Abstract
Structural variation (SV), involving deletions, duplications, inversions and translocations of DNA segments, is a major source of genetic variability in somatic cells and can dysregulate cancer-related pathways. However, discovering somatic SVs in single cells has been challenging, with copy-number-neutral and complex variants typically escaping detection. Here we describe single-cell tri-channel processing (scTRIP), a computational framework that integrates read depth, template strand and haplotype phase to comprehensively discover SVs in individual cells. We surveyed SV landscapes of 565 single cells, including transformed epithelial cells and patient-derived leukemic samples, to discover abundant SV classes including inversions, translocations and complex DNA rearrangements. Analysis of the leukemic samples revealed four times more somatic SVs than cytogenetic karyotyping, submicroscopic copy-number alterations, oncogenic copy-neutral rearrangements and a subclonal chromothripsis event. Advancing current methods, scTRIP can directly measure SV mutational processes in individual cells, such as breakage-fusion-bridge cycles, facilitating studies of clonal evolution, genetic mosaicism and SV formation mechanisms, which could improve disease classification for precision medicine.
Introduction
Cancer is a disease of the genome in which subclonal cell expansion is driven by mutation and selection. SVs represent the leading class of somatic driver mutation in many cancer types1,2. Comprising copy-number alterations (CNAs) and copy-neutral classes, SVs can amplify, disrupt and fuse genes or result in enhancer hijacking3–5. These variants can be inherited through the germline and be clonal, or can form de novo in somatic cells (in vivo or in culture) resulting in ‘somatic SVs’ present at subclonal cell fractions (CFs). Somatic SVs can lead to substantial genetic heterogeneity, can precipitate further rearrangements during periods of genomic instability, and contribute to disease development and therapy response6–9. A comprehensive understanding of the extent and nature of somatic SVs in single cells is imperative to elucidate clonal evolution and mutational processes acting in cancer and normal tissues10,11.
Important challenges have so far limited somatic SV studies. Current methods for discovering SVs depend on discordant paired-end or split read signatures that traverse breakpoints12. This requires ≥20-fold genome coverage for clonal, and vastly higher coverage for subclonal, SV detection13. The exception is read-depth analyses that can be pursued at lower depth, but are restricted to detecting only CNAs10. Somatic translocations, inversions and complex DNA rearrangements therefore largely escape detection in subclones, despite their known relevance in cancer and the relationship between complex SVs and poor disease prognosis2,5,14. While single cell analyses can overcome these limitations15, scalable methods for single cell SV detection are likewise only suited for somatic CNAs16–18. Discovering additional SV classes is constrained by requiring uniformly high coverage in each cell, and/or by using whole genome amplification (WGA) methods17 that lead to read chimera and confound SV calling. Although chimera can be filtered in deep coverage data19,20, SV surveys across hundreds of cells using these approaches are cost prohibitive.
Here we describe scTRIP (single cell tri-channel processing) and use it to comprehensively discover somatic SVs in individual cells. scTRIP leverages Strand-seq, a preamplification-free single cell technique that labels non-template DNA strands during normal replication21 and generates strand-specific reads for chromosome-length SNP haplotype phasing22. While Strand-seq has been used to identify polymorphic germline inversions21,23, efforts to exploit these data to characterize diverse SV classes and uncover somatic cell populations were lacking. scTRIP now unlocks the full potential of strand-specific sequencing, rendering a wide variety of disease-relevant SVs accessible to systematic single cell studies. It does so using a joint calling framework that integrates three separate layers of information - depth of coverage, read orientation, and haplotype phase - to build single cell SV landscapes and characterize subclonal SV heterogeneity.
Results
Discovering disease-relevant SV classes by scTRIP
The underlying rationale of scTRIP is that each SV class can be identified via a specific ‘diagnostic footprint’. These footprints capture the co-segregation patterns of rearranged DNA segments made discernible by sequencing single strands of each chromosome in a cell. Such strand-specific data is acquired using Strand-seq21, which exploits Bromodeoxyuridine (BrdU) to selectively remove one DNA strand (the nascent strand) during library preparation and thus only sequence the template DNA strand of each homolog (or ‘haplotype’) (Fig. 1a).Segregation patterns of all DNA segments can then be characterized for the cell, and assigned as Watson (‘W’) or Crick (‘C’) (Fig. 1b). For a cell sequenced with Strand-seq, we assign the haplotype phase to reads containing SNPs22 and jointly measure three data layers: (1.) the total number of reads in a region (‘depth’ layer), (2.) the relative proportion of W and C reads (‘strand’ layer) and (3.) the number of W and C reads assigned to one of the two haplotypes, denoted ‘H1’ or ‘H2’ (‘phase’ layer) (Fig. 1a,c). By integrating these three layers, scTRIP identifies and characterizes a wide variety of SV classes based on specific diagnostic footprints (Table S1).
Figure 1. Haplotype-aware discovery of SVs in single cells.
(a) Overview of the Strand-seq protocol used to preserve strand-orientation and homolog (haplotype) identity. BrdU (Bromodeoxyuridine) is incorporated into dividing cells, followed by removal of the BrdU-containing strands (dashed line) through nicking, and short read sequencing of the remaining (template, solid line) strand21. W, Watson strand (orange); C, Crick (blue); H, haplotype. Right panel: haplotagging approach that assigns individual Strand-seq reads to either haplotype 1 (H1) or H2. Red lollipops mark reads assigned to H1 based on overlapping SNPs; blue lollipops mark H2 reads. From this, three data layers are considered: 1. the total number of reads in a binned region are measured to calculate the ‘Depth’ layer, 2. the relative proportion of W and C reads are measured to calculate the ‘Strand’ layer, and 3. the number of W and C reads assigned to H1 or H2 are used to calculate the ‘Phase’ layer. (b) Scheme depicting how strands segregate during mitosis to reveal SVs in single cells. Del, deletion; Inv, inversion; Tr, translocation. Segments of derivative chromosomes share the same strand label during DNA replication and co-segregate. H1/H2 and h1/h2 designate haplotypes 1 and 2 for two different chromosomes. (c) scTRIP exploits read depth, strand ratio, and chromosome-length haplotype phase as data layers. Haplotype phase is assessed in a strand-aware fashion, with phased W reads shown as lollipops on left of ideogram and phased C reads shown to right (using the same haplotype colors as in (a)). An example “reference” state is shown, which contains 2N read depth, equal proportion of W:C reads and both haplotypes. Panels (d-f) depict diagnostic footprints for chromosomes where both haplotypes are labeled on different strands (‘WC/CW chromosomes’). Our framework also detects and scores equivalent footprints on CC and WW chromosomes (see Table S1). (d) Deletion (Del), detected as losses in read depth affecting a single haplotype, combined with unaltered read orientation. Duplication (Dup), detected as a haplotype-specific gain in depth with unaltered read orientation. (e) Balanced inversion (Inv), identified as haplotype-phased read orientation ‘flips’ with unaltered depth. Inverted duplication (InvDup), characterized by inverted reads detected for one haplotype coinciding with a read depth gain of the same haplotype. (f) Ploidy alterations can be detected as departures from diploid W and C segregation ratios (see also Table S2). (g) Balanced translocation show correlated template strand switches affecting the same paired genomic regions in cells harboring the SV.
The diagnostic footprint of a deletion (Del) is defined by a read depth loss affecting a single strand and haplotype, whereas a duplication (Dup) causes a haplotype-specific read depth gain, also with unaltered orientation (Fig. 1d). For balanced inversions (Inv), read orientation is reversed with the re-oriented reads mapping to a single haplotype, and if this co-locates with a read depth gain on the re-oriented haplotype it signifies an inverted duplication (InvDup; Fig. 1e). In the case of inter-chromosomal SVs, physically connected segments receive the same non-template strand label and hence co-segregate during mitosis (Fig. 1b). Thus segments showing correlating strand states in different cells without a change in depth characterize balanced translocations (Fig. 1f), whereas unbalanced translocations exhibit a similar footprint coupled with a read depth gain of the affected haplotype (Fig. S1). Altered cellular ploidy states also exhibit a unique diagnostic footprint (Fig. 1g, Fig. S2 and Table S2).
Using these principles, we developed a joint calling framework for SV discovery (Fig. 2, Fig. S3 and Methods). The framework first aligns, normalizes and places reads into genomic bins to assign template strand states and build chromosome-length haplotypes (Fig. 2a,b). It then infers SVs in the segmented data by employing a Bayesian model that estimates genotype likelihoods for each segment and each single cell (Fig. 2c, Fig. S4). This framework performs SV discovery in a haplotype-aware manner and combines signals across cells to sensitively detect SVs in a heterogeneous cell population (Fig. 2d,e). Finally, by analyzing adjacent SVs arising on the same haplotype it enables characterizing complex DNA rearrangements25,26. As a first benchmark, we performed simulation experiments (Supplementary Information) and observed excellent recall and precision after randomly placing somatic SVs into cell populations in silico, even down to a single cell (Fig. S5 and Fig. S6).
Figure 2. Analysis pipeline for locating somatic SVs in single cells.
