Abstract
Kinetochores assemble onto specialized centromeric CENP-A nucleosomes (CENP-ANuc) to mediate attachments between chromosomes and the mitotic spindle. We describe cryo-EM structures of the human inner kinetochore CCAN (Constitutive Centromere Associated Network) complex bound to CENP-ANuc reconstituted onto α-satellite DNA. CCAN forms edge-on contacts with CENP-ANuc, while a linker DNA segment of the α-satellite repeat emerges from the fully-wrapped end of the nucleosome to thread through the central CENP-LN channel that tightly grips the DNA. The CENP-TWSX histone-fold module further augments DNA binding and partially wraps the linker DNA in a manner reminiscent of canonical nucleosomes. Our study suggests that the topological entrapment of the linker DNA by CCAN provides a robust mechanism by which kinetochores withstand both pushing and pulling forces exerted by the mitotic spindle.
The centromere is a specialized genetic locus that interacts with the mitotic spindle to facilitate chromosome segregation. Centromeres of humans and other primates comprise multiple copies of the conserved ~171 bp α-satellite repeat that contains the histone H3 variant CENP-A (1, 2). Kinetochores assemble specifically onto centromeres to form large macromolecular machines that directly mediate chromosome segregation (3). A critical function of the kinetochore is to generate a load-bearing attachment between chromosomes and the spindle apparatus. How this function is achieved at the molecular level is unclear.
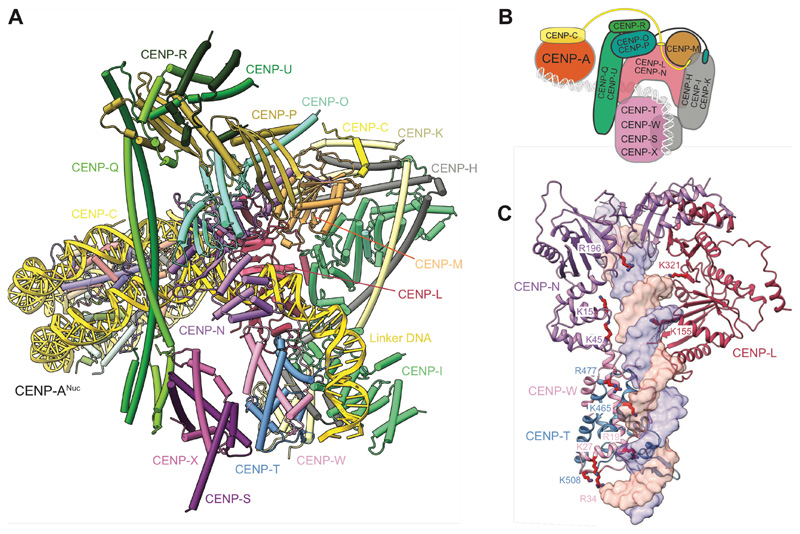
Throughout the cell cycle, CENP-A nucleosomes specify the constitutive association of the 16-subunit inner kinetochore CCAN complex with centromeric chromatin, whereas the ten-subunit KMN network of the outer kinetochore assembles onto CCAN during mitosis to couple CCAN to microtubules for spindle-mediated chromosome movement. Two human CCAN proteins, CENP-C and CENP-N, directly interact with CENP-ANuc (4, 5). CENP-C interacts with the C-terminus of CENP-A, and histones H2A and H2B (6), whereas CENP-N binds the L1 loop and adjacent DNA gyre of CENP-ANuc (7–11). The 17 bp B-box motif that interacts with CENP-B is the only DNA sequence within the α-satellite repeat that is specifically recognized by a centromere-associated protein.
In budding yeast, a single kinetochore complex associates with a 'point' centromere accommodating a sole CENP-A nucleosome. In contrast, the regional kinetochores of humans and other metazoans are generated from multiple copies of the CCAN and KMN network complexes to form large disk-like structures. Here, we determined the cryo-EM reconstruction of a CCAN-CENP-ANuc complex using a native 171 bp α-satellite repeat DNA, representing the repeating modular unit of a human inner kinetochore.
Results
CCAN is assembled from a network of interdependent modules
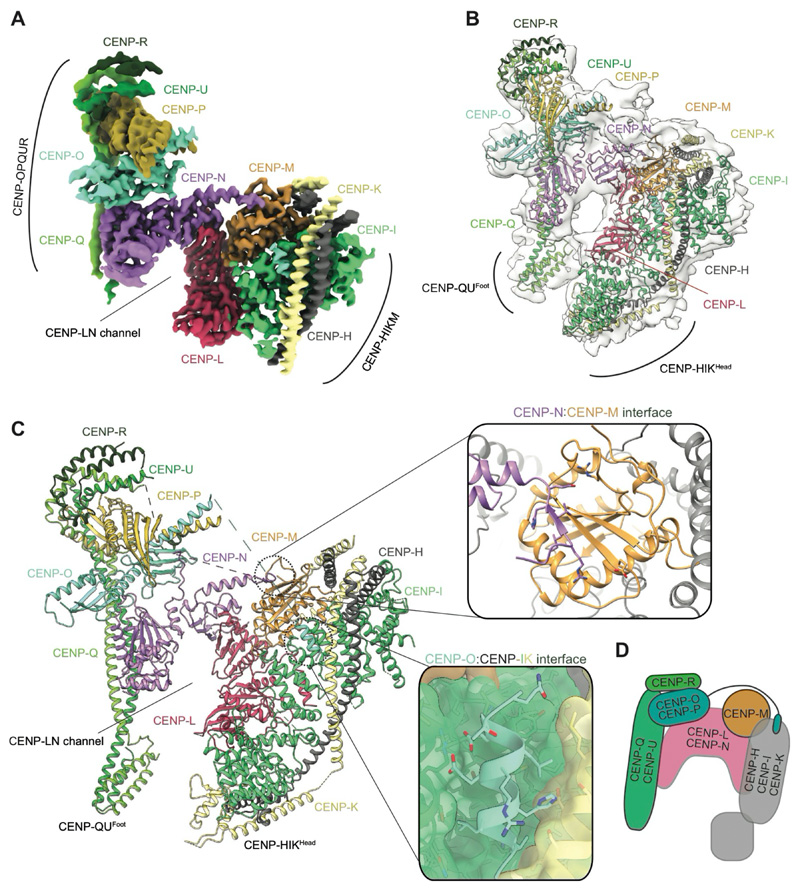
Human CCAN comprises four modules: CENP-LN, CENP-HIKM, CENP-OPQUR and CENP-TWS, that are additionally linked by the largely disordered CENP-C through its interactions with CENP-LN and CENP-HIKM (7, 12, 13). We expressed and purified CCAN sub-complexes, and used the N-terminal half of CENP-C (CENP-CN) that contains all known CCAN binding sites and the central domain essential for centromere localization (14, 15) (fig. S1A). Phosphoproteomic analysis indicated an interphase phosphorylation state of CCAN, as judged by the absence of CDK1-phosphorylation sites on CENP-T (3). Initially, we focused on the 11-subunit CCAN core complex (CENP-OPQUR-LN-HIKM) which could assemble without CENP-CN (termed CCANΔCT) (fig. S1B). We determined the cryo-EM structure of CCANΔCT at 3.2 Å resolution (Fig. 1, fig. S2A-D and Table S1). The cross-linked structure was identical to a non-cross-linked reconstruction obtained at lower resolution (fig. S2E). We also determined crystal structures of the CENP-OPQUR and CENP-HIKHead modules (fig. S2F and Table S2).
