Abstract
The Polycomb repressive system plays a fundamental role in controlling gene expression during mammalian development. To achieve this, Polycomb repressive complexes 1 and 2 (PRC1 and PRC2) bind target genes and use histone modification-dependent feedback mechanisms to form Polycomb chromatin domains and repress transcription. The interrelatedness of PRC1 and PRC2 activity at these sites has made it difficult to discover the specific components of Polycomb chromatin domains that drive gene repression and to understand mechanistically how this is achieved. Here, by exploiting rapid degron-based approaches and time-resolved genomics we kinetically dissect Polycomb-mediated repression and discover that PRC1 functions independently of PRC2 to counteract RNA polymerase II binding and transcription initiation. Using single-cell gene expression analysis, we reveal that PRC1 acts uniformly within the cell population, and that repression is achieved by controlling transcriptional burst frequency. These important new discoveries provide a mechanistic and conceptual framework for Polycomb-dependent transcriptional control.
Introduction
At the most basic level gene expression is controlled by DNA-binding transcription factors that shape how RNA polymerase functions at gene promoters. While this principle is shared across all clades of life, eukaryotes have evolved additional gene regulatory mechanisms that utilise chromatin and post-translational modification of histone proteins1,2. However, how chromatin-based systems control transcription to regulate expression remains poorly understood and a major gap in our understanding of gene regulation.
The importance of histone-modifying systems in gene regulation is exemplified by the vertebrate Polycomb repressive system3,4, whose perturbation causes catastrophic mis-regulation of gene expression and embryonic lethality5–8. The Polycomb system is composed of two multi-protein complexes, Polycomb repressive complex 1 (PRC1) and 2 (PRC2). PRC1 is an E3 ubiquitin ligase that monoubiquitylates histone H2A at lysine 119 (H2AK119ub1)9–11 and PRC2 is a histone methyltransferase that can mono-, di-, or trimethylate histone H3 at lysine 27 (H3K27me1/2/3)12–15. PRCs function at silent CpG island-associated gene promoters16–20 where they form Polycomb chromatin domains that are characterised by H2AK119ub1, H3K27me3, and high-level occupancy of Polycomb group proteins21,22. The formation of these domains relies on feedback mechanisms between PRC1 and PRC2. For example, H2AK119ub1 placed by PRC1 is recognised by PRC223–25 which deposits H3K27me3 and recruits more PRC112,26,27. To initiate Polycomb chromatin domain formation, PRC1 and PRC2 utilise DNA-binding activities to engage with, or sample, promoter-associated CpG islands16–20. These activities identify lowly transcribed or inactive genes to enable Polycomb chromatin domain formation which counteracts inappropriate gene expression and maintains normal cell identity3,28.
Despite an emerging view of how Polycomb chromatin domains form, we still have surprisingly little understanding of which components are required for gene repression and how the Polycomb system counteracts transcription. These fundamental questions have been difficult to answer due to the interrelated activities of PRC1 and PRC2 within Polycomb chromatin domains. For example, if one Polycomb complex is perturbed, this disrupts the targeting and activity of the other12,23,29, making it difficult to pinpoint the components of the system that repress transcription. Furthermore, previous approaches used to disrupt Polycomb proteins are slow and asynchronous29–31, making it impossible to study primary effects on RNA polymerase II (Pol II) activity and transcription. Therefore, despite the Polycomb system being a paradigm for chromatin-based gene regulation, we still have little mechanistic understanding of how it represses transcription to control normal gene expression and development.
To uncover the component(s) of the Polycomb system necessary for repression and to discover how it counteracts Pol II-driven transcription, we developed a degron system in mouse embryonic stem cells (ESCs) to remove PRC1 rapidly and synchronously via targeted proteolysis. By combining PRC1 depletion with time-resolved genomic analyses we discover that derepression of Polycomb target genes is remarkably quick and corresponds to active turnover of H2AK119ub1. Derepression occurs in the presence of H3K27me3, does not result from loss of PRC2 occupancy, and relies on new binding of Pol II and transcription initiation. Finally, single-cell analysis revealed that PRC1 counteracts low-level gene expression uniformly throughout the cell population by controlling the frequency of transcription bursts to maintain expression programmes and cell identity.
Results
Depletion of PRC1 reveals a rapid turnover of H2AK119ub1
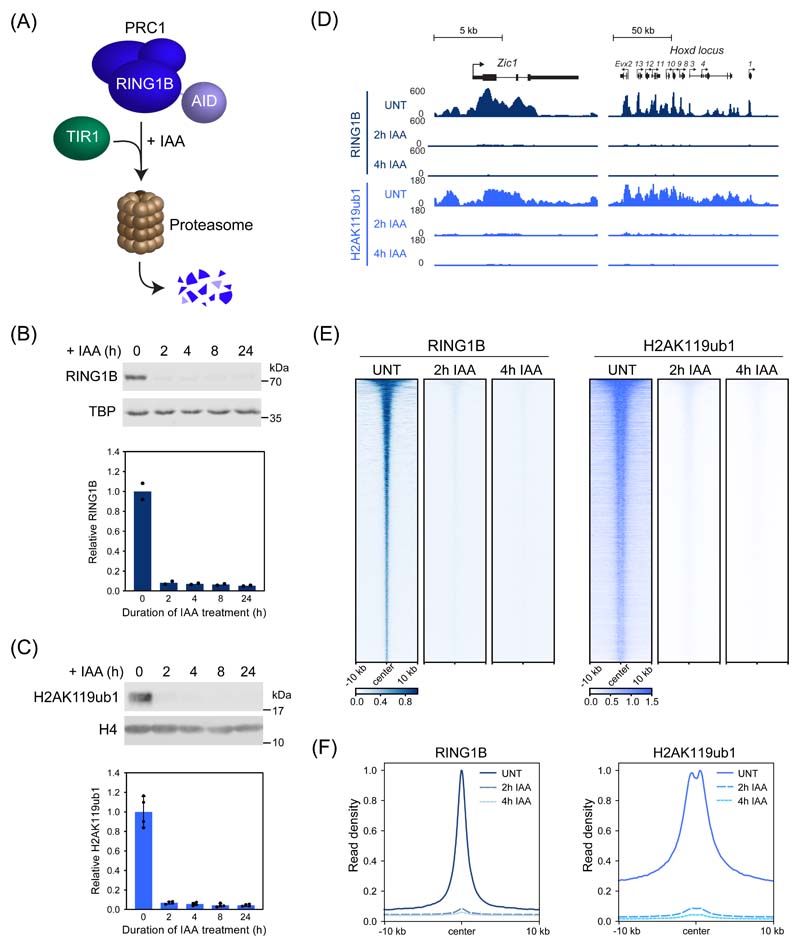
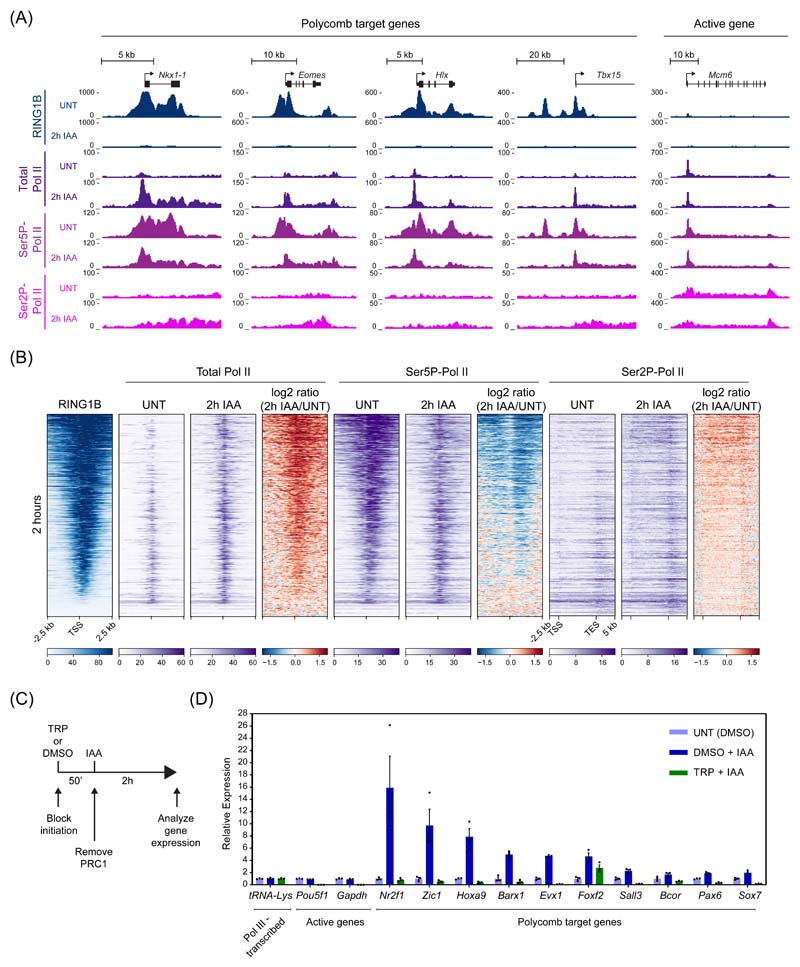
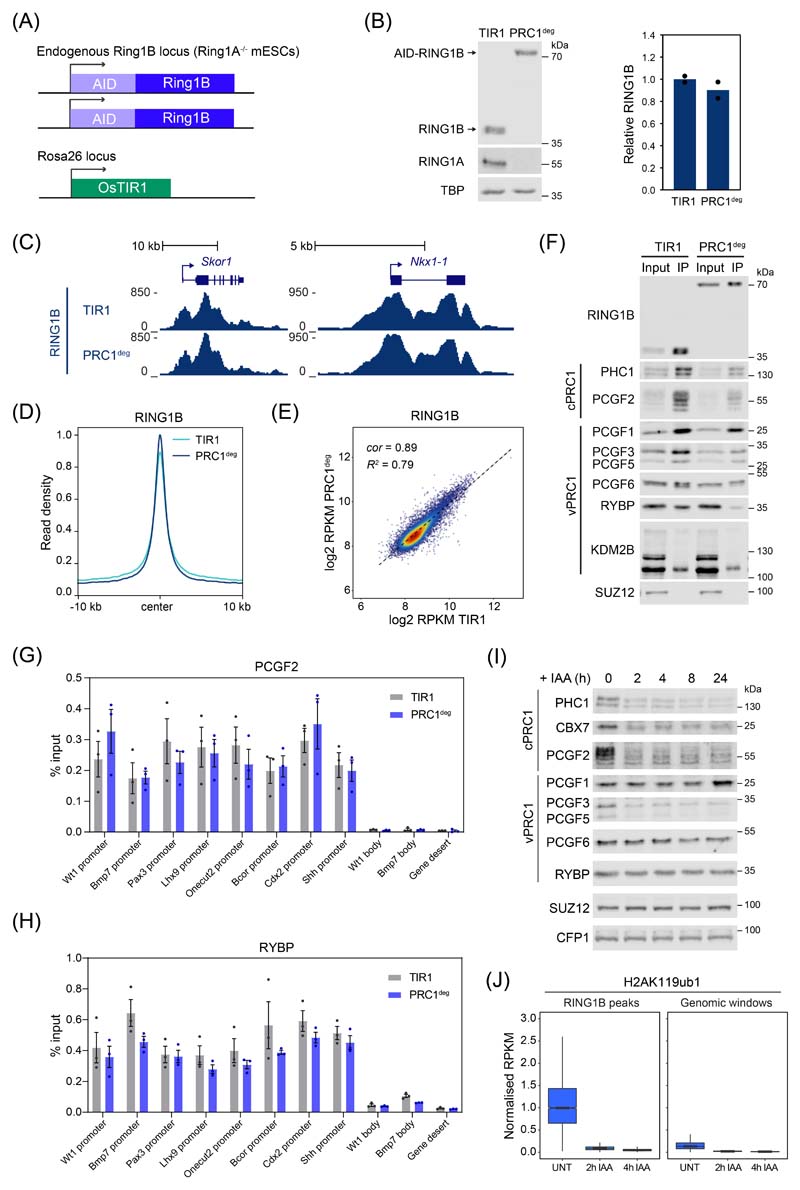
Conditional knockout studies in mouse ESCs have shown that PRC1 is important for Polycomb chromatin domain formation and its removal causes derepression of thousands of Polycomb target genes29–31. However, in these systems loss of PRC1 and Polycomb chromatin domains is slow and asynchronous, making it difficult to pinpoint the components required for gene repression and the primary mechanisms of Polycomb-mediated transcription inhibition. To overcome these limitations, we harnessed inducible proteolysis via an auxin-inducible degron (AID)32,33 to rapidly deplete PRC1 (PRC1deg) through removing its core structural and catalytic subunit, RING1B (Fig. 1A and Extended Data Fig. 1A)34. Endogenously AID-tagged RING1B is expressed at wild-type levels (Extended Data Fig. 1B), binds chromatin indistinguishably to untagged RING1B (Extended Data Fig. 1C-E), forms PRC1 complexes (Extended Data Fig. 1F-H), and is degraded within 2 hours of auxin treatment (Fig. 1B). In agreement with the requirement for RING1B in PRC1 complex formation30,35,36, its depletion caused a reduction in the levels of other PRC1 components (Extended Data Fig. 1I).
Figure 1. Acute depletion of PRC1 reveals a rapid turnover of H2AK119ub1.
(A) A schematic illustrating the PRC1deg system. Addition of auxin (IAA) induces proteasomal degradation of AID-RING1B.
(B) Western blot analysis (upper panel) and quantification (lower panel) of RING1B in PRC1deg cells treated with IAA for the indicated times. Shown are values and mean from n=2 independent experiments.
(C) Western blot analysis (upper panel) and quantification (lower panel) of H2AK119ub1 in PRC1deg cells treated with IAA for the indicated times. Shown are values and mean ± SD from n=4 independent experiments.
(D) Genomic snapshots of typical Polycomb target genes, showing cChIP-seq signal for RING1B and H2AK119ub1 in PRC1deg cells treated with IAA for the indicated times.
(E) Heatmap analysis of RING1B (left) and H2AK119ub1 (right) cChIP-seq at RING1B-bound sites in PRC1deg cells treated with IAA for the indicated times. Heatmaps were sorted by RING1B signal in untreated cells.
(F) Metaplot analysis of data shown in (E). Maximal read density in untreated cells was set to 1.
Using this system, we first set out to determine how removal of PRC1 affects H2AK119ub1. Western blotting revealed that H2AK119ub1 was barely detectable following 2 hours of auxin treatment (Fig. 1C) and calibrated chromatin immunoprecipitation coupled to massively parallel sequencing (cChIP-seq) demonstrated that RING1B and H2AK119ub1 were lost from PRC1 target sites and throughout the genome (Fig. 1D-F and Extended Data Fig. 1J). This indicates that H2AK119ub1 is rapidly turned over and dynamically regulated. Importantly, the rapid kinetics of this effect provided a new opportunity to dissect how PRC1 controls gene expression and to examine which Polycomb chromatin domain components enable gene repression.
Rapid target gene derepression following PRC1 removal
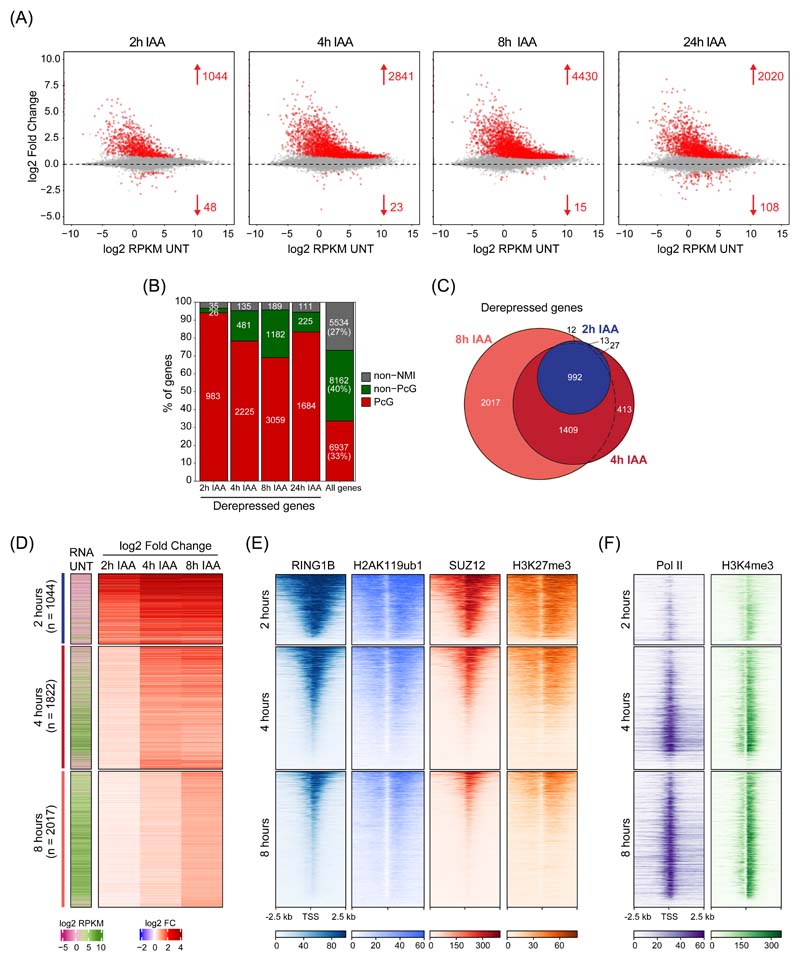
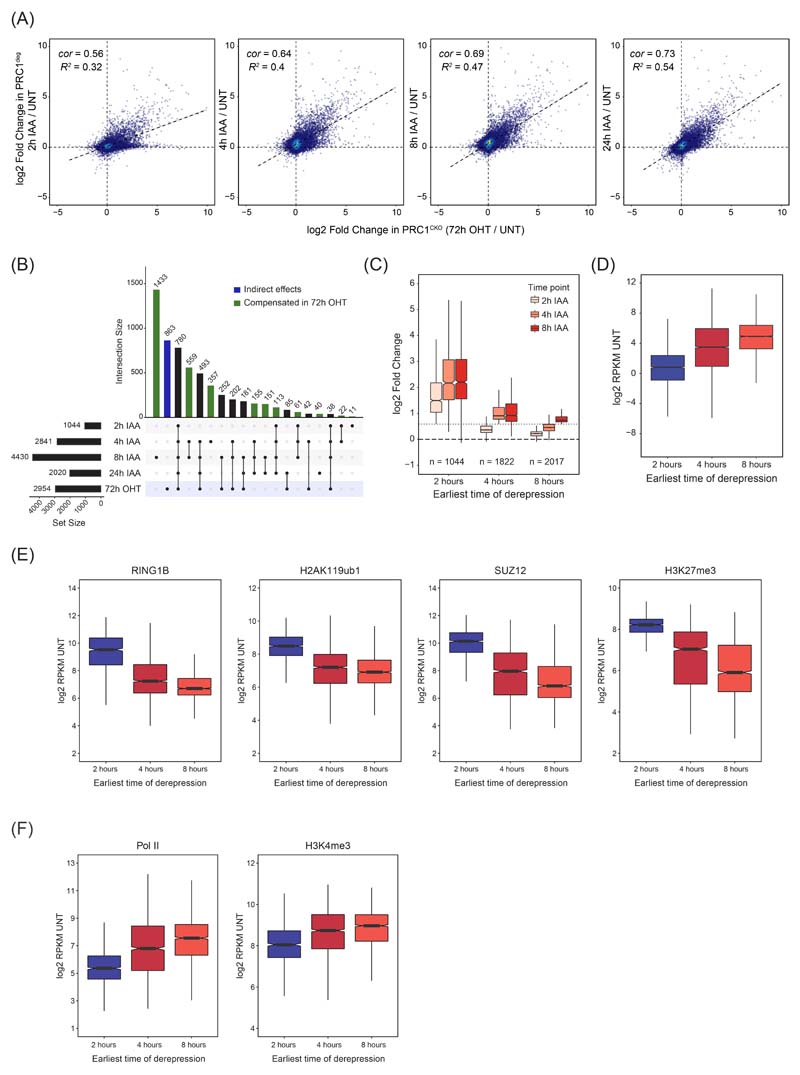
Our understanding of PRC1-mediated gene repression has largely been derived from end point gene expression analyses following prolonged PRC1 loss using conditional knockout systems 29–31. As such, these experiments do not distinguish between primary and secondary effects, nor do they capture the kinetics of gene expression alterations. Therefore, using our rapid PRC1deg system, we wanted to identify the primary target genes repressed by PRC1. To achieve this, we carried out calibrated nuclear RNA-sequencing (cnRNA-seq) in a time course experiment. Remarkably, after only 2 hours of auxin treatment roughly one thousand (1044) genes increased in expression (Fig. 2A). By 4 hours this number had more than doubled (2841) and plateaued at 8 hours (4430). 94% of genes with increased expression at 2 hours were Polycomb target genes and these genes also prominently featured at later time points (78% at 4 hours and 69% at 8 hours) (Fig. 2B). Genes exhibiting increased expression at 2, 4, and 8 hours showed a high degree of overlap (Fig. 2C), indicating that Polycomb target genes show different kinetics of derepression, with a subset becoming derepressed very rapidly. After 24 hours, the number of derepressed genes was lower (2020) (Fig. 2A), suggesting that gene expression adapts over time, which reinforces the importance of rapid depletion in identifying primary effects. In agreement with this, expression changes after 24 hours correlated more closely with the gene expression changes observed after long term conditional knockout of PRC1 (PRC1CKO)29 (Extended Data Fig. 2A). A detailed comparison of the gene expression alterations in the PRC1CKO and PRC1deg systems suggested that at later time points following PRC1 removal, indirect and compensatory effects may accumulate (Extended Data Fig. 2B). Therefore, our degron approach reveals primary Polycomb targets and identifies a subset of genes that are particularly reliant on PRC1 for repression.