Schematic shown for three single cells representing a heterogeneous population. (a) Single cell data are mapped to a reference genome (grey line) with Strand-seq reads (arrows) aligned in either the Watson (‘W’; orange) or Crick (‘C’; blue) direction. Left: reads are counted in 100 kb bins. Middle: Joint segmentation is performed on the binned data. Piecewise constant functions (black horizontal lines) are fitted to each segment and strand. Segmentation occurs across all cells based on change points in the fitted piecewise constant functions, to locate putative SV breakpoints between bin boundaries (vertical purple lines). Right: Heterozygous SNP positions are used to build consensus haplotypes using StrandphaseR22, resulting in SNPs assigned to chromosome-length haplotypes designated ‘H1’ (red) or ‘H2’ (blue). (b) Consensus haplotypes (now horizontal lines with SNP bubbles) are used to haplotype-tag (haplotag) individual Strand-seq reads in each cell. Any read overlapping a SNP is assigned to H1 (red lollipops) or H2 (blue lollipops) depending on the allele present in the read. Purple lines denote segment breakpoints. (c) Probabilistic model for SV calling. A multinomial distribution is used for the haplotagged read data (left panel). For each segment, the single cell data are considered as four different classes: C reads from H1 (C-H1), W reads from H1 (W-H1), C reads from H2 (C-H2), and W reads from H2 (W-H2). Random variables are represented by circles and parameter by boxes: N represents the true underlying copy-number (which we seek to infer) for each of these four categories, p the corresponding parameters of the multinomial distribution, and X represents the observed read counts in each category. A negative binomial (NB) distribution is used to model the total number of W and C reads (right panel). NB distributions for copy-numbers (CN) 0, 1, and 2 are depicted. Depending on the observed read counts (vertical dotted lines) for each segment, the likelihood of each CN is calculated. The full probabilistic graphical model is shown in Fig. S4. (d) Using this Bayesian model, the most probable SV type is assigned to each segment. In the schematic, two cells contain an inferred duplication on the H1 haplotype (Dup_H1; pink segment), and the other cell contains no SV (assignment to reference state; grey segment) (e) Example Strand-seq data analyzed with scTRIP for two RPE-1 cells and one C7 cell. RPE-1 cells exhibit a somatic duplication event (Dup_H1; chr3:60900000-62300000) absent in C7. Additional SVs called in Strand-seq data are shown in Fig. S8.
Surveying SV landscapes in single cells
To investigate single cell SV landscapes we generated Strand-seq libraries from telomerase-immortalized retinal pigment epithelial (RPE) cells. We used hTERT RPE cells (RPE-1) common to genomic instability research20,27–29, and C7 RPE cells showing anchorage-independent growth indicating cellular transformation30. Both lines originated from the same anonymous female donor. We generated 80 and 154 Strand-seq libraries for RPE-1 and C7, respectively (Methods), targeting more C7 cells to increase our power to uncover somatic SV heterogeneity in this transformed cell line. Libraries were sequenced to a median depth of 387,000 mapped non-duplicate fragments (Table S3), which amounts to ~0.017X coverage per cell.
We first searched for Dels, Dups, Invs and InvDups. Following normalization (Fig. S7), we identified 54 SVs in RPE-1 and 53 in C7 (Table S4). 25 SVs were present only in RPE-1, and 24 were only in C7 – these likely represent sample-specific somatic SVs that formed after the cell lines were derived, rather than corresponding to germline SVs (operationally defined as variants shared between both lines). Two representative somatic SVs include a 1.4 Megabase (Mb) Dup seen in RPE-1, and an 800 kilobase (kb) Del in C7 (Fig. S8). While all but three Del and Dup events were somatic and unique to RPE-1 or C7, Inv and InvDup events, including a 1.6 Mb Inv on 17p and a 900 kb InvDup on 17q (Fig. S8), were germline SVs mapping to known inversion polymorphisms23. We also identified previously-reported somatic chromosome arm-level CNAs, including deletion of 13q in C7, and duplication of a 10q region in RPE-1. These non-disomic regions enabled us to test our ploidy state footprints (Fig. S2 and Table S2). As predicted, the 13q-arm showed a 1:0 strand ratio diagnostic for monosomy, and the 10q region exhibited 2:1 and 3:0 strand ratios diagnostic for trisomy (Fig. S9).
We evaluated scTRIP by several means. First, we verified somatic SVs present with ≥30% CF by bulk whole genome sequencing (WGS), as CFs ≥30% are amenable to WGS-based SV calling13. This confirmed 9/9 (100%) of tested SVs in C7, and 8/9 (89%) in RPE-1 (Table S4). The single somatic SV not verified in RPE-1 partially overlapped a call in C7 and thus might actually represent a germline SV. Second, we examined sensitivity by using the Delly SV caller31 and read-depth analyses on bulk WGS data (Supplementary Information) to produce a curated test-set of SVs ≥200 kb for each line (Table S5). We successfully identified 82% of the test-set with scTRIP. We suspect many of the missed calls were Delly false-positives; all but one were copy-neutral (i.e. an SV class difficult to call from WGS data) and several involved template insertions26, which are small (<1 kb) DNA structure often mis-interpreted as large SVs in WGS data (Fig. S10). Third, by in silico cell mixing different proportions of C7 and RPE-1 cells (Supplementary Information), we tested scTRIP’s performance at varying subclone frequencies and found somatic SVs were detected at very low CF levels (<1% CF) including in individual cells (Fig. S11). Fourth, we compared scTRIP to a computational method tailored to single-cell CNA-profiling18, and found our approach was more accurate and sensitive (Fig. S12). Lastly, we verified scTRIP’s ability to identify altered cellular ploidy by sequencing 73 cells of the isogenic hyperploid RPE cell line C2928, and observed diagnostic strand ratios consistent with its near-tetraploid karyotype28 (Fig S9).
Discovering somatic translocations and novel fusion genes
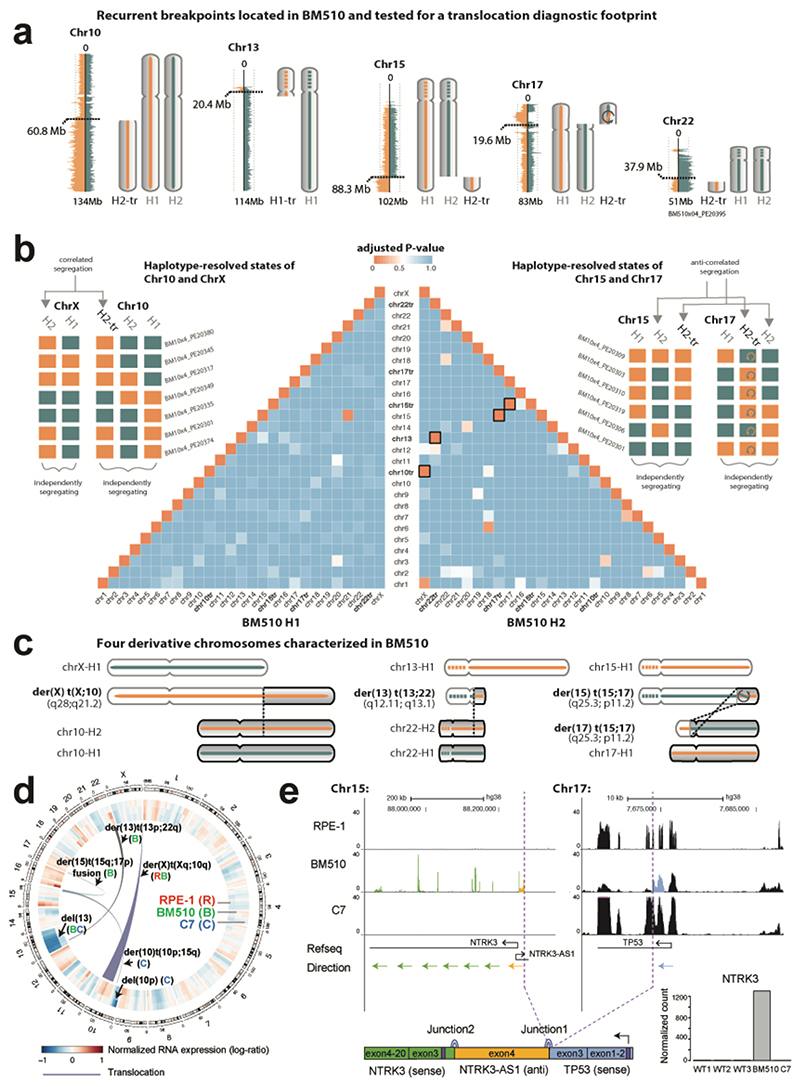
To explore whether scTRIP can detect a wider spectrum of somatic SV classes, we subjected RPE-1 cells to the CAST protocol28. By knocking-out TP53 and silencing the mitotic spindle machinery (Supplementary Information) we constructed the anchorage-independent line ‘BM510’ likely to exhibit genome instability. We sequenced 145 single BM510 cells and detected 67 Dels, Dups, Invs and InvDups (Table S4); 41 were germline SVs (i.e. shared with RPE-1), and 26 were somatic (i.e. unique to BM510 and formed during transformation). Notably, several DNA segments did not segregate with the respective chromosomes they originated from (Fig. 3a), indicating inter-chromosomal SV formation. We searched for co-segregation footprints (Supplementary Information) and identified four translocations in BM510, three of which were somatic (Fig. 3b,c). We then analyzed RPE-1 and C7 for translocations and identified one in each (Table S6). As no translocation was present in all three cell lines, they all constituted somatic events.
Figure 3. Translocation discovery in single cells.
(a) In BM510, segments from chromosomes 10, 13, 15, 17 and 22 failed to co-segregate with the respective chromosomes they originated from, suggesting putative involvement in translocations (use of ‘tr’, as in “H2-tr’ or ‘chr10tr’, denotes the candidate translocation status of these segments). (b) Independent and correlated segregation patterns reveal translocation partners. Schemes to the left and right show haplotype-resolved template strand states of chromosomal segments for six representative cells, exemplifying the segregation patterns of the Left: non-reciprocal der(X) t(X;10) translocation, and the Right: reciprocal t(15;17) translocation. Each colored box denotes the strand state for the segment as either Watson (orange) or Crick (blue). Grey arrows highlight pairwise ‘correlated segregation’ between segments on derivative chromosomes, which always exhibit the same strand state (e.g. chrX and chr10tr) or always exhibit inverse strand states (e.g. chr15tr and chr17; reflecting indirect orientations of these translocation partners). The inversion within the translocated portion of 17p is denoted with a circular arrow. Pyramid in the center: Unbiased analysis of translocations in BM510 (N=144 single cells). The pairwise heatmap depicts the template-strand correlation values for each haplotype segment (H1, left; H2, right), illustrating the co-segregation diagnostic footprint of translocations (Fig. 1f). Correlation values are here expressed as Benjamini-Hochberg adjusted P-values obtained from a two-sided Fisher’s exact test, where P=0 indicates prefect correlation (i.e. co-segregation) and P=1 indicates no correlation (i.e. independent segregation). Orange boxes with black outline depict significant (P<0.01) correlations found for four cases corresponding to the derivative chromosomes discovered in BM510. (c) Cartoon representation of the four inferred chromosomes (outlined boxes in pyramid b) with significant correlations, including: unbalanced der(X) t(X;10), showing chr10q H2-tr gain attached to chrXq H2 (adjusted correlation value P=2.26×10-32), unbalanced der(13) t(13;22), showing chr22q H2-tr gain attached to chr13p H2 (P=5.52×10-41), and balanced der(15) t(15;17) and der(17) t(15;17), showing reciprocal exchange of chr15q H2 and chr17p H2 (P=4.75×10-29, and P=3.93×10-30, respectively) (also see Table S6). Dashed lines within chromosomes (chr) correspond to unassembled regions at acrocentric chr13 and chr15. (d) Circos plot depicting translocations (internal links) and averaged gene expression values across genomic windows77, computed from RNA-seq data generated for BM510 (here denoted ‘B’), RPE-1 (‘R’) and C7 (‘C’). Fig. S16 resolves expression by haplotype. (e) Validation of gene fusion in BM510. RNA-seq based read depth for NTRK3 (green), NTRK3-AS1 (yellow) and TP53 (blue) depicted for C7, RPE-1 and BM510. Purple dashed lines: detected fusion junctions. Lower left corner: inferred fusion transcript. Purple boxes show start codon locations. Lower right corner: NTRK3 overexpression in BM510. WT1-3, RNA-seq replicates of RPE-1. Ex., exon.
