Abstract
Posttranslational modifications of the microtubule cytoskeleton have emerged as key regulators of cellular functions, and their perturbations have been linked to a growing number of human pathologies. Tubulin glycylation modifies microtubules specifically in cilia and flagella, but its functional and mechanistic roles remain unclear. In this study, we generated a mouse model entirely lacking tubulin glycylation. Male mice were subfertile owing to aberrant beat patterns of their sperm flagella, which impeded the straight swimming of sperm cells. Using cryo–electron tomography, we showed that lack of glycylation caused abnormal conformations of the dynein arms within sperm axonemes, providing the structural basis for the observed dysfunction. Our findings reveal the importance of microtubule glycylation for controlled flagellar beating, directional sperm swimming, and male fertility.
Microtubules (MTs) are ubiquitous cytoskeletal components that are structurally highly similar across most cell types and organisms. Their rich variety of posttranslational modifications (PTMs) has long been suggested to form a “tubulin code” that allows MTs to adapt to specific functions (1). However, clear-cut functional and mechanistic data verifying this concept are still scarce. Glycylation is among the least explored tubulin PTMs and has, so far, exclusively been found on axonemal MTs of a variety of species (2) but rarely on cytoplasmic MTs. Tubulin glycylation in mammals is catalyzed in two steps: the tubulin-tyrosine ligase-like (TTLL) glycylating enzymes TTLL3 and TTLL8 add the first glycine residues to unmodified tubulin, while TTLL10 exclusively elongates nascent glycine chains. Absence of elongated glycine chains owing to a constitutively inactive TTLL10 in humans is physiologically tolerated (3), whereas a complete loss of glycylation in mice has damaging effects: Knockout of Ttll3 in combination with short hairpin RNA (shRNA)–mediated knockdown of Ttll8 in mouse brain ventricles results in the loss of motile cilia from the affected ependymal cells (4). In retina and colon, two organs that do not express Ttll8, the knockout of Ttll3 alone leads, respectively, to a progressive shortening of the photoreceptors’ connecting cilia followed by retinal degeneration (5) and to the partial loss of primary cilia in the colon, accompanied by accelerated development of colorectal tumors (6). Finally, knocking down TTLL3 in cultured Madin-Darby canine kidney (MDCK) cells leads to a substantial shortening of primary cilia (7). Thus, glycylation appears to play a crucial role in ciliary integrity and function; however, its function in the most specialized mammalian cilium, the sperm flagellum, has remained elusive. Sperm flagella, similar to motile cilia, are extremely susceptible to alterations in the tubulin PTM polyglutamylation (8–11), but glycylation has only been shown to be essential for the integrity of the sperm flagellum in Drosophila melanogaster (3). The distinct assembly, structure, and size of Drosophila sperm flagella (12) do not allow direct conclusions to be drawn regarding mammalian sperm.
Loss of tubulin glycylation does not affect ciliogenesis
To understand the role of glycylation from the physiological to the ultrastructural, mechanistic level, we generated a double-knockout mouse for the two initiating glycylases Ttll3 and Ttll8 (Ttll3 −/− Ttll8 −/−; fig. S1). Ttll3 −/− Ttll8 −/− mice showed a complete absence of glycylation in motile ependymal cilia from brain ventricle walls (fig. S2, A and D), motile cilia in the trachea (fig. S2, B and D), and sperm flagella (Fig. 1, A and B), as well as the primary cilia from the kidney collecting tubules (fig. S2C). Cilia and flagella were still present in all tissues analyzed (Fig. 1A and fig. S2, A to C), and moreover, no gross defects were observed at the organism and tissue levels. For instance, the brain and kidneys appeared normal, and there were no signs of hydrocephaly (fig. S3A) or polycystic kidneys (fig. S3B), which are typical signs of ciliary dysfunctions (13). Trachea, too, showed no aberrations in the multiciliated epithelium (fig. S3C). The absence of ciliary defects in Ttll3 −/− Ttll8 −/− mice was unexpected. In particular, the loss-of-cilia phenotype in ependymal cells, which was induced by shRNA-mediated depletion of TTLL8 in a Ttll3−/− background (4), was not reproduced in Ttll3 −/− Ttll8 −/− mice, probably because of compensatory mechanisms during mouse development (14, 15).
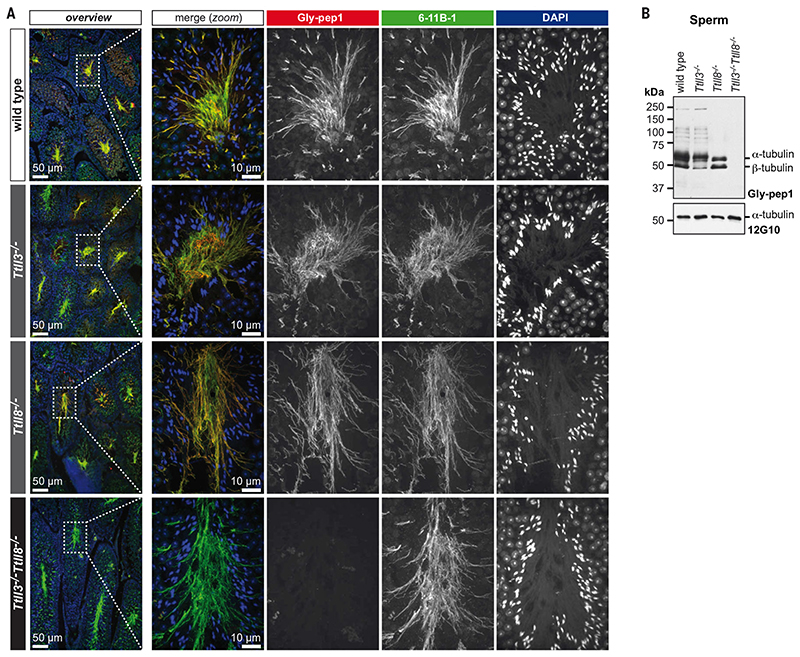
Fig. 1. Total absence of glycylation in Ttll3 −/− Ttll8 −/− mice.
((A) Paraffin-embedded testes sections stained for tubulin acetylation (6-11B-1; green), glycylation (Gly-pep1; red), and DNA [4′,6-diamidino-2-phenylindole (DAPI)]. Nuclear staining shows normal sperm head morphology and a normal arrangement of sperm heads of different developmental stages in the seminiferous tubules of wild-type, Ttll3 −/−, Ttll8 −/−, and Ttll3 −/− Ttll8 −/− testes. Sperm flagella are present in all genotypes (labeled by 6-11B-1), and are glycylated in wild-type and single-knockout but not in Ttll3 −/− Ttll8 −/− sperm tails. ((B) Immunoblot of sperm samples from wild-type, Ttll3 −/−, Ttll8 −/−, and Ttll3 −/− Ttll8 −/− mice. Note that in single-knockout mice (Ttll3 −/−, Ttll8 −/−), glycylation (Gly-pep1) is only partially abolished, while it is undetectable in Ttll3 −/− Ttll8 −/− sperm. The anti-α-tubulin antibody 12G10 confirmed equal tubulin load.
Absence of glycylation affects sperm motility and male fertility
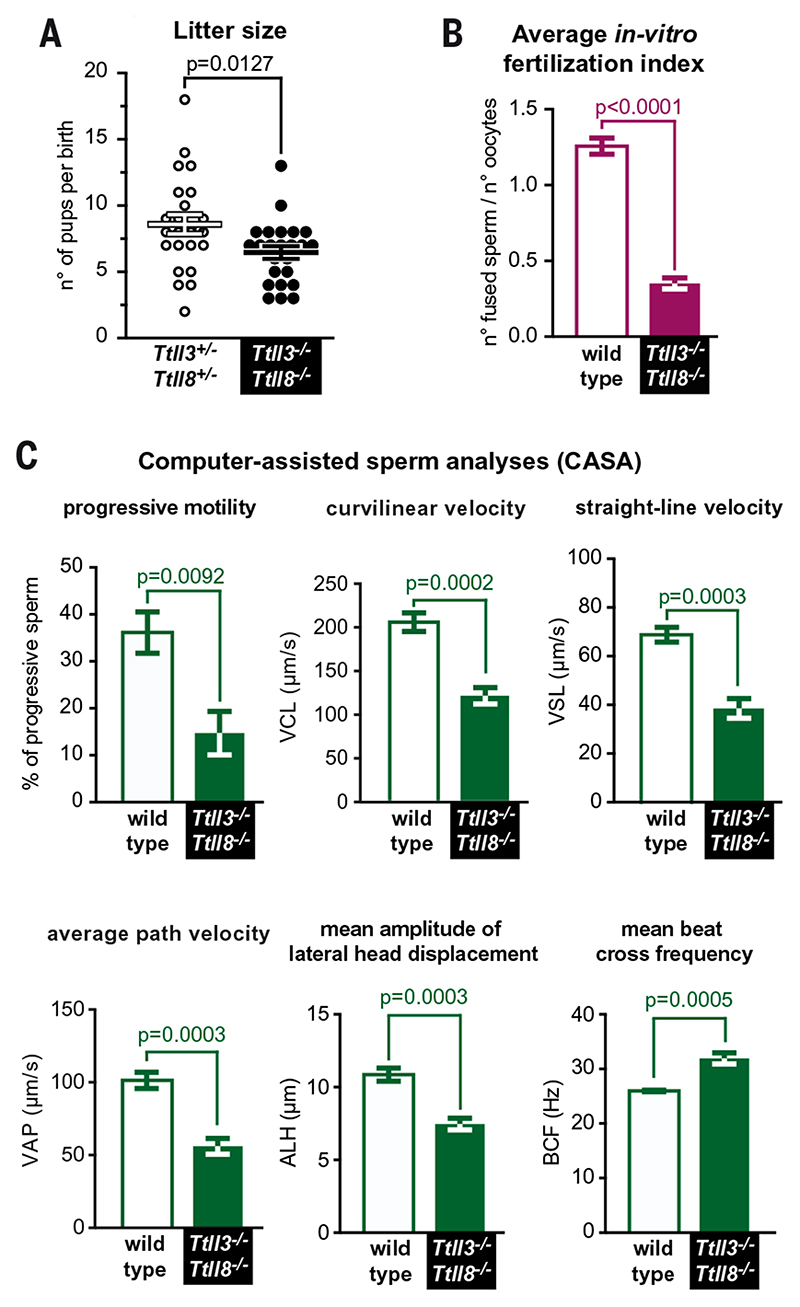
Although the overall tissue architecture of testes was not perturbed (fig. S3D), the decreased average litter size of Ttll3 −/− Ttll8 −/− mice (6.5 ± 2.4 versus 8.4 ± 3.9 for heterozygote controls) (Fig. 2A and fig. S3E) hinted at the possibility that mice were subfertile. To test specifically for male subfertility, we used in vitro fertilization assays, which provide a more sensitive readout than does in vivo fertilization (16). The fertilization index of Ttll3 −/− Ttll8 −/− sperm in vitro was about one-fourth that of wild type (Fig. 2B and fig. S4B), which establishes the subfertility of sperm lacking tubulin glycylation.
Fig. 2. Ttll3 −/− Ttll8 −/− mice show subfertility and reduced sperm motility.
(A) Analysis of litter size. Comparison of heterozygotes (Ttll3 +/− Ttll8 +/−) with Ttll3 −/−Ttll8 −/− mice shows a reduction in the average number of pups per litter (see fig. S3E for details). (B) In vitro fertilization assay with wild-type oocytes. The average fertilization index of five independent experiments (fig. S4B) shows a strong decrease of Ttll3 −/− Ttll8 −/− sperm fertility. (C) Computer-assisted sperm analyses (CASA) comparing wild-type and Ttll3 −/− Ttll8 −/− sperm. Ttll3 −/− Ttll8 −/− mice show a reduced proportion of progressive sperm. In particular, Ttll3 −/− Ttll8 −/− sperm had reduced velocity parameters: curvilinear velocity (VCL), straight-line velocity (VSL), and average path velocity (VAP), together with reduced amplitude of lateral head displacement (ALH). The beat-cross frequency (BCF), on the other hand, was higher in the Ttll3 −/− Ttll8 −/− sperm.