Figure 2. Perturbing PRC1 has immediate effects on gene expression.
(A) MA plots depicting gene expression changes in PRC1deg cells treated with IAA for the indicated times relative to untreated cells. Differentially expressed genes (padj < 0.05, fold change > 1.5) are labelled in red.
(B) Distribution of different gene classes among genes with increased expression at different times following IAA treatment. Non-NMI, genes lacking a non-methylated CGI; Non-PcG, non-Polycomb-occupied genes; PcG, Polycomb-occupied genes.
(C) A Venn diagram showing the overlap between significantly derepressed genes identified after treating cells with IAA for the indicated times.
(D) Heatmaps depicting gene expression (cnRNA-seq) in untreated cells and changes following IAA treatment for the three groups of genes defined by the earliest time of derepression. Heatmaps are sorted by RING1B cChIP-seq signal as in (E).
(E) Heatmaps showing cChIP-seq signal for Polycomb chromatin domain features (RING1B, H2AK119ub1, SUZ12, and H3K27me3) in untreated cells, at promoters of the gene groupings in (D). Heatmaps are sorted by RING1B cChIP-seq signal.
(F) As in (E) but for total Pol II and H3K4me3 cChIP-seq.
To understand why some genes rely heavily on PRC1 for their repression, we grouped genes based on the earliest time point they became derepressed (Fig. 2D and Extended Data Fig. 2C). Genes affected at 2 hours had the lowest expression in untreated cells (Fig. 2D and Extended Data Fig. 2D), the largest magnitude of derepression after PRC1 removal, and tended to reach maximal expression by 4 hours. In contrast, genes that displayed significant increases at either 4 or 8 hours showed higher initial expression and a substantially smaller magnitude of derepression. Moreover, genes that became derepressed at 2 hours had the highest levels of Polycomb chromatin domain features including RING1B, H2AK119ub1, SUZ12, and H3K27me3 in untreated cells (Fig. 2E and Extended Data Fig. 2E). They also had the lowest levels of Pol II and transcription-associated H3K4me3 (Fig. 2F and Extended Data Fig. 2F). Conversely, genes derepressed at 4 and 8 hours had lower levels of Polycomb features and higher Pol II and H3K4me3, indicating they were already partially transcribed. Therefore, time-resolved gene expression analysis revealed that PRC1 plays a central role in repressing Polycomb target genes with low occupancy of the transcriptional machinery, and that these genes become rapidly derepressed in the absence of PRC1.
PRC1-mediated repression does not rely on PRC2 or H3K27me3
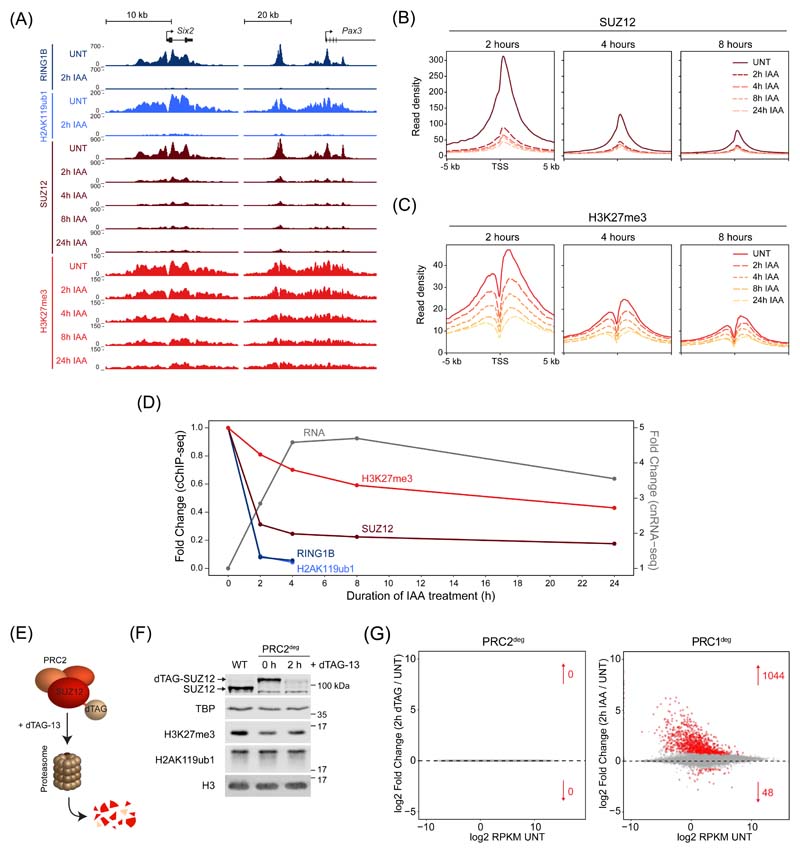
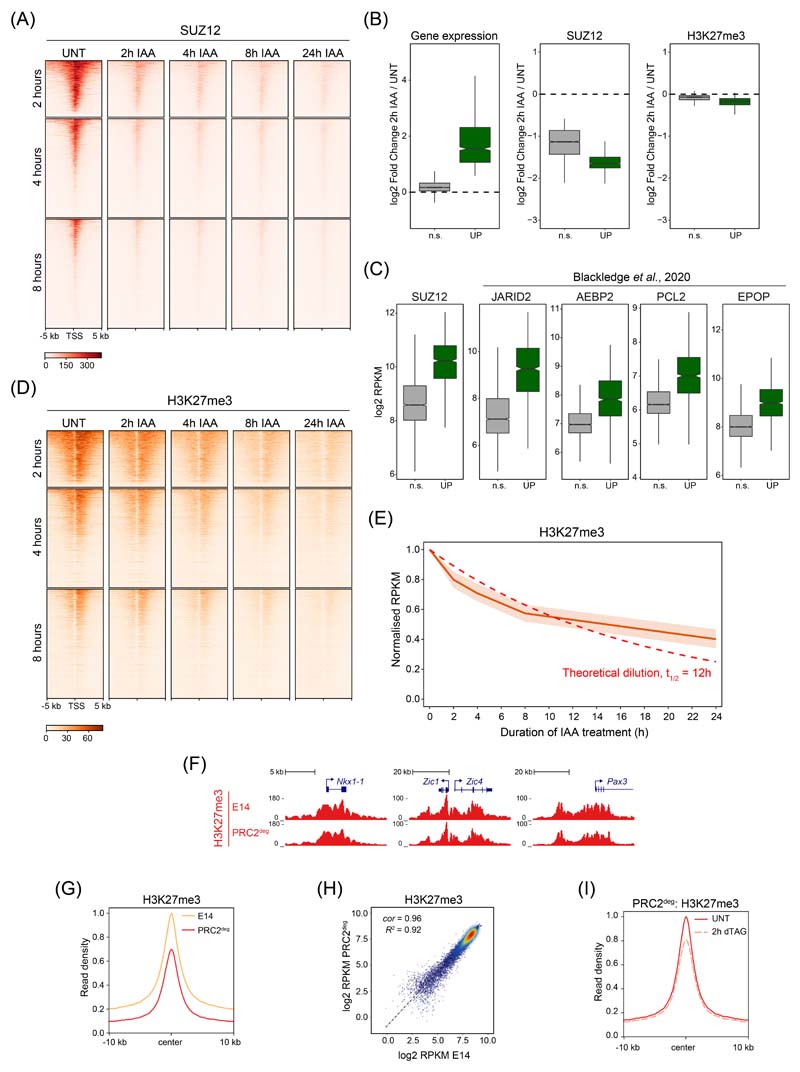
Having established that H2AK119ub1 is lost very rapidly following PRC1 removal, we set out to determine whether the rapidity of this turnover would provide an opportunity to dissect the relationship between other Polycomb chromatin domain features and gene repression. We first carried out time-resolved cChIP-seq analysis for SUZ12, a core structural component of PRC2. This revealed that PRC2 binding was majorly reduced at promoters of derepressed genes by 2 hours of PRC1 removal (Fig. 3A, B and Extended Data Fig. 3A). These reductions occurred in concert with loss of H2AK119ub1 and changed only modestly thereafter, demonstrating that PRC2 relies on H2AK119ub1 for occupancy in Polycomb chromatin domains23–25,37. Previous studies had indicated that displacement of PRC2 from chromatin could be caused by transcription and PRC2 binding to nascent RNA38–40. However, the effects we observed on PRC2 occupancy at 2 hours occurred regardless of whether genes displayed changes in expression (Extended Data Fig. 3B, C). Together this reveals that PRC2 binding to chromatin is highly dependent on PRC1/H2AK119ub1 and not simply dictated by effects on gene expression.
Figure 3. PRC1-mediated gene repression does not rely directly on PRC2 or H3K27me3.
(A) Genomic snapshots of typical Polycomb target genes, showing cChIP-seq signal for RING1B, H2AK119ub1, SUZ12 and H3K27me3 in PRC1deg cells treated with IAA for the indicated times.
(B) Metaplot analysis of SUZ12 cChIP-seq at promoters of the three groups of genes defined by the earliest time of derepression in PRC1deg cells treated with IAA for the indicated times.
(C) As in (B) for H3K27me3 cChIP-seq.
(D) Dynamics of gene expression changes in comparison with the reduction in cChIP-seq signal (TSS ± 2.5 kb) for Polycomb factors and associated histone modifications. Shown are median changes relative to untreated cells for genes derepressed after 2h of IAA treatment (n = 1044).
(E) Schematic illustration of the PRC2deg system. Endogenous SUZ12 is N-terminally tagged and degraded by the proteasome after addition of the dTAG-13 compound.
(F) Western blot analysis of SUZ12, H3K27me3 and H2AK119ub1 in wild type (WT) and PRC2deg cells (untreated and treated with dTAG-13 compound). Shown are representative results from n=4 independent experiments.
(G) MA plots depicting gene expression changes (cnRNA-seq) in PRC2deg cells treated with dTAG-13 compound for 2h (left) or PRC1deg cells treated with auxin for 2h (right). Arrows denote numbers of differentially expressed genes labelled in red (padj < 0.05, fold change > 1.5).
Based on rapid reductions in PRC2 binding in the absence of PRC1 and H2AK119ub1, we were curious to examine how this affected H3K27me3 and therefore carried out cChIP-seq for H3K27me3 over the same time course. Despite major reductions in PRC2 binding, there was only a modest effect on H3K27me3 at 2 hours and a slow reduction in H3K27me3 over the remainder of the time course (Fig. 3A, C and Extended Data Fig. 3D). The rate at which H3K27me3 eroded after PRC1 removal was consistent with an exponential decay model based on dilution during replication and cell division, not active turnover (Extended Data Fig. 3E). Importantly, as PRC2 protein levels are unaffected by PRC1 removal (Extended Data Fig. 1I), PRC2-H3K27me3 read-write mechanisms41–43 alone must be insufficient to reinstate H3K27me3 at Polycomb chromatin domains following DNA replication44. Instead, stabilisation of PRC2 at Polycomb chromatin domains through binding to H2AK119ub1 must contribute centrally to epigenetic maintenance of H3K27me3 in actively dividing cells.
These analyses revealed that derepression of Polycomb target genes at 2 hours occurred despite the fact that H3K27me3 was largely retained, suggesting at least in this context that PRC2 catalytic activity is not central to repression (Fig. 3D). However, PRC2 occupancy was majorly reduced, meaning a contribution of PRC2 binding to gene repression could not be excluded. To examine this possibility, we utilised a PRC2 degron system (PRC2deg) where treatment with the dTAG-13 compound for 2 hours results in the removal of SUZ12 (Fig. 3E, F)45,46. Using cnRNA-seq we examined gene expression before and 2 hours after PRC2 removal. This revealed no significant changes in gene expression, contrasting the dramatic effects caused by PRC1 removal at this same time point (Fig. 3G). Therefore, major reductions in PRC2 occupancy do not explain rapid derepression of Polycomb target genes caused by removal of PRC1. Although the PRC2deg cells have slightly lower starting levels of H3K27me3 than wild type cells (Extended Data Fig. 3F-H), rapid PRC2 removal caused only modest reductions in H3K27me3 (Extended Data Fig. 3I), again demonstrating that H3K27me3 turnover is slow in the absence of PRC2 occupancy. Therefore, we conclude that PRC2 occupancy at Polycomb chromatin domains is primarily defined by PRC1/H2AK119ub1 and that PRC1-mediated gene repression does not rely on PRC2 or H3K27me3.
