Abstract
Host-associated microbiomes harbor hundreds of bacterial species that co-occur creating the opportunity for manifold bacteria-bacteria interactions, which in turn contribute to the overall community structure. The mechanisms that underlie this self-organization among bacteria remain largely elusive. Here, we studied bacterial interactions in the phyllosphere microbiota. We screened for microbial interactions in planta by adding 200 endogenous strains individually to a 15-member synthetic community and tracking changes in community composition upon colonization of the model plant Arabidopsis. Ninety percent of the identified interactions in planta were negative, and phylogenetically closely related strains elicited consistent effects on the synthetic community, providing support for trait conservation. Community changes could be largely explained by binary interactions; however, we also identified a higher order interaction of more than two interacting strains. We further focused on a prominent interaction between two members of the Actinobacteria. In presence of Aeromicrobium Leaf245, the population of Nocardioides Leaf374 was reduced by almost two orders of magnitude. We identified a potent antimicrobial peptidase in Aeromicrobium Leaf245, which resulted in Nocardioides Leaf374 lysis. A respective Leaf245 mutant strain was necessary and sufficient to restore Nocardioides colonization in planta, demonstrating that direct bacteria-bacteria interactions were responsible for the population shift originally observed. Our study highlights the power of synthetic community screening and uncovers a strong microbial interaction that occurs despite a spatially heterogeneous environment.
Introduction
In the environment, bacteria often occur in multi-species communities. These may consist of few species or comprise hundreds of species as is often found in host-associated microbiomes1–3. Due to the complexity of these systems, it is particularly challenging to identify microbes that interact and thereby shape the community. Interactions among members of the microbiota are often studied in vitro4–6, but the observations are difficult to translate to the host-associated context due to differences in spatial and nutritional environments7.
The plant microbiome has been described for many species and the different plant compartments2,8–10. Members of the microbiota can provide beneficial functions to the plant host, such as growth promotion or increased resistance to biotic and abiotic stresses10–14. While there is variation in the composition of colonizing bacteria at the species level, the community is largely conserved at higher phylogenetic level over time and different locations10,15.
The mechanisms that govern microbial community assembly in the plant host only have started to be unraveled. Apart from host factors, microbe-microbe interactions also contribute to community assembly9,10,16–21, but little is known about the overall impact and specific mechanisms that prevail in planta. In general, interactions can be categorized based on the outcome for the interaction partners22,23. Positive interactions include commensalism and mutualism, in which one or both microbes benefit. Negative interactions include competition (negative for both microbes), amensalism (negative for one, neutral for the other microbe) and parasitism (negative for one and positive for the other microbe)24,25.
The genomes of bacterial phyllosphere and rhizosphere colonizers harbor a plethora of biosynthetic gene clusters. Therefore, the production of antimicrobial compounds to inhibit competitors is proposed to be one mechanism that shapes microbial communities6,20,26. Previous studies have also shown that the presence of fungi or oomycetes can alter bacterial community composition27–29. To date, most interactions have been described in the context of pathogen infection in diseased plants. Interactions among bacteria in healthy plants and hence the fundamental mechanisms driving community assembly are less studied.
Investigating bacteria-bacteria interactions directly in the host is challenging and usually relies on co-occurrence networks based on natural variation over time or between individuals29–31. The development of computational approaches applicable to microbiomes is a rapidly growing field and current limitations of the methods in interpreting species interactions in microbiome studies have been highlighted32. An inherent drawback of all co-occurrence network approaches is that they infer ecological associations based on abundance correlations and thus fall short in demonstrating direct interactions. Synthetic community experiments are attractive in this regard because: 1) the complexity of the system under study can be reduced and 2) individual bacteria can be removed or replaced to examine the community response to these controlled perturbations25,33. Over the last years, extensive culture collections have been established for several plant species, including Arabidopsis34, allowing synthetic communities to be assembled not only in vitro but also in planta33.
Here we describe a systematic in planta screen to identify positive and negative interactions of 200 Arabidopsis thaliana leaf bacteria with a synthetic 15-member focal community in a gnotobiotic model system. Mapping interactions of a broad range of bacteria in the phyllosphere allowed the identification of emerging patterns for groups of microbes as well as the identification of specific interactions. We then decipher an exemplary specific microbial interaction in planta to expand our mechanistic understanding of how these interactions work at the molecular level.
Results
Selection and characterization of the synthetic community
We aimed to dissect interactions among leaf strains during phyllosphere colonization of the model plant Arabidopsis thaliana. To this end, we selected 15 strains from the At-LSPHERE culture collection34 to assemble a focal community against which we probed 200 additional strains of the At-LSPHERE collection and a few selected model strains13,35,36 individually in drop-in synthetic communities (Fig. 1a, Supplementary Fig. 1). The use of a focal strain assembly, as opposed to testing binary interactions, provides a community context that potentially prevents evasion due to metabolic plasticity or spatial segregation23 and shifts the experimental design to a large, but still feasible set-up for testing bacterial interactions in planta. It also allows testing whether phylogenetically related strains exhibit similar interactions in the focal community and the emergence of higher order interactions.
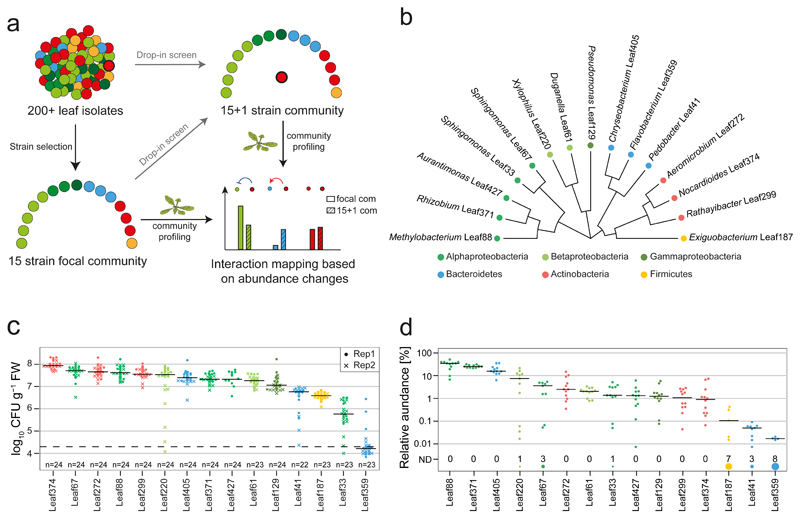
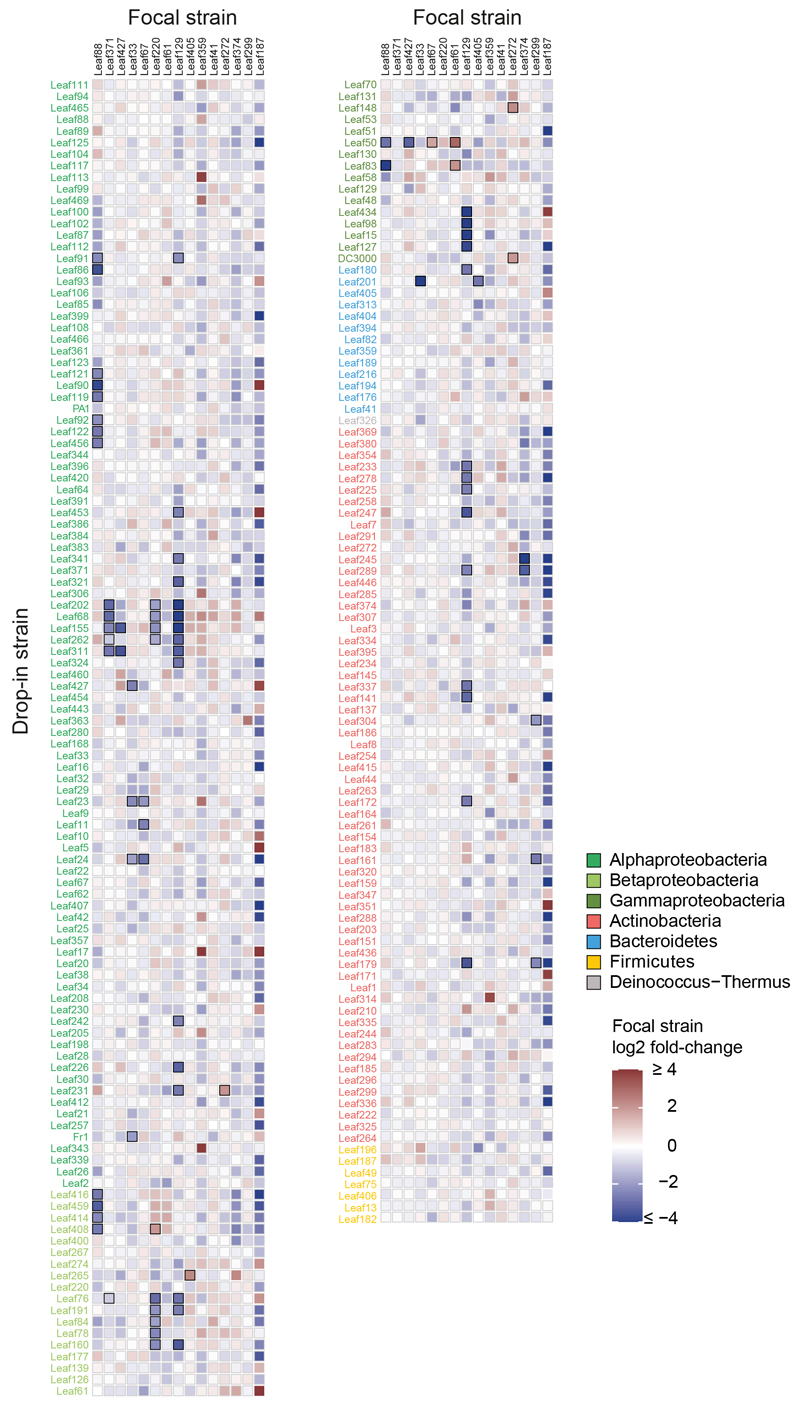
Fig. 1. A 15-strain focal SynCom to map bacteria-bacteria interactions.
a) Graphical representation of experimental design for interaction screening. b) Phylogenetic tree representing the selected strains based on full-length 16S rRNA gene sequences. c) Phyllosphere colonization by each focal strain in mono-association with A. thaliana Col-0 plants. Seedlings were inoculated with bacterial suspensions 7 days after planting and bacteria were enumerated 21 days later by CFU counting on R-2A+M agar. The limit of detection is indicated by a dashed line. The data across two independent experiments are shown. d) Relative focal community composition in the phyllosphere after 21 days based on 16S rRNA gene amplicon sequencing (n= 12). The number of replicates in which a particular strain was not detected (ND) are shown by the size of the circle and the count above the strain name. Colors throughout the figure correspond to the phylum, and in case of Proteobacteria, the respective class.
The focal community strains were selected to span the phylogenetic diversity of the At-LSPHERE culture collection34 and to include genera that are abundant in the Arabidopsis phyllosphere and part of the phyla Proteobacteria, Actinobacteria, Bacteroidetes and Firmicutes15,34 (Fig. 1b). Upon mono-association, most isolates colonized the phyllosphere in the range of 107 - 108 CFU g-1 plant fresh weight (FW) in a calcine clay based gnotobiotic model system (Fig. 1c). We also characterized the composition of the combined focal community in the phyllosphere by 16S rRNA gene sequencing by combining the strains in approximately equal ratio in the inoculum (Fig. 1d and Supplementary Fig. 2). The phyllosphere community was dominated by three strains that each contributed more than 10% relative abundance (Methylobacterium Leaf88, Rhizobium Leaf371 and Chryseobacterium Leaf405). Nine strains had relative abundances between 1-10% and three strains less than 0.1%.
Single strain drop-out and replacement experiments showed that the selected focal community is rather stable against perturbations within the community (see Supplementary Note), thus providing an ideal starting point to identify interactions directly induced by additional strains while still capturing potentially emerging higher order interactions.
Systematic mapping of interactions among At-LSPHERE isolates
Next, we mapped interactions of the focal community using a drop-in screen of 200 At-LSPHERE isolates in planta. We added the strains individually to the focal community, resulting in N (15 strains) +1 scenarios and used these for inoculation (see methods, Supplementary Fig. 3a and Supplementary Table 1). After community establishment for approximately three weeks, we harvested the plants and recorded relative abundance changes within the focal community compared to the unperturbed community control to map interactions (Fig. 2a, Extended Data Fig. 1, Supplementary Fig. 3, 4 and Supplementary Table 2). We also included conditions in which the focal strains themselves were added as a drop-in strain in the inoculum. These control conditions had no significant impact on the composition of the focal community except for the Leaf427 drop-in that affected Leaf 33 (log2FC: -2.52, padj: 0.0064). Thus, overall this result validated the robustness of the assembled focal community.
