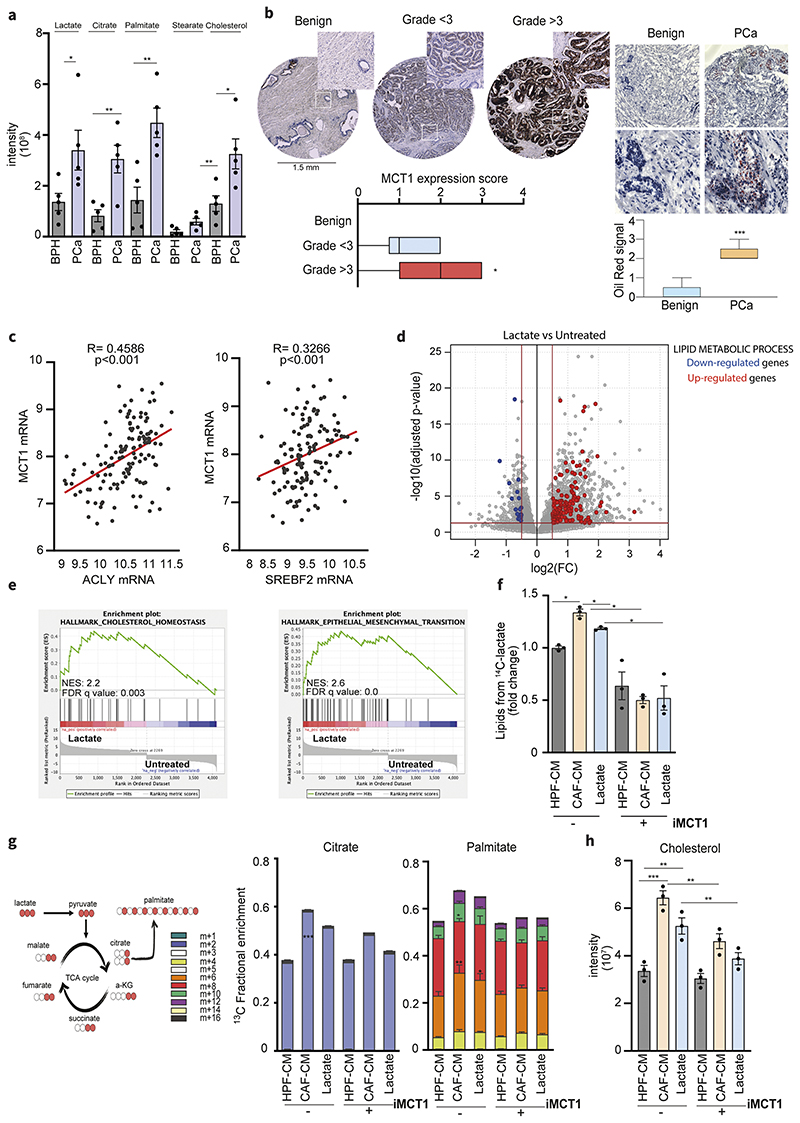
Intratumoral levels of lactate and the expression of the lactate transporter MCT1 (monocarboxylate transporter 1 also known as SLC16A1) have prognostic value in PCa and correlates with Gleason grade and cancer aggressiveness (33). Therefore, we hypothesized that higher lactate influx and subsequent lactate-dependent metabolites could be a feature of more aggressive cancers. By investigating a small cohort of PCas (Gleason grade >3) and benign tissues (i.e. benign hyperplasias, BPHs), we observed increased intratumoral levels of lactate in the cancer specimens paralleled by those of the TCA cycle intermediates and lipid species (palmitate, stearate and cholesterol) (Fig.1a and Suppl.Fig.1a). Then, we also analyzed a tissue microarray (TMA) containing tumor samples from PCa patients to correlate MCT1 protein expression with established grading scores. Representative tissue cores with high and low Gleason grades are shown in Fig.1b. Analysis of all cores using an MCT1 expression score revealed that MCT1 protein levels were increased in tumors with high Gleason grade, while low-graded and benign tissues display weak or negative signal, supporting an increasing MCT1 expression along the PCa progression. Strikingly, high Gleason scored PCa are characterized by an increased lipid content as revealed by Oil Red staining (Fig.1b). This correlation led us to investigate whether the increased ability of import lactate could support lipid accumulation in neoplastic tissues. The clinical relevance of this hypothesis is supported by in silico data analysis of PCa patient derived RNA sequencing (RNA-seq) data that revealed that ATP Citrate Lyase (ACLY) and Sterol Regulatory Element Binding Transcription Factor 2 (SREBF2), key players of the FA and cholesterol synthesis pathway respectively, are positively correlated with MCT1 expression (Fig.1c), supporting the interconnection between lactate trafficking and lipid metabolism in PCa.
Figure 1. Lactate reprograms lipid metabolism in PCa.
a) Levels of lactate, citrate, palmitate, stearate and cholesterol were measured in prostate carcinomas (PCa) and benign hyperplasia (BPH) (n=5). Data represent means ± SEMs. Student’s t test; *p < 0.05; **p < 0.01; ***p < 0.001 b) MCT1 expression in representative tissue cores from TMA (n=63, magnification 20X) including benign and tumor tissue samples with low (< 3) and high Gleason grade (≥3). Box-and-whiskers plots of MCT1 histoscores in tissue samples compared by Gleason grade. On the right, Oil Red staining and quantification in representative PCa and benign tissues (n=5, scale bar: 50μm, magnification 40X). c) Correlation analysis between ACLY/SREBF2 and MCT1 expression in primary tumor specimens of Taylor dataset. Each dot corresponds to an individual specimen. Pearson correlation r. d) Differential gene expression analysis between lactate-exposed (15 mM for 48h) and untreated (serum-starved) DU145 cells. The grey dots represent all the considered genes. Red and blue dots highlight, respectively, the up-regulated and down-regulated genes in the “lipid metabolic process” gene ontology pathway over a 0.5 |log2FC|, considering a p-value cut-off equal to 0.05. e) Enrichment plots of the Hallmark Cholesterol homeostasis and EMT pathways showing a positive association between these MSigDb datasets and the Lactate-exposed DU145 gene expression profile. NES, normalized enrichment score. f) DU145 cells were subjected to 14C lactate-containing medium, upon pre-exposure with HPF- or CAFCM and 15mM lactate, with or without AR-C155858 (iMCT1) inhibitor (40μM). Radioactive extracted lipids are shown. g) 13C-labeled lactate-derived fluxes and relative incorporation of 13C carbons derived from lactate in TCA cycle intermediates and palmitate in DU145 cells exposed to HPF- or CAF-CM and lactate (15mM). 13C carbons derived from lactate in citrate and palmitate in DU145 cells treated as indicated ± AR-C155858. h) Cholesterol steady-state levels detected in DU145 cells treated as indicated, ± AR-C155858. Data are represented as mean ± SEM of three independent experiments. Significance was determined one-way ANOVA with Tukey multiple-comparison analysis; *p < 0.05; **p < 0.01; ***p < 0.001.
