Abstract
Enhancers control the establishment of spatiotemporal gene expression patterns throughout development. Over the past decade, the development of new technologies has improved our capacity to link enhancers with their target genes based on their colocalization within the same topological domains. However, the mechanisms that regulate how enhancers specifically activate some genes but not others within a given domain remain unclear. In this Review, we discuss recent insights into the factors controlling enhancer specificity, including the genetic composition of enhancers and promoters, the linear and 3D distance between enhancers and their target genes, and cell-type specific chromatin landscapes. We also discuss how elucidating the molecular principles of enhancer specificity might help us to better understand and predict the pathological consequences of human genetic, epigenetic and structural variants.
Keywords: Enhanceropathies, Enhancers, Promoters, Specificity, Tethering elements
Introduction
Gene expression is mainly regulated through communication between two types of non-coding regulatory elements: promoters and enhancers. Historically, promoters were defined as regulatory elements that determine where gene transcription starts (i.e. the transcription start site, TSS) and serve as binding platforms for the transcriptional machinery and general co-factors. Enhancers are key regulatory elements that bind tissue-specific transcription factors (TFs) and specifically transmit regulatory information to their target gene promoters, thus dictating where and when genes are expressed. However, these classical definitions are ambiguous, and recent genomic studies show that both elements can overlap in their functions and properties (Andersson and Sandelin, 2020). In agreement with the importance of enhancers and promoters in controlling gene expression, the malfunction of these elements can lead to aberrant gene expression profiles and disease.
Since enhancers can activate gene transcription over long genomic distances (i.e. long-range) and do not always activate their closest target genes, uncovering how enhancers specifically communicate with their correct target gene and not others is still a major challenge in the field. The genome is organized into large megabase (Mb)-scale regulatory domains, such as topologically associating domains (TADs), insulated neighborhoods and contact domains (Beagan and Phillips-Cremins, 2020; Ibrahim and Mundlos, 2020; Szabo et al., 2019; de Wit, 2020). These domains are separated by topological boundaries that favor communication between enhancers and genes located in the same domain while limiting interactions between genes and enhancers located in neighboring domains. However, on a smaller scale, additional regulatory layers control enhancer-gene communication and contribute to the establishment of precise and specific gene expression programs.
In this Review, we discuss the genetic and molecular principles that jointly regulate enhancer specificity, including TAD organization, enhancer-promoter compatibility, linear genomic distance, tethering elements and cell type-specific chromatin landscapes. Finally, we discuss how modifications in these different regulatory layers contributing to enhancer-gene communication can ultimately lead to human disease.
3D genome organization, regulatory domains and TADs
The emergence of chromatin conformation capture (3C)-related methods, especially Hi-C, has dramatically changed our understanding of 3D genome organization. Hi-C studies uncovered that vertebrate genomes are organized in large (sub-Mb scale) and dynamic self-interacting domains (e.g. TADs, contact domains and insulated neighborhoods) that can regulate enhancer function (Dixon et al., 2012; Dowen et al., 2014; Nora et al., 2012; Rao et al., 2014). For the sake of simplicity, we broadly refer to these domains as TADs. TAD boundaries restrict the search space of the enhancers to loci located within the same domain. Consequently, TADs might represent fundamental regulatory units that favor communication between genes and enhancers located in the same domain, while TAD boundaries act as insulators that prevent enhancers from contacting non-target genes located in other domains. TAD formation and function is mainly achieved by the combined action of the cohesin complex and CCCTC-binding factor (CTCF) through a process referred to as loop extrusion (Mirny and Solovei, 2021). Briefly, cohesin is loaded into chromatin and progressively extrudes a loop until it encounters a barrier or is released by the cohesion-unloading factor Wapl (Fudenberg et al., 2016; Haarhuis et al., 2017; Sanborn et al., 2015; Valton et al., 2021 preprint). In vertebrates, the most common cohesin barrier is formed by convergent CTCF-binding sites (CBSs) (de Wit et al., 2015; Rao et al., 2014). However, TSSs and transcription termination sites as well as other proteins, including RNA polymerase II, might also act as cohesin barriers (Banigan et al., 2022 preprint; Valton et al., 2021 preprint). Loop extrusion can favor the interaction of loci located within the same TAD (Dekker and Mirny, 2016; Mach et al., 2022 preprint). Nevertheless, TADs are highly dynamic structures (Gabriele et al., 2022; Mach et al., 2022 preprint) and, depending on boundary strength and chromatin composition, inter-TAD communication can also occur (Bonev et al., 2017; Paulsen et al., 2019; Szabo et al., 2018).
The relevance of TADs to enhancer function is supported by several observations. First, enhancers and their target genes usually lie within the same TAD (Symmons et al., 2014). For example, major developmental genes and their cognate enhancers frequently occupy large genomic regions, previously termed genomic regulatory blocks (GRBs), which show remarkable overlap with TADs (Akalin et al., 2009; Kikuta et al., 2007; Ruf et al., 2011). In addition, disruption of TAD boundaries by structural variants (Fig. 1A) or mutations in CBSs can result in enhancer adoption/hijacking and ectopic gains in gene expression (Franke et al., 2016; Hanssen et al., 2017; Lupiáñez et al., 2015). For example, removal of a CBS located at the α-globin locus leads to fusion between the α-globin-TAD and its neighbor TAD, the establishment of ectopic interactions between the α-globin enhancers and the neighboring TAD, and the upregulation of genes located in the extended TAD (Hanssen et al., 2017). Similarly, deletion of two CBSs located between the Prdm14 super-enhancer (SE) and Slco5a1 causes a fusion between these neighboring TADs and an increase in Slco5a1 expression in mESCs (Vos et al., 2021).
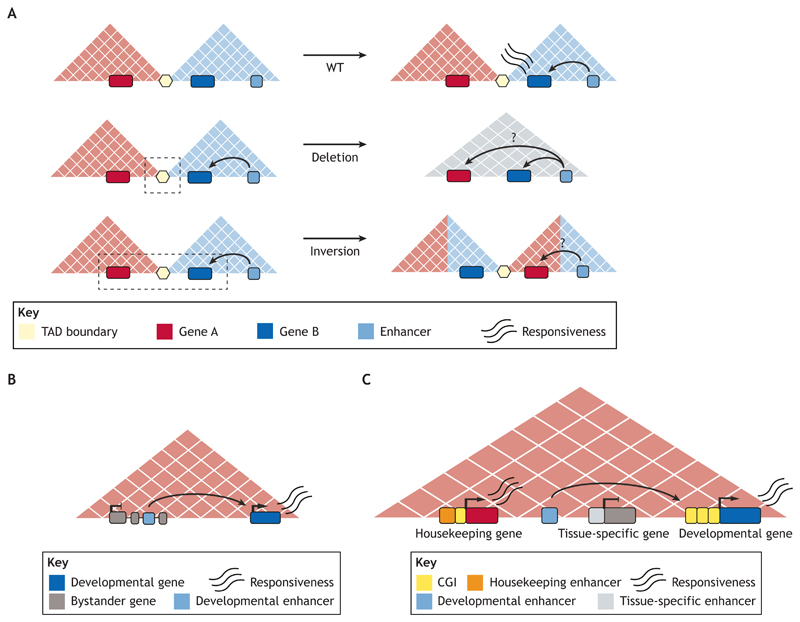
Fig. 1. The role of TADs in enhancer function and specificity.
(A) Enhancers and their target genes usually lie within the same TAD, enabling robust gene expression (top panel). However, structural variants can cause rearrangements of the genes and enhancers present within TADs. Deletions (middle panel) or inversions (lower panel) encompassing TAD boundaries can expose genes to new regulatory environments. Predicting the transcriptional effect of these rearrangements is not obvious, as genes do not always adopt enhancers located in the same TAD. (B) The majority of TADs contain several genes and enhancers. In these multi-gene TADs, enhancers associated with developmental genes can be found in the introns of neighboring genes with unrelated expression patterns (bystander genes). However, bystander genes do not respond to these enhancers, despite being within the same TAD, and are linked with their own regulatory elements. (C) The genetic diversity of promoters might explain the specificity of multi-gene TADs: housekeeping genes respond to housekeeping enhancers located within or close to the promoter regions; tissue-specific genes contain CpG-poor promoters and are responsive to proximal tissue-specific enhancers; while developmental genes contain promoters with large CpG island (CGI) clusters and respond to distal enhancers.
Nevertheless, TAD disruption does not always lead to enhancer rewiring. For example, only a minor set of genes show changes in their expression levels after most TADs are lost following CTCF or cohesin depletion (Nora et al., 2017; Rao et al., 2017). Similarly, highly rearranged chromosomes in Drosophila in which TAD organization is severely disrupted display minor gene expression changes (Ghavi-Helm et al., 2019). In mammals, deletion of a series of CBSs at the Sox9-Kcnj2 locus results in fusion of neighboring TADs, but without any major effects on gene expression (Despang et al., 2019). Furthermore, global analysis of how genes are distributed among TADs in the mouse limb, cortical neurons and ESCs revealed that only 12% of TADs contain a single gene (Ringel et al., 2021 preprint). Typically, these single-gene TADs contain developmental genes located within insulated regulatory landscapes as a mechanism that might ensure their specific expression (Wu et al., 2021). In contrast, the vast majority of TADs (88%) identified in these cell types contain on average 2.4 tissue-specific and 3.6 ubiquitously expressed genes (Ringel et al., 2021 preprint). Importantly, these multi-gene TADs can also contain major developmental genes and their cognate enhancers, together with additional genes (‘bystander’ genes) that do not seem to be regulated by those enhancers (Akalin et al., 2009; Kikuta et al., 2007) (Fig. 1B). Therefore, genes within a TAD are not always co-regulated, which strongly supports the existence of additional regulatory layers controlling the responsiveness between genes and enhancers and, thus, the establishment of specific gene expression profiles.
Promoter architecture and enhancer-gene compatibility
Eukaryotic promoters are composed of distinct core elements, including the TATA box, the initiator (Inr), the downstream promoter element (DPE), the downstream core element (DCE), the TFIIB-recognition element (BRE) and the motif ten element (MTE); these can appear in different combinations (reviewed by Haberle and Lenhard, 2016; Maston et al., 2006). A number of studies have examined how these various elements can influence enhancer-promoter specificity.
Since the original report showing that the 72 bp repeat of the SV40 remote viral sequence influences β-globin gene expression in a distance- and orientation-independent manner (Banerji et al., 1981), enhancers have been traditionally tested and validated according to their ability to influence transcription when placed immediately upstream of a minimal promoter using episomal vectors (i.e. reporter assays) (Banerji et al., 1983; Moreau et al., 1981; de Villiers et al., 1981). In addition, enhancer activity has been investigated using regulatory sensors consisting of a minimal promoter and a reporter gene that, once integrated into the genome, can detect the regulatory activity of nearby enhancers (Akhtar et al., 2014; Gierman et al., 2007; Ruf et al., 2011). Early studies using these two types of assays in Drosophila and mammals showed that, depending on their core elements, promoters can differ in their responsiveness to specific enhancers (Butler and Kadonaga, 2001; Merli et al., 1996; Ohtsuki et al., 1998; Simon et al., 1988; Wefald et al., 1990). In Drosophila, dpp enhancers regulate the expression of only dpp, despite being closer to other genes (i.e. Slh and oaf). Notably, replacing the oaf promoter with the dpp promoter makes oaf responsive to dpp enhancers (Merli et al., 1996). This model was further tested by generating Drosophila lines with random integrations of reporter genes containing a DPE- and TATA-motif promoter (Butler and Kadonaga, 2001). Selective excision of one of the two promoter motifs revealed that some enhancers are able to induce reporter gene expression only when they contain a specific promoter type.
