Abstract
In situ hybridization (ISH) methods remain the most popular approach for profiling the expression of a gene at high spatial resolution and have been broadly used to address many biological questions. One compelling application is in the field of evo-devo, where comparing gene expression patterns has offered insight into how vertebrate development has evolved. Gene expression profiling in the invertebrate chordate amphioxus (cephalochordate) has been particularly instrumental in this context: its key phylogenetic position as sister group to all other chordates makes it an ideal model system to compare with vertebrates and for reconstructing the ancestral condition of our phylum. However, while ISH methods have been developed extensively in vertebrate model systems to fluorescently detect the expression of multiple genes simultaneously at a cellular and subcellular resolution, amphioxus gene expression profiling is still based on single-gene nonfluorescent chromogenic methods, whose spatial resolution is often compromised by diffusion of the chromogenic product. This represents a serious limitation for reconciling gene expression dynamics between amphioxus and vertebrates and for molecularly identifying cell types, defined by their combinatorial code of gene expression, that may have played pivotal roles in evolutionary innovation. Herein we overcome these problems by describing a new protocol for application of the third-generation hybridization chain reaction (HCR) to the amphioxus, which permits fluorescent, multiplex, and quantitative detection of gene expression in situ, within the changing morphology of the developing embryo, and in adult tissues. A detailed protocol is herein provided for whole-mount preparations of embryos and vibratome sections of adult tissues.
Keywords: Amphioxus, Fluorophore-labeled, Gene expression profiling, HCR, In situ hybridization, Multiplex, Single-cell resolution
1. Introduction
In situ hybridization (ISH) methods have been utilized over the past several decades to describe spatial patterns of gene expression across tissues and across species. When combined with functional studies (e.g., gene expression inhibition) these methods have been particularly powerful for characterizing the roles and the regulatory interactions of specific genes in particular developmental processes. ISH methods have been, for example, key for defining genes and network interactions controlling vertebrate axial patterning [1, 2], vertebrate limb development [3, 4], and segmentation of the drosophila blastoderm [5, 6]. When combined with comparative developmental biology approaches, these methods have offered a means beyond morphological examination to define the homology of traits in different taxa, and therefore to define the origins of important evolutionary innovations. A remarkable discovery in this context was the observation that there is an evolutionarily conserved Hox code that patterns the anterior-posterior axis of most animals [7]. This sets the groundwork for comparisons between distantly related taxa, sharing homologous body parts (e.g., head, thorax, tail) that contain taxon-specific morphological specializations. Focusing on chordate evolution, gene expression profiling in the amphioxus has helped to resolve traits that are ancestral to the phylum, and those that are vertebrate innovations [8]. These include vertebrate traits, such as a complex brain and neural crest derivatives [9]. These studies have relied on the classic ISH protocol for amphioxus [10] which has been applied by researchers in the field for almost three decades [11].
While ISH methods have constantly evolved in other model systems to increase sensitivity, to target multiple genes simultaneously, and to achieve cellular and subcellular resolution, most of the gene expression profiling in amphioxus is still based on single-gene expression detection using nonfluorescent chromogenic methods. Chromogenic methods can be very sensitive, but they are not quantitative, as they depend on the nonlinear accumulation of a chromogenic product. Diffusion of this product can often compromise the spatial resolution of the signal, leading to loss of cellular and subcellular detail. Furthermore, chromogenic methods are limited in terms of multiplexing, usually restricted to three-color reactions (color per gene), and only two in amphioxus in very exceptional cases [12]. While possible, multiplex ISH is highly technically challenging, because it demands multiple chromogenic reactions to be performed in series without cross reaction. Many technologies have emerged recently that tackle some of these technical challenges in vertebrates and offer quantitative information by virtue of fluorescent RNA detection (e.g., TSA, RNAscope, HCR). However, because these have been inconsistently applied, reconciling gene expression dynamics between model systems is becoming increasingly challenging. Therefore, there is high demand for ISH technology that can be readily applied across model systems, and offers cellular resolution, multiplex potential, and fluorescent quantitative imaging.
To address these challenges, we have taken advantage of third-generation in situ hybridization chain reaction (HCR) technology, recently developed by Choi and collaborators [13], and adapted it to the particular requirements of amphioxus embryos and adult tissues. HCR version 3 (HCRv3) is based on the use of DNA probe pairs that bind in tandem to complementary sequences on the mRNA of interest. Split between each probe pair is an initiator sequence that triggers the focal binding and polymerization of metastable kinetically trapped DNA hairpins, each of which is conjugated to a fluorescent Alexa Fluor moiety. A complete adapter is formed where the probe pairs bind in tandem specifically to the mRNA sequence with certain separation. Only then can hairpins assemble through cooperative binding into tethered fluorescent amplification polymers, whose fluorescence intensity will scale linearly with the local density of mRNA molecules [14, 15]. Intrinsic to this design is automatic background suppression, in which the nonspecific off-site binding of a single probe will not accumulate fluorescent signal due to absence of a complete adapter sequence. This background suppression generates a significantly improved signal: noise ratio in HCRv3 imaging compared to previous versions, enabling highly sensitive imaging of even very weakly expressed genes. The specificity of adapter sequences and commercial availability of DNA hairpins conjugated to different fluorophores (Molecular Instruments) allow multiplexing probes to target up to five different genes in the same specimen per reaction [16]. The HCRv3 method is therefore multiplex, sensitive, and quantitative.