To determine whether lack of glycylation affects the fertilization potential of Ttll3 −/− Ttll8 −/− sperm, we investigated three major parameters that govern the quality of sperm isolated from the cauda epididymides: sperm cell count, morphology, and motility (17). Counts and viability of sperm from Ttll3 −/− Ttll8 −/− mice were nearly identical to those of controls (fig. S3, F and G, and table S1), and only some sperm cells showed morphological defects (fig. S3H). In contrast, most of the motility parameters assessed with computer-assisted sperm analyses (CASA) (fig. S4A) (18) were changed in Ttll3 −/− Ttll8 −/− sperm as compared with wild type (Fig. 2C, fig. S4C, and table S1). We found a clear decrease not only in the proportion of progressive sperm but also in curvilinear velocity (VCL), straight-line velocity (VSL), and average path velocity (VAP), three kinematic parameters assessing sperm velocity (Fig. 2C and fig. S4A). These results strongly suggested a reduced fertilization potential of Ttll3 −/− Ttll8 −/− sperm owing to reduced sperm motility (asthenozoospermia), a common cause of male infertility (19).
Defects in sperm motility are due to altered flagellar beat patterns
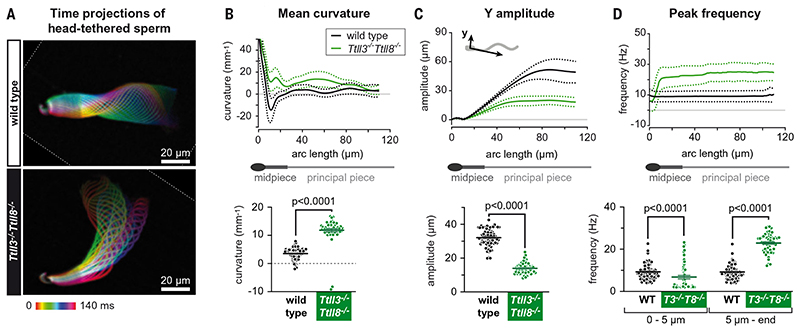
Following the observation of reduced motility of Ttll3 −/− Ttll8 −/− sperm, we further characterized the flagellar beat of those sperm. We first performed two-dimensional (2D) analyses of single sperm cells tethered with their heads at the glass surface. Wild-type sperm showed the characteristic symmetric flagellar beat, whereas flagella of sperm from Ttll3 −/− Ttll8 −/− mice were beating asymmetrically, thus inducing a rotatory swimming trajectory in one preferential direction (Fig. 3A and movie S1). Analyses of flagellar beat parameters (20) revealed that the mean flagellar curvature for Ttll3 −/− Ttll8 −/− sperm was more than three times higher than for wild type (Fig. 3B and fig. S5, A to C). Moreover, Ttll3 −/− Ttll8 −/− sperm flagella beat with less than half the amplitude of wild type, and their beat was biased toward the open-hook side of the sperm head, which could explain the asymmetric beat (Fig. 3C and fig. S5, C and D). Another major defect was found in the peak frequencies of the flagellar beat. Whereas a homogeneous peak frequency was detected along the entire length of wild-type flagella, Ttll3 −/− Ttll8 −/− flagella were beating with a lower frequency close to the sperm head but with a more than two times higher frequency in the rest of the flagellum compared with wild-type sperm (Fig. 3D and fig. S5E). Such modifications of beat patterns cannot be explained by premature hyperactivation, a sperm behavior that normally occurs during sperm transit in the female genital tract and ensures proper sperm progression toward the oocyte.
Fig. 3. Asymmetric beat of Ttll3 −/− Ttll8 −/− sperm flagella.
(A) Color-coded time projections of dark-field recordings of head-tethered mouse spermatozoa in two dimensions: The depicted color-coded time span (140 ms) corresponds to one beat cycle of wild-type sperm. The flagellar envelope of Ttll3 −/− Ttll8 −/− sperm cells is asymmetrically displaced to the open-hook side of the head, in contrast to wild-type sperm. See also movie S1. (B to D) Multiparameter motility analyses of the flagellar beat using the software SpermQ. In all line graphs, solid lines indicate the time-averaged values, and dotted lines the standard deviation for different arc-length positions along the flagellum. Scatter dot plots show the time- and arc-length-averaged values for individual sperm cells (dots) as well as the mean values ±SEM (bars). (B) Mean flagellar curvature. (C) Amplitude of flagellar beat in the direction perpendicular to the head-midpiece axis. (D) Representation of peak frequencies of the flagellar beat. Scatter plot shows the peak frequencies separately for the first 5 μm and for the rest of the flagellum.
Hyperactivation would result in a higher beat amplitude and lower beat frequencies and, accordingly, in a higher amplitude of lateral head displacement (ALH) and a lower beat-cross frequency (BCF) in CASA analyses (21–23). Given that ALH was persistently lower and BCF higher for Ttll3 −/− Ttll8 −/− sperm (Fig. 2C), we excluded the presence of premature hyperactivation.
Altered flagellar beat patterns result in anomalous sperm swim paths
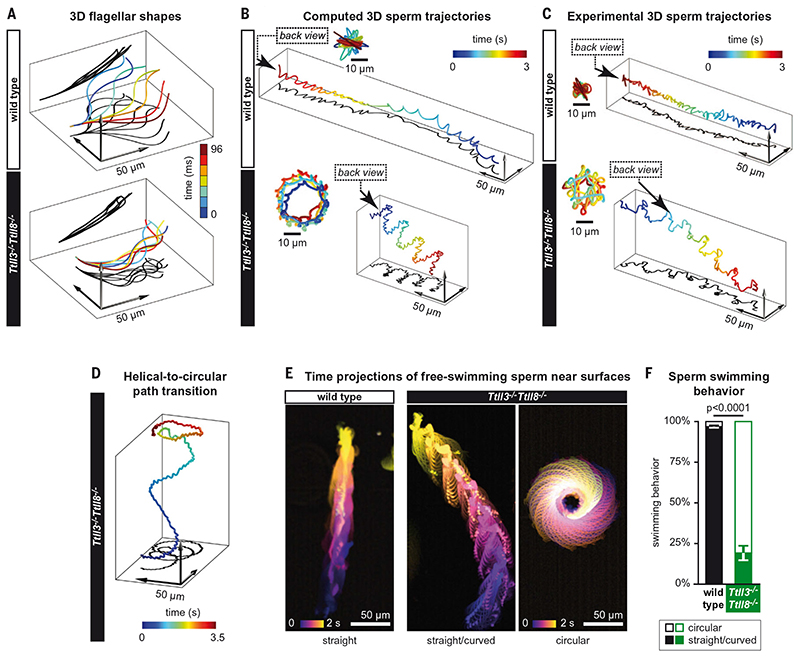
The altered beat patterns we observed for Ttll3 −/− Ttll8 −/− sperm were likely to translate into altered swim paths. We thus simulated the swim paths of sperm in three dimensions on the basis of the flagellar beat patterns we had determined from tethered sperm (Fig. 3), under the assumption that the flagellar beat is not perfectly planar (Fig. 4A). Our simulations showed that wild-type sperm should swim in a twisted ribbon trajectory (24), whereas Ttll3 −/− Ttll8 −/− sperm would adopt a helical swim path (Fig. 4B). To test these predictions, we directly determined the 3D trajectories of free-swimming sperm by in-line holographic microscopy (25). As predicted, wild-type sperm cells swam in a twisted ribbon trajectory, whereas Ttll3 −/− Ttll8 −/− sperm swam predominantly in helical paths (Fig. 4C and fig. S6). Both swimming patterns allow sperm to advance, however, when Ttll3 −/− Ttll8 −/− sperm reached the wall of the observation chamber, their swimming paths spontaneously switched from helical to circular swimming (Fig. 4D), which impedes progressive swimming. Circular swim patterns have been reported for sperm of marine invertebrates (26), but they are atypical for mammalian sperm (27). To test the propensity of Ttll3 −/− Ttll8 −/− sperm to switch to circular swimming, we recorded the swim paths of large numbers of free-swimming sperm cells near the glass surface of the recording chamber. Under these conditions, wild-type sperm swam almost exclusively along straight or curved paths, whereas most (~86%) of the Ttll3 −/− Ttll8 −/− sperm swam in circular paths (Fig. 4, E and F, and movie S2). Only a small fraction (~14%) of Ttll3 −/− Ttll8 −/− sperm showed incidental progressive movement, but even those switched intermittently to circular movements (movie S3). The predominantly circular swimming of Ttll3 −/− Ttll8 −/− spermatozoa near the glass surface can be explained by the increased average curvature of their flagella (Figs. 3B and 4A): Mesoscale hydrodynamics simulations predict that a high mean curvature of the flagellar beat contributes to the accumulation of sperm at the surface, prevents sperm rolling, and results in swimming along tight circles (28).
Fig. 4. Altered swimming behavior of Ttll3 −/− Ttll8 −/− sperm.
(A) 3D flagellar shapes used to compute the 3D swimming path of sperm cells. The shapes were extracted from experiments using tethered sperm (Fig. 3A). The flagellar shapes correspond to about half a beat period, but simulations were based on the complete time series. The z component was assumed to be a smooth arc of constant curvature along the flagellar arc length (κz = 5 × 10−3 μm−1; projections shown in black). (B) Computed swimming paths of sperm cells using resistive-force theory. Wild-type spermatozoa are predicted to swim along a twisted ribbon, whereas Ttll3 −/− Ttll8 −/− sperm cells are predicted to swim along a helical path. (C) Representative examples for reconstruction of 3D swimming paths from in-line holographic recordings of freely swimming sperm. Wild-type and Ttll3 −/− Ttll8 −/− sperm swim along a twisted ribbon and a helical path, respectively. Insets [(B) and (C)]: Back view of the path in the direction indicated by the arrow. See also fig. S6 for individual sperm trajectories. (D) 3D holographic trajectory of a Ttll3 −/− Ttll8 −/− sperm showing the transition from a helical to a circular swimming path when reaching the upper wall of the observation chamber. See also movie S3. (E) Color-coded time projections of representative dark-field recordings of freely swimming wild-type and Ttll3 −/− Ttll8 −/− sperm near the glass surface (see also movie S2). (F) Quantification of the swimming patterns observed in (E). The graph represents the mean (±SEM) of the different mice analyzed.
The main defect of Ttll3 −/− Ttll8 −/− sperm cells, their nonlinear swimming pattern, clearly hampered their capacity to reach the site of fertilization. To achieve progressive swimming, sperm require a symmetric flagellar beat, which Ttll3 −/− Ttll8 −/− sperm fail to generate. A higher beat asymmetry, as seen for Ttll3 −/− Ttll8 −/− sperm, is known to result in an increased rotational velocity perpendicular to the translational velocity of the sperm cell (20, 29, 30) and consequently induces swimming along helical or circular paths. Even helical swim paths can become circular when sperm cells encounter a surface, thus impeding progressive movement (28). Thus, while Ttll3 −/− Ttll8 −/− sperm can advance, and eventually proceed to fertilization, they are highly inefficient when compared with wild-type sperm. Therefore, it appears that the perturbed flagellar beat, which results in nonprogressive swim paths, is most likely the reason for the observed subfertility of the Ttll3 −/− Ttll8 −/− mice.
