Abstract
Single-cell genomics studies have decoded the immune-cell composition of several human prenatal organs but were limited in understanding the developing immune system as a distributed network across tissues. We profiled nine prenatal tissues combining single-cell RNA sequencing, antigen-receptor sequencing, and spatial transcriptomics to reconstruct the developing human immune system. This revealed the late acquisition of immune effector functions by myeloid and lymphoid cell subsets and the maturation of monocytes and T cells prior to peripheral tissue seeding. Moreover, we uncovered system-wide blood and immune cell development beyond primary hematopoietic organs, characterized human prenatal B1 cells, and shed light on the origin of unconventional T cells. Our atlas provides both valuable data resources and biological insights that will facilitate cell engineering, regenerative medicine, and disease understanding.
The human immune system develops across several anatomical sites throughout gestation. Immune cells differentiate initially from extra-embryonic yolk sac progenitors and subsequently from aorto-gonad-mesonephros-derived hematopoietic stem cells (HSCs), before liver and bone marrow take over as the primary sites of hematopoiesis (1, 2). Immune cells from these primary hematopoietic sites seed developing lymphoid organs such as the thymus, spleen, and lymph nodes, as well as peripheral non-lymphoid organs.
Recent advances in single-cell genomics technologies have revolutionized our understanding of the developing human organs (3–11). However, these studies have focused on one or a few organs, rather than reconstructing the entire immune system as a distributed network across all organs. Such a holistic understanding of the developing human immune system would have far-reaching implications for health and disease including cellular engineering, regenerative medicine, and a deeper mechanistic understanding of congenital disorders affecting the immune system.
Here we present a cross-tissue single-cell and spatial atlas of developing human immune cells across prenatal hematopoietic (yolk sac, liver, and bone marrow), lymphoid (thymus, spleen, and lymph node), and non-lymphoid peripheral organs (skin, kidney, and gut) to provide a detailed characterization of generic and tissue-specific properties of the developing immune system. We generated single-cell RNA sequencing (scRNA-seq) data from yolk sac, prenatal spleen, and skin, and integrated publicly available cell atlases of six additional organs, spanning weeks 4 to 17 post-conception (3, 4, 7, 8, 10, 11). We also generated single-cell γδ T cell receptor (γδTCR) sequencing data and additional αβ T cell receptor (αβTCR) and B cell receptor (BCR) sequencing data. Finally, we integrated the single-cell transcriptome profiles with in situ tissue location using spatial transcriptomics.
This study reveals the acquisition of immune effector functions of myeloid and lymphoid lineages from the second trimester, the maturation of developing monocytes and T cells prior to peripheral tissue seeding, and system-wide blood and immune cell development during human prenatal development. Moreover, we identify, characterize, and functionally validate the properties of human prenatal B1 cells and the origin of unconventional T cells.
Integrated cross-organ map of prenatal cell states in distinct tissue microenvironments
To systematically assess the heterogeneity of immune cell populations across human prenatal hematopoietic organs, lymphoid, and non-lymphoid tissues, we generated scRNA-seq data from prenatal spleen, yolk sac, and skin, which were integrated with a collection of publicly available single-cell datasets from the Human Developmental Cell Atlas initiative (3, 4, 7, 8, 10, 11). In total, our dataset comprised samples from 25 embryos/fetuses between 4 to 17 post-conception weeks (pcw) (Fig. 1A), profiled in 221 scRNA-seq libraries. For 65 of these libraries, paired antigen-receptor sequencing data was available for either αβTCR, γδTCR, or BCR (Fig. 1B). After mapping and preprocessing with a unified pipeline, a total of 908,178 cells were retained after quality control.
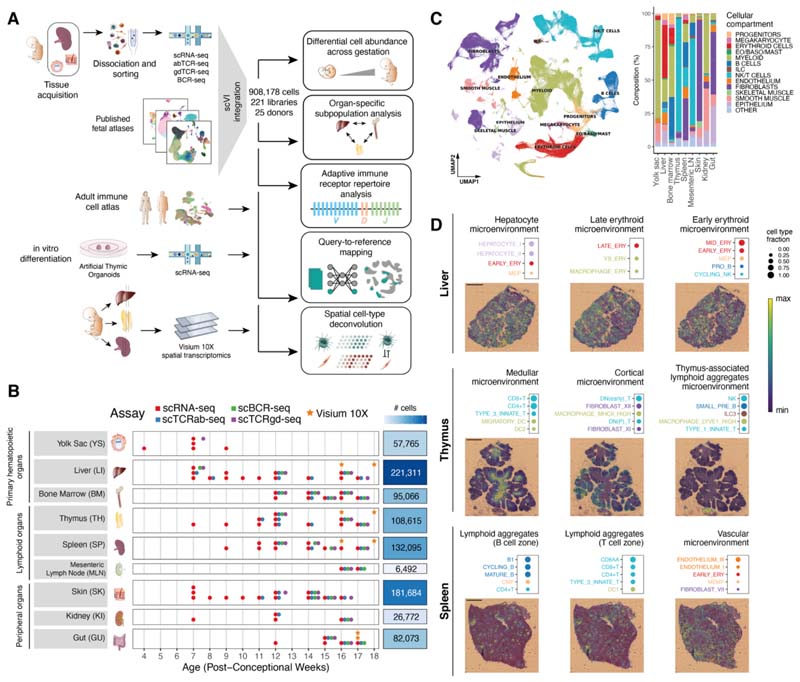
Fig. 1. Cross-tissue cellular atlas of the developing human immune system.
(A) Overview of study design and analysis pipeline. We generated scRNA-seq and scVDJ-seq data from prenatal spleen, yolk sac and skin which were integrated via scVI with a collection of publicly available single-cell RNA-seq datasets. This cell atlas was used for (i) differential abundance analysis across gestation and organs with Milo, (ii) antigen receptor repertoire analysis with scirpy and dandelion, (iii) comparison with adult immune cells and in vitro differentiated cells with scArches and CellTypist, and (iv) spatial cell type deconvolution on Visium 10X data of hematopoietic and lymphoid organs using cell2location. (B) Summary of analyzed samples by gestational stage (x-axis) and organ (y-axis). Colors denote the types of molecular assays performed for each sample. The side bar indicates the total number of cells collected for each organ (after quality control). (C) Left: UMAP embedding of scRNA-seq profiles in prenatal tissues (908,178 cells) colored by broad cellular compartments. Right: bar plot of percentage of cells assigned to each broad compartment for each of the profiled organs. Raw cell proportions are adjusted to account for FACS-based CD45 enrichment. The category “Other” denotes clusters annotated as low-quality cells. (Eo/Baso/Mast: eosinophils/basophils/mast cells; ILC: innate lymphoid cells; NK: natural killer cells). (D) Representative colocalization patterns identified with non-negative matrix factorization of spatial cell type abundances estimated with cell2location. For each annotated microenvironment, we show (top) a dot plot of relative contribution of cell types to microenvironment (dot size) and (bottom) spatial locations of microenvironments on tissue slides, with the color representing the weighted contribution of each microenvironment to each spot. Each scale bar indicates a length of 1 mm.
To facilitate joint analysis of the data, we integrated all libraries using scVI (12), minimizing protocol- and embryo-associated variation (fig. S1A) while retaining differences between organs.
In keeping with previous single-cell atlases of immune cells of prenatal and adult tissues (3, 11, 13), our data captured the emergence of myeloid and lymphoid lineages, as well as closely linked megakaryocytes (MK) and erythroid and non-neutrophilic granulocyte lineages from hematopoietic progenitors (Fig. 1C and figs. S1B to S3). Linking transcriptional phenotypes to paired antigen receptor sequence expression, we paired αβTCR sequences for 28,739 cells, paired γδTCR sequences for 813 cells, and paired BCR sequences for 14,506 cells (fig. S1C).
We repeated dimensionality reduction and clustering on subsets of cells from different lineages and used marker gene analysis and comparison with existing cell labels to comprehensively annotate cell types across tissues. In total, we defined 127 high-quality cell populations (figs. S4 and S5). Cross-tissue integration enabled the identification of cell populations that were too rare to be resolved through analysis of datasets from individual tissues, such as AXL and SIGLEC6-expressing dendritic cells (AS DC) (14) and plasma B cells (fig. S4). To facilitate the rapid reuse of our atlas for the analysis of newly collected samples, we made the weights from trained scVI models available to enable mapping of external scRNA-seq datasets using transfer learning with scArches (15).
To study the spatial localizations of the cell populations in an early hematopoietic tissue and lymphoid organs critical for B and T cell development, we profiled developing liver, thymus, and spleen from two donors at 16 and 18 pcw with spatial transcriptomics (10X Genomics Visium). Using our multi-organ scRNA-seq dataset as a reference, we performed spatial cell-type deconvolution with cell2location (16) to map cells in tissue (fig. S6). We used non-negative matrix factorization (NMF) of the cell-type-abundance estimates in tissue spots to identify microenvironments of colocalized cell types in the profiled tissues in an unbiased manner (Fig. 1D and figs. S7 to S10).
In the developing liver, we recovered expected signatures of tissue-specific parenchymal cells such as hepatocytes. In addition, we observed spatial segregation of early and late erythrocytes, suggesting distinctive hematopoietic zones (Fig. 1D and fig. S8). In the developing thymus, we recovered the localization of cell types in known histological structures. Developing T cells, for example, were largely localized to the thymic cortex, whereas mature T cells were consistently mapped to the thymic medulla. Furthermore, in two of the thymic tissue sections, we observed aggregates of lymphoid tissue (hereafter referred to as thymus-associated lymphoid aggregates). Within these, we mapped B cell subsets, innate lymphoid cells (ILCs), and macrophage subtypes (Fig. 1D and fig. S9). In the developing spleen, most of the tissue was highly vascularized. In addition, within splenic lymphoid aggregates, we were able to distinguish partially overlapping B cell zones and T cell zones (Fig. 1D and fig. S10).
Heterogeneity of prenatal myeloid cells across organs and gestation
We first examined the main compartments of immune cells in our multi-organ dataset to identify gestation-specific and organ-specific variability within cell populations.
The myeloid compartment captured the development from committed myeloid progenitors to neutrophils, monocytes, macrophages, and DCs (fig. S4, G and H). Our cross-tissue analysis distinguished three distinct subsets of monocytes, characterized by a differential distribution between prenatal bone marrow and peripheral tissues and by the expression of CXCR4, CCR2, or IL1B (17). Among macrophages, we identified eight broad macrophage groupings based on their transcriptome profile (fig. S4H): “LYVE1hi”, “iron-recycling”, “MHC class IIhi”, “Kupffer-like” (18), “microglia-like TREM2hi” (19), “osteoclasts” (11, 20), and “proliferating” macrophages. Assigning proliferating cells to the other identified subsets, we observed a high fraction of proliferating macrophages in the yolk sac and within the LYVE1hi subset across organs, suggesting an increased self-renewal potential (fig. S11).
We compared prenatal and adult immune-cell populations by mapping a cross-tissue adult dataset of immune cells (21) onto our prenatal myeloid reference (fig. S12, A and B). We found the transcriptional profiles of DC subsets were conserved between adult and prenatal counterparts (fig. S12C). Adult monocytes were most similar to the IL1Bhi and CCR2hi prenatal populations and no CXCR4hi monocytes in non-lymphoid adult tissues were observed (fig. S13). The majority of adult macrophages clustered separately from the prenatal macrophages, with the exception of erythrophagocytic macrophages (fig. S12, B and C). This population includes macrophages primarily from the spleen and liver that carry out iron-recycling functions (21).
