Summary
DNA polymerase epsilon (Polε) carries out high fidelity leading strand synthesis owing to its exonuclease activity. Polε polymerase and exonuclease activities are balanced, due to partitioning of nascent DNA strands between catalytic sites, so that net resection occurs when synthesis is impaired. In vivo, DNA synthesis stalling activates replication checkpoint kinases, which act to preserve the functional integrity of replication forks. We show that stalled Polε drives nascent strand resection causing fork functional collapse, averted via checkpoint-dependent phosphorylation. Polε catalytic subunit Pol2 is phosphorylated on serine 430, influencing partitioning between polymerase and exonuclease active sites. A phosphormimetic S430D change reduces exonucleolysis in vitro and counteracts fork collapse. Conversely, non-phosphorylatable pol2-S430A expression causes resection-driven stressed fork defects. Our findings reveal that checkpoint kinases switch Polε to an exonuclease-safe mode preventing nascent strand resection and stabilizing stalled replication forks. Elective partitioning suppression has implications for the diverse Polε roles in genome integrity maintenance.
Introduction
In eukaryotes, replication of the nuclear genome is carried out by two major replicases: DNA polymerase ε and δ (Burgers and Kunkel, 2017; Johansson and Dixon, 2013). Polε continuously synthetizes leading strands, while Polδ periodically extends Okazaki fragment primers, synthetized by DNA polymerase α, for the discontinuous synthesis of lagging strands (Lujan et al., 2016). Polε has a large subunit (Pol2 in budding yeast) carrying DNA polymerization and 3’ to 5’ exonucleolytic activities (Dua et al., 1999), as well as an essential non-catalytic (Dpb2) and two small non-essential (Dpb3 and Dpb4) subunits. Dpb2 mediates the association of Polε with the Cdc45-MCM-GINS (CMG) replicative helicase, while Dpb3 and Dpb4 contribute to DNA polymerization processivity and parental histone transfer toward nascent strands (Hogg and Johansson, 2012; Yu et al., 2018). Polδ holoenzyme is composed of a large catalytic subunit (budding yeast Pol3) bearing polymerization and exonucleolytic activities, and two accessory subunits (Pol31 and Pol32) (Gerik et al., 1998; Johnson et al., 2012).
Polε and Polδ act at branched replication fork structures, within large replisome complexes that coordinate chromosomal DNA unwinding and synthesis (Bell and Labib, 2016). Replisome progression along chromosomes can stall on account of events impairing helix unwinding or DNA synthesis, challenging genome integrity and globally termed replication stress, and is monitored by DNA damage response (DNA replication checkpoint in budding yeast) kinases (Berti and Vindigni, 2016; Giannattasio and Branzei, 2017; Jossen and Bermejo, 2013). Mec1ATR senses fork stalling upon association to ssDNA tracks, formed owing to helicase-polymerase uncoupling, and is thereby activated (Flynn and Zou, 2011; Pardo et al., 2017). Mec1 in turn phosphorylates and activates the Rad53CHK1 propagating checkpoint signaling to downstream targets. Checkpoint kinases counteract the genotoxic impact of replication stress through phosphorylation of diverse factors, thus delaying cell cycle progression, stabilizing replication forks, preventing firing of additional replication origins and upregulating dNTP pools and damage-inducible genes. In pre-malignant cells oncogene deregulation induces chronic replication stress and, in this context, the DNA damage response is thought to act as a barrier to transformation (Bartek et al., 2007).
Fork protection is thought to be the critical mechanism mediated by checkpoint kinases to counteract replication stress-induced genome instability and cell death (Jossen and Bermejo, 2013; Tercero et al., 2003). Treatment with the replication inhibitor hydroxyurea (HU) downregulates de novo dNTP synthesis inducing polymerase stalling, which causes irreversible fork arrest in checkpoint mutants. Checkpoint kinase defective cells display terminal loss of fork functionality along with structural transitions (nascent strand reannealing and nucleolytic resection), in a process termed fork collapse ultimately causing cell death (Branzei and Foiani, 2010; Cortez, 2015). The key events determining the functional and structural collapse of stalled forks that are counteracted by checkpoint kinases are not fully understood, though they likely involve deleterious nascent strand transitions (Bermejo et al., 2011; Hu et al., 2012; Pasero and Vindigni, 2017; Rossi et al., 2015). It was proposed that dissociation of replisomes from DNA causes terminal arrest of stalled forks in checkpoint mutants (Cobb et al., 2003; Lucca et al., 2004). However, structurally intact replisomes remain associated to a large fraction of terminal forks, suggesting that preservation of replication capacity might be achieved through direct regulation of replisome components (De Piccoli et al., 2012). Even if checkpoint-dependent phosphorylation of replisome components has been reported, specific events crucial for the maintenance of stalled fork integrity have not been characterized (Lanz et al., 2019).
Polε and Polδ bear 3’ to 5’ exonuclease activities that remove terminal nucleotides from nascent DNA chains, responsible for replication proofreading and involved in Okazaki fragment maturation and mismatch repair (Reha-Krantz, 2010). Exonucleolysis requires switching of nascent DNA 3’ ends from polymerase to exonuclease catalytic sites, which can occur intramolecularly through unpairing of primer strand nucleotides to cover the 40 Å distance separating the two active sites (Ganai et al., 2015; Hogg et al., 2014; Swan et al., 2009). As result of site partitioning, polymerase and exonuclease activities are in balance, which is normally shifted toward DNA synthesis because of a faster rate of polymerization in comparison to that of exonucleolysis (Reha-Krantz, 2010). When polymerization rates are impaired removal of terminal nucleotides dominates. This is proposed to contribute to polymerase proofreading, as misincorporations slow down the attachment of the subsequent nucleotide thus increasing the likelihood of exonucleolytic error correction (Hoekstra et al., 2017). In vitro, impaired DNA polymerization owing to limiting dNTPs shifts balance toward exonucleolysis, resulting in repeated removal events and processive degradation of nascent (primer) strands (Ganai et al., 2015; de Vega et al., 1996). To our knowledge, the in vivo behavior of replicative polymerase exonuclease activities upon fork stalling has not been addressed.
We report that the Polε is phosphorylated in a manner dependent on the Rad53 kinase, reflective of regulation by the DNA replication checkpoint in response to replication stress. Polε exonuclease activity drives nascent DNA strand resection at stalled forks in checkpoint kinase impaired cells, thus causing fork collapse and cell death. Phosphorylation of Pol2 on serine 430 within its exonuclease domain influences active site switching and nascent strand resection rates. Mimicking Pol2-S430 phosphorylation limits exonucleolysis by Polε in vitro and counteracts fork collapse in vivo, while precluding its phosphorylation results in resection and failure to stabilize stalled forks. These data suggest that, through phosphorylation of S430 in Pol2, nascent strands are prevented from gaining access to exonuclease sites, abnormally active when Polε stalls, thereby preserving stalled fork integrity.
Results
Replication checkpoint-dependent phosphorylation of DNA polymerase ε
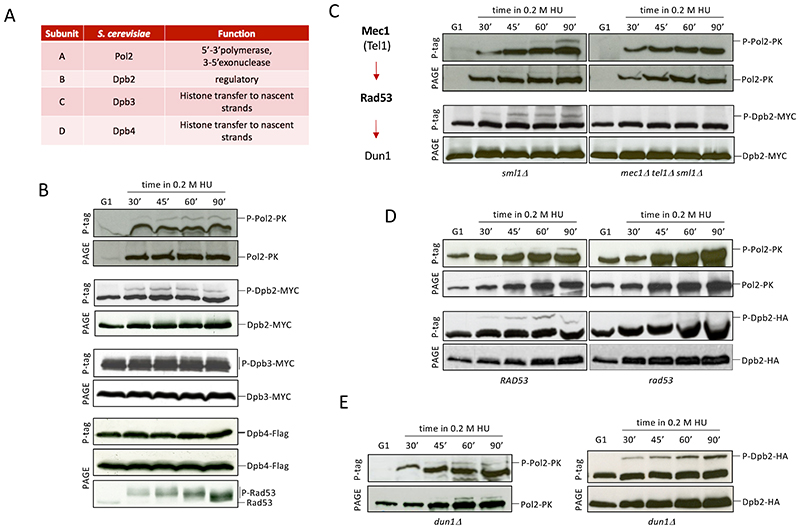
While searching for replisome factors targeted by checkpoint kinases to protect fork integrity, we investigated the phosphorylation status of the budding yeast replicases DNA polymerase ε and DNA polymerase δ. Cells expressing epitope-tagged subunits of Polε or Polδ (Figure 1A and S1A) were released into a synchronous S phase in the presence of 0.2M hydroxyurea (HU) to deplete dNTP pools and induce fork stalling. Polymerase subunits were immunodetected following polyacrylamide gel electrophoresis (PAGE) in the presence or absence of PhosTag, which retards the migration of phosphorylated proteins (Kinoshita et al., 2006). We observed band retardation in PhosTag gels for Pol2, Dpb2 and Dpb3 Polε subunits (Figure 1B) as well as of Polδ Pol3, Pol31 and Pol32 subunits (Figure S1B), pointing at phosphorylation of both holoenzymes. We analyzed the involvement of the DNA replication checkpoint in mediating these events by performing equivalent experiments with cells ablated for Mec1 and Tel1 apical kinases. Detection of Pol2 and Dpb2 phosphoisoforms, but not of Dpb3 or Polδ subunits, depended on checkpoint signaling (Figure 1C, S1C and S1D). Further dissecting downstream dependencies of checkpoint-mediated modifications, we found that phosphorylation of both Pol2 and Dpb2 was abolished in kinase-deficient rad53-K227A mutants (Figure 1D), but was observed in cells ablated for the downstream Dun1 kinase (Figure 1E), indicating that Polε phosphorylation is mediated through Rad53. Pol2 and Dpb2 Phos-Tag retarded bands were not detected in unperturbed replicating cells (Figure S1E). Hence, upon replication fork stalling the Rad53 checkpoint kinase mediates Polε phosphorylation on the essential catalytic and accessory Pol2 and Dpb2 subunits.
Figure 1. Pol2 and Dpb2 are phosphorylated in a checkpoint-dependent manner.
(A) DNA polymerase ε subunits. (B) Western blot (WB) of proteins from epitope-tagged Pol2, Dpb2, Dpb3 or Dpb4 cells released from an α-factor induced block (G1) into a synchronous S-phase in the presence of 0.2 M hydroxyurea (HU) subject to electrophoresis in the presence (P-tag) or absence (PAGE) of PhosTag. Checkpoint signaling was inferred from Rad53 phosphorylation status. (C) WB analysis of Pol2 and Dpb2 phosphorylation in mec1 tel1 cells. Schematic of the replication checkpoint signaling cascade. (D-E) WB analysis of Pol2 and Dpb2 phosphorylation in rad53-K227A (rad53) (D) and in dun1 (E) cells. See also Figure S1.