To validate this hypothesis and to gain molecular insights on the role of lactate in PCa, we exposed DU145 prostate cancer cell lines to exogenous lactate for 48 hours and addressed the transcriptional output through RNA-seq (Fig.1d). RNA-seq analysis identified 2151 genes that were differentially regulated (|log2FC| > 0.5 and adjusted P-value < 0.05). Importantly, a significant proportion of the genes that are upregulated by lactate exposure are listed as lipid metabolism involved genes, accordingly to Gene Ontology (red dots in volcano plot, Fig.1d). In addition, Gene Set Enrichment Analysis (GSEA) performed on Hallmark gene sets highlighted that lactate-induced genes in DU145 cells are positively associated with gene sets related to cholesterol homeostasis and epithelial-to-mesenchymal transition (EMT)-related gene sets, whereas lactate-induced genes are negatively associated with proliferation-related gene sets (Fig.1e, Suppl.Fig.1b-c). This is in line with previous findings showing that lactate-rich CAFs conditioning (CAF-CM, conditioned medium) leads to a highly motile and mitochondrially reprogrammed tumor phenotype (11). In particular, increased levels of citrate, which can be hydrolyzed in the cytoplasm to release acetyl-CoA, led us to hypothesize an activation of lipid biosynthesis. In keeping, tracing analysis showed that radioactive signal derived from uniformly labeled 14C-lactate was higher in the lipids extracted from PCa cells exposed to CAF-CM or exogenous lactate, with respect to the HPF-CM-treated cells. This enhanced lactate-derived carbons flux into lipids biosynthesis was significantly reduced by blocking lactate upload in PCa cells using MCT1 inhibitor (Fig.1f). No significant changes were observed in HPF-CM treated cells. To quantify the contribution of lactate to PCa cell metabolism, cells were cultured in medium containing uniformly (U) labeled 13C-lactate and the 13C-labeling of the TCA cycle intermediates (Suppl.Fig.1d, Suppl.Fig.2a) together with palmitate were analyzed by gas chromatography-mass spectrometry (GC-MS) (Fig.1g). 13C-tracing analysis revealed that lactate-derived carbons contributed to the relative TCA cycle metabolites levels (Suppl.Fig.1d) and that higher citrate as well as palmitate labeling was consistent with lactate-derived citrate carbons being used to synthesize FAs in CAF-reprogrammed PCa cells (Fig.1g). Taken together these data suggest that lactate induces a transcriptional and metabolic reprogramming in PCa cells that culminates in the enhancement of TCA cycle intermediates and subsequent synthesis of lipids (Fig.1f-h). Similarly, also cholesterol levels within PCa cells were reduced by MCT1 inhibition, suggesting that lactate provides acetyl-CoA that can be diverted into the cholesterol biosynthesis (Fig.1h).
2. CAF lactate-reprogrammed PCa cells store lipids into lipid droplets (LDs) to support mitochondrial metabolism
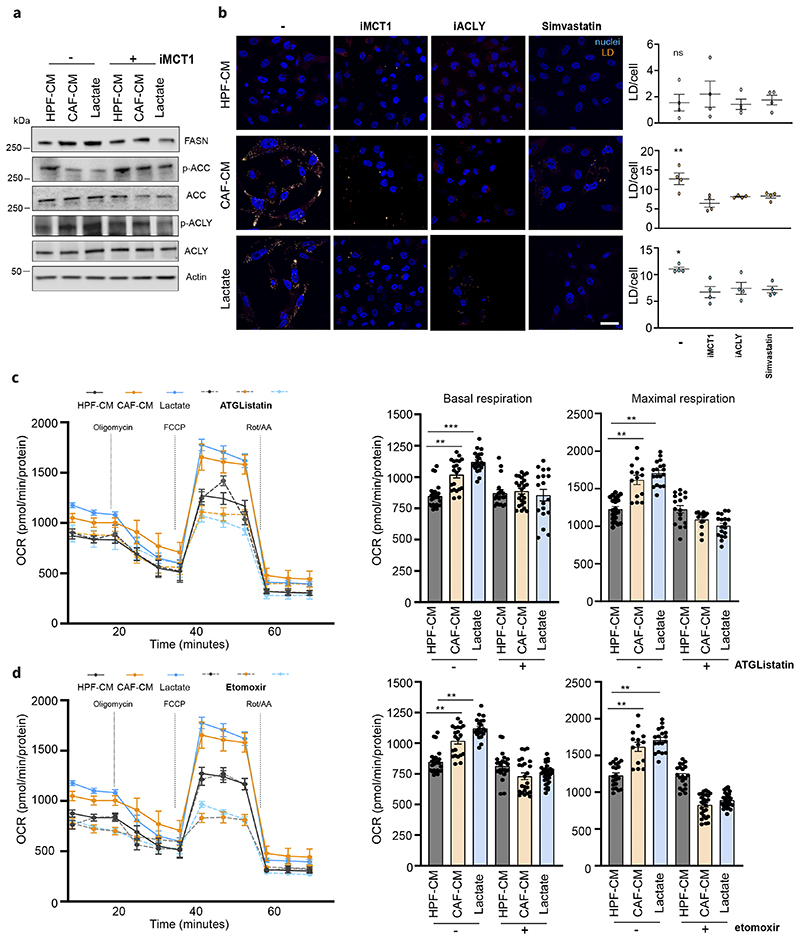
The ability of lactate to activate lipogenesis (16) in PCa cells is confirmed by immunoblot analysis showing that CAF-conditioned DU145 cells increased the expression and activation of ACLY, as shown by increased phosphorylation levels, the activation of acetyl-CoA carboxylase (ACC), as shown by reduced levels of ACC phosphorylation and increased expression of fatty acid synthase (FASN) (Fig.2a). To note, ACLY activity is crucial in the described tumor-stroma model as it allows lactate-dependent citrate to fuel lipid synthesis in DU145 cancer cells. Indeed, ACLY silencing reduced the lactate-derived lipogenesis as revealed by increased ACC phosphorylation (i.e. inactivation) (Suppl.Fig.3a) in ACLY-deficient tumor cells. Importantly, CAF-CM does not contain significant content of lipid species (Suppl.Fig.3b) strengthening the role of lactate as an essential source for lipid biosynthesis in CAF-exposed PCa cells. The lipid anabolism fueled by CAF-released lactate led us to identify whether the increase in lipid anabolism was paralleled by increased intracellular lipid deposition. LDs are intracellular organelles responsible for the storage of neutral lipids and cholesterol esters. Notably, LDs are significantly increased not only in AR- PCa cells but also in AR+ cells such as LNCap and 22Rv1, after exposure to CAF-CM and exogenous lactate (Fig.2b, Suppl.Fig.3c-d). Targeting MCT1 or impairing FAs and cholesterol biosynthesis, via ACLY or HMG-CoA reductase inhibition (simvastatin) respectively, reduced the LDs amount in CAF-CM and lactate-exposed PCa cells (Fig.2b).
Figure 2. Lactate promotes lipid droplets accumulation in PCa cells.
a) Immunoblot with the indicated antibodies is performed on DU145 cells treated with HPF- or CAF-CM and 15mM lactate for 48h. b) Representative pictures of BODIPY 493/503-stained DU145 cells treated as in a), ± iMCT1 (40μM), Simvastatin (50nM) and SB-204990 (25μM; iACLY). Lipid droplets were visualized as pale-yellow spots. Nuclei (blue) were stained with DAPI. Quantification of BODIPY spots/cell was reported. Scale bar: 10 μm c-d), Seahorse MitoStressTest and oxygen consumption rate (OCR) was monitored in presence of ATGListatin (25μM) and etomoxir (40μM) on DU145 cells, treated as indicted. Data are represented as mean ± SEM of three independent experiments (≥ 4 technical replicates). One-way ANOVA; Tukey’s corrected; *p < 0.05; **p < 0.01; ***p < 0.001.
Since LDs can store high amount of lipids, we speculated that LDs could act as a source of FAs to maintain and sustain the respiratory phenotype of PCa cells enhanced by CAF-derived lactate exposure. First, to directly monitor the energetic role of LDs we inhibited the lipid released from LDs by targeting adipose triglyceride lipase (ATGL) with ATGListatin. Oxygen Consumption Rate (OCR) levels upon ATGListatin administration in CAF-CM treated PCa cells are reduced when compared to cells that were exposed solely to CAF-CM, suggesting that oxidation of FAs released from LDs is needed to support stroma-rewired PCa cell oxidative metabolism (Fig.2c). This analysis was confirmed in CAF-CM treated PCa cells upon administration of etomoxir, an inhibitor of carnitine palmitoyl transferase CPT1, thus suggesting that the OCR levels were indeed sustained by lipid oxidation (Fig.2d).
