Summary
Autophagy is a conserved quality control pathway that mediates the degradation of cellular components by targeting them to the lysosomes or vacuoles1. Autophagy has been implicated in the regulation of some regulated cell death processes in animal systems2. However, its function in developmentally controlled programmed cell death (dPCD) in plants remains little studied and controversial3. Some studies reported autophagy pro-survival roles4,5, while others suggested pro-death functions for autophagy6,7, calling for further detailed investigations.
Here, we investigated the role of autophagy in dPCD using the Arabidopsis root cap as an accessible and genetically tractable model system8. In Arabidopsis, dPCD is an integral part of root cap differentiation, restricting root cap organ size to the root meristem9. The root cap consists of two distinct tissues: The proximally positioned columella that is located at the very root tip, and the lateral root cap (LRC) that flanks the root meristem up to its distal end at the start of the root elongation zone10.
We show that autophagic flux strongly increased prior to dPCD execution in both root cap tissues, and depends the key autophagy genes ATG2, ATG5, and ATG7. Systemic and organ-specific mutation of these genes show delayed PCD execution and lack of post-mortem corpse clearance in the columella, but no defects in dPCD execution or corpse clearance in the distal LRC. Our results reveal a high degree of cell-type specificity in autophagy functions, and suggest that autophagy roles in dPCD can considerably diverge between different cell types of the same plant organ.
Keywords: autophagic flux, columella, developmentally controlled cell death, lateral root cap, PCD, RCD, regulated cell death, rhizosphere, root
Results and Discussion
Arabidopsis root cap organ size is kept constant by a combination of cell death and cell sloughing8,11: When epidermal cells start to elongate at the distal end of the meristem, their neighbour cells of the distal LRC (Figure 1A) undergo a tightly controlled dPCD and corpse clearance process on the root surface9. Periodically, cell death advances proximally towards the columella creating distinct PCD sites that demark the respective edge of each LRC layer9. Finally, the columella cells, together with the adjacent proximal LRC cells, get shed as cell packages into the rhizosphere12, where they undergo dPCD shortly thereafter13.
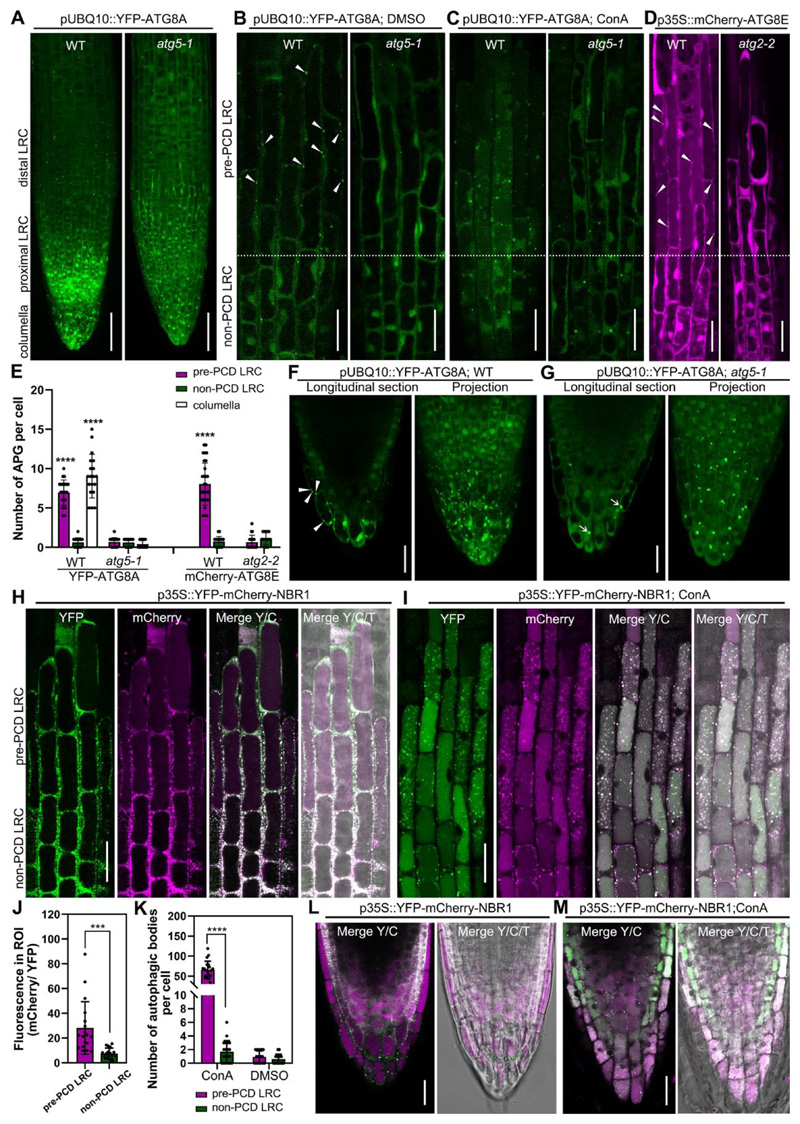
Figure 1. Visualization of the dynamics of autophagosomes/autophagy-related structures in root cap cells.
(A-C) Confocal laser scanning micrograph (CLSM) of lateral root cap (LRC) cells from seedlings at 4 days after germination (DAG) expressing pUBQ10::YFP-ATG8A in wild type and atg5-1 mutant, projection of the root tip (A), longitudinal section of LRC cells treated with DMSO as control (B), or treated with 1 μM ConA for 8 h (C). Scale bars are 50 μm (A), 20 μm (B-C).
(D) CLSM of LRC cells from 4 DAG seedlings of expressing p35S::mCherry-ATG8E in wild type and atg2-2 mutant. Scale bars are 20 μm.
(E) Quantification of autophagosomes (APG) in LRC and columella cells. Results are means ± SD. N = 18-30 cells from 5 roots. **** indicates a significant difference (t test, P < 0.0001).
(F-G) CLSM of columella cells from 4 DAG seedlings expressing pUBQ10::YFP-ATG8A in wild type (F) and atg5-1 mutant (G). Scale bars are 20 μm. White arrow heads indicate autophagosomes, white arrows indicate YFP aggregates.
(H-I) CLSM of LRC cells from 4 DAG seedlings expressing p35S::YFP-mCherry-NBR1 in wild type (H), treated with 1 μM ConA for 8 h (I). Scale bars are 20 μm.
(J) Quantification of the vacuolar import of NBR1 in LRC cells as shown in (H). Quantification was performed by dividing the average gray value in cytosol (YFP) with the average gray value in the vacuole (mCherry), using split channel images in ImageJ. Results are means ± SD. N = 18 cells from 5 roots. *** indicates a significant difference (t test, P < 0.001).
(K) Quantification of autophagic bodies in LRC cells treated with 1 μM ConA for 8 h as shown in (I). Results are means ± SD. N = 15-20 cells from 5 roots. **** indicates a significant difference (t test, P < 0.0001).
(L-M) CLSM of columella cells from 4 DAG seedlings expressing p35S::YFP-mCherry-NBR1 in wild type (L), treated with 1 μM ConA for 8 h (M). Scale bars are 20 μm.