Figure 1. CCAN is assembled from a network of interdependent modules.
(A, B) Cryo-EM density map (A) and molecular architecture (B) of the apo-CCANΔCT fitted into the cryo-EM map with resolved OPQURFoot and HIKHead. (C) Atomic model of the apo-CCANΔCT with details of the CENP-N-CENP-M interface as well as CENP-O binding to the CENP-HI pocket shown as insets. (D) Cartoon schematic of the CCANΔCT modules.
The architecture of CCANΔCT resembles budding yeast CCAN (ScCCAN) (16, 17) with CENP-OPQUR and CENP-HIKM 'lobes' assembled on either flank of a central CENP-LN module (Fig. 1). The arc-like CENP-LN module is the structural keystone of CCANΔCT, and generates a deep channel that is extended on one side by the N-terminal coiled-coil of CENP-QU. The RWD domains of CENP-O and CENP-P, with the C-terminal regions of CENP-QU and CENP-R, organize into a 'cap' domain above CENP-N. In human CCANΔCT, CENP-R partially substitutes for yeast Nkp1 and Nkp2. CENP-M, that is also unique to vertebrate CCAN, adopts a pseudo-GTPase-fold and forms a stable complex with CENP-HIK (18). CENP-M makes extensive contacts with the HEAT domain of CENP-I and the N-terminal helical bundle of CENP-H and CENP-K, and in the context of CCAN, with CENP-L (Fig. 1). CENP-M has a small deletion that removes the switch I region and an adjacent β-strand (18). A conserved β-strand of CENP-N occupies this position to augment the central β-sheet of CENP-M (Fig. 1C). An N-terminal α-helix of CENP-O, connected through a flexible linker to CENP-OPQUR, interacts with a conserved pocket formed by CENP-I and CENP-K (Fig. 1C). Disrupting this interaction by deleting the N-terminal 35 residues of CENP-O destabilizes CCANΔCT assembly, as does mutating residues on CENP-N that bind CENP-M (fig. S3A, B). Further supporting the human CCAN architecture described here is its consistency with the interdependent localization of CCAN subunits to centromeres (12, 13, 18, 19).
Two peripheral and conformationally flexible regions of CCAN are poorly resolved in the cryo-EM reconstruction: the CENP-HIKHead and N-terminal region of CENP-QU (CENP-QUFoot) (Fig. 1). Focused 3D classification of CENP-HIKHead identified multiple poses, two of which readily accommodate the CENP-HIKHead crystal structure (figs S2F, S3C).
Reconstitution of CCAN-CENP-ANuc onto a 171 bp α-satellite repeat
To assemble a complete CCANΔT-α-satellite nucleosome complex, we reconstituted CCANΔT complexes with CENP-CN and CENP-ANuc using 171 bp α-satellite repeat DNA (CENP-ANuc-171). The α-satellite DNA matches an α-satellite repeat from human chromosome 2, and also shares 93% sequence identity with at least 14 other human centromeres (20) (fig. S4A). We used two separate 171 bp α-satellite sequences, differing in register by 22 bp (termed AS1 and AS2) (fig. S4A). To define the exact nucleosome positioning of the AS2 sequence, we determined a cryo-EM structure of a CENP-ANuc-CENP-CN complex at 2.4 A resolution (fig. S4B-D and Table S1). The dyad axis of the nucleosome is equivalent to NCPs comprising X-chromosome α-satellite DNA, with which it shares 71% sequence identity (10, 21) (fig. S4A). This also matches nucleosome positions defined by in vivo ChIP micrococcal nuclease (MNase)-seq analyses of human centromeric chromatin (21, 22). Relative to histone H3 nucleosomes (H3Nuc), CENP-ANuc exhibits continuous conformational flexibility of its terminal DNA segments as shown by a 3D variability analysis. The DNA ends fluctuate between fully wrapped at both ends, to being unwrapped by ~22 bp at one end, with the majority of particles in intermediate states (fig. S4E and Movie S1). Unwrapped and flexible terminal DNA segments are seen in yeast and vertebrate CENP-ANuc structures (17, 23), and this correlates with a 110-120 bp nuclease-resistant DNA core (17, 23). These in vitro studies are also in concordance with CENP-ANuc protecting 110-150 bp of α-satellite DNA based on in vivo ChIP-MNase-seq studies of natural human centromeres and neocentromeres, as well as observations that CENP-ANuc is a canonical octameric nucleosome in cells (21, 24, 25). A structure of CENP-ANuc reconstituted with AS1 (from the CCANΔT-CENP-ANuc-AS1 cryo-EM data set) was positioned identically to CENP-ANuc-AS2, except the 3' unwrapped end is absent due to the 22 bp 5' register shift relative to AS2 (fig. S4A). In addition to resolving the CENP-ANuc, we were able to build the CENP-C central domain interacting with the C-terminus of CENP-A, and histones H2A/H2B, a structural feature of CENP-ANuc recognition conserved from yeast to human (6, 10, 11, 17).
The positively charged CCAN channel grips linker α-satellite DNA of CENP-ANuc
We then determined cryo-EM structures of CCANΔT-CENP-ANuc complexes using both the AS1 and AS2 DNA (figs S5, S6). These two structures were identical except for their DNA boundaries (figs S4A, S6B). Because the AS1 sequence produced a more stable complex, as judged by SEC, a larger cryo-EM data set was collected with this DNA (Table S1).
Cryo-EM micrographs of the CCANΔT-CENP-ANuc data sets and 2D-class averages showed particles corresponding to three species: (i) CENP-ANuc-CENP-CN, (ii) CCANΔT bound to free DNA and (iii) CCANΔT bound to CENP-ANuc (fig. S5A-C). The observation of one CCANΔT associated with CENP-ANuc is in agreement with an assessment of the oligomeric state of CCANΔT-CENP-ANuc in solution, indicating a single CCANΔT assembled onto CENP-ANuc (fig. S5D).
