Abstract
The antitumor action of immune checkpoint blockade (ICB) is primarily mediated by CD8+ T cells. How sensitivity to ICB varies across CD8+ T cell subsets and clonotypes and the relationship of these with clinical outcome is unclear. To explore this, we used single-cell V(D)J and RNA-sequencing to track gene expression changes elicited by ICB across individual peripheral CD8+ T cell clones, identify baseline markers of CD8+ T cell clonal sensitivity, and chart how CD8+ T cell transcriptional changes vary according to phenotypic subset and clonal size. We identified seven subsets of CD8+ T cells with divergent reactivity to ICB and found that the cytotoxic effector subset showed the greatest number of differentially expressed genes while remaining stable in clonal size after ICB. At the level of CD8+ T cell clonotypes, we found a relationship between transcriptional changes and clone size, with large clones showing a greater number of differentially regulated genes enriched for pathways including T cell receptor (TCR) signaling. Cytotoxic CD8+ effector clones were more likely to persist following ICB and were more likely to correspond with public tumor-infiltrating lymphocyte clonotypes. Last, we demonstrated that individuals whose CD8+ T cell pretreatment showed low cytotoxicity and had fewer expanded clones typically had worse outcomes after ICB treatment. This work further advances understanding of the molecular determinants of ICB response, assisting in the search for peripheral prognostic biomarkers and highlighting the importance of the baseline CD8+ immune landscape in determining ICB response in metastatic melanoma.
Introduction
Antibodies binding the checkpoint proteins CTLA-4 (cytotoxic T lymphocyte–associated protein-4) and PD-1 (programmed cell death protein-1), commonly referred to as immune checkpoint blockade (ICB), have revolutionized the treatment of metastatic melanoma (MM) and numerous other cancers (1, 2). There is, however, marked heterogeneity in ICB-induced clinical benefit (2) and insights into determinants of ICB activity and variation in clinical response remain limited (3–5). Similarly, the association of increased numbers of tumor-infiltrating lymphocytes (TILs) with favorable clinical outcome to ICB is long-established, although factors determining degree of T cell infiltration before and during treatment remain unresolved (6, 7). Crucially, it is increasingly apparent that general aspects of the peripheral immune landscape may play a critical, yet comparatively underinvestigated, role in determining tumor recognition and control (1, 2, 8).
CD8+ T cells are cardinal to the immune response toward MM (8, 9), with most research focusing on their presence within TILs (10, 11). However, recent analysis of non-melanoma skin cancer indicates the T cell response to ICB derives from a distinct repertoire of T cell clones, denoted by shared carriage of specific T cell receptors (TCRs), that migrate to the tumor upon treatment (12). In patients with MM, T cell clones are shared across tumor and blood compartments (11, 13–17), whereas the number of expanded clones (occupying >0.5% of the repertoire) in the periphery after the first cycle of ICB treatment associates with long-term clinical outcome (18). Thus, there is robust evidence that the peripheral clonal T cell response to ICB is clinically informative for patients with cancer and may allow for on-treatment prognostication. Most analyses are agnostic to CD8+ T cell phenotypes, and therefore the determinants of ICB activity across different CD8+ subsets, and at the level of individual clones, are unresolved. The CD8+ T cell ICB response has multiple components, including mitotic cell division (19) and increased cytotoxicity, as exemplified by interferon-γ (IFNγ) induction (20), with the relationship between these being undefined. The magnitude of the mitotic response is not strongly linked to clinical outcome (18). How these responses vary according to T cell phenotype (21), their heterogeneity across clones, and whether they are derived from de novo or preexisting clones are unknown.
To explore this, we used single-cell V(D)J and RNA sequencing (scRNA-seq) to assess transcriptomic responses of individual peripheral CD8+ T cell clones to ICB. In doing so, we described markers of clonal sensitivity to ICB and showed how phenotypic subset and clone size of CD8+ T cells similarly influence ICB response. We showed that large effector T cell clones were most sensitive to ICB and preferentially up-regulated genes involved in TCR signaling. We applied this observation to bulk CD8+ T cell RNA-seq data from a clinical dataset covering 131 pretreatment samples from patients with MM. Individuals with both low cytotoxicity and fewer expanded CD8+ T cell clones had less benefit from ICB treatment. Together, these data suggest that the peripheral CD8+ T cell repertoire pretreatment is a crucial determinant of the prognosis of patients with melanoma treated with ICB.
Results
Single-cell profiling of peripheral CD8+ T cells from patients with MM
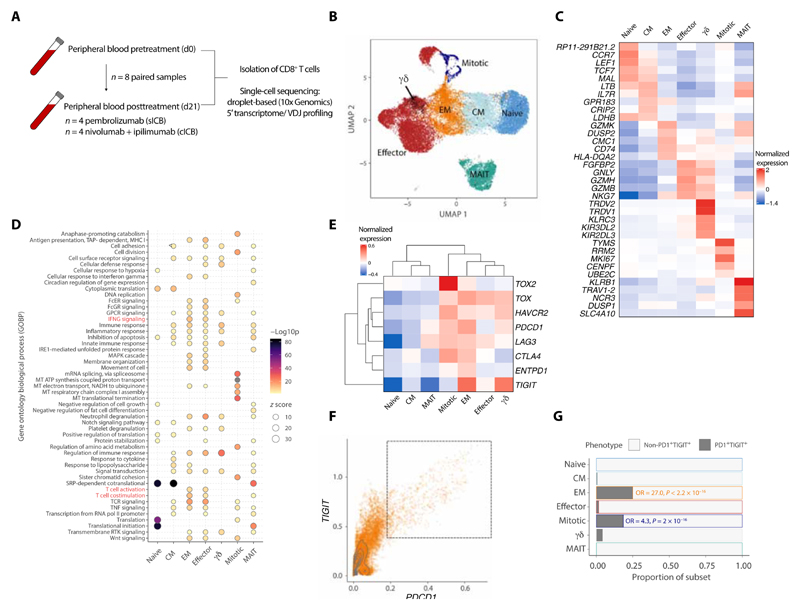
We generated paired baseline (day 0 = d0) and on-treatment (day 21 = d21) 5’ scRNA-seq and V(D)J profiles across peripheral CD8+ T cells from eight patients with MM undergoing treatment with ip-ilimumab plus nivolumab (cICB, n = 4) or pembrolizumab (sICB, n = 4) (Fig. 1A; table S2; and the “Study design” section). In total, 22,445 CD3/CD8 expressing cells passed quality control (QC; the “Data processing, QC, and clustering” section), of which 17,909 cells had TCR sequences. To identify distinct CD8+ T cell populations, we performed unsupervised clustering, observing globally similar uniform manifold approximation and project (UMAP) projection of cells from both time points (fig. S1A) and subsequently cross-referenced each cluster to identities from published CD8+ T cell scRNA-seq datasets (fig. S1, B to E), grouping cells aligned to each identity together. Seven distinct groups of CD8+ T cells were identified (Fig. 1, B and C): naïve, central memory (CM), effector memory (EM), and effector cells (ECs), which collectively represent most CD8+ T cells, as well as highly cycling cells (mitotic), γδ T cells (γδ), and KLRB1 expressing mucosal-associated invariant T cells, which formed a separate cluster from the rest of the cells. Mitotic cells had higher numbers of unique molecular identifiers (UMIs) and MKI67 expression per cell and a bias toward S and G2M cell cycle phases (fig. S1, F and G), consistent with previous descriptions (13, 22). Expression of TRDC was used to identify the γδ subset (23), which was enriched for KLRC3, KIR3DL2, and KIR3DL3 (fig. 1C). To distinguish the biological features of each subset, gene ontology biological process (GOBP) enrichment analysis was performed (Fig. 1D). This highlighted key immune pathways, including IFNG signaling and T cell activation/costimulation, which were enriched in EM and ECs, consistent with higher cytotoxicity (fig. S1H). In contrast, naïve/CM and mitotic subsets demonstrated up-regulated translational and mitochondrial machinery. We further examined the expression of T cell exhaustion markers across the cell subsets (Fig. 1E). Increased expression of TOX and HAVCR2 and most other exhaustion-related genes was found in mitotic, ECs, EM, and γδ cells compared with the remaining subsets. PDCD1 expression was the highest in EM and mitotic cells, indicating greater exhaustion within these subsets. The majority of PDCD1 + cells coexpressed TIGIT (Fig. 1F), and EM and mitotic cells, but not ECs, were enriched within this PDCD1+ TIGIT+ population (Fisher’s exact test; Fig. 1G). In summary, we identify seven distinct clusters of CD8+ T cells in the peripheral blood of patients with MM, which closely correspond with known cellular phenotypes.
Fig. 1. Identification of peripheral CD8+ T cell subsets in MM before and during ICB.
(A) Workflow for sample collection and processing for scRNA-seq. (B) UMAP clustering of n = 22,445 peripheral CD8+ T cells from n = 8 individuals sampled at pretreatment and after 21 days of ICB, across seven distinct subsets as represented by each color. (C) Expression heatmap showing distinct gene expression profiles for the identified subsets. Genes were selected as the five most significant DE genes present at both pretreatment and at d21 post-ICB for each time point. (D) Dot plot heatmap showing GOBP terms significantly enriched across each subset (adjusted P value < 0.05); selected pathways highlighted in red. MHC I, major histocompatibility complex class I; GPCR, G protein-coupled receptor. (E) Expression heatmap of selected T cell activation and exhaustion markers. (F) Dot plot of PDCD1 and TIGIT expression across CD8+ T cells, with the indicated PD1+ TIGIT+ peripheral exhausted population. (G) Subset proportions of nonexhausted and exhausted cells, showing an enrichment of EM and mitotic cells by Fisher’s exact test (odds ratio > 1).
