Abstract
The incretin hormone glucose-dependent insulinotropic polypeptide (GIP) augments glucose-dependent insulin secretion through its receptor expressed on islet β-cells. GIP also acts on adipose tissue; yet paradoxically, both enhanced and reduced GIP receptor (GIPR) signaling reduce adipose tissue mass and attenuate weight gain in response to nutrient excess. Moreover, the precise cellular localization of GIPR expression within white adipose tissue (WAT) remains uncertain. We used mouse genetics to target Gipr expression within adipocytes. Surprisingly, targeting Cre expression to adipocytes using the adiponectin (Adipoq) promoter did not produce meaningful reduction of WAT Gipr expression in Adipoq-Cre:Giprflx/flx mice. In contrast, adenoviral expression of Cre under the control of the cytomegalovirus promoter, or transgenic expression of Cre using nonadipocyte-selective promoters (Ap2/Fabp4 and Ubc) markedly attenuated WAT Gipr expression. Analysis of single-nucleus RNA-sequencing, adipose tissue data sets localized Gipr/GIPR expression predominantly to pericytes and mesothelial cells rather than to adipocytes. Together, these observations reveal that adipocytes are not the major GIPR+ cell type within WAT—findings with mechanistic implications for understanding how GIP and GIP-based co-agonists control adipose tissue biology.
Introduction
Incretin hormones are produced in specialized enteroendocrine cells and amplify meal-stimulated insulin release following food ingestion. The two incretins, glucose-dependent insulinotropic polypeptide (GIP) and glucagon-like peptide 1 (GLP-1), exert their actions through structurally related yet distinct receptors (1). The major target for incretin action is the islet β-cell, wherein GIP and GLP-1 potentiate insulin secretion through cyclic AMP-dependent and -independent pathways (1).
The molecular cloning of incretin receptor cDNAs enabled identification of extrapancreatic expression of both incretin receptors, consistent with characterization of multiple actions for GIP and GLP-1 beyond the β-cell (1,2). GLP-1 receptor mRNA transcripts have been identified within the central and enteric nervous systems, the heart, gastrointestinal tract, kidney, blood vessels, and immune cells (3–5), lending support for multiple direct actions of GLP-1 in peripheral tissues. Similarly, the tissue distribution of GIP receptor (GIPR) mRNA transcripts includes the brain, heart, gastrointestinal tract, blood vessels, and adipose tissue (6).
Among key differences in the extrapancreatic biology of GIP but not GLP-1 is that the former acts directly on adipose tissue (1,7). These actions of GIP have been studied using differentiated adipose tissue cell lines, primary adipocyte cultures, and experiments with animals and humans (8–14). Within white adipose tissue (WAT), GIP promotes both lipolysis and lipid accretion, glucose uptake, insulin sensitization, and adipokine expression (12,13,15–20). GIP also modifies adipose tissue biology through actions on blood vessels, and immune cells. For instance, GIP rapidly augments adipose tissue blood flow (21) and either augments or suppresses WAT inflammation through direct actions on immune cells (13,18,22,23).
Interpreting the actions of GIP on adipose tissue has been complicated by paradoxical observations that both sustained GIPR agonism and attenuation of GIPR signaling produce overlapping phenotypes in animals, including reduction of WAT inflammation, reduced WAT mass, resistance to weight gain, and improvement of insulin sensitivity (7,18,24–31). Understanding how gain or loss of GIP actions within unique adipose tissue GIPR+ cell types has been hampered in part by 1) the lack of highly specific validated antisera or labeled analogs for detection of the GIPR (32,33), and 2) a paucity of in situ hybridization or single-cell RNA sequencing (scRNA-seq) data for detection of the WAT GIPR. Hence, the cellular localization of GIPR in different adipose tissue depots remains incompletely understood.
In view of multiple studies linking expression of GIPR to adipocytes, we attempted to understand the metabolic consequences of targeting the adipocyte Gipr using mouse genetics. Surprisingly, although expression of Cre under control of the well-characterized adiponectin promoter recombined Gipr genomic DNA in WAT, we did not observe meaningful reduction of Gipr expression in WAT depots of Adipoq-Cre:Giprflx/flx mice. In contrast, expression of Cre recombinase under the control of the human adipocyte fatty acid binding protein (Ap2) Fabp promoter, or using Ubc-CREERT2 to direct widespread Cre expression, resulted in marked reduction of WAT Gipr mRNA transcripts in multiple WAT depots. Consistent with these findings, Gipr-directed reporter expression was not detected within the majority of adipocytes analyzed using a Gipr-Cre mouse to identify transcriptional domains of endogenous Gipr promoter activity. Finally, publicly available single-cell RNA-seq (scRNA-seq) data identified WAT Gipr/GIPR expression predominantly within pericytes and mesothelial cells (34). Taken together, these findings refine our understanding of WAT Gipr expression, providing insights for guiding interpretation of data linking GIP action to changes in adipose tissue biology.
Research Design and Methods
Animal Models
Animals were obtained from The Jackson Laboratory (Bar Harbor, ME). Giprflx/flx (35), Adipoq-Cre (36), AdipoqBAC-Cre (catalog no. 028020 [37]), Mip-Cre ERT (catalog no. 024709 [35,38]), αMHC-CreERT (32), Ubc-CreERT2 (catalog no. 008085 [39]), AdipoqBAC-CreERT (catalog no. 024671 [40]), and Fabp4(Ap2)-Cre (catalog 005069 [41]) mice have been previously described. Gipr-Cre (knock-in) mice were generated using CRISPR/Cas9, as previously described (42) and bred with Rosa26-LacZ [B6;129S4-Gt(ROSA)26Sortm1Sor/J (catalog no. 003309)] or Rosa29-mTmG [Gt(ROSA)26Sortm4(ACTBACTB-tdTomato,-EGFP)Luo/J (catalog no. 007576)] reporter mice. Male mice were used for all studies.
PCR Analysis
RNA isolation and quantitative PCR were carried out as previously described (35). PCR of genomic DNA was done using PrimeSTAR GXL DNA Polymerase (Takara Bio; catalog no. R050A). Primer sequences are described in Supplementary Table 1.
In Vivo Studies
All animal studies were conducted under protocols approved by the Animal Care Committees of the Toronto Centre for Phenogenomics, the Duke Molecular Physiology Department, and the University of Cambridge Animal Welfare and Ethical Review Body and conformed to the Animals (Scientific Procedures) Act 1986 Amendment Regulations (SI 2012/3039). Briefly, intraperitoneal and oral glucose tolerance tests (1.5 g/kg glucose) were performed in animals after a 5-h fast. For intraperitoneal glucose tolerance tests, animals were given PBS or d-Ala2 GIP (4 nmol/kg; Chi Scientific) intraperitoneally 10 min prior to glucose administration. Insulin tolerance was measured following administration of 0.7 units/kg fast-acting insulin (Humalog) in mice fasted for 5 h. The high-fat diet (HFD) feeding was a 45% fat diet from Research Diets (D12451). Body composition was measured with an EchoMRI device.
In Vivo Adenovirus Treatment
An adenoviral vector containing the human adenovirus type5 (dE1/E3) viral backbone encoding both Cre recombinase and GFP through separate cytomegalovirus (CMV) promoters was gifted by Dr. Andras Nagy (Lunenfeld-Tanenbaum Research Institute, Mt. Sinai Hospital, Toronto, ON, Canada). The virus was used at a titer of 1 × 1010 PFU/mL and a total of 50 μL was injected directly into a single inguinal fat pad in mice lightly anesthetized with isoflurane. Virus containing an empty vector was injected at the same amounts into the contralateral inguinal fat pad. Mice were sacrificed 72 h later and both inguinal fat pads were harvested, flash frozen, and stored at −80°C until used for RNA analysis.
