Abstract
Molecular profiling of small-molecules offers invaluable insights into the function of compounds and allows for hypothesis generation about small molecule direct targets and secondary effects. However, current profiling methods are either limited in the number of measurable parameters or throughput. Here, we developed a multiplexed, unbiased framework that, by linking genetic to drug-induced changes in nearly a thousand metabolites, allows for high-throughput functional annotation of compound libraries in Escherichia coli. First, we generated a reference map of metabolic changes from (CRISPR) interference with 352 genes in all major essential biological processes. Next, based on the comparison of genetic with 1342 drug-induced metabolic changes we made de novo predictions of compound functionality and revealed antibacterials with unconventional Modes of Action. We show that our framework, combining dynamic gene silencing with metabolomics, can be adapted as a general strategy for comprehensive high-throughput analysis of compound functionality, from bacteria to human cell lines.
Introduction
Due to the rapid emergence of antibiotic-resistant bacteria, there is an urgent need to speed up the discovery of antimicrobial compounds with new mechanisms of action1. However, the discovery of antimicrobials that can target new essential functions is extremely difficult 2 3 4. Despite significant progress in synthetic chemistry and the target-based design of biologically relevant molecules, high throughput growth inhibitory assays still represent a major pillar of drug discovery5. The lack of mechanistic knowledge on lead compound’s Modes of Action (MoA) is a major limitation, often leading to the rediscovery of conventional antibacterial compounds and hampering compound optimization, minimization of side effects, drug repurposing and rational design of combination therapies 6 7.
To fill this gap, there has been a continuous interest in reinventing phenotypic screens by integrating low-dimensional readouts (e.g. growth-rate, colony size) with high-content assays, such as image-based screening8 9 10, or molecular profiling11 12, simultaneously monitoring several consequences of small-molecule treatments. Comparison between multidimensional characteristics of small molecule effects is a powerful strategy to predict if a lead compound has a similar MoA and possibly bioactivity to drugs with already characterized mechanisms of action13 14. This approach has proven successful with a number of different molecular profiling technologies, like transcriptomics15 11, proteomics16, and metabolomics17 12 .
Compared to more mature omics profiling technologies, like transcriptomics and proteomics, non-targeted metabolomics is a cost-effective solution and still offers a throughput advantage, in that it can scale with the typical size of chemical libraries. Moreover, while most high-throughput technologies monitor growth-related phenotypes (e.g. colony size, morphology), by monitoring changes in thousands of cellular metabolites, current metabolomics platforms18 19 20 provide a rich multidimensional representation of drug effects that is largely independent from compounds’ growth inhibitory activity7. However, deconvolution of drug targets and mechanisms responsible for antibacterial activity remains challenging. Modern genetic tools21,22 to control expression of growth-essential genes and to systematically construct genome-wide mutant libraries opened new opportunities to expand the set of drug targets and to tackle fundamental bottlenecks in antimicrobial drug development and discovery22.
Here, by leveraging CRISPR technology23 and non-targeted metabolomics, we propose a combined computational/experimental strategy to perform high-throughput de novo functional annotations of antibacterials. We show that by comparing genetic to chemically induced metabolic changes is a scalable and general solution to gain mechanistic insights on small molecule functions and make experimentally testable hypotheses on direct and proximal drug targets in a single-pass screen. First, we validated the approach in Escherichia coli, Mycobacterium smegmatis and a lung cancer cell line on a set of antimicrobials and anticancer drugs with characterized MoAs. Then we applied our framework to a library of chemically diverse compounds (i.e. Prestwick library) and functionally annotate small-molecules with antimicrobial activity but unknown MoAs.
Results
Charting the metabolic landscape of essential genes
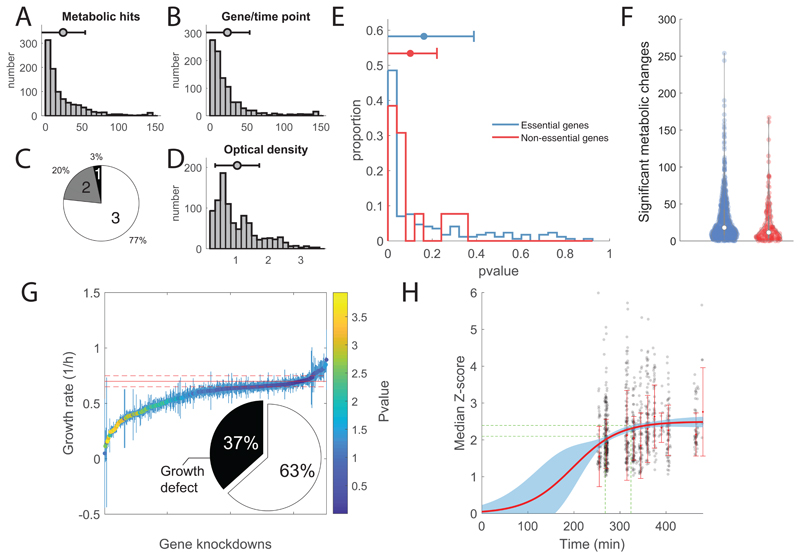
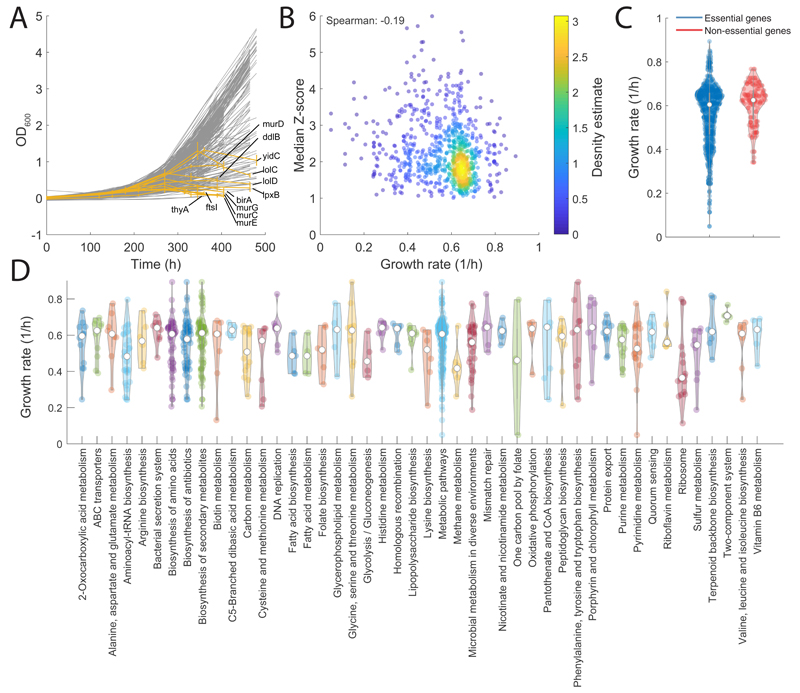
Small molecules exhibiting antimicrobial activity often act by inhibiting proteins essential for growth and disrupting metabolic homeostasis24 25. However, little is known on the metabolic consequences of limiting the expression of essential genes and whether genetic interference can mimic metabolic changes induced by chemical inhibition of essential proteins. Here we used a library of CRISPRi tunable knockdowns of essential genes in the model gram-negative bacterium E. coli 23. The arrayed strain library consists of mutant strains in which we can sterically hinder transcription at the sgRNA base-pairing genomic locus and ultimately induce selective knockdown of 376 gene targets, out of which 304 encodes for growth-essential proteins in glucose minimal medium (M9) (Supplementary Dataset 1). Each mutant strain was grown in glucose M9 for 12 hours before inoculation in glucose M9 with 1 mM IPTG, a concentration which induces ~10 fold repression of rfp 23. Because proteins are expressed and become limiting at different levels, samples were collected at multiple time points during mid-log growth phase between 3 to 7 hours after inoculation. Metabolic changes were measured by flow-injection time-of-flight mass spectrometry (FIA-TOFMS)18 (Supplementary Dataset 1). Overall, we detected 991 putatively annotated metabolites, and an average of 3 time points per mutant strain were profiled (Extended Data Figure 1). Raw mass spectrometry data were normalized by correcting for instrumental biases (e.g. plate effects) and systematic changes in cell numbers (i.e. optical density) (see Online Methods). Relative log2 fold-changes of metabolite levels were estimated for each mutant/time point with respect to wild-type levels7. A Z-score normalization was applied after estimating the average and standard deviation of fold-changes over the three replicates (Supplementary Dataset 1). Consistent with previous results22 26, a considerable fraction (63 %) of knockdown strains exhibited mild or no growth defects (pvalue>0.05) compared to wild-type (Extended Data Figure 1E), suggesting that growth of E. coli is robust to fluctuations in levels of essential proteins. For 12 genes, lowering gene expression caused a delayed drop in optical density (OD600) (Extended Data Figure 2). Consistent with cell lysis being a characteristic consequence of interfering with cell envelope, most of these genes (9 genes) are involved in cell wall biosynthesis and trafficking of lipoproteins (Extended Data Figure 2).
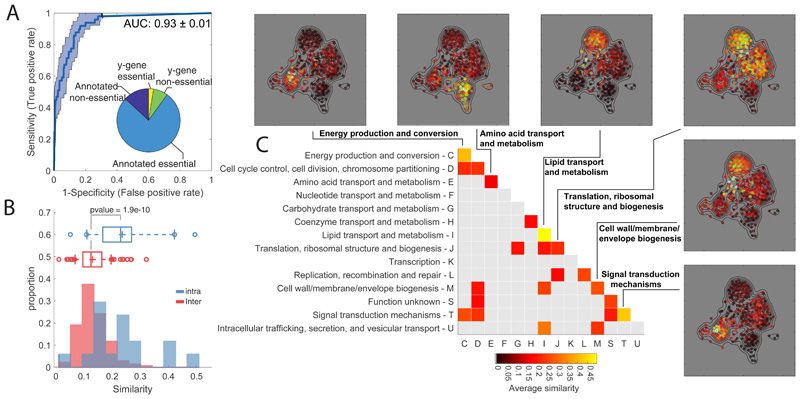
Figure 1. Metabolome profiling of essential gene knockdown.
A) ROC analysis of the similarity between metabolome profiles of non-essential enzyme knockout18 vs knockdown. The pie chart reports the fraction of non/essential genes in the CRISPRi mutant library with characterized vs non-characterized (i.e. y-genes) function. B) Histograms of the averaged similarities between genes from the same (blue) or different (red) functional groups (COG). The tops and bottoms of each box are the 25th and 75th percentiles, respectively, while the line in the middle of each box is the samples median. The lines extending above and below each box are the whiskers. Whiskers extend from the ends of the boxes delimited by the interquartile to the largest and smallest observations. Pvalue estimated by one-tailed t-test analysis. C) Heatmap of metabolome similarity between functional groups (i.e. COGs). For each pair of genes within the same functional group (diagonal) or between two different functional groups (off-diagonal), we estimated the average of similarity scores (iSim) and use a permutation test to estimate pvalue significance. COG names and identifiers are reported on the Y- and X-axis, respectively. Only similarities with pvalue≤0.05 are reported. The similarity between genes of the same functional group is illustrated for selected cases by a 2-dimensional projection of metabolome profiles. Metabolic profile similarities were calculated for all gene pairs by computing iSim. Each dot correspond to a gene knockdown/time point laid out using an edge-weighted, spring-embedded, network layout algorithm30. Genes sharing similar metabolic profiles are proximal to each other; less-similar genes are positioned farther apart. Each dot is colorcoded by the average distance to knockdowns from a specific functional group (green dots).
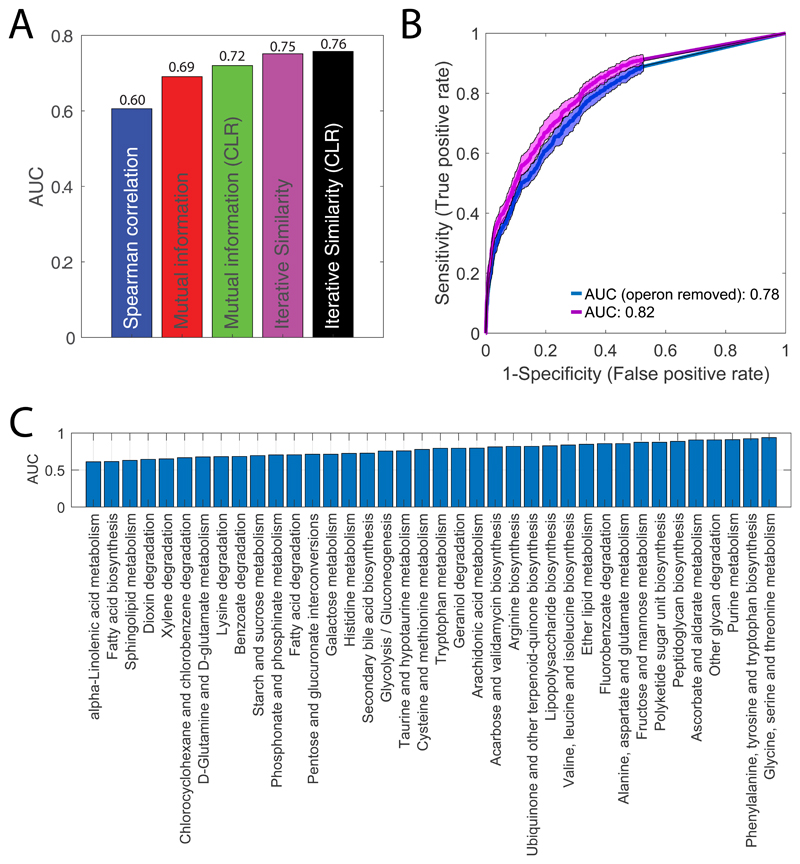
All 352 gene knockdowns exhibited at least one significant metabolic change during the course of the experiment (absolute Z-score ≥1 and pvalue ≤ 1e–5 Bonferroni-adjusted threshold) (Extended Data Figure 1). While the metabolic impact of gene knockdown varies with the targeted gene (Extended Data Figure 1), on average, after CRISPR induction metabolic changes increase with time until reaching a plateau approximately after 6 hours (Extended Data Figure 1). To test the specificity and reliability of the CRISPRi library and whether the protocol used for knockdown induction was sufficient to make protein availability limiting, we compared metabolic profiles between 49 non-essential gene knockdowns and their respective knockout strains27 (Supplementary Dataset 1). To measure the metabolic similarity between mutant strains, we perfected our previously developed similarity metric, we called iterative similarity7 (iSim). When assessing functional associations from metabolome profiles, iSim outperforms other similarity metrics – i.e. correlation metrics, such as Spearman, but also dependency measures, like mutual information (Extended Data Figure 3). Receiver operating characteristic (ROC) curve analysis showed that metabolic signatures observed in non-essential knockout mutant strains could be recapitulated by CRISPRi knockdown of the respective genes (Area Under the Curve - AUC = 0.93, Fig. 1A). Moreover, similar to knockout strains of non-essential enzymes27, knockdowns of essential enzymes tend to elicit metabolic changes in metabolites that are proximal within the metabolic network (Extended Data Figure 1). Altogether, the herein generated compendium of metabolome profiles provides an unprecedented reference set of metabolic signatures to interrogate the consequences of interfering with a largely diverse set of essential cellular functions.