More recently, the advent of next-generation sequencing has enabled the development of massive parallel reporter assays (MPRAs) in which the enhancer activity of libraries containing thousands of genomic fragments can be simultaneously measured (Arnold et al., 2013; Kheradpour et al., 2013; Neumayr et al., 2019). These libraries, which are tested in episomal plasmids (Arnold et al., 2013; Kheradpour et al., 2013; Neumayr et al., 2019) or randomly integrated in the genome (Dickel et al., 2014; Murtha et al., 2014), have revealed the striking selectivity of promoters as well as differences between Drosophila and mammalian promoters.
In Drosophila, one type of MPRA, referred to as self-transcribing active regulatory region sequencing (STARR-seq), was used to measure the enhancer activity of millions of non-coding sequences placed downstream of either housekeeping or developmental promoters (Zabidi et al., 2015). Remarkably, the identified enhancers showed clear preferences towards one of the two promoter classes. Furthermore, a variant of the STARR-seq method, termed self-transcribing active core promoter-sequencing (STAP-seq), determined a wide-range of responsiveness for thousands of Drosophila candidate core promoters tested against fixed enhancer sequences (Arnold et al., 2017). STAP-seq was also used to tether different co-activators to thousands of different Drosophila promoters (Haberle et al., 2019). In line with previous results, Drosophila promoters showed clear preferences for different co-activators, which could be connected to their highly variable enhancer responsiveness. Altogether, these MPRAs suggest that compatibility between promoters and enhancers in Drosophila is determined by both genetic and biochemical factors, which in turn can control the specific establishment of gene expression programs.
In the case of vertebrates, differences between core promoters were also observed, albeit not as clearly as in Drosophila. There are three main types of promoters in vertebrates that differ in the presence and density of sequences containing high frequency of CpG dinucleotides, i.e. CpG islands (CGIs) (Carninci et al., 2006; Saxonov et al., 2006) (Fig. 1C). Promoters associated with developmental genes contain large clusters of CGIs and are usually not associated with TATA elements (Lenhard et al., 2012). Promoters of housekeeping genes tend to have a single CGI and no TATA box, while tissue-specific promoters of genes expressed in terminally differentiated cells lack CGIs but are enriched in TATA elements (Lenhard et al., 2012). A recent MPRA combining different human promoters and enhancers in K562 cells showed that enhancers and promoters are broadly compatible, and combine their regulatory activities in a multiplicative manner (Bergman et al., 2022). Furthermore, this analysis showed that human housekeeping promoters are, in general, less responsive to enhancers, although this might be explained by the fact that core promoters already contain proximal regulatory elements that are intrinsically active (Bergman et al., 2022). On the other hand, context-specific/developmental promoters were shown to be generally more responsive to enhancers than housekeeping promoters, but without showing major preferences for any specific type of enhancer. Similarly, a recent MPRA study in which the activities of housekeeping and developmental core promoters inserted in different loci were measured in human K562 cell lines revealed that the intrinsic activity of different promoter classes is preserved across various genomic environments (Hong and Cohen, 2022). However, the genomic environment might scale the transcriptional strength of core promoters in a non-linear manner. That is, weak housekeeping or developmental core promoters might be more affected by the genomic environment than core promoters with strong transcriptional output (Hong and Cohen, 2022). Altogether, these results suggest that differences in enhancer-promoter compatibility are not as prevalent in human cells as in Drosophila. However, in another recent study using MPRAs in mouse ESCs (Martinez-Ara et al., 2022), the results were more similar to those previously reported in flies (Zabidi et al., 2015). Specifically, this approach combined all the distal regulatory elements (defined as DNAse I hypersensitive sites) and promoters from three different mouse loci and observed a wide range of compatibilities: some regulatory elements specifically activated or repressed a few promoters, whereas others were rather promiscuous (Martinez-Ara et al., 2022). The apparently contradictory results obtained in human, mouse and Drosophila cells might be explained by differences in experimental design (i.e. cell type, selection of candidate enhancers, the length of the promoter regions, etc.). For example, the lack of enhancer responsiveness reported for human housekeeping promoters is in stark contrast to previous findings in Drosophila. However, the core promoters used in the human MPRA were considerably longer than those used in the fly assays. In addition, most of the ‘housekeeping’ enhancers identified in flies overlap or are proximal (<200 bp) to TSSs. One possibility is that, at their endogenous loci, the ‘housekeeping’ enhancers described in Drosophila are part of extended housekeeping promoters that have been proposed for humans. Therefore, two major caveats of MPRAs are: (1) the inclusion of short promoter and enhancer sequences that do not contain the entire genetic information of the studied elements; and (2) investigation of the enhancers out of their endogenous genomic context, where the influence of the chromatin environment and the genetic distance between enhancers and promoters are neglected. Overall, MPRAs can offer highly valuable insights into enhancer function but, according to these recent and somehow contradictory results, they should be interpreted with caution.
Linear distance
Although enhancer function was initially considered to be distance independent, several studies have recently demonstrated that modifying the linear distance between enhancers and their target gene promoters can result in changes in gene expression (Fukaya et al., 2016; Pachano et al., 2021; Rinzema et al., 2021 preprint; Zuin et al., 2022). In Drosophila, positioning the sna enhancer at 6.5 or 9 kb from a reporter gene showed that transcription decreases as the linear enhancer-gene distance increases (Yokoshi et al., 2020). Notably, a recent study in which an enhancer was inserted around a reporter gene into more than 100 different genomic locations spanning ~300 kb strongly suggests that transcription decreases with increasing genomic distances (Zuin et al., 2022). Moreover, modeling of the resulting data suggests that the relationship between genetic distance, contact probability and transcription is non-linear (Zuin et al., 2022), which might be explained by a promoter being able to remain in an active transcriptional state for periods longer than the duration of its contact with an enhancer (i.e. promoter hysteresis) (Xiao et al., 2021; Zuin et al., 2022). Similarly, mobilization of the β-globin locus control region (LCR) with respect to a reporter gene containing the HBG1 promoter also showed that the linear distance between genes and enhancers inversely correlates with transcription (Rinzema et al., 2021 preprint). In addition, studies of the endogenous β-globin locus further illustrate the preference of the LCR for proximally located genes (Hanscombe et al., 1991; Peterson and Stamatoyannopoulos, 1993). Briefly, human β-globin genes are arranged in a 5′-ε-Gγ-Aγ-δ-β-3′ orientation downstream of the LCR. The LCR regulates the temporal expression of the globin genes during the embryonic (ε-globin), fetal (γ-globin) and adult (β-globin) stages (Hardison et al., 1997). The generation of transgenic animals in which the β-globin gene was inserted closer to the LCR led to higher gene expression (Dillon et al., 1997). Accordingly, an inversion switching the relative position of the β-globin genes with respect to the LCR resulted in preferential activation of the gene located more proximal to the LCR (Tanimoto et al., 1999). More recently, a tissue-specific gene (Gria1) was shown to be responsive to a developmental enhancer when both were placed in close proximity, but not when the same enhancer was placed 100 kb upstream of the gene within the same TAD (Pachano et al., 2021). In contrast, the activation of Shh in mouse ESCs through the forced recruitment of TFs to different enhancers is not significantly affected by linear distance (Kane et al., 2021 preprint). However, communication between the more-distal enhancers and the Shh promoter might be influenced by CTCF-mediated interactions, which in turn could buffer, in a cohesin-dependent manner, the negative effect that linear distance has on gene expression (Calderon et al., 2022; Kane et al., 2021 preprint; Rinzema et al., 2021 preprint). Interestingly, motif analyses suggest that tissue-specific genes with CpG-poor promoters, such as the β-globin genes and Gria1, often have their key regulatory elements in promoter-proximal regions (Roider et al., 2009), while developmental genes with CpG-rich promoters (e.g. Shh) are frequently regulated by distal enhancers (Lenhard et al., 2012).
Together with observations using MPRAs, these results indicate that: (1) housekeeping genes show a generally low responsiveness to enhancers, regardless of distance, because their regulatory elements are embedded within or close to their promoter regions (Bergman et al., 2022); (2) tissue-specific genes with CpG-poor promoters show high responsiveness to proximal but not to distal enhancers, probably reflecting that, endogenously, these promoters are typically regulated by proximal cis-regulatory elements (Roider et al., 2009); and (3) developmental genes, which in vertebrates have large CGI clusters in their promoter regions, are often regulated by and are highly responsive to distal enhancers, sometimes separated by >1 Mb (Benko et al., 2009; Lettice et al., 2002; Lim et al., 2018; Long et al., 2020) (Figs 1C and 2). Therefore, by placing genes and enhancers further away from each other within the same TAD, it might be possible to specifically activate some genes (e.g. developmental genes with CpG-rich promoters) but not others (e.g. housekeeping genes, tissue-specific genes with CpG-poor promoters) (Akalin et al., 2009; Engström et al., 2007; Kikuta et al., 2007).
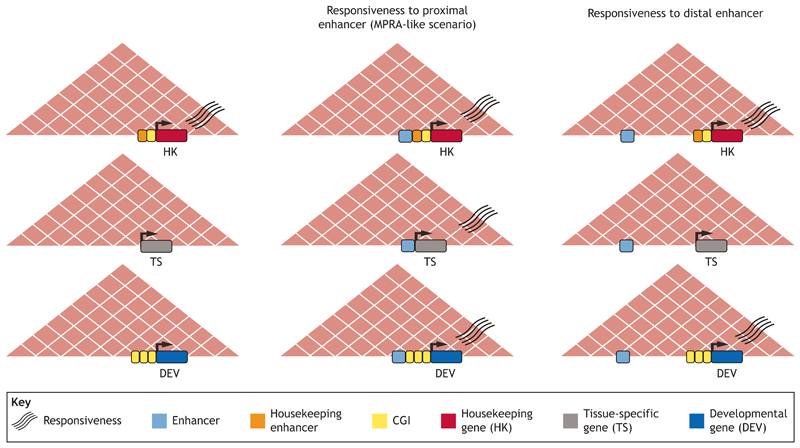
Fig. 2. Linear distance influences enhancer-gene regulatory specificity.
The responsiveness of different promoter types to enhancers can depend on linear distance. The promoters of housekeeping (HK) genes (top) are normally activated by proximal regulatory elements and show low responsiveness to proximal or distal enhancers. Tissue-specific (TS) genes (middle) also show low responsiveness to distal enhancers, but high responsiveness to proximal enhancers. In contrast, developmental genes (DEV; bottom) show high responsiveness to distal enhancers.
Factors that regulate enhancer-gene communication in 3D space
There are multiple examples of enhancers that can specifically control their target genes over long distances (Benko et al., 2009; Lettice et al., 2002; Long et al., 2020) or even on different chromosomes (Chang et al., 2004; Lim et al., 2018; Sanyal et al., 2012). Therefore, although genomic distance might contribute to enhancer-promoter specificity, there are additional mechanisms that enable distal enhancers to specifically encounter their target genes and execute their long-range regulatory function (Batut et al., 2022; Kane et al., 2021 preprint; Rinzema et al., 2021 preprint).