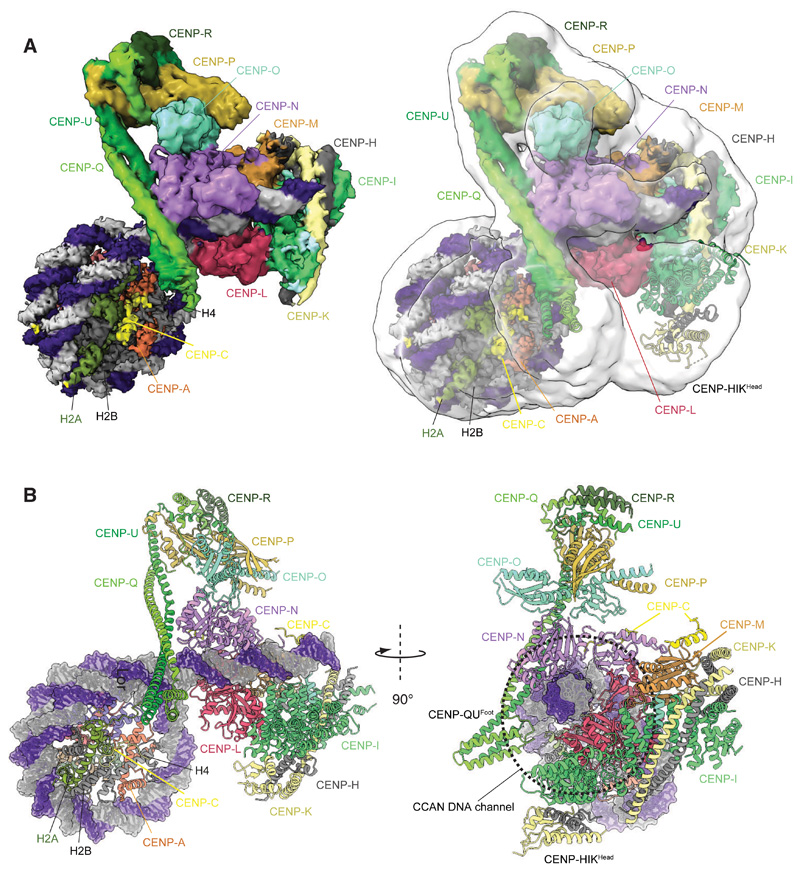
In the CCANΔT-DNA complex, determined at 4.5 Å resolution, EM density is visible for a linear ~24 bp DNA duplex threading through the CENP-LN channel (Figs 2A, 3A and fig. S5E). The CCANΔT-CENP-ANuc complexes exhibited conformational heterogeneity, limiting the resolution of a consensus cryo-EM reconstruction to 8.9 Å (Fig. 2A and figs S5, S6). Focused 3D classification and refinement of the CCANΔT-DNA component alone produced a 7.3 Å resolution reconstruction with a protein and DNA structure identical to that of the CCANΔT-DNA complex (fig. S5C, E). We therefore generated a composite CCANΔT-CENP-ANuc structure from the higher resolution CENP-ANuc-CENP-CN and CCANΔT-DNA reconstructions (Fig. 2A). In the CCANΔT-CENP-ANuc complex, the CENP-ANuc DNA is wrapped similarly to the free CENP-ANuc-CENP-CN (fig. S6).
Figure 2. Structure of the CCANΔT-CENP-ANuc complex.
(A) Right panel shows the consensus CCANΔT-5 CENP-ANuc cryo-EM map (transparent white) overlaid onto the composite CCANΔT-CENP-ANuc cryo-EM density map based on individual cryo-EM maps for the CENP-ANuc-CENP-CN and CCANΔT-DNA reconstructions. Left panel: composite map alone. (B) Two orthogonal views of the CCANΔT-CENP-ANuc complex depicted in cartoon representation for protein and space filling for DNA. CENP-ANuc has a diameter of 11 nm.
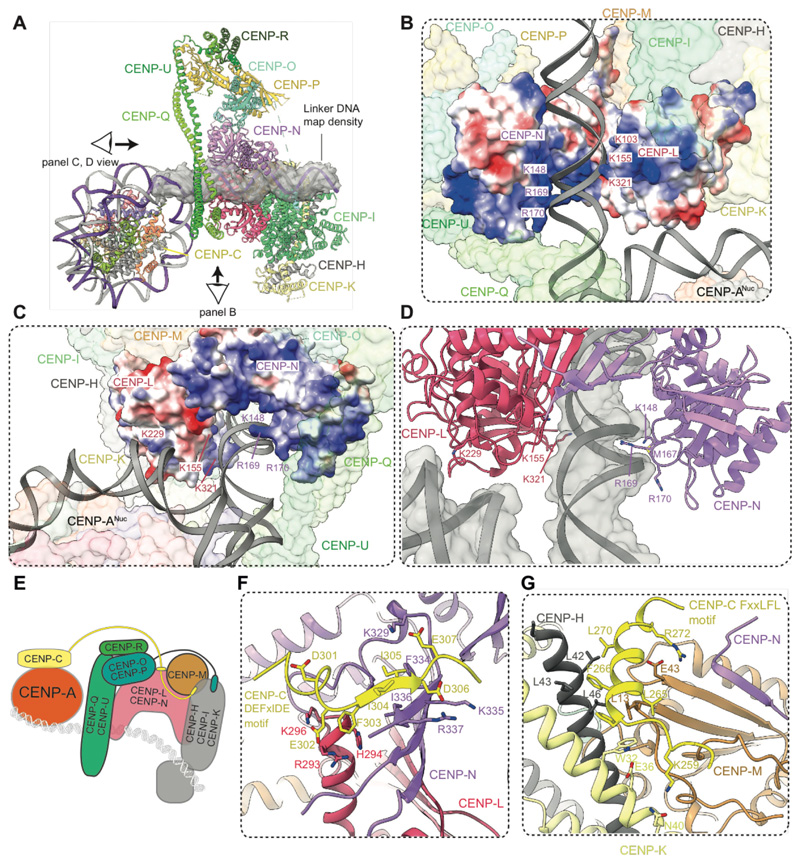
Figure 3. The extranucleosomal linker DNA is gripped by the CENP-LN channel.
(A) Cartoon representation of CCANΔT-CENP-ANuc with cryo-EM density shown for extranucleosomal linker DNA (from CCANΔT-DNA cryo-EM density map). (B) The CENP-LN module features a positively-charged channel that complements the DNA duplex. Electrostatic surface charge shown for CENP-LN. (C and D). Details of the CENP-LN channel-DNA interaction. R169 of CENP-N inserts into the DNA minor groove. CENP-LN surface charge shown in C. (E) Cartoon schematic of the CCANΔT modules showing DNA path. (F and G) CENP-C binds to two sites on CCAN. The DEFxIDE motif binds to CENP-LN (E), whereas the FxxLFL motif binds to CENP-HIKM (G).
A striking feature of the CCANΔT-CENP-ANuc complex is that CCANΔT forms few direct contacts with CENP-ANuc, but instead its primary contact with CENP-ANuc is through the extended extranucleosomal linker DNA. CENP-ANuc forms a small end-on contact, mediated through its DNA gyre, with the back-face of CCANΔT (Fig. 3B, C). This positions the extranucleosomal DNA to thread through the CENP-LN channel. Clearly resolved cryo-EM density for the DNA phosphate backbone shows it tightly gripped by conserved basic residues of the CENP-LN channel (fig. S5E). These create a dense, continuous positively-charged surface complementing the shape and charge of the 24 bp linker DNA duplex (Figs 2, 3). Interactions of the side chains of CENP-N residues K148, M167 and R169 with the DNA minor groove contribute to defining a fixed DNA register (Fig. 3C, D). In the DNA-bound state, the CENP-LN channel contracts, tightening its grip on the DNA duplex (fig. S5F), and the channel is extended by the apposition of CENP-HIKHead in the raised conformation (Fig. 3A, E).