To quantify changes in cellular composition across gestation, we used differential abundance analysis on cell neighborhoods with Milo (Fig. 2A and fig. S14A) (22). This analysis reaffirmed well-known compositional shifts that happen during gestation. For example, myeloid progenitor cells decreased in the liver but increased in the bone marrow, recapitulating the transition from liver to bone marrow hematopoiesis. DCs increased in proportional abundance across multiple tissues as previously described in the liver and bone marrow (3). Notably, for several cell populations, we found that some neighborhoods were enriched and others depleted across gestation, suggesting evolving transcriptional heterogeneity during development. This was especially evident in the macrophage compartment in the skin and liver (Fig. 2A) with a large fraction of neighborhoods overlapping the LYVE1hi and proliferating macrophages enriched in early gestation. Differential expression analysis revealed the upregulation of a proinflammatory gene signature with chemokines and cytokines specific to early stages in all macrophage subtypes across tissues (Fig. 2B, fig. S14B). TNF and NF-κB have been implicated in lymphoid tissue organogenesis (23) and the chemokines noted here have been associated with angiogenesis (24–27). Conversely, a large fraction of neighborhoods within the iron-recycling and MHCIIhi macrophage populations were enriched in later stages of gestation. We found that these subpopulations upregulated genes encoding for immune effector functions (Fig. 2B; fig. S14C; and table S1). In parallel to macrophages, we observed similar transcriptional heterogeneity during gestation in mast cells (Fig. 2A). Specifically, early mast cells in yolk sac, liver, and skin displayed a similar proinflammatory phenotype characterized by expression of TNF and NF-κB subunits, as well as chemokines associated with endothelial cell recruitment (CXCL3, CXCL2, and CXCL8) (26) (fig. S15).
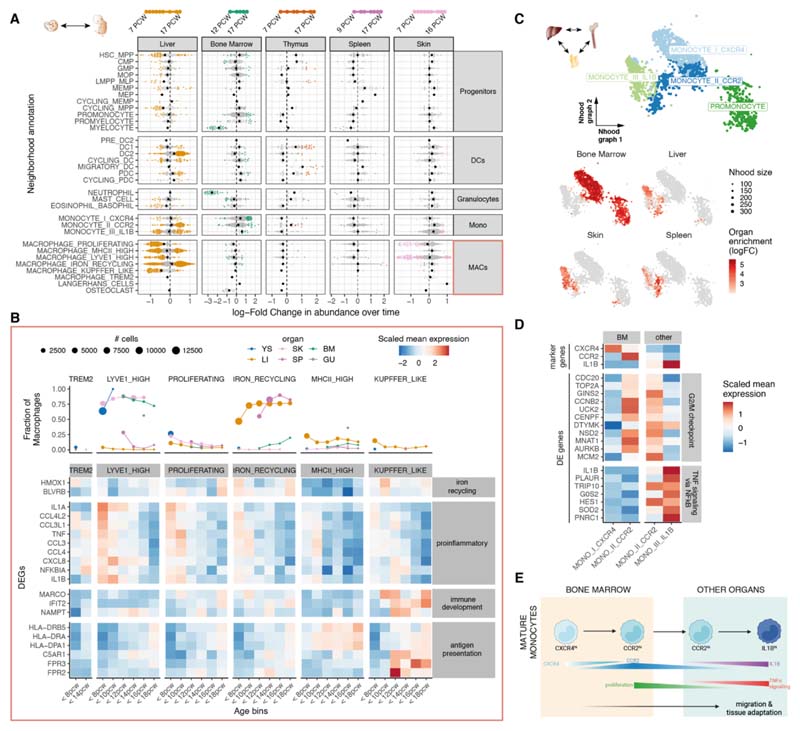
Fig. 2. Myeloid variation across time and tissues.
(A) Beeswarm plot of log-fold change (x-axis) in cell abundance across gestational stages in Milo neighborhoods of myeloid cells. Results from five organs are shown. Neighborhoods overlapping the same cell population are grouped together (y-axis), and colored if displaying significant differential abundance (DA) (SpatialFDR 10%). The black dot denotes the median log-fold change. The top bar denotes the range of gestational stages of the organ samples analyzed. (B) Heat map of average expression across time of a selection of markers of stage-specific macrophage neighborhoods. Mean log-normalized expression for each gene is scaled (z-score). Gestational ages are grouped in 5 age bins. Age bins where less than 30 cells of a given subset were present are not shown. The top panel shows the fraction of all macrophages belonging to the specified macrophage population in each time point and each organ (color). (C) Close-up view of monocytes on Milo neighborhood embedding of myeloid cells (subset from fig. S16). Top: neighborhoods are colored by overlapping cell population. Bottom: neighborhoods displaying significant DA (SpatialFDR 10%) are colored by log-fold change in abundance between the specified organ and all other organs. (D) Mean expression of a selection of differentially expressed genes between CCR2hi monocytes from bone marrow and other organs. Log-normalized expression for each gene is scaled (z-score). We show genes upregulated in bone marrow associated with G2/M checkpoint and genes downregulated in bone marrow associated with TNF signaling (from MSigDB Hallmark 2020 gene sets). (E) Schematic of the proposed process of monocyte egression from the bone marrow mediated by CXCR4 and CCR2 expression: CXCR4hi monocytes are retained in the bone marrow, until they switch to a proliferative state with increased expression of CCR2, mediating tissue egression. CCR2hi monocytes seed peripheral tissues and then mature further to the periphery-specific IL1B expressing subtype.
These findings suggest that early macrophages and mast cells may contribute to angiogenesis, tissue morphogenesis and homeostasis as previously reported (28–30), prior to adopting traditional immunological functions. Notably, acquisition of macrophage antigen-presentation properties (e.g., MHCII upregulation) between 10 and 12 pcw coincided with the expansion of adaptive lymphocytes (fig. S1E) and development of lymphatic vessels and lymph nodes (31).
Differential abundance analysis on cell neighborhoods to test for organ-specific enrichment (fig. S16A) revealed that CXCR4hi monocytes were enriched in bone marrow, and IL1Bhi monocytes in peripheral organs. Among CCR2hi monocytes, we distinguished bone marrow- and peripheral organ-specific subpopulations (Fig. 2C). Bone marrow CCR2hi monocytes expressed proliferation genes, whereas peripheral organ CCR2hi monocytes upregulated IL1B and other TNF-α signaling genes (Fig. 2D; fig. S16B; and table S2). This suggests that a CXCR4hi-to-CCR2hi transition accompanies monocyte egress from the bone marrow to seed peripheral tissues, and CCR2hi monocytes further mature in tissues into IL1Bhi monocytes (Fig. 2, D and E). In mouse bone marrow, interactions between monocyte CXCR4 and stromal cell CXCL12 retain monocytes in situ until CCR2-CCL2 interactions predominate, potentially enabling monocyte egress (17). Here, we observed CXCL12 expression in bone marrow fibroblasts and osteoblasts (fig. S16C). By contrast, the proportion of CXCR4hi monocytes in the developing liver was much lower (fig. S16D), in keeping with reports that alternative mechanisms of monocyte retention and release operate in the murine developing liver (32).
Heterogeneity of prenatal lymphoid cells across organs and gestation
The lymphoid compartment captures the development of B and T cells, together with ILC and natural killer (NK) cell subsets (fig. S4, I to L).
Mapping adult cells onto our prenatal lymphoid reference, NK cells and type 3 ILCs (ILC3) displayed high similarity between adult and prenatal counterparts (fig. S17, A and B). Among adult T cells, naive populations and regulatory T cells (Tregs) closely matched prenatal conventional T cells (CD4+ T, CD8+ T, and Tregs), whereas resident and effector memory T cells did not have a developmental equivalent (fig. S17, C and D), although we cannot exclude the possibility that memory T cells appear after 17 pcw as previously reported (33, 34). We did not find a clear matching between adult T cell subsets and prenatal unconventional T cells (type 1 and type 3 innate T and CD8AA in our annotation). All adult B cell progenitors, plasma B cells, naive B cells and memory B cells had prenatal counterparts, but no adult B cells were transcriptionally similar to prenatal putative B1 cells (fig. S17C).
Differential abundance analysis across gestation identified a broad shift from innate to adaptive immune populations (Fig. 3A and fig. S18A). ILCs and NK cells included cell neighborhoods both enriched and depleted across gestation. Genes involved in inflammatory responses, including TNF signaling, were overexpressed in <12 pcw liver and skin NK cells, although late splenic NK cells also expressed these genes. Conversely, late NK cells across organs overexpressed genes involved in cytokine signaling and granzyme genes (Fig. 3B; fig. S18, B and C; and table S3). As is the case for macrophages, these results suggest the progressive development of immune-effector function by NK cells.
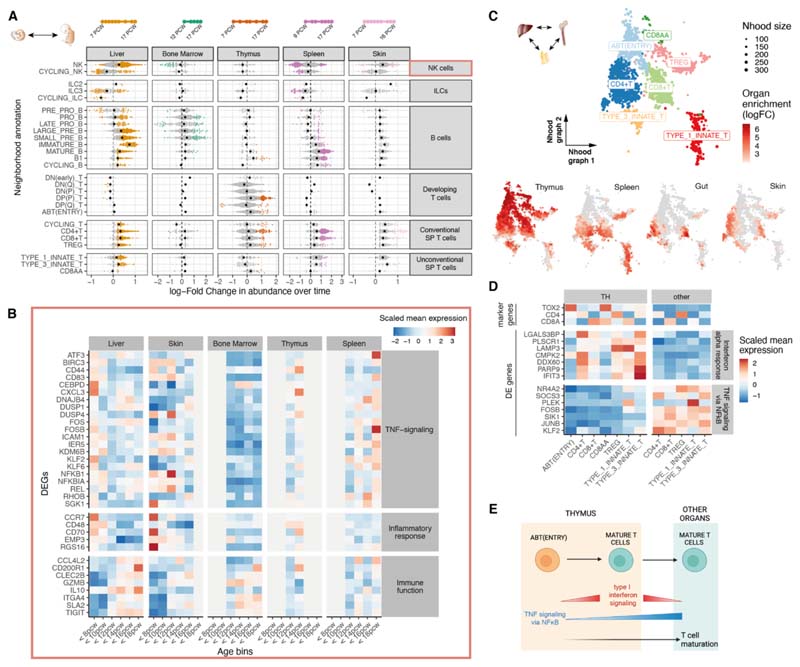
Fig. 3. Lymphoid variation across time and tissues.
(A) Beeswarm plot of log-fold change (x-axis) in cell abundance across gestational stages in Milo neighborhoods of lymphoid cells (as in fig. 2A). (B) Heat map showing average expression across time of a selection of genes identified as markers of early-specific and late-specific NK neighborhoods (as in fig. 2B): NK cells identified in liver and skin before 12 pcw express TNF proinflammatory genes, whereas expression of immune-effector genes such as cytokines, chemokines and granzyme genes increases after 12 pcw. Age bins where less than 30 NK cells were present in a given organ are grayed out. (C) Close-up view of single-positive T cells on Milo neighborhood embedding of lymphoid cells. Each point represents a neighborhood, the layout of points is determined by the position of the neighborhood index cell in the UMAP in fig. S4I. Top: neighborhoods are colored by the cell population they overlap. Bottom: neighborhoods are colored by their log-fold change in abundance between the specified organ and all other organs. Only neighborhoods displaying significant differential abundance (SpatialFDR 10%) are colored. (D) Mean expression of a selection of differentially expressed genes between single-positive T cells from thymus (TH) and other organs. We show genes downregulated in the thymus associated with TNF signaling (using MSigDB Hallmark 2020 gene sets) and genes upregulated in the thymus associated with an IFN-α response. (E) Schematic of the proposed mechanism of thymocyte maturation and egression from thymus mediated by type I interferon signaling and NF-κB signaling.
We next tested for organ-specific cell neighborhoods in the lymphoid compartment (fig. S19A). Although certain populations of mature T cells were exclusively enriched in the thymus (ABT(entry), CD8AA), we found that neighborhoods of conventional and unconventional T cells could be subdivided into a subset enriched in the thymus and other subsets enriched in peripheral organs (Fig. 3C). Thymic mature T cells overexpressed genes involved in interferon (IFN)-α signaling whereas peripheral mature T cells had higher expression of genes associated with TNF and NF-κB signaling (Fig. 3D; fig. S19B; and table S4). Both pathways have been implicated in last stages of functional maturation of murine T cells right before emigration out of the thymus (35, 36). In addition to the increase in type 1 IFN and NF-κB signaling accompanying ABT(entry) to thymic mature T cells, expression of NF-κB-signaling genes continued to increase when mature T cells migrate out to peripheral tissues (Fig. 3E).
