Abstract
Microbial mixed cultures are gaining increasing attention as biotechnological production systems, since they offer a large but untapped potential for future bioprocesses. Effects of secondary metabolite induction and advantages of labor division for the degradation of complex substrates offer new possibilities for process intensification. However, mixed cultures are highly complex, and, consequently, many biotic and abiotic parameters are required to be identified, characterized, and ideally controlled to establish a stable bioprocess. In this review, we discuss the advantages and disadvantages of existing measurement techniques for identifying, characterizing, monitoring, and controlling mixed cultures and highlight promising examples. Moreover, existing challenges and emerging technologies are discussed, which lay the foundation for novel analytical workflows to monitor mixed-culture bioprocesses.
Mixed Cultures: Opportunities and Challenges
An old tradition is revolutionizing modern biotechnology: microbial collaboration. Mixed-culture applications, such as wastewater treatment, composting, and a broad spectrum of fermentative food preparations, are the historical foundation of current biotechnology. Now, a targeted assembly of microorganisms to perform concerted bioproductions is forming a new cutting edge in biotechnology. Over the past decade, the research field of defined mixed cultures has gained increased attention due to their potential for process intensification and the chance to produce unknown secondary metabolites [1]. In the old-school mixed-culture approach, the driving force to develop mixed culture-specific (online) measurement techniques was very limited. Reasons for this might be the rather low dynamic of these processes and the strong closeness to natural processes and equilibria with no need for control. In other words, there was simply no need for a deeper understanding of these robust systems. However, this is now changing.
Since sustainability is currently the most important driving force in biotechnology, the number of published studies on mixed-culture consolidated bioprocesses (see Glossary) with plant biomass, waste streams, and gaseous waste substrates is increasing significantly [2–9]. The products of these processes are, in many cases, acids, alcohols, or enzymes for biomass pretreatment and hydrolysis [6,9]. Higher-value products of mixed cultures are secondary metabolites induced by interspecies communication [1,10–14]. Among these, antimicrobial substances, such as antibiotics, are of critical interest, especially in the context of increasingly multiresistant pathogens [15]. However, in-depth studies in this field are only possible with measurement tools for the identification and characterization of the microbial composition. To avoid the complexity of unknown natural microbial communities, synthetic co-culture systems allow defined combinations of microorganisms with beneficial metabolic pathways. Yet, in contrast to natural communities, which are typically stabilized by natural chemical, metabolic, and ecological equilibria, synthetic co-cultures have no natural equilibrium, which can result in highly dynamic and instable processes and makes monitoring and control especially necessary [16,17].
Glossary.
- Co-culture
defined mixture comprising a limited number of participants. Co-cultures are in general synthetic microbial systems.
- Consolidated bioprocess (CBP)
possible way for process intensification by a mixed or co-culture. Several normally individual process steps are combined in one reactor system. The term is often used in the context of conversion of complex substrates (e.g., lignocellulose) into higher valued products.
- Flow cytometry (FC)
technique used to detect and measure physical and chemical characteristics of single cells in a high-throughput flow channel.
- Invasive/non-invasive techniques
describes the degree of contact and possible influence of a measurement method on a sample.
- Microbial communities
groups of microorganisms that share a common living space. The number of participants as well as the composition is often unknown.
- Microfluidic systems
in the context of biotechnology, cultivation or cell-handling systems on a pico- or nanoliter scale.
- Mixed culture
culture that includes multiple species. The composition is known but the ratio between single microorganisms is undefined. The term is often used to describe natural microbial systems.
- rDNA sequencing
sequencing of ribosomal RNA genes and the internal transcribed spacers between them is used as a genetic barcode that allows the phylogenetic classification and identification of organisms.
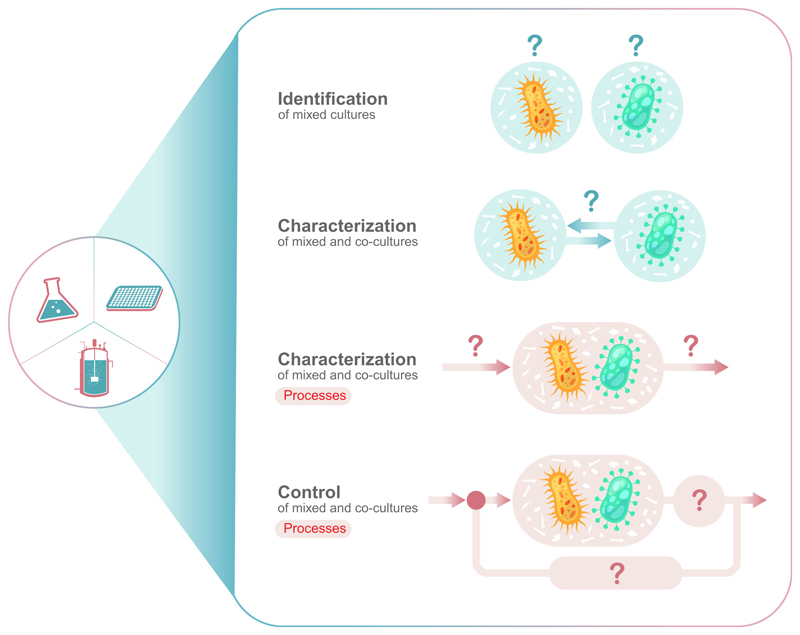
Compared with classic monoculture processes, the analysis of mixed and co-cultures can be structured into four fields with different requirements for the applied measurement technology (Figure 1, Key Figure): identification of species (mainly for mixed cultures); characterization of interactions; characterization of a process; and the control of that process. With respect to the necessary measurement technologies, there is a gradient in complexity from identification with only offline methods to process control with the necessity for online measured parameters (Box 1).
Figure 1. The four levels are identification, characterization, process development, and process control.
Key Figure Four Main Levels of Information to Investigate a Mixed or Co-Culture of Microorganisms
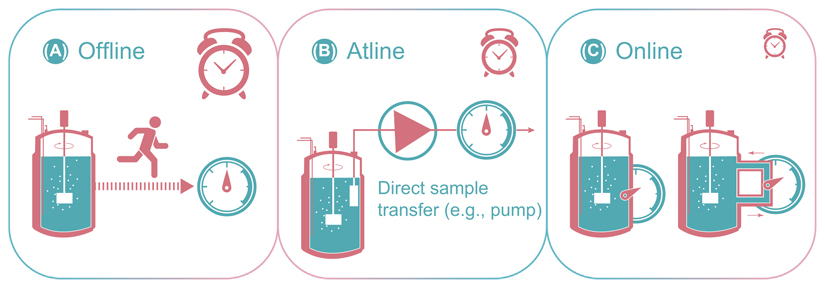
Box 1. Classification of Monitoring Techniques.
Monitoring techniques for bioprocess analysis and control can be classified in three categories (Figure I): offline, atline, and online. These categories are defined according to the location of the analytical system in relation to the bioreactor. Offline measurement systems (Figure IA) comprise manual or automatic sampling. Samples are afterwards analyzed in an external laboratory workflow and data are generally obtained with a temporal delay. Therefore, the information is not available for any bioreactor control strategies. In atline measurements (Figure IB), samples are analyzed by the side of the bioreactor. Data are typically available with an analysis-specific delay but generally more quickly compared with offline analyses. The third category is online analysis. For online measurements, in situ and bypass configurations are possible (Figure IC). In situ measurements are the preferred technology, since a sensor is directly located inside the bioreactor. All signals are directly available and, therefore, predestined for any kind of control. Alternatively, a sensor can also be located in a bypass, which increases the flexibility regarding size and geometry of the applied measurement technology. However, transfer of biomass in a bypass may cause changes in the physicochemical conditions of the sample, due to oxygen limitations and or heterogeneities caused by insufficient mixing.
Figure I. Schematic Illustration of Offline, Atline and Online Measurement Technologies for Bioprocess Control.
While all information about substrates, metabolic products, and cell morphologies is accessible as a summary parameter of all microbial activities via established offline methods, only a few techniques are available providing specific information about the mixed-culture composition and the individual-specific performance. Consequently, the in-depth characterization of an ongoing cultivation and directed optimization of process parameters via available online analysis technology are currently difficult to realize. Even more, with this lack of insight regarding the community and functional role, interactions, and so on, of specific subpopulation members, control of a specific mixed-culture composition is unachievable.
In our opinion, the lack of analytical tools to study processes, especially at larger scales, is a major reason for the discrepancy between the high scientific interest in mixed cultures but the limited number of successfully commercialized defined mixed and co-culture processes in biotechnology [18]. Therefore, the focus of this review is the discussion of advantages and disadvantages of existing measurement techniques to resolve and control population dynamics of mixed-culture processes. Researchers who are new in the field of mixed cultures could be guided by this article to find a suitable measurement technique to analyze their specific microbial community.
