Abstract
Major Depressive Disorder (MDD) often is associated with significant cognitive dysfunction. We conducted a meta-analysis of genome-wide interaction of MDD and cognitive function using data from 4 large European cohorts in a total of 3510 MDD cases and 6057 controls. In addition, we conducted analyses using polygenic risk scores (PRS) based on data from the Psychiatric Genomics Consortium (PGC) on the traits of MDD, Bipolar disorder (BD), Schizophrenia (SCZ), and mood instability (MIN). Functional exploration contained gene expression analyses and Ingenuity Pathway Analysis (IPA®). We identified a set of significantly interacting single nucleotide polymorphisms (SNPs) between MDD and the genome-wide association study (GWAS) of cognitive domains of executive function, processing speed, and global cognition. Several of these SNPs are located in genes expressed in brain, with important roles such as neuronal development (REST), oligodendrocyte maturation (TNFRSF21), and myelination (ARFGEF1). IPA® identified a set of core genes from our dataset that mapped to a wide range of canonical pathways and biological functions (MPO, FOXO1, PDE3A, TSLP, NLRP9, ADAMTS5, ROBO1, REST). Furthermore, IPA® identified upstream regulator molecules and causal networks impacting on the expression of dataset genes, providing a genetic basis for further clinical exploration (vitamin D receptor, beta-estradiol, tadalafil). PRS of MIN and meta-PRS of MDD, MIN and SCZ were significantly associated with all cognitive domains. Our results suggest several genes involved in physiological processes for the development and maintenance of cognition in MDD, as well as potential novel therapeutic agents that could be explored in patients with MDD associated cognitive dysfunction.
Keywords: cognitive function, MDD, GWAS, neurodevelopment
Introduction
Major Depressive Disorder (MDD) is an enormous health problem globally, with many years of life lived with disability1. In the 2017 Global Burden of Disease Study, MDD accounted for an estimated 32.8 million years lived with disability (YLDs)2. Cognitive dysfunction has been found to occur in over half of patients with MDD3, including deficits in memory, executive function, attention, and slower reaction time4, 5. Deficits in memory involve immediate memory6, verbal learning and memory7, visual memory, and working memory8.
Cognitive dysfunction observed in MDD is associated with impairment in functioning, including social and occupational functioning5, 9. Specifically, unemployment has been associated with cognitive dysfunction in both current and remitted MDD6. Furthermore, increased severity of MDD has been correlated with reduced cognitive performance in measures of executive function, processing speed, and episodic memory10. It has also been hypothesized that persistent cognitive dysfunction may be associated with a more disabling illness, including more frequent admissions to hospital 9 and non-response of depressive symptoms to pharmacotherapy11.
Importantly, cognitive dysfunction observed in MDD often persists, even after other symptoms of depression have remitted5. In addition to the cognitive dysfunction persisting, the impairment in psychosocial function can persist5, 9, 12. These clinical observations suggest that cognitive dysfunction is not only a state marker of MDD, but can present as a trait marker of MDD. Hence, there is a need to explore the underlying biology of cognitive function in MDD, including its genetic architecture, in more detail. While a number of novel treatments are showing promise in improving cognitive dysfunction in MDD, the research is generally in the early stages13. Further exploration of the underlying biology of cognitive dysfunction may enhance better targeting of treatment, and lead to the identification of novel molecular targets for treatments in patients with MDD5, 13.
Only few previous studies have specifically explored the genomic signature of cognitive performance in MDD patients and have produced heterogeneous results. A GWAS meta-analysis conducted in 24 independent cohorts as part of the Cognitive Genomics Consortium (COGENT) found genetic correlations between general cognitive performance and several psychiatric traits, but not for MDD14. In contrast, linkage disequilibrium (LD) score regression analyses using UK Biobank cognitive data found that MDD was genetically associated with slower reaction time15. In another study, healthy individuals with a higher MDD polygenic risk score (PRS) were found to show working memory activation patterns more like those seen in MDD16. Using data from over 7000 individuals participating in the Generation Scotland: the Scottish Family Health Study, Meijsen et al.17 confirmed significant deficits in those with MDD across a number of cognitive domains but found no single nucleotide polymorphism (SNP) associations with cognitive performance in patients.
These previous findings highlight common difficulties in this research area. First, both MDD and cognitive function are complex psychological concepts characterized by high levels of phenotypical heterogeneity, requiring very large samples to detect meaningful genetic associations. Second, the psychometric tools used to measure cognitive domains vary widely, and ‘composite’ cognition scores inherently introduce more variation. To achieve progress, studies are required in cohorts that are large and well enough characterized to break down cognitive function into recognized subdomains. Third, clinical data suggest that cognitive performance and MDD status interact with each other in complex ways. Therefore, analytic approaches are warranted that allow for the identification of such interactions on a genetic level.
To address these challenges, we aimed to investigate the phenotypic and genetic relationship between cognitive function and MDD through a genome wide interaction study, using several large European cohorts including MDD cases and healthy controls. Regular GWAS identifies the effect of a genetic variant on the phenotypes. The genome-wide interaction analysis aims to explore the modifying effect of an exposure variable on the genetic association. Due to the excellent clinical characterization of these cohorts, we were able to conduct separate analyses for global cognition as well as individual cognitive domains including executive function, immediate and delayed memory, and processing speed. We hypothesised that genetic associations with cognitive performance differ by MDD status.
Finally, to explore the biological functions of the loci identified in the genome-wide interaction study, we conducted analyses including psychiatric PRS, gene expression data, and Ingenuity Pathway Analysis (IPA®).
Methods
The overall sample consists of 9567 participants (3510 MDD cases and 6057 controls) with both genetic and other phenotypic data from four cohorts (Table 1). A detailed description of the cognitive tests in each cohort, including how each test is administered, appears in the supplementary material – including Table S1 (refer also to Supplementary Figure 1). A description of the method used to calculate z scores for each cognitive test within each cohort, for each cognitive domain, and then for each cohort (i.e. a global cognitive score) is also provided in the supplementary material.
Table 1. Sample description.
