Abstract
Orthogonal (O-) ribosome mediated translation of O-mRNAs enables the incorporation of up to three distinct non-canonical amino acids (ncAAs) into proteins in Escherichia coli. However, the general and efficient incorporation of multiple distinct ncAAs by O-ribosomes requires scalable strategies for both creating efficiently and specifically translated O-mRNAs, and the compact expression of multiple O-aminoacyl-tRNA synthetase (O-aaRS)/O-tRNA pairs. We automate the discovery of O-mRNAs that lead to up to 40-times more protein, and are up to 50-fold more orthogonal, than previous O-mRNAs; protein yields from our O-mRNAs match or exceed those from wild-type mRNAs. These advances enable a 33-fold increase in yield for incorporating three distinct ncAAs. We automate the creation of operons for O-tRNA genes, and develop operons for O-aaRS genes. Combining our advances creates a 68-codon, 24 amino acid genetic code to efficiently incorporate four distinct ncAAs into a single protein in response to four distinct quadruplet codons.
The ability to genetically encode the incorporation of multiple distinct non-canonical amino acids (ncAAs) into proteins will provide new opportunities for the engineering and directed evolution of protein function, will enable new strategies for biological discovery and understanding biological processes, and provides a foundation for the encoded cellular synthesis of non-canonical biopolymers1–3. We anticipate that the opportunities that arise from approaches to incorporate multiple distinct ncAAs will increase as the number of distinct ncAAs that can be incorporated increases.
Encoding multiple distinct ncAAs into proteins synthesized in cells requires orthogonal codons, beyond those used to encode natural protein synthesis in the same cell; these include quadruplet codons4–6, codons arising from sense codon compression3, 7, 8, and codons incorporating non-canonical bases9–12. Orthogonal codons must be assigned to ncAAs using engineered mutually orthogonal aminoacyl-tRNA synthetase (aaRS)/tRNA pairs. These pairs should be orthogonal in their aminoacylation specificity with respect to the synthetases and tRNAs used by the host organism for natural translation, and with respect to other orthogonal aaRSs and tRNAs used to direct ncAAs in the same cell; moreover, they should specifically recognize distinct ncAA monomers and decode distinct orthogonal codons4, 13–19.
Orthogonal ribosomes (O-ribosomes) are non-natural ribosomes that are directed towards an orthogonal mRNA (O-mRNA), which is not a substrate for wild-type (wt) ribosomes in Escherichia coli (E. coli) (Fig. 1). These ribosomes operate in parallel with natural ribosomes but contain alterations in their ribosomal RNA that direct them to an O-ribosome binding site (O-RBS) within the 5’ untranslated region (5’ UTR) of the orthogonal message20. Since O-ribosomes are not responsible for synthesizing the proteome, they can be engineered to perform new functions not accessed by natural ribosomes, including new decoding and new intrinsic polymerization functions4, 21, 22. O-riboQ1 (an evolved O-ribosome) efficiently decodes amber codons and quadruplet codons on O-mRNAs, using cognate tRNAs, and thus provides orthogonal codons that are selectively decoded on the orthogonal message4, 21.
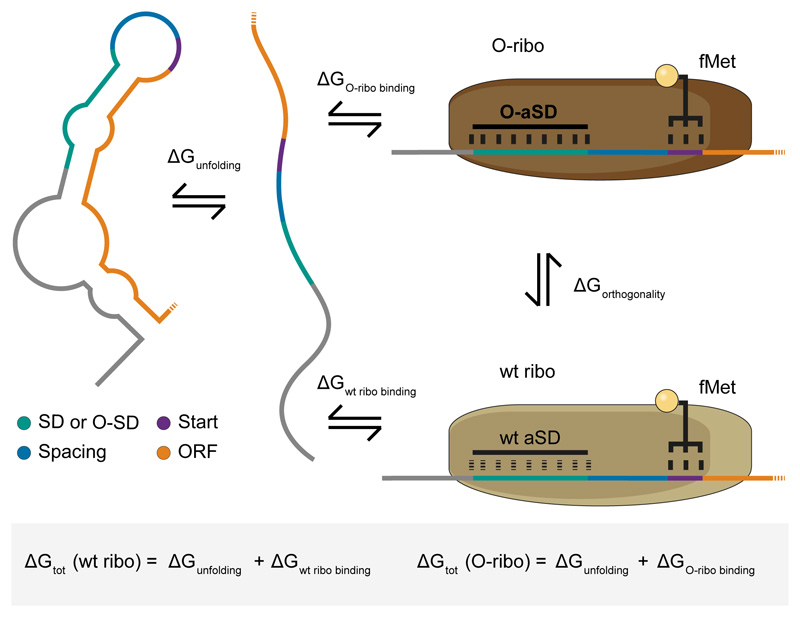
Figure 1. A thermodynamic model for the initiation of protein synthesis by wt and O-ribosomes on an mRNA.
The free energy for the formation of the initiation complex (ΔGtot) is the sum of the free energy required to unfold the mRNA (ΔGunfolding) and the free energy released (ΔGribo binding) when the mRNA forms the initiation complex through binding to a ribosomal 30S subunit and tRNAfMetCAU (black trident and yellow sphere). The 30S subunit of an O-ribosome (dark brown) contains an orthogonal anti-Shine Dalgarno (O-aSD) at the 3’ end of the O-16S rRNA, while the 30S subunit of the wt ribosome (light brown) contains a wt anti-Shine Dalgarno (wt aSD) at the 3’ end of its 16S rRNA. The free energy released on forming the initiation complex from unfolded mRNA with a wt and orthogonal 30S are ΔGwt ribo binding and ΔGO-ribo binding respectively. ORF open reading frame (orange), start codon (purple), SD/O-SD Shine-Dalgarno sequence or an orthogonal version, respectively (green), spacing between SD/O-SD and start codon (blue). The remainder of the 5’ UTR is shown in grey.
Engineered mutually orthogonal aaRS/tRNA pairs – which recognize distinct ncAAs and decode distinct codons – have been used to incorporate two or three distinct ncAAs into proteins4, 5, 15, 16, 19, 23. The homologous Methanosarcina mazei (Mm) or Methanosarcina barkeri (Mb) pyrrolysyl–tRNA synthetase (PylRS)/MmtRNAPylCUA or MbtRNAPylCUA pairs are the most widely used orthogonal aaRS/tRNA pairs for genetic code expansion2, 24. We recently investigated PylRS/tRNAPyl pairs from diverse organisms and discovered that natural PylRS and tRNAPyl sequences cluster into several subclasses with distinct specificities; this insight allowed us to engineer doubly and triply orthogonal PylRS/ tRNAPyl pairs that recognize distinct ncAAs and decode distinct codons15, 16.
By combining O-riboQ1-mediated translation of O(trans)-strepGFP(40TAG, 136AGGA, 150AGTA)His6 (an O-mRNA for a StrepGFPHis6 open reading frame (ORF) translated from a previously described 5’ UTR containing an O-ribosome binding site (O(trans)), and containing two quadruplet codons (AGGA and AGTA) and an amber codon (TAG)) with engineered triply orthogonal PylRS/tRNAPyl pairs, we demonstrated the incorporation of three ncAAs into recombinant StrepGFP(40BocK, 136NmH, 150CbzK)His616. However – as we noted15, 16 – the yield of protein from this expression system was low and un-optimized. Additional experiments – with O(trans)-strepGFPHis6 and a strepGFPHis6 open reading frame with a 5’ UTR containing a wt RBS – demonstrated that the translation of O(trans)-strepGFPHis6 by the O-ribosome leads to 31-fold less StrepGFPHis6 protein from their cognate mRNA than is produced by wt ribosomes. Moreover, transferring the O(trans) 5’ UTR to other ORFs also leads to substantially decreased levels of protein synthesis (Fig. 2 and Supplementary Fig. 1). The O(trans) 5’ UTR sequence was derived from constructs for producing GST fusion proteins, where it directed O-ribosome dependent translation at comparable levels to O-ribosome independent translation from a 5’ UTR containing a wt RBS4, 21. These observations demonstrated that – although the O(trans) sequence directs efficient orthogonal translation for some ORFs – it does not provide a general solution for the efficient translation of ORFs. This realization prompted us to investigate general solutions to the creation O-mRNAs that maximize protein yields in orthogonal translation.