Changes in CD8+ T cell subset composition during early ICB
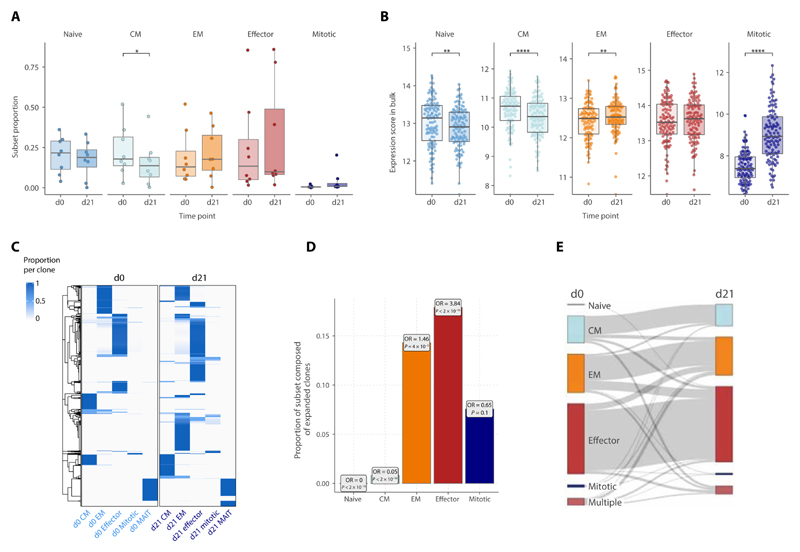
Focusing on the effect of ICB across key CD8+ T cell subsets (naïve, CM, EM, EC, and mitotic), we found that ICB only significantly reduced CM proportions in the single-cell data (Fig. 2A). To increase the power to detect generalized effects, we applied a composite expression signature for each single-cell subset across MM patient CD8+ T cell bulk RNA-seq data (n = 212 paired d0/d21 samples from 106 individuals) (18). We found significant correlation between score-inferred and actual single-cell proportions across all subsets, validating this approach (fig. S2A). Across the cohort, naïve, CM, and mitotic scores negatively correlated with age, whereas EM and EC scores positively correlated consistent with known aging effects (fig. S2B) (24). Changes to subset expression scores reflected the trends observed in the subset proportions from single-cell data, with a reduction in naïve and CM expression scores, increase in EM and mitotic scores, and no significant change in the EC score after ICB (Fig. 2, A and B). The increase in mitotic score after ICB remained sustained at d63+ of treatment (fig. S2C). Last, the decrease in naïve and CM score was positively correlated within individuals and inversely correlated with mitotic score increase (fig. S2D). Changes in EC score correlated with EM and, to a lesser extent, mitotic score, indicating shuttling from these subsets into the mitotic pool, potentially allowing for EC compartment replenishment after ICB.
Fig. 2. Dynamic immune composition changes upon ICB.
(A and B) Changes in the proportion of each cell subset between pretreatment (d0) and d21 post-ICB (d21) as determined by (A) scRNA-seq (n = 8, two-sided Wilcoxon signed-rank test) or (B) inferred from bulk RNA-seq data (n = 110, two-sided Wilcoxon signed-rank test). For the bulk RNA-seq inference, subset scores were calculated for each bulk RNA-seq sample based on a list of markers that distinguished each subset at both pretreatment and d21 post-ICB in the scRNA-seq data (the “Bulk RNA-seq” section; fig. 2, A and B). (C) TCR sharing heatmap between d0 and d21 CD8+ T cell subsets, showing T cell clones consisting of more than 1 detected clone and their relative presence and subset proportions at each time point. (D) Proportion within each subset at d21 of clones that have significantly expanded from d0 (boxes contain odds ratios of enrichment of clones expanded at d21 within each subset, P values by Fisher’s exact test). (E) Sankey plot tracking the cell subset phenotype of d0 clones at d21. For each clone, the subset in which most of its members were present in at a given time point was designated as the predominant subset for that clone; “multiple” indicates clones equally distributed across multiple subsets. The width of the connecting flowline represents the number of clones with the respective d0 and d21 subset phenotypes.
We next examined how TCR clones composed of more than one cell were distributed across subsets at d0 and d21 (Fig. 2C). Most of these clones were of an EC phenotype, being detected in this subset at both time points, and although there was an emergence of novel EM and EC clones at d21, few clones were shared across subsets at either time point (14, 22). To further characterize the mitotic cells, we tracked their clonal dynamics between d0 and d21. Most mitotic cells represented small clones, with the median clone size being similar to naïve and CM cells (fig. S2E) and not changing with treatment. Expanded clones were not enriched in the mitotic subset and were relatively depleted in naïve and CM subsets after treatment (Fig. 2D and table S5). In general, while there was limited TCR sharing between memory cells and ECs across time points, most resampled clones remained in the same subset over time (Fig. 2E), the exception being the mitotic subset where only 2.2% of d21 mitotic clones were present in any subset at d0. Together, the observed changes in subset proportion can be summarized by (i) a fall in the proportion of CM and, to a lesser extent, naïve subsets with a reciprocal and modest increase in EM cells (Fig. 2B), through both emergence and expansion of preexisting clones (Fig. 2, C and D); (ii) an increase in the mitotic subset, most of which were not sampled at both time points (Fig. 2, B and C); and (iii) stability in the EC compartment size (Fig. 2B), with an increase in the proportion of ECs composed of expanded clones (Fig. 2D).
Effector CD8+ T cells showed increased sensitivity to ICB
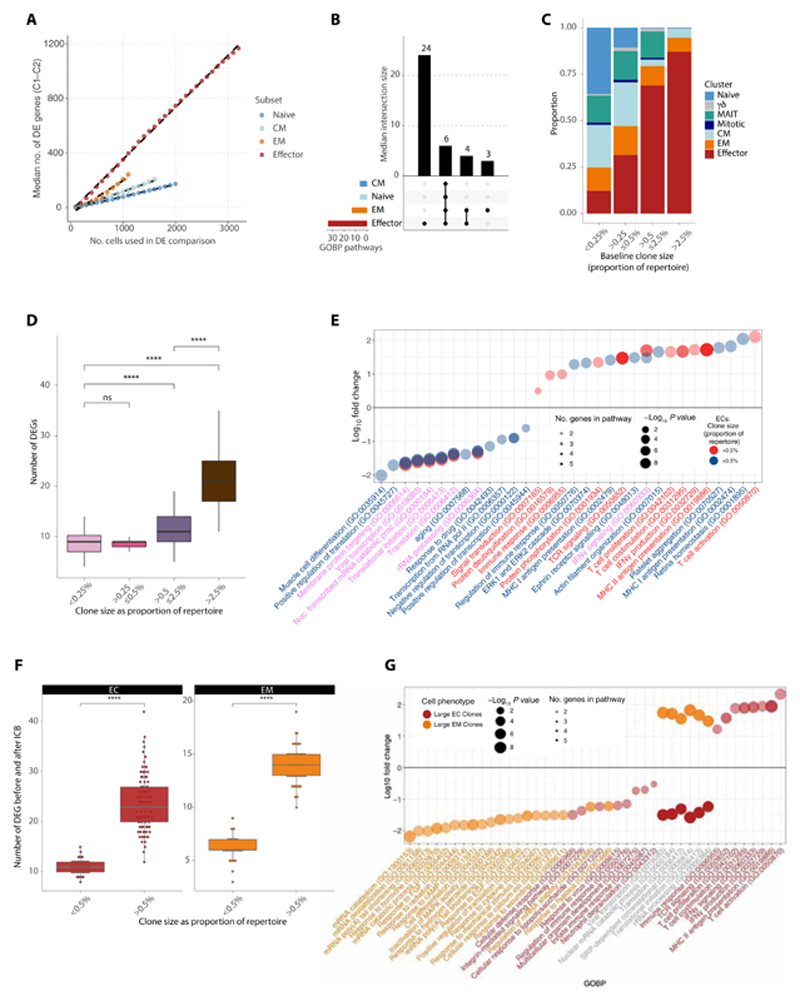
We further characterized responses to ICB between d0 and d21 at a CD8+ T cell subpopulation level, focusing on naïve, CM, EM, and ECs. We controlled for increased power to detect differentially expressed (DE) genes (comparing pre- with posttreatment) in larger subsets by subsampling varying numbers of cells across each subset (analysis bootstrapped 100 times) and used multiple DE analysis pipelines (the “Downstream expression analysis” section). Across the four CD8+ T cell subsets, ECs had more DE genes with treatment irrespective of subsample size (Fig. 3A). When we used a linear mixed-effects model to control for interindividual variation, we made similar observations (fig. S3A). To quantify these patterns of DE, we used a cutoff of n = 1100 cells per subsample and calculated the frequency of genes being detected as significant (adjusted P value below 0.05) across 100 bootstraps. Genes such as IL10RA and GZMA were consistently down- and up-regulated by treatment in all bootstraps in ECs but not in other subsets (fig. S3B and table S7). Comparing the DE genes in ECs versus other subsets, we observed that many were exclusively modulated in this subset (fig. S3C), suggesting that ICB has a distinct effect on these cells. Subset-wise GOBP analysis of the induced and suppressed genes demonstrated that the most modulated pathways were either conserved pathways represented across all CD8+ T cell subsets or specific to ECs (Fig. 3b). Specifically, pathways restricted to ECs encompassed pro-inflammatory and immune pathways including nuclear factor κB, mitogen-activated protein kinase, and tumor necrosis factor (TNF) signaling (fig. S3D), further supporting the profound immune-stimulatory effect of ICB on this subset. Together, these findings demonstrated that the EC subset has the greatest magnitude of DE genes after ICB and that the pathways modulated are unique to this subset.