| BiDirect | FOR2107 | Generation Scotland | SHIP-Trend | Total | |
|---|---|---|---|---|---|
| Total Sample (MDD and No MDD) | |||||
| Number | 1554 | 1254 | 6157 | 602 | 9567 |
| Sex: | |||||
| Male | 728 (46.8%) | 478 (38.1%) | 2399 (39.0%) | 282 (46.8%) | 3887(40.6%) |
| Female | 826 (53.2%) | 776 (61.9%) | 3758 (61.0%) | 320 (53.2%) | 5680(59.4%) |
| Age (years) | |||||
| Average | 51.1 | 34.8 | 47.9 | 48.8 | 46.75 |
| SD | 7.8 | 13.2 | 13.2 | 13.2 | 13.34 |
| Range | 35.1-66.1 | 18.0-69.0 | 18.0-93.0 | 22.0-80.0 | 18.0-93.0 |
| Edu (years) | |||||
| Average | 14.3 | 13.5 | 13.8 | 10.4 | 13.66 |
| SD | 2.7 | 2.6 | 3.4 | 1.2 | 3.23 |
| Range | 0.0-18.0 | 9.0-18.0 | 0.0-24+ | 8.0-12.0 | 0.0-24.5 |
| MDD Sample | |||||
| Number | 912 (58.7%) | 573 (45.7%) | 1877 (30.5%) | 148 (24.6%) | 3510 (36.7%) |
| Sex: Male | 391 | 223 | 538 | 40 | 1192 |
| Female | 521 | 350 | 1339 | 108 | 2318 |
| Age (years) | |||||
| Average | 49.98 | 37.55 | 46.23 | 48.95 | 45.91 |
| SD | 7.28 | 13.53 | 12.63 | 12.08 | 12.29 |
| Range | 35.08-66 | 18-69 | 18-84 | 22.0-80.0 | 18-84 |
| Edu (years) | |||||
| Average | 13.95 | 13.02 | 13.85 | 10.43 | 13.60 |
| SD | 2.70 | 2.72 | 3.41 | 1.18 | 3.15 |
| Range | 0-18 | 9-18 | 0-24.5 | 8.0-12.0 | 0-24.5 |
| Current MDD | 817 (89.6%) | 423 (73.8%) | 349 (18.6%) | 84 (56.8%) | 1673 (47.7%) |
| No MDD Sample | |||||
| Number | 642 (41.3%) | 681 (54.3%) | 4280 (69.5%) | 454 (75.4%) | 6057(63.3%) |
| Sex: Male | 305 | 255 | 1861 | 242 | 2695 |
| Female | 337 | 426 | 2419 | 212 | 3362 |
| (P=0.0002) | (P=0.634) | (P<2.2e-16) | (P=4.54e-08) | (P<2.2e-16) | |
| Age (years) | |||||
| Average | 52.56 | 32.56 | 48.63 | 48.74 | 47.25 |
| SD | 8.14 | 12.49 | 13.37 | 13.51 | 13.9 |
| Range | 35.19-66.09 | 18-65 | 18-93 | 22.0-80.0 | 18-93 |
| (P=2.03e-10) | (P=2.38e-11) | (P=2.02e-11) | (P=0.858) | (P=9.92e-07) | |
| Edu (years) | |||||
| Average | 14.84 | 13.92 | 13.82 | 10.44 | 13.68 |
| SD | 2.69 | 2.48 | 3.35 | 1.25 | 3.23 |
| Range | 0-18 | 9-18 | 2.5-24.5 | 8.0-12.0 | 0-24.5 |
| (P= 2.0e-10) | (P=1.97e-09) | (P=0.780) | (P=0.943) | (P=0.174) | |
MDD = Major Depressive Disorder (lifetime); SD = standard deviation; P-value in parenthesis is for comparison between MDD vs No MDD. T-test was used for comparison of age and education and chi-square test was done to test the association between sex and MDD status.
BiDirect study
BiDirect includes three different cohorts. The first cohort is comprised of individuals with a current episode of MDD at the time of recruitment, the second cohort consists of individuals with cardiovascular disease, and the third cohort is a reference cohort that was randomly sampled from the population18. Cognitive tests in the study assess executive function, processing speed, immediate memory, and delayed memory – with complete data available for close to 1600 participants.
FOR2107 cohort
The FOR2107 consortium investigates MDD, as well as Bipolar Disorder (BD), Schizoaffective Disorder, and Schizophrenia (SCZ)19. In addition to study participants meeting criteria for these disorders, the cohort includes participants at risk, as well as healthy controls, with a total of 2500 individuals19. Healthy controls are those without genetic risk (no relatives with MDD or BD) or environmental risk (no Childhood Trauma Questionnaire subscales meeting the maltreatment threshold)19. Executive function, processing speed, immediate and delayed memory are all measured in this cohort.
Generation Scotland cohort
Generation Scotland: Scottish Family Health Study (GS: SFHS) is a large community, family based study, with close to 24000 participants 20. The wide range of clinical information includes medical history, family history, as well as phenotypes of personality traits and mental health20. Cognitive tests measure processing speed, executive function, immediate and delayed memory.
SHIP Trend cohort
The Study of Health in Pomerania consists of two population-based independent cohorts (SHIP and SHIP-TREND)21. The SHIP-TREND study is the baseline examination of the second SHIP cohort, with data collected from 2008 to 201121. Complete data was available for 602 participants from SHIP Trend. Executive function and verbal episodic memory are assessed in the cohort.
Genome-wide association analysis
We performed the GWAS using SNP by MDD status interaction analysis based on three statistical tests, but summarised the main finding using the joint test of SNP and SNP by MDD interaction effect (2 degrees of freedom (2 df) test). This has been shown to be more powerful in detecting SNPs than either the marginal SNP or the pure SNP by MDD interaction test alone22. For each cohort and for each cognitive domain score, three genome-wide association tests, the marginal SNP effect, pure interaction effect of SNP with MDD status (SNPxMDD) and a joint test of both SNP and SNP by MDD status were performed using the GxEscan23 software. To account for confounding and population stratification issues, an additional set of covariates such as age, sex, total years of education and the first ten principal components were used in the regression models. Meta-analysis methods were used to combine each of the three GWAS results across the cohorts. Quality controlled GWAS results were meta-analysed for the marginal SNP effects and the interaction effect (SNPxMDD) using the METAL package24. Meta p-values for the joint effect were obtained using the sample size weighted linear combination of the joint effect 2df chi-square statistics25. The results were summarised based on the meta-analysis p-values of the joint test of SNP and SNP x MDD status (2 df) tests. The list of SNPs reported by the 2 df tests are either associated with cognitive function and/or differentially associated between the MDD subgroups. In other words, identified SNPs are associated with cognitive function domains, while also being moderated by MDD status. The GWAS p-value threshold was set at p<=5e-8, unadjusted for the number of traits as these traits are correlated.
The gene-based GWAS of the joint SNP and SNP x MDD (2df tests) were performed using MAGMA26 and the polygenic risk scores were generated using PRS-CS27 software. Regression analyses of the combined PRS score with relevant covariates were performed using R package v4.0.028. Additional details of the statistical analyses are provided in the supplementary material.
Functional analyses of GWAS findings
We conducted functional analyses of our GWAS findings using Qiagen’s Ingenuity Pathways Analysis software (IPA®, QIAGEN Redwood City, CA, USA, www.qiagen.com/ingenuity). Lists of genes for IPA® input were prepared using results from the genome-wide 2df tests and SNPxMDD interaction tests for all cognitive domains. For intergenic SNPs, the closest gene was added to the list. The input to IPA was an unranked list of these genes. IPA compares the proportion of input genes mapping to a biological pathway to the reference genes list in the ingenuity databases. The significance of the overrepresented canonical pathways and functional networks is determined using the right-tailed Fisher’s exact test and later adjusted for multiple testing using the Benjamini-Hochberg (BH) method. Significant results are determined at BH adjusted p-value <0.01.
Results
Study cohorts
A description of the study cohorts is provided in Table 1. Across all cohorts, there were a total of 9567 study participants. In all cohorts, there is a higher percentage of females (Table 1). Average age of participants is highest in the BiDirect cohort, with a similar average age in the BiDirect, Generation Scotland, and SHIP-Trend cohorts (51.1, 47.9, and 48.8 years respectively). The average age of the FOR2107 study was considerably lower at 34.8 years. The largest age range is in Generation Scotland (18 – 93 years), however only 58 participants were aged over 75 years in this cohort. Only 8 participants in SHIP-Trend were over the age of 75 years. Years of education were similar in the BiDirect, Generation Scotland, and FOR2107 groups (average 14.3, 13.8, and 13.5 years respectively).