Stalled fork resection is promoted by the exonuclease activity of DNA polymerase ε
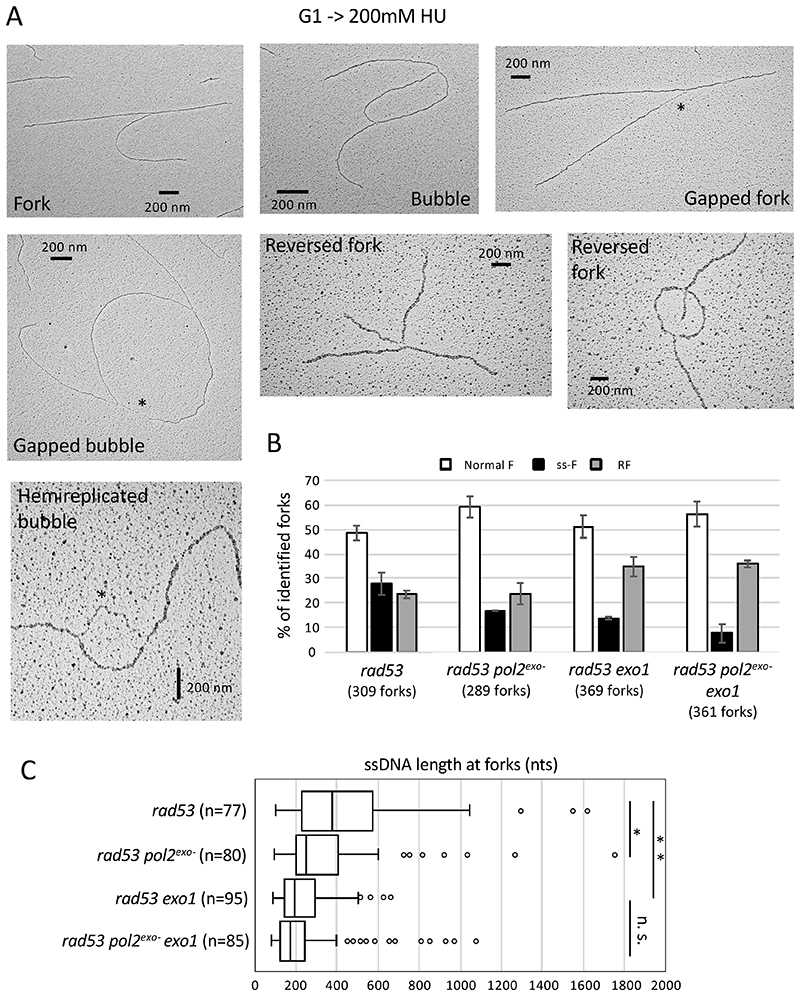
Checkpoint-dependent phosphorylation hinted at regulation of Polε function in response to replication stress. We reasoned that such regulation was likely exerted on its exonuclease activity that, similarly to what is observed in vitro, might degrade nascent DNA upon synthesis stalling. To address this hypothesis, we visualized psoralen-crosslinked replication intermediates from HU-treated checkpoint-deficient cells bearing exonuclease-inactivating mutations by electron microscopy (EM). Fork collapse in checkpoint mutants is characterized by the accumulation of reversed fork structures (RF), as well as gapped and hemi-replicated molecules with abnormally extended ssDNA stretches at fork branching points (ss-F) (Figure 2A), representing about 50% of replication intermediates in HU-treated rad53-K227A kinase-deficient cells (Figure 2B) (Sogo et al., 2002). Accumulation of forks containing extended ssDNA in rad53 cells partly depends on stalled nascent DNA strand resection by the Exo1 exonuclease (Cotta-Ramusino et al., 2005). We observed that expression of the pol2-4 exonuclease-dead allele (pol2exo-) (Shcherbakova and Pavlov, 1996) lead to a 40% reduction in the proportion of forks with extended ssDNA tracks and an equivalent increase in normal fork structures in checkpoint mutant cells, without altering reversed fork detection (Figure 2B). Consistent with previous studies, ablation of Exo1 also resulted in a (50%) reduction of the abundance of ssDNA intermediates in rad53 cells, further decreased (by 70%) in pol2exo- exo1 cells. ssDNA track lengths at fork junctions were drastically reduced in cells devoid of the exonuclease activities of Polε or Exo1 (with 120 and 180 nucleotide shorter median tracks, respectively) or lacking both (200 nucleotides shorter) (Figure 2C). These data indicate that the exonuclease activity of Polε is responsible for ssDNA accumulation at replication forks in absence of a functional checkpoint response, likely through resection of stalled nascent strands.
Figure 2. Polε exonuclease activity drives ssDNA accumulation at stalled replication forks in checkpoint-deficient cells.
(A) Representative transmission electron microscopy (TEM) pictures of in vivo psoralen-crosslinked normal or altered replication fork structures from rad53 cells 90 minutes after G1 release into 0.2M HU. Asterisks mark extended ssDNA tracks at fork junction points, the length of which is analyzed in C. Scale bars of 200nm correspond to roughly 555 base pairs. (B) Histogram plot representing means and SDs of the percentages of normal forks (bubbles and forks), ssDNA-containing forks (ss-F) (gapped forks, gapped bubbles and hemi-replicated bubbles) and reversed fork structures (RF) observed in rad53, rad53 pol2-4, rad53 exo1Δ and rad53 pol2-4 exo1Δ cells in two independent experiments. The total number of forks analyzed for each sample is reported. (C) Box plot showing length distributions of ssDNA gaps at fork branching points in ss-F molecules identified in B. Center line, median; box limits, 10th and 90th percentiles; whiskers, 1st and 99th percentiles; dots, outliers. * p < 0.05 and ** p<0.01 by two-tailed t test. N notes the number of ss-F molecules analyzed.
The exonuclease activity of DNA polymerase ε drives fork collapse
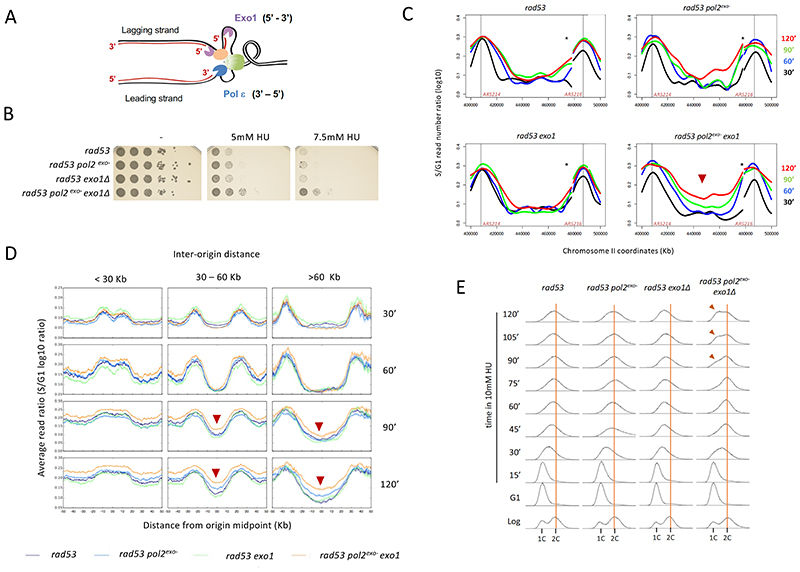
We examined the functional impact of Polε-mediated nascent strand exonucleolysis on stalled fork stability by evaluating sensitivity to replication stress. Due to its function in chromosome replication and position in the eukaryotic replisome, Polε contacts 3’ ends of stalled leading strands (Figure 3A). Conversely, Exo1 is thought to resect lagging nascent DNA, owing to its 5’ to 3’ exonuclease activity, using discontinuities between Okazaki fragments as entry points (Colosio et al., 2016). Of note, Rad53 phosphorylates Exo1 and this modification limits Exo1-mediated fork resection (Engels et al., 2011; Morafraile et al., 2020). We observed that counteracting Polε-mediated strand resection by introduction of a Pol2 exonuclease-dead allele fails to improve survival of rad53 mutants treated with HU (Figure 3B). As reported, Exo1 ablation also failed to supress the high HU sensitivity of checkpoint-deficient cells (Cotta-Ramusino et al., 2005; Segurado and Diffley, 2008). In contrast, combined abolishment of Polε and Exo1 exo activities synthetically rescued the viability of HU-treated rad53 cells (Figure 3B), indicating that these activities act in parallel to cause lethality, likely by resecting and inducing the collapse of stalled forks. Hence, fork protection by checkpoint kinases likely involves parallel suppression of Polε- and Exo1-mediated exonucleolysis of leading and lagging nascent strands.
Figure 3. Polε exonuclease activity drives fork collapse in checkpoint-deficient cells.
(A) Polε and Exo1 contacts with nascent strand ends. Exo1 and Polε exo polarities are shown. (B) Serial dilutions of rad53, rad53 pol2-4, rad53 exo1 and rad53 pol2-4 exo1 cells in absence or presence of 5mM and 7.5mM HU. (C) CGS analysis of fork progression in checkpoint and exonuclease deficient cells. Curves represent log10 S/G1 ratios of reads obtained from S-phase samples at the indicated times after G1 release in the presence of 10mM HU. A 120-Kb genomic region on chromosome II containing ARS214 and ARS216 early replication origins (black vertical lines) is shown. Asterisks mark a repetitive region rendering uninformative ratios. (D) Average read ratios across genomic regions categorized by inter-origin distance of the indicated cells along the time course in C. Red arrow heads mark time points with increased replication completion in rad53 pol2exo- exo1 cells. (E) Flow cytometry analysis of logarithmically growing (Log) cells, G1-blocked (G1) and released into S-phase in 10 mM HU. Vertical orange bars mark the 2C DNA content of G2/M cells. Red arrowheads mark 1C cells in a second cell cycle. See also Figure S2 and S3.
Combined ablation of Polε and Exo1 exo activities also improved MEC1 deleted cells survival in HU (Figure S2A), but failed to rescue the milder sensitivity of dun1 deleted cells (Figure S2B), consistent with Polε regulation occurring through Rad53 upstream of Dun1. We examined whether Polδ, in contact with nascent DNA at 3’ ends at lagging strands and bearing a 3’ to 5’ exo activity (Shcherbakova and Pavlov, 1996), contributed to fork collapse. We observed that the exo- pol3-01 (pol3exo-) allele failed to suppress rad53 cells HU sensitivity alone or in combination with pol2exo- (Figure S2C). We engineered a system for conditional Exo1 expression to test the combined effect of Exo1 elimination with Polε or Polδ exo inactivation. While Exo1 depletion suppressed of rad53 pol2exo- cells HU sensitivity (Figure S2D), it failed to rescue rad53 pol3exo- mutants, suggesting that the exonuclease activity of Polδ is not a critical fork collapse driver in this context.