Tethering elements and GAGA
Tethering elements are short DNA sequences found in promoters and/or enhancers that can bring both of these elements into 3D proximity. Tethering elements have been described in Drosophila and mammals, although they might differ across species in their genetic composition and the proteins that mediate their function (Batut et al., 2022; Pachano et al., 2021).
In Drosophila, tethering elements were first described for the Scr-ftz locus (Calhoun et al., 2002). This region contains the T1 enhancer, which specifically activates Scr even though it is located closer to ftz. The specific activation of Scr can be explained by a 450 bp region located next to the Scr TSS that was suggested to specifically bring the T1 enhancer in close proximity to the Scr promoter. Similarly, the IAB5 Drosophila enhancer, which is located equidistant from the abd-A and Abd-B genes, specifically interacts with Abd-B and not with abd-A (Akbari et al., 2007). The preferential physical association between IAB5 and Abd-B can be explained by the presence of a short 255 bp tethering element at the Abd-B TSS. Deletion of this element directs the IAB5 enhancer to the abd-A promoter and induces its expression. Similarly, in a transgenic configuration in which the IAB5 enhancer is placed between two genes, both genes are induced (Akbari et al., 2007).
However, placing the tethering element adjacent to the 3′ located gene results in preferential expression of this gene and no expression of the 5′ gene. These tethering elements frequently contain binding sites for the transcription factor GAGA (Gaf/Trl) (Mahmoudi et al., 2002). Once bound to both promoters and enhancers, GAGA can bring them into physical proximity due to its oligomerization capacity, thus facilitating enhancer function and gene activation (Mahmoudi et al., 2002). By mediating long-range enhancer-gene communication, these tethering elements enable the precise temporal transcription of developmental genes during early Drosophila embryogenesis (Batut et al., 2022). Interestingly, tethering elements are genetically and functionally distinct from TAD boundaries, the main function of which in Drosophila might be to insulate genes from spurious interactions with enhancers and silencers located in neighboring TADs (Batut et al., 2022).
CTCF-binding sites
Besides their roles in TAD boundaries and the insulation of regulatory landscapes, CBSs also occur at multiple mouse gene promoters, where they can facilitate physical interactions with distal enhancers (Kubo et al., 2021). For example, deletion of a CBS at the Vcan promoter impairs interactions with a distal enhancer and reduces its expression levels (Kubo et al., 2021). Vcan expression can be restored via the artificial tethering of CTCF to its promoter. In addition, deletion of the human TFF1 gene promoter can ‘release’ its cognate enhancer, which can then ectopically activate another gene from the same TAD that also contains a CBS on its promoter (Fig. 3A). Interestingly, the TFF1 promoter contains a CBS that, when disrupted, reproduces the effect of deleting the entire promoter (Oh et al., 2021). It is currently unclear whether this potential role of CTCF as a tethering element that facilitates enhancer function requires direct binding to both enhancers and promoters or whether binding to one of the regulatory elements is sufficient (Kubo et al., 2021; Oh et al., 2021). Recent work suggests that convergent CBSs flanking enhancer-gene pairs facilitate communication between genes and distal enhancers by favoring the formation of small contact domains (Rinzema et al., 2021 preprint) (Fig. 3B). In contrast, in the mouse Sox2 locus, CTCF binding to either the Sox2 promoter or a distal enhancer is dispensable for their interaction (Chakraborty et al., 2022 preprint; Taylor et al., 2022 preprint). It is important to note that, in the context of promoter regions, CTCF can function not only as a tethering element but also as an insulator and as a conventional transcriptional activator (Chernukhin et al., 2007; Paredes et al., 2013; Peña-Hernández et al., 2015), thus complicating the dissection of CTCF function in enhancer-driven gene expression.
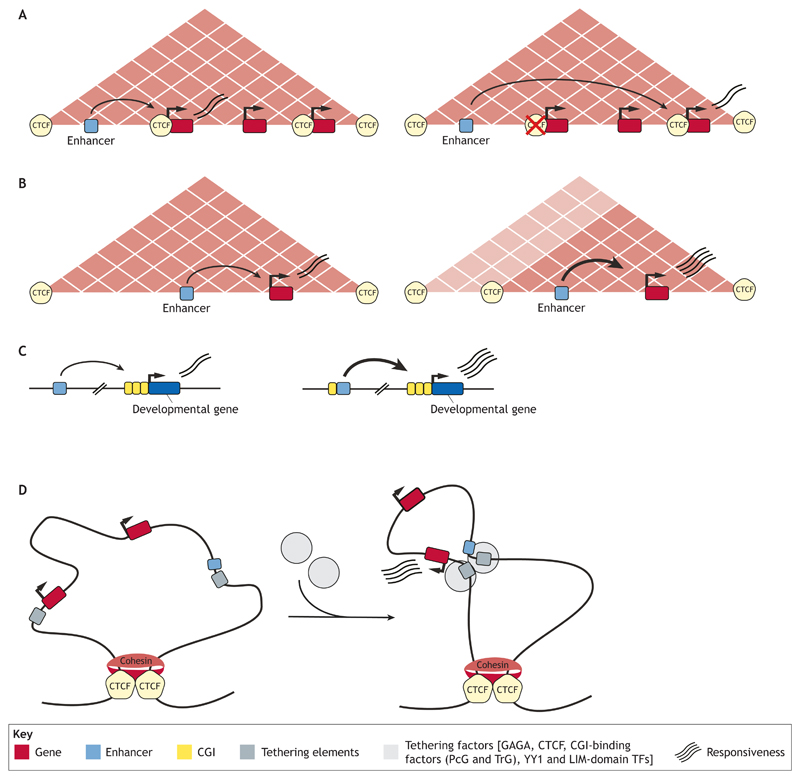
Fig. 3. CTCF-binding sites, CpG islands and tethering factors can modulate enhancer-gene communication in 3D space.
(A) The presence of a CTCF-binding site (CBS) can determine promoter choice by an enhancer. (B) The presence of a CBS can also favor the formation of smaller contact domains, leading to strong target gene induction. (C) CpG islands (CGIs) can boost the regulatory activity of distal developmental enhancers by conferring a permissive topological configuration that increases the responsiveness of target genes with CGI-associated promoters. (D) Schematic diagram illustrating the topological and regulatory role of tethering factors. Homotypic or heterotypic interactions between tethering elements facilitate physical communication between enhancers and their target gene promoters. This topological configuration might favor specific cis-activation of target genes.
CpG islands
In mammalian cells, a recent study identified a novel role for CGIs as tethering elements that facilitate the physical and functional communication between developmental genes and CpG-rich enhancers (also known as poised enhancers) (Pachano et al., 2021) (Fig. 3C). In this context, CGIs must be present at both promoters and enhancers to function as tethering elements. This results in homotypic intra-TAD interactions that enable enhancers to preferentially activate developmental genes (i.e. genes with CGI clusters in their promoters). Interestingly, the promoter from a CGI-poor gene (i.e. a tissue-specific gene) appears to be responsive to the CpG-rich enhancer when this is placed proximally but not distally (Pachano et al., 2021). This suggests that CGIs, and perhaps tethering elements in general, do not regulate the intrinsic compatibility between promoters and enhancers, but rather facilitate the long-range communication between enhancers and their target genes. As a corollary of this, tethering elements might not be necessary for proximal enhancers and their regulatory function might not be detectable using reporter assays.
Mechanistically, the interactions between CpG-rich enhancers and their target genes in pluripotent cells are likely to be mediated by Polycomb group (PcG) complexes, which are recruited to CGIs present at both enhancers and target promoters (Crispatzu et al., 2021; Cruz-Molina et al., 2017; Pachano et al., 2021). PcG complexes, particularly PRC1, can play important architectural roles and mediate long-range interactions (Blackledge and Klose, 2021; Cheutin and Cavalli, 2014; Entrevan et al., 2016; Pachano et al., 2019). It is currently unclear whether PcG complexes can still contribute to enhancer-gene communication once the CpG-rich enhancers and their target genes become active (Loubiere et al., 2020; Zhang et al., 2021) or whether, alternatively, other CGI-bound proteins (e.g. TrG) execute the tethering function in the active state (Xiang et al., 2020). Furthermore, regardless of whether enhancers contain CGI or not, promoters with large CGI clusters seem to be more responsive to distal enhancers than to CpG-poor promoters (Kraft et al., 2019; Pachano et al., 2021). In this case, the promoter CGIs might not act as tethering elements but rather confer a permissive chromatin state (e.g. low nucleosome occupancy and DNA hypomethylation) that increases enhancer responsiveness. Overall, mammalian CGIs and Drosophila tethering elements show remarkable functional similarities, and future work should elucidate whether this is achieved through either similar or distinct molecular mechanisms.
Transcription factors
There are several TFs that, as part of their trans-activating function, also contribute to the establishment of enhancer-gene contacts. One classical example is Sp1, which in mammals can bind to both enhancers and promoters, and, due to its oligomerization capacity, bring them into physical proximity (Mastrangelo et al., 1991). Although Sp1 can favor enhancer-gene communication over short distances (<2 kb), it is currently unknown whether it can also assist long-range enhancers. Another example of a TF intervening in enhancer-promoter looping is YY1 (Beagan et al., 2017; Weintraub et al., 2017): a zinc-finger protein that binds to active enhancers and promoters in mouse ESCs, and facilitates their physical and functional communication. Deletion of YY1-binding sites or degradation of YY1 leads to a loss of interactions between genes and enhancers and, consequently, to impaired gene expression. Moreover, the artificial tethering of YY1 to the Etv4 promoter bearing a mutation in a YY1-binding site rescues enhancer-promoter contacts and Etv4 expression (Weintraub et al., 2017). Another important group of TFs contributing to enhancer-gene communication in mammalian cells are the LIM-domain TFs, such as GATA1, LHX3 or ISL1. Previous work has shown that GATA1 binds to both the LCR and β-globin promoter in erythroid cells (Song et al., 2007; Tripic et al., 2009). Like other LIM-domain TFs, GATA1 interacts with LIM domain-binding protein 1 (LDB1), which mediates long-range interactions between the LCR and the β-globin gene due to its ability to form dimers through its SA domain (Song et al., 2007; Tripic et al., 2009). In support of the tethering role of LDB1, targeting LDB1 to the β-globin promoter leads to interactions with endogenous LDB1 present at the LCR and to the formation of a chromatin loop (Deng et al., 2012). Notably, this forced loop is sufficient to trigger β-globin gene induction in immature murine erythroblasts. Similarly, forced tethering of LDB1 or its SA domain to an otherwise silenced fetal γ-globin promoter was shown to activate gene expression in human primary adult erythroblasts (Bartman et al., 2016; Deng et al., 2014). Moreover, Chip, a Drosophila homolog of LDB1, also interacts with LIM domain-containing proteins and has been proposed to support enhancer-promoter interactions (Morcillo et al., 1997).
