Abstract
Early-onset conduct problems (CP) are a key predictor of adult criminality and poor mental health. While previous studies suggest that both genetic and environmental risks play an important role in the development of early-onset CP, little is known about potential biological processes underlying these associations. In this study, we examined prospective associations between DNA methylation (cord blood at birth) and trajectories of CP (4-13yrs), using data drawn from the Avon Longitudinal Study of Parents and Children (ALSPAC). Methylomic variation at seven loci across the genome (FDR<0.05) differentiated children who go on to develop early-onset (n = 174) vs low (n = 86) CP, including sites in the vicinity of MGLL (involved in endocannabinoid signaling and pain perception). Sub-threshold associations in the vicinity of three candidate genes for CP (MAOA, BDNF, and FKBP5) were also identified. Within the early-onset CP group, methylation levels of the identified sites did not distinguish children who will go on to persist vs desist in CP behavior over time. Overall, we found that several of the identified sites correlated with prenatal exposures, and none were linked to known genetic mQTLs. Findings contribute to a better understanding of epigenetic patterns associated with early-onset CP.
Keywords: DNA Methylation, genome-wide, ALSPAC, conduct problems, risk exposure
Introduction
Conduct problems (CP; e.g. fighting, stealing) are a major public health concern and a leading cause of youth treatment referral (Bywater, 2012). CP youth engage in antisocial and disruptive behaviors that result in significant distress to victims, impairment of life opportunities and economic burden on judicial, healthcare and social welfare services (Colman et al., 2009). In particular, an early-onset of CP (at or before the age of 10) designates a high-risk group of children, who show more severe and recidivistic patterns of antisocial behavior, higher levels of comorbid psychopathology (E. D. Barker, Oliver, & Maughan, 2010), increased risk for poor adult outcomes (Fergusson, John Horwood, & Ridder, 2005; Odgers et al., 2007), and who account for the majority of criminal offences within a given community (David P. Farrington, 2001). Consequently, early-onset CP are recognized as an important target for prevention and intervention efforts.
Like most complex phenotypes, early-onset CP are thought to result from the developmentally dynamic interplay of genetic and environmental influences. Studies have shown that the heritability of early-onset CP is moderate to high (~50-80%, see Barker, Cecil, Walton & Meehan, in press, for a review) and that this genetic vulnerability interacts with environmental risk exposure (e.g. Jaffee et al., 2005). Indeed, exposure to adversity during pregnancy (e.g. maternal smoking, stress and psychopathology) as well as the postnatal years (e.g. maltreatment, poverty) is frequent among this group of children, and thought to play an important role in the development and persistence of CP behavior (E. D. Barker & Maughan, 2009; T. E. Moffitt, 2006; Odgers et al., 2007; Odgers et al., 2008). Yet, little is known about the biological mechanisms through which these influences confer increased risk for early-onset CP.
In recent years, DNA methylation – an epigenetic mechanism that regulates gene expression (Jaenisch & Bird, 2003) – has emerged as a potential mechanism underlying geneenvironment interplay and disease susceptibility across the lifespan. Specifically, studies have shown that (i) DNAm patterns are under significant genetic control – as evidenced by the discovery of a large number of methylation quantitative trait loci (mQTL; Gaunt et al., 2016; Jones, Fejes, & Kobor, 2013); and (ii) DNAm is also sensitive to environmental influences (McGowan & Roth, 2015), including numerous nutritional, chemical, physical, and psychosocial exposures occurring in utero (e.g. Monk, Spicer, & Champagne, 2012; Richmond et al., 2015; Tobi et al., 2014) and postnatally (Lutz & Turecki, 2014; Szyf & Bick, 2013; Tyrka et al., 2015). In turn, DNAm patterns have been shown to play a central role in neurobiological and developmental processes (e.g. neurogenesis, synaptic plasticity, learning and memory; Murgatroyd & Spengler, 2011) and aberrations in DNAm have been implicated in a range of negative outcomes, including childhood psychopathology and stress-related psychiatric disorders (Klengel, Pape, Binder, & Mehta, 2014; van Mil et al., 2014; Weder et al., 2014). Consequently, it has been suggested that DNAm may represent a molecular pathway through which environmental exposures become translated into phenotypic variation, conferring increased susceptibility to mental and physical health problems (Kofink, Boks, Timmers, & Kas, 2013; Lewis & Olive, 2014).
For example, experimental studies in animals have shown that exposure to early adversity (e.g. prenatal stress, low postnatal care) can cause alterations in offspring DNAm (e.g. in HPA-axis genes), driving differences in the offspring’s behavioral response to future stressors (Weaver et al., 2004; Turecki & Meaney, 2016). Indeed, because DNAm marks can be passed on during cell division (and in some cases across generations), they can lead to long-term programming of gene expression, with downstream effects on stress sensitivity, behavior and disease risk (Rodgers & Bale, 2011; Zannas & West, 2014). In the context of CP, epigenetic alterations may thus represent a mechanism through which genetic and environmental factors intersect, influencing developmental trajectories of childhood aggression and antisocial behavior (Tremblay & Szyf, 2010).
So far, only a handful of studies in humans have examined associations between DNAm and CP-related phenotypes. Of these, the majority have been carried out by a research group comparing patterns of DNAm (extracted from peripheral blood samples collected at age 26-28) between adult males with a childhood history of chronic physical aggression (ages 6-15) vs controls who maintained low levels of physical aggression during the same time period, based on data from a longitudinal cohort (Booij et al., 2010; Guillemin et al., 2014; Provencal et al., 2013; Provencal et al., 2014; Wang et al., 2012). Using this case-control design, these studies reported that childhood aggression associated with (i) higher levels of SLC6A4 promoter methylation and lower in vivo levels of brain serotonin synthesis (Booij et al., 2010; Wang et al., 2012); (ii) DNAm levels in a set of genes involved in cytokine function and inflammation (Provencal et al., 2013); and (iii) across a large number of gene promoter regions in an epigenome-wide scan (Provencal et al., 2014) – a finding that was later extended to a small sample of adult females as well (Guillemin et al., 2014). Another set of studies based on a sample of antisocial boys diagnosed with oppositional defiant disorder or conduct disorder (4-16yrs) have also documented an association between severity of callous-unemotional traits (CU; e.g. low capacity for empathy, lack of guilt, shallow affect) and DNAm levels (obtained from peripheral blood) across several candidate genes, including lower DNAm of the Serotonin 1B Receptor gene (HTR1B; Moul, Dobson-Stone, Brennan, Hawes, & Dadds, 2015), and higher DNAm of the Oxytocin Receptor gene (OXTR; Dadds et al., 2014) – which in turn related to lower circulating oxytocin levels.
Despite these promising findings, research to date has been limited in a number of important ways. First, no study to our knowledge has specifically investigated DNAm patterns associated with early-onset CP, which designates a subgroup of children at high risk for negative adult outcomes. Second, the available research has been primarily cross-sectional or retrospective with regards to DNAm (i.e. associating behavioral phenotypes in childhood to DNAm later in development). As such, it is currently unclear whether observed alterations in DNAm patterns represent a risk factor for and/or a consequence of conduct problems. In other words, it has not been possible to establish whether DNAm is a prospective risk for CP, or alternatively, the biological correlate of a chronic antisocial lifestyle. Third, there is a lack of studies integrating measures of the environment, which has precluded the possibility of assessing the epigenome in relation to both risk exposure and psychiatric phenotypes.
To address the above gaps, research from our group has begun to assess prospective associations between environmental risks, DNAm at repeated time points (birth, childhood) and CP-related outcomes, using data from the Avon Longitudinal Study of Parents and Children (ALSPAC; Fraser et al., 2013). For example, in children with early-onset CP and low internalizing problems, we found that DNAm levels in the promoter region OXTR at birth (but not at later points in childhood) associated with higher risk exposure in utero, reduced experience of victimization during childhood (i.e. indicative of an ‘evocative epigenetic-environment correlation’) and more severe CU traits at age 13 (Cecil et al., 2014). Also in early-onset CP, we found that a sugar- and fat-rich diet during pregnancy prospectively associated with higher DNAm of the Intrauterine Growth Factor Gene 2 (IGF2) at birth (but not during childhood), which, in turn, associated with more severe co-occurring ADHD symptoms (Rijslasdaam et al., 2016a). The temporal specificity around birth observed across these and other studies (e.g. Cecil et al., 2016; Walton et al., 2016) may reflect a number of factors, including large-scale variability in DNAm patterns across developmental eras (Gaunt et al., 2016), differences between the tissues examined at each time point (i.e. cord blood at birth vs whole blood in childhood), as well as the influence of developmentally-specific genetic effects on symptom trajectories (e.g. Pingault, Rijsdijk, Zheng, Plomin & Viding, 2015). Additionally, findings may lend support for the developmental origins of health and disease hypothesis (DOHaD; D. J. Barker, 2007; Nigg, 2016), whereby methylation changes at birth may represent a more reliable proxy for intra-uterine risk exposures (compared to DNAm in childhood), and associated perturbations in fetal development. However, the extent to which DNAm patterns across the genome at birth may prospectively associate with an early-onset of CP has yet to be examined.