Fig. 3. Subset-specific and clone size-dependent transcriptome changes upon ICB.
(A) Median number of DE genes across 21 days of ICB in conventional T cell subsets. Each subset was subsampled to the indicated number of cells at d0 and d21 and tested for DE genes across 100 bootstraps. At each n value for number of cells, there was a significant difference across the subsets in the number of DE genes (P < 0.0001, Wilcoxon rank sum test). (B) Upset overlap plot of GOBP up-regulated in each subset after treatment (subsample of n = 1100 cells), based on the DE genes per bootstrap in (A). The median number of pathways in each intersecting set was calculated over the 100 bootstraps. (C) Clonal size bins at d0 filled by proportion of each phenotypic cluster. (D) Number of DE genes before and after treatment per clonal size bin. Each clonal size bin was subsampled to a constant size and the analysis bootstrapped 100x. (E) GOBP analysis demonstrating pathways up- and down-regulated in large versus small ECs before and after treatment. Large ECs are colored in bright red and small ECs in blue. Pathways where the text is in purple represent those that are shared between both groups. (F) Number of DE genes before and after treatment for large versus small ECs (red) and EMs (orange). Each cluster was subsampled to a constant size and the analysis bootstrapped 100x. (G) GOBP analysis demonstrating pathways up- and down-regulated in large ECs (red) versus large EMs (orange) before and after treatment. Pathways where the text is in gray represent those that are shared between both groups. For all analysis comparing numbers of DE genes, P values were determined by two-sided Wilcoxon rank sum test. ns, not significant.
CD8+ T cell clones differentially modulated gene expression after ICB as a function of their size
Given our findings linking the number of large clones with clinical outcome, we sought to understand the relationship between the size of a clone (the proportionate number of copies of that TCR clone compared with the entire repertoire in an individual patient) and its response to ICB. We grouped clones into four categories according to their size as a proportion of the total repertoire (Fig. 3C). As expected, most naïve cells were of the smallest sizes, whereas the larger clone categories were composed of ECs and, to a lesser extent, EMs (Fig. 3C). We next assessed the relationship between clonal size and ICB effect by performing category-wise DE analysis across treatment (including only clones present at both time points, agnostic to subset type), using sampling to control for variation in cell number across subpopulation and time point, bootstrapping analysis 100 times. This produced a response pattern of DE genes according to clone size, with small- and medium-sized clones having comparatively fewer DE genes than large and very large clones (Fig. 3D).
Given that this analysis is potentially confounded by differing subset proportions in each size category, we repeated this process restricted to EC and EM subsets, with clonal size simply dichotomized with a cutoff of 0.5% of the repertoire to denote large from small. The previously observed pattern was preserved within each subset, with larger clones again showing greater numbers of DE genes after ICB compared with smaller clones (Fig. 3E). These findings were replicated when analysis was performed sequentially, each time removing a different patient, suggesting that the findings were not reflecting skew from an outlier individual (fig. S3E).
To understand these effects at a functional level, we analyzed GOBP pathways corresponding to the DE gene sets within the EC and EM subsets (Fig. 3, F and G, and table S8). We again noted that large EC clones had a notable divergent gene expression profile after ICB compared with both small EC clones (Fig. 3F) and large EM clones (Fig. 3G), with pathways including T cell activation, proliferation and costimulation, TCR signaling, and IFNγ production uniquely up-regulated in large ECs. Together, clonal size influences both the magnitude of response to ICB and the types of genes regulated, with this phenomenon observed even within the same phenotypic subset. Larger clones display marked enhancement of immune function after ICB treatment, with large ECs uniquely up-regulating immune pathways critical to the ICB response.
Cytotoxic clones demonstrated a propensity to persist after ICB treatment
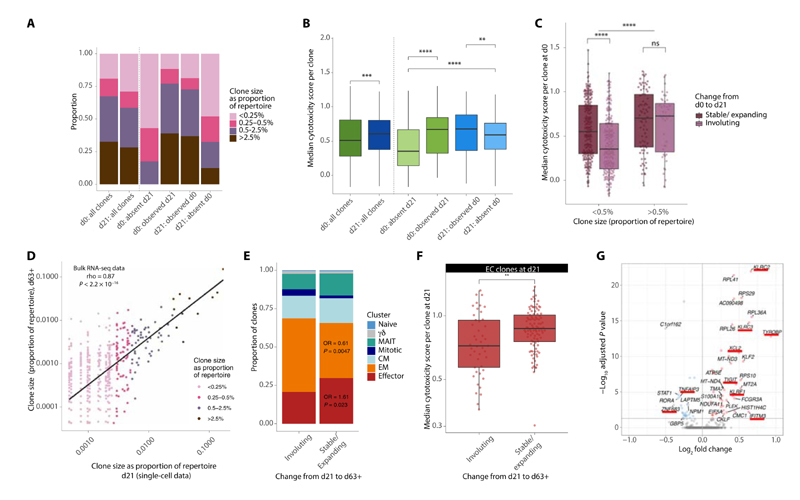
To further understand the temporal dynamics of clones, we tracked the presence or absence of each clone across d0 and d21, restricting the analysis to clones composed of two cells or more. Perhaps expectedly, large clones were more likely to be resampled, with clones only sampled at one time point predominantly grouped within smaller size categories (Fig. 4A). Genes correlated with IFNγ are informative as to cytotoxicity. We therefore adopted previous methods (13) to develop a cytotoxicity score for each cell based on key IFNγ-correlated genes. We found that although cytotoxicity was globally increased at d21, clones sampled at both time points or only at d21 had increased cytotoxicity over those present only at d0 (Fig. 4B). This finding was consistent across both small and large clones (defined by a 0.5% of repertoire size cutoff) (fig. S4A). To explore these observations further, we examined the d0 profile of clones, denoting those that significantly shrunk (as per a Fisher’s exact test, see the “Clonal definitions, size, and emergence versus involution” section) or were absent at d21 as involuting clones (shrinkage of the posttreatment cell population to the pre-expansion and pretreatment level). We compared these “involuting” clones with those clones that were found to expand or show no significant change in size from d0 to d21 (which we term “stable/expanding”). Small clones that were stable/expanding after ICB were significantly more cytotoxic at baseline than those involuting clones (Fig. 4C). This finding was also observed if clone size was determined by simple fold change (fig. S4B). This difference in baseline cytotoxicity across involuting and stable/expanding clones was not seen within larger clones, which were significantly more cytotoxic than smaller clones (Fig. 4C). Hence, these findings suggest that after ICB, there is an overall increase in cytotoxicity that is driven by a combination of the persistence of pretreatment cytotoxic T cell clones, involution of clones with low cytotoxicity, and the emergence of more cytotoxic clones.
Fig. 4. Cytotoxic clones can persist post-ICB treatment.
(A and B) Proportion of clonal size groupings (A) and median cytotoxicity per clone (B) for clones found at both d0 and d21 versus at just at one time point, compared with all clones at d0 and d21. (C) Median cytotoxicity per clone for those present at d0 that subsequently expand/remain stable (dark purple) or involute (light purple) by d21. Clones are separated based on size using a cutoff of 0.5% of the repertoire to denote large from small. The horizontal significance line demonstrates the significant difference between all large compared with all small clones. (D) Clone size at d21 compared with size at d63+; each point denotes a clone and is colored on the basis of size grouping at d21 (Spearman’s correlation). (E) Proportion of clones that are stable/expanding or involuting from d21 to d63+ colored by their phenotypic cluster; OR refers to the odds ratio for the proportion of ECs and EMs in the stable/expanding compared with the involuting group (Fisher’s exact test). (F) Median cytotoxicity score per clone across EC clones found at d21 that are stable/expanding or involuting by d63+. (G) Volcano plot of genes DE by ECs at d21 that are stable/expanding by d63+ compared with those that are involuting; genes of interest are denoted with an underscore. The direction is in favor of clones that are stable/expanding, i.e., those in red with a positive FC are up-regulated in clones that are stable/expanding. For all analyses, only clones with >1 cells were included, with Wilcoxon rank sum tests used for all comparisons unless otherwise stated. For (A) to (C), n = 8 individuals, and for (D) to (G), n = 6 individuals.