3. Histone acetylation is regulated by CAF lactate-dependent lipid reprogramming and supports invasiveness in PCa cells
Alterations in lipid metabolism have been documented to support TCA cycle in aggressive tumor models by regulating the abundance of acetyl-CoA under conditions of metabolic stress such as hypoxia or acidosis (34).
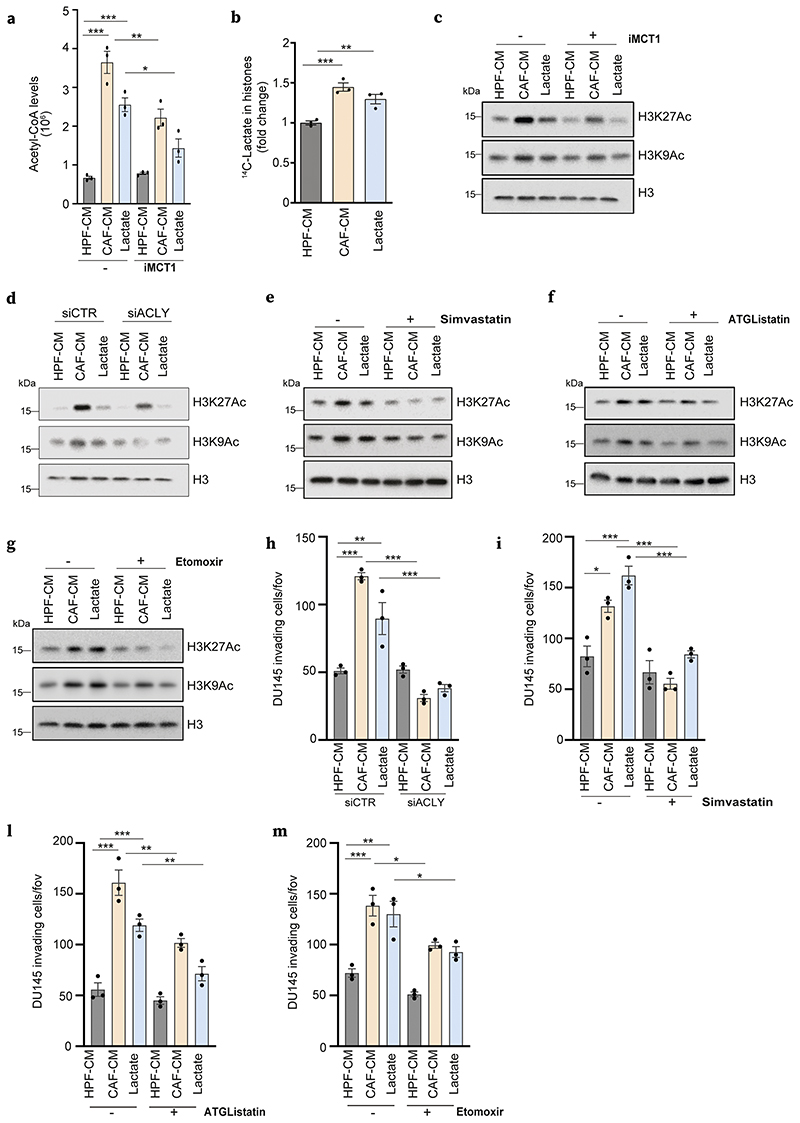
We therefore evaluated the contribution of CAF-CM and lactate conditioning to fuel acetyl-CoA in PCa cells and observed a significant increase of acetyl-CoA pool in DU145 cancer cells exposed to both stimuli (Fig.3a). Crucially, inhibition of MCT1 (i.e. blocking lactate entrance) and ACLY (i.e. blocking lipid synthesis from citrate) reduced acetyl-CoA intracellular levels, suggesting that CAF-derived lactate is the main source of acetyl-CoA in lipid-reprogrammed PCa cells (Fig.3a, Suppl.Fig.4a). Interestingly, in addition to sustain mitochondrial respiration, lipid-derived acetyl-CoA is a key source of carbon units for histone acetylation (15). In keeping, we found that radiolabeled carbon units derived from 14C-lactate are incorporated at higher levels in histone compartment in CAF-CM and lactate-conditioned PCa cells when compared to control DU145 cells (Fig.3b), suggesting a potential role of lactate in histones post translational modification. In particular, PCa cells exposed to CAF-CM and lactate showed increased levels of acetylation at H3K9 and H3K27, both markers of an ‘open’ chromatin state associated to active gene transcription (Fig.3c, Suppl.4b). Notably, MCT1 inhibition prevented histone acetylation, suggesting that lactate entry induces a transcriptional and metabolic reprogramming that involves epigenetic modifications, including those occurring on key residues of histones (Fig.3c, Suppl.4c).
Figure 3. Lactate-sustained lipid metabolism links to histone hyperacetylation in PCa cells.
a) Acetyl-CoA measured in DU145 cells treated with HPF- or CAF-CM and 15mM lactate ± iMCT1. b) Incorporation of 14C lactate into histones extracted as in Methods, from DU145 cells treated as indicated. c-g) Immunoblot using the indicated antibodies for DU145 cells treated as indicated ± iMCT1, simvastatin, ATGListatin and etomoxir. h-m) Invasion assays performed in DU145 cells with the same conditions and drug treatments as in Fig.2c-g.
In keeping with the contributions of CAF lactate-dependent lipid metabolic rewiring to generate acetyl-CoA, we investigated the impact of the inhibition of LDs formation and mobilization in the maintenance of histone acetylation in reprogrammed PCa cells. We provided evidence that both ACLY expression and activity are required for increased H3K9 and H3K27 acetylation observed in CAF-CM- and lactate-treated PCa cells (Fig.3d, Suppl. Fig.4d). Similarly, simvastatin, ATGListatin and etomoxir affected the acetylation of histone marks in PCa cells reprogrammed upon CAFs or lactate conditioning (Fig.3e-g, Suppl.Fig.4e), suggesting a crucial role for LDs as lipid storages to ensure acetyl-CoA supply for epigenetic modifications. Consistent with the link between lactate-induced metabolic and epigenetic remodeling observed in cancer cells, we hypothesized that this scenario defines a convergence point for the enhancement of aggressive features of PCa cells induced by stromal signals.
Intriguingly, emerging evidence showed that LDs abundance and increased lipid utilization in the mitochondria are linked to high metastatic potential in tumors (35,36). We examined whether lactate-dependent LDs could help potentiate the invasion ability of stroma-exposed PCa cells. To note, all the treatments that reduced lipogenesis and LD levels (i.e. siACLY, simvastatin, ATGListatin, etomoxir – Fig.2) impaired the invasive ability of PCa cells exposed to either CAF-CM or lactate (Fig.3h-m, Suppl.Fig.4f). Taken together, these data confirmed that lactate-derived lipid and acetyl-CoA availability participates in an epigenetic-dependent cell metabolic reprogramming (either generated ex novo or derived from LDs), conferring invasive ability to lactate-exposed cancer cells.