Figure 2. Automated design of O-mRNA sequences that are specifically and efficiently translated by O-ribosomes.
a, Algorithms developed to design O-mRNA sequences. Vol 1 generates a random 5’ UTR containing a wt SD sequence and predicts its ΔGtot (O-ribo). In an iterative process, a mutation is introduced into the 5’ UTR and the algorithm predicts a new orthogonal ΔGtotnew (O-ribo). If ΔGtotnew (O-ribo) is more negative than ΔGtot (O-ribo), the change is accepted; otherwise, the change is rejected with some conditional probability (see Methods). The algorithm terminates after 10,000 iterations. Vol 2 generates a random 5’ UTR containing an O-SD sequence at an optimal spacing from the start codon and predicts its ΔGtot (wt ribo) and ΔGtot (O-ribo). In an iterative process, a mutation is introduced into the 5’ UTR and the algorithm calculates new predicted values, ΔGtotnew (wt ribo) and ΔGtotnew (O-ribo). If ΔGtotnew (wt ribo) and ΔGtotnew (O-ribo) are more favourable than ΔGtot (wt ribo) and ΔGtot (O-ribo), the change is accepted; otherwise, the mutation is rejected with some conditional probability (see Methods). The algorithm terminates if 500 consecutive iterations fail to improve ΔGtot values. Vol 3 builds on vol 2, but has two notable differences: (1) Vol 3 starts with an ORF in which codons 2 to 12 are randomly exchanged with synonymous codons. (2) In the iterative process, both synonymous codon substitutions and mutations in the 5’ UTR are allowed. b, Discovering O-mRNA sequences that are specifically and efficiently translated by O-ribosomes. The y axis shows the production of strepGFPHis6 from O-mRNAs by O-ribosomes; the data is shown as a percentage of strepGFPHis6 produced by wt ribosomes from a wt message. The x axis shows the orthogonality of the O-mRNA; this is calculated as: strepGFPHis6 produced from the O-mRNA in the presence of O-ribosomes divided by strepGFPHis6 produced from the O-mRNA in the presence of wt ribosomes. Protein production levels are calculated from GFP absorption and fluorescence data; the wt system generates 30.6 ± 1.6 mg l-1 of strepGFPHis6. Each dot represents one O-mRNA. Trans (black dot) is O(trans)-strepGFPHis6. The coloured dots represent sequences from the indicated volume of the algorithm. c, d, Same as in b but done for mCherry (c) and E2Crimson (d) respectively. Bar charts representing the mean of three biological replicates ± s.d. for each measurement are provided in Supplementary Fig. 1.
Our understanding of the factors that determine protein yield for natural translation are incomplete: a design of experiment study suggests that only half the variance in observed protein yield can be explained by known parameters25. Nonetheless, initiation of protein synthesis is commonly the rate limiting step of translation26 and numerous studies suggest that RNA secondary structure in the 5’ UTR and the first 30 nt of the coding sequence are key determinants of translational initiation and protein yield25, 27. Indeed, thermodynamic models that predicts the total free energy change (ΔGtot (wt ribo)) from the free folded mRNA to a final ‘initiation competent’ state can be used to predict relative protein yields for natural translation28–30. Previous work – varying 35 nt in the 5’ UTR immediately upstream of the start codon – indicates that protein yields for a given ORF (interpreted as reflecting the rate of translational initiation) are proportional to the equilibrium constant (ie: proportional to the log of the ΔGtot (wt ribo)) for the formation of the initiation-competent state from the folded mRNA28, 31–34. ΔGtot (wt ribo) can be decomposed into mRNA unfolding (ΔGunfolding) and binding of the wt ribosome and tRNAfMetCAU, through base-pairing in the correct positions, to the mRNA (ΔGwt ribo binding) (Fig. 1).
Here we use a thermodynamic model of initiation and a simulated annealing optimization algorithm28 to automate the design of 5’ UTR sequences for orthogonal translation of ORFs. We develop the algorithm to explicitly select for messages that bind O-ribosomes, but not wt ribosomes, and increase the degrees of freedom in our search by exploring variation in both the 5’ UTR and the synonymous codons that encode amino acids 2 to 12 of the ORF. Automating the design of O-mRNAs leads to the discovery of sequences that provide up to 40-times more protein, and are up to 50-fold more orthogonal, than previous O-mRNAs; protein yields from our O-mRNAs match or exceed those from the wt mRNAs tested. These advances directly translate into a 33-fold increase in yield for incorporating three distinct ncAAs in response to an amber codon and two quadruplet codons using engineered triply orthogonal PylRS/tRNAPyl pairs. We automate the design of operons for the compact, scalable expression of distinct orthogonal tRNAs, and develop operons for expressing distinct orthogonal aaRSs. We develop compact operons expressing engineered triply orthogonal PylRS/tRNAPyl pairs and an Archaeoglobus fulgidus tyrosyl-tRNA synthetase (AfTyrRS)/tRNATyr derived pair, and demonstrate that all four pairs are mutually orthogonal. We combine our advances to create a 68-codon, 24 amino acid genetic code and efficiently incorporate four distinct ncAAs in response to four distinct quadruplet codons, via O-riboQ1-mediated translation of a designed O-mRNA.
Results
Automating 5’ UTR design for orthogonal translation
For our strepGFPHis6 ORF on a 5’ UTR containing a wt RBS the predicted ΔGtot (wt ribo) is -0.5 kcal/mol. In contrast, when we altered the anti-Shine Dalgarno sequence (aSD) used in the thermodynamic model to that of the O-ribosome the calculated free energy change for orthogonal translation (ΔGtot (O-ribo)) of O(trans)-StrepGFPHis6 was +3.5 kcal/mol. We decided to test whether an equilibrium model of initiation combined with a simulated annealing optimization algorithm, developed for wt translation28, could be adapted to design O-mRNA sequences that are more efficiently translated by O-ribosomes than O(trans)-strepGFPHis6 (Fig. 1 and Fig. 2a). We therefore varied the 5' UTR sequence between the +1 transcription site and the strepGFPHis6 ORF and searched – through a simulated annealing optimization algorithm28 – for sequences with highly favourable ΔGtot (O-ribo) for this ORF.
Using this algorithm (vol 1) we identified four new strepGFPHis6 constructs with optimised 5’ UTR regions (O1-strepGFPHis6 to O4-strepGFPHis6) for the production of strepGFPHis6 protein by the O-ribosome. The ΔGtot (O-ribo) for these constructs was: O1-strepGFPHis6 -5.8 kcal/mol, O2-strepGFPHis6 -4.9 kcal/mol, O3-strepGFPHis6 -5.1 kcal/mol, O4-strepGFPHis6 -6.6 kcal/mol. Thus, we predicted that these constructs may lead to higher protein levels than O(trans)-strepGFPHis6. We produced strepGFPHis6 from cells containing each construct and the O-ribosome. The optimised sequences (O1-strepGFPHis6 to O4-strepGFPHis6) led to large (11- to 31-fold) increases in protein production with orthogonal translation compared to O(trans)-strepGFPHis6 (Fig. 2b and Supplementary Fig. 1). The level of strepGFPHis6 protein produced from O1-strepGFPHis6 by the O-ribosome was comparable to that from the original construct containing a wt RBS and translated by the wt ribosome. ΔGtot (wt-ribo) (Fig. 1) for the new sequences was greater than +5 kcal/mol in all cases. Thus ΔGorthogonality (Fig. 1) predicts that these constructs will be selectively translated by the O-ribosome. Consistent with this prediction, additional experiments demonstrated that translation of strepGFPHis6 from each new 5’ UTR was O-ribosome dependent, and the orthogonality of the new sequences was 12- to 19-fold greater than that of O(trans)-strepGFPHis6 (Fig. 2b and Supplementary Fig. 1).
Automating 5’ UTR and ORF design for orthogonal translation
In an effort to fully automate the design of 5’ UTRs that do not direct efficient translation by wt ribosomes and direct maximal protein production by the O-ribosome, we created a new automated search (vol 2) (Fig. 2a). Our new search introduced an explicit penalty for 5’ UTR sequences that are predicted to be substrates for wt ribosomes and was biased towards sequences containing an optimally spaced canonical O-SD sequence.
The vol 2 search started from a 35 nt 5’ UTR which contained a 9 nt orthogonal SD (O-SD) sequence that is predicted to form perfect Watson-Crick base pairs with the orthogonal aSD sequence at the 3’ end of the O-16S rRNA. The spacing between the O-SD sequence and the start codon was set to 5 nucleotides and the sequence of the 5’ UTR, except the O-SD, was randomized. We then searched for sequences that maximize ΔGtot (O-ribo) but minimize ΔGtot (wt ribo). We disallowed mutations in the 5-nucleotide core of the O-SD site, which is predicted to base pair with the O-16S rRNA – but not the wt 16S rRNA – and thus determines orthogonality. Using the vol 2 algorithm we created new 5’ UTRs for strepGFPHis6 (O5- to O8-strepGFPHis6). These sequences had higher mean ΔGtot (O-ribo) (-7.7 ± 0.4 kcal/mol) than those derived from vol 1 (-5.6 ± 0.8 kcal/mol) (Supplementary Table 1). These sequences provided up to 18-fold more strepGFPHis6 protein than O(trans)-strepGFPHis6 (Fig. 2 and Supplementary Fig. 1). To investigate the generality of the vol 2 algorithm for enhancing protein production we investigated orthogonal translation of two additional ORFs, mCherry and E2Crimson. O(trans)-mCherry, and O(trans)-E2Crimson (in which the O(trans) 5’ UTR was placed between the +1 base of transcription and the ATG start codon) led to low levels of orthogonal translation. Applying the vol 2 algorithm led to mCherry expression constructs that are up to 10 times more active with the O-ribosome than O(trans)-mCherry, and also up to 8-fold more orthogonal (Fig. 2c and Supplementary Fig. 1). Similarly, applying the vol 2 algorithm led to E2Crimson production constructs that are up to 14-fold more active with the O-ribosome than O(trans)-E2Crimson, and up to 9-fold more orthogonal; E2Crimson was produced by the O-ribosome from O1-E2Crimson (designed using the vol 2 algorithm) at comparable levels to the levels produced from a wt RBS using a wt ribosome (Fig. 2d and Supplementary Fig. 1).