Removal of PRC1 causes rapid new binding of Pol II
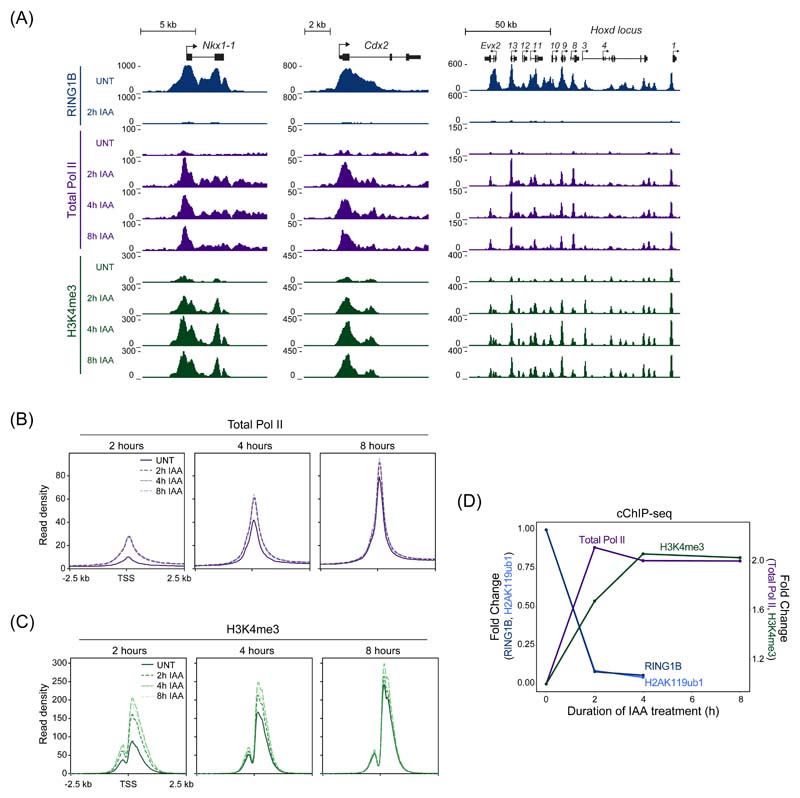
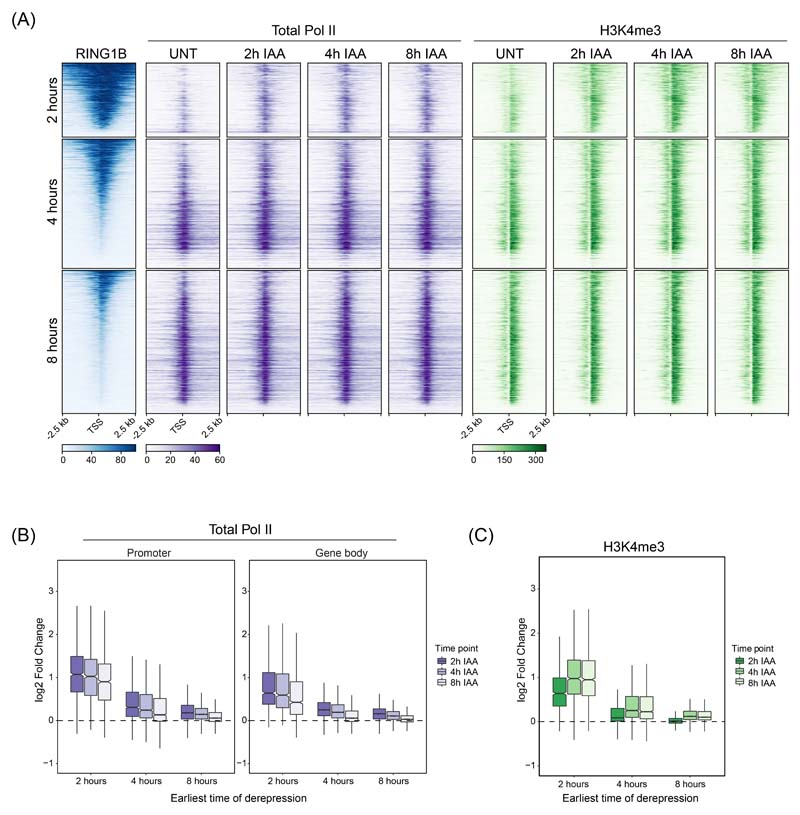
Our understanding of how the Polycomb system counteracts transcription to achieve repression has remained enigmatic. In fact, previous work has implicated the Polycomb system in regulating various and distinct phases of transcription, including initiation, pausing/priming, and elongation47–53. Having shown we can identify the earliest and most primary effects on gene expression after removing PRC1, we set out to discover how it counteracts the process of transcription at target genes. To achieve this, we carried out cChIP-seq for total Pol II in a time course experiment following removal of PRC1. This revealed rapid new binding of Pol II at Polycomb target gene promoters, which was most pronounced at genes with increased expression at 2 hours (Fig. 4A, B and Extended Data Fig. 4A). Despite expression continuing to increase for many of these genes throughout the time course, maximum levels of new Pol II binding were already reached at 2 hours (Fig. 4B and Extended Data Fig. 4B). In fact, even at genes where significant changes in expression did not manifest until 4 or 8 hours, maximal new promoter-proximal Pol II was evident at 2 hours. Therefore, the underlying defect in Polycomb chromatin domain function that enables new Pol II binding has already occurred by 2 hours, yet for many target genes significant expression changes are only detected at later time points, presumably after multiple rounds of transcription have occurred. In agreement with this, H3K4me3, a histone modification associated with active transcription54, accumulated until 4 hours (Fig. 4A, C) regardless of when the gene was first identified as significantly derepressed (Extended Data Fig. 4B, C). Therefore, we conclude that new binding of Pol II closely follows PRC1/H2AK119ub1 removal, with H3K4me3 accumulating more slowly as transcription proceeds (Fig. 4D), and that PRC1 functions to counteract Pol II binding at target gene promoters.
Figure 4. Removal of PRC1 causes a rapid new binding of Pol II and accumulation of H3K4me3.
(A) Genomic snapshots of typical Polycomb target genes, showing cChIP-seq signal for RING1B, total Pol II and H3K4me3 in PRC1deg cells treated with IAA for the indicated times.
(B) Metaplot analysis of total Pol II cChIP-seq at promoters of the three groups of genes defined by the earliest time of derepression in PRC1deg cells treated with IAA for the indicated times. The profiles represent the median signal over the shown genomic region.
(C) As in (B) but for H3K4me3 cChIP-seq.
(D) Dynamics of decreases in cChIP-seq signal for RING1B and H2AK119ub1 and increases in total Pol II and H3K4me3 signal at promoters (TSS ± 2.5 kb) of genes derepressed after 2h of IAA treatment (n = 1044). Shown are median changes relative to untreated cells.
New initiation underpins Polycomb target gene expression
The phosphorylation state of the C-terminal heptapeptide repeat domain (CTD) of Pol II characterises distinct stages of transcription. For example, phosphorylation of serine 5 (Ser5P) on the CTD is usually associated with initiated or promoter-proximal paused Pol II and serine 2 phosphorylation (Ser2P) with transcription elongation55. Previous studies examining Pol II phosphorylation in ESCs reported high levels of Ser5P-Pol II at Polycomb target gene promoters49,56 despite their lowly, or non-transcribed, state. Based on this it was proposed that the Polycomb system may repress genes by keeping Pol II in a poised state, and that release of poised Pol II was responsible for Polycomb target gene derepression in the absence of PRC149,56. However, subsequent analyses have suggested that Polycomb target genes have extremely low levels of transcriptionally engaged Pol II at their promoters57,58 and we found only low levels of total Pol II binding in our ChIP-seq analysis. These findings are therefore inconsistent with release of poised Pol II being sufficient to cause derepression of Polycomb target genes in the absence of PRC1.
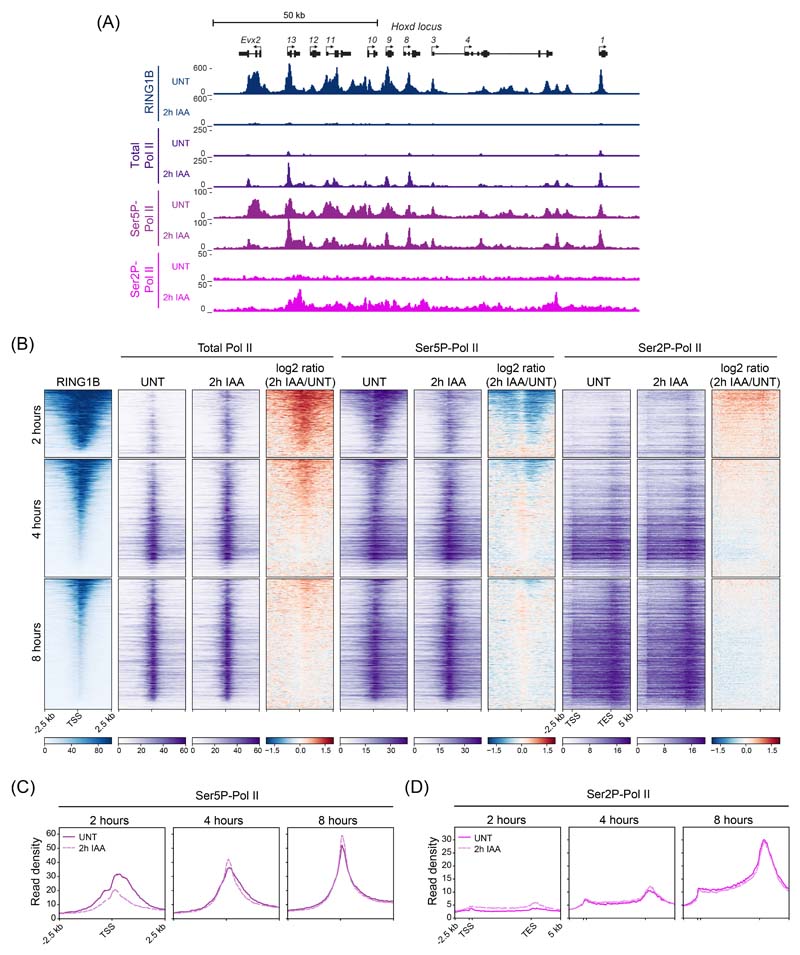
Given our capacity to capture the earliest events associated with Polycomb target gene derepression, we set out to examine how Ser5P-Pol II was affected by carrying out cChIP-seq in untreated cells and after removal of PRC1. As described previously, Ser5P-Pol II signal was present at Polycomb target genes in untreated cells49,56, despite extremely low levels of total Pol II (Fig. 5A and Extended Data Fig. 5A). However, compared to actively transcribed genes where Ser5P-Pol II occurs just downstream of the transcription start site, the distribution of Ser5P-Pol II at Polycomb target genes was highly atypical and mirrored the distribution of the underlying Polycomb chromatin domain, which in some instances extended tens of kilobases outside of the gene itself (Fig. 5A)49,56. This was evident at genes that are derepressed at 2 hours and have large Polycomb chromatin domains (Fig. 5B). Interestingly, after PRC1 removal, this atypical Ser5P-Pol II distribution was lost and Ser5P-Pol II signal was instead focused at transcription start site where new total Pol II binding was observed, presumably reflecting new transcription initiation (Fig. 5A,B and Extended Data Fig. 5A-C). We also observed small increases in Ser2P-Pol II levels in the body of derepressed genes, coincident with their increased transcription (Fig. 5A,B and Extended Data Fig. 5A, B, D). Together these observations are consistent with PRC1 removal causing new Pol II binding and transcription initiation, not release of a paused/poised Pol II. To test this more directly, we treated cells with triptolide, a small molecule inhibitor that disrupts initiation by Pol II, removed PRC1, and then asked if rapid derepression of Polycomb target genes could occur in the absence of new transcription initiation (Fig. 5C). Triptolide treatment did not affect the transcription of a RNA polymerase III (Pol III)-transcribed gene, but efficiently blocked active Pol II-transcribed genes. More importantly, derepression of almost all analysed Polycomb target genes was abolished in triptolide-treated cells (Figure 5D). We conclude that new transcription initiation, not pause release, explains the rapid derepression of Polycomb target genes in the absence of PRC1, and that PRC1 functions to counteract transcription initiation.
Figure 5. New initiation is required for Polycomb target gene derepression.
(A) Genomic snapshots of typical Polycomb target loci and one highly transcribed gene (Mcm6), showing cChIP-seq signal for RING1B, total Pol II, Ser5P-Pol II and Ser2P-Pol II in untreated PRC1deg cells and 2 hours after IAA addition.
(B) Heatmaps illustrating RING1B, total Pol II, Ser5P-Pol II and Ser2P-Pol II levels and changes in polymerase occupancy following 2h of IAA treatment for genes derepressed at that point (n = 1044). Heatmaps are sorted by RING1B signal in untreated cells. All derepressed Polycomb targets are shown in Extended Data Fig. 5B.
(C) Schematic of the experimental setup to test dependency of Polycomb target gene derepression on new initiation. 50 min prior to PRC1 depletion, cells are pre-treated with triptolide (TRP), an inhibitor of Pol II initiation, or DMSO. PRC1 removal is induced by IAA for 2 hours before collecting cells for the analysis.
(D) RT-qPCR analysis of gene expression changes in PRC1deg cells following 2h of IAA treatment, with or without 50 min pre-treatment with TRP. Gene expression is normalised to U6 snRNA and shown relative to the average expression in untreated cells. Data represents mean (n=3) ± SEM.
The Polycomb system constrains low-level gene expression
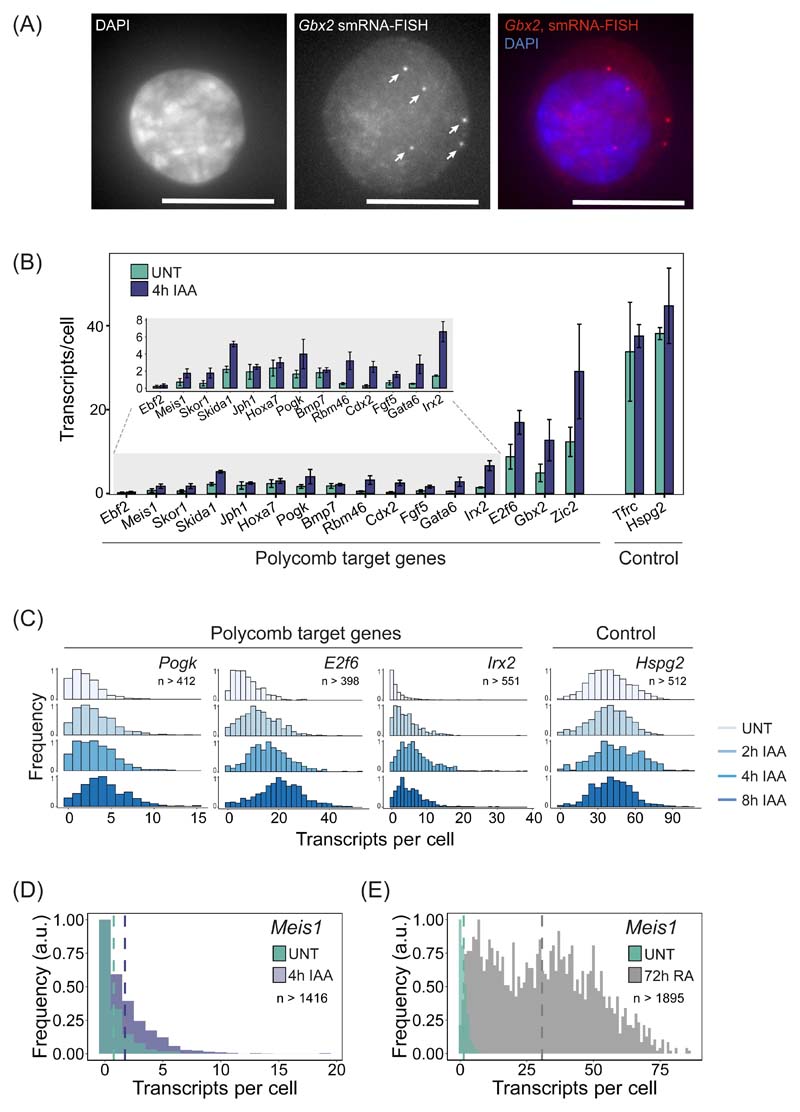
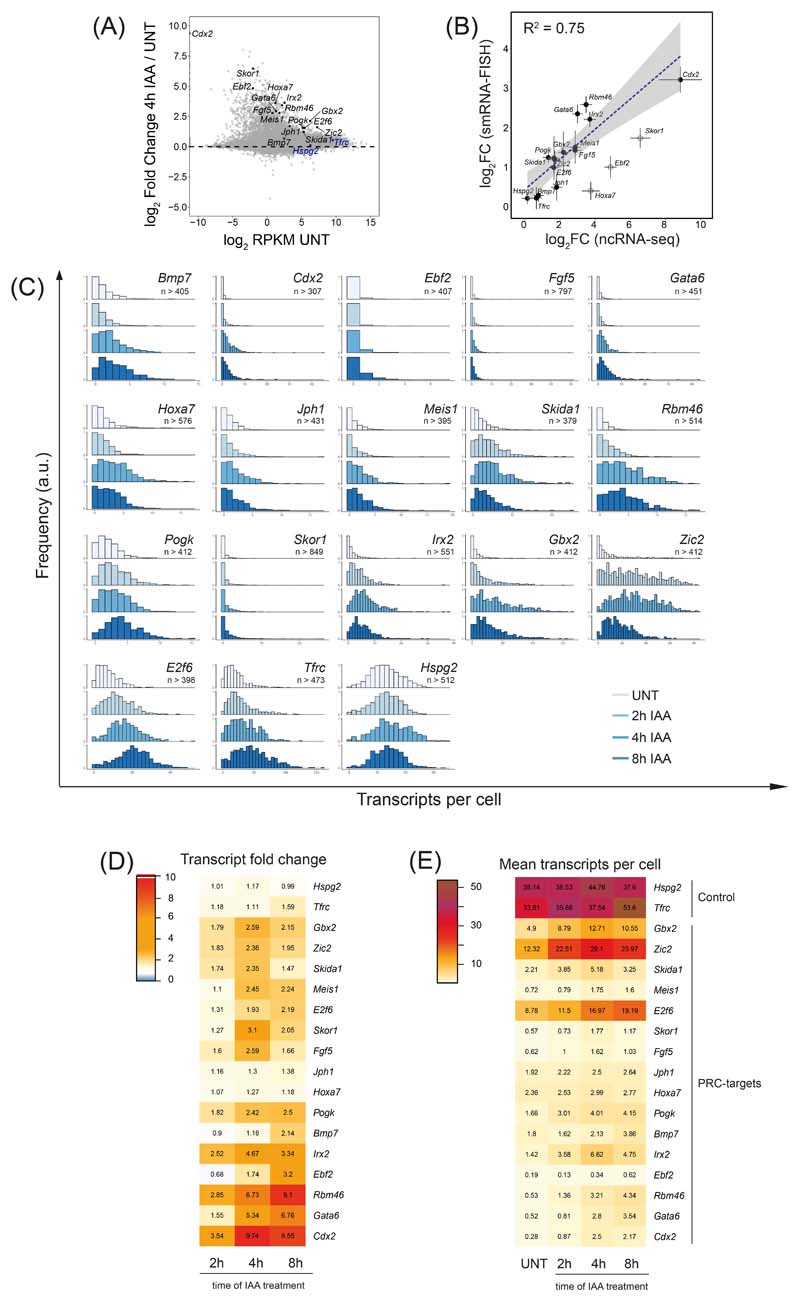
Our genomic experiments suggest that the Polycomb system functions through PRC1 and H2AK119ub1 to counteract transcription initiation. However, the ensemble nature of genomic experiments makes it impossible to understand if these effects occur in all cells in the population, or just in a subset of cells. Furthermore, they lack the resolution to define what aspect of transcription initiation is controlled by the Polycomb system. Therefore, to discover how the Polycomb system regulates gene expression in single cells, we employed single-molecule RNA fluorescent in situ hybridization (smRNA-FISH) (Fig. 6A) and developed a high-throughput imaging and analysis approach to count transcripts (Extended Data Fig. 6). We then studied the expression levels of 16 Polycomb target genes in single cells before and after PRC1 removal (Fig. 6B, Extended Data Fig. 7A,B). We found that all Polycomb target genes analysed were expressed to some degree, but most had very low transcript numbers (~1 transcript/cell) (Fig. 6B). A subset (E2f6, Gbx2, and Zic2) displayed higher average transcript numbers (~5 to ~12 transcripts/cell), but these were still much lower than actively transcribed control genes (~33 and ~38 transcripts/cell for Tfrc and Hspg2, respectively) (Fig. 6B). Therefore, Polycomb target genes are expressed, albeit very lowly, demonstrating that the Polycomb system is not an impervious block to gene transcription.
Figure 6. Single-cell analysis of Polycomb target gene expression.
