Abstract
Host-parasite species pairs can coevolve, but the impacts of diverse parasite communities on coevolution are unclear. By using experimental coevolution of a host bacterium and viral parasites, we revealed that diverse parasite communities accelerated host evolution and altered coevolutionary dynamics, resulting in enhanced host resistance and decreased parasite infectivity. Increases in parasite diversity drove shifts in the mode of selection from fluctuating (Red Queen dynamics) to predominately directional (Arms Race dynamics). The latter was characterized by selective sweeps of generalist resistance mutations in LPS biosynthesis genes, which caused faster molecular evolution within host populations and greater genetic divergence among populations. These results suggest that parasite communities can have profound effects on the rate and mechanism of host-parasite coevolution.
Coevolution between hosts and parasites is hypothesized as central to numerous biological phenomena, from rapid evolutionary change (1) to the maintenance of diversity (2) and sexual reproduction (3, 4). Host-parasite coevolution is traditionally investigated in a pairwise framework where a single parasite species infects one host species (5). However, in nature, these pairwise relationships rarely exist in isolation (6, 7): hosts are often under attack by multiple parasite species (8), and parasites must compete for hosts (9–12). Host-parasite coevolution thus operates within a network of species interactions (13). It nevertheless remains unknown whether and how diverse parasite communities can shape coevolutionary patterns and processes.
Coevolution with highly diverse parasite communities should impose stronger selection for host resistance if this diversity yields more infections and higher mortality than single parasites. This is an outcome that could ultimately drive faster host evolution and increased divergence among host populations. To test these predictions, we applied an experimental coevolution approach to an in vitro bacteria-phage system. We exposed the host bacterium, Pseudomoas aeruginosa, to communities of one to five lytic viral parasites (bacteriophages PEV2, LUZ19, LUZ7, 14-1 and LMA2) that are obligate lethal parasites. These parasites infect their hosts by each attaching to one of three specific cell surface receptors (lipopolysaccharides (LPS), Ton-B dependent receptors or Type IV pili) and hosts typically evolve resistance by modifying or deleting these attachment sites (14). Parasites, in turn, can evolve reciprocal adaptations to circumvent host resistance (15). Specifically, we experimentally coevolved host populations independently in cell culture tubes with replicates of a five-parasite community (High Diversity, N=30) alongside every possible pairwise parasite combination (Medium, N=30) and a single parasite treatment with replicates of each in monoculture (Low, N=30) alongside a parasite-free control (N=30). We tested for effects on the tempo and mode of host-parasite coevolution, conducting phenotypic assays of host resistance and parasite infectivity, as well as by using deep sequencing to measure changes in the genomic composition of host and parasite populations through time. Because we are able to experimentally manipulate parasite diversity, our work differs from the approaches used to investigate coevolution in natural communities, where diversity is not controlled exogenously (16, 17).