4. BET inhibition disrupts CAF lactate-sustained metabolic and epigenetic changes in PCa cells
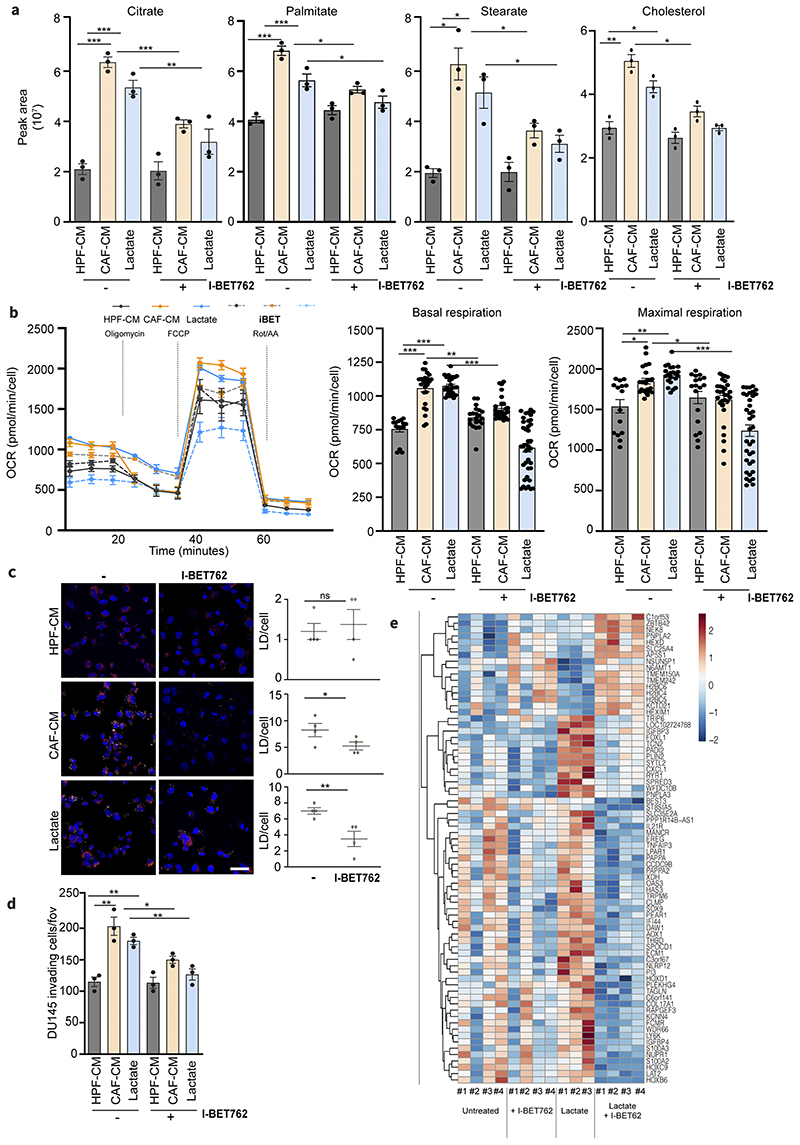
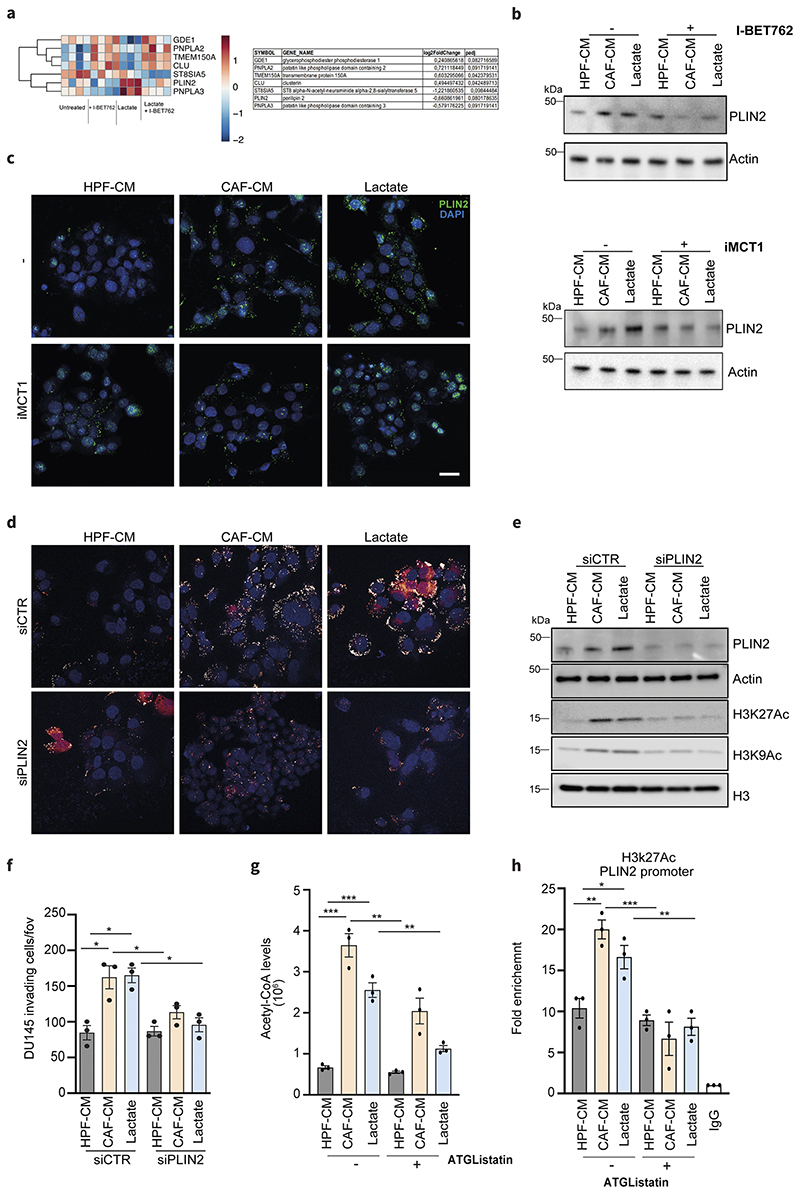
As lipids represent bona fide the major source of cellular acetyl-CoA destined for histone acetylation, we investigated whether targeting key players involved in the acetylation machinery can interfere with lactate-induced lipid reprogramming. Epigenetic inhibitors targeting BET proteins compete for binding to acetylated histones, showing a profound anticancer efficacy either alone or in combination with other drugs (37). Firstly, we investigated the impact of BET inhibition (I-BET762) in reprogramming of metabolism and invasion ability of CAF-CM and lactate-exposed PCa cells. The FDA-approved I-BET762 significantly reduced the mitochondrial-dependent metabolism of CAF-conditioned cancer cells, as monitored by the reduction of the intracellular levels of TCA cycle intermediates and relevant lipid species such as palmitate, stearate and cholesterol (Fig.4a, Suppl.Fig.5a) and, in parallel, of OCR levels (Fig.4b). Consequently, such metabolic effect was found to dampen the assembly of LDs (Fig.4c, Suppl.Fig.5b) and the invasive potential in lactate-reprogrammed PCa cells (Fig.4d). Interestingly, DU145 cells stably-silenced for BRD4, the most representative bromodomain in BET family, displayed a decreased content in LDs and ability to invade, when exposed to CAF-CM and lactate (Suppl.Fig.5c-d). To deeper explore the transcriptional regulation promoted by lactate-driven acetylation, we performed transcriptomic profiling using RNAseq on lactate-exposed PCa cells, with or without I-BET762 to discriminate the BET-dependent (i.e. acetylation-regulated) and -independent gene expression. Differential expression analysis revealed 74 genes that were differentially regulated (|log2FC| > 0.5 and adjusted P-value < 0.1) between I-BET762+Lactate and Lactate treated cells. Interestingly, we found that BET inhibition targets the expression of a series of LDs-associated genes including the patatin-like phospholipase 3 (PNPLA3), a gene involved in LDs hydrolysis (especially in hepatic steatosis) and the perilipin 2 (PLIN2), encoding for a protein located on the LDs surface responsible for their stability and formation (Fig.4e).
Figure 4. Lactate-induced metabolism and invasiveness are sensitive to BET inhibition in PCa cells.
a) Citrate, palmitate, stearate and cholesterol levels were detected in DU145 cells treated with HPF- or CAF-CM and 15mM lactate for 48h, ± I-BET762 (100nM). One-way ANOVA; Tukey’s corrected; *p < 0.05; **p < 0.01; ***p < 0.001 b) OCR from MitoStressTest analysis performed on DU145 cells treated as indicated ± I-BET762. Data are represented as mean ± SEM of three independent experiments (≥ 4 technical replicates). c) BODIPY 493/503 staining of DU145 cells treated as indicated ± I-BET762. Quantification of BODIPY spots per cell was reported. Scale bar: 10 μm d) Invasion assay of DU145 cells treated as indicated ± I-BET762. e) Heatmap showing the expression of differentially expressed genes between DU145 cells exposed to 15mM lactate for 48h and treated w/wo I-BET762.