In addition to advancing knowledge of potential biological differences between children with early-onset vs low CP, the study of DNAm may also help us to better understand why early-onset children themselves show considerable heterogeneity in their developmental trajectories of CP. Indeed, studies have found that – despite being considered a high-risk group as a whole - less than 50% of children with an early-onset of CP will actually continue to exhibit these problems into adolescence (i.e. early-onset persisting trajectory, EOP), while the rest will remit to near-typical levels of CP (i.e. childhood-limited trajectory, CL; E. D. Barker & Maughan, 2009; Odgers et al., 2008). This heterogeneity poses a challenge for accurate diagnostic screening, clinical classification and treatment formulation, as well as raising important questions about what factors may underlie these increasingly divergent trajectories of CP within early-onset children (i.e. in line with the concept of multifinality; Cicchetti & Rogosch, 1996). So far, evidence regarding early-life predictors of EOP vs CL trajectories has been somewhat mixed (see Moffitt et al., 2008, for a review), but generally suggests that the difference may be more quantitative than qualitative, with pre- and postnatal risk exposures found to be particularly high for EOP children, compared to their CL peers (i.e. EOP>CL>Low CP; E. D. Barker & Maughan, 2009). However, it is currently unknown whether such a ‘graded’ difference on the same risk factors may also exist at a biological level. The analysis of DNAm patterns between early-onsets who follow different developmental trajectories of CP (i.e. EOP vs CL), may therefore help to refine the phenotype and contribute to a better understanding of heterogeneity within this high-risk group of children.
The present study
In light of the above research gaps, we made use of data from the ALSPAC cohort spanning pregnancy to late childhood to address three key aims. First, we examined whether DNAm patterns at birth (measured at >450,000 sites across the genome) prospectively differentiate children who will go on to develop early-onset (n = 174) vs low CP (n = 86). Second, to explore heterogeneity in CP trajectories within the early-onset group, we tested whether these DNAm patterns further distinguish between children who will persist (EOP; n = 91) vs desist (CL; n = 83) in conduct problems over time. Third, we investigated potential genetic and environmental influences associated with the identified DNAm sites, including known genetic mQTLs and measures of prenatal adversity that have been previously linked to early-onset CP (e.g. maternal diet, smoking, and exposure to stressful events).
Methods
Sample
The Epigenetic Pathways to Conduct Problems Study consists of a subsample of youth (n=321; 50% female) drawn from the Avon Longitudinal Study of Parents and Children (ALSPAC) and partially overlapping with a larger study of DNA methylation, the Accessible Resource for Integrated Epigenomics Studies (ARIES, www.ariesepigenomics.org.uk; Relton et al., 2015). Specifically, children were included if they (i) have available measures of DNAm at two or more time points and (ii) follow previously established trajectories of conduct problems between 4 and 13 years of age (see measures section below; E. D. Barker & Maughan, 2009). Children in this study were white Caucasian (six removed from analyses due to missing ethnicity data).
ALSPAC is an ongoing epidemiological study of 14,541 pregnant women resident in Avon, UK with expected dates of delivery 1st April 1991 to 31st December 1992. Of these initial pregnancies, there was a total of 14,676 fetuses, resulting in 14,062 live births and 13,988 children who were alive at 1 year of age (Fraser et al., 2013). When compared to 1991 National Census Data, the ALSPAC sample was found to be broadly similar to the UK population as a whole (Boyd et al., 2013). The study website contains details of all the data that is available: http://www.bris.ac.uk/alspac/researchers/data-access/data-dictionary. Ethical approval for the study was obtained from the ALSPAC Law and Ethics Committee and the Local Research Ethics Committees.
Measures
Conduct Problems
CP trajectories were previously identified and validated in the whole ALSPAC sample using growth mixture models based on repeated measures of the Strengths and Difficulties Questionnaire ‘conduct problems’ subscale (ages 4,7,8,10,12,13; SDQ; Goodman, 2001), with boys and girls modeled in the same trajectories (E. D. Barker & Maughan, 2009). The original analysis identified four CP trajectories: (i) an early-onset persistent (EOP; 9%) group of children, who show high levels of CP early on and remain high across time; (ii) a childhood-limited (CL; 15%) group who also show an early-onset of CP, but desist around adolescence; (iii) an adolescent-onset group who only show CP behaviors later on (AO; 12%); and (iv) a low CP group, who maintain low levels of CP throughout (Low CP; 64%). The percentage of youth identified in the CP trajectories in ALSPAC is consistent with reported prevalence estimates in the general population (4-20%; Egger & Angold, 2006; Kim-Cohen et al., 2003). In contrast to the overall ALSPAC sample, our epigenetic subsample featured a near equal number of youth in each trajectory, and was therefore enriched for CP (EOP: n = 91, 28%; CL: n = 83, 26%; AO: n = 61, 19%; Low CP: n = 86, 27%). In this study, early-onset CP youth were defined as youth who follow either an EOP or CL trajectory. The adolescent-onset (AO) group was excluded from the main analyses for two reasons. First, although AO and Low children appear phenotypically similar in CP levels during the developmental period under investigation, they have been shown to differ in levels of environmental risk exposure and temperamental features (Barker & Maughan, 200), so that we decided against combining AOs within the Low (i.e. typical) group. Second, we did not include AOs as a separate comparison group in the epigenome-wide analyses due to concerns over statistical power and multiple testing burden resulting from the increase in number of comparisons.
DNA Extraction and Methylomic Profiling
DNA was obtained from cord blood samples at birth. 500ng genomic DNA was bisulfite converted using an EZ-DNA methylation kit (Zymo Research, Orange, CA, USA) and DNAm quantified using the Illumina HumanMethylation450 BeadChip (450K array; Illumina, USA) with arrays scanned using an Illumina iScan (software version 3.3.28). The Illumina 450K array comprises >485,000 probes, each quantifying DNAm at a specific CpG site, covering 99% of Reference Sequence (RefSeq) genes, with an average of 17 CpG sites per gene region (distributed across promoter, 5’UTR, first exon, gene body and 3’UTR regions). The BeadChip covers 96% of CpG islands, with additional coverage in island shores and their flanking regions. Initial quality control of data generated was conducted using GenomeStudio (version 2011.1) to determine the status of staining, extension, hybridization, target removal, bisulfite conversion, specificity, non-polymorphic and negative controls. Furthermore, multiple checks for sample mismatch were carried out to ascertain that all cord blood samples in the study reflected infant DNA as opposed to maternal DNA. Specifically, samples were checked by calculating concordance with (i) GWA data from the same participants, (ii) SNP probes on the Illumina 450k array across mother and child, and (iii) sex. Data were quantile normalized using the dasen function as part of the wateRmelon package (wateRmelon_1.0.3; Pidsley et al., 2013) within the R statistical analysis environment and batch corrected using the ComBat package (Johnson, Li, & Rabinovic, 2007). Probes previously reported to hybridize to multiple genomic regions or containing a SNP at the single base extension site were removed from subsequent analyses (Chen et al., 2013; Price et al., 2013), in addition to the 65 SNPs used for sample identification on the array (total probes removed 72,067). For each probe, DNAm levels were indexed by beta values – i.e. the ratio of methylated signal divided by the sum of the methylated and unmethylated signal (M/M+U).
Prenatal environment
Multiple environmental influences were included that have been previously linked to early-onset CP, including maternal prenatal diet, smoking, alcohol use and exposure to stressful events (E. D. Barker & Maughan, 2009). Information on dietary patterns was drawn from maternal ratings of the Food Frequency Questionnaire at 32 week gestation, where factor scores of ‘healthy’ (i.e. high on fish, non-meat protein and vegetables) and ‘unhealthy’ (i.e. high on processed and junk food) diet were extracted using confirmatory factor analysis, both of which showed acceptable model fit (for full details, see E. D. Barker, Kirkham, Ng, & Jensen, 2013). Maternal smoking and alcohol use during the first trimester of pregnancy were measured via maternal ratings, using a yes/no binary variable for smoking, and a 4-point scale for alcohol use (‘never’ to ‘daily’). With regards to stress exposure, we included cumulative risk scores of prenatal (18 to 32 weeks) adversity, based on 56 dichotomous items from the Life Events inventory (adapted in ALSPAC based on the work of Brown et al., 1973 and Barnett et al., 1983) and the Family Adversity Index (Wolke, Steer & Bowen, 2004). Briefly, items were organized into four conceptually distinct but related risk domains, and summed to create the following cumulative scores: (i) life events (22 items; e.g. death in family, accident); (ii) contextual risks (16 items; e.g. poor housing, financial problems); (iii) maternal risks (10 items; e.g. psychopathology, criminal behavior); and (iv) interpersonal risks (8 items; e.g. partner abuse, family conflict). The full list of items contained in these cumulative scores is provided in online supplement OS1, and further information is available elsewhere (Cecil et al., 2014b; Rijlaarsdam et al., 2016b).
Statistical Analyses
All analyses were performed within R (version 3.0.1; R Core Team, 2014) on DNAm data regressed for sex and cell-type proportions (CD8 T lymphocytes, CD4 T lymphocytes, natural killer cells, B lymphocytes, monocytes), estimated using the reference-based approach detailed in Houseman et al. (2012).
Step 1: Are there differences in neonatal patterns of DNAm between children who go on to develop early-onset vs low CP?
First, a methylome-wide association analysis was performed using the CpGAssoc package (Barfield, Kilaru, Smith, & Conneely, 2012) to identify DNAm sites at birth that differentiate children who go on to develop early-onset (i.e. EOP and CL) vs low CP. False discovery rate (FDR) correction was implemented across all probes (i.e. genome-wide significance threshold set at q < 0.05). Effect sizes were calculated and interpreted following Cohen’s guidelines, where an effect of 0.20 represents a small effect, and effect of 0.50 is a medium effect and an effect of 0.80 is a large effect (Cohen, 1988).