CENP-C interaction sites on CENP-LN and CENP-HIKM modules
CENP-C is required for the centromere localization of CENP-LN, CENP-T, CENP-O and the CENP-HIKM module (5, 7, 12, 13, 18). The CCAN-interacting region of CENP-C maps to the PEST sequence (CENP-CPEST), a 200-residue intrinsically disordered region N-terminal of the CENP-C central domain that features two highly conserved short linear sequence motifs (7, 13, 14). Cryo-EM maps of the CCANΔT-DNA and CCANΔT-CENP-ANuc complexes indicated two volumes of unassigned map density not present in apo-CCANΔCT and CCANΔC-DNA complexes (discussed below), associated with the CENP-HIKM and CENP-LN modules (fig. S6D), previously shown to interact with CENP-C (7, 12–14). To guide the interpretation of these densities, we used AlphaFold2 (26) to predict potential models of how CENP-CPEST would interact individually with CENP-LN, and CENP-HIKM. The first run predicted the conserved DEFxIDE motif (residues 301-307) (7, 12–14) of CENP-CPEST would form an edge β-strand with the CENP-N β-sheet (Fig. 3F). This position corresponded well to additional cryo-EM density associated with CENP-N (fig. S6D). Phe and Ile residues of the DEFxIDE motif dock into a hydrophobic pocket at the CENP-LN interface, whereas the flanking acidic residues form electrostatic interactions with conserved Arg and Lys residues of CENP-N, and CENP-L. In support of this model, a prior study mutating the EFxID residues of the CENP-C DEFxIDE motif abolished its interaction with CENP-LN in vitro and disrupted CENP-N recruitment to centromeres in vivo without perturbing CENP-C localization (7). For CENP-HIKM, AlphaFold2 also predicted that the CENP-CPEST FxxLFL motif (residues 262-267) (13) would bind as an α-helix to a hydrophobic site at the combined CENP-H-CENP-K-CENP-M interface (Fig. 3G). This prediction was validated by cryo-EM density at this location (fig. S6D), and is consistent with a previous report that mutating the LFL residues of this motif disrupts CENP-C interactions with CENP-HIKM, in vitro and in vivo (13). Both the LN and HIKM interaction sites are located on the back-face of CCAN, facing the CENP-ANuc, suggesting a mechanism for how CENP-C tethers kinetochores to CENP-ANuc (5) (Fig. 3E).
α-Satellite linker DNA is a crucial determinant of stable CCAN association with CENP-ANuc
Our CCANΔT-CENP-ANuc structure suggests that the interaction of the extranucleosomal DNA duplex with the CENP-LN channel is a major determinant of CCAN assembly onto a centromeric α-satellite-CENP-ANuc. To test this, we performed reconstitutions assessing the role of the extranucleosomal DNA, CENP-CN, basic residues lining the CENP-LN channel, CENP-HIKHead and CENP-QUFoot. We first tested the requirement of the extranucleosomal DNA. Using SEC, we found that CENP-ANuc wrapped with 147 bp of α-satellite DNA (10) did not form stable complexes with CCANΔT without CENP-CN, whereas a 171 bp α-satellite that included the extranucleosomal DNA formed stable complexes with CCANΔT independently of CENP-CN (fig. S7A, B). Interactions with CENP-ANuc reconstituted with a 171 bp α-satellite were equally stable in the presence or absence of the B-box, consistent with sequence-independent interactions with the DNA phosphate backbone (fig. S7C).
Having established a requirement for linker DNA in the assembly of CCANΔCT-CENP-ANuc complexes, we next assessed how modifications to the CENP-LN channel affected CCANΔT interactions with CENP-ANuc. In the absence of CENP-CN, mutating three positive patches of CENP-L (CENP-Lcm), that interact with linker DNA and the nucleosome gyre (Fig. 3B-D and fig. S5F), abolished binding of CCANΔCT to CENP-ANuc-171 without perturbing CCAN assembly (fig. S7D). Identical results were obtained by deleting either CENP-HIKHead, or CENP-QUFoot (fig. S8A, B). These deletions also moderately reduced CCAN binding to isolated DNA (fig. S8C). To assess the validity of the CCAN-CENP-ANuc model in a cellular context, we tested the effect of the CENP-Lcm mutant that disrupted CCANΔCT binding to CENP-ANuc-171 in vitro, on centromere localization of CENP-L in vivo. In HEK293 cells, localization of the CENP-Lcm mutant to kinetochores was substantially impaired compared to wild-type CENP-L (fig. S8D).
In previous structures of the CENP-N N-terminal domain (CENP-NNT) associated with CENP-ANuc (7, 8, 10), CENP-NNT interacts with the L1 loop of CENP-A (CENP-AL1 loop) and adjacent DNA gyre at SHL2-3. In agreement with these structures, we found that mutating CENP-AL1 loop disrupts the interaction of the isolated CENP-LN dimer with CENP-ANuc wrapped with 171bp α-satellite DNA (fig. S9A). By contrast, our structure of CCAN-CENP-ANuc-171 does not involve an interaction of CENP-NNT with CENP-AL1 loop, and consistent with this, mutating CENP-AL1 loop did not disrupt interactions of CENP-ANuc-171 with CCANΔCT, as assessed by both pulldown assays and during reconstitution on SEC (fig. S9A, B). These results, as well as previous structural information (7–10), suggest that the mode of interaction between CENP-LN and CENP-ANuc is different when CENP-LN is part of the full CCAN complex compared to CENP-LN alone. To further test this hypothesis, we performed competition assays where we added CENP-NNT, which binds specifically to CENP-AL1 loop, at a four-fold excess over the CCAN-CENP-ANuc complex (fig. S9C). The CCAN-CENP-ANuc complex readily accommodated the additional CENP-NNT, without evident diminution of CCAN binding, and this additional CENP-NNT incorporation was dependent on CENP-AL1 loop. This suggests that in a fully assembled CCAN-CENP-ANuc complex, CENP-AL1 loop is not occupied and therefore available for CENP-NNT binding (fig. S9D), consistent with our cryo-EM reconstruction. Modelling a CENP-NNT-based interaction of CCAN with CENP-AL1 loop shows that CENP-ANuc would clash extensively with the CENP-L, CENP-HIK and CENP-OPQUR modules (as well as CENP-TWSX module as discussed below) (fig. S9E). In both models, CENP-NNT forms the same interactions with the DNA backbone, although at SHL7-8 for CENP-NNT in CCAN (fig. S9F). Thus, previously described mutants at the CENP-NNT-DNA interface that disrupt CENP-N-centromere localization in vivo (4, 7, 8), are consistent with, but do not distinguish between, both modes of CENP-N-CENP-ANuc interaction.