Y/C indicates merge of the YFP channel and the mCherry channel; Y/C/T indicates merge of the YFP channel, the mCherry channel, and the transmission channel.
Autophagic flux is increased prior to dPCD in root cap cells
Macroautophagy, (here “autophagy”) is controlled by a highly diversified ensemble of autophagy-related genes (ATGs) in plants1. The Arabidopsis mutants atg7-214, atg5-115, and atg2-216 are deficient in autophagosome formation and autophagic flux to the vacuole. To test the relevance of autophagy for root cap dPCD, we imaged root cap cells of wild-type and atg mutant seedling expressing autophagy reporters. In the wild type, we detected pUBQ10::YFP-ATG8A17 in both the cytosol and in punctate foci (autophagosomes) in “pre-PCD LRC cells”, the last 1-2 cells from the distal root cap edge preparing for PCD. By contrast, we detected predominantly cytosolic YFP in more immature “non-PCD LRC cells”, which are 4 or more cells away from the distal root cap edge and not yet preparing for PCD (Figure 1A, B). In atg5-1 mutants15, we counted significantly less YFP-ATG8A foci in “pre-PCD” cells (Figure 1E), indicating autophagy activation prior to LRC cell death in an ATG5-dependent manner. These results were confirmed in seedlings expressing an alternative autophagy reporter, p35S::mCherry-ATG8E18 (Figure 1D, E).
In columella root cap cells we detected punctate pUBQ10::YFP-ATG8A in the outermost root cap layer in wild type roots (Figure 1F). By contrast, YFP-ATG8A was freely cytosolic and clustered in large cytoplasmic aggregates in atg5-1 mutants, as previously described17 (Figure 1G). The number of autophagosomes in mature columella cells was significantly increased compared to “non-PCD” cells (Figure 1E), suggesting that autophagic is activated prior to dPCD execution in the LRC and the columella.
Concanamycin A (ConA) is an established drug to elevate vacuolar pH allowing quantification of autophagic delivery to the vacuole19. After 8 hours of ConA treatment, numerous YFP-ATG8A-positive autophagic bodies accumulated in vacuoles of “pre-PCD” LRC cells (Figure 1C) and mature columella cells of wild type plants (Movie S1). Conversely, we detected no autophagic bodies in ConA-treated atg5-1 mutant (Figure 1C and Movie S2).
To independently quantify autophagic flux to the vacuole, we imaged root cap cells expressing a double-tagged autophagic cargo, YFP-mCherry-NBR1. After vacuolar delivery, low pH allows to differentiate between cytoplasmic to vacuolar fluorescence ratios comparing the pH-sensitive YFP and the less pH-sensitive mCherry20. We identified numerous NBR1 foci in the cytosol, and an increased vacuolar mCherry signal in “pre-PCD” LRC and columella cells (Figure 1H, J and L). When treated with ConA, in the central vacuole of both the LRC and the columella, we counted more NBR1-labelled autophagic bodies in “pre-PCD” root cap cells than in “non-PCD” root cap cells (Figure 1I, K and M). These results demonstrate that autophagic flux intensifies in different root cap cell types prior to PCD execution.
Autophagy is dispensable for dPCD execution and corpse clearance of distal LRC cells
To investigate the relevance of autophagy activation prior to dPCD execution, we performed a live-death assay applying fluorescein diacetate (FDA, live stain) and propidium iodide (PI, death stain)13 to roots of 5-day old wild type and atg mutant seedlings. The distal LRC PCD onset, indicated by the longitudinal root cap extent, was indistinguishable in atg mutants and the wild type (Figure S1A-B). Time-lapse imaging of the rapid clearance in PI-positive cell corpses on the root surface9 revealed no differences between the wild type and atg2-2 mutants (Figure S1E-F). This result is consistent with the absence of PI-stained cellular remnants of LRC cells in the elongation zone of atg mutants (Figure S1A) that are typical for corpse clearance mutants9,13. These results show that despite autophagy intensification prior to cell death, dPCD and post-mortem corpse clearance in the distal LRC occurs independently of autophagy. Our finding is in line with the observation that Arabidopsis atg mutants generally show mild or no phenotypes in several developmental processes involving dPCD, such as xylem tracheary element differentiation, seed development, or tapetum degeneration1. While autophagy has been implicated in dPCD promotion in Arabidopsis xylem21 and the suspensor of somatic embryos in Norway spruce (Piecea abies)7, in both systems early steps of cellular differentiation were affected. Secondary cell wall formation in the xylem, anisotropic cell expansion in the suspensor, and formation of large central vacuoles in both systems were affected6,7, suggesting that autophagy promotes cellular differentiation upstream of dPCD, and might affect dPCD execution in atg mutants only indirectly.
Autophagy is required for timely dPCD onset of proximal root cap cells
In contrast to the distal LRC, we observed a clear PCD phenotype in the columella and adjacent proximal LRC cells. While many of these cells in the wild type had already undergone PCD at the onset of the sloughing process, significantly more cells in atg mutants were still alive at this time point (Figure S1A, C-D). To investigate if general root cap development was affected in atg mutants, we visualized cell viability by FDA and cell outlines by FM4-64. Root cap architecture was indistinguishable in wild type and atg mutants (Figure S1G), and statolith production as a hallmark of columella differentiation22,23 was not affected in 5-day old atg mutants. While statoliths in atg mutants appeared to be smaller and more numerous than in the wild type, they disappeared prior to cell death in atg mutants and the wild type (Figure S1H-L), suggesting that autophagy is not involved in statolith degradation prior to columella dPCD. Seeds of a rice Osatg7-1 mutant are smaller and lower in starch content due to an abnormal activation of starch degradation in the endosperm during seed maturation24. Possibly, a similar pathway operates in the autophagy-deficient columella. Recently, autophagy has been shown to promote cell death processes associated with carbon starvation in tobacco BY-2 suspension culture cells25. Conceivably, carbon starvation is occurring and promoting PCD in detached root cap cells, but not in distal LRC cells, which would be in line with the different roles of autophagy we observed in these two cell types.
To follow columella viability after sloughing, we introduced a ubiquitously expressed tonoplast integrity marker (ToIM, co-expressing vacuolar RFP and cytoplasmic GFP)13 into the atg5-1 mutant. Twelve-hour time-lapse imaging of ToIM-expressing seedlings growing in imaging chambers did not reveal any aberration in the onset of sloughing of atg5-1 root cap cells. However, sloughed atg5-1 root cap cells remained viable significantly longer than the wild-type ones (Figures 2A-B). Even at 36 and 48 hours after shedding, living root cap cells could still be detected in atg5-1, which was rarely observed in wild-type plants. Interestingly, the vacuolar morphology was strikingly altered in these long-lived atg5-1 root cap cells; instead of a single large vacuole observed in the wild type, root cap cells showed two smaller vacuoles at either end of the cell (Figure 2A). Autophagy has been implicated in plant vacuole biogenesis before26,27, although to our knowledge, atg mutants have not been described to be affected in this process. Next, we investigated the ultrastructure in wild-type and atg2-2 mutant root cap cells by transmission electron microscopy (TEM). TEM confirmed the altered vacuolar structure in atg2-2 mutants (Figure S2J and K), but otherwise showed no obvious difference between wild type and atg mutant root cap cells prior to cell death (Figure S2D-F and J-L). Taken together, these results indicate that autophagy is neither critical for root cap development nor differentiation, but that autophagy promotes timely dPCD initiation in proximal LRC and columella cells.
