Abstract
Radiotherapy is one of the most effective approaches to achieve tumour control in cancer patients, although healthy tissue injury due to off-target radiation exposure can occur. In this study, we used a model of acute radiation injury to the lung in the context of cancer metastasis, to understand the biological link between tissue damage and cancer progression. We exposed healthy mouse lung tissue to radiation prior to the induction of metastasis and observed a strong enhancement of cancer cell growth. We found that locally activated neutrophils were key drivers of the tumour-supportive preconditioning of the lung microenvironment, governed by enhanced regenerative Notch signalling. Importantly, these tissue perturbations endowed arriving cancer cells with an augmented stemness phenotype. By preventing neutrophil-dependent Notch activation, via blocking degranulation, we were able to significantly offset the radiation-enhanced metastases. This work highlights a pro-tumorigenic activity of neutrophils, which is likely linked to their tissue regenerative functions.
Introduction
Radiotherapy (RT) is received by approximately 60% of all cancer patients, and remains one of the most successful non-surgical techniques to achieve local tumour control1. In recent decades, technological advances and increased sophistication in imaging have significantly improved the accuracy, efficacy and tolerability of RT. However, radiation-induced damage in non-target tissues can still occur, inducing a local injury1. A previous study from our laboratory identified the presence of a tissue regeneration program within the metastatic environment of pulmonary metastases from breast cancer2, generating the intriguing hypothesis that an injury could predispose the tissue for metastatic growth. In this study, we aimed to test whether acute lung injury triggered by radiation would set the stage for a perturbed tissue interaction with arriving cancer cells and foster metastasis. Both clinical and experimental studies have reported poor prognosis and increased metastasis associated with tumours occurring within a pre-irradiated site3,4. This has been attributed to both the release of pro-migratory factors from the irradiated local tissue5, and increased myeloid cell mobilisation induced by the primary tumour4. These studies, however, have not addressed whether an injury to healthy tissues induced by off-target radiation impacts subsequent metastatic growth within that organ. A 1973 study reported that radiation exposure influenced lung integrity and the seeding of metastatic cells6, while an earlier clinical study indeed suggested that lung metastases are increased after post-operative RT for breast cancer7. Here, analysis of women with clinically and pathologically comparable breast tumours revealed a significantly higher incidence of ipsilateral pulmonary metastasis (occurring on the same side as the breast cancer) in women who received post-operative RT compared to surgery alone. Of course the outdated RT technologies used in this study would have generated a large off-target radiation volume compared to the much more precise modern image-guided platforms7. Nonetheless, the biology behind the metastatic risk increase is not fully understood, and its uncovering may reveal important mechanistic connections between the processes of tissue injury and cancer.
In the present study, we set out to elucidate the injury response of healthy lung tissue to radiation exposure, and we report that radiation can generate a profoundly pro-metastatic microenvironment. Importantly, we found that lung neutrophils are key for this tumour promotion effect. Although neutrophils are vital for the host defence against pathogens, their infiltration and overzealous activation during acute inflammatory responses can exacerbate tissue damage8. However, an important role in the repair and regeneration of damaged epithelium in certain contexts is also emerging9-11. In cancer, neutrophils are well known to contribute to many aspects of tumour progression and metastasis and a plethora of pro-tumorigenic activities have been defined12. Here we describe an unexpected activity of neutrophils in the irradiated tissue, which, by influencing the responses of lung resident cells, bridges their tissue-repair functions with their tumour-supportive activity. Mechanistically, we identified Notch as the mediator of the neutrophil-dependent lung epithelial response to radiation injury, which is central to this tumour-supportive function. This surprising phenomenon of neutrophil tissue-priming following radiation exposure may have implications for the tissue response to radiation in patients, and warrants clinical studies to investigate neutrophil-driven perturbations in cancer patients undergoing RT.
Results
Radiation exposure in healthy lung tissue boosts metastasis
To examine the effects of off-target radiation exposure on healthy lung tissue, we delivered a single 13 Gy dose of focussed radiation specifically to the thoracic cavity of anaesthetised female BALB/c mice. We focused on the short-term tissue response to this acute lung injury, which in the long-term typically results in pneumonitis or fibrosis1. Mice were allowed to recover for four days, before being orthotopically injected with cancer cells into the mammary fat pad to induce primary tumours and test the onset of spontaneous metastasis (Fig. 1a). We first induced non-metastatic 4T07 primary tumours, where cancer cells can disseminate but are unable to initiate metastatic growth. Strikingly, we found an abundance of metastases within the lungs of mice previously irradiated, while only rare metastatic foci were detected in control lungs (sham-irradiated) (Fig. 1b,c). Similarly, when lungs were harvested from irradiated mice harbouring metastatic 4T1 tumours, we observed, at an early stage of tumour progression, a significant increase in metastatic burden compared to control mice while the primary tumour size was not affected (Extended Data Fig. 1a-c, Extended Data Fig. 2). We next confirmed our findings in an experimental metastasis model, in which metastases are induced via a single tail intravenous (IV) injection to drive cancer cell seeding in the lungs of mice which have received targeted lung irradiation 7 days earlier (Fig. 1d). Consistently, a profound increase in metastasis was observed following irradiation in FVB mice who received an IV injection of primary cancer cells isolated from tumours developed in mammary tumour virus-promoter middle tumour-antigen (MMTV-PyMT) mice crossed with actin-GFP transgenic mice (Extended Data Fig. 1d,e). This pro-metastatic effect of radiation was not unique for breast cancer cells, as a strong induction in metastasis was also observed in immune-compromised mice that were inoculated with the human oesophageal adenocarcinoma cell line Flo-1 following lung irradiation (Fig. 1e,f). Moreover, when injecting human non-small-cell lung cancer cell lines A549 and H460, we could only detect metastatic foci in pre-irradiated mice and not control animals (Extended Data Fig. 1f). Thus, radiation pre-exposure of healthy lung tissue strongly supports the growth of cancer cells across multiple mouse genetic backgrounds and induces an aggressive metastatic phenotype in poorly metastatic cells. Since cancer patients are typically treated with fractionated RT, whereby low doses of radiation are delivered over consecutive days, we employed fractionated dosing in our experimental setting (Fig. 1g). As with single-dose RT, a profound enhancement of 4T1 cancer cell metastasis was observed in BALB/c mice that received doses of 3 x 4 Gy compared to control mice (Fig. 1h,i), with the effect more pronounced with an additional 4 Gy fraction. We calculated the Biological Effective Dose (BED) in our different experimental settings, which measures the true biological dose delivered to a given tissue by a particular combination of dose-per-fraction and total dose, characterised by a specific α/β ratio13. We used an α/β of 10 in BED calculations for this early radiation-induced reaction14. Notably, the lowest fractionated regime (3x 4 Gy) delivered almost half the BED of a single high-dose 13 Gy treatment (BED 16.8 versus 29.9 Gy), yet still led to a strong boost in metastatic proficiency of the lung. Next, we used image-guided, focussed radiation to specifically target the right lung lobe of BALB/c mice, whilst leaving the left lung, heart and trachea largely unexposed (Fig. 1j). Given that the right is the larger side of the murine lung, more metastases are found on this side in both control and irradiated animals. Therefore, we measured if irradiation of the right side further increased the right/left ratio of metastases (Fig. 1j). Importantly, we observed a significant enrichment of 4T1 metastatic breast tumours in the lobe pre-exposed to either a medium (8 Gy) or high dose of radiation (12 Gy), when compared to the non-irradiated control mice (Fig. 1k). Of note, no changes in metastasis were observed in the non-irradiated left side of the lung between the experimental groups (Extended Data Fig. 1g), excluding systemic priming from irradiated to non-irradiated tissue in this setting. Thus, smaller irradiation volumes exert a similar effect to whole thoracic radiation, and the influence of radiation-induced cardiac damage on metastatic growth in our system can be excluded.
Fig. 1. Radiation exposure in healthy lung tissue enhances metastasis.
a, Experimental setup. Mice received targeted 13 Gy lung irradiation prior to orthotopic breast cancer transplantation. Spontaneous lung metastatic burden was assessed at day 21. b,c, Representative H&E images (b) and FACS quantification of GFP+ tumour cells (c) from control (UT) and irradiated BALB/c mice inoculated with 4T07 low-metastatic breast cancer cells. Primary tumour volume was equivalent between groups (Extended Data Fig. 1c) (n=12 control mice, n=10 irradiated; 2 independent experiments). Metastatic area is depicted with a dashed line. Scale bar, 250 μm. d, Experimental setup for experimental metastasis. Mice received 13Gy targeted lung irradiation prior to intravenous injection of cancer cells. e,f, Representative H&E images (e) and histology quantification (f) of metastatic lungs from control (UT) and irradiated NSG mice injected with human oesophageal adenocarcinoma Flo-1 cells (n=6 mice per group, one experiment). Metastatic area is depicted with a dashed line. Scale bar, 250 μm. g, Experimental setup for fractionated irradiation. BALB/c mice received a targeted 4Gy dose of lung irradiation over three or four consecutive days. Mice were injected intravenously with 4T1 breast cancer cells 7 days after the final dose. h,i, Representative H&E images (h) and tumour area quantification (i) for control (UT) mice and irradiated mice that received 3x 4Gy or 4x 4Gy (n=5 mice per group, one experiment). Scale bar, 250 μm. j,k Experimental setup (j) and histology quantification (k) for partial lung irradiation. Image-guided radiation at 8Gy or 12Gy was used to specifically target the right lung of BALB/c mice, one week prior to an intravenous injection of 4T1 breast cancer cells. The number of metastatic lesions in the right (irradiated) versus left (non-irradiated) lung lobes was quantified by H&E (n=6 control mice; n=5 8Gy mice; n=7 12Gy mice, one experiment). All data represented as mean ± s.e.m. Statistical analysis by non-parametric two-tailed Mann-Whitney test for (c) and (f), and one-way ANOVA with multiple comparisons for (i) and (k). UT, untreated; IR, irradiated. FACS gating strategies provided in Extended Data Fig. 2. Histology quantification rationale and process is outlined in Supplementary File 1. Source data.
Radiation induces neutrophil infiltration and activation
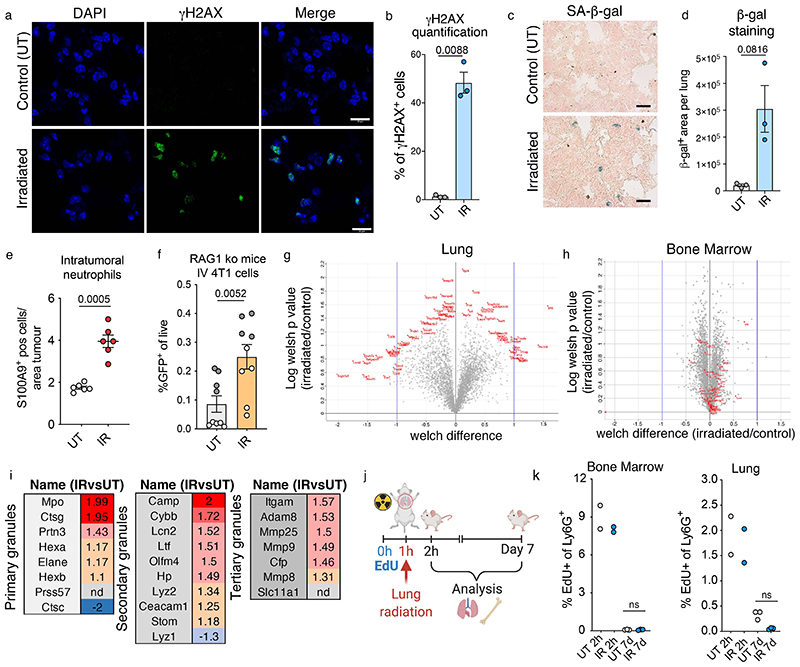
To probe how the tissue alterations induced by lung irradiation fuelled subsequent metastatic growth, we analysed lung tissue from BALB/c mice at day 7 following a single dose of 13 Gy irradiation, in the absence of cancer cell seeding. At this early time point, no overt alterations in the lungs were observed histologically (Fig. 2a), although, as expected, we detected radiation-induced DNA damage and senescence (Extended Data Fig 3a-d). When analysing immune infiltration in the lung at this time point, we observed a marked neutrophil infiltration compared to other immune subsets in the irradiated lungs (Fig. 2b,c). No significant alterations were detected within the adaptive immune compartment upon irradiation (Fig. 2c). This neutrophil increase was also reflected in the abundance of intra-tumoural neutrophil infiltration in the 4T1 metastatic lesions subsequently growing in pre-irradiated lungs compared to controls (Extended Data Fig. 3e). The pro-metastatic effect of radiation was maintained for murine cancer cells in immunodeficient mice, as we observed for human cell lines (Fig. 1e,f and Extended Data Fig. 1f and 3f).