We next investigated the impact of Polε and Exo1 exonuclease activities on stressed fork integrity monitoring chromosome duplication through copy number increase by Comparative Genome Sequencing (CGS) (Frattini et al., 2017). We inferred replication fork progression along chromosomes in checkpoint-deficient cells subject to mild stress (10 mM HU) (Figure 3C). In rad53 cells and single exo mutants, DNA synthesis slowly progressed away from replication origins for few (15-20) kilobases before arresting, likely as a result of fork collapse. In contrast, synthesis advanced steadily in cells devoid of both Polε and Exo1 exo activities, resulting in gradual replication completion most evident at genomic regions flanked by distant origins (as the 80 Kb-away ARS214 and ARS216 origins on chromosome II). Defects in Polε and Exo1 exo activities did not change the de-repression of late origins in rad53 mutants (Santocanale and Diffley, 1998; Shirahige et al., 1998), which also contribute to increased replication completion in rad53 pol2exo- exo1 cells (Figure S3A). Genomewide, more stable fork progression in Pol2 and Exo1 double exo deficient cells anticipated replication completion of regions flanked by close (less than 30 Kb away) origins, although a large fraction of rad53 and single exo- cells completed their replication with time (Figure 3D and S3B). In contrast, only rad53 pol2exo- exo1 cells significantly increased replication completion of larger regions (Figure 3D and S3C). Thus, suppression of Polε and Exo1 exo activities promotes stressed replication fork functional integrity in absence of an operative checkpoint. Taken together these observations suggest that unrestrained resection by Polε and Exo1 impairs stressed fork stable progression compromising chromosome replication completion, particularly at regions flanked by distant origins. Consistent with this conclusion, upon mild stress induction pol2exo- exo1 cells were able to complete bulk genome replication and undergo cell division, while rad53 and single exonuclease mutants arrested with a not fully replicated DNA complement (Figure 3E). These results uncover the exonuclease activity of Polε as a key fork collapse driver, acting along Exo1.
Polε is phosphorylated on Pol2 serine 430, which influences polymerase-exonuclease site partitioning and stalled fork integrity
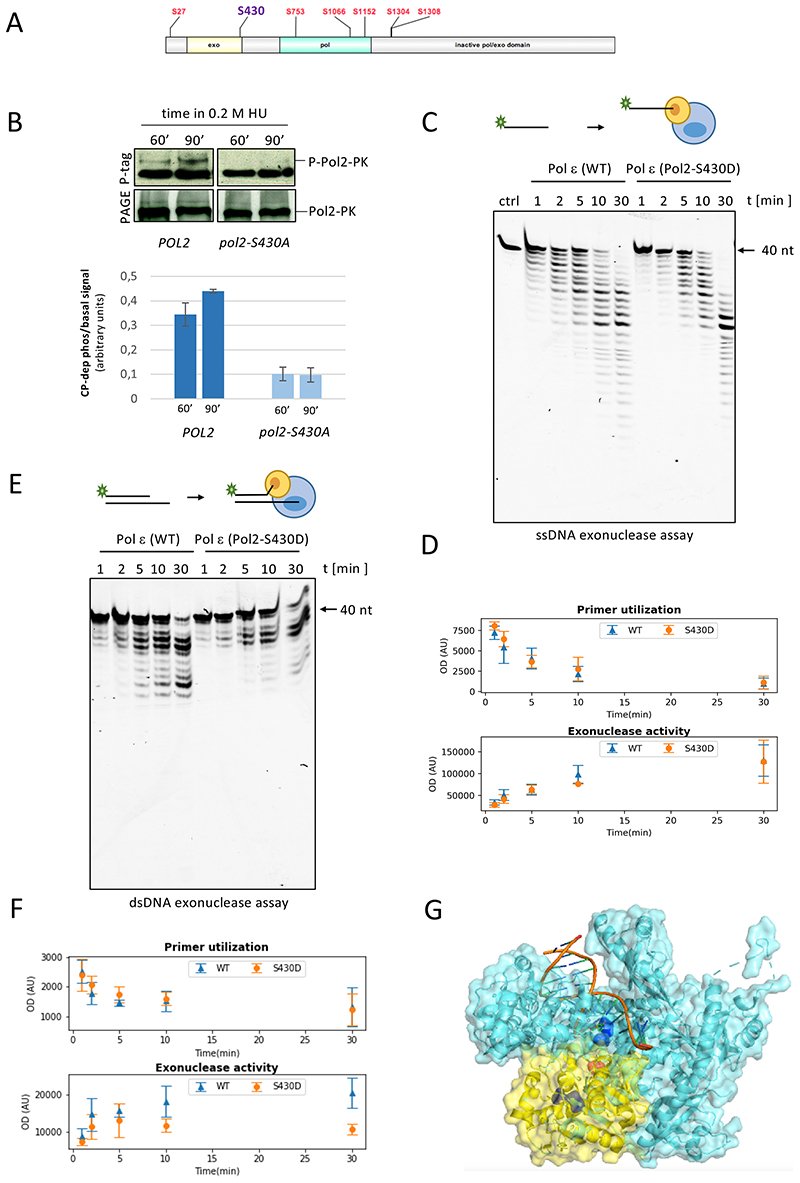
We reasoned that exonucleolysis by Polε might be suppressed via checkpoint-dependent phosphorylation. To address this point, we carried out mass spectrometric analyses and identified peptides containing phosphorylated serines and threonines in Pol2 and Dpb2 immunoprecipitates from cells experiencing replication stress (Figure S4A and S4B). Seven and four phospho-residues identified for Pol2 and Dpb2 proteins, respectively, matched the consensus motif of Rad53 targets (Smolka et al., 2007), with Pol2 phospho-residues clustered on the N-terminal half of the peptide sequence containing exonuclease and polymerase domains (Figure 4A). In vitro Rad53 phosphorylates Polε on the Pol2 and Dbp2 subunits, as well as Dpb4 and Dpb3 (Figure 4SC), raising the possibility that the checkpoint kinase directly targets the holoenzyme through the identified in vivo sites. We focused on phospho-serine 430 (Figure S4D), located at an extremity of the exonuclease domain sequence of Pol2. Substitution of Ser430 with a non-phosphorylatable alanine markedly decreased detection of Mec1/Rad53-dependent Pol2 phospho-isoforms in PhosTag gels (Figure 4B), suggesting that this residue accounts for a large fraction of checkpoint-driven phosphorylation in response to replication stress. To test the functional relevance of P-Ser430, we purified wild type Polε (Yeeles et al., 2015) along with a phosphomimetic variant in which Ser430 was replaced by aspartate (Pol2-S430D) and carried out in vitro exonuclease assays. We first performed reactions in which singled-stranded primers directly contact the exonuclease catalytic site. In these assays wild type and S430D Polε similarly degraded the (40 nt) primer substrate (Figure 4C and 4D), indicating that the phosphomimetic S430D change does not impart an exo catalytic defect or alter direct binding of primer (nascent) DNA to the exonuclease site. We performed similar assays using dsDNA substrates, in which annealed primer/template DNA interacts with the polymerase site first and primer strands must relocate to the exo site prior to undergoing resection. In these reactions, Polε-S430D showed a qualitatively different behaviour to wild type Polε, characterized by reduced exonucleolysis despite similar substrate utilization (Figure 4E and F), likely reflecting a defect in exonuclease processivity. Reduced activity on dsDNA assays suggests that the S430 phosphomimetic impairs switching between the pol and exo sites, so that the longer time required for primer DNA partitioning decreases the overall processivity of the exonuclease reaction. In order to analyse the balance between synthesis and exonucleolysis, we carried out polymerization/exonuclease-coupled assays in which the two activities compete (Figure S4E). Consistently, these reactions showed a similar reduction in the exo activity for Polε-S430D at low dNTP concentrations. Polε-S430D displayed decreased polymerization activity compared to the wild type protein in presence of limiting dNTPs, which was however abolished at high dNTP concentrations. Since high dNTPs limit switching to the exo active site, this further supports a defect in partitioning. In the crystal structure of the Pol2 catalytic core, Serine 430 lies between exonuclease and polymerase catalytic residues (Jain et al., 2014), in the path that nascent strands need to traverse upon displacement in order to reach the exonuclease catalytic cleft (Figure 4G and S4F). Hence, substitution by aspartate or attachment of a phosphate group to S430 likely limits nascent strand displacement from polymerase to nuclease active sites without directly affecting either.
Figure 4. Pol2 is phosphorylated on Serine 430, which influences partitioning between polymerase and exonuclease catalytic sites.
(A) Rad53 consensus phosphoresidue positions on Pol2 peptide sequence. (B) WB analysis of Pol2-S430A phosphorylation. Proteins extracted from cells expressing epitope-tagged Pol2 variants released from G1 block in 0.2 M hydroxyurea (HU) were run in the presence (P-tag) or absence (PAGE) of PhosTag. Histogram plots show mean and standard deviations of retarded (checkpoint-dependent phosphorylation) to basal Pol2 PhosTag band signal ratios from two independent experiments. (C-F) Single strand (C-D) and double strand (E-F) exonuclease assays performed with WT and S430D Polε. Means and standard deviations of quantified primer utilization and exonuclease activity for WT and S430D Polε in three independent experiments for each assay type are plotted. (G) Crystal structure of Pol2 catalytic core in ternary complex with 12-nt/16-nt DNA and an incoming nucleotide. Pol and exo domains depicted in light blue and yellow, respectively. Exo D290 and E292 and pol D640 and D877 catalytic residues are evidenced in dark blue and serine 430 in red. See also Figure S4.