Lack of glycylation leads to an altered distribution of axonemal dynein conformations
To understand the molecular basis of the perturbed beat patterns of sperm flagella that lack glycylation, we determined native 3D structures of axonemes in situ by plunge-freezing swimming sperm and imaging their flagella using cryo–electron tomography. We then applied subtomogram averaging to the straight segments of the distal part of the flagellar principal piece to generate high-resolution 3D electron density maps of the axonemal 96-nm repeat (31).
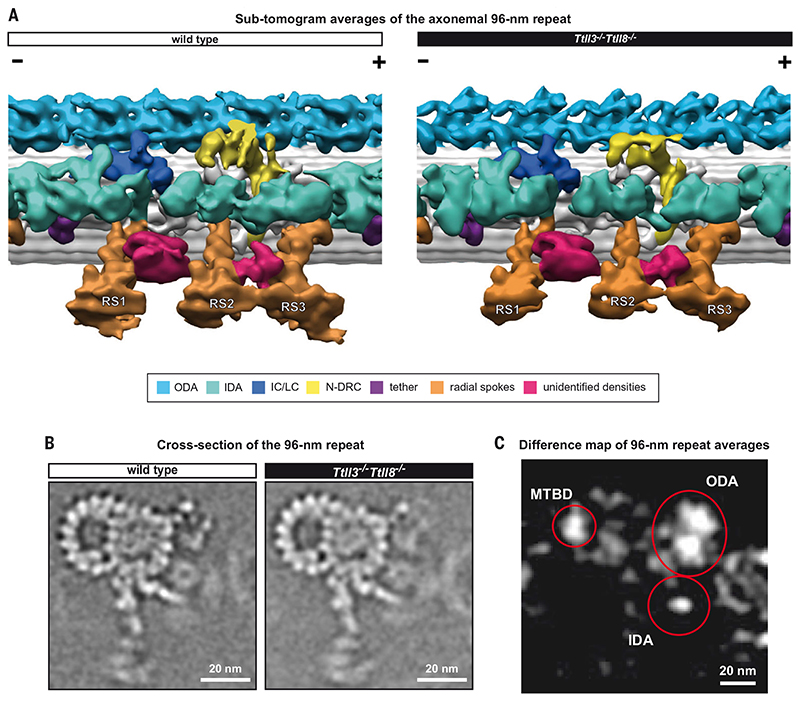
First, we observed that the absence of glycylation in Ttll3 −/− Ttll8 −/− axonemes affected neither the assembly of the MTs nor the overall macromolecular structure of the axonemes (Fig. 5, A and B). We also observed other axonemal features, such as a barrel-shaped structure between the radial spokes 1 and 2 and a protein structure linking the neck regions of radial spokes 2 and 3 (Fig. 5A). These structures, which were not seen in axonemes of other species, were present in both wild-type and Ttll3 −/− Ttll8 −/− mouse sperm flagella. The molecular identity of these barrel and linker structures remains unclear. However, the difference map between the 96-nm repeat models of wild-type and Ttll3 −/− Ttll8 −/− axonemes revealed structural alterations in the Ttll3 −/− Ttll8 −/− axo-nemal dynein arms and their MT-binding domains on the B tubule of each MT doublet (Fig. 5C).
Fig. 5. The assembly of the axonemal 96-nm repeat is not affected in Ttll3 −/− Ttll8 −/− sperm.
(A) 3D-isosurface rendering of the 96-nm repeats from active wild-type and Ttll3 −/− Ttll8 −/− sperm flagella after subtomogram averaging. All known components of the axonemal 96-nm repeat were identified in both wild type and Ttll3 −/− Ttll8 −/−, indicating that absence of glycylation did not affect their assembly. We further identified densities of structures that are not found in axonemes from other species (crimson-colored). A barrel-shaped structure between radial spokes 1 and 2 (RS1 and RS2) and a density that links the neck of radial spokes 2 and 3 (RS2 and RS3). ODA, outer dynein arm; IDA, inner dynein arm; IC/LC, dynein intermediate chain/light chain; N-DRC, nexin-dynein regulatory complex. (B) Slice through subtomogram averaged wild-type and Ttll3 −/− Ttll8 −/− 96-nm repeats. No evident modifications to the macromolecular assembly of the axoneme were found upon depletion of tubulin glycylation. (C) Difference map of wild-type and Ttll3 −/− Ttll8 −/− 96-nm repeat averages. Areas of the 96-nm repeat that present meaningful structural differences between the two averages are circled in red. These include ODAs, IDAs, and part of the external wall of the B-tubule. MTBD, microtubule-binding domain.
Axonemal dynein motors—the outer and inner dynein arms (ODAs and IDAs)—produce the force necessary to power flagellar motion and are organized in complexes that link together neighboring axonemal MT doublets (32). Functionally, ODAs are thought to control the beat amplitude and frequency, and IDAs the beat waveform (33). Consequently, loss or mislocalization of specific axonemal dynein isoforms in a flagellum can cause alterations of the flagellar beat. However, this appeared not to be the case in Ttll3 −/− Ttll8 −/− axonemes, because all ODA and IDA heavy chains were present in the 3D electron density maps (Figs. 5, A and C, and 6, A and B). Moreover, we confirmed that the two ODA heavy chains (DNAH17: β-heavy chain; DNAH8: γ-heavy chain; fig. S7A) and the IDA heavy and light chains (DNAH2: IDA-f b-heavy chain; DNALI1: light intermediate chain; fig. S7B) were present and correctly distributed along the entire length of the Ttll3 −/− Ttll8 −/− flagella. Thus, we reasoned that the structural alterations of Ttll3 −/− Ttll8 −/− axonemes that were highlighted by the difference map must have been of amore subtle nature.
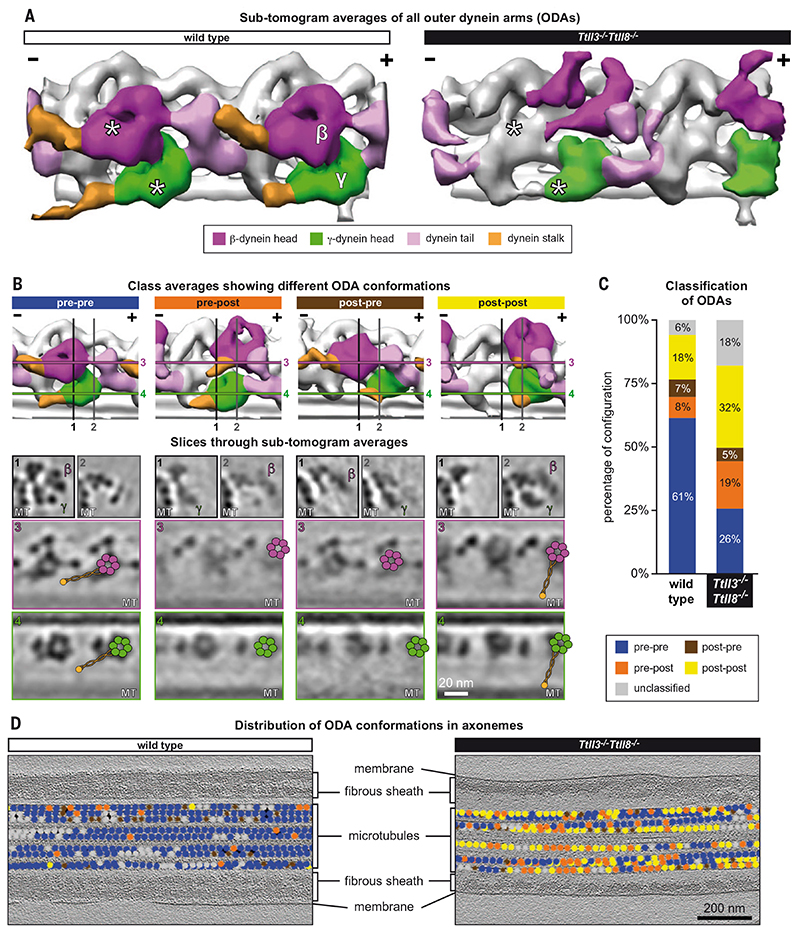
Fig. 6. Distribution of ODA conformations are perturbed in Ttll3 −/− Ttll8 −/− sperm flagella.
(A) Averages of all ODA subtomograms from wild-type and Ttll3 −/− Ttll8 −/− sperm flagella (from straight axonemal segments) highlight that the structures of the dynein heads (β-heavy chain, magenta; γ-heavy chain, green) are altered in Ttll3 −/− Ttll8 −/− sperm. Asterisks indicate the position of β- and γ-heavy chains in the wild-type structure and the corresponding coordinates in the Ttll3 −/− Ttll8 −/− average. The Ttll3 −/− Ttll8 −/− average shows a shift of both β- and γ-dynein heads toward the MT plus-end (+). (B) Representative class averages of the distinct ODA conformations identified in wild-type and Ttll3 −/− Ttll8 −/− flagella. Isosurface rendering (top panels) and representative orthogonal and longitudinal slices (lower panels) through β- and γ-heavy chains [color-coded as in (A)]. The numbered lines illustrate the slicing planes through the subtomogram averages in the lower panels. The superimposed schematic models show the positions of dynein β- and γ-AAA domains (magenta and green, respectively), the dynein stalks (orange lines), and the MT binding domains (orange dot). Note that pre-pre, pre-post, post-pre, and post-post refer to dynein γ- and β-heavy chain conformations, respectively (pre = pre-powerstroke, post = post-powerstroke). While the position of the stalk is clearly visible in the pre-pre and post-post conformations, as highlighted by the schematics, the stalks were not easily identified in the pre-post and post-pre conformations, probably because of their unstable conformation. (C) Incidence of the distinct ODA conformations in wild-type (n = 2691 particles) and Ttll3 −/− Ttll8 −/− (n = 3656 particles) axonemes. Ttll3 −/− Ttll8 −/− flagella show a particularly increased percentage of pre-post and post-post powerstroke conformations. (D) Distribution of the different ODAs conformations visualized in wild-type and Ttll3 −/− Ttll8 −/− axonemes shows the absence of a clear pattern of distribution in Ttll3 −/− Ttll8 −/− sperm [color coding of dots as in (B) and (C)]. The distribution in Ttll3 −/− Ttll8 −/− indicates a reduced ability of ODAs to coordinate their activity states.
Proper spatial and temporal coordination of dynein motor activities is crucial to drive flagellar motility. Experimental evidence from sea urchin sperm and Chlamydomonas flagella shows that in beating flagella, most ODAs adopt a pre-powerstroke conformation, whereas intermediate and post-powerstroke conformations are mostly found on subsets of MTs in the bent regions of axonemes (34). Regardless of the nature of the mechanism that initiates the beating of the flagellum—which is still debated in the field—a spatially clustered distribution of active and inactive dynein motors, correlating with the bending of the flagellum, is required for the propagation of the normal bending motion.
Our analysis of straight segments of active wild-type mouse sperm flagella showed that ODAs were predominantly in the pre-powerstroke conformation (Fig. 6A and movie S4, A and B), indicating that the distribution of dynein conformations, previously observed in echinoderms and algae (34), is also conserved in mammals. In contrast, analysis of Ttll3 −/− Ttll8 −/− flagella showed drastically altered structures of the β- and γ-heavy chains of ODAs (Fig. 6A and movie S4, A and B). The average of all Ttll3 −/− Ttll8 −/− ODAs from straight axoneme segments showed poorly resolved heavy chains (Fig. 6A), which is characteristic of flexible or conformationally heterogeneous structures that disappear in subtomogram averages. This observation indicated defects in the conformational arrangement of the dynein heavy chains in Ttll3 −/− Ttll8 −/− axonemes, which we confirmed with a subtomogram classification approach. Only 26% of Ttll3 −/− Ttll8 −/− ODAs exhibited the pre-powerstroke conformation, about 18% could not be classified, and the remaining 56% showed either the post-powerstroke state (32%) or mixed conformations of opposing states of the β- and γ-heavy chains, either as pre-post (19%) or post-pre conformations (5%) (Fig. 6, B and C, and movie S4C). Notably, the β-heavy chains were most affected by the absence of glycylation. The space distribution of the ODA conformations in the Ttll3 −/− Ttll8 −/− flagellar segments was considerably altered as compared with wild-type flagella (Fig. 6D), did not reveal a discernible pattern, and showed uncoordinated dynein activity. This altered conformational distribution of ODAs is presumably linked to an altered force-generation profile that results in an aberrant beating phenotype.