To extend this analysis across later time points, we examined bulk RNA-seq data from CD8+ T cells isolated 63 to 106 days after treatment (d63+), available for six of the eight individuals for which scRNA-seq data were available (the “Clonal definitions, size, and emergence versus involution” section). From the corresponding TCRs identified in these samples, we found 992 overlapping unique TCRβs associated with 4245 cells at d21 within the scRNA-seq data. Clone size at d21 and d63+ were highly correlated (Fig. 4D), indicating the relative preservation of clonal structure over this longer period. Comparison of d21 clones that were stable/expanding versus those that were involuting between d21 and d63+ highlighted an enrichment for ECs in the stable/expanding group, whereas there was underrepresentation of EMs (Fig. 4E). We subsequently constrained further analysis to the EC subset, analyzing d21 cytotoxicity of stable/expanding clones versus those involuting by d63+. Again, we found a significantly higher cytotoxicity in those stable/expanding clones (Fig. 4F), which was able to alternatively define changes to clone size based on fold change rather than by a Fisher’s test (fig. S4C). This trend persisted when clones were grouped on the basis of their size (fig. S4D) although was not statistically significant for the large clones. Last, by comparing expression of EC clones stable/expanding between d21 to d63+ with those that involuted, we identify genes linked to clonal stability. We found killer lectin receptor genes, including KLRF1 (killer cell lectin–like receptor F1), KLRC2/3, and the marker TIGIT, were significantly higher in stable/expanding ECs at d21, whereas STAT1 was significantly down-regulated (Fig. 4G and table S9), suggesting that these may form markers of clonal persistence. Collectively, these results indicate that preexisting cytotoxicity belies increased clonal persistence, with increased d0 to d21 persistence of clones that are more cytotoxic at baseline, whereas emerging clones, although small, also have higher levels of cytotoxicity. Similarly, within the EC compartment, d21 cytotoxic clones have increased likelihood to be found at d63+ of treatment.
Baseline and early on-treatment cytotoxicity was prognostic for clinical outcome during ICB
Given that baseline cytotoxicity delineates cells likely to persist or expand during ICB, potentially reflecting response to antigen, we explored the relationship between both pre- and on-treatment CD8+ T cell cytotoxicity and clinical response to ICB. We generated cytotoxicity scores for each bulk sequencing sample in our cohort (n = 131 at d0 and n = 109 at d21; the “Bulk RNA-seq” section), which we separately validated with flow cytometry across a subset of samples, finding a positive correlation of the cytotoxicity score to the expression of cytotoxic markers such as perforin-1 (fig. S5, A and B). Although cytotoxicity at d0 was positively correlated with d21 cytotoxicity across the cohort, indicative of the importance of the baseline response (fig. S5C), ICB tended to increase cytotoxicity, with a larger effect from cICB (fig. S5D).
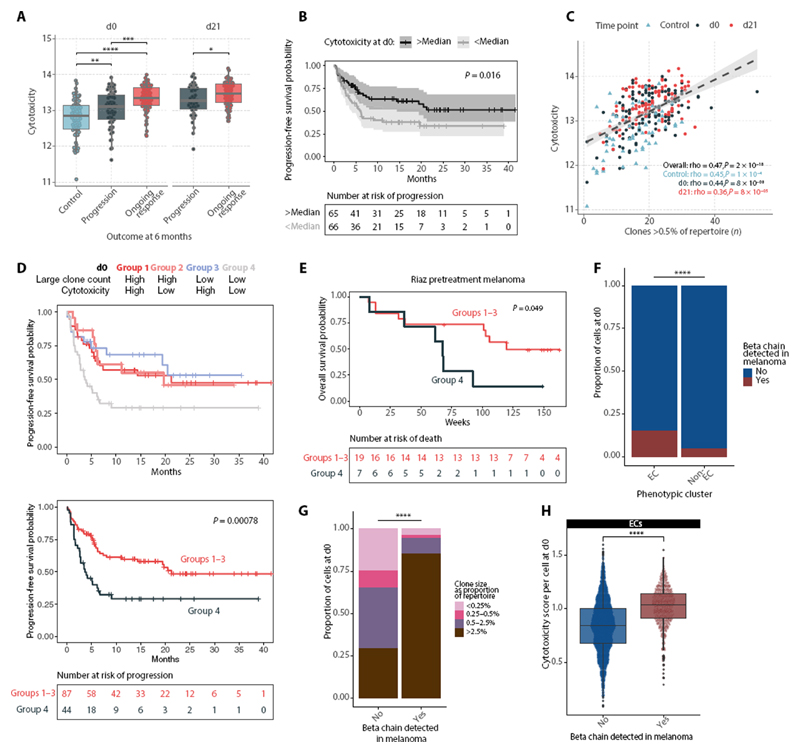
Patients had significantly higher CD8+ cytotoxicity pretreatment compared with healthy controls (Fig. 5A), reflecting the systemic immune effects of MM. In patients who continued to respond to treatment, both d0 and d21 cytotoxicity was higher than in those that had progressive disease 6 months after treatment. Using Kaplan-Meier estimates (log-rank test), we found a cytotoxicity score above the cohort median at either baseline or d21 positively correlated with increased progression-free survival (PFS) (Fig. 5B and fig. S5E), supporting the finding that baseline immune profile is important in determining clinical outcome and demonstrating that agnostic sampling of the peripheral CD8+ subset provides prognostic information. In contrast, we found no association between the mitotic score and PFS at either d0 or d21 (P = 0.78 and 0.69, respectively, log-rank test), suggesting that the degree of baseline CD8+ T cell division, or that induced by ICB, does not relate to clinical benefit.
Fig. 5. CD8+T cell cytotoxicity is associated with clinical response to ICB.
(A) Cytotoxicity scores per individual at d0 and d21, calculated in CD8+ bulk RNA-seq samples from healthy volunteers (control) and those who have do and do not have a response to treatment (“ongoing response” and “progression,” respectively; n = 69 controls, 112 patients). (B) Kaplan-Meier curves showing PFS in patients with baseline cytotoxicity above and below median (n = 131 patients) (two-sided log-rank test). (C) Correlation between large clone count and cytotoxicity score in the bulk cohort, across control individuals and patients at d0 and d21 (n = 69 controls, 137 patients, Spearman’s rank test). (D) PFS in patients separated by a combination of d0 large clone count and cytotoxicity (above/below median) (n = 131, two-sided log-rank test; group 1 versus group 4, P = 0.019; group 2 versus group 4, P = 0.023; group 3 versus group 4, P = 0.010). (E) Analysis of overall survival in patients with melanoma from Riaz et al. (25) based on intratumoral RNA-seq and TCRB-seq data. Patients were separated by a combination of pretreatment large clone count (TCRB-seq) and cytotoxicity (RNA-seq) [above/below median, as in (D)] (n = 26, two-sided log-rank test). (F) Proportion of ECs compared with non-ECs at d0 that carry a tumor-associated (TA)-TCRB chain, as identified by TCRseq of resected melanomas by Pruessman et al. (26) (Fisher’s exact test). (G) ECs at d0 that are either carrying a TA-TCRB or not, filled by clonal size; Fisher’s exact test for the proportion of large clones (>0.5%) compared with small clones carrying/not carrying a TA-TCRB. (H) Cytotoxicity scores of ECs at d0 categorized by whether they carry a TA-TCRB chain or not (Wilcoxon rank sum test).
As the number of large clones at d21 is associated with clinical response to ICB (18), we explored whether CD8+ cytotoxicity could be integrated with large clone count to stratify patients according to outcome before treatment. First, in this expanded cohort, we recapitulated the finding that d21 but not d0 large clone count positively associated with PFS, with increased significance in the cohort with a greater number of samples (fig. S5F). As expected, the CD8+ cytotoxicity score positively correlated with large clone count across healthy individuals and d0, and d21 samples from patients (Fig. 5C), whereas responding individuals had significantly higher cytotoxicity irrespective of large clone count when assessed with a linear model (fig. S5G). We next explored whether cytotoxicity and large clone count could additively predict 6-month patient outcome using a linear discriminant analysis (LDA) model. This yielded an area under the receiver operating characteristic curve of 0.69 (fig. S5H), which was minimally affected by removing cytotoxicity or large clone count in isolation but markedly reduced when both were removed (fig. S5I), indicating a compensatory interaction. To explore this, we categorized individuals into four groups based on whether large clone count and cytotoxicity were above or below median values and examined their PFS (Fig. 5F). We found that individuals with both submedian large clone count and submedian cytotoxicity (group 4) at baseline had significantly lower PFS compared with all other groups (Fig. 5D), which otherwise had comparable survival; this effect was also observed at d21 (fig. S5J). Hence, we further stratified a subset of patients with low pretreatment peripheral CD8+ T cell activity, with limited clonal expansion and low cytotoxicity, that receive minimal clinical benefit from treatment with ICB.
Validation using external datasets
To explore the prognostic value of cytotoxicity score in an alternative context, we looked to see whether our observations extended to the tumor environment. Using the Riaz et al. (25) dataset of paired RNA-seq and TCR beta chain sequencing data from melanoma samples before and during treatment with anti–PD-1, we devised an intratumoral CD8+ T cell cytotoxicity score by selecting genes used in our peripheral cytotoxicity analyses that showed the most significant positive correlation with CD3D, CD3E, CD8A, CD8B, IFNG, and PRF1 expression (fig. S6A) and subsequently identified the number of TCRB clones per sample with proportion above 0.5%, corresponding to the threshold for large clones. Despite the low number of individuals in this cohort, we found significantly lower overall survival for patients with below median large clone count and cytotoxicity (Fig. 5E) at baseline.