Here we describe a method for application of the HCRv3 technology to multiplex and quantitatively analyze gene expression in the amphioxus, both in embryos and adult sections. This protocol can be executed in 3 days in embryos or adult sections that had been previously fixed and dehydrated in methanol or ethanol. The sample preparation is, unsurprisingly, specific for amphioxus, but the hybridization and the amplification steps are essentially as described by Choi and colleagues [13], with some minor modifications. Below we briefly outline several key developments that have facilitated optimization of our protocol in relation to that described by Choi and colleagues [13], and the classic amphioxus ISH protocol from Holland and Holland [10]:
We have updated the protocol for amphioxus embryo fixation to improve the quality of the starting material (Subheading 3.1), thereby increasing the specificity and sensitivity of the HCR signal. In particular, the protocol described below differs from that previously published with respect to the fixative amount and the length of fixation [17, 18]. The handling of the embryos also differs since we do not include any centrifugation steps and all pipette tips and plates are siliconized. This latter step is motivated by our observation that amphioxus embryos are very sticky, especially at the early steps of fixation and at the beginning of the HCR protocol, meaning that they can get damaged very easily or lost during handling.
We describe a completely new protocol for generating thick vibratome sections of adult tissues (Subheading 3.2), which we illustrate using brain tissue (Fig. 1). This represents an improvement on methods for analyzing gene expression in adult amphioxus tissues, which to date were mostly dependent on paraffin embedding and on a very lengthy protocol for chromogenic detection [19, 20]. The classic protocol adopted these strategies since the high hybridization temperatures of the traditional amphioxus ISH protocol (60–63 °C) meant that the tissue would curl unless attached to the slide. By contrast, the milder hybridization conditions of the HCR protocol are permissive enough so the technique can be applied to thick floating vibratome sections with no curling or noticeable shrinkage. Consequently, we have been able to produce a protocol that can be applied more rapidly than the previous ISH on paraffin sections that allows a finer tuning depending on the gene of interest, and that is in general more robust (Fig. 1).
We introduced a bleaching step at the beginning of the HCR protocol (Subheading 3.4, step 3). Bleaching is a common step in the ISH protocols of many model systems, including flat-worms and most vertebrates [21–23]. In amphioxus, we have found that this step reduces the inherent autofluorescence of the tissue and completely eliminates the natural pigmentation of the eye and other photoreceptive spots. Thereafter, the tissue becomes more transparent, which permits imaging of entire embryos at single-cell resolution (Fig. 2).
We found that pretreatment with proteinase K is not necessary for HCR in amphioxus (Fig. 1), unlike when using DIG-labeled riboprobes, and indeed might only be useful when hybridizing very thick sections (Fig. 3). This is a major step forward, as imaging of gene expression is now possible in undigested tissues, showing a more normal morphology. Indeed, this protocol can be readily combined with immuno-histochemistry for morphological landmarks.
We propose a novel imaging approach for fluorescent labeling in amphioxus tissues using inverted confocal microscopy. This ensures that the specimens lie flat and within the working distance of the objective throughout the imaging process, even up to 100× optical magnification. Thus, Z-stacks can be acquired of entire specimens at single-cell resolution. This configuration is compatible with tile scanning of a large field of specimens in a multi-area time lapse, but also permits adjustment of orientation to acquire the same specimen from multiple views, or to repair small deviations of position after the initial mounting process. In principle, individual specimens can be retrieved after imaging with this method, bleached, and re-stained to further increase the number of channels in the HCR.
Fig. 1.
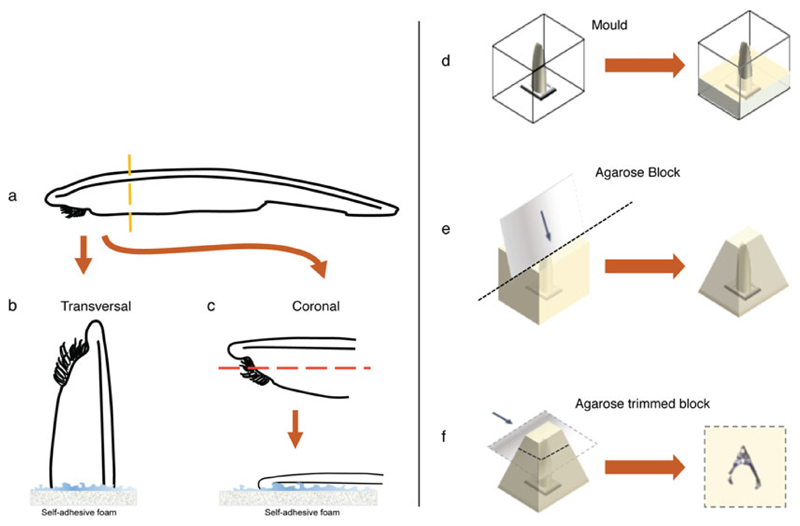
Embedding and vibratome sectioning of adult amphioxus brain tissues. Once the specimen has been anesthetized (a) the head is separated from the rest of the body and fixed for 24 h as indicated in Subheading 3.3, step 3. The separated head is then washed and glued into the self-adhesive foam either vertically (b) or horizontally (c), depending on the orientation needed. For coronal sections further dissection of the ventral side is recommended to properly align brain and neural tube in a same section. The foam with the attached head is then transferred into a peel-a-way square embedding mold and filled up with liquid low-melting agarose (d). Once the agarose has solidified the block is extracted from the mold and trimmed as a pyramid to provide better grip while sectioning (e). The wider part of the pyramid is then glued to the vibratome holder and the head is sectioned with the dorsal fin facing the blade (f)
Fig. 2.