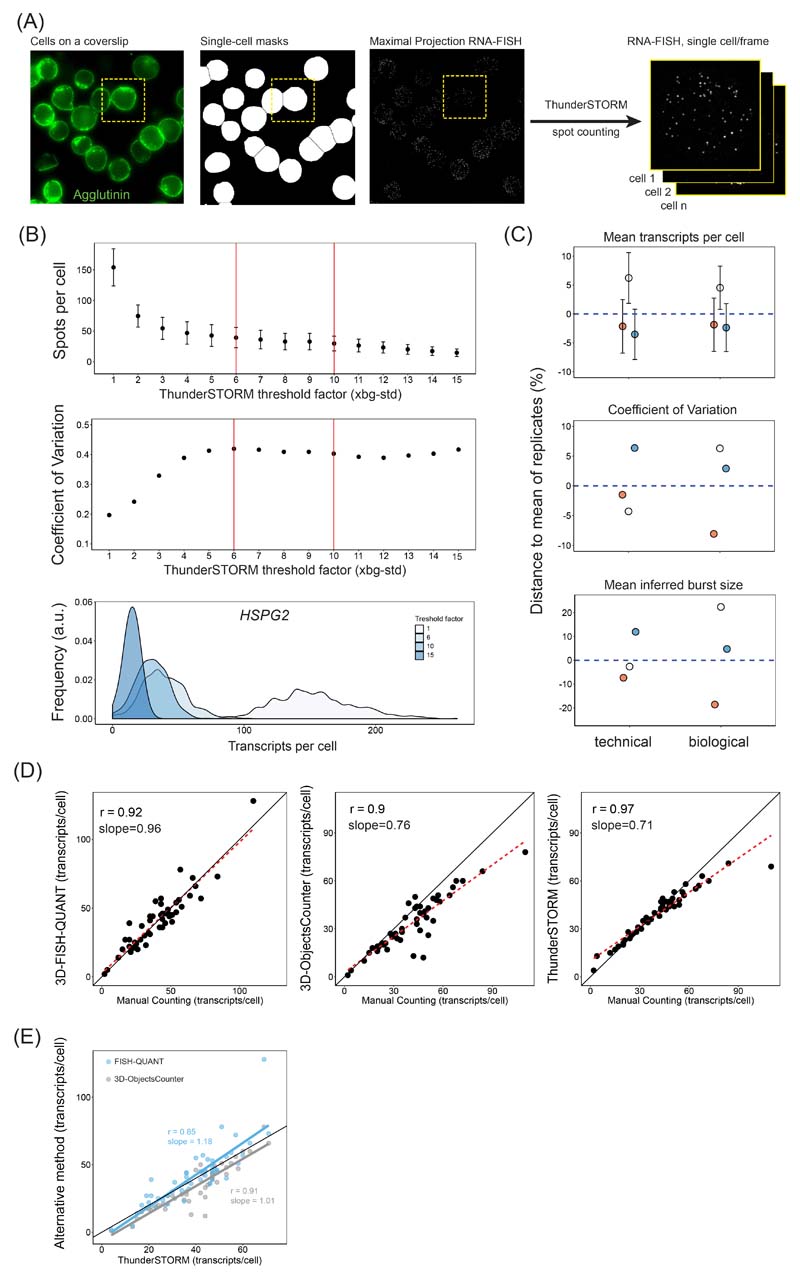
(A) A smRNA-FISH image of the Polycomb target gene Gbx2. Individual dots indicated with arrows correspond to single transcripts of Gbx2. DAPI is shown to indicate the nucleus (left panel). Maximal projections of 3D stacks are presented and the scale bar corresponds to 10 μm.
(B) A bar graph illustrating the mean number of transcripts per cell in PRC1deg cells before and after 4 hours of IAA treatment to remove PRC1 for 16 Polycomb target genes and two control genes. Data represents mean (n=3) ± SD.
(C) Normalised histograms illustrating the distribution of transcripts across the cell population in PRC1deg cells treated with IAA for the indicated times for selected Polycomb target genes and a control gene (n indicates minimum number of cells measured at any given time-point per gene dataset). All analysed genes are shown in Extended Data Fig. 7C.
(D) As in (C) but for Meis1 in untreated PRC1deg cells and after 4 hours of IAA treatment to remove PRC1. The vertical dashed lines correspond to the mean value of transcripts per cell for the respective condition.
(E) As in (D) but for Meis1 in untreated ESCs and after 72 hours of retinoic acid treatment (RA).
We then removed PRC1 and found that Polycomb target genes transitioned into a more expressed state (Fig. 6B). However, given their very low initial expression levels, even genes that showed a reasonably large increase in expression after 4 hours of auxin treatment (e.g. Irx2, ~5-fold) still only accumulated on average ~7 transcripts per cell (Fig. 6B). This modest effect in terms of absolute transcript number was evident for most genes across the time course (Extended Data Fig. 7D, E) and it was clear that almost all cells in the population displayed small increases in expression, as opposed to only a subset of cells switching into a highly expressed or actively transcribing state (Fig. 6C and Extended Data Fig. 7C). Therefore, we conclude that PRC1 counteracts low-level transcriptional signals that are present across the cell population, as opposed to blocking strong transcription signals that could activate gene expression either heterogeneously in single cells or more broadly.
In contrast to these observations, previous single-cell RNA-sequencing (scRNA-seq) had concluded that Polycomb target genes, and particularly those that were more highly expressed, show a bimodal distribution of transcript levels in the cell population59. It was proposed that this bimodality arose from some cells displaying very low Polycomb target gene expression and others existing in a highly expressed state. Furthermore, it was reported that increases in gene expression after removal of PRC1 resulted from lowly expressing cells switching into a highly expressed or active transcriptional state59. We did not observe bimodality for Polycomb target genes in untreated cells, nor did it emerge following removal of PRC1 (Fig. 6C and Extended Data Fig. 7C). This difference is likely explained by scRNA-seq lacking the capacity to accurately quantify lowly expressed genes60 and absolute transcript numbers. These limitations are overcome by smRNA-FISH which has single-transcript sensitivity. However, to further validate that removal of PRC1 did not result in lowly expressing cells switching into a highly expressed or active transcriptional state, we focussed on the Meis1 gene. When PRC1 was removed, Meis1 expression increased by 2.5-fold (0.7 to 1.75 transcripts/cell) (Fig. 6D). In contrast, when cells were treated with retinoic acid to induce differentiation, Meis1 expression increased more than 40-fold and switched into a highly expressed state with transcript numbers similar to other actively transcribed genes (~30 transcripts/cell) (Fig. 6E). This reinforces our conclusion that the Polycomb system functions to counteract low-level transcriptional signals present in most cells in the population, as opposed to protecting its target genes from switching into a highly expressed or active transcriptional state.
The Polycomb system limits transcription burst frequency
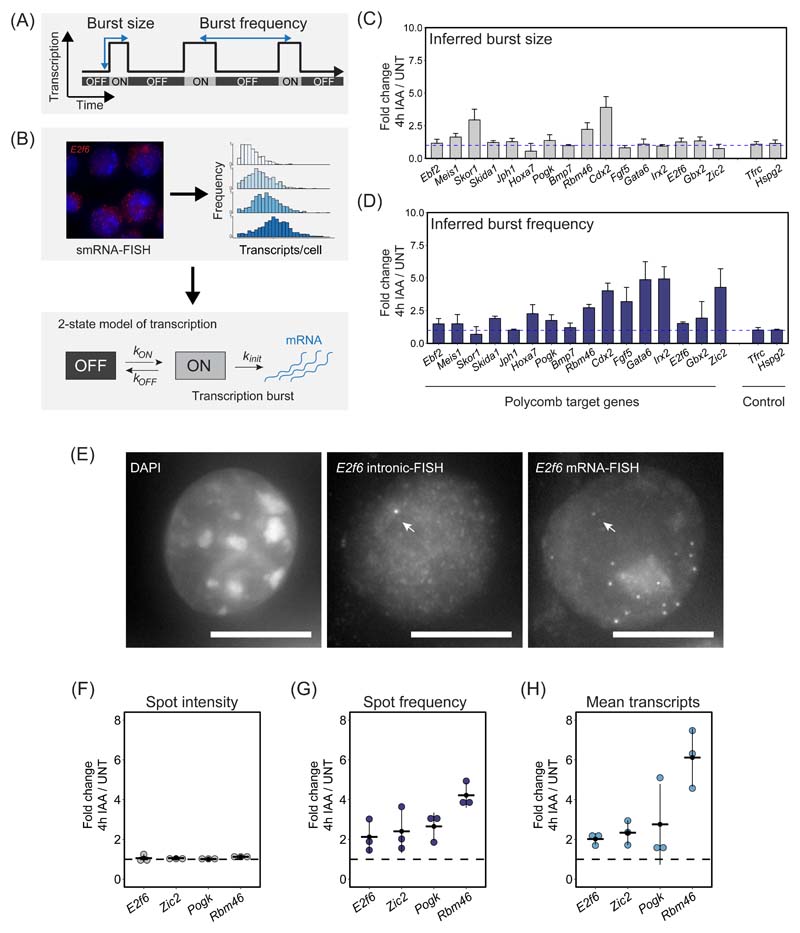
Our single-cell analysis revealed that Polycomb target genes are expressed, albeit at low levels, and that PRC1 functions to counteract expression in most cells in the population. This suggests that Polycomb target genes are constantly subject to signals that promote transcription and therefore PRC1 must function to counteract some fundamental aspect of transcription at their promoters. The process of transcription from gene promoters is pulsatile and stochastic61,62. During periods of activity, individual genes undergo bursts of transcription initiation and elongation, producing multiple transcripts. This is often followed by prolonged periods of inactivity. The frequency with which transcription bursts occur (burst frequency) and how many transcripts are produced during a burst (burst size) underpin gene expression (Fig. 7A).
Figure 7. The Polycomb system represses gene expression by limiting transcription burst frequency.
(A) A schematic illustrating the stochastic and pulsatile nature of gene transcription. Over time, a gene promoter can exist in an OFF state where no transcription is occurring or ON state where multiple transcripts are produced (a burst of transcription). The number of transcripts that are produced in a burst is referred to as the burst size and the burst frequency is the time between transcription bursts. Importantly, transcription burst sizes and their frequency are central determinants of gene expression.
(B) A schematic illustrating how population level transcript distributions from smRNA-FISH are used to inform a two state model of transcription and infer kinetic parameters of transcription.
(C) A bar graph illustrating the fold change in inferred transcription burst size for Polycomb target and control genes. Data represents mean (n=3) ± SD. The dashed horizontal line corresponds to no change.
(D) As in (C) but for inferred burst frequency.
(E) An example of a nascent smRNA-FISH (intronic FISH) image for E2f6, with an individual bright dot corresponding to a nascent transcription spot indicated with an arrow (middle panel). Standard smRNA-FISH (targeting exonic sequences) in the same cells illustrates individual transcripts and the nascent transcription spot (right panel). DAPI is shown to indicate the nucleus (left panel). The scale bar corresponds to 10 μm.
(F) Fold change in mean nascent transcription spot intensity for the indicated Polycomb target genes following 4h IAA treatment to remove PRC1. The dashed horizontal line corresponds to no change. Individual biological replicates are represented by dots, whereas the horizontal line represents the mean of the 3 biological replicates. A minimum of 887 cells was measured per biological replicate.
(G) As in (F) but fold change in nascent transcription spot frequency.
(H) As in (F) but fold change in the mean number of transcripts per cell.
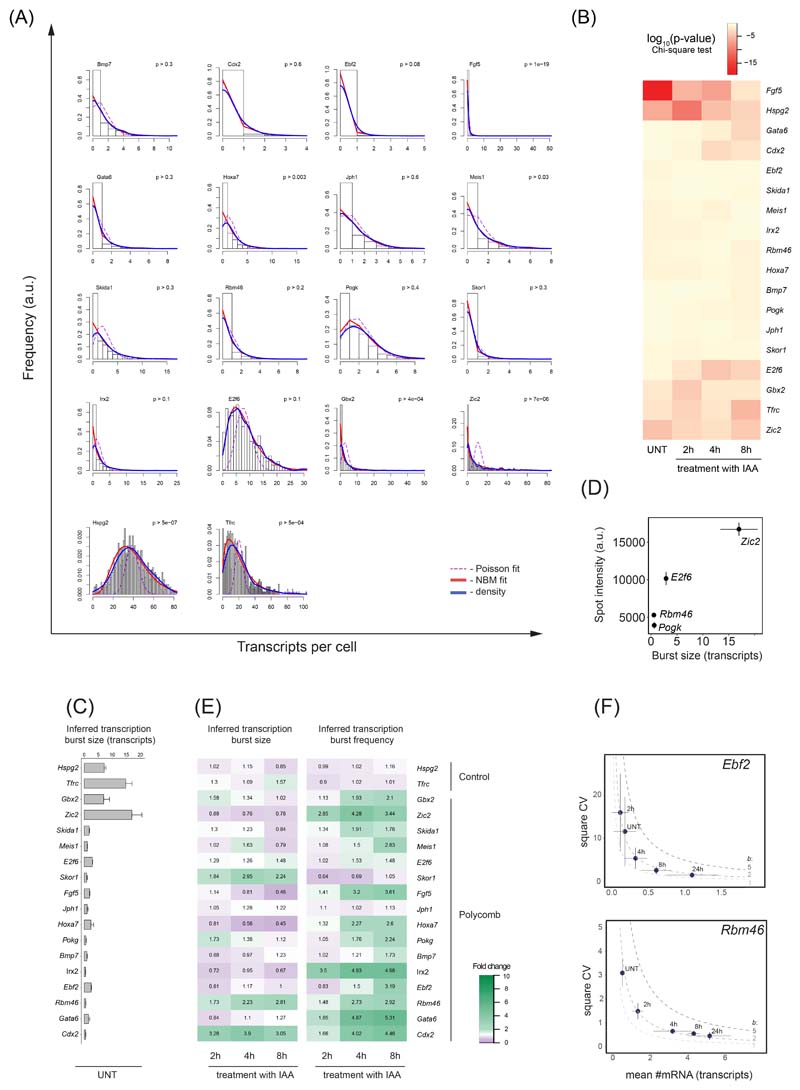
To characterise these features of Polycomb target gene transcription we used transcript distributions from our smRNA-FISH measurements and a 2-state model to infer kinetic parameters61,63–65 (Fig. 7B, Extended Data Fig. 8A, B). In untreated cells the majority of Polycomb target genes had very small transcription burst sizes (~1-3 transcripts) compared to expressed control genes (~7-15 transcripts) (Extended Data Fig. 8C). However, some Polycomb target genes with higher expression levels (Gbx2 and Zic2) also displayed larger burst sizes (~6-17 transcripts) (Extended Data Fig. 8C). Importantly, the burst size of most Polycomb target genes was unaffected by PRC1 removal (Fig. 7C and Extended Data Fig. 8E), suggesting that PRC1 does not primarily function to regulate transcription burst size.
To test the possibility that PRC1 may repress transcription by instead limiting burst frequency we revisited our smRNA-FISH measurements and the two-state model to estimate transcription burst initiation rate (kON), which can be used as a proxy for burst frequency. This revealed that the majority of Polycomb target genes displayed increases in burst frequency when PRC1 was removed (Fig. 7D), which was in line with the increased expression of these genes (Fig. 6B and Extended Data Fig. 7D). Therefore, modelling-based approaches suggest that PRC1 limits transcription burst frequency to repress gene expression.
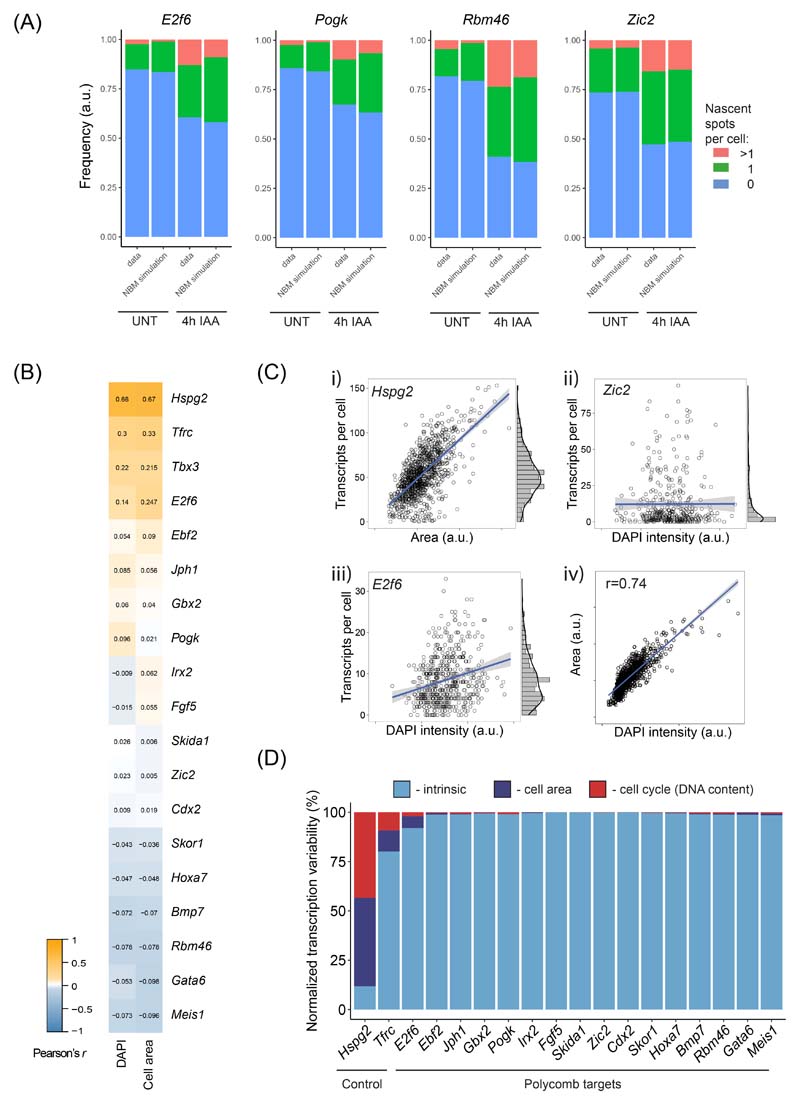
To test this possibility more directly, we used intronic smRNA-FISH that captures short-lived nascent pre-mRNAs at the site of active transcription and therefore more directly interrogates promoter activity (Fig. 7E). The number of nascent transcription spots in the cell population represents a measure of burst frequency, while the intensity of individual spots reflects transcription burst size66. Using this approach we examined two lowly expressed (Rbm46 and Pogk) and two more highly expressed (E2f6 and Zic2) target genes. The spot intensities for these genes in untreated cells scaled with inferred burst sizes (Extended Data Fig. 8D) and remained unchanged following removal of PRC1, in line with our conclusions from modelling of smRNA-FISH transcript distributions (Fig. 7F and Extended Data Fig. 8E). However, after removal of PRC1, the number of nascent transcription spots in the cell population increased in frequency for all four genes (Fig. 7G and Extended Data Fig. 9A) and this corresponded to increases in expression (Fig. 7H). Furthermore, the distribution of transcription spots in the cell population was consistent with independently transcribing alleles, as opposed to allelic-specific expression56, both before and after PRC1 removal (Extended Data Fig. 9A). Therefore, single-cell measurements reveal that PRC1 represses gene expression by limiting transcription burst frequency.