Genome-wide significant loci were then uploaded to the UCSC genome browser (GRCh37/hg19 assembly; Kent et al., 2002) to explore their potential functional relevance, by comparing their genomic location to that of key regulatory elements recorded in the Encyclopedia of DNA Elements (ENCODE) database (http://genome.ucsc.edu/ENCODE/). These included (i) transcription factor binding sites, i.e. DNA regions where one or more specific proteins responsible for regulating transcription bind to (data generated on 161 transcription factors in 91 cell types via ChIP-seq); (ii) DNase I hypersensitivity clusters, which tend to coincide with regulatory regions of the gene and indicate an open chromatin state that facilitates transcription (based on data from 125 cell types), and (iii) histone marks, i.e. chemical modifications that influence how tightly packaged the DNA is around histone protein, thereby regulating how accessible DNA is for transcription (data available for seven cell types; however, given that DNAm patterns can be tissue specific, we only selected here the three cell-types that are most relevant to cord blood; i.e. blood [GM12878, K562] and umbilical vein endothelial [HUVEC] cells). To identify enriched biological pathways, we also analyzed probes that were significant at p<0.001 using an optimized gene ontology method that corrects for multiple potential confounds, including background probe distribution and gene size (see OS2).
Step 2: Within the early-onset group, do these DNAm sites also differentiate children who will persist vs desist in CP over time?
As a second step, we examined whether the identified DNAm sites may help to explain heterogeneity in developmental trajectories within the early-onset CP group. Specifically, we used one-way Analysis of Covariance (ANCOVA) models to test whether DNAm levels across the sites identified in Step 1 also differentiate early-onsets who will go on to follow a persisting (i.e. EOP) vs desisting (i.e. CL) trajectory of CP behaviour over time, controlling for sex and cell-type proportions.
Step 3: Are the identified DNAm sites associated with genetic and environmental factors?
As a final step, we explored potential genetic and environmental factors that may influence the DNAm sites associated with early-onset CP (i.e. identified in Step 1). As our sample was underpowered to directly examine genetic polymorphisms (i.e. SNPs) affecting DNAm, we used the mQTLdb resource (http://www.mqtldb.org/) to search for known methylation quantitative trait loci (mQTLs) associated with our DNAm sites of interest. The mQTLdb database contains the results of a large-scale study based on the ARIES sample in ALSPAC (from which our subsample is derived), characterizing genome-wide significant cis effects (i.e. SNP within ±1000 base pairs of the DNAm site) and trans effects (i.e. ±1 million base pairs) on DNAm levels across Illumina 450k probes at five different life stages, including cord blood DNAm at birth (Gaunt et al., 2016). Here, we searched for mQTLs based on results from the conditional Genome-wide Complex Trait Analysis (GCTA), which was used to identify mQTLs with the most representative, independent effect on each DNAm site in order to account for linkage disequilibrium (Gaunt et al., 2016). With regards to environmental influences, we examined associations between DNAm levels in the identified sites and prenatal exposures (i.e. maternal diet, smoking, alcohol use and stressful events) using Pearson’s bivariate correlations. Again, the significance threshold was set at an FDR-corrected level of q<0.05 to adjust for multiple comparisons.
Results
Neonatal DNA methylation and early-onset CP
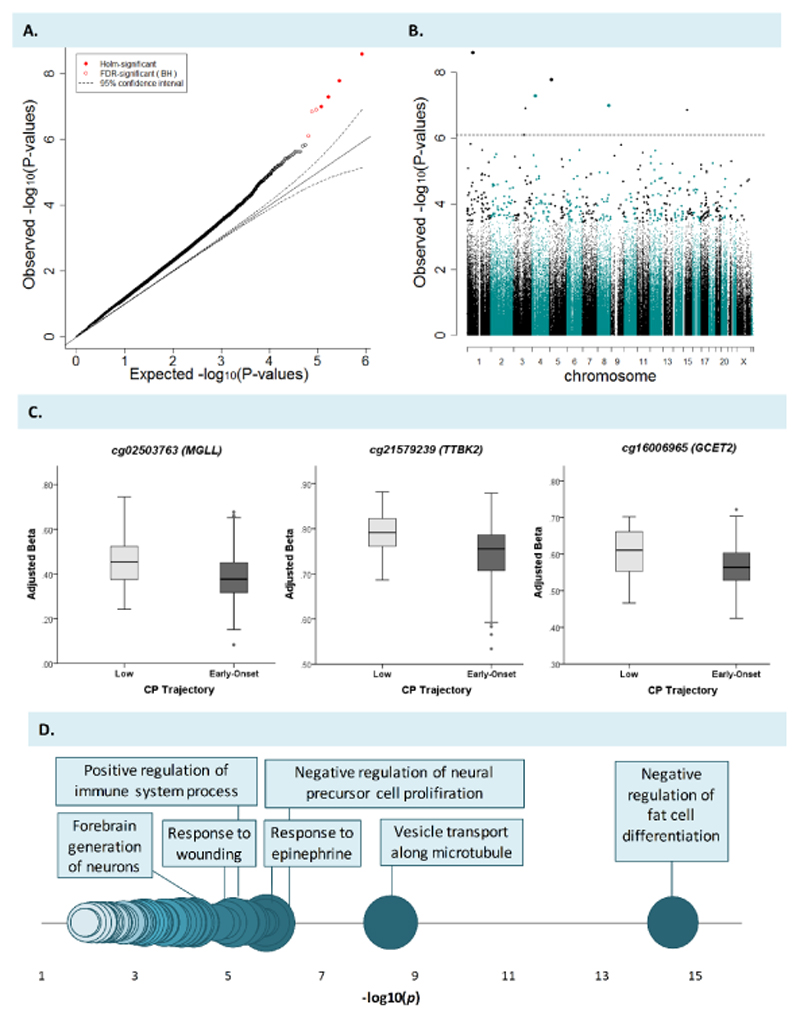
The Q-Q plot and Manhattan plot for the epigenome-wide association analysis at birth are displayed in Figure 1 (A-B). Table 1 shows the seven differentially methylated sites (q-value < 0.05) between early-onset vs low CP groups, as identified via our genome-wide analysis. Group differences were medium to large in effect size based on Cohen’s guidelines (d = 0.66 – 0.84), and ranged from 4% to 8% in terms of mean DNAm difference. Of note, all seven sites remained differentially methylated after additionally adjusting for gestational age. DNAm levels across these sites were all significantly inter-correlated (both positive and negative correlations observed; see OS3). Three of the sites had genic annotations (Figure 1C): (i) cg02503763, located in the promoter regulatory region of MGLL, a gene implicated in endocannabinoid signaling and nociperception (Iwasaki, Ishiguro, Higuchi, Onaivi, & Arinami, 2007); (ii) cg21579239, annotated to the 5'UTR region of TTBK2, a gene involved in tau protein phosphorylation, of which mutations have been linked to neurodegenerative disorders (Liao, Yang, Weng, Kuo, & Chang, 2015); and (iii) cg16006965, located in the promoter regulatory region of GCET2, a gene implicated in immune function (Pan et al., 2007). The other four differentially methylated sites identified at birth were distal to annotated transcripts (see OS4 for boxplots): cg12628061 located between AK127270 (-407kb) and PPAP2B (-507kb), cg02131674 located closest to BASP1 (-123kb), cg15384400 located between ARAP2 (-12kb) and DTHD1 (+26kb), and cg07128473 located between TRPS1 (-210kb) and EIF3H (+876kb). In all cases – except for probe cg15384400 – methylation of these sites was lower in the early-onset vs low CP groups. These seven sites were then viewed in Genome Browser for functional characterization, based on ENCODE data on regulatory elements. Four of the DNAm loci coincided with transcription factor binding sites, including MGLL cg02503763 and GCET2 cg16006965, and implicated a number of shared transcription factors, including MAX (4/4 loci), POLR2A, RUNX3 and EP300 (3/4 loci). The majority of DNAm sites also overlapped with histone marks (6/7 loci) and DNAse I hypersensitive clusters (4/7 loci), suggesting that they are located in genomic regions that are likely to play regulatory roles in transcription. Gene ontology analysis (n probes = 1101; n genes = 758) further indicated that DNAm sites (at birth) that differentiate between CP groups are annotated to genes involved in a range of biological processes, including regulation of fat cell differentiation and immune system process, response to epinephrine and wounding, as well as forebrain generation of neurons and CNS neuron differentiation (2.95E-15 <p< 6.66E-05, Figure 1D). GO results for the top 20 enriched biological pathways (including those displayed in Figure 1D) are provided in OS5.
Figure 1. Methylomic variation at birth prospectively associated with early-onset vs low conduct problems in childhood.
Epigenome-wide associations between DNA methylation at birth and early-onset vs low conduct problems (CP), including Q-Q plot (A), Manhattan plot (B) and scatterplots of the top 3 differentially methylated sites with genic annotations (C). Significantly enriched biological processes at birth are shown in (D), based on gene ontology (GO) analysis of genes annotated to probes that associate with early-onset vs low CP (p < 0.001). Circles represent GO terms that survive FDR-correction and contain at least two genes. The X axis represents -log(10) p-values. The opacity of the circles indicates level of significance (darker = more significant). The size of the circles indicates the percentage of genes in our results for a given pathway compared to the total number of genes in the same pathway (i.e. larger size = larger %).