Fig. 2. Radiation exposure induces the infiltration and local activation of lung neutrophils.
a,b, Representative H&E images of lungs (a) and S100A9 immunostaining for neutrophils (b) from control and irradiated mice at day 7 (n=6 mice per group, 2 independent experiments). Scale bars, 250 μm (a) and 100 μm (b). c, Immune cell frequencies in the lungs estimated by FACS, 7 days post-irradiation (n=7 mice per group, two independent experiments). Data is presented either as the frequency among live cells (for total CD45+ immune cells), or frequency among CD45+ cells. d,e, Representative immunofluorescent images (d) and quantification of nuclear segmentation (e) from sorted Ly6G+ lung neutrophils harvested from control and irradiated lungs at day 7 post-irradiation (n=7 mice per group, two independent experiments). DAPI staining was performed on fixed cells plated on poly-lysine coverslips (n=2 coverslips per mouse for each experiment). Each data point (n=30 UT, n=32 IR) represents the average segmentation across all cells within the field of view. Scale bars: main panel, 100 μm; enlarged insets, 10 μm. f, Experimental setup for quantitative mass-spectrometry based proteomic analysis of Ly6G+ positive cells. Cells were isolated from the lungs and from bone marrow extracted from the femur of control and irradiated BALB/c mice at day 7 by MACS sorting. g, Metacore pathway analysis of proteins upregulated by >1.2 fold in lung neutrophils from irradiated mice, compared to control mice (n=3 mice per group). h, Granule protein upregulation in the lungs of irradiated versus control mice (n=3 mice per group). Each dot represents an individual granule protein (n=24), red dots depict an enrichment in irradiated mice, blue dots represent downregulation. All data represented as mean ± s.e.m. Statistical analysis by a two-tailed t-test with Welch’s correction for (c) and unpaired two-tailed t-test for (e). Gating strategies for FACS analysis provided in Extended Data Fig. 2. Source data.
Interestingly, we found that neutrophils accumulating in irradiated lungs at this early time point homogenously acquired a distinctive hyper-segmented nuclear morphology, a hallmark feature of neutrophil activation (Fig. 2d,e). To further characterise this activated status, we performed mass-spectrometry based proteomic profiling of isolated lung and bone marrow neutrophils from irradiated and control mice (Fig. 2f). Striking differences were observed in the proteome of neutrophils harvested from irradiated lungs, while the proteome of bone marrow neutrophils isolated from distal femurs of the two groups were highly comparable (Extended Data Fig. 3g,h), although a level of radiation in the bone marrow of ribs cannot be excluded. Consistent with their hyper-segmented nuclear morphology (Fig. 2d,e), Metacore analysis of over-represented proteins revealed that neutrophils from irradiated lungs acquire a pro-inflammatory, activated phenotype with an increase in granule protein content (Fig. 2g,h and Extended Data Fig. 3i). Thus, radiation exposure in the lungs triggers a profound local activation of neutrophils. Recent evidence indicates that neutrophils adapt accordingly to tissue-derived signals and have a variable life time in mouse tissues, approximately 10.1h in the lung15. To test a direct effect of radiation exposure on neutrophil activation and their persistence in the tissue, we gave mice a pulse of EdU 1h prior to irradiation, and analysed neutrophils from lungs and bone marrow (Extended Data Fig. 3j). Flow cytometry analysis revealed negligible EdU positivity 7 days post-irradiation in neutrophils from both compartments, implying complete neutrophil turnover during this time (Extended Data Fig. 3k). We concluded that activated neutrophils observed 7 days post-irradiation were not directly exposed to radiation, and that neutrophil recruitment and activation is likely triggered by the local lung environment.
Radiation-primed neutrophils support metastatic colonization
Pro-inflammatory neutrophils with hyper-segmented nuclei have previously been linked with anti-cancer activity, including in the context of radiotherapy16,17. To test whether radiation-primed neutrophils alter the metastatic proficiency of irradiated lung tissue, we used daily injections of an anti-Ly6G antibody to deplete neutrophils from mice exposed to lung irradiation and 4T1-GFP+ cancer cells were intravenously inoculated 7 days post-irradiation (Fig. 3a). Strikingly, radiation-primed neutrophils strongly supported metastatic growth as their depletion dramatically reduced the tumour burden in the lung (Fig. 3b-d).
Fig. 3. Primed neutrophils support metastatic colonization in irradiated lungs.
a, Experimental setup. BALB/c mice were given daily injections of an anti-Ly6G neutrophil-depletion antibody or an IgG control antibody, beginning the day before lung radiation. One week later, mice received an intravenous injection of 4T1-GFP+ breast cancer cells. b, Representative H&E images of metastatic lungs from control (UT), irradiated and irradiated mice lacking neutrophils (n=7 mice per group, 2 independent experiments). The metastatic area is depicted with a dashed line. Scale bar, 250 μm. c, Ly6G+ neutrophil frequency among live cells by FACS (n=7 mice per group, 2 independent experiments). d, Metastatic burden as quantified by H&E staining (n=7 mice per group, 2 independent experiments). Mice from the two independent experiments are represented by different coloured dots. See Supplementary File 1 for quantification process. e, Schematic showing irradiations in neutropenic FVB G-CSF ko mice. Mice were treated with recombinant GCSF (rGCSF) or PBS every other day, beginning the day before irradiations, for a total of four injections. Mice were intravenously injected with MMTV-PyMT primary breast cancer cells two days after the final rGCSF injection. f,g, Representative immunofluorescent images (f) and quantification of nuclear segmentation (g) from sorted Ly6G+ lung neutrophils harvested from control (UT) and irradiated mice 24h after the final rGCSF treatment. DAPI staining was performed on Ly6G+ cells plated on poly-lysine coverslips (n=3 mice per group, 6 technical replicates quantified per mouse). Scale bars, 10 μm. h,i Quantification (h) and representative H&E images of metastatic lungs (i) from irradiated mice treated with either PBS or rGCSF (n=11 PBS-treated, n=9 rGCSF treated mice, 2 independent experiments). Scale bar, 500 μm. Data represented as mean ± s.e.m. Statistical analysis by One way ANOVA for (c), non-parametric Kruskal-Wallis test for (d), unpaired two-tailed t-test for (g) and (h). UT, untreated; IR, irradiated; px, pixals; Ko, knock-out; rGCSF, recombinant G-CSF. Gating strategies for FACS analysis provided in Extended Data Fig. 2. Histology quantification rationale and process is outlined in Supplementary File 1. Source data.
Many factors are at play when lungs are injured by radiation, for instance the integrity of the vasculature is compromised, which enhances extravasation. Indeed, cancer cells were increased 72h after seeding in previously irradiated lungs, a time point when extravasation is complete, however, this was unaffected by the absence of neutrophils (Extended Data Fig. 4a-c). NETosis has been increasingly reported as a mechanism mediating neutrophil pro-metastatic functions via inducing extracellular matrix remodelling and directly increasing cancer cell growth18-21. However, we did not detect NETosis in lung neutrophils at various timepoints following radiation exposure (Extended Data Fig. 4d).
We reasoned there might be various non-mutually exclusive mechanisms by which neutrophil recruitment and activation could favour metastatic growth in irradiated lungs12. Radiation-primed neutrophils could boost the growth of cancer cells directly or by orchestrating a tumour-supportive tissue response. To test if neutrophil activation directly influences their interaction with cancer cells, we performed in vitro co-cultures using a porous three-dimensional (3D) Alvetex™ scaffold (Extended Data Fig. 4e). We reported previously that lung neutrophils support cancer cell growth in this co-culture system2, but no further boost was observed when neutrophils were harvested from irradiated lungs (Extended Data Fig. 4f). We also did not detect an increase in cancer cells EdU incorporation following 2D co-culture with radiation-primed lung neutrophils compared to control neutrophils (Extended Data Fig. 4g,h). These results suggest radiation-primed lung neutrophils do not offer an additional direct growth-promoting advantage to cancer cells compared to control neutrophils.
This prompted us to test the alternative hypothesis that radiation-primed neutrophils boosted metastasis by perturbing the lung microenvironment prior to the arrival of cancer cells. Therefore we performed targeted lung irradiations in neutropenic G-csf ko mice that are unable to mobilise neutrophils from the bone marrow22. Irradiated G-csf ko mice were treated with recombinant G-CSF (rG-CSF) to permit neutrophil recruitment to the lungs (Fig. 3e), after which infiltrating neutrophils displayed a marked increase in nuclear segmentation indicating an activated phenotype similar to wild-type mice (Fig. 3f,g). rG-CSF treatment was stopped 48h before IV injection with MMTV-PyMT primary mammary cancer cells, by which time mice had returned to their original neutropenic state (Extended Data Fig. 5a,b). Importantly, long-term analysis revealed a significant increase in metastatic incidence in irradiated mice pre-treated with rG-CSF (Fig. 3h,i). Since the number of neutrophils was only increased prior to cancer cell seeding, this suggests that neutrophils may indirectly influence cancer cell growth in the irradiated lung by perturbing the tissue microenvironment.
Neutrophils influence the epithelial response to radiation
We next tested whether radiation-primed neutrophils alone were sufficient to drive a change in the healthy lung tissue microenvironment and influence metastatic receptiveness. We performed adoptive transfer of control or radiation-primed lung neutrophils into lungs of naïve recipient mice via a single IV injection (Fig. 4a). After four days of lung ‘conditioning’, at which time turnover of transferred neutrophils had occurred15, 4T1-GFP+ cancer cells were seeded to induce metastasis (Fig. 4a). Strikingly, mice whose lungs were conditioned with radiation-primed neutrophils had a marked increase in cancer cell growth compared with mice conditioned with control lung neutrophils (Fig. 4b-d). Thus, radiation-primed neutrophils can incite a tissue change in naïve lung tissue.
Fig. 4. Radiation-primed neutrophils perturb the lung tissue environment.
a, Experimental setup for the neutrophil adoptive transfer. Control or radiation-primed Ly6G+ lung neutrophils were MACS-sorted 7 days following irradiation and intravenously injected into naïve recipient BALB/c mice. Mice were given an intravenous injection of 4T1-GFP+ cancer cells 4 days later, and metastatic burden assessed after one week. b-d, Quantification of the entire lung (serial sectioning) (b) and representative images of GFP (c) and H&E stained lungs (d) (n=6 control, n=7 irradiated mice, two independent experiments). Metastatic foci are outlined and indicated with arrows. See methods for quantification details. Scale bar, 100 μm. e, Schematic depicting the experimental setup for bulk RNA-sequencing. BALB/c mice were given daily injections of anti-Ly6G to deplete neutrophils or a control IgG antibody, beginning the day before targeted lung irradiation. Flow cytometry was used to isolate CD45-CD31-Ter119-(Lin-)EpCAM+ lung epithelial and Lin-EpCAM- mesenchymal cells from control, irradiated and neutrophil-depleted irradiated mice 7 days post-irradiation (n=4 mice per group). f, Principle Component Analysis (PCA) of Lin-EpCAM+ epithelial cell signatures following RNA-seq analysis of control, irradiated and neutrophil-depleted irradiated lung samples. Each dot represents an individual mouse, with ovals enclosing the samples from each group to highlight their similarity in the PCA plot. g, Experimental setup for lung epithelial analysis. Lin-EpCAM+ lung epithelial cells harvested from control, irradiated and neutrophil-depleted irradiated BALB/c mice (7 days post-irradiation) were sorted by flow cytometry and co-cultured in Matrigel with MLg normal lung fibroblasts to generate lung organoids. h,i, Representative images (h) and quantification (i) of lung organoid co-cultures. Scale bar, 1000 μm. Quantification of organoid number is shown as the percentage reduction in organoids compared to the control group. Each dot represents an individual mouse, with the three independent experiments indicated by coloured dots (n=12 mice per group). Triplicate technical replicates were quantified for each mouse. Data represented as mean ± s.e.m. Statistical analysis by two-way ANOVA for (b) and non-parametric two-tailed Mann-Whitney test for (h). UT, untreated; IR, irradiation. Gating strategies for FACS sorting provided in Extended Data Fig. 2. Source data.
We then sought to uncover this neutrophil tissue-perturbation activity in the context of radiation exposure. We used flow cytometry to sort either epithelial (CD45-CD31-Ter119- EpCAM+) or mesenchymal (CD45-CD31-Ter119-EpCAM-) cells from control and irradiated lungs in the presence or absence of neutrophils (Fig. 4e). As expected, RNA-sequencing showed significant alterations in the transcriptome of both cell types 7 days after irradiation, while a profound influence of neutrophils also emerged (Fig. 4f and Extended Data Fig. 6a). Particularly in the lung epithelium, the absence of neutrophils resulted in a completely different irradiated cell cluster in the principle component analysis (PCA) (Fig. 4f). To test if the neutrophil-mediated transcriptional response was reflected in a functional difference in the epithelial compartment, we performed a lung organoid assay23. This is the gold-standard assay whereby lung progenitors are challenged to survive and grow organoid structures ex vivo. Lung epithelial cells were isolated from irradiated mice either in the presence or absence of neutrophils and co-cultured with lung fibroblasts in Matrigel (Fig. 4e,g). As expected, epithelial cells injured by radiation sharply reduced the overall number of organoids compared to non-injured cells24, yet strikingly epithelial cells irradiated in the absence of neutrophils were almost completely devoid of organoid forming ability (Fig. 4h,i). Thus, this data suggests that neutrophils are required to support lung epithelial fitness and progenitor function upon irradiation. To examine whether radiation-induced alterations in lung epithelial cells also functionally affects their interaction with cancer cells, we cultured them ex vivo on 3D Alvetex™ scaffolds with MMTV-PyMT-GFP+ cancer cells (as in Extended Data Fig. 4e). After confirming that the survival of the epithelial cells on the scaffolds was not affected by irradiation, we found that the growth advantage provided by epithelial cells was almost entirely abolished when the irradiation occurred in the absence of neutrophils (Extended Data Fig. 6b-e).