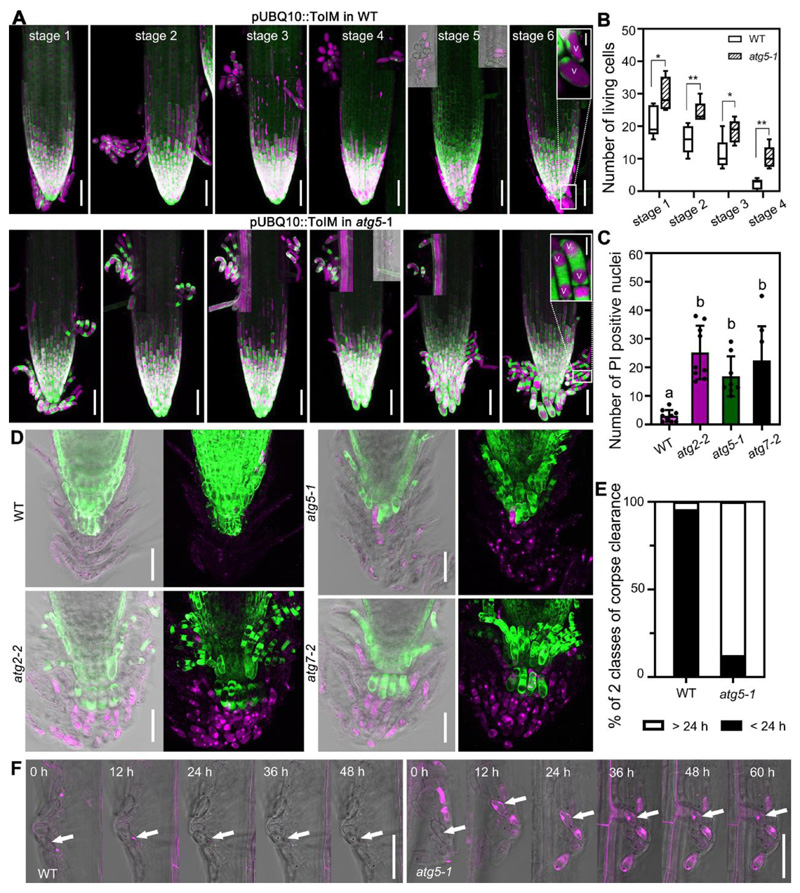
Figure 2. Autophagy is required for dPCD onset and postmortem nuclear degradation of proximal LRC and columella cells.
(A) pUBQ10::ToIM expression in wild type and atg5-1 was analyzed every 12 h (stages 1-6), with the visual onset of root cap shedding defined as stage 1. atg5-1 mutant shows longer living shed proximal LRC and columella cells compared with the wild type. EGFP signal is shown in green in the cytoplasm, and mRFP signal is shown in magenta in the vacuole. Scale bars are 50 μm. Close-up of cells are inserted in stage 6. V indicates the vacuole. Scale bars are 10 μm for inserted images.
(B) Quantification of living, shed cells from wild type and atg5-1 mutant at four stages after shedding shows that columella cells of the mutant lived longer compared with the wild type. Box plot whiskers show minimum and maximum values; cells were counted from 5 roots for each sample. * indicates a significant difference (t test, P < 0.05). ** indicates a significant difference (t test, P < 0.01).
(C) Quantification of PI-stained nuclei of wild type and atg mutants, shown in (D). Results are means ± SD. N = 8-9. Each mutant is significantly different from the wild type, as indicated by different letters (one-way ANOVA, Dunnett’s multiple comparison test, P < 0.05).
(D) CLSM of root tips from 14 DAG seedlings of wild type, atg2-2, atg5-1 and atg7-2, pulse labeled with FDA and PI. Scale bars are 50 μm.
(E) Quantification of corpse clearance of columella cells. Nuclei of atg5-1 took longer to be degraded after cell death compared with nuclei from the wild type. Box plots show data from at least 40 nuclei from at 6 different roots.
(F) Kymograph showing the delay of nuclear degradation in atg5-1 mutant compared with wild type. Cells were imaged every 12 h. PI is shown in magenta, and cells are shown with transmitted light. Scale bars are 50 μm.
See also Figure S1 and S2.
Autophagy is required for corpse clearance of proximal root cap cells
To test the involvement of autophagy in the corpse clearance of sloughed root cap cells, we cultivated atg mutants and wild type seedlings for 14 days on vertical agar plates. This method minimizes friction between the growing root tip and the medium, allowing root cap layers to accumulate. Using FDA-PI staining, we found an increased number of living root cap cells in atg mutants, confirming our results on younger seedlings (Figure S1D, Figure S2A-C). Additionally, we counted significantly more PI-stained columella cell corpses in atg mutants compared with the wild type (Figure 2C). This indicates that PI-stained cellular remains are rapidly degraded in the wild type, while cell corpse clearance is impaired in atg mutants. Time-lapse imaging confirmed that corpse clearance in atg5-1 mutant columella cells is strongly delayed, if not completely inhibited: While PI-stained nuclei were cleared within 24 hours in the wild type, nuclear remnants appeared practically unchanged as late as 60 hours after PI entry in atg5-1 mutants (Figure 2E-F). TEM imaging confirmed these results: In dead wild type root cap cells, cytosolic remnants were strongly condensed and organelles appeared to be degrading (Figure S2G, H and I). By contrast, in the atg2-2 mutant, columella cell corpses did not collapse, showed a continuous plasma membrane, and contained abundant cytoplasmic remains (Figure S2M, N and O).
Autophagy occurs independent of established dPCD gene regulatory networks
Loss of key autophagy regulators caused a columella longevity phenotype reminiscent of the double mutant ana046 anac087. These two NAC transcription factors jointly control the timely onset of PCD in the proximal root cap and regulate the expression of dPCD-associated genes13. Hence, we investigated whether autophagy is controlled by, or controlling, these transcription factors. As previously reported13, the ana046 anac087 mutant exhibits delayed corpse clearance in distal LRC cells (Figure S3F), and an increased number of living columella cells comparable to the one found in atg mutants (Figure S3F and G). Also, we detected a delay in corpse clearance in anac046 anac087 columella cells that had not been reported before (Figure S3H and I). However, direct comparison of anac046 anac087 and atg mutants revealed differences: The delay of corpse clearance in atg mutants (Figure 2) was much more pronounced than in anac046 anac087 mutants (Figure S3H and I). Also, the vacuolar morphology of anac046 anac087 is not similar to the one of atg mutants, but resembles the wild type (Figure S3J).