The single translocation shared between RPE-1 and BM510 involved the aforementioned gained 10q segment, which cosegregated with chromosome X (Fig. 3b and Fig. S13). Because no breakpoint was visible on chrX we leveraged sister chromatid exchanges21 to place the translocation to the tip of Xq (Supplementary Information), consistent with the published spectral karyotype27. Two somatic translocations in BM510 were formed through balanced reciprocal rearrangement of 15q and 17p (Fig. 3c). Notably, a somatic inversion was detected on the same 17p haplotype and shared one of its breakpoints with the reciprocal translocation (Fig. S14), suggesting these somatic SVs arose jointly, possibly involving a complex rearrangement process. In-depth analysis revealed the inversion encompassed the TP53 locus, which upon translocating fused the 5′ exons of TP53 to the NTRK3 oncogene32 (Fig. S14).
Again, bulk WGS and RNA-Seq analyses revealed excellent performance of our framework. We verified all translocations, with 4/5 recapitulated in WGS (Fig. 3d) and the remaining der(X) t(X;10) unbalanced translocation by the existing karyotype27. WGS failed to locate this translocation because the chrX breakpoint resides in highly repetitive telomeric DNA where read pair analysis is known to fail (Fig. S15); since scTRIP does not require breakpoint-traversing reads it is more sensitive than bulk WGS in such genomic regions. We also observed increased allele-specific expression of the duplicated haplotype predicted for the 10q segment, corroborating our haplotype placements (Fig. S16). Finally, we verified the complex rearrangement in BM510 by identifying TP53-NTRK3 fusion transcripts and along with extreme NTRK3 overexpression (Fig. 3e), which confirms scTRIP can discover novel fusion genes.
Direct measurements of complex DNA rearrangements
Cancer genomes frequently harbor complex DNA rearrangements that can facilitate accelerated tumor evolution33. One example are breakage-fusion-bridge cycles (BFBs) 34–39. BFBs initiate when the loss of a telomere causes replicated sister chromatids to fuse and form a dicentric chromosome. During anaphase, a chromosomal bridge forms that can lead to another DNA break to initiate another BFB cycle14. As a consequence, BFBs successively duplicate regions in inverted orientation (i.e. generate InvDups) adjacent to a terminal deletion (here called ‘DelTer’) on the same homolog. BFBs rising to high CF can be inferred from bulk WGS by locating ‘fold-back inversions’ from read-pair alignments34; however owing to high coverage requirements this cannot be systematically achieved in single cells. We reasoned that scTRIP could provide a new opportunity to directly study BFB formation in single cells.
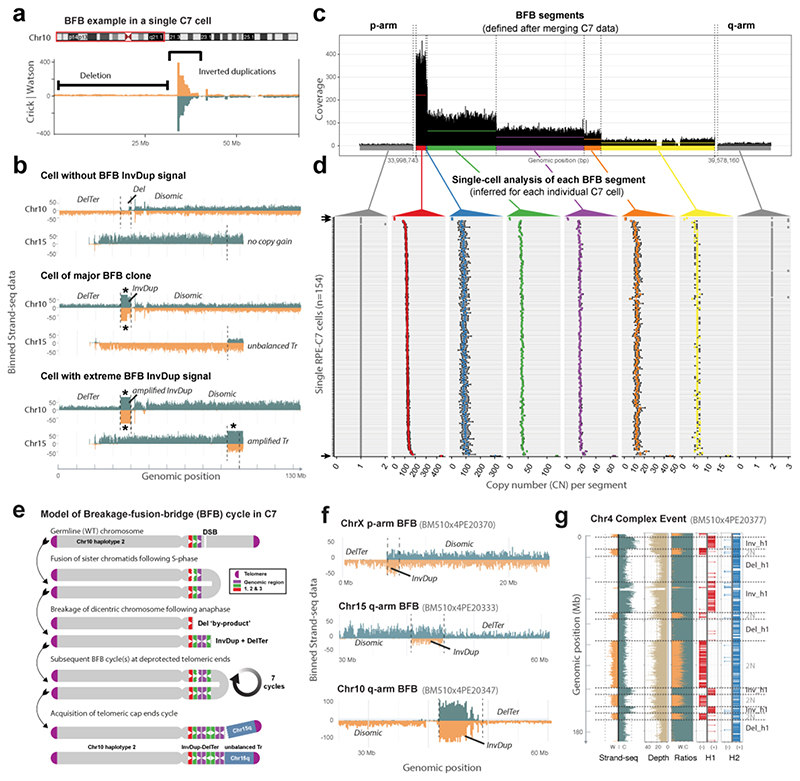
To investigate BFBs, we first interrogated C7, in which fold-back inversions were previously described28. scTRIP located a series of clustered InvDups on the 10p-arm, detected in 152/154 cells (Fig. 4). Closer analysis of 10p showed an amplicon containing ‘stepwise’ InvDups with an adjacent DelTer on the same haplotype, consistent with BFBs (Fig. 4a,b and Fig. S17). The remaining two cells lacked the InvDups but showed a larger DelTer affecting the same 10p segment (Fig. 4b). Upon aggregating reads across cells, we identified 8 discernable segments: the 10p amplicon comprising six step-wise copy-number changes, the adjacent 10p terminal deletion, and the centromere-proximal disomic region (Fig. 4c). We used these 8 segments to infer the cell-specific copy-number status for each cell (Fig. 4d, Table S7).). This revealed three genetically distinct subclones: (i) 151 cells (i.e. the ‘major clone’) showed ‘intermediate’ copy-numbers of 100-130 for the highest copy-number segment, (ii) two cells lost the corresponding 10p region through a DelTer, and (iii) one cell exhibited vastly higher copy-numbers (~440 copies) for this segment, suggesting it underwent additional BFBs (Fig. 4b and Fig. S18).
Figure 4. Single cell characterization of complex rearrangement processes.
(a) Strand-specific read depth of an example C7 cell showing a region of InvDup mediated amplification on 10p, with adjacent terminal deletion (DelTer) of the same haplotype, resulting from BFB cycles. (b) Depiction of three example C7 cells with different BFB statuses, based on estimated maximum copy-number (CN) of 1 (upper panel, cell without BFB), CN of ~110 (middle panel, major clone), and CN of ~440 (lower panel, amplified BFB) at the 10p amplicon region. These CN values correspond to the segment indicated in red, as defined in (c). For each cell, the corresponding gained segment on 15q is shown beneath, which scTRIP inferred to have undergone unbalanced translocation with the amplicon region. Note, the translocation is absent from cells lacking the 10p amplicon (upper panel). Read counts for W (orange) and C (blue) are capped at 50 (*, saturated read counts; see also Fig. S18, which uses a different y-axis scale). Tr, translocation. (c) Aggregated read data from 154 C7 cells to highlight the step-wise CN change for the 10p amplicon. Colours indicate six segments identified within the amplicon, with mean CN shown by horizontal lines (red=221; blue=151; green=65; purple=37; orange=28; yellow=13). Grey: regions flanking the amplicon (d) Genetic single cell diversity within the 10p amplicon. CN (x-axis) values are shown across each individual sequenced C7 cell (N=154; y-axis), to provide cell-by-cell estimates of CN for each segment defined in the merged data (shown in c) (see also Table S7). At least 3 different groups are readily discernible: high CN, intermediate CN, and loss of the 10p region (compare with panel b). Error bars reflect 95% confidence intervals. Arrows denote cells with CN=1 and CN of ~440 at the 10p amplicon. (e) Model of the mutational process leading to the observed structures seen for the ‘major clone’. Amplification via BFB cycles typically proceeds in 2n copy-number steps, suggesting ~7 successive BFB cycles occurred. According to our model, translocation of 15q terminal sequence stabilized 10p BFB. DSB, double strand break. (f) The scar of sporadic somatic BFBs, corresponding to InvDups flanked by DelTer on the same haplotype, identified in single BM510 cells. (g) Clustered rearrangements involving Dels and Invs on a single chr4 homolog of an individual BM510 cell. Shown is the binned read data (left) seperated into the three data channels typical to scTRIP.
Additional somatic SVs identified in C7 provided further insights into the BFB event. We detected an unbalanced translocation stitching a duplicated 15q segment to the 10p amplicon (Fig. 4b and Table S6). The duplicated segment encompassed the 15q telomere, which likely stabilized the amplicon to terminate the BFB process. In agreement, the unbalanced translocation was absent from the two cells harbouring the extended DelTer, and further amplified in the cell with extreme 10p copy-number (Fig. 4b). A model of the temporal rearrangement sequence leading to the major clone is shown in Fig. 4e. These data underscore the ability of scTRIP to characterize BFB-related mutational processes.