Mixed and Co-Culture Characterization
Natural and Synthetic Cellular Characteristics
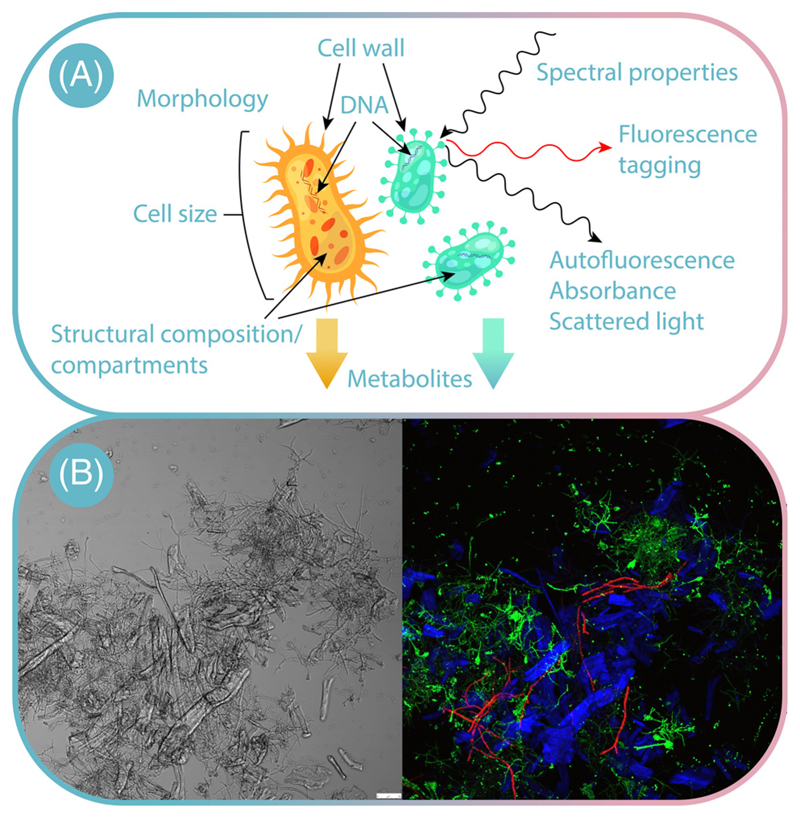
There are subtle structural, biochemical, and genetic differences between different species and strains of naturally occurring microorganisms (Figure 2). Whereas genetic differences are used for offline strain identification, differences in cell morphology and structural composition, such as compartments, can be used for online monitoring (Figure 2A). Differences within the metabolism lead to different biochemical characteristics, such as protein/metabolite composition and quantity. The infrared spectrum of these compounds gives a unique ‘fingerprint’ for different cells. Differences within fatty acid-related compounds, carbonyl residuals of proteins, the carboxylic groups of peptide, free amino acids and polysaccharides, as well as phospholipids, can be measured by different spectral regions [19,20]. Additionally, fluorescence properties of intracellular molecules and structures can be used as readout, leading to different autofluorescence spectra in organisms [21–24]. Furthermore, the difference within structural composition enables discrimination of cell types and strains by staining technique with specific dyes to create contrast between different species. Several staining protocols are currently in use and the reader is referred to existing literature in the field [25].
Figure 2. Differentiating between Co-Culture Members.
(A) Natural and synthetic cellular characteristics, which can be analyzed; (B) fluorescence image of a co-culture of Aspergillus terreus GFP1 (green) and Trichoderma reesei RFP1 (red) grown on cellulose (blue autofluorescence). Scale bar = 50 μm. Reproduced, with permission, from Ivan Schlembach.
Besides these natural differences, the rise of molecular biotechnology led to different methods that enable the engineering of not only existing, but also novel cellular characteristics. One example is the tagging of microorganisms or even metabolic pathways by different fluorescent reporters (Figure 2). Here, fluorescence proteins can be expressed to discriminate microorganisms [17,26,27]. The image in Figure 2B gives an example: it shows a high-resolution optical insight into a co-culture comprising Aspergillus terreus GFP1 (green) and Trichoderma reesei RFP1 (red) grown on cellulose (blue). Alternatively, fluorescence proteins can be used as reporters that visualize distinct metabolic traits and, thus, are only temporally expressed [28,29]. Compared with naturally occurring cellular differences, synthetic engineering of features is restricted to microorganisms that are genetically accessible [30].
Identification and Quantification of Culture Compositions
The identification of individual organisms in a microbial community is the fundamental beginning for understanding and learning from natural microbiomes [31,32]. Classically, the identification presupposes the isolation of the individual community members, which is invasive, involves cultivation, and thereby leads to a first selection between easy and difficult to cultivate microorganisms [33]. Currently, next-generation sequencing enables the characterization of the whole native community in a sample, with little, if any, selection of artifacts [34]. The entire genomic DNA of a community sample (i.e., the metagenome) can be directly sequenced, yielding simultaneously high-resolution qualitative as well as quantitative information on microbial composition and biosynthetic potential [35]. Even more, when including RNA-seq, the data can be used to discover metabolic interactions [36]. In case of microfluidic systems, a native environmental sample can be compartmentalized without the need for extended cultivation and enrichment into millions of droplets, each containing single cells, which can then be identified together with their metabolome by state-of-the-art technology, such as single cell DNA sequencing or mass spectrometry [37,38]. Once the members of a microbial mixed culture are identified and characterized in terms of their unique features, such as genetic, metabolic, morphological or optical differences (Figure 2), the dynamic change in the mixed population composition can be studied using a variety of methods.
Offline methods for mixed culture characterization are available with differing degrees of technological costs and effort (Table 1). For highly complex cultures containing tens or hundreds of different members, the single-species resolution can only be achieved using expensive DNA-sequencing techniques. However, when, for example, only the dynamics of certain subgenera or dominant species of a population are of interest, the population dynamics can be also estimated in a semiquantitative way using population fingerprinting methods, such as terminal restriction fragment length polymorphism (t-RFLP), lipid profiling, and quinone profiling [39–42]. Also, quantitative (q)PCR from genomic DNA extracts could be applied to enumerate the number of genomes of certain strains of interest using strain-specific primers [43].
Table 1. Offline Measurement Techniques for Mixed and Co-Culture Compositions for Different Microbial Systems.
| Method | Mixed-culture specification | Species | Refs |
|---|---|---|---|
| Plate counting | |||
| Plate counting | Bacterium and yeast | Lactobacillus lactis and Kluyveromyces marxianus | [44] |
| Yeast and yeast | Torulaspora delbrueckii and Saccharomyces cerevisiae | [45] | |
| Genetic techniques | |||
| t-RFLP | Three bacteria | Burkholderia cepacia, Staphylococcus aureus, and Pseudomonas aeruginosa | [41,42] |
| Pigment-DNA method | Fungus and fungus | Trichoderma reesei and Aspergillus wentii | [130] |
| 16S rDNA sequencing, RAPD analysis | Bacteria | Staphylococcus, Lactobacillus, Methanosarcinales, Methanomicrobiales, and Methanobacteriales | [46,47] |
| Quantitative real-time PCR and specific PCR analyses | Bacteria | Clostridium straminisolvens CSK1, Clostridium sp. strain FG4, Pseudoxanthomonas sp. strain M1-3, Brevibacillus sp. strain M1-5, and Bordetella sp. strain M1-6 | [43] |
| Full-length 16S rDNA sequencing | Microbiome | Undefined | [48] |
| Optical techniques | |||
| Reporter strains | Microbiome | Undefined | [61,62] |
| Flow cytometry and t-RFLP | Bacteria | B. cepacia, S. aureus, and P. aeruginosa | [51] |
| Flow cytometry with fluorescence-tagged strains | Yeast | Pichia pastoris (different strains expressing different pathways) | [58] |
| Bacteria | Different Escherichia coli strains | [59] | |
| Fluorescence in situ hybridization (FISH), oligonucleotide probes | Bacteria | Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Rhodobacter, Roseobacter, Rhodospirillum, and Rhodopseudomonas | [60] |
| Fluorescence proteins | Bacteria (biofilms) | Lactobacillus rhamnosus | [56] |
| Absorbance | Bacteria and yeasts | E. coli KO11, and S. cerevisiae D5A; Methylomicrobium buryatense 5GB1 and Scheffersomyces stipitis CBS 5773 | [55] |
| Autofluorescence | Fungi | Aspergillus flavus, Micosporum gypseum, Micosoprum canis, Trichophyton rubrum, and Trichophyton tonsurans | [23] |
| Microscopic image analysis with fluorescence-tagged strains | Bacterium and bacterium | Different E. coli strains | [27] |
| Particle size distribution | Eukaryote and bacteria | Tetrahymena pyriformis, E. coli, and Azotobacter vinelandii | [63] |
| Bacterium and yeast | L. lactis and K. marxianus | [52] | |
| Others | |||
| Cellulose consumption | Fungus and yeast | Trichoderma reesei and Ustilago maydis | [9] |
| Metabolic products (nisin and lactic acid) | Bacterium and yeast | L. lactis and S. cerevisiae | [53] |
| Measuring and -dependent O2 uptake | Bacteria | Nitrosomonas europaea and Nitrobacter winogradskyi | [54] |
Furthermore, two still-dominant methods for the offline quantification of complex mixed-culture systems are plate counting and 16S rDNA sequencing [44–48]. Classical plate counting appears outdated due to a huge amount of manual work as well as an unintended preselection of microorganisms growing on solid media. However, its combination with a liquid handling system as well as digital image analysis overcomes this disadvantage and it remains a robust and universal tool for most of the common microbial communities [33,45,49,50]. For lower-complexity mixed cultures and especially for defined co-cultures, various strain-specific features have been applied to resolve the population dynamics, which are often case specific. For example, bacteria–yeast co-cultures can be easily resolved using particle counting and size measurements [51,52]. When two co-culture members produce different metabolites, these metabolites can be taken as an indication of population ratios [53,54].