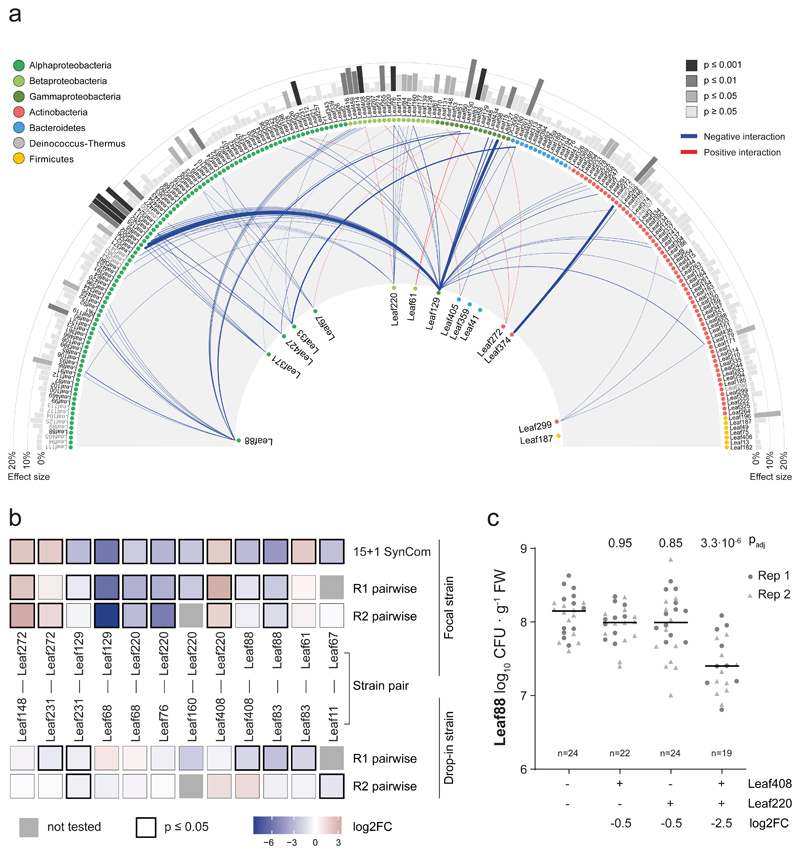
Fig. 2. Interactions of the At-LSPHERE collection with the focal SynCom in planta.
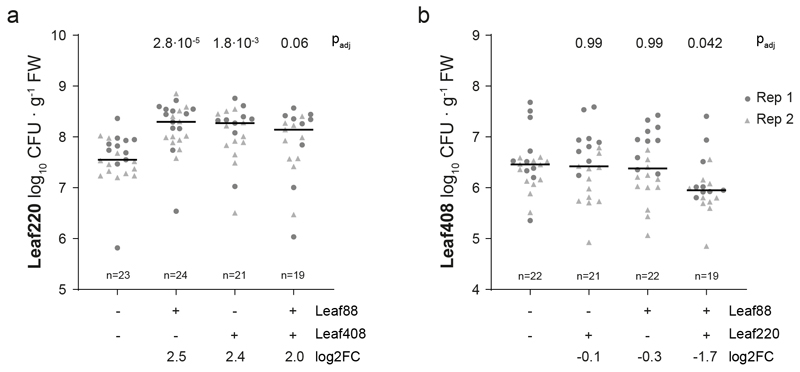
a) Interaction map showing all interactions identified between At-LSPHERE strains (top) and the focal community (bottom) based on log2-transformed abundance changes (DESeq2-normalized counts, Wald test, Benjamini-Hochberg corrected, padj ≤ 0.01, nfocal community = 16, ndrop-in = 3). Positive (red) and negative (blue) interactions are shown as connecting lines between drop-in strain (outside) and focal strain (center). Line thickness corresponds to fold-change. Bars in the outermost ring correspond to the overall effect size of the perturbation on the focal community (principle component analysis, PERMANOVA). Strains are ordered by phylogeny based on full-length 16S rRNA gene sequences and dots are colored by bacterial phyla or Proteobacteria class. Drop-in strains that belong to the same ASV as a focal strain are labelled in grey as for these no interactions with the focal strain of the same ASV can be determined. b) Pairwise strain inoculations of selected interacting strains identified in a). On top, the focal strain log2 fold-change observed in a) is shown as a reference for each strain pair. Below, the log2 fold-changes (pairwise inoculation vs mono-association) for the focal strain and drop-in strain are shown based on absolute abundances obtained by CFU enumeration for two biological replicates. The color of the boxes reflects the observed log2 fold-change and the black frames around the boxes indicate a significant difference compared to the mono-association condition (two-sided Wilcoxon rank sum test, p ≤ 0.05). Combinations that were not tested in a given experiment are marked with a grey box. Seedlings were inoculated at day 14 (replicate 1, R1) or day seven (replicate 2, R2) after planting. Exact p-values and number of replicates are provided in the source data. c) Phyllosphere colonization by Methylobacterium Leaf88 in mono-association or in combination with Xylophilus Leaf220, Methylophilus Leaf408 or both. Shown are the median and individual data points of log10-transformed CFU per g plant fresh weight across two independent experiments. Exact p-values (Kruskal-Wallis and post-hoc Dunn test, Bonferroni adjusted p) are indicated above and log2 fold changes below the graph. For the corresponding colonization levels of Leaf220 and Leaf408, see Extended Data Fig. 3.
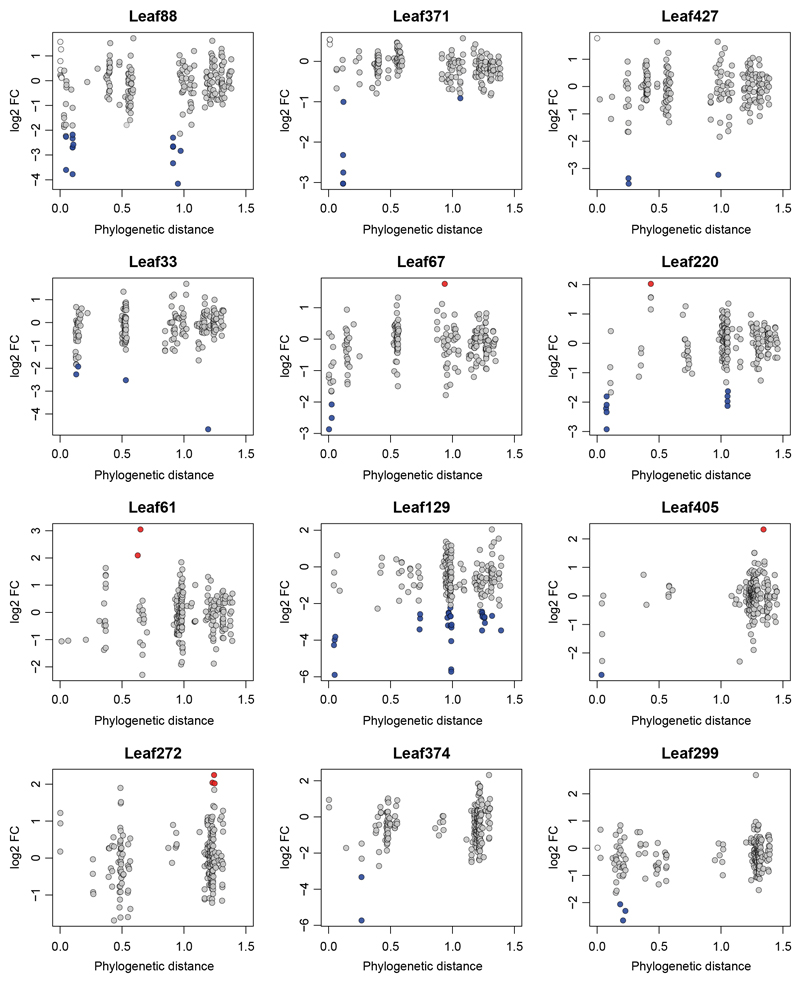
In total, we identified 84 interactions between strains (Wald test, Benjamini-Hochberg adjusted p ≤ 0.01) upon addition of 57 of the 200 strains tested. For the selected focal community, 90% of identified interactions were negative, indicating competitive interactions. We also observed that phylogenetically closely related isolates showed similar effects on the focal community, indicating trait conservation. The majority of negative interactions were observed with phylogenetically closely related strains, except for Pseudomonas Leaf 129, which also frequently interacted with distantly related strains (Extended Data Fig. 2). For focal strains that showed at least three negative interactions within the genus, we tested whether within genus interactions were more frequent than expected by chance (Fisher's exact test, Bonferroni adjusted p ≤ 0.05). This was the case for Rhizobium Leaf371, Sphingomonas Leaf67 and Methylobacterium Leaf88, suggesting competition among closely related isolates (Supplementary Table 3). Intriguingly, among the interactions found were some that have previously been identified in a larger 62 strain SynCom37 (e.g. Leaf88-Leaf416, Leaf129-Leaf233 and Leaf220-Leaf262), demonstrating that these interactions are also observed in even more complex communities.
We also estimated the impact of each added strain on the focal community by calculating the observed effect size based on PCA analysis (Fig. 2a, Supplementary Table 4). Except for Aeromicrobium Leaf245, all strains with an effect size larger than 20% interacted with two or more focal strains. Serratia Leaf50 interacted with four focal community strains - the maximum observed - and had an effect size of 27%, suggesting strong remodeling of the community. In fact, this strain and the plant pathogen Pseudomonas syringae pv. tomato DC3000 were the only strains that induced disease symptoms in some plants when present in the community (Supplementary Fig. 5). This is in line with the plant pathogenic behavior of Serratia Leaf5038 and may explain the strong impact on the focal community composition.
To test whether the identified interactions were binary or higher order interactions, and to independently validate the screen above using a complementary read-out allowing absolute quantification, we performed pairwise co-inoculations of Arabidopsis and compared colonization relative to mono-colonization using CFU counts. Notably, 7 out of 12 interactions tested could be recapitulated upon co-inoculation, indicating that the observed interactions do not require a community context (Fig. 2b). This complementary approach also allowed us to study the effect of the interaction on the drop-in strain during co-inoculation. For Rhizobium Leaf68 and Acidovorax Leaf76 and Leaf160, which all had negative effects on the interacting focal strains, no significant abundance changes were observed upon co-colonization compared to mono-association, indicating amensalism. For the interaction between Xanthomonas Leaf148 and the focal strain Aeromicrobium Leaf272, Leaf272 abundance increased when co-inoculated while Leaf148 abundance was not affected, suggesting commensalism.
Because higher order interactions would not manifest in pairwise inoculations, we also tested a trio in planta. We speculated that the addition of Xylophilus Leaf220 might be necessary to recapitulate the interaction of Methylobacterium Leaf88 and Methylophilus Leaf408, since Leaf408 also interacted with Xylophilus Leaf220 (Fig. 2a). Indeed, we were able to reproducibly measure a decrease of Methylobacterium Leaf88 abundance in the tripartite bacterial interaction (Fig. 2c) and the magnitude was comparable to the initial screen (log2FC: -2.7 and -2.5 in the screen and tripartite interaction, respectively). Interestingly, Xylophilus Leaf220 abundance was increased in binary combination with Methylophilus Leaf408 or Methylobacterium Leaf88 (Extended Data Fig. 3), suggesting a similar mechanism to support the growth of Leaf220, possibly linked to the methylotrophic lifestyle of both strains.
In summary, our screening approach allowed us to map bacterial interactions emerging due to targeted perturbation of a community by strain addition. These interactions are readily reproducible with a complementary read-out in minimal synthetic communities of two or three strains.