5. CAF lactate-dependent PLIN2 expression affects lipid-acetylation interplay in PCa cells
PLIN2 has a central role in the network that characterizes the interaction between the lactate-induced genes involved in lipid metabolism (Suppl.Fig.6a). Furthermore, PLIN2 is one of the most lactate-induced genes within lipid metabolic pathways whose expression is downregulated by I-BET762 (Fig.5a). PLIN2 expression was enhanced in CAF-CM- and lactate-exposed PCa cells and we confirmed that I-BET762 treatment as well as BRD4 silencing dramatically decreased PLIN2 levels both at mRNA (Suppl. Fig.6b) and protein (Fig.5b, Suppl.Fig.6c) levels. Importantly, targeting lactate entry with MCT1 inhibitor reduced PLIN2 expression and LDs levels (Fig.5c), suggesting a direct role of lactate in PLIN2 upregulation and LDs formation. To further ascertained the role of PLIN2 in our model, we silenced PLIN2 and found a significant decrease of the CAF-CM- and lactate-induced LDs accumulation (Fig.5d), histone acetylation (Fig.5e, Suppl.Fig.6d) and PCa invasiveness (Fig.5f). Since PLIN2 silencing significantly impairs histone acetylation, we hypothesized that a large proportion of acetyl-CoA availability could be dependent from LDs formation and utilization. In keeping, ATGListatin strongly interferes with lactate-induced acetyl-CoA levels (Fig.5g), corroborating the role of LDs mobilization to provide acetyl moieties for histone acetylation. Consequently, to uncover the role of PLIN2 as molecular linker between the lactate-dependent metabolic and epigenetic interplay in PCa cells we performed a ChIP-qPCR to detect the levels of H3K27ac on PLIN2 promoter. Strikingly, we found that PLIN2 promoter shows a higher enrichment in H3K27 acetylation, prevented by ATGListatin (Fig.5h), thus demonstrating a direct role of LDs-derived FAs in promoting PLIN2 acetylation and, consequently, its expression.
Figure 5. PLIN2 is a key player in metabolic-epigenetic interplay induced by lactate.
a) Lactate-dependent signature genes involved in lipid metabolism that are up- and down-modulated upon I-BET762 treatment in DU145 cells exposed to 15mM lactate for 48h. Relative log2FC and adjusted p-values are listed on the right. b) Immunoblot for PLIN2 levels in DU145 cells treated as above. c) Expression of PLIN2 evaluated by immunofluorescence or immunoblot in DU145 cells treated as indicated, ± iMCT1. Scale bar = 10 μm d-f) DU145 cells were silenced using the non-targeting control (siCTR) or PLIN2-specific siRNA and subsequently treated as indicated. LDs content analysis (d), immunoblot for H3K27ac and H3K9ac levels (e) and invasion assay (f) were performed in PLIN2-silenced DU145 cells using siCTR-treated cells as comparators. g) Acetyl-CoA levels were MS-evaluated in DU145 cells treated as indicated ± ATGListatin. e) ChIP-qPCR analysis of H3K27ac on PLIN2 promoters in DU145 cells in the indicated conditions. Enrichment as fold-enrichment relative to IgG is reported. One-way ANOVA; Tukey’s corrected; *p < 0.05; **p < 0.01; ***p < 0.001
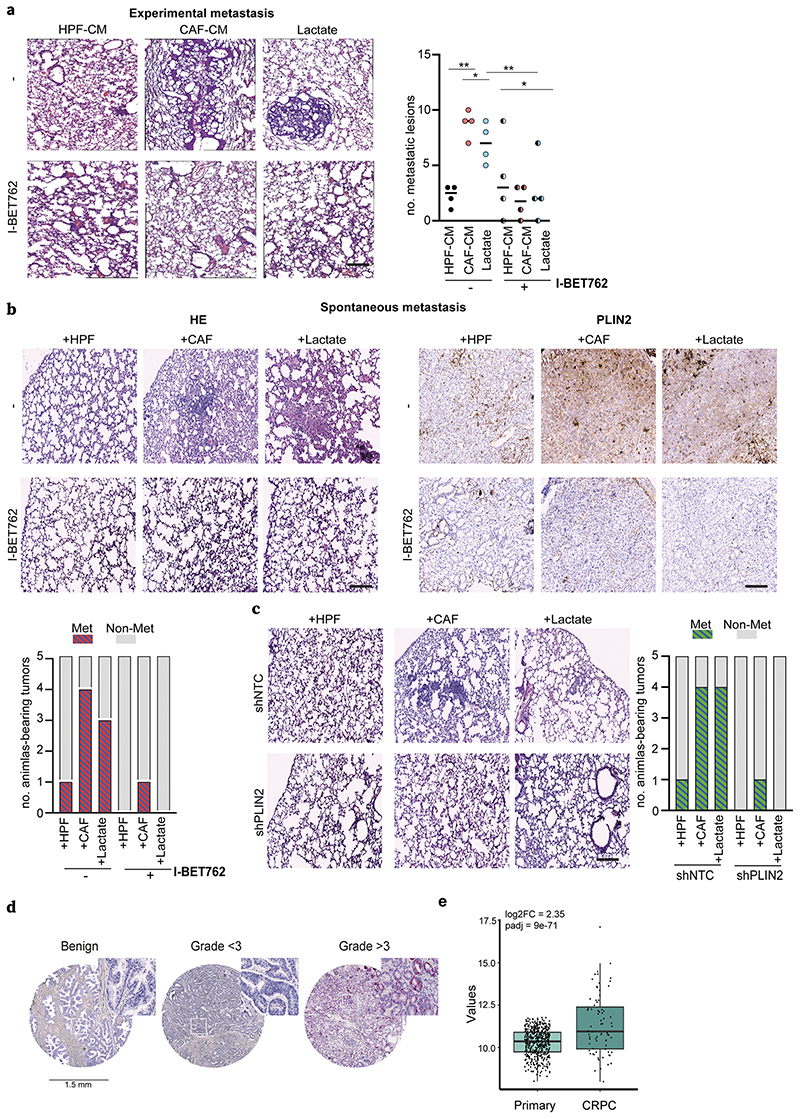
6. BET inhibitor suppresses the in vivo lactate-dependent lung colonization of PCa cells
To further examine the impact of I-BET762 on in vivo metastatic colonization of tumor, CAF-CM and lactate-treated DU145 cells were injected into the lateral tail vein of immunocompromised mice to get lung metastatic burden. We observed that the number and size of lung metastatic lesions were significantly larger in mice injected with CAF-CM and lactate-exposed PCa cells, indicating the lactate conditioning as a pro-metastatic event in PCa progression. Importantly, lung metastatic burden was dramatically reduced in I-BET762-treated mice injected with CAF-CM and lactate-conditioned cancer cells (Fig.6a). As expected, also tumor xenografts derived from CAFs:tumor cells co-injection as well as from lactate administration group displayed higher tumor mass compared with the healthy fibroblasts con-injected counterpart (Suppl.Fig.7a). Interestingly, I-BET762 treatment dramatically affects tumor metastatic incidence in mice bearing tumors sustained by CAF presence and lactate supplementation, also resulting in a strong reduction in PLIN2 expression as well as lipid accumulation (Fig.6b, Suppl.Fig.7b). To support the concept that the importance of lipid accumulation is associated to an aggressive PCa phenotype in vivo, we stably-silenced DU145 for PLIN2 (Suppl.Fig.7c) and we established tumor xenografts with HPFs/CAFs or administrating lactate to xenografted animals. In keeping, we observed that shPLIN2-derived tumors do not manifest any metastatic behavior, while only animals engrafting control (shNTC) tumor cells co-injected with CAFs or stimulated with lactate exhibit lung metastasis (Fig.6c, Suppl.Fig.8a-b). To note, the key role of PLIN2 in PCa progression is documented in human tumors by detecting that PLIN2 expression is predominantly associated with human high-Gleason grade PCa specimens (22/41, 50%) compared with the low-graded (3/10, 30%) and non-cancerous tissues (no positivity) in the TMA patients’ cohort (Fig.6d). Next, we found that the expression level of PLIN2 is higher in castration-resistant prostate cancer (CRPC) cohort of patients than the primary PCa samples (Fig.6e), indicating an important role of PLIN2 in aggressive tumors with high metastatic potential. Collectively, these results suggested that i) stromal lactate is determinant for PCa progression fostered by PLIN2-supported lipids storage in LDs and their subsequent exploitation as acetyl-CoA supplier for energetic and epigenetic purposes; ii) BET inhibition counteracts the effects of lactate-induced metabolic and epigenetic reprogramming and represents a potential therapeutic intervention to impair PCa metastatic spreading.