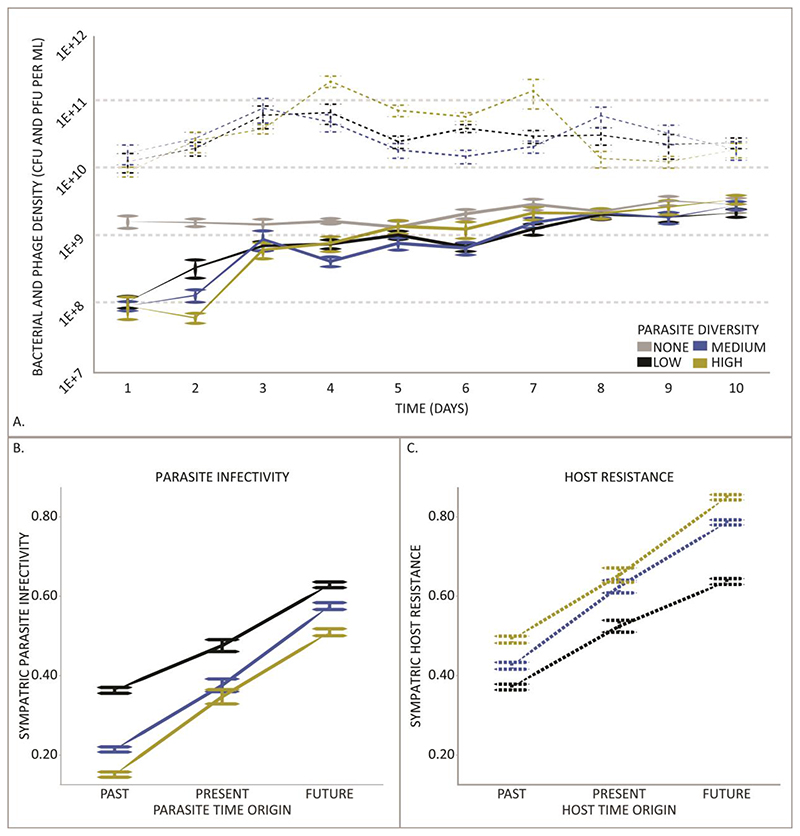
As with predators and prey, parasite densities are expected to track host densities producing time-lagged cycles in population sizes over time (18). Indeed, in these experiments, given that the hosts are initially susceptible, we might expect large amplitude oscillations in population size, unless host resistance rapidly evolves (19). By contrast, we observed that host densities increased throughout the experiment while parasite densities remained stable (Fig. 1.A, Supplementary Text), indicating that underlying coevolutionary dynamics interfered with the expected ecological dynamics (20).
Fig. 1. Ecological and Coevolutionary Dynamics of Host-Parasite Interactions.
(A) We tracked the population dynamics of hosts (solid lines) and parasites (dashed lines) for high (yellow), medium (blue) and low (black) diversity populations alongside the parasite free control (grey). All plotted points show the mean +/- standard error population density. Bacterial populations challenged with phage rapidly recovered their density. We did not observe cyclical oscillations in host and parasite densities. (B and C) To directly test for co-evolution, we used 13,500 time shift assays to measure changes in phage infectivity and bacterial resistance to phage. Lower levels or parasite infectivity (B) and higher levels of host resistance (C) evolve with increasing parasite diversity (F2,85 =9.7, P < 0.001). Furthermore, hosts are more susceptible to parasites from their future than their contemporary parasites and they are most resistant to parasites from their past. Likewise future parasite are more infective than past or contemporary parasites (F2,13044 =1766.21, P < 0.0001). Error bars show 1 standard error.
We determined whether coevolution took place in our experiment by performing 13,500 time-shift assays. This approach assesses the infectivity of parasites towards their past, present and future hosts, and reciprocally, the resistance of hosts to their past, present and future parasites (21). Resistance was measured using inhibition assays where individual host genotypes were challenged with parasite communities and resultant growth or inhibition was analysed to compare the mean levels of resistance among treatments. Infectivity and resistance varied widely across individual host-parasite communities over time (Fig. S1). Despite this complexity, two clear and significant patterns emerged. First, parasites from the future were more infective than parasites from the present or past (Fig. 1.B). Likewise, hosts evolved increased resistance over time (Fig. 1.C). This pattern is typical of Arms Race dynamics, commonly seen with in vitro bacteria-phage coevolution (5, 22). Second, bacterial resistance increased with parasite diversity, while parasite infectivity decreased (F2,85 =9.7, P < 0.001), suggesting that parasite communities impose stronger selection on their host than single parasites alone.
