Abstract
Oligodendrocytes that survive demyelination can remyelinate, including in Multiple Sclerosis (MS), but how they do so is unclear. Here, using zebrafish, we found that surviving oligodendrocytes make few new sheaths and frequently mistarget new myelin to neuronal cell bodies, a pathology we also found in MS. In contrast, oligodendrocytes generated after demyelination make abundant and correctly targeted sheaths, indicating that they likely also have a better regenerative potential in MS.
Introduction
Loss of myelin (demyelination) is a feature of debilitating disorders of the central nervous system (CNS), including Multiple Sclerosis (MS) 1 , but following demyelination, oligodendrocyte progenitor cells (OPCs) can produce new oligodendrocytes (OLs) that remyelinate axons 2 . Indeed therapeutic strategies aimed at promoting otherwise limited, variable, and age-depreciating remyelination in MS have focussed on enhancing oligodendrogenesis 1–3 . However, recent studies have indicated that oligodendrocytes that survive demyelination can also contribute to remyelination 2,4–6 , potentially offering additional opportunities to promote remyelination. Nonetheless, it remains unclear to what extent surviving oligodendrocytes can contribute to remyelination in vivo.
Results
A novel zebrafish model to study remyelination
To compare remyelination by surviving and newly-generated oligodendrocytes, we developed a zebrafish model of demyelination (Extended Data Figure 1 and Methods). To induce myelin damage we treated Tg(mbp:TRPV1-tagRFPt) animals with 10 μM capsaicin (csn) for 2 hours at 4 days post fertilisation (dpf). This led to extensive demyelination, as assessed by the transgenic reporter Tg(mbp:EGFP-CAAX) and transmission electron microscopy (TEM) (Extended Data Figure 1F and 2). TEM analyses revealed a 95% reduction in myelinated axon number in the dorsal tract of the spinal cord just 1 day post treatment (dpt) (Extended Data Figure 2A–F). Assessment of Tg(mbp:EGFP-CAAX), and quantification of TEM images indicated rapid remyelination, with no significant difference in myelinated axon number between control and csn-treated zebrafish by 3 days post treatment (3dpt) (Extended Data Figure 1F and 2G-J). The myelin present at 3dpt surrounded similarly sized axons in the same regions in control and csn-treated animals, confirming remyelination (Extended Data Figure 2G–J). Furthermore, axons at this timepoint showed both thick (>3 wraps) and thin (≤3 wraps) myelin, with those in the csn-treated animals having a higher proportion of thinly myelinated profiles (Extended Data Figure 2K–L), recapitulating a common feature of remyelination: thin myelin sheaths 2 . Despite the initial extensive (95%) loss of myelin, we saw a relatively modest reduction in total oligodendrocyte number, with only 33% fewer oligodendrocytes in csn-treated Tg(mbp:TRPV1-tagRFPt) animals at 3hpt (Extended Data Figure 2B and 3A–C), indicating that many oligodendrocytes survived demyelination. In line with ongoing remyelination, oligodendrocyte number increased over time (Extended Data Figure 3B–F). Therefore, this model allows investigation of remyelination by both surviving and newly-generated oligodendrocytes.
Surviving oligodendrocytes exhibit limited remyelination
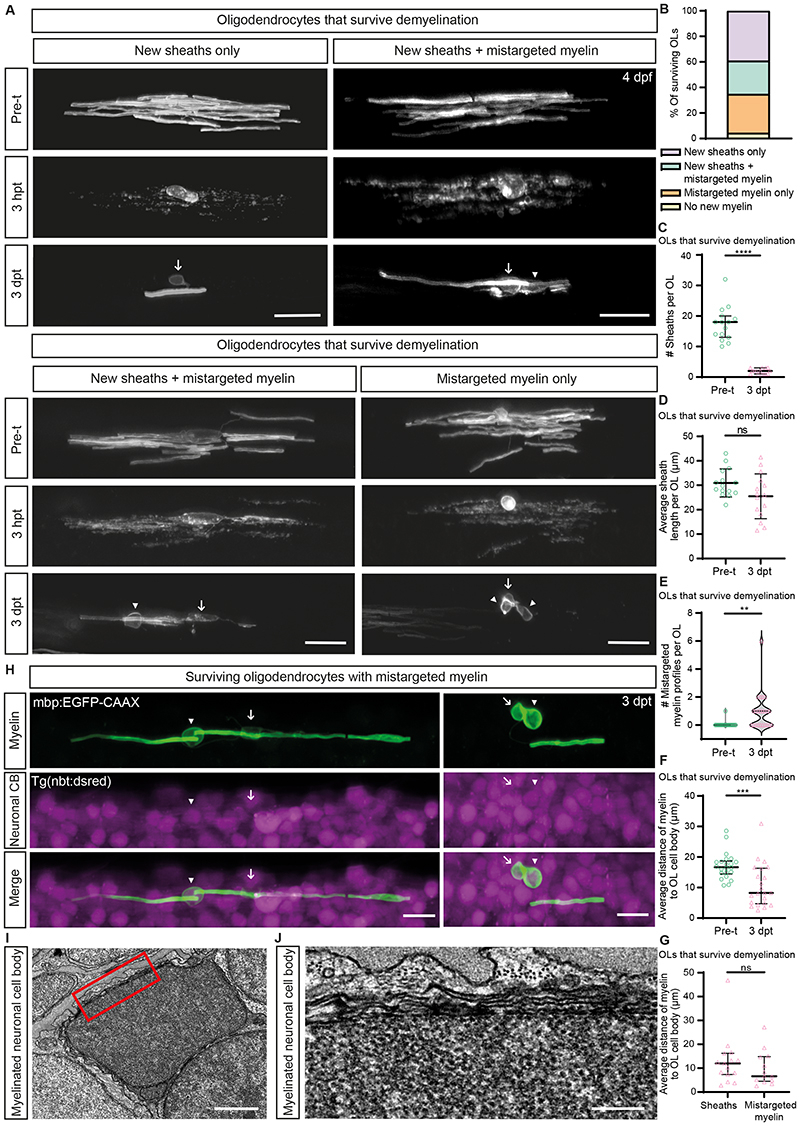
To follow the fate of individual surviving oligodendrocytes, we mosaically labelled them with mbp:EGFP-CAAX (Methods), and imaged them prior to demyelination, 3 hours post csn treatment (hpt), 1dpt, and after remyelination at 3dpt (Figure 1A). We imaged 37 oligodendrocytes with complete myelin sheath loss 3hpt, which led to production of myelin debris (Figure 1A and Extended Data Figure 4A) and an associated macrophage/microglial response (Extended Data Figure 4B–D). Of these 37 oligodendrocytes (analysed in 37 zebrafish), 14 cells underwent cell death, 12 of which died by 1dpt. The remaining 23 oligodendrocytes were present at 3dpt, confirming that oligodendrocytes can survive demyelination (Figure 1A). Of these surviving cells, 22 made myelin (Figure 1A–B), indicating that oligodendrocytes either rapidly die or make new myelin. Surviving oligodendrocytes made myelin sheaths of normal length, but made very few, producing only 2 per cell on average, compared with 18 prior to demyelination (Figure 1C–D). Furthermore, 15/22 surviving oligodendrocytes mistargeted newly-made myelin, as evidenced by the appearance of myelin profiles surrounding cell bodies (Figure 1A and H–I), as reported in other contexts 7,8 . In contrast, only one oligodendrocyte exhibited evidence of myelin mistargeting prior to demyelination (Figure 1E), indicating that extensive myelin mistargeting is a pathological feature of remyelination by surviving oligodendrocytes. We also observed that new sheaths and mistargeted myelin profiles were localised closer to the oligodendrocyte cell body than before demyelination (Figure 1F–G), suggesting that these cells have restricted process dynamics.
Figure 1. Remyelination by oligodendrocytes that survive demyelination in the Tg(mbp:TRPV1-tagRFPt) zebrafish model.
(A) Confocal images of single surviving oligodendrocytes followed over time from pre-treatment (Pre-t at 4dpf) to 3dpt. Scale bars, 20μm. (B) The percentage of different oligodendrocyte fates of the 23 oligodendrocytes that survived demyelination. (C) Quantification of the number of myelin sheaths produced by the same oligodendrocytes pre-treatment (median=18.00, IQR=13.00-20.00) and 3dpt (median=2.00, IQR=1.00-3.00), p<0.0001. Two-tailed Wilcoxon matched-pairs signed rank test. N=23 zebrafish. (D) Quantification of the average length of myelin sheaths produced by the same oligodendrocytes pre-treatment (mean=30.93±5.76SD) and 3dpt (mean=25.47±9.16SD), p=0.0655. Paired two-tailed t-test. N=15 zebrafish. (E) Quantification of the number of mistargeted myelin profiles produced by the same oligodendrocytes pre-treatment (median=0.00, IQR=0.00-0.00) and 3dpt (median=1.00, IQR=0.00-1.00), p=0.0012. Two-tailed Wilcoxon matched-pairs signed rank test. N=23 zebrafish. (F) Quantification of the average distance of myelin structures (sheaths and mistargeted myelin) to the oligodendrocyte cell body for the same oligodendrocytes pre-treatment (median=16.64, IQR=14.47-18.69) and 3dpt (median=8.24, IQR=4.73-16.33), p=0.0008. Two-tailed Wilcoxon matched-pairs signed rank test. N=22 zebrafish. (G) Quantification of the average distance of myelin structures to the oligodendrocyte cell body (μm) for myelin sheaths (median=11.97, IQR=7.36-16.24) and mistargeted myelin profiles (median=6.62, IQR=4.53-14.86) in surviving myelinating oligodendrocytes at 3dpt, p=0.3627. Two-tailed Mann-Whitney t-test. N=15 zebrafish (sheaths), N=13 zebrafish (mistargeted myelin). (H) Confocal images of single surviving oligodendrocytes at 3dpt which mistarget their myelin around neuronal cell bodies (CB). Scale bars, 10μm. (I-J’) Transmission electron microscopy images of a myelinated neuronal cell body surrounded by cell bodies which are not myelinated. (I) Scale bar=1μm. (J). Scale bar=0.2μm. In all figures, 1 oligodendrocyte was analysed per zebrafish. Data are presented as median with IQR in figures (C and E - G) and mean±SD in figure (D). In figure (A) and (H) Arrows show the position of oligodendrocyte cell bodies, and arrowheads show positions of mistargeted myelin.