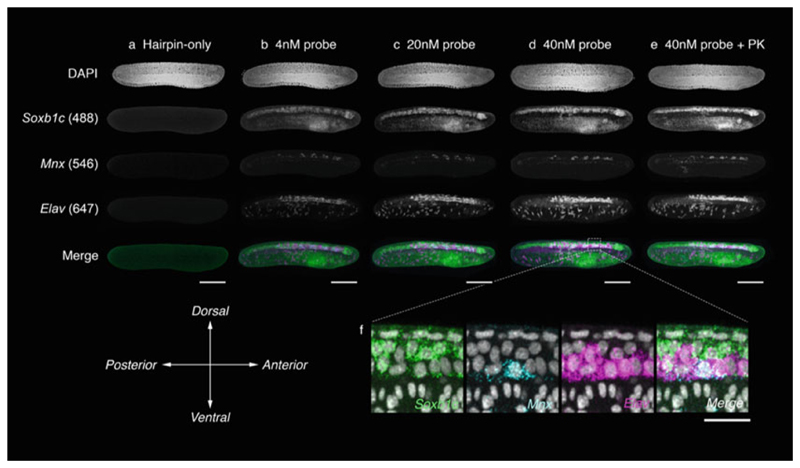
Triple HCR in whole-mount embryos of B. lanceolatum. HCR staining was performed on whole amphioxus embryos reared in our facility [24] to 24 hpf at 21 °C with varying concentrations of probe, and in the presence and absence of proteinase K digestion. For this combination, 20-pair probe sets were designed against Soxb1c for broad labeling of the neural tube, Mnx for labeling of motor neuron progenitors, and Elav for postmitotic neurons. Exposure to hairpins in the absence of probes generates channel-specific background fluorescence profiles (a). This is conspicuous in all channels, but most severe following excitation at a 488 nm wavelength. A 4 nM probe concentration, as suggested for use on zebrafish embryos, generates specific signal but with a poor signal:noise ratio, even after extensive washing (b). In the 488 channel, signal can be difficult to distinguish from background. Signal:noise is greatly improved with expose to 20 nM (c) and 40 nM (d) probe concentrations for common incubation times. At 40 nM, signal can readily be resolved from background. Proteinase K (PK) treatment is common in amphioxus ISH protocols to enhance embryo permeability. However, treatment with PK did not enhance HCR signal beyond that achieved with only permeabilization in TritonX-100 and DMSO (e). (f) Magnified view of parasagittal section through anterior neural tube of embryo in (d), revealing regional differences in expression profiles for each gene, and a single triple-positive cell. Scale bars measure 100 μm (a–e) and 20 μm (f)
Fig. 3.