The first 35 nucleotides of ORF sequence can contribute substantially to protein yields25, 27, 29. However, it remains controversial to what extent changing codons to their synonyms in this sequence influences translation through effects on mRNA secondary structure versus effects that result from the decoding of different synonyms with distinct isoacceptor tRNAs25–27, 35–37. We realized that varying codons within the first 35 nucleotides of the ORF to synonymous codons would provide additional degrees of freedom in the computational search for mRNAs that maximize ΔGtot (O-ribo) but minimize ΔGtot (wt ribo). And we hypothesized that, in some cases, this may allow us to discover mRNAs that are more efficiently translated by the O-ribosome and are more orthogonal with respect to translation by wt ribosomes. To investigate this hypothesis, we allowed codons 2 to 12 of each ORF to vary to their synonyms. We thereby created a third algorithm (vol 3), which builds on vol 2, to explore simultaneous variation in the ORF and 5’ UTR (Fig. 2a).
The vol 3 algorithm provided a notable increase in ΔGtot (O-ribo) (strepGFPHis6: -12.6 ± 0.2 kcal/mol; mCherry: -13.5 ± 0.3 kcal/mol; E2Crimson: -13.2 ± 0.0 kcal/mol) with respect to vol 2 (strepGFPHis6: -7.7 ± 0.4; mCherry: -9.6 ± 0.5 kcal/mol; E2Crimson: -8.9 ± 0.5 kcal/mol) and maintained the minimized ΔGtot (wt ribo) from vol 2. We discovered O-mRNA sequences for strepGFPHis6 and mCherry that are more orthogonal than those from the vol 2 algorithm and produce protein at levels higher than those produced by wt ribosomes from wt messages (Fig. 2b-d and Supplementary Fig. 1). Overall, our vol 2 and vol 3 algorithms provided protein yields that are 41-, 31- and 14-fold (for strepGFPHis6, mCherry, and E2Crimson, respectively) greater than when the O(trans) 5’ UTR was used with each ORF, and these yields match or exceed the yields from wt ribosomes on the wt message controls. The orthogonality of the best sequences we have discovered is 31-, 49- and 9-fold (for strepGFPHis6, mCherry, and E2Crimson, respectively) higher than when the O(trans) 5’ UTR was used with each ORF.
Optimized O-mRNAs increase yields for 3 distinct ncAAs
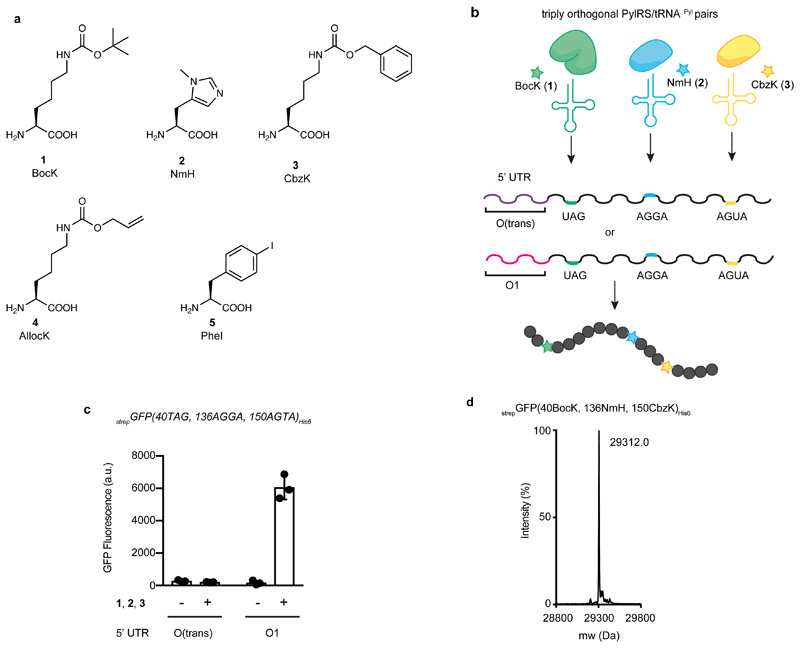
Next, we demonstrated that the increase in protein yields from optimized O-mRNAs enables an increase in the yield of protein containing three distinct ncAAs, via orthogonal translation. As this work proceeded in parallel with the algorithm development described above, we performed our experiments with the most active sequence available at the time, O1-strepGFPHis6, derived from the vol 1 algorithm (Fig. 2a,b). We created O1-strepGFP(40TAG, 136AGGA,150AGTA)His6 and translated this with O-riboQ1 in cells containing a triply orthogonal PylRS/tRNAPyl pair (composed of MmPylRS/ Methanosarcina spelaei (Mspe)tRNAPylCUA (which directs the incorporation of N6-(tert-butoxycarbonyl)-L-lysine (BocK) 1), Methanomassiliicoccus luminyensis 1 (Mlum)PylRS(NmH)/Methanomassiliicoccus intestinalis (Mint)tRNAPyl-A17VC10UCCU (L121M, L125I, Y126F, M129A, V168F mutant, which directs the incorporation of Nπ-methyl-L-histidine (NmH) 2) and Methanomethylophilus sp. 1R26 (M1r26)PylRS(CbzK)/Methanomethylophilus alvus (Malv)tRNAPyl-8UACU (Y126G, M129L mutant, which directs the incorporation of N6-((benzyloxy)carbonyl)-L-lysine (CbzK) 3). Full-length strepGFP(40BocK, 136NmH, 150CbzK)His6 was produced upon addition of BocK 1, NmH 2 and CbzK 3(Fig. 3a,b). Using this system, we synthesized 2.6±0.4 mg/L of strepGFP(40BocK, 136NmH, 150CbzK)His6. This yield is 33 times greater than the yield from O(trans)-strepGFP(40TAG, 136AGGA,150AGTA)His6 (Fig. 3c and Supplementary Table 2), corresponds to 9% of strepGFP(wt)His6 produced from O1-strepGFP(wt)His6, and to 11% of strepGFP(wt)His6 produced from strepGFP(wt)His6 translated from a wt RBS by wt ribosomes. The observed yields suggest a mean ncAA incorporation efficiency per step of 45%. Mass spectrometry confirmed the synthesis of the correct protein (Fig. 3d, Supplementary Fig. 2).
Figure 3. Efficient production of proteins containing three distinct ncAAs is enabled by new O-mRNAs.
a, Structures of the amino acids used in this work.N6-(tert-butoxycarbonyl)-L-lysine (BocK) 1; Nπ-methyl-L-histidine (NmH) 2; N6-((benzyloxy)carbonyl)-L-lysine (CbzK) 3; N6-((allyloxy)carbonyl)-L-lysine (AllocK) 4; (S)-2-amino-3-(4-iodophenyl)propanoic acid (PheI) 5. b, Engineered triply orthogonal pyrrolysyl-tRNA synthetase tRNA pairs for the incorporation of three distinct ncAAs using two different orthogonal messages. One message contains the O1-strepGFPHis6 5’UTR, generated by vol 1 of our algorithm, and the other message used the O-(trans) 5’UTR. c, Production of strepGFP(40BocK, 136NmH, 150CbzK)His6 from E. coli cells containing strepGFP(40TAG, 136 AGGA, 150 AGTA)His6 constructs with either the O(trans)- or O1-strepGFP-His6 5’UTRs. Cells also contained O-riboQ1 and the aaRS3/tRNA3 operons (encoding MmPylRS/MspetRNAPylCUA, MlumPylRS(NmH)/MinttRNAPyl-A17VC10UCCU and M1r26PylRS(CbzK)/MalvtRNAPyl-8UACU). ncAAs BocK 1, NmH 2, CbzK 3 were added to the cell. Each bar represents the mean of three biological replicates ± s.d. The individual data points are shown as dots. d, Results of positive electrospray TOF-MS of nickel-NTA purified strepGFP(40BocK, 136NmH, 150CbzK)His6 purified from cells. StrepGFP(40BocK, 136NmH, 150Cbz)His6 mass predicted: 29314.5, mass found: 29312.0. The experiment was performed three times with similar results.
Operon designs for quadruply orthogonal aaRS/tRNA pairs
Next, we aimed to build on the development of efficient O-mRNAs to enable the incorporation of four distinct ncAA into a single protein, with each ncAA encoded in response to a distinct quadruplet codon. This required four orthogonal aaRS/tRNA pairs that: (1) are mutually orthogonal in their aminoacylation specificity, (2) have four mutually orthogonal active sites, and (3) are assigned to four mutually orthogonal quadruplet codons. We chose a PylRS/tRNAPyl triplet – Methanomassiliicoccales archaeon RumEn M1 (Mrum)Pyl(NmH)RS/MinttRNAPyl-A17VC10UCCU (L121M, L125I, Y126F, M129A, V168F mutant, which directs the incorporation of NmH 2), Methanogenic archaeon ISO4-G1 (Mg1)Pyl(CbzK)RS/MalvtRNAPyl-8UACU (Y125G, M128L mutant, which directs the incorporation of CbzK 3) and MmPylRS/MspetRNAPyl-evolCUAG (which directs the incorporation of several ncAAs, including BocK 1 or N6-((allyloxy)carbonyl)-L-lysine (AllocK) 4) – as a starting point for our approach. We chose the AfTyrRS(PheI)/AftRNATyr-A01CUA, (Y36I, L69M, H74L, Q116E, D165T, I166G, F274V, L298G, D299R mutant, which directs the incorporation of (S)-2-amino-3-(4-iodophenyl)propanoic acid (PheI) 5) as the starting point for a fourth aaRS/tRNA pair; we have previously shown that this pair is orthogonal to several pyrrolysyl synthetases and tRNAPyls. Efforts to encode multiple ncAAs require strategies for the efficient and compact expression of the corresponding synthetases and tRNAs. We therefore established operon-based systems for the co-expression of the four exogenous tRNAs and the co-expression of their cognate synthetases.