Gene function-specific metabolic profiles
Because the CRISPRi mutant library consists of genes with largely diverse functions, we could systematically investigate whether knockdowns of genes with similar biological functions elicit similar metabolic changes. First, we used the Cluster of Orthologous Groups (COGs) classification which provides a broad classification of all genes in 26 functional groups28. For each group we estimated the average pairwise similarity and excluded gene pairs from the same operon to account for CRISPRi polarity22. We found that genes involved in the same essential process have significantly (pvalue<1e-9) higher similarity than between genes from different functional groups (Fig. 1B, Extended Data Figure 3 and Supplementary Dataset 1-2). We also found some notable exceptions and identified crosstalk between distinct functional groups (Fig. 1C). Some of the inter-similarity between functional groups are expected or obvious. For example, the similarity between ribosomal biogenesis and carbohydrate metabolism, reflecting the coordination between bacterial proteome and carbon metabolism29. In other cases, the origin of similarity is less obvious, suggesting highly coordinated regulation of functionally different processes like cell division and energy metabolism (Fig. 1C).
Next, we quantified the ability to retrieve specific gene functions using the Kyoto Encyclopedia of Genes and Genomes (KEGG annotation) (Supplementary Dataset 2). Even after accounting for CRISPRi polarity due to operon structure, comparing enrichment of KEGG pathways with known gene annotations revealed that metabolome-based similarity is highly predictive of gene functions (AUC>0.75) (Extended Data Figure 3). Hence, we demonstrated that despite interfering with major cellular hubs30, knocking down an essential gene induces metabolic changes predictive of its function. As expected, several of the most accurate resolved functions are directly related to metabolism, such as amino acids biosynthesis pathways (Extended Data Figure 3). Nevertheless, also metabolic changes induced by knockdowns of non-metabolic genes are highly specific of gene functions, e.g. ribosomal genes (AUC>0.8) (Extended Data Figure 2). Altogether, we showed that combining the CRISPR interference system with metabolome profiling can systematically reveal functional gene characteristics and new metabolic connections between essential cellular functions.
Testing the ability to predict antibiotic MoAs and targets
While we showed that interference with the expression of essential genes elicit function-specific metabolic changes, it remained to be investigated if chemical interference with the same gene products produces similar metabolic consequences. To this end, we profiled the metabolic responses of wild-type E. coli two hours after exposure to 63 commonly used antibiotics at 10μM (Supplementary Dataset 3) and combined this data with similar drug metabolic profiles previously acquired for 148 antimicrobials which are part of a chemically diverse library of 1279 small molecules applied at 100μM7 (i.e. Prestwick library). We considered five major antibiotic classes: protein, RNA, folic acid, cell wall biosynthesis, and DNA replication inhibitors (Supplementary Dataset 3). First, we estimated the pairwise metabolic similarity between gene knockdown and antimicrobial treatment. Next, we estimated the significance of the average similarity between genes in each COG and KEGG pathway and antibiotics in the five different classes. Remarkably, the strongest associations were found between antibiotic classes and gene knockdowns functionally related to antibiotic targets (Fig. 2A) - e.g. protein synthesis inhibitors tend to induce similar metabolic changes to silencing of genes involved in translation and ribosomal biogenesis, while antibiotics inhibiting DNA replication induced most similar metabolic changes to knockdowns of genes involved in DNA replication, recombination and repair. When zooming in at the KEGG pathway level, the associations became more accurate and specific (Fig. 2A). For example, we found that the strongest gene associations with inhibitors of folic acid biosynthesis were exclusively enriched for genes in folate synthesis (Fig. 2A and Supplementary Dataset 3).
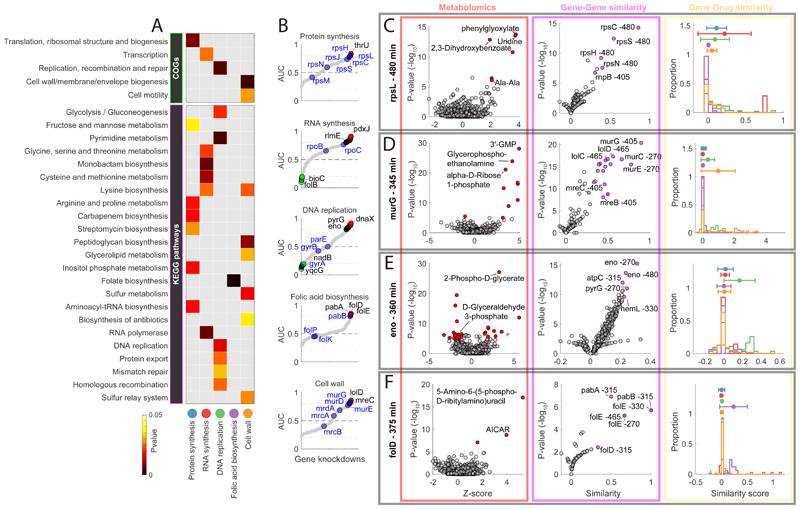
Figure 2. Functional associations between gene knockdowns and major antibiotic classes.
A) The heatmap highlights the functional groups of genes (i.e. COGs or KEGG pathways) that exhibit the most significant (pvalue≤0.05) averaged similarity to metabolic changes induced by antibiotics in 5 major classes. B) Sorted maximum AUC indexes estimated for each knockdown and antibiotic class (grey dots). Primary drug targets are highlighted in blue. Genes with an AUC greater than 0.8 or lower than 0.2 are marked in red and green respectively. C-D-E-F) Metabolic profiles (red panel), gene-gene similarity (purple panel) and associations with compounds in 5 major antibiotic classes (yellow) panel, for 4 gene knockdowns: rpsL (480 minutes after CRISPRi induction), murG (345 minutes after CRISPRi induction), eno (360 minutes after CRISPRi induction) and folD (375 minutes after CRISPRi induction). The volcano plots illustrate: the metabolic changes averaged over 3 biological replicates (i.e. Z-score and pvalue significance) for 991 putatively annotated metabolites (left, red panel); similarity between knockdown metabolic profiles (i.e. similarity and pvalue significance)(right, purple panel). Metabolites that are significantly (two-tailed t-test analysis) affected by the gene knockdown and genes that exhibit significant (hypergeometric test) metabolic similarities are highlighted in red and purple, respectively. Histograms (yellow panel) represent the distributions of similarities scores (iSim) with compounds in each major antibiotic class.
Overall, we showed that comparing metabolic consequences of silencing essential genes to the metabolic effects of antibiotic treatment is an effective and rapid strategy to identify the essential functions inhibited by small molecules. But can we identify primary drug targets? To address this question, for each gene knockdown and time point we assessed the specificity of metabolic similarities for antibiotics in the five distinct classes. For each knockdown, time point and antibiotic class, we calculated the Area Under the ROC Curve (AUC) index, i.e. the higher is the AUC the more specific and similar is the knockdown-induced metabolic signature to metabolic changes induced by drugs from one antibiotic class. Differently, a low AUC (e.g. AUC<0.2), means that the gene knockdown affected similar metabolites as the antibiotic, but in opposite directions (Supplementary Dataset 3). Remarkably, we identified gene knockdowns eliciting metabolic changes that are highly predictive (AUC>=0.8) of each antibiotic class (Fig. 2B). While several knockdowns of genes encoding for primary antibiotic targets, such as rpsL or murG(Fig. 2C-D), reproduced antibiotic-specific metabolic profiles, on average, we found that antibiotic-induced metabolic signatures most strongly associated with those of genes involved in the downstream effects of antibiotic treatment (Fig. 2E-F). Emblematic examples are inhibitors of folic acids biosynthesis, for which the most predictive knockdown folD (Fig. 2F) locates downstream of primary targets (e.g. folP).
Not surprisingly, we found that for antibiotic classes in which small molecules directly interfere with the catalytic activity of the target, such as ribosome, RNA or cell wall synthesis inhibitors, perturbing known antibiotic targets mimicked antibiotic-induced metabolic signatures (Fig. 2B). While antibiotics can act by blocking the overall catalytic functions of their targets (i.e. enzyme inhibition), antibacterial activity can also arise from modifying target catalytic functions. For example, quinolones increase the concentration of gyrase–DNA cleavage complexes, thereby turning gyrase into a toxic enzyme fragmenting the DNA and increasing levels of double strand breaks. We found that repressing expression of gyrA caused opposite metabolic changes to inhibitors of DNA replication (AUC<0.2) (Fig. 2B), with some notable exceptions like novobiocin, a competitive inhibitor of the ATPase reaction catalysed by GyrB. This result is consistent with the previously reported attempt of E. coli to buffer gyrase inhibitors effects by the overexpression of gyrA25. Hence, it is plausible that by inducing target overexpression, drug treatment partially caused metabolic effects opposite to gyrA knockdown. On the other hand, on average, DNA replication inhibitors induce metabolic changes that are similar to repressing essential genes involved in DNA replication (e.g. dnaB), cell cycle regulation (e.g. ftsZ), and non-essential genes in DNA synthesis (e.g. pyrG), revealing secondary drug effects and suggesting new potential targets to synergize with the drug mechanism of action (Extended Data Figure 6). Altogether, we demonstrate that phenocopying drugs’ action by genetic interference with their primary targets is not always possible. However, we show that for all current classes of antibiotics there exist genes functionally close to primary drug-targets that when knocked down feature similar metabolic consequences to drug treatment, thereby revealing mechanistic insights on compound functions. It is worth noting that function-specific metabolic signatures mimicking drug effects are not exclusive to metabolic genes or drugs with metabolic targets, but are also found for genes functionally distant from metabolism, e.g. gyrase or ribosomes.
From bacteria to human cells: a generalizable approach
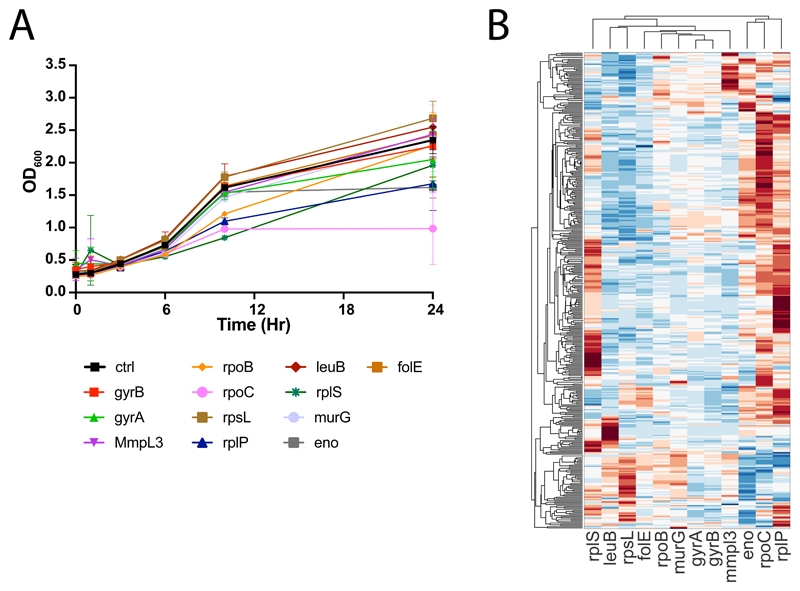
The next question we asked is whether the same principles and approach could be applied to other organisms. To this end, we first collected dynamic metabolic profiles of 12 essential gene knockdown in the acid-fast bacterium Mycobacterium smegmatis 31 (Supplementary Dataset 4). Based on the drug-gene similarity analysis in E. coli (Fig. 2B), we selected 10 representative genes (i.e. gyrA, gyrB, folE, mmpl3, murG, rplP, rplS, rlpS, rpoC, rpoB) and two enzymes that are functionally conserved between E. coli and M. smegmatis (i.e. eno, leuB). We profiled metabolic changes 3.5, 6, 10 and 24 hours after CRISPR induction (Extended Data Figure 4). Similar to E. coli, knockdowns of eno and leuB exhibit the characteristic accumulation (pvalue≤1e-5) of metabolic substrates (i.e. 2-phospho-D-glycerate and (2R,3S)-3-isopropylmalate, respectively) (Fig. 3A-B and Fig. 2E), confirming the ability to characterize the specific metabolic effect of interference with essential genes in largely diverse bacterial species (Supplementary Dataset 4). To further investigate whether metabolic profiles of knockdowns are able to mimic drug induced metabolic changes, we applied the same approach described for E. coli and used iSim to estimate the similarity between each pair of gene knockdowns and previously acquired metabolome profiles12 for 42 antimicrobials belonging to the 5 major antibiotic classes tested in E. coli (Fig 3C-L). Consistent with E. coli, we found that knockdowns of ribosomal, folate and cell wall biosynthesis related genes exhibit stronger metabolic similarity with inhibitors of the respective functions (Fig. 3C-L). Remarkably, the same opposite effects between knockdown of gyrA and inhibitors of DNA replication observed in E. coli (Fig. 2B) emerged also in M smegmatis (Fig. 3I). Moreover, similarly to E.coli, knockdown of rpoC featured a stronger similarity to inhibitors of RNA polymerases than rpoB knockdown (Fig. 3F-G and Fig. 2B). However, differently from E. coli, we observed a strong similarity between DNA replication inhibitors and rpoC knockdown (Fig. 3F), possibly reflecting the physical binding between the gyrase and RNA polymerase complexes in M. smegmatis 32.