Discussion
The interrelatedness of Polycomb repressive complex activities at target gene promoters has made identifying the mechanisms of gene repression extremely challenging. Using rapid depletion and time-resolved genomic analyses we now show that Polycomb target genes become immediately derepressed when PRC1 and H2AK119ub1 are lost from Polycomb chromatin domains, and that derepression occurs despite the presence of H3K27me3. Furthermore, rapid removal of PRC2 has no effect on Polycomb target gene repression. Therefore, these experiments narrow down the repressive activity of Polycomb chromatin domains to PRC1, but alone they do not define whether PRC1 complexes or H2AK119ub1 are responsible for transcriptional repression. Previously we and others developed ESCs systems where we could inducibly inactivate PRC1 catalysis, but leave PRC1 complexes intact and still able to bind target genes30,67. These experiments showed that catalysis by PRC1 is essential for gene repression, but these effects were confounded by the fact that H3K27me3 and PRC2 occupancy were also profoundly affected due to the aforementioned feedback mechanisms and the protracted time required to convert PRC1 into a catalytically deficient form. Our rapid perturbation experiments now enable us to exclude a central and direct role for PRC2 and H3K27me3 in PRC1-dependent gene repression, therefore strongly implicating H2AK119ub1 as a defining feature of Polycomb chromatin domains that enables repression in ESCs. These observations are in agreement with previous findings that canonical PRC1 complexes that bind to chromatin through H3K27me3, but which have limited capacity to deposit H2AK119ub123,68–70, contribute little to repression71–75. In contrast, variant PRC1 complexes that deposit almost all H2AK119ub1 are essential for gene repression29. Furthermore, when H2AK119ub1 is directly incorporated into chromatinised gene promoter templates in vitro, this specifically inhibits transcription48,76. Together this suggests that H2AK119ub1 is a primary mechanism through which the Polycomb systems mediates transcriptional repression in ESCs.
The contribution of PRC1 and its catalysis to regulation of mammalian gene expression has recently been examined in the context of neural cell differentiation77. In this case, catalysis was necessary for repression during early neurogenesis, although this dependency was less pronounced in more differentiated cells that were slowly proliferating. This raises the interesting possibility that H2AK119ub1 may represent a primary determinant of Polycomb-mediated repression in rapidly proliferating cells where the integrity of Polycomb chromatin domains is constantly challenged by DNA replication. In contrast, other Polycomb features may function to increase the fidelity of gene repression in contexts where rapid division has seceded and long-term stable repression must occur. In agreement with this possibility, the kinetics of H2AK119ub1 deposition (PD and RJK, unpublished observation) and removal appear to be very rapid, whereas PRC2-dependent H3K27me3 addition and removal is far slower78–82. Therefore, we speculate that PRC1 and H2AK119ub1 could function to rapidly respond to alterations in transcription to initiate repression and that feedback mechanisms that integrate PRC2 activity may function over longer time scales to increase the fidelity and stability of repression83.
How chromatin modifying systems regulate the process of transcription is poorly understood. Through defining the repressive features of Polycomb chromatin domains and combining genome-wide measurements of gene expression and Pol II binding with single-molecule single-cell transcription analysis we now gain the first detailed view of the mechanisms that enable Polycomb-mediated gene repression in ESCs. We show that the Polycomb system does not function as an absolute block to transcription. Instead, cells in the population display at least some expression of Polycomb target genes and low-level binding of Pol II at their promoters. Following PRC1 removal, most cells display increases in Polycomb target gene expression, indicating that the Polycomb system protects against ubiquitous low-level transcriptional signals, and does not actively repress genes that would otherwise become fully transcribed in its absence. As such, the Polycomb system appears to maintain gene expression patterns to support normal cell identity.
Our new findings raise the important question of how the Polycomb system mechanistically counteracts the process of transcription. By measuring transcription in single cells, we find that Polycomb-dependent gene repression works through regulating transcription burst frequency. This effect could in theory manifest from PRC1/H2AK119ub1 causing local chromatin compaction and formation of higher-order chromatin structures that are refractory to chromatin remodeling and active transcription as has been proposed previously84–88. However, we and others have shown that perturbation of the Polycomb system in ESCs does not profoundly influence target gene promoter accessibility89–91. We interpret this to mean that PRC1 and H2AK119ub1 must therefore have more direct roles in counteracting Pol II function. Indeed it had been suggested that the Polycomb system could limit the release of a poised or paused polymerase, based on the presence of Ser5-phosphorylated Pol II at Polycomb target gene promoters49,56. However, measurements of transcriptionally engaged Pol II suggest that Polycomb target gene promoters lack paused Pol II57,58 and we find that derepression of Polycomb target genes requires new transcription initiation. Furthermore, alterations to transcriptional pause release have been proposed to primarily affect transcription burst sizes92,93, not frequency as discovered here. Importantly, the frequency of transcription bursts can be increased by the action of transcriptional activators that bind to distal enhancer elements94–98 and influence Pol II activity at gene promoters99,100. Furthermore, histone acetylation has also been implicated in promoting burst frequency when found at enhancers and gene promoters98,101. Here we find that the Polycomb system elicits the opposite effect on burst frequency and does so through functioning directly at the gene promoter. As such, the Polycomb system may play a key role at promoters in counteracting low level or inappropriate transcription signals that emanate from regulatory elements, like enhancers, that if left unchecked could lead to bursts of transcription that would endanger maintenance of cell identity. Furthermore, we envisage that this Polycomb-dependent repressive activity could also play important roles during the early stages of gene induction to ensure appropriate and persistent activation signals are present before a gene can transition into an activated state, for example, during cell fate transitions3,28. Therefore, examining the influence of the Polycomb system on transcription dynamics during cellular differentiation will be important in the context of future work.
At a molecular level we envisage that PRC1 and H2AK119ub1 likely affect transcription burst frequency by controlling some key aspect of transcription initiation. This agrees with previous reports that the Polycomb system can interfere with the assembly or activity of the pre-initiation complex47,52. However, defining precisely how H2AK119ub1 influences transcription initiation will require more detailed consideration, and given the complexity of this process, it will be important to use highly defined in vitro systems where H2AK119ub1 has been shown to repress transcription48,76. Nevertheless, our detailed in vivo work provides a strong conceptual basis for Polycomb-mediated gene repression which places PRC1 and H2AK119ub1 as central components through which Polycomb chromatin domains control Pol II function and transcriptional burst frequency to regulate gene expression.
Methods
Cell lines, culturing and treatments
The control TIR1 and PRC1deg cell lines were previously derived from male E14 mouse embryonic stem cells (ESCs)34. In brief, the TIR1 coding sequence from the Oryza sativa was inserted into the Rosa26 locus by CRISPR-Cas9 engineering, generating the TIR1 control line. These cells were further engineered to create the PRC1deg cell line by introducing the full-length auxin inducible degron (AID) tag at the N-terminus of both Ring1B alleles, and constitutively deleting Ring1A by excising exons 1-3. The PRC2deg cell line was previously generated46 by inserting dTAG at the N-terminus of the endogenous Suz12 in the PCGF2-HaloTag genetic background. Wild type E14 mouse ESCs were used as a control.
Mouse ESCs were grown on gelatin-coated plates, in Dulbecco’s Modified Eagle Medium (DMEM, Life Technologies) supplemented with 15% fetal bovine serum (FBS, Labtech), 0.5 mM beta-mercaptoethanol (Life Technologies), 2 mM L-glutamine (Life Technologies), 1x penicillin-streptomycin (Life Technologies), 1x non-essential amino acids (Life Technologies) and 10 ng/mL leukemia inhibitory factor. Cells were cultured at 37°C with 5% CO2. For calibration of genomic experiments, we used either Drosophila S2 (SG4) cells, cultured at 25°C in Schneider’s Drosophila Medium (Life Technologies), supplemented with 10% heat-inactivated FBS and 1x penicillin-streptomycin, or human HEK293T cells, cultured at 37°C with 5% CO2, in DMEM supplemented with 10% FBS, 2 mM L-glutamine, 1x penicillin-streptomycin and 0.5 mM beta-mercaptoethanol. All cells were regularly tested for the presence of mycoplasma.
To induce AID-RING1B degradation, water-dissolved auxin (indole-3-acetic acid (IAA), Sigma) was mixed with cell medium to the final concentration of 500 μM and added to PRC1deg cells at designated times before harvesting by trypsinisation. To induce dTAG-SUZ12 degradation, PRC2deg cells were treated with 100 nM dTAG-13 compound45 for 2 hours. For the Pol II initiation inhibition experiments, PRC1deg cells were treated with 500 nM triptolide (Merck) or DMSO for 50 minutes before adding auxin for 2 hours, and harvested by scraping in the ice-cold PBS. For retinoic acid (RA) differentiation the cells were plated, allowed to attach and switched to a medium with 1 μM RA and without leukemia inhibitory factor105. Cells were grown for 72 hours with medium replaced every 24 hours before proceeding with smRNA-FISH.
Protein extraction and immunoblotting
To extract nuclear proteins, cell pellets were resuspended in 10 volumes of buffer A (10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.5 mM PMSF, and 1x cOmplete protease inhibitor (PIC, Roche)) and incubated for 10 min on ice. After centrifugation at 1500 g for 5 min, the cell pellets were resuspended in 3 volumes of buffer A supplemented with 0.1% NP-40. After mixing by inversion, nuclei were pelleted again at 1500 g for 5 min. Pelleted nuclei were resuspended in 1 volume of buffer C (250 mM NaCl, 5 mM HEPES pH 7.9, 26% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT and 1x PIC). The volume of the nuclear suspension was measured and concentration of NaCl raised to 400 mM by dropwise addition. Nuclei were rotated at 4°C for 1 h to extract nuclear proteins, which were recovered as the supernatant after centrifugation at 18,000 g for 20 min. Protein concentration was determined by Bradford assay (BioRad).
Histone extracts were prepared by washing pelleted cells in RSB (10 mM Tris HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2 and 20 mM N-ethylmaleimide (NEM)), followed by centrifugation at 240 g for 5 min and resuspension in RSB buffer supplemented with 0.5% NP-40. Following incubation on ice for 10 min, cells were centrifuged at 500 g for 5 minutes. After resuspending the nuclear pellet in 5 mM MgCl2, an equal volume of 0.8 M HCl was added, and histones were extracted for 20 min on ice and incubated on ice for 20 min to extract histones. The supernatant was taken after centrifugation for 20 min at 18,000 g, and histones precipitated by addition of TCA to 25% v/v and incubation on ice for 30 min. Histones were pelleted by centrifugation at 18,000 g for 15 min, and washed twice with cold acetone. The histone pellet was resuspended by vortexing in 1x SDS loading buffer (2% SDS, 100 mM Tris pH 6.8, 100 mM DTT, 10% glycerol and 0.1% bromophenol blue) and boiling at 95°C for 5 min. Histones were stained by Coomassie Brilliant Blue following SDS-PAGE to compare concentrations between samples.
For western blot analysis, nuclear or histone extracts were resolved by SDS-PAGE and transferred onto a nitrocellulose membrane using the Trans-Blot Turbo Transfer System (BioRad). Membranes were blocked in 5% milk in 1x PBS with 0.1% Tween 20 (PBST) for at least 30 min at RT. Membranes were incubated in the same buffer with primary antibodies (Supplementary Table 1) overnight at 4°C, washed 3x 5 min with PBST, and incubated with the IRDye secondary antibodies (LI-COR) in PBST for 60 min at RT. Following 2x 5 min PBST and 1x 5 min PBS wash, membranes were imaged using the Odyssey Fc system and analysed using Image Studio (LI-COR). Changes in bulk protein levels were quantified relative to the loading controls (TBP or CFP1 for nuclear extracts, H3 or H4 for histone extracts).
Co-immunoprecipitation
For co-immunoprecipitation experiments, 500 μg of nuclear extract from the control (TIR1) or PRC1deg cells was diluted to 550 μL with BC150 buffer (150 mM KCl, 10% glycerol, 50 mM HEPES (pH 7.9), 0.5 mM EDTA, 0.5 mM DTT, 1x PIC). A 50 μL aliquot was retained as input, and the rest was incubated overnight at 4°C with 5 μg of mouse monoclonal anti-RING1B antibody106. Protein A agarose beads (Repligen) were used to capture the immunoprecipitated material for 1 h at 4°C. Beads were pelleted at 1000 g, washed three times with BC150, resuspended in 120 μL of 1x SDS loading buffer and boiled at 95°C for 5 min. The supernatant was taken as the immunoprecipitate and analysed by western blotting along with the input, as described above.
Gene expression analysis by RT-qPCR
RNA was isolated from cell nuclei using either TRIzol reagent (Invitrogen) or RNeasy Mini Kit (Qiagen). Genomic DNA was digested with the TURBO DNA-free Kit (Invitrogen). cDNA was synthesized from 400 ng of RNA using ImProm-II Reverse Transcription system kit and random primers (Promega). Quantitative PCR was performed in technical duplicates using SensiMix SYBR mix (Bioline) and primers listed in Supplementary Table 2. The U6 snRNA gene was used as an internal control.
Calibrated nuclear RNA-sequencing (cnRNA-seq)
For cnRNA-seq, 2x107 mouse ESCs (untreated or treated with auxin or dTAG-13 compound for indicated times) were mixed with 8x106 Drosophila SG4 cells in PBS. Nuclei were released in 1 mL HS Lysis buffer (50 mM KCl, 10 mM MgSO4.7H20, 5 mM HEPES, 0.05% NP40, 1 mM PMSF, 3 mM DTT, 1x PIC) for 1 min at room temperature, and recovered by centrifugation at 1000 g for 5 min at 4°C, followed by three washes with ice-cold RSB buffer (10 mM NaCl, 10 mM Tris pH 7.4, 3 mM MgCl2). Pelleted nuclei were resuspended in 1 mL of TRIzol reagent and RNA was extracted according to the manufacturer’s protocol, followed by treatment with the TURBO DNA-free Kit. Quality of RNA was assessed using 2100 Bioanalyzer RNA 6000 Pico kit (Agilent). Next, rRNA was depleted using the NEBNext rRNA Depletion kit (NEB). RNA-seq libraries were prepared using the NEBNext Ultra (for PRC1deg) or Ultra II (for PRC2deg) Directional RNA Library Prep kit (NEB), indexed using NEBNext Multiplex Oligos (NEB), polled and sequenced as 80 bp (PRC1deg) or 40 bp (PRC2deg) paired-end reads on the Illumina NextSeq 500 in biological triplicates.
Preparation of chromatin for calibrated ChIP-seq (cChIP-seq)
All cChIP-seq experiments were performed in biological triplicates and calibrated using the whole-cell spike-in107–109.
For RING1B and SUZ12, we performed double-crosslinked cChIP-seq29. Briefly, 5x107 mouse ESCs were first crosslinked with 2 mM disuccinimidyl glutarate (DSG, Thermo Scientific) while rotating for 45 min at 25°C, and then with 1% formaldehyde (methanol-free, Thermo Scientific) for further 15 min. Reactions were quenched by addition of 125 mM glycine. Mouse ESCs were then mixed with 2x106 double-crosslinked HEK293T cells and incubated in lysis buffer (50 mM HEPES pH 7.9, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, 1x PIC) for 10 min at 4°C. The released nuclei were washed (10 mM Tris-HCl pH 8, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1x PIC) for 5 min at 4°C. Chromatin was then resuspended in 1 mL of sonication buffer (10 mM Tris-HCl pH 8, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium deoxycholate, 0.5% N-lauroylsarcosine, 1x PIC) and sonicated for 30 min using the BioRuptor Pico (Diagenode). Following sonication, Triton X-100 was added to a final concentration of 1%. After centrifugation at 20,000 g for 10 min at 4°C, supernatant was stored at -80°C until use.
For total and phosphorylated Pol II, we performed single-crosslinked cChIP-seq110. Briefly, 5x107 mouse ESCs were crosslinked in 1% formaldehyde for 10 min at 25°C and then quenched with 125 mM glycine. Mouse ECSs were mixed with 2x106 single-crosslinked HEK293T cells and incubated in FA-lysis buffer (50 mM HEPES pH 7.9, 150 mM NaCl, 2 mM EDTA, 0.5 mM EGTA, 0.5% NP-40, 0.1% sodium deoxycholate, 0.1% SDS, 10 mM NaF, 1 mM AEBSF, 1x PIC) for 10 min. Chromatin was sonicated for 30 min using the BioRuptor Pico, centrifuged at 20,000 g for 10 min at 4°C, and supernatant was stored at -80°C until use.
For H2AK119ub1, H3K27me3 and H3K4me3, we performed native cChIP-seq29. Briefly, 5x107 mouse ESCs were mixed with 2x107 Drosophila SG4 cells in PBS. The cells were pelleted and nuclei were released by resuspending in ice-cold lysis buffer (10 mM Tris-HCl pH 8, 10 mM NaCl, 3 mM MgCl2, 0.1% NP-40, 5 mM NEM). Nuclei were then washed, and resuspended in 1 mL of MNase digestion buffer (10 mM Tris-HCl pH 8, 10 mM NaCl, 3 mM MgCl2, 0.25 M sucrose, 3 mM CaCl2, 10 mM NEM, 1x PIC). Samples were digested with 150 units of MNase (Fermentas) for 5 min at 37°C, stopped by 4 mM EDTA. The supernatant (S1) was collected following centrifugation at 1500 g for 5 min at 4°C. The remaining pellet was incubated with 300 μl of nucleosome release buffer (10 mM Tris-HCl pH 7.5, 10 mM NaCl, 0.2 mM EDTA, 10 mM NEM, 1x PIC) at 4°C for 1 h, passed five times through a 27G needle, and spun at 1500 g for 5 min at 4°C. The second supernatant (S2) was collected and combined with corresponding S1 supernatant, and stored at -80°C until use. Digestion to predominantly mono-nucleosomal fragments was confirmed by agarose gel electrophoresies of purified DNA.