In E. coli, many tRNAs are transcribed in polycistronic operons, and the 5' and 3' ends of mature tRNAs are generated by post-transcriptional RNase processing38, 39. We created a program to automatically design synthetic tRNA operons in which the intergenic sequence between the exogenous tRNAs is derived from the sequence between E. coli tRNAs that are most similar to the exogenous tRNAs. The program first generates all possible orderings of the exogenous tRNAs. For each pair of adjacent exogenous tRNAs in an ordering, it identifies the adjacent natural tRNAs in the E. coli genome with the highest sequence identity to the exogenous pair. It then inserts the sequence of the intergenic region found between these natural tRNAs between the exogenous tRNAs. This process generates a synthetic operon sequence for each ordering of exogenous tRNAs. The program then compares the synthetic operons resulting from each tRNA order and ranks them based on the sum of the sequence identity between the exogenous tRNAs and the corresponding natural tRNAs used to define the intergenic regions in the operon.
We used our program for generating tRNA operons with AftRNATyr-A01, MspetRNAPyl-evol, MinttRNAPyl-A17VC10, and MalvtRNAPyl-8. Among the top ranked operons was: MinttRNAPyl-A17VC10UCCU - inter(glyX, glyY) - MalvtRNAPyl-8UACU- inter(glyW-cysT) – MspetRNAPyl-evolCUAG- inter(argY, argZ) - AftRNATyr-A01CUA, where inter(x, y) represents the intergenic spacer sequence between the E. coli tRNAs x and y. To adapt this operon for expressing tRNAs that decode four distinct quadruplet codons we replaced MspetRNAPyl-evolCUAG with MspetRNAPyl-evolUCUA (created by transplanting an anticodon stem that we have previously evolved in MbtRNAPyl into MspetRNAPyl) and AftRNATyr-A01CUA by AftRNATyr-A01CUAG (created by anticodon mutation of AftRNATyr-A01CUA). We named the resulting tRNA operon tRNA4(quad).
To identify operons that would allow high expression of the four exogenous aaRSs (MmPylRS, AfTyr(PheI)RS, Mg1(CbzK)PylRS and Mrum(NmH)PylRS) we first generated five optimized 5’ UTR regions for each synthetase gene, and then predicted the ΔGtot for going from the folded mRNA to the initiation competent translation complex for each 5’ UTR using any of the other three aaRS as 5’ sequence context. We chose two arrangements, RS4_1 and RS4_2, which had favorable ΔGtot for all four aaRS. We cloned each of the aaRS operons into a plasmid encoding tRNA4 to generate compact synthetase and tRNA expression modules (RS4_1/tRNA4 and RS4_2/tRNA4) (Supplementary Fig. 3). We tested the activity of each aaRS in each operon (Supplementary Fig. 4, Supplementary Table 3). These experiments led us to design an optimized chimeric aaRS operon in which we transplanted 150 nt upstream of the optimised 5’ UTR of Mrum(NmH)PylRS from RS4_2 into RS4_1, creating RS4_1-2. This operon combined the best properties of RS4_2 and RS4_1 (Supplementary Fig. 4c).
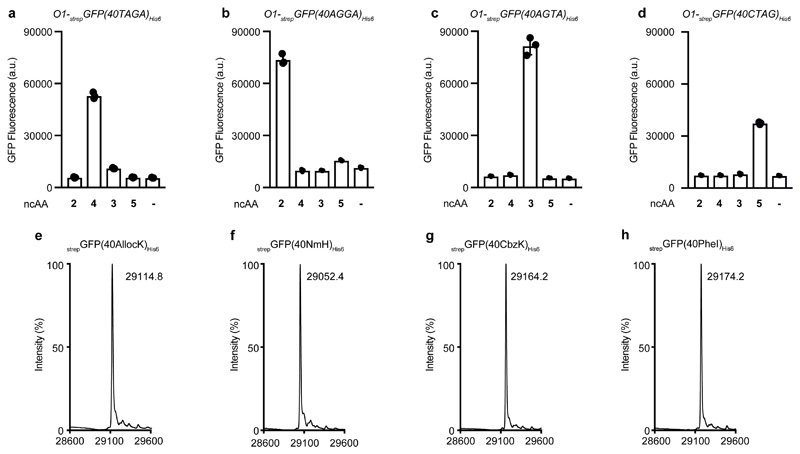
We combined the RS4_1-2 and tRNA4(quad) operons in a single vector (RS4_1-2/ tRNA4(quad)) and systematically tested the activity and orthogonality of each aaRS/tRNA pair produced by measuring the GFP fluorescence produced from O1-strepGFP(40XXX)His6, where XXXX stands for TAGA, AGGA, AGTA or CTAG. Cells contained O-riboQ1, each individual ncAA (NmH 2, CbzK 3, AllocK 4, PheI 5) or none, and RS4_1-2/tRNA4 (quad) (Fig. 4a-d). ESI-MS of strepGFP(40X)His6 (where X stands for NmH 2, CbzK 3, AllocK 4, PheI 5) produced by O-riboQ1 from O1-strepGFP(40XXXX)His6, (where XXXX stands for TAGA, AGGA, AGTA or TAGA) in the presence of RS4_1-2/ tRNA4(quad) and all four ncAAs (NmH 2, CbzK 3, PheI 5, AllocK 4) demonstrated that each aaRS, tRNA and codon are functionally orthogonal with respect to each other aaRS, tRNA and codon set (Fig 4e-h).
Figure 4. Four orthogonal aaRS/tRNA pairs decoding four orthogonal quadruplet codons are expressed from aaRS operons and computationally generated tRNA operons and are mutually orthogonal in their aminoacylation specificity, recognize distinct ncAAs, and decode distinct orthogonal codons.
a-d, Fluorescence from cells containing O1-strepGFP(40XXXX)His6, with XXXX being the codon at position 40 in sfGFP: TAGA, CTAG, AGGA or AGTA. E. coli also contained O-riboQ1 and the aaRS and tRNA operons (aaRS4_1-2/tRNA4(quad)); these operons expressed MmPylRS/MspetRNAPyl-evolUCUA, MrumPylRS(NmH)/MinttRNAPyl-A17VC10UCCU, AfTyrRS(PheI)/AftRNATyr-A01CUAG and Mg1PylRS(CbzK)/MalvtRNAPyl-8UACU. The indicated ncAAs: Nπ-methyl-L-histidine (NmH) 2, N6-((benzyloxy)carbonyl)-L-lysine (CbzK) 3, N6-((allyloxy)carbonyl)-L-lysine (AllocK) 4, (S)-2-amino-3-(4-iodophenyl)propanoic acid (PheI) 5 were added to cells or omitted (-). Each codon was only efficiently decoded in the presence of cognate ncAA of the aaRS/tRNA pair assigned to the respective quadruplet codon: (a) O1-strepGFP(40TAGA)His6 decoded by MmPylRS/MspetRNAPyl-evolUCUA, (b) O1-strepGFP(40AGGA)His6 decoded by MrumPylRS(NMH)/MinttRNAPyl-A17VC10UCCU, (c) O1-strepGFP(40AGTA)His6 decoded by Mg1PylRS(CbzK)/ MalvtRNAPyl-8UACU, and (d) O1-strepGFP(40CTAG)His6 decoded by AfTyrRS(PheI)/AftRNATyr-A01CUAG. Each bar represents the mean of three biological replicates ± s.d. The individual data points are shown as dots. e-h, Positive electrospray TOF-MS of nickel-NTA-purified strepGFPHis6, expressed from O1-strepGFP(40XXXX)His6, with XXXX being either TAGA (e), AGGA (f), AGTA (g) or CTAG (h), in the presence of NmH 2, CbzK 3, AllocK 4, PheI 5. Cells also contained O-riboQ1 and operon aaRS4_2-1/tRNA4(quad). strepGFP(40AllocK)His6: mass predicted 29113.2, mass found 29114.8. strepGFP(40NmH)His6: mass predicted 29052.1, mass found 29052.5. strepGFP(40CbzK)His6: mass predicted 29163.3, mass found 29164.2. strepGFP(40PheI)His6: mass predicted 29174.03, mass found 29174.2. The experiment was performed three times with similar results.