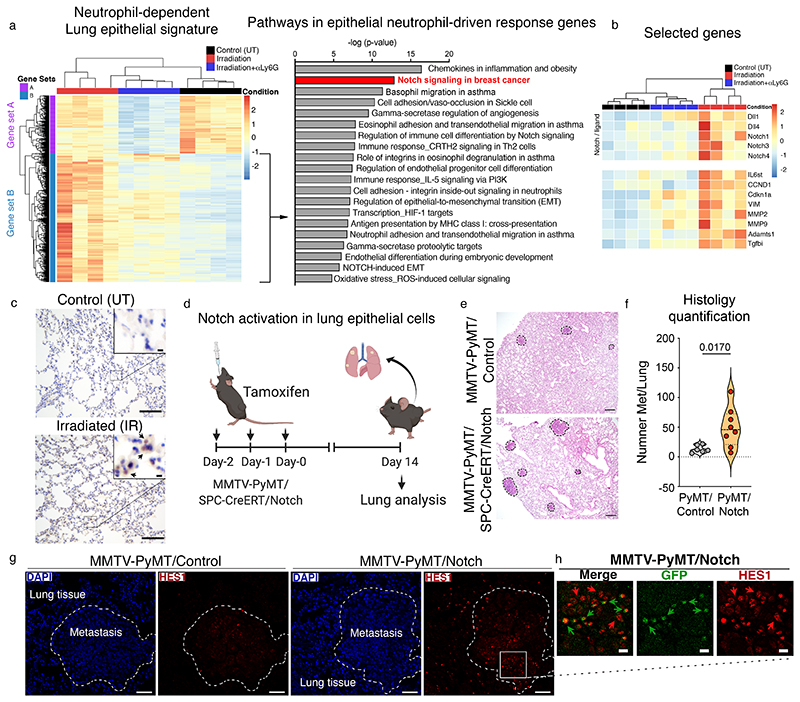
These functional differences in irradiated epithelial cells from neutrophil-depleted mice prompted us to further examine the transcriptional signatures of the different epithelial cell clusters we had distinguished by PCA analysis (Fig. 4f). We identified a distinctive cluster of genes whose expression was specifically enriched upon irradiation, and boosted by the presence of neutrophils (gene set B, Fig. 5a, left). In the mesenchymal compartment, a neutrophil-driven response was also evident, however this was less clear, and the presence of neutrophils appeared to have a greater impact on genes downregulated upon irradiation (gene set A, Extended Data Fig. 6f). Metacore pathway analysis of the neutrophil-promoted genes in epithelial cells (gene set B, Fig 5a, right) revealed an enrichment of Notch signalling, a critical regulator of stem cell proliferation and differentiation during lung development and tissue repair25, particularly in receptors Notch 1, 3 and 4 and the ligands Dll1 and Dll4 (Fig. 5b). The Notch signature was validated by reverse transcription with quantitative PCR (RT-qPCR), whereby a neutrophil-dependent induction of the canonical target genes Hes1 and Hey1 was also evident, as well as by immunofluorescence on isolated irradiated lung epithelial cells, where a reduction of intracellular activated Notch was detected in the absence of neutrophils (Extended Data Fig. 7a,b). Interestingly, in line with previous evidence of neutrophil-dependent vascular repair15, neutrophils also appeared to influence Notch genes in endothelial cells isolated from irradiated mice as shown by RT-qPCR (Extended Data Fig. 7c), although they were excluded during cell sorting for the RNA-seq analysis (Fig. 4e). Taken together, our findings suggest that the epithelial response to radiation-injury and their subsequent interaction with cancer cells is markedly influenced by the presence of neutrophils.
Fig. 5. Notch is activated in the lung epithelium and enhances spontaneous metastasis.
a, Heatmap of Lin-EpCAM+ lung epithelial cells from control, irradiated and neutrophil-depleted irradiated mice (left) (hierarchically clustered samples in columns and genes in rows, n=4 mice per group), and Metacore pathway analysis (right) of the genes triggered by radiation, but influenced by neutrophils (gene set B, indicated by the arrow). b, Heatmap showing selected genes from gene set B from (a) (hierarchically clustered samples in columns and selected genes in rows). c, Representative images for Hes1 immunostaining in lung tissue from control or irradiated mice at day 7. Positive cells are indicated by arrows in the enlarged inset. Scale bar, 100 μm (main image), 10 μm (inset) (n=4 mice per group, one experiment). d, Experiment setup for genetic Notch activation. PyMT/Notch mice (MMTV-PyMT/SPC-Cre-ERT2/Rosa26NICD-IRES-GFP) or PyMT/Control mice were administered tamoxifen by oral gavage (40 mg/kg) over three consecutive days to drive Cre expression in lung alveolar cells. Lungs were harvested 14 days after the last tamoxifen dose and assessed for metastatic burden. e,f, Representative H&E stained lungs (e) and histology quantification of lung metastases (f) from PyMT/Control and PyMT/Notch mice (n=6 PyMT/control mice, n=8 PyMT/Notch mice, spontaneous metastasis assessed over n=6 independent tamoxifen administrations). Scale bar, 250 μm g, Representative immunofluorescence images from DAPI (blue) and Hes1 (red) stained lung sections from PyMT/Control (left panel, n=5) and PyMT/Notch (right, n=7) mice harvested at experimental endpoint. Lung metastases are depicted with a dashed line. Scale bar, 50 μm. h, Enlargement from (g) showing Hes1 (red) and GFP (green) immunofluorescence staining in the PyMT/Notch metastatic lesion (n=7 mice). Positive cells are indicated by arrows. Scale bar, 10 μm. All data represented as mean ± s.e.m. Statistical analysis by a two-tailed t-test with Welch’s correction for (f). The violin plot displays the median, 25th and 75th percentiles as well as the density of data points. Dots represent individual mice. UT, untreated; IR, irradiation. Histology quantification process outlined in Supplementary File 1. Source data.
Radiation enhances Notch-signalling and cancer cell stemness
Given the enrichment in Notch signalling within the lung epithelium of irradiated mice (Fig. 5a-c), we explored whether genetic Notch activation in control mice, mimicking the activation observed downstream of radiation-primed neutrophils, could recapitulate the pro-metastatic effect of radiation. To test this, we forced the activation of Notch specifically in the lung epithelium of non-irradiated mice by crossing Rosa26-NICD-IRES-GFP mice26 with Sftpc-CreER mice that express Cre recombinase in lung alveolar type II cells. The resulting progeny were then crossed with the mammary tumour model MMTV-PyMT. Following the onset of mammary tumours, we administered tamoxifen to drive constitutive expression of the Notch intracellular domain in the lung alveoli in the absence of radiation exposure (Fig. 5d, Extended Data Fig. 8a). As expected, forced activation of Notch in the lung had no impact on primary tumour growth (Extended Data Fig. 8b). In contrast, when we analysed the lungs two weeks post-tamoxifen, we observed a striking enhancement of spontaneous metastasis in mice with activated Notch (PyMT/Notch) compared to control mice (Fig. 5e,f), indicating that cancer cells directly profit from enhanced Notch signals within the lung epithelium. Notch activation, marked by expression of the target gene Hes1, was clearly observed in the epithelium adjacent to metastatic foci in PyMT/Notch lungs, but not in control mice (Fig. 5g). Importantly, Hes1+ alveolar epithelial cells co-expressing GFP were readily detectable within the metastatic lesions of PyMT/Notch mice and, within the metastatic environment, adjacent cancer cells also showed staining (Figure 5h). This suggests a local induction of Notch signalling in metastatic cells. Indeed, there was a marked enrichment of Hes1 expression in the tumour cells themselves, compared to tumour cells from control mice (Extended Data Fig. 8c). This suggests that persistent Notch activation in epithelial cells within the metastatic microenvironment of the lung can foster the growth of arriving tumour cells. Of note, in this context, any potential influence of lung neutrophils would be independent from their radiation-primed activity.
The identification of Notch activation within the lung alveolar compartment of irradiated mice (Fig. 5a-c, and Extended Data Fig. 7a,b), together with the evidence that these cells are part of the early metastatic niche2, prompted us to assess whether Notch-activated alveolar cells could be detected in the metastatic niche of pre-irradiated lungs. To test this, we utilised Cherry-niche labelling 4T1 cancer cells, which are able to identify neighbouring non-cancerous cells by mCherry uptake27 (Fig. 6a). We isolated mCherry+ non-cancer lung niche cells (CD45-GFP-Cherry+) and Cherry− distant lung cells (CD45- GFP-Cherry-) from pre-irradiated or control lungs harbouring metastases (Extended Data Fig. 2) and performed single-cell RNA sequencing (scRNA-seq). Uniform Manifold Approximation and Projection (UMAP) analysis was used for the unbiased identification of epithelial, endothelial and mesenchymal cell clusters (Fig. 6b). We identified those clusters by the expression of EpCAM, CD31 (PECAM-1) and PDGFα/β, respectfully (Extended Data Fig. 8d). Excitingly, Notch signalling was enriched within the epithelial compartment of the metastatic niche (Cherry+) compared to the distal lung epithelial cells (Cherry-) of irradiated mice (Fig. 6c). Thus, the Notch-high environment we observed at day 7 (prior to cancer cell seeding) becomes positively selected within the metastatic niche of breast cancer cells 14 days post radiation-induced lung injury. Notably, the endothelial compartment (which also displayed Notch activation upon irradiation, Extended Data Fig. 7c) also contributes to the Notch-high environment within the metastatic niche, although we did not detect an enrichment compared to the distal lung (Fig. 6c). These findings, together with the enrichment of Notch in metastatic cells surrounded by Notch-activated lung epithelium (Fig. 5d-h), prompted us to examine Notch signalling within cancer cells growing in pre-irradiated lungs in the presence or absence of neutrophils (from Fig. 3a). Importantly, the nuclear localisation of the Notch downstream DNA-binding protein RBPJ and the expression of target gene Hes1 was strongly enriched in metastatic 4T1 cells growing in irradiated lungs compared to control tumours, and this was almost entirely abrogated by depleting neutrophils (Fig. 6d,e, Extended Data Fig. 8e), as well as in irradiated G-csf ko mice (from Fig. 3e, Extended Data Fig. 8f).
Fig. 6. Radiation exposure boosts Notch-signalling and stemness in metastatic cells.
a, Cherry-niche labelling tool. Irradiated BALB/c mice were intravenously injected with GFP+ 4T1-sLP-Cherry-labelling cancer cells 7 days post-irradiation. One week later, GFP-CD45-Ter119- mCherry+ (red) and GFP-CD45-Ter119-mCherry- cells (grey) were FACS-sorted from metastatic lungs, representing the labelled metastatic niche or distant lung cells, respectively. b, Combined Uniform Manifold Approximation and Projection (UMAP) plot of cells from the mCherry+ niche and mCherry- distant lung (n=10 mice, pooled). Clusters representing epithelial cells (EpCAM+), fibroblasts (Pdgfrα/β+) and endothelial cells (Pecam1(CD31)+) are outlined. c, Notch signalling (Reactome) score in irradiated Cherry+ niche (red) and Cherry- distant lung (blue) cells for the compartments in (b), calculated in VISION (methods). d,e, Quantification of RBPJ (d) and Hes1 (e) immunostaining in lung metastases from control, irradiated, and neutrophil-depleted irradiated mice at day 14 (n=7 mice per group, two independent experiments). Representative images in Extended Data Fig. 8e, quantification in methods. f, Cancer stemness assays. Metastatic lungs from control/irradiated BALB/c mice were harvested and the cell suspension plated to select for 4T1 metastatic cells (methods). The following day, equal numbers of cancer cells were plated in 2D or embedded in Matrigel. Colony/tumoroid formation was quantified after 7 days. (g) (left) Representative image of Giemsa-stained 6-well plate after seeding 1000 or 5000 cancer cells from control/irradiated lungs and (right) quantification (n=10 mice per group, 3 independent experiments). Scale bar, 1 cm. (h) (left) Representative image and (right) tumour organoid quantification (n=7 mice per group, 2 independent experiments). Scale bar, 50 μm. i,j, Quantification (i) and representative Sox9 immunostaining (j) in lung metastases from control, irradiated, and neutrophil-depleted irradiated BALB/c mice at day 14 (n=7 mice per group, two independent experiments). Scale bar, 100 μm (main), 10 μm (enlarged inset). All data represented as mean ± s.e.m. Statistical analysis by two-tailed non-parametric Mann-Whitney test for (g),one-way ANOVA for (d,e,i), and unpaired two-tailed t-test for (h). For box plots, boxes represent 25th/75th percentiles, the line represents the median and whiskers indicates the minimum/maximum values. Dots represent individual mice. UT, untreated; IR, irradiated. FACS gating strategies in Extended Data Fig. 2. Source data.
