Abstract
Proteomic biomarker discovery using formalin-fixed paraffin-embedded (FFPE) tissue requires robust workflows to support the analysis of large cohorts of patient samples. It also requires finding a reasonable balance between achieving high proteomic depth and limiting overall analysis time. To this end, we evaluated the merits of online coupling of single-use disposable trap column nano-flow liquid chromatography, high-field asymmetric-waveform ion-mobility spectrometry (FAIMS) and tandem mass spectrometry (nLC-FAIMS-MS/MS). The data shows that ≤ 600 ng of peptide digest should be loaded onto the chromatographic part of the system. Careful characterization of the FAIMS settings enabled the choice of optimal combinations of compensation voltages (CV) as a function of the employed LC gradient time. We found nLC-FAIMS-MS/MS to be on par with StageTip-based off-line basic pH reversed-phase fractionation in terms of proteomic depth and reproducibility of protein quantification (coefficient of variation ≤ 15 % for 90 % of all proteins) but requiring 50 % less sample and substantially reducing sample handling. Using FFPE material from lymph node, lung and prostate tissue as examples, we show that nLC-FAIMS-MS/MS can identify 5,000-6,000 proteins from the respective tissue within a total of 3 hours of analysis time.
Keywords: Quantitative proteomics, clinical proteomics, FFPE analysis, high field asymmetric waveform ion mobility spectrometry (FAIMS), LC-FAIMS-MS/MS
Introduction
Clinical specimen collected from patients are commonly archived as formalin fixed paraffin embedded (FFPE) tissue. FFPE not only preserves cellular morphology, but also many of the resident biomolecules permitting long-term, space- and energy-efficient storage. Countless samples are available in pathology departments and biobanks worldwide, frequently annotated with valuable clinical metadata (e.g., sex, age, diagnosis, response to treatment, survival, etc.) thus enabling many types of retrospective analysis to address numerous biomedical questions. While DNA and RNA-based analysis from FFPE tissue are already common today, proteomic investigations have only recently become feasible. An important advance in FFPE sample preparation was the development of the SP3 approach (single-pot, solid-phase-enhanced sample preparation for proteomics experiments) because it allows the use of sodium dodecyl sulfate (SDS) for efficient protein solubilization, and the use of organic solvents for removing small molecules, polymers and buffer components that reverse formaldehyde crosslinking from very limited sample quantities 1, 2.
As in other branches of proteomics, LC-MS/MS is also the method of choice for profiling the proteomes of FFPE material and all the major quantitative approaches such as classical data dependent acquisition (DDA), isobaric labeling using tandem mass tags (TMT) or data independent acquisition (DIA) have been implemented - each with its individual pros and cons 3–6. A challenge faced by any analytical proteomic approach is the sheer number of proteins present in a sample and the vast differences in expression between proteins in a tissue, which overwhelms even the most performant LC-MS/MS systems. One particular issue in the analysis of clinical specimen including FFPE material is sample-to-sample carry-over which may negatively affect results. An interesting new way to minimize carry-over between samples is the use of a nanoLC system with single-use disposable trap columns and the published literature suggests that such a system offers robust operation across large numbers of analysis while maintaining acceptable chromatographic performance 7, 8.
Simplifying and streamlining proteomic workflows particularly for clinical applications is a recent trend as this would reduce the number of sample processing steps during which variation may be encountered and may support higher throughput at the same time. In keeping with this idea, an alternative to separating peptides off-line prior to LC-MS/MS, is to do so in the gas phase ‘on-the-fly’. This can be achieved by ion mobility spectrometry and several variants of this approach have been implemented on commercial instruments 9–11.
High-field asymmetric waveform ion mobility spectrometry (FAIMS) is one of these approaches and has recently become more widely available12. FAIMS operates at atmospheric pressure and separates ions on the basis of differential mobility in low and high electric fields. FAIMS works on a millisecond time-scale which makes it well compatible with upstream LC-separations (second time scale) and downstream tandem MS (millisecond time-scale). The merits of FAIMS for proteomic applications have been investigated before and a substantial improvement in both peptide and protein identifications has been observed compared to standard analysis without FAIMS8, 13. In light of the above, coupling disposable trap column nanoLC separations with gas-phase fractionation using FAIMS and tandem mass spectrometry (nLC-FAIMS-MS/MS) appears particularly interesting for the analysis of FFPE material. However, to the best of our knowledge, such a setup has not yet been systematically evaluated for this purpose.
Experimental Section
Deparaffinization of FFPE tissue and protein extraction
The FFPE sample material used in this study, was healthy, freely available left-over patient material from routine diagnostics. Using a microtome, four 10 μm slices were cut from each FFPE tissue block (if not stated otherwise) and subsequently placed onto microscopy slides. To remove the bulk of the paraffin, slides were incubated at 60 °C for 30 min. Slices were fully deparaffinized using 100% xylene for 20 min followed by 1x100 %, 1x96 %, 1x70 % ethanol and 3x water for 10 min. An area of 5x5 mm of healthy tissue was micro-dissected using a scalpel from each slide. Protein extraction was performed by boiling in 260 μl extraction buffer (10 mM Dithiothreitol (DTT), 4 % SDS in 500 mM Tris-HCl, pH 9) for 1 h at 100 °C. Where noted, Tris concentrations of 100, 300 and 500 mM Tris as well as pH values of 8, 9 and 10 were used for optimization purposes. Extracts were sonicated for 25 cycles at 8 °C (60 sec on, 30 sec off), debris was pelleted by centrifugation at 17,000 x g for 10 min and the supernatant was further processed by SP3 digestion as described further below.
Preparation of HeLa peptides for nLC-FAIMS-MS/MS method optimization
Human HeLa cells (ATCC CCL-2) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% Fetal Bovine Serum at 37 °C and 5% CO2. Cells were lysed by incubation in lysis buffer (8 M urea, 1x EDTA free protease inhibitor in 40 mM Tris-HCl, pH 7.6) on ice for 5 min. The lysate was cleared by centrifugation for 30 min at 4 °C at 20,000 x g. Proteins were reduced with 10 mM DTT for 45 min at 37 °C and alkylated with 55 mM CAA (2-chloroacetamide) for 30 min at room temperature. Samples were diluted to 1 M urea using 40 mM Tris-HCl, pH 7.6. Initial digestion of proteins was performed by adding Trypsin and Lys-C (1:100 (wt/wt) enzyme-to-protein ratio) at 37 °C for 4 h. A second addition of the same amount of both proteases was performed and the digestion was allowed to continue overnight. Protease digestion was stopped by addition of formic acid (FA) to a final concentration of 1 %. The acidified peptides were centrifuged at 5,000 x g for 15 min, followed by desalting on a Sep-Pak C18 Cartridge. Peptide concentration was estimated by NanoDrop measurements, and the adequate amount of peptides was subsequently fractionated by self-packed StageTips or loaded on Evotips (Evosep).