Encoding four distinct ncAAs via quadruplet codons
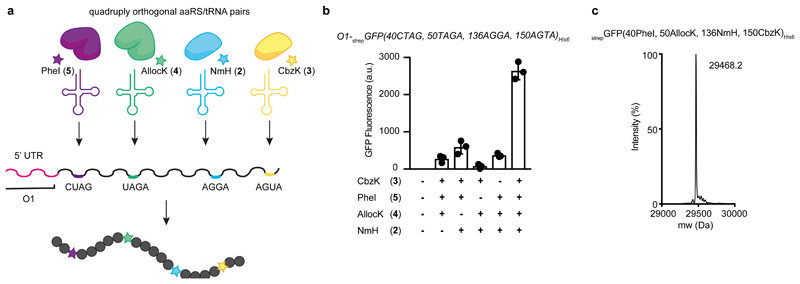
We combined our advances in generating aaRS/tRNA operons for orthogonal pairs with our advances in creating optimized O-mRNAs, which are efficiently read by O-riboQ1, to incorporate four distinct ncAAs into a single protein in response to four distinct quadruplet codons (Fig. 5a). We produced strepGFP(40PheI, 50AllocK, 136NmH, 150CbzK)His6 by O-riboQ1 mediated translation of O1-strepGFP(40CTAG, 50TAGA, 136AGGA, 150AGTA)His6 in cells that contained RS4_1-2/ tRNA4(quad) and were provided with all four ncAA substrates (NmH 2, CbzK 3, PheI 4, AllocK 5) (Fig. 5b). The production of strepGFP(40PheI, 50AllocK, 136NmH, 150CbzK)His6 was dependent upon the addition of all four ncAAs. We observed low levels of full-length protein in the absence of individual ncAAs. These observations are consistent with some orthogonal aaRSs using non-cognate ncAAs at a low level in the absence of their cognate substrate (Fig. 4a-d). In the presence of their cognate substrate these non-cognate interactions are out-competed (Fig. 4e-h, Fig. 5c, Supplementary Fig. 5). 0.41 ± 0.03 mg/L of the protein was produced (Supplementary Table 2). The observed yields suggest a mean ncAA incorporation efficiency per step of 38 %. Mass spectrometry confirmed the incorporation of all four ncAAs in response to four distinct quadruplet codons (Fig. 5c, Supplementary Fig. 5). In additional experiments we also demonstrated the incorporation of four distinct ncAAs in response to three quadruplet codons and the amber codon (Supplementary Fig. 6-8, Supplementary Table 2). The yield of protein containing four distinct ncAAs from translation of O-mRNAs containing four quadruplet codons by O-ribosomes is positively correlated with the yield of protein from the corresponding O-mRNAs without quadruplet codons (Supplementary Fig. 9). These data demonstrate that we can tune the yield of protein containing four distinct ncAAs by O-mRNA choice and further confirm that O-mRNA optimization leads directly to higher yields of proteins containing four distinct ncAAs.
Figure 5. Genetically encoding four distinct ncAAs into a protein using a 24 amino acid, 68 codon genetic code.
a, Schematic representation of four engineered mutually orthogonal aaRS/tRNA pairs used for the incorporation of four distinct ncAAs in response to four orthogonal quadruplet codons.
b, Efficient production of full length strepGFP(40PheI, 50AllocK, 136NmH, 150CbzK)His6 was dependent upon the addition of all four ncAAs (Nπ-methyl-L-histidine (NmH) 2, N6-((benzyloxy)carbonyl)-L-lysine (CbzK) 3, N6-((allyloxy)carbonyl)-L-lysine (AllocK) 4, (S)-2-amino-3-(4-iodophenyl)propanoic acid (PheI) 5). Fluorescence from cells containing O1-strepGFP(40CTAG, 50TAGA, 136AGGA, 150AGTA)His6, O-riboQ1, operon aaRS4/tRNA4(quad) (encoding MmPylRS/MspePyltRNAUCUA, MrumPylRS(NMH)/MintPyltRNA(A17,VC10)UCCU, AfTyrRS(PheI)/AftRNATyr-A01CUAG and Mg1PylRS(CbzK)/MalvPyltRNA(8)UACU) in presence or absence of a combination of NmH (2), CbzK (3), AllocK (4), PheI (5). Each bar represents the mean of three biological replicates ± s.d. The individual data points are shown as dots.
c, Positive electrospray TOF-MS of nickel-NTA purified strepGFP(40PheI, 50AllocK, 136NmH, 150CbzK)His6 from cells containing O1-strepGFP(40CTAG, 50TAGA, 136AGGA, 150AGTA)His6, O-riboQ1 and aaRS4_1-2/tRNA4(quad) in presence of the indicated ncAAs. Mass predicted 29470.4 mass found 29468.2. The experiment was performed three times with similar results.
Discussion
We have developed computational approaches to design O-mRNA sequences that are efficiently and selectively translated by O-ribosomes. The new O-mRNAs lead to up to 40-fold more protein, and are up to 50-fold more orthogonal, than O-mRNAs created by transplanting a previously used 5’ UTR containing the O-RBS in front of an ORF of interest. The O-mRNAs we created direct orthogonal protein production at levels comparable to – or greater than – those from the wt mRNAs we tested, which are translated by wt ribosomes. Our automated, rapid and scalable method for O-mRNA design will greatly accelerate the design and directed evolution of orthogonal translation systems that incorporate multiple ncAAs and polymerize new monomers3, 4, 20–22, as well as the creation and application of orthogonal gene expression systems40, 41.
Our O-mRNA optimization strategies include explicit selection for orthogonality and co-optimization of the 5’ UTR and ORF sequences. We found that co-optimizing the 5’ UTR and synonymous codon choices in the ORF led to O-mRNA sequences with predicted values for ΔGtot (O-ribo) that are larger (more negative) than those obtained through simply varying the 5’ UTR; these sequences also have large (positive) predicted values for ΔGtot (wt ribo). We discovered that co-optimization of the 5’ UTR sequence and synonymous codons within the ORF can improve protein yield, and testing four clones led to high levels of translation in each case tested. These observations are consistent with the view that mRNA folding is the major predictor – amongst known parameters – of protein yield25. We note that other parameters, including codon adaptation, may influence protein yield, and it will be interesting to see whether including these considerations in future iterations of the algorithm will lead to even greater predictive power. Future work may also explore the co-optimisation of 5’ UTR sequences and coding sequence to improve production of difficult-to-express proteins from wt ribosomes.
By combining our automated O-mRNA design with our previously developed triply orthogonal PylRS/tRNAPyl pairs, we increased the yield of a protein containing three distinct ncAA 33-fold. We established a pipeline for the efficient and compact co-expression of many exogenous aaRS and tRNAs. We developed a computational program to produce polycistronic tRNA operons which aims to mimic the endogenous transcription systems in E. coli. Our algorithm provides a general solution to producing multiple distinct tRNAs in E. coli under the same promoter, and may be readily adapted for other organisms. We also devised polycistronic aaRS operons for the efficient expression of four mutually orthogonal synthetases alongside the tRNA operon. We combined our advances to produce a protein consisting of 24 amino acids – the canonical 20 amino acids and 4 ncAAs – in vivo for the first time. Each ncAAs is encoded using quadruplet codons, which are selectively translated on the O-mRNA and not used in natural translation, creating an organism with a 68-codon genetic code.
We anticipate that emerging developments in creating mutually orthogonal aaRS/tRNA pairs that recognize distinct ncAAs and decode distinct quadruplet codons may allow an expansion of the quadruplet code. The efficiency of quadruplet decoding may be further improved by selecting ribosomes that no longer read triplet codons or developing quadruplet decoding in organisms with compressed genetic codes, where competing triplet decoding tRNAs are removed.3,7
Methods
Software implementation
he code for the O-mRNA design method and the tRNA operon designer are available at: https://www2.mrc-lmb.cam.ac.uk/research/technology-transfer/chinlab.
Thermodynamic model of translation initiation
The thermodynamic model has been described previously28. In brief, the model specifies the free energy difference, ΔGtot, of the predicted energy of the free folded mRNA, ΔGunfolding and an initiation-competent ribosome-bound state, ΔGribo_binding.
Here, ΔGunfolding is the energy required to unfold mRNA secondary structures. The free energy released on formation of the initiation-competent state. ΔGribo_binding, consists of four components.
ΔGmRNA−rRNA is the free energy of the predicted co-folded secondary structure of the last 9 nt of the 16S rRNA and the mRNA, in which the main contribution comes from the hybridization energy between the mRNA’s Shine Dalgarno (SD) or orthogonal Shine Dalgarno O-SD sequence and the 16S rRNA. mRNA folding downstream of the hybridization site is not permitted, reflecting the ribosomal footprint. ΔGstart is the energy released from the binding of the initiator tRNA to the start codon. ΔGspacing is an energy penalty for non-optimal spacing length between the SD site and the start codon. ΔGstandby is the energy required to unfold secondary structures that sequester the standby site, which is here defined as the four nucleotides upstream of the SD site.
Simulated annealing optimization algorithm for automated O-mRNA discovery
RNA secondary structure predictions were performed in the NuPACK suite using the ‘mfe’ algorithm. The calculations consider a window of at most 35 nt in the 5’ UTR and ORF; if longer sequences were used, only the 35 nt closest to the start codon were considered.