Notch signalling is critical for the regulation of stemness in cancer cells28, therefore we tested if metastases high in Notch activity also show an increase in stemness potential ex vivo. We isolated metastatic 4T1 breast cancer cells growing in the lungs of pre-irradiated (Notchhigh) or control mice (Notchlow), and analyzed the colony forming activity of equal numbers of tumour cells in 2D and 3D (Fig. 6f). In both experimental settings, tumour cells exposed to pre-irradiated lungs showed a striking increase in colony forming capacity, suggesting an augmented stemness potential (Fig 6g,h). In support of these findings, we observed a significant enrichment in the expression of the stem cell transcription factor Sox9 in tumour cells growing in irradiated lungs compared to control mice, which was reduced in metastases in neutrophil-depleted lungs (in which Notch expression is lower) (Fig. 6i,j). Interestingly, Sox9 has also been reported as a direct target gene of Notch129. Collectively, these data support the notion that Notch activation, which is part of the neutrophil-dependent radiation injury response, may be exploited by cancer cells seeding in the lung to enhance their stemness potential.
Radiation-enhanced metastasis requires degranulation
We have shown that radiation-activated neutrophils show a pro-inflammatory phenotype and an increase in granule proteins (Fig. 2g,h), therefore we tested if this tissue-perturbation effect was mediated by their degranulation. We performed targeted lung irradiation in the presence of a degranulation inhibitor Nexinhib2030 to prevent neutrophil exocytosis (Fig. 7a). Strikingly, this led to the abrogation of radiation-mediated metastatic enhancement and the reduction of Notch activation and Sox9 expression in metastatic 4T1 cells (Fig. 7b-d). To test the contribution of a single granule protein, Neutrophil elastase (Ela2), in the neutrophil-mediated tissue response, we irradiated the lungs of Ela2-ko mice and C57BL/6 littermates and intravenously injected E0771 breast cancer cells one week later (Fig. 7e). We observed a marked reduction in the metastatic proficiency of Ela2-ko lungs as well as a dampening of Notch activation (Fig. 7f-h). Thus, neutrophils strongly influence the response of lung epithelial cells to radiation injury, likely via the process of degranulation.
Fig. 7. Radiation-primed neutrophils boost lung metastasis via their degranulation activity.
a, Schematic depicting the treatment of control and irradiated mice with the neutrophil degranulation inhibitor Nexinhib20. Control or irradiated BALB/c mice were treated with Nexinhib20 or a vehicle control 3x per week (30 mg/kg), beginning the day before irradiation. GFP+ 4T1 breast cancer cells were intravenously injected at day 7 and metastatic burden assessed at day 14. b, Lung metastatic burden in control and irradiated mice treated with Nexinhib20 or vehicle, quantified as tumour area by H&E (see Supplementary File 1) (n=6 mice per group, 2 independent experiments). c,d, Quantification of Hes1 (c) and Sox9 (d) immunostaining in metastatic lesions from (b) (n=6 mice per group, two independent experiments) e, Experimental setup of Ela2ko mouse model. Ela2ko mice or C57BL/6J wild-type littermates received a 13 Gy dose of targeted whole-lung irradiation. Mice received an intravenous injection of E0771 breast cancer cells at day 7, and lungs were harvested at day 14 to assess metastatic load. f-h, Representative H&E staining (f) quantification of metastatic number (g) and Hes1 immunostaining in metastatic lesions (h) from the irradiated lungs of Ela2ko or wild-type control mice (n=6 mice per group, one experiment), Scale bar, 250 μm. All data represented as mean ± s.e.m. Statistical analysis by one-way ANOVA for (b-d), and a two-tailed t-test for (g) and (h). CTL, control; IR, irradiated; Veh, vehicle; Nex, Nexinhib20; IV, intravenous; Ko, knock-out. Metastases quantification process outlined in Supplementary File 1, immunostaining quantification in methods. Source data.
Finally, to confirm that Notch signalling activation in metastatic cells growing within irradiated lungs is responsible for enhancing their tumorigenicity, we treated control/irradiated mice with the γ-secretase inhibitor DAPT from the time of metastatic seeding (Fig. 8a). DAPT treatment led to a striking reduction in the growth of tumour cells in irradiated lungs (Fig. 8b,c), which coincided with a reduction in Hes1 expression (Fig. 8d). In line with our observation that the initial radiation-injury is required to amplify Notch activity, indeed, no effect was found following Notch inhibition in control mice (Fig. 8c). Taken together, these data show that tumour cells growing rapidly within irradiated lungs is predominantly due to enhanced Notch signalling, which is mediated by radiation-primed neutrophils influencing lung epithelial cell responses (Fig. 8e).
Fig. 8. Inhibition of Notch signalling attenuates the radiation-driven enhancement of metastatic growth in vivo.
a, Schematic depicting Notch signalling inhibition in vivo. Control or irradiated BALB/c mice were administered the γ-secretase inhibitor DAPT (10 mg/kg) or a vehicle control daily, beginning at the time of 4T1 cancer cell injection and continuing until the experimental endpoint one week later. b,c, Representative immunostaining for Ki67 (b) and quantification (c) of the number of Ki67+ metastatic lesions in control (UT) and irradiated mice treated with vehicle or DAPT (n=8 mice per group, two independent experiments). The quantification of Ki67+ metastatic foci is depicted as fold change, relative to the control mice treated with vehicle. Ki67+ metastatic foci are indicated with a dashed line. Scale bar, 250 μm. d, Quantification of Hes1 immunostaining in metastatic lesions from (b) (n=8 mice per group, two independent experiments) (see methods). All data represented as mean ± s.e.m. Statistical analysis by one-way ANOVA for (c), and a two-tailed t-test for (d). UT, untreated; IR, irradiated; Veh, vehicle. e, Proposed model for radiation-enhanced lung metastasis. Radiation exposure in healthy lung tissue leads to excessive neutrophil accumulation and activation, inducing an array of tissue perturbations such as Notch activation within epithelial cells. Together these alterations foster a pro-tumorigenic milieu within the irradiated lung, fuelling the subsequent growth of arriving metastatic cancer cells. Source data.
Discussion
A fundamental step in the metastatic cascade is the generation of a favourable metastatic niche31,32. We have recently discovered that the early response of lung tissue to breast cancer growth involves the activation of regeneration2. Therefore, in the present study, we probed whether the induction of a regenerative state in the tissue due to an injury would trigger a favourable environment for metastatic growth. Radiotherapy remains one of the most effective means of achieving curative outcomes for cancer patients with localized disease. However, in many patients the frequent development of metastasis significantly limits the long-term treatment success. Currently, the biological responses of healthy tissues to radiation-induced injury, the key inflammatory components involved, and the influence of this response on the metastatic proficiency of that organ are incompletely characterized.
To address these questions, we designed a preclinical study using targeted thoracic irradiation to generate an acute radiation injury to the lungs prior to the seeding of breast cancer cells. We found that radiation exposure to healthy lung induced a hospitable environment for metastatic growth. Most importantly, we show a key function of neutrophils recruited to the injured lung, which locally activate and influence the response of resident lung cells. We observed that neutrophils play a critical role particularly for the lung epithelium and directly support the progenitor function of alveolar cells when tested ex vivo.
We found a neutrophil-dependent enhancement of stem cell signalling including Notch in lung epithelial cells. In the adult lung, Notch is required for the regeneration of several airway cell types after chemical or infection-induced injury, including basal cells33, club cells34, alveolar type I cells35 and a population of lineage-negative epithelial progenitor cells36. Moreover, Notch signalling in response to irradiation was reported to promote survival of basal human and murine airway stem cells37. Here we identified an enrichment of Notch signaling within the metastatic niche of pre-irradiated lungs and demonstrate that the neutrophil presence results in Notch activation in metastatic cancer cells. Importantly, this induction of Notch signalling is a dominant effector enhancing tumorigenesis. In many types of human cancer including breast and lung, the Notch pathway is required for CSC self-renewal38-40. The transcription factor Sox9 is an essential regulator of self-renewal in CSCs, with its overexpression tightly correlated with metastasis formation and poor survival in multiple cancers41,42. Indeed, we observed the NotchhiSox9hi phenotype in breast cancer cells growing specifically within neutrophil-proficient irradiated lungs, along with their profoundly enhanced tumorigenicity. This suggests the induction of a regenerative program promoting cancer stemness.
We also observed a neutrophil-dependent boost in Notch signalling in irradiated endothelial cells, therefore we cannot exclude their involvement as an intermediary between the neutrophils and the epithelium. Indeed, neutrophils were shown to support lung vascular repair following whole-body irradiation15. In addition, the neutrophil-mediated change in the response of mesenchymal cells could also be relevant for metastatic outcome. Future studies are required to resolve the role of radiation-primed neutrophils in the perturbation of other cellular components, or determine if a similar activation occurs in other tissues exposed to radiation. Notably, as acute radiation injury can contribute to the onset of pneumonitis and fibrosis in the long-term, our data encourages further investigation into the role of radiation-primed neutrophils in these chronic pathologies.
Taken together, our study shows that a high level of radiation exposure in healthy lung tissue has profound effects on the tissue microenvironment that inadvertently increases its metastatic potential. Our findings place radiation-primed neutrophils as a key modulator of these pro-metastatic alterations.
Nowadays, the technologies used to administer conformal radiotherapy are highly effective in limiting both the dose and volume of tissue within the radiation field, making the treatment much safer for patients. We acknowledge that in our experimental setting, the volume of exposed tissue is considerably larger than in the clinic. Nonetheless, given the striking effects of radiation-primed neutrophils observed in this study using pre-clinical models, together with the abundant evidence of their multiple pro-tumour functions12, our work encourages closer attention to neutrophil responses in cancer patients receiving RT. Moreover, since the radiation-induced neutrophil activity is, at least in part, dependent on their degranulation, this study supports the interest in developing inhibitors of cancer cell exosome release for clinical use43,44.
Methods
All experiments in this study were approved by the Francis Crick Institute and University College London ethical review committees, and conducted according to UK Home Office regulations under the project license PPL80/2531 and PPL70/9032. Further information on research design, statistics and technical information is available in the Nature Research Reporting Summary linked to this article.
Statistics and reproducibility
All statistical analyses were performed using Prism (version 9.1.1, GraphPad Software). Graphic display was performed in Prism, and illustrative figures created with Biorender.com. A Kolmogorov-Smirnov normality test was performed before any other statistical test. After, if any of the comparative groups failed normality (or the number too low to estimate normality), a non-parametric Mann-Whitney test was performed. When groups showed a normal distribution, an unpaired two tailed t-test was performed. When groups showed a significant difference in the variance, we used a t-test with Welch’s correction. When assessing statistics of 3 or more groups, we performed one way ANOVA controlling for multiple comparison or non-parametric Kruskal-Wallis. 3D co-cultures on scaffolds were assessed via two-way ANOVA.
Litter mice were randomized prior to radiation exposure into control or irradiation groups. Since in vivo treatments were performed in pre-determined groups (control vs. irradiation), no further randomization could be performed. In vitro 2D/3D co-culture assays and organoid assays were blinded at quantification. For all histological analysis, quantification between different experimental groups was performed blinded. No statistical methods were used to pre-determine sample sizes, but these are similar to those reported in our previous publications32,45. All replicate/experiment numbers are clearly stated in the figure legends. No data was excluded.
Mouse strains
Breeding and all animal procedures were performed in accordance with UK Home Office regulations under project license PPL80/2531 and PPL70/9032. Female mice 6-12 weeks old were used in all experiments. Wild-type BALB/cJ, C57BL/6J, FVB/n, NSG and Nude mice from The Jackson Laboratory were provided by The Francis Crick Biological Research Facility. MMTV-PyMT mice on a FVB background were originally obtained from The Jackson Laboratory, actin-GFP mice and RAG1 deficient FVB/n mice were originally a gift from J. Huelsken laboratory (EPFL, Lausanne, Switzerland) and bred at the Francis Crick Institute. Gcsf knock-out mice were also a gift from the J. Huelsken laboratory and backcrossed to the FVB/n (N10) background. Sftpc-CreERT2 mice on a C57BL/6J background were a gift from the J-H Lee laboratory (MRC Cambridge Stem Cell Institute). Rosa26-NICD-IRES-GFP mice26 on a mixed background were purchased from The Jackson Laboratory (stock 008159). Ela2-Cre knock-in mice were originally purchased from European Mouse Mutant Archive (EMMA). For experiments in which mice developed a primary tumour, the maximum tumour size did not exceed 1.5 cm, in compliance with project license PPL80/2531.