Surviving oligodendrocytes exhibit limited dynamic process activity
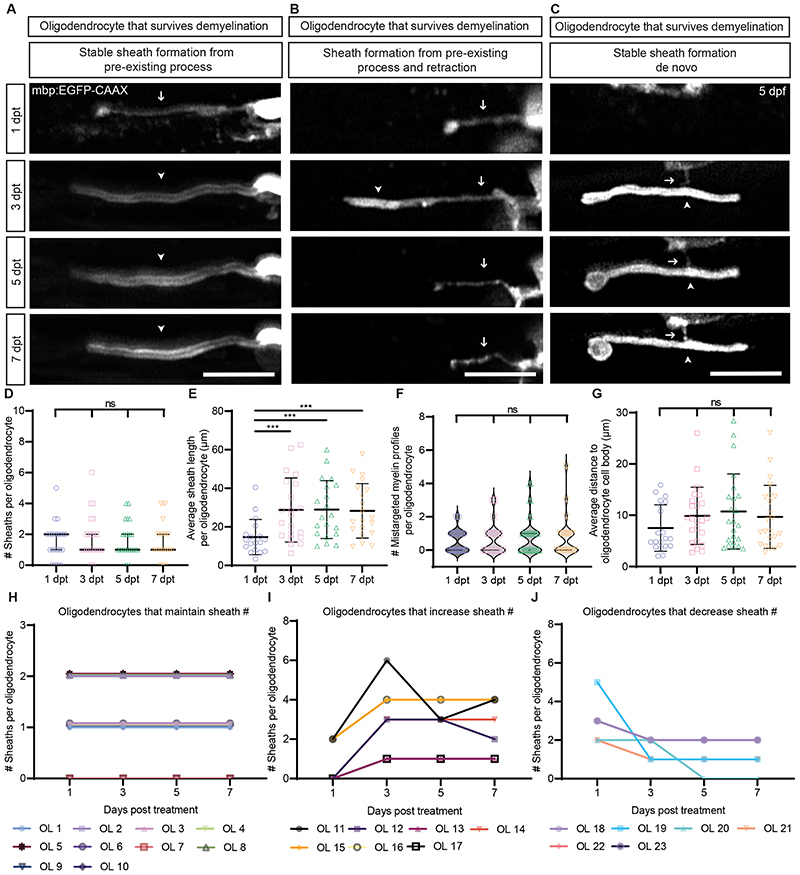
To study the dynamics of remyelination we imaged a further 23 surviving oligodendrocytes present at 1dpt in 23 zebrafish, at 3dpt, 5dpt and 7dpt. All 23 made myelin, with 9/23 (39%) making sheaths only, 12/23 (52%) sheaths and mistargeted myelin, and 2/23 (9%) mistargeted myelin only (Figure 2). The majority of bona fide sheaths were present at 1dpt, with sheath number per surviving cell remaining largely constant throughout the analyses (Figures 2D and H–J), reflecting previously documented rapid sheath formation by single oligodendrocytes 9 . However, 16 new sheaths were made across 7 cells after 1dpt, 11 of which formed from pre-existing processes (Figure 2A–B and I). Once formed, new sheaths extended along axons, completing their growth by 3dpt, in line with prior analyses of sheath elongation speeds in zebrafish 10 (Figure 2E). Similarly, most mistargeted myelin profiles were made by 1dpt, remaining stable thereafter, with only five oligodendrocytes producing mistargeted myelin at later time-points, all of which emerged from lingering processes (Figure 2F).
Figure 2. Myelin sheath dynamics of surviving oligodendrocytes following demyelination in the Tg(mbp:TRPV1-tagRFPt) zebrafish model.
(A-C) Confocal images of zoomed regions of oligodendrocytes that survive demyelination and form new sheaths from (A and B) pre-existing processes and (C) de novo sheath formation imaged over time at 1dpt, 3dpt, 5dpt and 7dpt. Arrows indicate locations of processes and arrow heads highlight the locations of newly formed myelin sheaths. Scale bars=10 μm. (D) Quantification of the number of sheaths produced per oligodendrocyte by the same cells following demyelination over time at 1dpt (median=2.00, IQR=1.00-2.00), 3dpt (median=1.00, IQR=1.00-2.00), 5dpt (median= 1.00, IQR = 1.00 - 2.00), 7dpt (median = 1.00, IQR = 1.00 - 2.00). 1dpt vs 3dpt p>0.9999, 1dpt vs 5dpt p>0.9999, 1dpt vs 7dpt p>0.9999, 3dpt vs 5dpt p>0.9999, 3dpt vs 7dpt p>0.9999, 5dpt vs 7dpt p>0.9999. Friedman test with Dunn’s multiple comparisons test. Data are presented as median with IQR. N=23 oligodendrocytes from 23 zebrafish. (E) Quantification of the average sheath length produced per oligodendrocyte by the same cells following demyelination over time at 1dpt (mean=14.68±9.12 SD), 3dpt (mean=28.75±16.62 SD), 5dpt (mean=28.98±14.98 SD), 7dpt (mean=28.32±14.12SD). 1dpt vs 3dpt p=0.0009, 1dpt vs 5dpt p=0.0006, 1dpt vs 7dpt p=0.0006, 3dpt vs 5dpt p=0.9296, 3dpt vs 7dpt p=0.9296, 5dpt vs 7dpt p=0.3654. Mixed-effects analysis with the Geisser-Greenhouse correction and Holm-Sidak’s multiple comparisons test. Data are presented as mean±SD. At 1dpt N=18 oligodendrocytes from 18 zebrafish, at 3dpt N=21 oligodendrocytes from 21 zebrafish, at 5 and 7dpt N=20 oligodendrocytes from 20 zebrafish. (F) Quantification of the number of mistargeted myelin profiles produced per oligodendrocyte following demyelination over time at 1dpt (median=0.00, IQR=0.00-1.00), 3dpt (median=0.00, IQR=0.00-1.00), 5dpt (median=1.00, IQR=0.00-1.00), 7dpt (median=0.00, IQR=0.00-1.00). 1dpt vs 3dpt p>0.9999, 1dpt vs 5dpt p>0.9999, 1dpt vs 7dpt p>0.9999, 3dpt vs 5dpt p>0.9999, 3dpt vs 7dpt p>0.9999, 5dpt vs 7dpt p>0.9999. Friedman test with Dunn’s multiple comparisons test. Data are presented as median with IQR. N=23 oligodendrocytes from 23 zebrafish. (G) Quantification of the average distance of myelin (sheaths and mistargeted myelin profiles) to the oligodendrocyte cell body (μm) per oligodendrocyte following demyelination over time at 1dpt (mean=7.52±4.53SD), 3dpt (mean= 9.89±5.58SD), 5dpt (mean=10.74±7.33SD), 7dpt (mean=9.69±6.16SD). 1dpt vs 3dpt p=0.1270, 1dpt vs 5dpt p=0.1643, 1dpt vs 7dpt p=0.2562, 3dpt vs 5dpt p=0.9067, 3dpt vs 7dpt p=0.9965, 5dpt vs 7dpt p=0.2543. Mixed-effects analysis, with Geisser-Greenhouse correction and Tukey’s multiple comparisons test. Data are presented as mean±SD. At 1dpt N=20 oligodendrocytes from 20 zebrafish, at 3, 5 and 7dpt N=23 oligodendrocytes from 23 zebrafish. (H) Quantification of the sheath dynamics of oligodendrocytes that survive demyelination and maintain their sheath number from 1 - 7dpt. N=10 oligodendrocytes from 10 zebrafish. (I) Quantification of the sheath dynamics of oligodendrocytes that survive demyelination and increase sheath number from 1 - 7dpt. N=7 oligodendrocytes from 7 zebrafish. (J) Quantification of the sheath dynamics of oligodendrocytes that survive demyelination and decrease sheath number from 1 - 7dpt. N=6 oligodendrocytes from 6 zebrafish.
Given that these analyses indicated limitations to process extension by surviving oligodendrocytes, we tried to promote sheath production by inhibiting Rho Kinase (ROCK) (Figure S5) 11,12 . We found that the ROCK inhibitor Y27632 increased the number of myelin sheaths produced by oligodendrocytes in control conditions (Extended Data Figure 5A and C), without increasing myelin mistargeting (Extended Data Figure 5E). Y27632 did not affect sheath number or length of surviving oligodendrocytes, but instead increased myelin mistargeting (Extended Data Figure 5B and F –G). Collectively our data indicate that surviving oligodendrocytes have a limited capacity to form new myelin sheaths in vivo.