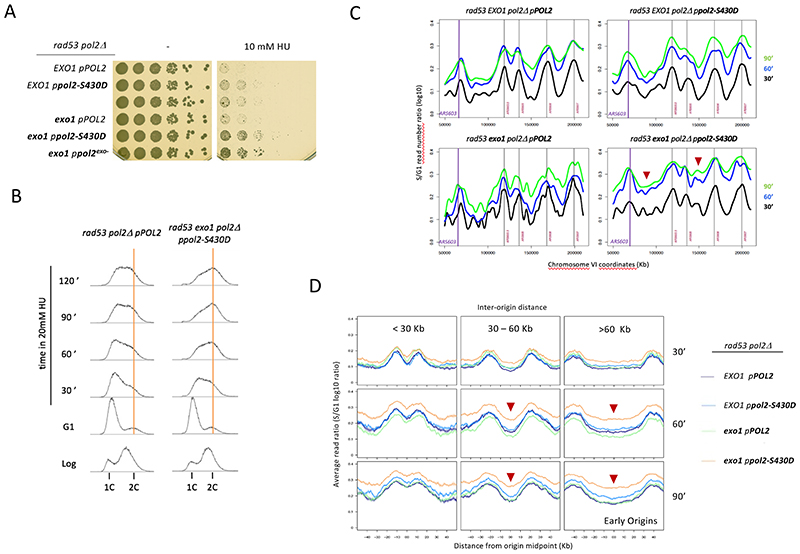
A reduced exonuclease activity owing to decreased partitioning would be expected to limit degradation of nascent strands also in vivo and hence promote the stability of stalling replication forks. To test this prediction, we provided plasmid-borne wild type or phospho-mimicking pol2-S430D as the only source of Pol2 expression in checkpoint-deficient cells combined, or not, with Exo1 ablation. Equivalently to pol2-4, pol2-S430D improved rad53 cells survival in HU only when combined with EXO1 deletion (Figure 5A). The fact that pol2-S430D phenocopies the synthetic rescue of the exo-deficient allele suggests that the phosphomimetic variant also counteracts fork collapse by limiting Polε-driven resection. In agreement with this notion, expression of pol2-S430D in combination with exo1 ablation improved bulk genome replication in checkpoint-deficient cells in presence of moderate stress (20-30 mM HU, Figure 5B and S5A), likely as a result of enhanced fork progression (Figure 5C and 5SB) that increases overall replication completion particularly at regions flanked by distant origins (Figure 5D, S5D and S5C). Expression of pol2-S430A did not alter replication progression profiles of rad53 cells (Figure S5E), consistent with inability to phosphorylate S430 not affecting fork stability in cells already impaired for the checkpoint kinase activity. Taken together, these results suggest that S430 phosphorylation curbs exonuclease activity when Pol2 stalls, protecting fork functional integrity if nascent strand resection by Exo1 is also prevented.
Figure 5. Phosphomimimetic modification of serine 430 of Pol2 counteracts Polε-driven stalled fork collapse.
(A) Serial dilutions of rad53 pPOL2, rad53 ppol2-S430D, rad53 ppol2-4, rad53 exo1 pPOL2, rad53 exo1 ppol2-S430D and rad53 exo1 ppol2-4 cells plated in absence (-) or presence of 10 mM HU. (B) Flow cytometry analysis of rad53 pPOL2 and rad53 exo1 ppol2-S430D cells released from G1 in 20 mM HU. Vertical orange bars mark the 2C DNA content of G2/M cells. (C) CGS analysis of fork progression. Curves represent log10 S/G1 ratios of reads obtained from S-phase cells at indicated time points after G1 release in the presence of 25mM HU. A 150-Kb genomic region on chromosome VI is shown. (D) Average read ratios across genomic regions categorized by inter-origin distance between early-firing origins corresponding to the experiment shown in C. Red arrow heads mark time points with increased replication completion in rad53 exo1 ppol2-S430D cells. See also Figure S5.
Phosphorylation of Pol2 on serine 430 counteracts Polε-driven nascent strand resection and stalled fork collapse
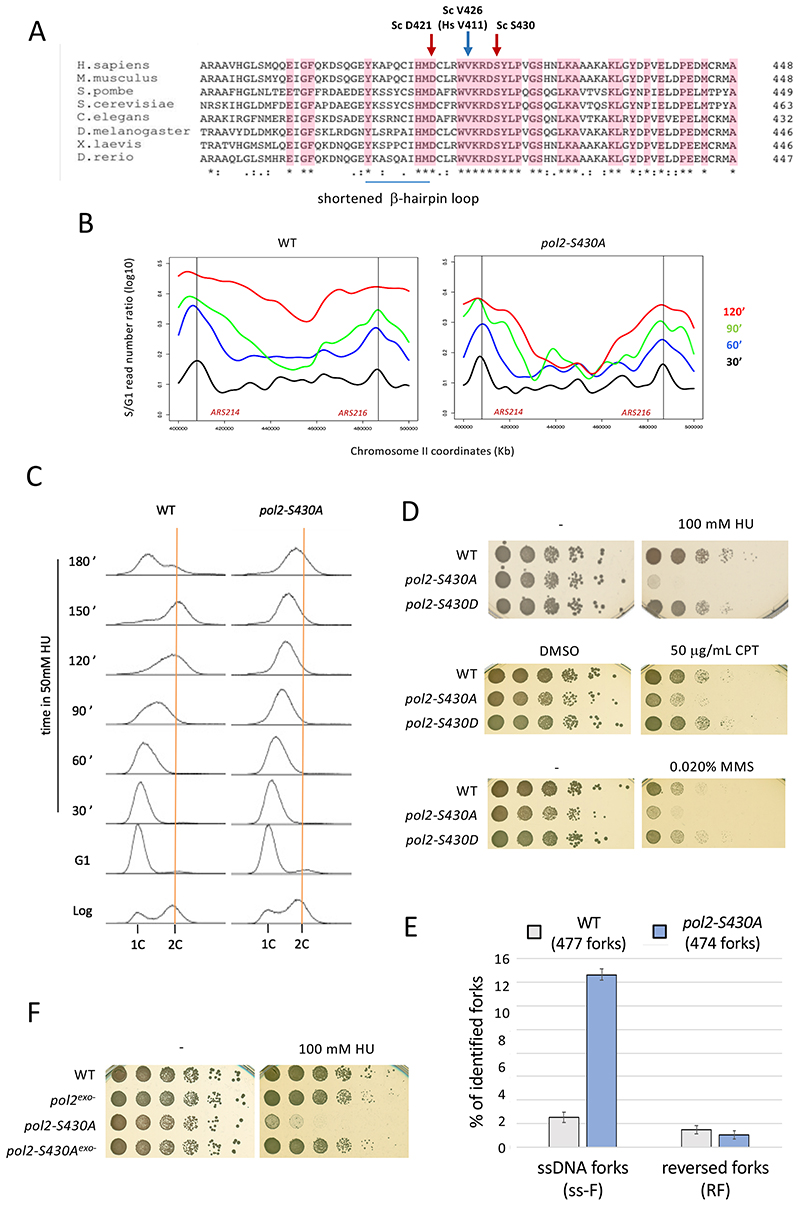
Pol2-S430 is situated next to a β-hairpin loop (Figure 6A), known to mediate switching of nascent DNA ends between polymerase and exo sites in B family polymerases though considerably shortened in Polε (Hogg et al., 2007), within a conserved structural element containing also valine 426 and aspartate 421 (Figure S4E and S4G). V426 is orthologous to POLE-V411, whose substitution to leucine is the second most common POLE mutation associated to human cancer (Henninger and Pursell, 2014). Aspartate 421 is also highly conserved in B-family polymerases and was shown to influence exo/pol partitioning in Phi29 polymerase (Del Prado et al., 2018). Hence, Pol2-S430 is strategically positioned to act as a molecular switch controlling nascent strand access to exo sites. Replacement of S430 by a non-phosphorylatable alanine does not impair Polε exonuclease activity in vitro (Figure S6) and may hence determine an inability to restrain fork resection in vivo upon Polε stalling despite checkpoint kinase activation. In agreement with this prediction, pol2-S430A cells exhibited markedly reduced fork progression under HU-induced stress, particularly failing to complete the replication of regions between most distant origins (Figure 6B, S7A and S7B). Replication fork defects in pol2-S430A mutants occur in a checkpoint signaling proficient context, as evidenced by retained repression of late/dormant origin firing (Figure S7C). Likely owing to fork progression impairment, pol2-S430A cells failed to complete bulk replication with a wild type timing in stress conditions (Figure 6C) and were sensitive to replication stalling agents HU, camptothecin or methane methyl sulphonate (Figure 6D). We addressed if fork defects in pol2-S430A cells related to nascent strand resection by inspecting replication intermediates upon treatment with (0.2M) HU by EM. Wild type cells exhibited almost exclusively normal forks, with only a marginal proportion of ssDNA-gapped (2%) or reversed (1%) forks (Figure 6E). In contrast, detection of replication forks with abnormally extended ssDNA tracks raised up to over 14% in pol2-S430A cells, without significant changes in reversed fork proportions (1%). Of note, the HU sensitivity of non-phosphorylatable pol2-S430A was suppressed intra-allelically by the pol2-4 exonuclease catalytic-dead mutation (Figure 6F), consistent with unrestrained resection underlying stalled fork instability. Hence, S430 phosphorylation is key for checkpoint-mediated fork protection by limiting unscheduled nascent strand resection upon Polε stalling.
Figure 6. Lack of phosphorylation on Pol2-S430 causes stalled fork resection and collapse in checkpoint-proficient cells.
(A) The exonuclease domains of Pol2 homologs aligned using Clustal Omega 1.2.0. Completely conserved residues in pink. S. cerevisiae D421, V426 and S430 are indicated. (B) CGS analysis of fork progression in wild type and pol2-S430A cells. S-phase time points were taken at the indicated times after G1 release in the presence of 50mM HU. A 120-Kb genomic region on chromosome II containing ARS214 and ARS216 early replication origins (vertical black lines) is shown. (C) Flow cytometry analysis of Log cells released from G1 in 50 mM HU. (D) Serial dilutions of wild-type, pol2-S430A and pol2-S430D cells plated in absence (-) or presence of 100 mM HU, 50 μg/mL camptothecin (CPT) or 0.015% methyl methansulphonate (MMS). 0.1% dimethyl sulfoxide (DMSO) plates serve as vector control for CPT. (E) TEM analysis of replication fork structures from wild type and pol2- S430A cells 90 minutes after G1 release into 200mM HU. Histogram plot representing means and SDs of the percentages of ssDNA-containing (ss-F) and reversed (RF) fork structures observed in two independent experiments. The total number of forks examined is shown. (F) Serial dilutions of WT, pol2-4, pol2-S430A and pol2-4-S430A cells plated in absence (-) or presence of 100 mM HU. See also Figure S6 and S7.
Our findings are consistent with a molecular mechanism for fork stabilization whereby upon DNA synthesis stalling checkpoint kinases promote Pol2 phosphorylation to switch Polε to a safe mode limiting nascent strand transfer to exonuclease active sites, thus preventing their deleterious resection.