HFD Feeding of Ap2-Cre Mice
Littermate, age-matched controls were weened at 3–4 weeks of age and maintained on standard rodent chow until 8 weeks of age. The diet was then switched to a 45% fat diet and the mice were maintained on this diet until study termination.
Whole-Mount β-Galactosidase Assay
Tissues were harvested from 11-month-old male mice that were hemizygous for Gipr-Cre and heterozygous for ROSA26-LacZ or heterozygous for ROSA26-LacZ (negative control). Tissues were rinsed in PBS and transferred to 6-well plates, where they were fixed for 2 h (in calcium- and magnesium-free PBS containing 1% paraformaldehyde, 0.2% glutaraldehyde, and 0.02% Nonidet P-40) at 4°C using an orbital shaker. Samples were then washed twice (20 min each) in PBS and incubated in the dark overnight (16 h) at 37°C in β-galactosidase substrate (calcium- and magnesium-free PBS containing 5 mmol/L potassium ferricyanide, 5 mmol/L potassium ferrocyanide, 2 mmol/L magnesium chloride, 0.02% NP-40, 0.01% sodium deoxycholate, and 1 mg/mL X-gal substrate). The following day, samples were rinsed twice in PBS as described, fixed in 10% neutral buffered formalin overnight at 4°C, and transferred to 70% ethanol until imaging. Whole-mount tissues were imaged using a Leica MZ6 stereomicroscope with an attached MC170 HD digital camera (Leica Microsystems Inc., Concord, ON, Canada).
Whole-Mount Confocal Microscopy
Adipose tissues were harvested from 14-week-old female mice that were hemizygous for Gipr-Cre and heterozygous for mTmG or heterozygous for mTmG (negative control), rinsed with PBS, cut into 0.5- to 1-cm pieces, transferred to 12-well plates, and fixed in 1% paraformaldehyde for 1 h at room temperature. Tissues were then washed three times (10 min each) in PBS containing 0.3% Triton X-100, followed by an additional three washes (10 min each) in PBS and then incubated in Lipidtox Deep Red (1:1,000 in PBS, catalog H34477; Thermo Fisher Scientific, Mississauga, ON, Canada) for 30 min at room temperature. Tissues were placed on a glass coverslip and saturated with DAPI-containing mounting media (Vectashield, catalog H-1200; Vector Laboratories Inc., Burlington, ON, Canada) and imaged using an inverted confocal laser microscope.
Single-Nucleus RNA-Seq Analysis
Detection of Gipr/GIPR and other class B, G protein–coupled receptor (GPCR) mRNA transcripts within mouse and human adipose tissue depots was accomplished using publicly available data derived from single-nucleus RNA-seq analyses, as previously described (34), accessed from the Broad Institute single-cell portal (https://singlecell.broadinstitute.org/single_cell).
Statistics
Data are presented as mean ± SEM. Differences were determined by Student t test or one- or two-way ANOVA, as appropriate. A Tukey test was used for post hoc analysis of ANOVAs, where appropriate. P < 0.05 was set as the criterion for statistically significant difference.
Data and Resource Availability
The original data in this article are available upon reasonable request from the authors. The Gipr floxed mice are available for collaborative sharing from the authors (contact D.J.D.). All of the other mouse lines and reagents described herein are available from commercial sources.
Results
Gipr mRNA Transcripts Are Not Reduced in Adipose Tissues From Adipoq-Cre:Giprflx/flx Mice
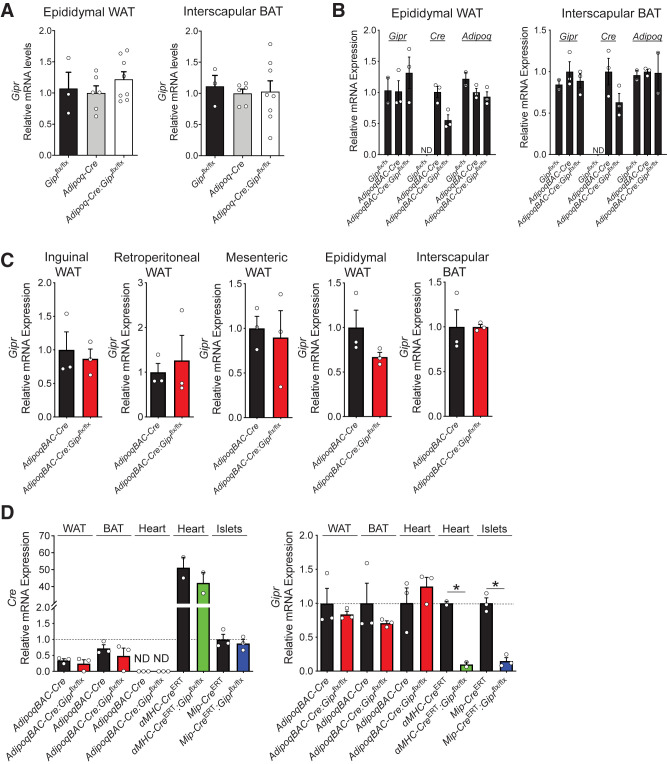
Several studies have reported targeting of murine Gipr expression in WAT; however, the extent of Gipr knockdown within adipocytes in vivo was not described (41,43). Nevertheless, based on reports localizing Gipr expression to adipocytes (43–45), we sought to inactivate adipocyte Gipr expression using the widely used adiponectin-Cre system (46). Accordingly, we generated Adipoq-Cre:Giprflx/flx mice by crossing Giprflx/flx mice (35,47) with mice expressing Cre driven by the Adipoq promoter (36). Surprisingly, levels of Gipr mRNA transcripts in WAT or brown adipose tissue (BAT) from Adipoq-Cre:Giprflx/flx mice were not reduced (Fig. 1A).
Figure 1.
Cre recombinase expression driven by the adiponectin promoter does not reduce Gipr mRNA transcripts in adipose tissue. A) Gipr expression in epididymal WAT and interscapular BAT. Cre expression is driven by transgenic expression of Adipoq-Cre (36) (n = 3–8). B) Gipr, Cre, and Adipoq expression in epididymal WAT and interscapular BAT. For reference, the average cycle threshold values for Gipr and Adipoq were 26.28 and 18.77, respectively, in WAT and 28.13 and 20.73, respectively, in BAT. Cre expression is driven by the AdipoqBAC promoter (AdipoqBAC-Cre [37]) (n = 3). C) Gipr expression in various adipose tissue depots in mice with the AdipoqBAC-Cre transgene (n = 3). D) Cre and Gipr expression in tissues, including epididymal WAT from various Cre recombinase models crossed with Giprflx/flx mice (n = 2–3). A–C) For relative RNA expression values, the values are normalized to expression in the Cre controls (Adipoq-Cre in A; AdipoqBAC-Cre in B and C). D) Values are normalized to levels for MIP-CreERT. *P < 0.05 vs control. ND, not detected.
We next generated a second mouse model using an independently generated Adipoq-Cre mouse that uses a BAC transgene containing the majority of the Adipoq regulatory elements (AdipoqBAC-Cre) (37), a mouse line successfully used by multiple groups to achieve adipocyte-selective gene recombination (48), including our own laboratory (49). Unexpectedly, AdipoqBAC-Cre:Giprflx/flx mice also failed to exhibit reduced Gipr expression in WAT and BAT (Fig. 1B and C) despite expressing Cre and Adipoq at levels similar to wild-type (WT) and AdipoqBAC-Cre control mice (Fig. 1B). Collectively these findings indirectly imply that the majority of adiponectin+ adipocytes do not express the Gipr within adipose tissue in vivo.