Myelin is mistargeted to neuronal cell bodies in MS
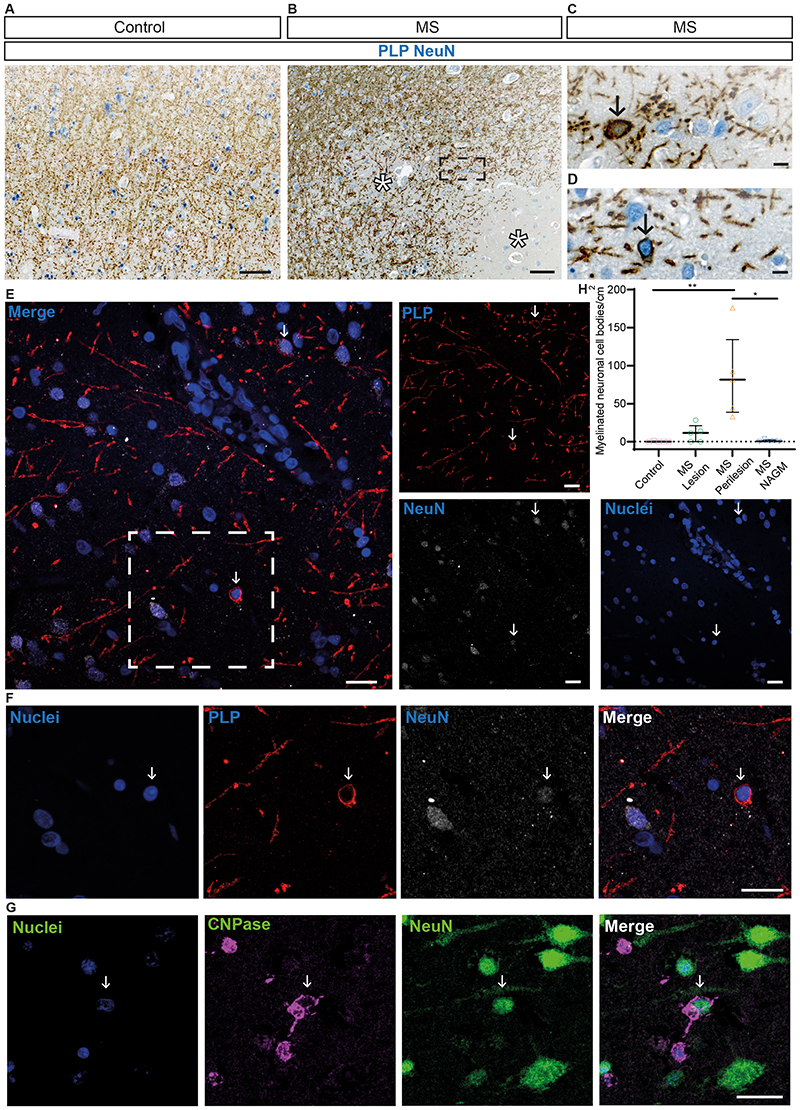
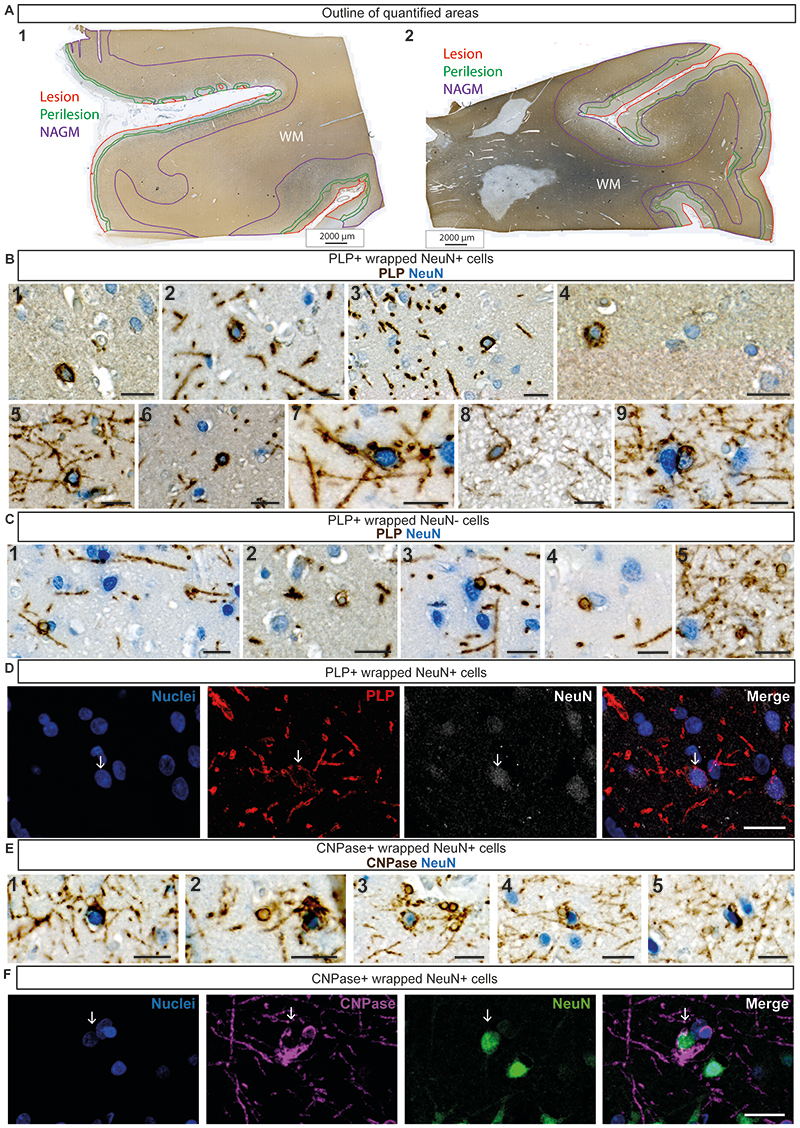
To assess whether myelin mistargeting is a feature of human disease we analysed post-mortem motor cortex samples from 5 people with, and 5 without MS (Figure 3 and Extended Data Figure 6). We focused on the grey matter because its lower density of myelin allows discernment of individual oligodendrocytes and myelin sheaths. We co-stained MS tissue with either PLP or CNPase to label oligodendrocytes and myelin sheaths and NeuN to label neurons and assessed subpial demyelinated lesions, perilesional areas and adjacent normal appearing grey matter (Figure 3 and Extended Data Figure 6) This revealed numerous aberrant-appearing myelinating profiles enwrapping NeuN-labelled neuronal cell bodies (Figure 3A–F’ and Extended Data Figure 6B), with the number of PLP+ myelinated neuronal cell bodies in MS perilesion sites, where remyelination is thought to take place 13 , almost 100-fold higher compared to control (non-MS) grey matter and normal appearing grey matter in MS (Figure 3H). Overall, these data indicate that myelin mistargeting is a previously unappreciated feature of MS grey matter pathology. Given the appearance of myelin mistargeting in areas of prospective remyelination, we asked whether this might be a general feature of remyelination or specific to oligodendrocytes that survive demyelination.
Figure 3. Mistargeted myelin profiles are present in remyelinating lesions in motor cortex tissue from people with MS.
(A and B) Low magnification images of chromogenic immunohistochemistry for proteolipid protein (PLP - brown) and NeuN (blue) in (A) human control (people without MS) motor cortex and (B) human MS motor cortex. Asterisks indicate lesion areas. Scale bars, 100μm. (C and D) High magnification images of myelin-wrapped (PLP - brown) neuronal cell bodies (NeuN - blue) in human MS motor cortex, at positions indicated by black arrows. (C) shows a high magnification view of the region highlighted in the black box in (B). Scale bars, 10μm. (E) Low magnification image of fluorescent immunohistochemistry for NeuN (white), PLP (red) and Hoechst (nuclei-blue) in human MS motor cortex. White arrows indicate locations of myelin wrapped (PLP+ve) neuronal (NeuN+ve) cell bodies (Hoechst+ve). Merged channel image scale bar, 20μm. Split channel scale bars, 10μm. (F) shows a high magnification view of the region highlighted in the white box in (E). (G) High magnification images of fluorescent immunohistochemistry for CNPase (magenta), NeuN (green) and Hoechst (nuclei-blue). White arrows indicate the location of myelin wrapped (CNPase+ve) neuronal (NeuN+ve) cell bodies (Hoechst+ve) close to the oligodendrocyte cell body. Scale bar, 20μm. H) Quantification of the number of myelinated neuronal cell bodies per grey matter area (cm2) in control grey matter (median=0.00, 25th percentile 0.00, 75th percentile 0.82), MS lesion area (median 11.65, IQR=0.00-21.06), MS perilesion area (median=81.63, IQR=38.65-134.30) and MS normal appearing grey matter (NAGM) (median = 0.61, IQR=0.00-2.32). Control versus MS lesion p>0.9999, control versus MS perilesion p=0.0076, control versus MS NAGM p>0.9999, MS lesion versus MS perilesion p=0.1735, MS lesion versus MS NAGM p>0.9999, MS perilesion versus MS NAGM p=0.0321. Kruskal-Wallis test with Dunn’s multiple comparisons test. Data are presented as median with IQR. All samples are from human motor cortex tissue: N=5 human tissue samples (from 5 different individuals) per condition.
Newly generated oligodendrocytes exhibit extensive remyelination
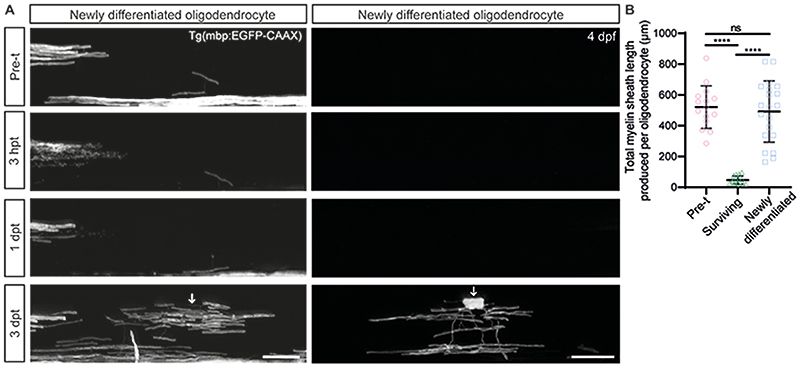
Given the limited contribution of surviving oligodendrocytes to the extensive remyelination observed by 3dpt, we next assessed the contribution of individual newly-generated oligodendrocytes to remyelination (Extended Data Figure 7 and see Methods). We found that they made an average of 25 sheaths per cell, thus over 10-fold more myelin than surviving cells (Extended Data Figure 7B). Furthermore, almost all sheaths made were correctly targeted to axons (509 correctly targeted sheaths versus 7 mistargeted myelin profiles across 20 newly-generated cells), demonstrating that myelin mistargeting is feature of remyelination by surviving oligodendrocytes. Together, our data indicate that the potential and accuracy for remyelination by newly-generated cells is far greater than that of oligodendrocytes that survive demyelination (Extended Data Figure 8).
Discussion
Our observation of limited remyelination by surviving oligodendrocytes is in line with recent evidence from studies of remyelination in the rodent cortex 6 , but it remains to be determined if surviving oligodendrocytes can be manipulated to exhibit more robust sheath formation, e.g. to reinstate dynamic process extension. Nonetheless, surviving cells can support the elongation of sheaths along axons in line with the ability of mature oligodendrocytes to support sheath growth 14–16 . Therefore, if demyelination results in the presence of lingering myelinating processes, significant remyelination may ensue. In contrast to surviving cells, newly generated oligodendrocytes have an extensive capacity for remyelination, as in mammals e.g. 6,17 , begging the question whether remyelination efficiency in MS might be related to oligodendrocyte diversity 18 , including the relative number of surviving and newly-made oligodendrocytes. The presence of surviving oligodendrocytes may even impair remyelination, given that cell death can stimulate the homeostatic generation of new oligodendrocytes from OPCs 19,20 . If oligodendrocyte death is indeed essential to triggering regeneration from OPCs, then it may be worth considering targeted destruction of surviving cells, to allow activation of OPCs with better remyelinating potential.
In summary, our analyses reveal limited and aberrant remyelination by oligodendrocytes that survive demyelination and suggest that therapeutic strategies to promote remyelination through the generation of new oligodendrocytes may remain the most promising approach for demyelinating disorders such as MS.
Methods
Zebrafish lines and maintenance
All zebrafish were maintained under standard conditions in the Queen’s Medical Research Institute CBS Aquatics facility at the University of Edinburgh. Studies were carried out with approval from the UK Home Office and according to its regulations, under project licenses 70/8436 and PP5258250. Adult animals were kept in a 14 hours light and 10 hours dark cycle. Embryos were kept at 28.5°C in 10 mM HEPES-buffered E3 embryo medium or conditioned aquarium water with methylene blue. Larval zebrafish were analysed between 4 - 9dpf, before the onset of sexual differentiation in zebrafish.