Next, we transformed a GFP-ATG8E autophagy reporter into the anac046 ana087 mutant, along with the Col-0 wild type and atg5-1 mutants as controls. GFP-ATG8E shows an ATG5-dependent increase of autophagic activity and autophagic flux to the vacuole in root cap cells preparing for dPCD (Figure S3A), confirming our earlier results (Figure 1). However, autophagy activation occured in anac046 anac087 mutant as in wild-type root cap cells (Figure 3A-C). These results indicate that the activation of autophagy in mature root cap cells occurs independently of ANA046 and ANAC087. Inducible misexpression of ANAC046 or SOMBRERO (SMB) leads to ectopic cell death in the entire plant13. To test if this ectopic cell death depends on autophagy, we transformed an estradiol-inducible XVE-ANAC046-GFP expression construct controlled by the pSMB promoter into atg5-1 mutants and the wild type. Two independent lines in each genotype were selected based GFP expression levels. Eight hours after induction, we observed a strong GFP signal and an increased number of PI-positive root cap cells in the wild type and the atg5-1 mutant (Figure 3G, upper row, and I). 24 hours after estradiol treatment this phenotype was exacerbated in both genotypes (Figure 3G, lower row, and I), suggesting that ANAC046-induced ectopic cell death does not depend on a functional autophagy pathway.
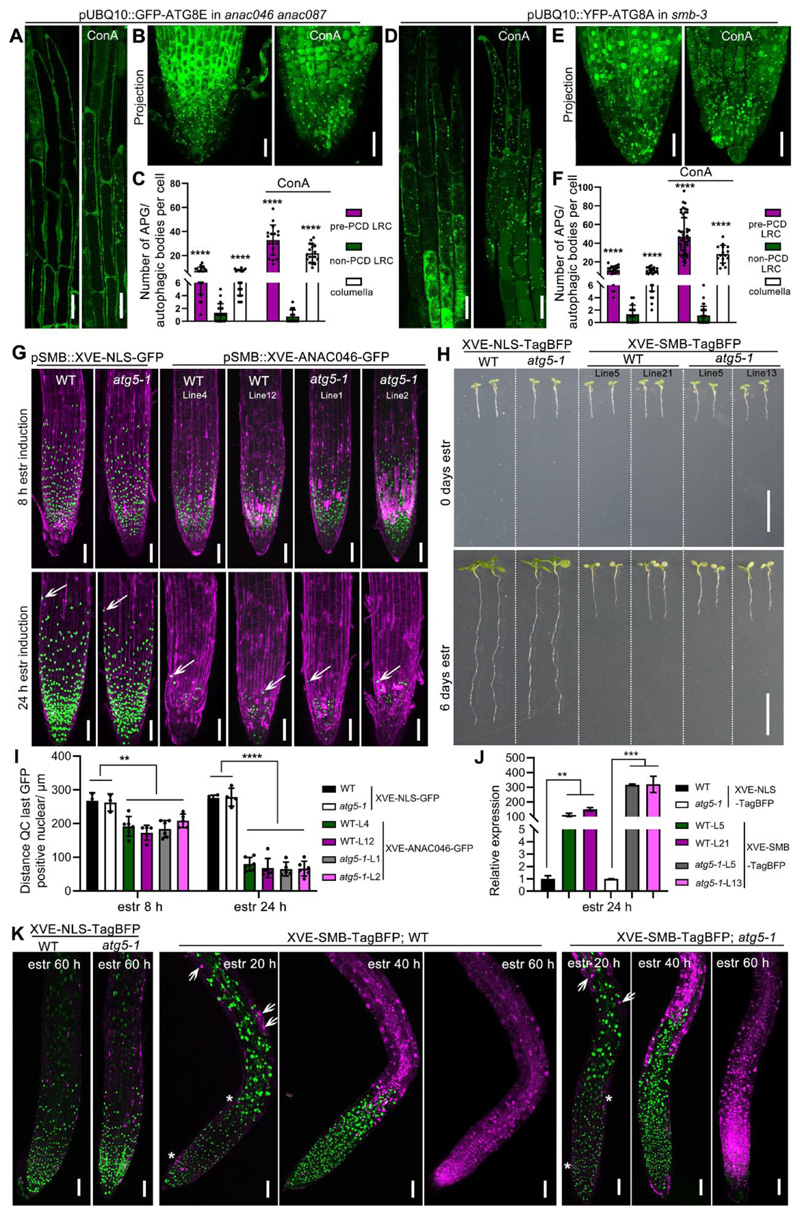
Figure 3. Autophagy occurs independently of established dPCD gene regulatory networks.
(A-C) CLSM of LRC cells from 5 DAG seedlings expressing pUBQ10::GFP-ATG8E in anac046 anac087 mutant, longitudinal section of LRC cells (A, left panel), treated with 1 μM ConA for 8 h (A, right panel), projection of root tip (B, left panel), treated with ConA for 8 h (B, right panel). Scale bars are 20 μm. C, Quantification of autophagosomes (APG) or autophagic bodies in LRC and columella cells shown in (A-B). Results are means ± SD. N = 16-22 cells from 6 roots. **** indicates a significant difference (t test, P < 0.0001).
(D-F) CLSM of LRC cells from 5 DAG seedlings expressing pUBQ10::YFP-ATG8A in smb-3 mutant, longitudinal section of LRC cells (D, left panel), treated with 1 μM ConA for 8 h (D, right panel), projection of root tip (E, left panel), treated with ConA (E, right panel). Scale bars are 20 μm. F, Quantification of autophagosomes (APG) or autophagic bodies in LRC and columella cells shown in (D-E). Results are means ± SD. N = 20-50 cells from 6 roots. **** indicates a significant difference (t test, P < 0.0001).
(G) CLSM of root tips from 5 DAG seedlings pSMB::XVE-NLS-GFP in wild type, in atg5-1, and pSMB::XVE-ANAC046-GFP in wild type and atg5-1 mutant. Two independent lines are shown, stained with PI (magenta). 8 h after estradiol induction (z-stack projection, up row), 24 h after estradiol induction (z-stack projection, down row). White arrows indicate the position of the last living root cap cells. Scale bars are 50 μm.
(H) Macroscopic appearance of estradiol-induction of SMB-TagBFP driven by promoter of pH3.3/HTR5 in wild type and atg5-1 mutant, estradiol-induction of NLS-TagBFP in wild type or atg5-1 mutant as control. Two independent lines are shown at 0 day (upper row) and 6 days (bottom row) after transfer to estradiol-containing medium. Overexpression of SMB resulted in root growth arrest in atg5-1 mutants similar to in the wild type, while the overexpression of NLS-TagBFP did not result in root growth arrest both in atg5-1 and wild type after 6 days of estradiol induction. Scale bars are 1 cm.
(I) Quantification of the root cap length of two independent overexpression lines of each XVE-ANA046-GFP and XVE-NLS-GFP shown in (G). At least 5 roots of each line were quantified. Results are means ± SD. Each line of XVE-ANAC046-GFP is significantly different from line of XVE-NLS-GFP, both in wild type and atg5-1 mutant, but there is no difference between wild type and atg5-1. (t test; ** P < 0.01, **** P < 0.0001).
(J) As indicated by RT-qPCR analysis, a 24 h induction by estradiol caused strong expression of SMB in two independent lines of XVE-TagBFP in wild type and atg5-1 mutant, XVE-NLS-TagBFP in wild type and atg5-1 as control. Results are means ± SD. N = 3 biological replicates. (t test; ** P < 0.01, *** P < 0.001).