Ratio of cases to controls was highest in the BiDirect cohort, with 912 cases (58.7% of participants) and 642 controls (Table 1). In addition, 801 (51.5%) of participants in this cohort had Center for Epidemiological Studies-Depression (CES-D)29 scores ≥ 16, a cut-off score used to indicate those at risk of depression. Of participants in the FOR2107 cohort with lifetime MDD, 13.5% and 2.3% had Hamilton Depression Rating Scale (HAM-D)30 scores in the moderate and severe range respectively. Both Generation Scotland (see suppl. Methods for case-control selection for this analysis) and SHIP-TREND are population samples, hence these cohorts have not specifically targeted MDD (and have a lower lifetime MDD case to control ratio, with 30.5% and 24.6% of study participants respectively meeting MDD criteria). There is significant difference between lifetime MDD cases and controls in age across the cohorts except in SHIP-Trend; in sex ratio except in FOR2107; and in education in BiDirect and FOR2107 (Table1).
Age, sex and education are significantly associated with cognitive domain scores across all the cohorts (Supplementary Table 2i). Associations of MDD status and severity of MDD with cognitive domains are not consistent across all the cohorts (Supplementary Tables 2i and 2j). The individual mean cognitive scores differ significantly across the cohorts (Supplementary Table 2k).
Genome-wide association analyses
i. GWAS of cognitive domains associated with MDD
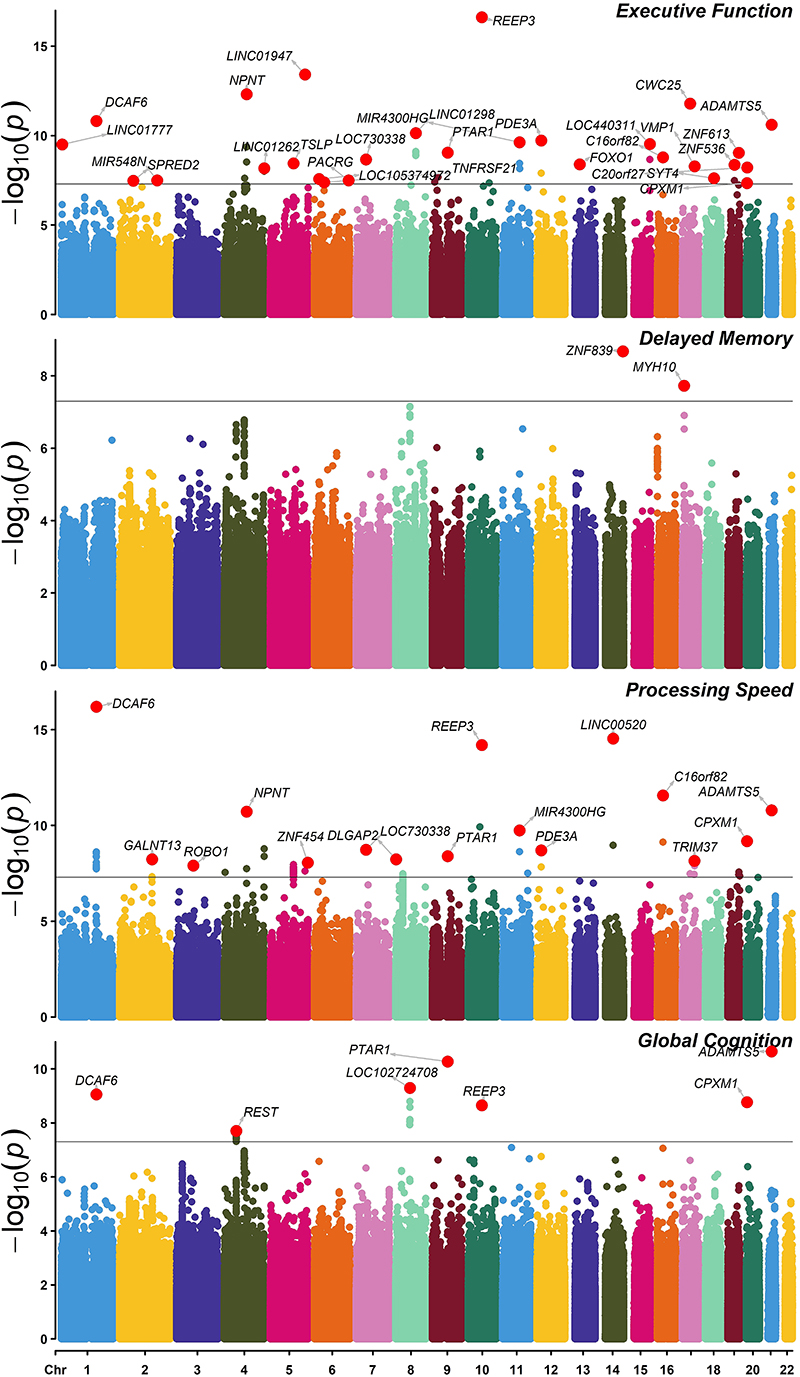
Manhattan plot of the p-values from the joint test of SNP and SNP x MDD interaction terms are presented in Figure 1. Presented SNPs are associated with cognitive function domains and/or also moderated by MDD status. The QQ plots are provided in Supplementary Figure 2. The marginal SNP, SNPxMDD and gene-based association results are provided in the supplementary text.
Figure 1. Manhattan plot for GWAS of SNP and SNP x MDD with cognitive domains.
Joint test of SNP and SNP x MDD interaction with cognitive domains. GWAS significant (p <= 5x10-8) loci are highlighted with the gene name closest to the top SNP. Identified SNPs are associated with cognitive function domains and/or moderated by MDD status.
The domain of executive function showed significant association with 48 SNPs. This included the SNP rs188552424 in TNFRSF21, a gene which has a role in the negative regulation of oligodendrocyte maturation31, and rs112979588 in DCAF6, a gene thought to be involved in stability of the neuromuscular junction32. Individual SNPs in TSLP (a gene involved in immune function33), REEP3 (involved in microtubule binding34, 35), and 2 SNPs in PDE3A (a gene implicated in cerebral endothelial dysfunction36) were also associated with executive function (Supplementary Table 2a).
In the domain of delayed memory, the SNPs rs117823280 (near ZNF839) and rs117688348 (near MYH10) were found to be significantly associated (Supplementary Table 2b). A point mutation of the MYH10 gene in mice is involved in developmental cardiac and brain defects37.
With processing speed 116 SNPs were found to be GWAS significant (Supplementary Table 2c). These included the SNPs in DCAF6, REEP3, and PDE3A associated with executive function, plus rs72635025 in ADAMTS5 (involved in regulation of reelin - an important protein for cortical development38), and rs114216628 in ROBO1 (a gene involved in axon guidance39).