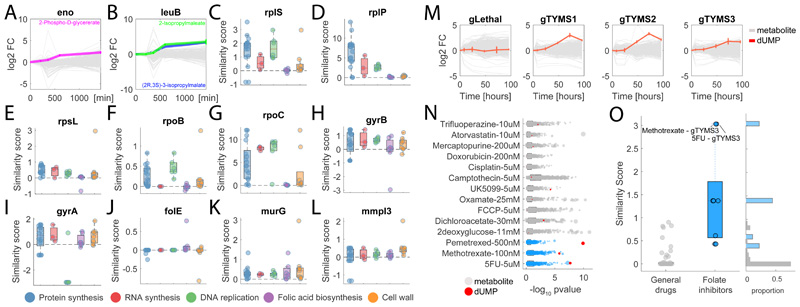
Figure 3. Metabolic profiling of chemical and genetic perturbations in M. smegmatis and A549 cancer cells.
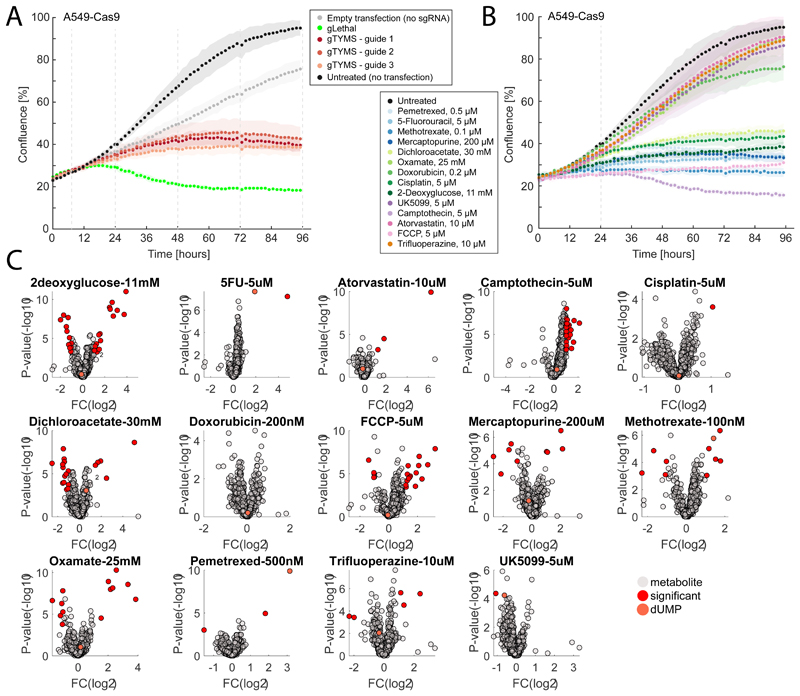
A-B) Time-course profiles of changes in intracellular metabolite abundances after CRISPR-mediated knockdown of enolase (eno) and 3-isopropylmalate dehydrogenase (leuB) in M. smegmatis. Reported data are average over 3 biological replicates. C-L) Each dot represents the metabolic similarity (i.e. similarity scores) between genetic and chemical perturbations in M. smegmatis. Antimicrobials are grouped in 5 major antibiotic classes and the distribution of similarities with gene knockdowns metabolic profiles are illustrated by box plots. The tops and bottoms of each box are the 25th and 75th percentiles, respectively, while the red line in the middle of each box is the samples median. The lines extending above and below each box are the whiskers. Whiskers extend from the ends of the boxes delimited by the interquartile to the largest and smallest observations. M) Time-course profiles of changes in intracellular metabolite abundances in A549 lung cancer cells after CRISPR-mediated knockout of thymidylate synthase (TYMS), using three individual guide RNAs targeting different gene exons and a separate guide RNA targeting multiple genomic regions (gLethal) is used to induce general growth inhibition (Extended Data Figure 5). Changes in dUMP are highlighted in red. Reported data are average ± standard deviation over 3 biological replicates. N) Metabolic changes induced by 14 drugs with diverse targets and MoAs. Each dot corresponds to a putatively annotated metabolite, the log2 fold-change is reflected in the dot size, and statistical significance (p-value estimated by by two-tailed t-test analysis) on the x-axis. Drugs targeting enzymes in folate metabolism are highlighted in blue, changes in dUMP abundance in red. O) Boxplot of metabolic similarity (i.e. similarity scores) between genetic and chemical perturbations in A549 human lung cancer cells. Similarities between genetic and chemical interference with TYMS (blue) are compared to all remaining pairwise similarities (grey). The tops and bottoms of each box are the 25th and 75th percentiles, respectively. The lines extending above and below each box are the whiskers. Whiskers extend from the ends of the boxes delimited by the interquartile to the largest and smallest observations.
To illustrate the relevance of this approach beyond antimicrobials and its applicability to virtually any type of organism or drug, we tested its performance in human cells. Specifically, we used a lung cancer cell line (A549) to interfere with the essential enzyme thymidylate synthase, target of several anticancer drugs. For the CRISPR-mediated knockout of thymidylate synthase (TYMS), we monitored the dynamic intracellular metabolic effects up to 96 hours of incubation with sgRNAs targeting three different gene sections (gTYMS1-3) in addition to a control sgRNA inducing cell death (gLethal) by targeting multiple sections in the genome. In parallel, we measured metabolic changes 48 hours after perturbation of cancer cells with 14 drugs (Extended Data Figure 5 and Supplementary Dataset 5), out of which 3 are targeting TYMS (methotrexate, pemetrexed and 5-fluorouracil), while the remaining drugs act on largely different cellular processes (Fig S5). Genetic interference with TYMS elicited a characteristic accumulation of the substrate of the thymidylate synthase reaction: deoxyuridine monophosphate (dUMP) (Fig. 3M). Remarkably, dUMP accumulation is observed only for drugs targeting TYMS (Fig. 3N) and resulted in a strong and selective metabolic similarity between genetic and chemical induced repression of TYMS (Fig. 3O). In conclusion, we provided a proof of principle that the same metabolome-based annotation of compound functionality is largely applicable to different systems, from bacteria to mammalian cell lines, and drug discovery fields, including anticancer discovery.
De novo prediction of antimicrobials MoA
By comparing small molecule-induced metabolic changes with the reference set of metabolic profiles from essential gene knockdowns we can rapidly identify cellular functions targeted by the small-molecule and, in the case of unconventional MoAs, make experimentally testable hypotheses on targets. We applied this approach in E. coli on a previously profiled drug library of 1279 chemically diverse small molecules (i.e., Prestwick Chemical Library)7, the majority (75%) consisting of human-targeted drugs. First, we estimated all pairwise similarities (iSim) between time-dependent metabolic profiles of gene-knockdowns and drug-induced metabolic changes and focused on the top 1% of gene-drug associations. In total, we found metabolic associations with at least one gene knockdown for 516 compounds (Fig. 4A). A major and significant fraction of these compounds (i.e. ~65%, pvalue≤1e-5) exhibits growth inhibitory activity against E. coli – i.e. 50% growth inhibitory concentration (GI50) below 100μM (Extended Data Figure 6). Metabolic similarities with essential genes such as lgt, ddlB, murG, lolD or lolC are the most frequent associations found among compounds in the library (Fig. 4A), suggesting that many of the compounds are interfering with the cell envelope.
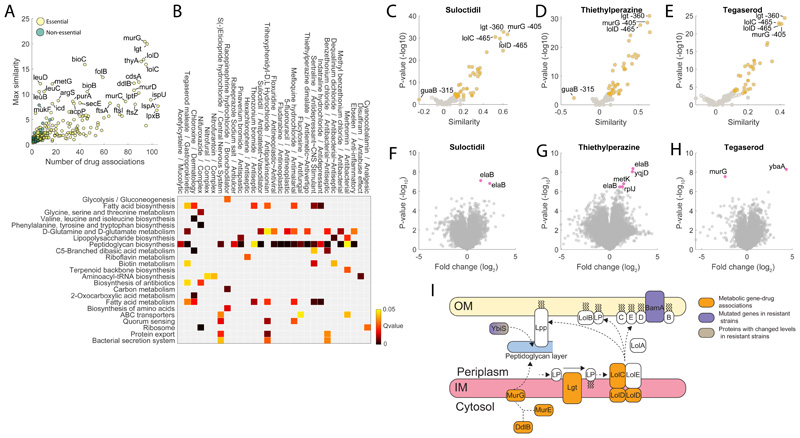
Figure 4. Chemical vs genetic interference.
A) Number of significant (top 1%) associations with drugs and the max similarity for each gene knockdown. B) The heatmap highlights drugs with a GI50 below 100μM that exhibit a significant (qvalue≤0.05) enrichment of gene associations with at least one KEGG pathway. C) Volcano plot of similarities between suloctidil and knockdowns metabolic profiles. D) Volcano plot of similarities between thietylperazine dimalate and knockdowns metabolic profiles. E) Volcano plot of similarities between tegaserod maleate and knockdowns metabolic profiles. Pvalues were estimated by hypergeometric test (see iSim). G) Volcano plots of limited proteolysis samples from E. coli lysates treated with suloctidil. Peptide mixes produced in the presence or absence of 500 μM suloctidil are compared. Fold changes (FC) in peptide abundance for treated and untreated samples are shown as a function of statistical significance. Significance cutoffs were pvalue = 3.3807e-07 (two-tailed t-test analysis, Bonferroni corrected) and FC = 1. Peptides passing both cutoffs are in purple and labeled by protein name (Supplementary Dataset 4). H) Volcano plots of limited proteolysis samples from E. coli lysates treated with thietylperazine dimalate. I) Volcano plots of limited proteolysis samples from E. coli lysates treated with tegaserod maleate. Data represent the average over 3 biological replicates. I) Schematics of metabolome derived gene-tegaserod associations and their relation to genetic and proteome changes in tegaserod resistant strains.
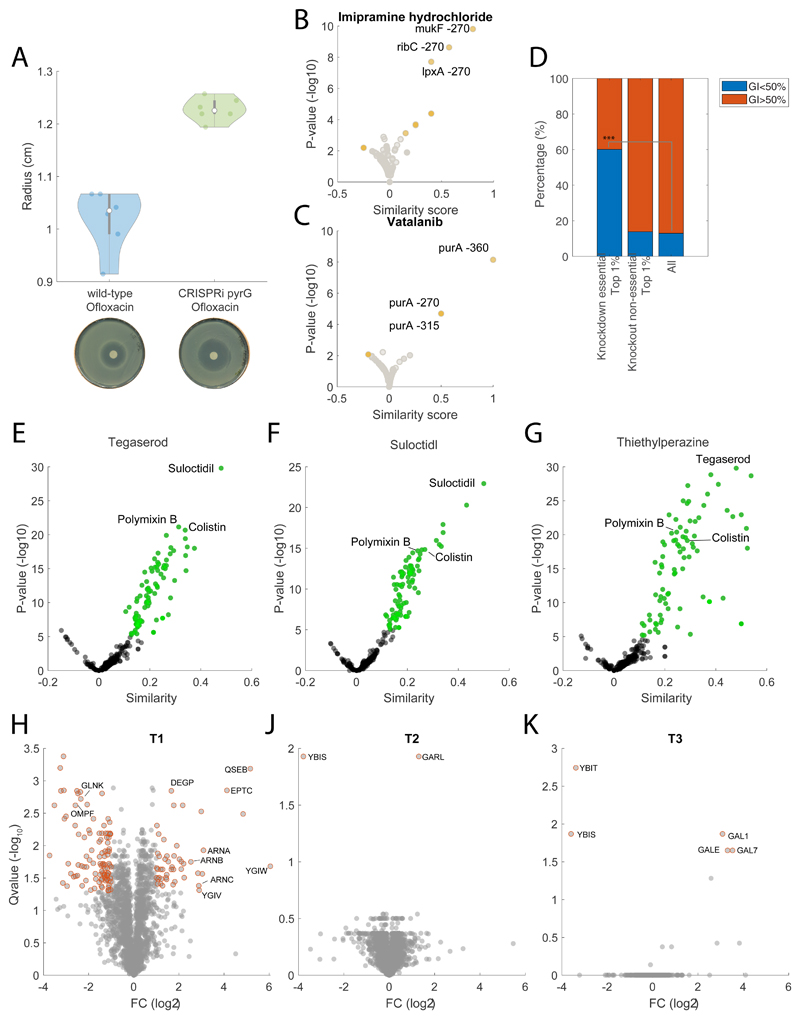
We restricted our analysis to the functional annotation of 39 compounds that exhibit growth inhibitory activity (i.e. GI50 below 100μM) and have a poorly characterized antibacterial mechanism of action. By performing KEGG enrichment analysis of metabolome-based similarity with gene knockdowns, we found significantly (qvalue≤0.05) enriched KEGG pathways for 29 out of the 39 selected drugs (Fig. 4B). A large fraction of the compounds (20 of 29) was predicted to interfere with the cell envelope, especially with periplasmic targets (e.g. Igt, LolC/E), possibly reflecting the difficulties of compounds in reaching essential targets in the cytoplasm. The three drugs exhibiting the largest metabolic similarities – i.e. suloctidil, thiethylperazine and tegaserod, induced metabolic changes similar to knockdowns of the lipoprotein release complex (lolCDE) which is required for trafficking of lipoproteins to the outer membrane33, phosphatidylglycerol—prolipoprotein diacylglyceryl transferase (lgt) and N-acetylglucosaminyl transferase (murG) (Fig. 4C, D, E). Consistent with metabolome-based predictions, suloctidil was previously shown to cause general perturbations of membrane, vesicle trafficking and lipid biosynthesis functions in yeast34, and phenothiazine derivatives, like thiethylperazine, were found to affect the cell envelope proteins of E. coli and to cause morphological responses similar to those produced by β-lactams35. Moreover, all three drugs exhibit metabolome profiles similar to colistin and polymixin B (Extended Data Figure 6), reinforcing the predicted role in interfering with the bacterial outer membrane.