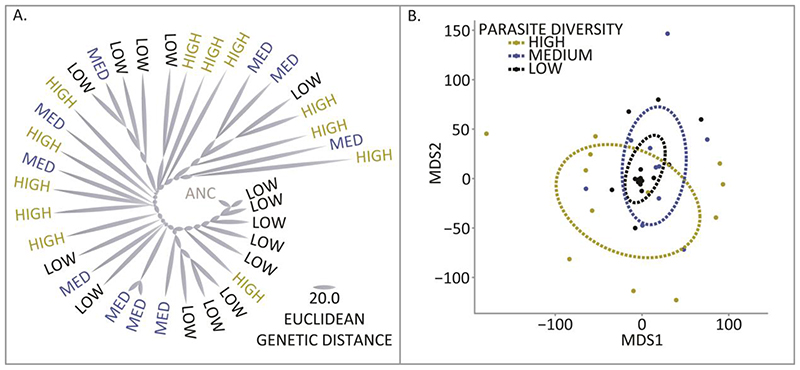
To test the hypothesis that parasite diversity accelerated coevolution at a molecular level, we sequenced entire host and parasite populations longitudinally at multiple time points (Ancestor, Midpoint and Endpoint) from the High (N=12), Medium (N=10), Low (N=15) and Control (N=10) treatments. We could thus detect any mutations that arose during coevolution and estimate their frequency changes within populations. In the hosts, we observed 474 non-synonymous and 75 synonymous polymorphisms across 173 genes and 133 intergenic mutations. Parallel evolution with mutations in the same genes or genes in the same pathways was common across all treatments: >50% of mutations occurred in <5% of these genes. Notably, we observed parallel evolution in genes that are known phage receptors (Table S1), including LPS (N=190 mutations), Type IV pili (N=69 mutations) and the TonB dependent receptor (N=55 mutations). To measure the rate of bacterial molecular evolution, we calculated the Euclidean genetic distance of each population from the ancestor (Fig. 2.A). We found that higher levels of parasite diversity drove greater host divergence from the ancestor, and consequently faster host evolution (Fig. S2; ANOVA F2,34 =10.5, P < 0.001; Post hoc Tukey test P < 0.05).
Fig. 2. Parasite diversity accelerates host evolution.
(A) Host allele frequencies after 10 days of coevolution from the high (yellow), medium (blue) and low (black) parasite diversity treatments were used to calculate pairwise Euclidean distances between the ancestral sequence (grey) and each coevolved population. Increasing parasite diversity accelerated the rate of host evolution. (B) The genetic distances between coevolved populations were ordinated by non-metric multidimensional scaling. The ellipses represent a 95% confidence bubble around the means for the different treatments. We found evidence of divergence between populations within treatments (ANOSIM R = 0.11 P < 0.01), and the greatest within treatment diversification was observed in the high parasite diversity treatment.
Two mechanisms could explain why diverse parasite communities accelerate the rate of host evolution. On one hand, diverse communities are more likely, by chance alone, to contain parasites that drive rapid evolution. On the other hand, it is possible that parasite diversity perse accelerates host evolution by increasing selection for resistance. To discriminate between these mechanisms, we calculated the contribution of parasite diversity to the speed of host evolution by adopting an approach developed to determine the contribution of biodiversity to ecosystem function, here represented by host evolutionary rate (23). Using this approach, we found that relative to phage monocultures, the combination of all five parasites contributed to faster host evolution overall (Dmax >0; Bonferroni corrected one-sample t-tests P < 0.05). Thus, the diversity of interactions between parasites within communities, and not merely the presence of a particular parasite, imposed stronger selection.
Whole genome sequencing also revealed profound changes in parasite communities. At an ecological level, the composition of communities changed markedly over time (Fig. S3). Parasite diversity decreased in both the medium and high diversity treatments, but diversity remained higher in the high diversity than in the medium diversity treatments (Fig. S4). At a genetic level, we found evidence for rapid molecular evolution of phage, which supports the results of our phenotypic assays. In total, we detected 533 SNPs across all five parasite types that reached >10% in frequency in their populations (Table S2). The proportions of non-synonymous SNPs were very high, exceeding 0.7 for each parasite (Table S3), and parallel evolution was common, implying that these SNPs were predominantly beneficial mutations. To investigate the role of parasite diversity in phage evolution, we focused our analysis on communities that included the parasite PEV2, as this phage was well-represented at all levels of parasite diversity, and compared with the other phages used here, the PEV2 genome is well-annotated. For example, all but two of the genes in the parasite 14-1 genome are hypothetical proteins. We detected 225 mutations in the PEV2 genome, and the rate of PEV2 evolution was independent of parasite diversity (Fig. S5). Thirty-nine percent (N=88) of the PEV2 mutations occurred in a single tail fibre gene, gp52. The tail fibers are the means by which the parasite recognizes and binds to its receptor and are known to undergo coevolution (24). We detected mutations in gp52 and at least one host LPS mutation (usually in wzz or migA) in every community containing PEV2 across all levels of parasite diversity (Fig. S6 and Fig. S7). Collectively, these results provide genetic confirmation for reciprocal co-evolution between host and parasite across all levels of parasite diversity, and they provide further evidence that parasite attachment to LPS is a key mechanism underpinning coevolution between PEV2 and P. aeruginosa.