Whole-lung targeted irradiation
Mice were anaesthetised with Fentanyl (0.05 mg/kg), Midazolam (5 mg/kg) and Medetomidine (0.5 mg/kg), then given a single 13 Gy dose of radiation (300 kV, 10 mA, 1 mm Cu filtration), targeted to the thoracic cavity. For fractionation experiments, 4Gy was delivered on 3 or 4 consecutive days. CT-based guidance was not used, but beam targeting to the lungs was achieved using a 7cm collimator (1 cm diameter) with a total field source distance of 20 cm. The machine was calibrated daily to achieve a consistent total dose. The dose was given at a rate of approximately 1.4 – 1.5 Gy/min each time, with a radiation time of approximately 8 - 9 minutes for 13 Gy and 2 – 3 minutes for 4 Gy. Probes confirmed radiation outside the collimator was negligible prior to experiments. The anaesthetic was reversed with Naloxone (1.2 mg/kg), Flumenzenil (0.5 mg/kg) and Atipam (2.5 mg/kg). Recovery was performed in warming chambers at 37°C. Control mice received a sham-irradiation, whereby they were placed under anaesthetic for the same length of time, and recovered in warming chambers.
Targeted Partial-lung irradiations
An Xstrahl Small Animal Radiation Research Platform (SARRP) S/N 525722 irradiator (225kV x-ray tube, half value layer 0.847 mm Cu), with 0.1 mm integral Be filtration, was used for mouse treatments. Treatment was performed at University College London under project license PPL70/9032. CBCT was performed before each treatment to confirm target position. Each mouse was anaesthetized with isoflurane and positioned for optimal right lung targeting on a 3D printed bed. The bed was rotated between the x-ray source and a digital flat-panel detector. The images were obtained at 60 kVp and 0.8 mA with 1 mm Al filtration of an uncollimated primary beam. During rotation, 360 projections were acquired (approx. 1° increments for each projection, approx. 0.01 Gy total radiation dose). The CBCT projections were rendered into a 3D image reconstruction, using the FDK® algorithm with a voxel size from between 0.01 to 5 mm. Muriplan software was used to set radiodensity thresholds for different tissues and enable treatment planning, setting an isocenter to target the right lung individually and avoid the left lung, heart and trachea. Dose calculation was computed using a Monte Carlo simulation superposition-convolution dose algorithm, similar to those used clinically.
Mice received a single treatment of either of 8 Gy or 12 Gy, delivered as one angled beam, targeted to maximise dose delivery to a single lung. The beam used was 220 kV and 13 mA, filtered with 0.15 mm Cu, dose rate 2.37 Gy/min under reference conditions. The beam was collimated, to match the treatment volume, with beam cross section up to 10x10mm. The time calculated to deliver 8Gy to individual mice varied between 222 and 229 seconds, and to deliver 12 Gy varied between 318 and 348 seconds.
Cell culture
All cell lines were provided by the Cell Services Unit of The Francis Crick Institute, where they were authenticated using Short-Tandom Repeat (STR) profiling and species-identification tests, and confirmed to be mycoplasma-free. Cells were cultured in DMEM (ThermoFisher Scientific) supplemented with 10% fetal bovine serum (FBS; Labtech) and 100 U/ml penicillin-streptomycin (ThermoFisher Scientific). MMTV-PyMT cells were isolated from late stage carcinomas and cultured on collagen-coated dishes in MEM medium (DMEM/F12 (ThermoFisher Scientific) with 2% FBS, 100 U/ml penicillin-streptomycin, 20 ng ml-1 EGF (ThermoFisher Scientific) and 10 μg/ml insulin (Merck Sigma-Aldrich)). Collagen solution contains 30 μg/mL PureCol collagen (Advanced Biomatrix), 0.1% bovine serum albumin (BSA) and 20 mM HEPES in HBSS (ThermoFisher Scientific). All cells were cultured at 37°C and 5% CO2.
Cherry Labelling tool
The Cherry labelling tool developed by our laboratory has been recently described in detail 2,27. A soluble peptide (SP) and a modified TAT peptide were cloned upstream of the mCherry cDNA, under the control of a mouse PGK promoter (sLP-Cherry). The sLP-Cherry sequence was then cloned into a pRRL lentiviral backbone. 4T1 cancer cells were stably infected with LP-Cherry and pLentiGFP lentiviral particles and subsequently sorted by flow cytometry to isolate 4T1-Cherry+GFP+ labelling cells.
Induction of metastasis
To induce spontaneous metastases, mice were given an orthotopic injection of 50μl growth factor-reduced (GFR) Matrigel (BD Biosciences) containing 1 × 106 4T1 or 4T07 breast cancer cells in the left mammary fat pad, using a 29-gauge insulin syringe. The injections were performed under anaesthesia (isoflurane).
To induce experimental metastasis, 4T1 breast cancer cells (0.5 × 106), MMTV-PyMT primary mammary tumour cells (1 × 106), E0771 breast cancer cells (0.5 × 106) were re-suspended in 100μl PBS and intravenously injected into BALB/c, Gcsf Ko or C57BL/6J mice, respectively. For human cancer cell metastasis, Flo-1 oesophageal cancer cells (1 × 106) were intravenously injected into NSG mice, and H460 and A549 lung NSCLC cells (1 × 106) were injected into Nude BALB/c mice. All animals were monitored daily for unexpected clinical signs following the project license PPL80/2531 guidelines and the principles set out in the NCRI Guidelines for the Welfare and Use of Animals in Cancer Research (UK). The rationale and process for histological quantification of lung metastatic burden is provided in Supplementary File 1.
In vivo treatments
For neutrophil depletion, rat anti-Ly6G antibody (BioXcell, clone 1A8, 12.5 μg/mouse) in PBS or rat IgG isotype control (Cell Services Unit, Francis Crick Institute) was administered daily to BALB/cJ female mice via intraperitoneal injection. Recombinant GCSF protein (Novoprotein, C002, 5 μg/mouse in PBS) was administered subcutaneously to Gcsf Ko mice every other day for a total of four injections. For Notch inhibition, BALB/cJ female mice received daily intraperitoneal injections of the γ-secretase inhibitor DAPT dissolved in corn oil (Sigma, 10 mg/kg body weight), or a vehicle control. To inhibit neutrophil degranulation, BALB/cJ female mice given an intraperitoneal injection of Nexinhib2030 (30 mg/kg in corn oil), or a vehicle control, three times per week for two weeks. For tamoxifen administration, tamoxifen (Sigma-Aldrich) was dissolved in corn oil in a 40 mg/ml stock solution. Three doses (0.2 mg per g body weight) were given via oral gavage over consecutive days, and mice were harvested two weeks after the final dose.
Tissue digestion for cell isolation or analysis
Lung tissue was minced manually with scissors and digested with Liberase TM and TH (Roche Diagnostics) and DNase I (Merck Sigma-Aldrich) in HBSS for 30 min at 37°C in a shaker at 180 rpm. Samples were passed through a 100 μm filter and centrifuged at 1250 rpm for 10 min. The cell pellet was incubated in Red Blood Cell Lysis buffer (Miltenyi Biotec) for 5 min at room temperature. After centrifugation, cells washed with MACS buffer (0.5% BSA and 250 mM EDTA in PBS) and passed through a 20 μm strainer-capped tube to generate a single cell suspension. Antibody staining was then performed for cell isolation (flow cytometry or magnetic-bead based cell separation), or for flow cytometry analysis.
FACS analysis and cell sorting
Mouse lung single-cell suspensions prepared as above were incubated with mouse FcR Blocking Reagent (Miltenyi Biotec) for 10 min at 4°C followed by incubation with fluorescently-conjugated antibodies for 30 min at 4°C (see Supplementary Table 1 for antibody list). Cells were washed twice with MACS buffer, then incubated with 4′,6-diamidino-2-phenylindole (DAPI) to stain dead cells. Flow cytometry analyses were performed on a BD LSR-Fortessa (BD Biosciences) and subsequently analysed using FlowJo 10.4.2 (FlowJO, LCC 2006-2018). Cell sorting was performed on a BD Influx cell sorter (BD Biosciences) using the BD FACS™ Sortware 1.2.0.142.
Magnetic-activated cell sorting (MACS)
EpCAM+ lung epithelial cells or Ly6G+ neutrophils were isolated from mouse single-cell suspensions via MACS. Cells were incubated with FcR Blocking Reagent for 10 min at 4°C followed by EpCAM-PE or Ly6G-PE (Supplementary Table 1) for 30 min at 4°C. After washing with MACS buffer, cells were incubated with PE-conjugated microbeads (Miltenyi Biotec) for 15 min at 4°C. Positive cell selection was performed using magnetic separation columns (Miltenyi Biotec).
Neutrophil adoptive transfer
Ly6G+ lung neutrophils were MACS-sorted from control or irradiated mice, 7 days post-irradiation (n=12 pooled lungs per group). Neutrophils were resuspended in PBS and 1.5 × 106 neutrophils intravenously injected into each naïve recipient mouse. After 4 days, recipient mice were intravenously injected with 0.3 × 106 4T1-GFP+ breast cancer cells.
Lungs were collected one week later, fixed in 4% PFA and embedded in paraffin. To quantify metastatic burden, 4μm serial sections from the entire lung were stained for haematoxylin & eosin (H&E). The total number of metastatic foci in each lung was counted manually, using a Nikon Eclipse 90i light microscope.
Lung organoid assay
Lung organoid co-culture assays have been previously described46. Lung epithelial cells (Epcam+CD45-CD31-Ter119-) from control BALB/cJ mice or irradiated mice were FACS sorted and resuspended in 3D organoid media (DMEM/F12 with 10% FBS, 100 U/ml penicillin-streptomycin, and insulin/transferrin/selenium (Merck Sigma-Aldrich)). Cells were mixed with murine normal lung fibroblast (MLg) cells and resuspended in GFR Matrigel at a ratio of 1:1. 100μl of this mixture was pipetted into a 24-well transwell insert with a 0.4 μm pore (Corning). 5,000 epithelial cells and 25,000 MLg cells were seeded in each insert. After incubating for 30 min at 37°C, 500μl organoid media was added to the lower chamber and media changed every other day. Bright-field images were acquired after 14 days using an EVOS microscope (ThermoFisher Scientific), and quantified using FiJi (version 2.0.0-rc-69/1.52r, ImageJ).
Cancer cell stemness assay
Control and irradiated lungs harbouring 4T1 metastases were dissociated to a single cell suspension as described above. Cells were plated overnight (>12h) in DMEM with 10% FBS and 100 U/ml penicillin-streptomycin. The following day, wells were washed 4x in PBS to remove non-adherent cells, trypsinised and counted. For all experiments, a metastasis-free normal lung was dissociated and plated overnight in the same conditions to confirm non-adherence of non-cancerous cells. For 2D assays, cancer cells were resuspended in DMEM/10% FBS at a density of 1 × 103 cells/ml or 5 × 103 cells/ml and cultured in 6-well plates for 7 days. At endpoint, cells were fixed in chilled acetone and methanol (1:1) and stained with Giemsa (Merck). For 3D assays, 5 × 104 cancer cells/ml were resuspended in GFR Matrigel and 20 μl droplets seeded into 24-well plates. After a 30 min incubation at 37°C, 500 μl 3D growth medium (DMEM/F12 supplemented with 100 U/ml penicillin-streptomycin, 20 ng/ml EGF, 20 ng/ml bFGF, 4 μg/ml Heparin and B27 (50x)) was added. Media was replaced every 48h and bright-field images acquired at day 7 using an EVOS microscope.
3D cell culture
GFP+MMTV-PyMT cells were seeded (5,000 cells/well) in a collagen-coated Alvetex™ Scaffold 96-well plate (ReproCELL). The following day, lungs were harvested from control or irradiated BALB/cJ mice (7 days post-irradiation), and EpCAM+ epithelial cells or Ly6G+ neutrophils were isolated by MACS sorting. Cells were seeded on top of the cancer cells (50,000 cells/well). The growth of GFP+ cells was monitored using the SteREO Lumar.V12 stereomicroscope (Zeiss), and images quantified using ImageJ. For quantification, the Li’s Minimum Cross Entropy thresholding algorithm was performed on image stacks.
EdU in vitro proliferation assay
GFP+MMTV-PyMT cells were seeded at a density of 10,000 cells/well into collagen-coated 6-well plates in MEM media. The following day, Ly6G+ lung neutrophils were isolated via MACS sorting from control or irradiated mice and added to the wells at a density of 100,000 cells/well. After 60h, wells were supplemented with 20 μM EdU (5-ethynyl-2’-deoxyuridine, Sigma-Aldrich). Cells were harvested 6h later, and EdU incorporation assessed using the Click-iT Plus EdU Flow Cytometry Assay Kit (ThermoFisher Scientific), according to the manufacturer’s instructions. Sample data were acquired on a BD LSR-Fortessa and analysed using FlowJo 10.4.2.