System-wide blood and immune cell development
While examining the distribution of various cell types across different organ systems, we were surprised to find that lineage-committed hematopoietic progenitors were present in non-hematopoietic organs. In particular, we detected B cell progenitors in almost all prenatal organs, megakaryocyte/erythroid progenitors in developing spleen and skin, and myeloid progenitors in the thymus, spleen, skin, and kidney (Fig. 4A). By contrast, T cell progenitors were restricted to the thymus, potentially reflecting more stringent niche requirements for T cell development and consistent with the observed absence of T cells in children with congenital athymia (37). This finding suggests that hematopoiesis is not restricted to developing liver and bone marrow between 7-17 pcw (38) and other organs can also support blood and immune cell differentiation during prenatal development.
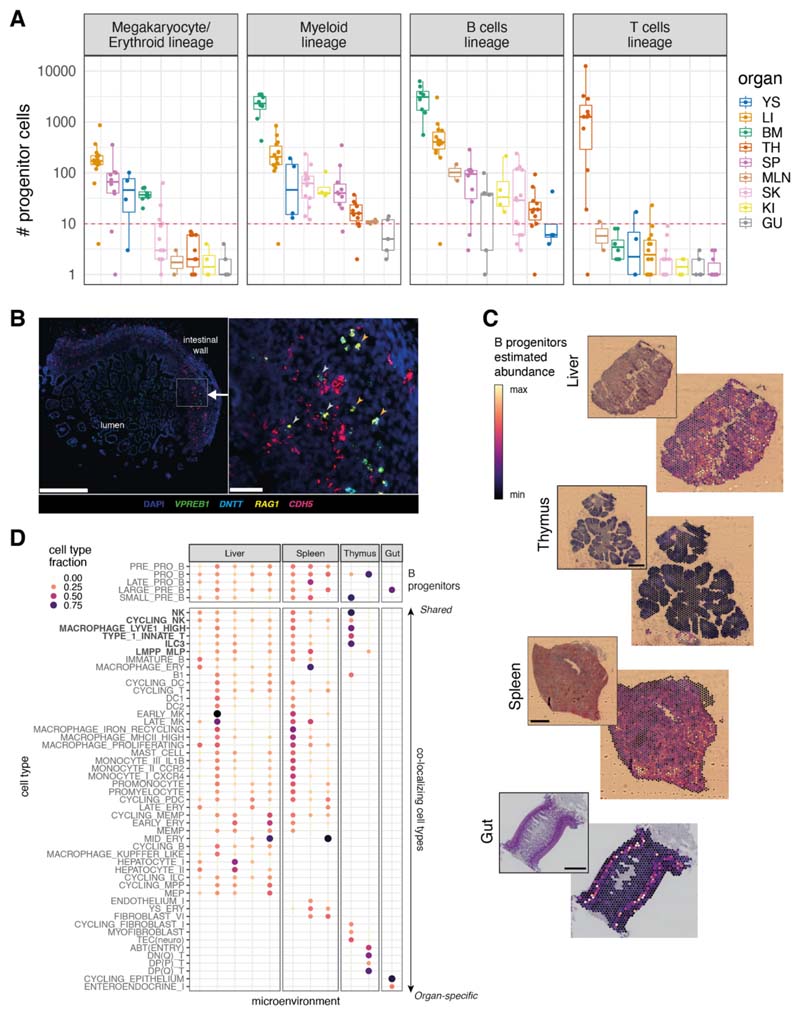
Fig. 4. System-wide blood and immune cell development.
(A) Boxplots of the number of progenitor cells in all donors across organs. Each point represents a donor, color-coded by organ (YS: yolk sac; LI: liver; BM: bone marrow; TH: thymus; SP: spleen; MLN: mesenteric lymph node; SK: skin; GU: gut; KI: kidney). The red dash line marks the threshold of 10 cells for potential technical artefacts. B_prog: B cell lineage progenitors; M/E_prog: megakaryocyte/erythroid progenitors; MYE_prog: myeloid progenitors; T_prog: T cell progenitors. Detailed cell types included in each lineage are shown in table S5. Boxes capture the first-to-third quartile of the cell number and whisks span a further 1.5X interquartile range on each side of the box. (B) Multiplex smFISH staining with DAPI, CDH5 for endothelial cells, and VPREB1, DNTT, RAG1 for B cell progenitors in the human prenatal intestine at 15 pcw. Left panel shows a zoomed-out view with the area of interest boxed in white (scale bar: 500 μm). Right panel shows a detailed view of the area of interest (scale bar: 50 μm). Gray arrows point to B cell progenitors associated with blood vessels and orange arrows point to B cell progenitors away from blood vessels. (C) Scaled sum of abundances of B progenitor cell types estimated with cell2location, shown on representative slides for each organ, with the corresponding H&E staining. Each scale bar represents the length of 1 mm. (D) Cell type contributions to microenvironments containing B cell progenitors in different organs identified with non-negative matrix factorization of spatial cell type abundances estimated with cell2location. The color and the size of the dots represent the relative fraction of cells of a type assigned to the microenvironment.
In addition, across progenitor lineages, cells of different developmental stages were simultaneously present in peripheral organs (fig. S20, A to D). Cell-fate-prediction analysis delineated a continuum of cells between HSCs and different lineage immune cells in multiple organs (fig. S20, C and D), supporting the conclusion that lineage-committed differentiation takes place within peripheral organs.
Single-molecule fluorescence in situ hybridization (smFISH) staining confirmed the existence of lineage-committed progenitors in multiple organs. Cells simultaneously expressing VPREB1, RAG1, and/or DNTT were present in the prenatal gut, spleen, and thymus (Fig. 4B and fig. S21, A to C), consistent with B cell progenitors. Although some B cell progenitors in the prenatal gut were associated with CDH5-expressing blood vessels, many could be detected extravascularly (Fig. 4B), further supporting the conclusion that B cells develop in prenatal peripheral organs. We also validated the presence of megakaryocyte/erythroid lineage progenitors in prenatal spleen and thymus (fig. S22, A to C), and that of myeloid lineage progenitors in prenatal gut and thymus (fig. S23, A to C).
Focusing on B lymphopoiesis given its widespread nature, we used cell2location (16) on Visium spatial transcriptomic data and found B cell progenitors were localized in the submucosa of the gut, in thymus-associated lymphoid aggregates, and proximal to lymphoid aggregates in the spleen (fig. S24B), in addition to their expected presence in the developing liver (Fig. 4C and fig. S24A). The widespread nature of B lymphopoiesis suggests that the cellular environments supporting B cell development are much more widely available than previously thought. Spatial transcriptomic data identified cells colocalizing with B cell progenitors across multiple organs, including ILC3, LYVE1hi macrophages, NK cells, type 1 innate T cells, and LMPP_MLP (see fig. S24C for predicted cell–cell interactions), whereas other colocalizing cell types were organ-specific (Fig. 4D).
Identification of putative prenatal B1 cells
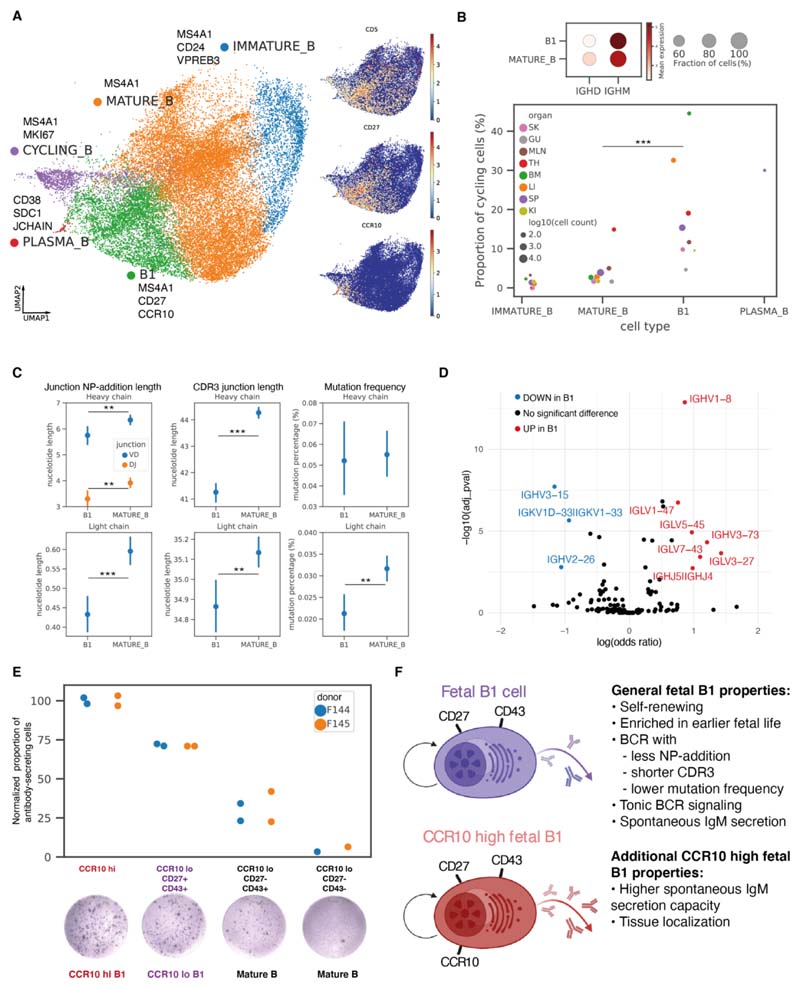
Among prenatal non-progenitor B cells that had productive BCR light chains and low IL7R expression (fig. S25A), we identified immature B, mature B, cycling B, plasma B cells, and putative B1 cells (Fig. 5A and fig. S25B). These putative B1 cells had the highest expression of CD5, CD27, and SPN(CD43), consistent with previously reported markers (39–41), as well as CCR10, a highly-specific marker that was expressed in a subset of B1 cells (Fig. 5A).
Fig. 5. Identification of putative prenatal B1 cells.
(A) Left: Close-up view of non-progenitor B cell populations on UMAP embedding of all lymphoid cells (fig. S4I), with marker genes listed next to each cell type. Right: expression of B1 marker genes on UMAP. (B) Top: dot plot of IGHM and IGHD expressions in B1 and mature B cells, with color of dots representing the mean expression and size representing the fraction of cells expressing the gene. Bottom: cycling cell proportions within each B cell subtype colored by organs, with dot size representing log10(cell count) and only dots with at least 10 cells shown. B1 cells had significantly higher cycling proportions than mature B cells in a logistic regression controlling for donors and organs. (C) Point plots of NP-addition length, CDR3 junction length, and mutation frequency in BCR heavy chains or light chains in B1 (N=2,357) and mature B (N=7,387) cells, with points representing the mean and lines representing 95% confidence intervals. Heavy chain VD and DJ junction NP-addition lengths are only calculated for cells with high-quality D gene mapping (B1: N=615, mature B: N=2,430). Difference in characteristics were tested with linear regressions controlling for donors and organs. (D) Volcano plot summarizing results of BCR heavy and light chain V, J gene segment usage comparison between B1 and mature B cells. The y-axis is the −log10(Benjamini–Hochberg adjusted P-value) and the x-axis is log(odds ratio) computed using logistic regression controlling for donors and organs. (E) Normalized proportions of antibody-secreting cells in different sorted fractions of the ELISpot experiments (raw counts in table S6), colored by donor. Each point represents a reaction well. The proportions of antibody-secreting cells were normalized against the average proportion in CCR10hi wells for each donor to remove donor-specific effects. A representative well image for each sorted fraction is shown on the bottom. (F) Schematic illustration summarizing the features of all human prenatal B1 cells, and additional features specific to CCR10hi prenatal B1 cells.