Coevolutionary dynamics are usually described as being one of two types. Arms Races occur when selection results in directional increases in host resistance and parasite infectivity (25, 26) and with Red Queen dynamics, negative frequency-dependent selection causes fluctuations in allele frequencies (27, 28). When interacting with this host in a pairwise fashion it has been previously shown that each parasite follows a different coevolutionary trajectory (29) (Fig. S8). Using a population genomic approach, we therefore sought to examine whether parasite diversity altered the mode of coevolution.
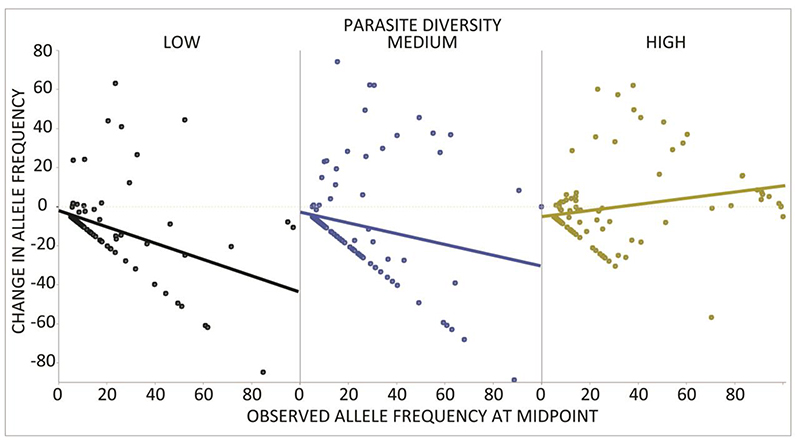
We examined the type of coevolution occurring in each community by comparing the change in frequencies of each host allele between the mid- and end-points of the experiment (29, 30). These timeframes were previously shown to be suitable for observing coevolutionary dynamics in P. aeruginosa-phage interactions (29). We focused our analysis on the host because changes in the frequency of phage alleles in diverse communities might additionally be driven by interspecific competition among parasites (14). On average, host alleles with higher frequencies tended to decline over time in the low and medium parasite diversity treatments, consistent with negative frequency-dependent selection on host resistance genes (Fig. 3.). This mode of selection becomes less predominant at higher levels of parasite diversity (ANCOVA F2,335 =12.64, P < 0.0001) suggesting that the occurrence of Red Queen dynamics was reduced in these communities.
Fig. 3.
Red Queen coevolution more common in pairwise host-parasite interactions. We tested for Red Queen dynamics by regressing the change in host allele frequency from day 5 to day 10 (y-axis) with observed frequency on day 5 (x-axis). We found evidence for negative frequency dependent selection on host alleles under low (black) and medium (blue) parasite diversity, but not at high parasite diversity (yellow). The string of points forming a straight downward slope from zero in all three panels are alleles observed on day 5 that had subsequently decreased in frequency to below our ability to detect them at day 10.