To support our metabolome-based predictions we probed for drug-induced protein structural alterations on a proteome-wide scale using limited proteolysis coupled to mass spectrometry (LiP-MS) 36. Protein extracts of E. coli harvested during exponential growth in M9 glucose without any drugs were incubated with suloctidil, thiethylperazine or tegaserod before conducting limited proteolysis. The analysis identified 29580 peptides mapping to 1504 unique proteins in E. coli (Supplementary Dataset 6). For each peptide, we quantified the difference in abundance between the treated and untreated proteome samples. Differences in peptide abundance reflected protein conformational changes induced by direct binding events or protein network rearrangements caused by the disruption of protein-protein interactions. (Fig. 4F-G-H and Supplementary Dataset 6). For all three drugs, LIP-MS analysis individuated only few protein structural alterations, that either directly supported our predictions, like in the case of tegaserod and its predicted effect on MurG (Fig. 4E), or identified proteins functionally linked to the predicted targets. Specifically, we found that tegaserod induced significant structural alterations (pvalue≤ 3.3807e-07 Bonferroni-corrected, and absolute log2 fold-changes≥1) in MurG, and YbaA, a nonessential protein with unknown function. Interestingly both thiethylperazine and suloctidil affected the conformation of ElaB, a small C-tail-anchored inner membrane protein also involved in membrane trafficking37. Moreover, in E. coli ElaB was found to physically interact with the penicillin-binding protein 1A (MrcA) 38 and inhibition of the LolCDE complex was shown to induce a strong (i.e. more than 10-fold) upregulation of elaB expression39. To further support our metabolome and LIP based evidence, we sought to identify mutations in E. coli strains that are resistant to tegaserod. Out of the six independent lineages of wild-type E. coli BW25133 that were allowed to evolve increasing resistance to tegaserod in LB medium, only 3 exhibited a mild but measurable level of resistance, between 2 (T1 mutant) and 1.3 times (T2 and T3 mutants) of the wildtype Minimal Inhibitory Concentration (MIC). Genome sequencing of a single isolate from each evolved population (i.e. T1, T2 and T3) revealed different mutations within the L,D-transpeptidase gene (ybiS) (Supplementary Dataset 7). YbiS is involved in the anchoring of the major outer membrane lipoprotein Lpp to the peptidoglycan layer, supporting metabolome-based similarities between tegaserod effects and knockdowns of lolCDE, lgt and murG, also indirectly involved in the structural anchoring between the outer membrane and the peptidoglycan (Fig. 4I). Moreover, whole proteomic analyses of mutant strains (Supplementary Dataset 8) revealed that T2 and T3 exhibit low levels of ybiS (Extended Data Figure 6), while in T1, point mutations (SNPs) in ybiS and the outer membrane protein assembly factor bamA associate with overexpression of several proteins involved in membrane stability (e.g. DegP, ArnA or EptC) (Extended Data Figure 6). Altogether, LIP-MS and genome sequencing analyses support metabolome-based MoA predictions, suggesting that tegaserod interferes with the essential structural anchoring between peptidoglycan and lipoproteins (Fig. 4I).
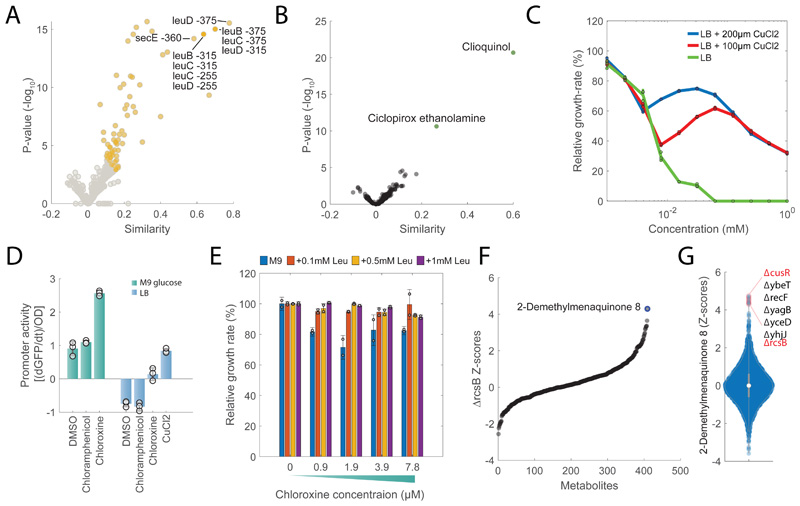
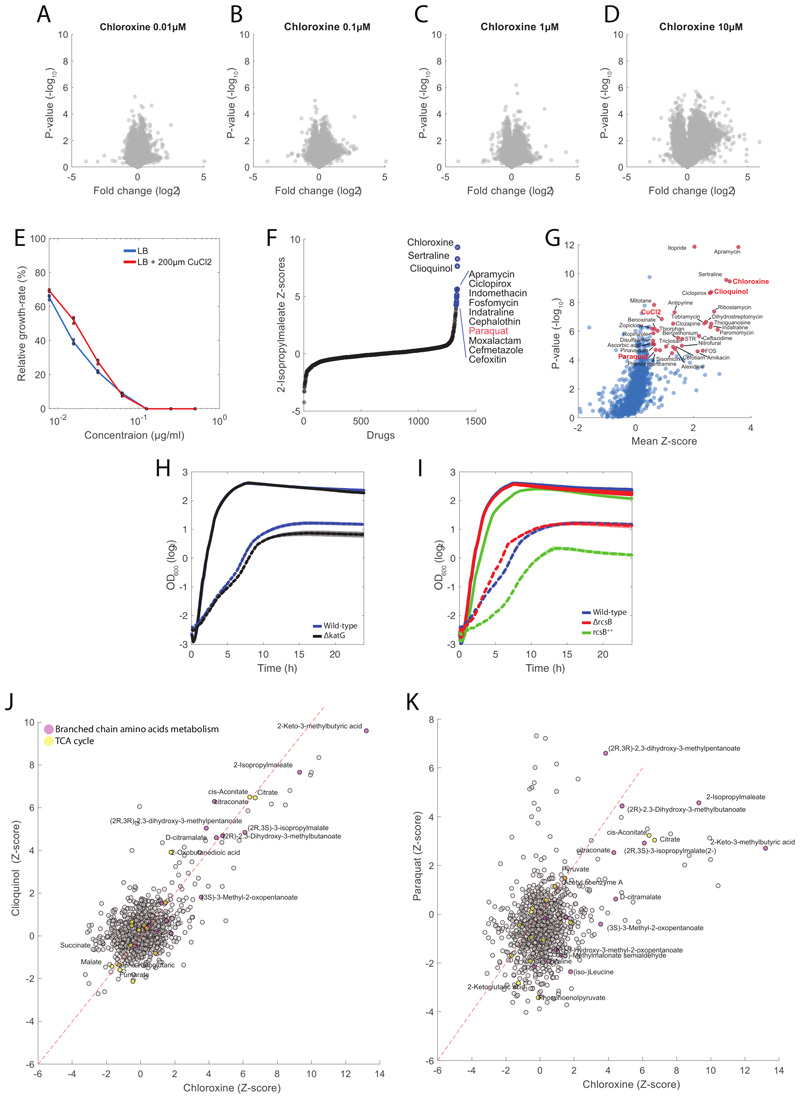
In addition to drugs targeting the cell wall, we also found drugs predicted to interfere with unconventional antibacterial targets, like chloroxine. Chloroxine has long been used as an antimicrobial in dermatology, but its mode of action is still unknown. Interestingly, we found that chloroxine induces a significant (pvalue≤1e-5) accumulation of intermediates in leucine biosynthesis (e.g. 2-Isopropylmaleate) and strongly associates to knockdowns of enzymes in the leucine pathway (e.g. LeuC, LeuD, Fig. 5A), raising the possibility that this compound interferes with branched chain amino acids (BCAA) biosynthesis. However, LiP-MS proteomics analysis did not detect any significant change in peptide patterns upon incubation of protein extracts with chloroxine (Extended Data Figure 7 and Supplementary Dataset 6), suggesting for an alternative MoA in which chloroxine acts without directly binding to proteins. Comparative analysis of chloroxine-induced metabolic changes revealed a striking similarity with clioquinol, another broad-spectrum antibiotic with unknown MoA (Fig. 5B). Both compounds have copper-chelating properties40 leading to the hypothesis that chloroxine acts primarily by interfering with copper availability. In agreement with this hypothesis, we verified that the MIC of chloroxine could be increased more than 5 fold with the supplementation of CuCl2 in the medium (Fig. 5C and Extended Data Figure 7). Surprisingly we found that CuCl2 supplementation doesn’t affect chloroxine inhibitory activity at low drug concentrations, and only partially rescued cells from chloroxine toxicity at concentrations above ~8μM (Fig. 5C). These results suggest that while chloroxine can inhibit bacterial growth by limiting copper availability, this is not the only growth inhibitory mechanism. Mounting evidence has shown that isopropylmalate isomerase (LeuC/LeuD) ), which featured a similar metabolic signature as chloroxine, is a primary target of reactive oxygen species41. Consistent with this evidence, the oxidative stress agent paraquat is among the 10 out of the 1342 chemical perturbations inducing the strongest accumulation of 2-Isopropylmaleate (Extended Data Figure 7). Hence, we hypothesized that chloroxine interferes with the redox cycling between the cuprous, Cu(I) and cupric Cu(II) oxidation states, responsible for the generation of highly reactive hydroxyl radicals42. To experimentally test this hypothesis, we treated cells with 7.8μM of chloroxine and monitored changes in the promoter activity of cueO, the enzyme responsible for oxidizing periplasmic oxidized copper (Cu(I)) to cuprous oxide (Cu(II)). Not only Chloroxine caused an overexpression of cueO (Fig. 5D). We also found that deletion of the catalase-peroxidase gene (katG) increases growth inhibitory activity of chloroxine at 7.8μM (Extended Data Figure 7), reinforcing the hypothesis of oxidative stress as a consequence of chloroxine treatment. Next, we asked whether chloroxine toxicity at low concentrations depends on the indirect inhibition of leucine biosynthesis. To this end, we measured growth inhibition in M9 glucose with and without 0.1, 0.5 or 1mM leucine. Consistent with a link between redox imbalance and isopropyl malate isomerase activity, we found that supplementing leucine in the medium almost completely abolished the inhibitory activity of chloroxine at concentrations below 8 μM (Fig. 5E).
Figure 5. Gene-Drug associations.
A) Volcano plot of similarities between chloroxine and knockdowns metabolic profiles at different time points. Chloroxine gene-drug associations that are among the top 1% are highlighted in yellow. Pvalues are estimated by hypergeometric test (see iSim). B) Volcano plot of similarities between chloroxine and the remaining 1341 drugs. Pvalues are estimated by hypergeometric test (see iSim). C) Growth rates at different concentrations of chloroxine relative to the untreated condition. Wild type E. coli was grown in LB medium with and without 100 or 200 μM of CuCl2 . Bar plot are mean ± standard deviation across three biological replicate. D) Promoter activity of cueO in M9 glucose medium and LB media with 7.8 μM chloroxine, 155 μM chloramphenicol and 185 μMCuCl2. Bar plots represent mean ± standard deviation of promoter activities in mid-log growth phase over 3 biological replicates. E) Growth rates at different concentrations of chloroxine relative to the untreated condition. Wild type E. coli was grown in M9 medium with and without 0.1/0.5/1 mM of leucine. Bar plot are mean ± standard deviation over two biological replicates. F) Relative metabolite z-score levels in ΔrcsB27. G) Relative z-score levels of 2-demethylmenaquinone across 3807 E. coli non-essential knockout strains27.