Sporadic BFB formation in transformed cells
How often BFBs form in somatic cells is unknown. We searched all 379 RPE-1, C7 and BM510 cells for evidence of a BFB (Methods) and identified 15 additional cells exhibiting the InvDup-DelTer signature (Table S8). Out of these, 11 displayed a ‘classical’ BFB event – an InvDup and DelTer with no other SV present (Fig. 4f and Fig. S19). The remaining four, further described below, showed additional SVs along with the InvDup-DelTer signature. We tested whether the InvDup-DelTer combination coincided by chance by asking whether an InvDup on one haplotype was ever adjacent to a DelTer on the other haplotype. Indeed, InvDup-DelTer structures always occurred on the same haplotype, consistent with the BFB model38. All 15 events were located in the transformed cell lines: 11 of them occurred in BM510 affecting 8% (11/145) of the cells, 4 occurred in C7 affecting 3% (4/154) of the cells, and none (0%; 0/80) were detected in RPE-1 cells. Copy-number estimates of the InvDup regions ranged from 3 to 9, indicating that up to three BFB cycles occurred (Fig. 4f). Finally, all were singleton events located in isolated cells and not shared between cells (Table S8), and therefore likely reflect sporadically formed (and potentially ongoing) BFB cycles.
We reasoned that SVs identified in individual cells can serve as a proxy for active mutational processes. Indeed, we identified 60 additional chromosomes in BM510 with evidence of mitotic errors40 involving somatic gains and losses of entire chromosome arms (35/60; 58%), terminal chromosome regions (17/60; 28%), and whole-chromosome aneuploidies (7/60; 12%). Moreover, nine cells showed multiple clustered rearrangements affecting the same haplotype, including the four cells harboring a sporadic BFB with additional SVs. By employing the infinite sites assumption37, we inferred the relative ordering of SVs occurring in these cells (Supplementary Information), and identified instances where the formation of an additional SV preceded the BFB, and cases where the SV succeeded the BFB (Fig. S20). This analysis also revealed a single cell exhibiting multiple reoriented and lost fragments on the same haplotype, resulting in 12 SV breakpoints that potentially arose through sporadic chromothripsis41,42 (Fig. 4g). Taken together, scTRIP enables the systematic detection of mitotic segregation errors, de novo SV formation and ongoing mutational processes acting in individual cells.
Karyotyping a patient sample from 41 single cells
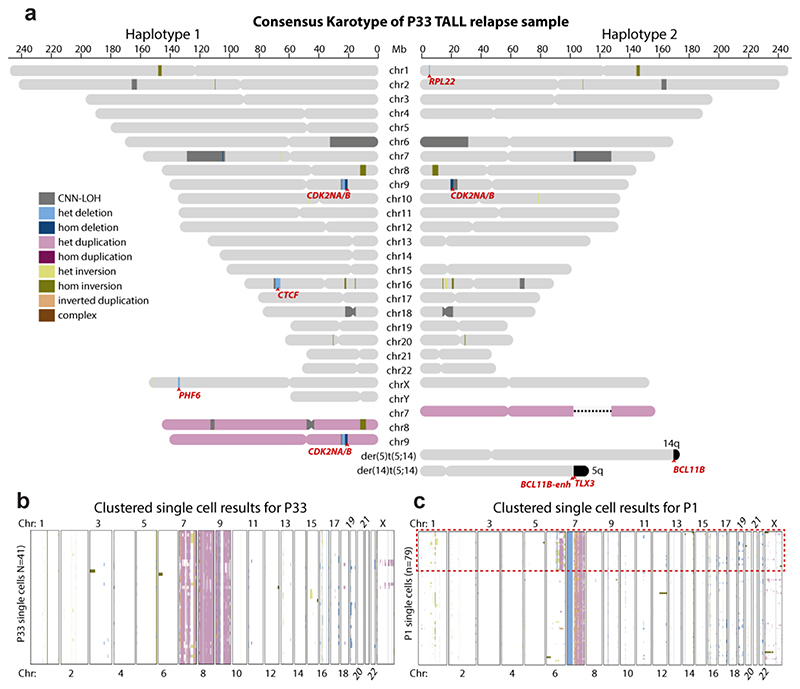
To evaluate the diagnostic value of scTRIP, we next analyzed leukemic samples. Both somatic balanced and complex SVs, which typically escape detection in single cells, are abundant in leukemia26,41,43. We characterized patient-derived xenograft (PDX)44 samples from two T-cell acute lymphoblastic leukaemia (T-ALL) patients. First focusing on P33, a T-ALL relapse of a juvenile patient with Klinefelter Syndrome, we sequenced 41 cells (Table S3). We used these to reconstruct a haplotype-resolved karyotype of the major clone to 200 kb resolution (Fig. 5a). We detected the typical XXY karyotype (Klinefelter Syndrome), trisomies of chromosomes 7, 8, and 9, along with 3 regions of copy-number neutral loss-of-heterozygosity (CNN-LOH) (Fig. S21 and Table S9). Furthermore, we observed 6 focal CNAs, 5 of which affected genes previously reported to be genetically altered in and/or ‘driving’ T-ALL43,45–47 – including PHF6, RPL2, CTCF, CDKN2A and CDKN2B (Fig. 5a, and Table S4). We also identified a t(5;14)(q35;q32) balanced translocation (Table S6) - a recurrent somatic SV in T-ALL known to target TLX3 for oncogenic dysregulation48. The majority of cells supported the karyotype of the major clone (Fig. 5b), with only few individual cells exhibiting karyotypic diversity (Fig. S22).
Figure 5. Single cell sequencing based karyotypes of PDX-derived T-ALL relapses.
(a) Haplotype-resolved consensus P33 karyotype constructed from 41 sequenced cells, using single cell sequencing based SV calls generated by scTRIP. Heterozygous SVs are depicted only on the haplotype they have been mapped to. Homozygous SVs (by definition) appear on both haplotypes. CNN-LOH, copy-neutral loss in heterozygosity (shown on both haplotypes)78. Chromosomes colored in pink reflect duplicated homologs. This T-ALL patient carries two chromosome X haplotypes as well as a Y chromosome, indicating transmission of an X and a Y chromosome from the father, whereas the mother contributed her X chromosome to the karyotype (Klinefelter or XXY syndrome). Affected leukemia-related genes are highlighted in red. ‘BCL11B-enh’ denotes a previously described enhancer region in 3′ of the BCL11B gene. (b) “Heatmap” of SVs arranged using Ward’s method for hierarchical clustering of SVs genotype likelihoods in P33, showing the presence of a single dominant clone and evidence of few additional somatic DNA alterations resulting in karyotypic diversity in this T-ALL relapse. (c) “Heatmap” of SV events called in an additional T-TALL sample, P1. Red dotted box outlines a clear subclonal population in the sample, represented by 25 cells.
We attempted to verify the major clone’s karyotype with classical cytogenetic karyotyping obtained during diagnosis - the current clinical standard to genetically characterize T-ALL. Although this verified the aneuploidies of chromosomes X, 7, 8 and 9, classical karyotyping missed all focal CNAs, and failed to capture the t(5;14)(q35;q32) translocation previously designated as ‘cryptic’ (i.e. ‘not detectable by karyotyping’)49. We next employed CNA profiling by bulk capture sequencing of P33 at diagnosis, remission and relapse50, as well as expression measurements (Supplementary Information). These experiments confirmed all (6/6; 100%) focal CNAs (Table S4), and verified TLX3 dysregulation (Fig. S23) supporting the t(5;14)(q35;q32) translocation. Thus, scTRIP’s haplotype-resolved karyotypes are highly accurate.
Novel and subclonal complex rearrangements uncovered in T-ALL
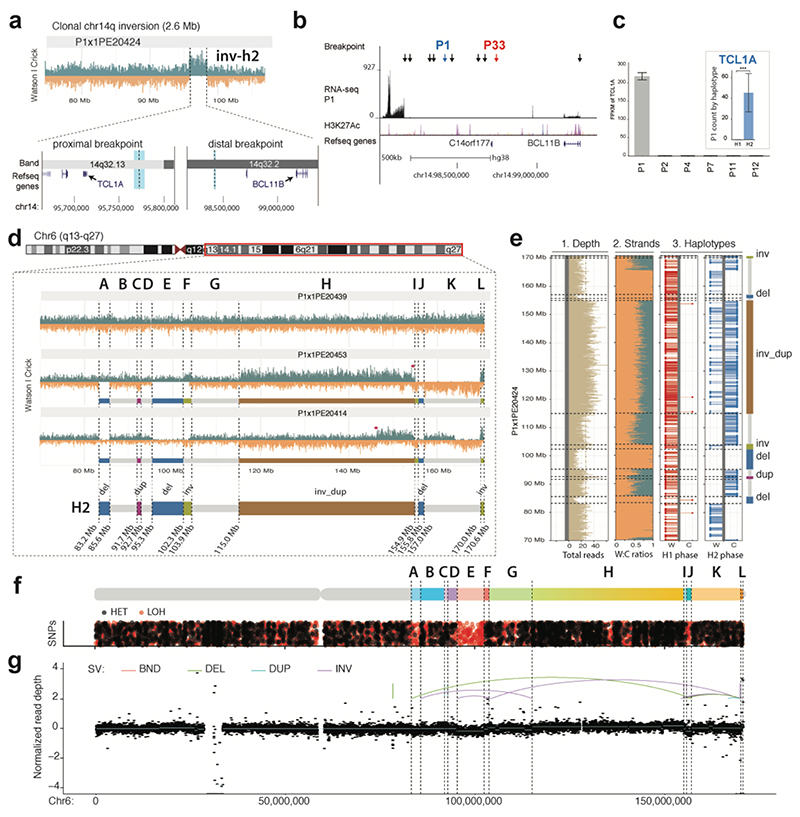
We next turned to a second T-ALL relapse sample obtained from a juvenile female patient (P1). We sequenced 79 cells (Table S3) and discovered two subclones, each represented by at least 25 cells (Fig. 5c and Table S4). First focusing on the clonal SVs, we found a novel 2.6 Mb balanced somatic inversion at 14q32 (Fig. 6a). Interestingly, one of the inversion breakpoints fell into the same 14q region affected by the P33 t(5;14)(q35;q32) translocation (Fig. 6b).