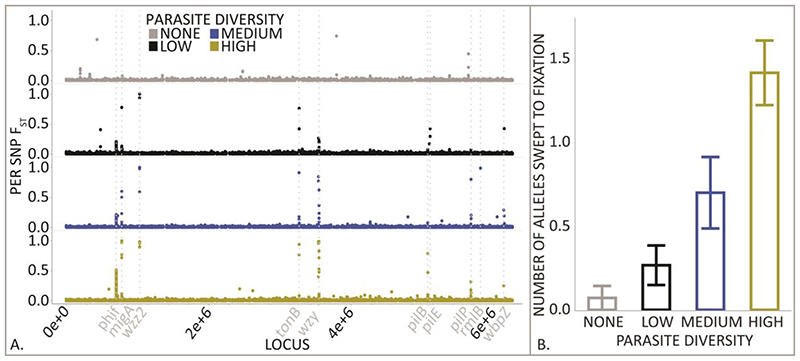
In contrast to Red Queen dynamics, Arms Races should result in recurrent selective sweeps of resistance mutations (5). The effects of directional selection in host-parasite interactions have been studied in a variety of systems and can be important for human disease coevolution (31–34). To test for selective sweeps in host populations, we calculated FST, which measures the genetic divergence per mutation between each coevolved host population and a sequenced ancestor (35), and we set a conservative threshold FST above which any mutation was deemed to be under strong directional selection (Fig. 4.A). Strong directional selection was more common as parasite diversity increased (X2= 20, df = 3, P <0.001) such that the number of fixed alleles in the high diversity treatment was over two-fold that observed under medium diversity (Fig. 4.B). Of the 29 alleles approaching fixation, 23 were in genes relating to LPS biosynthesis. LPS is very abundant on the bacterial cell surface, and phage may initially bind to LPS to anchor themselves to the cell before forming an irreversible attachment to a final receptor (36). Mutations that alter LPS biosynthesis may provide a general phage resistance mechanism. We have previously shown that mutations in LPS biosynthesis gene wzy provide resistance to four of the phages used in this study (14). Red Queen and Arms Race dynamics fall at two ends of a continuum (21), with tight interaction specificity required for the former, and for the latter, general host resistance mechanisms should evolve to allow future hosts to resist all previous parasite genotypes. Our data points to that the availability of generalist LPS biosynthesis resistance mutations as key to the transition from Red Queen to Arms Race dynamics as parasite diversity increases. That both dynamics can arise from host-parasite community coevolutionary interactions are consistent with studies of bacteria-phage systems conducted in natural communities (16, 17).
Fig. 4. Parasite diversity leads to more Arms Race coevolutionary dynamics.
(A) To test for Arms Race Dynamics, we calculated FST for all host SNPs across the P. aeruginosa genome across the high (yellow), medium (blue) and low (black) parasite diversity treatments and the parasite free control (grey). Each plotted point in the panel represents a single SNP and host genes that are known parasite targets are shown in italics, including genes involved in LPS biosynthesis, Type IV pili biosynthesis, and the Ton-B dependent receptor. (B) We used a conservative FST cutoff to identify selective sweeps of SNPs per host population (+/- standard error), and we found that increasing parasite diversity increased the rate of fixation of SNPs (X2= 20, df = 3, P <0.001).
Parasites have been shown to play a key role in evolutionary diversification within (37–39) and among host populations (40). Specifically, parasite-mediated selection against common host genotypes is predicted to stably maintain genetic diversity (31, 41). Host population genetic diversity should be greatest during pairwise coevolution where we observed Red Queen dynamics are relatively stronger. Surprisingly, we found that levels of genetic diversity within host populations were not impacted by parasite diversity (Fig. S9) despite the different modes of selection acting across treatments. Parasite diversity, conversely, had a profound impact on allopatric diversification among host populations (ANOSIM R = 0.11 P < 0.01). The greatest divergence was found in the high parasite diversity treatment (Fig. 2.B; ANOVA F2,213 =122.15, P < 0.001; Post-hoc Tukey test P < 0.05). The simplest explanation for this result is that rapid evolution within host populations leads to accelerated divergence among populations.
Our study reveals that parasite communities can form hotbeds of rapid antagonistic coevolution. Relative to single parasites, diverse parasite communities imposed stronger selection on host populations, causing faster selective sweeps of generalist resistance mutations and higher levels of host resistance. The multi-species coevolutionary process ultimately accelerated the rate of molecular evolution within host populations and increased the genomic divergence among populations. Host resistance to parasite infection can be specialized (26). However, diverse, broad-spectrum defence strategies which confer protection against different enemies are widespread in animals (42–44) and plants (45, 46), as well as bacteria (47–49). We should thus consider looking beyond the pairwise towards parasite communities as drivers of coevolution and rapid host evolution in nature. This focus could be particularly informative in biodiversity hotspots with elevated evolutionary and speciation rates (27): a multitude of parasites might be the fuel.