Figure 6. BET inhibition dampens lactate-sustained metastatization.
a) For experimental metastasis, CAF- and lactate-conditioned DU145 cells were i.v. injected into SCID mice (n = 4/group) and I-BET762 (20mg/kg) was administered daily. Representative HE pictures of lung metastatic lesions and relative quantification were shown. Scale bar: 50μm. b) DU145 cells were s.c. injected with HPFs, CAFs or alone (for lactate administration) in nude mice (n=5/group) and I-BET762 administered upon engrafting. Representative HE images of lung micrometastases and PLIN2 staining in primary tumors derived from the indicated animal cohorts were shown. Number of animals with metastasis is plotted. c) shNTC or shPLIN2 DU145 cells were s.c. injected as in b). Representative HE staining images of lung metastases were shown (magnification 10X, scale bars: 50μm). Number of animals with lung metastasis is plotted. d) Representative tissue cores for PLIN2 expression in TMA (magnification 40X). e) Expression level of PLIN2 in patients from CRPC (Fred Hutchinson Cancer Research Center and phs000909.v1.p1 - Trento/Cornell/Broad 2015) vs primary tumors (TCGA). Fold change and adjusted p-value were reported.
Discussion
Our study pinpoints lactate conditioning as driver of lipid metabolic rewiring fostered by stromal cues, to support the anabolic/catabolic needs of cancer cells. Lactate, previously considered a waste product of tumor fermentative metabolism, has currently been emancipated as a key nutrient in tumor microenvironment. Lung and pancreatic cancers use MCT1 to transport circulating lactate into the tumor cells, thus rerouting it into TCA cycle (8,9). To note, MCT1 has been highlighted as a discriminating point between inefficient and efficient metastasizer melanoma tumors, with the latter showing higher intratumoral TCA cycle intermediates levels (38). Coherently, in line with other models of CAF-tumor crosstalk (e.g. breast cancer) (39), we previously reported that PCa cells exposed to the CAF metabolic secretome prefer to exploit oxidative mitochondrial metabolism boosted by lactate entry, leading to a higher accumulation of TCA cycle intermediates (11). Here, we found that CAF-derived lactate stimulates lipogenesis by rerouting citrate to (i) activate ACLY and ACC, key enzymes in the FA synthesis, and (ii) cholesterol pathway. Similarly, lactate has been shown to induce lipid synthesis in CD4+ T cells recruited in inflammatory tissues (40), a well-known site characterized by high lactate concentration. However, de novo lipogenesis is not the exclusive source of lipids in cancer cells since it is reported that increased uptake of exogenous fatty acids is a known metabolic feature in several cancers (41,42). Notably, the deletion of the lipid transporter CD36 restricts not only FAs uptake from the microenvironment, but also reduces lipid biosynthesis, highlighting a wider rewiring of lipid metabolism in malignant prostate tissue (43), eventually culminating into the fueling of lipid pools necessary to sustain energy production (triglycerides and sterol lipids) and/or membrane synthesis (16). However, as lactate content increases in parallel with PCa grading (33), our data support the idea that MCT1-mediated transport of lactate used as carbon donor could be an important step in PCa progression. Moreover, as its abundance among microenvironmental nutrients, lactate can induce metabolic but also transcriptional changes. First, when glucose is deprived and lactate is accumulated, tumor cells can meet the demand of reducing power for proliferation or to handle oxidative stress. Thus, lactate can contribute to NADPH production by IDH1 (44) or through oxPPP (38). To note, NADPH generation is also advantageous for lipid biosynthesis. Secondly, metabolic plasticity can be accompanied to the transcriptional one. We observed a key role of lactate in modulating the expression of several lipid and cholesterol-related genes. Importantly, SREBP2, a master transcription regulator of lipid homeostasis, has been found to promote and sustain transcriptional upregulation of cholesterol pathway genes in response to tumor acidosis (45). Cholesterol esters together with newly-synthesized FAs are key components of LDs. We observed a LD phenotype in PCa cells, exposed to stromal lactate, independently on androgen receptor status. These cells also co-opt mobilization of LD-related triglycerides to promote cancer cells aggressiveness. However, the concomitant activation of lipogenesis to accumulate LDs and of lipolysis to release FAs for their subsequent oxidation has already been observed in other models as cancer-specific behavior under acidic conditions (34), where the acidity downregulates the mitochondrial ACC2 isoform disabling the negative feedback of CPT1 impairment (i.e. β-oxidation) by malonyl-CoA. Also, we highlighted that the lactate exploitation in PCa cells culminates in the formation of LDs acting as energy organelles used to support invasiveness. Indeed, we found that ATGL-dependent lipolytic activity allowing the breakdown of LDs as well as the oxidation of released FAs into mitochondria promote mitochondrial oxidative phosphorylation (and ATP synthesis) likely required for the invasive machinery of reprogrammed cancer cells. Although we observed that ATGL is a critical molecule to sustain metastatic potential, as reported in another tumor-stroma model (36) and in cancer-associated cachexia (46), other LD-related lipases could be involved (47,48). In addition, corroborating data revealed that extracellular acidosis modulates lipid metabolism and EMT-mediated invasive mechanisms (49). Together with our findings on the role of lactate in controlling LDs formation and cancer cell invasion, all the abovediscussed data strongly contribute to and integrate the highly complex and heterogeneous facets of tumor microenvironment.
Furthermore, our data support a model in which the entry and the exploitation of stromal-derived lactate in PCa cells promote lipid metabolism and increases acetyl-CoA availability, thereby leading to histone acetylation (and active gene expression). The interplay between nutrients and epigenome regulation has been widely reported (12) and, in particular, the readers of histones acetylation (e.g. BET proteins) have been emerged as druggable targets for the treatment of hematological malignancies and solid tumors, and their inhibitors showed remarkable efficacy in preclinical models and in clinical trials (37). Although the inhibition of the BET family has been found to impair cancer cell-stroma interaction (50,51) and prostate cancer stemness (17), the involvement of BET in the regulation of stromal-induced metabolic reprogramming is still unclear. Our results demonstrated that BET inhibition reduces the lactate-fueled TCA cycle intermediates and subsequent lipids accumulation in PCa cells. Accordingly, previous findings demonstrated that the BET inhibitor JQ1 reduces the expression of certain lipogenic enzymes (52). In addition, the effect of BET inhibitors is strictly dependent on acetyl-CoA levels (53) and suppressing acetyl-CoA–dependent processes impairs pancreatic tumor growth in vivo (54). This is in line with our data showing a BET-driven reduction of lactate-guided LDs formation, resulting in a consequent impairment of LDs-dependent metabolic/phenotypic behavior of PCa cells, and this is presumably dictated by BRD4. In this regard, we strikingly observed a BET (i.e. BRD4)-dependent lactate-induced transcriptional regulation of PLIN2, a crucial LDs-associated structural protein, proving the importance of LDs formation in PCa facing a lactate-rich environment. PLIN2 as well as other perilipins has been recognized as proteins required for adaptation to hypoxia and oxidative stress (55), that are conditions strictly associated with high levels of ambient lactate. We found that PLIN2 depletion has a double negative impact on histone acetylation in lactate-reprogrammed PCa cells in vitro and on lung PCa metastatization in vivo. Thus, PLIN2 sustains a metabolic signature associated with highly aggressive prostate carcinomas.