(K) CLSM of root tips from 5 DAG seedlings of pH3.3/HTR5::XVE-NLS-TagBFP and pH3.3/HTR5::XVE-SMB-TagBFP in wild type and atg5-1 mutant at different time points after estradiol induction, stained with the PI (magenta). 16 h after estradiol induction, ectopic cell death is detected and indicated by white arrows. PCD of root cap cells is indicated by white asterisks. 40 h after estradiol induction abundant cell death occurred above the root meristem. 60 h after estradiol induction the whole root tip is dead and stained with PI. The signal of TagBFP is shown in green. Scale bars are 50 μm.
See also Figure S3.
Next, we transformed an estradiol-inducible SMB-TagBFP construct driven by the ubiquitous pH3.3/HTR5 promoter28 into the wild type and atg5-1 mutants. As control, we transformed an inducible NLS-TagBFP construct under the same promoter. Two independent lines per construct and genotype were selected based on induction levels of SMB expression (Figure 3J). In both the wild type and the atg5-1 mutant, we observed ectopic cell death and root growth arrest followed by seedling death upon estradiol induction of SMB expression (Figure 3H, J, K). The NLS-TagBFP lines showed neither root growth arrest nor ectopic cell death after estradiol treatment (Figure 3H, K).
Finally, we investigated autophagic activity in smb-3 mutant root cap cells, that show a delayed and aberrant cell death devoid of cell corpse clearance9. We introgressed the YFP-ATG8A marker into the smb-3 mutant. The increased autophagic activity in mature LRC cells in comparison with “non-PCD” LRC cells occurred in smb-3 mutants (Figure 3D-F) in a fashion comparable to the wild type, suggesting that increased autophagic flux prior to root cap PCD is not regulated by SMB.
Taken together, autophagic flux increase in root cap cells preparing for dPCD occurs independently of the key PCD regulators SMB, ANAC046 and ANAC087, and the ectopic cell death caused by misexpression of these transcription factors does not depend on functional autophagy. These findings suggest that autophagy controls PCD in proximal root cap cells via parallel pathway that operates independently of the established PCD-promoting NAC transcription factor network.
Root cap-specific loss of function and complementation shows tissue-inherent functions of autophagy
As autophagic processes are active throughout the entire plant, we wondered whether the root cap phenotypes we observed were organ-specific, or possibly a secondary consequence of an organism-wide autophagy deficiency. To address this question, we first used a CRISPR-based tissue-specific knockout system, CRISPR-TSKO29, expressing a fluorescently tagged Cas9 exclusively in the root cap controlled by the pSMB promoter. Combined with guide-RNAs (gRNAs) targeting ATG2 or ATG5, we generated transgenic lines with root-cap specific autophagy deficiency. To verify the efficiency of our constructs, we transformed them into lines expressing the autophagy reporter p35S::mCherry-ATG8E. In most “pre-PCD” LRC cells, both constructs produced a significant reduction in the number of mCherry-labelled cytoplasmic foci and a clear reduction of vacuolar mCherry signal (Figure S4A-B), indicating a defect in autophagosome formation and delivery of ATG8E to the vacuole. Next, we transformed both constructs into the wild type and established four independent lines each. All lines showed significantly delayed corpse clearance in columella cells of 14-day old seedlings, comparable to the established atg mutants (Figure 4A-D). Additionally, FDA staining revealed a pronounced columella- and proximal LRC-cell longevity phenotype, similar to the one of the atg2-2 mutant (Figure S4C).
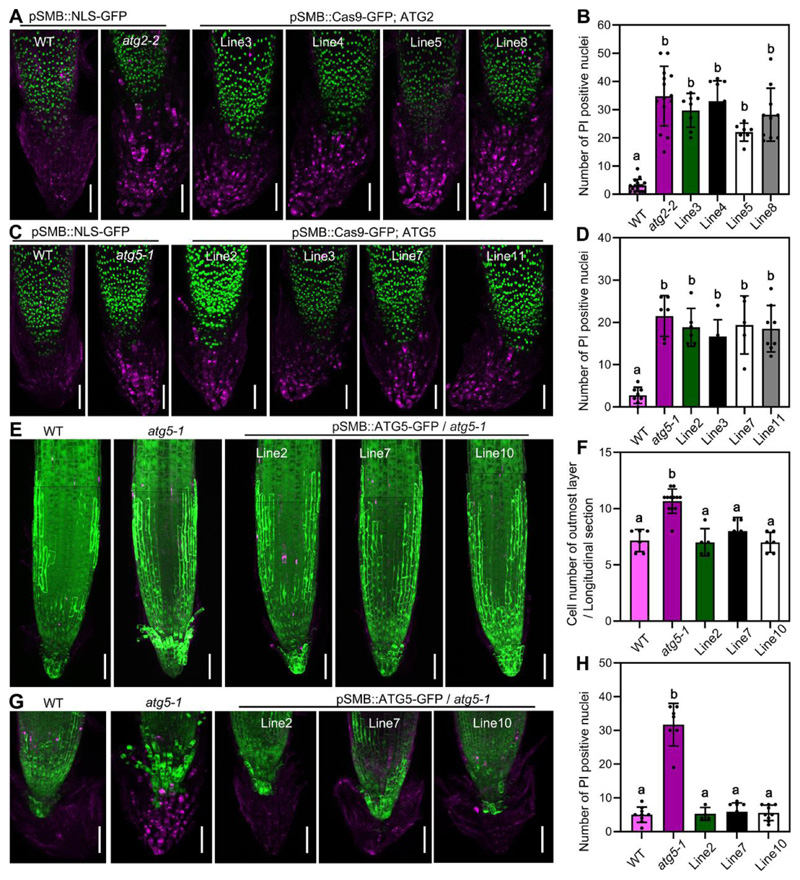
Figure 4. Autophagy controls cell death in a root-cap autonomous fashion.
(A) CLSM (z-stack projection) of root tips from 14 DAG seedlings of pSMB::NLS-GFP in wild type, in atg2-2, and four different lines of pSMB::Cas9-GFP;ATG2, pulse labeled with PI (magenta). Scale bars are 50 μm.
(B) Quantification of PI-stained nuclei of seedlings shown in (A). Results are means ± SD. N = 7-16 roots. Each mutant is significantly different from the wild type, as indicated by different letters (one-way ANOVA, Dunnett’s multiple comparison test, P < 0.05).
(C) CLSM (z-stack projection) of root tips from 14 DAG seedlings of pSMB::NLS-GFP in wild type, in atg5-1, and four different lines of pSMB::Cas9-GFP;ATG5, pulse labeled with PI (magenta). Scale bars are 50 μm.
(D) Quantification of PI-stained nuclei of seedlings shown in (C). Results are means ± SD. N = 7-8 roots. Each mutant is significantly different from the wild type, as indicated by different letters (one-way ANOVA, Dunnett’s multiple comparison test, P < 0.05).
(E) CLSM (z-stack) of root tips from 5 DAG seedlings of wild type, atg5-1 and three different root-cap specific complementation lines (pSMB::GFP-ATG5) in atg5-1 mutant, pulse labeled with FDA and PI. Scale bars are 50 μm.