Supplementary Material
One Sentence Summary.
Increasing parasite diversity accelerates coevolutionary arms races and leads to greater generalist host resistance.
Acknowledgments
We thank Michael Brockhurst, Dylan Dahan, Kevin Foster, Owen Lewis and Steve Paterson for their helpful comments. We also thank the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics funded by Wellcome Trust grant reference 090532/Z/09/Z and Medical Research Council Hub grant G0900747 91070 for generation of the high-throughput sequencing data. AB was supported by DPhil funding from the Biotechnology and Biological Sciences Research Council (BBSRC) [grant number BB/J014427/1]. R.C.M was funded by the Royal Society, European Research Council grant 281591, and Wellcome Trust Grant 106918/Z/15/Z. KCK was supported by a Leverhulme Research Project Grant RPG-2015-165.
The host and parasite sequences reported in this paper have been deposited in the National Center for Biotechnology (NCBI) database, www.ncbi.nlm.nih.gov (accession numbers PRJNA448325 and PRJNA448370 respectively). All other data are available in the supplementary materials.
AB, RCM, and KCK conceived and designed the study. AB and MZ collected the data. AB and CG conducted the data analysis. AB, RCM, and KCK drafted the article, with critical revisions provided by all authors.
References and Notes
- 1.Paterson S, et al. Antagonistic coevolution accelerates molecular evolution. Nature. 2010;464:275–8. doi: 10.1038/nature08798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haldane JBS. Disease and evolution. La Ric Sci. 1949;19:68–76. [Google Scholar]
- 3.Bell G. The masterpiece of nature : the evolution and genetics of sexuality. Croom Helm; London: 1982. [Google Scholar]
- 4.Hamilton WD. Sex versus Non-Sex versus Parasite. Oikos. 1980;35:282. [Google Scholar]
- 5.Brockhurst MA, et al. Running with the Red Queen: the role of biotic conflicts in evolution. Proc R Soc B. 2014;281:1–9. doi: 10.1098/rspb.2014.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcogliese DJ. Parasites of the superorganism: Are they indicators of ecosystem health? Int J Parasitol. 2005;35:705–716. doi: 10.1016/j.ijpara.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Hudson PJ, Dobson AP, Lafferty KD. Is a healthy ecosystem one that is rich in parasites? Trends Ecol Evol. 2006;21:381–385. doi: 10.1016/j.tree.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Telfer S, et al. Species interactions in a parasite community drive infection risk in a wildlife population. Science. 2010;330:243–246. doi: 10.1126/science.1190333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffiths EC, Pedersen AB, Fenton A, Petchey OL. The nature and consequences of coinfection in humans. J Infect. 2011;63:200–206. doi: 10.1016/j.jinf.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lello J. Coinfection: doing the math. Sci Transl Med. 2013;5:191fs24. doi: 10.1126/scitranslmed.3006565. [DOI] [PubMed] [Google Scholar]
- 11.Alizon S, de Roode JC, Michalakis Y. Multiple infections and the evolution of virulence. Ecol Lett. 2013;16:556–567. doi: 10.1111/ele.12076. [DOI] [PubMed] [Google Scholar]
- 12.Bashey F, Hawlena H, Lively CM. Alternative paths to success in a parasite community: Within-host competition can favor higher virulence or direct interference. Evolution. 2013;67:900–907. doi: 10.1111/j.1558-5646.2012.01825.x. [DOI] [PubMed] [Google Scholar]