Figure 6. Single cell sequencing of PDX-derived T-ALL relapse P1 reveals previously unrecognized SVs.
(a) Haplotype-resolved balanced 14q32 Inv inferred in P1 using scTRIP. The leftmost breakpoint (thick light blue line) resides close to TCL1A, whereas the rightmost breakpoint (thin light blue line) is in 3′ of BCL11B. (b) The rightmost Inv breakpoint falls into a “gene desert” region in 3′ BCL11B containing several enhancers. Black arrows show breakpoints of translocations resulting in T-ALL oncogene dysregulation from a recent study45. Colored arrows: 41 SV breakpoints in T-ALL donors P1 and P33. (c) Dysregulation of TCL1A in conjunction with 14q32 Inv. Larger barplot shows TCL1A overexpression in P1 compared to five arbitrarily chosen T-ALLs (FDR=2.3E22 two-sided Wald test with Benjamini-Hochberg correction). Inset barplot shows allele-specific RNA-seq analysis demonstrating TCL1A dysregulation occurs only on the inverted (H2) haplotype (FDR 6.68E-21 two-sided pairwise likelihood ratio test with Benjamini-Hochberg correction). The center values in the graph indicates mean of independent biological replicates (n=2 for P1, P4, P7, P11, P12; n=3 for P2). ***p<0.001. (d) Reconstruction of subclonal clustered DNA rearrangements at 6q via scTRIP. (e) Haplotype-resolved analysis of SVs clustered at 6q, all of which fall onto haplotype H2. (f) Detection of interspersed losses and retention of LOH in conjunction with the clustered SVs, indicative for a DNA rearrangements burst41. (LOH, signified by an abundance of red dots, was called as reported in the Methods. Regions with normal density of reference heterozygous SNPs (red), but with decreased density of additionally detected heterozygous SNPs (black), are indicative for LOH.) (g) Verification of subclonal clustered rearrangement burst at 6q, by bulk long-insert size paired-end sequencing75 to 165X physical coverage. Breakpoints inferred by scTRIP are shown as dotted lines, and scTRIP-inferred segments are denoted using the letters A to L. Colored breakpoint-connecting lines depict the paired-end mapping based rearrangement graph (i.e., deletion-type, tandem duplication-type, and inversion-type paired-ends). Using bulk whole-exome and mate-pair sequencing, read-depth shifts at these breakpoints were subtle and thus, this subclonal complex rearrangement escaped prior de novo SV detection efforts in bulk.
In-depth analysis of this locus revealed the 14q32 inversion in P1 juxtaposed an enhancer elementcontaining region 3′ of BCL11B48,51 into the immediate vicinity of the T-cell leukemia/lymphoma 1A (TCL1A) oncogene (Fig. 6a and Fig. S23). Prior studies reported different enhancer-juxtaposing rearrangements in T-cell leukemia or lymphoma resulting in oncogene overexpression43,51,52,53 (Fig. 6b). RNA-seq indeed confirmed TCL1A is the most highly overexpressed gene in P1, and showed >4000-fold increased expression over other T-ALL samples (Fig. 6c). We reasoned that if TCL1A dysregulation was driven by the inversion, then TCL1A overexpression should be restricted to the inverted haplotype, which was confirmed by allele-specific expression (Fig. 6c, inset). These data implicate a novel T-ALL inversion driving oncogene expression, likely involving enhancer hijacking. Further studies are needed to assess recurrence of this inversion in other T-cell malignancies, and the diversity of oncogene-dysregulating SVs involving the BCL11B enhancer region.
We next analyzed subclonal SVs in P1, and discovered a low frequency (CF=0.32) series of highly clustered rearrangements affecting a single 6q haplotype. These comprised two Invs, an InvDup, a Dup, and three Dels, resulting in 13 breakpoints spanning nearly 90 Mb (Fig. 6d,e). All cells in the subclone exhibited the full set of breakpoints, the copy-number profiles oscillated between only three states, and they displayed islands of retention and loss in heterozygosity (Fig. 6f) – patterns reminiscent of chromothripsis41,42. To corroborate this, we performed 4.9 kb insert size mate-pair sequencing in bulk to 165X physical coverage. These deep sequencing data confirmed all 13 subclonal SV breakpoints, verifying the existence of a DNA rearrangement burst consistent with chromothripsis (Fig. 6g), and underscoring the ability of scTRIP to uncover low-frequency complex SVs in cancer cells.
Discussion
scTRIP enables systematic SV detection in single cells by integrating three complementary data layers. We can now locate subclonal SVs at CF<1% and identify SV formation processes acting in single cells, addressing unmet needs10,13,26,55,56. The combined reagent costs are currently ~$15 USD per cell, and the protocol requires ~2 days to generate 96 libraries. Previous single cell studies investigating distinct SV classes involved deeply sequencing only few cells following WGA10,17,57, and prior SV detection efforts using Strand-seq were centered on germline inversions23. scTRIP, facilitated by our Bayesian calling framework, enables systematic discovery of a wide variety of disease-relevant somatic SV classes, including repeat-embedded SVs largely inaccessible to standard WGS in bulk. SVs detected by scTRIP are haplotype-resolved, which helps reduce false positive calls and facilitates allele-specific expression analyses57,58.
We showcase how scTRIP can infer complex mutational processes by identifying sporadic BFBs in up to 8% of transformed RPE cells, revealing that somatic SV formation via BFB cycles is markedly abundant. Indeed, BFB cycles represented the most common SV formation process identified after chromosomal arm-level and terminal loss/gain events, all of which can result from chromosome bridges40,59. BFB cycles have also been reported in cleavage-stage in vitro fertilization embryos (revealed by hybridization-based single cell assays)58 and occur in a wide variety of cancers14, can precipitate chromothripsis37, and correlate with disease prognosis60. An estimated 20% of somatic deletions and >50% of all somatic SVs in cancer genomes arise from complex rearrangements25,26. By directly measuring these events in single cells, scTRIP can facilitate investigating their role in cancer evolution.
Our study also exemplified a potential value for disease classification. We constructed a haplotype-resolved karyotype of a T-ALL sample at 200 kb resolution using 41 single cells, amounting to only 0.9X cumulative genomic coverage. This revealed submicroscopic CNAs and oncogenic rearrangements invisible to methods currently used in the clinic, and showed four times more leukemia-related somatic DNA alterations than the classical cytogenetic karyotype. Classical cytogenetics is typically pursued for only a limited number of metaphase spreads per patient, and thus can fail to capture subclonal karyotypic heterogeneity readily accessible to our approach. scTRIP uncovered a low-frequency chromothripsis event, highlighting utility for disease prognosis, considering chromothripsis is associated with dismal outcome61. Future studies of aberrant clonal expansions in healthy individuals10 and lineage tracing62 may be facilitated by scTRIP. Another potential application area is in rare disease genetics, where scTRIP may help resolve “unclear cases” by widening the spectrum of accessible SVs leading to somatic mosaicism56. Finally, scTRIP could be used to assess genome integrity in conjunction with cell therapy, gene therapy, and therapeutic CRISPR-Cas9 editing, which can result in unanticipated SVs63,64.
scTRIP is currently limited to Strand-seq, which requires labeling chromosomes during replication. Cells with incomplete BrdU labelling, or those that have undergone two rounds of labelling, must be excluded prior to analysis21,65. Non-dividing, apoptotic, or fixed cells cannot be studied. Nonetheless, many key cell types are naturally prone to divide or can be cultured, including fresh or frozen stem and progenitor cells, cancer cells, cells in regenerating or embryonic tissues, iPS cells, and cells from organoids.
Our approach enables studying somatic SV landscapes with much less sequence coverage than WGA-based methods. We demonstrated SV discovery using ~2000-fold less reads than required for read-pair or split-read based methods12. Single cell sequencing to deep coverage using WGA can map SVs <200 kb in size, and remains useful for detecting small CNAs or retrotransposons. However, WGA-based single cell SV analyses are subject to the limitations of paired-end analyses, allelic dropouts, low sensitivity in repetitive regions, and show limited scalability17. Low-depth and high-scale methods for CNA-profiling single cells exist and can detect CNAs of 1 to 5 Mb in size16,18. These show promise for investigating subclonal structure in non-dividing cancer cells harboring large CNAs, but miss key SV classes and fail to discriminate between SV formation processes.
In conclusion, scTRIP enables systematic SV landscape studies to decipher derivative chromosomes, karyotypic diversity, and to directly investigate SV formation in single cells. It provides important value over existing methods, and opens new avenues in single cell analysis.
Online Methods
Cell Lines and Culture
hTERT RPE-1 cells were purchased from ATCC (CRL-4000) and checked for mycoplasma contamination. The C29 hyperploid cell line was generated previously28. BM510 cells were generated newly using the CAST protocol and derived from the RPE-1 parental line (as previously-described28; see further detailed in the Supplementary Information). C7 cells were acquired from30. Cell lines were maintained in DMEM-F12 medium supplemented with 10% fetal bovine serum and antibiotics (Life Technologies).
Ethics Statement
The protocols used in this study received approval from the relevant institutional review boards and ethics committees. The T-ALL patient samples were approved by the University of Kiel ethics board, and obtained from clinical trials ALL-BFM 2000 (P33; age: 14 years at diagnosis) or AIEOP-BFM ALL 2009 (P1; age: 12 years at diagnosis). Written informed consent had been obtained from these patients, and experiments conformed to the principles set out in the WMA Declaration of Helsinki and the Department of Health and Human Services Belmont Report. The in vivo animal experiments were approved by the veterinary office of the Canton of Zurich, in compliance with ethical regulations for animal research.