Given this correlation between the peripheral and intratumoral T cell state, we used an alternative dataset of TCRB chains sequenced from melanoma biopsies (26). From the 199 samples, 85,528 unique TCRB were identified, of which 392 were found in our samples (subsequently referred to as tumor-associated TCRB, “TA-TCRB”). We assessed the characteristics of cells carrying one of these TA-TCRB at both time points and found that these cells contained a significantly increased proportion of ECs compared with other cells subsets (Fig. 5F and fig. S6B). Within these TA-TCRB ECs, there was additionally a significantly higher proportion of large clones (Fig. 5G and fig. S6D) with higher cytotoxicity score at both time points (Fig. 5H and fig. S6C). Extending these observations to our bulk cohort, we also observed greater clonal expansion of TA-TCRB matching T cell clones in patients compared with healthy controls (fig. S6E). When we compared this with the profile of cells matching public viral-reactive TCRs (27) (n = 149 cells), clonally expanded ECs were underrepresented, and there was no significant difference in cytotoxicity (fig. S6, F to H). Together, these data support our findings that large, peripheral EC clones that appear to be overrepresented within the tumor-infiltrating compartment positively correlate with patient response to ICB.
Discussion
Identifying the cells most responsive to ICB is vital to fully understand the mechanisms of ICB response and resistance. Here, we studied the effect of ICB on peripheral CD8+ T cells at a single-cell level, characterizing responses by subpopulations and by clones. We identified seven phenotypic subsets sharing conserved responses to ICB and displaying subset-specific changes, the greatest and most divergent being within the ECs. The EM subset expands intratumorally and peripherally (15, 28, 29) in response to ICB (15, 28, 29), and we observed increases in the peripheral EM score, inferring commensurate expansion of this subset. We showed though that although ICB does not change EC proportion, the EC compartment exhibits the greatest magnitude of clonal and gene expression changes and preferentially persists after ICB. Hence, we posit that in peripheral blood, the effector T cell compartment is most sensitive to ICB treatment. In keeping with this, recent work shows that clones common to blood and tumor are enriched for effector markers and are not peripherally exhausted in patients who respond to ICB (14).
The finding that large clones responded to ICB in a divergent manner to small clones provides insight into the mechanistic underpinnings of our previous observations linking large peripheral CD8+ T cell clone count with clinical outcome (18). Smaller clones down-regulated expression of immunological pathways and increased expression of ribosomal genes and translational activity after ICB. However, larger clones had the opposite response, consistent with the observation that translational suppression occurs in terminal effector T cells (30). It is unclear to what degree clonal size is a causal factor in the differential response to ICB or correlates with other factors. However, that large EC clones showed relative baseline up-regulation of TCR signaling, likely reflective of successful TCR ligation and subsequent expansion, suggests that the increased sensitivity to ICB reflects intrinsic activity of these clones.
The observation that ICB was associated with survival and expansion of CD8+ T cell clones displaying markers of cytotoxicity before treatment was supported by the inclusion of on-treatment bulk-sequencing data collated at later cycles of treatment. This highlighted that cytotoxic clones persisted across multiple cycles of ICB, consistent with previous descriptions of a relationship between cytotoxicity and response to PD-1 blockade in the context of nonsmall cell lung cancer (NSCLC) (31). Although it is difficult to definitively prove that ICB specifically elicits clonal survival, we found that ICB leads to increased clonal cytotoxicity. Given that persistent clones and expanding clones demonstrated higher cytotoxicity pretreatment or at d21, these data support this to be an action of ICB peripherally. The association of findings in pretreatment samples with outcome indicates the importance of preexisting tumor recognition by T cells and consequential effector T cell responses (32), with the absence of this linked to a poor clinical response (32). Thus, therapies targeted at overcoming further immune checkpoints may have relatively diminished clinical returns compared with those improving antigen presentation and priming.
Consistent with other studies, we found that ICB provided a marked stimulus to mitotic cell division in CD8+ T cells (8, 19, 33). The relevance of this response to outcome is less clear however. The mitotic subset showed increased expression of exhaustion markers, and most mitotic clones did not appear to become established at d21 or at later time points; only a small proportion of surveyed mitotic CD8+ T cells progressed to EC and EM cells. Although this implies dysfunction, whether these are classically exhausted or are in a state with features of exhaustion is unclear and will form the basis for future research. Conversely, we observed that ECs remained persistently expanded for long durations, and our work suggests that killer-lectin receptors may specifically denote long-lived ECs.
From a clinical outcome perspective, our work further demonstrates that analysis of peripheral CD8+ T cells both before and during treatment provides predictive information, reinforcing observations in other cancers as to the prognostic potential of baseline peripheral blood samples (34). The association of large clone count with response to ICB likely reflects the increased probability an individual will carry tumor reactive clones as this count rises. Nonetheless, a situation where a few specific anticancer clones are present could be beneficial but not register in total large clone count. These cells would be anticipated to be cytotoxic though, and here, we have shown that a high cytotoxicity score may compensate for low large clone count. Comparison of data from healthy individuals indicated that not only do healthy individuals have fewer large circulating clones but they also have diminished cytotoxicity. Last, we found no association between the magnitude of the mitotic response and clinical outcome, highlighting a disconnect between mitotic responses and those that are cytotoxic and therapeutically important.
It is unclear whether these peripheral CD8+ T cell clones are bystanders or whether they are enriched after exposure to melanoma antigens. By integration of our results with TCR sequencing of melanoma-resident T cells from Pruessman and colleagues (26), we demonstrated that public clones found in melanomas are predominantly large, and reactive clones show higher than average cytotoxicity. Conversely, although clones carrying TCR reactive to viral antigens are plentiful, they were not larger or more cytotoxic. These findings are consistent with recent descriptions of tumor-infiltrating clones that tend to have an effector-like phenotype in the peripheral blood in mouse models (17) and patients with MM (16).
A potential limitation of this study is the inability to definitively link the observed changes of gene expression in CD8+ T cells to ICB exposure and exclude the effects of cancer progression or other temporal factors. This is a drawback of this type of observational work in humans as it is difficult (and ethically challenging) to obtain samples from untreated individuals with cancer over time. Nonetheless, the changes we observed in patient profiles coupled to the frequently pronounced transformations in clinical performance argue that these are due to the effect of ICB. This is underlined by bulk RNA-seq data where cICB induced greater changes in cytotoxicity score than sICB, consistent with an effect of ICB on cytotoxicity. Likewise, baseline cytotoxicity was higher in patients than healthy controls and was higher in those that respond to treatment compared with those that do not, suggesting an important interaction between baseline immune state, the effect of ICB on cytotoxicity and response to treatment.
In summary, our work combined in-depth analysis of peripheral CD8+ T cells responses to ICB at the single-cell level, with observations corroborated and extended in a large dataset. The fact that most mitotic cells fail to expand into differentiated large effector clones indicates the limited clinical importance of magnitude of mitotic response to ICB. Instead, we find that enlarged clones within the effector subset are most sensitive to treatment. Clone size correlates with cytotoxicity and having larger and cytotoxic clones before treatment is prognostically favorable, reflecting that much of the effect of ICB relies upon preexistent CD8+ T cell responses. This work implicates failure of immune recognition as the major limiter of response to ICB and mechanisms to enhance baseline immune responses will be vital in improving sensitivity to ICB. Future work to assess the durability of tumor-reactive clones over longer time periods than covered here is important, as well as addressing the interplay with treatment-induced changes in tumor burden. Last, understanding the degree to which other non–tumor-related factors, such as germline genetics, affect the ICB response to cancer will be vital in further improving therapeutic outcomes.
Materials and Methods
Study design
The aim of this study was to understand peripheral CD8+ T cell responses to ICB at a single-cell level, particularly identifying key phenotypic or clonal features of response to therapy. Eight patients who were prescribed ICB for MM at the Churchill Hospital, Oxford University Hospitals NHS Foundation Trust, UK were prospectively and sequentially recruited. Four participants received single agent pembrolizumab (sICB) and four received combination ipilimumab/nivolumab (cICB). Characteristics of the participants are outlined in table S2. All participants provided written informed consent to donate samples to the Oxford Radcliffe Biobank (Oxford Centre for Histopathology Research ethical approval reference 19/SC/0173, project nos. 16/A019, 18/A064, and 19/A114). Thirty to 50 ml of blood was collected into EDTA tubes (BD Vacutainer System) taken immediately before treatment at d0 and d21 after 1 cycle of treatment (Fig. 1A). After single-cell isolation, transcriptome and V(D)J sequencing was undertaken with downstream analysis performed in R, as detailed below.
Sample collection
Peripheral blood mononuclear cells were immediately obtained from whole blood by density centrifugation (Ficoll Paque). CD8+ cell isolation was carried out by positive selection (Miltenyi Biotec) according to the manufacturer’s instructions, with all steps performed either at 4°C or on ice. Unless otherwise stated, patients receiving cICB (ipilimumab plus nivolumab, n = 4) or sICB (pembrolizumab, n = 4) were pooled for the purposes of analysis, based on previous work showing qualitatively similar genetic changes induced in peripheral CD8+ T cells by both cICB and sICB (18).
Single-cell sample preparation and sequencing
Magnetically separated CD8+ cells were immediately oil-partitioned into single-cell droplets, followed by cell lysis and a reverse transcription reaction using the 10x Genomics Chromium system. A total of 6000 CD8+ T cells were loaded onto each partitioned cassette. Single-cell 5’ RNA transcriptome and V(D)J libraries were constructed from the cDNA, according to the manufacturer’s instructions (https://10xgenomics.com/). Because of the sequential and prospective nature of sample collection, droplet generation and reverse transcription steps were performed on individual samples, but library generation and sequencing were performed in two batches of pooled samples (table S2). Sequencing was performed on an Illumina HiSeq4000: 75–base pair (bp) paired-end (PE) reads for the 5’ RNA libraries, 150-bp PE reads for the V(D)J libraries to a depth of about 50,000 reads per cell.