Single-pot solid-phase-enhanced sample preparation (SP3) protocol for FFPE samples
Prior to tryptic digestion, detergent was removed from lysates by SP3 clean-up, following the protocol first described by Hughes et al.1. Briefly, the lysate was mixed with SP3 beads and proteins were precipitated onto a 50:50 mixture of Sera-Mag Speed Beads types A and B (Thermo Fisher Scientific) in 70 % acetonitrile. Beads were washed twice with 80 % ethanol in water and once with acetonitrile. Disulfide bonds were reduced with 10 mM TCEP (tris(2-carboxyethyl)phosphine) for 45 min at 37 °C, followed by alkylation of cysteines with 55 mM CAA for 30 min at room temperature in 50 μl of digestion buffer (2 mM CaCl2 in 40 mM Tris-HCl, pH 7.8). Trypsin (1:50 (wt/wt) enzyme-to-protein ratio) was added and bead-precipitated proteins were digested at 37 °C for 3-4 hours. The same amount of Trypsin was added again and the digestion was allowed to continue overnight. The next day, beads were settled using a magnet and the supernatant was transferred to a new tube. Beads were washed by addition of 50 μl water, sonicated (3x 30 sec) and the supernatants were combined. Sample were acidified with formic acid to pH < 3. Subsequently, peptides were loaded onto Evotips. For the optimization of the extraction procedure, samples were desalted using a C18 SepPak 96-well desalting plate before measurement.
Off-line basic pH reversed-phase fractionation on self-packed StageTips
A StageTip14 containing Empore C18 disks was constructed. And the C18 material was conditioned with 0.1 % FA in 50 % acetonitrile in water and equilibrated with 0.1 % FA in water. 20 μg of HeLa peptides, dissolved in 0.1 % FA in water, were loaded twice onto the micro-column by centrifugation. The loaded peptides were washed twice with 0.1 % FA in water. 50 μl of 25 mM ammonium formate in water were added to the tip, the flow-through was reapplied and collected the second time. This was followed by sequential elution of the bound peptides using 5, 10, 15, 17.5 and 50 % acetonitrile in 25 mM ammonium formate resulting in 6 fractions. The 5 % and 50 % acetonitrile fractions were pooled and so were the the flow-through and 17.5 % acetonitrile fraction. The resulting four fractions were frozen and dried in a vacuum centrifuge, before loading onto Evotips.
nLC-MS/MS measurements for the optimization of the extraction procedure
Samples for the optimization of the extraction procedure were desalted prior to measurement using a C18 SepPak 96-well desalting plate (Waters). The C18 material was conditioned using 0.1 % FA in 40 % acetonitrile in water and equilibrated using 0.1 % FA in water. Acidified peptides were loaded onto the C18 material and washed twice with 0.1 % FA in water. Peptides were eluted with 40 % acetonitrile in 0.1 % FA in water followed by freezing and drying in a vacuum centrifuge. 50 ng of peptides were dissolved with 2 % acetonitrile, 0.1 % FA in water and analysed by nLC-MS/MS with a Dionex Ultimate3000 nano HPLC (Thermo Fisher Scientific) coupled online to a Q-Exactive HF-X mass spectrometer (Thermo Fisher Scientific). Peptides were delivered to a trap column (ReprosSil-pur C18-AQ, 5 μm, 20 mm x 75 μm, Dr. Maisch, Ammerbuch) and washed with 2 % solvent B (0.1 % formic acid, 5 % DMSO in acetonitrile) in solvent A (0.1 % formic acid, 5 % DMSO in HPLC grade water) at a flowrate of 5 μL/min for 10 min. Peptides were then separated on an analytical column (75 μm x 40 cm, packed in house with Reprosil-Gold C18 3 μm resin, Dr. Maisch, Ammerbuch) using a 50 min linear gradient ranging from 4-32 % solvent B with a flowrate of 300 nL/min.
The mass spectrometer was operated in positive ionization and data dependent acquisition mode, automatically switching between MS1 and MS2 spectra. MS1 spectra were acquired over a mass-to-charge (m/z) range of 360-1300 m/z at a resolution of 60,000 (at m/z 200) in the Orbitrap using a maximum injection time of 45 ms and an automatic gain control (AGC) target value of 3e6. Up to 18 peptide precursors were selected for fragmentation with an isolation width of 1.3 m/z, maximum injection time of 25 ms and an AGC value of 1e5, fragmented by HCD using 26 % normalized collision energy (NCE) and analysed in the Orbitrap at a resolution of 15,000 (at m/z 200). The dynamic exclusion duration of fragmented precursor ions was set to 25 s.
nLC-(FAIMS)-MS/MS analyses
Prior to measurement on the Evosep LC-system (Evosep), the desired amount of peptides was loaded onto Evotips (i. e. a single-use disposable trap column). The general loading procedure was according to manufacturer’s instructions with minor changes. All washing/loading volumes were increased from 20 to 100 μl such that Evotips were equilibrated in three steps: i) 0.1% FA in acetonitrile (EvoB); ii) 30 % EvoB in EvoA and iii) 0.1% FA in water (EvoA). Prepared tips are best stored in EvoA at 4 °C, preventing them from running dry. Peptides were eluted from Evotips into a long capillary using predefined low pressure gradient methods of 22 min (60 samples per day, SPD), 44 min (30 SPD) and 88 min (15 SPD) and flowrates of 1000, 500 and 220 nl/min, respectively. Using the high pressure pump, peptides were refocused and separated on an Evosep C18 analytical column (8 cm, 3 μm particle size, 100 μm ID for the 22 min and 15 cm, 1.9 μm particle size, 150 μm ID for the 44 and 88 min gradients).
The Orbitrap Exploris 480 mass spectrometer (Thermo Fisher Scientific) was operated in positive ionization mode (spray voltage of 2100 V and 2300 V without and with FAIMS, respectively) and data dependent acquisition mode, automatically switching between MS1 and MS2 spectra with a fixed cycle time. The cycle times chosen for different gradients with and without FAIMS are shown in Table 1. Briefly, cycle times for the three used gradients (1, 2 and 3 sec for the 21, 44 and 88 min gradient, respectively) was chosen based on the median respective chromatographic peak widths to ensure a sufficient number of data points (>10) across each peak for robust MS1 based label-free quantification. When more than one CV was used within an nLC-FAIMS-MS/MS run, the above cycle times were adjusted (by the number of internal CV values used) to keep the number of data points across the LC peak stable. We note that 0.4 sec is the minimum allowed cycle time. All actual CV values used in respective experiments are annotated in the figures, tabulated in Supplemental Table S1 and part of the raw file names.