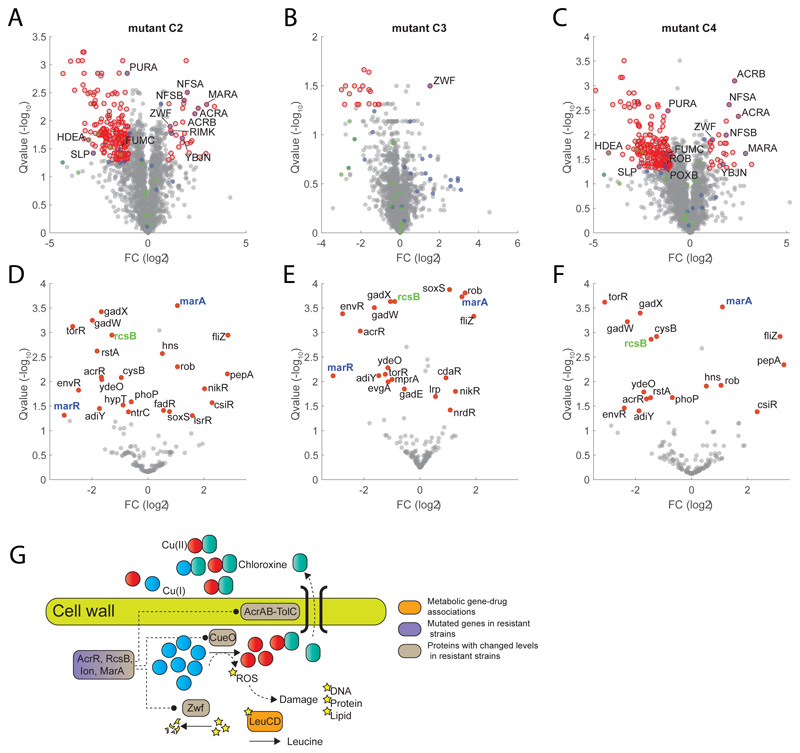
Overall, our results advocate for a scenario in which, by binding preferably to Cu(II), chloroxine not only limits copper availability but also disrupt intracellular copper homeostasis. The attempt of cells to maintain copper homeostasis by cueO overexpression to oxidize periplasmic Cu(I) to Cu(II) generates hydroxyl radicals in a process analogous to the Fenton reaction42, and ultimately contributes to chloroxine toxicity by inhibiting leucine biosynthesis. We sought to reinforce our metabolome-based predictions by sequencing 6 strains from 6 independent lineages that we evolved to acquire a 16-fold increase in chloroxine MIC. We found that most common mutations across the 6 independently evolved strains are mutations affecting transcriptional regulators (Supplementary Dataset 7). All 6 strains displayed disruptive insertions in the coding sequence of the acriflavine resistance repressor acrR, suggesting for a key role of AcrAB multidrug efflux pump in conferring resistance to chloroxine. 5 independent resistant strains also carry an insertion in the promoter region of the regulator capsule synthesis B (rcsB) gene and 4 of these lineages have an additional insertion upstream of lon protease, also regulating the capsule synthesis regulator (RcsAB) 43, suggesting for an additional key role of the Rcs system in mediating resistance to chloroxine. By analyzing previously acquired metabolome profiles for 3807 E. coli non-essential knockout strains27, we found that ΔrcsB exhibits a unique and strong accumulation of 2-demethylmenaquinone 8 (DMK-8), a metabolite involved in electron transfer and oxidative phosphorylation (Fig. 5F). Remarkably, out of 3807 E. coli non-essential knockout strains 27, the largest accumulation of DMK-8 occurs upon deletion of the key sensor of intracellular copper availability (cusR) (Fig. 5G). Such metabolic similarity indirectly links rcsB regulatory functions to those of cusR and further reinforce the predicted action of chloroxine on intracellular copper homeostasis. Moreover, at concentrations of chloroxine below 8 μM, deletion of rcsB is beneficial while its overexpression is detrimental (Extended Data Figure 7). To gain more insights on the regulatory events mediating chloroxine resistance, we performed proteome analysis of 3 chloroxine resistant strains: C2, C3 and C4 (Supplementary Dataset 7) (Fig. 6A-B-C and Supplementary Dataset 8). Notably, CueO and RcsB levels are significantly (qvalue≤0.05) reduced in C2 and C4 (Supplementary Dataset 8), while Zwf, a key enzyme mediating oxidative stress, is significantly (qvalue≤0.05) upregulated in all 3 resistant strains (Fig. Fig. 6A-B-C). Because changes in gene expression or protein levels of transcription factors (TFs) are a poor proxy of changes in activity, we estimated differences in TFs activity between wild type and chloroxine resistant strains directly from the combined protein levels of TF targets. To this end, for 163 TFs we estimated average and significance of changes in protein levels of regulated genes. We found that in addition to the expected reduced activity of RcsB in all three chloroxine resistant strains (Fig. 6D-E-F), the TF with the most significant changes in activity is MarA (qvalue≤1e-3). Expression of MarA is controlled by the multiple antibiotic resistance regulator and copper sensor MarR44 (Fig. 6D-E-F). Altogether experimental evidence suggest that chloroxine resistance is mediated by drug extrusion (e.g. acrA, acrB) and regulation of genes involved in copper homeostasis (e.g. cueO, marR) and redox balance (e.g. zwf, nfsA, nfsB), supporting the MoA initially predicated on the basis of the metabolome similarity between chloroxine, the copper chelator clioquinol and knockdown of the redox sensitive isopropylmalate isomerase (Fig. 6G).
Figure 6. Chloroxine resistance.
A-B-C) Volcano plot of proteome changes in three chloroxine resistant strains (Supplementary Dataset 7) with respect to wild-type E. coli (Supplementary Dataset 8). Significant changes (qvalue≤0.05 and |FC|≥1) are highlighted in red. Proteins that are transcriptionally regulated by marA or rcsB are highlighted in blue and green, respectively. Reported data are average over three biological replicates. Pvalues estimated from two-sided ttest analysis are corrected for multiple tests by q-value estimation49. D-E-F) Transcription Factor differential activity profiles. For each TF, we reported average fold-changes of regulated proteins and significance of the difference with respect to wild-type (qvalue) estimated using a permutation test (see Materials and Methods). TFs with significant changes in activity are highlighted in red (qvalue≤0.05). G) Schematics of metabolome derived gene-chloroxine associations and their relation to genetic and proteome changes in chloroxine resistant strains.
Discussion
Rapid advances in genome sequencing and editing technologies enabled the identification of a plethora of potential new promising targets. However, despite an ever-increasing throughput of phenotypic drug screening, chemical probes that can interfere with these targets are lagging behind. Our combined gene/drug metabolic profiling approach aims at closing this gap, providing a high-throughput platform for unbiased and rapid functional annotation of compound libraries. While there is not a single technology that provides a general solution to the problem of drug-target identification, metabolome profiling offers a complementary and sensitive readout orthogonal to directly probing protein-drug binding (e.g. LIP-MS), cell growth or morphological phenotypes. Directly measuring the metabolic consequences of drug treatment can unravel mechanistic insights on the MoA also of compounds that don’t directly bind to proteins (e.g. chloroxine) or have poor antibacterial activity. In our work, we have established principles for normalization and analysis of non-targeted metabolic profiling data that allow similarities and differences between drug- and genetic-induced metabolic changes to be measured. Combined with new genetic tools that allow tuning expression of individual genes, our approach offers the possibility to systematically investigate the consequences of selectively interfering with essential proteins. Because gene essentiality is condition-dependent, in vitro conditions that mimic in vivo environments will expand the inventory of essential bacterial functions that could become attractive antibacterial targets and improve the selection of lead compounds from susceptibility screening45. Here, we demonstrated that our approach can be applied in virtually any type of system or conditions and that, even if applied on a single condition, we were able to generate testable predictions for 29 out of 39 compounds that exhibit growth inhibitory activity and have an uncharacterized antibacterial MoA.
Despite perturbing major hubs in genetic networks30, we showed that limiting the abundance of essential proteins elicits specific metabolic signatures that are not only characteristic of protein function, but also similar to those induced by compounds targeting the same cellular function. This greatly accelerates hypothesis generation on drug-target interactions responsible for small-molecule growth inhibitory activity. The scalability of this framework, together with recent advances in CRISPR technology enabling genetic manipulation in non-model organism bacteria46, makes this approach of widespread use not only to dissect bacterial essential gene functions, but also to rapidly screen for compounds able to interfere with such functions, ultimately, mitigating the dereplication problem, i.e. the rediscovery of common antimicrobial compounds.
Combined with machine-learning tools 47 large scale metabolic profiling of chemical/genetic perturbations can open new opportunities for advancing in silico screening strategies and chemical optimization of lead compounds47. The presented computational/experimental framework to functionally annotate large compound libraries can be applied to largely diverse biological systems, from bacteria to human cells48, opening new opportunities to improve the selection of lead compounds and speed up drug discovery across many diverse therapeutic areas.
Materials and Method
E. coli CRISPRi library
The E. coli mutant library was generated in and shared by the group of Carol Gross at UCSF23. The library consists of arrayed BW25113 strains with an IPTG inducible, chromosomally integrated CRISPRi system (full details can be found here 23 and https://escholarship.org/uc/item/173451xm). The original library contained 540 strains expressing dCas9 constitutively as a part of a cassette integrated at Tn7att and marked with gentamicin resistance (Supplementary Dataset 1). Each strain contained sgRNA component of the CRISPRi targeting one single gene and the expression is under the control of an inducible promoter PLIacO-1. Because of technical issues which led to contamination of the strain library, deep-sequencing median reads/strain ~200,000) on all strains was performed. Only uncontaminated strains (genetic purity above 99.9%) were retained in our study, leading to a total number of 376 strains, out of which 24 did not grow in M9 glucose medium, 352 strains.
Physiology
Bacterial growth was measured using optical density (OD) at 600 nm in a TECAN spectrophotometer by diluting 50 μl of M9 bacteria culture in 150 μl in a 5 times diluted base salts solution (per liter): 7.52 g Na2HPO4-2H20, 3 g KH2PO4, 0.5 g NaCl, 2.5 g (NH4)2SO4). The growth rates were calculated as a slope of a linear regression of the logarithmically transformed growth curves. Samples were taken in parallel to metabolomics samples, from the same bacterial culture.
Culture media and growth of inducible-knockdown strains
Each strain was revived from frozen stock and incubated for 8 h in 500 μl selective LB medium (10 g/l yeast extract, 10 g/l tryptone, 5 g/l NaCl, 10 μg/ml gentamicin and 10 μg/ml chloramphenicol). 10 μl of culture were transferred to 690 μl of glucose minimal medium (M9) and grown over night. The medium consist of 5 g/l glucose, 7.52 g Na2HPO4-2H20, 3 g KH2PO4, 0.5 g NaCl, 2.5 g (NH4)2SO4, 14.7 mg CaCl2 2H2O, 246.5 mg MgSO4 7H2O, 16.2 mg FeCl3 6H2O, 180 μg ZnSO4 7H2O, 120 μg CuCl2 2H2O, 120 μg MnSO4 H2O, 180 μg CoCl2 6H2O and 1 mg thiamine HCl. Cells were grown in 96 well plates on a shaker at 37°C and 250 rpm. The day after, triplicates were prepared containing 10 μl of the overnight culture and 690 μl M9 supplemented with 1 mM IPTG. OD was measured at multiple times points and samples for metabolome extractions were collected at 3 time points with approximately 1 h time intervals. Because most proteins in E. coli are not actively degraded and each protein can be expressed and become limiting at different levels, we monitored dynamic changes across a time window in which most strains could undergo at least 2-3 duplications. This in order to observe the dynamic changes induced by the dilution of the knocked down protein. The procedure for metabolite extraction is described in 12 – i.e. extraction was performed at -20°C for 1 h diluting each sample in 1:4 extraction solution (50%(v/v) methanol and 50 %(v/v) acetonitrile, 25 μM phenyl hydrazine) in 160 μl total volume. The samples were then centrifuged at 4000 rpm for 5 min at 0°C and 100 μl of the supernatant were transferred into 96 well storage plates and stored at -80°C.
Metabolome profiling of the response to antimicrobials
Here we used the same protocol described in 7. E. coli overnight cultures growing on M9 minimal medium were diluted to an initial Optical Density (OD600) of 0.05. 700 μL cell cultures were distributed in 96 deep well plates and cells were grown at 37°C until exponential phase and an OD600 of 0.4 before exposure to the drug treatment by the addition of 7ul of a drug solution. The Prestwick library consisted mostly of human targeted drugs with unknown effects or penetration in gram negative bacteria like E. coli. The drug contraction of 100 μM was chosen because close to the estimated colon concentration range for many drugs50. On the contrary, the 63 newly screened compounds are well characterized antimicrobials. Hence we selected a lower concentration of 10μM at which most compounds already exhibit growth inhibitory activity. Samples for metabolomics profiling were taken after 2 hours from drug exposure. Previous analysis have shown that metabolic changes between 1 and 2 doublings (i.e. 1 or 2 hours for E. coli growing in M9 glucose) after drug treatment are the most consistent in time and maximize the predictive power to infer different classes of antibiotics12. 50 μl of whole cell broth were directly transferred to 120 μl extraction liquid solution containing 50% (v/v) methanol and 50% (v/v) acetonitrile at −20°C. The extraction was carried out by incubating the samples for 1 hour at −20°C. Samples were centrifuged for 5 minutes at 4000 RPM and 80 μl of the supernatant was transferred to 96 well storage plates and stored at −80°C. All treatments were measured in 3 biological replicates. Bacterial culture growth was estimated by measuring the optical density at 600 nm (OD600) in a plate reader. OD600measurements were acquired immediately before drug exposure and after 60, 120, 240 and 360 minutes. Growth rates were calculated as a slope of a linear fits to the logarithmically transformed growth curves. Only samples in which bacterial culture reached an optical density (OD600) of at least 0.2 were retained to have an adequate MS signal for further analysis. As a result, 24 gene knockdown mutants were excluded. The vast majority (22 genes) are encoding for metabolic enzymes, several of which catalyze intermediate steps in amino acids (8 genes) or purine biosynthesis (5 genes) (Supplementary Dataset 1).
Mass spectrometry analysis, ion annotation and ion abundance normalization
Cell extract samples were analyzed by flow-injection analysis time-of-flight mass spectrometry (FIA-TOFMS) on an Agilent 6550 iFunnel Q-TOF LC-MS System (Agilent Technologies, Santa Clara, CA, USA), as described in 18. Raw MS profiles were processed to align spectra and pick centroid ion masses using an in-house data processing environment in Matlab R2015b (The Mathworks, Natick). Measured ions were putatively annotated by matching mass-to-charge ratios to a reference list of calculated masses of metabolites listed E. coli K12 model iJO1366 51 within 0.003 amu mass accuracy. The reference mass list was generated from the respective sum formulae, considering deprotonation as the most prevalent mode of ionization in the chosen acquisition conditions. To allow for the annotation of α-keto acid derivatives formed in presence of phenyl hydrazine in the extraction solvent, sum formulae for the phenylhydrazone derivatives (+C6H8N2-H2O) of a total of 30 α-keto acid compounds (selected via KEGG SimComp search http://www.genome.jp/tools/simcomp/) were added to the metabolite list for annotation. Data were normalized using the multiple linear regression approach described in 12 and 7. Raw data normalization was performed by using a multiple linear regression approach. For each detected ion, measured intensity (i.e. ion counts) is considered to be a function of extracted biomass (measured by OD600 (O)), plate to plate variance (P), gene knockdown effect (G), and white noise (ε) , using the following model: Im = f(O,P,G) + ε. Function f is : , where Im,c,t is the measured intensity of ion m upon knockdown of gene g at time t after IPTG induction, Oc,t represents biomass (OD600) at the time of extraction, αm and βm represent quadratic and linear dependency between ion intensity and OD600. In our model the variance associated to plate effects across the large number of MS injections (~4000) is captured by the parameter γm,p. By assuming that overall, each ion concentration within the cell is “directly” affected only by few drugs, equivalently to assume sparsity of D, the proportionality factors α, β and γ could be determined by multiple least square fitting analysis performed on all collected samples (i.e. all muntants and time points), for each ion individually. The Matlab function fitlm using ordinary least squares, was used for the regression analysis. The relative difference between the model-derived expected intensity and the corresponding measured ion intensity under in each sample was used to estimate the gene knckdown effect at time t after IPTG induction: . Log 2 fold changes were calculated before Z-score normalization.