(F) Cell number quantification of the outmost root cap layer shown in (E) on longitudinal sections. Results are means ± SD. N = 5-12 roots. Wild type and complementation lines are significantly different from the mutant, as indicated by different letters (one-way ANOVA, Dunnett’s multiple comparison test, P < 0.0001).
(G) CLSM (z-stack projection) of root tips from 14 DAG seedlings of wild type, in atg5-1, and three different root-cap specific complementation lines (pSMB::GFP-ATG5) in atg5-1 mutant, pulse labeled with PI (magenta). Scale bars are 50 μm.
(H) Quantification of PI-stained nuclei of seedlings shown in (G). Results are means ± SD. N = 5-9 roots. Wild type and complementation lines are significantly different from the mutant, as indicated by different letters (one-way ANOVA, Dunnett’s multiple comparison test, P < 0.0001).
See also Figure S4.
Finally, we generated transgenic lines expressing a root-cap specific ATG5 complementation construct driven by pSMB in the atg5-1 mutant background. Three independent lines showed a complete rescue of the longevity and corpse clearance phenotypes (Figure 4E-H). These results demonstrate an autonomous regulation and function of autophagy in the root cap context.
More than a decade ago, cell-type specific approaches had been called for to address the specific roles of autophagy in different contexts30. The CRISPR-TSKO root-cap specific autophagy knock-out confirms that the PCD-promoting role of autophagy is controlled autonomously within the root cap. Cell-type specific approaches will be a powerful addition to the autophagy toolbox, and facilitate addressing the roles of autophagy in specific cell types, tissues, or organs while avoiding pleiotropic or secondary effects of conventional knockouts or pharmacological treatments.
STAR Methods
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Moritz Nowack (Moritz.Nowack@psb.vib-ugent.be).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Experimental Model and Subject Details
All Arabidopsis seedlings were grown vertically on 1/2 Murashige and Skoog (MS, Duchefa Biochemie) medium (0.1 g/L MES, pH 5.8 [KOH], and 0.8% plant agar) in a continuous light at 21°C before analysis, except where noted.
The Arabidopsis thaliana atg2-2 mutant allele (EMS, Gln803stop) was reported by16; atg5-1 (SAIL_129_B07) was reported by15; atg7-2 (GABI_655B06) was reported by14, smb-3 was reported by9, anac046 anac087 was reported by13. The Arabidopsis lines pUBQ10::YFP-ATG8A, p35S::mCherry-ATG8E18 and pSMB::NLS-GFP9 were introduced into atg5-1 and/or atg2-2, or smb-3 mutant by crossing, respectively. The line p35S::YFP-mCherry-NBR1 was reported by20. The pUBQ10::GFP-ATG8E was introduced into anac046 anac087 by dipping. pUBQ10::ToIM was reported previously13. Stable Arabidopsis transformations were produced using the floral dipping method described before31.
Method Details
Cloning
Golden Gate entry modules pGG-A-pSMB-B, pGG-B-Linker-C, pGG-C-Cas9-D, pGG-D-P2A-GFP-NLS-E, pGG-E-G7T-F, pGG-F-pATU6-26-AarI-G, were reported previously29. These entry modules were assembled in pFASTR-AG, resulting in the destination vector pFASTR-pSMB-Cas9-P2A-GFP-NLS-pATU6-26-AarI. 2 gRNAs (Table S1) were designed using CRISPOR32. Fragment gRNA1-pATU6-26-gRNA2 (ATG2 target) and gRNA1-pATU6-26-gRNA2 (ATG5 target) were amplified by PCR using the following primers, p43/p44 for ATG2, p41/42 for ATG5, and These purified PCR fragments were inserted into pFASTR-pSMB-Cas9-P2A-GFP-NLS-pATU6-26-AarI destination vector via a Golden Gate reaction. The resulting vectors were named pSMB-Cas9;ATG2 and pSMB-Cas9;ATG5.
Golden Gate entry modules pGG-A-pUBQ10-B, pGG-B-mGFP-C, pGG-C-ATG8E-D, pGG-D-Linker-E, pGG-E-tHSP18.2M-F, pGG-F-linkerII-G were collected from PSB plasmids stock. And these entry modules were assembled in pFASTRK-AG, resulting in the expression vector pFASTRK-pUBQ10::GFP-ATG8E.
Gateway entry modules L4-pSMB-R1, L1-ANAC046-L2, and R2-GFP-L3 were described previously9,13. L4-pSMB-XVE-R1, L4- pH3.3/HTR5 -XVE-R1, L1-NLS-GFP-L2, L1-NLS-TagBFP-L2, L1-SMB-L2 and R2-TagBFP-L3 were ordered from the PSB plasmid stock (https://gatewayvectors.vib.be). The ATG5 coding sequence was amplified by PCR with primers p214 and p215, using 4-day old seedling cDNA as template and inserted into pDONR221 via BP reaction, resulting in entry vector, L1-ATG5-L2. Entry vectors L4-pSMB-XVE-R1, L1-ANAC046-L2 and R2-GFP-L3 were assembled into pFASTRK-34GW destination vector via LR reaction. The result vector was pFASTRK-pSMB::XVE-ANAC046-GFP. L4-pSMB-XVE-R1, L1-NLS-GFP-L2 were assembled into pFASTRK-24GW destination vector via LR reaction. The resulting vector was pFASTRK-pSMB::XVE-NLS-GFP. L4- pH3.3/HTR5 -XVE-R1, L1-SMB-L2 and R2-TagBFP-L3 were assembled into pFASTGB-34GW destination vector via LR reaction. The resulting vector was pFASTGB- pH3.3/HTR5::XVE-SMB-TagBFP. L4- pH3.3/HTR5 -XVE-R1, L1-NLS-TagBFP-L2 were assembled into pFASTGB-24GW destination vector via LR reaction. The resulting vector was pFASTGB-pH3.3/HTR5::XVE-NLS-TagBFP. L4-pSMB-R1, L1-ATG5-L2 and R2-GFP-L3 were assembled into pFASTGB-34GW destination vector via LR reaction. The resulting vector was pFASTGB-pSMB::ATG5-GFP.
All PCRs for cloning was performed with Phusion high-fidelity DNA polymerase (Thermo Scientific). All entry vectors were sequenced by Eurofins Scientific using the Mix2Seq or TubeSeq services. All primers are listed in Table S1.
Staining and imaging
Confocal imaging was done on and LSM710 (Zeiss) using the Plan Apochromat 20× objective (numerical aperture 0.8), or an SP8X (Leica) using a 40x (HC PL APO CS2, NA=1.10) water immersion objective, unless stated otherwise.