Data processing, QC, and clustering
Single-cell reads were aligned, counted, and filtered for initial QC as previously described (12). In short, FASTQ files were generated from Illumina BCL outputs, used to produce gene expression and barcoding libraries, and then combined using the Cellranger mkfastq, count, and aggr functions respectively. For V(D)J data, Cellranger vdj was applied. For QC, the R package scater (35) was used to identify single-cell outliers as low-quality libraries and scran used to detect doublets that were removed from analysis. After alignment and data preprocessing, CD8+ T cells were filtered on detectable CD3D and either CD8A or CD8B reads while lacking CD14 expression. The R package Seurat (36) was used for further QC, data normalization, and identification of variable features; low-quality cells were excluded if they had less than 500 total UMIs, less than 300 detected features, and mitochondrial gene percentage above 20%. Genes expressed in <5 cells were also removed. The percentage of mitochondrial genes and cell cycle scores were regressed out upon scaling of the normalized data. Principal components analysis was performed on the scaled data, and the top 16 principal components were selected for dimensionality reduction and UMAP clustering as the most variable and biologically informative components. To determine an ideal resolution value, cell clusters generated using values between 0.1 and 2.5 were compared using the R package clustree (37) with values between 1.5 and 2.0, showing high cluster stability. There was no observed effect of batch on PC distribution or baseline cytotoxicity score.
Identification of cell subsets
To assign each cell cluster generated in Seurat to a functional T cell subset, we curated transcripts per million normalized counts for CD8+ T cells from previous T cell cancer scRNA-seq datasets (12, 13, 38–40). Using the R package SingleR (41), each individual cell was matched to one of the CD8+ T cell subsets present in the reference datasets based on global gene expression similarity, from which subset identity for each cluster was determined. Briefly, we noted cluster 12 (ECs) was grouped above the mitotic clusters despite being labeled as EC through SingleR and having high expression of FGFBP2 and other EC-characteristic genes; we detected a high level of expression of ribosomal genes in this subset relative to other ECs, explaining its projection on the UMAP despite being an EC cluster.
Clonal definitions, size, and emergence versus involution
T cell chains were filtered on the basis of called productivity and chain identity (TRA or TRB). Cells were selected as those that either contained a single TCRβ chain, a combination of one TCRα with one TCRβ chain or a group of three chains—two TCRα and TCRβ. Cells not meeting these criteria were excluded. A clone ID was defined by a concatenation of amino acid CDR3 chains present, with cells sharing identical clone IDs classed as members of the same clonotype. Clonal proportions were calculated by dividing the number of cells in each clone per sample by the total of number of cells (meeting the above criteria) per sample. Of 22,446 cells, which had transcriptome sequencing that had passed QC, 17,909 had matching V(D)J sequencing with required chain combinations as specified above, with these cells used for all clonal analyses.
For analyses examining subset phenotype of clones in terms of size and evolution (e.g., Fig. 2E and fig. S2F), cells were grouped by clone ID, individual, and time point, and proportion of its member cells within each subset was calculated; the highest proportion subset was used to describe the clone’s major subset. For global overview of clonal size at d0/d21 within each subset, clonal size was defined depending on the time point the clone was sampled at. Analysis of changes in subset identity per clone (Fig. 2D) used clones present at both time points only.
For analyses looking at clonal changes across time points, only clones present at both time points were included (e.g., Fig. 3, D to G). In this context, the size of the clone was defined on the basis of its d0 (pretreatment) size. For clones that are present at only one time point (e.g., Fig. 4A), the clone size was defined on the basis of its size when present. Similarly, for analyses only at one time point (e.g., Figs. 4, C to G, and 5, F to H), clonal size was defined on the basis of size at that time point.
For the analysis of stable/expanding versus involuting clones between d0 and d21, only clones present at d0 and minimally two cells in size (to reduce likelihood that clone disappearance was due to resampling chance alone) were analyzed (Fig. 4C). Clones found at both time points were designated to be involuting if the proportion at d21 was significantly (a nominal P value < 0.05, Fisher’s exact test) smaller than the proportion at d0 and vice versa for expanding. Clones that were found at d0 but not found at d21 were also classed as involuting. Expanding clones were grouped with those in which there was no significant difference between time points (P > 0.05, Fisher’s exact test) as the stable/expanding subset. We also performed an additional analysis where clones were classified as involuting based on a range of fold changes to size (fig. S4B). Here, involuting clones were defined as those where the proportion at d21 was either <0.6× or <0.4× the proportion at d0 and “stable or expanding” clones were all others.
Similarly, for analysis of expanding/stable versus involuting clones between d21 and d63+, only clones incorporating more than one cell and present at d21 were analyzed. Clones present at d63+ were identified on the basis of the presence of the TCRB amino acid sequence within bulk RNA-seq data of magnetically sorted CD8+ T cells for six of the same individuals. A clone was designated as being present at d21 and d63+ if that same TCRB was found at both time points (d21 in scRNA-seq, d63+ in bulk RNA-seq). The clonal size (as a proportion) at d63+ was calculated by dividing the number of reads for that TCRB by the total number of TCR reads in the sample. The designation of a clone into involution or expanding/stable followed the same methodology as described above, including a sensitivity analysis using fold change cutoffs.
Downstream expression analysis
Differential gene expression analysis with subsampling
Differential gene expression was performed in Seurat using the FindMarkers function for all genes present, unless otherwise indicated. Alternatively, to validate the FindMarkers results, a mixed linear model for all genes with variance of >0 in the subsampled cells was used, controlling for interindividual variation (fig. S3, A and D). Where specified, to control for differences in statistical power in detecting DE genes across groups with varying cell counts, the number of cells used for analysis was standardized to the number present in the smallest group and bootstrapped 100 times to account for sampling bias. For comparison of DE genes according to subset or clone size, 1100 or 245 cells were subsampled, respectively, as the minimum number of cells across each category (ECs and EMs) at both time points. This same methodology was repeated filtering for only ECs or EMs with cells grouped into large and small clones based on a 0.5% of repertoire cutoff, and subsampling was performed to the smallest group size.
Pathway analysis
Pathway enrichment analysis was performed using the R package XGR (42) with the GOBP database. Significantly induced or suppressed genes were assessed separately against a background of all detected genes using a hypergeometric test, ontology algorithm specified as “elimination,” and an elimination P value of 0.01. For comparisons of pathways modulated after ICB across subsets, pathways were assessed for DE gene from each subsample at n = 1100 cells, and the frequency of enriched pathways was used for comparison, with this approach used to maximize the number of DE genes/statistical power taken forward for pathway analysis in most subsets given the paucity of DE genes in naïve cells. For comparisons of pathways modulated after ICB in large versus small ECs/EMs, we included the minimum number of significant DE genes across all comparators, keeping the number of genes used in the analysis constant between groups.
Leave-one-out cross-validation
To explore the effect of interindividual variation, we repeated the differential gene expression with subsampling analysis eight times, with all cells from one individual excluded each time. This demonstrated consistency in results suggesting that the observed effect is not driven solely by cells from one donor.
Generation of cytotoxicity score
Cell scores for cytotoxicity were calculated using AddModuleScore in Seurat, based on the top 50 variable feature genes that significantly correlated with IFNG expression (see data file S2) (43). For PDCD1 and TIGIT expression analysis, the MAGIC R package was used to recover expression data (44). In most instances, and as indicated in the figures and figure legends, cytotoxicity score is reported on a clonal basis. Here, cells were grouped for each individual and time point based on clone ID, and the cytotoxicity score was calculated as a median of the cytotoxicity scores for all cells within that clone.
Bulk RNA-seq
Bulk RNA-seq was performed on CD8+ T cells from 69 healthy individuals and 140 patients receiving single or combination ICB for MM, with exclusion of adjuvant treated patients. Of 137 d0 and 113 d21 samples, 110 individuals had paired d0/d21 data with a further 49 having d63+ samples. Clinical follow-up was available for 132 d0 and 109 d21 samples. Gene expression and TCR analysis using the MiXCR package were performed as previously described; for full methods, please see (18). The ethical approval and consent process for the collection of these samples were the same as outlined above.
To generate subset scores from each bulk sample, hallmark genes for each subset were identified in the single-cell data using the FindAllMarkers function in Seurat across all cells from d0 and d21 samples separately. To minimize time point–specific genes, the top 20 genes at each time point were selected, and the overlapping genes were retained as robust subset-demarcating genes (table S3). Scores were then generated for each geneset using DESeq2-normalized expression data (45) to then calculate geometric mean expression for each group of subset genes. A similar approach was used for cytotoxic score but instead based on the top 50 genes used for single-cell cytotoxic scoring. All metadata are listed in table S12.
For clonal persistence at d63+, we used longitudinal bulk samples from six individuals for which single-cell data were obtained, four taken at day 63, one at day 84, and one at day 106 of treatment. Clones were defined within these samples using MiXCR (46) according to the beta chain and then compared with earlier scRNA-seq data with matching beta chain usage, as described above.