The following transgenic zebrafish (Danio rerio) lines were used in this study: Tg(mbp:TRPV1-tagRFPt) (generated for this study, see details below), Tg(mbp:EGFP), Tg(mbp:EGFP-CAAX) 21 Tg(nbt:dsred) and Tg(mpeg1:EGFP) 22 . Throughout the text and figures ‘Tg’ denotes stable, germline inserted transgenic line.
Generation of the Tg(mbp:TRPV1-tagRFPt) line
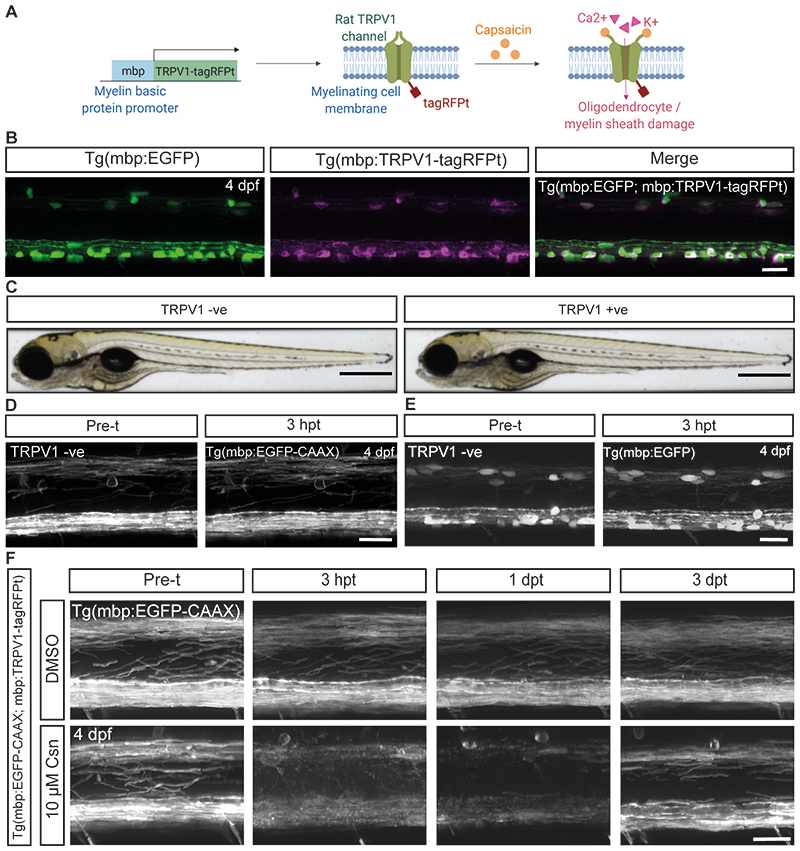
Given their advantages for longitudinal live imaging of myelination at high resolution in vivo 23 we designed a zebrafish model to follow the fate of single oligodendrocytes after demyelination. We wanted to generate a model in which we could induce extensive demyelination, but where a significant number of affected oligodendrocytes might survive. We reasoned that we might be able to induce primary disruption to myelin sheaths by generating a transgenic system in which we drove cationic influx into myelin. This was based on our observation that high amplitude, long duration Ca2+ transients prefigure myelin sheath retraction during development 24 , and that excitotoxic influx of cations can mimic myelin pathology in mammals 25,26 . To do so, we generated the Tg(mbp:TRPV1-tagRFPt) transgenic line, in which the cation-permeable TRPV1 channel of rats was expressed in myelinating oligodendrocytes downstream of the myelin basic protein (mbp) promoter of zebrafish (Extended Data Figure 1). Importantly, unlike the rat ortholog of the protein, zebrafish TRPV1 channels are not csn sensitive 27 , and treatment of zebrafish with csn alone had no observable adverse effects on myelin or oligodendrocytes (Extended Data Figure 1D–E): the principle of this system has previously been used to selectively ablate neurons in zebrafish 27 .
The mbp:TRPV1-tagRFPt plasmid was generated by first making a pME-TRPV1-tagRFPt entry vector plasmid. To do this, the TRPV1-tagRFPt sequence was amplified by PCR from the (islet1:GAL4VP16,4xUAS:TRPV1-RFPT) plasmid used by 27 , using the primers in Supplementary Table 1.
This PCR fragment was then inserted into the backbone pDONR221 by a BP reaction (BP Clonase II, Thermo Fisher Scientific). The entry vectors p5E-mbp-promoter, pME-TRPV1-tagRFPt, p3E-polyA, and pDest-Tol2pA2 were recombined by an LR reaction (LR Clonase II, Thermo Fisher Scientific) using the multisite Gateway system 28 to generate the final plasmid.
Finally, 1-2nl of 10 - 15ng/μl plasmid DNA was injected into wild-type eggs along with 25ng/μl tol2 transposase mRNA. Injected larvae were screened for mosaic transgene expression, and transgene positive larvae raised to adulthood (F0 generation). F0 generation zebrafish were then outcrossed and their progeny (F1 generation) screened for full expression of the transgene.
Capsaicin induction of demyelination
Capsaicin (csn) (Sigma-Aldrich) was prepared as a 20mM primary stock in 100% DMSO (Fisher Scientific) and stored at -80°C. Concentrations of csn were titrated to find an optimal dose to induce severe demyelination without affecting overall zebrafish health. 10μM csn 1% DMSO was selected as the lowest dose to induce severe demyelination in a 2 hour treatment period without inducing any visible behavioural response. We further tested the specificity of the 10 μM csn 1% DMSO 2 hour treatment on zebrafish which did not express rat TRPV1 channels. We observed no effect on oligodendrocytes or the myelin they produced in zebrafish which did not contain the Tg(mbp:TRPV1-tagRFPt) transgene (Extended Data Figure 1D–E).
Treatments of 10μM csn in 1% DMSO or 1% DMSO alone (vehicle control referred to as DMSO in figures) in E3 medium were applied by bath application to larval zebrafish for 2 hours at 28.5°C. Following treatment, to ensure no capsaicin or DMSO remained all solutions were replaced three times with fresh E3 embryo medium.
Y27632 treatment
Y27632 (Tocris) was prepared as a 200mM primary stock in 100% DMSO and stored at -80°C. To investigate the effect of ROCK inhibitor treatment on oligodendrocytes which had not undergone demyelination oligodendrocytes were treated with Y27632 100μM Y27632 in 1% DMSO or 1% DMSO alone (vehicle control referred to as DMSO in figures) in E3 medium by bath application to larval zebrafish between 2–4dpf. To investigate the effect of ROCK inhibitor treatment on demyelinated surviving oligodendrocytes zebrafish were demyelinated with capsaicin treatment as previously described at 4dpf. Immediately after washing out the csn treatment, 100μM Y27632 in 1% DMSO or 1% DMSO alone (vehicle control referred to as DMSO in figures) in E3 medium were applied by bath application to larval zebrafish between 4dpf (3hpt) to 7dpf (3dpt) with daily media changes. Zebrafish were kept at 28.5°C between imaging timepoints.
Transmission electron microscopy (TEM)
Tissue was prepared for TEM as previously described 29 . Briefly, zebrafish embryos were terminally anaesthetised in MS222 tricaine (Sigma-Aldrich) and incubated, with microwave stimulation, first in primary fixative (4% paraformaldehyde + 2% glutaraldehyde in 0.1M sodium cacodylate buffer) and then in secondary fixative (2% osmium tetroxide in 0.1M sodium cacodylate/imidazole buffer). Samples were then stained en bloc with saturated uranyl acetate solution and dehydrated in an ethanol and acetone series, both with microwave stimulation. Samples were embedded in EMbed-812 resin (Electron Microscopy Sciences) and sectioned using a Reichert Jung Ultracut Microtome. Sections were cut at comparable somite levels by inspection of blocks under a dissection microscope and stained in uranyl acetate and Sato lead stain. TEM images were taken with a Phillips CM120 Biotwin TEM. The Photomerge tool in Adobe Photoshop was used to automate image registration and tiling. To assess axon diameter (> 0.4μm), axonal areas were measured in ImageJ.
Single oligodendrocyte labelling
To mosaically label oligodendrocytes, fertilized eggs were injected with 1nl of 10ng/μl pTol2-mbp:EGFP-CAAX plasmid DNA and 50ng/μl tol2 transposase mRNA at the 1 cell stage. Animals were screened at 3 and 4dpf for isolated oligodendrocytes.
Live imaging
Larval zebrafish were live imaged after anesthetising them with MS222 tricaine and mounting them on glass coverslips, embedded in 1.5% low melting point agarose (Invitrogen). Z-stacks were acquired at relevant locations along the spinal cord using the LSM 880 confocal microscope equipped with Airyscan Fast and a 20X objective (Zeiss Plan-Apochromat 20X dry, NA=0.8). Z-stacks were acquired with an optimal z-step according to the experiment.
To follow the fate of oligodendrocytes, and myelination profiles over time, zebrafish were repeat imaged (4dpf pre-treatment, 3hpt, 1dpt, 3dpt, 5dpt and 7dpt). To do so zebrafish were carefully cut out from agarose following imaging and returned to E3 embryo medium, whilst being checked for signs of impaired health or swim behaviour. Zebrafish were maintained at 28.5°C in 10mM HEPES-buffered E3 embryo medium between imaging sessions.
To assess any developmental effects of the Tg(mbp:TRPV1-tagRFPt) transgene zebrafish were anesthetised with MS222 tricaine and imaged using the Vertebrate Automated Screening Technology BioImager system (Union Biometrica), which automated the positioning of larvae and image acquisition using a Zeiss spinning disk confocal microscope (VAST-SDCM) 30 .
Single oligodendrocyte imaging
Zebrafish were screened at 4dpf for isolated oligodendrocytes, and when identified were imaged over time by identifying their position along the spinal cord, relative to the nearest somite as well as their position relative to other oligodendrocytes in the surrounding area labelled with the Tg(mbp:TRPV1-tagRFPt) and Tg(mbp:EGFP-CAAX) transgenes. As not all oligodendrocytes die after initial demyelination, we were able to carefully assess relative positions using both tagRFPt and EGFP-CAAX reporters (Extended Data Figure 4A). Newly generated oligodendrocytes were carefully identified based on the appearance of oligodendrocytes labelled with the tagRFPt and EGFP-CAAX reporters in areas previously devoid of fluorescence.