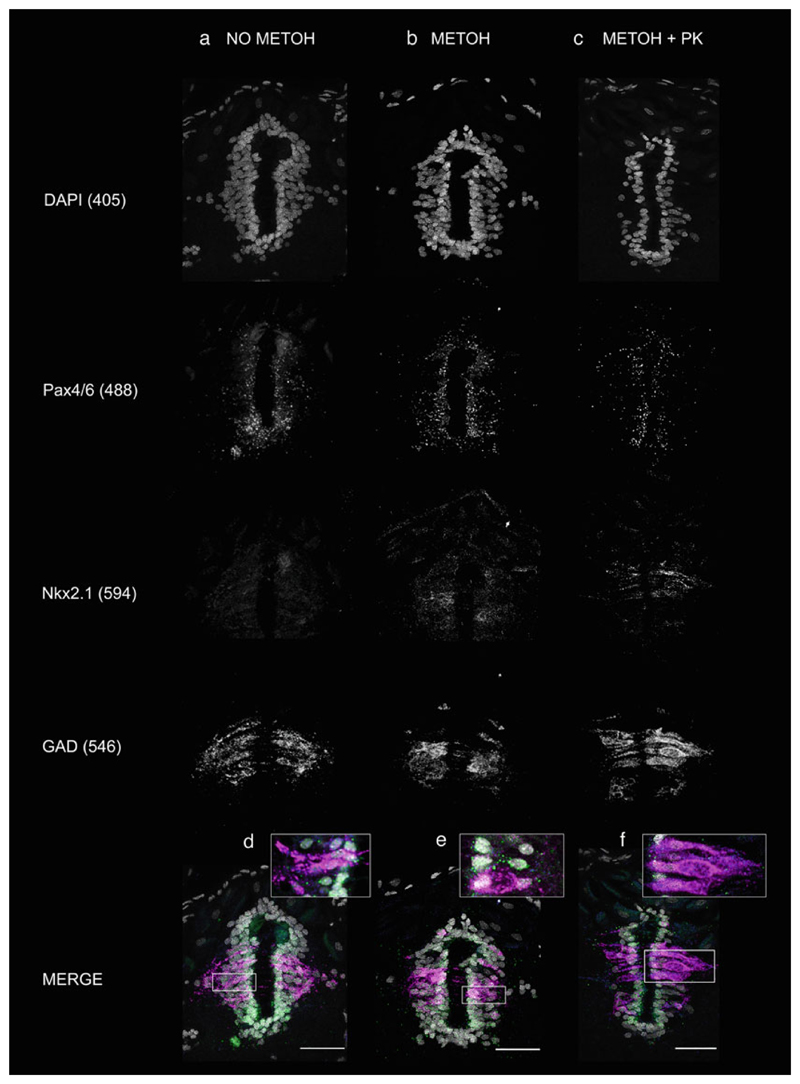
Triple HCR in B. lanceolatum brain vibratome sections. HCR staining was performed on floating vibratome sections of adult amphioxus maintained in our amphioxus facility [24]. Sections were stained with 20-pair probe sets designed against Pax4/6, used here as a neuronal marker, Nkx2.1, used here as a specifier of GABAergic fate and GAD, used here as a marker of GABAergic neurons. All probes were used at a concentration of 40 nM, as it was found to show the best signal:noise ratio for most of the genes in whole-mount preparations of embryos (see Fig. 2). Sections that had been preincubated in methanol after fixation (b) (see Note 1) showed a better signal:noise ratio than those that were not exposed to methanol (a). Low-expressed genes such as Nkx2.1 are for example hardly visible when sections are not preincubated in methanol (compare a and b). Additional pretreatment with proteinase K (c) improves the sharpness of the signal for all genes, including the low-level-expressing Nkx2.1, the mid-level-expressing Pax4/6, and the highly expressed GAD. Magnified views of merged images in d, e, and f show co-localization of the transcripts at a single-cell resolution
2. Materials
2.1. Reagents
Ethanol/methanol (for sample storage).
Split-initiator probe pairs (Molecular Instruments).
Fluorophore-labeled metastable DNA HCR hairpins (Molecular Instruments).
Probe hybridization buffer (Molecular Instruments).
Probe wash buffer (Molecular Instruments).
Amplification buffer (Molecular Instruments).
RNase AWAY (Ambion).
Proteinase K.
Glycerol or Aqua-Polymount (Polysciences).
2.2. Equipment
Siliconized or gelatinized p200 tips.
Sterile filter tips.
Siliconized or gelatinized 1.5 mL tubes.
Nunc untreated 4-well plates or siliconized/gelatinized 24-well plates.
Hybridization oven (e.g., Hybaid Shake ‘n’ Stack).
Orbital shaker.
Water bath.
Heat block.
Stereomicroscope.
Vibratome (Leica).
Vibratome blades.
Dissection Instruments (for adult tissues).
Peel-a-way square embedding molds (Sigma).
Self-adhesive foam stripes for window sealing.
Superglue (Loctite or Gorilla).
Parafilm.
0.2 μm pore size filters.
Glass-bottom dishes.
2.3. Buffers and Solutions
Tricaine solution: Tricaine, double-distilled water, 1 M Tris–HCl pH 9. Weigh 400 mg of tricaine and add this to 90 mL of double-distilled water. Once the tricaine has dissolved completely adjust the pH to 8.0–8.2 with 1 M Tris–HCl pH 9. Top up the solution to 100 mL with double-distilled water, filter through a 0.2 μm pore size filter, and aliquot in 2 mL doses. Store at −20 °C.
2× MOPS buffer pH 7.5–7.6: 0.1 M MOPS (free acid), 2 mM MgSO4, 1 mM EGTA, 0.5 M NaCl. Weigh all in powder and dissolve in DEPC-treated water or nuclease-free water. Adjust with NaOH pellets to a pH of 7.5–7.6 (for a volume of 250 mL add approximately 8 pellets). Filter through a 0.2 μm pore size filter and store at 4 °C. Discard after a month.
3.7% PFA-MOPS pH 7.5–7.6: Paraformaldehyde, 2× MOPS buffer pH 7.5–7.6. Weigh 1.85 g of paraformaldehyde and reserve in a 50 mL tube. Bring some DEPC-treated water or nuclease-free water to the boil in a microwave and add 20 mL of this to the paraformaldehyde reserved in the 50 mL tube. To the paraformaldehyde in water add approximately 50 μL of NaOH 10 M. Vortex until all paraformaldehyde is dissolved. If there are still particles of paraformaldehyde in suspension, leave the 50 mL tube in a water bath at 65 °C until everything is completely dissolved. Thereafter, add 25 mL of 2× MOPS buffer and confirm that pH is 7.5–7.6. Adjust the pH if it is not within range. Top up with DEPC-treated water or nuclease-free water to 50 mL. Filter through a 0.22 μm pore size filter and store at 4°C. For health and safety reasons it is recommended to perform all these steps, including weighing the paraformaldehyde, in a fume hood wearing protective goggles and gloves.