Nocardioides Leaf374 - Aeromicrobium Leaf245 interaction
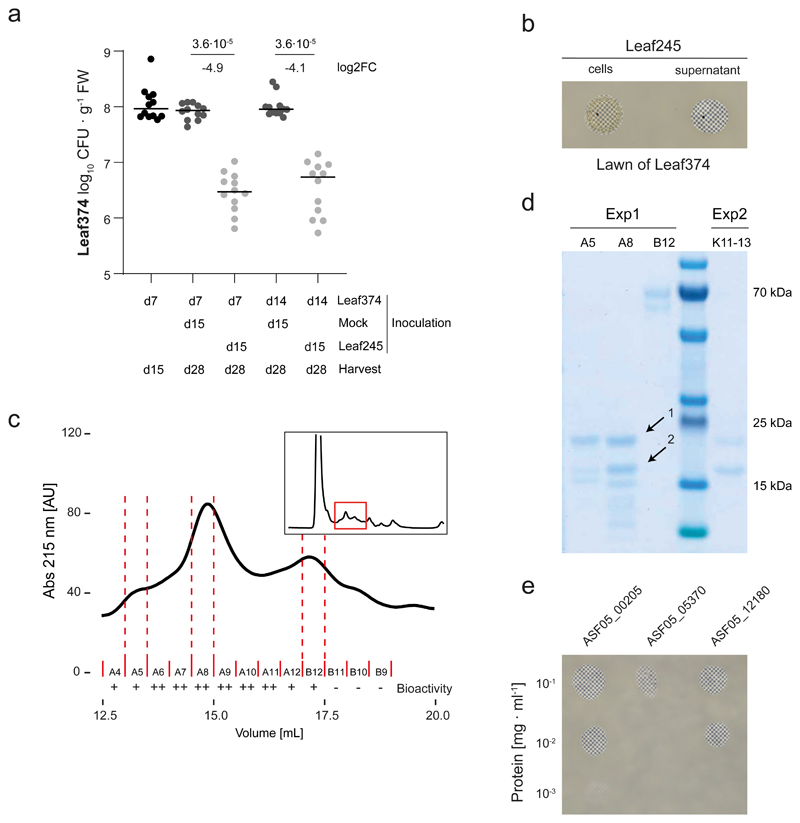
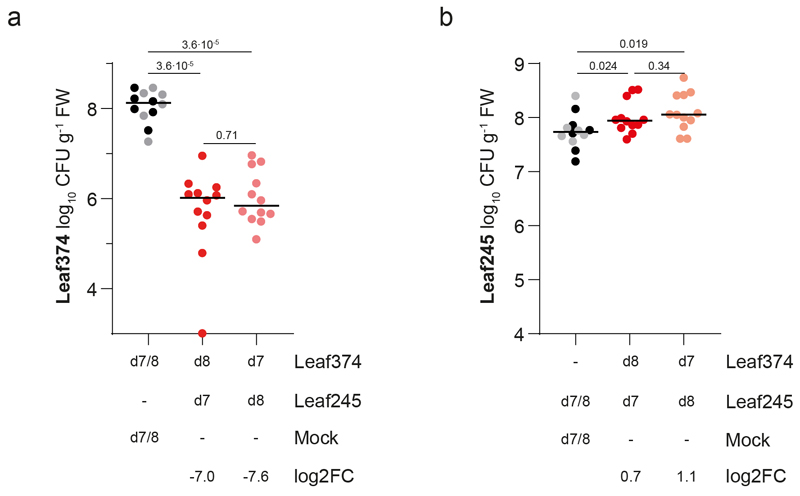
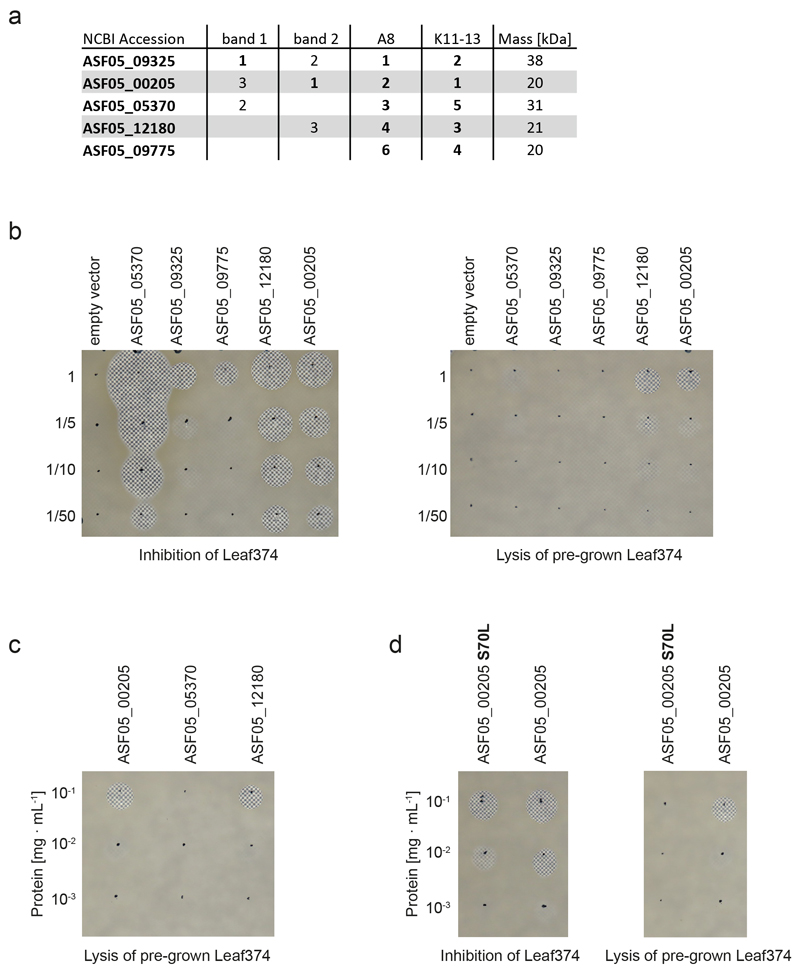
We had previously shown the potential of the leaf isolates to produce novel antimicrobial compounds6. Out of 18 inhibitory interactions observed in vitro between the tested strains two were also identified in planta (Supplementary Table 5), one of which was between Nocardioides Leaf374 (Leaf374) and Aeromicrobium Leaf245 (Leaf245). This interaction was not only conspicuous because of the overlap with previous in vitro data but, with a 50-fold reduction of relative abundance of Leaf374, was also particularly prominent in planta (Fig. 2a). We then further validated this strain pair by sequential phyllosphere inoculation. The order of inoculation did not matter (Extended Data Fig. 4) and Leaf245 also reduced an already established population of Leaf374 (Fig. 3a, Supplementary Fig. 6). The extent of Leaf374 reduction in the binary validation was two orders of magnitude (log2FC: -7.6 to -4.1), which is in a similar range to that in the in planta screen (log2FC: -5.7). To further characterize this conspicuous interaction, we reproduced it in vitro and found that also cell-free supernatant prepared from suspended Leaf245 cells was sufficient for inhibition of Leaf374 (see methods, Fig. 3b). Further, the supernatant not only inhibited growth of Leaf374, but also efficiently lysed the bacterial cells (Supplementary Video). Since the activity was lost upon boiling or addition of organic solvents (Supplementary Fig. 7), we speculated that the active molecule was likely a protein. Next, we fractionated the culture supernatant of Leaf245 and identified several protein candidates that could cause the observed inhibition (Fig. 3c,d, Extended Data Fig. 5a) and heterologously expressed these in E. coli (for details see Supplementary Note). Among these was a putative M23 family endopeptidase (ASF05_00205) which resulted in inhibition and lysis of the target cells (Fig. 3e, Extended Data Fig. 5b,c).
Fig. 3. Biochemical characterization of the interaction between Nocardioides Leaf374 and Aeromicrobium Leaf245.
a) Nocardioides Leaf374 colonization in mono-association or in combination with Leaf245. Strains were sequentially inoculated and harvested as indicated below. Leaf374 CFU were enumerated on MM maltose agar plates. Shown are the median and individual data points of log10-transformed CFU per gram fresh weight (FW) (n= 12). P-values (two-sided Wilcoxon rank sum test) and log2 fold-changes compared to mono-association are indicated. For colonization levels of Aeromicrobium Leaf245 see Supplementary Fig. 6. d: days after planting b) In vitro interaction between Leaf374 (in overlay agar) and Leaf245 live cells and cell free supernatant (spotted on top). To increase the contrast of the inhibition area, plates were imaged on top of a checkerboard background. c) Fractionation of Leaf245 supernatant by size exclusion chromatography. Shown is the absorption trace at 215 nm. Top right box shows trace of the complete run and marks the section that is enlarged in the main panel. Solid red lines indicate the collected fractions and the corresponding bioactivity is shown below. ++: complete-, +: partial- and -: no clearance of Leaf374 overlay agar. Dotted red lines indicate the fractions selected for SDS-PAGE. d) SDS-PAGE protein bands observed in the indicated fractions of Leaf245 supernatant (see panel c) and a second biological replicate. Arrows indicate protein bands that were identified by mass-spectrometry. e) In vitro activity of purified candidate proteins against Leaf374 in overlay agar. Proteins that could not be expressed in sufficient amounts are not shown.
To further substantiate that the observed in planta reduction of Leaf374 colonization was caused by the proteins found in the Leaf245 supernatant, we aimed to generate loss of function mutants. All attempts to genetically manipulate Leaf245 were unsuccessful (see Supplementary Note). We therefore used chemical mutagenesis and searched for mutants with loss of in vitro inhibition. We screened more than 10’000 clones from an ethyl methanesulfonate (EMS) treated population and identified 56 mutants with complete or partial loss of Leaf374 inhibition. We validated these mutants by comparing the activity of cell free supernatants to the wild type strain (Supplementary Fig. 8) and selected 33 clones that showed a maximum of 2% residual activity compared with the wild type for genome resequencing (Fig. 4a). Using this approach, we identified a gene locus, ASF05_00210 that was mutated in 40% of all clones analyzed. The locus encodes a 3 kb annotated transcriptional activator and is located directly upstream of the previously identified bacteriolytic protein ASF05_00205 (Fig. 4b). ASF05_00205 was also mutated in one clone (E_60, S70L mutation) and the corresponding purified mutant protein showed attenuated inhibition of Leaf374 (about tenfold less) compared to wild type protein. Furthermore, the altered protein failed to lyse Leaf374 cells grown in an agar pad (Extended Data Fig. 5d), thus identifying the S70L mutation as crucial for protein function. In the remaining identified clones, including those described above, no mutations in other predicted effector proteins (see Extended Data Fig. 5a) were found. We also identified two other clones (E_12 and E_48) with potential regulatory mutations and included these for further analyses. The analysis of supernatant fractions of these and the ASF05_00210 mutants showed that they no longer secreted the ASF05_00205 endopeptidase (Fig. 4c, for details see Supplementary Note).
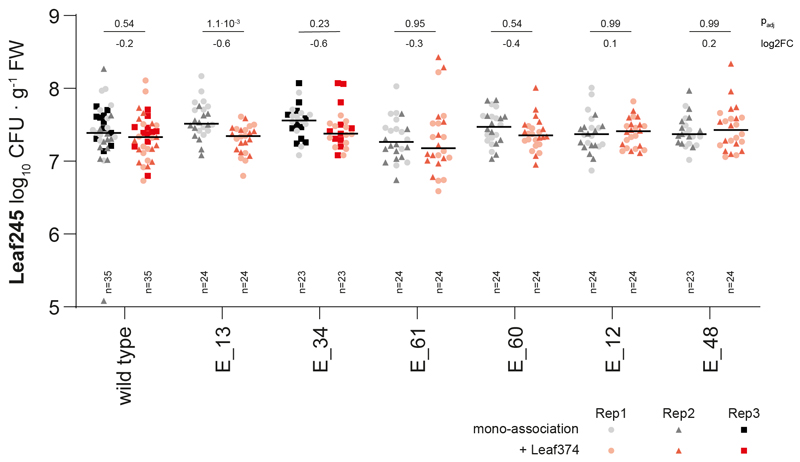
Fig. 4. Leaf374 phyllosphere colonization is less affected when co-inoculated with Leaf245 EMS mutants that no longer secrete ASF05_00205 compared to the wild type.