Metabolic comparison between CRISPRi mutants and knockout strains
We compared newly generated metabolic profiles in CRISPRi mediated gene knockdowns and previously generated metabolic profiles for gene deletion mutants27. We selected non-essential genes that when knockout exhibited a metabolic phenotype (i.e. at least one metabolite exhibit a Z-score≥5). Next, we used iSim to estimate pairwise similarity between knockdown and knockout metabolic profiles. Because knockdown profiles were measured at multiple time points, for each knockdown-knockout pair we selected the maximum similarity estimated at different time points after IPTG induction (Supplementary Dataset 1).
Pairwise metabolome comparisons between gene knockdowns and drug profiles (iSim)
We predicted gene function using a guilt by association scheme, in which two genes with similar metabolic profiles are predicted to have similar function. Similarly, we used drug-gene metabolic similarity to make predictions of functional annotations of small molecules. To compare the metabolome response between two different gene knockdowns we used an approach introduced by Campos et al7. Calculation of the similarity score consists of two main steps. First, for each pair of gene knockdowns A and B, we performed an exhaustive search over two parameters thrA and thrB, to maximize the significance of the overlap between metabolites that are significantly changed (|Zscore|≥thr) in A and in B.
Where ZGeneA, m is the z-score value of metabolite m in knockdown of gene A. The significance of the percentage of metabolites that exhibit consistent changes between the two drugs is estimated using a hypergeometric test:
where consistent changes in both gene knockdowns represent observed successes (s), the number of changes in one knockdown strain represents the maximum possible number of successes (K), the total number of metabolites represents the total population (N), and the number of changes in the second knockdown represents the number of events or draws (n). Pvalues were calculated in MATLAB using the hygecdf function. In the second step we generate one single similarity score (iSim) for each knockdown pair by multiplying the dot product dAvsB by the −log10 of the corresponding pvalue (PAvsB). To account for CRISPRi polarity effect we exclude pairs of gene belonging to the same operon according to the Operon Database v4 52.
Enrichment analysis of KEGG and COG functions
To predict the functional annotation of genes and cellular functions associated to drug MoA the iSim pair wise similarities between knockdown mutants or knockdown mutants and drug compounds were rank transformed. A statistical score that models the probability of the gene/drug functional annotation (see Tables S2) is based on the collective similarities of genes with known function (or drugs with known MoAs) following the approach described in 53. The significance of the rank distribution of all annotated gene knockdowns within the same functional groups or drugs with the same MoA is tested by means of an iterative hypergeometric test, indicating the statistical significance of, for example, all drugs targeting a common cellular process (e.g. protein biosynthesis) being distributed toward the top ranking ones. Pvalues were corrected for multiple tests by qvalues estimation.
Locality score
The procedure used here is also described in 1. A genome-scale network model of E. coli K12 model iJO1366 metabolism was used to determine the distance between each enzyme-metabolite pair. The resulting pairwise distance matrix (D) between metabolic enzymes and metabolites was estimated by means of the minimum number of reactions separating the two in a non-directional network. All highly connected metabolites – i.e. metabolites that participate in more than 20 reactions, were removed prior to calculation. Next we assessed whether largest metabolic changes were statistically more probable in the proximity of the repressed enzymes. To this end, we used a locality scoring function1, in which all metabolic changes are weighted by the respective distance to a tested enzyme, as follows:
where for each enzyme ei, a weighted mean over corresponding (metabolite m Z-score corresponding to drug d) is computed. Weights are functions of the inverse of the squared distance between the enzyme i and the metabolite m. We perform a permutation test by randomly shuffling the distance matrix D (Drand) K times (K=10000). For each gene we ensure to preserve original degree of connectivity in the stoichiometric network.
Disk diffusion assay
Filter paper circles of 6 mm diameter were impregnated with 5 μl of antibiotic solution (2.8 mM for oxfloxacin) and left to dry at room temperature. Bacteria in exponential phase grown in M9 liquid culture with 0.2 mM IPTG were plated on M9 glucose minimal medium agar plate. The plate was left to dry before applying the disk in the middle and incubating for 24 hours at 37°C.
Limited proteolysis (LiP) sample preparation
Cell pellets were resuspended in 200 μl of cold lysis buffer (100 mM HEPES, 1 mM MgCl2, 150 mM KCl, pH 7.5), supplemented with the same volume of acid-washed glass beads (Sigma) and disrupted at 4 °C by 3 consecutive rounds of beads-beating for 30 s with 4 min intercalating pauses in a FastPrep-24 5G instrument (MP Biomedicals). After centrifugation at 16,000 x g for 15 min at 4 °C, the supernatants were collected and the protein concentration was determined using a BCA assay (Thermo Fisher). The protein concentrations were adjusted to 1ug/ul of which 50ul was used for each LiP assay. For each sample, 1ul of the drug of interest (or DMSO used as mock treatment) was incubated with the 50 ul of protein extract for exactly 5 min at 25°C. Then 5ul containing 0.5ug of Proteinase K from Tritirachium album (Sigma Aldrich) was added to the LiP samples and incubated for 5 minutes at 25°C. Samples were then denatured by heating inactivation for 5 minutes at 99°C in a thermocycler followed. After 5 minutes cooling on ice 56ul of 10% sodium deoxycholate (Sigma) was added to each sample for a final concentration of 5%. The samples were then reduced with 5 mM TCEP (Thermo Fisher) for 30 minutes at 37 °C and alkylated with 40 mM iodoacetamide (Sigma) for 30 min at 25°C in the dark. The samples were then diluted with 100 mM ammonium bicarbonate to reach a final concentration of 1% sodium deoxycholate and finally digested by overnight incubation with 0.5 ug LysC (Wako Chemicals) and 0.5 ug trypsin (Promega) at 37°C under shaking. The digests were then acidified by addition of 50% formic acid to a final concentration of 2%. The peptide mixtures were passed through 0.2um PVDF filter plates (Corning) to remove the sodium deoxycholate precipitates. The filtrates were then desalted on a 96-well MicroSpin plate loaded with 300Å C18 silica material (The Nest Group). After elution with 50% acetonitrile/0.1% formic acid the peptides were dried in a speedvac, resolubilized with 25ul of 0.1 % formic acid, and transferred to MS vials for LC-MS analysis. Pipeline for data analysis can be found here: https://gitlab.ethz.ch/lgillet/mattia-zampieri-crispri.
Antibiotic evolutionary experiment
For each of the two drugs, tegaserod and chloroxine, five and six independent lineages were propagated in parallel, respectively. Serial passaging was performed in 96 deep-well plate cultivation (2 ml well volume, 500 μl culture volume), using the same protocol described in 54. Seven wells in a plate column were prepared with gradually increasing concentrations of the same antibiotic, and the last row of the plate served as a growth control and contained no drug. Every 48 h, OD600 was measured with a plate reader. 5 μl of the bacterial population that was able to grow (i.e. OD ≥ 0.5) at the highest of seven tested drug concentrations was used for the next passaging step. The number of generations during each passaging step was calculated by (i) measuring the final OD after a 48-h growth cycle (ODfin), (ii) 5 μl of selected evolved populations was reinoculated in 500 μl of fresh medium yielding a 1/100 dilution for the new starting OD. At the end of the 48-h growth cycle, OD was measured (OD*) and number of generation is calculated by the following formula: log2(OD*/(ODfin/100)). We reached ~60 generations when evolving resistance to tegaserod, and we were able to evolve mild but measurable level of resistance, from 0.015625 to 0.3125 mM, in one of the five populations. Already after ~40 generations we reached the solubility limit for chloroxine, from an MIC of 0.03125 mM in the wild type to 0.5 mM in all six evolved populations.
Genomic DNA extraction, whole-genome sequencing and mutations analyses
For each evolved strain, 1 ml of an over-night LB culture was pelleted and frozen at -80°C. Genomic DNA was then extracted using the GenElute Bacterial Genomic DNA kit (Sigma, NA2120-1KT). Genomic DNA integrity was monitored by 0.6% agar gel electrophoresis and gDNA was send to MiGS (Microbial Genome Sequencing Center), Pittsburgh, USA. PE151 sequencing runs were performed on NextSeq 2000 platform with a targeted coverage of c.a. 100X. Reads were mapped onto the genome of the reference Escherichia coli str. K-12 substr. MG1655 (NC_000913) using Breseq v.0.36.0 under default “consensus” mode settings with default settings (see supplements). Consistent genetic variations (as outputted by Breseq with these settings) were then analyzed and compared within the evolved resistant strains. Mutations conserved among all evolved strains were omitted from Supplementary Dataset 7. We used Breseq v.0.36.0 under default “consensus” mode settings. These include: --base-quality-cutoff=3, --require-match-fraction=0.9, --bowtie2-scoring=“--ma 1 --mp 3 --np 0 --rdg 2,3 --rfg 2,3 --ignore-quals”, --bowtie2-stage1=“--local -i S,1,0.25 --score-min L,1,0.9 -k 2000”,--bowtie2-stage2=“--local -i S,1,0.25 --score-min L,6,0.2 -k 2000”, --bowtie2-junction=“--local -i S,1,0.25 --score-min L,1,0.70 -k 2000”, --junction-minimum-candidates=100, --junction-candidate-length-factor=0.1, --junction-minimum-candidate-pos-hash-score=2, --junction-score-cutoff =3.0, --junction-minimum-pos-hash-score=3 , --junction-minimum-side-match=1, --junction-minimum-pr-no-read-start-per-position=0.1, --consensus-score-cutoff=10, --consensus-frequency-cutoff=0.8, --polymorphism-score-cutoff=10, --polymorphism-frequency-cutoff=0.2. SRA accession numbers for the ancestor E. coli strain, the 3 tegaserod and 6 chloroxine resistant strains: SRR16634183 Chloroxine 1, SRR16634182 Chloroxine 2, SRR16634181 Chloroxine 3, SRR16634191 Chloroxine 4, SRR16634190 Chloroxine 5, SRR16634189 Chloroxine 6, SRR16634192 Tegaserod 1, SRR16634188 Tegaserod 2, SRR16634187 Tegaserod 3, SRR16634193 Wt K-12 ancestor.
Whole proteome analysis of resistant mutants
Whole proteome of resistant mutants was generated following the Standard Operative Procedures (SOP v.2020.09.03) at the Proteomic Core Facility of the Biozentrum, Basel. A detailed stepwise description of the laboratory protocol and the analysis procedure is accessible in a recent methodologic tutorial article 55. Bacterial pellets of resistant strains grown in LB at 37°C under agitation for 16h and cells were harvest by centrifugations. Cell lysis was conducted in lysis buffer (1% sodium deoxycholate, 10 mM TCEP, 100 mM Tris-HCl, pH 8.5) with a microtube sonicator and a 10 minutes heat step at 95°C. Tryptic digestion of proteins was performed over 16h at 37°C in presence of chloroacetamide in order to stabilize cysteine residues. Peptides were then cleaned and purified with a solid-phase extraction cartridge (SDB-RPS). Eluted peptides were then dried and dissolved in liquid chromatography buffer A (0.15% formic acid, 2% acetonitrile) prior injection into a RP-HPLC column connected to a dual pressure LTQ-Orbitrap Elite mass spectrometer. Acquired peptide intensities were calculated using Progenesis QI (v2.0, Nonlinear Dynamics Limited) MaxQuant (Version 1.0.13.13) and MASCOT softwares and the SafeQuant R package v.2.3.4 (https://github.com/eahrne/SafeQuant/).
Measuring cueO promoter activity
We used GFP transcriptional reporter plasmids in which the promoter regions of cueO is fused to green fluorescent protein 56. Promoter activity was measured in M9 glucose minimal and LB media using a plate reader recording GFP intensity and optical density (OD600). Data were analysized using the protocol described in 56. E. coli was grown overnight in the respective media and 5μL of culture were inoculated in 200 μL of fresh media. Promoter activity over time was calculated by taking the time derivative of GFP divided by OD (dGFP/dt/OD). The analysis was performed using custom Matlab software. Negative promoter activity, such as in the case of E. coli exponentially growing in LB, reflects faster growth dilution than production of GFP. This is because CueO is transcribed more in inoculated starved cells than during exponential growth.
Estimation of Transcription Factors (TFs) differential activity
The network of TF-target genes reported in 57 was used to identify TFs regulating proteins with large difference between susceptible and resistant strains. For each TF, we estimated the average protein levels of the corresponding regulated genes. Significance was estimated using a permutation test (as above), where for each TF the same number of targets were randomized 100.000 times and the average protein levels compared to the true one.