Overall, in the current study we documented for the first time that lactate is a mandatory driver to promote metastatic spreading in PCa, a process that is primed by a metabolic-mediated epigenetic regulation (see Graphical abstract). Accordingly, the inhibition of BET proteins reduced the capacity of lactate-rewired PCa cells to form metastases and prevented the lipid accumulation in the metastatic lesions, in agreement with other findings showing that accumulation of dietary lipids in prostate tumors promotes a pro-metastatic program (56). Together, these results established a mechanistic link among aberrant lipogenesis, excess lipid accumulation and metastasis, thus highlighting the importance of integrating epigenetic with metabolic targeting approaches to prevent metastasis.
Supplementary Material
Significance.
This work shows that stromal-derived lactate induces accumulation of lipid droplets, stimulates epigenetic rewiring, and fosters metastatic potential in prostate cancer.
Acknowledgements
This research was funded by Associazione Italiana Ricerca sul Cancro (AIRC), and Fondazione Cassa di Risparmio di Firenze (grant #19515 to P.C. and A.M.), AIRC grants (#24731 to P.C. and #22941 to A.M.) and PRIN 2017-21 (grant to P.C.), Swiss Cancer League (KLS-4899-08-2019, to CVC) and Swiss National Science Foundation (SNSF-310030L_170182, to C.V.C). The data presented in the current study were in part generated using the equipment of the Facility di Medicina Molecolare, funded by “Ministero dell’Istruzione dell’Università e della Ricerca – Bando Dipartimenti di Eccellenza 2018-2022. L.I. is a Fondazione Umberto Veronesi fellow. N.L. is Annastaccatolisa association fellow. M.B. is a Pezcoller Foundation/SIC Prof.ssa De Gasperi Ronc. fellow. The illustration was created with BioRender.com.
Footnotes
Author contributions L.I. and G.C. designed and performed the experiments and analyzed the data; M.P. performed confocal imaging and GC-MS analysis; A.M. analyzed and interpreted the data and provided intellectual contributions; M.I., A.D., E.P., F.V. and L.F. provided technical support; F.B. performed in vivo experiment; N.L. and M.B. performed Seahorse analysis and participated to metabolic data interpretation; G.S. and C.V.C. performed gene expression, bioinformatic analyses and data interpretation; LM.G. and R.D. performed LC-MS analysis, P.S and S.S. provided human specimens; E.G. and P.C. conceived, designed, and supervised the study; L.I., P.C. and E.G. wrote the paper, with a critical feedback from A.M. All the authors reviewed the prepared manuscript.
Conflict of interest: The authors declare no potential conflicts of interest.
Data and code availability
The accession number for the RNA-seq data performed on DU145 cells treated with lactate ± I-BET762 reported in this paper is GEO: GSE195639.
References
- 1.Lyssiotis CA, Kimmelman AC. Metabolic Interactions in the Tumor Microenvironment. Trends Cell Biol. 2017;27:863–75. doi: 10.1016/j.tcb.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walenta S, Wetterling M, Lehrke M, Schwickert G, Sundfør K, Rofstad EK, et al. High lactate levels predict likelihood of metastases, tumor recurrence, and restricted patient survival in human cervical cancers. Cancer Res. 2000;60:916–21. [PubMed] [Google Scholar]
- 3.Brizel DM, Schroeder T, Scher RL, Walenta S, Clough RW, Dewhirst MW, et al. Elevated tumor lactate concentrations predict for an increased risk of metastases in head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2001;51:349–53. doi: 10.1016/s0360-3016(01)01630-3. [DOI] [PubMed] [Google Scholar]
- 4.Doyen J, Trastour C, Ettore F, Peyrottes I, Toussant N, Gal J, et al. Expression of the hypoxia-inducible monocarboxylate transporter MCT4 is increased in triple negative breast cancer and correlates independently with clinical outcome. Biochem Biophys Res Commun. 2014;451:54–61. doi: 10.1016/j.bbrc.2014.07.050. [DOI] [PubMed] [Google Scholar]
- 5.Sonveaux P, Végran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008;118:3930–42. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Végran F, Boidot R, Michiels C, Sonveaux P, Feron O. Lactate influx through the endothelial cell monocarboxylate transporter MCT1 supports an NF-κB/IL-8 pathway that drives tumor angiogenesis. Cancer Res. 2011;71:2550–60. doi: 10.1158/0008-5472.CAN-10-2828. [DOI] [PubMed] [Google Scholar]
- 7.Fiaschi T, Marini A, Giannoni E, Taddei ML, Gandellini P, De Donatis A, et al. Reciprocal metabolic reprogramming through lactate shuttle coordinately influences tumor-stroma interplay. Cancer Res. 2012;72:5130–40. doi: 10.1158/0008-5472.CAN-12-1949. [DOI] [PubMed] [Google Scholar]
- 8.Faubert B, Li KY, Cai L, Hensley CT, Kim J, Zacharias LG, et al. Lactate Metabolism in Human Lung Tumors. Cell. 2017;171:358–71.:e9. doi: 10.1016/j.cell.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, et al. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551:115–8. doi: 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comito G, Iscaro A, Bacci M, Morandi A, Ippolito L, Parri M, et al. Lactate modulates CD4. Oncogene. 2019 doi: 10.1038/s41388-019-0688-7. [DOI] [PubMed] [Google Scholar]
- 11.Ippolito L, Morandi A, Taddei ML, Parri M, Comito G, Iscaro A, et al. Cancer-associated fibroblasts promote prostate cancer malignancy via metabolic rewiring and mitochondrial transfer. Oncogene. 2019 doi: 10.1038/s41388-019-0805-7. [DOI] [PubMed] [Google Scholar]
- 12.Dai Z, Ramesh V, Locasale JW. The evolving metabolic landscape of chromatin biology and epigenetics. Nat Rev Genet. 2020;21:737–53. doi: 10.1038/s41576-020-0270-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan DG, Murphy MP, Frezza C, Prag HA, Chouchani ET, O’Neill LA, et al. Coupling Krebs cycle metabolites to signalling in immunity and cancer. Nat Metab. 2019;1:16–33. doi: 10.1038/s42255-018-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sivanand S, Viney I, Wellen KE. Spatiotemporal Control of Acetyl-CoA Metabolism in Chromatin Regulation. Trends Biochem Sci. 2018;43:61–74. doi: 10.1016/j.tibs.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonnell E, Crown SB, Fox DB, Kitir B, Ilkayeva OR, Olsen CA, et al. Lipids Reprogram Metabolism to Become a Major Carbon Source for Histone Acetylation. Cell Rep. 2016;17:1463–72. doi: 10.1016/j.celrep.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bacci M, Lorito N, Smiriglia A, Morandi A. Fat and Furious: Lipid Metabolism in Antitumoral Therapy Response and Resistance. Trends Cancer. 2020 doi: 10.1016/j.trecan.2020.10.004. [DOI] [PubMed] [Google Scholar]
- 17.Civenni G, Bosotti R, Timpanaro A, Vàzquez R, Merulla J, Pandit S, et al. Epigenetic Control of Mitochondrial Fission Enables Self-Renewal of Stem-like Tumor Cells in Human Prostate Cancer. Cell Metab. 2019;30:303–18.:e6. doi: 10.1016/j.cmet.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. [Online]. Available online at: http://www.bioinformatics.babraham.ac.uk/proiects/fastqc/
- 19.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. http://www.R-project.org/ [Google Scholar]
- 21.Oldham MC, Langfelder P, Horvath S. Network methods for describing sample relationships in genomic datasets: application to Huntington’s disease. BMC Syst Biol. 2012;6:63. doi: 10.1186/1752-0509-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolde RP. pheatmap: pretty heatmaps R package v 16. R Foundation for Statistical Computing; 2012. [Google Scholar]