Chromatin immunoprecipitation and massively parallel sequencing
For double-crosslinked ChIP, sonicated chromatin was diluted 10-fold in ChIP dilution buffer (1% Triton X-100, 1 mM EDTA, 20 mM Tris-HCl pH 8, 150 mM NaCl, 1x PIC) and pre-cleared for 1 h with either protein A agarose beads (Repligen, for RING1B ChIP) or protein A magnetic Dynabeads (Invitrogen, for SUZ12 ChIP) blocked with 1 mg/mL bovine serum albumin (BSA) and 1 mg/mL yeast tRNA. For each ChIP reaction, 1 mL of diluted and pre-cleared chromatin was incubated overnight with the appropriate antibody, either anti-RING1B (CST, D22F2, 3 μl), or anti-SUZ12 (CST, D39F6, 3 μl). Antibody-bound chromatin was captured using blocked protein A beads (agarose for RING1B, magnetic Dynabeads for SUZ12) for at least 2 h at 4°C, collected by centrifugation/on a magnetic rack and washed extensively. ChIP DNA was eluted in elution buffer (1% SDS, 0.1 M NaHCO3) and cross-linking was reversed overnight at 65°C with 200 mM NaCl and 2 μL RNase A (Sigma). A matched input sample (1/10 of original ChIP reaction) was treated identically. The following day, ChIP samples and inputs were incubated with Proteinase K (Sigma) for 1.5 h at 56°C and purified using ChIP DNA Clean and Concentrator Kit (Zymo Research).
For Pol II ChIP, 300 ug of chromatin was diluted to 1 ml in FA-lysis buffer and pre-cleared for 1 h with protein A agarose beads blocked with 1 mg/mL BSA and 1 mg/mL yeast tRNA. For each ChIP reaction, diluted and pre-cleared chromatin was incubated overnight with the appropriate antibody, anti-Rbp1-NTD (CST, D8L4Y, 15 μl) to detect total Pol II, anti-Rbp1-CTD-Ser5P (CST, D9N5I, 12.5 μl), or anti-Rbp1-CTD-Ser2P (CST, E1Z3G, 12.5 μl). Antibody-bound chromatin was isolated using blocked protein A agarose beads for 3 h at 4°C. Washes were performed with FA-Lysis buffer, FA-Lysis buffer containing 500 mM NaCl, DOC buffer (250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 2 mM EDTA, 10 mM Tris−HCl pH 8), followed by two washes with TE buffer pH 8. ChIP DNA was eluted, de-crosslinked and purified as described above, along with the matched input samples.
Native chromatin was diluted 10-fold in native ChIP incubation buffer (70 mM NaCl, 10 mM Tris−HCl pH 7.5, 2 mM MgCl2, 2 mM EDTA, 0.1% TritonX-100, 10 mM NEM, 1x PIC). For each ChIP reaction, 1 ml of diluted nucleosomes was incubated overnight at 4°C with the appropriate antibody, anti-H2AK119ub1 (CST, D27C4, 5 μL), anti-H3K27me3 (in-house, 5 μL) or anti-H3K4me3 (in-house, 3 μL). Antibody-bound nucleosomes were captured by incubation for 1 h at 4°C with protein A agarose beads, pre-blocked overnight in native ChIP incubation buffer supplemented with 1 mg/ml BSA and 1 mg/ml yeast tRNA. The beads were then washed four times with native ChIP wash buffer (20 mM Tris−HCl pH 7.5, 2 mM EDTA, 125 mM NaCl, 0.1% TritonX-100) and once with TE buffer pH 8. Immunoprecipitated DNA was eluted using 100 μl of elution buffer (1% SDS, 0.1 M NaHCO3) and purified using ChIP DNA Clean and Concentrator Kit. DNA from a matched input sample (corresponding to 10% of the original ChIP reaction) purified in the same way.
cChIP-seq libraries for both ChIP and input samples were prepared using NEBNext Ultra DNA Library Prep Kit for Illumina (double-crosslinked and native ChIP-seq) or NEBNext Ultra II DNA Library Prep Kit for Illumina (Pol II ChIP-seq), following manufacturer’s guidelines. Samples were indexed using NEBNext Multiplex Oligos. The average size and concentration of all libraries was analysed using the 2100 Bioanalyzer High Sensitivity DNA Kit (Agilent) followed by qPCR using SensiMix SYBR (Bioline, UK) and KAPA Illumina DNA standards (Roche). Libraries were sequenced as 40 bp paired-end reads on the Illumina NextSeq 500.
Chromatin immunoprecipitation followed by qPCR
For PCGF2 and RYBP ChIP-qPCR, double-crosslinked chromatin was prepared as described above. Chromatin was diluted and pre-cleared using protein A agarose beads (Repligen) and incubated overnight with either anti-PCGF2 (in house, 5 μl), or anti-RYBP (Millipore, AB3637, 5 μl) antibody. ChIP and Input DNA were purified as described above. ChIP-experiments were carried out in biological triplicates, and quantitative PCR was performed in technical duplicates using SensiMix SYBR mix (Bioline) and primers listed in Supplementary Table 2.
Single molecule RNA fluorescence in situ hybridization
smRNA-FISH probe sets comprised 48 individually labelled oligos that were 20-22 nt in length and designed using the online tool from the manufacturer (Stellaris). Mouse ESCs were harvested at designated times after treatment with IAA, fixed with 3.7 % formaldehyde for 10 min and then incubated in 70% (v/v) ethanol/PBS solution for at least 1 hour at 4°C. smRNA-FISH was carried out in suspension following the standard guidelines from the manufacturer (Stellaris). Briefly, the cells were pelleted and resuspended in buffer containing 10% formamide and 2x SSC (buffer A), followed by centrifugation at 300 g and further resuspension in hybridisation mixture comprising 20% dextran sulfate, 10% formamide, 2x SSC and probe sets specific to exons (conjugated with Q570) or introns (with Q670), in a total volume of 200 μl per cell pellet. After overnight incubation at 37°C, the cells were washed twice with buffer A. DNA was stained with DAPI dissolved in buffer A and cell membranes with Agglutinin conjugated with Alexa 488 (Thermofisher) in PBS. After a final wash and PBS aspiration the cell pellet’s volume was approximately 150 μl; 6 μl of the pellet was mixed with 10 μl of Vectashield (H-1000, Vectorlabs) and spread on a glass slide. The solution was covered with microscopy coverslip and pressed to remove excess mounting medium and ensure cell monolayer.
Polycomb target genes for study by smRNA-FISH were chosen based on transcript information from cnRNA-seq and fulfilled following criteria: they spanned a wide range of basal expression levels and had varying magnitude of derepression upon RING1B depletion (Extended Data Fig. 7A). Control genes (Tfrc and Hspg2) had moderate expression levels and were classified as non-PcG (see below). Whole-cell smRNA-FISH results were cross-validated with cnRNA-seq and were mostly in good agreement (Extended Data Fig. 7B).
Microscopy
Microscopy was carried out using Olympus IX-83 system and Cell Sens software, equipped with 63x 1.4 NA oil objective lens and 1200x1200px2 sCMOS camera (Photometrics), 91.5 nm pixel size. Fluorescence was excited with LED light (Coolled): 365 nm for DAPI, 488 nm for Alexa 488, 550 nm for Q570, and 635 nm for Q670. 3D-stacks contained 30 images and were acquired every 350 nm. Typically, 40 to 60 fields of view were acquired per sample, yielding between 1500 − 3000 cells in total from 3 biological replicates.
Quantification and Statistical Analysis
Sequencing data processing and normalization
For cnRNA-seq, paired-end reads were first aligned using Bowtie 2 (“−very-fast,” “−no-mixed” and “−no-discordant” options)111 against the concatenated mm10 and dm6 rDNA genomic sequence (GenBank: BK000964.3 and M21017.1) in order to discard the reads mapping to rDNA. The remaining unmapped reads were then aligned against the concatenated mm10 and dm6 genome using STAR112. Reads which failed to map using STAR were further aligned against the concatenated mm10 and dm6 genome using Bowtie 2 (“−sensitive-local,” “−no-mixed” and “−no-discordant” options), to improve the overall mapping of intronic sequences of nascent transcripts present in nuclear RNA fraction. Uniquely mapped reads from the two alignment steps were combined and PCR duplicates were removed using Sambamba113 before the further analysis.
For cChIP-seq, paired-end reads were aligned to the concatenated mouse (mm10) and spike-in (dm6 for native, hg19 for cross-linked cChIP-seq) genome sequences using Bowtie 2 (“−no-mixed” and “−no-discordant” options). Only uniquely mapped reads were kept for downstream analysis, after removal of PCR duplicates with Sambamba. All cnRNA-seq and cChIP-seq experiments performed in this study are listed in Supplementary Table 3 along with the number of uniquely aligned reads for both mouse and spike-in genomes.
For visualization of cChIP-seq sequencing data and annotation of genomic regions with read counts, uniquely mapped mouse reads were normalized using dm6 or hg19 spike-in29. Briefly, mouse reads were randomly subsampled based on the total number of spike-in (dm6 or hg19) reads in each sample. Additionally, to account for possible minor variations in spike-in cell mixing between cChIP-seq replicates, we corrected the subsampling factors by using the ratio of spike-in to mouse total read counts in the corresponding input samples. Prior to merging normalized replicates for visualization and analysis, read coverage across regions of interest (RING1B peaks for RING1B, SUZ12, H2AK119ub1 and H3K27me3, or TSS ± 2.5 kb for total Pol II, Ser5P-Pol II and H3K4me3, or gene bodies for Ser2P) was analyzed using multiBamSummary and plotCorrelation functions from deepTools (v3.1.1)114, confirming a high correlation between replicates (Pearson's correlation coefficient > 0.9) (Supplementary Table 4). Genome coverage tracks for cChIP-seq were generated using the pileup function from MACS2115 and visualized using the UCSC genome browser116. Differential genome coverage tracks (log2 ratio of the two conditions) were made by using bigwigCompare from deeptools.
Peak calling
To identify genomic regions bound by PRC1 we performed peak calling using MACS2 (“BAMPE” and “−broad” options) on the RING1B cChIP-seq data with corresponding input samples for background normalization. A set of peaks identified in all biological replicates of untreated PRC1deg cells was used for further analysis, after filtering out several peaks identified following 4 hours of auxin treatment (i.e. following loss of RING1B) and manually removing sequencing artefacts. In total, 7240 stringent RING1B peaks were identified.
Differential gene expression analysis and gene annotation
For differential gene expression analysis, we obtained read counts from the original bam files prior to spike-in normalization for a non-redundant mouse gene set using a SAMtools-based custom Perl script. The non-redundant mm10 gene set (n = 20633) was derived from mm10 refGene genes by filtering out very short genes with poor sequence mappability and highly similar transcripts. To identify significant gene expression changes following auxin treatment, we used a custom R script that incorporates spike-in calibration into DESeq2 analysis117 and uses “apeglm” method for LFC shrinkage118. Spike-in calibration was incorporated by using read counts from a set of unique dm6 refGene genes to calculate DESeq2 size factors for normalization of raw mm10 read counts, as previously described29,119. For a change to be considered significant, we applied a threshold of p-adj < 0.05 and fold change > 1.5. Based on the earliest time point when these thresholds were reached, derepressed genes were classified into three groups (“2 hours”, “4 hours” and “8 hours”). For visualization purposes, DESeq2-normalized read counts were averaged across the replicates and used to calculate RPKM. Log2 fold change values were visualized in R using ggplot2 to make MA plots and boxplots, and ComplexHeatmap120 for heatmap in Fig. 2D. Correlation in log2 fold change or log2 RPKM values was computed using cor.test function in R, expressed as Pearson correlation coefficient and visualized with ggplot2 using scatterplot colored by density with stat_density2d. Linear regression line was plotted using geom_smooth along with the R2 coefficient of determination.
Mouse genes from the custom non-redundant set (n = 20633) that was used for differential gene expression analysis were classified into three groups based on the overlap of their promoters (TSS ± 2500 bp) with RING1B-bound sites and non-methylated CpG islands (NMI). NMIs are regions enriched in non-methylated CpG dinucleotides, identified using MACS2 peak calling on BioCAP-seq data (n = 27047)121. Genes with promoters that do not overlap NMIs were classified as non-NMI (n = 5534). The rest of the genes were defined as Polycomb-occupied genes (PcG, n = 6937) if their promoters overlapped RINGB-bound sites identified in this study, and those that did not overlap as non-PcG genes (n = 8162). For genes with several alternative transcripts and promoters, we assigned all transcripts into the PcG group if at least one promoter overlapped a RINGB-bound site.
Read count quantitation and enrichment analysis for cChIP-seq
Metaplot and heatmap analysis of cChIP-seq read density at regions of interest was performed with computeMatrix and plotProfile/plotHeatmap from deepTools114. Metaplot profiles represented mean scores over sets of genomic regions, unless stated otherwise. Read coverages for genomic regions of interest from merged spike-in normalized replicates were computed with multiBamSummary from deeptools (“−outRawCounts”), and used for comparative boxplot analysis. To characterize low-level genomic blanket of H2AK119ub1, we counted reads in a set of 100 kb windows spanning the genome (n = 27282) defined by using the makewindows function from BEDtools (v2.17.0). RPKM values were calculated by dividing normalized read counts by a genomic interval size in kb and visualized with boxplots using custom R scripts and ggplot2. For boxplots comparing H2AK119ub1 cChIP-seq signal at RING1B peaks and 100 kb genomic windows, RPKM values are shown relative to the median signal at RING1B peaks in untreated cells.
Differential enrichment analysis of cChIP-seq data was performed similarly to the gene expression analysis described above, with a few differences. Namely, read counts from individual biological replicates prior to spike-in normalization were obtained with multiBamSummary from deeptools (“−outRawCounts”). Mouse reads were counted in the promoter regions (TSS ± 2.5 kb) of the custom non-redundant mm10 gene set, and over whole gene bodies (TSS to TES) for the analysis of total Pol II. Spike-in genomic reads were counted in the appropriate control regions: human (hg19) CpG islands obtained from UCSC Table Browser for RING1B and SUZ12, human (hg19) promoter regions (TSS ± 2.5 kb) of a custom unique gene set for Pol II, fly (dm6) promoter regions (TSS ± 2 kb) of a custom unique gene set for H3K4me3. Prior to quantification, spike-in reads were pre-normalized using the spike-in/mouse read ratio derived from the corresponding input sample in order to account for minor variations in spike-in cells mixing. For a change to be called significant, we applied a threshold of p-adj < 0.05 and fold change > 1.5.
Image Analysis
3D images were pre-processed using a custom ImageJ script which we call ThunderFISH. In brief, 3D smRNA-FISH images containing sparse transcripts appearing as diffraction limited spots were converted into 2D images through maximal projection. Due to low numbers of transcripts per cell this operation did not significantly affect the number of transcripts detected when compared with other spot counting methods relying on 3D detection (Extended Data Fig. 6D, E). Slight transcript undercounting was identified only in cells with more than approximately 70 transcripts/cell (Extended Data Fig. 6D). In contrast, most of the samples we imaged contained on average only a few transcripts per cell. In parallel, a threshold was applied on 2D average projections of DAPI and Agglutinin channels, and after converting into binary image and water-shedding, single cell masks were obtained (Extended Data Fig. 6A). The masks with low circularity, touching image boarders, and outside of a specified size range were discarded. Next, the remaining cell masks were used to extract single cell 2D smRNA-FISH images from the full field of view. Lastly, single cell images were converted to a data stack with each frame representing a single 2D smRNA-FISH image of an individual cell. This stack was used as input for diffraction-limited spot counting by the dedicated ImageJ plugin ThunderSTORM102. We used default settings, except for the threshold factor, which we adjusted to be 6-10x the background standard deviation. This range of values provided comparable transcripts per cell distributions (Extended Data Fig. 6B), hence we used it for our transcript detection. ThunderSTORM outputs the number of spots detected per frame (per cell). We used these numbers to produce density graphs of the transcript distributions using R and the ggplot2 package. The pre-processing by ThunderFISH allowed increased imaging and analysis throughput of up to 10,000 cells a day. This proved essential for accurately and efficiently counting Polycomb target gene transcripts, which are lowly expressed, and for extracting robust measures of expression change when the Polycomb system was disrupted.
Nascent RNA-FISH data were analysed using a custom-made ImageJ script utilizing 3D Objects counter104 that outputs number of nascent spots per cell with respective integrated intensity.
Two-state model
All transcript distributions were positively skewed and over-dispersed with a variance at least 2x larger than a mean, and poorly fitted a Poisson distribution (Extended Data Fig. 8A). In order to extract inferred mean transcription burst size (number of transcripts produced in a single transcription event), the transcript distribution in a cell population was approximated using a negative binomial distribution fit using the vcd package in R. This distribution is expected when the probability of burst initiation is constant in time and number of transcripts produced per burst is expected to follow exponential distribution. The mean burst size was calculated as b = (1-p)/p, where p denotes probability of promoter transition to the OFF state extracted from the individual distribution Maximum Likelihood estimation, similarly to as has been described previously101,122. The goodness of fit of the negative binomial distribution estimated p-values (χ2 test) in the majority of the cases exceeded 0.1 (Extended Data Fig. 8B) and were many orders of magnitude greater than if a Poisson distribution was assumed. Therefore, a negative binomial distribution was a better fit to the smRNA-FISH transcript-count distributions.
The frequency of transcription bursts, kON, for individual transcript distributions was computed using following equation123: kON = μƐ/b, where μ represents mean number of transcripts per cell, Ɛ corresponds to transcript half-life, and b corresponds to transcription burst size obtained from negative binomial fit. Since fold changes in kON were computed, transcript life-time Ɛ cancelled out as it was assumed constant and independent of auxin treatment.
Decomposition of sources of transcript heterogeneity
The two-state model assumes that transcript variability merely stems from an intrinsic stochastic nature of the studied promoter. In order to prove validity of this assumption we sought assessment of extrinsic sources of heterogeneity through decomposing overall transcript distribution heterogeneity. Previously it has been demonstrated that apart from intrinsic behaviour of the promoter, cell cycle and cell volume contribute most to the transcript heterogeneity in a cell population124. We used the previously published formula for cell volume-corrected noise measure N125 to estimate the contribution of cell area (proxy for cell volume) and DAPI fluorescence intensity (proxy for cell cycle) to overall measured noise expressed as squared coefficient of variation of transcript distribution CV2:
| (1) |
where <V> represents either average area or DAPI intensity, <m> is average number of transcripts, A and B correspond to the intercept and slope of a linear fit, respectively, of which the best line-fit for the relationship of transcripts and tested extrinsic parameter. Cov(m, V) stands for covariance between number of transcripts m and tested extrinsic parameter V. We note that for the most of the genes the linear slope B oscillates around 0, effectively cancelling the expression on the right side of the equation 1. Moreover, we note that cell area and DAPI intensity correlated well (Pearson’s r = 0.74, Extended Data Fig. 9C), likely both representing to some extent the same extrinsic cell parameter (likely the cell volume). Nucleus volume has been reported to increase in G2125, and since the nucleus constitutes the majority of the mouse ESC cell volume, correlation between DAPI intensity and total cell volume in ESCs can be expected, opposite to what was previously reported for another cell type125. Hence, our overall extrinsic noise estimation in spite of yielding miniscule values, is likely still a subject to overestimation. Nevertheless, intrinsic noise for all Polycomb target genes dominated total noise by >90%, and for vast majority of genes almost entirely (Extended Data Fig. 9D). Hence, raw transcript-per-cell distributions were used to infer transcription kinetics based on the two-state model.