A powerful and more general method for population analysis is microscopic image analysis. It can combine various modalities from other methods, such as fluorescence, absorbance, morphology, and size, while spatially separating cells, allowing separate unmixed signals to be assigned to each cell [27]. Besides microscopy, absorbance and fluorescence measurement are in general suitable techniques for the quantitative offline characterization of defined mixed-culture compositions [22,23,55–57]. Autofluorescence and absorbance are non-invasive and scalable measurement principles with applications from the pico-liter to cubic-meter level [21,33]. In combination with genetic modifications of the desired microorganisms by tagging them with fluorescent proteins, the mixed culture composition can be theoretically determined very precisely as long as the genetic tools are available for the specific strains involved [58–60]. Furthermore, reporter strains are suitable solutions to detect microbial activity. One example is the indirect analysis of unknown microbial communities for microorganisms that produce antimicrobial substances. These communities can be subcultured as single cell-derived colonies using droplet microfluidics and cultivated together with fluorescence-tagged reporter strains, which are killed if antimicrobial substances are produced [33,61,62].
A simple but powerful method to analyse and compare mixed cultures is the measurement of particle numbers and their size distributions. A first application of this technique was published in 1973 with a defined mixed culture of Tetrahymena pyriformis-consuming Escherichia coli and Azotobacter vinelandii [63]. There are two main limitations of this technique are mentioned: (i) the overlap of distributions of the bacterial strains; and (ii) changing cell volumes of A. vinelandii depending on culture conditions. The impact of both these limitations on the mixed culture analysis was shown more in detail and confirmed by Geinitz and colleagues, who investigated the mixed culture dynamics of Lactobacillus lactis and Kluyveromyces marxianus [52].
A related (and more expensive) way to distinguish organisms based in morphology is flow cytometry (FC), which additionally can be combined with fluorescence measurements. This technique has a long history in the investigation of population dynamics in mixed cultures and has become an essential tool for the quantitative study of microbial communities in ecosystems, biotechnology, and health research [64–66]. Recently, FC was applied to assess the viability of Staphylococcus aureus and Burkholderia cepacia in co-culture by membrane integrity analysis using SYBR® Green I and propidium iodide staining [67]. It was also used for the characterization of electrochemical performance and biofilm characteristics of a co-culture of Geobacter sulfurreducens and Shewanella oneidensis [68]. Lambrecht and colleagues used differences in autofluorescence to quantify and sort methanogenic Archaea from complex microbial communities [69].
In general, FC has the ability to generate population statistics because it can measure the relevant numbers and features of cells, identify subpopulations with similar properties, find (rare) events, and separate organisms of interest when expanded with a cell-sorting unit (fluorescence-activated cell sorting; FACS) [70].
Monitoring of Mixed and Co-Culture Processes
Since mixed cultures are significantly more complex than conventional pure culture systems, they are also less predictable and more dynamic. For establishing processes beyond fundamental research, it is necessary to react in time to these dynamics to control the process [17]. Therefore, online measurement techniques are essential not only to resolve, but also to steer microbial interaction patterns. However, despite their relevance, there is only a limited number of potential online techniques published and available, in contrast to the aforementioned offline techniques.
Table 2 illustrates the dominance of optical techniques along with the quantification of volatile metabolites via headspace mass spectrometry [71,72]. For example, Sovova and colleagues predicted population dynamics of different co-culture combinations of Serratia rubidaea, Serratia marcescens, and E. coli by measuring acetaldehyde, propanol, acetoin, and ethanol. Although other publications are available that use mass spectrometry to investigate mixed and co-cultures, most of them focus on the desired metabolic products, neglecting the population dynamics question.
Table 2. Online Measurement Techniques for Mixed and Co-Culture Compositions for Different Microbial Systems.
| Method | Mixed-culture specification | Species | Refs |
|---|---|---|---|
| Mass spectrometry | |||
| Mass spectrometry | Bacteria | Serratia rubidaea, Serratia marcescens, and Escherichia coli | [71] |
| Fungi | Eutypa lata and Botryosphaeria obtusa | [72] | |
| Scattered light and absorbance spectrum | |||
| Scattered light spectrum | Bacterium and yeast | Lactobacillus lactis and Kluyveromyces marxianus | [52] |
| Absorbance | Bacteria and yeasts | E. coli KO11 and Saccharomyces cerevisiae D5A; Methylomicrobium buryatense 5GB1 and Scheffersomyces stipitis CBS 5773 | [55] |
| Autofluorescence | |||
| Autofluorescence | Bacterium and yeast | Lactobacillus casei, Saccharomyces pastorianus Y275i, and Saccharomyces cerevisiae | [57] |
| Green algae and Cyanobacteria | Chlorella vulgaris and Spirulina | [24] | |
| Fluorescence-tagged strains | |||
| Flow cytometry with fluorescence-tagged strains | Yeasts | S. cerevisiae, Lachancea thermotolerans, Torulaspora delbrueckii, Wickerhamomyces anomalus, K. marxianus, and Hanseniaspora opuntiae | [26] |
| Fluorescent tagged strains + optical density measurements | Bacteria | Different E. coli strains | [104] |
| Microscopy with fluorescence-tagged strains | Bacteria | Corynebacterium glutamicum, E. coli, and Pseudomonas putida | [74] |
| E. coli and Salmonella typhimurium | [75] | ||
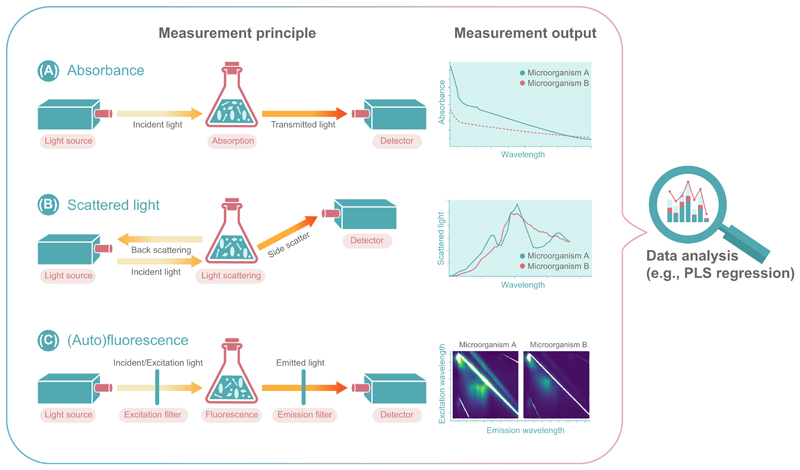
Optical measurement techniques are currently the most powerful tools that enable the monitoring of mixed cultures and will also have an integral role for active control of these systems (Table 2). Available technologies are based on fluorescence, absorbance, and scattered light at a single wavelength or with the advanced ability to determine the spectra (Box 2 and Figure 3) [33,52,55,73–75]. All technologies provide insight into the system without any relevant delay [76–78].
Box 2. Spectroscopic Techniques for Bioprocess Monitoring.
Spectroscopy is defined as the study of the interaction between matter and electromagnetic radiation [125]. The electromagnetic spectrum ranges from low-frequency radio waves to high-frequency γ-rays. The applications of spectroscopic methods for the analysis of processes in bioreactors are mainly the spectroscopic measurement of absorption, scattering light, and fluorescence spectroscopy. The theory behind each of these established methods is briefly illustrated and discussed herein. For instrumentation that is used for bioprocess monitoring, the reader is referred to Beutel and Henkel [126] and this is not discussed further here. Besides these established methods, NIR, MIR and Raman spectroscopy are also beginning to be used in bioprocess monitoring applications; however, because of their current under-representation in available strategies, these principles are not explained in detail here.
Absorbance Measurement/Spectroscopy
Absorbance measures the attenuation of light intensity after passage through a medium in relation to the wavelength of the light (see Figure 3A in the main text). In theory, an absorbance spectrum is defined exclusively by the light attenuation caused by resonant absorption (conversion of the light energy into heat).
In practice, however, conventional spectrophotometers measure the total light attenuation and cannot distinguish between absorption and scattering. Therefore, absorbance light spectra of cell suspensions are largely affected by scattered light in the UV/Vis part of the spectrum and it is uncommon in the literature to characterize nonpigmented microbial cells (see ‘Scattered Light Spectroscopy’ section). More common for cell suspensions is the IR absorbance spectroscopy, since IR light shows less scattering.
In absorbance measurements, light disappears following an encounter with a particle. Most prominent absorption measurements are UV/Vis measurements, which rely on the well-known Beer’s Law, which relates the amount of light absorbed by a sample to the amount of a chemical species present in that sample. The sample absorbs energy (i.e., photons) from the radiating field. The intensity of the absorption varies as a function of frequency, and this variation is the absorbance spectrum. For different microbes, different absorbance spectra can be found depending on the emission spectrum (see Figure 3A in the main text). More details regarding the physical principle and its implementation can be found elsewhere [127].
Scattered Light Measurement/Spectroscopy
Light scattering occurs when light interacts with small objects and thereby changes its direction. Hence, when characterizing cell suspensions by scattered light spectroscopy, the spectrum of the scattered light is not changed by the scattering event itself. It is rather the interplay between scattering and absorption that determines the scattered light spectra of cells. Thus, it can be seen as complementary to light absorbance. Different from the process of absorption, which assesses the amount of light absorbed, the amount of light that is scattered can be quantified at different positions. Thereby, the scattered light signal has a near- linear relationship with the cell concentration.