Subtomogram averaging of Ttll3−/−Ttll8−/− axonemes further revealed that IDAs also show an abnormal shift of their heavy chains toward the MT plus-end (fig. S8A), a conformation characteristic of immotile flagella (34). Subtomogram classification of IDAs from sperm flagella (fig. S8, B to D) revealed that in wild type, 42% of the IDAs were in pre-powerstroke and 22% in a post-powerstroke conformation (fig. S8, C and D). By contrast, only 20% of the Ttll3 −/− Ttll8 −/− IDAs were in pre-powerstroke and 45% IDAs were in post-powerstroke conformation (fig. S8, C and D). Among all IDAs analyzed, IDA-f showed the most prominent structural and positional difference between wild-type and Ttll3 −/− Ttll8 −/− sperm flagella (fig. S8B). The conformational reconfiguration of IDA-f occurred concurrently with an opposing tilting motion of the nexin-dynein regulatory complex (N-DRC) and the radial-spoke heads. This movement was perpendicular to the MT axis and associated with a displacement of the next MT doublet (movie S5). Similar movements have also been described for the nexindynein regulatory complex in Chlamydomonas axonemes (35) and for the MT doublets in sea urchin sperm flagella (34).
Mechanisms by which glycylation affects flagellar beat
Our structural data show that the lack of glycylation in Ttll3 −/− Ttll8 −/− sperm axonemes does not affect the binding and assembly of dyneins to A-tubules of the axonemal MT doublets (Fig. 6A, fig. S8A, and movie S4). Nevertheless, the lack of glycylation perturbed the coordination of motor activities. Thus, the affinity of the dynein MT-binding domain for the B tubule may depend on tubulin glycylation, potentially changing the dynamics of the “reloading” from the post-powerstroke into the pre-powerstroke state. Indeed, in vitro single-molecule experiments have shown that other tubulin PTMs can alter certain aspects of the interactions between dynein motors and MTs (36, 37). Perturbations of dynein–MT interactions are expected to affect the flagellar beat characteristics, such as the amplitude and the frequency, which is what we observed for Ttll3 −/− Ttll8 −/− sperm (Fig. 3, C and D). Yet it remains unclear why the flagellar beat of Ttll3 −/− Ttll8 −/− sperm features a higher mean curvature and why the midpiece and the principal piece displayed different beat frequencies. The observed increased mean curvature of Ttll3 −/− Ttll8 −/− flagella could either be caused by a differential distribution of glycylated tubulin across different MT doublets and the resulting effect of this asymmetry on motor activity, or it could be caused by the effect of a generalized increase in motor activity on an inherently asymmetric axonemal structure. Whatever the origin, such asymmetry would result in bending in one preferred direction and axonemal twist, which has been suggested to control the switching point of flagellar oscillations (38). The outer dense fibers and the mitochondrial sheath, structures that are present in the flagellar midpiece, could prevent flagellar twist at this location (39). This would result in the observed arc length–dependent beat frequencies of the strongly bent Ttll3 −/− Ttll8 −/− flagellum. In addition, the altered dynein activity we observed might result in local inhibition of MT sliding. Sliding inhibition, in turn, could result in secondary waves that emanate farther up in the flagellum, as observed in flagella lacking glycylation (movies S1 and S2). The increased frequency and reduced amplitude of these waves would be consistent with findings in demembranated sperm (40).
Our observation that tubulin glycylation is essential for the coordination of the power-stroke cycle of axonemal dynein motors does not exclude the possibility that additional processes sensitive to glycylation contribute to the observed flagellar beat phenotype. For instance, proper flagellar beat also involves the central-pair MTs that transmit mechanical signals via the radial spokes through the nexin-dynein regulatory complex, which control both IDAs and ODAs (41, 42). Indeed, central-pair MTs are also glycylated (43), and so we cannot exclude the possibility that the potential absence of glycylation on these MTs also contributes to a perturbed swimming pattern.
To verify whether the lack of glycylation also affects the beating of other motile cilia in Ttll3 −/− Ttll8 −/− mice, we recorded ependymal cilia of brain ventricles in isolated brain slices. Cilia from both wild-type and Ttll3 −/− Ttll8 −/− mice showed similar beat frequencies in three independent experiments (movie S6), suggesting no major beat defects. However, ependymal cilia are shorter than sperm flagella and might, similar to respiratory cilia in the trachea (44), contain different dynein heavy chains in their axonemes. Thus, the sensitivity of motile cilia to glycylation might be different from what we observed in sperm flagella.
While our data demonstrate a causative role of glycylation for the observed defects of Ttll3 −/− Ttll8 −/− sperm, other tubulin PTMs might also be involved. Earlier studies in Tetrahymena thermophila as well as in murine photoreceptors have revealed that absence of tubulin glycylation leads to an increase in tubulin polyglutamylation (5, 45, 46). This is intuitive, because both PTMs use overlapping modification sites on tubulin (47). Accordingly, we also found an increase in tubulin polyglutamylation in Ttll3 −/− Ttll8 −/− sperm (fig. S9). Nevertheless, it is rather unlikely that increased polyglutamylation participates in the observed defects in Ttll3 −/− Ttll8 −/− sperm, given that the defects in mice lacking the deglutamylases CCP1 (9, 11) or CCP5 (48) are substantially different from those found in Ttll3 −/− Ttll8 −/− mice.
Conclusion
Here we have elucidated the functional roles and molecular mechanisms of tubulin glycylation in ciliary and flagellar functions. The generation of a mouse model entirely lacking tubulin glycylation allowed us to reveal the importance of this PTM for mammalian sperm flagella. We demonstrated that the functional distribution of post- and pre-powerstroke conformations of axonemal dynein arms, which is an essential prerequisite for a physiological flagellar beat (33, 34, 42), depends on the presence of tubulin glycylation. Absence of glycylation in flagella of Ttll3 −/− Ttll8 −/− sperm led to severe alteration of the symmetry, amplitude, and frequency of the flagellar beat, which resulted in sperm failing to swim along a straight line. Straightline swimming, however, is a crucial requirement for sperm to reach the oocyte in the female reproductive tract. Although Ttll3 −/− Ttll8 −/− males were fertile, they produced smaller litter sizes and were revealed to be subfertile using in vitro fertilization assays. Human sperm are more susceptible to deficiencies than are mouse sperm(49), and sperm motility defects such as asthenozoospermia account for almost 80% of the sperm defects observed in infertile men (50). Thus, our findings could have implications for human fertility.
Our cryo–electron tomography of the mammalian sperm flagellum uncovers the molecular mechanism by which glycylation supports flagellar beating and in turn affects sperm swimming and male fertility. Considering the massive evolutionary pressure on sperm fitness (51, 52), our findings might explain why glycylation has, with only a few exceptions, been maintained throughout evolution. Finally, our work implies that a perturbation of tubulin glycylation could potentially provide a molecular cause of male infertility in humans.
Materials and methods
Transgenic mice
Generation of Ttll3−/− and Ttll8−/− mice
The Ttll3−/− mice were generated in the lab as previously described (5) (fig. S1A). The Ttll8flox/flox mouse was generated from the Ttll8tm1a(EUCOMM)Wtsi targeting vector obtained from EUCOMM consortium (IKMC Project #83158) at the Institut Clinique de la Souris, Strasbourg, France. The targeting vector was constructed as follows: the 5′ (4.7-kb) and 3′ (4.7-kb) regions of Ttll8, LacZ and inter-loxP (0.6 kb) fragments were amplified by polymerase chain reaction (PCR) and sequentially subcloned into an MCI proprietary vector containing the LoxP sites and a neomycin resistance cassette flanked by the flippase recognition target (FRT) sites (fig. S1B). The linearized construct was electroporated in 129S2/SvPas mouse embryonic stem (ES) cells. After selection, targeted clones were identified by PCR using external primers and further confirmed by Southern blot with 5′ and 3′ external probes. Two positive ES clones were injected into blastocysts, and derived male chimeras gave germline transmission. The excision of the neomycin resistance cassette was performed in vivo by breeding the chimeras with a Flp deleter line (C57BL/6N genetic background FLP under ACTB promoter) to generate the Ttll8flox/flox mice. To generate Ttll8 −/− mice, we excised exon 2 by crossing Ttll8flox/flox mice with mice expressing cre-recombinase under the control of a PGK promoter (53).
For generation of the double-knockout Ttll3 −/− Ttll8 −/− mice, the single-knockout mice were intercrossed to generate the heterozygotes Ttll3 +/− Ttll8 +/−, which were later crossed between each other to generate the homozygote Ttll3 −/− Ttll8 −/− mice. All animals were back-crossed at least seven generations to the C57BL/6N animals.
Mouse genotyping
DNA was extracted from ear fragments (collected during mouse identification) or tail fragments, using proteinase K (#193504, MP Biomedicals). PCR-based genotyping was performed using primers listed in fig. S1C and protocols listed in fig. S1D. Examples of genotyping results for all transgenes and deletions are shown in fig. S1E.
Mouse strain breeding
Animal care and use for this study were performed in accordance with the recommendations of the European Community (2010/63/UE) for the care and use of laboratory animals. Experimental procedures were specifically approved by the ethics committee of the Institut Curie CEEA-IC #118 (authorization no. 04395.03 given by National Authority) in compliance with the international guidelines.
Tissue histology
Six-month-old mice were anesthetized by intra-peritoneal application of ketamine and xylazine (100 mg per kilogram body weight for ketamine and 10 mg per kilogram body weight of xylazine) and subjected to cardiac perfusion with phosphate-buffered saline (PBS; pH 7.4) containing 4% paraformaldehyde (PFA). After perfusion, the mice were dissected, and the tissues were post-fixed with 4% PFA in PBS overnight at 4°C. The fixed tissues were washed and embedded in paraffin. Seven-micrometer sections were obtained using a microtome (#RM2245, Leica Biosystems). Sections were further analyzed using immunocytochemistry or hematoxylin-eosin staining.
Sperm isolation and Coomassie blue staining
Sperm from 3-month-old male mice were stained with Coomassie blue staining solution as described earlier (48). Briefly, the mice were cervically dislocated, and the epididymides isolated and lacerated in 1 ml PBS using 26G needles (0.45 mm by 13 mm; #613-5377, VWR) to allow the spermatozoa to swim out from the epididymides by incubating for 15 min at 37°C. The sperm suspensions obtained from both wild-type and Ttll3 −/− Ttll8 −/− mice were fixed with 4% PFA, centrifuged at 500g for 5 min at room temperature (RT), and resuspended in 100 mM ammonium acetate, and a smear was prepared with 50 to 70 μl of the sample on previously coated poly-l-lysine glass slides. Once the smear was dry, it was rehydrated and stained with 0.22% Coomassie blue (#35050, Serva Electrophoresis GmBH) for 2 min at RT. The slides were subsequently washed with water, mounted with Aquatex mounting medium (#108562, Merck Millipore), and analyzed.