5 × Gelatine stock for coating tips and plates: Gelatine, DEPC-treated water, formaldehyde 37%. For a volume of 50 mL of stock, weigh 0.25 g of gelatine. Dissolve by autoclaving in DEPC-treated water. Allow the solution to cool and add 250 μL of formaldehyde (37%). For health and safety reasons it is recommended to add the formaldehyde in a fume hood and wear protective goggles and gloves, as the solution might still be warm. To coat tips and plates, dilute the stock to 1 × with DEPC-treated water or nuclease-free water. After coating, dry plates and tips on a 65 °C oven before using them.
10× NPBS: 200 mM Phosphate buffer pH 7.4, 9% NaCl. Filter through a 0.22 μm pore size filter and store at room temperature. If stocked for long, filter before use.
NPBST: NPBS, 0.1% Tween 20.
Bleaching buffer: 5% Deionized formamide, 1.5% H2O2, 0.2× SSC in DEPC-treated water or nuclease-free water. Prepare fresh when needed. For health and safety reasons it is recommended to wear protective goggles and gloves while preparing this buffer.
3% LM-agarose: LM-agarose, DEPC-treated water or nuclease-free water. Weight 3 g of low-melting agarose for every 100 mL of DEPC-treated water or nuclease-free water. Add the 3 g of LM agarose to 80 mL of DEPC-treated water or nuclease-free water and boil in a microwave until completely dissolved. Add 10 mL of 10× NPBS and top up to 100 mL, if volume has decreased while boiling, with DEPC-treated water or nuclease-free water. The agarose can be kept at 4 °C and remelted when needed.
Permeabilization solution: 1% DMSO, 1% Triton. Dissolve in NPBS. Store at room temperature.
5× SSCT [13]: 5× SSC (pH 7), 0.1% Tween 20. For best results, filter just before use with a 0.22 μm pore size filter. Store at room temperature.
DAPI counterstain solution: 1 μg/mL DAPI, NPBST. Dilute DAPI (1 μg/mL) to 1:500 in NPBST.
3. Methods
Before starting it is recommended to clean all surfaces and equipment with RNase AWAY (Ambion). This is particularly relevant for the vibratome holder and tray, as the tissue can be especially vulnerable to RNases while sectioning. It is also recommended to autoclave all glassware and other autoclavable instruments used in the procedure. Use gloves throughout the entire procedure to protect yourself and to protect the samples from RNases.
3.1. Embryo Collection and Fixation
Collect the embryos in the center of the dish by concentrically moving the dish. Amphioxus embryos are very small, so it is necessary to observe this process under the stereomicroscope.
Pipette the embryos with a siliconized or gelatinized wide orifice p200 tip into siliconized 1.5 mL tubes.
Transfer the 1.5 mL tubes to an ice-cold rack and leave the embryos to pellet by gravity. When the pellet is formed, remove as much seawater as possible and fill the tube with ice-cold 3.7% PFA-MOPS buffer.
Leave the embryos to pellet by gravity. When the pellet is formed, remove as much liquid as possible and refill the tube with ice-cold 3.7% PFA-MOPS buffer.
Repeat step 4 and fix for 8–10 h (depending on the stage) at 4 °C.
Wash the embryos with 1 × MOPS buffer by pelleting the embryos by gravity, as indicated above, at least two times.
Wash the embryos in either ethanol or methanol, by pelleting the embryos by gravity, as indicated above, at least twice.
Store the embryos in either ethanol or methanol at −20 °C, or proceed to the amphioxus HCR ISH protocol (Subheading 3.3, step 2).
3.2. Adult Tissue Fixation, Embedding, and Sectioning
Anesthetize adult amphioxus for 30 min using 2 mL of tricaine solution per 50 mL of seawater.
Make a series of cuts to divide the animal into four segments of equal length. Remove excess seawater from the tissues and transfer into ice-cold 3.7% PFA-MOPS buffer.
Fix for 24 h at 4 °C.
Wash tissues with 1 × MOPS at least twice. The samples can at this point be archived for long-term storage in either ethanol or methanol at −20 °C. Otherwise, proceed to the following step to prepare samples for embedding.
Wash tissues with 1 × NPBS at least two times.
Transfer the tissues into pre-warmed low-melting agarose in a water or dry bath at 45 °C and leave to equilibrate for 30 min.