Scattered light spectra can be measured across different positions over a wide excitation spectrum. For different microorganisms, different scattered light spectra can be found depending on their pigmentation, cell wall characteristics, shape, and size. More details regarding the physical principle and its implementation can be found elsewhere [128]. An important form is Raman scattering, which can be used to not only characterize molecules, but also differentiate between microorganisms [129].
Fluorescence Measurement/Spectroscopy
When a fluorophore is excited by defined lights, it absorbs a photon and is lifted to a higher energy state. As the energy of the molecule relaxes to a lower energy state, it emits a photon at a different energy level and, thus, wavelength. This principle can be used to measure not only autofluorescence of samples, but also fluorescence based on genetically encoded biosensors or upon addition of fluorescent dyes. The range of frequencies of emitted photons from a particular excitation frequency is known as the emission spectrum. Typically, the wavelength varies between 180 and 800 nm, depending on the analyte of interest. Fluorescence profiles of co-cultures can be obtained by systematically testing the emission spectrum for a range of excitation frequencies.
Figure 3.
Optical Measurement Principles and Output of (A) Absorbance, (B) Scattered Light, and (C) (Auto)Fluorescence to Resolve Co-Culture Compositions and Dynamics. Abbreviation: PLS, partial least square.
Absorbance, Scattered Light, and Autofluorescence
The online measurements of absorbance, scattered light, and autofluorescence spectra (Figure 3) appear to be the most promising available methods to elucidate the population dynamics of single-species co-culture systems. An absorbance-based method was published by Stone and colleagues (Figure 3A) [55]. The authors demonstrated that it is possible to differentiate between Saccharomyces cerevisiae and E. coli cells. Although the method was only applied to offline samples, it would be simple to realize an online measurement of the absorbance spectra. Still, a general restriction of the absorbance signal is the limited linear correlation with biomass formation. For this reason, nearly every sample needs to be diluted before offline optical density determination. Therefore, absorbance measurements could be applied to co-cultures with low biomass concentrations and microbial communities containing pigments [79]. A beneficial property of pigments for online monitoring of mixed cultures is their strong difference of the absorbance spectra, depending on their specific physicochemical properties [80]. In addition to absorbance, pigments also have a clear influence on the scattered light spectrum.
The applicability of scattered light spectra detection for co-culture performance screening in microtiter plates was published by Geinitz and colleagues (Figure 3B), who measured the online trends of cell dry weight for the bacterium L. lactis and the yeast K. marxianus under different cultivation conditions [52]. Although both organisms differ in both size and morphology, there are only minor differences in the scattered light spectra (overlay >95%). Even with these subtle differences it is possible to distinguish both organisms. Interestingly, cell size appears to have only a minor impact on the scattered light spectra compared with differences in the structure of the cell wall between Gram-positive and Gram-negative bacteria. Therefore, the full potential of this method needs to be evaluated in further studies.
The highest information content is provided by measuring the autofluorescence spectra (Figure 3C). Since all microorganisms contain many different proteins, vitamins, and cofactors, the autofluorescence properties of these molecules can be used to gain information [21]. Moreover, it has become clear that the autofluorescence spectra of different microorganisms show detailed variations and could be used for identification and characterization of co-culture compositions [81]. A first application of this technique to distinguish between bacteria and yeast as well as different yeast strains was published by Bhatta and colleagues [57]. When analyzing the data, the authors focused on the relevant regions for tryptophan fluorescence. It was shown that minor differences between yeast and bacteria were detectable using the tryptophan spectrum, but only by comparing the results of monoseptic cultures and after washing the cells. This first example showed that the overall spectrum should be analyzed to identify fluorescence regions with larger differences between different microbial systems. The usage of complex media components, such as yeast extract with many autofluorescing constituents could significantly lower the precision of the measurement. To overcome this limitation and to maximize the output of information from UV/VIS spectra, the step size of extinction and emission should be as small as possible, which can be only realized by using stepless turning monochromators and, if necessary, by eliminating the effect of complex media components [82,83]. To condense and dissect the resulting large amount of data provided by full spectrum scans, mathematical tools such as partial least square (PLS) regression and principal component analysis (PCA) are essential to gain relevant information output [84,85].
An overview of advantages and disadvantages for the different optical measurement principles based on species-independent parameters is given in Table 3, guiding the reader to the most suitable method depending on strain-specific characteristics such as cell number, different cell sizes, different cell walls, pigments, and fluorescent metabolites. Nevertheless, all five parameters influence all three measurement parameters. Therefore, techniques marked with a ‘+’ give more precise and/or specific information compared with the other approaches and should be preferred (e.g., fluorescent metabolites can be also detected by absorbance but less specifically so).
Table 3. Overview of Advantages and Disadvantages for Optical Measurement Principles Based on Species-Independent Parameters to Identify a Suitable Techniquea.
| Absorbance | Scattered light | Autofluorescence | |
|---|---|---|---|
| High cell numbers | – | + | + |
| Different cell size | – | + | – |
| Different cell walls | – | + | + |
| Pigments | + | – | + |
| Fluorescent metabolites | – | – | + |
+ indicates a preferred principle.
The aforementioned optical methods focus on classic UV/VIS spectroscopy, but there is also a large potential for longer wavelength spectroscopy in the range of near-infrared (NIR) to Raman (Box 2) [86,87]. Beside a large number of medical applications, NIR is an established technique to monitor biomass, substrates, and metabolic products of well-known monoculture processes [88,89]. The direct determination of different microbial species in a mixed culture might be challenging by NIR, but indirect investigation of mixed culture (compositions) was shown by Grassi and colleagues, who cultured Lactobacillus bulgaricus and Streptococcus thermophilus for lactic acid formation [90]. A similar state-of-the-art can be observed in the field of mid-infrared (MIR) applications. Various publications are available for the online monitoring of monoculture processes [91–93], but only a few with established mixed-culture applications. For example, Goodacre and colleagues showed in 1996 that Fourier transform-infrared (FTIR) spectroscopy can be used to identify hospital isolates [94]. It was shown that the FTIR spectrum of, for example, Streptococcus pneumoniae differs from that of Streptococcus pyogenes, which should theoretically allow an online signal for each species in combination with a suitable chemometric method, comparable to the scattered light-based method published by Geinitz and colleagues [52]. A first step in this direction was published by Schawe and colleagues, who investigated the culture dynamic of a Pseudomonas putida and Rhodococcus ruber co-culture [95]. With respect to Raman spectroscopy, no publications elucidating a mixed-culture dynamic are currently available. Nevertheless, Raman measurement is a powerful and universal technique and, for example, García-Timermans and colleagues were able to identify different phenotypes in a microbial community by using Raman spectroscopy [96].
In any case, the robustness of the respective method has to be carefully evaluated in regard to spectral variations during axenic cultivations of the single strains. It is known that the spectral characteristics of a pure culture can change depending on age and the physiological conditions [97]. The autofluorescence (e.g., NADH) is highly dependent on the redox state and, therefore, can dynamically fluctuate without a change in cell number [98,99]. Moreover, cell size, cell wall composition, or pigmentation can also change depending on the culture conditions, leading to differences in absorption or scattered light spectra [100]. Thus, for a robust method, it is essential to find spectral regions that are invariant to external conditions. For yeast, for example, tryptophan fluorescence has been shown to be more invariant compared with NADH fluorescence [99].
Challenges for Enabling Online Optical Monitoring by Fluorescence Tagging
To enhance the power and resolution of the aforementioned optical methods, different fluorescent proteins can be expressed to artificially label the microorganisms and their metabolic activities. Given that fluorescence proteins typically have broad excitation and emission peaks, it is important to choose fluorescent proteins with minimal spectral overlap to avoid any crosstalk. Hence, experiments are typically restricted to a maximum of three fluorescent proteins to achieve acceptable spectral separation [101]. Nevertheless, theoretically, more complex communities involving more than three members could also be resolved using spectral unmixing algorithms [102]. Moreover, spectral unmixing is also essential to prevent crosstalk caused by autofluorescence [103]. Apart from these general spectroscopic challenges, the implementation of direct online monitoring of population dynamics solely by fluorescence measurement is not trivial and only a few studies using fluorescence tags have been published so far.
Main challenges arise from the fact that the relationship between fluorescence and biomass concentration is not constant but varies depending on how the fluorescence tag is integrated genetically (e.g., types of promotor, free or fused protein construct) and the growth phase [104]. The latter can be explained by a mismatch between fluorescence protein production rate and growth rate. At high growth rates, the fluorescent protein production rate cannot compensate for the volume growth by cell division and the fluorescence is diluted, reducing the fluorescence:biomass ratio [105]. Further problems may arise from pH fluctuations. The intracellular pH can be affected by many factors, such as growth rate, metabolism, and extracellular pH [106]. All fluorescent proteins have a certain pKa value for maximum fluorescence and any pH deviation will directly affect the fluorescence:biomass ratio. Therefore, the choice of a fluorescent protein with a suitable pKa value is vital.