Table 1. Cycle times set for different gradients and different numbers of internal CVs.
| cycle time per experiment [sec] | |||
|---|---|---|---|
| number of internal CVs | 21 min | 44 min | 88 min |
| 0 (FAIMS not mounted) | 1 | 2 | 3 |
| 1 | 1 | 2 | 3 |
| 2 | 0.5 | 1 | 1.5 |
| 3 | 0.4 | 0.66 | 1 |
| 4 | - | 0.5 | 0.75 |
| 5 | - | - | 0.6 |
| 6 | - | - | 0.5 |
| 7 | - | - | 0.42 |
For all experiments, MS1 spectra were acquired over a mass-to-charge (m/z) range of 360-1300 m/z at a resolution of 60,000 (at m/z 200) in the Orbitrap using a maximum injection time (maxIT) of 45 ms and a normalized automatic gain control (AGC) target value of 100% (1e6). The Monoisotopic Precursor Selection (MIPS) filter was activated and set to peptide mode. The charge state filter was set to 2-6. For MS2, precursors were isolated with a width of 1.3 Th, collected using a standard maxIT of 25 ms and a normalized AGC value of 100 % (1e5), fragmented by HCD at 28 % normalized collision energy (NCE) and spectra were acquired in the Orbitrap at a standard resolution of 15,000 (at m/z 200). For the dilution series of HeLa digests (Fig. S3E), MS2 resolution was ramped along with its matching maxIT (15k, 22 ms; 30k, 54 ms; 45k, 86 ms; 60k, 118 ms; 120k, 246 ms, each resolution at m/z 200). All other parameters were kept the same as specified above. The dynamic exclusion duration of fragmented precursor ions was set to 30, 45 and 90 s for the 21, 44 and 88 min gradient, respectively. The dynamic exclusion list was set to be shared across different experiments.
FFPE tissue material was analyzed as follows: 600 ng of each sample was measured twice (total: 1.2 μg) using an 88 min gradient (total: 176 min) with different sets of CVs per injection (Set1; - 30|-40|-50|-60|-70 V, Set2: -35|-45|-55|-65|-75 V). All other settings were the same as described above.
Database searching and data analysis
Raw MS data files were processed with MaxQuant (MQ) v1.6.17.015 using the integrated Andromeda Search engine and searched against a canonical human reference database (downloaded from Uniprot 15/03/2021; 75776 entries). Raw files with multiple internal FAIMS CVs were split into separate files based on the CV values of acquisition prior to MQ searches. These separated files were specified as different fractions, as for the bRP fractions, of the same experiment name in MQ. Multiple injections of the same sample were specified as the same experiment. Default MQ search parameters were used. Trypsin/P was specified as protease, allowing for up to a maximum of 2 missed cleavages. Carbamidomethylation of cysteine was specified as fixed modification, while oxidation of methionine and protein N-terminal acetylation were considered as variable modifications. The label free quantification (LFQ) algorithm, with a standard LFQ min. ratio count setting of 2, as well as the iBAQ algorithm, with log fit, was switched on where needed. Where used, the Match-Between-Runs (MBR) algorithm was switched on with default settings (0.7 min and 5 min for matching and retention time alignment window, respectively). The false discovery rate (FDR) was set to 1 % on protein and peptide-spectrum matches PSM level. For Prosit rescoring16, the MQ searches were reprocessed (partial processing from: “reading search engine results“) with a 100 % FDR cut-off at protein and PSM level. The respective MQ msms.txt files were rescored by Prosit. Peptides with q-values ≤ 0.01 were retained and proteins were grouped based on the picked FDR method. For both MQ and Prosit outputs, proteins for which no unique peptide was found were aggregated into protein groups. For simplicity, we refer to these only as proteins in throughput the manuscript. Data analysis and visualization was performed using R (v 4.1.0) in RStudio and Python.
Data repository
The mass spectrometry proteomics raw data and MaxQuant and Prosit search results have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD028136.
Results and Discussion
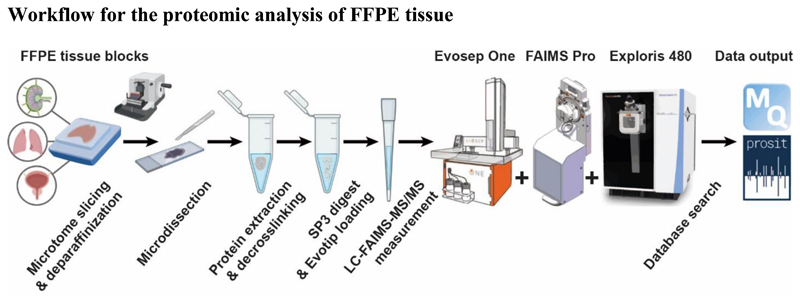
Workflow for the proteomic analysis of FFPE tissue
Figure 1 schematically shows the steps in the overall workflow and that are detailed in subsequent sections below. Briefly, archived FFPE tissue blocks are cut into slices using a microtome, deparaffinized and the area of interest is micro-dissected with a scalpel. For protein extraction and de-crosslinking, the tissue is boiled in SDS at alkaline pH, followed by detergent removal and protein digestion using the SP3 method. Tryptic peptides are loaded onto single-use disposable Evotips for desalting and interfacing to the nanoLC system. Each sample is analysed twice to accommodate extensive gas-phase fractionation by FAIMS prior to tandem mass spectrometry. Finally, peptide and protein identification and quantification are performed by MaxQuant15 and Prosit rescoring16.
Figure 1.
Schematic representation of the proteomic workflow for FFPE material evaluated in this study (created using BioRender). Drawing of the Evosep One LC courtesy of Evosep Biosystems. Drawing of the FAIMS Pro™ interface and Orbitrap Exploris™ 480 mass spectrometer courtesy of Thermo Fisher Scientific.
Evaluation of protein extraction and de-crosslinking
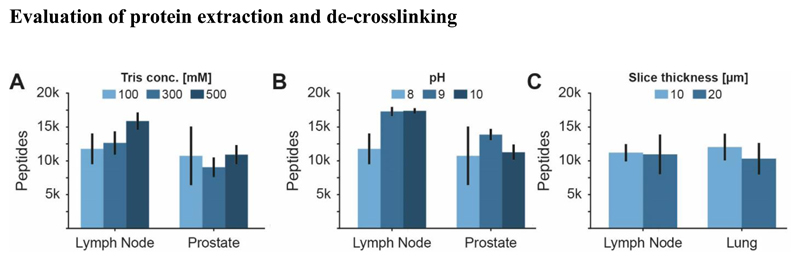
We briefly revisited an established sample preparation protocol that uses 10 μm tissue slices and extracts proteins from FFPE material in 100 mM Tris (pH 8) 17. Three human tissue types were selected for this evaluation, each having different characteristics. The lymph node is a dense and homogeneous tissue while prostate is more heterogeneous and lung has a looser texture. We observed that samples extracted at a higher ionic strength resulted in higher numbers of identified peptides for lymph node tissue (Fig. 2A; mean of 16,694 vs. 12,415 peptides at 500 mM vs. 100 mM Tris respectively). Similarly, raising the pH from 8 to 9 showed a benefit for lymph node and prostate tissue (Fig. 2B; mean of 11,787 vs 17,291 peptides at pH 8 and pH 9 respectively for lymph node). In contrast, slice thickness did not appear to have a strong influence (Fig. 2C). These results are in line with prior reports suggesting the use of higher Tris concentrations and more alkaline conditions18–20. The former may be explained by more efficient scavenging of the aldehydes that are liberated during de-crosslinking18 and the latter by more efficient hydrolysis of the formaldehyde cross links. Results obtained at the peptide level were mirrored at the protein level (Fig. S1) and thus, we settled on the use of 10 μm FFPE tissue slices and 500 mM Tris at pH 9 for all subsequent experiments.