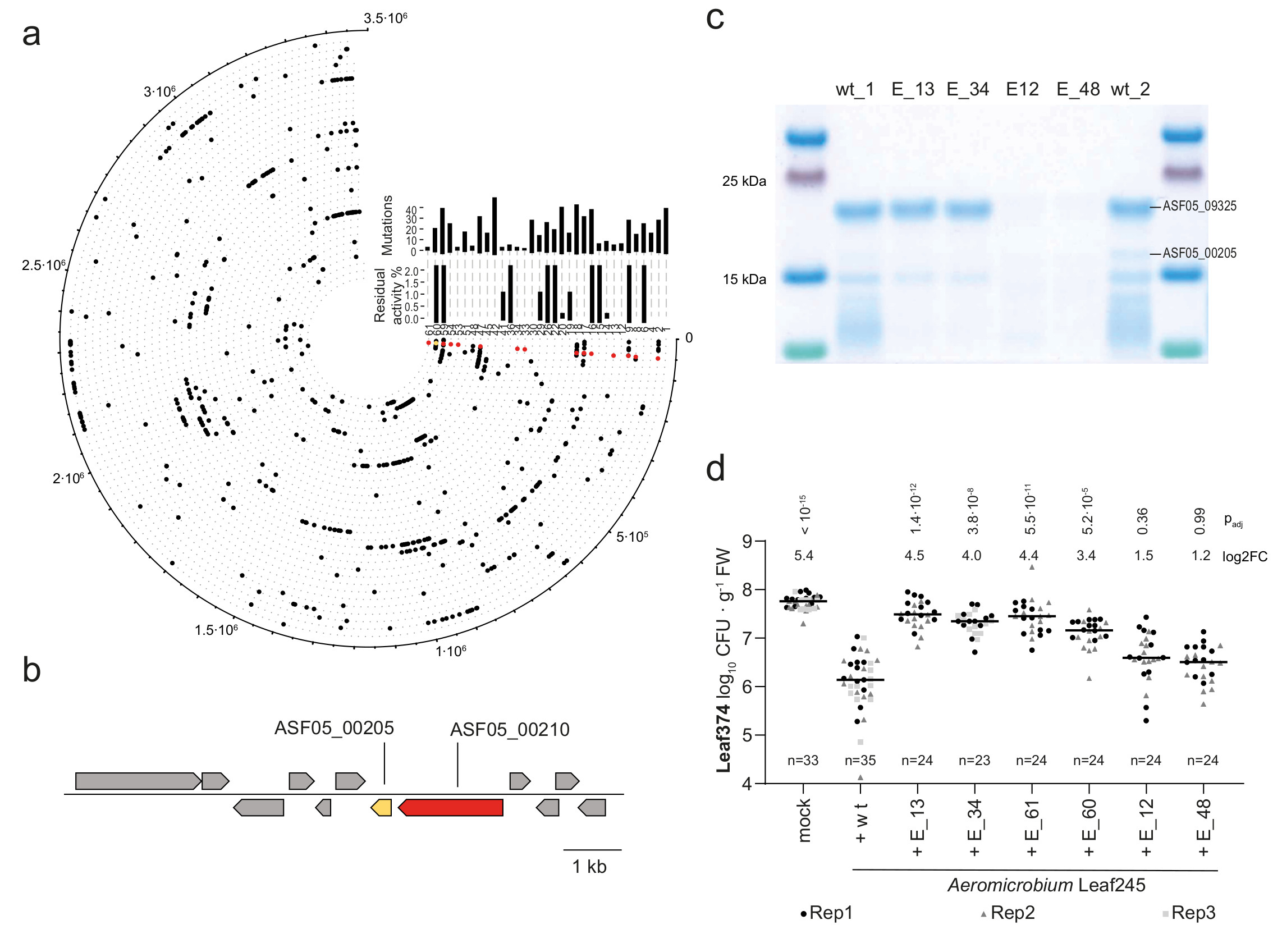
a) Graphic representation of mutations found in genome re-sequenced Leaf245 clones that no longer inhibit Leaf374 in vitro. Outermost circle indicates position on the Leaf245 genome and each circle with its corresponding number represents one clone. Total number of non-synonymous mutations and residual activity compared to the wild type strain are shown for each clone by the corresponding bar plots. Mutations in ASF05_00210 (red) and ASF05_00205 (yellow) are highlighted. b) Leaf245 genomic region comprising a gene pair encoding an identified regulator (red, ASF05_00210) and effector (yellow, ASF05_00205). c) SDS-PAGE protein bands observed in the active fraction of Leaf245 wild type and the corresponding fractions of selected Leaf245 EMS mutants. Protein bands with differing pattern between wild type and mutant strains are indicated with protein accession. A second biological replicate showed similar results. d) Leaf374 colonization of A. thaliana in mono association (mock) or in combination with Leaf245 wild-type or EMS mutants enumerated on MM maltose agar plates. The data of 2-3 biological replicates are shown. Each treatment was compared to Leaf374 colonization in combination with Leaf245 wild type, p-values (Kruskal-Wallis and post-hoc Dunn test, Bonferroni adjusted p) and log2 fold-changes are shown.
Next, we tested whether Leaf374 colonization in planta was no longer reduced when co-inoculated with Leaf245 EMS mutants that do not secrete the ASF05_00205 effector in vitro (E_12, E_13, E_34, E_48 and E_61) or secrete effector with attenuated activity (E_60, see Extended Data Fig. 5d and Supplementary Fig. 8). In mono-association studies, Leaf245 mutants reached similar colonization densities as the wild type indicating that they are not affected in plant colonization per se (Extended Data Fig. 6). Notably, Leaf374 reached 20-fold higher colonization levels in presence of the Leaf245 ASF05_00210 mutants (E_13, E_34 and E_61) and 10-fold higher levels in combination with clone E_60, carrying the ASF05_00205 S70G mutation, compared to in presence of wild type Leaf245 (Fig. 4d). This is in line with the observed supernatant activities in vitro and the partial loss of activity for the modified ASF05_00205 protein. In contrast, the E_12 and E_48 mutant clones did not significantly affect Leaf374 populations compared to the wild type, despite their loss of in vitro inhibition. This might be due to different regulation in planta compared to the tested in vitro conditions. Furthermore, increased Leaf374 levels did not impact colonization by Leaf245, as we did not detect a significant difference between the wild-type and mutant strains in combination with Leaf374 (Extended Data Fig. 6).
In summary we show that the reduction of Leaf374 colonization in planta by Leaf245 is caused by the putative M23 family endopeptidase ASF05_00205 that is regulated by ASF05_00210. We thus named the proteins Nocardioides lysis-associated peptidase (NlaP) and regulator (NlaR), respectively.
NlaP target range and prevalence in other species
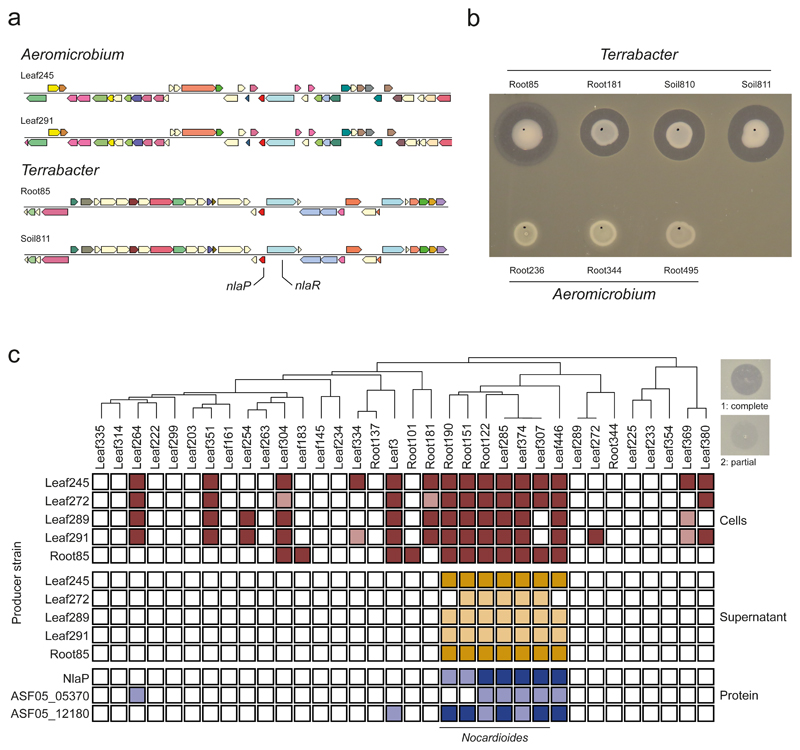
Having established the causative effect of NlaP, we wondered whether the protein was present in other bacterial species. For this purpose we conducted a search on the IMG database39, considering also synteny (gene's bidirectional best hits). This search yielded three other Aeromicrobium species of the At-LSPHERE collection, i.e. Leaf289, Leaf272 and Leaf291, and for one of these (Leaf289) a significant interaction with Leaf374 was also identified in the in planta screen above (Fig. 2a). Additionally, a group of Terrabacter spp. was identified that are part of the At-RSPHERE collection or related soil isolates34. These contained nlaP and nlaR, which are located next to each other in the genomes, but with opposite direction (Fig. 5a).
Fig. 5. Producer and target range of the Nocardioides lysis-associated peptidase NlaP.
a) Genomic region of Leaf245 (query) and exemplary strains containing similar gene neighborhoods compared to ASF05_00205 (nlaP, red) identified by IMG bidirectional best hit. The position of nlaP and nlaR are indicated in the genome map b) In vitro activity of candidate strains encoding nlaP and nlaR homologs and control Aeromicrobium strains not encoding a nlaP homolog. c) In vitro inhibition of a panel of At-SPHERE Actinobacteria by Aeromicrobium and Terrabacter strains (OD600= 1), the corresponding cell free supernatants (OD600 of suspended cells = 5) or by purified proteins (0.1 mg mL-1) indicated on the left side. Target strains are ordered by phylogeny (top) and were all normalized to an OD600 of 0.01 within the agar. Filled boxes indicate inhibition of the strain in the given condition, dark shade: complete clearance (1), light shade: partial clearance (2) as illustrated by the examples on the top right of the panel.
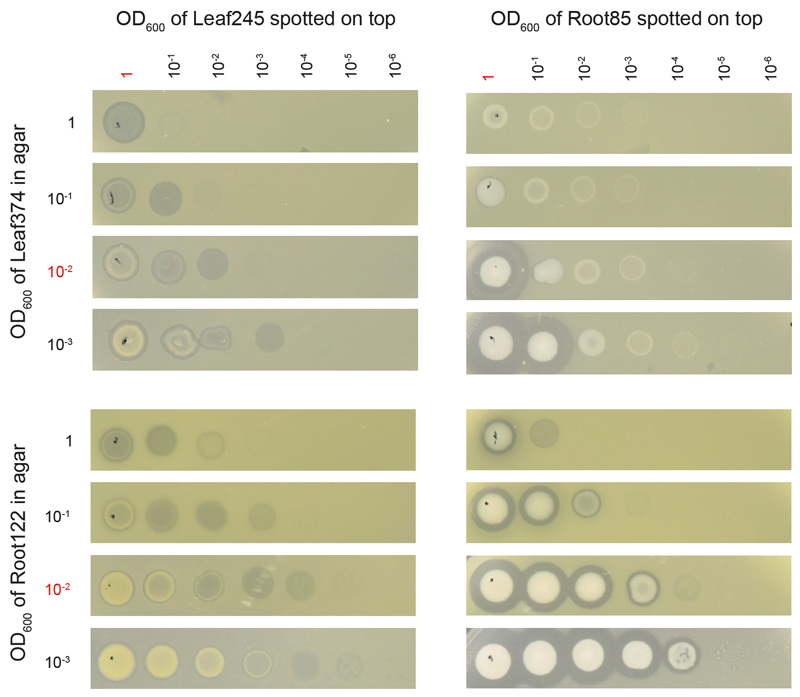
All Terrabacter spp. we tested showed strong inhibition of Leaf374 in vitro (Fig. 5b). Furthermore, we showed that Aeromicrobium spp. that lack nlaP, i.e. Root236, Root344 and Root495, did not inhibit Nocardioides Leaf374 in vitro (Fig. 5b), further supporting our finding of the relevance of NlaP.
Next, we investigated the target range of Aeromicrobium spp. with endopeptidase activity by selecting a panel of Actinobacteria from both the At-L- and At-R-SPHERE collections, which cover isolates from leaves and roots, and tested whether they were inhibited by living cells or cell-free supernatant of At-LSPHERE Aeromicrobium spp. or At-RSPHERE Terrabacter Root85 (Fig. 5c). Living cells consistently inhibited Nocardioides spp. from both the At-RSPHERE and At-LSPHERE collections and few other isolates. These inhibitions were density dependent as exemplarily shown for four strain pairs, especially if the target strain densities were high (Extended Data Fig. 7). The activity of cell-free supernatant on the other hand was restricted to Nocardioides spp. and a closely related Marmoricola sp. (Leaf446). Notably, the supernatants of Aeromicrobium Leaf245 and Terrabacter Leaf85 were more active than the other supernatants, suggesting that these strains release more protein or protein with higher activity. This may also explain the observed differences in interaction strength between Aeromicrobium spp. and Leaf374 in the in planta interaction screen.
Lastly, we also analyzed the spectrum of activity of the purified proteins NlaP, ASF05_05375 and ASF05_12180 that all inhibit Leaf374 (see Fig. 3e). These were also consistently active against Nocardioides spp. and Marmoricola sp. Leaf446 demonstrating the specificity of the endopeptidase-based negative interaction for this group of bacteria and providing a basis for future investigations regarding the target specificity within this so far poorly studied bacterial group.