The vol 1 algorithm is derived from a previously described simulated annealing optimization algorithm28, but using the final 9 nt of the orthogonal 16S rRNA (ATGGGATTA) instead of the canonical sequence (ACCTCCTTA) for the calculation of ΔGmRNA−rRNA. In brief, the algorithm starts from a random 5’ UTR sequence containing a canonical SD sequence. The ΔGtot (O−ribo) of the 5’ UTR and the ORF is evaluated using the thermodynamic model and compared to a target function ΔGtarget. In an iterative procedure, a mutation (either a single nucleotide change, an insertion or a deletion) is introduced into the 5’ UTR and a new ΔGtotnew (O−ribo) is calculated. If the mutated sequence invalidates one of the thermodynamic model’s assumptions28 or introduces a start codon the mutation is rejected. If the mutated sequence leads to a ΔGtotnew (O−ribo) closer to ΔGtarget, the mutation is accepted. If the ΔGtotnew (O−ribo) value is more different from ΔGtarget than the original ΔGtot (O−ribo), the mutation is accepted with a probability of . Here TSA, is the simulated annealing temperature, which is adjusted to maintain a 5-20 % acceptance rate. The algorithm terminates after 10,000 iterations and outputs the 5’ UTR and predicted ΔGtot (O−ribo).
The vol 2 algorithm builds on the vol 1 algorithm. The random starting 5’ UTR contains the 9 nucleotide O-SD site (TAATCCCAT) which is predicted to be perfectly complementary to the O-16S rRNA (ATGGGATTA) at an optimal spacing of 5 nucleotides from the ATG start codon. The ΔGtot (wt rbio) and ΔGtot (O−ribo) of the 5’ UTR and the ORF are evaluated using the thermodynamic model, and a hypothetical ΔGtot (opt) is calculated according to ΔGtot (opt) = ΔGtot (O ribo) − 0.5 * ΔGtot (wt ribo). In contrast to the vol 1 algorithm, no ΔGtarget value is specified. In an iterative procedure, a mutation (either a single nucleotide change, an insertion or a deletion) is introduced into the 5’ UTR and new ΔGtotnew values are calculated. If the mutated sequence violates sequence constraints or alters the 5 nucleotide core of the O-SD sequence (TCCCA), the mutation is rejected. If the mutated sequence leads to an improved (more negative) ΔGtotnew (opt) value, the mutation is accepted. If the ΔGtotnew (opt) value is greater (more positive) than the original ΔGtot (opt), the mutation is accepted with a probability of . If 500 consecutive iterations yield no improvements in ΔGtot (opt), the algorithm terminates and outputs the 5’ UTR and ΔGtot values. We typically run the algorithm multiple times and select sequences with the most favourable values ΔGtot values; we found this is computationally more efficient to identify highly translated 5’ UTRs than running the algorithm for more iterations per starting sequence. In this work, we chose 4 sequences out of 24 predicted 5’ UTRs.
The vol 3 algorithm builds on the vol 2 algorithm. In addition to the random starting 5’ UTR, the amino acids at positions 2 to 12 are encoded by a randomly selected choice of synonymous codons. Synonymous codon changes in positions 2 to 12 in the ORF, in addition to a single nucleotide change, insertion, or deletion in the 5’ UTR, are permitted as a mutation mechanism during the simulated annealing optimization.
tRNA operon designer
The program generates a list of all pairs of tRNAs in the host organism whose genes are adjacent to one another and on the same strand. It then extracts the gene sequences of these endogenous tRNA pairs as well as the corresponding intergenic sequences. Optionally, the user may specify minimum and maximum lengths of intergenic sequences to be considered by the program. For the tRNA operons used in this work, we used the E. coli strain K-12 substrain MG1655 genome (version U00096.3, last modified 24-Sep-2018) as the host genome, with minimum and maximum intergenic sequence lengths of 10 and 100 base pairs, respectively.
Next, the program generates all ordered pairs of the exogenous tRNAs. For each ordered pair of exogenous tRNAs, the acceptor stem sequences of these tRNAs are compared with the acceptor stem sequences of each endogenous tRNA pair. For consistency, we consider the first seven and last eight nucleotides of the tRNAs (excluding the CCA end), which comprise the canonical E. coli tRNA acceptor stem and discriminator base region. The program assigns each exogenous tRNA pair a score based on the sequence identity between the acceptor stems of the exogenous pair and its most similar endogenous tRNA pair.
Finally, the program generates all permutations of the exogenous tRNAs. Synthetic tRNA operons corresponding to each permutation are created by inserting the intergenic regions for endogenous tRNAs between each ordered pair of exogenous tRNA genes in the permutation. For each ordered exogenous pair, the intergenic region corresponding to the most similar endogenous tRNA pair is chosen. Each operon is assigned a score, calculated as the sum of the scores of all the ordered pairs in the permutation. The sequences and scores of the operons, along with information about the order of the tRNAs and the intergenic regions chosen, are presented as a ranked list of entries in an Office Open XML spreadsheet.
aaRS operon assembly
Details for the operon assembly are given in Supplementary Figure 3. All predicted 5’ UTRs with ΔGtot (wt ribo) for the alignments are given in Supplementary Table 3.
DNA constructs
Reporter genes (strepGFPHis6, mCherry and E2Crimson) were cloned by Gibson assembly into a p15A plasmid containing a tetracycline resistance cassette and were expressed from a lac promoter. Optimised 5’ UTRs were inserted between the +1 transcription site and the ORF by quick-change PCR Gibson assembly. Optimised 5’ UTRs and ORFs were inserted between the +1 transcription site and codon 13 by quick change PCR Gibson assembly. O(trans)-strepGFP(40TAG, 136AGGA, 150AGTA)His6 was expressed from a previously described p15A plasmid16. O1-strepGFP(40TAG, 136AGGA, 150AGTA)His6, O1-strepGFP(40TAG, 50CTAG, 136AGGA, 150AGTA)His6 and O1-strepGFP(40CTAG, 50TAGA, 136AGGA, 150AGTA)His6 were synthesized by IDT as gBlock double-stranded DNA fragments and cloned into the standard p15A reporter backbone by Gibson assembly.
Ribosomal RNA for O-ribosomes were encoded on previously described pRSF plasmids containing a kanamycin resistance cassette and were expressed from a trc promoter4, 5.
Synthetase operon RS3 and tRNA operon tRNA3 were encoded on a previously described pMB1 plasmid containing a spectinomycin resistance cassette16. Synthetase operons RS4_1 and RS4_2 were synthesized by IDT as gBlocks and inserted after the +1 transcription site of a glnS’ promoter by Gibson assembly16. RS4_1-2 was assembled by Gibson cloning of fragments from RS4_1 and RS4_2. tRNA operon tRNA4 was synthesized by IDT as a gBlock and assembled into the same pMB1 plasmid as the synthetase operons by Gibson cloning under control of a lpp promoter. tRNA4(quad) was assembled by quick change PCR Gibson assembly from tRNA4.
Measuring the activity and orthogonality of fluorescent reporters
To measure the activity and orthogonality of each fluorescent reporter (expressing strepGFPHis6, mCherry and E2Crimson from genes with a variety of 5’UTRs and coding sequences) we transformed 0.5 μL of p15A plasmids encoding the fluorescent reporter into 8 μL chemically competent E. coli DH10B cells bearing a pRSF plasmid encoding a copy of the O-ribosome rRNA or wt ribosome rRNA. We recovered the transformed cells for 1 h at 37°C and 750 rpm in 180 μL SOC medium in a 96-well microtiter plate format. 30 μL of the rescued cells were used to inoculate 500 μL selective 2xYT-kt (2xYT medium containing 50 μg/mL kanamycin, 12.5 μg/mL tetracycline) medium in a 1.2 mL 96-well plate format and the cultures were grown over night at 37°C and 750 rpm. 30 μL of the overnight cultures were used to inoculate 500 μL 2xYT-kt medium in a 1.2 mL 96-well plate format. Cells were grown for 2 h at 37°C and 750 rpm and production of fluorescent reporter as well as plasmid encoded rRNA was induced by addition of 10 μL 0.1 M IPTG to give a final concentration of 2 mM IPTG. Cells were grown for 18 h at 37°C and 750 rpm. 180 μL of each culture was transferred into 96-well flat bottom Costar plates and fluorescence and optical density were measured using a PHERAstar FS plate reader.
Incorporating three distinct ncAAs with O1-strepGFP(40TAG, 136AGGA, 150AGTA)His6 orO(trans)-strepGFP(40TAG, 136AGGA, 150AGTA)His6
To compare the efficiency of the incorporation of three distinct ncAAs into strepGFP(40TAG, 136AGGA, 150AGTA)His6 from reporters containing a transplanted or optimised orthogonal 5’UTR we transformed 0.4 μL pMB1 plasmid encoding operon RS3/tRNA3 together with 0.4 μL of p15A plasmid encoding O1-strepGFPHis6, O1-strepGFP(40TAG, 136AGGA, 150AGTA)His6 or O(trans)-strepGFP(40TAG, 136AGGA, 150AGTA)His6 into 8 μL chemically competent E. coli DH10B cells bearing a pRSF plasmid encoding a copy of O-riboQ1. We recovered the transformed cells for 1 h at 37°C and 750 rpm in 180 μL SOC medium in a 96-well microtiter plate format. 30 μL of the rescued cells were used to inoculate 500 μL 2xYT-kts medium (2xYT containing 25 μg/mL kanamycin, 12.5 μg/mL tetracyclin and 37.5 μg/mL spectinomycin) in a 1.2 mL 96-well plate format and the cultures were grown over night at 37°C and 750 rpm. 100 μL of the overnight cultures were used to inoculate 4 mL 2xYT-kts medium containing either 4 mM BocK 1, 4 mM NmH 2 and 2 mM CbzK 3 or no ncAA in a 10 mL 24-well plate format. Cells were grown for 2 h at 37°C and 220 rpm and production of strepGFPHis6 as well as O-riboQ1 was induced by addition of 8 μL 1 M IPTG to give a final concentration of 2 mM IPTG. Cells were grown for 18 h at 37°C and 750 rpm. 180 μL of each culture was transferred into 96-well flat bottom Costar plates and fluorescence and optical density were measured using PHERAstar FS. The rest of the cultures were centrifuged for 10 min at 3200 rcf and taken up in OD600 adjusted amounts of BugBuster containing Roche cOmplete proteinase inhibitor. Cells were lysed for 1 h under head-over-tail rotation at room temperature. The lysate was transferred into 1.5 mL Eppendorf tubes and spun down at 15000 rcf for 20 min. 180 μL of clarified cell lysate was transferred into 96-well flat bottom Costar plates and fluorescence and was measured using PHERAstar FS.