Discussion
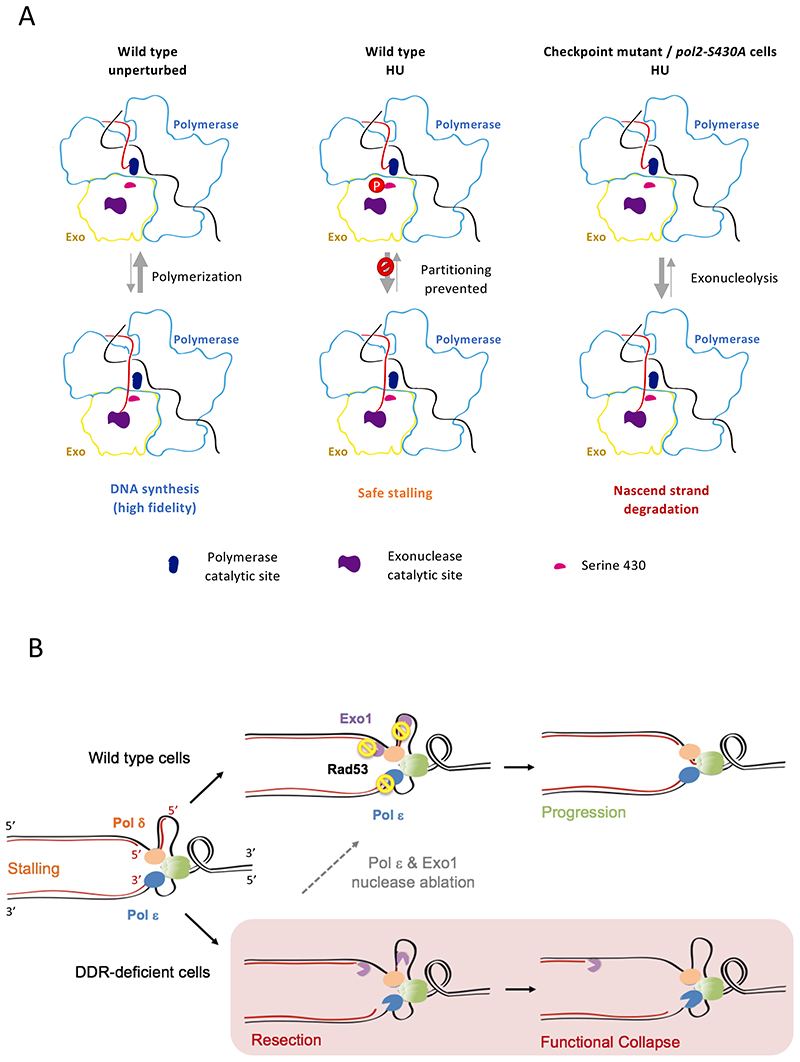
The results here presented indicate that the exonuclease activity of Polε can drive ssDNA accumulation at stalled replication forks, presumably as a result of unrestrained degradation of leading nascent strand 3’ ends that contact its catalytic core. Newly-synthesized DNA resection is most likely a consequence of a shifted balance in primer strand partitioning caused by the stalling of DNA synthesis (Figure 7A). During unperturbed replication polymerization is inherently favored while exonucleolysis has a marginal effect on nascent strands, only relevant in certain circumstances such as after nucleotide misincorporation for replication proofreading. Conversely, upon cessation of DNA synthesis recurrent transfer to the exo site promotes removal of terminal nucleotides, potentially resulting in processive nascent strand degradation. Our data suggest that upon fork stalling replication checkpoint kinases can counteract this shift in balance, which is controlled through the phosphorylation of Serine 430 of its catalytic subunit Pol2. This residue is located along the channel that the nascent strand needs to transverse in order to relocate from the polymerase to the exo catalytic site (Figure S4F). Attachment of a phosphate group to S430 might hence create a steric hindrance encumbering strand transfer. Alternatively, it might induce a conformational change involving the residue environment partly occluding the path normally navigated by nascent strands.
Figure 7. A model for checkpoint-dependent fork protection via limitation of nascent strand access to Polε exonuclease sites.
(A) Expected balances for polymerase-exonuclease strand partitioning in unperturbed and replication stress conditions, controlled via Pol2-S430 phosphorylation. Parental and nascent DNA strands are represented in black and red, respectively. Pol2 polymerase and exonuclease domains are outlined in light blue and yellow. Upon Polε stalling in HU, phosphorylation of S430 counteracts nascent strand switching to exonuclease active sites, thus lessening the partitioning unbalance toward net degradation. Cells deficient for checkpoint kinase activity or expressing non-phosphorylatable Pol2-S430A fail to curb partitioning and nascent strands are degraded. (B) Fork integrity protection by checkpoint kinases. A dual mechanism mediates inhibitory phosphorylation of Polε and Exo1, which counteracts exonucleolytic resection of both nascent strands. Pausing forks are switched to safe-mode favoring replication resumption when stress conditions are reverted. Cells deficient in the checkpoint response (DDR) fail to prevent fork resection which determines functional collapse. Genetic inactivation of Polε and Exo1 exonuclease activities mimics checkpoint-mediated control of nascent strand resection, partly restoring stalled fork integrity and counteracting deleterious outcomes related to fork collapse.
Pol2 Serine 430 is not solvent-exposed in the published Pol2 core structure. This suggests that conformational changes in response to stalling or during the Polε catalytic cycle facilitate its phosphorylation. Rad53-dependent in vivo phosphorylation is also observed on the Dpb2 non-catalytic subunit and additional Rad53 consensus motif phospho-residues are found for Pol2 in replication stress conditions. While Pol2-S430 plays a critical role controlling partitioning of nascent strands, it is possible that other phosphorylation events contribute, directly or indirectly, to counteract exonucleolytic nascent strand resection. In this respect, dpb2 mutations have been shown to increase mutagenesis rates in vivo (Jaszczur et al., 2008) and, hence, Dpb2 phosphorylation could contribute to curb Polε exo activity. However, we cannot exclude that the checkpoint modulates other aspects of Polε function in response to fork stalling through phosphorylation of these additional residues (Gan et al., 2017).
Our findings imply that, through phosphorylation of S430, the exo activity of Polε can be electively switched off in conditions in which exonucleolysis is deleterious. Equivalent mechanisms might be used for the control of DNA polymerase-driven exonucleolysis in diverse cellular contexts. For instance, during translesion synthesis (TLS), in which a proofreading polymerase takes over synthesis following TLS bypass by a specialized polymerase, 3’ exonucleolysis in the context of slowed polymerization could cause primer strand resection and futile cycles of translesion synthesis and degradation. Similarly, during DNA repair-associated gap-filling, which can occur outside of S-phase with low dNTP levels, a preponderance of the exonucleolytic activity might result in net resection of the repaired strand precluding repair and potentially extend original ssDNA gaps. Interestingly, there is evidence for activation of checkpoint kinases in both contexts (García - Rodríguez et al., 2018; Pagès et al., 2009). Elective suppression of the exo activity has been previously proposed for replicases that, as E. coli DNA polymerase III, bear polymerase and exonuclease in different subunits (Echols, 1991; Nick McElhinny et al., 2006). In the case of DNA polymerase III, the ε subunit proofreads errors made by the exo-deficient α subunit and it has been suggested that repression of the gene coding for the exonuclease subunit (DnaQ) may help the cell population to adapt to stressful conditions through hyper-mutagenesis because of replication by an exo- enzyme. For enzymes like Polε, carrying both activities in a single subunit, post-translational modification represents an easy and reversible means to curb the exonuclease activity.
B-family polymerases possess a β-hairpin loop structure that closely approaches nascent strand DNA at the polymerase catalytic site (Hogg et al., 2007). For this reason, it has been proposed to act as a wedge favoring nascent strand unpairing and translocation to the exonuclease active site. It was noted, however, that this β-hairpin loop is significantly shortened in Polε orthologs, and additional structures were proposed to take over this partitioning promoting function (Hogg et al., 2014). S430 lies within a structural element adjacent to the shortened β-hairpin loop, half way in between active sites (Figure S4F), which lines the path that the nascent strand needs to traverse to reach the exonuclease catalytic site during partitioning. Interestingly, this region harbors residue D421 orthologous to partitioning-relevant D211 in Phi29 polymerase. Hence, this element might bear the ability to influence switching between active sites as the β-hairpin loop does in other B-family polymerases.
Checkpoint kinases have been proposed to contribute to fork stabilization through phosphorylation of different factors, chiefly modulating nascent strand rearrangement, via inhibition of key nucleoporins and Pif1-family helicases, or limiting ssDNA generation by inhibiting CMG-driven unwinding of parental DNA (Bermejo et al., 2011; Devbhandari and Remus, 2020; Rossi et al., 2015). We observe stalled fork resection and progression defects in checkpoint-proficient cells expressing non-phosphorylatable Pol2-S430A, indicating that unrestrained Polε-driven exonucleolysis is sufficient to impair fork integrity even when other protective mechanisms are operative. Hence, leading strand resection by Polε represents a main potential driver of the functional collapse of stalled forks. Nonetheless, parallel suppression of Exo1-mediated resection is required for stressed fork stability and cell survival. Exo1 allegedly resects lagging nascent strands, likely acting as an opportunist engaging 5’ extremities abnormally exposed at the end of Okazaki fragments or at double-stranded ends of reversed forks (Colosio et al., 2016). This notion provides an explanation for the lack of suppression of checkpoint-mutants HU sensitivity previously observed upon Exo1 ablation (Cotta-Ramusino et al., 2005; Segurado and Diffley, 2008), as Polε-mediated leading strand resection likely masks the contribution to fork integrity of preventing lagging strand degradation. Our findings uncover that checkpoint kinases exert a dual role in stalled fork protection, parallelly counteracting the resection of leading and lagging strands through the inhibition of Polε and Exo1, respectively (Figure 7B).
DNA polymerase δ contacts 3’ ends of elongating Okazaki fragments at lagging strands. Like Polε, Polδ is expected to experience an imbalance toward exonucleolysis upon HU-induced stalling and checkpoint-dependent phosphorylation of different Polδ subunits hints at regulation in response to stress (Chen et al., 2010; Smolka et al., 2007). However, suppression of Polδ exo activity is not sufficient to restore checkpoint mutants’ viability in HU, even if combined with Polε exo-deficiency, suggesting that Exo1-mediated resection is the main fork collapse driver on lagging strands. Resection by Polδ might progress toward the fork branching point and terminate when reaching the 5’ end of an adjacent Okazaki fragment. Thus, Polδ-driven resection would be limited to few hundred base pairs, and Okazaki fragments may be readily re-primed once replication stress is overcome. In contrast, Exo1 is highly processive in chromosome resection and could advance away from the fork for longer distances generating kilobase-length ssDNA tracks (Mimitou and Symington, 2011). The precise molecular events through which unrestrained resection contributes to fork collapse remain to be determined. It is possible that extensive nascent strand degradation leads to excessive ssDNA accumulation making replication forks structurally unstable, for instance due to limiting amounts of ssDNA binding proteins (Toledo et al., 2013). Non-exclusive to this possibility, as resection proceeds away from the fork branching point efficient re-priming of leading strands might become progressively less efficient as a replication resumption mechanism.
Besides being the leading strand replicase, DNA polymerase ε plays crucial roles in replication origin firing, transmission of epigenetic information during replication and genome stability maintenance (Bellelli et al., 2018b, 2018a; Jain et al., 2018; Johansson and Dixon, 2013; Yu et al., 2018). Mutations in human POLE have been associated to different types of cancer, which often map to the exonuclease domain (Henninger and Pursell, 2014). Hence, Polε is thought to contribute to malignant transformation by promoting genome instability through increased mutagenesis owing to replication proofreading defects. Intriguingly, some of these mutations do not confer hyper-mutagenesis when transposed to budding yeast (Barbari et al., 2018), consistent with the notion that POLE mutations might promote cancer onset through diverse mechanisms (Meng et al., 2020; Xing et al., 2019). It is tantalizing to speculate that, besides promoting stalled fork integrity, exonuclease-curbing mechanisms may be relevant for other Polε functions impacting genome integrity.