Flow cytometry
Patient PBMCs frozen in 90% fetal calf serum + 10% dimethyl sulfoxide were thawed at 37°C and washed once in Hanks’ balanced salt solution (HBSS) before staining of 1 × 106 cells with LIVE/DEAD Fixable Near-IR (Invitrogen). Cells were washed and subsequently stained with antibodies against CD3, CD8, CD16, CD56, CD45RA, CD27, CX3CR1, and HLA-DR in 5% fetal calf serum + HBSS. After washing, cells were fixed for 10 min with 2% paraformaldehyde (Sigma-Aldrich), washed, and then permeabilized with 1× permeabilization buffer (eBioscience) for 10 min. The cells were then stained with perforin-1, CCL4, granzyme B, IFNγ, TNF-α, interleukin-32, and granulysin, washed with perm buffer, and stained with anti-rat immunoglobulin G2a secondary, before a final wash with perm buffer and acquisition on a BD LSRFortessa X-20. Data were analyzed in FlowJo (FlowJo LLC). Antibody details are shown in table S10 and gating is outlined in table S11, with representative plots found in fig. S7. All staining steps were performed for 30 min and at 4°C unless otherwise stated.
Analysis of external datasets
Melanoma tumor TCRB-seq and FPKM-normalized RNA-seq from Riaz et al. (25) were downloaded from https://github.com/riazn/bms038_analysis or from Gene Expression Omnibus: GSE91061, respectively. To generate an intratumoral cytotoxicity score, the 50 genes used for single-cell cytotoxic scoring were correlated against the expression of CD3D, CD3E, CD8A, CD8B, IFNG, and PRF1, and k-means were clustered to identify a module of genes most associated with CD8+ T cell cytotoxicity. Geometric mean expression of the eight genes was then calculated per sample and used as cytotoxic score. Large clone count per TCRB sample was calculated as for the bulk CD8+ sequencing data (i.e., the number of clones occupying greater than 0.5% of the repertoire). Survival analysis was conducted for the 52 samples with overlapping RNA-seq and TCRB data (26 pretreatment and 26 on-treatment) (table S13). Analysis of public melanoma and viral T cell clones was done using TCRB-seq data of 199 melanomas from Pruessman et al. (26) or viral-reactive TCR sequences downloaded from VDJdb (27, 47) with confidence scores above 0. Cells present in the scRNA-seq data were annotated as being found in melanoma or viral-reactive based on their ownership of a TCRB that was found in either the Pruessman or VDJdb dataset.
General statistical analysis
Statistical comparisons were performed as indicated in each figure legend and conducted in R. Paired tests were used for paired samples, usually using Wilcoxon signed-rank test as indicated. For unpaired samples, the Wilcoxon rank sum test was primarily used as per the figure legends. Cells and statistics from donor 6 were excluded from all analyses in Figs. 2 (C and D) and 4 and fig. S2E due to low numbers of cells at d0. For analysis of clonal persistence (Fig. 4, D to G), n = 6 as only six of the eight individuals profiled had samples at d63+. All analyses and statistical tests were performed in R using baseR or rstatix and lme4/lmerTest for linear effect models. LDA analysis was performed using MASS and Pi (48). Gene correlation analysis for phenotype scores was performed using DGCA (49). Survival analysis was performed using survival/survminer. UpSetR (50) networkD3 and ComplexHeatmap (51) were used for visualization. For all data, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Supplementary Material
Acknowledgments
We are very grateful to all patients who contributed samples and participated in the study. We thank all the staff of the Day Treatment Unit, Oxford Cancer Centre, and The Brodey Centre at the Horton General Hospital. We also thank M. Payne, N. Coupe, and R. Matin for assistance in collecting patient samples. We thank M. H. Al-Mossawi for discussion and advice.
Funding
This study was funded by a Wellcome Intermediate Clinical Fellowship to B.P.F. (no. 201488/Z/16/Z), additionally supporting A.V.d.l.A., E.A.M., and I.N. R.A.W. was a National Institute for Health Research (NIHR) Academic Clinical Fellow at the time of this project and was supported by a Cancer Research UK (CRUK) predoctoral fellowship (no. ANR00740), currently funded by a Wellcome Trust Doctoral Training Fellowship (no. BST00070). O.T. is supported by The Clarendon Fund, St Edmund Hall, and an Oxford Australia Scholarship. R.C. is supported by a CRUK Clinical Research Training Fellowship (S_3578). C.A.T. is funded by the Engineering and Physical Sciences Research Council and the Balliol Jowett Society (no. D4T00070). H.R. is funded by the Medical Research Council (MRC MC UU 00002/10) and the Lopez-Loreta Foundation. M.R.M. and B.P.F. are supported by the NIHR Oxford Biomedical Research Centre. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health.
Footnotes
Author contributions: R.A.W. and B.P.F. conceived the study; R.A.W., O.T., I.N., and B.P.F. performed the analysis; figures were generated by O.T. and R.A.W.; manuscript was drafted by R.A.W., O.T., and B.P.F.; R.A.W. and R.C. performed single-cell experiments; O.T. and P.K.S. performed flow cytometry experiments; R.A.W., E.A.M., R.C., C.A.T., P.K.S., A.V.d.l.A., and O.T. collected and prepared samples; H.R. provided expert statistical advice; M.R.M. contributed to discussions throughout the work; and B.P.F. oversaw the project. All authors read, edited, and approved the final manuscript.
Competing interests: R.A.W., O.T., R.C., C.A.T., A.V.d.l.A., P.K.S., E.A.M., H.R., I.N. declare that they have no competing interests. M.R.M. reports grants from Roche, grants from Astrazeneca, grants and personal fees from GSK, personal fees and other from Novartis, other from Millenium, personal fees and other from Immunocore, personal fees and other from BMS, personal fees and other from Eisai, other from Pfizer, personal fees, nonfinancial support and other from Merck/MSD, personal fees and other from Rigontec (acquired by MSD), other from Regeneron, personal fees and other from BiolineRx, personal fees and other from Array Biopharma (now Pfizer), nonfinancial support and other from Replimune, personal fees from Kineta, and personal fees from Silicon Therapeutics, outside the submitted work. B.P.F. received conference support from BMS.
Data and materials availability
Raw sequencing data (fastq files) has been deposited at the European Genome-Phenome Archive, which is hosted by the European Bioinformatics Institute and the Centre for Genomic Regulation under accession nos. EGAS00001005507 (for the scRNA-seq and VDJ sequencing data) and EGAS00001004081 (for the bulk RNA-seq data). Gene expression matrices and the Seurat object (which contains associated cell metadata for all cells that passed upstream QC) have been deposited on the Oxford University Research Archive and can be found at https://doi.org/10.5287/bodleian:2NVVop6Bz. Scripts used in data analysis and the generation of figures are available on the Fairfax group bitbucket account: https://bitbucket.org/bpfairfax/immune-checkpoint-blockade-sensitivity-and-progression-free/src/master/. All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials and deposited in online data.