EdU in vivo
Mice were treated with a single intraperitoneal injection of EdU dissolved in PBS (25 mg/kg body weight). Mice received targeted lung irradiation 1h later, and were culled either 1h or 7 days post-irradiation. Lungs and bone marrow were harvested, and EdU incorporation assessed in single cell suspensions stained for Ly6G-PE (BD Biosciences, clone 1A8) using the Click-iT Plus EdU Flow Cytometry Assay Kit. Data was acquired on a BD LSR-Fortessa and analysed using FlowJo 10.4.2.
Immunohistochemistry & Immunofluorescence
Mouse lungs were fixed overnight in 4% PFA and embedded in paraffin blocks. 4μm tissue sections were cut, deparaffinised and rehydrated using standard methods. Antigen retrieval was performed using pH 6.0 Citrate Buffer in a 900W microwave, with the exception of MPO and Cit-H3, for which the Dako Target Retrieval Solution, pH 9.0 was used (cat no. S2368). For immunohistochemistry, sections were blocked with 1% BSA, 5% Donkey serum in PBS for 1h at room temperature, then incubated overnight at 4°C with primary antibodies (see Supplementary Table 1). The following day, sections were incubated for 45 min at room temperature with biotinylated secondary antibodies (Vector Laboratories, Supplementary Table 1). Next, the VECTORSTAIN Elite ABC kit (Vector) and DAB substrate kit (Vector) was used according to the manufacturer’s instructions. Bright-field images were obtained on a Nikon Eclipse 90i light microscope. For immunofluorescence, samples were blocked with 1% BSA, 5% Donkey serum in PBS for 1h at room temperature, then incubated overnight with primary antibodies (Supplementary Table 1) at 4°C. The next day, slides were incubated with donkey anti-rabbit AlexaFluor 555 or donkey anti-goat 488 (1:500, ThermoFisher Scientific) for 1h at room temperature. Slides were mounted with Vectashield Mounting Medium with DAPI (Vector). Images were acquired using a Zeiss Upright710 confocal microscope with NIS-elements software (version 4.51, Nikon, Japan) and analysis performed with ImageJ.
Immunostaining quantification
To quantify Hes1, RBPJ and Sox9 expression within metastases, positive cells within each metastatic area were counted and normalised to tumour size by dividing the number of positive cells within each metastasis by the tumour area, as measured by ImageJ. A minimum of 20 images per lung were quantified. The intensity of RBPJ staining was determined using ImageJ. Following colour deconvolution, the mean grey value was measured specifically for the metastatic area within each image. The optical density was determined by the calculation OD = log10 (max grey value/mean grey value). The resulting optical density measurements were averaged across all images per sample, generating an average intensity of RBPJ staining per lung.
Immunofluorescent staining on coverslips
EpCAM+ epithelial cells were MACS-sorted from control, irradiated and neutrophil-depleted irradiated lungs, plated on poly-lysine glass coverslips for 15 min at room temperature, and fixed in 4% PFA in PBS for 10 min. After fixation, cells were permeabilized with 0.1% Triton-X-100 in PBS for 5 min and incubated with a blocking solution (1% BSA, 5% goat serum in PBS) for 1h at room temperature. Next, cells were incubated for 1h at room temperature with an anti-activated Notch1 antibody (Abcam, ab8925) diluted in blocking solution followed by a 1h incubation with a goat anti-rabbit AlexaFluor 488 (1:500, ThermoFisher Scientific) at room temperature. Coverslips were then mounted onto slides using Vectashield Mounting Medium with DAPI for imaging. Images were acquired using a Zeiss Upright710 confocal microscope, and quantified using the MeasureObjectIntensity function in CellProfiler. For analysis of neutrophil nuclear segmentation, Ly6G+ neutrophils were MACS-sorted from lung suspensions and plated on poly-lysine coverslips as described above. After fixation, coverslips were mounted onto slides using Vectashield Mounting Medium with DAPI and images captured with a Zeiss Upright710 confocal microscope. FiJi (version 2.0.0-rc-69/1.52r, ImageJ) was used to analyse fluorescence images.
Senescence-associated β-galactosidase staining
Senescence-associated β-galactosidase staining was performed on lung cryosections preserved in OCT freezing medium, using the Senescence β-galactosidase staining kit (cat#9860, Cell Signalling Technology), according to the manufacturer’s instructions. Cryosections of 10 μm were fixed in a 2% formaldehyde and 0.2% glutaraldehyde solution in PBS at room temperature for 5 min. Sections were washed 3 times with PBS, and incubated for 48 hrs at 37°C with the staining solution containing X-gal in N-N-dimethylformamide (pH 6.0). Sections were counterstained with Nuclear Fast Red (cat no. H-3403, Vector), mounted in VectoMount™ AQ (cat no. H-5501, Vector) and imaged on a Nikon Eclipse 90i microscope. For quantification, the area of SA-β-gal+ regions per lung was measured using ImageJ.
Quantitative Proteomics of neutrophils using TMT labelling
Ly6G+ lung or bone marrow neutrophils were isolated by MACS-sorting. Cells were lysed by adding three cell-pellet volumes of lysis buffer (8 M urea, 50 mM TEAB, protease inhibitors) followed by sonication. Insoluble material was removed by centrifugation at 16,000 × g for 10 minutes at 4°C. Following a Bicinchonic Acid (BCA) assay for protein quantification, 25 μg of total protein was transferred into 6 labelled tubes and adjusted to a final volume of 25 μl with 50 mM TEAB. Reduction and alkylation was performed using DTT and iodoacetamide in 50mM TEAB. Proteins were then precipitated overnight by adding six volumes of ice-cold acetone. The next day, proteins were resuspended in 25 μl of 50mM TEAB and digested by trypsin for 16 hours at 37°C (1:50 w:w). Peptides were then desalted using a C18 macrospin column (The Nest Group Inc) and re-solubilised in 50 mM TEAB and subsequently labelled using a 0.2mg TMT 10-plex kit (Thermo Scientific). Peptide labelling efficiency was assessed using a QExactive orbitrap (Thermo Scientific) mass spectrometer to ensure that all samples displayed >98% labelling efficiency. Next, 5 % hydroxylamine was added to each sample to sequester unreacted label and the samples were mixed. The 10-plex sample was desalted using a C18 macrospin column and peptides were eluted with 80 % acetonitrile and dried in a vacuum centrifuge. High pH reversed-phase fractionation (Pierce, UK) was employed with nine fraction collected and dried to completeness. Each fraction was analysed using a Fusion Lumos Tribrid orbitrap mass spectrometer coupled to an UltiMate 3000 HPLC system. The sample was loaded onto a C18 trap column (Acclaim PepMap 100; 75 μM × 2 cm) then transferred onto a C18 reversed-phase column (PepMap RSLC; 50 cm length, 75 μm inner diameter). Peptides were eluted with a linear gradient of 2–25 % buffer B (75 % acetonitrile, 20 % water, 0.1 % formic acid, 5 % DMSO) at a flow rate of 275 nl/min over 135 minutes followed by a second linear gradient of 25-40 % buffer B over 25 minutes. MS1 instrument settings: spectra were acquired in the orbitrap at 120,000 resolution with scan range of 350–1500 m/z; maximum injection time 50 milliseconds; AGC target value 4E5; 30 % RF lens setting; 90 second dynamic exclusion. MS2 instrument settings: precursor ions with charge states z = 2–7+ were selected for MS/MS higher-energy collision-induced dissociation (HCD) fragmentation; 38 % energy; acquired in the orbitrap at 60,000 resolution; maximum injection time 105 milliseconds; AGC target value 1E5. All data were analysed with MaxQuant software version 1.6.0.13. MS/MS spectra were searched against a UniProtKB Mus musculus database downloaded in July 2017 and a list of default contaminants. Reversed decoy sequences were used to estimate false discovery rates. Additional MaxQuant setting were; Trypsin with two missed cleavages, Carbamidomethylation of cysteine as a fixed modification, N-terminal protein acetylation and methionine oxidation as variable modifications. The MaxQuant output proteinGroups.txt file was imported to Perseus version 1.4.0.2. Proteins were filtered to common contaminants and decoy sequences. Intensity values were log 2 transformed and normalisation for each channel was performed by median subtraction.
Quantitative RT-PCR
RNA was isolated from sorted lung populations using the RNeasy Mini kit (Qiagen), with an on-column DNase treatment (Qiagen). cDNA was synthesized with random primers, using the SuperScript III First Strand Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. The PCR, data collection and data analysis was performed on a QuantStudio™ 3 Real-time PCR system (Thermo Fisher Scientific), using a PowerUp™ SYBR Green Master Mix (Thermo Fisher Scientific). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal expression reference. Primer sequences are provided in Supplementary Table 2.
RNA sequencing sample preparation
Single cell RNA sequencing
Single cell suspensions of metastatic lungs from control or irradiated mice (n=10 per group, pooled) were prepared as described above. CD45- Ter119- cells were sorted by flow cytometry following staining with anti-mouse CD45, Ter119 and DAPI. Library generation for 10x Genomics analysis were performed using the Chromium Single Cell 3’ Reagents Kits (10x Genomics), followed by sequencing on an Hiseq4000 (Illumina) to achieve an average of 50,000 reads per cell.
Bulk RNA sequencing
Lung CD45-CD31-Ter119-EpCAM+ and CD45-CD31-Ter119- EpCAM- cells were sorted from control, irradiated or neutrophil-depletion irradiated mice 7 days post-irradiation by flow cytometry. Total RNA was isolated using the miRNeasy Micro Kit (Qiagen, cat# 217084), according to the manufacturer’s instructions. Library generation was performed using the KAPA RNA HyperPrep with RiboErase (Roche), followed by sequencing on a HiSeq (Ilumina), to achieve an average of 30 million reads per sample.
Bioinformatic analysis
Bulk RNA sequencing
Raw fastq files were adapter trimmed using CutAdapt 1.547 and trimmed reads mapped to GRCm38 release 86 with associated ensemble transcript definitions using STAR 2.5.2a48 wrapped by RSEM 1.3.049 which was used to calculate estimated read counts per gene. Where necessary, bam files were merged using samtools 1.850. Estimated counts from samples in the Epithelial and Mesenchymal groups were normalised separately, with normalisation and differential expression of genes being called between genotype group using the R package DESeq2 1.12.3.51. Genes with an adjusted p-value less than or equal to 0.05 were said to be differentially expressed. Differentially expressed genes were further analysed for their pathway enrichments using Metacore (version 21.3, https://portal.genego.com).
Heatmaps were generated using the R package pheatmap 1.0.8 (https://cran.r-project.org/web/packages/pheatmap/index.html). All analyses were performed using R 3.3.1 (URL https://www.R-project.org/).
Single Cell RNA sequencing
Raw reads were first processed by the Cell Ranger v2.1.1 pipeline, using STAR (v2.5.1b) to align to the mm10 transcriptome, deconvolve reads to their cell of origin using the UMI tags and report cell-specific gene expression count estimates. All subsequent analyses were performed in R version 4.0.3 using the Seurat package (4.0.1)52. Datasets underwent initial filtering to remove genes expressed in fewer than 3 cells and cells with fewer than 200 detected genes. For the irradiated lung niche dataset (mCherry+ve), further filtering was performed by removing cells with: <500 genes, >3,000 genes, a mitochondrial gene content of >5.5% and a total number of reads >5300. Similarly, for the irradiated distal lung dataset, the cut-offs of <500 genes, >4000 genes, <6% mitochondrial gene content and <12000 total reads were used. Datasets were then normalised using the Seurat package SCTransform and integrated using the SelectIntegrationFeatures, PrepSCTIntegration, FindIntegrationAnchors and IntegrateData functions53. Dimensional reduction was performed using principal component analysis (PCA) on the integrated dataset. Unsupervised clustering was performed with the first 30 dimensions using the FindNeighbours and FindClusters functions and UMAP (Uniform Manifold Approximation and Projection) was used for cluster visualisation. Epithelial, endothelial and mesenchymal populations were identified based on expression of the markers Epcam, Pecam1 and Pdgfra and Pdgfrb respectively and FeaturePlot function was used for their visualisation. Cells with marker gene expression greater than 0.5 were selected for further analysis. Notch signalling pathway gene signature analysis was performed using the Reactome Signaling by Notch gene set within MSigDB. Single cell signature scores were calculated for niche and distal cells within each cell-type subset, using the Vision package v.2.1.054.