Table 1. DNA methylation sites at birth that differentiate early-onset vs low CP children (genome-wide significance; q<0.05).
| Probe | Gene | Chr | Genomic location | Position | p-value | q-value (FDR) | Direction | Early-onset CP Mean | Low CP Mean | % Diff | Cohen's d |
|---|---|---|---|---|---|---|---|---|---|---|---|
| cg12628061 | -- | 1 | -- | 56453730 | 2.47E-09 | 1.00E-03 | ↓ | 0.77 | 0.81 | 4% | 0.84 |
| cg02131674 | -- | 5 | -- | 17341065 | 1.64E-08 | 3.00E-03 | ↓ | 0.77 | 0.81 | 4% | 0.78 |
| cg15384400 | -- | 4 | -- | 36257279 | 5.09E-08 | 7.00E-03 | ↑ | 0.26 | 0.21 | 5% | 0.79 |
| cg07128473 | -- | 8 | -- | 116891579 | 1.00E-07 | 0.01 | ↓ | 0.83 | 0.86 | 3% | 0.76 |
| cg02503763 | MGLL | 3 | TSS1500 | 127542386 | 1.23E-07 | 0.01 | ↓ | 0.38 | 0.46 | 8% | 0.73 |
| cg21579239 | TTBK2 | 15 | 5'UTR | 43211292 | 1.40E-07 | 0.01 | ↓ | 0.74 | 0.78 | 4% | 0.73 |
| cg16006965 | GCET2 | 3 | TSS1500 | 111852325 | 7.85E-07 | 0.04 | ↓ | 0.53 | 0.6 | 7% | 0.66 |
Arrows represent the direction of methylation differences, where ↑ indicates higher methylation levels in early-onset vs low CP children, while ↓ indicates lower methylation levels in early-onset vs low CP children. Cohen’s d: .20 is a small effect size, .50 is a medium effect size, .80 is a large effect size.
In light of the above findings, we conducted two follow-up analyses to further investigate the relationship between DNAm at birth and early-onset CP.
Methylomic variation in MGLL, TTBK2 and GCET2
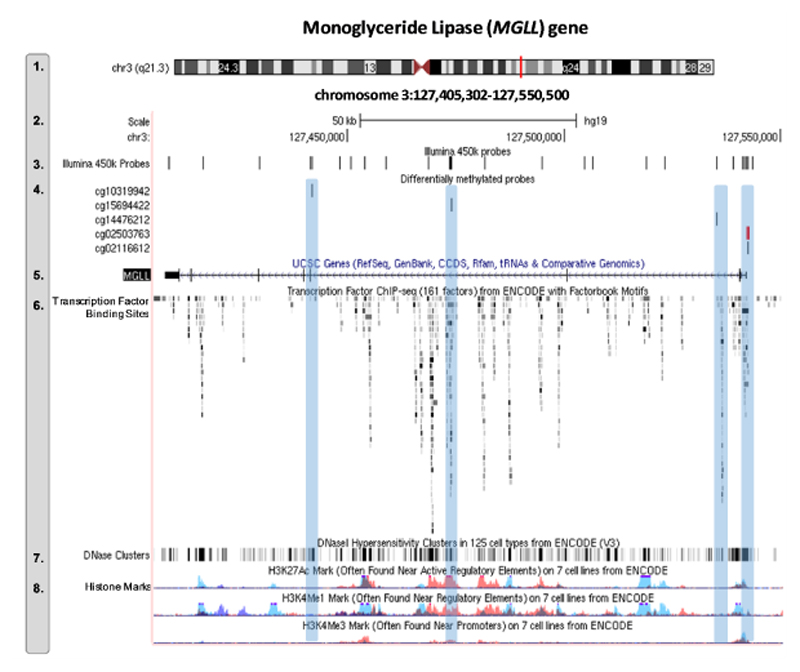
Our genome-wide analyses indicated that three of the seven sites identified as differentiating early-onset vs low CP children were annotated to genes, including MGLL, TTBK2 and GCET2 (one site each). As an additional step, we examined whether any other probes in the vicinity of these genes associated with early-onset CP at a nominal level. While none of the other probes in TTBK2 (n total = 10 probes) or GCET2 (n total = 19 probes) associated with early-onset vs low CP (p > 0.05), four additional probes in MGLL were differentially methylated between groups, including another probe in the TSS1500 region (cg02116612; p = 1.95E-05; significant at gene-level Bonferroni correction: 0.05/26 total annotated probes = 0.002) and three probes in the gene body region (cg14476212, p = 0.028; cg10319942, p = 0.034; and cg15694422, p = 0.048). The location of these probes and their relationship to ENCODE regulatory elements is displayed in Figure 2 (for more detailed information, see OS6). Of note, two of these additional probes correlated significantly with the genome-wide significant site MGLL cg02503763 – including a stronger association with the most proximal probe in the TSS1500 region (cg02116612; r = 0.46, p = 6.04E-15) and a weaker association with one of the probes in the gene body (cg10319942; r = 0.15, p = 0.01).
Figure 2. Functional characterization of MGLL DNA methylation sites associated with early-onset conduct problems.
Expanded views from the UCSC genome browser of the MGLL gene showing the position of the DNAm sites associated with early-onset CP relative to ENCODE regulatory elements. Track numbers are displayed on the left-hand site, and represent the following (1) genomic positon of MGLL in chromosome 3; (2) genomic coordinates and scale; (3) location of all Illumina 450k probes that map onto the MGLL gene (n = 26); (4) location of differentially methylated probes associated with early-onset vs low conduct problems (n = 5). These are highlighted in blue to facilitate comparison with the regulatory elements displayed in lower tracks (track 6-8). In red is the probe that survived genome-wide correction (MGLL cg02503763; all other probes significant at p<0.05); (5) schematic representation of the MGLL gene; (6) location of transcription factors (based on ChIP-seq data from 91 cell types), where darker shades indicate a stronger signal occupancy; (7) DNaseI hypersensitivity clusters (based on ChIP-seq data from 125 cell types), where darker shades also indicate a stronger signal; and (8) levels of enrichment of three histone marks (H3K27Ac, H3K4Me1, and H3K4Me3) across three cell-types, including blood (GM12878 [red], K562 [purple]) and umbilical vein endothelial (HUVEC [blue]) cells.
Methylomic variation in candidate genes for conduct problems
As shown in Table 1, none of the genome-wide significant loci identified resided in ‘traditional’ candidate genes for CP. In order to maximise comparability with existing molecular studies, we carried out a follow-up analysis to investigate DNAm variation in the vicinity of genes that have been previously implicated in CP-relevant phenotypes. The selection of candidate genes was informed by a recent systematic review (Waltes, Chiocchetti, & Freitag, 2015) and meta-analysis (Pappa et al., 2015) of genetic influences on aggression. Specifically, we selected a total of 15 genes (n total = 387 probes), across (i) dopaminergic (MAOA [14 probes], COMT [22 probes], SLC6A3 [52 probes],DRD2 [22 probes], DRD4 [21 probes]); (ii) serotonergic (SLC6A4 [14 probes], HTR1A [14 probes], HTR2A [25 probes], TPH1 [4 probes], TPH2 [18 probes]); and (iii) neuroendocrine and neurodevelopmental pathways (NR3C1 [35 probes], FKBP5 [32 probes], AVP [14 probes], OXTR [17 probes], BDNF [73 probes]). As done by Weder et al. (2014), we used a gene-level Bonferroni correction for these hypothesis-driven analyses, correcting for the total number of probes within each gene.
In total, 12% of probes were differentially methylated between groups at a nominal significance threshold (p<0.05), with at least one probe associating with early-onset CP for each gene (see OS7). However, only 4 of these loci survived gene-level Bonferroni-correction (see Table 2), including sites annotated to BDNF (cg01225698, cg18354203), FKBP5 (cg07061368) and the MAOA promoter region (i.e. TSS200; cg05443523). Effect sizes for these loci were small (d = 0.44-0.49), and mean group DNAm differences ranged from 1% to 3%. Based on ENCODE data, all four probes coincided with transcription factor binding sites and histone marks, with two of the probes additionally coinciding with DNase I hypersensitivity clusters. As with the loci identified via our epigenome-wide analysis, these four probes remained significantly associated with CP trajectory after controlling for gestational age.
Table 2. Candidate gene follow-up: Loci that differentiate between early-onset and low CP children (gene-level Bonferroni correction).
| Probe | Chr | Genomic Location | Position | p-value | q-value (genome-wide) | Bonferroni (gene-level) | Direction | Early-onset CP Mean | Low CP Mean | % Diff | Cohen's d |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BDNF | |||||||||||
| cg01225698 | 11 | Body; TSS1500 | 27742355 | 3.44E-04 | 0.30 | Significant | ↑ | 0.16 | 0.15 | 1% | 0.47 |
| cg18354203 | 11 | Body; 5'UTR | 27696004 | 4.97E-04 | 0.33 | Significant | ↓ | 0.72 | 0.75 | 3% | 0.49 |
| FKBP5 | |||||||||||
| cg07061368 | 6 | 5'UTR | 35631736 | 9.96E-04 | 0.37 | Significant | ↓ | 0.81 | 0.83 | 2% | 0.44 |
| MAOA | |||||||||||
| cg05443523 | X | TSS200 | 43515213 | 1.17E-03 | 0.38 | Significant | ↑ | 0.36 | 0.33 | 3% | 0.44 |
N.b. Arrows represent the direction of methylation differences, where ↑ indicates higher methylation levels in early-onset vs low CP children, while ↓ indicates lower methylation levels in early-onset vs low CP children. Cohen’s d: .20 is a small effect size, .50 is a medium effect size, .80 is a large effect size.
Comparison of top DNAm probes between early-onsets who persist vs desist in CP
Within the early-onset CP group, none of the DNAm sites identified in the previous step (i.e. 7 FDR-corrected genome-wide significant loci and 4 Bonferroni-corrected loci from the candidate gene follow-up analyses) were found to differentiate children who will go on to follow a persisting vs desisting trajectory of conduct problems over time (p > 0.05, see OS8 for full results). This lack of differences suggest that, while methylomic variation at birth prospectively associates with an early-onset of CP (i.e. distinguishing between high vs low CP early in childhood), this variation is not likely to contribute towards heterogeneity in developmental trajectories of conduct problems observed within early-onset children.