References and Notes
- 1.Postow MA, Callahan MK, Wolchok JD. Immune checkpoint blockade in cancer therapy. J Clin Oncol. 2015;33:1974–1982. doi: 10.1200/JCO.2014.59.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob J-J, Rutkowski P, Lao CD, Cowey CL, Schadendorf D, Wagstaff J, Dummer R, Ferrucci PF, et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2019;381:1535–1546. doi: 10.1056/NEJMoa1910836. [DOI] [PubMed] [Google Scholar]
- 3.Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, Kurzrock R. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim JY, Kronbichler A, Eisenhut M, Hong SH, van der Vliet HJ, Kang J, Shin JI, Gamerith G. Tumor mutational burden and efficacy of immune checkpoint inhibitors: A systematic review and meta-analysis. Cancer. 2019;11:1798. doi: 10.3390/cancers11111798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, Peters S. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann Oncol. 2019;30:44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnes TA, Amir E. HYPE or HOPE: The prognostic value of infiltrating immune cells in cancer. Br J Cancer. 2017;117:451–460. doi: 10.1038/bjc.2017.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maibach F, Sadozai H, Seyed Jafari SM, Hunger RE, Schenk M. Tumor-infiltrating lymphocytes and their prognostic value in cutaneous melanoma. Front Immunol. 2020;11:2105. doi: 10.3389/fimmu.2020.02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, Xu W, Harmon S, Giles JR, Wenz B, Adamow M, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017;545:60–65. doi: 10.1038/nature22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat Rev Immunol. 2020;20:651–668. doi: 10.1038/s41577-020-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas NE, Busam KJ, From L, Kricker A, Armstrong BK, Anton-Culver H, Gruber SB, Gallagher RP, Zanetti R, Rosso S, Dwyer T, et al. Tumor-infiltrating lymphocyte grade in primary melanomas is independently associated with melanoma-specific survival in the population-based genes, environment and melanoma study. J Clin Oncol. 2013;31:4252–4259. doi: 10.1200/JCO.2013.51.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJM, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, West AN, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yost KE, Satpathy AT, Wells DK, Qi Y, Wang C, Kageyama R, McNamara KL, Granja JM, Sarin KY, Brown RA, Gupta RK, et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med. 2019;25:1251–1259. doi: 10.1038/s41591-019-0522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu TD, Madireddi S, de Almeida PE, Banchereau R, Chen YJJ, Chitre AS, Chiang EY, Iftikhar H, O’Gorman WE, Au-Yeung A, Takahashi C, et al. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature. 2020;579:274–278. doi: 10.1038/s41586-020-2056-8. [DOI] [PubMed] [Google Scholar]
- 14.Lucca L, Axisa P-P, Lu B, Harnett B, Jessel S, Zhang L, Raddassi K, Zhang L, Olino K, Clune J, Singer M, et al. Circulating clonally expanded T cells reflect functions of tumor infiltrating T cells 2 3 running title 1 single-cell RNAseq of circulating TILs in melanoma 2 3. bioRxiv. 2020:2020.09.30.321240 [Google Scholar]
- 15.Valpione S, Galvani E, Tweedy J, Mundra PA, Banyard A, Middlehurst P, Barry J, Mills S, Salih Z, Weightman J, Gupta A, et al. Immune awakening revealed by peripheral T cell dynamics after one cycle of immunotherapy. Nat Can. 2020;1:210–221. doi: 10.1038/s43018-019-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucca LE, Axisa PP, Lu B, Harnett B, Jessel S, Zhang L, Raddassi K, Zhang L, Olino K, Clune J, Singer M, et al. Circulating clonally expanded T cells reflect functions of tumor-infiltrating T cells. J Exp Med. 2021;218:e20200921. doi: 10.1084/jem.20200921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pauken KE, Shahid O, Lagattuta KA, Mahuron KM, Luber JM, Lowe MM, Huang L, Delaney C, Long JM, Fung ME, Newcomer K, et al. Single-cell analyses identify circulating anti-tumor CD8 T cells and markers for their enrichment. J Exp Med. 2021;218:e20200920. doi: 10.1084/jem.20200920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fairfax BP, Taylor CA, Watson RA, Nassiri I, Danielli S, Fang H, Mahé EA, Cooper R, Woodcock V, Traill Z, Al-Mossawi MH, et al. Peripheral CD8+ T cell characteristics associated with durable responses to immune checkpoint blockade in patients with metastatic melanoma. Nat Med. 2020;26:193–199. doi: 10.1038/s41591-019-0734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim KH, Cho J, Ku BM, Koh J, Sun JM, Lee SH, Ahn JS, Cheon J, Min YJ, Park SH, Park K, et al. The first-week proliferative response of peripheral blood PD-1+CD8+T cells predicts the response to Anti-PD-1 therapy in solid tumors. Clin Cancer Res. 2019;25:2144–2154. doi: 10.1158/1078-0432.CCR-18-1449. [DOI] [PubMed] [Google Scholar]
- 20.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran V, Piha-Paul SA, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Investig. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szabo PA, Levitin HM, Miron M, Snyder ME, Senda T, Yuan J, Cheng YL, Bush EC, Dogra P, Thapa P, Farber DL, et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat Commun. 2019;10:4706. doi: 10.1038/s41467-019-12464-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pizzolato G, Kaminski H, Tosolini M, Franchini DM, Pont F, Martins F, Valle C, Labourdette D, Cadot S, Quillet-Mary A, Poupot M, et al. Single-cell RNA sequencing unveils the shared and the distinct cytotoxic hallmarks of human TCRVδ1 and TCRVδ2 γδ T lymphocytes. Proc Natl Acad Sci USA. 2019;116:11906–11915. doi: 10.1073/pnas.1818488116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aguirre-Gamboa R, Joosten I, Urbano PCM, van der Molen RG, van Rijssen E, van Cranenbroek B, Oosting M, Smeekens S, Jaeger M, Zorro M, Withoff S, et al. Differential effects of environmental and genetic factors on T and B cell immune traits. Cell Rep. 2016;17:2474–2487. doi: 10.1016/j.celrep.2016.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, Hodi FS, Martín-Algarra S, Mandal R, Sharfman WH, Bhatia S, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171:934–949.:e15. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruessmann W, Rytlewski J, Wilmott J, Mihm MC, Attrill GH, Dyring-Andersen B, Fields P, Zhan Q, Colebatch AJ, Ferguson PM, Thompson JF, et al. Molecular analysis of primary melanoma T cells identifies patients at risk for metastatic recurrence. Nat Can. 2020;1:197–209. doi: 10.1038/s43018-019-0019-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bagaev DV, Vroomans RMA, Samir J, Stervbo U, Rius C, Dolton G, Greenshields-Watson A, Attaf M, Egorov ES, Zvyagin IV, Babel N, et al. VDJdb in 2019: Database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Res. 2020;48:D1057–D1062. doi: 10.1093/nar/gkz874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enamorado M, Iborra S, Priego E, Cueto FJ, Quintana JA, Martýnez-Cano S, Mejyás-Perez E, Esteban M, Melero I, Hidalgo A, Sancho D. Enhanced anti-tumour immunity requires the interplay between resident and circulating memory CD8+ T cells. Nat Commun. 2017;8:16073. doi: 10.1038/ncomms16073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ribas A, Shin DS, Zaretsky J, Frederiksen J, Cornish A, Avramis E, Seja E, Kivork C, Siebert J, Kaplan-Lefko P, Wang X, et al. PD-1 blockade expands intratumoral memory T cells. Cancer Immunol Res. 2016;4:194–203. doi: 10.1158/2326-6066.CIR-15-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Araki K, Morita M, Bederman AG, Konieczny BT, Kissick HT, Sonenberg N, Ahmed R. Translation is actively regulated during the differentiation of CD8+ effector T cells. Nat Immunol. 2017;18:1046–1057. doi: 10.1038/ni.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwahori K, Shintani Y, Funaki S, Yamamoto Y, Matsumoto M, Yoshida T, Morimoto-Okazawa A, Kawashima A, Sato E, Gottschalk S, Okumura M, et al. Peripheral T cell cytotoxicity predicts T cell function in the tumor microenvironment. Sci Rep. 2019;9:2636. doi: 10.1038/s41598-019-39345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenkins RW, Barbie DA, Flaherty KT. Mechanisms of resistance to immune checkpoint inhibitors. Br J Cancer. 2018;118:9–16. doi: 10.1038/bjc.2017.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamphorst AO, Pillai RN, Yang S, Nasti TH, Akondy RS, Wieland A, Sica GL, Yu K, Koenig L, Patel NT, Behera M, et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc Natl Acad Sci USA. 2017;114:4993–4998. doi: 10.1073/pnas.1705327114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nabet BY, Esfahani MS, Moding EJ, Hamilton EG, Chabon JJ, Rizvi H, Steen CB, Chaudhuri AA, Liu CL, Hui AB, Almanza D, et al. Noninvasive early identification of therapeutic benefit from immune checkpoint inhibition. Cell. 2020;183:363–376.:e13. doi: 10.1016/j.cell.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarthy DJ, Campbell KR, Lun ATL, Wills QF. Scater: Pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zappia L, Oshlack A. Clustering trees: A visualization for evaluating clusterings at multiple resolutions. GigaScience. 2018;7:giy083. doi: 10.1093/gigascience/giy083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, Kang B, Liu Z, Jin L, Xing R, Gao R, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. 2018;24:978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 39.Savas P, Virassamy B, Ye C, Salim A, Mintoff CP, Caramia F, Salgado R, Byrne DJ, Teo ZL, Dushyanthen S, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med. 2018;24:986–993. doi: 10.1038/s41591-018-0078-7. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, Gao R, Kang B, Zhang Q, Huang JY, Konno H, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564:268–272. doi: 10.1038/s41586-018-0694-x. [DOI] [PubMed] [Google Scholar]
- 41.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, Butte AJ, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang H, Knezevic B, Burnham KL, Knight JC. XGR software for enhanced interpretation of genomic summary data, illustrated by application to immunological traits. Genome Med. 2016;8:129. doi: 10.1186/s13073-016-0384-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J, van den Oord J, et al. SCENIC: Single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, Burdziak C, Moon KR, Chaffer CL, Pattabiraman D, Bierie B, et al. Recovering gene interactions from single-cell data using data diffusion. Cell. 2018;174:716–729.:e27. doi: 10.1016/j.cell.2018.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, Chudakov DM. MiXCR: Software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 47.VDJdb. [accessed and downloaded April 2021]. https://vdjdb.cdr3.net/
- 48.Fang H, Beckmann G, Bountra C, Bowness P, Burgess-Brown N, Carpenter L, Chen L, Damerell D, Egner U, Fang H, Fujii R, et al. A genetics-led approach defines the drug target landscape of 30 immune-related traits. Nat Genet. 2019;51:1082–1091. doi: 10.1038/s41588-019-0456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKenzie AT, Katsyv I, Song WM, Wang M, Zhang B. DGCA: A comprehensive R package for differential gene correlation analysis. BMCSyst Biol. 2016;10:106. doi: 10.1186/s12918-016-0349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conway JR, Lex A, Gehlenborg N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33:2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data (fastq files) has been deposited at the European Genome-Phenome Archive, which is hosted by the European Bioinformatics Institute and the Centre for Genomic Regulation under accession nos. EGAS00001005507 (for the scRNA-seq and VDJ sequencing data) and EGAS00001004081 (for the bulk RNA-seq data). Gene expression matrices and the Seurat object (which contains associated cell metadata for all cells that passed upstream QC) have been deposited on the Oxford University Research Archive and can be found at https://doi.org/10.5287/bodleian:2NVVop6Bz. Scripts used in data analysis and the generation of figures are available on the Fairfax group bitbucket account: https://bitbucket.org/bpfairfax/immune-checkpoint-blockade-sensitivity-and-progression-free/src/master/. All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials and deposited in online data.