We next evaluated characteristics typical of murine B1 cells, including self-renewal (42, 43), high IgM and low IgD expression (44), emergence in early development (45), low levels of non-templated nucleotide BCR insertions (46, 47), tonic BCR signaling (39), and spontaneous antibody secretion (42).
With regards to B1 cell self-renewal, we calculated the percentage of cycling cells (as indicated by non-zero MKI67 expression) within immature B, mature B, B1, and plasma B cells, respectively (Fig. 5B and fig. S26A). The proportion of cycling B1 cells was significantly higher than cycling mature B cells, consistent with their capacity for self-renewal. B1 cells expressed lower levels of IGHD and higher levels of IGHM compared to mature B cells (Fig. 5B). Moreover, the highest frequency of B1 cells was found in early embryonic stages. These were gradually replaced by other subsets of non-progenitor B cells over time. The ratio of B1 to mature B cells showed a general decrease from the first to second trimester across most organs except the thymus (fig. S26B) where B1 cells persisted, consistent with a previous report of a shared phenotype between thymic B cells and B1 cells (48).
Analysis of non-templated nucleotide insertions in the BCR showed that both N/P-additions and CDR3 junctions in heavy and light chains were shorter in B1 cells compared to mature B cells (Fig. 5C). Moreover, a lower mutation frequency was observed in the light chains of the B1 cells compared to that in mature B cells, and the average mutation frequency was lower than that observed in adult B cells (21, 49). We next examined the V(D)J usage within different B cell subtypes along the developmental path (fig. S26C). Prenatal B1 and mature B cells both exhibited a varied BCR repertoire with minimal clonal expansion (fig. S26D) and had differing preferential usage of V(D)J segments (Fig. 5D).
Our putative B1 cells showed features of tonic BCR signaling, with higher B cell activation scores (fig. S26E), as well as higher transcription factor (TF) activity in the TNF-α and NF-κB signaling pathway (fig. S26F), which is downstream of BCR signaling (50), compared with mature B cells.
We assessed spontaneous antibody secretion capacity in B1 cells by flow-sorting B cell subsets, (fig. S26G) and assessing spontaneous IgM secretion by enzyme-linked immune absorbent spot (ELISpot) assay. The normalized antibody-secreting spot counts were higher in the two B1 fractions than the two mature B fractions, with the CCR10hi B1 fraction showing the highest spot counts (Fig. 5E). scRNA-seq of the sorted B cell fractions on a different sample using the same gating strategy further confirmed that the two sorted B1 fractions were indeed B1 cell-enriched compared to the mature B fractions (fig. S26H). We also explored the potential role of CCR10 in prenatal B1 cells and observed the expression of one of its ligands CCL28 in bone marrow stroma (chondrocyte and osteoblast), gut epithelium, and keratinocytes and melanocytes in skin (fig. S26I). Thus, CCR10 may play a role in the tissue localization of prenatal B1 cells.
Overall, our scRNA-seq, paired V(D)J sequencing data as well as the functional assay we have performed provide an extended characterization of human prenatal B1 cells (Fig. 5F).
Human unconventional T cells are trained by thymocyte-thymocyte selection
The mature T cell compartment consisted of conventional T cells (CD4+ T, CD8+ T and Tregs) and unconventional T cells. The origin of the latter in humans is poorly understood. Unconventional T cells expressed the key innate marker ZBTB16 (PLZF) (fig. S27A) (51) and could be further separated into three different subtypes: RORC- and CCR6- expressing type 3 innate T cells; EOMES- and TBX21- expressing type 1 innate T cells; and PDCD1- expressing and thymus-restricted CD8AA cells (figs. S2 and S4L), corresponding, respectively, to TH17-like cells, NKT-like cells, and CD8αα+ T cells (7).
The proportions of unconventional T cells among all mature T cells exhibited a decreasing trend from 7-9 pcw to 10-12 pcw across most of the organs surveyed here (fig. S27B). Type 1 and 3 innate T cells were almost negligible in postnatal thymus, whereas CD8AA T cell abundance rebounded in pediatric age groups before a further decline in adulthood (fig. S27B). Thus, type 1 and 3 innate T cells, but not CD8AA, appear to be developmental-specific unconventional T cells.
Spatially, we found that mature T cells segregated into two microenvironments in the thymic medulla (fig. S27C). Conventional CD4+ T and CD8+ T cells colocalized with mTECs close to the inner medulla, whereas CD8AA and type 1 innate T cells colocalized with type 1 DCs (DC1) near the cortico-medullary junction (fig. S27, D and E). Treg and type 3 innate T cells were located within both microenvironments. Thus, in contrast to conventional T cells, CD8AA and type I innate T cells likely undergo distinct negative selection processes mediated by DCs rather than mTECs and may also be involved in DC activation as previously suggested (7).
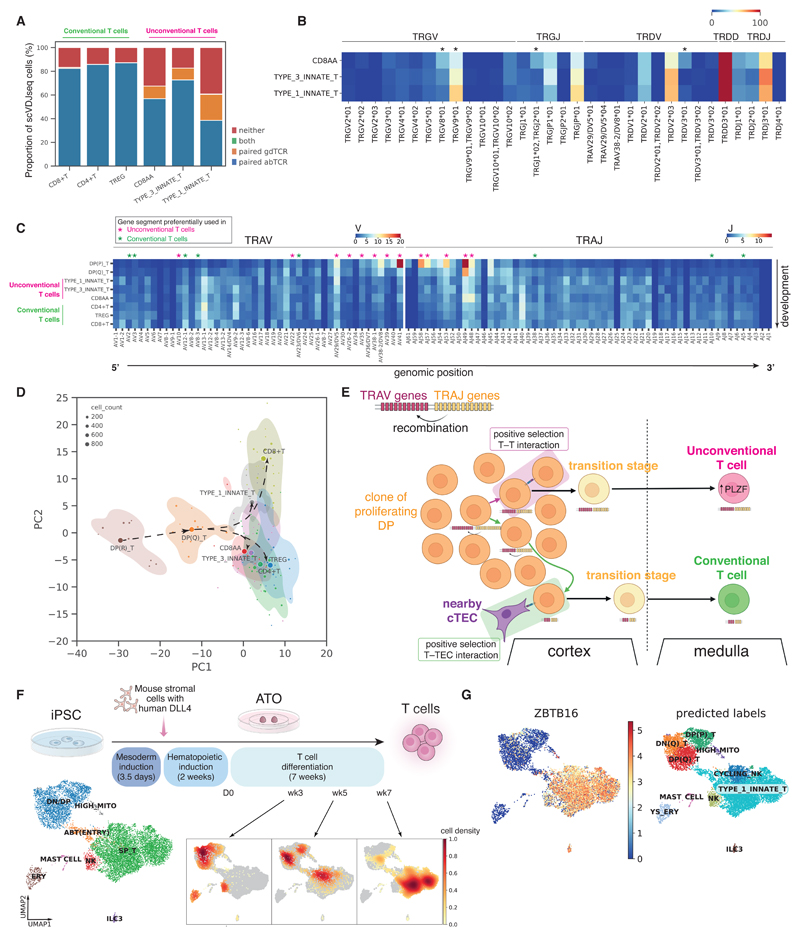
Single-cell sequencing of γδTCR and αβTCR was performed on a subset of samples to characterize antigen-receptor repertoires in unconventional T cells (Fig. 1B). The vast majority of unconventional T cells expressed paired αβTCR but some of these cells expressed paired γδTCR (Fig. 6A). The majority of the γδT cells expressed TRGV9 and TRDV2 (Fig. 6B), consistent with previous reports (52, 53). However, there was also a large proportion of γδT cells, particularly those of CD8AA and type 3 innate T cell subtypes, expressing TRGV8 or TRGV10 instead (Fig. 6B). Thus, the γδTCR showed a relatively restricted repertoire and substantial clonal expansion (fig. S28B).
Fig. 6. Deep characterization of human unconventional T cells.
(A) Proportions of cells expressing paired γδTCR, paired αβTCR, or both or neither. The proportions were calculated over cells that have had both single-cell αβTCR and γδTCR sequencing. The cells that expressed neither paired αβTCR nor paired γδTCR could be due to dropouts in single-cell TCR sequencing as over 50% of these contained orphan VDJ or VJ chains of αβTCR or γδTCR (fig. S28A). (B) Heat map showing the percentage of each γδTCR gene segment present in different T cell subtypes. Differential usage between cell subtypes was computed using the chi-squared test and gene segments with Benjamini–Hochberg adjusted P-values < 0.05 are marked with *. (C) Heat map showing the proportion of each TCRα gene segment present in different T cell subtypes. The gene segment usage in unconventional T cells and conventional T cells was compared using logistic regression, controlling for donors and organs. Gene segments with Benjamini–Hochberg adjusted P-value<0.05 are marked with magenta * (for preferential usage in unconventional T cells) and green * (for preferential usage in conventional T cells). (D) PCA plot summarizing TRAV, TRAJ, TRBV, TRBJ gene segment usage proportion in different T cell subtypes. Each dot represents a sample of at least 20 cells, with its size representing the cell count. The centroid of each cell type is shown as a filled circle, and 80% confidence contours are shown around the centroids. Arrows illustrate the proposed developmental trajectories. (E) Schematic illustration showing the T–T training origin of unconventional T cells in contrast to the T–TEC training origin of conventional T cells. (F) Top: schematic showing the experimental set-up of T cell differentiation from iPSCs in ATOs. Bottom left: UMAP visualization of different cell types in the ATO. Bottom right: density plots of cells from each time point over UMAP embedding. (G) Left: predicted annotations from logistic regression overlaid on the same UMAP plot as in (F); right: ZBTB16 expression pattern overlaid onto the same UMAP plot.
Prenatal unconventional T cells expressed a varied αβTCR repertoire (Fig. 6C and fig. S28C) with minimal clonal expansion (fig. S28D), unlike well-described unconventional T cell subsets (e.g., type I NKT and mucosal-associated invariant T (MAIT) cells) in adults (54). V-J gene usage in TCRα was previously observed to have a strong association with T cell developmental timing (7, 55). Specifically, double-positive (DP) T cells tend to use proximal TRAV, TRAJ gene segments, whereas mature T cells tend to use more distal pairs, governed by the processive depletion of proximal segments in V-J gene recombination (55). The V-J gene usage of αβTCR-expressing unconventional T cells lies between that of DP cells and conventional T cells, shown by the more proximal gene usage in unconventional T cells (Fig. 6C) and the principal component analysis (PCA) of TCR repertoire (Fig. 6D). This suggests that unconventional T cells are developmentally closer to DP cells (Fig. 6E) and undergo fewer recombinations before positive selection.
Previous studies have suggested that these PLZF-expressing unconventional T cells may originate from positive selection on neighboring T cells (51, 56–58) in contrast to conventional T cells arising from positive selection on cortical thymic epithelial cells (cTECs). Post β-selection, DP T cells undergo proliferation prior to recombination of TCRα (7, 59, 60). Each DP cell is hence surrounded by several neighboring DP cells from the same clone. It is therefore plausible that it requires less physical migration and hence is quicker for a DP T cell to receive positive signaling from a neighboring DP T cell rather than having to migrate to meet a nearby cTEC. Thus, the fact that unconventional T cells have a more similar TCR usage to DP cells agrees with the thymocyte–thymocyte (T–T) origin hypothesis.
To test our hypothesis for T–T-mediated selection of unconventional T cells, we differentiated induced pluripotent stem cells (iPSCs) into mature T cells using the artificial thymic organoid (ATO) (61). Importantly, there were no human thymic epithelial cells present in the ATO system (Fig. 6F). scRNA-seq analysis of differentiated cells harvested at weeks 3, 5, and 7 from two iPSC lines confirmed that the in vitro culture system recapitulated T cell development from double-negative (DN) and DP, to ABT(ENTRY) then to single-positive mature T cells (SP_T) (Fig. 6F, fig. S29A to C). SP_T cells differentiated in vitro were dominated by ZBTB16-expressing unconventional T cells (Fig. 6G). Both label transfer (Fig. 6G) and similarity scoring on merged embeddings (fig. S29D) showed that the in vitro SP_T were most similar to in vivo type 1 innate T cells. Thus, our in vitro experiments support the T–T origin hypothesis of unconventional T cells.