Thirty-two SNPs were significantly associated with global cognition (Supplementary Table 2d). Several of these SNPs were also associated in the domains of executive function and processing speed (SNP rs139747326 in PTAR1, rs148528269 in REEP3, rs112979588 in DCAF6, rs117658905 in CPXM1, and rs72635025 in ADAMTS5). No SNPs reached genome-wide significance for the domain of immediate memory (Supplementary Figure 3).
ii. Polygenic risk scores analyses
Association of PRSes of BD, MDD, SCZ, and mood instability (MIN) with cognitive domains were examined. PRS of SCZ and MIN were significantly associated with all the cognitive domains in our sample. BD PRS was associated with four cognitive domains (delayed memory, immediate memory, processing speed, global cognition), but not with executive function. MDD PRS had significant association with the cognitive domains of processing speed, immediate memory, and global cognition, but the effect was not in a consistent direction (Supplementary Table 2g, marginal model). Interestingly, none of the PRS had significant interaction with MDD status except the PRS of MDD. To understand the change in effect sizes, we have also examined the meta-PRS (MET3) of the three summary GWAS statistics (MDD, MIN, SCZ) with the cognitive domains. Although the MET3 is significantly associated with all the cognitive domains in consistent directions, the effect sizes are not bigger than the individual PRSes (Supplementary Table 2g, marginal).
Functional analyses
i. Gene expression analysis of significant genes
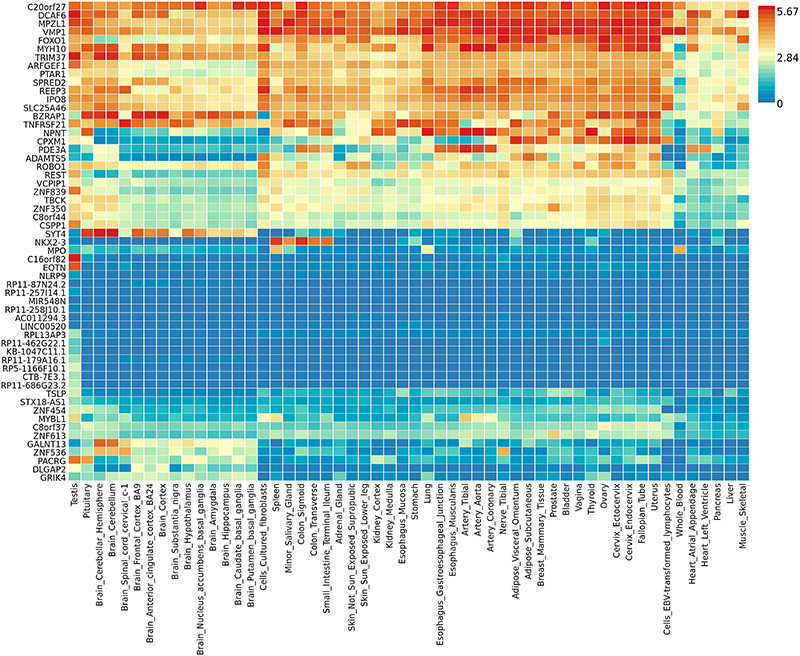
Expression pattern and tissue specific enrichment of the significantly associated genes (corresponding to the associated SNPs) across all cognitive domains were examined using the gene2func module of the FUMA software40 using the GTEx (https://gtexportal.org/home/) gene expression data. Tissue specific gene expression is displayed in Figure 2. A number of genes are expressed in brain, in particular the amygdala (TNFRSF21, DCAF6), anterior cingulate cortex (TNFRSF21), basal ganglia (MYH10, DCAF6), frontal cortex (TNFRSF21, DCAF6, VMP1), hippocampus (REEP3), hypothalamus (TNFRSF21, REEP3) and cerebellum (REEP3, TNFRSF21, DCAF6, VMP1, PTAR1) – but also in other body tissues. No tissue specific enrichment tests were found to be significant (Supplementary Figure 4; Supplementary Table 2h).
Figure 2. Tissue specific expression of top genes (p < 5.0 x 10-8) associated with cognitive function across all cognitive domains.
Tissue types are on the x-axis and gene symbols are on the y-axis. Scale bar on the right gives colour coding and level of gene expression.
ii. IPA® - Functional analyses of genes associated with cognitive phenotypes
We used IPA® to map genes implicated in the GWAS analysis to the service’s proprietary knowledge databases, which include canonical pathways, functional gene networks, upstream regulators, causal networks, diseases and bio-functions, toxicology functions, and toxicity lists. Detailed IPA® results for a summary gene list for all cognitive domains are provided in [Supplementary tables 3]. Overall, there is a relatively small number of genes that drive the IPA®-associations with various functional categories (canonical pathways, diseases and biofunctions etc.), including MPO, FOXO1, PDE3A, TSLP, NLRP9, ADAMTS5, ROBO1 and REST.
MPO was the dataset gene in the IPA® top canonical pathway, melatonin degradation, for the combined domains analysis and for processing speed, while FOXO1 and PDE3A drove the top canonical pathway for executive function (leptin signalling in obesity)[Supplementary tables 3].
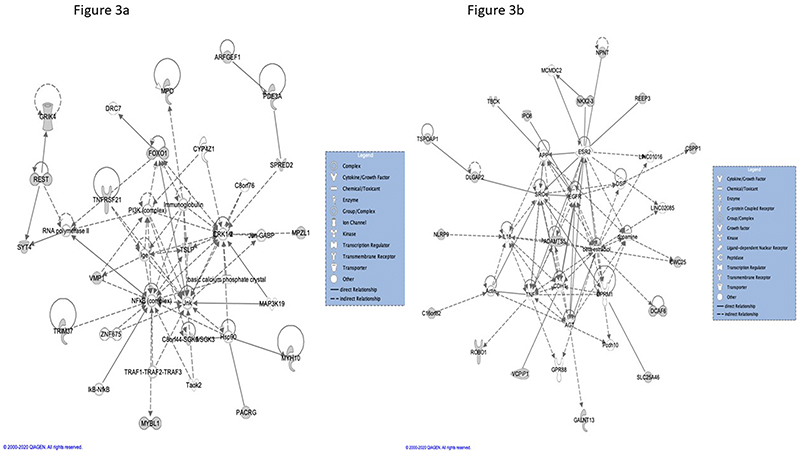
When all dataset genes where analysed, the two top IPA®-defined functional interaction networks implicated TSLP (network 1) and ADAMTS5, the latter together with beta-estradiol (network 2), as central functional nodes (Figure 3a and b). For executive function, the top interaction network centrally implicated the dataset gene NPNT, as well as the estrogen receptor 2 (ESR2), androgen receptor (AR), tumor protein 53 (TP 53), and amyloid precursor protein (APP). For processing speed, central connectivity was shown for the dataset gene VMP1 together with TP53, TGFB1, HNF4A, and the NFkB complex [Supplementary tables 3].
Figure 3. IPA® – functional networks 1 & 2 for all cognition-associated genes.
Further, IPA® identified upstream regulators and causal networks with associations to dataset genes. Amongst the top-listed molecules, the vitamin D receptor (VDR), beta-estradiol, the phosphodiesterase inhibitor tadalafil, and the protein kinase C inhibitor Go 6976 impact on several dataset genes and could therefore be of particular translational interest [Supplementary tables 3].
Discussion
We conducted a genome-wide interaction analysis of MDD with cognitive function in the BiDirect, FOR2107, Generation Scotland, and SHIP Trend cohorts. We observed a set of SNPs to be specifically associated with cognitive function, in the context of MDD. In other words, these SNPs became GWAS significant in the joint test of SNP and SNPxMDD, but were not marginally significant when the MDD status was not included in the analysis. The joint tests of SNP and SNPxMDD have improved power to find SNPs/ genes which would have been missing by the routine GWAS (our marginal test) because it looks for average effect among MDD vs non-MDD samples. Hence, MDD status has demonstrated a moderating effect on the association of these SNPs with cognitive domains.