Other TFs might also contribute to the specific communication between genes and enhancers through the establishment of homotypic interactions (Bouwman and De Laat, 2015; Denholtz et al., 2013; Giammartino et al., 2019). In mouse ESCs, regions bound by pluripotency TFs (i.e. KLF4, OCT4, SOX2 and NANOG) interact with each other and form distinct nuclear hubs (Bouwman and De Laat, 2015; Denholtz et al., 2013). However, these TFs seem to be necessary but not sufficient for long-range enhancer-gene interactions (Giammartino et al., 2019; Wei et al., 2013). This suggests that most TFs might recruit tethering (e.g. LDB1) or architectural (e.g. cohesin) proteins, which can then mediate specific communication between genes and enhancers (Dowen et al., 2014; Giammartino et al., 2019).
Identifying the genomic sequences that act as tethering elements and the proteins that are bound to them will be a key challenge in understanding how enhancers and genes can specifically and functionally communicate with each other (Fig. 3D). It is likely that additional tethering elements and their associated proteins are yet to be discovered. Moreover, tethering elements might also operate in a combinatorial manner, such that the communication between certain enhancer-gene pairs might involve more than one type of tethering element in order to increase specificity and/or gene expression levels.
Cell type-specific chromatin landscapes
Cell type-specific changes in chromatin modifications can, in principle, modulate the activity and function of the regulatory elements described above (i.e. promoters, enhancers and tethering elements). Assessing how such chromatin landscapes affect regulatory elements, however, is technically challenging. One limitation of episomal MPRAs, for example, is that they do not recapitulate the chromatin context in which regulatory elements function endogenously. Transgenic reporter assays, such as those frequently used in mouse or zebrafish embryos, can mitigate some of the limitations of episomal MPRAs by integrating the reporter constructs (i.e. enhancer, minimal promoter and reporter gene) into the genome. Using this type of reporter assay, the activity of the Pen enhancer, which is required for proper Pitx1 expression in the hindlimb, was investigated in mouse embryos (Kragesteen et al., 2018). Interestingly, the Pen enhancer was shown to be able to drive reporter expression not only in the hindlimb but also in the forelimb, where Pitx1 is not expressed. However, in the hindlimb, Pitx1 is in close physical proximity to the Pen enhancer and thus expressed, whereas, in the forelimb, Pitx1 is inactive, far from the Pen enhancer and associated with Neurog1. Both Neurog1 and Pitx1 are inactive in the forelimb and are marked with PcG, which can bring these two genes into physical proximity and create a chromatin environment refractory to Pen enhancer function (Kragesteen et al., 2018). This and other studies in which transgenic reporter assays were used illustrate how the chromatin environment can affect the cis-regulatory activity of enhancers (Akhtar et al., 2014; Ruf et al., 2011; Symmons et al., 2014). However, this advantage over MPRAs can also be a limitation, as enhancer responsiveness of the reporter gene can be severely affected by the genomic location of the integrations sites. These position effects in transgenic reporter assays can be mitigated by using safe harbor systems (Kvon et al., 2020).
The importance of chromatin context in enhancer-gene communication is also supported by a recent study showing that the divergent expression of Fat1 and Rex1, two genes located in the same TAD, involves cell type-specific chromatin changes that affect TAD organization and DNA methylation patterns (Ringel et al., 2021 preprint). In ESCs, both Fat1 and Rex1 are expressed, enabled by a TAD configuration in which these genes specifically interact with their cognate proximal enhancers. As differentiation progresses, Fat1 remains expressed while Rex1 is silenced. Remarkably, in differentiated cells, Fat1 is regulated by enhancers that are more proximal to Rex1 than to Fat1. The loss of Rex1 responsiveness to the Fat1 enhancers might be caused by DNA hypermethylation of its promoter region (Ringel et al., 2021 preprint). Interestingly, the Rex1 promoter belongs to the CpG-poor category, which might render it more susceptible to DNA hypermethylation in comparison with the CpG-rich Fat1 promoter (Blackledge and Klose, 2011; Deaton and Bird, 2011; Long et al., 2016).
Cell type-specific changes in DNA methylation patterns might also underlie the observed variability in TAD organization across cell types (McArthur and Capra, 2021), which seems more pronounced than originally proposed (Dixon et al., 2012). This variability in TAD organization could be caused by cell-type specific changes in CTCF occupancy, as the binding of this TF to its target sequences is sensitive to DNA methylation levels (Beagan et al., 2017; Wang et al., 2012).
The medical relevance of enhancer-gene specificity
Human disorders caused by enhancer malfunction are often referred to as enhanceropathies and can involve structural variants, single nucleotide polymorphisms and epigenetic alterations (Claringbould and Zaugg, 2021; Lupiáñez et al., 2016; Rickels and Shilatifard, 2018; Smith and Shilatifard, 2014). Although some enhanceropathies are caused by the direct disruption of enhancers (Haro et al., 2021; Long et al., 2020), others involve either gains (i.e. enhancer adoption/hijacking) or losses (i.e. enhancer disconnection) in long-range enhancer-gene communication (Franke et al., 2016; Gröschel et al., 2014; Laugsch et al., 2019; Lupiáñez et al., 2015).
Structural variants
Structural variants (SVs) include deletions, inversions, duplications, translocations and insertions. Deletions can result in either the direct loss of enhancers or changes in TAD structure. For example, the Pierre-Robert sequence craniofacial disorder is caused by deletions that eliminate enhancers located within the SOX9 TAD that are required for proper SOX9 expression in neural crest cells (Amarillo et al., 2013; Gordon et al., 2009; Long et al., 2020). Deletions spanning TAD boundaries can lead to the fusion of neighboring TADs, which in turn can lead to ectopic enhancer-gene interactions (i.e. enhancer adoption/hijacking) and, consequently, to pathological gains in gene expression without directly perturbing enhancer sequences (Franke et al., 2016; Gröschel et al., 2014; Lupiáñez et al., 2015) (Fig. 4A). Deletions can also affect tethering elements, as reported for a 12 kb homozygous deletion encompassing three CBSs located downstream of the limb-specific ZRS enhancer, within one of the Lmbrl introns (Ushiki et al., 2021). This deletion impairs communication between the Shh promoter and the ZRS enhancer, which reduces Shh expression in the limb and leads to a congenital limb truncation termed acheiropodia (Ushiki et al., 2021). On the other hand, inversions spanning TAD boundaries can place enhancers away from their target genes (i.e. enhancer disconnection) and/or in proximity of novel genes (i.e. enhancer adoption), which can result in either losses or gains in gene expression, respectively (Laugsch et al., 2019; Lupiáñez et al., 2016; Smith and Shilatifard, 2014; Spielmann et al., 2018) (Fig. 4A). For example, an inversion found in individuals with limb abnormalities relocates EPHA4 enhancers into the WNT6 TAD (Lupiáñez et al., 2015). Consequently, this leads to the ectopic and pathological expression of WNT6 in the limb. A recent study reported an individual with branchio-oculo-facial syndrome (BOFS) with a long heterozygous inversion that physically disconnects TFAP2A from its cognate neural crest enhancers (Laugsch et al., 2019). Notably, this results in the monoallelic and haploinsufficient expression of TFAP2A expression in the neural crest cells of the patient. Furthermore, the inversion also places novel genes in proximity to TFAP2A neural crest enhancers within a ‘shuffled’ TAD (Fig. 4A). However, none of these genes are responsive to the TFAP2A enhancers and enhancer adoption does not occur within this particular genomic context. Interestingly, although TFAP2A is a typical developmental gene with a large CGI cluster in its promoter region, none of the genes placed in the proximity of the neural crest enhancers due to the BOFS inversion display this type of promoter (Laugsch et al., 2019; Pachano et al., 2021). Thus, SVs that place enhancers and genes within the same TAD might not always lead to aberrant gene expression. In these cases, additional factors controlling enhancer-gene compatibility should be taken into consideration to better predict pathological changes in gene expression.
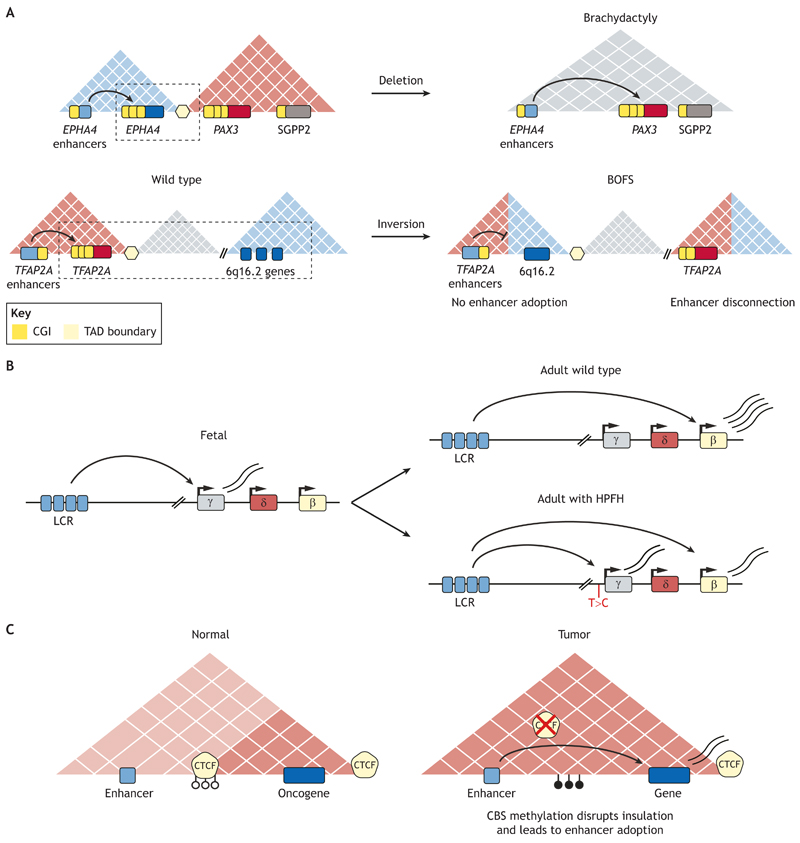
Fig. 4. The disruption of enhancer specificity can lead to human diseases.
(A) Structural variants (SVs) can rearrange genes and enhancers into different TADs, which in some cases can lead to either enhancer adoption or enhancer disconnection. Top panel: a deletion spanning a TAD boundary between the EPHA4-TAD and the PAX3-TAD is found in families with brachydactyly (Lupiáñez et al., 2015). As a result of this deletion, enhancers that are active in the developing limb and control EPHA4 expression in this tissue establish ectopic interactions with PAX3 and strongly induce its expression in the limb (i.e. enhancer adoption), a tissue in which this developmental gene is normally inactive. The SGGP2 gene, which is also located in the PAX3-TAD, does not respond to the EPHA4 enhancers. Bottom panel: an inversion that spans several TAD boundaries leads to TAD shuffling. The inversion places one of the TFAP2A alleles into a novel TAD and impairs its normal expression in neural crest cells due to physical disconnection from its cognate enhancers. The reduced expression of TFAP2A in neural crest cells leads to TFAP2A haploinsufficiency, which ultimately results in craniofacial abnormalities associated with the branchio-oculo-facial syndrome (BOFS). Moreover, this inversion also places novel genes originally found within the 6q16.2 locus in proximity to TFAP2A neural crest enhancers within a shuffled TAD. Importantly, none of the 6q16.2 genes is responsive to the TFAP2A neural crest enhancers (i.e. there is no enhancer adoption in neural crest cells) (Laugsch et al., 2019). (B) Individuals with hereditary persistence of fetal hemoglobin (HPFH) possess a single nucleotide polymorphism (SNP; T>C) at the γ-globin promoter, which increases its affinity for the locus control region (LCR). Consequently, this leads to persistent γ-globin expression in adulthood (Wienert et al., 2015). (C) Oncogenes can become overexpressed in tumor cells through the aberrant methylation of CTCF-binding sites (CBSs). Hypermethylation of CBSs can abrogate CTCF binding, which in turn can result in loss of insulation and ectopic communication between enhancers and responsive oncogenes (Flavahan et al., 2019). Pale-blue squares represent enhancers; dark-blue and red squares represent genes.