Culture of primary ependymal cells
Primary ependymal cells were cultured using the well-established protocol for isolation, culturing, and differentiation (54). Briefly, the brains of wild-type, Ttll3 −/−, Ttll8 −/−, or Ttll3 −/− Ttll8 −/− PN0-PN2 mouse pups were isolated and dissociated, and the cells from the subventricular zone were isolated by dissociating the ventricles using papain for 1 hour at 37°C. The cells were plated at high density in Dulbecco’s modified eagle’s medium (DMEM) containing 10% FBS in a 25-cm2 flask precoated with poly-l-lysine (P4832; Sigma-Aldrich). When cells reached confluency, the astroglial monolayer was replated onto precoated 12-mm or 24-mm glass coverslips respectively in 24-well or 6-well plates at a density of 104 cells/μl and maintained in serum-free medium to enable differentiation.
Dissection of lateral ventricles and whole-mount immunostaining
To analyze the ependymal cilia in the brain ventricle by immunocytochemistry, whole-mount sections of the ventricles were obtained as per the protocol described earlier (54). Briefly, PN15 pups from wild-type, Ttll3 −/−, Ttll8 −/−, or Ttll3 −/− Ttll8 −/− mice were cervically dislocated, and the whole brain was retrieved by carefully peeling off the skull without disturbing the brain tissue using spring scissors and forceps. The brain was dissected in PBS using a micro knife under a stereomicroscope. During the entire dissection procedure, the tissue was held by inserting the forceps above the corpus callosum and into the thalamus to ensure the ventricle wall was not disturbed. Once the two cortical hemispheres were separated, the hippocampus was carefully removed, followed by the median wall. Then, to ensure better access to the antibodies, the thalamus, choroid plexus, and corpus callosum were separated from the ventricle.
Once the lateral wall was obtained, it was permeabilized with 0.1% Triton X-100 in BRB80 (80 mM K-Pipes, pH 6.8, 5 mM Na-EGTA, 5 mM MgCl2) for 90 s, followed by fixing with 4% PFA in BRB80 for 20 min at RT. The tissue was then carefully washed three times with PBS, blocked for 1 hour with saturation buffer (PBS containing 10% FBS, 0.1% Triton X-100), and stained with 200 μl primary antibodies listed in table S2 in a 2-ml Eppendorf tube at 4°C overnight. After 3× washing off the primary antibodies with PBS containing 0.1% Triton X-100, the tissues were stained with the respective secondary antibodies, as indicated in table S2 for 1 hour at RT. All the antibodies were diluted in saturation buffer. Nuclei were visualized by staining for 5 min with DAPI (0.02 μg/ml; #D3571; Thermo Fisher Scientific).
After staining, the tissues were further dissected before mounting. First, the cortex around and under the lateral wall was excised, taking care of the curvature and depth of the lateral wall. The rostral part of the wall was cut prior to the caudal part. The tissue was held using forceps inserted into the thalamus. Once the thalamus was removed, the tissue was transferred to a clean slide and mounted using ProLong Gold (#P36934, Thermo Fisher Scientific), taking care not to exert excess pressure on the tissue while mounting.
Staining samples for immunofluorescence microscopy
Immunocytochemistry of testes, kidney, and trachea was carried out according to the established protocols (55). In brief, the tissue sections on the slides were hydrated with PBS, immersed in 10 mM Na-citrate pH 6.0, and boiled in a microwave for 7 min to enable antigen retrieval. The slides were subsequently cooled in ice for 1 hour, after which they were blocked with blocking buffer [PBS containing 10% normal goat serum (NGS), 0.3% Triton X-100]. Immunostaining of the different dyneins in spermatozoa was carried out by preparing smears of the wild-type and Ttll3 −/− Ttll8 −/− sperm on clean glass slides. The smears were fixed with 4% PFA for 10 min, followed by antigen retrieval and permeabilized with PBS containing 0.2% Triton X-100. The smears were then blocked for 1 hour at RT with buffer containing 1% BSA. The slides were then stained with the different primary antibodies as listed in table S2 overnight at 4°C in a humid chamber. After washing the slides 4 × 5 min with PBS containing 0.1% Triton X-100, the sections were stained with the respective secondary antibodies as listed in table S2 for 1 hour at RT. Most of the antibodies used were diluted in PBS containing 5% NGS, 0.3% Triton X-100, except the dynein antibodies that were diluted in PBS containing 1% BSA, 0.02% Triton X-100. Nuclei were visualized by staining for 5 min with DAPI (0.02 mg/ml; #D3571; Thermo Fisher Scientific). The slides were subsequently washed and mounted with ProLong Gold (#P36934, Thermo Fisher Scientific) or DAPI Vectashield (#H1200, Vector Laboratories).
Microscopy of histology, immunocytochemistry, and immunofluorescence
All immunofluorescence images apart from the whole-trachea section and sperm immunofluorescence (in figs. S2B and S7, respectively) were acquired using the Nikon Ti-E spinning disk inverted confocal laser microscope using either a 60× [oil immersion; numerical aperture (NA) 1.40] or a 100× (oil immersion; NA 1.49) objectives at RT. Images were acquired using the Prime95B sCMOS camera (Photometrics) and Leica MM AF imaging software. Multiple Z-stacks were acquired, and a maximum intensity projection was prepared in Fiji v2.1.0 (National Institute of Health) to generate the final image. Whole-trachea sections were imaged using the Structured Illumination Microscope (Optigrid/Leica systems) with a 5× (NA 0.15) objective at RT, an ORCA-Flash4.0 camera (Hamamatsu), and Leica MM AF imaging software and processed using Fiji v2.1.0. For the histology, hematoxylin-eosin stained tissue sections were imaged in the Axio Imager.M2 microscope (Zeiss) using either a 10× (NA 0.3), 20× (NA0.5), or a 63× (water immersion; NA1.2) objective. Multiple tiles of each tissue section were imaged. Tiles were subsequently stitched, background adjusted and processed using the ZEN v2.3 software (Zeiss). Sperm immunofluorescence was imaged using Zeiss Axiophot epifluorescence microscope with a 63× (oil immersion; NA 1.40) or 100× (oil immersion; NA 1.49) objective using a cooled charge-coupled device (CCD) camera (Hamamatsu) under identical instrument settings, with MetaMorph software (Molecular Devices).
Immunoblot analyses
Wild-type, Ttll3 −/−, Ttll8 −/−, or Ttll3 −/− Ttll8 −/− mice were sacrificed by cervical dislocation, and the trachea were harvested in 2-ml vials. The trachea were weighed and resuspended in lysis buffer (PBS pH 7.5, containing 25 mM HEPES, 200 mM NaCl, 5% glycerol, 1 mM dithiothreitol (DTT), 0.5% NP40 and protease inhibitor cocktail) at 100 μl per 100 mg of the tissue and homogenized. The homogenate was then centrifuged at 10,000g for 10 min at 4°C and the supernatant collected. The epididymides from these mice were lacerated in 1 ml PBS to allow the spermatozoa to swim out from the epididymides by incubating for 15 min at 37°C. The sperm suspension was centrifuged at 500g for 5 min at 4°C, after which the sperm pellets obtained were lysed in 100 μl of lysis buffer and centrifuged at 10,000g for 10 min at 4°C, and the supernatant collected. After estimating the total protein using the Pierce BCA protein estimation kit (#23227; Life Technologies), 1 to 2 μg of the total protein was run on 10% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gels. A specific protocol for SDS-PAGE gels was used to separate α- and β-tubulin (56). Proteins were transferred to nitrocellulose membranes with Trans-Blot Turbo using transfer packs (#1704159; Bio-Rad) and subjected to immunoblot analysis.
The ciliated cells, grown on six-well plates were harvested at different time intervals, with 500 μl of Laemmli buffer [400 mM Tris-HCl, pH 6.8 containing 450 mM DTT, 10% (w/v) SDS, 50% (v/v) glycerol, ~0.006% (w/v) bromo-phenol blue], heated to 95°C for 10 min and equal protein was run on 10% SDS-PAGE gels and analyzed by immunoblot analysis.
For all immunoblots, the membranes were blocked overnight with PBS containing 0.1% Tween 20 (PBS-T) and 5% nonfat milk and then incubated for 2 hours at RT with the different primary antibodies as listed in table S2. Antibodies were diluted in PBS-T containing 2.5% nonfat milk. Membranes were washed four times with PBS-T and then incubated for 1.5 hours with respective secondary antibodies conjugated to horseradish peroxidase, as listed in table S2, diluted to 1:10,000 in PBS-T. After washing, antibody labeling was revealed using Clarity Western ECL detection reagent (#1705061; Bio-Rad).
Ependymal cilia beating analyses
Image acquisition and generation of videos
Brain sections from PN20-PN30 wild-type and Ttll3 −/−,Ttll8 −/−, or Ttll3 −/−,Ttll8 −/− mice were collected and imaged as described in (57). Briefly, the dissected brains were sectioned into ~200-μm sagittal sections in Leibovitz L15 medium. The sections were then placed in 100 μl medium on a glass slide and covered with a 24 mm by 60 mm glass coverslip no. 1 by gently sealing it with vacuum grease. The sections were immediately imaged using the CLSM-SP5 confocal light microscope (Leica Biosystems) equipped with a resonance scanner for high-speed imaging, using a 60× (oil immersion; NA 1.4) objective at 37°C. The sections were imaged at 64 pixels by 48 pixels (15× zoom) at 100 frames per second (FPS) for 3 to 5 s using the LSM software, to obtain beating cilia with minimal tissue background. The time series obtained were exported as.avi videos, which were adjusted for their brightness and contrast using Fiji v2.1.0. For better visualization of the curvature of the ciliary beat, the videos were slowed down 20-fold, to run at 5 FPS.
Cilia beat frequency analysis
To analyze the ependymal cilia beat frequency (CBF), the videos obtained for both the wild-type and Ttll3 −/− Ttll8 −/− mice were analyzed using the CiliaFA plugin developed for ImageJ (58) that allowed fast, automated beat frequency analysis minimizing manual errors. The software works on extracting the pixel intensities, coupled with Fast Fourier Transformation in Microsoft Excel to provide the CBF. 256 frames of each video obtained from the wild-type or Ttll3 −/− Ttll8 −/− brain sections were analyzed in CiliaFA. The CBF obtained were tabulated and quantified using GraphPad prism version 8.
Sperm motility analyses
Sperm preparation for motility and holographic analyses of flagellar beat
Mouse sperm were collected by cutting off the cauda epididymites followed by a swim-out process in modified TYH medium (10 mM HEPES pH 7.4, 135 mM NaCl, 4.8 mM KCl, 2 mM CaCl2, 1.2 mM KH2PO4, 1 mM MgSO4, 5.6 mM glucose, 0.5 mM Na-pyruvate, 10 mM L-lactate) for 30 min at 37°C. Sperm were collected carefully without touching oil and tissue fragments.
Analysis of sperm motility and flagellar beating
Freely swimming and tethered sperm were studied in TYH medium containing 3 and 0.3 mg/ml BSA, respectively. Images were recorded using a high-speed camera (pco.dimax; PCO AG) and an inverted microscope (IX 71; Olympus). The temperature of the microscope was adjusted to 37°C using an incubator (Life Imaging Services, Switzerland). Wide-field images were recorded at 50 FPS using a 4× objective (UPLanFLN; Olympus) and custom-made observation chambers with a 100-μm depth. Illumination was achieved using a 660-nm LED. Dark-field and holographic recordings were achieved using a 10× objective (UPLSAPO10X; Olympus) and an additional 1.6× magnifying lens.
Dark-field images were recorded at 250 FPS using a M730L4 LED light source. For analysis, images were processed by a Gaussian blur (sigma: 0.5 pixels) and the Subtract Background method (radius: 5 pixels) in ImageJ. Next, the flagellar analysis was performed using SpermQ (20). Positive curvature refers to bending in the direction of the hook-shaped sperm head. The peak frequency was determined from the Fast Fourier Transform of the curvature values. Individual values for all the analyses are provided in table S3.