We previously achieved reduction of Gipr expression, using the same Giprflx/flx mice, in β-cells (35), cardiomyocytes (32), and BAT (47) using Mip-CreERT, αMHC-CreERT, and Myf5-Cre mice, respectively. To explain the lack of Gipr knockdown in WAT, we examined relative Cre expression across different mouse models. WAT Cre expression driven by the AdipoqBAC promoter was comparable to levels seen in primary islets, driven by Mip, but lower than levels detected in the heart, driven by αMHC. (Fig. 1D). Gipr mRNA transcript levels were reduced in heart and islet tissues of αMHC-Cre:Giprflx/flx and Mip-Cre:Giprflx/flx mice, respectively (Fig. 1D), consistent with previous observations (32,35). In contrast, Gipr mRNA transcripts were not reduced in adipose tissue depots from AdipoqBAC-Cre:Giprflx/flx mice (Fig. 1D).
Gipr Expression Can Be Reduced in WAT of Giprflx/flx Mice Through Nonadipocyte Selective Cre Expression
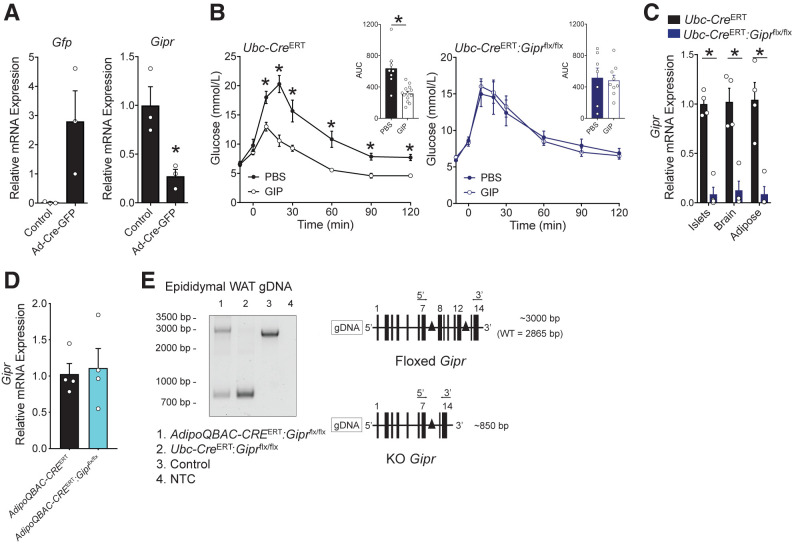
The failure to knock down WAT Gipr mRNA transcript levels using Adipoq-Cre raised several possibilities. First, we hypothesized that the adipocyte Gipr gene may be uniquely inaccessible to Cre within adipocytes, thereby preventing Cre-mediated recombination of the floxed alleles. Alternatively, we surmised that cellular Gipr expression may be inversely correlated to Adipoq expression, implying that the Gipr may not be expressed in the majority of white adipocytes. To examine these possibilities, we injected a Cre-expressing adenovirus directly into the inguinal adipose depot of Giprflx/flx mice as a means of producing widespread expression of Cre within WAT (50). Mice injected with Ad-CMV(GFP)-Cre exhibited a 70% knockdown in inguinal adipose tissue Gipr expression (Fig. 2A), illustrating the susceptibility of the Gipr allele within WAT of Giprflx/flx mice to recombination and subsequent reduction of WAT Gipr mRNA transcripts.
Figure 2.
Complementary genetic strategies enable reduction of Gipr expression in adipose tissue. A) An adenoviral vector expressing Cre and Gfp under control of the CMV promoter was injected into an inguinal adipose depot of Giprflx/flx mice; empty vector was administered into the contralateral depot as a control. Quantitative PCR was used to determine the expression of Gfp and Gipr in both depots (n = 3 mice.) B) Blood glucose levels during an intraperitoneal glucose tolerance test in which PBS or GIP (4 nmol/kg) was administered 10 min before glucose in control (Ubc-CreERT2; n = 8–13) or Ubc-CreERT2:Giprflx/flx (n = 7–9) mice. C) Gipr expression in primary islets, whole-brain, and epididymal adipose tissue (n = 4). D) Gipr expression in epididymal adipose tissue from AdipoqBAC-CreERT (control) and AdipoqBAC-CreERT:Giprflx/flx mice harvested 2 weeks following tamoxifen treatment, when mice were 10 weeks of age (n = 4). E) PCR analysis of genomic DNA (gDNA) from epididymal WAT from different genetic mouse models. Nonrecombined DNA produces a 2,865 base pair (bp) product in wild-type (WT) mice, and ∼3,000 bp product in Giprflx/flx mice. Recombination of the loxP alleles produces an 850-bp product. For relative mRNA values, expression in panel A was normalized to control; for panel C, values were normalized relative to Ubc-CreERT; and for panel D, levels were normalized to Adipo-CreERT. *P < 0.05 vs control. AUC, area under the curve; KO, knockout; NTC, no template control.
To obtain complementary evidence supporting these observations, we used Ubc-CreERT2 mice, which express Cre under the control of the human ubiquitin C promoter in most cell types (39). Consistent with loss of the insulin-stimulating actions of GIP in Giprβ-cell−/− mice (35), tamoxifen-treated Ubc-CreERT2:Giprflx/flx mice failed to exhibit reduction of glucose levels in response to exogenous GIP (Fig. 2B). Importantly, levels of Gipr mRNA transcripts were markedly reduced in the islets, brain, and WAT of Ubc-CreERT2:Giprflx/flx mice (Fig. 2C). Thus, both Ubc-CreERT2 and Ad-CMV-Cre can drive Cre expression enabling recombination of the Gipr gene and reduction of Gipr mRNA transcripts in WAT from Giprflx/flx mice.
AdipoqBAC-Cre Mice Exhibit Recombination of the Genomic Gipr Locus in WAT, Without Reduced Adipocyte Gipr Expression
Ubc-CreERT2 mice require tamoxifen to induce Cre recombinase activity, whereas the AdipoqBAC-Cre mice (Fig. 1B–D) exhibit constitutive expression of Cre recombinase, including during development. We wondered whether postnatal induction of Cre expression may confer preferential recombination of floxed adipocyte Gipr alleles. To assess this possibility, we crossed Giprflx/flx mice with AdipoqBAC-CreERT mice (40), which require tamoxifen for conditional induction of Cre activity. Following the same tamoxifen protocol used for the Ubc-CreERT2 model (Fig. 2B and C), treatment of AdipoqBAC-CreERT:Giprflx/flx mice with tamoxifen at 8 weeks of age failed to reduce epididymal Gipr expression (Fig. 2D). However, PCR analysis of epididymal adipose tissue DNA using primers that span both loxP sites demonstrated that the Cre recombinase effectively induced recombination of the Gipr genomic DNA (Fig. 2E). In control adipose tissue samples (WT or Giprflx/flx), only the full-length genomic DNA PCR product was amplified (no recombination). In contrast, both the full-length and Cre-generated products were amplified in genomic DNA from AdipoqBAC-CreERT:Giprflx/flx adipose tissue, and only the truncated Cre-generated PCR product was amplified in genomic DNA from Ubc-CreERT:Giprflx/flx adipose tissue (Fig. 2E). Thus, the Adipoq promoter is capable of generating sufficient Cre expression to permit recombination of the Gipr allele yet does not alter Gipr mRNA levels within WAT.