Overall, the interaction of free CENP-LN with CENP-ANuc is dependent on CENP-AL1 loop, but independent of the linker DNA, whereas CCAN interactions with CENP-ANuc are dependent on the linker DNA but independent of CENP-AL1 loop. These results are consistent with different modes of interaction between CENP-LN and CENP-ANuc when CENP-LN is incorporated into CCAN. This agrees with a previous observation that the L1 loop is not required for the in vitro assembly of a functional kinetochore (27).
CENP-C confers selectivity of assembled CCAN for CENP-A mono-nucleosomes
The only direct interaction between CCANΔT and the CENP-A histone in our structure is mediated by CENP-CN. To validate this finding, we produced human H3 nucleosomes reconstituted either with a 147 bp 601 sequence or a 601 sequence extended at the 5' end by 30 bp (177 bp) (fig. S4A). As expected, CCANΔT failed to bind H3Nuc reconstituted with the minimal 147 bp 601 sequence, whereas CCANΔT bound CENP-ANuc with the same DNA only in the presence of CENP-CN, similar to 147 bp α-satellite DNA (figs S7A and S10A). Consistent with our structure, binding was observed with the extended 177 bp sequence for both H3Nuc-177 and CENP-ANuc-177 (fig. S10B). The binding of CCANΔCT to CENP-ANuc-177 but not to H3Nuc-177, was enhanced four to five-fold by CENP-CN, consistent with selective recognition of CENP-ANuc by CENP-C (fig. S10C).
CENP-TWSX binds and partially wraps linker DNA
The CCANΔT-CENP-ANuc structure was reconstituted without the CENP-TWSX module due to our initial difficulties incorporating CENP-TWSX into CCAN. Removing all affinity tags from CENP-WSX enabled CENP-TWSX incorporation into CCAN (fig. S10D), and as judged by SEC, this 16-subunit holocomplex interacts with the 171 bp α-satellite-CENP-ANuc similarly to CCANΔT (fig. S10D), consistent with CENP-A, CENP-C and CENP-T comprising a single 20-subunit inner kinetochore complex (28). The full CCAN complex also requires extranucleosomal linker DNA to bind CENP-ANuc without CENP-C, suggesting that CENP-TWSX does not change the fundamental mechanism of CCAN assembly onto CENP-ANuc (fig. S10E). EMSA experiments revealed that CENP-TWSX increases the affinity of CCANΔCT for both CENP-ANuc and H3Nuc nucleosomes containing 177 bp 601 DNA approximately 3-fold, suggesting that it does not confer additional selectivity for CENP-ANuc over canonical H3Nuc (fig. S10 C, F).
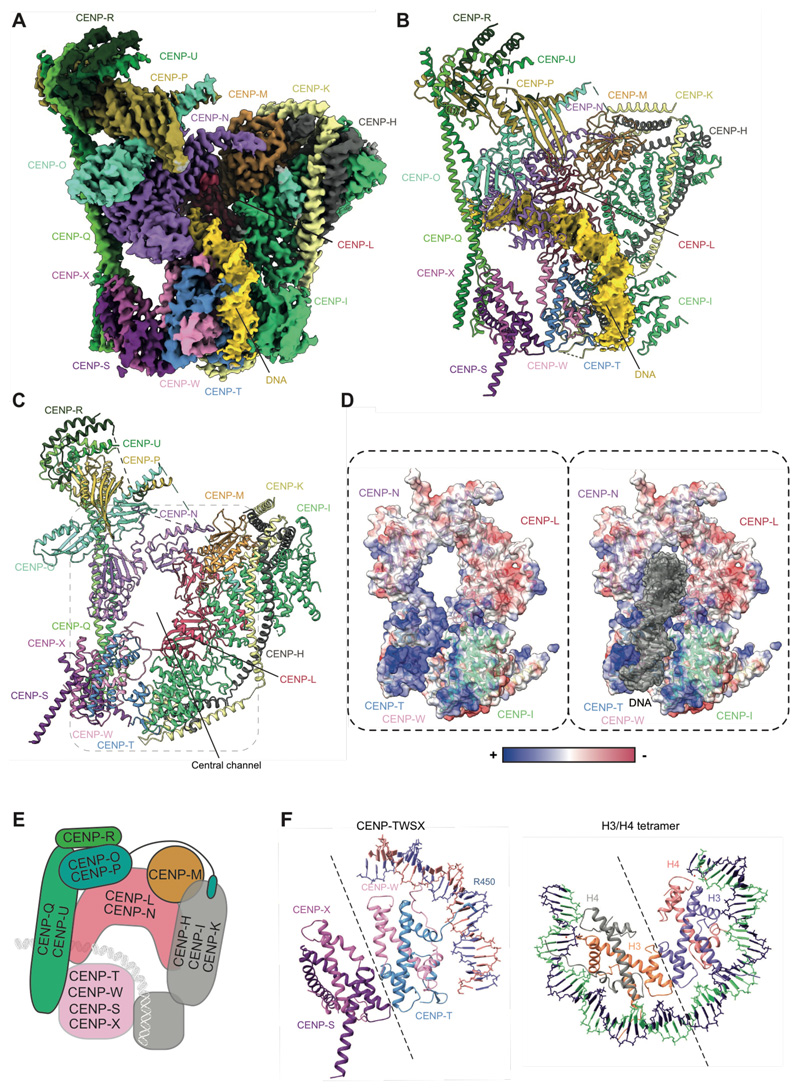
To understand how CENP-TWSX contributes to CCAN recognition of DNA and CENP-ANuc, we determined a cryo-EM structure of CCANΔC with a 53 bp DNA linker segment (fig. S4A). We obtained a well-resolved cryo-EM map at 2.8 Å resolution with all regions of CCAN and the DNA clearly defined (Fig. 4A, B, fig. S11, Table S1 and Movie S2).
Figure 4. Architecture of the full CCANΔC-DNA complex.
(A) Cryo-EM density map, and (B) molecular model of the CCANΔC-DNA complex with DNA map density shown in yellow. (C) CCAN assembles an enclosed chamber that topologically entraps linker DNA. (D) Electrostatic representation of the DNA-binding chamber formed by CENP-LN, CENP-TW and CENP-HIKHead modules shows a positively charged chamber that tightly grips DNA. (E) Schematic of CCAN showing DNA path through the CENP-LN, CENP-HIKM and CENP-TWSX modules. (F) The CENP-TWSX module resembles the canonical H3/H4 nucleosome tetramer, and partially wraps DNA.