Limitations
Because Pol2 is phosphorylated by Rad53 in vitro, and S430 matches the Rad53 target consensus and accounts for a large fraction of Mec1/Rad53-dependent Pol2 phosphorylation in vivo, we favor the hypothesis that this residue is directly targeted by the checkpoint kinase. However, we cannot formally exclude the existence of an interposed kinase, stimulated by checkpoint activation, that phosphorylates S430 to limit partitioning.
Star Methods
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Rodrigo Bermejo (rodrigo.bermejo@csic.es).
Materials Availability
Plasmids and strains generated in this study are available upon request.
Experimental Model and Subject Details
Yeast strains and growth conditions
All strains are RAD5+ derivatives from W303 and are listed in Table S1. For 10-fold serial dilution assays cells were grown on YPDA for 48 hours at 30ºC, unless otherwise stated. α-factor was used to a final concentration of 5 μg/ml to arrest cells in G1. Release into S-phase in the presence of HU was performed at the concentrations and for the times indicated in the figures. Plasmid-borne POL2 phosphosite mutants were generated by Quick Change mutagenesis on pRS415-POL2 and PAJ6 (yeast codon optimized POL2) plasmids (Williams et al., 2013; Yeeles et al., 2015) using pol2-S430DFw, pol2-S430DRev, pol2-S430AFw, pol2-S430DRev, pol2-S430DFw-oc and pol2-S430DRev-oc primers. Pol2 phosphosite mutagenesis on POL2 chromosomal loci were performed using a variation of delito perfetto technique (Storici and Resnick, 2006). In brief, pCORE cassettes were integrated on the POL2 locus and cells were transformed with pRS415-POL2 plasmids with the appropriate site mutagenized. Double strand breaks were generated at the pCORE cassette by I-SceI expression to induce repair with the mutated allele. Then the plasmid was eliminated by crossing, to leave the chromosomal allele as the only source of POL2 expression, and mutations were confirmed by Sanger sequencing. Oligos used to introduce point mutations are listed in the Key Resources Table. Gene deletion and protein tagging were performed by one-step PCR. Conditional expression of EXO1 was achieved by placing the chromosomal open reading frame under the control of the GAL1 promoter and by depletion of the protein via Auxin Inducible Degron (AID) tagging (Nishimura et al., 2009).
Method Details
Flow Cytometry analysis
For Flow Cytrometry analysis, 2 mL of culture were fixed using 70% ethanol/Tris-HCl 250 mM pH 7,6. Cells were treated with RNase A (1 mg/ml) in 50mM Tris-HCl pH 7.5 at 37°C, washed, resuspended in FACS Buffer (200 mM Tris-HCl pH 7.4, 200 mM NaCl, 80 mM MgCl2) containing 0,5 mg/mL Propidium Iodide and sonicated for analysis in a Beckton Dickinson fluorescence-activated cell analyser.
TEM analysis of replication intermediates
Transmission Electron Microscopy visualization of replication forks was performed as previously reported (Lopes, 2009). In brief, genomic DNA was in vivo crosslinked with psoralen and purified with commercial columns as described (Colosio et al., 2016). 10-20 μg of genomic DNA was partially digested with the PvuI and applied to 0.3 gr/mL BND cellulose or genomic tip G20 Qiagen columns (Zellweger and Lopes 2018) previously equilibrated with 10 mM Tris-HCl, pH 8.0, 300 mM NaCl. After washing with 10 mM Tris-HCl, pH 8.0, 1 mM NaCl, ssDNA-enriched DNA was eluted with 10 mM Tris-HCl, pH 8.0, 1 mM NaCl, 2% caffeine. Fractions of the samples were then spread onto a water surface in the presence of BAC (Benzalkonium Chloride) and the DNA molecules in the monomolecular layer were adsorbed on carbon-coated EM grids in the presence of uranyl acetate followed by platinum-based low angle rotatory shadowing and analyzed as described (Zellweger and Lopes, 2018). The thickness of DNA filaments distributed around 100 A (10 nm). EM pictures were acquired using a Tecnai 12 BIO TWIN G2 microscope operated at 120KV with a side-mounted GATAN Orius SC-1000 camera. Length measurements were performed using a conversion factor, expressed in nanometers/base pairs, calculated using a plasmid of known size as internal standard. The pixel size was corrected automatically at each magnification according to the internal calibrations of the electron microscope and camera.
Comparative Genome-wide Deep Sequencing (CGS)
CGS analysis was performed as previously reported (Frattini et al., 2017). In brief, genomic DNA was purified through glass bead breakage on Nuclei Isolation Buffer and subject to deep sequencing according to the manufacturer’s standard protocol (Applied Biosystems). Genomic DNA libraries were sequenced on Illumina HiSeq2500 platform using single-end sequencing strategy. Raw sequencing reads were aligned with reference yeast genome (S288C) using Bowtie2 software (Langmead and Salzberg, 2012). SAMtools (Li et al., 2009) and bedtools (Quinlan and Hall, 2010) were used to calculate genome wide read coverage. The total DNA replication origin list used were obtained from OriDB database (Siow et al., 2012) and we considered 410 confirmed origins for our analysis. Early and dormant/late origin lists were previously reported (Fachinetti et al., 2010). For generating replication profiles, log10 ratios of S and G1 sequencing reads around the intra and inter-origin regions were calculated using a 100 bp window. The extent of bulk genome replication was assessed by FACS analysis of each sample. A replication progression coefficient (the ratio of propidium iodide fluorenscence of S-phase and G1 cells modes or medians) was used for normalization of S/G1 ratios in respect to the replicated fraction of the genome. For ratio normalization and smoothing, we used the relevant functions from R package Repliscope (Müller et al., 2013). Median and associated standard error for all ratio (log10) values across each inter-origin region were calculated. Log10 S/G1 ratios in specified chromosomal coordinates were plotted using the plot function in R version 3.6.0. We used computeMatrix and plotProfile tools from deepTools (Ramírez et al., 2014) to plot the average read ratios across different region categories based on their inter-origin distances (less than 30 kilobases; greater than 30 but less than 60 kilobases and more than 60 kilobases) (Kent et al., 2010).
Protein phosphorylation analysis and Mass spectrometric phosphopeptide analysis
Phospho-tag polyacrylamide gels were prepared according to manufacturer instructions. Proteins were immunoblotted with antibodies specific to the corresponding epitope tag or with EL7 anti-Rad53 antibodies. ImageJ was used to quantify western blot signals. Lysates from HU-treated cells expressing HA-tagged Pol2 or Dpb2 subunits of Polε were and subjected to immunoprecipitation using anti-HA resin. The eluted protein was denatured, reduced, alkylated with iodoacetamide, and subjected to acetone precipitation. Precipitated proteins were resuspended in 2M urea / Tris solution and digested with trypsin. Phosphopeptides were subsequently enriched using home-made IMAC columns. Phosphopeptide fragmentations were acquired in a Q-exactive mass spectrometer as previously described (Lanz et al., 2018). PSM searches were conducted using Comet (integrated into TPP v5.2.0). All spectra Pol2 and Dpb2 were manually inspected.
DNA polymerase ε purification
Purification of Polε holoenzyme variants was performed as described (Yeeles et al., 2015). In brief, cells expressing Polε subunits from GAL1/10 promoters were grown in 2% raffinose and synchronized in G1 with 100 ng/ml of α-factor. Protein expression was induced by adding 2% galactose. Cells were harvested after 3 hours, washed with lysis buffer (25 mM Hepes-KOH pH7.6, 10% glycerol, 1 mM DTT) + 400 mM KOAc and resuspended in lysis buffer + 400 mM KOAc + Roche Complete protease inhibitors. Thawed cell powder was diluted in Lysis buffer supplemented with 400mM KOAc and one Roche Complete protease inhibitor tablet per 25 ml. After cell debris removal by centrifuging, CaCl2 was added to the supernatant to a final concertation of 2mM together with Calmodulin affinity resin and the lysate was rotated at 4ºC for 1 hour. Resin was collected in a 20 ml column and extensively washed in Lysis buffer + 400mM KOAc + 2 mM CaCl2 and eluted in 2 ml Lysis buffer + 400mM KOAc + 2 mM EDTA + 2 mM EGTA. The eluate was applied to a 5 mL Hi-trap heparin column previously equilibrated in Lysis buffer + 400mM KOAc. Polε was eluted with a 12 CV gradient from 450 mM - 1 M KOAc Lysis buffer after extensive washing with Lysis buffer + 400mM KOAc. Pool, concentration and separation of Polε heparin fractions were performed on a Superdex 200 column previously equilibrated in Lysis buffer + 500mM KOAc.
Exonuclease activity assays
The 3’-5’ exonuclease activity assay was performed in RQ reaction buffer containing 20mM Tris-HCl (PH 7.8), 100 μg/ml bovine serum albumin (BSA), and 1mM DTT. The reaction mixture contained 8mM MgAc2 and 200nM ssDNA (40 nt: 5’ Cy3- AGC CTG GAT TCT TAA CAC GAT TAT CAG CGG ACT GCT TAC C -3’) in RQ buffer, and the reaction was started by the addition of 5nM enzyme at 25°C. The reaction was terminated after the indicated time by adding ten microliters of stop solution (95% formamide, 20mM EDTA). Ten microliters of the reaction product were loaded onto a 10% denaturing polyacrylamide gel. The gel was scanned with a Typhoon Scanner. For exonuclease assays with dsDNA, the 40nt primer was pre-annealed with a 58nt template to form a duplex. 200nM dsDNA (40/58mer) was used as a substrate and the reaction was started by the addition of 20nM enzyme. Quantification was performed through densitometry using ImageJ.
Polymerization/exonuclease coupled assay
Exo-pol assays were performed in a reaction mixture containing 8mM MgAc2, 200nM dsDNA (40/70mer) and dNTP in RQ buffer. The ratio of each deoxynucleotide was based on the estimated in vivo dNTP concentration (dCTP=39μM, dTTP=66μM, dATP=22μM, dGTP=11μM), and the actual dNTP concentration used for each reaction is as indicated. The reactions were started by the addition of 20nM enzyme at 25°C and were terminated after the indicated time by adding ten microliters of stop solution. Products were resolved in denaturing polyacrylamide gels and visualized using a Typhoon imager.
In vitro kinase assays
The Rad53 kinase assays were performed using radiolabeled [γ-32P] adenosine tri-phosphate (ATP) (PerkinElmer NEG002A100UC) with polymerase epsilon as a substrate as described (Sun and Budde, 1999). Kinase reactions contained 100nM Rad53, 25mM HEPES (pH 7.5), 0.2μCi [γ-32P] ATP, 0.1mM ATP, 500nM polymerase epsilon, 10mM MgCl2, 10mM MnCl2, 1nM BSA, 0.5mM DTT, and 0.5mM activated sodium orthovanadate. After incubation at 30°C for 60 minutes, 10μl of the reaction was loaded on a 15% SDS-PAGE, dried, and scanned in a Typhoon FLA 9000 imager.