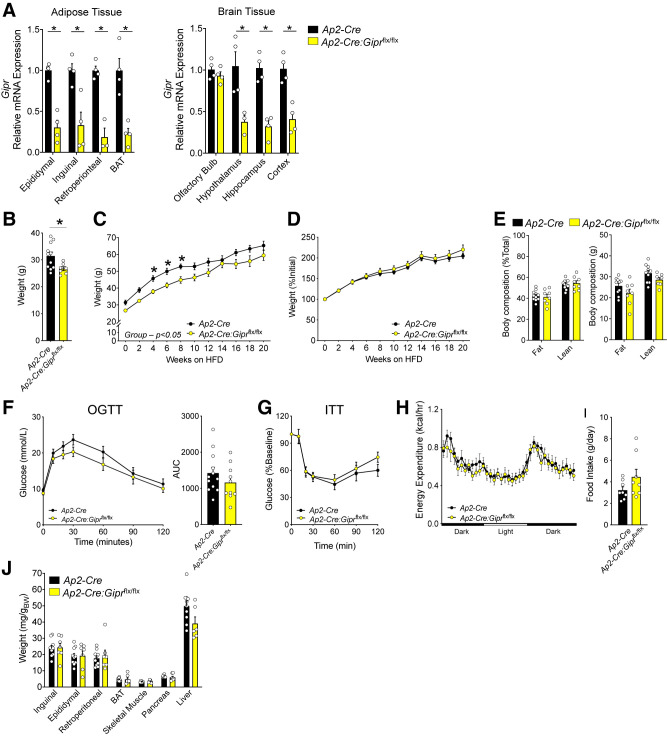
Ap2-Cre:Giprflx/flx Mice Exhibit Reduced Gipr Expression in Adipose Tissue and Brain
Several metabolic phenotypes were described for mice with Gipr knockdown in adipose tissue, generated using Ap2-Cre mice (41), including a modest reduction in body weight after HFD feeding, together with improved glucose tolerance, and reduced hepatic steatosis. Intriguingly, the reductions in body weight were driven by reduced lean mass, not fat mass. Although Gipr expression was markedly reduced in visceral and subcutaneous adipose tissue from Gipradipo−/− mice (41), Ap2/Fabp4 expression is not limited to adipocytes, because the Ap2/Fab4 promoter is transcriptionally active in heart, muscle, brain, macrophages, endothelium, and testis (46,51,52). We examined Gipr expression in adipose tissue depots and brain regions of independently generated Ap2-Cre:Giprflx/flx mice. Gipr transcripts were reduced in all adipose depots examined (Fig. 3A), as well as in the hypothalamus, hippocampus, and cortex (Fig. 3A). Ap2-Cre:Giprflx/flx mice weighed less at 8 weeks of age (Fig. 3B) and their weight remained below that of control mice throughout the HFD feeding period (Fig. 3C). However, the rate of weight gain between groups in response to HFD feeding was similar (Fig. 3D), as was body composition after 16 weeks of HFD feeding (Fig. 3E). Moreover, glucose tolerance (Fig. 3F), insulin tolerance (Fig. 3G), energy expenditure (Fig. 3H), food intake (Fig. 3I), and tissue weights (Fig. 3J) were similar, although liver weight trended lower (P = 0.055). Thus, although reduction of Gipr in adipose tissue depots with Ap2-Cre is associated with modest changes in body weight, these findings cannot be directly attributed to reductions in adipose tissue Gipr expression, because the concurrent reduction in brain Gipr expression confounds attribution of phenotypes to adipose tissue in this mouse model.
Figure 3.
Ap2-Cre expression results in Gipr knockdown in adipose and brain tissues. A) Gipr expression in adipose depots and brain tissues. (n = 4). B) Body weights (BWs) in 8-week-old mice prior to initiation of HFD feeding (n = 7–11). Absolute (C) and percentage increases (D) in BWs of mice fed a 45% HFD for several weeks (n = 7–11). E) Body composition expressed as a percentage of total (left) or absolute (right) weight in mice after 16 weeks of HFD feeding (n = 8–11). F) Blood glucose levels and area under the glucose curve (AUC) during an oral glucose tolerance test (OGTT) after 17 weeks of HFD feeding (n = 10–12). G) Blood glucose levels during an insulin tolerance test (ITT) after 6 weeks of HFD feeding (n = 10–12). H) Oxygen consumption after 20 weeks of HFD feeding (n = 8). I) Food intake (24 h) after 20 weeks of HFD feeding (n = 8). J) Tissues weights after 20 weeks of HFD feeding (n = 7–10). A) For relative mRNA expression, values were normalized to levels detected in RNA isolated from the same adipose tissue depots of Ap2-Cre mice. *P < 0.05 vs. control.
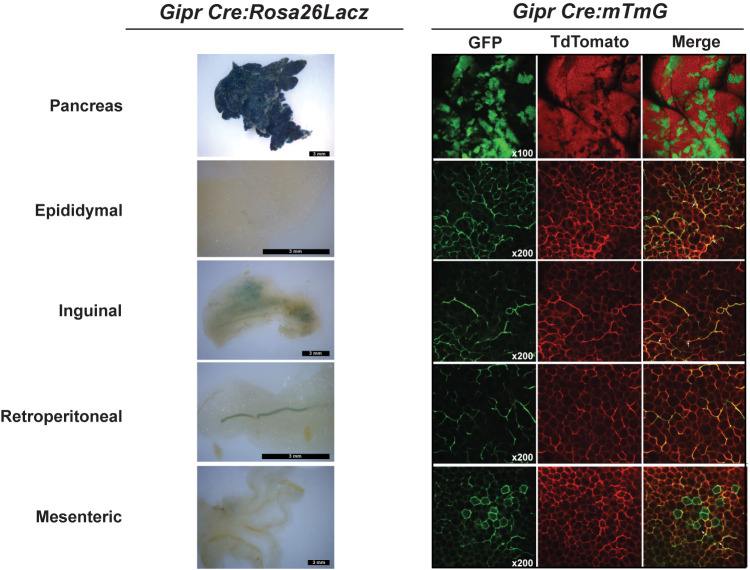
Expression of Gipr-Cre Demonstrates Heterogeneity of Reporter Protein Expression Within Adipose Tissue
To reconcile our inability to reduce WAT Gipr expression with multiple Adipoq-Cre driver lines, we used Gipr-Cre to direct reporter protein expression. Crossing Gipr-Cre mice with a Rosa26-LacZ reporter line (53) produced abundant β-galactosidase activity in the pancreas, yet with little activity detected in WAT depots, including staining localized to blood vessels (Fig. 4). We next crossed Gipr-Cre mice with a Rosa26-mT/mG reporter line (54). Confocal microscopy of tissues from these mice demonstrated Gipr promoter activity in the pancreas, in line with islet GIPR expression (Fig. 4). Gipr promoter activity within WAT was much more heterogeneous, with a definitive signal seen in only a small fraction of putative adipocytes within multiple WAT depots.
Figure 4.
Heterogeneous distribution of Gipr promoter activity in adipose depots. Representative whole-mount images of β-galactosidase activity (left column) and confocal fluorescence imaging (right column) of pancreas and adipose tissues from Gipr Cre:Rosa26Lacz and Gipr Cre:mTmG mice, respectively. GFP, Cre-positive green fluorescent protein; TdTomato, Cre-negative tomato red fluorescent protein.