Discussion
Identifying the nature of microbial interactions that manifest in complex microbiomes remains a challenge. Large numbers of colonizing bacteria create the possibility of numerous interactions, making the discovery of genotype-phenotype relationships difficult. In contrast to correlation-based interaction mapping, synthetic communities provide the opportunity to establish causal interactions by adding or removing strains33, and thus to empirically determine the consequence of inter-strain interactions on the remainder of the community. It also allows for the adjustment of community complexity, which is important because of the inherent complexity of the interaction network in which effects might superpose. There are, however, also drawbacks to the SynCom approach. While complexity reduction is favorable to identify individual interactions, fewer species combinations can be probed at once, and specific metabolic functions associated with only few strains may be absent in the community. Therefore, the size of the SynCom has to be carefully considered.
Here, we selected a focal community of 15 strains and mapped bacterial interactions observed when 200 leaf isolates were individually added during colonization of the Arabidopsis phyllosphere. We found that 90% of the observed interactions were negative (Fig. 2a). This suggests that interactions are mainly competitive in nature, which is consistent with previous studies5,37,40 and not surprising given the oligotrophic nature of the phyllosphere15. The constructed interaction map revealed high consistency of observed interactions for closely related isolates, suggesting similar functions of these within the community. This hints towards shared evolutionary history of these strains and phylogenetic trait conservation41. However, we also noted within genus interactions, which were all negative. It has been a longstanding question in ecology whether closely related species can co-exist in the same niche, due to phenotypic similarity and hence intense competition42. This hypothesis, termed "competition-relatedness hypothesis"43, was much debated in recent years43–47. In this study, we found that within genus negative interactions were more frequent than expected by chance. However, some strains (Methylobacterium Leaf88, Xylophilus Leaf220 and Pseudomonas Leaf129) also show negative interactions with distantly related groups (Extended Data Fig. 2).
To assess if the presence of the community is required to observe the identified interactions, we tested a subset of interactions in a binary inoculation model (Fig. 2b). Notably, we found, that two thirds of the interactions were reproducible without community background, meaning that these strains affect each other independently of other microbiota members. Further studies are necessary for each specific pair to demonstrate whether this is due to direct bacteria-bacteria interaction or due to indirect effects through host-modulation48, given that plants respond to most of its endogenous microbiota leaf members by inducing a conserved set of defense related genes49. Interactions between host-associated bacteria in binary and tertiary combinations have been characterized before5,40,50,51. However, most of the corresponding studies were conducted in vitro under conditions that are substantially different compared to the host-associated context. This is particularly pertinent when studying phyllosphere communities, since the leaf architecture provides a multitude of microenvironments that are spatially segregated and the oligotrophic nature of leaf surfaces is difficult to mimic in vitro52,53.
We also identified one higher order interaction54. A marked reduction of Methylobacterium Leaf88 was observed in the presence of at least two other strains that did not occur in the presence of either alone (Fig. 2c). Such higher order interactions are proposed to be important in stabilizing species coexistence in complex communities55; however, there is still little empirical evidence to assess their importance56. It was found that the presence of multiple strains can dampen the competition previously observed in a binary situation, which was attributed to higher order interactions57. Here, we found the opposite scenario, where a higher order interaction led to the emergence of a competitive interaction. Additional interactions observed in planta (Fig. 2a) might be the result of higher order interactions. In planta observed interactions that could not be validated using binary strain inoculations thus provide valuable starting points to test combinatorial permutations to uncover potentially underlying higher order interactions.
The patchy colonization observed on the leaf surface52,53,58, could also explain why higher order interactions in communities in the phyllosphere are indeed difficult to detect at the overall population level, as more than two strains have to co-localize for direct bacteria-bacteria interactions to occur. These events are less likely in spatially segregated environments. Similarly, binary interactions could also be affected as strains do not necessarily have a chance to encounter each other at the microscale, which mitigates the observed effect.
Spatial segregation may also explain why antibiotic interactions previously found in vitro6 were rarely observed in planta (Supplementary Table 5). Other reasons for the lack of antibiotic effects in planta, apart from leaf colonization patterns, include insufficient amounts or lack of production of the compound under the conditions found in situ59. While antibiotic production observed in vitro could not explain most interactions observed in planta, we found one prominent interaction based on antimicrobial production.
Intriguingly, the interaction is mediated by a protein, an endopeptidase termed NlaP. We hypothesize that proteins might be more stable in the phyllosphere compared to small molecules, which might be subject to rapid degradation due to exposure to UV irradiation or reactive oxygen species and hence explain the strong effect observed in planta. Moreover, proteins may directly impair the target cell. We speculate that NlaP cleaves Nocardioides peptidoglycan cross-linkages with high specificity, since other members of this protein family, including lysostaphin from Staphylococcus simulans60,61 and LasA from Pseudomonas aeruginosa62, have glycine-glycine endopeptidase activity. As observed for other bacteriolytic enzymes63–65, the target range of NlaP is taxonomically narrow (Fig. 5c) and potentially confined to strains with similar surface structure. Therefore, the production of NlaP likely provides a strategy to defend favorable niches on the leaf surface.
In summary, this study demonstrates the power of synthetic communities to systematically identify bacterial interactions in the phyllosphere. Since the microbes involved are all culturable, the identified interactions can be further characterized to decipher mechanisms that contribute to community assembly. Here we demonstrate a role that a bacteriolytic enzyme can play in community assembly. Further mechanistic understanding of how communities assemble will ultimately be critical to building lasting plant growth-promoting and disease suppressive microbial communities necessary for sustainable agriculture.
Methods
Strains and cultivation
Unless stated otherwise, At-SPHERE isolates were cultivated at room temperature on R-2A agar (Sigma-Aldrich) or at 28°C in R-2A broth (0.5 g yeast extract (Oxoid), 0.5 g Proteose peptone No. 3 (Becton, Dickinson and Company), 0.5 g Casamino acids (Becton, Dickinson and Company), 0.5 g D-glucose monohydrate (Sigma-Aldrich), 0.5 g starch (from potato, Fluka), 0.3 g sodium pyruvate (Sigma-Aldrich), 0.3 g K2HPO4 (AppliChem) and 0.05 g magnesium sulfate heptahydrate (Sigma-Aldrich) dissolved in 1 L deionized water), both supplemented after sterilization with 0.5% (v/v) methanol (R-2A+M). Minimal medium was prepared as described previously66 and supplemented with 5 mM maltose (Fluka). For Leaf374 cultivation, minimal medium agar was supplemented with vitamins (500 μg L-1 D-pantothenic acid hemi calcium salt, 100 μg L-1 biotin, 400 μg L-1 riboflavin, 400 μg L-1 thiamine HCl, 200 μg L-1 pyridoxal HCl 150 μg L-1 p-amino benzoic acid, 200 μg L-1 cobalamin, 50 μg L-1 lipoic acid, 150 μg L-1 nicotinic acid and 100 μg L-1 folic acid). For selective growth of Leaf272, R-2A+M was supplemented with 50 μg mL-1 rifampicin (Sigma-Aldrich), since this strain has a natural resistance towards this antibiotic.
E. coli BL21 (DE3) gold was cultivated in LB-Lennox at 37°C supplemented with 50 μg mL-1 kanamycin sulfate (AppliChem).
Plant growth conditions
Plants were cultivated as described previously37, with some modifications. In brief, Arabidopsis thaliana Columbia-0 (Col-0) seeds were surface-sterilized67 and stratified for 4 days at 4°C. Plants were cultivated in six- or twelve-well tissue culture plates (TechnoPlasticProducts) filled with 5 or 2 ml washed and heat-sterilized calcined clay and 2.5 or 1 ml half-strength Murashige & Skoog medium pH 5.8 including vitamins (½ MS, Duchefa), respectively. Individual seeds were placed in the center of each well. If seeds did not germinate, a seedling was transplanted to the corresponding well six days after seeding. Starting at day 6, each well was supplemented twice per week with 200 or 100 μL ½ MS, respectively. Plates were incubated in a growth chamber (Percival, CU41-L4) set to 22°C and 54% relative humidity with a 11 h light 13 hours dark regime fitted with full spectrum lights (Philips Master TL-D 18W/950 Graphica) and lights emitting a higher fraction of UVA and UVB (Sylvania Reptistar F18W/6500K). Combined light intensity was set to 200-210 µmol m-2 s-1 for wavelength 400-700 nm and 4-5 µmol m-2 s-1 for wavelength 280-400 nm.
Inoculation with bacteria
Bacteria were cultivated at 22 °C on R-2A+M agar plates for four days, re-streaked onto fresh R-2A+M plates and grown for another three days prior to inoculation. Inoculation suspensions were prepared by suspending individual strains in 10 mM MgCl2 solution. For the focal community, focal community drop-out and replacement as well as for binary and tripartite strain inoculations, the optical density at 600 nm (OD600) of each strain was adjusted to 0.2 and the strains were mixed at equal ratios. Finally, the strain mixtures were diluted to a final OD600 of 0.02 for inoculation. For strains added in the drop-in screen, similar volumes of cell paste were suspended in 10 mM MgCl2 solution corresponding to an OD600 of 0.1-0.3. Strains were diluted 20-fold when added to the premixed focal community (all strains adjusted to an OD600 of 0.2, mixed at equal ratio and then 10-fold diluted to OD600 0.02), but were present in approximately 10-fold excess compared to individual focal strains (Supplementary Table 1).
Unless otherwise indicated, 7 day old seedlings were inoculated with 50 μL final inoculation suspension by slowly dropping on the whole seedling. Axenic control plants were mock-treated with 10 mM MgCl2 instead of bacterial suspension. No statistical methods were used to pre-determine sample sizes for phyllosphere inoculation experiments, but sample sizes are similar to those previously reported19,37.
Phyllosphere harvest for 16S amplicon sequencing or CFU enumeration
The entire phyllosphere of 28 or 29 day old plants was harvested with sterilized tweezers and scalpels. Plants for community profiling by 16S rRNA gene amplicon sequencing were directly placed into lysis matrix E tubes (FastDNA SPIN Kit for Soil, MP Biomedicals), frozen in liquid nitrogen and stored at -80°C. For the in planta screen (Fig. 2a) two plants grown on separate plates were pooled for one biological replicate. In all other experiments, individual plants were analyzed.
Plants for colony-forming units (CFU) enumeration were harvested by two different protocols. The first protocol was performed as described previously13. The phyllosphere was placed in tubes containing 1.3 mL 100 mM phosphate buffer pH7 containing 0.2% Silwet-L77 (Leu+Gygax AG, Switzerland). Plant fresh weight was measured on an analytical balance (Mettler-Toledo) with an accuracy of 0.1 mg. Bacteria were subsequently washed off by shaking tubes for 15 min at 25 Hz with a TissueLyser II (Qiagen) followed by sonication (ultrasonic bath, Branson) for 5 min. Tubes containing wash-off solution were vortexed and 100 μL of the solution was transferred to a 96-well plate to prepare a 10-fold serial dilution in 100 mM phosphate buffer pH7.
The protocol was changed for experiments including Nocardioides Leaf374, since we observed that survival of the strain was reduced by the presence of Silwet-L77 in the wash solution. For harvest with the revised protocol, the phyllosphere was placed into tubes containing a metal bead (5 mm diameter) and 100 mM phosphate buffer pH 7 (200 µL). Plants were crushed with a TissueLyser II for 45 s at 30 Hz. Phosphate buffer (600 µL) was added to the crushed plant material and mixed by vortexing. Samples (100 µL) were transferred to a 96-well plate to prepare a 10-fold serial dilution in 100 mM phosphate buffer.
For dilution series prepared with either of the harvest protocols 4 µL of each dilution were spotted on R-2A+M agar square plates (Greiner) and dried under laminar flow. For samples inoculated with strain combinations or synthetic communities 50 µL of the appropriate dilutions were plated on round (9 cm) R-2A+M agar or appropriate selective agar plates to count individual strains based on morphology. Plates were incubated at room temperature and CFU were counted after 1-3 days on square plates and after 4-7 days on round plates. Strains in the combinations had different colony morphology to allow identification and quantification of each strain in the mix.
Phylogenetic tree construction
The phylogeny of the At-LSPHERE strains was based on full-length 16S rRNA gene sequences as described previously19. The phylogeny of At-L- and At-R-SPHERE Actinobacteria was based on AMPHORA genes as described in Bai et al34. The phylogenetic tree was divided with the keep.tip command of the R package ape 5.4-168. Leaf374 was placed manually in the tree with the command bind.tip in the R package phytools69 based on full-length 16S rRNA gene tree.