The four histone fold domains of CENP-TWSX resemble the histone H3-H4 tetramer, and are similar to crystal structures of isolated chicken CENP-TWSX (19). CENP-TWSX forms multiple interactions with neighboring CCAN subunits and DNA (Fig. 4B, C). Contacts with a slightly repositioned CENP-HIKHead are mediated by both CENP-T and the extended N terminus of CENP-W. CENP-W also directly contacts the CENP-N pyrin domain. A CENP-TWSX-mediated linkage of CENP-HIKHead to CENP-N and CENP-QUFoot creates an enclosed chamber that topologically entraps the extranucleosomal DNA (Fig. 4A-D). The chamber comprises CENP-LN, CENP-HIK and CENP-TW modules and is augmented and rigidified by all other CCAN subunits (Fig. 4E). As in the CCANΔT-DNA complex, the DNA duplex inserts into the deep CENP-LN channel, however CENP-TWSX and the repositioned CENP-HIKHead, induce a marked curvature of the DNA. Strikingly, the DNA starts to wrap around the CENP-TW histone fold domains as it emerges from the CENP-LN channel in a manner comparable to how a canonical H3-H4 tetramer wraps DNA (Fig. 4F). This DNA binding is augmented by CENP-HIKHead. Consequently, the DNA duplex engages a 50 A-long positively-charged groove at the interface of CENP-IHead and CENP-TW (Fig. 4D). Similar to nucleosomes, the N-terminus of the CENP-T (HFD) inserts into the minor groove of the extranucleosomal DNA positioning the highly conserved Arg450 to contact the DNA backbone (Fig. 4F). The CENP-LN channel and CENP-TW-CENP-IHead groove perfectly complement the DNA duplex, with the DNA phosphate backbone forming a continuous interface with CCAN, burying some 976 A2 of DNA surface area.
To complete our structural analysis, we determined cryo-EM structures of CCAN in complex with CENP-ANuc reconstituted with 171 bp α-satellite DNA (Fig. 5A, B and fig. S11C-E). CCAN assembles onto CENP-ANuc similarly to CCANΔT. The nucleosome associates with the back-face of CCAN, and the extranucleosomal DNA threads through the CENP-LN channel. The linker DNA of the α-satellite repeat is gently curved and interacts with CENP-TW-CENP-IHead reminiscent of the CCANΔC-DNA complex. We note that the AS2 α-satellite DNA of 171 bp in length is approximately 15 bp too short to wrap all the way around the CENP-TWSX module.
Figure 5. Structure of the complete CCAN-CENP-A inner kinetochore module.
(A) Atomic model of the CCAN-CENP-ANuc complex. (B) Cartoon schematic of the CCAN-CENP-ANuc complex. (C) Mutations of CENP-N and CENP-TW previously shown to impair centromere function contribute to the DNA-binding channel of the CCAN (4, 7–9). Residues implicated in DNA binding by CENP-L, and mutated in the current study, are also shown.
CENP-TW contributes to DNA binding via conserved basic residues that were previously implicated in mediating DNA binding in vitro, and for CENP-W, are required for mitotic progression, and its recruitment to kinetochores in vivo. These functions of CENP-TW, and CENP-TWSX's ability to supercoil DNA, led to proposals that CENP-TWSX is a DNA-binding nucleosome-like particle, consistent with the curvature of the DNA in our structure (19, 29). These residues of CENP-TW, together with basic residues of CENP-N, whose mutation disrupt CENP-N association with CENP-ANuc in vitro, and centromere localization in vivo (4, 7–9), form a continuous spine lining the conserved CCAN-DNA interface (Fig. 5C).
Many vertebrate cell lines are viable and exhibit normal chromosome segregation without the CENP-OPQUR module (12, 30), contrasting with our finding that deletion of the CENP-QUΔFoot abrogates CCANΔCT binding to CENP-ANuc (fig. S8B, C). A likely explanation for this is the absence of the CENP-TWSX module in CCANΔCT, that contributes to DNA binding (Figs 4 and 5A, B). Consistent with this, we found that in the presence of CENP-TWSX, removing the entire CENP-OPQUR module (CCANΔCO), caused only a small decrease in the affinity for both DNA and CENP-ANuc (fig. S12A, B). Additionally, CCANΔCO readily formed a complex with CENP-ANuc (fig. S12C). These in vitro results support the idea that CENP-OPQUR contributes relatively little to the attachment of vertebrate kinetochores to chromatin.
Discussion
The absence of both mitotic phosphorylation and the outer kinetochore suggests the structure we describe likely reflects an interphase kinetochore. A cryo-EM structure of human apo-CCAN (31) agrees with our CCAN reconstructions, and the proposed model of DNA engagement by the CENP-LN channel is shown from our CCAN-CENP-ANuc complex to represent extranucleosomal linker DNA. Defining the structure of the holo-kinetochore at mitosis remains an outstanding challenge, and it will be important to validate the cryo-EM structures in cells.
The remarkably similar features of DNA duplex recognition by the basic CENP-LN channel of HsCCAN and ScCCAN (17), suggests a common evolutionary origin of centromere-kinetochore assembly. In humans, however, the DNA duplex engaged by CCAN is the α-satellite linker DNA rather than the unwrapped CENP-ANuc DNA of budding yeast. CCAN engages ~40 bp of extranucleosomal DNA which could be facilitated either by unwrapping of ~16 bp of the upstream CENP-ANuc, consistent with our structures showing DNA unwrapping in CENP-ANuc, and/or by the sparse occupancy of CENP-ANuc on centromeric DNA (32). Such organization is also consistent with evidence that CENP-C and CENP-T bridge adjacent nucleosomes (28). The topological entrapment of linker DNA provides a molecular explanation for how the inner kinetochore can withstand strong pushing and pulling forces applied by the mitotic spindle, a central function of this large macromolecular assembly. Our work also confirms CENP-C as a key determinant of CCAN specificity for CENP-ANuc, providing a molecular explanation for how this essential CCAN component directly links the inner kinetochore to centromeric chromatin.
Our finding that CCAN can bind to either canonical or CENP-A nucleosomes bearing adequate linker DNA could provide an explanation for how the inner kinetochore maintains attachments at centromeres where H3Nuc vastly exceeds CENP-ANuc across the entire centromeric α-satellite repeats (33). Furthermore, it could explain why synthetic depletion of CENP-A once CCAN is assembled onto centromeres, nevertheless permits mitotic centromere function in human cells (34). Salt extraction of CENP-A does reduce CENP-N and CENP-C levels in centromeric chromatin (35), suggesting that once CCAN is assembled at centromeres in a CENP-A dependent process (36, 37), CENP-A is dispensable for localization. Lastly, assembly of CCAN onto linker DNA provides a possible explanation for how some organisms that lack CENP-A yet still depend on CCAN for chromosome segregation might function (38).
An emerging question from this study is how human CCAN localizes specifically to the centromere given its modest preference for CENP-ANuc mono-nucleosomes compared to H3Nuc (33). One explanation is that the CENP-LN module is selectively recruited to the centromere, through recognition of CENP-AL1 loop, during early stages of the cell cycle, and is then displaced to form a fully assembled CCAN, thus confining kinetochore assembly to a specific chromatin locus. CENP-LN levels at the centromere decline upon mitotic entry (10, 39), supporting this sequential and specific localization mechanism. CDK-mediated CENP-C phosphorylation (11, 40) and/or chromatin compaction (39) might regulate this process. CENP-B recognizes the B-box motifs of the α-satellite repeat and interacts with CENP-C, providing another possible mechanism for specific CCAN recruitment (41). Different degrees of wrapping of CENP-ANuc in centromeric arrays compared to H3Nuc might also provide selectivity for CCAN assembly, as could the H1 histone that would block access of CCAN to the linker DNA at H3Nuc. Our structure permits a model for the arrangement of CCAN within the centromeric arrays of regional kinetochores (fig. S12D).