Embedding: Cut a small rectangle of self-adhesive foam, large enough to hold the sample in the middle. Peel off the upside of the foam and add a small drop of superglue on top. With forceps extract the tissue that was equilibrating at 45 °C from the tube and place it in a suitable orientation on the top of the foam. Gently press the tissue so it gets properly attached to the foam. Transfer the foam rectangle with the attached tissue into the center of a peel-a-way square embedding mold. Fill the mold up with pre-warmed low-melting agarose (Fig. 1). Leave to cool and solidify at 4 °C.
Sectioning: Peel away the mold and glue the agarose block to the vibratome holder plate with superglue. Place at 4 °C while preparing the vibratome. Fill the tray of the vibratome with ice-cold RNase-free NPBS and the outer tray with ice. Assemble the vibratome blade following the instructions of the vibratome manufacturer. Attach the reserved holder plate containing the sample into the vibratome tray as indicated by the vibratome manufacturer. Place the hardest part of the tissue facing the blade (Fig. 1). If using a Leica VT100S the best sectioning conditions for amphioxus heads and trunks are as follows: speed 0.26 mm/s, frequency 50 Hz, and minimum thickness 50 μm. Collect the samples in gelatinized 24- or 96-well plates, to keep track of the sectioning order.
Sections can be stored at this point in either ethanol or methanol at −20 °C (see Note 1). Otherwise, proceed to the amphioxus HCR ISH protocol (Subheading 3.3, step 2).
3.3. Amphioxus HCR In Situ Hybridization (ISH) Steps
Embryo/section rehydration: If the embryos or sections are stored in ethanol it is best to transfer them to methanol for the rehydration steps. Thereafter, rehydrate through a methanol/water series, decreasing by 20% the proportion of methanol every 15 min. Perform all steps in gelatinized 4-well plates.
Wash embryos/sections twice in NPBST. Start the HCR protocol at this step if embryos and sections are already in NPBS or 1 × MOPS buffer.
Bleaching: Replace NPBST with 500 μL of bleaching solution. Incubate for 30–60 min, or as long as needed to remove the pigment spots, with light and reflective foil at the base of the plate. When embryos and tissues have reached translucency, wash twice in NPBST (see Note 2).
Permeabilization: Replace NPBST with 500 μL of permeabilization solution. Incubate for 3 h at room temperature (see Note 3).
Pre-hybridization: Remove permeabilization solution or NPBST (in case of the sections) and wash the embryos/sections in 5× SSCT for 5 min at room temperature. Wash the embryos/sections in pre-warmed hybridization buffer (without probes) at 37 °C to equilibrate the tissues. Replace with fresh pre-warmed hybridization buffer and pre-hybridize for at least 1 h at 37 °C (best results are obtained with longer pre-hybridization times).
Hybridization: Prepare the probe solution by diluting 1–10 μL of 2 μM probe mixed stock of each gene in 500 μL of pre-warmed hybridization solution. Replace the pre-hybridization buffer with the freshly made probe solution and incubate overnight at 37 °C. For adult sections, a minimum of 18-h incubation is recommended to ensure homogenous probe binding. For best results wrap the plate in parafilm to prevent evaporation during the hybridization time (see Note 4).
Pre-amplification: Remove the probe solution and wash the embryos/sections with pre-warmed probe wash buffer at 37 °C. Wash for a minimum of 20 min. In the last of the washes the plate can be transferred to an orbital shaker at room temperature, as the following steps are performed at room temperature. Replace the probe wash buffer with 5× SSCT and thoroughly wash embryos/sections three or four times, with each wash for a minimum of 1 h. Thereafter, pre-amplify by replacing the SSCT with 500 μL of pre-warmed amplification buffer at room temperature. Incubate in an orbital shaker at room temperature for a minimum of 1 h.
Preparation of the hairpins: Pipette 1 μL of each fluorescently labeled hairpin (of a 3 μM stock), per every 100 μL of amplification buffer to be used, into a fresh 1.5 mL tube. Heat up the hairpins by placing the 1.5 mL tube into a heat block at 95 ° C for 90 s. Cool the hairpins on ice, in the dark, for 30 min (see Note 5).
Amplification: In this step embryos and sections are transferred from the well plates to 1.5 mL tubes. In order to do this, concentrate the embryos/sections in the center of the well and use a wide-orifice siliconized p200 tip to collect them in no more of 20 μL of volume. Transfer the embryos/sections to a 1.5 mL tube containing 1 μL of each pre-cooled hairpin topped up to 80 μL with fresh amplification buffer. Protect the tubes from the light and incubate overnight in an orbital shaker at room temperature.
Washing: In this step embryos and sections are transferred back to a 4-well plate, so the washing volumes are bigger and therefore the excess of hairpins in the solution is removed more efficiently. To this aim, add 400 μL of 5 × SSCT to the 1.5 mL tube containing the embryos/sections and transfer the total volume of 500 μL to a 4-well plate. Add another 500 μL of 5 × SSCT to the tube, to ensure that all embryos and sections are recovered, and transfer these 500 μL to the 4-well plate. Wash three times at room temperature in 5× SSCT by replacing the buffer every 30 min. Wash a further four to five times at 4 °C in 5× SSCT, by replacing the buffer every hour. Best results are obtained with overnight washes at 4 °C (see Notes 6 and 7).