Figure 2. Influence of selected sample preparation parameters on the number of peptides identified from FFPE material.
(A) Ionic strength of the extraction buffer, (B) pH of the extraction buffer and (C) thickness of tissue slices. For each analysis, 50 ng of peptide digest were analysed by a 60 min nLC-MS/MS run. Error bars indicate the standard error of mean (SEM). For numbers of replicates see Supplemental Table S1.
Evaluation of the disposable trap nanoLC system
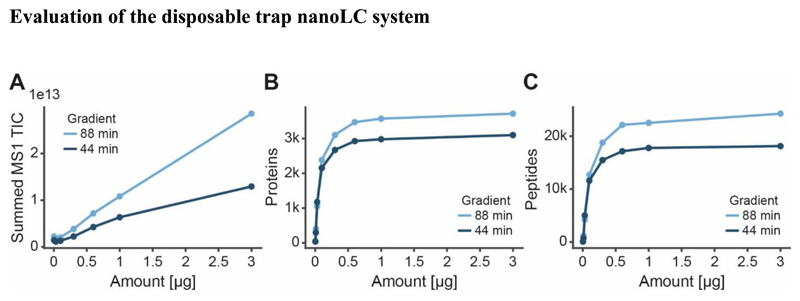
To characterize the nanoLC system, increasing amounts of peptides (HeLa cell line digest; 3 - 3,000 ng) were loaded onto Evotips and separated using 44 min and 88 min LC methods. The authors note that the Evosep One only supports certain LC methods and column dimensions corresponding to e. g. 30 and 15 samples per day for the 44 and 88 min method respectively. While the total ion current (TIC) of MS1 scans increased as a function of sample loading (Fig. 3A), the number of peptides and proteins that could be identified saturated at loadings of 600 ng or higher (Fig. 3B, 3C). As one would expect, using the longer LC gradient identified more peptides (+19 %) and proteins (+29 %). Because 600 ng of peptide yield is easily obtained from a single slice of micro-dissected tissue (4.8 μg median peptide yield from a 5x5 mm area of one 10 μm slice) and this loading is also in the linear range of the mass spectrometric signal, we used 600 ng for the subsequent experiments. The authors note that at low loadings (100 ng or less), a substantial loss of hydrophobic peptides was observed (Fig. S2), indicating that the workflow presented here may have to be adjusted (e. g. using smaller bed volumes of the disposable trap column or direct injection onto a conventional nanoLC system) to support the analysis of laser-capture micro-dissected tissue.
Figure 3. Results of the analysis of dilution series of Hela cell line digests using an Evosep One nLC system that operates at fixed gradients of either 44 or 88 min.
(A) Summed total ion current (TIC) for all MS1 scans, (B) number of identified protein groups and (C) number of identified peptides as a function of sample loading onto Evotips.
Characterization and optimization of FAIMS parameters
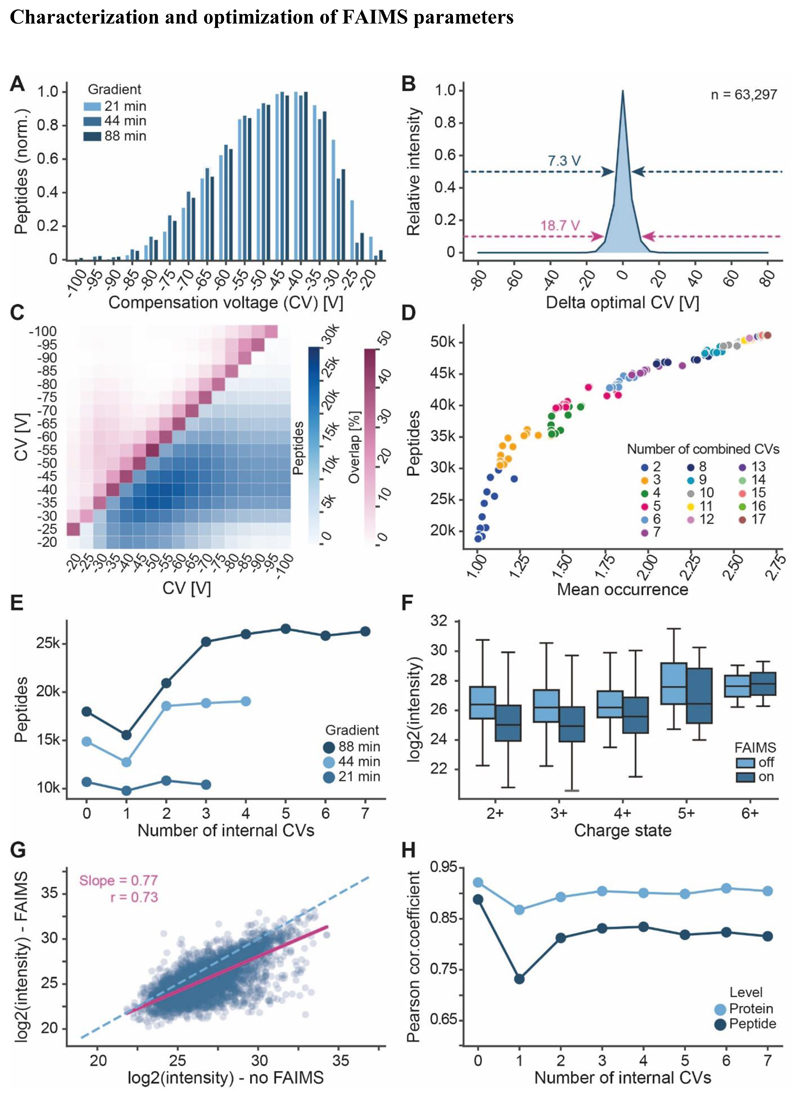
In order to characterize the gas phase fractionation of peptides by FAIMS, we investigated how many peptides can be identified from a complex digest as a function of the applied compensation voltage (CV), ranging from -100 V to - 20 V in 5 V intervals. The results showed a rather broad and skewed distribution which was independent of which LC gradient time was used (Fig. 4A; Fig. S3A-B). We next sought to estimate the separation power of FAIMS using the peptide identifications from the 88 min gradient. About 31% of all peptide features (n=63,297) were detected at a single CV only, another 29 % at 2 CVs, 25 % at 3 CVs and 15 % at more than 3 CVs (Fig. S3C). When analyzing the intensity distributions of these peptides across all CVs at which they were detected showed that, on average, 50 % of the intensity was contained within 7.3 V and 90 % within 18.7 V (Fig. 4B). These results indicate that multiple CVs should be used to maximize peptide coverage and quantitative information. Furthermore, multiple CVs should be spaced by at least 7.3 V to minimize redundant peptide identifications when combined in a single nLC-FAIMS-MS/MS run (see also below).