Significant SNPs from our GWAS were from various genes including LINC00520 (observed to promote tumour processes in glioma cells41), CPXM1 (also known as CPX-142, involved in adipogenesis43), VMP1 (thought to be important in releasing lipoproteins from the endoplasmic reticulum membrane44), and REEP3 (involved in microtubule binding34, 35 and possibly synaptic plasticity34). REEP3 is also involved in neural pathways linked to obsessive-compulsive disorder45 and has been proposed as a positional candidate gene for autism spectrum disorder46.
A number of significant SNPs were located in genes involved in negative regulation of oligodendrocyte maturation (TNFRSF21)31, axon guidance (ROBO1)39, and myelination (ARFGEF1)47. It is also notable that genes such as REEP3, TNFRSF21, and ARFGEF1 are all expressed in brain (Figure 2). Furthermore, SNPs from REEP3 and DCAF6 (also expressed in brain areas including the amygdala, basal ganglia, and frontal cortex) were specifically associated with multiple cognitive domains.
Several significant SNPs for global cognition are located in the REST gene (Supplementary Table 2d), which as a transcription repressor has an important role in the development of neurons48, 49, and also in regulating secretion of insulin from pancreatic β-cells49.
The functional analysis using IPA® software highlighted genes mapping to a high number of canonical pathways as well as to various disease- and biofunctions (MPO, FOXO1, PDE3A, TSLP, NLRP9, ADAMTS5, ROBO1 and REST). Several of these have previously been implicated in the neurobiology of cognitive function. For example, myeloperoxidase (MPO) is an enzyme highly expressed by neutrophils and is a primary mediator of neutrophils’ oxidative stress response. Elevated MPO levels have been implicated in the pathogenesis of Alzheimer’s disease, and mice with MPO deficiency were shown to exhibit superior cognitive performance50. Forkhead Box O (FOXO) transcription factor 1 is one of 4 isoforms which have previously been described as ‘guardians of neuronal integrity’ by inhibiting age‐progressive axonal degeneration in mammals through regulation of neuroprotective mechanisms under pro-inflammatory conditions51. In mice, depletion of neuronal FOXO 1, 3, and 4 initiates neurodegeneration and advances brain ageing51. ADAMTS5, at the centre of functional network 2, is a metalloprotease recently shown to play a role in cortical development through interactions with reelin and DISC1 38. Interestingly, a variant of TP53, which is implicated in the top functional networks for executive function and processing speed, has been described as a disease modifier in fronto-temporal dementia52.
The IPA -defined upstream regulator molecules and causal networks may provide a genetic rationale for further clinical evaluation and therapeutic strategies, in the context of MDD. These include beta-estradiol and the estrogen receptor, whose potential for cognitive enhancement has been demonstrated in a wide range of preclinical and clinical studies (for overview see Hamson et al.53). Signalling through the vitamin D receptor (VDR), another IPA® upstream regulator, has been proposed as a strategy for cognitive enhancement54 but has not been tested in depressed populations. Taurine supplementation has been observed to reduce MPO levels and boost the effects of exercise on cognition in women > 60 years55, but also does not appear to have been investigated as a therapeutic adjunct in MDD. Further, previous clinical and pre-clinical studies have suggested potential benefit of the upstream regulator tadalafil (a 5-phosphodiesterase inhibitor) on cognitive function56–58.
While only the PRS of MDD had significant interaction with MDD status (PRS associated with processing speed and executive function, although the effect was in an inconsistent direction), the PRS of SCZ and MIN were associated with all cognitive domains. The relationship between mood instability and psychiatric disorders has been previously investigated, with mood instability found to have a strong genetic correlation with MDD, and small but significant correlation with SCZ59. More specifically, mood instability and cognitive dysfunction are common in MDD, BD, and SCZ60, 61. These changes in affect regulation and cognitive function seen across diagnoses may relate to areas of the brain such as the prefrontal cortex. Specifically, reduced functional connectivity between the prefrontal cortex and amygdala, brain regions important in emotion regulation62, has been observed in BD63 and SCZ64. The prefrontal cortex is important not only in emotion regulation, but also in planning and other components of executive function65. With regard to MDD, altered functional connectivity has also been observed, with decreased resting state connectivity between prefrontal cortex and amygdala in adolescents, and increased connectivity between the amygdala and hippocampus in adults66.
No tissue specific enrichment tests of genes were significant. We also did not identify identical genome-wide significant SNPs found in previous GWA studies of cognitive function from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium67, the UK Biobank68, in meta-analyses of GWA studies from CHARGE69, or in a meta-analysis combining the UK Biobank, CHARGE, and Cognitive Genomics Consortium (COGENT) samples70. It is possible that biological pathways which may be involved in MDD, including inflammation, are associated with different genetic variants of cognitive traits.
Similarly, in our analyses, none of the genome-wide significant SNPs found in the recent Psychiatric Genomics Consortium GWA meta-analysis in MDD, which identified 44 significant loci71 or from the 23andMe MDD discovery data set72 were identified in the context of MDD and cognitive function. Reasons for this could extend beyond the smaller sample size of our cohorts, to include age, as well as MDD severity - with different genetic variants contributing to cognitive dysfunction during (compared with in between) depressive episodes.
There are strengths and limitations of our study. Strengths of this study include the number of cognitive tests performed and the coverage of a broad range of cognitive domains, covering multiple domains (for example the BiDirect and FOR2107 cohorts are rich in phenotypes, and assess MDD in a clinical sample). In addition, we conducted functional analyses of the genes associated with cognitive function, which we believe adds to the understanding of the neurobiology of cognitive dysfunction in MDD. Several limitations need to be considered when interpreting the results. First, the total sample size is relatively modest (particularly in comparison to the CHARGE Consortium, COGENT, and UK Biobank – which are all population studies). Hence, replications in other independent cohorts are important especially for those SNPs with low minor allele frequency (MAF). Second, although our GWAS covered a broad range of cognitive domains relevant to MDD, not all cohorts from our study contributed to the cognitive domains in the same way; hence, depending on the availability of individual tests in each cohort, different individual measures were used for a particular cognitive domain within the cohorts. Therefore, to address the heterogeneity of cognitive tests and best represent the relevant cognitive domain, we calculated z scores for each domain that was assessed by more than a single cognitive test. Any impact of this heterogeneity will therefore be more in a cohort where more than one cognitive measure was used within an individual domain. Third, cohorts included a mix of patients, with some tested during a major depressive episode, and some tested during remission. Hence, there may be SNPs associated with an acute episode of severe MDD and cognitive dysfunction that are different to those associated with persistent cognitive dysfunction following a major depressive episode. Fourth, the clinical and cognitive measures were obtained at a single time point only, hence the presented results are related to a trait of cognitive dysfunction rather than to changes in cognitive function over time. Fifth, some age-related impact on cognition in cohorts with participants over 75 years is possible, however, only a small number of participants were of this age. Sixth, databases for functional analysis such as IPA® are not biologically complete, and CNS processes are typically not as well covered as processes that can be studied in peripheral tissues such as blood. Therefore, it is possible that our functional analysis was unable to detect additional important pathways directly relevant to brain function.