Single cell DNA sequencing of RPE and T-ALL cells
RPE cells and PDX-derived T-ALL cells were cultured using previously established protocols28,67. We incorporated BrdU (40μM; Sigma, B5002) into growing cells for 18-48 hours, single nuclei were sorted into 96-well plates using the BD FACSMelody cell sorter, and strand-specific DNA sequencing libraries were generated using the previously described Strand-seq protocol21,65. Note, the BrdU concentration used was recently shown to have no measurable effect on sister chromatid exchanges24, a sensitive measure of DNA integrity and genomic instability24. To generate libraries at scale, the Strand-seq protocol was implemented on a Biomek FXP liquid handling robotic system, which requires two days to produce 96 barcoded single cell libraries. Libraries were sequenced on a NextSeq5000 (MID-mode, 75 bp paired-end protocol), demultiplexed and aligned to GRCh38 reference assembly (BWA 0.7.15).
Library selection for scTRIP analysis
High quality libraries (obtained from cells undergoing one complete round of DNA replication with BrdU incorporation) were selected as described in21,65. This is important because incomplete BrdU removal or incorporation could lead to false discovery SV calls. Libraries showing very low, uneven coverage, or an excess of ‘background reads’ yielding noisy single cell data were filtered prior to analysis. Cells with incomplete BrdU incorporation or cells undergoing more than one DNA synthesis phase under BrdU exposure are largely excluded during cell sorting and thus get only rarely sequenced during Strand-seq experiments21,65, typically contributing to less than 10% of sequenced cells. In a typical experiment, ~80% of cells yield high quality libraries reflecting efficient BrdU incorporation in exactly a single cell cycle, and thus ‘unusable libraries’ do not palpably contribute to experimental costs.
Chromosome-length haplotype phasing of heterozygous SNPs
Our SV discovery framework ‘MosaiCatcher’ phases template strands using StrandPhaseR22. The underlying rationale is that for ‘WC chromosomes’ (chromosomes where one parental homolog is inherited as W template strand and the other homolog is inherited as C template strand), heterozygous SNPs can be immediately phased into chromosome-length haplotypes (a feature unique to strand-specific DNA sequencing). To maximize the number of informative SNPs for full haplotype construction we aggregated reads from all single cell sequencing libraries and an internal 100 cell control and performed SNP discovery by re-genotyping the 1000 Genomes Project (1000GP) SNP sites68 using Freebayes69. All heterozygous SNPs with QUAL ≥10 where used for haplotype reconstruction and single cell haplotagging (described below). From a typical Strand-seq experiment (such as RPE-1, where N=80 libraries were analyzed) we observe ~1.4% of heterozygous positions sampled in any given cell, with ~78% of all SNPs in a given sample covered at least once (and ~18% are covered by more than one cell). (Fig. S24)
Discovery of somatic deletions, duplications, inversions and inverted duplications in single cells
We developed the core workflow of ‘MosaiCatcher’ to enable single cell discovery of Dup, Del, Inv, and InvDup SVs from strand-specific sequence data. Input data to the workflow are a set of single-cell BAM files from a donor sample, aligned to a reference genome. The core workflow performs binned read counting, normalization of coverage, segmentation, strand state and sister chromatid exchange (SCE) detection, and haplotype-aware SV classification. A brief description of each step is provided below, and for additional details see Supplementary Information.
Binned read counting
Reads for each individual cell, chromosome and strand were binned into 100 kb windows. PCR duplicates, improper pairs and reads with a low mapping quality (<10) were removed to count only unique, high-quality fragments.
Normalization of coverage
Normalization was performed to adjust for systematic read depth fluctuations. To derive suitable scaling factors, we performed an analysis of Strand-seq data from 1,058 single cells generated across nine 1000GP lymphoblastoid cell lines made available through the HGSVC project (http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/hgsv_sv_discovery/working/20151203_strand_seq/), and pursued normalization with a linear model used to infer a scaling factor for each genomic bin.
Joint segmentation of single cells in a population
Segmentation was performed by jointly processing strand-resolved binned read depth data across all single cells of a sample, used as multivariate input signal with a squared-error assumption70. Given a number of allowed change points k, a dynamic programming algorithm was employed to identify the discrete positions of k change points with a minimal sum of squared error. Analyzing all cells jointly in this way rendered even relatively small SVs (~200 kb) detectable once these are present with sufficient evidence in the single cell dataset (e.g. seen in enough cells). The number of breakpoints was chosen separately for each chromosome as the minimal k, such that using k+1 breakpoints would only yield a marginal improvement, operationalized as the difference of squared error terms being below a pre-selected threshold (Supplementary Information).
Strand-state and SCE detection in individual cells
The interpretation of strand-specific binned read counts relies on the knowledge of the underlying state of template strands for a given chromosome (WW, CC, or WC). These “ground states” stay constant over the length of each chromosome in each single cell, unless they are altered through SCEs21,71. To detect SCEs, we performed the same segmentation procedure described above in each cell separately (as opposed to jointly across all cells, as for the segmentation). We then inferred putative SCEs by identifying changes in strand state in individual cells that are otherwise incompatible with breakpoints uncovered by the joint segmentation (Supplementary Information). Leveraging these putative SCEs, we then assigned a ground state to each segment (Supplementary Information). To facilitate haplotype-resolved SV calling, we employed StrandPhaseR72 to distinguish segments with ground state WC, where Haplotype 1 is represented by Watson (W) reads and Haplotype 2 by Crick (C) reads, from ground state CW, where it is vice versa.
Haplotype-aware SV classification
We developed a Bayesian framework to compute posterior probabilities for each SV diagnostic footprint, and derive haplotype-resolved SV genotype likelihoods. To this end, we modeled strand-specific read counts using a negative binomial (NB) distribution, which captures the overdispersion typical for massively-parallel sequencing data54. The NB distribution has two parameters, p and r; the parameter p controls the relationship of mean and variance and was estimated jointly across all cells, while r is proportional to the mean and hence varies from cell to cell to reflect the different total read counts per single-cell library. After estimating p and r, we computed haplotype-aware SV genotype likelihoods for each segment in each single cell: For a given ground state (see above), each SV diagnostic footprint translates into the expected number of copies sequenced in W and C orientation contributing to the genomic segment (Table S1), which gives rise to a likelihood with respect to the NB model. The fact that our model distinguishes WC from CW ground states (see Strand-state and SCE detection above) renders our model implicitly whole-chromosome haplotype-aware - a key feature not met by any prior approach for somatic variant calling in single cells. In addition to this, we also incorporated the count of W or C reads assignable to a single haplotype via overlapping SNPs in the likelihood calculation, and refer to this procedure as “haplotagging” (since it involves reads “tagged” by a particular haplotype). We modeled the respective counts of tagged reads using a multinomial distribution (Supplementary Information). The output is a matrix of predicted SVs with probability scores for each single cell.
SV calling in a cell population
Our workflow estimates CF levels for each SV and uses them to define prior probabilities for each SV (Empirical Bayes). In this way, the framework benefits from observing SVs in more than one cell, which leads to an increased prior and hence to more confident SV discoveries. Our framework adjusts for the tradeoff between sensitively calling subclonal SVs, and accurately identifying SVs seen consistently among cells. We parameterized this tradeoff into a ‘strict’ and ‘lenient’ SV caller, whereby the ‘strict’ caller optimizes precision for SVs seen with CF ≥5%, and the ‘lenient’ caller targets all SVs including those present in a single cell only. Unless stated otherwise, SV calls presented in this study were generated using the ‘strict’ parameterization, to achieve a callset that minimises false positive SVs (Supplementary Information). We explored the limits of these parameterizations using simulations, by randomly implanting Dels, Dups and Invs into single cells in silico. We analyzed 200 single cells per simulation, applying coverage levels typical for Strand-seq21 (400,000 read fragments per cell). We observed excellent recall and precisions for SVs ≥1 Mb in size when present with >40% CF (Fig. S5). And while we detected a decrease in recall and precision for events present with lower CF, we were able to recover smaller SVs and those with lower CF down to individual cells (Fig. S5). When comparing SV profiles between samples, such as to determine which SVs were unique to a sample or shared between samples 50% reciprocal overlap tests were performed.
Single cell dissection of translocations
We discovered translocations in single cells by searching for segments exhibiting strand-states that are inconsistent with the chromosomes these segments originate from, while being consistent (correlated, or anti-correlated) in strand-state with another segment of the genome (i.e., their translocation partner) (Supplementary Information). To infer translocations, we determined the strand states of each chromosome in a homolog-resolved manner. In cases where strand states appeared to change across a haplotype (because this haplotype exhibited SVs or SCEs), we used the majority strand state (i.e. ‘ground state’, see above) to pursue translocation inference. We examined template strand co-segregation by generating contingency tables tallying the number of cells with equivalent strand states versus those not having equivalent strand states (see Fig. 3b). We employed Fisher’s exact test to infer the probability of the count distribution in the contingency table, followed by p-value adjustment73.
Characterization of breakage-fusion bridge (BFB) cycles in single cells
To infer and characterize BFB cycles in single cells, we first employed our framework with lenient parameterization to infer InvDups flanked by a DelTer event on the same homolog/haplotype. We tested whether InvDup-DelTer footprints resulting from BFB cycles may arise in single cells by chance, by searching for structures where an InvDup on one haplotype would be flanked by a DelTer on the other haplotype (for instance, an InvDup (H1)-DelTer (H2) event, where H1 and H2 denote different haplotypes). No such structures were detected, and InvDup-DelTer footprints thus always occurred on the same haplotype, consistent with BFB cycle formation. To ensure high sensitivity of our single cell based quantifications shown in Fig. S17, we additionally performed manual inspection of the single cell data for evidence of at least one of the following rearrangement classes: (i) an InvDup, (ii) a DelTer resulting in copy-number=1 on an otherwise disomic chromosome. These cells were inspected for InvDup-DelTer patterns indicative for BFBs, based on the diagnostic footprints defined in Fig. 1.