Activity and orthogonality assessment of aaRS/tRNA operons
To assess the activity and orthogonality of each aaRS/tRNA pair in our operons we transformed 0.4 μL pMB1 plasmids encoding operons (aaRS4_1/tRNA4, aaRS4_2/tRNA4, aaRS4_1-2/tRNA4 or aaRS4_1-2/tRNA(quad)) into 8 μL chemically competent E. coli DH10B cells harbouring a pRSF plasmid encoding a copy of O-riboQ1 as well as a p15A plasmid encoding O1-strepGFP(40XXXX)His6, where XXXX stands for either TAG (with all operons but aaRS4_1-2/tRNA4(quad)), TAGA (only with aaRS4_1-2/tRNA4(quad)), AGGA, AGTA or CTAG. We recovered the transformed cells for 1 h at 37°C and 750 rpm in 180 μL SOC medium in a 96-well microtiter plate format. 30 μL of the rescued cells were used to inoculate 500 μL of 2xYT-kts medium in a 1.2 mL 96-well plate format and the cultures were grown over night at 37°C and 750 rpm. 30 μL of the overnight cultures were used to inoculate 500 μL selective 2xYT-kts medium containing either 4 mM BocK 1, 4 mM NmH 2, 2 mM CbzK 3, 4 mM AllocK 4, 2 mM PheI 5 or no ncAA in a 1.2 mL 96-well plate format. Cells were grown for 2 h at 37°C and 750 rpm and expression of strepGFP(40XXXX)His6 as well as O-riboQ1 was induced by addition of 10 μL 0.1 M IPTG to give a final concentration of 2 mM IPTG. Cells were grown for 18 h at 37°C and 750 rpm. 180 μL of each culture was transferred into 96-well flat bottom Costar plates and fluorescence and optical density were measured using PHERAstar FS.
Production of strepGFP(40X)His6 for MS analysis
To isolate proteins for MS analysis to assess the orthogonality of the aaRS/tRNA operons, 0.4 μL pMB1 plasmid encoding operon RS4_1-2/tRNA4 or RS4_1-2/tRNA4(quad) together with 0.4 μL p15A plasmid encoding either O1-strepGFP(40XXXX)His6, where XXXX stands for TAG (only with RS4_1-2/tRNA4), TAGA (only with RS4_1-2/tRNA4(quad)), AGGA, AGTA and CTAG respectively were transformed into 50 μL chemically competent E. coli DH10B cells harbouring a pRSF plasmid encoding a copy of O-riboQ1. We recovered the transformed cells for 1 h at 37°C and 750 rpm in 400 μL SOC medium in a 1.5 mL Eppendorf tube. 100 μL of the rescued cells were used to inoculate 50 mL of 2xYT-kts medium in a 250 mL Erlenmeyer flask and the cultures were grown over night at 37°C, 220 rpm. 5 mL of the overnight cultures were used to inoculate 100 mL of 2xYT-kts medium containing a combination of ncAAs BocK 1, NmH 2, CbzK 3, AllocK 4 and PheI 5 according to the constructs used (RS4_1-2/tRNA4 with 1, 2, 3, 5 - RS4_1-2/tRNA4(quad) 2, 3, 4, 5). Cultures were grown for 2-3 h at 37°C and 220 rpm until an OD600 of 0.5, and then induced with 200 μL 1 M IPTG to a final concentration of 2 mM IPTG. Cells were grown at 37°C and 220 rpm for 18 h. Cells were centrifuged at 3200 rcf for 12 min, resuspended in 10 mL BugBuster containing Roche cOmplete proteinase inhibitor, sonicated for 1.5 min (2s on 2s off at 40% amplitude) and the lysate was centrifuged for 20 min at 15000 rcf at 4°C. The lysate was bound to 40 μL nickel NTA beads overnight. Beads were washed six times with 240 μL 20 mM imidazole in PBS. Proteins were eluted 9 times in 20 μL 250 mM imidazole. The buffer was exchanged for water using a 3 kDa Amicon ultra column for MS and MS/MS analysis.
Orthogonality and efficiency assessment of the incorporation of four distinct ncAAs in response to four distinct quadruplet codons from O1-strepGFP(40CTAG, 50TAGA, 136AGGA, 150AGTA)His6
To assess the efficiency and orthogonality of the incorporation of four distinct ncAAs into four distinct quadruplet codons we transformed 0.4 μL pMB1 plasmid encoding operon RS4_1-2/tRNA4(quad) together with 0.4 μL p15A plasmid encoding either O1-strepGFPHis6 or O1-strepGFP(40CTAG, 50TAGA, 136AGGA, 150AGTA)His6 into 8 μL chemically competent E. coli DH10B cells bearing a pRSF plasmid encoding a copy of O-riboQ1. We recovered the transformed cells for 1 h at 37 °C and 750 rpm in 180 μL SOC medium in a 96-well microtiter plate format. 30 μL of the rescued cells were used to inoculate 500 μL of 2xYT-kts medium in a 1.2 mL 96-well plate format and the cultures were grown over night at 37 °C and 750 rpm. 100 μL of the overnight cultures were used to inoculate 4 mL selective 2xYT-kts medium containing either each combination of three out of the four ncAAs: 4 mM NmH 2, 2 mM CbzK 3, 4 mM AllocK 4 and 2 mM PheI 5, all ncAAs or none (O1-strepGFPHis6 was only grown in presence of all ncAAs) in a 24-well plate format. Cells were grown for 2 h at 37°C and 220 rpm and production of strepGFPHis6 as well as O-riboQ1 was induced by addition of 8 μL 1 M IPTG to give a final concentration of 2 mM IPTG. Cells were grown for 18 h at 37°C and 750 rpm. 180 μL of each culture was transferred into 96-well flat bottom costar plates and fluorescence and optical density were measured using PHERAstar FS.
The same procedure was used for the orthogonality and efficiency assessment of the incorporation of four distinct ncAAs in response to one amber codon and three distinct quadruplet codons into O1-strepGFP(40TAG, 50CTAG, 136AGGA, 150AGTA)His6 However, RS4_1-2/tRNA4 was used to express the aaRS/tRNA pairs and O1-strepGFP(40TAG, 50CTAG, 136AGGA, 150AGTA)His6 as reporter for quadruplet incorporation. 4 mM BocK 1 was used instead of 4 mM AllocK 4.
Production of strepGFP(XXXX)His6 for MS analysis and determination of isolated yield of proteins containing three and four distinct ncAAs
An analogous procedure to that used for the production of proteins from O1-strepGFP(40TAG, 50CTAG, 136AGGA, 150AGTA)His6, O1-strepGFP(40CTAG, 50TAGA, 136AGGA, 150AGTA)His6 and O1-strepGFP(40TAG, 136AGGA, 150AGTA)His6 for mass spectrometry was used to produce and purify proteins containing three or four distinct ncAAs. The following combinations of reporters, operons and ncAAs were used to obtain proteins containing three or four distinct ncAAs. : O1-strepGFP(40TAG, 136AGGA, 150AGTA)His6 with RS3/tRNA3 and 4 mM BocK 1, 4 mM NmH 2, 2 mM CbzK 3 or O1-strepGFP(40TAG, 50CTAG, 136AGGA, 150AGTA)His6 with RS4_1-2/tRNA4 and 4 mM BocK 1, 4 mM NmH 2, 2 mM CbzK 3, 2 mM PheI 5 or O1-strepGFP(40CTAG, 50TAGA, 136AGGA, 150AGTA)His6 with RS4_1-2/tRNA4(quad) and 4 mM NmH 2, 2 mM CbzK 3, 4 mM AllocK 4, 2 mM PheI 5. To determine the isolated protein yield, the fluorescence of 180 μL isolated protein was measured using PHERAstar FS and the protein concentration was calculated based on a standard curve generated with a strepGFPHis6 standard. The buffer was exchanged for water using a 3 kDa Amicon ultra column for MS and MS/MS analysis.
Electrospray ionization mass spectrometry
Denatured protein samples (~10μM) were subjected to LC-MS analysis. Briefly, proteins were separated on a C4 BEH 1.7μm, 1.0 x 100mm UPLC column (Waters, UK) using a modified nanoAcquity (Waters, UK) to deliver a flow of approximately 50 μl/min. The column was developed over 20 minutes with a gradient of acetonitrile (2% v/v to 80% v/v) in 0.1% v/v formic acid. The analytical column outlet was directly interfaced via an electrospray ionisation source, with a hybrid quadrupole time-of-flight mass spectrometer (Xevo G2, Waters, UK). Data was acquired over a m/z range of 300–2000, in positive ion mode with a cone voltage of 30V. Scans were summed together manually and deconvoluted using MaxEnt1 (Masslynx, Waters, UK). The theoretical molecular weights of proteins with ncAAs was calculated by first computing the theoretical molecular weight of wild-type protein using an online tool (http://web.expasy.org/protparam/) and then manually correcting for the theoretical molecular weight of ncAAs.