Extended Data
Extended Data Fig. 1. Detailed characterisation of the PRC1deg.
(A) Schematic of the PRC1deg system. Both endogenous Ring1B alleles are N-terminally fused to the full-length AID tag in a Ring1A-/- background, while OsTIR1 is inserted in Rosa26 locus.
(B) Western blot analysis of RING1A and RING1B in the control cell line (TIR1) and PRC1deg cells (left panel) with a quantification of RING1B levels (right panel). Shown are values and mean from n=2 independent experiments.
(C) Genomic snapshots of typical Polycomb target genes showing cChIP-seq signal for RING1B in wild-type (TIR1) and PRC1deg cells before auxin treatment.
(D) Metaplot analysis of RING1B cChIP-seq at RING1B-bound sites in wild-type (TIR1) and PRC1deg cells before auxin treatment. Maximal read density in PRC1deg cells was set to 1.
(E) A scatterplot showing the relationship between RING1B cChIP-seq signal at RING1B-bound sites in wild-type (TIR1) and PRC1deg cells before auxin treatment. R2 is the coefficient of determination for linear regression and cor is Pearson correlation coefficient.
(F) Immunoprecipitation of RING1B from TIR1 and PRC1deg nuclear extracts followed by western blot analysis of PRC1 components, and SUZ12 (PRC2 component) as a negative control. Shown are representative results from the two independent experiments. vPRC1 − variant PRC1 components, cPRC1 − canonical PRC1 components.
(G) Chromatin occupancy of PCGF2 (a canonical PRC1 complex component) at Polycomb target gene promoters and control loci in wild-type (TIR1) and PRC1deg cells before auxin treatment assessed by ChIP-qPCR. Individual biological replicates (n = 3) are shown as dots along with mean and SEM.
(H) As in (G), but for RYBP (a variant PRC1 complex component).
(I) Western blot analysis of PRC1 components and SUZ12 in the nuclear extracts of the PRC1deg cells following IAA treatment. CFP1 serves as a loading control. Shown are representative results from the two independent experiments.
(J) Boxplots comparing H2AK119ub1 cChIP-seq signal before and after IAA treatment at RING1B-bound sites (n = 7240) and over 100 kb genomic windows (n = 27282). Read coverages represent merged spike-in normalised values from n=3 biologically independent experiments. All signal is normalised to the median RPKM value of RING1B-bound sites in untreated cells. Boxes show interquartile range, center line represents median, whiskers extend by 1.5x IQR, while notches extend by 1.58x IQR/sqrt(n), giving a roughly 95% confidence interval for comparing medians.
Extended Data Fig. 2. Expression and chromatin features of genes derepressed after PRC1 removal.
(A) Scatterplots comparing the log2 fold changes in gene expression (cnRNA-seq) at different time points after auxin treatment in PRC1deg cells with gene expression changes in PRC1CKO (72h OHT) cells. R2 is the coefficient of determination for linear regression and cor is Pearson correlation coefficient.
(B) An UpSet plot for genes with significantly increased expression (p-adj < 0.05, fold change > 1.5) following auxin treatment of PRC1deg cells at the indicated time points, or a 72h OHT treatment of PRC1CKO cells. The total number of genes in each set is shown in the bar chart on the left. Non-empty intersections were sorted by size, while excluding any intersection with less than 10 members for clarity. Intersections not included in the PRC1CKO (72h OHT) set are highlighted in green and may imply compensatory effects on expression, while the blue column denotes gene changes exclusive to the PRC1CKO and could correspond to secondary effects on expression.
(C) Boxplots comparing the log2 fold changes in expression of genes split into three groups based on the earliest time point when they became derepressed. Log2 fold changes are derived from n=3 biologically independent experiments. Dotted line represents 1.5-fold change. Boxes show interquartile range, center line represents median, whiskers extend by 1.5x IQR, while notches extend by 1.58x IQR/sqrt(n), giving a roughly 95% confidence interval for comparing medians.
(D) Boxplots comparing the gene expression levels in the untreated PRC1deg cells for the three groups of derepressed genes defined in (B). RPKM values represent mean of n=3 biologically independent experiments. Boxplots are defined as in (C).
(E) Boxplots comparing the promoter (TSS ±2.5 kb) cChIP-seq signal for RING1B, H2AK119ub1, SUZ12 and H3K27me3 in the untreated PRC1deg cells for the three groups of derepressed genes defined in (B). Read coverages represent merged spike-in normalised values from n=3 biologically independent experiments. Boxplots are defined as in (C).
(F) Boxplots comparing the promoter (TSS ± 2.5 kb) cChIP-seq signal for total Pol II and H3K4me3 in the untreated PRC1deg cells for the three groups of derepressed genes defined in (B). Read coverages represent merged spike-in normalised values from n=3 biologically independent experiments. Boxplots are defined as in (C).
Extended Data Fig. 3. Characterisation of PRC2 and H3K27me3 after PRC1 removal.
(A) Heatmap analysis of SUZ12 cChIP-seq at TSSs for the three groups of genes defined by the earliest time of derepression in PRC1deg cells treated with IAA for the indicated times. Heatmaps are sorted by RING1B cChIP-seq signal in untreated cells.
(B) Boxplots illustrating log2 fold changes in expression (cnRNA-seq, left panel), SUZ12 cChIP-seq signal (middle) or H3K27me3 cChIP-seq signal (right) at promoters (TSS ± 2.5 kb) of Polycomb target genes showing a significant reduction in SUZ12 levels following 2 hours of IAA treatment. Log2 fold changes are derived from n=3 biologically independent experiments. Genes are divided into Polycomb target genes that become derepressed (UP, n = 955) and those that do not change in expression (n.s., n = 3739) by 2 hours. SUZ12 binding was reduced in both groups, while H3K27me3 levels were only modestly affected. Boxes show interquartile range, center line represents median, whiskers extend by 1.5x IQR, while notches extend by 1.58x IQR/sqrt(n), giving a roughly 95% confidence interval for comparing medians.
(C) Boxplots comparing the promoter (TSS ± 2.5 kb) cChIP-seq signal of PRC2 components for the two groups of genes defined in (B), in the wild-type state. Polycomb target genes derepressed after 2 hours of PRC1 depletion are characterised by high levels of PRC2 components, in agreement with this group of genes being highly enriched in Polycomb chromatin domain features. SUZ12 cChIP-seq data is from this study (PRC1deg cells), while JARID2, AEBP2, PCL2 and EPOP are from 30. Read coverages represent merged spike-in normalised values from n=3 biologically independent experiments. Boxplots are defined as in (B).
(D) As in (A) but for H3K27me3 cChIP-seq.
(E) The dynamics of reduction in H3K27me3 cChIP-seq signal at RING1B-bound sites which show a significant reduction in H3K27me3 levels by 24 hours of IAA treatment (n = 5926). Central line represents median, with interquartile range shown as shaded area. A theoretical exponential decay function is shown as dashed line, assuming that H3K27me3 levels are halved with every cell cycle if maintenance is completely disrupted. The doubling time of mouse ESCs is approximately 12 hours.
(F) Genomic snapshots of typical Polycomb target genes showing cChIP-seq signal for H3K27me3 in wild-type (E14) and PRC2deg cells before dTAG-13 treatment.
(G) Metaplot analysis of H3K27me3 cChIP-seq at RING1B-bound sites in wild-type (E14) and PRC2deg cells before dTAG-13 treatment. Maximal read density in E14 cells was set to 1.
(H) A scatterplot showing the relationship between H3K27me3 cChIP-seq signal at RING1B-bound sites in wild-type (E14) and PRC2deg cells before dTAG-13 treatment. R2 is the coefficient of determination for linear regression and cor is Pearson correlation coefficient.
(I) Metaplot analysis of H3K27me3 cChIP-seq at RING1B-bound sites in PRC2deg cells before and after 2h dTAG-13 treatment. Maximal read density in untreated cells was set to 1.
Extended Data Fig. 4. Effects on total Pol II and H3K4me3 at genes after PRC1 removal.
(A) Heatmaps illustrating RING1B binding in untreated cells, and total Pol II and H3K4me3 cChIP-seq signal at TSSs of the three groups of genes defined by the earliest time of derepression in PRC1deg cells treated with IAA for the indicated times. Heatmaps are sorted by RING1B signal in untreated cells.
(B) Boxplots comparing the log2 fold changes in total Pol II cChIP-seq signal following IAA treatment for genes split into three groups defined by the earliest time of derepression (2 hours, n = 1044; 4 hours, n = 1822; 8 hours, n = 2017). Log2 fold changes are derived from n=3 biologically independent experiments. The analysis was done at promoters (TSS ± 2.5 kb) and over gene bodies (TSS to TES). Boxes show interquartile range, center line represents median, whiskers extend by 1.5x IQR, while notches extend by 1.58x IQR/sqrt(n), giving a roughly 95% confidence interval for comparing medians.
(C) Boxplots comparing the log2 fold changes in H3K4me3 cChIP-seq signal following IAA treatment at promoter regions (TSS ± 2.5 kb) for genes split into three groups defined by the earliest time of derepression (2 hours, n = 1044; 4 hours, n = 1822; 8 hours, n = 2017). Log2 fold changes are derived from n=3 biologically independent experiments. Boxplots are defined as in (B).
Extended Data Fig. 5. Effects on Pol II phosphorylation state after PRC1 removal.
(A) Genomic snapshot of Hoxd locus showing cChIP-seq signal for RING1B, total Pol II, Ser5P-Pol II and Ser2P-Pol II in untreated PRC1deg cells and 2 hours after IAA addition.
(B) Heatmaps illustrating RING1B, total Pol II, Ser5P-Pol II and Ser2P-Pol II levels and changes in polymerase occupancy following 2h of IAA treatment for three groups of genes defined by the earliest time of derepression. Heatmaps are sorted by RING1B signal in untreated cells.
(C) Metaplot analysis of Ser5P-Pol II cChIP-seq at promoters of the three groups of genes defined by the earliest time of derepression in untreated PRC1deg cells and 2 hours after IAA addition. The profiles represent the median signal over the shown genomic region.
(D) As in (C) but for Ser2P-Pol II cChIP-seq over gene bodies.
Extended Data Fig. 6. Detailed characterisation of the smRNA-FISH transcript counting approach.
A) A schematic illustrating our automated approach to analyse smRNA-FISH in a high throughput manner. This approach enables effective single-cell segmentation, conversion of the field of view into single-cell smRNA-FISH images, in which individual transcripts can then be counted using ThunderSTORM102.
B) An illustration of the effectiveness of ThunderSTORM threshold factor (TF) value identification for spot (transcript) detection in cells. An Hspg2 dataset is shown as an example. The TF unit is set at a standard deviation of the image background. Spots per cell (mean ± SD; top) and heterogeneity of detected transcript levels expressed as coefficient of variation (middle) are shown for a range of TF values. Vertical red lines indicate the range of TF values yielding similar spot-counting outcomes and which were ultimately employed in this study. Density plots (bottom) demonstrate transcript per cell distributions depending on the TF used. TFs from 6 to 10 yield comparable spot-counting values. Very large or very low TF values lead either to overcounting or undercounting of transcript signals.
C) An illustration of the reproducibility between technical and biological replicates for our smRNA-FISH transcript counting approach. Mean transcripts per cell (top), coefficient of variation (middle), and mean transcription burst size inferred from the 2-state model (bottom) are shown. Error bars in the top panel represent 10% of standard deviation of transcripts per cell distributions around the mean.
D) To ensure the robustness of our transcript counting approach, we compared it to other spot (transcript) counting methods and manual counting of transcripts in 50 cells. The methods compared are: 3D-FISH QUANT103 (left), 3D Objects Counter104 (middle), and technique used in this study (right). The right panel indicates that our technique can be prone to a slight undercounting when number of transcripts per cell exceeds 60-70, but otherwise performs comparably to other approaches. Pearson correlation coefficient (r) and slope derived from linear regression are presented.
E) Transcript counting using 3D-FISH Quant and 3D Objects Counter correlate well with the transcripts counting using our approach.
Extended Data Fig. 7. Effects on absolute transcript numbers in single cells after PRC1 removal.
A) An MA plot depicting gene expression (cnRNA-seq) changes following 4h IAA treatment in PRC1deg cells with candidate genes for smRNA-FISH highlighted. The genes chosen span a wide range of initial transcript levels and transcript increase upon RING1B removal. The control genes are highlighted in blue and Polycomb target genes in black.
B) Correlation of log2 fold changes between cnRNA-seq and smRNA-FISH after 4 hours of IAA treatment. Hollow dots represent genes excluded from the linear fit. Dots represent mean values while error bars represent standard error of 3 biological replicates.
C) Normalised histograms illustrating the distribution of transcripts per cell over the time course of RING1B removal for all genes studied with smRNA-FISH. Shown is a representative biological replicate of 3 independent experiments. n indicates minimum number of cells measured at any given time-point per gene dataset.
D) A heatmap of mean fold changes in transcripts per cell over the time course of RING1B removal. Numbers represent the mean of 3 biological replicates.
E) As in (D) but representing the mean number of transcripts per cell.
Extended Data Fig. 8. Analysis of transcription burst size and frequency after PRC1 removal.
A) Examples of model fits to cellular transcripts distributions for all the genes examined in untreated state. The negative binomial fit is in red and density in blue. The Poisson distribution fit is indicated for comparison as a dashed purple line.
B) A heatmap of the goodness of fit p-value (one-sided Chi-square test) throughout the time course of RING1B depletion. High p-value (light yellow) represents a good negative binomial fit to the data.
C) Barplots representing mean inferred transcription burst sizes for Polycomb target genes and control genes. Error bars correspond to standard error of 3 biological replicates.
D) The relationship between nascent spot intensity (active transcription site, measured using nascent smRNA-FISH targeting intronic sequences) and transcription burst size inferred from mRNA-FISH (targeting exonic sequences) reveals that genes with higher predicted burst size values have greater nascent spot intensity. Dots represent mean value while error bars correspond to standard deviation of 3 biological replicates.
E) Heatmaps illustrating the fold change in inferred transcription burst size (left) and burst frequency (right) over the time course of RING1B depletion for the panel of genes studied.
F) Examples of the relationship between square coefficient of variation (CV) and mean number of transcripts per cell demonstrate that Polycomb target genes derepressed upon PRC1 removal experience increase in transcript number while simultaneously retaining constant transcription burst size. Dashed lines represent theoretical relation between CV2 and the mean number of transcripts at constant burst size values (CV2=b/mean#mRNA) with changing burst frequencies. Dots represent mean values while error bars correspond to the standard deviation of 3 biological replicates.
Extended Data Fig. 9. Active allele distribution in the cell population and transcription noise decomposition.
A) Frequency of nascent spots representing active alleles in cell population (mean percentage of cells with 0, 1, or >1 nascent spot per cell obtained in 3 biological replicates). For comparison a simulated negative binomial distribution of active alleles per cell is shown assuming their expression is independent and random. The simulated distribution assumes the same frequency of spots as in the experimental data and 3 alleles per cell because a large proportion of ESCs will exist in S-phase at any given point. Importantly, the independence of allele expression is maintained 4h after removal of PRC1 by the addition of IAA. A minimum of 887 cells was measured per biological replicate.
B) Pearson’s correlation coefficient between number of transcripts per cell and either DAPI intensity per cell (a proxy for the cell cycle stage as G2 cells have generally 2x the genetic material of the cells in G1), or cell area (representing cell volume) − the two primary sources of extrinsic noise.
C) i − iii) Examples of area or DAPI signal per cell plotted against transcripts per cell for a control (Hspg2) or Polycomb target genes (Zic2, E2f6). Each data-point represents a single cell measurement. Shading around regression line represents 95% confidence interval. iv) Correlation between cell area and DAPI signal intensity (Pearson’s r = 0.74) suggests that volume of ESCs is strongly related to their cell cycle phase. We note that the cell nucleus in ESCs occupies a significant portion of the total cell volume.
D) Specific heterogeneity (noise) in transcripts per cell expressed as % of the total heterogeneity measured as square coefficient of variation of the transcripts per cell distributions. Cell area and DAPI signal intensity contribute very little to the overall variability in number of transcripts per cell in a population for Polycomb target genes. We note, that this measure of extrinsic variability likely suffers from overestimation as DAPI intensity and cell area are moderately correlated, and hence to some extent represent the same source of variability (see C).
Supplementary Material
Acknowledgements
Work in the Klose lab is supported by the Wellcome Trust (209400/Z/17/Z) (R.J.K.), the European Research Council (681440) (R.J.K.) and the Lister Institute of Preventive Medicine (R.J.K.). We are grateful to Amanda Williams at the Department of Zoology, University of Oxford, for sequencing support on the NextSeq 500. We express our gratitude to Darragh Ennis and Ilan Davis for their advice in RNA-FISH probe design. We thank Nadezda Fursova for her valuable advice on computational analysis. We are grateful to Nadezda Fursova, Neil Blackledge and Emilia Dimitrova for critical reading of the manuscript.
This version of the article has been accepted for publication, after peer review and is subject to Springer Nature’s AM terms of use, but is not the Version of Record and does not reflect post-acceptance improvements, or any corrections. The Version of Record is available online at: http://dx.doi.org/10.1038/s41594-021-00661-y
Footnotes
Author contributions
Conceptualization, P.D., A.S. and R.J.K.; Methodology, Investigation and Formal Analysis, P.D. and A.S.; Writing - Original Draft, P.D., A.S. and R.J.K.; Writing − Review and Editing, P.D., A.S. and R.J.K.; Funding Acquisition and Supervision, R.J.K.
Competing Interests
The authors declare no competing interests.