To measure the population composition in real time, the maturation time of the fluorescent protein is another critical factor. Delayed maturation will result in delayed delivery of the information. Hence, the maturation time should be a magnitude smaller than the doubling time of the tagged organism. An additional important factor in regard to most standard fluorescent protein maturation is the availability of oxygen for chromophore maturation. Maturation can be strongly delayed by oxygen-limited culture conditions. Therefore, recently developed fluorescent proteins without oxygen requirement should be used preferentially [107]. Otherwise, the application of oxygen-unlimited conditions is essential. Additionally, pigment formation can also interfere with fluorescent measurements because light absorption by pigments will affect both fluorescence excitation and emission.
Flow Cytometry-Based Online Analysis
FC, especially in combination with fluorescence-based measurements, is a powerful tool to follow population dynamics of complex cultures. Cells can be differentiated based on cell size, morphology, and fluorescence properties. Bessmer and colleagues used fully automated online FC to monitor microbial dynamics in groundwater. Besides the total cell concentration, the percentage of low nucleic acid (LNA) content bacteria was monitored over time through differences in the fluorescence fingerprints [108]. In a similar study, Buysschaert and colleagues additionally monitored diversity described by Hill number diversity indices [109]. The Hill number is an indicator of the richness and evenness of species within a mixed-culture population [110]. Recently, Liu and colleagues [111], used online FC in combination with sorting subpopulations for sequencing to follow the dynamics of cell populations of dozens of subgroups within a naturally mixed culture.
In recent years, additionally to classical FC, microfluidic systems have been developed and used in combination with automated microscopy for high-throughput image-based FC [microfluidic imaging FC (IFC)]. Here, in microfluidic channels, often parallelized individual cells can be monitored by imaging techniques. Combined with advanced image analysis tools, IFC provides more detailed information of the cell shape and structure compared with classical FC [112].
Standardization and Comparability
Spectroscopic data can be acquired at different scales with different instruments ranging from single cell-resolved FC, to microtiter plate readers up to large-scale stirred tank fermenters equipped with in situ optical probes. However, these data cannot be directly compared either between these scales or within one scale across labs, because the measurements are in arbitrary units that depend on the manufacturer of the device and the parameters adjusted by the user. For example, to make fluorescence measurements comparable, it is essential to standardize data. Thereby, different layers of standardization are necessary. First, the raw signals have to be converted into units, which can be universally compared across any device. This can be done by referencing arbitrary unit measurements from different instruments and scales to the signal intensity derived from defined standards, such as fluorescein [113]. Finally, these defined units can be correlated to a mass, such as the cell dry weight of biomass. A consequent calibration on cell dry weight would not only increase the comparability between different studies, but, even more importantly, enable mass and energy balances to validate the results on a fundamental level.
From Online Monitoring to Online Control
As illustrated in Figure 1, every control loop in a (bio)process comprises an input and an output variable, as well as an actuator to influence the current status of the co-culture. The input, in case of a co-culture, is its composition, which can be measured by any of the techniques discussed earlier. The actuator, which has to influence the population dynamic in the desired way, can be biotic and/or abiotic. A biotic way for controlling co-cultures is the establishment of a suitable self-regulating genetic interaction circuit. Such internal population control strategies have been realized in fundamental research work through, for example, obligatory cross-feeding interactions or tunable quorum-sensing circuits [114,115]. These strategies are not discussed further within this review and the reader is referred to recent reviews of biotic control strategies for further insights [116–118]. Instead, here we provide outlook on the possible implementation of external feedback control strategies, which could be enabled by the availability of online monitoring methods to precisely sense the population composition or differential performance parameters as control input. Up to now, such external control has been rarely realized experimentally. However, with the advance of online population measurement technologies, external control of co-cultures processes may also become more relevant and common in the future. Different control loop models and controllable actuators that could steer a co-culture are already available. In this way, abiotic and, therefore, often classic process parameters can be used as controls, which can be applied to many but not necessarily to every natural community or wild-type co-culture without the need for genetic engineering.
Possible actuators include all variables that influence the relative fitness of the participating organisms, such as pH, oxygen availability, or temperature. As an example, Krieger and colleagues dynamically controlled the community composition of a E. coli and P. putida co-culture by different temperatures [119]. Similarly, pH oscillations have been used to enable stable coexistence in a co-culture comprising E. coli and S. cerevisiae [120]. In another example, oxygen availability was found to influence population dynamics in a co-culture of lactic acid bacteria and T. reesei [4]. Besides these classical process parameters, the population could also be controlled by the addition of inducer compounds that regulate the expression of pathways for making obligate cross-feeding nutrients [121] or that induce cell lysis [114]. Another strategy could be the direct control of the growth rate via inducible expression of RNA polymerase [122].
With respect to the overall bioprocess control loop, there are also different options available. Established control loop techniques, which rely on high-frequency online measurement of input variables, could be applied in principle, such as proportional-integral-derivative controllers, which are reviewed elsewhere [123]. These techniques are not targeted to the specifics of mixed-culture processes. Interestingly, although not yet realized experimentally, it has been shown by computer models that population control can also work using more infrequent atline population measurements with the implementation of machine learning [124]. A benefit of this method is that the system automatically learns to fulfil the target objective, which could be either to establish a specified population ratio or to adapt the population ratio for maximization of product output [124]. One of the rare published examples of a full implementation of an external feedback control loop for population control was implemented by Sassi and colleagues, proposing the so-called ‘Segregostat’ concept, which comprises a FC that is coupled to a bioreactor to perform online FC analysis [73]. They used the setup for real-time monitoring and control of heterogeneity profiles of a continuous bioreactor process. By using propidium iodide as an effective fluorescent biomarker for cells adapting to nutrient limitation, the population of cells that switched their metabolic performance was controlled by the addition of glucose. Although this study was applied to control the subpopulation ratio of monocultures of E. coli and P. putida, the principle could be applied to co-culture processes. More progress in this field is clearly required to develop targeted mixed-culture control strategies.
Concluding Remarks and Future Perspectives
Microbial mixed cultures will greatly expand the biotechnological portfolio by opening novel strategies for the utilization of renewable biomass or waste materials and offering novel synthesis routes for the production of pharmacologically relevant compounds and high-value products. Currently, several high-performance analytical strategies have been developed to follow these complex biological processes. In this review, we have summarized and evaluated different offline, atline, and online methods, showing their advantages and disadvantages as well as the progress and challenges in the field. These analytical strategies lay the foundation and are prerequisites for the future development of operation control strategies for mixed-culture processes.
The review has shown that the resolution and control of mixed-culture population dynamics does not appear to be limited by the availability of suitable measurement principles but by their routine application and utilization of data (see Outstanding Questions). To overcome this limitation, data need to be standardized to generate ‘fingerprints’ for microorganisms. This task is significantly more difficult to realize compared with pure constituents, but should be possible by summarizing and mining all spectrometry data in a data collection. Another issue is a lack of commercialized equipment tailored for this specific application. Most of the studies discussed herein used special customized solutions, which are not available to the broader scientific community. Increasing implementation of optical monitoring and control technology for mixed cultures in basic research will reduce the difficulties associated with their handling and eventually boost the transfer of microbial mixed cultures from research to industrial biotechnology.
Outstanding Questions.
Are upcoming measurement technologies and control strategies for mixed cultures a game-changer for current single-species bioprocesses?
Will monoculture bioprocesses benefit from measurement strategies that are developed for co-culture analysis and control?
Are there suitable measurement techniques (utilizing novel physical principles) that could be adapted for mixed-culture monitoring in biotechnology?
Can the combination of absorbance, scattered light, and fluorescence result in a universal tool to resolve mixed and co-culture dynamics?
Can a bioinformatics combination of new spectroscopic information on mixed and co-culture composition with process performance/metabolomics data (e.g., from online sensors, online gas chromatography, or gas chromatography-mass spectrometry) be used to develop novel control strategies?
How will the application of deep learning and artificial intelligence approaches to complex bioprocess data analysis affect process control strategies?
Highlights.
Next-generation sequencing and microfluidic analysis can identify and characterize the complex native microbial community in a sample, with little, if any, selection of artifacts.
Optical measurement methods are currently the most promising fully scalable techniques for online monitoring and control of mixed-culture processes.
The potential of infrared techniques, Raman spectroscopy, and mass spectrometry currently appear to be underestimated and underused for all aspects of the identification, characterization, monitoring, and control of mixed and co-culture processes.
Acknowledgments
L.R. is supported by the Free State of Thuringia (project number 2019 FGR 0079). M.A.R. is partly supported by the German Research Foundation (Deutsche Forschungsgemeinschaft) under Germany’s Excellence Strategy (EXC 2051, –Project-ID 390713860) and the Priority Program 2170. She also has received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (Grant agreement No. 864669). A.G. is supported by the Priority Program 2170 of the German Research Foundation.
Footnotes
Declaration of Interests
There are no interests to declare.