Single cell based CNN-LOH discovery
For CNN-LOH detection, our framework first assembles consensus haplotypes for each sample, by analyzing all single cell Strand-seq libraries available for a sample using StrandPhaseR22. Each single cell is then compared to these consensus haplotypes in a disomic context, to identify discrepancies matching the CNN-LOH footprint. To detect clonally present CNN-LOH events, we used the 1000GP68 reference SNP panel to re-genotype aggregated single cell libraries in each sample. These re-genotyped (observed) SNPs were then compared to the 1000GP reference sets to identify genomic regions showing marked depletion in heterozygous SNPs indicative for CNN-LOH. To this end, we downsampled the 1000GP reference variants to the SNP numbers observed in the single cell data, and subsequently merged both data sets (observed and reference variants), sorting all SNPs by genomic position. We performed a sliding window search through these sorted SNPs, moving one SNP at a time, and compared the number of observed and reference SNPs in each window by computing the ratio R=observed SNPs/reference SNPs. In heterozygous disomic regions, R values of ~1 will be expected, whereas deviations are indicative of CNN-LOH. Window sizes (determined by the number of SNPs in a window) were defined as the median SNP count per 500 kb window. We employed circular binary segmentation (CBS)74 to detect changes in R, and assigned each segment a state based on the mean value of R. Segments ≥2 Mb in size exhibiting mean values R≤0.15 were reported as CNN-LOH.
Bulk genomic DNA sequencing
Genomic DNA was extracted using the DNA Blood Mini Kit (Qiagen, Hilden, Germany). 300 ng of high molecular weight genomic DNA was fragmented to 100 –700 bp (300 bp average size) with a Covaris S2 instrument (LGC Genomics) and cleaned up with Agencourt AMPure XP (Beckman Coulter, Brea, USA). DNA library preparation was performed using the NEBNext Ultra II DNA Library Prep Kit (New England Biolabs, Ipswich, USA). We employed 15ng of adapter ligated DNA and performed amplification with 10 cycles of PCR. DNA was size selected on a 0.75% agarose gel, by picking the length range between 400 and 500 bp. Library quantification and quality control was performed using a Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, USA) and a 2100 Bioanalyzer platform (Agilent Technologies, Santa Clara, USA). WGS was pursued using an Illumina HiSeq4000 (Illumina, San Diego, USA) platform, using 150 bp paired-end reads. Mate-pair sequencing with large insert size (~5 kb) was pursued as described previously75. SV detection in bulk DNA sequence data was pursued using Delly231. RPE-1 WGS data was sequenced to 32× coverage.
Bulk RNA-seq
Total RNA was extracted from RPE cells using the RNeasy MinElute Cleanup kit (Qiagen, Hilden, Germany). RNA quality control was performed using the 2100 Bioanalyzer platform (Agilent Technologies, Santa Clara, USA). Library preparation was pursued with a Beckman Biomek FX automated liquid handling system (Beckman Coulter, Brea, USA), with 200 ng starting material using TruSeq Stranded mRNA HT chemistry (Illumina, San Diego, USA). Samples were prepared with custom 6 base pair barcodes to enable pooling. Library quantification and quality control were performed using a Fragment Analyzer (Advanced Analytics Technologies, Ames, USA). RNA-Seq was pursued on an Illumina HiSeq 2500 platform (Illumina, San Diego, USA), using 50 base pair single reads. For RNA sequencing in T-ALL, total RNA was extracted using TRIzol (Invitrogen Life Technologies). The RNA was than treated with TURBO DNase (Thermo Fisher Scientific, Darmstadt, Germany) and purified using RNA Clean&Concentrator-5 (Zymo Research, Freiburg, Germany). We required a minimal RIN (RNA Integrity Number) of 7 as measured using a Bioanalyzer (Agilent, Santa Clara, CA) with the Agilent RNA 6000 Nano Kit. Cytoplasmic ribosomal RNA was depleted by Ribo-Zero rRNA Removal Kit (Illumina, San Diego, CA) and the libraries were prepared from 1 μg of RNA using TruSeq RNA Library Prep (Illumina, San Diego, CA). These samples were sequenced on a Illumina HiSeq 2000 lane as 75 bp single ends. Fusion junctions were detected using the STAR aligner76.
Quantitative real time PCR (qPCR)
RNA from PDX-derived T-ALL samples was extracted using a RNeasy Mini kit according to manufacturer’s instructions (cat 74106, Qiagen, Hombrechtikon, Switzerland), and cDNA was generated using High Capacity cDNA Reverse Transcription Kit (Applied BioSystems, Foster City, USA). qPCR was performed using a TaqMan Gene Expression Master Mix (Applied BioSystems) in triplicate using an ABI7900HT Analyzer with SDS Plate Utility (v2.2) software. Threshold cycle values were determined using the 2-ΔΔCT method, normalized to human-GAPDH (Hs02786624_g1, Applied BioSystems).
Statistical Analysis
For experiments with replicates, the results are shown as means ± s.d. with replicates from independent biological experiments, unless stated otherwise. For translocation analysis the correlation values were determined using a two-sided Fisher’s exact test adjusted using the Benjamini-Hochberg procedure for false discovery rate (FDR) control, and allele-specific RNA-seq analysis was tested using two-sided pairwise likelihood ratio test with Benjamini-Hochberg correction.
Supplementary Material
Acknowledgements
We thank Wolfgang Huber, Oliver Stegle, Francesco Marass, and Peter Lansdorp for discussions, and Tania Christiansen for software documentation. We thank Malte Paulsen (Flow Cytometry Core Facility) for assistance in sorting, and Cornelia Eckert for primary T-ALL samples for engraftment. JOK acknowledges funding from European Research Council (ERC) Starting (336045) and Consolidator Grants (773026) and the National Institutes of Health (3U41HG007497-04S1). Funding also came from the German Research Foundation (391137747 and 395192176) to TM, the José Carreras Foundation (DJCLS 06R/2016) to JOK, AEK and JBK, the Baden-Württemberg Stiftung (ID16) to AEK, and the Iten-Kohaut Stiftung to JPB. ADS and HY received postdoctoral fellowships through the Alexander von Humboldt Foundation.
Footnotes
Author contributions
Study conception: A.D.S., T.M., J.O.K. SV footprints: A.D.S., S.M., M.G., D.P., T.M., J.O.K. Strand-seq library preparation workflow: A.D.S., B.R., G.M.C.L., J.Z., V.B. BM510 generation: B.R.M., J.O.K. T-ALL samples. A.D.S., S.J., B.R., B.B., J.-P.B. MosaiCatcher tool for scTRIP data analysis: S.M., M.G., D.P., A.D.S., T.R, T.M., J.O.K. Bayesian framework: M.G., S.M., D.P., T.R., J.O.K., T.M. Cell mixing and simulations: S.M., T.R., D.B., T.M. Translocations: A.v.V., A.D.S., D.P., J.O.K. Clustered rearrangements: A.D.S., D.P., M.G., T.R., T.M., J.O.K. CNN-LOHs: D.P., A.D.S., T.M. Haplotagging: M.G., D.P., A.D.S., T.M. Bulk DNA sequencing: T.R., B.R. T-ALL clinical/cytogenetic data: P.R-P., J.B.K., M.S., A.K., B.B., J.-P.B. T-ALL expression analysis: H.J., P.R-P., J.B.K., S.J., B.B., B.R., J.-P.B., A.K. Manuscript: A.D.S., T.M., J.O.K wrote the manuscript, which was edited and approved by all authors.
Competing Interests statement
Disclosed patent application (EP19169090): A.D.S., J.O.K., T.M., D.P., S.M., M.G.
Reporting Summary. Further information on research design is available in the Life Sciences Reporting Summary.
Data Availability
Sequencing data from this study can be retrieved from the European Genome-phenome Archive (EGA), and the European Nucleotide Archive (ENA) [accessions: PRJEB30027, PRJEB30059, PRJEB8037, PRJEB33731, EGAS00001003248, EGAS00001003365]. Access to human patient data is governed by the EGA Data Access Committee.
Code Availability
The computational code of our analytical framework is hosted on GitHub (see https://github.com/friendsofstrandseq/mosaicatcher-pipeline, https://github.com/friendsofstrandseq/TranslocatoR, and https://github.com/friendsofstrandseq/mosaicatcher). All code is available freely for academic research.