Potential relevance of genetic and in utero environmental influences on the identified DNAm sites
The 11 DNAm sites associated with early-onset CP were carried forward to explore associations with potential genetic and environmental influences. Based on mQTLdb search, we found that none of the identified loci were associated with cis or trans mQTLs, suggesting that DNAm levels across these sites are not likely to be heavily influenced by known genetic polymorphisms. Bivariate correlations between the DNAm loci and prenatal exposures are shown in Table 3, controlling for sex and cell-type. Of the 11 probes examined, 7 were nominally correlated with one or more exposures (p < 0.05), but none of these associations remained significant after multiple correction (q > 0.05). The majority of associations involved maternal smoking and alcohol use during the first trimester of pregnancy. Around half of associations identified remained significant after additionally controlling for all other prenatal exposures (i.e. unique association, see Table 3).
Table 3. Associations between prenatal environmental exposures and loci that are differentially methylated in early-onset vs low CP children.
| Prenatal environment | ||||||||
|---|---|---|---|---|---|---|---|---|
| Healthy diet | Unhealthy diet | Maternal smoking | Maternal alcohol use | Life events | Contextual risks | Parental risks | Interpersonal risks | |
| Genome-wide analyses | ||||||||
| cg12628061 | 0.09 | -0.06 | -0.16 (p = 0.01) a | 0.17 (p = 0.01) | 0.09 | 0.02 | -0.09 | 0.07 |
| cg02131674 | 0.09 | -0.07 | -0.10 | 0.10 | 0.10 | 0.05 | -0.09 | 0.04 |
| cg15384400 | -0.07 | 0.07 | 0.09 | 0.00 | -0.05 | -0.01 | 0.11 | 0.05 |
| cg07128473 | 0.14 (p = 0.03) a | -0.08 | -0.10 | 0.12 | 0.05 | 0.05 | -0.07 | 0.06 |
| cg02503763 (MGLL) | 0.03 | -0.01 | -0.14 (p = 0.02) | 0.13 (p = 0.03) a | -0.05 | -0.04 | -0.08 | -0.02 |
| cg21579239 (TTBK2) | 0.08 | -0.07 | -0.16 (p = 0.01) | 0.09 | 0.09 | 0.04 | -0.12 | 0.02 |
| cg16006965 (GCET2) | 0.05 | 0.01 | -0.10 | 0.17 (p = 0.01) | 0.07 | 0.09 | -0.07 | 0.04 |
| Candidate gene follow-up analyses | ||||||||
| cg01225698 (BDNF) | -0.10 | 0.08 | 0.05 | -0.03 | -0.02 | 0.04 | 0.03 | 0.02 |
| cg18354203 (BDNF) | 0.06 | -0.12 | -0.09 | 0.00 | -0.01 | -0.03 | -0.06 | 0.07 |
| cg07061368 (FKBP5) | 0.00 | -0.03 | -0.08 | 0.20 (p = 0.001) a | 0.11 | 0.08 | -0.03 | 0.10 |
| cg05443523 (MAOA) | -0.09 | 0.10 | 0.03 | -0.09 | -0.13 (p = 0.001) a | -0.02 | -0.01 | -0.04 |
N.B. All associations control for sex and cell-type.
Association remains significant after additionally controlling for other exposures, p<0.05
Discussion
This is the first study to investigate the epigenetic landscape of early-onset conduct problems, using a prospective design. The study offers a number of strengths, including the examination of genome-wide data collected prior to the emergence of CP symptoms, the inclusion of the largest epigenetic CP sample to date, allowing for adequately-powered group comparisons (Tsai & Bell, 2015), as well as the integration of environmental, DNAm and phenotypic data. We highlight here three key findings: (i) methylomic variation across multiple loci at birth differentiated children who go on to develop early-onset vs low CP; (ii) DNAm levels across these loci were comparable for early-onsets who persist vs desist in CP over time; and (iii) several of the identified loci showed suggestive associations with prenatal exposures, while none were linked to known genetic mQTLs.
Genome-wide findings point to the potential importance of methylomic variation at birth
The use of a genome-wide, hypothesis-free approach enabled us to identify novel loci for CP, pointing to potential avenues for future investigation. Specifically, we found that seven loci at birth prospectively differentiated CP groups, showing a moderate-to-large effect size difference between children who go on to develop early-onset vs low CP. Functional characterization using ENCODE data indicated that the majority of the identified sites coincided with regulatory elements that play a key role in gene expression, including transcription factor binding sites, DNase I hypersensitivity clusters (i.e. regions of open chromatin) and histone marks. Six of the loci were significantly hypomethylated in early-onset vs low CP children, and three were annotated to genes. Of these, MGLL encodes a serine hydrolase that converts monoacylglycerides to free fatty acids and glycerol and acts as a major endocannabinoid metabolic enzyme (Iwasaki et al., 2007). Interestingly, previous studies have shown that MGLL activity associates with substance abuse (Hopfer et al., 2007), which is highly comorbid with CP (McAdams, Salekin, Marti, Lester, & Barker, 2014). Furthermore, CB1 cannabinoid receptor function (modulated indirectly by MGLL via 2-Arachidonoylglycerol [2-AG] activation) has been shown to mediate aggression in animal knockout studies (Rodriguez-Arias et al., 2013). Together, these findings support a link between MGLL function and CP-relevant phenotypes, although more work will be needed to delineate the functional effects of MGLL methylation on broader endocannabinoid signaling and downstream consequences on behavior. We do note that in follow-up analyses, a number of additional DNAm sites associated with early-onset CP across the promoter region and gene body of MGLL, as well as coinciding with key regulatory elements, which further supports a link between epigenetic regulation of this gene and CP.
Another DNAm site that showed CP-related DNAm at birth was annotated to TTBK2. This gene is highly expressed in subcortical brain structures (e.g. cerebellum, hippocampus) and is involved in the phosphorylation of tau and tubulin, two key microtubule stabilization proteins that are robustly associated with multiple nervous system pathologies, including Alzheimer’s disease (Liao et al., 2015). Although we are not aware of studies directly investigating the link between TTBK2 and CP-relevant phenotypes, it is noteworthy that associations between levels of tau phosphorylation in the brain and aggressive behavior have been previously reported in patients with Alzheimer’s disease (e.g., Guadagna, Esiri, Williams, & Francis, 2012). As such, it will be of interest in future to examine whether TTBK2 methylation may contribute to early-onset CP via microtubule alterations in the brain.
The third gene identified, GCET2, plays a role in immune processes, including cytokine function (Pan et al., 2007). While an increasing number of studies has documented a link between inflammation and psychiatric phenotypes, including childhood aggression (e.g. Provencal et al., 2013), little is known about the function of this gene in relation to CP. It is interesting; however, that a genome-wide study recently identified GCET2 expression levels as a potential biomarker for antidepressant treatment response (Hennings et al., 2015), pointing to a potential role of this gene in mood regulation.
DNA methylation of candidate genes: Maximizing comparability with previous studies
Consistent with what has been previously reported in the psychiatric epigenetic literature (e.g. Cecil, Walton, & Viding, 2015), we found that none of the DNAm sites identified via our hypothesis-free, epigenome-wide analysis were annotated to genes that are typically examined in hypothesis-driven, candidate gene studies. In order to maximize comparability with existing molecular studies, we carried out an additional analysis focusing on DNAm in the vicinity of 15 genes that have been extensively investigated in relation to CP and related phenotypes (e.g. aggression; Pappa et al., 2015; Waltes et al., 2015). It is important to note that these studies have primarily focused on genetic (as opposed to epigenetic) contributions to CP, so that a direct test of replication was not possible. Rather, this follow-up analysis was designed to explore the potential role of these genes in early-onset CP from an epigenetic perspective. Although none of the DNAm sites annotated to candidate genes passed genomewide correction, sub-threshold associations were identified across three genes, including BDNF (two probes), MAOA (one probe) and FKBP5 (one probe). Of note, effect sizes were small but not trivial (>0.44), suggesting that these DNAm differences may hold relevance for understanding biological vulnerability for early-onset CP.
BDNF encodes for a neutrophic protein that performs a wide range of functions, including regulation of neurodevelopment, synaptic plasticity and mood. While a link between aggression and BDNF activity has been documented by multiple lines of evidence – including findings from genetic, knock-out, endocrine and pharmacological studies (Waltes et al., 2015) – this is the first report, to our knowledge, of an association with BDNF DNAm levels. The role of MAOA activity on aggression has also received widespread attention from both the animal and human literature, due to its key role in the degradation of amine neurotransmitters in the brain, including dopamine, norepinephrine and serotonin (Nelson & Trainor, 2007). Consistent with our findings, a recent study reported that hypermethylation of the MAOA promoter region (with an average DNAm difference between groups of 3% - the same as observed in the present study) was associated with antisocial personality disorder in a forensic population, as well as decreased gene expression and dysregulation of serotonin blood serum levels (Checknita et al., 2015). Interestingly, MAOA promoter DNAm in blood has also been shown to robustly predict MAOA enzymatic activity in the brain (Shumay, Logan, Volkow, & Fowler, 2012), suggesting that peripheral MAOA DNAm status may be a promising biomarker for functional levels in live central tissue, which is most relevant to CP. Finally, the FKBP5 gene, which codes a co-chaperone of the glucocorticoid receptor that regulates its sensitivity, has been extensively investigated for its role in mediating stress, hormonal and immune responses to adverse experiences, particularly in the context of gene-environment interactions (Zannas & Binder, 2014). For example, FKBP5 polymorphisms have been found to interact with childhood trauma (e.g. maltreatment) to predict a range of psychiatric outcomes, including childhood aggression (Bryushkova et al., 2016). While emerging evidence suggests that FKBP5 methylation may mediate these observed GxE interactions (Zannas, Wiechmann, Gassen, & Binder, 2015), more work is needed to characterize its relevance to CP behaviour.