Discussion
Our study provides a comprehensive single-cell dataset of the developing human immune system, spanning over 900,000 single-cell profiles from nine tissues, encompassing over 100 cell states. Compared to previous multi-organ developmental atlases (9), we increased coverage of developmental organs and gestation stages, sequencing depth, and generated paired BCR, αβTCR, and γδTCR datasets. Moreover, we demonstrate the utility of scRNA-seq reference to delineate tissue organization and cellular communication in spatial transcriptomics, providing a proof-of-concept study of the localizations of immune cells across prenatal tissues. Our pre-processed data and pre-trained models (scVI and CellTypist models) will facilitate alignment of new data to our dataset and streamline future expansion and analysis of human developmental atlases.
Our cross-organ analysis reveals several important biological phenomena. First, human macrophages, mast cells and NK cells transcriptomically acquire immune effector functions between 10 and 12 pcw. Their transcriptomic signatures prior to this time-point suggest a role in tissue morphogenesis, in line with previous suggestion for murine macrophages (62) and may explain why these cells appear in early development. The coincidental development of the lymphatic system around 12 pcw (31) raises the possibility of its potential role in initiating this transcriptional switch. Second, there are conserved processes of proliferation and maturation for monocytes and T cells prior to their migration from the bone marrow and thymus respectively into peripheral tissues.
Third, in contrast to the previous dogma of hematopoiesis being restricted to the yolk sac, liver, and bone marrow during human development, system-wide blood and immune cell development takes place in peripheral organs, although at varying extents in different lineages. It is possible that hematopoiesis is supported to varying levels in prenatal organs, including the adrenal gland (9), prior to the onset of functional organ maturation as exemplified by the fetal liver, which transitions from a hematopoietic to a metabolic organ. The potential for other peripheral organs to support hematopoiesis is evidenced by the re-emergence of extramedullary hematopoiesis in adults, primarily in pathological settings (63–66), as well as the recent description of B lymphopoiesis in murine and non-human primate meninges (67–69).
Finally, this work identifies and functionally validates the properties of human prenatal innate-like B and T cells and provides an extensive characterization of human B1 cells. Our in vivo αβTCR V(D)J usage patterns and in vitro T cell differentiation data proposes T–T selection underpinning unconventional T cell development. Further studies are required to establish if B1 cells arise from different progenitors (lineage model) (70–72) or from the same progenitors but with different signaling (selection model) (73, 74), similar to the conventional and unconventional T cell model proposed here. Notably, both innate-like B and T cells were abundant during early development and their precise role at this developmental time-point warrants further investigations. Their reported debris-removal (41, 42), antigen-reactivity (41, 42, 54), and regulatory functions (42) may confer these prenatal innate-like B and T cells with tissue-homeostatic and important immunological roles.
In summary, this comprehensive atlas of the developing human immune system provides valuable resources and biological insights to facilitate in vitro cell engineering, regenerative medicine, and enhance our understanding of congenital disorders affecting the immune system.
Materials and Methods
A more detailed version of the materials and methods is provided in the Supplementary Materials.
Tissue acquisition and processing
Human developmental tissue samples (4 to 17 pcw, metadata in table S7) used for this study were obtained from the MRC–Wellcome Trust-funded Human Developmental Biology Resource (HDBR; http://www.hdbr.org) with written consent and approval from the Newcastle and North Tyneside NHS Health Authority Joint Ethics Committee (08/H0906/21+5). All tissues were digested into single-cell suspensions with 1.6 mg/ml of type IV collagenase (Worthington).
Single-cell RNA sequencing experiment
Dissociated cells were stained with anti-CD45 antibody and DAPI prior to sorting. For scRNA-seq experiments, either Chromium single cell 3′ reagent kit or Chromium single cell V(D)J reagent kits from 10X Genomics were used. Unsorted, or DAPI–CD45+, or DAPI–CD45– FACS-isolated cells were loaded onto each channel of the Chromium chip. Single-cell cDNA synthesis, amplification, gene expression (GEX) and targeted BCR and TCR libraries were generated. Targeted enrichment for γδTCR was performed following the TCR enrichment protocol from 10X with customized primers (table S8) (75). Sequencing was performed on the Illumina Novaseq 6000 system. The gene expression libraries were sequenced at a target depth of 50,000 reads per cell using the following parameters: Read1: 26 cycles, i7: 8 cycles, i5: 0 cycles; Read2: 91 cycles to generate 75-bp paired-end reads. BCR and TCR libraries were sequenced at a target depth of 5000 reads per cell.
ATO cell cultures
We followed the PSC-ATO protocol as previously described (61) (more details in the Supplementary Materials). Two iPSC lines HPSI0114i-kolf_2 (Kolf) and HPSI0514i-fiaj_1 (Fiaj) obtained from the Human Induced Pluripotent Stem Cell initiative (HipSci: www.hipsci.org) collection were used.
Visium
OCT-embedded freshly frozen samples (table S9) were used for 10X Genomics Visium. All tissues were sectioned with a thickness of 15 μm. Optimal 18-min permeabilization was selected for fetal spleen and liver, whereas a 24-min permeabilization was used for fetal thymus. The spatial gene expression library was then generated following the manufacturer's protocol. All images for this process were acquired with a Zeiss AxioImager (Carl Zeiss Microscopy) and a 20X air objective (0.8 NA) using either fluorescence (Zeiss Axiocam 503 monochrome camera) for optimization or brightfield mode (Zeiss Axiocam 105 color camera) for H&E imaging. ZEN (Blue edition) v.3.1 was used for acquisition and stitching of the image tiles.
Single-molecule fluorescence in situ hybridization (smFISH)
smFISH was performed on thymus, spleen, and gut sections, using the RNAScope 2.5 LS multiplex fluorescent assay (ACD, Bio-Techne) on the automated BOND RX system (Leica).
Slides were stained for DAPI (nuclei) and three or four probes of interest, with fluorophores atto 425, opal 520, opal 570, and opal 650. Positive and negative control probes were used to optimize staining conditions for all tissues.
For fetal gut and spleen, OCT-embedded freshly frozen 10 μm-thick sections were pretreated offline for 15 min with chilled 4% paraformaldehyde and dehydrated through an ethanol series (50%, 70%, 100%, and 100% ethanol), before processing on the Leica BOND RX with protease IV for 30 min at room temperature. The sections were imaged on a Perkin Elmer Opera Phenix High Content Screening System (16-bit sCMOS camera, PerkinElmer) with a 20X water objective (High NA, PerkinElmer). Due to high levels of endogenous autofluorescence, we imaged one of the spleen sections (fig. S21A) with a confocal microscope (Leica SP8) with a 40X 1.3NA oil immersion objective and SP8 Leica HyD and PMT detectors.
Due to the high cellular density in thymic sections, we used 3 μm-thick FFPE sections that were treated on the Leica Bond RX with epitope retrieval 2 for 15 min at 95°C and protease III for 15 min at 40°C. Imaging was performed on an Operetta CLS High Content Screening System (16-bit sCMOS camera, PerkinElmer) with a 40X water objective (High NA, PerkinElmer) and 2-μm z-steps.
scRNA-seq analysis
Preprocessing
The gene expression data was mapped with cellranger 3.0.2 to an Ensembl 93 based GRCh38 reference (10X-distributed 3.0.0 version). Ambient RNA was removed with cellbender v0.2.0 (76). Low-quality cells were filtered out (minimum number of reads = 2000, minimum number of genes = 500, Scrublet (v0.2.3) (77) doublet detection score <0.4). We identified possible maternal contamination using the souporcell pipeline for genotyping (v.2.4.0) (78) (see Supplementary Materials for details).
Data integration and annotation
Data normalization and preprocessing were performed using the Scanpy workflow (v1.8.1) (79). We normalized raw gene read counts by sequencing depth in each cell (scanpy.pp.normalize_per_cell, with parameters counts_per_cell_after=10e4) and performed ln(x) + 1 transformation. We then selected highly variable genes (HVG) for joint embedding by dispersion (scanpy.pp.highly_variable_genes with parameters min_mean = 0.001, max_mean = 10). We performed dimensionality reduction and batch correction using the scVI model (12) as implemented in scvi-tools (v0.14.5) (80), considering 10X chemistry (5′ and 3′) and the donor ID for each cell as the technical covariates to correct for (training parameters: dropout_rate = 0.2, n_layers = 2). The model was trained on raw counts of the 7500 most highly variable genes, excluding cell cycle genes and TCR/BCR genes (7) with 20 latent dimensions. To verify conservation of biological variation after integration, we collected and harmonized the available cell type labels from the published datasets (66% of cells) and quantified the agreement between labels across different datasets in the cell clusters identified post-integration, using the normalized mutual information (NMI) score, as implemented in scikit-learn (81). Unless otherwise specified, cell clustering was performed using the Leiden algorithm (82) (resolution = 1.5, n_neighbors = 30). To verify robustness to the choice of integration method, we performed in parallel integration using BBKNN (83) as previously described (7) (fig. S30A). We found that clustering post-integration both with scVI and BBKNN was consistent with previous annotations (fig. S30B).
To annotate fine cell populations across tissues, we clustered cells in the scVI latent space and preliminarily assigned cells to broad lineages, using expression of marker genes and previous annotations. For each broad lineage we repeated scVI integration and clustering as described above and defined further subsets (see hierarchy in fig. S5). Leiden clusters for the highest resolution subsets (stroma, megakaryocyte/erythroid, progenitors, lymphoid, myeloid) were annotated manually, using marker panels shown in fig. S4 (see Supplementary Materials for a more detailed description of annotation strategy). We verified that refined annotations were highly consistent with unsupervised clustering post-integration on the full dataset both with scVI and BBKNN (fig. S30C).
After full annotation, 23,156 cells (2.5% of total) were assigned to low quality clusters (doublet clusters, maternal contaminants clusters and clusters displaying a high percentage of reads from mitochondrial genes).
Differential abundance analysis
We tested for differences in cell abundances associated with gestational age or organ using the Milo framework for differential abundance testing (22), with the python implementation milopy (https://github.com/emdann/milopy). A more detailed description of this analysis can be found in the Supplementary Methods.
Briefly, we subsetted the dataset to cells from libraries obtained with CD45+ FACS, CD45– FACS or no FACS, excluding FACS-isolated samples for which we were not able to recover the true sorting fraction quantification. In total, we retained 228,731 lymphoid cells and 214,874 myeloid cells. To further minimize the differences in cell numbers driven by FACS efficiency, we calculated a FACS correction factor for each sample to use as confounding covariate in differential abundance testing (fig. S32, see Supplementary Methods). We constructed a KNN graph using similarity in the scVI embedding (k = 30 for test across gestation, k = 100 for test across tissues) and assigned cells to neighborhoods (milopy.core.make_nhoods, parameters: prop = 0.05). We then counted the number of cells belonging to each sample in each neighborhood (milopy.core.count_cells). We assigned each neighborhood a cell type label based on majority voting of the cells belonging to that neighborhood. We assigned a “Mixed” label if the most abundant label is present in less than 50% of cells within that neighborhood.