16S rRNA gene amplicon library preparation and sequencing
Frozen plant and inoculum samples were lyophilized (Christ Alpha 2-4 LD Plus) for 16-18 h and subsequently homogenized with a TissueLyser II for 2 min at 25 Hz. DNA was extracted with the FastDNA SPIN Kit for Soil (MP Biomedicals) according to the manufacturer's instructions. DNA was quantified (Promega QuantiFluor dsDNA, E2670) and normalized to a concentration of 1 ng µL-1. The 16S amplicon library was prepared as described before37. DNA amplification of the V5-V7 region of the 16S rRNA gene was performed in triplicate with primers 799F and 1193R with DFS Taq polymerase (Bioron) and 12.5 ng template DNA for 25 cycles with the following cycling parameters: Denaturation for 30 s at 94 °C, annealing for 30 s at 55 °C extension for 60 s at 72 °C. Triplicate samples were pooled and primers were removed by enzymatic digest with Antarctic phosphatase (NewEnglandBioLabs) and Exonuclease I (NewEnglandBioLabs). Barcoding PCR (10 cycles) was performed in triplicate with DFS Taq polymerase and plate specific forward and well specific reverse primers with the same cycling parameters as the first PCR. Triplicates were pooled and the amplification of each sample was verified by loading equal amounts (5 μL) on a 1.5% (w/v) agarose gel. Samples were pooled based on intensity of the obtained agarose gel band. The library was loaded on a 1.5% agarose gel and the band at a size of approximately 500 bp was cut out and DNA was cleaned up with the QIAquick gel extraction kit (Qiagen) according to manufacturer's recommendation. The library was cleaned twice by bead clean-up (AMPure XP, Beckman Coulter) with a ratio of 0.8:1 and finally with a ratio of 0.6:1 bead to sample ratio. The final length of the amplicon library was checked on a 2200 TapeStation (Agilent Technologies) using High sensitivity D1000 screen tape. Sequencing was performed on the Illumina MiSeq platform using a v3 cycle kit (2x 300 bp, paired-end) at the Genetic Diversity Center (ETH Zurich). The library was denatured and diluted to a final concentration of 12 pM with addition of 20% PhiX. Sequencing was performed with custom sequencing primers as described before34.
Data analysis
The 16S rRNA gene amplicon sequencing data was analyzed as described previously19,37. Based on V5-V7 16S rRNA gene sequences a list of amplicon sequencing variants (ASVs) included in the experiments was compiled and used as a reference database. When multiple strains existed with the same ASV they were assigned to one representative ASV for analysis, but could still be tracked as strains because they were not present with strains of the same ASV at the same time (except for few strains belonging to the same ASV as a focal strain).
Paired-end sequencing raw reads were processed using USEARCH70 v.11.0.667-i86 linux64. First, reads were merged with the -fastq_mergepairs command with minimum identity of 90% and minimum overlap of 16 bp. Next, the merged reads were filtered using -fastq_filter with a maximum expected error of 1 and a minimum length of 200 bp. The –otutab command was used to classify and count reads with 100% identity to the 16S rDNA reference database and assign them to individual samples, to generate an ASV table. The total number of sequences with a barcode corresponding to a sample but no match to the reference database were added as an additional line for sequencing depth estimation. Unclassified sequences were dereplicated and clustered with -fastx_uniques and -cluster_otus commands. The minimum cluster size was set to 1 and the identity threshold was set to 97%. Taxonomy was assigned to the de novo OTU clusters with the SILVA SSU Ref NR database71 (release132) to detect potential contaminations. Control samples of mock-inoculated plants and extraction/processing controls were used to detect possible systematic contaminations and were excluded for further analysis.
Further data analysis was performed in RStudio72 with R version 4.0.473 as described before37. The ASV table (Supplementary Data 1) was log-normalized and variance-stabilized using DESeq2 v.1.14.174 to account for different sequencing depths between samples. Log2 fold-changes and p-values (Wald test, Benjamini-Hochberg adjusted) were calculated with DESeq2 for each comparison between drop-in or drop-out condition compared to the unperturbed control. For the analysis of the screen, three replicates of each screening condition were compared to 16 replicates of the focal community. For the focal community control one sample was lost (S_58) and one sample was removed as an outlier (S_72), since the community composition was very different compared to other focal community samples (Supplementary Fig. 4). The drop-in strains Leaf72 and Leaf69 were excluded, since the former was neither detected in the inoculum nor in any of the plant samples and the latter was excluded due to contamination. Sample metadata can be found in Supplementary Data 1.
To assess the overall effect on the communities, PCA was performed with the transformed ASV table using the prcomp command. For PCA analysis only ASVs were included that were present in both conditions. The effect size represents the explained variance calculated on Euclidian distances and statistical significance was based on PERMANOVA with 50’000 permutations using the adonis command of the vegan v.2.5-7 package75.
Phylogenetic distance of all strains was calculated based on full-length 16S rRNA gene tree with the command cophenetic.phylo in package ape 5.4-168.
GraphPad Prism V9.2 was used to plot log10 transformed colonization data and to perform corresponding statistical tests.
Leaf245 supernatant preparation, fractionation and protein identification
Aeromicrobium Leaf245 was grown for three days on R-2A+M agar, re-streaked as a thin layer onto two square R-2A+M agar plates and grown for 24 h. The complete cell material was collected with a sterile inoculation loop and suspended in 800 μL sterile 100 mM ammonium bicarbonate buffer adjusted to pH 7.5 with HCl. Cells were vortex-mixed for 1 min and OD600 of cell suspension was measured for normalization purposes when different clones were compared. Cells were pelleted by centrifugation for 2 min at 8000 x g and the supernatant was passed through a 0.22 µm PES filter to remove residual cells. Finally, the supernatant was concentrated to a final volume of approximately 250 µL using an Amicon Ultra-4 spin filter tube (3 kDa, Merck).
A Superdex 75 column (GE Healthcare) fitted to an ÄKTA Purifier 10 FPLC system (GE Healthcare) was equilibrated with 100 mM ammonium bicarbonate buffer pH 7.5 before injecting 150 - 200 µL of concentrated supernatant. For fractionation, the flow rate was set to 0.2 mL min-1 for the first three mL and then increased to 0.5 mL min-1. Fractions (1 mL) were collected while the run was monitored by absorption measurement at 280 and 215 nm.
Single or pooled active fractions were concentrated with 3 kDa cutoff Amicon Ultra-4 spin filter tubes. Proteins were separated by SDS-PAGE on precast 10% acrylamide gels (Merck) and bands of interest were cut out or whole fractions were sent directly for protein identification by shotgun proteomics at the Functional Genomics Center Zurich.
Heterologous expression of effectors in E. coli
Plasmids for expression of selected candidate proteins were ordered from Twist Bioscience (USA). First, secretion signals of candidate proteins were predicted using SignalP-5.076 with default settings for Gram-positive bacteria. The protein sequences without signal sequence were subsequently reverse-translated and used to generate codon-optimized nucleotide sequences for expression in E. coli applying default settings of Twist. If necessary, spacer base pairs were added to allow in-frame cloning into the expression plasmid pET28a(+) (Novagen) using NcoI and XhoI restriction sites.
The nucleotide changes for the ASF05_00205S70L construct were introduced by quick-change mutagenesis with the primers 00205_S70L-F (CAATCGCGTAACGAAGCGGTGAGTGGCCGT CGC) and 00205_S70L-R (GCGACGGCCACTCACCGCTTCGTTACGCGATTG).
The expression plasmids were transformed into E. coli BL21 (DE3) Gold by electroporation and transformants were selected on LB agar supplemented with kanamycin. Pre-cultures in 20 mL LB supplemented with kanamycin were inoculated with cell material from several transformant colonies and were grown to mid-exponential phase. Bacteria were added to 100 or 200 mL main cultures at a starting OD600 of 0.01 and grown at 37°C. When cell cultures reached an OD600 of 0.5 they were shifted to 16 °C and expression was induced 30 min later by addition of isopropyl-β-D-thiogalactopyranosid (IPTG, Biosynth) at a final concentration of 0.1 mM. Expression was stopped 16 h after induction by placing shake-flasks on ice for 15 min. The cells were harvested by centrifugation (8000 x g, 15 min, 4°C), cell pellets were washed twice with 20 mM TRIS-HCl pH 7.5 containing 250 mM NaCl and were resuspended in in the same buffer supplemented with 40 mM imidazole. Cells were lysed by processing three times with a FrenchPressure cell (SLM AMINCO, SLM instruments Inc.) at a pressure of 1100 psi and the lysate was cleared by centrifugation for 1 h at 45'000 rpm in a Beckman Coulter LE-80 centrifuge equipped with a Ti50 rotor corresponding to approximately 130'000 x g. Cleared cell lysate was stored at 4°C.
Protein purification
Affinity purification was carried out on an ÄKTA Purifier 10 FPLC system (GE healthcare). Cleared cell lysate (4-6 mL) was passed through a 0.22 µm PES filter (TechnoPlasticProducts) and loaded on a HisTrap™ HP column (1 mL, Cytiva) equilibrated with 20 mM TRIS-HCl pH 7.5 containing 250 mM NaCl and 40 mM imidazole. The column was washed with 10 column volumes of the same buffer and bound proteins were eluted over a gradient of 10 mL to a final concentration of 500 mM imidazole. Elution of proteins was monitored by absorption at 215 and 280 nm and 1 mL fractions were collected. Based on SDS-PAGE gel band pattern, fractions containing purified protein were pooled and imidazole concentration was reduced by 20-fold dilution in buffer without imidazole followed by volume reduction with an Amicon Ultra-4 spin filter tube (3 kDa, Merck).
Protein quantification
The purified proteins were quantified by absorption measurement at 280 nm with a NanoDrop spectrophotometer (ThermoFisher) calibrated with extinction coefficients calculated for each purified protein with the Expasy ProtParam tool77.
Inhibition assay with living cells, supernatant and purified proteins
Nocardioides Leaf374 and other target strains were grown on R-2A+M agar plates for 2-4 days. Cells were suspended in 10 mM MgCl2 solution. To prepare overlay plates suspended bacteria were added to 3 mL R-2A top agar (43 °C, 1% agar) at a final OD600 of 0.01. The overlay agar was immediately poured on top of a round petri dish prefilled with 20 mL solidified R-2A agar and left to dry for 5 min under laminar flow. Larger square plates were prepared in the same way with 10 mL top agar and 30 mL prefilled agar. Suspended cells of producer strains (OD600 adjusted to 1, corresponding to approx. 5x109 CFU mL-1), cell free supernatants (from OD600 = 5 or 10 adjusted cell suspension) or purified proteins (adjusted to a concentration of 0.1 mg mL-1 protein) were spotted (2 μL) on top of overlay plates and dried for 5 min under laminar flow. Plates were incubated at room temperature and inhibitions that manifested by the appearance of halos were scored after 24 and 48 h.
Microscopy
Leaf245 was grown in R-2A+M broth and the supernatant was concentrated 200-fold with an Amicon Ultra-4 spin filter tube (100 kDa, Merck). Leaf374 grown on R-2A+M agar was resuspended in PBS pH7.4 (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl), spotted on a LB agar pad and dried. The concentrated supernatant of Leaf 245 was added (10 μL) and imaging was started one minute later. Phase contrast images were acquired on a Zeiss Axio Observer Z1 inverted microscope (Carl Zeiss AG) equipped with an EC Plan-Neofluar 100x/1.3 objective. Images were taken every 20 seconds over the course of 20 minutes and combined to a time lapse video using Fiji software78.
EMS mutagenesis of Leaf245 and identification of loss of function mutations
Aeromicrobium Leaf245 was grown on R-2A+M agar plates for three days and inoculated into 20 mL R-2A+M broth in a 100 mL baffled shake flask at a starting OD600 of approx. 0.02 and grown at 28°C to mid-exponential phase. Cells were harvested by centrifugation (8 min, 3320 x g, RT), washed with 30 mL 100 mM sodium phosphate buffer pH7 and were resuspended in the same buffer and adjusted to OD600 = 1. Suspended cells (400 µL) were mixed with 4% (v/v) ethyl methanesulfonate (EMS, Sigma-Aldrich), mixed vigorously and incubated for 1-5 hours at room temperature. Mutagenesis was stopped by adding 1.1 mL 0.16 M sodium thiosulfate (Sigma-Aldrich). Cells were pelleted by centrifugation (2 min, 8000 x g) and supernatant was completely removed. Cells were suspended in 1 mL 80 mM sodium phosphate buffer containing 32 mM sodium thiosulfate. Aliquots (200 µL) were mixed with the same amount of sterile 50% (v/v) glycerol and stored at -80°C. For each sample a 10-fold dilution series was prepared in 80 mM sodium phosphate buffer containing 32 mM sodium thiosulfate and 50 µL of dilutions 10-5 and 10-6 were plated on R-2A+M agar plates to estimate survival of cells. The sample with a survival rate of approximately 5% (reached after 3 hour EMS treatment) was chosen for further experiments.