Figure 4. Characterization of FAIMS parameters.
(A) Distribution of peptide identification at different compensation voltages (CV) and gradient lengths. (B) Approximation of FAIMS resolution by analysing the intensities of peptides detected at different CVs relative to the intensity at their optimal CV (88 min gradient data shown). Arrows and values indicate the full width at half maximum (dark blue) or the width of the distribution at 10% maximal intensity (purple). (C) Absolute number of identified peptides (blue scale) and relative number of overlapping peptides (purple scale) for any two combined CVs (88 min gradient data shown). (D) In-silico analysis of the number of unique peptides that may be identified by combing between 2-17 CV values (based 88 min gradient data). (E) Number of actually identified peptides when combining several CVs within an nLC-MS/MS run of different gradient lengths. (F) Precursor ion intensity distributions of peptides detected in different charge states with or without FAIMS (CV=-45 V). (G) Scatterplot of peptide precursor ion intensities with or without FAIMS (CV=-45 V). The dashed line marks the diagonal and the purple line depicts the linear regression (r=Pearson correlation coefficient). (H) Correlation of peptide and protein group intensities of nLC-MS/MS runs applying the indicated number of internal CVs to the nLC-MS/MS run without FAIMS (For 0, another replicate without FAIMS was used).
The total number of unique peptides identified from the 17 single CV experiments was 51,174 (5,239 proteins; 88 min gradients). Because performing that many runs is neither feasible nor desirable - particularly when sample quantities are limited -, we asked how many and which CVs should be fitted into a single nLC-FAIMS-MS/MS run to maximize peptide and protein coverage. To this end, we first simulated the number of unique peptides that may be detectable by combining results obtained from the single CV experiments in silico (Fig. 4C). The analysis showed that the best combination of two CVs (-40 and -55 V) may allow the identification of up to 30,218 peptides with less than 13 % overlap. Computing peptide identifications for combinations of 2-17 CVs showed that the relative gains become smaller the more CVs are combined. In addition, there is a substantial influence of which exact CVs are combined, this effect becomes smaller when many CVs are combined (Fig. 4D; Supplemental Table S1).
Next, we tested to which extent the above simulations may translate into practice. Combining 17 CVs into a single nLC-FAIMS-MS/MS run is not useful because switching CVs requires about 25 ms of time. This is about the same time as needed to record one MS/MS spectrum. Hence, a balance has to be found between using as many CVs as possible for peptide separation and recording as many MS/MS spectra as possible for peptide identification. The number of CV values that can be accommodated in a single run also depends on the available time which is determined by the chromatographic peak width. In other words, long gradients (i. e. wider LC peaks) should benefit more from combining multiple CVs than short gradients (i. e. sharp LC peaks). Using a gradient of 88 min and combining five CVs into a single run as an example, we found that only about 28,000 of the ~42,000 peptides expected from the simulation could actually be observed indicating that there was not enough time to sample all the peptides entering the mass spectrometer. As a result, there was also no noticeable difference between the five best combinations of five CVs (Fig. S3D). We note that these experiments already used the shortest tested MS2 scan time (15k resolution) as this provided the highest numbers of peptide and protein identifications for the sample loading of 600 ng used in this study. However, when less sample is available, higher resolution MS2 scans can be advantageous (Fig. S3E).
Next, we empirically tested the benefit of combining multiple CVs into nLC-FAIMS-MS/MS runs using different LC gradient times of 21, 44 and 88 min (Fig. 4E). At the peptide level, FAIMS with a single CV value invariably reduced the number of identified peptides but still improved the number of protein identifications (Fig. S3F). This has been observed for FAIMS before13 and is a typical fractionation effect also observed in the analysis of off-line HPLC separations, where different peptides of the same protein are distributed across fractions21. When analyzing just a single fraction (for FAIMS a single CV value), the mass spectrometer does not ‘see’ all of the most intense peptides at the same time. This allows the MS to also sample peptides of lower abundant proteins, thus sampling over a bigger dynamic range of the sample. Combining two CVs outperformed the number of peptide and protein identifications without FAIMS for all three gradients and the overall characteristics suggest that one, two and five CV values provide best results for 21, 44 and 88 min gradients respectively.
Peptide and protein quantification in nLC-FAIMS-MS/MS
As for any ion mobility system, an important parameter for peptide ion separation by FAIMS is their charge state. As tryptic peptides predominantly produce multiply charged ions, FAIMS is also an effective way to prevent singly charged background ions to enter the mass spectrometer. This keeps the instrument clean for longer times and can also facilitate the detection of low abundant multiply charged ions. However, the fact that the FAIMS device effectively blocks the entrance to the mass spectrometer, there is also the chance of losing ions produced by the electrospray source. In addition, and as shown above, only a part of the peptide ion signal is transmitted at a single CV. Both effects reduce the number of detectable ions and may negatively affect quantitative performance. This is illustrated in Fig. 4F where the intensities of identified peptides are shown as a function of their charge state and comparing their intensities without and with FAIMS (single CV of -45V). It is apparent, that the intensities of doubly and triply charged peptides (the vast majority of tryptic peptides) are substantially lower with FAIMS. When comparing the intensities of individual peptides measured with and without FAIMS, we observed a drop in Pearson correlation coefficient from 0.89 (two replicates without FAIMS) to 0.73 (no FAIMS vs. FAIMS with 1 CV) (Fig 2G-H, Fig. S4A). This is due to the aforementioned effect that a single CV is not optimal for many peptides. Even if it is, that single CV typically does not transmit the entire signal. Hence, the correlation improves to r=0.83 when three or more CVs are combined because more of the total signal of a given peptide can be detected. Still, this does not fully compensate for the absolute loss of ions by the FAIMS device. When rolling up peptide into protein intensities, the correlations between FAIMS and no FAIMS are much better (r=0.90) particularly when at least two CV values are included. Not only does this capture more of the signal of a particular peptide, it also increases the number of identified peptides per protein (Fig. 4H, Fig. S4B, S5).