Quantification and Statistical Analysis
Statistical analysis
For TEM replication intermediate identification data mean and standard deviation values for at least two independent experiments are represented. Statistical significance of gapped-molecules ssDNA track lengths was assessed by the two-tailed test.
Supplementary Material
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-Flag (M2) | Sigma | F1804 |
| anti HA (12CA5) | Roche | 11666606001 |
| Anti-Myc Tag (4A6) | Merck millipore | 05-724 |
| anti PK Anti-V5 Tag (SV5-Pk1) | Bionova | MCA1360 |
| anti-Mouse-HRP | Taper | T03.PI2000M001 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| RNase A | Sigma | R5503 |
| Shortcut RNase III | NEB | M0245 |
| Proteinase K | Roche | 03115852001 |
| Hydroxyurea | Ibian Technologies | HDU0250 |
| Metyl methanesulfonate | Sigma | 129925 |
| Camptothecin | Sigma | C9911 |
| Propidium iodide | Sigma | P4170 |
| Trimetilpsoralen | Sigma | P8399 |
| Phos-tag acrylamide | Rafer | 300-93523 |
| Complete Protease Inhibitor-EDTA free | Roche | 11873580001 |
| Alfa-factor Mating Pheromone | Insight Biotechnology | N/A |
| Spermine | Sigma | S1141 |
| Spermidine | Sigma | S2501 |
| QBT | QIAGEN | 19054 |
| QC | QIAGEN | 19055 |
| QF | QIAGEN | 19056 |
| QIAGEN Genomic-Tips 100/G | QIAGEN | 10243 |
| QIAGEN Genomic-Tips 20/G | QIAGEN | 10223 |
| Benzoylated Naphthoylated DEAE-Cellulose | Sigma | B6385 |
| Poly-Prep chromatography column | Biorad | 7311550 |
| Amicon Ultra-0.5 ml 100K | Merck millipore | UFC510096 |
| Deposited Data | ||
| CGS data | GEO | GSE156480 |
| Experimental Models: Organisms/Strains | ||
| A full list of yeast strains used in this study is provided in Table S1 | This paper | N/A |
| Oligonucleotides | ||
| 5’-GGGTGAAGCGTGATGACTATTTACCACAAGG-3’ | This paper | pol2-S430DFw |
| 5’-CCTTGTGGTAAATAGTCATCACGCTTCACCC-3’ | This paper | pol2-S430DRev |
| 5’-GGGTGAAGCGTGATGCTTATTTACCACAAGG-3’ | This paper | pol2-S430AFw |
| 5’-CCTTGTGGTAAATAAGCATCACGCTTCACCC-3’ | This paper | pol2-S430ARev |
| 5’-GGGTTAAGAGAGACGACTACTTGCCACAAGG-3’ | This paper | pol2-S430DFw-oc |
| 5’-CCTTGTGGCAAGTAGTCGTCTCTCTTAACCC-3’ | This paper | pol2-S430DRev-oc |
| 5’ GGGTTAAGAGACGACGCTTACTTGCCACAAGG 3’ | This paper | pol2-S430AFw-oc |
| 5’ CCTTGTGGCAAGTAAGCGCAGTCTCTTAACCC 3’ | This paper | pol2-S430ARev-oc |
| Recombinant DNA | ||
| pRS415-POL2 | Herr lab | N/A |
| pRS415-pol2-S430D | This paper | N/A |
| pRS415-pol2-S430A | This paper | N/A |
| pAJ6 | Yeeles lab | N/A |
| pAJ6-pol2-S430D | This paper | N/A |
| pAJ6-pol2-S430A | This paper | N/A |
| Software and Algorithms | ||
| R version 3.6.1 (2019-07-05) -- “Action of the Toes” Platform: x86_64-apple-darwin15.6.0 (64-bit) | R Core Team 2013 | N/A |
| Repliscope version ‘1.1.0’ | Muller at al. 2014 | N/A |
| Bowtie2 version 2.3.5.1 | Langmead and Salzberg 2012 | N/A |
| SAMtools version 1.9 (htslib 1.9) | Li et al. 2009 | N/A |
| Bedtools version 2.29.0 | Quinlan and Hall 2010 | N/A |
| Deeptools version 3.4.3 | Ramirez et al. 2014 | N/A |
| Bedgraphtobigwig version 2.8 (bbi version 4) | Kent et al. 2010 | N/A |
Acknowledgments
This work was supported by the Spanish Ministry of Ministry of Science, Innovation and Universities [BFU2014-52529-R & BFU2017-87013-R to R.B.], the Junta de Castilla y León [SA042P17 & SA103P20 to R.B.] the Spanish Formación del Personal Investigador (FPI) program [to S.V-H., G.P. and D.J], the Spanish Juan de la Cierva-Formación program [to M.A.M.] and a Beca Leonardo a Investigadores y Creadores Culturales 2018 from the BBVA foundation [to R.B.]. We thank T. Kunkel, A. Herr, M. Kanemaki and M. Foiani for strains, reagents and support. We are thankful to P. Bisht, I. Nebreda, S. Tamargo, M. Passari, M. García-Flores, A. Calzada and D. Batrakou for support and to other members of our labs for insightful discussions.
Footnotes
Authors Contributions
G.P. designed and conducted experiments. M.A.M. and K.S. performed deep sequencing and CGS analyses. S.V-H. and D.J. performed initial PhosTag gels and genomic experiments. M.C.L. and M.B.S. performed proteomic analyses. M.G. carried out EM analysis. X.Y. and M.G-D. designed and performed in vitro assays. J.Y. purified Polε variants. R.B. designed the project and wrote the paper.
Declaration of Interest
The authors declare no competing interests.
Inclusion and Diversity
One or more of the authors of this paper self-identifies as living with a disability.
Data and code availability
The CGS data generated in this study have been deposited in the GEO database under ID code GSE156480. The code generated for CGS data analysis is available at GitHub (https://github.com/MohammedAlMamun/RepProfs).
References
- Barbari SR, Kane DP, Moore EA, Shcherbakova PV. Functional Analysis of Cancer-Associated DNA Polymerase ε Variants in Saccharomyces cerevisiae. G3 (Bethesda, Md); Genes|Genomes|Genetics. 2018;8:g3.200042.2018. doi: 10.1534/g3.118.200042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- Bell SP, Labib K. Chromosome duplication in Saccharomyces cerevisiae. Genetics. 2016;203:1027–1067. doi: 10.1534/genetics.115.186452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellelli R, Belan O, Pye VE, Clement C, Maslen SL, Skehel JM, Cherepanov P, Almouzni G, Boulton SJ. POLE3-POLE4 Is a Histone H3-H4 Chaperone that Maintains Chromatin Integrity during DNA Replication. Mol Cell. 2018a:1–15. doi: 10.1016/j.molcel.2018.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellelli R, Borel V, Logan C, Jackson A, Boulton SJ, Bellelli R, Borel V, Logan C, Svendsen J, Cox DE, et al. Pol ε Instability Drives Replication Stress , Abnormal Article Pol ε Instability Drives Replication Stress , Abnormal Development , and Tumorigenesis. 2018b:1–15. doi: 10.1016/j.molcel.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Jossen R, Colosio A, Frattini C, Carotenuto W, Cocito A, Doksani Y, Klein H, Gómez-González B, et al. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell. 2011;146 doi: 10.1016/j.cell.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berti M, Vindigni A. Replication stress: Getting back on track. Nat Struct Mol Biol. 2016;23:103–109. doi: 10.1038/nsmb.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- Burgers PMJ, Kunkel TA. Eukaryotic DNA Replication Fork. Annu Rev Biochem. 2017;86:417–438. doi: 10.1146/annurev-biochem-061516-044709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SH, Albuquerque CP, Liang J, Suhandynata RT, Zhou H. A proteome-wide analysis of kinase-substrate network in the DNA damage response. J Biol Chem. 2010;285:12803–12812. doi: 10.1074/jbc.M110.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 2003;22:4325–4336. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosio A, Frattini C, Pellicanò G, Villa-Hernández S, Bermejo R. Nucleolytic processing of aberrant replication intermediates by an Exo1-Dna2-Sae2 axis counteracts fork collapse-driven chromosome instability. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkw858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D. Preventing replication fork collapse to maintain genome integrity. DNA Repair (Amst) 2015;32:149–157. doi: 10.1016/j.dnarep.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell. 2005;17:153–159. doi: 10.1016/j.molcel.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Devbhandari S, Remus D. Rad53 limits CMG helicase uncoupling from DNA synthesis at replication forks. Nat Struct Mol Biol. 2020:1–11. doi: 10.1038/s41594-020-0407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol ε and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 1999;274:22283–22288. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- Echols H. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- Engels K, Giannattasio M, Muzi-Falconi M, Lopes M, Ferrari S. 14-3-3 Proteins Regulate Exonuclease 1-Dependent Processing of Stalled Replication Forks. PLoS Genet. 2011;7:1–9. doi: 10.1371/journal.pgen.1001367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D, Bermejo R, Cocito A, Minardi S, Katou Y, Kanoh Y, Shirahige K, Azvolinsky A, Zakian VA, Foiani M. Replication Termination at Eukaryotic Chromosomes Is Mediated by Top2 and Occurs at Genomic Loci Containing Pausing Elements. Mol Cell. 2010;39 doi: 10.1016/j.molcel.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Zou L. ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem Sci. 2011;36:133–140. doi: 10.1016/j.tibs.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frattini C, Villa-Hernández S, Pellicanò G, Jossen R, Katou Y, Shirahige K, Bermejo R. Cohesin Ubiquitylation and Mobilization Facilitate Stalled Replication Fork Dynamics. Mol Cell. 2017;68:758–772.:e4. doi: 10.1016/j.molcel.2017.10.012. [DOI] [PubMed] [Google Scholar]
- Gan H, Yu C, Devbhandari S, Sharma S, Han J, Chabes A, Remus D, Zhang Z. Checkpoint Kinase Rad53 Couples Leading-and Lagging-Strand DNA Synthesis under Replication Stress. Mol Cell. 2017;68:446–455.:e3. doi: 10.1016/j.molcel.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganai RA, Bylund GO, Johansson E. Switching between polymerase and exonuclease sites in DNA polymerase ε. Nucleic Acids Res. 2015;43:932–942. doi: 10.1093/nar/gku1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Rodríguez N, Morawska M, Wong RP, Daigaku Y, Ulrich HD. Spatial separation between replisome‐ and template‐induced replication stress signaling. EMBO J. 2018;37:1–15. doi: 10.15252/embj.201798369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerik KJ, Li X, Pautz A, Burgers PMJ. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem. 1998;273:19747–19755. doi: 10.1074/jbc.273.31.19747. [DOI] [PubMed] [Google Scholar]
- Giannattasio M, Branzei D. S-phase checkpoint regulations that preserve replication and chromosome integrity upon dNTP depletion. Cell Mol Life Sci. 2017;74:2361–2380. doi: 10.1007/s00018-017-2474-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henninger EE, Pursell ZF. DNA polymerase ε and its roles in genome stability. IUBMB Life. 2014;66:339–351. doi: 10.1002/iub.1276. [DOI] [PubMed] [Google Scholar]