Single-Nucleus RNA-Seq Localization of Gipr/GIPR Expression in Adipose Tissue
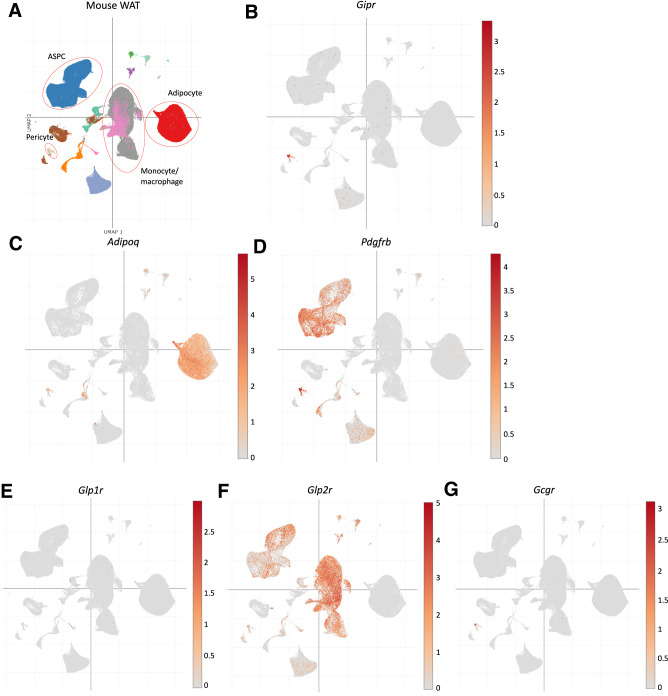
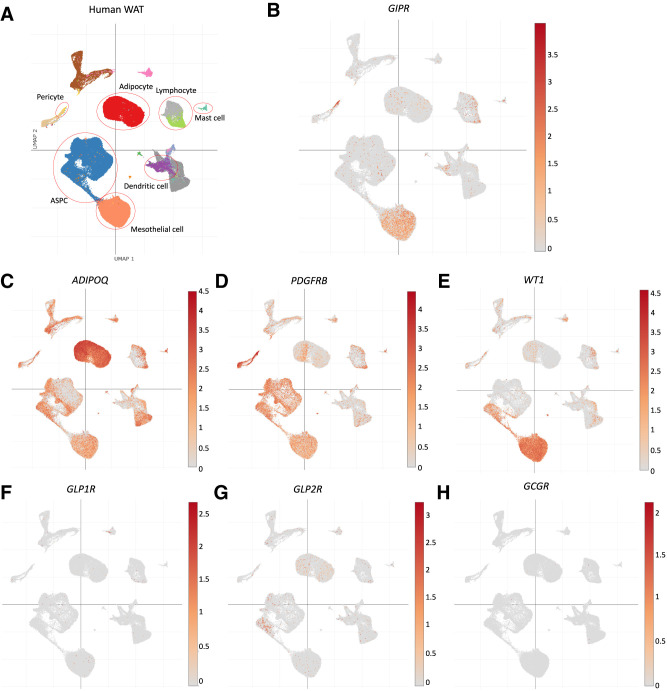
Collectively, the genetic findings in mice imply that Gipr expression within WAT is predominantly localized to nonadipocyte cell types. To further investigate this possibility, we analyzed Gipr/GIPR expression within distinct cell types of mouse and human inguinal and perigonadal adipose tissue using independently generated publicly available single nucleus RNA-seq data (34). Within mouse adipose tissue, Gipr mRNA was detected within Pdgfrbhigh pericytes, with minimal expression detected in Adipoq+ adipocytes (Fig. 5A–D). Glp1r was virtually absent in mouse adipose tissue (Fig. 5E); intriguingly, Glp2r was found in adipose stem and progenitor cells and macrophages (Fig. 5F). Gcgr showed an expression pattern similar to Gipr and was localized to pericytes (Fig. 5G). Similarly, scRNA-seq analysis of human subcutaneous and visceral adipose tissues detected GIPR primarily in PDGFRBhigh pericytes, followed by WT1+ mesothelial cells, but not in ADIPOQhigh adipocytes (Fig. 6A–E). Among the related class B, G protein–coupled receptors, only GLP2R, but not GLP1R and GCGR, was detected in some human adipose stem and progenitor cells and adipocytes (Fig. 6F–H). Hence, the available RNA-seq data are sufficiently useful for detection of GIPR and related class B, G protein–coupled receptor mRNA transcripts within various adipose tissue cell types, independently highlighting the lack of GIPR expression within the majority of mouse or human adipocytes.
Figure 5.
Mouse single-nucleus RNA-seq data were used to localize WAT Gipr expression to pericytes. A) Single-cell nucleus RNA-seq data (34) from a range of mouse adipose tissue cell types were analyzed for expression of B) Gipr; C) Adipoq; D) Pdgfrb, a gene expressed in endothelial/pericyte and adipose tissue progenitors; E) Glp1r; F) Glp2r; and G) Gcgr. ASPC, adipocyte stem progenitor cell.
Figure 6.
Human single-nucleus RNA-seq data were used to localize WAT GIPR expression to pericytes and mesothelial cells. A) Single-cell nucleus RNA-seq data (34) from a range of human adipose tissue cell types were analyzed for expression of B) GIPR; C) ADIPOQ; D) PDGFRB; E) WT1, a marker for the mesothelial cell lineage; F) GLP1R; G) GLP2R; and H) GCGR. ASPC, adipocyte stem progenitor cell.
Discussion
Our current findings have implications for interpreting studies of GIP biology in adipose tissue. First, using independent mouse lines, we found that the murine Gipr is not expressed within the majority of adipocytes. Second, consistent with these findings, expression of Cre recombinase under control of the Adiponectin promoter does not meaningfully reduce Gipr expression in multiple adipose tissue depots. Third, interpretation of data generated using nonadipocyte-selective promoters to target adipose tissue Gipr expression, exemplified by Ap2-Cre (41), may be confounded by reduction of Gipr expression in multiple nonadipocyte cell types, including immune, neuronal, and endothelial cells (46). Although relative levels of Gipr mRNA transcripts were reported as normal in the brain of Gipradipo−/− mice (41), our analyses, using the same Ap2 promoter to express Cre and inactivate the Gipr, revealed substantial reduction of Gipr mRNA transcripts in multiple regions of the murine central nervous system known to affect systemic metabolism. Taken together, these findings are consistent with a substantial proportion of adipose tissue GIPR expression arising within nonadipocyte lineages.
In agreement with interpretation of the data obtained using genetic approaches in vivo, the single-nucleus RNA-seq data provide further support for the concept that mouse and human adipocytes are not major sites of canonical Gipr/GIPR expression. Indeed, pericytes appear to be a putative GIPR-expressing cell type in both human and mouse WAT, and additional human WAT GIPR expression is identified in mesothelial cells. These findings have implications for interpretation of the existing literature describing mechanisms of GIP action in adipose tissue and may generate new hypotheses about the actions of GIPR within WAT cell types that contribute to the biology of adipose tissue development and function.
The importance of understanding the biology of the adipose tissue GIPR and its impact has accelerated in part due to translational interest in targeting the GIPR for the treatment of obesity and diabetes (7). Indeed, GIPR agonism reduces food intake, body weight, and fat mass in HFD-fed mice through mechanisms requiring central nervous system GIPR activation (55). A GIP–GLP-1 co-agonist LY3298716, subsequently renamed tirzepatide, robustly stimulated cAMP accumulation in adipocyte-like cells derived from progenitors differentiated ex vivo, reduced food intake and adipose tissue mass, and produced substantial weight loss in both preclinical and clinical studies (56). Moreover, tirzepatide augmented adipose tissue glucose uptake and enhanced insulin sensitivity in a GLP-1R–independent manner in mice (57). Remarkably, GIPR blockade with antibodies directed against the mouse or human GIPR also reduced fat (WAT) mass, blocked the actions of exogenous GIP on human adipocytes ex vivo, and attenuated weight gain, without changes in lean mass in mice and nonhuman primates (27,31). Reconciliation of how both gain and loss of function at the GIPR produce overlapping effects on body weight, WAT mass, and function requires a more detailed understanding of how GIP controls metabolism and adipose tissue biology.