To test for differential abundance across gestational age, we divided the sample ages into six equally sized bins (bin size = 2 pcw) and excluded samples from organs where less than three consecutive age bins were profiled (yolk sac, mesenteric lymph node, kidney, and gut). We model the cell count in neighborhoods as a negative binomial generalized linear model, using a log-linear model to model the effects of age on cell counts, while accounting for FACS correction factor and the total number of cells over all the neighborhoods. We control for multiple testing using the weighted BH correction as described in (22). To detect markers of early-specific neighborhoods (SpatialFDR < 0.1, logFC < 0) and/or late-specific neighborhoods (SpatialFDR < 0.1, logFC > 0) in cell type c and organ o, we tested for differential expression between cells from organ o assigned to the significant neighborhoods labeled as cell type c and cells belonging to all other neighborhoods labeled as cell type c. We used the t test implementation in scanpy (scanpy.tl.rank_genes_groups, method = "t-test_overestim_var"). Genes expressed in > 70% of tested cells were excluded. We considered genes as significantly overexpressed (i.e., markers) if the differential expression logFC > 1 and FDR < 0.1%. Gene set enrichment analysis was performed using the implementation of the EnrichR workflow (84) in the python package gseapy (https://gseapy.readthedocs.io/). The list of significantly overexpressed genes for all organs and cell types where differential expression testing was carried out can be found in tables S1 and S3.
To test for differential abundance between organs, we modeled the cell count in neighborhoods as above, using a log-linear model to model the effects of organ on cell counts, while accounting for FACS correction factor, library prep protocol, and the total number of cells over all the neighborhoods. We considered the neighborhoods where and SpatialFDR < 0.01 as cell subpopulations that show organ-specific transcriptional signatures.
Having identified a subset of neighborhoods overlapping a cell type that were enriched in a certain organ, we performed differential expression (DE) analysis between these cells and cells from the same cell type (see Supplementary Methods for a more detailed description). Briefly, we first aggregated single-cell expression profiles into pseudo-bulk expression profiles for each cell type and c sample s (as recommended by (85, 86)).
We then model the mRNA counts of gene g in pseudobulk p by a NB-GLM:
The expected count value μn,p is given by the following log-linear model
We estimated the log-fold change in expression in a given cell type for organ using the quasi-likelihood method (87) implemented in the R package glmGamPoi (85). We used the estimated logFC from the test on a set of control cell types (where we don't expect to see organ-specific differences) to filter out genes where differential expression is driven by technical differences in tissue processing. We provide the full results for the differential expression analysis between organs in mature T cells and monocytes in tables S2 and S4.
TCR analysis
Single-cell αβTCR sequencing data was mapped with cellranger-vdj (v.6.0.0). The output file filtered_contig_annotations.csv was used and analyzed with scirpy (v.0.6.0) (88). Single-cell γδTCR sequencing data was mapped with cellranger-vdj (v.4.0.0). All contigs deemed high-quality were selected, and re-annotated with igblastn (v.1.17.1) against IMGT reference sequences (last downloaded: 01/08/2021), via a workflow provided in dandelion (v0.2.0) (89) (https://github.com/zktuong/dandelion). The output file all_contig_dandelion.tsv was used and analyzed with scirpy (v0.6.0).
BCR analysis
Single-cell BCR data was initially processed with cellranger-vdj (v.6.0.0). BCR contigs contained in all_contigs.fasta and all_contig_annotations.csv were then processed further using dandelion (89) singularity container (v.0.2.0). BCR mutation frequencies were obtained using the observedMutations function in shazam (v.1.0.2) (90) with default settings.
B cell activation scoring
Gene Ontology B Cell Activation gene list was downloaded from Gene Set Enrichment Analysis website (http://www.gsea-msigdb.org/gsea/msigdb/genesets.jsp). Cells were scored according to expression values of all genes in this gene list apart from three genes that were not present in the dataset using scanpy.tl.score_genes() function.
Transcription factor activity inference
We used the DoRothEA Python package (v.1.0.5) (91) to infer TF activities in B1 and mature B cells. TFs that had higher activities (positive “meanchange”) in B1 cells were then ranked according to their adjusted P-values and only top 25 TFs are shown in fig. S26F.
Cell—cell interaction analysis
We used the CellPhoneDB Python package (v.3.0) (92, 93) to infer cell–cell interactions. The scRNA-seq dataset was split by organ and cell types with fewer than 20 cells in a given organ were filtered out. CellPhoneDB was run separately to infer cell–cell interactions in each organ, using default parameters. To explore cell–cell interactions between B cell progenitors and colocalizing cell types (fig. S24D), we aggregated the interactions predicted between each colocalizing cell type, by averaging the means and using the minimum of the P-values. We then filtered for the ligand-receptor pairs that were significant (P<0.05) across all three organs of liver, spleen and thymus, and ranked by the maximum aggregated means. Only the top 60 ligand-receptor pairs are shown in fig. S24C.
Query-to-reference mapping
We mapped query data to our prenatal data embeddings using online update of the scVI models following the scArches method (15), as implemented in the scvi-tools package (80). The model was trained for 200 epochs and setting weight_decay = 0, to ensure that the latent representation of the reference cells remained exactly the same. Reference genes missing in the query were set to 0, as recommended in (15). To generate a joint embedding of query and reference cells, we concatenated the latent dimensions learnt for query cells to the latent dimensions used for the reference embedding and computed the KNN graph and UMAP as described above. To assess that the mapping to the developmental reference conserves biological variation while minimising technical variation in the query data, we compared query cell type labels and batch labels with clusters obtained from Leiden clustering on the learnt latent dimensions, using the Normalized Mutual Information score (see fig. S33 for mapping of adult query data).
Annotation prediction using CellTypist
We used CellTypist v.0.1.9 Python package (21) to perform annotation prediction with logistic regression models. For prediction on cycling B cells, the rest of the non-progenitor B cells, including immature B, mature B, B1, and plasma B cells were used as training dataset. Default parameters were used for model building and prediction was made without majority voting for accurate enumeration of predicted B cell subtypes within cycling B cells.
Comparison with human adult immune cells
Single-cell RNA-seq data from adult immune cells was generated and preprocessed as described (21). The dataset including cell type annotations were provided by the authors. We mapped 264,929 adult lymphoid cells to the lymphoid embeddings of our developmental dataset and 54,047 adult myeloid cells to our myeloid embedding. In order to use cell annotations in our developmental dataset to predict adult cell types in the joint embedding, we adapted the KNN-classifier approach described in (15) and we calculated the similarity to prenatal cells labeled taking the Euclidean distance in the joint embedding, weighted by a Gaussian kernel.
Blood and immune cell progenitors scRNA-seq data analysis
For the cell-fate-prediction analysis shown in fig. S20, C and D, we used the Palantir method as implemented in CellRank (94, 95). Briefly, from the scVI embedding on all immune cells (fig. S20A), we selected cells belonging to progenitor populations and computed a KNN graph on scVI latent dimensions on these cells (k=30). Then transition probabilities were calculated using the ConnectivityKernel in the cellrank package. We computed coarse-grained macrostates with Generalized Perron Cluster Cluster Analysis, setting the number of macrostates to the number of annotated progenitor cell populations. We manually set the four target terminal states for each lineage (small pre B cells, DN(Q) T cells, early MKs, and promonocytes) and computed the probability of each cell to transition to one of the four terminal states. The fate simplex visualization in fig. S20, C and D, was generated using the function cellrank.pl.circular_projection.
Artificial thymic organoids scRNA-seq data analysis
Raw scRNA-seq reads were mapped with cellranger 3.0.2 with combined human reference of GRCh38.93 and mouse reference of mm10-3.1.0. Low quality cells were filtered out (minimum number of reads = 2000, minimum number of genes = 500, minimum Scrublet (77) doublet detection score <0.4). Cells where the percentage of counts from human genes was <90% were considered as mouse cells and excluded from downstream analysis. Cells were assigned to different cell lines (Kolf, Fiaj) using genotype prediction with souporcell (v.2.4.0) (78). We performed batch correction to minimize the differences between cells from different cell lines using scVI and clustered cells using the Leiden algorithm on the latent embedding as described above. We used CellTypist v.0.1.9 Python package (21) to perform annotation prediction with logistic regression using the whole in vivo scRNA-seq developmental dataset for training. For the in vivo-to-in vitro similarity analysis in fig. S29D, we mapped in vitro cells to the scVI model of lymphoid cells as described above. For each cell in the in vitro dataset, we calculated the Euclidean distance weighted by a Gaussian kernel to the closest in vivo cell from each in vivo cell population.
Spatial data analysis
Spatial transcriptomics data was mapped using spaceranger (v.1.2.1) and we used a custom image-processing script to identify regions overlapping tissues. To map cell types identified by scRNA-seq in the profiled spatial transcriptomics slides, we used the cell2location method (16) (see Supplementary Methods). Briefly, for the reference model training step, we excluded very lowly expressed genes using a recommended filtering strategy (16). Cell types where fewer than 20 cells were profiled in the organ of interest and cell types labeled as low-quality cells were excluded from the reference. For the analysis of unconventional T cell localization in thymus (fig. S27C), we trained a reference adding all the prenatal thymic epithelial cells from a thymus cell atlas (7) (data was downloaded from Zenodo (96)). For the spatial cell type deconvolution step, all slides representing a given organ were analyzed jointly. To identify microenvironments of colocalizing cell types, we used non-negative matrix factorization (NMF) on the matrix of estimated cell type abundances. Here latent factors correspond to tissue microenvironments defined by a set of colocalized cell types. We use the NMF implementation in scikit-learn (81), setting the number of factors d = 10. For downstream analysis, we excluded cell types where the 99% quantile of cell abundance across locations in every slide from the same organ was always below the detection threshold of 0.15. Unless otherwise specified, we consider a cell type to be part of a microenvironment if the cell type fraction was over 0.2.
For analysis of mature T cell localization in the thymic medulla (fig. S27, D and E), we retained factors where the sum of the cell type fractions for mature T cells (CD4+ T, CD8+ T, Treg, type 1 innate T, type 3 innate T, and CD8AA) was above 0.8. We assigned spots to the inner medulla or cortico-medullary microenvironment if the factor value in the spot was above the 90% quantile of all values in the slide. To annotate cortex and medulla from histology images, we extracted image features from the high resolution images of H&E staining using the python package squidpy (v1.1.2) (97) and performed Leiden clustering on image features. We then defined the cortico-medullary junction (CMJ) using spatial neighbor graph functionality in squidpy (see Supplementary Methods).
B1 functional validation experiment
Cryopreserved single-cell suspensions from F144 (17 pcw) and F145 (15 pcw) spleen samples were used for the ELISpot experiment. B cells were gated as singlet DAPI−CD3−CD20+ cells. Plasma cells should generally be CD20lo and therefore not included. To further exclude plasma cell contamination, we also gated out the top 1% of B cells expressing the highest level of CD38. The rest of the B cells were then sorted into four fractions: CCR10hi, CCR10loCD27+CD43+, CCR10loCD27–CD43+, and CCR10loCD27–CD43–. CD27 and CD43 gates were chosen based on fluorescence minus one (FMO) controls.
The ELISpot experiment was performed with Human IgM ELISpotBASIC kit (ALP) from Mabtech AB. Post sorting, 7000-8000 cells were added into ELISpot plate pre-coated with anti-IgM antibody and incubated at 37°C for 22 hours. The plate was then washed and incubated with biotinylated anti-IgM for 2 hours at room temperature, followed by a 1-hour incubation with streptavidin-ALP. The colored spots were developed with a 15-min incubation of BCIP/NBT substrate solution. Spots were counted with the AID ELISpot reader and iSpot software version 4.
In addition, we performed scRNA-seq of the sorted B cell fractions on a different donor (F149, 18 pcw fetal spleen), using the same gating strategy to further confirm the identity of sorted cells. The scRNA-seq data was preprocessed with scVI as above. Cell annotations were predicted using CellTypist v.0.1.9 (21).
Supplementary Material
One-Sentence Summary.
We present a human developing immune system atlas across time and multiple organs with important biological discoveries.