Mutagenized cells were thawed, diluted 103-105-fold in 100 mM sodium phosphate buffer pH7 and 50 µL aliquots plated on R-2A+M agar and incubated for 5 days. Individual clones were picked with sterile tooth picks and streaked on top of overlay agar plates containing Leaf374. Clones that showed complete loss of inhibition, smaller zone of inhibition or incomplete clearance were re-streaked twice on R-2A+M agar to obtain pure clones. A single colony was picked and grown in 20 mL R-2A+M broth. From this culture, cryo stocks were prepared and cell pellets were prepared and stored at -20°C for DNA extraction and subsequent genome sequencing.
Frozen cell pellets were thawed on ice. Cells were suspended in 50 µL freshly prepared 10 mM sodium phosphate buffer pH7 containing 3 mg mL-1 lysozyme (Carl Roth, Karlsruhe, Germany) and were incubated at 37°C for 30 min. After the lysozyme pre-treatment DNA was extracted with the Epicentre® MasterPure™ Complete DNA & RNA Purification kit (Lucigen, Middleton, WI, USA) according to the manufacturer's recommendation for bacterial cell cultures. DNA concentration was quantified using the Quantifluor® dsDNA System (Promega) and adjusted to 50 ng µL-1. Library preparation and Illumina sequencing was performed by Novogene (UK sequencing center). DNA was randomly fragmented by sonication, end-polished, A-tailed and ligated with the full-length adapters for Illumina sequencing. Adaptor ligated fragments were PCR amplified with P5 and indexed P7 oligos and purified with AMPure XP beads. The size distribution of the libraries was checked with a 2100 Bioanalyzer (Agilent Technologies) and quantified by real-time PCR. The combined library was sequenced on a NovaSeq 6000 sequencing system (2x 150 bp, paired-end). Reads containing adaptors, unassigned bases (N > 10%) or low quality bases (Qscore ≤ 5 for > 50% of bases) were removed from the raw data by the sequencing center.
Illumina short-reads were adapter-trimmed (ktrim=r, k=23, mink=11, hdist=1, tpe, tbo, ftm=5), contaminant-filtered (k=31, hdist=1) and quality-filtered (trimq=14, maq=20, minlength=36, maxns=0, qtrim=r) using BBDuk of BBTools79 (v.38.87, sourceforge.net/projects/bbmap). Mutations in EMS clones of Leaf245 were identified with Breseq80 (v.0.35.4). Variations also observed in the wild-type resequenced strain were removed from the list.
Sequencing of Leaf245 with Nanopore long reads and Illumina read polishing
Leaf245 wild-type DNA was also sequenced using ONT long-read sequencing technology to close the genome prior to EMS mutant identification. DNA was tagmented and rapid barcoded (RB03) with the SKT-RBK004 kit (Nanopore) and pooled with other tagmented DNA samples (not included here) according to the manufacturer's instructions. The tagmented DNA pool was concentrated with SPEED beads and half of the mixture was size selected with a 4 kb high-pass cutoff on a BluePippin (Sage Science) with a BLF-7510 cassette. The size-selected DNA was cleaned and concentrated using an equal volume of SPEED beads prior to elution in 10 mM Tris-HCl pH 8 containing 50 mM NaCl. Nanopore sequencing adapters were added according to the SKT-RBK004 kit instructions prior to sequencing on a MinION with a FLO-MIN106 flow cell. Base calling and demultiplexing was done post-sequencing using Guppy (v.3.2.4+d9ed22f, Nanopore) and reads from two sequencing runs/flow cells were combined. Adapters were trimmed and possible chimeras removed using porechop (v.0.2.4, https://github.com/rrwick/Porechop). The resulting long-reads were assembled with Flye81 (v.2.8-b1674) into a single circular chromosome, followed by five rounds of Illumina short-read polishing with Pilon82 (v.1.23). One circular chromosome of 3463989 bp was obtained and annotated using Prokka83 (v.1.13.3).
Extended Data
Extended Data Figure 1. Changes in focal community strain abundances upon strain drop-in.
Heatmap showing log2-transformed fold changes (treatment vs control) for each focal community strain (top) upon strain drop-in (left). Significant changes (DESeq2-normalized counts, Wald test, Benjamini-Hochberg corrected, padj ≤ 0.01, nfocal community = 16, ndrop-in = 3) are indicated with a black frame. Drop-in strains are colored by phylum or Proteobacteria class. Exact p-values are provided in Supplementary Table 2.
Extended Data Figure 2. Phylogenetic distance of interacting strains.
Focal strains that showed ≥ 1 interaction are shown. The phylogenetic distance of the focal strain to all added strains is indicated on the x-axis. Log2-fold changes (log2FC) are shown on the y-axis. Significant fold-changes are highlighted with blue (negative interactions) or red (positive interactions) fill color. Strains that belong to the same ASV as a focal strain have a light grey fill color.
Extended Data Figure 3. Phyllosphere colonization of Leaf220 and Leaf408.
Phyllosphere colonization of a) Leaf220 or b) Leaf408 in mono-association or in combination with other strains (indicated below). Shown are the median and individual data points of log10-transformed CFU per gram fresh weight across two independent experiments. Colors/shapes refer to experiment. Exact p-values (Krusksal-Wallis test with post-hoc Dunn test, Bonferroni adjusted p) and log2 fold-changes (log2FC) for comparisons to the mono-association condition are indicated above or below the graph, respectively. For corresponding Leaf88 colonization levels see Fig. 2c.
Extended Data Figure 4. Validation of Leaf374-Leaf245 interaction by sequential inoculation of A. thaliana.
Phyllosphere colonization of a) Nocardioides Leaf374 and b) Aeromicrobium Leaf245 in mono-association or in combination with the other strain. Shown are the median and individual data points of log10-transformed CFU per gram fresh weight recovered after plant colonization (n= 12). CFU were enumerated on MM maltose agar plates and R2A+M for Nocardioides Leaf374 and Aeromicrobium Leaf245, respectively. The timepoint of inoculation with Leaf245 or Leaf374 is indicated below the graph as days (d) after planting. For mono-association controls half of the replicates were inoculated on day7 (black) or day8 (grey) and mock-treated with 10 mM MgCl2 on the other day. Exact p-values (two-sided Wilcoxon rank sum test) and log2 fold changes (log2FC) compared to the mono-association control are indicated above or below the graph, respectively.
Extended Data Figure 5. Characterization of selected proteins identified in the supernatant of Leaf245.
a) List of proteins identified in the supernatant of Leaf245 and selected for heterologous expression in E. coli. For each gel band in Fig. 3d as well as the most active fractions from two independent experiments the rank abundance of the proteins based on total ion count as identified by mass spectrometry are shown. b, c, d) Leaf374 inhibition and lysis by heterologously expressed candidate proteins. b) Activity of cleared cell lysate after expression of proteins (top label) in E. coli BL21 DE3 gold. Lysate was applied on Leaf374 overlay plates directly after preparation (left panel) or on Leaf374 overlay plates that were pre-incubated for 24 h to assess lysis (right panel). Lysate concentration was normalized to the final OD600 that each expression culture reached. Up to 50-fold dilutions in the buffer used for lysis (see methods) were prepared as indicated on the left. c) Activity of purified proteins against 24 h pre-grown Leaf374. d) Activity of ASF05_00205 native and mutant (S70L) protein against Leaf374. Concentrations of both purified proteins were normalized (0.1 mg mL-1) and a 10- and 100- fold dilution was prepared and assayed on a Leaf374 overlay plate directly after preparation (left panel) or after the overlay was pre-incubated for 24 h (right panel) to test for growth inhibition and cell lysis, respectively. Data shown for ASF05_00205 native protein is the same as shown in Fig. 3e and panel c) of this figure.
Extended Data Figure 6. Aeromicrobium Leaf245 wild type and EMS mutant colonization level.
Phyllosphere colonization of Leaf245 in mono-association or in combination with Leaf374. Shown are the median and individual data points of log10-transformed CFU per g fresh weight across 2-3 independent experiments. Log2 fold changes and exact p-values (two-sided Wilcoxon rank sum test) between Leaf245 mono-association and co-colonization treatments with Leaf374 are shown above the graph. For the corresponding Leaf374 colonization see Fig. 4d.
Extended Data Figure 7. In vitro inhibition assay with different producer and target cell densities.
The concentrations of the NlaP producer strains (Leaf245 and Root85) spotted on top (2 µL) and target strains (Leaf374 and Root122) within the agar are indicated on top and on the left of the graph, respectively. Pictures were taken 36 hours after overlay preparation and spotting. The standard concentrations used for all other assays performed in this study are highlighted in red.
Supplementary Material
Acknowledgements
We thank Dr. Aria Minder and Silvia Kobel at the Genetic Diversity Center Zurich (GDC, ETH Zurich) for MiSeq amplicon sequencing service, Dr. Christopher Field (Institute of Microbiology, ETH Zurich) for amplicon raw read processing, the Functional Genomics Center Zurich (FGCZ) for protein identification service and Prof. Dr. Paul Schulze-Lefert (Max Planck Institute for Plant Breeding Research, Cologne, Germany) for sharing At-RSPHERE isolates. This work was supported by the Swiss National Science Foundation through NRP72 (no. 407240_167051) and as part of NCCR Microbiomes, a National Centre of Competence in Research (no. 51NF40_180575) and a European Research Council Advanced Grant (PhyMo; no. 668991).
Footnotes
Author contributions
M.S. and J.A.V. designed the research. M.S., M.B.-M. and C.M.V. conducted the in planta interaction screen and binary competition experiments. M.S. prepared the amplicon sequencing libraries and analyzed amplicon sequencing data, performed biochemical analyses of Leaf245 supernatants and heterologous expression of proteins. M.M conducted microscopy time course experiment to monitor Leaf374 lysis. M.S. prepared and M.S. and C.M.V screened the Leaf245 EMS library. C.M.V. closed the Leaf245 genome and mapped point mutations of genome re-sequenced EMS clones. M.S. screened the inhibition spectrum of NlaP producer strains, performed statistical analyses and visualized the data. M.S. and J.V. wrote the manuscript with contributions from all authors.
Competing interests
The authors declare no competing interests.
Data availability
The 16S rRNA gene amplicon sequencing reads and the reads obtained from Leaf245 EMS mutant genome sequencing are available online at the European nucleotide archive (https://www.ebi.ac.uk/ena/browser/) under the accession numbers PRJEB50894 and PRJEB47688, respectively. The SILVA SSU Ref NR database (release 132) can be found under www.arb-silva.de/download/archive. Source data are provided with this article.
Code availability
The code used for analysis of 16S rRNA gene amplicon data can be found in the package phylloR available on GitHub (https://github.com/MicrobiologyETHZ/phylloR/). No unpublished algorithms were used.