Three-dimensional holographic images were recorded at 500 or 1000 FPS in observation chambers with a 150-mm depth. Coherent illumination was produced by a 510-nm laser (LDH-D-C serials, PicoQuant GmbH) and the corresponding controller (Sepia II Multichannel Processor, PicoQuant GmbH). Holograms were analyzed as previously described (25). In brief: The background was calculated by averaging all the holograms from each recording, and a background-free hologram was obtained by dividing the original hologram by the background. The Rayleigh-Sommerfeld back-propagation method (59) was used to numerically refocus each background-free hologram, resulting in a refocused stack. A filter based on the Gouy phase anomaly (60) was applied on the refocused stack, and the 3D coordinates of the sperm flagellum were extracted by localizing the brightest shape in the refocused stack. The head x and y coordinates were estimated by calculating the center of mass of the scattering pattern from the sperm head. To determine robustly the z head coordinates, we fitted a plane to the basal end of the flagellum (about 11 μm from the head center). The head z coordinate was obtained by substituting the head x and y coordinates into the plane equation. All the analysis was done in the open-source software ImageJ combined with custom-made Java plugins.
Three-dimensional trajectories predicted from flagellar beat patterns were computed using resistive-force theory, as previously described (25, 29), using the 2D flagellar beat pattern from tethered sperm (Fig. 3A) and assuming a constant z = z(s) component characterized by an arc of curvature κz = 5 × 10−3 μm−1 along the full flagellar arc length s.
Generating color-coded projections
Color-coded videos and projections were generated using custom ImageJ macros. The macros and code are open source and freely available at https://github.com/hansenjn/ColorStackByTimeAndProject.
Computer-assisted sperm analyses (CASA)
Sperm motility was assessed by computer-assisted sperm analysis (CASA) using the CEROS II apparatus (Hamilton Thorne, Beverly, MA, USA). Briefly, mouse sperm cells expelled from the cauda epididymis were recovered in M2 medium (Sigma-Aldrich, Saint-Louis, MO, USA) and incubated for 10 min at 37°C in 5% CO2. The suspension was collected, and total sperm count determined. At least 500 sperm cells per sample were analyzed in 20 μm chambers (Leja Products B.V., Netherlands) with a Zeiss AX10 Lab. A1 microscope (10× objective) and using HT CASAII software.
The settings were as follows: acquisition rate, 60 Hz; number of frames, 45; minimum head brightness, 175; minimum tail brightness, 80; minimum head size, 10 μm2; minimum elongation gate, 1%; maximum elongation gate, 100%; objective magnification factor, 1.2.
The principal motility parameters measured were curvilinear velocity (VCL), average path velocity (VAP), straight-line velocity (VSL), beat/cross frequency (BCF), amplitude of lateral head displacement (ALH). Progressive sperm cells were characterized by average path velocity (VAP) >45 μm/s and straightness (STR = VSL/ VAP) >45%, respectively.
Sperm viability analyses
Sperm viability was assessed by eosin-nigrosin staining as previously described (61). Briefly, 10 μl of sperm suspension was mixed with 20 μl of eosin and 30 μl nigrosin, and a 20-μl droplet was smeared onto a clean glass slide. After drying, a minimum of 200 cells per genotype were counted.
In vitro fertilization analyses
Preparation of oocytes
C75B6L/N female mice (6 to 8 weeks old, Janvier Labs, France) were superovulated with 5 IU of pregnant mare serum gonado-tropin (PMSG) and 5 IU human chorionic gonadotropin (hCG; Intervet, France) 48 hours apart. About 14 hours after hCG injection, the animals were sacrificed by cervical dislocation. Cumulus oophorus were collected by tearing the ampulla wall of the oviduct, placed in Ferticult medium (#FECU100; FertiPro N.V., Belgium) supplemented with 3% BSA (#A3311; Sigma-Aldrich). To remove the zona pellucida (ZP), cumulus cells were removed by a brief exposure to hyaluronidase IV-S (1 mg/ml; #H3884, Sigma-Aldrich) and ZP was then dissolved with acidic Tyrode’s (AT) solution, pH 2.5 (#T1788; Sigma-Aldrich) under visual monitoring. The zona-free eggs were rapidly washed five times with Ferticult medium and kept at 37°C under 5% CO2 for 2 to 3 hours to recover their fertilization ability.
Capacitated sperm preparation
Mouse spermatozoa were obtained from the cauda epididymides of wild-type and Ttll3 −/− Ttll8 −/− male mice (8- to 10-weekold) and capacitated at 37°C under 5% CO2 for 90 min in a 500-μl drop Ferticult medium supplemented with 3% BSA, under mineral oil. The total sperm count was obtained, and 105 sperm per ml from either wild-type or Ttll3 −/− Ttll8 −/− mice were used for each experiment.
In vitro fertilization
Zona-free eggs were inseminated with capacitated spermatozoa for 3 hours in a 100-μl drop of Ferticult medium 3% BSA at a final concentration of 105/ml. Then, they were washed and directly mounted in Vectashield/DAPI (Vector Laboratories, CA, USA) for observation under ultra-violet light (Nikon Eclipse E600 microscope). Only oocytes showing at least one fluorescent decondensed sperm head within their cytoplasm were considered fertilized. To assess the fertilization index (FI), the number of decondensed sperm heads per oocyte was counted.
Cryo-EM analyses
Cauda epididymides were extracted, lacerated in PBS, and kept for 10 min at 37°C. Swimming sperm cells were collected and rapidly plunge-frozen on Quantifoil Holey Carbon grids using a Leica Automatic Plunge Freezer EM GP. Samples were imaged with a FEI Titan Halo (Thermo Fisher Scientific) operated at 300 keV using a Gatan energy filter and a Gatan K2 Summit direct electron detector. SerialEM software was used to operate the microscope during acquisition of tomographic tilt series. Tomographic series were acquired at angles from −26° to 40° and −28° to −40° using a 2° tilt increments. Each image was acquired in dose fractionation mode with about 2e/Å2 total electron dose. Image frames were further processed with K2Align software (62) to generate each image of a tilt series. Generated tilt series were processed with etomo from IMOD software package (63) to reconstruct tomograms by weighted back-projection using fiducial-based aligned, CTF-corrected, dose-weighted filtered image stacks (64). Tomograms were further filtered using nonlinear anisotropic diffusion (63) before tomographic inspection and annotation. We chose straight segments of the sperm tails for our analyses to optimize the conditions for the tomographic reconstruction. The positions of the 96-nm axonemal repeats and the associated ODAs within unfiltered tomograms were modeled using 3dmod. Initially, 983 96-nm repeats and 3166 ODA particles were extracted for wild type, while for Ttll3 −/− Ttll8 −/−, 61796-nm repeats and 3656 ODA particles were extracted. All particles from each MT doublet were manually aligned to a common reference before fine automatic alignments and subtomogram averaging using PEET (65). For the final averages of the 96-nm repeat, 900 wild-type and 540 Ttll3 −/− Ttll8 −/− particles were used (fig. S10). For the ODA-related analysis, only particles from MT doublet pairs showing no mechanical disruption were included for further conformational analysis. The number of ODA particles used in this analysis was 2691 for wild type and 3656 for Ttll3 −/− Ttll8 −/−. Each ODA particle was classified according to the combined result of principal component analysis and k-means clustering (65) of particle regions containing either β- or γ-heavy chains. IDAs were classified with principal component analysis of independently aligned wild-type and Ttll3 −/− Ttll8 −/− sperm flagella. This allowed their classification into pre- and post-powerstroke configurations. 3dmod was used to inspect the resulting averages and generate sliced views, while further 3D rendering and structure fitting for clearer visualization was carried out using Chimera (66). For the complete cryo-analysis, sperm samples from one wild-type mouse were used, while in the Ttll3 −/− Ttll8 −/− case, the samples used came from two different mice.
Statistical analyses
All the quantifications are listed in table S1. P values were calculated by unpaired Student’s t test.
2D and 3D motility analyses
To determine the swimming patterns, ~250 sperm from six different wild-type and five different Ttll3 −/− Ttll8 −/− mice were analyzed. For all the flagellar beat parameter analyses, 76 spermatozoa from six different wild-type mice and 58 spermatozoa from four different Ttll3 −/− Ttll8 −/− mice were analyzed.
Ependymal cilia beat frequency analyses
To determine the CBF, sections from three individual wild-type or Ttll3 −/− Ttll8 −/− mice were analyzed. From each section, 13 to 26 individual cells were imaged for their ciliary beat and CBF was determined using CiliaFA plugin for ImageJ. Scatter plots from each experiment were plotted using GraphPad Prism version 8. The lines indicate the mean (± SEM).
CASA analyses
To determine the motility parameters, sperm from five wild-type and six Ttll3 −/− Ttll8 −/− mice were analyzed. The graphs for each of the individual motility parameters were plotted using GraphPad Prism version 8. Mean values of individual experiments were plotted as bar graphs with standard error of mean (±SEM) between individual experiments.
In vitro fertilization assay
To analyze the fertilization index of the sperm with in vitro fertilization tests, the experiments were performed five times, using five individual wild-type and Ttll3 −/− Ttll8 −/− mice. The fertilization index is plotted as a ratio of the total number of fused sperm to the total number of oocytes used per experiment. The graphs were plotted using GraphPad Prism version 8. Mean values of individual experiments were plotted in bar graphs with standard error of mean (±SEM) between individual experiments.
Image and video processing
Immunofluorescence and histology images were treated with Adobe Photoshop. Intensities were adjusted exclusively in a linear manner, and no additional image treatments were performed.
For mounting the composite videos, the still panels were prepared in Adobe Illustrator and then subsequently mounted onto the individual video compositions in Adobe After Effects. Each composition was rendered as a QuickTime movie with Adobe Media Encoder and exported as 1280 pixel by 720 pixel videos with QuickTime Player v10.4.
Supplementary Material
Research Article Summary.
Introduction
Microtubules are key components of the eukaryotic cytoskeleton. Although they are involved in a wide variety of functions, microtubules are structurally highly similar across most cell types and organisms. It was suggested that a “tubulin code,” formed by combinations of tubulin posttranslational modifications, adapts individual microtubules to specific functions within living cells. However, clear-cut functional and mechanistic data verifying this concept are still scarce. Glycylation is among the least explored posttranslational modifications of tubulin and has, so far, exclusively been found on microtubules of cilia and flagella from a variety of species. Previous work has suggested that glycylation might be essential for cilia and flagella, but mechanistic insight remains lacking.
Rationale
Two enzymes from the tubulintyrosine ligase-like (TTLL) family, TTLL3 and TTLL8, are essential to initiate glycylation of tubulin in mammals. To entirely abolish glycylation at the organism level and to determine its physiological function, we generated a double-knockout mouse lacking both glycylating enzymes (Ttll3 −/− Ttll8 −/−). Inactivation of these two enzymes led to a lack of glycylation in all analyzed cilia and flagella. This allowed us to investigate the role of glycylation in the function of these organelles.