Deletions of promoter sequences can also lead to oncogenic gains in gene expression through long-range regulatory mechanisms (Cho et al., 2018; Oh et al., 2021). In this case, deletions can impair physical communication between enhancers and their endogenous target genes (Cho et al., 2018; Oh et al., 2021), which in turn enables the enhancers to interact with and activate neighboring oncogenes located within the same TAD. For example, in the absence of the α-globin gene, its cognate LCR makes contact with and induces the expression of an unrelated gene, NME4, which is located ~300 kb away (Lower et al., 2009). Interestingly, there are up to five additional genes between the α-globin LCR and NME4, the expression of which is not affected by α-globin deletion, further highlighting the importance of factors contributing to enhancer-gene specificity. Moreover, the previous examples illustrate how active promoters can also act as insulators that restrict enhancer activity, which might involve the formation of topological boundaries (Dixon et al., 2012) and/or promoter competition (Calhoun et al., 2002).
Single nucleotide polymorphisms
Genome-wide association studies (GWAS) have revealed that most single nucleotide polymorphisms (SNPs) associated with complex diseases occur within non-coding regulatory regions (Maurano et al., 2012). These SNPs often disrupt TF-binding sites within promoters or enhancer elements, resulting in quantitative changes in gene expression that can be either detrimental or beneficial (Rockman and Kruglyak, 2006). Below, we summarize some of these examples and direct the reader to excellent reviews on this topic for additional data (Carullo and Day, 2019; Claringbould and Zaugg, 2021; Rickels and Shilatifard, 2018; Smith and Shilatifard, 2014).
Individuals with hereditary persistence of fetal hemoglobin (HPFH) carry a SNP that reduces the symptoms of β-thalassemia and sickle cell anemia (Wilber et al., 2011). During development, a postnatal switch in the affinity of the LCR from the γ-globin promoter towards the β-globin promoter results in the downregulation of fetal hemoglobin in adulthood (Wilber et al., 2011) (Fig. 4B). However, individuals with HPFH show higher levels of fetal hemoglobin in adulthood because of an SNP in the γ-globin promoter that increases its affinity for the LCR (Fig. 4B). Consequently, fetal hemoglobin expression persists during adulthood, while the β-globin genes become downregulated (Wienert et al., 2017). One of the SNPs associated with HPFH (A198 T>C) generates a new binding site for the erythroid-specific TF KLF1 (Martyn et al., 2019; Wienert et al., 2015). Other β-thalassemia cases are caused by a SNP found within a non-coding region at the α-globin locus. This SNP, which is located between the α-globin genes and the LCR, creates a new promoter that interferes with α-globin transcription (Bozhilov et al., 2021). The postnatal switch from γ-globin expression towards β-globin expression is also regulated by BCL11A, a TF that represses γ-globin expression. Individuals with a SNP located at the BCL11A enhancer do not express BCL11A in adulthood and present HPFH (Bauer et al., 2015). Interestingly, CRISPR-Cas9 targeting of the BCL11A enhancer in individuals with β-thalassemia and sickle cell anemia resulted in an increase in fetal hemoglobin and reduced morbidity and mortality (Frangoul et al., 2020).
A SNP in the HNE-F2 enhancer, located at the Sox9 TAD, is implicated in the etiology of the Pierre Robin sequence condition (Benko et al., 2009). This SNP disrupts a binding site for MSX1 within the HNE-F2 enhancer and downregulates Sox9 expression in the mandibular and maxillary mesenchymal tissues (Benko et al., 2009). Several SNPs within the limb-specific ZRS enhancer in humans and other vertebrates have been shown to drive ectopic Shh in the anterior limb mesenchyme, causing a polydactyly phenotype (Hill and Lettice, 2013; Lettice et al., 2003; VanderMeer and Ahituv, 2011). Finally, GWAS revealed that almost all SNPs associated with Alzheimer’s disease (AD) lie within intronic or intergenic regions (Kikuchi et al., 2019). However, only 30% of these SNPs map to enhancers, highlighting the possibility that these SNPs alter the function of other types of non-coding regulatory elements. Interestingly, some of these SNPs (e.g. rs1476679 and rs1990620) overlap with CBSs and can affect CTCF recruitment, the formation of chromatin loops and the expression of genes associated with AD (Gallagher et al., 2017; Kikuchi et al., 2019).
Epigenetic alterations
DNA methylation profiles are frequently altered in cancer, which can disrupt the binding of CTCF to its cognate elements (Bell and Felsenfeld, 2000; Hark et al., 2000; Liu et al., 2016). For example, recent studies indicate that DNA hypermethylation in different cancer types can block the recruitment of CTCF to key insulators, resulting in enhancer adoption and oncogenic gains in gene expression (Flavahan et al., 2016; 2019) (Fig. 4C).
Altogether, the above examples illustrate the prevalence of human enhanceropathies and the complexity of their associated pathomechanisms. Therefore, revealing the regulatory factors that control the specific communication between genes and enhancers is an essential step towards a complete and predictive understanding of human disease.
Conclusions
During the past few decades, the field of enhancer biology has vastly expanded through the identification of enhancers and their association with target genes. Despite this tremendous progress, it is still unclear how enhancers communicate specifically with their target genes. Recent MPRAs and genomic engineering approaches are starting to reveal a number of complex and diverse mechanisms that control enhancer specificity. While TAD structures might favor specific communication between genes and their enhancers, other regulatory layers further refine these interactions. One of the most striking examples is the observed lack of compatibility between developmental and housekeeping regulatory elements in Drosophila (Haberle et al., 2019; Zabidi et al., 2015; Zabidi and Stark, 2016). However, these compatibility rules do not seem to be as clear cut in mammalian cells (Bergman et al., 2022; Martinez-Ara et al., 2022), with chromatin context, genetic distance and tethering elements representing additional and important regulators of enhancer specificity.
In the case of tethering elements, recent reports have discovered novel roles for genetic elements (e.g. CGIs and GAGA elements) and proteins (e.g. CTCFs, LDB1 and YY1) that favor specific communication between enhancers and their target genes (Calhoun et al., 2002; Giammartino et al., 2019; Kubo et al., 2021; Oh et al., 2021; Pachano et al., 2021; Weintraub et al., 2017). Similar to its role in TAD boundary formation and enhancer insulation, CTCF tethering function is likely connected to its capacity to block cohesin and loop extrusion. However, the extent to which CTCF is required for enhancer-gene communication is still unclear, as either the deletion of CBSs or the depletion of CTCF does not always affect the topology or the expression of genes containing CBSs at their promoters (Chakraborty et al., 2022 preprint; Despang et al., 2019; Taylor et al., 2022 preprint). Moreover, the molecular mechanisms by which other types of tethering elements (e.g. CGIs, GAGA-elements) control enhancer specificity and function are still poorly understood. It is currently unclear whether the regulatory function of tethering elements is cohesion dependent and generally involves loop extrusion or whether alternative mechanisms (e.g. polymerization and liquid-liquid phase separation) might be also involved. Therefore, a future goal should be to identify the full repertoire of tethering elements that contribute to enhancer function and to compare their mechanisms of action.
More broadly, there are still fundamental questions regarding the mechanisms of enhancer function that remain to be solved. Uncovering the biochemical processes whereby enhancers activate their target gene might help us to better understand the compatibility rules between enhancers and promoters, and how tethering elements work. We anticipate that understanding the principles that regulate enhancer specificity will help us to better predict the effect of genetic variation in human diseases. As illustrated by the examples discussed above, disease-causing genetic variants do not necessarily disrupt enhancer or gene sequences, but instead can affect the ability of enhancers to specifically activate their target genes. Therefore, elucidating the molecular and genetic principles controlling enhancer specificity will certainly help us to uncover and predict the etiological basis of human disease.
Acknowledgements
We thank all the Rada-Iglesias lab members for useful discussions and ideas while preparing this Review.
Funding
Work in the Rada-Iglesias laboratory is funded by the Ministerio de Ciencia e Innovación and the Agencia Española de Investigación; by the European Regional Development Fund (PGC2018-095301-B-I00 and RED2018-102553-T); by the European Research Council (862022); and by the European Commission (H2020-MSCA-ITN-2019-860002).
Footnotes
Competing interests
The authors declare no competing or financial interests.