References
- 1.Flemming HC, Wuertz S. Bacteria and archaea on Earth and their abundance in biofilms. Nat Rev Microbiol. 2019;17:247–260. doi: 10.1038/s41579-019-0158-9. [DOI] [PubMed] [Google Scholar]
- 2.Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venturelli OS, et al. Deciphering microbial interactions in synthetic human gut microbiome communities. Mol Syst Biol. 2018;14:e8157. doi: 10.15252/msb.20178157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster KR, Bell T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr Biol. 2012;22:1845–1850. doi: 10.1016/j.cub.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Helfrich EJN, et al. Bipartite interactions, antibiotic production and biosynthetic potential of the Arabidopsis leaf microbiome. Nat Microbiol. 2018;3:909–919. doi: 10.1038/s41564-018-0200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coyte KZ, Rakoff-Nahoum S. Understanding competition and cooperation within the mammalian gut microbiome. Curr Biol. 2019;29:538–544. doi: 10.1016/j.cub.2019.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner TR, et al. Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 2013;7:2248–2258. doi: 10.1038/ismej.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK. Plant-microbiome interactions: from community assembly to plant health. Nat Rev Microbiol. 2020;18:607–621. doi: 10.1038/s41579-020-0412-1. [DOI] [PubMed] [Google Scholar]
- 10.Müller DB, Vogel C, Bai Y, Vorholt JA. The plant microbiota: Systems-level insights and perspectives. Annu Rev Genet. 2016;50:211–234. doi: 10.1146/annurev-genet-120215-034952. [DOI] [PubMed] [Google Scholar]
- 11.Lugtenberg B, Kamilova F. Plant-growth-promoting Rhizobacteria. Annu Rev Microbiol. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 12.Berendsen RL, Pieterse CMJ, Bakker PAHM. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Innerebner G, Knief C, Vorholt JA. Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl Environ Microb. 2011;77:3202–3210. doi: 10.1128/AEM.00133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shekhawat K, et al. Root endophyte induced plant thermotolerance by constitutive chromatin modification at heat stress memory gene loci. EMBO Rep. 2021;22:e51049. doi: 10.15252/embr.202051049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vorholt JA. Microbial life in the phyllosphere. Nat Rev Microbiol. 2012;10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 16.Bodenhausen N, Bortfeld-Miller M, Ackermann M, Vorholt JA. A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. Plos Genetics. 2014;10:e1004283. doi: 10.1371/journal.pgen.1004283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reisberg EE, Hildebrandt U, Riederer M, Hentschel U. Distinct phyllosphere bacterial communities on Arabidopsis wax mutant leaves. Plos One. 2013;8:e78613. doi: 10.1371/journal.pone.0078613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kniskern JM, Traw MB, Bergelson J. Salicylic acid and jasmonic acid signaling defense pathways reduce natural bacterial diversity on Arabidopsis thaliana . Mol Plant Microbe In. 2007;20:1512–1522. doi: 10.1094/MPMI-20-12-1512. [DOI] [PubMed] [Google Scholar]
- 19.Pfeilmeier S, et al. The plant NADPH oxidase RBOHD is required for microbiota homeostasis in leaves. Nat Microbiol. 2021;6:852–864. doi: 10.1038/s41564-021-00929-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen T, et al. A plant genetic network for preventing dysbiosis in the phyllosphere. Nature. 2020;580:653–657. doi: 10.1038/s41586-020-2185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassani MA, Duran P, Hacquard S. Microbial interactions within the plant holobiont. Microbiome. 2018;6:58. doi: 10.1186/s40168-018-0445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lidicker WZ. Clarification of interactions in ecological systems. Bioscience. 1979;29:475–477. [Google Scholar]
- 23.Rudolf O, Schlechter MM, Mitja NP. Remus-Emsermann. Driving factors of epiphytic bacterial communities: A review. J Adv Res. 2019;19:57–65. doi: 10.1016/j.jare.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbio. 2012;10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 25.Grosskopf T, Soyer OS. Synthetic microbial communities. Curr Opin Microbiol. 2014;18:72–77. doi: 10.1016/j.mib.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blair PM, et al. Exploration of the biosynthetic potential of the Populus microbiome. mSystems. 2018;3:e00045. doi: 10.1128/mSystems.00045-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suda W, Nagasaki A, Shishido M. Powdery mildew-infection changes bacterial community composition in the phyllosphere. Microbes Environ. 2009;24:217–223. doi: 10.1264/jsme2.me09114. [DOI] [PubMed] [Google Scholar]
- 28.Manching HC, Balint-Kurti PJ, Stapleton AE. Southern leaf blight disease severity is correlated with decreased maize leaf epiphytic bacterial species richness and the phyllosphere bacterial diversity decline is enhanced by nitrogen fertilization. Front Plant Sci. 2014;5:403. doi: 10.3389/fpls.2014.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agler MT, et al. hub taxa link host and abiotic factors to plant microbiome variation. Plos Biol. 2016;14:100235. doi: 10.1371/journal.pbio.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Layeghifard M, Hwang DM, Guttman DS. Disentangling interactions in the microbiome: A network perspective. Trends in Microbiology. 2017;25:217–228. doi: 10.1016/j.tim.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faust K. Microbial co-occurrence relationships in the human microbiome. Plos Comput Biol. 2012;8:e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carr A, Diener C, Baliga NS, Gibbons SM. Use and abuse of correlation analyses in microbial ecology. ISME J. 2019;13:2647–2655. doi: 10.1038/s41396-019-0459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vorholt JA, Vogel C, Carlström CI, Müller DB. Establishing causality: Opportunities of synthetic communities for plant microbiome research. Cell Host Microbe. 2017;22:142–155. doi: 10.1016/j.chom.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Bai Y, et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature. 2015;528:364–369. doi: 10.1038/nature16192. [DOI] [PubMed] [Google Scholar]
- 35.Knief C, Frances L, Vorholt JA. Competitiveness of diverse Methylobacterium strains in the phyllosphere of Arabidopsis thaliana and identification of representative models, including M. extorquens PA1. Microb Ecol. 2010;60:440–452. doi: 10.1007/s00248-010-9725-3. [DOI] [PubMed] [Google Scholar]
- 36.Fan J, Crooks C, Lamb C. High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J. 2008;53:393–399. doi: 10.1111/j.1365-313X.2007.03303.x. [DOI] [PubMed] [Google Scholar]
- 37.Carlström CI, et al. Synthetic microbiota reveal priority effects and keystone strains in the Arabidopsis phyllosphere. Nat Ecol Evol. 2019;3:1445–1454. doi: 10.1038/s41559-019-0994-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogel CM, Potthoff DM, Schäfer M, Barandun N, Vorholt JA. Protective role of the Arabidopsis leaf microbiota against a bacterial pathogen. Nat Microbiol. 2021;6:1537–1548. doi: 10.1038/s41564-021-00997-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen I-MA, et al. The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucl Acids Res. 2020;49:751–763. doi: 10.1093/nar/gkaa939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ortiz A, Vega NM, Ratzke C, Gore J. Interspecies bacterial competition regulates community assembly in the C. elegans intestine. ISME J. 2021;15:2131–2145. doi: 10.1038/s41396-021-00910-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goberna M, Verdú M. Predicting microbial traits with phylogenies. ISME J. 2016;10:959–967. doi: 10.1038/ismej.2015.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. Phylogenies and community ecology. Annu Rev Ecol Syst. 2002;33:475–505. [Google Scholar]
- 43.Cahill JF, Kembel SW, Lamb EG, Keddy PA. Does phylogenetic relatedness influence the strength of competition among vascular plants? Perspect Plant Ecol. 2008;10:41–50. [Google Scholar]
- 44.Maherali H, Klironomos JN. Influence of phylogeny on fungal community assembly and ecosystem functioning. Science. 2007;316:1746–1748. doi: 10.1126/science.1143082. [DOI] [PubMed] [Google Scholar]
- 45.Duncan RP, Williams PA. Ecology - Darwin's naturalization hypothesis challenged. Nature. 2002;417:608–609. doi: 10.1038/417608a. [DOI] [PubMed] [Google Scholar]
- 46.Slingsby JA, Verboom GA. Phylogenetic relatedness limits co-occurrence at fine spatial scales: Evidence from the schoenoid sedges (Cyperaceae : Schoeneae) of the Cape Floristic Region, South Africa. Am Nat. 2006;168:14–27. doi: 10.1086/505158. [DOI] [PubMed] [Google Scholar]
- 47.Mayfield MM, Levine JM. Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecol Lett. 2010;13:1085–1093. doi: 10.1111/j.1461-0248.2010.01509.x. [DOI] [PubMed] [Google Scholar]
- 48.Teixeira PJPL, Colaianni NR, Fitzpatrick CR, Dangl JL. Beyond pathogens: microbiota interactions with the plant immune system. Curr Opin Microbiol. 2019;49:7–17. doi: 10.1016/j.mib.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 49.Maier BA, et al. A general non-self response as part of plant immunity. Nat Plants. 2021;7:696–705. doi: 10.1038/s41477-021-00913-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friedman J, Higgins LM, Gore J. Community structure follows simple assembly rules in microbial microcosms. Nat Ecol Evol. 2017;1:0109. doi: 10.1038/s41559-017-0109. [DOI] [PubMed] [Google Scholar]
- 51.Kehe J, et al. Positive interactions are common among culturable bacteria. Sci Adv. 2021;7 doi: 10.1126/sciadv.abi7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindow SE, Brandl MT. Microbiology of the phyllosphere. Appl Environ Microb. 2003;69:1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Remus-Emsermann MNP, et al. Spatial distribution analyses of natural phyllosphere-colonizing bacteria on Arabidopsis thaliana revealed by fluorescence in situ hybridization. Environ Microbiol. 2014;16:2329–2340. doi: 10.1111/1462-2920.12482. [DOI] [PubMed] [Google Scholar]
- 54.Billick I, Case TJ. Higher-order interactions in ecological communities - What are they and how can they be detected. Ecology. 1994;75:1529–1543. [Google Scholar]
- 55.Grilli J, Barabas G, Michalska-Smith MJ, Allesina S. Higher-order interactions stabilize dynamics in competitive network models. Nature. 2017;548:210–213. doi: 10.1038/nature23273. [DOI] [PubMed] [Google Scholar]
- 56.Levine JM, Bascompte J, Adler PB, Allesina S. Beyond pairwise mechanisms of species coexistence in complex communities. Nature. 2017;546:56–64. doi: 10.1038/nature22898. [DOI] [PubMed] [Google Scholar]
- 57.Sundarraman D, et al. Higher-order interactions dampen pairwise competition in the zebrafish gut microbiome. Mbio. 2020;11:e01667-01620. doi: 10.1128/mBio.01667-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morris C. Encyclopedia for life sciences. National Publishing Group; London, UK: 2002. Phyllosphere. [Google Scholar]
- 59.Raaijmakers JM, Mazzola M. Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu Rev Phytopathol. 2012;50:403–424. doi: 10.1146/annurev-phyto-081211-172908. [DOI] [PubMed] [Google Scholar]
- 60.Iversen OJ, Grov A. Studies on lysostaphin - separation and characterization of 3 enzymes. Eur J Biochem. 1973;38:293–300. doi: 10.1111/j.1432-1033.1973.tb03061.x. [DOI] [PubMed] [Google Scholar]
- 61.Recsei PA, Gruss AD, Novick RP. Cloning, sequence, and expression of the lysostaphin gene from Staphylococcus simulans. Proc Natl Acad Sci USA. 1987;84:1127–1131. doi: 10.1073/pnas.84.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kessler E, Safrin M, Abrams WR, Rosenbloom J, Ohman DE. Inhibitors and specificity of Pseudomonas aeruginosa LasA. J Biol Chem. 1997;272:9884–9889. doi: 10.1074/jbc.272.15.9884. [DOI] [PubMed] [Google Scholar]
- 63.Trayer HR, Buckley CE. Molecular properties of lysostaphin, a bacteriolytic agent specific for Staphylococcus aureus. J Biol Chem. 1970;245:4842–4846. [PubMed] [Google Scholar]
- 64.Heymer B, Schmidt WC. Purification and characterization of a Streptomyces albus endo-N-acetylmuramidase lytic for group A and other beta hemolytic Streptococci. Microbios. 1975;12:51–66. [PubMed] [Google Scholar]
- 65.Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 66.Peyraud R, et al. Demonstration of the ethylmalonyl-CoA pathway by using C-13 metabolomics. Proc Natl Acad Sci USA. 2009;106:4846–4851. doi: 10.1073/pnas.0810932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schlesier B, Breton F, Mock HP. A hydroponic culture system for growing Arabidopsis thaliana plantlets under sterile conditions. Plant Mol Biol Rep. 2003;21:449–456. [Google Scholar]
- 68.Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 69.Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3:217–223. [Google Scholar]
- 70.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 71.Pruesse E, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucl Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.RStudio: Integrated development environment for R. R Studio; PBC Boston, MA USA: 2020. [Google Scholar]
- 73.R: A language and environment for statistical computing. R foundation for statistical computing; Vienna, Austria: 2021. [Google Scholar]
- 74.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.vegan: community ecology package v. 2.57 (2020)
- 76.Armenteros JJA, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nature Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 77.Gasteiger E, et al. The proteomics protocols handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]
- 78.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bushnell B. BBMap: short read aligner, and other bioinformatic tools. Available from https://sourceforge.net/projects/bbmap.
- 80.Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 82.Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA gene amplicon sequencing reads and the reads obtained from Leaf245 EMS mutant genome sequencing are available online at the European nucleotide archive (https://www.ebi.ac.uk/ena/browser/) under the accession numbers PRJEB50894 and PRJEB47688, respectively. The SILVA SSU Ref NR database (release 132) can be found under www.arb-silva.de/download/archive. Source data are provided with this article.
The code used for analysis of 16S rRNA gene amplicon data can be found in the package phylloR available on GitHub (https://github.com/MicrobiologyETHZ/phylloR/). No unpublished algorithms were used.