- 13.Betts A, Rafaluk C, King KC. Host and Parasite Evolution in a Tangled Bank. Trends Parasitol. 2016;32:863–873. doi: 10.1016/j.pt.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Betts A, Gifford DR, MacLean RC, King KC. Parasite diversity drives rapid host dynamics and evolution of resistance in a bacteria-phage system. Evolution. 2016;70:969–978. doi: 10.1111/evo.12909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samson JE, Magadán AH, Sabri M, Moineau S. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol. 2013;11:675–87. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- 16.Koskella B, Parr N. The evolution of bacterial resistance against bacteriophages in the horse chestnut phyllosphere is general across both space and time. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140297. doi: 10.1098/rstb.2014.0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gómez P, Buckling A. Bacteria-phage antagonistic coevolution in soil. Science. 2011;332:106–109. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- 18.Hudson PJ, Dobson AP, Newborn D. Prevention of Population Cycles by Parasite Removal. Science. 1998;282:2256–2258. doi: 10.1126/science.282.5397.2256. [DOI] [PubMed] [Google Scholar]
- 19.Mougi A, Iwasa Y. Evolution towards oscillation or stability in a predator-prey system. Proc R Soc B. 2010;277:3163–71. doi: 10.1098/rspb.2010.0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cortez MH, Weitz JS. Coevolution can reverse predator-prey cycles. Proc Natl Acad Sci. 2014;111:7486–91. doi: 10.1073/pnas.1317693111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gandon S, Buckling A, Decaestecker E, Day T. Host-parasite coevolution and patterns of adaptation across time and space. J Evol Biol. 2008;21:1861–6. doi: 10.1111/j.1420-9101.2008.01598.x. [DOI] [PubMed] [Google Scholar]
- 22.Buckling A, Wei Y, Massey RC, Brockhurst MA, Hochberg ME. Antagonistic coevolution with parasites increases the cost of host deleterious mutations. Proc R Soc B. 2006;273:45–49. doi: 10.1098/rspb.2005.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loreau M, Hector A. Biodiversity and Ecosystem Functioning : Current Knowledge and Future Challenges. Science. 2001;294:804–809. doi: 10.1126/science.1064088. [DOI] [PubMed] [Google Scholar]
- 24.Weitz JS, Hartman H, Levin SA. Coevolutionary arms races between bacteria and bacteriophage. Proc Natl Acad Sci. 2005;102:9535–9540. doi: 10.1073/pnas.0504062102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buckling A, Rainey PB. Antagonistic coevolution between a bacterium and a bacteriophage. Proc R Soc B. 2002;269:931–936. doi: 10.1098/rspb.2001.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agrawal A, Lively C. Infection genetics: gene-for-gene versus matching-alleles models and all points in between. Evol Ecol Res. 2002;4:79–90. [Google Scholar]
- 27.Pirie MD, et al. The biodiversity hotspot as evolutionary hot-bed: spectacular radiation of Erica in the Cape Floristic Region. BMC Evol Biol. 2016;16:190. doi: 10.1186/s12862-016-0764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Decaestecker E, et al. Host-parasite “Red Queen” dynamics archived in pond sediment. Nature. 2007;450:870–873. doi: 10.1038/nature06291. [DOI] [PubMed] [Google Scholar]
- 29.Betts A, Kaltz O, Hochberg MEE. Contrasted coevolutionary dynamics between a bacterial pathogen and its bacteriophages. Proc Natl Acad Sci. 2014;111:11109–11114. doi: 10.1073/pnas.1406763111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koskella B, Lively CM. Evidence for negative frequency-dependent selection during experimental coevolution of a freshwater snail and a sterilizing trematode. Evolution. 2009;63:2213–21. doi: 10.1111/j.1558-5646.2009.00711.x. [DOI] [PubMed] [Google Scholar]
- 31.Schulte RD, Makus C, Hasert B, Michiels NK, Schulenburg H. Multiple reciprocal adaptations and rapid genetic change upon experimental coevolution of an animal host and its microbial parasite. Proc Natl Acad Sci. 2010;107:7359–64. doi: 10.1073/pnas.1003113107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilfert L, Jiggins FM. Disease association mapping in Drosophila can be replicated in the wild. Biol Lett. 2010;6:666–668. doi: 10.1098/rsbl.2010.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roche B, et al. Might Interspecific Interactions between Pathogens Drive Host Evolution? The Case of Plasmodium Species and Duffy-Negativity in Human Populations. Trends Parasitol. 2017;33:21–29. doi: 10.1016/j.pt.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 34.Eizaguirre C, Lenz TL, Kalbe M, Milinski M. Divergent selection on locally adapted major histocompatibility complex immune genes experimentally proven in the field. Ecol Lett. 2012;15:723–731. doi: 10.1111/j.1461-0248.2012.01791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wright S. Evolution and the Genetics of Populations. Vol. 2. University of Chicago Press; Chicago, IL: 1969. pp. 1–295. [Google Scholar]