Metabolic profiling of genetic-/chemical perturbations in human cells
A549 human lung cancer cells stably expressing Cas9 nuclease (A549-Cas) were purchased from Horizon Discovery (cat. no. HD Cas9-001) and maintained in RPMI-1640 cell culture medium (Thermo Fisher Scientific, cat. no. 21870076) supplemented with 5% dialyzed fetal bovine serum (FBS), 2 mM glutamine (Thermo Fisher Scientific, cat. no. A2916801) and 1% penicillin-streptomycin solution (Thermo Fisher Scientific, cat. no. 15140122). On the day before transfection, A549-Cas9 cells were seeded in 96-well plates (Thermo Fisher Scientific, cat. no. 167008) at a cell density of 2.3e4 cells/mL (150 μL per well) in RPMI-1640 medium without antibiotics supplementation. The cells were incubated over-night at 37°C in 5% CO2 atmosphere to allow cell attachment. On the next day, cells were treated in triplicates with either genetic- or drug perturbations as follows. For CRISPR-mediated knockouts of thymidylate synthase (TYMS), sgRNAs targeting three different gene sections were purchased from Horizon Discovery (Edit-R synthetic guide RNAs, cat. no. SG-004717-01-0002, SG-004717-02-0002 and SG-004717-03-0002), in addition to a control sgRNA inducing cell death (lethal control, gLethal) by targeting multiple sections in the genome (Horizon Discovery, cat. no. U-008000-02-02). The sgRNAs were separately introduced into A549-Cas9 cells using lipid-mediated transfection. To that end, 0.2 μL/well transfection reagent (DharmaFECT I, Horizon Discovery, cat. no. T-2001-01) was prepared in serum- and antibiotics-free RPMI-1640 medium, and mixed with sgRNA solutions in serum- and antibiotics-free RPMI-1640 medium. After 15 min incubation, the transfection mix was spiked into the supernatants of A549-Cas9 cells with a final sgRNA concentration of 25 nM. Three wells were treated only with transfection reagent without sgRNAs as a negative control. In parallel to the sgRNA transfections, separate triplicate cell cultures were supplemented with 14 drugs with variable metabolic and non-metabolic targets, including three inhibitors acting on folate metabolism. The treatment concentrations for each drug were chosen based on prior or literature data: 2-Deoxyglucose (11 mM, cat. no. D8375, Sigma-Aldrich), 5-fluorouracil (5 μM, cat. no. F8423, Sigma-Aldrich), Atorvastatin (10 μM, cat. no. PHR1422, Sigma-Aldrich), Camptothecin (5 μM, cat. no. C9911, Sigma-Aldrich), Cisplatin (5 μM, cat. no. P4394, Sigma-Aldrich), Dichloroacetate (30 mM, cat. no. 347796, Sigma-Aldrich), Doxorubicin (0.2 μM, cat. no. D1515, Sigma-Aldrich), Carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP, 5 μM, cat. no. C2920, Sigma-Aldrich), Mercaptopurine (200 μM, cat. no. 38171, Sigma-Aldrich), Methotrexate (0.1 μM, cat. no. M9929, Sigma-Aldrich), Oxamate (25 mM, cat. no. O2751, Sigma-Aldrich), Pemetrexed (0.5 μM, cat. no. Y0001539, Sigma Aldrich), Trifluoperazine (10 μM, cat. no. T8516, Sigma-Aldrich), UK5099 (5 μM, cat. no. PZ0160, Sigma-Aldrich). Metabolite extraction was carried out at 48 hours after treatment as described in detail previously 48. Metabolite extracts were stored at -80°C, and transferred to fresh 96-well plates immediately prior to metabolomics measurements (FIA-TOFMS, see above). Measured metabolites were annotated to endogenous metabolites listed in HMDBv4 58and Recon3D59 based on accurate mass (putative annotation). We detected 1593 putatively annotated metabolites. The MS profiles were normalized to account for differences in cell confluence in each sample following the procedure described procedure described above for E. coli and previously 48, yielding log2 fold-changes and statistical significance (p-value) for each putatively annotated metabolite in each treatment as compared to untreated cells (Supplementary Dataset 5).
Metabolic profiling of M. smegmatis
We selected 12 strains from a collection of M. smegmatis strains with inducible CRISPR-interference system targeting essential genes: murG, mmpl3, gyrA, gyrB, folE, eno, leuB, rpoB, rpoC, rpsL, rplP, rplS31. 100 μl of culture from frozen glycerol stocks were used to inoculate inkwell bottles containing 5 ml Middlebrook 7H9 broth (BD Difco™) supplemented with 0.2% (v/v) glycerol, 0.05% (v/v) tyloxapol and 20 μg/ml kanamycin. The same media was used throughout the course of the experiment. Starter cultures were incubated for ~24 h at 37°C with shaking at 100 rpm then 75 μl was transferred to 5 ml fresh media and incubated for ~15 h overnight at 37°C with shaking at 150 rpm. Cell cultures were diluted to an OD600 of 0.3 with fresh media and 700 μl of each strain distributed into triplicate wells in a 96-well 2 ml square well V-bottom deep-well block. A vector only control strain and media only controls were also included on the plate. The plate was incubated at 37°C with shaking at 300 rpm and after 1 h incubation OD600 was measured and cultures adjusted back to OD600 ~0.3-0.4 using pre-warmed media. Anhydrotetracycline hydrochloride (Acros Organics) in DMSO was then added at a final concentration of 100 ng/ml to induce CRIPSR-mediated gene knockdown and plates returned to the incubator. Cultures were sampled for metabolomics profiling and OD measurement after 3.5, 6, 10 and 24 h from induction of gene knockdown. All strains and controls were measured as biological triplicates. At each time point, 40 μl of whole-cell broth was transferred to 120 μl medium on microtiter plates and OD600 was measured on a FLUOstar Omega plate reader (BMG Labtech). In parallel, 40 μl culture was transferred to 120 μl of extraction liquid solution containing 50% (v/v) methanol and 50% (v/v) acetonitrile at –20°C. The extraction was carried out by incubating the samples for 1 h at –20°C. Samples were centrifuged for 5 min at 3900 rpm and 50 μl of the supernatant was transferred to replicate 96-well storage plates and stored at −80°C. We detected 942 putatively annotated metabolites. The MS data was normalized to account for differences in cell confluence in each sample following the procedure described above for E. coli and previously 48, yielding log2 fold-changes and statistical significance (p-value) for each putatively annotated metabolite in each treatment as compared to untreated cells (Supplementary Dataset 4).
1. Extended Data
Extended Data Fig. 1. Metabolome profiles of CRISPRi mutant library.
A) Distribution of significant metabolic changes per gene knockdowns/time points. On average, 24 metabolites underwent significant changes (absolute Z-score ≥ 1 and p value ≤ 1e–5) per mutant and time point, and most of the metabolites (95%) exhibited a significant response in at least one gene knockdown. B) Distribution of significant changes for each metabolite across knockdowns/time points. C) Relative distribution of collected time points per gene knockdown. D) Distribution of Optical Densities (OD600) at sampling. E) Distribution of locality scores for essential (blue) and non-essential knockdowns (red). F) Distribution of the number of significant affected metabolites by essential (blue) and non-essential (red) gene knockdowns in each time point. G) Estimates of growth rates of CRISPRi mutants grown in M9 glucose + 1mM IPTG. Each dot corresponds to mean ± standard deviation (SD) across three biological replicates. Average Wild-type growth rate ± SD are represented by solid and dashed red lines, respectively. Each dot is color-coded according to the significance in growth rate difference with respect to wild-type (i.e. t-test analysis). Pie chart reports on the fraction of knockdown strains growing significantly slower than wild type (pvalue≤0.01). H) Median absolute Z-score of significantly affected metabolites (|Z-score| ≥ 1 and p value ≤ 1e–5) in each mutant/time point. Median values for each timepoint across all mutant strains are used to fit a sigmoidal curve (red curve). The blue shaded area identifies the confidence region (95% confidence interval). Green dashed lines report the 95% confidence interval for the time necessary to reach 90% of the maximum estimated changes (i.e. plateau).
Extended Data Fig. 2. Analysis of growth phenotypes.
A) Growth dynamics of each individual mutant. Each line corresponds to the OD averaged across 3 biological replicates. In orange, mutants exhibiting a sudden drop in OD are highlighted. B) Median absolute Z-score of significantly affected metabolites in each mutant/time point vs estimated growth rate. As expected the slower the growth rate the larger is the impact of gene knockdowns on metabolism7, although the dependency is mild: Spearman correlation of -0.19. C) Distribution of growth rates for essential and non-essential gene knockdowns. D) Distribution of growth rates for gene knockdowns across KEGG pathways.
Extended Data Fig. 3. Metabolome-based predictions of functional gene-gene similarity.
A) ROC analysis comparing the performance of different similarity metrics in detecting pair of genes that are either encoding for subunits of the same protein complex or for iso-enzymes. We used previously published data27 profiling the metabolome across 3873 gene knockout mutants. We selected only genes encoding for proteins complexes and isoenzymes with at least one significant metabolic changes as defined in 27 (|Z-score|>5). Area Under the Curve (AUC) is reported for Spearman correlation, mutual information, context likelihood of relatedness (CLR) 60 applied on mutual information, iterative similarity and CLR applied on iterative similarity (iSim). The best performance was obtained with iSim (Table S1). To cope with the fact that similarity metrics, like mutual information, can be biased by hidden global patterns in the data (in our case likely to reflect indirect and general type of effects (e.g. growth rates)), and to take into account the typical patterns of interaction of multivariate datasets, the authors in 60 developed an effective and simple approach to normalize pair wise mutual information. The CLR algorithm applies an adaptive background correction step to the matrix of pair-wise similarity scores to eliminate indirect global similarities between drug/gene knockdown metabolome profiles. After computing the similarity between drug/gene pairs, the algorithm compares the similarity between drug/gene A and drug/gene B to the background distribution of similarity scores calculated for all possible drug/gene pairs that include either A or B. The pairs with the most probable functional associations are those whose similarity scores is larger than the background distribution of similarity scores. This step, when applied on mutual information, improves predictions by eliminating “promiscuous” cases, where one gene weakly co-varies with a large numbers of genes. The improvement of CLR applied on iSim are minor. Hence, in this work iSim is used instead of iSim+CLR. B) For each gene, we ranked gene-gene metabolic similarity and performed KEGG-functional enrichment analysis -i.e. KEGG pathways that exhibit a significant (qvalue≤0.01) enrichment of gene knockdowns exhibiting similar metabolic profiles. In blue the ROC curve obtained by considering only similarities between gene pairs from different operons. In purple, we report ROC analysis of KEGG functional enrichment without accounting for operon structure. D) Each corresponds to a KEGG metabolic pathway and the respective AUC values estimated from gene-gene similarity. Only KEGG pathways with an AUC≥0.6 are reported.
Extended Data Fig. 4. Metabolic profiles of genetic perturbations in Mycobacterium Smegmatis.
A) Growth dynamics of each individual M. smegmatis CRISPR mutant after ATC induction. Each line and errorbar corresponds to the OD600 average and standard deviation across 3 biological replicates. B) Heatmap of metabolome profiles 24 hours after CRISPR induction.
Extended Data Fig. 5. Metabolic profiles of chemical perturbations in A549 cancer cells.
A) Dynamic monitoring of cell confluence (% well area covered by cells) in A549 cancer cell line in the presence of sgRNAs targeting three different sections of thymidylate synthase, in addition to a control sgRNA inducing cell death (lethal control, gLethal) by targeting multiple sections in the genome. B) Dynamic monitoring of cell confluence in A549 cancer cell line treated with different drugs. C) The volcano plots illustrate metabolic changes (i.e. log2 fold-change and p-value significance) for 1593 putatively annotated metabolites in A549 cells 48 hours after treatment with 14 different drugs. Pvalues are estimated by two-sided t-test analysis. 3 drugs are targeting TYMS (methotrexate, pemetrexed and 5-fluorouracil). The remaining drugs act on largely different cellular processes, such as central metabolism (oxamate, dichloroacetate, carbonyl cyanide, 2-deoxyglucose, UK5099), nucleotide biosynthesis (mercaptopurine), mevalonate pathway (atorvastatin), DNA repair (camptothecin, cisplatin, doxorubicin) and dopamine receptors (trifluoperazine).
Extended Data Fig. 6. Gene-drug associations.
A) Disk diffusion assay testing the susceptibility to ofloxacin (10 μg) in wild-type E. coli and pyrG knockdown. Violin plot represents mean ± SD across 3 biological replicates. Two representative petri dishes are reported on the lower panel. B) Imipramine gene-drug associations. Volcano plot of similarities between drug and knockdowns time dependent metabolic profiles. Notably, we found genes exhibiting more rare but strong associations to chemically induced metabolic changes, such as the association between the gene mediating chromosome partitioning during division mukF, and imipramine, a drug typically used as antidepressant, potentially hinting at the toxic side effects of imipramine as DNA damaging agent; C) Vatalanib gene-drug associations. Vatalanib, a tyrosine kinase inhibitor that interferes with the ATP-binding site of vascular endothelial growth factor receptors (VEGFR1-3), induces similar metabolic changes to purA knockdown, a key gene in the adenosine ribonucleotides de novo biosynthesis. D) fraction of compounds inhibiting at least 50% of the growth-rate at 100μM for (i) compounds within the top 1% of drug-gene knockdown similarities, (ii) compounds within the top 1% of drug-gene knockout similarities (data reported in27), (iii) all tested compounds. We found that drugs with strong similar metabolic profiles to at least one essential gene knockdown (i.e. top 1% of all drug-genes associations) are significantly enriched (pvalue = 2.3071e-183 ≤ 0.001 ***, from 13% to 60%) for drugs inducing Growth Inhibition (GI) greater than 50%. On the contrary when selecting the top 1% of similarities between drugs and the ~3800 non-essential gene knockout7 27, we found no significant enrichment and the majority of drugs exhibits relatively low growth inhibition (GI<50%). E-F-G) Volcano plot of similarities between tegaserod, suloctidil, thiethylperazine and the remaining 1341 drugs. Pvalues are estimated by hypergeometric test analysis. H-J-K) Volcano plot of proteome changes in three tegaserod resistant strains (Table 1 of the main text) with respect to the ancestor wild-type E. coli (Table S7). Proteins with significant (qvalue≤0.05 and | FC|≥1) changes with respect to the wild-type are highlighted in red. Pvalues are estimated by two-sided t-test analysis and corrected for multiple tests.
Extended Data Fig. 7. Chloroxine induce copper toxicity.
A-B-C-D) Results from limited proteolysis analysis. Each dot in the volcano plot represents the relative difference in peptide abundance between the treated and untreated proteome extracts. None of the 13797 detected peptides exhibit significant (pvalue<7.2480e-07, Bonferroni corrected) conformational changes between untreated and whole proteome extracts treated with 4 different concentrations of chloroxine (Table S6). Pvalues are estimated by twosided t-test analysis. E) Relative growth rates at different concentrations of chloramphenicol with respect to the untreated condition. Wild type E. coli was grown in LB medium with and without 200 μM of CuCl2. Growth inhibitory activity for different chloramphenicol concentrations were measured in triplicates. Errorbars are mean ± standard deviation across three biological replicates. F) 2-Isopropylmaleate z-score levels across the 1342 tested compounds. G) Average Z-score across metabolites in the branched chain amino acids biosynthesis pathway as defined in KEGG. Significance (i.e. pvalue estimated by permutation test) of enrichment analysis is testing for each compound an average increase of metabolite levels. Out of the 1342 tested compounds (i.e. Prestwick library + antibiotics), only 35 (less than 3%) induced significant changes in the levels of BCAA intermediates (pvalue≤5e-4 Bonferroni corrected and mean Z-score≥0.5). In addition to chloroxine, clioquinol, CuCl2 and paraquat, we observed that several of the 35 compounds inhibit protein synthesis (e.g. apramacyn, tobramycin, ribostamacyn...). This is likely reflecting the indirect accumulation of amino acids, including BCAA intermediates, as a results of the inhibition of protein synethesis4,18. H) Growth of wild-type E.coli (blue) and ΔkatG (black) without and with (dashed lines) 7.8μM of chloroxine. H) Growth of wild-type E.coli (blue), ΔrcsB (black) and rcsB++ (expression of rcsB is under the control of an IPTG-inducible promoter, here we used 1mM of IPTG) without and with (dashed lines) 7.8μM of chloroxine. J) Scatter plot of metabolic changes induced by chloroxine vs clioquinol. K) Scatter plot of metabolic changes induced by chloroxine vs paraquat. Some of the largest metabolic changes induced by paraquat involved metabolic intermediates of branched chain amino acids.