The results of several previous studies examining GIP action in adipose tissue have yielded conflicting results, with some studies demonstrating that GIP acts directly on WAT and other experiments invoking a role for GIP as an insulin sensitizer on adipocytes, using cells differentiated from adipocyte progenitors ex vivo (11,58,59). Indeed, the very slow kinetics of the adipose tissue response to GIP (59) have prompted the suggestion that GIP might act indirectly on adipocyte lipid metabolism, through one or more downstream mediators such as insulin or resistin (35,60,61). Nevertheless, substantial data suggest that adipocyte-like cells studied ex vivo express a functional GIPR coupled to cAMP accumulation and fatty acid uptake (43,57).
Previous studies using mouse genetics to interrogate the role of the adipocyte GIPR have been partially inconclusive. For example, transgenic targeting of GIPR expression to WAT of Gipr−/− mice using the Ap2/Fabp4 promoter produced weight gain independent of changes in fat mass, without any meaningfully evident metabolic phenotypes (62). Conversely, reduction of WAT Gipr mRNA transcripts using the Ap2/Fabp4 promoter to direct Cre expression to several cell types, including adipocytes, reduced WAT Gipr expression in mice with lower body weight and lean body mass, yet without change in fat mass (41). More recent studies using Adipoq-Cre to target the mouse adipocyte Gipr revealed loss of Gipr expression in adipocytes differentiated ex vivo, together with reduced GIP-stimulated cAMP accumulation and decreased fatty acid uptake (43). Notably, however, the levels of Gipr mRNA within WAT depots from GiprAdipo−/− mice were not reported.
The growing importance in understanding the actions of GIP in WAT is further augmented by interest in the mechanisms of action of tirzepatide (63). Administration of tirzepatide or a long-acting GIPR agonist improved insulin sensitivity, associated with enhanced glucose uptake into WAT (57). Interestingly, however, RNA-seq analysis of WAT from mice treated with tirzepatide or a long-acting GIPR agonist showed no changes in metabolic gene expression within WAT depots, whereas a GIPR agonist and tirzepatide differentially regulated >1,000 genes within BAT (57). These latter findings are consistent with a functional role for the canonical murine GIPR in regulation of genes important for thermogenesis, lipid metabolism, and cytokine expression in BAT (47,61).
Limitations and Future Perspectives
Our data require interpretation with caution due to a number of important limitations. First, we focused almost entirely on mRNA expression, because of the lack of suitably validated antisera for detection of the mouse GIPR (32,33). Data from experiments using reporter genes to infer expression should be considered with caveats, because the readouts may reflect activation of transcriptional sequences in one or more early adipose tissue lineages that subsequently give rise to differentiated adipocytes. Hence, whether reporter gene expression within a few adipocytes coincides with simultaneous coexpression of the Gipr mRNA transcript or protein in the same differentiated cell remains uncertain. Although the scRNA-seq and gene-targeting data align with the concept that mouse adipocyte Gipr expression is uncommon, we did not study adipose tissue depots from a wide range of mice with metabolic perturbations. For example, animals with diabetes, insulin deficiency or resistance, or obesity, might exhibit upregulation of adipocyte Gipr expression—scenarios that require additional investigation.
It is also worth noting that low-level adipocyte expression of class B GPCRs such as Glp1r or Gipr might not easily be detected using thresholds set for scRNA-seq, although the same analyses successfully detected Gipr and Glp2r mRNAs in nonadipocyte cell types within adipose tissue (34). Finally, our data do not rule out an important role for GIPR activity in adipose tissue. The small fraction of mature GIPR+ adipocytes identified by scRNA-seq could represent a key subset of cells that contribute to regulation of overall adipose tissue function through paracrine or endocrine processes. Alternatively, the colocalization of Gipr/GIPR with Pdgfrb/PDGFRB populations may represent early precursor cells, potentially suggesting a role for GIP in preadipocyte function. Finally, there is much less information available on the cellular localization of GIPR in human adipose tissue depots across the life span in people living with diabetes or obesity, hence the putative importance of adipocyte expression of the human GIPR requires greater scrutiny. In summary, our data introduce further complexity in conceptualizing how gain or loss of GIPR signaling affects adipose tissue biology and adipocytes in vivo.
Article Information
Acknowledgments. The authors thank Jackie Koehler (Lunenfeld Tanenbaum Research Institute) and Ju Hee Lee (The Hospital for Sick Children, Toronto, Canada) for their assistance with whole-mount β-galactosidase activity and confocal microscopy studies, respectively. The authors also thank Ilona Zvetkova from the Metabolic Research Laboratories–Genome Engineering Core (grant MRC_MC_UU_12012/5) and Debbie Drage from University of Cambridge Central Biomedical Services for their assistance in generating GIPR-Cre mice.
Funding. J.E.C. previously received fellowships from the Banting and Best Diabetes Centre, University of Toronto, Canadian Institutes of Health Research, and the American Diabetes Association (1-18-JDF-017); is currently funded by a career development award from the American Diabetes Association (1-18-JDF-017) and by funding from the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (grants DK123075 and DK125353); and is a Borden Scholar. J.L.B. has received fellowship funding from Diabetes Canada and the Banting and Best Diabetes Centre and is currently funded by Natural Sciences and Engineering Research Council of Canada Discovery early career grants (RGPIN-2021-03439 and DGECR-2021-00388) and a Connaught New Researcher award (NR-2020-21). J.R.U. has received fellowships from Canadian Institutes of Health Research and the Alberta Innovates Health Solutions and is supported by operating grants from the Canadian Institutes of Health Research (CIHR), the Heart and Stroke Foundation of Canada, and the Canada Research Chairs program. D.A.D. is supported by grants from the NIH (DK101991) and Veterans Administration (CX001401). Research in the Reimann/Gribble laboratories is supported by the Wellcome Trust (grants 106262/Z/14/Z and 106263/Z/14/Z) and the Medical Research Council (grant MRC_MC_UU_12012/3). D.J.D. is supported by a Banting and Best Diabetes Centre–Novo Nordisk Chair in Incretin Biology, the Sinai Health Novo Nordisk Foundation in Regulatory Peptides, and CIHR Foundation (grant 154321). Mt. Sinai Hospital receives funding for incretin biology and obesity research in the Drucker laboratory from Novo Nordisk.
Duality of Interest. D.J.D. has served as an advisor or speaker within the past 12 months to Altimmune, Applied Molecular Transport, Kallyope, Eli Lilly and Co., Merck Research Laboratories, Novo Nordisk, and Pfizer Inc. Neither D.J.D. nor his family members hold stock in these companies. D.A.D. is a consultant to Eli Lilly and Co., Merck, and Novo Nordisk and received grants from Merck and Ligand during the conduct of the study. J.E.C. has served as an advisor or speaker within the past 12 months to Altimmune, Eli Lilly and Co., and ShouTi and receives funding for preclinical studies from Eli Lilly and Co. and Novo Nordisk. F.G. and F.R. receive funding from Astra Zeneca, Eli Lilly and Co., and LGC Ltd. for preclinical studies. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. J.E.C., J.L.B., and D.J.D. contributed to the project design. J.E.C., J.L.B., B.S., L.L.B., A.N.G., J.R.U., and C.K.W. contributed to the experimental investigation. J.E.C. and J.L.B. conducted the formal analysis. J.E.C. and D.J.D. supervised the project and wrote the original manuscript draft. J.E.C., J.L.B., B.S., L.L.B., J.R.U., C.K.W., D.A.D., and D.J.D. reviewed and edited the manuscript. J.E.C., F.M.G., D.A.D., F.R., and D.J.D. contributed to funding acquisition and project administration. D.J.D. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
J.R.U. currently is affiliated with the Faculty of Pharmacy and Pharmaceutical Sciences, University of Alberta, Edmonton, Alberta, Canada.