Image analysis
Image processing and analysis was performed in Fiji (ImageJ). Figure panels were produced using Fiji and Adobe Illustrator. For figures, maximum-intensity projections of z-stacks were made, and a representative x-y area was cropped. All zebrafish images represent a lateral view of the spinal cord, anterior to the left and dorsal on top. For most images, processing included only global change of brightness and contrast; further processing and analysis is as follows.
Cell counts
Mbp:EGFP cell bodies filled with cytoplasmic EGFP were counted through z-stacks which encompassed the depth of the spinal cord using the cell counter plugin in Fiji. The same area of the spinal cord was imaged and counted for analysis in different zebrafish (5 somite section). Dorsal and ventral oligodendrocytes were counted separately and combined for total analysis. All images were blinded to treatment condition during analysis.
Single oligodendrocyte morphology analysis
Oligodendrocytes were analysed for sheath length, sheath number and the distance of the myelin sheath/ mistargeted myelin profile from the cell body using the segmented line tracing tool in Fiji (Image J) through z-stacks which encompassed the depth of the oligodendrocyte and its myelin. Regions of interest were saved per oligodendrocyte. Cell body counts were carried out manually using the full z-stack of each oligodendrocyte. Oligodendrocytes with too much overlapping myelin, from oligodendrocytes in the surrounding area, to confidently trace the source of the myelin sheath were excluded from analysis.
Human data
Post-mortem brain tissue (motor cortices) from MS patients and controls without neurological defects were provided by a UK prospective donor scheme with full ethical approval from the UK Multiple Sclerosis Society Tissue Bank (MREC/02/2/39). MS diagnosis was confirmed by neuropathological means by F. Roncaroli (Imperial College London) and clinical history was provided by R. Nicholas (Imperial College London). Supplementary Table 2 includes details on samples used. Tissue blocks of 2cm x 2cm x 1cm were collected, fixed, dehydrated and embedded in paraffin blocks. 4μm sequential sections were cut and stored at room temperature. Grey matter MS lesions were identified using anti-Proteolipid Protein (PLP) immunostaining.
Human post-mortem brain tissue immunohistochemistry
Paraffin sections were rehydrated, washed in PBS and microwaved for 15 minutes in Vector Unmasking Solution for antigen retrieval (H-3300, Vector). For colorimetric immunohistochemistry, endogenous peroxidase and alkaline phosphatase activities were blocked for 10 minutes using Bioxal solution (SP-6000, Vector). Sections were then blocked with 2.5% normal horse serum (S-2012, Vector) for 1 hour at room temperature. Primary antibodies were incubated in antibody diluent solution (003118, Thermo Fisher Scientific), overnight at 4°C in a humidified chamber. Horse peroxidase or alkaline phosphatase-conjugated secondary antibodies (Vector) were applied for an hour at room temperature. Staining development was performed using either DAB HRP substrate kit or Vector Blue substrate kit (both from Vector) according to the manufacturer’s instructions.
Human post-mortem brain tissue immunofluorescence
For immunofluorescence, sections were incubated with Autofluorescent Eliminator Reagent (2160, MERCK-Millipore) for 1 minute and briefly washed in 70% ethanol after antigen retrieval. The sections were subsequently incubated with Image-iT® FX Signal Enhancer (I36933, Thermo Fisher Scientific) for 30 minutes at room temperature, washed and blocked for 1 hour with 10% normal horse serum, 0.3% Triton-X in PBS. Primary antibodies were diluted in antibody diluent solution (as above) and placed on sections overnight at 4°C in a humidified chamber. The next day the sections were incubated with Alexa Fluor secondary antibodies (Thermo Fischer Scientific, 1:1000) for 1 hour at room temperature and counterstained with Hoechst for the visualization of the nuclei.
Primary antibodies used: rabbit polyclonal IgG antibody to NeuN (104225, Abcam, 1:100), mouse monoclonal IgG2A antibody to myelin Proteolipid Protein (clone PLPC1, MAB388, MERCK-Millipore, 1:100) and mouse monoclonal IgG2b antibody to CNPase (2’,3’-cyclic nucleotide 3’ phosphodiesterase,CL2887, AMAb91072, Atlas antibodies, 1:1000). All slides were mounted using Mowiol mounting medium (475904, MERCK- Millipore).
Secondary antibodies used: Goat anti-Mouse IgG2a Cross-Adsorbed Secondary Antibody, Alexa Fluor 568, A-21134, RRID: AB_2535773 Thermo Fisher Scientific Goat anti-Mouse IgG2b Cross-Adsorbed Secondary Antibody, Alexa Fluor 568, A-21144, RRID:AB_2535780 Thermo Fisher Scientific Goat anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor® 488 conjugate, A-11008, RRID: AB_143165 Thermo Fisher Scientific ImmPRESS® HRP Horse Anti-Mouse IgG Polymer Detection Kit, Peroxidase (MP-7402-15) Vector Laboratories ImmPRESS-AP Anti-Rabbit Ig (alkaline phosphatase) Polymer Detection Kit (MP-5401) Vector Laboratories. All listed secondary antibodies were ready to use antibody kits and no specific dilution was used.
For chromogenic IHC signal detection the following were used:
DAB Substrate Kit, Peroxidase (HRP), with Nickel, (3,3’-diaminobenzidine) SK-4100 Vector Laboratories Vector® Blue Substrate Kit, Alkaline Phosphatase (AP) SK-5300 Vector Laboratories.
Entire sections were imaged using the Zeiss AxioScan Slide scanner and all quantifications were performed using Zeiss Zen lite imaging software. For all cases, the whole grey matter area of the section was investigated focusing on perilesion areas in MS cases and in corresponding cortical layers in control tissue. Cell densities are presented as immune-positive cells per cm2. Z-stack images of the fluorescent-labelled samples were acquired with the LSM 880 confocal microscope equipped with Airyscanner and a 20X objective (Zeiss Plan-Apochromat 20X dry, NA=0.8). Z-stacks were acquired with an optimal z-step according to the experiment.
Reproducibility and sample size selection
For zebrafish data no statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications 7 . Larval zebrafish were grown up in the same incubator to avoid any differences due to developmental stage between experiments. Following screening to identify transgene positive zebrafish, larvae were randomly assigned to treatment or control groups. During live imaging analyses, experimental and control animals were imaged in an alternating pattern (per experiment) to ensure no confounding effects of developmental stage between groups.
For human data no statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications 31,32 . For human data samples were randomly chosen for each group. We used donor tissue from both sexes randomly distributed in each group (see Supplementary Table 2). The age of the donors the post-mortem interval and in the case of MS samples disease duration was also randomly chosen. The selected MS samples were selected based on the presence of at least one demyelinated lesion in the grey matter. Data collection and analysis could not be performed blind to the conditions of the experiments but in the case of human IHC analysis the whole tissue section was analysed so as not to introduce any regional bias. The results were validated by two independent researchers. No points were excluded.
Statistics and reproducibility
Graphs and statistical tests were carried out using GraphPad Prism. Following screening to identify transgene positive zebrafish, larvae were randomly assigned to treatment or control groups. Analysis of in vivo data was carried out blinded before treatment group was revealed. Data were averaged per biological replicate (N represents number of zebrafish or humans) unless stated otherwise in figure legends. All single oligodendrocytes which were imaged over a time course were analysed to make sure the same cell was captured over time, where this was not the case the oligodendrocyte was excluded for analysis. No data points were excluded from analysis due to variability.
For oligodendrocyte counts, and electron microscopy analysis findings are reported from a single experimental run, from multiple zebrafish per condition. Data from Figures 2 and 3 characterising single surviving oligodendrocytes are taken from at least 3 experimental runs, data documenting oligodendrocytes which did not undergo demyelination in Extended Data Figure 5 is from 2 experimental runs.
Data was tested for normal distribution using the D’Agostino & Pearson test or the Shapiro-Wilk test. The variance of the data was assessed using the F test for variance. Indicated p values are from two-tailed unpaired t-tests, two-tailed unpaired t-test with Welch’s correction, two-tailed paired t-tests, Wilcoxon signed-rank tests, or Mann-Whitney tests. To compare more than 2 groups a one-way ANOVA with Tukey’s multiple comparisons test, a Kruskal-Wallis test with Dunn’s multiple comparisons test, a mixed-effects analysis or a Friedman test was used. A difference was considered statistically significant when p < 0.05. Throughout the figures p values are indicated as follows: not significant or ‘ns’ p > 0.05, ‘*’ p < 0.05, ‘**’ p < 0.01, ‘***’p < 0.001. All data are shown as mean ± standard deviation (SD) where data is normally distributed or median with interquartile range (IQR) (25th percentile and 75th percentile) where data is not normally distributed. Details of statistical test used, precise p values and N values for each comparison are detailed in figure legends.
Extended Data
Extended Data Figure 1. Characterisation of the Tg(mbp:TRPV1-tagRFPt) zebrafish model.
(A) Schematic illustrating the Tg(mbp:TRPV1-tagRFPt) demyelination model made using Biorender. The rat ortholog of the TRPV1 channel is expressed in myelinating oligodendrocytes and is activated by addition of csn which drives cation influx. Zebrafish TRPV1 channels are insensitive to csn, therefore csn treatment specifically results in damage to myelinating glia which express the rat ortholog of the TRPV1 channel. (B) Confocal images of myelinating oligodendrocytes in the Tg(mbp:EGFP; mbp:TRPV1- tagRFPt) zebrafish line at 4dpf showing oligodendrocytes co-expressing EGFP and tagRFPt in the merged image. Scale bar, 20 µm. (C) Brightfield images of a zebrafish containing the Tg(mbp:TRPV1-tagRFPt) transgene (TRPV1+ve), or wildtype siblings without the Tg(mbp:TRPV1-tagRFPt) transgene (TRPV1-ve) which show no developmental differences at 4dpf. Scale bars, 500µm. (D and E) Confocal images of the (D) Tg(mbp:EGFP-CAAX) line and (E) the Tg(mbp:EGFP) line pre-treatment (indicated here as pre-t) at 4dpf, and 3hpt. Zebrafish not containing the Tg(mbp:TRPV1-tagRFPt) transgene show no disruption to myelin or oligodendrocytes following a 2 hour treatment of 10µM csn. Scale bars, 20µm. (F) Confocal images of myelin visualised in Tg(mbp:EGFP-CAAX; mbp:TRPV1-tagRFPt) zebrafish, with control (DMSO) and csn treated animals in top and bottom panels respectively pre-treatment (pre-t) at 4dpf, 3hpt, 1dpt and 3dpt. Scale bar, 20µm.