References
- 1.Ciriello G, et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mertens F, Johansson B, Fioretos T, Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15:371–381. doi: 10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- 3.Northcott PA, et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547:311–317. doi: 10.1038/nature22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beroukhim R, Zhang X, Meyerson M. Copy number alterations unmasked as enhancer hijackers. Nat Genet. 2016;49:5–6. doi: 10.1038/ng.3754. [DOI] [PubMed] [Google Scholar]
- 5.Northcott PA, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim C, et al. Chemoresistance Evolution in Triple-Negative Breast Cancer Delineated by Single-Cell Sequencing. Cell. 2018;173:879–893.:e13. doi: 10.1016/j.cell.2018.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turajlic S, et al. Tracking Cancer Evolution Reveals Constrained Routes to Metastases: TRACERx Renal. Cell. 2018;173:581–594.:e12. doi: 10.1016/j.cell.2018.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sottoriva A, et al. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47:209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aparicio S, Caldas C. The implications of clonal genome evolution for cancer medicine. N Engl J Med. 2013;368:842–851. doi: 10.1056/NEJMra1204892. [DOI] [PubMed] [Google Scholar]
- 10.Forsberg LA, Gisselsson D, Dumanski JP. Mosaicism in health and disease - clones picking up speed. Nat Rev Genet. 2017;18:128–142. doi: 10.1038/nrg.2016.145. [DOI] [PubMed] [Google Scholar]
- 11.Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–1558. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 12.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leibowitz ML, Zhang C-Z, Pellman D. Chromothripsis: A New Mechanism for Rapid Karyotype Evolution. Annu Rev Genet. 2015;49:183–211. doi: 10.1146/annurev-genet-120213-092228. [DOI] [PubMed] [Google Scholar]
- 15.Navin NE. Cancer genomics: one cell at a time. Genome Biol. 2014;15:452. doi: 10.1186/s13059-014-0452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zahn H, et al. Scalable whole-genome single-cell library preparation without preamplification. Nat Methods. 2017;14:167–173. doi: 10.1038/nmeth.4140. [DOI] [PubMed] [Google Scholar]
- 17.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 18.Bakker B, et al. Single-cell sequencing reveals karyotype heterogeneity in murine and human malignancies. Genome Biol. 2016;17:115. doi: 10.1186/s13059-016-0971-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voet T, et al. Single-cell paired-end genome sequencing reveals structural variation per cell cycle. Nucleic Acids Res. 2013;41:6119–6138. doi: 10.1093/nar/gkt345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang CZ, et al. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falconer E, et al. DNA template strand sequencing of single-cells maps genomic rearrangements at high resolution. Nat Methods. 2012;9:1107–1112. doi: 10.1038/nmeth.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porubsky D, et al. Dense and accurate whole-chromosome haplotyping of individual genomes. Nat Commun. 2017;8:1293. doi: 10.1038/s41467-017-01389-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanders AD, et al. Characterizing polymorphic inversions in human genomes by single-cell sequencing. Genome Res. 2016;26:1575–1587. doi: 10.1101/gr.201160.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Wietmarschen N, Lansdorp PM. Bromodeoxyuridine does not contribute to sister chromatid exchange events in normal or Bloom syndrome cells. Nucleic Acids Res. 2016;44:6787–6793. doi: 10.1093/nar/gkw422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang L, et al. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell. 2013;153:919–929. doi: 10.1016/j.cell.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, et al. Patterns of structural variation in human cancer, bioRxiv. bioRxiv. 2017:181339. doi: 10.1101/181339. [DOI] [Google Scholar]
- 27.Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 28.Mardin BR, et al. A cell-based model system links chromothripsis with hyperploidy. Mol Syst Biol. 2015;11:828. doi: 10.15252/msb.20156505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maciejowski J, Li Y, Bosco N, Campbell PJ, de Lange T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell. 2015;163:1641–1654. doi: 10.1016/j.cell.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riches A, et al. Neoplastic transformation and cytogenetic changes after Gamma irradiation of human epithelial cells expressing telomerase. Radiat Res. 2001;155:222–229. doi: 10.1667/0033-7587(2001)155[0222:ntacca]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 31.Rausch T, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amatu A, Sartore-Bianchi A, Siena S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open. 2016;1:e000023. doi: 10.1136/esmoopen-2015-000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C-Z, Leibowitz ML, Pellman D. Chromothripsis and beyond: rapid genome evolution from complex chromosomal rearrangements. Genes Dev. 2013;27:2513–2530. doi: 10.1101/gad.229559.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rode A, Maass KK, Willmund KV, Lichter P, Ernst A. Chromothripsis in cancer cells: An update. Int J Cancer. 2016;138:2322–2333. doi: 10.1002/ijc.29888. [DOI] [PubMed] [Google Scholar]
- 36.Selvarajah S, et al. The breakage-fusion-bridge (BFB) cycle as a mechanism for generating genetic heterogeneity in osteosarcoma. Chromosoma. 2006;115:459–467. doi: 10.1007/s00412-006-0074-4. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, et al. Constitutional and somatic rearrangement of chromosome 21 in acute lymphoblastic leukaemia. Nature. 2014;508:98–102. doi: 10.1038/nature13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McClintock B. The Stability of Broken Ends of Chromosomes in Zea Mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gisselsson D, et al. Chromosomal breakage-fusion-bridge events cause genetic intratumor heterogeneity. Proc Natl Acad Sci U S A. 2000;97:5357–5362. doi: 10.1073/pnas.090013497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Curr Biol. 2010;20:R285–95. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 43.Girardi T, Vicente C, Cools J, De Keersmaecker K. The genetics and molecular biology of T-ALL. Blood. 2017;129:1113–1123. doi: 10.1182/blood-2016-10-706465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richter-Pechańska P, et al. PDX models recapitulate the genetic and epigenetic landscape of pediatric T-cell leukemia. EMBO Mol Med. 2018:e9443. doi: 10.15252/emmm.201809443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49:1211–1218. doi: 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Q, et al. Mutations of PHF6 are associated with mutations of NOTCH1, JAK1 and rearrangement of SET-NUP214 in T-cell acute lymphoblastic leukemia. Haematologica. 2011;96:1808–1814. doi: 10.3324/haematol.2011.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rao S, et al. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood. 2012;120:3764–3773. doi: 10.1182/blood-2012-03-415349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagel S, et al. Activation of TLX3 and NKX2-5 in t(5;14)(q35;q32) T-cell acute lymphoblastic leukemia by remote 3’-BCL11B enhancers and coregulation by PU.1 and HMGA1. Cancer Res. 2007;67:1461–1471. doi: 10.1158/0008-5472.CAN-06-2615. [DOI] [PubMed] [Google Scholar]
- 49.Bernard OA, et al. A new recurrent and specific cryptic translocation, t(5;14)(q35;q32), is associated with expression of the Hox11L2 gene in T acute lymphoblastic leukemia. Leukemia. 2001;15:1495–1504. doi: 10.1038/sj.leu.2402249. [DOI] [PubMed] [Google Scholar]
- 50.Kunz JB, et al. Pediatric T-cell lymphoblastic leukemia evolves into relapse by clonal selection, acquisition of mutations and promoter hypomethylation. Haematologica. 2015;100:1442–1450. doi: 10.3324/haematol.2015.129692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li L, et al. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood. 2013;122:902–911. doi: 10.1182/blood-2012-08-447839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugimoto K-J, et al. T-cell lymphoblastic leukemia/lymphoma with t(7;14)(p15;q32) [TCRγ-TCL1A translocation]: a case report and a review of the literature. Int J Clin Exp Pathol. 2014;7:2615–2623. [PMC free article] [PubMed] [Google Scholar]
- 53.Virgilio L, et al. Deregulated expression of TCL1 causes T cell leukemia in mice. Proc Natl Acad Sci U S A. 1998;95:3885–3889. doi: 10.1073/pnas.95.7.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campbell IM, Shaw CA, Stankiewicz P, Lupski JR. Somatic mosaicism: implications for disease and transmission genetics. Trends Genet. 2015;31:382–392. doi: 10.1016/j.tig.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dou Y, Gold HD, Luquette LJ, Park PJ. Detecting Somatic Mutations in Normal Cells. Trends Genet. 2018;34:545–557. doi: 10.1016/j.tig.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voet T, et al. Breakage-fusion-bridge cycles leading to inv dup del occur in human cleavage stage embryos. Hum Mutat. 2011;32:783–793. doi: 10.1002/humu.21502. [DOI] [PubMed] [Google Scholar]
- 59.Bakhoum SF, et al. The mitotic origin of chromosomal instability. Curr Biol. 2014;24:R148–9. doi: 10.1016/j.cub.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang YK, et al. Genomic consequences of aberrant DNA repair mechanisms stratify ovarian cancer histotypes. Nat Genet. 2017;49:856–865. doi: 10.1038/ng.3849. [DOI] [PubMed] [Google Scholar]
- 61.Rücker FG, et al. Chromothripsis is linked to TP53 alteration, cell cycle impairment, and dismal outcome in acute myeloid leukemia with complex karyotype. Haematologica. 2018;103:e17–e20. doi: 10.3324/haematol.2017.180497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Navin NE, Hicks J. Tracing the tumor lineage. Mol Oncol. 2010;4:267–283. doi: 10.1016/j.molonc.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee H, Kim J-S. Unexpected CRISPR on-target effects. Nat Biotechnol. 2018;36:703–704. doi: 10.1038/nbt.4207. [DOI] [PubMed] [Google Scholar]
- 64.Yoshihara M, Hayashizaki Y, Murakawa Y. Genomic Instability of iPSCs: Challenges Towards Their Clinical Applications. Stem Cell Rev. 2017;13:7–16. doi: 10.1007/s12015-016-9680-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanders AD, Falconer E, Hills M, Spierings DCJ, Lansdorp PM. Single-cell template strand sequencing by Strand-seq enables the characterization of individual homologs. Nat Protoc. 2017;12:1151–1176. doi: 10.1038/nprot.2017.029. [DOI] [PubMed] [Google Scholar]
- 66.Mooijman D, Dey SS, Boisset JC, Crosetto N, van Oudenaarden A. Single-cell 5hmC sequencing reveals chromosome-wide cell-to-cell variability and enables lineage reconstruction. Nat Biotechnol. 2016;34:852–856. doi: 10.1038/nbt.3598. [DOI] [PubMed] [Google Scholar]
- 67.Frismantas V, et al. Ex vivo drug response profiling detects recurrent sensitivity patterns in drug-resistant acute lymphoblastic leukemia. Blood. 2017;129:e26–e37. doi: 10.1182/blood-2016-09-738070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.1000-Genomes-Project-Consortium et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv [q-bioGN] 2012 [Google Scholar]
- 70.Huber W, Toedling J, Steinmetz LM. Transcript mapping with high-density oligonucleotide tiling arrays. Bioinformatics. 2006;22:1963–1970. doi: 10.1093/bioinformatics/btl289. [DOI] [PubMed] [Google Scholar]
- 71.Claussin C, et al. Genome-wide mapping of sister chromatid exchange events in single yeast cells using Strand-seq. Elife. 2017;6 doi: 10.7554/eLife.30560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Porubsky D, et al. Direct chromosome-length haplotyping by single-cell sequencing. Genome Res. 2016;26:1565–1574. doi: 10.1101/gr.209841.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 74.Klambauer G, et al. cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res. 2012;40:e69. doi: 10.1093/nar/gks003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rausch T, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fan J, et al. Linking transcriptional and genetic tumor heterogeneity through allele analysis of single-cell RNA-seq data. Genome Res. 2018;28:1217–1227. doi: 10.1101/gr.228080.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lapunzina P, Monk D. The consequences of uniparental disomy and copy number neutral loss-of-heterozygosity during human development and cancer. Biol Cell. 2011;103:303–317. doi: 10.1042/BC20110013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data from this study can be retrieved from the European Genome-phenome Archive (EGA), and the European Nucleotide Archive (ENA) [accessions: PRJEB30027, PRJEB30059, PRJEB8037, PRJEB33731, EGAS00001003248, EGAS00001003365]. Access to human patient data is governed by the EGA Data Access Committee.
The computational code of our analytical framework is hosted on GitHub (see https://github.com/friendsofstrandseq/mosaicatcher-pipeline, https://github.com/friendsofstrandseq/TranslocatoR, and https://github.com/friendsofstrandseq/mosaicatcher). All code is available freely for academic research.