References
- 1.Bertrand S, et al. Metabolite induction via microorganism co-culture: a potential way to enhance chemical diversity for drug discovery. Biotechnol Adv. 2014;32:1180–1204. doi: 10.1016/j.biotechadv.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Zuroff TR, et al. Consortia-mediated bioprocessing of cellulose to ethanol with a symbiotic Clostridium phytofermentans/yeast co-culture. Biotechno Biofuels. 2013;6:59. doi: 10.1186/1754-6834-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minty JJ, et al. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass. Proc Natl Acad Sci U S A. 2013;110:14592–14597. doi: 10.1073/pnas.1218447110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shahab RL, et al. Consolidated bioprocessing of lignocellulosic biomass to lactic acid by a synthetic fungal-bacterial consortium. Biotechnol Bioeng. 2018;115:1207–1215. doi: 10.1002/bit.26541. [DOI] [PubMed] [Google Scholar]
- 5.Brethauer S, Studer MH. Consolidated bioprocessing of lignocellulose by a microbial consortium. Energy Environ Sci. 2014;7:1446–1453. [Google Scholar]
- 6.Zhao C, et al. Consolidated bioprocessing of lignocellulosic biomass to itaconic acid by metabolically engineering Neurospora crassa. Appl Microbiol Biotechnol. 2018;102:9577–9584. doi: 10.1007/s00253-018-9362-1. [DOI] [PubMed] [Google Scholar]
- 7.Shahab RL, et al. A heterogeneous microbial consortium producing short-chain fatty acids from lignocellulose. Science. 2020;369:eabb1214. doi: 10.1126/science.abb1214. [DOI] [PubMed] [Google Scholar]
- 8.Catur Utomo RN, et al. Defined microbial mixed culture for utilization of polyurethane monomers. ACS Sustain Chem Eng. 2020;8:17466–17474. [Google Scholar]
- 9.Schlembach I, et al. Consolidated bioprocessing of cellulose to itaconic acid by a co-culture of Trichoderma reesei and Ustilago maydis. Biotechnol Biofuels. 2020;13:207. doi: 10.1186/s13068-020-01835-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keller L, Surette MG. Communication in bacteria: an ecological and evolutionary perspective. Nat Rev Micro. 2006;4:249–258. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- 11.Adnani N, et al. Symbiosis-inspired approaches to antibiotic discovery. Nat Prod Rep. 2017;34:784–814. doi: 10.1039/c7np00009j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benndorf R, et al. Natural products from actinobacteria associated with fungus-growing termites. Anttbiotics (Base) 2018;7:83. doi: 10.3390/antibiotics7030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khalil ZG, et al. Inter-Kingdom beach warfare: microbial chemical communication activates natural chemica defences. ISME J. 2019;13:147–158. doi: 10.1038/s41396-018-0265-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arp J, et al. Synergistic activity of cosecreted natural products from amoebae-associated bacteria. PNAS. 2018;115:3758–3763. doi: 10.1073/pnas.1721790115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molloy EM, Hertweck C. Antimicrobial discovery inspired by ecological interactions. Curr Opin Microbiol. 2017;39:121–127. doi: 10.1016/j.mib.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 16.Johns NI, et al. Principles for designing synthetic microbial communities. Curr Opin Microbiol. 2016;31:146–153. doi: 10.1016/j.mib.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goers L, et al. Co-culture systems and technologies: taking synthetic biology to the next level. J R Soc Interface. 2014;11 doi: 10.1098/rsif.2014.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabra W, Zeng AP. In: Industrial Biocatalysis. Grunwald P, editor. Pan Stanford Publishing; 2014. Mixed microbial cultures for industrial biotechnology: success, chance, and challenges; pp. 201–233. [Google Scholar]
- 19.Maity JP, et al. Identification and discrimination of bacteria using Fourier transform infrared spectroscopy. Spectrochim Acta A Mol Biomol Spectrosc. 2013;116:478–484. doi: 10.1016/j.saa.2013.07.062. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y-D, et al. Synchrotron infrared spectral regions as signatures for foodborne bacterial typing. Nucl Sci Tech. 2019;30:25 [Google Scholar]
- 21.Marose S, et al. Two-dimensional fluorescence spectroscopy: a new tool for on-line bioprocess monitoring. Biotechnol Prog. 1998;14:63–74. doi: 10.1021/bp970124o. [DOI] [PubMed] [Google Scholar]
- 22.Wen S-H, et al. Confocal autofluorescence identification of bacteria, fungi, and acanthamoeba in infected porcine cornea models. Optik. 2018;168:384–389. [Google Scholar]
- 23.Lin S-J, et al. Multiphoton autofluorescence spectral analysis for fungus imaging and identification. Appl Phys Lett. 2009;95:043703 [Google Scholar]
- 24.Shin Y-H, et al. A hand-held fluorescent sensor platform for selectively estimating green algae and cyanobacteria biomass. Sensors Actuators B Chem. 2018;262:938–946. [Google Scholar]
- 25.Kiernan JA. Dyes and other colorants in microtechnique and biomedical research. Color Techno. 2006;122:1–21. [Google Scholar]
- 26.Conacher CG, et al. Real-time monitoring of population dynamics and physical interactions in a synthetic yeast ecosystem by use of multicolour flow cytometry. Appl Microbiol Biotechnol. 2020;104:5547–5562. doi: 10.1007/s00253-020-10607-x. [DOI] [PubMed] [Google Scholar]
- 27.Stephens K, et al. Bacterial co-culture with cell signaling translator and growth controller modules for autonomously regulated culture composition. Nat Commun. 2019;10:4129. doi: 10.1038/s41467-019-12027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costantini LM, et al. A palette of fluorescent proteins optimized for diverse cellular environments. Nat Commun. 2015;6:7670. doi: 10.1038/ncomms8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tebo AG, Gautier A. A spl it fluorescent reporter with rapid and reversible complementation. Nat Commun. 2019;10:2822. doi: 10.1038/s41467-019-10855-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farkas JA, et al. Genetic techniques for the Archaea. Annu Rev Genet. 2013;47:539–561. doi: 10.1146/annurev-genet-111212-133225. [DOI] [PubMed] [Google Scholar]
- 31.Woloszynek S, et al. Exploring thematic structure and predicted functionality of 16S rRNA amplicon data. PLoS ONE. 2019;14:e0219235. doi: 10.1371/journal.pone.0219235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber KP, Legge RL. Community-level physiological profiling. Methods Mol Biol. 2010;599:263–281. doi: 10.1007/978-1-60761-439-5_16. [DOI] [PubMed] [Google Scholar]
- 33.Mahler L, et al. Highly parallelized microfluidic droplet cultivation and prioritization on antibiotic producers from complex natural microbial communities. bioRxiv. 2019;2019:2019.2012.2018.877530 [Google Scholar]
- 34.Cao Y, et al. A review on the applications of next generation sequencing technologies as applied to food-related microbiome studies. Front Microbiol. 2017;8:1829. doi: 10.3389/fmicb.2017.01829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayo B, et al. Impact of next generation sequencing techniques in food microbiology. Curr Genomics. 2014;15:293–309. doi: 10.2174/1389202915666140616233211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenthal AZ, et al. RNA-seq reveals cooperative metabolic interactions between two termite-gut spirochete species in co-culture. ISME J. 2011;5:1133–1142. doi: 10.1038/ismej.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lan F, et al. Single-cell genome sequencing at ultra-high-throughput with microfluidic droplet barcoding. Nat Biotechnol. 2017;35:640–646. doi: 10.1038/nbt.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terekhov SS, et al. Microfluidic droplet platform for ultrahigh-throughput single-cell screening of biodiversity. PNAS. 2017;114:2550–2555. doi: 10.1073/pnas.1621226114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spiegelman D, et al. A survey of the methods for the characterization of microbial consortia and communities. Can J Microbiol. 2005;51:355–386. doi: 10.1139/w05-003. [DOI] [PubMed] [Google Scholar]
- 40.Sabra W, et al. Biosystems analysis and engineering of microbial consortia for industrial biotechnology. Eng Life Sci. 2010;10:407–421. [Google Scholar]
- 41.Schmidt JK, et al. A novel concept combining experimental and mathematical analysis for the identification of unknown interspecies effects in a mixed culture. Biotechnol Bioeng. 2011;108:1900–1911. doi: 10.1002/bit.23126. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt JK, et al. Characterization of a three bacteria mixed culture in a chemostat: evaluation and application of a quantitative terminal-restriction fragment length polymorphism (T-RFLP) analysis for absolute and species specific cell enumeration. Biotechnol Bioeng. 2007;96:738–756. doi: 10.1002/bit.21147. [DOI] [PubMed] [Google Scholar]
- 43.Kato S, et al. Stable coexistence of five bacterial strains as a cellulose-degrading community. Appl Environ Microbiol. 2005;71:7099–7106. doi: 10.1128/AEM.71.11.7099-7106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimizu H, et al. Nisin Production by a Mixed-Culture System Consisting of Lactococcus lactis and Kluyveromyces marxianus. Appl Environ Microbiol. 1999;65:3134–3141. doi: 10.1128/aem.65.7.3134-3141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brou P, et al. Mixed culture fermentation using Torulaspora delbrueckii and Saccharomyces cerevisiae with direct and indirect contact: impact of anaerobic growth factors. Eur Food Res Techno. 2018;244:1699–1710. [Google Scholar]