Gas-phase vs basic pH reversed-phase fractionation
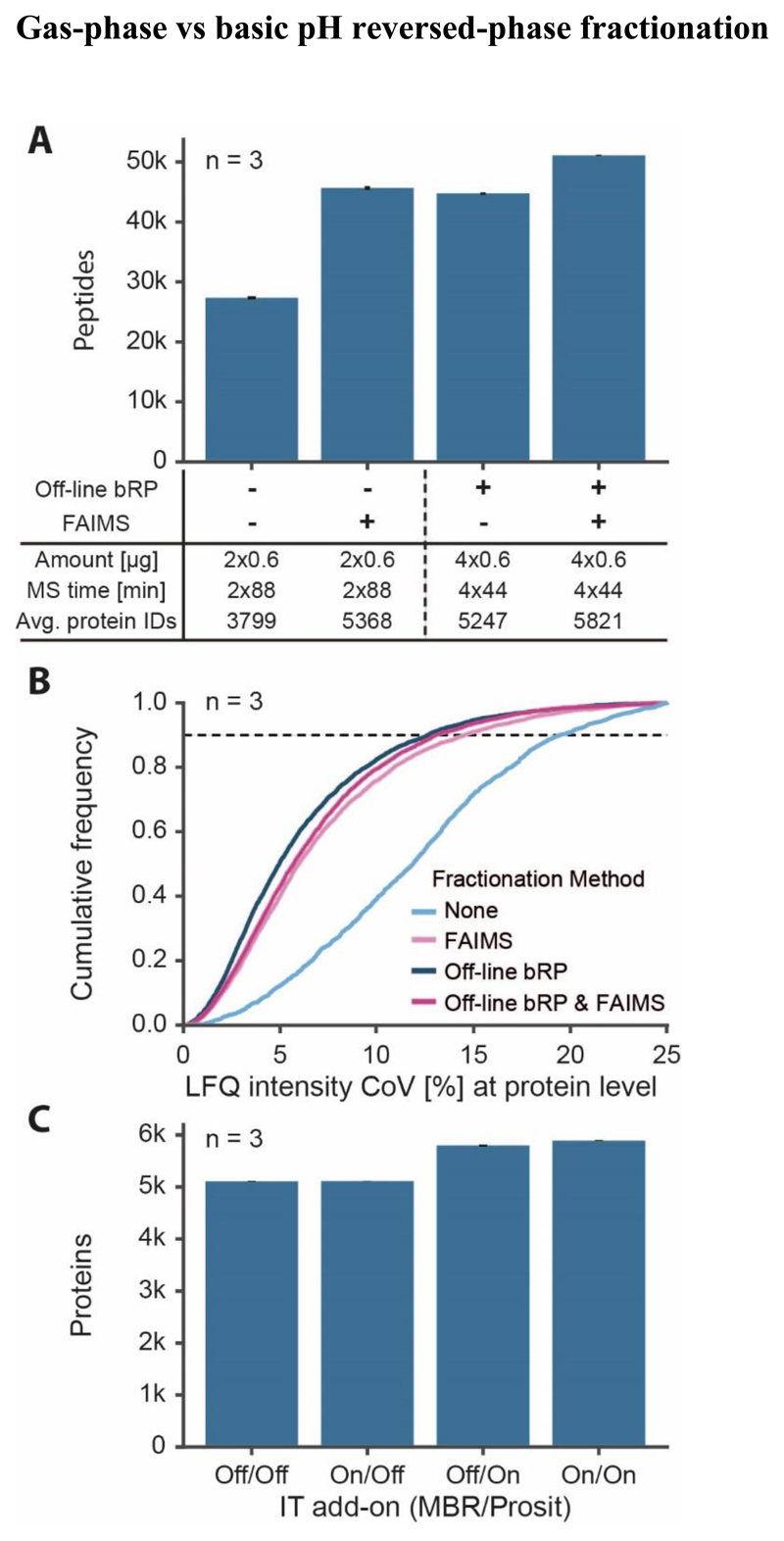
One popular approach to obtain deeper proteome coverage from a given sample is fractionation by basic pH reversed-phase (bRP) chromatography on ‘StageTips’ because this is partially orthogonal to online coupled low pH chromatography14, 22. Although this step may be automated on liquid handling platforms to compensate for the additional time needed, a disadvantage is that some sample loss is inevitably incurred. Therefore, nLC-FAIMS-MS/MS may be an attractive alternative as it does not require the laboratory step and sample losses may be minimized at the same time. To test this hypothesis, a series of experiments was performed as outlined in Fig. 5A (more details in Fig. S6A). To remove the influence of sample losses during bRP, a large quantity (20 μg) of HeLa digest was separated on StageTips into four fractions. 600 ng peptides per fraction were loaded on Evotips and each fraction was measured using the 44 min gradient with or without FAIMS (2 CVs). This led to the identification of, on average, 5,247 and 5,821 proteins respectively (n=3). To analyse the same digest without fractionation but using the same amount of nLC-MS/MS time, 600 ng of peptides were loaded onto Evotips and analysed twice using an 88 min gradient with or without FAIMS (2 sets of 5 CVs per run; total of 10 different CVs). On average, this led to the identification of, 3,799 and 5,368 proteins respectively (n=3) showing that nLC-FAIMS-MS/MS can reach a similar proteomic depth as bRP fractionation but using half of the amount of sample and not requiring additional laboratory steps. Replicate analysis of the above experiments (n=3) showed that nLC-FAIMS-MS/MS was similarly reproducible as bRP fractionation nLC-MS/MS, both achieving a coefficient of variation (CoV) of less than 15 % for more than 90 % of the quantified proteins (Pearson correlation coefficient r>0.95 for intensities of replicates) and performing substantially better than the analysis without any fractionation (Fig. 5B; Fig. S6B). Finally, applying the ‘match-between-run’ feature of MaxQuant/Andromeda and/or Prosit rescoring of database search engine results16, 23 led to nearly 5,900 proteins that can be detected from 1.2 μg of HeLa peptides within 3 hours of nLC-FAIMS-MS/MS time (Fig. 5C).
Figure 5. Comparison of proteomic depth obtained by FAIMS or off-line fractionation.
(A) Peptides identified (with SEM) with or without FAIMS and with or without basic pH reversed-phase fractionation (bRP; four fractions). Two sets of five CVs were combined for 88 min separations and 2 CVs were used for 44 min separations. (B) Empirical cumulative distribution function (ECDF) of the quantitative reproducibility described by the coefficient of variation (CoV) based on protein group intensities of three replicates (same data as for A). (C) Effect of using the ‘match between runs’ (MBR) feature of MaxQuant and/or Prosit rescoring on the number of identified proteins (with SEM, 88 min gradient with double injection using FAIMS with two sets of 5 CVs).