Acknowledgements
We thank the Cooks lab (A. Montel-Hagen, S. Lopez, and G. Cooks) for their kind help in setting up the ATO experiments, R. Lindeboom, C. Talavera-Lopez, and K. Kanemaru for helpful discussions, and J. Eliasova, A. Garcia, and BioRender.com for graphical illustrations. We gratefully acknowledge the Sanger Flow Cytometry Facility, Newcastle University Flow Cytometry Core Facility, Sanger Cellular Generation and Phenotyping (CGaP) Core Facility, and Sanger Core Sequencing pipeline for support with sample processing and sequencing library preparation. The human embryonic and fetal material was provided by the MRC-Wellcome Trust-funded Human Developmental Biology Resource (HDBR; http://www.hdbr.org). We are grateful to the donors and donor families for granting access to the tissue samples. This publication is part of the Human Cell Atlas: www.humancellatlas.org/publications. We acknowledge Wellcome Trust Sanger Institute as the source of HPSI0114i-kolf_2 and HPSI0514i-fiaj_1 human induced pluripotent cell lines which were generated under the Human Induced Pluripotent Stem Cell Initiative funded by a grant from the Wellcome Trust and Medical Research Council, supported by the Wellcome Trust (WT098051) and the NIHR/Wellcome Trust Clinical Research Facility, and acknowledge Life Science Technologies Corporation as the provider of Cytotune.
Funding
We acknowledge funding from the Wellcome Human Cell Atlas Strategic Science Support (WT211276/Z/18/Z), CZI Seed Networks for the Human Cell Atlas (Thymus award), MRC Human Cell Atlas award and Wellcome Human Developmental Biology Initiative. M.H. is funded by Wellcome (WT107931/Z/15/Z), The Lister Institute for Preventive Medicine and NIHR and Newcastle Biomedical Research Centre. S.A.T. is funded by Wellcome (WT206194), the ERC Consolidator Grant ThDEFINE. C.S. is supported by a Wellcome Trust Ph.D. Fellowship for Clinicians. Z.K.T and M.R.C are supported by a Medical Research Council Research Project Grant (MR/S035842/1). M.R.C. was supported by an NIHR Research Professorship (RP-2017-08-ST2-002) and a Wellcome Investigator Award (220268/Z/20/Z).
Footnotes
Author contributions: Conceptualization: S.A.T., M.H., M.R.C., C.S., and E.D. Data curation: C.S., E.D., and I.G. Formal analysis: E.D., C.S., I.G., L.J., J.E.P., V.K., Z.K.T., K.P., C.X., N.Y., R.E., C.D.C., P.H., C.M., and J.C.M. Funding acquisition: S.A.T. and M.H. Methodology: C.S., I.G., R.A.B., E.S., J.E., M.M., and A.S.S. Project administration: S.A.T., M.H., C.S., E.D., and I.G. Software: E.D., K.P., Z.K.T., C.X., M.P., P.M., and D.H. Supervision: S.A.T., M.H., and M.R.C. Validation: C.S., S.P., N.Y., and O.S. Visualization: C.S., E.D., and N.Y. Writing - original draft: C.S., E.D., M.H., L.J., I.G., V.K., N.Y., K.P., Z.K.T., and S.P. Writing - review and editing: all authors.
Competing interests: In the past 3 years, S.A.T. has consulted for Genentech and Roche and sits on Scientific Advisory Boards for Qiagen, Foresite Labs, Biogen, and GlaxoSmithKline and is a co-founder and equity holder of Transition Bio. R.E. is a paid consultant of Foresite Capital. The remaining authors declare no competing interests.
Data and materials availability
Raw sequencing data for newly generated sequencing libraries have been deposited in ArrayExpress (scRNA-seq libraries: accession number E-MTAB-11343; scVDJ-seq libraries: accession number E-MTAB-11388; Visium 10X libraries: accession number E-MTAB-11341). Processed data objects are available for online visualization and download in AnnData format (98) at https://developmental.cellatlas.io/fetal-immune, as well as trained scVI models for query to reference mapping and trained Celltypist models for cell annotation. All code scripts and notebooks for analysis presented in the manuscript are available at https://github.com/Teichlab/Pan_fetal_immune (99). This publication is part of the Human Cell Atlas (www.humancellatlas.org/publications).
References
- 1.Park J-E, Jardine L, Gottgens B, Teichmann SA, Haniffa M. Prenatal development of human immunity. Science. 2020;368:600–603. doi: 10.1126/science.aaz9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jagannathan-Bogdan M, Zon LI. Hematopoiesis. Dev Camb Engl. 2013;140:2463–2467. doi: 10.1242/dev.083147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Popescu D-M, et al. Decoding human fetal liver haematopoiesis. Nature. 2019;574:365–371. doi: 10.1038/s41586-019-1652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stewart BJ, et al. Spatiotemporal immune zonation of the human kidney. Science. 2019;365:1461–1466. doi: 10.1126/science.aat5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng Y, et al. Tracing the first hematopoietic stem cell generation in human embryo by single-cell RNA sequencing. Cell Res. 2019;29:881–894. doi: 10.1038/s41422-019-0228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng Y, et al. Single-Cell RNA Sequencing Resolves Spatiotemporal Development of Pre-thymic Lymphoid Progenitors and Thymus Organogenesis in Human Embryos. Immunity. 2019;51:930–948.:e6. doi: 10.1016/j.immuni.2019.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Park J-E, et al. A cell atlas of human thymic development defines T cell repertoire formation. Science. 2020;367 doi: 10.1126/science.aay3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elmentaite R, et al. Single-Cell Sequencing of Developing Human Gut Reveals Transcriptional Links to Childhood Crohn’s Disease. Dev Cell. 2020;55:771–785.:e5. doi: 10.1016/j.devcel.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao J, et al. A human cell atlas of fetal gene expression. Science. 2020;370 doi: 10.1126/science.aba7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reynolds G, et al. Developmental cell programs are co-opted in inflammatory skin disease. Science. 2021;371:eaba6500. doi: 10.1126/science.aba6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jardine L, et al. Blood and immune development in human fetal bone marrow and Down syndrome. Nature. 2021:1–5. doi: 10.1038/s41586-021-03929-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez R, Regier J, Cole MB, Jordan MI, Yosef N. Deep generative modeling for single-cell transcriptomics. Nat Methods. 2018;15:1053–1058. doi: 10.1038/s41592-018-0229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pellin D, et al. A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat Commun. 2019;10:2395. doi: 10.1038/s41467-019-10291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villani A-C, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lotfollahi M, et al. Mapping single-cell data to reference atlases by transfer learning. Nat Biotechnol. 2021:1–10. doi: 10.1038/s41587-021-01001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleshchevnikov V, et al. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat Biotechnol. 2022:1–11. doi: 10.1038/s41587-021-01139-4. [DOI] [PubMed] [Google Scholar]
- 17.Chong SZ, et al. CXCR4 identifies transitional bone marrow premonocytes that replenish the mature monocyte pool for peripheral responses. J Exp Med. 2016;213:2293–2314. doi: 10.1084/jem.20160800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacParland SA, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. 2018;9:4383. doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerrits E, Heng Y, Boddeke EWGM, Eggen BJL, et al. Transcriptional profiling of microglia; current state of the art and future perspectives. Glia. 2020;68:740–755. doi: 10.1002/glia.23767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toor SM, Wani S, Albagha OME, et al. Comprehensive Transcriptomic Profiling of Murine Osteoclast Differentiation Reveals Novel Differentially Expressed Genes and LncRNAs. Front Genet. 2021;12 doi: 10.3389/fgene.2021.781272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conde CD, et al. Cross-tissue immune cell analysis reveals tissue-specific adaptations and clonal architecture in humans. 2021:2021.04.28.441762. doi: 10.1101/2021.04.28.441762. [DOI] [Google Scholar]
- 22.Dann E, Henderson NC, Teichmann SA, Morgan MD, Marioni JC. Differential abundance testing on single-cell data using k-nearest neighbor graphs. Nat Biotechnol. 2021:1–9. doi: 10.1038/s41587-021-01033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeucken KCM, Koning JJ, Mebius RE, Tas SW. The Role of Endothelial Cells and TNF-Receptor Superfamily Members in Lymphoid Organogenesis and Function During Health and Inflammation. Front Immunol. 2019;10:2700. doi: 10.3389/fimmu.2019.02700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X, et al. Essential contribution of a chemokine, CCL3, and its receptor, CCR1, to hepatocellular carcinoma progression. Int J Cancer. 2006;118:1869–1876. doi: 10.1002/ijc.21596. [DOI] [PubMed] [Google Scholar]
- 25.Hua F, Tian Y. CCL4 promotes the cell proliferation, invasion and migration of endometrial carcinoma by targeting the VEGF-A signal pathway. Int J Clin Exp Pathol. 2017;10:11288–11299. [PMC free article] [PubMed] [Google Scholar]
- 26.Keeley EC, Mehrad B, Strieter RM. CXC chemokines in cancer angiogenesis and metastases. Adv Cancer Res. 2010;106:91–111. doi: 10.1016/S0065-230X(10)06003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heidemann J, et al. Angiogenic effects of interleukin 8 (CXCL8) in human intestinal microvascular endothelial cells are mediated by CXCR2. J Biol Chem. 2003;278:8508–8515. doi: 10.1074/jbc.M208231200. [DOI] [PubMed] [Google Scholar]
- 28.Norrby K. Mast cells and angiogenesis. APMIS Acta Pathol Microbiol Immunol Scand. 2002;110:355–371. doi: 10.1034/j.1600-0463.2002.100501.x. [DOI] [PubMed] [Google Scholar]
- 29.Ribatti D, Crivellato E. The role of mast cell in tissue morphogenesis. Thymus, duodenum, and mammary gland as examples. Exp Cell Res. 2016;341:105–109. doi: 10.1016/j.yexcr.2015.11.022. [DOI] [PubMed] [Google Scholar]
- 30.Wood W, Martin P. Macrophage Functions in Tissue Patterning and Disease: New Insights from the Fly. Dev Cell. 2017;40:221–233. doi: 10.1016/j.devcel.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoorweg K, Cupedo T. Development of human lymph nodes and Peyer's patches. Semin Immunol. 2008;20:164–170. doi: 10.1016/j.smim.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Rantakari P, et al. Fetal liver endothelium regulates the seeding of tissue-resident macrophages. Nature. 2016;538:392–396. doi: 10.1038/nature19814. [DOI] [PubMed] [Google Scholar]
- 33.Li N, et al. Memory CD4+ T cells are generated in the human fetal intestine. Nat Immunol. 2019;20:301–312. doi: 10.1038/s41590-018-0294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mishra A, et al. Microbial exposure during early human development primes fetal immune cells. Cell. 2021;184:3394–3409.:e20. doi: 10.1016/j.cell.2021.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xing Y, Wang X, Jameson SC, Hogquist KA. Late stages of T cell maturation in the thymus involve NF-κB and tonic type I interferon signaling. Nat Immunol. 2016;17:565–573. doi: 10.1038/ni.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Webb LV, Ley SC, Seddon B. TNF activation of NF-κB is essential for development of single-positive thymocytes. J Exp Med. 2016;213:1399–1407. doi: 10.1084/jem.20151604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collins C, Sharpe E, Silber A, Kulke S, Hsieh EWY. Congenital Athymia: Genetic Etiologies, Clinical Manifestations, Diagnosis, and Treatment. J Clin Immunol. 2021;41:881–895. doi: 10.1007/s10875-021-01059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holt PG, Jones CA. The development of the immune system during pregnancy and early life. Allergy. 2000;55:688–697. doi: 10.1034/j.1398-9995.2000.00118.x. [DOI] [PubMed] [Google Scholar]
- 39.Griffin DO, Holodick NE, Rothstein TL. Human B1 cells in umbilical cord and adult peripheral blood express the novel phenotype CD20+CD27+CD43+CD70−. J Exp Med. 2011;208:67–80. doi: 10.1084/jem.20101499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffin D, Rothstein T. Human B1 Cell Frequency: Isolation and Analysis of Human B1 Cells. Front Immunol. 2012;3:122. doi: 10.3389/fimmu.2012.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rothstein TL, Griffin DO, Holodick NE, Quach TD, Kaku H. Human B-1 cells take the stage. Ann NY Acad Sci. 2013;1285:97–114. doi: 10.1111/nyas.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baumgarth N. The double life of a B-1 cell: self-reactivity selects for protective effector functions. Nat Rev Immunol. 2011;11:34–46. doi: 10.1038/nri2901. [DOI] [PubMed] [Google Scholar]