- 46.Raskin L, et al. Characterization of microbial communities in anaerobic bioreactors using molecular probes. Antonie Van Leeuwenhoek. 1995;68:297–308. doi: 10.1007/BF00874140. [DOI] [PubMed] [Google Scholar]
- 47.Rebecchi A, et al. Physiological and molecular techniques for the study of bacterial community development in sausage fermentation. J Appl Microbiol. 1998;84:1043–1049. doi: 10.1046/j.1365-2672.1998.00442.x. [DOI] [PubMed] [Google Scholar]
- 48.Dethlefsen L, et al. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taniguchi M, et al. Production of a mixture of antimicrobial organic acids from lactose by co-culture of Bifidobacterium longum and Propionibacterium freudenreichii. Biosci Biotechnol Biochem. 1998;62:1522–1527. doi: 10.1271/bbb.62.1522. [DOI] [PubMed] [Google Scholar]
- 50.Shekhawat K, et al. Impact of oxygenation on the performance of three non-Saccharomyces yeasts in co-fermentation with Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2017;101:2479–2491. doi: 10.1007/s00253-016-8001-y. [DOI] [PubMed] [Google Scholar]
- 51.Rüger M, et al. Species-specific viability analysis of Pseudomonas aeruginosa, Burkholderia cepacia and Staphylococcus aureus in mixed culture by flow cytometry. BMC Microbiol. 2014;14:15. doi: 10.1186/1471-2180-14-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geinitz B, et al. Noninvasive tool for optical online monitoring of individual biomass concentrations in a defined coculture. Biotech Bioeng. 2020;117:999–1011. doi: 10.1002/bit.27256. [DOI] [PubMed] [Google Scholar]
- 53.Liu C, et al. Stimulation of Nisin production from whey by a mixed culture of Lactococcus lactis and Saccharomyces cerevisiae. Appl Biochem Biotechnol. 2006;131:751–761. doi: 10.1385/ABAB:131:1:751. [DOI] [PubMed] [Google Scholar]
- 54.Pérez J, et al. Interactions of Nitrosomonas europaea and Nitrobacter winogradskyi grown in co-culture. Arch Microbiol. 2015;197:79–89. doi: 10.1007/s00203-014-1056-1. [DOI] [PubMed] [Google Scholar]
- 55.Stone KA, et al. A novel soft sensor approach for estimating individual biomass in mixed cultures. Biotechnol Prog. 2017;33:347–354. doi: 10.1002/btpr.2453. [DOI] [PubMed] [Google Scholar]
- 56.Spacova I, et al. Expression of fluorescent proteins in Lactobacillus rhamnosus to study host-microbe and microbe-microbe interactions. Microb Biotechnol. 2018;11:317–331. doi: 10.1111/1751-7915.12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhatta H, et al. Use of fluorescence spectroscopy to differentiate yeast and bacterial cells. Appl Microbiol Biotechnol. 2006;71:121–126. doi: 10.1007/s00253-005-0309-y. [DOI] [PubMed] [Google Scholar]
- 58.Liu Y, et al. Engineered monoculture and co-culture of methylotrophic yeast for de novo production of monacolin J and lovastatin from methanol. Metab Eng. 2018;45:189–199. doi: 10.1016/j.ymben.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 59.Dinh CV, et al. Development of a quorum-sensing based circuit for control of coculture population composition in a naringenin production system. ACS Synth Biol. 2020;9:590–597. doi: 10.1021/acssynbio.9b00451. [DOI] [PubMed] [Google Scholar]
- 60.Fradinho JC, et al. Photosynthetic mixed culture polyhydroxyalkanoate (PHA) production from individual and mixed volatile fatty acids (VFAs): substrate preferences and co-substrate uptake. J Biotechnol. 2014;185:19–27. doi: 10.1016/j.jbiotec.2014.05.035. [DOI] [PubMed] [Google Scholar]
- 61.Mahler L, et al. Detection of antibiotics synthetized in microfluidic picolitre-droplets by various actinobacteria. Sci Rep. 2018;8:13087. doi: 10.1038/s41598-018-31263-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mahler L, et al. Publisher Correction: Detection of antibiotics synthetized in microfluidic picolitre-droplets by various actinobacteria. Sci Rep. 2018;8:15859. doi: 10.1038/s41598-018-34069-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Drake JF, Tsuchiya HM. Differential counting in mixed cultures with coulter counters. Appl Microbiol. 1973;26:9–13. doi: 10.1128/am.26.1.9-13.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Z, et al. Ecological Stability properties of microbial communities assessed by flow cytometry. mSphere. 2018;3:e00564-00517. doi: 10.1128/mSphere.00564-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koch C, Müller S. Personalized microbiome dynamics – cytometric fingerprints for routine diagnostics. Mol Asp Med. 2018;59:123–134. doi: 10.1016/j.mam.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 66.Díaz M, et al. Application of flow cytometry to industrial microbial bioprocesses. Biochem Eng J. 2010;48:385–407. [Google Scholar]
- 67.Rüger M, et al. A flow cytometric method for viability assessment of Staphylococcus aureus and Burkholderia cepacia in mixed culture. Cytometry A. 2012;81A:1055–1066. doi: 10.1002/cyto.a.22219. [DOI] [PubMed] [Google Scholar]
- 68.Engel C, et al. Long-term behavior of defined mixed cultures of Geobacter sulfurreducens and Shewanella oneidensis in bioelectrochemical systems. Front Bioeng Biotechnol. 2019;7:60. doi: 10.3389/fbioe.2019.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lambrecht J, et al. Flow cytometric quantification, sorting and sequencing of methanogenic archaea based on F420 autofluorescence. Microb Ceii Factories. 2017;16:180. doi: 10.1186/s12934-017-0793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mattanovich D, Borth N. Applications of cell sorting in biotechnology. Microb Cell Factories. 2006;5:12. doi: 10.1186/1475-2859-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sovová K, et al. Real time monitoring of population dynamics in concurrent bacterial growth using SIFT-MS quantification of volatile metabolites. Analyst. 2013;138:4795–4801. doi: 10.1039/c3an00472d. [DOI] [PubMed] [Google Scholar]
- 72.Azzollini A, Boggia L, Boccard J, Sgorbini B, Lecoultre N, Allard P-M, Rubiolo P, Rudaz S, Gindro K, Bicchi C, Wolfender J-L. Dynamics of metabolite induction in fungal co-cultures by metabolomics at both volatile and non-volatile levels. Front Microbiol. 2018;9(72) doi: 10.3389/fmicb.2018.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sassi H, et al. Segregostat: a novel concept to control phenotypic diversification dynamics on the example of Gram-negative bacteria. Microb Biotechnol. 2019;12:1064–1075. doi: 10.1111/1751-7915.13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burmeister A, et al. A microfluidic co-cultivation platform to investigate microbial interactions at defined microenvironments. Lab Chip. 2019;19:98–110. doi: 10.1039/c8lc00977e. [DOI] [PubMed] [Google Scholar]
- 75.Scott SR, et al. A stabilized microbial ecosystem of self-limiting bacteria using synthetic quorum-regulated lysis. Nat Microbiol. 2017;2:17083. doi: 10.1038/nmicrobiol.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bajracharya S, et al. Long-term operation of microbial electrosynthesis cell reducing CO2 to multi-carbon chemicals with a mixed culture avoiding methanogenesis. Bioelectrochemistry. 2017;113:26–34. doi: 10.1016/j.bioelechem.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 77.Stierle AA, et al. The berkeleylactones, antibiotic macrolides from fungal coculture. J Nat Prod. 2017;80:1150–1160. doi: 10.1021/acs.jnatprod.7b00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rubbens P, et al. Flow cytometric single-cell identification of populations in synthetic bacterial communities. PLoS ONE. 2017;12:e0169754. doi: 10.1371/journal.pone.0169754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dufossé L. In: Natural and Artificial Flavoring Agents and Food Dyes. Grumezescu AM, Holban AM, editors. Academic Press; 2018. Microbial pigments from bacteria, yeasts, fungi, and microalgae for the food and feed industries; pp. 113–132. [Google Scholar]
- 80.Thrane J-E, et al. Spectrophotometric analysis of pigments: a critical assessment of a high-throughput method for analysis of algal pigment mixtures by spectral deconvolution. PLoS ONE. 2015;10:e0137645. doi: 10.1371/journal.pone.0137645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ku H-M, et al. Using autofluorescence to detect bacterial contamination in root fractures. J Dent. 2019;86:27–32. doi: 10.1016/j.jdent.2019.05.024. [DOI] [PubMed] [Google Scholar]
- 82.Ladner T, et al. Parallel online multi-wavelength (2D) fluorescence spectroscopy in each well of a continuously shaken microtiter plate. Biotechnol J. 2016;11:1605–1616. doi: 10.1002/biot.201600515. [DOI] [PubMed] [Google Scholar]
- 83.Siepert E-M, et al. Short-chain fluorescent tryptophan tags for on-line detection of functional recombinant proteins. BMC Biotechnol. 2012;12:65. doi: 10.1186/1472-6750-12-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Giana HE, et al. Rapid identification of bacterial species by fluorescence spectroscopy and classification through principal components analysis. J Fluoresc. 2003;13:489–493. [Google Scholar]
- 85.Faassen SM, Hitzmann B. Fluorescence spectroscopy and chemometric modeling for bioprocess monitoring. Sensors (Basel) 2015;15:10271–10291. doi: 10.3390/s150510271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mazarevica G, et al. On-line fermentation monitoring by mid-infrared spectroscopy. Appl Spectrosc. 2004;58:804–810. doi: 10.1366/0003702041389229. [DOI] [PubMed] [Google Scholar]