Results
Despite the absence of glycylation in Ttll3 −/− Ttll8 −/− mice, no gross defects were observed at the organism and tissue levels. Motile ependymal cilia in brain ventricles as well as motile cilia in the respiratory tract were present and appeared normal. Sperm flagella were also assembled normally, and sperm were able to swim. However, in vitro fertility assays showed that male Ttll3 −/− Ttll8 −/− mice were subfertile. Computer-assisted sperm analyses revealed motility defects of Ttll3 −/− Ttll8 −/− sperm. Further analyses showed that lack of glycylation leads to perturbed flagellar beat patterns, causing Ttll3 −/− Ttll8 −/− sperm to swim predominantly along circular paths. This is highly unusual for mammalian sperm and interferes with their ability to reach the oocyte for fertilization. To determine the molecular mechanisms underlying this aberrant flagellar beat, we used cryo–electron tomography. The three-dimensional structure of the 96-nm repeat of the Ttll3 −/− Ttll8 −/− sperm axoneme showed no aberrations in its overall assembly. By contrast, the structure of both outer and inner dynein arms (ODAs and IDAs) was perturbed in Ttll3 −/− Ttll8 −/− flagella. Classification analysis showed that the incidence and distribution of pre-powerstroke and post-powerstroke conformations of ODAs and IDAs were altered in Ttll3 −/− Ttll8 −/− sperm. These ultrastructural findings indicate that glycylation is required to efficiently control the dynein powerstroke cycle, which is essential for the generation of a physiological flagellar beat.
Conclusion
Our work shows that tubulin glycylation regulates the beat of mammalian flagella by modulating axonemal dynein motor activity. Lack of glycylation leads to perturbed sperm motility and male subfertility in mice. Considering that human sperm are more susceptible than mouse sperm to deficiencies in sperm motility, our findings imply that a perturbation of tubulin glycylation could underlie some forms of male infertility in humans.
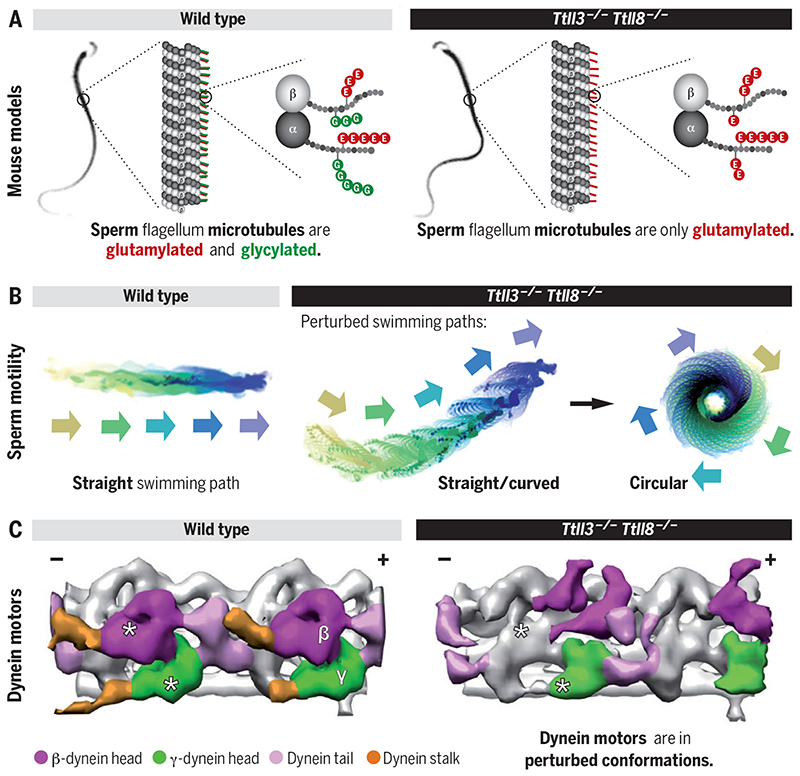
Tubulin glycylation controls sperm motility.
(A) Microtubules in sperm flagella are rich in tubulin posttranslational modifications. Mice deficient for the glycylating enzymes TTLL3 and TTLL8 lack glycylation. (B) Mammalian sperm swim in linear paths. In the absence of glycylation, abnormal, mostly circular swimming patterns are observed, which impede progressive swimming. (C) Absence of glycylation leads to perturbed distribution of axonemal dynein conformations in Ttll3 −/− Ttll8 −/− flagella, which impedes normal flagellar beating.
Acknowledgments
We thank C. Arnoult (University of Grenoble, France), S. Bodakuntla, M. M. Magiera (Institut Curie), E. Feraille (University of Geneva, Switzerland), T. Stünker, J. Wistuba (University Clinics Münster, Germany), and D. Wachten (University of Bonn, Germany) for critical analysis and insightful discussions. We also thank N. Spassky and N. Delgehyr (Ecole Normale Superieur, Paris) for technical assistance with the mouse primary ependymal cell culture and mouse ventricle whole-mount staining experiments. We acknowledge the Mouse Clinical Institute (PHENOMIN-Institut Clinique de la Souris, MCI/ICS) for generating the transgenic Ttll8 mice in the Genetic Engineering and Model Validation Department by M.-C. Birling with funds from the CNRS. We thank J. Helppi [Biomedical Services of Max Planck Institute for Molecular Cell Biology and Genetics (MPI-CBG)] for managing the genetically modified mice and for helping with tissue dissection. We thank the Electron Microscopy facility of the MPI-CBG for technical support. We further thank C. Alberti, E. Belloir, F. Bertrand, V. Dangles-Marie, I. Grandjean, H. Hermange, C. Caspersen, M. M. Magiera, S. Bodakuntla, N.-L.J. Nguyen, C. Rollin, C. Serieyssol, A. Thadal, and L. Vaslin (Institut Curie) for technical assistance. We are grateful to M.-N. Soler, C. Lovo, and L. Besse from the imaging platform PICT-IBiSA@Orsay (Institut Curie, Orsay) for technical assistance in the use of the Leica Structured Illumination Microscope (Optigrid) and the Nikon Eclipse Ti-E inverted microscope systems. We also thank B. Friedrich for his support using resistive-force theory.
Funding
This work was supported by the ANR-10-IDEX-0001-02, the LabEx Cell(n)Scale ANR-11-LBX-0038. C.J. is supported by the Institut Curie, the French National Research Agency (ANR) awards ANR-12-BSV2-0007 and ANR-17-CE13-0021, the Institut National du Cancer (INCA) grant 2014-PL BIO-11-ICR-1, and the Fondation pour la Recherche Médicale (FRM) grant DEQ. 20170336756. S.G. received a 1-year postdoctoral fellowship from LabEx Cell(n)Scale ANR-11-LBX-0038. G.P., G.A.V., and A.K. are supported by the Max Planck Society and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant 819826) to G.P. J.N.H. holds a PhD Fellowship by the Boehringer Ingelheim Fonds. The work of L.A. was supported by the Deutsche Forschungsgemeinschaft via the priority program SPP 1726 “Microswimmers.” M.W. and A.T. are supported by the French National Research Agency (ANR) awards DIVERCIL-17-CE13-0023 and ANR FLAGELOME 19-CE17-0014.
Footnotes
Author contributions: Conceptualization: S.G., L.A., G.P., and C.J. Methodology, analyses, and validation: Mouse generation, genotyping, ependymal cell culture, immunoblot, immunocytochemistry, and histological analyses: S.G., S.L., M.W., and C.J. CASA and in vitro fertility analyses: A.T., M.W., A.Z., C.I.-R., S.G., and C.J. 2D motility and 3D in-line holography analyses: J.N.H., A.G., and L.A. Computation of ideal trajectories using resistive-force theory: A.G. and L.A. Sample preparation for cryo–electron tomography: G.A.V., A.K., and S.G. Cryo–electron tomography, subtomogram averaging, particle classification, 3D model rendering, and interpretation: G.A.V., A.K., and G.P. Writing: original draft: S.G. and C.J.; review and editing: S.G., G.A.V., J.N.H., A.G., A.T., A.Z., L.A., G.P. and C.J. Visualization: S.G., G.A.V., J.N.H., A.G., A.K., A.T., M.W., A.Z., L.A., G.P., and C.J. Supervision: L.A., G.P., A.T., and C.J. Project administration: C.J. Funding acquisition: S.G., L.A., G.P., A.T. and C.J.
Competing interests: The authors declare no competing financial interests.
Data and materials availability
The 3D averaged structures of the 96-nm repeats of the wild-type and TTLL3 −/− TTLL8 −/− sperm flagella as well as all major classes of the ODAs and IDAs have been deposited in the Electron Microscopy Data Bank (EMDB) under IDs EMD-11942 (pre-pre-powerstroke conformation of the ODAs), EMD-11943 (pre-post-powerstroke conformation of the ODAs), EMD-11944 (post-pre-powerstroke conformation of the ODAs), EMD-11945 (post-post-powerstroke conformation of the ODAs), EMD-11946 (axonemal 96-nm repeat from active wild-type sperm flagella), EMD-11947 (axonemal 96-nm repeat from active Ttll3 −/− Ttll8 −/− sperm flagella), EMD-11948 (axonemal 96-nm repeat from active sperm flagella showing IDAs in pre-powerstroke conformations), and EMD-11949 (axonemal 96-nm repeat from active sperm flagella showing IDAs in post-powerstroke conformations). All other data are available in the manuscript or the supplementary material.