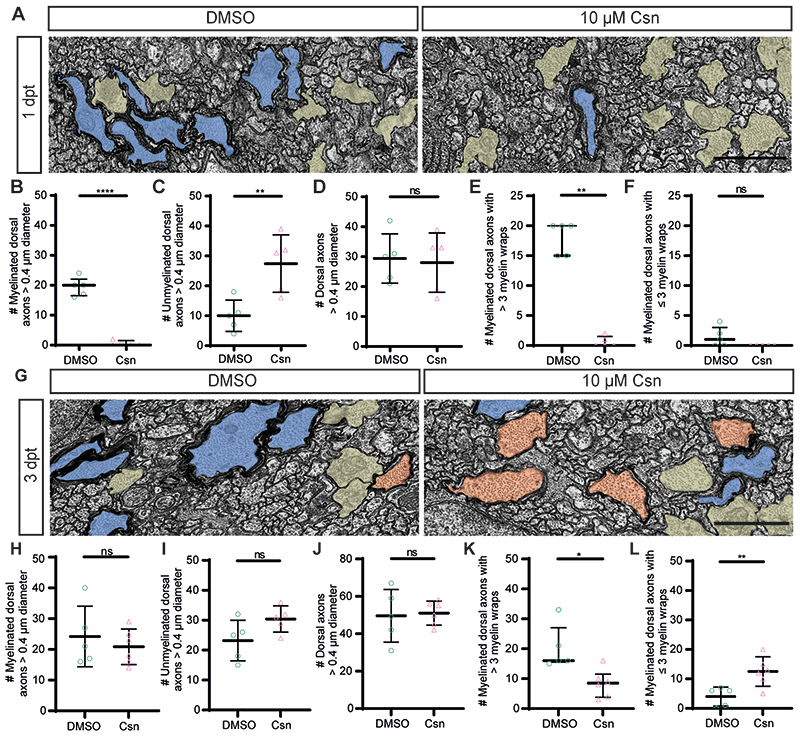
Extended Data Figure 2. Csn treatment induces severe demyelination in the Tg(mbp:TRPV1-tagRFPt) zebrafish model.
(A) Transmission electron microscopy images of DMSO and csn treated Tg(mbp:TRPV1-tagRFPt) zebrafish at 1dpt show numerous large calibre myelinated axons (blue) in DMSO treated animals and numerous large calibre unmyelinated axons (yellow) in csn treated animals. Scale bar, 1µm. (B) Quantification of the number of myelinated axons>0.4µm diameter in the dorsal spinal cord at 1dpt in DMSO (median=20.00, IQR=16.50-22.00) versus csn (median = 0.00, IQR=0.00-1.50) treated zebrafish, p<0.0001. Unpaired two-tailed t-test with Welch’s correction. N=5 zebrafish per condition. Data are presented as median with IQR. (C) Quantification of the number of unmyelinated axons>0.4µm diameter in the dorsal spinal cord at 1dpt in DMSO (mean=10.00±5.24SD) versus csn (mean=27.40±9.61SD) treated zebrafish, p=0.0075. Unpaired two-tailed t-test. N=5 zebrafish per condition. Data are presented as mean±SEM. (D) Quantification of the number of axons>0.4µm diameter in the dorsal spinal cord at 1dpt in DMSO (mean=29.40±8.26SD) versus csn (mean=28.00±9.95SD) treated zebrafish, p=0.8148. Unpaired two-tailed t-test. N=5 zebrafish per condition. Data are presented as mean±SEM. (E) Quantification of the number of axons with>3 myelin wraps in the dorsal spinal cord at 1dpt in DMSO (median=20.00, IQR=15.00-20.00) versus csn (median=0.00,IQR=0.00-1.50) treated zebrafish, p=0.0079. Two-tailed Mann-Whitney t-test. N=5 zebrafish per condition. Data are presented as median with IQR (F) Quantification of the number of axons with≤3 myelin wraps in the dorsal spinal cord at 1dpt in DMSO (median=1.00, IQR=0.00-3.00) versus csn (median=0.00, IQR=0.00-0.00) treated zebrafish, p=0.1667. Two-tailed Kolmogorov-Smirnov test. N=5 zebrafish per condition. Data is presented as median with IQR.(G) Transmission electron microscopy images of DMSO and csn treated Tg(mbp:TRPV1-tagRFPt) zebrafish at 3dpt show numerous large calibre myelinated axons (>3 myelin wraps highlighted in blue, ≤3 myelin wraps highlighted in orange) and unmyelinated axons (highlighted in yellow) in DMSO and csn treated animals. Scale bar, 1µm. (H) Quantification of the number of myelinated axons >0.4µm diameter in the dorsal spinal cord at 3dpt in DMSO (mean=24.20±9.83SD) versus csn (mean=20.83±5.78SD) treated zebrafish, p=0.4963. Unpaired two-tailed t-test. N=5 DMSO treated zebrafish, N=6 csn treated zebrafish. Data are presented as mean±SEM.(I) Quantification of the number of unmyelinated axons>0.4µm diameter in the dorsal spinal cord at 3dpt in DMSO (mean=23.20±6.76SD) versus csn (mean=30.40±4.39SD) treated zebrafish, p=0.0809. Unpaired two-tailed t-test. N=5 DMSO treated zebrafish, N=6 csn treated zebrafish. Data are presented as mean±SEM.(J) Quantification of the number of axons>0.4µm diameter in the dorsal spinal cord at 3dpt in DMSO (mean=47.60±10.92SD) versus csn (mean=51.00±6.45SD) treated zebrafish, p=0.5359. Unpaired two-tailed t-test. N=5 DMSO treated zebrafish, N=6 csn treated zebrafish. (K) Quantification of the number of axons with>3 myelin wraps in the dorsal spinal cord at 3dpt in DMSO (median=16.00, IQR=15.50-27.00) versus csn (median=8.50, IQR=3.75-11.50) treated zebrafish, p=0.0152. Two-tailed Mann-Whitney t-test. N=5 DMSO treated zebrafish, N=6 csn treated zebrafish. (L) Quantification of the number of axons with≤3 myelin wraps in the dorsal spinal cord at 3dpt in DMSO (mean=4.00±3.24SD) versus csn (mean=12.50±5.01SD) treated zebrafish, p=0.0099. Unpaired two-tailed t-test. N=5 DMSO treated zebrafish, N=6 csn treated zebrafish.
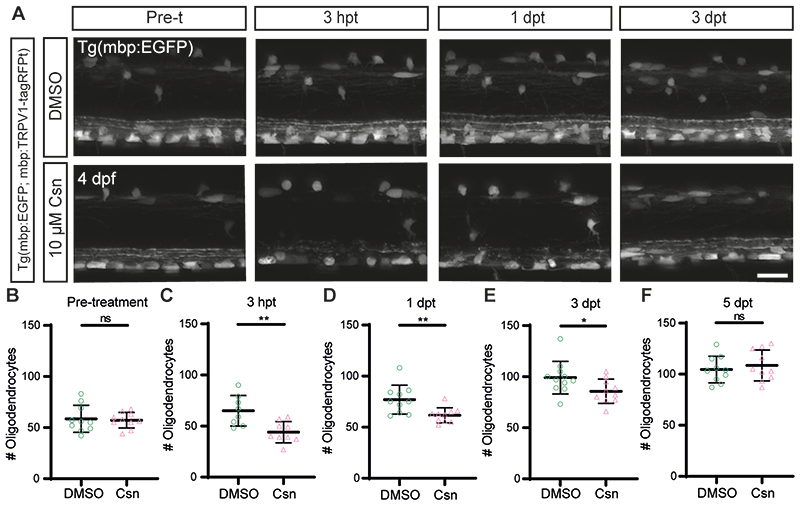
Extended Data Figure 3. Csn treatment induces minimal oligodendrocyte loss in the Tg(mbp:TRPV1-tagRFPt) model.
(A) Confocal images of myelinating oligodendrocytes visualised in Tg(mbp:EGFP; mbp:TRPV1-tagRFPt) zebrafish, with control (DMSO) and csn treated animals in top and bottom panels respectively pre-treatment (pre-t) at 4dpf, 3hpt, 1dpt and 3dpt. Scale bar, 20 µm. (B-F) Quantification of myelinating oligodendrocyte number in DMSO and csn treated Tg(mbp:EGFP; mbp:TRPV1-tagRFPt) zebrafish over time. (B) Pre-treatment DMSO (mean=58.60±13.23SD) versus csn (mean=57.20±7.64SD), p=0.7753. Data are presented as mean±SEM. (C) 3hpt DMSO (mean=65.00±14.93SD) versus csn (mean=44.11±10.36SD), p=0.0041. Data are presented as mean±SEM. (D) 1dpt DMSO (mean=76.90±14.14SD) versus csn (mean=61.70±7.39SD), p=0.0075. Data are presented as mean±SEM. (E) 3dpt DMSO (mean=99.00±15.93SD) versus csn (mean=85.70±11.83SD), p=0.0444. Data are presented as mean±SEM. (F) 5dpt DMSO (mean=104.5±12.95SD) versus csn (mean=108.5±15.10SD), p=0.5328. (B-F) Unpaired two-tailed t-tests. Pre-treatment N=10 zebrafish per treatment group, 3hpt N=8 (DMSO) and 9 (csn) treated zebrafish, 1dpt N=10zebrafish per treatment group, 3dpt N=11 zebrafish (DMSO) and 10 zebrafish (DMSO), 5 dpt N=10 zebrafish per treatment group. Data are presented as mean±SEM. Each data point represents total (dorsal + ventral) oligodendrocyte number analysed per imaged area per zebrafish.
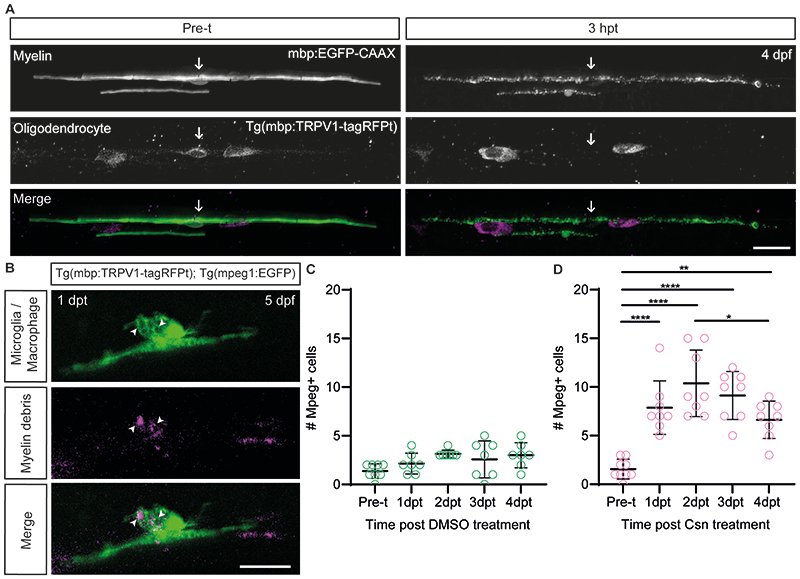
Extended Data Figure 4. Characterisation of single oligodendrocyte loss and myelin debris phagocytosis following demyelination in the Tg(mbp:TRPV1-tagRFPt) zebrafish model.