Heterogeneity in developmental trajectories within the early-onset CP group
Whereas the identified DNAm sites at birth differentiated children who develop early-onset vs low CP, these same sites did not further discriminate between early-onsets who go on to follow a persisting (i.e. EOP) vs desisting (i.e. CL) trajectory of CP into adolescence. This lack of associations may reflect a number of possibilities. For example, it is possible that, at birth, EOP and CL children may be phenotypically and biologically similar. In other words, while they may both be at increased risk for early-onset CP relative to typical children, it may not yet be possible to separate those who will persist vs desist in CP behavior over time. Given that EOP vs CL children start to diverge in levels of CP around late childhood and predict different outcomes in adulthood (David P Farrington, Gallagher, Morley, Ledger, & West, 1988; Terrie E Moffitt, Caspi, Harrington, & Milne, 2002), it will be of interest in future to extend epigenetic analyses later in development, so as to investigate whether these behavioural differences may coincide with underlying epigenetic differences. In addition, the examination of DNAm at later time points would enable one to test potential associations with postnatal environmental influences, which are likely to play an important role in the divergence between EOP and CL trajectories. It is also possible that statistically, our analyses were underpowered to examine within-group differences in DNAm patterns among the early-onset group. Larger longitudinal studies will be needed in future to enable a more fined-tuned comparison of DNAm patterns between EOP and CL children across multiple developmental periods.
Genetic and prenatal influences on DNAm patterns associated with early-onset CP
As a final step, we investigated whether DNAm loci at birth that differentiated between early-onset vs low CP children (both at the genome-wide and candidate-gene level) were also associated with potential genetic and environmental influences relevant to CP (E. D. Barker & Maughan, 2009; Salvatore & Dick, 2016). Based on findings from a large-scale study of genetic effects on DNAm (Gaunt et al., 2016), we found that none of the identified loci associated with known cis or trans mQTLs. While this suggests that the identified loci were unlikely to be heavily influenced by genetic structure, it is important to note that the heritability of DNAm patterns is greater than what can currently be explained using known mQTLs (Gaunt et al., 2016). As such, it is still possible that the identified loci may have been influenced by polygenic effects, involving many mQTL that each explain too little variance in isolation to be detected with currently available samples.
With regards to environmental influences, we identified suggestive associations between the identified DNAm sites and prenatal exposures. Of note, we found that proximal exposures (e.g. maternal smoking and alcohol use) correlated with DNAm more strongly than more distal exposures (e.g. life events). Albeit preliminary, these findings are consistent with previous studies showing that DNAm is associated with environmental influences (e.g. Lewis & Olive, 2014), with smoking-related effects, in particular, being amongst the most replicated across epigenetic studies (Gao, Jia, Zhang, Breitling, & Brenner, 2015). For example, we found that prenatal smoking correlated with hypomethylation of the MGLL probe, which in turn prospectively associated with early-onset CP. This is of interest given that activity of this gene has been shown to influence nicotine withdrawal in animals and humans (Muldoon et al., 2015), as well as interacting with environmental exposures to predict cannabis dependence symptoms and amygdala function (Carey et al., 2015). However, it is important to note that the associations identified in the present study were based on correlational analyses and did not survive multiple correction. Consequently, they should be interpreted with caution and considered more as well-grounded hypotheses for further examination in larger longitudinal studies.
Limitations
Findings should be interpreted in light of a number of limitations. First, because results from this study were based on DNAm collected from peripheral samples, the extent to which they may be tissue-specific is unclear. More research will be needed to assess the relevance of the identified DNAm sites to brain tissue, as well as characterizing their potential role in CP-related brain-based phenotypes. Furthermore, because we did not have access to RNA, functional characterization of the identified loci was performed using recorded ENCODE data. However, integration of transcriptomic data will mark an important step toward establishing the functional significance and downstream effects of the observed DNAm changes. Second, the current study was based on a community sample of children with relatively low rates of CP. In future, it will be important to replicate our findings in high-risk populations who show more severe externalising problems (e.g. young offenders, psychiatric inpatients). Furthermore, the use of a larger number of cases vs controls will make it possible to examine potential moderators in the relationship between DNAm and early-onset CP (e.g. sex). Finally, although we used a prospective design where DNAm was collected prior to the emergence of CP symptoms, the results from the study are correlational and cannot be taken to reflect causal pathways. As such, findings should be considered as hypothesis-generating and in need of replication. In future, the application of advanced inference methods (e.g. two-step Mendelian randomization; Pingault, Cecil, Murray, Munafo & Viding, 2016; Relton & Davey Smith, 2012), will offer unique opportunities to establish causal relationship between prenatal environmental risk, DNAm and CP.
Conclusion
In this study, we find that DNA methylation at multiple loci at birth prospectively associated with early-onset vs low conduct problems. These findings highlight the neonatal period as a potentially important window of biological vulnerability for conduct problems, as well as pinpointing novel potential risk markers for future investigation. Suggestive evidence of associations between the identified loci and prenatal exposures also lend preliminary support for a link between the early environment, DNA methylation and the development of early-onset conduct problems.
Supplementary Material
Acknowledgements
We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. With regard to the ALSPAC DNA methylation, we thank all involved, particularly the laboratory scientists and bioinformaticians who contributed considerable time and expertise to the data in this paper. The UK Medical Research Council and the Wellcome Trust (Grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. This publication is the work of the authors who will serve as guarantors for the contents of this paper. This research was specifically supported by the National Institute of Child and Human Development grant (E. D.B.; grant ref: R01HD068437). C.C. is supported by the Economic and Social Research Council (grant ref: ES/N001273/1).
References
- Barfield RT, Kilaru V, Smith AK, Conneely KN. CpGassoc: an R function for analysis of DNA methylation microarray data. Bioinformatics. 2012;28(9):1280–1281. doi: 10.1093/bioinformatics/bts124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett BE, Hanna B, Parker G. Life event scales for obstetric groups. Journal of psychosomatic research. 1983;27(4):313–320. doi: 10.1016/0022-3999(83)90054-5. [DOI] [PubMed] [Google Scholar]
- Barker DJ. The origins of the developmental origins theory. Journal of internal medicine. 2007;261(5):412–417. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- Barker ED, Cecil CAM, Walton E, Meehan A. In: The Wiley Handbook of Disruptive and Impulse-Control Disorders. Lochman J, Matthys W, editors. New York: Wiley; 2017. Genetic contributions to disruptive and impulse-control disorders. [Google Scholar]
- Barker ED, Kirkham N, Ng J, Jensen SK. Prenatal maternal depression symptoms and nutrition, and child cognitive function. Br J Psychiatry. 2013;203(6):417421. doi: 10.1192/bjp.bp.113.129486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker ED, Maughan B. Differentiating early-onset persistent versus childhoodlimited conduct problem youth. Am J Psychiatry. 2009;166(8):900–908. doi: 10.1176/appi.ajp.2009.08121770. [DOI] [PubMed] [Google Scholar]
- Barker ED, Oliver BR, Maughan B. Co-occurring problems of early onset persistent, childhood limited, and adolescent onset conduct problem youth. J Child Psychol Psychiatry. 2010;51(11):1217–1226. doi: 10.1111/j.1469-7610.2010.02240.x. [DOI] [PubMed] [Google Scholar]
- Bergman Y, Cedar H. DNA methylation dynamics in health and disease. Nat Struct Mol Biol. 2013;20(3):274–281. doi: 10.1038/nsmb.2518. [DOI] [PubMed] [Google Scholar]
- Booij L, Tremblay RE, Leyton M, Seguin JR, Vitaro F, Gravel P, et al. Diksic M. Brain serotonin synthesis in adult males characterized by physical aggression during childhood: a 21-year longitudinal study. PLoS ONE. 2010;5(6):e11255. doi: 10.1371/journal.pone.0011255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, et al. Davey Smith G. Cohort Profile: the 'children of the 90s'--the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2013;42(1):111–127. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GW, Sklair F, Harris TO, Birley JLT. Life-events and psychiatric disorders Part 1: some methodological issues. Psychological Medicine. 1973;3(01):74–87. doi: 10.1017/s0033291700046365. [DOI] [PubMed] [Google Scholar]
- Bryushkova L, Zai C, Chen S, Pappa I, Mileva V, Tiemeier H, et al. Beitchman JH. FKBP5 Interacts with Maltreatment in a Sample of Children with Extreme, Pervasive, and Persistent Aggression. Psychiatry research. 2016 doi: 10.1016/j.psychres.2015.09.052. [DOI] [PubMed] [Google Scholar]