Data Availability
The high-throughput sequencing datasets generated for this study are available in GEO database under the accession number GSE159400. Published data used in this study include BioCAP-seq (GEO: GSE43512) from 121, cnRNA-seq in Ring1a-/-; Ring1bfl/fl (PRC1CKO)(GEO: GSE119619) from 29 and cChIP-seq for JARID2, AEBP2, PCL2 and EPOP in untreated (wt) Ring1aI50A/D53K; Ring1b(WT->I53A/D56K)fl/fl ESCs (GEO: GSE132754) from 30. For cnRNA-seq processing we used mm10 (GenBank: BK000964.3, https://www.ncbi.nlm.nih.gov/nuccore/bk000964) and dm6 (GenBank: M21017.1, https://www.ncbi.nlm.nih.gov/nuccore/M21017.1) rDNA genomic datasets.
Code availability
All R and Perl scripts used for genomic data analysis in this study are available at https://github.com/pauladobrinic/PRC1degron-2021. A custom-made ImageJ script for pre-processing 3D images (ThunderFISH) is publicly available with detailed manual of sample preparation and script usage at https://github.com/aleks-szczure/ThunderFISH.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackledge NP, Rose NR, Klose RJ. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol. 2015;16:643–649. doi: 10.1038/nrm4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuettengruber B, Bourbon H-M, Di Croce L, Cavalli G. Genome regulation by polycomb and trithorax: 70 years and counting. Cell. 2017;171:34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Voncken JW, et al. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci USA. 2003;100:2468–2473. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faust C, Schumacher A, Holdener B, Magnuson T. The eed mutation disrupts anterior mesoderm production in mice. Development. 1995;121:273–285. doi: 10.1242/dev.121.2.273. [DOI] [PubMed] [Google Scholar]
- 7.Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Carroll D, et al. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, et al. Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J Biol Chem. 2006;281:20643–20649. doi: 10.1074/jbc.M602461200. [DOI] [PubMed] [Google Scholar]
- 11.Buchwald G, et al. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 13.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Müller J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 15.Czermin B, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 16.Farcas AM, et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife. 2012;1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He J, et al. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol. 2013;15:373–384. doi: 10.1038/ncb2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, et al. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature. 2017;549:287–291. doi: 10.1038/nature23881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perino M, et al. MTF2 recruits Polycomb Repressive Complex 2 by helical-shape-selective DNA binding. Nat Genet. 2018;50:1002–1010. doi: 10.1038/s41588-018-0134-8. [DOI] [PubMed] [Google Scholar]
- 20.Wu X, Johansen JV, Helin K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell. 2013;49:1134–1146. doi: 10.1016/j.molcel.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 21.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ku M, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blackledge NP, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper S, et al. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 2014;7:1456–1470. doi: 10.1016/j.celrep.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalb R, et al. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol. 2014;21:569–571. doi: 10.1038/nsmb.2833. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, et al. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Min J, Zhang Y, Xu R-M. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klose RJ, Cooper S, Farcas AM, Blackledge NP, Brockdorff N. Chromatin sampling--an emerging perspective on targeting polycomb repressor proteins. PLoS Genet. 2013;9:e1003717. doi: 10.1371/journal.pgen.1003717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fursova NA, et al. Synergy between Variant PRC1 Complexes Defines Polycomb-Mediated Gene Repression. Mol Cell. 2019;74:1020–1036.:e8. doi: 10.1016/j.molcel.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blackledge NP, et al. PRC1 catalytic activity is central to polycomb system function. Mol Cell. 2020;77:857–874.:e9. doi: 10.1016/j.molcel.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Endoh M, et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135:1513–1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- 32.Natsume T, Kiyomitsu T, Saga Y, Kanemaki MT. Rapid Protein Depletion in Human Cells by Auxin-Inducible Degron Tagging with Short Homology Donors. Cell Rep. 2016;15:210–218. doi: 10.1016/j.celrep.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 34.Rhodes JDP, et al. Cohesin Disrupts Polycomb-Dependent Chromosome Interactions in Embryonic Stem Cells. Cell Rep. 2020;30:820–835.:e10. doi: 10.1016/j.celrep.2019.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujimura Y, et al. Distinct roles of Polycomb group gene products in transcriptionally repressed and active domains of Hoxb8. Development. 2006;133:2371–2381. doi: 10.1242/dev.02405. [DOI] [PubMed] [Google Scholar]
- 36.Leeb M, Wutz A. Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J Cell Biol. 2007;178:219–229. doi: 10.1083/jcb.200612127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cooper S, et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat Commun. 2016;7:13661. doi: 10.1038/ncomms13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beltran M, et al. G-tract RNA removes Polycomb repressive complex 2 from genes. Nat Struct Mol Biol. 2019;26:899–909. doi: 10.1038/s41594-019-0293-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, et al. Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat Struct Mol Biol. 2017;24:1028–1038. doi: 10.1038/nsmb.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu C, et al. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2) Proc Natl Acad Sci USA. 2010;107:19266–19271. doi: 10.1073/pnas.1008937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hansen KH, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 44.Laprell F, Finkl K, Müller J. Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science. 2017;356:85–88. doi: 10.1126/science.aai8266. [DOI] [PubMed] [Google Scholar]
- 45.Nabet B, et al. The dTAG system for immediate and target-specific protein degradation. Nat Chem Biol. 2018;14:431–441. doi: 10.1038/s41589-018-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huseyin MK, Klose RJ. Live-cell single particle tracking of PRC1 reveals a highly dynamic system with low target site occupancy. BioRxiv. 2020 doi: 10.1101/2020.04.25.061358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehmann L, et al. Polycomb repressive complex 1 (PRC1) disassembles RNA polymerase II preinitiation complexes. J Biol Chem. 2012;287:35784–35794. doi: 10.1074/jbc.M112.397430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakagawa T, et al. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008;22:37–49. doi: 10.1101/gad.1609708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stock JK, et al. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 50.Zhou W, et al. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chopra VS, et al. The polycomb group mutant esc leads to augmented levels of paused Pol II in the Drosophila embryo. Mol Cell. 2011;42:837–844. doi: 10.1016/j.molcel.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dellino GI, et al. Polycomb silencing blocks transcription initiation. Mol Cell. 2004;13:887–893. doi: 10.1016/s1097-2765(04)00128-5. [DOI] [PubMed] [Google Scholar]
- 53.King IFG, Francis NJ, Kingston RE. Native and recombinant polycomb group complexes establish a selective block to template accessibility to repress transcription in vitro. Mol Cell Biol. 2002;22:7919–7928. doi: 10.1128/MCB.22.22.7919-7928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hughes AL, Kelley JR, Klose RJ. Understanding the interplay between CpG island-associated gene promoters and H3K4 methylation. Biochim Biophys Acta Gene Regul Mech. 2020;1863:194567. doi: 10.1016/j.bbagrm.2020.194567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harlen KM, Churchman LS. The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat Rev Mol Cell Biol. 2017;18:263–273. doi: 10.1038/nrm.2017.10. [DOI] [PubMed] [Google Scholar]
- 56.Brookes E, et al. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell. 2012;10:157–170. doi: 10.1016/j.stem.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Min IM, et al. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams LH, et al. Pausing of RNA polymerase II regulates mammalian developmental potential through control of signaling networks. Mol Cell. 2015;58:311–322. doi: 10.1016/j.molcel.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kar G, et al. Flipping between Polycomb repressed and active transcriptional states introduces noise in gene expression. Nat Commun. 2017;8:36. doi: 10.1038/s41467-017-00052-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Torre E, et al. Rare Cell Detection by Single-Cell RNA Sequencing as Guided by Single-Molecule RNA FISH. Cell Syst. 2018;6:171–179.:e5. doi: 10.1016/j.cels.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodriguez J, Larson DR. Transcription in living cells: molecular mechanisms of bursting. Annu Rev Biochem. 2020;89:189–212. doi: 10.1146/annurev-biochem-011520-105250. [DOI] [PubMed] [Google Scholar]
- 63.Peccoud J, Ycart B. Markovian Modeling of Gene-Product Synthesis. Theor Popul Biol. 1995;48:222–234. [Google Scholar]
- 64.Zoller B, Little SC, Gregor T. Diverse Spatial Expression Patterns Emerge from Unified Kinetics of Transcriptional Bursting. Cell. 2018;175:835–847.:e25. doi: 10.1016/j.cell.2018.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fritzsch C, et al. Estrogen-dependent control and cell-to-cell variability of transcriptional bursting. Mol Syst Biol. 2018;14:e7678. doi: 10.15252/msb.20177678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Levesque MJ, Raj A. Single-chromosome transcriptional profiling reveals chromosomal gene expression regulation. Nat Methods. 2013;10:246–248. doi: 10.1038/nmeth.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tamburri S, et al. Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression. Mol Cell. 2020;77:840–856.:e5. doi: 10.1016/j.molcel.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gao Z, et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taherbhoy AM, Huang OW, Cochran AG. BMI1-RING1B is an autoinhibited RING E3 ubiquitin ligase. Nat Commun. 2015;6:7621. doi: 10.1038/ncomms8621. [DOI] [PubMed] [Google Scholar]
- 70.Rose NR, et al. RYBP stimulates PRC1 to shape chromatin-based communication between Polycomb repressive complexes. Elife. 2016;5 doi: 10.7554/eLife.18591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morey L, et al. Polycomb Regulates Mesoderm Cell Fate-Specification in Embryonic Stem Cells through Activation and Repression Mechanisms. Cell Stem Cell. 2015;17:300–315. doi: 10.1016/j.stem.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 72.Morey L, et al. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 2012;10:47–62. doi: 10.1016/j.stem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 73.Kundu S, et al. Polycomb Repressive Complex 1 Generates Discrete Compacted Domains that Change during Differentiation. Mol Cell. 2017;65:432–446.:e5. doi: 10.1016/j.molcel.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zepeda-Martinez JA, et al. Parallel PRC2/cPRC1 and vPRC1 pathways silence lineage-specific genes and maintain self-renewal in mouse embryonic stem cells. Sci Adv. 2020;6:eaax5692. doi: 10.1126/sciadv.aax5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cohen I, Bar C, Ezhkova E. Activity of PRC1 and histone H2AK119 monoubiquitination: revising popular misconceptions. Bioessays. 2020;42:e1900192. doi: 10.1002/bies.201900192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aihara H, et al. Histone H2A T120 phosphorylation promotes oncogenic transformation via upregulation of cyclin D1. Mol Cell. 2016;64:176–188. doi: 10.1016/j.molcel.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 77.Tsuboi M, et al. Ubiquitination-Independent Repression of PRC1 Targets during Neuronal Fate Restriction in the Developing Mouse Neocortex. Dev Cell. 2018;47:758–772.:e5. doi: 10.1016/j.devcel.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 78.Højfeldt JW, et al. Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nat Struct Mol Biol. 2018;25:225–232. doi: 10.1038/s41594-018-0036-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alabert C, et al. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 2015;29:585–590. doi: 10.1101/gad.256354.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oksuz O, et al. Capturing the Onset of PRC2-Mediated Repressive Domain Formation. Mol Cell. 2018;70:1149–1162.:e5. doi: 10.1016/j.molcel.2018.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reverón-Gómez N, et al. Accurate Recycling of Parental Histones Reproduces the Histone Modification Landscape during DNA Replication. Mol Cell. 2018;72:239–249.:e5. doi: 10.1016/j.molcel.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alabert C, et al. Domain model explains propagation dynamics and stability of histone H3K27 and H3K36 methylation landscapes. Cell Rep. 2020;30:1223–1234.:e8. doi: 10.1016/j.celrep.2019.12.060. [DOI] [PubMed] [Google Scholar]
- 83.Miller SA, Damle M, Kim J, Kingston RE. Full methylation of H3K27 by PRC2 is dispensable for initial embryoid body formation but required to maintain differentiated cell identity. Development. 2021;148 doi: 10.1242/dev.196329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Francis NJ, Saurin AJ, Shao Z, Kingston RE. Reconstitution of a functional core polycomb repressive complex. Mol Cell. 2001;8:545–556. doi: 10.1016/s1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- 85.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 86.Shao Z, et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 87.Isono K, et al. SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev Cell. 2013;26:565–577. doi: 10.1016/j.devcel.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 88.Lavigne M, Francis NJ, King IFG, Kingston RE. Propagation of Silencing. Mol Cell. 2004;13:415–425. doi: 10.1016/s1097-2765(04)00006-1. [DOI] [PubMed] [Google Scholar]
- 89.King HW, Fursova NA, Blackledge NP, Klose RJ. Polycomb repressive complex 1 shapes the nucleosome landscape but not accessibility at target genes. Genome Res. 2018;28:1494–1507. doi: 10.1101/gr.237180.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lavarone E, Barbieri CM, Pasini D. Dissecting the role of H3K27 acetylation and methylation in PRC2 mediated control of cellular identity. Nat Commun. 2019;10:1679. doi: 10.1038/s41467-019-09624-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hodges HC, et al. Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nat Struct Mol Biol. 2018;25:61–72. doi: 10.1038/s41594-017-0007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bartman CR, et al. Transcriptional burst initiation and polymerase pause release are key control points of transcriptional regulation. Mol Cell. 2019;73:519–532.:e4. doi: 10.1016/j.molcel.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ochiai H, et al. Genome-wide kinetic properties of transcriptional bursting in mouse embryonic stem cells. Sci Adv. 2020;6:eaaz6699. doi: 10.1126/sciadv.aaz6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Larsson AJM, et al. Genomic encoding of transcriptional burst kinetics. Nature. 2019;565:251–254. doi: 10.1038/s41586-018-0836-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bartman CR, Hsu SC, Hsiung CC-S, Raj A, Blobel GA. Enhancer regulation of transcriptional bursting parameters revealed by forced chromatin looping. Mol Cell. 2016;62:237–247. doi: 10.1016/j.molcel.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mermet J, et al. Clock-dependent chromatin topology modulates circadian transcription and behavior. Genes Dev. 2018;32:347–358. doi: 10.1101/gad.312397.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fukaya T, Lim B, Levine M. Enhancer control of transcriptional bursting. Cell. 2016;166:358–368. doi: 10.1016/j.cell.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen L-F, et al. Enhancer Histone Acetylation Modulates Transcriptional Bursting Dynamics of Neuronal Activity-Inducible Genes. Cell Rep. 2019;26:1174–1188.:e5. doi: 10.1016/j.celrep.2019.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Larson DR, et al. Direct observation of frequency modulated transcription in single cells using light activation. Elife. 2013;2:e00750. doi: 10.7554/eLife.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Senecal A, et al. Transcription factors modulate c-Fos transcriptional bursts. Cell Rep. 2014;8:75–83. doi: 10.1016/j.celrep.2014.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nicolas D, Zoller B, Suter DM, Naef F. Modulation of transcriptional burst frequency by histone acetylation. Proc Natl Acad Sci USA. 2018;115:7153–7158. doi: 10.1073/pnas.1722330115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ovesný M, Křížek P, Borkovec J, Svindrych Z, Hagen GM. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mueller F, et al. FISH-quant: automatic counting of transcripts in 3D FISH images. Nat Methods. 2013;10:277–278. doi: 10.1038/nmeth.2406. [DOI] [PubMed] [Google Scholar]
- 104.Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 105.Dimitrova E, et al. FBXL19 recruits CDK-Mediator to CpG islands of developmental genes priming them for activation during lineage commitment. Elife. 2018;7 doi: 10.7554/eLife.37084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Atsuta T, et al. Production of monoclonal antibodies against mammalian Ring1B proteins. Hybridoma. 2001;20:43–46. doi: 10.1089/027245701300060427. [DOI] [PubMed] [Google Scholar]
- 107.Bonhoure N, et al. Quantifying ChIP-seq data: a spiking method providing an internal reference for sample-to-sample normalization. Genome Res. 2014;24:1157–1168. doi: 10.1101/gr.168260.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hu B, et al. Biological chromodynamics: a general method for measuring protein occupancy across the genome by calibrating ChIP-seq. Nucleic Acids Res. 2015;43:e132. doi: 10.1093/nar/gkv670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Orlando DA, et al. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep. 2014;9:1163–1170. doi: 10.1016/j.celrep.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 110.Turberfield AH, et al. KDM2 proteins constrain transcription from CpG island gene promoters independently of their histone demethylase activity. Nucleic Acids Res. 2019;47:9005–9023. doi: 10.1093/nar/gkz607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032–2034. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ramírez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–5. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Love MI, Huber W, Anders S. Moderated estimation of fold change and’ ’ dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhu A, Ibrahim JG, Love MI. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics. 2019;35:2084–2092. doi: 10.1093/bioinformatics/bty895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Taruttis F, et al. External calibration with Drosophila whole-cell spike-ins delivers absolute mRNA fold changes from human RNA-Seq and qPCR data. BioTechniques. 2017;62:53–61. doi: 10.2144/000114514. [DOI] [PubMed] [Google Scholar]
- 120.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 121.Long HK, et al. Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. Elife. 2013;2:e00348. doi: 10.7554/eLife.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Singer ZS, et al. Dynamic heterogeneity and DNA methylation in embryonic stem cells. Mol Cell. 2014;55:319–331. doi: 10.1016/j.molcel.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Singh A, Razooky B, Cox CD, Simpson ML, Weinberger LS. Transcriptional bursting from the HIV-1 promoter is a significant source of stochastic noise in HIV-1 gene expression. Biophys J. 2010;98:L32–4. doi: 10.1016/j.bpj.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Foreman R, Wollman R. Mammalian gene expression variability is explained by underlying cell state. Mol Syst Biol. 2020;16:e9146. doi: 10.15252/msb.20199146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Padovan-Merhar O, et al. Single mammalian cells compensate for differences in cellular volume and DNA copy number through independent global transcriptional mechanisms. Mol Cell. 2015;58:339–352. doi: 10.1016/j.molcel.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The high-throughput sequencing datasets generated for this study are available in GEO database under the accession number GSE159400. Published data used in this study include BioCAP-seq (GEO: GSE43512) from 121, cnRNA-seq in Ring1a-/-; Ring1bfl/fl (PRC1CKO)(GEO: GSE119619) from 29 and cChIP-seq for JARID2, AEBP2, PCL2 and EPOP in untreated (wt) Ring1aI50A/D53K; Ring1b(WT->I53A/D56K)fl/fl ESCs (GEO: GSE132754) from 30. For cnRNA-seq processing we used mm10 (GenBank: BK000964.3, https://www.ncbi.nlm.nih.gov/nuccore/bk000964) and dm6 (GenBank: M21017.1, https://www.ncbi.nlm.nih.gov/nuccore/M21017.1) rDNA genomic datasets.
All R and Perl scripts used for genomic data analysis in this study are available at https://github.com/pauladobrinic/PRC1degron-2021. A custom-made ImageJ script for pre-processing 3D images (ThunderFISH) is publicly available with detailed manual of sample preparation and script usage at https://github.com/aleks-szczure/ThunderFISH.