- 36.Abedon ST. The Bacteriophages. Vol. 2 Oxford: 2006. [Google Scholar]
- 37.King KC, Delph LF, Jokela J, Lively CM. The Geographic Mosaic of Sex and the Red Queen. Curr Biol. 2009;19:1438–1441. doi: 10.1016/j.cub.2009.06.062. [DOI] [PubMed] [Google Scholar]
- 38.Marston MF, et al. Rapid diversification of coevolving marine Synechococcus and a virus. Proc Natl Acad Sci. 2012;109:4544–4549. doi: 10.1073/pnas.1120310109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morran LT, Schmidt OG, Gelarden IA, Parrish RC, Lively CM. Running with the Red Queen: host-parasite coevolution selects for biparental sex. Science. 2011;333:216–218. doi: 10.1126/science.1206360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson J. Geographic Mosaic of Coevolution. The University of Chicago Press; Chicago, IL: 2005. [Google Scholar]
- 41.D’Costa VM, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–61. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 42.Klein J, O’Huigin C. MHC Polymorphism and Parasites. Philos Trans R Soc B Biol Sci. 1994;346:351–358. doi: 10.1098/rstb.1994.0152. [DOI] [PubMed] [Google Scholar]
- 43.Duncan AB, Fellous S, Kaltz O. Reverse Evolution : Selection Against Costly Resistance In Disease-Free Microcosm Populations Of Paramecium Caudatum . Evolution. 2011;65:3462–3474. doi: 10.1111/j.1558-5646.2011.01388.x. [DOI] [PubMed] [Google Scholar]
- 44.Tetreau G, Stalinski R, David J, Després L. Increase In Larval Gut Proteolytic Activities And Bti Resistance In The Dengue Fever Mosquito. Arch Insect Biochem Physiol. 2013;82:71–83. doi: 10.1002/arch.21076. [DOI] [PubMed] [Google Scholar]
- 45.Nicaise V. Crop immunity against viruses: outcomes and future challenges. Front Plant Sci. 2014;5 doi: 10.3389/fpls.2014.00660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barrett LG, Encinas-viso F, Burdon JJ, Thrall PH. Specialization for resistance in wild host-pathogen interaction networks. Front Plant Sci. 2015;6:1–13. doi: 10.3389/fpls.2015.00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez-Valera F, et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 48.Avrani S, Wurtzel O, Sharon I, Sorek R, Lindell D. Genomic island variability facilitates Prochlorococcusvirus coexistence. Nature. 2011;474:604–608. doi: 10.1038/nature10172. [DOI] [PubMed] [Google Scholar]
- 49.Doron S, et al. Systematic discovery of antiphage defense systems in the microbial pangenome. Science. 2018;359 doi: 10.1126/science.aar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R. R Development Core Team, R. D. C. Team. R: A Language and Environment for Statistical Computing. R Found Stat Comput. 2011;1:409. Ed. [Google Scholar]
- 51.Winsor GL, et al. Pseudomonas Genome Database : improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 2011;39:596–600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kofler R, et al. PoPoolation2 : identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq) Bioinformatics. 2011;27:3435–3436. doi: 10.1093/bioinformatics/btr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Popescu AA, Huber KT, Paradis E. Ape 3.0: New tools for distance-based phylogenetics and evolutionary analysis in R. Bioinformatics. 2012;28:1536–1537. doi: 10.1093/bioinformatics/bts184. [DOI] [PubMed] [Google Scholar]
- 55.Loreau M. Separating sampling and other effects in biodiversity experiments. Oikos. 1998;82:600–602. [Google Scholar]
- 56.Sheldon BC, Verhulst S. Ecological immunology - costly parasite defenses and trade-offs in evolutionary ecology. Trends Ecol Evol. 1996;11:317–321. doi: 10.1016/0169-5347(96)10039-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.