Extended Data
Extended Data Fig. 1. Radiation exposure in healthy lung tissue enhances metastasis.
a, Representative H&E images of metastatic lungs from control (UT) and irradiated BALB/c mice orthotopically injected with 4T1 breast cancer cells to generate a primary tumour. The metastatic area is depicted with a dashed line (n=4 mice per group, 2 independent experiments). Scale bar, 250 μm. b,c, FACS quantification of GFP+ tumour cells in the metastatic lung (b) and primary tumour volume (c) from control and irradiated mice (n=4 mice per group, one experiment). d,e, Representative H&E images (d) and FACS quantification of GFP+ tumour cells (e) from metastatic lungs from control (UT) or irradiated FVB mice intravenously injected with GFP+ MMTV-PyMT primary mammary tumour cells at day 7 (n=5 per group, one experiment). Scale bar, 100 μm. f, Table and representative immunostaining of human-specific Lamin A/C to detect human lung cancer cells growing in BALB/c Nude mice (n=4 mice per group for each cell line tested). Mice were intravenously injected with H460 or A549 human NSCLC cells 7 days following targeted lung irradiation (or sham-irradiation for control mice) and lungs harvested 3 weeks later. The table depicts the number of mice in which h-Lamin A/C+ cancer cells were present in the lungs. No foci were detected in control mice from either experimental group. The representative images show an example of a LaminA/C+ metastatic foci within an irradiated lung, for each cell line. The H460 metastatic lesion (left) is indicated with the arrow. Scale bar, 250 μm. g, Violin plot showing the number of metastatic foci in the non-irradiated left lung lobe from mice that received image-guided targeted radiation to the right lung (n=5 for 8Gy mice; n=7 for 12Gy mice), compared with control mice (n=6). The violin plot displays the median, 25th and 75th percentiles as well as the density of data points. Dots represent individual mice. All data represented as mean ± s.e.m. Statistical analysis by non-parametric two-tailed Mann-Whitney test for (b), (c) and (e) and a one-way ANOVA with multiple comparisons for (g). UT, untreated. IR, irradiated. Gating strategies for FACS analysis provided in Extended Data Fig. 2. Histology quantification process outlined in Supplementary File 1. Source data.
Extended Data Fig. 2. FACS gating strategy.
a-c, Example of FACS gating strategy to determine the frequency of (a) GFP+ cancer cells or (b) Ly6G+ neutrophils in the lung tissue of control/irradiated mice, or (c) Lineage–/EpCAM+ epithelial cells in the irradiated metastatic niche (Cherry-labelled, left), or unlabelled distant lung (Cherry-negative, right). All samples were gated to exclude debris and doublets, followed by live cell discrimination by DAPI staining. All gates were set based on fluorescence-minus-one (FMO) controls, containing all antibodies minus the one of interest, to determine the background signal. Importantly, lung tissue displays a high level of autofluorescence, which needs to be considered when excluding dead cells (an FMO-DAPI is critical for this).
Extended Data Fig. 3. Radiation exposure induces lung perturbations including neutrophil infiltration and activation.
a,b, Representative immunofluorescent images (a) and quantification (b) of phospho-Histone H2A.X (Ser139) (green) and DAPI (blue) stained lungs from control/irradiated mice, 7 days post-irradiation (n=3 mice per group, one experiment). Scale bar, 10 μm. 6 fields of view were quantified per mouse. c,d, Representative images of senescence-associated β-galactosidase (SA-β-gal) staining on lung cryosections (c) and quantification (d) from control/irradiated mice, 7 days post-irradiation (n=3 mice per group, one experiment). Scale bar, 25 μm. e, Quantification of immunostaining for S100A9+ neutrophils in metastases from control and irradiated lungs at day 14 (7 days post-IV, Figure 1d) (n=6 mice per group, 2 independent experiments). The number of neutrophils within the metastatic area was normalised to tumour area. f, FACS quantification of GFP+ cancer cells in control/irradiated lungs from RAG1-ko mice at day 14 (7 days post-IV). n=9 mice per group, two independent experiments, grey dots C57BL/6J and white dots FVB background. g,h, Volcano plots showing protein expression from irradiated versus control lung (g) and bone marrow (h) neutrophils. A selection of differentially expressed proteins in the lung are depicted in red, with the same proteins shown in bone marrow samples (n=3 mice per group). i, Table of granule proteins within primary, secondary or tertiary granules with their fold change in irradiated lung neutrophils (IR) vs untreated control (UT). Nd = not detected. Highly enriched proteins are highlighted in red. j, Mice received an intraperitoneal EdU injection (25 mg/kg) 1h prior to lung irradiation. Lungs/bone marrow were harvested 1h or 7 days later, and EdU incorporation assessed by FACS. k, EdU incorporation in Ly6G+ neutrophils from bone marrow (left) and lungs (right) from control (UT) and irradiated (IR) mice (n=2 control/irradiated mice at 2h, n=3 control/irradiated mice at day 7). A representative of two independent experiments is shown (total for both experiments n=5 control/irradiated at 2h,, n=6 control/irradiated mice at day 7) All data represented as mean ± s.e.m. Statistical analysis by two-tailed t-test with Welch’s correction for (d) and (e), two-tailed non-parametric Mann-Whitney test for (f) and (k) and one-sample t-test (for value different from 2) for (b). UT, untreated; IR, irradiated. FACS gating strategies provided in Extended Data Fig. 2. Source data.
Extended Data Fig. 4. Radiation-primed neutrophil fuel metastatic growth independently of NETosis and extravasation.
a, Experimental setup. Irradiated mice were given daily injections of an anti-Ly6G neutrophil depletion antibody or IgG control, beginning the day before an IV injection of 4T1-GFP+ cancer cells. Mice were collected 72h post-IV. b,c, Frequency of Ly6G+ neutrophils (b) and GFP+ cancer cells (c) among live cells by FACS (n=7 mice per group, 2 independent experiments). d, Representative immunofluorescent images of myeloperoxidase (MPO, green), citrullinated histone-H3 (Cit-H3, red) and DAPI (blue) stained lungs at 2h, 24h and 7 days post-irradiation (n=3 mice per group, each timepoint). The positive control represents lung tissue from a mouse infected intratracheally with C.albicans and harvested 24h later. Scale bar, 100 μm. e, Schematic of 3D co-cultures. GFP+MMTV-PyMT cancer cells were seeded in Alvetex™ Scaffold 96-well plates with MACS-sorted Ly6G+ neutrophils from control or irradiated mice, harvested 7 days after irradiation. f, GFP signal quantification. Cancer cell growth on the scaffold is shown as the fold change compared to cancer cells alone (n=3 mice, each with at least 3 technical replicates, see methods). g, Schematic of GFP+MMTV-PyMT cancer cell proliferation in 2D co-culture with MACS-sorted Ly6G+ lung neutrophils from control (UT) or irradiated mice, harvested 7 days after irradiation (n=6 mice per group). Cells were treated with EdU (20 μM) and incorporation was assessed by FACS 6h later. h, Quantification of EdU+ cells among GFP+ cancer cells. PyMT cancer cells co-cultured with neutrophils isolated from control (UT) or irradiated (IR) lungs (n=6 mice per group) were compared to PyMT cells cultured alone (n=2 replicates). All data represented as mean ± s.e.m. Statistical analysis by one-way ANOVA for (b) and (c), two-way ANOVA for (f) and an unpaired two-tailed t-test for (h). UT, untreated; IR, irradiated; NT, neutrophils. Gating strategies for FACS analysis provided in Extended Data Fig. 2. Source data.
Extended Data Fig. 5. Recombinant G-CSF treatment permits controlled neutrophil recruitment.
a, Experimental setup for rG-CSF time course. Mice were given a subcutaneous injection of rGCSF every second day for a total of 4 doses, beginning the day before irradiations. On day 0, 2, 4, 6, 7 and 8, a blood sample was taken from each mouse to quantify Ly6G+ neutrophils. Mice were harvested at day 8. b, FACS analysis of Ly6G+CD11b+ neutrophils among CD45+ cells in the blood (n=6 mice per group). The day of rGCSF injections is depicted in blue and indicated by an arrow. The day of irradiation (day 0), the day of cancer cell injection (day 7) and the day of lung seeding (day 8) are indicated. Each dot represents an individual mouse, treated with either rGCSF (blue dots) or PBS (black dots). IV, intravenous; rG-CSF, recombinant G-CSF; ko, knock-out. Source data.
Extended Data Fig. 6. Primed neutrophils influence the lung epithelial cell response to radiation-induced injury.
a, Principle Component Analysis (PCA) of Lin-EpCAM- mesenchymal cell signatures following RNA-seq analysis of control, irradiated and neutrophil-depleted irradiated lungs. Each dot represents an individual mouse, ovals enclose samples from each group to highlight their similarity in the PCA plot (n=4 mice per group). b,c, 3D co-culture of GFP+MMTV-PyMT+ cancer cells on Alvetex™ Scaffolds with EpCAM+ lung epithelial cells Isolated from control (UT), irradiated (IR) and neutrophil-depleted irradiated (IR + α-Ly6G) mice, 7 days after irradiation with (b) showing GFP signal quantification at day 4, normalised to cancer cell growth alone and (c) displaying representative images of GFP intensity at day 4 (n=9 mice total per group, 3 independent experiments). Each dot in (b) represents the average of n=3 mice for an independent experiment, with at least 3 technical replicates quantified per mouse in each experiment. Scale bar, 400 μm. d,e, 3D culture of GFP+EpCAM+ epithelial cells to assess survival on the scaffold, with (d) showing representative images of GFP intensity on the scaffold at day 4 (n=3 mice per group, at least 3 technical replicates per mouse) and (e) showing GFP signal quantification (n=3 mice per group, at least 3 technical replicates per mouse). Each dot represents an individual mouse. GFP+EpCAM+ cells were sorted from the lungs of control, irradiated and neutrophil-depleted irradiated actin-GFP mice 7 days after irradiation, and seeded in Alvetex™ Scaffolds. f, Heatmap of Lin-EpCAM- mesenchymal cells from control, irradiated and neutrophil-depleted irradiated lungs (hierarchically clustered samples in columns and genes in rows) (n=4 mice per group). All data represented as mean ± s.e.m. Statistical analysis by oneway ANOVA for (b). UT, untreated; IR, irradiated; Ep, epithelial. Source data.
Extended Data Fig. 7. Radiation-induced Notch signalling in the lung environment is boosted by the presence of neutrophils.
a, Representative images and quantification of immunofluorescent staining for the Notch Intracellular Domain (NCID) in irradiated EpCAM+ lung epithelial cells that were MACS-sorted and seeded on coverslips. Cells were harvested at day 7 from irradiated mice that were treated daily with an anti-Ly6G neutrophil depletion antibody or an IgG control antibody. Cells were quantified using CellProfiler, each dot represents the intensity of nuclear NCID staining in an individual cell (n=130 IgG cells, n=205 α-L·y6G cells, see methods). n=3 mice per group, 3 technical replicates quantified per mouse. Scale bar, 5 μm. b, Quantitative RT-PCR validation of differentially-expressed genes identified by RNA-sequencing in sorted Lin-EpCAM+ lung epithelial cells. Lungs were harvested from an independent cohort of control, irradiated and neutrophil-depleted irradiated mice (n=3 mice per group). c, Quantitative RT-PCR expression of Notch genes in sorted CD31+ lung endothelial cells from control, irradiated and neutrophil-depleted irradiated mice (n=4 mice per group). Gapdh was used as a housekeeper gene for normalisation in (b) and (c). All data represented as mean ± s.e.m. Statistical analysis by unpaired two-tailed t-test for (a) and one-way ANOVA for (b) and (c). UT, untreated; IR, irradiated; Ep, epithelial. Source data.
Extended Data Fig. 8. Radiation-exposure boosts Notch-signalling and stemness in metastatic cells.
a, Quantitative RT-PCR analysis of Notch1 and target gene Hes1 in lineage-traced lung alveolar type 2 cells from SPC-Cre-ERT2 control mice and SPC-Cre-ERT2/Rosa26NICD-IRES-GFP Notch-activated mice. Mice were administered tamoxifen by oral gavage (40 mg/kg) over three consecutive days. Lungs were harvested 14 days after the last tamoxifen dose and GFP+ cells were sorted by flow cytometry (n=3 control mice, n=4 Notch mice). b, Primary tumour weight from PyMT/Control (n=6) and PyMT/Notch mice (n=8) harvested two weeks post-tamoxifen induction. c, Quantification and representative images of Hes1 immunostaining in lung metastases from PyMT/Control (n=5) and PyMT/Notch mice (n=7) mice (metastatic lungs harvested over n=5 independent tamoxifen administrations, immunostaining quantification in methods). The enlarged inset shows nuclear localisation. Scale bar, 100 μm (main image), 10 μm (enlarged inset). d, Combined Uniform Manifold Approximation and Projection (UMAP) plot of cells from the mCherry+ niche and mCherry- distant lung from irradiated mice (n=10 mice, pooled). The expression level of EpCAM (distinguishing epithelial cells), Pdgfrα and Pdgfrβ (fibroblasts) and Pecam1(CD31) (endothelial cells) is indicated in blue. e, Representative immunostaining for RBPJ (top panel) and Hes1 (lower panel) in metastatic lungs from control, irradiated, and neutrophil-depleted irradiated mice harvested at day 14 (n=7 mice per group, 2 independent experiments). Quantification shown in Figure 6d,e. The enlarged inset shows nuclear localisation within tumour cells. Hes1+ cells are indicated by arrows. Scale bar, 100 μm (main image), 10 μm (enlarged inset). f, Representative immunostaining and quantification of RBPJ staining intensity in metastatic lungs from irradiated FVB mice pre-treated with rGCSF or PBS prior to injection of MMTV-PyMT cancer cells (Figure 3e, n=5 mice per group). Staining intensity within the metastatic area was measured using ImageJ (see methods). Scale bar, 100 μm. All data represented as mean ± s.e.m. Statistical analysis by non-parametric two-tailed Mann-Whitney test for (a), a two-tailed unpaired t-test for (b), (c) and (f). UT, untreated; IR, irradiated. The violin plot displays the median, 25th and 75th percentiles as well as the density of data points. Dots represent individual mice. Gating strategies for FACS sorting for (d) provided in Extended Data Fig. 2. Source data.