Conclusions
We find a set of SNPs to be specifically associated with cognitive function, in the context of MDD. Many of these SNPs are expressed in brain, and functional analysis of the results point to central physiological processes involved in neuronal development, neuroprotection, and maintenance of optimal cognition, thereby offering putative therapeutic targets. Potentially this cognitive phenotype - if confirmed in future analyses - represents a subgroup in MDD, with unique biological characteristics.
Supplementary Material
Acknowledgements
Generation Scotland received core support from the Chief Scientist Office of the Scottish Government Health Directorates [CZD/16/6] and the Scottish Funding Council [HR03006] and is currently supported by the Wellcome Trust [216767/Z/19/Z]. Genotyping of the GS:SFHS samples was carried out by the Genetics Core Laboratory at the Edinburgh Clinical Research Facility, University of Edinburgh, Scotland and was funded by the Medical Research Council UK and the Wellcome Trust (Wellcome Trust Strategic Award “STratifying Resilience and Depression Longitudinally” (STRADL) Reference 104036/Z/14/Z). SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs and the Social Ministry of the Federal State of Mecklenburg-West Pomerania. This study was further supported by the EU-JPND Funding for BRIDGET (FKZ:01ED1615).
Footnotes
Conflicts of Interest
HJG has received travel grants and speaker honoraria from Fresenius Medical Care, Neuraxpharm, Servier and Janssen Cilag as well as research funding from Fresenius Medical Care. KOS reports having received lecture fees and research funding from Janssen Australia, research funding from Lundbeck Otsuka, and research funding from Gilead. BTB reports having received speaker and consultation fees from AstraZeneca, Lundbeck, Pfizer, Takeda, Servier, Bristol Myers Squibb, Otsuka, LivaNova and Janssen-Cilag.
Author Contributions
KB (Klaus Berger), HM, DG, UD, SM, NO, JR, MG, IN, FS, KB (Katharina Brosch), TM, JP, AJF, PH, MMN, SW, MR, TK, MA, AMM, DJP, IJD, CH, AC, HJG, AT (Alexander Teumer), GH, SV contributed clinical and GWAS data, commented on results, and reviewed and approved the final manuscript. BTB conceived the study. AT (Anbupalam Thalamuthu), KOS, NTM, and BTB developed the design and analysis plan. AT (Anbupalam Thalamuthu) and KOS conducted the analyses. AT (Anbupalam Thalamuthu), KOS, NTM, and BTB interpreted the results, wrote and edited the manuscript. All authors approved the final manuscript.
References
- 1.Whiteford HA, Degenhardt L, Rehm J, Baxter AJ, Ferrari AJ, Erskine HE, et al. Global burden of disease attributable to mental and substance use disorders: findings from the Global Burden of Disease Study 2010. Lancet. 2013;382(9904):1575–1586. doi: 10.1016/S0140-6736(13)61611-6. [DOI] [PubMed] [Google Scholar]
- 2.Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1789–1858. doi: 10.1016/S0140-6736(18)32279-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butters MA, Whyte EM, Nebes RD, Begley AE, Dew MA, Mulsant BH, et al. The nature and determinants of neuropsychological functioning in late-life depression. Arch Gen Psychiatry. 2004;61(6):587–595. doi: 10.1001/archpsyc.61.6.587. [DOI] [PubMed] [Google Scholar]
- 4.Beblo T, Sinnamon G, Baune BT. Specifying the Neuropsychology of Affective Disorders: Clinical, Demographic and Neurobiological Factors. Neuropsychol Rev. 2011;21(4):337–359. doi: 10.1007/s11065-011-9171-0. [DOI] [PubMed] [Google Scholar]
- 5.Rock PL, Roiser JP, Riedel WJ, Blackwell AD. Cognitive impairment in depression: a systematic review and meta-analysis. Psychol Med. 2014;44(10):2029–2040. doi: 10.1017/S0033291713002535. [DOI] [PubMed] [Google Scholar]
- 6.Baune BT, Miller R, McAfoose J, Johnson M, Quirk F, Mitchell D. The role of cognitive impairment in general functioning in major depression. Psychiatry Res. 2010;176:183–189. doi: 10.1016/j.psychres.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Davis MT, DellaGioia N, Matuskey D, Harel B, Maruff P, Pietrzak RH, et al. Preliminary evidence concerning the pattern and magnitude of cognitive dysfunction in major depressive disorder using cogstate measures. J Affect Disord. 2017;218:82–85. doi: 10.1016/j.jad.2017.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shiroma PR, Albott CS, Johns B, Thuras P, Wels J, Lim KO. Neurocognitive performance and serial intravenous subanesthetic ketamine in treatment-resistant depression. Int J Neuropsychopharmacol. 2014;17(11):1805–1813. doi: 10.1017/S1461145714001011. [DOI] [PubMed] [Google Scholar]
- 9.Jaeger J, Berns S, Uzelac S, Davis-Conway S. Neurocognitive deficits and disability in major depressive disorder. Psychiatry Res. 2006;145(1):39–48. doi: 10.1016/j.psychres.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 10.McDermott LM, Ebmeier KP. A meta-analysis of depression severity and cognitive function. J Affect Disord. 2009;119:1–8. doi: 10.1016/j.jad.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 11.Mandelli L, Serretti A, Colombo C, Florita M, Santoro A, Rossini D, et al. Improvement of cognitive functioning in mood disorder patients with depressive symptomatic recovery during treatment: an exploratory analysis. Psychiatry Clin Neurosci. 2006;60(5):598–604. doi: 10.1111/j.1440-1819.2006.01564.x. [DOI] [PubMed] [Google Scholar]
- 12.Withall A, Harris LM, Cumming SR. The relationship between cognitive function and clinical and functional outcomes in major depressive disorder. Psychol Med. 2009;39(3):393–402. doi: 10.1017/S0033291708003620. [DOI] [PubMed] [Google Scholar]
- 13.Knight MJ, Mills NT, Baune BT. Contemporary methods of improving cognitive dysfunction in clinical depression. Expert Rev Neurother. 2019;19(5):431–443. doi: 10.1080/14737175.2019.1610395. [DOI] [PubMed] [Google Scholar]
- 14.Trampush JW, Yang ML, Yu J, Knowles E, Davies G, Liewald DC, et al. GWAS meta-analysis reveals novel loci and genetic correlates for general cognitive function: a report from the COGENT consortium. Mol Psychiatry. 2017;22(3):336–345. doi: 10.1038/mp.2016.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagenaars SP, Harris SE, Davies G, Hill WD, Liewald DCM, Ritchie SJ, et al. Shared genetic aetiology between cognitive functions and physical and mental health in UK Biobank (N=112151) and 24 GWAS consortia. Mol Psychiatry. 2016;21(11):1624–1632. doi: 10.1038/mp.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuksel D, Dietsche B, Forstner AJ, Witt SH, Maier R, Rietschel M, et al. Polygenic risk for depression and the neural correlates of working memory in healthy subjects. Prog Neuropsychopharmacol Biol Psychiatry. 2017;79:67–76. doi: 10.1016/j.pnpbp.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 17.Meijsen JJ, Campbell A, Hayward C, Porteous DJ, Deary IJ, Marioni RE, et al. Phenotypic and genetic analysis of cognitive performance in Major Depressive Disorder in the Generation Scotland: Scottish Family Health Study. Transl Psychiatry. 2018;8(1):63. doi: 10.1038/s41398-018-0111-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teismann H, Wersching H, Nagel M, Arolt V, Heindel W, Baune BT, et al. Establishing the bidirectional relationship between depression and subclinical arteriosclerosis - rationale, design, and characteristics of the BiDirect Study. BMC Psychiatry. 2014;14:174. doi: 10.1186/1471-244X-14-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kircher T, Wohr M, Nenadic I, Schwarting R, Schratt G, Alferink J, et al. Neurobiology of the major psychoses: a translational perspective on brain structure and function-the FOR2107 consortium. Eur Arch Psychiatry Clin Neurosci. 2019;269(8):949–962. doi: 10.1007/s00406-018-0943-x. [DOI] [PubMed] [Google Scholar]