This article contains supplementary material online at https://doi.org/10.2337/figshare.19184462.
References
- 1. Campbell JE, Drucker DJ. Pharmacology, physiology, and mechanisms of incretin hormone action. Cell Metab 2013;17:819–837 [DOI] [PubMed] [Google Scholar]
- 2. Gallwitz B. Extra-pancreatic effects of incretin-based therapies. Endocrine 2014;47:360–371 [DOI] [PubMed] [Google Scholar]
- 3. Bullock BP, Heller RS, Habener JF. Tissue distribution of messenger ribonucleic acid encoding the rat glucagon-like peptide-1 receptor. Endocrinology 1996;137:2968–2978 [DOI] [PubMed] [Google Scholar]
- 4. Wei Y, Mojsov S. Tissue-specific expression of the human receptor for glucagon-like peptide-I: brain, heart and pancreatic forms have the same deduced amino acid sequences. FEBS Lett 1995;358:219–224 [DOI] [PubMed] [Google Scholar]
- 5. McLean BA, Wong CK, Campbell JE, Hodson DJ, Trapp S, Drucker DJ. Revisiting the complexity of GLP-1 action from sites of synthesis to receptor activation. Endocr Rev 2021;42:101–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Usdin TB, Mezey E, Button DC, Brownstein MJ, Bonner TI. Gastric inhibitory polypeptide receptor, a member of the secretin-vasoactive intestinal peptide receptor family, is widely distributed in peripheral organs and the brain. Endocrinology 1993;133:2861–2870 [DOI] [PubMed] [Google Scholar]
- 7. Campbell JE. Targeting the GIPR for obesity: to agonize or antagonize? Potential mechanisms. Mol Metab 2021;46:101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yip RG, Boylan MO, Kieffer TJ, Wolfe MM. Functional GIP receptors are present on adipocytes. Endocrinology 1998;139:4004–4007 [DOI] [PubMed] [Google Scholar]
- 9. Timper K, Grisouard J, Radimerski T, et al. Glucose-dependent insulinotropic polypeptide (GIP) induces calcitonin gene-related peptide (CGRP)-I and procalcitonin (Pro-CT) production in human adipocytes. J Clin Endocrinol Metab 2011;96:E297–E303 [DOI] [PubMed] [Google Scholar]
- 10. Omar B, Banke E, Guirguis E, et al. Regulation of the pro-inflammatory cytokine osteopontin by GIP in adipocytes--a role for the transcription factor NFAT and phosphodiesterase 3B. Biochem Biophys Res Commun 2012;425:812–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ceperuelo-Mallafré V, Duran X, Pachón G, et al. Disruption of GIP/GIPR axis in human adipose tissue is linked to obesity and insulin resistance. J Clin Endocrinol Metab 2014;99:E908–E919 [DOI] [PubMed] [Google Scholar]
- 12. Thondam SK, Daousi C, Wilding JP, et al. Glucose-dependent insulinotropic polypeptide promotes lipid deposition in subcutaneous adipocytes in obese type 2 diabetes patients: a maladaptive response. Am J Physiol Endocrinol Metab 2017;312:E224–E233 [DOI] [PubMed] [Google Scholar]
- 13. Gögebakan Ö, Osterhoff MA, Schüler R, et al. GIP increases adipose tissue expression and blood levels of MCP-1 in humans and links high energy diets to inflammation: a randomised trial. Diabetologia 2015;58:1759–1768 [DOI] [PubMed] [Google Scholar]
- 14. Rudovich N, Kaiser S, Engeli S, et al. GIP receptor mRNA expression in different fat tissue depots in postmenopausal non-diabetic women. Regul Pept 2007;142:138–145 [DOI] [PubMed] [Google Scholar]
- 15. Yip RG, Wolfe MM. GIP biology and fat metabolism. Life Sci 2000;66:91–103 [DOI] [PubMed] [Google Scholar]
- 16. Kim SJ, Nian C, McIntosh CH. GIP increases human adipocyte LPL expression through CREB and TORC2-mediated trans-activation of the LPL gene. J Lipid Res 2010;51:3145–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lamont BJ, Drucker DJ. Differential antidiabetic efficacy of incretin agonists versus DPP-4 inhibition in high fat fed mice. Diabetes 2008;57:190–198 [DOI] [PubMed] [Google Scholar]
- 18. Varol C, Zvibel I, Spektor L, et al. Long-acting glucose-dependent insulinotropic polypeptide ameliorates obesity-induced adipose tissue inflammation. J Immunol 2014;193:4002–4009 [DOI] [PubMed] [Google Scholar]
- 19. Ben-Shlomo S, Zvibel I, Varol C, et al. Role of glucose-dependent insulinotropic polypeptide in adipose tissue inflammation of dipeptidylpeptidase 4-deficient rats. Obesity (Silver Spring) 2013;21:2331–2341 [DOI] [PubMed] [Google Scholar]
- 20. Ahlqvist E, Osmark P, Kuulasmaa T, et al. Link between GIP and osteopontin in adipose tissue and insulin resistance. Diabetes 2013;62:2088–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Asmar M, Simonsen L, Madsbad S, Stallknecht B, Holst JJ, Bülow J. Glucose-dependent insulinotropic polypeptide may enhance fatty acid re-esterification in subcutaneous abdominal adipose tissue in lean humans. Diabetes 2010;59:2160–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen S, Okahara F, Osaki N, Shimotoyodome A. Increased GIP signaling induces adipose inflammation via a HIF-1α-dependent pathway and impairs insulin sensitivity in mice. Am J Physiol Endocrinol Metab 2015;308:E414–E425 [DOI] [PubMed] [Google Scholar]
- 23. Mantelmacher FD, Zvibel I, Cohen K, et al. GIP regulates inflammation and body weight by restraining myeloid-cell-derived S100A8/A9. Nat Metab 2019;1:58–69 [DOI] [PubMed] [Google Scholar]
- 24. Nasteska D, Harada N, Suzuki K, et al. Chronic reduction of GIP secretion alleviates obesity and insulin resistance under high-fat diet conditions. Diabetes 2014;63:2332–2343 [DOI] [PubMed] [Google Scholar]
- 25. Miyawaki K, Yamada Y, Ban N, et al. Inhibition of gastric inhibitory polypeptide signaling prevents obesity. Nat Med 2002;8:738–742 [DOI] [PubMed] [Google Scholar]
- 26. Boylan MO, Glazebrook PA, Tatalovic M, Wolfe MM. Gastric inhibitory polypeptide immunoneutralization attenuates development of obesity in mice. Am J Physiol Endocrinol Metab 2015;309:E1008–E1018 [DOI] [PubMed] [Google Scholar]
- 27. Killion EA, Wang J, Yie J, et al. Anti-obesity effects of GIPR antagonists alone and in combination with GLP-1R agonists in preclinical models. Sci Transl Med 2018;10:eaat3392. [DOI] [PubMed] [Google Scholar]
- 28. Szalowska E, Meijer K, Kloosterhuis N, Razaee F, Priebe M, Vonk RJ. Sub-chronic administration of stable GIP analog in mice decreases serum LPL activity and body weight. Peptides 2011;32:938–945 [DOI] [PubMed] [Google Scholar]
- 29. Kim SJ, Nian C, Karunakaran S, Clee SM, Isales CM, McIntosh CH. GIP-overexpressing mice demonstrate reduced diet-induced obesity and steatosis, and improved glucose homeostasis. PLoS One 2012;7:e40156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mroz PA, Finan B, Gelfanov V, et al. Optimized GIP analogs promote body weight lowering in mice through GIPR agonism not antagonism. Mol Metab 2019;20:51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Svendsen B, Capozzi ME, Nui J, et al. Pharmacological antagonism of the incretin system protects against diet-induced obesity. Mol Metab 2020;32:44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ussher JR, Campbell JE, Mulvihill EE, et al. Inactivation of the glucose-dependent insulinotropic polypeptide receptor improves outcomes following experimental myocardial infarction. Cell Metab 2018;27:450–460.e6 [DOI] [PubMed] [Google Scholar]