- 43.Lalor PA, Herzenberg LA, Adams S, Stall AM. Feedback regulation of murine Ly-1 B cell development. Eur J Immunol. 1989;19:507–513. doi: 10.1002/eji.1830190315. [DOI] [PubMed] [Google Scholar]
- 44.Hayakawa K, Hardy RR, Parks DR, Herzenberg LA. The “Ly-1 B” cell subpopulation in normal immunodefective, and autoimmune mice. J Exp Med. 1983;157:202–218. doi: 10.1084/jem.157.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montecino-Rodriguez E, Dorshkind K. B-1 B Cell Development in the Fetus and Adult. Immunity. 2012;36:13–21. doi: 10.1016/j.immuni.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kantor AB, Merrill CE, Herzenberg LA, Hillson JL. An unbiased analysis of V(H)-D-J(H) sequences from B-1a, B-1b, and conventional B cells. J Immunol Baltim Md 1950. 1997;158:1175–1186. [PubMed] [Google Scholar]
- 47.Tornberg UC, Holmberg D. B-1a, B-1b and B-2 B cells display unique VHDJH repertoires formed at different stages of ontogeny and under different selection pressures. EMBO J. 1995;14:1680–1689. doi: 10.1002/j.1460-2075.1995.tb07157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyama-Inaba M, et al. Unusual phenotype of B cells in the thymus of normal mice. J Exp Med. 1988;168:811–816. doi: 10.1084/jem.168.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elmentaite R, et al. Cells of the human intestinal tract mapped across space and time. Nature. 2021;597:250–255. doi: 10.1038/s41586-021-03852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schulze-Luehrmann J, Ghosh S. Antigen-receptor signaling to nuclear factor kappa B. Immunity. 2006;25:701–715. doi: 10.1016/j.immuni.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 51.Alonzo ES, Sant’Angelo DB. Development of PLZF-expressing innate T cells. Curr Opin Immunol. 2011;23:220–227. doi: 10.1016/j.coi.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dimova T, et al. Effector Vγ9Vδ2 T cells dominate the human fetal γδ T-cell repertoire. Proc Natl Acad Sci. 2015;112:E556–E565. doi: 10.1073/pnas.1412058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan L, et al. A fetal wave of human type 3 effector γδ cells with restricted TCR diversity persists into adulthood. Sci Immunol. 2021;6:eabf0125. doi: 10.1126/sciimmunol.abf0125. [DOI] [PubMed] [Google Scholar]
- 54.Mayassi T, Barreiro LB, Rossjohn J, Jabri B. A multilayered immune system through the lens of unconventional T cells. Nature. 2021;595:501–510. doi: 10.1038/s41586-021-03578-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carico ZM, Roy Choudhury K, Zhang B, Zhuang Y, Krangel MS. Tcrd Rearrangement Redirects a Processive Tcra Recombination Program to Expand the Tcra Repertoire. Cell Rep. 2017;19:2157–2173. doi: 10.1016/j.celrep.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee YJ, et al. Generation of PLZF+ CD4+ T cells via MHC class II-dependent thymocyte–thymocyte interaction is a physiological process in humans. J Exp Med. 2010;207:237–246. doi: 10.1084/jem.20091519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho H, Bediako Y, Xu H, Choi H-J, Wang C-R. Positive selecting cell type determines the phenotype of MHC class Ib-restricted CD8+ T cells. Proc Natl Acad Sci. 2011;108:13241–13246. doi: 10.1073/pnas.1105118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Georgiev H, Peng C, Huggins MA, Jameson SC, Hogquist KA. Classical MHC expression by DP thymocytes impairs the selection of non-classical MHC restricted innate-like T cells. Nat Commun. 2021;12:2308. doi: 10.1038/s41467-021-22589-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoffman ES, et al. Productive T-cell receptor beta-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev. 1996;10:948–962. doi: 10.1101/gad.10.8.948. [DOI] [PubMed] [Google Scholar]
- 60.Spits H. Development of αβ T cells in the human thymus. Nat Rev Immunol. 2002;2:760–772. doi: 10.1038/nri913. [DOI] [PubMed] [Google Scholar]
- 61.Montel-Hagen A, et al. Organoid-induced differentiation of conventional T cells from human pluripotent stem cells. Cell Stem Cell. 2019;24:376–389.:e8. doi: 10.1016/j.stem.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mass E, et al. Specification of tissue-resident macrophages during organogenesis. Science. 2016;353:aaf4238. doi: 10.1126/science.aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mende N, Laurenti E. Hematopoietic stem and progenitor cells outside the bone marrow: where, when, and why. Exp Hematol. 2021;104:9–16. doi: 10.1016/j.exphem.2021.10.002. [DOI] [PubMed] [Google Scholar]
- 64.Mende N, et al. Blood. doi: 10.1182/blood.2021013450. in press. [DOI] [Google Scholar]
- 65.Krishnan S, et al. Hematopoietic stem and progenitor cells are present in healthy gingiva tissue. J Exp Med. 2021;218:e20200737. doi: 10.1084/jem.20200737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim H. Homeostatic and pathogenic extramedullary hematopoiesis. J Blood Med. 2010;1:13–19. doi: 10.2147/JBM.S7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brioschi S, et al. Heterogeneity of meningeal B cells reveals a lymphopoietic niche at the CNS borders. Science. 2021;373:eabf9277. doi: 10.1126/science.abf9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schafflick D, et al. Single-cell profiling of CNS border compartment leukocytes reveals that B cells and their progenitors reside in non-diseased meninges. Nat Neurosci. 2021;24:1225–1234. doi: 10.1038/s41593-021-00880-y. [DOI] [PubMed] [Google Scholar]
- 69.Wang Y, et al. Early developing B cells undergo negative selection by central nervous system-specific antigens in the meninges. Immunity. 2021;54:2784–2794.:e6. doi: 10.1016/j.immuni.2021.09.016. [DOI] [PubMed] [Google Scholar]
- 70.Montecino-Rodriguez E, Leathers H, Dorshkind K. Identification of a B-1 B cell–specified progenitor. Nat Immunol. 2006;7:293–301. doi: 10.1038/ni1301. [DOI] [PubMed] [Google Scholar]
- 71.Esplin BL, Welner RS, Zhang Q, Borghesi LA, Kincade PW. A differentiation pathway for B1 cells in adult bone marrow. Proc Natl Acad Sci. 2009;106:5773–5778. doi: 10.1073/pnas.0811632106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshimoto M, et al. Embryonic day 9 yolk sac and intra-embryonic hemogenic endothelium independently generate a B-1 and marginal zone progenitor lacking B-2 potential. Proc Natl Acad Sci U S A. 2011;108:1468–1473. doi: 10.1073/pnas.1015841108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kreslavsky T, Wong JB, Fischer M, Skok JA, Busslinger M. Control of B-1a cell development by instructive BCR signaling. Curr Opin Immunol. 2018;51:24–31. doi: 10.1016/j.coi.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Graf R, et al. BCR-dependent lineage plasticity in mature B cells. Science. 2019;363:748–753. doi: 10.1126/science.aau8475. [DOI] [PubMed] [Google Scholar]
- 75.Mimitou EP, et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat Methods. 2019;16:409–412. doi: 10.1038/s41592-019-0392-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fleming SJ, Marioni JC, Babadi M. CellBender remove-background: a deep generative model for unsupervised removal of background noise from scRNA-seq datasets. 2019:791699 [Google Scholar]
- 77.Wolock SL, Lopez R, Klein AM. Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst. 2019;8:281–291.:e9. doi: 10.1016/j.cels.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heaton H, et al. Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat Methods. 2020;17:615–620. doi: 10.1038/s41592-020-0820-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gayoso A, et al. A Python library for probabilistic analysis of single-cell omics data. Nat Biotechnol. 2022;40:163–166. doi: 10.1038/s41587-021-01206-w. [DOI] [PubMed] [Google Scholar]
- 81.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 82.Traag VA, Waltman L, van Eck NJ. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep. 2019;9:5233. doi: 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Polański K, et al. BBKNN: fast batch alignment of single cell transcriptomes. Bioinforma Oxf Engl. 2020;36:964–965. doi: 10.1093/bioinformatics/btz625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ahlmann-Eltze C, Huber W. glmGamPoi: fitting Gamma-Poisson generalized linear models on single cell count data. Bioinformatics. 2020;36:5701–5702. doi: 10.1093/bioinformatics/btaa1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Squair JW, et al. Confronting false discoveries in single-cell differential expression. Nat Commun. 2021;12:5692. doi: 10.1038/s41467-021-25960-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sturm G, et al. Scirpy: a Scanpy extension for analyzing single-cell T-cell receptor-sequencing data. Bioinformatics. 2020;36:4817–4818. doi: 10.1093/bioinformatics/btaa611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stephenson E, et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat Med. 2021;27:904–916. doi: 10.1038/s41591-021-01329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gupta NT, et al. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015;31:3356–3358. doi: 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Holland CH, et al. Robustness and applicability of transcription factor and pathway analysis tools on single-cell RNA-seq data. Genome Biol. 2020;21:36. doi: 10.1186/s13059-020-1949-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 93.Garcia-Alonso L, et al. Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro. Nat Genet. 2021;53:1698–1711. doi: 10.1038/s41588-021-00972-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Setty M, et al. Characterization of cell fate probabilities in single-cell data with Palantir. Nat Biotechnol. 2019;37:451–460. doi: 10.1038/s41587-019-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lange M, et al. CellRank for directed single-cell fate mapping. Nat Methods. 2022:1–12. doi: 10.1038/s41592-021-01346-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Park J-E, Teichmann S, Haniffa M, Taghon T. Collection of codes and annotated matrix for the paper “A cell atlas of human thymic development defines T cell repertoire formation”. 2021 doi: 10.5281/zenodo.5500511. [DOI] [Google Scholar]
- 97.Palla G, et al. Squidpy: a scalable framework for spatial omics analysis. Nat Methods. 2022;19:171–178. doi: 10.1038/s41592-021-01358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Virshup I, Rybakov S, Theis FJ, Angerer P, Wolf FA. anndata: Annotated data. 2021:2021.12.16.473007. doi: 10.1101/2021.12.16.473007. [DOI] [Google Scholar]
- 99.Dann E, Suo C, Goh I, Kleshchevnikov V. Teichlab/Pan_fetal_immune: Analysis code for publication. Zenodo; 2022. [DOI] [Google Scholar]
- 100.Dick SA, et al. Three tissue resident macrophage subsets coexist across organs with conserved origins and life cycles. Sci Immunol. 2022;7:eabf7777. doi: 10.1126/sciimmunol.abf7777. [DOI] [PubMed] [Google Scholar]
- 101.Gadala-Maria D, Yaari G, Uduman M, Kleinstein SH. Automated analysis of high-throughput B-cell sequencing data reveals a high frequency of novel immunoglobulin V gene segment alleles. Proc Natl Acad Sci. 2015;112:E862–E870. doi: 10.1073/pnas.1417683112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Egawa T, et al. The earliest stages of B cell development require a chemokine stromal cell-derived factor/pre-B cell growth-stimulating factor. Immunity. 2001;15:323–334. doi: 10.1016/s1074-7613(01)00185-6. [DOI] [PubMed] [Google Scholar]
- 103.Nagasawa T, et al. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382:635–638. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data for newly generated sequencing libraries have been deposited in ArrayExpress (scRNA-seq libraries: accession number E-MTAB-11343; scVDJ-seq libraries: accession number E-MTAB-11388; Visium 10X libraries: accession number E-MTAB-11341). Processed data objects are available for online visualization and download in AnnData format (98) at https://developmental.cellatlas.io/fetal-immune, as well as trained scVI models for query to reference mapping and trained Celltypist models for cell annotation. All code scripts and notebooks for analysis presented in the manuscript are available at https://github.com/Teichlab/Pan_fetal_immune (99). This publication is part of the Human Cell Atlas (www.humancellatlas.org/publications).