DAPI staining: Wash the embryos/sections twice in NPBST. Replace with fresh DAPI counterstain solution. Incubate overnight protected from light at 4 °C (see Note 8).
Wash four to five times in NPBST, replacing the buffer every 15 min.
3.4. Whole-Mount Imaging
Replace NPBST with 100% glycerol and allow the embryos/sections to equilibrate.
Fill the base of a glass-bottomed dish suitable for imaging on an inverted confocal microscope with 100% glycerol.
Transfer the embryos/sections to the glass-bottomed dish using a wide-orifice siliconized p200 pipette tip. Use an eyelash wand to push the specimens to the bottom of the dish, such that they lie flat in direct context with the glass in the correct orientation for imaging. Embryos can be organized into rows and columns based on variations in state, treatment, and HCR gene combination. Adult sections should be manually flattened against the glass with the eyelash wand to remove folds and curvatures (see Note 9).
Leave the glass-bottomed dish in the dark at 4 °C for at least half an hour prior to imaging to prevent drifting during the imaging process.
Image whole embryos and sections using a multi-area time-lapse function on an inverted confocal microscope (see Note 10).
After imaging, retrieve specimens from the glass-bottomed dish for long-term storage in PBS.
Acknowledgments
The authors would like to thank Ben Steventon for encouraging us to develop the HCR protocol in amphioxus; to Christo Christov for technical support to our lab and amphioxus facility, the latter supported by a Sir Isaac Newton Trust Research Grant (Ref. 15.07 (r)); to everybody in the histopathology and imaging facilities at the CRUK-CI; and to Matt Wayland in the imaging facilities at the Department of Zoology, which are supported by a Sir Isaac Newton Trust Research Grant (Ref. 18.07ii(c)). We also acknowledge support from CRUK (C9545/A29580) to EBG, Wellcome Trust Grant (203806/Z/16/A) to TGA, and the Claire Barnes Trust to GG.
4 Notes
Methanol incubation. To maximize signal-to-background ratio in adult sections an incubation of at least 12 h in 100% MeOH is strongly recommended. This step has proved to be particularly useful for transcripts present at low levels, while patterns of highly expressed genes can be identified even in sections that are not treated with MeOH (see Fig. 3).
Bleaching solution. This solution can also be used to remove the chorion of early amphioxus embryos.
Pretreatment with proteinase K. For very thick sections incubate with proteinase K (PK) at a concentration of 1 μg/mL, for up to 8 min at 37 °C. Thereafter, thoroughly wash in NPBST and postfix with 3.7% PFA-PBS for 30 min at room temperature. Then wash thoroughly at least three times in NPBST.
Hybridization time. Hybridization time is critical for thick sections, as probes take longer to penetrate in the tissue. At least 18 h of incubation is recommended.
Amplifier label. Even with bleaching, endogenous autofluorescence in amphioxus is still particularly visible at 488 nm. We therefore advise using Alexa Fluor 488 hairpins (Molecular Instruments) for strongly expressed genes, where signal intensity is expected to exceed the autofluorescence levels. If this is not possible, background subtraction might be required during image acquisition or registration to enhance the signal of the probes.
Background removal. Although the V3 method ensures a high signal:noise ratio by preventing off-site binding, in amphioxus we find that background can still be an issue, particularly for weakly expressed genes where laser powers must be high to visualize the signal. However, we have found background to be reduced by extending probe washes and hairpin washes overnight at 4 °C. If after an overnight wash background is still high, washes can be extended for a further 2–3 days at 4 °C.
Combined immunohistochemistry. HCR imaging can be readily coupled to immunohistochemistry to label specific subcellular structures or morphological landmarks. For this, wash specimens out of the hairpin solution and then proceed directly to the immunoblock and subsequent steps of primary and secondary antibody incubation. All steps from here on should be in the dark at 4 °C to preserve HCR fluorescence. However, this is very robust, and we find no severe quenching during the immunohistochemistry protocol.
Dapi staining. This step can also be done by incubating at a higher concentration (1:200) for 3 h at 4 °C.
Vibratome sections mounting for microscopy. The use of Aqua Polymount might be more appropriate for vibratome sections of big specimens. This mounting media solidifies at application, thereby preventing the sections from moving or bend during image acquisition. This mounting media can also be used for embryos if high magnification objectives, with a shorter working distance, are used (e.g., 40×, 60×, 100×). In this case, use a wide-orifice siliconized p200 pipette tip to transfer sections to a glass-bottom dish, then remove NPBST, and apply Aqua-Polymount to fill the dish. Use tungsten needles to gently arrange sections ensuring that they are completely flat and lying at the bottom of the dish, and then leave to harden at 4 °C for at least 30 min.
Cross-talk suppression. This method permits imaging five fluorophores simultaneously in the same specimen. However, we have found cross-talk between fluorophores when detection windows are too wide. To compensate for this, detection windows for each channel should be narrowed to sit exclusively on the emission peak of the fluorophore of interest.