References
- Akalin A, Fredman D, Arner E, Dong X, Bryne JC, Suzuki H, Daub CO, Hayashizaki Y, Lenhard B. Transcriptional features of genomic regulatory blocks. Genome Biol. 2009;10:R38. doi: 10.1186/gb-2009-10-4-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari OS, Bae E, Johnsen H, Villaluz A, Wong D, Drewell RA. A novel promoter-tethering element regulates enhancer-driven gene expression at the bithorax complex in the Drosophila embryo. Development. 2007;135:123–131. doi: 10.1242/dev.010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar W, Pindyurin AV, De Jong J, Pagie L, Ten Hoeve J, Berns A, Wessels LFA, Van Steensel B, Van Lohuizen M. Using TRIP for genome-wide position effect analysis in cultured cells. Nat Protoc. 2014;9:1255–1281. doi: 10.1038/nprot.2014.072. [DOI] [PubMed] [Google Scholar]
- Amarillo IE, Dipple KM, Quintero-Rivera F. Familial microdeletion of 17q24.3 upstream of SOX9 is associated with isolated pierre robin sequence due to position effect. Am J Med Genet A. 2013;161:1167–1172. doi: 10.1002/ajmg.a.35847. [DOI] [PubMed] [Google Scholar]
- Andersson R, Sandelin A. Determinants of enhancer and promoter activities of regulatory elements. Nat Rev Genet. 2020;21:71–87. doi: 10.1038/s41576-019-0173-8. [DOI] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Stelzer C, Boryń ŁM, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- Arnold CD, Zabidi MA, Pagani M, Rath M, Schernhuber K, Kazmar T, Stark A. Genome-wide assessment of sequence-intrinsic enhancer responsiveness at single-base-pair resolution. Nat Biotechnol. 2017;35:136–144. doi: 10.1038/nbt.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J, Olson L, Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983;33:729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Banerji J, Rusconi S, Schaffner W. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-X. [DOI] [PubMed] [Google Scholar]
- Banigan EJ, Tang W, van den Berg AA, Stocsits RR, Wutz G, Brandão HB, Busslinger GA, Peters J-M, Mirny LA. Transcription shapes 3D chromatin organization by interacting with loop-extruding cohesin complexes. BioRxiv. 2022:2022.01.07.475367. doi: 10.1101/2022.01.07.475367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartman CR, Hsu SC, Hsiung CC-S, Raj A, Blobel GA. Enhancer regulation of transcriptional bursting parameters revealed by forced chromatin looping. Mol Cell. 2016;62:237–247. doi: 10.1016/j.molcel.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batut PJ, Bing XY, Sisco Z, Raimundo J, Levo M, Levine MS. Genome organization controls transcriptional dynamics during development. Science. 2022;375:566–570. doi: 10.1126/science.abi7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DE, Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, et al. Crispr-Cas9 saturating mutagenesis reveals an achilles heel in the BCL11A erythroid enhancer for fetal hemoglobin induction (by Genome Editing) Blood. 2015;126:638. doi: 10.1182/blood.V126.23.638.638. [DOI] [Google Scholar]
- Beagan JA, Phillips-Cremins JE. On the existence and functionality of topologically associating domains. Nat Genet. 2022;52:8–16. doi: 10.1038/s41588-019-0561-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagan JA, Duong MT, Titus KR, Zhou L, Cao Z, Ma J, Lachanski CV, Gillis DR, Phillips-Cremins JE. YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res. 2017;27:1139–1152. doi: 10.1101/gr.215160.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Benko S, Fantes JA, Amiel J, Kleinjan D-J, Thomas S, Ramsay J, Jamshidi N, Essafi A, Heaney S, Gordon CT, et al. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat Genet. 2009;41:359–364. doi: 10.1038/ng.329. [DOI] [PubMed] [Google Scholar]
- Bergman DT, Jones TR, Liu V, Ray J, Jagoda E, Siraj L, Kang HY, Nasser J, Kane M, Rios A, et al. Compatibility rules of human enhancer and promoter sequences. Nature. 2022 doi: 10.1038/s41586-022-04877-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Klose R. CpG island chromatin. Epigenetics. 2011;6:147–152. doi: 10.4161/epi.6.2.13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Klose RJ. The molecular principles of gene regulation by Polycomb repressive complexes. Nat Rev Mol Cell Biol. 2021;22:815–833. doi: 10.1038/s41580-021-00398-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Cohen NM, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot J-P, Tanay A, et al. Multiscale 3D genome rewiring during mouse neural development. Cell. 2017;171:557–572.:e24. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman BA, De Laat W. Getting the genome in shape: the formation of loops, domains and compartments. Genome Biol. 2015;16:154. doi: 10.1186/s13059-015-0730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozhilov YK, Downes DJ, Telenius J, Oudelaar AM, Olivier EN, Mountford JC, Hughes JR, Gibbons RJ, Higgs DR. A gain-of-function single nucleotide variant creates a new promoter which acts as an orientation-dependent enhancer-blocker. Nat Commun. 2021;12:3806. doi: 10.1038/s41467-021-23980-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JEF, Kadonaga JT. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon L, Weiss FD, Beagan JA, Oliveira MS, Georgieva R, Wang Y-F, Carroll TS, Dharmalingam G, Gong W, Tossell K, et al. Cohesin-dependence of neuronal gene expression relates to chromatin loop length. eLife. 2022;11:e76539. doi: 10.7554/eLife.76539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VC, Stathopoulos A, Levine M. Promoter-proximal tethering elements regulate enhancer-promoter specificity in the Drosophila Antennapedia complex. Proc Natl Acad Sci USA. 2002;99:9243–9247. doi: 10.1073/pnas.142291299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CAM, Taylor MS, Engström PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Carullo NVN, Day JJ. Genomic enhancers in brain health and disease. Genes. 2019;10:43. doi: 10.3390/genes10010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Kopitchinski N, Eraso A, Awasthi P, Chari R, Rocha PP. High affinity enhancer-promoter interactions can bypass CTCF/cohesin-mediated insulation and contribute to phenotypic robustness. BioRxiv. 2022:2021.12.30.474562. doi: 10.1101/2021.12.30.474562. [DOI] [Google Scholar]
- Chang TH-T, Primig M, Hadchouel J, Tajbakhsh S, Rocancourt D, Fernandez A, Kappler R, Scherthan H, Buckingham M. An enhancer directs differential expression of the linked Mrf4 and Myf5 myogenic regulatory genes in the mouse. Dev Biol. 2004;269:595–608. doi: 10.1016/j.ydbio.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Chernukhin I, Shamsuddin S, Kang SY, Bergström R, Kwon Y-W, Yu W, Whitehead J, Mukhopadhyay R, Docquier F, Farrar D, et al. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol. 2007;27:1631–1648. doi: 10.1128/MCB.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin T, Cavalli G. Polycomb silencing: from linear chromatin domains to 3D chromosome folding. Curr Opin Genet Dev. 2014;25:30–37. doi: 10.1016/j.gde.2013.11.016. [DOI] [PubMed] [Google Scholar]
- Cho SW, Xu J, Sun R, Mumbach MR, Carter AC, Chen YG, Yost KE, Kim J, He J, Nevins SA, et al. Promoter of lncRNA gene PVT1 is a tumor-suppressor DNA boundary element. Cell. 2018;173:1398–1412.:e22. doi: 10.1016/j.cell.2018.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claringbould A, Zaugg JB. Enhancers in disease: molecular basis and emerging treatment strategies. Trends Mol Med. 2021;27:1060–1073. doi: 10.1016/j.molmed.2021.07.012. [DOI] [PubMed] [Google Scholar]
- Crispatzu G, Rehimi R, Pachano T, Bleckwehl T, Cruz-Molina S, Xiao C, Mahabir E, Bazzi H, Rada-Iglesias A. The chromatin, topological and regulatory properties of pluripotency-associated poised enhancers are conserved in vivo. Nat Commun. 2021;12:4344. doi: 10.1038/s41467-021-24641-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Molina S, Respuela P, Tebartz C, Kolovos P, Nikolic M, Fueyo R, Van Ijcken WFJ, Grosveld F, Frommolt P, Bazzi H, et al. PRC2 Facilitates the regulatory topology required for poised enhancer function during pluripotent stem cell differentiation. Cell Stem Cell. 2017;20:689–705.:e9. doi: 10.1016/j.stem.2017.02.004. [DOI] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Mirny L. The 3D genome as moderator of chromosomal communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Rupon JW, Krivega I, Breda L, Motta I, Jahn KS, Reik A, Gregory PD, Rivella S, Dean A, et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158:849–860. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denholtz M, Bonora G, Chronis C, Splinter E, De Laat W, Ernst J, Pellegrini M, Plath K. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell. 2013;13:602–616. doi: 10.1016/j.stem.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despang A, Schöpflin R, Franke M, Ali S, Jerkoví I, Paliou C, Chan W-L, Timmermann B, Wittler L, Vingron M, et al. Functional dissection of the Sox9–Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat Genet. 2019;51:1263–1271. doi: 10.1038/s41588-019-0466-z. [DOI] [PubMed] [Google Scholar]
- de Wit E. TADs as the caller calls them. J Mol Biol. 2020;432:638–642. doi: 10.1016/j.jmb.2019.09.026. [DOI] [PubMed] [Google Scholar]
- de Wit E, Vos ESM, Holwerda SJB, Valdes-Quezada C, Verstegen MJAM, Teunissen H, Splinter E, Wijchers PJ, Krijger PHL, de Laat W. CTCF binding polarity determines chromatin looping. Mol Cell. 2015;60:676–684. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- De Villiers EM, Gissmann L, Zur Hausen H. Molecular cloning of viral DNA from human genital warts. J Virol. 1981;40:932–935. doi: 10.1128/jvi.40.3.932-935.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickel DE, Zhu Y, Nord AS, Wylie JN, Akiyama JA, Afzal V, Plajzer-Frick I, Kirkpatrick A, Göttgens B, Bruneau BG, et al. Function-based identification of mammalian enhancers using site-specific integration. Nat Methods. 2014;11:566–571. doi: 10.1038/nmeth.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon N, Trimborn T, Strouboulis J, Fraser P, Grosveld F. The Effect of Distance on Long-Range Chromatin Interactions. Mol Cell. 1997;1:131–139. doi: 10.1016/S1097-2765(00)80014-3. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström PG, Sui SJH, Drivenes Ø, Becker TS, Lenhard B. Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome Res. 2007;17:1898–1908. doi: 10.1101/gr.6669607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entrevan M, Schuettengruber B, Cavalli G. Regulation of genome architecture and function by polycomb proteins. Trends Cell Biol. 2016;26:511–525. doi: 10.1016/j.tcb.2016.04.009. [DOI] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Johnstone SE, Hemming ML, Tarjan DR, Hegazi E, Shareef SJ, Javed NM, Raut CP, Eschle BK, et al. Altered chromosomal topology drives oncogenic programs in SDH-deficient GISTs. Nature. 2019;575:229–233. doi: 10.1038/s41586-019-1668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangoul H, Altshuler D, Cappellini MD, Chen Y-S, Domm J, Eustace BK, Foell J, de la Fuente J, Grupp S, Handgretinger R, et al. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N Engl J Med. 2020;384:252–260. doi: 10.1056/NEJMoa2031054. [DOI] [PubMed] [Google Scholar]
- Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schöpflin R, Kraft K, Kempfer R, Jerković I, Chan W-L, et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature. 2016;538:265–269. doi: 10.1038/nature19800. [DOI] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T, Lim B, Levine M. Enhancer control of transcriptional bursting. Cell. 2016;166:358–368. doi: 10.1016/j.cell.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriele M, Brandão HB, Grosse-Holz S, Jha A, Dailey GM, Cattoglio C, Hsieh T-HS, Mirny L, Zechner C, Hansen AS. Dynamics of CTCF and cohesin mediated chromatin looping revealed by live-cell imaging. Science. 2022;376:496–501. doi: 10.1126/science.abn6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher MD, Posavi M, Huang P, Unger TL, Berlyand Y, Gruenewald L, Chesi A, Manduchi E, Wells AD, Grant SFA, et al. A dementia-associated risk variant near TMEM106B alters chromatin architecture and gene expression. Am J Hum Genet. 2017;101:643–663. doi: 10.1016/j.ajhg.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Jankowski A, Meiers S, Viales RR, Korbel JO, Furlong EEM. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat Genet. 2019;51:1272–1282. doi: 10.1038/s41588-019-0462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giammartino DCD, Kloetgen A, Polyzos A, Liu Y, Kim D, Murphy D, Abuhashem A, Cavaliere P, Aronson B, Shah V, et al. KLF4 is involved in the organization and regulation of pluripotency-associated three-dimensional enhancer networks. Nat Cell Biol. 2019;21:1179–1190. doi: 10.1038/s41556-019-0390-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierman HJ, Indemans MHG, Koster J, Goetze S, Seppen J, Geerts D, Van Driel R, Versteeg R. Domain-wide regulation of gene expression in the human genome. Genome Res. 2007;17:1286–1295. doi: 10.1101/gr.6276007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CT, Tan TY, Benko S, FitzPatrick D, Lyonnet S, Farlie PG. Long-range regulation at the SOX9 locus in development and disease. J Med Genet. 2009;46:649. doi: 10.1136/jmg.2009.068361. [DOI] [PubMed] [Google Scholar]
- Gröschel S, Sanders MA, Hoogenboezem R, De Wit E, Bouwman AM, Erpelinck C, Van Der Velden VHJ, Havermans M, Avellino R, van Lom K, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–381. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- Haarhuis JHI, Van Der Weide RH, Blomen VA, Yáñez-Cuna JO, Amendola M, Van Ruiten MS, Krijger PHL, Teunissen H, Medema RH, Van Steensel B, et al. The cohesin release factor WAPL restricts chromatin loop extension. Cell. 2017;169:693–707.:e14. doi: 10.1016/j.cell.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V, Lenhard B. Promoter architectures and developmental gene regulation. Semin Cell Dev Biol. 2016;5711 doi: 10.1016/j.semcdb.2016.01.014. [DOI] [PubMed] [Google Scholar]
- Haberle V, Arnold CD, Pagani M, Rath M, Schernhuber K, Stark A. Transcriptional cofactors display specificity for distinct types of core promoters. Nature. 2019;570:122–126. doi: 10.1038/s41586-019-1210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanscombe O, Whyatt D, Fraser P, Yannoutsos N, Greaves D, Dillon N, Grosveld F. Importance of globin gene order for correct developmental expression. Genes Dev. 1991;5:1387–1394. doi: 10.1101/gad.5.8.1387. [DOI] [PubMed] [Google Scholar]
- Hanssen LLP, Kassouf MT, Oudelaar AM, Biggs D, Preece C, Downes DJ, Gosden M, Sharpe JA, Sloane-Stanley JA, Hughes JR, et al. Tissue-specific CTCF/Cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat Cell Biol. 2017;19:952–961. doi: 10.1038/ncb3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison R, Slightom JL, Gumucio DL, Goodman M, Stojanovic N, Miller W. Locus control regions of mammalian β-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene. 1997;205:73–94. doi: 10.1016/S0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Haro E, Petit F, Pira CU, Spady CD, Lucas-Toca S, Yorozuya LI, Gray AL, Escande F, Jourdain A-S, Nguyen A, et al. Identification of limb-specific Lmx1b auto-regulatory modules with Nail-patella syndrome pathogenicity. Nat Commun. 2021;12:5533. doi: 10.1038/s41467-021-25844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RE, Lettice LA. Alterations to the remote control of Shh gene expression cause congenital abnormalities. Philos Trans R Soc B Biol Sci. 2013;368:20120357. doi: 10.1098/rstb.2012.0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CKY, Cohen BA. Genomic environments scale the activities of diverse core promoters. Genome Res. 2022;32:85–96. doi: 10.1101/gr.276025.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim DM, Mundlos S. The role of 3D chromatin domains in gene regulation: a multi-facetted view on genome organization. Curr Opin Genet Dev. 2020;61:1–8. doi: 10.1016/j.gde.2020.02.015. [DOI] [PubMed] [Google Scholar]
- Kane L, Williamson I, Flyamer IM, Kumar Y, Hill RE, Lettice LA, Bickmore WA. Cohesin is required for long-range enhancer action. BioRxiv. 2021:2021.06.24.449812. doi: 10.1101/2021.06.24.449812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradpour P, Ernst J, Melnikov A, Rogov P, Wang L, Zhang X, Alston J, Mikkelsen TS, Kellis M. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 2013;23:800–811. doi: 10.1101/gr.144899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi M, Hara N, Hasegawa M, Miyashita A, Kuwano R, Ikeuchi T, Nakaya A. Enhancer variants associated with Alzheimer's disease affect gene expression via chromatin looping. BMC Medical Genomics. 2019;12:128. doi: 10.1186/s12920-019-0574-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuta H, Laplante M, Navratilova P, Komisarczuk AZ, Engström PG, Fredman D, Akalin A, Caccamo M, Sealy I, Howe K, et al. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 2007;17:545–555. doi: 10.1101/gr.6086307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft K, Magg A, Heinrich V, Riemenschneider C, Schöpflin R, Markowski J, Ibrahim DM, Acuna-Hidalgo R, Despang A, Andrey G, et al. Serial genomic inversions induce tissue-specific architectural stripes gene misexpression and congenital malformations. Nat Cell Biol. 2019;21:305–310. doi: 10.1038/s41556-019-0273-x. [DOI] [PubMed] [Google Scholar]
- Kragesteen BK, Spielmann M, Paliou C, Heinrich V, Schoöpflin R, Esposito A, Annunziatella C, Bianco S, Chiariello AM, Jerkovicć I, et al. Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis. Nat Genet. 2018;50:1463–1473. doi: 10.1038/s41588-018-0221-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo N, Ishii H, Xiong X, Bianco S, Meitinger F, Hu R, Hocker JD, Conte M, Gorkin D, Yu M, et al. Promoter-proximal CTCF-binding promotes long-range-enhancer dependent gene activation. Nat Struct Mol Biol. 2021;28:152–161. doi: 10.1038/s41594-020-00539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Zhu Y, Kelman G, Novak CS, Plajzer-Frick I, Kato M, Garvin TH, Pham Q, Harrington AN, Hunter RD, et al. Comprehensive In Vivo interrogation reveals phenotypic impact of human enhancer variants. Cell. 2020;180:1262–1271.:e15. doi: 10.1016/j.cell.2020.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugsch M, Bartusel M, Rehimi R, Alirzayeva H, Karaolidou A, Crispatzu G, Zentis P, Nikolic M, Bleckwehl T, Kolovos P, et al. Modeling the pathological long-range regulatory effects of human structural variation with patient-specific hiPSCs. Cell Stem Cell. 2019;24:736–752.:e12. doi: 10.1016/j.stem.2019.03.004. [DOI] [PubMed] [Google Scholar]
- Lenhard B, Sandelin A, Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat Rev Genet. 2012;13:233–245. doi: 10.1038/nrg3163. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Horikoshi T, Heaney SJH, Van Baren MJ, Van Der Linde HC, Breedveld GJ, Joosse M, Akarsu N, Oostra BA, Endo N, et al. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc Natl Acad Sci USA. 2002;99:7548–7553. doi: 10.1073/pnas.112212199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJH, Purdie LA, Li L, De Beer P, Oostra BA, Goode D. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- Lim B, Heist T, Levine M, Fukaya T. Visualization of transvection in living Drosophila embryos. Mol Cell. 2018;70:287–296.:e6. doi: 10.1016/j.molcel.2018.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. Editing DNA methylation in the mammalian genome. Cell. 2016;167:233–247.:e1710. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, King HW, Patient RK, Odom DT, Klose RJ. Protection of CpG islands from DNA methylation is DNA-encoded and evolutionarily conserved. Nucleic Acids Res. 2016;44:6693–6706. doi: 10.1093/nar/gkw258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Osterwalder M, Welsh IC, Hansen K, Davies JOJ, Liu YE, Koska M, Adams AT, Aho R, Arora N, et al. Loss of extreme long-range enhancers in human neural crest drives a craniofacial disorder. Cell Stem Cell. 2020;27:765–783.:e1410. doi: 10.1016/j.stem.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loubiere V, Papadopoulos GL, Szabo Q, Martinez A-M, Cavalli G. Widespread activation of developmental gene expression characterized by PRC1-dependent chromatin looping. Sci Adv. 2020;6:eaax4001. doi: 10.1126/sciadv.aax4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower KM, Hughes JR, Gobbi MD, Henderson S, Viprakasit V, Fisher C, Goriely A, Ayyub H, Sloane-Stanley J, Vernimmen D, et al. Adventitious changes in long-range gene expression caused by polymorphic structural variation and promoter competition. Proc Natl Acad Sci USA. 2009;106:21771–21776. doi: 10.1073/pnas.0909331106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Spielmann M, Mundlos S. Breaking TADs: how alterations of chromatin domains result in disease. Trends Genet. 2016;32:225–237. doi: 10.1016/j.tig.2016.01.003. [DOI] [PubMed] [Google Scholar]
- Mach P, Kos PI, Zhan Y, Cramard J, Gaudin S, Tünnermann J, Marchi E, Eglinger J, Zuin J, Kryzhanovska M, et al. Live-cell imaging and physical modeling reveal control of chromosome folding dynamics by cohesin and CTCF. BioRxiv. 2022:2022.03.03.482826. doi: 10.1101/2022.03.03.482826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi T, Katsani KR, Verrijzer CP. GAGA can mediate enhancer function in trans by linking two separate DNA molecules. EMBO J. 2022;21:1775–1781. doi: 10.1093/emboj/21.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Ara M, Comoglio F, van Arensbergen J, van Steensel B. Systematic analysis of intrinsic enhancer-promoter compatibility in the mouse genome. Mol Cell. 2022 doi: 10.1016/j.molcel.2022.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyn GE, Wienert B, Kurita R, Nakamura Y, Quinlan KGR, Crossley M. A natural regulatory mutation in the proximal promoter elevates fetal globin expression by creating a de novo GATA1 site. Blood. 2019;133:852–856. doi: 10.1182/blood-2018-07-863951. [DOI] [PubMed] [Google Scholar]
- Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Genomics Hum Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- Mastrangelo IA, Courey AJ, Wall JS, Jackson SP, Hough PV. DNA looping and Sp1 multimer links: a mechanism for transcriptional synergism and enhancement. Proc Natl Acad Sci USA. 1991;88:5670–5674. doi: 10.1073/pnas.88.13.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcarthur E, Capra JA. Topologically associating domain boundaries that are stable across diverse cell types are evolutionarily constrained and enriched for heritability. The American Journal of Human Genetics. 2021;108:269–283. doi: 10.1016/j.ajhg.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merli C, Bergstrom DE, Cygan JA, Blackman RK. Promoter specificity mediates the independent regulation of neighboring genes. Genes Dev. 1996;11:1260–1270. doi: 10.1101/gad.10.10.1260. [DOI] [PubMed] [Google Scholar]
- Mirny LA, Solovei I. Keeping chromatin in the loop(s) Nat Rev Mol Cell Biol. 2021;22:439–440. doi: 10.1038/s41580-021-00337-x. [DOI] [PubMed] [Google Scholar]