Application of nLC-FAIMS-MS/MS to the analysis of FFPE tissue material
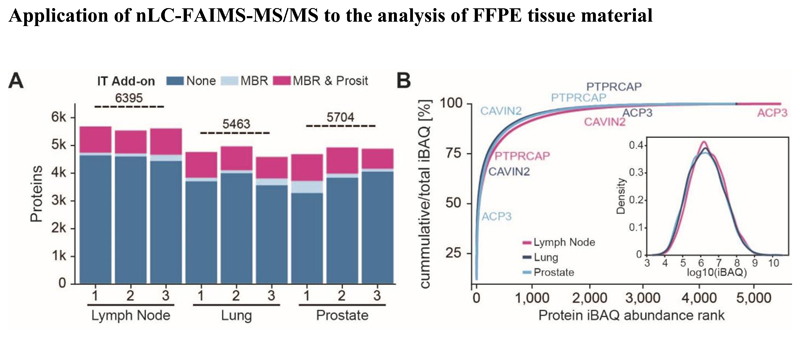
In light of the above technical evaluation, we conclude that nLC-FAIMS-MS/MS can yield good quantitative performance for a large number of proteins from a limited amount of FFPE sample and using an acceptable amount of time per sample. This makes it a viable option for the analysis of clinical FFPE material. To show that this is indeed the case, we deployed the workflow shown in Fig. 1 to the proteomic profiling of three exemplary FFPE tissues (n=3 each). Combining the results of each individual tissue, a total number of 6,395, 5,463 and 5,704 proteins could be identified from lymph node, lung and prostate, respectively (Fig. 5A, Fig. S7). The dynamic range of protein abundance as measured by the iBAQ approach24 spanned nearly 7 orders of magnitude for all three tissues (Fig. 6B). Transforming this data into a cumulative abundance plot, confirmed yet again that the vast majority of the total protein content of a tissue is comprised of few proteins and that this characteristic is tissue-dependent. For example, prostatic acid phosphatase (ACP3) is highly abundant in prostate but positioned at the lower end of the abundance distribution in the other two tissues. Similarly, caveolae-associated protein 2 (CAVIN2) is highly expressed in the lung, moderately expressed in prostate and of low abundance in lymph node. In contrast, protein tyrosine phosphatase receptor type C-associated protein (PTPRCAP), a CD-45 associated protein, is high the lymph node but low in the other two tissues.
Figure 6.
Analysis of three exemplary human FFPE tissues (A) Number of proteins identified from lymph node, lung and prostate (n=3 each) by standard database searching, match-between-runs enabled and re-scored by Prosit. Horizontal lines depict the total number of proteins identified in each tissue. (B) Cumulative protein abundance distributions of the three human tissues. The inset shows a density plot of protein abundance measured by the iBAQ approach (intensity-based absolute protein quantification). Tissues are color coded and examples for proteins detected in all three tissues but in very different quantities are marked.
Conclusions
The results obtained above for the analysis of FFPE tissue proteomes compare well to published work in this area. For instance, Coscia et al.3 analysed several tumor entities and identified ~5,000 proteins without fractionation in two hours of LC-MS/MS or ~7,000 proteins in the same time when using data independent analysis (DIA). Pozniak et al.25 analysed a cohort of breast cancer patients and quantified ~1700-7,700 proteins per patient from 12 FFPE slices followed by chromatographic fractionation and requiring ~24 hours of time per sample. Buczak et al.26 used tandem mass tags27 for sample multiplexing and basic pH off-line fractionation and report the identification of 5,000-6,000 proteins from single FFPE slices of hepatocellular carcinoma patients requiring the equivalent of 4 hours of time per patient. All these approaches clearly have merit. Some are rather laborious, some require expensive stable isotope reagents and some consume larger amounts of sample and/or time. The workflow advocated in this study aimed to balance many of these aspects to enable the measurement of large sample cohorts. This is important to ensure that e. g. biomarker identification campaigns are statistically well powered. Experience from work in the authors’ laboratory (>1,000 FFPE samples from different tumor entities) suggests that disposable trap column nLC-FAIMS-MS/MS provides the necessary throughput and robustness to achieve this. The workflow also offers substantial potential for downscaling e. g. to samples obtained by laser-capture micro-dissection. Given that disposable trap nanoLCs and FAIMS are becoming more widely available, we anticipate that the workflow presented in this study will find ample application in the field.
Supplementary Material
The following supporting information is available free of charge on the ACS website (http://pubs.acs.org)
Supplemental Figure S1-S7. Additional results and data analysis (PDF)
Supplemental Table S1. Numbers of replicates for Fig. 2, CV blender output for Fig. 4D and optimal internal CVs used for Fig. 4E (XLS)
Acknowledgment
This work was partially supported by the German Science foundation (DFG-SFB1309), the ERC Advanced Grant (TOPAS; 833710) and the Federal Ministry of Education and Research (Clinspect-M; FKZ161L0214A). We are grateful to Miriam Abele for the acquisition of mass spectrometry data shown in Figure 2. We wish to thank all members of the Kuster group for technical assistance and fruitful discussions.
Abbreviations
- FFPE
formalin-fixed paraffin-embedded
- FAIMS
high field asymmetric waveform ion mobility spectrometry
- LC-MS
liquid chromatography-mass spectrometry
- CV
compensation voltage
- HPLC
high performance liquid chromatography
- bRP
basic pH reversed-phase
- CoV
coefficient of variation
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Bernhard Kuster is co-founder and shareholder of OmicScouts and MSAID. He has no operational role in either company. The authors have no competing financial interest to declare. The mass spectrometry proteomics raw data and MaxQuant and Prosit search results have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD028136.