References
- 1.Graham A, Papalopulu N, Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989;57(3):367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- 2.Dubrulle J, Pourquié O. fgf8 mRNA decay establishes a gradient that couples axia elongation to patterning in the vertebrate embryo. Nature. 2004;427:419–422. doi: 10.1038/nature02216. [DOI] [PubMed] [Google Scholar]
- 3.Riddle RD, Johnson RL, Laufer E, et al. Sonic Hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75(7):1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- 4.Nelson C, Morgan B, Burke AC, et al. Analysis of Hox gene expression in the chick limb bud. Development. 1996;122:1449–1466. doi: 10.1242/dev.122.5.1449. [DOI] [PubMed] [Google Scholar]
- 5.Akam M, Martinez-Arias A. The distribution of Ultrabithorax transcripts in Drosophila embryos. EMBO. 1985;4:1689–1700. doi: 10.1002/j.1460-2075.1985.tb03838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker NE. Molecular cloning oi sequences from wingless, a segment polarity gene in Drosophila: the spatial distribution o a transcript in embryos. EMBO. 1987;6:1765–1773. doi: 10.1002/j.1460-2075.1987.tb02429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 8.Benito-Gutiérrez È. In: Amphioxus as a model for mechanisms in vertebrate development. eLS, editor. 2011. [DOI] [Google Scholar]
- 9.Meuleman D, Bronner-Fraser M. Gene-Regulatory Interactions in neural crest evolution and development. Dev Cell. 2004;7:291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Holland ND, Holland LZ. In: Essential developmental biology, a practical approach. Stern CD, Holland PWH, editors. IRL Press; Oxford: 1993. Embryos and larvae of invertebrate deuterostomes; pp. 21–32. [Google Scholar]
- 11.Holland PW, Holland LZ, Williams NA, et al. An amphioxus homeobox gene: sequence conservation, spatial expression during development and insights into vertebrate evolution. Development. 1992;116:653–661. doi: 10.1242/dev.116.3.653. [DOI] [PubMed] [Google Scholar]
- 12.Irimia M, Pineiro C, Maeso I, et al. Conserved developmental expression of Fezf in chordates and Drosophila and the origin of the Zona Limitans Intrathalamica (ZLI) brain organizer. EvoDevo. 2010;1:7. doi: 10.1186/2041-9139-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi HMT, Schwarzkopf M, Fornace ME, et al. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development. 2018;145:dev165753. doi: 10.1242/dev.165753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi HMT, Chang JY, Trinh LA, et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat Bio-technol. 2010;28:1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi HMT, Beck VA, Pierce NA. Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability. ACS Nano. 2014;8:4284–4294. doi: 10.1021/nn405717p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi HMT, Calvert CR, Husain N, et al. Mapping a multiplexed zoo of mRNA expression. Development. 2016;143:3632–3637. doi: 10.1242/dev.140137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu JK, Holland LZ. Amphioxus whole- mount in situ hybridization. Cold Spring Harb Protoc. 2009;2009:pdb.prot5286. doi: 10.1101/pdb.prot5286. [DOI] [PubMed] [Google Scholar]
- 18.Yu JK, Holland LZ. Amphioxus (Bran- chiostoma floridae) spawning and embryo collection. Cold Spring Harb Protoc. 2009;2009:pdb.prot5285. doi: 10.1101/pdb.prot5285. [DOI] [PubMed] [Google Scholar]
- 19.Benito-Gutierrez E, Nake C, Llovera M, et al. The single AmphiTrk receptor highlights increased complexity of neurotrophin signalling in vertebrates and suggests an early role in developing sensory neuroepidermal cells. Development. 2005;132:2191–2202. doi: 10.1242/dev.01803. [DOI] [PubMed] [Google Scholar]
- 20.Benito-Gutiérrez È, Stemmer M, Rohr SD, et al. Patterning of a telencephalon-like region in the adult brain of amphioxus. bioRxiv. 2018:307629 [Google Scholar]
- 21.Rybak-Wolf A, Solana J. In: Whole-Mount in situ hybridization using DIG-labelled probes in planarian in in situ hybridization protocols, methods in molecular biology. Nielsen BS, editor. Vol. 1211. Springer; New York: 2014. pp. 41–50. [DOI] [PubMed] [Google Scholar]
- 22.Fuentes R, Fernández J. In: In situ hybridization protocols, methods in molecular biology. Nielsen BS, editor. Vol. 1211. Springer; New York: 2014. Fixation/per-meabilization procedure for mRNA in situ hybridisation of zebrafish whole-mount oocytes, embryos and larvae; pp. 1–12. [DOI] [PubMed] [Google Scholar]
- 23.Saint-Jeannet JP. In: Cold Spring Harb Protoc. Sive Hazel L., editor. 2017. Whole-mount in situ hybridization of xenopus embryos. From the xenopus collection. [DOI] [PubMed] [Google Scholar]
- 24.Benito-Gutiérrez E, Weber H, Bryant DV, et al. Methods for generating year-round access to amphioxus in the laboratory. PLoS One. 2013;8:e71599. doi: 10.1371/journal.pone.0071599. [DOI] [PMC free article] [PubMed] [Google Scholar]