- 87.Claßen J, et al. Spectroscopic sensors for in-line bioprocess monitoring in research and pharmaceutical industrial application. Anal Bioanal Chem. 2017;409:651–666. doi: 10.1007/s00216-016-0068-x. [DOI] [PubMed] [Google Scholar]
- 88.Sandor M, et al. NIR-spectroscopy for bioprocess monitoring & control. BMC Proc. 2013;7:P29 [Google Scholar]
- 89.Zimmerleiter R, et al. Probeless non-invasive near-infrared spectroscopic bioprocess monitoring using microspectrometer technology. Anal Bioanal Chem. 2020;412:2103–2109. doi: 10.1007/s00216-019-02227-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grassi S, et al. Monitoring of lactic acid fermentation process using fourier transform near infrared spectroscopy. J Near Infrared Spectrosc. 2013;21:417–425. [Google Scholar]
- 91.Kornmann H, et al. Monitoring and control of Gluconacetobacter xylinus fed-batch cultures using in situ mid-IR spectroscopy. J Biotechnol. 2004;113:231–245. doi: 10.1016/j.jbiotec.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 92.Koch C, et al. Multi-analyte quantification in bioprocesses by Fourier-transform-infrared spectroscopy by partial least squares regression and multivariate curve resolution. Anal Chim Acta. 2014;807:103–110. doi: 10.1016/j.aca.2013.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Narayana S, et al. Mid-infrared spectroscopy and multivariate analysis to characterize Lactobacillus acidophilus fermentation processes. Appl Spectrosc. 2019;73:1087–1098. doi: 10.1177/0003702819848486. [DOI] [PubMed] [Google Scholar]
- 94.Goodacre R, et al. Rapid identification of Streptococcus and Enterococcus species using diffuse reflectance-absorbance Fourier transform infrared spectroscopy and artificial neural networks. FEMS Microbiol Let. 1996;140:233–239. doi: 10.1016/0378-1097(96)00186-3. [DOI] [PubMed] [Google Scholar]
- 95.Schäwe R, et al. Evaluation of FT-IR spectroscopy as a tool to quantify bacteria in binary mixed cultures. J Microbiol Methods. 2011;86:182–187. doi: 10.1016/j.mimet.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 96.García-Timermans C, et al. Raman spectroscopy-based measurements of single-cell phenotypic diversity in microbial populations. mSphere. 2020;5:e00806-00820. doi: 10.1128/mSphere.00806-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Boehl D, et al. Chemometric modelling with two-dimensional fluorescence data for Claviceps purpurea bioprocess characterization. J Biotechnol. 2003;105:179–188. doi: 10.1016/s0168-1656(03)00189-5. [DOI] [PubMed] [Google Scholar]
- 98.Lara AR, et al. Effect of the oxygen transfer rate on oxygen-limited production of plasmid DNA by Escherichia coli. Biochem Eng J. 2019;150:107303 [Google Scholar]
- 99.Horvath JJ, et al. In situ fluorescence cell mass measurements of Saccharomyces cerevisiae using cellular tryptophan. Biotechnol Prog. 1993;9:666–670. doi: 10.1021/bp00024a016. [DOI] [PubMed] [Google Scholar]
- 100.Shi K, et al. Pigment fingerprint profile during extractive fermentation with Monascus anka GIM 3.592. BMC Biotechnol. 2017;17:46. doi: 10.1186/s12896-017-0366-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Heins A-L, et al. Development and characterization of Escherichia coli triple reporter strains for investigation of population heterogeneity in bioprocesses. Microb Ceil Factories. 2020;19:14. doi: 10.1186/s12934-020-1283-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McRae TD, et al. Robust blind spectral unmixing for fluorescence microscopy using unsupervised learning. PLoS ONE. 2019;14:e0225410. doi: 10.1371/journal.pone.0225410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lichten CA, et al. Unmixing of fluorescence spectra to resolve quantitative time-series measurements of gene expression in plate readers. BMC Biotechnol. 2014;14:11. doi: 10.1186/1472-6750-14-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Noonan AJC, et al. CRAGE-mediated insertion of fluorescent chromosomal markers for accurate and scalable measurement of co-culture dynamics in Escherichia coli. Synth Biol. 2020;5 doi: 10.1093/synbio/ysaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nordholt N, et al. Effects of growth rate and promoter activity on single-cell protein expression. Sci Rep. 2017;7:6299. doi: 10.1038/s41598-017-05871-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Moussa R, et al. An evaluation of genetically encoded FRET-based biosensors for quantitative metabolite analyses in vivo. J Biotechnol. 2014;191:250–259. doi: 10.1016/j.jbiotec.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 107.Drepper T, et al. Flavin mononucleotide-based fluorescent reporter proteins outperform green fluorescent protein-like proteins as quantitative in vivo real-time reporters. Appl Environ Microbiol. 2010;76:5990–5994. doi: 10.1128/AEM.00701-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Besmer MD, et al. Online flow cytometry reveals microbial dynamics influenced by concurrent natural and operational events in groundwater used for drinking water treatment. Sci Rep. 2016;6:38462. doi: 10.1038/srep38462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Buysschaert B, et al. Online flow cytometric monitoring of microbial water quality in a full-scale water treatment plant. NPJ Clean Water. 2018;1:16 [Google Scholar]
- 110.Alberdi A, Gilbert MTP. A guide to the application of Hill numbers to DNA-based diversity analyses. Mol Ecol Resour. 2019;19:804–817. doi: 10.1111/1755-0998.13014. [DOI] [PubMed] [Google Scholar]
- 111.Liu Z, et al. Neutral mechanisms and niche differentiation in steady-state insular microbial communities revealed by single cell analysis. Environ Microbiol. 2019;21:164–181. doi: 10.1111/1462-2920.14437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stavrakis S, et al. A. deMello, High-throughput microfluidic imaging flow cytometry. Curr Opin Biotechnol. 2019;55:36–43. doi: 10.1016/j.copbio.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 113.Fedorec AJH, et al. FlopR: an open source software package for calibration and normalization of plate reader and flow cytometry data. ACS Synth Biol. 2020;9:2258–2266. doi: 10.1021/acssynbio.0c00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Miano A, et al. Inducible cell-to-cell signaling for tunable dynamics in microbial communities. Nat Commun. 2020;11:1193. doi: 10.1038/s41467-020-15056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shou W, et al. Synthetic cooperation in engineered yeast populations. PNAS. 2007;104:1877–1882. doi: 10.1073/pnas.0610575104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brenner K, et al. Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol. 2008;26:483–489. doi: 10.1016/j.tibtech.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 117.Shong J, et al. Towards synthetic microbial consortia for bioprocessing. Curr Opin Biotechnol. 2012;23:798–802. doi: 10.1016/j.copbio.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 118.Rollié S, et al. Designing biological systems: systems engineering meets synthetic biology. Chem Eng Sci. 2012;69:1–29. [Google Scholar]
- 119.Krieger A, et al. Temperature regulation as a tool to program synthetic microbial community composition. bioRxiv. 2020;2020:2020.2002.2014.944090. doi: 10.1002/bit.27662. [DOI] [PubMed] [Google Scholar]
- 120.Davison BH, Stephanopoulos G. Effect of pH oscillations on a competing mixed culture. Biotechnol Bioeng. 1986;28:1127–1137. doi: 10.1002/bit.260280802. [DOI] [PubMed] [Google Scholar]
- 121.Kerner A, et al. A programmable Escherichia coli consortium via tunable symbiosis. PLoS ONE. 2012;7:e34032. doi: 10.1371/journal.pone.0034032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Izard J, et al. A synthetic growth switch based on controlled expression of RNA polymerase. Mol Syst Biol. 2015;11:840. doi: 10.15252/msb.20156382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Simutis R, Lübbert A. Bioreactor control improves bioprocess performance. Biotechnol J. 2015;10:1115–1130. doi: 10.1002/biot.201500016. [DOI] [PubMed] [Google Scholar]
- 124.Treloar NJ, et al. Deep reinforcement learning for the control of microbial co-cultures in bioreactors. PLoS Comput Biol. 2020;16:e1007783. doi: 10.1371/journal.pcbi.1007783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dopfer O. Schmidt Werner. ChemPhysChem. Vol. 7. 2006. Optical spectroscopy in chemistry and life sciences. An introduction; p. 1598. [Google Scholar]
- 126.Beutel S, Henkel S. In situ sensor techniques in modern bioprocess monitoring. Appl Microbiol Biotechnol. 2011;91:1493–1505. doi: 10.1007/s00253-011-3470-5. [DOI] [PubMed] [Google Scholar]
- 127.Burgess C. In: UV-Visible Spectrophotometry of Water and Wastewater. 2nd edn. Thomas O, Burgess C, editors. Elsevier; 2017. The basis for good spectrophotometric UV-visible measurements; pp. 1–35. [Google Scholar]
- 128.Campbell ID, Dwek RA. Biological Spectroscopy. Benjamin/Cummings Publishing Company; 1994. [Google Scholar]
- 129.Kotanen CN, et al. Surface enhanced Raman scattering spectroscopy for detection and identification of microbial pathogens isolated from human serum. Sensing Bio Sensing Res. 2016;8:20–26. [Google Scholar]
- 130.Panda T, et al. Method to estimate growth of Trichoderma reesei and Aspergillus wentii in mixed culture on cellulosic substrates. Appl Environ Microbiol. 1989;55:1044–1046. doi: 10.1128/aem.55.4.1044-1046.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]