References and Notes
- 1.Janke C, Magiera MM. The tubulin code and its role in controlling microtubule properties and functions. Nat Rev Mol Cell Biol. 2020;21:307–326. doi: 10.1038/s41580-020-0214-3. [DOI] [PubMed] [Google Scholar]
- 2.Bré MH, et al. Axonemal tubulin polyglycylation probed with two monoclonal antibodies: Widespread evolutionary distribution, appearance during spermatozoan maturation and possible function in motility. J Cell Sci. 1996;109:727–738. doi: 10.1242/jcs.109.4.727. [DOI] [PubMed] [Google Scholar]
- 3.Rogowski K, et al. Evolutionary divergence of enzymatic mechanisms for posttranslational polyglycylation. Cell. 2009;137:1076–1087. doi: 10.1016/j.cell.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 4.Bosch Grau M, et al. Tubulin glycylases and glutamylases have distinct functions in stabilization and motility of ependymal cilia. J Cell Biol. 2013;202:441–451. doi: 10.1083/jcb.201305041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch Grau M, et al. Alterations in the balance of tubulin glycylation and glutamylation in photoreceptors leads to retinal degeneration. J Cell Sci. 2017;130:938–949. doi: 10.1242/jcs.199091. [DOI] [PubMed] [Google Scholar]
- 6.Rocha C, et al. Tubulin glycylases are required for primary cilia, control of cell proliferation and tumor development in colon. EMBO J. 2014;33:2247–2260. doi: 10.15252/embj.201488466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gadadhar S, et al. Tubulin glycylation controls primary cilia length. J Cell Biol. 2017;216:2701–2713. doi: 10.1083/jcb.201612050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konno A, et al. Ttll9 −/− mice sperm flagella show shortening of doublet 7, reduction of doublet 5 polyglutamylation and a stall in beating. Cell J Sci. 2016;129:2757–2766. doi: 10.1242/jcs.185983. [DOI] [PubMed] [Google Scholar]
- 9.Mullen RJ, Eicher EM, Sidman RL. Purkinje cell degeneration, a new neurological mutation in the mouse. Proc Natl Acad Sci USA. 1976;73:208–212. doi: 10.1073/pnas.73.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogel P, Hansen G, Fontenot G, Read R. Tubulin tyrosine ligase-like 1 deficiency results in chronic rhinosinusitis and abnormal development of spermatid flagella in mice. Vet Pathol. 2010;47:703–712. doi: 10.1177/0300985810363485. [DOI] [PubMed] [Google Scholar]
- 11.Wu H-Y, Wei P, Morgan JI. Role of cytosolic carboxypeptidase 5 in neuronal survival and spermatogenesis. Sci Rep. 2017;7:41428. doi: 10.1038/srep41428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mencarelli C, Lupetti P, Dallai R. New insights into the cell biology of insect axonemes. Int Rev Cell Mol Biol. 2008;268:95–145. doi: 10.1016/S1937-6448(08)00804-6. [DOI] [PubMed] [Google Scholar]
- 13.Reiter JF, Leroux MR. Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol. 2017;18:533–547. doi: 10.1038/nrm.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Brolosy MA, Stainier DYR. Genetic compensation: A phenomenon in search of mechanisms. PLOS Genet. 2017;13:e1006780. doi: 10.1371/journal.pgen.1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Brolosy MA, et al. Genetic compensation triggered by mutant mRNA degradation. Nature. 2019;568:193–197. doi: 10.1038/s41586-019-1064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talevi R, Gualtieri R. In vivo versus in vitro fertilization. Eur J Obstet Gynecol Reprod Biol. 2004;115(suppl. 1):S68–S71. doi: 10.1016/j.ejogrb.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Pusch HH. The importance of sperm motility for the fertilization of human oocytes in vivo and in vitro. Andrologia. 1987;19:514–527. doi: 10.1111/j.1439-0272.1987.tb01891.x. [DOI] [PubMed] [Google Scholar]
- 18.Amann RP, Waberski D. Computer-assisted sperm analysis (CASA): Capabilities and potential developments. Theriogenology. 2014;81(5–17):e1–3. doi: 10.1016/j.theriogenology.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Heidary Z, Saliminejad K, Zaki-Dizaji M, Khorram HR. Khorshid, Genetic aspects of idiopathic asthenozoospermia as a cause of male infertility. Hum Fertil. 2020;23:83–92. doi: 10.1080/14647273.2018.1504325. [DOI] [PubMed] [Google Scholar]
- 20.Hansen JN, Rassmann S, Jikeli JF, Wachten D. SpermQ-Asimple analysis software to comprehensively study flagellar beating and sperm steering. Cells. 2018;8:10. doi: 10.3390/cells8010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mortimer ST, Mortimer D. Kinematics of human spermatozoa incubated under capacitating conditions. J Androl. 1990;11:195–203. [PubMed] [Google Scholar]
- 22.Neill JM, Olds-Clarke P. A computer-assisted assay for mouse sperm hyperactivation demonstrates that bicarbonate but not bovine serum albumin is required. Gamete Res. 1987;18:121–140. doi: 10.1002/mrd.1120180204. [DOI] [PubMed] [Google Scholar]
- 23.Marquez B, Ignotz G, Suarez SS. Contributions of extracellular and intracellular Ca2+to regulation of sperm motility: Release of intracellular stores can hyperactivate CatSperl and CatSper2 null sperm. Dev Biol. 2007;303:214–221. doi: 10.1016/j.ydbio.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su T-W, et al. Sperm trajectories form chiral ribbons. Sci Rep. 2013;3:1664. doi: 10.1038/srep01664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jikeli JF, et al. Sperm navigation along helical paths in 3D chemoattractant landscapes. Nat Commun. 2015;6:7985. doi: 10.1038/ncomms8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller RL. In: Biology of Fertilization. Metz CB, Monroy A, editors. Vol. 2. Academic Press; 1985. Sperm chemo-orientation in the metazoa; pp. 275–337. [Google Scholar]
- 27.Su T-W, Xue L, Ozcan A. High-throughput lensfree 3D tracking of human sperms reveals rare statistics of helical trajectories. Proc Natl Acad Sci USA. 2012;109:16018–16022. doi: 10.1073/pnas.1212506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elgeti J, Kaupp UB, Gompper G. Hydrodynamics of sperm cells near surfaces. Biophys J. 2010;99:1018–1026. doi: 10.1016/j.bpj.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong A, et al. The steering gaits of sperm. Philos Trans R Soc London Ser B. 2020;375:20190149. doi: 10.1098/rstb.2019.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crenshaw HC. A new look at locomotion in microorganisms: Rotating and translating. Am Zool. 1996;36:608–618. doi: 10.1093/icb/36.6.608. [DOI] [Google Scholar]
- 31.Nicastro D, et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 32.Viswanadha R, Sale WS, Porter ME. Ciliary motility: Regulation of axonemal dynein motors. Cold Spring Harbor Perspect Biol. 2017;9:a018325. doi: 10.1101/cshperspect.a018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Porter ME, Sale WS. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J Cell Biol. 2000;151:F37–F42. doi: 10.1083/jcb.151.5.F37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin J, Nicastro D. Asymmetric distribution and spatial switching of dynein activity generates ciliary motility. Science. 2018;360:eaar1968. doi: 10.1126/science.aar1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gui L, et al. Scaffold subunits support associated subunit assembly in theChlamydomonasciliary nexin-dynein regulatory complex. Proc Natl Acad Sci USA. 2019;116:23152–23162. doi: 10.1073/pnas.1910960116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alper JD, Tovar M, Howard J. Displacement-weighted velocity analysis of gliding assays reveals thatChlamydomonasaxonemal dynein preferentially moves conspecific microtubules. Biophys J. 2013;104:1989–1998. doi: 10.1016/j.bpj.2013.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenney RJ, Huynh W, Vale RD, Sirajuddin M. Tyrosination of a-tubulin controls the initiation of processive dynein-dynactin motility. EMBO J. 2016;35:1175–1185. doi: 10.15252/embj.201593071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sartori P, Geyer VF, Howard J, Jülicher F. Curvature regulation of the ciliary beat through axonemal twist. Phys Rev. 2016;E94:042426. doi: 10.1103/PhysRevE.94.042426. [DOI] [PubMed] [Google Scholar]
- 39.Lindemann CB, Lesich KA. Functional anatomy of the mammalian sperm flagellum. Cytoskeleton. 2016;73:652–669. doi: 10.1002/cm.21338. [DOI] [PubMed] [Google Scholar]
- 40.Fujimura M, Okuno M. Requirement of the fixed end for spontaneous beating in flagella. J Exp Biol. 2006;209:1336–1343. doi: 10.1242/jeb.02131. [DOI] [PubMed] [Google Scholar]
- 41.Smith EF, Yang P. The radial spokes and central apparatus: Mechano-chemical transducers that regulate flagellar motility. Cell Motil Cytoskeleton. 2004;57:8–17. doi: 10.1002/cm.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kubo T, Hou Y, Cochran DA, Witman GB, Oda T. A microtubule-dynein tethering complex regulates the axonemal inner dyneinf(I1) Mol Biol Cell. 2018;29:1060–1074. doi: 10.1091/mbc.E17-11-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orbach R, Howard J. The dynamic and structural properties of axonemal tubulins support the high length stability of cilia. Nat Commun. 2019;10:1838. doi: 10.1038/s41467-019-09779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishikawa T. Axoneme structure from motile cilia. Cold Spring Harbor Perspect Biol. 2017;9:a028076. doi: 10.1101/cshperspect.a028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wloga D, et al. TTLL3 Is a tubulin glycine ligase that regulates the assembly of cilia. Dev Cell. 2009;16:867–876. doi: 10.1016/j.devcel.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 46.Wloga D, et al. Hyperglutamylation of tubulin can either stabilize or destabilize microtubules in the same cell. Eukaryot Cell. 2010;9:184–193. doi: 10.1128/EC.00176-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Janke C. The tubulin code: Molecular components, readout mechanisms, and functions. J Cell Biol. 2014;206:461–472. doi: 10.1083/jcb.201406055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giordano T, et al. Loss of the deglutamylase CCP5 perturbs multiple steps of spermatogenesis and leads to male infertility. J Cell Sci. 2019;132:jcs226951. doi: 10.1242/jcs.226951. [DOI] [PubMed] [Google Scholar]
- 49.Kherraf Z-E, et al. SPINK2 deficiency causes infertility by inducing sperm defects in heterozygotes and azoospermia in homozygotes. EMBO Mol Med. 2017;9:1132–1149. doi: 10.15252/emmm.201607461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Curi SM, et al. Asthenozoospermia: Analysis of a large population. Arch Androl. 2003;49:343–349. doi: 10.1080/01485010390219656. [DOI] [PubMed] [Google Scholar]
- 51.Birkhead TR, Hosken DJ, Pitnick S. In: Sperm Biology: An Evolutionary Perspective. Birkhead TR, Hosken DJ, Pitnick S, editors. Academic Press; 2009. pp. vii–viii. [Google Scholar]
- 52.Alvarez L. The tailored sperm cell. J Plant Res. 2017;130:455–464. doi: 10.1007/s10265-017-0936-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lallemand Y, Luria V, Haffner-Krausz R, Lonai P. Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res. 1998;7:105–112. doi: 10.1023/A:1008868325009. [DOI] [PubMed] [Google Scholar]
- 54.Delgehyr N, et al. Ependymal cell differentiation, from monociliated to multiciliated cells. Methods Cell Biol. 2015;127:19–35. doi: 10.1016/bs.mcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 55.Hussaini SMQ, et al. Heat-induced antigen retrieval: An effective method to detect and identify progenitor cell types during adult hippocampal neurogenesis. J Vis Exp. 2013;78:e50769. doi: 10.3791/50769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Magiera MM, Janke C. In: Microtubules, In Vitro of Methods Cell Biology. Correia JJ, Wilson L, editors. Vol. 115. Academic Press; 2013. Investigating tubulin posttranslational modifications with specific antibodies; pp. 247–267. [DOI] [PubMed] [Google Scholar]
- 57.AlOmran AJ, Saternos HC, Liu T, Nauli SM, AbouAlaiwi WA. Live imaging of the ependymal cilia in the lateral ventricles of the mouse brain. J Vis Exp. 2015;100:e52853. doi: 10.3791/52853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith CM, et al. ciliaFA: A research tool for automated, high-throughput measurement of ciliary beat frequency using freely available software. Cilia. 2012;1:14. doi: 10.1186/2046-2530-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee S-H, Grier DG. Holographic microscopy of holographically trapped three-dimensional structures. Opt Express. 2007;15:1505–1512. doi: 10.1364/OE.15.001505. [DOI] [PubMed] [Google Scholar]
- 60.Wilson L, Zhang R., 3rd Localization of weak scatterers in digital holographic microscopy using Rayleigh-Sommerfeld back-propagation. Opt Express. 2012;20:16735. doi: 10.1364/OE.20.016735. [DOI] [Google Scholar]
- 61.Björndahl L, Söderlund I, Kvist U. Evaluation of the one-step eosin-nigrosin staining technique for human sperm vitality assessment. Hum Reprod. 2003;18:813–816. doi: 10.1093/humrep/deg199. [DOI] [PubMed] [Google Scholar]
- 62.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 64.Xiong Q, Morphew MK, Schwartz CL, Hoenger AH, Mastronarde DN. CTF determination and correction for low dose tomographic tilt series. J Struct Biol. 2009;168:378–387. doi: 10.1016/j.jsb.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heumann JM, Hoenger A, Mastronarde DN. Clustering and variance maps for cryo-electron tomography using wedge-masked differences. J Struct Biol. 2011;175:288–299. doi: 10.1016/j.jsb.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 3D averaged structures of the 96-nm repeats of the wild-type and TTLL3 −/− TTLL8 −/− sperm flagella as well as all major classes of the ODAs and IDAs have been deposited in the Electron Microscopy Data Bank (EMDB) under IDs EMD-11942 (pre-pre-powerstroke conformation of the ODAs), EMD-11943 (pre-post-powerstroke conformation of the ODAs), EMD-11944 (post-pre-powerstroke conformation of the ODAs), EMD-11945 (post-post-powerstroke conformation of the ODAs), EMD-11946 (axonemal 96-nm repeat from active wild-type sperm flagella), EMD-11947 (axonemal 96-nm repeat from active Ttll3 −/− Ttll8 −/− sperm flagella), EMD-11948 (axonemal 96-nm repeat from active sperm flagella showing IDAs in pre-powerstroke conformations), and EMD-11949 (axonemal 96-nm repeat from active sperm flagella showing IDAs in post-powerstroke conformations). All other data are available in the manuscript or the supplementary material.