- Bywater TJ. Perspectives on the Incredible Years programme: psychological management of conduct disorder. Br J Psychiatry. 2012;201:85–87. doi: 10.1192/bjp.bp.111.107920. [DOI] [PubMed] [Google Scholar]
- Carey CE, Agrawal A, Zhang B, Conley ED, Degenhardt L, Heath AC, et al. Bogdan R. Monoacylglycerol lipase (MGLL) polymorphism rs604300 interacts with childhood adversity to predict cannabis dependence symptoms and amygdala habituation: Evidence from an endocannabinoid system-level analysis. J Abnorm Psychol. 2015;124(4):860–877. doi: 10.1037/abn0000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecil CA, Lysenko LJ, Jaffee SR, Pingault JB, Smith RG, Relton CL, et al. Barker ED. Environmental risk, Oxytocin Receptor Gene (OXTR) methylation and youth callous-unemotional traits: a 13-year longitudinal study. Mol Psychiatry. 2014;19(10):1071–1077. doi: 10.1038/mp.2014.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecil CAM, Walton E, Smith RG, Viding E, McCrory EJ, Relton CL, et al. Barker ED. DNA methylation and substance-use risk: a prospective, genome-wide study spanning gestation to adolescence. Translational Psychiatry. 2016;6(12):e976. doi: 10.1038/tp.2016.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecil CM, Walton E, Viding E. DNA Methylation, Substance Use and Addiction: a Systematic Review of Recent Animal and Human Research from a Developmental Perspective. Current Addiction Reports. 2015;2(4):331–346. doi: 10.1007/s40429-015-0072-9. [DOI] [Google Scholar]
- Checknita D, Maussion G, Labonte B, Comai S, Tremblay RE, Vitaro F, et al. Turecki G. Monoamine oxidase A gene promoter methylation and transcriptional downregulation in an offender population with antisocial personality disorder. Br J Psychiatry. 2015;206(3):216–222. doi: 10.1192/bjp.bp.114.144964. [DOI] [PubMed] [Google Scholar]
- Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, et al. Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8(2):203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchetti D, Rogosch FA. Equifinality and multifinality in developmental psychopathology. Dev Psychopathol. 1996;8(04):597–600. [Google Scholar]
- Cohen J. Statistical Power Analysis for the Behavioral Sciences. 2nd edn. Hillsdale, New Jersey: L: Erlbaum; 1988. [Google Scholar]
- Colman I, Murray J, Abbott RA, Maughan B, Kuh D, Croudace TJ, Jones PB. Outcomes of conduct problems in adolescence: 40 year follow-up of national cohort. 2009;338 doi: 10.1136/bmj.a2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadds MR, Moul C, Cauchi A, Dobson-Stone C, Hawes DJ, Brennan J, Ebstein RE. Methylation of the oxytocin receptor gene and oxytocin blood levels in the development of psychopathy. Dev Psychopathol. 2014;26(1):33–40. doi: 10.1017/s0954579413000497. [DOI] [PubMed] [Google Scholar]
- Egger HL, Angold A. Common emotional and behavioral disorders in preschool children: presentation, nosology, and epidemiology. Journal of Child Psychology and Psychiatry. 2006;47(3–4):313–337. doi: 10.1111/j.1469-7610.2006.01618.x. [DOI] [PubMed] [Google Scholar]
- Farrington DP. In: Clinical assessment of dangerousness: Empirical contributions. Pinard Georges-Franck, Pagani Linda., editors. 2001. Predicting adult official and self-reported violence. 2001. [Google Scholar]
- Farrington DP, Gallagher B, Morley L, Ledger RJS, West DJ. Are there any successful men from criminogenic backgrounds? Psychiatry. 1988;51(2):116–130. [PubMed] [Google Scholar]
- Fergusson DM, John Horwood L, Ridder EM. Show me the child at seven: the consequences of conduct problems in childhood for psychosocial functioning in adulthood. Journal of Child Psychology and Psychiatry. 2005;46(8):837–849. doi: 10.1111/j.1469-7610.2004.00387.x. [DOI] [PubMed] [Google Scholar]
- Fraser A, Macdonald-Wallis C, Tilling K, Boyd A, Golding J, Davey Smith G, et al. Lawlor DA. Cohort Profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int J Epidemiol. 2013;42(1):97–110. doi: 10.1093/ije/dys066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Jia M, Zhang Y, Breitling LP, Brenner H. DNA methylation changes of whole blood cells in response to active smoking exposure in adults: a systematic review of DNA methylation studies. Clin Epigenetics. 2015;7:113. doi: 10.1186/s13148-015-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt TR, Shihab HA, Hemani G, Min JL, Woodward G, Lyttleton O, et al. Ho K. Systematic identification of genetic influences on methylation across the human life course. Genome biology. 2016;17(1):1. doi: 10.1186/s13059-016-0926-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman R. Psychometric properties of the strengths and difficulties questionnaire. J Am Acad Child Adolesc Psychiatry. 2001;40(11):1337–1345. doi: 10.1097/00004583-200111000-00015. [DOI] [PubMed] [Google Scholar]
- Guadagna S, Esiri MM, Williams RJ, Francis PT. Tau phosphorylation in human brain: relationship to behavioral disturbance in dementia. Neurobiology of Aging. 2012;33(12):2798–2806. doi: 10.1016/j.neurobiolaging.2012.01.015. [DOI] [PubMed] [Google Scholar]
- Guillemin C, Provencal N, Suderman M, Cote SM, Vitaro F, Hallett M, et al. Szyf M. DNA methylation signature of childhood chronic physical aggression in T cells of both men and women. PLoS ONE. 2014;9(1):e86822. doi: 10.1371/journal.pone.0086822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennings JM, Uhr M, Klengel T, Weber P, Putz B, Touma C, et al. Lucae S. RNA expression profiling in depressed patients suggests retinoid-related orphan receptor alpha as a biomarker for antidepressant response. TranslPsychiatry. 2015;5:e538. doi: 10.1038/tp.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfer CJ, Lessem JM, Hartman CA, Stallings MC, Cherny SS, Corley RP, et al. Crowley TJ. A genome-wide scan for loci influencing adolescent cannabis dependence symptoms: evidence for linkage on chromosomes 3 and 9. Drug Alcohol Depend. 2007;89(1):34–41. doi: 10.1016/j.drugalcdep.2006.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Ishiguro H, Higuchi S, Onaivi ES, Arinami T. Association study between alcoholism and endocannabinoid metabolic enzyme genes encoding fatty acid amide hydrolase and monoglyceride lipase in a Japanese population. Psychiatr Genet. 2007;17(4):215–220. doi: 10.1097/YPG.0b013e32809913d8. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jaffee SR, Caspi A, Moffitt TE, Dodge KA, Rutter M, Taylor A, Tully LA. Nature X nurture: genetic vulnerabilities interact with physical maltreatment to promote conduct problems. Dev Psychopathol. 2005;17(1):67–84. doi: 10.1017/s0954579405050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Jones MJ, Fejes AP, Kobor MS. DNA methylation, genotype and gene expression: who is driving and who is along for the ride? Genome Biol. 2013;14(7):126. doi: 10.1186/gb-2013-14-7-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Cohen J, Caspi A, Moffitt TE, Harrington H, Milne BJ, Poulton R. Prior juvenile diagnoses in adults with mental disorder: developmental follow-back of a prospective-longitudinal cohort. Archives of general psychiatry. 2003;60(7):709–717. doi: 10.1001/archpsyc.60.7.709. [DOI] [PubMed] [Google Scholar]
- Klengel T, Pape J, Binder EB, Mehta D. The role of DNA methylation in stress-related psychiatric disorders. Neuropharmacology. 2014;80:115–132. doi: 10.1016/j.neuropharm.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Kofink D, Boks MP, Timmers HT, Kas MJ. Epigenetic dynamics in psychiatric disorders: environmental programming of neurodevelopmental processes. Neurosci Biobehav Rev. 2013;37(5):831–845. doi: 10.1016/j.neubiorev.2013.03.020. [DOI] [PubMed] [Google Scholar]
- Lewis CR, Olive MF. Early-life stress interactions with the epigenome: potential mechanisms driving vulnerability toward psychiatric illness. Behav Pharmacol. 2014;25(5–6):341–351. doi: 10.1097/fbp.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JC, Yang TT, Weng RR, Kuo CT, Chang CW. TTBK2: a tau protein kinase beyond tau phosphorylation. Biomed Res Int. 2015;2015:575170. doi: 10.1155/2015/575170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz PE, Turecki G. DNA methylation and childhood maltreatment: from animal models to human studies. Neuroscience. 2014;264:142–156. doi: 10.1016/j.neuroscience.2013.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams TA, Salekin RD, Marti CN, Lester WS, Barker ED. Cooccurrence of antisocial behavior and substance use: testing for sex differences in the impact of older male friends, low parental knowledge and friends' delinquency. Journal of adolescence. 2014;37:247–256. doi: 10.1016/j.adolescence.2014.01.001. [DOI] [PubMed] [Google Scholar]
- McGowan PO, Roth TL. Epigenetic pathways through which experiences become linked with biology. Dev Psychopathol. 2015;27(2):637–648. doi: 10.1017/s0954579415000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt TE. In: Risk, Disorder, and Adaptation. C Cicchetti D, editor. Vol. 3. New York: Wiley and Sons; 2006. Life-course persistent versus adolescence-limited antisocial behaviour, in Developmental Psycholopathology; pp. 570–598. [Google Scholar]
- Moffitt TE, Arseneault L, Jaffee SR, Kim-Cohen J, Koenen KC, Odgers CL, et al. Viding E. Research Review: DSM-V conduct disorder: research needs for an evidence base. Journal of Child Psychology and Psychiatry. 2008;49(1):3–33. doi: 10.1111/j.1469-7610.2007.01823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt TE, Caspi A, Harrington H, Milne BJ. Males on the life-course-persistent and adolescence-limited antisocial pathways: Follow-up at age 26 years. Dev Psychopathol. 2002;14(01):179–207. doi: 10.1017/s0954579402001104. [DOI] [PubMed] [Google Scholar]