References
- 1.Hughes CS, Moggridge S, Müller T, Sorensen PH, Morin GB, Krijgsveld J. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nature Protocols. 2019;14(1):68–85. doi: 10.1038/s41596-018-0082-x. [DOI] [PubMed] [Google Scholar]
- 2.Müller T, Kalxdorf M, Longuespée R, Kazdal DN, Stenzinger A, Krijgsveld J. Automated sample preparation with SP3 for low-input clinical proteomics. Molecular Systems Biology. 2020;16(1):e9111. doi: 10.15252/msb.20199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coscia F, Doll S, Bech JM, Schweizer L, Mund A, Lengyel E, Lindebjerg J, Madsen GI, Moreira JMA, Mann M. A streamlined mass spectrometry–based proteomics workflow for large-scale FFPE tissue analysis. The Journal of Pathology. 2020;251(1):100–112. doi: 10.1002/path.5420. [DOI] [PubMed] [Google Scholar]
- 4.Hughes CS, McConechy MK, Cochrane DR, Nazeran T, Karnezis AN, Huntsman DG, Morin GB. Quantitative Profiling of Single Formalin Fixed Tumour Sections: proteomics for translational research. Scientific Reports. 2016;6(1):34949. doi: 10.1038/srep34949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedrich C, Schallenberg S, Kirchner M, Ziehm M, Niquet S, Haji M, Beier C, Neudecker J, Klauschen F, Mertins P. Comprehensive micro-scaled proteome and phosphoproteome characterization of archived retrospective cancer repositories. Nature Communications. 2021;12(1):3576. doi: 10.1038/s41467-021-23855-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim YJ, Sweet SMM, Egertson JD, Sedgewick AJ, Woo S, Liao WL, Merrihew GE, Searle BC, Vaske C, Heaton R, MacCoss MJ, et al. Data-Independent Acquisition Mass Spectrometry To Quantify Protein Levels in FFPE Tumor Biopsies for Molecular Diagnostics. J Proteome Res. 2019;18(1):426–435. doi: 10.1021/acs.jproteome.8b00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bache N, Geyer PE, Bekker-Jensen DB, Hoerning O, Falkenby L, Treit PV, Doll S, Paron I, Méller JB, Meier F, Olsen JV, et al. A Novel LC System Embeds Analytes in Pre-formed Gradients for Rapid Ultra-robust Proteomics*. Molecular & Cellular Proteomics. 2018;17(11):2284–2296. doi: 10.1074/mcp.TIR118.000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bekker-Jensen DB, Martínez-Val A, Steigerwald S, Rüther P, Fort KL, Arrey TN, Harder A, Makarov A, Olsen JV. A Compact Quadrupole-Orbitrap Mass Spectrometer with FAIMS Interface Improves Proteome Coverage in Short LC Gradients. Mol Cell Proteomics. 2020;19(4):716–729. doi: 10.1074/mcp.TIR119.001906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helm D, Vissers JPC, Hughes CJ, Hahne H, Ruprecht B, Pachl F, Grzyb A, Richardson K, Wildgoose J, Maier SK, Marx H, et al. Ion mobility tandem mass spectrometry enhances performance of bottom-up proteomics. Mol Cell Proteomics. 2014;13(12):3709–3715. doi: 10.1074/mcp.M114.041038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez-Lima F, Kaplan DA, Suetering J, Park MA. Gas-phase separation using a trapped ion mobility spectrometer. International Journal for Ion Mobility Spectrometry. 2011;14(2):93–98. doi: 10.1007/s12127-011-0067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonneil E, Pfammatter S, Thibault P. Enhancement of mass spectrometry performance for proteomic analyses using high-field asymmetric waveform ion mobility spectrometry (FAIMS) Journal of Mass Spectrometry. 2015;50(11):1181–1195. doi: 10.1002/jms.3646. [DOI] [PubMed] [Google Scholar]
- 12.Pfammatter S, Bonneil E, McManus FP, Prasad S, Bailey DJ, Belford M, Dunyach J-J, Thibault P. A Novel Differential Ion Mobility Device Expands the Depth of Proteome Coverage and the Sensitivity of Multiplex Proteomic Measurements. Mol Cell Proteomics. 2018;17(10):2051–2067. doi: 10.1074/mcp.TIR118.000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hebert AS, Prasad S, Belford MW, Bailey DJ, McAlister GC, Abbatiello SE, Huguet R, Wouters ER, Dunyach J-J, Brademan DR, Westphall MS, et al. Comprehensive Single-Shot Proteomics with FAIMS on a Hybrid Orbitrap Mass Spectrometer. Analytical Chemistry. 2018;90(15):9529–9537. doi: 10.1021/acs.analchem.8b02233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature Protocols. 2007;2(8):1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 15.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 16.Gessulat S, Schmidt T, Zolg DP, Samaras P, Schnatbaum K, Zerweck J, Knaute T, Rechenberger J, Delanghe B, Huhmer A, Reimer U, et al. Prosit: proteome-wide prediction of peptide tandem mass spectra by deep learning. Nature Methods. 2019;16(6):509–518. doi: 10.1038/s41592-019-0426-7. [DOI] [PubMed] [Google Scholar]
- 17.Tyanova S, Albrechtsen R, Kronqvist P, Cox J, Mann M, Geiger T. Proteomic maps of breast cancer subtypes. Nature Communications. 2016;7(1):10259. doi: 10.1038/ncomms10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawashima Y, Kodera Y, Singh A, Matsumoto M, Matsumoto H. Efficient extraction of proteins from formalin-fixed paraffin-embedded tissues requires higher concentration of tris(hydroxymethyl)aminomethane. Clin Proteomics. 2014;11(1):4. doi: 10.1186/1559-0275-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magdeldin S, Yamamoto T. Toward deciphering proteomes of formalin-fixed paraffin-embedded (FFPE) tissues. Proteomics. 2012;12(7):1045–58. doi: 10.1002/pmic.201100550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Y, Weiss T, Zhang Q, Sun R, Wang B, Yi X, Wu Z, Gao H, Cai X, Ruan G, Zhu T, et al. High-throughput proteomic analysis of FFPE tissue samples facilitates tumor stratification. Mol Oncol. 2019;13(11):2305–2328. doi: 10.1002/1878-0261.12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulak NA, Geyer PE, Mann M. Loss-less Nano-fractionator for High Sensitivity, High Coverage Proteomics*. Molecular & Cellular Proteomics. 2017;16(4):694–705. doi: 10.1074/mcp.O116.065136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruprecht B, Zecha J, Zolg DP, Kuster B. High pH Reversed-Phase Micro-Columns for Simple, Sensitive, and Efficient Fractionation of Proteome and (TMT labeled) Phosphoproteome Digests. Methods Mol Biol. 2017;1550:83–98. doi: 10.1007/978-1-4939-6747-6_8. [DOI] [PubMed] [Google Scholar]
- 23.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nature Protocols. 2016;11(12):2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 24.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 25.Pozniak Y, Balint-Lahat N, Rudolph Jan D, Lindskog C, Katzir R, Avivi C, Pontén F, Ruppin E, Barshack I, Geiger T. System-wide Clinical Proteomics of Breast Cancer Reveals Global Remodeling of Tissue Homeostasis. Cell Systems. 2016;2(3):172–184. doi: 10.1016/j.cels.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Buczak K, Kirkpatrick JM, Truckenmueller F, Santinha D, Ferreira L, Roessler S, Singer S, Beck M, Ori A. Spatially resolved analysis of FFPE tissue proteomes by quantitative mass spectrometry. Nature Protocols. 2020;15(9):2956–2979. doi: 10.1038/s41596-020-0356-y. [DOI] [PubMed] [Google Scholar]
- 27.Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Hamon C. Tandem Mass Tags: A Novel Quantification Strategy for Comparative Analysis of Complex Protein Mixtures by MS/MS. Analytical Chemistry. 2003;75(8):1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1-S7. Additional results and data analysis (PDF)
Supplemental Table S1. Numbers of replicates for Fig. 2, CV blender output for Fig. 4D and optimal internal CVs used for Fig. 4E (XLS)