- 20.Smith BH, Campbell A, Linksted P, Fitzpatrick B, Jackson C, Kerr SM, et al. Cohort Profile: Generation Scotland: Scottish Family Health Study (GS:SFHS). The study, its participants and their potential for genetic research on health and illness. Int J Epidemiol. 2013;42(3):689–700. doi: 10.1093/ije/dys084. [DOI] [PubMed] [Google Scholar]
- 21.Volzke H, Alte D, Schmidt CO, Radke D, Lorbeer R, Friedrich N, et al. Cohort Profile: The Study of Health in Pomerania. Int J Epidemiol. 2011;40(2):294–307. doi: 10.1093/ije/dyp394. [DOI] [PubMed] [Google Scholar]
- 22.Kraft P, Yen YC, Stram DO, Morrison J, Gauderman WJ. Exploiting gene-environment interaction to detect genetic associations. Hum Hered. 2007;63(2):111–119. doi: 10.1159/000099183. [DOI] [PubMed] [Google Scholar]
- 23.Zhang P, Lewinger JP, Conti D, Morrison JL, Gauderman WJ. Detecting Gene-Environment Interactions for a Quantitative Trait in a Genome-Wide Association Study. Genet Epidemiol. 2016;40(5):394–403. doi: 10.1002/gepi.21977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duchesne P, De Micheaux PL. Computing the distribution of quadratic forms: Further comparisons between the Liu-Tang-Zhang approximation and exact methods. Comput Stat Data Anal. 2010;54(4):858–862. [Google Scholar]
- 26.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11(4):e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ge T, Chen CY, Ni Y, Feng YA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10(1):1776. doi: 10.1038/s41467-019-09718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2020. [Google Scholar]
- 29.Radloff L. The CES-D scale: a self-report depression scale for research in the general population. Appl Psychol Meas. 1977;1:385–401. [Google Scholar]
- 30.Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 1960;23(1):56–62. doi: 10.1136/jnnp.23.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mi S, Lee X, Hu Y, Ji B, Shao Z, Yang W, et al. Death receptor 6 negatively regulates oligodendrocyte survival, maturation and myelination. Nat Med. 2011;17(7):816–821. doi: 10.1038/nm.2373. [DOI] [PubMed] [Google Scholar]
- 32.Chen HH, Tsai LK, Liao KY, Wu TC, Huang YH, Huang YC, et al. Muscle-restricted nuclear receptor interaction protein knockout causes motor neuron degeneration through down-regulation of myogenin at the neuromuscular junction. J Cachexia Sarcopenia Muscle. 2018;9(4):771–785. doi: 10.1002/jcsm.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito T, Liu YJ, Arima K. Cellular and molecular mechanisms of TSLP function in human allergic disorders--TSLP programs the “Th2 code” in dendritic cells. Allergol Int. 2012;61(1):35–43. doi: 10.2332/allergolint.11-RAI-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blackstone C, O’Kane CJ, Reid E. Hereditary spastic paraplegias: membrane traffic and the motor pathway. Nat Rev Neurosci. 2011;12(1):31–42. doi: 10.1038/nrn2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park SH, Zhu PP, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest. 2010;120(4):1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yasmeen S, Kaur S, Mirza AH, Brodin B, Pociot F, Kruuse C. miRNA-27a-3p and miRNA-222-3p as Novel Modulators of Phosphodiesterase 3a (PDE3A) in Cerebral Microvascular Endothelial Cells. Mol Neurobiol. 2019;56(8):5304–5314. doi: 10.1007/s12035-018-1446-5. [DOI] [PubMed] [Google Scholar]
- 37.Ridge LA, Mitchell K, Al-Anbaki A, Shaikh Qureshi WM, Stephen LA, Tenin G, et al. Non-muscle myosin IIB (Myh10) is required for epicardial function and coronary vessel formation during mammalian development. PLoS Genet. 2017;13(10):e1007068. doi: 10.1371/journal.pgen.1007068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bradshaw NJ, Trossbach SV, Köber S, Walter S, Prikulis I, Weggen S, et al. Disrupted in Schizophrenia 1 regulates the processing of reelin in the perinatal cortex. Schizophr Res. 2020;215:506–513. doi: 10.1016/j.schres.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Gao Y, Xu X, Shi R, Liu J, Yao W, et al. Slit2/Robo1 promotes synaptogenesis and functional recovery of spinal cord injury. Neuroreport. 2017;28(2):75–81. doi: 10.1097/WNR.0000000000000715. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Yang C, Liu X, Zheng J, Zhang F, Wang D, et al. Transcription factor AP-4 (TFAP4)-upstream ORF coding 66 aa inhibits the malignant behaviors of glioma cells by suppressing the TFAP4/long noncoding RNA 00520/microRNA-520f-3p feedback loop. Cancer Sci. 2020;111(3):891–906. doi: 10.1111/cas.14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar A, Bandapalli OR, Paramasivam N, Giangiobbe S, Diquigiovanni C, Bonora E, et al. Familial Cancer Variant Prioritization Pipeline version 2 (FCVPPv2) applied to a papillary thyroid cancer family. Sci Rep. 2018;8(1):11635. doi: 10.1038/s41598-018-29952-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim YH, Barclay JL, He J, Luo X, O’Neill HM, Keshvari S, et al. Identification of carboxypeptidase X (CPX)-1 as a positive regulator of adipogenesis. FASEB J. 2016;30(7):2528–2540. doi: 10.1096/fj.201500107R. [DOI] [PubMed] [Google Scholar]
- 44.Morishita H, Zhao YG, Tamura N, Nishimura T, Kanda Y, Sakamaki Y, et al. A critical role of VMP1 in lipoprotein secretion. eLife. 2019;8 doi: 10.7554/eLife.48834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noh HJ, Tang R, Flannick J, O’Dushlaine C, Swofford R, Howrigan D, et al. Integrating evolutionary and regulatory information with a multispecies approach implicates genes and pathways in obsessive-compulsive disorder. Nat Commun. 2017;8(1):774. doi: 10.1038/s41467-017-00831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castermans D, Vermeesch JR, Fryns JP, Steyaert JG, Van de Ven WJ, Creemers JW, et al. Identification and characterization of the TRIP8 and REEP3 genes on chromosome 10q21.3 as novel candidate genes for autism. Eur J Hum Genet. 2007;15(4):422–431. doi: 10.1038/sj.ejhg.5201785. [DOI] [PubMed] [Google Scholar]