This study also highlights the common mechanisms of topological entrapment used by biological systems responsible for accurately mediating chromosome segregation, such as the cohesin complex that, likewise resisting vast forces exerted by the spindle, maintains cohesion between sister chromatids.
Supplementary Material
3D variability analysis demonstrates a continuous conformational heterogeneity of the DNA termini of the CENP-ANuc.
Overview of the cryo-EM reconstruction and molecular model of the CCANΔC-DNA complex.
One-Sentence Summary.
The human inner kinetochore CCAN complex tightly grips the linker DNA of the α-satellite CENP-A nucleosome.
Acknowledgments
We are grateful to the LMB and eBIC EM facilities, and Diamond Light Source for help with the EM and X-ray data collection, J. Grimmett and T. Darling for computing, A. Burt for assistance with 3D variability analysis, H. Kramer and F. Begum for mass spectrometry analysis, and J. Shi for help with insect cell expression.
Funding
UKRI/Medical Research Council MC_UP_1201/6 (DB)
Cancer Research UK C576/A14109 (DB)
Boehringer Ingleheim Fonds Fellowship (SY)
Footnotes
Author contributions:
D.B., K.W.M. and S.Y. designed the study and experiments. K.W.M. and S.Y. cloned constructs except H3 nucleosome, which was cloned by Z.Z. Z.Z. produced and purified all DNA substrates. S.Y. and K.W.M. purified all proteins with help from J.Y., Z.Z. and M.P. S.Y. and K.W.M. performed all biochemical experiments, prepared EM grids, collected and processed the cryo-EM data and built atomic models together with D.B. T.D. helped with cryo-EM data processing. T.T. performed all in vivo experiments. D.Be. crystallized OPQURFoot and collected crystallography data and assisted with crystallography data processing. S.H.M. performed SEC-MALS experiments. D.B., K.W.M. and S.Y. wrote the manuscript with input from all authors.
Competing interests: Authors declare that they have no competing interests.
Data and materials availability
PDB and cryo-EM maps have been deposited with RCSB and EMDB, respectively. Accession numbers are listed in Tables S1 and S2.
References and Notes
- 1.Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 2.Logsdon GA, Vollger MR, Hsieh P, Mao Y, Liskovykh MA, Koren S, Nurk S, Mercuri L, Dishuck PC, Rhie A, de Lima LG, et al. The structure, function and evolution of a complete human chromosome 8. Nature. 2021;593:101–107. doi: 10.1038/s41586-021-03420-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Navarro AP, Cheeseman IM. Kinetochore assembly throughout the cell cycle. Semin Cell Dev Biol. 2021 doi: 10.1016/j.semcdb.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nature cell biology. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. The Journal of cell biology. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 2013;340:1110–1113. doi: 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pentakota S, Zhou K, Smith C, Maffini S, Petrovic A, Morgan GP, Weir JR, Vetter IR, Musacchio A, Luger K. Decoding the centromeric nucleosome through CENP-N. eLife. 2017;6 doi: 10.7554/eLife.33442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chittori S, Hong J, Saunders H, Feng H, Ghirlando R, Kelly AE, Bai Y, Subramaniam S. Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N. Science. 2018;359:339–343. doi: 10.1126/science.aar2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian T, Li X, Liu Y, Wang C, Liu X, Bi G, Zhang X, Yao X, Zhou ZH, Zang J. Molecular basis for CENP-N recognition of CENP-A nucleosome on the human kinetochore. Cell Res. 2018;28:374–378. doi: 10.1038/cr.2018.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allu PK, Dawicki-McKenna JM, Van Eeuwen T, Slavin M, Braitbard M, Xu C, Kalisman N, Murakami K, Black BE. Structure of the Human Core Centromeric Nucleosome Complex. Current biology: CB. 2019;29:2625–2639.:e2625. doi: 10.1016/j.cub.2019.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ariyoshi M, Makino F, Watanabe R, Nakagawa R, Kato T, Namba K, Arimura Y, Fujita R, Kurumizaka H, Okumura EI, Hara M, et al. Cryo-EM structure of the CENP-A nucleosome in complex with phosphorylated CENP-C. The EMBO journal. 2021;40:e105671. doi: 10.15252/embj.2020105671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKinley KL, Sekulic N, Guo LY, Tsinman T, Black BE, Cheeseman IM. The CENP-L-N Complex Forms a Critical Node in an Integrated Meshwork of Interactions at the Centromere-Kinetochore Interface. Molecular cell. 2015;60:886–898. doi: 10.1016/j.molcel.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klare K, Weir JR, Basilico F, Zimniak T, Massimiliano L, Ludwigs N, Herzog F, Musacchio A. CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. The Journal of cell biology. 2015;210:11–22. doi: 10.1083/jcb.201412028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo LY, Allu PK, Zandarashvili L, McKinley KL, Sekulic N, Dawicki-McKenna JM, Fachinetti D, Logsdon GA, Jamiolkowski RM, Cleveland DW, Cheeseman IM, et al. Centromeres are maintained by fastening CENP-A to DNA and directing an arginine anchor-dependent nucleosome transition. Nature communications. 2017;8:15775. doi: 10.1038/ncomms15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pesenti ME, Prumbaum D, Auckland P, Smith CM, Faesen AC, Petrovic A, Erent M, Maffini S, Pentakota S, Weir JR, Lin YC, et al. Reconstitution of a 26-Subunit Human Kinetochore Reveals Cooperative Microtubule Binding by CENP-OPQUR and NDC80. Molecular cell. 2018;71:923–939.:e910. doi: 10.1016/j.molcel.2018.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinshaw SM, Harrison SC. The structure of the Ctf19c/CCAN from budding yeast. eLife. 2019;8 doi: 10.7554/eLife.44239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan K, Yang J, Zhang Z, McLaughlin SH, Chang L, Fasci D, Ehrenhofer-Murray AE, Heck AJR, Barford D. Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome. Nature. 2019;574:278–282. doi: 10.1038/s41586-019-1609-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basilico F, Maffini S, Weir JR, Prumbaum D, Rojas AM, Zimniak T, De Antoni A, Jeganathan S, Voss B, van Gerwen S, Krenn V, et al. The pseudo GTPase CENP-M drives human kinetochore assembly. eLife. 2014;3:e02978. doi: 10.7554/eLife.02978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishino T, Takeuchi K, Gascoigne KE, Suzuki A, Hori T, Oyama T, Morikawa K, Cheeseman IM, Fukagawa T. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 2012;148:487–501. doi: 10.1016/j.cell.2011.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka Y, Tachiwana H, Yoda K, Masumoto H, Okazaki T, Kurumizaka H, Yokoyama S. Human centromere protein B induces translational positioning of nucleosomes on alpha-satellite sequences. The Journal of biological chemistry. 2005;280:41609–41618. doi: 10.1074/jbc.M509666200. [DOI] [PubMed] [Google Scholar]