- 33. Ast J, Broichhagen J, Hodson DJ. Reagents and models for detecting endogenous GLP1R and GIPR. EBioMedicine 2021;74:103739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emont MP, Jacobs C, Essene AL, et al. A single-cell atlas of human and mouse white adipose tissue. Nature 2022;603:926–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Campbell JE, Ussher JR, Mulvihill EE, et al. TCF1 links GIPR signaling to the control of beta cell function and survival. Nat Med 2016;22:84–90 [DOI] [PubMed] [Google Scholar]
- 36. Wang ZV, Deng Y, Wang QA, Sun K, Scherer PE. Identification and characterization of a promoter cassette conferring adipocyte-specific gene expression. Endocrinology 2010;151:2933–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eguchi J, Wang X, Yu S, et al. Transcriptional control of adipose lipid handling by IRF4. Cell Metab 2011;13:249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tamarina NA, Roe MW, Philipson L. Characterization of mice expressing Ins1 gene promoter driven CreERT recombinase for conditional gene deletion in pancreatic β-cells. Islets 2014;6:e27685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ruzankina Y, Pinzon-Guzman C, Asare A, et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 2007;1:113–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jeffery E, Church CD, Holtrup B, Colman L, Rodeheffer MS. Rapid depot-specific activation of adipocyte precursor cells at the onset of obesity. Nat Cell Biol 2015;17:376–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Joo E, Harada N, Yamane S, et al. Inhibition of gastric inhibitory polypeptide receptor signaling in adipose tissue reduces insulin resistance and hepatic steatosis in high-fat diet-fed mice. Diabetes 2017;66:868–879 [DOI] [PubMed] [Google Scholar]
- 42. Adriaenssens AE, Biggs EK, Darwish T, et al. Glucose-Dependent Insulinotropic Polypeptide Receptor-Expressing Cells in the Hypothalamus Regulate Food Intake. Cell Metab 2019;30:987–996.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Killion EA, Chen M, Falsey JR, et al. Chronic glucose-dependent insulinotropic polypeptide receptor (GIPR) agonism desensitizes adipocyte GIPR activity mimicking functional GIPR antagonism. Nat Commun 2020;11:4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McIntosh CH, Widenmaier S, Kim SJ. Glucose-dependent insulinotropic polypeptide signaling in pancreatic β-cells and adipocytes. J Diabetes Investig 2012;3:96–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Weaver RE, Donnelly D, Wabitsch M, Grant PJ, Balmforth AJ. Functional expression of glucose-dependent insulinotropic polypeptide receptors is coupled to differentiation in a human adipocyte model. Int J Obes 2008;32:1705–1711 [DOI] [PubMed] [Google Scholar]
- 46. Lee KY, Russell SJ, Ussar S, et al. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes 2013;62:864–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beaudry JL, Kaur KD, Varin EM, et al. Physiological roles of the GIP receptor in murine brown adipose tissue. Mol Metab 2019;28:14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jeffery E, Berry R, Church CD, et al. Characterization of Cre recombinase models for the study of adipose tissue. Adipocyte 2014;3:206–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Varin EM, Mulvihill EE, Beaudry JL, et al. Circulating levels of soluble dipeptidyl peptidase-4 are dissociated from inflammation and induced by enzymatic DPP4 inhibition. Cell Metab 2019;29:320–334.e5 [DOI] [PubMed] [Google Scholar]
- 50. Prost S, Sheahan S, Rannie D, Harrison DJ. Adenovirus-mediated Cre deletion of floxed sequences in primary mouse cells is an efficient alternative for studies of gene deletion. Nucleic Acids Res 2001;29:E80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Martens K, Bottelbergs A, Baes M. Ectopic recombination in the central and peripheral nervous system by aP2/FABP4-Cre mice: implications for metabolism research. FEBS Lett 2010;584:1054–1058 [DOI] [PubMed] [Google Scholar]
- 52. Mullican SE, Tomaru T, Gaddis CA, Peed LC, Sundaram A, Lazar MA. A novel adipose-specific gene deletion model demonstrates potential pitfalls of existing methods. Mol Endocrinol 2013;27:127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 1999;21:70–71 [DOI] [PubMed] [Google Scholar]
- 54. Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis 2007;45:593–605 [DOI] [PubMed] [Google Scholar]
- 55. Zhang Q, Delessa CT, Augustin R, et al. The glucose-dependent insulinotropic polypeptide (GIP) regulates body weight and food intake via CNS-GIPR signaling. Cell Metab 2021;33:833–844.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Coskun T, Sloop KW, Loghin C, et al. LY3298176, a novel dual GIP and GLP-1 receptor agonist for the treatment of type 2 diabetes mellitus: from discovery to clinical proof of concept. Mol Metab 2018;18:3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Samms RJ, Christe ME, Collins KA, et al. GIPR agonism mediates weight-independent insulin sensitization by tirzepatide in obese mice. J Clin Invest 2021;131:e146353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mohammad S, Ramos LS, Buck J, Levin LR, Rubino F, McGraw TE. Gastric inhibitory peptide controls adipose insulin sensitivity via activation of cAMP-response element-binding protein and p110β isoform of phosphatidylinositol 3-kinase. J Biol Chem 2011;286:43062–43070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kim SJ, Nian C, McIntosh CH. Activation of lipoprotein lipase by glucose-dependent insulinotropic polypeptide in adipocytes. A role for a protein kinase B, LKB1, and AMP-activated protein kinase cascade. J Biol Chem 2007;282:8557–8567 [DOI] [PubMed] [Google Scholar]
- 60. Kim SJ, Nian C, McIntosh CH. Resistin is a key mediator of glucose-dependent insulinotropic polypeptide (GIP) stimulation of lipoprotein lipase (LPL) activity in adipocytes. J Biol Chem 2007;282:34139–34147 [DOI] [PubMed] [Google Scholar]
- 61. Hansotia T, Maida A, Flock G, et al. Extrapancreatic incretin receptors modulate glucose homeostasis, body weight, and energy expenditure. J Clin Invest 2007;117:143–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ugleholdt R, Pedersen J, Bassi MR, et al. Transgenic rescue of adipocyte glucose-dependent insulinotropic polypeptide receptor expression restores high fat diet-induced body weight gain. J Biol Chem 2011;286:44632–44645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Baggio LL, Drucker DJ. Glucagon-like peptide-1 receptor co-agonists for treating metabolic disease. Mol Metab 2021;46:101090. [DOI] [PMC free article] [PubMed] [Google Scholar]