(A) Confocal images of a single oligodendrocyte labelled with mbp:EGFP-CAAX in the Tg(mbp:TRPV1-tagRFPt) line pre-treatment (pre-t) at 4dpf, and 3hpt. An example of oligodendrocyte cell death is demonstrated here by the disappearance of a tagRFPt+ve cell body following csn treatment in the same zebrafish before and after demyelination, whilst 2 tagRFPt+ve oligodendrocytes which survive demyelination are seen neighbouring it at 3hpt. Arrows indicate the location of the oligodendrocyte cell body which undergoes cell death, or where it was prior to demyelination. Scale bar, 20µm. (B) Confocal images of microglia / macrophage engulfment of myelin debris following demyelination at 1dpt. Arrowheads highlight the location of myelin debris engulfment. Scale bar, 20µm. (C) Quantification of the number of mpeg+ve cells (macrophages / microglia) in a 4-somite section of the spinal cord at pre-treatment (mean=1.38±0.74SD), 1dpt (mean=2.14±1.07SD), 2dpt (mean=3.14±0.38SD), 3dpt (mean=2.57±1.90SD) and 4dpt (mean=3.00±1.29SD) (where treatment was a DMSO control). Pre-t vs 1dpt p=0.7196, pre-t vs 2dpt p=0.0505, pre-t vs 3dpt p=0.3106, pre-t vs 4dpt p=0.0844, 1dpt vs 2dpt p=0.5190, 1dpt vs 3dpt p=0.9597, 1dpt vs 4dpt p=0.6588, 2dpt vs 3dpt p=0.8930, 2dpt vs 4dpt p=0.9994, 3dpt vs 4dpt p=0.9597. Ordinary one-way ANOVA with Tukey’s multiple comparison test. Pre-treatment N=8 zebrafish , 1–4dpt N=7 zebrafish. Data are presented as mean±SEM. (D) Quantification of the number of mpeg+ve cells (macrophages / microglia) in a 4-somite section of the spinal cord at pre-treatment (mean=1.56±1.01SD), 1dpt (mean=7.88±2.75SD), 2dpt (mean=10.38±3.42SD), 3dpt (mean=9.12±2.48SD) and 4dpt (mean=6.63±1.92SD) (where treatment was a demyelinating csn treatment). Pre-t vs 1dpt p<0.0001, pre-t vs 2dpt p<0.0001, pre-t vs 3dpt p<0.0001, pre-t vs 4dpt p=0.0011, 1dpt vs 2dpt p=0.2586, 1dpt vs 3dpt p=0.8394, 1dpt vs 4dpt p=0.8394, 2dpt vs 3dpt p=0.8394, 2dpt vs 4dpt p=0.0294, 3dpt vs 4dpt p=0.2586. Ordinary one-way ANOVA with Tukey’s multiple comparison test. Pre-treatment N=9 zebrafish, 1–4dpt N=8 zebrafish. Data are presented as mean±SEM.
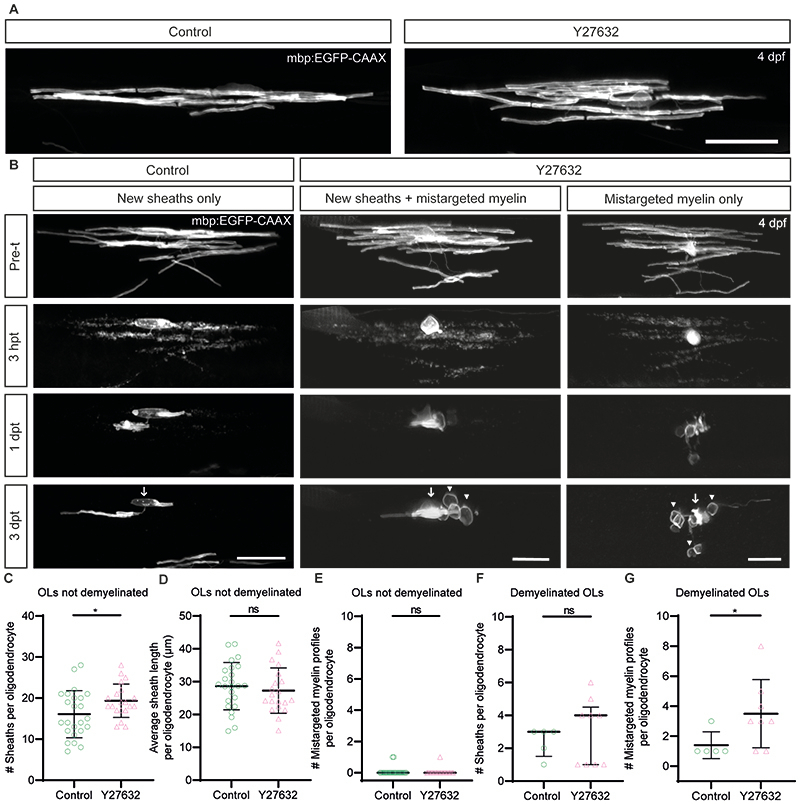
Extended Data Figure 5. ROCK inhibitor treatment further increases myelin mistargeting by surviving oligodendrocytes in the Tg(mbp:TRPV1-tagRFPt) zebrafish model.
(A) Confocal images of single oligodendrocytes which have not undergone demyelination treated with DMSO (control) or Y27632 ROCK inhibitor and imaged at 4dpf. Scale bar, 20µm. (B) Confocal images of single surviving oligodendrocytes followed over time from prior to demyelination (csn treatment) at 4dpf through to 3dpt. Following demyelination oligodendrocytes were treated with either DMSO (control) or Y27632 ROCK inhibitor. Scale bars, 20µm. (C) Quantification of the number of sheaths produced in oligodendrocytes which have not been demyelinated in control (mean=16.08±5.73SD) and Y27632 ROCK inhibitor (mean=19.35±4.08SD) treated zebrafish, p=0.0386. Unpaired two-tailed t-test. N=24 oligodendrocytes from 24 zebrafish (control), N=20 oligodendrocytes from 20 zebrafish (Y27632). Data are presented as mean±SEM. (D) Quantification of the average sheath length (µm) produced in oligodendrocytes which have not been demyelinated in control (mean=28.62±7.18SD) and Y27632 ROCK inhibitor (mean=27.26±6.91SD) treated zebrafish, p=0.5308. Unpaired two-tailed t-test. N=24 oligodendrocytes from 24 zebrafish (control), N=20 oligodendrocytes from 20 zebrafish (Y27632). Data are presented as mean±SEM.(E) Quantification of the number of mistargeted myelin profiles produced in oligodendrocytes which have not been demyelinated in control (median=0.00, IQR=0.00-0.00) and Y27632 ROCK inhibitor (median=0.00, IQR=0.00-0.00) treated zebrafish, p>0.9999. Two-tailed Mann-Whitney test. N=24 oligodendrocytes from 24 zebrafish (control), N=20 oligodendrocytes from 20 zebrafish (Y27632). Data are presented as median with IQR.(F) Quantification of the number of sheaths produced per oligodendrocyte following demyelination in control (median = 3.00, 25th percentile 1.50, 75th percentile 3.00) and Y27632 ROCK inhibitor (median=4.00, IQR=1.00-4.50) treated zebrafish, p=0.5964. Two-tailed Mann-Whitney t-test. N=5 oligodendrocytes from 5 zebrafish (control), N=9 oligodendrocytes from 9 zebrafish (Y27632). Data are presented as median with IQR. (G) Quantification of the number of mistargeted myelin profiles produced per oligodendrocyte following demyelination in control (median=1.00, IQR=1.00-2.00) and Y27632 ROCK inhibitor (median=3.00, IQR=1.50-4.75) treated zebrafish, p=0.0414. Unpaired two tailed t-test with Welch’s correction . N=5 oligodendrocytes from 5 zebrafish (control), N=9 oligodendrocytes from 9 zebrafish (Y27632). Data are presented as mean±SEM.
Extended Data Figure 6. Mistargeted myelin profiles are present in remyelinating lesions in motor cortex tissue from people with MS.
(A) Low magnification image of chromogenic immunohistochemistry for proteolipid protein (PLP - brown) and NeuN (blue) in human MS motor cortex. Outline of quantified areas shown with lesion area highlighted in red, perilesion area highlighted in green, normal appearing grey matter (NAGM) in purple and white matter indicated by ‘WM’ in white. Images 1 and 2 show examples of quantified areas in 2 different human MS motor cortex samples. Scale bars, 2000µm. (B and C) High magnification images of chromogenic immunohistochemistry for proteolipid protein (PLP - brown) and NeuN (blue) in human MS motor cortex. (B) Images 1-9 show example images of PLP+ve wrapped NeuN+ve cells (myelinated neuronal cell bodies). Images 1 and 3-9 scale bars, 20µm. Image 2 scale bar, 10µm. (C) Images 1-5 show example images of PLP+ve wrapped NeuN-ve cells (oligodendrocytes). Scale bars, 20µm. (D) Fluorescent immunohistochemistry for NeuN (white), PLP (red) and Hoechst (nuclei-blue) in human MS motor cortex. Arrows indicate the location of PLP+ve wrapped NeuN+veHoechst+ve cells (myelinated neuronal cell body). Scale bar, 20µm. (E) Images 1-5 show example images of CNPase+ve wrapped NeuN+ve cells (myelinated neuronal cell bodies). Scale bars, 20µm. (F) Fluorescent immunohistochemistry for NeuN (green), CNPase (magenta) and Hoechst (nuclei-blue) in human MS motor cortex. Arrows indicate the location of CNPase+ve wrapped NeuN+veHoechst+ve cells. Scale bar, 20µ
Extended Data Figure 7. Extensive remyelination by newly generated oligodendrocytes in the Tg(mbp:TRPV1-tagRFPt) zebrafish model.