- 21.Hasson D, Panchenko T, Salimian KJ, Salman MU, Sekulic N, Alonso A, Warburton PE, Black BE. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nature structural & molecular biology. 2013;20:687–695. doi: 10.1038/nsmb.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henikoff JG, Thakur J, Kasinathan S, Henikoff S. A unique chromatin complex occupies young alpha-satellite arrays of human centromeres. Sci Adv. 2015;1 doi: 10.1126/sciadv.1400234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, Kimura H, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 24.Nechemia-Arbely Y, Fachinetti D, Miga KH, Sekulic N, Soni GV, Kim DH, Wong AK, Lee AY, Nguyen K, Dekker C, Ren B, et al. Human centromeric CENP-A chromatin is a homotypic, octameric nucleosome at all cell cycle points. The Journal of cell biology. 2017;216:607–621. doi: 10.1083/jcb.201608083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Padeganeh A, Ryan J, Boisvert J, Ladouceur AM, Dorn JF, Maddox PS. Octameric CENP-A nucleosomes are present at human centromeres throughout the cell cycle. Current biology : CB. 2013;23:764–769. doi: 10.1016/j.cub.2013.03.037. [DOI] [PubMed] [Google Scholar]
- 26.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, Bridgland A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021 doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thakur J, Henikoff S. CENPT bridges adjacent CENPA nucleosomes on young human alpha-satellite dimers. Genome Res. 2016;26:1178–1187. doi: 10.1101/gr.204784.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, Cheeseman IM, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 30.Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, Desai A, 3rd, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nature cell biology. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 31.Pesenti ME, Raisch T, Conti D, Hoffmann I, Vogt D, Prumbaum D, Vetter IR, Raunser S, Musacchio A. Structure of the human inner kinetochore CCAN complex and its significance for human centromere organization. BioRxiv. 2022:2022.01.06.475204. doi: 10.1101/2022.01.06.475204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann S, Izquierdo HM, Gamba R, Chardon F, Dumont M, Keizer V, Herve S, McNulty SM, Sullivan BA, Manel N, Fachinetti D. A genetic memory initiates the epigenetic loop necessary to preserve centromere position. The EMBO journal. 2020;39:e105505. doi: 10.15252/embj.2020105505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, Jansen LE. The quantitative architecture of centromeric chromatin. eLife. 2014;3:e02137. doi: 10.7554/eLife.02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmann S, Dumont M, Barra V, Ly P, Nechemia-Arbely Y, McMahon MA, Herve S, Cleveland DW, Fachinetti D. CENP-A Is Dispensable for Mitotic Centromere Function after Initial Centromere/Kinetochore Assembly. Cell reports. 2016;17:2394–2404. doi: 10.1016/j.celrep.2016.10.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao S, Zhou K, Zhang Z, Luger K, Straight AF. Constitutive centromere-associated network contacts confer differential stability on CENP-A nucleosomes in vitro and in the cell. Molecular biology of the cell. 2018;29:751–762. doi: 10.1091/mbc.E17-10-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LE, Cleveland DW. A two-step mechanism for epigenetic specification of centromere identity and function. Nature cell biology. 2013;15:1056–1066. doi: 10.1038/ncb2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 38.Cortes-Silva N, Ulmer J, Kiuchi T, Hsieh E, Cornilleau G, Ladid I, Dingli F, Loew D, Katsuma S, Drinnenberg IA. CenH3-Independent Kinetochore Assembly in Lepidoptera Requires CCAN, Including CENP-T. Current biology: CB. 2020;30:561–572.:e510. doi: 10.1016/j.cub.2019.12.014. [DOI] [PubMed] [Google Scholar]
- 39.Fang J, Liu Y, Wei Y, Deng W, Yu Z, Huang L, Teng Y, Yao T, You Q, Ruan H, Chen P, et al. Structural transitions of centromeric chromatin regulate the cell cycle-dependent recruitment of CENP-N. Genes & development. 2015;29:1058–1073. doi: 10.1101/gad.259432.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe R, Hara M, Okumura EI, Herve S, Fachinetti D, Ariyoshi M, Fukagawa T. CDK1-mediated CENP-C phosphorylation modulates CENP-A binding and mitotic kinetochore localization. The Journal of cell biology. 2019;218:4042–4062. doi: 10.1083/jcb.201907006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, Cleveland DW. DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function. Developmental cell. 2015;33:314–327. doi: 10.1016/j.devcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, Yang J, Barford D. Recombinant expression and reconstitution of multiprotein complexes by the USER cloning method in the insect cell-baculovirus expression system. Methods. 2016;95:13–25. doi: 10.1016/j.ymeth.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 43.Yang TP, Hansen SK, Oishi KK, Ryder OA, Hamkalo BA. Characterization of a cloned repetitive DNA sequence concentrated on the human X chromosome. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:6593–6597. doi: 10.1073/pnas.79.21.6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou BR, Yadav KNS, Borgnia M, Hong J, Cao B, Olins AL, Olins DE, Bai Y, Zhang P. Atomic resolution cryo-EM structure of a native-like CENP-A nucleosome aided by an antibody fragment. Nature communications. 2019;10:2301. doi: 10.1038/s41467-019-10247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 46.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta crystallographica. Section D, Biological crystallography. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta crystallographica Section D, Biological crystallography. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 48.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nature protocols. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 51.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7 doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagner T, Merino F, Stabrin M, Moriya T, Antoni C, Apelbaum A, Hagel P, Sitsel O, Raisch T, Prumbaum D, Quentin D, et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun Biol. 2019;2:218. doi: 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tegunov D, Cramer P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods. 2019;16:1146–1152. doi: 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 59.Wilkinson ME, Kumar A, Casanal A. Methods for merging data sets in electron cryo-microscopy. Acta crystallographica. Section D, Structural biology. 2019;75:782–791. doi: 10.1107/S2059798319010519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Punjani A, Zhang H, Fleet DJ. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat Methods. 2020;17:1214–1221. doi: 10.1038/s41592-020-00990-8. [DOI] [PubMed] [Google Scholar]
- 61.Tunyasuvunakool K, Adler J, Wu Z, Green T, Zielinski M, Zidek A, Bridgland A, Cowie A, Meyer C, Laydon A, Velankar S, et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021 doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein science : a publication of the Protein Society. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
3D variability analysis demonstrates a continuous conformational heterogeneity of the DNA termini of the CENP-ANuc.
Overview of the cryo-EM reconstruction and molecular model of the CCANΔC-DNA complex.
Data Availability Statement
PDB and cryo-EM maps have been deposited with RCSB and EMDB, respectively. Accession numbers are listed in Tables S1 and S2.