Supplementary Material
Acknowledgements
We are grateful to N. Osborne from the Safety, Health and Sustainability Team at the Francis Crick Institute for his invaluable support during this work. We thank E. Nye from the Experimental Histopathology Unit at the Crick Institute for histological processing and advice and D. Barry from the Advanced Light Microscopy Facility at the Crick Institute for image analysis advice. We are grateful to P. Chakravarty from the Bioinformatics & Biostatistics Facility at the Crick Institute for bioinformatics support. We thank B. Snijders from the Proteomics Facility at the Crick Institute and R. Goldstone and A. Edwards from the Advanced Sequencing Facility at the Crick Institute for their technical support. We are also grateful for support from the Flow Cytometry Unit, the Cell Services Unit and the Biological Resources Unit at the Francis Crick Institute, particularly we thank Daiva Poniskaitiene and Tim Zverev specifically for their extended contributions to welfare monitoring of experimental mice and for Tim performing oral gavages. We are grateful to S. Quezada for his support for the image-guided focussed irradiations. We thank Dr Anna Wilkins, Clinician Scientist at the ICR and Honorary Consultant at the Royal Marsden, London, for providing critical input on the manuscript sections discussing the clinical relevance of the study. We thank R. Ferreira for critical reading of the manuscript. This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001112), the UK Medical Research Council (FC001112), and the Wellcome Trust (FC001112) and the European Research Council grant (ERC CoG-H2020-725492). The work at UCL was supported by the Radiation Research Unit at the Cancer Research UK City of London Centre Award (C7893/A28990).
Footnotes
Author Contributions:
E.N. designed and performed most of the experiments, analysed and interpreted the data. V.B. managed colony breeding and generated experimental mice, supported the animal work and provided technical support and discussion. L.O. developed the sLP-Cherry labelling tool, and provided valuable discussion. A.K and N.R. performed the single cell RNA analysis. C.S. performed the γ-H2AX immunofluorescence and SA-β-gal staining. M.V.D performed the injections and harvested mice for the image-guided partial-lung irradiation. S.H. performed bioinformatic analysis. F.R. provided technical support. P.F. performed the neutrophil proteomics. R.C. planned and performed the image-guided, focussed partial-lung radiation. L.O., A.K., N.R. and V.B. critically reviewed the manuscript. I.M. designed and supervised the study and interpreted the data. I.M and E.N. wrote the manuscript.
Competing Interests Statement:
The authors’ declare no competing interests.
Data Availability
The bulk RNA sequencing datasets (GSE180823) and the single cell RNA sequencing datasets (GSE181306) are deposited in the Gene Expression Omnibus (GEO, NCBI) repository. The proteomic datasets are deposited in PRoteomics IDEntifications (PRIDE) repository (PXD027628). Source data for Figures 1-8, Extended Data Figure 1 and Extended Data Figures 3-8 are provided as source data files.
References
- 1.Wirsdörfer F, Jendrossek V. The Role of Lymphocytes in Radiotherapy-Induced Adverse Late Effects in the Lung. Front Immunol. 2016;7:207–20. doi: 10.3389/fimmu.2016.00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ombrato L, et al. Metastatic-niche labelling reveals parenchymal cells with stem features. Nature. 2019;572:603–608. doi: 10.1038/s41586-019-1487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vicini FA, Kestin L, Huang R, Martinez A. Does local recurrence affect the rate of distant metastases and survival in patients with early-stage breast carcinoma treated with breast-conserving therapy? Cancer. 2003;97:910–919. doi: 10.1002/cncr.11143. [DOI] [PubMed] [Google Scholar]
- 4.Kuonen F, Secondini C, Rüegg C. Molecular pathways: emerging pathways mediating growth, invasion, and metastasis of tumors progressing in an irradiated microenvironment. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:5196–5202. doi: 10.1158/1078-0432.CCR-11-1758. [DOI] [PubMed] [Google Scholar]
- 5.Bouchard G, et al. Pre-irradiation of mouse mammary gland stimulates cancer cell migration and development of lung metastases. Br J Cancer. 2013;109:1829–1838. doi: 10.1038/bjc.2013.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown JM. The effect of lung irradiation on the incidence of pulmonary metastases in mice. BJR. 1973;46:613–618. doi: 10.1259/0007-1285-46-548-613. [DOI] [PubMed] [Google Scholar]
- 7.Dao TL, Kovaric J. Incidence of pulmonary and skin metastases in women with breast cancer who received postoperative irradiation. surgery. 1962;52:203–212. [Google Scholar]
- 8.Wang J. Neutrophils in tissue injury and repair. Cell Tissue Res. 2018;371:531–539. doi: 10.1007/s00441-017-2785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and in ammation. Nat Rev Immunol. 2013;13:159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- 10.Paris AJ, et al. Neutrophils promote alveolar epithelial regeneration by enhancing type II pneumocyte proliferation in a model of acid-induced acute lung injury. AJP: Lung Cellular and Molecular Physiology. 2016;311:L1062–L1075. doi: 10.1152/ajplung.00327.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zemans RL, et al. Neutrophil transmigration triggers repair of the lung epithelium via β-catenin signaling. Proc Natl Acad Sci USA. 2011;108:15990–15995. doi: 10.1073/pnas.1110144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedrick CC, Malanchi I. Neutrophils in cancer: heterogeneous and multifaceted. Nat Rev Immunol. 2021:1–15. doi: 10.1038/s41577-021-00571-6. [DOI] [PubMed] [Google Scholar]
- 13.Jones B, Dale RG, Deehan C, Hopkins KI, Morgan DA. The role of biologically effective dose (BED) in clinical oncology. Clin Oncol (R Coll Radiol) 2001;13:71–81. doi: 10.1053/clon.2001.9221. [DOI] [PubMed] [Google Scholar]
- 14.Emami B, et al. Tolerance of normal tissue to therapeutic irradiation. Int J Radiat Oncol Biol Phys. 1991;21:109–122. doi: 10.1016/0360-3016(91)90171-y. [DOI] [PubMed] [Google Scholar]
- 15.Ballesteros I, et al. Co-option of Neutrophil Fates by Tissue Environments. Cell. 2020 doi: 10.1016/j.cell.2020.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Granot Z, et al. Tumor entrained neutrophils inhibit seeding in the premetastatic lung. Cancer Cell. 2011;20:300–314. doi: 10.1016/j.ccr.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeshima T, et al. Key role for neutrophils in radiation-induced antitumor immune responses: Potentiation with G-CSF. Proc Natl Acad Sci USA. 2016;113:11300–11305. doi: 10.1073/pnas.1613187113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El Rayes T, et al. Lung inflammation promotes metastasis through neutrophil protease-mediated degradation of Tsp-1. Proc Natl Acad Sci USA. 2015;112:16000–16005. doi: 10.1073/pnas.1507294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park J, et al. Cancer cells induce metastasis-supporting neutrophil extracellular DNA traps. Sci Transl Med. 2016;8:361ra138. doi: 10.1126/scitranslmed.aag1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albrengues J, et al. Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science. 2018;361:eaao4227. doi: 10.1126/science.aao4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang L, et al. DNA of neutrophil extracellular traps promotes cancer metastasis via CCDC25. Nature. 2020;583:1–30. doi: 10.1038/s41586-020-2394-6. [DOI] [PubMed] [Google Scholar]
- 22.Lieschke GJ, et al. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood. 1994;84:1737–1746. [PubMed] [Google Scholar]
- 23.Lee J-H, et al. Anatomically and Functionally Distinct Lung Mesenchymal Populations Marked by Lgr5 and Lgr6. Cell. 2017;170:1149–1156.:e12. doi: 10.1016/j.cell.2017.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McConnell AM, Konda B, Kirsch DG, Stripp BR. Distal airway epithelial progenitor cells are radiosensitive to High-LET radiation. Sci Rep. 2016;6:33455–11. doi: 10.1038/srep33455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Sato C, Cerletti M, Wagers A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr Top Dev Biol. 2010;92:367–409. doi: 10.1016/S0070-2153(10)92012-7. [DOI] [PubMed] [Google Scholar]
- 26.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci USA. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ombrato L, et al. Generation of neighbor-labeling cells to study intercellular interactions in vivo. Nature Protocols. 2021;16:872–892. doi: 10.1038/s41596-020-00438-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Sullenger BA, Rich JN. Notch signaling in cancer stem cells. Adv Exp Med Biol. 2012;727:174–185. doi: 10.1007/978-1-4614-0899-4_13. [DOI] [PubMed] [Google Scholar]
- 29.Meier-Stiegen F, et al. Activated Notch1 Target Genes during Embryonic Cell Differentiation Depend on the Cellular Context and Include Lineage Determinants and Inhibitors. PLoS ONE. 2010;5:e11481–18. doi: 10.1371/journal.pone.0011481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson JL, et al. Identification of Neutrophil Exocytosis Inhibitors (Nexinhibs), Small Molecule Inhibitors of Neutrophil Exocytosis and Inflammation. J Biol Chem. 2016;291:25965–25982. doi: 10.1074/jbc.M116.741884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malanchi I, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2011;481:85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- 32.del P Martin Y, et al. Mesenchymal Cancer Cell-Stroma Crosstalk Promotes Niche Activation, Epithelial Reversion, and Metastatic Colonization. CellReports. 2015;22:2456–2469. doi: 10.1016/j.celrep.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rock JR, et al. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell. 2011;8:639–648. doi: 10.1016/j.stem.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xing Y, Li A, Borok Z, Li C, Minoo P. NOTCH1 is required for regeneration of Clara cells during repair of airway injury. Stem Cells. 2012;30:946–955. doi: 10.1002/stem.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finn J, et al. Dlk1-Mediated Temporal Regulation of Notch Signaling Is Required for Differentiation of Alveolar Type II to Type I Cells during Repair. CellReports. 2019;26:2942–2954.:e5. doi: 10.1016/j.celrep.2019.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaughan AE, et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 2015;517:621–625. doi: 10.1038/nature14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giuranno L, et al. NOTCH signaling promotes the survival of irradiated basal airway stem cells. Am J Physiol-lung Cell Mol Physiol. 2019;317:L414–L423. doi: 10.1152/ajplung.00197.2019. [DOI] [PubMed] [Google Scholar]
- 38.Fan X, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoey T, et al. DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell. 2009;5:168–177. doi: 10.1016/j.stem.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 40.Zheng Y, et al. A rare population of CD24(+)ITGB4(+)Notch(hi) cells drives tumor propagation in NSCLC and requires Notch3 for self-renewal. Cancer Cell. 2013;24:59–74. doi: 10.1016/j.ccr.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mamun MA, Mannoor K, Cao J, Qadri F, Song X. SOX2 in cancer stemness: tumor malignancy and therapeutic potentials. J Mol Cell Biol. 2020;12:85–98. doi: 10.1093/jmcb/mjy080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aguilar-Medina M, et al. SOX9 Stem-Cell Factor: Clinical and Functional Relevance in Cancer. Journal of Oncology. 2019;2019:6754040. doi: 10.1155/2019/6754040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H, Lu J, Liu J, Zhang G, Lu A. Advances in the discovery of exosome inhibitors in cancer. J Enzyme Inhib Med Chem. 2020;35:1322–1330. doi: 10.1080/14756366.2020.1754814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polverino E, Rosales-Mayor E, Dale GE, Dembowsky K, Torres A. The Role of Neutrophil Elastase Inhibitors in Lung Diseases. Chest. 2017;152:249–262. doi: 10.1016/j.chest.2017.03.056. [DOI] [PubMed] [Google Scholar]
- 45.Wculek SK, Malanchi I. Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature. 2015;528(7582):413–417. doi: 10.1038/nature16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J-H, et al. Lung Stem Cell Differentiation in Mice Directed by Endothelial Cells via a BMP4-NFATc1-Thrombospondin-1 Axis. Cell. 2014;156:440–455. doi: 10.1016/j.cell.2013.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17:10–12. [Google Scholar]
- 48.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hao Y, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biology. 2019;20(1):296–15. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DeTomaso D, Jones MG, Subramaniam M, Ashuach T, Ye CJ, Yosef N. Functional interpretation of single cell similarity maps. Nature Communications. 2019;10(1):4376–11. doi: 10.1038/s41467-019-12235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bulk RNA sequencing datasets (GSE180823) and the single cell RNA sequencing datasets (GSE181306) are deposited in the Gene Expression Omnibus (GEO, NCBI) repository. The proteomic datasets are deposited in PRoteomics IDEntifications (PRIDE) repository (PXD027628). Source data for Figures 1-8, Extended Data Figure 1 and Extended Data Figures 3-8 are provided as source data files.