(A) Confocal images of csn treated zebrafish with oligodendrocytes newly generated after demyelination. Arrows show position of oligodendrocyte cell bodies. Scale bars, 20µm. (B) Quantification of the total myelin produced per oligodendrocyte (calculated by multiplying number of sheaths per oligodendrocyte by the average sheath length per oligodendrocyte) (mean=521.5±138.30SD), versus the same oligodendrocytes 3dpt (mean=47.18±26.57SD) and by newly differentiated oligodendrocytes at 3dpt (mean=491.80±199.10SD). Pre-treatment versus surviving p<0.0001, pre-treatment versus newly differentiated p=0.8271, surviving versus newly differentiated p<0.0001. Ordinary one-way ANOVA with Tukey’s multiple comparison test. N=15 oligodendrocytes from 15 zebrafish (pre-treatment and surviving). N=20 oligodendrocytes from 11 zebrafish (newly differentiated). Data are presented as mean±SEM.
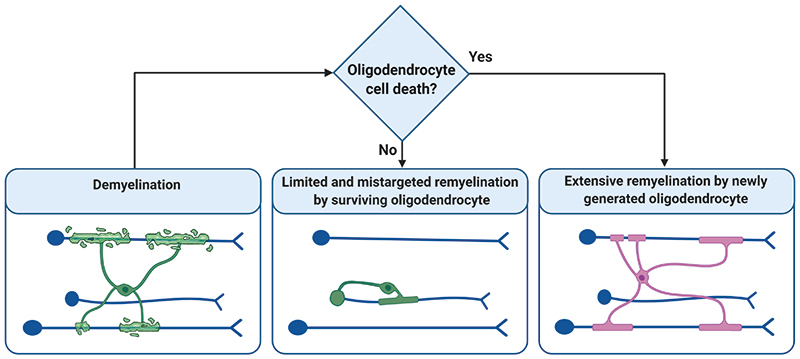
Extended Data Figure 8. Summary Schematic.
Summary schematic outlining the responses of oligodendrocytes which survive demyelination and those newly generated after demyelination made using Biorender.
Supplementary Material
Acknowledgements
We thank Charles ffrench-Constant, Ethan Hughes and the Lyons lab for feedback, the BVS aquatics facility for fish care, Stephen Mitchell for electron microscopy assistance, and Carmen Melendez-Vasquez for suggesting the ROCK experiment. This work was supported by Wellcome Trust Senior Research Fellowships (102836/Z/13/Z and 214244/Z/18/Z), a Medical Research Council Project Grant (MR/P006272/1) and an MS Society Innovative Grant (95) to DAL. SAN and JMW were supported by a Wellcome Trust Four-Year Ph.D. Program in Tissue Repair (Grant 108906/Z/15/Z) and JMW by a University of Edinburgh Ph.D. Tissue Repair Studentship Award (MRC Doctoral Training Partnership MR/K501293/1). LZ and AW were supported by MS Society UK Centre grant.
Footnotes
Author Contributions statement
SAN, JMW and DAL conceived the project. SAN, JMW, AK and JJE designed and performed the in vivo experiments. LZ and AW designed and performed the human tissue experiments. SAN and DAL co-wrote the manuscript, edited by all. DAL managed the project.
Competing Interests Statement
The authors declare no competing interests.
Data analysis
The following programs were used for data analysis: Fiji Image J version 1.52p and 1.53c, Microsoft Excel 365, Graph Pad Prism9, Adobe Illustrator 2021, Adobe Photoshop 2021.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Code Availability
No code was used in this manuscript.
References
- 1.Thompson AJ, Baranzini SE, Geurts J, Hemmer B, Ciccarelli O. Multiple sclerosis. Lancet. 2018;391:1622–1636. doi: 10.1016/S0140-6736(18)30481-1. [DOI] [PubMed] [Google Scholar]
- 2.Franklin RJM, Frisén J, Lyons DA. Revisiting remyelination: Towards a consensus on the regeneration of CNS myelin. Semin Cell Dev Biol. 2020;116:3–9. doi: 10.1016/j.semcdb.2020.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Cole KLH, Early JJ, Lyons DA. Drug discovery for remyelination and treatment of MS. Glia. 2017;65:1565–1589. doi: 10.1002/glia.23166. [DOI] [PubMed] [Google Scholar]
- 4.Yeung MSY, et al. Dynamics of oligodendrocyte generation in multiple sclerosis. Nature. 2019;566:538–542. doi: 10.1038/s41586-018-0842-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duncan ID, et al. The adult oligodendrocyte can participate in remyelination. Proc Natl Acad Sci U S A. 2018;115:E11807–E11816. doi: 10.1073/pnas.1808064115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacmeister CM, et al. Motor learning promotes remyelination via new and surviving oligodendrocytes. Nat Neurosci. 2020;23:819–831. doi: 10.1038/s41593-020-0637-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klingseisen A, et al. Oligodendrocyte Neurofascin Independently Regulates Both Myelin Targeting and Sheath Growth in the CNS Article Oligodendrocyte Neurofascin Independently Regulates Both Myelin Targeting and Sheath Growth in the CNS. Dev Cell. 2019;51:730–744. doi: 10.1016/j.devcel.2019.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Almeida RG, et al. Myelination of Neuronal Cell Bodies when Myelin Supply Exceeds Axonal Demand. Curr Biol. 2018;28:1296–1305. doi: 10.1016/j.cub.2018.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czopka T, ffrench-Constant C, Lyons DA. Individual oligodendrocytes have only a few hours in which to generate new myelin sheaths invivo. Dev Cell. 2013;25:599–609. doi: 10.1016/j.devcel.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auer F, Vagionitis S, Czopka T. Evidence for Myelin Sheath Remodeling in the CNS Revealed by In Vivo Imaging. Curr Biol. 2018;28:549–559. doi: 10.1016/j.cub.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Harboe M, Torvund-Jensen J, Kjaer-Sorensen K, Laursen LS. Ephrin-A1-EphA4 signaling negatively regulates myelination in the central nervous system. Glia. 2018;66:934–950. doi: 10.1002/glia.23293. [DOI] [PubMed] [Google Scholar]
- 12.Wolf RM, Wilkes JJ, Chao MV, Resh MD. Tyrosine Phosphorylation of p190 RhoGAP by Fyn Regulates Oligodendrocyte Differentiation. J Neurobiol. 2001;49:62–78. doi: 10.1002/neu.1066. [DOI] [PubMed] [Google Scholar]
- 13.Albert M, Antel J, Brück W, Stadelmann C. Extensive Cortical Remyelination in Patients with Chronic Multiple Sclerosis. Brain Pathol. 2007;17:129–138. doi: 10.1111/j.1750-3639.2006.00043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snaidero N, et al. Myelin membrane wrapping of CNS axons by PI(3,4,5)P3-dependent polarized growth at the inner tongue. Cell. 2014;156:277–290. doi: 10.1016/j.cell.2013.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes EG, Orthmann-Murphy JL, Langseth AJ, Bergles DE. Myelin remodeling through experience-dependent oligodendrogenesis in the adult somatosensory cortex. Nat Neurosci. 2018;21:696–708. doi: 10.1038/s41593-018-0121-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill RA, Patel KD, Goncalves CM, Grutzendler J, Nishiyama A. Modulation of oligodendrocyte generation during a critical temporal window after NG2 cell division. Nat Neurosci. 2014;17:1518–1529. doi: 10.1038/nn.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orthmann-Murphy J, et al. Remyelination alters the pattern of myelin in the cerebral cortex. Elife. 2020;9:1–32. doi: 10.7554/eLife.56621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jäkel S, et al. Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature. 2019;566:543–547. doi: 10.1038/s41586-019-0903-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirby BB, et al. In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. 2006;9:1506–1511. doi: 10.1038/nn1803. [DOI] [PubMed] [Google Scholar]
- 20.Hughes EG, Kang SH, Fukaya M, Bergles DE. Oligodendrocyte progenitors balance growth with self-repulsion to achieve homeostasis in the adult brain. Nat Neurosci. 2013;16:668–679. doi: 10.1038/nn.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Almeida RG, Czopka T, ffrench-Constant C, Lyons DA. Individual axons regulate the myelinating potential of single oligodendrocytes in vivo. Development. 2011;138:4443–4450. doi: 10.1242/dev.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. Phagocytes, Granulocytes, and Myelopoiesis mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood. 2011;27:e49–e56. doi: 10.1182/blood-2010-10-314120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bin JM, Lyons DA. Imaging Myelination In Vivo Using Transparent Animal Models. Brain Plast. 2016;2:3–29. doi: 10.3233/BPL-160029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baraban M, Koudelka S, Lyons DA. Ca 2+ activity signatures of myelin sheath formation and growth in vivo. Nat Neurosci. 2018;21:19–25. doi: 10.1038/s41593-017-0040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamilton nicolaB, Kolodziejczyk K, Kougioumtzidou E, Attwell D. Proton-gated Ca 2+-permeable TRP channels damage myelin in conditions mimicking ischaemia. Nature. 2016;529:523–527. doi: 10.1038/nature16519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paez PM, Lyons DA. Annual Review of Neuroscience Calcium Signaling in the Oligodendrocyte Lineage: Regulators and Consequences. Annu Rev Neurosci. 2020;43:163–186. doi: 10.1146/annurev-neuro-100719-093305. (2020) [DOI] [PubMed] [Google Scholar]
- 27.Chen S, Chiu CN, McArthur KL, Fetcho JR, Prober DA. TRP channel mediated neuronal activation and ablation in freely behaving zebrafish. Nat Methods. 2016;13:147–150. doi: 10.1038/nmeth.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwan KM, et al. The Tol2kit: A Multisite Gateway-Based Construction Kit for Tol2 Transposon Transgenesis Constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 29.Karttunen MJ, et al. Regeneration of myelin sheaths of normal length and thickness in the zebrafish CNS correlates with growth of axons in caliber. PLoS One. 2017;12:1–27. doi: 10.1371/journal.pone.0178058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Early JJ, et al. An automated high-resolution in vivo screen in zebrafish to identify chemical regulators of myelination. Elife. 2018;7:1–31. doi: 10.7554/eLife.35136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicaise AM, et al. Cellular senescence in progenitor cells contributes to diminished remyelination potential in progressive multiple sclerosis. Proc Natl Acad Sci. 2019;116:9030–9039. doi: 10.1073/pnas.1818348116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lloyd AF, et al. Central nervous system regeneration is driven by microglia necroptosis and repopulation. Nat Neurosci. 2019;22:1046–1052. doi: 10.1038/s41593-019-0418-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
No code was used in this manuscript.