Tandem MS/MS analysis
Proteins were run on 4-12% NuPAGE Bis-Tris gel (Invitrogen) with MES buffer and briefly stained using InstantBlue (Expedeon). The bands were excised and stored in water. Tryptic digestion and tandem MS/MS analyses were done by Mark Skehel (Biological Mass Spectrometry and Proteomics Laboratory, MRC Laboratory of Molecular Biology).
Supplementary Material
Acknowledgements
This work was supported by the UK Medical Research Council (MRC; MC_U105181009 and MC_UP_A024_1008) and an ERC Advanced Grant SGCR (all to J.W.C.). D.L.D. and S.B.O. were supported by the Boehringer Ingelheim Fonds. We thank M. Skehel at the MRC-LMB mass spectrometry facility for performing mass spectrometry.
Footnotes
Author Contributions Statement
D.L.D., S.B.O. and J.W.C. conceived the study. D.L.D. performed all wet-lab experiments and managed data. S.B.O. developed the automated orthogonal mRNA design, with input from D.L.D.. D.L.D. developed aaRS operons. A.T.B. developed the tRNA operon generator and analysed the MS/MS data. D.L.D., S.B.O. and J.W.C. wrote the paper with input from A.T.B..
Competing interests
The authors declare no competing interests.
Data availability
Source data for main figures is provided in Source Data Files 2 to 5. All other relevant data are included in the article and its Supplementary Information. Materials generated or analysed in this study are available from the corresponding author upon reasonable request.
References
- 1.Chin JW. Expanding and reprogramming the genetic code. Nature. 2017;550:53–60. doi: 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- 2.de la Torre D, Chin JW. Reprogramming the genetic code. Nat Rev Genet. 2021;22:169–184. doi: 10.1038/s41576-020-00307-7. [DOI] [PubMed] [Google Scholar]
- 3.Robertson WE, et al. Sense codon reassignment enables viral resistance and encoded polymer synthesis. Science. 2021;372:1057–1062. doi: 10.1126/science.abg3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 5.Wang K, et al. Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nat Chem. 2014;6:393–403. doi: 10.1038/nchem.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson JC, et al. An expanded genetic code with a functional quadruplet codon. Proc Natl Acad Sci U S A. 2004;101:7566–7571. doi: 10.1073/pnas.0401517101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fredens J, et al. Total synthesis of Escherichia coli with a recoded genome. Nature. 2019;569:514–518. doi: 10.1038/s41586-019-1192-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang K, et al. Defining synonymous codon compression schemes by genome recoding. Nature. 2016;539:59–64. doi: 10.1038/nature20124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malyshev DA, et al. A semi-synthetic organism with an expanded genetic alphabet. Nature. 2014;509:385–388. doi: 10.1038/nature13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, et al. A semi-synthetic organism that stores and retrieves increased genetic information. Nature. 2017;551:644–647. doi: 10.1038/nature24659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. A semisynthetic organism engineered for the stable expansion of the genetic alphabet. Proc Natl Acad Sci U S A. 2017;114:1317–1322. doi: 10.1073/pnas.1616443114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer EC, et al. New codons for efficient production of unnatural proteins in a semisynthetic organism. Nat Chem Biol. 2020;16:570–576. doi: 10.1038/s41589-020-0507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neumann H, Slusarczyk AL, Chin JW. De Novo Generation of Mutually Orthogonal Aminoacyl-tRNA Synthetase/tRNA Pairs. J Am Chem Soc. 2010;132:2142–2144. doi: 10.1021/ja9068722. [DOI] [PubMed] [Google Scholar]
- 14.Chatterjee A, Sun SB, Furman JL, Xiao H, Schultz PG. A Versatile Platform for Single- and Multiple-Unnatural Amino Acid Mutagenesis in Escherichia coli. Biochemistry. 2013;52:1828–1837. doi: 10.1021/bi4000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willis JCW, Chin JW. Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs. Nat Chem. 2018;10:831–837. doi: 10.1038/s41557-018-0052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunkelmann DL, Willis JCW, Beattie AT, Chin JW. Engineered triply orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs enable the genetic encoding of three distinct non-canonical amino acids. Nat Chem. 2020;12:535–544. doi: 10.1038/s41557-020-0472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cervettini D, et al. Rapid discovery and evolution of orthogonal aminoacyl-tRNA synthetase-tRNA pairs. Nat Biotechnol. 2020;38:989–999. doi: 10.1038/s41587-020-0479-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang MS, et al. Biosynthesis and genetic encoding of phosphothreonine through parallel selection and deep sequencing. Nat Methods. 2017;14:729–736. doi: 10.1038/nmeth.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Italia J, et al. Mutually Orthogonal Nonsense-Suppression Systems and Conjugation Chemistries for Precise Protein Labeling at up to Three Distinct Sites. J Am Chem Soc. 2019;141:6204–6212. doi: 10.1021/jacs.8b12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rackham O, Chin JW. A network of orthogonal ribosome·mRNA pairs. Nat Chem Biol. 2005;1:159–166. doi: 10.1038/nchembio719. [DOI] [PubMed] [Google Scholar]
- 21.Wang K, Neumann H, Peak-Chew SY, Chin JW. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat Biotechnol. 2007;25:770–777. doi: 10.1038/nbt1314. [DOI] [PubMed] [Google Scholar]
- 22.Schmied WH, et al. Controlling orthogonal ribosome subunit interactions enables evolution of new function. Nature. 2018;564:444–448. doi: 10.1038/s41586-018-0773-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venkat S, et al. Genetically Incorporating Two Distinct Post-translational Modifications into One Protein Simultaneously. ACS Synth Biol. 2018;7:689–695. doi: 10.1021/acssynbio.7b00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chin JW. Expanding and Reprogramming the Genetic Code of Cells and Animals. Annu Rev Biochem. 2014;83:379–408. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 25.Cambray G, Guimaraes JC, Arkin AP. Evaluation of 244,000 synthetic sequences reveals design principles to optimize translation in Escherichia coli. Nat Biotechnol. 2018;36:1005–1015. doi: 10.1038/nbt.4238. [DOI] [PubMed] [Google Scholar]
- 26.Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuller T, Zur H. Multiple roles of the coding sequence 5' end in gene expression regulation. Nucleic Acids Res. 2015;43:13–28. doi: 10.1093/nar/gku1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Na D, Lee S, Lee D. Mathematical modeling of translation initiation for the estimation of its efficiency to computationally design mRNA sequences with desired expression levels in prokaryotes. BMC Syst Biol. 2010;4:1–16. doi: 10.1186/1752-0509-4-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seo SW, et al. Predictive design of mRNA translation initiation region to control prokaryotic translation efficiency. Metab Eng. 2013;15:67–74. doi: 10.1016/j.ymben.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 31.Salis HM. Methods in Enzymology. Vol. 498. Academic Press; Cambridge, MA USA: 2011. pp. 19–42. [DOI] [PubMed] [Google Scholar]
- 32.Espah Borujeni A, Channarasappa AS, Salis HM. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 2014;42:2646–2659. doi: 10.1093/nar/gkt1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Espah Borujeni A, Salis HM. Translation Initiation is Controlled by RNA Folding Kinetics via a Ribosome Drafting Mechanism. J Am Chem Soc. 2016;138:7016–7023. doi: 10.1021/jacs.6b01453. [DOI] [PubMed] [Google Scholar]
- 34.Espah Borujeni A, et al. Precise quantification of translation inhibition by mRNA structures that overlap with the ribosomal footprint in N-terminal coding sequences. Nucleic Acids Res. 2017;45:5437–5448. doi: 10.1093/nar/gkx061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-Sequence Determinants of Gene Expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allert M, Cox JC, Hellinga HW. Multifactorial Determinants of Protein Expression in Prokaryotic Open Reading Frames. J Mol Biol. 2010;402:905–918. doi: 10.1016/j.jmb.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodman DB, Church GM, Kosuri S. Causes and Effects of N-Terminal Codon Bias in Bacterial Genes. Science. 2013;342:475–479. doi: 10.1126/science.1241934. [DOI] [PubMed] [Google Scholar]
- 38.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El Yacoubi B, Bailly M, de Crécy-Lagard V. Biosynthesis and Function of Posttranscriptional Modifications of Transfer RNAs. Annu Rev Genet. 2012;46:69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- 40.An W, Chin JW. Synthesis of orthogonal transcription-translation networks. Proc Natl Acad Sci U S A. 2009;106:8477–8482. doi: 10.1073/pnas.0900267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darlington APS, Kim J, Jiménez JI, Bates DG. Dynamic allocation of orthogonal ribosomes facilitates uncoupling of co-expressed genes. Nat Commun. 2018;9:1–12. doi: 10.1038/s41467-018-02898-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for main figures is provided in Source Data Files 2 to 5. All other relevant data are included in the article and its Supplementary Information. Materials generated or analysed in this study are available from the corresponding author upon reasonable request.