- Hoekstra TP, Depken M, Lin SN, Cabanas-Danés J, Gross P, Dame RT, Peterman EJG, Wuite GJL. Switching between Exonucleolysis and Replication by T7 DNA Polymerase Ensures High Fidelity. Biophys J. 2017;112:575–583. doi: 10.1016/j.bpj.2016.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg M, Johansson E. In: The Eukaryotic Replisome: A Guide to Protein Structure and Function. MacNeill S, editor. Springer Netherlands; Dordrecht: 2012. DNA Polymerase $ε$ pp. 237–257. [Google Scholar]
- Hogg M, Aller P, Konigsberg W, Wallace SS, Doublie S. Structural and biochemical investigation of the role in proofreading of a β hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family. J Biol Chem. 2007;282:1432–1444. doi: 10.1074/jbc.M605675200. [DOI] [PubMed] [Google Scholar]
- Hogg M, Osterman P, Bylund GO, Ganai RA, Lundström E-B, Sauer-Eriksson AE, Johansson E. Structural basis for processive DNA synthesis by yeast DNA polymerase ɛ. Nat Struct Mol Biol. 2014;21:49–55. doi: 10.1038/nsmb.2712. [DOI] [PubMed] [Google Scholar]
- Hu J, Sun L, Shen F, Chen Y, Hua Y, Liu Y, Zhang M, Hu Y, Wang Q, Xu W, et al. The intra-S phase checkpoint targets Dna2 to prevent stalled replication forks from reversing. Cell. 2012;149:1221–1232. doi: 10.1016/j.cell.2012.04.030. [DOI] [PubMed] [Google Scholar]
- Jain R, Rajashankar KR, Buku A, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Crystal structure of yeast DNA polymerase ε catalytic domain. PLoS One. 2014;9 doi: 10.1371/journal.pone.0094835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Aggarwal AK, Rechkoblit O. Eukaryotic DNA polymerases. Curr Opin Struct Biol. 2018;53:77–87. doi: 10.1016/j.sbi.2018.06.003. [DOI] [PubMed] [Google Scholar]
- Jaszczur M, Flis K, Rudzka J, Kraszewska J, Budd ME, Polaczek P, Campbell JL, Jonczyk P, Fijalkowska IJ. Dpb2p, a noncatalytic subunit of DNA polymerase ε, contributes to the fidelity of DNA replication in Saccharomyces cerevisiae. Genetics. 2008;178:633–647. doi: 10.1534/genetics.107.082818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson E, Dixon N. Replicative DNA polymerases. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Prakash L, Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase are also essential subunits of DNA polymerase. Proc Natl Acad Sci. 2012;109:12455–12460. doi: 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossen R, Bermejo R. The DNA damage checkpoint response to replication stress: A game of forks. Front Genet. 2013;4 doi: 10.3389/fgene.2013.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz MC, Oberly S, Sanford EJ, Sharma S, Chabes A, Smolka MB. Separable roles for Mec1/ATR in genome maintenance, DNA replication, and checkpoint signaling. Genes Dev. 2018;32:822–835. doi: 10.1101/gad.308148.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz MC, Dibitetto D, Smolka MB. DNA damage kinase signaling: checkpoint and repair at 30 years. EMBO J. 2019;38 doi: 10.15252/embj.2019101801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M. Electron microscopy methods for studying in vivo DNA replication intermediates. Methods Mol Biol. 2009;521:605–631. doi: 10.1007/978-1-60327-815-7_34. [DOI] [PubMed] [Google Scholar]
- Lucca C, Vanoli F, Cotta-Ramusino C, Pellicioli A, Liberi G, Haber J, Foiani M. Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene. 2004;23:1206–1213. doi: 10.1038/sj.onc.1207199. [DOI] [PubMed] [Google Scholar]
- Lujan SA, Williams JS, Kunkel TA. DNA Polymerases Divide the Labor of Genome Replication. Trends Cell Biol. 2016;26:640–654. doi: 10.1016/j.tcb.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Wei L, Devbhandari S, Zhang T, Xiang J, Remus D, Zhao X. DNA polymerase ε relies on a unique domain for efficient replisome assembly and strand synthesis. Nat Commun. 2020;11 doi: 10.1038/s41467-020-16095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. DNA end resection--unraveling the tail. DNA Repair (Amst) 2011;10:344–348. doi: 10.1016/j.dnarep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morafraile EC, Bugallo A, Carreira R, Fernández M, Martín-Castellanos C, Blanco MG, Segurado M. Exo1 phosphorylation inhibits exonuclease activity and prevents fork collapse in rad53 mutants independently of the 14-3-3 proteins. Nucleic Acids Res. 2020;48:3053–3070. doi: 10.1093/nar/gkaa054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller CA, Hawkins M, Retkute R, Malla S, Wilson R, Blythe MJ, Nakato R, Komata M, Shirahige K, De Moura APS, et al. The dynamics of genome replication using deep sequencing. Nucleic Acids Res. 2013;42:e3. doi: 10.1093/nar/gkt878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Pavlov YI, Kunkel TA. Evidence for extrinsic exonucleolytic proofreading. Cell Cycle. 2006;5:958–962. doi: 10.4161/cc.5.9.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Pagès V, Santa Maria SR, Prakash L, Prakash S. Role of DNA damage-induced replication checkpoint in promoting lesion bypass by translesion synthesis in yeast. Genes Dev. 2009;23:1438–1449. doi: 10.1101/gad.1793409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo B, Crabbé L, Pasero P. Signaling pathways of replication stress in yeast. FEMS Yeast Res. 2017;17:1–11. doi: 10.1093/femsyr/fow101. [DOI] [PubMed] [Google Scholar]
- Pasero P, Vindigni A. Nucleases Acting at Stalled Forks: How to Reboot the Replication Program with a Few Shortcuts. Annu Rev Genet. 2017;51:477–499. doi: 10.1146/annurev-genet-120116-024745. [DOI] [PubMed] [Google Scholar]
- De Piccoli G, Katou Y, Itoh T, Nakato R, Shirahige K, Labib K. Replisome Stability at Defective DNA Replication Forks Is Independent of S Phase Checkpoint Kinases. Mol Cell. 2012;45:696–704. doi: 10.1016/j.molcel.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Del Prado A, Franco-Echevarría E, González B, Blanco L, Salas M, De Vega M. Noncatalytic aspartate at the exonuclease domain of proofreading DNA polymerases regulates both degradative and synthetic activities. Proc Natl Acad Sci U S A. 2018;115:E2921–E2929. doi: 10.1073/pnas.1718787115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–91. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reha-Krantz LJ. DNA polymerase proofreading: Multiple roles maintain genome stability. Biochim Biophys Acta - Proteins Proteomics. 2010;1804:1049–1063. doi: 10.1016/j.bbapap.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Rossi SE, Ajazi A, Carotenuto W, Foiani M, Giannattasio M. Rad53-Mediated Regulation of Rrm3 and Pif1 DNA Helicases Contributes to Prevention of Aberrant Fork Transitions under Replication Stress. Cell Rep. 2015:1–13. doi: 10.1016/j.celrep.2015.08.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF. A Mec1-and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- Segurado M, Diffley JFX. Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev. 2008;22:1816–1827. doi: 10.1101/gad.477208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova PV, Pavlov YI. 3′ → 5′ Exonucleases of DNA polymerases ε and δ correct base analog induced DNA replication errors on opposite DNA strands in Saccharomyces cerevisiae. Genetics. 1996;142:717–726. doi: 10.1093/genetics/142.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- Siow CC, Nieduszynska SR, Müller CA, Nieduszynski CA. OriDB, the DNA replication origin database updated and extended. Nucleic Acids Res. 2012;40:D682–6. doi: 10.1093/nar/gkr1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci U S A. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- Storici F, Resnick MA. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 2006;409:329–345. doi: 10.1016/S0076-6879(05)09019-1. [DOI] [PubMed] [Google Scholar]
- Sun G, Budde RJA. Substitution studies of the second divalent metal cation requirement of protein tyrosine kinase CSK. Biochemistry. 1999;38:5659–5665. doi: 10.1021/bi982793w. [DOI] [PubMed] [Google Scholar]
- Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat Struct Mol Biol. 2009;16:979–986. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JA, Longhese MP, Diffley JFX. A central role for DNA replication forks in checkpoint activation and response. Mol Cell. 2003;11:1323–1336. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- Toledo LI, Altmeyer M, Rask M-B, Lukas C, Larsen DH, Povlsen LK, Bekker-Jensen S, Mailand N, Bartek J, Lukas J. ATR Prohibits Replication Catastrophe by Preventing Global Exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- de Vega M, Lazaro JM, Salas M, Blanco L. Primer-terminus stabilization at the 3′-5′ exonuclease active site of phi29 DNA polymerase. Involvement of two amino acid residues highly conserved in proofreading DNA polymerases. EMBO J. 1996;15:1182–1192. [PMC free article] [PubMed] [Google Scholar]
- Williams LN, Herr AJ, Preston BD. Emergence of DNA polymerase ε antimutators that escape error-induced extinction in yeast. Genetics. 2013;193:751–770. doi: 10.1534/genetics.112.146910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing X, Kane DP, Bulock CR, Moore EA, Sharma S, Chabes A, Shcherbakova PV. A recurrent cancer-associated substitution in DNA polymerase ε produces a hyperactive enzyme. Nat Commun. 2019;10 doi: 10.1038/s41467-018-08145-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JTP, Deegan TD, Janska A, Early A, Diffley JFX. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519:431–435. doi: 10.1038/nature14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Gan H, Serra-Cardona A, Zhang L, Gan S, Sharma S, Johansson E, Chabes A, Xu RM, Zhang Z. A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science (80- ) 2018;361:1386–1389. doi: 10.1126/science.aat8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger R, Lopes M. Dynamic Architecture of Eukaryotic DNA Replication Forks In Vivo, Visualized by Electron Microscopy. Methods Mol Biol. 2018;1672:261–294. doi: 10.1007/978-1-4939-7306-4_19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The CGS data generated in this study have been deposited in the GEO database under ID code GSE156480. The code generated for CGS data analysis is available at GitHub (https://github.com/MohammedAlMamun/RepProfs).