- 47.Miyamoto Y, Torii T, Tago K, Tanoue A, Takashima S, Yamauchi J. BIG1/Arfgef1 and Arf1 regulate the initiation of myelination by Schwann cells in mice. Sci Adv. 2018;4:eaar4471. doi: 10.1126/sciadv.aar4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia-Manteiga JM, D’Alessandro R, Meldolesi J. News about the Role of the Transcription Factor REST in Neurons: From Physiology to Pathology. Int J Mol Sci. 2019;21(1) doi: 10.3390/ijms21010235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thiel G, Ekici M, Rössler OG. RE-1 silencing transcription factor (REST): a regulator of neuronal development and neuronal/endocrine function. Cell Tissue Res. 2015;359(1):99–109. doi: 10.1007/s00441-014-1963-0. [DOI] [PubMed] [Google Scholar]
- 50.Volkman R, Ben-Zur T, Kahana A, Garty BZ, Offen D. Myeloperoxidase Deficiency Inhibits Cognitive Decline in the 5XFAD Mouse Model of Alzheimer’s Disease. Front Neurosci. 2019;13:990. doi: 10.3389/fnins.2019.00990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hwang I, Oh H, Santo E, Kim DY, Chen JW, Bronson RT, et al. FOXO protects against age-progressive axonal degeneration. Aging Cell. 2018;17(1):e12701. doi: 10.1111/acel.12701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosas I, Martínez C, Coto E, Clarimón J, Lleó A, Illán-Gala I, et al. Genetic variation in APOE, GRN, and TP53 are phenotype modifiers in frontotemporal dementia. Neurobiol Aging. 2021;99:99.e15–99.e22. doi: 10.1016/j.neurobiolaging.2020.08.018. [DOI] [PubMed] [Google Scholar]
- 53.Hamson DK, Roes MM, Galea LA. Sex Hormones and Cognition: Neuroendocrine Influences on Memory and Learning. Compr Physiol. 2016;6(3):1295–1337. doi: 10.1002/cphy.c150031. [DOI] [PubMed] [Google Scholar]
- 54.Goodwill AM, Szoeke C. A Systematic Review and Meta-Analysis of The Effect of Low Vitamin D on Cognition. J Am Geriatr Soc. 2017;65(10):2161–2168. doi: 10.1111/jgs.15012. [DOI] [PubMed] [Google Scholar]
- 55.Chupel MU, Minuzzi LG, Furtado GE, Santos ML, Ferreira JP, Filaire E, et al. Taurine supplementation reduces myeloperoxidase and matrix-metalloproteinase-9 levels and improves the effects of exercise in cognition and physical fitness in older women. Amino acids. 2021;53(3):333–345. doi: 10.1007/s00726-021-02952-6. [DOI] [PubMed] [Google Scholar]
- 56.Urios A, Ordoño F, García-García R, Mangas-Losada A, Leone P, JoséGallego J, et al. Tadalafil Treatment Improves Inflammation, Cognitive Function, And Mismatch Negativity Of Patients With Low Urinary Tract Symptoms And Erectile Dysfunction. Sci Rep. 2019;9(1):17119. doi: 10.1038/s41598-019-53136-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi JB, Cho KJ, Kim JC, Kim CH, Chung YA, Jeong HS, et al. The Effect of Daily Low Dose Tadalafil on Cerebral Perfusion and Cognition in Patients with Erectile Dysfunction and Mild Cognitive Impairment. Clin Psychopharmacol Neurosci. 2019;17(3):432–437. doi: 10.9758/cpn.2019.17.3.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Al-Amin MM, Hasan SM, Alam T, Hasan AT, Hossain I, Didar RR, et al. Tadalafil enhances working memory, and reduces hippocampal oxidative stress in both young and aged mice. Eur J Pharmacol. 2014;745:84–90. doi: 10.1016/j.ejphar.2014.10.026. [DOI] [PubMed] [Google Scholar]
- 59.Ward J, Strawbridge RJ, Bailey MES, Graham N, Ferguson A, Lyall DM, et al. Genome-wide analysis in UK Biobank identifies four loci associated with mood instability and genetic correlation with major depressive disorder, anxiety disorder and schizophrenia. Transl Psychiatry. 2017;7(11):1264. doi: 10.1038/s41398-017-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Broome MR, Saunders KE, Harrison PJ, Marwaha S. Mood instability: significance, definition and measurement. Br J Psychiatry. 2015;207(4):283–285. doi: 10.1192/bjp.bp.114.158543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu Y, Womer FY, Leng H, Chang M, Yin Z, Wei Y, et al. The Relationship Between Cognitive Dysfunction and Symptom Dimensions Across Schizophrenia, Bipolar Disorder, and Major Depressive Disorder. Front Psychiatry. 2019;10:253. doi: 10.3389/fpsyt.2019.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Phillips ML, Ladouceur CD, Drevets WC. A neural model of voluntary and automatic emotion regulation: implications for understanding the pathophysiology and neurodevelopment of bipolar disorder. Mol Psychiatry. 2008;13(9):833–857. doi: 10.1038/mp.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Radaelli D, Sferrazza Papa G, Vai B, Poletti S, Smeraldi E, Colombo C, et al. Fronto-limbic disconnection in bipolar disorder. Eur Psychiatry. 2015;30(1):82–88. doi: 10.1016/j.eurpsy.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 64.Liu H, Tang Y, Womer F, Fan G, Lu T, Driesen N, et al. Differentiating patterns of amygdala-frontal functional connectivity in schizophrenia and bipolar disorder. Schizophr Bull. 2014;40(2):469–477. doi: 10.1093/schbul/sbt044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Logue SF, Gould TJ. The neural and genetic basis of executive function: attention, cognitive flexibility, and response inhibition. Pharmacol Biochem Behav. 2014;123:45–54. doi: 10.1016/j.pbb.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang S, Lu L, Zhang L, Hu X, Bu X, Li H, et al. Abnormal amygdala resting-state functional connectivity in adults and adolescents with major depressive disorder: A comparative meta-analysis. EBioMedicine. 2018;36:436–445. doi: 10.1016/j.ebiom.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ibrahim-Verbaas CA, Bressler J, Debette S, Schuur M, Smith AV, Bis JC, et al. GWAS for executive function and processing speed suggests involvement of the CADM2 gene. Mol Psychiatry. 2016;21(2):189–197. doi: 10.1038/mp.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davies G, Marioni RE, Liewald DC, Hill WD, Hagenaars SP, Harris SE, et al. Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N=112 151) Mol Psychiatry. 2016;21(6):758–767. doi: 10.1038/mp.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davies G, Armstrong N, Bis JC, Bressler J, Chouraki V, Giddaluru S, et al. Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N=53949) Mol Psychiatry. 2015;20(2):183–192. doi: 10.1038/mp.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davies G, Lam M, Harris SE, Trampush JW, Luciano M, Hill WD, et al. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat Commun. 2018;9(1):2098. doi: 10.1038/s41467-018-04362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50(5):668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hyde CL, Nagle MW, Tian C, Chen X, Paciga SA, Wendland JR, et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet. 2016;48(9):1031–1036. doi: 10.1038/ng.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.