Supplementary Material
Acknowledgments
we thank Uwe Sauer for supporting this work and providing laboratory facilities, Dr. Natalie de Souza for helpful feedback and discussions, the Gross and KC Huang groups at UCSF and Stanford for discussion and sharing the E. coli CRISPRi library ahead of publication, Timothy J de Wet and Digby F Warner at the University of Cape Town for sharing the CRISPRi mutants in M. smegmatis.
Funding
This work was supported by a NCCR AntiResist project funding to M. Z., U. J. and P. P. (1-006425); P. P. and L. G. were additionally funded by the European Research Council (grant agreement no. 866004) and through a Personalized Health and Related Technologies grant (PHRT-506). C.M. and M.B. were supported by NIH grant AI133191.
Footnotes
Author Contributions Statement: M. Z. designed the project. M. G. and G. H. performed the metabolic profiling of the E. coli mutant library. A. C. performed the drug metabolome profiles. K. O. performed the metabolic profiling in human cell lines. C. M. performed the metabolic profiling of M. smegmatis under the supervision of M. B. P. M. performed the genome sequencing and proteome profiling of resistant strains under the supervision of U. J. L. G. performed the limited proteolysis experiments and analyses under P. P. supervision. M. G., G. H. and M. Z. performed data analysis. All authors contributed to preparing the manuscript.
Competing interests: P. P. is advisor at Biognosys AG (Schlieren, Switzerland), the company that licenses the LiP technology.
Data and materials availability
All data generated or analyzed during this study are included in this published article as Supplementary Data. A detailed description of all data analysis steps is published in this article in the Methods section and Supplementary Information. Matlab code is available for download at http://www.imsb.ethz.ch/research/zampieri-group/resources.html. Metabolome data and results from data analysis can be found as Supplementary Tables. The mass spectrometry proteomics raw data and analysis results have been deposited through the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD024133. Genome sequence data have been deposited through SRA Bioproject: PRJNA776168 (also see Supplementary Notes).
References
- 1.Tacconelli E, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 2.Cox G, et al. A Common Platform for Antibiotic Dereplication and Adjuvant Discovery. Cell Chem Biol. 2017;24:98–109. doi: 10.1016/j.chembiol.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 3.Eckhardt M, Hultquist JF, Kaake RM, Hüttenhain R, Krogan NJ. A systems approach to infectious disease. Nat Rev Genet. 2020;21:339–354. doi: 10.1038/s41576-020-0212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imai Y, et al. A new antibiotic selectively kills Gram-negative pathogens. Nature. 2019;576:459–464. doi: 10.1038/s41586-019-1791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson EO, et al. Large-scale chemical-genetics yields new M. tuberculosis inhibitor classes. Nature. 2019;571:72–78. doi: 10.1038/s41586-019-1315-z. [DOI] [PubMed] [Google Scholar]
- 6.Mechanism matters. Nat Med. 2010;16:347–347. doi: 10.1038/nm0410-347. [DOI] [PubMed] [Google Scholar]
- 7.Campos AI, Zampieri M. Metabolomics-Driven Exploration of the Chemical Drug Space to Predict Combination Antimicrobial Therapies. Mol Cell. 2019;74:1291–1303.:e6. doi: 10.1016/j.molcel.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nonejuie P, Burkart M, Pogliano K, Pogliano J. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc Natl Acad Sci. 2013;110:16169–16174. doi: 10.1073/pnas.1311066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stokes JM, et al. A multiplexable assay for screening antibiotic lethality against drug-tolerant bacteria. Nat Methods. 2019;16:303–306. doi: 10.1038/s41592-019-0333-y. [DOI] [PubMed] [Google Scholar]
- 10.French S, Coutts BE, Brown ED. Open-Source High-Throughput Phenomics of Bacterial Promoter-Reporter Strains. Cell Syst. 2018;7:339–346.:e3. doi: 10.1016/j.cels.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Gardner TS, Bernardo D di, Lorenz D, Collins JJ. Inferring Genetic Networks and Identifying Compound Mode of Action via Expression Profiling. Science. 2003;301:102–105. doi: 10.1126/science.1081900. [DOI] [PubMed] [Google Scholar]
- 12.Zampieri M, et al. High-throughput metabolomic analysis predicts mode of action of uncharacterized antimicrobial compounds. Sci Transl Med. 2018;10:eaal3973. doi: 10.1126/scitranslmed.aal3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zampieri M. From the metabolic profiling of drug response to drug mode of action. Curr Opin Syst Biol. 2018;10:26–33. [Google Scholar]
- 14.Iorio F, et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc Natl Acad Sci. 2010;107:14621–14626. doi: 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Rourke A, et al. Mechanism-of-Action Classification of Antibiotics by Global Transcriptome Profiling. Antimicrob Agents Chemother. 2020;64 doi: 10.1128/AAC.01207-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brötz-Oesterhelt H, Bandow JE, Labischinski H. Bacterial proteomics and its role in antibacterial drug discovery. Mass Spectrom Rev. 2005;24:549–565. doi: 10.1002/mas.20030. [DOI] [PubMed] [Google Scholar]
- 17.Vincent IM, Ehmann DE, Mills SD, Perros M, Barrett MP. Untargeted Metabolomics To Ascertain Antibiotic Modes of Action. Antimicrob Agents Chemother. 2016;60:2281–2291. doi: 10.1128/AAC.02109-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuhrer T, Heer D, Begemann B, Zamboni N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal Chem. 2011;83:7074–7080. doi: 10.1021/ac201267k. [DOI] [PubMed] [Google Scholar]
- 19.Mülleder M, et al. Functional Metabolomics Describes the Yeast Biosynthetic Regulome. Cell. 2016;167:553–565.:e12. doi: 10.1016/j.cell.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarvin B, et al. Fast and sensitive flow-injection mass spectrometry metabolomics by analyzing sample-specific ion distributions. Nat Commun. 2020;11:3186. doi: 10.1038/s41467-020-17026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters JM, et al. A Comprehensive, CRISPR-based Functional Analysis of Essential Genes in Bacteria. Cell. 2016;165:1493–1506. doi: 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silvis MR, et al. Morphological and Transcriptional Responses to CRISPRi Knockdown of Essential Genes in Escherichia coli. mBio. 0:e02561-21. doi: 10.1128/mBio.02561-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belenky P, et al. Bactericidal Antibiotics Induce Toxic Metabolic Perturbations that Lead to Cellular Damage. Cell Rep. 2015;13:968–980. doi: 10.1016/j.celrep.2015.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zampieri M, Zimmermann M, Claassen M, Sauer U. Nontargeted Metabolomics Reveals the Multilevel Response to Antibiotic Perturbations. Cell Rep. 2017;19:1214–1228. doi: 10.1016/j.celrep.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Donati S, et al. Multi-omics Analysis of CRISPRi-Knockdowns Identifies Mechanisms that Buffer Decreases of Enzymes in E. coli Metabolism. CellSyst. 2020 doi: 10.1016/j.cels.2020.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Fuhrer T, Zampieri M, Sévin DC, Sauer U, Zamboni N. Genomewide landscape of gene-metabolome associations in Escherichia coli. Mol Syst Biol. 2017;13:907. doi: 10.15252/msb.20167150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.You C, et al. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature. 2013;500:301–306. doi: 10.1038/nature12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costanzo M, et al. The Genetic Landscape of a Cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Wet TJ, Winkler KR, Mhlanga M, Mizrahi V, Warner DF. Arrayed CRISPRi and quantitative imaging describe the morphotypic landscape of essential mycobacterial genes. eLife. 2020;9:e60083. doi: 10.7554/eLife.60083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta R, China A, Manjunatha UH, Ponnanna NM, Nagaraja V. A complex of DNA gyrase and RNA polymerase fosters transcription in Mycobacterium smegmatis. Biochem Biophys Res Commun. 2006;343:1141–1145. doi: 10.1016/j.bbrc.2006.02.195. [DOI] [PubMed] [Google Scholar]
- 33.Nayar AS, et al. Novel Antibacterial Targets and Compounds Revealed by a High-Throughput Cell Wall Reporter Assay. J Bacteriol. 2015;197:1726–1734. doi: 10.1128/JB.02552-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spitzer M, et al. Cross-species discovery of syncretic drug combinations that potentiate the antifungal fluconazole. Mol Syst Biol. 2011;7:499. doi: 10.1038/msb.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amaral L, Lorian V. Effects of chlorpromazine on the cell envelope proteins of Escherichia coli. Antimicrob Agents Chemother. 1991;35:1923–1924. doi: 10.1128/aac.35.9.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng Y, et al. Global analysis of protein structural changes in complex proteomes. Nat Biotechnol. 2014;32:1036–1044. doi: 10.1038/nbt.2999. [DOI] [PubMed] [Google Scholar]
- 37.Guo Y, et al. Tail-Anchored Inner Membrane Protein ElaB Increases Resistance to Stress While Reducing Persistence in Escherichia coli. J Bacteriol. 2017;199 doi: 10.1128/JB.00057-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu P, et al. Global Functional Atlas of Escherichia coli Encompassing Previously Uncharacterized Proteins. PLOS Biol. 2009;7:e1000096. doi: 10.1371/journal.pbio.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lorenz C, Dougherty TJ, Lory S. Transcriptional Responses of Escherichia coli to a Small-Molecule Inhibitor of LolCDE, an Essential Component of the Lipoprotein Transport Pathway. J Bacteriol. 2016;198:3162–3175. doi: 10.1128/JB.00502-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Říha M, et al. Novel method for rapid copper chelation assessment confirmed low affinity of D-penicillamine for copper in comparison with trientine and 8-hydroxyquinolines. J Inorg Biochem. 2013;123:80–87. doi: 10.1016/j.jinorgbio.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 41.Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol. 2013;11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A. 2009;106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torres-Cabassa AS, Gottesman S. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J Bacteriol. 1987;169:981–989. doi: 10.1128/jb.169.3.981-989.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hao Z, et al. The multiple antibiotic resistance regulator MarR is a copper sensor in Escherichia coli. Nat Chem Biol. 2014;10:21–28. doi: 10.1038/nchembio.1380. [DOI] [PubMed] [Google Scholar]
- 45.Kirn TJ, Onyeaso E, Syed M, Weinstein MP. Systematic evaluation of commercial susceptibility testing methods for determining the in vitro activity of daptomycin versus Staphylococcus aureus and Enterococci. J Clin Microbiol. 2014;52:1877–1882. doi: 10.1128/JCM.03439-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peters JM, et al. Enabling genetic analysis of diverse bacteria with Mobile-CRISPRi. Nat Microbiol. 2019;4:244–250. doi: 10.1038/s41564-018-0327-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stokes JM, et al. A Deep Learning Approach to Antibiotic Discovery. Cell. 2020;180:688–702.:e13. doi: 10.1016/j.cell.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dubuis S, Ortmayr K, Zampieri M. A framework for large-scale metabolome drug profiling links coenzyme A metabolism to the toxicity of anti-cancer drug dichloroacetate. Commun Biol. 2018;1:101. doi: 10.1038/s42003-018-0111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Storey JD. A Direct Approach to False Discovery Rates. J R Stat Soc Ser B. 2002;64:479–498. [Google Scholar]
- 50.Maier L, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555:623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orth JD, et al. A comprehensive genome-scale reconstruction of Escherichia coli metabolism-2011. Mol Syst Biol. 2011;7:535. doi: 10.1038/msb.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okuda S, Yoshizawa AC. ODB: a database for operon organizations, 2011 update. Nucleic Acids Res. 2011;39:D552-555. doi: 10.1093/nar/gkq1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Konig R, et al. A probability-based approach for the analysis of large-scale RNAi screens. Nat Meth. 2007;4:847–849. doi: 10.1038/nmeth1089. [DOI] [PubMed] [Google Scholar]
- 54.Zampieri M, et al. Metabolic constraints on the evolution of antibiotic resistance. Mol Syst Biol. 2017;13:917. doi: 10.15252/msb.20167028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manfredi P, et al. Defining Proteomic Signatures to Predict Multidrug Persistence in Pseudomonas aeruginosa. Methods Mol Biol Clifton NJ. 2021;2357:161–175. doi: 10.1007/978-1-0716-1621-5_11. [DOI] [PubMed] [Google Scholar]
- 56.Zaslaver A, et al. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods. 2006;3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- 57.Fang X, et al. Global transcriptional regulatory network for Escherichia coli robustly connects gene expression to transcription factor activities. Proc Natl Acad Sci U S A. 2017;114:10286–10291. doi: 10.1073/pnas.1702581114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wishart DS, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brunk E, et al. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat Biotechnol. 2018;36:272–281. doi: 10.1038/nbt.4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Faith JJ, et al. Large-Scale Mapping and Validation of Escherichia coli Transcriptional Regulation from a Compendium of Expression Profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article as Supplementary Data. A detailed description of all data analysis steps is published in this article in the Methods section and Supplementary Information. Matlab code is available for download at http://www.imsb.ethz.ch/research/zampieri-group/resources.html. Metabolome data and results from data analysis can be found as Supplementary Tables. The mass spectrometry proteomics raw data and analysis results have been deposited through the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD024133. Genome sequence data have been deposited through SRA Bioproject: PRJNA776168 (also see Supplementary Notes).