- 26.H W. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York: 2016. [Google Scholar]
- 27.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Millard P, Delépine B, Guionnet M, Heuillet M, Bellvert F, Létisse F. IsoCor: isotope correction for high-resolution MS labeling experiments. Bioinformatics. 2019;35:4484–7. doi: 10.1093/bioinformatics/btz209. [DOI] [PubMed] [Google Scholar]
- 30.Speziale R, Montesano C, De Leonibus ML, Bonelli F, Fezzardi P, Beconi MG, et al. Determination of acetyl coenzyme A in human whole blood by ultra-performance liquid chromatography-mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2018;1083:57–62. doi: 10.1016/j.jchromb.2018.02.039. [DOI] [PubMed] [Google Scholar]
- 31.Mehlem A, Hagberg CE, Muhl L, Eriksson U, Falkevall A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nat Protoc. 2013;8:1149–54. doi: 10.1038/nprot.2013.055. [DOI] [PubMed] [Google Scholar]
- 32.Cortazar AR, Torrano V, Martín-Martín N, Caro-Maldonado A, Camacho L, Hermanova I, et al. CANCERTOOL: A Visualization and Representation Interface to Exploit Cancer Datasets. Cancer Res. 2018;78:6320–8. doi: 10.1158/0008-5472.CAN-18-1669. [DOI] [PubMed] [Google Scholar]
- 33.Granlund KL, Tee SS, Vargas HA, Lyashchenko SK, Reznik E, Fine S, et al. Hyperpolarized MRI of Human Prostate Cancer Reveals Increased Lactate with Tumor Grade Driven by Monocarboxylate Transporter 1. Cell Metab. 2020;31:105–14.:e3. doi: 10.1016/j.cmet.2019.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corbet C, Pinto A, Martherus R, Santiago de Jesus JP, Polet F, Feron O. Acidosis Drives the Reprogramming of Fatty Acid Metabolism in Cancer Cells through Changes in Mitochondrial and Histone Acetylation. Cell Metab. 2016;24:311–23. doi: 10.1016/j.cmet.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Wright HJ, Hou J, Xu B, Cortez M, Potma EO, Tromberg BJ, et al. CDCP1 drives triple-negative breast cancer metastasis through reduction of lipid-droplet abundance and stimulation of fatty acid oxidation. Proc Natl Acad Sci U S A. 2017;114:E6556–E65. doi: 10.1073/pnas.1703791114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang YY, Attané C, Milhas D, Dirat B, Dauvillier S, Guerard A, et al. Mammary adipocytes stimulate breast cancer invasion through metabolic remodeling of tumor cells. JCI Insight. 2017;2:e87489. doi: 10.1172/jci.insight.87489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doroshow DB, Eder JP, LoRusso PM. BET inhibitors: a novel epigenetic approach. Ann Oncol. 2017;28:1776–87. doi: 10.1093/annonc/mdx157. [DOI] [PubMed] [Google Scholar]
- 38.Tasdogan A, Faubert B, Ramesh V, Ubellacker JM, Shen B, Solmonson A, et al. Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature. 2020;577:115–20. doi: 10.1038/s41586-019-1847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonuccelli G, Tsirigos A, Whitaker-Menezes D, Pavlides S, Pestell RG, Chiavarina B, et al. Ketones and lactate “fuel” tumor growth and metastasis: Evidence that epithelial cancer cells use oxidative mitochondrial metabolism. Cell Cycle. 2010;9:3506–14. doi: 10.4161/cc.9.17.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pucino V, Certo M, Bulusu V, Cucchi D, Goldmann K, Pontarini E, et al. Lactate Buildup at the Site of Chronic Inflammation Promotes Disease by Inducing CD4. Cell Metab. 2019;30:1055–74.:e8. doi: 10.1016/j.cmet.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ladanyi A, Mukherjee A, Kenny HA, Johnson A, Mitra AK, Sundaresan S, et al. Adipocyte-induced CD36 expression drives ovarian cancer progression and metastasis. Oncogene. 2018;37:2285–301. doi: 10.1038/s41388-017-0093-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pascual G, Avgustinova A, Mejetta S, Martín M, Castellanos A, Attolini CS, et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017;541:41–5. doi: 10.1038/nature20791. [DOI] [PubMed] [Google Scholar]
- 43.Watt MJ, Clark AK, Selth LA, Haynes VR, Lister N, Rebello R, et al. Suppressing fatty acid uptake has therapeutic effects in preclinical models of prostate cancer. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aau5758. [DOI] [PubMed] [Google Scholar]
- 44.Ying M, You D, Zhu X, Cai L, Zeng S, Hu X. Lactate and glutamine support NADPH generation in cancer cells under glucose deprived conditions. Redox Biol. 2021;46:102065. doi: 10.1016/j.redox.2021.102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kondo A, Yamamoto S, Nakaki R, Shimamura T, Hamakubo T, Sakai J, et al. Extracellular Acidic pH Activates the Sterol Regulatory Element-Binding Protein 2 to Promote Tumor Progression. Cell Rep. 2017;18:2228–42. doi: 10.1016/j.celrep.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Das SK, Eder S, Schauer S, Diwoky C, Temmel H, Guertl B, et al. Adipose triglyceride lipase contributes to cancer-associated cachexia. Science. 2011;333:233–8. doi: 10.1126/science.1198973. [DOI] [PubMed] [Google Scholar]
- 47.Rozeveld CN, Johnson KM, Zhang L, Razidlo GL. KRAS Controls Pancreatic Cancer Cell Lipid Metabolism and Invasive Potential through the Lipase HSL. Cancer Res. 2020;80:4932–45. doi: 10.1158/0008-5472.CAN-20-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corbet C, Bastien E, Santiago de Jesus JP, Dierge E, Martherus R, Vander Linden C, et al. TGFβ2-induced formation of lipid droplets supports acidosis-driven EMT and the metastatic spreading of cancer cells. Nat Commun. 2020;11:454. doi: 10.1038/s41467-019-14262-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sherman MH, Yu RT, Tseng TW, Sousa CM, Liu S, Truitt ML, et al. Stromal cues regulate the pancreatic cancer epigenome and metabolome. Proc Natl Acad Sci U S A. 2017;114:1129–34. doi: 10.1073/pnas.1620164114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yin M, Guo Y, Hu R, Cai WL, Li Y, Pei S, et al. Potent BRD4 inhibitor suppresses cancer cell-macrophage interaction. Nat Commun. 2020;11:1833. doi: 10.1038/s41467-020-15290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tonini C, Colardo M, Colella B, Di Bartolomeo S, Berardinelli F, Caretti G, et al. Inhibition of Bromodomain and Extraterminal Domain (BET) Proteins by JQ1 Unravels a Novel Epigenetic Modulation to Control Lipid Homeostasis. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21041297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang Y, Hu T, Wang T, Shi X, Kitano A, Eagle K, et al. AMP-activated protein kinase links acetyl-CoA homeostasis to BRD4 recruitment in acute myeloid leukemia. Blood. 2019;134:2183–94. doi: 10.1182/blood.2019001076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carrer A, Trefely S, Zhao S, Campbell SL, Norgard RJ, Schultz KC, et al. Acetyl-CoA Metabolism Supports Multistep Pancreatic Tumorigenesis. Cancer Discov. 2019;9:416–35. doi: 10.1158/2159-8290.CD-18-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bailey AP, Koster G, Guillermier C, Hirst EM, MacRae JI, Lechene CP, et al. Antioxidant Role for Lipid Droplets in a Stem Cell Niche of Drosophila. Cell. 2015;163:340–53. doi: 10.1016/j.cell.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen M, Zhang J, Sampieri K, Clohessy JG, Mendez L, Gonzalez-Billalabeitia E, et al. An aberrant SREBP-dependent lipogenic program promotes metastatic prostate cancer. Nat Genet. 2018;50:206–18. doi: 10.1038/s41588-017-0027-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-seq data performed on DU145 cells treated with lactate ± I-BET762 reported in this paper is GEO: GSE195639.