For the FDA-PI viability staining, seedlings were mounted on a glass slide in FDA solution (1 μL dissolved FDA stock solution [2 mg in 1 ml acetone] in 1 ml 0.5 1/2 MS) supplemented with 10 μg/ml PI. FDA and PI were imaged simultaneously on the LSM710 (Zeiss) using the 488-nm laser line to excite FDA and the 561-nm laser line to excite PI. Emission was detected between 500 and 550 nm and between 600 and 700 nm for FDA and PI, respectively. For the FDA-FM4-64 staining, seedlings were mounted on a glass slide in FDA solution (1 μL dissolved FDA [2 mg in 1 ml acetone] in 1 ml 1/2 MS) supplemented with 4 μg/ml FM4-64. FM4-64 was imaged on the LSM710 (Zeiss) using the 561-nm laser line to excite FM4-64. Emission was detected between 600 and 700 nm for FM4-64.
In the inducible overexpression lines, seedlings were sprayed with 50 μg/ml estradiol or DMSO (mock) and then mounted on a glass slide in 1/2 MS supplemented with 10 μg/ml PI at different timepoints. GFP/TagBFP and PI were detected simultaneously in the same track on the LSM710 (Zeiss). GFP was excited by the 488-nm line of the argon laser and detected between 500 and 550 nm in channel 1, or TagBFP was excited by 405-nm line and detected between 450 and 510 nm in channel 1, whereas PI was excited by the 561-nm line and detected between 590 and 700 nm in channel 2. For the overnight time course, 20 hours induced seedlings were transferred to a Lab-Tek chamber and covered with an agar slab (1/2 MS) supplemented with 10 μg/ml estradiol and 10 μg/ml PI. Roots were imaged at different timepoints.
For the reporter lines of pUBQ10::YFP-ATG8A, pUBQ10::GFP-ATG8E, YFP-mCherry-NBR1, seedlings were mounted on a glass slide in 1/2 MS medium and imaged on SP8 (Leica). YFP was excited by the 514-nm line of the argon laser and detected between 525 and 580 nm. GFP was excited by the 488-nm line of the argon laser and detected between 500 and 550 nm. The signal of mCherry was excited by the 561-nm line and detected between 600 nm and 700 nm. Imaging of ToIM was performed as described before13.
For starch staining, 5 days old seedlings were stained with Lugol solution (Sigma-Aldrich) for 5 min, washed in distilled water for 1 min and then mounted with clearing solution (chloral hydrate: glycerol: water=8:1:3). Samples were imaged on an Olympus BX51 microscope with a 40× DIC objective.
All images were processed and analyzed using Fiji (https://fiji.sc/)33.
Transmission Electron Microcopy (TEM)
Root tips of 5 days old and 14 days old seedlings of Arabidopsis thaliana Col-0 and atg2-2 were excised, immersed in 20% (w/v) BSA and frozen immediately in a high-pressure freezer (Leica EM ICE; Leica Microsystems, Vienna, Austria). Freeze substitution was carried out using a Leica EM AFS (Leica Microsystems) in dry acetone containing 1% (w/v) OsO4 and 0.2% glutaraldehyde over a 4-days period as follows: - 90°C for 54 hours, 2°C per hour increase for 15 hours, -60°C for 8 hours, 2°C per hour increase for 15 hours, and -30°C for 8 hours. Samples were then slowly warmed up to 4°C, infiltrated stepwise over 3 days at 4°C in Spurr’s resin and embedded in capsules. The polymerization was performed at 70°C for 16 h. Ultrathin sections were made using an ultra-microtome (Leica EM UC6) and post-stained in in a Leica EM AC20 for 40 min in uranyl acetate at 20°C and for 10 min in lead stain at 20°C. Sections were collected on formvar-coated copper slot grids.
Grids were viewed with a JEM1400plus transmission electron microscope (JEOL, Tokyo, Japan) operating at 80 kV.
Quantification and Statistical Analysis
The statistical details of experiments can be found in the corresponding Figure legends. The results of statistical tests can be found in the corresponding Results section. Statistical tests were carried out using GraphPad Prism 9.0.0 for Windows.
Supplementary Material
The root tip of 5 days old wild type expressing pUBQ10::YFP-ATG8A treated with ConA for 8 h. Scale bar is 20 μm.
The root tip an atg5-1 mutant seedling at 5 DAG expressing pUBQ10-YFP-ATG8A treated with ConA for 8 h. Scale bar is 20 μm.
Highlights.
Autophagy intensifies prior to developmental cell death in the Arabidopsis root cap
The role of autophagy in cell death is divergent depending on the root cap cell type
Autophagy is dispensable for cell death and corpse clearance in the lateral root cap
Autophagy deficiency delays cell death and blocks corpse clearance in the columella
eTOC Blurb.
Does autophagy play a role in developmentally controlled programmed cell death in plants? Feng et al. show that cell death at the distal edge of the Arabidopsis lateral root cap is entirely independent of autophagy, while autophagy is required for timely cell death execution and corpse clearance in the proximal columella root cap.
Acknowledgments
We thank Peter Bozhkov (Department of Chemistry and Biotechnology, Uppsala BioCenter, Swedish University of Agricultural Sciences, Sweden) for the seeds of atg5-1, atg7-2 and GFP-ATG8A in atg5-1; Celine Masclaux-Daubresse for seeds of p35S::mCherry-ATG8E (originally) from Romain Lebars; Matyas Fendrych for the seeds of pUBQ10::YFP-ATG8A in Col-0. This research was financially supported by the European Research Council (ERC) StG PROCELLDEATH 639234 and CoG EXECUT.ER 864952 (M.K.N.), by the Austrian Science Fund (FWF- SFB F79) and the Vienna Science and Technology Fund (WWTF, LS17-047) to Y.D., and by a postdoctoral fellowship (01P06118) of the University of Ghent Bijzonder Onderzoeksfonds (BOF) (Q.F.).
Footnotes
Author contributions
Q.F. performed the experiments; Y.D. provided monitoring autophagy support; R.D.R. provided TEM imaging support; Q.F., M.K.N., and Y.D. analyzed the data, designed the experiments and wrote the article.
Declaration of interests
The authors declare no competing interests.