- Monk C, Spicer J, Champagne FA. Linking prenatal maternal adversity to developmental outcomes in infants: the role of epigenetic pathways. Dev Psychopathol. 2012;24(4):1361. doi: 10.1017/S0954579412000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moul C, Dobson-Stone C, Brennan J, Hawes DJ, Dadds MR. Serotonin 1B Receptor Gene (HTR1B) Methylation as a Risk Factor for Callous-Unemotional Traits in Antisocial Boys. PLoS ONE. 2015;10(5):e0126903. doi: 10.1371/journal.pone.0126903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muldoon PP, Chen J, Harenza JL, Abdullah RA, Sim-Selley LJ, Cravatt BF, et al. Damaj MI. Inhibition of monoacylglycerol lipase reduces nicotine withdrawal. Br J Pharmacol. 2015;172(3):869–882. doi: 10.1111/bph.12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C, Spengler D. Epigenetics of early child development. Frontiers in Psychiatry. 2011;2:16. doi: 10.3389/fpsyt.2011.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RJ, Trainor BC. Neural mechanisms of aggression. Nature Reviews Neuroscience. 2007;8(7):536–546. doi: 10.1038/nrn2174. [DOI] [PubMed] [Google Scholar]
- Nigg JT. Where Do Epigenetics and Developmental Origins Take the Field of Developmental Psychopathology? Journal of abnormal child psychology. 2016;44(3):405–419. doi: 10.1007/s10802-015-0121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odgers CL, Caspi A, Broadbent JM, Dickson N, Hancox RJ, Harrington H, et al. Moffitt TE. Prediction of differential adult health burden by conduct problem subtypes in males. Arch Gen Psychiatry. 2007;64(4):476–484. doi: 10.1001/archpsyc.64.4.476. [DOI] [PubMed] [Google Scholar]
- Odgers CL, Moffitt TE, Broadbent JM, Dickson N, Hancox RJ, Harrington H, et al. Caspi A. Female and male antisocial trajectories: from childhood origins to adult outcomes. DevPsychopathol. 2008;20(2):673–716. doi: 10.1017/S0954579408000333. [DOI] [PubMed] [Google Scholar]
- Pan Z, Shen Y, Ge B, Du C, McKeithan T, Chan WC. Studies of a germinal centre B-cell expressed gene, GCET2, suggest its role as a membrane associated adapter protein. Br J Haematol. 2007;137(6):578–590. doi: 10.1111/j.1365-2141.2007.06597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappa I, St Pourcain B, Benke K, Cavadino A, Hakulinen C, Nivard MG, et al. Tiemeier H. A genome-wide approach to children's aggressive behavior: The EAGLE consortium. Am J Med Genet B Neuropsychiatr Genet. 2015 doi: 10.1002/ajmg.b.32333. [DOI] [PubMed] [Google Scholar]
- Pidsley R, CC YW, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingault JB, Cecil CAM, Murray J, Munafo MR, Viding E. Causal inference in psychopathology: A systematic review of Mendelian randomisation studies aiming to identify environmental risk factors for psychopathology. Psychopathology Review. 2016 [Google Scholar]
- Pingault J-B, Rijsdijk F, Zheng Y, Plomin R, Viding E. Developmentally dynamic genome: Evidence of genetic influences on increases and decreases in conduct problems from early childhood to adolescence. Scientific reports. 2015;5 doi: 10.1038/srep10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price ME, Cotton AM, Lam LL, Farre P, Emberly E, Brown CJ, et al. Kobor MS. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin. 2013;6(1):4. doi: 10.1186/1756-8935-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencal N, Suderman MJ, Caramaschi D, Wang D, Hallett M, Vitaro F, et al. Szyf M. Differential DNA methylation regions in cytokine and transcription factor genomic loci associate with childhood physical aggression. PLoS ONE. 2013;8(8):e71691. doi: 10.1371/journal.pone.0071691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencal N, Suderman MJ, Guillemin C, Vitaro F, Cote SM, Hallett M, et al. Szyf M. Association of Childhood Chronic Physical Aggression with a DNA Methylation Signature in Adult Human T Cells. PLoS ONE. 2014;9(4):e89839. doi: 10.1371/journal.pone.0089839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2014. Retrieved from http://www.R-proiect.org/ [Google Scholar]
- Relton CL, Davey Smith G. Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. Int J Epidemiol. 2012;41(1):161–176. doi: 10.1093/ije/dyr233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relton CL, Gaunt T, McArdle W, Ho K, Duggirala A, Shihab H, et al. Davey Smith G. Data Resource Profile: Accessible Resource for Integrated Epigenomic Studies (ARIES) Int J Epidemiol. 2015 doi: 10.1093/ije/dyv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond RC, Simpkin AJ, Woodward G, Gaunt TR, Lyttleton O, McArdle WL, et al. Tilling K. Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: findings from the Avon Longitudinal Study of Parents and Children (ALSPAC) Human molecular genetics. 2015;24(8):2201–2217. doi: 10.1093/hmg/ddu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijlaarsdam J, Cecil C, Walton E, Chapman Mesirow MS, Relton C, Gaunt TR, et al. Barker E. Prenatal unhealthy diet, insulin-like growth factor 2 gene (IGF2) methylation and attention deficit hyperactivity disorder (ADHD) symptoms for early-onset conduct problem youth: Prenatal unhealthy diet, IGF2 methylation and ADHD. Journal of Child Psychology and Psychiatry. 2016a doi: 10.1111/jcpp.12589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijlaarsdam J, Pappa I, Walton E, Bakermans-Kranenburg MJ, Mileva-Seitz VR, Rippe RC, et al. van IMH. An Epigenome-Wide Association Metaanalysis of Prenatal Maternal Stress in Neonates: A Model Approach for Replication. Epigenetics. 2016b doi: 10.1080/15592294.2016.1145329. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers AB, Bale TL. Germ cell origins of posttraumatic stress disorder risk: the transgenerational impact of parental stress experience. Biological psychiatry. 2015;78(5):307–314. doi: 10.1016/j.biopsych.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Arias M, Navarrete F, Daza-Losada M, Navarro D, Aguilar MA, Berbel P, et al. Manzanares J. CB1 cannabinoid receptor-mediated aggressive behavior. Neuropharmacology. 2013;75:172–180. doi: 10.1016/j.neuropharm.2013.07.013. [DOI] [PubMed] [Google Scholar]
- Salvatore JE, Dick DM. Genetic Influences on Conduct Disorder. Neuroscience & Biobehavioral Reviews. 2016 doi: 10.1016/j.neubiorev.2016.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumay E, Logan J, Volkow ND, Fowler JS. Evidence that the methylation state of the monoamine oxidase A (MAOA) gene predicts brain activity of MAO A enzyme in healthy men. Epigenetics. 2012;7(10):1151–1160. doi: 10.4161/epi.21976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M, Bick J. DNA methylation: a mechanism for embedding early life experiences in the genome. Child Dev. 2013;84(1):49–57. doi: 10.1111/j.1467-8624.2012.01793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobi EW, Goeman JJ, Monajemi R, Gu H, Putter H, Zhang Y, et al. Müller F. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nature communications. 2014;5 doi: 10.1038/ncomms6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay RE, Szyf M. Developmental origins of chronic physical aggression and epigenetics. Epigenomics. 2010;2(4):495–499. doi: 10.2217/epi.10.40. [DOI] [PubMed] [Google Scholar]
- Tsai PC, Bell JT. Power and sample size estimation for epigenome-wide association scans to detect differential DNA methylation. Int J Epidemiol. 2015 doi: 10.1093/ije/dyv041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turecki G, Meaney MJ. Effects of the social environment and stress on glucocorticoid receptor gene methylation: a systematic review. Biological psychiatry. 2016;79(2):87–96. doi: 10.1016/j.biopsych.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrka AR, Parade SH, Eslinger NM, Marsit CJ, Lesseur C, Armstrong DA, et al. Seifer R. Methylation of exons 1 D, 1 F, and 1 H of the glucocorticoid receptor gene promoter and exposure to adversity in preschool-aged children. Dev Psychopalhol. 2015;27(02):577–585. doi: 10.1017/S0954579415000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mil NH, Steegers-Theunissen RP, Bouwland-Both MI, Verbiest MM, Rijlaarsdam J, Hofman A, et al. Verhulst FC. DNA methylation profiles at birth and child ADHD symptoms. J Psychiatr Res. 2014;49:51–59. doi: 10.1016/j.jpsychires.2013.10.017. [DOI] [PubMed] [Google Scholar]
- Waltes R, Chiocchetti AG, Freitag CM. The neurobiological basis of human aggression: A review on genetic and epigenetic mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2015 doi: 10.1002/ajmg.b.32388. [DOI] [PubMed] [Google Scholar]
- Walton E, Pingault J-B, Cecil C, Gaunt T, Relton C, Mill J, Barker E. Epigenetic profiling of ADHD symptoms trajectories: a prospective, methylome-wide study. Molecular psychiatry. 2016 doi: 10.1038/mp.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Szyf M, Benkelfat C, Provencal N, Turecki G, Caramaschi D, et al. Booij L. Peripheral SLC6A4 DNA methylation is associated with in vivo measures of human brain serotonin synthesis and childhood physical aggression. PLoS ONE. 2012;7(6):e39501. doi: 10.1371/journal.pone.0039501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, et al. Meaney MJ. Epigenetic programming by maternal behavior. Nature neuroscience. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weder N, Zhang H, Jensen K, Yang BZ, Simen A, Jackowski A, et al. Kaufman J. Child abuse, depression, and methylation in genes involved with stress, neural plasticity, and brain circuitry. J Am Acad Child Adolesc Psychiatry. 2014;53(4):417–424.:e415. doi: 10.1016/j.jaac.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolke D, Steer C, Bowen E. The ALSPAC Family Adversity Index (FAI): Background and Development. ALSPAC data documentation. 2004 [Google Scholar]
- Zannas A, Binder E. Gene-environment interactions at the FKBP5 locus: Sensitive periods, mechanisms and pleiotropism. Genes, Brain and Behavior. 2014;73(1):25–37. doi: 10.1111/gbb.12104. [DOI] [PubMed] [Google Scholar]
- Zannas AS, West AE. Epigenetics and the regulation of stress vulnerability and resilience. Neuroscience. 2014;264:157–170. doi: 10.1016/j.neuroscience.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannas AS, Wiechmann T, Gassen NC, Binder EB. Gene-Stress-Epigenetic Regulation of FKBP5: Clinical and Translational Implications. Neuropsychopharmacology. 2015 doi: 10.1038/npp.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.