- Morcillo P, Rosen C, Baylies MK, Dorsett D. Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer inDrosophila. Genes Dev. 1997;11:2729–2740. doi: 10.1101/gad.11.20.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau P, Hen R, Wasylyk B, Everett R, Gaub MP, Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981;9:6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtha M, Tokcaer-Keskin Z, Tang Z, Strino F, Chen X, Wang Y, Xi X, Basilico C, Brown S, Bonneau R, et al. FIREWACh: high-throughput functional detection of transcriptional regulatory modules in mammalian cells. Nat Methods. 2014;11:559–565. doi: 10.1038/nmeth.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumayr C, Pagani M, Stark A, Arnold CD. STARR–seq and UMI–STARR–seq: assessing enhancer activities for genome-wide–, high–, and low–complexity candidate libraries. Curr Protoc Mol Biol. 2019;128:e105. doi: 10.1002/cpmb.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, Van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton A-L, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell. 2017;169:930–944.:e22. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Shao J, Mitra J, Xiong F, D’antonio M, Wang R, Garcia-Bassets I, Ma Q, Zhu X, Lee J-H, et al. Enhancer release and retargeting activates disease-susceptibility genes. Nature. 2021;595:735–740. doi: 10.1038/s41586-021-03577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S, Levine M, Cai HN. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 1998;12:547–556. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachano T, Crispatzu G, Rada-Iglesias A. Polycomb proteins as organizers of 3D genome architecture in embryonic stem cells. Briefings in Functional Genomics. 2019;18:358–366. doi: 10.1093/bfgp/elz022. [DOI] [PubMed] [Google Scholar]
- Pachano T, Sánchez-Gaya V, Ealo T, Mariner-Faulí M, Bleckwehl T, Asenjo HG, Respuela P, Cruz-Molina S, Muñoz-San Martín M, Haro E, et al. Orphan CpG islands amplify poised enhancer regulatory activity and determine target gene responsiveness. Nat Genet. 2021;53:1036–1049. doi: 10.1038/s41588-021-00888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes SH, Melgar MF, Sethupathy P. Promoter-proximal CCCTC-factor binding is associated with an increase in the transcriptional pausing index. Bioinformatics. 2013;29:1485–1487. doi: 10.1093/bioinformatics/bts596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen J, Ali TML, Nekrasov M, Delbarre E, Baudement M-O, Kurscheid S, Tremethick D, Collas P. Long-range interactions between topologically associating domains shape the four-dimensional genome during differentiation. Nat Genet. 2019;51:835–843. doi: 10.1038/s41588-019-0392-0. [DOI] [PubMed] [Google Scholar]
- Peña-Hernández R, Marques M, Hilmi K, Zhao T, Saad A, Alaoui-Jamali MA, Del Rincon SV, Ashworth T, Roy AL, Emerson BM, et al. Genome-wide targeting of the epigenetic regulatory protein CTCF to gene promoters by the transcription factor TFII-I. Proc Natl Acad Sci U S A. 2015;112:E677–E686. doi: 10.1073/pnas.1416674112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KR, Stamatoyannopoulos G. Role of gene order in developmental control of human gamma- and beta-globin gene expression. Mol Cell Biol. 1993;13:4836–4843. doi: 10.1128/mcb.13.8.4836-4843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang S-C, Hilaire BGS, Engreitz JM, Perez EM, Kieffer-Kwon K-R, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. Cohesin loss eliminates all loop domains. Cell. 2017;171:305–320.:e24. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickels R, Shilatifard A. Enhancer logic and mechanics in development and disease. Trends Cell Biol. 2018;28:608–630. doi: 10.1016/j.tcb.2018.04.003. [DOI] [PubMed] [Google Scholar]
- Ringel AR, Szabo Q, Chiariello AM, Chudzik K, Schöpflin R, Rothe P, Mattei AL, Zehnder T, Harnett D, Laupert V, et al. Promoter repression and 3D-restructuring resolves divergent developmental gene expression in TADs. BioRxiv. 2021:2021.10.08.463672. doi: 10.2139/ssrn.3947354. [DOI] [Google Scholar]
- Rinzema NJ, Sofiadis K, Tjalsma SJD, Verstegen MJAM, Oz Y, Valdes-Quezada C, Felder A-K, Filipovska T, Van Der Elst S, de Andrade dos Ramos Z, et al. Building regulatory landscapes: enhancer recruits cohesin to create contact domains, engage CTCF sites and activate distant genes. BioRxiv. 2021:2021.10.05.463209. doi: 10.1101/2021.10.05.463209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L. Genetics of global gene expression. Nat Rev Genet. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- Roider HG, Lenhard B, Kanhere A, Haas SA, Vingron M. CpG-depleted promoters harbor tissue-specific transcription factor binding signals—implications for motif overrepresentation analyses. Nucleic Acids Res. 2009;37:6305–6315. doi: 10.1093/nar/gkp682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S, Symmons O, Uslu VV, Dolle D, Hot C, Ettwiller L, Spitz F. Large-scale analysis of the regulatory architecture of the mouse genome with a transposon-associated sensor. Nat Genet. 2011;43:379–386. doi: 10.1038/ng.790. [DOI] [PubMed] [Google Scholar]
- Sanborn AL, Rao SSP, Huang S-C, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci USA. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MC, Fisch TM, Benecke BJ, Nevins JR, Heintz N. Definition of multiple, functionally distinct TATA elements, one of which is a target in the hsp70 promoter for E1A regulation. Cell. 1988;52:723–729. doi: 10.1016/0092-8674(88)90410-2. [DOI] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. Enhancer biology and enhanceropathies. Nat Struct Mol Biol. 2014;21:210–219. doi: 10.1038/nsmb.2784. [DOI] [PubMed] [Google Scholar]
- Song S-H, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range β-globin locus control region function. Mol Cell. 2007;28:810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmann M, Lupiáñez DG, Mundlos S. Structural variation in the 3D genome. Nat Rev Genet. 2018;19:453–467. doi: 10.1038/s41576-018-0007-0. [DOI] [PubMed] [Google Scholar]
- Symmons O, Uslu VV, Tsujimura T, Ruf S, Nassari S, Schwarzer W, Ettwiller L, Spitz F. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014;24:390–400. doi: 10.1101/gr.163519.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Q, Jost D, Chang J-M, Cattoni DI, Papadopoulos GL, Bonev B, Sexton T, Gurgo J, Jacquier C, Nollmann M, et al. TADs are 3D structural units of higher-order chromosome organization in Drosophila. Sci Adv. 2018;4:eaar8082. doi: 10.1126/sciadv.aar8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Q, Bantignies F, Cavalli G. Principles of genome folding into topologically associating domains. Sci Adv. 2019;5:eaaw1668. doi: 10.1126/sciadv.aaw1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto K, Liu Q, Bungert J, Engel JD. Effects of altered gene order or orientation of the locus control region on human β-globin gene expression in mice. Nature. 1999;398:344–348. doi: 10.1038/18698. [DOI] [PubMed] [Google Scholar]
- Taylor T, Sikorska N, Shchuka VM, Chahar S, Ji C, Macpherson NN, Moorthy SD, Mitchell JA, Sexton T. Transcriptional regulation and chromatin architecture maintenance are decoupled modular functions at the Sox2 locus. BioRxiv. 2022:2022.02.09.479674. doi: 10.1101/2022.02.09.479674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripic T, Deng W, Cheng Y, Zhang Y, Vakoc CR, Gregory GD, Hardison RC, Blobel GA. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood. 2009;113:2191–2201. doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushiki A, Zhang Y, Xiong C, Zhao J, Georgakopoulos-Soares I, Kane L, Jamieson K, Bamshad MJ, Nickerson DA, Shen Y, et al. Deletion of CTCF sites in the SHH locus alters enhancer–promoter interactions and leads to acheiropodia. Nat Commun. 2021;12:2282. doi: 10.1038/s41467-021-22470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valton A-L, Valton SV, Mair B, Khokhar E, Tong AHY, Usaj M, Chan KSK, Pai AA, Moffat J, Dekker J. A cohesin traffic pattern genetically linked to gene regulation. BioRxiv. 2021:2021.07.29.454218. doi: 10.1101/2021.07.29.454218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandermeer JE, Ahituv N. cis–regulatory mutations are a genetic cause of human limb malformations. Dev Dyn. 2011;240:920–930. doi: 10.1002/dvdy.22535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos ESM, Valdes-Quezada C, Huang Y, Allahyar A, Verstegen MJAM, Felder A-K, Van Der Vegt F, Uijttewaal ECH, Krijger PHL, De Laat W. Interplay between CTCF boundaries and a super enhancer controls cohesin extrusion trajectories and gene expression. Mol Cell. 2021;81:3082–3095.:e6. doi: 10.1016/j.molcel.2021.06.008. [DOI] [PubMed] [Google Scholar]
- Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wefald FC, Devlin BH, Williams RS. Functional heterogeneity of mammalian TATA-box sequences revealed by interaction with a cell-specific enhancer. Nature. 1990;344:260–262. doi: 10.1038/344260a0. [DOI] [PubMed] [Google Scholar]
- Wei Z, Gao F, Kim S, Yang H, Lyu J, An W, Wang K, Lu W. Klf4 Organizes Long-Range Chromosomal Interactions with the Oct4 Locus in Reprogramming and Pluripotency. Cell Stem Cell. 2013;13:36–47. doi: 10.1016/j.stem.2013.05.010. [DOI] [PubMed] [Google Scholar]
- Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, Abraham BJ, Cohen MA, Nabet B, Buckley DL, et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell. 2017;171:1573–1588.:e28. doi: 10.1016/j.cell.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienert B, Funnell APW, Norton LJ, Pearson RCM, Wilkinson-White LE, Lester K, Vadolas J, Porteus MH, Matthews JM, Quinlan KGR, et al. Editing the genome to introduce a beneficial naturally occurring mutation associated with increased fetal globin. Nat Commun. 2015;6:7085. doi: 10.1038/ncomms8085. [DOI] [PubMed] [Google Scholar]
- Wienert B, Martyn GE, Kurita R, Nakamura Y, Quinlan KGR, Crossley M. KLF1 drives the expression of fetal hemoglobin in British HPFH. Blood. 2017;130:803–807. doi: 10.1182/blood-2017-02-767400. [DOI] [PubMed] [Google Scholar]
- Wilber A, Nienhuis AW, Persons DA. Transcriptional regulation of fetal to adult hemoglobin switching: new therapeutic opportunities. Blood. 2011;117:3945–3953. doi: 10.1182/blood-2010-11-316893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H-J, Landshammer A, Stamenova EK, Bolondi A, Kretzmer H, Meissner A, Michor F. Topological isolation of developmental regulators in mammalian genomes. Nat Commun. 2021;12:4897. doi: 10.1038/s41467-021-24951-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhang Y, Xu Q, Zhou C, Liu B, Du Z, Zhang K, Zhang B, Wang X, Gayen S, et al. Epigenomic analysis of gastrulation identifies a unique chromatin state for primed pluripotency. Nat Genet. 2020;52:95–105. doi: 10.1038/s41588-019-0545-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao JY, Hafner A, Boettiger AN. How subtle changes in 3D structure can create large changes in transcription. ELife. 2021;10:e64320. doi: 10.7554/eLife.64320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoshi M, Segawa K, Fukaya T. Visualizing the role of boundary elements in enhancer-promoter communication. Mol Cell. 2020;78:224–235.:e5. doi: 10.1016/j.molcel.2020.02.007. [DOI] [PubMed] [Google Scholar]
- Zabidi MA, Stark A. Regulatory enhancer–core-promoter communication via transcription factors and cofactors. Trends Genet. 2016;32:801–814. doi: 10.1016/j.tig.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, Stark A. Enhancer–core-promoter specificity separates developmental and housekeeping gene regulation. Nature. 2015;518:556–559. doi: 10.1038/nature13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Yuan F, Garcia-Martinez L, Lee KD, Stransky S, Sidoli S, Verdun RE, Zhang Y, Wang Z, et al. The Polycomb protein RING1B enables estrogen-mediated gene expression by promoting enhancer–promoter interaction and R-loop formation. Nucleic Acids Res. 2021;49:9768–9782. doi: 10.1093/nar/gkab723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J, Roth G, Zhan Y, Cramard J, Redolfi J, Piskadlo E, Mach P, Kryzhanovska M, Tihanyi G, Kohler H, et al. Nonlinear control of transcription through enhancer–promoter interactions. Nature. 2022;604 doi: 10.1038/s41586-022-04570-y. [DOI] [PMC free article] [PubMed] [Google Scholar]