Data and code availability
Plasmids generated in this study have been deposited to the VIB-UGent plasmid repository (https://gatewayvectors.vib.be) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. All data generated or analyzed during this study are included in this published article (and its supplementary information files). All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Marshall RS, Vierstra RD. Autophagy: The Master of Bulk and Selective Recycling. Annual review of plant biology. 2018;69:173–208. doi: 10.1146/annurev-arplant-042817-040606. [DOI] [PubMed] [Google Scholar]
- 2.Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, Alnemri ES, Altucci L, Amelio I, Andrews DW, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell death and differentiation. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ustun S, Hafren A, Hofius D. Autophagy as a mediator of life and death in plants. Current opinion in plant biology. 2017;40:122–130. doi: 10.1016/j.pbi.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Dauphinee AN, Denbigh GL, Rollini A, Fraser M, Lacroix CR, Gunawardena A. The Function of Autophagy in Lace Plant Programmed Cell Death. Frontiers in plant science. 2019;10:1198. doi: 10.3389/fpls.2019.01198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. The Plant cell. 2009;21:2914–2927. doi: 10.1105/tpc.109.068635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwon SI, Cho HJ, Park OK. Role of Arabidopsis RabG3b and autophagy in tracheary element differentiation. Autophagy. 2010;6:1187–1189. doi: 10.4161/auto.6.8.13429. [DOI] [PubMed] [Google Scholar]
- 7.Minina EA, Filonova LH, Fukada K, Savenkov EI, Gogvadze V, Clapham D, Sanchez-Vera V, Suarez MF, Zhivotovsky B, Daniel G, et al. Autophagy and metacaspase determine the mode of cell death in plants. J Cell Biol. 2013;203:917–927. doi: 10.1083/jcb.201307082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumpf RP, Nowack MK. The root cap: a short story of life and death. Journal of experimental botany. 2015;66:5651–5662. doi: 10.1093/jxb/erv295. [DOI] [PubMed] [Google Scholar]
- 9.Fendrych M, Van Hautegem T, Van Durme M, Olvera-Carrillo Y, Huysmans M, Karimi M, Lippens S, Guerin CJ, Krebs M, Schumacher K, Nowack MK. Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Current biology: CB. 2014;24:931–940. doi: 10.1016/j.cub.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 10.Rost TL. The organization of roots of dicotyledonous plants and the positions of control points. Annals of botany. 2011;107:1213–1222. doi: 10.1093/aob/mcq229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennett T, van den Toorn A, Sanchez-Perez GF, Campilho A, Willemsen V, Snel B, Scheres B. SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis. The Plant cell. 2010;22:640–654. doi: 10.1105/tpc.109.072272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi CL, von Wangenheim D, Herrmann U, Wildhagen M, Kulik I, Kopf A, Ishida T, Olsson V, Anker MK, Albert M, et al. The dynamics of root cap sloughing in Arabidopsis is regulated by peptide signalling. Nature plants. 2018;4:596–604. doi: 10.1038/s41477-018-0212-z. [DOI] [PubMed] [Google Scholar]
- 13.Huysmans M, Andrade Buono R, Skorzinski N, Cubria Radio M, De Winter F, Parizot B, Mertens J, Karimi M, Fendrych M, Nowack MK. ANAC087 and ANAC046 control distinct aspects of programmed cell death in the Arabidopsis columella and lateral root cap. The Plant cell. 2018 doi: 10.1105/tpc.18.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O, Jorgensen LB, Jones JD, Mundy J, Petersen M. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009;137:773–783. doi: 10.1016/j.cell.2009.02.036. [DOI] [PubMed] [Google Scholar]
- 15.Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant physiology. 2005;138:2097–2110. doi: 10.1104/pp.105.060673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Nishimura MT, Zhao T, Tang D. ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. The Plant journal: for cell and molecular biology. 2011;68:74–87. doi: 10.1111/j.1365-313X.2011.04669.x. [DOI] [PubMed] [Google Scholar]
- 17.Li F, Chung T, Pennington JG, Federico ML, Kaeppler HF, Kaeppler SM, Otegui MS, Vierstra RD. Autophagic recycling plays a central role in maize nitrogen remobilization. The Plant cell. 2015;27:1389–1408. doi: 10.1105/tpc.15.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Bars R, Marion J, Le Borgne R, Satiat-Jeunemaitre B, Bianchi MW. ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nature communications. 2014;5:4121. doi: 10.1038/ncomms5121. [DOI] [PubMed] [Google Scholar]
- 19.Tamura K, Shimada T, Ono E, Tanaka Y, Nagatani A, Higashi SI, Watanabe M, Nishimura M, Hara-Nishimura I. Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. The Plant journal: for cell and molecular biology. 2003;35:545–555. doi: 10.1046/j.1365-313x.2003.01822.x. [DOI] [PubMed] [Google Scholar]
- 20.Svenning S, Lamark T, Krause K, Johansen T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy. 2014;7:993–1010. doi: 10.4161/auto.7.9.16389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon SI, Cho HJ, Jung JH, Yoshimoto K, Shirasu K, Park OK. The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. The Plant journal: for cell and molecular biology. 2010;64:151–164. doi: 10.1111/j.1365-313X.2010.04315.x. [DOI] [PubMed] [Google Scholar]
- 22.Hong JH, Chu H, Zhang C, Ghosh D, Gong X, Xu J. A quantitative analysis of stem cell homeostasis in the Arabidopsis columella root cap. Frontiers in plant science. 2015;6:206. doi: 10.3389/fpls.2015.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature. 1997;390:287–289. doi: 10.1038/36856. [DOI] [PubMed] [Google Scholar]
- 24.Sera Y, Hanamata S, Sakamoto S, Ono S, Kaneko K, Mitsui Y, Koyano T, Fujita N, Sasou A, Masumura T, et al. Essential roles of autophagy in metabolic regulation in endosperm development during rice seed maturation. Sci Rep. 2019;9:18544. doi: 10.1038/s41598-019-54361-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teper-Bamnolker P, Danieli R, Peled-Zehavi H, Belausov E, Abu-Abied M, Avin-Wittenberg T, Sadot E, Eshel D. Vacuolar processing enzyme translocates to the vacuole through the autophagy pathway to induce programmed cell death. Autophagy. 2021;17:3109–3123. doi: 10.1080/15548627.2020.1856492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolb C, Nagel MK, Kalinowska K, Hagmann J, Ichikawa M, Anzenberger F, Alkofer A, Sato MH, Braun P, Isono E. FYVE1 is essential for vacuole biogenesis and intracellular trafficking in Arabidopsis. Plant physiology. 2015;167:1361–1373. doi: 10.1104/pp.114.253377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machado SR, Rodrigues TM. Autophagy and vacuolar biogenesis during the nectary development. Planta. 2019;250:519–533. doi: 10.1007/s00425-019-03190-7. [DOI] [PubMed] [Google Scholar]
- 28.Ingouff M, Selles B, Michaud C, Vu TM, Berger F, Schorn AJ, Autran D, Van Durme M, Nowack MK, Martienssen RA, Grimanelli D. Live-cell analysis of DNA methylation during sexual reproduction in Arabidopsis reveals context and sex-specific dynamics controlled by noncanonical RdDM. Genes Dev. 2017;31:72–83. doi: 10.1101/gad.289397.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Decaestecker W, Buono RA, Pfeiffer ML, Vangheluwe N, Jourquin J, Karimi M, Van Isterdael G, Beeckman T, Nowack MK, Jacobs TB. CRISPR-TSKO: A Technique for Efficient Mutagenesis in Specific Cell Types, Tissues, or Organs in Arabidopsis. The Plant cell. 2019;31:2868–2887. doi: 10.1105/tpc.19.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, Makino A. Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant physiology. 2009;149:885–893. doi: 10.1104/pp.108.130013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clough SJ, Bent AF. Floral dip: a simplified method fo Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant journal: for cell and molecular biology. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 32.Concordet JP, Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46:W242–W245. doi: 10.1093/nar/gky354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nature methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The root tip of 5 days old wild type expressing pUBQ10::YFP-ATG8A treated with ConA for 8 h. Scale bar is 20 μm.
The root tip an atg5-1 mutant seedling at 5 DAG expressing pUBQ10-YFP-ATG8A treated with ConA for 8 h. Scale bar is 20 μm.
Data Availability Statement
Plasmids generated in this study have been deposited to the VIB-UGent plasmid repository (https://gatewayvectors.vib.be) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. All data generated or analyzed during this study are included in this published article (and its supplementary information files). All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.